G2C::Proteomics
Comparative analysis of human and mouse postsynaptic proteomes identifies high compositional conservation and abundance differences for key synaptic proteins
Àlex Bayés1,2,3*,Mark O Collins4*, Mike DR Croning2,3, Louie N van de Lagemaat2,3, Jyoti S Choudhary4 and Seth GN Grant2,3*
Corresponding authors: Seth.Grant@ed.ac.uk, ABayesP@santpau.cat
- Molecular Physiology of the Synapse Laboratory, Institut de Recerca de l’Hospital de la Santa Creu i Sant Pau, UAB. C/ Sant Antoni Maria Claret 167, 08025 Barcelona, Catalonia, Spain
- Genes to Cognition Programme, The Wellcome Trust Sanger Institute, Genome Campus, Hinxton, Cambridgeshire, CB10 1SA, UK
- Centre for Clinical Brain Sciences and Centre for Neuroregeneration, The University of Edinburgh, Chancellors Building, 47 Little France Crescent, Edinburgh EH16 4SB, UK
- Proteomic Mass Spectrometry. The Wellcome Trust Sanger Institute, Hinxton, Cambridgeshire, CB10 1SA, UK
Overview
Direct comparison of protein components from human and mouse excitatory synapses is important for determining the suitability of mice as models of human brain disease and to understand the evolution of the mammalian brain. The postsynaptic density (PSD) is a highly complex set of proteins organized into molecular networks that play a central role in behavior and disease. We report the first direct comparison of the proteome of triplicate isolates of mouse and human cortical PSDs. The mouse PSD (mPSD) comprised 1556 proteins and the human PSD (hPSD) 1461.
A large compositional overlap was observed; more than 70% of hPSD proteins were also observed in the mPSD. Quantitative analysis of PSD components in both species indicates a broadly similar profile of abundance but also shows that there is higher abundance variation between species than within species.
Well-known PSD components are generally more abundant in the mouse PSD. Significant inter-species abundance differences exist in some families of key PSD proteins including glutamatergic neurotransmitter receptors and adaptor proteins. We have identified a closely interacting set of molecules enriched in the human PSD that could be involved in dendrite and spine structural plasticity. Understanding synapse proteome diversity within and between species will be important to further our understanding of brain complexity and disease.
The full results and analyses of the study can be sorted, searched and downloaded
Figures
- Figure 1.
- Validation of fractionation method and quantitative comparison of mouse and human cortical PSDs.
- Figure 2.
- Analysis of evolutionary conservation of PSD proteins in mouse and humans.
- Figure 3.
- Inter-species differences in postsynaptic density proteins.
- Figure 4.
- Bioinformatic functional analysis of proteins enriched in the human PSD.
- Figure 5.
- Predicted functional protein groups with the mammalian cortical PSD.
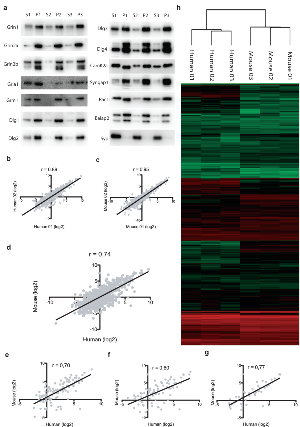
Figure 1. Validation of fractionation method and quantitative comparison of mouse and human cortical PSDs.
a. Immunoblots of known postsynaptic density components. Mouse synaptosomes (S) and PSD (P) fractions are analysed for each of the three replicates. All proteins show enrichment in the PSD compared to synaptosomes. The presynaptic marker synaptophysin (Syn) is only detected in the synaptosomal fraction.
Only genes identified in triplicate in both species were considered for the quantitative analysis. Correlations are calculated based on linear models. Coefficients of correlations are represented by r.
b. Intra-species protein abundance correlation, human PSD first replica against second
c. Intra-species protein abundance correlation, mouse PSD first replica against second.
d. Plot of average values of PSD proteins abundance from human and mouse cortex.
e. Protein abundance correlation of PSD components found in the NMDA receptor complex [3];
f. Protein abundance correlation of PSD components found in the PSD-95 complex [26].
g.Protein abundance correlation of PSD components found in the mGluR5 complex [27].
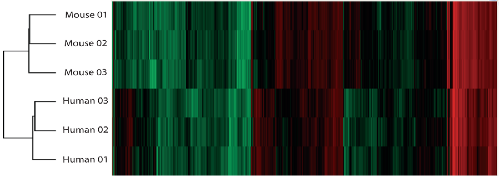
h. Clustering of PSD protein abundance values for human and mouse replicates.
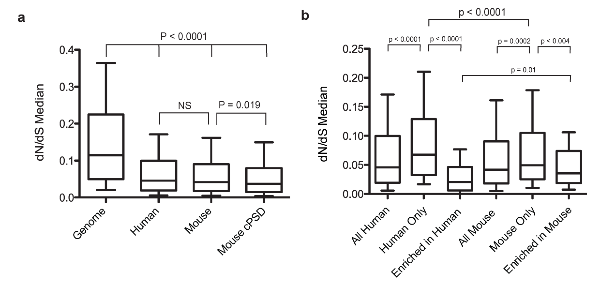
Figure 2. Analysis of evolutionary conservation of PSD proteins in mouse and humans.
a.Median dN/dS of proteins identified in the human PSD, mouse PSD and mouse consensus set (cPSD) compared with the median genomic value.
b.Comparison of dN/dS values in proteins exclusive or enriched in one species with all PSD proteins.
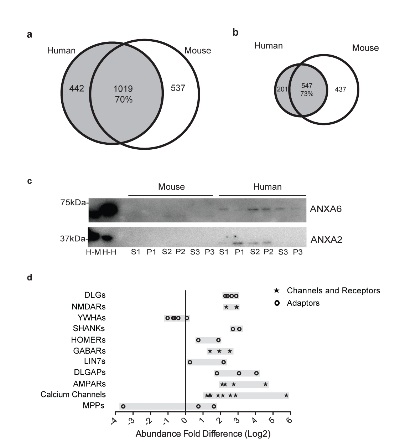
Figure 3. Inter-species differences in postsynaptic density proteins.
a.Venn diagrams showing the total amount of proteins in mPSD and hPSD as well as the overlap between them relative to the hPSD.
b.Venn diagrams showing the proteins identified in triplicate for the mPSD and hPSD, the overlap relative to hPSD between the two sets is also shown.
c.Immunoblot validation of proteins identified only in the hPSD by mass spectrometry. The following samples were analysed: H-M, mouse brain homogenate; H-H, human brain homogenate; and SX, each of the three synaptosomal fractions, PX, each of the three postsynaptic density fractions for both species.
d.The abundance fold difference (AFD) between mouse and human is shown for members of adaptors, channels and neurotransmitter receptors protein families. AFD is calculated as the mouse to human ratio of normalized and log2 transformed average abundance values. For each protein family the number of proteins displayed is given in brackets.
Figure 4. Bioinformatic functional analysis of proteins enriched in the human PSD.
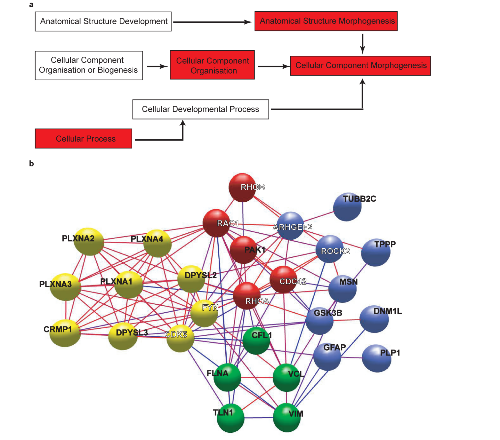
a.Ontology hierarchical tree leading to the Biological Process ‘Cellular Component Morphogenesis’. In red are the terms significantly enriched in proteins abundant in the human PSD.
b.Molecular interaction map of proteins from the human PSD that belong to the Biological Process ‘Cellular Component Morphogenesis’ (CCM) or are known to be involved in semaphorin signaling. Only CCM proteins enriched in the human PSD are considered. Some proteins related to semaphorin signaling are not enriched in either species, these are identified with a white name. Proteins highly interconnected are represented in clusters of different colors. Clustering was done using the Markov Clustering Algorithm (MCL) in BioLayout Express 3D [24]. Node edges are coloured according to a confidence score, from red (100% confidence) to blue (40% confidence).
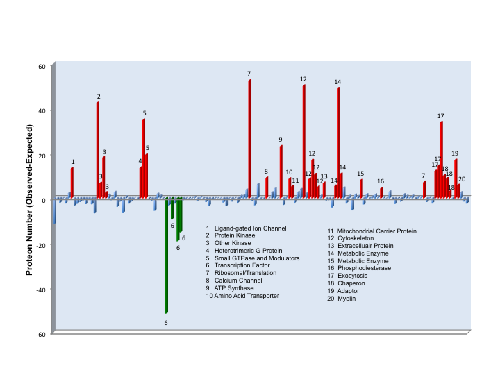
Figure 5. Predicted functional protein groups with the mammalian cortical PSD.
The mammalian cortical PSD was defined by combining all proteins identified in mouse and human. This set of PSD proteins were then functionally classified using the Panther Classification descriptor ‘Protein Class’. Enrichment analysis was done comparing the number of PSD proteins per class with the genome. The Panther Protein Class ontology is hierarchically organized, only end-branch classes are displayed to remove redundancy from the graph (all data in Table S8). Protein classes that did not show difference with the genome after correction for multiple testing, are shown in blue. Classes overrepresented are shown in red and classes underrepresented are shown in green.
Tables
- Table 1.
- Significantly enriched protein classes of species-specific PSD proteins.
- Table 2.
- Abundance of canonical PSD components as defined by studies with rodent brains.
Table 1. Significantly enriched protein classes of species-specific PSD proteins.
| PANTHER Protein Class | Proteins Identified | p-value | Proteins Identified | p-value | |
|---|---|---|---|---|---|
| PANTHER Protein Class | PSD Proteins | Consensus PSD Proteins | Enriched In | ||
| Cytoskeletal protein | 36 | 1.30E-09 | 16 | 3.60E-07 | Human |
| Actin family cytoskeletal protein | 22 | 7.60E-08 | 10 | 3.40E-05 | Human |
| Oxidoreductase | 39 | 5.20E-09 | 13 | 5.10E-04 | Human |
| Dehydrogenase | 22 | 4.50E-08 | 8 | 8.90E-04 | Human |
| Non-motor actin binding protein | 14 | 3.40E-06 | 6 | 1.40E-03 | Human |
| Reductase | 8 | 1.10E-03 | 4 | 1.40E-02 | Human |
| Transferase | 70 | 1.40E-08 | 18 | 2.50E-02 | Human |
| Microtubule family cytoskeletal protein | 11 | 5.90E-03 | 5 | 3.30E-02 | Human |
| Transfer/carrier protein | 14 | 8.50E-03 | 6 | 4.30E-02 | Human |
| Membrane trafficking regulatory protein | 16 | 4.70E-08 | 4 | 4.30E-02 | Human |
| Nucleotide kinase | 10 | 3.40E-06 | 3 | 4.30E-02 | Human |
| Membrane traffic protein | 27 | 4.70E-08 | 5 | 3.00E-01 | Human |
| Kinase | 40 | 1.20E-07 | 8 | 3.00E-01 | Human |
| Protein kinase | 25 | 1.20E-03 | 5 | 7.60E-01 | Human |
| Enzyme modulator | 33 | 6.50E-03 | 7 | 7.90E-01 | Human |
| G-protein modulator | 15 | 8.50E-03 | 1 | 1.00E+00 | Human |
| Ribosomal protein | 29 | 8.00E-13 | 16 | 2.30E-09 | Mouse |
| Calcium channel | 7 | 2.90E-04 | 6 | 3.90E-05 | Mouse |
| RNA binding protein | 60 | 1.20E-13 | 20 | 3.90E-04 | Mouse |
| Ion channel | 31 | 3.70E-08 | 12 | 2.10E-03 | Mouse |
| Nucleic acid binding | 78 | 6.70E-09 | 28 | 2.50E-03 | Mouse |
| SNARE protein | 8 | 1.10E-04 | 4 | 9.40E-03 | Mouse |
| Transporter | 57 | 4.10E-06 | 19 | 8.80E-02 | Mouse |
| Ligand-gated ion channel | 11 | 9.60E-04 | 4 | 2.20E-01 | Mouse |
Table 2. Abundance of canonical PSD components as defined by studies with rodent brains.
Mouse to human abundance difference of proteins known to be key components of the postsynaptic density, as derived from studies on mouse and rat brains. Abundance is expressed as the fold of mouse to human average abundance values.
| Appr Gene (m) | Appr Gene (h) | Abundance Fold Difference (Mouse/Human) | Significantly Enriched in | Appr Gene (m) | Appr Gene (h) | Abundance Fold Difference (Mouse/Human) | Significantly Enriched |
|---|---|---|---|---|---|---|---|
| Ablim1 | ABLIM1 | 1.3 | Gria3 | GRIA3 | 4.3 | Mouse | |
| Baiap2 | BAIAP2 | 3.1 | Gria4 | GRIA4 | 23.8 | Mouse | |
| Begain | BEGAIN | 10.4 | Mouse | Grin1 | GRIN1 | 7.7 | Mouse |
| Cacng2 | CACNG2 | 2.6 | Grin2a | GRIN2A | 5 | Mouse | |
| Camk2a | CAMK2A | 1 | Grin2b | GRIN2B | 7.5 | ||
| Camk2b | CAMK2B | 3.4 | Homer1 | HOMER1 | 3.7 | Mouse | |
| Camk2d | CAMK2D | 0.9 | Homer2 | HOMER2 | 1.7 | ||
| Capza2 | CAPZA2 | 1.6 | Iqsec1 | IQSEC1 | 2.4 | ||
| Cntnap1 | CNTNAP1 | 0.5 | Iqsec2 | IQSEC2 | 2.7 | Mouse | |
| Cntnap2 | CNTNAP2 | 2.1 | Kalrn | KALRN | 8.3 | Mouse | |
| Dlg1 | DLG1 | 5.3 | Mouse | Kcnab2 | KCNAB2 | 4.4 | |
| Dlg2 | DLG2 | 6.3 | Mouse | Kcnq2 | KCNQ2 | 0.9 | |
| Dlg3 | DLG3 | 7.5 | Mouse | Lgi1 | LGI1 | 11 | Mouse |
| Dlg4 | DLG4 | 4.9 | Magi2 | MAGI2 | 0.5 | ||
| Dlgap1 | DLGAP1 | 3.5 | Mouse | Nlgn2 | NLGN2 | 4.4 | |
| Dlgap2 | DLGAP2 | 8.3 | Mouse | Nrxn1 | NRXN1 | 1.5 | |
| Dlgap3 | DLGAP3 | 8.3 | Mouse | Nrxn3 | NRXN3 | 3.8 | Mouse |
| Dlgap4 | DLGAP4 | 16.6 | Mouse | Rac1 | RAC1 | 1.7 | |
| Gabbr1 | GABBR1 | 5.8 | Shank2 | SHANK2 | 8.4 | Mouse | |
| Gabbr2 | GABBR2 | 3.8 | Mouse | Shank3 | SHANK3 | 6.5 | Mouse |
| Gabra1 | GABRA1 | 2.6 | Syngap1 | SYNGAP1 | 5.9 | Mouse | |
| Gnas | GNAS | 1.1 | Ywhae | YWHAE | 1 | ||
| Gria1 | GRIA1 | 6.7 | Ywhah | YWHAH | 0.5 | Human | |
| Gria2 | GRIA2 | 4.8 | Mouse | Ywhaq | YWHAQ | 0.6 |
Supplementary Tables
- Supplementary Table 1.
- Protein Identifications and Proteomic Data.
- Supplementary Table 2a.
- PSD protein abundance (all)
- Supplementary Table 2b.
- PSD protein abundance (enriched in human)
- Supplementary Table 2c.
- PSD protein abundance (enriched in mouse)
- Supplementary Table 2d.
- PSD protein abundance (PSD complexes)
- Supplementary Table 3a.
- Proteins only detected in human PSD
- Supplementary Table 3b.
- Proteins only detected in mouse PSD
- Supplementary Table 4a.
- Functional comparison of human and mouse PSD proteins (all)
- Supplementary Table 4b.
- Functional comparison of human and mouse PSD proteins (consensus)
- Supplementary Table 5.
- Pathways enriched in human and mouse unique or enriched proteins.
- Supplementary Table 6.
- GO Biological Process term enrichment in human and mouse unique or enriched proteins.
- Supplementary Table 7.
- Proteins related to axon guidance or cell morphology.
- Supplementary Table 8.
- Functional comparison of all PSD proteins identified in human and mouse.
- Supplementary Table 9a.
- Pathway analysis of all PSD proteins identified in human and mouse.
- Supplementary Table 9b.
- Pathway analysis summary of all PSD proteins identified in human and mouse.
- Supplementary Table 10.
- Analysis of dN/dS values from mouse and human PSD proteins.
Supplementary Table 1. Protein Identifications and Proteomic Data.
For each identified protein several identification (ID) numbers from biological databases are given. The number of total and uniquely identified peptides for each replicate is also provided. Proteins found with two or more peptides in all replicates are classified as members of the consensus mouse PSD. Human orthologues to mouse proteins are provided. Data regarding human PSD proteins is from: Bayes A et al. Nat Neurosci. 2011 Jan;14(1):19-21.
| Internal ID | Detected Mouse | Detected Human (PMID:21170055) | Appr Gene (m) | MGI ID | Appr Gene (h) | HGNC ID | Ensembl (m) | Ensembl (h) | Entrez Gene (m) | Entrez Gene (h) | Cluster Number | Gene (m) | Gene (h) | IPI (m) | UP (m) | IPI (h) | UP (h) | Shared peptides | mPSD 010203 | mPSD01 | mPSD02 | mPSD03 | Present in 3 replicas (m) | Present in 3 replicas (h) (PMID:21170055) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ab16moc_919c | AMBIGUOUS | AMBIGUOUS | Calm3 | MGI:103249 | CALM3 | HGNC:1449 | ENSMUSG00000019370 | ENSG00000160014 | 12315 | 808 | Cluster 451 | Calm1|Calm2|Calm3 | CALM1|CALM2|CALM3 | IPI00761696 | P62204 | IPI00386621 | P62158 | Shared peptides with CALM1|CALM2|CALM3 | 4 | 1 | 1 | 4 | ||
| ab16moc_919a | AMBIGUOUS | AMBIGUOUS | Calm1 | MGI:88251 | CALM1 | HGNC:1442 | ENSMUSG00000001175 | ENSG00000160014 | 12313 | 801 | Cluster 451 | Calm1|Calm2|Calm3 | CALM1|CALM2|CALM3 | IPI00761696 | P62204 | IPI00386621 | P62158 | Shared peptides with CALM1|CALM2|CALM3 | 4 | 1 | 1 | 4 | ||
| ab16moc_919b | AMBIGUOUS | AMBIGUOUS | Calm2 | MGI:103250 | CALM2 | HGNC:1445 | ENSMUSG00000036438 | ENSG00000143933 | 12314 | 805 | Cluster 451 | Calm1|Calm2|Calm3 | CALM1|CALM2|CALM3 | IPI00761696 | P62204 | IPI00386621 | P62158 | Shared peptides with CALM1|CALM2|CALM3 | 4 | 1 | 1 | 4 | ||
| ab16moc_920 | YES | AMBIGUOUS | Arf3 | MGI:99432 | ARF3 | HGNC:654 | ENSMUSG00000051853 | ENSG00000134287 | 11842 | 377 | Cluster 382 | Arf3 | ARF3 | IPI00221614 | P61205 | IPI00215917 | P61204 | Shared peptides with ARF1 | 4 | 4 | 3 | 3 | YES | YES |
| ab16moc_925 | YES | AMBIGUOUS | Arf1 | MGI:99431 | ARF1 | HGNC:652 | ENSMUSG00000048076 | ENSG00000143761 | 11840 | 375 | Cluster 382 | Arf1 | ARF1 | IPI00221613 | P84078 | IPI00215914 | P84077 | Shared peptides with ARF3 | 4 | 4 | 3 | 3 | YES | YES |
| ab16moc_505 | YES | AMBIGUOUS | Hspa1l | MGI:96231 | HSPA1L | HGNC:5234 | ENSMUSG00000007033 | ENSG00000204390 | 15482 | 3305 | Cluster 307 | Hspa1l | HSPA1L | IPI00133208 | P16627 | IPI00301277 | P34931 | Shared peptides with Hspa1a|Hspa1b | 9 | 9 | 6 | 8 | YES | YES |
| ab16moc_465a | YES | AMBIGUOUS | Hspa1a | MGI:96244 | HSPA1A | HGNC:5232 | ENSMUSG00000067283 | ENSG00000204389 | 193740 | 3303 | Cluster 307 | Hspa1a|Hspa1b | HSPA1A|HSPA1B | IPI00798482 | Q61696 | IPI00304925 | P08107 | Shared peptides with HSPA1L | 10 | 9 | 6 | 9 | YES | YES |
| ab16moc_465b | AMBIGUOUS | Hspa1b | MGI:99517 | HSPA1B | HGNC:5233 | ENSMUSG00000067284 | ENSG00000204388 | 15511 | 3304 | Cluster 307 | Hspa1a|Hspa1b | HSPA1A|HSPA1B | IPI00798482 | Q61696 | IPI00304925 | P08107 | Shared peptides with HSPA1L | 10 | 9 | 6 | 9 | YES | YES | |
| ab16moc_1868 | AMBIGUOUS | Tubg2 | MGI:2144208 | TUBG2 | HGNC:12419 | ENSMUSG00000045007 | ENSG00000037042 | 103768 | 27175 | Cluster 2218 | TUBG2 | IPI00299830 | Q9NRH3 | Shared peptides with TUBG1 | ||||||||||
| ab16moc_1867 | AMBIGUOUS | Tubg1 | MGI:101834 | TUBG1 | HGNC:12417 | ENSMUSG00000035198 | ENSG00000131462 | 103733 | 7283 | Cluster 2218 | TUBG1 | IPI00295081 | P23258 | Shared peptides with TUBG2 | ||||||||||
| ab16moc_1050 | YES | YES | Mrps36 | MGI:1913378 | MRPS36 | HGNC:16631 | ENSMUSG00000061474 | ENSG00000134056 | 66128 | 92259 | Cluster 372 | Mrps36 | MRPS36 | IPI00315808 | Q9CQX8 | IPI00020495 | P82909 | 3 | 2 | 1 | 2 | |||
| ab16moc_1307 | YES | YES | Prr7 | MGI:3487246 | PRR7 | HGNC:28130 | ENSMUSG00000034686 | ENSG00000131188 | 432763 | 80758 | Cluster 1663 | Prr7 | PRR7 | IPI00132812 | Q3V0I2 | IPI00152142 | Q8TB68 | 2 | 2 | 2 | 2 | YES | ||
| ab16moc_1313 | YES | YES | Usmg5 | MGI:1891435 | USMG5 | HGNC:30889 | ENSMUSG00000071528 | ENSG00000173915 | 66477 | 84833 | Cluster 463 | Usmg5 | USMG5 | IPI00624653 | Q78IK2 | IPI00063903 | Q96IX5 | 2 | 2 | 2 | ||||
| ab16moc_514 | YES | YES | Cend1 | MGI:1929898 | CEND1 | HGNC:24153 | ENSMUSG00000060240 | ENSG00000184524 | 57754 | 51286 | Cluster 77 | Cend1 | CEND1 | IPI00122826 | Q9JKC6 | IPI00295601 | Q8N111 | 9 | 8 | 7 | 7 | YES | YES | |
| ab16moc_560 | YES | YES | N28178 | MGI:2140712 | KIAA1045 | HGNC:29180 | ENSMUSG00000078351 | ENSG00000122733 | 230085 | 23349 | Cluster 728 | KIAA1045 | N28178 | IPI00647986 | Q80TL4 | IPI00414898 | Q9UPV7 | 8 | 3 | 7 | 3 | YES | YES | |
| ab16moc_597 | YES | YES | Ccdc127 | MGI:1914683 | CCDC127 | HGNC:30520 | ENSMUSG00000021578 | ENSG00000164366 | 67433 | 133957 | Cluster 512 | Ccdc127 | CCDC127 | IPI00131843 | Q3TC33 | IPI00060148 | Q96BQ5 | 8 | 8 | 4 | 3 | YES | ||
| ab16moc_713 | YES | YES | Calcoco1 | MGI:1914738 | CALCOCO1 | HGNC:29306 | ENSMUSG00000023055 | ENSG00000012822 | 67488 | 57658 | Cluster 1537 | Calcoco1 | CALCOCO1 | IPI00461826 | Q5DTX0 | IPI00011232 | Q6FI59 | 6 | 3 | 4 | 5 | YES | ||
| ab16moc_931 | YES | YES | 1810074P20Rik | MGI:1914740 | KIAA0776 | HGNC:23039 | ENSMUSG00000040359 | ENSG00000014123 | 67490 | 23376 | Cluster 2090 | KIAA0776 | KIAA0776 | IPI00317684 | Q8CCJ3 | IPI00844000 | O94874 | 4 | 4 | |||||
| ab16moc_155 | YES | YES | Ckap4 | MGI:2444926 | CKAP4 | HGNC:16991 | ENSMUSG00000046841 | ENSG00000136026 | 216197 | 10970 | Cluster 193 | Ckap4 | CKAP4 | IPI00223047 | Q8BMK4 | IPI00433214 | Q6NWZ1 | 27 | 24 | 17 | 21 | YES | YES | |
| ab16moc_362 | YES | YES | 2010300C02Rik | MGI:1919347 | C2orf55 | HGNC:33454 | ENSMUSG00000026090 | ENSG00000196872 | 72097 | 343990 | "Cluster 1084, Cluster 2471" | 2010300C02Rik | C2ORF55 | IPI00454053 | Q6GQT3 | IPI00410436 | Q6NV74 | 14 | 12 | 11 | 4 | YES | ||
| ab16moc_857 | YES | YES | 2310022B05Rik | MGI:1916801 | C1orf198 | HGNC:25900 | ENSMUSG00000031983 | ENSG00000119280 | 69551 | 84886 | "Cluster 831, Cluster 1477" | 2310022B05Rik | C1ORF198 | IPI00225267 | Q8C3W1 | IPI00013912 | Q9H425 | 5 | 2 | 2 | 5 | YES | ||
| ab16moc_1315 | YES | YES | Agpat5 | MGI:1196345 | AGPAT5 | HGNC:20886 | ENSMUSG00000031467 | ENSG00000155189 | 52123 | 55326 | Cluster 2049 | Agpat5 | AGPAT5 | IPI00221486 | Q9D1E8 | IPI00028491 | Q9NUQ2 | 2 | 2 | 1 | ||||
| ab16moc_426 | YES | YES | Plcb1 | MGI:97613 | PLCB1 | HGNC:15917 | ENSMUSG00000051177 | ENSG00000182621 | 18795 | 23236 | Cluster 595 | Plcb1 | PLCB1 | IPI00468121 | Q9Z1B3 | IPI00395561 | Q9NQ66 | 11 | 6 | 8 | 4 | YES | YES | |
| ab16moc_831 | YES | YES | Plcb4 | MGI:107464 | PLCB4 | HGNC:9059 | ENSMUSG00000039943 | ENSG00000101333 | 18798 | 5332 | Cluster 1985 | Plcb4 | PLCB4 | IPI00625848 | Q8BXH1 | IPI00014897 | Q15147 | 5 | 4 | 2 | 3 | YES | ||
| ab16moc_54 | YES | YES | Cnp | MGI:88437 | CNP | HGNC:2158 | ENSMUSG00000006782 | ENSG00000173786 | 12799 | 1267 | Cluster 11 | Cnp1 | CNP | IPI00319602 | P16330 | IPI00306667 | P09543 | 46 | 36 | 40 | 40 | YES | YES |
Supplementary Table 2a. PSD protein abundance (all)
Individual and average abundance values (iBAQ) for mouse and human PSD proteins are given. Data was normalized and log2 transformed. Normalization was achieved by dividing abundance data points by its species abundance average. Abundance fold difference was defined as the ratio of mouse to human average abundance values. To measure significant abundance differences between species a Student’s t-test was used. Proteins significantly enriched in one species are shown in separate sheets. The abundance of proteins from the postsynaptic protein complexes shown in figure 1 are also shown in a separate sheet.
Individual and median abundance values (iBAQ) for mouse and human PSD proteins are given. Normalisation was done by dividing abundace by each species abundance median. Abundance fold difference is defined as the ratio of mouse by human normalised abundance values.
| Appr Gene (m) | MGI ID | Appr Gene (h) | HGNC ID | Ensembl (m) | Ensembl (h) | human ibaq protein group id | mouse ibaq protein group id | iBAQ mPSD01 (Log2, normalised) | iBAQ mPSD02 (Log2, normalised) | iBAQ mPSD03 (Log2, normalised) | iBAQ hPSD01 (Log2, normalised) | iBAQ hPSD02 (Log2, normalised) | iBAQ hPSD03 (Log2, normalised) | iBAQ hPSD average (Log2, normalised) | iBAQ mPSD average (Log2, normalised) | Average fold change (mPSD from hDB/hPSD from mDB) | t-test Significant (Perm 0.01) | t-test p value |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sept2 | MGI:97298 | SEPT2 | HGNC:7729 | ENSMUSG00000026276 | ENSG00000168385 | 485 | 207 | 3.336 | 3.186 | 4.176 | 3.687 | 5.141 | 4.505 | 4.445 | 3.566 | 0.544 | 0.1674 | |
| Sept3 | MGI:1345148 | SEPT3 | HGNC:10750 | ENSMUSG00000022456 | ENSG00000100167 | 1858 | 2331 | 5.443 | 5.234 | 5.717 | 4.861 | 5.084 | 5.635 | 5.194 | 5.465 | 1.207 | 0.3712 | |
| Sept4 | MGI:1270156 | SEPT4 | HGNC:9165 | ENSMUSG00000020486 | ENSG00000181013 | 406 | 2170 | 1.193 | 0.978 | 1.011 | 0.019 | -0.254 | -0.014 | -0.083 | 1.061 | 2.209 | + | 0.0005 |
| Sept5 | MGI:1195461 | SEPT5 | HGNC:9164 | ENSMUSG00000072214 | ENSG00000184702 | 2308 | 2408 | 7.316 | 7.516 | 7.626 | -1.426 | 1.365 | -0.138 | -0.066 | 7.486 | 187.678 | + | 0.0007 |
| Sept6 | MGI:1888939 | SEPT6 | HGNC:15848 | ENSMUSG00000050379 | ENSG00000125354 | 1337 | 1133 | 3.242 | 4.874 | 4.896 | 2.798 | 3.562 | 1.913 | 2.758 | 4.338 | 2.989 | 0.0952 | |
| Sept7 | MGI:1335094 | SEPT7 | HGNC:1717 | ENSMUSG00000001833 | ENSG00000122545 | 1022 | 1098 | 7.574 | 7.457 | 7.264 | 6.964 | 7.466 | 6.894 | 7.108 | 7.432 | 1.251 | 0.1836 | |
| Sept8 | MGI:894310 | SEPT8 | HGNC:16511 | ENSMUSG00000018398 | ENSG00000164402 | 2128 | 2371 | 3.632 | 3.218 | 3.575 | 2.884 | 3.254 | 2.449 | 2.862 | 3.475 | 1.529 | 0.0829 | |
| Sept9 | MGI:1858222 | SEPT9 | HGNC:7323 | ENSMUSG00000059248 | ENSG00000184640 | 2294 | 2083 | 2.196 | 2.197 | 2.125 | 2.599 | 3.303 | 2.898 | 2.934 | 2.172 | 0.590 | 0.0208 | |
| Sept10 | MGI:1918110 | SEPT10 | HGNC:14349 | ENSMUSG00000019917 | ENSG00000186522 | 1828 | 1317 | 1.608 | 2.869 | 2.417 | -1.801 | -0.201 | -1.419 | -1.141 | 2.298 | 10.843 | + | 0.0048 |
| Sept11 | MGI:1277214 | SEPT11 | HGNC:25589 | ENSMUSG00000058013 | ENSG00000138758 | 2456 | 1862 | 7.728 | 7.159 | 7.480 | 7.955 | 7.946 | 8.179 | 8.027 | 7.456 | 0.673 | 0.0348 | |
| 1810074P20Rik | MGI:1914740 | KIAA0776 | HGNC:23039 | ENSMUSG00000040359 | ENSG00000014123 | 2334 | 2114 | 1.207 | 1.899 | 1.711 | -0.702 | -0.632 | 0.671 | -0.221 | 1.606 | 3.547 | 0.0206 | |
| 2400001E08Rik | MGI:1913758 | LAMTOR1 | HGNC:26068 | ENSMUSG00000030842 | ENSG00000149357 | 553 | 1421 | 0.113 | -0.713 | -0.906 | -0.413 | -0.142 | -1.146 | -0.567 | -0.502 | 1.046 | 0.8875 | |
| 2400003C14Rik | MGI:1919205 | KIAA0174 | HGNC:28977 | ENSMUSG00000031729 | ENSG00000182149 | 781 | 66 | 0.690 | 1.032 | 0.450 | 0.762 | 1.384 | 2.047 | 1.398 | 0.724 | 0.627 | 0.1736 | |
| Aak1 | MGI:1098687 | AAK1 | HGNC:19679 | ENSMUSG00000057230 | ENSG00000115977 | 2421 | 1690 | 2.670 | 2.174 | 2.792 | 0.914 | 0.893 | 1.461 | 1.090 | 2.545 | 2.743 | + | 0.0054 |
| Aars | MGI:2384560 | AARS | HGNC:20 | ENSMUSG00000031960 | ENSG00000090861 | 879 | 1490 | 2.670 | -1.091 | -4.910 | -0.852 | -1.116 | -0.430 | -0.799 | -1.110 | 0.806 | 0.8943 | |
| Abcd2 | MGI:1349467 | ABCD2 | HGNC:66 | ENSMUSG00000055782 | ENSG00000173208 | 107 | 1364 | -0.642 | -1.128 | -2.418 | -0.778 | -0.382 | -0.581 | -0.580 | -1.396 | 0.568 | 0.2066 | |
| Abcd3 | MGI:1349216 | ABCD3 | HGNC:67 | ENSMUSG00000028127 | ENSG00000117528 | 94 | 864 | -1.540 | -1.040 | -1.266 | 0.217 | -0.929 | -0.275 | -0.329 | -1.282 | 0.517 | 0.0581 | |
| Abhd16a | MGI:99476 | ABHD16A | HGNC:13921 | ENSMUSG00000007036 | ENSG00000204427 | 1024 | 694 | -2.434 | -1.152 | -2.737 | -1.349 | -0.819 | -0.405 | -0.858 | -2.108 | 0.420 | 0.0883 | |
| Abi1 | MGI:104913 | ABI1 | HGNC:11320 | ENSMUSG00000058835 | ENSG00000136754 | 2011 | 1927 | 4.389 | 4.853 | 4.938 | 2.519 | 3.334 | 4.331 | 3.395 | 4.726 | 2.517 | 0.0731 | |
| Abi2 | MGI:106913 | ABI2 | HGNC:24011 | ENSMUSG00000026782 | ENSG00000138443 | 2030 | 1734 | 2.829 | 3.260 | 3.213 | 1.594 | 2.988 | 3.315 | 2.632 | 3.100 | 1.383 | 0.4389 | |
| Ablim1 | MGI:1194500 | ABLIM1 | HGNC:78 | ENSMUSG00000078103 | ENSG00000099204 | 1776 | 1688 | 0.902 | 1.554 | 1.489 | -0.062 | 1.860 | 1.144 | 0.981 | 1.315 | 1.261 | 0.6061 | |
| Ablim2 | MGI:2385758 | ABLIM2 | HGNC:19195 | ENSMUSG00000029095 | ENSG00000163995 | 2017 | 1136 | -2.949 | -1.597 | -1.847 | -2.017 | -0.338 | -0.640 | -0.998 | -2.131 | 0.456 | 0.1627 | |
| Ablim3 | MGI:2442582 | ABLIM3 | HGNC:29132 | ENSMUSG00000032735 | ENSG00000173210 | 1305 | 1989 | -1.000 | -0.658 | -0.596 | -3.797 | -2.034 | -2.183 | -2.672 | -0.751 | 3.785 | 0.0294 | |
| Abr | MGI:107771 | ABR | HGNC:81 | ENSMUSG00000017631 | ENSG00000159842 | 962 | 1762 | -2.507 | -4.425 | -3.230 | -3.250 | -0.571 | -0.829 | -1.550 | -3.387 | 0.280 | 0.1461 | |
| Acat1 | MGI:87870 | ACAT1 | HGNC:93 | ENSMUSG00000032047 | ENSG00000075239 | 961 | 956 | -2.814 | -3.035 | -1.048 | -2.587 | -1.578 | -3.925 | -2.697 | -2.299 | 1.318 | 0.6894 |
Supplementary Table 2b. PSD protein abundance (enriched in human)
Individual and average abundance values (iBAQ) for mouse and human PSD proteins are given. Data was normalized and log2 transformed. Normalization was achieved by dividing abundance data points by its species abundance average. Abundance fold difference was defined as the ratio of mouse to human average abundance values. To measure significant abundance differences between species a Student’s t-test was used. Proteins significantly enriched in one species are shown in separate sheets. The abundance of proteins from the postsynaptic protein complexes shown in figure 1 are also shown in a separate sheet.
Individual and median abundance values (iBAQ) for mouse and human PSD proteins are given. Normalisation was done by dividing abundace by each species abundance median. Abundance fold difference is defined as the ratio of mouse by human normalised abundance values.
| Appr Gene (m) | MGI ID | Appr Gene (h) | HGNC ID | Ensembl (m) | Ensembl (h) | human ibaq protein group id | mouse ibaq protein group id | iBAQ mPSD01 (Log2, normalised) | iBAQ mPSD02 (Log2, normalised) | iBAQ mPSD03 (Log2, normalised) | iBAQ hPSD01 (Log2, normalised) | iBAQ hPSD02 (Log2, normalised) | iBAQ hPSD03 (Log2, normalised) | iBAQ hPSD average (Log2, normalised) | iBAQ mPSD average (Log2, normalised) | Average fold change (mPSD from hDB/hPSD from mDB) | t-test Significant (Permutation 0.01) | t-test p value |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Stxbp1 | MGI:107363 | STXBP1 | HGNC:11444 | ENSMUSG00000026797 | ENSG00000136854 | 1098 | 1849 | -2.846 | -2.331 | -2.836 | 5.526 | 6.183 | 6.722 | 6.144 | -2.671 | 0.002 | + | 0.0000 |
| Vim | MGI:98932 | VIM | HGNC:12692 | ENSMUSG00000026728 | ENSG00000026025 | 1987 | 1156 | -0.116 | 0.132 | -0.019 | 5.177 | 6.275 | 5.295 | 5.583 | -0.001 | 0.021 | + | 0.0001 |
| Gdi1 | MGI:99846 | GDI1 | HGNC:4226 | ENSMUSG00000015291 | ENSG00000203879 | 357 | 1518 | -2.370 | -0.363 | -2.290 | 3.486 | 3.794 | 4.054 | 3.778 | -1.674 | 0.023 | + | 0.0013 |
| Gfap | MGI:95697 | GFAP | HGNC:4235 | ENSMUSG00000020932 | ENSG00000131095 | 818 | 272 | 1.695 | 1.634 | 1.823 | 6.280 | 7.516 | 6.863 | 6.886 | 1.717 | 0.028 | + | 0.0001 |
| Ppia | MGI:97749 | PPIA | HGNC:9253 | ENSMUSG00000071866 | ENSG00000196262 | 1994 | 2096 | -1.795 | -3.107 | -2.893 | 2.085 | 3.053 | 2.375 | 2.505 | -2.598 | 0.029 | + | 0.0005 |
| Dpysl3 | MGI:1349762 | DPYSL3 | HGNC:3015 | ENSMUSG00000024501 | ENSG00000113657 | 2369 | 443 | -4.083 | -3.283 | -4.568 | 0.937 | 0.551 | 1.464 | 0.984 | -3.978 | 0.032 | + | 0.0004 |
| Msn | MGI:97167 | MSN | HGNC:7373 | ENSMUSG00000031207 | ENSG00000147065 | 1426 | 91 | -2.338 | -2.133 | -2.721 | 2.050 | 2.761 | 2.120 | 2.311 | -2.397 | 0.038 | + | 0.0001 |
| Ldhb | MGI:96763 | LDHB | HGNC:6541 | ENSMUSG00000030246 | ENSG00000111716 | 1420 | 1203 | -2.648 | -2.299 | -3.636 | 2.522 | 1.676 | 1.184 | 1.794 | -2.861 | 0.040 | + | 0.0011 |
| Hspa1a | MGI:96244 | HSPA1A | HGNC:5232 | ENSMUSG00000067283 | ENSG00000204389 | 1712 | 1666 | -1.911 | -2.758 | -1.297 | 2.447 | 2.934 | 2.209 | 2.530 | -1.989 | 0.044 | + | 0.0007 |
| Hspa1b | MGI:99517 | HSPA1B | HGNC:5233 | ENSMUSG00000067284 | ENSG00000204388 | 1712 | 1666 | -1.911 | -2.758 | -1.297 | 2.447 | 2.934 | 2.209 | 2.530 | -1.989 | 0.044 | + | 0.0007 |
| Glipr2 | MGI:1917770 | GLIPR2 | HGNC:18007 | ENSMUSG00000028480 | ENSG00000122694 | 245 | 1853 | -0.304 | -0.957 | -2.347 | 3.452 | 3.101 | 3.092 | 3.215 | -1.203 | 0.047 | + | 0.0020 |
| Mapre2 | MGI:106271 | MAPRE2 | HGNC:6891 | ENSMUSG00000024277 | ENSG00000166974 | 2418 | 2383 | -4.583 | -2.726 | -3.024 | 0.961 | 0.353 | 1.591 | 0.969 | -3.444 | 0.047 | + | 0.0029 |
| Vps45 | MGI:891965 | VPS45 | HGNC:14579 | ENSMUSG00000015747 | ENSG00000136631 | 1100 | 516 | -2.319 | -2.146 | -2.311 | 1.450 | 2.695 | 2.086 | 2.077 | -2.259 | 0.050 | + | 0.0003 |
| Eno2 | MGI:95394 | ENO2 | HGNC:3353 | ENSMUSG00000004267 | ENSG00000111674 | 1339 | 1617 | -1.499 | -3.182 | -2.390 | 2.521 | 1.477 | 1.817 | 1.938 | -2.357 | 0.051 | + | 0.0017 |
| Aco2 | MGI:87880 | ACO2 | HGNC:118 | ENSMUSG00000022477 | ENSG00000100412 | 582 | 240 | -2.417 | -1.370 | -2.372 | 1.862 | 2.760 | 1.613 | 2.078 | -2.053 | 0.057 | + | 0.0011 |
| Uba1 | MGI:98890 | UBA1 | HGNC:12469 | ENSMUSG00000001924 | ENSG00000130985 | 2198 | 481 | -2.037 | -1.669 | -2.973 | 1.236 | 1.751 | 2.514 | 1.834 | -2.226 | 0.060 | + | 0.0016 |
| Fscn1 | MGI:1352745 | FSCN1 | HGNC:11148 | ENSMUSG00000029581 | ENSG00000075618 | 1180 | 1682 | -2.239 | -2.102 | -1.853 | 0.621 | 2.772 | 2.383 | 1.925 | -2.065 | 0.063 | + | 0.0040 |
| Plp1 | MGI:97623 | PLP1 | HGNC:9086 | ENSMUSG00000031425 | ENSG00000123560 | 1440 | 406 | 0.036 | 0.609 | -0.353 | 3.607 | 3.870 | 4.168 | 3.882 | 0.097 | 0.073 | + | 0.0003 |
| Pdia3 | MGI:95834 | PDIA3 | HGNC:4606 | ENSMUSG00000027248 | ENSG00000167004 | 809 | 1233 | -3.881 | -4.253 | -4.748 | -0.797 | -0.357 | -0.559 | -0.571 | -4.294 | 0.076 | + | 0.0002 |
| Ckb | MGI:88407 | CKB | HGNC:1991 | ENSMUSG00000001270 | ENSG00000166165 | 732 | 889 | 2.355 | 3.096 | 3.526 | 6.245 | 6.176 | 6.965 | 6.462 | 2.992 | 0.090 | + | 0.0012 |
| Ube2v2 | MGI:1917870 | UBE2V2 | HGNC:12495 | ENSMUSG00000022674 | ENSG00000169139 | 622 | 1782 | -0.197 | 0.273 | -0.657 | 3.338 | 2.918 | 3.550 | 3.269 | -0.194 | 0.091 | + | 0.0004 |
| Pip4k2a | MGI:1298206 | PIP4K2A | HGNC:8997 | ENSMUSG00000026737 | ENSG00000150867 | 338 | 229 | -0.253 | -0.522 | -0.001 | 3.539 | 2.440 | 3.619 | 3.199 | -0.259 | 0.091 | + | 0.0011 |
| Pacsin1 | MGI:1345181 | PACSIN1 | HGNC:8570 | ENSMUSG00000040276 | ENSG00000124507 | 404 | 493 | -0.664 | -1.285 | -0.778 | 1.704 | 2.581 | 3.170 | 2.485 | -0.909 | 0.095 | + | 0.0019 |
| Rras2 | MGI:1914172 | RRAS2 | HGNC:17271 | ENSMUSG00000055723 | ENSG00000133818 | 432 | 1534 | -1.115 | -1.052 | -1.926 | 2.329 | 1.161 | 2.194 | 1.895 | -1.364 | 0.104 | + | 0.0022 |
| Lrrc47 | MGI:1920196 | LRRC47 | HGNC:29207 | ENSMUSG00000029028 | ENSG00000130764 | 1224 | 917 | -5.811 | -4.686 | -4.074 | -2.047 | -1.352 | -1.625 | -1.675 | -4.857 | 0.110 | + | 0.0044 |
Supplementary Table 2c. PSD protein abundance (enriched in mouse)
Individual and average abundance values (iBAQ) for mouse and human PSD proteins are given. Data was normalized and log2 transformed. Normalization was achieved by dividing abundance data points by its species abundance average. Abundance fold difference was defined as the ratio of mouse to human average abundance values. To measure significant abundance differences between species a Student’s t-test was used. Proteins significantly enriched in one species are shown in separate sheets. The abundance of proteins from the postsynaptic protein complexes shown in figure 1 are also shown in a separate sheet.
Individual and median abundance values (iBAQ) for mouse and human PSD proteins are given. Normalisation was done by dividing abundace by each species abundance median. Abundance fold difference is defined as the ratio of mouse by human normalised abundance values.
| Appr Gene (m) | MGI ID | Appr Gene (h) | HGNC ID | Ensembl (m) | Ensembl (h) | human ibaq protein group id | mouse ibaq protein group id | iBAQ mPSD01 (Log2, normalised) | iBAQ mPSD02 (Log2, normalised) | iBAQ mPSD03 (Log2, normalised) | iBAQ hPSD01 (Log2, normalised) | iBAQ hPSD02 (Log2, normalised) | iBAQ hPSD03 (Log2, normalised) | iBAQ hPSD average (Log2, normalised) | iBAQ mPSD average (Log2, normalised) | Average fold change (mPSD from hDB/hPSD from mDB) | t-test Significant (Permutation 0.01) | t-test p value |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sept5 | MGI:1195461 | SEPT5 | HGNC:9164 | ENSMUSG00000072214 | ENSG00000184702 | 2308 | 2408 | 7.316 | 7.516 | 7.626 | -1.426 | 1.365 | -0.138 | -0.066 | 7.486 | 187.678 | + | 0.0007 |
| Cacna2d3 | MGI:1338890 | CACNA2D3 | HGNC:15460 | ENSMUSG00000021991 | ENSG00000157445 | 1687 | 689 | 1.365 | 0.648 | 0.570 | -3.585 | -4.646 | -6.482 | -4.905 | 0.861 | 54.406 | + | 0.0028 |
| Mdh1 | MGI:97051 | MDH1 | HGNC:6970 | ENSMUSG00000020321 | ENSG00000014641 | 2420 | 1622 | 7.557 | 5.877 | 7.426 | 2.417 | 1.993 | 0.587 | 1.666 | 6.953 | 39.060 | + | 0.0024 |
| Camk2g | MGI:88259 | CAMK2G | HGNC:1463 | ENSMUSG00000021820 | ENSG00000148660 | 2403 | 526 | 2.747 | 3.748 | 3.277 | -2.436 | -2.479 | -0.822 | -1.913 | 3.257 | 36.001 | + | 0.0011 |
| Gria4 | MGI:95811 | GRIA4 | HGNC:4574 | ENSMUSG00000025892 | ENSG00000152578 | 2229 | 740 | 0.372 | 0.537 | 1.152 | -4.956 | -3.320 | -3.386 | -3.887 | 0.687 | 23.829 | + | 0.0014 |
| Dbn1 | MGI:1931838 | DBN1 | HGNC:2695 | ENSMUSG00000034675 | ENSG00000113758 | 2318 | 858 | 4.026 | 3.929 | 4.859 | -0.970 | 0.726 | -0.340 | -0.195 | 4.271 | 22.101 | + | 0.0015 |
| Anks1b | MGI:1924781 | ANKS1B | HGNC:24600 | ENSMUSG00000058589 | ENSG00000185046 | 2042 | 2205 | 5.526 | 5.941 | 5.593 | 0.740 | 1.673 | 1.688 | 1.367 | 5.687 | 19.975 | + | 0.0002 |
| Dlgap4 | MGI:2138865 | DLGAP4 | HGNC:24476 | ENSMUSG00000061689 | ENSG00000080845 | 253 | 2249 | 1.425 | 1.299 | 1.402 | -1.971 | -2.190 | -3.878 | -2.680 | 1.375 | 16.621 | + | 0.0026 |
| Mal2 | MGI:2146021 | MAL2 | HGNC:13634 | ENSMUSG00000024479 | ENSG00000147676 | 1056 | 1036 | 4.980 | 5.122 | 5.302 | 0.742 | 2.150 | 0.751 | 1.214 | 5.135 | 15.140 | + | 0.0012 |
| Rpl6 | MGI:108057 | RPL6 | HGNC:10362 | ENSMUSG00000029614 | ENSG00000089009 | 1775 | 1401 | 1.121 | 0.051 | 0.614 | -2.119 | -3.970 | -3.098 | -3.062 | 0.595 | 12.616 | + | 0.0041 |
| D3Bwg0562e | MGI:106530 | NOT_FOUND | NOT_FOUND | ENSMUSG00000044667 | ENSG00000117600 | 1818 | 1870 | 1.797 | 1.459 | 1.301 | -2.533 | -1.840 | -1.690 | -2.021 | 1.519 | 11.631 | + | 0.0003 |
| Shisa7 | MGI:3605641 | SHISA7 | HGNC:35409 | ENSMUSG00000053550 | ENSG00000187902 | 2422 | 2377 | 1.996 | 2.393 | 2.150 | -2.083 | -0.354 | -1.425 | -1.287 | 2.180 | 11.056 | + | 0.0026 |
| Lgi1 | MGI:1861691 | LGI1 | HGNC:6572 | ENSMUSG00000067242 | ENSG00000108231 | 661 | 384 | 1.607 | 2.344 | 2.308 | -1.292 | -2.008 | -0.797 | -1.366 | 2.086 | 10.943 | + | 0.0013 |
| Sept10 | MGI:1918110 | SEPT10 | HGNC:14349 | ENSMUSG00000019917 | ENSG00000186522 | 1828 | 1317 | 1.608 | 2.869 | 2.417 | -1.801 | -0.201 | -1.419 | -1.141 | 2.298 | 10.843 | + | 0.0048 |
| Begain | MGI:3044626 | BEGAIN | HGNC:24163 | ENSMUSG00000040867 | ENSG00000183092 | 386 | 1732 | 0.708 | 1.320 | 0.943 | -2.243 | -1.662 | -3.271 | -2.392 | 0.991 | 10.430 | + | 0.0025 |
| Ppfia4 | MGI:1915757 | PPFIA4 | HGNC:9248 | ENSMUSG00000026458 | ENSG00000143847 | 2183 | 2406 | 0.713 | 0.673 | 0.462 | -2.257 | -3.308 | -2.397 | -2.654 | 0.616 | 9.645 | + | 0.0006 |
| Pura | MGI:103079 | PURA | HGNC:9701 | ENSMUSG00000043991 | ENSG00000185129 | 754 | 312 | 4.891 | 4.600 | 4.791 | 1.275 | 1.381 | 2.042 | 1.566 | 4.760 | 9.156 | + | 0.0002 |
| Shank2 | MGI:2671987 | SHANK2 | HGNC:14295 | ENSMUSG00000037541 | ENSG00000162105 | 2357 | 1673 | 3.188 | 2.773 | 3.377 | -0.412 | 0.432 | 0.081 | 0.034 | 3.113 | 8.450 | + | 0.0005 |
| Sh3pxd2a | MGI:1298393 | SH3PXD2A | HGNC:23664 | ENSMUSG00000053617 | ENSG00000107957 | 1924 | 1329 | -2.363 | -2.577 | -2.296 | -5.278 | -6.411 | -4.737 | -5.475 | -2.412 | 8.359 | + | 0.0036 |
| Dlgap2 | MGI:2443181 | DLGAP2 | HGNC:2906 | ENSMUSG00000047495 | ENSG00000198010 | 1025 | 1045 | 1.773 | 1.997 | 1.976 | -1.389 | -0.682 | -1.366 | -1.145 | 1.916 | 8.345 | + | 0.0002 |
| Dlgap3 | MGI:3039563 | DLGAP3 | HGNC:30368 | ENSMUSG00000042388 | ENSG00000116544 | 1429 | 847 | 3.011 | 3.335 | 3.164 | -0.426 | 0.553 | 0.204 | 0.110 | 3.170 | 8.339 | + | 0.0005 |
| Kalrn | MGI:2685385 | KALRN | HGNC:4814 | ENSMUSG00000061751 | ENSG00000160145 | 909 | 2119 | 1.127 | 0.865 | 1.453 | -2.497 | -0.952 | -2.262 | -1.904 | 1.148 | 8.293 | + | 0.0039 |
| Dpp6 | MGI:94921 | DPP6 | HGNC:3010 | ENSMUSG00000061576 | ENSG00000130226 | 1520 | 2378 | -1.423 | -1.620 | -0.973 | -4.494 | -4.200 | -4.371 | -4.355 | -1.338 | 8.091 | + | 0.0001 |
| Grin1 | MGI:95819 | GRIN1 | HGNC:4584 | ENSMUSG00000026959 | ENSG00000176884 | 2090 | 2247 | 3.412 | 3.695 | 3.018 | -0.085 | 0.503 | 0.891 | 0.437 | 3.375 | 7.666 | + | 0.0010 |
| Rimbp2 | MGI:2443235 | RIMBP2 | HGNC:30339 | ENSMUSG00000029420 | ENSG00000060709 | 1290 | 2325 | 0.647 | 0.446 | 0.292 | -2.149 | -2.088 | -3.116 | -2.451 | 0.462 | 7.528 | + | 0.0011 |
Supplementary Table 2d. PSD protein abundance (PSD complexes)
Individual and average abundance values (iBAQ) for mouse and human PSD proteins are given. Data was normalized and log2 transformed. Normalization was achieved by dividing abundance data points by its species abundance average. Abundance fold difference was defined as the ratio of mouse to human average abundance values. To measure significant abundance differences between species a Student’s t-test was used. Proteins significantly enriched in one species are shown in separate sheets. The abundance of proteins from the postsynaptic protein complexes shown in figure 1 are also shown in a separate sheet.
Individual and median abundance values (iBAQ) for mouse and human PSD proteins are given. Normalisation was done by dividing abundace by each species abundance median. Abundance fold difference is defined as the ratio of mouse by human normalised abundance values.
| MGI ID | Appr Gene (h) | PSD95 Complex | NR Complex | mGluR5 Complex | human ibaq protein group id | mouse ibaq protein group id | iBAQ mPSD01 (Log2, normalised) | iBAQ mPSD02 (Log2, normalised) | iBAQ mPSD03 (Log2, normalised) | iBAQ hPSD01 (Log2, normalised) | iBAQ hPSD02 (Log2, normalised) | iBAQ hPSD03 (Log2, normalised) | iBAQ hPSD average (Log2, normalised) | iBAQ mPSD average (Log2, normalised) | Average fold change (mPSD from hDB/hPSD from mDB) | t-test Significant (Perm 0.01) | t-test p value |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MGI:95820 | GRIN2A | YES | YES | YES | 951 | 309 | 1.027 | 0.953 | 1.121 | -1.310 | -1.009 | -1.563 | -1.294 | 1.034 | 5.020 | + | 0.000 |
| MGI:104560 | NSF | YES | YES | YES | 222 | 2190 | 3.051 | 4.066 | 3.771 | 3.361 | 4.806 | 4.163 | 4.110 | 3.629 | 0.717 | 0.404 | |
| MGI:2388633 | CLTC | YES | YES | YES | 767 | 982 | 4.540 | 3.592 | 4.005 | 3.970 | 5.206 | 4.683 | 4.620 | 4.046 | 0.672 | 0.272 | |
| MGI:1353495 | SLC25A4 | YES | YES | NO | 729 | 219 | -0.786 | -2.634 | -4.733 | 3.294 | 2.646 | 1.863 | 2.601 | -2.718 | 0.025 | 0.012 | |
| MGI:99432 | ARF3 | YES | YES | NO | 1321 | 1026 | -0.599 | -2.604 | -5.061 | 0.756 | 0.564 | -1.215 | 0.035 | -2.755 | 0.145 | 0.124 | |
| MGI:107363 | STXBP1 | YES | YES | NO | 1098 | 1849 | -2.846 | -2.331 | -2.836 | 5.526 | 6.183 | 6.722 | 6.144 | -2.671 | 0.002 | + | 0.000 |
| MGI:87880 | ACO2 | YES | YES | NO | 582 | 240 | -2.417 | -1.370 | -2.372 | 1.862 | 2.760 | 1.613 | 2.078 | -2.053 | 0.057 | + | 0.001 |
| MGI:1346858 | MAPK1 | YES | YES | NO | -1.803 | -1.043 | -2.232 | 0.992 | -0.670 | 2.196 | 0.839 | -1.693 | 0.173 | ||||
| MGI:99523 | PRDX1 | YES | YES | NO | 60 | 427 | -1.416 | -0.599 | -0.677 | 1.335 | 1.851 | -0.986 | 0.733 | -0.897 | 0.323 | 0.148 | |
| MGI:1316660 | CACNG2 | YES | YES | NO | 82 | 783 | -0.456 | -0.267 | 0.385 | -0.950 | -2.174 | -1.317 | -1.481 | -0.113 | 2.581 | 0.037 | |
| MGI:97623 | PLP1 | YES | YES | NO | 1440 | 406 | 0.036 | 0.609 | -0.353 | 3.607 | 3.870 | 4.168 | 3.882 | 0.097 | 0.073 | + | 0.000 |
| MGI:1915339 | ARPC4 | YES | YES | NO | 2155 | 926 | 0.743 | 0.321 | 0.083 | 0.534 | 1.201 | -0.431 | 0.435 | 0.382 | 0.964 | 0.923 | |
| MGI:101931 | SLC1A2 | YES | YES | NO | 1672 | 2058 | 1.999 | 1.442 | 0.701 | -0.287 | -0.456 | 0.228 | -0.172 | 1.380 | 2.932 | 0.022 | |
| MGI:1194500 | ABLIM1 | YES | YES | NO | 1776 | 1688 | 0.902 | 1.554 | 1.489 | -0.062 | 1.860 | 1.144 | 0.981 | 1.315 | 1.261 | 0.606 | |
| MGI:2685385 | KALRN | YES | YES | NO | 909 | 2119 | 1.127 | 0.865 | 1.453 | -2.497 | -0.952 | -2.262 | -1.904 | 1.148 | 8.293 | + | 0.004 |
| MGI:107164 | PPP3CA | YES | YES | NO | 1271 | 421 | 1.732 | 2.067 | 1.521 | 1.824 | 2.069 | 2.455 | 2.116 | 1.773 | 0.789 | 0.231 | |
| MGI:102520 | PHB2 | YES | YES | NO | 868 | 1498 | 2.754 | 2.279 | 1.455 | 3.710 | 3.109 | 1.994 | 2.938 | 2.163 | 0.584 | 0.286 | |
| MGI:1861691 | LGI1 | YES | YES | NO | 661 | 384 | 1.607 | 2.344 | 2.308 | -1.292 | -2.008 | -0.797 | -1.366 | 2.086 | 10.943 | + | 0.001 |
| MGI:95739 | GLUL | YES | YES | NO | 354 | 2150 | 2.507 | 3.041 | 2.607 | 2.763 | 1.865 | 1.264 | 1.964 | 2.719 | 1.687 | 0.180 | |
| MGI:1346065 | DLGAP1 | YES | YES | NO | 691 | 2169 | 2.606 | 3.020 | 3.270 | 1.189 | 1.626 | 0.688 | 1.167 | 2.965 | 3.477 | + | 0.006 |
| MGI:95821 | GRIN2B | YES | YES | NO | 1639 | 1491 | 2.991 | 2.992 | 3.007 | -1.201 | 0.707 | 0.758 | 0.088 | 2.996 | 7.507 | 0.011 | |
| MGI:107231 | DLG1 | YES | YES | NO | 959 | 1834 | 3.016 | 2.871 | 2.839 | 0.433 | 0.355 | 0.734 | 0.507 | 2.909 | 5.282 | + | 0.000 |
| MGI:1353496 | SLC25A5 | YES | YES | NO | 257 | 608 | 4.122 | 3.584 | 2.337 | 0.979 | -0.645 | -1.403 | -0.356 | 3.348 | 13.034 | 0.014 | |
| MGI:95819 | GRIN1 | YES | YES | NO | 2090 | 2247 | 3.412 | 3.695 | 3.018 | -0.085 | 0.503 | 0.891 | 0.437 | 3.375 | 7.666 | + | 0.001 |
| MGI:95640 | GAPDH | YES | YES | NO | 1408 | 1310 | 3.378 | 3.400 | 3.480 | 3.873 | 3.807 | 4.643 | 4.108 | 3.419 | 0.620 | 0.063 |
Supplementary Table 3a. Proteins only detected in human PSD
Proteins only found in human or mouse are shown in separate sheets. For each protein several identification (ID) numbers from biological databases are given.
For each identified protein several identification (ID) numbers from biological databases are given.
Supplementary Table 3b. Proteins only detected in mouse PSD
Proteins only found in human or mouse are shown in separate sheets. For each protein several identification (ID) numbers from biological databases are given.
For each identified protein several identification (ID) numbers from biological databases are given.
Supplementary Table 4a. All PSD
Proteins from the total PSD exclusive to human or mouse were classified independently using the Panther 'Protein Class' descriptor. Enrichment analysis was done to determine Protein Classes overrepresented in each species set of exclusive molecules. The Benjamini-Hochberg procedure was used to correct for multiple testing. The column 'observed' retrieves the number of proteins identified in each 'Protein Class' while the column 'expected', the number that would have been identified by chance. Over or under-representations are shown by a (+) or (-) symbol respectively. A second sheet contains the same analysis but done only for the molecules from the consensus PSD.
Human and mouse PSD proteins not found in the other specie were classified independenly using the Panther 'Protein Class' descriptor. An enrichment analysis was done to determine Protein Classes overrepresented in each species as compared with the human genome. P-values were corrected for multiple testing using the Benjamini-Hochberg procedure. The colum 'observed' retrives the number of proteins identified in each 'Protein Class' while the column 'expected', the number which would have been identified by chance. Over or underrepresntations are shown by a (+) or (-) symbol respectively.
| Protein classes in human PSD as compared with human genome | Protein classes in mouse PSD as compared with human genome | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PANTHER Protein Class | Homo sapiens genes | Observed | Expected | over/under | Corrected P-value | Observed | Expected | over/under | P-value | Corrected P-value | |||
| cytoskeletal protein | 441 | 36 | 9.17 | + | 1.30E-09 | 14 | 10.85 | + | 2.03E-01 | 6.18E-01 | |||
| oxidoreductase | 550 | 39 | 11.44 | + | 5.20E-09 | 24 | 13.54 | + | 5.68E-03 | 5.22E-02 | |||
| transcription factor | 2067 | 9 | 42.98 | - | 5.26E-09 | 11 | 50.87 | - | 2.59E-12 | 1.67E-10 | |||
| transferase | 1512 | 70 | 31.44 | + | 1.44E-08 | 40 | 37.21 | + | 3.41E-01 | 7.86E-01 | |||
| dehydrogenase | 210 | 22 | 4.37 | + | 4.52E-08 | 13 | 5.17 | + | 2.51E-03 | 3.22E-02 | |||
| membrane traffic protein | 321 | 27 | 6.67 | + | 4.74E-08 | 18 | 7.9 | + | 1.26E-03 | 2.03E-02 | |||
| membrane trafficking regulatory protein | 107 | 16 | 2.22 | + | 4.74E-08 | 4 | 2.63 | + | 2.71E-01 | 7.42E-01 | |||
| actin family cytoskeletal protein | 222 | 22 | 4.62 | + | 7.62E-08 | 6 | 5.46 | + | 4.65E-01 | 8.19E-01 | |||
| kinase | 679 | 40 | 14.12 | + | 1.25E-07 | 25 | 16.71 | + | 3.20E-02 | 1.82E-01 | |||
| nucleotide kinase | 51 | 10 | 1.06 | + | 3.36E-06 | 4 | 1.26 | + | 3.85E-02 | 2.12E-01 | |||
| non-motor actin binding protein | 114 | 14 | 2.37 | + | 3.39E-06 | 2 | 2.81 | - | 4.68E-01 | 8.19E-01 | |||
| zinc finger transcription factor | 803 | 1 | 16.7 | - | 1.17E-05 | 3 | 19.76 | - | 2.92E-06 | 7.04E-05 | |||
| Unclassified | 6763 | 104 | 140.62 | - | 9.26E-04 | 162 | 166.43 | - | 3.55E-01 | 8.03E-01 | |||
| reductase | 65 | 8 | 1.35 | + | 1.10E-03 | 5 | 1.6 | + | 2.34E-02 | 1.51E-01 | |||
| protein kinase | 510 | 25 | 10.6 | + | 1.15E-03 | 16 | 12.55 | + | 1.95E-01 | 6.13E-01 | |||
| KRAB box transcription factor | 552 | 1 | 11.48 | - | 1.36E-03 | 0 | 13.58 | - | 1.04E-06 | 2.87E-05 | |||
| microtubule family cytoskeletal protein | 156 | 11 | 3.24 | + | 5.89E-03 | 6 | 3.84 | + | 1.90E-01 | 6.13E-01 | |||
| enzyme modulator | 857 | 33 | 17.82 | + | 6.53E-03 | 34 | 21.09 | + | 4.89E-03 | 4.97E-02 | |||
| transfer/carrier protein | 248 | 14 | 5.16 | + | 8.54E-03 | 3 | 6.1 | - | 1.41E-01 | 5.04E-01 | |||
| G-protein modulator | 278 | 15 | 5.78 | + | 8.54E-03 | 16 | 6.84 | + | 1.77E-03 | 2.58E-02 | |||
| transaminase | 25 | 4 | 0.52 | + | 1.83E-02 | 0 | 0.62 | - | 5.40E-01 | 8.19E-01 | |||
| peroxidase | 27 | 4 | 0.56 | + | 2.31E-02 | 0 | 0.66 | - | 5.14E-01 | 8.19E-01 | |||
| G-protein | 206 | 11 | 4.28 | + | 3.74E-02 | 7 | 5.07 | + | 2.47E-01 | 7.12E-01 | |||
| ion channel | 341 | 1 | 7.09 | - | 5.18E-02 | 31 | 8.39 | + | 9.63E-10 | 3.72E-08 | |||
| acetyltransferase | 105 | 7 | 2.18 | + | 5.40E-02 | 3 | 2.58 | + | 4.78E-01 | 8.19E-01 | |||
| aminoacyl-tRNA synthetase | 20 | 3 | 0.42 | + | 6.50E-02 | 1 | 0.49 | + | 3.89E-01 | 8.19E-01 | |||
| ligase | 260 | 12 | 5.41 | + | 6.60E-02 | 10 | 6.4 | + | 1.13E-01 | 4.54E-01 | |||
| isomerase | 94 | 6 | 1.95 | + | 1.01E-01 | 0 | 2.31 | - | 9.84E-02 | 4.13E-01 | |||
| small GTPase | 158 | 8 | 3.29 | + | 1.25E-01 | 6 | 3.89 | + | 1.97E-01 | 6.13E-01 | |||
| chaperone | 130 | 7 | 2.7 | + | 1.25E-01 | 2 | 3.2 | - | 3.79E-01 | 8.19E-01 | |||
| transaldolase | 1 | 1 | 0.02 | + | 1.25E-01 | 0 | 0.02 | - | 9.76E-01 | 1.00E+00 | |||
| receptor | 1076 | 13 | 22.37 | - | 1.25E-01 | 34 | 26.48 | + | 8.40E-02 | 3.85E-01 | |||
| lyase | 104 | 6 | 2.16 | + | 1.33E-01 | 5 | 2.56 | + | 1.16E-01 | 4.57E-01 | |||
| mRNA processing factor | 179 | 0 | 3.72 | - | 1.35E-01 | 11 | 4.41 | + | 5.50E-03 | 5.22E-02 | |||
| vesicle coat protein | 30 | 3 | 0.62 | + | 1.38E-01 | 1 | 0.74 | + | 5.22E-01 | 8.19E-01 | |||
| non-motor microtubule binding protein | 53 | 4 | 1.1 | + | 1.38E-01 | 4 | 1.3 | + | 4.33E-02 | 2.23E-01 | |||
| dehydratase | 31 | 3 | 0.64 | + | 1.44E-01 | 0 | 0.76 | - | 4.66E-01 | 8.19E-01 | |||
| phosphorylase | 13 | 2 | 0.27 | + | 1.52E-01 | 0 | 0.32 | - | 7.26E-01 | 8.87E-01 | |||
| transcription cofactor | 255 | 1 | 5.3 | - | 1.52E-01 | 3 | 6.28 | - | 1.27E-01 | 4.84E-01 | |||
| transmembrane receptor regulatory/adaptor protein | 84 | 5 | 1.75 | + | 1.56E-01 | 1 | 2.07 | - | 3.88E-01 | 8.19E-01 | |||
| G-protein coupled receptor | 447 | 4 | 9.29 | - | 2.08E-01 | 13 | 11 | + | 3.10E-01 | 7.86E-01 | |||
| antibacterial response protein | 18 | 2 | 0.37 | + | 2.49E-01 | 0 | 0.44 | - | 6.42E-01 | 8.60E-01 | |||
| hydrolase | 454 | 15 | 9.44 | + | 2.49E-01 | 12 | 11.17 | + | 4.42E-01 | 8.19E-01 | |||
| nucleic acid binding | 1466 | 22 | 30.48 | - | 2.70E-01 | 78 | 36.08 | + | 1.38E-10 | 6.66E-09 | |||
| basic helix-loop-helix transcription factor | 120 | 0 | 2.5 | - | 3.51E-01 | 1 | 2.95 | - | 2.05E-01 | 6.18E-01 | |||
| guanyl-nucleotide exchange factor | 79 | 4 | 1.64 | + | 3.54E-01 | 3 | 1.94 | + | 3.08E-01 | 7.86E-01 | |||
| tubulin | 25 | 2 | 0.52 | + | 3.95E-01 | 1 | 0.62 | + | 4.60E-01 | 8.19E-01 | |||
| transketolase | 5 | 1 | 0.1 | + | 3.97E-01 | 0 | 0.12 | - | 8.84E-01 | 9.69E-01 | |||
| phosphodiesterase | 27 | 2 | 0.56 | + | 4.25E-01 | 3 | 0.66 | + | 2.99E-02 | 1.79E-01 | |||
| cysteine protease | 121 | 5 | 2.52 | + | 4.25E-01 | 2 | 2.98 | - | 4.28E-01 | 8.19E-01 | |||
| kinase modulator | 103 | 0 | 2.14 | - | 4.43E-01 | 6 | 2.53 | + | 4.39E-02 | 2.23E-01 | |||
| lipase | 57 | 3 | 1.19 | + | 4.43E-01 | 3 | 1.4 | + | 1.67E-01 | 5.65E-01 | |||
| ligand-gated ion channel | 101 | 0 | 2.1 | - | 4.44E-01 | 11 | 2.49 | + | 5.47E-05 | 9.60E-04 | |||
| peptide hormone | 169 | 1 | 3.51 | - | 4.70E-01 | 3 | 4.16 | - | 4.02E-01 | 8.19E-01 | |||
| DNA binding protein | 476 | 6 | 9.9 | - | 4.70E-01 | 11 | 11.71 | - | 4.94E-01 | 8.19E-01 | |||
| homeobox transcription factor | 233 | 2 | 4.84 | - | 4.72E-01 | 0 | 5.73 | - | 3.13E-03 | 3.55E-02 | |||
| helix-turn-helix transcription factor | 233 | 2 | 4.84 | - | 4.72E-01 | 0 | 5.73 | - | 3.13E-03 | 3.55E-02 | |||
| amino acid transporter | 98 | 4 | 2.04 | + | 4.96E-01 | 3 | 2.41 | + | 4.34E-01 | 8.19E-01 | |||
| potassium channel | 89 | 0 | 1.85 | - | 5.14E-01 | 8 | 2.19 | + | 1.87E-03 | 2.58E-02 | |||
| actin binding motor protein | 9 | 1 | 0.19 | + | 5.47E-01 | 0 | 0.22 | - | 8.01E-01 | 9.37E-01 | |||
| heterotrimeric G-protein | 36 | 2 | 0.75 | + | 5.47E-01 | 1 | 0.89 | + | 5.88E-01 | 8.20E-01 | |||
| phospholipase | 36 | 2 | 0.75 | + | 5.47E-01 | 3 | 0.89 | + | 6.04E-02 | 2.84E-01 | |||
| cytokine receptor | 213 | 2 | 4.43 | - | 5.51E-01 | 6 | 5.24 | + | 4.27E-01 | 8.19E-01 | |||
| intermediate filament binding protein | 10 | 1 | 0.21 | + | 5.67E-01 | 2 | 0.25 | + | 2.57E-02 | 1.60E-01 | |||
| Hsp90 family chaperone | 10 | 1 | 0.21 | + | 5.67E-01 | 0 | 0.25 | - | 7.82E-01 | 9.26E-01 | |||
| chromatin/chromatin-binding protein | 74 | 3 | 1.54 | + | 5.85E-01 | 2 | 1.82 | + | 5.44E-01 | 8.19E-01 | |||
| intermediate filament | 11 | 1 | 0.23 | + | 5.85E-01 | 2 | 0.27 | + | 3.06E-02 | 1.79E-01 | |||
| phosphatase | 230 | 7 | 4.78 | + | 5.85E-01 | 8 | 5.66 | + | 2.10E-01 | 6.23E-01 | |||
| Hsp70 family chaperone | 13 | 1 | 0.27 | + | 6.63E-01 | 0 | 0.32 | - | 7.26E-01 | 8.87E-01 | |||
| cell junction protein | 67 | 0 | 1.39 | - | 6.78E-01 | 0 | 1.65 | - | 1.92E-01 | 6.13E-01 | |||
| microtubule binding motor protein | 46 | 2 | 0.96 | + | 6.78E-01 | 1 | 1.13 | - | 6.87E-01 | 8.70E-01 | |||
| mutase | 14 | 1 | 0.29 | + | 6.78E-01 | 0 | 0.34 | - | 7.08E-01 | 8.70E-01 | |||
| RNA binding protein | 727 | 12 | 15.12 | - | 6.78E-01 | 60 | 17.89 | + | 6.16E-16 | 1.19E-13 | |||
| glucosidase | 15 | 1 | 0.31 | + | 6.83E-01 | 0 | 0.37 | - | 6.91E-01 | 8.70E-01 | |||
| storage protein | 15 | 1 | 0.31 | + | 6.83E-01 | 0 | 0.37 | - | 6.91E-01 | 8.70E-01 | |||
| calcium-binding protein | 63 | 0 | 1.31 | - | 6.83E-01 | 1 | 1.55 | - | 5.41E-01 | 8.19E-01 | |||
| acyltransferase | 88 | 3 | 1.83 | + | 6.94E-01 | 1 | 2.17 | - | 3.62E-01 | 8.03E-01 | |||
| cation transporter | 178 | 2 | 3.7 | - | 6.99E-01 | 7 | 4.38 | + | 1.53E-01 | 5.37E-01 | |||
| methyltransferase | 130 | 4 | 2.7 | + | 6.99E-01 | 2 | 3.2 | - | 3.79E-01 | 8.19E-01 | |||
| ubiquitin-protein ligase | 132 | 4 | 2.74 | + | 7.04E-01 | 6 | 3.25 | + | 1.10E-01 | 4.52E-01 | |||
| DNA methyltransferase | 17 | 1 | 0.35 | + | 7.04E-01 | 0 | 0.42 | - | 6.58E-01 | 8.70E-01 | |||
| glycosyltransferase | 229 | 3 | 4.76 | - | 7.04E-01 | 6 | 5.64 | + | 4.95E-01 | 8.19E-01 | |||
| translation factor | 56 | 2 | 1.16 | + | 7.56E-01 | 5 | 1.38 | + | 1.33E-02 | 9.51E-02 | |||
| histone | 54 | 0 | 1.12 | - | 7.56E-01 | 5 | 1.33 | + | 1.15E-02 | 8.54E-02 | |||
| chaperonin | 20 | 1 | 0.42 | + | 7.72E-01 | 0 | 0.49 | - | 6.11E-01 | 8.30E-01 | |||
| signaling molecule | 961 | 22 | 19.98 | + | 7.85E-01 | 23 | 23.65 | - | 5.00E-01 | 8.19E-01 | |||
| cysteine protease inhibitor | 21 | 1 | 0.44 | + | 7.85E-01 | 0 | 0.52 | - | 5.96E-01 | 8.20E-01 | |||
| structural protein | 280 | 7 | 5.82 | + | 8.00E-01 | 10 | 6.89 | + | 1.57E-01 | 5.41E-01 | |||
| immunoglobulin receptor superfamily | 155 | 2 | 3.22 | - | 8.00E-01 | 5 | 3.81 | + | 3.35E-01 | 7.86E-01 | |||
| replication origin binding protein | 47 | 0 | 0.98 | - | 8.00E-01 | 2 | 1.16 | + | 3.22E-01 | 7.86E-01 | |||
| phosphatase modulator | 63 | 2 | 1.31 | + | 8.00E-01 | 1 | 1.55 | - | 5.41E-01 | 8.19E-01 | |||
| nuclear hormone receptor | 46 | 0 | 0.96 | - | 8.05E-01 | 1 | 1.13 | - | 6.87E-01 | 8.70E-01 | |||
| defense/immunity protein | 107 | 3 | 2.22 | + | 8.05E-01 | 1 | 2.63 | - | 2.60E-01 | 7.27E-01 | |||
| kinase activator | 45 | 0 | 0.94 | - | 8.05E-01 | 2 | 1.11 | + | 3.04E-01 | 7.86E-01 | |||
| damaged DNA-binding protein | 45 | 0 | 0.94 | - | 8.05E-01 | 0 | 1.11 | - | 3.30E-01 | 7.86E-01 | |||
| esterase | 25 | 1 | 0.52 | + | 8.16E-01 | 0 | 0.62 | - | 5.40E-01 | 8.19E-01 | |||
| protein phosphatase | 111 | 3 | 2.31 | + | 8.16E-01 | 3 | 2.73 | + | 5.15E-01 | 8.19E-01 | |||
| cell adhesion molecule | 93 | 1 | 1.93 | - | 8.35E-01 | 2 | 2.29 | - | 5.99E-01 | 8.20E-01 | |||
| transporter | 1098 | 24 | 22.83 | + | 8.38E-01 | 57 | 27.02 | + | 1.27E-07 | 4.09E-06 | |||
| phosphatase inhibitor | 28 | 1 | 0.58 | + | 8.48E-01 | 0 | 0.69 | - | 5.02E-01 | 8.19E-01 | |||
| growth factor | 165 | 4 | 3.43 | + | 8.48E-01 | 1 | 4.06 | - | 8.64E-02 | 3.85E-01 | |||
| actin and actin related protein | 31 | 1 | 0.64 | + | 8.48E-01 | 1 | 0.76 | + | 5.34E-01 | 8.19E-01 | |||
| membrane-bound signaling molecule | 133 | 2 | 2.77 | - | 8.48E-01 | 3 | 3.27 | - | 5.86E-01 | 8.20E-01 | |||
| nucleotidyltransferase | 84 | 1 | 1.75 | - | 8.48E-01 | 2 | 2.07 | - | 6.58E-01 | 8.70E-01 | |||
| nucleotide phosphatase | 35 | 0 | 0.73 | - | 8.48E-01 | 3 | 0.86 | + | 5.65E-02 | 2.73E-01 | |||
| DNA helicase | 80 | 2 | 1.66 | + | 8.48E-01 | 0 | 1.97 | - | 1.39E-01 | 5.04E-01 | |||
| immunoglobulin | 33 | 1 | 0.69 | + | 8.48E-01 | 0 | 0.81 | - | 4.44E-01 | 8.19E-01 | |||
| intracellular calcium-sensing protein | 33 | 0 | 0.69 | - | 8.48E-01 | 1 | 0.81 | + | 5.56E-01 | 8.19E-01 | |||
| endoribonuclease | 33 | 0 | 0.69 | - | 8.48E-01 | 0 | 0.81 | - | 4.44E-01 | 8.19E-01 | |||
| calmodulin | 33 | 0 | 0.69 | - | 8.48E-01 | 1 | 0.81 | + | 5.56E-01 | 8.19E-01 | |||
| myelin protein | 33 | 0 | 0.69 | - | 8.48E-01 | 4 | 0.81 | + | 9.48E-03 | 7.35E-02 | |||
| serine protease inhibitor | 79 | 1 | 1.64 | - | 8.48E-01 | 1 | 1.94 | - | 4.21E-01 | 8.19E-01 | |||
| carbohydrate kinase | 35 | 1 | 0.73 | + | 8.48E-01 | 0 | 0.86 | - | 4.22E-01 | 8.19E-01 | |||
| nuclease | 35 | 1 | 0.73 | + | 8.48E-01 | 2 | 0.86 | + | 2.13E-01 | 6.23E-01 | |||
| calcium channel | 31 | 0 | 0.64 | - | 8.48E-01 | 7 | 0.76 | + | 1.48E-05 | 2.86E-04 | |||
| type I cytokine receptor | 31 | 0 | 0.64 | - | 8.48E-01 | 1 | 0.76 | + | 5.34E-01 | 8.19E-01 | |||
| interleukin superfamily | 36 | 1 | 0.75 | + | 8.48E-01 | 0 | 0.89 | - | 4.12E-01 | 8.19E-01 | |||
| protease | 476 | 10 | 9.9 | + | 8.48E-01 | 6 | 11.71 | - | 5.16E-02 | 2.55E-01 | |||
| ribosomal protein | 184 | 4 | 3.83 | + | 8.48E-01 | 29 | 4.53 | + | 8.24E-15 | 7.95E-13 | |||
| tight junction | 30 | 0 | 0.62 | - | 8.48E-01 | 0 | 0.74 | - | 4.78E-01 | 8.19E-01 | |||
| anion channel | 37 | 1 | 0.77 | + | 8.48E-01 | 1 | 0.91 | + | 5.98E-01 | 8.20E-01 | |||
| SNARE protein | 37 | 1 | 0.77 | + | 8.48E-01 | 8 | 0.91 | + | 5.01E-06 | 1.07E-04 | |||
| oxygenase | 74 | 1 | 1.54 | - | 8.48E-01 | 2 | 1.82 | + | 5.44E-01 | 8.19E-01 | |||
| ATP-binding cassette (ABC) transporter | 74 | 1 | 1.54 | - | 8.48E-01 | 2 | 1.82 | + | 5.44E-01 | 8.19E-01 | |||
| major histocompatibility complex antigen | 28 | 0 | 0.58 | - | 8.48E-01 | 1 | 0.69 | + | 4.98E-01 | 8.19E-01 | |||
| neuropeptide | 28 | 0 | 0.58 | - | 8.48E-01 | 0 | 0.69 | - | 5.02E-01 | 8.19E-01 | |||
| extracellular matrix protein | 72 | 1 | 1.5 | - | 8.48E-01 | 6 | 1.77 | + | 9.52E-03 | 7.35E-02 | |||
| RNA helicase | 71 | 1 | 1.48 | - | 8.48E-01 | 6 | 1.75 | + | 8.93E-03 | 7.35E-02 | |||
| cytokine | 159 | 3 | 3.31 | - | 8.48E-01 | 1 | 3.91 | - | 9.73E-02 | 4.13E-01 | |||
| glycosidase | 26 | 0 | 0.54 | - | 8.48E-01 | 0 | 0.64 | - | 5.27E-01 | 8.19E-01 | |||
| apolipoprotein | 42 | 1 | 0.87 | + | 8.48E-01 | 0 | 1.03 | - | 3.55E-01 | 8.03E-01 | |||
| immunoglobulin superfamily cell adhesion molecule | 25 | 0 | 0.52 | - | 8.48E-01 | 1 | 0.62 | + | 4.60E-01 | 8.19E-01 | |||
| hydroxylase | 44 | 1 | 0.91 | + | 8.48E-01 | 0 | 1.08 | - | 3.38E-01 | 7.86E-01 | |||
| HMG box transcription factor | 44 | 1 | 0.91 | + | 8.48E-01 | 0 | 1.08 | - | 3.38E-01 | 7.86E-01 | |||
| protease inhibitor | 109 | 2 | 2.27 | - | 8.48E-01 | 1 | 2.68 | - | 2.51E-01 | 7.12E-01 | |||
| serine protease | 153 | 3 | 3.18 | - | 8.48E-01 | 2 | 3.77 | - | 2.73E-01 | 7.42E-01 | |||
| exoribonuclease | 24 | 0 | 0.5 | - | 8.48E-01 | 0 | 0.59 | - | 5.54E-01 | 8.19E-01 | |||
| basic leucine zipper transcription factor | 24 | 0 | 0.5 | - | 8.48E-01 | 0 | 0.59 | - | 5.54E-01 | 8.19E-01 | |||
| CREB transcription factor | 24 | 0 | 0.5 | - | 8.48E-01 | 0 | 0.59 | - | 5.54E-01 | 8.19E-01 | |||
| carbohydrate transporter | 46 | 1 | 0.96 | + | 8.48E-01 | 0 | 1.13 | - | 3.22E-01 | 7.86E-01 | |||
| cyclase | 23 | 0 | 0.48 | - | 8.48E-01 | 3 | 0.57 | + | 1.98E-02 | 1.32E-01 | |||
| chemokine | 48 | 1 | 1 | + | 8.48E-01 | 0 | 1.18 | - | 3.06E-01 | 7.86E-01 | |||
| tumor necrosis factor receptor | 22 | 0 | 0.46 | - | 8.48E-01 | 0 | 0.54 | - | 5.82E-01 | 8.20E-01 | |||
| RNA methyltransferase | 22 | 0 | 0.46 | - | 8.48E-01 | 0 | 0.54 | - | 5.82E-01 | 8.20E-01 | |||
| ribonucleoprotein | 61 | 1 | 1.27 | - | 8.48E-01 | 3 | 1.5 | + | 1.91E-01 | 6.13E-01 | |||
| metalloprotease | 145 | 3 | 3.01 | - | 8.48E-01 | 1 | 3.57 | - | 1.28E-01 | 4.84E-01 | |||
| gap junction | 21 | 0 | 0.44 | - | 8.48E-01 | 0 | 0.52 | - | 5.96E-01 | 8.20E-01 | |||
| adenylate cyclase | 21 | 0 | 0.44 | - | 8.48E-01 | 3 | 0.52 | + | 1.56E-02 | 1.08E-01 | |||
| endodeoxyribonuclease | 20 | 0 | 0.42 | - | 8.54E-01 | 0 | 0.49 | - | 6.11E-01 | 8.30E-01 | |||
| sodium channel | 20 | 0 | 0.42 | - | 8.54E-01 | 2 | 0.49 | + | 8.78E-02 | 3.85E-01 | |||
| oxidase | 57 | 1 | 1.19 | - | 8.54E-01 | 1 | 1.4 | - | 5.91E-01 | 8.20E-01 | |||
| extracellular matrix glycoprotein | 55 | 1 | 1.14 | - | 8.67E-01 | 6 | 1.35 | + | 2.67E-03 | 3.22E-02 | |||
| centromere DNA-binding protein | 18 | 0 | 0.37 | - | 8.68E-01 | 0 | 0.44 | - | 6.42E-01 | 8.60E-01 | |||
| mitochondrial carrier protein | 17 | 0 | 0.35 | - | 8.76E-01 | 3 | 0.42 | + | 8.91E-03 | 7.35E-02 | |||
| interferon superfamily | 17 | 0 | 0.35 | - | 8.76E-01 | 0 | 0.42 | - | 6.58E-01 | 8.70E-01 | |||
| decarboxylase | 17 | 0 | 0.35 | - | 8.76E-01 | 1 | 0.42 | + | 3.42E-01 | 7.86E-01 | |||
| ATP synthase | 51 | 1 | 1.06 | - | 8.76E-01 | 4 | 1.26 | + | 3.85E-02 | 2.12E-01 | |||
| galactosidase | 16 | 0 | 0.33 | - | 8.76E-01 | 0 | 0.39 | - | 6.74E-01 | 8.70E-01 | |||
| kinase inhibitor | 15 | 0 | 0.31 | - | 8.89E-01 | 0 | 0.37 | - | 6.91E-01 | 8.70E-01 | |||
| surfactant | 15 | 0 | 0.31 | - | 8.89E-01 | 0 | 0.37 | - | 6.91E-01 | 8.70E-01 | |||
| DNA-directed RNA polymerase | 15 | 0 | 0.31 | - | 8.89E-01 | 0 | 0.37 | - | 6.91E-01 | 8.70E-01 | |||
| aspartic protease | 14 | 0 | 0.29 | - | 8.90E-01 | 1 | 0.34 | + | 2.92E-01 | 7.83E-01 | |||
| deaminase | 12 | 0 | 0.25 | - | 9.22E-01 | 0 | 0.3 | - | 7.44E-01 | 8.97E-01 | |||
| DNA glycosylase | 12 | 0 | 0.25 | - | 9.22E-01 | 0 | 0.3 | - | 7.44E-01 | 8.97E-01 | |||
| tumor necrosis factor family member | 11 | 0 | 0.23 | - | 9.31E-01 | 0 | 0.27 | - | 7.63E-01 | 9.09E-01 | |||
| DNA-directed DNA polymerase | 10 | 0 | 0.21 | - | 9.44E-01 | 0 | 0.25 | - | 7.82E-01 | 9.26E-01 | |||
| extracellular matrix structural protein | 9 | 0 | 0.19 | - | 9.58E-01 | 0 | 0.22 | - | 8.01E-01 | 9.37E-01 | |||
| carbohydrate phosphatase | 9 | 0 | 0.19 | - | 9.58E-01 | 0 | 0.22 | - | 8.01E-01 | 9.37E-01 | |||
| annexin | 9 | 0 | 0.19 | - | 9.58E-01 | 0 | 0.22 | - | 8.01E-01 | 9.37E-01 | |||
| protein kinase receptor | 8 | 0 | 0.17 | - | 9.62E-01 | 0 | 0.2 | - | 8.21E-01 | 9.38E-01 | |||
| neurotrophic factor | 8 | 0 | 0.17 | - | 9.62E-01 | 0 | 0.2 | - | 8.21E-01 | 9.38E-01 | |||
| epimerase/racemase | 7 | 0 | 0.15 | - | 9.68E-01 | 0 | 0.17 | - | 8.42E-01 | 9.50E-01 | |||
| viral protein | 7 | 0 | 0.15 | - | 9.68E-01 | 0 | 0.17 | - | 8.42E-01 | 9.50E-01 | |||
| viral coat protein | 7 | 0 | 0.15 | - | 9.68E-01 | 0 | 0.17 | - | 8.42E-01 | 9.50E-01 | |||
| DNA strand-pairing protein | 7 | 0 | 0.15 | - | 9.68E-01 | 0 | 0.17 | - | 8.42E-01 | 9.50E-01 | |||
| amylase | 6 | 0 | 0.12 | - | 9.68E-01 | 0 | 0.15 | - | 8.63E-01 | 9.52E-01 | |||
| DNA ligase | 6 | 0 | 0.12 | - | 9.68E-01 | 1 | 0.15 | + | 1.37E-01 | 5.04E-01 | |||
| helicase | 5 | 0 | 0.1 | - | 9.77E-01 | 0 | 0.12 | - | 8.84E-01 | 9.69E-01 | |||
| amino acid kinase | 5 | 0 | 0.1 | - | 9.77E-01 | 0 | 0.12 | - | 8.84E-01 | 9.69E-01 | |||
| pyrophosphatase | 5 | 0 | 0.1 | - | 9.77E-01 | 0 | 0.12 | - | 8.84E-01 | 9.69E-01 | |||
| metalloprotease inhibitor | 4 | 0 | 0.08 | - | 9.81E-01 | 0 | 0.1 | - | 9.06E-01 | 9.71E-01 | |||
| aldolase | 4 | 0 | 0.08 | - | 9.81E-01 | 0 | 0.1 | - | 9.06E-01 | 9.71E-01 | |||
| DNA topoisomerase | 4 | 0 | 0.08 | - | 9.81E-01 | 0 | 0.1 | - | 9.06E-01 | 9.71E-01 | |||
| deacetylase | 2 | 0 | 0.04 | - | 1.00E+00 | 0 | 0.05 | - | 9.52E-01 | 1.00E+00 | |||
| calsequestrin | 2 | 0 | 0.04 | - | 1.00E+00 | 0 | 0.05 | - | 9.52E-01 | 1.00E+00 | |||
| TGF-beta receptor | 2 | 0 | 0.04 | - | 1.00E+00 | 0 | 0.05 | - | 9.52E-01 | 1.00E+00 | |||
| DNA polymerase processivity factor | 2 | 0 | 0.04 | - | 1.00E+00 | 0 | 0.05 | - | 9.52E-01 | 1.00E+00 | |||
| DNA photolyase | 2 | 0 | 0.04 | - | 1.00E+00 | 0 | 0.05 | - | 9.52E-01 | 1.00E+00 | |||
| exodeoxyribonuclease | 1 | 0 | 0.02 | - | 1.00E+00 | 0 | 0.02 | - | 9.76E-01 | 1.00E+00 | |||
| voltage-gated ion channel | 1 | 0 | 0.02 | - | 1.00E+00 | 0 | 0.02 | - | 9.76E-01 | 1.00E+00 | |||
| primase | 1 | 0 | 0.02 | - | 1.00E+00 | 0 | 0.02 | - | 9.76E-01 | 1.00E+00 | |||
| phosphatase activator | 1 | 0 | 0.02 | - | 1.00E+00 | 0 | 0.02 | - | 9.76E-01 | 1.00E+00 | |||
| reverse transcriptase | 1 | 0 | 0.02 | - | 1.00E+00 | 0 | 0.02 | - | 9.76E-01 | 1.00E+00 | |||
Supplementary Table 4b. Consensus PSD
Proteins from the total PSD exclusive to human or mouse were classified independently using the Panther 'Protein Class' descriptor. Enrichment analysis was done to determine Protein Classes overrepresented in each species set of exclusive molecules. The Benjamini-Hochberg procedure was used to correct for multiple testing. The column 'observed' retrieves the number of proteins identified in each 'Protein Class' while the column 'expected', the number that would have been identified by chance. Over or under-representations are shown by a (+) or (-) symbol respectively. A second sheet contains the same analysis but done only for the molecules from the consensus PSD.
