G2C::Press Releases
Evolution of NMDA receptor cytoplasmic interaction domains
A recent Genes to Cognition study in BMC Neuroscience has brought an evolutionary perspective to bear on NMDA receptor signalling, providing a new avenue into our understanding of vertebrate brain evolution (Ryan et al '08).
NMDA receptors have long been known to be essential for learning and memory. They are a unique class of neurotransmitter receptor that are specifically activated by persistent stimulation. This property allows them to strengthen the connections between neurons following coincident activity. Therefore they are widely believed to underlie the cellular mechanisms for memory formation in the brain. NMDA receptors have been heavily studied in the mammalian brain, and more recently have been shown to be important for learning and memory in Drosophila (Xia et al '05).
Though NMDA receptors can rightly be regarded as an ancient type of receptor fundamental to learning in all animal species, Ryan et al's study identifies a dramatic elaboration in the signalling capability of vertebrate NMDA receptors. This adaptation in synaptic complexity is likely to have contributed to the evolution of higher cognitive function.
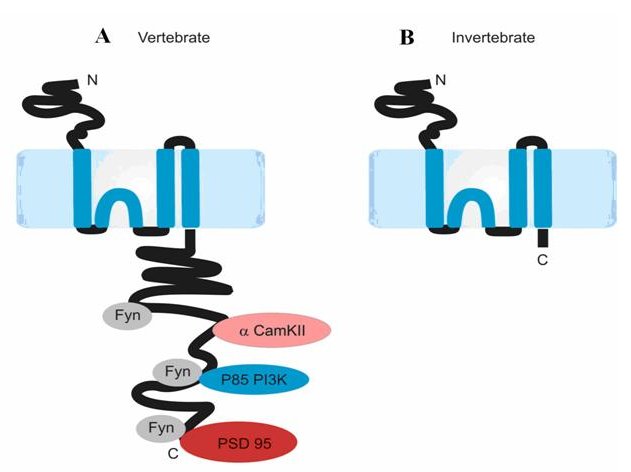
NMDA receptors are calcium ion channels that function at the postsynaptic side of excitatory synapses in the central nervous system. NMDA receptors are thought to be tetrameric in structure, composed of two NR1 subunits and two NR2 subunits. It is established that the NMDA receptor's function in synaptic plasticity is largely mediated by the NR2 intracellular C-terminal domain, which interacts directly and indirectly with numerous postsynaptic signalling molecules (Sprengel et al '98, Migaud et al '98, Husi et al '00). The purpose of this study was to address questions of evolutionary conservation of the NR2 C-terminal domain and structural properties.
Ryan et al's study provides two main insights. The first was based on comparisons of the length of the NR2 C-terminus between paralogues and across species. Through simple amino acid sequence alignments of NR2 orthologues it became clear that the NR2 C-terminus profoundly differs in size between vertebrates and invertebrates, with vertebrates possessing C-termini of up 500 amino acids larger than all sequenced invertebrates. This suggests that vertebrate NR2 subunits have a greater set of protein interactions than invertebrate NR2. Moreover, within the vertebrate clade, alignments of the four NR2 paralogues show that that while the N-terminal regions of the four NR2 subunits are highly conserved, they have diverged significantly in their C-terminal domains, implying that NR2 subunits functionally differ primarily at the level of intracellular signalling. Secondly, Ryan et al applied computational tools to predict the structure of the vertebrate NR2 C-terminus. In doing so it became apparent that despite being very distinct at the amino acid sequence level, each of the NR2 C-termini domains share a common intrinsically disordered structure. Moreover, NR2A and NR2B have a similar potential secondary structure.
Taken together, these results infer an evolutionary scenario where the vertebrate NMDA receptor has evolved a large NR2 C-terminal domain with the capacity to function as a multi-protein hub. The NR2 gene duplicated in a common ancestor of all vertebrates, creating four NR2 paralogues that have diverged in their C-terminal domains to allow differential signalling and/or synaptic localisation. Nevertheless the four NR2 subunits maintained the common ability to function as a postsynaptic signalling hub. Consistent with at this model, a larger Genes to Cognition study has inferred a significant difference in the molecular complexity of postsynaptic proteomes between vertebrates and invertebrates, with greater complexity at vertebrates synapses (Emes et al '08). Thus, the molecular evolution of the NMDA receptor and associated signalling complexes is likely to have contributed to the evolution of higher physiological and behavioural properties in vertebrates. On-going transgenic studies will test these hypotheses.
References
- Ryan TJ, Emes RD, Grant SG, Komiyama NH. BMC Neuroscience 2008, Jan 15;9:6
- Xia S, Miyashita T, Fu TF, Lin WY, Wu CL, Pyzocha L, Lin IR, Saitoe M, Tully T, Chiang AS. Current Biology 2005 Apr 12;15(7):603-15
- Sprengel R, Suchanek B, Amico C, Brusa R, Burnashev N, Rozov A, Hvalby O, Jensen V, Paulsen O, Andersen P, Kim JJ, Thompson RF, Sun W, Webster LC, Grant SG, Eilers J, Konnerth A, Li J, McNamara JO, Seeburg PH. Cell 1998 Jan 23;92(2):279-89
- Migaud M, Charlesworth P, Dempster M, Webster LC, Watabe AM, Makhinson M, He Y, Ramsay MF, Morris RG, Morrison JH, O'Dell TJ, Grant SG. Nature 1998 Dec 3;396(6710):433-9.
- Husi H, Ward MA, Choudhary JS, Blackstock WP, Grant SG. Nature Neuroscience 2000 Jul 3(7):661-9
- Richard D. Emes, Andrew J. Pocklington, Christopher N.G. Anderson, Alex Bayes, Mark O. Collins, Catherine A. Vickers, Mike D.R. Croning, Bilal R. Malik, Jyoti S. Choudhary, J. Douglas Armstrong, Seth G.N. Grant. Nature Neuroscience 2008 (in press)
