G2C::Proteomics
Zebrafish synapse proteome complexity, evolution and ultrastructure.
Àlex Bayés1,2*, Mark O Collins3, Rita Reig-Viader1,2, Gemma Gou1,2, David Goulding4, Abril Izquierdo5, Jyoti S Choudhary6, Richard D Emes5,7, and Seth GN Grant8*
*Corresponding author: abayesp@santpau.cat
*Corresponding author: Seth.Grant@ed.ac.uk
- Molecular Physiology of the Synapse Laboratory, Biomedical Research Institute Sant Pau (IIB Sant Pau) Sant Antoni Ma Claret 167, 08025 Barcelona, Catalonia, Spain
- Universitat Autònoma de Barcelona, 08193 Bellaterra, Cerdanyola del Vallès, Spain
- Department of Biomedical Science, The Centre for Membrane Interactions and Dynamics, University of Sheffield, Western Bank, Sheffield S10 2TN, UK
- Pathogen Genomics, Wellcome Trust Sanger Institute, Hinxton, CB101SA
- School of Veterinary Medicine and Science, University of Nottingham. Sutton Bonington Campus, Leicestershire, LE12 5RD UK
- Proteomic Mass Spectrometry, The Wellcome Trust Sanger Institute, Hinxton, Cambridgeshire, UK
- Advanced Data Analysis Centre, University of Nottingham. Sutton Bonnington Campus, Leicestershire, LE12 5RD UK
- Genes to Cognition Programme, Molecular Neuroscience, Centre for Clinical Brain Science and Centre for Neuroregeneration, University of Edinburgh, Edinburgh EH16 4SB, UK
Overview
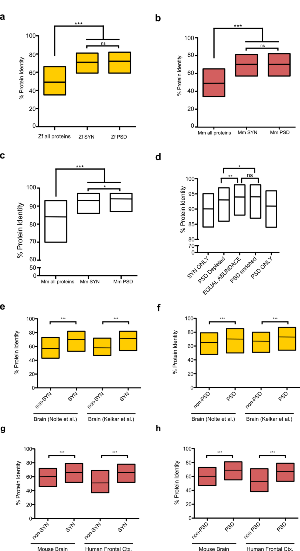
Zebrafish are a widely used model organism for studying the biology and diseases of the vertebrate nervous system. Mammalian synapse proteomes are highly complex and disrupted by many mutations causing developmental, neurological and psychiatric disorders. Here we report the characterisation of zebrafish synapse ultrastructure, proteome composition and complexity and analyse the importance of the teleost-specific whole genome duplication. The complexity of the zebrafish postsynaptic density proteome was lower than in mammals with fewer protein families comprised of more gene duplicates. A highly conserved vertebrate PSD proteome spanning the last ~450 million years of evolution contained ~1000 proteins. Lineage-specific PSD proteins conferred functional specialization to vertebrate species. Synapse ultrastructure showed conserved and species specific features. These datasets are a valuable resource for studying the biology of zebrafish and other vertebrates and have important implications for their use as models of human disease.
a. Transmission electron microscopy image of a characteristic asymmetric synapse from the zebrafish cerebellar
corpus. The presynaptic bouton, filled with sinaptic vesicles (red astreisk), and the opposing postsyanptic density (red
arrow) are clearly visiable. Noticieably the presynaptic bouton (highlighted in purple) greatly surronds the postsynaptic
spine a feature that is not observed in mammalian cerebellar asymmetric synapses. Scale bar 200nm.
b. PSD protein sequence identity between mouse and zebrafish for proteins found in both species (vertebrate PSD)
or exclusively in one of them. Vertebrate PSD proteins show higher levels of conservation than proteins esclusive to one
species. Red lines represent the median % of identitiy for two zebrafish brain proteomes and one mouse brain proteome.
The full results and analyses of the study can be sorted, searched and downloaded
Figures
- Figure 1.
- Transmission electron microscopy of zebrafish asymmetric synapses in four different brain areas.
- Figure 2.
- Mouse and zebrafish synaptic protoeme.
- Figure 3.
- Expansion and retention of synaptic proteins after teleost-specific whole genome duplication.
- Figure 4.
- Functional similarity between mouse and zebrafish synaptic proteomes.
- Figure 5.
- Differential expression at the mouse PSD of proteins involved in intracellular vesicle biogenesis.
- Figure 6.
- Conservation of the core vertebrate PSD machinery.
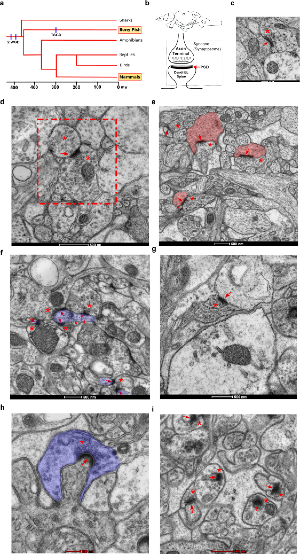
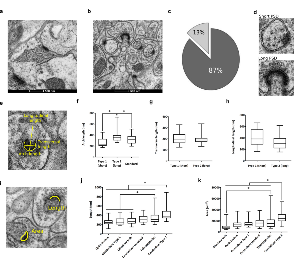
Figure 1. Transmission electron microscopy of zebrafish asymmetric synapses in four different brain areas.
a. Evolutionary tree of the vertebrate lineage with timescale in million years (my). The occurrence of the two whole genome duplication events common to all vertebrates and specific to teleost (TSGD) are indicated by blue lines.
b. Schematic representation of the zebrafish brain with the four regions studied (o: olfactory bulb, t: telencephalon, ot: optic tectum and cc: cerebellar corpus) and of an excitatory synapse. Synaptosomes are formed by the axon terminal and the dendritic spine, which are separated from their corresponding neurons during tissue processing. The location of the postsynaptic density (PSD) is also indicated.
c. Asymmetric synapse from the olfactory bulb. A red asterisk and a red arrow indicate the location of presynaptic vesicles and the PSD respectively. Scale bar 200 nm.
d. An asymmetric dendro-dendritic synapse of the olfactory bulb (framed by a red dotted square) is shown. Asterisks indicate pre- and post-synaptic vesicles. The PSD is indicated by a red arrow. Scale bar 500 nm.
e. Asymmetric synapses from the telencephalon. Red asterisks and arrows indicate the location of presynaptic vesicles and PSDs respectively. The area corresponding to postsynaptic spine-like structures is filled with pink. Scale bar 500 nm.
f. Asymmetric synapses from the optic tectum. Red asterisks and arrows indicate the location of presynaptic vesicles and the PSD respectively. Red arrowheads indicate microtubule location within thin dendritic-like projection. The area of a thin dendritic-like projection, where synapses are form, is filled with purple. Scale bar 500 nm.
g. Flat (standard) asymmetric synapse from the cerebellar corpus. A red asterisk and a red arrow indicate the location of presynaptic vesicles and the PSD respectively. Scale bar 500 nm.
h. Asymmetric synapse from the medial part of the cerebellar corpus showing the extent at which the presynaptic element (highlighted in purple) surrounds the dendritic spine. Scale bar 200 nm.
i. Micrograph displaying the morphology of most abundant asymmetric synapses from the cerebellar corpus. Red asterisks and arrows indicate the location of presynaptic vesicles and the PSD respectively. Scale bar 500 nm.
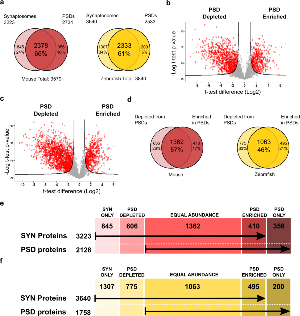
Figure 2. Mouse and zebrafish synaptic protoeme.
a. Venn diagrams of mouse (red) and zebrafish (yellow) proteins identified in synaptosomal and PSD preparations indicating the percentage of proteins found in both fractions. The total number of protein identified in each species is also indicated.
b/c. Volcano plots showing quantitative enrichment and depletion of proteins between synaptosomes and postsynaptic densities purified from mouse (b) and zebrafish brain (c). Enriched or depleted proteins were identified from statistical analysis of triplicate PSD and synaptosome datasets for each species using t-testing and a Permutation based False discovery rate of 0.05.
d. Venn diagrams of mouse (red) and zebrafish (yellow) proteins found depleted or enriched in the PSD when compared with the synaptosomal fraction. The percentage of proteins found with equal abundance at the synaptosomal and PSD fraction is also indicated.
e. Scheme indicating the number of mouse synaptosomal and PSD proteins found only in one of the two fractions, depleted or enriched at the PSD or found in equal abundance in both fractions.
f. Scheme indicating the number of zebrafish synaptosomal and PSD proteins found only in one of the two fractions, depleted or enriched at the PSD or found in equal abundance in both fractions.
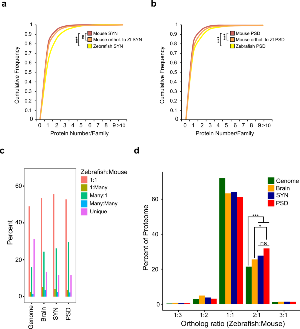
Figure 3. Expansion and retention of synaptic proteins after teleost-specific whole genome duplication.
a. Cumulative frequency plot of proteins found per Ensemble Family amongst mouse, zebrafish and mouse orthologs of zebrafish SYN proteins. Kruskal-Wallis Test used to calculate significance between distributions (** p < 0.01 and *** p < 0.0001).
b. Cumulative frequency plot of proteins found per Ensemble Family amongst mouse, zebrafish and mouse orthologs of zebrafish PSD proteins. Kruskal-Wallis Test used to calculate significance between distributions (** p < 0.01 and *** p < 0.0001).
c. Distribution of orthology types between zebrafish and mouse. For each protein in the proteome, the corresponding gene was identified and orthologous genes between species were determined using the biomaRt bioconductor package1. For each gene the zebrafish:mouse ratio of orthologs was determined and characterised as 1:1, 1:Many, Many:1, Many:Many or unique to mouse or zebrafish.
d. Density of proteome by ortholog ratio (zebrafish:mouse). Statistically significant comparisons between pairs of distributions (two-tailed Kolmogorov–Smirnov test applied to distributions) are shown. *** p < 0.001 and * p < 0.05, non significant comparisons are not shown.
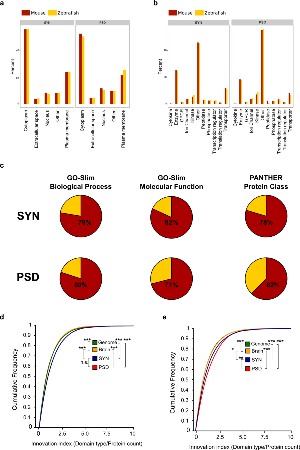
Figure 4. Functional similarity between mouse and zebrafish synaptic proteomes.
a. Fraction of mouse (red) and zebrafish (yellow) synaptic proteins annotated to IPA cell location categories.
b. Fraction of mouse (red) and zebrafish (yellow) synaptic proteins annotated to IPA molecular function categories.
c. Overlap between Biological Process and Molecular Function GO-slim terms and PANTHER Protein Classes enriched in mouse and zebrafish. Top row shows data for synaptosomes and bottom row for PSD proteomes. The exact percentage of overlap is indicated.
d. Cumulative frequency distribution plots of individual protein innovation index (number of unique domain types per protein) for each proteome in mouse. Statistically significant comparisons between pairs of distributions (two-tailed Kolmogorov–Smirnov test applied to distributions) are shown at the legend. *** p < 0.001, ** p < 0.01, * p < 0.05 non significant comparisons are not shown.
e. Cumulative frequency distribution plots of individual protein innovation index (number of unique domain types per protein) for each proteome in zebrafish. Statistically significant comparisons between pairs of distributions (two-tailed Kolmogorov–Smirnov test applied to distributions) are shown at the legend. *** p < 0.001, ** p < 0.01, * p < 0.05 non significant comparisons are not shown.
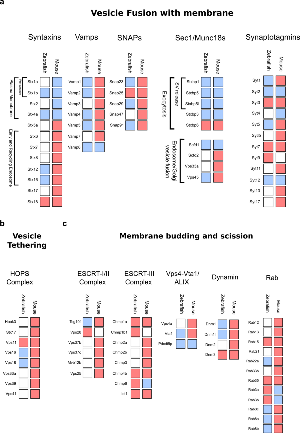
Figure 5. Differential expression at the mouse PSD of proteins involved in intracellular vesicle biogenesis.
For each protein the mouse ortholog gene name is given. A white squares denote that a protein has not been found in a given species, a blue square identifies proteins found in synaptosomes but not at the level of postsynaptic densities, and a red square identifies proteins found at the PSD.
a. Proteins involved in vesicle fusion with membranes, including SNARE complex proteins: Syntaxins, Vamps and SNAPs, syntaxin binding proteins (Sec1/Munc18) and synaptotagmins.
b. Proteins form the HOPS complex, involved in membrane vesicle tethering to membranes.
c. Proteins involved in membrane bending/budding and scission to form vesicles, including proteins from ESCRT complexes (ESCRT I, II, III and Vps4-Vta1/ALIX), dynamins and small GTPases from the Rab family.
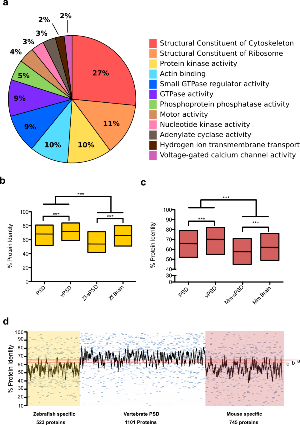
Figure 6. Conservation of the core vertebrate PSD machinery.
a. Pie chart of main GO-Slim Molecular Function categories enriched amongst vPSD proteins.
b. Box plots for the percentage of protein identity since the last common ancestor between zebrafish and mouse of zebrafish PSD proteins, PSD components found in both species (vPSD), zebrafish-specific PSD proteins (Zf-sPSD) and a zebrafish brain proteom. Distributions compared using the Mann-Whitney U test (*** p < 0.001).
c. Box plots for the percentage of protein identity since the last common ancestor between zebrafish and mouse of mouse PSD proteins, PSD components found in both species (vPSD), mouse-specific PSD proteins (Mm-sPSD) and a mouse brain proteome. Distributions compared using the Mann-Whitney U test (*** p < 0.001).
d. Percentage of protein identity for individual proteins found only in zebrafish PSD (yellow), in both species PSD (white) and only in mouse PSD (red). Red lines represent the median percentage of protein identity for two zebrafish (a and b) and a mouse (c) whole brain proteomes.
Tables
- Table 1.
- Protein Families among mouse and zebrafish SYN and PSD proteins.
- Table 2.
- Number of proteins in families characteristic of the synapse.
Table 1. Protein Families among mouse and zebrafish SYN and PSD proteins.
| Proteins | Ensembl Families | Families/Protein | |
|---|---|---|---|
| Mouse SYN | 3223 | 2552 | 0.79 |
| Zebrafish SYN | 3640 | 2354 | 0.65 |
| Mouse orthologues of Zebrafish SYN | 3138 | 2410 | 0.77 |
| Mouse PSD | 2128 | 1688 | 0.79 |
| Zebrafish PSD | 1758 | *1089 | 0.62 |
| Mouse orthologues of Zebrafish PSD | 1556 | ***1128 | 0.72 |
* p < 0.05
*** p < 0.0001
Table 2. Number of proteins in families characteristic of the synapse.
*Fold enrichment of observed proteins per GO term.
#Binomial statistics p-value.
| Human | Rat | Mouse | ||||
|---|---|---|---|---|---|---|
| *Fold Change | #P-value | Fold Change | P-value | Fold Change | P-value | |
| Related to vesicle-mediated protein trafficking and localization | ||||||
| regulation of vesicle-mediated transport (GO:0060627) | 3.3 | 1.3E-04 | 3.4 | 1.7E-03 | 2.3 | 3.0E-04 |
| vesicle-mediated transport (GO:0016192) | 2.9 | 5.8E-10 | 2.4 | 7.1E-03 | 2.2 | 1.7E-12 |
| cytoplasmic transport (GO:0016482) | 2.9 | 5.0E-06 | 2.6 | 1.9E-02 | 2.0 | 1.2E-04 |
| establishment of protein localization (GO:0045184) | 2.9 | 1.2E-12 | 2.2 | 1.7E-03 | 2.2 | 2.5E-15 |
| protein transport (GO:0015031) | 2.8 | 2.7E-10 | 2.3 | 4.1E-03 | 2.2 | 3.5E-14 |
| intracellular transport (GO:0046907) | 2.7 | 9.9E-10 | 2.5 | 6.3E-06 | 2.2 | 4.0E-15 |
| endosome (GO:0005768) | 2.2 | 3.3E-02 | 2.5 | 3.2E-03 | 2.3 | 6.8E-10 |
| Related to protein location at plasma membrane | ||||||
| extrinsic component of cytoplasmic side of plasma membrane (GO:0031234) | 5.8 | 2.8E-04 | 5.9 | 5.2E-03 | 3.5 | 9.4E-04 |
| extrinsic component of plasma membrane (GO:0019897) | 5.1 | 1.1E-04 | 5.3 | 8.1E-04 | 3.3 | 6.5E-05 |
| cytoplasmic side of plasma membrane (GO:0009898) | 4.5 | 5.3E-04 | 6.1 | 2.6E-06 | 3.7 | 1.1E-07 |
| cytoplasmic side of membrane (GO:0098562) | 4.2 | 1.4E-03 | 5.7 | 7.7E-06 | 3.5 | 2.1E-07 |
| extrinsic component of membrane (GO:0019898) | 3.3 | 6.3E-03 | 3.6 | 1.1E-02 | 2.5 | 2.8E-04 |
| Related to actin filaments function and dynamics | ||||||
| regulation of actin filament length (GO:0030832) | 4.6 | 2.8E-03 | 4.8 | 1.7E-02 | 2.9 | 9.7E-03 |
| actin cytoskeleton organization (GO:0030036) | 4.0 | 1.8E-08 | 3.3 | 3.7E-03 | 2.4 | 7.9E-06 |
| actin filament-based process (GO:0030029) | 4.0 | 1.3E-09 | 3.4 | 5.9E-04 | 2.6 | 3.8E-08 |
| regulation of cytoskeleton organization (GO:0051493) | 3.4 | 8.1E-06 | 3.2 | 3.9E-03 | 2.7 | 3.0E-09 |
| Related to Ion channels | ||||||
| regulation of cation transmembrane transport (GO:1904062) | 4.4 | 1.1E-04 | 4.6 | 1.2E-03 | 2.7 | 2.7E-03 |
| regulation of metal ion transport (GO:0010959) | 3.0 | 2.3E-02 | 3.4 | 1.7E-02 | 2.2 | 4.2E-03 |
| transmembrane transporter complex (GO:1902495) | 3.0 | 2.8E-02 | 3.4 | 1.1E-02 | 2.7 | 5.0E-06 |
| transporter complex (GO:1990351) | 2.9 | 3.8E-02 | 3.3 | 1.5E-02 | 2.7 | 1.2E-06 |
| Related to Cell Junction | ||||||
| adherens junction (GO:0005912) | 3.9 | 1.8E-09 | 3.3 | 2.1E-04 | 2.2 | 1.3E-04 |
| cell-substrate junction (GO:0030055) | 3.9 | 1.1E-07 | 3.7 | 3.8E-05 | 2.0 | 1.7E-02 |
| anchoring junction (GO:0070161) | 3.7 | 6.3E-09 | 3.3 | 1.3E-04 | 2.2 | 9.8E-05 |
| cell junction (GO:0030054) | 2.9 | 2.0E-13 | 3.0 | 3.0E-12 | 2.4 | 2.1E-22 |
| Related to Microtubules and supramolecular fibers | ||||||
| microtubule (GO:0005874) | 4.6 | 9.9E-13 | 2.9 | 2.0E-02 | 3.5 | 1.1E-19 |
| polymeric cytoskeletal fiber (GO:0099513) | 4.2 | 4.9E-17 | 2.5 | 2.7E-02 | 3.1 | 5.0E-23 |
| supramolecular fiber (GO:0099512) | 4.2 | 9.9E-17 | 2.4 | 3.4E-02 | 3.1 | 1.7E-22 |
Supplementary Figures
- Supplementary Figure 1.
- Ultrastructure of Zebrafish PSD morphologies.
- Supplementary Figure 2.
- Morphological characteristics of zebrafish PSDs.
- Supplementary Figure 3.
- PSD morphological metrics.
- Supplementary Figure 4.
- Isolation of postsynaptic densities from mouse and zebrafish brains.
- Supplementary Figure 5.
- Label Free Quantification (LFQ) intensities of zebrafish and mouse synaptic proteins.
- Supplementary Figure 6.
- Conservation of SYN and PSD proteins along vertebrate evolution.
Supplementary Figure 1. Ultrastructure of Zebrafish PSD morphologies.
Transmission electron microscopy images taken from sections of four different zebrafish brain regions.
|
|
|
|
|
|
a. Olfactory bulb: (a') Schematic representation of the zebrafish brain. The black line indicates the position of the olfactory bulb. (a1-a8) PSDs found in different areas of the olfactory bulb. Arrowheads indicate PSDs from dendro-dendritic synapses.
b. Telencephalon: (b') Schematic representation of the zebrafish brain. The black line indicates the position of the telencephalon. (b'') Coronal semi-thin section through telencephalon stained with toluidine blue. (b1-b8) PSDs found in different areas of the telencephalon.
c. Optic tectum: (c') Schematic representation of the zebrafish brain. The black line indicates the position of the midbrain. (c'') Coronal semi-thin section through midbrain stained with toluidine blue. The red encircled area corresponds to the optic tectum. The orange line indicates the area where asymmetric synapses were identified. (c1-c8) PSDs found in different sections of the optic tectum. Arrowheads denote PSDs.
d. Cerebellar corpus: (d') Schematic representation of the zebrafish brain. The black line indicates the position of the hindbrain. (d'') Coronal semi-thin section through hindbrain stained with toluidine blue. The red encircled area corresponds to the cerebellar corpus. The orange line indicates the area where asymmetric synapses were identified. (d1-d8) PSDs found in different sections of the cerebellum. Arrows indicate PSDs showing the curved morphology with the presynaptic element surrounding the postsynaptic spine.
Supplementary Figure 2. Morphological characteristics of zebrafish PSDs.
a. Synapses from the cerebellar corpus with morphological characteristics similar to those observed in mammals and other zebrafish brain regions, including flat PSDs.
b. Synapses from the cerebellar corpus with presynaptic elements engulfing the postsynaptic terminal and highly curved PSDs.
c. Proportion of flat (light grey) and curved PSDs found at the cerebellar corpus of zebrafish brain.
d. Representation of the three measures taken on cerebellar synapses as in (b): PSD arch length, PSD transversal length and spine longitudinal length.
e. Representative images of the two types of PSD found amongst curved ones: type 1 (left image) and Type 2 (right image).
f. PSD arch length differences between the three types of PSDs found at the cerebellar corpus (p < 0.05). Type 1 n = 24; type 2 n = 40; standard n = 10.
g. PSD transversal length differences between the three types of PSDs found at the cerebellar corpus. Type 1 n = 24; type 2 n = 40; standard n = 10.
h. Spine longitudinal length differences between the three types of PSDs found at the cerebellar corpus. Type 1 n = 24; type 2 n = 40; standard n = 10.
i. Representation of the two measures taken on PSD from all brain areas: length and area.
j. Differences in PSD length between brain regions (p < 0.05). Optic tectum n = 55; Cerebellum type 1 n = 25; Olfactory bulb n = 52; Cerebellum standard n = 10; Telencephalon n = 86; Cerebellum type 2 n = 40.
k. Differences in PSD area between brain regions (p < 0.05). Olfactory bulb = 53; Optic tectum = 53; Cerebellum type 1 = 26; Cerebellum standard = 10; Telencephalon = 87; Cerebellum type 2 = 37.
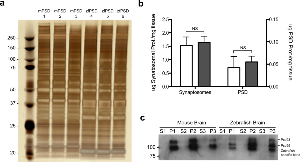
Supplementary Figure 4. Isolation of postsynaptic densities from mouse and zebrafish brains.
a. SDS-PAGE electrophoresis of the three PSD fractions isolated from mouse brain (lanes 1-3) and zebrafish brain (lanes 4-6).
b. Protein yield for synaptosomal (SYN) preparations from mouse and zebrafish brains.
c. Protein yield for postsynaptic density (PSD) preparations from mouse and zebrafish brains.
d. Immunoblot of synaptosomal (S) and PSD (P) fractions for PSD95 protein in triplicate samples from mouse and zebrafish brains. Representative image of 3 western blots.
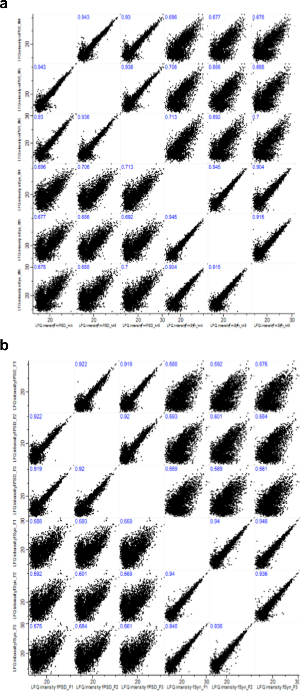
Supplementary Figure 5. Label Free Quantification (LFQ) intensities of zebrafish and mouse synaptic proteins.
a. Multiscatter plot of protein LFQ intensities calculated from data obtained from proteomic analysis of Zebrafish synaptosome and PSD samples. Pearson correlation coefficients are shown for each comparison and were >0.90 for biological replicates in each group.
b. Multiscatter plot of protein LFQ intensities calculated from data obtained from proteomic analysis of mouse synaptosome and PSD samples. Pearson correlation coefficients are shown for each comparison and were >0.90 for biological replicates in each group.
Supplementary Figure 6. Conservation of SYN and PSD proteins along vertebrate evolution.
a. Box plots for the percentage of protein identity since the last common ancestor between zebrafish and mouse in the following zebrafish protein groups: all proteins encoded in the genome, brain synaptosomes (Zf-SYN) and brain postsynaptic densities (Zf-PSD).
b. Box plots for the percentage of protein identity since the last common ancestor between zebrafish and mouse in the following mouse protein groups: all proteins encoded in the genome, brain synaptosomes (Mm-SYN) and brain postsynaptic densities (Mm-PSD).
c. Box plots for the percentage of protein identity since the last common ancestor between mouse and human in the following protein groups: all mouse proteins encoded in the genome, mouse synaptosomal (Mm-SYN) and postsynaptic density (Mm-PSD) proteins.
d. Box plots for the percentage of protein identity since the last common ancestor between mouse and human of of proteins found only in the mouse synaptosomal fraction, with equal abundance in both fractions or depleted, enriched or only found at the PSD.
e. Box plots for the percentage of protein identity since the last common ancestor between zebrafish and mouse of proteins identified in the zebrafish brain divided into those found (SYN) or absent (non-SYN) in our synaptosomal preparation. Data from two independent zebrafish brain proteomes are used2,5.
f. Box plots for the percentage of protein identity since the last common ancestor between zebrafish and mouse of proteins identified in the zebrafish brain divided into those found (PSD) or absent (non-PSD) in our postsynaptic density preparation. Data from two independent zebrafish brain proteomes are used2,5.
g. Box plots for the percentage of protein identity since the last common ancestor between zebrafish and mouse of proteins identified in a mouse brain proteome3 and a human frontal cortex proteome6 divided into those found (SYN) or absent (non-SYN) in our synaptosomal preparation.
h. Box plots for the percentage of protein identity since the last common ancestor between zebrafish and mouse of proteins identified in a mouse brain proteome3 and a human frontal cortex proteome6 divided into those found (PSD) or absent (non-PSD) in our postsynaptic density preparation.
All distribution compared using the Mann-Whitney U test (*** p < 0.001; ** p < 0.01 and * p< 0.05).
- Supplementary Table 1a.
- Synaptosomal proteins identified in mouse.
- Supplementary Table 1b.
- Postsynaptic density proteins identified in mouse.
- Supplementary Table 2a.
- Synaptosomal proteins identified in zebrafish.
- Supplementary Table 2b.
- Postsynaptic density proteins identified in zebrafish.
- Supplementary Table 3a.
- IPA cell location and molecular function categories in mouse SYN and PSD proteins.
- Supplementary Table 3b.
- IPA cell location and molecular function categories in zebrafish SYN and PSD proteins.
- Supplementary Table 4a.
- Ensemble Families from mouse synaptosomal proteins.
- Supplementary Table 4b.
- Ensemble Families from mouse postsynaptic density proteins.
- Supplementary Table 4c.
- Ensemble Families from zebrafish synaptosomal proteins.
- Supplementary Table 4d.
- Ensemble Families from mouse postsynaptic density proteins.
- Supplementary Table 5.
- Domains in mouse and zebrafish synaptic proteins
- Supplementary Table 6a.
- GO analysis of mouse specific PSD proteins
- Supplementary Table 6b.
- GO analysis of zebrafish specific PSD proteins.
- Supplementary Table 7a.
- Mouse SYN orthologues in zebrafish SYN.
- Supplementary Table 7b.
- Mouse PSD orthologues in zebrafish PSD.
- Supplementary Table 7c.
- Zebrafish SYN orthologues in Mouse SYN
- Supplementary Table 7d.
- Zebrafish PSD orthologues in mouse PSD
Supplementary Table 1a. Synaptosomal proteins identified in mouse.
Proteins identified in mouse whole brain synaptosomal preparations. For each protein the following database identifiers are given: Ensembl Gene, Associated Gene Name, Description and MGI ID.
| ENSEBLE ID | Associated Gene Name | Description | MGI ID |
|---|---|---|---|
| ENSMUSG00000057230 | Aak1 | AP2 associated kinase 1 | MGI:1098687 |
| ENSMUSG00000023938 | Aars2 | alanyl-tRNA synthetase 2, mitochondrial (putative) | MGI:2681839 |
| ENSMUSG00000031960 | Aars | alanyl-tRNA synthetase | MGI:2384560 |
| ENSMUSG00000025375 | Aatk | apoptosis-associated tyrosine kinase | MGI:1197518 |
| ENSMUSG00000057880 | Abat | 4-aminobutyrate aminotransferase | MGI:2443582 |
| ENSMUSG00000015243 | Abca1 | ATP-binding cassette, sub-family A (ABC1), member 1 | MGI:99607 |
| ENSMUSG00000026944 | Abca2 | ATP-binding cassette, sub-family A (ABC1), member 2 | MGI:99606 |
| ENSMUSG00000024130 | Abca3 | ATP-binding cassette, sub-family A (ABC1), member 3 | MGI:1351617 |
| ENSMUSG00000018800 | Abca5 | ATP-binding cassette, sub-family A (ABC1), member 5 | MGI:2386607 |
| ENSMUSG00000035722 | Abca7 | ATP-binding cassette, sub-family A (ABC1), member 7 | MGI:1351646 |
| ENSMUSG00000041797 | Abca9 | ATP-binding cassette, sub-family A (ABC1), member 9 | MGI:2386796 |
| ENSMUSG00000031974 | Abcb10 | ATP-binding cassette, sub-family B (MDR/TAP), member 10 | MGI:1860508 |
| ENSMUSG00000040584 | Abcb1a | ATP-binding cassette, sub-family B (MDR/TAP), member 1A | MGI:97570 |
| ENSMUSG00000026198 | Abcb6 | ATP-binding cassette, sub-family B (MDR/TAP), member 6 | MGI:1921354 |
| ENSMUSG00000031333 | Abcb7 | ATP-binding cassette, sub-family B (MDR/TAP), member 7 | MGI:109533 |
| ENSMUSG00000028973 | Abcb8 | ATP-binding cassette, sub-family B (MDR/TAP), member 8 | MGI:1351667 |
| ENSMUSG00000029408 | Abcb9 | ATP-binding cassette, sub-family B (MDR/TAP), member 9 | MGI:1861729 |
| ENSMUSG00000032842 | Abcc10 | ATP-binding cassette, sub-family C (CFTR/MRP), member 10 | MGI:2386976 |
| ENSMUSG00000022822 | Abcc5 | ATP-binding cassette, sub-family C (CFTR/MRP), member 5 | MGI:1351644 |
| ENSMUSG00000055782 | Abcd2 | ATP-binding cassette, sub-family D (ALD), member 2 | MGI:1349467 |
| ENSMUSG00000028127 | Abcd3 | ATP-binding cassette, sub-family D (ALD), member 3 | MGI:1349216 |
| ENSMUSG00000029802 | Abcg2 | ATP-binding cassette, sub-family G (WHITE), member 2 | MGI:1347061 |
| ENSMUSG00000032046 | Abhd12 | abhydrolase domain containing 12 | MGI:1923442 |
| ENSMUSG00000007036 | Abhd16a | abhydrolase domain containing 16A | MGI:99476 |
| ENSMUSG00000047368 | Abhd17b | abhydrolase domain containing 17B | MGI:1917816 |
| ENSMUSG00000002475 | Abhd3 | abhydrolase domain containing 3 | MGI:2147183 |
| ENSMUSG00000025277 | Abhd6 | abhydrolase domain containing 6 | MGI:1913332 |
| ENSMUSG00000058835 | Abi1 | abl-interactor 1 | MGI:104913 |
| ENSMUSG00000026782 | Abi2 | abl-interactor 2 | MGI:106913 |
| ENSMUSG00000026596 | Abl2 | v-abl Abelson murine leukemia viral oncogene 2 (arg, Abelson-related gene) | MGI:87860 |
| ENSMUSG00000025085 | Ablim1 | actin-binding LIM protein 1 | MGI:1194500 |
| ENSMUSG00000032735 | Ablim3 | actin binding LIM protein family, member 3 | MGI:2442582 |
| ENSMUSG00000017631 | Abr | active BCR-related gene | MGI:107771 |
| ENSMUSG00000036138 | Acaa1a | acetyl-Coenzyme A acyltransferase 1A | MGI:2148491 |
| ENSMUSG00000036880 | Acaa2 | acetyl-Coenzyme A acyltransferase 2 (mitochondrial 3-oxoacyl-Coenzyme A thiolase) | MGI:1098623 |
| ENSMUSG00000020532 | Acaca | acetyl-Coenzyme A carboxylase alpha | MGI:108451 |
| ENSMUSG00000090150 | Acad11 | acyl-Coenzyme A dehydrogenase family, member 11 | MGI:2143169 |
| ENSMUSG00000042647 | Acad12 | acyl-Coenzyme A dehydrogenase family, member 12 | MGI:2443320 |
| ENSMUSG00000031969 | Acad8 | acyl-Coenzyme A dehydrogenase family, member 8 | MGI:1914198 |
| ENSMUSG00000027710 | Acad9 | acyl-Coenzyme A dehydrogenase family, member 9 | MGI:1914272 |
| ENSMUSG00000026003 | Acadl | acyl-Coenzyme A dehydrogenase, long-chain | MGI:87866 |
| ENSMUSG00000062908 | Acadm | acyl-Coenzyme A dehydrogenase, medium chain | MGI:87867 |
| ENSMUSG00000030861 | Acadsb | acyl-Coenzyme A dehydrogenase, short/branched chain | MGI:1914135 |
| ENSMUSG00000018574 | Acadvl | acyl-Coenzyme A dehydrogenase, very long chain | MGI:895149 |
| ENSMUSG00000030607 | Acan | aggrecan | MGI:99602 |
| ENSMUSG00000049076 | Acap2 | ArfGAP with coiled-coil, ankyrin repeat and PH domains 2 | MGI:1925868 |
| ENSMUSG00000029033 | Acap3 | ArfGAP with coiled-coil, ankyrin repeat and PH domains 3 | MGI:2153589 |
| ENSMUSG00000032047 | Acat1 | acetyl-Coenzyme A acetyltransferase 1 | MGI:87870 |
| ENSMUSG00000023832 | Acat2 | acetyl-Coenzyme A acetyltransferase 2 | MGI:87871 |
| ENSMUSG00000026499 | Acbd3 | acyl-Coenzyme A binding domain containing 3 | MGI:2181074 |
| ENSMUSG00000020681 | Ace | angiotensin I converting enzyme (peptidyl-dipeptidase A) 1 | MGI:87874 |
| ENSMUSG00000023328 | Ache | acetylcholinesterase | MGI:87876 |
| ENSMUSG00000020917 | Acly | ATP citrate lyase | MGI:103251 |
| ENSMUSG00000028405 | Aco1 | aconitase 1 | MGI:87879 |
| ENSMUSG00000022477 | Aco2 | aconitase 2, mitochondrial | MGI:87880 |
| ENSMUSG00000034853 | Acot11 | acyl-CoA thioesterase 11 | MGI:1913736 |
| ENSMUSG00000006717 | Acot13 | acyl-CoA thioesterase 13 | MGI:1914084 |
| ENSMUSG00000028937 | Acot7 | acyl-CoA thioesterase 7 | MGI:1917275 |
| ENSMUSG00000017307 | Acot8 | acyl-CoA thioesterase 8 | MGI:2158201 |
| ENSMUSG00000025287 | Acot9 | acyl-CoA thioesterase 9 | MGI:1928939 |
| ENSMUSG00000020777 | Acox1 | acyl-Coenzyme A oxidase 1, palmitoyl | MGI:1330812 |
| ENSMUSG00000029098 | Acox3 | acyl-Coenzyme A oxidase 3, pristanoyl | MGI:1933156 |
| ENSMUSG00000002103 | Acp2 | acid phosphatase 2, lysosomal | MGI:87882 |
| ENSMUSG00000028093 | Acp6 | acid phosphatase 6, lysophosphatidic | MGI:1931010 |
| ENSMUSG00000032281 | Acsbg1 | acyl-CoA synthetase bubblegum family member 1 | MGI:2385656 |
| ENSMUSG00000076435 | Acsf2 | acyl-CoA synthetase family member 2 | MGI:2388287 |
| ENSMUSG00000015016 | Acsf3 | acyl-CoA synthetase family member 3 | MGI:2182591 |
| ENSMUSG00000018796 | Acsl1 | acyl-CoA synthetase long-chain family member 1 | MGI:102797 |
| ENSMUSG00000032883 | Acsl3 | acyl-CoA synthetase long-chain family member 3 | MGI:1921455 |
| ENSMUSG00000031278 | Acsl4 | acyl-CoA synthetase long-chain family member 4 | MGI:1354713 |
| ENSMUSG00000024981 | Acsl5 | acyl-CoA synthetase long-chain family member 5 | MGI:1919129 |
| ENSMUSG00000020333 | Acsl6 | acyl-CoA synthetase long-chain family member 6 | MGI:894291 |
| ENSMUSG00000031972 | Acta1 | actin, alpha 1, skeletal muscle | MGI:87902 |
| ENSMUSG00000029580 | Actb | actin, beta | MGI:87904 |
| ENSMUSG00000055194 | Actbl2 | actin, beta-like 2 | MGI:2444552 |
| ENSMUSG00000062825 | Actg1 | actin, gamma, cytoplasmic 1 | MGI:87906 |
| ENSMUSG00000052374 | Actn2 | actinin alpha 2 | MGI:109192 |
| ENSMUSG00000054808 | Actn4 | actinin alpha 4 | MGI:1890773 |
| ENSMUSG00000021076 | Actr10 | ARP10 actin-related protein 10 | MGI:1891654 |
| ENSMUSG00000025228 | Actr1a | ARP1 actin-related protein 1A, centractin alpha | MGI:1858964 |
| ENSMUSG00000037351 | Actr1b | ARP1 actin-related protein 1B, centractin beta | MGI:1917446 |
| ENSMUSG00000020152 | Actr2 | ARP2 actin-related protein 2 | MGI:1913963 |
| ENSMUSG00000026341 | Actr3 | ARP3 actin-related protein 3 | MGI:1921367 |
| ENSMUSG00000056367 | Actr3b | ARP3 actin-related protein 3B | MGI:2661120 |
| ENSMUSG00000023262 | Acy1 | aminoacylase 1 | MGI:87913 |
| ENSMUSG00000008822 | Acyp1 | acylphosphatase 1, erythrocyte (common) type | MGI:1913454 |
| ENSMUSG00000060923 | Acyp2 | acylphosphatase 2, muscle type | MGI:1922822 |
| ENSMUSG00000054693 | Adam10 | a disintegrin and metallopeptidase domain 10 | MGI:109548 |
| ENSMUSG00000020926 | Adam11 | a disintegrin and metallopeptidase domain 11 | MGI:1098667 |
| ENSMUSG00000025964 | Adam23 | a disintegrin and metallopeptidase domain 23 | MGI:1345162 |
| ENSMUSG00000031555 | Adam9 | a disintegrin and metallopeptidase domain 9 (meltrin gamma) | MGI:105376 |
| ENSMUSG00000056413 | Adap1 | ArfGAP with dual PH domains 1 | MGI:2442201 |
| ENSMUSG00000021044 | Adck1 | aarF domain containing kinase 1 | MGI:1919363 |
| ENSMUSG00000020431 | Adcy1 | adenylate cyclase 1 | MGI:99677 |
| ENSMUSG00000021536 | Adcy2 | adenylate cyclase 2 | MGI:99676 |
| ENSMUSG00000020654 | Adcy3 | adenylate cyclase 3 | MGI:99675 |
| ENSMUSG00000022840 | Adcy5 | adenylate cyclase 5 | MGI:99673 |
| ENSMUSG00000022994 | Adcy6 | adenylate cyclase 6 | MGI:87917 |
| ENSMUSG00000022376 | Adcy8 | adenylate cyclase 8 | MGI:1341110 |
| ENSMUSG00000005580 | Adcy9 | adenylate cyclase 9 | MGI:108450 |
| ENSMUSG00000029778 | Adcyap1r1 | adenylate cyclase activating polypeptide 1 receptor 1 | MGI:108449 |
| ENSMUSG00000030000 | Add2 | adducin 2 (beta) | MGI:87919 |
| ENSMUSG00000025026 | Add3 | adducin 3 (gamma) | MGI:1351615 |
| ENSMUSG00000034730 | Adgrb1 | adhesion G protein-coupled receptor B1 | MGI:1933736 |
| ENSMUSG00000028782 | Adgrb2 | adhesion G protein-coupled receptor B2 | MGI:2451244 |
| ENSMUSG00000033569 | Adgrb3 | adhesion G protein-coupled receptor B3 | MGI:2441837 |
| ENSMUSG00000031785 | Adgrg1 | adhesion G protein-coupled receptor G1 | MGI:1340051 |
| ENSMUSG00000013033 | Adgrl1 | adhesion G protein-coupled receptor L1 | MGI:1929461 |
| ENSMUSG00000028184 | Adgrl2 | adhesion G protein-coupled receptor L2 | MGI:2139714 |
| ENSMUSG00000037605 | Adgrl3 | adhesion G protein-coupled receptor L3 | MGI:2441950 |
| ENSMUSG00000028138 | Adh5 | alcohol dehydrogenase 5 (class III), chi polypeptide | MGI:87929 |
| ENSMUSG00000026457 | Adipor1 | adiponectin receptor 1 | MGI:1919924 |
| ENSMUSG00000025236 | Adpgk | ADP-dependent glucokinase | MGI:1919391 |
| ENSMUSG00000033717 | Adra2a | adrenergic receptor, alpha 2a | MGI:87934 |
| ENSMUSG00000024858 | Adrbk1 | adrenergic receptor kinase, beta 1 | MGI:87940 |
| ENSMUSG00000022407 | Adsl | adenylosuccinate lyase | MGI:103202 |
| ENSMUSG00000039313 | AF529169 | cDNA sequence AF529169 | MGI:2667167 |
| ENSMUSG00000024527 | Afg3l2 | AFG3-like AAA ATPase 2 | MGI:1916847 |
| ENSMUSG00000055013 | Agap1 | ArfGAP with GTPase domain, ankyrin repeat and PH domain 1 | MGI:2653690 |
| ENSMUSG00000025422 | Agap2 | ArfGAP with GTPase domain, ankyrin repeat and PH domain 2 | MGI:3580016 |
| ENSMUSG00000023353 | Agap3 | ArfGAP with GTPase domain, ankyrin repeat and PH domain 3 | MGI:2183446 |
| ENSMUSG00000026159 | Agfg1 | ArfGAP with FG repeats 1 | MGI:1333754 |
| ENSMUSG00000029916 | Agk | acylglycerol kinase | MGI:1917173 |
| ENSMUSG00000033400 | Agl | amylo-1,6-glucosidase, 4-alpha-glucanotransferase | MGI:1924809 |
| ENSMUSG00000036698 | Ago2 | argonaute RISC catalytic subunit 2 | MGI:2446632 |
| ENSMUSG00000034254 | Agpat1 | 1-acylglycerol-3-phosphate O-acyltransferase 1 (lysophosphatidic acid acyltransferase, alpha) | MGI:1932075 |
| ENSMUSG00000001211 | Agpat3 | 1-acylglycerol-3-phosphate O-acyltransferase 3 | MGI:1336186 |
| ENSMUSG00000023827 | Agpat4 | 1-acylglycerol-3-phosphate O-acyltransferase 4 (lysophosphatidic acid acyltransferase, delta) | MGI:1915512 |
| ENSMUSG00000031467 | Agpat5 | 1-acylglycerol-3-phosphate O-acyltransferase 5 (lysophosphatidic acid acyltransferase, epsilon) | MGI:1196345 |
| ENSMUSG00000042410 | Agps | alkylglycerone phosphate synthase | MGI:2443065 |
| ENSMUSG00000041936 | Agrn | agrin | MGI:87961 |
| ENSMUSG00000027893 | Ahcyl1 | S-adenosylhomocysteine hydrolase-like 1 | MGI:2385184 |
| ENSMUSG00000029772 | Ahcyl2 | S-adenosylhomocysteine hydrolase-like 2 | MGI:1921590 |
| ENSMUSG00000019986 | Ahi1 | Abelson helper integration site 1 | MGI:87971 |
| ENSMUSG00000021037 | Ahsa1 | AHA1, activator of heat shock protein ATPase 1 | MGI:2387603 |
| ENSMUSG00000032313 | AI118078 | expressed sequence AI118078 | MGI:2142980 |
| ENSMUSG00000050812 | AI314180 | expressed sequence AI314180 | MGI:2140220 |
| ENSMUSG00000042901 | Aida | axin interactor, dorsalization associated | MGI:1919737 |
| ENSMUSG00000036932 | Aifm1 | apoptosis-inducing factor, mitochondrion-associated 1 | MGI:1349419 |
| ENSMUSG00000022763 | Aifm3 | apoptosis-inducing factor, mitochondrion-associated 3 | MGI:1919418 |
| ENSMUSG00000028029 | Aimp1 | aminoacyl tRNA synthetase complex-interacting multifunctional protein 1 | MGI:102774 |
| ENSMUSG00000029610 | Aimp2 | aminoacyl tRNA synthetase complex-interacting multifunctional protein 2 | MGI:2385237 |
| ENSMUSG00000026817 | Ak1 | adenylate kinase 1 | MGI:87977 |
| ENSMUSG00000028792 | Ak2 | adenylate kinase 2 | MGI:87978 |
| ENSMUSG00000024782 | Ak3 | adenylate kinase 3 | MGI:1860835 |
| ENSMUSG00000028527 | Ak4 | adenylate kinase 4 | MGI:87979 |
| ENSMUSG00000039058 | Ak5 | adenylate kinase 5 | MGI:2677491 |
| ENSMUSG00000047804 | Akap10 | A kinase (PRKA) anchor protein 10 | MGI:1890218 |
| ENSMUSG00000038587 | Akap12 | A kinase (PRKA) anchor protein (gravin) 12 | MGI:1932576 |
| ENSMUSG00000021057 | Akap5 | A kinase (PRKA) anchor protein 5 | MGI:2685104 |
| ENSMUSG00000039166 | Akap7 | A kinase (PRKA) anchor protein 7 | MGI:1859150 |
| ENSMUSG00000028692 | Akr1a1 | aldo-keto reductase family 1, member A1 (aldehyde reductase) | MGI:1929955 |
| ENSMUSG00000061758 | Akr1b10 | aldo-keto reductase family 1, member B10 (aldose reductase) | MGI:1915111 |
| ENSMUSG00000001642 | Akr1b3 | aldo-keto reductase family 1, member B3 (aldose reductase) | MGI:1353494 |
| ENSMUSG00000028743 | Akr7a5 | aldo-keto reductase family 7, member A5 (aflatoxin aldehyde reductase) | MGI:107796 |
| ENSMUSG00000028393 | Alad | aminolevulinate, delta-, dehydratase | MGI:96853 |
| ENSMUSG00000029368 | Alb | albumin | MGI:87991 |
| ENSMUSG00000022636 | Alcam | activated leukocyte cell adhesion molecule | MGI:1313266 |
| ENSMUSG00000025007 | Aldh18a1 | aldehyde dehydrogenase 18 family, member A1 | MGI:1888908 |
| ENSMUSG00000053279 | Aldh1a1 | aldehyde dehydrogenase family 1, subfamily A1 | MGI:1353450 |
| ENSMUSG00000035561 | Aldh1b1 | aldehyde dehydrogenase 1 family, member B1 | MGI:1919785 |
| ENSMUSG00000030088 | Aldh1l1 | aldehyde dehydrogenase 1 family, member L1 | MGI:1340024 |
| ENSMUSG00000029455 | Aldh2 | aldehyde dehydrogenase 2, mitochondrial | MGI:99600 |
| ENSMUSG00000010025 | Aldh3a2 | aldehyde dehydrogenase family 3, subfamily A2 | MGI:1353452 |
| ENSMUSG00000024885 | Aldh3b1 | aldehyde dehydrogenase 3 family, member B1 | MGI:1914939 |
| ENSMUSG00000028737 | Aldh4a1 | aldehyde dehydrogenase 4 family, member A1 | MGI:2443883 |
| ENSMUSG00000035936 | Aldh5a1 | aldhehyde dehydrogenase family 5, subfamily A1 | MGI:2441982 |
| ENSMUSG00000021238 | Aldh6a1 | aldehyde dehydrogenase family 6, subfamily A1 | MGI:1915077 |
| ENSMUSG00000053644 | Aldh7a1 | aldehyde dehydrogenase family 7, member A1 | MGI:108186 |
| ENSMUSG00000026687 | Aldh9a1 | aldehyde dehydrogenase 9, subfamily A1 | MGI:1861622 |
| ENSMUSG00000030695 | Aldoa | aldolase A, fructose-bisphosphate | MGI:87994 |
| ENSMUSG00000017390 | Aldoc | aldolase C, fructose-bisphosphate | MGI:101863 |
| ENSMUSG00000039740 | Alg2 | asparagine-linked glycosylation 2 (alpha-1,3-mannosyltransferase) | MGI:1914731 |
| ENSMUSG00000028766 | Alpl | alkaline phosphatase, liver/bone/kidney | MGI:87983 |
| ENSMUSG00000026024 | Als2 | amyotrophic lateral sclerosis 2 (juvenile) | MGI:1921268 |
| ENSMUSG00000021986 | Amer2 | APC membrane recruitment 2 | MGI:1919375 |
| ENSMUSG00000031751 | Amfr | autocrine motility factor receptor | MGI:1345634 |
| ENSMUSG00000050947 | Amigo1 | adhesion molecule with Ig like domain 1 | MGI:2653612 |
| ENSMUSG00000041688 | Amot | angiomotin | MGI:108440 |
| ENSMUSG00000027889 | Ampd2 | adenosine monophosphate deaminase 2 | MGI:88016 |
| ENSMUSG00000005686 | Ampd3 | adenosine monophosphate deaminase 3 | MGI:1096344 |
| ENSMUSG00000021314 | Amph | amphiphysin | MGI:103574 |
| ENSMUSG00000032826 | Ank2 | ankyrin 2, brain | MGI:88025 |
| ENSMUSG00000069601 | Ank3 | ankyrin 3, epithelial | MGI:88026 |
| ENSMUSG00000040351 | Ankib1 | ankyrin repeat and IBR domain containing 1 | MGI:1918047 |
| ENSMUSG00000022265 | Ank | progressive ankylosis | MGI:3045421 |
| ENSMUSG00000048307 | Ankrd46 | ankyrin repeat domain 46 | MGI:1916089 |
| ENSMUSG00000078137 | Ankrd63 | ankyrin repeat domain 63 | MGI:2686183 |
| ENSMUSG00000058589 | Anks1b | ankyrin repeat and sterile alpha motif domain containing 1B | MGI:1924781 |
| ENSMUSG00000037949 | Ano10 | anoctamin 10 | MGI:2143103 |
| ENSMUSG00000034863 | Ano8 | anoctamin 8 | MGI:2687327 |
| ENSMUSG00000039062 | Anpep | alanyl (membrane) aminopeptidase | MGI:5000466 |
| ENSMUSG00000032231 | Anxa2 | annexin A2 | MGI:88246 |
| ENSMUSG00000029994 | Anxa4 | annexin A4 | MGI:88030 |
| ENSMUSG00000027712 | Anxa5 | annexin A5 | MGI:106008 |
| ENSMUSG00000018340 | Anxa6 | annexin A6 | MGI:88255 |
| ENSMUSG00000021814 | Anxa7 | annexin A7 | MGI:88031 |
| ENSMUSG00000009090 | Ap1b1 | adaptor protein complex AP-1, beta 1 subunit | MGI:1096368 |
| ENSMUSG00000031731 | Ap1g1 | adaptor protein complex AP-1, gamma 1 subunit | MGI:101919 |
| ENSMUSG00000003033 | Ap1m1 | adaptor-related protein complex AP-1, mu subunit 1 | MGI:102776 |
| ENSMUSG00000004849 | Ap1s1 | adaptor protein complex AP-1, sigma 1 | MGI:1098244 |
| ENSMUSG00000031367 | Ap1s2 | adaptor-related protein complex 1, sigma 2 subunit | MGI:1889383 |
| ENSMUSG00000060279 | Ap2a1 | adaptor-related protein complex 2, alpha 1 subunit | MGI:101921 |
| ENSMUSG00000002957 | Ap2a2 | adaptor-related protein complex 2, alpha 2 subunit | MGI:101920 |
| ENSMUSG00000035152 | Ap2b1 | adaptor-related protein complex 2, beta 1 subunit | MGI:1919020 |
| ENSMUSG00000022841 | Ap2m1 | adaptor-related protein complex 2, mu 1 subunit | MGI:1298405 |
| ENSMUSG00000008036 | Ap2s1 | adaptor-related protein complex 2, sigma 1 subunit | MGI:2141861 |
| ENSMUSG00000062444 | Ap3b2 | adaptor-related protein complex 3, beta 2 subunit | MGI:1100869 |
| ENSMUSG00000020198 | Ap3d1 | adaptor-related protein complex 3, delta 1 subunit | MGI:107734 |
| ENSMUSG00000031539 | Ap3m2 | adaptor-related protein complex 3, mu 2 subunit | MGI:1929214 |
| ENSMUSG00000024480 | Ap3s1 | adaptor-related protein complex 3, sigma 1 subunit | MGI:1337062 |
| ENSMUSG00000005871 | Apc | adenomatosis polyposis coli | MGI:88039 |
| ENSMUSG00000032590 | Apeh | acylpeptide hydrolase | MGI:88041 |
| ENSMUSG00000035960 | Apex1 | apurinic/apyrimidinic endonuclease 1 | MGI:88042 |
| ENSMUSG00000006651 | Aplp1 | amyloid beta (A4) precursor-like protein 1 | MGI:88046 |
| ENSMUSG00000031996 | Aplp2 | amyloid beta (A4) precursor-like protein 2 | MGI:88047 |
| ENSMUSG00000033096 | Apmap | adipocyte plasma membrane associated protein | MGI:1919131 |
| ENSMUSG00000032083 | Apoa1 | apolipoprotein A-I | MGI:88049 |
| ENSMUSG00000028070 | Apoa1bp | apolipoprotein A-I binding protein | MGI:2180167 |
| ENSMUSG00000022548 | Apod | apolipoprotein D | MGI:88056 |
| ENSMUSG00000002985 | Apoe | apolipoprotein E | MGI:88057 |
| ENSMUSG00000079508 | Apoo | apolipoprotein O | MGI:1915566 |
| ENSMUSG00000025525 | Apool | apolipoprotein O-like | MGI:1915367 |
| ENSMUSG00000022892 | App | amyloid beta (A4) precursor protein | MGI:88059 |
| ENSMUSG00000040760 | Appl1 | adaptor protein, phosphotyrosine interaction, PH domain and leucine zipper containing 1 | MGI:1920243 |
| ENSMUSG00000020263 | Appl2 | adaptor protein, phosphotyrosine interaction, PH domain and leucine zipper containing 2 | MGI:2384914 |
| ENSMUSG00000004655 | Aqp1 | aquaporin 1 | MGI:103201 |
| ENSMUSG00000024411 | Aqp4 | aquaporin 4 | MGI:107387 |
| ENSMUSG00000032096 | Arcn1 | archain 1 | MGI:2387591 |
| ENSMUSG00000048076 | Arf1 | ADP-ribosylation factor 1 | MGI:99431 |
| ENSMUSG00000021877 | Arf4 | ADP-ribosylation factor 4 | MGI:99433 |
| ENSMUSG00000020440 | Arf5 | ADP-ribosylation factor 5 | MGI:99434 |
| ENSMUSG00000044147 | Arf6 | ADP-ribosylation factor 6 | MGI:99435 |
| ENSMUSG00000027575 | Arfgap1 | ADP-ribosylation factor GTPase activating protein 1 | MGI:2183559 |
| ENSMUSG00000027255 | Arfgap2 | ADP-ribosylation factor GTPase activating protein 2 | MGI:1924288 |
| ENSMUSG00000030881 | Arfip2 | ADP-ribosylation factor interacting protein 2 | MGI:1924182 |
| ENSMUSG00000027247 | Arhgap1 | Rho GTPase activating protein 1 | MGI:2445003 |
| ENSMUSG00000049807 | Arhgap23 | Rho GTPase activating protein 23 | MGI:3697726 |
| ENSMUSG00000036452 | Arhgap26 | Rho GTPase activating protein 26 | MGI:1918552 |
| ENSMUSG00000041444 | Arhgap32 | Rho GTPase activating protein 32 | MGI:2450166 |
| ENSMUSG00000036882 | Arhgap33 | Rho GTPase activating protein 33 | MGI:2673998 |
| ENSMUSG00000033697 | Arhgap39 | Rho GTPase activating protein 39 | MGI:107858 |
| ENSMUSG00000033389 | Arhgap44 | Rho GTPase activating protein 44 | MGI:2144423 |
| ENSMUSG00000035133 | Arhgap5 | Rho GTPase activating protein 5 | MGI:1332637 |
| ENSMUSG00000025132 | Arhgdia | Rho GDP dissociation inhibitor (GDI) alpha | MGI:2178103 |
| ENSMUSG00000041977 | Arhgef11 | Rho guanine nucleotide exchange factor (GEF) 11 | MGI:2441869 |
| ENSMUSG00000059495 | Arhgef12 | Rho guanine nucleotide exchange factor (GEF) 12 | MGI:1916882 |
| ENSMUSG00000028059 | Arhgef2 | rho/rac guanine nucleotide exchange factor (GEF) 2 | MGI:103264 |
| ENSMUSG00000054901 | Arhgef33 | Rho guanine nucleotide exchange factor (GEF) 33 | MGI:2685787 |
| ENSMUSG00000031511 | Arhgef7 | Rho guanine nucleotide exchange factor (GEF7) | MGI:1860493 |
| ENSMUSG00000042348 | Arl15 | ADP-ribosylation factor-like 15 | MGI:2442308 |
| ENSMUSG00000060904 | Arl1 | ADP-ribosylation factor-like 1 | MGI:99436 |
| ENSMUSG00000035199 | Arl6ip5 | ADP-ribosylation factor-like 6 interacting protein 5 | MGI:1929501 |
| ENSMUSG00000026426 | Arl8a | ADP-ribosylation factor-like 8A | MGI:1915974 |
| ENSMUSG00000030105 | Arl8b | ADP-ribosylation factor-like 8B | MGI:1914416 |
| ENSMUSG00000038525 | Armc10 | armadillo repeat containing 10 | MGI:1914461 |
| ENSMUSG00000027599 | Armc1 | armadillo repeat containing 1 | MGI:1921502 |
| ENSMUSG00000032468 | Armc8 | armadillo repeat containing 8 | MGI:1921375 |
| ENSMUSG00000033436 | Armcx2 | armadillo repeat containing, X-linked 2 | MGI:1914666 |
| ENSMUSG00000049047 | Armcx3 | armadillo repeat containing, X-linked 3 | MGI:1918953 |
| ENSMUSG00000029621 | Arpc1a | actin related protein 2/3 complex, subunit 1A | MGI:1928896 |
| ENSMUSG00000006304 | Arpc2 | actin related protein 2/3 complex, subunit 2 | MGI:1923959 |
| ENSMUSG00000029465 | Arpc3 | actin related protein 2/3 complex, subunit 3 | MGI:1928375 |
| ENSMUSG00000079426 | Arpc4 | actin related protein 2/3 complex, subunit 4 | MGI:1915339 |
| ENSMUSG00000008475 | Arpc5 | actin related protein 2/3 complex, subunit 5 | MGI:1915021 |
| ENSMUSG00000026755 | Arpc5l | actin related protein 2/3 complex, subunit 5-like | MGI:1921442 |
| ENSMUSG00000018909 | Arrb1 | arrestin, beta 1 | MGI:99473 |
| ENSMUSG00000022620 | Arsa | arylsulfatase A | MGI:88077 |
| ENSMUSG00000000325 | Arvcf | armadillo repeat gene deleted in velo-cardio-facial syndrome | MGI:109620 |
| ENSMUSG00000048040 | Arxes2 | adipocyte-related X-chromosome expressed sequence 2 | MGI:1924226 |
| ENSMUSG00000031591 | Asah1 | N-acylsphingosine amidohydrolase 1 | MGI:1277124 |
| ENSMUSG00000024887 | Asah2 | N-acylsphingosine amidohydrolase 2 | MGI:1859310 |
| ENSMUSG00000052456 | Asna1 | arsA arsenite transporter, ATP-binding, homolog 1 (bacterial) | MGI:1928379 |
| ENSMUSG00000028207 | Asph | aspartate-beta-hydroxylase | MGI:1914186 |
| ENSMUSG00000029348 | Asphd2 | aspartate beta-hydroxylase domain containing 2 | MGI:1920148 |
| ENSMUSG00000024654 | Asrgl1 | asparaginase like 1 | MGI:1913764 |
| ENSMUSG00000026587 | Astn1 | astrotactin 1 | MGI:1098567 |
| ENSMUSG00000028373 | Astn2 | astrotactin 2 | MGI:1889277 |
| ENSMUSG00000013662 | Atad1 | ATPase family, AAA domain containing 1 | MGI:1915229 |
| ENSMUSG00000029036 | Atad3a | ATPase family, AAA domain containing 3A | MGI:1919214 |
| ENSMUSG00000024426 | Atat1 | alpha tubulin acetyltransferase 1 | MGI:1913869 |
| ENSMUSG00000034958 | Atcay | ataxia, cerebellar, Cayman type homolog (human) | MGI:2448730 |
| ENSMUSG00000022663 | Atg3 | autophagy related 3 | MGI:1915091 |
| ENSMUSG00000033124 | Atg9a | autophagy related 9A | MGI:2138446 |
| ENSMUSG00000026192 | Atic | 5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase/IMP cyclohydrolase | MGI:1351352 |
| ENSMUSG00000021066 | Atl1 | atlastin GTPase 1 | MGI:1921241 |
| ENSMUSG00000059811 | Atl2 | atlastin GTPase 2 | MGI:1929492 |
| ENSMUSG00000024759 | Atl3 | atlastin GTPase 3 | MGI:1924270 |
| ENSMUSG00000062949 | Atp11c | ATPase, class VI, type 11C | MGI:1859661 |
| ENSMUSG00000031862 | Atp13a1 | ATPase type 13A1 | MGI:2180801 |
| ENSMUSG00000036622 | Atp13a2 | ATPase type 13A2 | MGI:1922022 |
| ENSMUSG00000038094 | Atp13a4 | ATPase type 13A4 | MGI:1924456 |
| ENSMUSG00000033161 | Atp1a1 | ATPase, Na+/K+ transporting, alpha 1 polypeptide | MGI:88105 |
| ENSMUSG00000007097 | Atp1a2 | ATPase, Na+/K+ transporting, alpha 2 polypeptide | MGI:88106 |
| ENSMUSG00000040907 | Atp1a3 | ATPase, Na+/K+ transporting, alpha 3 polypeptide | MGI:88107 |
| ENSMUSG00000007107 | Atp1a4 | ATPase, Na+/K+ transporting, alpha 4 polypeptide | MGI:1351335 |
| ENSMUSG00000026576 | Atp1b1 | ATPase, Na+/K+ transporting, beta 1 polypeptide | MGI:88108 |
| ENSMUSG00000041329 | Atp1b2 | ATPase, Na+/K+ transporting, beta 2 polypeptide | MGI:88109 |
| ENSMUSG00000032412 | Atp1b3 | ATPase, Na+/K+ transporting, beta 3 polypeptide | MGI:107788 |
| ENSMUSG00000029467 | Atp2a2 | ATPase, Ca++ transporting, cardiac muscle, slow twitch 2 | MGI:88110 |
| ENSMUSG00000020788 | Atp2a3 | ATPase, Ca++ transporting, ubiquitous | MGI:1194503 |
| ENSMUSG00000019943 | Atp2b1 | ATPase, Ca++ transporting, plasma membrane 1 | MGI:104653 |
| ENSMUSG00000030302 | Atp2b2 | ATPase, Ca++ transporting, plasma membrane 2 | MGI:105368 |
| ENSMUSG00000031376 | Atp2b3 | ATPase, Ca++ transporting, plasma membrane 3 | MGI:1347353 |
| ENSMUSG00000026463 | Atp2b4 | ATPase, Ca++ transporting, plasma membrane 4 | MGI:88111 |
| ENSMUSG00000032570 | Atp2c1 | ATPase, Ca++-sequestering | MGI:1889008 |
| ENSMUSG00000025428 | Atp5a1 | ATP synthase, H+ transporting, mitochondrial F1 complex, alpha subunit 1 | MGI:88115 |
| ENSMUSG00000025393 | Atp5b | ATP synthase, H+ transporting mitochondrial F1 complex, beta subunit | MGI:107801 |
| ENSMUSG00000025781 | Atp5c1 | ATP synthase, H+ transporting, mitochondrial F1 complex, gamma polypeptide 1 | MGI:1261437 |
| ENSMUSG00000003072 | Atp5d | ATP synthase, H+ transporting, mitochondrial F1 complex, delta subunit | MGI:1913293 |
| ENSMUSG00000016252 | Atp5e | ATP synthase, H+ transporting, mitochondrial F1 complex, epsilon subunit | MGI:1855697 |
| ENSMUSG00000000563 | Atp5f1 | ATP synthase, H+ transporting, mitochondrial F0 complex, subunit B1 | MGI:1100495 |
| ENSMUSG00000034566 | Atp5h | ATP synthase, H+ transporting, mitochondrial F0 complex, subunit D | MGI:1918929 |
| ENSMUSG00000038690 | Atp5j2 | ATP synthase, H+ transporting, mitochondrial F0 complex, subunit F2 | MGI:1927558 |
| ENSMUSG00000022890 | Atp5j | ATP synthase, H+ transporting, mitochondrial F0 complex, subunit F | MGI:107777 |
| ENSMUSG00000050856 | Atp5k | ATP synthase, H+ transporting, mitochondrial F1F0 complex, subunit E | MGI:106636 |
| ENSMUSG00000038717 | Atp5l | ATP synthase, H+ transporting, mitochondrial F0 complex, subunit G | MGI:1351597 |
| ENSMUSG00000022956 | Atp5o | ATP synthase, H+ transporting, mitochondrial F1 complex, O subunit | MGI:106341 |
| ENSMUSG00000019087 | Atp6ap1 | ATPase, H+ transporting, lysosomal accessory protein 1 | MGI:109629 |
| ENSMUSG00000031007 | Atp6ap2 | ATPase, H+ transporting, lysosomal accessory protein 2 | MGI:1917745 |
| ENSMUSG00000019302 | Atp6v0a1 | ATPase, H+ transporting, lysosomal V0 subunit A1 | MGI:103286 |
| ENSMUSG00000024121 | Atp6v0c | ATPase, H+ transporting, lysosomal V0 subunit C | MGI:88116 |
| ENSMUSG00000013160 | Atp6v0d1 | ATPase, H+ transporting, lysosomal V0 subunit D1 | MGI:1201778 |
| ENSMUSG00000052459 | Atp6v1a | ATPase, H+ transporting, lysosomal V1 subunit A | MGI:1201780 |
| ENSMUSG00000006273 | Atp6v1b2 | ATPase, H+ transporting, lysosomal V1 subunit B2 | MGI:109618 |
| ENSMUSG00000022295 | Atp6v1c1 | ATPase, H+ transporting, lysosomal V1 subunit C1 | MGI:1913585 |
| ENSMUSG00000021114 | Atp6v1d | ATPase, H+ transporting, lysosomal V1 subunit D | MGI:1921084 |
| ENSMUSG00000019210 | Atp6v1e1 | ATPase, H+ transporting, lysosomal V1 subunit E1 | MGI:894326 |
| ENSMUSG00000004285 | Atp6v1f | ATPase, H+ transporting, lysosomal V1 subunit F | MGI:1913394 |
| ENSMUSG00000039105 | Atp6v1g1 | ATPase, H+ transporting, lysosomal V1 subunit G1 | MGI:1913540 |
| ENSMUSG00000024403 | Atp6v1g2 | ATPase, H+ transporting, lysosomal V1 subunit G2 | MGI:1913487 |
| ENSMUSG00000033793 | Atp6v1h | ATPase, H+ transporting, lysosomal V1 subunit H | MGI:1914864 |
| ENSMUSG00000037685 | Atp8a1 | ATPase, aminophospholipid transporter (APLT), class I, type 8A, member 1 | MGI:1330848 |
| ENSMUSG00000021983 | Atp8a2 | ATPase, aminophospholipid transporter-like, class I, type 8A, member 2 | MGI:1354710 |
| ENSMUSG00000027546 | Atp9a | ATPase, class II, type 9A | MGI:1330826 |
| ENSMUSG00000024566 | Atp9b | ATPase, class II, type 9B | MGI:1354757 |
| ENSMUSG00000027312 | Atrn | attractin | MGI:1341628 |
| ENSMUSG00000016541 | Atxn10 | ataxin 10 | MGI:1859293 |
| ENSMUSG00000028830 | AU040320 | expressed sequence AU040320 | MGI:2140475 |
| ENSMUSG00000021460 | Auh | AU RNA binding protein/enoyl-coenzyme A hydratase | MGI:1338011 |
| ENSMUSG00000071649 | B3gat3 | beta-1,3-glucuronyltransferase 3 (glucuronosyltransferase I) | MGI:1919977 |
| ENSMUSG00000006731 | B4galnt1 | beta-1,4-N-acetyl-galactosaminyl transferase 1 | MGI:1342057 |
| ENSMUSG00000047379 | B4gat1 | beta-1,4-glucuronyltransferase 1 | MGI:1919680 |
| ENSMUSG00000052364 | B630019K06Rik | RIKEN cDNA B630019K06 gene | MGI:2147918 |
| ENSMUSG00000042215 | Bag2 | BCL2-associated athanogene 2 | MGI:1891254 |
| ENSMUSG00000049792 | Bag5 | BCL2-associated athanogene 5 | MGI:1917619 |
| ENSMUSG00000024392 | Bag6 | BCL2-associated athanogene 6 | MGI:1919439 |
| ENSMUSG00000025372 | Baiap2 | brain-specific angiogenesis inhibitor 1-associated protein 2 | MGI:2137336 |
| ENSMUSG00000024844 | Banf1 | barrier to autointegration factor 1 | MGI:1346330 |
| ENSMUSG00000045763 | Basp1 | brain abundant, membrane attached signal protein 1 | MGI:1917600 |
| ENSMUSG00000006010 | BC003331 | cDNA sequence BC003331 | MGI:2385108 |
| ENSMUSG00000067722 | BC003965 | cDNA sequence BC003965 | MGI:2670966 |
| ENSMUSG00000033488 | BC026585 | cDNA sequence BC026585 | MGI:2448516 |
| ENSMUSG00000036948 | BC037034 | cDNA sequence BC037034 | MGI:2385896 |
| ENSMUSG00000002980 | Bcam | basal cell adhesion molecule | MGI:1929940 |
| ENSMUSG00000004892 | Bcan | brevican | MGI:1096385 |
| ENSMUSG00000020650 | Bcap29 | B cell receptor associated protein 29 | MGI:101917 |
| ENSMUSG00000002015 | Bcap31 | B cell receptor associated protein 31 | MGI:1350933 |
| ENSMUSG00000059439 | Bcas3 | breast carcinoma amplified sequence 3 | MGI:2385848 |
| ENSMUSG00000030268 | Bcat1 | branched chain aminotransferase 1, cytosolic | MGI:104861 |
| ENSMUSG00000030802 | Bckdk | branched chain ketoacid dehydrogenase kinase | MGI:1276121 |
| ENSMUSG00000009112 | Bcl2l13 | BCL2-like 13 (apoptosis facilitator) | MGI:2136959 |
| ENSMUSG00000007659 | Bcl2l1 | BCL2-like 1 | MGI:88139 |
| ENSMUSG00000009681 | Bcr | breakpoint cluster region | MGI:88141 |
| ENSMUSG00000026172 | Bcs1l | BCS1-like (yeast) | MGI:1914071 |
| ENSMUSG00000046598 | Bdh1 | 3-hydroxybutyrate dehydrogenase, type 1 | MGI:1919161 |
| ENSMUSG00000102226 | Begain | brain-enriched guanylate kinase-associated | MGI:3044626 |
| ENSMUSG00000032757 | Bet1 | blocked early in transport 1 homolog (S. cerevisiae) | MGI:1343104 |
| ENSMUSG00000003452 | Bicd1 | bicaudal D homolog 1 (Drosophila) | MGI:1101760 |
| ENSMUSG00000024381 | Bin1 | bridging integrator 1 | MGI:108092 |
| ENSMUSG00000098112 | Bin2 | bridging integrator 2 | MGI:3611448 |
| ENSMUSG00000020840 | Blmh | bleomycin hydrolase | MGI:1345186 |
| ENSMUSG00000040466 | Blvrb | biliverdin reductase B (flavin reductase (NADPH)) | MGI:2385271 |
| ENSMUSG00000021796 | Bmpr1a | bone morphogenetic protein receptor, type 1A | MGI:1338938 |
| ENSMUSG00000067336 | Bmpr2 | bone morphogenetic protein receptor, type II (serine/threonine kinase) | MGI:1095407 |
| ENSMUSG00000024191 | Bnip1 | BCL2/adenovirus E1B interacting protein 1 | MGI:109328 |
| ENSMUSG00000015943 | Bola1 | bolA-like 1 (E. coli) | MGI:1916418 |
| ENSMUSG00000038286 | Bphl | biphenyl hydrolase-like (serine hydrolase, breast epithelial mucin-associated antigen) | MGI:1915271 |
| ENSMUSG00000026617 | Bpnt1 | bisphosphate 3'-nucleotidase 1 | MGI:1338800 |
| ENSMUSG00000037905 | Bri3bp | Bri3 binding protein | MGI:1924059 |
| ENSMUSG00000028351 | Brinp1 | bone morphogenic protein/retinoic acid inducible neural specific 1 | MGI:1928478 |
| ENSMUSG00000004031 | Brinp2 | bone morphogenic protein/retinoic acid inducible neural-specific 2 | MGI:2443333 |
| ENSMUSG00000033940 | Brk1 | BRICK1, SCAR/WAVE actin-nucleating complex subunit | MGI:1915406 |
| ENSMUSG00000035390 | Brsk1 | BR serine/threonine kinase 1 | MGI:2685946 |
| ENSMUSG00000053046 | Brsk2 | BR serine/threonine kinase 2 | MGI:1923020 |
| ENSMUSG00000023175 | Bsg | basigin | MGI:88208 |
| ENSMUSG00000032589 | Bsn | bassoon | MGI:1277955 |
| ENSMUSG00000020042 | Btbd11 | BTB (POZ) domain containing 11 | MGI:1921257 |
| ENSMUSG00000000202 | Btbd17 | BTB (POZ) domain containing 17 | MGI:1919264 |
| ENSMUSG00000036905 | C1qb | complement component 1, q subcomponent, beta polypeptide | MGI:88224 |
| ENSMUSG00000018446 | C1qbp | complement component 1, q subcomponent binding protein | MGI:1194505 |
| ENSMUSG00000040794 | C1qtnf4 | C1q and tumor necrosis factor related protein 4 | MGI:1914695 |
| ENSMUSG00000032120 | C2cd2l | C2 calcium-dependent domain containing 2-like | MGI:1919014 |
| ENSMUSG00000045912 | C2cd4c | C2 calcium-dependent domain containing 4C | MGI:2685084 |
| ENSMUSG00000036707 | Cab39 | calcium binding protein 39 | MGI:107438 |
| ENSMUSG00000015488 | Cacfd1 | calcium channel flower domain containing 1 | MGI:1924317 |
| ENSMUSG00000034656 | Cacna1a | calcium channel, voltage-dependent, P/Q type, alpha 1A subunit | MGI:109482 |