Human and mouse PSD proteins not found in the other specie were classified independenly using the Panther 'Protein Class' descriptor. An enrichment analysis was done to determine Protein Classes overrepresented in each species as compared with the human genome. P-values were corrected for multiple testing using the Benjamini-Hochberg procedure. The colum 'observed' retrives the number of proteins identified in each 'Protein Class' while the column 'expected', the number which would have been identified by chance. Over or underrepresntations are shown by a (+) or (-) symbol respectively.
| Protein classes in human PSD as compared with human genome | Protein classes in mouse PSD as compared with human genome | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PANTHER Protein Class | Homo sapiens genes | Observed | Expected | over/under | Corrected P-value | Observed | Expected | over/under | P-value | Corrected P-value | |||
| cytoskeletal protein | 441 | 16 | 2.35 | + | 3.61E-07 | 5 | 3.85 | + | 3.42E-01 | 1.00E+00 | |||
| actin family cytoskeletal protein | 222 | 10 | 1.18 | + | 3.45E-05 | 2 | 1.94 | + | 5.79E-01 | 1.00E+00 | |||
| oxidoreductase | 550 | 13 | 2.93 | + | 5.06E-04 | 10 | 4.81 | + | 2.36E-02 | 3.25E-01 | |||
| dehydrogenase | 210 | 8 | 1.12 | + | 8.88E-04 | 6 | 1.84 | + | 1.09E-02 | 2.10E-01 | |||
| non-motor actin binding protein | 114 | 6 | 0.61 | + | 1.42E-03 | 1 | 1 | + | 6.32E-01 | 1.00E+00 | |||
| reductase | 65 | 4 | 0.35 | + | 1.39E-02 | 3 | 0.57 | + | 1.99E-02 | 2.95E-01 | |||
| transcription factor | 2067 | 2 | 11 | - | 2.18E-02 | 3 | 18.06 | - | 8.16E-06 | 3.94E-04 | |||
| transferase | 1512 | 18 | 8.05 | + | 2.48E-02 | 14 | 13.21 | + | 4.51E-01 | 1.00E+00 | |||
| microtubule family cytoskeletal protein | 156 | 5 | 0.83 | + | 3.32E-02 | 2 | 1.36 | + | 3.96E-01 | 1.00E+00 | |||
| transfer/carrier protein | 248 | 6 | 1.32 | + | 4.27E-02 | 0 | 2.17 | - | 1.13E-01 | 7.40E-01 | |||
| nucleotide kinase | 51 | 3 | 0.27 | + | 4.31E-02 | 2 | 0.45 | + | 7.40E-02 | 6.80E-01 | |||
| membrane trafficking regulatory protein | 107 | 4 | 0.57 | + | 4.31E-02 | 2 | 0.94 | + | 2.40E-01 | 1.00E+00 | |||
| transaminase | 25 | 2 | 0.13 | + | 1.20E-01 | 0 | 0.22 | - | 8.04E-01 | 1.00E+00 | |||
| dehydratase | 31 | 2 | 0.17 | + | 1.63E-01 | 0 | 0.27 | - | 7.63E-01 | 1.00E+00 | |||
| zinc finger transcription factor | 803 | 0 | 4.27 | - | 1.63E-01 | 2 | 7.02 | - | 2.70E-02 | 3.26E-01 | |||
| lyase | 104 | 3 | 0.55 | + | 2.23E-01 | 0 | 0.91 | - | 4.02E-01 | 1.00E+00 | |||
| kinase | 679 | 8 | 3.61 | + | 3.02E-01 | 11 | 5.93 | + | 3.72E-02 | 3.99E-01 | |||
| membrane traffic protein | 321 | 5 | 1.71 | + | 3.02E-01 | 7 | 2.81 | + | 2.36E-02 | 3.25E-01 | |||
| methyltransferase | 130 | 3 | 0.69 | + | 3.02E-01 | 0 | 1.14 | - | 3.20E-01 | 1.00E+00 | |||
| chaperone | 130 | 3 | 0.69 | + | 3.02E-01 | 0 | 1.14 | - | 3.20E-01 | 1.00E+00 | |||
| non-motor microtubule binding protein | 53 | 2 | 0.28 | + | 3.02E-01 | 1 | 0.46 | + | 3.71E-01 | 1.00E+00 | |||
| actin binding motor protein | 9 | 1 | 0.05 | + | 4.11E-01 | 0 | 0.08 | - | 9.24E-01 | 1.00E+00 | |||
| KRAB box transcription factor | 552 | 0 | 2.94 | - | 4.26E-01 | 0 | 4.82 | - | 7.51E-03 | 1.61E-01 | |||
| Unclassified | 6763 | 28 | 36 | - | 4.81E-01 | 54 | 59.1 | - | 2.32E-01 | 1.00E+00 | |||
| Hsp70 family chaperone | 13 | 1 | 0.07 | + | 5.12E-01 | 0 | 0.11 | - | 8.93E-01 | 1.00E+00 | |||
| phosphorylase | 13 | 1 | 0.07 | + | 5.12E-01 | 0 | 0.11 | - | 8.93E-01 | 1.00E+00 | |||
| transmembrane receptor regulatory/adaptor protein | 84 | 2 | 0.45 | + | 5.12E-01 | 1 | 0.73 | + | 5.21E-01 | 1.00E+00 | |||
| storage protein | 15 | 1 | 0.08 | + | 5.12E-01 | 0 | 0.13 | - | 8.77E-01 | 1.00E+00 | |||
| DNA binding protein | 476 | 0 | 2.53 | - | 5.12E-01 | 3 | 4.16 | - | 4.00E-01 | 1.00E+00 | |||
| acyltransferase | 88 | 2 | 0.47 | + | 5.18E-01 | 1 | 0.77 | + | 5.37E-01 | 1.00E+00 | |||
| DNA methyltransferase | 17 | 1 | 0.09 | + | 5.39E-01 | 0 | 0.15 | - | 8.62E-01 | 1.00E+00 | |||
| G-protein | 206 | 3 | 1.1 | + | 5.91E-01 | 4 | 1.8 | + | 1.08E-01 | 7.40E-01 | |||
| cysteine protease inhibitor | 21 | 1 | 0.11 | + | 6.20E-01 | 0 | 0.18 | - | 8.32E-01 | 1.00E+00 | |||
| protease inhibitor | 109 | 2 | 0.58 | + | 6.53E-01 | 0 | 0.95 | - | 3.85E-01 | 1.00E+00 | |||
| protein kinase | 510 | 5 | 2.72 | + | 7.55E-01 | 6 | 4.46 | + | 2.88E-01 | 1.00E+00 | |||
| actin and actin related protein | 31 | 1 | 0.17 | + | 7.94E-01 | 1 | 0.27 | + | 2.37E-01 | 1.00E+00 | |||
| ion channel | 341 | 0 | 1.82 | - | 7.94E-01 | 12 | 2.98 | + | 5.37E-05 | 2.07E-03 | |||
| receptor | 1076 | 3 | 5.73 | - | 7.94E-01 | 11 | 9.4 | + | 3.41E-01 | 1.00E+00 | |||
| carbohydrate kinase | 35 | 1 | 0.19 | + | 7.94E-01 | 0 | 0.31 | - | 7.36E-01 | 1.00E+00 | |||
| enzyme modulator | 857 | 7 | 4.56 | + | 7.94E-01 | 14 | 7.49 | + | 1.89E-02 | 2.95E-01 | |||
| heterotrimeric G-protein | 36 | 1 | 0.19 | + | 7.94E-01 | 1 | 0.31 | + | 2.70E-01 | 1.00E+00 | |||
| interleukin superfamily | 36 | 1 | 0.19 | + | 7.94E-01 | 0 | 0.31 | - | 7.30E-01 | 1.00E+00 | |||
| phospholipase | 36 | 1 | 0.19 | + | 7.94E-01 | 0 | 0.31 | - | 7.30E-01 | 1.00E+00 | |||
| metalloprotease | 145 | 2 | 0.77 | + | 7.94E-01 | 1 | 1.27 | - | 6.38E-01 | 1.00E+00 | |||
| apolipoprotein | 42 | 1 | 0.22 | + | 8.58E-01 | 0 | 0.37 | - | 6.93E-01 | 1.00E+00 | |||
| small GTPase | 158 | 2 | 0.84 | + | 8.58E-01 | 3 | 1.38 | + | 1.61E-01 | 9.42E-01 | |||
| HMG box transcription factor | 44 | 1 | 0.23 | + | 8.58E-01 | 0 | 0.38 | - | 6.80E-01 | 1.00E+00 | |||
| microtubule binding motor protein | 46 | 1 | 0.24 | + | 8.73E-01 | 0 | 0.4 | - | 6.69E-01 | 1.00E+00 | |||
| extracellular matrix glycoprotein | 55 | 1 | 0.29 | + | 1.00E+00 | 1 | 0.48 | + | 3.82E-01 | 1.00E+00 | |||
| lipase | 57 | 1 | 0.3 | + | 1.00E+00 | 0 | 0.5 | - | 6.07E-01 | 1.00E+00 | |||
| phosphatase modulator | 63 | 1 | 0.34 | + | 1.00E+00 | 0 | 0.55 | - | 5.76E-01 | 1.00E+00 | |||
| homeobox transcription factor | 233 | 0 | 1.24 | - | 1.00E+00 | 0 | 2.04 | - | 1.29E-01 | 8.03E-01 | |||
| helix-turn-helix transcription factor | 233 | 0 | 1.24 | - | 1.00E+00 | 0 | 2.04 | - | 1.29E-01 | 8.03E-01 | |||
| glycosyltransferase | 229 | 0 | 1.22 | - | 1.00E+00 | 2 | 2 | - | 6.76E-01 | 1.00E+00 | |||
| transporter | 1098 | 4 | 5.85 | - | 1.00E+00 | 19 | 9.6 | + | 3.64E-03 | 8.78E-02 | |||
| hydrolase | 454 | 1 | 2.42 | - | 1.00E+00 | 1 | 3.97 | - | 9.14E-02 | 6.81E-01 | |||
| G-protein coupled receptor | 447 | 1 | 2.38 | - | 1.00E+00 | 4 | 3.91 | + | 5.50E-01 | 1.00E+00 | |||
| RNA helicase | 71 | 1 | 0.38 | + | 1.00E+00 | 1 | 0.62 | + | 4.63E-01 | 1.00E+00 | |||
| extracellular matrix protein | 72 | 1 | 0.38 | + | 1.00E+00 | 1 | 0.63 | + | 4.68E-01 | 1.00E+00 | |||
| nucleic acid binding | 1466 | 6 | 7.8 | - | 1.00E+00 | 28 | 12.81 | + | 7.88E-05 | 2.53E-03 | |||
| serine protease inhibitor | 79 | 1 | 0.42 | + | 1.00E+00 | 0 | 0.69 | - | 5.01E-01 | 1.00E+00 | |||
| mRNA processing factor | 179 | 0 | 0.95 | - | 1.00E+00 | 0 | 1.56 | - | 2.08E-01 | 1.00E+00 | |||
| cation transporter | 178 | 0 | 0.95 | - | 1.00E+00 | 1 | 1.56 | - | 5.39E-01 | 1.00E+00 | |||
| isomerase | 94 | 1 | 0.5 | + | 1.00E+00 | 0 | 0.82 | - | 4.39E-01 | 1.00E+00 | |||
| signaling molecule | 961 | 6 | 5.12 | + | 1.00E+00 | 13 | 8.4 | + | 7.97E-02 | 6.81E-01 | |||
| amino acid transporter | 98 | 1 | 0.52 | + | 1.00E+00 | 0 | 0.86 | - | 4.24E-01 | 1.00E+00 | |||
| structural protein | 280 | 2 | 1.49 | + | 1.00E+00 | 3 | 2.45 | + | 4.43E-01 | 1.00E+00 | |||
| protein phosphatase | 111 | 1 | 0.59 | + | 1.00E+00 | 1 | 0.97 | + | 6.22E-01 | 1.00E+00 | |||
| protease | 476 | 3 | 2.53 | + | 1.00E+00 | 3 | 4.16 | - | 4.00E-01 | 1.00E+00 | |||
| membrane-bound signaling molecule | 133 | 0 | 0.71 | - | 1.00E+00 | 2 | 1.16 | + | 3.24E-01 | 1.00E+00 | |||
| ubiquitin-protein ligase | 132 | 1 | 0.7 | + | 1.00E+00 | 0 | 1.15 | - | 3.14E-01 | 1.00E+00 | |||
| cysteine protease | 121 | 0 | 0.64 | - | 1.00E+00 | 1 | 1.06 | - | 7.15E-01 | 1.00E+00 | |||
| basic helix-loop-helix transcription factor | 120 | 0 | 0.64 | - | 1.00E+00 | 0 | 1.05 | - | 3.49E-01 | 1.00E+00 | |||
| RNA binding protein | 727 | 4 | 3.87 | + | 1.00E+00 | 20 | 6.35 | + | 6.78E-06 | 3.94E-04 | |||
| serine protease | 153 | 1 | 0.81 | + | 1.00E+00 | 0 | 1.34 | - | 2.61E-01 | 1.00E+00 | |||
| G-protein modulator | 278 | 1 | 1.48 | - | 1.00E+00 | 6 | 2.43 | + | 3.64E-02 | 3.99E-01 | |||
| immunoglobulin receptor superfamily | 155 | 1 | 0.83 | + | 1.00E+00 | 1 | 1.35 | - | 6.07E-01 | 1.00E+00 | |||
| defense/immunity protein | 107 | 0 | 0.57 | - | 1.00E+00 | 0 | 0.94 | - | 3.92E-01 | 1.00E+00 | |||
| acetyltransferase | 105 | 0 | 0.56 | - | 1.00E+00 | 1 | 0.92 | + | 6.01E-01 | 1.00E+00 | |||
| cytokine | 159 | 1 | 0.85 | + | 1.00E+00 | 0 | 1.39 | - | 2.48E-01 | 1.00E+00 | |||
| kinase modulator | 103 | 0 | 0.55 | - | 1.00E+00 | 3 | 0.9 | + | 6.24E-02 | 6.02E-01 | |||
| ligand-gated ion channel | 101 | 0 | 0.54 | - | 1.00E+00 | 4 | 0.88 | + | 1.24E-02 | 2.18E-01 | |||
| growth factor | 165 | 1 | 0.88 | + | 1.00E+00 | 0 | 1.44 | - | 2.35E-01 | 1.00E+00 | |||
| peptide hormone | 169 | 1 | 0.9 | + | 1.00E+00 | 2 | 1.48 | + | 4.35E-01 | 1.00E+00 | |||
| ligase | 260 | 1 | 1.38 | - | 1.00E+00 | 1 | 2.27 | - | 3.35E-01 | 1.00E+00 | |||
| transcription cofactor | 255 | 1 | 1.36 | - | 1.00E+00 | 0 | 2.23 | - | 1.06E-01 | 7.40E-01 | |||
| cell adhesion molecule | 93 | 0 | 0.5 | - | 1.00E+00 | 1 | 0.81 | + | 5.57E-01 | 1.00E+00 | |||
| potassium channel | 89 | 0 | 0.47 | - | 1.00E+00 | 2 | 0.78 | + | 1.83E-01 | 1.00E+00 | |||
| ribosomal protein | 184 | 1 | 0.98 | + | 1.00E+00 | 16 | 1.61 | + | 1.18E-11 | 2.28E-09 | |||
| nucleotidyltransferase | 84 | 0 | 0.45 | - | 1.00E+00 | 0 | 0.73 | - | 4.79E-01 | 1.00E+00 | |||
| phosphatase | 230 | 1 | 1.22 | - | 1.00E+00 | 3 | 2.01 | + | 3.26E-01 | 1.00E+00 | |||
| DNA helicase | 80 | 0 | 0.43 | - | 1.00E+00 | 0 | 0.7 | - | 4.96E-01 | 1.00E+00 | |||
| guanyl-nucleotide exchange factor | 79 | 0 | 0.42 | - | 1.00E+00 | 1 | 0.69 | + | 4.99E-01 | 1.00E+00 | |||
| chromatin/chromatin-binding protein | 74 | 0 | 0.39 | - | 1.00E+00 | 1 | 0.65 | + | 4.77E-01 | 1.00E+00 | |||
| oxygenase | 74 | 0 | 0.39 | - | 1.00E+00 | 1 | 0.65 | + | 4.77E-01 | 1.00E+00 | |||
| ATP-binding cassette (ABC) transporter | 74 | 0 | 0.39 | - | 1.00E+00 | 1 | 0.65 | + | 4.77E-01 | 1.00E+00 | |||
| cytokine receptor | 213 | 1 | 1.13 | - | 1.00E+00 | 1 | 1.86 | - | 4.43E-01 | 1.00E+00 | |||
| cell junction protein | 67 | 0 | 0.36 | - | 1.00E+00 | 0 | 0.59 | - | 5.56E-01 | 1.00E+00 | |||
| calcium-binding protein | 63 | 0 | 0.34 | - | 1.00E+00 | 0 | 0.55 | - | 5.76E-01 | 1.00E+00 | |||
| ribonucleoprotein | 61 | 0 | 0.32 | - | 1.00E+00 | 0 | 0.53 | - | 5.86E-01 | 1.00E+00 | |||
| oxidase | 57 | 0 | 0.3 | - | 1.00E+00 | 0 | 0.5 | - | 6.07E-01 | 1.00E+00 | |||
| translation factor | 56 | 0 | 0.3 | - | 1.00E+00 | 2 | 0.49 | + | 8.68E-02 | 6.81E-01 | |||
| histone | 54 | 0 | 0.29 | - | 1.00E+00 | 0 | 0.47 | - | 6.23E-01 | 1.00E+00 | |||
| ATP synthase | 51 | 0 | 0.27 | - | 1.00E+00 | 1 | 0.45 | + | 3.60E-01 | 1.00E+00 | |||
| chemokine | 48 | 0 | 0.26 | - | 1.00E+00 | 0 | 0.42 | - | 6.57E-01 | 1.00E+00 | |||
| replication origin binding protein | 47 | 0 | 0.25 | - | 1.00E+00 | 0 | 0.41 | - | 6.63E-01 | 1.00E+00 | |||
| carbohydrate transporter | 46 | 0 | 0.24 | - | 1.00E+00 | 0 | 0.4 | - | 6.69E-01 | 1.00E+00 | |||
| nuclear hormone receptor | 46 | 0 | 0.24 | - | 1.00E+00 | 0 | 0.4 | - | 6.69E-01 | 1.00E+00 | |||
| kinase activator | 45 | 0 | 0.24 | - | 1.00E+00 | 1 | 0.39 | + | 3.25E-01 | 1.00E+00 | |||
| damaged DNA-binding protein | 45 | 0 | 0.24 | - | 1.00E+00 | 0 | 0.39 | - | 6.75E-01 | 1.00E+00 | |||
| hydroxylase | 44 | 0 | 0.23 | - | 1.00E+00 | 0 | 0.38 | - | 6.80E-01 | 1.00E+00 | |||
| SNARE protein | 37 | 0 | 0.2 | - | 1.00E+00 | 4 | 0.32 | + | 3.42E-04 | 9.43E-03 | |||
| anion channel | 37 | 0 | 0.2 | - | 1.00E+00 | 0 | 0.32 | - | 7.24E-01 | 1.00E+00 | |||
| nucleotide phosphatase | 35 | 0 | 0.19 | - | 1.00E+00 | 1 | 0.31 | + | 2.64E-01 | 1.00E+00 | |||
| nuclease | 35 | 0 | 0.19 | - | 1.00E+00 | 1 | 0.31 | + | 2.64E-01 | 1.00E+00 | |||
| myelin protein | 33 | 0 | 0.18 | - | 1.00E+00 | 1 | 0.29 | + | 2.51E-01 | 1.00E+00 | |||
| intracellular calcium-sensing protein | 33 | 0 | 0.18 | - | 1.00E+00 | 0 | 0.29 | - | 7.49E-01 | 1.00E+00 | |||
| endoribonuclease | 33 | 0 | 0.18 | - | 1.00E+00 | 0 | 0.29 | - | 7.49E-01 | 1.00E+00 | |||
| immunoglobulin | 33 | 0 | 0.18 | - | 1.00E+00 | 0 | 0.29 | - | 7.49E-01 | 1.00E+00 | |||
| calmodulin | 33 | 0 | 0.18 | - | 1.00E+00 | 0 | 0.29 | - | 7.49E-01 | 1.00E+00 | |||
| calcium channel | 31 | 0 | 0.17 | - | 1.00E+00 | 6 | 0.27 | + | 4.02E-07 | 3.88E-05 | |||
| type I cytokine receptor | 31 | 0 | 0.17 | - | 1.00E+00 | 0 | 0.27 | - | 7.63E-01 | 1.00E+00 | |||
| vesicle coat protein | 30 | 0 | 0.16 | - | 1.00E+00 | 1 | 0.26 | + | 2.31E-01 | 1.00E+00 | |||
| tight junction | 30 | 0 | 0.16 | - | 1.00E+00 | 0 | 0.26 | - | 7.69E-01 | 1.00E+00 | |||
| major histocompatibility complex antigen | 28 | 0 | 0.15 | - | 1.00E+00 | 0 | 0.24 | - | 7.83E-01 | 1.00E+00 | |||
| phosphatase inhibitor | 28 | 0 | 0.15 | - | 1.00E+00 | 0 | 0.24 | - | 7.83E-01 | 1.00E+00 | |||
| neuropeptide | 28 | 0 | 0.15 | - | 1.00E+00 | 0 | 0.24 | - | 7.83E-01 | 1.00E+00 | |||
| phosphodiesterase | 27 | 0 | 0.14 | - | 1.00E+00 | 0 | 0.24 | - | 7.90E-01 | 1.00E+00 | |||
| peroxidase | 27 | 0 | 0.14 | - | 1.00E+00 | 0 | 0.24 | - | 7.90E-01 | 1.00E+00 | |||
| glycosidase | 26 | 0 | 0.14 | - | 1.00E+00 | 0 | 0.23 | - | 7.97E-01 | 1.00E+00 | |||
| immunoglobulin superfamily cell adhesion molecule | 25 | 0 | 0.13 | - | 1.00E+00 | 1 | 0.22 | + | 1.96E-01 | 1.00E+00 | |||
| tubulin | 25 | 0 | 0.13 | - | 1.00E+00 | 1 | 0.22 | + | 1.96E-01 | 1.00E+00 | |||
| esterase | 25 | 0 | 0.13 | - | 1.00E+00 | 0 | 0.22 | - | 8.04E-01 | 1.00E+00 | |||
| exoribonuclease | 24 | 0 | 0.13 | - | 1.00E+00 | 0 | 0.21 | - | 8.11E-01 | 1.00E+00 | |||
| basic leucine zipper transcription factor | 24 | 0 | 0.13 | - | 1.00E+00 | 0 | 0.21 | - | 8.11E-01 | 1.00E+00 | |||
| CREB transcription factor | 24 | 0 | 0.13 | - | 1.00E+00 | 0 | 0.21 | - | 8.11E-01 | 1.00E+00 | |||
| cyclase | 23 | 0 | 0.12 | - | 1.00E+00 | 0 | 0.2 | - | 8.18E-01 | 1.00E+00 | |||
| tumor necrosis factor receptor | 22 | 0 | 0.12 | - | 1.00E+00 | 0 | 0.19 | - | 8.25E-01 | 1.00E+00 | |||
| RNA methyltransferase | 22 | 0 | 0.12 | - | 1.00E+00 | 0 | 0.19 | - | 8.25E-01 | 1.00E+00 | |||
| gap junction | 21 | 0 | 0.11 | - | 1.00E+00 | 0 | 0.18 | - | 8.32E-01 | 1.00E+00 | |||
| adenylate cyclase | 21 | 0 | 0.11 | - | 1.00E+00 | 0 | 0.18 | - | 8.32E-01 | 1.00E+00 | |||
| endodeoxyribonuclease | 20 | 0 | 0.11 | - | 1.00E+00 | 0 | 0.17 | - | 8.40E-01 | 1.00E+00 | |||
| chaperonin | 20 | 0 | 0.11 | - | 1.00E+00 | 0 | 0.17 | - | 8.40E-01 | 1.00E+00 | |||
| aminoacyl-tRNA synthetase | 20 | 0 | 0.11 | - | 1.00E+00 | 0 | 0.17 | - | 8.40E-01 | 1.00E+00 | |||
| sodium channel | 20 | 0 | 0.11 | - | 1.00E+00 | 0 | 0.17 | - | 8.40E-01 | 1.00E+00 | |||
| centromere DNA-binding protein | 18 | 0 | 0.1 | - | 1.00E+00 | 0 | 0.16 | - | 8.54E-01 | 1.00E+00 | |||
| antibacterial response protein | 18 | 0 | 0.1 | - | 1.00E+00 | 0 | 0.16 | - | 8.54E-01 | 1.00E+00 | |||
| mitochondrial carrier protein | 17 | 0 | 0.09 | - | 1.00E+00 | 0 | 0.15 | - | 8.62E-01 | 1.00E+00 | |||
| interferon superfamily | 17 | 0 | 0.09 | - | 1.00E+00 | 0 | 0.15 | - | 8.62E-01 | 1.00E+00 | |||
| decarboxylase | 17 | 0 | 0.09 | - | 1.00E+00 | 0 | 0.15 | - | 8.62E-01 | 1.00E+00 | |||
| galactosidase | 16 | 0 | 0.09 | - | 1.00E+00 | 0 | 0.14 | - | 8.69E-01 | 1.00E+00 | |||
| kinase inhibitor | 15 | 0 | 0.08 | - | 1.00E+00 | 0 | 0.13 | - | 8.77E-01 | 1.00E+00 | |||
| glucosidase | 15 | 0 | 0.08 | - | 1.00E+00 | 0 | 0.13 | - | 8.77E-01 | 1.00E+00 | |||
| surfactant | 15 | 0 | 0.08 | - | 1.00E+00 | 0 | 0.13 | - | 8.77E-01 | 1.00E+00 | |||
| DNA-directed RNA polymerase | 15 | 0 | 0.08 | - | 1.00E+00 | 0 | 0.13 | - | 8.77E-01 | 1.00E+00 | |||
| aspartic protease | 14 | 0 | 0.07 | - | 1.00E+00 | 1 | 0.12 | + | 1.15E-01 | 7.40E-01 | |||
| mutase | 14 | 0 | 0.07 | - | 1.00E+00 | 0 | 0.12 | - | 8.85E-01 | 1.00E+00 | |||
| deaminase | 12 | 0 | 0.06 | - | 1.00E+00 | 0 | 0.1 | - | 9.00E-01 | 1.00E+00 | |||
| DNA glycosylase | 12 | 0 | 0.06 | - | 1.00E+00 | 0 | 0.1 | - | 9.00E-01 | 1.00E+00 | |||
| intermediate filament | 11 | 0 | 0.06 | - | 1.00E+00 | 1 | 0.1 | + | 9.17E-02 | 6.81E-01 | |||
| tumor necrosis factor family member | 11 | 0 | 0.06 | - | 1.00E+00 | 0 | 0.1 | - | 9.08E-01 | 1.00E+00 | |||
| intermediate filament binding protein | 10 | 0 | 0.05 | - | 1.00E+00 | 1 | 0.09 | + | 8.37E-02 | 6.81E-01 | |||
| Hsp90 family chaperone | 10 | 0 | 0.05 | - | 1.00E+00 | 0 | 0.09 | - | 9.16E-01 | 1.00E+00 | |||
| DNA-directed DNA polymerase | 10 | 0 | 0.05 | - | 1.00E+00 | 0 | 0.09 | - | 9.16E-01 | 1.00E+00 | |||
| extracellular matrix structural protein | 9 | 0 | 0.05 | - | 1.00E+00 | 0 | 0.08 | - | 9.24E-01 | 1.00E+00 | |||
| carbohydrate phosphatase | 9 | 0 | 0.05 | - | 1.00E+00 | 0 | 0.08 | - | 9.24E-01 | 1.00E+00 | |||
| annexin | 9 | 0 | 0.05 | - | 1.00E+00 | 0 | 0.08 | - | 9.24E-01 | 1.00E+00 | |||
| protein kinase receptor | 8 | 0 | 0.04 | - | 1.00E+00 | 0 | 0.07 | - | 9.32E-01 | 1.00E+00 | |||
| neurotrophic factor | 8 | 0 | 0.04 | - | 1.00E+00 | 0 | 0.07 | - | 9.32E-01 | 1.00E+00 | |||
| epimerase/racemase | 7 | 0 | 0.04 | - | 1.00E+00 | 0 | 0.06 | - | 9.41E-01 | 1.00E+00 | |||
| viral protein | 7 | 0 | 0.04 | - | 1.00E+00 | 0 | 0.06 | - | 9.41E-01 | 1.00E+00 | |||
| viral coat protein | 7 | 0 | 0.04 | - | 1.00E+00 | 0 | 0.06 | - | 9.41E-01 | 1.00E+00 | |||
| DNA strand-pairing protein | 7 | 0 | 0.04 | - | 1.00E+00 | 0 | 0.06 | - | 9.41E-01 | 1.00E+00 | |||
| DNA ligase | 6 | 0 | 0.03 | - | 1.00E+00 | 1 | 0.05 | + | 5.11E-02 | 5.19E-01 | |||
| amylase | 6 | 0 | 0.03 | - | 1.00E+00 | 0 | 0.05 | - | 9.49E-01 | 1.00E+00 | |||
| helicase | 5 | 0 | 0.03 | - | 1.00E+00 | 0 | 0.04 | - | 9.57E-01 | 1.00E+00 | |||
| amino acid kinase | 5 | 0 | 0.03 | - | 1.00E+00 | 0 | 0.04 | - | 9.57E-01 | 1.00E+00 | |||
| pyrophosphatase | 5 | 0 | 0.03 | - | 1.00E+00 | 0 | 0.04 | - | 9.57E-01 | 1.00E+00 | |||
| transketolase | 5 | 0 | 0.03 | - | 1.00E+00 | 0 | 0.04 | - | 9.57E-01 | 1.00E+00 | |||
| metalloprotease inhibitor | 4 | 0 | 0.02 | - | 1.00E+00 | 0 | 0.03 | - | 9.66E-01 | 1.00E+00 | |||
| aldolase | 4 | 0 | 0.02 | - | 1.00E+00 | 0 | 0.03 | - | 9.66E-01 | 1.00E+00 | |||
| DNA topoisomerase | 4 | 0 | 0.02 | - | 1.00E+00 | 0 | 0.03 | - | 9.66E-01 | 1.00E+00 | |||
| deacetylase | 2 | 0 | 0.01 | - | 1.00E+00 | 0 | 0.02 | - | 9.83E-01 | 1.00E+00 | |||
| calsequestrin | 2 | 0 | 0.01 | - | 1.00E+00 | 0 | 0.02 | - | 9.83E-01 | 1.00E+00 | |||
| TGF-beta receptor | 2 | 0 | 0.01 | - | 1.00E+00 | 0 | 0.02 | - | 9.83E-01 | 1.00E+00 | |||
| DNA polymerase processivity factor | 2 | 0 | 0.01 | - | 1.00E+00 | 0 | 0.02 | - | 9.83E-01 | 1.00E+00 | |||
| DNA photolyase | 2 | 0 | 0.01 | - | 1.00E+00 | 0 | 0.02 | - | 9.83E-01 | 1.00E+00 | |||
| exodeoxyribonuclease | 1 | 0 | 0.01 | - | 1.00E+00 | 0 | 0.01 | - | 9.91E-01 | 1.00E+00 | |||
| voltage-gated ion channel | 1 | 0 | 0.01 | - | 1.00E+00 | 0 | 0.01 | - | 9.91E-01 | 1.00E+00 | |||
| primase | 1 | 0 | 0.01 | - | 1.00E+00 | 0 | 0.01 | - | 9.91E-01 | 1.00E+00 | |||
| phosphatase activator | 1 | 0 | 0.01 | - | 1.00E+00 | 0 | 0.01 | - | 9.91E-01 | 1.00E+00 | |||
| transaldolase | 1 | 0 | 0.01 | - | 1.00E+00 | 0 | 0.01 | - | 9.91E-01 | 1.00E+00 | |||
| reverse transcriptase | 1 | 0 | 0.01 | - | 1.00E+00 | 0 | 0.01 | - | 9.91E-01 | 1.00E+00 | |||
Supplementary Table 5. Pathways enriched in human and mouse unique or enriched proteins.
Proteins from the consensus PSD exclusive or significantly enriched in human or mouse were classified independently using the Panther 'Pathway' descriptor. Enrichment analysis was done to determine Pathways overrepresented in each species set of specific proteins. The Benjamini-Hochberg procedure was used to correct for multiple testing. The column 'observed' retrieves the number of proteins identified in each 'Pathway' while the column 'expected', the number that would have been identified by chance. Over or under-representations are shown by a (+) or (-) symbol respectively.
| Pathway (Human) | Homo sapiens genes | Observed | Expected | (over/under) | Corrected P-value | Pathway (Mouse) | Observed | Expected | (over/under) | P-value | Corrected P-value |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Unclassified | 17270 | 121 | 148.32 | - | 5.47E-06 | Ionotropic glutamate receptor pathway | 15 | .73 | + | 2.65E-15 | 4.08E-13 |
| Pyrimidine Metabolism | 15 | 4 | .13 | + | 7.70E-04 | Metabotropic glutamate receptor group III pathway | 11 | .95 | + | 5.19E-09 | 7.99E-07 |
| Parkinson disease | 97 | 7 | .83 | + | 1.25E-03 | Metabotropic glutamate receptor group II pathway | 7 | .64 | + | 4.67E-06 | 7.19E-04 |
| Axon guidance mediated by semaphorins | 37 | 4 | .32 | + | 1.23E-02 | Synaptic_vesicle_trafficking | 6 | .50 | + | 1.41E-05 | 2.17E-03 |
| Apoptosis signaling pathway | 119 | 6 | 1.02 | + | 1.93E-02 | Metabotropic glutamate receptor group I pathway | 5 | .35 | + | 3.34E-05 | 5.14E-03 |
| Glycolysis | 24 | 3 | .21 | + | 3.16E-02 | GABA-B_receptor_II_signaling | 5 | .54 | + | 2.47E-04 | 3.80E-02 |
| De novo purine biosynthesis | 33 | 3 | .28 | + | 6.67E-02 | Unclassified | 214 | 235.05 | - | 2.69E-04 | 4.14E-02 |
| EGF receptor signaling pathway | 130 | 5 | 1.12 | + | 1.07E-01 | Heterotrimeric G-protein signaling pathway-Gq alpha and Go alpha mediated pathway | 8 | 1.82 | + | 5.74E-04 | 8.84E-02 |
| Integrin signalling pathway | 196 | 6 | 1.68 | + | 1.25E-01 | Thyrotropin-releasing hormone receptor signaling pathway | 5 | .84 | + | 1.73E-03 | 2.66E-01 |
| TCA cycle | 16 | 2 | .14 | + | 1.32E-01 | Endothelin signaling pathway | 5 | 1.20 | + | 7.53E-03 | 1.00E+00 |
| Cytoskeletal regulation by Rho GTPase | 98 | 4 | .84 | + | 1.45E-01 | Heterotrimeric G-protein signaling pathway-Gi alpha and Gs alpha mediated pathway | 7 | 2.23 | + | 7.75E-03 | 1.00E+00 |
| 5-Hydroxytryptamine degredation | 19 | 2 | .16 | + | 1.45E-01 | Muscarinic acetylcholine receptor 2 and 4 signaling pathway | 4 | .84 | + | 1.07E-02 | 1.00E+00 |
| Toll receptor signaling pathway | 56 | 3 | .48 | + | 1.45E-01 | Beta2 adrenergic receptor signaling pathway | 3 | .60 | + | 2.29E-02 | 1.00E+00 |
| Huntington disease | 161 | 5 | 1.38 | + | 1.45E-01 | Beta1 adrenergic receptor signaling pathway | 3 | .60 | + | 2.29E-02 | 1.00E+00 |
| Ubiquitin proteasome pathway | 69 | 3 | .59 | + | 2.07E-01 | Alpha adrenergic receptor signaling pathway | 2 | .31 | + | 3.98E-02 | 1.00E+00 |
| VEGF signaling pathway | 69 | 3 | .59 | + | 2.07E-01 | Endogenous_cannabinoid_signaling | 2 | .33 | + | 4.29E-02 | 1.00E+00 |
| FGF signaling pathway | 124 | 4 | 1.06 | + | 2.07E-01 | Nicotinic acetylcholine receptor signaling pathway | 4 | 1.31 | + | 4.33E-02 | 1.00E+00 |
| Phenylalanine biosynthesis | 3 | 1 | .03 | + | 2.17E-01 | Cytoskeletal regulation by Rho GTPase | 4 | 1.33 | + | 4.61E-02 | 1.00E+00 |
| Tyrosine biosynthesis | 4 | 1 | .03 | + | 2.74E-01 | Beta3 adrenergic receptor signaling pathway | 2 | .35 | + | 4.95E-02 | 1.00E+00 |
| Synaptic_vesicle_trafficking | 37 | 2 | .32 | + | 3.08E-01 | Muscarinic acetylcholine receptor 1 and 3 signaling pathway | 3 | .83 | + | 5.16E-02 | 1.00E+00 |
| Serine glycine biosynthesis | 5 | 1 | .04 | + | 3.08E-01 | Proline biosynthesis | 1 | .05 | + | 5.30E-02 | 1.00E+00 |
| Asparagine and aspartate biosynthesis | 5 | 1 | .04 | + | 3.08E-01 | Hedgehog signaling pathway | 2 | .38 | + | 5.64E-02 | 1.00E+00 |
| Angiogenesis | 176 | 4 | 1.51 | + | 4.26E-01 | Cortocotropin releasing factor receptor signaling pathway | 2 | .41 | + | 6.37E-02 | 1.00E+00 |
| Pentose phosphate pathway | 8 | 1 | .07 | + | 4.26E-01 | 5HT4 type receptor mediated signaling pathway | 2 | .42 | + | 6.74E-02 | 1.00E+00 |
| Arginine biosynthesis | 8 | 1 | .07 | + | 4.26E-01 | Huntington disease | 5 | 2.19 | + | 7.08E-02 | 1.00E+00 |
| Phenylethylamine degradation | 10 | 1 | .09 | + | 4.87E-01 | Adrenaline and noradrenaline biosynthesis | 2 | .44 | + | 7.12E-02 | 1.00E+00 |
| Inflammation mediated by chemokine and cytokine signaling pathway | 272 | 5 | 2.34 | + | 4.94E-01 | EGF receptor signaling pathway | 4 | 1.77 | + | 1.03E-01 | 1.00E+00 |
| Ras Pathway | 75 | 2 | .64 | + | 7.48E-01 | Integrin signalling pathway | 5 | 2.67 | + | 1.31E-01 | 1.00E+00 |
| Dopamine receptor mediated signaling pathway | 80 | 2 | .69 | + | 8.02E-01 | Notch signaling pathway | 2 | .64 | + | 1.35E-01 | 1.00E+00 |
| T cell activation | 88 | 2 | .76 | + | 8.98E-01 | Transcription regulation by bZIP transcription factor | 2 | .73 | + | 1.68E-01 | 1.00E+00 |
| Interferon-gamma signaling pathway | 29 | 1 | .25 | + | 1.00E+00 | TCA cycle | 1 | .22 | + | 1.96E-01 | 1.00E+00 |
| Wnt signaling pathway | 318 | 1 | 2.73 | - | 1.00E+00 | Oxytocin receptor mediated signaling pathway | 2 | .82 | + | 1.97E-01 | 1.00E+00 |
| Insulin/IGF pathway-mitogen activated protein kinase kinase/MAP kinase cascade | 34 | 1 | .29 | + | 1.00E+00 | 5HT3 type receptor mediated signaling pathway | 1 | .24 | + | 2.17E-01 | 1.00E+00 |
| FAS signaling pathway | 36 | 1 | .31 | + | 1.00E+00 | VEGF signaling pathway | 2 | .94 | + | 2.42E-01 | 1.00E+00 |
| Alzheimer disease-presenilin pathway | 122 | 2 | 1.05 | + | 1.00E+00 | 5HT2 type receptor mediated signaling pathway | 2 | .94 | + | 2.42E-01 | 1.00E+00 |
| Angiotensin II-stimulated signaling through G proteins and beta-arrestin | 40 | 1 | .34 | + | 1.00E+00 | Ras Pathway | 2 | 1.02 | + | 2.72E-01 | 1.00E+00 |
| Heterotrimeric G-protein signaling pathway-Gq alpha and Go alpha mediated pathway | 134 | 0 | 1.15 | - | 1.00E+00 | Histamine H2 receptor mediated signaling pathway | 1 | .34 | + | 2.89E-01 | 1.00E+00 |
| p38 MAPK pathway | 46 | 1 | .40 | + | 1.00E+00 | Axon guidance mediated by Slit/Robo | 1 | .34 | + | 2.89E-01 | 1.00E+00 |
| PDGF signaling pathway | 148 | 2 | 1.27 | + | 1.00E+00 | Dopamine receptor mediated signaling pathway | 2 | 1.09 | + | 2.97E-01 | 1.00E+00 |
| TGF-beta signaling pathway | 149 | 2 | 1.28 | + | 1.00E+00 | Insulin/IGF pathway-protein kinase B signaling cascade | 0 | 1.20 | - | 3.01E-01 | 1.00E+00 |
| Alzheimer disease-amyloid secretase pathway | 67 | 1 | .58 | + | 1.00E+00 | Inflammation mediated by chemokine and cytokine signaling pathway | 5 | 3.70 | + | 3.13E-01 | 1.00E+00 |
| B cell activation | 73 | 1 | .63 | + | 1.00E+00 | Cadherin signaling pathway | 3 | 1.99 | + | 3.20E-01 | 1.00E+00 |
| Insulin/IGF pathway-protein kinase B signaling cascade | 88 | 0 | .76 | - | 1.00E+00 | TGF-beta signaling pathway | 3 | 2.03 | + | 3.31E-01 | 1.00E+00 |
| Endothelin signaling pathway | 88 | 1 | .76 | + | 1.00E+00 | Opioid prodynorphin pathway | 1 | .42 | + | 3.44E-01 | 1.00E+00 |
| Metabotropic glutamate receptor group III pathway | 70 | 0 | .60 | - | 1.00E+00 | Opioid proenkephalin pathway | 1 | .42 | + | 3.44E-01 | 1.00E+00 |
| 5HT2 type receptor mediated signaling pathway | 69 | 0 | .59 | - | 1.00E+00 | Opioid proopiomelanocortin pathway | 1 | .44 | + | 3.53E-01 | 1.00E+00 |
| Nicotinic acetylcholine receptor signaling pathway | 96 | 1 | .82 | + | 1.00E+00 | Interleukin signaling pathway | 1 | 2.16 | - | 3.62E-01 | 1.00E+00 |
| Thyrotropin-releasing hormone receptor signaling pathway | 62 | 0 | .53 | - | 1.00E+00 | Wnt signaling pathway | 3 | 4.33 | - | 3.70E-01 | 1.00E+00 |
| Muscarinic acetylcholine receptor 2 and 4 signaling pathway | 62 | 0 | .53 | - | 1.00E+00 | Axon guidance mediated by netrin | 1 | .48 | + | 3.79E-01 | 1.00E+00 |
| Heterotrimeric G-protein signaling pathway-Gi alpha and Gs alpha mediated pathway | 164 | 1 | 1.41 | - | 1.00E+00 | Parkinson disease | 2 | 1.32 | + | 3.81E-01 | 1.00E+00 |
| Muscarinic acetylcholine receptor 1 and 3 signaling pathway | 61 | 0 | .52 | - | 1.00E+00 | Ubiquitin proteasome pathway | 0 | .94 | - | 3.90E-01 | 1.00E+00 |
| Oxytocin receptor mediated signaling pathway | 60 | 0 | .52 | - | 1.00E+00 | PDGF signaling pathway | 1 | 2.01 | - | 4.01E-01 | 1.00E+00 |
| Interleukin signaling pathway | 159 | 1 | 1.37 | - | 1.00E+00 | Alzheimer disease-amyloid secretase pathway | 0 | .91 | - | 4.01E-01 | 1.00E+00 |
| Oxidative stress response | 56 | 0 | .48 | - | 1.00E+00 | Heterotrimeric G-protein signaling pathway-rod outer segment phototransduction | 1 | .53 | + | 4.12E-01 | 1.00E+00 |
| p53 pathway | 114 | 1 | .98 | + | 1.00E+00 | 5HT1 type receptor mediated signaling pathway | 1 | .60 | + | 4.51E-01 | 1.00E+00 |
| Transcription regulation by bZIP transcription factor | 54 | 0 | .46 | - | 1.00E+00 | Toll receptor signaling pathway | 0 | .76 | - | 4.66E-01 | 1.00E+00 |
| Ionotropic glutamate receptor pathway | 54 | 0 | .46 | - | 1.00E+00 | Oxidative stress response | 0 | .76 | - | 4.66E-01 | 1.00E+00 |
| p53 pathway feedback loops 2 | 52 | 0 | .45 | - | 1.00E+00 | PI3 kinase pathway | 2 | 1.59 | + | 4.73E-01 | 1.00E+00 |
| Cadherin signaling pathway | 146 | 1 | 1.25 | - | 1.00E+00 | FGF signaling pathway | 2 | 1.69 | + | 5.04E-01 | 1.00E+00 |
| Blood coagulation | 48 | 0 | .41 | - | 1.00E+00 | Alzheimer disease-presenilin pathway | 1 | 1.66 | - | 5.05E-01 | 1.00E+00 |
| Notch signaling pathway | 47 | 0 | .40 | - | 1.00E+00 | p53 pathway feedback loops 2 | 1 | .71 | + | 5.08E-01 | 1.00E+00 |
| Metabotropic glutamate receptor group II pathway | 47 | 0 | .40 | - | 1.00E+00 | Apoptosis signaling pathway | 1 | 1.62 | - | 5.18E-01 | 1.00E+00 |
| Histamine H1 receptor mediated signaling pathway | 47 | 0 | .40 | - | 1.00E+00 | Blood coagulation | 0 | .65 | - | 5.20E-01 | 1.00E+00 |
| Beta2 adrenergic receptor signaling pathway | 44 | 0 | .38 | - | 1.00E+00 | Histamine H1 receptor mediated signaling pathway | 0 | .64 | - | 5.27E-01 | 1.00E+00 |
| Beta1 adrenergic receptor signaling pathway | 44 | 0 | .38 | - | 1.00E+00 | p38 MAPK pathway | 0 | .63 | - | 5.34E-01 | 1.00E+00 |
| 5HT1 type receptor mediated signaling pathway | 44 | 0 | .38 | - | 1.00E+00 | p53 pathway | 1 | 1.55 | - | 5.40E-01 | 1.00E+00 |
| GABA-B_receptor_II_signaling | 40 | 0 | .34 | - | 1.00E+00 | Angiogenesis | 2 | 2.40 | - | 5.70E-01 | 1.00E+00 |
| Heterotrimeric G-protein signaling pathway-rod outer segment phototransduction | 39 | 0 | .33 | - | 1.00E+00 | Angiotensin II-stimulated signaling through G proteins and beta-arrestin | 0 | .54 | - | 5.80E-01 | 1.00E+00 |
| General transcription regulation | 37 | 0 | .32 | - | 1.00E+00 | Axon guidance mediated by semaphorins | 0 | .50 | - | 6.04E-01 | 1.00E+00 |
| PI3 kinase pathway | 117 | 1 | 1.00 | - | 1.00E+00 | General transcription regulation | 0 | .50 | - | 6.04E-01 | 1.00E+00 |
| Axon guidance mediated by netrin | 35 | 0 | .30 | - | 1.00E+00 | FAS signaling pathway | 0 | .49 | - | 6.12E-01 | 1.00E+00 |
| Enkephalin release | 33 | 0 | .28 | - | 1.00E+00 | Insulin/IGF pathway-mitogen activated protein kinase kinase/MAP kinase cascade | 0 | .46 | - | 6.29E-01 | 1.00E+00 |
| Adrenaline and noradrenaline biosynthesis | 32 | 0 | .27 | - | 1.00E+00 | B cell activation | 1 | .99 | + | 6.30E-01 | 1.00E+00 |
| Opioid proopiomelanocortin pathway | 32 | 0 | .27 | - | 1.00E+00 | De novo purine biosynthesis | 0 | .45 | - | 6.38E-01 | 1.00E+00 |
| Hypoxia response via HIF activation | 32 | 0 | .27 | - | 1.00E+00 | Enkephalin release | 0 | .45 | - | 6.38E-01 | 1.00E+00 |
| Opioid prodynorphin pathway | 31 | 0 | .27 | - | 1.00E+00 | Hypoxia response via HIF activation | 0 | .44 | - | 6.47E-01 | 1.00E+00 |
| Opioid proenkephalin pathway | 31 | 0 | .27 | - | 1.00E+00 | T cell activation | 1 | 1.20 | - | 6.63E-01 | 1.00E+00 |
| 5HT4 type receptor mediated signaling pathway | 31 | 0 | .27 | - | 1.00E+00 | Interferon-gamma signaling pathway | 0 | .39 | - | 6.74E-01 | 1.00E+00 |
| Cortocotropin releasing factor receptor signaling pathway | 30 | 0 | .26 | - | 1.00E+00 | Nicotine degradation | 0 | .34 | - | 7.11E-01 | 1.00E+00 |
| Hedgehog signaling pathway | 28 | 0 | .24 | - | 1.00E+00 | p53 pathway by glucose deprivation | 0 | .34 | - | 7.11E-01 | 1.00E+00 |
| Metabotropic glutamate receptor group I pathway | 26 | 0 | .22 | - | 1.00E+00 | Glycolysis | 0 | .33 | - | 7.21E-01 | 1.00E+00 |
| Beta3 adrenergic receptor signaling pathway | 26 | 0 | .22 | - | 1.00E+00 | Cell cycle | 0 | .30 | - | 7.41E-01 | 1.00E+00 |
| Nicotine degradation | 25 | 0 | .21 | - | 1.00E+00 | DNA replication | 0 | .29 | - | 7.51E-01 | 1.00E+00 |
| p53 pathway by glucose deprivation | 25 | 0 | .21 | - | 1.00E+00 | General transcription by RNA polymerase I | 0 | .27 | - | 7.62E-01 | 1.00E+00 |
| Histamine H2 receptor mediated signaling pathway | 25 | 0 | .21 | - | 1.00E+00 | 5-Hydroxytryptamine degredation | 0 | .26 | - | 7.72E-01 | 1.00E+00 |
| Axon guidance mediated by Slit/Robo | 25 | 0 | .21 | - | 1.00E+00 | Androgen/estrogene/progesterone biosynthesis | 0 | .26 | - | 7.72E-01 | 1.00E+00 |
| Endogenous_cannabinoid_signaling | 24 | 0 | .21 | - | 1.00E+00 | De novo pyrmidine ribonucleotides biosythesis | 0 | .24 | - | 7.83E-01 | 1.00E+00 |
| Alpha adrenergic receptor signaling pathway | 23 | 0 | .20 | - | 1.00E+00 | Plasminogen activating cascade | 0 | .24 | - | 7.83E-01 | 1.00E+00 |
| Cell cycle | 22 | 0 | .19 | - | 1.00E+00 | De novo pyrimidine deoxyribonucleotide biosynthesis | 0 | .24 | - | 7.83E-01 | 1.00E+00 |
| DNA replication | 21 | 0 | .18 | - | 1.00E+00 | JAK/STAT signaling pathway | 0 | .23 | - | 7.93E-01 | 1.00E+00 |
| General transcription by RNA polymerase I | 20 | 0 | .17 | - | 1.00E+00 | Pyrimidine Metabolism | 0 | .20 | - | 8.15E-01 | 1.00E+00 |
| Androgen/estrogene/progesterone biosynthesis | 19 | 0 | .16 | - | 1.00E+00 | Salvage pyrimidine ribonucleotides | 0 | .19 | - | 8.26E-01 | 1.00E+00 |
| De novo pyrmidine ribonucleotides biosythesis | 18 | 0 | .15 | - | 1.00E+00 | Pyruvate metabolism | 0 | .19 | - | 8.26E-01 | 1.00E+00 |
| Plasminogen activating cascade | 18 | 0 | .15 | - | 1.00E+00 | Heme biosynthesis | 0 | .18 | - | 8.38E-01 | 1.00E+00 |
| De novo pyrimidine deoxyribonucleotide biosynthesis | 18 | 0 | .15 | - | 1.00E+00 | Cholesterol biosynthesis | 0 | .18 | - | 8.38E-01 | 1.00E+00 |
| 5HT3 type receptor mediated signaling pathway | 18 | 0 | .15 | - | 1.00E+00 | Fructose galactose metabolism | 0 | .16 | - | 8.49E-01 | 1.00E+00 |
| JAK/STAT signaling pathway | 17 | 0 | .15 | - | 1.00E+00 | Vitamin D metabolism and pathway | 0 | .16 | - | 8.49E-01 | 1.00E+00 |
| Salvage pyrimidine ribonucleotides | 14 | 0 | .12 | - | 1.00E+00 | Vasopressin synthesis | 0 | .16 | - | 8.49E-01 | 1.00E+00 |
| Pyruvate metabolism | 14 | 0 | .12 | - | 1.00E+00 | Circadian clock system | 0 | .16 | - | 8.49E-01 | 1.00E+00 |
| Heme biosynthesis | 13 | 0 | .11 | - | 1.00E+00 | Phenylethylamine degradation | 0 | .14 | - | 8.73E-01 | 1.00E+00 |
| Cholesterol biosynthesis | 13 | 0 | .11 | - | 1.00E+00 | Formyltetrahydroformate biosynthesis | 0 | .14 | - | 8.73E-01 | 1.00E+00 |
| Fructose galactose metabolism | 12 | 0 | .10 | - | 1.00E+00 | mRNA splicing | 0 | .12 | - | 8.85E-01 | 1.00E+00 |
| Vitamin D metabolism and pathway | 12 | 0 | .10 | - | 1.00E+00 | Pentose phosphate pathway | 0 | .11 | - | 8.97E-01 | 1.00E+00 |
| Vasopressin synthesis | 12 | 0 | .10 | - | 1.00E+00 | Arginine biosynthesis | 0 | .11 | - | 8.97E-01 | 1.00E+00 |
| Circadian clock system | 12 | 0 | .10 | - | 1.00E+00 | Adenine and hypoxanthine salvage pathway | 0 | .11 | - | 8.97E-01 | 1.00E+00 |
| Formyltetrahydroformate biosynthesis | 10 | 0 | .09 | - | 1.00E+00 | ATP synthesis | 0 | .11 | - | 8.97E-01 | 1.00E+00 |
| mRNA splicing | 9 | 0 | .08 | - | 1.00E+00 | 2-arachidonoylglycerol biosynthesis | 0 | .10 | - | 9.09E-01 | 1.00E+00 |
| Adenine and hypoxanthine salvage pathway | 8 | 0 | .07 | - | 1.00E+00 | N-acetylglucosamine metabolism | 0 | .10 | - | 9.09E-01 | 1.00E+00 |
| ATP synthesis | 8 | 0 | .07 | - | 1.00E+00 | Mannose metabolism | 0 | .10 | - | 9.09E-01 | 1.00E+00 |
| 2-arachidonoylglycerol biosynthesis | 7 | 0 | .06 | - | 1.00E+00 | P53 pathway feedback loops 1 | 0 | .10 | - | 9.09E-01 | 1.00E+00 |
| N-acetylglucosamine metabolism | 7 | 0 | .06 | - | 1.00E+00 | Purine metabolism | 0 | .08 | - | 9.22E-01 | 1.00E+00 |
| Mannose metabolism | 7 | 0 | .06 | - | 1.00E+00 | Folate biosynthesis | 0 | .08 | - | 9.22E-01 | 1.00E+00 |
| P53 pathway feedback loops 1 | 7 | 0 | .06 | - | 1.00E+00 | Coenzyme A biosynthesis | 0 | .08 | - | 9.22E-01 | 1.00E+00 |
| Purine metabolism | 6 | 0 | .05 | - | 1.00E+00 | Gamma-aminobutyric acid synthesis | 0 | .08 | - | 9.22E-01 | 1.00E+00 |
| Folate biosynthesis | 6 | 0 | .05 | - | 1.00E+00 | 5-Hydroxytryptamine biosynthesis | 0 | .08 | - | 9.22E-01 | 1.00E+00 |
| Coenzyme A biosynthesis | 6 | 0 | .05 | - | 1.00E+00 | Serine glycine biosynthesis | 0 | .07 | - | 9.34E-01 | 1.00E+00 |
| Gamma-aminobutyric acid synthesis | 6 | 0 | .05 | - | 1.00E+00 | Asparagine and aspartate biosynthesis | 0 | .07 | - | 9.34E-01 | 1.00E+00 |
| 5-Hydroxytryptamine biosynthesis | 6 | 0 | .05 | - | 1.00E+00 | Tyrosine biosynthesis | 0 | .05 | - | 9.47E-01 | 1.00E+00 |
| Xanthine and guanine salvage pathway | 4 | 0 | .03 | - | 1.00E+00 | Xanthine and guanine salvage pathway | 0 | .05 | - | 9.47E-01 | 1.00E+00 |
| Salvage pyrimidine deoxyribonucleotides | 4 | 0 | .03 | - | 1.00E+00 | Salvage pyrimidine deoxyribonucleotides | 0 | .05 | - | 9.47E-01 | 1.00E+00 |
| Proline biosynthesis | 4 | 0 | .03 | - | 1.00E+00 | O-antigen biosynthesis | 0 | .05 | - | 9.47E-01 | 1.00E+00 |
| O-antigen biosynthesis | 4 | 0 | .03 | - | 1.00E+00 | Methylmalonyl pathway | 0 | .05 | - | 9.47E-01 | 1.00E+00 |
| Methylmalonyl pathway | 4 | 0 | .03 | - | 1.00E+00 | Methylcitrate cycle | 0 | .05 | - | 9.47E-01 | 1.00E+00 |
| Methylcitrate cycle | 4 | 0 | .03 | - | 1.00E+00 | Glutamine glutamate conversion | 0 | .05 | - | 9.47E-01 | 1.00E+00 |
| Glutamine glutamate conversion | 4 | 0 | .03 | - | 1.00E+00 | Ascorbate degradation | 0 | .05 | - | 9.47E-01 | 1.00E+00 |
| Ascorbate degradation | 4 | 0 | .03 | - | 1.00E+00 | Phenylalanine biosynthesis | 0 | .04 | - | 9.60E-01 | 1.00E+00 |
| Vitamin B6 metabolism | 3 | 0 | .03 | - | 1.00E+00 | Vitamin B6 metabolism | 0 | .04 | - | 9.60E-01 | 1.00E+00 |
| Valine biosynthesis | 3 | 0 | .03 | - | 1.00E+00 | Valine biosynthesis | 0 | .04 | - | 9.60E-01 | 1.00E+00 |
| S adenosyl methionine biosynthesis | 3 | 0 | .03 | - | 1.00E+00 | S adenosyl methionine biosynthesis | 0 | .04 | - | 9.60E-01 | 1.00E+00 |
| Ornithine degradation | 3 | 0 | .03 | - | 1.00E+00 | Ornithine degradation | 0 | .04 | - | 9.60E-01 | 1.00E+00 |
| Isoleucine biosynthesis | 3 | 0 | .03 | - | 1.00E+00 | Isoleucine biosynthesis | 0 | .04 | - | 9.60E-01 | 1.00E+00 |
| Acetate utilization | 3 | 0 | .03 | - | 1.00E+00 | Acetate utilization | 0 | .04 | - | 9.60E-01 | 1.00E+00 |
| Threonine biosynthesis | 2 | 0 | .02 | - | 1.00E+00 | Threonine biosynthesis | 0 | .03 | - | 9.73E-01 | 1.00E+00 |
| Thiamine metabolism | 2 | 0 | .02 | - | 1.00E+00 | Thiamine metabolism | 0 | .03 | - | 9.73E-01 | 1.00E+00 |
| Sulfate assimilation | 2 | 0 | .02 | - | 1.00E+00 | Sulfate assimilation | 0 | .03 | - | 9.73E-01 | 1.00E+00 |
| Succinate to proprionate conversion | 2 | 0 | .02 | - | 1.00E+00 | Succinate to proprionate conversion | 0 | .03 | - | 9.73E-01 | 1.00E+00 |
| Pyridoxal phosphate salvage pathway | 2 | 0 | .02 | - | 1.00E+00 | Pyridoxal phosphate salvage pathway | 0 | .03 | - | 9.73E-01 | 1.00E+00 |
| PLP biosynthesis | 2 | 0 | .02 | - | 1.00E+00 | PLP biosynthesis | 0 | .03 | - | 9.73E-01 | 1.00E+00 |
| Lysine biosynthesis | 2 | 0 | .02 | - | 1.00E+00 | Lysine biosynthesis | 0 | .03 | - | 9.73E-01 | 1.00E+00 |
| Lipoate_biosynthesis | 2 | 0 | .02 | - | 1.00E+00 | Lipoate_biosynthesis | 0 | .03 | - | 9.73E-01 | 1.00E+00 |
| Leucine biosynthesis | 2 | 0 | .02 | - | 1.00E+00 | Leucine biosynthesis | 0 | .03 | - | 9.73E-01 | 1.00E+00 |
| Carnitine metabolism | 2 | 0 | .02 | - | 1.00E+00 | Carnitine metabolism | 0 | .03 | - | 9.73E-01 | 1.00E+00 |
| Carnitine and CoA metabolism | 2 | 0 | .02 | - | 1.00E+00 | Carnitine and CoA metabolism | 0 | .03 | - | 9.73E-01 | 1.00E+00 |
| Histamine synthesis | 2 | 0 | .02 | - | 1.00E+00 | Histamine synthesis | 0 | .03 | - | 9.73E-01 | 1.00E+00 |
| Aminobutyrate degradation | 2 | 0 | .02 | - | 1.00E+00 | Aminobutyrate degradation | 0 | .03 | - | 9.73E-01 | 1.00E+00 |
| Alanine biosynthesis | 2 | 0 | .02 | - | 1.00E+00 | Alanine biosynthesis | 0 | .03 | - | 9.73E-01 | 1.00E+00 |
| Triacylglycerol metabolism | 1 | 0 | .01 | - | 1.00E+00 | Triacylglycerol metabolism | 0 | .01 | - | 9.86E-01 | 1.00E+00 |
| Bupropion_degradation | 1 | 0 | .01 | - | 1.00E+00 | Bupropion_degradation | 0 | .01 | - | 9.86E-01 | 1.00E+00 |
| Anandamide_degradation | 1 | 0 | .01 | - | 1.00E+00 | Anandamide_degradation | 0 | .01 | - | 9.86E-01 | 1.00E+00 |
| Methionine biosynthesis | 1 | 0 | .01 | - | 1.00E+00 | Methionine biosynthesis | 0 | .01 | - | 9.86E-01 | 1.00E+00 |
| Flavin biosynthesis | 1 | 0 | .01 | - | 1.00E+00 | Flavin biosynthesis | 0 | .01 | - | 9.86E-01 | 1.00E+00 |
| Cysteine biosynthesis | 1 | 0 | .01 | - | 1.00E+00 | Cysteine biosynthesis | 0 | .01 | - | 9.86E-01 | 1.00E+00 |
| Cobalamin biosynthesis | 1 | 0 | .01 | - | 1.00E+00 | Cobalamin biosynthesis | 0 | .01 | - | 9.86E-01 | 1.00E+00 |
| Allantoin degradation | 1 | 0 | .01 | - | 1.00E+00 | Allantoin degradation | 0 | .01 | - | 9.86E-01 | 1.00E+00 |
| 1.00E+00 | 1.00E+00 |
Supplementary Table 6. GO Biological Process term enrichment in human and mouse unique or enriched proteins.
Proteins from the consensus PSD exclusive or significantly enriched in human or mouse were classified independently using the Gene Ontology (GO) term 'Biological Process'. Enrichment analysis was done to determine Biological Processes overrepresented in each species set of specific proteins. The Benjamini-Hochberg procedure was used to correct for multiple testing. The column 'observed' retrieves the number of proteins identified in each 'Biological Process' while the column 'expected', the number that would have been identified by chance. Over or under-representations are shown by a (+) or (-) symbol respectively.
| Biological Process | Homo sapiens genes | Observed | Expected | (over/under) | Corrected P-value | Biological Process | Observed | Expected | (over/under) | P-value | Corrected P-value |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Unclassified | 7108 | 25 | 61.05 | - | 1.36E-07 | neurological system process | 49 | 15.72 | + | 1.62E-12 | 2.93E-10 |
| protein folding | 197 | 14 | 1.69 | + | 2.61E-07 | synaptic transmission | 30 | 6.29 | + | 2.75E-12 | 2.93E-10 |
| transcription | 1993 | 1 | 17.12 | - | 1.48E-05 | system process | 52 | 19.19 | + | 5.21E-11 | 3.70E-09 |
| transcription from RNA polymerase II promoter | 1987 | 1 | 17.06 | - | 1.48E-05 | cell communication | 94 | 49.42 | + | 9.05E-11 | 4.82E-09 |
| protein transport | 1254 | 30 | 10.77 | + | 1.48E-05 | nerve-nerve synaptic transmission | 13 | 1.14 | + | 2.49E-10 | 1.06E-08 |
| intracellular protein transport | 1254 | 30 | 10.77 | + | 1.48E-05 | "nucleobase, nucleoside, nucleotide and nucleic acid metabolic process" | 12 | 44.87 | - | 6.11E-10 | 2.17E-08 |
| regulation of transcription from RNA polymerase II promoter | 1563 | 0 | 13.42 | - | 2.58E-05 | cellular process | 118 | 72.49 | + | 1.96E-09 | 5.96E-08 |
| cellular component morphogenesis | 734 | 19 | 6.3 | + | 4.64E-04 | regulation of transcription from RNA polymerase II promoter | 1 | 21.27 | - | 5.76E-09 | 1.53E-07 |
| anatomical structure morphogenesis | 734 | 19 | 6.3 | + | 4.64E-04 | transport | 64 | 32.24 | + | 5.90E-08 | 1.40E-06 |
| response to stress | 265 | 11 | 2.28 | + | 4.64E-04 | cell-cell signaling | 32 | 11.73 | + | 3.53E-07 | 7.52E-06 |
| protein metabolic process | 2803 | 44 | 24.07 | + | 8.04E-04 | transcription | 6 | 27.13 | - | 4.73E-07 | 9.02E-06 |
| vesicle-mediated transport | 801 | 19 | 6.88 | + | 1.20E-03 | transcription from RNA polymerase II promoter | 6 | 27.04 | - | 5.08E-07 | 9.02E-06 |
| cellular component organization | 989 | 20 | 8.49 | + | 5.82E-03 | signal transduction | 80 | 46.97 | + | 5.55E-07 | 9.09E-06 |
| protein complex assembly | 73 | 5 | 0.63 | + | 6.59E-03 | vesicle-mediated transport | 29 | 10.9 | + | 2.12E-06 | 3.23E-05 |
| transport | 2369 | 36 | 20.35 | + | 6.59E-03 | protein transport | 36 | 17.07 | + | 2.12E-05 | 3.01E-04 |
| metabolic process | 7511 | 85 | 64.51 | + | 1.24E-02 | intracellular protein transport | 36 | 17.07 | + | 2.12E-05 | 3.01E-04 |
| intracellular signaling cascade | 1017 | 19 | 8.73 | + | 1.59E-02 | neurotransmitter secretion | 13 | 3.17 | + | 2.40E-05 | 3.01E-04 |
| "nucleobase, nucleoside, nucleotide and nucleic acid metabolic process" | 3297 | 15 | 28.32 | - | 2.90E-02 | exocytosis | 14 | 3.77 | + | 3.42E-05 | 4.05E-04 |
| cellular process | 5326 | 63 | 45.74 | + | 2.90E-02 | translation | 16 | 4.82 | + | 3.61E-05 | 4.05E-04 |
| glycolysis | 72 | 4 | 0.62 | + | 3.88E-02 | protein amino acid phosphorylation | 22 | 8.96 | + | 1.19E-04 | 1.27E-03 |
| cell motion | 448 | 10 | 3.85 | + | 5.76E-02 | neuronal action potential propagation | 5 | 0.65 | + | 5.62E-04 | 5.63E-03 |
| primary metabolic process | 7131 | 77 | 61.24 | + | 7.86E-02 | response to stimulus | 4 | 15.18 | - | 5.86E-04 | 5.63E-03 |
| monosaccharide metabolic process | 151 | 5 | 1.3 | + | 9.45E-02 | immune system process | 9 | 22.82 | - | 6.08E-04 | 5.63E-03 |
| endocytosis | 353 | 8 | 3.03 | + | 9.99E-02 | Unclassified | 72 | 96.74 | - | 8.54E-04 | 7.58E-03 |
| generation of precursor metabolites and energy | 285 | 7 | 2.45 | + | 9.99E-02 | synaptic vesicle exocytosis | 8 | 1.99 | + | 9.93E-04 | 8.46E-03 |
| pyrimidine base metabolic process | 55 | 3 | 0.47 | + | 9.99E-02 | ion transport | 19 | 8.55 | + | 1.13E-03 | 9.26E-03 |
| G-protein coupled receptor protein signaling pathway | 680 | 1 | 5.84 | - | 1.45E-01 | ferredoxin metabolic process | 2 | 0.08 | + | 3.15E-03 | 2.49E-02 |
| synaptic transmission | 462 | 9 | 3.97 | + | 1.45E-01 | cation transport | 15 | 6.89 | + | 4.40E-03 | 3.35E-02 |
| system development | 1206 | 4 | 10.36 | - | 1.48E-01 | DNA metabolic process | 0 | 5.08 | - | 5.95E-03 | 4.37E-02 |
| tricarboxylic acid cycle | 26 | 2 | 0.22 | + | 1.48E-01 | intracellular signaling cascade | 24 | 13.84 | + | 6.72E-03 | 4.77E-02 |
| cell-cell adhesion | 442 | 0 | 3.8 | - | 1.48E-01 | primary metabolic process | 78 | 97.06 | - | 8.59E-03 | 5.80E-02 |
| neuromuscular synaptic transmission | 27 | 2 | 0.23 | + | 1.53E-01 | RNA metabolic process | 1 | 6.72 | - | 8.72E-03 | 5.80E-02 |
| cell surface receptor linked signal transduction | 1628 | 7 | 13.98 | - | 1.74E-01 | immune response | 1 | 6.47 | - | 1.10E-02 | 7.10E-02 |
| fatty acid metabolic process | 204 | 5 | 1.75 | + | 2.02E-01 | dorsal/ventral axis specification | 3 | 0.5 | + | 1.46E-02 | 9.15E-02 |
| reproduction | 368 | 0 | 3.16 | - | 2.51E-01 | cell adhesion | 20 | 11.76 | + | 1.55E-02 | 9.43E-02 |
| protein modification process | 1177 | 16 | 10.11 | + | 2.79E-01 | metabolic process | 85 | 102.23 | - | 1.71E-02 | 1.01E-01 |
| tRNA aminoacylation for protein translation | 42 | 2 | 0.36 | + | 2.90E-01 | proteolysis | 6 | 13.43 | - | 1.79E-02 | 1.03E-01 |
| neurotransmitter secretion | 233 | 5 | 2 | + | 2.90E-01 | protein metabolic process | 51 | 38.15 | + | 1.83E-02 | 1.03E-01 |
| gamete generation | 335 | 0 | 2.88 | - | 2.97E-01 | protein modification process | 25 | 16.02 | + | 1.92E-02 | 1.05E-01 |
| purine base metabolic process | 100 | 3 | 0.86 | + | 2.97E-01 | muscle contraction | 9 | 4.1 | + | 2.34E-02 | 1.22E-01 |
| response to stimulus | 1115 | 15 | 9.58 | + | 2.98E-01 | cell surface receptor linked signal transduction | 32 | 22.16 | + | 2.35E-02 | 1.22E-01 |
| protein amino acid phosphorylation | 658 | 10 | 5.65 | + | 2.98E-01 | G-protein coupled receptor protein signaling pathway | 16 | 9.26 | + | 2.52E-02 | 1.28E-01 |
| system process | 1410 | 18 | 12.11 | + | 2.98E-01 | anion transport | 4 | 1.22 | + | 3.56E-02 | 1.76E-01 |
| pentose-phosphate shunt | 8 | 1 | 0.07 | + | 3.21E-01 | asymmetric protein localization | 2 | 0.3 | + | 3.67E-02 | 1.78E-01 |
| RNA metabolic process | 494 | 1 | 4.24 | - | 3.40E-01 | protein localization | 2 | 0.3 | + | 3.67E-02 | 1.78E-01 |
| carbohydrate metabolic process | 687 | 10 | 5.9 | + | 3.40E-01 | phosphate metabolic process | 5 | 1.91 | + | 4.39E-02 | 2.03E-01 |
| mRNA processing | 298 | 0 | 2.56 | - | 3.40E-01 | protein targeting | 5 | 1.96 | + | 4.85E-02 | 2.20E-01 |
| respiratory electron transport chain | 262 | 5 | 2.25 | + | 3.40E-01 | tricarboxylic acid cycle | 2 | 0.35 | + | 4.95E-02 | 2.20E-01 |
| cellular amino acid metabolic process | 266 | 5 | 2.28 | + | 3.50E-01 | "nuclear mRNA splicing, via spliceosome" | 0 | 2.97 | - | 5.06E-02 | 2.20E-01 |
| cellular amino acid and derivative metabolic process | 266 | 5 | 2.28 | + | 3.50E-01 | cytokinesis | 4 | 1.39 | + | 5.20E-02 | 2.22E-01 |
| ion transport | 628 | 2 | 5.39 | - | 3.76E-01 | calcium-mediated signaling | 5 | 2.15 | + | 6.65E-02 | 2.78E-01 |
| exocytosis | 277 | 5 | 2.38 | + | 3.76E-01 | DNA repair | 0 | 2.61 | - | 7.24E-02 | 2.97E-01 |
| cellular defense response | 264 | 0 | 2.27 | - | 4.07E-01 | nitric oxide biosynthetic process | 1 | 0.08 | + | 7.84E-02 | 3.15E-01 |
| receptor-mediated endocytosis | 131 | 3 | 1.13 | + | 4.07E-01 | nitric oxide mediated signal transduction | 1 | 0.08 | + | 7.84E-02 | 3.15E-01 |
| cytokine-mediated signaling pathway | 261 | 0 | 2.24 | - | 4.07E-01 | mRNA processing | 1 | 4.06 | - | 8.60E-02 | 3.33E-01 |
| mesoderm development | 754 | 3 | 6.48 | - | 4.15E-01 | mitosis | 9 | 5.31 | + | 8.80E-02 | 3.35E-01 |
| nervous system development | 738 | 3 | 6.34 | - | 4.44E-01 | macrophage activation | 0 | 2.29 | - | 1.01E-01 | 3.77E-01 |
| muscle contraction | 301 | 5 | 2.59 | + | 4.44E-01 | mesoderm development | 6 | 10.26 | - | 1.10E-01 | 4.04E-01 |
| neurological system process | 1155 | 14 | 9.92 | + | 4.44E-01 | DNA replication | 0 | 2.11 | - | 1.20E-01 | 4.16E-01 |
| developmental process | 2093 | 23 | 17.98 | + | 4.61E-01 | reproduction | 2 | 5.01 | - | 1.22E-01 | 4.16E-01 |
| cell adhesion | 864 | 4 | 7.42 | - | 4.61E-01 | response to stress | 1 | 3.61 | - | 1.23E-01 | 4.16E-01 |
| oxidative phosphorylation | 76 | 2 | 0.65 | + | 4.78E-01 | cellular defense response | 1 | 3.59 | - | 1.25E-01 | 4.16E-01 |
| "nuclear mRNA splicing, via spliceosome" | 218 | 0 | 1.87 | - | 5.14E-01 | general transcription from RNA polymerase II promoter | 2 | 0.61 | + | 1.26E-01 | 4.16E-01 |
| pattern specification process | 207 | 0 | 1.78 | - | 5.51E-01 | monosaccharide metabolic process | 0 | 2.06 | - | 1.27E-01 | 4.16E-01 |
| DNA metabolic process | 373 | 1 | 3.2 | - | 5.51E-01 | cytokine-mediated signaling pathway | 1 | 3.55 | - | 1.29E-01 | 4.16E-01 |
| coenzyme metabolic process | 92 | 2 | 0.79 | + | 5.95E-01 | organelle organization | 1 | 3.55 | - | 1.29E-01 | 4.16E-01 |
| cation transport | 506 | 2 | 4.35 | - | 5.95E-01 | segment specification | 0 | 2.03 | - | 1.31E-01 | 4.16E-01 |
| translation | 354 | 5 | 3.04 | + | 5.95E-01 | oxygen and reactive oxygen species metabolic process | 2 | 0.69 | + | 1.54E-01 | 4.82E-01 |
| immune system process | 1677 | 18 | 14.4 | + | 5.99E-01 | transmembrane receptor protein tyrosine kinase signaling pathway | 1 | 3.29 | - | 1.58E-01 | 4.88E-01 |
| spermatogenesis | 187 | 0 | 1.61 | - | 6.06E-01 | gamete generation | 2 | 4.56 | - | 1.65E-01 | 4.98E-01 |
| nuclear transport | 98 | 2 | 0.84 | + | 6.18E-01 | MAPKKK cascade | 6 | 3.7 | + | 1.69E-01 | 4.98E-01 |
| skeletal system development | 181 | 0 | 1.55 | - | 6.21E-01 | establishment or maintenance of chromatin architecture | 1 | 3.2 | - | 1.70E-01 | 4.98E-01 |
| cytokinesis | 102 | 2 | 0.88 | + | 6.39E-01 | carbohydrate metabolic process | 6 | 9.35 | - | 1.72E-01 | 4.98E-01 |
| angiogenesis | 172 | 0 | 1.48 | - | 6.53E-01 | "nucleobase, nucleoside, nucleotide and nucleic acid transport" | 2 | 0.75 | + | 1.73E-01 | 4.98E-01 |
| mitosis | 390 | 5 | 3.35 | + | 6.90E-01 | localization | 2 | 0.76 | + | 1.78E-01 | 5.06E-01 |
| cell communication | 3631 | 35 | 31.18 | + | 6.90E-01 | B cell mediated immunity | 0 | 1.7 | - | 1.81E-01 | 5.07E-01 |
| RNA localization | 34 | 1 | 0.29 | + | 6.90E-01 | lipid transport | 4 | 2.26 | + | 1.92E-01 | 5.31E-01 |
| fatty acid biosynthetic process | 34 | 1 | 0.29 | + | 6.90E-01 | generation of precursor metabolites and energy | 6 | 3.88 | + | 1.95E-01 | 5.33E-01 |
| blood coagulation | 158 | 0 | 1.36 | - | 6.90E-01 | carbohydrate transport | 2 | 0.87 | + | 2.17E-01 | 5.84E-01 |
| response to external stimulus | 158 | 0 | 1.36 | - | 6.90E-01 | nitrogen compound metabolic process | 1 | 0.24 | + | 2.17E-01 | 5.84E-01 |
| DNA replication | 155 | 0 | 1.33 | - | 6.92E-01 | cellular amino acid catabolic process | 2 | 0.88 | + | 2.22E-01 | 5.84E-01 |
| embryonic development | 151 | 0 | 1.3 | - | 7.07E-01 | phospholipid metabolic process | 1 | 2.8 | - | 2.29E-01 | 5.95E-01 |
| segment specification | 149 | 0 | 1.28 | - | 7.11E-01 | fatty acid metabolic process | 1 | 2.78 | - | 2.34E-01 | 5.96E-01 |
| ectoderm development | 827 | 5 | 7.1 | - | 7.15E-01 | extracellular transport | 0 | 1.42 | - | 2.42E-01 | 5.96E-01 |
| cell cycle | 1152 | 12 | 9.89 | + | 7.19E-01 | protein amino acid glycosylation | 1 | 2.72 | - | 2.43E-01 | 5.96E-01 |
| lipid metabolic process | 836 | 9 | 7.18 | + | 7.23E-01 | natural killer cell activation | 0 | 1.4 | - | 2.45E-01 | 5.96E-01 |
| cell-cell signaling | 862 | 9 | 7.4 | + | 7.91E-01 | induction of apoptosis | 5 | 3.36 | + | 2.48E-01 | 5.96E-01 |
| B cell mediated immunity | 125 | 0 | 1.07 | - | 8.21E-01 | protein folding | 1 | 2.68 | - | 2.51E-01 | 5.96E-01 |
| organelle organization | 261 | 1 | 2.24 | - | 8.21E-01 | steroid metabolic process | 1 | 2.68 | - | 2.51E-01 | 5.96E-01 |
| protein targeting | 144 | 2 | 1.24 | + | 8.31E-01 | hemopoiesis | 0 | 1.37 | - | 2.52E-01 | 5.96E-01 |
| synaptic vesicle exocytosis | 146 | 2 | 1.25 | + | 8.36E-01 | purine base metabolic process | 0 | 1.36 | - | 2.56E-01 | 5.99E-01 |
| induction of apoptosis | 247 | 1 | 2.12 | - | 8.40E-01 | nuclear transport | 0 | 1.33 | - | 2.63E-01 | 6.09E-01 |
| "nucleobase, nucleoside, nucleotide and nucleic acid transport" | 55 | 1 | 0.47 | + | 8.40E-01 | spermatogenesis | 1 | 2.55 | - | 2.77E-01 | 6.30E-01 |
| localization | 56 | 1 | 0.48 | + | 8.40E-01 | sulfur metabolic process | 0 | 1.28 | - | 2.77E-01 | 6.30E-01 |
| tRNA metabolic process | 56 | 1 | 0.48 | + | 8.40E-01 | visual perception | 5 | 3.54 | + | 2.81E-01 | 6.30E-01 |
| transmembrane receptor protein tyrosine kinase signaling pathway | 242 | 1 | 2.08 | - | 8.40E-01 | coenzyme metabolic process | 0 | 1.25 | - | 2.85E-01 | 6.32E-01 |
| visual perception | 260 | 3 | 2.23 | + | 8.40E-01 | response to interferon-gamma | 0 | 1.24 | - | 2.89E-01 | 6.35E-01 |
| establishment or maintenance of chromatin architecture | 235 | 1 | 2.02 | - | 8.40E-01 | muscle organ development | 1 | 2.45 | - | 2.96E-01 | 6.37E-01 |
| heart development | 105 | 0 | 0.9 | - | 8.40E-01 | JAK-STAT cascade | 0 | 1.21 | - | 2.97E-01 | 6.37E-01 |
| extracellular transport | 104 | 0 | 0.89 | - | 8.40E-01 | cellular component morphogenesis | 12 | 9.99 | + | 3.00E-01 | 6.37E-01 |
| natural killer cell activation | 103 | 0 | 0.88 | - | 8.40E-01 | anatomical structure morphogenesis | 12 | 9.99 | + | 3.00E-01 | 6.37E-01 |
| immune response | 475 | 3 | 4.08 | - | 8.40E-01 | homeostatic process | 0 | 1.18 | - | 3.05E-01 | 6.37E-01 |
| lipid transport | 166 | 2 | 1.43 | + | 8.40E-01 | neuromuscular synaptic transmission | 1 | 0.37 | + | 3.08E-01 | 6.37E-01 |
| hemopoiesis | 101 | 0 | 0.87 | - | 8.40E-01 | transcription initiation from RNA polymerase II promoter | 0 | 1.16 | - | 3.14E-01 | 6.43E-01 |
| signal transduction | 3451 | 31 | 29.64 | + | 8.40E-01 | cell-matrix adhesion | 0 | 1.14 | - | 3.18E-01 | 6.43E-01 |
| polysaccharide metabolic process | 225 | 1 | 1.93 | - | 8.40E-01 | response to toxin | 0 | 1.13 | - | 3.22E-01 | 6.43E-01 |
| cellular amino acid biosynthetic process | 64 | 1 | 0.55 | + | 8.40E-01 | system development | 14 | 16.41 | - | 3.23E-01 | 6.43E-01 |
| cellular amino acid catabolic process | 65 | 1 | 0.56 | + | 8.40E-01 | meiosis | 2 | 1.21 | + | 3.42E-01 | 6.75E-01 |
| JNK cascade | 98 | 0 | 0.84 | - | 8.40E-01 | endocytosis | 6 | 4.8 | + | 3.49E-01 | 6.75E-01 |
| blood circulation | 96 | 0 | 0.82 | - | 8.45E-01 | DNA recombination | 0 | 1.05 | - | 3.50E-01 | 6.75E-01 |
| anion transport | 90 | 0 | 0.77 | - | 8.45E-01 | rRNA metabolic process | 0 | 1.05 | - | 3.50E-01 | 6.75E-01 |
| JAK-STAT cascade | 89 | 0 | 0.76 | - | 8.45E-01 | oxidative phosphorylation | 0 | 1.03 | - | 3.55E-01 | 6.75E-01 |
| complement activation | 73 | 1 | 0.63 | + | 8.45E-01 | blood coagulation | 1 | 2.15 | - | 3.66E-01 | 6.79E-01 |
| apoptosis | 681 | 5 | 5.85 | - | 8.45E-01 | response to external stimulus | 1 | 2.15 | - | 3.66E-01 | 6.79E-01 |
| phospholipid metabolic process | 206 | 1 | 1.77 | - | 8.45E-01 | negative regulation of apoptosis | 1 | 2.14 | - | 3.69E-01 | 6.79E-01 |
| homeostatic process | 87 | 0 | 0.75 | - | 8.45E-01 | protein complex assembly | 0 | 0.99 | - | 3.70E-01 | 6.79E-01 |
| proteolysis | 987 | 9 | 8.48 | + | 8.45E-01 | glycolysis | 0 | 0.98 | - | 3.75E-01 | 6.83E-01 |
| transcription initiation from RNA polymerase II promoter | 85 | 0 | 0.73 | - | 8.45E-01 | antigen processing and presentation | 0 | 0.98 | - | 3.75E-01 | 6.83E-01 |
| DNA recombination | 77 | 1 | 0.66 | + | 8.45E-01 | embryonic development | 1 | 2.06 | - | 3.90E-01 | 6.98E-01 |
| I-kappaB kinase/NF-kappaB cascade | 84 | 0 | 0.72 | - | 8.45E-01 | RNA catabolic process | 0 | 0.93 | - | 3.96E-01 | 7.03E-01 |
| nerve-nerve synaptic transmission | 84 | 0 | 0.72 | - | 8.45E-01 | fertilization | 0 | 0.93 | - | 3.96E-01 | 7.03E-01 |
| protein amino acid glycosylation | 200 | 1 | 1.72 | - | 8.45E-01 | polysaccharide metabolic process | 2 | 3.06 | - | 4.08E-01 | 7.07E-01 |
| steroid metabolic process | 197 | 1 | 1.69 | - | 8.45E-01 | cell motion | 7 | 6.1 | + | 4.10E-01 | 7.07E-01 |
| vitamin transport | 79 | 0 | 0.68 | - | 8.45E-01 | lipid metabolic process | 10 | 11.38 | - | 4.12E-01 | 7.07E-01 |
| anterior/posterior axis specification | 79 | 0 | 0.68 | - | 8.45E-01 | angiogenesis | 3 | 2.34 | + | 4.15E-01 | 7.07E-01 |
| DNA repair | 192 | 1 | 1.65 | - | 8.45E-01 | apoptosis | 8 | 9.27 | - | 4.18E-01 | 7.07E-01 |
| response to toxin | 83 | 1 | 0.71 | + | 8.45E-01 | nervous system development | 11 | 10.04 | + | 4.23E-01 | 7.09E-01 |
| cell-matrix adhesion | 84 | 1 | 0.72 | + | 8.45E-01 | skeletal system development | 3 | 2.46 | + | 4.47E-01 | 7.44E-01 |
| rRNA metabolic process | 77 | 0 | 0.66 | - | 8.45E-01 | tRNA metabolic process | 0 | 0.76 | - | 4.66E-01 | 7.69E-01 |
| glycogen metabolic process | 85 | 1 | 0.73 | + | 8.45E-01 | pyrimidine base metabolic process | 0 | 0.75 | - | 4.73E-01 | 7.75E-01 |
| female gamete generation | 76 | 0 | 0.65 | - | 8.45E-01 | respiratory electron transport chain | 4 | 3.57 | + | 4.78E-01 | 7.75E-01 |
| meiosis | 89 | 1 | 0.76 | + | 8.57E-01 | amino acid transport | 1 | 0.65 | + | 4.80E-01 | 7.75E-01 |
| antigen processing and presentation | 72 | 0 | 0.62 | - | 8.57E-01 | chromosome segregation | 1 | 1.74 | - | 4.80E-01 | 7.75E-01 |
| muscle organ development | 180 | 1 | 1.55 | - | 8.57E-01 | cellular amino acid metabolic process | 4 | 3.62 | + | 4.90E-01 | 7.78E-01 |
| response to interferon-gamma | 91 | 1 | 0.78 | + | 8.57E-01 | cellular amino acid and derivative metabolic process | 4 | 3.62 | + | 4.90E-01 | 7.78E-01 |
| cholesterol metabolic process | 93 | 1 | 0.8 | + | 8.60E-01 | cell cycle | 15 | 15.68 | - | 4.97E-01 | 7.78E-01 |
| sulfur metabolic process | 94 | 1 | 0.81 | + | 8.60E-01 | developmental process | 28 | 28.49 | - | 5.11E-01 | 7.92E-01 |
| RNA catabolic process | 68 | 0 | 0.58 | - | 8.60E-01 | regulation of biological process | 0 | 0.67 | - | 5.13E-01 | 7.92E-01 |
| fertilization | 68 | 0 | 0.58 | - | 8.60E-01 | regulation of vasoconstriction | 0 | 0.67 | - | 5.13E-01 | 7.92E-01 |
| macrophage activation | 168 | 1 | 1.44 | - | 8.70E-01 | cellular component organization | 13 | 13.46 | - | 5.21E-01 | 7.93E-01 |
| carbohydrate transport | 64 | 0 | 0.55 | - | 8.70E-01 | receptor-mediated endocytosis | 2 | 1.78 | + | 5.33E-01 | 7.98E-01 |
| sensory perception | 384 | 3 | 3.3 | - | 8.70E-01 | cyclic nucleotide metabolic process | 0 | 0.63 | - | 5.34E-01 | 7.98E-01 |
| MAPKKK cascade | 272 | 2 | 2.34 | - | 8.73E-01 | pattern specification process | 3 | 2.82 | + | 5.36E-01 | 7.98E-01 |
| calcium-mediated signaling | 158 | 1 | 1.36 | - | 8.95E-01 | cellular calcium ion homeostasis | 0 | 0.61 | - | 5.42E-01 | 8.00E-01 |
| negative regulation of apoptosis | 157 | 1 | 1.35 | - | 8.95E-01 | ectoderm development | 11 | 11.26 | - | 5.48E-01 | 8.00E-01 |
| oxygen and reactive oxygen species metabolic process | 51 | 0 | 0.44 | - | 9.36E-01 | vitamin metabolic process | 0 | 0.6 | - | 5.49E-01 | 8.00E-01 |
| regulation of biological process | 49 | 0 | 0.42 | - | 9.36E-01 | transmembrane receptor protein serine/threonine kinase signaling pathway | 0 | 0.6 | - | 5.49E-01 | 8.00E-01 |
| regulation of vasoconstriction | 49 | 0 | 0.42 | - | 9.36E-01 | RNA elongation from RNA polymerase II promoter | 0 | 0.59 | - | 5.57E-01 | 8.00E-01 |
| phosphate metabolic process | 140 | 1 | 1.2 | - | 9.36E-01 | defense response to bacterium | 0 | 0.59 | - | 5.57E-01 | 8.00E-01 |
| neuronal action potential propagation | 48 | 0 | 0.41 | - | 9.36E-01 | tRNA aminoacylation for protein translation | 0 | 0.57 | - | 5.64E-01 | 8.00E-01 |
| amino acid transport | 48 | 0 | 0.41 | - | 9.36E-01 | phagocytosis | 0 | 0.56 | - | 5.72E-01 | 8.00E-01 |
| cyclic nucleotide metabolic process | 46 | 0 | 0.4 | - | 9.36E-01 | sensory perception | 5 | 5.23 | - | 5.76E-01 | 8.00E-01 |
| general transcription from RNA polymerase II promoter | 45 | 0 | 0.39 | - | 9.36E-01 | sensory perception of sound | 0 | 0.54 | - | 5.80E-01 | 8.00E-01 |
| cellular calcium ion homeostasis | 45 | 0 | 0.39 | - | 9.36E-01 | heart development | 1 | 1.43 | - | 5.81E-01 | 8.00E-01 |
| vitamin metabolic process | 44 | 0 | 0.38 | - | 9.36E-01 | cellular amino acid biosynthetic process | 1 | 0.87 | + | 5.82E-01 | 8.00E-01 |
| transmembrane receptor protein serine/threonine kinase signaling pathway | 44 | 0 | 0.38 | - | 9.36E-01 | mRNA 3'-end processing | 0 | 0.53 | - | 5.88E-01 | 8.03E-01 |
| RNA elongation from RNA polymerase II promoter | 43 | 0 | 0.37 | - | 9.36E-01 | cell-cell adhesion | 6 | 6.02 | - | 6.04E-01 | 8.18E-01 |
| defense response to bacterium | 43 | 0 | 0.37 | - | 9.36E-01 | sensory perception of pain | 0 | 0.5 | - | 6.04E-01 | 8.18E-01 |
| chromosome segregation | 128 | 1 | 1.1 | - | 9.36E-01 | JNK cascade | 1 | 1.33 | - | 6.15E-01 | 8.18E-01 |
| phagocytosis | 41 | 0 | 0.35 | - | 9.36E-01 | antigen processing and presentation of peptide or polysaccharide antigen via MHC class II | 0 | 0.48 | - | 6.21E-01 | 8.18E-01 |
| sensory perception of sound | 40 | 0 | 0.34 | - | 9.38E-01 | blood circulation | 1 | 1.31 | - | 6.24E-01 | 8.18E-01 |
| mRNA 3'-end processing | 39 | 0 | 0.33 | - | 9.40E-01 | RNA localization | 0 | 0.46 | - | 6.29E-01 | 8.18E-01 |
| sensory perception of pain | 37 | 0 | 0.32 | - | 9.51E-01 | fatty acid biosynthetic process | 0 | 0.46 | - | 6.29E-01 | 8.18E-01 |
| dorsal/ventral axis specification | 37 | 0 | 0.32 | - | 9.51E-01 | complement activation | 1 | 0.99 | + | 6.30E-01 | 8.18E-01 |
| antigen processing and presentation of peptide or polysaccharide antigen via MHC class II | 35 | 0 | 0.3 | - | 9.55E-01 | cellular glucose homeostasis | 0 | 0.45 | - | 6.38E-01 | 8.20E-01 |
| cellular glucose homeostasis | 33 | 0 | 0.28 | - | 9.66E-01 | cholesterol metabolic process | 1 | 1.27 | - | 6.39E-01 | 8.20E-01 |
| mRNA polyadenylation | 32 | 0 | 0.27 | - | 9.69E-01 | mRNA polyadenylation | 0 | 0.44 | - | 6.47E-01 | 8.25E-01 |
| gluconeogenesis | 30 | 0 | 0.26 | - | 9.79E-01 | gluconeogenesis | 0 | 0.41 | - | 6.65E-01 | 8.43E-01 |
| mRNA transcription | 29 | 0 | 0.25 | - | 9.79E-01 | mRNA transcription | 0 | 0.39 | - | 6.74E-01 | 8.45E-01 |
| mammary gland development | 29 | 0 | 0.25 | - | 9.79E-01 | mammary gland development | 0 | 0.39 | - | 6.74E-01 | 8.45E-01 |
| endoderm development | 28 | 0 | 0.24 | - | 9.79E-01 | glycogen metabolic process | 1 | 1.16 | - | 6.78E-01 | 8.45E-01 |
| protein amino acid acetylation | 27 | 0 | 0.23 | - | 9.82E-01 | I-kappaB kinase/NF-kappaB cascade | 1 | 1.14 | - | 6.83E-01 | 8.46E-01 |
| mitochondrion organization | 26 | 0 | 0.22 | - | 9.82E-01 | endoderm development | 0 | 0.38 | - | 6.83E-01 | 8.46E-01 |
| mitochondrial transport | 26 | 0 | 0.22 | - | 9.82E-01 | protein amino acid acetylation | 0 | 0.37 | - | 6.92E-01 | 8.46E-01 |
| fatty acid beta-oxidation | 25 | 0 | 0.21 | - | 9.82E-01 | mitochondrion organization | 0 | 0.35 | - | 7.02E-01 | 8.46E-01 |
| acyl-CoA metabolic process | 24 | 0 | 0.21 | - | 9.85E-01 | mitochondrial transport | 0 | 0.35 | - | 7.02E-01 | 8.46E-01 |
| porphyrin metabolic process | 22 | 0 | 0.19 | - | 9.88E-01 | vitamin transport | 1 | 1.08 | - | 7.08E-01 | 8.46E-01 |
| asymmetric protein localization | 22 | 0 | 0.19 | - | 9.88E-01 | anterior/posterior axis specification | 1 | 1.08 | - | 7.08E-01 | 8.46E-01 |
| protein localization | 22 | 0 | 0.19 | - | 9.88E-01 | fatty acid beta-oxidation | 0 | 0.34 | - | 7.11E-01 | 8.46E-01 |
| vitamin biosynthetic process | 21 | 0 | 0.18 | - | 9.88E-01 | acyl-CoA metabolic process | 0 | 0.33 | - | 7.21E-01 | 8.51E-01 |
| protein amino acid lipidation | 21 | 0 | 0.18 | - | 9.88E-01 | female gamete generation | 1 | 1.03 | - | 7.23E-01 | 8.51E-01 |
| termination of RNA polymerase II transcription | 19 | 0 | 0.16 | - | 9.93E-01 | porphyrin metabolic process | 0 | 0.3 | - | 7.41E-01 | 8.67E-01 |
| lysosomal transport | 18 | 0 | 0.15 | - | 9.93E-01 | vitamin biosynthetic process | 0 | 0.29 | - | 7.51E-01 | 8.74E-01 |
| nitrogen compound metabolic process | 18 | 0 | 0.15 | - | 9.93E-01 | protein amino acid lipidation | 0 | 0.29 | - | 7.51E-01 | 8.74E-01 |
| sex determination | 17 | 0 | 0.15 | - | 9.93E-01 | termination of RNA polymerase II transcription | 0 | 0.26 | - | 7.72E-01 | 8.89E-01 |
| regulation of carbohydrate metabolic process | 17 | 0 | 0.15 | - | 9.93E-01 | lysosomal transport | 0 | 0.24 | - | 7.83E-01 | 8.97E-01 |
| phosphate transport | 16 | 0 | 0.14 | - | 9.93E-01 | sex determination | 0 | 0.23 | - | 7.93E-01 | 9.03E-01 |
| gut mesoderm development | 15 | 0 | 0.13 | - | 9.95E-01 | regulation of carbohydrate metabolic process | 0 | 0.23 | - | 7.93E-01 | 9.03E-01 |
| peroxisomal transport | 14 | 0 | 0.12 | - | 9.95E-01 | phosphate transport | 0 | 0.22 | - | 8.04E-01 | 9.06E-01 |
| "regulation of nucleobase, nucleoside, nucleotide and nucleic acid metabolic process" | 14 | 0 | 0.12 | - | 9.95E-01 | gut mesoderm development | 0 | 0.2 | - | 8.15E-01 | 9.14E-01 |
| sensory perception of chemical stimulus | 12 | 0 | 0.1 | - | 9.95E-01 | peroxisomal transport | 0 | 0.19 | - | 8.26E-01 | 9.21E-01 |
| protein amino acid methylation | 10 | 0 | 0.09 | - | 9.95E-01 | "regulation of nucleobase, nucleoside, nucleotide and nucleic acid metabolic process" | 0 | 0.19 | - | 8.26E-01 | 9.21E-01 |
| protein amino acid ADP-ribosylation | 10 | 0 | 0.09 | - | 9.95E-01 | sensory perception of chemical stimulus | 0 | 0.16 | - | 8.49E-01 | 9.37E-01 |
| DNA catabolic process | 10 | 0 | 0.09 | - | 9.95E-01 | protein amino acid methylation | 0 | 0.14 | - | 8.73E-01 | 9.52E-01 |
| sensory perception of taste | 10 | 0 | 0.09 | - | 9.95E-01 | protein amino acid ADP-ribosylation | 0 | 0.14 | - | 8.73E-01 | 9.52E-01 |
| regulation of liquid surface tension | 9 | 0 | 0.08 | - | 9.95E-01 | DNA catabolic process | 0 | 0.14 | - | 8.73E-01 | 9.52E-01 |
| response to pheromone | 7 | 0 | 0.06 | - | 9.95E-01 | sensory perception of taste | 0 | 0.14 | - | 8.73E-01 | 9.52E-01 |
| pinocytosis | 7 | 0 | 0.06 | - | 9.95E-01 | regulation of liquid surface tension | 0 | 0.12 | - | 8.85E-01 | 9.52E-01 |
| "transcription, RNA-dependent" | 6 | 0 | 0.05 | - | 9.95E-01 | pentose-phosphate shunt | 0 | 0.11 | - | 8.97E-01 | 9.60E-01 |
| nitric oxide biosynthetic process | 6 | 0 | 0.05 | - | 9.95E-01 | response to pheromone | 0 | 0.1 | - | 9.09E-01 | 9.68E-01 |
| regulation of amino acid metabolic process | 6 | 0 | 0.05 | - | 9.95E-01 | pinocytosis | 0 | 0.1 | - | 9.09E-01 | 9.68E-01 |
| ferredoxin metabolic process | 6 | 0 | 0.05 | - | 9.95E-01 | "transcription, RNA-dependent" | 0 | 0.08 | - | 9.22E-01 | 9.70E-01 |
| unsaturated fatty acid biosynthetic process | 6 | 0 | 0.05 | - | 9.95E-01 | regulation of amino acid metabolic process | 0 | 0.08 | - | 9.22E-01 | 9.70E-01 |
| nitric oxide mediated signal transduction | 6 | 0 | 0.05 | - | 9.95E-01 | unsaturated fatty acid biosynthetic process | 0 | 0.08 | - | 9.22E-01 | 9.70E-01 |
| disaccharide metabolic process | 5 | 0 | 0.04 | - | 9.95E-01 | disaccharide metabolic process | 0 | 0.07 | - | 9.34E-01 | 9.70E-01 |
| polyphosphate catabolic process | 4 | 0 | 0.03 | - | 9.99E-01 | polyphosphate catabolic process | 0 | 0.05 | - | 9.47E-01 | 9.79E-01 |
| mRNA capping | 3 | 0 | 0.03 | - | 1.00E+00 | mRNA capping | 0 | 0.04 | - | 9.60E-01 | 9.88E-01 |
| regulation of phosphate metabolic process | 3 | 0 | 0.03 | - | 1.00E+00 | regulation of phosphate metabolic process | 0 | 0.04 | - | 9.60E-01 | 9.88E-01 |
| sensory perception of smell | 2 | 0 | 0.02 | - | 1.00E+00 | sensory perception of smell | 0 | 0.03 | - | 9.73E-01 | 9.92E-01 |
| nitrogen utilization | 2 | 0 | 0.02 | - | 1.00E+00 | nitrogen utilization | 0 | 0.03 | - | 9.73E-01 | 9.92E-01 |
| vitamin catabolic process | 1 | 0 | 0.01 | - | 1.00E+00 | vitamin catabolic process | 0 | 0.01 | - | 9.86E-01 | 9.95E-01 |
| catabolic process | 1 | 0 | 0.01 | - | 1.00E+00 | catabolic process | 0 | 0.01 | - | 9.86E-01 | 9.95E-01 |
| bile acid metabolic process | 1 | 0 | 0.01 | - | 1.00E+00 | bile acid metabolic process | 0 | 0.01 | - | 9.86E-01 | 9.95E-01 |
| 1 | 1.00E+00 |
Supplementary Table 7. Proteins related to axon guidance or cell morphology.
