G2C::Genetics
Confirmed Rare Copy Number Variants Implicate Novel Genes in Schizophrenia
Gloria W. C. Tam1*, Louie N. van de Lagemaat1*, Richard R. Redon1, Mary P. Malloy2, Walter J. Muir2, Ben S. Pickard3, Karen E. Strathdee1, Mike D. R. Croning1, Ian J. Deary4, Douglas H.R. Blackwood2, Nigel P. Carter1 and Seth G. N. Grant1
Author email: sg3@sanger.ac.uk * - These authors contributed equally to this work
- Wellcome Trust Sanger Institute, Wellcome Trust Genome Campus, Hinxton, Cambridge, CB10 1SA, UK
- Division of Psychiatry, University of Edinburgh, Royal Edinburgh Hospital, Morningside Park, Edinburgh, EH10 5HF, UK
- Strathclyde Institute of Pharmacy and Biomedical Sciences (SIPBS), University of Strathclyde, Glasgow G4 0NR
- Department of Psychology, University of Edinburgh, EH8 9JZ
Introduction
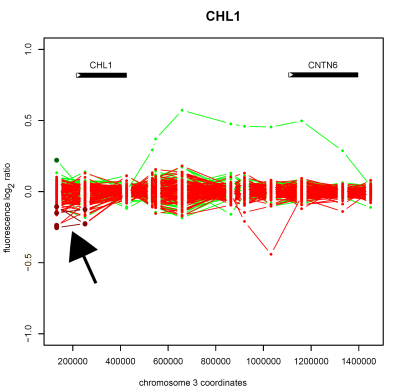
Detection of copy number deletion on chromosome 3 (arrowed) in the cell-recognition protein encoding gene <em>CHL1</em> in schizophrenic patient samples, as detected by whole-genome tiling path array CGH.
Understanding how cognitive processes including learning, memory, decision making and ideation are encoded by the genome is a key question in biology. Identification of sets of genes underlying human cognitive disorders is one path that may ultimately give insight to the biochemical and cellular mechanisms of cognition. Schizophrenia is a common disease with cognitive symptoms, high heritability and complex genetics ( 19748074).
Here we identify genes involved with schizophrenia by measuring differences in DNA copy number across the entire genome in 91 schizophrenia cases and 92 controls in the Scottish population. Comparing this data with >3000 cases in the public domain confirms loci demonstrating a bias for mutation in schizophrenia.
In addition to identifying novel loci we found copy number variants in phosphodiesterase PDE10A, the FMR1 interacting protein CYFIP1, potassium channel genes KCNE1 and KCNE2, the Down Syndrome Critical Region 1 gene RCAN1, cell-recognition protein CHL1, the transcription factor SP4 and a histone deacetylase HDAC9 amongst others. Integrating the function of these many genes into a coherent model of schizophrenia and cognition is a major unanswered challenge.
Data resources
1. What are copy number variants (CNVs)?
Copy number variations are spontaneous mutations in the genome that result in duplications or deletions of the genomic sequence. Duplications can produce extra copies of a gene, deletions can remove it altogether. CNVs are an interesting biological phenomenon because they are not inherited from ones parents. Rather, they are acquired de novo when certain sequences of genetic code fail to copy properly. Acquisition of CNVs is unpredictable, random, and spontaneous. Smaller variants are probably very common. Each one of us may have one or two (taken from G2COnline).
For full review see: 19748074
2. What is schizophrenia?
Schizophrenia is a psychiatric disorder characterized by abnormalities in the perception or expression of reality. It most commonly manifests as auditory hallucinations, paranoid or bizarre delusions, or disorganized speech and thinking with significant social or occupational dysfunction. See also Schizophrenia at Wikipedia.
3. What is the evidence CNVs are associated with schizophrenia?
Recent studies based on genome-wide study of CNVs have detected novel recurrent submicroscopic copy number changes, including recurrent deletions at a number of loci.
Available evidence is fully reviewed in: 19748074
4.How do we assay an indivdual's genome for CNVs?
Copy number variation can be asssayed by PCR and DNA array-based approaches. In the present study we utilised whole-genome tiling path array comparative genome hybridisation (array CGH). See e.g. methodological description by Catherine Shaffer.
5. What is the Lothian Birth Cohort (LBC)?
The Lothian Birth Cohort was obtained from the Scottish Mental Survey of 1947 (SMS1947; N = 70,805) examining individuals born in the year 1936 and residing in the Edinburgh area (Lothian) of Scotland. See 14717632 for full description.
6. What is CNVFinder?
CNVFinder is a programme designed to detect copy number variants (CNVs) in human population from large-insert clone DNA microarray data covering the entire human genome in tiling path resolution (WGTP platform). More information on the CNV Project is available on the Sanger website.
The algorithm is describes in 17122085
Supplementary Table 1. Technical confirmations of CNV calls.
Shown are the patient ID in which the CNV was detected, locus of the variation on the NCBI36 human genome assembly, whether predicted as a loss or gain by CNVFinder, result of confirmation by qPCR, Nimblegen result, and ISC coordinates of the variation.
Supplementary Table 2. Genic case-specific ISC CNVs confirmed by CNVFinder.
Shown are HGNC-approved gene symbols and IDs, locus of the variation on the NCBI36 human genome assembly, detection in the various patient and control cohorts, and whether the variation in an insertion or deletion.
References
-
Schizophrenia and Autism - Opposite Ends of the Same Spectrum?
Connolly, J