| ENSMUSG00000004113 | Cacna1b | calcium channel, voltage-dependent, N type, alpha 1B subunit | MGI:88296 |
| ENSMUSG00000051331 | Cacna1c | calcium channel, voltage-dependent, L type, alpha 1C subunit | MGI:103013 |
| ENSMUSG00000004110 | Cacna1e | calcium channel, voltage-dependent, R type, alpha 1E subunit | MGI:106217 |
| ENSMUSG00000022416 | Cacna1i | calcium channel, voltage-dependent, alpha 1I subunit | MGI:2178051 |
| ENSMUSG00000040118 | Cacna2d1 | calcium channel, voltage-dependent, alpha2/delta subunit 1 | MGI:88295 |
| ENSMUSG00000010066 | Cacna2d2 | calcium channel, voltage-dependent, alpha 2/delta subunit 2 | MGI:1929813 |
| ENSMUSG00000021991 | Cacna2d3 | calcium channel, voltage-dependent, alpha2/delta subunit 3 | MGI:1338890 |
| ENSMUSG00000020882 | Cacnb1 | calcium channel, voltage-dependent, beta 1 subunit | MGI:102522 |
| ENSMUSG00000003352 | Cacnb3 | calcium channel, voltage-dependent, beta 3 subunit | MGI:103307 |
| ENSMUSG00000017412 | Cacnb4 | calcium channel, voltage-dependent, beta 4 subunit | MGI:103301 |
| ENSMUSG00000019146 | Cacng2 | calcium channel, voltage-dependent, gamma subunit 2 | MGI:1316660 |
| ENSMUSG00000066189 | Cacng3 | calcium channel, voltage-dependent, gamma subunit 3 | MGI:1859165 |
| ENSMUSG00000053395 | Cacng8 | calcium channel, voltage-dependent, gamma subunit 8 | MGI:1932376 |
| ENSMUSG00000014226 | Cacybp | calcyclin binding protein | MGI:1270839 |
| ENSMUSG00000013629 | Cad | carbamoyl-phosphate synthetase 2, aspartate transcarbamylase, and dihydroorotase | MGI:1916969 |
| ENSMUSG00000032076 | Cadm1 | cell adhesion molecule 1 | MGI:1889272 |
| ENSMUSG00000064115 | Cadm2 | cell adhesion molecule 2 | MGI:2442722 |
| ENSMUSG00000005338 | Cadm3 | cell adhesion molecule 3 | MGI:2137858 |
| ENSMUSG00000054793 | Cadm4 | cell adhesion molecule 4 | MGI:2449088 |
| ENSMUSG00000054423 | Cadps | Ca2+-dependent secretion activator | MGI:1350922 |
| ENSMUSG00000028222 | Calb1 | calbindin 1 | MGI:88248 |
| ENSMUSG00000003657 | Calb2 | calbindin 2 | MGI:101914 |
| ENSMUSG00000023055 | Calcoco1 | calcium binding and coiled coil domain 1 | MGI:1914738 |
| ENSMUSG00000019370 | Calm3 | calmodulin 3 | MGI:103249 |
| ENSMUSG00000003814 | Calr | calreticulin | MGI:88252 |
| ENSMUSG00000024617 | Camk2a | calcium/calmodulin-dependent protein kinase II alpha | MGI:88256 |
| ENSMUSG00000021820 | Camk2g | calcium/calmodulin-dependent protein kinase II gamma | MGI:88259 |
| ENSMUSG00000032936 | Camkv | CaM kinase-like vesicle-associated | MGI:2384296 |
| ENSMUSG00000021501 | Caml | calcium modulating ligand | MGI:104728 |
| ENSMUSG00000041570 | Camsap2 | calmodulin regulated spectrin-associated protein family, member 2 | MGI:1922434 |
| ENSMUSG00000044433 | Camsap3 | calmodulin regulated spectrin-associated protein family, member 3 | MGI:1916947 |
| ENSMUSG00000020114 | Cand1 | cullin associated and neddylation disassociated 1 | MGI:1261820 |
| ENSMUSG00000020368 | Canx | calnexin | MGI:88261 |
| ENSMUSG00000028656 | Cap1 | CAP, adenylate cyclase-associated protein 1 (yeast) | MGI:88262 |
| ENSMUSG00000021373 | Cap2 | CAP, adenylate cyclase-associated protein, 2 (yeast) | MGI:1914502 |
| ENSMUSG00000035547 | Capn5 | calpain 5 | MGI:1100859 |
| ENSMUSG00000001794 | Capns1 | calpain, small subunit 1 | MGI:88266 |
| ENSMUSG00000027184 | Caprin1 | cell cycle associated protein 1 | MGI:1858234 |
| ENSMUSG00000070372 | Capza1 | capping protein (actin filament) muscle Z-line, alpha 1 | MGI:106227 |
| ENSMUSG00000015733 | Capza2 | capping protein (actin filament) muscle Z-line, alpha 2 | MGI:106222 |
| ENSMUSG00000028745 | Capzb | capping protein (actin filament) muscle Z-line, beta | MGI:104652 |
| ENSMUSG00000056158 | Car10 | carbonic anhydrase 10 | MGI:1919855 |
| ENSMUSG00000038526 | Car14 | carbonic anhydrase 14 | MGI:1344341 |
| ENSMUSG00000027562 | Car2 | carbonic anhydrase 2 | MGI:88269 |
| ENSMUSG00000000805 | Car4 | carbonic anhydrase 4 | MGI:1096574 |
| ENSMUSG00000031505 | Carkd | carbohydrate kinase domain containing | MGI:1913353 |
| ENSMUSG00000060227 | Casc4 | cancer susceptibility candidate 4 | MGI:2443129 |
| ENSMUSG00000033597 | Caskin1 | CASK interacting protein 1 | MGI:2442952 |
| ENSMUSG00000027187 | Cat | catalase | MGI:88271 |
| ENSMUSG00000038498 | Catsper1 | cation channel, sperm associated 1 | MGI:2179947 |
| ENSMUSG00000031654 | Cbln1 | cerebellin 1 precursor protein | MGI:88281 |
| ENSMUSG00000051483 | Cbr1 | carbonyl reductase 1 | MGI:88284 |
| ENSMUSG00000031641 | Cbr4 | carbonyl reductase 4 | MGI:2384567 |
| ENSMUSG00000036686 | Cc2d1a | coiled-coil and C2 domain containing 1A | MGI:2384831 |
| ENSMUSG00000021578 | Ccdc127 | coiled-coil domain containing 127 | MGI:1914683 |
| ENSMUSG00000001376 | Ccdc132 | coiled-coil domain containing 132 | MGI:1920538 |
| ENSMUSG00000029769 | Ccdc136 | coiled-coil domain containing 136 | MGI:1918128 |
| ENSMUSG00000062961 | Ccdc177 | coiled-coil domain containing 177 | MGI:2686414 |
| ENSMUSG00000031143 | Ccdc22 | coiled-coil domain containing 22 | MGI:1859608 |
| ENSMUSG00000059554 | Ccdc28a | coiled-coil domain containing 28A | MGI:2443508 |
| ENSMUSG00000078622 | Ccdc47 | coiled-coil domain containing 47 | MGI:1914413 |
| ENSMUSG00000025645 | Ccdc51 | coiled-coil domain containing 51 | MGI:1913908 |
| ENSMUSG00000075229 | Ccdc58 | coiled-coil domain containing 58 | MGI:2146423 |
| ENSMUSG00000032740 | Ccdc88a | coiled coil domain containing 88A | MGI:1925177 |
| ENSMUSG00000030613 | Ccdc90b | coiled-coil domain containing 90B | MGI:1913615 |
| ENSMUSG00000024286 | Ccny | cyclin Y | MGI:1915224 |
| ENSMUSG00000070871 | Ccnyl1 | cyclin Y-like 1 | MGI:2138614 |
| ENSMUSG00000034024 | Cct2 | chaperonin containing Tcp1, subunit 2 (beta) | MGI:107186 |
| ENSMUSG00000001416 | Cct3 | chaperonin containing Tcp1, subunit 3 (gamma) | MGI:104708 |
| ENSMUSG00000007739 | Cct4 | chaperonin containing Tcp1, subunit 4 (delta) | MGI:104689 |
| ENSMUSG00000022234 | Cct5 | chaperonin containing Tcp1, subunit 5 (epsilon) | MGI:107185 |
| ENSMUSG00000029447 | Cct6a | chaperonin containing Tcp1, subunit 6a (zeta) | MGI:107943 |
| ENSMUSG00000030007 | Cct7 | chaperonin containing Tcp1, subunit 7 (eta) | MGI:107184 |
| ENSMUSG00000025613 | Cct8 | chaperonin containing Tcp1, subunit 8 (theta) | MGI:107183 |
| ENSMUSG00000029617 | Ccz1 | CCZ1 vacuolar protein trafficking and biogenesis associated | MGI:2141070 |
| ENSMUSG00000022661 | Cd200 | CD200 antigen | MGI:1196990 |
| ENSMUSG00000016494 | Cd34 | CD34 antigen | MGI:88329 |
| ENSMUSG00000002944 | Cd36 | CD36 antigen | MGI:107899 |
| ENSMUSG00000029084 | Cd38 | CD38 antigen | MGI:107474 |
| ENSMUSG00000055447 | Cd47 | CD47 antigen (Rh-related antigen, integrin-associated signal transducer) | MGI:96617 |
| ENSMUSG00000037706 | Cd81 | CD81 antigen | MGI:1096398 |
| ENSMUSG00000027215 | Cd82 | CD82 antigen | MGI:104651 |
| ENSMUSG00000035776 | Cd99l2 | CD99 antigen-like 2 | MGI:2177151 |
| ENSMUSG00000030342 | Cd9 | CD9 antigen | MGI:88348 |
| ENSMUSG00000019471 | Cdc37 | cell division cycle 37 | MGI:109531 |
| ENSMUSG00000026490 | Cdc42bpa | CDC42 binding protein kinase alpha | MGI:2441841 |
| ENSMUSG00000021279 | Cdc42bpb | CDC42 binding protein kinase beta | MGI:2136459 |
| ENSMUSG00000006699 | Cdc42 | cell division cycle 42 | MGI:106211 |
| ENSMUSG00000049521 | Cdc42ep1 | CDC42 effector protein (Rho GTPase binding) 1 | MGI:1929763 |
| ENSMUSG00000041598 | Cdc42ep4 | CDC42 effector protein (Rho GTPase binding) 4 | MGI:1929760 |
| ENSMUSG00000052298 | Cdc42se2 | CDC42 small effector 2 | MGI:1919979 |
| ENSMUSG00000022321 | Cdh10 | cadherin 10 | MGI:107436 |
| ENSMUSG00000031673 | Cdh11 | cadherin 11 | MGI:99217 |
| ENSMUSG00000031841 | Cdh13 | cadherin 13 | MGI:99551 |
| ENSMUSG00000040420 | Cdh18 | cadherin 18 | MGI:1344366 |
| ENSMUSG00000024304 | Cdh2 | cadherin 2 | MGI:88355 |
| ENSMUSG00000039385 | Cdh6 | cadherin 6 | MGI:107435 |
| ENSMUSG00000004071 | Cdip1 | cell death inducing Trp53 target 1 | MGI:1913876 |
| ENSMUSG00000030682 | Cdipt | CDP-diacylglycerol--inositol 3-phosphatidyltransferase (phosphatidylinositol synthase) | MGI:105491 |
| ENSMUSG00000020015 | Cdk17 | cyclin-dependent kinase 17 | MGI:97517 |
| ENSMUSG00000026437 | Cdk18 | cyclin-dependent kinase 18 | MGI:97518 |
| ENSMUSG00000028969 | Cdk5 | cyclin-dependent kinase 5 | MGI:101765 |
| ENSMUSG00000090071 | Cdk5r2 | cyclin-dependent kinase 5, regulatory subunit 2 (p39) | MGI:1330828 |
| ENSMUSG00000018669 | Cdk5rap3 | CDK5 regulatory subunit associated protein 3 | MGI:1933126 |
| ENSMUSG00000031292 | Cdkl5 | cyclin-dependent kinase-like 5 | MGI:1278336 |
| ENSMUSG00000029330 | Cds1 | CDP-diacylglycerol synthase 1 | MGI:1921846 |
| ENSMUSG00000058793 | Cds2 | CDP-diacylglycerol synthase (phosphatidate cytidylyltransferase) 2 | MGI:1332236 |
| ENSMUSG00000032803 | Cdv3 | carnitine deficiency-associated gene expressed in ventricle 3 | MGI:2448759 |
| ENSMUSG00000094626 | Cecr6 | cat eye syndrome chromosome region, candidate 6 | MGI:2136977 |
| ENSMUSG00000002107 | Celf2 | CUGBP, Elav-like family member 2 | MGI:1338822 |
| ENSMUSG00000068740 | Celsr2 | cadherin, EGF LAG seven-pass G-type receptor 2 (flamingo homolog, Drosophila) | MGI:1858235 |
| ENSMUSG00000060240 | Cend1 | cell cycle exit and neuronal differentiation 1 | MGI:1929898 |
| ENSMUSG00000026605 | Cenpf | centromere protein F | MGI:1313302 |
| ENSMUSG00000072825 | Cep170b | centrosomal protein 170B | MGI:2145043 |
| ENSMUSG00000057335 | Cep170 | centrosomal protein 170 | MGI:1918348 |
| ENSMUSG00000022604 | Cep97 | centrosomal protein 97 | MGI:1921451 |
| ENSMUSG00000087408 | Cers1 | ceramide synthase 1 | MGI:2136690 |
| ENSMUSG00000015714 | Cers2 | ceramide synthase 2 | MGI:1924143 |
| ENSMUSG00000031347 | Cetn2 | centrin 2 | MGI:1347085 |
| ENSMUSG00000056201 | Cfl1 | cofilin 1, non-muscle | MGI:101757 |
| ENSMUSG00000062929 | Cfl2 | cofilin 2, muscle | MGI:101763 |
| ENSMUSG00000053768 | Chchd3 | coiled-coil-helix-coiled-coil-helix domain containing 3 | MGI:1913325 |
| ENSMUSG00000030086 | Chchd6 | coiled-coil-helix-coiled-coil-helix domain containing 6 | MGI:1913348 |
| ENSMUSG00000025512 | Chid1 | chitinase domain containing 1 | MGI:1915288 |
| ENSMUSG00000030077 | Chl1 | cell adhesion molecule with homology to L1CAM | MGI:1098266 |
| ENSMUSG00000000743 | Chmp1a | charged multivesicular body protein 1A | MGI:1920159 |
| ENSMUSG00000033916 | Chmp2a | charged multivesicular body protein 2A | MGI:1916203 |
| ENSMUSG00000004843 | Chmp2b | charged multivesicular body protein 2B | MGI:1916192 |
| ENSMUSG00000053119 | Chmp3 | charged multivesicular body protein 3 | MGI:1913950 |
| ENSMUSG00000038467 | Chmp4b | charged multivesicular body protein 4B | MGI:1922858 |
| ENSMUSG00000025371 | Chmp6 | charged multivesicular body protein 6 | MGI:3583942 |
| ENSMUSG00000014077 | Chp1 | calcineurin-like EF hand protein 1 | MGI:1927185 |
| ENSMUSG00000060002 | Chpt1 | choline phosphotransferase 1 | MGI:2384841 |
| ENSMUSG00000032773 | Chrm1 | cholinergic receptor, muscarinic 1, CNS | MGI:88396 |
| ENSMUSG00000046159 | Chrm3 | cholinergic receptor, muscarinic 3, cardiac | MGI:88398 |
| ENSMUSG00000040495 | Chrm4 | cholinergic receptor, muscarinic 4 | MGI:88399 |
| ENSMUSG00000045193 | Cirbp | cold inducible RNA binding protein | MGI:893588 |
| ENSMUSG00000037710 | Cisd1 | CDGSH iron sulfur domain 1 | MGI:1261855 |
| ENSMUSG00000028165 | Cisd2 | CDGSH iron sulfur domain 2 | MGI:1914256 |
| ENSMUSG00000029516 | Cit | citron | MGI:105313 |
| ENSMUSG00000046841 | Ckap4 | cytoskeleton-associated protein 4 | MGI:2444926 |
| ENSMUSG00000040549 | Ckap5 | cytoskeleton associated protein 5 | MGI:1923036 |
| ENSMUSG00000001270 | Ckb | creatine kinase, brain | MGI:88407 |
| ENSMUSG00000000308 | Ckmt1 | creatine kinase, mitochondrial 1, ubiquitous | MGI:99441 |
| ENSMUSG00000064302 | Clasp1 | CLIP associating protein 1 | MGI:1923957 |
| ENSMUSG00000033392 | Clasp2 | CLIP associating protein 2 | MGI:1923749 |
| ENSMUSG00000027884 | Clcc1 | chloride channel CLIC-like 1 | MGI:2385186 |
| ENSMUSG00000022843 | Clcn2 | chloride channel 2 | MGI:105061 |
| ENSMUSG00000004319 | Clcn3 | chloride channel 3 | MGI:103555 |
| ENSMUSG00000000605 | Clcn4-2 | chloride channel 4-2 | MGI:104571 |
| ENSMUSG00000029016 | Clcn6 | chloride channel 6 | MGI:1347049 |
| ENSMUSG00000036636 | Clcn7 | chloride channel 7 | MGI:1347048 |
| ENSMUSG00000022132 | Cldn10 | claudin 10 | MGI:1913101 |
| ENSMUSG00000037625 | Cldn11 | claudin 11 | MGI:106925 |
| ENSMUSG00000046798 | Cldn12 | claudin 12 | MGI:1929288 |
| ENSMUSG00000079598 | Clec2l | C-type lectin domain family 2, member L | MGI:2141402 |
| ENSMUSG00000037242 | Clic4 | chloride intracellular channel 4 (mitochondrial) | MGI:1352754 |
| ENSMUSG00000006169 | Clint1 | clathrin interactor 1 | MGI:2144243 |
| ENSMUSG00000049550 | Clip1 | CAP-GLY domain containing linker protein 1 | MGI:1928401 |
| ENSMUSG00000063146 | Clip2 | CAP-GLY domain containing linker protein 2 | MGI:1313136 |
| ENSMUSG00000013921 | Clip3 | CAP-GLY domain containing linker protein 3 | MGI:1923936 |
| ENSMUSG00000021097 | Clmn | calmin | MGI:2136957 |
| ENSMUSG00000032024 | Clmp | CXADR-like membrane protein | MGI:1918816 |
| ENSMUSG00000001829 | Clpb | ClpB caseinolytic peptidase B | MGI:1100517 |
| ENSMUSG00000002660 | Clpp | caseinolytic mitochondrial matrix peptidase proteolytic subunit | MGI:1858213 |
| ENSMUSG00000002981 | Clptm1 | cleft lip and palate associated transmembrane protein 1 | MGI:1927155 |
| ENSMUSG00000021610 | Clptm1l | CLPTM1-like | MGI:2442892 |
| ENSMUSG00000039953 | Clstn1 | calsyntenin 1 | MGI:1929895 |
| ENSMUSG00000008153 | Clstn3 | calsyntenin 3 | MGI:2178323 |
| ENSMUSG00000028478 | Clta | clathrin, light polypeptide (Lca) | MGI:894297 |
| ENSMUSG00000047547 | Cltb | clathrin, light polypeptide (Lcb) | MGI:1921575 |
| ENSMUSG00000047126 | Cltc | clathrin, heavy polypeptide (Hc) | MGI:2388633 |
| ENSMUSG00000022037 | Clu | clusterin | MGI:88423 |
| ENSMUSG00000025545 | Clybl | citrate lyase beta like | MGI:1916884 |
| ENSMUSG00000030282 | Cmas | cytidine monophospho-N-acetylneuraminic acid synthetase | MGI:1337124 |
| ENSMUSG00000039163 | Cmc1 | COX assembly mitochondrial protein 1 | MGI:1915149 |
| ENSMUSG00000034390 | Cmip | c-Maf inducing protein | MGI:1921690 |
| ENSMUSG00000028719 | Cmpk1 | cytidine monophosphate (UMP-CMP) kinase 1 | MGI:1913838 |
| ENSMUSG00000096188 | Cmtm4 | CKLF-like MARVEL transmembrane domain containing 4 | MGI:2142888 |
| ENSMUSG00000040759 | Cmtm5 | CKLF-like MARVEL transmembrane domain containing 5 | MGI:2447164 |
| ENSMUSG00000024644 | Cndp2 | CNDP dipeptidase 2 (metallopeptidase M20 family) | MGI:1913304 |
| ENSMUSG00000025658 | Cnksr2 | connector enhancer of kinase suppressor of Ras 2 | MGI:2661175 |
| ENSMUSG00000053931 | Cnn3 | calponin 3, acidic | MGI:1919244 |
| ENSMUSG00000025189 | Cnnm1 | cyclin M1 | MGI:1891366 |
| ENSMUSG00000064105 | Cnnm2 | cyclin M2 | MGI:2151054 |
| ENSMUSG00000001138 | Cnnm3 | cyclin M3 | MGI:2151055 |
| ENSMUSG00000003135 | Cnot11 | CCR4-NOT transcription complex, subunit 11 | MGI:106580 |
| ENSMUSG00000006782 | Cnp | 2',3'-cyclic nucleotide 3' phosphodiesterase | MGI:88437 |
| ENSMUSG00000025381 | Cnpy2 | canopy 2 homolog (zebrafish) | MGI:1928477 |
| ENSMUSG00000023973 | Cnpy3 | canopy 3 homolog (zebrafish) | MGI:1919279 |
| ENSMUSG00000044288 | Cnr1 | cannabinoid receptor 1 (brain) | MGI:104615 |
| ENSMUSG00000044629 | Cnrip1 | cannabinoid receptor interacting protein 1 | MGI:1917505 |
| ENSMUSG00000038949 | Cnst | consortin, connexin sorting protein | MGI:2445141 |
| ENSMUSG00000028444 | Cntfr | ciliary neurotrophic factor receptor | MGI:99605 |
| ENSMUSG00000055022 | Cntn1 | contactin 1 | MGI:105980 |
| ENSMUSG00000053024 | Cntn2 | contactin 2 | MGI:104518 |
| ENSMUSG00000030075 | Cntn3 | contactin 3 | MGI:99534 |
| ENSMUSG00000064293 | Cntn4 | contactin 4 | MGI:1095737 |
| ENSMUSG00000039488 | Cntn5 | contactin 5 | MGI:3042287 |
| ENSMUSG00000030092 | Cntn6 | contactin 6 | MGI:1858223 |
| ENSMUSG00000017167 | Cntnap1 | contactin associated protein-like 1 | MGI:1858201 |
| ENSMUSG00000039419 | Cntnap2 | contactin associated protein-like 2 | MGI:1914047 |
| ENSMUSG00000031772 | Cntnap4 | contactin associated protein-like 4 | MGI:2183572 |
| ENSMUSG00000070695 | Cntnap5a | contactin associated protein-like 5A | MGI:3643623 |
| ENSMUSG00000017188 | Coa3 | cytochrome C oxidase assembly factor 3 | MGI:1098757 |
| ENSMUSG00000048351 | Coa7 | cytochrome c oxidase assembly factor 7 | MGI:1917143 |
| ENSMUSG00000001755 | Coasy | Coenzyme A synthase | MGI:1918993 |
| ENSMUSG00000042705 | Commd10 | COMM domain containing 10 | MGI:1916706 |
| ENSMUSG00000051355 | Commd1 | COMM domain containing 1 | MGI:109474 |
| ENSMUSG00000036513 | Commd2 | COMM domain containing 2 | MGI:1098806 |
| ENSMUSG00000051154 | Commd3 | COMM domain containing 3 | MGI:88218 |
| ENSMUSG00000027163 | Commd9 | COMM domain containing 9 | MGI:1923751 |
| ENSMUSG00000000326 | Comt | catechol-O-methyltransferase | MGI:88470 |
| ENSMUSG00000021773 | Comtd1 | catechol-O-methyltransferase domain containing 1 | MGI:1916406 |
| ENSMUSG00000026553 | Copa | coatomer protein complex subunit alpha | MGI:1334462 |
| ENSMUSG00000030754 | Copb1 | coatomer protein complex, subunit beta 1 | MGI:1917599 |
| ENSMUSG00000032458 | Copb2 | coatomer protein complex, subunit beta 2 (beta prime) | MGI:1354962 |
| ENSMUSG00000030058 | Copg1 | coatomer protein complex, subunit gamma 1 | MGI:1858696 |
| ENSMUSG00000027206 | Cops2 | COP9 (constitutive photomorphogenic) homolog, subunit 2 (Arabidopsis thaliana) | MGI:1330276 |
| ENSMUSG00000019373 | Cops3 | COP9 (constitutive photomorphogenic) homolog, subunit 3 (Arabidopsis thaliana) | MGI:1349409 |
| ENSMUSG00000035297 | Cops4 | COP9 (constitutive photomorphogenic) homolog, subunit 4 (Arabidopsis thaliana) | MGI:1349414 |
| ENSMUSG00000019494 | Cops6 | COP9 (constitutive photomorphogenic) homolog, subunit 6 (Arabidopsis thaliana) | MGI:1349439 |
| ENSMUSG00000030127 | Cops7a | COP9 (constitutive photomorphogenic) homolog, subunit 7a (Arabidopsis thaliana) | MGI:1349400 |
| ENSMUSG00000034432 | Cops8 | COP9 (constitutive photomorphogenic) homolog, subunit 8 (Arabidopsis thaliana) | MGI:1915363 |
| ENSMUSG00000060992 | Copz1 | coatomer protein complex, subunit zeta 1 | MGI:1929063 |
| ENSMUSG00000028247 | Coq3 | coenzyme Q3 homolog, methyltransferase (yeast) | MGI:101813 |
| ENSMUSG00000030652 | Coq7 | demethyl-Q 7 | MGI:107207 |
| ENSMUSG00000031782 | Coq9 | coenzyme Q9 homolog (yeast) | MGI:1915164 |
| ENSMUSG00000030707 | Coro1a | coronin, actin binding protein 1A | MGI:1345961 |
| ENSMUSG00000024835 | Coro1b | coronin, actin binding protein 1B | MGI:1345963 |
| ENSMUSG00000004530 | Coro1c | coronin, actin binding protein 1C | MGI:1345964 |
| ENSMUSG00000028337 | Coro2a | coronin, actin binding protein 2A | MGI:1345966 |
| ENSMUSG00000041729 | Coro2b | coronin, actin binding protein, 2B | MGI:2444283 |
| ENSMUSG00000031827 | Cotl1 | coactosin-like 1 (Dictyostelium) | MGI:1919292 |
| ENSMUSG00000031818 | Cox4i1 | cytochrome c oxidase subunit IV isoform 1 | MGI:88473 |
| ENSMUSG00000000088 | Cox5a | cytochrome c oxidase subunit Va | MGI:88474 |
| ENSMUSG00000061518 | Cox5b | cytochrome c oxidase subunit Vb | MGI:88475 |
| ENSMUSG00000041697 | Cox6a1 | cytochrome c oxidase subunit VIa polypeptide 1 | MGI:103099 |
| ENSMUSG00000036751 | Cox6b1 | cytochrome c oxidase, subunit VIb polypeptide 1 | MGI:107460 |
| ENSMUSG00000014313 | Cox6c | cytochrome c oxidase subunit VIc | MGI:104614 |
| ENSMUSG00000032330 | Cox7a2 | cytochrome c oxidase subunit VIIa 2 | MGI:1316715 |
| ENSMUSG00000024248 | Cox7a2l | cytochrome c oxidase subunit VIIa polypeptide 2-like | MGI:106015 |
| ENSMUSG00000017778 | Cox7c | cytochrome c oxidase subunit VIIc | MGI:103226 |
| ENSMUSG00000020841 | Cpd | carboxypeptidase D | MGI:107265 |
| ENSMUSG00000020300 | Cpeb4 | cytoplasmic polyadenylation element binding protein 4 | MGI:1914829 |
| ENSMUSG00000037852 | Cpe | carboxypeptidase E | MGI:101932 |
| ENSMUSG00000033615 | Cplx1 | complexin 1 | MGI:104727 |
| ENSMUSG00000020183 | Cpm | carboxypeptidase M | MGI:1917824 |
| ENSMUSG00000024008 | Cpne5 | copine V | MGI:2385908 |
| ENSMUSG00000022212 | Cpne6 | copine VI | MGI:1334445 |
| ENSMUSG00000055531 | Cpsf6 | cleavage and polyadenylation specific factor 6 | MGI:1913948 |
| ENSMUSG00000024900 | Cpt1a | carnitine palmitoyltransferase 1a, liver | MGI:1098296 |
| ENSMUSG00000007783 | Cpt1c | carnitine palmitoyltransferase 1c | MGI:2446526 |
| ENSMUSG00000028607 | Cpt2 | carnitine palmitoyltransferase 2 | MGI:109176 |
| ENSMUSG00000016481 | Cr1l | complement component (3b/4b) receptor 1-like | MGI:88513 |
| ENSMUSG00000026853 | Crat | carnitine acetyltransferase | MGI:109501 |
| ENSMUSG00000030284 | Creld1 | cysteine-rich with EGF-like domains 1 | MGI:2152539 |
| ENSMUSG00000006356 | Crip2 | cysteine rich protein 2 | MGI:1915587 |
| ENSMUSG00000017776 | Crk | v-crk sarcoma virus CT10 oncogene homolog (avian) | MGI:88508 |
| ENSMUSG00000029121 | Crmp1 | collapsin response mediator protein 1 | MGI:107793 |
| ENSMUSG00000040860 | Crocc | ciliary rootlet coiled-coil, rootletin | MGI:3529431 |
| ENSMUSG00000003623 | Crot | carnitine O-octanoyltransferase | MGI:1921364 |
| ENSMUSG00000042401 | Crtac1 | cartilage acidic protein 1 | MGI:1920082 |
| ENSMUSG00000032060 | Cryab | crystallin, alpha B | MGI:88516 |
| ENSMUSG00000030905 | Crym | crystallin, mu | MGI:102675 |
| ENSMUSG00000005683 | Cs | citrate synthase | MGI:88529 |
| ENSMUSG00000068823 | Csde1 | cold shock domain containing E1, RNA binding | MGI:92356 |
| ENSMUSG00000002718 | Cse1l | chromosome segregation 1-like (S. cerevisiae) | MGI:1339951 |
| ENSMUSG00000060924 | Csmd1 | CUB and Sushi multiple domains 1 | MGI:2137383 |
| ENSMUSG00000028804 | Csmd2 | CUB and Sushi multiple domains 2 | MGI:2386401 |
| ENSMUSG00000024576 | Csnk1a1 | casein kinase 1, alpha 1 | MGI:1934950 |
| ENSMUSG00000025162 | Csnk1d | casein kinase 1, delta | MGI:1355272 |
| ENSMUSG00000022433 | Csnk1e | casein kinase 1, epsilon | MGI:1351660 |
| ENSMUSG00000032384 | Csnk1g1 | casein kinase 1, gamma 1 | MGI:2660884 |
| ENSMUSG00000003345 | Csnk1g2 | casein kinase 1, gamma 2 | MGI:1920014 |
| ENSMUSG00000073563 | Csnk1g3 | casein kinase 1, gamma 3 | MGI:1917675 |
| ENSMUSG00000074698 | Csnk2a1 | casein kinase 2, alpha 1 polypeptide | MGI:88543 |
| ENSMUSG00000046707 | Csnk2a2 | casein kinase 2, alpha prime polypeptide | MGI:88547 |
| ENSMUSG00000024387 | Csnk2b | casein kinase 2, beta polypeptide | MGI:88548 |
| ENSMUSG00000032911 | Cspg4 | chondroitin sulfate proteoglycan 4 | MGI:2153093 |
| ENSMUSG00000032482 | Cspg5 | chondroitin sulfate proteoglycan 5 | MGI:1352747 |
| ENSMUSG00000026421 | Csrp1 | cysteine and glycine-rich protein 1 | MGI:88549 |
| ENSMUSG00000027447 | Cst3 | cystatin C | MGI:102519 |
| ENSMUSG00000021000 | Ctage5 | CTAGE family, member 5 | MGI:1346056 |
| ENSMUSG00000037373 | Ctbp1 | C-terminal binding protein 1 | MGI:1201685 |
| ENSMUSG00000026176 | Ctdsp1 | CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 1 | MGI:2654470 |
| ENSMUSG00000037815 | Ctnna1 | catenin (cadherin associated protein), alpha 1 | MGI:88274 |
| ENSMUSG00000063063 | Ctnna2 | catenin (cadherin associated protein), alpha 2 | MGI:88275 |
| ENSMUSG00000006932 | Ctnnb1 | catenin (cadherin associated protein), beta 1 | MGI:88276 |
| ENSMUSG00000022240 | Ctnnd2 | catenin (cadherin associated protein), delta 2 | MGI:1195966 |
| ENSMUSG00000021939 | Ctsb | cathepsin B | MGI:88561 |
| ENSMUSG00000007891 | Ctsd | cathepsin D | MGI:88562 |
| ENSMUSG00000021477 | Ctsl | cathepsin L | MGI:88564 |
| ENSMUSG00000000416 | Cttnbp2 | cortactin binding protein 2 | MGI:1353467 |
| ENSMUSG00000031078 | Cttn | cortactin | MGI:99695 |
| ENSMUSG00000029686 | Cul1 | cullin 1 | MGI:1349658 |
| ENSMUSG00000004364 | Cul3 | cullin 3 | MGI:1347360 |
| ENSMUSG00000031446 | Cul4a | cullin 4A | MGI:1914487 |
| ENSMUSG00000031095 | Cul4b | cullin 4B | MGI:1919834 |
| ENSMUSG00000032030 | Cul5 | cullin 5 | MGI:1922967 |
| ENSMUSG00000031778 | Cx3cl1 | chemokine (C-X3-C motif) ligand 1 | MGI:1097153 |
| ENSMUSG00000022865 | Cxadr | coxsackie virus and adenovirus receptor | MGI:1201679 |
| ENSMUSG00000024646 | Cyb5a | cytochrome b5 type A (microsomal) | MGI:1926952 |
| ENSMUSG00000031924 | Cyb5b | cytochrome b5 type B | MGI:1913677 |
| ENSMUSG00000026456 | Cyb5r1 | cytochrome b5 reductase 1 | MGI:1919267 |
| ENSMUSG00000018042 | Cyb5r3 | cytochrome b5 reductase 3 | MGI:94893 |
| ENSMUSG00000027015 | Cybrd1 | cytochrome b reductase 1 | MGI:2654575 |
| ENSMUSG00000022551 | Cyc1 | cytochrome c-1 | MGI:1913695 |
| ENSMUSG00000030447 | Cyfip1 | cytoplasmic FMR1 interacting protein 1 | MGI:1338801 |
| ENSMUSG00000020340 | Cyfip2 | cytoplasmic FMR1 interacting protein 2 | MGI:1924134 |
| ENSMUSG00000061740 | Cyp2d22 | cytochrome P450, family 2, subfamily d, polypeptide 22 | MGI:1929474 |
| ENSMUSG00000015224 | Cyp2j9 | cytochrome P450, family 2, subfamily j, polypeptide 9 | MGI:1921769 |
| ENSMUSG00000021259 | Cyp46a1 | cytochrome P450, family 46, subfamily a, polypeptide 1 | MGI:1341877 |
| ENSMUSG00000001467 | Cyp51 | cytochrome P450, family 51 | MGI:106040 |
| ENSMUSG00000003269 | Cyth2 | cytohesin 2 | MGI:1334255 |
| ENSMUSG00000053329 | D10Jhu81e | DNA segment, Chr 10, Johns Hopkins University 81 expressed | MGI:1351861 |
| ENSMUSG00000068373 | D430041D05Rik | RIKEN cDNA D430041D05 gene | MGI:2181743 |
| ENSMUSG00000063455 | D630045J12Rik | RIKEN cDNA D630045J12 gene | MGI:2669829 |
| ENSMUSG00000034574 | Daam1 | dishevelled associated activator of morphogenesis 1 | MGI:1914596 |
| ENSMUSG00000040260 | Daam2 | dishevelled associated activator of morphogenesis 2 | MGI:1923691 |
| ENSMUSG00000028519 | Dab1 | disabled 1 | MGI:108554 |
| ENSMUSG00000026883 | Dab2ip | disabled 2 interacting protein | MGI:1916851 |
| ENSMUSG00000022174 | Dad1 | defender against cell death 1 | MGI:101912 |
| ENSMUSG00000039952 | Dag1 | dystroglycan 1 | MGI:101864 |
| ENSMUSG00000035735 | Dagla | diacylglycerol lipase, alpha | MGI:2677061 |
| ENSMUSG00000042096 | Dao | D-amino acid oxidase | MGI:94859 |
| ENSMUSG00000026356 | Dars | aspartyl-tRNA synthetase | MGI:2442544 |
| ENSMUSG00000026385 | Dbi | diazepam binding inhibitor | MGI:94865 |
| ENSMUSG00000034675 | Dbn1 | drebrin 1 | MGI:1931838 |
| ENSMUSG00000020476 | Dbnl | drebrin-like | MGI:700006 |
| ENSMUSG00000000340 | Dbt | dihydrolipoamide branched chain transacylase E2 | MGI:105386 |
| ENSMUSG00000020935 | Dcakd | dephospho-CoA kinase domain containing | MGI:1915337 |
| ENSMUSG00000060534 | Dcc | deleted in colorectal carcinoma | MGI:94869 |
| ENSMUSG00000028078 | Dclk2 | doublecortin-like kinase 2 | MGI:1918012 |
| ENSMUSG00000032040 | Dcps | decapping enzyme, scavenger | MGI:1916555 |
| ENSMUSG00000031865 | Dctn1 | dynactin 1 | MGI:107745 |
| ENSMUSG00000025410 | Dctn2 | dynactin 2 | MGI:107733 |
| ENSMUSG00000028447 | Dctn3 | dynactin 3 | MGI:1859251 |
| ENSMUSG00000024603 | Dctn4 | dynactin 4 | MGI:1914915 |
| ENSMUSG00000030868 | Dctn5 | dynactin 5 | MGI:1891689 |
| ENSMUSG00000048787 | Dcun1d3 | DCN1, defective in cullin neddylation 1, domain containing 3 (S. cerevisiae) | MGI:2679003 |
| ENSMUSG00000028194 | Ddah1 | dimethylarginine dimethylaminohydrolase 1 | MGI:1916469 |
| ENSMUSG00000007039 | Ddah2 | dimethylarginine dimethylaminohydrolase 2 | MGI:1859016 |
| ENSMUSG00000024740 | Ddb1 | damage specific DNA binding protein 1 | MGI:1202384 |
| ENSMUSG00000028757 | Ddost | dolichyl-di-phosphooligosaccharide-protein glycotransferase | MGI:1194508 |
| ENSMUSG00000068290 | Ddrgk1 | DDRGK domain containing 1 | MGI:1924256 |
| ENSMUSG00000001666 | Ddt | D-dopachrome tautomerase | MGI:1298381 |
| ENSMUSG00000055065 | Ddx17 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 17 | MGI:1914290 |
| ENSMUSG00000037149 | Ddx1 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 1 | MGI:2144727 |
| ENSMUSG00000000787 | Ddx3x | DEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide 3, X-linked | MGI:103064 |
| ENSMUSG00000069045 | Ddx3y | DEAD (Asp-Glu-Ala-Asp) box polypeptide 3, Y-linked | MGI:1349406 |
| ENSMUSG00000020719 | Ddx5 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 5 | MGI:105037 |
| ENSMUSG00000032097 | Ddx6 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 6 | MGI:104976 |
| ENSMUSG00000028223 | Decr1 | 2,4-dienoyl CoA reductase 1, mitochondrial | MGI:1914710 |
| ENSMUSG00000038633 | Degs1 | degenerative spermatocyte homolog 1 (Drosophila) | MGI:1097711 |
| ENSMUSG00000035392 | Dennd1a | DENN/MADD domain containing 1A | MGI:2442794 |
| ENSMUSG00000042404 | Dennd4b | DENN/MADD domain containing 4B | MGI:2446201 |
| ENSMUSG00000022365 | Derl1 | Der1-like domain family, member 1 | MGI:1915069 |
| ENSMUSG00000036095 | Dgkb | diacylglycerol kinase, beta | MGI:2442474 |
| ENSMUSG00000000276 | Dgke | diacylglycerol kinase, epsilon | MGI:1889276 |
| ENSMUSG00000022861 | Dgkg | diacylglycerol kinase, gamma | MGI:105060 |
| ENSMUSG00000040479 | Dgkz | diacylglycerol kinase zeta | MGI:1278339 |
| ENSMUSG00000031730 | Dhodh | dihydroorotate dehydrogenase | MGI:1928378 |
| ENSMUSG00000020834 | Dhrs13 | dehydrogenase/reductase (SDR family) member 13 | MGI:1917701 |
| ENSMUSG00000002332 | Dhrs1 | dehydrogenase/reductase (SDR family) member 1 | MGI:1196314 |
| ENSMUSG00000022210 | Dhrs4 | dehydrogenase/reductase (SDR family) member 4 | MGI:90169 |
| ENSMUSG00000042569 | Dhrs7b | dehydrogenase/reductase (SDR family) member 7B | MGI:2384931 |
| ENSMUSG00000042426 | Dhx29 | DEAH (Asp-Glu-Ala-His) box polypeptide 29 | MGI:2145374 |
| ENSMUSG00000032480 | Dhx30 | DEAH (Asp-Glu-Ala-His) box polypeptide 30 | MGI:1920081 |
| ENSMUSG00000029433 | Diablo | diablo homolog (Drosophila) | MGI:1913843 |
| ENSMUSG00000023026 | Dip2b | DIP2 disco-interacting protein 2 homolog B (Drosophila) | MGI:2145977 |
| ENSMUSG00000043670 | Diras1 | DIRAS family, GTP-binding RAS-like 1 | MGI:2183442 |
| ENSMUSG00000047842 | Diras2 | DIRAS family, GTP-binding RAS-like 2 | MGI:1915453 |
| ENSMUSG00000022848 | Dirc2 | disrupted in renal carcinoma 2 (human) | MGI:2387188 |
| ENSMUSG00000040035 | Disp2 | dispatched homolog 2 (Drosophila) | MGI:2388733 |
| ENSMUSG00000000168 | Dlat | dihydrolipoamide S-acetyltransferase (E2 component of pyruvate dehydrogenase complex) | MGI:2385311 |
| ENSMUSG00000020664 | Dld | dihydrolipoamide dehydrogenase | MGI:107450 |
| ENSMUSG00000022770 | Dlg1 | discs, large homolog 1 (Drosophila) | MGI:107231 |
| ENSMUSG00000052572 | Dlg2 | discs, large homolog 2 (Drosophila) | MGI:1344351 |
| ENSMUSG00000020886 | Dlg4 | discs, large homolog 4 (Drosophila) | MGI:1277959 |
| ENSMUSG00000003279 | Dlgap1 | discs, large (Drosophila) homolog-associated protein 1 | MGI:1346065 |
| ENSMUSG00000047495 | Dlgap2 | discs, large (Drosophila) homolog-associated protein 2 | MGI:2443181 |
| ENSMUSG00000042388 | Dlgap3 | discs, large (Drosophila) homolog-associated protein 3 | MGI:3039563 |
| ENSMUSG00000061689 | Dlgap4 | discs, large homolog-associated protein 4 (Drosophila) | MGI:2138865 |
| ENSMUSG00000004789 | Dlst | dihydrolipoamide S-succinyltransferase (E2 component of 2-oxo-glutarate complex) | MGI:1926170 |
| ENSMUSG00000045103 | Dmd | dystrophin, muscular dystrophy | MGI:94909 |
| ENSMUSG00000022099 | Dmtn | dematin actin binding protein | MGI:99670 |
| ENSMUSG00000030410 | Dmwd | dystrophia myotonica-containing WD repeat motif | MGI:94907 |
| ENSMUSG00000033987 | Dnah17 | dynein, axonemal, heavy chain 17 | MGI:1917176 |
| ENSMUSG00000028410 | Dnaja1 | DnaJ (Hsp40) homolog, subfamily A, member 1 | MGI:1270129 |
| ENSMUSG00000031701 | Dnaja2 | DnaJ (Hsp40) homolog, subfamily A, member 2 | MGI:1931882 |
| ENSMUSG00000004069 | Dnaja3 | DnaJ (Hsp40) homolog, subfamily A, member 3 | MGI:1933786 |
| ENSMUSG00000032285 | Dnaja4 | DnaJ (Hsp40) homolog, subfamily A, member 4 | MGI:1927638 |
| ENSMUSG00000004460 | Dnajb11 | DnaJ (Hsp40) homolog, subfamily B, member 11 | MGI:1915088 |
| ENSMUSG00000020109 | Dnajb12 | DnaJ (Hsp40) homolog, subfamily B, member 12 | MGI:1931881 |
| ENSMUSG00000074212 | Dnajb14 | DnaJ (Hsp40) homolog, subfamily B, member 14 | MGI:1917854 |
| ENSMUSG00000005483 | Dnajb1 | DnaJ (Hsp40) homolog, subfamily B, member 1 | MGI:1931874 |
| ENSMUSG00000026203 | Dnajb2 | DnaJ (Hsp40) homolog, subfamily B, member 2 | MGI:1928739 |
| ENSMUSG00000029131 | Dnajb6 | DnaJ (Hsp40) homolog, subfamily B, member 6 | MGI:1344381 |
| ENSMUSG00000027006 | Dnajc10 | DnaJ (Hsp40) homolog, subfamily C, member 10 | MGI:1914111 |
| ENSMUSG00000039768 | Dnajc11 | DnaJ (Hsp40) homolog, subfamily C, member 11 | MGI:2443386 |
| ENSMUSG00000032560 | Dnajc13 | DnaJ (Hsp40) homolog, subfamily C, member 13 | MGI:2676368 |
| ENSMUSG00000040697 | Dnajc16 | DnaJ (Hsp40) homolog, subfamily C, member 16 | MGI:2442146 |
| ENSMUSG00000027679 | Dnajc19 | DnaJ (Hsp40) homolog, subfamily C, member 19 | MGI:1914963 |
| ENSMUSG00000022136 | Dnajc3 | DnaJ (Hsp40) homolog, subfamily C, member 3 | MGI:107373 |
| ENSMUSG00000000826 | Dnajc5 | DnaJ (Hsp40) homolog, subfamily C, member 5 | MGI:892995 |
| ENSMUSG00000028528 | Dnajc6 | DnaJ (Hsp40) homolog, subfamily C, member 6 | MGI:1919935 |
| ENSMUSG00000036766 | Dner | delta/notch-like EGF-related receptor | MGI:2152889 |
| ENSMUSG00000026825 | Dnm1 | dynamin 1 | MGI:107384 |
| ENSMUSG00000022789 | Dnm1l | dynamin 1-like | MGI:1921256 |
| ENSMUSG00000033335 | Dnm2 | dynamin 2 | MGI:109547 |
| ENSMUSG00000040265 | Dnm3 | dynamin 3 | MGI:1341299 |
| ENSMUSG00000026209 | Dnpep | aspartyl aminopeptidase | MGI:1278328 |
| ENSMUSG00000058325 | Dock1 | dedicator of cytokinesis 1 | MGI:2429765 |
| ENSMUSG00000039716 | Dock3 | dedicator of cyto-kinesis 3 | MGI:2429763 |
| ENSMUSG00000035954 | Dock4 | dedicator of cytokinesis 4 | MGI:1918006 |
| ENSMUSG00000028556 | Dock7 | dedicator of cytokinesis 7 | MGI:1914549 |
| ENSMUSG00000025558 | Dock9 | dedicator of cytokinesis 9 | MGI:106321 |
| ENSMUSG00000035640 | Dos | downstream of Stk11 | MGI:1354170 |
| ENSMUSG00000041035 | Dpcd | deleted in primary ciliary dyskinesia | MGI:1924407 |
| ENSMUSG00000078919 | Dpm1 | dolichol-phosphate (beta-D) mannosyltransferase 1 | MGI:1330239 |
| ENSMUSG00000063904 | Dpp3 | dipeptidylpeptidase 3 | MGI:1922471 |
| ENSMUSG00000061576 | Dpp6 | dipeptidylpeptidase 6 | MGI:94921 |
| ENSMUSG00000022048 | Dpysl2 | dihydropyrimidinase-like 2 | MGI:1349763 |
| ENSMUSG00000024501 | Dpysl3 | dihydropyrimidinase-like 3 | MGI:1349762 |
| ENSMUSG00000025478 | Dpysl4 | dihydropyrimidinase-like 4 | MGI:1349764 |
| ENSMUSG00000029168 | Dpysl5 | dihydropyrimidinase-like 5 | MGI:1929772 |
| ENSMUSG00000020537 | Drg2 | developmentally regulated GTP binding protein 2 | MGI:1342307 |
| ENSMUSG00000103888 | Dst | dystonin | MGI:104627 |
| ENSMUSG00000015932 | Dstn | destrin | MGI:1929270 |
| ENSMUSG00000027430 | Dtd1 | D-tyrosyl-tRNA deacylase 1 | MGI:1913294 |
| ENSMUSG00000024302 | Dtna | dystrobrevin alpha | MGI:106039 |
| ENSMUSG00000042662 | Dusp15 | dual specificity phosphatase-like 15 | MGI:1934928 |
| ENSMUSG00000003518 | Dusp3 | dual specificity phosphatase 3 (vaccinia virus phosphatase VH1-related) | MGI:1919599 |
| ENSMUSG00000021791 | Dydc2 | DPY30 domain containing 2 | MGI:1918450 |
| ENSMUSG00000018707 | Dync1h1 | dynein cytoplasmic 1 heavy chain 1 | MGI:103147 |
| ENSMUSG00000029757 | Dync1i1 | dynein cytoplasmic 1 intermediate chain 1 | MGI:107743 |
| ENSMUSG00000027012 | Dync1i2 | dynein cytoplasmic 1 intermediate chain 2 | MGI:107750 |
| ENSMUSG00000032435 | Dync1li1 | dynein cytoplasmic 1 light intermediate chain 1 | MGI:2135610 |
| ENSMUSG00000035770 | Dync1li2 | dynein, cytoplasmic 1 light intermediate chain 2 | MGI:107738 |
| ENSMUSG00000009013 | Dynll1 | dynein light chain LC8-type 1 | MGI:1861457 |
| ENSMUSG00000020483 | Dynll2 | dynein light chain LC8-type 2 | MGI:1915347 |
| ENSMUSG00000047459 | Dynlrb1 | dynein light chain roadblock-type 1 | MGI:1914318 |
| ENSMUSG00000037259 | Dzank1 | double zinc ribbon and ankyrin repeat domains 1 | MGI:2139080 |
| ENSMUSG00000037172 | E330009J07Rik | RIKEN cDNA E330009J07 gene | MGI:2444256 |
| ENSMUSG00000022350 | E430025E21Rik | RIKEN cDNA E430025E21 gene | MGI:2146110 |
| ENSMUSG00000031168 | Ebp | phenylalkylamine Ca2+ antagonist (emopamil) binding protein | MGI:107822 |
| ENSMUSG00000026247 | Ecel1 | endothelin converting enzyme-like 1 | MGI:1343461 |
| ENSMUSG00000053898 | Ech1 | enoyl coenzyme A hydratase 1, peroxisomal | MGI:1858208 |
| ENSMUSG00000025465 | Echs1 | enoyl Coenzyme A hydratase, short chain, 1, mitochondrial | MGI:2136460 |
| ENSMUSG00000024132 | Eci1 | enoyl-Coenzyme A delta isomerase 1 | MGI:94871 |
| ENSMUSG00000021417 | Eci2 | enoyl-Coenzyme A delta isomerase 2 | MGI:1346064 |
| ENSMUSG00000066839 | Ecsit | ECSIT homolog (Drosophila) | MGI:1349469 |
| ENSMUSG00000015092 | Edf1 | endothelial differentiation-related factor 1 | MGI:1891227 |
| ENSMUSG00000034488 | Edil3 | EGF-like repeats and discoidin I-like domains 3 | MGI:1329025 |
| ENSMUSG00000036499 | Eea1 | early endosome antigen 1 | MGI:2442192 |
| ENSMUSG00000037742 | Eef1a1 | eukaryotic translation elongation factor 1 alpha 1 | MGI:1096881 |
| ENSMUSG00000016349 | Eef1a2 | eukaryotic translation elongation factor 1 alpha 2 | MGI:1096317 |
| ENSMUSG00000025967 | Eef1b2 | eukaryotic translation elongation factor 1 beta 2 | MGI:1929520 |
| ENSMUSG00000055762 | Eef1d | eukaryotic translation elongation factor 1 delta (guanine nucleotide exchange protein) | MGI:1913906 |
| ENSMUSG00000001707 | Eef1e1 | eukaryotic translation elongation factor 1 epsilon 1 | MGI:1913393 |
| ENSMUSG00000071644 | Eef1g | eukaryotic translation elongation factor 1 gamma | MGI:1914410 |
| ENSMUSG00000034994 | Eef2 | eukaryotic translation elongation factor 2 | MGI:95288 |
| ENSMUSG00000036611 | Eepd1 | endonuclease/exonuclease/phosphatase family domain containing 1 | MGI:1914734 |
| ENSMUSG00000040659 | Efhd2 | EF hand domain containing 2 | MGI:106504 |
| ENSMUSG00000028039 | Efna3 | ephrin A3 | MGI:106644 |
| ENSMUSG00000031217 | Efnb1 | ephrin B1 | MGI:102708 |
| ENSMUSG00000003934 | Efnb3 | ephrin B3 | MGI:109196 |
| ENSMUSG00000015002 | Efr3a | EFR3 homolog A (S. cerevisiae) | MGI:1923990 |
| ENSMUSG00000020658 | Efr3b | EFR3 homolog B (S. cerevisiae) | MGI:2444851 |
| ENSMUSG00000020122 | Egfr | epidermal growth factor receptor | MGI:95294 |
| ENSMUSG00000042302 | Ehbp1 | EH domain binding protein 1 | MGI:2667252 |
| ENSMUSG00000024772 | Ehd1 | EH-domain containing 1 | MGI:1341878 |
| ENSMUSG00000024065 | Ehd3 | EH-domain containing 3 | MGI:1928900 |
| ENSMUSG00000027293 | Ehd4 | EH-domain containing 4 | MGI:1919619 |
| ENSMUSG00000062762 | Ei24 | etoposide induced 2.4 mRNA | MGI:108090 |
| ENSMUSG00000067194 | Eif1ax | eukaryotic translation initiation factor 1A, X-linked | MGI:1913485 |
| ENSMUSG00000021116 | Eif2s1 | eukaryotic translation initiation factor 2, subunit 1 alpha | MGI:95299 |
| ENSMUSG00000035150 | Eif2s3x | eukaryotic translation initiation factor 2, subunit 3, structural gene X-linked | MGI:1349431 |
| ENSMUSG00000024991 | Eif3a | eukaryotic translation initiation factor 3, subunit A | MGI:95301 |
| ENSMUSG00000033047 | Eif3l | eukaryotic translation initiation factor 3, subunit L | MGI:2386251 |
| ENSMUSG00000022884 | Eif4a2 | eukaryotic translation initiation factor 4A2 | MGI:106906 |
| ENSMUSG00000025580 | Eif4a3 | eukaryotic translation initiation factor 4A3 | MGI:1923731 |
| ENSMUSG00000045983 | Eif4g1 | eukaryotic translation initiation factor 4, gamma 1 | MGI:2384784 |
| ENSMUSG00000028760 | Eif4g3 | eukaryotic translation initiation factor 4 gamma, 3 | MGI:1923935 |
| ENSMUSG00000040731 | Eif4h | eukaryotic translation initiation factor 4H | MGI:1341822 |
| ENSMUSG00000078812 | Eif5a | eukaryotic translation initiation factor 5A | MGI:106248 |
| ENSMUSG00000021282 | Eif5 | eukaryotic translation initiation factor 5 | MGI:95309 |
| ENSMUSG00000040028 | Elavl1 | ELAV (embryonic lethal, abnormal vision)-like 1 (Hu antigen R) | MGI:1100851 |
| ENSMUSG00000008489 | Elavl2 | ELAV (embryonic lethal, abnormal vision, Drosophila)-like 2 (Hu antigen B) | MGI:1100887 |
| ENSMUSG00000028546 | Elavl4 | ELAV (embryonic lethal, abnormal vision, Drosophila)-like 4 (Hu antigen D) | MGI:107427 |
| ENSMUSG00000048988 | Elfn1 | leucine rich repeat and fibronectin type III, extracellular 1 | MGI:2442479 |
| ENSMUSG00000043460 | Elfn2 | leucine rich repeat and fibronectin type III, extracellular 2 | MGI:3608416 |
| ENSMUSG00000041112 | Elmo1 | engulfment and cell motility 1 | MGI:2153044 |
| ENSMUSG00000017670 | Elmo2 | engulfment and cell motility 2 | MGI:2153045 |
| ENSMUSG00000041986 | Elmod1 | ELMO/CED-12 domain containing 1 | MGI:3583900 |
| ENSMUSG00000006390 | Elovl1 | elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 1 | MGI:1858959 |
| ENSMUSG00000032349 | Elovl5 | ELOVL family member 5, elongation of long chain fatty acids (yeast) | MGI:1916051 |
| ENSMUSG00000021728 | Emb | embigin | MGI:95321 |
| ENSMUSG00000008140 | Emc10 | ER membrane protein complex subunit 10 | MGI:1916933 |
| ENSMUSG00000078517 | Emc1 | ER membrane protein complex subunit 1 | MGI:2443696 |
| ENSMUSG00000022337 | Emc2 | ER membrane protein complex subunit 2 | MGI:1913986 |
| ENSMUSG00000030286 | Emc3 | ER membrane protein complex subunit 3 | MGI:1913337 |
| ENSMUSG00000027131 | Emc4 | ER membrane protein complex subunit 4 | MGI:1915282 |
| ENSMUSG00000047260 | Emc6 | ER membrane protein complex subunit 6 | MGI:1913298 |
| ENSMUSG00000055943 | Emc7 | ER membrane protein complex subunit 7 | MGI:1920274 |
| ENSMUSG00000031819 | Emc8 | ER membrane protein complex subunit 8 | MGI:1343095 |
| ENSMUSG00000001964 | Emd | emerin | MGI:108117 |
| ENSMUSG00000058070 | Eml1 | echinoderm microtubule associated protein like 1 | MGI:1915769 |
| ENSMUSG00000022995 | Enah | enabled homolog (Drosophila) | MGI:108360 |
| ENSMUSG00000037419 | Endod1 | endonuclease domain containing 1 | MGI:1919196 |
| ENSMUSG00000059040 | Eno1b | enolase 1B, retrotransposed | MGI:3648653 |
| ENSMUSG00000004267 | Eno2 | enolase 2, gamma neuronal | MGI:95394 |
| ENSMUSG00000038173 | Enpp6 | ectonucleotide pyrophosphatase/phosphodiesterase 6 | MGI:2445171 |
| ENSMUSG00000038619 | Ensa | endosulfine alpha | MGI:1891189 |
| ENSMUSG00000048120 | Entpd1 | ectonucleoside triphosphate diphosphohydrolase 1 | MGI:102805 |
| ENSMUSG00000015085 | Entpd2 | ectonucleoside triphosphate diphosphohydrolase 2 | MGI:1096863 |
| ENSMUSG00000041608 | Entpd3 | ectonucleoside triphosphate diphosphohydrolase 3 | MGI:1321386 |
| ENSMUSG00000033068 | Entpd6 | ectonucleoside triphosphate diphosphohydrolase 6 | MGI:1202295 |
| ENSMUSG00000027624 | Epb4.1l1 | erythrocyte protein band 4.1 like 1 | MGI:103010 |
| ENSMUSG00000019978 | Epb4.1l2 | erythrocyte protein band 4.1 like 2 | MGI:103009 |
| ENSMUSG00000024044 | Epb4.1l3 | erythrocyte protein band 4.1 like 3 | MGI:103008 |
| ENSMUSG00000023216 | Epb4.2 | erythrocyte protein band 4.2 | MGI:95402 |
| ENSMUSG00000002808 | Epdr1 | ependymin related protein 1 (zebrafish) | MGI:2145369 |
| ENSMUSG00000026235 | Epha4 | Eph receptor A4 | MGI:98277 |
| ENSMUSG00000028664 | Ephb2 | Eph receptor B2 | MGI:99611 |
| ENSMUSG00000005958 | Ephb3 | Eph receptor B3 | MGI:104770 |
| ENSMUSG00000038776 | Ephx1 | epoxide hydrolase 1, microsomal | MGI:95405 |
| ENSMUSG00000035203 | Epn1 | epsin 1 | MGI:1333763 |
| ENSMUSG00000010080 | Epn3 | epsin 3 | MGI:1919139 |
| ENSMUSG00000026615 | Eprs | glutamyl-prolyl-tRNA synthetase | MGI:97838 |
| ENSMUSG00000028552 | Eps15 | epidermal growth factor receptor pathway substrate 15 | MGI:104583 |
| ENSMUSG00000006276 | Eps15l1 | epidermal growth factor receptor pathway substrate 15-like 1 | MGI:104582 |
| ENSMUSG00000015766 | Eps8 | epidermal growth factor receptor pathway substrate 8 | MGI:104684 |
| ENSMUSG00000075703 | Ept1 | ethanolaminephosphotransferase 1 (CDP-ethanolamine-specific) | MGI:107898 |
| ENSMUSG00000021583 | Erap1 | endoplasmic reticulum aminopeptidase 1 | MGI:1933403 |
| ENSMUSG00000021709 | Erbb2ip | Erbb2 interacting protein | MGI:1890169 |
| ENSMUSG00000062209 | Erbb4 | v-erb-a erythroblastic leukemia viral oncogene homolog 4 (avian) | MGI:104771 |
| ENSMUSG00000030172 | Erc1 | ELKS/RAB6-interacting/CAST family member 1 | MGI:2151013 |
| ENSMUSG00000040640 | Erc2 | ELKS/RAB6-interacting/CAST family member 2 | MGI:1098749 |
| ENSMUSG00000001576 | Ergic1 | endoplasmic reticulum-golgi intermediate compartment (ERGIC) 1 | MGI:1914708 |
| ENSMUSG00000020311 | Erlec1 | endoplasmic reticulum lectin 1 | MGI:1914003 |
| ENSMUSG00000025198 | Erlin1 | ER lipid raft associated 1 | MGI:2387613 |
| ENSMUSG00000031483 | Erlin2 | ER lipid raft associated 2 | MGI:2387215 |
| ENSMUSG00000026830 | Ermn | ermin, ERM-like protein | MGI:1925017 |
| ENSMUSG00000046324 | Ermp1 | endoplasmic reticulum metallopeptidase 1 | MGI:106250 |
| ENSMUSG00000021831 | Ero1l | ERO1-like (S. cerevisiae) | MGI:1354385 |
| ENSMUSG00000029616 | Erp29 | endoplasmic reticulum protein 29 | MGI:1914647 |
| ENSMUSG00000028343 | Erp44 | endoplasmic reticulum protein 44 | MGI:1923549 |
| ENSMUSG00000025366 | Esyt1 | extended synaptotagmin-like protein 1 | MGI:1344426 |
| ENSMUSG00000021171 | Esyt2 | extended synaptotagmin-like protein 2 | MGI:1261845 |
| ENSMUSG00000024360 | Etf1 | eukaryotic translation termination factor 1 | MGI:2385071 |
| ENSMUSG00000032314 | Etfa | electron transferring flavoprotein, alpha polypeptide | MGI:106092 |
| ENSMUSG00000004610 | Etfb | electron transferring flavoprotein, beta polypeptide | MGI:106098 |
| ENSMUSG00000027809 | Etfdh | electron transferring flavoprotein, dehydrogenase | MGI:106100 |
| ENSMUSG00000036617 | Etl4 | enhancer trap locus 4 | MGI:95454 |
| ENSMUSG00000019232 | Etnppl | ethanolamine phosphate phospholyase | MGI:1919010 |
| ENSMUSG00000035104 | Eva1a | eva-1 homolog A (C. elegans) | MGI:2385247 |
| ENSMUSG00000011832 | Evi5l | ecotropic viral integration site 5 like | MGI:2442167 |
| ENSMUSG00000021262 | Evl | Ena-vasodilator stimulated phosphoprotein | MGI:1194884 |
| ENSMUSG00000032705 | Exd2 | exonuclease 3'-5' domain containing 2 | MGI:1922485 |
| ENSMUSG00000036435 | Exoc1 | exocyst complex component 1 | MGI:2445020 |
| ENSMUSG00000021357 | Exoc2 | exocyst complex component 2 | MGI:1913732 |
| ENSMUSG00000034152 | Exoc3 | exocyst complex component 3 | MGI:2443972 |
| ENSMUSG00000029763 | Exoc4 | exocyst complex component 4 | MGI:1096376 |
| ENSMUSG00000061244 | Exoc5 | exocyst complex component 5 | MGI:2145645 |
| ENSMUSG00000033769 | Exoc6b | exocyst complex component 6B | MGI:1923164 |
| ENSMUSG00000020792 | Exoc7 | exocyst complex component 7 | MGI:1859270 |
| ENSMUSG00000074030 | Exoc8 | exocyst complex component 8 | MGI:2142527 |
| ENSMUSG00000042787 | Exog | endo/exonuclease (5'-3'), endonuclease G-like | MGI:2143333 |
| ENSMUSG00000052397 | Ezr | ezrin | MGI:98931 |
| ENSMUSG00000028128 | F3 | coagulation factor III | MGI:88381 |
| ENSMUSG00000034171 | Faah | fatty acid amide hydrolase | MGI:109609 |
| ENSMUSG00000028773 | Fabp3 | fatty acid binding protein 3, muscle and heart | MGI:95476 |
| ENSMUSG00000027533 | Fabp5 | fatty acid binding protein 5, epidermal | MGI:101790 |
| ENSMUSG00000019874 | Fabp7 | fatty acid binding protein 7, brain | MGI:101916 |
| ENSMUSG00000010663 | Fads1 | fatty acid desaturase 1 | MGI:1923517 |
| ENSMUSG00000025873 | Faf2 | Fas associated factor family member 2 | MGI:1923827 |
| ENSMUSG00000045316 | Fahd1 | fumarylacetoacetate hydrolase domain containing 1 | MGI:1915886 |
| ENSMUSG00000027371 | Fahd2a | fumarylacetoacetate hydrolase domain containing 2A | MGI:1915376 |
| ENSMUSG00000030630 | Fah | fumarylacetoacetate hydrolase | MGI:95482 |
| ENSMUSG00000023011 | Faim2 | Fas apoptotic inhibitory molecule 2 | MGI:1919643 |
| ENSMUSG00000038014 | Fam120a | family with sequence similarity 120, member A | MGI:2446163 |
| ENSMUSG00000025262 | Fam120c | family with sequence similarity 120, member C | MGI:2387687 |
| ENSMUSG00000028995 | Fam126a | family with sequence similarity 126, member A | MGI:2149839 |
| ENSMUSG00000038174 | Fam126b | family with sequence similarity 126, member B | MGI:1098784 |
| ENSMUSG00000029861 | Fam131b | family with sequence similarity 131, member B | MGI:1923406 |
| ENSMUSG00000049339 | Fam134a | family with sequence similarity 134, member A | MGI:2388278 |
| ENSMUSG00000017802 | Fam134c | family with sequence similarity 134, member C | MGI:1915248 |
| ENSMUSG00000057497 | Fam136a | family with sequence similarity 136, member A | MGI:1913738 |
| ENSMUSG00000003955 | Fam162a | family with sequence similarity 162, member A | MGI:1917436 |
| ENSMUSG00000009216 | Fam163b | family with sequence similarity 163, member B | MGI:1926106 |
| ENSMUSG00000050530 | Fam171a1 | family with sequence similarity 171, member A1 | MGI:2442917 |
| ENSMUSG00000034685 | Fam171a2 | family with sequence similarity 171, member A2 | MGI:2448496 |
| ENSMUSG00000048388 | Fam171b | family with sequence similarity 171, member B | MGI:2444579 |
| ENSMUSG00000061111 | Fam195b | family with sequence similarity 195, member B | MGI:2384752 |
| ENSMUSG00000038121 | Fam210a | family with sequence similarity 210, member A | MGI:1914000 |
| ENSMUSG00000021792 | Fam213a | family with sequence similarity 213, member A | MGI:1917814 |
| ENSMUSG00000029059 | Fam213b | family with sequence similarity 213, member B | MGI:1913719 |
| ENSMUSG00000028439 | Fam219a | family with sequence similarity 219, member A | MGI:1919151 |
| ENSMUSG00000020589 | Fam49a | family with sequence similarity 49, member A | MGI:1261783 |
| ENSMUSG00000022378 | Fam49b | family with sequence similarity 49, member B | MGI:1923520 |
| ENSMUSG00000058966 | Fam57b | family with sequence similarity 57, member B | MGI:1916202 |
| ENSMUSG00000042444 | Fam63b | family with sequence similarity 63, member B | MGI:2443086 |
| ENSMUSG00000054942 | Fam73a | family with sequence similarity 73, member A | MGI:1924567 |
| ENSMUSG00000026858 | Fam73b | family with sequence similarity 73, member B | MGI:1922035 |
| ENSMUSG00000032224 | Fam81a | family with sequence similarity 81, member A | MGI:1924136 |
| ENSMUSG00000027349 | Fam98b | family with sequence similarity 98, member B | MGI:1915465 |
| ENSMUSG00000025555 | Farp1 | FERM, RhoGEF (Arhgef) and pleckstrin domain protein 1 (chondrocyte-derived) | MGI:2446173 |
| ENSMUSG00000003808 | Farsa | phenylalanyl-tRNA synthetase, alpha subunit | MGI:1913840 |
| ENSMUSG00000026245 | Farsb | phenylalanyl-tRNA synthetase, beta subunit | MGI:1346035 |
| ENSMUSG00000025153 | Fasn | fatty acid synthase | MGI:95485 |
| ENSMUSG00000070047 | Fat1 | FAT tumor suppressor homolog 1 (Drosophila) | MGI:109168 |
| ENSMUSG00000055333 | Fat2 | FAT tumor suppressor homolog 2 (Drosophila) | MGI:2685369 |
| ENSMUSG00000074505 | Fat3 | FAT tumor suppressor homolog 3 (Drosophila) | MGI:2444314 |
| ENSMUSG00000038274 | Fau | Finkel-Biskis-Reilly murine sarcoma virus (FBR-MuSV) ubiquitously expressed (fox derived) | MGI:102547 |
| ENSMUSG00000025738 | Fbxl16 | F-box and leucine-rich repeat protein 16 | MGI:2448488 |
| ENSMUSG00000020883 | Fbxl20 | F-box and leucine-rich repeat protein 20 | MGI:1919444 |
| ENSMUSG00000041556 | Fbxo2 | F-box protein 2 | MGI:2446216 |
| ENSMUSG00000047013 | Fbxo41 | F-box protein 41 | MGI:1261912 |
| ENSMUSG00000058715 | Fcer1g | Fc receptor, IgE, high affinity I, gamma polypeptide | MGI:95496 |
| ENSMUSG00000059743 | Fdps | farnesyl diphosphate synthetase | MGI:104888 |
| ENSMUSG00000079677 | Fdx1l | ferredoxin 1-like | MGI:1915415 |
| ENSMUSG00000018861 | Fdxr | ferredoxin reductase | MGI:104724 |
| ENSMUSG00000024588 | Fech | ferrochelatase | MGI:95513 |
| ENSMUSG00000022523 | Fgf12 | fibroblast growth factor 12 | MGI:109183 |
| ENSMUSG00000040242 | Fgfr1op2 | FGFR1 oncogene partner 2 | MGI:1914779 |
| ENSMUSG00000054252 | Fgfr3 | fibroblast growth factor receptor 3 | MGI:95524 |
| ENSMUSG00000026526 | Fh1 | fumarate hydratase 1 | MGI:95530 |
| ENSMUSG00000024911 | Fibp | fibroblast growth factor (acidic) intracellular binding protein | MGI:1926233 |
| ENSMUSG00000019054 | Fis1 | fission 1 (mitochondrial outer membrane) homolog (yeast) | MGI:1913687 |
| ENSMUSG00000066151 | Fkbp15 | FK506 binding protein 15 | MGI:2444782 |
| ENSMUSG00000032966 | Fkbp1a | FK506 binding protein 1a | MGI:95541 |
| ENSMUSG00000020635 | Fkbp1b | FK506 binding protein 1b | MGI:1336205 |
| ENSMUSG00000056629 | Fkbp2 | FK506 binding protein 2 | MGI:95542 |
| ENSMUSG00000020949 | Fkbp3 | FK506 binding protein 3 | MGI:1353460 |
| ENSMUSG00000030357 | Fkbp4 | FK506 binding protein 4 | MGI:95543 |
| ENSMUSG00000019428 | Fkbp8 | FK506 binding protein 8 | MGI:1341070 |
| ENSMUSG00000002812 | Flii | flightless I homolog (Drosophila) | MGI:1342286 |
| ENSMUSG00000059714 | Flot1 | flotillin 1 | MGI:1100500 |
| ENSMUSG00000061981 | Flot2 | flotillin 2 | MGI:103309 |
| ENSMUSG00000047414 | Flrt2 | fibronectin leucine rich transmembrane protein 2 | MGI:3603594 |
| ENSMUSG00000028354 | Fmn2 | formin 2 | MGI:1859252 |
| ENSMUSG00000055805 | Fmnl1 | formin-like 1 | MGI:1888994 |
| ENSMUSG00000036053 | Fmnl2 | formin-like 2 | MGI:1918659 |
| ENSMUSG00000000838 | Fmr1 | fragile X mental retardation syndrome 1 | MGI:95564 |
| ENSMUSG00000075415 | Fnbp1 | formin binding protein 1 | MGI:109606 |
| ENSMUSG00000039735 | Fnbp1l | formin binding protein 1-like | MGI:1925642 |
| ENSMUSG00000001773 | Folh1 | folate hydrolase 1 | MGI:1858193 |
| ENSMUSG00000045589 | Frrs1l | ferric-chelate reductase 1 like | MGI:2442704 |
| ENSMUSG00000020170 | Frs2 | fibroblast growth factor receptor substrate 2 | MGI:1100860 |
| ENSMUSG00000023266 | Frs3 | fibroblast growth factor receptor substrate 3 | MGI:2135965 |
| ENSMUSG00000056602 | Fry | furry homolog (Drosophila) | MGI:2443895 |
| ENSMUSG00000029581 | Fscn1 | fascin homolog 1, actin bundling protein (Strongylocentrotus purpuratus) | MGI:1352745 |
| ENSMUSG00000011589 | Fsd1 | fibronectin type 3 and SPRY domain-containing protein | MGI:1934858 |
| ENSMUSG00000024661 | Fth1 | ferritin heavy chain 1 | MGI:95588 |
| ENSMUSG00000025040 | Fundc1 | FUN14 domain containing 1 | MGI:1919268 |
| ENSMUSG00000031198 | Fundc2 | FUN14 domain containing 2 | MGI:1914641 |
| ENSMUSG00000025466 | Fuom | fucose mutarotase | MGI:1916314 |
| ENSMUSG00000027680 | Fxr1 | fragile X mental retardation gene 1, autosomal homolog | MGI:104860 |
| ENSMUSG00000018765 | Fxr2 | fragile X mental retardation, autosomal homolog 2 | MGI:1346074 |
| ENSMUSG00000036570 | Fxyd1 | FXYD domain-containing ion transport regulator 1 | MGI:1889273 |
| ENSMUSG00000066705 | Fxyd6 | FXYD domain-containing ion transport regulator 6 | MGI:1890226 |
| ENSMUSG00000036578 | Fxyd7 | FXYD domain-containing ion transport regulator 7 | MGI:1889006 |
| ENSMUSG00000019843 | Fyn | Fyn proto-oncogene | MGI:95602 |
| ENSMUSG00000029405 | G3bp2 | GTPase activating protein (SH3 domain) binding protein 2 | MGI:2442040 |
| ENSMUSG00000031400 | G6pdx | glucose-6-phosphate dehydrogenase X-linked | MGI:105979 |
| ENSMUSG00000025579 | Gaa | glucosidase, alpha, acid | MGI:95609 |
| ENSMUSG00000031950 | Gabarapl2 | gamma-aminobutyric acid (GABA) A receptor-associated protein-like 2 | MGI:1890602 |
| ENSMUSG00000024462 | Gabbr1 | gamma-aminobutyric acid (GABA) B receptor, 1 | MGI:1860139 |
| ENSMUSG00000039809 | Gabbr2 | gamma-aminobutyric acid (GABA) B receptor, 2 | MGI:2386030 |
| ENSMUSG00000010803 | Gabra1 | gamma-aminobutyric acid (GABA) A receptor, subunit alpha 1 | MGI:95613 |
| ENSMUSG00000000560 | Gabra2 | gamma-aminobutyric acid (GABA) A receptor, subunit alpha 2 | MGI:95614 |
| ENSMUSG00000031343 | Gabra3 | gamma-aminobutyric acid (GABA) A receptor, subunit alpha 3 | MGI:95615 |
| ENSMUSG00000029211 | Gabra4 | gamma-aminobutyric acid (GABA) A receptor, subunit alpha 4 | MGI:95616 |
| ENSMUSG00000020428 | Gabra6 | gamma-aminobutyric acid (GABA) A receptor, subunit alpha 6 | MGI:95618 |
| ENSMUSG00000029212 | Gabrb1 | gamma-aminobutyric acid (GABA) A receptor, subunit beta 1 | MGI:95619 |
| ENSMUSG00000007653 | Gabrb2 | gamma-aminobutyric acid (GABA) A receptor, subunit beta 2 | MGI:95620 |
| ENSMUSG00000033676 | Gabrb3 | gamma-aminobutyric acid (GABA) A receptor, subunit beta 3 | MGI:95621 |
| ENSMUSG00000020436 | Gabrg2 | gamma-aminobutyric acid (GABA) A receptor, subunit gamma 2 | MGI:95623 |
| ENSMUSG00000070880 | Gad1 | glutamate decarboxylase 1 | MGI:95632 |
| ENSMUSG00000026787 | Gad2 | glutamic acid decarboxylase 2 | MGI:95634 |
| ENSMUSG00000062234 | Gak | cyclin G associated kinase | MGI:2442153 |
| ENSMUSG00000021003 | Galc | galactosylceramidase | MGI:95636 |
| ENSMUSG00000071650 | Ganab | alpha glucosidase 2 alpha neutral subunit | MGI:1097667 |
| ENSMUSG00000047261 | Gap43 | growth associated protein 43 | MGI:95639 |
| ENSMUSG00000057666 | Gapdh | glyceraldehyde-3-phosphate dehydrogenase | MGI:95640 |
| ENSMUSG00000029777 | Gars | glycyl-tRNA synthetase | MGI:2449057 |
| ENSMUSG00000033066 | Gas7 | growth arrest specific 7 | MGI:1202388 |
| ENSMUSG00000028048 | Gba | glucosidase, beta, acid | MGI:95665 |
| ENSMUSG00000029432 | Gbas | glioblastoma amplified sequence | MGI:1278343 |
| ENSMUSG00000022707 | Gbe1 | glucan (1,4-alpha-), branching enzyme 1 | MGI:1921435 |
| ENSMUSG00000003809 | Gcdh | glutaryl-Coenzyme A dehydrogenase | MGI:104541 |
| ENSMUSG00000041638 | Gcn1l1 | GCN1 general control of amino-acid synthesis 1-like 1 (yeast) | MGI:2444248 |
| ENSMUSG00000058624 | Gda | guanine deaminase | MGI:95678 |
| ENSMUSG00000025777 | Gdap1 | ganglioside-induced differentiation-associated-protein 1 | MGI:1338002 |
| ENSMUSG00000017943 | Gdap1l1 | ganglioside-induced differentiation-associated protein 1-like 1 | MGI:2385163 |
| ENSMUSG00000033917 | Gde1 | glycerophosphodiester phosphodiesterase 1 | MGI:1891827 |
| ENSMUSG00000015291 | Gdi1 | guanosine diphosphate (GDP) dissociation inhibitor 1 | MGI:99846 |
| ENSMUSG00000021218 | Gdi2 | guanosine diphosphate (GDP) dissociation inhibitor 2 | MGI:99845 |
| ENSMUSG00000061666 | Gdpd1 | glycerophosphodiester phosphodiesterase domain containing 1 | MGI:1913819 |
| ENSMUSG00000020932 | Gfap | glial fibrillary acidic protein | MGI:95697 |
| ENSMUSG00000040888 | Gfer | growth factor, erv1 (S. cerevisiae)-like (augmenter of liver regeneration) | MGI:107757 |
| ENSMUSG00000027774 | Gfm1 | G elongation factor, mitochondrial 1 | MGI:107339 |
| ENSMUSG00000027603 | Ggt7 | gamma-glutamyltransferase 7 | MGI:1913385 |
| ENSMUSG00000041028 | Ghitm | growth hormone inducible transmembrane protein | MGI:1913342 |
| ENSMUSG00000029714 | Gigyf1 | GRB10 interacting GYF protein 1 | MGI:1888677 |
| ENSMUSG00000048000 | Gigyf2 | GRB10 interacting GYF protein 2 | MGI:2138584 |
| ENSMUSG00000019433 | Gipc1 | GIPC PDZ domain containing family, member 1 | MGI:1926252 |
| ENSMUSG00000011877 | Git1 | G protein-coupled receptor kinase-interactor 1 | MGI:1927140 |
| ENSMUSG00000050953 | Gja1 | gap junction protein, alpha 1 | MGI:95713 |
| ENSMUSG00000040055 | Gjb6 | gap junction protein, beta 6 | MGI:107588 |
| ENSMUSG00000043448 | Gjc2 | gap junction protein, gamma 2 | MGI:2153060 |
| ENSMUSG00000056966 | Gjc3 | gap junction protein, gamma 3 | MGI:2153041 |
| ENSMUSG00000029638 | Glcci1 | glucocorticoid induced transcript 1 | MGI:2179717 |
| ENSMUSG00000003316 | Glg1 | golgi apparatus protein 1 | MGI:104967 |