Proteins shown are either members of the group of molecules involved in 'axon guidance mediated by semaphorins', as defined by Panther, or are known to be involved in this process (literature), or belong to the Biological Process: "cellular component morphogenesis'. Of all molecules belonging to the later group only those that are unique or significantly enriched in human PSD were considered.
| Gene Name | HGNC ID | MGI ID | Panther Pathway: Axon guidance mediated by Semaphorins | Panther Biological Process Cellular Compoenet Morphogenesis | Involved in Axonguidance mediated by sempahorins acording to Literature | Abundance Fold Difference (AFD) Mouse/Human | Significantly Enriched |
|---|---|---|---|---|---|---|---|
| CRMP1 | HGNC:2365 | MGI:107793 | YES | YES | 0.141 | Human | |
| DPYSL2 | HGNC:3014 | MGI:1349763 | YES | YES | 0.122 | Human | |
| DPYSL3 | HGNC:3015 | MGI:1349762 | YES | YES | 0.032 | Human | |
| CDK5 | HGNC:1774 | MGI:101765 | YES | YES | 0.733 | NOT | |
| PLXNA1 | HGNC:9099 | MGI:107685 | YES | YES | 0.302 | NOT | |
| RAC1 | HGNC:9801 | MGI:97845 | YES | YES | 1.736 | NOT | |
| RHOA | HGNC:667 | MGI:1096342 | YES | YES | 1.071 | NOT | |
| PAK1 | HGNC:8590 | MGI:1339975 | YES | YES | 0.154 | Human | |
| ARHGEF2 | HGNC:682 | MGI:103264 | YES | 1.331 | NOT | ||
| FYN | HGNC:4037 | MGI:95602 | YES | 0.813 | NOT | ||
| RHOB | HGNC:668 | MGI:107949 | YES | 2.387 | NOT | ||
| ROCK2 | HGNC:10252 | MGI:107926 | YES | 1.988 | NOT | ||
| PLXNA2 | HGNC:9100 | MGI:107684 | YES | Human Only | |||
| PLXNA3 | HGNC:9101 | MGI:107683 | YES | Human Only | |||
| CDC42 | HGNC:1736 | MGI:106211 | YES | 1.530 | NOT | ||
| GSK3B | HGNC:4617 | MGI:1861437 | YES | 0.936 | NOT | ||
| PLXNA4 | HGNC:9102 | MGI:2179061 | YES | 0.484 | NOT | ||
| CFL1 | HGNC:1874 | MGI:101757 | YES | 0.134 | Human | ||
| DNM1L | HGNC:2973 | MGI:1921256 | YES | 0.246 | Human | ||
| GFAP | HGNC:4235 | MGI:95697 | YES | 0.028 | Human | ||
| MAPRE2 | HGNC:6891 | MGI:106271 | YES | 0.047 | Human | ||
| MSN | HGNC:7373 | MGI:97167 | YES | 0.038 | Human | ||
| PLP1 | HGNC:9086 | MGI:97623 | YES | 0.073 | Human | ||
| VIM | HGNC:12692 | MGI:98932 | YES | 0.021 | Human | ||
| TUBB2C | HGNC:20771 | MGI:1915472 | YES | 0.484 | Human | ||
| TPPP | HGNC:24164 | MGI:1920198 | YES | 0.201 | Human | ||
| MAP4 | HGNC:6862 | MGI:97178 | YES | 0.167 | Human | ||
| DYNC1LI2 | HGNC:2966 | MGI:107738 | 0.262 | Human | |||
| CAPG | HGNC:1474 | MGI:1098259 | YES | Human Only | |||
| KIF21A | HGNC:19349 | MGI:109188 | YES | Human Only | |||
| TBCB | HGNC:1989 | MGI:1913661 | YES | Human Only | |||
| VCL | HGNC:12665 | MGI:98927 | YES | Human Only | |||
| TLN1 | HGNC:11845 | MGI:1099832 | YES | Human Only | |||
| FLNA | HGNC:3754 | MGI:95556 | YES | Human Only |
Supplementary Table 8. Functional comparison of all PSD proteins identified in human and mouse.
All human and mouse PSD proteins were classified together using the Panther 'Protein Class' descriptor. An enrichment analysis was done to determine Protein Classes overrepresented compared with the human genome. The Benjamini-Hochberg procedure was used to correct for multiple testing. The column 'observed' retrieves the number of proteins identified in each 'Protein Class' while the column 'expected', the number that would have been identified by chance. Over or under-representations are shown by a (+) or (-) symbol respectively.
| PANTHER Protein Class | Homo sapiens genes | Observed | Expected | Over/Under | P-value | Corrected P-value |
|---|---|---|---|---|---|---|
| Cytoskeletal protein | 441 | 163 | 41.91 | + | 2.0E-47 | 3.9E-45 |
| Transcription factor | 2067 | 41 | 196.41 | - | 1.9E-44 | 1.9E-42 |
| Membrane traffic protein | 321 | 123 | 30.5 | + | 2.3E-37 | 1.5E-35 |
| Actin family cytoskeletal protein | 222 | 96 | 21.1 | + | 2.6E-33 | 1.3E-31 |
| Zinc finger transcription factor | 803 | 7 | 76.3 | - | 6.5E-25 | 2.1E-23 |
| Enzyme modulator | 857 | 183 | 81.43 | + | 1.7E-23 | 4.6E-22 |
| G-protein | 206 | 68 | 19.57 | + | 6.5E-18 | 1.1E-16 |
| G-protein modulator | 278 | 79 | 26.42 | + | 5.7E-17 | 9.2E-16 |
| Oxidoreductase | 550 | 121 | 52.26 | + | 9.4E-17 | 1.4E-15 |
| Chaperone | 130 | 50 | 12.35 | + | 5.0E-16 | 6.9E-15 |
| Transporter | 1098 | 193 | 104.34 | + | 5.9E-16 | 7.6E-15 |
| Microtubule family cytoskeletal protein | 156 | 55 | 14.82 | + | 6.4E-16 | 7.8E-15 |
| Kinase | 679 | 137 | 64.52 | + | 7.1E-16 | 8.0E-15 |
| Cation transporter | 178 | 57 | 16.91 | + | 1.1E-14 | 1.1E-13 |
| Unclassified | 6763 | 508 | 642.64 | - | 1.6E-11 | 1.4E-10 |
| RNA binding protein | 727 | 129 | 69.08 | + | 3.0E-11 | 2.5E-10 |
| Transferase | 1512 | 215 | 143.67 | + | 3.9E-09 | 3.0E-08 |
| Helix-turn-helix transcription factor | 233 | 3 | 22.14 | - | 4.6E-07 | 2.8E-06 |
| Ion channel | 341 | 58 | 32.4 | + | 2.7E-05 | 1.4E-04 |
| DNA binding protein | 476 | 25 | 45.23 | - | 6.7E-04 | 2.8E-03 |
| Intermediate filament | 11 | 6 | 1.05 | + | 7.4E-04 | 3.0E-03 |
| Nucleic acid binding | 1466 | 173 | 139.3 | + | 2.3E-03 | 8.7E-03 |
| Hydrolase | 454 | 63 | 43.14 | + | 2.4E-03 | 8.9E-03 |
| Cytokine | 159 | 5 | 15.11 | - | 2.5E-03 | 9.2E-03 |
| Peptide hormone | 169 | 6 | 16.06 | - | 3.8E-03 | 1.3E-02 |
| Signaling molecule | 961 | 68 | 91.32 | - | 5.6E-03 | 1.9E-02 |
| Lyase | 104 | 19 | 9.88 | + | 6.3E-03 | 2.1E-02 |
| Protease inhibitor | 109 | 3 | 10.36 | - | 7.8E-03 | 2.5E-02 |
| Protease | 476 | 30 | 45.23 | - | 9.9E-03 | 3.1E-02 |
| Extracellular matrix protein | 72 | 14 | 6.84 | + | 1.1E-02 | 3.2E-02 |
| Ligase | 260 | 37 | 24.71 | + | 1.2E-02 | 3.6E-02 |
| Structural protein | 280 | 37 | 26.61 | + | 3.2E-02 | 8.2E-02 |
| Receptor | 1076 | 85 | 102.24 | - | 4.1E-02 | 1.0E-01 |
| Defense/immunity protein | 107 | 5 | 10.17 | - | 6.1E-02 | 1.4E-01 |
| Phosphatase | 230 | 29 | 21.86 | + | 8.1E-02 | 1.7E-01 |
| Growth factor | 165 | 10 | 15.68 | - | 8.8E-02 | 1.8E-01 |
| Isomerase | 94 | 13 | 8.93 | + | 1.2E-01 | 2.3E-01 |
| Calcium-binding protein | 63 | 9 | 5.99 | + | 1.5E-01 | 2.7E-01 |
| Intracellular calcium-sensing protein | 33 | 5 | 3.14 | + | 2.1E-01 | 3.4E-01 |
| Transfer/carrier protein | 248 | 26 | 23.57 | + | 3.3E-01 | 4.8E-01 |
| Cell adhesion molecule | 93 | 7 | 8.84 | - | 3.4E-01 | 4.8E-01 |
| Cytokine receptor | 213 | 18 | 20.24 | - | 3.6E-01 | 4.9E-01 |
| Cyclase | 23 | 3 | 2.19 | + | 3.7E-01 | 5.0E-01 |
| Lipase | 57 | 6 | 5.42 | + | 4.6E-01 | 5.6E-01 |
| Viral protein | 7 | 0 | 0.67 | - | 5.1E-01 | 5.8E-01 |
| Kinase modulator | 103 | 10 | 9.79 | + | 5.2E-01 | 5.8E-01 |
| Cell junction protein | 67 | 6 | 6.37 | - | 5.5E-01 | 6.1E-01 |
| Phosphatase modulator | 63 | 6 | 5.99 | + | 5.5E-01 | 6.1E-01 |
| Non-motor actin binding protein | 114 | 61 | 10.83 | + | 3.2E-26 | 1.2E-24 |
| KRAB box transcription factor | 552 | 1 | 52.45 | - | 4.4E-22 | 1.0E-20 |
| Ribosomal protein | 184 | 70 | 17.48 | + | 1.3E-21 | 2.8E-20 |
| Dehydrogenase | 210 | 69 | 19.95 | + | 4.7E-18 | 9.0E-17 |
| Membrane trafficking regulatory protein | 107 | 44 | 10.17 | + | 2.9E-15 | 3.1E-14 |
| ATP synthase | 51 | 28 | 4.85 | + | 4.2E-13 | 4.0E-12 |
| Small GTPase | 158 | 50 | 15.01 | + | 6.8E-13 | 6.2E-12 |
| Nucleotide kinase | 51 | 23 | 4.85 | + | 2.0E-09 | 1.6E-08 |
| Non-motor microtubule binding protein | 53 | 22 | 5.04 | + | 1.9E-08 | 1.4E-07 |
| Protein kinase | 510 | 91 | 48.46 | + | 2.0E-08 | 1.4E-07 |
| Guanyl-nucleotide exchange factor | 79 | 27 | 7.51 | + | 2.7E-08 | 1.9E-07 |
| SNARE protein | 37 | 18 | 3.52 | + | 3.6E-08 | 2.4E-07 |
| Heterotrimeric G-protein | 36 | 17 | 3.42 | + | 1.3E-07 | 8.4E-07 |
| Vesicle coat protein | 30 | 15 | 2.85 | + | 3.5E-07 | 2.1E-06 |
| Homeobox transcription factor | 233 | 3 | 22.14 | - | 4.6E-07 | 2.8E-06 |
| Hsp70 family chaperone | 13 | 10 | 1.24 | + | 7.3E-07 | 4.2E-06 |
| Chaperonin | 20 | 12 | 1.9 | + | 7.9E-07 | 4.4E-06 |
| Transmembrane receptor regulatory/adaptor protein | 84 | 25 | 7.98 | + | 1.1E-06 | 5.6E-06 |
| Tubulin | 25 | 11 | 2.38 | + | 3.9E-05 | 2.0E-04 |
| Microtubule binding motor protein | 46 | 15 | 4.37 | + | 5.2E-05 | 2.6E-04 |
| Calcium channel | 31 | 12 | 2.95 | + | 5.9E-05 | 2.8E-04 |
| Ligand-gated ion channel | 101 | 23 | 9.6 | + | 1.6E-04 | 7.4E-04 |
| Aminoacyl-tRNA synthetase | 20 | 9 | 1.9 | + | 1.6E-04 | 7.4E-04 |
| Reductase | 65 | 17 | 6.18 | + | 2.4E-04 | 1.1E-03 |
| Transcription cofactor | 255 | 9 | 24.23 | - | 3.4E-04 | 1.5E-03 |
| Intermediate filament binding protein | 10 | 6 | 0.95 | + | 4.5E-04 | 1.9E-03 |
| Basic helix-loop-helix transcription factor | 120 | 2 | 11.4 | - | 8.4E-04 | 3.4E-03 |
| Mitochondrial carrier protein | 17 | 7 | 1.62 | + | 1.4E-03 | 5.5E-03 |
| Carbohydrate kinase | 35 | 10 | 3.33 | + | 2.3E-03 | 8.7E-03 |
| Peroxidase | 27 | 8 | 2.57 | + | 4.9E-03 | 1.7E-02 |
| Myelin protein | 33 | 9 | 3.14 | + | 5.0E-03 | 1.7E-02 |
| Amino acid transporter | 98 | 18 | 9.31 | + | 7.3E-03 | 2.4E-02 |
| Extracellular matrix glycoprotein | 55 | 12 | 5.23 | + | 7.5E-03 | 2.4E-02 |
| Amino acid kinase | 5 | 3 | 0.48 | + | 1.3E-02 | 3.7E-02 |
| Acetyltransferase | 105 | 18 | 9.98 | + | 1.4E-02 | 4.0E-02 |
| Phosphodiesterase | 27 | 7 | 2.57 | + | 1.6E-02 | 4.6E-02 |
| Hsp90 family chaperone | 10 | 4 | 0.95 | + | 1.6E-02 | 4.6E-02 |
| Serine protease inhibitor | 79 | 2 | 7.51 | - | 2.0E-02 | 5.6E-02 |
| RNA helicase | 71 | 13 | 6.75 | + | 2.1E-02 | 5.7E-02 |
| Anion channel | 37 | 8 | 3.52 | + | 2.7E-02 | 7.4E-02 |
| Deaminase | 12 | 4 | 1.14 | + | 2.9E-02 | 7.7E-02 |
| Actin and actin related protein | 31 | 7 | 2.95 | + | 3.1E-02 | 8.1E-02 |
| Membrane-bound signaling molecule | 133 | 6 | 12.64 | - | 3.2E-02 | 8.2E-02 |
| G-protein coupled receptor | 447 | 31 | 42.48 | - | 3.9E-02 | 1.0E-01 |
| Endoribonuclease | 33 | 0 | 3.14 | - | 4.3E-02 | 1.1E-01 |
| Serine protease | 153 | 8 | 14.54 | - | 4.7E-02 | 1.1E-01 |
| Oxidase | 57 | 10 | 5.42 | + | 4.9E-02 | 1.2E-01 |
| Actin binding motor protein | 9 | 3 | 0.86 | + | 5.6E-02 | 1.3E-01 |
| Aldolase | 4 | 2 | 0.38 | + | 5.6E-02 | 1.3E-01 |
| Storage protein | 15 | 4 | 1.43 | + | 5.7E-02 | 1.3E-01 |
| Nuclear hormone receptor | 46 | 1 | 4.37 | - | 6.8E-02 | 1.5E-01 |
| Neuropeptide | 28 | 0 | 2.66 | - | 7.0E-02 | 1.6E-01 |
| Damaged DNA-binding protein | 45 | 1 | 4.28 | - | 7.3E-02 | 1.6E-01 |
| Methyltransferase | 130 | 7 | 12.35 | - | 7.5E-02 | 1.6E-01 |
| HMG box transcription factor | 44 | 1 | 4.18 | - | 7.9E-02 | 1.7E-01 |
| Oxygenase | 74 | 3 | 7.03 | - | 8.0E-02 | 1.7E-01 |
| Transketolase | 5 | 2 | 0.48 | + | 8.3E-02 | 1.7E-01 |
| Glycosidase | 26 | 0 | 2.47 | - | 8.4E-02 | 1.8E-01 |
| Transaldolase | 1 | 1 | 0.1 | + | 9.1E-02 | 1.8E-01 |
| Translation factor | 56 | 9 | 5.32 | + | 9.1E-02 | 1.8E-01 |
| Immunoglobulin superfamily cell adhesion molecule | 25 | 5 | 2.38 | + | 9.3E-02 | 1.8E-01 |
| Basic leucine zipper transcription factor | 24 | 0 | 2.28 | - | 1.0E-01 | 2.0E-01 |
| CREB transcription factor | 24 | 0 | 2.28 | - | 1.0E-01 | 2.0E-01 |
| Nucleotide phosphatase | 35 | 6 | 3.33 | + | 1.2E-01 | 2.3E-01 |
| Tumor necrosis factor receptor | 22 | 0 | 2.09 | - | 1.2E-01 | 2.3E-01 |
| RNA methyltransferase | 22 | 0 | 2.09 | - | 1.2E-01 | 2.3E-01 |
| DNA helicase | 80 | 4 | 7.6 | - | 1.2E-01 | 2.3E-01 |
| Interleukin superfamily | 36 | 1 | 3.42 | - | 1.4E-01 | 2.6E-01 |
| Endodeoxyribonuclease | 20 | 0 | 1.9 | - | 1.5E-01 | 2.7E-01 |
| Chemokine | 48 | 2 | 4.56 | - | 1.7E-01 | 3.0E-01 |
| ATP-binding cassette (ABC) transporter | 74 | 4 | 7.03 | - | 1.7E-01 | 3.0E-01 |
| Immunoglobulin | 33 | 1 | 3.14 | - | 1.8E-01 | 3.1E-01 |
| Centromere DNA-binding protein | 18 | 0 | 1.71 | - | 1.8E-01 | 3.1E-01 |
| Metalloprotease | 145 | 10 | 13.78 | - | 1.9E-01 | 3.3E-01 |
| Interferon superfamily | 17 | 0 | 1.62 | - | 2.0E-01 | 3.4E-01 |
| Kinase activator | 45 | 2 | 4.28 | - | 2.0E-01 | 3.4E-01 |
| Type I cytokine receptor | 31 | 1 | 2.95 | - | 2.1E-01 | 3.4E-01 |
| Calmodulin | 33 | 5 | 3.14 | + | 2.1E-01 | 3.4E-01 |
| Extracellular matrix structural protein | 9 | 2 | 0.86 | + | 2.1E-01 | 3.4E-01 |
| Annexin | 9 | 2 | 0.86 | + | 2.1E-01 | 3.4E-01 |
| Hydroxylase | 44 | 2 | 4.18 | - | 2.1E-01 | 3.4E-01 |
| Transaminase | 25 | 4 | 2.38 | + | 2.2E-01 | 3.4E-01 |
| Galactosidase | 16 | 0 | 1.52 | - | 2.2E-01 | 3.4E-01 |
| Potassium channel | 89 | 11 | 8.46 | + | 2.3E-01 | 3.6E-01 |
| Apolipoprotein | 42 | 2 | 3.99 | - | 2.4E-01 | 3.7E-01 |
| Kinase inhibitor | 15 | 0 | 1.43 | - | 2.4E-01 | 3.7E-01 |
| Surfactant | 15 | 0 | 1.43 | - | 2.4E-01 | 3.7E-01 |
| DNA-directed RNA polymerase | 15 | 0 | 1.43 | - | 2.4E-01 | 3.7E-01 |
| Major histocompatibility complex antigen | 28 | 1 | 2.66 | - | 2.6E-01 | 3.9E-01 |
| Phospholipase | 36 | 5 | 3.42 | + | 2.6E-01 | 3.9E-01 |
| Immunoglobulin receptor superfamily | 155 | 17 | 14.73 | + | 3.1E-01 | 4.6E-01 |
| DNA glycosylase | 12 | 0 | 1.14 | - | 3.2E-01 | 4.7E-01 |
| Adenylate cyclase | 21 | 3 | 2 | + | 3.2E-01 | 4.7E-01 |
| Glycosyltransferase | 229 | 19 | 21.76 | - | 3.2E-01 | 4.7E-01 |
| Exoribonuclease | 24 | 1 | 2.28 | - | 3.4E-01 | 4.8E-01 |
| Dehydratase | 31 | 4 | 2.95 | + | 3.4E-01 | 4.8E-01 |
| Phosphorylase | 13 | 2 | 1.24 | + | 3.5E-01 | 4.9E-01 |
| Tumor necrosis factor family member | 11 | 0 | 1.05 | - | 3.5E-01 | 4.9E-01 |
| Ribonucleoprotein | 61 | 7 | 5.8 | + | 3.6E-01 | 4.9E-01 |
| Protein phosphatase | 111 | 12 | 10.55 | + | 3.7E-01 | 5.0E-01 |
| Aspartic protease | 14 | 2 | 1.33 | + | 3.8E-01 | 5.1E-01 |
| Mutase | 14 | 2 | 1.33 | + | 3.8E-01 | 5.1E-01 |
| DNA-directed DNA polymerase | 10 | 0 | 0.95 | - | 3.9E-01 | 5.1E-01 |
| Cysteine protease | 121 | 10 | 11.5 | - | 4.0E-01 | 5.3E-01 |
| Nucleotidyltransferase | 84 | 9 | 7.98 | + | 4.1E-01 | 5.3E-01 |
| Cysteine protease inhibitor | 21 | 1 | 2 | - | 4.1E-01 | 5.3E-01 |
| Gap junction | 21 | 1 | 2 | - | 4.1E-01 | 5.3E-01 |
| Esterase | 25 | 3 | 2.38 | + | 4.2E-01 | 5.4E-01 |
| Carbohydrate phosphatase | 9 | 0 | 0.86 | - | 4.3E-01 | 5.4E-01 |
| Amylase | 6 | 1 | 0.57 | + | 4.4E-01 | 5.4E-01 |
| DNA ligase | 6 | 1 | 0.57 | + | 4.4E-01 | 5.4E-01 |
| mRNA processing factor | 179 | 18 | 17.01 | + | 4.4E-01 | 5.4E-01 |
| Carbohydrate transporter | 46 | 5 | 4.37 | + | 4.4E-01 | 5.5E-01 |
| Tight junction | 30 | 2 | 2.85 | - | 4.6E-01 | 5.6E-01 |
| Protein kinase receptor | 8 | 0 | 0.76 | - | 4.7E-01 | 5.7E-01 |
| Neurotrophic factor | 8 | 0 | 0.76 | - | 4.7E-01 | 5.7E-01 |
| Decarboxylase | 17 | 2 | 1.62 | + | 4.8E-01 | 5.8E-01 |
| DNA methyltransferase | 17 | 2 | 1.62 | + | 4.8E-01 | 5.8E-01 |
| Ubiquitin-protein ligase | 132 | 13 | 12.54 | + | 4.9E-01 | 5.8E-01 |
| Phosphatase inhibitor | 28 | 2 | 2.66 | - | 5.0E-01 | 5.8E-01 |
| Antibacterial response protein | 18 | 2 | 1.71 | + | 5.1E-01 | 5.8E-01 |
| Epimerase/racemase | 7 | 0 | 0.67 | - | 5.1E-01 | 5.8E-01 |
| Viral coat protein | 7 | 0 | 0.67 | - | 5.1E-01 | 5.8E-01 |
| DNA strand-pairing protein | 7 | 0 | 0.67 | - | 5.1E-01 | 5.8E-01 |
| Replication origin binding protein | 47 | 4 | 4.47 | - | 5.4E-01 | 6.1E-01 |
| Acyltransferase | 88 | 8 | 8.36 | - | 5.4E-01 | 6.1E-01 |
| Sodium channel | 20 | 2 | 1.9 | + | 5.7E-01 | 6.3E-01 |
| Nuclease | 35 | 3 | 3.33 | - | 5.8E-01 | 6.3E-01 |
| Glucosidase | 15 | 1 | 1.43 | - | 5.8E-01 | 6.4E-01 |
| Histone | 54 | 5 | 5.13 | - | 5.9E-01 | 6.4E-01 |
| Chromatin/chromatin-binding protein | 74 | 7 | 7.03 | - | 5.9E-01 | 6.4E-01 |
| Helicase | 5 | 0 | 0.48 | - | 6.2E-01 | 6.7E-01 |
| Pyrophosphatase | 5 | 0 | 0.48 | - | 6.2E-01 | 6.7E-01 |
| Metalloprotease inhibitor | 4 | 0 | 0.38 | - | 6.8E-01 | 7.3E-01 |
| DNA topoisomerase | 4 | 0 | 0.38 | - | 6.8E-01 | 7.3E-01 |
| Deacetylase | 2 | 0 | 0.19 | - | 8.3E-01 | 8.7E-01 |
| Calsequestrin | 2 | 0 | 0.19 | - | 8.3E-01 | 8.7E-01 |
| TGF-beta receptor | 2 | 0 | 0.19 | - | 8.3E-01 | 8.7E-01 |
| DNA polymerase processivity factor | 2 | 0 | 0.19 | - | 8.3E-01 | 8.7E-01 |
| DNA photolyase | 2 | 0 | 0.19 | - | 8.3E-01 | 8.7E-01 |
| Exodeoxyribonuclease | 1 | 0 | 0.1 | - | 9.1E-01 | 9.3E-01 |
| Voltage-gated ion channel | 1 | 0 | 0.1 | - | 9.1E-01 | 9.3E-01 |
| Primase | 1 | 0 | 0.1 | - | 9.1E-01 | 9.3E-01 |
| Phosphatase activator | 1 | 0 | 0.1 | - | 9.1E-01 | 9.3E-01 |
| Reverse transcriptase | 1 | 0 | 0.1 | - | 9.1E-01 | 9.3E-01 |
Supplementary Table 9a. Pathway analysis of all PSD proteins identified in human and mouse.
All human and mouse PSD proteins were classified together using the Panther 'Pathway' descriptor. An enrichment analysis was done to determine Pathways overrepresented compared with the human genome. The Benjamini-Hochberg procedure was used to correct for multiple testing. The column 'observed' retrieves the number of proteins identified in each 'Pathway' while the column 'expected', the number that would have been identified by chance. Over or under-representations are shown by a (+) or (-) symbol respectively.
Human and mouse PSD proteins were classified using the Panther 'Pathway' descriptor. An enrichment analysis was done to determine Pathways overrepresented as compared with the human genome, a set of mouse brain expressed proteins and a set of genes highly expressed inhuman neurons. P-values were corrected for multiple testing using the Benjamini-Hochberg procedure. The colum 'observed' retrives the number of proteins identified in each 'Pathway' while the column 'expected', the number which would have been identified by chance. Over or underrepresntations are shown by a (+) or (-) symbol respectively.
| PSD vs Human Genome |
|---|
| Pathway | Observed | Expected | over/under | P-value | Corrected P-value |
|---|---|---|---|---|---|
| Metabotropic glutamate receptor group III pathway | 44 | 6.94 | + | 3.09E-21 | 4.51E-19 |
| Metabotropic glutamate receptor group II pathway | 34 | 4.85 | + | 4.99E-18 | 3.64E-16 |
| Muscarinic acetylcholine receptor 1 and 3 signaling pathway | 35 | 5.8 | + | 1.44E-16 | 7.01E-15 |
| Huntington disease | 58 | 15.87 | + | 2.00E-16 | 7.30E-15 |
| Ionotropic glutamate receptor pathway | 32 | 5.13 | + | 1.18E-15 | 3.43E-14 |
| Thyrotropin-releasing hormone receptor signaling pathway | 34 | 5.89 | + | 1.41E-15 | 3.43E-14 |
| Synaptic_vesicle_trafficking | 27 | 3.61 | + | 2.81E-15 | 5.86E-14 |
| Parkinson disease | 42 | 9.5 | + | 6.06E-15 | 1.11E-13 |
| Heterotrimeric G-protein signaling pathway-Gq alpha and Go alpha mediated pathway | 48 | 12.73 | + | 2.52E-14 | 4.09E-13 |
| Oxytocin receptor mediated signaling pathway | 31 | 5.7 | + | 1.14E-13 | 1.66E-12 |
| 5HT2 type receptor mediated signaling pathway | 33 | 6.56 | + | 1.51E-13 | 2.00E-12 |
| Muscarinic acetylcholine receptor 2 and 4 signaling pathway | 30 | 5.89 | + | 1.41E-12 | 1.72E-11 |
| Histamine H1 receptor mediated signaling pathway | 26 | 4.47 | + | 2.39E-12 | 2.68E-11 |
| Beta2 adrenergic receptor signaling pathway | 25 | 4.18 | + | 3.57E-12 | 3.72E-11 |
| Beta1 adrenergic receptor signaling pathway | 25 | 4.18 | + | 3.57E-12 | 3.72E-11 |
| Inflammation mediated by chemokine and cytokine signaling pathway | 69 | 26.89 | + | 5.42E-12 | 4.95E-11 |
| Cortocotropin releasing factor receptor signaling pathway | 20 | 2.85 | + | 3.20E-11 | 2.75E-10 |
| Heterotrimeric G-protein signaling pathway-Gi alpha and Gs alpha mediated pathway | 48 | 15.77 | + | 4.02E-11 | 3.26E-10 |
| Cytoskeletal regulation by Rho GTPase | 35 | 9.31 | + | 8.18E-11 | 6.29E-10 |
| 5HT1 type receptor mediated signaling pathway | 23 | 4.18 | + | 1.27E-10 | 9.27E-10 |
| 5HT4 type receptor mediated signaling pathway | 19 | 2.95 | + | 3.90E-10 | 2.71E-09 |
| GABA-B_receptor_II_signaling | 21 | 3.8 | + | 7.37E-10 | 4.89E-09 |
| Beta3 adrenergic receptor signaling pathway | 17 | 2.47 | + | 1.24E-09 | 7.87E-09 |
| EGF receptor signaling pathway | 39 | 12.83 | + | 2.66E-09 | 1.62E-08 |
| Metabotropic glutamate receptor group I pathway | 19 | 3.42 | + | 4.28E-09 | 2.50E-08 |
| Unclassified | 1560 | 1647.41 | - | 5.90E-09 | 3.31E-08 |
| Alpha adrenergic receptor signaling pathway | 17 | 3.04 | + | 2.50E-08 | 1.35E-07 |
| Histamine H2 receptor mediated signaling pathway | 15 | 2.38 | + | 3.47E-08 | 1.81E-07 |
| Nicotinic acetylcholine receptor signaling pathway | 30 | 9.22 | + | 4.11E-08 | 2.07E-07 |
| Endogenous_cannabinoid_signaling | 14 | 2.28 | + | 1.37E-07 | 6.67E-07 |
| Glycolysis | 14 | 2.38 | + | 2.23E-07 | 1.05E-06 |
| Axon guidance mediated by semaphorins | 18 | 4.09 | + | 3.20E-07 | 1.46E-06 |
| Endothelin signaling pathway | 27 | 8.65 | + | 4.20E-07 | 1.86E-06 |
| Heterotrimeric G-protein signaling pathway-rod outer segment phototransduction | 17 | 4.28 | + | 2.62E-06 | 1.13E-05 |
| FGF signaling pathway | 30 | 11.78 | + | 5.78E-06 | 2.41E-05 |
| TCA cycle | 10 | 1.62 | + | 7.62E-06 | 3.09E-05 |
| Integrin signalling pathway | 38 | 17.2 | + | 9.08E-06 | 3.58E-05 |
| T cell activation | 26 | 9.69 | + | 9.80E-06 | 3.77E-05 |
| Wnt signaling pathway | 56 | 30.12 | + | 1.33E-05 | 4.98E-05 |
| PI3 kinase pathway | 27 | 10.93 | + | 2.74E-05 | 1.00E-04 |
| B cell activation | 21 | 7.79 | + | 6.27E-05 | 2.23E-04 |
| Ras Pathway | 20 | 7.51 | + | 1.08E-04 | 3.75E-04 |
| Cadherin signaling pathway | 30 | 13.97 | + | 1.22E-04 | 4.14E-04 |
| De novo purine biosynthesis | 12 | 3.23 | + | 1.38E-04 | 4.58E-04 |
| Hedgehog signaling pathway | 10 | 2.38 | + | 1.83E-04 | 5.94E-04 |
| Adrenaline and noradrenaline biosynthesis | 11 | 3.04 | + | 3.22E-04 | 1.02E-03 |
| 5-Hydroxytryptamine degredation | 8 | 1.9 | + | 7.88E-04 | 2.45E-03 |
| Apoptosis signaling pathway | 24 | 11.69 | + | 1.01E-03 | 3.07E-03 |
| Angiogenesis | 33 | 18.15 | + | 1.03E-03 | 3.07E-03 |
| ATP synthesis | 5 | 0.76 | + | 1.13E-03 | 3.30E-03 |
| 5HT3 type receptor mediated signaling pathway | 7 | 1.71 | + | 1.93E-03 | 5.53E-03 |
| VEGF signaling pathway | 16 | 7.13 | + | 2.80E-03 | 7.86E-03 |
| Pyrimidine Metabolism | 6 | 1.43 | + | 3.47E-03 | 9.46E-03 |
| PDGF signaling pathway | 27 | 15.11 | + | 3.50E-03 | 9.46E-03 |
| Ubiquitin proteasome pathway | 15 | 6.65 | + | 3.58E-03 | 9.50E-03 |
| Glutamine glutamate conversion | 3 | 0.38 | + | 6.89E-03 | 1.80E-02 |
| Pentose phosphate pathway | 4 | 0.76 | + | 7.62E-03 | 1.95E-02 |
| Axon guidance mediated by Slit/Robo | 6 | 1.9 | + | 1.32E-02 | 3.32E-02 |
| Phenylethylamine degradation | 4 | 0.95 | + | 1.61E-02 | 3.98E-02 |
| Adenine and hypoxanthine salvage pathway | 4 | 0.95 | + | 1.61E-02 | 3.98E-02 |
| Alzheimer disease-amyloid secretase pathway | 13 | 6.75 | + | 2.07E-02 | 4.95E-02 |
| De novo pyrmidine ribonucleotides biosythesis | 5 | 1.71 | + | 3.02E-02 | 7.11E-02 |
| General transcription regulation | 0 | 3.42 | - | 3.26E-02 | 7.55E-02 |
| Phenylalanine biosynthesis | 2 | 0.29 | + | 3.37E-02 | 7.69E-02 |
| Arginine biosynthesis | 3 | 0.76 | + | 4.18E-02 | 9.39E-02 |
| Pyruvate metabolism | 4 | 1.33 | + | 4.61E-02 | 1.02E-01 |
| Circadian clock system | 3 | 0.86 | + | 5.56E-02 | 1.21E-01 |
| Tyrosine biosynthesis | 2 | 0.38 | + | 5.63E-02 | 1.21E-01 |
| Proline biosynthesis | 2 | 0.38 | + | 5.63E-02 | 1.21E-01 |
| FAS signaling pathway | 7 | 3.42 | + | 5.92E-02 | 1.23E-01 |
| Insulin/IGF pathway-protein kinase B signaling cascade | 4 | 8.46 | - | 7.58E-02 | 1.55E-01 |
| Oxidative stress response | 2 | 5.7 | - | 7.64E-02 | 1.55E-01 |
| Serine glycine biosynthesis | 2 | 0.48 | + | 8.27E-02 | 1.65E-01 |
| Asparagine and aspartate biosynthesis | 2 | 0.48 | + | 8.27E-02 | 1.65E-01 |
| Triacylglycerol metabolism | 1 | 0.1 | + | 9.07E-02 | 1.77E-01 |
| Anandamide_degradation | 1 | 0.1 | + | 9.07E-02 | 1.77E-01 |
| Fructose galactose metabolism | 3 | 1.14 | + | 1.08E-01 | 2.05E-01 |
| Gamma-aminobutyric acid synthesis | 2 | 0.57 | + | 1.12E-01 | 2.10E-01 |
| Heme biosynthesis | 3 | 1.24 | + | 1.28E-01 | 2.37E-01 |
| General transcription by RNA polymerase I | 0 | 1.9 | - | 1.49E-01 | 2.72E-01 |
| Axon guidance mediated by netrin | 5 | 2.85 | + | 1.60E-01 | 2.88E-01 |
| Interleukin signaling pathway | 11 | 15.3 | - | 1.65E-01 | 2.92E-01 |
| Blood coagulation | 2 | 4.56 | - | 1.66E-01 | 2.92E-01 |
| Sulfate assimilation | 1 | 0.19 | + | 1.73E-01 | 2.97E-01 |
| Pyridoxal phosphate salvage pathway | 1 | 0.19 | + | 1.73E-01 | 2.97E-01 |
| Aminobutyrate degradation | 1 | 0.19 | + | 1.73E-01 | 2.97E-01 |
| Purine metabolism | 2 | 0.76 | + | 1.77E-01 | 2.97E-01 |
| Hypoxia response via HIF activation | 1 | 3.04 | - | 1.93E-01 | 3.20E-01 |
| Toll receptor signaling pathway | 8 | 5.89 | + | 2.41E-01 | 3.95E-01 |
| Vitamin B6 metabolism | 1 | 0.29 | + | 2.48E-01 | 4.02E-01 |
| De novo pyrimidine deoxyribonucleotide biosynthesis | 3 | 1.81 | + | 2.71E-01 | 4.35E-01 |
| Alzheimer disease-presenilin pathway | 14 | 11.59 | + | 2.76E-01 | 4.36E-01 |
| TGF-beta signaling pathway | 11 | 13.78 | - | 2.78E-01 | 4.36E-01 |
| Formyltetrahydroformate biosynthesis | 2 | 1.05 | + | 2.81E-01 | 4.36E-01 |
| Androgen/estrogene/progesterone biosynthesis | 3 | 1.9 | + | 2.96E-01 | 4.53E-01 |
| Interferon-gamma signaling pathway | 4 | 2.76 | + | 2.98E-01 | 4.53E-01 |
| p53 pathway by glucose deprivation | 1 | 2.38 | - | 3.14E-01 | 4.71E-01 |
| Methylmalonyl pathway | 1 | 0.38 | + | 3.16E-01 | 4.71E-01 |
| Ascorbate degradation | 1 | 0.38 | + | 3.16E-01 | 4.71E-01 |
| Cell cycle | 3 | 2.09 | + | 3.48E-01 | 5.06E-01 |
| Cholesterol biosynthesis | 2 | 1.24 | + | 3.50E-01 | 5.06E-01 |
| p53 pathway feedback loops 2 | 6 | 4.94 | + | 3.74E-01 | 5.35E-01 |
| Salvage pyrimidine ribonucleotides | 2 | 1.33 | + | 3.84E-01 | 5.44E-01 |
| p53 pathway | 12 | 10.74 | + | 3.89E-01 | 5.46E-01 |
| DNA replication | 1 | 2 | - | 4.07E-01 | 5.66E-01 |
| mRNA splicing | 0 | 0.86 | - | 4.25E-01 | 5.77E-01 |
| Insulin/IGF pathway-mitogen activated protein kinase kinase/MAP kinase cascade | 4 | 3.33 | + | 4.25E-01 | 5.77E-01 |
| JAK/STAT signaling pathway | 1 | 1.9 | - | 4.33E-01 | 5.77E-01 |
| Transcription regulation by bZIP transcription factor | 4 | 5.04 | - | 4.34E-01 | 5.77E-01 |
| Xanthine and guanine salvage pathway | 1 | 0.57 | + | 4.35E-01 | 5.77E-01 |
| Coenzyme A biosynthesis | 1 | 0.57 | + | 4.35E-01 | 5.77E-01 |
| 2-arachidonoylglycerol biosynthesis | 1 | 0.67 | + | 4.86E-01 | 6.34E-01 |
| Plasminogen activating cascade | 2 | 1.71 | + | 5.10E-01 | 6.58E-01 |
| N-acetylglucosamine metabolism | 0 | 0.67 | - | 5.14E-01 | 6.58E-01 |
| Mannose metabolism | 0 | 0.67 | - | 5.14E-01 | 6.58E-01 |
| P53 pathway feedback loops 1 | 0 | 0.67 | - | 5.14E-01 | 6.58E-01 |
| Notch signaling pathway | 4 | 4.47 | - | 5.38E-01 | 6.71E-01 |
| Folate biosynthesis | 0 | 0.57 | - | 5.65E-01 | 6.99E-01 |
| 5-Hydroxytryptamine biosynthesis | 0 | 0.57 | - | 5.65E-01 | 6.99E-01 |
| Vitamin D metabolism and pathway | 1 | 1.33 | - | 6.16E-01 | 7.49E-01 |
| Salvage pyrimidine deoxyribonucleotides | 0 | 0.38 | - | 6.84E-01 | 8.25E-01 |
| O-antigen biosynthesis | 0 | 0.38 | - | 6.84E-01 | 8.25E-01 |
| Methylcitrate cycle | 0 | 0.38 | - | 6.84E-01 | 8.25E-01 |
| Vasopressin synthesis | 1 | 1.14 | - | 6.84E-01 | 8.25E-01 |
| Valine biosynthesis | 0 | 0.29 | - | 7.52E-01 | 8.78E-01 |
| S adenosyl methionine biosynthesis | 0 | 0.29 | - | 7.52E-01 | 8.78E-01 |
| Ornithine degradation | 0 | 0.29 | - | 7.52E-01 | 8.78E-01 |
| Isoleucine biosynthesis | 0 | 0.29 | - | 7.52E-01 | 8.78E-01 |
| Acetate utilization | 0 | 0.29 | - | 7.52E-01 | 8.78E-01 |
| Threonine biosynthesis | 0 | 0.19 | - | 8.27E-01 | 9.29E-01 |
| Thiamine metabolism | 0 | 0.19 | - | 8.27E-01 | 9.29E-01 |
| Succinate to proprionate conversion | 0 | 0.19 | - | 8.27E-01 | 9.29E-01 |
| PLP biosynthesis | 0 | 0.19 | - | 8.27E-01 | 9.29E-01 |
| Lysine biosynthesis | 0 | 0.19 | - | 8.27E-01 | 9.29E-01 |
| Lipoate_biosynthesis | 0 | 0.19 | - | 8.27E-01 | 9.29E-01 |
| Leucine biosynthesis | 0 | 0.19 | - | 8.27E-01 | 9.29E-01 |
| Carnitine metabolism | 0 | 0.19 | - | 8.27E-01 | 9.29E-01 |
| Carnitine and CoA metabolism | 0 | 0.19 | - | 8.27E-01 | 9.29E-01 |
| Histamine synthesis | 0 | 0.19 | - | 8.27E-01 | 9.29E-01 |
| Alanine biosynthesis | 0 | 0.19 | - | 8.27E-01 | 9.29E-01 |
| Bupropion_degradation | 0 | 0.1 | - | 9.09E-01 | 9.41E-01 |
| Methionine biosynthesis | 0 | 0.1 | - | 9.09E-01 | 9.41E-01 |
| Flavin biosynthesis | 0 | 0.1 | - | 9.09E-01 | 9.41E-01 |
| Cysteine biosynthesis | 0 | 0.1 | - | 9.09E-01 | 9.41E-01 |
| Cobalamin biosynthesis | 0 | 0.1 | - | 9.09E-01 | 9.41E-01 |
| Allantoin degradation | 0 | 0.1 | - | 9.09E-01 | 9.41E-01 |
Supplementary Table 9b. Pathway analysis summary of all PSD proteins identified in human and mouse.
All human and mouse PSD proteins were classified together using the Panther 'Pathway' descriptor. An enrichment analysis was done to determine Pathways overrepresented compared with the human genome. The Benjamini-Hochberg procedure was used to correct for multiple testing. The column 'observed' retrieves the number of proteins identified in each 'Pathway' while the column 'expected', the number that would have been identified by chance. Over or under-representations are shown by a (+) or (-) symbol respectively.
Human and mouse PSD proteins were classified using the Panther 'Pathway' descriptor. An enrichment analysis was done to determine Pathways overrepresented as compared with the human genome, a set of mouse brain expressed proteins and a set of genes highly expressed inhuman neurons. P-values were corrected for multiple testing using the Benjamini-Hochberg procedure. The colum 'observed' retrives the number of proteins identified in each 'Pathway' while the column 'expected', the number which would have been identified by chance. Over or underrepresntations are shown by a (+) or (-) symbol respectively.
| Pathway | Corrected P-value vs Genome | Corrected P-value vs Mouse Brain | Corrected P-value vs Neuron | Significantly overreprresented against the 3 neural lists |
|---|---|---|---|---|
| Metabotropic glutamate receptor group III pathway | 4.51E-19 | 2.13E-06 | 2.41E-03 | YES |
| Metabotropic glutamate receptor group II pathway | 3.64E-16 | 3.51E-05 | 4.34E-04 | YES |
| Muscarinic acetylcholine receptor 1 and 3 signaling pathway | 7.01E-15 | 2.10E-05 | 1.93E-06 | YES |
| Huntington disease | 7.30E-15 | 4.35E-04 | 9.66E-07 | YES |
| Ionotropic glutamate receptor pathway | 3.43E-14 | 6.33E-06 | 4.24E-03 | YES |
| Thyrotropin-releasing hormone receptor signaling pathway | 3.43E-14 | 2.10E-05 | 4.34E-04 | YES |
| Synaptic_vesicle_trafficking | 5.86E-14 | 1.42E-04 | 1.21E-02 | YES |
| Parkinson disease | 1.11E-13 | 8.85E-03 | 1.04E-07 | YES |
| Heterotrimeric G-protein signaling pathway-Gq alpha and Go alpha mediated pathway | 4.09E-13 | 3.25E-05 | 9.10E-06 | YES |
| Oxytocin receptor mediated signaling pathway | 1.66E-12 | 8.75E-05 | 2.99E-03 | YES |
| 5HT2 type receptor mediated signaling pathway | 2.00E-12 | 7.41E-05 | 5.96E-03 | YES |
| Muscarinic acetylcholine receptor 2 and 4 signaling pathway | 1.72E-11 | 2.87E-03 | 3.52E-05 | YES |
| Histamine H1 receptor mediated signaling pathway | 2.68E-11 | 7.41E-05 | 9.04E-04 | YES |
| Beta2 adrenergic receptor signaling pathway | 3.72E-11 | 1.55E-03 | 3.21E-02 | YES |
| Beta1 adrenergic receptor signaling pathway | 3.72E-11 | 1.55E-03 | 3.21E-02 | YES |
| Inflammation mediated by chemokine and cytokine signaling pathway | 4.95E-11 | 2.12E-05 | 5.95E-07 | YES |
| Cortocotropin releasing factor receptor signaling pathway | 2.75E-10 | 1.33E-02 | 2.40E-04 | YES |
| Heterotrimeric G-protein signaling pathway-Gi alpha and Gs alpha mediated pathway | 3.26E-10 | 2.87E-03 | 9.79E-05 | YES |
| Cytoskeletal regulation by Rho GTPase | 6.29E-10 | 1.75E-04 | 1.96E-03 | YES |
| 5HT1 type receptor mediated signaling pathway | 9.27E-10 | 5.24E-03 | 7.43E-03 | YES |
| 5HT4 type receptor mediated signaling pathway | 2.71E-09 | 1.56E-02 | 3.07E-03 | YES |
| EGF receptor signaling pathway | 1.62E-08 | 2.23E-02 | 4.34E-04 | YES |
| Metabotropic glutamate receptor group I pathway | 2.50E-08 | 3.54E-05 | 3.00E-02 | YES |
| Histamine H2 receptor mediated signaling pathway | 1.81E-07 | 2.86E-02 | 4.97E-02 | YES |
| Nicotinic acetylcholine receptor signaling pathway | 2.07E-07 | 3.79E-04 | 3.52E-05 | YES |
| Glycolysis | 1.05E-06 | 2.53E-03 | 1.05E-04 | YES |
| Heterotrimeric G-protein signaling pathway-rod outer segment phototransduction | 1.13E-05 | 1.29E-02 | 5.85E-04 | YES |
| TCA cycle | 3.09E-05 | 1.66E-02 | 3.42E-06 | YES |
| Wnt signaling pathway | 4.98E-05 | 2.87E-03 | 1.36E-02 | YES |
| PI3 kinase pathway | 1.00E-04 | 4.71E-03 | 1.21E-02 | YES |
| Cadherin signaling pathway | 4.14E-04 | 1.70E-05 | 1.23E-02 | YES |
| Hedgehog signaling pathway | 5.94E-04 | 1.66E-02 | 1.07E-02 | YES |
| 5-Hydroxytryptamine degredation | 2.45E-03 | 4.90E-02 | 0.00E+00 | YES |
| Glutamine glutamate conversion | 1.80E-02 | 0.00E+00 | 0.00E+00 | YES |
| GABA-B_receptor_II_signaling | 4.89E-09 | 1.17E-02 | 1.83E-01 | |
| Beta3 adrenergic receptor signaling pathway | 7.87E-09 | 5.14E-02 | 1.34E-02 | |
| Unclassified | 3.31E-08 | 4.91E-01 | 4.76E-01 | |
| Alpha adrenergic receptor signaling pathway | 1.35E-07 | 1.98E-03 | 8.71E-02 | |
| Endogenous_cannabinoid_signaling | 6.67E-07 | 5.21E-02 | 1.83E-01 | |
| Axon guidance mediated by semaphorins | 1.46E-06 | 1.85E-03 | 5.27E-02 | |
| Endothelin signaling pathway | 1.86E-06 | 2.87E-03 | 2.12E-01 | |
| FGF signaling pathway | 2.41E-05 | 9.87E-02 | 2.00E-01 | |
| Integrin signalling pathway | 3.58E-05 | 5.21E-02 | 2.15E-03 | |
| T cell activation | 3.77E-05 | 1.70E-02 | 4.79E-01 | |
| B cell activation | 2.23E-04 | 7.26E-03 | 1.83E-01 | |
| Ras Pathway | 3.75E-04 | 4.91E-01 | 4.53E-01 | |
| De novo purine biosynthesis | 4.58E-04 | 7.28E-02 | 3.96E-05 | |
| Adrenaline and noradrenaline biosynthesis | 1.02E-03 | 5.21E-02 | 2.32E-02 | |
| Apoptosis signaling pathway | 3.07E-03 | 5.49E-01 | 4.72E-01 | |
| Angiogenesis | 3.07E-03 | 1.81E-01 | 4.76E-01 | |
| ATP synthesis | 3.30E-03 | 3.72E-01 | 1.89E-02 | |
| 5HT3 type receptor mediated signaling pathway | 5.53E-03 | 1.03E-01 | 2.65E-02 | |
| VEGF signaling pathway | 7.86E-03 | 3.09E-01 | 4.79E-01 | |
| Pyrimidine Metabolism | 9.46E-03 | 4.27E-01 | 4.16E-01 | |
| PDGF signaling pathway | 9.46E-03 | 5.21E-02 | 8.88E-02 | |
| Ubiquitin proteasome pathway | 9.50E-03 | 6.13E-01 | 4.97E-01 | |
| Pentose phosphate pathway | 1.95E-02 | 5.80E-02 | 7.19E-02 | |
| Axon guidance mediated by Slit/Robo | 3.32E-02 | 1.23E-01 | 2.47E-01 | |
| Phenylethylamine degradation | 3.98E-02 | 1.72E-01 | 0.00E+00 | |
| Adenine and hypoxanthine salvage pathway | 3.98E-02 | 8.85E-03 | 7.19E-02 | |
| Alzheimer disease-amyloid secretase pathway | 4.95E-02 | 3.09E-01 | 8.71E-02 | |
| De novo pyrmidine ribonucleotides biosythesis | 7.11E-02 | 1.62E-02 | 1.81E-01 | |
| General transcription regulation | 7.55E-02 | 4.71E-03 | 9.08E-03 | |
| Phenylalanine biosynthesis | 7.69E-02 | 2.04E-01 | 4.16E-01 | |
| Arginine biosynthesis | 9.39E-02 | 4.97E-02 | 2.10E-01 | |
| Pyruvate metabolism | 1.02E-01 | 6.29E-01 | 3.35E-01 | |
| Circadian clock system | 1.21E-01 | 3.72E-01 | 2.10E-01 | |
| Tyrosine biosynthesis | 1.21E-01 | 2.04E-01 | 4.16E-01 | |
| Proline biosynthesis | 1.21E-01 | 6.13E-01 | 0.00E+00 | |
| FAS signaling pathway | 1.23E-01 | 5.49E-01 | 4.60E-01 | |
| Insulin/IGF pathway-protein kinase B signaling cascade | 1.55E-01 | 6.13E-01 | 2.80E-01 | |
| Oxidative stress response | 1.55E-01 | 1.03E-02 | 9.83E-04 | |
| Serine glycine biosynthesis | 1.65E-01 | 2.04E-01 | 0.00E+00 | |
| Asparagine and aspartate biosynthesis | 1.65E-01 | 2.04E-01 | 4.16E-01 | |
| Triacylglycerol metabolism | 1.77E-01 | 5.70E-01 | 0.00E+00 | |
| Anandamide_degradation | 1.77E-01 | 5.70E-01 | 0.00E+00 | |
| Fructose galactose metabolism | 2.05E-01 | 6.34E-01 | 4.79E-01 | |
| Gamma-aminobutyric acid synthesis | 2.10E-01 | 6.29E-01 | 4.16E-01 | |
| Heme biosynthesis | 2.37E-01 | 6.34E-01 | 0.00E+00 | |
| General transcription by RNA polymerase I | 2.72E-01 | 2.30E-01 | 1.83E-01 | |
| Axon guidance mediated by netrin | 2.88E-01 | 3.72E-01 | 5.22E-01 | |
| Interleukin signaling pathway | 2.92E-01 | 4.90E-01 | 3.31E-01 | |
| Blood coagulation | 2.92E-01 | 6.29E-01 | 6.34E-01 | |
| Sulfate assimilation | 2.97E-01 | 0.00E+00 | 7.24E-01 | |
| Pyridoxal phosphate salvage pathway | 2.97E-01 | 7.24E-01 | 0.00E+00 | |
| Aminobutyrate degradation | 2.97E-01 | 7.24E-01 | 7.24E-01 | |
| Purine metabolism | 2.97E-01 | 6.64E-01 | 4.16E-01 | |
| Hypoxia response via HIF activation | 3.20E-01 | 2.27E-01 | 1.89E-02 | |
| Toll receptor signaling pathway | 3.95E-01 | 6.34E-01 | 3.69E-01 | |
| Vitamin B6 metabolism | 4.02E-01 | 6.34E-01 | 0.00E+00 | |
| De novo pyrimidine deoxyribonucleotide biosynthesis | 4.35E-01 | 4.97E-02 | 2.10E-01 | |
| Alzheimer disease-presenilin pathway | 4.36E-01 | 1.66E-01 | 2.70E-01 | |
| TGF-beta signaling pathway | 4.36E-01 | 5.16E-01 | 5.37E-01 | |
| Formyltetrahydroformate biosynthesis | 4.36E-01 | 6.13E-01 | 4.16E-01 | |
| Androgen/estrogene/progesterone biosynthesis | 4.53E-01 | 5.18E-01 | 4.79E-01 | |
| Interferon-gamma signaling pathway | 4.53E-01 | 6.29E-01 | 5.43E-01 | |
| p53 pathway by glucose deprivation | 4.71E-01 | 5.21E-02 | 7.24E-01 | |
| Methylmalonyl pathway | 4.71E-01 | 7.24E-01 | 7.24E-01 | |
| Ascorbate degradation | 4.71E-01 | 5.70E-01 | 0.00E+00 | |
| Cell cycle | 5.06E-01 | 4.91E-01 | 4.79E-01 | |
| Cholesterol biosynthesis | 5.06E-01 | 6.64E-01 | 4.16E-01 | |
| p53 pathway feedback loops 2 | 5.35E-01 | 5.81E-01 | 1.84E-01 | |
| Salvage pyrimidine ribonucleotides | 5.44E-01 | 5.49E-01 | 4.16E-01 | |
| p53 pathway | 5.46E-01 | 5.81E-01 | 8.69E-02 | |
| DNA replication | 5.66E-01 | 3.16E-01 | 7.24E-01 | |
| mRNA splicing | 5.77E-01 | 1.62E-01 | 4.16E-01 | |
| Insulin/IGF pathway-mitogen activated protein kinase kinase/MAP kinase cascade | 5.77E-01 | 4.92E-01 | 3.35E-01 | |
| JAK/STAT signaling pathway | 5.77E-01 | 6.34E-01 | 7.24E-01 | |
| Transcription regulation by bZIP transcription factor | 5.77E-01 | 2.04E-01 | 2.80E-01 | |
| Xanthine and guanine salvage pathway | 5.77E-01 | 5.70E-01 | 7.24E-01 | |
| Coenzyme A biosynthesis | 5.77E-01 | 6.34E-01 | 2.31E-01 | |
| 2-arachidonoylglycerol biosynthesis | 6.34E-01 | 0.00E+00 | 7.24E-01 | |
| Plasminogen activating cascade | 6.58E-01 | 2.04E-01 | 0.00E+00 | |
| N-acetylglucosamine metabolism | 6.58E-01 | 2.30E-01 | 4.16E-01 | |
| Mannose metabolism | 6.58E-01 | 1.00E-01 | #N/A | |
| P53 pathway feedback loops 1 | 6.58E-01 | 6.34E-01 | #N/A | |
| Notch signaling pathway | 6.71E-01 | 5.50E-01 | 5.22E-01 | |
| Folate biosynthesis | 6.99E-01 | 6.34E-01 | #N/A | |
| 5-Hydroxytryptamine biosynthesis | 6.99E-01 | 5.18E-01 | #N/A | |
| Vitamin D metabolism and pathway | 7.49E-01 | 6.34E-01 | 7.24E-01 | |
| Salvage pyrimidine deoxyribonucleotides | 8.25E-01 | #N/A | #N/A | |
| O-antigen biosynthesis | 8.25E-01 | 6.34E-01 | 4.16E-01 | |
| Methylcitrate cycle | 8.25E-01 | #N/A | #N/A | |
| Vasopressin synthesis | 8.25E-01 | 2.27E-01 | 1.07E-01 | |
| Valine biosynthesis | 8.78E-01 | 3.66E-01 | #N/A | |
| S adenosyl methionine biosynthesis | 8.78E-01 | 6.34E-01 | 4.16E-01 | |
| Ornithine degradation | 8.78E-01 | #N/A | #N/A | |
| Isoleucine biosynthesis | 8.78E-01 | 3.66E-01 | #N/A | |
| Acetate utilization | 8.78E-01 | 6.34E-01 | #N/A | |
| Threonine biosynthesis | 9.29E-01 | 5.18E-01 | #N/A | |
| Thiamine metabolism | 9.29E-01 | 5.18E-01 | 4.16E-01 | |
| Succinate to proprionate conversion | 9.29E-01 | 6.34E-01 | 4.16E-01 | |
| PLP biosynthesis | 9.29E-01 | 5.18E-01 | #N/A | |
| Lysine biosynthesis | 9.29E-01 | 6.34E-01 | #N/A | |
| Lipoate_biosynthesis | 9.29E-01 | 6.34E-01 | #N/A | |
| Leucine biosynthesis | 9.29E-01 | 5.18E-01 | #N/A | |
| Carnitine metabolism | 9.29E-01 | 6.34E-01 | 4.16E-01 | |
| Carnitine and CoA metabolism | 9.29E-01 | 6.34E-01 | 4.16E-01 | |
| Histamine synthesis | 9.29E-01 | 6.34E-01 | #N/A | |
| Alanine biosynthesis | 9.29E-01 | 5.18E-01 | #N/A | |
| Bupropion_degradation | 9.41E-01 | #N/A | #N/A | |
| Methionine biosynthesis | 9.41E-01 | #N/A | 4.16E-01 | |
| Flavin biosynthesis | 9.41E-01 | 6.34E-01 | 4.16E-01 | |
| Cysteine biosynthesis | 9.41E-01 | 6.34E-01 | #N/A | |
| Cobalamin biosynthesis | 9.41E-01 | 6.34E-01 | #N/A | |
| Allantoin degradation | 9.41E-01 | #N/A | #N/A |
Supplementary Table 10. Analysis of dN/dS values from mouse and human PSD proteins.
dN, dS, dN/dS and median dN/dS values are given for proteins found in human and mouse cortical PSD, and proteins exclusive or significantly enriched to each species’ PSD. All values were obtained from Ensembl 62.
