G2C::Electrophysiology::Brain Slice
Contemporary proteomic studies demonstrate a complex structure of the brain synapse with over a 1000 different proteins being associated with the postsynaptic density. However, the function of about 80% of these proteins is still poorly understood and there is an urgent need in experimental platforms that would permit their expedient characterisation.
Electrophysiological methods offer a straightforward quantitative way to assess consequences of the loss, mutation or over-expression of a particular protein since it is plausible that many of the neurone-specific proteins would be important for synaptic transmission. Classical electrophysiological approaches produce extremely useful sets of data however they are not particularly amenable for increased throughput required for description of a multitude of novel proteins.
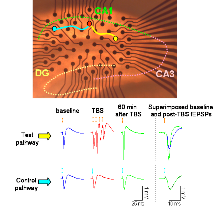
Figure 1: a thin (350 m thick) hippocampal slice rests on top of a multi-electrode array containing 60 electrodes.
An ideal way of synaptic proteome profiling must also take into account the fact that magnitude of signalling between neurones is an activity-dependent variable. In vitro models recapitulating short- and long-term plastic properties of synaptic transmission have become an extremely popular way to assess functions of proteins following their pharmacological and/or genetic manipulation. Long-term potentiation (LTP) of synaptic transmission evoked by trains of patterned stimulation represents one of the most widely used models to study plastic properties of the synapse. Its application to the versatile hippocampal slice preparation has made LTP a quintessential element of many studies that aimed to elucidate functional roles of brain proteins.
In our attempt to systematically describe roles of different postsynaptic proteins, we study synaptic transmission and its plasticity in CA1 area of hippocampal slices prepared from brains of mutant mice generated within G2C Programme. An important part of this large-scale screening effort is maximisation of throughput and standardisation of experimental conditions. For this purpose, we employ novel multi-electrode array (MEA) technology that offers excellent multiplexing capabilities and sets clear spatial landmarks necessary for stereotypic arrangement of stimulation and recording sites.
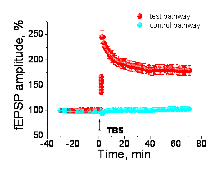
Figure 2 illustrates a typical LTP summary graph (mean ± s.e.m.) for wild-type littermates of one of the mutant strains studied. Normalised test pathway and control pathway fEPSPs are plotted in red and blue symbols respectively
In Figure 1 above, a thin (350 m thick) hippocampal slice rests on top of a multi-electrode array containing 60 electrodes. Field excitatory postsynaptic potentials (fEPSPs) are recorded by electrodes in the upper part of the array, including the principal recording electrode marked by the red circle. fEPSPs are evoked by successive stimulations of the test pathway (yellow electrode) and control pathway (blue electrode). Following theta-burst stimulation of the test (yellow) pathway, consistently larger responses are observed in that pathway (as shown in the overlaid baseline and post-TBS fEPSP traces), whereas the amplitude of the fEPSP in the control (blue) pathway stays the same. Time scale of superimposed recordings was enlarged for convenience.
Currently, we use four Multi Channel Systems MCS GmbH MEA set-ups and analyse up to 4 slices per animal per day. A typical experimental cohort includes 5 mutant and 5 litter-matched wild-type mice. Therefore, characterisation of one strain typically involves two weeks (10 working days) and yields data that are averages of up to 20 experimental points. Our plan is to expand the number of set-ups, in order to cut phenotyping period up to one week.
Our studies will help to shed light on hitherto unknown neuronal functions of many synaptic proteins by assaying effects of their absence (knock-out) or modification (domain deletion, knock-in) on basal properties of synaptic transmission and its plasticity. Combined with results of other electrophysiological studies (network activity in cell cultures) as well as biochemical and behavioural experiments, slice electrophysiology data will be an important element of G2C multifaceted characterisation of postsynaptic proteome.
Phenotyping Protocols
| Protocol | Description | View |
|---|---|---|
| P00000013 | Hippocampal slice electrophysiology | View |