| ENSMUSG00000028480 | Glipr2 | GLI pathogenesis-related 2 | MGI:1917770 |
| ENSMUSG00000024026 | Glo1 | glyoxalase 1 | MGI:95742 |
| ENSMUSG00000017286 | Glod4 | glyoxalase domain containing 4 | MGI:1914451 |
| ENSMUSG00000000263 | Glra1 | glycine receptor, alpha 1 subunit | MGI:95747 |
| ENSMUSG00000018196 | Glrx2 | glutaredoxin 2 (thioltransferase) | MGI:1916617 |
| ENSMUSG00000031068 | Glrx3 | glutaredoxin 3 | MGI:1353653 |
| ENSMUSG00000021102 | Glrx5 | glutaredoxin 5 homolog (S. cerevisiae) | MGI:1920296 |
| ENSMUSG00000021591 | Glrx | glutaredoxin | MGI:2135625 |
| ENSMUSG00000026103 | Gls | glutaminase | MGI:95752 |
| ENSMUSG00000011884 | Gltp | glycolipid transfer protein | MGI:1929253 |
| ENSMUSG00000036568 | Gltscr1l | GLTSCR1-like | MGI:2673855 |
| ENSMUSG00000021794 | Glud1 | glutamate dehydrogenase 1 | MGI:95753 |
| ENSMUSG00000026473 | Glul | glutamate-ammonia ligase (glutamine synthetase) | MGI:95739 |
| ENSMUSG00000057262 | Gm10020 | predicted pseudogene 10020 | MGI:3642192 |
| ENSMUSG00000058064 | Gm10036 | predicted gene 10036 | MGI:3642334 |
| ENSMUSG00000058927 | Gm10053 | predicted gene 10053 | MGI:3704493 |
| ENSMUSG00000062382 | Gm10116 | predicted pseudogene 10116 | MGI:3779109 |
| ENSMUSG00000069117 | Gm10260 | predicted gene 10260 | MGI:3642298 |
| ENSMUSG00000042744 | Gm15800 | predicted gene 15800 | MGI:3647820 |
| ENSMUSG00000043192 | Gm1840 | predicted gene 1840 | MGI:3037698 |
| ENSMUSG00000078193 | Gm2000 | predicted gene 2000 | MGI:3780170 |
| ENSMUSG00000091228 | Gm20390 | predicted gene 20390 | MGI:5141855 |
| ENSMUSG00000090639 | Gm20425 | predicted gene 20425 | MGI:5141890 |
| ENSMUSG00000021139 | Gm20498 | predicted gene 20498 | MGI:5141963 |
| ENSMUSG00000100725 | Gm28062 | predicted gene 28062 | MGI:5578768 |
| ENSMUSG00000100679 | Gm28778 | predicted gene 28778 | MGI:5579484 |
| ENSMUSG00000000594 | Gm2a | GM2 ganglioside activator protein | MGI:95762 |
| ENSMUSG00000048087 | Gm4737 | predicted gene 4737 | MGI:3643647 |
| ENSMUSG00000066487 | Gm5786 | predicted pseudogene 5786 | MGI:3645003 |
| ENSMUSG00000094685 | Gm5900 | predicted pseudogene 5900 | MGI:3643907 |
| ENSMUSG00000063696 | Gm8730 | predicted pseudogene 8730 | MGI:3644565 |
| ENSMUSG00000044424 | Gm9493 | predicted gene 9493 | MGI:3779903 |
| ENSMUSG00000042165 | Gm9774 | predicted pseudogene 9774 | MGI:3642386 |
| ENSMUSG00000045455 | Gm9797 | predicted pseudogene 9797 | MGI:3704349 |
| ENSMUSG00000029419 | Gm996 | predicted gene 996 | MGI:2685842 |
| ENSMUSG00000062014 | Gmfb | glia maturation factor, beta | MGI:1927133 |
| ENSMUSG00000000253 | Gmpr | guanosine monophosphate reductase | MGI:1913605 |
| ENSMUSG00000034781 | Gna11 | guanine nucleotide binding protein, alpha 11 | MGI:95766 |
| ENSMUSG00000000149 | Gna12 | guanine nucleotide binding protein, alpha 12 | MGI:95767 |
| ENSMUSG00000020611 | Gna13 | guanine nucleotide binding protein, alpha 13 | MGI:95768 |
| ENSMUSG00000057614 | Gnai1 | guanine nucleotide binding protein (G protein), alpha inhibiting 1 | MGI:95771 |
| ENSMUSG00000032562 | Gnai2 | guanine nucleotide binding protein (G protein), alpha inhibiting 2 | MGI:95772 |
| ENSMUSG00000000001 | Gnai3 | guanine nucleotide binding protein (G protein), alpha inhibiting 3 | MGI:95773 |
| ENSMUSG00000024524 | Gnal | guanine nucleotide binding protein, alpha stimulating, olfactory type | MGI:95774 |
| ENSMUSG00000031748 | Gnao1 | guanine nucleotide binding protein, alpha O | MGI:95775 |
| ENSMUSG00000024639 | Gnaq | guanine nucleotide binding protein, alpha q polypeptide | MGI:95776 |
| ENSMUSG00000027523 | Gnas | GNAS (guanine nucleotide binding protein, alpha stimulating) complex locus | MGI:95777 |
| ENSMUSG00000040009 | Gnaz | guanine nucleotide binding protein, alpha z subunit | MGI:95780 |
| ENSMUSG00000029064 | Gnb1 | guanine nucleotide binding protein (G protein), beta 1 | MGI:95781 |
| ENSMUSG00000029713 | Gnb2 | guanine nucleotide binding protein (G protein), beta 2 | MGI:95784 |
| ENSMUSG00000020372 | Gnb2l1 | guanine nucleotide binding protein (G protein), beta polypeptide 2 like 1 | MGI:101849 |
| ENSMUSG00000023439 | Gnb3 | guanine nucleotide binding protein (G protein), beta 3 | MGI:95785 |
| ENSMUSG00000027669 | Gnb4 | guanine nucleotide binding protein (G protein), beta 4 | MGI:104581 |
| ENSMUSG00000032192 | Gnb5 | guanine nucleotide binding protein (G protein), beta 5 | MGI:101848 |
| ENSMUSG00000036402 | Gng12 | guanine nucleotide binding protein (G protein), gamma 12 | MGI:1336171 |
| ENSMUSG00000025739 | Gng13 | guanine nucleotide binding protein (G protein), gamma 13 | MGI:1925616 |
| ENSMUSG00000043004 | Gng2 | guanine nucleotide binding protein (G protein), gamma 2 | MGI:102705 |
| ENSMUSG00000071658 | Gng3 | guanine nucleotide binding protein (G protein), gamma 3 | MGI:102704 |
| ENSMUSG00000021303 | Gng4 | guanine nucleotide binding protein (G protein), gamma 4 | MGI:102703 |
| ENSMUSG00000024429 | Gnl1 | guanine nucleotide binding protein-like 1 | MGI:95764 |
| ENSMUSG00000031985 | Gnpat | glyceronephosphate O-acyltransferase | MGI:1343460 |
| ENSMUSG00000002546 | Golga2 | golgi autoantigen, golgin subfamily a, 2 | MGI:2139395 |
| ENSMUSG00000029502 | Golga3 | golgi autoantigen, golgin subfamily a, 3 | MGI:96958 |
| ENSMUSG00000021192 | Golga5 | golgi autoantigen, golgin subfamily a, 5 | MGI:1351475 |
| ENSMUSG00000042532 | Golga7b | golgi autoantigen, golgin subfamily a, 7B | MGI:1918396 |
| ENSMUSG00000015341 | Golga7 | golgi autoantigen, golgin subfamily a, 7 | MGI:1931029 |
| ENSMUSG00000034243 | Golgb1 | golgi autoantigen, golgin subfamily b, macrogolgin 1 | MGI:1099447 |
| ENSMUSG00000030245 | Golt1b | golgi transport 1 homolog B (S. cerevisiae) | MGI:1914214 |
| ENSMUSG00000010392 | Gosr1 | golgi SNAP receptor complex member 1 | MGI:1858260 |
| ENSMUSG00000020946 | Gosr2 | golgi SNAP receptor complex member 2 | MGI:1927204 |
| ENSMUSG00000025190 | Got1 | glutamic-oxaloacetic transaminase 1, soluble | MGI:95791 |
| ENSMUSG00000031672 | Got2 | glutamatic-oxaloacetic transaminase 2, mitochondrial | MGI:95792 |
| ENSMUSG00000022561 | Gpaa1 | GPI anchor attachment protein 1 | MGI:1202392 |
| ENSMUSG00000034220 | Gpc1 | glypican 1 | MGI:1194891 |
| ENSMUSG00000031119 | Gpc4 | glypican 4 | MGI:104902 |
| ENSMUSG00000022112 | Gpc5 | glypican 5 | MGI:1194894 |
| ENSMUSG00000023019 | Gpd1 | glycerol-3-phosphate dehydrogenase 1 (soluble) | MGI:95679 |
| ENSMUSG00000050627 | Gpd1l | glycerol-3-phosphate dehydrogenase 1-like | MGI:1289257 |
| ENSMUSG00000026827 | Gpd2 | glycerol phosphate dehydrogenase 2, mitochondrial | MGI:99778 |
| ENSMUSG00000047454 | Gphn | gephyrin | MGI:109602 |
| ENSMUSG00000036427 | Gpi1 | glucose phosphate isomerase 1 | MGI:95797 |
| ENSMUSG00000031517 | Gpm6a | glycoprotein m6a | MGI:107671 |
| ENSMUSG00000031342 | Gpm6b | glycoprotein m6b | MGI:107672 |
| ENSMUSG00000036357 | Gpr101 | G protein-coupled receptor 101 | MGI:2685211 |
| ENSMUSG00000045967 | Gpr158 | G protein-coupled receptor 158 | MGI:2441697 |
| ENSMUSG00000038390 | Gpr162 | G protein-coupled receptor 162 | MGI:1315214 |
| ENSMUSG00000008734 | Gprc5b | G protein-coupled receptor, family C, group 5, member B | MGI:1927596 |
| ENSMUSG00000069227 | Gprin1 | G protein-regulated inducer of neurite outgrowth 1 | MGI:1349455 |
| ENSMUSG00000045441 | Gprin3 | GPRIN family member 3 | MGI:1924785 |
| ENSMUSG00000025156 | Gps1 | G protein pathway suppressor 1 | MGI:2384801 |
| ENSMUSG00000026930 | Gpsm1 | G-protein signalling modulator 1 (AGS3-like, C. elegans) | MGI:1915089 |
| ENSMUSG00000034786 | Gpsm3 | G-protein signalling modulator 3 (AGS3-like, C. elegans) | MGI:2146785 |
| ENSMUSG00000063856 | Gpx1 | glutathione peroxidase 1 | MGI:104887 |
| ENSMUSG00000075706 | Gpx4 | glutathione peroxidase 4 | MGI:104767 |
| ENSMUSG00000040111 | Gramd1b | GRAM domain containing 1B | MGI:1925037 |
| ENSMUSG00000035900 | Gramd4 | GRAM domain containing 4 | MGI:2676308 |
| ENSMUSG00000059923 | Grb2 | growth factor receptor bound protein 2 | MGI:95805 |
| ENSMUSG00000072772 | Grcc10 | gene rich cluster, C10 gene | MGI:1315201 |
| ENSMUSG00000035637 | Grhpr | glyoxylate reductase/hydroxypyruvate reductase | MGI:1923488 |
| ENSMUSG00000020524 | Gria1 | glutamate receptor, ionotropic, AMPA1 (alpha 1) | MGI:95808 |
| ENSMUSG00000033981 | Gria2 | glutamate receptor, ionotropic, AMPA2 (alpha 2) | MGI:95809 |
| ENSMUSG00000001986 | Gria3 | glutamate receptor, ionotropic, AMPA3 (alpha 3) | MGI:95810 |
| ENSMUSG00000025892 | Gria4 | glutamate receptor, ionotropic, AMPA4 (alpha 4) | MGI:95811 |
| ENSMUSG00000041078 | Grid1 | glutamate receptor, ionotropic, delta 1 | MGI:95812 |
| ENSMUSG00000071424 | Grid2 | glutamate receptor, ionotropic, delta 2 | MGI:95813 |
| ENSMUSG00000056073 | Grik2 | glutamate receptor, ionotropic, kainate 2 (beta 2) | MGI:95815 |
| ENSMUSG00000001985 | Grik3 | glutamate receptor, ionotropic, kainate 3 | MGI:95816 |
| ENSMUSG00000059003 | Grin2a | glutamate receptor, ionotropic, NMDA2A (epsilon 1) | MGI:95820 |
| ENSMUSG00000030209 | Grin2b | glutamate receptor, ionotropic, NMDA2B (epsilon 2) | MGI:95821 |
| ENSMUSG00000002771 | Grin2d | glutamate receptor, ionotropic, NMDA2D (epsilon 4) | MGI:95823 |
| ENSMUSG00000031153 | Gripap1 | GRIP1 associated protein 1 | MGI:1859616 |
| ENSMUSG00000074886 | Grk6 | G protein-coupled receptor kinase 6 | MGI:1347078 |
| ENSMUSG00000019828 | Grm1 | glutamate receptor, metabotropic 1 | MGI:1351338 |
| ENSMUSG00000023192 | Grm2 | glutamate receptor, metabotropic 2 | MGI:1351339 |
| ENSMUSG00000003974 | Grm3 | glutamate receptor, metabotropic 3 | MGI:1351340 |
| ENSMUSG00000063239 | Grm4 | glutamate receptor, metabotropic 4 | MGI:1351341 |
| ENSMUSG00000049583 | Grm5 | glutamate receptor, metabotropic 5 | MGI:1351342 |
| ENSMUSG00000056755 | Grm7 | glutamate receptor, metabotropic 7 | MGI:1351344 |
| ENSMUSG00000024211 | Grm8 | glutamate receptor, metabotropic 8 | MGI:1351345 |
| ENSMUSG00000029198 | Grpel1 | GrpE-like 1, mitochondrial | MGI:1334417 |
| ENSMUSG00000057177 | Gsk3a | glycogen synthase kinase 3 alpha | MGI:2152453 |
| ENSMUSG00000022812 | Gsk3b | glycogen synthase kinase 3 beta | MGI:1861437 |
| ENSMUSG00000026879 | Gsn | gelsolin | MGI:95851 |
| ENSMUSG00000031584 | Gsr | glutathione reductase | MGI:95804 |
| ENSMUSG00000032348 | Gsta4 | glutathione S-transferase, alpha 4 | MGI:1309515 |
| ENSMUSG00000029864 | Gstk1 | glutathione S-transferase kappa 1 | MGI:1923513 |
| ENSMUSG00000058135 | Gstm1 | glutathione S-transferase, mu 1 | MGI:95860 |
| ENSMUSG00000004032 | Gstm5 | glutathione S-transferase, mu 5 | MGI:1309466 |
| ENSMUSG00000060803 | Gstp1 | glutathione S-transferase, pi 1 | MGI:95865 |
| ENSMUSG00000021033 | Gstz1 | glutathione transferase zeta 1 (maleylacetoacetate isomerase) | MGI:1341859 |
| ENSMUSG00000060261 | Gtf2i | general transcription factor II I | MGI:1202722 |
| ENSMUSG00000042535 | Gtpbp1 | GTP binding protein 1 | MGI:109443 |
| ENSMUSG00000041624 | Gucy1a2 | guanylate cyclase 1, soluble, alpha 2 | MGI:2660877 |
| ENSMUSG00000028005 | Gucy1b3 | guanylate cyclase 1, soluble, beta 3 | MGI:1860604 |
| ENSMUSG00000020444 | Guk1 | guanylate kinase 1 | MGI:95871 |
| ENSMUSG00000025059 | Gyk | glycerol kinase | MGI:106594 |
| ENSMUSG00000051839 | Gypa | glycophorin A | MGI:95880 |
| ENSMUSG00000090523 | Gypc | glycophorin C | MGI:1098566 |
| ENSMUSG00000019188 | H13 | histocompatibility 13 | MGI:95886 |
| ENSMUSG00000073422 | H2-Ke6 | H2-K region expressed gene 6 | MGI:95911 |
| ENSMUSG00000060743 | H3f3a | H3 histone, family 3A | MGI:1097686 |
| ENSMUSG00000033629 | Hacd3 | 3-hydroxyacyl-CoA dehydratase 3 | MGI:1889341 |
| ENSMUSG00000021884 | Hacl1 | 2-hydroxyacyl-CoA lyase 1 | MGI:1929657 |
| ENSMUSG00000025745 | Hadha | hydroxyacyl-Coenzyme A dehydrogenase/3-ketoacyl-Coenzyme A thiolase/enoyl-Coenzyme A hydratase (trifunctional protein), alpha subunit | MGI:2135593 |
| ENSMUSG00000059447 | Hadhb | hydroxyacyl-Coenzyme A dehydrogenase/3-ketoacyl-Coenzyme A thiolase/enoyl-Coenzyme A hydratase (trifunctional protein), beta subunit | MGI:2136381 |
| ENSMUSG00000027984 | Hadh | hydroxyacyl-Coenzyme A dehydrogenase | MGI:96009 |
| ENSMUSG00000024158 | Hagh | hydroxyacyl glutathione hydrolase | MGI:95745 |
| ENSMUSG00000021613 | Hapln1 | hyaluronan and proteoglycan link protein 1 | MGI:1337006 |
| ENSMUSG00000007594 | Hapln4 | hyaluronan and proteoglycan link protein 4 | MGI:2679531 |
| ENSMUSG00000069919 | Hba-a1 | hemoglobin alpha, adult chain 1 | MGI:96015 |
| ENSMUSG00000073940 | Hbb-bt | hemoglobin, beta adult t chain | MGI:5474850 |
| ENSMUSG00000031386 | Hcfc1 | host cell factor C1 | MGI:105942 |
| ENSMUSG00000021730 | Hcn1 | hyperpolarization-activated, cyclic nucleotide-gated K+ 1 | MGI:1096392 |
| ENSMUSG00000020331 | Hcn2 | hyperpolarization-activated, cyclic nucleotide-gated K+ 2 | MGI:1298210 |
| ENSMUSG00000028051 | Hcn3 | hyperpolarization-activated, cyclic nucleotide-gated K+ 3 | MGI:1298211 |
| ENSMUSG00000032338 | Hcn4 | hyperpolarization-activated, cyclic nucleotide-gated K+ 4 | MGI:1298209 |
| ENSMUSG00000034245 | Hdac11 | histone deacetylase 11 | MGI:2385252 |
| ENSMUSG00000030532 | Hddc3 | HD domain containing 3 | MGI:1915945 |
| ENSMUSG00000025421 | Hdhd2 | haloacid dehalogenase-like hydrolase domain containing 2 | MGI:1924237 |
| ENSMUSG00000038422 | Hdhd3 | haloacid dehalogenase-like hydrolase domain containing 3 | MGI:1919998 |
| ENSMUSG00000034088 | Hdlbp | high density lipoprotein (HDL) binding protein | MGI:99256 |
| ENSMUSG00000039414 | Heatr5b | HEAT repeat containing 5B | MGI:2444098 |
| ENSMUSG00000042770 | Hebp1 | heme binding protein 1 | MGI:1333880 |
| ENSMUSG00000021301 | Hecw1 | HECT, C2 and WW domain containing E3 ubiquitin protein ligase 1 | MGI:2444115 |
| ENSMUSG00000042807 | Hecw2 | HECT, C2 and WW domain containing E3 ubiquitin protein ligase 2 | MGI:2685817 |
| ENSMUSG00000046240 | Hepacam | hepatocyte cell adhesion molecule | MGI:1920177 |
| ENSMUSG00000025232 | Hexa | hexosaminidase A | MGI:96073 |
| ENSMUSG00000021665 | Hexb | hexosaminidase B | MGI:96074 |
| ENSMUSG00000025793 | Hgs | HGF-regulated tyrosine kinase substrate | MGI:104681 |
| ENSMUSG00000032523 | Hhatl | hedgehog acyltransferase-like | MGI:1922020 |
| ENSMUSG00000029776 | Hibadh | 3-hydroxyisobutyrate dehydrogenase | MGI:1889802 |
| ENSMUSG00000041426 | Hibch | 3-hydroxyisobutyryl-Coenzyme A hydrolase | MGI:1923792 |
| ENSMUSG00000038412 | Higd1a | HIG1 domain family, member 1A | MGI:1930666 |
| ENSMUSG00000020267 | Hint1 | histidine triad nucleotide binding protein 1 | MGI:1321133 |
| ENSMUSG00000028470 | Hint2 | histidine triad nucleotide binding protein 2 | MGI:1916167 |
| ENSMUSG00000019791 | Hint3 | histidine triad nucleotide binding protein 3 | MGI:1914097 |
| ENSMUSG00000039959 | Hip1 | huntingtin interacting protein 1 | MGI:1099804 |
| ENSMUSG00000000915 | Hip1r | huntingtin interacting protein 1 related | MGI:1352504 |
| ENSMUSG00000060081 | Hist1h2aa | histone cluster 1, H2aa | MGI:2448285 |
| ENSMUSG00000069308 | Hist1h2bp | histone cluster 1, H2bp | MGI:2448409 |
| ENSMUSG00000069266 | Hist1h4b | histone cluster 1, H4b | MGI:2448420 |
| ENSMUSG00000037012 | Hk1 | hexokinase 1 | MGI:96103 |
| ENSMUSG00000000628 | Hk2 | hexokinase 2 | MGI:1315197 |
| ENSMUSG00000028672 | Hmgcl | 3-hydroxy-3-methylglutaryl-Coenzyme A lyase | MGI:96158 |
| ENSMUSG00000021670 | Hmgcr | 3-hydroxy-3-methylglutaryl-Coenzyme A reductase | MGI:96159 |
| ENSMUSG00000093930 | Hmgcs1 | 3-hydroxy-3-methylglutaryl-Coenzyme A synthase 1 | MGI:107592 |
| ENSMUSG00000004070 | Hmox2 | heme oxygenase 2 | MGI:109373 |
| ENSMUSG00000046434 | Hnrnpa1 | heterogeneous nuclear ribonucleoprotein A1 | MGI:104820 |
| ENSMUSG00000004980 | Hnrnpa2b1 | heterogeneous nuclear ribonucleoprotein A2/B1 | MGI:104819 |
| ENSMUSG00000059005 | Hnrnpa3 | heterogeneous nuclear ribonucleoprotein A3 | MGI:1917171 |
| ENSMUSG00000020358 | Hnrnpab | heterogeneous nuclear ribonucleoprotein A/B | MGI:1330294 |
| ENSMUSG00000000568 | Hnrnpd | heterogeneous nuclear ribonucleoprotein D | MGI:101947 |
| ENSMUSG00000021546 | Hnrnpk | heterogeneous nuclear ribonucleoprotein K | MGI:99894 |
| ENSMUSG00000059208 | Hnrnpm | heterogeneous nuclear ribonucleoprotein M | MGI:1926465 |
| ENSMUSG00000007617 | Homer1 | homer homolog 1 (Drosophila) | MGI:1347345 |
| ENSMUSG00000025813 | Homer2 | homer homolog 2 (Drosophila) | MGI:1347354 |
| ENSMUSG00000003573 | Homer3 | homer homolog 3 (Drosophila) | MGI:1347359 |
| ENSMUSG00000037234 | Hook3 | hook homolog 3 (Drosophila) | MGI:2443554 |
| ENSMUSG00000028785 | Hpca | hippocalcin | MGI:1336200 |
| ENSMUSG00000071379 | Hpcal1 | hippocalcin-like 1 | MGI:1855689 |
| ENSMUSG00000046093 | Hpcal4 | hippocalcin-like 4 | MGI:2157521 |
| ENSMUSG00000025630 | Hprt | hypoxanthine guanine phosphoribosyl transferase | MGI:96217 |
| ENSMUSG00000025499 | Hras | Harvey rat sarcoma virus oncogene | MGI:96224 |
| ENSMUSG00000022323 | Hrsp12 | heat-responsive protein 12 | MGI:1095401 |
| ENSMUSG00000031839 | Hsbp1 | heat shock factor binding protein 1 | MGI:1915446 |
| ENSMUSG00000025260 | Hsd17b10 | hydroxysteroid (17-beta) dehydrogenase 10 | MGI:1333871 |
| ENSMUSG00000029311 | Hsd17b11 | hydroxysteroid (17-beta) dehydrogenase 11 | MGI:2149821 |
| ENSMUSG00000027195 | Hsd17b12 | hydroxysteroid (17-beta) dehydrogenase 12 | MGI:1926967 |
| ENSMUSG00000024507 | Hsd17b4 | hydroxysteroid (17-beta) dehydrogenase 4 | MGI:105089 |
| ENSMUSG00000034189 | Hsdl1 | hydroxysteroid dehydrogenase like 1 | MGI:1919802 |
| ENSMUSG00000028383 | Hsdl2 | hydroxysteroid dehydrogenase like 2 | MGI:1919729 |
| ENSMUSG00000021270 | Hsp90aa1 | heat shock protein 90, alpha (cytosolic), class A member 1 | MGI:96250 |
| ENSMUSG00000023944 | Hsp90ab1 | heat shock protein 90 alpha (cytosolic), class B member 1 | MGI:96247 |
| ENSMUSG00000020048 | Hsp90b1 | heat shock protein 90, beta (Grp94), member 1 | MGI:98817 |
| ENSMUSG00000025092 | Hspa12a | heat shock protein 12A | MGI:1920692 |
| ENSMUSG00000074793 | Hspa12b | heat shock protein 12B | MGI:1919880 |
| ENSMUSG00000032932 | Hspa13 | heat shock protein 70 family, member 13 | MGI:1309463 |
| ENSMUSG00000091971 | Hspa1a | heat shock protein 1A | MGI:96244 |
| ENSMUSG00000059970 | Hspa2 | heat shock protein 2 | MGI:96243 |
| ENSMUSG00000020361 | Hspa4 | heat shock protein 4 | MGI:1342292 |
| ENSMUSG00000025757 | Hspa4l | heat shock protein 4 like | MGI:107422 |
| ENSMUSG00000026864 | Hspa5 | heat shock protein 5 | MGI:95835 |
| ENSMUSG00000015656 | Hspa8 | heat shock protein 8 | MGI:105384 |
| ENSMUSG00000024359 | Hspa9 | heat shock protein 9 | MGI:96245 |
| ENSMUSG00000004951 | Hspb1 | heat shock protein 1 | MGI:96240 |
| ENSMUSG00000025980 | Hspd1 | heat shock protein 1 (chaperonin) | MGI:96242 |
| ENSMUSG00000073676 | Hspe1 | heat shock protein 1 (chaperonin 10) | MGI:104680 |
| ENSMUSG00000029657 | Hsph1 | heat shock 105kDa/110kDa protein 1 | MGI:105053 |
| ENSMUSG00000049511 | Htr1b | 5-hydroxytryptamine (serotonin) receptor 1B | MGI:96274 |
| ENSMUSG00000006205 | Htra1 | HtrA serine peptidase 1 | MGI:1929076 |
| ENSMUSG00000068329 | Htra2 | HtrA serine peptidase 2 | MGI:1928676 |
| ENSMUSG00000029104 | Htt | huntingtin | MGI:96067 |
| ENSMUSG00000025261 | Huwe1 | HECT, UBA and WWE domain containing 1 | MGI:1926884 |
| ENSMUSG00000032115 | Hyou1 | hypoxia up-regulated 1 | MGI:108030 |
| ENSMUSG00000026618 | Iars2 | isoleucine-tRNA synthetase 2, mitochondrial | MGI:1919586 |
| ENSMUSG00000037851 | Iars | isoleucine-tRNA synthetase | MGI:2145219 |
| ENSMUSG00000049287 | Iba57 | IBA57, iron-sulfur cluster assembly homolog (S. cerevisiae) | MGI:3041174 |
| ENSMUSG00000032174 | Icam5 | intercellular adhesion molecule 5, telencephalin | MGI:109430 |
| ENSMUSG00000018858 | Ict1 | immature colon carcinoma transcript 1 | MGI:1915822 |
| ENSMUSG00000025950 | Idh1 | isocitrate dehydrogenase 1 (NADP+), soluble | MGI:96413 |
| ENSMUSG00000030541 | Idh2 | isocitrate dehydrogenase 2 (NADP+), mitochondrial | MGI:96414 |
| ENSMUSG00000032279 | Idh3a | isocitrate dehydrogenase 3 (NAD+) alpha | MGI:1915084 |
| ENSMUSG00000027406 | Idh3b | isocitrate dehydrogenase 3 (NAD+) beta | MGI:2158650 |
| ENSMUSG00000002010 | Idh3g | isocitrate dehydrogenase 3 (NAD+), gamma | MGI:1099463 |
| ENSMUSG00000032816 | Igdcc4 | immunoglobulin superfamily, DCC subclass, member 4 | MGI:1858497 |
| ENSMUSG00000005533 | Igf1r | insulin-like growth factor I receptor | MGI:96433 |
| ENSMUSG00000023830 | Igf2r | insulin-like growth factor 2 receptor | MGI:96435 |
| ENSMUSG00000076617 | Ighm | immunoglobulin heavy constant mu | MGI:96448 |
| ENSMUSG00000013367 | Iglon5 | IgLON family member 5 | MGI:2686277 |
| ENSMUSG00000040972 | Igsf21 | immunoglobulin superfamily, member 21 | MGI:2681842 |
| ENSMUSG00000042035 | Igsf3 | immunoglobulin superfamily, member 3 | MGI:1926158 |
| ENSMUSG00000000159 | Igsf5 | immunoglobulin superfamily, member 5 | MGI:1919308 |
| ENSMUSG00000038034 | Igsf8 | immunoglobulin superfamily, member 8 | MGI:2154090 |
| ENSMUSG00000022514 | Il1rap | interleukin 1 receptor accessory protein | MGI:104975 |
| ENSMUSG00000052372 | Il1rapl1 | interleukin 1 receptor accessory protein-like 1 | MGI:2687319 |
| ENSMUSG00000021756 | Il6st | interleukin 6 signal transducer | MGI:96560 |
| ENSMUSG00000040612 | Ildr2 | immunoglobulin-like domain containing receptor 2 | MGI:1196370 |
| ENSMUSG00000032763 | Ilvbl | ilvB (bacterial acetolactate synthase)-like | MGI:1351911 |
| ENSMUSG00000052337 | Immt | inner membrane protein, mitochondrial | MGI:1923864 |
| ENSMUSG00000027531 | Impa1 | inositol (myo)-1(or 4)-monophosphatase 1 | MGI:1933158 |
| ENSMUSG00000024423 | Impact | impact, RWD domain protein | MGI:1098233 |
| ENSMUSG00000066324 | Impad1 | inositol monophosphatase domain containing 1 | MGI:1915720 |
| ENSMUSG00000034336 | Ina | internexin neuronal intermediate filament protein, alpha | MGI:96568 |
| ENSMUSG00000037679 | Inf2 | inverted formin, FH2 and WH2 domain containing | MGI:1917685 |
| ENSMUSG00000026102 | Inpp1 | inositol polyphosphate-1-phosphatase | MGI:104848 |
| ENSMUSG00000026113 | Inpp4a | inositol polyphosphate-4-phosphatase, type I | MGI:1931123 |
| ENSMUSG00000025477 | Inpp5a | inositol polyphosphate-5-phosphatase A | MGI:2686961 |
| ENSMUSG00000030662 | Ipo5 | importin 5 | MGI:1917822 |
| ENSMUSG00000066232 | Ipo7 | importin 7 | MGI:2152414 |
| ENSMUSG00000034312 | Iqsec1 | IQ motif and Sec7 domain 1 | MGI:1196356 |
| ENSMUSG00000041115 | Iqsec2 | IQ motif and Sec7 domain 2 | MGI:3528396 |
| ENSMUSG00000040797 | Iqsec3 | IQ motif and Sec7 domain 3 | MGI:2677208 |
| ENSMUSG00000046879 | Irgm1 | immunity-related GTPase family M member 1 | MGI:107567 |
| ENSMUSG00000021241 | Isca2 | iron-sulfur cluster assembly 2 homolog (S. cerevisiae) | MGI:1921566 |
| ENSMUSG00000051243 | Islr2 | immunoglobulin superfamily containing leucine-rich repeat 2 | MGI:2444277 |
| ENSMUSG00000086784 | Isoc2a | isochorismatase domain containing 2a | MGI:3609243 |
| ENSMUSG00000031729 | Ist1 | increased sodium tolerance 1 homolog (yeast) | MGI:1919205 |
| ENSMUSG00000031703 | Itfg1 | integrin alpha FG-GAP repeat containing 1 | MGI:106419 |
| ENSMUSG00000027111 | Itga6 | integrin alpha 6 | MGI:96605 |
| ENSMUSG00000030786 | Itgam | integrin alpha M | MGI:96607 |
| ENSMUSG00000027087 | Itgav | integrin alpha V | MGI:96608 |
| ENSMUSG00000025809 | Itgb1 | integrin beta 1 (fibronectin receptor beta) | MGI:96610 |
| ENSMUSG00000000290 | Itgb2 | integrin beta 2 | MGI:96611 |
| ENSMUSG00000020689 | Itgb3 | integrin beta 3 | MGI:96612 |
| ENSMUSG00000022817 | Itgb5 | integrin beta 5 | MGI:96614 |
| ENSMUSG00000026971 | Itgb6 | integrin beta 6 | MGI:96615 |
| ENSMUSG00000025321 | Itgb8 | integrin beta 8 | MGI:1338035 |
| ENSMUSG00000006522 | Itih3 | inter-alpha trypsin inhibitor, heavy chain 3 | MGI:96620 |
| ENSMUSG00000022108 | Itm2b | integral membrane protein 2B | MGI:1309517 |
| ENSMUSG00000026223 | Itm2c | integral membrane protein 2C | MGI:1927594 |
| ENSMUSG00000074797 | Itpa | inosine triphosphatase (nucleoside triphosphate pyrophosphatase) | MGI:96622 |
| ENSMUSG00000027296 | Itpka | inositol 1,4,5-trisphosphate 3-kinase A | MGI:1333822 |
| ENSMUSG00000030102 | Itpr1 | inositol 1,4,5-trisphosphate receptor 1 | MGI:96623 |
| ENSMUSG00000030287 | Itpr2 | inositol 1,4,5-triphosphate receptor 2 | MGI:99418 |
| ENSMUSG00000042644 | Itpr3 | inositol 1,4,5-triphosphate receptor 3 | MGI:96624 |
| ENSMUSG00000020640 | Itsn2 | intersectin 2 | MGI:1338049 |
| ENSMUSG00000028530 | Jak1 | Janus kinase 1 | MGI:96628 |
| ENSMUSG00000063646 | Jakmip1 | janus kinase and microtubule interacting protein 1 | MGI:1923321 |
| ENSMUSG00000024502 | Jakmip2 | janus kinase and microtubule interacting protein 2 | MGI:1923467 |
| ENSMUSG00000056856 | Jakmip3 | janus kinase and microtubule interacting protein 3 | MGI:1921254 |
| ENSMUSG00000053062 | Jam2 | junction adhesion molecule 2 | MGI:1933820 |
| ENSMUSG00000031990 | Jam3 | junction adhesion molecule 3 | MGI:1933825 |
| ENSMUSG00000042686 | Jph1 | junctophilin 1 | MGI:1891495 |
| ENSMUSG00000025318 | Jph3 | junctophilin 3 | MGI:1891497 |
| ENSMUSG00000001552 | Jup | junction plakoglobin | MGI:96650 |
| ENSMUSG00000031948 | Kars | lysyl-tRNA synthetase | MGI:1934754 |
| ENSMUSG00000040606 | Kazn | kazrin, periplakin interacting protein | MGI:1918779 |
| ENSMUSG00000047976 | Kcna1 | potassium voltage-gated channel, shaker-related subfamily, member 1 | MGI:96654 |
| ENSMUSG00000040724 | Kcna2 | potassium voltage-gated channel, shaker-related subfamily, member 2 | MGI:96659 |
| ENSMUSG00000047959 | Kcna3 | potassium voltage-gated channel, shaker-related subfamily, member 3 | MGI:96660 |
| ENSMUSG00000042604 | Kcna4 | potassium voltage-gated channel, shaker-related subfamily, member 4 | MGI:96661 |
| ENSMUSG00000038077 | Kcna6 | potassium voltage-gated channel, shaker-related, subfamily, member 6 | MGI:96663 |
| ENSMUSG00000027827 | Kcnab1 | potassium voltage-gated channel, shaker-related subfamily, beta member 1 | MGI:109155 |
| ENSMUSG00000028931 | Kcnab2 | potassium voltage-gated channel, shaker-related subfamily, beta member 2 | MGI:109239 |
| ENSMUSG00000018470 | Kcnab3 | potassium voltage-gated channel, shaker-related subfamily, beta member 3 | MGI:1336208 |
| ENSMUSG00000050556 | Kcnb1 | potassium voltage gated channel, Shab-related subfamily, member 1 | MGI:96666 |
| ENSMUSG00000058975 | Kcnc1 | potassium voltage gated channel, Shaw-related subfamily, member 1 | MGI:96667 |
| ENSMUSG00000062785 | Kcnc3 | potassium voltage gated channel, Shaw-related subfamily, member 3 | MGI:96669 |
| ENSMUSG00000009731 | Kcnd1 | potassium voltage-gated channel, Shal-related family, member 1 | MGI:96671 |
| ENSMUSG00000060882 | Kcnd2 | potassium voltage-gated channel, Shal-related family, member 2 | MGI:102663 |
| ENSMUSG00000040896 | Kcnd3 | potassium voltage-gated channel, Shal-related family, member 3 | MGI:1928743 |
| ENSMUSG00000059742 | Kcnh7 | potassium voltage-gated channel, subfamily H (eag-related), member 7 | MGI:2159566 |
| ENSMUSG00000029088 | Kcnip4 | Kv channel interacting protein 4 | MGI:1933131 |
| ENSMUSG00000044708 | Kcnj10 | potassium inwardly-rectifying channel, subfamily J, member 10 | MGI:1194504 |
| ENSMUSG00000043301 | Kcnj6 | potassium inwardly-rectifying channel, subfamily J, member 6 | MGI:104781 |
| ENSMUSG00000033998 | Kcnk1 | potassium channel, subfamily K, member 1 | MGI:109322 |
| ENSMUSG00000063142 | Kcnma1 | potassium large conductance calcium-activated channel, subfamily M, alpha member 1 | MGI:99923 |
| ENSMUSG00000054477 | Kcnn2 | potassium intermediate/small conductance calcium-activated channel, subfamily N, member 2 | MGI:2153182 |
| ENSMUSG00000016346 | Kcnq2 | potassium voltage-gated channel, subfamily Q, member 2 | MGI:1309503 |
| ENSMUSG00000028033 | Kcnq5 | potassium voltage-gated channel, subfamily Q, member 5 | MGI:1924937 |
| ENSMUSG00000098557 | Kctd12 | potassium channel tetramerisation domain containing 12 | MGI:2145823 |
| ENSMUSG00000051401 | Kctd16 | potassium channel tetramerisation domain containing 16 | MGI:1914659 |
| ENSMUSG00000037653 | Kctd8 | potassium channel tetramerisation domain containing 8 | MGI:2443804 |
| ENSMUSG00000036333 | Kidins220 | kinase D-interacting substrate 220 | MGI:1924730 |
| ENSMUSG00000060012 | Kif13b | kinesin family member 13B | MGI:1098265 |
| ENSMUSG00000014602 | Kif1a | kinesin family member 1A | MGI:108391 |
| ENSMUSG00000022629 | Kif21a | kinesin family member 21A | MGI:109188 |
| ENSMUSG00000021693 | Kif2a | kinesin family member 2A | MGI:108390 |
| ENSMUSG00000074657 | Kif5a | kinesin family member 5A | MGI:109564 |
| ENSMUSG00000006740 | Kif5b | kinesin family member 5B | MGI:1098268 |
| ENSMUSG00000026764 | Kif5c | kinesin family member 5C | MGI:1098269 |
| ENSMUSG00000026585 | Kifap3 | kinesin-associated protein 3 | MGI:107566 |
| ENSMUSG00000032036 | Kirrel3 | kin of IRRE like 3 (Drosophila) | MGI:1914953 |
| ENSMUSG00000005672 | Kit | kit oncogene | MGI:96677 |
| ENSMUSG00000021288 | Klc1 | kinesin light chain 1 | MGI:107978 |
| ENSMUSG00000024862 | Klc2 | kinesin light chain 2 | MGI:107953 |
| ENSMUSG00000021929 | Kpna3 | karyopherin (importin) alpha 3 | MGI:1100863 |
| ENSMUSG00000003731 | Kpna6 | karyopherin (importin) alpha 6 | MGI:1100836 |
| ENSMUSG00000001440 | Kpnb1 | karyopherin (importin) beta 1 | MGI:107532 |
| ENSMUSG00000030265 | Kras | v-Ki-ras2 Kirsten rat sarcoma viral oncogene homolog | MGI:96680 |
| ENSMUSG00000075402 | Krt76 | keratin 76 | MGI:1924305 |
| ENSMUSG00000031391 | L1cam | L1 cell adhesion molecule | MGI:96721 |
| ENSMUSG00000020988 | L2hgdh | L-2-hydroxyglutarate dehydrogenase | MGI:2384968 |
| ENSMUSG00000032370 | Lactb | lactamase, beta | MGI:1933395 |
| ENSMUSG00000032796 | Lama1 | laminin, alpha 1 | MGI:99892 |
| ENSMUSG00000031447 | Lamp1 | lysosomal-associated membrane protein 1 | MGI:96745 |
| ENSMUSG00000016534 | Lamp2 | lysosomal-associated membrane protein 2 | MGI:96748 |
| ENSMUSG00000027270 | Lamp5 | lysosomal-associated membrane protein family, member 5 | MGI:1923411 |
| ENSMUSG00000030842 | Lamtor1 | late endosomal/lysosomal adaptor, MAPK and MTOR activator 1 | MGI:1913758 |
| ENSMUSG00000028062 | Lamtor2 | late endosomal/lysosomal adaptor, MAPK and MTOR activator 2 | MGI:1932697 |
| ENSMUSG00000050552 | Lamtor4 | late endosomal/lysosomal adaptor, MAPK and MTOR activator 4 | MGI:1913346 |
| ENSMUSG00000026000 | Lancl1 | LanC (bacterial lantibiotic synthetase component C)-like 1 | MGI:1336997 |
| ENSMUSG00000062190 | Lancl2 | LanC (bacterial lantibiotic synthetase component C)-like 2 | MGI:1919085 |
| ENSMUSG00000039682 | Lap3 | leucine aminopeptidase 3 | MGI:1914238 |
| ENSMUSG00000004383 | Large | like-glycosyltransferase | MGI:1342270 |
| ENSMUSG00000037331 | Larp1 | La ribonucleoprotein domain family, member 1 | MGI:1890165 |
| ENSMUSG00000035202 | Lars2 | leucyl-tRNA synthetase, mitochondrial | MGI:2142973 |
| ENSMUSG00000024493 | Lars | leucyl-tRNA synthetase | MGI:1913808 |
| ENSMUSG00000038366 | Lasp1 | LIM and SH3 protein 1 | MGI:109656 |
| ENSMUSG00000054469 | Lclat1 | lysocardiolipin acyltransferase 1 | MGI:2684937 |
| ENSMUSG00000037669 | Ldah | lipid droplet associated hydrolase | MGI:1916082 |
| ENSMUSG00000063229 | Ldha | lactate dehydrogenase A | MGI:96759 |
| ENSMUSG00000030246 | Ldhb | lactate dehydrogenase B | MGI:96763 |
| ENSMUSG00000005299 | Letm1 | leucine zipper-EF-hand containing transmembrane protein 1 | MGI:1932557 |
| ENSMUSG00000042363 | Lgalsl | lectin, galactoside binding-like | MGI:1916114 |
| ENSMUSG00000067242 | Lgi1 | leucine-rich repeat LGI family, member 1 | MGI:1861691 |
| ENSMUSG00000039252 | Lgi2 | leucine-rich repeat LGI family, member 2 | MGI:2180196 |
| ENSMUSG00000033595 | Lgi3 | leucine-rich repeat LGI family, member 3 | MGI:2182619 |
| ENSMUSG00000106379 | Lhfpl3 | lipoma HMGIC fusion partner-like 3 | MGI:1925076 |
| ENSMUSG00000023022 | Lima1 | LIM domain and actin binding 1 | MGI:1920992 |
| ENSMUSG00000037736 | Limch1 | LIM and calponin homology domains 1 | MGI:1924819 |
| ENSMUSG00000040699 | Limd2 | LIM domain containing 2 | MGI:1915053 |
| ENSMUSG00000019906 | Lin7a | lin-7 homolog A (C. elegans) | MGI:2135609 |
| ENSMUSG00000003872 | Lin7b | lin-7 homolog B (C. elegans) | MGI:1330858 |
| ENSMUSG00000027162 | Lin7c | lin-7 homolog C (C. elegans) | MGI:1330839 |
| ENSMUSG00000049556 | Lingo1 | leucine rich repeat and Ig domain containing 1 | MGI:1915522 |
| ENSMUSG00000045083 | Lingo2 | leucine rich repeat and Ig domain containing 2 | MGI:2442298 |
| ENSMUSG00000020536 | Llgl1 | lethal giant larvae homolog 1 (Drosophila) | MGI:102682 |
| ENSMUSG00000021484 | Lman2 | lectin, mannose-binding 2 | MGI:1914140 |
| ENSMUSG00000001143 | Lman2l | lectin, mannose-binding 2-like | MGI:2443010 |
| ENSMUSG00000073725 | Lmbrd1 | LMBR1 domain containing 1 | MGI:1915671 |
| ENSMUSG00000039704 | Lmbrd2 | LMBR1 domain containing 2 | MGI:2444173 |
| ENSMUSG00000028063 | Lmna | lamin A | MGI:96794 |
| ENSMUSG00000024590 | Lmnb1 | lamin B1 | MGI:96795 |
| ENSMUSG00000033060 | Lmo7 | LIM domain only 7 | MGI:1353586 |
| ENSMUSG00000038970 | Lmtk2 | lemur tyrosine kinase 2 | MGI:3036247 |
| ENSMUSG00000062044 | Lmtk3 | lemur tyrosine kinase 3 | MGI:3039582 |
| ENSMUSG00000023845 | Lnpep | leucyl/cystinyl aminopeptidase | MGI:2387123 |
| ENSMUSG00000009207 | Lnp | limb and neural patterns | MGI:1918115 |
| ENSMUSG00000042992 | Loh12cr1 | loss of heterozygosity, 12, chromosomal region 1 homolog (human) | MGI:1915024 |
| ENSMUSG00000041168 | Lonp1 | lon peptidase 1, mitochondrial | MGI:1921392 |
| ENSMUSG00000047866 | Lonp2 | lon peptidase 2, peroxisomal | MGI:1914137 |
| ENSMUSG00000021608 | Lpcat1 | lysophosphatidylcholine acyltransferase 1 | MGI:2384812 |
| ENSMUSG00000027134 | Lpcat4 | lysophosphatidylcholine acyltransferase 4 | MGI:2138993 |
| ENSMUSG00000026623 | Lpgat1 | lysophosphatidylglycerol acyltransferase 1 | MGI:2446186 |
| ENSMUSG00000063446 | Lppr1 | lipid phosphate phosphatase-related protein type 1 | MGI:2445015 |
| ENSMUSG00000035835 | Lppr3 | lipid phosphate phosphatase-related protein type 3 | MGI:2388640 |
| ENSMUSG00000044667 | Lppr4 | lipid phosphate phosphatase-related protein type 4 | MGI:106530 |
| ENSMUSG00000030600 | Lrfn1 | leucine rich repeat and fibronectin type III domain containing 1 | MGI:2136810 |
| ENSMUSG00000040490 | Lrfn2 | leucine rich repeat and fibronectin type III domain containing 2 | MGI:1917780 |
| ENSMUSG00000036957 | Lrfn3 | leucine rich repeat and fibronectin type III domain containing 3 | MGI:2442512 |
| ENSMUSG00000045045 | Lrfn4 | leucine rich repeat and fibronectin type III domain containing 4 | MGI:2385612 |
| ENSMUSG00000035653 | Lrfn5 | leucine rich repeat and fibronectin type III domain containing 5 | MGI:2144814 |
| ENSMUSG00000030029 | Lrig1 | leucine-rich repeats and immunoglobulin-like domains 1 | MGI:107935 |
| ENSMUSG00000049252 | Lrp1b | low density lipoprotein-related protein 1B (deleted in tumors) | MGI:2151136 |
| ENSMUSG00000040249 | Lrp1 | low density lipoprotein receptor-related protein 1 | MGI:96828 |
| ENSMUSG00000029103 | Lrpap1 | low density lipoprotein receptor-related protein associated protein 1 | MGI:96829 |
| ENSMUSG00000024120 | Lrpprc | leucine-rich PPR-motif containing | MGI:1919666 |
| ENSMUSG00000021338 | Lrrc16a | leucine rich repeat containing 16A | MGI:1915982 |
| ENSMUSG00000063052 | Lrrc40 | leucine rich repeat containing 40 | MGI:1914394 |
| ENSMUSG00000029028 | Lrrc47 | leucine rich repeat containing 47 | MGI:1920196 |
| ENSMUSG00000047085 | Lrrc4b | leucine rich repeat containing 4B | MGI:3027390 |
| ENSMUSG00000050587 | Lrrc4c | leucine rich repeat containing 4C | MGI:2442636 |
| ENSMUSG00000027286 | Lrrc57 | leucine rich repeat containing 57 | MGI:1913856 |
| ENSMUSG00000020869 | Lrrc59 | leucine rich repeat containing 59 | MGI:2138133 |
| ENSMUSG00000071073 | Lrrc73 | leucine rich repeat containing 73 | MGI:2684934 |
| ENSMUSG00000028176 | Lrrc7 | leucine rich repeat containing 7 | MGI:2676665 |
| ENSMUSG00000007476 | Lrrc8a | leucine rich repeat containing 8A | MGI:2652847 |
| ENSMUSG00000070639 | Lrrc8b | leucine rich repeat containing 8 family, member B | MGI:2141353 |
| ENSMUSG00000054720 | Lrrc8c | leucine rich repeat containing 8 family, member C | MGI:2140839 |
| ENSMUSG00000046079 | Lrrc8d | leucine rich repeat containing 8D | MGI:1922368 |
| ENSMUSG00000034648 | Lrrn1 | leucine rich repeat protein 1, neuronal | MGI:106038 |
| ENSMUSG00000060780 | Lrrtm1 | leucine rich repeat transmembrane neuronal 1 | MGI:2389173 |
| ENSMUSG00000071862 | Lrrtm2 | leucine rich repeat transmembrane neuronal 2 | MGI:2389174 |
| ENSMUSG00000061080 | Lsamp | limbic system-associated membrane protein | MGI:1261760 |
| ENSMUSG00000044847 | Lsm11 | U7 snRNP-specific Sm-like protein LSM11 | MGI:1919540 |
| ENSMUSG00000020922 | Lsm12 | LSM12 homolog (S. cerevisiae) | MGI:1919592 |
| ENSMUSG00000015889 | Lta4h | leukotriene A4 hydrolase | MGI:96836 |
| ENSMUSG00000047557 | Lxn | latexin | MGI:107633 |
| ENSMUSG00000022577 | Ly6h | lymphocyte antigen 6 complex, locus H | MGI:1346030 |
| ENSMUSG00000042228 | Lyn | Yamaguchi sarcoma viral (v-yes-1) oncogene homolog | MGI:96892 |
| ENSMUSG00000026344 | Lypd1 | Ly6/Plaur domain containing 1 | MGI:1919835 |
| ENSMUSG00000025903 | Lypla1 | lysophospholipase 1 | MGI:1344588 |
| ENSMUSG00000046573 | Lyrm4 | LYR motif containing 4 | MGI:2683538 |
| ENSMUSG00000036306 | Lzts1 | leucine zipper, putative tumor suppressor 1 | MGI:2684762 |
| ENSMUSG00000037703 | Lzts3 | leucine zipper, putative tumor suppressor family member 3 | MGI:2656976 |
| ENSMUSG00000007458 | M6pr | mannose-6-phosphate receptor, cation dependent | MGI:96904 |
| ENSMUSG00000036278 | Macrod1 | MACRO domain containing 1 | MGI:2147583 |
| ENSMUSG00000040687 | Madd | MAP-kinase activating death domain | MGI:2444672 |
| ENSMUSG00000045095 | Magi1 | membrane associated guanylate kinase, WW and PDZ domain containing 1 | MGI:1203522 |
| ENSMUSG00000040003 | Magi2 | membrane associated guanylate kinase, WW and PDZ domain containing 2 | MGI:1354953 |
| ENSMUSG00000036634 | Mag | myelin-associated glycoprotein | MGI:96912 |
| ENSMUSG00000024479 | Mal2 | mal, T cell differentiation protein 2 | MGI:2146021 |
| ENSMUSG00000032295 | Man2c1 | mannosidase, alpha, class 2C, member 1 | MGI:1920994 |
| ENSMUSG00000032575 | Manf | mesencephalic astrocyte-derived neurotrophic factor | MGI:1922090 |
| ENSMUSG00000025037 | Maoa | monoamine oxidase A | MGI:96915 |
| ENSMUSG00000040147 | Maob | monoamine oxidase B | MGI:96916 |
| ENSMUSG00000027254 | Map1a | microtubule-associated protein 1 A | MGI:1306776 |
| ENSMUSG00000052727 | Map1b | microtubule-associated protein 1B | MGI:1306778 |
| ENSMUSG00000027602 | Map1lc3a | microtubule-associated protein 1 light chain 3 alpha | MGI:1915661 |
| ENSMUSG00000031812 | Map1lc3b | microtubule-associated protein 1 light chain 3 beta | MGI:1914693 |
| ENSMUSG00000019261 | Map1s | microtubule-associated protein 1S | MGI:2443304 |
| ENSMUSG00000004936 | Map2k1 | mitogen-activated protein kinase kinase 1 | MGI:1346866 |
| ENSMUSG00000033352 | Map2k4 | mitogen-activated protein kinase kinase 4 | MGI:1346869 |
| ENSMUSG00000015222 | Map2 | microtubule-associated protein 2 | MGI:97175 |
| ENSMUSG00000026074 | Map4k4 | mitogen-activated protein kinase kinase kinase kinase 4 | MGI:1349394 |
| ENSMUSG00000032479 | Map4 | microtubule-associated protein 4 | MGI:97178 |
| ENSMUSG00000041205 | Map6d1 | MAP6 domain containing 1 | MGI:3607784 |
| ENSMUSG00000055407 | Map6 | microtubule-associated protein 6 | MGI:1201690 |
| ENSMUSG00000028849 | Map7d1 | MAP7 domain containing 1 | MGI:2384297 |
| ENSMUSG00000041020 | Map7d2 | MAP7 domain containing 2 | MGI:1917474 |
| ENSMUSG00000046709 | Mapk10 | mitogen-activated protein kinase 10 | MGI:1346863 |
| ENSMUSG00000063358 | Mapk1 | mitogen-activated protein kinase 1 | MGI:1346858 |
| ENSMUSG00000063065 | Mapk3 | mitogen-activated protein kinase 3 | MGI:1346859 |
| ENSMUSG00000024163 | Mapk8ip3 | mitogen-activated protein kinase 8 interacting protein 3 | MGI:1353598 |
| ENSMUSG00000027479 | Mapre1 | microtubule-associated protein, RP/EB family, member 1 | MGI:891995 |
| ENSMUSG00000024277 | Mapre2 | microtubule-associated protein, RP/EB family, member 2 | MGI:106271 |
| ENSMUSG00000029166 | Mapre3 | microtubule-associated protein, RP/EB family, member 3 | MGI:2140967 |
| ENSMUSG00000018411 | Mapt | microtubule-associated protein tau | MGI:97180 |
| ENSMUSG00000073481 | Marc2 | mitochondrial amidoxime reducing component 2 | MGI:1914497 |
| ENSMUSG00000023307 | March5 | membrane-associated ring finger (C3HC4) 5 | MGI:1915207 |
| ENSMUSG00000047945 | Marcksl1 | MARCKS-like 1 | MGI:97143 |
| ENSMUSG00000069662 | Marcks | myristoylated alanine rich protein kinase C substrate | MGI:96907 |
| ENSMUSG00000026620 | Mark1 | MAP/microtubule affinity regulating kinase 1 | MGI:2664902 |
| ENSMUSG00000024969 | Mark2 | MAP/microtubule affinity regulating kinase 2 | MGI:99638 |
| ENSMUSG00000007411 | Mark3 | MAP/microtubule affinity regulating kinase 3 | MGI:1341865 |
| ENSMUSG00000030397 | Mark4 | MAP/microtubule affinity regulating kinase 4 | MGI:1920955 |
| ENSMUSG00000040354 | Mars | methionine-tRNA synthetase | MGI:1345633 |
| ENSMUSG00000053907 | Mat2a | methionine adenosyltransferase II, alpha | MGI:2443731 |
| ENSMUSG00000004933 | Matk | megakaryocyte-associated tyrosine kinase | MGI:99259 |
| ENSMUSG00000037236 | Matr3 | matrin 3 | MGI:1298379 |
| ENSMUSG00000037523 | Mavs | mitochondrial antiviral signaling protein | MGI:2444773 |
| ENSMUSG00000051065 | Mb21d2 | Mab-21 domain containing 2 | MGI:1917028 |
| ENSMUSG00000051098 | Mblac2 | metallo-beta-lactamase domain containing 2 | MGI:1920102 |
| ENSMUSG00000035596 | Mboat7 | membrane bound O-acyltransferase domain containing 7 | MGI:1924832 |
| ENSMUSG00000041607 | Mbp | myelin basic protein | MGI:96925 |
| ENSMUSG00000032135 | Mcam | melanoma cell adhesion molecule | MGI:1933966 |
| ENSMUSG00000048755 | Mcat | malonyl CoA:ACP acyltransferase (mitochondrial) | MGI:2388651 |
| ENSMUSG00000027709 | Mccc1 | methylcrotonoyl-Coenzyme A carboxylase 1 (alpha) | MGI:1919289 |
| ENSMUSG00000021646 | Mccc2 | methylcrotonoyl-Coenzyme A carboxylase 2 (beta) | MGI:1925288 |
| ENSMUSG00000031442 | Mcf2l | mcf.2 transforming sequence-like | MGI:103263 |
| ENSMUSG00000004567 | Mcoln1 | mucolipin 1 | MGI:1890498 |
| ENSMUSG00000021596 | Mctp1 | multiple C2 domains, transmembrane 1 | MGI:1926021 |
| ENSMUSG00000009647 | Mcu | mitochondrial calcium uniporter | MGI:3026965 |
| ENSMUSG00000021371 | Mcur1 | mitochondrial calcium uniporter regulator 1 | MGI:1923387 |
| ENSMUSG00000034912 | Mdga2 | MAM domain containing glycosylphosphatidylinositol anchor 2 | MGI:2444706 |
| ENSMUSG00000020321 | Mdh1 | malate dehydrogenase 1, NAD (soluble) | MGI:97051 |
| ENSMUSG00000019179 | Mdh2 | malate dehydrogenase 2, NAD (mitochondrial) | MGI:97050 |
| ENSMUSG00000032418 | Me1 | malic enzyme 1, NADP(+)-dependent, cytosolic | MGI:97043 |
| ENSMUSG00000024556 | Me2 | malic enzyme 2, NAD(+)-dependent, mitochondrial | MGI:2147351 |
| ENSMUSG00000030621 | Me3 | malic enzyme 3, NADP(+)-dependent, mitochondrial | MGI:1916679 |
| ENSMUSG00000028910 | Mecr | mitochondrial trans-2-enoyl-CoA reductase | MGI:1349441 |
| ENSMUSG00000014361 | Mertk | c-mer proto-oncogene tyrosine kinase | MGI:96965 |
| ENSMUSG00000038503 | Mesdc2 | mesoderm development candidate 2 | MGI:1891421 |
| ENSMUSG00000054619 | Mettl7a1 | methyltransferase like 7A1 | MGI:1916523 |
| ENSMUSG00000026150 | Mff | mitochondrial fission factor | MGI:1922984 |
| ENSMUSG00000030605 | Mfge8 | milk fat globule-EGF factor 8 protein | MGI:102768 |
| ENSMUSG00000027668 | Mfn1 | mitofusin 1 | MGI:1914664 |
| ENSMUSG00000029020 | Mfn2 | mitofusin 2 | MGI:2442230 |
| ENSMUSG00000041439 | Mfsd6 | major facilitator superfamily domain containing 6 | MGI:1922925 |
| ENSMUSG00000033174 | Mgll | monoglyceride lipase | MGI:1346042 |
| ENSMUSG00000008540 | Mgst1 | microsomal glutathione S-transferase 1 | MGI:1913850 |
| ENSMUSG00000026688 | Mgst3 | microsomal glutathione S-transferase 3 | MGI:1913697 |
| ENSMUSG00000056050 | Mia3 | melanoma inhibitory activity 3 | MGI:2443183 |
| ENSMUSG00000003178 | Mical3 | microtubule associated monooxygenase, calponin and LIM domain containing 3 | MGI:2442733 |
| ENSMUSG00000020111 | Micu1 | mitochondrial calcium uptake 1 | MGI:2384909 |
| ENSMUSG00000039478 | Micu3 | mitochondrial calcium uptake family, member 3 | MGI:1925756 |
| ENSMUSG00000033307 | Mif | macrophage migration inhibitory factor | MGI:96982 |
| ENSMUSG00000020827 | Mink1 | misshapen-like kinase 1 (zebrafish) | MGI:1355329 |
| ENSMUSG00000042447 | Mios | missing oocyte, meiosis regulator, homolog (Drosophila) | MGI:2182066 |
| ENSMUSG00000035805 | Mlc1 | megalencephalic leukoencephalopathy with subcortical cysts 1 homolog (human) | MGI:2157910 |
| ENSMUSG00000048578 | Mlec | malectin | MGI:1924015 |
| ENSMUSG00000030120 | Mlf2 | myeloid leukemia factor 2 | MGI:1353554 |
| ENSMUSG00000032355 | Mlip | muscular LMNA-interacting protein | MGI:1916892 |
| ENSMUSG00000068036 | Mllt4 | myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila) translocated to, 4 | MGI:1314653 |
| ENSMUSG00000074064 | Mlycd | malonyl-CoA decarboxylase | MGI:1928485 |
| ENSMUSG00000037022 | Mmaa | methylmalonic aciduria (cobalamin deficiency) type A | MGI:1923805 |
| ENSMUSG00000029575 | Mmab | methylmalonic aciduria (cobalamin deficiency) type B homolog (human) | MGI:1924947 |
| ENSMUSG00000061273 | Mmgt1 | membrane magnesium transporter 1 | MGI:2384305 |
| ENSMUSG00000025979 | Mob4 | MOB family member 4, phocein | MGI:104899 |
| ENSMUSG00000076439 | Mog | myelin oligodendrocyte glycoprotein | MGI:97435 |
| ENSMUSG00000030036 | Mogs | mannosyl-oligosaccharide glucosidase | MGI:1929872 |
| ENSMUSG00000032583 | Mon1a | MON1 homolog A (yeast) | MGI:1920075 |
| ENSMUSG00000078908 | Mon1b | MON1 homolog b (yeast) | MGI:1923231 |
| ENSMUSG00000034602 | Mon2 | MON2 homolog (yeast) | MGI:1914324 |
| ENSMUSG00000023861 | Mpc1 | mitochondrial pyruvate carrier 1 | MGI:1915240 |
| ENSMUSG00000026568 | Mpc2 | mitochondrial pyruvate carrier 2 | MGI:1917706 |
| ENSMUSG00000028402 | Mpdz | multiple PDZ domain protein | MGI:1343489 |
| ENSMUSG00000031402 | Mpp1 | membrane protein, palmitoylated | MGI:105941 |
| ENSMUSG00000017314 | Mpp2 | membrane protein, palmitoylated 2 (MAGUK p55 subfamily member 2) | MGI:1858257 |
| ENSMUSG00000052373 | Mpp3 | membrane protein, palmitoylated 3 (MAGUK p55 subfamily member 3) | MGI:1328354 |
| ENSMUSG00000038388 | Mpp6 | membrane protein, palmitoylated 6 (MAGUK p55 subfamily member 6) | MGI:1927340 |
| ENSMUSG00000005417 | Mprip | myosin phosphatase Rho interacting protein | MGI:1349438 |
| ENSMUSG00000071711 | Mpst | mercaptopyruvate sulfurtransferase | MGI:2179733 |
| ENSMUSG00000107283 | Mpv17 | MpV17 mitochondrial inner membrane protein | MGI:97138 |
| ENSMUSG00000032470 | Mras | muscle and microspikes RAS | MGI:1100856 |
| ENSMUSG00000024902 | Mrpl11 | mitochondrial ribosomal protein L11 | MGI:2137215 |
| ENSMUSG00000039640 | Mrpl12 | mitochondrial ribosomal protein L12 | MGI:1926273 |
| ENSMUSG00000020514 | Mrpl22 | mitochondrial ribosomal protein L22 | MGI:1333794 |
| ENSMUSG00000037772 | Mrpl23 | mitochondrial ribosomal protein L23 | MGI:1196612 |
| ENSMUSG00000028622 | Mrpl37 | mitochondrial ribosomal protein L37 | MGI:1926268 |
| ENSMUSG00000022889 | Mrpl39 | mitochondrial ribosomal protein L39 | MGI:1351620 |
| ENSMUSG00000022706 | Mrpl40 | mitochondrial ribosomal protein L40 | MGI:1332635 |
| ENSMUSG00000025208 | Mrpl43 | mitochondrial ribosomal protein L43 | MGI:2137229 |
| ENSMUSG00000030612 | Mrpl46 | mitochondrial ribosomal protein L46 | MGI:1914558 |
| ENSMUSG00000030706 | Mrpl48 | mitochondrial ribosomal protein L48 | MGI:1289321 |
| ENSMUSG00000007338 | Mrpl49 | mitochondrial ribosomal protein L49 | MGI:108180 |
| ENSMUSG00000030037 | Mrpl53 | mitochondrial ribosomal protein L53 | MGI:1915749 |
| ENSMUSG00000034729 | Mrps10 | mitochondrial ribosomal protein S10 | MGI:1928139 |
| ENSMUSG00000054312 | Mrps21 | mitochondrial ribosomal protein S21 | MGI:1913542 |
| ENSMUSG00000032459 | Mrps22 | mitochondrial ribosomal protein S22 | MGI:1928137 |
| ENSMUSG00000023723 | Mrps23 | mitochondrial ribosomal protein S23 | MGI:1928138 |
| ENSMUSG00000037740 | Mrps26 | mitochondrial ribosomal protein S26 | MGI:1333830 |
| ENSMUSG00000041632 | Mrps27 | mitochondrial ribosomal protein S27 | MGI:1919064 |
| ENSMUSG00000040269 | Mrps28 | mitochondrial ribosomal protein S28 | MGI:1913480 |
| ENSMUSG00000040112 | Mrps35 | mitochondrial ribosomal protein S35 | MGI:2385255 |
| ENSMUSG00000061474 | Mrps36 | mitochondrial ribosomal protein S36 | MGI:1913378 |
| ENSMUSG00000026887 | Mrrf | mitochondrial ribosome recycling factor | MGI:1915121 |
| ENSMUSG00000031207 | Msn | moesin | MGI:97167 |
| ENSMUSG00000054733 | Msra | methionine sulfoxide reductase A | MGI:106916 |
| ENSMUSG00000064356 | mt-Atp8 | mitochondrially encoded ATP synthase 8 | MGI:99926 |
| ENSMUSG00000024012 | Mtch1 | mitochondrial carrier homolog 1 (C. elegans) | MGI:1929261 |
| ENSMUSG00000027282 | Mtch2 | mitochondrial carrier homolog 2 (C. elegans) | MGI:1929260 |
| ENSMUSG00000064351 | mt-Co1 | mitochondrially encoded cytochrome c oxidase I | MGI:102504 |
| ENSMUSG00000064354 | mt-Co2 | mitochondrially encoded cytochrome c oxidase II | MGI:102503 |
| ENSMUSG00000064358 | mt-Co3 | mitochondrially encoded cytochrome c oxidase III | MGI:102502 |
| ENSMUSG00000022255 | Mtdh | metadherin | MGI:1914404 |
| ENSMUSG00000004748 | Mtfp1 | mitochondrial fission process 1 | MGI:1916686 |
| ENSMUSG00000046671 | Mtfr1l | mitochondrial fission regulator 1-like | MGI:1924074 |
| ENSMUSG00000040675 | Mthfd1l | methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1-like | MGI:1924836 |
| ENSMUSG00000021048 | Mthfd1 | methylenetetrahydrofolate dehydrogenase (NADP+ dependent), methenyltetrahydrofolate cyclohydrolase, formyltetrahydrofolate synthase | MGI:1342005 |
| ENSMUSG00000015214 | Mtmr1 | myotubularin related protein 1 | MGI:1858271 |
| ENSMUSG00000031918 | Mtmr2 | myotubularin related protein 2 | MGI:1924366 |
| ENSMUSG00000064341 | mt-Nd1 | mitochondrially encoded NADH dehydrogenase 1 | MGI:101787 |
| ENSMUSG00000064345 | mt-Nd2 | mitochondrially encoded NADH dehydrogenase 2 | MGI:102500 |
| ENSMUSG00000064363 | mt-Nd4 | mitochondrially encoded NADH dehydrogenase 4 | MGI:102498 |
| ENSMUSG00000064367 | mt-Nd5 | mitochondrially encoded NADH dehydrogenase 5 | MGI:102496 |
| ENSMUSG00000028991 | Mtor | mechanistic target of rapamycin (serine/threonine kinase) | MGI:1928394 |
| ENSMUSG00000024234 | Mtpap | mitochondrial poly(A) polymerase | MGI:1914690 |
| ENSMUSG00000029840 | Mtpn | myotrophin | MGI:99445 |
| ENSMUSG00000033763 | Mtss1l | metastasis suppressor 1-like | MGI:3039591 |
| ENSMUSG00000029651 | Mtus2 | microtubule associated tumor suppressor candidate 2 | MGI:1915388 |
| ENSMUSG00000064068 | Mtx1 | metaxin 1 | MGI:103025 |
| ENSMUSG00000027099 | Mtx2 | metaxin 2 | MGI:1859652 |
| ENSMUSG00000021704 | Mtx3 | metaxin 3 | MGI:2686040 |
| ENSMUSG00000023921 | Mut | methylmalonyl-Coenzyme A mutase | MGI:97239 |
| ENSMUSG00000038740 | Mvb12b | multivesicular body subunit 12B | MGI:1919793 |
| ENSMUSG00000068566 | Myadm | myeloid-associated differentiation marker | MGI:1355332 |
| ENSMUSG00000033004 | Mycbp2 | MYC binding protein 2 | MGI:2179432 |
| ENSMUSG00000019579 | Mydgf | myeloid derived growth factor | MGI:2156020 |
| ENSMUSG00000020900 | Myh10 | myosin, heavy polypeptide 10, non-muscle | MGI:1930780 |
| ENSMUSG00000018830 | Myh11 | myosin, heavy polypeptide 11, smooth muscle | MGI:102643 |
| ENSMUSG00000030739 | Myh14 | myosin, heavy polypeptide 14 | MGI:1919210 |
| ENSMUSG00000033196 | Myh2 | myosin, heavy polypeptide 2, skeletal muscle, adult | MGI:1339710 |
| ENSMUSG00000040752 | Myh6 | myosin, heavy polypeptide 6, cardiac muscle, alpha | MGI:97255 |
| ENSMUSG00000022443 | Myh9 | myosin, heavy polypeptide 9, non-muscle | MGI:107717 |
| ENSMUSG00000034868 | Myl12b | myosin, light chain 12B, regulatory | MGI:107494 |
| ENSMUSG00000061816 | Myl1 | myosin, light polypeptide 1 | MGI:97269 |
| ENSMUSG00000039824 | Myl6b | myosin, light polypeptide 6B | MGI:1917789 |
| ENSMUSG00000090841 | Myl6 | myosin, light polypeptide 6, alkali, smooth muscle and non-muscle | MGI:109318 |
| ENSMUSG00000022836 | Mylk | myosin, light polypeptide kinase | MGI:894806 |
| ENSMUSG00000000631 | Myo18a | myosin XVIIIA | MGI:2667185 |
| ENSMUSG00000018417 | Myo1b | myosin IB | MGI:107752 |
| ENSMUSG00000017774 | Myo1c | myosin IC | MGI:106612 |
| ENSMUSG00000035441 | Myo1d | myosin ID | MGI:107728 |
| ENSMUSG00000034593 | Myo5a | myosin VA | MGI:105976 |
| ENSMUSG00000041794 | Myrip | myosin VIIA and Rab interacting protein | MGI:2384407 |
| ENSMUSG00000022671 | Mzt2 | mitotic spindle organizing protein 2 | MGI:1922845 |
| ENSMUSG00000036062 | N28178 | expressed sequence N28178 | MGI:2140712 |
| ENSMUSG00000022698 | Naa50 | N(alpha)-acetyltransferase 50, NatE catalytic subunit | MGI:1919367 |
| ENSMUSG00000041073 | Nacad | NAC alpha domain containing | MGI:3603030 |
| ENSMUSG00000022253 | Nadk2 | NAD kinase 2, mitochondrial | MGI:1915896 |
| ENSMUSG00000028334 | Nans | N-acetylneuraminic acid synthase (sialic acid synthase) | MGI:2149820 |
| ENSMUSG00000058799 | Nap1l1 | nucleosome assembly protein 1-like 1 | MGI:1855693 |
| ENSMUSG00000059119 | Nap1l4 | nucleosome assembly protein 1-like 4 | MGI:1316687 |
| ENSMUSG00000006024 | Napa | N-ethylmaleimide sensitive fusion protein attachment protein alpha | MGI:104563 |
| ENSMUSG00000027438 | Napb | N-ethylmaleimide sensitive fusion protein attachment protein beta | MGI:104562 |
| ENSMUSG00000024581 | Napg | N-ethylmaleimide sensitive fusion protein attachment protein gamma | MGI:104561 |
| ENSMUSG00000024587 | Nars | asparaginyl-tRNA synthetase | MGI:1917473 |
| ENSMUSG00000009418 | Nav1 | neuron navigator 1 | MGI:2183683 |
| ENSMUSG00000020576 | Nbas | neuroblastoma amplified sequence | MGI:1918419 |
| ENSMUSG00000027799 | Nbea | neurobeachin | MGI:1347075 |
| ENSMUSG00000051359 | Ncald | neurocalcin delta | MGI:1196326 |
| ENSMUSG00000039542 | Ncam1 | neural cell adhesion molecule 1 | MGI:97281 |
| ENSMUSG00000022762 | Ncam2 | neural cell adhesion molecule 2 | MGI:97282 |
| ENSMUSG00000002341 | Ncan | neurocan | MGI:104694 |
| ENSMUSG00000028833 | Ncdn | neurochondrin | MGI:1347351 |
| ENSMUSG00000027698 | Nceh1 | neutral cholesterol ester hydrolase 1 | MGI:2443191 |
| ENSMUSG00000027002 | Nckap1 | NCK-associated protein 1 | MGI:1355333 |
| ENSMUSG00000032598 | Nckipsd | NCK interacting protein with SH3 domain | MGI:1931834 |
| ENSMUSG00000020238 | Ncln | nicalin homolog (zebrafish) | MGI:1926081 |
| ENSMUSG00000026234 | Ncl | nucleolin | MGI:97286 |
| ENSMUSG00000062661 | Ncs1 | neuronal calcium sensor 1 | MGI:109166 |
| ENSMUSG00000003458 | Ncstn | nicastrin | MGI:1891700 |
| ENSMUSG00000028614 | Ndc1 | NDC1 transmembrane nucleoporin | MGI:1920037 |
| ENSMUSG00000018736 | Ndel1 | nuclear distribution gene E-like homolog 1 (A. nidulans) | MGI:1932915 |
| ENSMUSG00000053253 | Ndfip2 | Nedd4 family interacting protein 2 | MGI:1923523 |
| ENSMUSG00000005125 | Ndrg1 | N-myc downstream regulated gene 1 | MGI:1341799 |
| ENSMUSG00000004558 | Ndrg2 | N-myc downstream regulated gene 2 | MGI:1352498 |
| ENSMUSG00000027634 | Ndrg3 | N-myc downstream regulated gene 3 | MGI:1352499 |
| ENSMUSG00000036564 | Ndrg4 | N-myc downstream regulated gene 4 | MGI:2384590 |
| ENSMUSG00000026260 | Ndufa10 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex 10 | MGI:1914523 |
| ENSMUSG00000002379 | Ndufa11 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex 11 | MGI:1917125 |
| ENSMUSG00000020022 | Ndufa12 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 12 | MGI:1913664 |
| ENSMUSG00000036199 | Ndufa13 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 13 | MGI:1914434 |
| ENSMUSG00000016427 | Ndufa1 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 1 | MGI:1929511 |
| ENSMUSG00000014294 | Ndufa2 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 2 | MGI:1343103 |
| ENSMUSG00000035674 | Ndufa3 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 3 | MGI:1913341 |
| ENSMUSG00000029632 | Ndufa4 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 4 | MGI:107686 |
| ENSMUSG00000023089 | Ndufa5 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 5 | MGI:1915452 |
| ENSMUSG00000022450 | Ndufa6 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 6 (B14) | MGI:1914380 |
| ENSMUSG00000041881 | Ndufa7 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 7 (B14.5a) | MGI:1913666 |
| ENSMUSG00000026895 | Ndufa8 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 8 | MGI:1915625 |
| ENSMUSG00000000399 | Ndufa9 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 9 | MGI:1913358 |
| ENSMUSG00000030869 | Ndufab1 | NADH dehydrogenase (ubiquinone) 1, alpha/beta subcomplex, 1 | MGI:1917566 |
| ENSMUSG00000068184 | Ndufaf2 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, assembly factor 2 | MGI:1922847 |
| ENSMUSG00000070283 | Ndufaf3 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, assembly factor 3 | MGI:1913956 |
| ENSMUSG00000028261 | Ndufaf4 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, assembly factor 4 | MGI:1915743 |
| ENSMUSG00000050323 | Ndufaf6 | NADH dehydrogenase (ubiquinone) complex I, assembly factor 6 | MGI:1924197 |
| ENSMUSG00000040048 | Ndufb10 | NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 10 | MGI:1915592 |
| ENSMUSG00000031059 | Ndufb11 | NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 11 | MGI:1349919 |
| ENSMUSG00000002416 | Ndufb2 | NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 2 | MGI:1915448 |
| ENSMUSG00000026032 | Ndufb3 | NADH dehydrogenase (ubiquinone) 1 beta subcomplex 3 | MGI:1913745 |
| ENSMUSG00000022820 | Ndufb4 | NADH dehydrogenase (ubiquinone) 1 beta subcomplex 4 | MGI:1915444 |
| ENSMUSG00000027673 | Ndufb5 | NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 5 | MGI:1913296 |
| ENSMUSG00000071014 | Ndufb6 | NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 6 | MGI:2684983 |
| ENSMUSG00000033938 | Ndufb7 | NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 7 | MGI:1914166 |
| ENSMUSG00000025204 | Ndufb8 | NADH dehydrogenase (ubiquinone) 1 beta subcomplex 8 | MGI:1914514 |
| ENSMUSG00000022354 | Ndufb9 | NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 9 | MGI:1913468 |
| ENSMUSG00000030647 | Ndufc2 | NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 2 | MGI:1344370 |
| ENSMUSG00000025968 | Ndufs1 | NADH dehydrogenase (ubiquinone) Fe-S protein 1 | MGI:2443241 |
| ENSMUSG00000013593 | Ndufs2 | NADH dehydrogenase (ubiquinone) Fe-S protein 2 | MGI:2385112 |
| ENSMUSG00000005510 | Ndufs3 | NADH dehydrogenase (ubiquinone) Fe-S protein 3 | MGI:1915599 |
| ENSMUSG00000021764 | Ndufs4 | NADH dehydrogenase (ubiquinone) Fe-S protein 4 | MGI:1343135 |
| ENSMUSG00000028648 | Ndufs5 | NADH dehydrogenase (ubiquinone) Fe-S protein 5 | MGI:1890889 |
| ENSMUSG00000021606 | Ndufs6 | NADH dehydrogenase (ubiquinone) Fe-S protein 6 | MGI:107932 |
| ENSMUSG00000020153 | Ndufs7 | NADH dehydrogenase (ubiquinone) Fe-S protein 7 | MGI:1922656 |
| ENSMUSG00000059734 | Ndufs8 | NADH dehydrogenase (ubiquinone) Fe-S protein 8 | MGI:2385079 |