| Ensembl Gene ID/ | Mouse Ensembl Gene ID/ | dN | dS | dN/dS | Median dN/dS | Ensembl Gene ID/ | Human Ensembl Gene ID/ | dN | dS | dN/dS | Median dN/dS | Ensembl Gene ID/ | Mouse Ensembl Gene ID/ | dN/ | dS | dN/dS | Median dN/dS | Ensembl Gene ID/ | Mouse Ensembl Gene ID/ | dN | dS | dN/dS | Median dN/dS | Ensembl Gene ID/ | Human Ensembl Gene ID/ | dN/ | dS | dN/dS | Median dN/dS | Ensembl Gene ID/ | Human Ensembl Gene ID/ | dN/ | dS | dN/dS | Median dN/dS |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Human PSD (Nat Neurosci. 2011 Jan;14(1):19-21) | Mouse PSD | Identified only in human PSD | Identified only in mouse PSD | Enriched in human PSD | Enriched in mouse PSD | ||||||||||||||||||||||||||||||
| ENSG00000121083 | ENSMUSG00000020483 | 0 | 0.0817 | 0 | 0.045432099 | ENSMUSG00000020483 | ENSG00000121083 | 0 | 0.0817 | 0 | 0.041538096 | ENSG00000108518 | ENSMUSG00000018293 | 0.0188 | 0.2138 | 0.087932647 | 0.067476115 | ENSG00000123091 | ENSMUSG00000028557 | 0.003 | 0.1131 | 0.026525199 | 0.049200877 | ENSMUSG00000004849 | ENSG00000106367 | 0 | 0.6521 | 0 | 0.02032421 | ENSMUSG00000023944 | ENSG00000096384 | 0.0018 | 0.7489 | 0.0024 | 0.0352 |
| ENSG00000197111 | ENSMUSG00000056851 | 0 | 0.1043 | 0 | ENSMUSG00000056851 | ENSG00000197111 | 0 | 0.1043 | 0 | ENSG00000108671 | ENSMUSG00000017428 | 0.0012 | 0.2141 | 0.005604858 | ENSG00000112531 | ENSMUSG00000062078 | 0 | 0.1417 | 0 | ENSMUSG00000020149 | ENSG00000138069 | 0 | 0.1842 | 0 | ENSMUSG00000004455 | ENSG00000186298 | 0.0014 | 0.4273 | 0.0033 | ||||||
| ENSG00000124785 | ENSMUSG00000039114 | 0.0127 | 0.1058 | 0.120037807 | ENSMUSG00000039114 | ENSG00000124785 | 0.0127 | 0.1058 | 0.120037807 | ENSG00000123728 | ENSMUSG00000050029 | 0 | 0.2257 | 0 | ENSG00000146006 | ENSMUSG00000071862 | 0.0085 | 0.1585 | 0.05362776 | ENSMUSG00000022884 | ENSG00000156976 | 0 | 0.2833 | 0 | ENSMUSG00000021820 | ENSG00000148660 | 0.0009 | 0.2708 | 0.0033 | ||||||
| ENSG00000123560 | ENSMUSG00000031425 | 0 | 0.1147 | 0 | ENSMUSG00000028557 | ENSG00000123091 | 0.003 | 0.1131 | 0.026525199 | ENSG00000175130 | ENSMUSG00000047945 | 0.0202 | 0.2348 | 0.086030664 | ENSG00000198492 | ENSMUSG00000040025 | 0.0017 | 0.1666 | 0.010204082 | ENSMUSG00000026797 | ENSG00000136854 | 0 | 0.5646 | 0 | ENSMUSG00000072214 | ENSG00000184702 | 0.0044 | 1.1790 | 0.0037 | ||||||
| ENSG00000107758 | ENSMUSG00000021816 | 0.0035 | 0.1603 | 0.021834061 | ENSMUSG00000031425 | ENSG00000123560 | 0 | 0.1147 | 0 | ENSG00000153310 | ENSMUSG00000022378 | 0.0014 | 0.2568 | 0.005451713 | ENSG00000154001 | ENSMUSG00000021051 | 0.001 | 0.1675 | 0.005970149 | ENSMUSG00000027438 | ENSG00000125814 | 0 | 0.5154 | 0 | ENSMUSG00000025892 | ENSG00000152578 | 0.0015 | 0.3846 | 0.0039 | ||||||
| ENSG00000070831 | ENSMUSG00000006699 | 0 | 0.1723 | 0 | ENSMUSG00000062078 | ENSG00000112531 | 0 | 0.1417 | 0 | ENSG00000141279 | ENSMUSG00000001441 | 0.0108 | 0.2652 | 0.040723982 | ENSG00000102081 | ENSMUSG00000000838 | 0.0136 | 0.1939 | 0.070139247 | ENSMUSG00000031425 | ENSG00000123560 | 0 | 0.1147 | 0 | ENSMUSG00000043991 | ENSG00000185129 | 0.0012 | 0.2722 | 0.0044 | ||||||
| ENSG00000138069 | ENSMUSG00000020149 | 0 | 0.1842 | 0 | ENSMUSG00000071862 | ENSG00000146006 | 0.0085 | 0.1585 | 0.05362776 | ENSG00000099246 | ENSMUSG00000073639 | 0.0045 | 0.2669 | 0.016860247 | ENSG00000073921 | ENSMUSG00000039361 | 0.0114 | 0.2191 | 0.052031036 | ENSMUSG00000054459 | ENSG00000163032 | 0 | 0.5023 | 0 | ENSMUSG00000026959 | ENSG00000176884 | 0.0041 | 0.7495 | 0.0055 | ||||||
| ENSG00000102069 | ENSMUSG00000059585 | 0.068 | 0.1853 | 0.366972477 | ENSMUSG00000021816 | ENSG00000107758 | 0.0035 | 0.1603 | 0.021834061 | ENSG00000156976 | ENSMUSG00000022884 | 0 | 0.2833 | 0 | ENSG00000114416 | ENSMUSG00000027680 | 0.0086 | 0.2251 | 0.038205242 | ENSMUSG00000055723 | ENSG00000133818 | 0 | 0.3523 | 0 | ENSMUSG00000015143 | ENSG00000072110 | 0.0039 | 0.5925 | 0.0066 | ||||||
| ENSG00000147044 | ENSMUSG00000031012 | 0.0005 | 0.1924 | 0.002598753 | ENSMUSG00000040025 | ENSG00000198492 | 0.0017 | 0.1666 | 0.010204082 | ENSG00000138430 | ENSMUSG00000027108 | 0.0024 | 0.3022 | 0.00794176 | ENSG00000116667 | ENSMUSG00000032666 | 0.0193 | 0.2275 | 0.084835165 | ENSMUSG00000036752 | ENSG00000188229 | 0.0008 | 0.7997 | 0.001000375 | ENSMUSG00000035390 | ENSG00000160469 | 0.0040 | 0.4793 | 0.0083 | ||||||
| ENSG00000125675 | ENSMUSG00000001986 | 0.0032 | 0.194 | 0.016494845 | ENSMUSG00000021051 | ENSG00000154001 | 0.001 | 0.1675 | 0.005970149 | ENSG00000174238 | ENSMUSG00000017781 | 0.005 | 0.3078 | 0.016244314 | ENSG00000134108 | ENSMUSG00000030105 | 0 | 0.2387 | 0 | ENSMUSG00000037742 | ENSG00000156508 | 0.001 | 0.4075 | 0.002453988 | ENSMUSG00000066392 | ENSG00000021645 | 0.0046 | 0.5253 | 0.0088 | ||||||
| ENSG00000165410 | ENSMUSG00000062929 | 0.0027 | 0.2026 | 0.013326752 | ENSMUSG00000006699 | ENSG00000070831 | 0 | 0.1723 | 0 | ENSG00000102144 | ENSMUSG00000062070 | 0.0108 | 0.3088 | 0.034974093 | ENSG00000139190 | ENSMUSG00000030337 | 0.0083 | 0.255 | 0.03254902 | ENSMUSG00000059970 | ENSG00000126803 | 0.0019 | 0.7296 | 0.002604167 | ENSMUSG00000061751 | ENSG00000160145 | 0.0049 | 0.4947 | 0.0099 | ||||||
| ENSG00000070756 | ENSMUSG00000022283 | 0.0014 | 0.2082 | 0.006724304 | ENSMUSG00000020149 | ENSG00000138069 | 0 | 0.1842 | 0 | ENSG00000112335 | ENSMUSG00000019804 | 0 | 0.3102 | 0 | ENSG00000151846 | ENSMUSG00000022283 | 0.0376 | 0.2588 | 0.145285935 | ENSMUSG00000020114 | ENSG00000111530 | 0.0027 | 0.5551 | 0.004863988 | ENSMUSG00000000881 | ENSG00000082458 | 0.0033 | 0.3224 | 0.0102 | ||||||
| ENSG00000084733 | ENSMUSG00000020671 | 0 | 0.213 | 0 | ENSMUSG00000031012 | ENSG00000147044 | 0.0005 | 0.1924 | 0.002598753 | ENSG00000131779 | ENSMUSG00000028102 | 0.0394 | 0.3106 | 0.126851256 | ENSG00000048740 | ENSMUSG00000002107 | 0.0361 | 0.2596 | 0.139060092 | ENSMUSG00000034160 | ENSG00000147162 | 0.0018 | 0.3689 | 0.004879371 | ENSMUSG00000039809 | ENSG00000136928 | 0.0088 | 0.7527 | 0.0117 | ||||||
| ENSG00000108518 | ENSMUSG00000018293 | 0.0188 | 0.2138 | 0.087932647 | ENSMUSG00000000838 | ENSG00000102081 | 0.0136 | 0.1939 | 0.070139247 | ENSG00000175073 | ENSMUSG00000045210 | 0.0217 | 0.3113 | 0.069707677 | ENSG00000149970 | ENSMUSG00000025658 | 0.0054 | 0.2634 | 0.020501139 | ENSMUSG00000022378 | ENSG00000153310 | 0.0014 | 0.2568 | 0.005451713 | ENSMUSG00000043460 | ENSG00000166897 | 0.0170 | 1.2052 | 0.0141 | ||||||
| ENSG00000108671 | ENSMUSG00000017428 | 0.0012 | 0.2141 | 0.005604858 | ENSMUSG00000001986 | ENSG00000125675 | 0.0032 | 0.194 | 0.016494845 | ENSG00000088833 | ENSMUSG00000027455 | 0.0182 | 0.3124 | 0.058258643 | ENSG00000196792 | ENSMUSG00000020954 | 0.0189 | 0.265 | 0.071320755 | ENSMUSG00000061981 | ENSG00000132589 | 0.0032 | 0.5667 | 0.005646727 | ENSMUSG00000024743 | ENSG00000011347 | 0.0082 | 0.5541 | 0.0148 | ||||||
| ENSG00000135316 | ENSMUSG00000032423 | 0.0088 | 0.2221 | 0.039621792 | ENSMUSG00000062929 | ENSG00000165410 | 0.0027 | 0.2026 | 0.013326752 | ENSG00000068366 | ENSMUSG00000031278 | 0.0147 | 0.3126 | 0.047024952 | ENSG00000165527 | ENSMUSG00000044147 | 0 | 0.2657 | 0 | ENSMUSG00000021270 | ENSG00000080824 | 0.0036 | 0.61 | 0.005901639 | ENSMUSG00000040640 | ENSG00000187672 | 0.0084 | 0.5661 | 0.0148 | ||||||
| ENSG00000132639 | ENSMUSG00000027273 | 0 | 0.2223 | 0 | ENSMUSG00000006456 | ENSG00000239306 | 0.0087 | 0.2036 | 0.042730845 | ENSG00000110344 | ENSMUSG00000059890 | 0.0153 | 0.3173 | 0.048219351 | ENSG00000113558 | ENSMUSG00000036309 | 0.0027 | 0.2673 | 0.01010101 | ENSMUSG00000039615 | ENSG00000103266 | 0.0098 | 1 | 0.007915994 | ENSMUSG00000041115 | ENSG00000124313 | 0.0047 | 0.3121 | 0.0151 | ||||||
| ENSG00000177879 | ENSMUSG00000074154 | 0.0096 | 0.2239 | 0.042876284 | ENSMUSG00000022283 | ENSG00000070756 | 0.0014 | 0.2082 | 0.006724304 | ENSG00000079332 | ENSMUSG00000020088 | 0.0023 | 0.3187 | 0.007216818 | ENSG00000046653 | ENSMUSG00000031342 | 0.0084 | 0.2705 | 0.031053604 | ENSMUSG00000018965 | ENSG00000128245 | 0.0036 | 0.3628 | 0.009922822 | ENSMUSG00000001986 | ENSG00000125675 | 0.0032 | 0.1940 | 0.0165 | ||||||
| ENSG00000123728 | ENSMUSG00000050029 | 0 | 0.2257 | 0 | ENSMUSG00000020671 | ENSG00000084733 | 0 | 0.213 | 0 | ENSG00000147164 | ENSMUSG00000046032 | 0.0055 | 0.3204 | 0.017166042 | ENSG00000156709 | ENSMUSG00000036932 | 0.043 | 0.2744 | 0.156705539 | ENSMUSG00000057322 | ENSG00000172809 | 0.0065 | 0.6395 | 0.010164191 | ENSMUSG00000017412 | ENSG00000182389 | 0.0080 | 0.4756 | 0.0168 | ||||||
| ENSG00000177879 | ENSMUSG00000024480 | 0.0002 | 0.2275 | 0.000879121 | ENSMUSG00000039361 | ENSG00000073921 | 0.0114 | 0.2191 | 0.052031036 | ENSG00000162734 | ENSMUSG00000013698 | 0.0032 | 0.3234 | 0.009894867 | ENSG00000108654 | ENSMUSG00000020719 | 0.0036 | 0.289 | 0.012456747 | ENSMUSG00000030246 | ENSG00000111716 | 0.0101 | 0.8754 | 0.011537583 | ENSMUSG00000071073 | ENSG00000204052 | 0.0058 | 0.3194 | 0.0182 | ||||||
| ENSG00000101266 | ENSMUSG00000074698 | 0.0059 | 0.2277 | 0.025911287 | ENSMUSG00000032423 | ENSG00000135316 | 0.0088 | 0.2221 | 0.039621792 | ENSG00000111540 | ENSMUSG00000000711 | 0 | 0.3237 | 0 | ENSG00000177301 | ENSMUSG00000040724 | 0.0027 | 0.2894 | 0.009329648 | ENSMUSG00000029581 | ENSG00000075618 | 0.0146 | 1 | 0.012005592 | ENSMUSG00000021991 | ENSG00000157445 | 0.0093 | 0.5089 | 0.0183 | ||||||
| ENSG00000119782 | ENSMUSG00000020635 | 0.0129 | 0.2297 | 0.056160209 | ENSMUSG00000027273 | ENSG00000132639 | 0 | 0.2223 | 0 | ENSG00000158882 | ENSMUSG00000005674 | 0.0108 | 0.3249 | 0.033240997 | ENSG00000126950 | ENSMUSG00000033578 | 0.0143 | 0.2916 | 0.049039781 | ENSMUSG00000059714 | ENSG00000137312 | 0.0074 | 0.6061 | 0.012209206 | ENSMUSG00000047013 | ENSG00000163013 | 0.0133 | 0.7097 | 0.0187 | ||||||
| ENSG00000171723 | ENSMUSG00000047454 | 0.0061 | 0.234 | 0.026068376 | ENSMUSG00000027680 | ENSG00000114416 | 0.0086 | 0.2251 | 0.038205242 | ENSG00000095139 | ENSMUSG00000032096 | 0.0046 | 0.33 | 0.013939394 | ENSG00000109171 | ENSMUSG00000036087 | 0.0309 | 0.2934 | 0.105316973 | ENSMUSG00000026737 | ENSG00000150867 | 0.0078 | 0.6142 | 0.012699446 | ENSMUSG00000018765 | ENSG00000129245 | 0.0077 | 0.3774 | 0.0204 | ||||||
| ENSG00000175130 | ENSMUSG00000047945 | 0.0202 | 0.2348 | 0.086030664 | ENSMUSG00000024480 | ENSG00000177879 | 0.0002 | 0.2275 | 0.000879121 | ENSG00000101367 | ENSMUSG00000027479 | 0.0164 | 0.3311 | 0.049531863 | ENSG00000188215 | ENSMUSG00000048787 | 0.0104 | 0.2941 | 0.035362122 | ENSMUSG00000031207 | ENSG00000147065 | 0.006 | 0.4717 | 0.012719949 | ENSMUSG00000030172 | ENSG00000082805 | 0.0128 | 0.5522 | 0.0232 | ||||||
| ENSG00000169139 | ENSMUSG00000060811 | 0.0652 | 0.2353 | 0.277093073 | ENSMUSG00000032666 | ENSG00000116667 | 0.0193 | 0.2275 | 0.084835165 | ENSG00000091164 | ENSMUSG00000024583 | 0.0064 | 0.337 | 0.018991098 | ENSG00000184083 | ENSMUSG00000025262 | 0.0395 | 0.2958 | 0.133536173 | ENSMUSG00000022048 | ENSG00000092964 | 0.0056 | 0.4314 | 0.012980992 | ENSMUSG00000022763 | ENSG00000183773 | 0.0183 | 0.7800 | 0.0235 | ||||||
| ENSG00000213639 | ENSMUSG00000014956 | 0 | 0.2364 | 0 | ENSMUSG00000074698 | ENSG00000101266 | 0.0059 | 0.2277 | 0.025911287 | ENSG00000169891 | ENSMUSG00000040855 | 0.0625 | 0.3379 | 0.184965966 | ENSG00000142864 | ENSMUSG00000036371 | 0.0266 | 0.2964 | 0.08974359 | ENSMUSG00000014602 | ENSG00000130294 | 0.0111 | 0.7975 | 0.013918495 | ENSMUSG00000033981 | ENSG00000120251 | 0.0083 | 0.3467 | 0.0239 | ||||||
| ENSG00000070770 | ENSMUSG00000046707 | 0.005 | 0.2404 | 0.020798669 | ENSMUSG00000020635 | ENSG00000119782 | 0.0129 | 0.2297 | 0.056160209 | ENSG00000134444 | ENSMUSG00000026319 | 0.0367 | 0.3411 | 0.107593081 | ENSG00000170471 | ENSMUSG00000027652 | 0.0123 | 0.2985 | 0.04120603 | ENSMUSG00000022789 | ENSG00000087470 | 0.006 | 0.4124 | 0.014548982 | ENSMUSG00000024969 | ENSG00000072518 | 0.0079 | 0.3270 | 0.0242 | ||||||
| ENSG00000126861 | ENSMUSG00000049612 | 0.0632 | 0.2412 | 0.262023217 | ENSMUSG00000047454 | ENSG00000171723 | 0.0061 | 0.234 | 0.026068376 | ENSG00000170624 | ENSMUSG00000020354 | 0.0273 | 0.347 | 0.078674352 | ENSG00000122566 | ENSMUSG00000004980 | 0.0027 | 0.2986 | 0.009042197 | ENSMUSG00000024501 | ENSG00000113657 | 0.0072 | 0.483 | 0.014906832 | ENSMUSG00000067629 | ENSG00000197283 | 0.0111 | 0.4581 | 0.0242 | ||||||
| ENSG00000101558 | ENSMUSG00000024091 | 0.013 | 0.2413 | 0.053874845 | ENSMUSG00000014956 | ENSG00000213639 | 0 | 0.2364 | 0 | ENSG00000205302 | ENSMUSG00000034484 | 0.0098 | 0.3472 | 0.028225806 | ENSG00000196338 | ENSMUSG00000031302 | 0.0045 | 0.3041 | 0.014797764 | ENSMUSG00000054423 | ENSG00000163618 | 0.0065 | 0.4227 | 0.015377336 | ENSMUSG00000020827 | ENSG00000141503 | 0.0118 | 0.4760 | 0.0248 | ||||||
| ENSG00000115233 | ENSMUSG00000026914 | 0 | 0.2442 | 0 | ENSMUSG00000030105 | ENSG00000134108 | 0 | 0.2387 | 0 | ENSG00000159348 | ENSMUSG00000026456 | 0.0428 | 0.3474 | 0.123200921 | ENSG00000144320 | ENSMUSG00000009207 | 0.0745 | 0.3047 | 0.24450279 | ENSMUSG00000015291 | ENSG00000203879 | 0.0092 | 0.5061 | 0.018178226 | ENSMUSG00000038453 | ENSG00000017373 | 0.0170 | 0.6555 | 0.0259 | ||||||
| ENSG00000110367 | ENSMUSG00000032097 | 0.0108 | 0.2455 | 0.043991853 | ENSMUSG00000046707 | ENSG00000070770 | 0.005 | 0.2404 | 0.020798669 | ENSG00000119711 | ENSMUSG00000021238 | 0.0246 | 0.3506 | 0.070165431 | ENSG00000155744 | ENSMUSG00000038174 | 0.0238 | 0.3078 | 0.077322937 | ENSMUSG00000004267 | ENSG00000111674 | 0.0083 | 0.4236 | 0.019593957 | ENSMUSG00000047454 | ENSG00000171723 | 0.0061 | 0.2340 | 0.0261 | ||||||
| ENSG00000169139 | ENSMUSG00000022674 | 0.0059 | 0.254 | 0.023228346 | ENSMUSG00000049612 | ENSG00000126861 | 0.0632 | 0.2412 | 0.262023217 | ENSG00000124486 | ENSMUSG00000031010 | 0.0072 | 0.3512 | 0.020501139 | ENSG00000204469 | ENSMUSG00000024393 | 0.0545 | 0.3079 | 0.177005521 | ENSMUSG00000041329 | ENSG00000129244 | 0.0121 | 0.5747 | 0.021054463 | ENSMUSG00000020612 | ENSG00000108946 | 0.0160 | 0.6073 | 0.0263 | ||||||
| ENSG00000153310 | ENSMUSG00000022378 | 0.0014 | 0.2568 | 0.005451713 | ENSMUSG00000024091 | ENSG00000101558 | 0.013 | 0.2413 | 0.053874845 | ENSG00000134001 | ENSMUSG00000021116 | 0.0042 | 0.3513 | 0.011955594 | ENSG00000146147 | ENSMUSG00000032355 | 0.1882 | 0.309 | 0.609061489 | ENSMUSG00000001270 | ENSG00000166165 | 0.0184 | 0.8655 | 0.021259388 | ENSMUSG00000029516 | ENSG00000122966 | 0.0152 | 0.5522 | 0.0275 | ||||||
| ENSG00000067560 | ENSMUSG00000007815 | 0.0025 | 0.2593 | 0.009641342 | ENSMUSG00000026914 | ENSG00000115233 | 0 | 0.2442 | 0 | ENSG00000130227 | ENSMUSG00000022100 | 0.0027 | 0.3518 | 0.007674815 | ENSG00000186479 | ENSMUSG00000021719 | 0.0249 | 0.3106 | 0.080167418 | ENSMUSG00000022674 | ENSG00000169139 | 0.0059 | 0.254 | 0.023228346 | ENSMUSG00000007617 | ENSG00000152413 | 0.0111 | 0.3996 | 0.0278 | ||||||
| ENSG00000143376 | ENSMUSG00000028136 | 0.0099 | 0.2608 | 0.037960123 | ENSMUSG00000032097 | ENSG00000110367 | 0.0108 | 0.2455 | 0.043991853 | ENSG00000162368 | ENSMUSG00000028719 | 0.0422 | 0.3535 | 0.119377652 | ENSG00000167258 | ENSMUSG00000003119 | 0.0426 | 0.3111 | 0.136933462 | ENSMUSG00000026728 | ENSG00000026025 | 0.0111 | 0.4681 | 0.023712882 | ENSMUSG00000030376 | ENSG00000118160 | 0.0254 | 0.9112 | 0.0279 | ||||||
| ENSG00000092841 | ENSMUSG00000060680 | 0.0203 | 0.2612 | 0.077718224 | ENSMUSG00000022674 | ENSG00000169139 | 0.0059 | 0.254 | 0.023228346 | ENSG00000157500 | ENSMUSG00000040760 | 0.0078 | 0.3564 | 0.021885522 | ENSG00000184831 | ENSMUSG00000079508 | 0.1011 | 0.3127 | 0.32331308 | ENSMUSG00000040276 | ENSG00000124507 | 0.0201 | 0.84 | 0.023928571 | ENSMUSG00000037703 | ENSG00000088899 | 0.0210 | 0.7118 | 0.0295 | ||||||
| ENSG00000105372 | ENSMUSG00000072528 | 0.01 | 0.2613 | 0.038270188 | ENSMUSG00000030337 | ENSG00000139190 | 0.0083 | 0.255 | 0.03254902 | ENSG00000025772 | ENSMUSG00000018322 | 0.0585 | 0.3608 | 0.16213969 | ENSG00000184831 | ENSMUSG00000049233 | 0.1011 | 0.3127 | 0.32331308 | ENSMUSG00000029121 | ENSG00000072832 | 0.0157 | 0.6366 | 0.024662268 | ENSMUSG00000044667 | ENSG00000117600 | 0.0180 | 0.5861 | 0.0307 | ||||||
| ENSG00000116473 | ENSMUSG00000068798 | 0 | 0.2642 | 0 | ENSMUSG00000022283 | ENSG00000151846 | 0.0376 | 0.2588 | 0.145285935 | ENSG00000174373 | ENSMUSG00000021027 | 0.0252 | 0.3646 | 0.06911684 | ENSG00000182134 | ENSMUSG00000041912 | 0.0465 | 0.3134 | 0.148372687 | ENSMUSG00000021573 | ENSG00000171368 | 0.0369 | 1 | 0.024831763 | ENSMUSG00000000202 | ENSG00000204347 | 0.0347 | 1.1077 | 0.0313 | ||||||
| ENSG00000141279 | ENSMUSG00000001441 | 0.0108 | 0.2652 | 0.040723982 | ENSMUSG00000007815 | ENSG00000067560 | 0.0025 | 0.2593 | 0.009641342 | ENSG00000115866 | ENSMUSG00000026356 | 0.0183 | 0.3647 | 0.050178229 | ENSG00000048828 | ENSMUSG00000038014 | 0.0157 | 0.3191 | 0.049200877 | ENSMUSG00000031865 | ENSG00000204843 | 0.0145 | 0.502 | 0.028884462 | ENSMUSG00000003279 | ENSG00000170579 | 0.0178 | 0.5573 | 0.0319 | ||||||
| ENSG00000163466 | ENSMUSG00000006304 | 0.003 | 0.2663 | 0.01126549 | ENSMUSG00000002107 | ENSG00000048740 | 0.0361 | 0.2596 | 0.139060092 | ENSG00000100591 | ENSMUSG00000021037 | 0.0249 | 0.3649 | 0.068237873 | ENSG00000204842 | ENSMUSG00000042605 | 0.0564 | 0.3203 | 0.17608492 | ENSMUSG00000024277 | ENSG00000166974 | 0.0125 | 0.4283 | 0.029185151 | ENSMUSG00000061689 | ENSG00000080845 | 0.0184 | 0.5601 | 0.0329 | ||||||
| ENSG00000099246 | ENSMUSG00000073639 | 0.0045 | 0.2669 | 0.016860247 | ENSMUSG00000028136 | ENSG00000143376 | 0.0099 | 0.2608 | 0.037960123 | ENSG00000205544 | ENSMUSG00000070394 | 0.0628 | 0.3697 | 0.16986746 | ENSG00000085832 | ENSMUSG00000028552 | 0.0585 | 0.3215 | 0.181959565 | ENSMUSG00000022477 | ENSG00000100412 | 0.0158 | 0.5347 | 0.02954928 | ENSMUSG00000022623 | ENSG00000251322 | 0.0231 | 0.6816 | 0.0339 | ||||||
| ENSG00000119396 | ENSMUSG00000026878 | 0 | 0.2681 | 0 | ENSMUSG00000060680 | ENSG00000092841 | 0.0203 | 0.2612 | 0.077718224 | ENSG00000100462 | ENSMUSG00000023110 | 0.0095 | 0.3702 | 0.025661804 | ENSG00000162374 | ENSMUSG00000028546 | 0.0104 | 0.3215 | 0.032348367 | ENSMUSG00000025139 | ENSG00000078902 | 0.0337 | 1 | 0.02994757 | ENSMUSG00000034593 | ENSG00000197535 | 0.0148 | 0.4356 | 0.0340 | ||||||
| ENSG00000109919 | ENSMUSG00000027282 | 0.0376 | 0.2689 | 0.139828933 | ENSMUSG00000025658 | ENSG00000149970 | 0.0054 | 0.2634 | 0.020501139 | ENSG00000168807 | ENSMUSG00000041308 | 0.0171 | 0.3711 | 0.046079224 | ENSG00000198947 | ENSMUSG00000045103 | 0.0439 | 0.3215 | 0.136547434 | ENSMUSG00000035770 | ENSG00000135720 | 0.0146 | 0.3996 | 0.036536537 | ENSMUSG00000047495 | ENSG00000198010 | 0.0519 | 1.4223 | 0.0365 | ||||||
| ENSG00000156482 | ENSMUSG00000058600 | 0 | 0.2707 | 0 | ENSMUSG00000068798 | ENSG00000116473 | 0 | 0.2642 | 0 | ENSG00000204463 | ENSMUSG00000024392 | 0.0294 | 0.3717 | 0.079096045 | ENSG00000138757 | ENSMUSG00000029405 | 0.0074 | 0.3226 | 0.022938624 | ENSMUSG00000015747 | ENSG00000136631 | 0.0145 | 0.3764 | 0.038522848 | ENSMUSG00000024077 | ENSG00000115808 | 0.0161 | 0.4409 | 0.0365 | ||||||
| ENSG00000148660 | ENSMUSG00000021820 | 0.0009 | 0.2708 | 0.003323486 | ENSMUSG00000020954 | ENSG00000196792 | 0.0189 | 0.265 | 0.071320755 | ENSG00000198648 | ENSMUSG00000027030 | 0.021 | 0.3719 | 0.056466792 | ENSG00000092847 | ENSMUSG00000041530 | 0.0003 | 0.3254 | 0.000921942 | ENSMUSG00000037601 | ENSG00000239672 | 0.0295 | 0.716 | 0.041201117 | ENSMUSG00000053550 | ENSG00000187902 | 0.0336 | 0.8709 | 0.0386 | ||||||
| ENSG00000105372 | ENSMUSG00000071476 | 0.0133 | 0.2721 | 0.048879089 | ENSMUSG00000044147 | ENSG00000165527 | 0 | 0.2657 | 0 | ENSG00000035403 | ENSMUSG00000021823 | 0.0031 | 0.3723 | 0.008326618 | ENSG00000155970 | ENSMUSG00000039478 | 0.038 | 0.3281 | 0.115818348 | ENSMUSG00000066235 | ENSG00000144647 | 0.0302 | 0.7156 | 0.042202348 | ENSMUSG00000059003 | ENSG00000183454 | 0.0229 | 0.5876 | 0.0390 | ||||||
| ENSG00000105372 | ENSMUSG00000072548 | 0.0133 | 0.2721 | 0.048879089 | ENSMUSG00000006304 | ENSG00000163466 | 0.003 | 0.2663 | 0.01126549 | ENSG00000132142 | ENSMUSG00000020532 | 0.0113 | 0.3727 | 0.030319292 | ENSG00000131089 | ENSMUSG00000025656 | 0.009 | 0.3313 | 0.027165711 | ENSMUSG00000062444 | ENSG00000103723 | 0.0243 | 0.5369 | 0.045259825 | ENSMUSG00000037605 | ENSG00000150471 | 0.0190 | 0.4742 | 0.0401 | ||||||
| ENSG00000185129 | ENSMUSG00000043991 | 0.0012 | 0.2722 | 0.004408523 | ENSMUSG00000036309 | ENSG00000113558 | 0.0027 | 0.2673 | 0.01010101 | ENSG00000165795 | ENSMUSG00000004558 | 0.0227 | 0.3811 | 0.059564419 | ENSG00000109606 | ENSMUSG00000029169 | 0.0017 | 0.3342 | 0.005086774 | ENSMUSG00000020932 | ENSG00000131095 | 0.0372 | 0.7595 | 0.048979592 | ENSMUSG00000014932 | ENSG00000176105 | 0.0173 | 0.4174 | 0.0414 | ||||||
| ENSG00000122545 | ENSMUSG00000001833 | 0.0039 | 0.2747 | 0.014197306 | ENSMUSG00000026878 | ENSG00000119396 | 0 | 0.2681 | 0 | ENSG00000099942 | ENSMUSG00000006134 | 0.0159 | 0.3832 | 0.041492693 | ENSG00000147010 | ENSMUSG00000040990 | 0.0278 | 0.3349 | 0.083009854 | ENSMUSG00000001924 | ENSG00000130985 | 0.0228 | 0.4641 | 0.049127343 | ENSMUSG00000061576 | ENSG00000130226 | 0.0356 | 0.8467 | 0.0420 | ||||||
| ENSG00000099783 | ENSMUSG00000059208 | 0.0116 | 0.2747 | 0.042227885 | ENSMUSG00000027282 | ENSG00000109919 | 0.0376 | 0.2689 | 0.139828933 | ENSG00000060237 | ENSMUSG00000045962 | 0.0761 | 0.3833 | 0.198539003 | ENSG00000121774 | ENSMUSG00000028790 | 0.0274 | 0.3353 | 0.081717865 | ENSMUSG00000028480 | ENSG00000122694 | 0.0331 | 0.5882 | 0.056273376 | ENSMUSG00000031292 | ENSG00000008086 | 0.0208 | 0.4894 | 0.0425 | ||||||
| ENSG00000075391 | ENSMUSG00000070565 | 0.0145 | 0.2753 | 0.052669815 | ENSMUSG00000031342 | ENSG00000046653 | 0.0084 | 0.2705 | 0.031053604 | ENSG00000119787 | ENSMUSG00000059811 | 0.0113 | 0.3845 | 0.029388817 | ENSG00000137094 | ENSMUSG00000036052 | 0.0038 | 0.3356 | 0.011323004 | ENSMUSG00000053398 | ENSG00000092621 | 0.0302 | 0.5289 | 0.057099641 | ENSMUSG00000067242 | ENSG00000108231 | 0.0144 | 0.3247 | 0.0443 | ||||||
| ENSG00000147649 | ENSMUSG00000022255 | 0.0428 | 0.2753 | 0.155466764 | ENSMUSG00000058600 | ENSG00000156482 | 0 | 0.2707 | 0 | ENSG00000120265 | ENSMUSG00000019795 | 0.0209 | 0.3856 | 0.054201245 | ENSG00000033170 | ENSMUSG00000021065 | 0.0163 | 0.3361 | 0.048497471 | ENSMUSG00000002504 | ENSG00000065054 | 0.0395 | 0.6812 | 0.057985907 | ENSMUSG00000020321 | ENSG00000014641 | 0.0188 | 0.3945 | 0.0477 | ||||||
| ENSG00000135486 | ENSMUSG00000046434 | 0.0013 | 0.2756 | 0.004716981 | ENSMUSG00000021820 | ENSG00000148660 | 0.0009 | 0.2708 | 0.003323486 | ENSG00000167004 | ENSMUSG00000027248 | 0.0363 | 0.3862 | 0.09399275 | ENSG00000135823 | ENSMUSG00000026470 | 0.0305 | 0.3376 | 0.090343602 | ENSMUSG00000032382 | ENSG00000028528 | 0.0249 | 0.4228 | 0.058893094 | ENSMUSG00000063160 | ENSG00000105245 | 0.0222 | 0.4593 | 0.0483 | ||||||
| ENSG00000065135 | ENSMUSG00000000001 | 0.0079 | 0.2774 | 0.028478731 | ENSMUSG00000043991 | ENSG00000185129 | 0.0012 | 0.2722 | 0.004408523 | ENSG00000089006 | ENSMUSG00000027423 | 0.0081 | 0.3916 | 0.020684372 | ENSG00000156113 | ENSMUSG00000063142 | 0.0074 | 0.344 | 0.021511628 | ENSMUSG00000073838 | ENSG00000178952 | 0.0349 | 0.5743 | 0.060769633 | ENSMUSG00000022770 | ENSG00000075711 | 0.0213 | 0.4152 | 0.0513 | ||||||
| ENSG00000092841 | ENSMUSG00000090841 | 0 | 0.2779 | 0 | ENSMUSG00000036932 | ENSG00000156709 | 0.043 | 0.2744 | 0.156705539 | ENSG00000117592 | ENSMUSG00000026701 | 0.0671 | 0.3975 | 0.168805031 | ENSG00000135387 | ENSMUSG00000027184 | 0.014 | 0.3442 | 0.040674027 | ENSMUSG00000029028 | ENSG00000130764 | 0.0678 | 1 | 0.067476115 | ENSMUSG00000070565 | ENSG00000075391 | 0.0145 | 0.2753 | 0.0527 | ||||||
| ENSG00000168710 | ENSMUSG00000027893 | 0 | 0.2832 | 0 | ENSMUSG00000001833 | ENSG00000122545 | 0.0039 | 0.2747 | 0.014197306 | ENSG00000187118 | ENSMUSG00000039163 | 0.0515 | 0.398 | 0.129396985 | ENSG00000136950 | ENSMUSG00000026755 | 0.0152 | 0.3442 | 0.044160372 | ENSMUSG00000005981 | ENSG00000126602 | 0.059 | 0.8247 | 0.071541166 | ENSMUSG00000042388 | ENSG00000116544 | 0.0164 | 0.3100 | 0.0529 | ||||||
| ENSG00000156976 | ENSMUSG00000022884 | 0 | 0.2833 | 0 | ENSMUSG00000059208 | ENSG00000099783 | 0.0116 | 0.2747 | 0.042227885 | ENSG00000167699 | ENSMUSG00000017286 | 0.0448 | 0.4002 | 0.111944028 | ENSG00000112697 | ENSMUSG00000032328 | 0.0604 | 0.345 | 0.175072464 | ENSMUSG00000030774 | ENSG00000149269 | 0.0323 | 0.4086 | 0.079050416 | ENSMUSG00000026991 | ENSG00000144283 | 0.0172 | 0.3142 | 0.0547 | ||||||
| ENSG00000138443 | ENSMUSG00000026782 | 0.0041 | 0.2871 | 0.014280738 | ENSMUSG00000070565 | ENSG00000075391 | 0.0145 | 0.2753 | 0.052669815 | ENSG00000169764 | ENSMUSG00000001891 | 0.0053 | 0.4002 | 0.013243378 | ENSG00000147140 | ENSMUSG00000031311 | 0.004 | 0.345 | 0.011594203 | ENSMUSG00000027248 | ENSG00000167004 | 0.0363 | 0.3862 | 0.09399275 | ENSMUSG00000022103 | ENSG00000168546 | 0.0279 | 0.5026 | 0.0555 | ||||||
| ENSG00000108953 | ENSMUSG00000020849 | 0 | 0.2889 | 0 | ENSMUSG00000022255 | ENSG00000147649 | 0.0428 | 0.2753 | 0.155466764 | ENSG00000165434 | ENSMUSG00000030729 | 0.0314 | 0.4018 | 0.078148333 | ENSG00000092531 | ENSMUSG00000027287 | 0.0654 | 0.3459 | 0.189071986 | ENSMUSG00000028249 | ENSG00000137575 | 0.0504 | 0.5001 | 0.100779844 | ENSMUSG00000040867 | ENSG00000183092 | 0.0559 | 0.9731 | 0.0574 | ||||||
| ENSG00000105372 | ENSMUSG00000040952 | 0.0033 | 0.2923 | 0.011289771 | ENSMUSG00000046434 | ENSG00000135486 | 0.0013 | 0.2756 | 0.004716981 | ENSG00000140600 | ENSMUSG00000030638 | 0.0748 | 0.4024 | 0.185884692 | ENSG00000166579 | ENSMUSG00000018736 | 0.0192 | 0.3462 | 0.055459272 | ENSMUSG00000038173 | ENSG00000164303 | 0.0738 | 0.6733 | 0.109609387 | ENSMUSG00000021357 | ENSG00000112685 | 0.0334 | 0.5696 | 0.0586 | ||||||
| ENSG00000178741 | ENSMUSG00000000088 | 0.0742 | 0.2928 | 0.253415301 | ENSMUSG00000000001 | ENSG00000065135 | 0.0079 | 0.2774 | 0.028478731 | ENSG00000106771 | ENSMUSG00000055296 | 0.0396 | 0.4049 | 0.097801926 | ENSG00000102225 | ENSMUSG00000031065 | 0.0124 | 0.3478 | 0.035652674 | ENSMUSG00000030884 | ENSG00000140740 | 0.077 | 0.4502 | 0.171035096 | ENSMUSG00000025422 | ENSG00000135439 | 0.0240 | 0.4026 | 0.0596 | ||||||
| ENSG00000156052 | ENSMUSG00000024639 | 0.0012 | 0.2934 | 0.00408998 | ENSMUSG00000027893 | ENSG00000168710 | 0 | 0.2832 | 0 | ENSG00000205339 | ENSMUSG00000066232 | 0.0026 | 0.4057 | 0.006408676 | ENSG00000135298 | ENSMUSG00000033569 | 0.0078 | 0.3497 | 0.022304833 | ENSMUSG00000061518 | ENSG00000135940 | 0.0994 | 0.3084 | 0.32230869 | ENSMUSG00000053617 | ENSG00000107957 | 0.0443 | 0.7077 | 0.0626 | ||||||
| ENSG00000082701 | ENSMUSG00000022812 | 0.0045 | 0.2939 | 0.01531133 | ENSMUSG00000026782 | ENSG00000138443 | 0.0041 | 0.2871 | 0.014280738 | ENSG00000147853 | ENSMUSG00000024782 | 0.0381 | 0.4072 | 0.093565815 | ENSG00000011677 | ENSMUSG00000031343 | 0.0196 | 0.3498 | 0.056032018 | ENSMUSG00000026458 | ENSG00000143847 | 0.0365 | 0.4980 | 0.0733 | |||||||||||
| ENSG00000198898 | ENSMUSG00000015733 | 0.0078 | 0.2941 | 0.026521591 | ENSMUSG00000020849 | ENSG00000108953 | 0 | 0.2889 | 0 | ENSG00000067900 | ENSMUSG00000024290 | 0.015 | 0.4076 | 0.036800785 | ENSG00000150625 | ENSMUSG00000031517 | 0.0052 | 0.3507 | 0.014827488 | ENSMUSG00000020640 | ENSG00000198399 | 0.0330 | 0.4453 | 0.0741 | |||||||||||
| ENSG00000115091 | ENSMUSG00000026341 | 0.0011 | 0.2991 | 0.0036777 | ENSMUSG00000020719 | ENSG00000108654 | 0.0036 | 0.289 | 0.012456747 | ENSG00000198513 | ENSMUSG00000021066 | 0.0165 | 0.4112 | 0.040126459 | ENSG00000198722 | ENSMUSG00000028456 | 0.0222 | 0.351 | 0.063247863 | ENSMUSG00000062257 | ENSG00000183715 | 0.0346 | 0.4475 | 0.0773 | |||||||||||
| ENSG00000128463 | ENSMUSG00000027131 | 0.0025 | 0.2991 | 0.008358409 | ENSMUSG00000040724 | ENSG00000177301 | 0.0027 | 0.2894 | 0.009329648 | ENSG00000169398 | ENSMUSG00000022607 | 0.0128 | 0.4129 | 0.031000242 | ENSG00000133818 | ENSMUSG00000055723 | 0 | 0.3523 | 0 | ENSMUSG00000052572 | ENSG00000150672 | 0.0308 | 0.3930 | 0.0784 | |||||||||||
| ENSG00000171475 | ENSMUSG00000038013 | 0.0387 | 0.2993 | 0.129301704 | ENSMUSG00000033578 | ENSG00000126950 | 0.0143 | 0.2916 | 0.049039781 | ENSG00000136152 | ENSMUSG00000034893 | 0.0202 | 0.4131 | 0.048898572 | ENSG00000169306 | ENSMUSG00000052372 | 0.0066 | 0.3532 | 0.018686297 | ENSMUSG00000031626 | ENSG00000154556 | 0.0368 | 0.4678 | 0.0787 | |||||||||||
| ENSG00000104381 | ENSMUSG00000025777 | 0.0384 | 0.2997 | 0.128128128 | ENSMUSG00000020894 | ENSG00000220205 | 0.0038 | 0.2921 | 0.013009243 | ENSG00000080371 | ENSMUSG00000020132 | 0.0108 | 0.4142 | 0.02607436 | ENSG00000137942 | ENSMUSG00000039735 | 0.0103 | 0.3546 | 0.029046813 | ENSMUSG00000029614 | ENSG00000089009 | 0.0595 | 0.7275 | 0.0818 | |||||||||||
| ENSG00000122705 | ENSMUSG00000028478 | 0.0061 | 0.3012 | 0.020252324 | ENSMUSG00000040952 | ENSG00000105372 | 0.0033 | 0.2923 | 0.011289771 | ENSG00000137076 | ENSMUSG00000028465 | 0.0063 | 0.4148 | 0.015188042 | ENSG00000204131 | ENSMUSG00000079481 | 0.1067 | 0.3547 | 0.300817592 | ENSMUSG00000050908 | ENSG00000166676 | 0.0618 | 0.7007 | 0.0882 | |||||||||||
| ENSG00000138430 | ENSMUSG00000027108 | 0.0024 | 0.3022 | 0.00794176 | ENSMUSG00000000088 | ENSG00000178741 | 0.0742 | 0.2928 | 0.253415301 | ENSG00000163344 | ENSMUSG00000027952 | 0.0907 | 0.4165 | 0.217767107 | ENSG00000170515 | ENSMUSG00000025364 | 0.0048 | 0.356 | 0.013483146 | ENSMUSG00000035863 | ENSG00000099864 | 0.1249 | 1.3825 | 0.0903 | |||||||||||
| ENSG00000168958 | ENSMUSG00000026150 | 0.0242 | 0.303 | 0.079867987 | ENSMUSG00000024639 | ENSG00000156052 | 0.0012 | 0.2934 | 0.00408998 | ENSG00000213281 | ENSMUSG00000027852 | 0.0024 | 0.4173 | 0.005751258 | ENSG00000123124 | ENSMUSG00000041058 | 0.045 | 0.36 | 0.125 | ENSMUSG00000020486 | ENSG00000108387 | 0.0369 | 0.4058 | 0.0909 | |||||||||||
| ENSG00000118473 | ENSMUSG00000028524 | 0.0323 | 0.304 | 0.10625 | ENSMUSG00000036087 | ENSG00000109171 | 0.0309 | 0.2934 | 0.105316973 | ENSG00000109846 | ENSMUSG00000032060 | 0.0106 | 0.4183 | 0.025340665 | ENSG00000183723 | ENSMUSG00000051578 | 0.0219 | 0.3606 | 0.060732113 | ENSMUSG00000029420 | ENSG00000060709 | 0.0722 | 0.7701 | 0.0938 | |||||||||||
| ENSG00000114030 | ENSMUSG00000022905 | 0.0103 | 0.3045 | 0.033825944 | ENSMUSG00000015733 | ENSG00000198898 | 0.0078 | 0.2941 | 0.026521591 | ENSG00000028528 | ENSMUSG00000032382 | 0.0249 | 0.4228 | 0.058893094 | ENSG00000089818 | ENSMUSG00000030327 | 0.0351 | 0.3618 | 0.097014925 | ENSMUSG00000025964 | ENSG00000114948 | 0.0481 | 0.4999 | 0.0962 | |||||||||||
| ENSG00000182890 | ENSMUSG00000091316 | 0.0329 | 0.3047 | 0.107975057 | ENSMUSG00000048787 | ENSG00000188215 | 0.0104 | 0.2941 | 0.035362122 | ENSG00000101557 | ENSMUSG00000047879 | 0.0158 | 0.4229 | 0.037361078 | ENSG00000133961 | ENSMUSG00000021224 | 0.0337 | 0.3634 | 0.092735278 | ENSMUSG00000062044 | ENSG00000142235 | 0.0507 | 0.5098 | 0.0995 | |||||||||||
| ENSG00000182890 | ENSMUSG00000021794 | 0.0329 | 0.3047 | 0.107975057 | ENSMUSG00000025262 | ENSG00000184083 | 0.0395 | 0.2958 | 0.133536173 | ENSG00000156467 | ENSMUSG00000021520 | 0.068 | 0.4249 | 0.160037656 | ENSG00000077063 | ENSMUSG00000000416 | 0.0782 | 0.3643 | 0.214658249 | ENSMUSG00000058589 | ENSG00000185046 | 0.0421 | 0.4145 | 0.1016 | |||||||||||
| ENSG00000005022 | ENSMUSG00000016319 | 0.0081 | 0.3048 | 0.026574803 | ENSMUSG00000036371 | ENSG00000142864 | 0.0266 | 0.2964 | 0.08974359 | ENSG00000133812 | ENSMUSG00000038371 | 0.03 | 0.4257 | 0.070472163 | ENSG00000167371 | ENSMUSG00000045114 | 0.1324 | 0.3646 | 0.363137685 | ENSMUSG00000022577 | ENSG00000176956 | 0.0532 | 0.5035 | 0.1057 | |||||||||||
| ENSG00000165637 | ENSMUSG00000021771 | 0.0295 | 0.3048 | 0.096784777 | ENSMUSG00000027652 | ENSG00000170471 | 0.0123 | 0.2985 | 0.04120603 | ENSG00000122359 | ENSMUSG00000021866 | 0.0339 | 0.4273 | 0.079335362 | ENSG00000114850 | ENSMUSG00000027828 | 0.0051 | 0.3672 | 0.013888889 | ENSMUSG00000059974 | ENSG00000182667 | 0.0655 | 0.6133 | 0.1068 | |||||||||||
| ENSG00000123416 | ENSMUSG00000023004 | 0 | 0.3058 | 0 | ENSMUSG00000004980 | ENSG00000122566 | 0.0027 | 0.2986 | 0.009042197 | ENSG00000100934 | ENSMUSG00000020986 | 0.0048 | 0.4276 | 0.011225444 | ENSG00000172733 | ENSMUSG00000049184 | 0.0723 | 0.3682 | 0.196360674 | ENSMUSG00000057230 | ENSG00000115977 | 0.0361 | 0.3276 | 0.1102 | |||||||||||
| ENSG00000172020 | ENSMUSG00000047261 | 0.1247 | 0.3059 | 0.407649559 | ENSMUSG00000026341 | ENSG00000115091 | 0.0011 | 0.2991 | 0.0036777 | ENSG00000166974 | ENSMUSG00000024277 | 0.0125 | 0.4283 | 0.029185151 | ENSG00000111218 | ENSMUSG00000030350 | 0.0067 | 0.3685 | 0.018181818 | ENSMUSG00000041670 | ENSG00000079841 | 0.0597 | 0.4994 | 0.1195 | |||||||||||
| ENSG00000105372 | ENSMUSG00000069391 | 0.0965 | 0.3072 | 0.314127604 | ENSMUSG00000027131 | ENSG00000128463 | 0.0025 | 0.2991 | 0.008358409 | ENSG00000107679 | ENSMUSG00000040268 | 0.0515 | 0.4284 | 0.120214753 | ENSG00000157680 | ENSMUSG00000038665 | 0.0188 | 0.3704 | 0.05075594 | ENSMUSG00000037541 | ENSG00000162105 | 0.0774 | 0.5993 | 0.1292 | |||||||||||
| ENSG00000174238 | ENSMUSG00000017781 | 0.005 | 0.3078 | 0.016244314 | ENSMUSG00000038013 | ENSG00000171475 | 0.0387 | 0.2993 | 0.129301704 | ENSG00000165219 | ENSMUSG00000026867 | 0.0102 | 0.4329 | 0.023562024 | ENSG00000120049 | ENSMUSG00000025221 | 0.0146 | 0.3794 | 0.038481813 | ENSMUSG00000034675 | ENSG00000113758 | 0.0792 | 0.5821 | 0.1361 | |||||||||||
| ENSG00000107566 | ENSMUSG00000025198 | 0.0252 | 0.3081 | 0.081791626 | ENSMUSG00000025777 | ENSG00000104381 | 0.0384 | 0.2997 | 0.128128128 | ENSG00000151929 | ENSMUSG00000030847 | 0.0798 | 0.4333 | 0.184168013 | ENSG00000104177 | ENSMUSG00000027201 | 0.0422 | 0.3802 | 0.110994214 | ENSMUSG00000019917 | ENSG00000186522 | 0.0761 | 0.5381 | 0.1414 | |||||||||||
| ENSG00000135940 | ENSMUSG00000061518 | 0.0994 | 0.3084 | 0.32230869 | ENSMUSG00000028478 | ENSG00000122705 | 0.0061 | 0.3012 | 0.020252324 | ENSG00000123338 | ENSMUSG00000022488 | 0.0255 | 0.4368 | 0.058379121 | ENSG00000076716 | ENSMUSG00000031119 | 0.0241 | 0.3846 | 0.062662507 | ENSMUSG00000029711 | ENSG00000130427 | 0.1276 | 0.6737 | 0.1894 | |||||||||||
| ENSG00000102144 | ENSMUSG00000062070 | 0.0108 | 0.3088 | 0.034974093 | ENSMUSG00000026150 | ENSG00000168958 | 0.0242 | 0.303 | 0.079867987 | ENSG00000130725 | ENSMUSG00000005575 | 0 | 0.4368 | 0 | ENSG00000125249 | ENSMUSG00000051615 | 0.0022 | 0.3854 | 0.005708355 | ENSMUSG00000068373 | ENSG00000110427 | 0.1466 | 0.6921 | 0.2118 | |||||||||||
| ENSG00000116544 | ENSMUSG00000042388 | 0.0164 | 0.31 | 0.052903226 | ENSMUSG00000028524 | ENSG00000118473 | 0.0323 | 0.304 | 0.10625 | ENSG00000172164 | ENSMUSG00000060429 | 0.0325 | 0.4378 | 0.07423481 | ENSG00000163155 | ENSMUSG00000053769 | 0.0934 | 0.3882 | 0.24059763 | ||||||||||||||||
| ENSG00000112335 | ENSMUSG00000019804 | 0 | 0.3102 | 0 | ENSMUSG00000031302 | ENSG00000196338 | 0.0045 | 0.3041 | 0.014797764 | ENSG00000166340 | ENSMUSG00000030894 | 0.0637 | 0.4385 | 0.145267959 | ENSG00000081026 | ENSMUSG00000052539 | 0.0724 | 0.3884 | 0.186405767 | ||||||||||||||||
| ENSG00000131779 | ENSMUSG00000028102 | 0.0394 | 0.3106 | 0.126851256 | ENSMUSG00000022905 | ENSG00000114030 | 0.0103 | 0.3045 | 0.033825944 | ENSG00000110514 | ENSMUSG00000040687 | 0.0258 | 0.4406 | 0.058556514 | ENSG00000139197 | ENSMUSG00000005069 | 0.0316 | 0.3889 | 0.081254821 | ||||||||||||||||
| ENSG00000126858 | ENSMUSG00000017686 | 0.0099 | 0.3107 | 0.031863534 | ENSMUSG00000021794 | ENSG00000182890 | 0.0329 | 0.3047 | 0.107975057 | ENSG00000180228 | ENSMUSG00000002731 | 0.0106 | 0.4413 | 0.024019941 | ENSG00000128683 | ENSMUSG00000070880 | 0.0126 | 0.3894 | 0.032357473 | ||||||||||||||||
| ENSG00000175073 | ENSMUSG00000045210 | 0.0217 | 0.3113 | 0.069707677 | ENSMUSG00000009207 | ENSG00000144320 | 0.0745 | 0.3047 | 0.24450279 | ENSG00000031698 | ENSMUSG00000068739 | 0.0207 | 0.4427 | 0.046758527 | ENSG00000147234 | ENSMUSG00000042425 | 0.0659 | 0.3914 | 0.168369954 | ||||||||||||||||
| ENSG00000010810 | ENSMUSG00000019843 | 0.0026 | 0.3115 | 0.008346709 | ENSMUSG00000016319 | ENSG00000005022 | 0.0081 | 0.3048 | 0.026574803 | ENSG00000166501 | ENSMUSG00000052889 | 0.0054 | 0.4495 | 0.012013348 | ENSG00000153317 | ENSMUSG00000022377 | 0.0188 | 0.392 | 0.047959184 | ||||||||||||||||
| ENSG00000160551 | ENSMUSG00000017291 | 0.0028 | 0.3118 | 0.008980115 | ENSMUSG00000021771 | ENSG00000165637 | 0.0295 | 0.3048 | 0.096784777 | ENSG00000103316 | ENSMUSG00000030905 | 0.0634 | 0.4512 | 0.140514184 | ENSG00000163288 | ENSMUSG00000029212 | 0.0087 | 0.3924 | 0.022171254 | ||||||||||||||||
| ENSG00000124313 | ENSMUSG00000041115 | 0.0047 | 0.3121 | 0.015059276 | ENSMUSG00000023004 | ENSG00000123416 | 0 | 0.3058 | 0 | ENSG00000159023 | ENSMUSG00000028906 | 0.045 | 0.4525 | 0.099447514 | ENSG00000128683 | ENSMUSG00000090665 | 0.0245 | 0.3926 | 0.062404483 | ||||||||||||||||
| ENSG00000088833 | ENSMUSG00000027455 | 0.0182 | 0.3124 | 0.058258643 | ENSMUSG00000047261 | ENSG00000172020 | 0.1247 | 0.3059 | 0.407649559 | ENSG00000119471 | ENSMUSG00000028383 | 0.0697 | 0.453 | 0.153863135 | ENSG00000089091 | ENSMUSG00000037259 | 0.1332 | 0.3953 | 0.336959271 | ||||||||||||||||
| ENSG00000068366 | ENSMUSG00000031278 | 0.0147 | 0.3126 | 0.047024952 | ENSMUSG00000038174 | ENSG00000155744 | 0.0238 | 0.3078 | 0.077322937 | ENSG00000166986 | ENSMUSG00000040354 | 0.0587 | 0.4592 | 0.12783101 | ENSG00000108733 | ENSMUSG00000018733 | 0.0646 | 0.3957 | 0.163254991 | ||||||||||||||||
| ENSG00000144283 | ENSMUSG00000026991 | 0.0172 | 0.3142 | 0.054742202 | ENSMUSG00000024393 | ENSG00000204469 | 0.0545 | 0.3079 | 0.177005521 | ENSG00000114520 | ENSMUSG00000022808 | 0.0322 | 0.46 | 0.07 | ENSG00000100568 | ENSMUSG00000021124 | 0.0378 | 0.3987 | 0.094808126 | ||||||||||||||||
| ENSG00000116489 | ENSMUSG00000070372 | 0.0177 | 0.3145 | 0.056279809 | ENSMUSG00000025198 | ENSG00000107566 | 0.0252 | 0.3081 | 0.081791626 | ENSG00000198171 | ENSMUSG00000068290 | 0.0933 | 0.4605 | 0.202605863 | ENSG00000115355 | ENSMUSG00000032740 | 0.0254 | 0.3998 | 0.063531766 | ||||||||||||||||
| ENSG00000176986 | ENSMUSG00000039367 | 0.0397 | 0.3148 | 0.126111817 | ENSMUSG00000061518 | ENSG00000135940 | 0.0994 | 0.3084 | 0.32230869 | ENSG00000176148 | ENSMUSG00000027175 | 0.0822 | 0.4615 | 0.178114843 | ENSG00000138073 | ENSMUSG00000045302 | 0.0646 | 0.4006 | 0.161258113 | ||||||||||||||||
| ENSG00000110344 | ENSMUSG00000059890 | 0.0153 | 0.3173 | 0.048219351 | ENSMUSG00000032355 | ENSG00000146147 | 0.1882 | 0.309 | 0.609061489 | ENSG00000082898 | ENSMUSG00000020290 | 0.0091 | 0.4631 | 0.019650184 | ENSG00000117640 | ENSMUSG00000046671 | 0.0202 | 0.4009 | 0.05038663 | ||||||||||||||||
| ENSG00000079332 | ENSMUSG00000020088 | 0.0023 | 0.3187 | 0.007216818 | ENSMUSG00000042388 | ENSG00000116544 | 0.0164 | 0.31 | 0.052903226 | ENSG00000083312 | ENSMUSG00000009470 | 0.0016 | 0.4638 | 0.003449763 | ENSG00000047188 | ENSMUSG00000034653 | 0.0135 | 0.4024 | 0.033548708 | ||||||||||||||||
| ENSG00000204052 | ENSMUSG00000071073 | 0.0058 | 0.3194 | 0.018159048 | ENSMUSG00000021719 | ENSG00000186479 | 0.0249 | 0.3106 | 0.080167418 | ENSG00000135924 | ENSMUSG00000026203 | 0.0687 | 0.4639 | 0.148092261 | ENSG00000068971 | ENSMUSG00000024777 | 0.0054 | 0.4026 | 0.013412817 | ||||||||||||||||
| ENSG00000100030 | ENSMUSG00000063358 | 0.0053 | 0.3202 | 0.016552155 | ENSMUSG00000017686 | ENSG00000126858 | 0.0099 | 0.3107 | 0.031863534 | ENSG00000187164 | ENSMUSG00000041362 | 0.0455 | 0.4643 | 0.097996985 | ENSG00000111262 | ENSMUSG00000047976 | 0.0069 | 0.4036 | 0.017096135 | ||||||||||||||||
| ENSG00000147164 | ENSMUSG00000046032 | 0.0055 | 0.3204 | 0.017166042 | ENSMUSG00000003119 | ENSG00000167258 | 0.0426 | 0.3111 | 0.136933462 | ENSG00000119321 | ENSMUSG00000066151 | 0.123 | 0.4645 | 0.264800861 | ENSG00000104177 | ENSMUSG00000049230 | 0.0598 | 0.4044 | 0.147873393 | ||||||||||||||||
| ENSG00000185565 | ENSMUSG00000061080 | 0.0699 | 0.3219 | 0.217148183 | ENSMUSG00000019843 | ENSG00000010810 | 0.0026 | 0.3115 | 0.008346709 | ENSG00000138685 | ENSMUSG00000037225 | 0.0206 | 0.4669 | 0.044120797 | ENSG00000146826 | ENSMUSG00000036948 | 0.0178 | 0.405 | 0.043950617 | ||||||||||||||||
| ENSG00000082458 | ENSMUSG00000000881 | 0.0033 | 0.3224 | 0.010235732 | ENSMUSG00000017291 | ENSG00000160551 | 0.0028 | 0.3118 | 0.008980115 | ENSG00000136051 | ENSMUSG00000034560 | 0.0156 | 0.4679 | 0.033340457 | ENSG00000146826 | ENSMUSG00000091964 | 0.0178 | 0.405 | 0.043950617 | ||||||||||||||||
| ENSG00000111052 | ENSMUSG00000019906 | 0.0021 | 0.3233 | 0.006495515 | ENSMUSG00000041115 | ENSG00000124313 | 0.0047 | 0.3121 | 0.015059276 | ENSG00000166925 | ENSMUSG00000029723 | 0.0862 | 0.4702 | 0.183326244 | ENSG00000204228 | ENSMUSG00000073422 | 0.0647 | 0.4059 | 0.159398867 | ||||||||||||||||
| ENSG00000162734 | ENSMUSG00000013698 | 0.0032 | 0.3234 | 0.009894867 | ENSMUSG00000079508 | ENSG00000184831 | 0.1011 | 0.3127 | 0.32331308 | ENSG00000090013 | ENSMUSG00000040466 | 0.0389 | 0.472 | 0.082415254 | ENSG00000126016 | ENSMUSG00000041688 | 0.0597 | 0.4066 | 0.146827349 | ||||||||||||||||
| ENSG00000067141 | ENSMUSG00000032340 | 0.0283 | 0.3236 | 0.087453646 | ENSMUSG00000041912 | ENSG00000182134 | 0.0465 | 0.3134 | 0.148372687 | ENSG00000125971 | ENSMUSG00000047459 | 0.0136 | 0.472 | 0.028813559 | ENSG00000166206 | ENSMUSG00000033676 | 0.0329 | 0.4092 | 0.080400782 | ||||||||||||||||
| ENSG00000111540 | ENSMUSG00000000711 | 0 | 0.3237 | 0 | ENSMUSG00000026991 | ENSG00000144283 | 0.0172 | 0.3142 | 0.054742202 | ENSG00000138674 | ENSMUSG00000035325 | 0.0432 | 0.4748 | 0.090985678 | ENSG00000143195 | ENSMUSG00000040612 | 0.0438 | 0.4103 | 0.106751158 | ||||||||||||||||
| ENSG00000187391 | ENSMUSG00000040003 | 0.0285 | 0.324 | 0.087962963 | ENSMUSG00000070372 | ENSG00000116489 | 0.0177 | 0.3145 | 0.056279809 | ENSG00000175198 | ENSMUSG00000041650 | 0.0574 | 0.4767 | 0.12041116 | ENSG00000063601 | ENSMUSG00000015214 | 0.0335 | 0.4107 | 0.081568055 | ||||||||||||||||
| ENSG00000108231 | ENSMUSG00000067242 | 0.0144 | 0.3247 | 0.04434863 | ENSMUSG00000039367 | ENSG00000176986 | 0.0397 | 0.3148 | 0.126111817 | ENSG00000154229 | ENSMUSG00000050965 | 0.0027 | 0.4785 | 0.005642633 | ENSG00000151012 | ENSMUSG00000027737 | 0.0572 | 0.4114 | 0.139037433 | ||||||||||||||||
| ENSG00000158882 | ENSMUSG00000005674 | 0.0108 | 0.3249 | 0.033240997 | ENSMUSG00000038014 | ENSG00000048828 | 0.0157 | 0.3191 | 0.049200877 | ENSG00000004660 | ENSMUSG00000020785 | 0.0317 | 0.4835 | 0.065563599 | ENSG00000163686 | ENSMUSG00000025277 | 0.0281 | 0.4117 | 0.068253583 | ||||||||||||||||
| ENSG00000136754 | ENSMUSG00000058835 | 0.0089 | 0.3252 | 0.027367774 | ENSMUSG00000071073 | ENSG00000204052 | 0.0058 | 0.3194 | 0.018159048 | ENSG00000023734 | ENSMUSG00000030224 | 0.0105 | 0.4861 | 0.021600494 | ENSG00000143575 | ENSMUSG00000027944 | 0.1024 | 0.4118 | 0.2486644 | ||||||||||||||||
| ENSG00000156482 | ENSMUSG00000062582 | 0 | 0.3258 | 0 | ENSMUSG00000063358 | ENSG00000100030 | 0.0053 | 0.3202 | 0.016552155 | ENSG00000117868 | ENSMUSG00000021171 | 0.039 | 0.487 | 0.080082136 | ENSG00000123360 | ENSMUSG00000022489 | 0.02 | 0.4128 | 0.048449612 | ||||||||||||||||
| ENSG00000173915 | ENSMUSG00000071528 | 0.0481 | 0.3267 | 0.147229875 | ENSMUSG00000042605 | ENSG00000204842 | 0.0564 | 0.3203 | 0.17608492 | ENSG00000170248 | ENSMUSG00000032504 | 0.0271 | 0.4888 | 0.055441899 | ENSG00000176454 | ENSMUSG00000027134 | 0.0346 | 0.4133 | 0.083716429 | ||||||||||||||||
| ENSG00000072518 | ENSMUSG00000024969 | 0.0079 | 0.327 | 0.024159021 | ENSMUSG00000028546 | ENSG00000162374 | 0.0104 | 0.3215 | 0.032348367 | ENSG00000064042 | ENSMUSG00000037736 | 0.0685 | 0.4899 | 0.139824454 | ENSG00000198739 | ENSMUSG00000042846 | 0.011 | 0.4146 | 0.026531597 | ||||||||||||||||
| ENSG00000115977 | ENSMUSG00000057230 | 0.0361 | 0.3276 | 0.11019536 | ENSMUSG00000045103 | ENSG00000198947 | 0.0439 | 0.3215 | 0.136547434 | ENSG00000116161 | ENSMUSG00000014226 | 0.0305 | 0.4899 | 0.062257604 | ENSG00000142676 | ENSMUSG00000058064 | 0.0052 | 0.4152 | 0.012524085 | ||||||||||||||||
| ENSG00000006125 | ENSMUSG00000035152 | 0.0005 | 0.3289 | 0.001520219 | ENSMUSG00000028552 | ENSG00000085832 | 0.0585 | 0.3215 | 0.181959565 | ENSG00000105197 | ENSMUSG00000003438 | 0.0421 | 0.4904 | 0.085848287 | ENSG00000182220 | ENSMUSG00000031007 | 0.034 | 0.4176 | 0.081417625 | ||||||||||||||||
| ENSG00000004059 | ENSMUSG00000020440 | 0 | 0.3294 | 0 | ENSMUSG00000021794 | ENSG00000148672 | 0.0118 | 0.3218 | 0.036668738 | ENSG00000167578 | ENSMUSG00000053291 | 0.0042 | 0.4904 | 0.008564437 | ENSG00000122406 | ENSMUSG00000058558 | 0.0075 | 0.4179 | 0.017946877 | ||||||||||||||||
| ENSG00000095139 | ENSMUSG00000032096 | 0.0046 | 0.33 | 0.013939394 | ENSMUSG00000061080 | ENSG00000185565 | 0.0699 | 0.3219 | 0.217148183 | ENSG00000107020 | ENSMUSG00000016495 | 0.0919 | 0.4952 | 0.185581583 | ENSG00000166862 | ENSMUSG00000019146 | 0.0051 | 0.4179 | 0.012203877 | ||||||||||||||||
| ENSG00000101367 | ENSMUSG00000027479 | 0.0164 | 0.3311 | 0.049531863 | ENSMUSG00000000881 | ENSG00000082458 | 0.0033 | 0.3224 | 0.010235732 | ENSG00000084774 | ENSMUSG00000013629 | 0.0236 | 0.4962 | 0.047561467 | ENSG00000137216 | ENSMUSG00000036026 | 0.0082 | 0.4203 | 0.019509874 | ||||||||||||||||
| ENSG00000167552 | ENSMUSG00000072235 | 0 | 0.332 | 0 | ENSMUSG00000029405 | ENSG00000138757 | 0.0074 | 0.3226 | 0.022938624 | ENSG00000136193 | ENSMUSG00000019124 | 0.0618 | 0.4963 | 0.124521459 | ENSG00000154146 | ENSMUSG00000053310 | 0.0175 | 0.4213 | 0.041538096 | ||||||||||||||||
| ENSG00000162188 | ENSMUSG00000071658 | 0 | 0.3355 | 0 | ENSMUSG00000019906 | ENSG00000111052 | 0.0021 | 0.3233 | 0.006495515 | ENSG00000131462 | ENSMUSG00000035198 | 0.0047 | 0.4967 | 0.009462452 | ENSG00000127483 | ENSMUSG00000028759 | 0.0512 | 0.4217 | 0.121413327 | ||||||||||||||||
| ENSG00000138814 | ENSMUSG00000028161 | 0.0018 | 0.336 | 0.005357143 | ENSMUSG00000032340 | ENSG00000067141 | 0.0283 | 0.3236 | 0.087453646 | ENSG00000115252 | ENSMUSG00000059173 | 0.038 | 0.4975 | 0.07638191 | ENSG00000108588 | ENSMUSG00000078622 | 0.009 | 0.4225 | 0.021301775 | ||||||||||||||||
| ENSG00000087258 | ENSMUSG00000031748 | 0.0086 | 0.3362 | 0.025580012 | ENSMUSG00000040003 | ENSG00000187391 | 0.0285 | 0.324 | 0.087962963 | ENSG00000138279 | ENSMUSG00000021814 | 0.0394 | 0.4984 | 0.07905297 | ENSG00000078725 | ENSMUSG00000028351 | 0.0052 | 0.4229 | 0.012296051 | ||||||||||||||||
| ENSG00000135940 | ENSMUSG00000079941 | 0.0998 | 0.3363 | 0.296758846 | ENSMUSG00000067242 | ENSG00000108231 | 0.0144 | 0.3247 | 0.04434863 | ENSG00000135655 | ENSMUSG00000020124 | 0.0111 | 0.4995 | 0.022222222 | ENSG00000066926 | ENSMUSG00000024588 | 0.063 | 0.4231 | 0.148900969 | ||||||||||||||||
| ENSG00000091164 | ENSMUSG00000024583 | 0.0064 | 0.337 | 0.018991098 | ENSMUSG00000058835 | ENSG00000136754 | 0.0089 | 0.3252 | 0.027367774 | ENSG00000060971 | ENSMUSG00000010651 | 0.0789 | 0.5 | 0.1578 | ENSG00000158987 | ENSMUSG00000037533 | 0.0425 | 0.4236 | 0.1003305 | ||||||||||||||||
| ENSG00000164924 | ENSMUSG00000022285 | 0.0019 | 0.3375 | 0.00562963 | ENSMUSG00000041530 | ENSG00000092847 | 0.0003 | 0.3254 | 0.000921942 | ENSG00000198677 | ENSMUSG00000033991 | 0.0856 | 0.5002 | 0.171131547 | ENSG00000157005 | ENSMUSG00000004366 | 0.0162 | 0.4283 | 0.037823955 | ||||||||||||||||
| ENSG00000169891 | ENSMUSG00000040855 | 0.0625 | 0.3379 | 0.184965966 | ENSMUSG00000071528 | ENSG00000173915 | 0.0481 | 0.3267 | 0.147229875 | ENSG00000112640 | ENSMUSG00000059409 | 0.0136 | 0.5043 | 0.026968075 | ENSG00000101349 | ENSMUSG00000039913 | 0.0291 | 0.4288 | 0.067863806 | ||||||||||||||||
| ENSG00000169564 | ENSMUSG00000051695 | 0 | 0.3383 | 0 | ENSMUSG00000024969 | ENSG00000072518 | 0.0079 | 0.327 | 0.024159021 | ENSG00000141252 | ENSMUSG00000017288 | 0.0158 | 0.5049 | 0.031293325 | ENSG00000001631 | ENSMUSG00000000600 | 0.0277 | 0.4293 | 0.064523643 | ||||||||||||||||
| ENSG00000134444 | ENSMUSG00000026319 | 0.0367 | 0.3411 | 0.107593081 | ENSMUSG00000057230 | ENSG00000115977 | 0.0361 | 0.3276 | 0.11019536 | ENSG00000117448 | ENSMUSG00000028692 | 0.0356 | 0.5052 | 0.070467142 | ENSG00000154380 | ENSMUSG00000022995 | 0.0542 | 0.4306 | 0.125870878 | ||||||||||||||||
| ENSG00000143653 | ENSMUSG00000038936 | 0.0721 | 0.3414 | 0.211189221 | ENSMUSG00000039478 | ENSG00000155970 | 0.038 | 0.3281 | 0.115818348 | ENSG00000071967 | ENSMUSG00000027015 | 0.1663 | 0.5054 | 0.3290463 | ENSG00000144895 | ENSMUSG00000027810 | 0.0331 | 0.4307 | 0.076851637 | ||||||||||||||||
| ENSG00000069345 | ENSMUSG00000031701 | 0.0023 | 0.3432 | 0.006701632 | ENSMUSG00000035152 | ENSG00000006125 | 0.0005 | 0.3289 | 0.001520219 | ENSG00000160007 | ENSMUSG00000058230 | 0.0109 | 0.5054 | 0.021567076 | ENSG00000059758 | ENSMUSG00000020015 | 0.0063 | 0.4341 | 0.014512785 | ||||||||||||||||
| ENSG00000115310 | ENSMUSG00000020458 | 0.1372 | 0.3439 | 0.398953184 | ENSMUSG00000020440 | ENSG00000004059 | 0 | 0.3294 | 0 | ENSG00000151835 | ENSMUSG00000048279 | 0.0663 | 0.5055 | 0.13115727 | ENSG00000151834 | ENSMUSG00000000560 | 0.0114 | 0.4344 | 0.026243094 | ||||||||||||||||
| ENSG00000182934 | ENSMUSG00000032042 | 0.0119 | 0.3461 | 0.034383126 | ENSMUSG00000025656 | ENSG00000131089 | 0.009 | 0.3313 | 0.027165711 | ENSG00000112855 | ENSMUSG00000019143 | 0.0912 | 0.5056 | 0.180379747 | ENSG00000121753 | ENSMUSG00000028782 | 0.0175 | 0.4358 | 0.040156035 | ||||||||||||||||
| ENSG00000108424 | ENSMUSG00000001440 | 0.0039 | 0.3466 | 0.011252164 | ENSMUSG00000072235 | ENSG00000167552 | 0 | 0.332 | 0 | ENSG00000137106 | ENSMUSG00000035637 | 0.0764 | 0.5064 | 0.150868878 | ENSG00000166225 | ENSMUSG00000020170 | 0.0216 | 0.4369 | 0.049439231 | ||||||||||||||||
| ENSG00000120251 | ENSMUSG00000033981 | 0.0083 | 0.3467 | 0.023940006 | ENSMUSG00000029169 | ENSG00000109606 | 0.0017 | 0.3342 | 0.005086774 | ENSG00000151552 | ENSMUSG00000015806 | 0.0319 | 0.51 | 0.06254902 | ENSG00000172375 | ENSMUSG00000032120 | 0.0469 | 0.4369 | 0.107347219 | ||||||||||||||||
| ENSG00000178127 | ENSMUSG00000024099 | 0.0358 | 0.3468 | 0.103229527 | ENSMUSG00000040990 | ENSG00000147010 | 0.0278 | 0.3349 | 0.083009854 | ENSG00000135046 | ENSMUSG00000024659 | 0.0626 | 0.5101 | 0.122721035 | ENSG00000114942 | ENSMUSG00000025967 | 0.0293 | 0.44 | 0.066590909 | ||||||||||||||||
| ENSG00000170624 | ENSMUSG00000020354 | 0.0273 | 0.347 | 0.078674352 | ENSMUSG00000028790 | ENSG00000121774 | 0.0274 | 0.3353 | 0.081717865 | ENSG00000073417 | ENSMUSG00000025584 | 0.1218 | 0.5117 | 0.238030096 | ENSG00000106636 | ENSMUSG00000002741 | 0.0354 | 0.4404 | 0.080381471 | ||||||||||||||||
| ENSG00000205302 | ENSMUSG00000034484 | 0.0098 | 0.3472 | 0.028225806 | ENSMUSG00000071658 | ENSG00000162188 | 0 | 0.3355 | 0 | ENSG00000197386 | ENSMUSG00000029104 | 0.049 | 0.5122 | 0.095665756 | ENSG00000115808 | ENSMUSG00000024077 | 0.0161 | 0.4409 | 0.036516217 | ||||||||||||||||
| ENSG00000159348 | ENSMUSG00000026456 | 0.0428 | 0.3474 | 0.123200921 | ENSMUSG00000036052 | ENSG00000137094 | 0.0038 | 0.3356 | 0.011323004 | ENSG00000211455 | ENSMUSG00000001630 | 0.0111 | 0.514 | 0.021595331 | ENSG00000113231 | ENSMUSG00000021684 | 0.0205 | 0.4411 | 0.046474722 | ||||||||||||||||
| ENSG00000198814 | ENSMUSG00000025059 | 0.0127 | 0.3477 | 0.036525741 | ENSMUSG00000028161 | ENSG00000138814 | 0.0018 | 0.336 | 0.005357143 | ENSG00000060971 | ENSMUSG00000036138 | 0.0832 | 0.5145 | 0.161710398 | ENSG00000179091 | ENSMUSG00000022551 | 0.0525 | 0.4416 | 0.11888587 | ||||||||||||||||
| ENSG00000134308 | ENSMUSG00000076432 | 0.0018 | 0.3482 | 0.005169443 | ENSMUSG00000021065 | ENSG00000033170 | 0.0163 | 0.3361 | 0.048497471 | ENSG00000002549 | ENSMUSG00000039682 | 0.0478 | 0.5163 | 0.092581832 | ENSG00000034693 | ENSMUSG00000019809 | 0.0293 | 0.4424 | 0.066229656 | ||||||||||||||||
| ENSG00000059915 | ENSMUSG00000037126 | 0.0407 | 0.3504 | 0.116152968 | ENSMUSG00000031748 | ENSG00000087258 | 0.0086 | 0.3362 | 0.025580012 | ENSG00000148341 | ENSMUSG00000026860 | 0.0149 | 0.5166 | 0.028842431 | ENSG00000072501 | ENSMUSG00000041133 | 0.0007 | 0.4427 | 0.001581206 | ||||||||||||||||
| ENSG00000119711 | ENSMUSG00000021238 | 0.0246 | 0.3506 | 0.070165431 | ENSMUSG00000022285 | ENSG00000164924 | 0.0019 | 0.3375 | 0.00562963 | ENSG00000120053 | ENSMUSG00000025190 | 0.0468 | 0.5173 | 0.090469747 | ENSG00000125037 | ENSMUSG00000030286 | 0.0018 | 0.4433 | 0.004060456 | ||||||||||||||||
| ENSG00000147224 | ENSMUSG00000031432 | 0 | 0.3507 | 0 | ENSMUSG00000026470 | ENSG00000135823 | 0.0305 | 0.3376 | 0.090343602 | ENSG00000125166 | ENSMUSG00000031672 | 0.025 | 0.5176 | 0.048299845 | ENSG00000173517 | ENSMUSG00000074305 | 0.0735 | 0.4451 | 0.165131431 | ||||||||||||||||
| ENSG00000173611 | ENSMUSG00000035236 | 0.0145 | 0.3509 | 0.041322314 | ENSMUSG00000051695 | ENSG00000169564 | 0 | 0.3383 | 0 | ENSG00000164904 | ENSMUSG00000053644 | 0.064 | 0.5181 | 0.123528276 | ENSG00000145864 | ENSMUSG00000007653 | 0.001 | 0.4457 | 0.002243662 | ||||||||||||||||
| ENSG00000124486 | ENSMUSG00000031010 | 0.0072 | 0.3512 | 0.020501139 | ENSMUSG00000038936 | ENSG00000143653 | 0.0721 | 0.3414 | 0.211189221 | ENSG00000133742 | ENSMUSG00000027556 | 0.134 | 0.5197 | 0.257841062 | ENSG00000184983 | ENSMUSG00000022450 | 0.0855 | 0.4458 | 0.19179004 | ||||||||||||||||
| ENSG00000134001 | ENSMUSG00000021116 | 0.0042 | 0.3513 | 0.011955594 | ENSMUSG00000031701 | ENSG00000069345 | 0.0023 | 0.3432 | 0.006701632 | ENSG00000109654 | ENSMUSG00000027993 | 0.0041 | 0.5205 | 0.007877041 | ENSG00000006715 | ENSMUSG00000041236 | 0.0122 | 0.4462 | 0.027341999 | ||||||||||||||||
| ENSG00000130227 | ENSMUSG00000022100 | 0.0027 | 0.3518 | 0.007674815 | ENSMUSG00000020458 | ENSG00000115310 | 0.1372 | 0.3439 | 0.398953184 | ENSG00000164961 | ENSMUSG00000022350 | 0.0204 | 0.5205 | 0.039193084 | ENSG00000152092 | ENSMUSG00000026587 | 0.0096 | 0.4484 | 0.021409456 | ||||||||||||||||
| ENSG00000117758 | ENSMUSG00000028879 | 0.0359 | 0.3522 | 0.101930721 | ENSMUSG00000063142 | ENSG00000156113 | 0.0074 | 0.344 | 0.021511628 | ENSG00000067992 | ENSMUSG00000035232 | 0.0151 | 0.5213 | 0.028966046 | ENSG00000159784 | ENSMUSG00000029861 | 0.0192 | 0.4492 | 0.042742654 | ||||||||||||||||
| ENSG00000162368 | ENSMUSG00000028719 | 0.0422 | 0.3535 | 0.119377652 | ENSMUSG00000027184 | ENSG00000135387 | 0.014 | 0.3442 | 0.040674027 | ENSG00000042493 | ENSMUSG00000056737 | 0.0341 | 0.522 | 0.06532567 | ENSG00000144290 | ENSMUSG00000026904 | 0.0105 | 0.4503 | 0.023317788 | ||||||||||||||||
| ENSG00000167535 | ENSMUSG00000003352 | 0.0036 | 0.3539 | 0.010172365 | ENSMUSG00000026755 | ENSG00000136950 | 0.0152 | 0.3442 | 0.044160372 | ENSG00000139641 | ENSMUSG00000025366 | 0.063 | 0.5224 | 0.120597243 | ENSG00000116560 | ENSMUSG00000028820 | 0.0364 | 0.4532 | 0.080317741 | ||||||||||||||||
| ENSG00000144036 | ENSMUSG00000033769 | 0.0046 | 0.3541 | 0.012990681 | ENSMUSG00000031311 | ENSG00000147140 | 0.004 | 0.345 | 0.011594203 | ENSG00000108465 | ENSMUSG00000018669 | 0.0586 | 0.5235 | 0.111938873 | ENSG00000198838 | ENSMUSG00000057378 | 0.021 | 0.4535 | 0.046306505 | ||||||||||||||||
| ENSG00000091428 | ENSMUSG00000049044 | 0.0134 | 0.3547 | 0.037778404 | ENSMUSG00000032328 | ENSG00000112697 | 0.0604 | 0.345 | 0.175072464 | ENSG00000185722 | ENSMUSG00000020790 | 0.0268 | 0.5246 | 0.051086542 | ENSG00000143702 | ENSMUSG00000057335 | 0.0508 | 0.4536 | 0.111992945 | ||||||||||||||||
| ENSG00000044574 | ENSMUSG00000026864 | 0.007 | 0.3553 | 0.019701661 | ENSMUSG00000020152 | ENSG00000138071 | 0 | 0.3455 | 0 | ENSG00000106125 | ENSMUSG00000038022 | 0.1503 | 0.5249 | 0.286340255 | ENSG00000113805 | ENSMUSG00000030075 | 0.0429 | 0.4542 | 0.094451783 | ||||||||||||||||
| ENSG00000144834 | ENSMUSG00000022658 | 0.0044 | 0.3563 | 0.012349144 | ENSMUSG00000027287 | ENSG00000092531 | 0.0654 | 0.3459 | 0.189071986 | ENSG00000151475 | ENSMUSG00000069041 | 0.0592 | 0.5261 | 0.112526136 | ENSG00000155974 | ENSMUSG00000034813 | 0.0188 | 0.4543 | 0.041382346 | ||||||||||||||||
| ENSG00000157500 | ENSMUSG00000040760 | 0.0078 | 0.3564 | 0.021885522 | ENSMUSG00000032042 | ENSG00000182934 | 0.0119 | 0.3461 | 0.034383126 | ENSG00000091483 | ENSMUSG00000026526 | 0.0361 | 0.5267 | 0.068539966 | ENSG00000104723 | ENSMUSG00000039530 | 0.0122 | 0.4546 | 0.02683678 | ||||||||||||||||
| ENSG00000173726 | ENSMUSG00000058779 | 0.0032 | 0.3571 | 0.008961075 | ENSMUSG00000018736 | ENSG00000166579 | 0.0192 | 0.3462 | 0.055459272 | ENSG00000197043 | ENSMUSG00000018340 | 0.0283 | 0.5271 | 0.053690002 | ENSG00000103404 | ENSMUSG00000063317 | 0.0436 | 0.4557 | 0.09567698 | ||||||||||||||||
| ENSG00000095637 | ENSMUSG00000025006 | 0.0595 | 0.3583 | 0.166061959 | ENSMUSG00000001440 | ENSG00000108424 | 0.0039 | 0.3466 | 0.011252164 | ENSG00000057608 | ENSMUSG00000021218 | 0.0178 | 0.529 | 0.033648393 | ENSG00000136805 | ENSMUSG00000045589 | 0.0409 | 0.4557 | 0.08975203 | ||||||||||||||||
| ENSG00000117450 | ENSMUSG00000048164 | 0.0478 | 0.3585 | 0.133333333 | ENSMUSG00000033981 | ENSG00000120251 | 0.0083 | 0.3467 | 0.023940006 | ENSG00000130340 | ENSMUSG00000002365 | 0.0452 | 0.5294 | 0.085379675 | ENSG00000136040 | ENSMUSG00000074785 | 0.0494 | 0.456 | 0.108333333 | ||||||||||||||||
| ENSG00000215301 | ENSMUSG00000000787 | 0.0069 | 0.3605 | 0.019140083 | ENSMUSG00000024099 | ENSG00000178127 | 0.0358 | 0.3468 | 0.103229527 | ENSG00000159176 | ENSMUSG00000026421 | 0.0024 | 0.5303 | 0.00452574 | ENSG00000077157 | ENSMUSG00000073557 | 0.0628 | 0.4561 | 0.137689103 | ||||||||||||||||
| ENSG00000025772 | ENSMUSG00000018322 | 0.0585 | 0.3608 | 0.16213969 | ENSMUSG00000025059 | ENSG00000198814 | 0.0127 | 0.3477 | 0.036525741 | ENSG00000184752 | ENSMUSG00000020022 | 0.0632 | 0.5313 | 0.11895351 | ENSG00000173208 | ENSMUSG00000055782 | 0.0302 | 0.4578 | 0.065967671 | ||||||||||||||||
| ENSG00000023228 | ENSMUSG00000025968 | 0.028 | 0.3609 | 0.077583818 | ENSMUSG00000031065 | ENSG00000102225 | 0.0124 | 0.3478 | 0.035652674 | ENSG00000127481 | ENSMUSG00000066036 | 0.0151 | 0.5326 | 0.028351483 | ENSG00000170049 | ENSMUSG00000018470 | 0.0345 | 0.4611 | 0.07482108 | ||||||||||||||||
| ENSG00000164587 | ENSMUSG00000024608 | 0 | 0.3621 | 0 | ENSMUSG00000076432 | ENSG00000134308 | 0.0018 | 0.3482 | 0.005169443 | ENSG00000009335 | ENSMUSG00000039000 | 0.0333 | 0.5335 | 0.062417994 | ENSG00000155506 | ENSMUSG00000037331 | 0.0524 | 0.4615 | 0.113542795 | ||||||||||||||||
| ENSG00000132305 | ENSMUSG00000052337 | 0.059 | 0.3624 | 0.162803532 | ENSMUSG00000033569 | ENSG00000135298 | 0.0078 | 0.3497 | 0.022304833 | ENSG00000100412 | ENSMUSG00000022477 | 0.0158 | 0.5347 | 0.02954928 | ENSG00000151532 | ENSMUSG00000024983 | 0.0235 | 0.4617 | 0.050898852 | ||||||||||||||||
| ENSG00000131236 | ENSMUSG00000028656 | 0.0285 | 0.3627 | 0.078577337 | ENSMUSG00000031343 | ENSG00000011677 | 0.0196 | 0.3498 | 0.056032018 | ENSG00000004478 | ENSMUSG00000030357 | 0.0525 | 0.5355 | 0.098039216 | ENSG00000134909 | ENSMUSG00000041444 | 0.0672 | 0.4619 | 0.145486036 | ||||||||||||||||
| ENSG00000128245 | ENSMUSG00000018965 | 0.0036 | 0.3628 | 0.009922822 | ENSMUSG00000037126 | ENSG00000059915 | 0.0407 | 0.3504 | 0.116152968 | ENSG00000134516 | ENSMUSG00000020143 | 0.0235 | 0.5363 | 0.043818758 | ENSG00000075413 | ENSMUSG00000007411 | 0.0149 | 0.462 | 0.032251082 | ||||||||||||||||
| ENSG00000114867 | ENSMUSG00000045983 | 0.0438 | 0.3633 | 0.120561519 | ENSMUSG00000031432 | ENSG00000147224 | 0 | 0.3507 | 0 | ENSG00000198130 | ENSMUSG00000041426 | 0.0968 | 0.5368 | 0.180327869 | ENSG00000170348 | ENSMUSG00000021248 | 0.0335 | 0.4625 | 0.072432432 | ||||||||||||||||
| ENSG00000119689 | ENSMUSG00000004789 | 0.0411 | 0.3643 | 0.112819105 | ENSMUSG00000031517 | ENSG00000150625 | 0.0052 | 0.3507 | 0.014827488 | ENSG00000197872 | ENSMUSG00000020589 | 0.0014 | 0.5381 | 0.002601747 | ENSG00000143183 | ENSMUSG00000052428 | 0 | 0.4633 | 0 | ||||||||||||||||
| ENSG00000174373 | ENSMUSG00000021027 | 0.0252 | 0.3646 | 0.06911684 | ENSMUSG00000035236 | ENSG00000173611 | 0.0145 | 0.3509 | 0.041322314 | ENSG00000009844 | ENSMUSG00000019868 | 0.0393 | 0.5383 | 0.073007617 | ENSG00000145247 | ENSMUSG00000029153 | 0.0783 | 0.4644 | 0.168604651 | ||||||||||||||||
| ENSG00000115866 | ENSMUSG00000026356 | 0.0183 | 0.3647 | 0.050178229 | ENSMUSG00000028456 | ENSG00000198722 | 0.0222 | 0.351 | 0.063247863 | ENSG00000136167 | ENSMUSG00000021998 | 0.0152 | 0.5394 | 0.028179459 | ENSG00000177034 | ENSMUSG00000021704 | 0.0536 | 0.4673 | 0.114701477 | ||||||||||||||||
| ENSG00000100591 | ENSMUSG00000021037 | 0.0249 | 0.3649 | 0.068237873 | ENSMUSG00000028879 | ENSG00000117758 | 0.0359 | 0.3522 | 0.101930721 | ENSG00000187231 | ENSMUSG00000042272 | 0.0086 | 0.5394 | 0.015943641 | ENSG00000169221 | ENSMUSG00000042492 | 0.0517 | 0.4677 | 0.110540945 | ||||||||||||||||
| ENSG00000088538 | ENSMUSG00000039716 | 0.0091 | 0.3653 | 0.024911032 | ENSMUSG00000055723 | ENSG00000133818 | 0 | 0.3523 | 0 | ENSG00000124198 | ENSMUSG00000074582 | 0.0219 | 0.5399 | 0.040563067 | ENSG00000156026 | ENSMUSG00000009647 | 0.0196 | 0.4678 | 0.041898247 | ||||||||||||||||
| ENSG00000169862 | ENSMUSG00000022240 | 0.0135 | 0.3657 | 0.036915505 | ENSMUSG00000052372 | ENSG00000169306 | 0.0066 | 0.3532 | 0.018686297 | ENSG00000140575 | ENSMUSG00000030536 | 0.0213 | 0.5419 | 0.039306145 | ENSG00000147123 | ENSMUSG00000031059 | 0.1345 | 0.4682 | 0.287270397 | ||||||||||||||||
| ENSG00000117450 | ENSMUSG00000028691 | 0.0285 | 0.3669 | 0.077677841 | ENSMUSG00000003352 | ENSG00000167535 | 0.0036 | 0.3539 | 0.010172365 | ENSG00000006747 | ENSMUSG00000002565 | 0.0465 | 0.5432 | 0.085603829 | ENSG00000198216 | ENSMUSG00000004110 | 0.0156 | 0.4688 | 0.033276451 | ||||||||||||||||
| ENSG00000197971 | ENSMUSG00000041607 | 0.0656 | 0.367 | 0.178746594 | ENSMUSG00000033769 | ENSG00000144036 | 0.0046 | 0.3541 | 0.012990681 | ENSG00000150054 | ENSMUSG00000057440 | 0.0356 | 0.5448 | 0.065345081 | ENSG00000107560 | ENSMUSG00000040022 | 0.0413 | 0.4692 | 0.088022165 | ||||||||||||||||
| ENSG00000115944 | ENSMUSG00000024248 | 0.0849 | 0.368 | 0.230706522 | ENSMUSG00000039735 | ENSG00000137942 | 0.0103 | 0.3546 | 0.029046813 | ENSG00000166947 | ENSMUSG00000023216 | 0.1688 | 0.5456 | 0.309384164 | ENSG00000145730 | ENSMUSG00000026335 | 0.0475 | 0.4706 | 0.100934977 | ||||||||||||||||
| ENSG00000147162 | ENSMUSG00000034160 | 0.0018 | 0.3689 | 0.004879371 | ENSMUSG00000049044 | ENSG00000091428 | 0.0134 | 0.3547 | 0.037778404 | ENSG00000125447 | ENSMUSG00000020740 | 0.0668 | 0.546 | 0.122344322 | ENSG00000140937 | ENSMUSG00000031673 | 0.0126 | 0.4711 | 0.026745914 | ||||||||||||||||
| ENSG00000175216 | ENSMUSG00000040549 | 0.0248 | 0.3696 | 0.067099567 | ENSMUSG00000026864 | ENSG00000044574 | 0.007 | 0.3553 | 0.019701661 | ENSG00000116266 | ENSMUSG00000027882 | 0.0413 | 0.5461 | 0.075627175 | ENSG00000110076 | ENSMUSG00000033768 | 0.0078 | 0.4736 | 0.016469595 | ||||||||||||||||
| ENSG00000205544 | ENSMUSG00000070394 | 0.0628 | 0.3697 | 0.16986746 | ENSMUSG00000025364 | ENSG00000170515 | 0.0048 | 0.356 | 0.013483146 | ENSG00000100003 | ENSMUSG00000003585 | 0.033 | 0.5466 | 0.060373216 | ENSG00000144369 | ENSMUSG00000048388 | 0.0587 | 0.4741 | 0.123813541 | ||||||||||||||||
| ENSG00000100462 | ENSMUSG00000023110 | 0.0095 | 0.3702 | 0.025661804 | ENSMUSG00000022658 | ENSG00000144834 | 0.0044 | 0.3563 | 0.012349144 | ENSG00000124207 | ENSMUSG00000002718 | 0.0048 | 0.5475 | 0.008767123 | ENSG00000147123 | ENSMUSG00000061633 | 0.2271 | 0.4749 | 0.478205938 | ||||||||||||||||
| ENSG00000168807 | ENSMUSG00000041308 | 0.0171 | 0.3711 | 0.046079224 | ENSMUSG00000058779 | ENSG00000173726 | 0.0032 | 0.3571 | 0.008961075 | ENSG00000037042 | ENSMUSG00000045007 | 0.0104 | 0.5504 | 0.018895349 | ENSG00000066044 | ENSMUSG00000040028 | 0.0069 | 0.475 | 0.014526316 | ||||||||||||||||
| ENSG00000111737 | ENSMUSG00000029518 | 0.0004 | 0.3712 | 0.001077586 | ENSMUSG00000025006 | ENSG00000095637 | 0.0595 | 0.3583 | 0.166061959 | ENSG00000124942 | ENSMUSG00000069833 | 0.0905 | 0.5535 | 0.163504968 | ENSG00000151726 | ENSMUSG00000018796 | 0.0807 | 0.4753 | 0.169787503 | ||||||||||||||||