| ENSMUSG00000037916 | Ndufv1 | NADH dehydrogenase (ubiquinone) flavoprotein 1 | MGI:107851 |
| ENSMUSG00000024099 | Ndufv2 | NADH dehydrogenase (ubiquinone) flavoprotein 2 | MGI:1920150 |
| ENSMUSG00000024038 | Ndufv3 | NADH dehydrogenase (ubiquinone) flavoprotein 3 | MGI:1890894 |
| ENSMUSG00000053702 | Nebl | nebulette | MGI:1921353 |
| ENSMUSG00000031837 | Necab2 | N-terminal EF-hand calcium binding protein 2 | MGI:2152211 |
| ENSMUSG00000030327 | Necap1 | NECAP endocytosis associated 1 | MGI:1914852 |
| ENSMUSG00000024589 | Nedd4l | neural precursor cell expressed, developmentally down-regulated gene 4-like | MGI:1933754 |
| ENSMUSG00000032216 | Nedd4 | neural precursor cell expressed, developmentally down-regulated 4 | MGI:97297 |
| ENSMUSG00000010376 | Nedd8 | neural precursor cell expressed, developmentally down-regulated gene 8 | MGI:97301 |
| ENSMUSG00000020396 | Nefh | neurofilament, heavy polypeptide | MGI:97309 |
| ENSMUSG00000022055 | Nefl | neurofilament, light polypeptide | MGI:97313 |
| ENSMUSG00000022054 | Nefm | neurofilament, medium polypeptide | MGI:97314 |
| ENSMUSG00000040037 | Negr1 | neuronal growth regulator 1 | MGI:2444846 |
| ENSMUSG00000022454 | Nell2 | NEL-like 2 | MGI:1858510 |
| ENSMUSG00000037499 | Nenf | neuron derived neurotrophic factor | MGI:1913458 |
| ENSMUSG00000032340 | Neo1 | neogenin | MGI:1097159 |
| ENSMUSG00000020716 | Nf1 | neurofibromatosis 1 | MGI:97306 |
| ENSMUSG00000026442 | Nfasc | neurofascin | MGI:104753 |
| ENSMUSG00000027618 | Nfs1 | nitrogen fixation gene 1 (S. cerevisiae) | MGI:1316706 |
| ENSMUSG00000026259 | Ngef | neuronal guanine nucleotide exchange factor | MGI:1858414 |
| ENSMUSG00000079481 | Nhsl2 | NHS-like 2 | MGI:3645090 |
| ENSMUSG00000026036 | Nif3l1 | Ngg1 interacting factor 3-like 1 (S. pombe) | MGI:1929485 |
| ENSMUSG00000047037 | Nipa1 | non imprinted in Prader-Willi/Angelman syndrome 1 homolog (human) | MGI:2442058 |
| ENSMUSG00000034285 | Nipsnap1 | 4-nitrophenylphosphatase domain and non-neuronal SNAP25-like protein homolog 1 (C. elegans) | MGI:1278344 |
| ENSMUSG00000015247 | Nipsnap3b | nipsnap homolog 3B (C. elegans) | MGI:1913786 |
| ENSMUSG00000021910 | Nisch | nischarin | MGI:1928323 |
| ENSMUSG00000021772 | Nkiras1 | NFKB inhibitor interacting Ras-like protein 1 | MGI:1916971 |
| ENSMUSG00000017837 | Nkiras2 | NFKB inhibitor interacting Ras-like protein 2 | MGI:1919216 |
| ENSMUSG00000029112 | Nkx1-1 | NK1 transcription factor related, locus 1 (Drosophila) | MGI:109346 |
| ENSMUSG00000063887 | Nlgn1 | neuroligin 1 | MGI:2179435 |
| ENSMUSG00000051790 | Nlgn2 | neuroligin 2 | MGI:2681835 |
| ENSMUSG00000031302 | Nlgn3 | neuroligin 3 | MGI:2444609 |
| ENSMUSG00000021710 | Nln | neurolysin (metallopeptidase M3 family) | MGI:1923055 |
| ENSMUSG00000032109 | Nlrx1 | NLR family member X1 | MGI:2429611 |
| ENSMUSG00000037601 | Nme1 | NME/NM23 nucleoside diphosphate kinase 1 | MGI:97355 |
| ENSMUSG00000073435 | Nme3 | NME/NM23 nucleoside diphosphate kinase 3 | MGI:1930182 |
| ENSMUSG00000025453 | Nnt | nicotinamide nucleotide transhydrogenase | MGI:109279 |
| ENSMUSG00000030835 | Nomo1 | nodal modulator 1 | MGI:2385850 |
| ENSMUSG00000031311 | Nono | non-POU-domain-containing, octamer binding protein | MGI:1855692 |
| ENSMUSG00000029361 | Nos1 | nitric oxide synthase 1, neuronal | MGI:97360 |
| ENSMUSG00000021047 | Nova1 | neuro-oncological ventral antigen 1 | MGI:104297 |
| ENSMUSG00000024413 | Npc1 | Niemann-Pick type C1 | MGI:1097712 |
| ENSMUSG00000015094 | Npdc1 | neural proliferation, differentiation and control 1 | MGI:1099802 |
| ENSMUSG00000001441 | Npepps | aminopeptidase puromycin sensitive | MGI:1101358 |
| ENSMUSG00000032558 | Nphp3 | nephronophthisis 3 (adolescent) | MGI:1921275 |
| ENSMUSG00000057113 | Npm1 | nucleophosmin 1 | MGI:106184 |
| ENSMUSG00000032336 | Nptn | neuroplastin | MGI:108077 |
| ENSMUSG00000025582 | Nptx1 | neuronal pentraxin 1 | MGI:107811 |
| ENSMUSG00000022421 | Nptxr | neuronal pentraxin receptor | MGI:1920590 |
| ENSMUSG00000027852 | Nras | neuroblastoma ras oncogene | MGI:97376 |
| ENSMUSG00000020598 | Nrcam | neuronal cell adhesion molecule | MGI:104750 |
| ENSMUSG00000039114 | Nrn1 | neuritin 1 | MGI:1915654 |
| ENSMUSG00000025810 | Nrp1 | neuropilin 1 | MGI:106206 |
| ENSMUSG00000025969 | Nrp2 | neuropilin 2 | MGI:1100492 |
| ENSMUSG00000024109 | Nrxn1 | neurexin I | MGI:1096391 |
| ENSMUSG00000033768 | Nrxn2 | neurexin II | MGI:1096362 |
| ENSMUSG00000066392 | Nrxn3 | neurexin III | MGI:1096389 |
| ENSMUSG00000031349 | Nsdhl | NAD(P) dependent steroid dehydrogenase-like | MGI:1099438 |
| ENSMUSG00000027455 | Nsfl1c | NSFL1 (p97) cofactor (p47) | MGI:3042273 |
| ENSMUSG00000034187 | Nsf | N-ethylmaleimide sensitive fusion protein | MGI:104560 |
| ENSMUSG00000029126 | Nsg1 | neuron specific gene family member 1 | MGI:109149 |
| ENSMUSG00000020297 | Nsg2 | neuron specific gene family member 2 | MGI:1202070 |
| ENSMUSG00000006476 | Nsmf | NMDA receptor synaptonuclear signaling and neuronal migration factor | MGI:1861755 |
| ENSMUSG00000020736 | Nt5c | 5',3'-nucleotidase, cytosolic | MGI:1354954 |
| ENSMUSG00000054027 | Nt5dc3 | 5'-nucleotidase domain containing 3 | MGI:3513266 |
| ENSMUSG00000032420 | Nt5e | 5' nucleotidase, ecto | MGI:99782 |
| ENSMUSG00000032615 | Nt5m | 5',3'-nucleotidase, mitochondrial | MGI:1917127 |
| ENSMUSG00000059974 | Ntm | neurotrimin | MGI:2446259 |
| ENSMUSG00000059857 | Ntng1 | netrin G1 | MGI:1934028 |
| ENSMUSG00000055254 | Ntrk2 | neurotrophic tyrosine kinase, receptor, type 2 | MGI:97384 |
| ENSMUSG00000059146 | Ntrk3 | neurotrophic tyrosine kinase, receptor, type 3 | MGI:97385 |
| ENSMUSG00000030824 | Nucb1 | nucleobindin 1 | MGI:97388 |
| ENSMUSG00000030659 | Nucb2 | nucleobindin 2 | MGI:1858179 |
| ENSMUSG00000028851 | Nudc | nuclear distribution gene C homolog (Aspergillus) | MGI:106014 |
| ENSMUSG00000045211 | Nudt18 | nudix (nucleoside diphosphate linked moiety X)-type motif 18 | MGI:2385853 |
| ENSMUSG00000028443 | Nudt2 | nudix (nucleoside diphosphate linked moiety X)-type motif 2 | MGI:1913651 |
| ENSMUSG00000024213 | Nudt3 | nudix (nucleotide diphosphate linked moiety X)-type motif 3 | MGI:1928484 |
| ENSMUSG00000063160 | Numbl | numb-like | MGI:894702 |
| ENSMUSG00000021224 | Numb | numb gene homolog (Drosophila) | MGI:107423 |
| ENSMUSG00000054976 | Nyap2 | neuronal tyrosine-phophorylated phosphoinositide 3-kinase adaptor 2 | MGI:2443135 |
| ENSMUSG00000030934 | Oat | ornithine aminotransferase | MGI:97394 |
| ENSMUSG00000029152 | Ociad1 | OCIA domain containing 1 | MGI:1915345 |
| ENSMUSG00000029153 | Ociad2 | OCIA domain containing 2 | MGI:1916377 |
| ENSMUSG00000001173 | Ocrl | oculocerebrorenal syndrome of Lowe | MGI:109589 |
| ENSMUSG00000021913 | Ogdhl | oxoglutarate dehydrogenase-like | MGI:3616088 |
| ENSMUSG00000020456 | Ogdh | oxoglutarate (alpha-ketoglutarate) dehydrogenase (lipoamide) | MGI:1098267 |
| ENSMUSG00000034160 | Ogt | O-linked N-acetylglucosamine (GlcNAc) transferase (UDP-N-acetylglucosamine:polypeptide-N-acetylglucosaminyl transferase) | MGI:1339639 |
| ENSMUSG00000027108 | Ola1 | Obg-like ATPase 1 | MGI:1914309 |
| ENSMUSG00000026833 | Olfm1 | olfactomedin 1 | MGI:1860437 |
| ENSMUSG00000032172 | Olfm2 | olfactomedin 2 | MGI:3045350 |
| ENSMUSG00000027965 | Olfm3 | olfactomedin 3 | MGI:2387329 |
| ENSMUSG00000049612 | Omg | oligodendrocyte myelin glycoprotein | MGI:106586 |
| ENSMUSG00000038084 | Opa1 | optic atrophy 1 | MGI:1921393 |
| ENSMUSG00000052214 | Opa3 | optic atrophy 3 | MGI:2686271 |
| ENSMUSG00000050121 | Opalin | oligodendrocytic myelin paranodal and inner loop protein | MGI:2657025 |
| ENSMUSG00000062257 | Opcml | opioid binding protein/cell adhesion molecule-like | MGI:97397 |
| ENSMUSG00000038150 | Ormdl3 | ORM1-like 3 (S. cerevisiae) | MGI:1913862 |
| ENSMUSG00000040462 | Os9 | amplified in osteosarcoma | MGI:1924301 |
| ENSMUSG00000040875 | Osbpl10 | oxysterol binding protein-like 10 | MGI:1921736 |
| ENSMUSG00000020189 | Osbpl8 | oxysterol binding protein-like 8 | MGI:2443807 |
| ENSMUSG00000024687 | Osbp | oxysterol binding protein | MGI:97447 |
| ENSMUSG00000024767 | Otub1 | OTU domain, ubiquitin aldehyde binding 1 | MGI:2147616 |
| ENSMUSG00000022186 | Oxct1 | 3-oxoacid CoA transferase 1 | MGI:1914291 |
| ENSMUSG00000022307 | Oxr1 | oxidation resistance 1 | MGI:2179326 |
| ENSMUSG00000036353 | P2ry12 | purinergic receptor P2Y, G-protein coupled 12 | MGI:1918089 |
| ENSMUSG00000025130 | P4hb | prolyl 4-hydroxylase, beta polypeptide | MGI:97464 |
| ENSMUSG00000025364 | Pa2g4 | proliferation-associated 2G4 | MGI:894684 |
| ENSMUSG00000022283 | Pabpc1 | poly(A) binding protein, cytoplasmic 1 | MGI:1349722 |
| ENSMUSG00000024855 | Pacs1 | phosphofurin acidic cluster sorting protein 1 | MGI:1277113 |
| ENSMUSG00000040276 | Pacsin1 | protein kinase C and casein kinase substrate in neurons 1 | MGI:1345181 |
| ENSMUSG00000016664 | Pacsin2 | protein kinase C and casein kinase substrate in neurons 2 | MGI:1345153 |
| ENSMUSG00000027257 | Pacsin3 | protein kinase C and casein kinase substrate in neurons 3 | MGI:1891410 |
| ENSMUSG00000028927 | Padi2 | peptidyl arginine deiminase, type II | MGI:1338892 |
| ENSMUSG00000020745 | Pafah1b1 | platelet-activating factor acetylhydrolase, isoform 1b, subunit 1 | MGI:109520 |
| ENSMUSG00000003131 | Pafah1b2 | platelet-activating factor acetylhydrolase, isoform 1b, subunit 2 | MGI:108415 |
| ENSMUSG00000005447 | Pafah1b3 | platelet-activating factor acetylhydrolase, isoform 1b, subunit 3 | MGI:108414 |
| ENSMUSG00000027508 | Pag1 | phosphoprotein associated with glycosphingolipid microdomains 1 | MGI:2443160 |
| ENSMUSG00000029247 | Paics | phosphoribosylaminoimidazole carboxylase, phosphoribosylaminoribosylaminoimidazole, succinocarboxamide synthetase | MGI:1914304 |
| ENSMUSG00000030774 | Pak1 | p21 protein (Cdc42/Rac)-activated kinase 1 | MGI:1339975 |
| ENSMUSG00000039913 | Pak7 | p21 protein (Cdc42/Rac)-activated kinase 7 | MGI:1920334 |
| ENSMUSG00000089945 | Pakap | paralemmin A kinase anchor protein | MGI:5141924 |
| ENSMUSG00000090053 | Palm2 | paralemmin 2 | MGI:1934601 |
| ENSMUSG00000047986 | Palm3 | paralemmin 3 | MGI:1921587 |
| ENSMUSG00000035863 | Palm | paralemmin | MGI:1261814 |
| ENSMUSG00000014301 | Pam16 | presequence translocase-asssociated motor 16 homolog (S. cerevisiae) | MGI:1913699 |
| ENSMUSG00000026335 | Pam | peptidylglycine alpha-amidating monooxygenase | MGI:97475 |
| ENSMUSG00000037514 | Pank2 | pantothenate kinase 2 | MGI:1921700 |
| ENSMUSG00000058441 | Panx2 | pannexin 2 | MGI:1890615 |
| ENSMUSG00000028964 | Park7 | Parkinson disease (autosomal recessive, early onset) 7 | MGI:2135637 |
| ENSMUSG00000034981 | Parm1 | prostate androgen-regulated mucin-like protein 1 | MGI:2443349 |
| ENSMUSG00000043572 | Pars2 | prolyl-tRNA synthetase (mitochondrial)(putative) | MGI:2386296 |
| ENSMUSG00000051695 | Pcbp1 | poly(rC) binding protein 1 | MGI:1345635 |
| ENSMUSG00000056851 | Pcbp2 | poly(rC) binding protein 2 | MGI:108202 |
| ENSMUSG00000001120 | Pcbp3 | poly(rC) binding protein 3 | MGI:1890470 |
| ENSMUSG00000041650 | Pcca | propionyl-Coenzyme A carboxylase, alpha polypeptide | MGI:97499 |
| ENSMUSG00000032527 | Pccb | propionyl Coenzyme A carboxylase, beta polypeptide | MGI:1914154 |
| ENSMUSG00000049100 | Pcdh10 | protocadherin 10 | MGI:1338042 |
| ENSMUSG00000035566 | Pcdh17 | protocadherin 17 | MGI:2684924 |
| ENSMUSG00000051323 | Pcdh19 | protocadherin 19 | MGI:2685563 |
| ENSMUSG00000051375 | Pcdh1 | protocadherin 1 | MGI:104692 |
| ENSMUSG00000029108 | Pcdh7 | protocadherin 7 | MGI:1860487 |
| ENSMUSG00000036422 | Pcdh8 | protocadherin 8 | MGI:1306800 |
| ENSMUSG00000055421 | Pcdh9 | protocadherin 9 | MGI:1306801 |
| ENSMUSG00000102697 | Pcdhac2 | protocadherin alpha subfamily C, 2 | MGI:1891443 |
| ENSMUSG00000102918 | Pcdhgc3 | protocadherin gamma subfamily C, 3 | MGI:1935201 |
| ENSMUSG00000102543 | Pcdhgc5 | protocadherin gamma subfamily C, 5 | MGI:1935205 |
| ENSMUSG00000040618 | Pck2 | phosphoenolpyruvate carboxykinase 2 (mitochondrial) | MGI:1860456 |
| ENSMUSG00000061601 | Pclo | piccolo (presynaptic cytomatrix protein) | MGI:1349390 |
| ENSMUSG00000019795 | Pcmt1 | protein-L-isoaspartate (D-aspartate) O-methyltransferase 1 | MGI:97502 |
| ENSMUSG00000038370 | Pcp4l1 | Purkinje cell protein 4-like 1 | MGI:1913675 |
| ENSMUSG00000039278 | Pcsk1n | proprotein convertase subtilisin/kexin type 1 inhibitor | MGI:1353431 |
| ENSMUSG00000027419 | Pcsk2 | proprotein convertase subtilisin/kexin type 2 | MGI:97512 |
| ENSMUSG00000024892 | Pcx | pyruvate carboxylase | MGI:97520 |
| ENSMUSG00000024579 | Pcyox1l | prenylcysteine oxidase 1 like | MGI:3606062 |
| ENSMUSG00000029998 | Pcyox1 | prenylcysteine oxidase 1 | MGI:1914131 |
| ENSMUSG00000035246 | Pcyt1b | phosphate cytidylyltransferase 1, choline, beta isoform | MGI:2147987 |
| ENSMUSG00000029623 | Pdap1 | PDGFA associated protein 1 | MGI:2448536 |
| ENSMUSG00000027835 | Pdcd10 | programmed cell death 10 | MGI:1928396 |
| ENSMUSG00000032504 | Pdcd6ip | programmed cell death 6 interacting protein | MGI:1333753 |
| ENSMUSG00000021576 | Pdcd6 | programmed cell death 6 | MGI:109283 |
| ENSMUSG00000051007 | Pddc1 | Parkinson disease 7 domain containing 1 | MGI:2387178 |
| ENSMUSG00000023868 | Pde10a | phosphodiesterase 10A | MGI:1345143 |
| ENSMUSG00000059173 | Pde1a | phosphodiesterase 1A, calmodulin-dependent | MGI:1201792 |
| ENSMUSG00000022489 | Pde1b | phosphodiesterase 1B, Ca2+-calmodulin dependent | MGI:97523 |
| ENSMUSG00000030653 | Pde2a | phosphodiesterase 2A, cGMP-stimulated | MGI:2446107 |
| ENSMUSG00000028525 | Pde4b | phosphodiesterase 4B, cAMP specific | MGI:99557 |
| ENSMUSG00000025584 | Pde8a | phosphodiesterase 8A | MGI:1277116 |
| ENSMUSG00000029231 | Pdgfra | platelet derived growth factor receptor, alpha polypeptide | MGI:97530 |
| ENSMUSG00000024620 | Pdgfrb | platelet derived growth factor receptor, beta polypeptide | MGI:97531 |
| ENSMUSG00000031299 | Pdha1 | pyruvate dehydrogenase E1 alpha 1 | MGI:97532 |
| ENSMUSG00000021748 | Pdhb | pyruvate dehydrogenase (lipoamide) beta | MGI:1915513 |
| ENSMUSG00000010914 | Pdhx | pyruvate dehydrogenase complex, component X | MGI:1351627 |
| ENSMUSG00000027248 | Pdia3 | protein disulfide isomerase associated 3 | MGI:95834 |
| ENSMUSG00000025823 | Pdia4 | protein disulfide isomerase associated 4 | MGI:104864 |
| ENSMUSG00000020571 | Pdia6 | protein disulfide isomerase associated 6 | MGI:1919103 |
| ENSMUSG00000050890 | Pdik1l | PDLIM1 interacting kinase 1 like | MGI:2385213 |
| ENSMUSG00000006494 | Pdk1 | pyruvate dehydrogenase kinase, isoenzyme 1 | MGI:1926119 |
| ENSMUSG00000038967 | Pdk2 | pyruvate dehydrogenase kinase, isoenzyme 2 | MGI:1343087 |
| ENSMUSG00000035232 | Pdk3 | pyruvate dehydrogenase kinase, isoenzyme 3 | MGI:2384308 |
| ENSMUSG00000028273 | Pdlim5 | PDZ and LIM domain 5 | MGI:1927489 |
| ENSMUSG00000049225 | Pdp1 | pyruvate dehyrogenase phosphatase catalytic subunit 1 | MGI:2685870 |
| ENSMUSG00000024122 | Pdpk1 | 3-phosphoinositide dependent protein kinase 1 | MGI:1338068 |
| ENSMUSG00000033624 | Pdpr | pyruvate dehydrogenase phosphatase regulatory subunit | MGI:2442188 |
| ENSMUSG00000032788 | Pdxk | pyridoxal (pyridoxine, vitamin B6) kinase | MGI:1351869 |
| ENSMUSG00000015668 | Pdzd11 | PDZ domain containing 11 | MGI:1919871 |
| ENSMUSG00000074746 | Pdzd8 | PDZ domain containing 8 | MGI:2677270 |
| ENSMUSG00000013698 | Pea15a | phosphoprotein enriched in astrocytes 15A | MGI:104799 |
| ENSMUSG00000032959 | Pebp1 | phosphatidylethanolamine binding protein 1 | MGI:1344408 |
| ENSMUSG00000028102 | Pex11b | peroxisomal biogenesis factor 11 beta | MGI:1338882 |
| ENSMUSG00000018733 | Pex12 | peroxisomal biogenesis factor 12 | MGI:2144177 |
| ENSMUSG00000028975 | Pex14 | peroxisomal biogenesis factor 14 | MGI:1927868 |
| ENSMUSG00000019809 | Pex3 | peroxisomal biogenesis factor 3 | MGI:1929646 |
| ENSMUSG00000027674 | Pex5l | peroxisomal biogenesis factor 5-like | MGI:1916672 |
| ENSMUSG00000005069 | Pex5 | peroxisomal biogenesis factor 5 | MGI:1098808 |
| ENSMUSG00000006412 | Pfdn2 | prefoldin 2 | MGI:1276111 |
| ENSMUSG00000052033 | Pfdn4 | prefoldin 4 | MGI:1923512 |
| ENSMUSG00000001289 | Pfdn5 | prefoldin 5 | MGI:1928753 |
| ENSMUSG00000024309 | Pfdn6 | prefoldin subunit 6 | MGI:95908 |
| ENSMUSG00000020277 | Pfkl | phosphofructokinase, liver, B-type | MGI:97547 |
| ENSMUSG00000033065 | Pfkm | phosphofructokinase, muscle | MGI:97548 |
| ENSMUSG00000021196 | Pfkp | phosphofructokinase, platelet | MGI:1891833 |
| ENSMUSG00000027805 | Pfn2 | profilin 2 | MGI:97550 |
| ENSMUSG00000011752 | Pgam1 | phosphoglycerate mutase 1 | MGI:97552 |
| ENSMUSG00000029500 | Pgam5 | phosphoglycerate mutase family member 5 | MGI:1919792 |
| ENSMUSG00000073678 | Pgap1 | post-GPI attachment to proteins 1 | MGI:2443342 |
| ENSMUSG00000028961 | Pgd | phosphogluconate dehydrogenase | MGI:97553 |
| ENSMUSG00000062070 | Pgk1 | phosphoglycerate kinase 1 | MGI:97555 |
| ENSMUSG00000031807 | Pgls | 6-phosphogluconolactonase | MGI:1913421 |
| ENSMUSG00000030729 | Pgm2l1 | phosphoglucomutase 2-like 1 | MGI:1918224 |
| ENSMUSG00000025791 | Pgm2 | phosphoglucomutase 2 | MGI:97565 |
| ENSMUSG00000043445 | Pgp | phosphoglycolate phosphatase | MGI:1914328 |
| ENSMUSG00000006373 | Pgrmc1 | progesterone receptor membrane component 1 | MGI:1858305 |
| ENSMUSG00000049940 | Pgrmc2 | progesterone receptor membrane component 2 | MGI:1918054 |
| ENSMUSG00000017715 | Pgs1 | phosphatidylglycerophosphate synthase 1 | MGI:1921701 |
| ENSMUSG00000054728 | Phactr1 | phosphatase and actin regulator 1 | MGI:2659021 |
| ENSMUSG00000004264 | Phb2 | prohibitin 2 | MGI:102520 |
| ENSMUSG00000038845 | Phb | prohibitin | MGI:97572 |
| ENSMUSG00000053398 | Phgdh | 3-phosphoglycerate dehydrogenase | MGI:1355330 |
| ENSMUSG00000010760 | Phlda2 | pleckstrin homology-like domain, family A, member 2 | MGI:1202307 |
| ENSMUSG00000048537 | Phldb1 | pleckstrin homology-like domain, family B, member 1 | MGI:2143230 |
| ENSMUSG00000036504 | Phpt1 | phosphohistidine phosphatase 1 | MGI:1922704 |
| ENSMUSG00000037747 | Phyhipl | phytanoyl-CoA hydroxylase interacting protein-like | MGI:1918161 |
| ENSMUSG00000003469 | Phyhip | phytanoyl-CoA hydroxylase interacting protein | MGI:1860417 |
| ENSMUSG00000025178 | Pi4k2a | phosphatidylinositol 4-kinase type 2 alpha | MGI:1934031 |
| ENSMUSG00000041720 | Pi4ka | phosphatidylinositol 4-kinase, catalytic, alpha polypeptide | MGI:2448506 |
| ENSMUSG00000039361 | Picalm | phosphatidylinositol binding clathrin assembly protein | MGI:2385902 |
| ENSMUSG00000039047 | Pigk | phosphatidylinositol glycan anchor biosynthesis, class K | MGI:1913863 |
| ENSMUSG00000041958 | Pigs | phosphatidylinositol glycan anchor biosynthesis, class S | MGI:2687325 |
| ENSMUSG00000017721 | Pigt | phosphatidylinositol glycan anchor biosynthesis, class T | MGI:1926178 |
| ENSMUSG00000033628 | Pik3c3 | phosphoinositide-3-kinase, class 3 | MGI:2445019 |
| ENSMUSG00000032462 | Pik3cb | phosphatidylinositol 3-kinase, catalytic, beta polypeptide | MGI:1922019 |
| ENSMUSG00000032571 | Pik3r4 | phosphatidylinositol 3 kinase, regulatory subunit, polypeptide 4, p150 | MGI:1922919 |
| ENSMUSG00000032171 | Pin1 | protein (peptidyl-prolyl cis/trans isomerase) NIMA-interacting 1 | MGI:1346036 |
| ENSMUSG00000079480 | Pin4 | protein (peptidyl-prolyl cis/trans isomerase) NIMA-interacting, 4 (parvulin) | MGI:1916963 |
| ENSMUSG00000026737 | Pip4k2a | phosphatidylinositol-5-phosphate 4-kinase, type II, alpha | MGI:1298206 |
| ENSMUSG00000018547 | Pip4k2b | phosphatidylinositol-5-phosphate 4-kinase, type II, beta | MGI:1934234 |
| ENSMUSG00000034902 | Pip5k1c | phosphatidylinositol-4-phosphate 5-kinase, type 1 gamma | MGI:1298224 |
| ENSMUSG00000023452 | Pisd | phosphatidylserine decarboxylase | MGI:2445114 |
| ENSMUSG00000017781 | Pitpna | phosphatidylinositol transfer protein, alpha | MGI:99887 |
| ENSMUSG00000040430 | Pitpnc1 | phosphatidylinositol transfer protein, cytoplasmic 1 | MGI:1919045 |
| ENSMUSG00000024851 | Pitpnm1 | phosphatidylinositol transfer protein, membrane-associated 1 | MGI:1197524 |
| ENSMUSG00000029406 | Pitpnm2 | phosphatidylinositol transfer protein, membrane-associated 2 | MGI:1336192 |
| ENSMUSG00000021193 | Pitrm1 | pitrilysin metallepetidase 1 | MGI:1916867 |
| ENSMUSG00000027499 | Pkia | protein kinase inhibitor, alpha | MGI:104747 |
| ENSMUSG00000032294 | Pkm | pyruvate kinase, muscle | MGI:97591 |
| ENSMUSG00000041957 | Pkp2 | plakophilin 2 | MGI:1914701 |
| ENSMUSG00000026991 | Pkp4 | plakophilin 4 | MGI:109281 |
| ENSMUSG00000060675 | Pla2g16 | phospholipase A2, group XVI | MGI:2179715 |
| ENSMUSG00000050211 | Pla2g4e | phospholipase A2, group IVE | MGI:1919144 |
| ENSMUSG00000051177 | Plcb1 | phospholipase C, beta 1 | MGI:97613 |
| ENSMUSG00000039943 | Plcb4 | phospholipase C, beta 4 | MGI:107464 |
| ENSMUSG00000010660 | Plcd1 | phospholipase C, delta 1 | MGI:97614 |
| ENSMUSG00000029055 | Plch2 | phospholipase C, eta 2 | MGI:2443078 |
| ENSMUSG00000038910 | Plcl2 | phospholipase C-like 2 | MGI:1352756 |
| ENSMUSG00000020828 | Pld2 | phospholipase D2 | MGI:892877 |
| ENSMUSG00000003363 | Pld3 | phospholipase D family, member 3 | MGI:1333782 |
| ENSMUSG00000022565 | Plec | plectin | MGI:1277961 |
| ENSMUSG00000030231 | Plekha5 | pleckstrin homology domain containing, family A member 5 | MGI:1923802 |
| ENSMUSG00000041757 | Plekha6 | pleckstrin homology domain containing, family A member 6 | MGI:2388662 |
| ENSMUSG00000030701 | Plekhb1 | pleckstrin homology domain containing, family B (evectins) member 1 | MGI:1351469 |
| ENSMUSG00000066438 | Plekhd1 | pleckstrin homology domain containing, family D (with coiled-coil domains) member 1 | MGI:3036228 |
| ENSMUSG00000040624 | Plekhg1 | pleckstrin homology domain containing, family G (with RhoGef domain) member 1 | MGI:2676551 |
| ENSMUSG00000016495 | Plgrkt | plasminogen receptor, C-terminal lysine transmembrane protein | MGI:1915009 |
| ENSMUSG00000031775 | Pllp | plasma membrane proteolipid | MGI:1915051 |
| ENSMUSG00000031425 | Plp1 | proteolipid protein (myelin) 1 | MGI:97623 |
| ENSMUSG00000031146 | Plp2 | proteolipid protein 2 | MGI:1298382 |
| ENSMUSG00000030084 | Plxna1 | plexin A1 | MGI:107685 |
| ENSMUSG00000026640 | Plxna2 | plexin A2 | MGI:107684 |
| ENSMUSG00000029765 | Plxna4 | plexin A4 | MGI:2179061 |
| ENSMUSG00000053646 | Plxnb1 | plexin B1 | MGI:2154238 |
| ENSMUSG00000036606 | Plxnb2 | plexin B2 | MGI:2154239 |
| ENSMUSG00000074785 | Plxnc1 | plexin C1 | MGI:1890127 |
| ENSMUSG00000030123 | Plxnd1 | plexin D1 | MGI:2154244 |
| ENSMUSG00000042251 | Pm20d1 | peptidase M20 domain containing 1 | MGI:2442939 |
| ENSMUSG00000026926 | Pmpca | peptidase (mitochondrial processing) alpha | MGI:1918568 |
| ENSMUSG00000004565 | Pnpla6 | patatin-like phospholipase domain containing 6 | MGI:1354723 |
| ENSMUSG00000036257 | Pnpla8 | patatin-like phospholipase domain containing 8 | MGI:1914702 |
| ENSMUSG00000033152 | Podxl2 | podocalyxin-like 2 | MGI:2442488 |
| ENSMUSG00000034064 | Poglut1 | protein O-glucosyltransferase 1 | MGI:2444232 |
| ENSMUSG00000021018 | Polr2h | polymerase (RNA) II (DNA directed) polypeptide H | MGI:2384309 |
| ENSMUSG00000066235 | Pomgnt2 | protein O-linked mannose beta 1,4-N-acetylglucosaminyltransferase 2 | MGI:2143424 |
| ENSMUSG00000039254 | Pomt1 | protein-O-mannosyltransferase 1 | MGI:2138994 |
| ENSMUSG00000032667 | Pon2 | paraoxonase 2 | MGI:106687 |
| ENSMUSG00000005514 | Por | P450 (cytochrome) oxidoreductase | MGI:97744 |
| ENSMUSG00000020089 | Ppa1 | pyrophosphatase (inorganic) 1 | MGI:97831 |
| ENSMUSG00000028013 | Ppa2 | pyrophosphatase (inorganic) 2 | MGI:1922026 |
| ENSMUSG00000028517 | Ppap2b | phosphatidic acid phosphatase type 2B | MGI:1915166 |
| ENSMUSG00000037519 | Ppfia1 | protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 1 | MGI:1924750 |
| ENSMUSG00000053825 | Ppfia2 | protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2 | MGI:2443834 |
| ENSMUSG00000003863 | Ppfia3 | protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 3 | MGI:1924037 |
| ENSMUSG00000026458 | Ppfia4 | protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 4 | MGI:1915757 |
| ENSMUSG00000071866 | Ppia | peptidylprolyl isomerase A | MGI:97749 |
| ENSMUSG00000032383 | Ppib | peptidylprolyl isomerase B | MGI:97750 |
| ENSMUSG00000027804 | Ppid | peptidylprolyl isomerase D (cyclophilin D) | MGI:1914988 |
| ENSMUSG00000021868 | Ppif | peptidylprolyl isomerase F (cyclophilin F) | MGI:2145814 |
| ENSMUSG00000039457 | Ppl | periplakin | MGI:1194898 |
| ENSMUSG00000021096 | Ppm1a | protein phosphatase 1A, magnesium dependent, alpha isoform | MGI:99878 |
| ENSMUSG00000034613 | Ppm1h | protein phosphatase 1H (PP2C domain containing) | MGI:2442087 |
| ENSMUSG00000030718 | Ppme1 | protein phosphatase methylesterase 1 | MGI:1919840 |
| ENSMUSG00000040385 | Ppp1ca | protein phosphatase 1, catalytic subunit, alpha isoform | MGI:103016 |
| ENSMUSG00000014956 | Ppp1cb | protein phosphatase 1, catalytic subunit, beta isoform | MGI:104871 |
| ENSMUSG00000004455 | Ppp1cc | protein phosphatase 1, catalytic subunit, gamma isoform | MGI:104872 |
| ENSMUSG00000019907 | Ppp1r12a | protein phosphatase 1, regulatory (inhibitor) subunit 12A | MGI:1309528 |
| ENSMUSG00000033819 | Ppp1r16a | protein phosphatase 1, regulatory (inhibitor) subunit 16A | MGI:1920312 |
| ENSMUSG00000037754 | Ppp1r16b | protein phosphatase 1, regulatory (inhibitor) subunit 16B | MGI:2151841 |
| ENSMUSG00000061718 | Ppp1r1b | protein phosphatase 1, regulatory (inhibitor) subunit 1B | MGI:94860 |
| ENSMUSG00000034709 | Ppp1r21 | protein phosphatase 1, regulatory subunit 21 | MGI:1921075 |
| ENSMUSG00000026275 | Ppp1r7 | protein phosphatase 1, regulatory (inhibitor) subunit 7 | MGI:1913635 |
| ENSMUSG00000032827 | Ppp1r9a | protein phosphatase 1, regulatory (inhibitor) subunit 9A | MGI:2442401 |
| ENSMUSG00000038976 | Ppp1r9b | protein phosphatase 1, regulatory subunit 9B | MGI:2387581 |
| ENSMUSG00000020349 | Ppp2ca | protein phosphatase 2 (formerly 2A), catalytic subunit, alpha isoform | MGI:1321159 |
| ENSMUSG00000007564 | Ppp2r1a | protein phosphatase 2, regulatory subunit A, alpha | MGI:1926334 |
| ENSMUSG00000022052 | Ppp2r2a | protein phosphatase 2, regulatory subunit B, alpha | MGI:1919228 |
| ENSMUSG00000029120 | Ppp2r2c | protein phosphatase 2, regulatory subunit B, gamma | MGI:2442660 |
| ENSMUSG00000039515 | Ppp2r4 | protein phosphatase 2A activator, regulatory subunit B | MGI:1346006 |
| ENSMUSG00000026626 | Ppp2r5a | protein phosphatase 2, regulatory subunit B', alpha | MGI:2388479 |
| ENSMUSG00000024777 | Ppp2r5b | protein phosphatase 2, regulatory subunit B', beta | MGI:2388480 |
| ENSMUSG00000059409 | Ppp2r5d | protein phosphatase 2, regulatory subunit B', delta | MGI:2388481 |
| ENSMUSG00000021051 | Ppp2r5e | protein phosphatase 2, regulatory subunit B', epsilon | MGI:1349473 |
| ENSMUSG00000028161 | Ppp3ca | protein phosphatase 3, catalytic subunit, alpha isoform | MGI:107164 |
| ENSMUSG00000021816 | Ppp3cb | protein phosphatase 3, catalytic subunit, beta isoform | MGI:107163 |
| ENSMUSG00000033953 | Ppp3r1 | protein phosphatase 3, regulatory subunit B, alpha isoform (calcineurin B, type I) | MGI:107172 |
| ENSMUSG00000003099 | Ppp5c | protein phosphatase 5, catalytic subunit | MGI:102666 |
| ENSMUSG00000028657 | Ppt1 | palmitoyl-protein thioesterase 1 | MGI:1298204 |
| ENSMUSG00000031149 | Praf2 | PRA1 domain family 2 | MGI:1859607 |
| ENSMUSG00000028691 | Prdx1 | peroxiredoxin 1 | MGI:99523 |
| ENSMUSG00000005161 | Prdx2 | peroxiredoxin 2 | MGI:109486 |
| ENSMUSG00000024997 | Prdx3 | peroxiredoxin 3 | MGI:88034 |
| ENSMUSG00000025289 | Prdx4 | peroxiredoxin 4 | MGI:1859815 |
| ENSMUSG00000024953 | Prdx5 | peroxiredoxin 5 | MGI:1859821 |
| ENSMUSG00000026701 | Prdx6 | peroxiredoxin 6 | MGI:894320 |
| ENSMUSG00000045302 | Preb | prolactin regulatory element binding | MGI:1355326 |
| ENSMUSG00000019849 | Prep | prolyl endopeptidase | MGI:1270863 |
| ENSMUSG00000030020 | Prickle2 | prickle homolog 2 (Drosophila) | MGI:1925144 |
| ENSMUSG00000050697 | Prkaa1 | protein kinase, AMP-activated, alpha 1 catalytic subunit | MGI:2145955 |
| ENSMUSG00000005469 | Prkaca | protein kinase, cAMP dependent, catalytic, alpha | MGI:97592 |
| ENSMUSG00000005034 | Prkacb | protein kinase, cAMP dependent, catalytic, beta | MGI:97594 |
| ENSMUSG00000067713 | Prkag1 | protein kinase, AMP-activated, gamma 1 non-catalytic subunit | MGI:108411 |
| ENSMUSG00000028944 | Prkag2 | protein kinase, AMP-activated, gamma 2 non-catalytic subunit | MGI:1336153 |
| ENSMUSG00000020612 | Prkar1a | protein kinase, cAMP dependent regulatory, type I, alpha | MGI:104878 |
| ENSMUSG00000025855 | Prkar1b | protein kinase, cAMP dependent regulatory, type I beta | MGI:97759 |
| ENSMUSG00000032601 | Prkar2a | protein kinase, cAMP dependent regulatory, type II alpha | MGI:108025 |
| ENSMUSG00000002997 | Prkar2b | protein kinase, cAMP dependent regulatory, type II beta | MGI:97760 |
| ENSMUSG00000050965 | Prkca | protein kinase C, alpha | MGI:97595 |
| ENSMUSG00000052889 | Prkcb | protein kinase C, beta | MGI:97596 |
| ENSMUSG00000021948 | Prkcd | protein kinase C, delta | MGI:97598 |
| ENSMUSG00000045038 | Prkce | protein kinase C, epsilon | MGI:97599 |
| ENSMUSG00000078816 | Prkcg | protein kinase C, gamma | MGI:97597 |
| ENSMUSG00000003402 | Prkcsh | protein kinase C substrate 80K-H | MGI:107877 |
| ENSMUSG00000029334 | Prkg2 | protein kinase, cGMP-dependent, type II | MGI:108173 |
| ENSMUSG00000052429 | Prmt1 | protein arginine N-methyltransferase 1 | MGI:107846 |
| ENSMUSG00000023110 | Prmt5 | protein arginine N-methyltransferase 5 | MGI:1351645 |
| ENSMUSG00000030350 | Prmt8 | protein arginine N-methyltransferase 8 | MGI:3043083 |
| ENSMUSG00000079037 | Prnp | prion protein | MGI:97769 |
| ENSMUSG00000003526 | Prodh | proline dehydrogenase | MGI:97770 |
| ENSMUSG00000029086 | Prom1 | prominin 1 | MGI:1100886 |
| ENSMUSG00000092305 | Prps1l1 | phosphoribosyl pyrophosphate synthetase 1-like 1 | MGI:1922706 |
| ENSMUSG00000015869 | Prpsap1 | phosphoribosyl pyrophosphate synthetase-associated protein 1 | MGI:1915013 |
| ENSMUSG00000020528 | Prpsap2 | phosphoribosyl pyrophosphate synthetase-associated protein 2 | MGI:2384838 |
| ENSMUSG00000064125 | Prr36 | proline rich 36 | MGI:3605626 |
| ENSMUSG00000024393 | Prrc2a | proline-rich coiled-coil 2A | MGI:1915467 |
| ENSMUSG00000040225 | Prrc2c | proline-rich coiled-coil 2C | MGI:1913754 |
| ENSMUSG00000015476 | Prrt1 | proline-rich transmembrane protein 1 | MGI:1932118 |
| ENSMUSG00000045114 | Prrt2 | proline-rich transmembrane protein 2 | MGI:1916267 |
| ENSMUSG00000045009 | Prrt3 | proline-rich transmembrane protein 3 | MGI:2444810 |
| ENSMUSG00000071519 | Prss3 | protease, serine 3 | MGI:102758 |
| ENSMUSG00000004207 | Psap | prosaposin | MGI:97783 |
| ENSMUSG00000024640 | Psat1 | phosphoserine aminotransferase 1 | MGI:2183441 |
| ENSMUSG00000024347 | Psd2 | pleckstrin and Sec7 domain containing 2 | MGI:1921252 |
| ENSMUSG00000030465 | Psd3 | pleckstrin and Sec7 domain containing 3 | MGI:1918215 |
| ENSMUSG00000037126 | Psd | pleckstrin and Sec7 domain containing | MGI:1920978 |
| ENSMUSG00000019969 | Psen1 | presenilin 1 | MGI:1202717 |
| ENSMUSG00000028484 | Psip1 | PC4 and SFRS1 interacting protein 1 | MGI:2142116 |
| ENSMUSG00000030751 | Psma1 | proteasome (prosome, macropain) subunit, alpha type 1 | MGI:1347005 |
| ENSMUSG00000015671 | Psma2 | proteasome (prosome, macropain) subunit, alpha type 2 | MGI:104885 |
| ENSMUSG00000060073 | Psma3 | proteasome (prosome, macropain) subunit, alpha type 3 | MGI:104883 |
| ENSMUSG00000032301 | Psma4 | proteasome (prosome, macropain) subunit, alpha type 4 | MGI:1347060 |
| ENSMUSG00000068749 | Psma5 | proteasome (prosome, macropain) subunit, alpha type 5 | MGI:1347009 |
| ENSMUSG00000021024 | Psma6 | proteasome (prosome, macropain) subunit, alpha type 6 | MGI:1347006 |
| ENSMUSG00000027566 | Psma7 | proteasome (prosome, macropain) subunit, alpha type 7 | MGI:1347070 |
| ENSMUSG00000014769 | Psmb1 | proteasome (prosome, macropain) subunit, beta type 1 | MGI:104884 |
| ENSMUSG00000028837 | Psmb2 | proteasome (prosome, macropain) subunit, beta type 2 | MGI:1347045 |
| ENSMUSG00000069744 | Psmb3 | proteasome (prosome, macropain) subunit, beta type 3 | MGI:1347014 |
| ENSMUSG00000005779 | Psmb4 | proteasome (prosome, macropain) subunit, beta type 4 | MGI:1098257 |
| ENSMUSG00000022193 | Psmb5 | proteasome (prosome, macropain) subunit, beta type 5 | MGI:1194513 |
| ENSMUSG00000018286 | Psmb6 | proteasome (prosome, macropain) subunit, beta type 6 | MGI:104880 |
| ENSMUSG00000021178 | Psmc1 | protease (prosome, macropain) 26S subunit, ATPase 1 | MGI:106054 |
| ENSMUSG00000028932 | Psmc2 | proteasome (prosome, macropain) 26S subunit, ATPase 2 | MGI:109555 |
| ENSMUSG00000002102 | Psmc3 | proteasome (prosome, macropain) 26S subunit, ATPase 3 | MGI:1098754 |
| ENSMUSG00000030603 | Psmc4 | proteasome (prosome, macropain) 26S subunit, ATPase, 4 | MGI:1346093 |
| ENSMUSG00000020708 | Psmc5 | protease (prosome, macropain) 26S subunit, ATPase 5 | MGI:105047 |
| ENSMUSG00000021832 | Psmc6 | proteasome (prosome, macropain) 26S subunit, ATPase, 6 | MGI:1914339 |
| ENSMUSG00000017428 | Psmd11 | proteasome (prosome, macropain) 26S subunit, non-ATPase, 11 | MGI:1916327 |
| ENSMUSG00000020720 | Psmd12 | proteasome (prosome, macropain) 26S subunit, non-ATPase, 12 | MGI:1914247 |
| ENSMUSG00000025487 | Psmd13 | proteasome (prosome, macropain) 26S subunit, non-ATPase, 13 | MGI:1345192 |
| ENSMUSG00000026914 | Psmd14 | proteasome (prosome, macropain) 26S subunit, non-ATPase, 14 | MGI:1913284 |
| ENSMUSG00000026229 | Psmd1 | proteasome (prosome, macropain) 26S subunit, non-ATPase, 1 | MGI:1917497 |
| ENSMUSG00000006998 | Psmd2 | proteasome (prosome, macropain) 26S subunit, non-ATPase, 2 | MGI:1096584 |
| ENSMUSG00000017221 | Psmd3 | proteasome (prosome, macropain) 26S subunit, non-ATPase, 3 | MGI:98858 |
| ENSMUSG00000005625 | Psmd4 | proteasome (prosome, macropain) 26S subunit, non-ATPase, 4 | MGI:1201670 |
| ENSMUSG00000026869 | Psmd5 | proteasome (prosome, macropain) 26S subunit, non-ATPase, 5 | MGI:1914248 |
| ENSMUSG00000021737 | Psmd6 | proteasome (prosome, macropain) 26S subunit, non-ATPase, 6 | MGI:1913663 |
| ENSMUSG00000039067 | Psmd7 | proteasome (prosome, macropain) 26S subunit, non-ATPase, 7 | MGI:1351511 |
| ENSMUSG00000021938 | Pspc1 | paraspeckle protein 1 | MGI:1913895 |
| ENSMUSG00000028134 | Ptbp2 | polypyrimidine tract binding protein 2 | MGI:1860489 |
| ENSMUSG00000021518 | Ptdss1 | phosphatidylserine synthase 1 | MGI:1276575 |
| ENSMUSG00000015090 | Ptgds | prostaglandin D2 synthase (brain) | MGI:99261 |
| ENSMUSG00000026820 | Ptges2 | prostaglandin E synthase 2 | MGI:1917592 |
| ENSMUSG00000027864 | Ptgfrn | prostaglandin F2 receptor negative regulator | MGI:1277114 |
| ENSMUSG00000047250 | Ptgs1 | prostaglandin-endoperoxide synthase 1 | MGI:97797 |
| ENSMUSG00000023972 | Ptk7 | PTK7 protein tyrosine kinase 7 | MGI:1918711 |
| ENSMUSG00000026238 | Ptma | prothymosin alpha | MGI:97803 |
| ENSMUSG00000030122 | Ptms | parathymosin | MGI:1916452 |
| ENSMUSG00000026064 | Ptp4a1 | protein tyrosine phosphatase 4a1 | MGI:1277096 |
| ENSMUSG00000028788 | Ptp4a2 | protein tyrosine phosphatase 4a2 | MGI:1277117 |
| ENSMUSG00000063235 | Ptpmt1 | protein tyrosine phosphatase, mitochondrial 1 | MGI:1913711 |
| ENSMUSG00000027540 | Ptpn1 | protein tyrosine phosphatase, non-receptor type 1 | MGI:97805 |
| ENSMUSG00000030854 | Ptpn5 | protein tyrosine phosphatase, non-receptor type 5 | MGI:97807 |
| ENSMUSG00000032290 | Ptpn9 | protein tyrosine phosphatase, non-receptor type 9 | MGI:1928376 |
| ENSMUSG00000027303 | Ptpra | protein tyrosine phosphatase, receptor type, A | MGI:97808 |
| ENSMUSG00000026395 | Ptprc | protein tyrosine phosphatase, receptor type, C | MGI:97810 |
| ENSMUSG00000028399 | Ptprd | protein tyrosine phosphatase, receptor type, D | MGI:97812 |
| ENSMUSG00000041836 | Ptpre | protein tyrosine phosphatase, receptor type, E | MGI:97813 |
| ENSMUSG00000033295 | Ptprf | protein tyrosine phosphatase, receptor type, F | MGI:102695 |
| ENSMUSG00000021745 | Ptprg | protein tyrosine phosphatase, receptor type, G | MGI:97814 |
| ENSMUSG00000025314 | Ptprj | protein tyrosine phosphatase, receptor type, J | MGI:104574 |
| ENSMUSG00000056553 | Ptprn2 | protein tyrosine phosphatase, receptor type, N polypeptide 2 | MGI:107418 |
| ENSMUSG00000026204 | Ptprn | protein tyrosine phosphatase, receptor type, N | MGI:102765 |
| ENSMUSG00000013236 | Ptprs | protein tyrosine phosphatase, receptor type, S | MGI:97815 |
| ENSMUSG00000053141 | Ptprt | protein tyrosine phosphatase, receptor type, T | MGI:1321152 |
| ENSMUSG00000068748 | Ptprz1 | protein tyrosine phosphatase, receptor type Z, polypeptide 1 | MGI:97816 |
| ENSMUSG00000072582 | Ptrh2 | peptidyl-tRNA hydrolase 2 | MGI:2444848 |
| ENSMUSG00000043991 | Pura | purine rich element binding protein A | MGI:103079 |
| ENSMUSG00000094483 | Purb | purine rich element binding protein B | MGI:1338779 |
| ENSMUSG00000049184 | Purg | purine-rich element binding protein G | MGI:1922279 |
| ENSMUSG00000005716 | Pvalb | parvalbumin | MGI:97821 |
| ENSMUSG00000032012 | Pvrl1 | poliovirus receptor-related 1 | MGI:1926483 |
| ENSMUSG00000022656 | Pvrl3 | poliovirus receptor-related 3 | MGI:1930171 |
| ENSMUSG00000026520 | Pycr2 | pyrroline-5-carboxylate reductase family, member 2 | MGI:1277956 |
| ENSMUSG00000022571 | Pycrl | pyrroline-5-carboxylate reductase-like | MGI:1913444 |
| ENSMUSG00000033059 | Pygb | brain glycogen phosphorylase | MGI:97828 |
| ENSMUSG00000032648 | Pygm | muscle glycogen phosphorylase | MGI:97830 |
| ENSMUSG00000030359 | Pzp | pregnancy zone protein | MGI:87854 |
| ENSMUSG00000032604 | Qars | glutaminyl-tRNA synthetase | MGI:1915851 |
| ENSMUSG00000015806 | Qdpr | quinoid dihydropteridine reductase | MGI:97836 |
| ENSMUSG00000062078 | Qk | quaking | MGI:97837 |
| ENSMUSG00000020671 | Rab10 | RAB10, member RAS oncogene family | MGI:105066 |
| ENSMUSG00000077450 | Rab11b | RAB11B, member RAS oncogene family | MGI:99425 |
| ENSMUSG00000040022 | Rab11fip2 | RAB11 family interacting protein 2 (class I) | MGI:1922248 |
| ENSMUSG00000051343 | Rab11fip5 | RAB11 family interacting protein 5 (class I) | MGI:1098586 |
| ENSMUSG00000023460 | Rab12 | RAB12, member RAS oncogene family | MGI:894284 |
| ENSMUSG00000027935 | Rab13 | RAB13, member RAS oncogene family | MGI:1927232 |
| ENSMUSG00000026878 | Rab14 | RAB14, member RAS oncogene family | MGI:1915615 |
| ENSMUSG00000021062 | Rab15 | RAB15, member RAS oncogene family | MGI:1916865 |
| ENSMUSG00000073639 | Rab18 | RAB18, member RAS oncogene family | MGI:102790 |
| ENSMUSG00000024870 | Rab1b | RAB1B, member RAS oncogene family | MGI:1923558 |
| ENSMUSG00000020149 | Rab1 | RAB1, member RAS oncogene family | MGI:97842 |
| ENSMUSG00000020132 | Rab21 | RAB21, member RAS oncogene family | MGI:894308 |
| ENSMUSG00000027519 | Rab22a | RAB22A, member RAS oncogene family | MGI:105072 |
| ENSMUSG00000004768 | Rab23 | RAB23, member RAS oncogene family | MGI:99833 |
| ENSMUSG00000034789 | Rab24 | RAB24, member RAS oncogene family | MGI:105065 |
| ENSMUSG00000024511 | Rab27b | RAB27B, member RAS oncogene family | MGI:1931295 |
| ENSMUSG00000047187 | Rab2a | RAB2A, member RAS oncogene family | MGI:1928750 |
| ENSMUSG00000022159 | Rab2b | RAB2B, member RAS oncogene family | MGI:1923588 |
| ENSMUSG00000030643 | Rab30 | RAB30, member RAS oncogene family | MGI:1923235 |
| ENSMUSG00000056515 | Rab31 | RAB31, member RAS oncogene family | MGI:1914603 |
| ENSMUSG00000027739 | Rab33b | RAB33B, member RAS oncogene family | MGI:1330805 |
| ENSMUSG00000029518 | Rab35 | RAB35, member RAS oncogene family | MGI:1924657 |
| ENSMUSG00000020732 | Rab37 | RAB37, member RAS oncogene family | MGI:1929945 |
| ENSMUSG00000031202 | Rab39b | RAB39B, member RAS oncogene family | MGI:1915040 |
| ENSMUSG00000055069 | Rab39 | RAB39, member RAS oncogene family | MGI:2442855 |
| ENSMUSG00000031840 | Rab3a | RAB3A, member RAS oncogene family | MGI:97843 |
| ENSMUSG00000003411 | Rab3b | RAB3B, member RAS oncogene family | MGI:1917158 |
| ENSMUSG00000021700 | Rab3c | RAB3C, member RAS oncogene family | MGI:1914545 |
| ENSMUSG00000019066 | Rab3d | RAB3D, member RAS oncogene family | MGI:97844 |
| ENSMUSG00000036104 | Rab3gap1 | RAB3 GTPase activating protein subunit 1 | MGI:2445001 |
| ENSMUSG00000039318 | Rab3gap2 | RAB3 GTPase activating protein subunit 2 | MGI:1916043 |
| ENSMUSG00000019478 | Rab4a | RAB4A, member RAS oncogene family | MGI:105069 |
| ENSMUSG00000053291 | Rab4b | RAB4B, member RAS oncogene family | MGI:105071 |
| ENSMUSG00000017831 | Rab5a | RAB5A, member RAS oncogene family | MGI:105926 |
| ENSMUSG00000000711 | Rab5b | RAB5B, member RAS oncogene family | MGI:105938 |
| ENSMUSG00000019173 | Rab5c | RAB5C, member RAS oncogene family | MGI:105306 |
| ENSMUSG00000030704 | Rab6a | RAB6A, member RAS oncogene family | MGI:894313 |
| ENSMUSG00000032549 | Rab6b | RAB6B, member RAS oncogene family | MGI:107283 |
| ENSMUSG00000079477 | Rab7 | RAB7, member RAS oncogene family | MGI:105068 |
| ENSMUSG00000003037 | Rab8a | RAB8A, member RAS oncogene family | MGI:96960 |
| ENSMUSG00000036943 | Rab8b | RAB8B, member RAS oncogene family | MGI:2442982 |
| ENSMUSG00000043463 | Rab9b | RAB9B, member RAS oncogene family | MGI:2442454 |
| ENSMUSG00000079316 | Rab9 | RAB9, member RAS oncogene family | MGI:1890695 |
| ENSMUSG00000020817 | Rabep1 | rabaptin, RAB GTPase binding effector protein 1 | MGI:1860236 |
| ENSMUSG00000015087 | Rabl6 | RAB, member RAS oncogene family-like 6 | MGI:2442633 |
| ENSMUSG00000001847 | Rac1 | RAS-related C3 botulinum substrate 1 | MGI:97845 |
| ENSMUSG00000018012 | Rac3 | RAS-related C3 botulinum substrate 3 | MGI:2180784 |
| ENSMUSG00000008859 | Rala | v-ral simian leukemia viral oncogene homolog A (ras related) | MGI:1927243 |
| ENSMUSG00000004451 | Ralb | v-ral simian leukemia viral oncogene homolog B (ras related) | MGI:1927244 |
| ENSMUSG00000021027 | Ralgapa1 | Ral GTPase activating protein, alpha subunit 1 | MGI:1931050 |
| ENSMUSG00000027652 | Ralgapb | Ral GTPase activating protein, beta subunit (non-catalytic) | MGI:2444531 |
| ENSMUSG00000029430 | Ran | RAN, member RAS oncogene family | MGI:1333112 |
| ENSMUSG00000068798 | Rap1a | RAS-related protein-1a | MGI:97852 |
| ENSMUSG00000052681 | Rap1b | RAS related protein 1b | MGI:894315 |
| ENSMUSG00000041351 | Rap1gap | Rap1 GTPase-activating protein | MGI:109338 |
| ENSMUSG00000028149 | Rap1gds1 | RAP1, GTP-GDP dissociation stimulator 1 | MGI:2385189 |
| ENSMUSG00000051615 | Rap2a | RAS related protein 2a | MGI:97855 |
| ENSMUSG00000036894 | Rap2b | RAP2B, member of RAS oncogene family | MGI:1921262 |
| ENSMUSG00000050029 | Rap2c | RAP2C, member of RAS oncogene family | MGI:1919315 |
| ENSMUSG00000062232 | Rapgef2 | Rap guanine nucleotide exchange factor (GEF) 2 | MGI:2659071 |
| ENSMUSG00000049044 | Rapgef4 | Rap guanine nucleotide exchange factor (GEF) 4 | MGI:1917723 |
| ENSMUSG00000026014 | Raph1 | Ras association (RalGDS/AF-6) and pleckstrin homology domains 1 | MGI:1924550 |
| ENSMUSG00000018848 | Rars | arginyl-tRNA synthetase | MGI:1914297 |
| ENSMUSG00000031453 | Rasa3 | RAS p21 protein activator 3 | MGI:1197013 |
| ENSMUSG00000070565 | Rasal2 | RAS protein activator like 2 | MGI:2443881 |
| ENSMUSG00000032356 | Rasgrf1 | RAS protein-specific guanine nucleotide-releasing factor 1 | MGI:99694 |
| ENSMUSG00000021708 | Rasgrf2 | RAS protein-specific guanine nucleotide-releasing factor 2 | MGI:109137 |
| ENSMUSG00000025907 | Rb1cc1 | RB1-inducible coiled-coil 1 | MGI:1341850 |
| ENSMUSG00000022400 | Rbx1 | ring-box 1 | MGI:1891829 |
| ENSMUSG00000032320 | Rcn2 | reticulocalbin 2 | MGI:1349765 |
| ENSMUSG00000066441 | Rdh11 | retinol dehydrogenase 11 | MGI:102581 |
| ENSMUSG00000008435 | Rdh13 | retinol dehydrogenase 13 (all-trans and 9-cis) | MGI:1918732 |
| ENSMUSG00000020621 | Rdh14 | retinol dehydrogenase 14 (all-trans and 9-cis) | MGI:1920402 |
| ENSMUSG00000032050 | Rdx | radixin | MGI:97887 |
| ENSMUSG00000052852 | Reep1 | receptor accessory protein 1 | MGI:1098827 |
| ENSMUSG00000038555 | Reep2 | receptor accessory protein 2 | MGI:2385070 |
| ENSMUSG00000005873 | Reep5 | receptor accessory protein 5 | MGI:1270152 |
| ENSMUSG00000044024 | Rell2 | RELT-like 2 | MGI:1918044 |
| ENSMUSG00000029048 | Rer1 | RER1 retention in endoplasmic reticulum 1 homolog (S. cerevisiae) | MGI:1915080 |
| ENSMUSG00000032026 | Rexo2 | REX2, RNA exonuclease 2 homolog (S. cerevisiae) | MGI:1888981 |
| ENSMUSG00000039316 | Rftn1 | raftlin lipid raft linker 1 | MGI:1923688 |
| ENSMUSG00000025978 | Rftn2 | raftlin family member 2 | MGI:1921263 |
| ENSMUSG00000070509 | Rgma | repulsive guidance molecule family member A | MGI:2679262 |
| ENSMUSG00000029101 | Rgs12 | regulator of G-protein signaling 12 | MGI:1918979 |
| ENSMUSG00000052087 | Rgs14 | regulator of G-protein signaling 14 | MGI:1859709 |
| ENSMUSG00000019775 | Rgs17 | regulator of G-protein signaling 17 | MGI:1927469 |
| ENSMUSG00000002459 | Rgs20 | regulator of G-protein signaling 20 | MGI:1929866 |
| ENSMUSG00000021219 | Rgs6 | regulator of G-protein signaling 6 | MGI:1354730 |
| ENSMUSG00000021719 | Rgs7bp | regulator of G-protein signalling 7 binding protein | MGI:106334 |
| ENSMUSG00000026527 | Rgs7 | regulator of G protein signaling 7 | MGI:1346089 |
| ENSMUSG00000042671 | Rgs8 | regulator of G-protein signaling 8 | MGI:108408 |
| ENSMUSG00000020599 | Rgs9 | regulator of G-protein signaling 9 | MGI:1338824 |
| ENSMUSG00000028945 | Rheb | Ras homolog enriched in brain | MGI:97912 |
| ENSMUSG00000007815 | Rhoa | ras homolog gene family, member A | MGI:1096342 |
| ENSMUSG00000054364 | Rhob | ras homolog gene family, member B | MGI:107949 |
| ENSMUSG00000029449 | Rhof | ras homolog gene family, member f | MGI:1345629 |
| ENSMUSG00000073982 | Rhog | ras homolog gene family, member G | MGI:1928370 |
| ENSMUSG00000024143 | Rhoq | ras homolog gene family, member Q | MGI:1931553 |
| ENSMUSG00000017686 | Rhot1 | ras homolog gene family, member T1 | MGI:1926078 |
| ENSMUSG00000025733 | Rhot2 | ras homolog gene family, member T2 | MGI:2384892 |
| ENSMUSG00000025257 | Ribc1 | RIB43A domain with coiled-coils 1 | MGI:1913861 |
| ENSMUSG00000025485 | Ric8 | resistance to inhibitors of cholinesterase 8 homolog (C. elegans) | MGI:2141866 |
| ENSMUSG00000029420 | Rimbp2 | RIMS binding protein 2 | MGI:2443235 |
| ENSMUSG00000041670 | Rims1 | regulating synaptic membrane exocytosis 1 | MGI:2152971 |
| ENSMUSG00000037386 | Rims2 | regulating synaptic membrane exocytosis 2 | MGI:2152972 |
| ENSMUSG00000032890 | Rims3 | regulating synaptic membrane exocytosis 3 | MGI:2443331 |
| ENSMUSG00000024883 | Rin1 | Ras and Rab interactor 1 | MGI:2385695 |
| ENSMUSG00000028229 | Rmdn1 | regulator of microtubule dynamics 1 | MGI:1913552 |
| ENSMUSG00000070730 | Rmdn3 | regulator of microtubule dynamics 3 | MGI:1915059 |
| ENSMUSG00000019763 | Rmnd1 | required for meiotic nuclear division 1 homolog (S. cerevisiae) | MGI:1913334 |
| ENSMUSG00000017144 | Rnd3 | Rho family GTPase 3 | MGI:1921444 |
| ENSMUSG00000035890 | Rnf126 | ring finger protein 126 | MGI:1917544 |
| ENSMUSG00000013878 | Rnf170 | ring finger protein 170 | MGI:1924983 |
| ENSMUSG00000038650 | Rnh1 | ribonuclease/angiogenin inhibitor 1 | MGI:1195456 |
| ENSMUSG00000009535 | Rnmt | RNA (guanine-7-) methyltransferase | MGI:1915147 |
| ENSMUSG00000041926 | Rnpep | arginyl aminopeptidase (aminopeptidase B) | MGI:2384902 |
| ENSMUSG00000022883 | Robo1 | roundabout homolog 1 (Drosophila) | MGI:1274781 |
| ENSMUSG00000052516 | Robo2 | roundabout homolog 2 (Drosophila) | MGI:1890110 |
| ENSMUSG00000020580 | Rock2 | Rho-associated coiled-coil containing protein kinase 2 | MGI:107926 |
| ENSMUSG00000022540 | Rogdi | rogdi homolog (Drosophila) | MGI:1913299 |
| ENSMUSG00000060090 | Rp2h | retinitis pigmentosa 2 homolog (human) | MGI:1277953 |
| ENSMUSG00000038510 | Rpf2 | ribosome production factor 2 homolog (S. cerevisiae) | MGI:1914489 |
| ENSMUSG00000029608 | Rph3a | rabphilin 3A | MGI:102788 |
| ENSMUSG00000037805 | Rpl10a | ribosomal protein L10A | MGI:1343877 |
| ENSMUSG00000058443 | Rpl10-ps3 | ribosomal protein L10, pseudogene 3 | MGI:3704336 |
| ENSMUSG00000038900 | Rpl12 | ribosomal protein L12 | MGI:98002 |
| ENSMUSG00000074129 | Rpl13a | ribosomal protein L13A | MGI:1351455 |
| ENSMUSG00000000740 | Rpl13 | ribosomal protein L13 | MGI:105922 |
| ENSMUSG00000025794 | Rpl14 | ribosomal protein L14 | MGI:1914365 |
| ENSMUSG00000062328 | Rpl17 | ribosomal protein L17 | MGI:2448270 |
| ENSMUSG00000045128 | Rpl18a | ribosomal protein L18A | MGI:1924058 |
| ENSMUSG00000059070 | Rpl18 | ribosomal protein L18 | MGI:98003 |
| ENSMUSG00000017404 | Rpl19 | ribosomal protein L19 | MGI:98020 |
| ENSMUSG00000041453 | Rpl21 | ribosomal protein L21 | MGI:1278340 |
| ENSMUSG00000039221 | Rpl22l1 | ribosomal protein L22 like 1 | MGI:1915278 |
| ENSMUSG00000028936 | Rpl22 | ribosomal protein L22 | MGI:99262 |
| ENSMUSG00000058546 | Rpl23a | ribosomal protein L23A | MGI:3040672 |
| ENSMUSG00000071415 | Rpl23 | ribosomal protein L23 | MGI:1929455 |
| ENSMUSG00000098274 | Rpl24 | ribosomal protein L24 | MGI:1915443 |
| ENSMUSG00000060938 | Rpl26 | ribosomal protein L26 | MGI:106022 |
| ENSMUSG00000046364 | Rpl27a | ribosomal protein L27A | MGI:1347076 |
| ENSMUSG00000073640 | Rpl27-ps3 | ribosomal protein L27, pseudogene 3 | MGI:3646174 |
| ENSMUSG00000030432 | Rpl28 | ribosomal protein L28 | MGI:101839 |
| ENSMUSG00000058600 | Rpl30 | ribosomal protein L30 | MGI:98037 |
| ENSMUSG00000073702 | Rpl31 | ribosomal protein L31 | MGI:2149632 |
| ENSMUSG00000057841 | Rpl32 | ribosomal protein L32 | MGI:98038 |
| ENSMUSG00000062006 | Rpl34 | ribosomal protein L34 | MGI:1915686 |
| ENSMUSG00000057863 | Rpl36 | ribosomal protein L36 | MGI:1860603 |
| ENSMUSG00000046330 | Rpl37a | ribosomal protein L37a | MGI:98068 |
| ENSMUSG00000057322 | Rpl38 | ribosomal protein L38 | MGI:1914921 |
| ENSMUSG00000060036 | Rpl3 | ribosomal protein L3 | MGI:1351605 |
| ENSMUSG00000032399 | Rpl4 | ribosomal protein L4 | MGI:1915141 |
| ENSMUSG00000058558 | Rpl5 | ribosomal protein L5 | MGI:102854 |
| ENSMUSG00000029614 | Rpl6 | ribosomal protein L6 | MGI:108057 |
| ENSMUSG00000062647 | Rpl7a | ribosomal protein L7A | MGI:1353472 |
| ENSMUSG00000043716 | Rpl7 | ribosomal protein L7 | MGI:98073 |
| ENSMUSG00000003970 | Rpl8 | ribosomal protein L8 | MGI:1350927 |
| ENSMUSG00000047215 | Rpl9 | ribosomal protein L9 | MGI:1298373 |
| ENSMUSG00000067274 | Rplp0 | ribosomal protein, large, P0 | MGI:1927636 |
| ENSMUSG00000007892 | Rplp1 | ribosomal protein, large, P1 | MGI:1927099 |
| ENSMUSG00000025508 | Rplp2 | ribosomal protein, large P2 | MGI:1914436 |
| ENSMUSG00000030062 | Rpn1 | ribophorin I | MGI:98084 |
| ENSMUSG00000027642 | Rpn2 | ribophorin II | MGI:98085 |
| ENSMUSG00000052146 | Rps10 | ribosomal protein S10 | MGI:1914347 |
| ENSMUSG00000003429 | Rps11 | ribosomal protein S11 | MGI:1351329 |
| ENSMUSG00000067038 | Rps12-ps3 | ribosomal protein S12, pseudogene 3 | MGI:3704503 |
| ENSMUSG00000090862 | Rps13 | ribosomal protein S13 | MGI:1915302 |
| ENSMUSG00000024608 | Rps14 | ribosomal protein S14 | MGI:98107 |
| ENSMUSG00000008683 | Rps15a | ribosomal protein S15A | MGI:2389091 |
| ENSMUSG00000063457 | Rps15 | ribosomal protein S15 | MGI:98117 |
| ENSMUSG00000037563 | Rps16 | ribosomal protein S16 | MGI:98118 |
| ENSMUSG00000061787 | Rps17 | ribosomal protein S17 | MGI:1309526 |
| ENSMUSG00000040952 | Rps19 | ribosomal protein S19 | MGI:1333780 |
| ENSMUSG00000028234 | Rps20 | ribosomal protein S20 | MGI:1914677 |
| ENSMUSG00000025290 | Rps24 | ribosomal protein S24 | MGI:98147 |
| ENSMUSG00000009927 | Rps25 | ribosomal protein S25 | MGI:1922867 |
| ENSMUSG00000059775 | Rps26-ps1 | ribosomal protein S26, pseudogene 1 | MGI:3704322 |
| ENSMUSG00000020460 | Rps27a | ribosomal protein S27A | MGI:1925544 |
| ENSMUSG00000050621 | Rps27rt | ribosomal protein S27, retrogene | MGI:3704345 |
| ENSMUSG00000067288 | Rps28 | ribosomal protein S28 | MGI:1859516 |
| ENSMUSG00000034892 | Rps29 | ribosomal protein S29 | MGI:107681 |
| ENSMUSG00000028081 | Rps3a1 | ribosomal protein S3A1 | MGI:1202063 |
| ENSMUSG00000030744 | Rps3 | ribosomal protein S3 | MGI:1350917 |
| ENSMUSG00000012848 | Rps5 | ribosomal protein S5 | MGI:1097682 |
| ENSMUSG00000028495 | Rps6 | ribosomal protein S6 | MGI:98159 |
| ENSMUSG00000047675 | Rps8 | ribosomal protein S8 | MGI:98166 |
| ENSMUSG00000006333 | Rps9 | ribosomal protein S9 | MGI:1924096 |
| ENSMUSG00000032518 | Rpsa | ribosomal protein SA | MGI:105381 |
| ENSMUSG00000055723 | Rras2 | related RAS viral (r-ras) oncogene homolog 2 | MGI:1914172 |
| ENSMUSG00000038387 | Rras | Harvey rat sarcoma oncogene, subgroup R | MGI:98179 |
| ENSMUSG00000027422 | Rrbp1 | ribosome binding protein 1 | MGI:1932395 |
| ENSMUSG00000001783 | Rtcb | RNA 2',3'-cyclic phosphate and 5'-OH ligase | MGI:106379 |
| ENSMUSG00000021087 | Rtn1 | reticulon 1 | MGI:1933947 |
| ENSMUSG00000024758 | Rtn3 | reticulon 3 | MGI:1339970 |
| ENSMUSG00000019864 | Rtn4ip1 | reticulon 4 interacting protein 1 | MGI:2178759 |
| ENSMUSG00000020458 | Rtn4 | reticulon 4 | MGI:1915835 |
| ENSMUSG00000050896 | Rtn4rl2 | reticulon 4 receptor-like 2 | MGI:2669796 |
| ENSMUSG00000029291 | Rufy3 | RUN and FYVE domain containing 3 | MGI:106484 |
| ENSMUSG00000006575 | Rundc3a | RUN domain containing 3A | MGI:1858752 |
| ENSMUSG00000003868 | Ruvbl2 | RuvB-like protein 2 | MGI:1342299 |
| ENSMUSG00000021313 | Ryr2 | ryanodine receptor 2, cardiac | MGI:99685 |
| ENSMUSG00000074457 | S100a16 | S100 calcium binding protein A16 | MGI:1915110 |
| ENSMUSG00000044080 | S100a1 | S100 calcium binding protein A1 | MGI:1338917 |
| ENSMUSG00000033208 | S100b | S100 protein, beta polypeptide, neural | MGI:98217 |
| ENSMUSG00000045092 | S1pr1 | sphingosine-1-phosphate receptor 1 | MGI:1096355 |
| ENSMUSG00000025240 | Sacm1l | SAC1 (suppressor of actin mutations 1, homolog)-like (S. cerevisiae) | MGI:1933169 |
| ENSMUSG00000048279 | Sacs | sacsin | MGI:1354724 |
| ENSMUSG00000022437 | Samm50 | sorting and assembly machinery component 50 homolog (S. cerevisiae) | MGI:1915903 |
| ENSMUSG00000020088 | Sar1a | SAR1 gene homolog A (S. cerevisiae) | MGI:98230 |
| ENSMUSG00000050132 | Sarm1 | sterile alpha and HEAT/Armadillo motif containing 1 | MGI:2136419 |
| ENSMUSG00000068739 | Sars | seryl-aminoacyl-tRNA synthetase | MGI:102809 |
| ENSMUSG00000036529 | Sbf1 | SET binding factor 1 | MGI:1925230 |
| ENSMUSG00000035236 | Scai | suppressor of cancer cell invasion | MGI:2443716 |
| ENSMUSG00000021687 | Scamp1 | secretory carrier membrane protein 1 | MGI:1349480 |
| ENSMUSG00000040188 | Scamp2 | secretory carrier membrane protein 2 | MGI:1346518 |
| ENSMUSG00000028049 | Scamp3 | secretory carrier membrane protein 3 | MGI:1346346 |
| ENSMUSG00000078441 | Scamp4 | secretory carrier membrane protein 4 | MGI:1928947 |
| ENSMUSG00000040722 | Scamp5 | secretory carrier membrane protein 5 | MGI:1928948 |
| ENSMUSG00000029426 | Scarb2 | scavenger receptor class B, member 2 | MGI:1196458 |
| ENSMUSG00000038936 | Sccpdh | saccharopine dehydrogenase (putative) | MGI:1924486 |
| ENSMUSG00000020952 | Scfd1 | Sec1 family domain containing 1 | MGI:1924233 |
| ENSMUSG00000062110 | Scfd2 | Sec1 family domain containing 2 | MGI:2443446 |
| ENSMUSG00000050711 | Scg2 | secretogranin II | MGI:103033 |
| ENSMUSG00000064329 | Scn1a | sodium channel, voltage-gated, type I, alpha | MGI:98246 |
| ENSMUSG00000019194 | Scn1b | sodium channel, voltage-gated, type I, beta | MGI:98247 |
| ENSMUSG00000075318 | Scn2a1 | sodium channel, voltage-gated, type II, alpha 1 | MGI:98248 |
| ENSMUSG00000070304 | Scn2b | sodium channel, voltage-gated, type II, beta | MGI:106921 |
| ENSMUSG00000057182 | Scn3a | sodium channel, voltage-gated, type III, alpha | MGI:98249 |
| ENSMUSG00000049281 | Scn3b | sodium channel, voltage-gated, type III, beta | MGI:1918882 |
| ENSMUSG00000046480 | Scn4b | sodium channel, type IV, beta | MGI:2687406 |
| ENSMUSG00000023033 | Scn8a | sodium channel, voltage-gated, type VIII, alpha | MGI:103169 |
| ENSMUSG00000069844 | Sco1 | SCO cytochrome oxidase deficient homolog 1 (yeast) | MGI:106362 |
| ENSMUSG00000091780 | Sco2 | SCO cytochrome oxidase deficient homolog 2 (yeast) | MGI:3818630 |
| ENSMUSG00000028603 | Scp2 | sterol carrier protein 2, liver | MGI:98254 |
| ENSMUSG00000022568 | Scrib | scribbled homolog (Drosophila) | MGI:2145950 |
| ENSMUSG00000019124 | Scrn1 | secernin 1 | MGI:1917188 |
| ENSMUSG00000069539 | Scyl2 | SCY1-like 2 (S. cerevisiae) | MGI:1289172 |
| ENSMUSG00000025743 | Sdc3 | syndecan 3 | MGI:1349163 |
| ENSMUSG00000017009 | Sdc4 | syndecan 4 | MGI:1349164 |
| ENSMUSG00000028249 | Sdcbp | syndecan binding protein | MGI:1337026 |
| ENSMUSG00000022769 | Sdf2l1 | stromal cell-derived factor 2-like 1 | MGI:2149842 |
| ENSMUSG00000002064 | Sdf2 | stromal cell derived factor 2 | MGI:108019 |
| ENSMUSG00000074211 | Sdhaf1 | succinate dehydrogenase complex assembly factor 1 | MGI:1915582 |
| ENSMUSG00000021577 | Sdha | succinate dehydrogenase complex, subunit A, flavoprotein (Fp) | MGI:1914195 |
| ENSMUSG00000009863 | Sdhb | succinate dehydrogenase complex, subunit B, iron sulfur (Ip) | MGI:1914930 |
| ENSMUSG00000058076 | Sdhc | succinate dehydrogenase complex, subunit C, integral membrane protein | MGI:1913302 |
| ENSMUSG00000000171 | Sdhd | succinate dehydrogenase complex, subunit D, integral membrane protein | MGI:1914175 |
| ENSMUSG00000041592 | Sdk2 | sidekick homolog 2 (chicken) | MGI:2443847 |
| ENSMUSG00000022223 | Sdr39u1 | short chain dehydrogenase/reductase family 39U, member 1 | MGI:1916876 |
| ENSMUSG00000025724 | Sec11a | SEC11 homolog A (S. cerevisiae) | MGI:1929464 |
| ENSMUSG00000024516 | Sec11c | SEC11 homolog C (S. cerevisiae) | MGI:1913536 |
| ENSMUSG00000027879 | Sec22b | SEC22 vesicle trafficking protein homolog B (S. cerevisiae) | MGI:1338759 |
| ENSMUSG00000020986 | Sec23a | SEC23A (S. cerevisiae) | MGI:1349635 |
| ENSMUSG00000001052 | Sec24b | Sec24 related gene family, member B (S. cerevisiae) | MGI:2139764 |
| ENSMUSG00000039367 | Sec24c | Sec24 related gene family, member C (S. cerevisiae) | MGI:1919746 |
| ENSMUSG00000035325 | Sec31a | Sec31 homolog A (S. cerevisiae) | MGI:1916412 |
| ENSMUSG00000053317 | Sec61b | Sec61 beta subunit | MGI:1913462 |
| ENSMUSG00000078974 | Sec61g | SEC61, gamma subunit | MGI:1202066 |
| ENSMUSG00000019802 | Sec63 | SEC63-like (S. cerevisiae) | MGI:2155302 |
| ENSMUSG00000020964 | Sel1l | sel-1 suppressor of lin-12-like (C. elegans) | MGI:1329016 |
| ENSMUSG00000042682 | Selk | selenoprotein K | MGI:1931466 |
| ENSMUSG00000075702 | Selm | selenoprotein M | MGI:2149786 |
| ENSMUSG00000075700 | Selt | selenoprotein T | MGI:1916477 |
| ENSMUSG00000028064 | Sema4a | sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A | MGI:107560 |
| ENSMUSG00000030539 | Sema4b | sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4B | MGI:107559 |
| ENSMUSG00000021451 | Sema4d | sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4D | MGI:109244 |
| ENSMUSG00000000627 | Sema4f | sema domain, immunoglobulin domain (Ig), TM domain, and short cytoplasmic domain | MGI:1340055 |
| ENSMUSG00000001227 | Sema6b | sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6B | MGI:1202889 |
| ENSMUSG00000038264 | Sema7a | sema domain, immunoglobulin domain (Ig), and GPI membrane anchor, (semaphorin) 7A | MGI:1306826 |
| ENSMUSG00000037072 | Sep15 | selenoprotein | MGI:1927947 |
| ENSMUSG00000058013 | Sept11 | septin 11 | MGI:1277214 |
| ENSMUSG00000026276 | Sept2 | septin 2 | MGI:97298 |
| ENSMUSG00000022456 | Sept3 | septin 3 | MGI:1345148 |
| ENSMUSG00000020486 | Sept4 | septin 4 | MGI:1270156 |
| ENSMUSG00000072214 | Sept5 | septin 5 | MGI:1195461 |
| ENSMUSG00000050379 | Sept6 | septin 6 | MGI:1888939 |
| ENSMUSG00000001833 | Sept7 | septin 7 | MGI:1335094 |
| ENSMUSG00000018398 | Sept8 | septin 8 | MGI:894310 |
| ENSMUSG00000059248 | Sept9 | septin 9 | MGI:1858222 |
| ENSMUSG00000019877 | Serinc1 | serine incorporator 1 | MGI:1926228 |
| ENSMUSG00000058207 | Serpina3k | serine (or cysteine) peptidase inhibitor, clade A, member 3K | MGI:98377 |
| ENSMUSG00000044734 | Serpinb1a | serine (or cysteine) peptidase inhibitor, clade B, member 1a | MGI:1913472 |
| ENSMUSG00000060147 | Serpinb6a | serine (or cysteine) peptidase inhibitor, clade B, member 6a | MGI:103123 |
| ENSMUSG00000026249 | Serpine2 | serine (or cysteine) peptidase inhibitor, clade E, member 2 | MGI:101780 |
| ENSMUSG00000070436 | Serpinh1 | serine (or cysteine) peptidase inhibitor, clade H, member 1 | MGI:88283 |
| ENSMUSG00000042272 | Sestd1 | SEC14 and spectrin domains 1 | MGI:1916262 |
| ENSMUSG00000030683 | Sez6l2 | seizure related 6 homolog like 2 | MGI:2385295 |
| ENSMUSG00000058153 | Sez6l | seizure related 6 homolog like | MGI:1935121 |
| ENSMUSG00000006527 | Sfmbt1 | Scm-like with four mbt domains 1 | MGI:1859609 |
| ENSMUSG00000028820 | Sfpq | splicing factor proline/glutamine rich (polypyrimidine tract binding protein associated) | MGI:1918764 |
| ENSMUSG00000021474 | Sfxn1 | sideroflexin 1 | MGI:2137677 |
| ENSMUSG00000025212 | Sfxn3 | sideroflexin 3 | MGI:2137679 |
| ENSMUSG00000033720 | Sfxn5 | sideroflexin 5 | MGI:2137681 |
| ENSMUSG00000028524 | Sgip1 | SH3-domain GRB2-like (endophilin) interacting protein 1 | MGI:1920344 |
| ENSMUSG00000020097 | Sgpl1 | sphingosine phosphate lyase 1 | MGI:1261415 |
| ENSMUSG00000004937 | Sgta | small glutamine-rich tetratricopeptide repeat (TPR)-containing, alpha | MGI:1098703 |
| ENSMUSG00000032261 | Sh3bgrl2 | SH3 domain binding glutamic acid-rich protein like 2 | MGI:1915350 |
| ENSMUSG00000028843 | Sh3bgrl3 | SH3 domain binding glutamic acid-rich protein-like 3 | MGI:1920973 |
| ENSMUSG00000022436 | Sh3bp1 | SH3-domain binding protein 1 | MGI:104603 |
| ENSMUSG00000003200 | Sh3gl1 | SH3-domain GRB2-like 1 | MGI:700010 |
| ENSMUSG00000028488 | Sh3gl2 | SH3-domain GRB2-like 2 | MGI:700009 |
| ENSMUSG00000037062 | Sh3glb1 | SH3-domain GRB2-like B1 (endophilin) | MGI:1859730 |
| ENSMUSG00000026860 | Sh3glb2 | SH3-domain GRB2-like endophilin B2 | MGI:2385131 |
| ENSMUSG00000040990 | Sh3kbp1 | SH3-domain kinase binding protein 1 | MGI:1889583 |
| ENSMUSG00000041889 | Shisa4 | shisa family member 4 | MGI:1924802 |
| ENSMUSG00000053930 | Shisa6 | shisa family member 6 | MGI:2685725 |
| ENSMUSG00000053550 | Shisa7 | shisa family member 7 | MGI:3605641 |
| ENSMUSG00000022494 | Shisa9 | shisa family member 9 | MGI:1919805 |
| ENSMUSG00000025403 | Shmt2 | serine hydroxymethyltransferase 2 (mitochondrial) | MGI:1277989 |
| ENSMUSG00000024976 | Shoc2 | soc-2 (suppressor of clear) homolog (C. elegans) | MGI:1927197 |
| ENSMUSG00000034135 | Sik3 | SIK family kinase 3 | MGI:2446296 |
| ENSMUSG00000042700 | Sipa1l1 | signal-induced proliferation-associated 1 like 1 | MGI:2443679 |
| ENSMUSG00000037902 | Sirpa | signal-regulatory protein alpha | MGI:108563 |
| ENSMUSG00000015149 | Sirt2 | sirtuin 2 | MGI:1927664 |
| ENSMUSG00000036309 | Skp1a | S-phase kinase-associated protein 1A | MGI:103575 |
| ENSMUSG00000036087 | Slain2 | SLAIN motif family, member 2 | MGI:1923241 |
| ENSMUSG00000029219 | Slc10a4 | solute carrier family 10 (sodium/bile acid cotransporter family), member 4 | MGI:3606480 |
| ENSMUSG00000024597 | Slc12a2 | solute carrier family 12, member 2 | MGI:101924 |
| ENSMUSG00000017740 | Slc12a5 | solute carrier family 12, member 5 | MGI:1862037 |
| ENSMUSG00000027130 | Slc12a6 | solute carrier family 12, member 6 | MGI:2135960 |
| ENSMUSG00000037344 | Slc12a9 | solute carrier family 12 (potassium/chloride transporters), member 9 | MGI:1933532 |
| ENSMUSG00000018459 | Slc13a3 | solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 3 | MGI:2149635 |
| ENSMUSG00000020805 | Slc13a5 | solute carrier family 13 (sodium-dependent citrate transporter), member 5 | MGI:3037150 |
| ENSMUSG00000059336 | Slc14a1 | solute carrier family 14 (urea transporter), member 1 | MGI:1351654 |
| ENSMUSG00000032902 | Slc16a1 | solute carrier family 16 (monocarboxylic acid transporters), member 1 | MGI:106013 |
| ENSMUSG00000030500 | Slc17a6 | solute carrier family 17 (sodium-dependent inorganic phosphate cotransporter), member 6 | MGI:2156052 |
| ENSMUSG00000070570 | Slc17a7 | solute carrier family 17 (sodium-dependent inorganic phosphate cotransporter), member 7 | MGI:1920211 |
| ENSMUSG00000025094 | Slc18a2 | solute carrier family 18 (vesicular monoamine), member 2 | MGI:106677 |
| ENSMUSG00000024935 | Slc1a1 | solute carrier family 1 (neuronal/epithelial high affinity glutamate transporter, system Xag), member 1 | MGI:105083 |
| ENSMUSG00000005089 | Slc1a2 | solute carrier family 1 (glial high affinity glutamate transporter), member 2 | MGI:101931 |
| ENSMUSG00000005360 | Slc1a3 | solute carrier family 1 (glial high affinity glutamate transporter), member 3 | MGI:99917 |
| ENSMUSG00000020142 | Slc1a4 | solute carrier family 1 (glutamate/neutral amino acid transporter), member 4 | MGI:2135601 |
| ENSMUSG00000005357 | Slc1a6 | solute carrier family 1 (high affinity aspartate/glutamate transporter), member 6 | MGI:1096331 |
| ENSMUSG00000027397 | Slc20a1 | solute carrier family 20, member 1 | MGI:108392 |
| ENSMUSG00000037656 | Slc20a2 | solute carrier family 20, member 2 | MGI:97851 |
| ENSMUSG00000022199 | Slc22a17 | solute carrier family 22 (organic cation transporter), member 17 | MGI:1926225 |
| ENSMUSG00000038267 | Slc22a23 | solute carrier family 22, member 23 | MGI:1920352 |
| ENSMUSG00000020334 | Slc22a4 | solute carrier family 22 (organic cation transporter), member 4 | MGI:1353479 |
| ENSMUSG00000027340 | Slc23a2 | solute carrier family 23 (nucleobase transporters), member 2 | MGI:1859682 |
| ENSMUSG00000037996 | Slc24a2 | solute carrier family 24 (sodium/potassium/calcium exchanger), member 2 | MGI:1923626 |
| ENSMUSG00000041771 | Slc24a4 | solute carrier family 24 (sodium/potassium/calcium exchanger), member 4 | MGI:2447362 |
| ENSMUSG00000025792 | Slc25a10 | solute carrier family 25 (mitochondrial carrier, dicarboxylate transporter), member 10 | MGI:1353497 |
| ENSMUSG00000014606 | Slc25a11 | solute carrier family 25 (mitochondrial carrier oxoglutarate carrier), member 11 | MGI:1915113 |
| ENSMUSG00000027010 | Slc25a12 | solute carrier family 25 (mitochondrial carrier, Aralar), member 12 | MGI:1926080 |
| ENSMUSG00000015112 | Slc25a13 | solute carrier family 25 (mitochondrial carrier, adenine nucleotide translocator), member 13 | MGI:1354721 |
| ENSMUSG00000031105 | Slc25a14 | solute carrier family 25 (mitochondrial carrier, brain), member 14 | MGI:1330823 |
| ENSMUSG00000004902 | Slc25a18 | solute carrier family 25 (mitochondrial carrier), member 18 | MGI:1919053 |
| ENSMUSG00000003528 | Slc25a1 | solute carrier family 25 (mitochondrial carrier, citrate transporter), member 1 | MGI:1345283 |
| ENSMUSG00000032602 | Slc25a20 | solute carrier family 25 (mitochondrial carnitine/acylcarnitine translocase), member 20 | MGI:1928738 |
| ENSMUSG00000019082 | Slc25a22 | solute carrier family 25 (mitochondrial carrier, glutamate), member 22 | MGI:1915517 |
| ENSMUSG00000046329 | Slc25a23 | solute carrier family 25 (mitochondrial carrier phosphate carrier), member 23 | MGI:1914222 |
| ENSMUSG00000026819 | Slc25a25 | solute carrier family 25 (mitochondrial carrier, phosphate carrier), member 25 | MGI:1915913 |
| ENSMUSG00000023912 | Slc25a27 | solute carrier family 25, member 27 | MGI:1921261 |
| ENSMUSG00000018740 | Slc25a35 | solute carrier family 25, member 35 | MGI:1919248 |
| ENSMUSG00000061904 | Slc25a3 | solute carrier family 25 (mitochondrial carrier, phosphate carrier), member 3 | MGI:1353498 |
| ENSMUSG00000054099 | Slc25a40 | solute carrier family 25, member 40 | MGI:2442486 |
| ENSMUSG00000002346 | Slc25a42 | solute carrier family 25, member 42 | MGI:1920345 |
| ENSMUSG00000024259 | Slc25a46 | solute carrier family 25, member 46 | MGI:1914703 |
| ENSMUSG00000031633 | Slc25a4 | solute carrier family 25 (mitochondrial carrier, adenine nucleotide translocator), member 4 | MGI:1353495 |
| ENSMUSG00000045973 | Slc25a51 | solute carrier family 25, member 51 | MGI:2684984 |
| ENSMUSG00000016319 | Slc25a5 | solute carrier family 25 (mitochondrial carrier, adenine nucleotide translocator), member 5 | MGI:1353496 |
| ENSMUSG00000031808 | Slc27a1 | solute carrier family 27 (fatty acid transporter), member 1 | MGI:1347098 |
| ENSMUSG00000059316 | Slc27a4 | solute carrier family 27 (fatty acid transporter), member 4 | MGI:1347347 |
| ENSMUSG00000023942 | Slc29a1 | solute carrier family 29 (nucleoside transporters), member 1 | MGI:1927073 |
| ENSMUSG00000024891 | Slc29a2 | solute carrier family 29 (nucleoside transporters), member 2 | MGI:1345278 |
| ENSMUSG00000036298 | Slc2a13 | solute carrier family 2 (facilitated glucose transporter), member 13 | MGI:2146030 |
| ENSMUSG00000028645 | Slc2a1 | solute carrier family 2 (facilitated glucose transporter), member 1 | MGI:95755 |
| ENSMUSG00000003153 | Slc2a3 | solute carrier family 2 (facilitated glucose transporter), member 3 | MGI:95757 |
| ENSMUSG00000037434 | Slc30a1 | solute carrier family 30 (zinc transporter), member 1 | MGI:1345281 |
| ENSMUSG00000029151 | Slc30a3 | solute carrier family 30 (zinc transporter), member 3 | MGI:1345280 |
| ENSMUSG00000029221 | Slc30a9 | solute carrier family 30 (zinc transporter), member 9 | MGI:1923690 |
| ENSMUSG00000037771 | Slc32a1 | solute carrier family 32 (GABA vesicular transporter), member 1 | MGI:1194488 |
| ENSMUSG00000033272 | Slc35a4 | solute carrier family 35, member A4 | MGI:1915093 |
| ENSMUSG00000038602 | Slc35f1 | solute carrier family 35, member F1 | MGI:2139810 |
| ENSMUSG00000070287 | Slc35g2 | solute carrier family 35, member G2 | MGI:2685365 |
| ENSMUSG00000010064 | Slc38a3 | solute carrier family 38, member 3 | MGI:1923507 |
| ENSMUSG00000025986 | Slc39a10 | solute carrier family 39 (zinc transporter), member 10 | MGI:1914515 |
| ENSMUSG00000024270 | Slc39a6 | solute carrier family 39 (metal ion transporter), member 6 | MGI:2147279 |
| ENSMUSG00000024327 | Slc39a7 | solute carrier family 39 (zinc transporter), member 7 | MGI:95909 |
| ENSMUSG00000010095 | Slc3a2 | solute carrier family 3 (activators of dibasic and neutral amino acid transport), member 2 | MGI:96955 |
| ENSMUSG00000038178 | Slc43a2 | solute carrier family 43, member 2 | MGI:2442746 |
| ENSMUSG00000028412 | Slc44a1 | solute carrier family 44, member 1 | MGI:2140592 |
| ENSMUSG00000057193 | Slc44a2 | solute carrier family 44, member 2 | MGI:1915932 |
| ENSMUSG00000039838 | Slc45a1 | solute carrier family 45, member 1 | MGI:2653235 |
| ENSMUSG00000026904 | Slc4a10 | solute carrier family 4, sodium bicarbonate cotransporter-like, member 10 | MGI:2150150 |
| ENSMUSG00000006574 | Slc4a1 | solute carrier family 4 (anion exchanger), member 1 | MGI:109393 |
| ENSMUSG00000006576 | Slc4a3 | solute carrier family 4 (anion exchanger), member 3 | MGI:109350 |
| ENSMUSG00000060961 | Slc4a4 | solute carrier family 4 (anion exchanger), member 4 | MGI:1927555 |
| ENSMUSG00000021733 | Slc4a7 | solute carrier family 4, sodium bicarbonate cotransporter, member 7 | MGI:2443878 |
| ENSMUSG00000023032 | Slc4a8 | solute carrier family 4 (anion exchanger), member 8 | MGI:1928745 |
| ENSMUSG00000089774 | Slc5a3 | solute carrier family 5 (inositol transporters), member 3 | MGI:1858226 |
| ENSMUSG00000030307 | Slc6a11 | solute carrier family 6 (neurotransmitter transporter, GABA), member 11 | MGI:95630 |
| ENSMUSG00000027894 | Slc6a17 | solute carrier family 6 (neurotransmitter transporter), member 17 | MGI:2442535 |
| ENSMUSG00000030310 | Slc6a1 | solute carrier family 6 (neurotransmitter transporter, GABA), member 1 | MGI:95627 |
| ENSMUSG00000021609 | Slc6a3 | solute carrier family 6 (neurotransmitter transporter, dopamine), member 3 | MGI:94862 |
| ENSMUSG00000020838 | Slc6a4 | solute carrier family 6 (neurotransmitter transporter, serotonin), member 4 | MGI:96285 |
| ENSMUSG00000039728 | Slc6a5 | solute carrier family 6 (neurotransmitter transporter, glycine), member 5 | MGI:105090 |
| ENSMUSG00000052026 | Slc6a7 | solute carrier family 6 (neurotransmitter transporter, L-proline), member 7 | MGI:2147363 |
| ENSMUSG00000028542 | Slc6a9 | solute carrier family 6 (neurotransmitter transporter, glycine), member 9 | MGI:95760 |
| ENSMUSG00000027737 | Slc7a11 | solute carrier family 7 (cationic amino acid transporter, y+ system), member 11 | MGI:1347355 |
| ENSMUSG00000069072 | Slc7a14 | solute carrier family 7 (cationic amino acid transporter, y+ system), member 14 | MGI:3040688 |
| ENSMUSG00000041313 | Slc7a1 | solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 | MGI:88117 |
| ENSMUSG00000031596 | Slc7a2 | solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 | MGI:99828 |
| ENSMUSG00000040010 | Slc7a5 | solute carrier family 7 (cationic amino acid transporter, y+ system), member 5 | MGI:1298205 |
| ENSMUSG00000031904 | Slc7a6 | solute carrier family 7 (cationic amino acid transporter, y+ system), member 6 | MGI:2142598 |
| ENSMUSG00000022180 | Slc7a8 | solute carrier family 7 (cationic amino acid transporter, y+ system), member 8 | MGI:1355323 |
| ENSMUSG00000054640 | Slc8a1 | solute carrier family 8 (sodium/calcium exchanger), member 1 | MGI:107956 |
| ENSMUSG00000030376 | Slc8a2 | solute carrier family 8 (sodium/calcium exchanger), member 2 | MGI:107996 |
| ENSMUSG00000079055 | Slc8a3 | solute carrier family 8 (sodium/calcium exchanger), member 3 | MGI:107976 |
| ENSMUSG00000028854 | Slc9a1 | solute carrier family 9 (sodium/hydrogen exchanger), member 1 | MGI:102462 |
| ENSMUSG00000020733 | Slc9a3r1 | solute carrier family 9 (sodium/hydrogen exchanger), member 3 regulator 1 | MGI:1349482 |
| ENSMUSG00000002504 | Slc9a3r2 | solute carrier family 9 (sodium/hydrogen exchanger), member 3 regulator 2 | MGI:1890662 |
| ENSMUSG00000060681 | Slc9a6 | solute carrier family 9 (sodium/hydrogen exchanger), member 6 | MGI:2443511 |
| ENSMUSG00000037341 | Slc9a7 | solute carrier family 9 (sodium/hydrogen exchanger), member 7 | MGI:2444530 |
| ENSMUSG00000025020 | Slit1 | slit homolog 1 (Drosophila) | MGI:1315203 |
| ENSMUSG00000075478 | Slitrk1 | SLIT and NTRK-like family, member 1 | MGI:2679446 |
| ENSMUSG00000048304 | Slitrk3 | SLIT and NTRK-like family, member 3 | MGI:2679447 |
| ENSMUSG00000046699 | Slitrk4 | SLIT and NTRK-like family, member 4 | MGI:2442509 |
| ENSMUSG00000025060 | Slk | STE20-like kinase | MGI:103241 |
| ENSMUSG00000021870 | Slmap | sarcolemma associated protein | MGI:1933549 |
| ENSMUSG00000026155 | Smap1 | small ArfGAP 1 | MGI:2138261 |
| ENSMUSG00000078350 | Smim1 | small integral membrane protein 1 | MGI:1916109 |
| ENSMUSG00000061461 | Smim20 | small integral membrane protein 20 | MGI:1913528 |
| ENSMUSG00000031906 | Smpd3 | sphingomyelin phosphodiesterase 3, neutral | MGI:1927578 |
| ENSMUSG00000027287 | Snap23 | synaptosomal-associated protein 23 | MGI:109356 |
| ENSMUSG00000027273 | Snap25 | synaptosomal-associated protein 25 | MGI:98331 |
| ENSMUSG00000022765 | Snap29 | synaptosomal-associated protein 29 | MGI:1914724 |
| ENSMUSG00000009894 | Snap47 | synaptosomal-associated protein, 47 | MGI:1915076 |
| ENSMUSG00000033419 | Snap91 | synaptosomal-associated protein 91 | MGI:109132 |
| ENSMUSG00000001018 | Snapin | SNAP-associated protein | MGI:1333745 |
| ENSMUSG00000025889 | Snca | synuclein, alpha | MGI:1277151 |
| ENSMUSG00000034891 | Sncb | synuclein, beta | MGI:1889011 |
| ENSMUSG00000023064 | Sncg | synuclein, gamma | MGI:1298397 |
| ENSMUSG00000001424 | Snd1 | staphylococcal nuclease and tudor domain containing 1 | MGI:1929266 |
| ENSMUSG00000037972 | Snn | stannin | MGI:1276549 |
| ENSMUSG00000027457 | Snph | syntaphilin | MGI:2139270 |
| ENSMUSG00000020018 | Snrpf | small nuclear ribonucleoprotein polypeptide F | MGI:1917128 |
| ENSMUSG00000027488 | Snta1 | syntrophin, acidic 1 | MGI:101772 |
| ENSMUSG00000060429 | Sntb1 | syntrophin, basic 1 | MGI:101781 |
| ENSMUSG00000046032 | Snx12 | sorting nexin 12 | MGI:1919331 |
| ENSMUSG00000027534 | Snx16 | sorting nexin 16 | MGI:1921968 |
| ENSMUSG00000032382 | Snx1 | sorting nexin 1 | MGI:1928395 |
| ENSMUSG00000038291 | Snx25 | sorting nexin 25 | MGI:2142610 |
| ENSMUSG00000028136 | Snx27 | sorting nexin family member 27 | MGI:1923992 |
| ENSMUSG00000034484 | Snx2 | sorting nexin 2 | MGI:1915054 |
| ENSMUSG00000028385 | Snx30 | sorting nexin family member 30 | MGI:2443882 |
| ENSMUSG00000019804 | Snx3 | sorting nexin 3 | MGI:1860188 |
| ENSMUSG00000022808 | Snx4 | sorting nexin 4 | MGI:1916400 |
| ENSMUSG00000027423 | Snx5 | sorting nexin 5 | MGI:1916428 |
| ENSMUSG00000005656 | Snx6 | sorting nexin 6 | MGI:1919433 |
| ENSMUSG00000022982 | Sod1 | superoxide dismutase 1, soluble | MGI:98351 |
| ENSMUSG00000006818 | Sod2 | superoxide dismutase 2, mitochondrial | MGI:98352 |
| ENSMUSG00000038916 | Soga3 | SOGA family member 3 | MGI:1914662 |
| ENSMUSG00000025006 | Sorbs1 | sorbin and SH3 domain containing 1 | MGI:700014 |
| ENSMUSG00000029093 | Sorcs2 | sortilin-related VPS10 domain containing receptor 2 | MGI:1932289 |
| ENSMUSG00000049313 | Sorl1 | sortilin-related receptor, LDLR class A repeats-containing | MGI:1202296 |
| ENSMUSG00000068747 | Sort1 | sortilin 1 | MGI:1338015 |
| ENSMUSG00000020859 | Spag9 | sperm associated antigen 9 | MGI:1918084 |
| ENSMUSG00000029309 | Sparcl1 | SPARC-like 1 | MGI:108110 |
| ENSMUSG00000018593 | Sparc | secreted acidic cysteine rich glycoprotein | MGI:98373 |
| ENSMUSG00000051934 | Spats2 | spermatogenesis associated, serine-rich 2 | MGI:1919822 |
| ENSMUSG00000035227 | Spcs2 | signal peptidase complex subunit 2 homolog (S. cerevisiae) | MGI:1913874 |
| ENSMUSG00000042331 | Specc1 | sperm antigen with calponin homology and coiled-coil domains 1 | MGI:2442356 |
| ENSMUSG00000000738 | Spg7 | spastic paraplegia 7 homolog (human) | MGI:2385906 |
| ENSMUSG00000026163 | Sphkap | SPHK1 interactor, AKAP domain containing | MGI:1924879 |
| ENSMUSG00000024533 | Spire1 | spire homolog 1 (Drosophila) | MGI:1915416 |
| ENSMUSG00000056222 | Spock1 | sparc/osteonectin, cwcv and kazal-like domains proteoglycan 1 | MGI:105371 |
| ENSMUSG00000027351 | Spred1 | sprouty protein with EVH-1 domain 1, related sequence | MGI:2150016 |
| ENSMUSG00000045671 | Spred2 | sprouty-related, EVH1 domain containing 2 | MGI:2150019 |
| ENSMUSG00000033735 | Spr | sepiapterin reductase | MGI:103078 |
| ENSMUSG00000022114 | Spry2 | sprouty homolog 2 (Drosophila) | MGI:1345138 |
| ENSMUSG00000021930 | Spryd7 | SPRY domain containing 7 | MGI:1913924 |
| ENSMUSG00000026532 | Spta1 | spectrin alpha, erythrocytic 1 | MGI:98385 |
| ENSMUSG00000057738 | Sptan1 | spectrin alpha, non-erythrocytic 1 | MGI:98386 |
| ENSMUSG00000020315 | Sptbn1 | spectrin beta, non-erythrocytic 1 | MGI:98388 |
| ENSMUSG00000067889 | Sptbn2 | spectrin beta, non-erythrocytic 2 | MGI:1313261 |
| ENSMUSG00000011751 | Sptbn4 | spectrin beta, non-erythrocytic 4 | MGI:1890574 |
| ENSMUSG00000021061 | Sptb | spectrin beta, erythrocytic | MGI:98387 |
| ENSMUSG00000021468 | Sptlc1 | serine palmitoyltransferase, long chain base subunit 1 | MGI:1099431 |
| ENSMUSG00000005803 | Sqrdl | sulfide quinone reductase-like (yeast) | MGI:1929899 |
| ENSMUSG00000015837 | Sqstm1 | sequestosome 1 | MGI:107931 |
| ENSMUSG00000038453 | Srcin1 | SRC kinase signaling inhibitor 1 | MGI:1933179 |
| ENSMUSG00000027646 | Src | Rous sarcoma oncogene | MGI:98397 |
| ENSMUSG00000026425 | Srgap2 | SLIT-ROBO Rho GTPase activating protein 2 | MGI:109605 |
| ENSMUSG00000030257 | Srgap3 | SLIT-ROBO Rho GTPase activating protein 3 | MGI:2152938 |
| ENSMUSG00000003161 | Sri | sorcin | MGI:98419 |
| ENSMUSG00000009549 | Srp14 | signal recognition particle 14 | MGI:107169 |
| ENSMUSG00000020780 | Srp68 | signal recognition particle 68 | MGI:1917447 |
| ENSMUSG00000062604 | Srpk2 | serine/arginine-rich protein specific kinase 2 | MGI:1201408 |
| ENSMUSG00000032042 | Srpr | signal recognition particle receptor ('docking protein') | MGI:1914648 |
| ENSMUSG00000001323 | Srr | serine racemase | MGI:1351636 |
| ENSMUSG00000071172 | Srsf3 | serine/arginine-rich splicing factor 3 | MGI:98285 |
| ENSMUSG00000029911 | Ssbp1 | single-stranded DNA binding protein 1 | MGI:1920040 |
| ENSMUSG00000021427 | Ssr1 | signal sequence receptor, alpha | MGI:105082 |
| ENSMUSG00000027828 | Ssr3 | signal sequence receptor, gamma | MGI:1914687 |
| ENSMUSG00000002014 | Ssr4 | signal sequence receptor, delta | MGI:1099464 |
| ENSMUSG00000022403 | St13 | suppression of tumorigenicity 13 | MGI:1917606 |
| ENSMUSG00000009588 | St6galnac1 | ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 1 | MGI:1341826 |
| ENSMUSG00000056812 | St8sia3 | ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 3 | MGI:106019 |
| ENSMUSG00000026718 | Stam | signal transducing adaptor molecule (SH3 domain and ITAM motif) 1 | MGI:1329014 |
| ENSMUSG00000003062 | Stard3nl | STARD3 N-terminal like | MGI:1923455 |
| ENSMUSG00000026389 | Steap3 | STEAP family member 3 | MGI:1915678 |
| ENSMUSG00000030987 | Stim1 | stromal interaction molecule 1 | MGI:107476 |
| ENSMUSG00000039156 | Stim2 | stromal interaction molecule 2 | MGI:2151156 |
| ENSMUSG00000024966 | Stip1 | stress-induced phosphoprotein 1 | MGI:109130 |
| ENSMUSG00000026213 | Stk11ip | serine/threonine kinase 11 interacting protein | MGI:1918978 |
| ENSMUSG00000063410 | Stk24 | serine/threonine kinase 24 | MGI:2385007 |
| ENSMUSG00000028832 | Stmn1 | stathmin 1 | MGI:96739 |
| ENSMUSG00000027500 | Stmn2 | stathmin-like 2 | MGI:98241 |
| ENSMUSG00000027581 | Stmn3 | stathmin-like 3 | MGI:1277137 |
| ENSMUSG00000028455 | Stoml2 | stomatin (Epb7.2)-like 2 | MGI:1913842 |
| ENSMUSG00000026880 | Stom | stomatin | MGI:95403 |
| ENSMUSG00000030224 | Strap | serine/threonine kinase receptor associated protein | MGI:1329037 |
| ENSMUSG00000014601 | Strip1 | striatin interacting protein 1 | MGI:2443884 |
| ENSMUSG00000020954 | Strn3 | striatin, calmodulin binding protein 3 | MGI:2151064 |
| ENSMUSG00000030374 | Strn4 | striatin, calmodulin binding protein 4 | MGI:2142346 |
| ENSMUSG00000024077 | Strn | striatin, calmodulin binding protein | MGI:1333757 |
| ENSMUSG00000032116 | Stt3a | STT3, subunit of the oligosaccharyltransferase complex, homolog A (S. cerevisiae) | MGI:105124 |
| ENSMUSG00000032437 | Stt3b | STT3, subunit of the oligosaccharyltransferase complex, homolog B (S. cerevisiae) | MGI:1915542 |
| ENSMUSG00000039615 | Stub1 | STIP1 homology and U-Box containing protein 1 | MGI:1891731 |
| ENSMUSG00000028879 | Stx12 | syntaxin 12 | MGI:1931027 |
| ENSMUSG00000027522 | Stx16 | syntaxin 16 | MGI:1923396 |
| ENSMUSG00000061455 | Stx17 | syntaxin 17 | MGI:1914977 |
| ENSMUSG00000029125 | Stx18 | syntaxin 18 | MGI:1918366 |
| ENSMUSG00000007207 | Stx1a | syntaxin 1A (brain) | MGI:109355 |
| ENSMUSG00000030806 | Stx1b | syntaxin 1B | MGI:1930705 |
| ENSMUSG00000029428 | Stx2 | syntaxin 2 | MGI:108059 |
| ENSMUSG00000030805 | Stx4a | syntaxin 4A (placental) | MGI:893577 |
| ENSMUSG00000010110 | Stx5a | syntaxin 5A | MGI:1928483 |
| ENSMUSG00000026470 | Stx6 | syntaxin 6 | MGI:1926235 |
| ENSMUSG00000019998 | Stx7 | syntaxin 7 | MGI:1858210 |
| ENSMUSG00000020903 | Stx8 | syntaxin 8 | MGI:1890156 |
| ENSMUSG00000026797 | Stxbp1 | syntaxin binding protein 1 | MGI:107363 |
| ENSMUSG00000027882 | Stxbp3 | syntaxin binding protein 3 | MGI:107362 |
| ENSMUSG00000022829 | Stxbp5l | syntaxin binding protein 5-like | MGI:2443815 |
| ENSMUSG00000019790 | Stxbp5 | syntaxin binding protein 5 (tomosyn) | MGI:1926058 |
| ENSMUSG00000046314 | Stxbp6 | syntaxin binding protein 6 (amisyn) | MGI:2384963 |
| ENSMUSG00000022110 | Sucla2 | succinate-Coenzyme A ligase, ADP-forming, beta subunit | MGI:1306775 |
| ENSMUSG00000052738 | Suclg1 | succinate-CoA ligase, GDP-forming, alpha subunit | MGI:1927234 |
| ENSMUSG00000022024 | Sugt1 | SGT1, suppressor of G2 allele of SKP1 (S. cerevisiae) | MGI:1915205 |
| ENSMUSG00000020265 | Sumo3 | SMT3 suppressor of mif two 3 homolog 3 (yeast) | MGI:1336201 |
| ENSMUSG00000049858 | Suox | sulfite oxidase | MGI:2446117 |
| ENSMUSG00000014867 | Surf4 | surfeit gene 4 | MGI:98445 |
| ENSMUSG00000038486 | Sv2a | synaptic vesicle glycoprotein 2 a | MGI:1927139 |
| ENSMUSG00000053025 | Sv2b | synaptic vesicle glycoprotein 2 b | MGI:1927338 |
| ENSMUSG00000051111 | Sv2c | synaptic vesicle glycoprotein 2c | MGI:1922459 |
| ENSMUSG00000074093 | Svip | small VCP/p97-interacting protein | MGI:1922994 |
| ENSMUSG00000042078 | Svop | SV2 related protein | MGI:1915916 |
| ENSMUSG00000022340 | Sybu | syntabulin (syntaxin-interacting) | MGI:2442392 |
| ENSMUSG00000037217 | Syn1 | synapsin I | MGI:98460 |
| ENSMUSG00000009394 | Syn2 | synapsin II | MGI:103020 |
| ENSMUSG00000059602 | Syn3 | synapsin III | MGI:1351334 |
| ENSMUSG00000032423 | Syncrip | synaptotagmin binding, cytoplasmic RNA interacting protein | MGI:1891690 |
| ENSMUSG00000074736 | Syndig1 | synapse differentiation inducing 1 | MGI:3702158 |
| ENSMUSG00000096054 | Syne1 | spectrin repeat containing, nuclear envelope 1 | MGI:1927152 |
| ENSMUSG00000067629 | Syngap1 | synaptic Ras GTPase activating protein 1 homolog (rat) | MGI:3039785 |
| ENSMUSG00000022415 | Syngr1 | synaptogyrin 1 | MGI:1328323 |
| ENSMUSG00000007021 | Syngr3 | synaptogyrin 3 | MGI:1341881 |
| ENSMUSG00000022973 | Synj1 | synaptojanin 1 | MGI:1354961 |
| ENSMUSG00000043079 | Synpo | synaptopodin | MGI:1099446 |
| ENSMUSG00000020570 | Sypl | synaptophysin-like protein | MGI:108081 |
| ENSMUSG00000031144 | Syp | synaptophysin | MGI:98467 |
| ENSMUSG00000068923 | Syt11 | synaptotagmin XI | MGI:1859547 |
| ENSMUSG00000049303 | Syt12 | synaptotagmin XII | MGI:2159601 |
| ENSMUSG00000058420 | Syt17 | synaptotagmin XVII | MGI:104966 |
| ENSMUSG00000035864 | Syt1 | synaptotagmin I | MGI:99667 |
| ENSMUSG00000026452 | Syt2 | synaptotagmin II | MGI:99666 |
| ENSMUSG00000030731 | Syt3 | synaptotagmin III | MGI:99665 |
| ENSMUSG00000024261 | Syt4 | synaptotagmin IV | MGI:101759 |
| ENSMUSG00000004961 | Syt5 | synaptotagmin V | MGI:1926368 |
| ENSMUSG00000024743 | Syt7 | synaptotagmin VII | MGI:1859545 |
| ENSMUSG00000065954 | Tacc1 | transforming, acidic coiled-coil containing protein 1 | MGI:2443510 |
| ENSMUSG00000022658 | Tagln3 | transgelin 3 | MGI:1926784 |
| ENSMUSG00000025503 | Taldo1 | transaldolase 1 | MGI:1274789 |
| ENSMUSG00000030316 | Tamm41 | TAM41, mitochondrial translocator assembly and maintenance protein, homolog (S. cerevisiae) | MGI:1916221 |
| ENSMUSG00000035168 | Tanc1 | tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1 | MGI:1914110 |
| ENSMUSG00000053580 | Tanc2 | tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2 | MGI:2444121 |
| ENSMUSG00000017291 | Taok1 | TAO kinase 1 | MGI:1914490 |
| ENSMUSG00000059981 | Taok2 | TAO kinase 2 | MGI:1915919 |
| ENSMUSG00000046985 | Tapt1 | transmembrane anterior posterior transformation 1 | MGI:2683537 |
| ENSMUSG00000041459 | Tardbp | TAR DNA binding protein | MGI:2387629 |
| ENSMUSG00000030515 | Tarsl2 | threonyl-tRNA synthetase-like 2 | MGI:2444486 |
| ENSMUSG00000022241 | Tars | threonyl-tRNA synthetase | MGI:106314 |
| ENSMUSG00000042492 | Tbc1d10b | TBC1 domain family, member 10b | MGI:1915699 |
| ENSMUSG00000038520 | Tbc1d17 | TBC1 domain family, member 17 | MGI:2449973 |
| ENSMUSG00000036473 | Tbc1d24 | TBC1 domain family, member 24 | MGI:2443456 |
| ENSMUSG00000005374 | Tbl2 | transducin (beta)-like 2 | MGI:1351652 |
| ENSMUSG00000079658 | Tceb1 | transcription elongation factor B (SIII), polypeptide 1 | MGI:1915173 |
| ENSMUSG00000055839 | Tceb2 | transcription elongation factor B (SIII), polypeptide 2 | MGI:1914923 |
| ENSMUSG00000068039 | Tcp1 | t-complex protein 1 | MGI:98535 |
| ENSMUSG00000041912 | Tdrkh | tudor and KH domain containing protein | MGI:1919884 |
| ENSMUSG00000066621 | Tecpr1 | tectonin beta-propeller repeat containing 1 | MGI:1917631 |
| ENSMUSG00000031708 | Tecr | trans-2,3-enoyl-CoA reductase | MGI:1915408 |
| ENSMUSG00000016150 | Tenm1 | teneurin transmembrane protein 1 | MGI:1345185 |
| ENSMUSG00000049336 | Tenm2 | teneurin transmembrane protein 2 | MGI:1345184 |
| ENSMUSG00000031561 | Tenm3 | teneurin transmembrane protein 3 | MGI:1345183 |
| ENSMUSG00000048078 | Tenm4 | teneurin transmembrane protein 4 | MGI:2447063 |
| ENSMUSG00000029359 | Tesc | tescalcin | MGI:1930803 |
| ENSMUSG00000009670 | Tex11 | testis expressed gene 11 | MGI:1933237 |
| ENSMUSG00000040813 | Tex264 | testis expressed gene 264 | MGI:1096570 |
| ENSMUSG00000040548 | Tex2 | testis expressed gene 2 | MGI:102465 |
| ENSMUSG00000003923 | Tfam | transcription factor A, mitochondrial | MGI:107810 |
| ENSMUSG00000022797 | Tfrc | transferrin receptor | MGI:98822 |
| ENSMUSG00000056429 | Tgoln1 | trans-golgi network protein | MGI:105080 |
| ENSMUSG00000028145 | Them4 | thioesterase superfamily member 4 | MGI:1923028 |
| ENSMUSG00000056665 | Them6 | thioesterase superfamily member 6 | MGI:1925301 |
| ENSMUSG00000048550 | Thnsl1 | threonine synthase-like 1 (bacterial) | MGI:2139347 |
| ENSMUSG00000004929 | Thop1 | thimet oligopeptidase 1 | MGI:1354165 |
| ENSMUSG00000032625 | Thsd7a | thrombospondin, type I, domain containing 7A | MGI:2685683 |
| ENSMUSG00000032011 | Thy1 | thymus cell antigen 1, theta | MGI:98747 |
| ENSMUSG00000038028 | Tigar | Trp53 induced glycolysis repulatory phosphatase | MGI:2442752 |
| ENSMUSG00000089847 | Timm10b | translocase of inner mitochondrial membrane 10B | MGI:1315196 |
| ENSMUSG00000027076 | Timm10 | translocase of inner mitochondrial membrane 10 | MGI:1353429 |
| ENSMUSG00000020219 | Timm13 | translocase of inner mitochondrial membrane 13 | MGI:1353432 |
| ENSMUSG00000062580 | Timm17a | translocase of inner mitochondrial membrane 17a | MGI:1343131 |
| ENSMUSG00000031158 | Timm17b | translocase of inner mitochondrial membrane 17b | MGI:1343176 |
| ENSMUSG00000020843 | Timm22 | translocase of inner mitochondrial membrane 22 | MGI:1929742 |
| ENSMUSG00000013701 | Timm23 | translocase of inner mitochondrial membrane 23 | MGI:1858317 |
| ENSMUSG00000002949 | Timm44 | translocase of inner mitochondrial membrane 44 | MGI:1343262 |
| ENSMUSG00000003438 | Timm50 | translocase of inner mitochondrial membrane 50 | MGI:1913775 |
| ENSMUSG00000039016 | Timm8b | translocase of inner mitochondrial membrane 8B | MGI:1353424 |
| ENSMUSG00000021079 | Timm9 | translocase of inner mitochondrial membrane 9 | MGI:1353436 |
| ENSMUSG00000030516 | Tjp1 | tight junction protein 1 | MGI:98759 |
| ENSMUSG00000024812 | Tjp2 | tight junction protein 2 | MGI:1341872 |
| ENSMUSG00000031397 | Tktl1 | transketolase-like 1 | MGI:1933244 |
| ENSMUSG00000021957 | Tkt | transketolase | MGI:105992 |
| ENSMUSG00000028465 | Tln1 | talin 1 | MGI:1099832 |
| ENSMUSG00000052698 | Tln2 | talin 2 | MGI:1917799 |
| ENSMUSG00000025544 | Tm9sf2 | transmembrane 9 superfamily member 2 | MGI:1915309 |
| ENSMUSG00000025016 | Tm9sf3 | transmembrane 9 superfamily member 3 | MGI:1914262 |
| ENSMUSG00000068040 | Tm9sf4 | transmembrane 9 superfamily protein member 4 | MGI:2139220 |
| ENSMUSG00000030126 | Tmcc1 | transmembrane and coiled coil domains 1 | MGI:2442368 |
| ENSMUSG00000042066 | Tmcc2 | transmembrane and coiled-coil domains 2 | MGI:1916125 |
| ENSMUSG00000052428 | Tmco1 | transmembrane and coiled-coil domains 1 | MGI:1921173 |
| ENSMUSG00000021248 | Tmed10 | transmembrane emp24-like trafficking protein 10 (yeast) | MGI:1915831 |
| ENSMUSG00000029390 | Tmed2 | transmembrane emp24 domain trafficking protein 2 | MGI:1929269 |
| ENSMUSG00000004394 | Tmed4 | transmembrane emp24 protein transport domain containing 4 | MGI:1915070 |
| ENSMUSG00000033184 | Tmed7 | transmembrane emp24 protein transport domain containing 7 | MGI:1913926 |
| ENSMUSG00000058569 | Tmed9 | transmembrane emp24 protein transport domain containing 9 | MGI:1914761 |
| ENSMUSG00000028347 | Tmeff1 | transmembrane protein with EGF-like and two follistatin-like domains 1 | MGI:1926810 |
| ENSMUSG00000026109 | Tmeff2 | transmembrane protein with EGF-like and two follistatin-like domains 2 | MGI:1861735 |
| ENSMUSG00000029571 | Tmem106b | transmembrane protein 106B | MGI:1919150 |
| ENSMUSG00000034659 | Tmem109 | transmembrane protein 109 | MGI:1915789 |
| ENSMUSG00000043284 | Tmem11 | transmembrane protein 11 | MGI:2144726 |
| ENSMUSG00000030615 | Tmem126a | transmembrane protein 126A | MGI:1913521 |
| ENSMUSG00000024736 | Tmem132a | transmembrane protein 132A | MGI:2147810 |
| ENSMUSG00000070498 | Tmem132b | transmembrane protein 132B | MGI:3609245 |
| ENSMUSG00000020701 | Tmem132e | transmembrane protein 132E | MGI:2685490 |
| ENSMUSG00000039428 | Tmem135 | transmembrane protein 135 | MGI:1920009 |
| ENSMUSG00000043843 | Tmem145 | transmembrane protein 145 | MGI:3607779 |
| ENSMUSG00000021361 | Tmem14c | transmembrane protein 14C | MGI:1913404 |
| ENSMUSG00000026347 | Tmem163 | transmembrane protein 163 | MGI:1919410 |
| ENSMUSG00000029234 | Tmem165 | transmembrane protein 165 | MGI:894407 |
| ENSMUSG00000013495 | Tmem175 | transmembrane protein 175 | MGI:1919642 |
| ENSMUSG00000057716 | Tmem178b | transmembrane protein 178B | MGI:3647581 |
| ENSMUSG00000025521 | Tmem192 | transmembrane protein 192 | MGI:1920317 |
| ENSMUSG00000040883 | Tmem205 | transmembrane protein 205 | MGI:3045495 |
| ENSMUSG00000028857 | Tmem222 | transmembrane protein 222 | MGI:1098568 |
| ENSMUSG00000048022 | Tmem229a | transmembrane protein 229A | MGI:2442812 |
| ENSMUSG00000070394 | Tmem256 | transmembrane protein 256 | MGI:1916436 |
| ENSMUSG00000036372 | Tmem258 | transmembrane protein 258 | MGI:1916288 |
| ENSMUSG00000013858 | Tmem259 | transmembrane protein 259 | MGI:2177957 |
| ENSMUSG00000032328 | Tmem30a | transmembrane protein 30A | MGI:106402 |
| ENSMUSG00000037720 | Tmem33 | transmembrane protein 33 | MGI:1915128 |
| ENSMUSG00000033578 | Tmem35 | transmembrane protein 35 | MGI:1914814 |
| ENSMUSG00000031791 | Tmem38a | transmembrane protein 38A | MGI:1921416 |
| ENSMUSG00000030095 | Tmem43 | transmembrane protein 43 | MGI:1921372 |
| ENSMUSG00000025666 | Tmem47 | transmembrane protein 47 | MGI:2177570 |
| ENSMUSG00000028221 | Tmem55a | transmembrane protein 55A | MGI:1919769 |
| ENSMUSG00000035953 | Tmem55b | transmembrane protein 55b | MGI:2448501 |
| ENSMUSG00000036026 | Tmem63b | transmembrane protein 63b | MGI:2387609 |
| ENSMUSG00000034145 | Tmem63c | transmembrane protein 63c | MGI:2444386 |
| ENSMUSG00000062373 | Tmem65 | transmembrane protein 65 | MGI:1922118 |
| ENSMUSG00000028232 | Tmem68 | transmembrane protein 68 | MGI:1919348 |
| ENSMUSG00000033808 | Tmem87a | transmembrane protein 87A | MGI:2441844 |
| ENSMUSG00000031021 | Tmem9b | TMEM9 domain family, member B | MGI:1915254 |
| ENSMUSG00000026411 | Tmem9 | transmembrane protein 9 | MGI:1913491 |
| ENSMUSG00000028328 | Tmod1 | tropomodulin 1 | MGI:98775 |
| ENSMUSG00000032186 | Tmod2 | tropomodulin 2 | MGI:1355335 |
| ENSMUSG00000058587 | Tmod3 | tropomodulin 3 | MGI:1355315 |
| ENSMUSG00000028958 | Tmub1 | transmembrane and ubiquitin-like domain containing 1 | MGI:1923764 |
| ENSMUSG00000021072 | Tmx1 | thioredoxin-related transmembrane protein 1 | MGI:1919986 |
| ENSMUSG00000050043 | Tmx2 | thioredoxin-related transmembrane protein 2 | MGI:1914208 |
| ENSMUSG00000024614 | Tmx3 | thioredoxin-related transmembrane protein 3 | MGI:2442418 |
| ENSMUSG00000034723 | Tmx4 | thioredoxin-related transmembrane protein 4 | MGI:106558 |
| ENSMUSG00000028364 | Tnc | tenascin C | MGI:101922 |
| ENSMUSG00000023915 | Tnfrsf21 | tumor necrosis factor receptor superfamily, member 21 | MGI:2151075 |
| ENSMUSG00000027692 | Tnik | TRAF2 and NCK interacting kinase | MGI:1916264 |
| ENSMUSG00000015829 | Tnr | tenascin R | MGI:99516 |
| ENSMUSG00000025139 | Tollip | toll interacting protein | MGI:1891808 |
| ENSMUSG00000000538 | Tom1l2 | target of myb1-like 2 (chicken) | MGI:2443306 |
| ENSMUSG00000042870 | Tom1 | target of myb1 homolog (chicken) | MGI:1338026 |
| ENSMUSG00000093904 | Tomm20 | translocase of outer mitochondrial membrane 20 homolog (yeast) | MGI:1915202 |
| ENSMUSG00000022427 | Tomm22 | translocase of outer mitochondrial membrane 22 homolog (yeast) | MGI:2450248 |
| ENSMUSG00000018322 | Tomm34 | translocase of outer mitochondrial membrane 34 | MGI:1914395 |
| ENSMUSG00000002984 | Tomm40 | translocase of outer mitochondrial membrane 40 homolog (yeast) | MGI:1858259 |
| ENSMUSG00000022752 | Tomm70a | translocase of outer mitochondrial membrane 70 homolog A (yeast) | MGI:106295 |
| ENSMUSG00000059323 | Tonsl | tonsoku-like, DNA repair protein | MGI:1919999 |
| ENSMUSG00000035274 | Tpbg | trophoblast glycoprotein | MGI:1341264 |
| ENSMUSG00000027506 | Tpd52 | tumor protein D52 | MGI:107749 |
| ENSMUSG00000023456 | Tpi1 | triosephosphate isomerase 1 | MGI:98797 |
| ENSMUSG00000032366 | Tpm1 | tropomyosin 1, alpha | MGI:98809 |
| ENSMUSG00000027940 | Tpm3 | tropomyosin 3, gamma | MGI:1890149 |
| ENSMUSG00000041763 | Tpp2 | tripeptidyl peptidase II | MGI:102724 |
| ENSMUSG00000014846 | Tppp3 | tubulin polymerization-promoting protein family member 3 | MGI:1915221 |
| ENSMUSG00000021573 | Tppp | tubulin polymerization promoting protein | MGI:1920198 |
| ENSMUSG00000029030 | Tprgl | transformation related protein 63 regulated like | MGI:1915058 |
| ENSMUSG00000060126 | Tpt1 | tumor protein, translationally-controlled 1 | MGI:104890 |
| ENSMUSG00000015363 | Trabd | TraB domain containing | MGI:1915226 |
| ENSMUSG00000021277 | Traf3 | TNF receptor-associated factor 3 | MGI:108041 |
| ENSMUSG00000005981 | Trap1 | TNF receptor-associated protein 1 | MGI:1915265 |
| ENSMUSG00000000374 | Trappc10 | trafficking protein particle complex 10 | MGI:1336209 |
| ENSMUSG00000020628 | Trappc12 | trafficking protein particle complex 12 | MGI:2445089 |
| ENSMUSG00000028847 | Trappc3 | trafficking protein particle complex 3 | MGI:1351486 |
| ENSMUSG00000032112 | Trappc4 | trafficking protein particle complex 4 | MGI:1926211 |
| ENSMUSG00000040236 | Trappc5 | trafficking protein particle complex 5 | MGI:1913932 |
| ENSMUSG00000020993 | Trappc6b | trafficking protein particle complex 6B | MGI:1925482 |
| ENSMUSG00000033382 | Trappc8 | trafficking protein particle complex 8 | MGI:2443008 |
| ENSMUSG00000047921 | Trappc9 | trafficking protein particle complex 9 | MGI:1923760 |
| ENSMUSG00000050663 | Trhde | TRH-degrading enzyme | MGI:2384311 |
| ENSMUSG00000021712 | Trim23 | tripartite motif-containing 23 | MGI:1933161 |
| ENSMUSG00000027993 | Trim2 | tripartite motif-containing 2 | MGI:1933163 |
| ENSMUSG00000051675 | Trim32 | tripartite motif-containing 32 | MGI:1917057 |
| ENSMUSG00000036989 | Trim3 | tripartite motif-containing 3 | MGI:1860040 |
| ENSMUSG00000042766 | Trim46 | tripartite motif-containing 46 | MGI:2673000 |
| ENSMUSG00000021071 | Trim9 | tripartite motif-containing 9 | MGI:2137354 |
| ENSMUSG00000022263 | Trio | triple functional domain (PTPRF interacting) | MGI:1927230 |
| ENSMUSG00000055963 | Triqk | triple QxxK/R motif containing | MGI:3650048 |
| ENSMUSG00000038812 | Trmt112 | tRNA methyltransferase 11-2 | MGI:1914924 |
| ENSMUSG00000068735 | Trp53i11 | transformation related protein 53 inducible protein 11 | MGI:2670995 |
| ENSMUSG00000052387 | Trpm3 | transient receptor potential cation channel, subfamily M, member 3 | MGI:2443101 |
| ENSMUSG00000018507 | Trpv2 | transient receptor potential cation channel, subfamily V, member 2 | MGI:1341836 |
| ENSMUSG00000029723 | Tsc22d4 | TSC22 domain family, member 4 | MGI:1926079 |
| ENSMUSG00000002496 | Tsc2 | tuberous sclerosis 2 | MGI:102548 |
| ENSMUSG00000040521 | Tsfm | Ts translation elongation factor, mitochondrial | MGI:1913649 |
| ENSMUSG00000014402 | Tsg101 | tumor susceptibility gene 101 | MGI:106581 |
| ENSMUSG00000026374 | Tsn | translin | MGI:109263 |
| ENSMUSG00000027858 | Tspan2 | tetraspanin 2 | MGI:1917997 |
| ENSMUSG00000058254 | Tspan7 | tetraspanin 7 | MGI:1298407 |
| ENSMUSG00000030352 | Tspan9 | tetraspanin 9 | MGI:1924558 |
| ENSMUSG00000044986 | Tst | thiosulfate sulfurtransferase, mitochondrial | MGI:98852 |
| ENSMUSG00000041278 | Ttc1 | tetratricopeptide repeat domain 1 | MGI:1914077 |
| ENSMUSG00000033530 | Ttc7b | tetratricopeptide repeat domain 7B | MGI:2144724 |
| ENSMUSG00000007944 | Ttc9b | tetratricopeptide repeat domain 9B | MGI:1920282 |
| ENSMUSG00000071660 | Ttc9c | tetratricopeptide repeat domain 9C | MGI:1917637 |
| ENSMUSG00000051747 | Ttn | titin | MGI:98864 |
| ENSMUSG00000061808 | Ttr | transthyretin | MGI:98865 |
| ENSMUSG00000030428 | Ttyh1 | tweety homolog 1 (Drosophila) | MGI:1889007 |
| ENSMUSG00000036565 | Ttyh3 | tweety homolog 3 (Drosophila) | MGI:1925589 |
| ENSMUSG00000072235 | Tuba1a | tubulin, alpha 1A | MGI:98869 |
| ENSMUSG00000026202 | Tuba4a | tubulin, alpha 4A | MGI:1095410 |
| ENSMUSG00000058672 | Tubb2a | tubulin, beta 2A class IIA | MGI:107861 |
| ENSMUSG00000045136 | Tubb2b | tubulin, beta 2B class IIB | MGI:1920960 |
| ENSMUSG00000062380 | Tubb3 | tubulin, beta 3 class III | MGI:107813 |
| ENSMUSG00000062591 | Tubb4a | tubulin, beta 4A class IVA | MGI:107848 |
| ENSMUSG00000036752 | Tubb4b | tubulin, beta 4B class IVB | MGI:1915472 |
| ENSMUSG00000001525 | Tubb5 | tubulin, beta 5 class I | MGI:107812 |
| ENSMUSG00000001473 | Tubb6 | tubulin, beta 6 class V | MGI:1915201 |
| ENSMUSG00000045007 | Tubg2 | tubulin, gamma 2 | MGI:2144208 |
| ENSMUSG00000025474 | Tubgcp2 | tubulin, gamma complex associated protein 2 | MGI:1921487 |
| ENSMUSG00000073838 | Tufm | Tu translation elongation factor, mitochondrial | MGI:1923686 |
| ENSMUSG00000039530 | Tusc3 | tumor suppressor candidate 3 | MGI:1933134 |
| ENSMUSG00000022451 | Twf1 | twinfilin, actin-binding protein, homolog 1 (Drosophila) | MGI:1100520 |
| ENSMUSG00000023277 | Twf2 | twinfilin, actin-binding protein, homolog 2 (Drosophila) | MGI:1346078 |
| ENSMUSG00000028367 | Txn1 | thioredoxin 1 | MGI:98874 |
| ENSMUSG00000028567 | Txndc12 | thioredoxin domain containing 12 (endoplasmic reticulum) | MGI:1913323 |
| ENSMUSG00000038991 | Txndc5 | thioredoxin domain containing 5 | MGI:2145316 |
| ENSMUSG00000024583 | Txnl1 | thioredoxin-like 1 | MGI:1860078 |
| ENSMUSG00000020250 | Txnrd1 | thioredoxin reductase 1 | MGI:1354175 |
| ENSMUSG00000075704 | Txnrd2 | thioredoxin reductase 2 | MGI:1347023 |
| ENSMUSG00000001924 | Uba1 | ubiquitin-like modifier activating enzyme 1 | MGI:98890 |
| ENSMUSG00000032557 | Uba5 | ubiquitin-like modifier activating enzyme 5 | MGI:1913913 |
| ENSMUSG00000042520 | Ubap2l | ubiquitin-associated protein 2-like | MGI:1921633 |
| ENSMUSG00000042345 | Ubash3a | ubiquitin associated and SH3 domain containing, A | MGI:1926074 |
| ENSMUSG00000032020 | Ubash3b | ubiquitin associated and SH3 domain containing, B | MGI:1920078 |
| ENSMUSG00000028277 | Ube2j1 | ubiquitin-conjugating enzyme E2J 1 | MGI:1926245 |
| ENSMUSG00000038965 | Ube2l3 | ubiquitin-conjugating enzyme E2L 3 | MGI:109240 |
| ENSMUSG00000005575 | Ube2m | ubiquitin-conjugating enzyme E2M | MGI:108278 |
| ENSMUSG00000074781 | Ube2n | ubiquitin-conjugating enzyme E2N | MGI:1934835 |
| ENSMUSG00000020802 | Ube2o | ubiquitin-conjugating enzyme E2O | MGI:2444266 |
| ENSMUSG00000078923 | Ube2v1 | ubiquitin-conjugating enzyme E2 variant 1 | MGI:1913839 |
| ENSMUSG00000022674 | Ube2v2 | ubiquitin-conjugating enzyme E2 variant 2 | MGI:1917870 |
| ENSMUSG00000025326 | Ube3a | ubiquitin protein ligase E3A | MGI:105098 |
| ENSMUSG00000059890 | Ube4a | ubiquitination factor E4A | MGI:2154580 |
| ENSMUSG00000001687 | Ubl3 | ubiquitin-like 3 | MGI:1344373 |
| ENSMUSG00000015290 | Ubl4 | ubiquitin-like 4 | MGI:95049 |
| ENSMUSG00000005312 | Ubqln1 | ubiquilin 1 | MGI:1860276 |
| ENSMUSG00000050148 | Ubqln2 | ubiquilin 2 | MGI:1860283 |
| ENSMUSG00000066036 | Ubr4 | ubiquitin protein ligase E3 component n-recognin 4 | MGI:1916366 |
| ENSMUSG00000026353 | Ubxn4 | UBX domain protein 4 | MGI:1915062 |
| ENSMUSG00000019578 | Ubxn6 | UBX domain protein 6 | MGI:1913780 |
| ENSMUSG00000029223 | Uchl1 | ubiquitin carboxy-terminal hydrolase L1 | MGI:103149 |
| ENSMUSG00000018189 | Uchl5 | ubiquitin carboxyl-terminal esterase L5 | MGI:1914848 |
| ENSMUSG00000062963 | Ufc1 | ubiquitin-fold modifier conjugating enzyme 1 | MGI:1913405 |
| ENSMUSG00000040359 | Ufl1 | UFM1 specific ligase 1 | MGI:1914740 |
| ENSMUSG00000027746 | Ufm1 | ubiquitin-fold modifier 1 | MGI:1915140 |
| ENSMUSG00000031634 | Ufsp2 | UFM1-specific peptidase 2 | MGI:1913679 |
| ENSMUSG00000037470 | Uggt1 | UDP-glucose glycoprotein glucosyltransferase 1 | MGI:2443162 |
| ENSMUSG00000001891 | Ugp2 | UDP-glucose pyrophosphorylase 2 | MGI:2183447 |
| ENSMUSG00000032854 | Ugt8a | UDP galactosyltransferase 8A | MGI:109522 |
| ENSMUSG00000019951 | Uhrf1bp1l | UHRF1 (ICBP90) binding protein 1-like | MGI:2442888 |
| ENSMUSG00000062151 | Unc13c | unc-13 homolog C (C. elegans) | MGI:2149021 |
| ENSMUSG00000055567 | Unc80 | unc-80 homolog (C. elegans) | MGI:2652882 |
| ENSMUSG00000058301 | Upf1 | UPF1 regulator of nonsense transcripts homolog (yeast) | MGI:107995 |
| ENSMUSG00000005882 | Uqcc1 | ubiquinol-cytochrome c reductase complex assembly factor 1 | MGI:1929472 |
| ENSMUSG00000024208 | Uqcc2 | ubiquinol-cytochrome c reductase complex assembly factor 2 | MGI:1914517 |
| ENSMUSG00000071654 | Uqcc3 | ubiquinol-cytochrome c reductase complex assembly factor 3 | MGI:2147553 |
| ENSMUSG00000059534 | Uqcr10 | ubiquinol-cytochrome c reductase, complex III subunit X | MGI:1913402 |
| ENSMUSG00000021520 | Uqcrb | ubiquinol-cytochrome c reductase binding protein | MGI:1914780 |
| ENSMUSG00000025651 | Uqcrc1 | ubiquinol-cytochrome c reductase core protein 1 | MGI:107876 |
| ENSMUSG00000030884 | Uqcrc2 | ubiquinol cytochrome c reductase core protein 2 | MGI:1914253 |
| ENSMUSG00000038462 | Uqcrfs1 | ubiquinol-cytochrome c reductase, Rieske iron-sulfur polypeptide 1 | MGI:1913944 |
| ENSMUSG00000063882 | Uqcrh | ubiquinol-cytochrome c reductase hinge protein | MGI:1913826 |
| ENSMUSG00000044894 | Uqcrq | ubiquinol-cytochrome c reductase, complex III subunit VII | MGI:107807 |
| ENSMUSG00000028684 | Urod | uroporphyrinogen decarboxylase | MGI:98916 |
| ENSMUSG00000002395 | Use1 | unconventional SNARE in the ER 1 homolog (S. cerevisiae) | MGI:1914273 |
| ENSMUSG00000071528 | Usmg5 | upregulated during skeletal muscle growth 5 | MGI:1891435 |
| ENSMUSG00000029407 | Uso1 | USO1 vesicle docking factor | MGI:1929095 |
| ENSMUSG00000031066 | Usp11 | ubiquitin specific peptidase 11 | MGI:2384312 |
| ENSMUSG00000047879 | Usp14 | ubiquitin specific peptidase 14 | MGI:1928898 |
| ENSMUSG00000029592 | Usp30 | ubiquitin specific peptidase 30 | MGI:2140991 |
| ENSMUSG00000063317 | Usp31 | ubiquitin specific peptidase 31 | MGI:1923429 |
| ENSMUSG00000054814 | Usp46 | ubiquitin specific peptidase 46 | MGI:1916977 |
| ENSMUSG00000038429 | Usp5 | ubiquitin specific peptidase 5 (isopeptidase T) | MGI:1347343 |
| ENSMUSG00000022710 | Usp7 | ubiquitin specific peptidase 7 | MGI:2182061 |
| ENSMUSG00000031010 | Usp9x | ubiquitin specific peptidase 9, X chromosome | MGI:894681 |
| ENSMUSG00000019820 | Utrn | utrophin | MGI:104631 |
| ENSMUSG00000010936 | Vac14 | Vac14 homolog (S. cerevisiae) | MGI:2157980 |
| ENSMUSG00000030337 | Vamp1 | vesicle-associated membrane protein 1 | MGI:1313276 |
| ENSMUSG00000020894 | Vamp2 | vesicle-associated membrane protein 2 | MGI:1313277 |
| ENSMUSG00000028955 | Vamp3 | vesicle-associated membrane protein 3 | MGI:1321389 |
| ENSMUSG00000026696 | Vamp4 | vesicle-associated membrane protein 4 | MGI:1858730 |
| ENSMUSG00000073002 | Vamp5 | vesicle-associated membrane protein 5 | MGI:1858622 |
| ENSMUSG00000051412 | Vamp7 | vesicle-associated membrane protein 7 | MGI:1096399 |
| ENSMUSG00000050732 | Vamp8 | vesicle-associated membrane protein 8 | MGI:1336882 |
| ENSMUSG00000024091 | Vapa | vesicle-associated membrane protein, associated protein A | MGI:1353561 |
| ENSMUSG00000054455 | Vapb | vesicle-associated membrane protein, associated protein B and C | MGI:1928744 |
| ENSMUSG00000007029 | Vars | valyl-tRNA synthetase | MGI:90675 |
| ENSMUSG00000046844 | Vat1l | vesicle amine transport protein 1 homolog-like (T. californica) | MGI:2142534 |
| ENSMUSG00000034993 | Vat1 | vesicle amine transport protein 1 homolog (T californica) | MGI:1349450 |
| ENSMUSG00000027962 | Vcam1 | vascular cell adhesion molecule 1 | MGI:98926 |
| ENSMUSG00000021614 | Vcan | versican | MGI:102889 |
| ENSMUSG00000021823 | Vcl | vinculin | MGI:98927 |
| ENSMUSG00000045210 | Vcpip1 | valosin containing protein (p97)/p47 complex interacting protein 1 | MGI:1917925 |
| ENSMUSG00000028452 | Vcp | valosin containing protein | MGI:99919 |
| ENSMUSG00000020402 | Vdac1 | voltage-dependent anion channel 1 | MGI:106919 |
| ENSMUSG00000021771 | Vdac2 | voltage-dependent anion channel 2 | MGI:106915 |
| ENSMUSG00000008892 | Vdac3 | voltage-dependent anion channel 3 | MGI:106922 |
| ENSMUSG00000037428 | Vgf | VGF nerve growth factor inducible | MGI:1343180 |
| ENSMUSG00000075701 | Vimp | VCP-interacting membrane protein | MGI:95994 |
| ENSMUSG00000026728 | Vim | vimentin | MGI:98932 |
| ENSMUSG00000024924 | Vldlr | very low density lipoprotein receptor | MGI:98935 |
| ENSMUSG00000073131 | Vma21 | VMA21 vacuolar H+-ATPase homolog (S. cerevisiae) | MGI:1914298 |
| ENSMUSG00000018171 | Vmp1 | vacuole membrane protein 1 | MGI:1923159 |
| ENSMUSG00000032127 | Vps11 | vacuolar protein sorting 11 (yeast) | MGI:1918982 |
| ENSMUSG00000046230 | Vps13a | vacuolar protein sorting 13A (yeast) | MGI:2444304 |
| ENSMUSG00000035284 | Vps13c | vacuolar protein sorting 13C (yeast) | MGI:2444207 |
| ENSMUSG00000020220 | Vps13d | vacuolar protein sorting 13 D (yeast) | MGI:2448530 |
| ENSMUSG00000027411 | Vps16 | vacuolar protein sorting 16 (yeast) | MGI:2136772 |
| ENSMUSG00000034216 | Vps18 | vacuolar protein sorting 18 (yeast) | MGI:2443626 |
| ENSMUSG00000078656 | Vps25 | vacuolar protein sorting 25 (yeast) | MGI:106354 |
| ENSMUSG00000020078 | Vps26a | vacuolar protein sorting 26 homolog A (yeast) | MGI:1353654 |
| ENSMUSG00000031988 | Vps26b | vacuolar protein sorting 26 homolog B (yeast) | MGI:1917656 |
| ENSMUSG00000029462 | Vps29 | vacuolar protein sorting 29 (S. pombe) | MGI:1928344 |
| ENSMUSG00000029434 | Vps33a | vacuolar protein sorting 33A (yeast) | MGI:1924823 |
| ENSMUSG00000031696 | Vps35 | vacuolar protein sorting 35 | MGI:1890467 |
| ENSMUSG00000048832 | Vps37c | vacuolar protein sorting 37C (yeast) | MGI:2147661 |
| ENSMUSG00000027291 | Vps39 | vacuolar protein sorting 39 (yeast) | MGI:2443189 |
| ENSMUSG00000041236 | Vps41 | vacuolar protein sorting 41 (yeast) | MGI:1929215 |
| ENSMUSG00000015747 | Vps45 | vacuolar protein sorting 45 (yeast) | MGI:891965 |
| ENSMUSG00000024797 | Vps51 | vacuolar protein sorting 51 homolog (S. cerevisiae) | MGI:1915755 |
| ENSMUSG00000024319 | Vps52 | vacuolar protein sorting 52 (yeast) | MGI:1330304 |
| ENSMUSG00000017288 | Vps53 | vacuolar protein sorting 53 (yeast) | MGI:1915549 |
| ENSMUSG00000001062 | Vps9d1 | VPS9 domain containing 1 | MGI:1914143 |
| ENSMUSG00000054459 | Vsnl1 | visinin-like 1 | MGI:1349453 |
| ENSMUSG00000019868 | Vta1 | Vps20-associated 1 homolog (S. cerevisiae) | MGI:1913451 |
| ENSMUSG00000024983 | Vti1a | vesicle transport through interaction with t-SNAREs 1A | MGI:1855699 |
| ENSMUSG00000021124 | Vti1b | vesicle transport through interaction with t-SNAREs 1B | MGI:1855688 |
| ENSMUSG00000023186 | Vwa5a | von Willebrand factor A domain containing 5A | MGI:1915026 |
| ENSMUSG00000028753 | Vwa5b1 | von Willebrand factor A domain containing 5B1 | MGI:1922968 |
| ENSMUSG00000058997 | Vwa8 | von Willebrand factor A domain containing 8 | MGI:1919008 |
| ENSMUSG00000021266 | Wars | tryptophanyl-tRNA synthetase | MGI:104630 |
| ENSMUSG00000019831 | Wasf1 | WAS protein family, member 1 | MGI:1890563 |
| ENSMUSG00000028868 | Wasf2 | WAS protein family, member 2 | MGI:1098641 |
| ENSMUSG00000029636 | Wasf3 | WAS protein family, member 3 | MGI:2658986 |
| ENSMUSG00000024101 | Wash1 | WAS protein family homolog 1 | MGI:1916017 |
| ENSMUSG00000029684 | Wasl | Wiskott-Aldrich syndrome-like (human) | MGI:1920428 |
| ENSMUSG00000034341 | Wbp2 | WW domain binding protein 2 | MGI:104709 |
| ENSMUSG00000034040 | Wbscr17 | Williams-Beuren syndrome chromosome region 17 homolog (human) | MGI:2137594 |
| ENSMUSG00000073643 | Wdfy1 | WD repeat and FYVE domain containing 1 | MGI:1916618 |
| ENSMUSG00000043940 | Wdfy3 | WD repeat and FYVE domain containing 3 | MGI:1096875 |
| ENSMUSG00000005103 | Wdr1 | WD repeat domain 1 | MGI:1337100 |
| ENSMUSG00000037957 | Wdr20 | WD repeat domain 20 | MGI:1916891 |
| ENSMUSG00000021147 | Wdr37 | WD repeat domain 37 | MGI:1920393 |
| ENSMUSG00000025173 | Wdr45b | WD repeat domain 45B | MGI:1914090 |
| ENSMUSG00000040389 | Wdr47 | WD repeat domain 47 | MGI:2139593 |
| ENSMUSG00000032512 | Wdr48 | WD repeat domain 48 | MGI:1914811 |
| ENSMUSG00000039828 | Wdr70 | WD repeat domain 70 | MGI:1921020 |
| ENSMUSG00000040560 | Wdr7 | WD repeat domain 7 | MGI:1860197 |
| ENSMUSG00000039474 | Wfs1 | Wolfram syndrome 1 homolog (human) | MGI:1328355 |
| ENSMUSG00000064030 | Wibg | within bgcn homolog (Drosophila) | MGI:1925678 |
| ENSMUSG00000086040 | Wipf3 | WAS/WASL interacting protein family, member 3 | MGI:3044681 |
| ENSMUSG00000045962 | Wnk1 | WNK lysine deficient protein kinase 1 | MGI:2442092 |
| ENSMUSG00000037989 | Wnk2 | WNK lysine deficient protein kinase 2 | MGI:1922857 |
| ENSMUSG00000041245 | Wnk3 | WNK lysine deficient protein kinase 3 | MGI:2652875 |
| ENSMUSG00000004637 | Wwox | WW domain-containing oxidoreductase | MGI:1931237 |
| ENSMUSG00000027022 | Xirp2 | xin actin-binding repeat containing 2 | MGI:2685198 |
| ENSMUSG00000051951 | Xkr4 | X Kell blood group precursor related family member 4 | MGI:3528744 |
| ENSMUSG00000035067 | Xkr6 | X Kell blood group precursor related family member 6 homolog | MGI:2447765 |
| ENSMUSG00000025027 | Xpnpep1 | X-prolyl aminopeptidase (aminopeptidase P) 1, soluble | MGI:2180003 |
| ENSMUSG00000020290 | Xpo1 | exportin 1, CRM1 homolog (yeast) | MGI:2144013 |
| ENSMUSG00000022100 | Xpo7 | exportin 7 | MGI:1929705 |
| ENSMUSG00000026469 | Xpr1 | xenotropic and polytropic retrovirus receptor 1 | MGI:97932 |
| ENSMUSG00000022792 | Yars2 | tyrosyl-tRNA synthetase 2 (mitochondrial) | MGI:1917370 |
| ENSMUSG00000028811 | Yars | tyrosyl-tRNA synthetase | MGI:2147627 |
| ENSMUSG00000014932 | Yes1 | Yamaguchi sarcoma viral (v-yes) oncogene homolog 1 | MGI:99147 |
| ENSMUSG00000071074 | Yipf3 | Yip1 domain family, member 3 | MGI:106280 |
| ENSMUSG00000024487 | Yipf5 | Yip1 domain family, member 5 | MGI:1914430 |
| ENSMUSG00000002741 | Ykt6 | YKT6 homolog (S. Cerevisiae) | MGI:1927550 |
| ENSMUSG00000026775 | Yme1l1 | YME1-like 1 (S. cerevisiae) | MGI:1351651 |
| ENSMUSG00000018326 | Ywhab | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, beta polypeptide | MGI:1891917 |
| ENSMUSG00000020849 | Ywhae | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, epsilon polypeptide | MGI:894689 |
| ENSMUSG00000051391 | Ywhag | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, gamma polypeptide | MGI:108109 |
| ENSMUSG00000018965 | Ywhah | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, eta polypeptide | MGI:109194 |
| ENSMUSG00000076432 | Ywhaq | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta polypeptide | MGI:891963 |
| ENSMUSG00000022285 | Ywhaz | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide | MGI:109484 |
| ENSMUSG00000034173 | Zbed5 | zinc finger, BED type containing 5 | MGI:1919220 |
| ENSMUSG00000043542 | Zc2hc1a | zinc finger, C2HC-type containing 1A | MGI:1914556 |
| ENSMUSG00000035798 | Zdhhc17 | zinc finger, DHHC domain containing 17 | MGI:2445110 |
| ENSMUSG00000021969 | Zdhhc20 | zinc finger, DHHC domain containing 20 | MGI:1923215 |
| ENSMUSG00000034075 | Zdhhc5 | zinc finger, DHHC domain containing 5 | MGI:1923573 |
| ENSMUSG00000037855 | Zfp365 | zinc finger protein 365 | MGI:2143676 |
| ENSMUSG00000032264 | Zw10 | zw10 kinetochore protein | MGI:1349478 |
| ENSMUSG00000019923 | Zwint | ZW10 interactor | MGI:1289227 |
| ENSMUSG00000021252 | 0610007P14Rik | RIKEN cDNA 0610007P14 gene | MGI:1915571 |
| ENSMUSG00000025731 | 0610011F06Rik | RIKEN cDNA 0610011F06 gene | MGI:1915597 |
| ENSMUSG00000087651 | 1500009L16Rik | RIKEN cDNA 1500009L16 gene | MGI:1917034 |
| ENSMUSG00000032666 | 1700025G04Rik | RIKEN cDNA 1700025G04 gene | MGI:1916649 |
| ENSMUSG00000094103 | 1700047I17Rik2 | RIKEN cDNA 1700047I17 gene 2 | MGI:3714351 |
| ENSMUSG00000048429 | 1810026J23Rik | RIKEN cDNA 1810026J23 gene | MGI:1917023 |
| ENSMUSG00000054091 | 1810037I17Rik | RIKEN cDNA 1810037I17 gene | MGI:1914954 |
| ENSMUSG00000062376 | 2010012O05Rik | RIKEN cDNA 2010012O05 gene | MGI:1913689 |
| ENSMUSG00000021290 | 2010107E04Rik | RIKEN cDNA 2010107E04 gene | MGI:1917507 |
| ENSMUSG00000020083 | 2010107G23Rik | RIKEN cDNA 2010107G23 gene | MGI:1917144 |
| ENSMUSG00000026090 | 2010300C02Rik | RIKEN cDNA 2010300C02 gene | MGI:1919347 |
| ENSMUSG00000075605 | 2300005B03Rik | RIKEN cDNA 2300005B03 gene | MGI:1916712 |
| ENSMUSG00000031983 | 2310022B05Rik | RIKEN cDNA 2310022B05 gene | MGI:1916801 |
| ENSMUSG00000026319 | 2310035C23Rik | RIKEN cDNA 2310035C23 gene | MGI:1922832 |
| ENSMUSG00000045176 | 2310047M10Rik | RIKEN cDNA 2310047M10 gene | MGI:1919173 |
| ENSMUSG00000050705 | 2310061I04Rik | RIKEN cDNA 2310061I04 gene | MGI:1916912 |
| ENSMUSG00000049760 | 2410015M20Rik | RIKEN cDNA 2410015M20 gene | MGI:2442174 |
| ENSMUSG00000010277 | 2610507B11Rik | RIKEN cDNA 2610507B11 gene | MGI:1919753 |
| ENSMUSG00000021807 | 2700060E02Rik | RIKEN cDNA 2700060E02 gene | MGI:1915295 |
| ENSMUSG00000024410 | 3110002H16Rik | RIKEN cDNA 3110002H16 gene | MGI:1916528 |
| ENSMUSG00000037270 | 4932438A13Rik | RIKEN cDNA 4932438A13 gene | MGI:2444631 |
| ENSMUSG00000027942 | 4933434E20Rik | RIKEN cDNA 4933434E20 gene | MGI:1914027 |
| ENSMUSG00000040412 | 5330417C22Rik | RIKEN cDNA 5330417C22 gene | MGI:1923930 |
| ENSMUSG00000053963 | 6330403A02Rik | RIKEN cDNA 6330403A02 gene | MGI:2138735 |
| ENSMUSG00000031824 | 6430548M08Rik | RIKEN cDNA 6430548M08 gene | MGI:2443793 |
| ENSMUSG00000037977 | 6430571L13Rik | RIKEN cDNA 6430571L13 gene | MGI:2445137 |
| ENSMUSG00000030207 | 8430419L09Rik | RIKEN cDNA 8430419L09 gene | MGI:1921775 |
| ENSMUSG00000056004 | 9330182L06Rik | RIKEN cDNA 9330182L06 gene | MGI:2443264 |
| ENSMUSG00000025971 | 9430016H08Rik | RIKEN cDNA 9430016H08 gene | MGI:1915365 |
| ENSMUSG00000034560 | A230046K03Rik | RIKEN cDNA A230046K03 gene | MGI:2441787 |
| ENSMUSG00000050875 | A730017C20Rik | RIKEN cDNA A730017C20 gene | MGI:2442934 |
| ENSMUSG00000044060 | A830010M20Rik | RIKEN cDNA A830010M20 gene | MGI:2445097 |
Supplementary Table 1b. Postsynaptic density proteins identified in mouse.