| ENSG00000204463 | ENSMUSG00000024392 | 0.0294 | 0.3717 | 0.079096045 | ENSMUSG00000041058 | ENSG00000123124 | 0.045 | 0.36 | 0.125 | ENSG00000129515 | ENSMUSG00000005656 | 0.0022 | 0.5535 | 0.003974706 | ENSG00000164751 | ENSMUSG00000040374 | 0.0652 | 0.4758 | 0.137032367 | ||||||||||||||||
| ENSG00000198648 | ENSMUSG00000027030 | 0.021 | 0.3719 | 0.056466792 | ENSMUSG00000051578 | ENSG00000183723 | 0.0219 | 0.3606 | 0.060732113 | ENSG00000171202 | ENSMUSG00000030615 | 0.1702 | 0.5536 | 0.307442197 | ENSG00000134369 | ENSMUSG00000009418 | 0.0173 | 0.479 | 0.03611691 | ||||||||||||||||
| ENSG00000166887 | ENSMUSG00000027291 | 0.0144 | 0.372 | 0.038709677 | ENSMUSG00000025968 | ENSG00000023228 | 0.028 | 0.3609 | 0.077583818 | ENSG00000204351 | ENSMUSG00000040356 | 0.0339 | 0.5542 | 0.061169253 | ENSG00000163462 | ENSMUSG00000042766 | 0.0102 | 0.4807 | 0.021219056 | ||||||||||||||||
| ENSG00000035403 | ENSMUSG00000021823 | 0.0031 | 0.3723 | 0.008326618 | ENSMUSG00000030327 | ENSG00000089818 | 0.0351 | 0.3618 | 0.097014925 | ENSG00000132842 | ENSMUSG00000021686 | 0.0717 | 0.5554 | 0.129096147 | ENSG00000153933 | ENSMUSG00000000276 | 0.0456 | 0.4811 | 0.094782789 | ||||||||||||||||
| ENSG00000132142 | ENSMUSG00000020532 | 0.0113 | 0.3727 | 0.030319292 | ENSMUSG00000024608 | ENSG00000164587 | 0 | 0.3621 | 0 | ENSG00000149541 | ENSMUSG00000071649 | 0.0209 | 0.5588 | 0.037401575 | ENSG00000065665 | ENSMUSG00000025816 | 0.0005 | 0.4829 | 0.001035411 | ||||||||||||||||
| ENSG00000166128 | ENSMUSG00000036943 | 0.0064 | 0.3746 | 0.017084891 | ENSMUSG00000052337 | ENSG00000132305 | 0.059 | 0.3624 | 0.162803532 | ENSG00000161204 | ENSMUSG00000003234 | 0.0171 | 0.5595 | 0.030563003 | ENSG00000111110 | ENSMUSG00000034613 | 0.0158 | 0.4837 | 0.032664875 | ||||||||||||||||
| ENSG00000005249 | ENSMUSG00000002997 | 0.0156 | 0.3756 | 0.041533546 | ENSMUSG00000028656 | ENSG00000131236 | 0.0285 | 0.3627 | 0.078577337 | ENSG00000130638 | ENSMUSG00000016541 | 0.0988 | 0.5604 | 0.176302641 | ENSG00000075151 | ENSMUSG00000028760 | 0.0712 | 0.4842 | 0.147046675 | ||||||||||||||||
| ENSG00000153707 | ENSMUSG00000028399 | 0.0062 | 0.3758 | 0.016498137 | ENSMUSG00000018965 | ENSG00000128245 | 0.0036 | 0.3628 | 0.009922822 | ENSG00000065613 | ENSMUSG00000025060 | 0.0932 | 0.564 | 0.165248227 | ENSG00000100226 | ENSMUSG00000042535 | 0.0127 | 0.4842 | 0.026228831 | ||||||||||||||||
| ENSG00000112290 | ENSMUSG00000019831 | 0.0081 | 0.3758 | 0.021554018 | ENSMUSG00000045983 | ENSG00000114867 | 0.0438 | 0.3633 | 0.120561519 | ENSG00000068120 | ENSMUSG00000001755 | 0.0692 | 0.5641 | 0.122673285 | ENSG00000166924 | ENSMUSG00000045348 | 0.0396 | 0.4852 | 0.081615829 | ||||||||||||||||
| ENSG00000078668 | ENSMUSG00000008892 | 0.0082 | 0.376 | 0.021808511 | ENSMUSG00000021224 | ENSG00000133961 | 0.0337 | 0.3634 | 0.092735278 | ENSG00000108861 | ENSMUSG00000003518 | 0.0323 | 0.5669 | 0.056976539 | ENSG00000141376 | ENSMUSG00000059439 | 0.0151 | 0.4863 | 0.031050792 | ||||||||||||||||
| ENSG00000136631 | ENSMUSG00000015747 | 0.0145 | 0.3764 | 0.038522848 | ENSMUSG00000004789 | ENSG00000119689 | 0.0411 | 0.3643 | 0.112819105 | ENSG00000164830 | ENSMUSG00000022307 | 0.0919 | 0.5683 | 0.161710364 | ENSG00000117523 | ENSMUSG00000040225 | 0.0702 | 0.4869 | 0.144177449 | ||||||||||||||||
| ENSG00000129245 | ENSMUSG00000018765 | 0.0077 | 0.3774 | 0.020402756 | ENSMUSG00000000416 | ENSG00000077063 | 0.0782 | 0.3643 | 0.214658249 | ENSG00000115685 | ENSMUSG00000026275 | 0.0272 | 0.5688 | 0.047819972 | ENSG00000101365 | ENSMUSG00000027406 | 0.0311 | 0.4874 | 0.063807961 | ||||||||||||||||
| ENSG00000165795 | ENSMUSG00000004558 | 0.0227 | 0.3811 | 0.059564419 | ENSMUSG00000045114 | ENSG00000167371 | 0.1324 | 0.3646 | 0.363137685 | ENSG00000124181 | ENSMUSG00000016933 | 0.019 | 0.5689 | 0.033397785 | ENSG00000172915 | ENSMUSG00000027799 | 0.0125 | 0.4914 | 0.025437525 | ||||||||||||||||
| ENSG00000099942 | ENSMUSG00000006134 | 0.0159 | 0.3832 | 0.041492693 | ENSMUSG00000039716 | ENSG00000088538 | 0.0091 | 0.3653 | 0.024911032 | ENSG00000217930 | ENSMUSG00000014301 | 0.0228 | 0.57 | 0.04 | ENSG00000071054 | ENSMUSG00000026074 | 0.0072 | 0.4916 | 0.014646054 | ||||||||||||||||
| ENSG00000060237 | ENSMUSG00000045962 | 0.0761 | 0.3833 | 0.198539003 | ENSMUSG00000022240 | ENSG00000169862 | 0.0135 | 0.3657 | 0.036915505 | ENSG00000041982 | ENSMUSG00000028364 | 0.0676 | 0.5717 | 0.118243834 | ENSG00000152402 | ENSMUSG00000041624 | 0.0501 | 0.4942 | 0.101375961 | ||||||||||||||||
| ENSG00000211456 | ENSMUSG00000025240 | 0.0234 | 0.3834 | 0.061032864 | ENSMUSG00000020716 | ENSG00000196712 | 0.008 | 0.3658 | 0.021869874 | ENSG00000149313 | ENSMUSG00000025894 | 0.0417 | 0.5718 | 0.072927597 | ENSG00000132563 | ENSMUSG00000038555 | 0.0331 | 0.4986 | 0.06638588 | ||||||||||||||||
| ENSG00000136238 | ENSMUSG00000001847 | 0 | 0.3842 | 0 | ENSMUSG00000028691 | ENSG00000117450 | 0.0285 | 0.3669 | 0.077677841 | ENSG00000182199 | ENSMUSG00000025403 | 0.0316 | 0.5725 | 0.055196507 | ENSG00000040933 | ENSMUSG00000026113 | 0.0159 | 0.4998 | 0.031812725 | ||||||||||||||||
| ENSG00000132535 | ENSMUSG00000020886 | 0.0105 | 0.3844 | 0.027315297 | ENSMUSG00000041607 | ENSG00000197971 | 0.0656 | 0.367 | 0.178746594 | ENSG00000160752 | ENSMUSG00000059743 | 0.0855 | 0.5731 | 0.149188623 | ENSG00000100997 | ENSMUSG00000032046 | 0.0572 | 0.5006 | 0.114262885 | ||||||||||||||||
| ENSG00000119787 | ENSMUSG00000059811 | 0.0113 | 0.3845 | 0.029388817 | ENSMUSG00000027828 | ENSG00000114850 | 0.0051 | 0.3672 | 0.013888889 | ENSG00000151632 | ENSMUSG00000021214 | 0.1846 | 0.5738 | 0.321714883 | ENSG00000116641 | ENSMUSG00000028556 | 0.0084 | 0.5016 | 0.016746411 | ||||||||||||||||
| ENSG00000152578 | ENSMUSG00000025892 | 0.0015 | 0.3846 | 0.003900156 | ENSMUSG00000024248 | ENSG00000115944 | 0.0849 | 0.368 | 0.230706522 | ENSG00000182718 | ENSMUSG00000032231 | 0.0121 | 0.5743 | 0.021069128 | ENSG00000129219 | ENSMUSG00000020828 | 0.0526 | 0.502 | 0.104780876 | ||||||||||||||||
| ENSG00000067191 | ENSMUSG00000020882 | 0.0083 | 0.385 | 0.021558442 | ENSMUSG00000049184 | ENSG00000172733 | 0.0723 | 0.3682 | 0.196360674 | ENSG00000103043 | ENSMUSG00000010936 | 0.0265 | 0.5747 | 0.046111014 | ENSG00000011021 | ENSMUSG00000029016 | 0.0146 | 0.5021 | 0.029077873 | ||||||||||||||||
| ENSG00000006451 | ENSMUSG00000008859 | 0.0022 | 0.3853 | 0.005709836 | ENSMUSG00000030350 | ENSG00000111218 | 0.0067 | 0.3685 | 0.018181818 | ENSG00000189058 | ENSMUSG00000022548 | 0.1916 | 0.5761 | 0.332581149 | ENSG00000168546 | ENSMUSG00000022103 | 0.0279 | 0.5026 | 0.055511341 | ||||||||||||||||
| ENSG00000128872 | ENSMUSG00000032186 | 0.0264 | 0.3853 | 0.068518038 | ENSMUSG00000034160 | ENSG00000147162 | 0.0018 | 0.3689 | 0.004879371 | ENSG00000133943 | ENSMUSG00000021185 | 0.1624 | 0.5769 | 0.281504594 | ENSG00000106803 | ENSMUSG00000092116 | 0.0152 | 0.5028 | 0.030230708 | ||||||||||||||||
| ENSG00000149357 | ENSMUSG00000030842 | 0.0028 | 0.3854 | 0.007265179 | ENSMUSG00000040549 | ENSG00000175216 | 0.0248 | 0.3696 | 0.067099567 | ENSG00000164434 | ENSMUSG00000019874 | 0.0754 | 0.5773 | 0.130608003 | ENSG00000131771 | ENSMUSG00000061718 | 0.0664 | 0.5038 | 0.131798333 | ||||||||||||||||
| ENSG00000120265 | ENSMUSG00000019795 | 0.0209 | 0.3856 | 0.054201245 | ENSMUSG00000046434 | ENSG00000139675 | 0.0144 | 0.3699 | 0.03892944 | ENSG00000184277 | ENSMUSG00000078681 | 0.0789 | 0.5773 | 0.136670708 | ENSG00000101098 | ENSMUSG00000035226 | 0.0032 | 0.5045 | 0.006342914 | ||||||||||||||||
| ENSG00000158864 | ENSMUSG00000013593 | 0.0413 | 0.3856 | 0.107105809 | ENSMUSG00000038665 | ENSG00000157680 | 0.0188 | 0.3704 | 0.05075594 | ENSG00000158710 | ENSMUSG00000026547 | 0.0156 | 0.5787 | 0.026956973 | ENSG00000130779 | ENSMUSG00000049550 | 0.0538 | 0.5045 | 0.106640238 | ||||||||||||||||
| ENSG00000135250 | ENSMUSG00000062604 | 0.0393 | 0.3859 | 0.101839855 | ENSMUSG00000029518 | ENSG00000111737 | 0.0004 | 0.3712 | 0.001077586 | ENSG00000137992 | ENSMUSG00000000340 | 0.0654 | 0.5801 | 0.112739183 | ENSG00000151498 | ENSMUSG00000031969 | 0.0554 | 0.5049 | 0.109724698 | ||||||||||||||||
| ENSG00000167004 | ENSMUSG00000027248 | 0.0363 | 0.3862 | 0.09399275 | ENSMUSG00000027291 | ENSG00000166887 | 0.0144 | 0.372 | 0.038709677 | ENSG00000196305 | ENSMUSG00000037851 | 0.0493 | 0.5806 | 0.08491216 | ENSG00000181418 | ENSMUSG00000059213 | 0.1404 | 0.5051 | 0.277964759 | ||||||||||||||||
| ENSG00000138029 | ENSMUSG00000059447 | 0.0464 | 0.3881 | 0.119556815 | ENSMUSG00000036943 | ENSG00000166128 | 0.0064 | 0.3746 | 0.017084891 | ENSG00000115839 | ENSMUSG00000036104 | 0.036 | 0.5807 | 0.061994145 | ENSG00000213593 | ENSMUSG00000050043 | 0.0362 | 0.5052 | 0.07165479 | ||||||||||||||||
| ENSG00000108100 | ENSMUSG00000024286 | 0.0094 | 0.3884 | 0.024201854 | ENSMUSG00000002997 | ENSG00000005249 | 0.0156 | 0.3756 | 0.041533546 | ENSG00000083720 | ENSMUSG00000022186 | 0.0404 | 0.5812 | 0.069511356 | ENSG00000132718 | ENSMUSG00000068923 | 0.0212 | 0.5057 | 0.041922088 | ||||||||||||||||
| ENSG00000044115 | ENSMUSG00000037815 | 0.0031 | 0.3889 | 0.007971201 | ENSMUSG00000028399 | ENSG00000153707 | 0.0062 | 0.3758 | 0.016498137 | ENSG00000147526 | ENSMUSG00000065954 | 0.1486 | 0.5821 | 0.255282597 | ENSG00000148219 | ENSMUSG00000028373 | 0.0121 | 0.5057 | 0.02392723 | ||||||||||||||||
| ENSG00000196465 | ENSMUSG00000039824 | 0.0403 | 0.3892 | 0.103545735 | ENSMUSG00000019831 | ENSG00000112290 | 0.0081 | 0.3758 | 0.021554018 | ENSG00000076201 | ENSMUSG00000036057 | 0.0438 | 0.5844 | 0.074948665 | ENSG00000072778 | ENSMUSG00000018574 | 0.0917 | 0.5059 | 0.181261119 | ||||||||||||||||
| ENSG00000100220 | ENSMUSG00000001783 | 0.0028 | 0.3895 | 0.007188703 | ENSMUSG00000008892 | ENSG00000078668 | 0.0082 | 0.376 | 0.021808511 | ENSG00000100884 | ENSMUSG00000022212 | 0.0097 | 0.5853 | 0.016572698 | ENSG00000110881 | ENSMUSG00000023017 | 0.0083 | 0.5065 | 0.016386969 | ||||||||||||||||
| ENSG00000158195 | ENSMUSG00000028868 | 0.0454 | 0.3902 | 0.116350589 | ENSMUSG00000015747 | ENSG00000136631 | 0.0145 | 0.3764 | 0.038522848 | ENSG00000077279 | ENSMUSG00000031285 | 0.0025 | 0.5857 | 0.004268397 | ENSG00000185008 | ENSMUSG00000052516 | 0.0207 | 0.5067 | 0.040852575 | ||||||||||||||||
| ENSG00000155189 | ENSMUSG00000031467 | 0.1094 | 0.3902 | 0.280369042 | ENSMUSG00000033940 | ENSG00000254999 | 0.0111 | 0.3769 | 0.029450783 | ENSG00000134121 | ENSMUSG00000030077 | 0.0814 | 0.5857 | 0.138978999 | ENSG00000162951 | ENSMUSG00000060780 | 0.0138 | 0.5088 | 0.027122642 | ||||||||||||||||
| ENSG00000116459 | ENSMUSG00000000563 | 0.1043 | 0.3906 | 0.26702509 | ENSMUSG00000018765 | ENSG00000129245 | 0.0077 | 0.3774 | 0.020402756 | ENSG00000101400 | ENSMUSG00000027488 | 0.038 | 0.5859 | 0.064857484 | ENSG00000072134 | ENSMUSG00000001036 | 0.0586 | 0.5089 | 0.115150324 | ||||||||||||||||
| ENSG00000089006 | ENSMUSG00000027423 | 0.0081 | 0.3916 | 0.020684372 | ENSMUSG00000025221 | ENSG00000120049 | 0.0146 | 0.3794 | 0.038481813 | ENSG00000101997 | ENSMUSG00000031143 | 0.0641 | 0.5878 | 0.109050698 | ENSG00000157445 | ENSMUSG00000021991 | 0.0093 | 0.5089 | 0.01827471 | ||||||||||||||||
| ENSG00000127824 | ENSMUSG00000026202 | 0 | 0.3919 | 0 | ENSMUSG00000027201 | ENSG00000104177 | 0.0422 | 0.3802 | 0.110994214 | ENSG00000138182 | ENSMUSG00000024795 | 0.1617 | 0.5879 | 0.275046777 | ENSG00000164588 | ENSMUSG00000021730 | 0.0183 | 0.5097 | 0.035903473 | ||||||||||||||||
| ENSG00000065609 | ENSMUSG00000033419 | 0.0399 | 0.3921 | 0.101759755 | ENSMUSG00000025240 | ENSG00000211456 | 0.0234 | 0.3834 | 0.061032864 | ENSG00000138095 | ENSMUSG00000024120 | 0.156 | 0.5881 | 0.26526101 | ENSG00000006116 | ENSMUSG00000066189 | 0.0057 | 0.5112 | 0.011150235 | ||||||||||||||||
| ENSG00000062485 | ENSMUSG00000046934 | 0.0526 | 0.3928 | 0.133910387 | ENSMUSG00000001847 | ENSG00000136238 | 0 | 0.3842 | 0 | ENSG00000158186 | ENSMUSG00000032470 | 0.0157 | 0.5888 | 0.026664402 | ENSG00000198822 | ENSMUSG00000003974 | 0.0152 | 0.5129 | 0.029635407 | ||||||||||||||||
| ENSG00000150672 | ENSMUSG00000052572 | 0.0308 | 0.393 | 0.078371501 | ENSMUSG00000020886 | ENSG00000132535 | 0.0105 | 0.3844 | 0.027315297 | ENSG00000148468 | ENSMUSG00000050530 | 0.0397 | 0.589 | 0.067402377 | ENSG00000011566 | ENSMUSG00000024242 | 0.0198 | 0.5131 | 0.038588969 | ||||||||||||||||
| ENSG00000064651 | ENSMUSG00000024597 | 0.0284 | 0.3933 | 0.072209509 | ENSMUSG00000025892 | ENSG00000152578 | 0.0015 | 0.3846 | 0.003900156 | ENSG00000154027 | ENSMUSG00000039058 | 0.0445 | 0.5896 | 0.075474898 | ENSG00000169851 | ENSMUSG00000029108 | 0.0183 | 0.5132 | 0.035658613 | ||||||||||||||||
| ENSG00000079819 | ENSMUSG00000019978 | 0.0932 | 0.3933 | 0.236969235 | ENSMUSG00000031119 | ENSG00000076716 | 0.0241 | 0.3846 | 0.062662507 | ENSG00000105875 | ENSMUSG00000058486 | 0.0387 | 0.5904 | 0.06554878 | ENSG00000135525 | ENSMUSG00000019996 | 0.1154 | 0.5141 | 0.224469947 | ||||||||||||||||
| ENSG00000213619 | ENSMUSG00000005510 | 0.0726 | 0.3937 | 0.184404369 | ENSMUSG00000020882 | ENSG00000067191 | 0.0083 | 0.385 | 0.021558442 | ENSG00000171246 | ENSMUSG00000025582 | 0.0162 | 0.5904 | 0.027439024 | ENSG00000196090 | ENSMUSG00000053141 | 0.0067 | 0.5148 | 0.013014763 | ||||||||||||||||
| ENSG00000008056 | ENSMUSG00000037217 | 0.022 | 0.3939 | 0.055851739 | ENSMUSG00000008859 | ENSG00000006451 | 0.0022 | 0.3853 | 0.005709836 | ENSG00000090861 | ENSMUSG00000031960 | 0.0222 | 0.5914 | 0.037538045 | ENSG00000109016 | ENSMUSG00000042569 | 0.1285 | 0.5157 | 0.249175877 | ||||||||||||||||
| ENSG00000165283 | ENSMUSG00000028455 | 0.0282 | 0.3944 | 0.071501014 | ENSMUSG00000032186 | ENSG00000128872 | 0.0264 | 0.3853 | 0.068518038 | ENSG00000204590 | ENSMUSG00000024429 | 0.0221 | 0.5949 | 0.037149101 | ENSG00000080854 | ENSMUSG00000034275 | 0.0189 | 0.5163 | 0.036606624 | ||||||||||||||||
| ENSG00000014641 | ENSMUSG00000020321 | 0.0188 | 0.3945 | 0.04765526 | ENSMUSG00000051615 | ENSG00000125249 | 0.0022 | 0.3854 | 0.005708355 | ENSG00000187134 | ENSMUSG00000021214 | 0.1729 | 0.5957 | 0.290246769 | ENSG00000155849 | ENSMUSG00000041112 | 0.0019 | 0.5164 | 0.003679318 | ||||||||||||||||
| ENSG00000164209 | ENSMUSG00000024259 | 0.0572 | 0.3956 | 0.144590495 | ENSMUSG00000030842 | ENSG00000149357 | 0.0028 | 0.3854 | 0.007265179 | ENSG00000157483 | ENSMUSG00000032220 | 0.0194 | 0.5965 | 0.032523051 | ENSG00000114646 | ENSMUSG00000032482 | 0.0705 | 0.517 | 0.136363636 | ||||||||||||||||
| ENSG00000022355 | ENSMUSG00000010803 | 0.0061 | 0.3962 | 0.015396265 | ENSMUSG00000013593 | ENSG00000158864 | 0.0413 | 0.3856 | 0.107105809 | ENSG00000002746 | ENSMUSG00000021301 | 0.0629 | 0.5967 | 0.105413105 | ENSG00000106803 | ENSMUSG00000067657 | 0.0315 | 0.5188 | 0.060717039 | ||||||||||||||||
| ENSG00000114302 | ENSMUSG00000032601 | 0.0903 | 0.3968 | 0.227570565 | ENSMUSG00000062604 | ENSG00000135250 | 0.0393 | 0.3859 | 0.101839855 | ENSG00000142657 | ENSMUSG00000028961 | 0.0313 | 0.5972 | 0.052411253 | ENSG00000033867 | ENSMUSG00000021733 | 0.0318 | 0.5198 | 0.061177376 | ||||||||||||||||
| ENSG00000070367 | ENSMUSG00000061244 | 0.0076 | 0.3972 | 0.019133938 | ENSMUSG00000059447 | ENSG00000138029 | 0.0464 | 0.3881 | 0.119556815 | ENSG00000075142 | ENSMUSG00000003161 | 0.0196 | 0.5977 | 0.032792371 | ENSG00000168539 | ENSMUSG00000032773 | 0.0048 | 0.5228 | 0.009181331 | ||||||||||||||||
| ENSG00000033122 | ENSMUSG00000028176 | 0.022 | 0.3974 | 0.055359839 | ENSMUSG00000053769 | ENSG00000163155 | 0.0934 | 0.3882 | 0.24059763 | ENSG00000170584 | ENSMUSG00000020328 | 0.0028 | 0.5989 | 0.004675238 | ENSG00000177600 | ENSMUSG00000091018 | 0.0088 | 0.5233 | 0.016816358 | ||||||||||||||||
| ENSG00000117592 | ENSMUSG00000026701 | 0.0671 | 0.3975 | 0.168805031 | ENSMUSG00000024286 | ENSG00000108100 | 0.0094 | 0.3884 | 0.024201854 | ENSG00000147471 | ENSMUSG00000031485 | 0.0775 | 0.6006 | 0.129037629 | ENSG00000106803 | ENSMUSG00000053317 | 0.005 | 0.5236 | 0.009549274 | ||||||||||||||||
| ENSG00000115484 | ENSMUSG00000007739 | 0.0158 | 0.3979 | 0.039708469 | ENSMUSG00000052539 | ENSG00000081026 | 0.0724 | 0.3884 | 0.186405767 | ENSG00000181789 | ENSMUSG00000030058 | 0.0126 | 0.6014 | 0.020951114 | ENSG00000138623 | ENSMUSG00000038264 | 0.0528 | 0.5243 | 0.100705703 | ||||||||||||||||
| ENSG00000187118 | ENSMUSG00000039163 | 0.0515 | 0.398 | 0.129396985 | ENSMUSG00000037815 | ENSG00000044115 | 0.0031 | 0.3889 | 0.007971201 | ENSG00000134202 | ENSMUSG00000004032 | 0.0661 | 0.6021 | 0.109782428 | ENSG00000142655 | ENSMUSG00000028975 | 0.0338 | 0.5244 | 0.064454615 | ||||||||||||||||
| ENSG00000122218 | ENSMUSG00000026553 | 0.0074 | 0.3982 | 0.018583626 | ENSMUSG00000005069 | ENSG00000139197 | 0.0316 | 0.3889 | 0.081254821 | ENSG00000112304 | ENSMUSG00000006717 | 0.0991 | 0.6022 | 0.164563268 | ENSG00000111726 | ENSMUSG00000030282 | 0.0322 | 0.5266 | 0.061146981 | ||||||||||||||||
| ENSG00000077380 | ENSMUSG00000027012 | 0.0099 | 0.3989 | 0.02481825 | ENSMUSG00000039824 | ENSG00000196465 | 0.0403 | 0.3892 | 0.103545735 | ENSG00000111731 | ENSMUSG00000030279 | 0.0187 | 0.6047 | 0.030924425 | ENSG00000176204 | ENSMUSG00000052581 | 0.0228 | 0.5273 | 0.043239143 | ||||||||||||||||
| ENSG00000152413 | ENSMUSG00000007617 | 0.0111 | 0.3996 | 0.027777778 | ENSMUSG00000070880 | ENSG00000128683 | 0.0126 | 0.3894 | 0.032357473 | ENSG00000164111 | ENSMUSG00000027712 | 0.0374 | 0.6054 | 0.061777337 | ENSG00000110917 | ENSMUSG00000048578 | 0.0453 | 0.5295 | 0.085552408 | ||||||||||||||||
| ENSG00000135720 | ENSMUSG00000035770 | 0.0146 | 0.3996 | 0.036536537 | ENSMUSG00000001783 | ENSG00000100220 | 0.0028 | 0.3895 | 0.007188703 | ENSG00000119705 | ENSMUSG00000021040 | 0.1906 | 0.6069 | 0.314055034 | ENSG00000162522 | ENSMUSG00000050390 | 0.1061 | 0.5301 | 0.200150915 | ||||||||||||||||
| ENSG00000198561 | ENSMUSG00000034101 | 0.0116 | 0.3998 | 0.029014507 | ENSMUSG00000028868 | ENSG00000158195 | 0.0454 | 0.3902 | 0.116350589 | ENSG00000143870 | ENSMUSG00000020571 | 0.0276 | 0.6075 | 0.045432099 | ENSG00000165478 | ENSMUSG00000046240 | 0.0246 | 0.5304 | 0.04638009 | ||||||||||||||||
| ENSG00000165914 | ENSMUSG00000033530 | 0.0157 | 0.3999 | 0.039259815 | ENSMUSG00000031467 | ENSG00000155189 | 0.1094 | 0.3902 | 0.280369042 | ENSG00000187134 | ENSMUSG00000033715 | 0.2061 | 0.6091 | 0.338368084 | ENSG00000114331 | ENSMUSG00000049076 | 0.0178 | 0.5313 | 0.033502729 | ||||||||||||||||
| ENSG00000169764 | ENSMUSG00000001891 | 0.0053 | 0.4002 | 0.013243378 | ENSMUSG00000000563 | ENSG00000116459 | 0.1043 | 0.3906 | 0.26702509 | ENSG00000122778 | ENSMUSG00000063455 | 0.1824 | 0.6095 | 0.29926169 | ENSG00000176049 | ENSMUSG00000024502 | 0.0098 | 0.5322 | 0.01841413 | ||||||||||||||||
| ENSG00000167699 | ENSMUSG00000017286 | 0.0448 | 0.4002 | 0.111944028 | ENSMUSG00000042425 | ENSG00000147234 | 0.0659 | 0.3914 | 0.168369954 | ENSG00000137486 | ENSMUSG00000018909 | 0.0064 | 0.6096 | 0.010498688 | ENSG00000137843 | ENSMUSG00000074923 | 0.0359 | 0.5326 | 0.067405182 | ||||||||||||||||
| ENSG00000100554 | ENSMUSG00000021114 | 0.0077 | 0.401 | 0.019201995 | ENSMUSG00000026202 | ENSG00000127824 | 0 | 0.3919 | 0 | ENSG00000138363 | ENSMUSG00000026192 | 0.0476 | 0.6118 | 0.077803204 | ENSG00000154822 | ENSMUSG00000038910 | 0.0109 | 0.5346 | 0.020389076 | ||||||||||||||||
| ENSG00000165434 | ENSMUSG00000030729 | 0.0314 | 0.4018 | 0.078148333 | ENSMUSG00000022377 | ENSG00000153317 | 0.0188 | 0.392 | 0.047959184 | ENSG00000116977 | ENSMUSG00000057554 | 0.1274 | 0.6123 | 0.208067941 | ENSG00000119777 | ENSMUSG00000038828 | 0.1035 | 0.5362 | 0.193024991 | ||||||||||||||||
| ENSG00000158560 | ENSMUSG00000029757 | 0.0096 | 0.4019 | 0.023886539 | ENSMUSG00000033419 | ENSG00000065609 | 0.0399 | 0.3921 | 0.101759755 | ENSG00000120899 | ENSMUSG00000059456 | 0.0221 | 0.6142 | 0.035981765 | ENSG00000143862 | ENSMUSG00000026426 | 0.0023 | 0.5373 | 0.004280663 | ||||||||||||||||
| ENSG00000143776 | ENSMUSG00000026490 | 0.0196 | 0.402 | 0.048756219 | ENSMUSG00000029212 | ENSG00000163288 | 0.0087 | 0.3924 | 0.022171254 | ENSG00000108561 | ENSMUSG00000018446 | 0.0713 | 0.6147 | 0.115991541 | ENSG00000102218 | ENSMUSG00000060090 | 0.0605 | 0.5378 | 0.112495351 | ||||||||||||||||
| ENSG00000140600 | ENSMUSG00000030638 | 0.0748 | 0.4024 | 0.185884692 | ENSMUSG00000052572 | ENSG00000150672 | 0.0308 | 0.393 | 0.078371501 | ENSG00000112186 | ENSMUSG00000021373 | 0.0584 | 0.6148 | 0.094990241 | ENSG00000163214 | ENSMUSG00000035051 | 0.0781 | 0.5384 | 0.145059435 | ||||||||||||||||
| ENSG00000135439 | ENSMUSG00000025422 | 0.024 | 0.4026 | 0.059612519 | ENSMUSG00000024597 | ENSG00000064651 | 0.0284 | 0.3933 | 0.072209509 | ENSG00000128564 | ENSMUSG00000037428 | 0.0702 | 0.615 | 0.114146341 | ENSG00000185477 | ENSMUSG00000045441 | 0.3075 | 0.5403 | 0.569128262 | ||||||||||||||||
| ENSG00000133318 | ENSMUSG00000024758 | 0.2087 | 0.4031 | 0.517737534 | ENSMUSG00000019978 | ENSG00000079819 | 0.0932 | 0.3933 | 0.236969235 | ENSG00000213366 | ENSMUSG00000040562 | 0.1219 | 0.6156 | 0.198018194 | ENSG00000072195 | ENSMUSG00000026207 | 0.0441 | 0.5432 | 0.081185567 | ||||||||||||||||
| ENSG00000116871 | ENSMUSG00000028849 | 0.0688 | 0.4033 | 0.170592611 | ENSMUSG00000005510 | ENSG00000213619 | 0.0726 | 0.3937 | 0.184404369 | ENSG00000128609 | ENSMUSG00000023089 | 0.0967 | 0.616 | 0.156980519 | ENSG00000155858 | ENSMUSG00000044847 | 0.0664 | 0.5433 | 0.122216087 | ||||||||||||||||
| ENSG00000165280 | ENSMUSG00000028452 | 0.0004 | 0.4043 | 0.000989364 | ENSMUSG00000037217 | ENSG00000008056 | 0.022 | 0.3939 | 0.055851739 | ENSG00000165566 | ENSMUSG00000021986 | 0.1048 | 0.6182 | 0.169524426 | ENSG00000105767 | ENSMUSG00000054793 | 0.0083 | 0.5453 | 0.015220979 | ||||||||||||||||
| ENSG00000100764 | ENSMUSG00000021178 | 0.0004 | 0.4044 | 0.00098912 | ENSMUSG00000028455 | ENSG00000165283 | 0.0282 | 0.3944 | 0.071501014 | ENSG00000151632 | ENSMUSG00000033715 | 0.2123 | 0.6193 | 0.342806394 | ENSG00000092051 | ENSMUSG00000022208 | 0.0194 | 0.546 | 0.035531136 | ||||||||||||||||
| ENSG00000106771 | ENSMUSG00000055296 | 0.0396 | 0.4049 | 0.097801926 | ENSMUSG00000020321 | ENSG00000014641 | 0.0188 | 0.3945 | 0.04765526 | ENSG00000186197 | ENSMUSG00000039309 | 0.1439 | 0.6197 | 0.232209133 | ENSG00000186468 | ENSMUSG00000073559 | 0.0203 | 0.5483 | 0.037023527 | ||||||||||||||||
| ENSG00000141367 | ENSMUSG00000047126 | 0.0005 | 0.4052 | 0.001233959 | ENSMUSG00000037259 | ENSG00000089091 | 0.1332 | 0.3953 | 0.336959271 | ENSG00000152137 | ENSMUSG00000041548 | 0.0286 | 0.6201 | 0.046121593 | ENSG00000004777 | ENSMUSG00000036882 | 0.0433 | 0.5488 | 0.078899417 | ||||||||||||||||
| ENSG00000196405 | ENSMUSG00000021262 | 0.0297 | 0.4054 | 0.073260977 | ENSMUSG00000024259 | ENSG00000164209 | 0.0572 | 0.3956 | 0.144590495 | ENSG00000156453 | ENSMUSG00000051375 | 0.0329 | 0.6215 | 0.052936444 | ENSG00000130956 | ENSMUSG00000021476 | 0.0857 | 0.549 | 0.156102004 | ||||||||||||||||
| ENSG00000205339 | ENSMUSG00000066232 | 0.0026 | 0.4057 | 0.006408676 | ENSMUSG00000018733 | ENSG00000108733 | 0.0646 | 0.3957 | 0.163254991 | ENSG00000157020 | ENSMUSG00000030298 | 0.0141 | 0.6215 | 0.022687047 | ENSG00000036530 | ENSMUSG00000021259 | 0.0236 | 0.5504 | 0.042877907 | ||||||||||||||||
| ENSG00000179915 | ENSMUSG00000024109 | 0.0051 | 0.4068 | 0.012536873 | ENSMUSG00000010803 | ENSG00000022355 | 0.0061 | 0.3962 | 0.015396265 | ENSG00000103415 | ENSMUSG00000004070 | 0.0548 | 0.6257 | 0.087581908 | ENSG00000121057 | ENSMUSG00000018428 | 0.19 | 0.5512 | 0.344702467 | ||||||||||||||||
| ENSG00000069329 | ENSMUSG00000031696 | 0.003 | 0.407 | 0.007371007 | ENSMUSG00000032601 | ENSG00000114302 | 0.0903 | 0.3968 | 0.227570565 | ENSG00000198721 | ENSMUSG00000021417 | 0.1493 | 0.6274 | 0.23796621 | ENSG00000154127 | ENSMUSG00000032020 | 0.0115 | 0.5518 | 0.020840884 | ||||||||||||||||
| ENSG00000147853 | ENSMUSG00000024782 | 0.0381 | 0.4072 | 0.093565815 | ENSMUSG00000061244 | ENSG00000070367 | 0.0076 | 0.3972 | 0.019133938 | ENSG00000066557 | ENSMUSG00000063052 | 0.0996 | 0.6275 | 0.1587251 | ENSG00000134115 | ENSMUSG00000030092 | 0.054 | 0.5524 | 0.09775525 | ||||||||||||||||
| ENSG00000156508 | ENSMUSG00000037742 | 0.001 | 0.4075 | 0.002453988 | ENSMUSG00000028176 | ENSG00000033122 | 0.022 | 0.3974 | 0.055359839 | ENSG00000100401 | ENSMUSG00000022391 | 0.0574 | 0.629 | 0.091255962 | ENSG00000164199 | ENSMUSG00000069170 | 0.1138 | 0.5551 | 0.205008107 | ||||||||||||||||
| ENSG00000067900 | ENSMUSG00000024290 | 0.015 | 0.4076 | 0.036800785 | ENSMUSG00000007739 | ENSG00000115484 | 0.0158 | 0.3979 | 0.039708469 | ENSG00000213088 | ENSMUSG00000037872 | 0.3473 | 0.6352 | 0.546756927 | ENSG00000162300 | ENSMUSG00000024792 | 0.0386 | 0.5566 | 0.069349623 | ||||||||||||||||
| ENSG00000106299 | ENSMUSG00000029684 | 0.0195 | 0.4078 | 0.047817558 | ENSMUSG00000026553 | ENSG00000122218 | 0.0074 | 0.3982 | 0.018583626 | ENSG00000068903 | ENSMUSG00000015149 | 0.0681 | 0.6381 | 0.106723084 | ENSG00000107816 | ENSMUSG00000035342 | 0.0376 | 0.5571 | 0.067492371 | ||||||||||||||||
| ENSG00000149269 | ENSMUSG00000030774 | 0.0323 | 0.4086 | 0.079050416 | ENSMUSG00000021124 | ENSG00000100568 | 0.0378 | 0.3987 | 0.094808126 | ENSG00000075539 | ENSMUSG00000070733 | 0.0266 | 0.6381 | 0.041686256 | ENSG00000197969 | ENSMUSG00000046230 | 0.0856 | 0.5577 | 0.153487538 | ||||||||||||||||
| ENSG00000104490 | ENSMUSG00000051359 | 0 | 0.4093 | 0 | ENSMUSG00000027012 | ENSG00000077380 | 0.0099 | 0.3989 | 0.02481825 | ENSG00000102606 | ENSMUSG00000031511 | 0.0266 | 0.6396 | 0.041588493 | ENSG00000124222 | ENSMUSG00000027522 | 0.0322 | 0.5613 | 0.057366827 | ||||||||||||||||
| ENSG00000047849 | ENSMUSG00000032479 | 0.2619 | 0.4097 | 0.63924823 | ENSMUSG00000007617 | ENSG00000152413 | 0.0111 | 0.3996 | 0.027777778 | ENSG00000162688 | ENSMUSG00000033400 | 0.0409 | 0.6401 | 0.063896266 | ENSG00000123908 | ENSMUSG00000036698 | 0.0036 | 0.5629 | 0.006395452 | ||||||||||||||||
| ENSG00000134287 | ENSMUSG00000051853 | 0 | 0.41 | 0 | ENSMUSG00000035770 | ENSG00000135720 | 0.0146 | 0.3996 | 0.036536537 | ENSG00000125633 | ENSMUSG00000026339 | 0.0403 | 0.6421 | 0.06276281 | ENSG00000161267 | ENSMUSG00000046598 | 0.0722 | 0.5645 | 0.127900797 | ||||||||||||||||
| ENSG00000198513 | ENSMUSG00000021066 | 0.0165 | 0.4112 | 0.040126459 | ENSMUSG00000034101 | ENSG00000198561 | 0.0116 | 0.3998 | 0.029014507 | ENSG00000105223 | ENSMUSG00000003363 | 0.0301 | 0.644 | 0.04673913 | ENSG00000177600 | ENSMUSG00000025508 | 0.0044 | 0.5649 | 0.007788989 | ||||||||||||||||
| ENSG00000087470 | ENSMUSG00000022789 | 0.006 | 0.4124 | 0.014548982 | ENSMUSG00000032740 | ENSG00000115355 | 0.0254 | 0.3998 | 0.063531766 | ENSG00000177963 | ENSMUSG00000025485 | 0.0647 | 0.6446 | 0.100372324 | ENSG00000149658 | ENSMUSG00000038848 | 0.0309 | 0.565 | 0.054690265 | ||||||||||||||||
| ENSG00000142507 | ENSMUSG00000018286 | 0.0335 | 0.4127 | 0.081172765 | ENSMUSG00000033530 | ENSG00000165914 | 0.0157 | 0.3999 | 0.039259815 | ENSG00000175220 | ENSMUSG00000027247 | 0.0176 | 0.6471 | 0.027198269 | ENSG00000161647 | ENSMUSG00000052373 | 0.0448 | 0.5651 | 0.079278004 | ||||||||||||||||
| ENSG00000169398 | ENSMUSG00000022607 | 0.0128 | 0.4129 | 0.031000242 | ENSMUSG00000045302 | ENSG00000138073 | 0.0646 | 0.4006 | 0.161258113 | ENSG00000115520 | ENSMUSG00000025981 | 0.0822 | 0.6477 | 0.126910607 | ENSG00000175497 | ENSMUSG00000036815 | 0.0625 | 0.5655 | 0.110521662 | ||||||||||||||||
| ENSG00000136152 | ENSMUSG00000034893 | 0.0202 | 0.4131 | 0.048898572 | ENSMUSG00000046671 | ENSG00000117640 | 0.0202 | 0.4009 | 0.05038663 | ENSG00000144908 | ENSMUSG00000030088 | 0.0395 | 0.6484 | 0.060919186 | ENSG00000163637 | ENSMUSG00000030020 | 0.0209 | 0.5697 | 0.036685975 | ||||||||||||||||
| ENSG00000145819 | ENSMUSG00000036452 | 0.0141 | 0.4133 | 0.034115654 | ENSMUSG00000021114 | ENSG00000100554 | 0.0077 | 0.401 | 0.019201995 | ENSG00000148175 | ENSMUSG00000026880 | 0.0726 | 0.6487 | 0.11191614 | ENSG00000119523 | ENSMUSG00000039740 | 0.1188 | 0.5698 | 0.208494208 | ||||||||||||||||
| ENSG00000080371 | ENSMUSG00000020132 | 0.0108 | 0.4142 | 0.02607436 | ENSMUSG00000029757 | ENSG00000158560 | 0.0096 | 0.4019 | 0.023886539 | ENSG00000116266 | ENSMUSG00000071640 | 0.0789 | 0.649 | 0.121571649 | ENSG00000136531 | ENSMUSG00000075318 | 0.0103 | 0.5714 | 0.018025901 | ||||||||||||||||
| ENSG00000185046 | ENSMUSG00000058589 | 0.0421 | 0.4145 | 0.101568154 | ENSMUSG00000026490 | ENSG00000143776 | 0.0196 | 0.402 | 0.048756219 | ENSG00000050405 | ENSMUSG00000023022 | 0.1394 | 0.6493 | 0.214692746 | ENSG00000149256 | ENSMUSG00000048078 | 0.0126 | 0.5715 | 0.022047244 | ||||||||||||||||
| ENSG00000137076 | ENSMUSG00000028465 | 0.0063 | 0.4148 | 0.015188042 | ENSMUSG00000034653 | ENSG00000047188 | 0.0135 | 0.4024 | 0.033548708 | ENSG00000156931 | ENSMUSG00000033653 | 0.0376 | 0.6509 | 0.05776617 | ENSG00000103018 | ENSMUSG00000031924 | 0.1457 | 0.5729 | 0.254320126 | ||||||||||||||||
| ENSG00000075711 | ENSMUSG00000022770 | 0.0213 | 0.4152 | 0.051300578 | ENSMUSG00000024777 | ENSG00000068971 | 0.0054 | 0.4026 | 0.013412817 | ENSG00000102189 | ENSMUSG00000036499 | 0.0842 | 0.6518 | 0.12918073 | ENSG00000150995 | ENSMUSG00000030102 | 0.0053 | 0.576 | 0.009201389 | ||||||||||||||||
| ENSG00000163344 | ENSMUSG00000027952 | 0.0907 | 0.4165 | 0.217767107 | ENSMUSG00000025422 | ENSG00000135439 | 0.024 | 0.4026 | 0.059612519 | ENSG00000132932 | ENSMUSG00000021983 | 0.0291 | 0.6525 | 0.044597701 | ENSG00000128524 | ENSMUSG00000004285 | 0.0072 | 0.5789 | 0.012437381 | ||||||||||||||||
| ENSG00000138138 | ENSMUSG00000013662 | 0 | 0.417 | 0 | ENSMUSG00000024758 | ENSG00000133318 | 0.2087 | 0.4031 | 0.517737534 | ENSG00000177156 | ENSMUSG00000025503 | 0.0273 | 0.6536 | 0.041768666 | ENSG00000176406 | ENSMUSG00000037386 | 0.0526 | 0.579 | 0.090846287 | ||||||||||||||||
| ENSG00000109107 | ENSMUSG00000017390 | 0.0106 | 0.417 | 0.025419664 | ENSMUSG00000028849 | ENSG00000116871 | 0.0688 | 0.4033 | 0.170592611 | ENSG00000121053 | ENSMUSG00000052234 | 0.0558 | 0.6549 | 0.085203848 | ENSG00000147403 | ENSMUSG00000008682 | 0 | 0.5791 | 0 | ||||||||||||||||
| ENSG00000213281 | ENSMUSG00000027852 | 0.0024 | 0.4173 | 0.005751258 | ENSMUSG00000047976 | ENSG00000111262 | 0.0069 | 0.4036 | 0.017096135 | ENSG00000163346 | ENSMUSG00000042613 | 0.1718 | 0.6559 | 0.261930172 | ENSG00000147403 | ENSMUSG00000058443 | 0 | 0.5791 | 0 | ||||||||||||||||
| ENSG00000196220 | ENSMUSG00000030257 | 0.006 | 0.4174 | 0.014374701 | ENSMUSG00000028452 | ENSG00000165280 | 0.0004 | 0.4043 | 0.000989364 | ENSG00000168589 | ENSMUSG00000034467 | 0.0097 | 0.6574 | 0.014755096 | ENSG00000130724 | ENSMUSG00000033916 | 0.0019 | 0.5797 | 0.003277557 | ||||||||||||||||
| ENSG00000176105 | ENSMUSG00000014932 | 0.0173 | 0.4174 | 0.041447053 | ENSMUSG00000021178 | ENSG00000100764 | 0.0004 | 0.4044 | 0.00098912 | ENSG00000147459 | ENSMUSG00000044447 | 0.0305 | 0.6582 | 0.046338499 | ENSG00000151846 | ENSMUSG00000046173 | 0.1326 | 0.5842 | 0.226977063 | ||||||||||||||||
| ENSG00000106028 | ENSMUSG00000029911 | 0.1028 | 0.4174 | 0.246286536 | ENSMUSG00000036948 | ENSG00000146826 | 0.0178 | 0.405 | 0.043950617 | ENSG00000171564 | ENSMUSG00000033831 | 0.0916 | 0.6627 | 0.138222423 | ENSG00000005108 | ENSMUSG00000032625 | 0.0467 | 0.5845 | 0.079897348 | ||||||||||||||||
| ENSG00000109846 | ENSMUSG00000032060 | 0.0106 | 0.4183 | 0.025340665 | ENSMUSG00000047126 | ENSG00000141367 | 0.0005 | 0.4052 | 0.001233959 | ENSG00000182054 | ENSMUSG00000030541 | 0.0218 | 0.6628 | 0.032890766 | ENSG00000089693 | ENSMUSG00000030120 | 0.0125 | 0.5845 | 0.0213858 | ||||||||||||||||
| ENSG00000106278 | ENSMUSG00000068748 | 0.097 | 0.4203 | 0.230787533 | ENSMUSG00000021262 | ENSG00000196405 | 0.0297 | 0.4054 | 0.073260977 | ENSG00000132612 | ENSMUSG00000031913 | 0.0041 | 0.6648 | 0.006167268 | ENSG00000184368 | ENSMUSG00000041020 | 0.1463 | 0.5857 | 0.24978658 | ||||||||||||||||
| ENSG00000075239 | ENSMUSG00000032047 | 0.086 | 0.4204 | 0.204567079 | ENSMUSG00000020486 | ENSG00000108387 | 0.0369 | 0.4058 | 0.090931493 | ENSG00000100266 | ENSMUSG00000016664 | 0.0308 | 0.6693 | 0.046018228 | ENSG00000198055 | ENSMUSG00000074886 | 0.0199 | 0.5865 | 0.033930094 | ||||||||||||||||
| ENSG00000068796 | ENSMUSG00000021693 | 0.0106 | 0.4211 | 0.025172168 | ENSMUSG00000073422 | ENSG00000204228 | 0.0647 | 0.4059 | 0.159398867 | ENSG00000075624 | ENSMUSG00000029580 | 0.0022 | 0.6694 | 0.003286525 | ENSG00000099968 | ENSMUSG00000009112 | 0.1496 | 0.5871 | 0.254811787 | ||||||||||||||||
| ENSG00000108774 | ENSMUSG00000019173 | 0.0063 | 0.4212 | 0.014957265 | ENSMUSG00000041688 | ENSG00000126016 | 0.0597 | 0.4066 | 0.146827349 | ENSG00000214253 | ENSMUSG00000019054 | 0.0202 | 0.6698 | 0.030158256 | ENSG00000078070 | ENSMUSG00000027709 | 0.0855 | 0.5883 | 0.145334013 | ||||||||||||||||
| ENSG00000175166 | ENSMUSG00000006998 | 0.0063 | 0.4226 | 0.014907714 | ENSMUSG00000024109 | ENSG00000179915 | 0.0051 | 0.4068 | 0.012536873 | ENSG00000104267 | ENSMUSG00000027562 | 0.1117 | 0.6729 | 0.165997919 | ENSG00000138650 | ENSMUSG00000049100 | 0.0161 | 0.589 | 0.027334465 | ||||||||||||||||
| ENSG00000163618 | ENSMUSG00000054423 | 0.0065 | 0.4227 | 0.015377336 | ENSMUSG00000007440 | ENSG00000243232 | 0.0356 | 0.4068 | 0.087512291 | ENSG00000115415 | ENSMUSG00000026104 | 0.0336 | 0.6756 | 0.04973357 | ENSG00000196230 | ENSMUSG00000001525 | 0 | 0.5904 | 0 | ||||||||||||||||
| ENSG00000028528 | ENSMUSG00000032382 | 0.0249 | 0.4228 | 0.058893094 | ENSMUSG00000031696 | ENSG00000069329 | 0.003 | 0.407 | 0.007371007 | ENSG00000137168 | ENSMUSG00000024007 | 0.0084 | 0.678 | 0.012389381 | ENSG00000141404 | ENSMUSG00000024524 | 0.0402 | 0.5909 | 0.068031816 | ||||||||||||||||
| ENSG00000101557 | ENSMUSG00000047879 | 0.0158 | 0.4229 | 0.037361078 | ENSMUSG00000037742 | ENSG00000156508 | 0.001 | 0.4075 | 0.002453988 | ENSG00000187134 | ENSMUSG00000021211 | 0.2088 | 0.6793 | 0.307375239 | ENSG00000169282 | ENSMUSG00000027827 | 0.0842 | 0.5909 | 0.1424945 | ||||||||||||||||
| ENSG00000111674 | ENSMUSG00000004267 | 0.0083 | 0.4236 | 0.019593957 | ENSMUSG00000029684 | ENSG00000106299 | 0.0195 | 0.4078 | 0.047817558 | ENSG00000187134 | ENSMUSG00000021213 | 0.2224 | 0.6807 | 0.326722492 | ENSG00000119401 | ENSMUSG00000051675 | 0.0208 | 0.5942 | 0.035005049 | ||||||||||||||||
| ENSG00000156467 | ENSMUSG00000021520 | 0.068 | 0.4249 | 0.160037656 | ENSMUSG00000030774 | ENSG00000149269 | 0.0323 | 0.4086 | 0.079050416 | ENSG00000065054 | ENSMUSG00000002504 | 0.0395 | 0.6812 | 0.057985907 | ENSG00000147642 | ENSMUSG00000022340 | 0.0788 | 0.5951 | 0.13241472 | ||||||||||||||||
| ENSG00000118705 | ENSMUSG00000027642 | 0.0421 | 0.425 | 0.099058824 | ENSMUSG00000033676 | ENSG00000166206 | 0.0329 | 0.4092 | 0.080400782 | ENSG00000102178 | ENSMUSG00000015290 | 0.0519 | 0.6824 | 0.0760551 | ENSG00000151846 | ENSMUSG00000051732 | 0.1459 | 0.5952 | 0.245127688 | ||||||||||||||||
| ENSG00000171497 | ENSMUSG00000027804 | 0.0294 | 0.4253 | 0.069127675 | ENSMUSG00000051359 | ENSG00000104490 | 0 | 0.4093 | 0 | ENSG00000187134 | ENSMUSG00000054757 | 0.1593 | 0.6827 | 0.233338216 | ENSG00000153820 | ENSMUSG00000026163 | 0.1635 | 0.5961 | 0.274282838 | ||||||||||||||||
| ENSG00000115419 | ENSMUSG00000026103 | 0.0522 | 0.4254 | 0.122708039 | ENSMUSG00000051853 | ENSG00000134287 | 0 | 0.41 | 0 | ENSG00000215642 | ENSMUSG00000059854 | 0.1013 | 0.6834 | 0.148229441 | ENSG00000138669 | ENSMUSG00000029334 | 0.0153 | 0.599 | 0.025542571 | ||||||||||||||||
| ENSG00000133812 | ENSMUSG00000038371 | 0.03 | 0.4257 | 0.070472163 | ENSMUSG00000040612 | ENSG00000143195 | 0.0438 | 0.4103 | 0.106751158 | ENSG00000102882 | ENSMUSG00000063065 | 0.0124 | 0.6846 | 0.018112767 | ENSG00000119013 | ENSMUSG00000026032 | 0.1361 | 0.6023 | 0.225967126 | ||||||||||||||||
| ENSG00000087274 | ENSMUSG00000029106 | 0.0394 | 0.4258 | 0.092531705 | ENSMUSG00000015214 | ENSG00000063601 | 0.0335 | 0.4107 | 0.081568055 | ENSG00000139116 | ENSMUSG00000022629 | 0.0291 | 0.6856 | 0.042444574 | ENSG00000111664 | ENSMUSG00000023439 | 0.0141 | 0.6037 | 0.023355972 | ||||||||||||||||
| ENSG00000151729 | ENSMUSG00000031633 | 0.0207 | 0.4262 | 0.048568747 | ENSMUSG00000027737 | ENSG00000151012 | 0.0572 | 0.4114 | 0.139037433 | ENSG00000213366 | ENSMUSG00000004035 | 0.0893 | 0.6867 | 0.130042231 | ENSG00000103502 | ENSMUSG00000030682 | 0.0165 | 0.6038 | 0.027326929 | ||||||||||||||||
| ENSG00000170606 | ENSMUSG00000020361 | 0.02 | 0.4267 | 0.046871338 | ENSMUSG00000025277 | ENSG00000163686 | 0.0281 | 0.4117 | 0.068253583 | ENSG00000213366 | ENSMUSG00000068762 | 0.1386 | 0.6914 | 0.200462829 | ENSG00000184408 | ENSMUSG00000060882 | 0.0044 | 0.6038 | 0.007287181 | ||||||||||||||||
| ENSG00000186298 | ENSMUSG00000004455 | 0.0014 | 0.4273 | 0.003276387 | ENSMUSG00000027944 | ENSG00000143575 | 0.1024 | 0.4118 | 0.2486644 | ENSG00000138768 | ENSMUSG00000029407 | 0.0749 | 0.6955 | 0.107692308 | ENSG00000061918 | ENSMUSG00000028005 | 0.0053 | 0.6052 | 0.008757436 | ||||||||||||||||
| ENSG00000090621 | ENSMUSG00000011257 | 0.0109 | 0.4273 | 0.02550901 | ENSMUSG00000022789 | ENSG00000087470 | 0.006 | 0.4124 | 0.014548982 | ENSG00000013364 | ENSMUSG00000030681 | 0.045 | 0.696 | 0.064655172 | ENSG00000196497 | ENSMUSG00000002319 | 0.1035 | 0.6073 | 0.170426478 | ||||||||||||||||
| ENSG00000122359 | ENSMUSG00000021866 | 0.0339 | 0.4273 | 0.079335362 | ENSMUSG00000018286 | ENSG00000142507 | 0.0335 | 0.4127 | 0.081172765 | ENSG00000198721 | ENSMUSG00000021416 | 0.2112 | 0.6961 | 0.303404683 | ENSG00000118733 | ENSMUSG00000027965 | 0.024 | 0.6078 | 0.039486673 | ||||||||||||||||
| ENSG00000136888 | ENSMUSG00000039105 | 0.0258 | 0.4274 | 0.060364998 | ENSMUSG00000022489 | ENSG00000123360 | 0.02 | 0.4128 | 0.048449612 | ENSG00000076356 | ENSMUSG00000026640 | 0.0167 | 0.6968 | 0.023966705 | ENSG00000177272 | ENSMUSG00000047959 | 0.0124 | 0.608 | 0.020394737 | ||||||||||||||||
| ENSG00000100934 | ENSMUSG00000020986 | 0.0048 | 0.4276 | 0.011225444 | ENSMUSG00000036452 | ENSG00000145819 | 0.0141 | 0.4133 | 0.034115654 | ENSG00000151632 | ENSMUSG00000054757 | 0.1659 | 0.6987 | 0.237440962 | ENSG00000165802 | ENSMUSG00000006476 | 0.0261 | 0.609 | 0.042857143 | ||||||||||||||||
| ENSG00000155008 | ENSMUSG00000025525 | 0.1334 | 0.4276 | 0.311973807 | ENSMUSG00000027134 | ENSG00000176454 | 0.0346 | 0.4133 | 0.083716429 | ENSG00000108828 | ENSMUSG00000034993 | 0.0463 | 0.6988 | 0.06625644 | ENSG00000071553 | ENSMUSG00000019087 | 0.0777 | 0.6121 | 0.126940042 | ||||||||||||||||
| ENSG00000084764 | ENSMUSG00000029166 | 0.0032 | 0.428 | 0.007476636 | ENSMUSG00000058589 | ENSG00000185046 | 0.0421 | 0.4145 | 0.101568154 | ENSG00000173599 | ENSMUSG00000024892 | 0.0151 | 0.6994 | 0.021589934 | ENSG00000168481 | ENSMUSG00000033595 | 0.0156 | 0.6148 | 0.025374105 | ||||||||||||||||
| ENSG00000161203 | ENSMUSG00000022841 | 0 | 0.4282 | 0 | ENSMUSG00000042846 | ENSG00000198739 | 0.011 | 0.4146 | 0.026531597 | ENSG00000151632 | ENSMUSG00000021211 | 0.2109 | 0.6996 | 0.301457976 | ENSG00000006071 | ENSMUSG00000040136 | 0.0229 | 0.6173 | 0.037097035 | ||||||||||||||||
| ENSG00000070808 | ENSMUSG00000024617 | 0.0009 | 0.4282 | 0.002101822 | ENSMUSG00000022770 | ENSG00000075711 | 0.0213 | 0.4152 | 0.051300578 | ENSG00000151632 | ENSMUSG00000021213 | 0.225 | 0.7002 | 0.321336761 | ENSG00000157654 | ENSMUSG00000089945 | 0.1013 | 0.6212 | 0.163071475 | ||||||||||||||||
| ENSG00000166974 | ENSMUSG00000024277 | 0.0125 | 0.4283 | 0.029185151 | ENSMUSG00000013662 | ENSG00000138138 | 0 | 0.417 | 0 | ENSG00000125844 | ENSMUSG00000027422 | 0.0765 | 0.704 | 0.108664773 | ENSG00000166689 | ENSMUSG00000045659 | 0.0611 | 0.622 | 0.098231511 | ||||||||||||||||
| ENSG00000107679 | ENSMUSG00000040268 | 0.0515 | 0.4284 | 0.120214753 | ENSMUSG00000017390 | ENSG00000109107 | 0.0106 | 0.417 | 0.025419664 | ENSG00000171557 | ENSMUSG00000033860 | 0.0947 | 0.7046 | 0.134402498 | ENSG00000093100 | ENSMUSG00000003178 | 0.0549 | 0.6221 | 0.088249478 | ||||||||||||||||
| ENSG00000034713 | ENSMUSG00000031950 | 0 | 0.4287 | 0 | ENSMUSG00000030257 | ENSG00000196220 | 0.006 | 0.4174 | 0.014374701 | ENSG00000187134 | ENSMUSG00000025955 | 0.1779 | 0.706 | 0.251983003 | ENSG00000103035 | ENSMUSG00000039067 | 0.0166 | 0.6257 | 0.026530286 | ||||||||||||||||
| ENSG00000111667 | ENSMUSG00000038429 | 0.0077 | 0.4289 | 0.017952903 | ENSMUSG00000014932 | ENSG00000176105 | 0.0173 | 0.4174 | 0.041447053 | ENSG00000151632 | ENSMUSG00000071551 | 0.1806 | 0.7062 | 0.255734919 | ENSG00000148719 | ENSMUSG00000020109 | 0.0273 | 0.6303 | 0.043312708 | ||||||||||||||||
| ENSG00000065427 | ENSMUSG00000031948 | 0.0397 | 0.4295 | 0.092433062 | ENSMUSG00000029911 | ENSG00000106028 | 0.1028 | 0.4174 | 0.246286536 | ENSG00000076864 | ENSMUSG00000041351 | 0.0149 | 0.7072 | 0.021069005 | ENSG00000105894 | ENSMUSG00000029838 | 0.0134 | 0.6304 | 0.021256345 | ||||||||||||||||
| ENSG00000149084 | ENSMUSG00000027195 | 0.1113 | 0.4298 | 0.258957655 | ENSMUSG00000023036 | ENSG00000240764 | 0.0377 | 0.4175 | 0.090299401 | ENSG00000107957 | ENSMUSG00000053617 | 0.0443 | 0.7077 | 0.062597146 | ENSG00000108298 | ENSMUSG00000017404 | 0 | 0.6307 | 0 | ||||||||||||||||
| ENSG00000120438 | ENSMUSG00000068039 | 0.0182 | 0.4307 | 0.042256791 | ENSMUSG00000031007 | ENSG00000182220 | 0.034 | 0.4176 | 0.081417625 | ENSG00000183735 | ENSMUSG00000020115 | 0.0307 | 0.7089 | 0.043306531 | ENSG00000008988 | ENSMUSG00000028234 | 0 | 0.6308 | 0 | ||||||||||||||||
| ENSG00000091140 | ENSMUSG00000020664 | 0.0243 | 0.4311 | 0.056367432 | ENSMUSG00000058558 | ENSG00000122406 | 0.0075 | 0.4179 | 0.017946877 | ENSG00000133315 | ENSMUSG00000036278 | 0.0735 | 0.7103 | 0.103477404 | ENSG00000188089 | ENSMUSG00000050211 | 0.1452 | 0.6314 | 0.229965157 | ||||||||||||||||
| ENSG00000092964 | ENSMUSG00000022048 | 0.0056 | 0.4314 | 0.012980992 | ENSMUSG00000071415 | ENSG00000125691 | 0 | 0.4184 | 0 | ENSG00000112294 | ENSMUSG00000035936 | 0.0867 | 0.7109 | 0.121958081 | ENSG00000134313 | ENSMUSG00000036333 | 0.0297 | 0.6315 | 0.047030879 | ||||||||||||||||
| ENSG00000173210 | ENSMUSG00000032735 | 0.0091 | 0.432 | 0.021064815 | ENSMUSG00000001175 | ENSG00000198668 | 0 | 0.4188 | 0 | ENSG00000151632 | ENSMUSG00000025955 | 0.1894 | 0.7113 | 0.266273021 | ENSG00000196361 | ENSMUSG00000003410 | 0.0023 | 0.6316 | 0.003641545 | ||||||||||||||||
| ENSG00000197157 | ENSMUSG00000001424 | 0.0122 | 0.4321 | 0.028234205 | ENSMUSG00000036026 | ENSG00000137216 | 0.0082 | 0.4203 | 0.019509874 | ENSG00000204128 | ENSMUSG00000026227 | 0.1803 | 0.7135 | 0.252697968 | ENSG00000141452 | ENSMUSG00000024410 | 0.0255 | 0.6335 | 0.040252565 | ||||||||||||||||
| ENSG00000008277 | ENSMUSG00000040537 | 0.0444 | 0.4321 | 0.102753992 | ENSMUSG00000068748 | ENSG00000106278 | 0.097 | 0.4203 | 0.230787533 | ENSG00000096696 | ENSMUSG00000054889 | 0.0232 | 0.7152 | 0.032438479 | ENSG00000176749 | ENSMUSG00000048895 | 0.0067 | 0.6357 | 0.010539563 | ||||||||||||||||
| ENSG00000165219 | ENSMUSG00000026867 | 0.0102 | 0.4329 | 0.023562024 | ENSMUSG00000032047 | ENSG00000075239 | 0.086 | 0.4204 | 0.204567079 | ENSG00000005381 | ENSMUSG00000009350 | 0.0775 | 0.7161 | 0.108225108 | ENSG00000131323 | ENSMUSG00000021277 | 0.017 | 0.6361 | 0.026725358 | ||||||||||||||||
| ENSG00000151929 | ENSMUSG00000030847 | 0.0798 | 0.4333 | 0.184168013 | ENSMUSG00000021693 | ENSG00000068796 | 0.0106 | 0.4211 | 0.025172168 | ENSG00000187134 | ENSMUSG00000071551 | 0.1769 | 0.7165 | 0.246894627 | ENSG00000073670 | ENSMUSG00000020926 | 0.0305 | 0.6376 | 0.047835634 | ||||||||||||||||
| ENSG00000074319 | ENSMUSG00000014402 | 0.0255 | 0.4334 | 0.058837102 | ENSMUSG00000019173 | ENSG00000108774 | 0.0063 | 0.4212 | 0.014957265 | ENSG00000161618 | ENSMUSG00000007833 | 0.1104 | 0.7177 | 0.153824718 | ENSG00000117114 | ENSMUSG00000028184 | 0.0213 | 0.6402 | 0.033270853 | ||||||||||||||||
| ENSG00000100387 | ENSMUSG00000022400 | 0 | 0.4341 | 0 | ENSMUSG00000053310 | ENSG00000154146 | 0.0175 | 0.4213 | 0.041538096 | ENSG00000105254 | ENSMUSG00000006095 | 0.0558 | 0.7186 | 0.077650988 | ENSG00000135824 | ENSMUSG00000042671 | 0.03 | 0.6404 | 0.046845721 | ||||||||||||||||
| ENSG00000149182 | ENSMUSG00000027255 | 0.0363 | 0.4342 | 0.083602027 | ENSMUSG00000028759 | ENSG00000127483 | 0.0512 | 0.4217 | 0.121413327 | ENSG00000132613 | ENSMUSG00000033763 | 0.0226 | 0.7192 | 0.031423804 | ENSG00000185274 | ENSMUSG00000034040 | 0.0067 | 0.644 | 0.010403727 | ||||||||||||||||
| ENSG00000127838 | ENSMUSG00000026179 | 0.0192 | 0.4354 | 0.044097382 | ENSMUSG00000078622 | ENSG00000108588 | 0.009 | 0.4225 | 0.021301775 | ENSG00000071127 | ENSMUSG00000005103 | 0.0227 | 0.7209 | 0.031488417 | ENSG00000213023 | ENSMUSG00000030731 | 0.0167 | 0.6464 | 0.025835396 | ||||||||||||||||
| ENSG00000197535 | ENSMUSG00000034593 | 0.0148 | 0.4356 | 0.033976125 | ENSMUSG00000006998 | ENSG00000175166 | 0.0063 | 0.4226 | 0.014907714 | ENSG00000089220 | ENSMUSG00000047104 | 0.0911 | 0.7227 | 0.126055071 | ENSG00000079691 | ENSMUSG00000021338 | 0.056 | 0.6471 | 0.086539947 | ||||||||||||||||
| ENSG00000120063 | ENSMUSG00000020611 | 0.017 | 0.4358 | 0.03900872 | ENSMUSG00000054423 | ENSG00000163618 | 0.0065 | 0.4227 | 0.015377336 | ENSG00000159231 | ENSMUSG00000022947 | 0.0796 | 0.7241 | 0.109929568 | ENSG00000110697 | ENSMUSG00000024851 | 0.0325 | 0.6473 | 0.050208559 | ||||||||||||||||
| ENSG00000147684 | ENSMUSG00000022354 | 0.0802 | 0.4361 | 0.183902775 | ENSMUSG00000028351 | ENSG00000078725 | 0.0052 | 0.4229 | 0.012296051 | ENSG00000116288 | ENSMUSG00000028964 | 0.0495 | 0.7255 | 0.068228808 | ENSG00000119421 | ENSMUSG00000026895 | 0.0786 | 0.6481 | 0.121277581 | ||||||||||||||||
| ENSG00000130830 | ENSMUSG00000031402 | 0.0381 | 0.4365 | 0.087285223 | ENSMUSG00000024588 | ENSG00000066926 | 0.063 | 0.4231 | 0.148900969 | ENSG00000174791 | ENSMUSG00000024883 | 0.1283 | 0.7291 | 0.175970374 | ENSG00000100284 | ENSMUSG00000042870 | 0.0561 | 0.6503 | 0.086267876 | ||||||||||||||||
| ENSG00000130725 | ENSMUSG00000005575 | 0 | 0.4368 | 0 | ENSMUSG00000004267 | ENSG00000111674 | 0.0083 | 0.4236 | 0.019593957 | ENSG00000004939 | ENSMUSG00000006574 | 0.103 | 0.73 | 0.14109589 | ENSG00000100284 | ENSMUSG00000037827 | 0.0561 | 0.6503 | 0.086267876 | ||||||||||||||||
| ENSG00000123338 | ENSMUSG00000022488 | 0.0255 | 0.4368 | 0.058379121 | ENSMUSG00000037533 | ENSG00000158987 | 0.0425 | 0.4236 | 0.1003305 | ENSG00000068976 | ENSMUSG00000032648 | 0.0139 | 0.7326 | 0.018973519 | ENSG00000189127 | ENSMUSG00000045034 | 0.1044 | 0.6524 | 0.160024525 | ||||||||||||||||
| ENSG00000069535 | ENSMUSG00000040147 | 0.0575 | 0.4372 | 0.131518756 | ENSMUSG00000027642 | ENSG00000118705 | 0.0421 | 0.425 | 0.099058824 | ENSG00000110931 | ENSMUSG00000029471 | 0.0545 | 0.7363 | 0.074018742 | ENSG00000196730 | ENSMUSG00000021559 | 0.0238 | 0.6527 | 0.036463919 | ||||||||||||||||
| ENSG00000172164 | ENSMUSG00000060429 | 0.0325 | 0.4378 | 0.07423481 | ENSMUSG00000027804 | ENSG00000171497 | 0.0294 | 0.4253 | 0.069127675 | ENSG00000187134 | ENSMUSG00000021207 | 0.1679 | 0.7402 | 0.226830586 | ENSG00000143811 | ENSMUSG00000026520 | 0.0377 | 0.6544 | 0.057610024 | ||||||||||||||||
| ENSG00000163539 | ENSMUSG00000033392 | 0.019 | 0.438 | 0.043378995 | ENSMUSG00000026103 | ENSG00000115419 | 0.0522 | 0.4254 | 0.122708039 | ENSG00000100092 | ENSMUSG00000022436 | 0.1038 | 0.7427 | 0.139760334 | ENSG00000108370 | ENSMUSG00000020599 | 0.0451 | 0.6546 | 0.068897036 | ||||||||||||||||
| ENSG00000166340 | ENSMUSG00000030894 | 0.0637 | 0.4385 | 0.145267959 | ENSMUSG00000029106 | ENSG00000087274 | 0.0394 | 0.4258 | 0.092531705 | ENSG00000138796 | ENSMUSG00000027984 | 0.0516 | 0.7449 | 0.069271043 | ENSG00000169213 | ENSMUSG00000003411 | 0.0121 | 0.6558 | 0.018450747 | ||||||||||||||||
| ENSG00000127603 | ENSMUSG00000028649 | 0.1143 | 0.4386 | 0.260601915 | ENSMUSG00000031633 | ENSG00000151729 | 0.0207 | 0.4262 | 0.048568747 | ENSG00000148356 | ENSMUSG00000026792 | 0.0637 | 0.7472 | 0.085251606 | ENSG00000104427 | ENSMUSG00000043542 | 0.0684 | 0.6588 | 0.103825137 | ||||||||||||||||
| ENSG00000125354 | ENSMUSG00000050379 | 0.0187 | 0.439 | 0.042596811 | ENSMUSG00000020361 | ENSG00000170606 | 0.02 | 0.4267 | 0.046871338 | ENSG00000168439 | ENSMUSG00000024966 | 0.0128 | 0.7489 | 0.017091735 | ENSG00000074416 | ENSMUSG00000033174 | 0.0864 | 0.6651 | 0.129905277 | ||||||||||||||||
| ENSG00000166794 | ENSMUSG00000032383 | 0.0294 | 0.439 | 0.066970387 | ENSMUSG00000004455 | ENSG00000186298 | 0.0014 | 0.4273 | 0.003276387 | ENSG00000187134 | ENSMUSG00000021210 | 0.1443 | 0.7499 | 0.192425657 | ENSG00000089154 | ENSMUSG00000041638 | 0.031 | 0.6682 | 0.046393295 | ||||||||||||||||
| ENSG00000142945 | ENSMUSG00000028678 | 0.0642 | 0.4395 | 0.146075085 | ENSMUSG00000011257 | ENSG00000090621 | 0.0109 | 0.4273 | 0.02550901 | ENSG00000213366 | ENSMUSG00000027890 | 0.1278 | 0.751 | 0.170173103 | ENSG00000138326 | ENSMUSG00000025290 | 0.0106 | 0.6687 | 0.015851652 | ||||||||||||||||
| ENSG00000110514 | ENSMUSG00000040687 | 0.0258 | 0.4406 | 0.058556514 | ENSMUSG00000039105 | ENSG00000136888 | 0.0258 | 0.4274 | 0.060364998 | ENSG00000010278 | ENSMUSG00000030342 | 0.0606 | 0.7533 | 0.080446037 | ENSG00000110108 | ENSMUSG00000034659 | 0.1588 | 0.6688 | 0.237440191 | ||||||||||||||||
| ENSG00000061676 | ENSMUSG00000027002 | 0.0028 | 0.4409 | 0.006350646 | ENSMUSG00000025525 | ENSG00000155008 | 0.1334 | 0.4276 | 0.311973807 | ENSG00000106992 | ENSMUSG00000026817 | 0.0574 | 0.7554 | 0.075986232 | ENSG00000103056 | ENSMUSG00000031906 | 0.0438 | 0.67 | 0.065373134 | ||||||||||||||||
| ENSG00000180228 | ENSMUSG00000002731 | 0.0106 | 0.4413 | 0.024019941 | ENSMUSG00000029166 | ENSG00000084764 | 0.0032 | 0.428 | 0.007476636 | ENSG00000151632 | ENSMUSG00000021207 | 0.1673 | 0.7571 | 0.220974772 | ENSG00000073910 | ENSMUSG00000056602 | 0.0156 | 0.6716 | 0.023228112 | ||||||||||||||||
| ENSG00000134769 | ENSMUSG00000024302 | 0.0164 | 0.4414 | 0.037154508 | ENSMUSG00000022841 | ENSG00000161203 | 0 | 0.4282 | 0 | ENSG00000111344 | ENSMUSG00000029602 | 0.0738 | 0.7601 | 0.097092488 | ENSG00000161835 | ENSMUSG00000000531 | 0.0425 | 0.6724 | 0.063206425 | ||||||||||||||||
| ENSG00000104412 | ENSMUSG00000022337 | 0.0118 | 0.4419 | 0.026702874 | ENSMUSG00000024617 | ENSG00000070808 | 0.0009 | 0.4282 | 0.002101822 | ENSG00000103496 | ENSMUSG00000030805 | 0.0454 | 0.7624 | 0.059548793 | ENSG00000100243 | ENSMUSG00000018042 | 0.0588 | 0.6755 | 0.087046632 | ||||||||||||||||
| ENSG00000175203 | ENSMUSG00000025410 | 0.0204 | 0.442 | 0.046153846 | ENSMUSG00000004366 | ENSG00000157005 | 0.0162 | 0.4283 | 0.037823955 | ENSG00000151632 | ENSMUSG00000021210 | 0.1532 | 0.7634 | 0.200681163 | ENSG00000122574 | ENSMUSG00000086040 | 0.1499 | 0.6774 | 0.221287275 | ||||||||||||||||
| ENSG00000088367 | ENSMUSG00000027624 | 0.0218 | 0.4422 | 0.04929896 | ENSMUSG00000031950 | ENSG00000034713 | 0 | 0.4287 | 0 | ENSG00000143799 | ENSMUSG00000026496 | 0.0373 | 0.7647 | 0.048777298 | ENSG00000152128 | ENSMUSG00000026347 | 0.0533 | 0.6781 | 0.078601976 | ||||||||||||||||
| ENSG00000140416 | ENSMUSG00000032366 | 0.0955 | 0.4424 | 0.215867993 | ENSMUSG00000039913 | ENSG00000101349 | 0.0291 | 0.4288 | 0.067863806 | ENSG00000196924 | ENSMUSG00000031328 | 0.0138 | 0.7686 | 0.017954723 | ENSG00000068383 | ENSMUSG00000025477 | 0.029 | 0.6795 | 0.04267844 | ||||||||||||||||
| ENSG00000031698 | ENSMUSG00000068739 | 0.0207 | 0.4427 | 0.046758527 | ENSMUSG00000038429 | ENSG00000111667 | 0.0077 | 0.4289 | 0.017952903 | ENSG00000167460 | ENSMUSG00000031799 | 0.1017 | 0.771 | 0.131906615 | ENSG00000184588 | ENSMUSG00000028525 | 0.0905 | 0.6815 | 0.132795304 | ||||||||||||||||
| ENSG00000138411 | ENSMUSG00000042807 | 0.0241 | 0.4432 | 0.054377256 | ENSMUSG00000000600 | ENSG00000001631 | 0.0277 | 0.4293 | 0.064523643 | ENSG00000110711 | ENSMUSG00000024847 | 0.0254 | 0.7783 | 0.032635231 | ENSG00000086289 | ENSMUSG00000002808 | 0.1132 | 0.6823 | 0.165909424 | ||||||||||||||||
| ENSG00000186469 | ENSMUSG00000043004 | 0 | 0.444 | 0 | ENSMUSG00000031948 | ENSG00000065427 | 0.0397 | 0.4295 | 0.092433062 | ENSG00000121769 | ENSMUSG00000028773 | 0.0788 | 0.7823 | 0.100728621 | ENSG00000067248 | ENSMUSG00000042426 | 0.0477 | 0.6825 | 0.06989011 | ||||||||||||||||
| ENSG00000092108 | ENSMUSG00000020952 | 0.0175 | 0.4441 | 0.039405539 | ENSMUSG00000027195 | ENSG00000149084 | 0.1113 | 0.4298 | 0.258957655 | ENSG00000181061 | ENSMUSG00000038412 | 0.0705 | 0.7946 | 0.088723886 | ENSG00000173175 | ENSMUSG00000022840 | 0.0197 | 0.6839 | 0.028805381 | ||||||||||||||||
| ENSG00000164398 | ENSMUSG00000020333 | 0.0585 | 0.4443 | 0.131667792 | ENSMUSG00000052146 | ENSG00000124614 | 0.0058 | 0.4299 | 0.01349151 | ENSG00000105379 | ENSMUSG00000004610 | 0.0886 | 0.7979 | 0.111041484 | ENSG00000122026 | ENSMUSG00000060566 | 0.0244 | 0.6842 | 0.035662087 | ||||||||||||||||
| ENSG00000128512 | ENSMUSG00000035954 | 0.0075 | 0.4453 | 0.016842578 | ENSMUSG00000022995 | ENSG00000154380 | 0.0542 | 0.4306 | 0.125870878 | ENSG00000141556 | ENSMUSG00000039230 | 0.1059 | 0.7989 | 0.132557266 | ENSG00000196072 | ENSMUSG00000057506 | 0.0253 | 0.6866 | 0.036848238 | ||||||||||||||||
| ENSG00000198399 | ENSMUSG00000020640 | 0.033 | 0.4453 | 0.074107343 | ENSMUSG00000068039 | ENSG00000120438 | 0.0182 | 0.4307 | 0.042256791 | ENSG00000159423 | ENSMUSG00000028737 | 0.0406 | 0.7993 | 0.050794445 | ENSG00000131469 | ENSMUSG00000063316 | 0.0007 | 0.6875 | 0.001018182 | ||||||||||||||||
| ENSG00000108528 | ENSMUSG00000014606 | 0.0165 | 0.4454 | 0.037045352 | ENSMUSG00000027810 | ENSG00000144895 | 0.0331 | 0.4307 | 0.076851637 | ENSG00000197448 | ENSMUSG00000029864 | 0.1658 | 0.8 | 0.20725 | ENSG00000180096 | ENSMUSG00000000486 | 0.0197 | 0.6884 | 0.028617083 | ||||||||||||||||
| ENSG00000083799 | ENSMUSG00000036712 | 0.0245 | 0.4456 | 0.054982047 | ENSMUSG00000020664 | ENSG00000091140 | 0.0243 | 0.4311 | 0.056367432 | ENSG00000170634 | ENSMUSG00000060923 | 0.0908 | 0.8036 | 0.112991538 | ENSG00000165238 | ENSMUSG00000037989 | 0.1176 | 0.6931 | 0.169672486 | ||||||||||||||||
| ENSG00000103423 | ENSMUSG00000004069 | 0.0704 | 0.4459 | 0.157882933 | ENSMUSG00000022048 | ENSG00000092964 | 0.0056 | 0.4314 | 0.012980992 | ENSG00000186907 | ENSMUSG00000050896 | 0.0212 | 0.8065 | 0.026286423 | ENSG00000170473 | ENSMUSG00000064030 | 0.0501 | 0.6956 | 0.072024152 | ||||||||||||||||
| ENSG00000144040 | ENSMUSG00000033720 | 0.0155 | 0.4461 | 0.034745573 | ENSMUSG00000032735 | ENSG00000173210 | 0.0091 | 0.432 | 0.021064815 | ENSG00000133731 | ENSMUSG00000027531 | 0.073 | 0.8171 | 0.08934035 | ENSG00000099889 | ENSMUSG00000000325 | 0.0434 | 0.6987 | 0.062115357 | ||||||||||||||||
| ENSG00000188846 | ENSMUSG00000025794 | 0.0773 | 0.4462 | 0.173240699 | ENSMUSG00000001424 | ENSG00000197157 | 0.0122 | 0.4321 | 0.028234205 | ENSG00000168490 | ENSMUSG00000003469 | 0.0025 | 0.8175 | 0.003058104 | ENSG00000142549 | ENSMUSG00000013367 | 0.0089 | 0.701 | 0.012696148 | ||||||||||||||||
| ENSG00000134318 | ENSMUSG00000020580 | 0.0159 | 0.4467 | 0.035594359 | ENSMUSG00000040537 | ENSG00000008277 | 0.0444 | 0.4321 | 0.102753992 | ENSG00000149823 | ENSMUSG00000024797 | 0.0194 | 0.8226 | 0.023583759 | ENSG00000151491 | ENSMUSG00000015766 | 0.055 | 0.7059 | 0.077914719 | ||||||||||||||||
| ENSG00000183715 | ENSMUSG00000062257 | 0.0346 | 0.4475 | 0.077318436 | ENSMUSG00000014402 | ENSG00000074319 | 0.0255 | 0.4334 | 0.058837102 | ENSG00000213366 | ENSMUSG00000091578 | 0.1129 | 0.8274 | 0.136451535 | ENSG00000137154 | ENSMUSG00000028495 | 0.0007 | 0.7072 | 0.000989819 | ||||||||||||||||
| ENSG00000204681 | ENSMUSG00000024462 | 0.0057 | 0.4478 | 0.012728897 | ENSMUSG00000022400 | ENSG00000100387 | 0 | 0.4341 | 0 | ENSG00000144712 | ENSMUSG00000030319 | 0.0501 | 0.8325 | 0.06018018 | ENSG00000163517 | ENSMUSG00000034245 | 0.0398 | 0.7085 | 0.056175018 | ||||||||||||||||
| ENSG00000167085 | ENSMUSG00000038845 | 0.0017 | 0.4479 | 0.00379549 | ENSMUSG00000020015 | ENSG00000059758 | 0.0063 | 0.4341 | 0.014512785 | ENSG00000163931 | ENSMUSG00000021957 | 0.0265 | 0.8431 | 0.031431621 | ENSG00000067829 | ENSMUSG00000002010 | 0.0252 | 0.7095 | 0.03551797 | ||||||||||||||||
| ENSG00000004864 | ENSMUSG00000015112 | 0.0228 | 0.448 | 0.050892857 | ENSMUSG00000027255 | ENSG00000149182 | 0.0363 | 0.4342 | 0.083602027 | ENSG00000213366 | ENSMUSG00000058135 | 0.1363 | 0.8457 | 0.161168263 | ENSG00000185774 | ENSMUSG00000029088 | 0.0741 | 0.7101 | 0.1043515 | ||||||||||||||||
| ENSG00000166501 | ENSMUSG00000052889 | 0.0054 | 0.4495 | 0.012013348 | ENSMUSG00000000560 | ENSG00000151834 | 0.0114 | 0.4344 | 0.026243094 | ENSG00000117115 | ENSMUSG00000028927 | 0.0347 | 0.8459 | 0.041021397 | ENSG00000007047 | ENSMUSG00000030397 | 0.0506 | 0.7135 | 0.07091801 | ||||||||||||||||
| ENSG00000138190 | ENSMUSG00000053799 | 0.0354 | 0.4502 | 0.078631719 | ENSMUSG00000026179 | ENSG00000127838 | 0.0192 | 0.4354 | 0.044097382 | ENSG00000132702 | ENSMUSG00000004894 | 0.0411 | 0.8487 | 0.048427006 | ENSG00000144647 | ENSMUSG00000066235 | 0.0302 | 0.7156 | 0.042202348 | ||||||||||||||||
| ENSG00000140740 | ENSMUSG00000030884 | 0.077 | 0.4502 | 0.171035096 | ENSMUSG00000034593 | ENSG00000197535 | 0.0148 | 0.4356 | 0.033976125 | ENSG00000167378 | ENSMUSG00000041037 | 0.109 | 0.8487 | 0.128431719 | ENSG00000161682 | ENSMUSG00000034685 | 0.0256 | 0.7157 | 0.035769177 | ||||||||||||||||
| ENSG00000110955 | ENSMUSG00000025393 | 0.0195 | 0.4505 | 0.043285239 | ENSMUSG00000020611 | ENSG00000120063 | 0.017 | 0.4358 | 0.03900872 | ENSG00000171224 | ENSMUSG00000020083 | 0.0366 | 0.857 | 0.042707118 | ENSG00000106078 | ENSMUSG00000020173 | 0.2314 | 0.7176 | 0.322463768 | ||||||||||||||||
| ENSG00000132912 | ENSMUSG00000024603 | 0.0156 | 0.4506 | 0.034620506 | ENSMUSG00000028782 | ENSG00000121753 | 0.0175 | 0.4358 | 0.040156035 | ENSG00000103197 | ENSMUSG00000002496 | 0.0467 | 0.8606 | 0.054264467 | ENSG00000108669 | ENSMUSG00000017132 | 0.0068 | 0.7229 | 0.009406557 | ||||||||||||||||
| ENSG00000182901 | ENSMUSG00000026527 | 0.0079 | 0.4508 | 0.017524401 | ENSMUSG00000022354 | ENSG00000147684 | 0.0802 | 0.4361 | 0.183902775 | ENSG00000143727 | ENSMUSG00000044573 | 0.1248 | 0.8613 | 0.144897248 | ENSG00000148408 | ENSMUSG00000004113 | 0.0336 | 0.7256 | 0.046306505 | ||||||||||||||||
| ENSG00000103316 | ENSMUSG00000030905 | 0.0634 | 0.4512 | 0.140514184 | ENSMUSG00000031402 | ENSG00000130830 | 0.0381 | 0.4365 | 0.087285223 | ENSG00000079435 | ENSMUSG00000003123 | 0.0941 | 0.8634 | 0.108987723 | ENSG00000122547 | ENSMUSG00000036611 | 0.0502 | 0.726 | 0.069146006 | ||||||||||||||||
| ENSG00000020129 | ENSMUSG00000028833 | 0.0091 | 0.4514 | 0.020159504 | ENSMUSG00000020170 | ENSG00000166225 | 0.0216 | 0.4369 | 0.049439231 | ENSG00000160211 | ENSMUSG00000031400 | 0.0296 | 0.864 | 0.034259259 | ENSG00000117118 | ENSMUSG00000009863 | 0.047 | 0.7292 | 0.064454196 | ||||||||||||||||
| ENSG00000168036 | ENSMUSG00000006932 | 0.0006 | 0.4516 | 0.001328609 | ENSMUSG00000032120 | ENSG00000172375 | 0.0469 | 0.4369 | 0.107347219 | ENSG00000213366 | ENSMUSG00000004038 | 0.1395 | 0.8655 | 0.16117851 | ENSG00000149575 | ENSMUSG00000070304 | 0.0378 | 0.7327 | 0.05159001 | ||||||||||||||||
| ENSG00000174437 | ENSMUSG00000029467 | 0.0055 | 0.452 | 0.012168142 | ENSMUSG00000040147 | ENSG00000069535 | 0.0575 | 0.4372 | 0.131518756 | ENSG00000111275 | ENSMUSG00000029455 | 0.0255 | 0.8733 | 0.029199588 | ENSG00000137766 | ENSMUSG00000062151 | 0.0528 | 0.7339 | 0.071944407 | ||||||||||||||||
| ENSG00000164061 | ENSMUSG00000032589 | 0.0559 | 0.452 | 0.123672566 | ENSMUSG00000033392 | ENSG00000163539 | 0.019 | 0.438 | 0.043378995 | ENSG00000104946 | ENSMUSG00000038520 | 0.0536 | 0.8749 | 0.061264144 | ENSG00000171488 | ENSMUSG00000054720 | 0.0147 | 0.7345 | 0.020013615 | ||||||||||||||||
| ENSG00000087460 | ENSMUSG00000027523 | 0.2024 | 0.452 | 0.447787611 | ENSMUSG00000028649 | ENSG00000127603 | 0.1143 | 0.4386 | 0.260601915 | ENSG00000100994 | ENSMUSG00000033059 | 0.0203 | 0.8789 | 0.023097053 | ENSG00000196072 | ENSMUSG00000025201 | 0.0251 | 0.7346 | 0.034168255 | ||||||||||||||||
| ENSG00000117528 | ENSMUSG00000028127 | 0.023 | 0.4522 | 0.05086245 | ENSMUSG00000050379 | ENSG00000125354 | 0.0187 | 0.439 | 0.042596811 | ENSG00000055332 | ENSMUSG00000024079 | 0.3148 | 0.8795 | 0.357930642 | ENSG00000187688 | ENSMUSG00000018507 | 0.1113 | 0.7394 | 0.150527455 | ||||||||||||||||
| ENSG00000159023 | ENSMUSG00000028906 | 0.045 | 0.4525 | 0.099447514 | ENSMUSG00000032383 | ENSG00000166794 | 0.0294 | 0.439 | 0.066970387 | ENSG00000185359 | ENSMUSG00000025793 | 0.0344 | 0.8852 | 0.038861274 | ENSG00000179841 | ENSMUSG00000021057 | 0.254 | 0.7406 | 0.342965163 | ||||||||||||||||
| ENSG00000119471 | ENSMUSG00000028383 | 0.0697 | 0.453 | 0.153863135 | ENSMUSG00000028678 | ENSG00000142945 | 0.0642 | 0.4395 | 0.146075085 | ENSG00000014216 | ENSMUSG00000024942 | 0.0522 | 0.8853 | 0.058963063 | ENSG00000149091 | ENSMUSG00000040479 | 0.0457 | 0.7437 | 0.061449509 | ||||||||||||||||
| ENSG00000111237 | ENSMUSG00000029462 | 0.0028 | 0.4533 | 0.006176925 | ENSMUSG00000025967 | ENSG00000114942 | 0.0293 | 0.44 | 0.066590909 | ENSG00000100033 | ENSMUSG00000003526 | 0.0973 | 0.8868 | 0.109720343 | ENSG00000188803 | ENSMUSG00000053930 | 0.0497 | 0.7442 | 0.066783123 | ||||||||||||||||
| ENSG00000074054 | ENSMUSG00000064302 | 0.0099 | 0.4533 | 0.021839841 | ENSMUSG00000002741 | ENSG00000106636 | 0.0354 | 0.4404 | 0.080381471 | ENSG00000123143 | ENSMUSG00000057672 | 0.0434 | 0.8869 | 0.048934491 | ENSG00000111696 | ENSMUSG00000054027 | 0.038 | 0.745 | 0.051006711 | ||||||||||||||||
| ENSG00000180834 | ENSMUSG00000041205 | 0.1478 | 0.4536 | 0.325837743 | ENSMUSG00000027002 | ENSG00000061676 | 0.0028 | 0.4409 | 0.006350646 | ENSG00000148834 | ENSMUSG00000025068 | 0.1754 | 0.8878 | 0.19756702 | ENSG00000151148 | ENSMUSG00000029577 | 0.0395 | 0.7467 | 0.052899424 | ||||||||||||||||
| ENSG00000084754 | ENSMUSG00000025745 | 0.0772 | 0.4537 | 0.170156491 | ENSMUSG00000024077 | ENSG00000115808 | 0.0161 | 0.4409 | 0.036516217 | ENSG00000127445 | ENSMUSG00000032171 | 0.0232 | 0.8882 | 0.026120243 | ENSG00000069712 | ENSMUSG00000044060 | 0.2828 | 0.7575 | 0.373333333 | ||||||||||||||||
| ENSG00000138107 | ENSMUSG00000025228 | 0 | 0.454 | 0 | ENSMUSG00000021684 | ENSG00000113231 | 0.0205 | 0.4411 | 0.046474722 | ENSG00000187091 | ENSMUSG00000010660 | 0.0486 | 0.894 | 0.054362416 | ENSG00000159788 | ENSMUSG00000029101 | 0.0808 | 0.7595 | 0.10638578 | ||||||||||||||||
| ENSG00000164076 | ENSMUSG00000032936 | 0.025 | 0.4544 | 0.055017606 | ENSMUSG00000040389 | ENSG00000085433 | 0.023 | 0.4412 | 0.052130553 | ENSG00000095321 | ENSMUSG00000026853 | 0.0459 | 0.9014 | 0.05092079 | ENSG00000157103 | ENSMUSG00000030310 | 0.0093 | 0.7661 | 0.012139407 | ||||||||||||||||
| ENSG00000062485 | ENSMUSG00000005683 | 0.0316 | 0.4544 | 0.069542254 | ENSMUSG00000024302 | ENSG00000134769 | 0.0164 | 0.4414 | 0.037154508 | ENSG00000171840 | ENSMUSG00000041377 | 0.1589 | 0.9018 | 0.176203149 | ENSG00000124159 | ENSMUSG00000016995 | 0.0485 | 0.7724 | 0.0627913 | ||||||||||||||||
| ENSG00000058404 | ENSMUSG00000057897 | 0.0064 | 0.4547 | 0.014075214 | ENSMUSG00000022551 | ENSG00000179091 | 0.0525 | 0.4416 | 0.11888587 | ENSG00000130827 | ENSMUSG00000031398 | 0.0282 | 0.907 | 0.03109151 | ENSG00000170989 | ENSMUSG00000045092 | 0.0273 | 0.775 | 0.035225806 | ||||||||||||||||
| ENSG00000123983 | ENSMUSG00000032883 | 0.0362 | 0.4565 | 0.079299014 | ENSMUSG00000022337 | ENSG00000104412 | 0.0118 | 0.4419 | 0.026702874 | ENSG00000180900 | ENSMUSG00000022568 | 0.0661 | 0.9091 | 0.072709273 | ENSG00000165757 | ENSMUSG00000033960 | 0.3317 | 0.7751 | 0.427944781 | ||||||||||||||||
| ENSG00000012822 | ENSMUSG00000023055 | 0.0561 | 0.4568 | 0.122810858 | ENSMUSG00000025410 | ENSG00000175203 | 0.0204 | 0.442 | 0.046153846 | ENSG00000160211 | ENSMUSG00000089992 | 0.0638 | 0.9123 | 0.069933136 | ENSG00000134780 | ENSMUSG00000035735 | 0.0127 | 0.7828 | 0.016223812 | ||||||||||||||||
| ENSG00000197283 | ENSMUSG00000067629 | 0.0111 | 0.4581 | 0.024230517 | ENSMUSG00000027624 | ENSG00000088367 | 0.0218 | 0.4422 | 0.04929896 | ENSG00000116791 | ENSMUSG00000028199 | 0.1114 | 0.9239 | 0.12057582 | ENSG00000116039 | ENSMUSG00000006269 | 0.0316 | 0.7838 | 0.040316407 | ||||||||||||||||