Proteins identified in mouse whole brain postsynaptic density preparations. For each protein the following database identifiers are given: Ensembl Gene, Associated Gene Name, Description and MGI ID.
| ENSEBLE ID | Associated Gene Name | Description | MGI ID |
|---|---|---|---|
| ENSMUSG00000057230 | Aak1 | AP2 associated kinase 1 | MGI:1098687 |
| ENSMUSG00000040584 | Abcb1a | ATP-binding cassette, sub-family B (MDR/TAP), member 1A | MGI:97570 |
| ENSMUSG00000031333 | Abcb7 | ATP-binding cassette, sub-family B (MDR/TAP), member 7 | MGI:109533 |
| ENSMUSG00000028973 | Abcb8 | ATP-binding cassette, sub-family B (MDR/TAP), member 8 | MGI:1351667 |
| ENSMUSG00000055782 | Abcd2 | ATP-binding cassette, sub-family D (ALD), member 2 | MGI:1349467 |
| ENSMUSG00000028127 | Abcd3 | ATP-binding cassette, sub-family D (ALD), member 3 | MGI:1349216 |
| ENSMUSG00000028953 | Abcf2 | ATP-binding cassette, sub-family F (GCN20), member 2 | MGI:1351657 |
| ENSMUSG00000032046 | Abhd12 | abhydrolase domain containing 12 | MGI:1923442 |
| ENSMUSG00000007036 | Abhd16a | abhydrolase domain containing 16A | MGI:99476 |
| ENSMUSG00000025277 | Abhd6 | abhydrolase domain containing 6 | MGI:1913332 |
| ENSMUSG00000058835 | Abi1 | abl-interactor 1 | MGI:104913 |
| ENSMUSG00000026782 | Abi2 | abl-interactor 2 | MGI:106913 |
| ENSMUSG00000026596 | Abl2 | v-abl Abelson murine leukemia viral oncogene 2 (arg, Abelson-related gene) | MGI:87860 |
| ENSMUSG00000025085 | Ablim1 | actin-binding LIM protein 1 | MGI:1194500 |
| ENSMUSG00000029095 | Ablim2 | actin-binding LIM protein 2 | MGI:2385758 |
| ENSMUSG00000032735 | Ablim3 | actin binding LIM protein family, member 3 | MGI:2442582 |
| ENSMUSG00000036880 | Acaa2 | acetyl-Coenzyme A acyltransferase 2 (mitochondrial 3-oxoacyl-Coenzyme A thiolase) | MGI:1098623 |
| ENSMUSG00000090150 | Acad11 | acyl-Coenzyme A dehydrogenase family, member 11 | MGI:2143169 |
| ENSMUSG00000042647 | Acad12 | acyl-Coenzyme A dehydrogenase family, member 12 | MGI:2443320 |
| ENSMUSG00000031969 | Acad8 | acyl-Coenzyme A dehydrogenase family, member 8 | MGI:1914198 |
| ENSMUSG00000030861 | Acadsb | acyl-Coenzyme A dehydrogenase, short/branched chain | MGI:1914135 |
| ENSMUSG00000030607 | Acan | aggrecan | MGI:99602 |
| ENSMUSG00000049076 | Acap2 | ArfGAP with coiled-coil, ankyrin repeat and PH domains 2 | MGI:1925868 |
| ENSMUSG00000032047 | Acat1 | acetyl-Coenzyme A acetyltransferase 1 | MGI:87870 |
| ENSMUSG00000026499 | Acbd3 | acyl-Coenzyme A binding domain containing 3 | MGI:2181074 |
| ENSMUSG00000017307 | Acot8 | acyl-CoA thioesterase 8 | MGI:2158201 |
| ENSMUSG00000025287 | Acot9 | acyl-CoA thioesterase 9 | MGI:1928939 |
| ENSMUSG00000032883 | Acsl3 | acyl-CoA synthetase long-chain family member 3 | MGI:1921455 |
| ENSMUSG00000031278 | Acsl4 | acyl-CoA synthetase long-chain family member 4 | MGI:1354713 |
| ENSMUSG00000031972 | Acta1 | actin, alpha 1, skeletal muscle | MGI:87902 |
| ENSMUSG00000029580 | Actb | actin, beta | MGI:87904 |
| ENSMUSG00000055194 | Actbl2 | actin, beta-like 2 | MGI:2444552 |
| ENSMUSG00000062825 | Actg1 | actin, gamma, cytoplasmic 1 | MGI:87906 |
| ENSMUSG00000052374 | Actn2 | actinin alpha 2 | MGI:109192 |
| ENSMUSG00000054808 | Actn4 | actinin alpha 4 | MGI:1890773 |
| ENSMUSG00000021076 | Actr10 | ARP10 actin-related protein 10 | MGI:1891654 |
| ENSMUSG00000025228 | Actr1a | ARP1 actin-related protein 1A, centractin alpha | MGI:1858964 |
| ENSMUSG00000037351 | Actr1b | ARP1 actin-related protein 1B, centractin beta | MGI:1917446 |
| ENSMUSG00000020152 | Actr2 | ARP2 actin-related protein 2 | MGI:1913963 |
| ENSMUSG00000026341 | Actr3 | ARP3 actin-related protein 3 | MGI:1921367 |
| ENSMUSG00000056367 | Actr3b | ARP3 actin-related protein 3B | MGI:2661120 |
| ENSMUSG00000020926 | Adam11 | a disintegrin and metallopeptidase domain 11 | MGI:1098667 |
| ENSMUSG00000025964 | Adam23 | a disintegrin and metallopeptidase domain 23 | MGI:1345162 |
| ENSMUSG00000056413 | Adap1 | ArfGAP with dual PH domains 1 | MGI:2442201 |
| ENSMUSG00000020431 | Adcy1 | adenylate cyclase 1 | MGI:99677 |
| ENSMUSG00000021536 | Adcy2 | adenylate cyclase 2 | MGI:99676 |
| ENSMUSG00000022994 | Adcy6 | adenylate cyclase 6 | MGI:87917 |
| ENSMUSG00000029106 | Add1 | adducin 1 (alpha) | MGI:87918 |
| ENSMUSG00000030000 | Add2 | adducin 2 (beta) | MGI:87919 |
| ENSMUSG00000025026 | Add3 | adducin 3 (gamma) | MGI:1351615 |
| ENSMUSG00000025475 | Adgra1 | adhesion G protein-coupled receptor A1 | MGI:1277167 |
| ENSMUSG00000034730 | Adgrb1 | adhesion G protein-coupled receptor B1 | MGI:1933736 |
| ENSMUSG00000028782 | Adgrb2 | adhesion G protein-coupled receptor B2 | MGI:2451244 |
| ENSMUSG00000033569 | Adgrb3 | adhesion G protein-coupled receptor B3 | MGI:2441837 |
| ENSMUSG00000013033 | Adgrl1 | adhesion G protein-coupled receptor L1 | MGI:1929461 |
| ENSMUSG00000028184 | Adgrl2 | adhesion G protein-coupled receptor L2 | MGI:2139714 |
| ENSMUSG00000037605 | Adgrl3 | adhesion G protein-coupled receptor L3 | MGI:2441950 |
| ENSMUSG00000033717 | Adra2a | adrenergic receptor, alpha 2a | MGI:87934 |
| ENSMUSG00000024858 | Adrbk1 | adrenergic receptor kinase, beta 1 | MGI:87940 |
| ENSMUSG00000024527 | Afg3l2 | AFG3-like AAA ATPase 2 | MGI:1916847 |
| ENSMUSG00000055013 | Agap1 | ArfGAP with GTPase domain, ankyrin repeat and PH domain 1 | MGI:2653690 |
| ENSMUSG00000025422 | Agap2 | ArfGAP with GTPase domain, ankyrin repeat and PH domain 2 | MGI:3580016 |
| ENSMUSG00000023353 | Agap3 | ArfGAP with GTPase domain, ankyrin repeat and PH domain 3 | MGI:2183446 |
| ENSMUSG00000026159 | Agfg1 | ArfGAP with FG repeats 1 | MGI:1333754 |
| ENSMUSG00000029916 | Agk | acylglycerol kinase | MGI:1917173 |
| ENSMUSG00000041530 | Ago1 | argonaute RISC catalytic subunit 1 | MGI:2446630 |
| ENSMUSG00000036698 | Ago2 | argonaute RISC catalytic subunit 2 | MGI:2446632 |
| ENSMUSG00000034254 | Agpat1 | 1-acylglycerol-3-phosphate O-acyltransferase 1 (lysophosphatidic acid acyltransferase, alpha) | MGI:1932075 |
| ENSMUSG00000031467 | Agpat5 | 1-acylglycerol-3-phosphate O-acyltransferase 5 (lysophosphatidic acid acyltransferase, epsilon) | MGI:1196345 |
| ENSMUSG00000019986 | Ahi1 | Abelson helper integration site 1 | MGI:87971 |
| ENSMUSG00000072812 | Ahnak2 | AHNAK nucleoprotein 2 | MGI:2144831 |
| ENSMUSG00000069833 | Ahnak | AHNAK nucleoprotein (desmoyokin) | MGI:1316648 |
| ENSMUSG00000021037 | Ahsa1 | AHA1, activator of heat shock protein ATPase 1 | MGI:2387603 |
| ENSMUSG00000046312 | AI464131 | expressed sequence AI464131 | MGI:2140300 |
| ENSMUSG00000036932 | Aifm1 | apoptosis-inducing factor, mitochondrion-associated 1 | MGI:1349419 |
| ENSMUSG00000022763 | Aifm3 | apoptosis-inducing factor, mitochondrion-associated 3 | MGI:1919418 |
| ENSMUSG00000028029 | Aimp1 | aminoacyl tRNA synthetase complex-interacting multifunctional protein 1 | MGI:102774 |
| ENSMUSG00000029610 | Aimp2 | aminoacyl tRNA synthetase complex-interacting multifunctional protein 2 | MGI:2385237 |
| ENSMUSG00000039546 | Ajap1 | adherens junction associated protein 1 | MGI:2685419 |
| ENSMUSG00000078139 | AK157302 | cDNA sequence AK157302 | MGI:3574096 |
| ENSMUSG00000026817 | Ak1 | adenylate kinase 1 | MGI:87977 |
| ENSMUSG00000024782 | Ak3 | adenylate kinase 3 | MGI:1860835 |
| ENSMUSG00000028527 | Ak4 | adenylate kinase 4 | MGI:87979 |
| ENSMUSG00000039058 | Ak5 | adenylate kinase 5 | MGI:2677491 |
| ENSMUSG00000021057 | Akap5 | A kinase (PRKA) anchor protein 5 | MGI:2685104 |
| ENSMUSG00000061758 | Akr1b10 | aldo-keto reductase family 1, member B10 (aldose reductase) | MGI:1915111 |
| ENSMUSG00000010025 | Aldh3a2 | aldehyde dehydrogenase family 3, subfamily A2 | MGI:1353452 |
| ENSMUSG00000024885 | Aldh3b1 | aldehyde dehydrogenase 3 family, member B1 | MGI:1914939 |
| ENSMUSG00000030695 | Aldoa | aldolase A, fructose-bisphosphate | MGI:87994 |
| ENSMUSG00000039740 | Alg2 | asparagine-linked glycosylation 2 (alpha-1,3-mannosyltransferase) | MGI:1914731 |
| ENSMUSG00000028766 | Alpl | alkaline phosphatase, liver/bone/kidney | MGI:87983 |
| ENSMUSG00000021986 | Amer2 | APC membrane recruitment 2 | MGI:1919375 |
| ENSMUSG00000041688 | Amot | angiomotin | MGI:108440 |
| ENSMUSG00000027889 | Ampd2 | adenosine monophosphate deaminase 2 | MGI:88016 |
| ENSMUSG00000005686 | Ampd3 | adenosine monophosphate deaminase 3 | MGI:1096344 |
| ENSMUSG00000032826 | Ank2 | ankyrin 2, brain | MGI:88025 |
| ENSMUSG00000069601 | Ank3 | ankyrin 3, epithelial | MGI:88026 |
| ENSMUSG00000040351 | Ankib1 | ankyrin repeat and IBR domain containing 1 | MGI:1918047 |
| ENSMUSG00000022265 | Ank | progressive ankylosis | MGI:3045421 |
| ENSMUSG00000078137 | Ankrd63 | ankyrin repeat domain 63 | MGI:2686183 |
| ENSMUSG00000058589 | Anks1b | ankyrin repeat and sterile alpha motif domain containing 1B | MGI:1924781 |
| ENSMUSG00000039062 | Anpep | alanyl (membrane) aminopeptidase | MGI:5000466 |
| ENSMUSG00000021866 | Anxa11 | annexin A11 | MGI:108481 |
| ENSMUSG00000032231 | Anxa2 | annexin A2 | MGI:88246 |
| ENSMUSG00000029994 | Anxa4 | annexin A4 | MGI:88030 |
| ENSMUSG00000021814 | Anxa7 | annexin A7 | MGI:88031 |
| ENSMUSG00000009090 | Ap1b1 | adaptor protein complex AP-1, beta 1 subunit | MGI:1096368 |
| ENSMUSG00000003033 | Ap1m1 | adaptor-related protein complex AP-1, mu subunit 1 | MGI:102776 |
| ENSMUSG00000004849 | Ap1s1 | adaptor protein complex AP-1, sigma 1 | MGI:1098244 |
| ENSMUSG00000031367 | Ap1s2 | adaptor-related protein complex 1, sigma 2 subunit | MGI:1889383 |
| ENSMUSG00000060279 | Ap2a1 | adaptor-related protein complex 2, alpha 1 subunit | MGI:101921 |
| ENSMUSG00000002957 | Ap2a2 | adaptor-related protein complex 2, alpha 2 subunit | MGI:101920 |
| ENSMUSG00000035152 | Ap2b1 | adaptor-related protein complex 2, beta 1 subunit | MGI:1919020 |
| ENSMUSG00000022841 | Ap2m1 | adaptor-related protein complex 2, mu 1 subunit | MGI:1298405 |
| ENSMUSG00000008036 | Ap2s1 | adaptor-related protein complex 2, sigma 1 subunit | MGI:2141861 |
| ENSMUSG00000062444 | Ap3b2 | adaptor-related protein complex 3, beta 2 subunit | MGI:1100869 |
| ENSMUSG00000020198 | Ap3d1 | adaptor-related protein complex 3, delta 1 subunit | MGI:107734 |
| ENSMUSG00000021824 | Ap3m1 | adaptor-related protein complex 3, mu 1 subunit | MGI:1929212 |
| ENSMUSG00000031539 | Ap3m2 | adaptor-related protein complex 3, mu 2 subunit | MGI:1929214 |
| ENSMUSG00000024480 | Ap3s1 | adaptor-related protein complex 3, sigma 1 subunit | MGI:1337062 |
| ENSMUSG00000063801 | Ap3s2 | adaptor-related protein complex 3, sigma 2 subunit | MGI:1337060 |
| ENSMUSG00000005871 | Apc | adenomatosis polyposis coli | MGI:88039 |
| ENSMUSG00000032081 | Apoc3 | apolipoprotein C-III | MGI:88055 |
| ENSMUSG00000025525 | Apool | apolipoprotein O-like | MGI:1915367 |
| ENSMUSG00000020263 | Appl2 | adaptor protein, phosphotyrosine interaction, PH domain and leucine zipper containing 2 | MGI:2384914 |
| ENSMUSG00000004655 | Aqp1 | aquaporin 1 | MGI:103201 |
| ENSMUSG00000001127 | Araf | v-raf murine sarcoma 3611 viral oncogene homolog | MGI:88065 |
| ENSMUSG00000022602 | Arc | activity regulated cytoskeletal-associated protein | MGI:88067 |
| ENSMUSG00000032096 | Arcn1 | archain 1 | MGI:2387591 |
| ENSMUSG00000048076 | Arf1 | ADP-ribosylation factor 1 | MGI:99431 |
| ENSMUSG00000020440 | Arf5 | ADP-ribosylation factor 5 | MGI:99434 |
| ENSMUSG00000044147 | Arf6 | ADP-ribosylation factor 6 | MGI:99435 |
| ENSMUSG00000027575 | Arfgap1 | ADP-ribosylation factor GTPase activating protein 1 | MGI:2183559 |
| ENSMUSG00000027255 | Arfgap2 | ADP-ribosylation factor GTPase activating protein 2 | MGI:1924288 |
| ENSMUSG00000054277 | Arfgap3 | ADP-ribosylation factor GTPase activating protein 3 | MGI:1913501 |
| ENSMUSG00000036591 | Arhgap21 | Rho GTPase activating protein 21 | MGI:1918685 |
| ENSMUSG00000049807 | Arhgap23 | Rho GTPase activating protein 23 | MGI:3697726 |
| ENSMUSG00000036452 | Arhgap26 | Rho GTPase activating protein 26 | MGI:1918552 |
| ENSMUSG00000022799 | Arhgap31 | Rho GTPase activating protein 31 | MGI:1333857 |
| ENSMUSG00000041444 | Arhgap32 | Rho GTPase activating protein 32 | MGI:2450166 |
| ENSMUSG00000036882 | Arhgap33 | Rho GTPase activating protein 33 | MGI:2673998 |
| ENSMUSG00000033697 | Arhgap39 | Rho GTPase activating protein 39 | MGI:107858 |
| ENSMUSG00000033389 | Arhgap44 | Rho GTPase activating protein 44 | MGI:2144423 |
| ENSMUSG00000025132 | Arhgdia | Rho GDP dissociation inhibitor (GDI) alpha | MGI:2178103 |
| ENSMUSG00000041977 | Arhgef11 | Rho guanine nucleotide exchange factor (GEF) 11 | MGI:2441869 |
| ENSMUSG00000019467 | Arhgef25 | Rho guanine nucleotide exchange factor (GEF) 25 | MGI:1277173 |
| ENSMUSG00000028059 | Arhgef2 | rho/rac guanine nucleotide exchange factor (GEF) 2 | MGI:103264 |
| ENSMUSG00000054901 | Arhgef33 | Rho guanine nucleotide exchange factor (GEF) 33 | MGI:2685787 |
| ENSMUSG00000031511 | Arhgef7 | Rho guanine nucleotide exchange factor (GEF7) | MGI:1860493 |
| ENSMUSG00000025656 | Arhgef9 | CDC42 guanine nucleotide exchange factor (GEF) 9 | MGI:2442233 |
| ENSMUSG00000026426 | Arl8a | ADP-ribosylation factor-like 8A | MGI:1915974 |
| ENSMUSG00000038525 | Armc10 | armadillo repeat containing 10 | MGI:1914461 |
| ENSMUSG00000027599 | Armc1 | armadillo repeat containing 1 | MGI:1921502 |
| ENSMUSG00000033436 | Armcx2 | armadillo repeat containing, X-linked 2 | MGI:1914666 |
| ENSMUSG00000049804 | Armcx4 | armadillo repeat containing, X-linked 4 | MGI:2147887 |
| ENSMUSG00000029621 | Arpc1a | actin related protein 2/3 complex, subunit 1A | MGI:1928896 |
| ENSMUSG00000006304 | Arpc2 | actin related protein 2/3 complex, subunit 2 | MGI:1923959 |
| ENSMUSG00000029465 | Arpc3 | actin related protein 2/3 complex, subunit 3 | MGI:1928375 |
| ENSMUSG00000079426 | Arpc4 | actin related protein 2/3 complex, subunit 4 | MGI:1915339 |
| ENSMUSG00000026755 | Arpc5l | actin related protein 2/3 complex, subunit 5-like | MGI:1921442 |
| ENSMUSG00000000325 | Arvcf | armadillo repeat gene deleted in velo-cardio-facial syndrome | MGI:109620 |
| ENSMUSG00000031591 | Asah1 | N-acylsphingosine amidohydrolase 1 | MGI:1277124 |
| ENSMUSG00000023017 | Asic1 | acid-sensing (proton-gated) ion channel 1 | MGI:1194915 |
| ENSMUSG00000028207 | Asph | aspartate-beta-hydroxylase | MGI:1914186 |
| ENSMUSG00000029348 | Asphd2 | aspartate beta-hydroxylase domain containing 2 | MGI:1920148 |
| ENSMUSG00000026587 | Astn1 | astrotactin 1 | MGI:1098567 |
| ENSMUSG00000028373 | Astn2 | astrotactin 2 | MGI:1889277 |
| ENSMUSG00000013662 | Atad1 | ATPase family, AAA domain containing 1 | MGI:1915229 |
| ENSMUSG00000029036 | Atad3a | ATPase family, AAA domain containing 3A | MGI:1919214 |
| ENSMUSG00000024426 | Atat1 | alpha tubulin acetyltransferase 1 | MGI:1913869 |
| ENSMUSG00000034958 | Atcay | ataxia, cerebellar, Cayman type homolog (human) | MGI:2448730 |
| ENSMUSG00000034566 | Atp5h | ATP synthase, H+ transporting, mitochondrial F0 complex, subunit D | MGI:1918929 |
| ENSMUSG00000050856 | Atp5k | ATP synthase, H+ transporting, mitochondrial F1F0 complex, subunit E | MGI:106636 |
| ENSMUSG00000022956 | Atp5o | ATP synthase, H+ transporting, mitochondrial F1 complex, O subunit | MGI:106341 |
| ENSMUSG00000019087 | Atp6ap1 | ATPase, H+ transporting, lysosomal accessory protein 1 | MGI:109629 |
| ENSMUSG00000031007 | Atp6ap2 | ATPase, H+ transporting, lysosomal accessory protein 2 | MGI:1917745 |
| ENSMUSG00000019302 | Atp6v0a1 | ATPase, H+ transporting, lysosomal V0 subunit A1 | MGI:103286 |
| ENSMUSG00000024121 | Atp6v0c | ATPase, H+ transporting, lysosomal V0 subunit C | MGI:88116 |
| ENSMUSG00000013160 | Atp6v0d1 | ATPase, H+ transporting, lysosomal V0 subunit D1 | MGI:1201778 |
| ENSMUSG00000052459 | Atp6v1a | ATPase, H+ transporting, lysosomal V1 subunit A | MGI:1201780 |
| ENSMUSG00000006273 | Atp6v1b2 | ATPase, H+ transporting, lysosomal V1 subunit B2 | MGI:109618 |
| ENSMUSG00000022295 | Atp6v1c1 | ATPase, H+ transporting, lysosomal V1 subunit C1 | MGI:1913585 |
| ENSMUSG00000021114 | Atp6v1d | ATPase, H+ transporting, lysosomal V1 subunit D | MGI:1921084 |
| ENSMUSG00000019210 | Atp6v1e1 | ATPase, H+ transporting, lysosomal V1 subunit E1 | MGI:894326 |
| ENSMUSG00000004285 | Atp6v1f | ATPase, H+ transporting, lysosomal V1 subunit F | MGI:1913394 |
| ENSMUSG00000039105 | Atp6v1g1 | ATPase, H+ transporting, lysosomal V1 subunit G1 | MGI:1913540 |
| ENSMUSG00000024403 | Atp6v1g2 | ATPase, H+ transporting, lysosomal V1 subunit G2 | MGI:1913487 |
| ENSMUSG00000033793 | Atp6v1h | ATPase, H+ transporting, lysosomal V1 subunit H | MGI:1914864 |
| ENSMUSG00000027546 | Atp9a | ATPase, class II, type 9A | MGI:1330826 |
| ENSMUSG00000016541 | Atxn10 | ataxin 10 | MGI:1859293 |
| ENSMUSG00000042605 | Atxn2 | ataxin 2 | MGI:1277223 |
| ENSMUSG00000021460 | Auh | AU RNA binding protein/enoyl-coenzyme A hydratase | MGI:1338011 |
| ENSMUSG00000071649 | B3gat3 | beta-1,3-glucuronyltransferase 3 (glucuronosyltransferase I) | MGI:1919977 |
| ENSMUSG00000006731 | B4galnt1 | beta-1,4-N-acetyl-galactosaminyl transferase 1 | MGI:1342057 |
| ENSMUSG00000052364 | B630019K06Rik | RIKEN cDNA B630019K06 gene | MGI:2147918 |
| ENSMUSG00000042215 | Bag2 | BCL2-associated athanogene 2 | MGI:1891254 |
| ENSMUSG00000049792 | Bag5 | BCL2-associated athanogene 5 | MGI:1917619 |
| ENSMUSG00000025372 | Baiap2 | brain-specific angiogenesis inhibitor 1-associated protein 2 | MGI:2137336 |
| ENSMUSG00000047507 | Baiap3 | BAI1-associated protein 3 | MGI:2685783 |
| ENSMUSG00000024844 | Banf1 | barrier to autointegration factor 1 | MGI:1346330 |
| ENSMUSG00000045763 | Basp1 | brain abundant, membrane attached signal protein 1 | MGI:1917600 |
| ENSMUSG00000006464 | Bbs1 | Bardet-Biedl syndrome 1 (human) | MGI:1277215 |
| ENSMUSG00000036948 | BC037034 | cDNA sequence BC037034 | MGI:2385896 |
| ENSMUSG00000004892 | Bcan | brevican | MGI:1096385 |
| ENSMUSG00000020650 | Bcap29 | B cell receptor associated protein 29 | MGI:101917 |
| ENSMUSG00000002015 | Bcap31 | B cell receptor associated protein 31 | MGI:1350933 |
| ENSMUSG00000013523 | Bcas1 | breast carcinoma amplified sequence 1 | MGI:1924210 |
| ENSMUSG00000059439 | Bcas3 | breast carcinoma amplified sequence 3 | MGI:2385848 |
| ENSMUSG00000030802 | Bckdk | branched chain ketoacid dehydrogenase kinase | MGI:1276121 |
| ENSMUSG00000009681 | Bcr | breakpoint cluster region | MGI:88141 |
| ENSMUSG00000102226 | Begain | brain-enriched guanylate kinase-associated | MGI:3044626 |
| ENSMUSG00000015943 | Bola1 | bolA-like 1 (E. coli) | MGI:1916418 |
| ENSMUSG00000037905 | Bri3bp | Bri3 binding protein | MGI:1924059 |
| ENSMUSG00000028351 | Brinp1 | bone morphogenic protein/retinoic acid inducible neural specific 1 | MGI:1928478 |
| ENSMUSG00000004031 | Brinp2 | bone morphogenic protein/retinoic acid inducible neural-specific 2 | MGI:2443333 |
| ENSMUSG00000033940 | Brk1 | BRICK1, SCAR/WAVE actin-nucleating complex subunit | MGI:1915406 |
| ENSMUSG00000035390 | Brsk1 | BR serine/threonine kinase 1 | MGI:2685946 |
| ENSMUSG00000053046 | Brsk2 | BR serine/threonine kinase 2 | MGI:1923020 |
| ENSMUSG00000032589 | Bsn | bassoon | MGI:1277955 |
| ENSMUSG00000020042 | Btbd11 | BTB (POZ) domain containing 11 | MGI:1921257 |
| ENSMUSG00000000202 | Btbd17 | BTB (POZ) domain containing 17 | MGI:1919264 |
| ENSMUSG00000036905 | C1qb | complement component 1, q subcomponent, beta polypeptide | MGI:88224 |
| ENSMUSG00000018446 | C1qbp | complement component 1, q subcomponent binding protein | MGI:1194505 |
| ENSMUSG00000036896 | C1qc | complement component 1, q subcomponent, C chain | MGI:88225 |
| ENSMUSG00000040794 | C1qtnf4 | C1q and tumor necrosis factor related protein 4 | MGI:1914695 |
| ENSMUSG00000045912 | C2cd4c | C2 calcium-dependent domain containing 4C | MGI:2685084 |
| ENSMUSG00000030279 | C2cd5 | C2 calcium-dependent domain containing 5 | MGI:1921991 |
| ENSMUSG00000036377 | C530008M17Rik | RIKEN cDNA C530008M17 gene | MGI:2444817 |
| ENSMUSG00000050390 | C77080 | expressed sequence C77080 | MGI:2140651 |
| ENSMUSG00000036707 | Cab39 | calcium binding protein 39 | MGI:107438 |
| ENSMUSG00000034656 | Cacna1a | calcium channel, voltage-dependent, P/Q type, alpha 1A subunit | MGI:109482 |
| ENSMUSG00000004113 | Cacna1b | calcium channel, voltage-dependent, N type, alpha 1B subunit | MGI:88296 |
| ENSMUSG00000040118 | Cacna2d1 | calcium channel, voltage-dependent, alpha2/delta subunit 1 | MGI:88295 |
| ENSMUSG00000010066 | Cacna2d2 | calcium channel, voltage-dependent, alpha 2/delta subunit 2 | MGI:1929813 |
| ENSMUSG00000021991 | Cacna2d3 | calcium channel, voltage-dependent, alpha2/delta subunit 3 | MGI:1338890 |
| ENSMUSG00000020882 | Cacnb1 | calcium channel, voltage-dependent, beta 1 subunit | MGI:102522 |
| ENSMUSG00000057914 | Cacnb2 | calcium channel, voltage-dependent, beta 2 subunit | MGI:894644 |
| ENSMUSG00000003352 | Cacnb3 | calcium channel, voltage-dependent, beta 3 subunit | MGI:103307 |
| ENSMUSG00000017412 | Cacnb4 | calcium channel, voltage-dependent, beta 4 subunit | MGI:103301 |
| ENSMUSG00000019146 | Cacng2 | calcium channel, voltage-dependent, gamma subunit 2 | MGI:1316660 |
| ENSMUSG00000066189 | Cacng3 | calcium channel, voltage-dependent, gamma subunit 3 | MGI:1859165 |
| ENSMUSG00000053395 | Cacng8 | calcium channel, voltage-dependent, gamma subunit 8 | MGI:1932376 |
| ENSMUSG00000023055 | Calcoco1 | calcium binding and coiled coil domain 1 | MGI:1914738 |
| ENSMUSG00000029761 | Cald1 | caldesmon 1 | MGI:88250 |
| ENSMUSG00000019370 | Calm3 | calmodulin 3 | MGI:103249 |
| ENSMUSG00000024617 | Camk2a | calcium/calmodulin-dependent protein kinase II alpha | MGI:88256 |
| ENSMUSG00000057897 | Camk2b | calcium/calmodulin-dependent protein kinase II, beta | MGI:88257 |
| ENSMUSG00000053819 | Camk2d | calcium/calmodulin-dependent protein kinase II, delta | MGI:1341265 |
| ENSMUSG00000021820 | Camk2g | calcium/calmodulin-dependent protein kinase II gamma | MGI:88259 |
| ENSMUSG00000029471 | Camkk2 | calcium/calmodulin-dependent protein kinase kinase 2, beta | MGI:2444812 |
| ENSMUSG00000032936 | Camkv | CaM kinase-like vesicle-associated | MGI:2384296 |
| ENSMUSG00000041570 | Camsap2 | calmodulin regulated spectrin-associated protein family, member 2 | MGI:1922434 |
| ENSMUSG00000044433 | Camsap3 | calmodulin regulated spectrin-associated protein family, member 3 | MGI:1916947 |
| ENSMUSG00000021373 | Cap2 | CAP, adenylate cyclase-associated protein, 2 (yeast) | MGI:1914502 |
| ENSMUSG00000035547 | Capn5 | calpain 5 | MGI:1100859 |
| ENSMUSG00000001794 | Capns1 | calpain, small subunit 1 | MGI:88266 |
| ENSMUSG00000027184 | Caprin1 | cell cycle associated protein 1 | MGI:1858234 |
| ENSMUSG00000070372 | Capza1 | capping protein (actin filament) muscle Z-line, alpha 1 | MGI:106227 |
| ENSMUSG00000015733 | Capza2 | capping protein (actin filament) muscle Z-line, alpha 2 | MGI:106222 |
| ENSMUSG00000028745 | Capzb | capping protein (actin filament) muscle Z-line, beta | MGI:104652 |
| ENSMUSG00000056158 | Car10 | carbonic anhydrase 10 | MGI:1919855 |
| ENSMUSG00000000805 | Car4 | carbonic anhydrase 4 | MGI:1096574 |
| ENSMUSG00000060227 | Casc4 | cancer susceptibility candidate 4 | MGI:2443129 |
| ENSMUSG00000031012 | Cask | calcium/calmodulin-dependent serine protein kinase (MAGUK family) | MGI:1309489 |
| ENSMUSG00000033597 | Caskin1 | CASK interacting protein 1 | MGI:2442952 |
| ENSMUSG00000034471 | Caskin2 | CASK-interacting protein 2 | MGI:2157062 |
| ENSMUSG00000031654 | Cbln1 | cerebellin 1 precursor protein | MGI:88281 |
| ENSMUSG00000040380 | Cbln3 | cerebellin 3 precursor protein | MGI:1889286 |
| ENSMUSG00000051483 | Cbr1 | carbonyl reductase 1 | MGI:88284 |
| ENSMUSG00000022428 | Cby1 | chibby homolog 1 (Drosophila) | MGI:1920989 |
| ENSMUSG00000036686 | Cc2d1a | coiled-coil and C2 domain containing 1A | MGI:2384831 |
| ENSMUSG00000031150 | Ccdc120 | coiled-coil domain containing 120 | MGI:1859619 |
| ENSMUSG00000021578 | Ccdc127 | coiled-coil domain containing 127 | MGI:1914683 |
| ENSMUSG00000029769 | Ccdc136 | coiled-coil domain containing 136 | MGI:1918128 |
| ENSMUSG00000062961 | Ccdc177 | coiled-coil domain containing 177 | MGI:2686414 |
| ENSMUSG00000059554 | Ccdc28a | coiled-coil domain containing 28A | MGI:2443508 |
| ENSMUSG00000078622 | Ccdc47 | coiled-coil domain containing 47 | MGI:1914413 |
| ENSMUSG00000025645 | Ccdc51 | coiled-coil domain containing 51 | MGI:1913908 |
| ENSMUSG00000041609 | Ccdc64 | coiled-coil domain containing 64 | MGI:1922915 |
| ENSMUSG00000032878 | Ccdc85a | coiled-coil domain containing 85A | MGI:2445069 |
| ENSMUSG00000032740 | Ccdc88a | coiled coil domain containing 88A | MGI:1925177 |
| ENSMUSG00000037979 | Ccdc92 | coiled-coil domain containing 92 | MGI:106485 |
| ENSMUSG00000000378 | Ccm2 | cerebral cavernous malformation 2 | MGI:2384924 |
| ENSMUSG00000024286 | Ccny | cyclin Y | MGI:1915224 |
| ENSMUSG00000031971 | Ccsap | centriole, cilia and spindle associated protein | MGI:1920670 |
| ENSMUSG00000034024 | Cct2 | chaperonin containing Tcp1, subunit 2 (beta) | MGI:107186 |
| ENSMUSG00000001416 | Cct3 | chaperonin containing Tcp1, subunit 3 (gamma) | MGI:104708 |
| ENSMUSG00000007739 | Cct4 | chaperonin containing Tcp1, subunit 4 (delta) | MGI:104689 |
| ENSMUSG00000022234 | Cct5 | chaperonin containing Tcp1, subunit 5 (epsilon) | MGI:107185 |
| ENSMUSG00000029447 | Cct6a | chaperonin containing Tcp1, subunit 6a (zeta) | MGI:107943 |
| ENSMUSG00000030007 | Cct7 | chaperonin containing Tcp1, subunit 7 (eta) | MGI:107184 |
| ENSMUSG00000025613 | Cct8 | chaperonin containing Tcp1, subunit 8 (theta) | MGI:107183 |
| ENSMUSG00000029617 | Ccz1 | CCZ1 vacuolar protein trafficking and biogenesis associated | MGI:2141070 |
| ENSMUSG00000055447 | Cd47 | CD47 antigen (Rh-related antigen, integrin-associated signal transducer) | MGI:96617 |
| ENSMUSG00000037706 | Cd81 | CD81 antigen | MGI:1096398 |
| ENSMUSG00000026490 | Cdc42bpa | CDC42 binding protein kinase alpha | MGI:2441841 |
| ENSMUSG00000021279 | Cdc42bpb | CDC42 binding protein kinase beta | MGI:2136459 |
| ENSMUSG00000049521 | Cdc42ep1 | CDC42 effector protein (Rho GTPase binding) 1 | MGI:1929763 |
| ENSMUSG00000041598 | Cdc42ep4 | CDC42 effector protein (Rho GTPase binding) 4 | MGI:1929760 |
| ENSMUSG00000052298 | Cdc42se2 | CDC42 small effector 2 | MGI:1919979 |
| ENSMUSG00000023932 | Cdc5l | cell division cycle 5-like (S. pombe) | MGI:1918952 |
| ENSMUSG00000022321 | Cdh10 | cadherin 10 | MGI:107436 |
| ENSMUSG00000031673 | Cdh11 | cadherin 11 | MGI:99217 |
| ENSMUSG00000031841 | Cdh13 | cadherin 13 | MGI:99551 |
| ENSMUSG00000040420 | Cdh18 | cadherin 18 | MGI:1344366 |
| ENSMUSG00000050840 | Cdh20 | cadherin 20 | MGI:1346069 |
| ENSMUSG00000024304 | Cdh2 | cadherin 2 | MGI:88355 |
| ENSMUSG00000000305 | Cdh4 | cadherin 4 | MGI:99218 |
| ENSMUSG00000039385 | Cdh6 | cadherin 6 | MGI:107435 |
| ENSMUSG00000026312 | Cdh7 | cadherin 7, type 2 | MGI:2442792 |
| ENSMUSG00000025370 | Cdh9 | cadherin 9 | MGI:107433 |
| ENSMUSG00000030682 | Cdipt | CDP-diacylglycerol--inositol 3-phosphatidyltransferase (phosphatidylinositol synthase) | MGI:105491 |
| ENSMUSG00000020015 | Cdk17 | cyclin-dependent kinase 17 | MGI:97517 |
| ENSMUSG00000026437 | Cdk18 | cyclin-dependent kinase 18 | MGI:97518 |
| ENSMUSG00000028969 | Cdk5 | cyclin-dependent kinase 5 | MGI:101765 |
| ENSMUSG00000090071 | Cdk5r2 | cyclin-dependent kinase 5, regulatory subunit 2 (p39) | MGI:1330828 |
| ENSMUSG00000018669 | Cdk5rap3 | CDK5 regulatory subunit associated protein 3 | MGI:1933126 |
| ENSMUSG00000031292 | Cdkl5 | cyclin-dependent kinase-like 5 | MGI:1278336 |
| ENSMUSG00000094626 | Cecr6 | cat eye syndrome chromosome region, candidate 6 | MGI:2136977 |
| ENSMUSG00000002107 | Celf2 | CUGBP, Elav-like family member 2 | MGI:1338822 |
| ENSMUSG00000060240 | Cend1 | cell cycle exit and neuronal differentiation 1 | MGI:1929898 |
| ENSMUSG00000072825 | Cep170b | centrosomal protein 170B | MGI:2145043 |
| ENSMUSG00000057335 | Cep170 | centrosomal protein 170 | MGI:1918348 |
| ENSMUSG00000022604 | Cep97 | centrosomal protein 97 | MGI:1921451 |
| ENSMUSG00000031347 | Cetn2 | centrin 2 | MGI:1347085 |
| ENSMUSG00000056201 | Cfl1 | cofilin 1, non-muscle | MGI:101757 |
| ENSMUSG00000062929 | Cfl2 | cofilin 2, muscle | MGI:101763 |
| ENSMUSG00000053768 | Chchd3 | coiled-coil-helix-coiled-coil-helix domain containing 3 | MGI:1913325 |
| ENSMUSG00000030086 | Chchd6 | coiled-coil-helix-coiled-coil-helix domain containing 6 | MGI:1913348 |
| ENSMUSG00000027350 | Chgb | chromogranin B | MGI:88395 |
| ENSMUSG00000000743 | Chmp1a | charged multivesicular body protein 1A | MGI:1920159 |
| ENSMUSG00000033916 | Chmp2a | charged multivesicular body protein 2A | MGI:1916203 |
| ENSMUSG00000004843 | Chmp2b | charged multivesicular body protein 2B | MGI:1916192 |
| ENSMUSG00000053119 | Chmp3 | charged multivesicular body protein 3 | MGI:1913950 |
| ENSMUSG00000038467 | Chmp4b | charged multivesicular body protein 4B | MGI:1922858 |
| ENSMUSG00000014077 | Chp1 | calcineurin-like EF hand protein 1 | MGI:1927185 |
| ENSMUSG00000032773 | Chrm1 | cholinergic receptor, muscarinic 1, CNS | MGI:88396 |
| ENSMUSG00000046159 | Chrm3 | cholinergic receptor, muscarinic 3, cardiac | MGI:88398 |
| ENSMUSG00000040495 | Chrm4 | cholinergic receptor, muscarinic 4 | MGI:88399 |
| ENSMUSG00000046691 | Chtf8 | CTF8, chromosome transmission fidelity factor 8 | MGI:2443370 |
| ENSMUSG00000045193 | Cirbp | cold inducible RNA binding protein | MGI:893588 |
| ENSMUSG00000037710 | Cisd1 | CDGSH iron sulfur domain 1 | MGI:1261855 |
| ENSMUSG00000078695 | Cisd3 | CDGSH iron sulfur domain 3 | MGI:101788 |
| ENSMUSG00000029516 | Cit | citron | MGI:105313 |
| ENSMUSG00000046841 | Ckap4 | cytoskeleton-associated protein 4 | MGI:2444926 |
| ENSMUSG00000040549 | Ckap5 | cytoskeleton associated protein 5 | MGI:1923036 |
| ENSMUSG00000000308 | Ckmt1 | creatine kinase, mitochondrial 1, ubiquitous | MGI:99441 |
| ENSMUSG00000064302 | Clasp1 | CLIP associating protein 1 | MGI:1923957 |
| ENSMUSG00000033392 | Clasp2 | CLIP associating protein 2 | MGI:1923749 |
| ENSMUSG00000022843 | Clcn2 | chloride channel 2 | MGI:105061 |
| ENSMUSG00000029016 | Clcn6 | chloride channel 6 | MGI:1347049 |
| ENSMUSG00000022132 | Cldn10 | claudin 10 | MGI:1913101 |
| ENSMUSG00000037625 | Cldn11 | claudin 11 | MGI:106925 |
| ENSMUSG00000046798 | Cldn12 | claudin 12 | MGI:1929288 |
| ENSMUSG00000006169 | Clint1 | clathrin interactor 1 | MGI:2144243 |
| ENSMUSG00000049550 | Clip1 | CAP-GLY domain containing linker protein 1 | MGI:1928401 |
| ENSMUSG00000063146 | Clip2 | CAP-GLY domain containing linker protein 2 | MGI:1313136 |
| ENSMUSG00000013921 | Clip3 | CAP-GLY domain containing linker protein 3 | MGI:1923936 |
| ENSMUSG00000021097 | Clmn | calmin | MGI:2136957 |
| ENSMUSG00000001829 | Clpb | ClpB caseinolytic peptidase B | MGI:1100517 |
| ENSMUSG00000028478 | Clta | clathrin, light polypeptide (Lca) | MGI:894297 |
| ENSMUSG00000047547 | Cltb | clathrin, light polypeptide (Lcb) | MGI:1921575 |
| ENSMUSG00000022037 | Clu | clusterin | MGI:88423 |
| ENSMUSG00000030282 | Cmas | cytidine monophospho-N-acetylneuraminic acid synthetase | MGI:1337124 |
| ENSMUSG00000034390 | Cmip | c-Maf inducing protein | MGI:1921690 |
| ENSMUSG00000028719 | Cmpk1 | cytidine monophosphate (UMP-CMP) kinase 1 | MGI:1913838 |
| ENSMUSG00000025658 | Cnksr2 | connector enhancer of kinase suppressor of Ras 2 | MGI:2661175 |
| ENSMUSG00000053931 | Cnn3 | calponin 3, acidic | MGI:1919244 |
| ENSMUSG00000036550 | Cnot1 | CCR4-NOT transcription complex, subunit 1 | MGI:2442402 |
| ENSMUSG00000006782 | Cnp | 2',3'-cyclic nucleotide 3' phosphodiesterase | MGI:88437 |
| ENSMUSG00000028444 | Cntfr | ciliary neurotrophic factor receptor | MGI:99605 |
| ENSMUSG00000055022 | Cntn1 | contactin 1 | MGI:105980 |
| ENSMUSG00000053024 | Cntn2 | contactin 2 | MGI:104518 |
| ENSMUSG00000030075 | Cntn3 | contactin 3 | MGI:99534 |
| ENSMUSG00000064293 | Cntn4 | contactin 4 | MGI:1095737 |
| ENSMUSG00000039488 | Cntn5 | contactin 5 | MGI:3042287 |
| ENSMUSG00000030092 | Cntn6 | contactin 6 | MGI:1858223 |
| ENSMUSG00000017167 | Cntnap1 | contactin associated protein-like 1 | MGI:1858201 |
| ENSMUSG00000017188 | Coa3 | cytochrome C oxidase assembly factor 3 | MGI:1098757 |
| ENSMUSG00000058897 | Col25a1 | collagen, type XXV, alpha 1 | MGI:1924268 |
| ENSMUSG00000036103 | Colec12 | collectin sub-family member 12 | MGI:2152907 |
| ENSMUSG00000034807 | Colgalt1 | collagen beta(1-O)galactosyltransferase 1 | MGI:1924348 |
| ENSMUSG00000042705 | Commd10 | COMM domain containing 10 | MGI:1916706 |
| ENSMUSG00000051355 | Commd1 | COMM domain containing 1 | MGI:109474 |
| ENSMUSG00000051154 | Commd3 | COMM domain containing 3 | MGI:88218 |
| ENSMUSG00000027163 | Commd9 | COMM domain containing 9 | MGI:1923751 |
| ENSMUSG00000021773 | Comtd1 | catechol-O-methyltransferase domain containing 1 | MGI:1916406 |
| ENSMUSG00000026553 | Copa | coatomer protein complex subunit alpha | MGI:1334462 |
| ENSMUSG00000030058 | Copg1 | coatomer protein complex, subunit gamma 1 | MGI:1858696 |
| ENSMUSG00000019373 | Cops3 | COP9 (constitutive photomorphogenic) homolog, subunit 3 (Arabidopsis thaliana) | MGI:1349409 |
| ENSMUSG00000035297 | Cops4 | COP9 (constitutive photomorphogenic) homolog, subunit 4 (Arabidopsis thaliana) | MGI:1349414 |
| ENSMUSG00000030127 | Cops7a | COP9 (constitutive photomorphogenic) homolog, subunit 7a (Arabidopsis thaliana) | MGI:1349400 |
| ENSMUSG00000034432 | Cops8 | COP9 (constitutive photomorphogenic) homolog, subunit 8 (Arabidopsis thaliana) | MGI:1915363 |
| ENSMUSG00000060992 | Copz1 | coatomer protein complex, subunit zeta 1 | MGI:1929063 |
| ENSMUSG00000024835 | Coro1b | coronin, actin binding protein 1B | MGI:1345963 |
| ENSMUSG00000004530 | Coro1c | coronin, actin binding protein 1C | MGI:1345964 |
| ENSMUSG00000028337 | Coro2a | coronin, actin binding protein 2A | MGI:1345966 |
| ENSMUSG00000041729 | Coro2b | coronin, actin binding protein, 2B | MGI:2444283 |
| ENSMUSG00000023020 | Cox14 | cytochrome c oxidase assembly protein 14 | MGI:1913629 |
| ENSMUSG00000031818 | Cox4i1 | cytochrome c oxidase subunit IV isoform 1 | MGI:88473 |
| ENSMUSG00000041697 | Cox6a1 | cytochrome c oxidase subunit VIa polypeptide 1 | MGI:103099 |
| ENSMUSG00000024248 | Cox7a2l | cytochrome c oxidase subunit VIIa polypeptide 2-like | MGI:106015 |
| ENSMUSG00000039652 | Cpeb3 | cytoplasmic polyadenylation element binding protein 3 | MGI:2443075 |
| ENSMUSG00000020300 | Cpeb4 | cytoplasmic polyadenylation element binding protein 4 | MGI:1914829 |
| ENSMUSG00000020183 | Cpm | carboxypeptidase M | MGI:1917824 |
| ENSMUSG00000034361 | Cpne2 | copine II | MGI:2387578 |
| ENSMUSG00000032564 | Cpne4 | copine IV | MGI:1921270 |
| ENSMUSG00000024008 | Cpne5 | copine V | MGI:2385908 |
| ENSMUSG00000022212 | Cpne6 | copine VI | MGI:1334445 |
| ENSMUSG00000030270 | Cpne9 | copine family member IX | MGI:2443052 |
| ENSMUSG00000055531 | Cpsf6 | cleavage and polyadenylation specific factor 6 | MGI:1913948 |
| ENSMUSG00000034820 | Cpsf7 | cleavage and polyadenylation specific factor 7 | MGI:1917826 |
| ENSMUSG00000024900 | Cpt1a | carnitine palmitoyltransferase 1a, liver | MGI:1098296 |
| ENSMUSG00000006356 | Crip2 | cysteine rich protein 2 | MGI:1915587 |
| ENSMUSG00000017776 | Crk | v-crk sarcoma virus CT10 oncogene homolog (avian) | MGI:88508 |
| ENSMUSG00000040860 | Crocc | ciliary rootlet coiled-coil, rootletin | MGI:3529431 |
| ENSMUSG00000042401 | Crtac1 | cartilage acidic protein 1 | MGI:1920082 |
| ENSMUSG00000032060 | Cryab | crystallin, alpha B | MGI:88516 |
| ENSMUSG00000068823 | Csde1 | cold shock domain containing E1, RNA binding | MGI:92356 |
| ENSMUSG00000024576 | Csnk1a1 | casein kinase 1, alpha 1 | MGI:1934950 |
| ENSMUSG00000025162 | Csnk1d | casein kinase 1, delta | MGI:1355272 |
| ENSMUSG00000022433 | Csnk1e | casein kinase 1, epsilon | MGI:1351660 |
| ENSMUSG00000032384 | Csnk1g1 | casein kinase 1, gamma 1 | MGI:2660884 |
| ENSMUSG00000003345 | Csnk1g2 | casein kinase 1, gamma 2 | MGI:1920014 |
| ENSMUSG00000073563 | Csnk1g3 | casein kinase 1, gamma 3 | MGI:1917675 |
| ENSMUSG00000074698 | Csnk2a1 | casein kinase 2, alpha 1 polypeptide | MGI:88543 |
| ENSMUSG00000046707 | Csnk2a2 | casein kinase 2, alpha prime polypeptide | MGI:88547 |
| ENSMUSG00000024387 | Csnk2b | casein kinase 2, beta polypeptide | MGI:88548 |
| ENSMUSG00000026421 | Csrp1 | cysteine and glycine-rich protein 1 | MGI:88549 |
| ENSMUSG00000027447 | Cst3 | cystatin C | MGI:102519 |
| ENSMUSG00000037373 | Ctbp1 | C-terminal binding protein 1 | MGI:1201685 |
| ENSMUSG00000037815 | Ctnna1 | catenin (cadherin associated protein), alpha 1 | MGI:88274 |
| ENSMUSG00000063063 | Ctnna2 | catenin (cadherin associated protein), alpha 2 | MGI:88275 |
| ENSMUSG00000060843 | Ctnna3 | catenin (cadherin associated protein), alpha 3 | MGI:2661445 |
| ENSMUSG00000006932 | Ctnnb1 | catenin (cadherin associated protein), beta 1 | MGI:88276 |
| ENSMUSG00000022240 | Ctnnd2 | catenin (cadherin associated protein), delta 2 | MGI:1195966 |
| ENSMUSG00000000416 | Cttnbp2 | cortactin binding protein 2 | MGI:1353467 |
| ENSMUSG00000062127 | Cttnbp2nl | CTTNBP2 N-terminal like | MGI:1933137 |
| ENSMUSG00000031078 | Cttn | cortactin | MGI:99695 |
| ENSMUSG00000029686 | Cul1 | cullin 1 | MGI:1349658 |
| ENSMUSG00000004364 | Cul3 | cullin 3 | MGI:1347360 |
| ENSMUSG00000031446 | Cul4a | cullin 4A | MGI:1914487 |
| ENSMUSG00000032030 | Cul5 | cullin 5 | MGI:1922967 |
| ENSMUSG00000031924 | Cyb5b | cytochrome b5 type B | MGI:1913677 |
| ENSMUSG00000027015 | Cybrd1 | cytochrome b reductase 1 | MGI:2654575 |
| ENSMUSG00000022551 | Cyc1 | cytochrome c-1 | MGI:1913695 |
| ENSMUSG00000030447 | Cyfip1 | cytoplasmic FMR1 interacting protein 1 | MGI:1338801 |
| ENSMUSG00000020340 | Cyfip2 | cytoplasmic FMR1 interacting protein 2 | MGI:1924134 |
| ENSMUSG00000036712 | Cyld | cylindromatosis (turban tumor syndrome) | MGI:1921506 |
| ENSMUSG00000021259 | Cyp46a1 | cytochrome P450, family 46, subfamily a, polypeptide 1 | MGI:1341877 |
| ENSMUSG00000017132 | Cyth1 | cytohesin 1 | MGI:1334257 |
| ENSMUSG00000003269 | Cyth2 | cytohesin 2 | MGI:1334255 |
| ENSMUSG00000018001 | Cyth3 | cytohesin 3 | MGI:1335107 |
| ENSMUSG00000053329 | D10Jhu81e | DNA segment, Chr 10, Johns Hopkins University 81 expressed | MGI:1351861 |
| ENSMUSG00000068373 | D430041D05Rik | RIKEN cDNA D430041D05 gene | MGI:2181743 |
| ENSMUSG00000063455 | D630045J12Rik | RIKEN cDNA D630045J12 gene | MGI:2669829 |
| ENSMUSG00000034574 | Daam1 | dishevelled associated activator of morphogenesis 1 | MGI:1914596 |
| ENSMUSG00000040260 | Daam2 | dishevelled associated activator of morphogenesis 2 | MGI:1923691 |
| ENSMUSG00000028519 | Dab1 | disabled 1 | MGI:108554 |
| ENSMUSG00000022150 | Dab2 | disabled 2, mitogen-responsive phosphoprotein | MGI:109175 |
| ENSMUSG00000026883 | Dab2ip | disabled 2 interacting protein | MGI:1916851 |
| ENSMUSG00000078794 | Dact3 | dapper homolog 3, antagonist of beta-catenin (xenopus) | MGI:3654828 |
| ENSMUSG00000035735 | Dagla | diacylglycerol lipase, alpha | MGI:2677061 |
| ENSMUSG00000042096 | Dao | D-amino acid oxidase | MGI:94859 |
| ENSMUSG00000021559 | Dapk1 | death associated protein kinase 1 | MGI:1916885 |
| ENSMUSG00000034974 | Dapk3 | death-associated protein kinase 3 | MGI:1203520 |
| ENSMUSG00000026356 | Dars | aspartyl-tRNA synthetase | MGI:2442544 |
| ENSMUSG00000034675 | Dbn1 | drebrin 1 | MGI:1931838 |
| ENSMUSG00000020476 | Dbnl | drebrin-like | MGI:700006 |
| ENSMUSG00000000340 | Dbt | dihydrolipoamide branched chain transacylase E2 | MGI:105386 |
| ENSMUSG00000049354 | Dcaf7 | DDB1 and CUL4 associated factor 7 | MGI:1919083 |
| ENSMUSG00000020935 | Dcakd | dephospho-CoA kinase domain containing | MGI:1915337 |
| ENSMUSG00000027797 | Dclk1 | doublecortin-like kinase 1 | MGI:1330861 |
| ENSMUSG00000028078 | Dclk2 | doublecortin-like kinase 2 | MGI:1918012 |
| ENSMUSG00000025410 | Dctn2 | dynactin 2 | MGI:107733 |
| ENSMUSG00000028447 | Dctn3 | dynactin 3 | MGI:1859251 |
| ENSMUSG00000024603 | Dctn4 | dynactin 4 | MGI:1914915 |
| ENSMUSG00000030868 | Dctn5 | dynactin 5 | MGI:1891689 |
| ENSMUSG00000024740 | Ddb1 | damage specific DNA binding protein 1 | MGI:1202384 |
| ENSMUSG00000059213 | Ddn | dendrin | MGI:108101 |
| ENSMUSG00000028757 | Ddost | dolichyl-di-phosphooligosaccharide-protein glycotransferase | MGI:1194508 |
| ENSMUSG00000068290 | Ddrgk1 | DDRGK domain containing 1 | MGI:1924256 |
| ENSMUSG00000053289 | Ddx10 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 10 | MGI:1924841 |
| ENSMUSG00000055065 | Ddx17 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 17 | MGI:1914290 |
| ENSMUSG00000037149 | Ddx1 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 1 | MGI:2144727 |
| ENSMUSG00000000787 | Ddx3x | DEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide 3, X-linked | MGI:103064 |
| ENSMUSG00000069045 | Ddx3y | DEAD (Asp-Glu-Ala-Asp) box polypeptide 3, Y-linked | MGI:1349406 |
| ENSMUSG00000020719 | Ddx5 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 5 | MGI:105037 |
| ENSMUSG00000032097 | Ddx6 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 6 | MGI:104976 |
| ENSMUSG00000028223 | Decr1 | 2,4-dienoyl CoA reductase 1, mitochondrial | MGI:1914710 |
| ENSMUSG00000035392 | Dennd1a | DENN/MADD domain containing 1A | MGI:2442794 |
| ENSMUSG00000036095 | Dgkb | diacylglycerol kinase, beta | MGI:2442474 |
| ENSMUSG00000000276 | Dgke | diacylglycerol kinase, epsilon | MGI:1889276 |
| ENSMUSG00000022861 | Dgkg | diacylglycerol kinase, gamma | MGI:105060 |
| ENSMUSG00000038665 | Dgki | diacylglycerol kinase, iota | MGI:2443430 |
| ENSMUSG00000040479 | Dgkz | diacylglycerol kinase zeta | MGI:1278339 |
| ENSMUSG00000020834 | Dhrs13 | dehydrogenase/reductase (SDR family) member 13 | MGI:1917701 |
| ENSMUSG00000022210 | Dhrs4 | dehydrogenase/reductase (SDR family) member 4 | MGI:90169 |
| ENSMUSG00000042569 | Dhrs7b | dehydrogenase/reductase (SDR family) member 7B | MGI:2384931 |
| ENSMUSG00000029169 | Dhx15 | DEAH (Asp-Glu-Ala-His) box polypeptide 15 | MGI:1099786 |
| ENSMUSG00000032480 | Dhx30 | DEAH (Asp-Glu-Ala-His) box polypeptide 30 | MGI:1920081 |
| ENSMUSG00000040620 | Dhx33 | DEAH (Asp-Glu-Ala-His) box polypeptide 33 | MGI:2445102 |
| ENSMUSG00000042699 | Dhx9 | DEAH (Asp-Glu-Ala-His) box polypeptide 9 | MGI:108177 |
| ENSMUSG00000023026 | Dip2b | DIP2 disco-interacting protein 2 homolog B (Drosophila) | MGI:2145977 |
| ENSMUSG00000043670 | Diras1 | DIRAS family, GTP-binding RAS-like 1 | MGI:2183442 |
| ENSMUSG00000047842 | Diras2 | DIRAS family, GTP-binding RAS-like 2 | MGI:1915453 |
| ENSMUSG00000020664 | Dld | dihydrolipoamide dehydrogenase | MGI:107450 |
| ENSMUSG00000022770 | Dlg1 | discs, large homolog 1 (Drosophila) | MGI:107231 |
| ENSMUSG00000052572 | Dlg2 | discs, large homolog 2 (Drosophila) | MGI:1344351 |
| ENSMUSG00000000881 | Dlg3 | discs, large homolog 3 (Drosophila) | MGI:1888986 |
| ENSMUSG00000020886 | Dlg4 | discs, large homolog 4 (Drosophila) | MGI:1277959 |
| ENSMUSG00000003279 | Dlgap1 | discs, large (Drosophila) homolog-associated protein 1 | MGI:1346065 |
| ENSMUSG00000047495 | Dlgap2 | discs, large (Drosophila) homolog-associated protein 2 | MGI:2443181 |
| ENSMUSG00000042388 | Dlgap3 | discs, large (Drosophila) homolog-associated protein 3 | MGI:3039563 |
| ENSMUSG00000061689 | Dlgap4 | discs, large homolog-associated protein 4 (Drosophila) | MGI:2138865 |
| ENSMUSG00000004789 | Dlst | dihydrolipoamide S-succinyltransferase (E2 component of 2-oxo-glutarate complex) | MGI:1926170 |
| ENSMUSG00000022099 | Dmtn | dematin actin binding protein | MGI:99670 |
| ENSMUSG00000030410 | Dmwd | dystrophia myotonica-containing WD repeat motif | MGI:94907 |
| ENSMUSG00000028410 | Dnaja1 | DnaJ (Hsp40) homolog, subfamily A, member 1 | MGI:1270129 |
| ENSMUSG00000031701 | Dnaja2 | DnaJ (Hsp40) homolog, subfamily A, member 2 | MGI:1931882 |
| ENSMUSG00000004069 | Dnaja3 | DnaJ (Hsp40) homolog, subfamily A, member 3 | MGI:1933786 |
| ENSMUSG00000004460 | Dnajb11 | DnaJ (Hsp40) homolog, subfamily B, member 11 | MGI:1915088 |
| ENSMUSG00000020109 | Dnajb12 | DnaJ (Hsp40) homolog, subfamily B, member 12 | MGI:1931881 |
| ENSMUSG00000074212 | Dnajb14 | DnaJ (Hsp40) homolog, subfamily B, member 14 | MGI:1917854 |
| ENSMUSG00000005483 | Dnajb1 | DnaJ (Hsp40) homolog, subfamily B, member 1 | MGI:1931874 |
| ENSMUSG00000026203 | Dnajb2 | DnaJ (Hsp40) homolog, subfamily B, member 2 | MGI:1928739 |
| ENSMUSG00000028035 | Dnajb4 | DnaJ (Hsp40) homolog, subfamily B, member 4 | MGI:1914285 |
| ENSMUSG00000036052 | Dnajb5 | DnaJ (Hsp40) homolog, subfamily B, member 5 | MGI:1930018 |
| ENSMUSG00000029131 | Dnajb6 | DnaJ (Hsp40) homolog, subfamily B, member 6 | MGI:1344381 |
| ENSMUSG00000027006 | Dnajc10 | DnaJ (Hsp40) homolog, subfamily C, member 10 | MGI:1914111 |
| ENSMUSG00000039768 | Dnajc11 | DnaJ (Hsp40) homolog, subfamily C, member 11 | MGI:2443386 |
| ENSMUSG00000040697 | Dnajc16 | DnaJ (Hsp40) homolog, subfamily C, member 16 | MGI:2442146 |
| ENSMUSG00000024350 | Dnajc18 | DnaJ (Hsp40) homolog, subfamily C, member 18 | MGI:1923844 |
| ENSMUSG00000027679 | Dnajc19 | DnaJ (Hsp40) homolog, subfamily C, member 19 | MGI:1914963 |
| ENSMUSG00000026825 | Dnm1 | dynamin 1 | MGI:107384 |
| ENSMUSG00000022789 | Dnm1l | dynamin 1-like | MGI:1921256 |
| ENSMUSG00000033335 | Dnm2 | dynamin 2 | MGI:109547 |
| ENSMUSG00000040265 | Dnm3 | dynamin 3 | MGI:1341299 |
| ENSMUSG00000038608 | Dock10 | dedicator of cytokinesis 10 | MGI:2146320 |
| ENSMUSG00000058325 | Dock1 | dedicator of cytokinesis 1 | MGI:2429765 |
| ENSMUSG00000039716 | Dock3 | dedicator of cyto-kinesis 3 | MGI:2429763 |
| ENSMUSG00000035954 | Dock4 | dedicator of cytokinesis 4 | MGI:1918006 |
| ENSMUSG00000044447 | Dock5 | dedicator of cytokinesis 5 | MGI:2652871 |
| ENSMUSG00000028556 | Dock7 | dedicator of cytokinesis 7 | MGI:1914549 |
| ENSMUSG00000025558 | Dock9 | dedicator of cytokinesis 9 | MGI:106321 |
| ENSMUSG00000078919 | Dpm1 | dolichol-phosphate (beta-D) mannosyltransferase 1 | MGI:1330239 |
| ENSMUSG00000001229 | Dpp9 | dipeptidylpeptidase 9 | MGI:2443967 |
| ENSMUSG00000020537 | Drg2 | developmentally regulated GTP binding protein 2 | MGI:1342307 |
| ENSMUSG00000103888 | Dst | dystonin | MGI:104627 |
| ENSMUSG00000042662 | Dusp15 | dual specificity phosphatase-like 15 | MGI:1934928 |
| ENSMUSG00000003518 | Dusp3 | dual specificity phosphatase 3 (vaccinia virus phosphatase VH1-related) | MGI:1919599 |
| ENSMUSG00000021791 | Dydc2 | DPY30 domain containing 2 | MGI:1918450 |
| ENSMUSG00000029757 | Dync1i1 | dynein cytoplasmic 1 intermediate chain 1 | MGI:107743 |
| ENSMUSG00000027012 | Dync1i2 | dynein cytoplasmic 1 intermediate chain 2 | MGI:107750 |
| ENSMUSG00000032435 | Dync1li1 | dynein cytoplasmic 1 light intermediate chain 1 | MGI:2135610 |
| ENSMUSG00000035770 | Dync1li2 | dynein, cytoplasmic 1 light intermediate chain 2 | MGI:107738 |
| ENSMUSG00000009013 | Dynll1 | dynein light chain LC8-type 1 | MGI:1861457 |
| ENSMUSG00000020483 | Dynll2 | dynein light chain LC8-type 2 | MGI:1915347 |
| ENSMUSG00000022897 | Dyrk1a | dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1a | MGI:1330299 |
| ENSMUSG00000037259 | Dzank1 | double zinc ribbon and ankyrin repeat domains 1 | MGI:2139080 |
| ENSMUSG00000022350 | E430025E21Rik | RIKEN cDNA E430025E21 gene | MGI:2146110 |
| ENSMUSG00000026247 | Ecel1 | endothelin converting enzyme-like 1 | MGI:1343461 |
| ENSMUSG00000025465 | Echs1 | enoyl Coenzyme A hydratase, short chain, 1, mitochondrial | MGI:2136460 |
| ENSMUSG00000066839 | Ecsit | ECSIT homolog (Drosophila) | MGI:1349469 |
| ENSMUSG00000015092 | Edf1 | endothelial differentiation-related factor 1 | MGI:1891227 |
| ENSMUSG00000034488 | Edil3 | EGF-like repeats and discoidin I-like domains 3 | MGI:1329025 |
| ENSMUSG00000037742 | Eef1a1 | eukaryotic translation elongation factor 1 alpha 1 | MGI:1096881 |
| ENSMUSG00000016349 | Eef1a2 | eukaryotic translation elongation factor 1 alpha 2 | MGI:1096317 |
| ENSMUSG00000055762 | Eef1d | eukaryotic translation elongation factor 1 delta (guanine nucleotide exchange protein) | MGI:1913906 |
| ENSMUSG00000001707 | Eef1e1 | eukaryotic translation elongation factor 1 epsilon 1 | MGI:1913393 |
| ENSMUSG00000071644 | Eef1g | eukaryotic translation elongation factor 1 gamma | MGI:1914410 |
| ENSMUSG00000036611 | Eepd1 | endonuclease/exonuclease/phosphatase family domain containing 1 | MGI:1914734 |
| ENSMUSG00000028039 | Efna3 | ephrin A3 | MGI:106644 |
| ENSMUSG00000015002 | Efr3a | EFR3 homolog A (S. cerevisiae) | MGI:1923990 |
| ENSMUSG00000020658 | Efr3b | EFR3 homolog B (S. cerevisiae) | MGI:2444851 |
| ENSMUSG00000024772 | Ehd1 | EH-domain containing 1 | MGI:1341878 |
| ENSMUSG00000024065 | Ehd3 | EH-domain containing 3 | MGI:1928900 |
| ENSMUSG00000027810 | Eif2a | eukaryotic translation initiation factor 2A | MGI:1098684 |
| ENSMUSG00000035150 | Eif2s3x | eukaryotic translation initiation factor 2, subunit 3, structural gene X-linked | MGI:1349431 |
| ENSMUSG00000031029 | Eif3f | eukaryotic translation initiation factor 3, subunit F | MGI:1913335 |
| ENSMUSG00000033047 | Eif3l | eukaryotic translation initiation factor 3, subunit L | MGI:2386251 |
| ENSMUSG00000022884 | Eif4a2 | eukaryotic translation initiation factor 4A2 | MGI:106906 |
| ENSMUSG00000025580 | Eif4a3 | eukaryotic translation initiation factor 4A3 | MGI:1923731 |
| ENSMUSG00000058655 | Eif4b | eukaryotic translation initiation factor 4B | MGI:95304 |
| ENSMUSG00000045983 | Eif4g1 | eukaryotic translation initiation factor 4, gamma 1 | MGI:2384784 |
| ENSMUSG00000005610 | Eif4g2 | eukaryotic translation initiation factor 4, gamma 2 | MGI:109207 |
| ENSMUSG00000008489 | Elavl2 | ELAV (embryonic lethal, abnormal vision, Drosophila)-like 2 (Hu antigen B) | MGI:1100887 |
| ENSMUSG00000003410 | Elavl3 | ELAV (embryonic lethal, abnormal vision, Drosophila)-like 3 (Hu antigen C) | MGI:109157 |
| ENSMUSG00000028546 | Elavl4 | ELAV (embryonic lethal, abnormal vision, Drosophila)-like 4 (Hu antigen D) | MGI:107427 |
| ENSMUSG00000048988 | Elfn1 | leucine rich repeat and fibronectin type III, extracellular 1 | MGI:2442479 |
| ENSMUSG00000043460 | Elfn2 | leucine rich repeat and fibronectin type III, extracellular 2 | MGI:3608416 |
| ENSMUSG00000041112 | Elmo1 | engulfment and cell motility 1 | MGI:2153044 |
| ENSMUSG00000017670 | Elmo2 | engulfment and cell motility 2 | MGI:2153045 |
| ENSMUSG00000041986 | Elmod1 | ELMO/CED-12 domain containing 1 | MGI:3583900 |
| ENSMUSG00000008140 | Emc10 | ER membrane protein complex subunit 10 | MGI:1916933 |
| ENSMUSG00000078517 | Emc1 | ER membrane protein complex subunit 1 | MGI:2443696 |
| ENSMUSG00000022337 | Emc2 | ER membrane protein complex subunit 2 | MGI:1913986 |
| ENSMUSG00000030286 | Emc3 | ER membrane protein complex subunit 3 | MGI:1913337 |
| ENSMUSG00000027131 | Emc4 | ER membrane protein complex subunit 4 | MGI:1915282 |
| ENSMUSG00000055943 | Emc7 | ER membrane protein complex subunit 7 | MGI:1920274 |
| ENSMUSG00000031819 | Emc8 | ER membrane protein complex subunit 8 | MGI:1343095 |
| ENSMUSG00000001964 | Emd | emerin | MGI:108117 |
| ENSMUSG00000022995 | Enah | enabled homolog (Drosophila) | MGI:108360 |
| ENSMUSG00000015337 | Endog | endonuclease G | MGI:1261433 |
| ENSMUSG00000023960 | Enpp5 | ectonucleotide pyrophosphatase/phosphodiesterase 5 | MGI:1933830 |
| ENSMUSG00000038173 | Enpp6 | ectonucleotide pyrophosphatase/phosphodiesterase 6 | MGI:2445171 |
| ENSMUSG00000033068 | Entpd6 | ectonucleoside triphosphate diphosphohydrolase 6 | MGI:1202295 |
| ENSMUSG00000027624 | Epb4.1l1 | erythrocyte protein band 4.1 like 1 | MGI:103010 |
| ENSMUSG00000024044 | Epb4.1l3 | erythrocyte protein band 4.1 like 3 | MGI:103008 |
| ENSMUSG00000028434 | Epb4.1l4b | erythrocyte protein band 4.1 like 4b | MGI:1859149 |
| ENSMUSG00000023216 | Epb4.2 | erythrocyte protein band 4.2 | MGI:95402 |
| ENSMUSG00000002808 | Epdr1 | ependymin related protein 1 (zebrafish) | MGI:2145369 |
| ENSMUSG00000026235 | Epha4 | Eph receptor A4 | MGI:98277 |
| ENSMUSG00000010080 | Epn3 | epsin 3 | MGI:1919139 |
| ENSMUSG00000006276 | Eps15l1 | epidermal growth factor receptor pathway substrate 15-like 1 | MGI:104582 |
| ENSMUSG00000015766 | Eps8 | epidermal growth factor receptor pathway substrate 8 | MGI:104684 |
| ENSMUSG00000021709 | Erbb2ip | Erbb2 interacting protein | MGI:1890169 |
| ENSMUSG00000030172 | Erc1 | ELKS/RAB6-interacting/CAST family member 1 | MGI:2151013 |
| ENSMUSG00000040640 | Erc2 | ELKS/RAB6-interacting/CAST family member 2 | MGI:1098749 |
| ENSMUSG00000021131 | Erh | enhancer of rudimentary homolog (Drosophila) | MGI:108089 |
| ENSMUSG00000025198 | Erlin1 | ER lipid raft associated 1 | MGI:2387613 |
| ENSMUSG00000031483 | Erlin2 | ER lipid raft associated 2 | MGI:2387215 |
| ENSMUSG00000026830 | Ermn | ermin, ERM-like protein | MGI:1925017 |
| ENSMUSG00000021171 | Esyt2 | extended synaptotagmin-like protein 2 | MGI:1261845 |
| ENSMUSG00000024360 | Etf1 | eukaryotic translation termination factor 1 | MGI:2385071 |
| ENSMUSG00000036617 | Etl4 | enhancer trap locus 4 | MGI:95454 |
| ENSMUSG00000035104 | Eva1a | eva-1 homolog A (C. elegans) | MGI:2385247 |
| ENSMUSG00000011832 | Evi5l | ecotropic viral integration site 5 like | MGI:2442167 |
| ENSMUSG00000021262 | Evl | Ena-vasodilator stimulated phosphoprotein | MGI:1194884 |
| ENSMUSG00000009079 | Ewsr1 | Ewing sarcoma breakpoint region 1 | MGI:99960 |
| ENSMUSG00000032705 | Exd2 | exonuclease 3'-5' domain containing 2 | MGI:1922485 |
| ENSMUSG00000036435 | Exoc1 | exocyst complex component 1 | MGI:2445020 |
| ENSMUSG00000021357 | Exoc2 | exocyst complex component 2 | MGI:1913732 |
| ENSMUSG00000034152 | Exoc3 | exocyst complex component 3 | MGI:2443972 |
| ENSMUSG00000029763 | Exoc4 | exocyst complex component 4 | MGI:1096376 |
| ENSMUSG00000061244 | Exoc5 | exocyst complex component 5 | MGI:2145645 |
| ENSMUSG00000033769 | Exoc6b | exocyst complex component 6B | MGI:1923164 |
| ENSMUSG00000053799 | Exoc6 | exocyst complex component 6 | MGI:1351611 |
| ENSMUSG00000020792 | Exoc7 | exocyst complex component 7 | MGI:1859270 |
| ENSMUSG00000074030 | Exoc8 | exocyst complex component 8 | MGI:2142527 |
| ENSMUSG00000042787 | Exog | endo/exonuclease (5'-3'), endonuclease G-like | MGI:2143333 |
| ENSMUSG00000027963 | Extl2 | exostoses (multiple)-like 2 | MGI:1889574 |
| ENSMUSG00000028128 | F3 | coagulation factor III | MGI:88381 |
| ENSMUSG00000034171 | Faah | fatty acid amide hydrolase | MGI:109609 |
| ENSMUSG00000045316 | Fahd1 | fumarylacetoacetate hydrolase domain containing 1 | MGI:1915886 |
| ENSMUSG00000023011 | Faim2 | Fas apoptotic inhibitory molecule 2 | MGI:1919643 |
| ENSMUSG00000021750 | Fam107a | family with sequence similarity 107, member A | MGI:3041256 |
| ENSMUSG00000049119 | Fam110b | family with sequence similarity 110, member B | MGI:1916593 |
| ENSMUSG00000038014 | Fam120a | family with sequence similarity 120, member A | MGI:2446163 |
| ENSMUSG00000025262 | Fam120c | family with sequence similarity 120, member C | MGI:2387687 |
| ENSMUSG00000028995 | Fam126a | family with sequence similarity 126, member A | MGI:2149839 |
| ENSMUSG00000038174 | Fam126b | family with sequence similarity 126, member B | MGI:1098784 |
| ENSMUSG00000029861 | Fam131b | family with sequence similarity 131, member B | MGI:1923406 |
| ENSMUSG00000003955 | Fam162a | family with sequence similarity 162, member A | MGI:1917436 |
| ENSMUSG00000009216 | Fam163b | family with sequence similarity 163, member B | MGI:1926106 |
| ENSMUSG00000050530 | Fam171a1 | family with sequence similarity 171, member A1 | MGI:2442917 |
| ENSMUSG00000034685 | Fam171a2 | family with sequence similarity 171, member A2 | MGI:2448496 |
| ENSMUSG00000048388 | Fam171b | family with sequence similarity 171, member B | MGI:2444579 |
| ENSMUSG00000061111 | Fam195b | family with sequence similarity 195, member B | MGI:2384752 |
| ENSMUSG00000073805 | Fam196a | family with sequence similarity 196, member A | MGI:3605068 |
| ENSMUSG00000021792 | Fam213a | family with sequence similarity 213, member A | MGI:1917814 |
| ENSMUSG00000029059 | Fam213b | family with sequence similarity 213, member B | MGI:1913719 |
| ENSMUSG00000058966 | Fam57b | family with sequence similarity 57, member B | MGI:1916202 |
| ENSMUSG00000054942 | Fam73a | family with sequence similarity 73, member A | MGI:1924567 |
| ENSMUSG00000032224 | Fam81a | family with sequence similarity 81, member A | MGI:1924136 |
| ENSMUSG00000002017 | Fam98a | family with sequence similarity 98, member A | MGI:1919972 |
| ENSMUSG00000027349 | Fam98b | family with sequence similarity 98, member B | MGI:1915465 |
| ENSMUSG00000030759 | Far1 | fatty acyl CoA reductase 1 | MGI:1914670 |
| ENSMUSG00000025555 | Farp1 | FERM, RhoGEF (Arhgef) and pleckstrin domain protein 1 (chondrocyte-derived) | MGI:2446173 |
| ENSMUSG00000003808 | Farsa | phenylalanyl-tRNA synthetase, alpha subunit | MGI:1913840 |
| ENSMUSG00000026245 | Farsb | phenylalanyl-tRNA synthetase, beta subunit | MGI:1346035 |
| ENSMUSG00000070047 | Fat1 | FAT tumor suppressor homolog 1 (Drosophila) | MGI:109168 |
| ENSMUSG00000055333 | Fat2 | FAT tumor suppressor homolog 2 (Drosophila) | MGI:2685369 |
| ENSMUSG00000074505 | Fat3 | FAT tumor suppressor homolog 3 (Drosophila) | MGI:2444314 |
| ENSMUSG00000038274 | Fau | Finkel-Biskis-Reilly murine sarcoma virus (FBR-MuSV) ubiquitously expressed (fox derived) | MGI:102547 |
| ENSMUSG00000041556 | Fbxo2 | F-box protein 2 | MGI:2446216 |
| ENSMUSG00000047013 | Fbxo41 | F-box protein 41 | MGI:1261912 |
| ENSMUSG00000058715 | Fcer1g | Fc receptor, IgE, high affinity I, gamma polypeptide | MGI:95496 |
| ENSMUSG00000041685 | Fcho2 | FCH domain only 2 | MGI:3505790 |
| ENSMUSG00000024588 | Fech | ferrochelatase | MGI:95513 |
| ENSMUSG00000028001 | Fga | fibrinogen alpha chain | MGI:1316726 |
| ENSMUSG00000022523 | Fgf12 | fibroblast growth factor 12 | MGI:109183 |
| ENSMUSG00000036585 | Fgf1 | fibroblast growth factor 1 | MGI:95515 |
| ENSMUSG00000040242 | Fgfr1op2 | FGFR1 oncogene partner 2 | MGI:1914779 |
| ENSMUSG00000033860 | Fgg | fibrinogen gamma chain | MGI:95526 |
| ENSMUSG00000026841 | Fibcd1 | fibrinogen C domain containing 1 | MGI:2138953 |
| ENSMUSG00000024911 | Fibp | fibroblast growth factor (acidic) intracellular binding protein | MGI:1926233 |
| ENSMUSG00000020635 | Fkbp1b | FK506 binding protein 1b | MGI:1336205 |
| ENSMUSG00000019428 | Fkbp8 | FK506 binding protein 8 | MGI:1341070 |
| ENSMUSG00000002812 | Flii | flightless I homolog (Drosophila) | MGI:1342286 |
| ENSMUSG00000059714 | Flot1 | flotillin 1 | MGI:1100500 |
| ENSMUSG00000061981 | Flot2 | flotillin 2 | MGI:103309 |
| ENSMUSG00000047414 | Flrt2 | fibronectin leucine rich transmembrane protein 2 | MGI:3603594 |
| ENSMUSG00000051379 | Flrt3 | fibronectin leucine rich transmembrane protein 3 | MGI:1918686 |
| ENSMUSG00000028354 | Fmn2 | formin 2 | MGI:1859252 |
| ENSMUSG00000055805 | Fmnl1 | formin-like 1 | MGI:1888994 |
| ENSMUSG00000036053 | Fmnl2 | formin-like 2 | MGI:1918659 |
| ENSMUSG00000000838 | Fmr1 | fragile X mental retardation syndrome 1 | MGI:95564 |
| ENSMUSG00000026657 | Frmd4a | FERM domain containing 4A | MGI:1919850 |
| ENSMUSG00000042425 | Frmpd3 | FERM and PDZ domain containing 3 | MGI:3646547 |
| ENSMUSG00000045589 | Frrs1l | ferric-chelate reductase 1 like | MGI:2442704 |
| ENSMUSG00000020170 | Frs2 | fibroblast growth factor receptor substrate 2 | MGI:1100860 |
| ENSMUSG00000023266 | Frs3 | fibroblast growth factor receptor substrate 3 | MGI:2135965 |
| ENSMUSG00000056602 | Fry | furry homolog (Drosophila) | MGI:2443895 |
| ENSMUSG00000011589 | Fsd1 | fibronectin type 3 and SPRY domain-containing protein | MGI:1934858 |
| ENSMUSG00000024661 | Fth1 | ferritin heavy chain 1 | MGI:95588 |
| ENSMUSG00000025040 | Fundc1 | FUN14 domain containing 1 | MGI:1919268 |
| ENSMUSG00000031198 | Fundc2 | FUN14 domain containing 2 | MGI:1914641 |
| ENSMUSG00000030795 | Fus | fused in sarcoma | MGI:1353633 |
| ENSMUSG00000021065 | Fut8 | fucosyltransferase 8 | MGI:1858901 |
| ENSMUSG00000027680 | Fxr1 | fragile X mental retardation gene 1, autosomal homolog | MGI:104860 |
| ENSMUSG00000018765 | Fxr2 | fragile X mental retardation, autosomal homolog 2 | MGI:1346074 |
| ENSMUSG00000036570 | Fxyd1 | FXYD domain-containing ion transport regulator 1 | MGI:1889273 |
| ENSMUSG00000025241 | Fyco1 | FYVE and coiled-coil domain containing 1 | MGI:107277 |
| ENSMUSG00000019843 | Fyn | Fyn proto-oncogene | MGI:95602 |
| ENSMUSG00000018583 | G3bp1 | GTPase activating protein (SH3 domain) binding protein 1 | MGI:1351465 |
| ENSMUSG00000029405 | G3bp2 | GTPase activating protein (SH3 domain) binding protein 2 | MGI:2442040 |
| ENSMUSG00000030161 | Gabarapl1 | gamma-aminobutyric acid (GABA) A receptor-associated protein-like 1 | MGI:1914980 |
| ENSMUSG00000031950 | Gabarapl2 | gamma-aminobutyric acid (GABA) A receptor-associated protein-like 2 | MGI:1890602 |
| ENSMUSG00000024462 | Gabbr1 | gamma-aminobutyric acid (GABA) B receptor, 1 | MGI:1860139 |
| ENSMUSG00000039809 | Gabbr2 | gamma-aminobutyric acid (GABA) B receptor, 2 | MGI:2386030 |
| ENSMUSG00000010803 | Gabra1 | gamma-aminobutyric acid (GABA) A receptor, subunit alpha 1 | MGI:95613 |
| ENSMUSG00000000560 | Gabra2 | gamma-aminobutyric acid (GABA) A receptor, subunit alpha 2 | MGI:95614 |
| ENSMUSG00000031343 | Gabra3 | gamma-aminobutyric acid (GABA) A receptor, subunit alpha 3 | MGI:95615 |
| ENSMUSG00000029211 | Gabra4 | gamma-aminobutyric acid (GABA) A receptor, subunit alpha 4 | MGI:95616 |
| ENSMUSG00000020428 | Gabra6 | gamma-aminobutyric acid (GABA) A receptor, subunit alpha 6 | MGI:95618 |
| ENSMUSG00000029212 | Gabrb1 | gamma-aminobutyric acid (GABA) A receptor, subunit beta 1 | MGI:95619 |
| ENSMUSG00000007653 | Gabrb2 | gamma-aminobutyric acid (GABA) A receptor, subunit beta 2 | MGI:95620 |
| ENSMUSG00000033676 | Gabrb3 | gamma-aminobutyric acid (GABA) A receptor, subunit beta 3 | MGI:95621 |
| ENSMUSG00000020436 | Gabrg2 | gamma-aminobutyric acid (GABA) A receptor, subunit gamma 2 | MGI:95623 |
| ENSMUSG00000033751 | Gadd45gip1 | growth arrest and DNA-damage-inducible, gamma interacting protein 1 | MGI:1914947 |
| ENSMUSG00000062234 | Gak | cyclin G associated kinase | MGI:2442153 |
| ENSMUSG00000021003 | Galc | galactosylceramidase | MGI:95636 |
| ENSMUSG00000047261 | Gap43 | growth associated protein 43 | MGI:95639 |
| ENSMUSG00000057666 | Gapdh | glyceraldehyde-3-phosphate dehydrogenase | MGI:95640 |
| ENSMUSG00000034201 | Gas2l1 | growth arrest-specific 2 like 1 | MGI:1926176 |
| ENSMUSG00000033066 | Gas7 | growth arrest specific 7 | MGI:1202388 |
| ENSMUSG00000029432 | Gbas | glioblastoma amplified sequence | MGI:1278343 |
| ENSMUSG00000003809 | Gcdh | glutaryl-Coenzyme A dehydrogenase | MGI:104541 |