| ENSG00000170390 | ENSMUSG00000028078 | 0.0299 | 0.4582 | 0.065255347 | ENSMUSG00000019809 | ENSG00000034693 | 0.0293 | 0.4424 | 0.066229656 | ENSG00000167363 | ENSMUSG00000025175 | 0.0658 | 0.9293 | 0.070805983 | ENSG00000106638 | ENSMUSG00000005374 | 0.0594 | 0.7897 | 0.075218437 | ||||||||||||||||
| ENSG00000086061 | ENSMUSG00000028410 | 0.0022 | 0.4583 | 0.004800349 | ENSMUSG00000032366 | ENSG00000140416 | 0.0955 | 0.4424 | 0.215867993 | ENSG00000161714 | ENSMUSG00000020937 | 0.059 | 0.9294 | 0.063481816 | ENSG00000141837 | ENSMUSG00000034656 | 0.0474 | 0.7899 | 0.060007596 | ||||||||||||||||
| ENSG00000166986 | ENSMUSG00000040354 | 0.0587 | 0.4592 | 0.12783101 | ENSMUSG00000041133 | ENSG00000072501 | 0.0007 | 0.4427 | 0.001581206 | ENSG00000106211 | ENSMUSG00000004951 | 0.0803 | 0.9334 | 0.086029569 | ENSG00000162738 | ENSMUSG00000026556 | 0.0025 | 0.7899 | 0.003164958 | ||||||||||||||||
| ENSG00000132294 | ENSMUSG00000015002 | 0.0132 | 0.4593 | 0.028739386 | ENSMUSG00000042807 | ENSG00000138411 | 0.0241 | 0.4432 | 0.054377256 | ENSG00000160209 | ENSMUSG00000032788 | 0.07 | 0.9384 | 0.074595055 | ENSG00000130559 | ENSMUSG00000026933 | 0.0842 | 0.7915 | 0.106380291 | ||||||||||||||||
| ENSG00000105245 | ENSMUSG00000063160 | 0.0222 | 0.4593 | 0.048334422 | ENSMUSG00000030286 | ENSG00000125037 | 0.0018 | 0.4433 | 0.004060456 | ENSG00000165752 | ENSMUSG00000015981 | 0.0423 | 0.9443 | 0.044795086 | ENSG00000165959 | ENSMUSG00000021097 | 0.1807 | 0.7918 | 0.228214196 | ||||||||||||||||
| ENSG00000180979 | ENSMUSG00000027286 | 0.0226 | 0.4594 | 0.049194602 | ENSMUSG00000043004 | ENSG00000186469 | 0 | 0.444 | 0 | ENSG00000205531 | ENSMUSG00000059119 | 0.0246 | 0.9462 | 0.025998732 | ENSG00000090971 | ENSMUSG00000035285 | 0.0245 | 0.7955 | 0.03079824 | ||||||||||||||||
| ENSG00000157827 | ENSMUSG00000036053 | 0.0346 | 0.4596 | 0.075282855 | ENSMUSG00000020952 | ENSG00000092108 | 0.0175 | 0.4441 | 0.039405539 | ENSG00000135709 | ENSMUSG00000031824 | 0.0503 | 0.9539 | 0.052730894 | ENSG00000141543 | ENSMUSG00000060782 | 0.0144 | 0.797 | 0.018067754 | ||||||||||||||||
| ENSG00000114520 | ENSMUSG00000022808 | 0.0322 | 0.46 | 0.07 | ENSMUSG00000020333 | ENSG00000164398 | 0.0585 | 0.4443 | 0.131667792 | ENSG00000104419 | ENSMUSG00000005125 | 0.0282 | 0.96 | 0.029375 | ENSG00000099904 | ENSMUSG00000060166 | 0.0415 | 0.7986 | 0.05196594 | ||||||||||||||||
| ENSG00000198171 | ENSMUSG00000068290 | 0.0933 | 0.4605 | 0.202605863 | ENSMUSG00000074305 | ENSG00000173517 | 0.0735 | 0.4451 | 0.165131431 | ENSG00000124126 | ENSMUSG00000039621 | 0.0404 | 0.9659 | 0.041826276 | ENSG00000131148 | ENSMUSG00000031819 | 0.0322 | 0.8012 | 0.040189715 | ||||||||||||||||
| ENSG00000112851 | ENSMUSG00000021709 | 0.0504 | 0.4606 | 0.109422492 | ENSMUSG00000035954 | ENSG00000128512 | 0.0075 | 0.4453 | 0.016842578 | ENSG00000108064 | ENSMUSG00000003923 | 0.2531 | 0.9679 | 0.261493956 | ENSG00000136156 | ENSMUSG00000022108 | 0.0217 | 0.8014 | 0.027077614 | ||||||||||||||||
| ENSG00000151025 | ENSMUSG00000045967 | 0.0687 | 0.4607 | 0.149120903 | ENSMUSG00000020640 | ENSG00000198399 | 0.033 | 0.4453 | 0.074107343 | ENSG00000211899 | ENSMUSG00000076617 | 0.2497 | 0.9696 | 0.257528878 | ENSG00000137825 | ENSMUSG00000027296 | 0.0371 | 0.8021 | 0.046253584 | ||||||||||||||||
| ENSG00000162616 | ENSMUSG00000028035 | 0.0285 | 0.4608 | 0.061848958 | ENSMUSG00000014606 | ENSG00000108528 | 0.0165 | 0.4454 | 0.037045352 | ENSG00000125730 | ENSMUSG00000024164 | 0.1335 | 0.972 | 0.137345679 | ENSG00000089250 | ENSMUSG00000029361 | 0.0295 | 0.806 | 0.036600496 | ||||||||||||||||
| ENSG00000137100 | ENSMUSG00000028447 | 0.031 | 0.4614 | 0.067186823 | ENSMUSG00000036712 | ENSG00000083799 | 0.0245 | 0.4456 | 0.054982047 | ENSG00000133313 | ENSMUSG00000024644 | 0.0449 | 1.0031 | 0.04476124 | ENSG00000144559 | ENSMUSG00000030316 | 0.1255 | 0.8077 | 0.155379473 | ||||||||||||||||
| ENSG00000176148 | ENSMUSG00000027175 | 0.0822 | 0.4615 | 0.178114843 | ENSMUSG00000007653 | ENSG00000145864 | 0.001 | 0.4457 | 0.002243662 | ENSG00000131016 | ENSMUSG00000038587 | 0.3012 | 1.0045 | 0.299850672 | ENSG00000054793 | ENSMUSG00000027546 | 0.0088 | 0.8081 | 0.010889741 | ||||||||||||||||
| ENSG00000104067 | ENSMUSG00000030516 | 0.0465 | 0.4619 | 0.100671141 | ENSMUSG00000022450 | ENSG00000184983 | 0.0855 | 0.4458 | 0.19179004 | ENSG00000130764 | ENSMUSG00000029028 | 0.0678 | 1.0048 | 0.067476115 | ENSG00000164929 | ENSMUSG00000022296 | 0.2638 | 0.8112 | 0.325197239 | ||||||||||||||||
| ENSG00000173166 | ENSMUSG00000026014 | 0.0502 | 0.4627 | 0.108493624 | ENSMUSG00000004069 | ENSG00000103423 | 0.0704 | 0.4459 | 0.157882933 | ENSG00000203485 | ENSMUSG00000037679 | 0.1279 | 1.0263 | 0.12462243 | ENSG00000147509 | ENSMUSG00000002459 | 0.2541 | 0.8122 | 0.312853977 | ||||||||||||||||
| ENSG00000082898 | ENSMUSG00000020290 | 0.0091 | 0.4631 | 0.019650184 | ENSMUSG00000033720 | ENSG00000144040 | 0.0155 | 0.4461 | 0.034745573 | ENSG00000103202 | ENSMUSG00000024177 | 0.0872 | 1.0328 | 0.084430674 | ENSG00000139174 | ENSMUSG00000036158 | 0.0342 | 0.8131 | 0.042061247 | ||||||||||||||||
| ENSG00000172260 | ENSMUSG00000040037 | 0.0249 | 0.4634 | 0.053733276 | ENSMUSG00000041236 | ENSG00000006715 | 0.0122 | 0.4462 | 0.027341999 | ENSG00000167037 | ENSMUSG00000042216 | 0.0276 | 1.0363 | 0.026633214 | ENSG00000181467 | ENSMUSG00000036894 | 0 | 0.8131 | 0 | ||||||||||||||||
| ENSG00000091129 | ENSMUSG00000020598 | 0.0354 | 0.4634 | 0.076391886 | ENSMUSG00000025794 | ENSG00000188846 | 0.0773 | 0.4462 | 0.173240699 | ENSG00000142347 | ENSMUSG00000024300 | 0.03 | 1.0404 | 0.028835063 | ENSG00000102119 | ENSMUSG00000001964 | 0.1396 | 0.8184 | 0.170576735 | ||||||||||||||||
| ENSG00000083312 | ENSMUSG00000009470 | 0.0016 | 0.4638 | 0.003449763 | ENSMUSG00000020580 | ENSG00000134318 | 0.0159 | 0.4467 | 0.035594359 | ENSG00000159189 | ENSMUSG00000036896 | 0.1503 | 1.0613 | 0.141618769 | ENSG00000100364 | ENSMUSG00000036046 | 0.052 | 0.8308 | 0.062590274 | ||||||||||||||||
| ENSG00000135924 | ENSMUSG00000026203 | 0.0687 | 0.4639 | 0.148092261 | ENSMUSG00000062257 | ENSG00000183715 | 0.0346 | 0.4475 | 0.077318436 | ENSG00000179604 | ENSMUSG00000041598 | 0.1522 | 1.064 | 0.143045113 | ENSG00000099800 | ENSMUSG00000020219 | 0.0252 | 0.8409 | 0.029967892 | ||||||||||||||||
| ENSG00000130985 | ENSMUSG00000001924 | 0.0228 | 0.4641 | 0.049127343 | ENSMUSG00000079426 | ENSG00000241553 | 0 | 0.4478 | 0 | ENSG00000186111 | ENSMUSG00000034902 | 0.0307 | 1.0714 | 0.028654097 | ENSG00000090975 | ENSMUSG00000029406 | 0.0463 | 0.8495 | 0.054502649 | ||||||||||||||||
| ENSG00000187164 | ENSMUSG00000041362 | 0.0455 | 0.4643 | 0.097996985 | ENSMUSG00000024462 | ENSG00000204681 | 0.0057 | 0.4478 | 0.012728897 | ENSG00000169710 | ENSMUSG00000025153 | 0.1023 | 1.0734 | 0.095304639 | ENSG00000100427 | ENSMUSG00000035805 | 0.0588 | 0.8511 | 0.069087064 | ||||||||||||||||
| ENSG00000204427 | ENSMUSG00000007036 | 0.0193 | 0.4644 | 0.041559001 | ENSMUSG00000038845 | ENSG00000167085 | 0.0017 | 0.4479 | 0.00379549 | ENSG00000110090 | ENSMUSG00000024900 | 0.0684 | 1.0786 | 0.063415539 | ENSG00000141543 | ENSMUSG00000063692 | 0.0122 | 0.8532 | 0.014299109 | ||||||||||||||||
| ENSG00000214338 | ENSMUSG00000038916 | 0.0454 | 0.4644 | 0.097760551 | ENSMUSG00000015112 | ENSG00000004864 | 0.0228 | 0.448 | 0.050892857 | ENSG00000103160 | ENSMUSG00000034189 | 0.0688 | 1.0883 | 0.063217863 | ENSG00000197380 | ENSMUSG00000078794 | 0.0597 | 0.8595 | 0.069458988 | ||||||||||||||||
| ENSG00000119321 | ENSMUSG00000066151 | 0.123 | 0.4645 | 0.264800861 | ENSMUSG00000026587 | ENSG00000152092 | 0.0096 | 0.4484 | 0.021409456 | ENSG00000213366 | ENSMUSG00000091561 | 0.1669 | 1.0971 | 0.152128338 | ENSG00000154096 | ENSMUSG00000032011 | 0.2074 | 0.8602 | 0.241106719 | ||||||||||||||||
| ENSG00000157823 | ENSMUSG00000063801 | 0 | 0.4651 | 0 | ENSMUSG00000029861 | ENSG00000159784 | 0.0192 | 0.4492 | 0.042742654 | ENSG00000172992 | ENSMUSG00000020935 | 0.0431 | 1.1234 | 0.038365676 | ENSG00000105679 | ENSMUSG00000061099 | 0.1098 | 0.8671 | 0.126628993 | ||||||||||||||||
| ENSG00000138698 | ENSMUSG00000028149 | 0.0242 | 0.4654 | 0.051998281 | ENSMUSG00000053799 | ENSG00000138190 | 0.0354 | 0.4502 | 0.078631719 | ENSG00000197256 | ENSMUSG00000032194 | 0.0889 | 1.1647 | 0.076328668 | ENSG00000085733 | ENSMUSG00000031078 | 0.0375 | 0.8682 | 0.043192813 | ||||||||||||||||
| ENSG00000110171 | ENSMUSG00000036989 | 0.007 | 0.466 | 0.015021459 | ENSMUSG00000030884 | ENSG00000140740 | 0.077 | 0.4502 | 0.171035096 | ENSG00000160360 | ENSMUSG00000026930 | 0.0358 | 1.1868 | 0.03016515 | ENSG00000165912 | ENSMUSG00000027257 | 0.0262 | 0.8731 | 0.030008017 | ||||||||||||||||
| ENSG00000067715 | ENSMUSG00000035864 | 0.0112 | 0.4661 | 0.024029178 | ENSMUSG00000026904 | ENSG00000144290 | 0.0105 | 0.4503 | 0.023317788 | ENSG00000072958 | ENSMUSG00000003033 | 0.003 | 1.189 | 0.002523129 | ENSG00000103154 | ENSMUSG00000031837 | 0.075 | 0.8762 | 0.085596896 | ||||||||||||||||
| ENSG00000138685 | ENSMUSG00000037225 | 0.0206 | 0.4669 | 0.044120797 | ENSMUSG00000025393 | ENSG00000110955 | 0.0195 | 0.4505 | 0.043285239 | ENSG00000099795 | ENSMUSG00000033938 | 0.096 | 1.1901 | 0.08066549 | ENSG00000154429 | ENSMUSG00000031971 | 0.1766 | 0.8773 | 0.201299441 | ||||||||||||||||
| ENSG00000154556 | ENSMUSG00000031626 | 0.0368 | 0.4678 | 0.078666097 | ENSMUSG00000024603 | ENSG00000132912 | 0.0156 | 0.4506 | 0.034620506 | ENSG00000176595 | ENSMUSG00000055675 | 0.0981 | 1.239 | 0.079176755 | ENSG00000171603 | ENSMUSG00000039953 | 0.0433 | 0.8834 | 0.049015169 | ||||||||||||||||
| ENSG00000171132 | ENSMUSG00000045038 | 0.0104 | 0.4679 | 0.022226972 | ENSMUSG00000026527 | ENSG00000182901 | 0.0079 | 0.4508 | 0.017524401 | ENSG00000126934 | ENSMUSG00000035027 | 0.0246 | 1.2561 | 0.019584428 | ENSG00000075399 | ENSMUSG00000001062 | 0.093 | 0.8837 | 0.105239335 | ||||||||||||||||
| ENSG00000136051 | ENSMUSG00000034560 | 0.0156 | 0.4679 | 0.033340457 | ENSMUSG00000028833 | ENSG00000020129 | 0.0091 | 0.4514 | 0.020159504 | ENSG00000127948 | ENSMUSG00000005514 | 0.0356 | 1.297 | 0.027447957 | ENSG00000040608 | ENSMUSG00000043811 | 0.0622 | 0.8844 | 0.070330167 | ||||||||||||||||
| ENSG00000164885 | ENSMUSG00000028969 | 0.0016 | 0.4681 | 0.003418073 | ENSMUSG00000006932 | ENSG00000168036 | 0.0006 | 0.4516 | 0.001328609 | ENSG00000065000 | ENSMUSG00000020198 | 0.0442 | 1.3155 | 0.033599392 | ENSG00000187630 | ENSMUSG00000022210 | 0.1963 | 0.8955 | 0.219207147 | ||||||||||||||||
| ENSG00000026025 | ENSMUSG00000026728 | 0.0111 | 0.4681 | 0.023712882 | ENSMUSG00000029467 | ENSG00000174437 | 0.0055 | 0.452 | 0.012168142 | ENSG00000143774 | ENSMUSG00000020444 | 0.0625 | 1.4153 | 0.044160249 | ENSG00000128011 | ENSMUSG00000030600 | 0.0207 | 0.8982 | 0.023046092 | ||||||||||||||||
| ENSG00000154174 | ENSMUSG00000022752 | 0.0429 | 0.4694 | 0.091393268 | ENSMUSG00000032589 | ENSG00000164061 | 0.0559 | 0.452 | 0.123672566 | ENSG00000204388 | ENSMUSG00000091971 | 0.0183 | 1.4379 | 0.012726893 | ENSG00000174672 | ENSMUSG00000053046 | 0.0098 | 0.899 | 0.010901001 | ||||||||||||||||
| ENSG00000144381 | ENSMUSG00000025980 | 0.0121 | 0.4699 | 0.02575016 | ENSMUSG00000027523 | ENSG00000087460 | 0.2024 | 0.452 | 0.447787611 | ENSG00000204388 | ENSMUSG00000090877 | 0.0183 | 1.4565 | 0.012564367 | ENSG00000161671 | ENSMUSG00000008140 | 0.0788 | 0.9021 | 0.087351735 | ||||||||||||||||
| ENSG00000166925 | ENSMUSG00000029723 | 0.0862 | 0.4702 | 0.183326244 | ENSMUSG00000028127 | ENSG00000117528 | 0.023 | 0.4522 | 0.05086245 | ENSG00000196365 | ENSMUSG00000041168 | 0.0621 | 1.466 | 0.042360164 | ENSG00000161970 | ENSMUSG00000059150 | 0.0412 | 0.9028 | 0.0456358 | ||||||||||||||||
| ENSG00000105993 | ENSMUSG00000029131 | 0.089 | 0.4705 | 0.189160468 | ENSMUSG00000028820 | ENSG00000116560 | 0.0364 | 0.4532 | 0.080317741 | ENSG00000007080 | ENSMUSG00000007721 | 0.0423 | 1.4903 | 0.028383547 | ENSG00000168298 | ENSMUSG00000051627 | 0.029 | 0.9049 | 0.03204774 | ||||||||||||||||
| ENSG00000070961 | ENSMUSG00000019943 | 0.0023 | 0.4708 | 0.004885302 | ENSMUSG00000029462 | ENSG00000111237 | 0.0028 | 0.4533 | 0.006176925 | ENSG00000167671 | ENSMUSG00000019578 | 0.1087 | 1.5206 | 0.07148494 | ENSG00000154118 | ENSMUSG00000025318 | 0.0319 | 0.9092 | 0.03508579 | ||||||||||||||||
| ENSG00000145681 | ENSMUSG00000021613 | 0.0181 | 0.4711 | 0.038420717 | ENSMUSG00000064302 | ENSG00000074054 | 0.0099 | 0.4533 | 0.021839841 | ENSG00000130193 | ENSMUSG00000056665 | 0.0614 | 1.5821 | 0.038809178 | ENSG00000123104 | ENSMUSG00000030287 | 0.0205 | 0.9164 | 0.022370144 | ||||||||||||||||
| ENSG00000006530 | ENSMUSG00000029916 | 0.0419 | 0.4711 | 0.088940777 | ENSMUSG00000057378 | ENSG00000198838 | 0.021 | 0.4535 | 0.046306505 | ENSG00000110047 | ENSMUSG00000024772 | 0.0022 | 1.6009 | 0.001374227 | ENSG00000076641 | ENSMUSG00000027508 | 0.1093 | 0.9167 | 0.119232028 | ||||||||||||||||
| ENSG00000168959 | ENSMUSG00000049583 | 0.0179 | 0.4715 | 0.037963945 | ENSMUSG00000057335 | ENSG00000143702 | 0.0508 | 0.4536 | 0.111992945 | ENSG00000103024 | ENSMUSG00000073435 | 0.0578 | 1.7278 | 0.033452946 | ENSG00000141543 | ENSMUSG00000025580 | 0.0033 | 0.9217 | 0.003580341 | ||||||||||||||||
| ENSG00000215021 | ENSMUSG00000004264 | 0.0005 | 0.4716 | 0.001060221 | ENSMUSG00000041205 | ENSG00000180834 | 0.1478 | 0.4536 | 0.325837743 | ENSG00000176619 | ENSMUSG00000062075 | 0.0867 | 2.2395 | 0.038713999 | ENSG00000143858 | ENSMUSG00000026452 | 0.0093 | 0.9239 | 0.010066024 | ||||||||||||||||
| ENSG00000147065 | ENSMUSG00000031207 | 0.006 | 0.4717 | 0.012719949 | ENSMUSG00000025745 | ENSG00000084754 | 0.0772 | 0.4537 | 0.170156491 | ENSG00000170373 | ENSMUSG00000027442 | 0.9662 | 3.142 | 0.307511139 | ENSG00000162551 | ENSMUSG00000028766 | 0.0569 | 0.9249 | 0.061520164 | ||||||||||||||||
| ENSG00000132002 | ENSMUSG00000005483 | 0.0211 | 0.4718 | 0.04472234 | ENSMUSG00000025228 | ENSG00000138107 | 0 | 0.454 | 0 | ENSG00000170373 | ENSMUSG00000036924 | 13.4209 | 11.1545 | 1.203182572 | ENSG00000105520 | ENSMUSG00000040563 | 0.0681 | 0.9269 | 0.073470709 | ||||||||||||||||
| ENSG00000125971 | ENSMUSG00000047459 | 0.0136 | 0.472 | 0.028813559 | ENSMUSG00000030075 | ENSG00000113805 | 0.0429 | 0.4542 | 0.094451783 | ENSG00000085063 | ENSMUSG00000068686 | 0.5046 | 34.08 | 0.014806338 | ENSG00000184985 | ENSMUSG00000029093 | 0.0968 | 0.9435 | 0.102596714 | ||||||||||||||||
| ENSG00000090013 | ENSMUSG00000040466 | 0.0389 | 0.472 | 0.082415254 | ENSMUSG00000034813 | ENSG00000155974 | 0.0188 | 0.4543 | 0.041382346 | ENSG00000066248 | ENSMUSG00000026259 | 0.0729 | 0.9538 | 0.076431118 | |||||||||||||||||||||
| ENSG00000137343 | ENSMUSG00000024426 | 0.046 | 0.4724 | 0.097375106 | ENSMUSG00000032936 | ENSG00000164076 | 0.025 | 0.4544 | 0.055017606 | ENSG00000004139 | ENSMUSG00000050132 | 0.0697 | 0.9598 | 0.072619296 | |||||||||||||||||||||
| ENSG00000122733 | ENSMUSG00000036062 | 0.0501 | 0.4731 | 0.105897273 | ENSMUSG00000005683 | ENSG00000062485 | 0.0316 | 0.4544 | 0.069542254 | ENSG00000184009 | ENSMUSG00000062825 | 0.001 | 0.9622 | 0.001039285 | |||||||||||||||||||||
| ENSG00000090621 | ENSMUSG00000062093 | 0.0139 | 0.4737 | 0.029343466 | ENSMUSG00000039530 | ENSG00000104723 | 0.0122 | 0.4546 | 0.02683678 | ENSG00000116991 | ENSMUSG00000001995 | 0.0419 | 0.9637 | 0.043478261 | |||||||||||||||||||||
| ENSG00000171885 | ENSMUSG00000024411 | 0.0367 | 0.4737 | 0.077475195 | ENSMUSG00000057897 | ENSG00000058404 | 0.0064 | 0.4547 | 0.014075214 | ENSG00000140511 | ENSMUSG00000030606 | 0.1096 | 0.9773 | 0.112145708 | |||||||||||||||||||||
| ENSG00000150471 | ENSMUSG00000037605 | 0.019 | 0.4742 | 0.040067482 | ENSMUSG00000045589 | ENSG00000136805 | 0.0409 | 0.4557 | 0.08975203 | ENSG00000104980 | ENSMUSG00000002949 | 0.0521 | 0.9819 | 0.053060393 | |||||||||||||||||||||
| ENSG00000161791 | ENSMUSG00000023008 | 0.0226 | 0.4743 | 0.047649167 | ENSMUSG00000074785 | ENSG00000136040 | 0.0494 | 0.456 | 0.108333333 | ENSG00000185340 | ENSMUSG00000034201 | 0.0827 | 0.9828 | 0.084147334 | |||||||||||||||||||||
| ENSG00000138674 | ENSMUSG00000035325 | 0.0432 | 0.4748 | 0.090985678 | ENSMUSG00000073557 | ENSG00000077157 | 0.0628 | 0.4561 | 0.137689103 | ENSG00000096433 | ENSMUSG00000042644 | 0.0208 | 0.9861 | 0.021093195 | |||||||||||||||||||||
| ENSG00000116106 | ENSMUSG00000026235 | 0.0069 | 0.4754 | 0.014514093 | ENSMUSG00000032883 | ENSG00000123983 | 0.0362 | 0.4565 | 0.079299014 | ENSG00000161970 | ENSMUSG00000063754 | 0.0065 | 0.9896 | 0.00656831 | |||||||||||||||||||||
| ENSG00000125868 | ENSMUSG00000015932 | 0.0188 | 0.4755 | 0.039537329 | ENSMUSG00000023055 | ENSG00000012822 | 0.0561 | 0.4568 | 0.122810858 | ENSG00000138311 | ENSMUSG00000037855 | 0.2516 | 0.996 | 0.252610442 | |||||||||||||||||||||
| ENSG00000182389 | ENSMUSG00000017412 | 0.008 | 0.4756 | 0.016820858 | ENSMUSG00000055782 | ENSG00000173208 | 0.0302 | 0.4578 | 0.065967671 | ENSG00000158445 | ENSMUSG00000050556 | 0.0293 | 1.0026 | 0.029224018 | |||||||||||||||||||||
| ENSG00000152556 | ENSMUSG00000033065 | 0.0103 | 0.4757 | 0.021652302 | ENSMUSG00000067629 | ENSG00000197283 | 0.0111 | 0.4581 | 0.024230517 | ENSG00000158792 | ENSMUSG00000033594 | 0.0849 | 1.0277 | 0.082611657 | |||||||||||||||||||||
| ENSG00000141503 | ENSMUSG00000020827 | 0.0118 | 0.476 | 0.024789916 | ENSMUSG00000028078 | ENSG00000170390 | 0.0299 | 0.4582 | 0.065255347 | ENSG00000135916 | ENSMUSG00000026223 | 0.0291 | 1.0311 | 0.028222287 | |||||||||||||||||||||
| ENSG00000175198 | ENSMUSG00000041650 | 0.0574 | 0.4767 | 0.12041116 | ENSMUSG00000028410 | ENSG00000086061 | 0.0022 | 0.4583 | 0.004800349 | ENSG00000170889 | ENSMUSG00000006333 | 0 | 1.0328 | 0 | |||||||||||||||||||||
| ENSG00000135821 | ENSMUSG00000026473 | 0.0267 | 0.4769 | 0.05598658 | ENSMUSG00000015002 | ENSG00000132294 | 0.0132 | 0.4593 | 0.028739386 | ENSG00000172247 | ENSMUSG00000040794 | 0.021 | 1.039 | 0.020211742 | |||||||||||||||||||||
| ENSG00000115944 | ENSMUSG00000068537 | 0.1855 | 0.4771 | 0.388807378 | ENSMUSG00000063160 | ENSG00000105245 | 0.0222 | 0.4593 | 0.048334422 | ENSG00000117245 | ENSMUSG00000028758 | 0.1058 | 1.0469 | 0.101060273 | |||||||||||||||||||||
| ENSG00000077549 | ENSMUSG00000028745 | 0.0497 | 0.4775 | 0.10408377 | ENSMUSG00000027286 | ENSG00000180979 | 0.0226 | 0.4594 | 0.049194602 | ENSG00000143740 | ENSMUSG00000009894 | 0.1995 | 1.0484 | 0.190289966 | |||||||||||||||||||||
| ENSG00000215305 | ENSMUSG00000027411 | 0.0154 | 0.4777 | 0.032237806 | ENSMUSG00000036053 | ENSG00000157827 | 0.0346 | 0.4596 | 0.075282855 | ENSG00000124831 | ENSMUSG00000026305 | 0.3234 | 1.0524 | 0.307297605 | |||||||||||||||||||||
| ENSG00000154229 | ENSMUSG00000050965 | 0.0027 | 0.4785 | 0.005642633 | ENSMUSG00000021709 | ENSG00000112851 | 0.0504 | 0.4606 | 0.109422492 | ENSG00000185989 | ENSMUSG00000031453 | 0.0226 | 1.0637 | 0.021246592 | |||||||||||||||||||||
| ENSG00000105926 | ENSMUSG00000038388 | 0.0077 | 0.4788 | 0.016081871 | ENSMUSG00000045967 | ENSG00000151025 | 0.0687 | 0.4607 | 0.149120903 | ENSG00000175274 | ENSMUSG00000068735 | 0.0414 | 1.071 | 0.038655462 | |||||||||||||||||||||
| ENSG00000166963 | ENSMUSG00000027254 | 0.1106 | 0.4788 | 0.230994152 | ENSMUSG00000028035 | ENSG00000162616 | 0.0285 | 0.4608 | 0.061848958 | ENSG00000197530 | ENSMUSG00000029060 | 0.0377 | 1.0822 | 0.034836444 | |||||||||||||||||||||
| ENSG00000136267 | ENSMUSG00000036095 | 0.0155 | 0.479 | 0.032359081 | ENSMUSG00000018470 | ENSG00000170049 | 0.0345 | 0.4611 | 0.07482108 | ENSG00000196358 | ENSMUSG00000035513 | 0.0256 | 1.096 | 0.023357664 | |||||||||||||||||||||
| ENSG00000160469 | ENSMUSG00000035390 | 0.004 | 0.4793 | 0.008345504 | ENSMUSG00000028447 | ENSG00000137100 | 0.031 | 0.4614 | 0.067186823 | ENSG00000188191 | ENSMUSG00000025855 | 0.0274 | 1.0984 | 0.024945375 | |||||||||||||||||||||
| ENSG00000131100 | ENSMUSG00000019210 | 0.0061 | 0.4794 | 0.012724239 | ENSMUSG00000024983 | ENSG00000151532 | 0.0235 | 0.4617 | 0.050898852 | ENSG00000157911 | ENSMUSG00000029047 | 0.1056 | 1.1063 | 0.095453313 | |||||||||||||||||||||
| ENSG00000135624 | ENSMUSG00000030007 | 0.0219 | 0.4797 | 0.045653533 | ENSMUSG00000030516 | ENSG00000104067 | 0.0465 | 0.4619 | 0.100671141 | ENSG00000187664 | ENSMUSG00000007594 | 0.0456 | 1.1147 | 0.040907868 | |||||||||||||||||||||
| ENSG00000149273 | ENSMUSG00000030744 | 0.0019 | 0.4799 | 0.003959158 | ENSMUSG00000041444 | ENSG00000134909 | 0.0672 | 0.4619 | 0.145486036 | ENSG00000171700 | ENSMUSG00000002458 | 0.0405 | 1.1504 | 0.035205146 | |||||||||||||||||||||
| ENSG00000007402 | ENSMUSG00000010066 | 0.0155 | 0.4799 | 0.032298395 | ENSMUSG00000007411 | ENSG00000075413 | 0.0149 | 0.462 | 0.032251082 | ENSG00000116663 | ENSMUSG00000055401 | 0.1966 | 1.1544 | 0.17030492 | |||||||||||||||||||||
| ENSG00000151239 | ENSMUSG00000022451 | 0.0256 | 0.48 | 0.053333333 | ENSMUSG00000021248 | ENSG00000170348 | 0.0335 | 0.4625 | 0.072432432 | ENSG00000105088 | ENSMUSG00000032172 | 0.0277 | 1.1615 | 0.023848472 | |||||||||||||||||||||
| ENSG00000101333 | ENSMUSG00000039943 | 0.0221 | 0.4806 | 0.045984186 | ENSMUSG00000026014 | ENSG00000173166 | 0.0502 | 0.4627 | 0.108493624 | ENSG00000161970 | ENSMUSG00000060938 | 0 | 1.1622 | 0 | |||||||||||||||||||||
| ENSG00000135318 | ENSMUSG00000032420 | 0.0752 | 0.4812 | 0.156275977 | ENSMUSG00000052428 | ENSG00000143183 | 0 | 0.4633 | 0 | ENSG00000099308 | ENSMUSG00000031833 | 0.0552 | 1.1683 | 0.047248138 | |||||||||||||||||||||
| ENSG00000156642 | ENSMUSG00000032336 | 0.0968 | 0.4823 | 0.200704955 | ENSMUSG00000040037 | ENSG00000172260 | 0.0249 | 0.4634 | 0.053733276 | ENSG00000073150 | ENSMUSG00000058441 | 0.0321 | 1.1734 | 0.0273564 | |||||||||||||||||||||
| ENSG00000005810 | ENSMUSG00000033004 | 0.0131 | 0.4827 | 0.02713901 | ENSMUSG00000020598 | ENSG00000091129 | 0.0354 | 0.4634 | 0.076391886 | ENSG00000184992 | ENSMUSG00000037905 | 0.1016 | 1.1931 | 0.085156315 | |||||||||||||||||||||
| ENSG00000167792 | ENSMUSG00000037916 | 0.0195 | 0.4828 | 0.040389395 | ENSMUSG00000001924 | ENSG00000130985 | 0.0228 | 0.4641 | 0.049127343 | ENSG00000125912 | ENSMUSG00000020238 | 0.0321 | 1.2 | 0.02675 | |||||||||||||||||||||
| ENSG00000113657 | ENSMUSG00000024501 | 0.0072 | 0.483 | 0.014906832 | ENSMUSG00000007036 | ENSG00000204427 | 0.0193 | 0.4644 | 0.041559001 | ENSG00000197177 | ENSMUSG00000025475 | 0.0905 | 1.2028 | 0.075241104 | |||||||||||||||||||||
| ENSG00000066084 | ENSMUSG00000023026 | 0.0099 | 0.4831 | 0.020492652 | ENSMUSG00000038916 | ENSG00000255330 | 0.0454 | 0.4644 | 0.097760551 | ENSG00000061337 | ENSMUSG00000036306 | 0.0466 | 1.205 | 0.038672199 | |||||||||||||||||||||
| ENSG00000004660 | ENSMUSG00000020785 | 0.0317 | 0.4835 | 0.065563599 | ENSMUSG00000038916 | ENSG00000214338 | 0.0454 | 0.4644 | 0.097760551 | ENSG00000051128 | ENSMUSG00000003573 | 0.0564 | 1.2201 | 0.046225719 | |||||||||||||||||||||
| ENSG00000074696 | ENSMUSG00000033629 | 0.0415 | 0.4835 | 0.085832472 | ENSMUSG00000029153 | ENSG00000145247 | 0.0783 | 0.4644 | 0.168604651 | ENSG00000152969 | ENSMUSG00000063646 | 0.0349 | 1.2232 | 0.02853172 | |||||||||||||||||||||
| ENSG00000136143 | ENSMUSG00000022110 | 0.0458 | 0.4842 | 0.094589013 | ENSMUSG00000063801 | ENSG00000157823 | 0 | 0.4651 | 0 | ENSG00000130813 | ENSMUSG00000038884 | 0.0208 | 1.2248 | 0.016982364 | |||||||||||||||||||||
| ENSG00000138246 | ENSMUSG00000032560 | 0.0159 | 0.4846 | 0.032810565 | ENSMUSG00000028149 | ENSG00000138698 | 0.0242 | 0.4654 | 0.051998281 | ENSG00000176108 | ENSMUSG00000025371 | 0.0306 | 1.2719 | 0.024058495 | |||||||||||||||||||||
| ENSG00000168291 | ENSMUSG00000021748 | 0.0313 | 0.4852 | 0.064509481 | ENSMUSG00000036989 | ENSG00000110171 | 0.007 | 0.466 | 0.015021459 | ENSG00000164114 | ENSMUSG00000033900 | 0.209 | 1.304 | 0.160276074 | |||||||||||||||||||||
| ENSG00000176658 | ENSMUSG00000035441 | 0.0108 | 0.4856 | 0.022240527 | ENSMUSG00000035864 | ENSG00000067715 | 0.0112 | 0.4661 | 0.024029178 | ENSG00000142408 | ENSMUSG00000053395 | 0.013 | 1.3586 | 0.009568674 | |||||||||||||||||||||
| ENSG00000183775 | ENSMUSG00000051401 | 0.017 | 0.486 | 0.034979424 | ENSMUSG00000021704 | ENSG00000177034 | 0.0536 | 0.4673 | 0.114701477 | ENSG00000099822 | ENSMUSG00000020331 | 0.0276 | 1.3785 | 0.020021763 | |||||||||||||||||||||
| ENSG00000023734 | ENSMUSG00000030224 | 0.0105 | 0.4861 | 0.021600494 | ENSMUSG00000042492 | ENSG00000169221 | 0.0517 | 0.4677 | 0.110540945 | ENSG00000118898 | ENSMUSG00000039457 | 0.0625 | 1.3948 | 0.044809292 | |||||||||||||||||||||
| ENSG00000117868 | ENSMUSG00000021171 | 0.039 | 0.487 | 0.080082136 | ENSMUSG00000009647 | ENSG00000156026 | 0.0196 | 0.4678 | 0.041898247 | ENSG00000174775 | ENSMUSG00000025499 | 0 | 1.4086 | 0 | |||||||||||||||||||||
| ENSG00000140992 | ENSMUSG00000024122 | 0.0244 | 0.4876 | 0.050041017 | ENSMUSG00000031626 | ENSG00000154556 | 0.0368 | 0.4678 | 0.078666097 | ENSG00000119242 | ENSMUSG00000037979 | 0.068 | 1.5494 | 0.043887957 | |||||||||||||||||||||
| ENSG00000124164 | ENSMUSG00000054455 | 0.0488 | 0.4876 | 0.100082034 | ENSMUSG00000045038 | ENSG00000171132 | 0.0104 | 0.4679 | 0.022226972 | ENSG00000125741 | ENSMUSG00000052214 | 0.1685 | 1.7863 | 0.09432906 | |||||||||||||||||||||
| ENSG00000000419 | ENSMUSG00000078919 | 0.0536 | 0.4881 | 0.109813563 | ENSMUSG00000028969 | ENSG00000164885 | 0.0016 | 0.4681 | 0.003418073 | ENSG00000124575 | ENSMUSG00000052565 | 0.0729 | 1.8636 | 0.039117836 | |||||||||||||||||||||
| ENSG00000149925 | ENSMUSG00000063129 | 0.0371 | 0.4886 | 0.075931232 | ENSMUSG00000026728 | ENSG00000026025 | 0.0111 | 0.4681 | 0.023712882 | ENSG00000177733 | ENSMUSG00000007836 | 0.0322 | 1.9088 | 0.016869237 | |||||||||||||||||||||
| ENSG00000170248 | ENSMUSG00000032504 | 0.0271 | 0.4888 | 0.055441899 | ENSMUSG00000031059 | ENSG00000147123 | 0.1345 | 0.4682 | 0.287270397 | ENSG00000142444 | ENSMUSG00000048429 | 0.0959 | 2.1873 | 0.043844009 | |||||||||||||||||||||
| ENSG00000100201 | ENSMUSG00000055065 | 0.0057 | 0.4889 | 0.011658826 | ENSMUSG00000004110 | ENSG00000198216 | 0.0156 | 0.4688 | 0.033276451 | ENSG00000141933 | ENSMUSG00000020308 | 0.0736 | 2.203 | 0.033408988 | |||||||||||||||||||||
| ENSG00000008086 | ENSMUSG00000031292 | 0.0208 | 0.4894 | 0.042501022 | ENSMUSG00000040022 | ENSG00000107560 | 0.0413 | 0.4692 | 0.088022165 | ENSG00000198576 | ENSMUSG00000022602 | 0.0324 | 2.2847 | 0.014181293 | |||||||||||||||||||||
| ENSG00000078018 | ENSMUSG00000015222 | 0.0406 | 0.4898 | 0.082890976 | ENSMUSG00000022752 | ENSG00000154174 | 0.0429 | 0.4694 | 0.091393268 | ENSG00000176022 | ENSMUSG00000050796 | 0.0793 | 2.3042 | 0.034415415 | |||||||||||||||||||||
| ENSG00000181013 | ENSMUSG00000090107 | 0.3441 | 0.4898 | 0.702531646 | ENSMUSG00000025980 | ENSG00000144381 | 0.0121 | 0.4699 | 0.02575016 | ENSG00000187837 | ENSMUSG00000036181 | 0.061 | 2.3826 | 0.025602283 | |||||||||||||||||||||
| ENSG00000116161 | ENSMUSG00000014226 | 0.0305 | 0.4899 | 0.062257604 | ENSMUSG00000029131 | ENSG00000105993 | 0.089 | 0.4705 | 0.189160468 | ENSG00000129951 | ENSMUSG00000035835 | 0.0523 | 2.477 | 0.021114251 | |||||||||||||||||||||
| ENSG00000064042 | ENSMUSG00000037736 | 0.0685 | 0.4899 | 0.139824454 | ENSMUSG00000026335 | ENSG00000145730 | 0.0475 | 0.4706 | 0.100934977 | ENSG00000115268 | ENSMUSG00000063457 | 0.0028 | 2.8141 | 0.00099499 | |||||||||||||||||||||
| ENSG00000104093 | ENSMUSG00000041268 | 0.0368 | 0.49 | 0.075102041 | ENSMUSG00000019943 | ENSG00000070961 | 0.0023 | 0.4708 | 0.004885302 | ENSG00000115268 | ENSMUSG00000090538 | 0.0664 | 3.4555 | 0.019215743 | |||||||||||||||||||||
| ENSG00000115365 | ENSMUSG00000026000 | 0.0442 | 0.4901 | 0.090185676 | ENSMUSG00000031673 | ENSG00000140937 | 0.0126 | 0.4711 | 0.026745914 | ENSG00000130479 | ENSMUSG00000019261 | 0.1929 | 4.4571 | 0.043279262 | |||||||||||||||||||||
| ENSG00000148943 | ENSMUSG00000027162 | 0 | 0.4904 | 0 | ENSMUSG00000021613 | ENSG00000145681 | 0.0181 | 0.4711 | 0.038420717 | ENSG00000184357 | ENSMUSG00000058773 | 0.0421 | 6.1729 | 0.006820133 | |||||||||||||||||||||
| ENSG00000167578 | ENSMUSG00000053291 | 0.0042 | 0.4904 | 0.008564437 | ENSMUSG00000029916 | ENSG00000006530 | 0.0419 | 0.4711 | 0.088940777 | ENSG00000184357 | ENSMUSG00000091230 | 0.1728 | 7.0301 | 0.02458002 | |||||||||||||||||||||
| ENSG00000105197 | ENSMUSG00000003438 | 0.0421 | 0.4904 | 0.085848287 | ENSMUSG00000049583 | ENSG00000168959 | 0.0179 | 0.4715 | 0.037963945 | ENSG00000178695 | ENSMUSG00000041633 | 0.2745 | 26.69 | 0.010284751 | |||||||||||||||||||||
| ENSG00000144566 | ENSMUSG00000017831 | 0.0083 | 0.4923 | 0.016859638 | ENSMUSG00000004264 | ENSG00000215021 | 0.0005 | 0.4716 | 0.001060221 | ||||||||||||||||||||||||||
| ENSG00000129562 | ENSMUSG00000022174 | 0 | 0.4926 | 0 | ENSMUSG00000031207 | ENSG00000147065 | 0.006 | 0.4717 | 0.012719949 | ||||||||||||||||||||||||||
| ENSG00000153956 | ENSMUSG00000040118 | 0.0192 | 0.4929 | 0.038953135 | ENSMUSG00000005483 | ENSG00000132002 | 0.0211 | 0.4718 | 0.04472234 | ||||||||||||||||||||||||||
| ENSG00000079785 | ENSMUSG00000037149 | 0.0118 | 0.4935 | 0.023910841 | ENSMUSG00000024426 | ENSG00000137343 | 0.046 | 0.4724 | 0.097375106 | ||||||||||||||||||||||||||
| ENSG00000106554 | ENSMUSG00000053768 | 0.0514 | 0.4943 | 0.103985434 | ENSMUSG00000033768 | ENSG00000110076 | 0.0078 | 0.4736 | 0.016469595 | ||||||||||||||||||||||||||
| ENSG00000113013 | ENSMUSG00000024359 | 0.0076 | 0.4944 | 0.015372168 | ENSMUSG00000024411 | ENSG00000171885 | 0.0367 | 0.4737 | 0.077475195 | ||||||||||||||||||||||||||
| ENSG00000126777 | ENSMUSG00000021843 | 0.0896 | 0.4946 | 0.18115649 | ENSMUSG00000048388 | ENSG00000144369 | 0.0587 | 0.4741 | 0.123813541 | ||||||||||||||||||||||||||
| ENSG00000160145 | ENSMUSG00000061751 | 0.0049 | 0.4947 | 0.009904993 | ENSMUSG00000037605 | ENSG00000150471 | 0.019 | 0.4742 | 0.040067482 | ||||||||||||||||||||||||||
| ENSG00000163468 | ENSMUSG00000001416 | 0.0137 | 0.4951 | 0.027671178 | ENSMUSG00000023008 | ENSG00000161791 | 0.0226 | 0.4743 | 0.047649167 | ||||||||||||||||||||||||||
| ENSG00000107020 | ENSMUSG00000016495 | 0.0919 | 0.4952 | 0.185581583 | ENSMUSG00000040028 | ENSG00000066044 | 0.0069 | 0.475 | 0.014526316 | ||||||||||||||||||||||||||
| ENSG00000159082 | ENSMUSG00000022973 | 0.0522 | 0.4954 | 0.105369398 | ENSMUSG00000018796 | ENSG00000151726 | 0.0807 | 0.4753 | 0.169787503 | ||||||||||||||||||||||||||
| ENSG00000143933 | ENSMUSG00000036438 | 0 | 0.496 | 0 | ENSMUSG00000026235 | ENSG00000116106 | 0.0069 | 0.4754 | 0.014514093 | ||||||||||||||||||||||||||
| ENSG00000084774 | ENSMUSG00000013629 | 0.0236 | 0.4962 | 0.047561467 | ENSMUSG00000015932 | ENSG00000125868 | 0.0188 | 0.4755 | 0.039537329 | ||||||||||||||||||||||||||
| ENSG00000136193 | ENSMUSG00000019124 | 0.0618 | 0.4963 | 0.124521459 | ENSMUSG00000017412 | ENSG00000182389 | 0.008 | 0.4756 | 0.016820858 | ||||||||||||||||||||||||||
| ENSG00000127463 | ENSMUSG00000078517 | 0.0337 | 0.4965 | 0.067875126 | ENSMUSG00000033065 | ENSG00000152556 | 0.0103 | 0.4757 | 0.021652302 | ||||||||||||||||||||||||||
| ENSG00000131462 | ENSMUSG00000035198 | 0.0047 | 0.4967 | 0.009462452 | ENSMUSG00000040374 | ENSG00000164751 | 0.0652 | 0.4758 | 0.137032367 | ||||||||||||||||||||||||||
| ENSG00000047249 | ENSMUSG00000033793 | 0.0065 | 0.4968 | 0.013083736 | ENSMUSG00000031320 | ENSG00000198034 | 0 | 0.476 | 0 | ||||||||||||||||||||||||||
| ENSG00000136044 | ENSMUSG00000020263 | 0.0399 | 0.4969 | 0.080297847 | ENSMUSG00000020827 | ENSG00000141503 | 0.0118 | 0.476 | 0.024789916 | ||||||||||||||||||||||||||
| ENSG00000115252 | ENSMUSG00000059173 | 0.038 | 0.4975 | 0.07638191 | ENSMUSG00000026473 | ENSG00000135821 | 0.0267 | 0.4769 | 0.05598658 | ||||||||||||||||||||||||||
| ENSG00000143847 | ENSMUSG00000026458 | 0.0365 | 0.498 | 0.073293173 | ENSMUSG00000028745 | ENSG00000077549 | 0.0497 | 0.4775 | 0.10408377 | ||||||||||||||||||||||||||
| ENSG00000114391 | ENSMUSG00000091285 | 0.0662 | 0.4982 | 0.132878362 | ENSMUSG00000027411 | ENSG00000215305 | 0.0154 | 0.4777 | 0.032237806 | ||||||||||||||||||||||||||
| ENSG00000138279 | ENSMUSG00000021814 | 0.0394 | 0.4984 | 0.07905297 | ENSMUSG00000029621 | ENSG00000241685 | 0.0074 | 0.4784 | 0.015468227 | ||||||||||||||||||||||||||
| ENSG00000136717 | ENSMUSG00000024381 | 0.02 | 0.4994 | 0.040048058 | ENSMUSG00000038388 | ENSG00000105926 | 0.0077 | 0.4788 | 0.016081871 | ||||||||||||||||||||||||||
| ENSG00000079841 | ENSMUSG00000041670 | 0.0597 | 0.4994 | 0.119543452 | ENSMUSG00000027254 | ENSG00000166963 | 0.1106 | 0.4788 | 0.230994152 | ||||||||||||||||||||||||||
| ENSG00000135655 | ENSMUSG00000020124 | 0.0111 | 0.4995 | 0.022222222 | ENSMUSG00000036095 | ENSG00000136267 | 0.0155 | 0.479 | 0.032359081 | ||||||||||||||||||||||||||
| ENSG00000114948 | ENSMUSG00000025964 | 0.0481 | 0.4999 | 0.096219244 | ENSMUSG00000009418 | ENSG00000134369 | 0.0173 | 0.479 | 0.03611691 | ||||||||||||||||||||||||||
| ENSG00000102003 | ENSMUSG00000031144 | 0.0216 | 0.5 | 0.0432 | ENSMUSG00000035390 | ENSG00000160469 | 0.004 | 0.4793 | 0.008345504 | ||||||||||||||||||||||||||
| ENSG00000060971 | ENSMUSG00000010651 | 0.0789 | 0.5 | 0.1578 | ENSMUSG00000019210 | ENSG00000131100 | 0.0061 | 0.4794 | 0.012724239 | ||||||||||||||||||||||||||
| ENSG00000137575 | ENSMUSG00000028249 | 0.0504 | 0.5001 | 0.100779844 | ENSMUSG00000030007 | ENSG00000135624 | 0.0219 | 0.4797 | 0.045653533 | ||||||||||||||||||||||||||
| ENSG00000198677 | ENSMUSG00000033991 | 0.0856 | 0.5002 | 0.171131547 | ENSMUSG00000030744 | ENSG00000149273 | 0.0019 | 0.4799 | 0.003959158 | ||||||||||||||||||||||||||
| ENSG00000166913 | ENSMUSG00000018326 | 0.0056 | 0.5013 | 0.011170956 | ENSMUSG00000010066 | ENSG00000007402 | 0.0155 | 0.4799 | 0.032298395 | ||||||||||||||||||||||||||
| ENSG00000153904 | ENSMUSG00000028194 | 0.0317 | 0.5016 | 0.063197767 | ENSMUSG00000022451 | ENSG00000151239 | 0.0256 | 0.48 | 0.053333333 | ||||||||||||||||||||||||||
| ENSG00000159164 | ENSMUSG00000038486 | 0.0056 | 0.502 | 0.011155378 | ENSMUSG00000039943 | ENSG00000101333 | 0.0221 | 0.4806 | 0.045984186 | ||||||||||||||||||||||||||
| ENSG00000204843 | ENSMUSG00000031865 | 0.0145 | 0.502 | 0.028884462 | ENSMUSG00000042766 | ENSG00000163462 | 0.0102 | 0.4807 | 0.021219056 | ||||||||||||||||||||||||||
| ENSG00000064787 | ENSMUSG00000013523 | 0.2772 | 0.5022 | 0.551971326 | ENSMUSG00000000276 | ENSG00000153933 | 0.0456 | 0.4811 | 0.094782789 | ||||||||||||||||||||||||||
| ENSG00000163032 | ENSMUSG00000054459 | 0 | 0.5023 | 0 | ENSMUSG00000032420 | ENSG00000135318 | 0.0752 | 0.4812 | 0.156275977 | ||||||||||||||||||||||||||
| ENSG00000065150 | ENSMUSG00000030662 | 0.0082 | 0.5029 | 0.016305429 | ENSMUSG00000032336 | ENSG00000156642 | 0.0968 | 0.4823 | 0.200704955 | ||||||||||||||||||||||||||
| ENSG00000213672 | ENSMUSG00000032598 | 0.054 | 0.5029 | 0.107377212 | ENSMUSG00000022601 | ENSG00000066422 | 0.0575 | 0.4825 | 0.119170984 | ||||||||||||||||||||||||||
| ENSG00000116688 | ENSMUSG00000029020 | 0.0238 | 0.5035 | 0.047269116 | ENSMUSG00000033004 | ENSG00000005810 | 0.0131 | 0.4827 | 0.02713901 | ||||||||||||||||||||||||||
| ENSG00000176956 | ENSMUSG00000022577 | 0.0532 | 0.5035 | 0.105660377 | ENSMUSG00000037916 | ENSG00000167792 | 0.0195 | 0.4828 | 0.040389395 | ||||||||||||||||||||||||||
| ENSG00000136842 | ENSMUSG00000028328 | 0.0143 | 0.5037 | 0.028389915 | ENSMUSG00000025816 | ENSG00000065665 | 0.0005 | 0.4829 | 0.001035411 | ||||||||||||||||||||||||||
| ENSG00000170759 | ENSMUSG00000006740 | 0.0143 | 0.5038 | 0.028384279 | ENSMUSG00000024501 | ENSG00000113657 | 0.0072 | 0.483 | 0.014906832 | ||||||||||||||||||||||||||
| ENSG00000112640 | ENSMUSG00000059409 | 0.0136 | 0.5043 | 0.026968075 | ENSMUSG00000023026 | ENSG00000066084 | 0.0099 | 0.4831 | 0.020492652 | ||||||||||||||||||||||||||
| ENSG00000141252 | ENSMUSG00000017288 | 0.0158 | 0.5049 | 0.031293325 | ENSMUSG00000033629 | ENSG00000074696 | 0.0415 | 0.4835 | 0.085832472 | ||||||||||||||||||||||||||
| ENSG00000117448 | ENSMUSG00000028692 | 0.0356 | 0.5052 | 0.070467142 | ENSMUSG00000034613 | ENSG00000111110 | 0.0158 | 0.4837 | 0.032664875 | ||||||||||||||||||||||||||
| ENSG00000160007 | ENSMUSG00000058230 | 0.0109 | 0.5054 | 0.021567076 | ENSMUSG00000042535 | ENSG00000100226 | 0.0127 | 0.4842 | 0.026228831 | ||||||||||||||||||||||||||
| ENSG00000071967 | ENSMUSG00000027015 | 0.1663 | 0.5054 | 0.3290463 | ENSMUSG00000022110 | ENSG00000136143 | 0.0458 | 0.4842 | 0.094589013 | ||||||||||||||||||||||||||
| ENSG00000165629 | ENSMUSG00000025781 | 0.044 | 0.5055 | 0.087042532 | ENSMUSG00000028760 | ENSG00000075151 | 0.0712 | 0.4842 | 0.147046675 | ||||||||||||||||||||||||||
| ENSG00000151835 | ENSMUSG00000048279 | 0.0663 | 0.5055 | 0.13115727 | ENSMUSG00000032560 | ENSG00000138246 | 0.0159 | 0.4846 | 0.032810565 | ||||||||||||||||||||||||||
| ENSG00000112855 | ENSMUSG00000019143 | 0.0912 | 0.5056 | 0.180379747 | ENSMUSG00000021748 | ENSG00000168291 | 0.0313 | 0.4852 | 0.064509481 | ||||||||||||||||||||||||||
| ENSG00000134684 | ENSMUSG00000028811 | 0.0176 | 0.5058 | 0.034796362 | ENSMUSG00000045348 | ENSG00000166924 | 0.0396 | 0.4852 | 0.081615829 | ||||||||||||||||||||||||||
| ENSG00000203879 | ENSMUSG00000015291 | 0.0092 | 0.5061 | 0.018178226 | ENSMUSG00000035441 | ENSG00000176658 | 0.0108 | 0.4856 | 0.022240527 | ||||||||||||||||||||||||||
| ENSG00000137106 | ENSMUSG00000035637 | 0.0764 | 0.5064 | 0.150868878 | ENSMUSG00000051401 | ENSG00000183775 | 0.017 | 0.486 | 0.034979424 | ||||||||||||||||||||||||||
| ENSG00000099204 | ENSMUSG00000025085 | 0.052 | 0.5067 | 0.102624827 | ENSMUSG00000059439 | ENSG00000141376 | 0.0151 | 0.4863 | 0.031050792 | ||||||||||||||||||||||||||
| ENSG00000107897 | ENSMUSG00000026781 | 0.1025 | 0.5087 | 0.201494004 | ENSMUSG00000040225 | ENSG00000117523 | 0.0702 | 0.4869 | 0.144177449 | ||||||||||||||||||||||||||
| ENSG00000168172 | ENSMUSG00000037234 | 0.0102 | 0.5088 | 0.02004717 | ENSMUSG00000027406 | ENSG00000101365 | 0.0311 | 0.4874 | 0.063807961 | ||||||||||||||||||||||||||
| ENSG00000090863 | ENSMUSG00000003316 | 0.0171 | 0.509 | 0.033595285 | ENSMUSG00000024122 | ENSG00000140992 | 0.0244 | 0.4876 | 0.050041017 | ||||||||||||||||||||||||||
| ENSG00000007237 | ENSMUSG00000033066 | 0.0126 | 0.5091 | 0.024749558 | ENSMUSG00000054455 | ENSG00000124164 | 0.0488 | 0.4876 | 0.100082034 | ||||||||||||||||||||||||||
| ENSG00000155511 | ENSMUSG00000020524 | 0.0056 | 0.5092 | 0.010997643 | ENSMUSG00000078919 | ENSG00000000419 | 0.0536 | 0.4881 | 0.109813563 | ||||||||||||||||||||||||||
| ENSG00000142235 | ENSMUSG00000062044 | 0.0507 | 0.5098 | 0.099450765 | ENSMUSG00000055065 | ENSG00000100201 | 0.0057 | 0.4889 | 0.011658826 | ||||||||||||||||||||||||||
| ENSG00000150768 | ENSMUSG00000000168 | 0.0823 | 0.5099 | 0.161404197 | ENSMUSG00000031292 | ENSG00000008086 | 0.0208 | 0.4894 | 0.042501022 | ||||||||||||||||||||||||||
| ENSG00000151552 | ENSMUSG00000015806 | 0.0319 | 0.51 | 0.06254902 | ENSMUSG00000015222 | ENSG00000078018 | 0.0406 | 0.4898 | 0.082890976 | ||||||||||||||||||||||||||
| ENSG00000135046 | ENSMUSG00000024659 | 0.0626 | 0.5101 | 0.122721035 | ENSMUSG00000041268 | ENSG00000104093 | 0.0368 | 0.49 | 0.075102041 | ||||||||||||||||||||||||||
| ENSG00000066032 | ENSMUSG00000063063 | 0.0005 | 0.5102 | 0.000980008 | ENSMUSG00000026000 | ENSG00000115365 | 0.0442 | 0.4901 | 0.090185676 | ||||||||||||||||||||||||||
| ENSG00000171914 | ENSMUSG00000052698 | 0.0062 | 0.5102 | 0.012152097 | ENSMUSG00000027162 | ENSG00000148943 | 0 | 0.4904 | 0 | ||||||||||||||||||||||||||
| ENSG00000058272 | ENSMUSG00000019907 | 0.0429 | 0.5106 | 0.084018801 | ENSMUSG00000027799 | ENSG00000172915 | 0.0125 | 0.4914 | 0.025437525 | ||||||||||||||||||||||||||
| ENSG00000197959 | ENSMUSG00000040265 | 0.0099 | 0.5111 | 0.019369986 | ENSMUSG00000026074 | ENSG00000071054 | 0.0072 | 0.4916 | 0.014646054 | ||||||||||||||||||||||||||
| ENSG00000073417 | ENSMUSG00000025584 | 0.1218 | 0.5117 | 0.238030096 | ENSMUSG00000017831 | ENSG00000144566 | 0.0083 | 0.4923 | 0.016859638 | ||||||||||||||||||||||||||
| ENSG00000158856 | ENSMUSG00000022099 | 0.0202 | 0.5122 | 0.03943772 | ENSMUSG00000022174 | ENSG00000129562 | 0 | 0.4926 | 0 | ||||||||||||||||||||||||||
| ENSG00000197386 | ENSMUSG00000029104 | 0.049 | 0.5122 | 0.095665756 | ENSMUSG00000037149 | ENSG00000079785 | 0.0118 | 0.4935 | 0.023910841 | ||||||||||||||||||||||||||
| ENSG00000080493 | ENSMUSG00000060961 | 0.0154 | 0.5123 | 0.030060511 | ENSMUSG00000041624 | ENSG00000152402 | 0.0501 | 0.4942 | 0.101375961 | ||||||||||||||||||||||||||
| ENSG00000145349 | ENSMUSG00000053819 | 0.0088 | 0.5124 | 0.017174083 | ENSMUSG00000053768 | ENSG00000106554 | 0.0514 | 0.4943 | 0.103985434 | ||||||||||||||||||||||||||
| ENSG00000109158 | ENSMUSG00000029211 | 0.0479 | 0.5126 | 0.093445181 | ENSMUSG00000024359 | ENSG00000113013 | 0.0076 | 0.4944 | 0.015372168 | ||||||||||||||||||||||||||
| ENSG00000131558 | ENSMUSG00000029763 | 0.02 | 0.5129 | 0.038993956 | ENSMUSG00000021843 | ENSG00000126777 | 0.0896 | 0.4946 | 0.18115649 | ||||||||||||||||||||||||||
| ENSG00000211455 | ENSMUSG00000001630 | 0.0111 | 0.514 | 0.021595331 | ENSMUSG00000061751 | ENSG00000160145 | 0.0049 | 0.4947 | 0.009904993 | ||||||||||||||||||||||||||
| ENSG00000155097 | ENSMUSG00000022295 | 0.0089 | 0.5141 | 0.017311807 | ENSMUSG00000001416 | ENSG00000163468 | 0.0137 | 0.4951 | 0.027671178 | ||||||||||||||||||||||||||
| ENSG00000164070 | ENSMUSG00000025757 | 0.0382 | 0.5143 | 0.074275715 | ENSMUSG00000022973 | ENSG00000159082 | 0.0522 | 0.4954 | 0.105369398 | ||||||||||||||||||||||||||
| ENSG00000060971 | ENSMUSG00000036138 | 0.0832 | 0.5145 | 0.161710398 | ENSMUSG00000036438 | ENSG00000143933 | 0 | 0.496 | 0 | ||||||||||||||||||||||||||
| ENSG00000147416 | ENSMUSG00000006273 | 0.0056 | 0.5148 | 0.010878011 | ENSMUSG00000078517 | ENSG00000127463 | 0.0337 | 0.4965 | 0.067875126 | ||||||||||||||||||||||||||
| ENSG00000125814 | ENSMUSG00000027438 | 0 | 0.5154 | 0 | ENSMUSG00000033793 | ENSG00000047249 | 0.0065 | 0.4968 | 0.013083736 | ||||||||||||||||||||||||||
| ENSG00000137824 | ENSMUSG00000070730 | 0.0822 | 0.5157 | 0.159394997 | ENSMUSG00000020263 | ENSG00000136044 | 0.0399 | 0.4969 | 0.080297847 | ||||||||||||||||||||||||||
| ENSG00000132153 | ENSMUSG00000032480 | 0.0115 | 0.516 | 0.022286822 | ENSMUSG00000026458 | ENSG00000143847 | 0.0365 | 0.498 | 0.073293173 | ||||||||||||||||||||||||||
| ENSG00000002549 | ENSMUSG00000039682 | 0.0478 | 0.5163 | 0.092581832 | ENSMUSG00000038555 | ENSG00000132563 | 0.0331 | 0.4986 | 0.06638588 | ||||||||||||||||||||||||||
| ENSG00000148341 | ENSMUSG00000026860 | 0.0149 | 0.5166 | 0.028842431 | ENSMUSG00000024381 | ENSG00000136717 | 0.02 | 0.4994 | 0.040048058 | ||||||||||||||||||||||||||
| ENSG00000205726 | ENSMUSG00000022957 | 0.0247 | 0.5166 | 0.047812621 | ENSMUSG00000041670 | ENSG00000079841 | 0.0597 | 0.4994 | 0.119543452 | ||||||||||||||||||||||||||
| ENSG00000169032 | ENSMUSG00000004936 | 0.0046 | 0.5169 | 0.008899207 | ENSMUSG00000026113 | ENSG00000040933 | 0.0159 | 0.4998 | 0.031812725 | ||||||||||||||||||||||||||
| ENSG00000120053 | ENSMUSG00000025190 | 0.0468 | 0.5173 | 0.090469747 | ENSMUSG00000025964 | ENSG00000114948 | 0.0481 | 0.4999 | 0.096219244 | ||||||||||||||||||||||||||
| ENSG00000105953 | ENSMUSG00000020456 | 0.0216 | 0.5175 | 0.04173913 | ENSMUSG00000031144 | ENSG00000102003 | 0.0216 | 0.5 | 0.0432 | ||||||||||||||||||||||||||
| ENSG00000204655 | ENSMUSG00000076439 | 0.0831 | 0.5175 | 0.16057971 | ENSMUSG00000028249 | ENSG00000137575 | 0.0504 | 0.5001 | 0.100779844 | ||||||||||||||||||||||||||
| ENSG00000125166 | ENSMUSG00000031672 | 0.025 | 0.5176 | 0.048299845 | ENSMUSG00000032046 | ENSG00000100997 | 0.0572 | 0.5006 | 0.114262885 | ||||||||||||||||||||||||||
| ENSG00000134278 | ENSMUSG00000024533 | 0.0359 | 0.5176 | 0.069358578 | ENSMUSG00000018326 | ENSG00000166913 | 0.0056 | 0.5013 | 0.011170956 | ||||||||||||||||||||||||||
| ENSG00000166411 | ENSMUSG00000032279 | 0.0117 | 0.5181 | 0.022582513 | ENSMUSG00000028556 | ENSG00000116641 | 0.0084 | 0.5016 | 0.016746411 | ||||||||||||||||||||||||||
| ENSG00000164904 | ENSMUSG00000053644 | 0.064 | 0.5181 | 0.123528276 | ENSMUSG00000028194 | ENSG00000153904 | 0.0317 | 0.5016 | 0.063197767 | ||||||||||||||||||||||||||
| ENSG00000164506 | ENSMUSG00000019790 | 0.0223 | 0.5182 | 0.043033578 | ENSMUSG00000038486 | ENSG00000159164 | 0.0056 | 0.502 | 0.011155378 | ||||||||||||||||||||||||||
| ENSG00000014123 | ENSMUSG00000040359 | 0.0497 | 0.5188 | 0.095797995 | ENSMUSG00000031865 | ENSG00000204843 | 0.0145 | 0.502 | 0.028884462 | ||||||||||||||||||||||||||
| ENSG00000149930 | ENSMUSG00000059981 | 0.0388 | 0.519 | 0.074759152 | ENSMUSG00000020828 | ENSG00000129219 | 0.0526 | 0.502 | 0.104780876 | ||||||||||||||||||||||||||
| ENSG00000116147 | ENSMUSG00000015829 | 0.0334 | 0.5191 | 0.064342131 | ENSMUSG00000029016 | ENSG00000011021 | 0.0146 | 0.5021 | 0.029077873 | ||||||||||||||||||||||||||
| ENSG00000155816 | ENSMUSG00000028354 | 0.1126 | 0.5191 | 0.216913889 | ENSMUSG00000013523 | ENSG00000064787 | 0.2772 | 0.5022 | 0.551971326 | ||||||||||||||||||||||||||
| ENSG00000213585 | ENSMUSG00000020402 | 0.0065 | 0.5192 | 0.01251926 | ENSMUSG00000054459 | ENSG00000163032 | 0 | 0.5023 | 0 | ||||||||||||||||||||||||||
| ENSG00000149925 | ENSMUSG00000030695 | 0.0101 | 0.5192 | 0.019453005 | ENSMUSG00000022103 | ENSG00000168546 | 0.0279 | 0.5026 | 0.055511341 | ||||||||||||||||||||||||||
| ENSG00000130396 | ENSMUSG00000068036 | 0.031 | 0.5195 | 0.059672762 | ENSMUSG00000030662 | ENSG00000065150 | 0.0082 | 0.5029 | 0.016305429 | ||||||||||||||||||||||||||
| ENSG00000133742 | ENSMUSG00000027556 | 0.134 | 0.5197 | 0.257841062 | ENSMUSG00000032598 | ENSG00000213672 | 0.054 | 0.5029 | 0.107377212 | ||||||||||||||||||||||||||
| ENSG00000150086 | ENSMUSG00000030209 | 0.0058 | 0.5199 | 0.011155992 | ENSMUSG00000029020 | ENSG00000116688 | 0.0238 | 0.5035 | 0.047269116 | ||||||||||||||||||||||||||
| ENSG00000109654 | ENSMUSG00000027993 | 0.0041 | 0.5205 | 0.007877041 | ENSMUSG00000022577 | ENSG00000176956 | 0.0532 | 0.5035 | 0.105660377 | ||||||||||||||||||||||||||
| ENSG00000164961 | ENSMUSG00000022350 | 0.0204 | 0.5205 | 0.039193084 | ENSMUSG00000028328 | ENSG00000136842 | 0.0143 | 0.5037 | 0.028389915 | ||||||||||||||||||||||||||
| ENSG00000067992 | ENSMUSG00000035232 | 0.0151 | 0.5213 | 0.028966046 | ENSMUSG00000006740 | ENSG00000170759 | 0.0143 | 0.5038 | 0.028384279 | ||||||||||||||||||||||||||
| ENSG00000139970 | ENSMUSG00000021087 | 0.107 | 0.5217 | 0.205098716 | ENSMUSG00000061718 | ENSG00000131771 | 0.0664 | 0.5038 | 0.131798333 | ||||||||||||||||||||||||||
| ENSG00000139220 | ENSMUSG00000053825 | 0.0105 | 0.5218 | 0.020122652 | ENSMUSG00000035226 | ENSG00000101098 | 0.0032 | 0.5045 | 0.006342914 | ||||||||||||||||||||||||||
| ENSG00000162729 | ENSMUSG00000038034 | 0.0782 | 0.5218 | 0.149865849 | ENSMUSG00000049550 | ENSG00000130779 | 0.0538 | 0.5045 | 0.106640238 | ||||||||||||||||||||||||||
| ENSG00000042493 | ENSMUSG00000056737 | 0.0341 | 0.522 | 0.06532567 | ENSMUSG00000031969 | ENSG00000151498 | 0.0554 | 0.5049 | 0.109724698 | ||||||||||||||||||||||||||
| ENSG00000139641 | ENSMUSG00000025366 | 0.063 | 0.5224 | 0.120597243 | ENSMUSG00000059213 | ENSG00000181418 | 0.1404 | 0.5051 | 0.277964759 | ||||||||||||||||||||||||||
| ENSG00000078053 | ENSMUSG00000021314 | 0.086 | 0.5226 | 0.164561806 | ENSMUSG00000050043 | ENSG00000213593 | 0.0362 | 0.5052 | 0.07165479 | ||||||||||||||||||||||||||
| ENSG00000148180 | ENSMUSG00000026879 | 0.0363 | 0.5227 | 0.069447102 | ENSMUSG00000025781 | ENSG00000165629 | 0.044 | 0.5055 | 0.087042532 | ||||||||||||||||||||||||||
| ENSG00000115677 | ENSMUSG00000034088 | 0.011 | 0.5235 | 0.021012416 | ENSMUSG00000028373 | ENSG00000148219 | 0.0121 | 0.5057 | 0.02392723 | ||||||||||||||||||||||||||
| ENSG00000108465 | ENSMUSG00000018669 | 0.0586 | 0.5235 | 0.111938873 | ENSMUSG00000068923 | ENSG00000132718 | 0.0212 | 0.5057 | 0.041922088 | ||||||||||||||||||||||||||
| ENSG00000114573 | ENSMUSG00000052459 | 0.0099 | 0.5239 | 0.018896736 | ENSMUSG00000028811 | ENSG00000134684 | 0.0176 | 0.5058 | 0.034796362 | ||||||||||||||||||||||||||
| ENSG00000185722 | ENSMUSG00000020790 | 0.0268 | 0.5246 | 0.051086542 | ENSMUSG00000018574 | ENSG00000072778 | 0.0917 | 0.5059 | 0.181261119 | ||||||||||||||||||||||||||
| ENSG00000132141 | ENSMUSG00000020698 | 0.0985 | 0.5247 | 0.18772632 | ENSMUSG00000015291 | ENSG00000203879 | 0.0092 | 0.5061 | 0.018178226 | ||||||||||||||||||||||||||
| ENSG00000106125 | ENSMUSG00000038022 | 0.1503 | 0.5249 | 0.286340255 | ENSMUSG00000023017 | ENSG00000110881 | 0.0083 | 0.5065 | 0.016386969 | ||||||||||||||||||||||||||
| ENSG00000134982 | ENSMUSG00000005871 | 0.0467 | 0.5251 | 0.088935441 | ENSMUSG00000052516 | ENSG00000185008 | 0.0207 | 0.5067 | 0.040852575 | ||||||||||||||||||||||||||
| ENSG00000175115 | ENSMUSG00000024855 | 0.0119 | 0.5252 | 0.022658035 | ENSMUSG00000026781 | ENSG00000107897 | 0.1025 | 0.5087 | 0.201494004 | ||||||||||||||||||||||||||
| ENSG00000021645 | ENSMUSG00000066392 | 0.0046 | 0.5253 | 0.008756901 | ENSMUSG00000037234 | ENSG00000168172 | 0.0102 | 0.5088 | 0.02004717 | ||||||||||||||||||||||||||
| ENSG00000151475 | ENSMUSG00000069041 | 0.0592 | 0.5261 | 0.112526136 | ENSMUSG00000060780 | ENSG00000162951 | 0.0138 | 0.5088 | 0.027122642 | ||||||||||||||||||||||||||
| ENSG00000091483 | ENSMUSG00000026526 | 0.0361 | 0.5267 | 0.068539966 | ENSMUSG00000021991 | ENSG00000157445 | 0.0093 | 0.5089 | 0.01827471 | ||||||||||||||||||||||||||
| ENSG00000143761 | ENSMUSG00000048076 | 0 | 0.5271 | 0 | ENSMUSG00000001036 | ENSG00000072134 | 0.0586 | 0.5089 | 0.115150324 | ||||||||||||||||||||||||||
| ENSG00000197043 | ENSMUSG00000018340 | 0.0283 | 0.5271 | 0.053690002 | ENSMUSG00000003316 | ENSG00000090863 | 0.0171 | 0.509 | 0.033595285 | ||||||||||||||||||||||||||
| ENSG00000116675 | ENSMUSG00000028528 | 0.0359 | 0.5271 | 0.068108518 | ENSMUSG00000033066 | ENSG00000007237 | 0.0126 | 0.5091 | 0.024749558 | ||||||||||||||||||||||||||
| ENSG00000107295 | ENSMUSG00000028488 | 0.0051 | 0.5276 | 0.009666414 | ENSMUSG00000020524 | ENSG00000155511 | 0.0056 | 0.5092 | 0.010997643 | ||||||||||||||||||||||||||
| ENSG00000143153 | ENSMUSG00000026576 | 0.0306 | 0.5283 | 0.057921635 | ENSMUSG00000021730 | ENSG00000164588 | 0.0183 | 0.5097 | 0.035903473 | ||||||||||||||||||||||||||
| ENSG00000101421 | ENSMUSG00000038467 | 0.0019 | 0.5288 | 0.003593041 | ENSMUSG00000062044 | ENSG00000142235 | 0.0507 | 0.5098 | 0.099450765 | ||||||||||||||||||||||||||
| ENSG00000121905 | ENSMUSG00000028785 | 0 | 0.5289 | 0 | ENSMUSG00000000168 | ENSG00000150768 | 0.0823 | 0.5099 | 0.161404197 | ||||||||||||||||||||||||||
| ENSG00000092621 | ENSMUSG00000053398 | 0.0302 | 0.5289 | 0.057099641 | ENSMUSG00000063063 | ENSG00000066032 | 0.0005 | 0.5102 | 0.000980008 | ||||||||||||||||||||||||||
| ENSG00000154917 | ENSMUSG00000032549 | 0 | 0.529 | 0 | ENSMUSG00000052698 | ENSG00000171914 | 0.0062 | 0.5102 | 0.012152097 | ||||||||||||||||||||||||||
| ENSG00000057608 | ENSMUSG00000021218 | 0.0178 | 0.529 | 0.033648393 | ENSMUSG00000019907 | ENSG00000058272 | 0.0429 | 0.5106 | 0.084018801 | ||||||||||||||||||||||||||
| ENSG00000100714 | ENSMUSG00000021048 | 0.0357 | 0.5292 | 0.067460317 | ENSMUSG00000040265 | ENSG00000197959 | 0.0099 | 0.5111 | 0.019369986 | ||||||||||||||||||||||||||
| ENSG00000130340 | ENSMUSG00000002365 | 0.0452 | 0.5294 | 0.085379675 | ENSMUSG00000066189 | ENSG00000006116 | 0.0057 | 0.5112 | 0.011150235 | ||||||||||||||||||||||||||
| ENSG00000134056 | ENSMUSG00000021631 | 0.0744 | 0.5296 | 0.140483384 | ENSMUSG00000022099 | ENSG00000158856 | 0.0202 | 0.5122 | 0.03943772 | ||||||||||||||||||||||||||
| ENSG00000134056 | ENSMUSG00000061474 | 0.0748 | 0.5302 | 0.141078838 | ENSMUSG00000060961 | ENSG00000080493 | 0.0154 | 0.5123 | 0.030060511 | ||||||||||||||||||||||||||
| ENSG00000173171 | ENSMUSG00000064068 | 0.1537 | 0.5302 | 0.289890607 | ENSMUSG00000053819 | ENSG00000145349 | 0.0088 | 0.5124 | 0.017174083 | ||||||||||||||||||||||||||
| ENSG00000159176 | ENSMUSG00000026421 | 0.0024 | 0.5303 | 0.00452574 | ENSMUSG00000029211 | ENSG00000109158 | 0.0479 | 0.5126 | 0.093445181 | ||||||||||||||||||||||||||
| ENSG00000110436 | ENSMUSG00000005089 | 0.0227 | 0.5306 | 0.042781757 | ENSMUSG00000003974 | ENSG00000198822 | 0.0152 | 0.5129 | 0.029635407 | ||||||||||||||||||||||||||
| ENSG00000075785 | ENSMUSG00000079477 | 0 | 0.531 | 0 | ENSMUSG00000029763 | ENSG00000131558 | 0.02 | 0.5129 | 0.038993956 | ||||||||||||||||||||||||||
| ENSG00000184752 | ENSMUSG00000020022 | 0.0632 | 0.5313 | 0.11895351 | ENSMUSG00000024242 | ENSG00000011566 | 0.0198 | 0.5131 | 0.038588969 | ||||||||||||||||||||||||||
| ENSG00000174444 | ENSMUSG00000032399 | 0.0305 | 0.5323 | 0.057298516 | ENSMUSG00000029108 | ENSG00000169851 | 0.0183 | 0.5132 | 0.035658613 | ||||||||||||||||||||||||||
| ENSG00000127481 | ENSMUSG00000066036 | 0.0151 | 0.5326 | 0.028351483 | ENSMUSG00000022295 | ENSG00000155097 | 0.0089 | 0.5141 | 0.017311807 | ||||||||||||||||||||||||||
| ENSG00000038427 | ENSMUSG00000021614 | 0.2379 | 0.5329 | 0.44642522 | ENSMUSG00000019996 | ENSG00000135525 | 0.1154 | 0.5141 | 0.224469947 | ||||||||||||||||||||||||||
| ENSG00000009335 | ENSMUSG00000039000 | 0.0333 | 0.5335 | 0.062417994 | ENSMUSG00000025757 | ENSG00000164070 | 0.0382 | 0.5143 | 0.074275715 | ||||||||||||||||||||||||||
| ENSG00000010256 | ENSMUSG00000025651 | 0.0656 | 0.5345 | 0.122731525 | ENSMUSG00000006273 | ENSG00000147416 | 0.0056 | 0.5148 | 0.010878011 | ||||||||||||||||||||||||||
| ENSG00000100412 | ENSMUSG00000022477 | 0.0158 | 0.5347 | 0.02954928 | ENSMUSG00000053141 | ENSG00000196090 | 0.0067 | 0.5148 | 0.013014763 | ||||||||||||||||||||||||||
| ENSG00000163399 | ENSMUSG00000033161 | 0.0158 | 0.5353 | 0.029516159 | ENSMUSG00000027438 | ENSG00000125814 | 0 | 0.5154 | 0 | ||||||||||||||||||||||||||
| ENSG00000004478 | ENSMUSG00000030357 | 0.0525 | 0.5355 | 0.098039216 | ENSMUSG00000070730 | ENSG00000137824 | 0.0822 | 0.5157 | 0.159394997 | ||||||||||||||||||||||||||
| ENSG00000169992 | ENSMUSG00000051790 | 0.0079 | 0.5359 | 0.014741556 | ENSMUSG00000042569 | ENSG00000109016 | 0.1285 | 0.5157 | 0.249175877 | ||||||||||||||||||||||||||
| ENSG00000134516 | ENSMUSG00000020143 | 0.0235 | 0.5363 | 0.043818758 | ENSMUSG00000032480 | ENSG00000132153 | 0.0115 | 0.516 | 0.022286822 | ||||||||||||||||||||||||||
| ENSG00000162869 | ENSMUSG00000034709 | 0.0431 | 0.5363 | 0.080365467 | ENSMUSG00000034275 | ENSG00000080854 | 0.0189 | 0.5163 | 0.036606624 | ||||||||||||||||||||||||||
| ENSG00000198130 | ENSMUSG00000041426 | 0.0968 | 0.5368 | 0.180327869 | ENSMUSG00000041112 | ENSG00000155849 | 0.0019 | 0.5164 | 0.003679318 | ||||||||||||||||||||||||||
| ENSG00000103723 | ENSMUSG00000062444 | 0.0243 | 0.5369 | 0.045259825 | ENSMUSG00000022957 | ENSG00000205726 | 0.0247 | 0.5166 | 0.047812621 | ||||||||||||||||||||||||||
| ENSG00000018189 | ENSMUSG00000029291 | 0.0343 | 0.5373 | 0.063837707 | ENSMUSG00000004936 | ENSG00000169032 | 0.0046 | 0.5169 | 0.008899207 | ||||||||||||||||||||||||||
| ENSG00000006740 | ENSMUSG00000033389 | 0.0324 | 0.5374 | 0.060290287 | ENSMUSG00000032482 | ENSG00000114646 | 0.0705 | 0.517 | 0.136363636 | ||||||||||||||||||||||||||
| ENSG00000128641 | ENSMUSG00000018417 | 0.0103 | 0.5375 | 0.019162791 | ENSMUSG00000020456 | ENSG00000105953 | 0.0216 | 0.5175 | 0.04173913 | ||||||||||||||||||||||||||
| ENSG00000168385 | ENSMUSG00000026276 | 0.0063 | 0.5378 | 0.011714392 | ENSMUSG00000076439 | ENSG00000204655 | 0.0831 | 0.5175 | 0.16057971 | ||||||||||||||||||||||||||
| ENSG00000171533 | ENSMUSG00000055407 | 0.1828 | 0.538 | 0.339776952 | ENSMUSG00000024533 | ENSG00000134278 | 0.0359 | 0.5176 | 0.069358578 | ||||||||||||||||||||||||||
| ENSG00000197872 | ENSMUSG00000020589 | 0.0014 | 0.5381 | 0.002601747 | ENSMUSG00000032279 | ENSG00000166411 | 0.0117 | 0.5181 | 0.022582513 | ||||||||||||||||||||||||||
| ENSG00000186522 | ENSMUSG00000019917 | 0.0761 | 0.5381 | 0.141423527 | ENSMUSG00000019790 | ENSG00000164506 | 0.0223 | 0.5182 | 0.043033578 | ||||||||||||||||||||||||||
| ENSG00000009844 | ENSMUSG00000019868 | 0.0393 | 0.5383 | 0.073007617 | ENSMUSG00000040359 | ENSG00000014123 | 0.0497 | 0.5188 | 0.095797995 | ||||||||||||||||||||||||||
| ENSG00000129625 | ENSMUSG00000005873 | 0.027 | 0.5391 | 0.050083472 | ENSMUSG00000059981 | ENSG00000149930 | 0.0388 | 0.519 | 0.074759152 | ||||||||||||||||||||||||||
| ENSG00000187231 | ENSMUSG00000042272 | 0.0086 | 0.5394 | 0.015943641 | ENSMUSG00000015829 | ENSG00000116147 | 0.0334 | 0.5191 | 0.064342131 | ||||||||||||||||||||||||||
| ENSG00000136167 | ENSMUSG00000021998 | 0.0152 | 0.5394 | 0.028179459 | ENSMUSG00000028354 | ENSG00000155816 | 0.1126 | 0.5191 | 0.216913889 | ||||||||||||||||||||||||||
| ENSG00000124198 | ENSMUSG00000074582 | 0.0219 | 0.5399 | 0.040563067 | ENSMUSG00000020402 | ENSG00000213585 | 0.0065 | 0.5192 | 0.01251926 | ||||||||||||||||||||||||||
| ENSG00000185666 | ENSMUSG00000059602 | 0.0387 | 0.5401 | 0.071653398 | ENSMUSG00000030695 | ENSG00000149925 | 0.0101 | 0.5192 | 0.019453005 | ||||||||||||||||||||||||||
| ENSG00000100592 | ENSMUSG00000034574 | 0.0138 | 0.5404 | 0.02553664 | ENSMUSG00000068036 | ENSG00000130396 | 0.031 | 0.5195 | 0.059672762 | ||||||||||||||||||||||||||
| ENSG00000124406 | ENSMUSG00000037685 | 0.0089 | 0.5405 | 0.016466235 | ENSMUSG00000021733 | ENSG00000033867 | 0.0318 | 0.5198 | 0.061177376 | ||||||||||||||||||||||||||
| ENSG00000107819 | ENSMUSG00000025212 | 0.0313 | 0.5413 | 0.057823758 | ENSMUSG00000030209 | ENSG00000150086 | 0.0058 | 0.5199 | 0.011155992 | ||||||||||||||||||||||||||
| ENSG00000140575 | ENSMUSG00000030536 | 0.0213 | 0.5419 | 0.039306145 | ENSMUSG00000021087 | ENSG00000139970 | 0.107 | 0.5217 | 0.205098716 | ||||||||||||||||||||||||||
| ENSG00000149294 | ENSMUSG00000039542 | 0.0278 | 0.5432 | 0.051178203 | ENSMUSG00000053825 | ENSG00000139220 | 0.0105 | 0.5218 | 0.020122652 | ||||||||||||||||||||||||||
| ENSG00000006747 | ENSMUSG00000002565 | 0.0465 | 0.5432 | 0.085603829 | ENSMUSG00000038034 | ENSG00000162729 | 0.0782 | 0.5218 | 0.149865849 | ||||||||||||||||||||||||||
| ENSG00000111669 | ENSMUSG00000023456 | 0.0244 | 0.5433 | 0.044910731 | ENSMUSG00000021314 | ENSG00000078053 | 0.086 | 0.5226 | 0.164561806 | ||||||||||||||||||||||||||
| ENSG00000108262 | ENSMUSG00000011877 | 0.0106 | 0.5442 | 0.019478133 | ENSMUSG00000026879 | ENSG00000148180 | 0.0363 | 0.5227 | 0.069447102 | ||||||||||||||||||||||||||
| ENSG00000108797 | ENSMUSG00000017167 | 0.0339 | 0.5446 | 0.062247521 | ENSMUSG00000032773 | ENSG00000168539 | 0.0048 | 0.5228 | 0.009181331 | ||||||||||||||||||||||||||
| ENSG00000150054 | ENSMUSG00000057440 | 0.0356 | 0.5448 | 0.065345081 | ENSMUSG00000034088 | ENSG00000115677 | 0.011 | 0.5235 | 0.021012416 | ||||||||||||||||||||||||||
| ENSG00000145362 | ENSMUSG00000032826 | 0.0419 | 0.5448 | 0.076908957 | ENSMUSG00000053317 | ENSG00000106803 | 0.005 | 0.5236 | 0.009549274 | ||||||||||||||||||||||||||
| ENSG00000124194 | ENSMUSG00000017943 | 0.0093 | 0.5451 | 0.01706109 | ENSMUSG00000052459 | ENSG00000114573 | 0.0099 | 0.5239 | 0.018896736 | ||||||||||||||||||||||||||
| ENSG00000073969 | ENSMUSG00000034187 | 0.0089 | 0.5455 | 0.016315307 | ENSMUSG00000038264 | ENSG00000138623 | 0.0528 | 0.5243 | 0.100705703 | ||||||||||||||||||||||||||
| ENSG00000166947 | ENSMUSG00000023216 | 0.1688 | 0.5456 | 0.309384164 | ENSMUSG00000028975 | ENSG00000142655 | 0.0338 | 0.5244 | 0.064454615 | ||||||||||||||||||||||||||
| ENSG00000186472 | ENSMUSG00000061601 | 0.0926 | 0.5459 | 0.169628137 | ENSMUSG00000020698 | ENSG00000132141 | 0.0985 | 0.5247 | 0.18772632 | ||||||||||||||||||||||||||
| ENSG00000125447 | ENSMUSG00000020740 | 0.0668 | 0.546 | 0.122344322 | ENSMUSG00000005871 | ENSG00000134982 | 0.0467 | 0.5251 | 0.088935441 | ||||||||||||||||||||||||||
| ENSG00000116266 | ENSMUSG00000027882 | 0.0413 | 0.5461 | 0.075627175 | ENSMUSG00000024855 | ENSG00000175115 | 0.0119 | 0.5252 | 0.022658035 | ||||||||||||||||||||||||||
| ENSG00000113643 | ENSMUSG00000018848 | 0.044 | 0.5462 | 0.080556573 | ENSMUSG00000066392 | ENSG00000021645 | 0.0046 | 0.5253 | 0.008756901 | ||||||||||||||||||||||||||
| ENSG00000100167 | ENSMUSG00000022456 | 0.028 | 0.5463 | 0.05125389 | ENSMUSG00000030282 | ENSG00000111726 | 0.0322 | 0.5266 | 0.061146981 | ||||||||||||||||||||||||||
| ENSG00000131473 | ENSMUSG00000020917 | 0.0088 | 0.5464 | 0.016105417 | ENSMUSG00000048076 | ENSG00000143761 | 0 | 0.5271 | 0 | ||||||||||||||||||||||||||
| ENSG00000100003 | ENSMUSG00000003585 | 0.033 | 0.5466 | 0.060373216 | ENSMUSG00000028528 | ENSG00000116675 | 0.0359 | 0.5271 | 0.068108518 | ||||||||||||||||||||||||||
| ENSG00000124207 | ENSMUSG00000002718 | 0.0048 | 0.5475 | 0.008767123 | ENSMUSG00000052581 | ENSG00000176204 | 0.0228 | 0.5273 | 0.043239143 | ||||||||||||||||||||||||||
| ENSG00000163541 | ENSMUSG00000052738 | 0.0396 | 0.548 | 0.072262774 | ENSMUSG00000028488 | ENSG00000107295 | 0.0051 | 0.5276 | 0.009666414 | ||||||||||||||||||||||||||
| ENSG00000126432 | ENSMUSG00000024953 | 0.1489 | 0.5481 | 0.271665754 | ENSMUSG00000026576 | ENSG00000143153 | 0.0306 | 0.5283 | 0.057921635 | ||||||||||||||||||||||||||
| ENSG00000120549 | ENSMUSG00000036617 | 0.113 | 0.5482 | 0.20612915 | ENSMUSG00000038467 | ENSG00000101421 | 0.0019 | 0.5288 | 0.003593041 | ||||||||||||||||||||||||||
| ENSG00000197555 | ENSMUSG00000042700 | 0.0195 | 0.5492 | 0.035506191 | ENSMUSG00000028785 | ENSG00000121905 | 0 | 0.5289 | 0 | ||||||||||||||||||||||||||
| ENSG00000115306 | ENSMUSG00000020315 | 0.0077 | 0.5493 | 0.014017841 | ENSMUSG00000053398 | ENSG00000092621 | 0.0302 | 0.5289 | 0.057099641 | ||||||||||||||||||||||||||
| ENSG00000099365 | ENSMUSG00000030806 | 0 | 0.5497 | 0 | ENSMUSG00000032549 | ENSG00000154917 | 0 | 0.529 | 0 | ||||||||||||||||||||||||||
| ENSG00000058668 | ENSMUSG00000026463 | 0.0884 | 0.5502 | 0.160668848 | ENSMUSG00000021048 | ENSG00000100714 | 0.0357 | 0.5292 | 0.067460317 | ||||||||||||||||||||||||||
| ENSG00000037042 | ENSMUSG00000045007 | 0.0104 | 0.5504 | 0.018895349 | ENSMUSG00000048578 | ENSG00000110917 | 0.0453 | 0.5295 | 0.085552408 | ||||||||||||||||||||||||||
| ENSG00000189043 | ENSMUSG00000029632 | 0.057 | 0.5509 | 0.103467054 | ENSMUSG00000050390 | ENSG00000162522 | 0.1061 | 0.5301 | 0.200150915 | ||||||||||||||||||||||||||
| ENSG00000079215 | ENSMUSG00000005360 | 0.0156 | 0.5511 | 0.028307022 | ENSMUSG00000061474 | ENSG00000134056 | 0.0748 | 0.5302 | 0.141078838 | ||||||||||||||||||||||||||
| ENSG00000141720 | ENSMUSG00000018547 | 0.0041 | 0.5521 | 0.007426191 | ENSMUSG00000064068 | ENSG00000173171 | 0.1537 | 0.5302 | 0.289890607 | ||||||||||||||||||||||||||
| ENSG00000082805 | ENSMUSG00000030172 | 0.0128 | 0.5522 | 0.023180007 | ENSMUSG00000046240 | ENSG00000165478 | 0.0246 | 0.5304 | 0.04638009 | ||||||||||||||||||||||||||
| ENSG00000122966 | ENSMUSG00000029516 | 0.0152 | 0.5522 | 0.027526259 | ENSMUSG00000005089 | ENSG00000110436 | 0.0227 | 0.5306 | 0.042781757 | ||||||||||||||||||||||||||
| ENSG00000152234 | ENSMUSG00000025428 | 0.0123 | 0.5525 | 0.022262443 | ENSMUSG00000079477 | ENSG00000075785 | 0 | 0.531 | 0 | ||||||||||||||||||||||||||
| ENSG00000129515 | ENSMUSG00000005656 | 0.0022 | 0.5535 | 0.003974706 | ENSMUSG00000049076 | ENSG00000114331 | 0.0178 | 0.5313 | 0.033502729 | ||||||||||||||||||||||||||
| ENSG00000124942 | ENSMUSG00000069833 | 0.0905 | 0.5535 | 0.163504968 | ENSMUSG00000024502 | ENSG00000176049 | 0.0098 | 0.5322 | 0.01841413 | ||||||||||||||||||||||||||
| ENSG00000171202 | ENSMUSG00000030615 | 0.1702 | 0.5536 | 0.307442197 | ENSMUSG00000020621 | ENSG00000240857 | 0.0506 | 0.5322 | 0.095077039 | ||||||||||||||||||||||||||
| ENSG00000115840 | ENSMUSG00000027010 | 0.0167 | 0.5537 | 0.030160737 | ENSMUSG00000032399 | ENSG00000174444 | 0.0305 | 0.5323 | 0.057298516 | ||||||||||||||||||||||||||
| ENSG00000152932 | ENSMUSG00000021700 | 0.0124 | 0.5538 | 0.022390755 | ENSMUSG00000074923 | ENSG00000137843 | 0.0359 | 0.5326 | 0.067405182 | ||||||||||||||||||||||||||
| ENSG00000011347 | ENSMUSG00000024743 | 0.0082 | 0.5541 | 0.014798773 | ENSMUSG00000021614 | ENSG00000038427 | 0.2379 | 0.5329 | 0.44642522 | ||||||||||||||||||||||||||