| ENSMUSG00000041638 | Gcn1l1 | GCN1 general control of amino-acid synthesis 1-like 1 (yeast) | MGI:2444248 |
| ENSMUSG00000025777 | Gdap1 | ganglioside-induced differentiation-associated-protein 1 | MGI:1338002 |
| ENSMUSG00000017943 | Gdap1l1 | ganglioside-induced differentiation-associated protein 1-like 1 | MGI:2385163 |
| ENSMUSG00000025858 | Get4 | golgi to ER traffic protein 4 homolog (S. cerevisiae) | MGI:1914854 |
| ENSMUSG00000020932 | Gfap | glial fibrillary acidic protein | MGI:95697 |
| ENSMUSG00000022103 | Gfra2 | glial cell line derived neurotrophic factor family receptor alpha 2 | MGI:1195462 |
| ENSMUSG00000027573 | Gid8 | GID complex subunit 8 homolog (S. cerevisiae) | MGI:1923675 |
| ENSMUSG00000029714 | Gigyf1 | GRB10 interacting GYF protein 1 | MGI:1888677 |
| ENSMUSG00000048000 | Gigyf2 | GRB10 interacting GYF protein 2 | MGI:2138584 |
| ENSMUSG00000019433 | Gipc1 | GIPC PDZ domain containing family, member 1 | MGI:1926252 |
| ENSMUSG00000011877 | Git1 | G protein-coupled receptor kinase-interactor 1 | MGI:1927140 |
| ENSMUSG00000050953 | Gja1 | gap junction protein, alpha 1 | MGI:95713 |
| ENSMUSG00000040055 | Gjb6 | gap junction protein, beta 6 | MGI:107588 |
| ENSMUSG00000043448 | Gjc2 | gap junction protein, gamma 2 | MGI:2153060 |
| ENSMUSG00000021552 | Gkap1 | G kinase anchoring protein 1 | MGI:1891694 |
| ENSMUSG00000028480 | Glipr2 | GLI pathogenesis-related 2 | MGI:1917770 |
| ENSMUSG00000000263 | Glra1 | glycine receptor, alpha 1 subunit | MGI:95747 |
| ENSMUSG00000028020 | Glrb | glycine receptor, beta subunit | MGI:95751 |
| ENSMUSG00000031068 | Glrx3 | glutaredoxin 3 | MGI:1353653 |
| ENSMUSG00000021591 | Glrx | glutaredoxin | MGI:2135625 |
| ENSMUSG00000011884 | Gltp | glycolipid transfer protein | MGI:1929253 |
| ENSMUSG00000036568 | Gltscr1l | GLTSCR1-like | MGI:2673855 |
| ENSMUSG00000021794 | Glud1 | glutamate dehydrogenase 1 | MGI:95753 |
| ENSMUSG00000057262 | Gm10020 | predicted pseudogene 10020 | MGI:3642192 |
| ENSMUSG00000058064 | Gm10036 | predicted gene 10036 | MGI:3642334 |
| ENSMUSG00000069117 | Gm10260 | predicted gene 10260 | MGI:3642298 |
| ENSMUSG00000042744 | Gm15800 | predicted gene 15800 | MGI:3647820 |
| ENSMUSG00000078193 | Gm2000 | predicted gene 2000 | MGI:3780170 |
| ENSMUSG00000091228 | Gm20390 | predicted gene 20390 | MGI:5141855 |
| ENSMUSG00000090639 | Gm20425 | predicted gene 20425 | MGI:5141890 |
| ENSMUSG00000021139 | Gm20498 | predicted gene 20498 | MGI:5141963 |
| ENSMUSG00000100725 | Gm28062 | predicted gene 28062 | MGI:5578768 |
| ENSMUSG00000066487 | Gm5786 | predicted pseudogene 5786 | MGI:3645003 |
| ENSMUSG00000094685 | Gm5900 | predicted pseudogene 5900 | MGI:3643907 |
| ENSMUSG00000050490 | Gm8394 | predicted gene 8394 | MGI:3647964 |
| ENSMUSG00000063696 | Gm8730 | predicted pseudogene 8730 | MGI:3644565 |
| ENSMUSG00000044424 | Gm9493 | predicted gene 9493 | MGI:3779903 |
| ENSMUSG00000045455 | Gm9797 | predicted pseudogene 9797 | MGI:3704349 |
| ENSMUSG00000049230 | Gm9833 | predicted gene 9833 | MGI:3641855 |
| ENSMUSG00000029419 | Gm996 | predicted gene 996 | MGI:2685842 |
| ENSMUSG00000062014 | Gmfb | glia maturation factor, beta | MGI:1927133 |
| ENSMUSG00000000149 | Gna12 | guanine nucleotide binding protein, alpha 12 | MGI:95767 |
| ENSMUSG00000057614 | Gnai1 | guanine nucleotide binding protein (G protein), alpha inhibiting 1 | MGI:95771 |
| ENSMUSG00000032562 | Gnai2 | guanine nucleotide binding protein (G protein), alpha inhibiting 2 | MGI:95772 |
| ENSMUSG00000000001 | Gnai3 | guanine nucleotide binding protein (G protein), alpha inhibiting 3 | MGI:95773 |
| ENSMUSG00000024524 | Gnal | guanine nucleotide binding protein, alpha stimulating, olfactory type | MGI:95774 |
| ENSMUSG00000031748 | Gnao1 | guanine nucleotide binding protein, alpha O | MGI:95775 |
| ENSMUSG00000024639 | Gnaq | guanine nucleotide binding protein, alpha q polypeptide | MGI:95776 |
| ENSMUSG00000027523 | Gnas | GNAS (guanine nucleotide binding protein, alpha stimulating) complex locus | MGI:95777 |
| ENSMUSG00000040009 | Gnaz | guanine nucleotide binding protein, alpha z subunit | MGI:95780 |
| ENSMUSG00000029713 | Gnb2 | guanine nucleotide binding protein (G protein), beta 2 | MGI:95784 |
| ENSMUSG00000023439 | Gnb3 | guanine nucleotide binding protein (G protein), beta 3 | MGI:95785 |
| ENSMUSG00000027669 | Gnb4 | guanine nucleotide binding protein (G protein), beta 4 | MGI:104581 |
| ENSMUSG00000032192 | Gnb5 | guanine nucleotide binding protein (G protein), beta 5 | MGI:101848 |
| ENSMUSG00000036402 | Gng12 | guanine nucleotide binding protein (G protein), gamma 12 | MGI:1336171 |
| ENSMUSG00000025739 | Gng13 | guanine nucleotide binding protein (G protein), gamma 13 | MGI:1925616 |
| ENSMUSG00000043004 | Gng2 | guanine nucleotide binding protein (G protein), gamma 2 | MGI:102705 |
| ENSMUSG00000021303 | Gng4 | guanine nucleotide binding protein (G protein), gamma 4 | MGI:102703 |
| ENSMUSG00000031985 | Gnpat | glyceronephosphate O-acyltransferase | MGI:1343460 |
| ENSMUSG00000042532 | Golga7b | golgi autoantigen, golgin subfamily a, 7B | MGI:1918396 |
| ENSMUSG00000015341 | Golga7 | golgi autoantigen, golgin subfamily a, 7 | MGI:1931029 |
| ENSMUSG00000034243 | Golgb1 | golgi autoantigen, golgin subfamily b, macrogolgin 1 | MGI:1099447 |
| ENSMUSG00000010392 | Gosr1 | golgi SNAP receptor complex member 1 | MGI:1858260 |
| ENSMUSG00000034220 | Gpc1 | glypican 1 | MGI:1194891 |
| ENSMUSG00000022112 | Gpc5 | glypican 5 | MGI:1194894 |
| ENSMUSG00000047454 | Gphn | gephyrin | MGI:109602 |
| ENSMUSG00000045967 | Gpr158 | G protein-coupled receptor 158 | MGI:2441697 |
| ENSMUSG00000038390 | Gpr162 | G protein-coupled receptor 162 | MGI:1315214 |
| ENSMUSG00000043384 | Gprasp1 | G protein-coupled receptor associated sorting protein 1 | MGI:1917418 |
| ENSMUSG00000008734 | Gprc5b | G protein-coupled receptor, family C, group 5, member B | MGI:1927596 |
| ENSMUSG00000069227 | Gprin1 | G protein-regulated inducer of neurite outgrowth 1 | MGI:1349455 |
| ENSMUSG00000071531 | Gprin2 | G protein regulated inducer of neurite outgrowth 2 | MGI:2444560 |
| ENSMUSG00000045441 | Gprin3 | GPRIN family member 3 | MGI:1924785 |
| ENSMUSG00000026930 | Gpsm1 | G-protein signalling modulator 1 (AGS3-like, C. elegans) | MGI:1915089 |
| ENSMUSG00000034786 | Gpsm3 | G-protein signalling modulator 3 (AGS3-like, C. elegans) | MGI:2146785 |
| ENSMUSG00000063856 | Gpx1 | glutathione peroxidase 1 | MGI:104887 |
| ENSMUSG00000075706 | Gpx4 | glutathione peroxidase 4 | MGI:104767 |
| ENSMUSG00000035900 | Gramd4 | GRAM domain containing 4 | MGI:2676308 |
| ENSMUSG00000000531 | Grasp | GRP1 (general receptor for phosphoinositides 1)-associated scaffold protein | MGI:1860303 |
| ENSMUSG00000072772 | Grcc10 | gene rich cluster, C10 gene | MGI:1315201 |
| ENSMUSG00000020524 | Gria1 | glutamate receptor, ionotropic, AMPA1 (alpha 1) | MGI:95808 |
| ENSMUSG00000033981 | Gria2 | glutamate receptor, ionotropic, AMPA2 (alpha 2) | MGI:95809 |
| ENSMUSG00000001986 | Gria3 | glutamate receptor, ionotropic, AMPA3 (alpha 3) | MGI:95810 |
| ENSMUSG00000025892 | Gria4 | glutamate receptor, ionotropic, AMPA4 (alpha 4) | MGI:95811 |
| ENSMUSG00000071424 | Grid2 | glutamate receptor, ionotropic, delta 2 | MGI:95813 |
| ENSMUSG00000010825 | Grid2ip | glutamate receptor, ionotropic, delta 2 (Grid2) interacting protein 1 | MGI:2176213 |
| ENSMUSG00000056073 | Grik2 | glutamate receptor, ionotropic, kainate 2 (beta 2) | MGI:95815 |
| ENSMUSG00000001985 | Grik3 | glutamate receptor, ionotropic, kainate 3 | MGI:95816 |
| ENSMUSG00000003378 | Grik5 | glutamate receptor, ionotropic, kainate 5 (gamma 2) | MGI:95818 |
| ENSMUSG00000026959 | Grin1 | glutamate receptor, ionotropic, NMDA1 (zeta 1) | MGI:95819 |
| ENSMUSG00000059003 | Grin2a | glutamate receptor, ionotropic, NMDA2A (epsilon 1) | MGI:95820 |
| ENSMUSG00000030209 | Grin2b | glutamate receptor, ionotropic, NMDA2B (epsilon 2) | MGI:95821 |
| ENSMUSG00000020734 | Grin2c | glutamate receptor, ionotropic, NMDA2C (epsilon 3) | MGI:95822 |
| ENSMUSG00000002771 | Grin2d | glutamate receptor, ionotropic, NMDA2D (epsilon 4) | MGI:95823 |
| ENSMUSG00000034813 | Grip1 | glutamate receptor interacting protein 1 | MGI:1921303 |
| ENSMUSG00000030098 | Grip2 | glutamate receptor interacting protein 2 | MGI:2681173 |
| ENSMUSG00000031153 | Gripap1 | GRIP1 associated protein 1 | MGI:1859616 |
| ENSMUSG00000074886 | Grk6 | G protein-coupled receptor kinase 6 | MGI:1347078 |
| ENSMUSG00000023192 | Grm2 | glutamate receptor, metabotropic 2 | MGI:1351339 |
| ENSMUSG00000003974 | Grm3 | glutamate receptor, metabotropic 3 | MGI:1351340 |
| ENSMUSG00000063239 | Grm4 | glutamate receptor, metabotropic 4 | MGI:1351341 |
| ENSMUSG00000049583 | Grm5 | glutamate receptor, metabotropic 5 | MGI:1351342 |
| ENSMUSG00000056755 | Grm7 | glutamate receptor, metabotropic 7 | MGI:1351344 |
| ENSMUSG00000024211 | Grm8 | glutamate receptor, metabotropic 8 | MGI:1351345 |
| ENSMUSG00000046182 | Gsg1l | GSG1-like | MGI:2685483 |
| ENSMUSG00000022812 | Gsk3b | glycogen synthase kinase 3 beta | MGI:1861437 |
| ENSMUSG00000026879 | Gsn | gelsolin | MGI:95851 |
| ENSMUSG00000032348 | Gsta4 | glutathione S-transferase, alpha 4 | MGI:1309515 |
| ENSMUSG00000060261 | Gtf2i | general transcription factor II I | MGI:1202722 |
| ENSMUSG00000042535 | Gtpbp1 | GTP binding protein 1 | MGI:109443 |
| ENSMUSG00000041624 | Gucy1a2 | guanylate cyclase 1, soluble, alpha 2 | MGI:2660877 |
| ENSMUSG00000028005 | Gucy1b3 | guanylate cyclase 1, soluble, beta 3 | MGI:1860604 |
| ENSMUSG00000020444 | Guk1 | guanylate kinase 1 | MGI:95871 |
| ENSMUSG00000025059 | Gyk | glycerol kinase | MGI:106594 |
| ENSMUSG00000051839 | Gypa | glycophorin A | MGI:95880 |
| ENSMUSG00000090523 | Gypc | glycophorin C | MGI:1098566 |
| ENSMUSG00000060743 | H3f3a | H3 histone, family 3A | MGI:1097686 |
| ENSMUSG00000035376 | Hacd2 | 3-hydroxyacyl-CoA dehydratase 2 | MGI:1918007 |
| ENSMUSG00000033629 | Hacd3 | 3-hydroxyacyl-CoA dehydratase 3 | MGI:1889341 |
| ENSMUSG00000025745 | Hadha | hydroxyacyl-Coenzyme A dehydrogenase/3-ketoacyl-Coenzyme A thiolase/enoyl-Coenzyme A hydratase (trifunctional protein), alpha subunit | MGI:2135593 |
| ENSMUSG00000059447 | Hadhb | hydroxyacyl-Coenzyme A dehydrogenase/3-ketoacyl-Coenzyme A thiolase/enoyl-Coenzyme A hydratase (trifunctional protein), beta subunit | MGI:2136381 |
| ENSMUSG00000006930 | Hap1 | huntingtin-associated protein 1 | MGI:1261831 |
| ENSMUSG00000021613 | Hapln1 | hyaluronan and proteoglycan link protein 1 | MGI:1337006 |
| ENSMUSG00000004894 | Hapln2 | hyaluronan and proteoglycan link protein 2 | MGI:2137300 |
| ENSMUSG00000007594 | Hapln4 | hyaluronan and proteoglycan link protein 4 | MGI:2679531 |
| ENSMUSG00000031386 | Hcfc1 | host cell factor C1 | MGI:105942 |
| ENSMUSG00000034245 | Hdac11 | histone deacetylase 11 | MGI:2385252 |
| ENSMUSG00000042807 | Hecw2 | HECT, C2 and WW domain containing E3 ubiquitin protein ligase 2 | MGI:2685817 |
| ENSMUSG00000046240 | Hepacam | hepatocyte cell adhesion molecule | MGI:1920177 |
| ENSMUSG00000021665 | Hexb | hexosaminidase B | MGI:96074 |
| ENSMUSG00000020267 | Hint1 | histidine triad nucleotide binding protein 1 | MGI:1321133 |
| ENSMUSG00000039959 | Hip1 | huntingtin interacting protein 1 | MGI:1099804 |
| ENSMUSG00000000915 | Hip1r | huntingtin interacting protein 1 related | MGI:1352504 |
| ENSMUSG00000060081 | Hist1h2aa | histone cluster 1, H2aa | MGI:2448285 |
| ENSMUSG00000069308 | Hist1h2bp | histone cluster 1, H2bp | MGI:2448409 |
| ENSMUSG00000069266 | Hist1h4b | histone cluster 1, H4b | MGI:2448420 |
| ENSMUSG00000037012 | Hk1 | hexokinase 1 | MGI:96103 |
| ENSMUSG00000000628 | Hk2 | hexokinase 2 | MGI:1315197 |
| ENSMUSG00000078249 | Hmga1-rs1 | high mobility group AT-hook I, related sequence 1 | MGI:96161 |
| ENSMUSG00000046434 | Hnrnpa1 | heterogeneous nuclear ribonucleoprotein A1 | MGI:104820 |
| ENSMUSG00000004980 | Hnrnpa2b1 | heterogeneous nuclear ribonucleoprotein A2/B1 | MGI:104819 |
| ENSMUSG00000059005 | Hnrnpa3 | heterogeneous nuclear ribonucleoprotein A3 | MGI:1917171 |
| ENSMUSG00000020358 | Hnrnpab | heterogeneous nuclear ribonucleoprotein A/B | MGI:1330294 |
| ENSMUSG00000060373 | Hnrnpc | heterogeneous nuclear ribonucleoprotein C | MGI:107795 |
| ENSMUSG00000000568 | Hnrnpd | heterogeneous nuclear ribonucleoprotein D | MGI:101947 |
| ENSMUSG00000021546 | Hnrnpk | heterogeneous nuclear ribonucleoprotein K | MGI:99894 |
| ENSMUSG00000015165 | Hnrnpl | heterogeneous nuclear ribonucleoprotein L | MGI:104816 |
| ENSMUSG00000059208 | Hnrnpm | heterogeneous nuclear ribonucleoprotein M | MGI:1926465 |
| ENSMUSG00000007617 | Homer1 | homer homolog 1 (Drosophila) | MGI:1347345 |
| ENSMUSG00000025813 | Homer2 | homer homolog 2 (Drosophila) | MGI:1347354 |
| ENSMUSG00000003573 | Homer3 | homer homolog 3 (Drosophila) | MGI:1347359 |
| ENSMUSG00000037234 | Hook3 | hook homolog 3 (Drosophila) | MGI:2443554 |
| ENSMUSG00000028759 | Hp1bp3 | heterochromatin protein 1, binding protein 3 | MGI:109369 |
| ENSMUSG00000028785 | Hpca | hippocalcin | MGI:1336200 |
| ENSMUSG00000071379 | Hpcal1 | hippocalcin-like 1 | MGI:1855689 |
| ENSMUSG00000046093 | Hpcal4 | hippocalcin-like 4 | MGI:2157521 |
| ENSMUSG00000025499 | Hras | Harvey rat sarcoma virus oncogene | MGI:96224 |
| ENSMUSG00000031839 | Hsbp1 | heat shock factor binding protein 1 | MGI:1915446 |
| ENSMUSG00000025260 | Hsd17b10 | hydroxysteroid (17-beta) dehydrogenase 10 | MGI:1333871 |
| ENSMUSG00000027195 | Hsd17b12 | hydroxysteroid (17-beta) dehydrogenase 12 | MGI:1926967 |
| ENSMUSG00000024507 | Hsd17b4 | hydroxysteroid (17-beta) dehydrogenase 4 | MGI:105089 |
| ENSMUSG00000034189 | Hsdl1 | hydroxysteroid dehydrogenase like 1 | MGI:1919802 |
| ENSMUSG00000028383 | Hsdl2 | hydroxysteroid dehydrogenase like 2 | MGI:1919729 |
| ENSMUSG00000025092 | Hspa12a | heat shock protein 12A | MGI:1920692 |
| ENSMUSG00000074793 | Hspa12b | heat shock protein 12B | MGI:1919880 |
| ENSMUSG00000091971 | Hspa1a | heat shock protein 1A | MGI:96244 |
| ENSMUSG00000059970 | Hspa2 | heat shock protein 2 | MGI:96243 |
| ENSMUSG00000015656 | Hspa8 | heat shock protein 8 | MGI:105384 |
| ENSMUSG00000004951 | Hspb1 | heat shock protein 1 | MGI:96240 |
| ENSMUSG00000006205 | Htra1 | HtrA serine peptidase 1 | MGI:1929076 |
| ENSMUSG00000037851 | Iars | isoleucine-tRNA synthetase | MGI:2145219 |
| ENSMUSG00000018858 | Ict1 | immature colon carcinoma transcript 1 | MGI:1915822 |
| ENSMUSG00000002010 | Idh3g | isocitrate dehydrogenase 3 (NAD+), gamma | MGI:1099463 |
| ENSMUSG00000076617 | Ighm | immunoglobulin heavy constant mu | MGI:96448 |
| ENSMUSG00000013367 | Iglon5 | IgLON family member 5 | MGI:2686277 |
| ENSMUSG00000040972 | Igsf21 | immunoglobulin superfamily, member 21 | MGI:2681842 |
| ENSMUSG00000038034 | Igsf8 | immunoglobulin superfamily, member 8 | MGI:2154090 |
| ENSMUSG00000034275 | Igsf9b | immunoglobulin superfamily, member 9B | MGI:2685354 |
| ENSMUSG00000052372 | Il1rapl1 | interleukin 1 receptor accessory protein-like 1 | MGI:2687319 |
| ENSMUSG00000020279 | Il9r | interleukin 9 receptor | MGI:96564 |
| ENSMUSG00000040612 | Ildr2 | immunoglobulin-like domain containing receptor 2 | MGI:1196370 |
| ENSMUSG00000001016 | Ilf2 | interleukin enhancer binding factor 2 | MGI:1915031 |
| ENSMUSG00000032763 | Ilvbl | ilvB (bacterial acetolactate synthase)-like | MGI:1351911 |
| ENSMUSG00000052337 | Immt | inner membrane protein, mitochondrial | MGI:1923864 |
| ENSMUSG00000034336 | Ina | internexin neuronal intermediate filament protein, alpha | MGI:96568 |
| ENSMUSG00000026113 | Inpp4a | inositol polyphosphate-4-phosphatase, type I | MGI:1931123 |
| ENSMUSG00000025477 | Inpp5a | inositol polyphosphate-5-phosphatase A | MGI:2686961 |
| ENSMUSG00000026925 | Inpp5e | inositol polyphosphate-5-phosphatase E | MGI:1927753 |
| ENSMUSG00000066232 | Ipo7 | importin 7 | MGI:2152414 |
| ENSMUSG00000034312 | Iqsec1 | IQ motif and Sec7 domain 1 | MGI:1196356 |
| ENSMUSG00000041115 | Iqsec2 | IQ motif and Sec7 domain 2 | MGI:3528396 |
| ENSMUSG00000040797 | Iqsec3 | IQ motif and Sec7 domain 3 | MGI:2677208 |
| ENSMUSG00000046879 | Irgm1 | immunity-related GTPase family M member 1 | MGI:107567 |
| ENSMUSG00000031729 | Ist1 | increased sodium tolerance 1 homolog (yeast) | MGI:1919205 |
| ENSMUSG00000031703 | Itfg1 | integrin alpha FG-GAP repeat containing 1 | MGI:106419 |
| ENSMUSG00000024187 | Itfg3 | integrin alpha FG-GAP repeat containing 3 | MGI:2146854 |
| ENSMUSG00000006522 | Itih3 | inter-alpha trypsin inhibitor, heavy chain 3 | MGI:96620 |
| ENSMUSG00000022108 | Itm2b | integral membrane protein 2B | MGI:1309517 |
| ENSMUSG00000026223 | Itm2c | integral membrane protein 2C | MGI:1927594 |
| ENSMUSG00000027296 | Itpka | inositol 1,4,5-trisphosphate 3-kinase A | MGI:1333822 |
| ENSMUSG00000030102 | Itpr1 | inositol 1,4,5-trisphosphate receptor 1 | MGI:96623 |
| ENSMUSG00000030287 | Itpr2 | inositol 1,4,5-triphosphate receptor 2 | MGI:99418 |
| ENSMUSG00000020640 | Itsn2 | intersectin 2 | MGI:1338049 |
| ENSMUSG00000028530 | Jak1 | Janus kinase 1 | MGI:96628 |
| ENSMUSG00000063646 | Jakmip1 | janus kinase and microtubule interacting protein 1 | MGI:1923321 |
| ENSMUSG00000024502 | Jakmip2 | janus kinase and microtubule interacting protein 2 | MGI:1923467 |
| ENSMUSG00000056856 | Jakmip3 | janus kinase and microtubule interacting protein 3 | MGI:1921254 |
| ENSMUSG00000031990 | Jam3 | junction adhesion molecule 3 | MGI:1933825 |
| ENSMUSG00000042686 | Jph1 | junctophilin 1 | MGI:1891495 |
| ENSMUSG00000025318 | Jph3 | junctophilin 3 | MGI:1891497 |
| ENSMUSG00000022208 | Jph4 | junctophilin 4 | MGI:2443113 |
| ENSMUSG00000001552 | Jup | junction plakoglobin | MGI:96650 |
| ENSMUSG00000061751 | Kalrn | kalirin, RhoGEF kinase | MGI:2685385 |
| ENSMUSG00000040606 | Kazn | kazrin, periplakin interacting protein | MGI:1918779 |
| ENSMUSG00000047976 | Kcna1 | potassium voltage-gated channel, shaker-related subfamily, member 1 | MGI:96654 |
| ENSMUSG00000040724 | Kcna2 | potassium voltage-gated channel, shaker-related subfamily, member 2 | MGI:96659 |
| ENSMUSG00000047959 | Kcna3 | potassium voltage-gated channel, shaker-related subfamily, member 3 | MGI:96660 |
| ENSMUSG00000042604 | Kcna4 | potassium voltage-gated channel, shaker-related subfamily, member 4 | MGI:96661 |
| ENSMUSG00000038077 | Kcna6 | potassium voltage-gated channel, shaker-related, subfamily, member 6 | MGI:96663 |
| ENSMUSG00000027827 | Kcnab1 | potassium voltage-gated channel, shaker-related subfamily, beta member 1 | MGI:109155 |
| ENSMUSG00000050556 | Kcnb1 | potassium voltage gated channel, Shab-related subfamily, member 1 | MGI:96666 |
| ENSMUSG00000062785 | Kcnc3 | potassium voltage gated channel, Shaw-related subfamily, member 3 | MGI:96669 |
| ENSMUSG00000009731 | Kcnd1 | potassium voltage-gated channel, Shal-related family, member 1 | MGI:96671 |
| ENSMUSG00000060882 | Kcnd2 | potassium voltage-gated channel, Shal-related family, member 2 | MGI:102663 |
| ENSMUSG00000040896 | Kcnd3 | potassium voltage-gated channel, Shal-related family, member 3 | MGI:1928743 |
| ENSMUSG00000053519 | Kcnip1 | Kv channel-interacting protein 1 | MGI:1917607 |
| ENSMUSG00000029088 | Kcnip4 | Kv channel interacting protein 4 | MGI:1933131 |
| ENSMUSG00000044216 | Kcnj4 | potassium inwardly-rectifying channel, subfamily J, member 4 | MGI:104743 |
| ENSMUSG00000054477 | Kcnn2 | potassium intermediate/small conductance calcium-activated channel, subfamily N, member 2 | MGI:2153182 |
| ENSMUSG00000016346 | Kcnq2 | potassium voltage-gated channel, subfamily Q, member 2 | MGI:1309503 |
| ENSMUSG00000098557 | Kctd12 | potassium channel tetramerisation domain containing 12 | MGI:2145823 |
| ENSMUSG00000051401 | Kctd16 | potassium channel tetramerisation domain containing 16 | MGI:1914659 |
| ENSMUSG00000037653 | Kctd8 | potassium channel tetramerisation domain containing 8 | MGI:2443804 |
| ENSMUSG00000028790 | Khdrbs1 | KH domain containing, RNA binding, signal transduction associated 1 | MGI:893579 |
| ENSMUSG00000003779 | Kif20a | kinesin family member 20A | MGI:1201682 |
| ENSMUSG00000021693 | Kif2a | kinesin family member 2A | MGI:108390 |
| ENSMUSG00000020668 | Kif3c | kinesin family member 3C | MGI:107979 |
| ENSMUSG00000006740 | Kif5b | kinesin family member 5B | MGI:1098268 |
| ENSMUSG00000026764 | Kif5c | kinesin family member 5C | MGI:1098269 |
| ENSMUSG00000026585 | Kifap3 | kinesin-associated protein 3 | MGI:107566 |
| ENSMUSG00000032036 | Kirrel3 | kin of IRRE like 3 (Drosophila) | MGI:1914953 |
| ENSMUSG00000021288 | Klc1 | kinesin light chain 1 | MGI:107978 |
| ENSMUSG00000024862 | Klc2 | kinesin light chain 2 | MGI:107953 |
| ENSMUSG00000066129 | Kndc1 | kinase non-catalytic C-lobe domain (KIND) containing 1 | MGI:1923734 |
| ENSMUSG00000030265 | Kras | v-Ki-ras2 Kirsten rat sarcoma viral oncogene homolog | MGI:96680 |
| ENSMUSG00000000600 | Krit1 | KRIT1, ankyrin repeat containing | MGI:1930618 |
| ENSMUSG00000075402 | Krt76 | keratin 76 | MGI:1924305 |
| ENSMUSG00000021843 | Ktn1 | kinectin 1 | MGI:109153 |
| ENSMUSG00000031391 | L1cam | L1 cell adhesion molecule | MGI:96721 |
| ENSMUSG00000032370 | Lactb | lactamase, beta | MGI:1933395 |
| ENSMUSG00000030842 | Lamtor1 | late endosomal/lysosomal adaptor, MAPK and MTOR activator 1 | MGI:1913758 |
| ENSMUSG00000087260 | Lamtor5 | late endosomal/lysosomal adaptor, MAPK and MTOR activator 5 | MGI:1915826 |
| ENSMUSG00000026000 | Lancl1 | LanC (bacterial lantibiotic synthetase component C)-like 1 | MGI:1336997 |
| ENSMUSG00000062190 | Lancl2 | LanC (bacterial lantibiotic synthetase component C)-like 2 | MGI:1919085 |
| ENSMUSG00000020585 | Laptm4a | lysosomal-associated protein transmembrane 4A | MGI:108017 |
| ENSMUSG00000037331 | Larp1 | La ribonucleoprotein domain family, member 1 | MGI:1890165 |
| ENSMUSG00000024493 | Lars | leucyl-tRNA synthetase | MGI:1913808 |
| ENSMUSG00000038366 | Lasp1 | LIM and SH3 protein 1 | MGI:109656 |
| ENSMUSG00000037353 | Letmd1 | LETM1 domain containing 1 | MGI:1915864 |
| ENSMUSG00000067242 | Lgi1 | leucine-rich repeat LGI family, member 1 | MGI:1861691 |
| ENSMUSG00000039252 | Lgi2 | leucine-rich repeat LGI family, member 2 | MGI:2180196 |
| ENSMUSG00000033595 | Lgi3 | leucine-rich repeat LGI family, member 3 | MGI:2182619 |
| ENSMUSG00000036560 | Lgi4 | leucine-rich repeat LGI family, member 4 | MGI:2180197 |
| ENSMUSG00000106379 | Lhfpl3 | lipoma HMGIC fusion partner-like 3 | MGI:1925076 |
| ENSMUSG00000042873 | Lhfpl4 | lipoma HMGIC fusion partner-like protein 4 | MGI:3057108 |
| ENSMUSG00000023022 | Lima1 | LIM domain and actin binding 1 | MGI:1920992 |
| ENSMUSG00000037736 | Limch1 | LIM and calponin homology domains 1 | MGI:1924819 |
| ENSMUSG00000040699 | Limd2 | LIM domain containing 2 | MGI:1915053 |
| ENSMUSG00000019906 | Lin7a | lin-7 homolog A (C. elegans) | MGI:2135609 |
| ENSMUSG00000003872 | Lin7b | lin-7 homolog B (C. elegans) | MGI:1330858 |
| ENSMUSG00000027162 | Lin7c | lin-7 homolog C (C. elegans) | MGI:1330839 |
| ENSMUSG00000051067 | Lingo3 | leucine rich repeat and Ig domain containing 3 | MGI:3609246 |
| ENSMUSG00000022802 | Lmln | leishmanolysin-like (metallopeptidase M8 family) | MGI:2444736 |
| ENSMUSG00000028063 | Lmna | lamin A | MGI:96794 |
| ENSMUSG00000024590 | Lmnb1 | lamin B1 | MGI:96795 |
| ENSMUSG00000062075 | Lmnb2 | lamin B2 | MGI:96796 |
| ENSMUSG00000033060 | Lmo7 | LIM domain only 7 | MGI:1353586 |
| ENSMUSG00000038970 | Lmtk2 | lemur tyrosine kinase 2 | MGI:3036247 |
| ENSMUSG00000062044 | Lmtk3 | lemur tyrosine kinase 3 | MGI:3039582 |
| ENSMUSG00000047866 | Lonp2 | lon peptidase 2, peroxisomal | MGI:1914137 |
| ENSMUSG00000026623 | Lpgat1 | lysophosphatidylglycerol acyltransferase 1 | MGI:2446186 |
| ENSMUSG00000040563 | Lppr2 | lipid phosphate phosphatase-related protein type 2 | MGI:2384575 |
| ENSMUSG00000035835 | Lppr3 | lipid phosphate phosphatase-related protein type 3 | MGI:2388640 |
| ENSMUSG00000044667 | Lppr4 | lipid phosphate phosphatase-related protein type 4 | MGI:106530 |
| ENSMUSG00000068015 | Lrch1 | leucine-rich repeats and calponin homology (CH) domain containing 1 | MGI:2443390 |
| ENSMUSG00000022801 | Lrch3 | leucine-rich repeats and calponin homology (CH) domain containing 3 | MGI:1917394 |
| ENSMUSG00000030600 | Lrfn1 | leucine rich repeat and fibronectin type III domain containing 1 | MGI:2136810 |
| ENSMUSG00000036957 | Lrfn3 | leucine rich repeat and fibronectin type III domain containing 3 | MGI:2442512 |
| ENSMUSG00000045045 | Lrfn4 | leucine rich repeat and fibronectin type III domain containing 4 | MGI:2385612 |
| ENSMUSG00000063052 | Lrrc40 | leucine rich repeat containing 40 | MGI:1914394 |
| ENSMUSG00000050587 | Lrrc4c | leucine rich repeat containing 4C | MGI:2442636 |
| ENSMUSG00000027286 | Lrrc57 | leucine rich repeat containing 57 | MGI:1913856 |
| ENSMUSG00000020869 | Lrrc59 | leucine rich repeat containing 59 | MGI:2138133 |
| ENSMUSG00000071073 | Lrrc73 | leucine rich repeat containing 73 | MGI:2684934 |
| ENSMUSG00000046417 | Lrrc75a | leucine rich repeat containing 75A | MGI:2682293 |
| ENSMUSG00000028176 | Lrrc7 | leucine rich repeat containing 7 | MGI:2676665 |
| ENSMUSG00000007476 | Lrrc8a | leucine rich repeat containing 8A | MGI:2652847 |
| ENSMUSG00000070639 | Lrrc8b | leucine rich repeat containing 8 family, member B | MGI:2141353 |
| ENSMUSG00000054720 | Lrrc8c | leucine rich repeat containing 8 family, member C | MGI:2140839 |
| ENSMUSG00000046079 | Lrrc8d | leucine rich repeat containing 8D | MGI:1922368 |
| ENSMUSG00000060780 | Lrrtm1 | leucine rich repeat transmembrane neuronal 1 | MGI:2389173 |
| ENSMUSG00000071862 | Lrrtm2 | leucine rich repeat transmembrane neuronal 2 | MGI:2389174 |
| ENSMUSG00000042846 | Lrrtm3 | leucine rich repeat transmembrane neuronal 3 | MGI:2389177 |
| ENSMUSG00000052581 | Lrrtm4 | leucine rich repeat transmembrane neuronal 4 | MGI:2389180 |
| ENSMUSG00000055003 | Lrtm2 | leucine-rich repeats and transmembrane domains 2 | MGI:2141485 |
| ENSMUSG00000061080 | Lsamp | limbic system-associated membrane protein | MGI:1261760 |
| ENSMUSG00000044847 | Lsm11 | U7 snRNP-specific Sm-like protein LSM11 | MGI:1919540 |
| ENSMUSG00000018819 | Lsp1 | lymphocyte specific 1 | MGI:96832 |
| ENSMUSG00000001089 | Luzp1 | leucine zipper protein 1 | MGI:107629 |
| ENSMUSG00000063297 | Luzp2 | leucine zipper protein 2 | MGI:1889615 |
| ENSMUSG00000043807 | Ly6g5b | lymphocyte antigen 6 complex, locus G5B | MGI:2385809 |
| ENSMUSG00000042228 | Lyn | Yamaguchi sarcoma viral (v-yes-1) oncogene homolog | MGI:96892 |
| ENSMUSG00000026344 | Lypd1 | Ly6/Plaur domain containing 1 | MGI:1919835 |
| ENSMUSG00000036306 | Lzts1 | leucine zipper, putative tumor suppressor 1 | MGI:2684762 |
| ENSMUSG00000037703 | Lzts3 | leucine zipper, putative tumor suppressor family member 3 | MGI:2656976 |
| ENSMUSG00000025151 | Maged1 | melanoma antigen, family D, 1 | MGI:1930187 |
| ENSMUSG00000045095 | Magi1 | membrane associated guanylate kinase, WW and PDZ domain containing 1 | MGI:1203522 |
| ENSMUSG00000040003 | Magi2 | membrane associated guanylate kinase, WW and PDZ domain containing 2 | MGI:1354953 |
| ENSMUSG00000052539 | Magi3 | membrane associated guanylate kinase, WW and PDZ domain containing 3 | MGI:1923484 |
| ENSMUSG00000024479 | Mal2 | mal, T cell differentiation protein 2 | MGI:2146021 |
| ENSMUSG00000032295 | Man2c1 | mannosidase, alpha, class 2C, member 1 | MGI:1920994 |
| ENSMUSG00000027254 | Map1a | microtubule-associated protein 1 A | MGI:1306776 |
| ENSMUSG00000052727 | Map1b | microtubule-associated protein 1B | MGI:1306778 |
| ENSMUSG00000027602 | Map1lc3a | microtubule-associated protein 1 light chain 3 alpha | MGI:1915661 |
| ENSMUSG00000031812 | Map1lc3b | microtubule-associated protein 1 light chain 3 beta | MGI:1914693 |
| ENSMUSG00000019261 | Map1s | microtubule-associated protein 1S | MGI:2443304 |
| ENSMUSG00000004936 | Map2k1 | mitogen-activated protein kinase kinase 1 | MGI:1346866 |
| ENSMUSG00000033352 | Map2k4 | mitogen-activated protein kinase kinase 4 | MGI:1346869 |
| ENSMUSG00000002948 | Map2k7 | mitogen-activated protein kinase kinase 7 | MGI:1346871 |
| ENSMUSG00000026074 | Map4k4 | mitogen-activated protein kinase kinase kinase kinase 4 | MGI:1349394 |
| ENSMUSG00000041205 | Map6d1 | MAP6 domain containing 1 | MGI:3607784 |
| ENSMUSG00000055407 | Map6 | microtubule-associated protein 6 | MGI:1201690 |
| ENSMUSG00000028849 | Map7d1 | MAP7 domain containing 1 | MGI:2384297 |
| ENSMUSG00000041020 | Map7d2 | MAP7 domain containing 2 | MGI:1917474 |
| ENSMUSG00000019996 | Map7 | microtubule-associated protein 7 | MGI:1328328 |
| ENSMUSG00000033900 | Map9 | microtubule-associated protein 9 | MGI:2442208 |
| ENSMUSG00000046709 | Mapk10 | mitogen-activated protein kinase 10 | MGI:1346863 |
| ENSMUSG00000063358 | Mapk1 | mitogen-activated protein kinase 1 | MGI:1346858 |
| ENSMUSG00000063065 | Mapk3 | mitogen-activated protein kinase 3 | MGI:1346859 |
| ENSMUSG00000024163 | Mapk8ip3 | mitogen-activated protein kinase 8 interacting protein 3 | MGI:1353598 |
| ENSMUSG00000027479 | Mapre1 | microtubule-associated protein, RP/EB family, member 1 | MGI:891995 |
| ENSMUSG00000024277 | Mapre2 | microtubule-associated protein, RP/EB family, member 2 | MGI:106271 |
| ENSMUSG00000029166 | Mapre3 | microtubule-associated protein, RP/EB family, member 3 | MGI:2140967 |
| ENSMUSG00000018411 | Mapt | microtubule-associated protein tau | MGI:97180 |
| ENSMUSG00000073481 | Marc2 | mitochondrial amidoxime reducing component 2 | MGI:1914497 |
| ENSMUSG00000023307 | March5 | membrane-associated ring finger (C3HC4) 5 | MGI:1915207 |
| ENSMUSG00000026620 | Mark1 | MAP/microtubule affinity regulating kinase 1 | MGI:2664902 |
| ENSMUSG00000024969 | Mark2 | MAP/microtubule affinity regulating kinase 2 | MGI:99638 |
| ENSMUSG00000007411 | Mark3 | MAP/microtubule affinity regulating kinase 3 | MGI:1341865 |
| ENSMUSG00000030397 | Mark4 | MAP/microtubule affinity regulating kinase 4 | MGI:1920955 |
| ENSMUSG00000053693 | Mast1 | microtubule associated serine/threonine kinase 1 | MGI:1861901 |
| ENSMUSG00000031833 | Mast3 | microtubule associated serine/threonine kinase 3 | MGI:2683541 |
| ENSMUSG00000034751 | Mast4 | microtubule associated serine/threonine kinase family member 4 | MGI:1918885 |
| ENSMUSG00000004933 | Matk | megakaryocyte-associated tyrosine kinase | MGI:99259 |
| ENSMUSG00000016995 | Matn4 | matrilin 4 | MGI:1328314 |
| ENSMUSG00000037236 | Matr3 | matrin 3 | MGI:1298379 |
| ENSMUSG00000041607 | Mbp | myelin basic protein | MGI:96925 |
| ENSMUSG00000027709 | Mccc1 | methylcrotonoyl-Coenzyme A carboxylase 1 (alpha) | MGI:1919289 |
| ENSMUSG00000021646 | Mccc2 | methylcrotonoyl-Coenzyme A carboxylase 2 (beta) | MGI:1925288 |
| ENSMUSG00000031442 | Mcf2l | mcf.2 transforming sequence-like | MGI:103263 |
| ENSMUSG00000009647 | Mcu | mitochondrial calcium uniporter | MGI:3026965 |
| ENSMUSG00000043557 | Mdga1 | MAM domain containing glycosylphosphatidylinositol anchor 1 | MGI:1922012 |
| ENSMUSG00000034912 | Mdga2 | MAM domain containing glycosylphosphatidylinositol anchor 2 | MGI:2444706 |
| ENSMUSG00000018076 | Med13l | mediator complex subunit 13-like | MGI:2670178 |
| ENSMUSG00000070462 | Mesdc1 | mesoderm development candidate 1 | MGI:1891420 |
| ENSMUSG00000026150 | Mff | mitochondrial fission factor | MGI:1922984 |
| ENSMUSG00000030605 | Mfge8 | milk fat globule-EGF factor 8 protein | MGI:102768 |
| ENSMUSG00000026688 | Mgst3 | microsomal glutathione S-transferase 3 | MGI:1913697 |
| ENSMUSG00000003178 | Mical3 | microtubule associated monooxygenase, calponin and LIM domain containing 3 | MGI:2442733 |
| ENSMUSG00000020111 | Micu1 | mitochondrial calcium uptake 1 | MGI:2384909 |
| ENSMUSG00000039478 | Micu3 | mitochondrial calcium uptake family, member 3 | MGI:1925756 |
| ENSMUSG00000020827 | Mink1 | misshapen-like kinase 1 (zebrafish) | MGI:1355329 |
| ENSMUSG00000042447 | Mios | missing oocyte, meiosis regulator, homolog (Drosophila) | MGI:2182066 |
| ENSMUSG00000035805 | Mlc1 | megalencephalic leukoencephalopathy with subcortical cysts 1 homolog (human) | MGI:2157910 |
| ENSMUSG00000048578 | Mlec | malectin | MGI:1924015 |
| ENSMUSG00000030120 | Mlf2 | myeloid leukemia factor 2 | MGI:1353554 |
| ENSMUSG00000032355 | Mlip | muscular LMNA-interacting protein | MGI:1916892 |
| ENSMUSG00000068036 | Mllt4 | myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila) translocated to, 4 | MGI:1314653 |
| ENSMUSG00000074064 | Mlycd | malonyl-CoA decarboxylase | MGI:1928485 |
| ENSMUSG00000037022 | Mmaa | methylmalonic aciduria (cobalamin deficiency) type A | MGI:1923805 |
| ENSMUSG00000061273 | Mmgt1 | membrane magnesium transporter 1 | MGI:2384305 |
| ENSMUSG00000029436 | Mmp17 | matrix metallopeptidase 17 | MGI:1346076 |
| ENSMUSG00000025979 | Mob4 | MOB family member 4, phocein | MGI:104899 |
| ENSMUSG00000032517 | Mobp | myelin-associated oligodendrocytic basic protein | MGI:108511 |
| ENSMUSG00000076439 | Mog | myelin oligodendrocyte glycoprotein | MGI:97435 |
| ENSMUSG00000030036 | Mogs | mannosyl-oligosaccharide glucosidase | MGI:1929872 |
| ENSMUSG00000032583 | Mon1a | MON1 homolog A (yeast) | MGI:1920075 |
| ENSMUSG00000078908 | Mon1b | MON1 homolog b (yeast) | MGI:1923231 |
| ENSMUSG00000026568 | Mpc2 | mitochondrial pyruvate carrier 2 | MGI:1917706 |
| ENSMUSG00000028402 | Mpdz | multiple PDZ domain protein | MGI:1343489 |
| ENSMUSG00000031402 | Mpp1 | membrane protein, palmitoylated | MGI:105941 |
| ENSMUSG00000017314 | Mpp2 | membrane protein, palmitoylated 2 (MAGUK p55 subfamily member 2) | MGI:1858257 |
| ENSMUSG00000052373 | Mpp3 | membrane protein, palmitoylated 3 (MAGUK p55 subfamily member 3) | MGI:1328354 |
| ENSMUSG00000021112 | Mpp5 | membrane protein, palmitoylated 5 (MAGUK p55 subfamily member 5) | MGI:1927339 |
| ENSMUSG00000005417 | Mprip | myosin phosphatase Rho interacting protein | MGI:1349438 |
| ENSMUSG00000032470 | Mras | muscle and microspikes RAS | MGI:1100856 |
| ENSMUSG00000024902 | Mrpl11 | mitochondrial ribosomal protein L11 | MGI:2137215 |
| ENSMUSG00000039640 | Mrpl12 | mitochondrial ribosomal protein L12 | MGI:1926273 |
| ENSMUSG00000023939 | Mrpl14 | mitochondrial ribosomal protein L14 | MGI:1333864 |
| ENSMUSG00000020514 | Mrpl22 | mitochondrial ribosomal protein L22 | MGI:1333794 |
| ENSMUSG00000037772 | Mrpl23 | mitochondrial ribosomal protein L23 | MGI:1196612 |
| ENSMUSG00000022706 | Mrpl40 | mitochondrial ribosomal protein L40 | MGI:1332635 |
| ENSMUSG00000025208 | Mrpl43 | mitochondrial ribosomal protein L43 | MGI:2137229 |
| ENSMUSG00000044018 | Mrpl50 | mitochondrial ribosomal protein L50 | MGI:107329 |
| ENSMUSG00000030037 | Mrpl53 | mitochondrial ribosomal protein L53 | MGI:1915749 |
| ENSMUSG00000034729 | Mrps10 | mitochondrial ribosomal protein S10 | MGI:1928139 |
| ENSMUSG00000054312 | Mrps21 | mitochondrial ribosomal protein S21 | MGI:1913542 |
| ENSMUSG00000032459 | Mrps22 | mitochondrial ribosomal protein S22 | MGI:1928137 |
| ENSMUSG00000023723 | Mrps23 | mitochondrial ribosomal protein S23 | MGI:1928138 |
| ENSMUSG00000041632 | Mrps27 | mitochondrial ribosomal protein S27 | MGI:1919064 |
| ENSMUSG00000040269 | Mrps28 | mitochondrial ribosomal protein S28 | MGI:1913480 |
| ENSMUSG00000035772 | Mrps2 | mitochondrial ribosomal protein S2 | MGI:2153089 |
| ENSMUSG00000021731 | Mrps30 | mitochondrial ribosomal protein S30 | MGI:1926237 |
| ENSMUSG00000061474 | Mrps36 | mitochondrial ribosomal protein S36 | MGI:1913378 |
| ENSMUSG00000026887 | Mrrf | mitochondrial ribosome recycling factor | MGI:1915121 |
| ENSMUSG00000021339 | Mrs2 | MRS2 magnesium homeostasis factor homolog (S. cerevisiae) | MGI:2685748 |
| ENSMUSG00000069769 | Msi2 | musashi RNA-binding protein 2 | MGI:1923876 |
| ENSMUSG00000064356 | mt-Atp8 | mitochondrially encoded ATP synthase 8 | MGI:99926 |
| ENSMUSG00000024012 | Mtch1 | mitochondrial carrier homolog 1 (C. elegans) | MGI:1929261 |
| ENSMUSG00000027282 | Mtch2 | mitochondrial carrier homolog 2 (C. elegans) | MGI:1929260 |
| ENSMUSG00000064354 | mt-Co2 | mitochondrially encoded cytochrome c oxidase II | MGI:102503 |
| ENSMUSG00000022255 | Mtdh | metadherin | MGI:1914404 |
| ENSMUSG00000046671 | Mtfr1l | mitochondrial fission regulator 1-like | MGI:1924074 |
| ENSMUSG00000021048 | Mthfd1 | methylenetetrahydrofolate dehydrogenase (NADP+ dependent), methenyltetrahydrofolate cyclohydrolase, formyltetrahydrofolate synthase | MGI:1342005 |
| ENSMUSG00000015214 | Mtmr1 | myotubularin related protein 1 | MGI:1858271 |
| ENSMUSG00000031918 | Mtmr2 | myotubularin related protein 2 | MGI:1924366 |
| ENSMUSG00000028991 | Mtor | mechanistic target of rapamycin (serine/threonine kinase) | MGI:1928394 |
| ENSMUSG00000024234 | Mtpap | mitochondrial poly(A) polymerase | MGI:1914690 |
| ENSMUSG00000029840 | Mtpn | myotrophin | MGI:99445 |
| ENSMUSG00000033763 | Mtss1l | metastasis suppressor 1-like | MGI:3039591 |
| ENSMUSG00000029651 | Mtus2 | microtubule associated tumor suppressor candidate 2 | MGI:1915388 |
| ENSMUSG00000064068 | Mtx1 | metaxin 1 | MGI:103025 |
| ENSMUSG00000027099 | Mtx2 | metaxin 2 | MGI:1859652 |
| ENSMUSG00000021704 | Mtx3 | metaxin 3 | MGI:2686040 |
| ENSMUSG00000038740 | Mvb12b | multivesicular body subunit 12B | MGI:1919793 |
| ENSMUSG00000068566 | Myadm | myeloid-associated differentiation marker | MGI:1355332 |
| ENSMUSG00000033004 | Mycbp2 | MYC binding protein 2 | MGI:2179432 |
| ENSMUSG00000020900 | Myh10 | myosin, heavy polypeptide 10, non-muscle | MGI:1930780 |
| ENSMUSG00000018830 | Myh11 | myosin, heavy polypeptide 11, smooth muscle | MGI:102643 |
| ENSMUSG00000030739 | Myh14 | myosin, heavy polypeptide 14 | MGI:1919210 |
| ENSMUSG00000022443 | Myh9 | myosin, heavy polypeptide 9, non-muscle | MGI:107717 |
| ENSMUSG00000034868 | Myl12b | myosin, light chain 12B, regulatory | MGI:107494 |
| ENSMUSG00000061816 | Myl1 | myosin, light polypeptide 1 | MGI:97269 |
| ENSMUSG00000061086 | Myl4 | myosin, light polypeptide 4 | MGI:97267 |
| ENSMUSG00000039824 | Myl6b | myosin, light polypeptide 6B | MGI:1917789 |
| ENSMUSG00000090841 | Myl6 | myosin, light polypeptide 6, alkali, smooth muscle and non-muscle | MGI:109318 |
| ENSMUSG00000022836 | Mylk | myosin, light polypeptide kinase | MGI:894806 |
| ENSMUSG00000000631 | Myo18a | myosin XVIIIA | MGI:2667185 |
| ENSMUSG00000018417 | Myo1b | myosin IB | MGI:107752 |
| ENSMUSG00000017774 | Myo1c | myosin IC | MGI:106612 |
| ENSMUSG00000035441 | Myo1d | myosin ID | MGI:107728 |
| ENSMUSG00000032220 | Myo1e | myosin IE | MGI:106621 |
| ENSMUSG00000024300 | Myo1f | myosin IF | MGI:107711 |
| ENSMUSG00000034593 | Myo5a | myosin VA | MGI:105976 |
| ENSMUSG00000033577 | Myo6 | myosin VI | MGI:104785 |
| ENSMUSG00000039585 | Myo9a | myosin IXa | MGI:107735 |
| ENSMUSG00000022671 | Mzt2 | mitotic spindle organizing protein 2 | MGI:1922845 |
| ENSMUSG00000058799 | Nap1l1 | nucleosome assembly protein 1-like 1 | MGI:1855693 |
| ENSMUSG00000059119 | Nap1l4 | nucleosome assembly protein 1-like 4 | MGI:1316687 |
| ENSMUSG00000035285 | Nat14 | N-acetyltransferase 14 | MGI:3039561 |
| ENSMUSG00000009418 | Nav1 | neuron navigator 1 | MGI:2183683 |
| ENSMUSG00000002341 | Ncan | neurocan | MGI:104694 |
| ENSMUSG00000027002 | Nckap1 | NCK-associated protein 1 | MGI:1355333 |
| ENSMUSG00000032598 | Nckipsd | NCK interacting protein with SH3 domain | MGI:1931834 |
| ENSMUSG00000020238 | Ncln | nicalin homolog (zebrafish) | MGI:1926081 |
| ENSMUSG00000026234 | Ncl | nucleolin | MGI:97286 |
| ENSMUSG00000062661 | Ncs1 | neuronal calcium sensor 1 | MGI:109166 |
| ENSMUSG00000018736 | Ndel1 | nuclear distribution gene E-like homolog 1 (A. nidulans) | MGI:1932915 |
| ENSMUSG00000020022 | Ndufa12 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 12 | MGI:1913664 |
| ENSMUSG00000036199 | Ndufa13 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 13 | MGI:1914434 |
| ENSMUSG00000016427 | Ndufa1 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 1 | MGI:1929511 |
| ENSMUSG00000014294 | Ndufa2 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 2 | MGI:1343103 |
| ENSMUSG00000029632 | Ndufa4 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 4 | MGI:107686 |
| ENSMUSG00000022450 | Ndufa6 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 6 (B14) | MGI:1914380 |
| ENSMUSG00000041881 | Ndufa7 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 7 (B14.5a) | MGI:1913666 |
| ENSMUSG00000026895 | Ndufa8 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 8 | MGI:1915625 |
| ENSMUSG00000000399 | Ndufa9 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 9 | MGI:1913358 |
| ENSMUSG00000070283 | Ndufaf3 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, assembly factor 3 | MGI:1913956 |
| ENSMUSG00000028261 | Ndufaf4 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, assembly factor 4 | MGI:1915743 |
| ENSMUSG00000050323 | Ndufaf6 | NADH dehydrogenase (ubiquinone) complex I, assembly factor 6 | MGI:1924197 |
| ENSMUSG00000040048 | Ndufb10 | NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 10 | MGI:1915592 |
| ENSMUSG00000031059 | Ndufb11 | NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 11 | MGI:1349919 |
| ENSMUSG00000002416 | Ndufb2 | NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 2 | MGI:1915448 |
| ENSMUSG00000026032 | Ndufb3 | NADH dehydrogenase (ubiquinone) 1 beta subcomplex 3 | MGI:1913745 |
| ENSMUSG00000022820 | Ndufb4 | NADH dehydrogenase (ubiquinone) 1 beta subcomplex 4 | MGI:1915444 |
| ENSMUSG00000027673 | Ndufb5 | NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 5 | MGI:1913296 |
| ENSMUSG00000071014 | Ndufb6 | NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 6 | MGI:2684983 |
| ENSMUSG00000033938 | Ndufb7 | NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 7 | MGI:1914166 |
| ENSMUSG00000025204 | Ndufb8 | NADH dehydrogenase (ubiquinone) 1 beta subcomplex 8 | MGI:1914514 |
| ENSMUSG00000022354 | Ndufb9 | NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 9 | MGI:1913468 |
| ENSMUSG00000030647 | Ndufc2 | NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 2 | MGI:1344370 |
| ENSMUSG00000005510 | Ndufs3 | NADH dehydrogenase (ubiquinone) Fe-S protein 3 | MGI:1915599 |
| ENSMUSG00000021606 | Ndufs6 | NADH dehydrogenase (ubiquinone) Fe-S protein 6 | MGI:107932 |
| ENSMUSG00000020153 | Ndufs7 | NADH dehydrogenase (ubiquinone) Fe-S protein 7 | MGI:1922656 |
| ENSMUSG00000024099 | Ndufv2 | NADH dehydrogenase (ubiquinone) flavoprotein 2 | MGI:1920150 |
| ENSMUSG00000024038 | Ndufv3 | NADH dehydrogenase (ubiquinone) flavoprotein 3 | MGI:1890894 |
| ENSMUSG00000053702 | Nebl | nebulette | MGI:1921353 |
| ENSMUSG00000031837 | Necab2 | N-terminal EF-hand calcium binding protein 2 | MGI:2152211 |
| ENSMUSG00000030327 | Necap1 | NECAP endocytosis associated 1 | MGI:1914852 |
| ENSMUSG00000024589 | Nedd4l | neural precursor cell expressed, developmentally down-regulated gene 4-like | MGI:1933754 |
| ENSMUSG00000010376 | Nedd8 | neural precursor cell expressed, developmentally down-regulated gene 8 | MGI:97301 |
| ENSMUSG00000020396 | Nefh | neurofilament, heavy polypeptide | MGI:97309 |
| ENSMUSG00000022055 | Nefl | neurofilament, light polypeptide | MGI:97313 |
| ENSMUSG00000022054 | Nefm | neurofilament, medium polypeptide | MGI:97314 |
| ENSMUSG00000040037 | Negr1 | neuronal growth regulator 1 | MGI:2444846 |
| ENSMUSG00000050321 | Neto1 | neuropilin (NRP) and tolloid (TLL)-like 1 | MGI:2180216 |
| ENSMUSG00000036902 | Neto2 | neuropilin (NRP) and tolloid (TLL)-like 2 | MGI:1921763 |
| ENSMUSG00000034000 | Neu4 | sialidase 4 | MGI:2661364 |
| ENSMUSG00000039103 | Nexn | nexilin | MGI:1916060 |
| ENSMUSG00000020716 | Nf1 | neurofibromatosis 1 | MGI:97306 |
| ENSMUSG00000009073 | Nf2 | neurofibromatosis 2 | MGI:97307 |
| ENSMUSG00000027618 | Nfs1 | nitrogen fixation gene 1 (S. cerevisiae) | MGI:1316706 |
| ENSMUSG00000026259 | Ngef | neuronal guanine nucleotide exchange factor | MGI:1858414 |
| ENSMUSG00000079481 | Nhsl2 | NHS-like 2 | MGI:3645090 |
| ENSMUSG00000032606 | Nicn1 | nicolin 1 | MGI:1913507 |
| ENSMUSG00000047037 | Nipa1 | non imprinted in Prader-Willi/Angelman syndrome 1 homolog (human) | MGI:2442058 |
| ENSMUSG00000034285 | Nipsnap1 | 4-nitrophenylphosphatase domain and non-neuronal SNAP25-like protein homolog 1 (C. elegans) | MGI:1278344 |
| ENSMUSG00000021910 | Nisch | nischarin | MGI:1928323 |
| ENSMUSG00000078532 | Nkain1 | Na+/K+ transporting ATPase interacting 1 | MGI:1914399 |
| ENSMUSG00000021772 | Nkiras1 | NFKB inhibitor interacting Ras-like protein 1 | MGI:1916971 |
| ENSMUSG00000017837 | Nkiras2 | NFKB inhibitor interacting Ras-like protein 2 | MGI:1919216 |
| ENSMUSG00000051790 | Nlgn2 | neuroligin 2 | MGI:2681835 |
| ENSMUSG00000031302 | Nlgn3 | neuroligin 3 | MGI:2444609 |
| ENSMUSG00000073435 | Nme3 | NME/NM23 nucleoside diphosphate kinase 3 | MGI:1930182 |
| ENSMUSG00000031311 | Nono | non-POU-domain-containing, octamer binding protein | MGI:1855692 |
| ENSMUSG00000021047 | Nova1 | neuro-oncological ventral antigen 1 | MGI:104297 |
| ENSMUSG00000057113 | Npm1 | nucleophosmin 1 | MGI:106184 |
| ENSMUSG00000056209 | Npm3 | nucleoplasmin 3 | MGI:894653 |
| ENSMUSG00000025582 | Nptx1 | neuronal pentraxin 1 | MGI:107811 |
| ENSMUSG00000060275 | Nrg2 | neuregulin 2 | MGI:1098246 |
| ENSMUSG00000044287 | Nrn1l | neuritin 1-like | MGI:2443642 |
| ENSMUSG00000039114 | Nrn1 | neuritin 1 | MGI:1915654 |
| ENSMUSG00000066392 | Nrxn3 | neurexin III | MGI:1096389 |
| ENSMUSG00000029126 | Nsg1 | neuron specific gene family member 1 | MGI:109149 |
| ENSMUSG00000020297 | Nsg2 | neuron specific gene family member 2 | MGI:1202070 |
| ENSMUSG00000006476 | Nsmf | NMDA receptor synaptonuclear signaling and neuronal migration factor | MGI:1861755 |
| ENSMUSG00000032420 | Nt5e | 5' nucleotidase, ecto | MGI:99782 |
| ENSMUSG00000059974 | Ntm | neurotrimin | MGI:2446259 |
| ENSMUSG00000059857 | Ntng1 | netrin G1 | MGI:1934028 |
| ENSMUSG00000035513 | Ntng2 | netrin G2 | MGI:2159341 |
| ENSMUSG00000045211 | Nudt18 | nudix (nucleoside diphosphate linked moiety X)-type motif 18 | MGI:2385853 |
| ENSMUSG00000031754 | Nudt21 | nudix (nucleoside diphosphate linked moiety X)-type motif 21 | MGI:1915469 |
| ENSMUSG00000063160 | Numbl | numb-like | MGI:894702 |
| ENSMUSG00000021224 | Numb | numb gene homolog (Drosophila) | MGI:107423 |
| ENSMUSG00000036992 | Nxt1 | NTF2-related export protein 1 | MGI:1929619 |
| ENSMUSG00000045348 | Nyap1 | neuronal tyrosine-phosphorylated phosphoinositide 3-kinase adaptor 1 | MGI:2443880 |
| ENSMUSG00000054976 | Nyap2 | neuronal tyrosine-phophorylated phosphoinositide 3-kinase adaptor 2 | MGI:2443135 |
| ENSMUSG00000029152 | Ociad1 | OCIA domain containing 1 | MGI:1915345 |
| ENSMUSG00000021913 | Ogdhl | oxoglutarate dehydrogenase-like | MGI:3616088 |
| ENSMUSG00000020456 | Ogdh | oxoglutarate (alpha-ketoglutarate) dehydrogenase (lipoamide) | MGI:1098267 |
| ENSMUSG00000034160 | Ogt | O-linked N-acetylglucosamine (GlcNAc) transferase (UDP-N-acetylglucosamine:polypeptide-N-acetylglucosaminyl transferase) | MGI:1339639 |
| ENSMUSG00000026833 | Olfm1 | olfactomedin 1 | MGI:1860437 |
| ENSMUSG00000032172 | Olfm2 | olfactomedin 2 | MGI:3045350 |
| ENSMUSG00000027965 | Olfm3 | olfactomedin 3 | MGI:2387329 |
| ENSMUSG00000049612 | Omg | oligodendrocyte myelin glycoprotein | MGI:106586 |
| ENSMUSG00000052214 | Opa3 | optic atrophy 3 | MGI:2686271 |
| ENSMUSG00000062257 | Opcml | opioid binding protein/cell adhesion molecule-like | MGI:97397 |
| ENSMUSG00000044252 | Osbpl1a | oxysterol binding protein-like 1A | MGI:1927551 |
| ENSMUSG00000024767 | Otub1 | OTU domain, ubiquitin aldehyde binding 1 | MGI:2147616 |
| ENSMUSG00000025364 | Pa2g4 | proliferation-associated 2G4 | MGI:894684 |
| ENSMUSG00000022283 | Pabpc1 | poly(A) binding protein, cytoplasmic 1 | MGI:1349722 |
| ENSMUSG00000024855 | Pacs1 | phosphofurin acidic cluster sorting protein 1 | MGI:1277113 |
| ENSMUSG00000027257 | Pacsin3 | protein kinase C and casein kinase substrate in neurons 3 | MGI:1891410 |
| ENSMUSG00000020745 | Pafah1b1 | platelet-activating factor acetylhydrolase, isoform 1b, subunit 1 | MGI:109520 |
| ENSMUSG00000003131 | Pafah1b2 | platelet-activating factor acetylhydrolase, isoform 1b, subunit 2 | MGI:108415 |
| ENSMUSG00000027508 | Pag1 | phosphoprotein associated with glycosphingolipid microdomains 1 | MGI:2443160 |
| ENSMUSG00000029247 | Paics | phosphoribosylaminoimidazole carboxylase, phosphoribosylaminoribosylaminoimidazole, succinocarboxamide synthetase | MGI:1914304 |
| ENSMUSG00000030774 | Pak1 | p21 protein (Cdc42/Rac)-activated kinase 1 | MGI:1339975 |
| ENSMUSG00000030602 | Pak4 | p21 protein (Cdc42/Rac)-activated kinase 4 | MGI:1917834 |
| ENSMUSG00000074923 | Pak6 | p21 protein (Cdc42/Rac)-activated kinase 6 | MGI:2679420 |
| ENSMUSG00000039913 | Pak7 | p21 protein (Cdc42/Rac)-activated kinase 7 | MGI:1920334 |
| ENSMUSG00000089945 | Pakap | paralemmin A kinase anchor protein | MGI:5141924 |
| ENSMUSG00000090053 | Palm2 | paralemmin 2 | MGI:1934601 |
| ENSMUSG00000047986 | Palm3 | paralemmin 3 | MGI:1921587 |
| ENSMUSG00000035863 | Palm | paralemmin | MGI:1261814 |
| ENSMUSG00000014301 | Pam16 | presequence translocase-asssociated motor 16 homolog (S. cerevisiae) | MGI:1913699 |
| ENSMUSG00000026335 | Pam | peptidylglycine alpha-amidating monooxygenase | MGI:97475 |
| ENSMUSG00000058441 | Panx2 | pannexin 2 | MGI:1890615 |
| ENSMUSG00000027233 | Patl2 | protein associated with topoisomerase II homolog 2 (yeast) | MGI:1914828 |
| ENSMUSG00000051695 | Pcbp1 | poly(rC) binding protein 1 | MGI:1345635 |
| ENSMUSG00000056851 | Pcbp2 | poly(rC) binding protein 2 | MGI:108202 |
| ENSMUSG00000001120 | Pcbp3 | poly(rC) binding protein 3 | MGI:1890470 |
| ENSMUSG00000049100 | Pcdh10 | protocadherin 10 | MGI:1338042 |
| ENSMUSG00000102697 | Pcdhac2 | protocadherin alpha subfamily C, 2 | MGI:1891443 |
| ENSMUSG00000051242 | Pcdhb9 | protocadherin beta 9 | MGI:2136744 |
| ENSMUSG00000102543 | Pcdhgc5 | protocadherin gamma subfamily C, 5 | MGI:1935205 |
| ENSMUSG00000061601 | Pclo | piccolo (presynaptic cytomatrix protein) | MGI:1349390 |
| ENSMUSG00000031592 | Pcm1 | pericentriolar material 1 | MGI:1277958 |
| ENSMUSG00000019795 | Pcmt1 | protein-L-isoaspartate (D-aspartate) O-methyltransferase 1 | MGI:97502 |
| ENSMUSG00000054874 | Pcnxl3 | pecanex-like 3 (Drosophila) | MGI:1861733 |
| ENSMUSG00000021140 | Pcnx | pecanex homolog (Drosophila) | MGI:1891924 |
| ENSMUSG00000025137 | Pcyt2 | phosphate cytidylyltransferase 2, ethanolamine | MGI:1915921 |
| ENSMUSG00000029623 | Pdap1 | PDGFA associated protein 1 | MGI:2448536 |
| ENSMUSG00000027835 | Pdcd10 | programmed cell death 10 | MGI:1928396 |
| ENSMUSG00000021576 | Pdcd6 | programmed cell death 6 | MGI:109283 |
| ENSMUSG00000022489 | Pde1b | phosphodiesterase 1B, Ca2+-calmodulin dependent | MGI:97523 |
| ENSMUSG00000030653 | Pde2a | phosphodiesterase 2A, cGMP-stimulated | MGI:2446107 |
| ENSMUSG00000028525 | Pde4b | phosphodiesterase 4B, cAMP specific | MGI:99557 |
| ENSMUSG00000021699 | Pde4d | phosphodiesterase 4D, cAMP specific | MGI:99555 |
| ENSMUSG00000025584 | Pde8a | phosphodiesterase 8A | MGI:1277116 |
| ENSMUSG00000021684 | Pde8b | phosphodiesterase 8B | MGI:2443999 |
| ENSMUSG00000041119 | Pde9a | phosphodiesterase 9A | MGI:1277179 |
| ENSMUSG00000031299 | Pdha1 | pyruvate dehydrogenase E1 alpha 1 | MGI:97532 |
| ENSMUSG00000021748 | Pdhb | pyruvate dehydrogenase (lipoamide) beta | MGI:1915513 |
| ENSMUSG00000010914 | Pdhx | pyruvate dehydrogenase complex, component X | MGI:1351627 |
| ENSMUSG00000050890 | Pdik1l | PDLIM1 interacting kinase 1 like | MGI:2385213 |
| ENSMUSG00000035232 | Pdk3 | pyruvate dehydrogenase kinase, isoenzyme 3 | MGI:2384308 |
| ENSMUSG00000024122 | Pdpk1 | 3-phosphoinositide dependent protein kinase 1 | MGI:1338068 |
| ENSMUSG00000015668 | Pdzd11 | PDZ domain containing 11 | MGI:1919871 |
| ENSMUSG00000002006 | Pdzd4 | PDZ domain containing 4 | MGI:2443483 |
| ENSMUSG00000013698 | Pea15a | phosphoprotein enriched in astrocytes 15A | MGI:104799 |
| ENSMUSG00000074305 | Peak1 | pseudopodium-enriched atypical kinase 1 | MGI:2442366 |
| ENSMUSG00000042275 | Pelo | pelota homolog (Drosophila) | MGI:2145154 |
| ENSMUSG00000035395 | Pet2 | plasmacytoma expressed transcript 2 | MGI:101758 |
| ENSMUSG00000028102 | Pex11b | peroxisomal biogenesis factor 11 beta | MGI:1338882 |
| ENSMUSG00000018733 | Pex12 | peroxisomal biogenesis factor 12 | MGI:2144177 |
| ENSMUSG00000028975 | Pex14 | peroxisomal biogenesis factor 14 | MGI:1927868 |
| ENSMUSG00000019809 | Pex3 | peroxisomal biogenesis factor 3 | MGI:1929646 |
| ENSMUSG00000005069 | Pex5 | peroxisomal biogenesis factor 5 | MGI:1098808 |
| ENSMUSG00000020277 | Pfkl | phosphofructokinase, liver, B-type | MGI:97547 |
| ENSMUSG00000033065 | Pfkm | phosphofructokinase, muscle | MGI:97548 |
| ENSMUSG00000021196 | Pfkp | phosphofructokinase, platelet | MGI:1891833 |
| ENSMUSG00000011752 | Pgam1 | phosphoglycerate mutase 1 | MGI:97552 |
| ENSMUSG00000029500 | Pgam5 | phosphoglycerate mutase family member 5 | MGI:1919792 |
| ENSMUSG00000073678 | Pgap1 | post-GPI attachment to proteins 1 | MGI:2443342 |
| ENSMUSG00000017715 | Pgs1 | phosphatidylglycerophosphate synthase 1 | MGI:1921701 |
| ENSMUSG00000054728 | Phactr1 | phosphatase and actin regulator 1 | MGI:2659021 |
| ENSMUSG00000004264 | Phb2 | prohibitin 2 | MGI:102520 |
| ENSMUSG00000038845 | Phb | prohibitin | MGI:97572 |
| ENSMUSG00000048537 | Phldb1 | pleckstrin homology-like domain, family B, member 1 | MGI:2143230 |
| ENSMUSG00000003469 | Phyhip | phytanoyl-CoA hydroxylase interacting protein | MGI:1860417 |
| ENSMUSG00000041720 | Pi4ka | phosphatidylinositol 4-kinase, catalytic, alpha polypeptide | MGI:2448506 |
| ENSMUSG00000041958 | Pigs | phosphatidylinositol glycan anchor biosynthesis, class S | MGI:2687325 |
| ENSMUSG00000017721 | Pigt | phosphatidylinositol glycan anchor biosynthesis, class T | MGI:1926178 |
| ENSMUSG00000041417 | Pik3r1 | phosphatidylinositol 3-kinase, regulatory subunit, polypeptide 1 (p85 alpha) | MGI:97583 |
| ENSMUSG00000032571 | Pik3r4 | phosphatidylinositol 3 kinase, regulatory subunit, polypeptide 4, p150 | MGI:1922919 |
| ENSMUSG00000032171 | Pin1 | protein (peptidyl-prolyl cis/trans isomerase) NIMA-interacting 1 | MGI:1346036 |
| ENSMUSG00000026737 | Pip4k2a | phosphatidylinositol-5-phosphate 4-kinase, type II, alpha | MGI:1298206 |
| ENSMUSG00000018547 | Pip4k2b | phosphatidylinositol-5-phosphate 4-kinase, type II, beta | MGI:1934234 |
| ENSMUSG00000028126 | Pip5k1a | phosphatidylinositol-4-phosphate 5-kinase, type 1 alpha | MGI:107929 |
| ENSMUSG00000034902 | Pip5k1c | phosphatidylinositol-4-phosphate 5-kinase, type 1 gamma | MGI:1298224 |
| ENSMUSG00000023452 | Pisd | phosphatidylserine decarboxylase | MGI:2445114 |
| ENSMUSG00000017781 | Pitpna | phosphatidylinositol transfer protein, alpha | MGI:99887 |
| ENSMUSG00000029406 | Pitpnm2 | phosphatidylinositol transfer protein, membrane-associated 2 | MGI:1336192 |
| ENSMUSG00000041957 | Pkp2 | plakophilin 2 | MGI:1914701 |
| ENSMUSG00000026991 | Pkp4 | plakophilin 4 | MGI:109281 |
| ENSMUSG00000050211 | Pla2g4e | phospholipase A2, group IVE | MGI:1919144 |
| ENSMUSG00000024960 | Plcb3 | phospholipase C, beta 3 | MGI:104778 |
| ENSMUSG00000039943 | Plcb4 | phospholipase C, beta 4 | MGI:107464 |
| ENSMUSG00000029055 | Plch2 | phospholipase C, eta 2 | MGI:2443078 |
| ENSMUSG00000038910 | Plcl2 | phospholipase C-like 2 | MGI:1352756 |
| ENSMUSG00000027695 | Pld1 | phospholipase D1 | MGI:109585 |
| ENSMUSG00000003363 | Pld3 | phospholipase D family, member 3 | MGI:1333782 |
| ENSMUSG00000022565 | Plec | plectin | MGI:1277961 |
| ENSMUSG00000030231 | Plekha5 | pleckstrin homology domain containing, family A member 5 | MGI:1923802 |
| ENSMUSG00000041757 | Plekha6 | pleckstrin homology domain containing, family A member 6 | MGI:2388662 |
| ENSMUSG00000045659 | Plekha7 | pleckstrin homology domain containing, family A member 7 | MGI:2445094 |
| ENSMUSG00000030701 | Plekhb1 | pleckstrin homology domain containing, family B (evectins) member 1 | MGI:1351469 |
| ENSMUSG00000066438 | Plekhd1 | pleckstrin homology domain containing, family D (with coiled-coil domains) member 1 | MGI:3036228 |
| ENSMUSG00000040624 | Plekhg1 | pleckstrin homology domain containing, family G (with RhoGef domain) member 1 | MGI:2676551 |
| ENSMUSG00000016495 | Plgrkt | plasminogen receptor, C-terminal lysine transmembrane protein | MGI:1915009 |
| ENSMUSG00000031775 | Pllp | plasma membrane proteolipid | MGI:1915051 |
| ENSMUSG00000035383 | Pmch | pro-melanin-concentrating hormone | MGI:97629 |
| ENSMUSG00000026179 | Pnkd | paroxysmal nonkinesiogenic dyskinesia | MGI:1930773 |
| ENSMUSG00000045731 | Pnoc | prepronociceptin | MGI:105308 |
| ENSMUSG00000004565 | Pnpla6 | patatin-like phospholipase domain containing 6 | MGI:1354723 |
| ENSMUSG00000066235 | Pomgnt2 | protein O-linked mannose beta 1,4-N-acetylglucosaminyltransferase 2 | MGI:2143424 |
| ENSMUSG00000039254 | Pomt1 | protein-O-mannosyltransferase 1 | MGI:2138994 |
| ENSMUSG00000028517 | Ppap2b | phosphatidic acid phosphatase type 2B | MGI:1915166 |
| ENSMUSG00000040105 | Ppapdc2 | phosphatidic acid phosphatase type 2 domain containing 2 | MGI:1921661 |
| ENSMUSG00000037519 | Ppfia1 | protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 1 | MGI:1924750 |
| ENSMUSG00000053825 | Ppfia2 | protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2 | MGI:2443834 |
| ENSMUSG00000003863 | Ppfia3 | protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 3 | MGI:1924037 |
| ENSMUSG00000026458 | Ppfia4 | protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 4 | MGI:1915757 |
| ENSMUSG00000032383 | Ppib | peptidylprolyl isomerase B | MGI:97750 |
| ENSMUSG00000027804 | Ppid | peptidylprolyl isomerase D (cyclophilin D) | MGI:1914988 |
| ENSMUSG00000039457 | Ppl | periplakin | MGI:1194898 |
| ENSMUSG00000026181 | Ppm1f | protein phosphatase 1F (PP2C domain containing) | MGI:1918464 |
| ENSMUSG00000034613 | Ppm1h | protein phosphatase 1H (PP2C domain containing) | MGI:2442087 |
| ENSMUSG00000030718 | Ppme1 | protein phosphatase methylesterase 1 | MGI:1919840 |
| ENSMUSG00000040385 | Ppp1ca | protein phosphatase 1, catalytic subunit, alpha isoform | MGI:103016 |
| ENSMUSG00000014956 | Ppp1cb | protein phosphatase 1, catalytic subunit, beta isoform | MGI:104871 |
| ENSMUSG00000004455 | Ppp1cc | protein phosphatase 1, catalytic subunit, gamma isoform | MGI:104872 |
| ENSMUSG00000019907 | Ppp1r12a | protein phosphatase 1, regulatory (inhibitor) subunit 12A | MGI:1309528 |
| ENSMUSG00000073557 | Ppp1r12b | protein phosphatase 1, regulatory (inhibitor) subunit 12B | MGI:1916417 |
| ENSMUSG00000037754 | Ppp1r16b | protein phosphatase 1, regulatory (inhibitor) subunit 16B | MGI:2151841 |
| ENSMUSG00000032827 | Ppp1r9a | protein phosphatase 1, regulatory (inhibitor) subunit 9A | MGI:2442401 |
| ENSMUSG00000038976 | Ppp1r9b | protein phosphatase 1, regulatory subunit 9B | MGI:2387581 |
| ENSMUSG00000020349 | Ppp2ca | protein phosphatase 2 (formerly 2A), catalytic subunit, alpha isoform | MGI:1321159 |
| ENSMUSG00000007564 | Ppp2r1a | protein phosphatase 2, regulatory subunit A, alpha | MGI:1926334 |
| ENSMUSG00000022052 | Ppp2r2a | protein phosphatase 2, regulatory subunit B, alpha | MGI:1919228 |
| ENSMUSG00000029120 | Ppp2r2c | protein phosphatase 2, regulatory subunit B, gamma | MGI:2442660 |
| ENSMUSG00000039515 | Ppp2r4 | protein phosphatase 2A activator, regulatory subunit B | MGI:1346006 |
| ENSMUSG00000024777 | Ppp2r5b | protein phosphatase 2, regulatory subunit B', beta | MGI:2388480 |
| ENSMUSG00000017843 | Ppp2r5c | protein phosphatase 2, regulatory subunit B', gamma | MGI:1349475 |
| ENSMUSG00000021051 | Ppp2r5e | protein phosphatase 2, regulatory subunit B', epsilon | MGI:1349473 |
| ENSMUSG00000028657 | Ppt1 | palmitoyl-protein thioesterase 1 | MGI:1298204 |
| ENSMUSG00000028691 | Prdx1 | peroxiredoxin 1 | MGI:99523 |
| ENSMUSG00000005161 | Prdx2 | peroxiredoxin 2 | MGI:109486 |
| ENSMUSG00000025289 | Prdx4 | peroxiredoxin 4 | MGI:1859815 |
| ENSMUSG00000045302 | Preb | prolactin regulatory element binding | MGI:1355326 |
| ENSMUSG00000030020 | Prickle2 | prickle homolog 2 (Drosophila) | MGI:1925144 |
| ENSMUSG00000005469 | Prkaca | protein kinase, cAMP dependent, catalytic, alpha | MGI:97592 |
| ENSMUSG00000005034 | Prkacb | protein kinase, cAMP dependent, catalytic, beta | MGI:97594 |
| ENSMUSG00000020612 | Prkar1a | protein kinase, cAMP dependent regulatory, type I, alpha | MGI:104878 |
| ENSMUSG00000025855 | Prkar1b | protein kinase, cAMP dependent regulatory, type I beta | MGI:97759 |
| ENSMUSG00000032601 | Prkar2a | protein kinase, cAMP dependent regulatory, type II alpha | MGI:108025 |
| ENSMUSG00000002997 | Prkar2b | protein kinase, cAMP dependent regulatory, type II beta | MGI:97760 |
| ENSMUSG00000050965 | Prkca | protein kinase C, alpha | MGI:97595 |
| ENSMUSG00000052889 | Prkcb | protein kinase C, beta | MGI:97596 |
| ENSMUSG00000078816 | Prkcg | protein kinase C, gamma | MGI:97597 |
| ENSMUSG00000029334 | Prkg2 | protein kinase, cGMP-dependent, type II | MGI:108173 |
| ENSMUSG00000002731 | Prkra | protein kinase, interferon inducible double stranded RNA dependent activator | MGI:1344375 |
| ENSMUSG00000052429 | Prmt1 | protein arginine N-methyltransferase 1 | MGI:107846 |
| ENSMUSG00000030350 | Prmt8 | protein arginine N-methyltransferase 8 | MGI:3043083 |
| ENSMUSG00000023484 | Prph | peripherin | MGI:97774 |
| ENSMUSG00000092305 | Prps1l1 | phosphoribosyl pyrophosphate synthetase 1-like 1 | MGI:1922706 |
| ENSMUSG00000079104 | Prps1l3 | phosphoribosyl pyrophosphate synthetase 1-like 3 | MGI:3779453 |
| ENSMUSG00000015869 | Prpsap1 | phosphoribosyl pyrophosphate synthetase-associated protein 1 | MGI:1915013 |
| ENSMUSG00000020528 | Prpsap2 | phosphoribosyl pyrophosphate synthetase-associated protein 2 | MGI:2384838 |
| ENSMUSG00000064125 | Prr36 | proline rich 36 | MGI:3605626 |
| ENSMUSG00000034686 | Prr7 | proline rich 7 (synaptic) | MGI:3487246 |
| ENSMUSG00000024393 | Prrc2a | proline-rich coiled-coil 2A | MGI:1915467 |
| ENSMUSG00000040225 | Prrc2c | proline-rich coiled-coil 2C | MGI:1913754 |
| ENSMUSG00000015476 | Prrt1 | proline-rich transmembrane protein 1 | MGI:1932118 |
| ENSMUSG00000071519 | Prss3 | protease, serine 3 | MGI:102758 |
| ENSMUSG00000030465 | Psd3 | pleckstrin and Sec7 domain containing 3 | MGI:1918215 |
| ENSMUSG00000037126 | Psd | pleckstrin and Sec7 domain containing | MGI:1920978 |
| ENSMUSG00000019969 | Psen1 | presenilin 1 | MGI:1202717 |
| ENSMUSG00000028484 | Psip1 | PC4 and SFRS1 interacting protein 1 | MGI:2142116 |
| ENSMUSG00000060073 | Psma3 | proteasome (prosome, macropain) subunit, alpha type 3 | MGI:104883 |
| ENSMUSG00000068749 | Psma5 | proteasome (prosome, macropain) subunit, alpha type 5 | MGI:1347009 |