| ENSG00000204351 | ENSMUSG00000040356 | 0.0339 | 0.5542 | 0.061169253 | ENSMUSG00000025651 | ENSG00000010256 | 0.0656 | 0.5345 | 0.122731525 | ||||||||||||||||||||||||||
| ENSG00000105568 | ENSMUSG00000007564 | 0.0008 | 0.5548 | 0.001441961 | ENSMUSG00000038910 | ENSG00000154822 | 0.0109 | 0.5346 | 0.020389076 | ||||||||||||||||||||||||||
| ENSG00000189221 | ENSMUSG00000025037 | 0.0711 | 0.555 | 0.128108108 | ENSMUSG00000033161 | ENSG00000163399 | 0.0158 | 0.5353 | 0.029516159 | ||||||||||||||||||||||||||
| ENSG00000111530 | ENSMUSG00000020114 | 0.0027 | 0.5551 | 0.004863988 | ENSMUSG00000051790 | ENSG00000169992 | 0.0079 | 0.5359 | 0.014741556 | ||||||||||||||||||||||||||
| ENSG00000132842 | ENSMUSG00000021686 | 0.0717 | 0.5554 | 0.129096147 | ENSMUSG00000038828 | ENSG00000119777 | 0.1035 | 0.5362 | 0.193024991 | ||||||||||||||||||||||||||
| ENSG00000110435 | ENSMUSG00000010914 | 0.0838 | 0.5554 | 0.150882247 | ENSMUSG00000034709 | ENSG00000162869 | 0.0431 | 0.5363 | 0.080365467 | ||||||||||||||||||||||||||
| ENSG00000182621 | ENSMUSG00000051177 | 0.0126 | 0.556 | 0.022661871 | ENSMUSG00000062444 | ENSG00000103723 | 0.0243 | 0.5369 | 0.045259825 | ||||||||||||||||||||||||||
| ENSG00000133835 | ENSMUSG00000024507 | 0.0735 | 0.5563 | 0.132122955 | ENSMUSG00000026426 | ENSG00000143862 | 0.0023 | 0.5373 | 0.004280663 | ||||||||||||||||||||||||||
| ENSG00000069966 | ENSMUSG00000032192 | 0.0034 | 0.5567 | 0.006107419 | ENSMUSG00000029291 | ENSG00000018189 | 0.0343 | 0.5373 | 0.063837707 | ||||||||||||||||||||||||||
| ENSG00000170579 | ENSMUSG00000003279 | 0.0178 | 0.5573 | 0.031939709 | ENSMUSG00000033389 | ENSG00000006740 | 0.0324 | 0.5374 | 0.060290287 | ||||||||||||||||||||||||||
| ENSG00000078369 | ENSMUSG00000029064 | 0.0006 | 0.5577 | 0.001075847 | ENSMUSG00000018417 | ENSG00000128641 | 0.0103 | 0.5375 | 0.019162791 | ||||||||||||||||||||||||||
| ENSG00000149541 | ENSMUSG00000071649 | 0.0209 | 0.5588 | 0.037401575 | ENSMUSG00000026276 | ENSG00000168385 | 0.0063 | 0.5378 | 0.011714392 | ||||||||||||||||||||||||||
| ENSG00000103647 | ENSMUSG00000041729 | 0.008 | 0.5591 | 0.01430871 | ENSMUSG00000060090 | ENSG00000102218 | 0.0605 | 0.5378 | 0.112495351 | ||||||||||||||||||||||||||
| ENSG00000161204 | ENSMUSG00000003234 | 0.0171 | 0.5595 | 0.030563003 | ENSMUSG00000055407 | ENSG00000171533 | 0.1828 | 0.538 | 0.339776952 | ||||||||||||||||||||||||||
| ENSG00000166136 | ENSMUSG00000025204 | 0.0979 | 0.5596 | 0.17494639 | ENSMUSG00000019917 | ENSG00000186522 | 0.0761 | 0.5381 | 0.141423527 | ||||||||||||||||||||||||||
| ENSG00000174903 | ENSMUSG00000024870 | 0.0043 | 0.5598 | 0.007681315 | ENSMUSG00000035051 | ENSG00000163214 | 0.0781 | 0.5384 | 0.145059435 | ||||||||||||||||||||||||||
| ENSG00000080845 | ENSMUSG00000061689 | 0.0184 | 0.5601 | 0.032851277 | ENSMUSG00000005873 | ENSG00000129625 | 0.027 | 0.5391 | 0.050083472 | ||||||||||||||||||||||||||
| ENSG00000130638 | ENSMUSG00000016541 | 0.0988 | 0.5604 | 0.176302641 | ENSMUSG00000059602 | ENSG00000185666 | 0.0387 | 0.5401 | 0.071653398 | ||||||||||||||||||||||||||
| ENSG00000136153 | ENSMUSG00000033060 | 0.1163 | 0.561 | 0.207308378 | ENSMUSG00000045441 | ENSG00000185477 | 0.3075 | 0.5403 | 0.569128262 | ||||||||||||||||||||||||||
| ENSG00000019144 | ENSMUSG00000048537 | 0.0283 | 0.5618 | 0.050373799 | ENSMUSG00000034574 | ENSG00000100592 | 0.0138 | 0.5404 | 0.02553664 | ||||||||||||||||||||||||||
| ENSG00000070756 | ENSMUSG00000051732 | 0.1049 | 0.5619 | 0.186688023 | ENSMUSG00000037685 | ENSG00000124406 | 0.0089 | 0.5405 | 0.016466235 | ||||||||||||||||||||||||||
| ENSG00000146676 | ENSMUSG00000091207 | 0.0076 | 0.5626 | 0.01350871 | ENSMUSG00000025212 | ENSG00000107819 | 0.0313 | 0.5413 | 0.057823758 | ||||||||||||||||||||||||||
| ENSG00000054116 | ENSMUSG00000028847 | 0.0079 | 0.5627 | 0.014039453 | ENSMUSG00000004748 | ENSG00000242114 | 0.0658 | 0.5414 | 0.121536757 | ||||||||||||||||||||||||||
| ENSG00000033627 | ENSMUSG00000019302 | 0.0187 | 0.5628 | 0.033226724 | ENSMUSG00000039542 | ENSG00000149294 | 0.0278 | 0.5432 | 0.051178203 | ||||||||||||||||||||||||||
| ENSG00000105193 | ENSMUSG00000037563 | 0 | 0.5629 | 0 | ENSMUSG00000026207 | ENSG00000072195 | 0.0441 | 0.5432 | 0.081185567 | ||||||||||||||||||||||||||
| ENSG00000215301 | ENSMUSG00000039224 | 0.0221 | 0.5631 | 0.039247025 | ENSMUSG00000023456 | ENSG00000111669 | 0.0244 | 0.5433 | 0.044910731 | ||||||||||||||||||||||||||
| ENSG00000156011 | ENSMUSG00000030465 | 0.1148 | 0.5639 | 0.203582195 | ENSMUSG00000044847 | ENSG00000155858 | 0.0664 | 0.5433 | 0.122216087 | ||||||||||||||||||||||||||
| ENSG00000166226 | ENSMUSG00000034024 | 0.0114 | 0.564 | 0.020212766 | ENSMUSG00000011877 | ENSG00000108262 | 0.0106 | 0.5442 | 0.019478133 | ||||||||||||||||||||||||||
| ENSG00000065613 | ENSMUSG00000025060 | 0.0932 | 0.564 | 0.165248227 | ENSMUSG00000017167 | ENSG00000108797 | 0.0339 | 0.5446 | 0.062247521 | ||||||||||||||||||||||||||
| ENSG00000068120 | ENSMUSG00000001755 | 0.0692 | 0.5641 | 0.122673285 | ENSMUSG00000032826 | ENSG00000145362 | 0.0419 | 0.5448 | 0.076908957 | ||||||||||||||||||||||||||
| ENSG00000160014 | ENSMUSG00000019370 | 0 | 0.5644 | 0 | ENSMUSG00000017943 | ENSG00000124194 | 0.0093 | 0.5451 | 0.01706109 | ||||||||||||||||||||||||||
| ENSG00000136854 | ENSMUSG00000026797 | 0 | 0.5646 | 0 | ENSMUSG00000054793 | ENSG00000105767 | 0.0083 | 0.5453 | 0.015220979 | ||||||||||||||||||||||||||
| ENSG00000116120 | ENSMUSG00000026245 | 0.036 | 0.5649 | 0.063728093 | ENSMUSG00000034187 | ENSG00000073969 | 0.0089 | 0.5455 | 0.016315307 | ||||||||||||||||||||||||||
| ENSG00000169021 | ENSMUSG00000038462 | 0.0652 | 0.5649 | 0.115418658 | ENSMUSG00000011752 | ENSG00000226784 | 0.0215 | 0.5459 | 0.039384503 | ||||||||||||||||||||||||||
| ENSG00000143850 | ENSMUSG00000041757 | 0.0671 | 0.5649 | 0.118782085 | ENSMUSG00000061601 | ENSG00000186472 | 0.0926 | 0.5459 | 0.169628137 | ||||||||||||||||||||||||||
| ENSG00000136628 | ENSMUSG00000026615 | 0.057 | 0.5657 | 0.10076012 | ENSMUSG00000022208 | ENSG00000092051 | 0.0194 | 0.546 | 0.035531136 | ||||||||||||||||||||||||||
| ENSG00000187672 | ENSMUSG00000040640 | 0.0084 | 0.5661 | 0.014838368 | ENSMUSG00000018848 | ENSG00000113643 | 0.044 | 0.5462 | 0.080556573 | ||||||||||||||||||||||||||
| ENSG00000135905 | ENSMUSG00000038608 | 0.0312 | 0.5663 | 0.055094473 | ENSMUSG00000022456 | ENSG00000100167 | 0.028 | 0.5463 | 0.05125389 | ||||||||||||||||||||||||||
| ENSG00000132589 | ENSMUSG00000061981 | 0.0032 | 0.5667 | 0.005646727 | ENSMUSG00000020917 | ENSG00000131473 | 0.0088 | 0.5464 | 0.016105417 | ||||||||||||||||||||||||||
| ENSG00000108861 | ENSMUSG00000003518 | 0.0323 | 0.5669 | 0.056976539 | ENSMUSG00000052738 | ENSG00000163541 | 0.0396 | 0.548 | 0.072262774 | ||||||||||||||||||||||||||
| ENSG00000068793 | ENSMUSG00000030447 | 0.0043 | 0.5683 | 0.007566426 | ENSMUSG00000024953 | ENSG00000126432 | 0.1489 | 0.5481 | 0.271665754 | ||||||||||||||||||||||||||
| ENSG00000164830 | ENSMUSG00000022307 | 0.0919 | 0.5683 | 0.161710364 | ENSMUSG00000036882 | ENSG00000004777 | 0.0433 | 0.5488 | 0.078899417 | ||||||||||||||||||||||||||
| ENSG00000204394 | ENSMUSG00000007029 | 0.0331 | 0.5686 | 0.058213155 | ENSMUSG00000021476 | ENSG00000130956 | 0.0857 | 0.549 | 0.156102004 | ||||||||||||||||||||||||||
| ENSG00000115685 | ENSMUSG00000026275 | 0.0272 | 0.5688 | 0.047819972 | ENSMUSG00000042700 | ENSG00000197555 | 0.0195 | 0.5492 | 0.035506191 | ||||||||||||||||||||||||||
| ENSG00000133083 | ENSMUSG00000027797 | 0.0363 | 0.5688 | 0.063818565 | ENSMUSG00000020315 | ENSG00000115306 | 0.0077 | 0.5493 | 0.014017841 | ||||||||||||||||||||||||||
| ENSG00000124181 | ENSMUSG00000016933 | 0.019 | 0.5689 | 0.033397785 | ENSMUSG00000030806 | ENSG00000099365 | 0 | 0.5497 | 0 | ||||||||||||||||||||||||||
| ENSG00000083845 | ENSMUSG00000012848 | 0.0044 | 0.5691 | 0.007731506 | ENSMUSG00000026463 | ENSG00000058668 | 0.0884 | 0.5502 | 0.160668848 | ||||||||||||||||||||||||||
| ENSG00000112992 | ENSMUSG00000025453 | 0.0322 | 0.5693 | 0.056560689 | ENSMUSG00000021259 | ENSG00000036530 | 0.0236 | 0.5504 | 0.042877907 | ||||||||||||||||||||||||||
| ENSG00000148700 | ENSMUSG00000025026 | 0.0393 | 0.5694 | 0.069020021 | ENSMUSG00000029632 | ENSG00000189043 | 0.057 | 0.5509 | 0.103467054 | ||||||||||||||||||||||||||
| ENSG00000112685 | ENSMUSG00000021357 | 0.0334 | 0.5696 | 0.05863764 | ENSMUSG00000005360 | ENSG00000079215 | 0.0156 | 0.5511 | 0.028307022 | ||||||||||||||||||||||||||
| ENSG00000141551 | ENSMUSG00000025162 | 0.0011 | 0.5699 | 0.001930163 | ENSMUSG00000018428 | ENSG00000121057 | 0.19 | 0.5512 | 0.344702467 | ||||||||||||||||||||||||||
| ENSG00000217930 | ENSMUSG00000014301 | 0.0228 | 0.57 | 0.04 | ENSMUSG00000032020 | ENSG00000154127 | 0.0115 | 0.5518 | 0.020840884 | ||||||||||||||||||||||||||
| ENSG00000091157 | ENSMUSG00000040560 | 0.0253 | 0.5704 | 0.044354839 | ENSMUSG00000011752 | ENSG00000171314 | 0.0017 | 0.552 | 0.00307971 | ||||||||||||||||||||||||||
| ENSG00000041982 | ENSMUSG00000028364 | 0.0676 | 0.5717 | 0.118243834 | ENSMUSG00000018547 | ENSG00000141720 | 0.0041 | 0.5521 | 0.007426191 | ||||||||||||||||||||||||||
| ENSG00000149313 | ENSMUSG00000025894 | 0.0417 | 0.5718 | 0.072927597 | ENSMUSG00000030172 | ENSG00000082805 | 0.0128 | 0.5522 | 0.023180007 | ||||||||||||||||||||||||||
| ENSG00000164919 | ENSMUSG00000014313 | 0.1536 | 0.572 | 0.268531469 | ENSMUSG00000029516 | ENSG00000122966 | 0.0152 | 0.5522 | 0.027526259 | ||||||||||||||||||||||||||
| ENSG00000047056 | ENSMUSG00000021147 | 0.0157 | 0.5722 | 0.027437959 | ENSMUSG00000038525 | ENSG00000170632 | 0.1319 | 0.5522 | 0.238862731 | ||||||||||||||||||||||||||
| ENSG00000182199 | ENSMUSG00000025403 | 0.0316 | 0.5725 | 0.055196507 | ENSMUSG00000030092 | ENSG00000134115 | 0.054 | 0.5524 | 0.09775525 | ||||||||||||||||||||||||||
| ENSG00000160752 | ENSMUSG00000059743 | 0.0855 | 0.5731 | 0.149188623 | ENSMUSG00000025428 | ENSG00000152234 | 0.0123 | 0.5525 | 0.022262443 | ||||||||||||||||||||||||||
| ENSG00000151632 | ENSMUSG00000021214 | 0.1846 | 0.5738 | 0.321714883 | ENSMUSG00000027010 | ENSG00000115840 | 0.0167 | 0.5537 | 0.030160737 | ||||||||||||||||||||||||||
| ENSG00000127527 | ENSMUSG00000006276 | 0.0404 | 0.574 | 0.070383275 | ENSMUSG00000021700 | ENSG00000152932 | 0.0124 | 0.5538 | 0.022390755 | ||||||||||||||||||||||||||
| ENSG00000105270 | ENSMUSG00000013921 | 0.0068 | 0.5742 | 0.011842564 | ENSMUSG00000024743 | ENSG00000011347 | 0.0082 | 0.5541 | 0.014798773 | ||||||||||||||||||||||||||
| ENSG00000182718 | ENSMUSG00000032231 | 0.0121 | 0.5743 | 0.021069128 | ENSMUSG00000007564 | ENSG00000105568 | 0.0008 | 0.5548 | 0.001441961 | ||||||||||||||||||||||||||
| ENSG00000178952 | ENSMUSG00000073838 | 0.0349 | 0.5743 | 0.060769633 | ENSMUSG00000025037 | ENSG00000189221 | 0.0711 | 0.555 | 0.128108108 | ||||||||||||||||||||||||||
| ENSG00000184524 | ENSMUSG00000060240 | 0.1118 | 0.5743 | 0.194671774 | ENSMUSG00000020114 | ENSG00000111530 | 0.0027 | 0.5551 | 0.004863988 | ||||||||||||||||||||||||||
| ENSG00000137710 | ENSMUSG00000032050 | 0.007 | 0.5745 | 0.012184508 | ENSMUSG00000069170 | ENSG00000164199 | 0.1138 | 0.5551 | 0.205008107 | ||||||||||||||||||||||||||
| ENSG00000172053 | ENSMUSG00000032604 | 0.0547 | 0.5746 | 0.095196659 | ENSMUSG00000010914 | ENSG00000110435 | 0.0838 | 0.5554 | 0.150882247 | ||||||||||||||||||||||||||
| ENSG00000129244 | ENSMUSG00000041329 | 0.0121 | 0.5747 | 0.021054463 | ENSMUSG00000051177 | ENSG00000182621 | 0.0126 | 0.556 | 0.022661871 | ||||||||||||||||||||||||||
| ENSG00000103043 | ENSMUSG00000010936 | 0.0265 | 0.5747 | 0.046111014 | ENSMUSG00000024507 | ENSG00000133835 | 0.0735 | 0.5563 | 0.132122955 | ||||||||||||||||||||||||||
| ENSG00000131711 | ENSMUSG00000052727 | 0.0494 | 0.5754 | 0.085853319 | ENSMUSG00000024792 | ENSG00000162300 | 0.0386 | 0.5566 | 0.069349623 | ||||||||||||||||||||||||||
| ENSG00000170558 | ENSMUSG00000024304 | 0.0165 | 0.576 | 0.028645833 | ENSMUSG00000032192 | ENSG00000069966 | 0.0034 | 0.5567 | 0.006107419 | ||||||||||||||||||||||||||
| ENSG00000189058 | ENSMUSG00000022548 | 0.1916 | 0.5761 | 0.332581149 | ENSMUSG00000035342 | ENSG00000107816 | 0.0376 | 0.5571 | 0.067492371 | ||||||||||||||||||||||||||
| ENSG00000133943 | ENSMUSG00000021185 | 0.1624 | 0.5769 | 0.281504594 | ENSMUSG00000003279 | ENSG00000170579 | 0.0178 | 0.5573 | 0.031939709 | ||||||||||||||||||||||||||
| ENSG00000164434 | ENSMUSG00000019874 | 0.0754 | 0.5773 | 0.130608003 | ENSMUSG00000029064 | ENSG00000078369 | 0.0006 | 0.5577 | 0.001075847 | ||||||||||||||||||||||||||
| ENSG00000184277 | ENSMUSG00000078681 | 0.0789 | 0.5773 | 0.136670708 | ENSMUSG00000046230 | ENSG00000197969 | 0.0856 | 0.5577 | 0.153487538 | ||||||||||||||||||||||||||
| ENSG00000108829 | ENSMUSG00000020869 | 0.0207 | 0.5784 | 0.035788382 | ENSMUSG00000041729 | ENSG00000103647 | 0.008 | 0.5591 | 0.01430871 | ||||||||||||||||||||||||||
| ENSG00000137409 | ENSMUSG00000024012 | 0.0083 | 0.5785 | 0.01434745 | ENSMUSG00000025204 | ENSG00000166136 | 0.0979 | 0.5596 | 0.17494639 | ||||||||||||||||||||||||||
| ENSG00000100280 | ENSMUSG00000009090 | 0.0142 | 0.5787 | 0.024537757 | ENSMUSG00000024870 | ENSG00000174903 | 0.0043 | 0.5598 | 0.007681315 | ||||||||||||||||||||||||||
| ENSG00000158710 | ENSMUSG00000026547 | 0.0156 | 0.5787 | 0.026956973 | ENSMUSG00000061689 | ENSG00000080845 | 0.0184 | 0.5601 | 0.032851277 | ||||||||||||||||||||||||||
| ENSG00000137992 | ENSMUSG00000000340 | 0.0654 | 0.5801 | 0.112739183 | ENSMUSG00000041720 | ENSG00000241973 | 0.012 | 0.5606 | 0.021405637 | ||||||||||||||||||||||||||
| ENSG00000178952 | ENSMUSG00000030735 | 0.0361 | 0.5802 | 0.062219924 | ENSMUSG00000049807 | ENSG00000225485 | 0.0459 | 0.5609 | 0.081832769 | ||||||||||||||||||||||||||
| ENSG00000175662 | ENSMUSG00000000538 | 0.0264 | 0.5805 | 0.045478036 | ENSMUSG00000033060 | ENSG00000136153 | 0.1163 | 0.561 | 0.207308378 | ||||||||||||||||||||||||||
| ENSG00000075673 | ENSMUSG00000022229 | 0.0721 | 0.5805 | 0.124203273 | ENSMUSG00000027522 | ENSG00000124222 | 0.0322 | 0.5613 | 0.057366827 | ||||||||||||||||||||||||||
| ENSG00000196305 | ENSMUSG00000037851 | 0.0493 | 0.5806 | 0.08491216 | ENSMUSG00000048537 | ENSG00000019144 | 0.0283 | 0.5618 | 0.050373799 | ||||||||||||||||||||||||||
| ENSG00000115839 | ENSMUSG00000036104 | 0.036 | 0.5807 | 0.061994145 | ENSMUSG00000051732 | ENSG00000070756 | 0.1049 | 0.5619 | 0.186688023 | ||||||||||||||||||||||||||
| ENSG00000083720 | ENSMUSG00000022186 | 0.0404 | 0.5812 | 0.069511356 | ENSMUSG00000028847 | ENSG00000054116 | 0.0079 | 0.5627 | 0.014039453 | ||||||||||||||||||||||||||
| ENSG00000132434 | ENSMUSG00000062190 | 0.0582 | 0.5812 | 0.100137646 | ENSMUSG00000019302 | ENSG00000033627 | 0.0187 | 0.5628 | 0.033226724 | ||||||||||||||||||||||||||
| ENSG00000113758 | ENSMUSG00000034675 | 0.0792 | 0.5821 | 0.136059096 | ENSMUSG00000037563 | ENSG00000105193 | 0 | 0.5629 | 0 | ||||||||||||||||||||||||||
| ENSG00000147526 | ENSMUSG00000065954 | 0.1486 | 0.5821 | 0.255282597 | ENSMUSG00000036698 | ENSG00000123908 | 0.0036 | 0.5629 | 0.006395452 | ||||||||||||||||||||||||||
| ENSG00000174469 | ENSMUSG00000039419 | 0.0322 | 0.5834 | 0.055193692 | ENSMUSG00000022820 | ENSG00000065518 | 0.179 | 0.563 | 0.317939609 | ||||||||||||||||||||||||||
| ENSG00000117266 | ENSMUSG00000026437 | 0.04 | 0.5835 | 0.068551842 | ENSMUSG00000034024 | ENSG00000166226 | 0.0114 | 0.564 | 0.020212766 | ||||||||||||||||||||||||||
| ENSG00000214021 | ENSMUSG00000030276 | 0.1469 | 0.584 | 0.251541096 | ENSMUSG00000019370 | ENSG00000160014 | 0 | 0.5644 | 0 | ||||||||||||||||||||||||||
| ENSG00000076201 | ENSMUSG00000036057 | 0.0438 | 0.5844 | 0.074948665 | ENSMUSG00000046598 | ENSG00000161267 | 0.0722 | 0.5645 | 0.127900797 | ||||||||||||||||||||||||||
| ENSG00000164082 | ENSMUSG00000023192 | 0.0119 | 0.5848 | 0.020348837 | ENSMUSG00000026797 | ENSG00000136854 | 0 | 0.5646 | 0 | ||||||||||||||||||||||||||
| ENSG00000100884 | ENSMUSG00000022212 | 0.0097 | 0.5853 | 0.016572698 | ENSMUSG00000025508 | ENSG00000177600 | 0.0044 | 0.5649 | 0.007788989 | ||||||||||||||||||||||||||
| ENSG00000077279 | ENSMUSG00000031285 | 0.0025 | 0.5857 | 0.004268397 | ENSMUSG00000026245 | ENSG00000116120 | 0.036 | 0.5649 | 0.063728093 | ||||||||||||||||||||||||||
| ENSG00000134121 | ENSMUSG00000030077 | 0.0814 | 0.5857 | 0.138978999 | ENSMUSG00000038462 | ENSG00000169021 | 0.0652 | 0.5649 | 0.115418658 | ||||||||||||||||||||||||||
| ENSG00000157087 | ENSMUSG00000030302 | 0.0049 | 0.5859 | 0.008363202 | ENSMUSG00000041757 | ENSG00000143850 | 0.0671 | 0.5649 | 0.118782085 | ||||||||||||||||||||||||||
| ENSG00000101400 | ENSMUSG00000027488 | 0.038 | 0.5859 | 0.064857484 | ENSMUSG00000038848 | ENSG00000149658 | 0.0309 | 0.565 | 0.054690265 | ||||||||||||||||||||||||||
| ENSG00000102934 | ENSMUSG00000031775 | 0.0572 | 0.5859 | 0.097627581 | ENSMUSG00000052373 | ENSG00000161647 | 0.0448 | 0.5651 | 0.079278004 | ||||||||||||||||||||||||||
| ENSG00000117600 | ENSMUSG00000044667 | 0.018 | 0.5861 | 0.030711483 | ENSMUSG00000036815 | ENSG00000175497 | 0.0625 | 0.5655 | 0.110521662 | ||||||||||||||||||||||||||
| ENSG00000198836 | ENSMUSG00000038084 | 0.0189 | 0.5863 | 0.032236057 | ENSMUSG00000031149 | ENSG00000243279 | 0.0462 | 0.5656 | 0.081683168 | ||||||||||||||||||||||||||
| ENSG00000183454 | ENSMUSG00000059003 | 0.0229 | 0.5876 | 0.03897209 | ENSMUSG00000026615 | ENSG00000136628 | 0.057 | 0.5657 | 0.10076012 | ||||||||||||||||||||||||||
| ENSG00000101997 | ENSMUSG00000031143 | 0.0641 | 0.5878 | 0.109050698 | ENSMUSG00000040640 | ENSG00000187672 | 0.0084 | 0.5661 | 0.014838368 | ||||||||||||||||||||||||||
| ENSG00000070756 | ENSMUSG00000046173 | 0.1005 | 0.5879 | 0.17094744 | ENSMUSG00000038608 | ENSG00000135905 | 0.0312 | 0.5663 | 0.055094473 | ||||||||||||||||||||||||||
| ENSG00000138182 | ENSMUSG00000024795 | 0.1617 | 0.5879 | 0.275046777 | ENSMUSG00000061981 | ENSG00000132589 | 0.0032 | 0.5667 | 0.005646727 | ||||||||||||||||||||||||||
| ENSG00000138095 | ENSMUSG00000024120 | 0.156 | 0.5881 | 0.26526101 | ENSMUSG00000030447 | ENSG00000068793 | 0.0043 | 0.5683 | 0.007566426 | ||||||||||||||||||||||||||
| ENSG00000122694 | ENSMUSG00000028480 | 0.0331 | 0.5882 | 0.056273376 | ENSMUSG00000007029 | ENSG00000204394 | 0.0331 | 0.5686 | 0.058213155 | ||||||||||||||||||||||||||
| ENSG00000156049 | ENSMUSG00000024697 | 0.0152 | 0.5885 | 0.025828377 | ENSMUSG00000027797 | ENSG00000133083 | 0.0363 | 0.5688 | 0.063818565 | ||||||||||||||||||||||||||
| ENSG00000158186 | ENSMUSG00000032470 | 0.0157 | 0.5888 | 0.026664402 | ENSMUSG00000012848 | ENSG00000083845 | 0.0044 | 0.5691 | 0.007731506 | ||||||||||||||||||||||||||
| ENSG00000148468 | ENSMUSG00000050530 | 0.0397 | 0.589 | 0.067402377 | ENSMUSG00000025453 | ENSG00000112992 | 0.0322 | 0.5693 | 0.056560689 | ||||||||||||||||||||||||||
| ENSG00000154027 | ENSMUSG00000039058 | 0.0445 | 0.5896 | 0.075474898 | ENSMUSG00000025026 | ENSG00000148700 | 0.0393 | 0.5694 | 0.069020021 | ||||||||||||||||||||||||||
| ENSG00000148798 | ENSMUSG00000034336 | 0.0277 | 0.5903 | 0.046925292 | ENSMUSG00000021357 | ENSG00000112685 | 0.0334 | 0.5696 | 0.05863764 | ||||||||||||||||||||||||||
| ENSG00000171246 | ENSMUSG00000025582 | 0.0162 | 0.5904 | 0.027439024 | ENSMUSG00000030020 | ENSG00000163637 | 0.0209 | 0.5697 | 0.036685975 | ||||||||||||||||||||||||||
| ENSG00000105875 | ENSMUSG00000058486 | 0.0387 | 0.5904 | 0.06554878 | ENSMUSG00000039740 | ENSG00000119523 | 0.1188 | 0.5698 | 0.208494208 | ||||||||||||||||||||||||||
| ENSG00000165264 | ENSMUSG00000071014 | 0.2153 | 0.5905 | 0.364606266 | ENSMUSG00000025162 | ENSG00000141551 | 0.0011 | 0.5699 | 0.001930163 | ||||||||||||||||||||||||||
| ENSG00000106976 | ENSMUSG00000026825 | 0.0069 | 0.5908 | 0.011679079 | ENSMUSG00000040560 | ENSG00000091157 | 0.0253 | 0.5704 | 0.044354839 | ||||||||||||||||||||||||||
| ENSG00000144635 | ENSMUSG00000032435 | 0.0277 | 0.5908 | 0.046885579 | ENSMUSG00000075318 | ENSG00000136531 | 0.0103 | 0.5714 | 0.018025901 | ||||||||||||||||||||||||||
| ENSG00000090861 | ENSMUSG00000031960 | 0.0222 | 0.5914 | 0.037538045 | ENSMUSG00000014313 | ENSG00000164919 | 0.1536 | 0.572 | 0.268531469 | ||||||||||||||||||||||||||
| ENSG00000140945 | ENSMUSG00000031841 | 0.049 | 0.5914 | 0.082854244 | ENSMUSG00000021147 | ENSG00000047056 | 0.0157 | 0.5722 | 0.027437959 | ||||||||||||||||||||||||||
| ENSG00000182149 | ENSMUSG00000031729 | 0.1346 | 0.5914 | 0.227595536 | ENSMUSG00000031924 | ENSG00000103018 | 0.1457 | 0.5729 | 0.254320126 | ||||||||||||||||||||||||||
| ENSG00000132692 | ENSMUSG00000004892 | 0.0928 | 0.5917 | 0.156836235 | ENSMUSG00000006276 | ENSG00000127527 | 0.0404 | 0.574 | 0.070383275 | ||||||||||||||||||||||||||
| ENSG00000108515 | ENSMUSG00000060600 | 0.0104 | 0.5918 | 0.017573505 | ENSMUSG00000013921 | ENSG00000105270 | 0.0068 | 0.5742 | 0.011842564 | ||||||||||||||||||||||||||
| ENSG00000072110 | ENSMUSG00000015143 | 0.0039 | 0.5925 | 0.006582278 | ENSMUSG00000073838 | ENSG00000178952 | 0.0349 | 0.5743 | 0.060769633 | ||||||||||||||||||||||||||
| ENSG00000128487 | ENSMUSG00000042331 | 0.0844 | 0.5929 | 0.142351155 | ENSMUSG00000060240 | ENSG00000184524 | 0.1118 | 0.5743 | 0.194671774 | ||||||||||||||||||||||||||
| ENSG00000204590 | ENSMUSG00000024429 | 0.0221 | 0.5949 | 0.037149101 | ENSMUSG00000032050 | ENSG00000137710 | 0.007 | 0.5745 | 0.012184508 | ||||||||||||||||||||||||||
| ENSG00000007923 | ENSMUSG00000039768 | 0.0154 | 0.5956 | 0.025856279 | ENSMUSG00000032604 | ENSG00000172053 | 0.0547 | 0.5746 | 0.095196659 | ||||||||||||||||||||||||||
| ENSG00000187134 | ENSMUSG00000021214 | 0.1729 | 0.5957 | 0.290246769 | ENSMUSG00000041329 | ENSG00000129244 | 0.0121 | 0.5747 | 0.021054463 | ||||||||||||||||||||||||||
| ENSG00000157483 | ENSMUSG00000032220 | 0.0194 | 0.5965 | 0.032523051 | ENSMUSG00000052727 | ENSG00000131711 | 0.0494 | 0.5754 | 0.085853319 | ||||||||||||||||||||||||||
| ENSG00000143878 | ENSMUSG00000054364 | 0 | 0.5967 | 0 | ENSMUSG00000030102 | ENSG00000150995 | 0.0053 | 0.576 | 0.009201389 | ||||||||||||||||||||||||||
| ENSG00000002746 | ENSMUSG00000021301 | 0.0629 | 0.5967 | 0.105413105 | ENSMUSG00000024304 | ENSG00000170558 | 0.0165 | 0.576 | 0.028645833 | ||||||||||||||||||||||||||
| ENSG00000142657 | ENSMUSG00000028961 | 0.0313 | 0.5972 | 0.052411253 | ENSMUSG00000020869 | ENSG00000108829 | 0.0207 | 0.5784 | 0.035788382 | ||||||||||||||||||||||||||
| ENSG00000075142 | ENSMUSG00000003161 | 0.0196 | 0.5977 | 0.032792371 | ENSMUSG00000024012 | ENSG00000137409 | 0.0083 | 0.5785 | 0.01434745 | ||||||||||||||||||||||||||
| ENSG00000170584 | ENSMUSG00000020328 | 0.0028 | 0.5989 | 0.004675238 | ENSMUSG00000009090 | ENSG00000100280 | 0.0142 | 0.5787 | 0.024537757 | ||||||||||||||||||||||||||
| ENSG00000162105 | ENSMUSG00000037541 | 0.0774 | 0.5993 | 0.129150676 | ENSMUSG00000004285 | ENSG00000128524 | 0.0072 | 0.5789 | 0.012437381 | ||||||||||||||||||||||||||
| ENSG00000147471 | ENSMUSG00000031485 | 0.0775 | 0.6006 | 0.129037629 | ENSMUSG00000037386 | ENSG00000176406 | 0.0526 | 0.579 | 0.090846287 | ||||||||||||||||||||||||||
| ENSG00000149925 | ENSMUSG00000059343 | 0.0315 | 0.6008 | 0.052430093 | ENSMUSG00000008682 | ENSG00000147403 | 0 | 0.5791 | 0 | ||||||||||||||||||||||||||
| ENSG00000181789 | ENSMUSG00000030058 | 0.0126 | 0.6014 | 0.020951114 | ENSMUSG00000033916 | ENSG00000130724 | 0.0019 | 0.5797 | 0.003277557 | ||||||||||||||||||||||||||
| ENSG00000101473 | ENSMUSG00000017307 | 0.0799 | 0.6017 | 0.132790427 | ENSMUSG00000000538 | ENSG00000175662 | 0.0264 | 0.5805 | 0.045478036 | ||||||||||||||||||||||||||
| ENSG00000124140 | ENSMUSG00000017740 | 0.0139 | 0.6018 | 0.023097375 | ENSMUSG00000022229 | ENSG00000075673 | 0.0721 | 0.5805 | 0.124203273 | ||||||||||||||||||||||||||
| ENSG00000128050 | ENSMUSG00000029247 | 0.025 | 0.602 | 0.041528239 | ENSMUSG00000062190 | ENSG00000132434 | 0.0582 | 0.5812 | 0.100137646 | ||||||||||||||||||||||||||
| ENSG00000134202 | ENSMUSG00000004032 | 0.0661 | 0.6021 | 0.109782428 | ENSMUSG00000034675 | ENSG00000113758 | 0.0792 | 0.5821 | 0.136059096 | ||||||||||||||||||||||||||
| ENSG00000131828 | ENSMUSG00000031299 | 0.0108 | 0.6022 | 0.017934241 | ENSMUSG00000039419 | ENSG00000174469 | 0.0322 | 0.5834 | 0.055193692 | ||||||||||||||||||||||||||
| ENSG00000112304 | ENSMUSG00000006717 | 0.0991 | 0.6022 | 0.164563268 | ENSMUSG00000026437 | ENSG00000117266 | 0.04 | 0.5835 | 0.068551842 | ||||||||||||||||||||||||||
| ENSG00000115275 | ENSMUSG00000030036 | 0.0782 | 0.6034 | 0.129598939 | ENSMUSG00000046173 | ENSG00000151846 | 0.1326 | 0.5842 | 0.226977063 | ||||||||||||||||||||||||||
| ENSG00000079950 | ENSMUSG00000019998 | 0.0298 | 0.604 | 0.049337748 | ENSMUSG00000030120 | ENSG00000089693 | 0.0125 | 0.5845 | 0.0213858 | ||||||||||||||||||||||||||
| ENSG00000154277 | ENSMUSG00000029223 | 0.0213 | 0.6044 | 0.035241562 | ENSMUSG00000032625 | ENSG00000005108 | 0.0467 | 0.5845 | 0.079897348 | ||||||||||||||||||||||||||
| ENSG00000111731 | ENSMUSG00000030279 | 0.0187 | 0.6047 | 0.030924425 | ENSMUSG00000023192 | ENSG00000164082 | 0.0119 | 0.5848 | 0.020348837 | ||||||||||||||||||||||||||
| ENSG00000090989 | ENSMUSG00000036435 | 0.0129 | 0.6049 | 0.021325839 | ENSMUSG00000041020 | ENSG00000184368 | 0.1463 | 0.5857 | 0.24978658 | ||||||||||||||||||||||||||
| ENSG00000069424 | ENSMUSG00000028931 | 0.0047 | 0.6052 | 0.007766028 | ENSMUSG00000030302 | ENSG00000157087 | 0.0049 | 0.5859 | 0.008363202 | ||||||||||||||||||||||||||
| ENSG00000164111 | ENSMUSG00000027712 | 0.0374 | 0.6054 | 0.061777337 | ENSMUSG00000031775 | ENSG00000102934 | 0.0572 | 0.5859 | 0.097627581 | ||||||||||||||||||||||||||
| ENSG00000137312 | ENSMUSG00000059714 | 0.0074 | 0.6061 | 0.012209206 | ENSMUSG00000044667 | ENSG00000117600 | 0.018 | 0.5861 | 0.030711483 | ||||||||||||||||||||||||||
| ENSG00000075415 | ENSMUSG00000061904 | 0.0598 | 0.6065 | 0.098598516 | ENSMUSG00000038084 | ENSG00000198836 | 0.0189 | 0.5863 | 0.032236057 | ||||||||||||||||||||||||||
| ENSG00000119705 | ENSMUSG00000021040 | 0.1906 | 0.6069 | 0.314055034 | ENSMUSG00000074886 | ENSG00000198055 | 0.0199 | 0.5865 | 0.033930094 | ||||||||||||||||||||||||||
| ENSG00000156261 | ENSMUSG00000025613 | 0.018 | 0.6072 | 0.029644269 | ENSMUSG00000009112 | ENSG00000099968 | 0.1496 | 0.5871 | 0.254811787 | ||||||||||||||||||||||||||
| ENSG00000108946 | ENSMUSG00000020612 | 0.016 | 0.6073 | 0.026346122 | ENSMUSG00000059003 | ENSG00000183454 | 0.0229 | 0.5876 | 0.03897209 | ||||||||||||||||||||||||||
| ENSG00000143870 | ENSMUSG00000020571 | 0.0276 | 0.6075 | 0.045432099 | ENSMUSG00000046173 | ENSG00000070756 | 0.1005 | 0.5879 | 0.17094744 | ||||||||||||||||||||||||||
| ENSG00000186642 | ENSMUSG00000030653 | 0.0306 | 0.6083 | 0.050304126 | ENSMUSG00000028480 | ENSG00000122694 | 0.0331 | 0.5882 | 0.056273376 | ||||||||||||||||||||||||||
| ENSG00000070718 | ENSMUSG00000031539 | 0.0079 | 0.6085 | 0.012982744 | ENSMUSG00000027709 | ENSG00000078070 | 0.0855 | 0.5883 | 0.145334013 | ||||||||||||||||||||||||||
| ENSG00000147475 | ENSMUSG00000031483 | 0.0121 | 0.6086 | 0.019881696 | ENSMUSG00000024697 | ENSG00000156049 | 0.0152 | 0.5885 | 0.025828377 | ||||||||||||||||||||||||||
| ENSG00000084710 | ENSMUSG00000020658 | 0.0115 | 0.6091 | 0.018880315 | ENSMUSG00000049100 | ENSG00000138650 | 0.0161 | 0.589 | 0.027334465 | ||||||||||||||||||||||||||
| ENSG00000187134 | ENSMUSG00000033715 | 0.2061 | 0.6091 | 0.338368084 | ENSMUSG00000034336 | ENSG00000148798 | 0.0277 | 0.5903 | 0.046925292 | ||||||||||||||||||||||||||
| ENSG00000122778 | ENSMUSG00000063455 | 0.1824 | 0.6095 | 0.29926169 | ENSMUSG00000001525 | ENSG00000196230 | 0 | 0.5904 | 0 | ||||||||||||||||||||||||||
| ENSG00000137486 | ENSMUSG00000018909 | 0.0064 | 0.6096 | 0.010498688 | ENSMUSG00000071014 | ENSG00000165264 | 0.2153 | 0.5905 | 0.364606266 | ||||||||||||||||||||||||||
| ENSG00000102879 | ENSMUSG00000030707 | 0.0268 | 0.6098 | 0.043948836 | ENSMUSG00000026825 | ENSG00000106976 | 0.0069 | 0.5908 | 0.011679079 | ||||||||||||||||||||||||||
| ENSG00000171992 | ENSMUSG00000043079 | 0.0903 | 0.6098 | 0.148081338 | ENSMUSG00000032435 | ENSG00000144635 | 0.0277 | 0.5908 | 0.046885579 | ||||||||||||||||||||||||||
| ENSG00000080824 | ENSMUSG00000021270 | 0.0036 | 0.61 | 0.005901639 | ENSMUSG00000024524 | ENSG00000141404 | 0.0402 | 0.5909 | 0.068031816 | ||||||||||||||||||||||||||
| ENSG00000038382 | ENSMUSG00000022263 | 0.0161 | 0.6108 | 0.026358874 | ENSMUSG00000027827 | ENSG00000169282 | 0.0842 | 0.5909 | 0.1424945 | ||||||||||||||||||||||||||
| ENSG00000072210 | ENSMUSG00000010025 | 0.0879 | 0.6114 | 0.1437684 | ENSMUSG00000024319 | ENSG00000223501 | 0.0098 | 0.5914 | 0.016570849 | ||||||||||||||||||||||||||
| ENSG00000138363 | ENSMUSG00000026192 | 0.0476 | 0.6118 | 0.077803204 | ENSMUSG00000042228 | ENSG00000254087 | 0.0175 | 0.5914 | 0.029590801 | ||||||||||||||||||||||||||
| ENSG00000158528 | ENSMUSG00000032827 | 0.0496 | 0.6123 | 0.081006043 | ENSMUSG00000031841 | ENSG00000140945 | 0.049 | 0.5914 | 0.082854244 | ||||||||||||||||||||||||||
| ENSG00000116977 | ENSMUSG00000057554 | 0.1274 | 0.6123 | 0.208067941 | ENSMUSG00000031729 | ENSG00000182149 | 0.1346 | 0.5914 | 0.227595536 | ||||||||||||||||||||||||||
| ENSG00000148303 | ENSMUSG00000070778 | 0.0236 | 0.6126 | 0.038524323 | ENSMUSG00000004892 | ENSG00000132692 | 0.0928 | 0.5917 | 0.156836235 | ||||||||||||||||||||||||||
| ENSG00000182667 | ENSMUSG00000059974 | 0.0655 | 0.6133 | 0.106799283 | ENSMUSG00000060600 | ENSG00000108515 | 0.0104 | 0.5918 | 0.017573505 | ||||||||||||||||||||||||||
| ENSG00000018625 | ENSMUSG00000007097 | 0.004 | 0.6139 | 0.006515719 | ENSMUSG00000015143 | ENSG00000072110 | 0.0039 | 0.5925 | 0.006582278 | ||||||||||||||||||||||||||
| ENSG00000150867 | ENSMUSG00000026737 | 0.0078 | 0.6142 | 0.012699446 | ENSMUSG00000042331 | ENSG00000128487 | 0.0844 | 0.5929 | 0.142351155 | ||||||||||||||||||||||||||
| ENSG00000120899 | ENSMUSG00000059456 | 0.0221 | 0.6142 | 0.035981765 | ENSMUSG00000051675 | ENSG00000119401 | 0.0208 | 0.5942 | 0.035005049 | ||||||||||||||||||||||||||
| ENSG00000165868 | ENSMUSG00000025092 | 0.016 | 0.6143 | 0.026045906 | ENSMUSG00000022340 | ENSG00000147642 | 0.0788 | 0.5951 | 0.13241472 | ||||||||||||||||||||||||||
| ENSG00000196296 | ENSMUSG00000030730 | 0.0173 | 0.6144 | 0.028157552 | ENSMUSG00000051732 | ENSG00000151846 | 0.1459 | 0.5952 | 0.245127688 | ||||||||||||||||||||||||||
| ENSG00000108561 | ENSMUSG00000018446 | 0.0713 | 0.6147 | 0.115991541 | ENSMUSG00000039768 | ENSG00000007923 | 0.0154 | 0.5956 | 0.025856279 | ||||||||||||||||||||||||||
| ENSG00000112186 | ENSMUSG00000021373 | 0.0584 | 0.6148 | 0.094990241 | ENSMUSG00000026163 | ENSG00000153820 | 0.1635 | 0.5961 | 0.274282838 | ||||||||||||||||||||||||||
| ENSG00000128564 | ENSMUSG00000037428 | 0.0702 | 0.615 | 0.114146341 | ENSMUSG00000054364 | ENSG00000143878 | 0 | 0.5967 | 0 | ||||||||||||||||||||||||||
| ENSG00000114742 | ENSMUSG00000032512 | 0.0049 | 0.6151 | 0.007966184 | ENSMUSG00000029334 | ENSG00000138669 | 0.0153 | 0.599 | 0.025542571 | ||||||||||||||||||||||||||
| ENSG00000055163 | ENSMUSG00000020340 | 0.0006 | 0.6155 | 0.000974817 | ENSMUSG00000037541 | ENSG00000162105 | 0.0774 | 0.5993 | 0.129150676 | ||||||||||||||||||||||||||
| ENSG00000177105 | ENSMUSG00000073982 | 0 | 0.6156 | 0 | ENSMUSG00000017307 | ENSG00000101473 | 0.0799 | 0.6017 | 0.132790427 | ||||||||||||||||||||||||||
| ENSG00000213366 | ENSMUSG00000040562 | 0.1219 | 0.6156 | 0.198018194 | ENSMUSG00000017740 | ENSG00000124140 | 0.0139 | 0.6018 | 0.023097375 | ||||||||||||||||||||||||||
| ENSG00000128609 | ENSMUSG00000023089 | 0.0967 | 0.616 | 0.156980519 | ENSMUSG00000029247 | ENSG00000128050 | 0.025 | 0.602 | 0.041528239 | ||||||||||||||||||||||||||
| ENSG00000168280 | ENSMUSG00000026764 | 0.0076 | 0.6162 | 0.012333658 | ENSMUSG00000031299 | ENSG00000131828 | 0.0108 | 0.6022 | 0.017934241 | ||||||||||||||||||||||||||
| ENSG00000116141 | ENSMUSG00000026620 | 0.028 | 0.6167 | 0.045402951 | ENSMUSG00000026032 | ENSG00000119013 | 0.1361 | 0.6023 | 0.225967126 | ||||||||||||||||||||||||||
| ENSG00000118873 | ENSMUSG00000039318 | 0.0577 | 0.6172 | 0.093486714 | ENSMUSG00000030036 | ENSG00000115275 | 0.0782 | 0.6034 | 0.129598939 | ||||||||||||||||||||||||||
| ENSG00000159685 | ENSMUSG00000030086 | 0.2027 | 0.618 | 0.327993528 | ENSMUSG00000023439 | ENSG00000111664 | 0.0141 | 0.6037 | 0.023355972 | ||||||||||||||||||||||||||
| ENSG00000052126 | ENSMUSG00000030231 | 0.0859 | 0.6182 | 0.138951796 | ENSMUSG00000060882 | ENSG00000184408 | 0.0044 | 0.6038 | 0.007287181 | ||||||||||||||||||||||||||
| ENSG00000165566 | ENSMUSG00000021986 | 0.1048 | 0.6182 | 0.169524426 | ENSMUSG00000030682 | ENSG00000103502 | 0.0165 | 0.6038 | 0.027326929 | ||||||||||||||||||||||||||
| ENSG00000198242 | ENSMUSG00000058546 | 0 | 0.6187 | 0 | ENSMUSG00000019998 | ENSG00000079950 | 0.0298 | 0.604 | 0.049337748 | ||||||||||||||||||||||||||
| ENSG00000159842 | ENSMUSG00000017631 | 0.0166 | 0.6189 | 0.026821781 | ENSMUSG00000029223 | ENSG00000154277 | 0.0213 | 0.6044 | 0.035241562 | ||||||||||||||||||||||||||
| ENSG00000151632 | ENSMUSG00000033715 | 0.2123 | 0.6193 | 0.342806394 | ENSMUSG00000036435 | ENSG00000090989 | 0.0129 | 0.6049 | 0.021325839 | ||||||||||||||||||||||||||
| ENSG00000186197 | ENSMUSG00000039309 | 0.1439 | 0.6197 | 0.232209133 | ENSMUSG00000028931 | ENSG00000069424 | 0.0047 | 0.6052 | 0.007766028 | ||||||||||||||||||||||||||
| ENSG00000040731 | ENSMUSG00000022321 | 0.0167 | 0.6199 | 0.026939829 | ENSMUSG00000028005 | ENSG00000061918 | 0.0053 | 0.6052 | 0.008757436 | ||||||||||||||||||||||||||
| ENSG00000108852 | ENSMUSG00000017314 | 0.012 | 0.62 | 0.019354839 | ENSMUSG00000059714 | ENSG00000137312 | 0.0074 | 0.6061 | 0.012209206 | ||||||||||||||||||||||||||
| ENSG00000152137 | ENSMUSG00000041548 | 0.0286 | 0.6201 | 0.046121593 | ENSMUSG00000061904 | ENSG00000075415 | 0.0598 | 0.6065 | 0.098598516 | ||||||||||||||||||||||||||
| ENSG00000155980 | ENSMUSG00000074657 | 0.0098 | 0.6205 | 0.015793715 | ENSMUSG00000025613 | ENSG00000156261 | 0.018 | 0.6072 | 0.029644269 | ||||||||||||||||||||||||||
| ENSG00000133612 | ENSMUSG00000023353 | 0.0191 | 0.6205 | 0.030781628 | ENSMUSG00000020612 | ENSG00000108946 | 0.016 | 0.6073 | 0.026346122 | ||||||||||||||||||||||||||
| ENSG00000157654 | ENSMUSG00000089945 | 0.1013 | 0.6212 | 0.163071475 | ENSMUSG00000002319 | ENSG00000196497 | 0.1035 | 0.6073 | 0.170426478 | ||||||||||||||||||||||||||
| ENSG00000157020 | ENSMUSG00000030298 | 0.0141 | 0.6215 | 0.022687047 | ENSMUSG00000027965 | ENSG00000118733 | 0.024 | 0.6078 | 0.039486673 | ||||||||||||||||||||||||||
| ENSG00000156453 | ENSMUSG00000051375 | 0.0329 | 0.6215 | 0.052936444 | ENSMUSG00000047959 | ENSG00000177272 | 0.0124 | 0.608 | 0.020394737 | ||||||||||||||||||||||||||
| ENSG00000198242 | ENSMUSG00000078126 | 0.0029 | 0.6225 | 0.004658635 | ENSMUSG00000030653 | ENSG00000186642 | 0.0306 | 0.6083 | 0.050304126 | ||||||||||||||||||||||||||
| ENSG00000117480 | ENSMUSG00000034171 | 0.0895 | 0.6231 | 0.143636655 | ENSMUSG00000031539 | ENSG00000070718 | 0.0079 | 0.6085 | 0.012982744 | ||||||||||||||||||||||||||
| ENSG00000101298 | ENSMUSG00000027457 | 0.0426 | 0.6239 | 0.068280173 | ENSMUSG00000031483 | ENSG00000147475 | 0.0121 | 0.6086 | 0.019881696 | ||||||||||||||||||||||||||
| ENSG00000103942 | ENSMUSG00000025813 | 0.0242 | 0.6251 | 0.038713806 | ENSMUSG00000006476 | ENSG00000165802 | 0.0261 | 0.609 | 0.042857143 | ||||||||||||||||||||||||||
| ENSG00000103415 | ENSMUSG00000004070 | 0.0548 | 0.6257 | 0.087581908 | ENSMUSG00000020658 | ENSG00000084710 | 0.0115 | 0.6091 | 0.018880315 | ||||||||||||||||||||||||||
| ENSG00000167114 | ENSMUSG00000059316 | 0.0399 | 0.626 | 0.063738019 | ENSMUSG00000030707 | ENSG00000102879 | 0.0268 | 0.6098 | 0.043948836 | ||||||||||||||||||||||||||
| ENSG00000104722 | ENSMUSG00000022054 | 0.0413 | 0.6263 | 0.065942839 | ENSMUSG00000043079 | ENSG00000171992 | 0.0903 | 0.6098 | 0.148081338 | ||||||||||||||||||||||||||
| ENSG00000120254 | ENSMUSG00000040675 | 0.0652 | 0.6263 | 0.104103465 | ENSMUSG00000021270 | ENSG00000080824 | 0.0036 | 0.61 | 0.005901639 | ||||||||||||||||||||||||||
| ENSG00000198721 | ENSMUSG00000021417 | 0.1493 | 0.6274 | 0.23796621 | ENSMUSG00000003178 | ENSG00000243156 | 0.0483 | 0.6107 | 0.079089569 | ||||||||||||||||||||||||||
| ENSG00000066557 | ENSMUSG00000063052 | 0.0996 | 0.6275 | 0.1587251 | ENSMUSG00000022263 | ENSG00000038382 | 0.0161 | 0.6108 | 0.026358874 | ||||||||||||||||||||||||||
| ENSG00000167815 | ENSMUSG00000005161 | 0.0305 | 0.6278 | 0.048582351 | ENSMUSG00000010025 | ENSG00000072210 | 0.0879 | 0.6114 | 0.1437684 | ||||||||||||||||||||||||||
| ENSG00000100401 | ENSMUSG00000022391 | 0.0574 | 0.629 | 0.091255962 | ENSMUSG00000019087 | ENSG00000071553 | 0.0777 | 0.6121 | 0.126940042 | ||||||||||||||||||||||||||
| ENSG00000163995 | ENSMUSG00000029095 | 0.0407 | 0.6307 | 0.064531473 | ENSMUSG00000032827 | ENSG00000158528 | 0.0496 | 0.6123 | 0.081006043 | ||||||||||||||||||||||||||
| ENSG00000151150 | ENSMUSG00000069601 | 0.0274 | 0.6311 | 0.043416257 | ENSMUSG00000059974 | ENSG00000182667 | 0.0655 | 0.6133 | 0.106799283 | ||||||||||||||||||||||||||
| ENSG00000146731 | ENSMUSG00000029447 | 0.0641 | 0.6315 | 0.101504355 | ENSMUSG00000007097 | ENSG00000018625 | 0.004 | 0.6139 | 0.006515719 | ||||||||||||||||||||||||||
| ENSG00000109971 | ENSMUSG00000015656 | 0.0007 | 0.633 | 0.001105845 | ENSMUSG00000026737 | ENSG00000150867 | 0.0078 | 0.6142 | 0.012699446 | ||||||||||||||||||||||||||
| ENSG00000196586 | ENSMUSG00000033577 | 0.0153 | 0.6336 | 0.024147727 | ENSMUSG00000025092 | ENSG00000165868 | 0.016 | 0.6143 | 0.026045906 | ||||||||||||||||||||||||||
| ENSG00000175084 | ENSMUSG00000026208 | 0.0099 | 0.6349 | 0.015593007 | ENSMUSG00000030730 | ENSG00000196296 | 0.0173 | 0.6144 | 0.028157552 | ||||||||||||||||||||||||||
| ENSG00000213088 | ENSMUSG00000037872 | 0.3473 | 0.6352 | 0.546756927 | ENSMUSG00000033595 | ENSG00000168481 | 0.0156 | 0.6148 | 0.025374105 | ||||||||||||||||||||||||||
| ENSG00000072832 | ENSMUSG00000029121 | 0.0157 | 0.6366 | 0.024662268 | ENSMUSG00000032512 | ENSG00000114742 | 0.0049 | 0.6151 | 0.007966184 | ||||||||||||||||||||||||||
| ENSG00000119139 | ENSMUSG00000024812 | 0.0674 | 0.6369 | 0.10582509 | ENSMUSG00000020340 | ENSG00000055163 | 0.0006 | 0.6155 | 0.000974817 | ||||||||||||||||||||||||||
| ENSG00000103507 | ENSMUSG00000030802 | 0.0236 | 0.637 | 0.037048666 | ENSMUSG00000073982 | ENSG00000177105 | 0 | 0.6156 | 0 | ||||||||||||||||||||||||||
| ENSG00000131188 | ENSMUSG00000034686 | 0.0126 | 0.6372 | 0.019774011 | ENSMUSG00000026764 | ENSG00000168280 | 0.0076 | 0.6162 | 0.012333658 | ||||||||||||||||||||||||||
| ENSG00000198242 | ENSMUSG00000063556 | 0 | 0.6375 | 0 | ENSMUSG00000026620 | ENSG00000116141 | 0.028 | 0.6167 | 0.045402951 | ||||||||||||||||||||||||||
| ENSG00000075539 | ENSMUSG00000070733 | 0.0266 | 0.6381 | 0.041686256 | ENSMUSG00000039318 | ENSG00000118873 | 0.0577 | 0.6172 | 0.093486714 | ||||||||||||||||||||||||||
| ENSG00000068903 | ENSMUSG00000015149 | 0.0681 | 0.6381 | 0.106723084 | ENSMUSG00000040136 | ENSG00000006071 | 0.0229 | 0.6173 | 0.037097035 | ||||||||||||||||||||||||||
| ENSG00000172809 | ENSMUSG00000057322 | 0.0065 | 0.6395 | 0.010164191 | ENSMUSG00000030086 | ENSG00000159685 | 0.2027 | 0.618 | 0.327993528 | ||||||||||||||||||||||||||
| ENSG00000102606 | ENSMUSG00000031511 | 0.0266 | 0.6396 | 0.041588493 | ENSMUSG00000030231 | ENSG00000052126 | 0.0859 | 0.6182 | 0.138951796 | ||||||||||||||||||||||||||
| ENSG00000107863 | ENSMUSG00000036591 | 0.0742 | 0.6397 | 0.115991871 | ENSMUSG00000058546 | ENSG00000198242 | 0 | 0.6187 | 0 | ||||||||||||||||||||||||||
| ENSG00000162688 | ENSMUSG00000033400 | 0.0409 | 0.6401 | 0.063896266 | ENSMUSG00000017631 | ENSG00000159842 | 0.0166 | 0.6189 | 0.026821781 | ||||||||||||||||||||||||||
| ENSG00000186868 | ENSMUSG00000018411 | 0.0469 | 0.6411 | 0.073155514 | ENSMUSG00000022321 | ENSG00000040731 | 0.0167 | 0.6199 | 0.026939829 | ||||||||||||||||||||||||||
| ENSG00000125633 | ENSMUSG00000026339 | 0.0403 | 0.6421 | 0.06276281 | ENSMUSG00000017314 | ENSG00000108852 | 0.012 | 0.62 | 0.019354839 | ||||||||||||||||||||||||||
| ENSG00000184144 | ENSMUSG00000053024 | 0.0495 | 0.6432 | 0.076958955 | ENSMUSG00000074657 | ENSG00000155980 | 0.0098 | 0.6205 | 0.015793715 | ||||||||||||||||||||||||||
| ENSG00000136848 | ENSMUSG00000026883 | 0.0192 | 0.6433 | 0.029846106 | ENSMUSG00000023353 | ENSG00000133612 | 0.0191 | 0.6205 | 0.030781628 | ||||||||||||||||||||||||||
| ENSG00000105223 | ENSMUSG00000003363 | 0.0301 | 0.644 | 0.04673913 | ENSMUSG00000045659 | ENSG00000166689 | 0.0611 | 0.622 | 0.098231511 | ||||||||||||||||||||||||||
| ENSG00000090372 | ENSMUSG00000030374 | 0.0261 | 0.6442 | 0.040515368 | ENSMUSG00000003178 | ENSG00000093100 | 0.0549 | 0.6221 | 0.088249478 | ||||||||||||||||||||||||||
| ENSG00000182902 | ENSMUSG00000004902 | 0.0638 | 0.6443 | 0.099022195 | ENSMUSG00000034171 | ENSG00000117480 | 0.0895 | 0.6231 | 0.143636655 | ||||||||||||||||||||||||||
| ENSG00000177963 | ENSMUSG00000025485 | 0.0647 | 0.6446 | 0.100372324 | ENSMUSG00000027457 | ENSG00000101298 | 0.0426 | 0.6239 | 0.068280173 | ||||||||||||||||||||||||||
| ENSG00000175220 | ENSMUSG00000027247 | 0.0176 | 0.6471 | 0.027198269 | ENSMUSG00000025813 | ENSG00000103942 | 0.0242 | 0.6251 | 0.038713806 | ||||||||||||||||||||||||||
| ENSG00000115520 | ENSMUSG00000025981 | 0.0822 | 0.6477 | 0.126910607 | ENSMUSG00000039067 | ENSG00000103035 | 0.0166 | 0.6257 | 0.026530286 | ||||||||||||||||||||||||||
| ENSG00000088387 | ENSMUSG00000025558 | 0.0254 | 0.6482 | 0.039185437 | ENSMUSG00000047675 | ENSG00000142937 | 0 | 0.6259 | 0 | ||||||||||||||||||||||||||
| ENSG00000104325 | ENSMUSG00000028223 | 0.0988 | 0.6483 | 0.152398581 | ENSMUSG00000059316 | ENSG00000167114 | 0.0399 | 0.626 | 0.063738019 | ||||||||||||||||||||||||||
| ENSG00000144908 | ENSMUSG00000030088 | 0.0395 | 0.6484 | 0.060919186 | ENSMUSG00000022054 | ENSG00000104722 | 0.0413 | 0.6263 | 0.065942839 | ||||||||||||||||||||||||||
| ENSG00000148175 | ENSMUSG00000026880 | 0.0726 | 0.6487 | 0.11191614 | ENSMUSG00000040675 | ENSG00000120254 | 0.0652 | 0.6263 | 0.104103465 | ||||||||||||||||||||||||||
| ENSG00000116266 | ENSMUSG00000071640 | 0.0789 | 0.649 | 0.121571649 | ENSMUSG00000005161 | ENSG00000167815 | 0.0305 | 0.6278 | 0.048582351 | ||||||||||||||||||||||||||
| ENSG00000050405 | ENSMUSG00000023022 | 0.1394 | 0.6493 | 0.214692746 | ENSMUSG00000020109 | ENSG00000148719 | 0.0273 | 0.6303 | 0.043312708 | ||||||||||||||||||||||||||
| ENSG00000204314 | ENSMUSG00000015476 | 0.0146 | 0.6498 | 0.022468452 | ENSMUSG00000029838 | ENSG00000105894 | 0.0134 | 0.6304 | 0.021256345 | ||||||||||||||||||||||||||
| ENSG00000163902 | ENSMUSG00000030062 | 0.0243 | 0.6499 | 0.037390368 | ENSMUSG00000017404 | ENSG00000108298 | 0 | 0.6307 | 0 | ||||||||||||||||||||||||||
| ENSG00000156931 | ENSMUSG00000033653 | 0.0376 | 0.6509 | 0.05776617 | ENSMUSG00000029095 | ENSG00000163995 | 0.0407 | 0.6307 | 0.064531473 | ||||||||||||||||||||||||||
| ENSG00000102189 | ENSMUSG00000036499 | 0.0842 | 0.6518 | 0.12918073 | ENSMUSG00000028234 | ENSG00000008988 | 0 | 0.6308 | 0 | ||||||||||||||||||||||||||
| ENSG00000133026 | ENSMUSG00000020900 | 0.0041 | 0.652 | 0.006288344 | ENSMUSG00000069601 | ENSG00000151150 | 0.0274 | 0.6311 | 0.043416257 | ||||||||||||||||||||||||||
| ENSG00000106367 | ENSMUSG00000004849 | 0 | 0.6521 | 0 | ENSMUSG00000050211 | ENSG00000188089 | 0.1452 | 0.6314 | 0.229965157 | ||||||||||||||||||||||||||
| ENSG00000132932 | ENSMUSG00000021983 | 0.0291 | 0.6525 | 0.044597701 | ENSMUSG00000036333 | ENSG00000134313 | 0.0297 | 0.6315 | 0.047030879 | ||||||||||||||||||||||||||
| ENSG00000177156 | ENSMUSG00000025503 | 0.0273 | 0.6536 | 0.041768666 | ENSMUSG00000029447 | ENSG00000146731 | 0.0641 | 0.6315 | 0.101504355 | ||||||||||||||||||||||||||
| ENSG00000173786 | ENSMUSG00000006782 | 0.0728 | 0.6544 | 0.111246944 | ENSMUSG00000003410 | ENSG00000196361 | 0.0023 | 0.6316 | 0.003641545 | ||||||||||||||||||||||||||
| ENSG00000105402 | ENSMUSG00000006024 | 0.0074 | 0.6545 | 0.011306341 | ENSMUSG00000015656 | ENSG00000109971 | 0.0007 | 0.633 | 0.001105845 | ||||||||||||||||||||||||||
| ENSG00000197694 | ENSMUSG00000057738 | 0.0075 | 0.6547 | 0.011455629 | ENSMUSG00000078695 | ENSG00000230055 | 0.0959 | 0.6331 | 0.15147686 | ||||||||||||||||||||||||||
| ENSG00000121053 | ENSMUSG00000052234 | 0.0558 | 0.6549 | 0.085203848 | ENSMUSG00000024410 | ENSG00000141452 | 0.0255 | 0.6335 | 0.040252565 | ||||||||||||||||||||||||||
| ENSG00000182473 | ENSMUSG00000020792 | 0.0163 | 0.6551 | 0.024881697 | ENSMUSG00000033577 | ENSG00000196586 | 0.0153 | 0.6336 | 0.024147727 | ||||||||||||||||||||||||||
| ENSG00000029534 | ENSMUSG00000031543 | 0.0478 | 0.6551 | 0.072965959 | ENSMUSG00000004285 | ENSG00000226138 | 0.0579 | 0.6348 | 0.09120983 | ||||||||||||||||||||||||||
| ENSG00000116337 | ENSMUSG00000027889 | 0.014 | 0.6553 | 0.021364261 | ENSMUSG00000026208 | ENSG00000175084 | 0.0099 | 0.6349 | 0.015593007 | ||||||||||||||||||||||||||
| ENSG00000017373 | ENSMUSG00000038453 | 0.017 | 0.6555 | 0.025934401 | ENSMUSG00000048895 | ENSG00000176749 | 0.0067 | 0.6357 | 0.010539563 | ||||||||||||||||||||||||||
| ENSG00000163346 | ENSMUSG00000042613 | 0.1718 | 0.6559 | 0.261930172 | ENSMUSG00000021277 | ENSG00000131323 | 0.017 | 0.6361 | 0.026725358 | ||||||||||||||||||||||||||
| ENSG00000168589 | ENSMUSG00000034467 | 0.0097 | 0.6574 | 0.014755096 | ENSMUSG00000029121 | ENSG00000072832 | 0.0157 | 0.6366 | 0.024662268 | ||||||||||||||||||||||||||
| ENSG00000142949 | ENSMUSG00000033295 | 0.0199 | 0.6578 | 0.030252356 | ENSMUSG00000024812 | ENSG00000119139 | 0.0674 | 0.6369 | 0.10582509 | ||||||||||||||||||||||||||
| ENSG00000147459 | ENSMUSG00000044447 | 0.0305 | 0.6582 | 0.046338499 | ENSMUSG00000030802 | ENSG00000103507 | 0.0236 | 0.637 | 0.037048666 | ||||||||||||||||||||||||||
| ENSG00000134265 | ENSMUSG00000024581 | 0.0091 | 0.6585 | 0.013819286 | ENSMUSG00000034686 | ENSG00000131188 | 0.0126 | 0.6372 | 0.019774011 | ||||||||||||||||||||||||||
| ENSG00000115159 | ENSMUSG00000026827 | 0.0359 | 0.6586 | 0.054509566 | ENSMUSG00000020926 | ENSG00000073670 | 0.0305 | 0.6376 | 0.047835634 | ||||||||||||||||||||||||||
| ENSG00000116584 | ENSMUSG00000028059 | 0.0603 | 0.6601 | 0.091349795 | ENSMUSG00000057322 | ENSG00000172809 | 0.0065 | 0.6395 | 0.010164191 | ||||||||||||||||||||||||||
| ENSG00000144867 | ENSMUSG00000032553 | 0.045 | 0.6603 | 0.068150841 | ENSMUSG00000036591 | ENSG00000107863 | 0.0742 | 0.6397 | 0.115991871 | ||||||||||||||||||||||||||
| ENSG00000114353 | ENSMUSG00000032562 | 0.0083 | 0.6606 | 0.012564335 | ENSMUSG00000028184 | ENSG00000117114 | 0.0213 | 0.6402 | 0.033270853 | ||||||||||||||||||||||||||
| ENSG00000042753 | ENSMUSG00000008036 | 0 | 0.6616 | 0 | ENSMUSG00000042671 | ENSG00000135824 | 0.03 | 0.6404 | 0.046845721 | ||||||||||||||||||||||||||
| ENSG00000067225 | ENSMUSG00000032294 | 0.0387 | 0.662 | 0.058459215 | ENSMUSG00000018411 | ENSG00000186868 | 0.0469 | 0.6411 | 0.073155514 | ||||||||||||||||||||||||||
| ENSG00000134419 | ENSMUSG00000067351 | 0.031 | 0.6627 | 0.046778331 | ENSMUSG00000053024 | ENSG00000184144 | 0.0495 | 0.6432 | 0.076958955 | ||||||||||||||||||||||||||
| ENSG00000171564 | ENSMUSG00000033831 | 0.0916 | 0.6627 | 0.138222423 | ENSMUSG00000026883 | ENSG00000136848 | 0.0192 | 0.6433 | 0.029846106 | ||||||||||||||||||||||||||
| ENSG00000182054 | ENSMUSG00000030541 | 0.0218 | 0.6628 | 0.032890766 | ENSMUSG00000034040 | ENSG00000185274 | 0.0067 | 0.644 | 0.010403727 | ||||||||||||||||||||||||||
| ENSG00000077522 | ENSMUSG00000052374 | 0.0033 | 0.6637 | 0.004972126 | ENSMUSG00000030374 | ENSG00000090372 | 0.0261 | 0.6442 | 0.040515368 | ||||||||||||||||||||||||||
| ENSG00000172531 | ENSMUSG00000040385 | 0.0013 | 0.6645 | 0.001956358 | ENSMUSG00000004902 | ENSG00000182902 | 0.0638 | 0.6443 | 0.099022195 | ||||||||||||||||||||||||||
| ENSG00000132612 | ENSMUSG00000031913 | 0.0041 | 0.6648 | 0.006167268 | ENSMUSG00000030731 | ENSG00000213023 | 0.0167 | 0.6464 | 0.025835396 | ||||||||||||||||||||||||||
| ENSG00000198910 | ENSMUSG00000031391 | 0.0592 | 0.6655 | 0.088955672 | ENSMUSG00000021338 | ENSG00000079691 | 0.056 | 0.6471 | 0.086539947 | ||||||||||||||||||||||||||
| ENSG00000131844 | ENSMUSG00000021646 | 0.0623 | 0.6656 | 0.09359976 | ENSMUSG00000024851 | ENSG00000110697 | 0.0325 | 0.6473 | 0.050208559 | ||||||||||||||||||||||||||
| ENSG00000113319 | ENSMUSG00000021708 | 0.0752 | 0.6659 | 0.112929869 | ENSMUSG00000026895 | ENSG00000119421 | 0.0786 | 0.6481 | 0.121277581 | ||||||||||||||||||||||||||
| ENSG00000185236 | ENSMUSG00000077450 | 0.0039 | 0.6664 | 0.005852341 | ENSMUSG00000025558 | ENSG00000088387 | 0.0254 | 0.6482 | 0.039185437 | ||||||||||||||||||||||||||
| ENSG00000117394 | ENSMUSG00000028645 | 0.0156 | 0.6669 | 0.023391813 | ENSMUSG00000028223 | ENSG00000104325 | 0.0988 | 0.6483 | 0.152398581 | ||||||||||||||||||||||||||
| ENSG00000132970 | ENSMUSG00000029636 | 0.0318 | 0.6677 | 0.047626179 | ENSMUSG00000015476 | ENSG00000204314 | 0.0146 | 0.6498 | 0.022468452 | ||||||||||||||||||||||||||
| ENSG00000116906 | ENSMUSG00000031985 | 0.1116 | 0.6678 | 0.167115903 | ENSMUSG00000030062 | ENSG00000163902 | 0.0243 | 0.6499 | 0.037390368 | ||||||||||||||||||||||||||
| ENSG00000100266 | ENSMUSG00000016664 | 0.0308 | 0.6693 | 0.046018228 | ENSMUSG00000042870 | ENSG00000100284 | 0.0561 | 0.6503 | 0.086267876 | ||||||||||||||||||||||||||
| ENSG00000075624 | ENSMUSG00000029580 | 0.0022 | 0.6694 | 0.003286525 | ENSMUSG00000025204 | ENSG00000255339 | 0.1511 | 0.6505 | 0.232282859 | ||||||||||||||||||||||||||
| ENSG00000214253 | ENSMUSG00000019054 | 0.0202 | 0.6698 | 0.030158256 | ENSMUSG00000020900 | ENSG00000133026 | 0.0041 | 0.652 | 0.006288344 | ||||||||||||||||||||||||||
| ENSG00000157985 | ENSMUSG00000055013 | 0.0106 | 0.671 | 0.015797317 | ENSMUSG00000004849 | ENSG00000106367 | 0 | 0.6521 | 0 | ||||||||||||||||||||||||||
| ENSG00000139180 | ENSMUSG00000000399 | 0.1431 | 0.6714 | 0.213136729 | ENSMUSG00000045034 | ENSG00000189127 | 0.1044 | 0.6524 | 0.160024525 | ||||||||||||||||||||||||||
| ENSG00000154654 | ENSMUSG00000022762 | 0.0311 | 0.6715 | 0.046314222 | ENSMUSG00000021559 | ENSG00000196730 | 0.0238 | 0.6527 | 0.036463919 | ||||||||||||||||||||||||||
| ENSG00000148303 | ENSMUSG00000068579 | 0.0105 | 0.6723 | 0.015618028 | ENSMUSG00000078137 | ENSG00000230778 | 0.0419 | 0.6527 | 0.064194883 | ||||||||||||||||||||||||||
| ENSG00000148303 | ENSMUSG00000062758 | 0.0035 | 0.6724 | 0.005205235 | ENSMUSG00000026520 | ENSG00000143811 | 0.0377 | 0.6544 | 0.057610024 | ||||||||||||||||||||||||||
| ENSG00000175416 | ENSMUSG00000047547 | 0.0228 | 0.6725 | 0.033903346 | ENSMUSG00000006782 | ENSG00000173786 | 0.0728 | 0.6544 | 0.111246944 | ||||||||||||||||||||||||||
| ENSG00000104267 | ENSMUSG00000027562 | 0.1117 | 0.6729 | 0.165997919 | ENSMUSG00000006024 | ENSG00000105402 | 0.0074 | 0.6545 | 0.011306341 | ||||||||||||||||||||||||||
| ENSG00000164303 | ENSMUSG00000038173 | 0.0738 | 0.6733 | 0.109609387 | ENSMUSG00000020599 | ENSG00000108370 | 0.0451 | 0.6546 | 0.068897036 | ||||||||||||||||||||||||||
| ENSG00000130427 | ENSMUSG00000029711 | 0.1276 | 0.6737 | 0.189401811 | ENSMUSG00000057738 | ENSG00000197694 | 0.0075 | 0.6547 | 0.011455629 | ||||||||||||||||||||||||||
| ENSG00000115073 | ENSMUSG00000037351 | 0.0047 | 0.674 | 0.006973294 | ENSMUSG00000020792 | ENSG00000182473 | 0.0163 | 0.6551 | 0.024881697 | ||||||||||||||||||||||||||
| ENSG00000183783 | ENSMUSG00000037653 | 0.0211 | 0.6743 | 0.03129171 | ENSMUSG00000031543 | ENSG00000029534 | 0.0478 | 0.6551 | 0.072965959 | ||||||||||||||||||||||||||
| ENSG00000198053 | ENSMUSG00000037902 | 0.2395 | 0.6746 | 0.3550252 | ENSMUSG00000027889 | ENSG00000116337 | 0.014 | 0.6553 | 0.021364261 | ||||||||||||||||||||||||||
| ENSG00000115415 | ENSMUSG00000026104 | 0.0336 | 0.6756 | 0.04973357 | ENSMUSG00000038453 | ENSG00000017373 | 0.017 | 0.6555 | 0.025934401 | ||||||||||||||||||||||||||
| ENSG00000150753 | ENSMUSG00000022234 | 0.0208 | 0.6757 | 0.030782892 | ENSMUSG00000003411 | ENSG00000169213 | 0.0121 | 0.6558 | 0.018450747 | ||||||||||||||||||||||||||
| ENSG00000148090 | ENSMUSG00000021460 | 0.0435 | 0.6771 | 0.064244572 | ENSMUSG00000033295 | ENSG00000142949 | 0.0199 | 0.6578 | 0.030252356 | ||||||||||||||||||||||||||
| ENSG00000137168 | ENSMUSG00000024007 | 0.0084 | 0.678 | 0.012389381 | ENSMUSG00000024581 | ENSG00000134265 | 0.0091 | 0.6585 | 0.013819286 | ||||||||||||||||||||||||||
| ENSG00000131378 | ENSMUSG00000039316 | 0.1897 | 0.679 | 0.279381443 | ENSMUSG00000026827 | ENSG00000115159 | 0.0359 | 0.6586 | 0.054509566 | ||||||||||||||||||||||||||
| ENSG00000187134 | ENSMUSG00000021211 | 0.2088 | 0.6793 | 0.307375239 | ENSMUSG00000043542 | ENSG00000104427 | 0.0684 | 0.6588 | 0.103825137 | ||||||||||||||||||||||||||
| ENSG00000106789 | ENSMUSG00000028337 | 0.0761 | 0.6803 | 0.111862414 | ENSMUSG00000028059 | ENSG00000116584 | 0.0603 | 0.6601 | 0.091349795 | ||||||||||||||||||||||||||
| ENSG00000104529 | ENSMUSG00000055762 | 0.0799 | 0.6803 | 0.117448185 | ENSMUSG00000032553 | ENSG00000144867 | 0.045 | 0.6603 | 0.068150841 | ||||||||||||||||||||||||||
| ENSG00000187134 | ENSMUSG00000021213 | 0.2224 | 0.6807 | 0.326722492 | ENSMUSG00000032562 | ENSG00000114353 | 0.0083 | 0.6606 | 0.012564335 | ||||||||||||||||||||||||||
| ENSG00000197150 | ENSMUSG00000028973 | 0.0932 | 0.6808 | 0.136897767 | ENSMUSG00000008036 | ENSG00000042753 | 0 | 0.6616 | 0 | ||||||||||||||||||||||||||
| ENSG00000163531 | ENSMUSG00000026442 | 0.0157 | 0.6811 | 0.023050947 | ENSMUSG00000032294 | ENSG00000067225 | 0.0387 | 0.662 | 0.058459215 | ||||||||||||||||||||||||||
| ENSG00000065054 | ENSMUSG00000002504 | 0.0395 | 0.6812 | 0.057985907 | ENSMUSG00000052374 | ENSG00000077522 | 0.0033 | 0.6637 | 0.004972126 | ||||||||||||||||||||||||||
| ENSG00000110880 | ENSMUSG00000004530 | 0.0122 | 0.6823 | 0.017880698 | ENSMUSG00000040385 | ENSG00000172531 | 0.0013 | 0.6645 | 0.001956358 | ||||||||||||||||||||||||||
| ENSG00000102178 | ENSMUSG00000015290 | 0.0519 | 0.6824 | 0.0760551 | ENSMUSG00000033174 | ENSG00000074416 | 0.0864 | 0.6651 | 0.129905277 | ||||||||||||||||||||||||||
| ENSG00000187134 | ENSMUSG00000054757 | 0.1593 | 0.6827 | 0.233338216 | ENSMUSG00000031391 | ENSG00000198910 | 0.0592 | 0.6655 | 0.088955672 | ||||||||||||||||||||||||||
| ENSG00000215642 | ENSMUSG00000059854 | 0.1013 | 0.6834 | 0.148229441 | ENSMUSG00000021646 | ENSG00000131844 | 0.0623 | 0.6656 | 0.09359976 | ||||||||||||||||||||||||||
| ENSG00000102882 | ENSMUSG00000063065 | 0.0124 | 0.6846 | 0.018112767 | ENSMUSG00000021708 | ENSG00000113319 | 0.0752 | 0.6659 | 0.112929869 | ||||||||||||||||||||||||||
| ENSG00000148303 | ENSMUSG00000070289 | 0.0105 | 0.6853 | 0.015321757 | ENSMUSG00000077450 | ENSG00000185236 | 0.0039 | 0.6664 | 0.005852341 | ||||||||||||||||||||||||||
| ENSG00000139116 | ENSMUSG00000022629 | 0.0291 | 0.6856 | 0.042444574 | ENSMUSG00000028645 | ENSG00000117394 | 0.0156 | 0.6669 | 0.023391813 | ||||||||||||||||||||||||||
| ENSG00000135631 | ENSMUSG00000051343 | 0.0675 | 0.686 | 0.098396501 | ENSMUSG00000029636 | ENSG00000132970 | 0.0318 | 0.6677 | 0.047626179 | ||||||||||||||||||||||||||
| ENSG00000213366 | ENSMUSG00000004035 | 0.0893 | 0.6867 | 0.130042231 | ENSMUSG00000031985 | ENSG00000116906 | 0.1116 | 0.6678 | 0.167115903 | ||||||||||||||||||||||||||
| ENSG00000103226 | ENSMUSG00000030835 | 0.0363 | 0.6876 | 0.052792321 | ENSMUSG00000041638 | ENSG00000089154 | 0.031 | 0.6682 | 0.046393295 | ||||||||||||||||||||||||||
| ENSG00000146729 | ENSMUSG00000029432 | 0.0399 | 0.6884 | 0.057960488 | ENSMUSG00000025290 | ENSG00000138326 | 0.0106 | 0.6687 | 0.015851652 | ||||||||||||||||||||||||||
| ENSG00000049245 | ENSMUSG00000028955 | 0.0331 | 0.6885 | 0.048075527 | ENSMUSG00000034659 | ENSG00000110108 | 0.1588 | 0.6688 | 0.237440191 | ||||||||||||||||||||||||||
| ENSG00000177380 | ENSMUSG00000003863 | 0.0089 | 0.6891 | 0.012915397 | ENSMUSG00000031906 | ENSG00000103056 | 0.0438 | 0.67 | 0.065373134 | ||||||||||||||||||||||||||
| ENSG00000155265 | ENSMUSG00000042532 | 0.005 | 0.6892 | 0.007254788 | ENSMUSG00000055013 | ENSG00000157985 | 0.0106 | 0.671 | 0.015797317 | ||||||||||||||||||||||||||
| ENSG00000148303 | ENSMUSG00000062358 | 0.0053 | 0.6905 | 0.007675597 | ENSMUSG00000000399 | ENSG00000139180 | 0.1431 | 0.6714 | 0.213136729 | ||||||||||||||||||||||||||
| ENSG00000196535 | ENSMUSG00000000631 | 0.0241 | 0.6905 | 0.034902245 | ENSMUSG00000022762 | ENSG00000154654 | 0.0311 | 0.6715 | 0.046314222 | ||||||||||||||||||||||||||
| ENSG00000213366 | ENSMUSG00000068762 | 0.1386 | 0.6914 | 0.200462829 | ENSMUSG00000056602 | ENSG00000073910 | 0.0156 | 0.6716 | 0.023228112 | ||||||||||||||||||||||||||
| ENSG00000110427 | ENSMUSG00000068373 | 0.1466 | 0.6921 | 0.211819101 | ENSMUSG00000000531 | ENSG00000161835 | 0.0425 | 0.6724 | 0.063206425 | ||||||||||||||||||||||||||
| ENSG00000213923 | ENSMUSG00000022433 | 0.0051 | 0.6924 | 0.007365685 | ENSMUSG00000047547 | ENSG00000175416 | 0.0228 | 0.6725 | 0.033903346 | ||||||||||||||||||||||||||
| ENSG00000104888 | ENSMUSG00000070570 | 0.0062 | 0.6934 | 0.008941448 | ENSMUSG00000038173 | ENSG00000164303 | 0.0738 | 0.6733 | 0.109609387 | ||||||||||||||||||||||||||
| ENSG00000132681 | ENSMUSG00000007107 | 0.099 | 0.6935 | 0.142754146 | ENSMUSG00000029711 | ENSG00000130427 | 0.1276 | 0.6737 | 0.189401811 | ||||||||||||||||||||||||||
| ENSG00000126583 | ENSMUSG00000078816 | 0.0096 | 0.6942 | 0.013828868 | ENSMUSG00000037351 | ENSG00000115073 | 0.0047 | 0.674 | 0.006973294 | ||||||||||||||||||||||||||
| ENSG00000159720 | ENSMUSG00000013160 | 0.0013 | 0.6943 | 0.001872389 | ENSMUSG00000037653 | ENSG00000183783 | 0.0211 | 0.6743 | 0.03129171 | ||||||||||||||||||||||||||
| ENSG00000101336 | ENSMUSG00000003283 | 0.0502 | 0.6948 | 0.072251007 | ENSMUSG00000037902 | ENSG00000198053 | 0.2395 | 0.6746 | 0.3550252 | ||||||||||||||||||||||||||
| ENSG00000138768 | ENSMUSG00000029407 | 0.0749 | 0.6955 | 0.107692308 | ENSMUSG00000018042 | ENSG00000100243 | 0.0588 | 0.6755 | 0.087046632 | ||||||||||||||||||||||||||
| ENSG00000013364 | ENSMUSG00000030681 | 0.045 | 0.696 | 0.064655172 | ENSMUSG00000022234 | ENSG00000150753 | 0.0208 | 0.6757 | 0.030782892 | ||||||||||||||||||||||||||
| ENSG00000198721 | ENSMUSG00000021416 | 0.2112 | 0.6961 | 0.303404683 | ENSMUSG00000021460 | ENSG00000148090 | 0.0435 | 0.6771 | 0.064244572 | ||||||||||||||||||||||||||
| ENSG00000076356 | ENSMUSG00000026640 | 0.0167 | 0.6968 | 0.023966705 | ENSMUSG00000026347 | ENSG00000152128 | 0.0533 | 0.6781 | 0.078601976 | ||||||||||||||||||||||||||
| ENSG00000164402 | ENSMUSG00000018398 | 0.0097 | 0.6978 | 0.013900831 | ENSMUSG00000039316 | ENSG00000131378 | 0.1897 | 0.679 | 0.279381443 | ||||||||||||||||||||||||||
| ENSG00000148303 | ENSMUSG00000066452 | 0.0018 | 0.6987 | 0.002576213 | ENSMUSG00000025477 | ENSG00000068383 | 0.029 | 0.6795 | 0.04267844 | ||||||||||||||||||||||||||
| ENSG00000151632 | ENSMUSG00000054757 | 0.1659 | 0.6987 | 0.237440962 | ENSMUSG00000028337 | ENSG00000106789 | 0.0761 | 0.6803 | 0.111862414 | ||||||||||||||||||||||||||
| ENSG00000108828 | ENSMUSG00000034993 | 0.0463 | 0.6988 | 0.06625644 | ENSMUSG00000055762 | ENSG00000104529 | 0.0799 | 0.6803 | 0.117448185 | ||||||||||||||||||||||||||
| ENSG00000173599 | ENSMUSG00000024892 | 0.0151 | 0.6994 | 0.021589934 | ENSMUSG00000028973 | ENSG00000197150 | 0.0932 | 0.6808 | 0.136897767 | ||||||||||||||||||||||||||
| ENSG00000167191 | ENSMUSG00000008734 | 0.068 | 0.6994 | 0.097226194 | ENSMUSG00000026442 | ENSG00000163531 | 0.0157 | 0.6811 | 0.023050947 | ||||||||||||||||||||||||||
| ENSG00000151632 | ENSMUSG00000021211 | 0.2109 | 0.6996 | 0.301457976 | ENSMUSG00000028757 | ENSG00000244038 | 0.0253 | 0.6815 | 0.037123991 | ||||||||||||||||||||||||||
| ENSG00000120694 | ENSMUSG00000029657 | 0.0347 | 0.6998 | 0.049585596 | ENSMUSG00000028525 | ENSG00000184588 | 0.0905 | 0.6815 | 0.132795304 | ||||||||||||||||||||||||||
| ENSG00000184117 | ENSMUSG00000034285 | 0.0295 | 0.7001 | 0.042136838 | ENSMUSG00000022623 | ENSG00000251322 | 0.0231 | 0.6816 | 0.033890845 | ||||||||||||||||||||||||||
| ENSG00000151632 | ENSMUSG00000021213 | 0.225 | 0.7002 | 0.321336761 | ENSMUSG00000004530 | ENSG00000110880 | 0.0122 | 0.6823 | 0.017880698 | ||||||||||||||||||||||||||
| ENSG00000166676 | ENSMUSG00000050908 | 0.0618 | 0.7007 | 0.088197517 | ENSMUSG00000002808 | ENSG00000086289 | 0.1132 | 0.6823 | 0.165909424 | ||||||||||||||||||||||||||
| ENSG00000125844 | ENSMUSG00000027422 | 0.0765 | 0.704 | 0.108664773 | ENSMUSG00000042426 | ENSG00000067248 | 0.0477 | 0.6825 | 0.06989011 | ||||||||||||||||||||||||||
| ENSG00000171557 | ENSMUSG00000033860 | 0.0947 | 0.7046 | 0.134402498 | ENSMUSG00000022840 | ENSG00000173175 | 0.0197 | 0.6839 | 0.028805381 | ||||||||||||||||||||||||||
| ENSG00000197102 | ENSMUSG00000018707 | 0.0036 | 0.7054 | 0.005103487 | ENSMUSG00000051343 | ENSG00000135631 | 0.0675 | 0.686 | 0.098396501 | ||||||||||||||||||||||||||
| ENSG00000187134 | ENSMUSG00000025955 | 0.1779 | 0.706 | 0.251983003 | ENSMUSG00000057506 | ENSG00000196072 | 0.0253 | 0.6866 | 0.036848238 | ||||||||||||||||||||||||||
| ENSG00000151632 | ENSMUSG00000071551 | 0.1806 | 0.7062 | 0.255734919 | ENSMUSG00000063316 | ENSG00000131469 | 0.0007 | 0.6875 | 0.001018182 | ||||||||||||||||||||||||||
| ENSG00000186716 | ENSMUSG00000009681 | 0.026 | 0.7067 | 0.036790717 | ENSMUSG00000030835 | ENSG00000103226 | 0.0363 | 0.6876 | 0.052792321 | ||||||||||||||||||||||||||
| ENSG00000076864 | ENSMUSG00000041351 | 0.0149 | 0.7072 | 0.021069005 | ENSMUSG00000000486 | ENSG00000180096 | 0.0197 | 0.6884 | 0.028617083 | ||||||||||||||||||||||||||
| ENSG00000095713 | ENSMUSG00000042401 | 0.0664 | 0.7072 | 0.093891403 | ENSMUSG00000029432 | ENSG00000146729 | 0.0399 | 0.6884 | 0.057960488 | ||||||||||||||||||||||||||
| ENSG00000107957 | ENSMUSG00000053617 | 0.0443 | 0.7077 | 0.062597146 | ENSMUSG00000028955 | ENSG00000049245 | 0.0331 | 0.6885 | 0.048075527 | ||||||||||||||||||||||||||
| ENSG00000148303 | ENSMUSG00000062647 | 0.0018 | 0.7085 | 0.002540579 | ENSMUSG00000003863 | ENSG00000177380 | 0.0089 | 0.6891 | 0.012915397 | ||||||||||||||||||||||||||
| ENSG00000156515 | ENSMUSG00000037012 | 0.0478 | 0.7085 | 0.067466478 | ENSMUSG00000042532 | ENSG00000155265 | 0.005 | 0.6892 | 0.007254788 | ||||||||||||||||||||||||||
| ENSG00000152767 | ENSMUSG00000025555 | 0.0396 | 0.7088 | 0.055869074 | ENSMUSG00000000631 | ENSG00000196535 | 0.0241 | 0.6905 | 0.034902245 | ||||||||||||||||||||||||||
| ENSG00000148303 | ENSMUSG00000071052 | 0.0071 | 0.7089 | 0.010015517 | ENSMUSG00000029765 | ENSG00000221866 | 0.01 | 0.6911 | 0.014469686 | ||||||||||||||||||||||||||
| ENSG00000183735 | ENSMUSG00000020115 | 0.0307 | 0.7089 | 0.043306531 | ENSMUSG00000030835 | ENSG00000185164 | 0.0363 | 0.6911 | 0.05252496 | ||||||||||||||||||||||||||
| ENSG00000163013 | ENSMUSG00000047013 | 0.0133 | 0.7097 | 0.018740313 | ENSMUSG00000068373 | ENSG00000110427 | 0.1466 | 0.6921 | 0.211819101 | ||||||||||||||||||||||||||
| ENSG00000173230 | ENSMUSG00000034243 | 0.1541 | 0.7098 | 0.217103409 | ENSMUSG00000022433 | ENSG00000213923 | 0.0051 | 0.6924 | 0.007365685 | ||||||||||||||||||||||||||
| ENSG00000133315 | ENSMUSG00000036278 | 0.0735 | 0.7103 | 0.103477404 | ENSMUSG00000037989 | ENSG00000165238 | 0.1176 | 0.6931 | 0.169672486 | ||||||||||||||||||||||||||
| ENSG00000112294 | ENSMUSG00000035936 | 0.0867 | 0.7109 | 0.121958081 | ENSMUSG00000070570 | ENSG00000104888 | 0.0062 | 0.6934 | 0.008941448 | ||||||||||||||||||||||||||
| ENSG00000151632 | ENSMUSG00000025955 | 0.1894 | 0.7113 | 0.266273021 | ENSMUSG00000007107 | ENSG00000132681 | 0.099 | 0.6935 | 0.142754146 | ||||||||||||||||||||||||||
| ENSG00000067842 | ENSMUSG00000031376 | 0.0088 | 0.7116 | 0.012366498 | ENSMUSG00000078816 | ENSG00000126583 | 0.0096 | 0.6942 | 0.013828868 | ||||||||||||||||||||||||||
| ENSG00000088899 | ENSMUSG00000037703 | 0.021 | 0.7118 | 0.029502669 | ENSMUSG00000013160 | ENSG00000159720 | 0.0013 | 0.6943 | 0.001872389 | ||||||||||||||||||||||||||
| ENSG00000082397 | ENSMUSG00000024044 | 0.0644 | 0.7119 | 0.090462144 | ENSMUSG00000003283 | ENSG00000101336 | 0.0502 | 0.6948 | 0.072251007 | ||||||||||||||||||||||||||
| ENSG00000204128 | ENSMUSG00000026227 | 0.1803 | 0.7135 | 0.252697968 | ENSMUSG00000064030 | ENSG00000170473 | 0.0501 | 0.6956 | 0.072024152 | ||||||||||||||||||||||||||
| ENSG00000002834 | ENSMUSG00000038366 | 0.019 | 0.715 | 0.026573427 | ENSMUSG00000018398 | ENSG00000164402 | 0.0097 | 0.6978 | 0.013900831 | ||||||||||||||||||||||||||
| ENSG00000096696 | ENSMUSG00000054889 | 0.0232 | 0.7152 | 0.032438479 | ENSMUSG00000030835 | ENSG00000103512 | 0.0354 | 0.6978 | 0.050730868 | ||||||||||||||||||||||||||
| ENSG00000162407 | ENSMUSG00000028517 | 0.0232 | 0.716 | 0.032402235 | ENSMUSG00000000325 | ENSG00000099889 | 0.0434 | 0.6987 | 0.062115357 | ||||||||||||||||||||||||||
| ENSG00000005381 | ENSMUSG00000009350 | 0.0775 | 0.7161 | 0.108225108 | ENSMUSG00000008734 | ENSG00000167191 | 0.068 | 0.6994 | 0.097226194 | ||||||||||||||||||||||||||
| ENSG00000187134 | ENSMUSG00000071551 | 0.1769 | 0.7165 | 0.246894627 | ENSMUSG00000029657 | ENSG00000120694 | 0.0347 | 0.6998 | 0.049585596 | ||||||||||||||||||||||||||
| ENSG00000141385 | ENSMUSG00000024527 | 0.0364 | 0.7166 | 0.050795423 | ENSMUSG00000034285 | ENSG00000184117 | 0.0295 | 0.7001 | 0.042136838 | ||||||||||||||||||||||||||
| ENSG00000100075 | ENSMUSG00000003528 | 0.0256 | 0.7173 | 0.035689391 | ENSMUSG00000050908 | ENSG00000166676 | 0.0618 | 0.7007 | 0.088197517 | ||||||||||||||||||||||||||
| ENSG00000161618 | ENSMUSG00000007833 | 0.1104 | 0.7177 | 0.153824718 | ENSMUSG00000013367 | ENSG00000142549 | 0.0089 | 0.701 | 0.012696148 | ||||||||||||||||||||||||||
| ENSG00000105254 | ENSMUSG00000006095 | 0.0558 | 0.7186 | 0.077650988 | ENSMUSG00000018707 | ENSG00000197102 | 0.0036 | 0.7054 | 0.005103487 | ||||||||||||||||||||||||||
| ENSG00000150760 | ENSMUSG00000058325 | 0.017 | 0.7208 | 0.023584906 | ENSMUSG00000015766 | ENSG00000151491 | 0.055 | 0.7059 | 0.077914719 | ||||||||||||||||||||||||||
| ENSG00000071127 | ENSMUSG00000005103 | 0.0227 | 0.7209 | 0.031488417 | ENSMUSG00000009681 | ENSG00000186716 | 0.026 | 0.7067 | 0.036790717 | ||||||||||||||||||||||||||
| ENSG00000138758 | ENSMUSG00000058013 | 0.0043 | 0.7227 | 0.00594991 | ENSMUSG00000028495 | ENSG00000137154 | 0.0007 | 0.7072 | 0.000989819 | ||||||||||||||||||||||||||
| ENSG00000089220 | ENSMUSG00000047104 | 0.0911 | 0.7227 | 0.126055071 | ENSMUSG00000042401 | ENSG00000095713 | 0.0664 | 0.7072 | 0.093891403 | ||||||||||||||||||||||||||
| ENSG00000103966 | ENSMUSG00000027293 | 0.0146 | 0.723 | 0.020193638 | ENSMUSG00000062647 | ENSG00000148303 | 0.0018 | 0.7085 | 0.002540579 | ||||||||||||||||||||||||||
| ENSG00000174996 | ENSMUSG00000024862 | 0.0205 | 0.7236 | 0.028330569 | ENSMUSG00000034245 | ENSG00000163517 | 0.0398 | 0.7085 | 0.056175018 | ||||||||||||||||||||||||||
| ENSG00000159231 | ENSMUSG00000022947 | 0.0796 | 0.7241 | 0.109929568 | ENSMUSG00000037012 | ENSG00000156515 | 0.0478 | 0.7085 | 0.067466478 | ||||||||||||||||||||||||||
| ENSG00000075340 | ENSMUSG00000030000 | 0.0351 | 0.7255 | 0.048380427 | ENSMUSG00000025555 | ENSG00000152767 | 0.0396 | 0.7088 | 0.055869074 | ||||||||||||||||||||||||||
| ENSG00000116288 | ENSMUSG00000028964 | 0.0495 | 0.7255 | 0.068228808 | ENSMUSG00000002010 | ENSG00000067829 | 0.0252 | 0.7095 | 0.03551797 | ||||||||||||||||||||||||||
| ENSG00000131018 | ENSMUSG00000019769 | 0.0735 | 0.7274 | 0.101044817 | ENSMUSG00000047013 | ENSG00000163013 | 0.0133 | 0.7097 | 0.018740313 | ||||||||||||||||||||||||||
| ENSG00000089009 | ENSMUSG00000029614 | 0.0595 | 0.7275 | 0.081786942 | ENSMUSG00000029088 | ENSG00000185774 | 0.0741 | 0.7101 | 0.1043515 | ||||||||||||||||||||||||||
| ENSG00000105726 | ENSMUSG00000031862 | 0.0255 | 0.7278 | 0.035037098 | ENSMUSG00000031376 | ENSG00000067842 | 0.0088 | 0.7116 | 0.012366498 | ||||||||||||||||||||||||||
| ENSG00000105443 | ENSMUSG00000003269 | 0.0032 | 0.728 | 0.004395604 | ENSMUSG00000037703 | ENSG00000088899 | 0.021 | 0.7118 | 0.029502669 | ||||||||||||||||||||||||||
| ENSG00000109756 | ENSMUSG00000062232 | 0.0328 | 0.7285 | 0.045024022 | ENSMUSG00000024044 | ENSG00000082397 | 0.0644 | 0.7119 | 0.090462144 | ||||||||||||||||||||||||||
| ENSG00000174791 | ENSMUSG00000024883 | 0.1283 | 0.7291 | 0.175970374 | ENSMUSG00000030397 | ENSG00000007047 | 0.0506 | 0.7135 | 0.07091801 | ||||||||||||||||||||||||||
| ENSG00000126803 | ENSMUSG00000059970 | 0.0019 | 0.7296 | 0.002604167 | ENSMUSG00000038366 | ENSG00000002834 | 0.019 | 0.715 | 0.026573427 | ||||||||||||||||||||||||||
| ENSG00000184779 | ENSMUSG00000050978 | 0.0775 | 0.7299 | 0.106178929 | ENSMUSG00000066235 | ENSG00000144647 | 0.0302 | 0.7156 | 0.042202348 | ||||||||||||||||||||||||||
| ENSG00000004939 | ENSMUSG00000006574 | 0.103 | 0.73 | 0.14109589 | ENSMUSG00000034685 | ENSG00000161682 | 0.0256 | 0.7157 | 0.035769177 | ||||||||||||||||||||||||||
| ENSG00000146122 | ENSMUSG00000040260 | 0.0401 | 0.7303 | 0.054908942 | ENSMUSG00000028517 | ENSG00000162407 | 0.0232 | 0.716 | 0.032402235 | ||||||||||||||||||||||||||
| ENSG00000105220 | ENSMUSG00000036427 | 0.0596 | 0.7309 | 0.081543303 | ENSMUSG00000037601 | ENSG00000239672 | 0.0295 | 0.716 | 0.041201117 | ||||||||||||||||||||||||||
| ENSG00000068976 | ENSMUSG00000032648 | 0.0139 | 0.7326 | 0.018973519 | ENSMUSG00000024527 | ENSG00000141385 | 0.0364 | 0.7166 | 0.050795423 | ||||||||||||||||||||||||||
| ENSG00000196961 | ENSMUSG00000060279 | 0.0062 | 0.7345 | 0.008441116 | ENSMUSG00000003528 | ENSG00000100075 | 0.0256 | 0.7173 | 0.035689391 | ||||||||||||||||||||||||||
| ENSG00000184922 | ENSMUSG00000055805 | 0.0601 | 0.736 | 0.081657609 | ENSMUSG00000020173 | ENSG00000106078 | 0.2314 | 0.7176 | 0.322463768 | ||||||||||||||||||||||||||
| ENSG00000110931 | ENSMUSG00000029471 | 0.0545 | 0.7363 | 0.074018742 | ENSMUSG00000022210 | ENSG00000157326 | 0.121 | 0.7189 | 0.1683127 | ||||||||||||||||||||||||||
| ENSG00000197444 | ENSMUSG00000021913 | 0.0325 | 0.7382 | 0.044026009 | ENSMUSG00000058325 | ENSG00000150760 | 0.017 | 0.7208 | 0.023584906 | ||||||||||||||||||||||||||
| ENSG00000168003 | ENSMUSG00000010095 | 0.1946 | 0.7392 | 0.263257576 | ENSMUSG00000058013 | ENSG00000138758 | 0.0043 | 0.7227 | 0.00594991 | ||||||||||||||||||||||||||
| ENSG00000187134 | ENSMUSG00000021207 | 0.1679 | 0.7402 | 0.226830586 | ENSMUSG00000017132 | ENSG00000108669 | 0.0068 | 0.7229 | 0.009406557 | ||||||||||||||||||||||||||
| ENSG00000131899 | ENSMUSG00000020536 | 0.0541 | 0.7409 | 0.073019301 | ENSMUSG00000027293 | ENSG00000103966 | 0.0146 | 0.723 | 0.020193638 | ||||||||||||||||||||||||||
| ENSG00000169258 | ENSMUSG00000069227 | 0.3717 | 0.7413 | 0.501416431 | ENSMUSG00000024862 | ENSG00000174996 | 0.0205 | 0.7236 | 0.028330569 | ||||||||||||||||||||||||||
| ENSG00000124772 | ENSMUSG00000024008 | 0.0087 | 0.7424 | 0.01171875 | ENSMUSG00000030000 | ENSG00000075340 | 0.0351 | 0.7255 | 0.048380427 | ||||||||||||||||||||||||||
| ENSG00000100092 | ENSMUSG00000022436 | 0.1038 | 0.7427 | 0.139760334 | ENSMUSG00000004113 | ENSG00000148408 | 0.0336 | 0.7256 | 0.046306505 | ||||||||||||||||||||||||||
| ENSG00000138796 | ENSMUSG00000027984 | 0.0516 | 0.7449 | 0.069271043 | ENSMUSG00000036611 | ENSG00000122547 | 0.0502 | 0.726 | 0.069146006 | ||||||||||||||||||||||||||
| ENSG00000072071 | ENSMUSG00000013033 | 0.0071 | 0.7464 | 0.009512326 | ENSMUSG00000019769 | ENSG00000131018 | 0.0735 | 0.7274 | 0.101044817 | ||||||||||||||||||||||||||
| ENSG00000139719 | ENSMUSG00000029434 | 0.0102 | 0.7471 | 0.013652791 | ENSMUSG00000029614 | ENSG00000089009 | 0.0595 | 0.7275 | 0.081786942 | ||||||||||||||||||||||||||
| ENSG00000148356 | ENSMUSG00000026792 | 0.0637 | 0.7472 | 0.085251606 | ENSMUSG00000031862 | ENSG00000105726 | 0.0255 | 0.7278 | 0.035037098 | ||||||||||||||||||||||||||
| ENSG00000131626 | ENSMUSG00000037519 | 0.0211 | 0.7474 | 0.028231201 | ENSMUSG00000003269 | ENSG00000105443 | 0.0032 | 0.728 | 0.004395604 | ||||||||||||||||||||||||||
| ENSG00000169020 | ENSMUSG00000050856 | 0.0622 | 0.7477 | 0.083188445 | ENSMUSG00000062232 | ENSG00000109756 | 0.0328 | 0.7285 | 0.045024022 | ||||||||||||||||||||||||||
| ENSG00000168439 | ENSMUSG00000024966 | 0.0128 | 0.7489 | 0.017091735 | ENSMUSG00000009863 | ENSG00000117118 | 0.047 | 0.7292 | 0.064454196 | ||||||||||||||||||||||||||
| ENSG00000134333 | ENSMUSG00000063229 | 0.0332 | 0.7492 | 0.044313935 | ENSMUSG00000059970 | ENSG00000126803 | 0.0019 | 0.7296 | 0.002604167 | ||||||||||||||||||||||||||
| ENSG00000176884 | ENSMUSG00000026959 | 0.0041 | 0.7495 | 0.005470314 | ENSMUSG00000040260 | ENSG00000146122 | 0.0401 | 0.7303 | 0.054908942 | ||||||||||||||||||||||||||
| ENSG00000132024 | ENSMUSG00000036686 | 0.0928 | 0.7498 | 0.123766338 | ENSMUSG00000036427 | ENSG00000105220 | 0.0596 | 0.7309 | 0.081543303 | ||||||||||||||||||||||||||
| ENSG00000187134 | ENSMUSG00000021210 | 0.1443 | 0.7499 | 0.192425657 | ENSMUSG00000070304 | ENSG00000149575 | 0.0378 | 0.7327 | 0.05159001 | ||||||||||||||||||||||||||
| ENSG00000176894 | ENSMUSG00000029499 | 0.1378 | 0.7509 | 0.183513118 | ENSMUSG00000062151 | ENSG00000137766 | 0.0528 | 0.7339 | 0.071944407 | ||||||||||||||||||||||||||
| ENSG00000213366 | ENSMUSG00000027890 | 0.1278 | 0.751 | 0.170173103 | ENSMUSG00000060279 | ENSG00000196961 | 0.0062 | 0.7345 | 0.008441116 | ||||||||||||||||||||||||||
| ENSG00000205981 | ENSMUSG00000027679 | 0.1616 | 0.7521 | 0.214865045 | ENSMUSG00000054720 | ENSG00000171488 | 0.0147 | 0.7345 | 0.020013615 | ||||||||||||||||||||||||||
| ENSG00000100285 | ENSMUSG00000020396 | 0.0925 | 0.7524 | 0.122939926 | ENSMUSG00000055805 | ENSG00000184922 | 0.0601 | 0.736 | 0.081657609 | ||||||||||||||||||||||||||
| ENSG00000136928 | ENSMUSG00000039809 | 0.0088 | 0.7527 | 0.011691245 | ENSMUSG00000021913 | ENSG00000197444 | 0.0325 | 0.7382 | 0.044026009 | ||||||||||||||||||||||||||
| ENSG00000010278 | ENSMUSG00000030342 | 0.0606 | 0.7533 | 0.080446037 | ENSMUSG00000010095 | ENSG00000168003 | 0.1946 | 0.7392 | 0.263257576 | ||||||||||||||||||||||||||
| ENSG00000142875 | ENSMUSG00000005034 | 0.0402 | 0.7537 | 0.053336871 | ENSMUSG00000018507 | ENSG00000187688 | 0.1113 | 0.7394 | 0.150527455 | ||||||||||||||||||||||||||
| ENSG00000117154 | ENSMUSG00000040972 | 0.0298 | 0.7553 | 0.039454521 | ENSMUSG00000021057 | ENSG00000179841 | 0.254 | 0.7406 | 0.342965163 | ||||||||||||||||||||||||||
| ENSG00000106992 | ENSMUSG00000026817 | 0.0574 | 0.7554 | 0.075986232 | ENSMUSG00000020536 | ENSG00000131899 | 0.0541 | 0.7409 | 0.073019301 | ||||||||||||||||||||||||||
| ENSG00000197879 | ENSMUSG00000017774 | 0.0183 | 0.7564 | 0.024193548 | ENSMUSG00000069227 | ENSG00000169258 | 0.3717 | 0.7413 | 0.501416431 | ||||||||||||||||||||||||||
| ENSG00000110700 | ENSMUSG00000090862 | 0 | 0.7565 | 0 | ENSMUSG00000024008 | ENSG00000124772 | 0.0087 | 0.7424 | 0.01171875 | ||||||||||||||||||||||||||
| ENSG00000110700 | ENSMUSG00000069972 | 0 | 0.7565 | 0 | ENSMUSG00000040479 | ENSG00000149091 | 0.0457 | 0.7437 | 0.061449509 | ||||||||||||||||||||||||||
| ENSG00000151632 | ENSMUSG00000021207 | 0.1673 | 0.7571 | 0.220974772 | ENSMUSG00000060244 | ENSG00000183684 | 0.0465 | 0.744 | 0.0625 | ||||||||||||||||||||||||||
| ENSG00000131437 | ENSMUSG00000018395 | 0.01 | 0.7572 | 0.01320655 | ENSMUSG00000053930 | ENSG00000188803 | 0.0497 | 0.7442 | 0.066783123 | ||||||||||||||||||||||||||
| ENSG00000161681 | ENSMUSG00000038738 | 0.0297 | 0.7582 | 0.039171723 | ENSMUSG00000054027 | ENSG00000111696 | 0.038 | 0.745 | 0.051006711 | ||||||||||||||||||||||||||
| ENSG00000104142 | ENSMUSG00000034216 | 0.0194 | 0.7583 | 0.025583542 | ENSMUSG00000013033 | ENSG00000072071 | 0.0071 | 0.7464 | 0.009512326 | ||||||||||||||||||||||||||
| ENSG00000167863 | ENSMUSG00000061992 | 0.1563 | 0.7583 | 0.20611895 | ENSMUSG00000029577 | ENSG00000151148 | 0.0395 | 0.7467 | 0.052899424 | ||||||||||||||||||||||||||
| ENSG00000131095 | ENSMUSG00000020932 | 0.0372 | 0.7595 | 0.048979592 | ENSMUSG00000029434 | ENSG00000139719 | 0.0102 | 0.7471 | 0.013652791 | ||||||||||||||||||||||||||
| ENSG00000110700 | ENSMUSG00000066362 | 0 | 0.7601 | 0 | ENSMUSG00000037519 | ENSG00000131626 | 0.0211 | 0.7474 | 0.028231201 | ||||||||||||||||||||||||||
| ENSG00000111344 | ENSMUSG00000029602 | 0.0738 | 0.7601 | 0.097092488 | ENSMUSG00000050856 | ENSG00000169020 | 0.0622 | 0.7477 | 0.083188445 | ||||||||||||||||||||||||||
| ENSG00000100241 | ENSMUSG00000036529 | 0.0265 | 0.7612 | 0.034813452 | ENSMUSG00000023944 | ENSG00000096384 | 0.0018 | 0.7489 | 0.002403525 | ||||||||||||||||||||||||||
| ENSG00000089009 | ENSMUSG00000057605 | 0.0666 | 0.7615 | 0.087458963 | ENSMUSG00000063229 | ENSG00000134333 | 0.0332 | 0.7492 | 0.044313935 | ||||||||||||||||||||||||||
| ENSG00000103496 | ENSMUSG00000030805 | 0.0454 | 0.7624 | 0.059548793 | ENSMUSG00000026959 | ENSG00000176884 | 0.0041 | 0.7495 | 0.005470314 | ||||||||||||||||||||||||||
| ENSG00000173898 | ENSMUSG00000067889 | 0.0255 | 0.7634 | 0.033403196 | ENSMUSG00000036686 | ENSG00000132024 | 0.0928 | 0.7498 | 0.123766338 | ||||||||||||||||||||||||||
| ENSG00000151632 | ENSMUSG00000021210 | 0.1532 | 0.7634 | 0.200681163 | ENSMUSG00000027679 | ENSG00000205981 | 0.1616 | 0.7521 | 0.214865045 | ||||||||||||||||||||||||||
| ENSG00000089009 | ENSMUSG00000091086 | 0.0791 | 0.7637 | 0.103574702 | ENSMUSG00000020396 | ENSG00000100285 | 0.0925 | 0.7524 | 0.122939926 | ||||||||||||||||||||||||||
| ENSG00000143799 | ENSMUSG00000026496 | 0.0373 | 0.7647 | 0.048777298 | ENSMUSG00000039809 | ENSG00000136928 | 0.0088 | 0.7527 | 0.011691245 | ||||||||||||||||||||||||||
| ENSG00000018236 | ENSMUSG00000055022 | 0.0222 | 0.7658 | 0.028989292 | ENSMUSG00000005034 | ENSG00000142875 | 0.0402 | 0.7537 | 0.053336871 | ||||||||||||||||||||||||||
| ENSG00000100316 | ENSMUSG00000068262 | 0.0129 | 0.7661 | 0.016838533 | ENSMUSG00000026520 | ENSG00000255835 | 0.1846 | 0.7539 | 0.244860061 | ||||||||||||||||||||||||||
| ENSG00000127946 | ENSMUSG00000039959 | 0.0578 | 0.7663 | 0.075427378 | ENSMUSG00000040972 | ENSG00000117154 | 0.0298 | 0.7553 | 0.039454521 | ||||||||||||||||||||||||||
| ENSG00000177731 | ENSMUSG00000002812 | 0.0221 | 0.7666 | 0.028828594 | ENSMUSG00000017774 | ENSG00000197879 | 0.0183 | 0.7564 | 0.024193548 | ||||||||||||||||||||||||||
| ENSG00000149260 | ENSMUSG00000035547 | 0.036 | 0.7682 | 0.046862796 | ENSMUSG00000018395 | ENSG00000131437 | 0.01 | 0.7572 | 0.01320655 | ||||||||||||||||||||||||||
| ENSG00000196924 | ENSMUSG00000031328 | 0.0138 | 0.7686 | 0.017954723 | ENSMUSG00000044060 | ENSG00000069712 | 0.2828 | 0.7575 | 0.373333333 | ||||||||||||||||||||||||||
| ENSG00000198626 | ENSMUSG00000021313 | 0.0227 | 0.7696 | 0.029495842 | ENSMUSG00000038738 | ENSG00000161681 | 0.0297 | 0.7582 | 0.039171723 | ||||||||||||||||||||||||||
| ENSG00000128266 | ENSMUSG00000040009 | 0.0068 | 0.7701 | 0.008830022 | ENSMUSG00000034216 | ENSG00000104142 | 0.0194 | 0.7583 | 0.025583542 | ||||||||||||||||||||||||||
| ENSG00000060709 | ENSMUSG00000029420 | 0.0722 | 0.7701 | 0.093754058 | ENSMUSG00000020932 | ENSG00000131095 | 0.0372 | 0.7595 | 0.048979592 | ||||||||||||||||||||||||||
| ENSG00000167460 | ENSMUSG00000031799 | 0.1017 | 0.771 | 0.131906615 | ENSMUSG00000029101 | ENSG00000159788 | 0.0808 | 0.7595 | 0.10638578 | ||||||||||||||||||||||||||
| ENSG00000198752 | ENSMUSG00000021279 | 0.0403 | 0.7723 | 0.052181795 | ENSMUSG00000066362 | ENSG00000110700 | 0 | 0.7601 | 0 | ||||||||||||||||||||||||||
| ENSG00000100316 | ENSMUSG00000060036 | 0.0075 | 0.7726 | 0.009707481 | ENSMUSG00000036529 | ENSG00000100241 | 0.0265 | 0.7612 | 0.034813452 | ||||||||||||||||||||||||||
| ENSG00000105409 | ENSMUSG00000040907 | 0.0054 | 0.7731 | 0.006984866 | ENSMUSG00000067889 | ENSG00000173898 | 0.0255 | 0.7634 | 0.033403196 | ||||||||||||||||||||||||||
| ENSG00000133030 | ENSMUSG00000005417 | 0.0448 | 0.7735 | 0.057918552 | ENSMUSG00000055022 | ENSG00000018236 | 0.0222 | 0.7658 | 0.028989292 | ||||||||||||||||||||||||||
| ENSG00000127955 | ENSMUSG00000057614 | 0 | 0.7738 | 0 | ENSMUSG00000030310 | ENSG00000157103 | 0.0093 | 0.7661 | 0.012139407 | ||||||||||||||||||||||||||
| ENSG00000203685 | ENSMUSG00000053963 | 0.0088 | 0.7745 | 0.011362169 | ENSMUSG00000039959 | ENSG00000127946 | 0.0578 | 0.7663 | 0.075427378 | ||||||||||||||||||||||||||
| ENSG00000110711 | ENSMUSG00000024847 | 0.0254 | 0.7783 | 0.032635231 | ENSMUSG00000002812 | ENSG00000177731 | 0.0221 | 0.7666 | 0.028828594 | ||||||||||||||||||||||||||
| ENSG00000183773 | ENSMUSG00000022763 | 0.0183 | 0.78 | 0.023461538 | ENSMUSG00000035547 | ENSG00000149260 | 0.036 | 0.7682 | 0.046862796 | ||||||||||||||||||||||||||
| ENSG00000181790 | ENSMUSG00000034730 | 0.0263 | 0.7801 | 0.033713626 | ENSMUSG00000021313 | ENSG00000198626 | 0.0227 | 0.7696 | 0.029495842 | ||||||||||||||||||||||||||
| ENSG00000105701 | ENSMUSG00000019428 | 0.0327 | 0.7803 | 0.041906959 | ENSMUSG00000040009 | ENSG00000128266 | 0.0068 | 0.7701 | 0.008830022 | ||||||||||||||||||||||||||
| ENSG00000151914 | ENSMUSG00000026131 | 0.067 | 0.7804 | 0.085853409 | ENSMUSG00000029420 | ENSG00000060709 | 0.0722 | 0.7701 | 0.093754058 | ||||||||||||||||||||||||||
| ENSG00000123384 | ENSMUSG00000040249 | 0.0089 | 0.782 | 0.011381074 | ENSMUSG00000021279 | ENSG00000198752 | 0.0403 | 0.7723 | 0.052181795 | ||||||||||||||||||||||||||
| ENSG00000121769 | ENSMUSG00000028773 | 0.0788 | 0.7823 | 0.100728621 | ENSMUSG00000016995 | ENSG00000124159 | 0.0485 | 0.7724 | 0.0627913 | ||||||||||||||||||||||||||
| ENSG00000167863 | ENSMUSG00000034566 | 0.1217 | 0.7834 | 0.155348481 | ENSMUSG00000060036 | ENSG00000100316 | 0.0075 | 0.7726 | 0.009707481 | ||||||||||||||||||||||||||
| ENSG00000117791 | ENSMUSG00000073481 | 0.1399 | 0.7844 | 0.178352881 | ENSMUSG00000040907 | ENSG00000105409 | 0.0054 | 0.7731 | 0.006984866 | ||||||||||||||||||||||||||
| ENSG00000136026 | ENSMUSG00000046841 | 0.0852 | 0.7854 | 0.108479756 | ENSMUSG00000005417 | ENSG00000133030 | 0.0448 | 0.7735 | 0.057918552 | ||||||||||||||||||||||||||
| ENSG00000100347 | ENSMUSG00000022437 | 0.0216 | 0.786 | 0.027480916 | ENSMUSG00000057614 | ENSG00000127955 | 0 | 0.7738 | 0 | ||||||||||||||||||||||||||
| ENSG00000070182 | ENSMUSG00000021061 | 0.0449 | 0.786 | 0.057124682 | ENSMUSG00000053963 | ENSG00000203685 | 0.0088 | 0.7745 | 0.011362169 | ||||||||||||||||||||||||||
| ENSG00000105376 | ENSMUSG00000032174 | 0.0812 | 0.788 | 0.103045685 | ENSMUSG00000045092 | ENSG00000170989 | 0.0273 | 0.775 | 0.035225806 | ||||||||||||||||||||||||||
| ENSG00000130558 | ENSMUSG00000026833 | 0.0448 | 0.7883 | 0.056831156 | ENSMUSG00000033960 | ENSG00000165757 | 0.3317 | 0.7751 | 0.427944781 | ||||||||||||||||||||||||||
| ENSG00000079805 | ENSMUSG00000033335 | 0.0079 | 0.7914 | 0.00998231 | ENSMUSG00000022763 | ENSG00000183773 | 0.0183 | 0.78 | 0.023461538 | ||||||||||||||||||||||||||
| ENSG00000105675 | ENSMUSG00000005553 | 0.0099 | 0.7919 | 0.012501578 | ENSMUSG00000034730 | ENSG00000181790 | 0.0263 | 0.7801 | 0.033713626 | ||||||||||||||||||||||||||
| ENSG00000181061 | ENSMUSG00000038412 | 0.0705 | 0.7946 | 0.088723886 | ENSMUSG00000019428 | ENSG00000105701 | 0.0327 | 0.7803 | 0.041906959 | ||||||||||||||||||||||||||
| ENSG00000131495 | ENSMUSG00000014294 | 0.0665 | 0.7972 | 0.083416959 | ENSMUSG00000026131 | ENSG00000151914 | 0.067 | 0.7804 | 0.085853409 | ||||||||||||||||||||||||||
| ENSG00000130294 | ENSMUSG00000014602 | 0.0111 | 0.7975 | 0.013918495 | ENSMUSG00000040249 | ENSG00000123384 | 0.0089 | 0.782 | 0.011381074 | ||||||||||||||||||||||||||
| ENSG00000105379 | ENSMUSG00000004610 | 0.0886 | 0.7979 | 0.111041484 | ENSMUSG00000035735 | ENSG00000134780 | 0.0127 | 0.7828 | 0.016223812 | ||||||||||||||||||||||||||
| ENSG00000167863 | ENSMUSG00000068706 | 0.123 | 0.7986 | 0.154019534 | ENSMUSG00000034566 | ENSG00000167863 | 0.1217 | 0.7834 | 0.155348481 | ||||||||||||||||||||||||||
| ENSG00000141556 | ENSMUSG00000039230 | 0.1059 | 0.7989 | 0.132557266 | ENSMUSG00000006269 | ENSG00000116039 | 0.0316 | 0.7838 | 0.040316407 | ||||||||||||||||||||||||||
| ENSG00000159423 | ENSMUSG00000028737 | 0.0406 | 0.7993 | 0.050794445 | ENSMUSG00000073481 | ENSG00000117791 | 0.1399 | 0.7844 | 0.178352881 | ||||||||||||||||||||||||||
| ENSG00000188229 | ENSMUSG00000036752 | 0.0008 | 0.7997 | 0.001000375 | ENSMUSG00000046841 | ENSG00000136026 | 0.0852 | 0.7854 | 0.108479756 | ||||||||||||||||||||||||||
| ENSG00000197448 | ENSMUSG00000029864 | 0.1658 | 0.8 | 0.20725 | ENSMUSG00000022437 | ENSG00000100347 | 0.0216 | 0.786 | 0.027480916 | ||||||||||||||||||||||||||
| ENSG00000183020 | ENSMUSG00000002957 | 0.0117 | 0.8013 | 0.014601273 | ENSMUSG00000021061 | ENSG00000070182 | 0.0449 | 0.786 | 0.057124682 | ||||||||||||||||||||||||||
| ENSG00000126267 | ENSMUSG00000036751 | 0.0696 | 0.8014 | 0.086848016 | ENSMUSG00000037902 | ENSG00000089012 | 0.3809 | 0.7874 | 0.483743967 | ||||||||||||||||||||||||||
| ENSG00000177542 | ENSMUSG00000019082 | 0.0195 | 0.8019 | 0.024317247 | ENSMUSG00000032174 | ENSG00000105376 | 0.0812 | 0.788 | 0.103045685 | ||||||||||||||||||||||||||
| ENSG00000164715 | ENSMUSG00000038970 | 0.1289 | 0.8023 | 0.160663094 | ENSMUSG00000026833 | ENSG00000130558 | 0.0448 | 0.7883 | 0.056831156 | ||||||||||||||||||||||||||
| ENSG00000062598 | ENSMUSG00000017670 | 0.0566 | 0.8029 | 0.070494458 | ENSMUSG00000005374 | ENSG00000106638 | 0.0594 | 0.7897 | 0.075218437 | ||||||||||||||||||||||||||
| ENSG00000213760 | ENSMUSG00000024403 | 0.0228 | 0.8032 | 0.028386454 | ENSMUSG00000026556 | ENSG00000162738 | 0.0025 | 0.7899 | 0.003164958 | ||||||||||||||||||||||||||
| ENSG00000170634 | ENSMUSG00000060923 | 0.0908 | 0.8036 | 0.112991538 | ENSMUSG00000034656 | ENSG00000141837 | 0.0474 | 0.7899 | 0.060007596 | ||||||||||||||||||||||||||
| ENSG00000186907 | ENSMUSG00000050896 | 0.0212 | 0.8065 | 0.026286423 | ENSMUSG00000033335 | ENSG00000079805 | 0.0079 | 0.7914 | 0.00998231 | ||||||||||||||||||||||||||
| ENSG00000172725 | ENSMUSG00000024835 | 0.0284 | 0.8084 | 0.035131123 | ENSMUSG00000026933 | ENSG00000130559 | 0.0842 | 0.7915 | 0.106380291 | ||||||||||||||||||||||||||
| ENSG00000185800 | ENSMUSG00000030410 | 0.0379 | 0.8105 | 0.046761258 | ENSMUSG00000021097 | ENSG00000165959 | 0.1807 | 0.7918 | 0.228214196 | ||||||||||||||||||||||||||
| ENSG00000164466 | ENSMUSG00000021474 | 0.0325 | 0.8115 | 0.040049291 | ENSMUSG00000005553 | ENSG00000105675 | 0.0099 | 0.7919 | 0.012501578 | ||||||||||||||||||||||||||
| ENSG00000145494 | ENSMUSG00000021606 | 0.0777 | 0.8129 | 0.095583713 | ENSMUSG00000035285 | ENSG00000090971 | 0.0245 | 0.7955 | 0.03079824 | ||||||||||||||||||||||||||
| ENSG00000133731 | ENSMUSG00000027531 | 0.073 | 0.8171 | 0.08934035 | ENSMUSG00000014294 | ENSG00000131495 | 0.0665 | 0.7972 | 0.083416959 | ||||||||||||||||||||||||||
| ENSG00000105220 | ENSMUSG00000043192 | 0.0765 | 0.8172 | 0.093612335 | ENSMUSG00000014602 | ENSG00000130294 | 0.0111 | 0.7975 | 0.013918495 | ||||||||||||||||||||||||||
| ENSG00000168490 | ENSMUSG00000003469 | 0.0025 | 0.8175 | 0.003058104 | ENSMUSG00000060166 | ENSG00000099904 | 0.0415 | 0.7986 | 0.05196594 | ||||||||||||||||||||||||||
| ENSG00000141200 | ENSMUSG00000046755 | 0.1201 | 0.8178 | 0.146857422 | ENSMUSG00000036752 | ENSG00000188229 | 0.0008 | 0.7997 | 0.001000375 | ||||||||||||||||||||||||||
| ENSG00000167774 | ENSMUSG00000041881 | 0.0503 | 0.8185 | 0.061453879 | ENSMUSG00000031819 | ENSG00000131148 | 0.0322 | 0.8012 | 0.040189715 | ||||||||||||||||||||||||||
| ENSG00000149823 | ENSMUSG00000024797 | 0.0194 | 0.8226 | 0.023583759 | ENSMUSG00000002957 | ENSG00000183020 | 0.0117 | 0.8013 | 0.014601273 | ||||||||||||||||||||||||||
| ENSG00000164366 | ENSMUSG00000021578 | 0.0951 | 0.8238 | 0.115440641 | ENSMUSG00000022108 | ENSG00000136156 | 0.0217 | 0.8014 | 0.027077614 | ||||||||||||||||||||||||||
| ENSG00000159692 | ENSMUSG00000037373 | 0.005 | 0.8246 | 0.006063546 | ENSMUSG00000036751 | ENSG00000126267 | 0.0696 | 0.8014 | 0.086848016 | ||||||||||||||||||||||||||
| ENSG00000126602 | ENSMUSG00000005981 | 0.059 | 0.8247 | 0.071541166 | ENSMUSG00000019082 | ENSG00000177542 | 0.0195 | 0.8019 | 0.024317247 | ||||||||||||||||||||||||||
| ENSG00000180104 | ENSMUSG00000034152 | 0.0189 | 0.8253 | 0.022900763 | ENSMUSG00000027296 | ENSG00000137825 | 0.0371 | 0.8021 | 0.046253584 | ||||||||||||||||||||||||||
| ENSG00000213366 | ENSMUSG00000091578 | 0.1129 | 0.8274 | 0.136451535 | ENSMUSG00000038970 | ENSG00000164715 | 0.1289 | 0.8023 | 0.160663094 | ||||||||||||||||||||||||||
| ENSG00000137285 | ENSMUSG00000045136 | 0 | 0.8292 | 0 | ENSMUSG00000017670 | ENSG00000062598 | 0.0566 | 0.8029 | 0.070494458 | ||||||||||||||||||||||||||
| ENSG00000130414 | ENSMUSG00000026260 | 0.1407 | 0.8297 | 0.169579366 | ENSMUSG00000024403 | ENSG00000213760 | 0.0228 | 0.8032 | 0.028386454 | ||||||||||||||||||||||||||
| ENSG00000144712 | ENSMUSG00000030319 | 0.0501 | 0.8325 | 0.06018018 | ENSMUSG00000029361 | ENSG00000089250 | 0.0295 | 0.806 | 0.036600496 | ||||||||||||||||||||||||||
| ENSG00000136279 | ENSMUSG00000020476 | 0.0775 | 0.8369 | 0.092603656 | ENSMUSG00000030316 | ENSG00000144559 | 0.1255 | 0.8077 | 0.155379473 | ||||||||||||||||||||||||||
| ENSG00000124507 | ENSMUSG00000040276 | 0.0201 | 0.84 | 0.023928571 | ENSMUSG00000027546 | ENSG00000054793 | 0.0088 | 0.8081 | 0.010889741 | ||||||||||||||||||||||||||
| ENSG00000179295 | ENSMUSG00000043733 | 0.0024 | 0.8403 | 0.002856123 | ENSMUSG00000024835 | ENSG00000172725 | 0.0284 | 0.8084 | 0.035131123 | ||||||||||||||||||||||||||
| ENSG00000169783 | ENSMUSG00000049556 | 0.0027 | 0.8426 | 0.003204367 | ENSMUSG00000030410 | ENSG00000185800 | 0.0379 | 0.8105 | 0.046761258 | ||||||||||||||||||||||||||
| ENSG00000163931 | ENSMUSG00000021957 | 0.0265 | 0.8431 | 0.031431621 | ENSMUSG00000022296 | ENSG00000164929 | 0.2638 | 0.8112 | 0.325197239 | ||||||||||||||||||||||||||
| ENSG00000178950 | ENSMUSG00000062234 | 0.0985 | 0.8448 | 0.116595644 | ENSMUSG00000021474 | ENSG00000164466 | 0.0325 | 0.8115 | 0.040049291 | ||||||||||||||||||||||||||
| ENSG00000160460 | ENSMUSG00000011751 | 0.0205 | 0.8451 | 0.024257484 | ENSMUSG00000002459 | ENSG00000147509 | 0.2541 | 0.8122 | 0.312853977 | ||||||||||||||||||||||||||
| ENSG00000213366 | ENSMUSG00000058135 | 0.1363 | 0.8457 | 0.161168263 | ENSMUSG00000021606 | ENSG00000145494 | 0.0777 | 0.8129 | 0.095583713 | ||||||||||||||||||||||||||
| ENSG00000117115 | ENSMUSG00000028927 | 0.0347 | 0.8459 | 0.041021397 | ENSMUSG00000036894 | ENSG00000181467 | 0 | 0.8131 | 0 | ||||||||||||||||||||||||||
| ENSG00000032444 | ENSMUSG00000004565 | 0.0178 | 0.8466 | 0.021025278 | ENSMUSG00000036158 | ENSG00000139174 | 0.0342 | 0.8131 | 0.042061247 | ||||||||||||||||||||||||||
| ENSG00000130226 | ENSMUSG00000061576 | 0.0356 | 0.8467 | 0.042045589 | ENSMUSG00000029434 | ENSG00000256861 | 0.0194 | 0.8134 | 0.023850504 | ||||||||||||||||||||||||||
| ENSG00000089169 | ENSMUSG00000029608 | 0.0526 | 0.8476 | 0.062057574 | ENSMUSG00000046755 | ENSG00000141200 | 0.1201 | 0.8178 | 0.146857422 | ||||||||||||||||||||||||||
| ENSG00000144711 | ENSMUSG00000034312 | 0.0272 | 0.8483 | 0.032064128 | ENSMUSG00000014313 | ENSG00000255792 | 0.2161 | 0.8179 | 0.264213229 | ||||||||||||||||||||||||||
| ENSG00000132702 | ENSMUSG00000004894 | 0.0411 | 0.8487 | 0.048427006 | ENSMUSG00000001964 | ENSG00000102119 | 0.1396 | 0.8184 | 0.170576735 | ||||||||||||||||||||||||||
| ENSG00000167378 | ENSMUSG00000041037 | 0.109 | 0.8487 | 0.128431719 | ENSMUSG00000041881 | ENSG00000167774 | 0.0503 | 0.8185 | 0.061453879 | ||||||||||||||||||||||||||
| ENSG00000171224 | ENSMUSG00000020083 | 0.0366 | 0.857 | 0.042707118 | ENSMUSG00000022055 | ENSG00000256228 | 0.0182 | 0.8189 | 0.022224936 | ||||||||||||||||||||||||||
| ENSG00000137267 | ENSMUSG00000058672 | 0.0009 | 0.8588 | 0.001047974 | ENSMUSG00000021578 | ENSG00000164366 | 0.0951 | 0.8238 | 0.115440641 | ||||||||||||||||||||||||||
| ENSG00000103197 | ENSMUSG00000002496 | 0.0467 | 0.8606 | 0.054264467 | ENSMUSG00000037373 | ENSG00000159692 | 0.005 | 0.8246 | 0.006063546 | ||||||||||||||||||||||||||
| ENSG00000143727 | ENSMUSG00000044573 | 0.1248 | 0.8613 | 0.144897248 | ENSMUSG00000005981 | ENSG00000126602 | 0.059 | 0.8247 | 0.071541166 | ||||||||||||||||||||||||||
| ENSG00000076826 | ENSMUSG00000044433 | 0.0469 | 0.8631 | 0.054339011 | ENSMUSG00000034152 | ENSG00000180104 | 0.0189 | 0.8253 | 0.022900763 | ||||||||||||||||||||||||||
| ENSG00000079435 | ENSMUSG00000003123 | 0.0941 | 0.8634 | 0.108987723 | ENSMUSG00000045136 | ENSG00000137285 | 0 | 0.8292 | 0 | ||||||||||||||||||||||||||
| ENSG00000197122 | ENSMUSG00000027646 | 0.0039 | 0.8637 | 0.004515457 | ENSMUSG00000026260 | ENSG00000130414 | 0.1407 | 0.8297 | 0.169579366 | ||||||||||||||||||||||||||
| ENSG00000160211 | ENSMUSG00000031400 | 0.0296 | 0.864 | 0.034259259 | ENSMUSG00000036046 | ENSG00000100364 | 0.052 | 0.8308 | 0.062590274 | ||||||||||||||||||||||||||
| ENSG00000105255 | ENSMUSG00000011589 | 0.0358 | 0.8641 | 0.04143039 | ENSMUSG00000020476 | ENSG00000136279 | 0.0775 | 0.8369 | 0.092603656 | ||||||||||||||||||||||||||
| ENSG00000166165 | ENSMUSG00000001270 | 0.0184 | 0.8655 | 0.021259388 | ENSMUSG00000037902 | ENSG00000101307 | 0.3737 | 0.8369 | 0.446528856 | ||||||||||||||||||||||||||
| ENSG00000213366 | ENSMUSG00000004038 | 0.1395 | 0.8655 | 0.16117851 | ENSMUSG00000040276 | ENSG00000124507 | 0.0201 | 0.84 | 0.023928571 | ||||||||||||||||||||||||||
| ENSG00000129990 | ENSMUSG00000004961 | 0.0444 | 0.867 | 0.051211073 | ENSMUSG00000043733 | ENSG00000179295 | 0.0024 | 0.8403 | 0.002856123 | ||||||||||||||||||||||||||
| ENSG00000187902 | ENSMUSG00000053550 | 0.0336 | 0.8709 | 0.038580779 | ENSMUSG00000020219 | ENSG00000099800 | 0.0252 | 0.8409 | 0.029967892 | ||||||||||||||||||||||||||
| ENSG00000166170 | ENSMUSG00000049792 | 0.049 | 0.8718 | 0.056205552 | ENSMUSG00000049556 | ENSG00000169783 | 0.0027 | 0.8426 | 0.003204367 | ||||||||||||||||||||||||||
| ENSG00000111275 | ENSMUSG00000029455 | 0.0255 | 0.8733 | 0.029199588 | ENSMUSG00000062234 | ENSG00000178950 | 0.0985 | 0.8448 | 0.116595644 | ||||||||||||||||||||||||||
| ENSG00000104946 | ENSMUSG00000038520 | 0.0536 | 0.8749 | 0.061264144 | ENSMUSG00000004565 | ENSG00000032444 | 0.0178 | 0.8466 | 0.021025278 | ||||||||||||||||||||||||||
| ENSG00000111716 | ENSMUSG00000030246 | 0.0101 | 0.8754 | 0.011537583 | ENSMUSG00000061576 | ENSG00000130226 | 0.0356 | 0.8467 | 0.042045589 | ||||||||||||||||||||||||||
| ENSG00000136802 | ENSMUSG00000007476 | 0.0045 | 0.8757 | 0.005138746 | ENSMUSG00000029608 | ENSG00000089169 | 0.0526 | 0.8476 | 0.062057574 | ||||||||||||||||||||||||||
| ENSG00000100994 | ENSMUSG00000033059 | 0.0203 | 0.8789 | 0.023097053 | ENSMUSG00000034312 | ENSG00000144711 | 0.0272 | 0.8483 | 0.032064128 | ||||||||||||||||||||||||||
| ENSG00000055332 | ENSMUSG00000024079 | 0.3148 | 0.8795 | 0.357930642 | ENSMUSG00000029406 | ENSG00000090975 | 0.0463 | 0.8495 | 0.054502649 | ||||||||||||||||||||||||||
| ENSG00000185359 | ENSMUSG00000025793 | 0.0344 | 0.8852 | 0.038861274 | ENSMUSG00000035805 | ENSG00000100427 | 0.0588 | 0.8511 | 0.069087064 | ||||||||||||||||||||||||||
| ENSG00000014216 | ENSMUSG00000024942 | 0.0522 | 0.8853 | 0.058963063 | ENSMUSG00000053375 | ENSG00000250565 | 0.122 | 0.8535 | 0.142940832 | ||||||||||||||||||||||||||
| ENSG00000100033 | ENSMUSG00000003526 | 0.0973 | 0.8868 | 0.109720343 | ENSMUSG00000058672 | ENSG00000137267 | 0.0009 | 0.8588 | 0.001047974 | ||||||||||||||||||||||||||
| ENSG00000123143 | ENSMUSG00000057672 | 0.0434 | 0.8869 | 0.048934491 | ENSMUSG00000078794 | ENSG00000197380 | 0.0597 | 0.8595 | 0.069458988 | ||||||||||||||||||||||||||
| ENSG00000148834 | ENSMUSG00000025068 | 0.1754 | 0.8878 | 0.19756702 | ENSMUSG00000032011 | ENSG00000154096 | 0.2074 | 0.8602 | 0.241106719 | ||||||||||||||||||||||||||
| ENSG00000127445 | ENSMUSG00000032171 | 0.0232 | 0.8882 | 0.026120243 | ENSMUSG00000044433 | ENSG00000076826 | 0.0469 | 0.8631 | 0.054339011 | ||||||||||||||||||||||||||
| ENSG00000168924 | ENSMUSG00000005299 | 0.1041 | 0.8928 | 0.116599462 | ENSMUSG00000027646 | ENSG00000197122 | 0.0039 | 0.8637 | 0.004515457 | ||||||||||||||||||||||||||
| ENSG00000187091 | ENSMUSG00000010660 | 0.0486 | 0.894 | 0.054362416 | ENSMUSG00000011589 | ENSG00000105255 | 0.0358 | 0.8641 | 0.04143039 | ||||||||||||||||||||||||||
| ENSG00000172380 | ENSMUSG00000036402 | 0.0128 | 0.8982 | 0.014250724 | ENSMUSG00000001270 | ENSG00000166165 | 0.0184 | 0.8655 | 0.021259388 | ||||||||||||||||||||||||||
| ENSG00000146701 | ENSMUSG00000019179 | 0.025 | 0.8983 | 0.027830346 | ENSMUSG00000022956 | ENSG00000241837 | 0.1025 | 0.8657 | 0.118401294 | ||||||||||||||||||||||||||
| ENSG00000063660 | ENSMUSG00000034220 | 0.0533 | 0.8995 | 0.059255142 | ENSMUSG00000004961 | ENSG00000129990 | 0.0444 | 0.867 | 0.051211073 | ||||||||||||||||||||||||||
| ENSG00000095321 | ENSMUSG00000026853 | 0.0459 | 0.9014 | 0.05092079 | ENSMUSG00000061099 | ENSG00000105679 | 0.1098 | 0.8671 | 0.126628993 | ||||||||||||||||||||||||||
| ENSG00000171840 | ENSMUSG00000041377 | 0.1589 | 0.9018 | 0.176203149 | ENSMUSG00000031078 | ENSG00000085733 | 0.0375 | 0.8682 | 0.043192813 | ||||||||||||||||||||||||||
| ENSG00000106089 | ENSMUSG00000007207 | 0.0097 | 0.9028 | 0.010744351 | ENSMUSG00000053550 | ENSG00000187902 | 0.0336 | 0.8709 | 0.038580779 | ||||||||||||||||||||||||||
| ENSG00000114450 | ENSMUSG00000027669 | 0.0155 | 0.9059 | 0.017110056 | ENSMUSG00000049792 | ENSG00000166170 | 0.049 | 0.8718 | 0.056205552 | ||||||||||||||||||||||||||
| ENSG00000130827 | ENSMUSG00000031398 | 0.0282 | 0.907 | 0.03109151 | ENSMUSG00000027257 | ENSG00000165912 | 0.0262 | 0.8731 | 0.030008017 | ||||||||||||||||||||||||||
| ENSG00000180900 | ENSMUSG00000022568 | 0.0661 | 0.9091 | 0.072709273 | ENSMUSG00000030246 | ENSG00000111716 | 0.0101 | 0.8754 | 0.011537583 | ||||||||||||||||||||||||||
| ENSG00000118160 | ENSMUSG00000030376 | 0.0254 | 0.9112 | 0.027875329 | ENSMUSG00000007476 | ENSG00000136802 | 0.0045 | 0.8757 | 0.005138746 | ||||||||||||||||||||||||||
| ENSG00000123159 | ENSMUSG00000019433 | 0.0159 | 0.9119 | 0.017436122 | ENSMUSG00000031837 | ENSG00000103154 | 0.075 | 0.8762 | 0.085596896 | ||||||||||||||||||||||||||
| ENSG00000161016 | ENSMUSG00000003970 | 0.0009 | 0.9123 | 0.000986518 | ENSMUSG00000031971 | ENSG00000154429 | 0.1766 | 0.8773 | 0.201299441 | ||||||||||||||||||||||||||
| ENSG00000160211 | ENSMUSG00000089992 | 0.0638 | 0.9123 | 0.069933136 | ENSMUSG00000067288 | ENSG00000233927 | 0 | 0.8803 | 0 | ||||||||||||||||||||||||||
| ENSG00000105464 | ENSMUSG00000002771 | 0.0103 | 0.9129 | 0.011282725 | ENSMUSG00000039953 | ENSG00000171603 | 0.0433 | 0.8834 | 0.049015169 | ||||||||||||||||||||||||||
| ENSG00000167971 | ENSMUSG00000033597 | 0.0498 | 0.9154 | 0.054402447 | ENSMUSG00000001062 | ENSG00000075399 | 0.093 | 0.8837 | 0.105239335 | ||||||||||||||||||||||||||
| ENSG00000138834 | ENSMUSG00000024163 | 0.0317 | 0.9165 | 0.034588107 | ENSMUSG00000043811 | ENSG00000040608 | 0.0622 | 0.8844 | 0.070330167 | ||||||||||||||||||||||||||
| ENSG00000130402 | ENSMUSG00000054808 | 0.0063 | 0.9179 | 0.006863493 | ENSMUSG00000054277 | ENSG00000242247 | 0.1012 | 0.8861 | 0.114208329 | ||||||||||||||||||||||||||
| ENSG00000151640 | ENSMUSG00000025478 | 0.0319 | 0.9203 | 0.03466261 | ENSMUSG00000005299 | ENSG00000168924 | 0.1041 | 0.8928 | 0.116599462 | ||||||||||||||||||||||||||
| ENSG00000141959 | ENSMUSG00000020277 | 0.0289 | 0.9206 | 0.03139257 | ENSMUSG00000022210 | ENSG00000187630 | 0.1963 | 0.8955 | 0.219207147 | ||||||||||||||||||||||||||
| ENSG00000120885 | ENSMUSG00000022037 | 0.1295 | 0.9207 | 0.14065385 | ENSMUSG00000036402 | ENSG00000172380 | 0.0128 | 0.8982 | 0.014250724 | ||||||||||||||||||||||||||
| ENSG00000142534 | ENSMUSG00000003429 | 0 | 0.9212 | 0 | ENSMUSG00000030600 | ENSG00000128011 | 0.0207 | 0.8982 | 0.023046092 | ||||||||||||||||||||||||||
| ENSG00000116791 | ENSMUSG00000028199 | 0.1114 | 0.9239 | 0.12057582 | ENSMUSG00000019179 | ENSG00000146701 | 0.025 | 0.8983 | 0.027830346 | ||||||||||||||||||||||||||
| ENSG00000167363 | ENSMUSG00000025175 | 0.0658 | 0.9293 | 0.070805983 | ENSMUSG00000053046 | ENSG00000174672 | 0.0098 | 0.899 | 0.010901001 | ||||||||||||||||||||||||||
| ENSG00000161714 | ENSMUSG00000020937 | 0.059 | 0.9294 | 0.063481816 | ENSMUSG00000034220 | ENSG00000063660 | 0.0533 | 0.8995 | 0.059255142 | ||||||||||||||||||||||||||
| ENSG00000106211 | ENSMUSG00000004951 | 0.0803 | 0.9334 | 0.086029569 | ENSMUSG00000008140 | ENSG00000161671 | 0.0788 | 0.9021 | 0.087351735 | ||||||||||||||||||||||||||
| ENSG00000130787 | ENSMUSG00000000915 | 0.0444 | 0.9341 | 0.047532384 | ENSMUSG00000007207 | ENSG00000106089 | 0.0097 | 0.9028 | 0.010744351 | ||||||||||||||||||||||||||
| ENSG00000130287 | ENSMUSG00000002341 | 0.2121 | 0.9356 | 0.226699444 | ENSMUSG00000051627 | ENSG00000168298 | 0.029 | 0.9049 | 0.03204774 | ||||||||||||||||||||||||||
| ENSG00000160209 | ENSMUSG00000032788 | 0.07 | 0.9384 | 0.074595055 | ENSMUSG00000027669 | ENSG00000114450 | 0.0155 | 0.9059 | 0.017110056 | ||||||||||||||||||||||||||
| ENSG00000099814 | ENSMUSG00000072825 | 0.1133 | 0.9393 | 0.12062174 | ENSMUSG00000025318 | ENSG00000154118 | 0.0319 | 0.9092 | 0.03508579 | ||||||||||||||||||||||||||
| ENSG00000005007 | ENSMUSG00000058301 | 0.0054 | 0.9394 | 0.00574835 | ENSMUSG00000030376 | ENSG00000118160 | 0.0254 | 0.9112 | 0.027875329 | ||||||||||||||||||||||||||
| ENSG00000116661 | ENSMUSG00000041556 | 0.0613 | 0.9421 | 0.065067403 | ENSMUSG00000019433 | ENSG00000123159 | 0.0159 | 0.9119 | 0.017436122 | ||||||||||||||||||||||||||
| ENSG00000184640 | ENSMUSG00000059248 | 0.0449 | 0.9425 | 0.047639257 | ENSMUSG00000003970 | ENSG00000161016 | 0.0009 | 0.9123 | 0.000986518 | ||||||||||||||||||||||||||
| ENSG00000075043 | ENSMUSG00000016346 | 0.0219 | 0.9436 | 0.023208987 | ENSMUSG00000002771 | ENSG00000105464 | 0.0103 | 0.9129 | 0.011282725 | ||||||||||||||||||||||||||
| ENSG00000165752 | ENSMUSG00000015981 | 0.0423 | 0.9443 | 0.044795086 | ENSMUSG00000022210 | ENSG00000225766 | 0.2198 | 0.9134 | 0.240639369 | ||||||||||||||||||||||||||
| ENSG00000205531 | ENSMUSG00000059119 | 0.0246 | 0.9462 | 0.025998732 | ENSMUSG00000033597 | ENSG00000167971 | 0.0498 | 0.9154 | 0.054402447 | ||||||||||||||||||||||||||
| ENSG00000135709 | ENSMUSG00000031824 | 0.0503 | 0.9539 | 0.052730894 | ENSMUSG00000030287 | ENSG00000123104 | 0.0205 | 0.9164 | 0.022370144 | ||||||||||||||||||||||||||
| ENSG00000170027 | ENSMUSG00000051391 | 0 | 0.9543 | 0 | ENSMUSG00000024163 | ENSG00000138834 | 0.0317 | 0.9165 | 0.034588107 | ||||||||||||||||||||||||||
| ENSG00000173801 | ENSMUSG00000001552 | 0.0068 | 0.9569 | 0.007106281 | ENSMUSG00000027508 | ENSG00000076641 | 0.1093 | 0.9167 | 0.119232028 | ||||||||||||||||||||||||||
| ENSG00000104419 | ENSMUSG00000005125 | 0.0282 | 0.96 | 0.029375 | ENSMUSG00000054808 | ENSG00000130402 | 0.0063 | 0.9179 | 0.006863493 | ||||||||||||||||||||||||||
| ENSG00000124126 | ENSMUSG00000039621 | 0.0404 | 0.9659 | 0.041826276 | ENSMUSG00000025478 | ENSG00000151640 | 0.0319 | 0.9203 | 0.03466261 | ||||||||||||||||||||||||||
| ENSG00000105695 | ENSMUSG00000036634 | 0.0365 | 0.966 | 0.037784679 | ENSMUSG00000020277 | ENSG00000141959 | 0.0289 | 0.9206 | 0.03139257 | ||||||||||||||||||||||||||
| ENSG00000108064 | ENSMUSG00000003923 | 0.2531 | 0.9679 | 0.261493956 | ENSMUSG00000022037 | ENSG00000120885 | 0.1295 | 0.9207 | 0.14065385 | ||||||||||||||||||||||||||
| ENSG00000211899 | ENSMUSG00000076617 | 0.2497 | 0.9696 | 0.257528878 | ENSMUSG00000003429 | ENSG00000142534 | 0 | 0.9212 | 0 | ||||||||||||||||||||||||||
| ENSG00000135406 | ENSMUSG00000023484 | 0.0198 | 0.9702 | 0.020408163 | ENSMUSG00000025580 | ENSG00000141543 | 0.0033 | 0.9217 | 0.003580341 | ||||||||||||||||||||||||||
| ENSG00000125730 | ENSMUSG00000024164 | 0.1335 | 0.972 | 0.137345679 | ENSMUSG00000026452 | ENSG00000143858 | 0.0093 | 0.9239 | 0.010066024 | ||||||||||||||||||||||||||
| ENSG00000183092 | ENSMUSG00000040867 | 0.0559 | 0.9731 | 0.057445278 | ENSMUSG00000028766 | ENSG00000162551 | 0.0569 | 0.9249 | 0.061520164 | ||||||||||||||||||||||||||
| ENSG00000114554 | ENSMUSG00000030084 | 0.0177 | 0.9755 | 0.018144541 | ENSMUSG00000040563 | ENSG00000105520 | 0.0681 | 0.9269 | 0.073470709 | ||||||||||||||||||||||||||
| ENSG00000130477 | ENSMUSG00000034799 | 0.0193 | 0.978 | 0.019734151 | ENSMUSG00000000915 | ENSG00000130787 | 0.0444 | 0.9341 | 0.047532384 | ||||||||||||||||||||||||||
| ENSG00000104863 | ENSMUSG00000003872 | 0.0086 | 0.9789 | 0.008785371 | ENSMUSG00000002341 | ENSG00000130287 | 0.2121 | 0.9356 | 0.226699444 | ||||||||||||||||||||||||||
| ENSG00000013016 | ENSMUSG00000024065 | 0.0115 | 0.9795 | 0.011740684 | ENSMUSG00000072825 | ENSG00000099814 | 0.1133 | 0.9393 | 0.12062174 | ||||||||||||||||||||||||||
| ENSG00000118680 | ENSMUSG00000034868 | 0 | 0.9801 | 0 | ENSMUSG00000058301 | ENSG00000005007 | 0.0054 | 0.9394 | 0.00574835 | ||||||||||||||||||||||||||
| ENSG00000126214 | ENSMUSG00000021288 | 0.0184 | 0.9803 | 0.018769764 | ENSMUSG00000041556 | ENSG00000116661 | 0.0613 | 0.9421 | 0.065067403 | ||||||||||||||||||||||||||
| ENSG00000067057 | ENSMUSG00000021196 | 0.0657 | 0.9898 | 0.066377046 | ENSMUSG00000059248 | ENSG00000184640 | 0.0449 | 0.9425 | 0.047639257 | ||||||||||||||||||||||||||
| ENSG00000198755 | ENSMUSG00000061988 | 0.048 | 0.9903 | 0.048470161 | ENSMUSG00000029093 | ENSG00000184985 | 0.0968 | 0.9435 | 0.102596714 | ||||||||||||||||||||||||||
| ENSG00000111229 | ENSMUSG00000029465 | 0.01 | 0.9927 | 0.010073537 | ENSMUSG00000016346 | ENSG00000075043 | 0.0219 | 0.9436 | 0.023208987 | ||||||||||||||||||||||||||
| ENSG00000204390 | ENSMUSG00000007033 | 0.0277 | 0.9936 | 0.027878422 | ENSMUSG00000026259 | ENSG00000066248 | 0.0729 | 0.9538 | 0.076431118 | ||||||||||||||||||||||||||
| ENSG00000167434 | ENSMUSG00000000805 | 0.34 | 0.9936 | 0.342190016 | ENSMUSG00000051391 | ENSG00000170027 | 0 | 0.9543 | 0 | ||||||||||||||||||||||||||
| ENSG00000008256 | ENSMUSG00000018001 | 0.0042 | 0.997 | 0.004212638 | ENSMUSG00000001552 | ENSG00000173801 | 0.0068 | 0.9569 | 0.007106281 | ||||||||||||||||||||||||||
| ENSG00000133313 | ENSMUSG00000024644 | 0.0449 | 1.0031 | 0.04476124 | ENSMUSG00000050132 | ENSG00000004139 | 0.0697 | 0.9598 | 0.072619296 | ||||||||||||||||||||||||||
| ENSG00000131016 | ENSMUSG00000038587 | 0.3012 | 1.0045 | 0.299850672 | ENSMUSG00000062825 | ENSG00000184009 | 0.001 | 0.9622 | 0.001039285 | ||||||||||||||||||||||||||
| ENSG00000130764 | ENSMUSG00000029028 | 0.0678 | 1.0048 | 0.067476115 | ENSMUSG00000036634 | ENSG00000105695 | 0.0365 | 0.966 | 0.037784679 | ||||||||||||||||||||||||||
| ENSG00000173020 | ENSMUSG00000024858 | 0.0054 | 1.0073 | 0.005360866 | ENSMUSG00000023484 | ENSG00000135406 | 0.0198 | 0.9702 | 0.020408163 | ||||||||||||||||||||||||||
| ENSG00000167632 | ENSMUSG00000047921 | 0.0384 | 1.0073 | 0.038121712 | ENSMUSG00000040867 | ENSG00000183092 | 0.0559 | 0.9731 | 0.057445278 | ||||||||||||||||||||||||||
| ENSG00000100345 | ENSMUSG00000022443 | 0.0128 | 1.011 | 0.012660732 | ENSMUSG00000030084 | ENSG00000114554 | 0.0177 | 0.9755 | 0.018144541 | ||||||||||||||||||||||||||
| ENSG00000092820 | ENSMUSG00000052397 | 0.0153 | 1.0119 | 0.015120071 | ENSMUSG00000030606 | ENSG00000140511 | 0.1096 | 0.9773 | 0.112145708 | ||||||||||||||||||||||||||
| ENSG00000179115 | ENSMUSG00000003808 | 0.0392 | 1.012 | 0.038735178 | ENSMUSG00000003872 | ENSG00000104863 | 0.0086 | 0.9789 | 0.008785371 | ||||||||||||||||||||||||||
| ENSG00000104524 | ENSMUSG00000022571 | 0.1072 | 1.017 | 0.105408063 | ENSMUSG00000024065 | ENSG00000013016 | 0.0115 | 0.9795 | 0.011740684 | ||||||||||||||||||||||||||
| ENSG00000175866 | ENSMUSG00000025372 | 0.0244 | 1.0195 | 0.023933301 | ENSMUSG00000004952 | ENSG00000105808 | 0.0722 | 0.9799 | 0.073680988 | ||||||||||||||||||||||||||
| ENSG00000203485 | ENSMUSG00000037679 | 0.1279 | 1.0263 | 0.12462243 | ENSMUSG00000034868 | ENSG00000118680 | 0 | 0.9801 | 0 | ||||||||||||||||||||||||||
| ENSG00000196872 | ENSMUSG00000026090 | 0.2563 | 1.0272 | 0.24951324 | ENSMUSG00000021288 | ENSG00000126214 | 0.0184 | 0.9803 | 0.018769764 | ||||||||||||||||||||||||||
| ENSG00000103202 | ENSMUSG00000024177 | 0.0872 | 1.0328 | 0.084430674 | ENSMUSG00000002949 | ENSG00000104980 | 0.0521 | 0.9819 | 0.053060393 | ||||||||||||||||||||||||||
| ENSG00000167037 | ENSMUSG00000042216 | 0.0276 | 1.0363 | 0.026633214 | ENSMUSG00000034201 | ENSG00000185340 | 0.0827 | 0.9828 | 0.084147334 | ||||||||||||||||||||||||||
| ENSG00000142347 | ENSMUSG00000024300 | 0.03 | 1.0404 | 0.028835063 | ENSMUSG00000004952 | ENSG00000170667 | 0.0735 | 0.9848 | 0.074634444 | ||||||||||||||||||||||||||
| ENSG00000131143 | ENSMUSG00000031818 | 0.1136 | 1.06 | 0.107169811 | ENSMUSG00000042644 | ENSG00000096433 | 0.0208 | 0.9861 | 0.021093195 | ||||||||||||||||||||||||||
| ENSG00000159189 | ENSMUSG00000036896 | 0.1503 | 1.0613 | 0.141618769 | ENSMUSG00000021196 | ENSG00000067057 | 0.0657 | 0.9898 | 0.066377046 | ||||||||||||||||||||||||||
| ENSG00000165023 | ENSMUSG00000047842 | 0.004 | 1.0622 | 0.003765769 | ENSMUSG00000029465 | ENSG00000111229 | 0.01 | 0.9927 | 0.010073537 | ||||||||||||||||||||||||||
| ENSG00000119280 | ENSMUSG00000031983 | 0.1155 | 1.0624 | 0.108716114 | ENSMUSG00000007033 | ENSG00000204390 | 0.0277 | 0.9936 | 0.027878422 | ||||||||||||||||||||||||||
| ENSG00000179604 | ENSMUSG00000041598 | 0.1522 | 1.064 | 0.143045113 | ENSMUSG00000000805 | ENSG00000167434 | 0.34 | 0.9936 | 0.342190016 | ||||||||||||||||||||||||||
| ENSG00000120645 | ENSMUSG00000040797 | 0.0692 | 1.0711 | 0.064606479 | ENSMUSG00000037855 | ENSG00000138311 | 0.2516 | 0.996 | 0.252610442 | ||||||||||||||||||||||||||
| ENSG00000186111 | ENSMUSG00000034902 | 0.0307 | 1.0714 | 0.028654097 | ENSMUSG00000018001 | ENSG00000008256 | 0.0042 | 0.997 | 0.004212638 | ||||||||||||||||||||||||||
| ENSG00000169710 | ENSMUSG00000025153 | 0.1023 | 1.0734 | 0.095304639 | ENSMUSG00000050556 | ENSG00000158445 | 0.0293 | 1.0026 | 0.029224018 | ||||||||||||||||||||||||||
| ENSG00000110090 | ENSMUSG00000024900 | 0.0684 | 1.0786 | 0.063415539 | ENSMUSG00000024858 | ENSG00000173020 | 0.0054 | 1.0073 | 0.005360866 | ||||||||||||||||||||||||||
| ENSG00000105649 | ENSMUSG00000031840 | 0.0037 | 1.0814 | 0.003421491 | ENSMUSG00000047921 | ENSG00000167632 | 0.0384 | 1.0073 | 0.038121712 | ||||||||||||||||||||||||||
| ENSG00000133392 | ENSMUSG00000018830 | 0.0166 | 1.0852 | 0.015296719 | ENSMUSG00000022443 | ENSG00000100345 | 0.0128 | 1.011 | 0.012660732 | ||||||||||||||||||||||||||
| ENSG00000182809 | ENSMUSG00000006356 | 0.0321 | 1.0883 | 0.029495544 | ENSMUSG00000052397 | ENSG00000092820 | 0.0153 | 1.0119 | 0.015120071 | ||||||||||||||||||||||||||
| ENSG00000103160 | ENSMUSG00000034189 | 0.0688 | 1.0883 | 0.063217863 | ENSMUSG00000003808 | ENSG00000179115 | 0.0392 | 1.012 | 0.038735178 | ||||||||||||||||||||||||||
| ENSG00000175793 | ENSMUSG00000047281 | 0.0133 | 1.0953 | 0.012142792 | ENSMUSG00000022571 | ENSG00000104524 | 0.1072 | 1.017 | 0.105408063 | ||||||||||||||||||||||||||
| ENSG00000213366 | ENSMUSG00000091561 | 0.1669 | 1.0971 | 0.152128338 | ENSMUSG00000025372 | ENSG00000175866 | 0.0244 | 1.0195 | 0.023933301 | ||||||||||||||||||||||||||
| ENSG00000198053 | ENSMUSG00000078783 | 0.5762 | 1.1022 | 0.522772637 | ENSMUSG00000026090 | ENSG00000196872 | 0.2563 | 1.0272 | 0.24951324 | ||||||||||||||||||||||||||
| ENSG00000130203 | ENSMUSG00000002985 | 0.1621 | 1.1109 | 0.145917724 | ENSMUSG00000033594 | ENSG00000158792 | 0.0849 | 1.0277 | 0.082611657 | ||||||||||||||||||||||||||
| ENSG00000105426 | ENSMUSG00000013236 | 0.028 | 1.1169 | 0.025069388 | ENSMUSG00000026223 | ENSG00000135916 | 0.0291 | 1.0311 | 0.028222287 | ||||||||||||||||||||||||||
| ENSG00000172992 | ENSMUSG00000020935 | 0.0431 | 1.1234 | 0.038365676 | ENSMUSG00000006333 | ENSG00000170889 | 0 | 1.0328 | 0 | ||||||||||||||||||||||||||
| ENSG00000078902 | ENSMUSG00000025139 | 0.0337 | 1.1253 | 0.02994757 | ENSMUSG00000040794 | ENSG00000172247 | 0.021 | 1.039 | 0.020211742 | ||||||||||||||||||||||||||
| ENSG00000198053 | ENSMUSG00000074677 | 0.3661 | 1.1364 | 0.322157691 | ENSMUSG00000028758 | ENSG00000117245 | 0.1058 | 1.0469 | 0.101060273 | ||||||||||||||||||||||||||
| ENSG00000130304 | ENSMUSG00000031808 | 0.0504 | 1.1428 | 0.044102205 | ENSMUSG00000009894 | ENSG00000143740 | 0.1995 | 1.0484 | 0.190289966 | ||||||||||||||||||||||||||
| ENSG00000140983 | ENSMUSG00000025733 | 0.0816 | 1.1552 | 0.070637119 | ENSMUSG00000026305 | ENSG00000124831 | 0.3234 | 1.0524 | 0.307297605 | ||||||||||||||||||||||||||
| ENSG00000106665 | ENSMUSG00000063146 | 0.0402 | 1.1555 | 0.034790134 | ENSMUSG00000031818 | ENSG00000131143 | 0.1136 | 1.06 | 0.107169811 | ||||||||||||||||||||||||||
| ENSG00000197256 | ENSMUSG00000032194 | 0.0889 | 1.1647 | 0.076328668 | ENSMUSG00000047842 | ENSG00000165023 | 0.004 | 1.0622 | 0.003765769 | ||||||||||||||||||||||||||
| ENSG00000198053 | ENSMUSG00000078780 | 0.5605 | 1.172 | 0.478242321 | ENSMUSG00000029500 | ENSG00000247077 | 0.0685 | 1.0624 | 0.064476657 | ||||||||||||||||||||||||||
| ENSG00000147799 | ENSMUSG00000033697 | 0.0512 | 1.1761 | 0.043533713 | ENSMUSG00000031983 | ENSG00000119280 | 0.1155 | 1.0624 | 0.108716114 | ||||||||||||||||||||||||||
| ENSG00000184702 | ENSMUSG00000072214 | 0.0044 | 1.179 | 0.003731976 | ENSMUSG00000031453 | ENSG00000185989 | 0.0226 | 1.0637 | 0.021246592 | ||||||||||||||||||||||||||
| ENSG00000198053 | ENSMUSG00000078781 | 0.3648 | 1.1834 | 0.308264323 | ENSMUSG00000022956 | ENSG00000249209 | 0.4239 | 1.0697 | 0.396279331 | ||||||||||||||||||||||||||
| ENSG00000162065 | ENSMUSG00000036473 | 0.0347 | 1.1848 | 0.029287643 | ENSMUSG00000068735 | ENSG00000175274 | 0.0414 | 1.071 | 0.038655462 | ||||||||||||||||||||||||||
| ENSG00000160360 | ENSMUSG00000026930 | 0.0358 | 1.1868 | 0.03016515 | ENSMUSG00000040797 | ENSG00000120645 | 0.0692 | 1.0711 | 0.064606479 | ||||||||||||||||||||||||||
| ENSG00000072958 | ENSMUSG00000003033 | 0.003 | 1.189 | 0.002523129 | ENSMUSG00000031840 | ENSG00000105649 | 0.0037 | 1.0814 | 0.003421491 | ||||||||||||||||||||||||||
| ENSG00000099795 | ENSMUSG00000033938 | 0.096 | 1.1901 | 0.08066549 | ENSMUSG00000029060 | ENSG00000197530 | 0.0377 | 1.0822 | 0.034836444 | ||||||||||||||||||||||||||
| ENSG00000067836 | ENSMUSG00000022540 | 0.0256 | 1.2029 | 0.021281902 | ENSMUSG00000018830 | ENSG00000133392 | 0.0166 | 1.0852 | 0.015296719 | ||||||||||||||||||||||||||
| ENSG00000166897 | ENSMUSG00000043460 | 0.017 | 1.2052 | 0.014105543 | ENSMUSG00000006356 | ENSG00000182809 | 0.0321 | 1.0883 | 0.029495544 | ||||||||||||||||||||||||||
| ENSG00000122873 | ENSMUSG00000037710 | 0.0756 | 1.2145 | 0.062247839 | ENSMUSG00000047281 | ENSG00000175793 | 0.0133 | 1.0953 | 0.012142792 | ||||||||||||||||||||||||||
| ENSG00000075618 | ENSMUSG00000029581 | 0.0146 | 1.2161 | 0.012005592 | ENSMUSG00000035513 | ENSG00000196358 | 0.0256 | 1.096 | 0.023357664 | ||||||||||||||||||||||||||
| ENSG00000198053 | ENSMUSG00000044138 | 0.363 | 1.227 | 0.295843521 | ENSMUSG00000025855 | ENSG00000188191 | 0.0274 | 1.0984 | 0.024945375 | ||||||||||||||||||||||||||
| ENSG00000167468 | ENSMUSG00000075706 | 0.1129 | 1.233 | 0.091565288 | ENSMUSG00000029047 | ENSG00000157911 | 0.1056 | 1.1063 | 0.095453313 | ||||||||||||||||||||||||||
| ENSG00000103266 | ENSMUSG00000039615 | 0.0098 | 1.238 | 0.007915994 | ENSMUSG00000000202 | ENSG00000204347 | 0.0347 | 1.1077 | 0.031326171 | ||||||||||||||||||||||||||
| ENSG00000176595 | ENSMUSG00000055675 | 0.0981 | 1.239 | 0.079176755 | ENSMUSG00000002985 | ENSG00000130203 | 0.1621 | 1.1109 | 0.145917724 | ||||||||||||||||||||||||||
| ENSG00000126934 | ENSMUSG00000035027 | 0.0246 | 1.2561 | 0.019584428 | ENSMUSG00000007594 | ENSG00000187664 | 0.0456 | 1.1147 | 0.040907868 | ||||||||||||||||||||||||||
| ENSG00000111640 | ENSMUSG00000063885 | 0.2797 | 1.2636 | 0.221351694 | ENSMUSG00000013236 | ENSG00000105426 | 0.028 | 1.1169 | 0.025069388 | ||||||||||||||||||||||||||
| ENSG00000105357 | ENSMUSG00000030739 | 0.0334 | 1.2706 | 0.026286794 | ENSMUSG00000025139 | ENSG00000078902 | 0.0337 | 1.1253 | 0.02994757 | ||||||||||||||||||||||||||
| ENSG00000109501 | ENSMUSG00000039474 | 0.061 | 1.2797 | 0.047667422 | ENSMUSG00000031808 | ENSG00000130304 | 0.0504 | 1.1428 | 0.044102205 | ||||||||||||||||||||||||||
| ENSG00000198211 | ENSMUSG00000062380 | 0.0013 | 1.289 | 0.001008534 | ENSMUSG00000055401 | ENSG00000116663 | 0.1966 | 1.1544 | 0.17030492 | ||||||||||||||||||||||||||
| ENSG00000141985 | ENSMUSG00000003200 | 0.025 | 1.2899 | 0.019381347 | ENSMUSG00000025733 | ENSG00000140983 | 0.0816 | 1.1552 | 0.070637119 | ||||||||||||||||||||||||||
| ENSG00000127948 | ENSMUSG00000005514 | 0.0356 | 1.297 | 0.027447957 | ENSMUSG00000063146 | ENSG00000106665 | 0.0402 | 1.1555 | 0.034790134 | ||||||||||||||||||||||||||
| ENSG00000101460 | ENSMUSG00000027602 | 0.0736 | 1.3014 | 0.05655448 | ENSMUSG00000032172 | ENSG00000105088 | 0.0277 | 1.1615 | 0.023848472 | ||||||||||||||||||||||||||
| ENSG00000065000 | ENSMUSG00000020198 | 0.0442 | 1.3155 | 0.033599392 | ENSMUSG00000060938 | ENSG00000161970 | 0 | 1.1622 | 0 | ||||||||||||||||||||||||||
| ENSG00000176788 | ENSMUSG00000045763 | 0.1544 | 1.3343 | 0.115716106 | ENSMUSG00000031833 | ENSG00000099308 | 0.0552 | 1.1683 | 0.047248138 | ||||||||||||||||||||||||||
| ENSG00000167658 | ENSMUSG00000034994 | 0.0043 | 1.348 | 0.003189911 | ENSMUSG00000058441 | ENSG00000073150 | 0.0321 | 1.1734 | 0.0273564 | ||||||||||||||||||||||||||
| ENSG00000179242 | ENSMUSG00000000305 | 0.0384 | 1.3552 | 0.028335301 | ENSMUSG00000033697 | ENSG00000147799 | 0.0512 | 1.1761 | 0.043533713 | ||||||||||||||||||||||||||
| ENSG00000183186 | ENSMUSG00000045912 | 0.0433 | 1.3797 | 0.031383634 | ENSMUSG00000072214 | ENSG00000184702 | 0.0044 | 1.179 | 0.003731976 | ||||||||||||||||||||||||||
| ENSG00000099864 | ENSMUSG00000035863 | 0.1249 | 1.3825 | 0.09034358 | ENSMUSG00000036473 | ENSG00000162065 | 0.0347 | 1.1848 | 0.029287643 | ||||||||||||||||||||||||||
| ENSG00000101210 | ENSMUSG00000016349 | 0.0014 | 1.3919 | 0.001005819 | ENSMUSG00000037905 | ENSG00000184992 | 0.1016 | 1.1931 | 0.085156315 | ||||||||||||||||||||||||||
| ENSG00000204389 | ENSMUSG00000091971 | 0.0183 | 1.3928 | 0.013139001 | ENSMUSG00000020238 | ENSG00000125912 | 0.0321 | 1.2 | 0.02675 | ||||||||||||||||||||||||||
| ENSG00000204389 | ENSMUSG00000090877 | 0.0182 | 1.4097 | 0.012910548 | ENSMUSG00000025475 | ENSG00000197177 | 0.0905 | 1.2028 | 0.075241104 | ||||||||||||||||||||||||||
| ENSG00000143774 | ENSMUSG00000020444 | 0.0625 | 1.4153 | 0.044160249 | ENSMUSG00000022540 | ENSG00000067836 | 0.0256 | 1.2029 | 0.021281902 | ||||||||||||||||||||||||||
| ENSG00000198010 | ENSMUSG00000047495 | 0.0519 | 1.4223 | 0.036490192 | ENSMUSG00000029036 | ENSG00000160072 | 0.0743 | 1.2043 | 0.061695591 | ||||||||||||||||||||||||||
| ENSG00000140990 | ENSMUSG00000040048 | 0.1058 | 1.426 | 0.074193548 | ENSMUSG00000036306 | ENSG00000061337 | 0.0466 | 1.205 | 0.038672199 | ||||||||||||||||||||||||||
| ENSG00000204388 | ENSMUSG00000091971 | 0.0183 | 1.4379 | 0.012726893 | ENSMUSG00000043460 | ENSG00000166897 | 0.017 | 1.2052 | 0.014105543 | ||||||||||||||||||||||||||
| ENSG00000204388 | ENSMUSG00000090877 | 0.0183 | 1.4565 | 0.012564367 | ENSMUSG00000008348 | ENSG00000170315 | 0.0039 | 1.213 | 0.003215169 | ||||||||||||||||||||||||||
| ENSG00000116903 | ENSMUSG00000074030 | 0.0299 | 1.4609 | 0.020466836 | ENSMUSG00000037710 | ENSG00000122873 | 0.0756 | 1.2145 | 0.062247839 | ||||||||||||||||||||||||||
| ENSG00000196365 | ENSMUSG00000041168 | 0.0621 | 1.466 | 0.042360164 | ENSMUSG00000029581 | ENSG00000075618 | 0.0146 | 1.2161 | 0.012005592 | ||||||||||||||||||||||||||
| ENSG00000171368 | ENSMUSG00000021573 | 0.0369 | 1.486 | 0.024831763 | ENSMUSG00000003573 | ENSG00000051128 | 0.0564 | 1.2201 | 0.046225719 | ||||||||||||||||||||||||||
| ENSG00000115286 | ENSMUSG00000020153 | 0.0606 | 1.4872 | 0.040747714 | ENSMUSG00000063646 | ENSG00000152969 | 0.0349 | 1.2232 | 0.02853172 | ||||||||||||||||||||||||||
| ENSG00000007080 | ENSMUSG00000007721 | 0.0423 | 1.4903 | 0.028383547 | ENSMUSG00000038884 | ENSG00000130813 | 0.0208 | 1.2248 | 0.016982364 | ||||||||||||||||||||||||||
| ENSG00000167671 | ENSMUSG00000019578 | 0.1087 | 1.5206 | 0.07148494 | ENSMUSG00000075706 | ENSG00000167468 | 0.1129 | 1.233 | 0.091565288 | ||||||||||||||||||||||||||
| ENSG00000142634 | ENSMUSG00000040659 | 0.0249 | 1.549 | 0.016074887 | ENSMUSG00000039615 | ENSG00000103266 | 0.0098 | 1.238 | 0.007915994 | ||||||||||||||||||||||||||
| ENSG00000127561 | ENSMUSG00000007021 | 0.0288 | 1.5509 | 0.018569863 | ENSMUSG00000029036 | ENSG00000215915 | 0.1602 | 1.2527 | 0.127883771 | ||||||||||||||||||||||||||
| ENSG00000168309 | ENSMUSG00000021750 | 0.0416 | 1.565 | 0.02658147 | ENSMUSG00000000399 | ENSG00000255639 | 0.2092 | 1.2606 | 0.165952721 | ||||||||||||||||||||||||||
| ENSG00000130193 | ENSMUSG00000056665 | 0.0614 | 1.5821 | 0.038809178 | ENSMUSG00000038690 | ENSG00000241468 | 0.1134 | 1.2649 | 0.089651356 | ||||||||||||||||||||||||||
| ENSG00000178209 | ENSMUSG00000022565 | 0.0299 | 1.595 | 0.018746082 | ENSMUSG00000030739 | ENSG00000105357 | 0.0334 | 1.2706 | 0.026286794 | ||||||||||||||||||||||||||
| ENSG00000110047 | ENSMUSG00000024772 | 0.0022 | 1.6009 | 0.001374227 | ENSMUSG00000029036 | ENSG00000197785 | 0.0468 | 1.2715 | 0.036806921 | ||||||||||||||||||||||||||
| ENSG00000172270 | ENSMUSG00000023175 | 0.2229 | 1.6011 | 0.139216788 | ENSMUSG00000025371 | ENSG00000176108 | 0.0306 | 1.2719 | 0.024058495 | ||||||||||||||||||||||||||
| ENSG00000103024 | ENSMUSG00000073435 | 0.0578 | 1.7278 | 0.033452946 | ENSMUSG00000039474 | ENSG00000109501 | 0.061 | 1.2797 | 0.047667422 | ||||||||||||||||||||||||||
| ENSG00000166136 | ENSMUSG00000091471 | 0.3498 | 1.7419 | 0.200815202 | ENSMUSG00000062380 | ENSG00000198211 | 0.0013 | 1.289 | 0.001008534 | ||||||||||||||||||||||||||
| ENSG00000104833 | ENSMUSG00000062591 | 0.0018 | 1.8334 | 0.000981782 | ENSMUSG00000003200 | ENSG00000141985 | 0.025 | 1.2899 | 0.019381347 | ||||||||||||||||||||||||||
| ENSG00000099624 | ENSMUSG00000003072 | 0.0757 | 2.0109 | 0.037644836 | ENSMUSG00000027602 | ENSG00000101460 | 0.0736 | 1.3014 | 0.05655448 | ||||||||||||||||||||||||||
| ENSG00000176619 | ENSMUSG00000062075 | 0.0867 | 2.2395 | 0.038713999 | ENSMUSG00000033900 | ENSG00000164114 | 0.209 | 1.304 | 0.160276074 | ||||||||||||||||||||||||||
| ENSG00000180155 | ENSMUSG00000075605 | 0.4135 | 2.4725 | 0.167239636 | ENSMUSG00000055013 | ENSG00000204172 | 0.2136 | 1.3086 | 0.163227877 | ||||||||||||||||||||||||||
| ENSG00000170373 | ENSMUSG00000027442 | 0.9662 | 3.142 | 0.307511139 | ENSMUSG00000055013 | ENSG00000198035 | 0.2138 | 1.3102 | 0.163181194 | ||||||||||||||||||||||||||
| ENSG00000130255 | ENSMUSG00000057863 | 0.0043 | 4.2774 | 0.001005284 | ENSMUSG00000045763 | ENSG00000176788 | 0.1544 | 1.3343 | 0.115716106 | ||||||||||||||||||||||||||
| ENSG00000088256 | ENSMUSG00000034781 | 0.0089 | 4.6581 | 0.00191065 | ENSMUSG00000034994 | ENSG00000167658 | 0.0043 | 1.348 | 0.003189911 | ||||||||||||||||||||||||||
| ENSG00000170373 | ENSMUSG00000036924 | 13.4209 | 11.1545 | 1.203182572 | ENSMUSG00000053395 | ENSG00000142408 | 0.013 | 1.3586 | 0.009568674 | ||||||||||||||||||||||||||
| ENSG00000178695 | ENSMUSG00000041633 | 0.2745 | 26.69 | 0.010284751 | ENSMUSG00000020331 | ENSG00000099822 | 0.0276 | 1.3785 | 0.020021763 | ||||||||||||||||||||||||||
| ENSG00000085063 | ENSMUSG00000068686 | 0.5046 | 34.08 | 0.014806338 | ENSMUSG00000045912 | ENSG00000183186 | 0.0433 | 1.3797 | 0.031383634 | ||||||||||||||||||||||||||
| ENSMUSG00000035863 | ENSG00000099864 | 0.1249 | 1.3825 | 0.09034358 | |||||||||||||||||||||||||||||||
| ENSMUSG00000016349 | ENSG00000101210 | 0.0014 | 1.3919 | 0.001005819 | |||||||||||||||||||||||||||||||
| ENSMUSG00000039457 | ENSG00000118898 | 0.0625 | 1.3948 | 0.044809292 | |||||||||||||||||||||||||||||||
| ENSMUSG00000025499 | ENSG00000174775 | 0 | 1.4086 | 0 | |||||||||||||||||||||||||||||||
| ENSMUSG00000047495 | ENSG00000198010 | 0.0519 | 1.4223 | 0.036490192 | |||||||||||||||||||||||||||||||
| ENSMUSG00000048240 | ENSG00000176533 | 0.0263 | 1.4239 | 0.018470398 | |||||||||||||||||||||||||||||||
| ENSMUSG00000040048 | ENSG00000140990 | 0.1058 | 1.426 | 0.074193548 | |||||||||||||||||||||||||||||||
| ENSMUSG00000055013 | ENSG00000204169 | 0.2432 | 1.4276 | 0.170355842 | |||||||||||||||||||||||||||||||
| ENSMUSG00000029419 | ENSG00000232434 | 0.0678 | 1.4407 | 0.047060457 | |||||||||||||||||||||||||||||||
| ENSMUSG00000055013 | ENSG00000174194 | 0.2407 | 1.4521 | 0.165759934 | |||||||||||||||||||||||||||||||
| ENSMUSG00000074030 | ENSG00000116903 | 0.0299 | 1.4609 | 0.020466836 | |||||||||||||||||||||||||||||||
| ENSMUSG00000055013 | ENSG00000188234 | 0.2414 | 1.4623 | 0.165082404 | |||||||||||||||||||||||||||||||
| ENSMUSG00000021573 | ENSG00000171368 | 0.0369 | 1.486 | 0.024831763 | |||||||||||||||||||||||||||||||
| ENSMUSG00000020153 | ENSG00000115286 | 0.0606 | 1.4872 | 0.040747714 | |||||||||||||||||||||||||||||||
| ENSMUSG00000055013 | ENSG00000172650 | 0.2629 | 1.5077 | 0.174371559 | |||||||||||||||||||||||||||||||
| ENSMUSG00000055013 | ENSG00000204149 | 0.2716 | 1.5174 | 0.178990378 | |||||||||||||||||||||||||||||||
| ENSMUSG00000040659 | ENSG00000142634 | 0.0249 | 1.549 | 0.016074887 | |||||||||||||||||||||||||||||||
| ENSMUSG00000037979 | ENSG00000119242 | 0.068 | 1.5494 | 0.043887957 | |||||||||||||||||||||||||||||||
| ENSMUSG00000007021 | ENSG00000127561 | 0.0288 | 1.5509 | 0.018569863 | |||||||||||||||||||||||||||||||
| ENSMUSG00000021750 | ENSG00000168309 | 0.0416 | 1.565 | 0.02658147 | |||||||||||||||||||||||||||||||
| ENSMUSG00000022565 | ENSG00000178209 | 0.0299 | 1.595 | 0.018746082 | |||||||||||||||||||||||||||||||
| ENSMUSG00000023175 | ENSG00000172270 | 0.2229 | 1.6011 | 0.139216788 | |||||||||||||||||||||||||||||||
| ENSMUSG00000052214 | ENSG00000125741 | 0.1685 | 1.7863 | 0.09432906 | |||||||||||||||||||||||||||||||
| ENSMUSG00000062591 | ENSG00000104833 | 0.0018 | 1.8334 | 0.000981782 | |||||||||||||||||||||||||||||||
| ENSMUSG00000052565 | ENSG00000124575 | 0.0729 | 1.8636 | 0.039117836 | |||||||||||||||||||||||||||||||
| ENSMUSG00000007836 | ENSG00000177733 | 0.0322 | 1.9088 | 0.016869237 | |||||||||||||||||||||||||||||||
| ENSMUSG00000036199 | ENSG00000186010 | 0.0818 | 1.989 | 0.041126194 | |||||||||||||||||||||||||||||||
| ENSMUSG00000003072 | ENSG00000099624 | 0.0757 | 2.0109 | 0.037644836 | |||||||||||||||||||||||||||||||
| ENSMUSG00000048429 | ENSG00000142444 | 0.0959 | 2.1873 | 0.043844009 | |||||||||||||||||||||||||||||||
| ENSMUSG00000020308 | ENSG00000141933 | 0.0736 | 2.203 | 0.033408988 | |||||||||||||||||||||||||||||||
| ENSMUSG00000022602 | ENSG00000198576 | 0.0324 | 2.2847 | 0.014181293 | |||||||||||||||||||||||||||||||
| ENSMUSG00000050796 | ENSG00000176022 | 0.0793 | 2.3042 | 0.034415415 | |||||||||||||||||||||||||||||||
| ENSMUSG00000036181 | ENSG00000187837 | 0.061 | 2.3826 | 0.025602283 | |||||||||||||||||||||||||||||||
| ENSMUSG00000020331 | ENSG00000256952 | 0.0238 | 2.4115 | 0.009869376 | |||||||||||||||||||||||||||||||
| ENSMUSG00000035835 | ENSG00000129951 | 0.0523 | 2.477 | 0.021114251 | |||||||||||||||||||||||||||||||
| ENSMUSG00000064125 | ENSG00000183248 | 0.7099 | 2.7757 | 0.255755305 | |||||||||||||||||||||||||||||||
| ENSMUSG00000063457 | ENSG00000115268 | 0.0028 | 2.8141 | 0.00099499 | |||||||||||||||||||||||||||||||
| ENSMUSG00000057863 | ENSG00000130255 | 0.0043 | 4.2774 | 0.001005284 | |||||||||||||||||||||||||||||||
| ENSMUSG00000019261 | ENSG00000130479 | 0.1929 | 4.4571 | 0.043279262 | |||||||||||||||||||||||||||||||
| ENSMUSG00000034781 | ENSG00000088256 | 0.0089 | 4.6581 | 0.00191065 | |||||||||||||||||||||||||||||||
| ENSMUSG00000046691 | ENSG00000178928 | 1.09 | 5.3777 | 0.202688882 | |||||||||||||||||||||||||||||||
| ENSMUSG00000058773 | ENSG00000184357 | 0.0421 | 6.1729 | 0.006820133 | |||||||||||||||||||||||||||||||
| ENSMUSG00000041633 | ENSG00000178695 | 0.2745 | 26.69 | 0.010284751 | |||||||||||||||||||||||||||||||
| ENSMUSG00000024403 | ENSG00000254870 | 0.3693 | 59.4828 | 0.006208517 | |||||||||||||||||||||||||||||||