| ENSMUSG00000021024 | Psma6 | proteasome (prosome, macropain) subunit, alpha type 6 | MGI:1347006 |
| ENSMUSG00000027566 | Psma7 | proteasome (prosome, macropain) subunit, alpha type 7 | MGI:1347070 |
| ENSMUSG00000014769 | Psmb1 | proteasome (prosome, macropain) subunit, beta type 1 | MGI:104884 |
| ENSMUSG00000005779 | Psmb4 | proteasome (prosome, macropain) subunit, beta type 4 | MGI:1098257 |
| ENSMUSG00000018286 | Psmb6 | proteasome (prosome, macropain) subunit, beta type 6 | MGI:104880 |
| ENSMUSG00000028932 | Psmc2 | proteasome (prosome, macropain) 26S subunit, ATPase 2 | MGI:109555 |
| ENSMUSG00000002102 | Psmc3 | proteasome (prosome, macropain) 26S subunit, ATPase 3 | MGI:1098754 |
| ENSMUSG00000030603 | Psmc4 | proteasome (prosome, macropain) 26S subunit, ATPase, 4 | MGI:1346093 |
| ENSMUSG00000020708 | Psmc5 | protease (prosome, macropain) 26S subunit, ATPase 5 | MGI:105047 |
| ENSMUSG00000021832 | Psmc6 | proteasome (prosome, macropain) 26S subunit, ATPase, 6 | MGI:1914339 |
| ENSMUSG00000017428 | Psmd11 | proteasome (prosome, macropain) 26S subunit, non-ATPase, 11 | MGI:1916327 |
| ENSMUSG00000020720 | Psmd12 | proteasome (prosome, macropain) 26S subunit, non-ATPase, 12 | MGI:1914247 |
| ENSMUSG00000025487 | Psmd13 | proteasome (prosome, macropain) 26S subunit, non-ATPase, 13 | MGI:1345192 |
| ENSMUSG00000026914 | Psmd14 | proteasome (prosome, macropain) 26S subunit, non-ATPase, 14 | MGI:1913284 |
| ENSMUSG00000017221 | Psmd3 | proteasome (prosome, macropain) 26S subunit, non-ATPase, 3 | MGI:98858 |
| ENSMUSG00000021737 | Psmd6 | proteasome (prosome, macropain) 26S subunit, non-ATPase, 6 | MGI:1913663 |
| ENSMUSG00000039067 | Psmd7 | proteasome (prosome, macropain) 26S subunit, non-ATPase, 7 | MGI:1351511 |
| ENSMUSG00000021938 | Pspc1 | paraspeckle protein 1 | MGI:1913895 |
| ENSMUSG00000006498 | Ptbp1 | polypyrimidine tract binding protein 1 | MGI:97791 |
| ENSMUSG00000028134 | Ptbp2 | polypyrimidine tract binding protein 2 | MGI:1860489 |
| ENSMUSG00000021518 | Ptdss1 | phosphatidylserine synthase 1 | MGI:1276575 |
| ENSMUSG00000063235 | Ptpmt1 | protein tyrosine phosphatase, mitochondrial 1 | MGI:1913711 |
| ENSMUSG00000028399 | Ptprd | protein tyrosine phosphatase, receptor type, D | MGI:97812 |
| ENSMUSG00000021745 | Ptprg | protein tyrosine phosphatase, receptor type, G | MGI:97814 |
| ENSMUSG00000056553 | Ptprn2 | protein tyrosine phosphatase, receptor type, N polypeptide 2 | MGI:107418 |
| ENSMUSG00000013236 | Ptprs | protein tyrosine phosphatase, receptor type, S | MGI:97815 |
| ENSMUSG00000053141 | Ptprt | protein tyrosine phosphatase, receptor type, T | MGI:1321152 |
| ENSMUSG00000028909 | Ptpru | protein tyrosine phosphatase, receptor type, U | MGI:1321151 |
| ENSMUSG00000068748 | Ptprz1 | protein tyrosine phosphatase, receptor type Z, polypeptide 1 | MGI:97816 |
| ENSMUSG00000072582 | Ptrh2 | peptidyl-tRNA hydrolase 2 | MGI:2444848 |
| ENSMUSG00000043991 | Pura | purine rich element binding protein A | MGI:103079 |
| ENSMUSG00000094483 | Purb | purine rich element binding protein B | MGI:1338779 |
| ENSMUSG00000049184 | Purg | purine-rich element binding protein G | MGI:1922279 |
| ENSMUSG00000026520 | Pycr2 | pyrroline-5-carboxylate reductase family, member 2 | MGI:1277956 |
| ENSMUSG00000022571 | Pycrl | pyrroline-5-carboxylate reductase-like | MGI:1913444 |
| ENSMUSG00000030359 | Pzp | pregnancy zone protein | MGI:87854 |
| ENSMUSG00000032604 | Qars | glutaminyl-tRNA synthetase | MGI:1915851 |
| ENSMUSG00000062078 | Qk | quaking | MGI:97837 |
| ENSMUSG00000040022 | Rab11fip2 | RAB11 family interacting protein 2 (class I) | MGI:1922248 |
| ENSMUSG00000051343 | Rab11fip5 | RAB11 family interacting protein 5 (class I) | MGI:1098586 |
| ENSMUSG00000023460 | Rab12 | RAB12, member RAS oncogene family | MGI:894284 |
| ENSMUSG00000027935 | Rab13 | RAB13, member RAS oncogene family | MGI:1927232 |
| ENSMUSG00000021062 | Rab15 | RAB15, member RAS oncogene family | MGI:1916865 |
| ENSMUSG00000020132 | Rab21 | RAB21, member RAS oncogene family | MGI:894308 |
| ENSMUSG00000027519 | Rab22a | RAB22A, member RAS oncogene family | MGI:105072 |
| ENSMUSG00000027739 | Rab33b | RAB33B, member RAS oncogene family | MGI:1330805 |
| ENSMUSG00000029518 | Rab35 | RAB35, member RAS oncogene family | MGI:1924657 |
| ENSMUSG00000019066 | Rab3d | RAB3D, member RAS oncogene family | MGI:97844 |
| ENSMUSG00000003037 | Rab8a | RAB8A, member RAS oncogene family | MGI:96960 |
| ENSMUSG00000036943 | Rab8b | RAB8B, member RAS oncogene family | MGI:2442982 |
| ENSMUSG00000020817 | Rabep1 | rabaptin, RAB GTPase binding effector protein 1 | MGI:1860236 |
| ENSMUSG00000040472 | Rabggta | Rab geranylgeranyl transferase, a subunit | MGI:1860443 |
| ENSMUSG00000001847 | Rac1 | RAS-related C3 botulinum substrate 1 | MGI:97845 |
| ENSMUSG00000018012 | Rac3 | RAS-related C3 botulinum substrate 3 | MGI:2180784 |
| ENSMUSG00000022246 | Rai14 | retinoic acid induced 14 | MGI:1922896 |
| ENSMUSG00000008859 | Rala | v-ral simian leukemia viral oncogene homolog A (ras related) | MGI:1927243 |
| ENSMUSG00000024096 | Ralbp1 | ralA binding protein 1 | MGI:108466 |
| ENSMUSG00000004451 | Ralb | v-ral simian leukemia viral oncogene homolog B (ras related) | MGI:1927244 |
| ENSMUSG00000021027 | Ralgapa1 | Ral GTPase activating protein, alpha subunit 1 | MGI:1931050 |
| ENSMUSG00000027652 | Ralgapb | Ral GTPase activating protein, beta subunit (non-catalytic) | MGI:2444531 |
| ENSMUSG00000029430 | Ran | RAN, member RAS oncogene family | MGI:1333112 |
| ENSMUSG00000068798 | Rap1a | RAS-related protein-1a | MGI:97852 |
| ENSMUSG00000052681 | Rap1b | RAS related protein 1b | MGI:894315 |
| ENSMUSG00000041351 | Rap1gap | Rap1 GTPase-activating protein | MGI:109338 |
| ENSMUSG00000062232 | Rapgef2 | Rap guanine nucleotide exchange factor (GEF) 2 | MGI:2659071 |
| ENSMUSG00000049044 | Rapgef4 | Rap guanine nucleotide exchange factor (GEF) 4 | MGI:1917723 |
| ENSMUSG00000026014 | Raph1 | Ras association (RalGDS/AF-6) and pleckstrin homology domains 1 | MGI:1924550 |
| ENSMUSG00000028292 | Rars2 | arginyl-tRNA synthetase 2, mitochondrial | MGI:1923596 |
| ENSMUSG00000031453 | Rasa3 | RAS p21 protein activator 3 | MGI:1197013 |
| ENSMUSG00000029602 | Rasal1 | RAS protein activator like 1 (GAP1 like) | MGI:1330842 |
| ENSMUSG00000070565 | Rasal2 | RAS protein activator like 2 | MGI:2443881 |
| ENSMUSG00000030134 | Rasgef1a | RasGEF domain family, member 1A | MGI:1917977 |
| ENSMUSG00000032356 | Rasgrf1 | RAS protein-specific guanine nucleotide-releasing factor 1 | MGI:99694 |
| ENSMUSG00000021708 | Rasgrf2 | RAS protein-specific guanine nucleotide-releasing factor 2 | MGI:109137 |
| ENSMUSG00000027347 | Rasgrp1 | RAS guanyl releasing protein 1 | MGI:1314635 |
| ENSMUSG00000032946 | Rasgrp2 | RAS, guanyl releasing protein 2 | MGI:1333849 |
| ENSMUSG00000006456 | Rbm14 | RNA binding motif protein 14 | MGI:1929092 |
| ENSMUSG00000031167 | Rbm3 | RNA binding motif protein 3 | MGI:1099460 |
| ENSMUSG00000031134 | Rbmx | RNA binding motif protein, X chromosome | MGI:1343044 |
| ENSMUSG00000035469 | Rcbtb1 | regulator of chromosome condensation (RCC1) and BTB (POZ) domain containing protein 1 | MGI:1918580 |
| ENSMUSG00000032320 | Rcn2 | reticulocalbin 2 | MGI:1349765 |
| ENSMUSG00000008435 | Rdh13 | retinol dehydrogenase 13 (all-trans and 9-cis) | MGI:1918732 |
| ENSMUSG00000038555 | Reep2 | receptor accessory protein 2 | MGI:2385070 |
| ENSMUSG00000022176 | Rem2 | rad and gem related GTP binding protein 2 | MGI:2155260 |
| ENSMUSG00000039316 | Rftn1 | raftlin lipid raft linker 1 | MGI:1923688 |
| ENSMUSG00000070509 | Rgma | repulsive guidance molecule family member A | MGI:2679262 |
| ENSMUSG00000019775 | Rgs17 | regulator of G-protein signaling 17 | MGI:1927469 |
| ENSMUSG00000002458 | Rgs19 | regulator of G-protein signaling 19 | MGI:1915153 |
| ENSMUSG00000002459 | Rgs20 | regulator of G-protein signaling 20 | MGI:1929866 |
| ENSMUSG00000021219 | Rgs6 | regulator of G-protein signaling 6 | MGI:1354730 |
| ENSMUSG00000021719 | Rgs7bp | regulator of G-protein signalling 7 binding protein | MGI:106334 |
| ENSMUSG00000026527 | Rgs7 | regulator of G protein signaling 7 | MGI:1346089 |
| ENSMUSG00000042671 | Rgs8 | regulator of G-protein signaling 8 | MGI:108408 |
| ENSMUSG00000020599 | Rgs9 | regulator of G-protein signaling 9 | MGI:1338824 |
| ENSMUSG00000028945 | Rheb | Ras homolog enriched in brain | MGI:97912 |
| ENSMUSG00000029449 | Rhof | ras homolog gene family, member f | MGI:1345629 |
| ENSMUSG00000073982 | Rhog | ras homolog gene family, member G | MGI:1928370 |
| ENSMUSG00000024143 | Rhoq | ras homolog gene family, member Q | MGI:1931553 |
| ENSMUSG00000017686 | Rhot1 | ras homolog gene family, member T1 | MGI:1926078 |
| ENSMUSG00000025257 | Ribc1 | RIB43A domain with coiled-coils 1 | MGI:1913861 |
| ENSMUSG00000025485 | Ric8 | resistance to inhibitors of cholinesterase 8 homolog (C. elegans) | MGI:2141866 |
| ENSMUSG00000029420 | Rimbp2 | RIMS binding protein 2 | MGI:2443235 |
| ENSMUSG00000041670 | Rims1 | regulating synaptic membrane exocytosis 1 | MGI:2152971 |
| ENSMUSG00000037386 | Rims2 | regulating synaptic membrane exocytosis 2 | MGI:2152972 |
| ENSMUSG00000032890 | Rims3 | regulating synaptic membrane exocytosis 3 | MGI:2443331 |
| ENSMUSG00000035226 | Rims4 | regulating synaptic membrane exocytosis 4 | MGI:2674366 |
| ENSMUSG00000024883 | Rin1 | Ras and Rab interactor 1 | MGI:2385695 |
| ENSMUSG00000050357 | Rltpr | RGD motif, leucine rich repeats, tropomodulin domain and proline-rich containing | MGI:2685431 |
| ENSMUSG00000070730 | Rmdn3 | regulator of microtubule dynamics 3 | MGI:1915059 |
| ENSMUSG00000019763 | Rmnd1 | required for meiotic nuclear division 1 homolog (S. cerevisiae) | MGI:1913334 |
| ENSMUSG00000010086 | Rnf112 | ring finger protein 112 | MGI:106611 |
| ENSMUSG00000042790 | Rnf214 | ring finger protein 214 | MGI:2444451 |
| ENSMUSG00000009535 | Rnmt | RNA (guanine-7-) methyltransferase | MGI:1915147 |
| ENSMUSG00000022883 | Robo1 | roundabout homolog 1 (Drosophila) | MGI:1274781 |
| ENSMUSG00000052516 | Robo2 | roundabout homolog 2 (Drosophila) | MGI:1890110 |
| ENSMUSG00000022540 | Rogdi | rogdi homolog (Drosophila) | MGI:1913299 |
| ENSMUSG00000060090 | Rp2h | retinitis pigmentosa 2 homolog (human) | MGI:1277953 |
| ENSMUSG00000038510 | Rpf2 | ribosome production factor 2 homolog (S. cerevisiae) | MGI:1914489 |
| ENSMUSG00000029608 | Rph3a | rabphilin 3A | MGI:102788 |
| ENSMUSG00000037805 | Rpl10a | ribosomal protein L10A | MGI:1343877 |
| ENSMUSG00000058443 | Rpl10-ps3 | ribosomal protein L10, pseudogene 3 | MGI:3704336 |
| ENSMUSG00000059291 | Rpl11 | ribosomal protein L11 | MGI:1914275 |
| ENSMUSG00000038900 | Rpl12 | ribosomal protein L12 | MGI:98002 |
| ENSMUSG00000074129 | Rpl13a | ribosomal protein L13A | MGI:1351455 |
| ENSMUSG00000000740 | Rpl13 | ribosomal protein L13 | MGI:105922 |
| ENSMUSG00000025794 | Rpl14 | ribosomal protein L14 | MGI:1914365 |
| ENSMUSG00000062328 | Rpl17 | ribosomal protein L17 | MGI:2448270 |
| ENSMUSG00000045128 | Rpl18a | ribosomal protein L18A | MGI:1924058 |
| ENSMUSG00000059070 | Rpl18 | ribosomal protein L18 | MGI:98003 |
| ENSMUSG00000017404 | Rpl19 | ribosomal protein L19 | MGI:98020 |
| ENSMUSG00000041453 | Rpl21 | ribosomal protein L21 | MGI:1278340 |
| ENSMUSG00000039221 | Rpl22l1 | ribosomal protein L22 like 1 | MGI:1915278 |
| ENSMUSG00000028936 | Rpl22 | ribosomal protein L22 | MGI:99262 |
| ENSMUSG00000058546 | Rpl23a | ribosomal protein L23A | MGI:3040672 |
| ENSMUSG00000071415 | Rpl23 | ribosomal protein L23 | MGI:1929455 |
| ENSMUSG00000098274 | Rpl24 | ribosomal protein L24 | MGI:1915443 |
| ENSMUSG00000060938 | Rpl26 | ribosomal protein L26 | MGI:106022 |
| ENSMUSG00000046364 | Rpl27a | ribosomal protein L27A | MGI:1347076 |
| ENSMUSG00000073640 | Rpl27-ps3 | ribosomal protein L27, pseudogene 3 | MGI:3646174 |
| ENSMUSG00000030432 | Rpl28 | ribosomal protein L28 | MGI:101839 |
| ENSMUSG00000048758 | Rpl29 | ribosomal protein L29 | MGI:99687 |
| ENSMUSG00000058600 | Rpl30 | ribosomal protein L30 | MGI:98037 |
| ENSMUSG00000073702 | Rpl31 | ribosomal protein L31 | MGI:2149632 |
| ENSMUSG00000057841 | Rpl32 | ribosomal protein L32 | MGI:98038 |
| ENSMUSG00000062006 | Rpl34 | ribosomal protein L34 | MGI:1915686 |
| ENSMUSG00000060636 | Rpl35a | ribosomal protein L35A | MGI:1928894 |
| ENSMUSG00000057863 | Rpl36 | ribosomal protein L36 | MGI:1860603 |
| ENSMUSG00000046330 | Rpl37a | ribosomal protein L37a | MGI:98068 |
| ENSMUSG00000057322 | Rpl38 | ribosomal protein L38 | MGI:1914921 |
| ENSMUSG00000060036 | Rpl3 | ribosomal protein L3 | MGI:1351605 |
| ENSMUSG00000032399 | Rpl4 | ribosomal protein L4 | MGI:1915141 |
| ENSMUSG00000058558 | Rpl5 | ribosomal protein L5 | MGI:102854 |
| ENSMUSG00000029614 | Rpl6 | ribosomal protein L6 | MGI:108057 |
| ENSMUSG00000062647 | Rpl7a | ribosomal protein L7A | MGI:1353472 |
| ENSMUSG00000043716 | Rpl7 | ribosomal protein L7 | MGI:98073 |
| ENSMUSG00000003970 | Rpl8 | ribosomal protein L8 | MGI:1350927 |
| ENSMUSG00000047215 | Rpl9 | ribosomal protein L9 | MGI:1298373 |
| ENSMUSG00000067274 | Rplp0 | ribosomal protein, large, P0 | MGI:1927636 |
| ENSMUSG00000007892 | Rplp1 | ribosomal protein, large, P1 | MGI:1927099 |
| ENSMUSG00000025508 | Rplp2 | ribosomal protein, large P2 | MGI:1914436 |
| ENSMUSG00000030062 | Rpn1 | ribophorin I | MGI:98084 |
| ENSMUSG00000027642 | Rpn2 | ribophorin II | MGI:98085 |
| ENSMUSG00000052146 | Rps10 | ribosomal protein S10 | MGI:1914347 |
| ENSMUSG00000003429 | Rps11 | ribosomal protein S11 | MGI:1351329 |
| ENSMUSG00000067038 | Rps12-ps3 | ribosomal protein S12, pseudogene 3 | MGI:3704503 |
| ENSMUSG00000090862 | Rps13 | ribosomal protein S13 | MGI:1915302 |
| ENSMUSG00000024608 | Rps14 | ribosomal protein S14 | MGI:98107 |
| ENSMUSG00000008683 | Rps15a | ribosomal protein S15A | MGI:2389091 |
| ENSMUSG00000063457 | Rps15 | ribosomal protein S15 | MGI:98117 |
| ENSMUSG00000037563 | Rps16 | ribosomal protein S16 | MGI:98118 |
| ENSMUSG00000061787 | Rps17 | ribosomal protein S17 | MGI:1309526 |
| ENSMUSG00000040952 | Rps19 | ribosomal protein S19 | MGI:1333780 |
| ENSMUSG00000028234 | Rps20 | ribosomal protein S20 | MGI:1914677 |
| ENSMUSG00000049517 | Rps23 | ribosomal protein S23 | MGI:1913725 |
| ENSMUSG00000025290 | Rps24 | ribosomal protein S24 | MGI:98147 |
| ENSMUSG00000009927 | Rps25 | ribosomal protein S25 | MGI:1922867 |
| ENSMUSG00000059775 | Rps26-ps1 | ribosomal protein S26, pseudogene 1 | MGI:3704322 |
| ENSMUSG00000020460 | Rps27a | ribosomal protein S27A | MGI:1925544 |
| ENSMUSG00000036781 | Rps27l | ribosomal protein S27-like | MGI:1915191 |
| ENSMUSG00000050621 | Rps27rt | ribosomal protein S27, retrogene | MGI:3704345 |
| ENSMUSG00000067288 | Rps28 | ribosomal protein S28 | MGI:1859516 |
| ENSMUSG00000034892 | Rps29 | ribosomal protein S29 | MGI:107681 |
| ENSMUSG00000028081 | Rps3a1 | ribosomal protein S3A1 | MGI:1202063 |
| ENSMUSG00000030744 | Rps3 | ribosomal protein S3 | MGI:1350917 |
| ENSMUSG00000031320 | Rps4x | ribosomal protein S4, X-linked | MGI:98158 |
| ENSMUSG00000012848 | Rps5 | ribosomal protein S5 | MGI:1097682 |
| ENSMUSG00000003644 | Rps6ka1 | ribosomal protein S6 kinase polypeptide 1 | MGI:104558 |
| ENSMUSG00000028495 | Rps6 | ribosomal protein S6 | MGI:98159 |
| ENSMUSG00000047675 | Rps8 | ribosomal protein S8 | MGI:98166 |
| ENSMUSG00000006333 | Rps9 | ribosomal protein S9 | MGI:1924096 |
| ENSMUSG00000055723 | Rras2 | related RAS viral (r-ras) oncogene homolog 2 | MGI:1914172 |
| ENSMUSG00000038387 | Rras | Harvey rat sarcoma oncogene, subgroup R | MGI:98179 |
| ENSMUSG00000027422 | Rrbp1 | ribosome binding protein 1 | MGI:1932395 |
| ENSMUSG00000050079 | Rspry1 | ring finger and SPRY domain containing 1 | MGI:1914860 |
| ENSMUSG00000000339 | Rtca | RNA 3'-terminal phosphate cyclase | MGI:1913618 |
| ENSMUSG00000001783 | Rtcb | RNA 2',3'-cyclic phosphate and 5'-OH ligase | MGI:106379 |
| ENSMUSG00000050896 | Rtn4rl2 | reticulon 4 receptor-like 2 | MGI:2669796 |
| ENSMUSG00000043811 | Rtn4r | reticulon 4 receptor | MGI:2136886 |
| ENSMUSG00000029291 | Rufy3 | RUN and FYVE domain containing 3 | MGI:106484 |
| ENSMUSG00000006575 | Rundc3a | RUN domain containing 3A | MGI:1858752 |
| ENSMUSG00000021313 | Ryr2 | ryanodine receptor 2, cardiac | MGI:99685 |
| ENSMUSG00000057378 | Ryr3 | ryanodine receptor 3 | MGI:99684 |
| ENSMUSG00000074457 | S100a16 | S100 calcium binding protein A16 | MGI:1915110 |
| ENSMUSG00000045092 | S1pr1 | sphingosine-1-phosphate receptor 1 | MGI:1096355 |
| ENSMUSG00000025240 | Sacm1l | SAC1 (suppressor of actin mutations 1, homolog)-like (S. cerevisiae) | MGI:1933169 |
| ENSMUSG00000048279 | Sacs | sacsin | MGI:1354724 |
| ENSMUSG00000022437 | Samm50 | sorting and assembly machinery component 50 homolog (S. cerevisiae) | MGI:1915903 |
| ENSMUSG00000020088 | Sar1a | SAR1 gene homolog A (S. cerevisiae) | MGI:98230 |
| ENSMUSG00000050132 | Sarm1 | sterile alpha and HEAT/Armadillo motif containing 1 | MGI:2136419 |
| ENSMUSG00000068739 | Sars | seryl-aminoacyl-tRNA synthetase | MGI:102809 |
| ENSMUSG00000035236 | Scai | suppressor of cancer cell invasion | MGI:2443716 |
| ENSMUSG00000040722 | Scamp5 | secretory carrier membrane protein 5 | MGI:1928948 |
| ENSMUSG00000038936 | Sccpdh | saccharopine dehydrogenase (putative) | MGI:1924486 |
| ENSMUSG00000062110 | Scfd2 | Sec1 family domain containing 2 | MGI:2443446 |
| ENSMUSG00000050711 | Scg2 | secretogranin II | MGI:103033 |
| ENSMUSG00000091780 | Sco2 | SCO cytochrome oxidase deficient homolog 2 (yeast) | MGI:3818630 |
| ENSMUSG00000022568 | Scrib | scribbled homolog (Drosophila) | MGI:2145950 |
| ENSMUSG00000019124 | Scrn1 | secernin 1 | MGI:1917188 |
| ENSMUSG00000025743 | Sdc3 | syndecan 3 | MGI:1349163 |
| ENSMUSG00000028249 | Sdcbp | syndecan binding protein | MGI:1337026 |
| ENSMUSG00000002064 | Sdf2 | stromal cell derived factor 2 | MGI:108019 |
| ENSMUSG00000025724 | Sec11a | SEC11 homolog A (S. cerevisiae) | MGI:1929464 |
| ENSMUSG00000024516 | Sec11c | SEC11 homolog C (S. cerevisiae) | MGI:1913536 |
| ENSMUSG00000027879 | Sec22b | SEC22 vesicle trafficking protein homolog B (S. cerevisiae) | MGI:1338759 |
| ENSMUSG00000020986 | Sec23a | SEC23A (S. cerevisiae) | MGI:1349635 |
| ENSMUSG00000027429 | Sec23b | SEC23B (S. cerevisiae) | MGI:1350925 |
| ENSMUSG00000001052 | Sec24b | Sec24 related gene family, member B (S. cerevisiae) | MGI:2139764 |
| ENSMUSG00000039367 | Sec24c | Sec24 related gene family, member C (S. cerevisiae) | MGI:1919746 |
| ENSMUSG00000025816 | Sec61a2 | Sec61, alpha subunit 2 (S. cerevisiae) | MGI:1931071 |
| ENSMUSG00000053317 | Sec61b | Sec61 beta subunit | MGI:1913462 |
| ENSMUSG00000042682 | Selk | selenoprotein K | MGI:1931466 |
| ENSMUSG00000021451 | Sema4d | sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4D | MGI:109244 |
| ENSMUSG00000000627 | Sema4f | sema domain, immunoglobulin domain (Ig), TM domain, and short cytoplasmic domain | MGI:1340055 |
| ENSMUSG00000038264 | Sema7a | sema domain, immunoglobulin domain (Ig), and GPI membrane anchor, (semaphorin) 7A | MGI:1306826 |
| ENSMUSG00000058013 | Sept11 | septin 11 | MGI:1277214 |
| ENSMUSG00000026276 | Sept2 | septin 2 | MGI:97298 |
| ENSMUSG00000022456 | Sept3 | septin 3 | MGI:1345148 |
| ENSMUSG00000020486 | Sept4 | septin 4 | MGI:1270156 |
| ENSMUSG00000072214 | Sept5 | septin 5 | MGI:1195461 |
| ENSMUSG00000050379 | Sept6 | septin 6 | MGI:1888939 |
| ENSMUSG00000001833 | Sept7 | septin 7 | MGI:1335094 |
| ENSMUSG00000018398 | Sept8 | septin 8 | MGI:894310 |
| ENSMUSG00000059248 | Sept9 | septin 9 | MGI:1858222 |
| ENSMUSG00000036371 | Serbp1 | serpine1 mRNA binding protein 1 | MGI:1914120 |
| ENSMUSG00000026249 | Serpine2 | serine (or cysteine) peptidase inhibitor, clade E, member 2 | MGI:101780 |
| ENSMUSG00000070436 | Serpinh1 | serine (or cysteine) peptidase inhibitor, clade H, member 1 | MGI:88283 |
| ENSMUSG00000042272 | Sestd1 | SEC14 and spectrin domains 1 | MGI:1916262 |
| ENSMUSG00000006527 | Sfmbt1 | Scm-like with four mbt domains 1 | MGI:1859609 |
| ENSMUSG00000028820 | Sfpq | splicing factor proline/glutamine rich (polypyrimidine tract binding protein associated) | MGI:1918764 |
| ENSMUSG00000025212 | Sfxn3 | sideroflexin 3 | MGI:2137679 |
| ENSMUSG00000004631 | Sgce | sarcoglycan, epsilon | MGI:1329042 |
| ENSMUSG00000028524 | Sgip1 | SH3-domain GRB2-like (endophilin) interacting protein 1 | MGI:1920344 |
| ENSMUSG00000020097 | Sgpl1 | sphingosine phosphate lyase 1 | MGI:1261415 |
| ENSMUSG00000032261 | Sh3bgrl2 | SH3 domain binding glutamic acid-rich protein like 2 | MGI:1915350 |
| ENSMUSG00000040990 | Sh3kbp1 | SH3-domain kinase binding protein 1 | MGI:1889583 |
| ENSMUSG00000053617 | Sh3pxd2a | SH3 and PX domains 2A | MGI:1298393 |
| ENSMUSG00000022623 | Shank3 | SH3/ankyrin domain gene 3 | MGI:1930016 |
| ENSMUSG00000053930 | Shisa6 | shisa family member 6 | MGI:2685725 |
| ENSMUSG00000053550 | Shisa7 | shisa family member 7 | MGI:3605641 |
| ENSMUSG00000025403 | Shmt2 | serine hydroxymethyltransferase 2 (mitochondrial) | MGI:1277989 |
| ENSMUSG00000024976 | Shoc2 | soc-2 (suppressor of clear) homolog (C. elegans) | MGI:1927197 |
| ENSMUSG00000034135 | Sik3 | SIK family kinase 3 | MGI:2446296 |
| ENSMUSG00000042700 | Sipa1l1 | signal-induced proliferation-associated 1 like 1 | MGI:2443679 |
| ENSMUSG00000001995 | Sipa1l2 | signal-induced proliferation-associated 1 like 2 | MGI:2676970 |
| ENSMUSG00000030583 | Sipa1l3 | signal-induced proliferation-associated 1 like 3 | MGI:1921456 |
| ENSMUSG00000036309 | Skp1a | S-phase kinase-associated protein 1A | MGI:103575 |
| ENSMUSG00000036087 | Slain2 | SLAIN motif family, member 2 | MGI:1923241 |
| ENSMUSG00000024597 | Slc12a2 | solute carrier family 12, member 2 | MGI:101924 |
| ENSMUSG00000059336 | Slc14a1 | solute carrier family 14 (urea transporter), member 1 | MGI:1351654 |
| ENSMUSG00000005360 | Slc1a3 | solute carrier family 1 (glial high affinity glutamate transporter), member 3 | MGI:99917 |
| ENSMUSG00000025792 | Slc25a10 | solute carrier family 25 (mitochondrial carrier, dicarboxylate transporter), member 10 | MGI:1353497 |
| ENSMUSG00000014606 | Slc25a11 | solute carrier family 25 (mitochondrial carrier oxoglutarate carrier), member 11 | MGI:1915113 |
| ENSMUSG00000027010 | Slc25a12 | solute carrier family 25 (mitochondrial carrier, Aralar), member 12 | MGI:1926080 |
| ENSMUSG00000004902 | Slc25a18 | solute carrier family 25 (mitochondrial carrier), member 18 | MGI:1919053 |
| ENSMUSG00000003528 | Slc25a1 | solute carrier family 25 (mitochondrial carrier, citrate transporter), member 1 | MGI:1345283 |
| ENSMUSG00000019082 | Slc25a22 | solute carrier family 25 (mitochondrial carrier, glutamate), member 22 | MGI:1915517 |
| ENSMUSG00000023912 | Slc25a27 | solute carrier family 25, member 27 | MGI:1921261 |
| ENSMUSG00000054099 | Slc25a40 | solute carrier family 25, member 40 | MGI:2442486 |
| ENSMUSG00000002346 | Slc25a42 | solute carrier family 25, member 42 | MGI:1920345 |
| ENSMUSG00000024259 | Slc25a46 | solute carrier family 25, member 46 | MGI:1914703 |
| ENSMUSG00000031633 | Slc25a4 | solute carrier family 25 (mitochondrial carrier, adenine nucleotide translocator), member 4 | MGI:1353495 |
| ENSMUSG00000016319 | Slc25a5 | solute carrier family 25 (mitochondrial carrier, adenine nucleotide translocator), member 5 | MGI:1353496 |
| ENSMUSG00000031808 | Slc27a1 | solute carrier family 27 (fatty acid transporter), member 1 | MGI:1347098 |
| ENSMUSG00000059316 | Slc27a4 | solute carrier family 27 (fatty acid transporter), member 4 | MGI:1347347 |
| ENSMUSG00000028645 | Slc2a1 | solute carrier family 2 (facilitated glucose transporter), member 1 | MGI:95755 |
| ENSMUSG00000024270 | Slc39a6 | solute carrier family 39 (metal ion transporter), member 6 | MGI:2147279 |
| ENSMUSG00000024327 | Slc39a7 | solute carrier family 39 (zinc transporter), member 7 | MGI:95909 |
| ENSMUSG00000027075 | Slc43a1 | solute carrier family 43, member 1 | MGI:1931352 |
| ENSMUSG00000006574 | Slc4a1 | solute carrier family 4 (anion exchanger), member 1 | MGI:109393 |
| ENSMUSG00000002504 | Slc9a3r2 | solute carrier family 9 (sodium/hydrogen exchanger), member 3 regulator 2 | MGI:1890662 |
| ENSMUSG00000048304 | Slitrk3 | SLIT and NTRK-like family, member 3 | MGI:2679447 |
| ENSMUSG00000021870 | Slmap | sarcolemma associated protein | MGI:1933549 |
| ENSMUSG00000026155 | Smap1 | small ArfGAP 1 | MGI:2138261 |
| ENSMUSG00000041133 | Smc1a | structural maintenance of chromosomes 1A | MGI:1344345 |
| ENSMUSG00000024974 | Smc3 | structural maintenance of chromosomes 3 | MGI:1339795 |
| ENSMUSG00000078350 | Smim1 | small integral membrane protein 1 | MGI:1916109 |
| ENSMUSG00000061461 | Smim20 | small integral membrane protein 20 | MGI:1913528 |
| ENSMUSG00000027287 | Snap23 | synaptosomal-associated protein 23 | MGI:109356 |
| ENSMUSG00000027273 | Snap25 | synaptosomal-associated protein 25 | MGI:98331 |
| ENSMUSG00000022765 | Snap29 | synaptosomal-associated protein 29 | MGI:1914724 |
| ENSMUSG00000009894 | Snap47 | synaptosomal-associated protein, 47 | MGI:1915076 |
| ENSMUSG00000001424 | Snd1 | staphylococcal nuclease and tudor domain containing 1 | MGI:1929266 |
| ENSMUSG00000027457 | Snph | syntaphilin | MGI:2139270 |
| ENSMUSG00000021039 | Snw1 | SNW domain containing 1 | MGI:1913604 |
| ENSMUSG00000028136 | Snx27 | sorting nexin family member 27 | MGI:1923992 |
| ENSMUSG00000038916 | Soga3 | SOGA family member 3 | MGI:1914662 |
| ENSMUSG00000025006 | Sorbs1 | sorbin and SH3 domain containing 1 | MGI:700014 |
| ENSMUSG00000031626 | Sorbs2 | sorbin and SH3 domain containing 2 | MGI:1924574 |
| ENSMUSG00000029093 | Sorcs2 | sortilin-related VPS10 domain containing receptor 2 | MGI:1932289 |
| ENSMUSG00000044352 | Sowaha | sosondowah ankyrin repeat domain family member A | MGI:2687280 |
| ENSMUSG00000018593 | Sparc | secreted acidic cysteine rich glycoprotein | MGI:98373 |
| ENSMUSG00000024068 | Spast | spastin | MGI:1858896 |
| ENSMUSG00000033594 | Spata2l | spermatogenesis associated 2-like | MGI:1926029 |
| ENSMUSG00000051934 | Spats2 | spermatogenesis associated, serine-rich 2 | MGI:1919822 |
| ENSMUSG00000021917 | Spcs1 | signal peptidase complex subunit 1 homolog (S. cerevisiae) | MGI:1916269 |
| ENSMUSG00000035227 | Spcs2 | signal peptidase complex subunit 2 homolog (S. cerevisiae) | MGI:1913874 |
| ENSMUSG00000042331 | Specc1 | sperm antigen with calponin homology and coiled-coil domains 1 | MGI:2442356 |
| ENSMUSG00000026207 | Speg | SPEG complex locus | MGI:109282 |
| ENSMUSG00000026163 | Sphkap | SPHK1 interactor, AKAP domain containing | MGI:1924879 |
| ENSMUSG00000024533 | Spire1 | spire homolog 1 (Drosophila) | MGI:1915416 |
| ENSMUSG00000027351 | Spred1 | sprouty protein with EVH-1 domain 1, related sequence | MGI:2150016 |
| ENSMUSG00000045671 | Spred2 | sprouty-related, EVH1 domain containing 2 | MGI:2150019 |
| ENSMUSG00000022114 | Spry2 | sprouty homolog 2 (Drosophila) | MGI:1345138 |
| ENSMUSG00000021930 | Spryd7 | SPRY domain containing 7 | MGI:1913924 |
| ENSMUSG00000026532 | Spta1 | spectrin alpha, erythrocytic 1 | MGI:98385 |
| ENSMUSG00000057738 | Sptan1 | spectrin alpha, non-erythrocytic 1 | MGI:98386 |
| ENSMUSG00000020315 | Sptbn1 | spectrin beta, non-erythrocytic 1 | MGI:98388 |
| ENSMUSG00000067889 | Sptbn2 | spectrin beta, non-erythrocytic 2 | MGI:1313261 |
| ENSMUSG00000011751 | Sptbn4 | spectrin beta, non-erythrocytic 4 | MGI:1890574 |
| ENSMUSG00000021061 | Sptb | spectrin beta, erythrocytic | MGI:98387 |
| ENSMUSG00000005803 | Sqrdl | sulfide quinone reductase-like (yeast) | MGI:1929899 |
| ENSMUSG00000015837 | Sqstm1 | sequestosome 1 | MGI:107931 |
| ENSMUSG00000038453 | Srcin1 | SRC kinase signaling inhibitor 1 | MGI:1933179 |
| ENSMUSG00000027646 | Src | Rous sarcoma oncogene | MGI:98397 |
| ENSMUSG00000026425 | Srgap2 | SLIT-ROBO Rho GTPase activating protein 2 | MGI:109605 |
| ENSMUSG00000030257 | Srgap3 | SLIT-ROBO Rho GTPase activating protein 3 | MGI:2152938 |
| ENSMUSG00000009549 | Srp14 | signal recognition particle 14 | MGI:107169 |
| ENSMUSG00000079108 | Srp54b | signal recognition particle 54B | MGI:3714357 |
| ENSMUSG00000020780 | Srp68 | signal recognition particle 68 | MGI:1917447 |
| ENSMUSG00000062604 | Srpk2 | serine/arginine-rich protein specific kinase 2 | MGI:1201408 |
| ENSMUSG00000001323 | Srr | serine racemase | MGI:1351636 |
| ENSMUSG00000071172 | Srsf3 | serine/arginine-rich splicing factor 3 | MGI:98285 |
| ENSMUSG00000029911 | Ssbp1 | single-stranded DNA binding protein 1 | MGI:1920040 |
| ENSMUSG00000027828 | Ssr3 | signal sequence receptor, gamma | MGI:1914687 |
| ENSMUSG00000002014 | Ssr4 | signal sequence receptor, delta | MGI:1099464 |
| ENSMUSG00000004366 | Sst | somatostatin | MGI:98326 |
| ENSMUSG00000056812 | St8sia3 | ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 3 | MGI:106019 |
| ENSMUSG00000039536 | Stau1 | staufen (RNA binding protein) homolog 1 (Drosophila) | MGI:1338864 |
| ENSMUSG00000039156 | Stim2 | stromal interaction molecule 2 | MGI:2151156 |
| ENSMUSG00000063410 | Stk24 | serine/threonine kinase 24 | MGI:2385007 |
| ENSMUSG00000027581 | Stmn3 | stathmin-like 3 | MGI:1277137 |
| ENSMUSG00000028455 | Stoml2 | stomatin (Epb7.2)-like 2 | MGI:1913842 |
| ENSMUSG00000026880 | Stom | stomatin | MGI:95403 |
| ENSMUSG00000030224 | Strap | serine/threonine kinase receptor associated protein | MGI:1329037 |
| ENSMUSG00000020954 | Strn3 | striatin, calmodulin binding protein 3 | MGI:2151064 |
| ENSMUSG00000030374 | Strn4 | striatin, calmodulin binding protein 4 | MGI:2142346 |
| ENSMUSG00000024077 | Strn | striatin, calmodulin binding protein | MGI:1333757 |
| ENSMUSG00000032116 | Stt3a | STT3, subunit of the oligosaccharyltransferase complex, homolog A (S. cerevisiae) | MGI:105124 |
| ENSMUSG00000032437 | Stt3b | STT3, subunit of the oligosaccharyltransferase complex, homolog B (S. cerevisiae) | MGI:1915542 |
| ENSMUSG00000039615 | Stub1 | STIP1 homology and U-Box containing protein 1 | MGI:1891731 |
| ENSMUSG00000028879 | Stx12 | syntaxin 12 | MGI:1931027 |
| ENSMUSG00000027522 | Stx16 | syntaxin 16 | MGI:1923396 |
| ENSMUSG00000061455 | Stx17 | syntaxin 17 | MGI:1914977 |
| ENSMUSG00000029125 | Stx18 | syntaxin 18 | MGI:1918366 |
| ENSMUSG00000010110 | Stx5a | syntaxin 5A | MGI:1928483 |
| ENSMUSG00000026470 | Stx6 | syntaxin 6 | MGI:1926235 |
| ENSMUSG00000019998 | Stx7 | syntaxin 7 | MGI:1858210 |
| ENSMUSG00000020903 | Stx8 | syntaxin 8 | MGI:1890156 |
| ENSMUSG00000046314 | Stxbp6 | syntaxin binding protein 6 (amisyn) | MGI:2384963 |
| ENSMUSG00000052738 | Suclg1 | succinate-CoA ligase, GDP-forming, alpha subunit | MGI:1927234 |
| ENSMUSG00000022024 | Sugt1 | SGT1, suppressor of G2 allele of SKP1 (S. cerevisiae) | MGI:1915205 |
| ENSMUSG00000022340 | Sybu | syntabulin (syntaxin-interacting) | MGI:2442392 |
| ENSMUSG00000037217 | Syn1 | synapsin I | MGI:98460 |
| ENSMUSG00000009394 | Syn2 | synapsin II | MGI:103020 |
| ENSMUSG00000059602 | Syn3 | synapsin III | MGI:1351334 |
| ENSMUSG00000032423 | Syncrip | synaptotagmin binding, cytoplasmic RNA interacting protein | MGI:1891690 |
| ENSMUSG00000074736 | Syndig1 | synapse differentiation inducing 1 | MGI:3702158 |
| ENSMUSG00000096054 | Syne1 | spectrin repeat containing, nuclear envelope 1 | MGI:1927152 |
| ENSMUSG00000067629 | Syngap1 | synaptic Ras GTPase activating protein 1 homolog (rat) | MGI:3039785 |
| ENSMUSG00000007021 | Syngr3 | synaptogyrin 3 | MGI:1341881 |
| ENSMUSG00000043079 | Synpo | synaptopodin | MGI:1099446 |
| ENSMUSG00000068923 | Syt11 | synaptotagmin XI | MGI:1859547 |
| ENSMUSG00000027220 | Syt13 | synaptotagmin XIII | MGI:1933945 |
| ENSMUSG00000058420 | Syt17 | synaptotagmin XVII | MGI:104966 |
| ENSMUSG00000035864 | Syt1 | synaptotagmin I | MGI:99667 |
| ENSMUSG00000030731 | Syt3 | synaptotagmin III | MGI:99665 |
| ENSMUSG00000004961 | Syt5 | synaptotagmin V | MGI:1926368 |
| ENSMUSG00000024743 | Syt7 | synaptotagmin VII | MGI:1859545 |
| ENSMUSG00000054453 | Sytl5 | synaptotagmin-like 5 | MGI:2668451 |
| ENSMUSG00000065954 | Tacc1 | transforming, acidic coiled-coil containing protein 1 | MGI:2443510 |
| ENSMUSG00000022658 | Tagln3 | transgelin 3 | MGI:1926784 |
| ENSMUSG00000030316 | Tamm41 | TAM41, mitochondrial translocator assembly and maintenance protein, homolog (S. cerevisiae) | MGI:1916221 |
| ENSMUSG00000035168 | Tanc1 | tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1 | MGI:1914110 |
| ENSMUSG00000053580 | Tanc2 | tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2 | MGI:2444121 |
| ENSMUSG00000017291 | Taok1 | TAO kinase 1 | MGI:1914490 |
| ENSMUSG00000059981 | Taok2 | TAO kinase 2 | MGI:1915919 |
| ENSMUSG00000041459 | Tardbp | TAR DNA binding protein | MGI:2387629 |
| ENSMUSG00000034412 | Tbc1d10a | TBC1 domain family, member 10a | MGI:2144164 |
| ENSMUSG00000042492 | Tbc1d10b | TBC1 domain family, member 10b | MGI:1915699 |
| ENSMUSG00000036473 | Tbc1d24 | TBC1 domain family, member 24 | MGI:2443456 |
| ENSMUSG00000020115 | Tbk1 | TANK-binding kinase 1 | MGI:1929658 |
| ENSMUSG00000038517 | Tbkbp1 | TBK1 binding protein 1 | MGI:1920424 |
| ENSMUSG00000005374 | Tbl2 | transducin (beta)-like 2 | MGI:1351652 |
| ENSMUSG00000079658 | Tceb1 | transcription elongation factor B (SIII), polypeptide 1 | MGI:1915173 |
| ENSMUSG00000055839 | Tceb2 | transcription elongation factor B (SIII), polypeptide 2 | MGI:1914923 |
| ENSMUSG00000068039 | Tcp1 | t-complex protein 1 | MGI:98535 |
| ENSMUSG00000022019 | Tdrd3 | tudor domain containing 3 | MGI:2444023 |
| ENSMUSG00000041912 | Tdrkh | tudor and KH domain containing protein | MGI:1919884 |
| ENSMUSG00000031708 | Tecr | trans-2,3-enoyl-CoA reductase | MGI:1915408 |
| ENSMUSG00000049336 | Tenm2 | teneurin transmembrane protein 2 | MGI:1345184 |
| ENSMUSG00000031561 | Tenm3 | teneurin transmembrane protein 3 | MGI:1345183 |
| ENSMUSG00000009670 | Tex11 | testis expressed gene 11 | MGI:1933237 |
| ENSMUSG00000003923 | Tfam | transcription factor A, mitochondrial | MGI:107810 |
| ENSMUSG00000056665 | Them6 | thioesterase superfamily member 6 | MGI:1925301 |
| ENSMUSG00000032011 | Thy1 | thymus cell antigen 1, theta | MGI:98747 |
| ENSMUSG00000038028 | Tigar | Trp53 induced glycolysis repulatory phosphatase | MGI:2442752 |
| ENSMUSG00000020219 | Timm13 | translocase of inner mitochondrial membrane 13 | MGI:1353432 |
| ENSMUSG00000031158 | Timm17b | translocase of inner mitochondrial membrane 17b | MGI:1343176 |
| ENSMUSG00000020843 | Timm22 | translocase of inner mitochondrial membrane 22 | MGI:1929742 |
| ENSMUSG00000002949 | Timm44 | translocase of inner mitochondrial membrane 44 | MGI:1343262 |
| ENSMUSG00000030516 | Tjp1 | tight junction protein 1 | MGI:98759 |
| ENSMUSG00000024812 | Tjp2 | tight junction protein 2 | MGI:1341872 |
| ENSMUSG00000042066 | Tmcc2 | transmembrane and coiled-coil domains 2 | MGI:1916125 |
| ENSMUSG00000052428 | Tmco1 | transmembrane and coiled-coil domains 1 | MGI:1921173 |
| ENSMUSG00000021248 | Tmed10 | transmembrane emp24-like trafficking protein 10 (yeast) | MGI:1915831 |
| ENSMUSG00000058569 | Tmed9 | transmembrane emp24 protein transport domain containing 9 | MGI:1914761 |
| ENSMUSG00000028347 | Tmeff1 | transmembrane protein with EGF-like and two follistatin-like domains 1 | MGI:1926810 |
| ENSMUSG00000026109 | Tmeff2 | transmembrane protein with EGF-like and two follistatin-like domains 2 | MGI:1861735 |
| ENSMUSG00000034659 | Tmem109 | transmembrane protein 109 | MGI:1915789 |
| ENSMUSG00000043284 | Tmem11 | transmembrane protein 11 | MGI:2144726 |
| ENSMUSG00000030615 | Tmem126a | transmembrane protein 126A | MGI:1913521 |
| ENSMUSG00000070498 | Tmem132b | transmembrane protein 132B | MGI:3609245 |
| ENSMUSG00000026347 | Tmem163 | transmembrane protein 163 | MGI:1919410 |
| ENSMUSG00000057716 | Tmem178b | transmembrane protein 178B | MGI:3647581 |
| ENSMUSG00000038828 | Tmem214 | transmembrane protein 214 | MGI:1916046 |
| ENSMUSG00000038079 | Tmem237 | transmembrane protein 237 | MGI:2138365 |
| ENSMUSG00000084845 | Tmem240 | transmembrane protein 240 | MGI:3648074 |
| ENSMUSG00000070394 | Tmem256 | transmembrane protein 256 | MGI:1916436 |
| ENSMUSG00000036372 | Tmem258 | transmembrane protein 258 | MGI:1916288 |
| ENSMUSG00000032328 | Tmem30a | transmembrane protein 30A | MGI:106402 |
| ENSMUSG00000037720 | Tmem33 | transmembrane protein 33 | MGI:1915128 |
| ENSMUSG00000033578 | Tmem35 | transmembrane protein 35 | MGI:1914814 |
| ENSMUSG00000025666 | Tmem47 | transmembrane protein 47 | MGI:2177570 |
| ENSMUSG00000028221 | Tmem55a | transmembrane protein 55A | MGI:1919769 |
| ENSMUSG00000028826 | Tmem57 | transmembrane protein 57 | MGI:1913396 |
| ENSMUSG00000036026 | Tmem63b | transmembrane protein 63b | MGI:2387609 |
| ENSMUSG00000028232 | Tmem68 | transmembrane protein 68 | MGI:1919348 |
| ENSMUSG00000014353 | Tmem87b | transmembrane protein 87B | MGI:1919727 |
| ENSMUSG00000031021 | Tmem9b | TMEM9 domain family, member B | MGI:1915254 |
| ENSMUSG00000028328 | Tmod1 | tropomodulin 1 | MGI:98775 |
| ENSMUSG00000032186 | Tmod2 | tropomodulin 2 | MGI:1355335 |
| ENSMUSG00000058587 | Tmod3 | tropomodulin 3 | MGI:1355315 |
| ENSMUSG00000019961 | Tmpo | thymopoietin | MGI:106920 |
| ENSMUSG00000049775 | Tmsb4x | thymosin, beta 4, X chromosome | MGI:99510 |
| ENSMUSG00000028364 | Tnc | tenascin C | MGI:101922 |
| ENSMUSG00000027692 | Tnik | TRAF2 and NCK interacting kinase | MGI:1916264 |
| ENSMUSG00000015829 | Tnr | tenascin R | MGI:99516 |
| ENSMUSG00000025139 | Tollip | toll interacting protein | MGI:1891808 |
| ENSMUSG00000000538 | Tom1l2 | target of myb1-like 2 (chicken) | MGI:2443306 |
| ENSMUSG00000042870 | Tom1 | target of myb1 homolog (chicken) | MGI:1338026 |
| ENSMUSG00000093904 | Tomm20 | translocase of outer mitochondrial membrane 20 homolog (yeast) | MGI:1915202 |
| ENSMUSG00000022427 | Tomm22 | translocase of outer mitochondrial membrane 22 homolog (yeast) | MGI:2450248 |
| ENSMUSG00000005674 | Tomm40l | translocase of outer mitochondrial membrane 40 homolog-like (yeast) | MGI:3589112 |
| ENSMUSG00000002984 | Tomm40 | translocase of outer mitochondrial membrane 40 homolog (yeast) | MGI:1858259 |
| ENSMUSG00000078713 | Tomm5 | translocase of outer mitochondrial membrane 5 homolog (yeast) | MGI:1915762 |
| ENSMUSG00000033475 | Tomm6 | translocase of outer mitochondrial membrane 6 homolog (yeast) | MGI:1913369 |
| ENSMUSG00000022752 | Tomm70a | translocase of outer mitochondrial membrane 70 homolog A (yeast) | MGI:106295 |
| ENSMUSG00000059323 | Tonsl | tonsoku-like, DNA repair protein | MGI:1919999 |
| ENSMUSG00000000296 | Tpd52l1 | tumor protein D52-like 1 | MGI:1298386 |
| ENSMUSG00000000827 | Tpd52l2 | tumor protein D52-like 2 | MGI:1913564 |
| ENSMUSG00000027506 | Tpd52 | tumor protein D52 | MGI:107749 |
| ENSMUSG00000020308 | Tpgs1 | tubulin polyglutamylase complex subunit 1 | MGI:106618 |
| ENSMUSG00000023456 | Tpi1 | triosephosphate isomerase 1 | MGI:98797 |
| ENSMUSG00000032366 | Tpm1 | tropomyosin 1, alpha | MGI:98809 |
| ENSMUSG00000027940 | Tpm3 | tropomyosin 3, gamma | MGI:1890149 |
| ENSMUSG00000021573 | Tppp | tubulin polymerization promoting protein | MGI:1920198 |
| ENSMUSG00000048707 | Tprn | taperin | MGI:2139535 |
| ENSMUSG00000021277 | Traf3 | TNF receptor-associated factor 3 | MGI:108041 |
| ENSMUSG00000000374 | Trappc10 | trafficking protein particle complex 10 | MGI:1336209 |
| ENSMUSG00000028847 | Trappc3 | trafficking protein particle complex 3 | MGI:1351486 |
| ENSMUSG00000040236 | Trappc5 | trafficking protein particle complex 5 | MGI:1913932 |
| ENSMUSG00000020993 | Trappc6b | trafficking protein particle complex 6B | MGI:1925482 |
| ENSMUSG00000033382 | Trappc8 | trafficking protein particle complex 8 | MGI:2443008 |
| ENSMUSG00000047921 | Trappc9 | trafficking protein particle complex 9 | MGI:1923760 |
| ENSMUSG00000050663 | Trhde | TRH-degrading enzyme | MGI:2384311 |
| ENSMUSG00000021712 | Trim23 | tripartite motif-containing 23 | MGI:1933161 |
| ENSMUSG00000027993 | Trim2 | tripartite motif-containing 2 | MGI:1933163 |
| ENSMUSG00000036989 | Trim3 | tripartite motif-containing 3 | MGI:1860040 |
| ENSMUSG00000042766 | Trim46 | tripartite motif-containing 46 | MGI:2673000 |
| ENSMUSG00000021071 | Trim9 | tripartite motif-containing 9 | MGI:2137354 |
| ENSMUSG00000022263 | Trio | triple functional domain (PTPRF interacting) | MGI:1927230 |
| ENSMUSG00000021188 | Trip11 | thyroid hormone receptor interactor 11 | MGI:1924393 |
| ENSMUSG00000055963 | Triqk | triple QxxK/R motif containing | MGI:3650048 |
| ENSMUSG00000026510 | Trp53bp2 | transformation related protein 53 binding protein 2 | MGI:2138319 |
| ENSMUSG00000068735 | Trp53i11 | transformation related protein 53 inducible protein 11 | MGI:2670995 |
| ENSMUSG00000018507 | Trpv2 | transient receptor potential cation channel, subfamily V, member 2 | MGI:1341836 |
| ENSMUSG00000029723 | Tsc22d4 | TSC22 domain family, member 4 | MGI:1926079 |
| ENSMUSG00000002496 | Tsc2 | tuberous sclerosis 2 | MGI:102548 |
| ENSMUSG00000014402 | Tsg101 | tumor susceptibility gene 101 | MGI:106581 |
| ENSMUSG00000027858 | Tspan2 | tetraspanin 2 | MGI:1917997 |
| ENSMUSG00000033530 | Ttc7b | tetratricopeptide repeat domain 7B | MGI:2144724 |
| ENSMUSG00000007944 | Ttc9b | tetratricopeptide repeat domain 9B | MGI:1920282 |
| ENSMUSG00000071660 | Ttc9c | tetratricopeptide repeat domain 9C | MGI:1917637 |
| ENSMUSG00000042734 | Ttc9 | tetratricopeptide repeat domain 9 | MGI:1916730 |
| ENSMUSG00000051747 | Ttn | titin | MGI:98864 |
| ENSMUSG00000043091 | Tuba1c | tubulin, alpha 1C | MGI:1095409 |
| ENSMUSG00000026202 | Tuba4a | tubulin, alpha 4A | MGI:1095410 |
| ENSMUSG00000030137 | Tuba8 | tubulin, alpha 8 | MGI:1858225 |
| ENSMUSG00000058672 | Tubb2a | tubulin, beta 2A class IIA | MGI:107861 |
| ENSMUSG00000045136 | Tubb2b | tubulin, beta 2B class IIB | MGI:1920960 |
| ENSMUSG00000062380 | Tubb3 | tubulin, beta 3 class III | MGI:107813 |
| ENSMUSG00000062591 | Tubb4a | tubulin, beta 4A class IVA | MGI:107848 |
| ENSMUSG00000036752 | Tubb4b | tubulin, beta 4B class IVB | MGI:1915472 |
| ENSMUSG00000001525 | Tubb5 | tubulin, beta 5 class I | MGI:107812 |
| ENSMUSG00000001473 | Tubb6 | tubulin, beta 6 class V | MGI:1915201 |
| ENSMUSG00000045007 | Tubg2 | tubulin, gamma 2 | MGI:2144208 |
| ENSMUSG00000000759 | Tubgcp3 | tubulin, gamma complex associated protein 3 | MGI:2183752 |
| ENSMUSG00000073838 | Tufm | Tu translation elongation factor, mitochondrial | MGI:1923686 |
| ENSMUSG00000039530 | Tusc3 | tumor suppressor candidate 3 | MGI:1933134 |
| ENSMUSG00000022451 | Twf1 | twinfilin, actin-binding protein, homolog 1 (Drosophila) | MGI:1100520 |
| ENSMUSG00000023277 | Twf2 | twinfilin, actin-binding protein, homolog 2 (Drosophila) | MGI:1346078 |
| ENSMUSG00000028567 | Txndc12 | thioredoxin domain containing 12 (endoplasmic reticulum) | MGI:1913323 |
| ENSMUSG00000024583 | Txnl1 | thioredoxin-like 1 | MGI:1860078 |
| ENSMUSG00000034485 | Uaca | uveal autoantigen with coiled-coil domains and ankyrin repeats | MGI:1919815 |
| ENSMUSG00000042520 | Ubap2l | ubiquitin-associated protein 2-like | MGI:1921633 |
| ENSMUSG00000074781 | Ube2n | ubiquitin-conjugating enzyme E2N | MGI:1934835 |
| ENSMUSG00000052981 | Ube2ql1 | ubiquitin-conjugating enzyme E2Q family-like 1 | MGI:1924230 |
| ENSMUSG00000078923 | Ube2v1 | ubiquitin-conjugating enzyme E2 variant 1 | MGI:1913839 |
| ENSMUSG00000022674 | Ube2v2 | ubiquitin-conjugating enzyme E2 variant 2 | MGI:1917870 |
| ENSMUSG00000001687 | Ubl3 | ubiquitin-like 3 | MGI:1344373 |
| ENSMUSG00000015290 | Ubl4 | ubiquitin-like 4 | MGI:95049 |
| ENSMUSG00000009741 | Ubp1 | upstream binding protein 1 | MGI:104889 |
| ENSMUSG00000066036 | Ubr4 | ubiquitin protein ligase E3 component n-recognin 4 | MGI:1916366 |
| ENSMUSG00000020923 | Ubtf | upstream binding transcription factor, RNA polymerase I | MGI:98512 |
| ENSMUSG00000026353 | Ubxn4 | UBX domain protein 4 | MGI:1915062 |
| ENSMUSG00000019578 | Ubxn6 | UBX domain protein 6 | MGI:1913780 |
| ENSMUSG00000062963 | Ufc1 | ubiquitin-fold modifier conjugating enzyme 1 | MGI:1913405 |
| ENSMUSG00000040359 | Ufl1 | UFM1 specific ligase 1 | MGI:1914740 |
| ENSMUSG00000032854 | Ugt8a | UDP galactosyltransferase 8A | MGI:109522 |
| ENSMUSG00000034799 | Unc13a | unc-13 homolog A (C. elegans) | MGI:3051532 |
| ENSMUSG00000028456 | Unc13b | unc-13 homolog B (C. elegans) | MGI:1342278 |
| ENSMUSG00000062151 | Unc13c | unc-13 homolog C (C. elegans) | MGI:2149021 |
| ENSMUSG00000021198 | Unc79 | unc-79 homolog (C. elegans) | MGI:2684729 |
| ENSMUSG00000055567 | Unc80 | unc-80 homolog (C. elegans) | MGI:2652882 |
| ENSMUSG00000058301 | Upf1 | UPF1 regulator of nonsense transcripts homolog (yeast) | MGI:107995 |
| ENSMUSG00000024208 | Uqcc2 | ubiquinol-cytochrome c reductase complex assembly factor 2 | MGI:1914517 |
| ENSMUSG00000044894 | Uqcrq | ubiquinol-cytochrome c reductase, complex III subunit VII | MGI:107807 |
| ENSMUSG00000039929 | Urb1 | URB1 ribosome biogenesis 1 homolog (S. cerevisiae) | MGI:2146468 |
| ENSMUSG00000071528 | Usmg5 | upregulated during skeletal muscle growth 5 | MGI:1891435 |
| ENSMUSG00000029592 | Usp30 | ubiquitin specific peptidase 30 | MGI:2140991 |
| ENSMUSG00000063317 | Usp31 | ubiquitin specific peptidase 31 | MGI:1923429 |
| ENSMUSG00000054814 | Usp46 | ubiquitin specific peptidase 46 | MGI:1916977 |
| ENSMUSG00000039701 | Usp53 | ubiquitin specific peptidase 53 | MGI:2139607 |
| ENSMUSG00000027363 | Usp8 | ubiquitin specific peptidase 8 | MGI:1934029 |
| ENSMUSG00000019820 | Utrn | utrophin | MGI:104631 |
| ENSMUSG00000010936 | Vac14 | Vac14 homolog (S. cerevisiae) | MGI:2157980 |
| ENSMUSG00000030337 | Vamp1 | vesicle-associated membrane protein 1 | MGI:1313276 |
| ENSMUSG00000020894 | Vamp2 | vesicle-associated membrane protein 2 | MGI:1313277 |
| ENSMUSG00000028955 | Vamp3 | vesicle-associated membrane protein 3 | MGI:1321389 |
| ENSMUSG00000026696 | Vamp4 | vesicle-associated membrane protein 4 | MGI:1858730 |
| ENSMUSG00000073002 | Vamp5 | vesicle-associated membrane protein 5 | MGI:1858622 |
| ENSMUSG00000051412 | Vamp7 | vesicle-associated membrane protein 7 | MGI:1096399 |
| ENSMUSG00000026556 | Vangl2 | vang-like 2 (van gogh, Drosophila) | MGI:2135272 |
| ENSMUSG00000024091 | Vapa | vesicle-associated membrane protein, associated protein A | MGI:1353561 |
| ENSMUSG00000054455 | Vapb | vesicle-associated membrane protein, associated protein B and C | MGI:1928744 |
| ENSMUSG00000007029 | Vars | valyl-tRNA synthetase | MGI:90675 |
| ENSMUSG00000030403 | Vasp | vasodilator-stimulated phosphoprotein | MGI:109268 |
| ENSMUSG00000021614 | Vcan | versican | MGI:102889 |
| ENSMUSG00000021823 | Vcl | vinculin | MGI:98927 |
| ENSMUSG00000020402 | Vdac1 | voltage-dependent anion channel 1 | MGI:106919 |
| ENSMUSG00000021771 | Vdac2 | voltage-dependent anion channel 2 | MGI:106915 |
| ENSMUSG00000008892 | Vdac3 | voltage-dependent anion channel 3 | MGI:106922 |
| ENSMUSG00000037428 | Vgf | VGF nerve growth factor inducible | MGI:1343180 |
| ENSMUSG00000075701 | Vimp | VCP-interacting membrane protein | MGI:95994 |
| ENSMUSG00000026728 | Vim | vimentin | MGI:98932 |
| ENSMUSG00000032127 | Vps11 | vacuolar protein sorting 11 (yeast) | MGI:1918982 |
| ENSMUSG00000027411 | Vps16 | vacuolar protein sorting 16 (yeast) | MGI:2136772 |
| ENSMUSG00000034216 | Vps18 | vacuolar protein sorting 18 (yeast) | MGI:2443626 |
| ENSMUSG00000078656 | Vps25 | vacuolar protein sorting 25 (yeast) | MGI:106354 |
| ENSMUSG00000031988 | Vps26b | vacuolar protein sorting 26 homolog B (yeast) | MGI:1917656 |
| ENSMUSG00000029462 | Vps29 | vacuolar protein sorting 29 (S. pombe) | MGI:1928344 |
| ENSMUSG00000029434 | Vps33a | vacuolar protein sorting 33A (yeast) | MGI:1924823 |
| ENSMUSG00000066278 | Vps37b | vacuolar protein sorting 37B (yeast) | MGI:1916724 |
| ENSMUSG00000048832 | Vps37c | vacuolar protein sorting 37C (yeast) | MGI:2147661 |
| ENSMUSG00000027291 | Vps39 | vacuolar protein sorting 39 (yeast) | MGI:2443189 |
| ENSMUSG00000041236 | Vps41 | vacuolar protein sorting 41 (yeast) | MGI:1929215 |
| ENSMUSG00000015747 | Vps45 | vacuolar protein sorting 45 (yeast) | MGI:891965 |
| ENSMUSG00000031913 | Vps4a | vacuolar protein sorting 4a (yeast) | MGI:1890520 |
| ENSMUSG00000024797 | Vps51 | vacuolar protein sorting 51 homolog (S. cerevisiae) | MGI:1915755 |
| ENSMUSG00000024319 | Vps52 | vacuolar protein sorting 52 (yeast) | MGI:1330304 |
| ENSMUSG00000017288 | Vps53 | vacuolar protein sorting 53 (yeast) | MGI:1915549 |
| ENSMUSG00000001062 | Vps9d1 | VPS9 domain containing 1 | MGI:1914143 |
| ENSMUSG00000054459 | Vsnl1 | visinin-like 1 | MGI:1349453 |
| ENSMUSG00000019868 | Vta1 | Vps20-associated 1 homolog (S. cerevisiae) | MGI:1913451 |
| ENSMUSG00000024983 | Vti1a | vesicle transport through interaction with t-SNAREs 1A | MGI:1855699 |
| ENSMUSG00000021124 | Vti1b | vesicle transport through interaction with t-SNAREs 1B | MGI:1855688 |
| ENSMUSG00000058997 | Vwa8 | von Willebrand factor A domain containing 8 | MGI:1919008 |
| ENSMUSG00000019831 | Wasf1 | WAS protein family, member 1 | MGI:1890563 |
| ENSMUSG00000028868 | Wasf2 | WAS protein family, member 2 | MGI:1098641 |
| ENSMUSG00000029636 | Wasf3 | WAS protein family, member 3 | MGI:2658986 |
| ENSMUSG00000024101 | Wash1 | WAS protein family homolog 1 | MGI:1916017 |
| ENSMUSG00000029684 | Wasl | Wiskott-Aldrich syndrome-like (human) | MGI:1920428 |
| ENSMUSG00000034040 | Wbscr17 | Williams-Beuren syndrome chromosome region 17 homolog (human) | MGI:2137594 |
| ENSMUSG00000037957 | Wdr20 | WD repeat domain 20 | MGI:1916891 |
| ENSMUSG00000021147 | Wdr37 | WD repeat domain 37 | MGI:1920393 |
| ENSMUSG00000040389 | Wdr47 | WD repeat domain 47 | MGI:2139593 |
| ENSMUSG00000032512 | Wdr48 | WD repeat domain 48 | MGI:1914811 |
| ENSMUSG00000040560 | Wdr7 | WD repeat domain 7 | MGI:1860197 |
| ENSMUSG00000039474 | Wfs1 | Wolfram syndrome 1 homolog (human) | MGI:1328355 |
| ENSMUSG00000064030 | Wibg | within bgcn homolog (Drosophila) | MGI:1925678 |
| ENSMUSG00000038013 | Wipf2 | WAS/WASL interacting protein family, member 2 | MGI:1924462 |
| ENSMUSG00000086040 | Wipf3 | WAS/WASL interacting protein family, member 3 | MGI:3044681 |
| ENSMUSG00000037989 | Wnk2 | WNK lysine deficient protein kinase 2 | MGI:1922857 |
| ENSMUSG00000021994 | Wnt5a | wingless-type MMTV integration site family, member 5A | MGI:98958 |
| ENSMUSG00000004637 | Wwox | WW domain-containing oxidoreductase | MGI:1931237 |
| ENSMUSG00000031930 | Wwp2 | WW domain containing E3 ubiquitin protein ligase 2 | MGI:1914144 |
| ENSMUSG00000027022 | Xirp2 | xin actin-binding repeat containing 2 | MGI:2685198 |
| ENSMUSG00000051951 | Xkr4 | X Kell blood group precursor related family member 4 | MGI:3528744 |
| ENSMUSG00000035067 | Xkr6 | X Kell blood group precursor related family member 6 homolog | MGI:2447765 |
| ENSMUSG00000022401 | Xpnpep3 | X-prolyl aminopeptidase (aminopeptidase P) 3, putative | MGI:2445217 |
| ENSMUSG00000022792 | Yars2 | tyrosyl-tRNA synthetase 2 (mitochondrial) | MGI:1917370 |
| ENSMUSG00000028811 | Yars | tyrosyl-tRNA synthetase | MGI:2147627 |
| ENSMUSG00000028639 | Ybx1 | Y box protein 1 | MGI:99146 |
| ENSMUSG00000014932 | Yes1 | Yamaguchi sarcoma viral (v-yes) oncogene homolog 1 | MGI:99147 |
| ENSMUSG00000002741 | Ykt6 | YKT6 homolog (S. Cerevisiae) | MGI:1927550 |
| ENSMUSG00000026775 | Yme1l1 | YME1-like 1 (S. cerevisiae) | MGI:1351651 |
| ENSMUSG00000018326 | Ywhab | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, beta polypeptide | MGI:1891917 |
| ENSMUSG00000020849 | Ywhae | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, epsilon polypeptide | MGI:894689 |
| ENSMUSG00000051391 | Ywhag | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, gamma polypeptide | MGI:108109 |
| ENSMUSG00000018965 | Ywhah | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, eta polypeptide | MGI:109194 |
| ENSMUSG00000076432 | Ywhaq | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta polypeptide | MGI:891963 |
| ENSMUSG00000022285 | Ywhaz | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide | MGI:109484 |
| ENSMUSG00000034173 | Zbed5 | zinc finger, BED type containing 5 | MGI:1919220 |
| ENSMUSG00000043542 | Zc2hc1a | zinc finger, C2HC-type containing 1A | MGI:1914556 |
| ENSMUSG00000022390 | Zc3h7b | zinc finger CCCH type containing 7B | MGI:1328310 |
| ENSMUSG00000034075 | Zdhhc5 | zinc finger, DHHC domain containing 5 | MGI:1923573 |
| ENSMUSG00000060166 | Zdhhc8 | zinc finger, DHHC domain containing 8 | MGI:1338012 |
| ENSMUSG00000037855 | Zfp365 | zinc finger protein 365 | MGI:2143676 |
| ENSMUSG00000032264 | Zw10 | zw10 kinetochore protein | MGI:1349478 |
| ENSMUSG00000019923 | Zwint | ZW10 interactor | MGI:1289227 |
| ENSMUSG00000045414 | 1190002N15Rik | RIKEN cDNA 1190002N15 gene | MGI:1916111 |
| ENSMUSG00000087651 | 1500009L16Rik | RIKEN cDNA 1500009L16 gene | MGI:1917034 |
| ENSMUSG00000043629 | 1700019D03Rik | RIKEN cDNA 1700019D03 gene | MGI:1914330 |
| ENSMUSG00000032666 | 1700025G04Rik | RIKEN cDNA 1700025G04 gene | MGI:1916649 |
| ENSMUSG00000021290 | 2010107E04Rik | RIKEN cDNA 2010107E04 gene | MGI:1917507 |
| ENSMUSG00000026090 | 2010300C02Rik | RIKEN cDNA 2010300C02 gene | MGI:1919347 |
| ENSMUSG00000031983 | 2310022B05Rik | RIKEN cDNA 2310022B05 gene | MGI:1916801 |
| ENSMUSG00000026319 | 2310035C23Rik | RIKEN cDNA 2310035C23 gene | MGI:1922832 |
| ENSMUSG00000045411 | 2410002F23Rik | RIKEN cDNA 2410002F23 gene | MGI:1914226 |
| ENSMUSG00000049760 | 2410015M20Rik | RIKEN cDNA 2410015M20 gene | MGI:2442174 |
| ENSMUSG00000021807 | 2700060E02Rik | RIKEN cDNA 2700060E02 gene | MGI:1915295 |
| ENSMUSG00000024410 | 3110002H16Rik | RIKEN cDNA 3110002H16 gene | MGI:1916528 |
| ENSMUSG00000036046 | 5031439G07Rik | RIKEN cDNA 5031439G07 gene | MGI:2444899 |
| ENSMUSG00000053963 | 6330403A02Rik | RIKEN cDNA 6330403A02 gene | MGI:2138735 |
| ENSMUSG00000033960 | 9430020K01Rik | RIKEN cDNA 9430020K01 gene | MGI:2685174 |
| ENSMUSG00000034560 | A230046K03Rik | RIKEN cDNA A230046K03 gene | MGI:2441787 |
| ENSMUSG00000044060 | A830010M20Rik | RIKEN cDNA A830010M20 gene | MGI:2445097 |
Supplementary Table 2a. Synaptosomal proteins identified in zebrafish.
Proteins identified in zebrafish whole brain synaptosomal preparations. For each protein the following database identifiers are given: Ensembl Gene, Associated Gene Name, Description and MGI ID.
| ENSEMBL ID | Associated Gene Name | Description | EntrezGene ID |
|---|---|---|---|
| ENSDARG00000041685 | A2ML1 (3 of 13) | alpha-2-macroglobulin-like 1 | 100006782 |
| ENSDARG00000056314 | a2ml | alpha-2-macroglobulin-like | 100006972 |
| ENSDARG00000011855 | aak1a | AP2 associated kinase 1a | 568999 |
| ENSDARG00000077686 | aak1b | AP2 associated kinase 1b | 100534954 |
| ENSDARG00000069142 | aars | alanyl-tRNA synthetase | 324940 |
| ENSDARG00000051816 | aass | aminoadipate-semialdehyde synthase | 556229 |
| ENSDARG00000006031 | abat | 4-aminobutyrate aminotransferase | 378968 |
| ENSDARG00000013500 | abca2 | ATP-binding cassette, sub-family A (ABC1), member 2 | 100006090 |
| ENSDARG00000057169 | abca4a | ATP-binding cassette, sub-family A (ABC1), member 4a | 798993 |
| ENSDARG00000074041 | abca5 | ATP-binding cassette, sub-family A (ABC1), member 5 | 100151075 |
| ENSDARG00000061591 | abcb10 | ATP-binding cassette, sub-family B (MDR/TAP), member 10 | 100003744 |
| ENSDARG00000010936 | abcb4 | ATP-binding cassette, sub-family B (MDR/TAP), member 4 | 100136865 |
| ENSDARG00000063297 | abcb6a | ATP-binding cassette, sub-family B (MDR/TAP), member 6a | 564067 |
| ENSDARG00000062795 | abcb7 | ATP-binding cassette, sub-family B (MDR/TAP), member 7 | 566515 |
| ENSDARG00000056672 | abcb8 | ATP-binding cassette, sub-family B (MDR/TAP), member 8 | 548340 |
| ENSDARG00000014031 | abcc2 | ATP-binding cassette, sub-family C (CFTR/MRP), member 2 | 393561 |
| ENSDARG00000051879 | abcc8 | ATP-binding cassette, sub-family C (CFTR/MRP), member 8 | 553281 |
| ENSDARG00000074876 | abcd1 | ATP-binding cassette, sub-family D (ALD), member 1 | 566367 |
| ENSDARG00000059409 | ABCD2 (1 of 2) | ATP-binding cassette, sub-family D (ALD), member 2 | 100537809 |
| ENSDARG00000104085 | abcd3a | ATP-binding cassette, sub-family D (ALD), member 3a | 406803 |
| ENSDARG00000007216 | abce1 | ATP-binding cassette, sub-family E (OABP), member 1 | 406324 |
| ENSDARG00000031795 | abcf1 | ATP-binding cassette, sub-family F (GCN20), member 1 | 406467 |
| ENSDARG00000100075 | abcg2a | ATP-binding cassette, sub-family G (WHITE), member 2a | 735312 |
| ENSDARG00000058574 | abcg2c | ATP-binding cassette, sub-family G (WHITE), member 2c | 569858 |
| ENSDARG00000069501 | abhd11 | abhydrolase domain containing 11 | 446169 |
| ENSDARG00000071004 | abhd12 | abhydrolase domain containing 12 | 767657 |
| ENSDARG00000058367 | abhd14a | abhydrolase domain containing 14A | 404598 |
| ENSDARG00000100531 | abhd14b | abhydrolase domain containing 14B | 393338 |
| ENSDARG00000078929 | abhd16a | abhydrolase domain containing 16A | 564613 |
| ENSDARG00000091433 | abhd17aa | abhydrolase domain containing 17Aa | 322121 |
| ENSDARG00000010155 | abi1a | abl-interactor 1a | 393711 |
| ENSDARG00000062991 | abi1b | abl-interactor 1b | 767738 |
| ENSDARG00000058207 | abi2a | abl-interactor 2a | 767646 |
| ENSDARG00000014875 | abi2b | abl-interactor 2b | 692331 |
| ENSDARG00000060072 | abi3a | ABI family, member 3a | 767807 |
| ENSDARG00000063283 | abi3b | ABI family, member 3b | 566059 |
| ENSDARG00000013841 | abl2 | c-abl oncogene 2, non-receptor tyrosine kinase | 570636 |
| ENSDARG00000060149 | ablim1a | actin binding LIM protein 1a | 100003558 |
| ENSDARG00000055223 | ablim2 | actin binding LIM protein family, member 2 | 568454 |
| ENSDARG00000004687 | acaa1 | acetyl-CoA acyltransferase 1 | 431754 |
| ENSDARG00000099056 | ACAA2 (2 of 2) | acetyl-CoA acyltransferase 2 | 0 |
| ENSDARG00000038881 | acaa2 | acetyl-CoA acyltransferase 2 | 406325 |
| ENSDARG00000078512 | acaca | acetyl-CoA carboxylase alpha | 559403 |
| ENSDARG00000045411 | acad11 | acyl-CoA dehydrogenase family, member 11 | 393147 |
| ENSDARG00000042658 | acad8 | acyl-CoA dehydrogenase family, member 8 | 394130 |
| ENSDARG00000055620 | acad9 | acyl-CoA dehydrogenase family, member 9 | 724002 |
| ENSDARG00000088357 | acadl | acyl-CoA dehydrogenase, long chain | 394156 |
| ENSDARG00000038900 | acadm | acyl-CoA dehydrogenase, C-4 to C-12 straight chain | 406283 |
| ENSDARG00000030781 | acads | acyl-CoA dehydrogenase, C-2 to C-3 short chain | 445288 |
| ENSDARG00000076780 | acadsb | acyl-CoA dehydrogenase, short/branched chain | 0 |
| ENSDARG00000016687 | acadvl | acyl-CoA dehydrogenase, very long chain | 573723 |
| ENSDARG00000045799 | acanb | aggrecan b | 559593 |
| ENSDARG00000045888 | acat1 | acetyl-CoA acetyltransferase 1 | 445290 |
| ENSDARG00000007127 | acat2 | acetyl-CoA acetyltransferase 2 | 30643 |
| ENSDARG00000034883 | acbd5a | acyl-CoA binding domain containing 5a | 553674 |
| ENSDARG00000094730 | acbd7 | acyl-CoA binding domain containing 7 | 619256 |
| ENSDARG00000079166 | ace | angiotensin I converting enzyme (peptidyl-dipeptidase A) 1 | 565980 |
| ENSDARG00000031796 | ache | acetylcholinesterase | 114549 |
| ENSDARG00000099079 | aclya | ATP citrate lyase a | 436922 |
| ENSDARG00000026376 | aco1 | aconitase 1, soluble | 568448 |
| ENSDARG00000007294 | aco2 | aconitase 2, mitochondrial | 322670 |
| ENSDARG00000061728 | acot13 | acyl-CoA thioesterase 13 | 564010 |
| ENSDARG00000018653 | acot7 | acyl-CoA thioesterase 7 | 447878 |
| ENSDARG00000058352 | acot9.1 | acyl-CoA thioesterase 9, tandem duplicate 1 | 798073 |
| ENSDARG00000014727 | acox1 | acyl-CoA oxidase 1, palmitoyl | 449662 |
| ENSDARG00000038865 | acox3 | acyl-CoA oxidase 3, pristanoyl | 406421 |
| ENSDARG00000020149 | acoxl | acyl-CoA oxidase-like | 569104 |
| ENSDARG00000076486 | acp1 | acid phosphatase 1, soluble | 541489 |
| ENSDARG00000007244 | ACP2 | acid phosphatase 2, lysosomal | 0 |
| ENSDARG00000040064 | acp6 | acid phosphatase 6, lysophosphatidic | 558758 |
| ENSDARG00000030514 | acsl1a | acyl-CoA synthetase long-chain family member 1a | 555308 |
| ENSDARG00000003854 | acsl1b | acyl-CoA synthetase long-chain family member 1b | 445175 |
| ENSDARG00000014674 | acsl3b | acyl-CoA synthetase long-chain family member 3b | 559656 |
| ENSDARG00000004078 | acsl4a | acyl-CoA synthetase long-chain family member 4a | 393622 |
| ENSDARG00000099648 | ACSL6 | acyl-CoA synthetase long-chain family member 6 | 0 |
| ENSDARG00000044142 | acss1 | acyl-CoA synthetase short-chain family member 1 | 541435 |
| ENSDARG00000037746 | actb1 | actin, beta 1 | 57934 |
| ENSDARG00000037870 | actb2 | actin, beta 2 | 57935 |
| ENSDARG00000099786 | ACTN1 (2 of 3) | actinin, alpha 1 | 0 |
| ENSDARG00000007219 | actn1 | actinin, alpha 1 | 560400 |
| ENSDARG00000038432 | actr10 | actin-related protein 10 homolog (S. cerevisiae) | 393139 |
| ENSDARG00000011611 | actr1 | ARP1 actin-related protein 1, centractin (yeast) | 406681 |
| ENSDARG00000091138 | ACTR3 (1 of 2) | ARP3 actin-related protein 3 homolog (yeast) | 0 |
| ENSDARG00000100510 | ACTR3 (2 of 2) | ARP3 actin-related protein 3 homolog (yeast) | 0 |
| ENSDARG00000008790 | actr3b | ARP3 actin-related protein 3 homolog B (yeast) | 100038776 |
| ENSDARG00000060126 | acyp1 | acylphosphatase 1, erythrocyte (common) type | 0 |
| ENSDARG00000053468 | adam10a | ADAM metallopeptidase domain 10a | 566044 |
| ENSDARG00000015502 | adam10b | ADAM metallopeptidase domain 10b | 0 |
| ENSDARG00000079204 | adam11 | ADAM metallopeptidase domain 11 | 567145 |
| ENSDARG00000060532 | adam22 | ADAM metallopeptidase domain 22 | 100126805 |
| ENSDARG00000062323 | adam23a | ADAM metallopeptidase domain 23a | 794175 |
| ENSDARG00000010070 | adam9 | ADAM metallopeptidase domain 9 | 447940 |
| ENSDARG00000003544 | adarb1b | adenosine deaminase, RNA-specific, B1b | 558759 |
| ENSDARG00000062561 | adck1 | aarF domain containing kinase 1 | 100144553 |
| ENSDARG00000073923 | adck2 | aarF domain containing kinase 2 | 572087 |
| ENSDARG00000020123 | adck3 | aarF domain containing kinase 3 | 437001 |
| ENSDARG00000101130 | adck4 | aarF domain containing kinase 4 | 799071 |
| ENSDARG00000069026 | adcy1a | adenylate cyclase 1a | 557026 |
| ENSDARG00000088634 | adcy1b | adenylate cyclase 1b | 0 |
| ENSDARG00000077145 | ADCY3 | adenylate cyclase 3 | 571825 |
| ENSDARG00000061445 | adcy6a | adenylate cyclase 6a | 0 |
| ENSDARG00000103847 | ADCY9 (2 of 2) | adenylate cyclase 9 | 0 |
| ENSDARG00000100869 | adcy9 | adenylate cyclase 9 | 100333702 |
| ENSDARG00000027740 | adcyap1b | adenylate cyclase activating polypeptide 1b | 335625 |
| ENSDARG00000105201 | adcyap1r1a | adenylate cyclase activating polypeptide 1a (pituitary) receptor type I | 0 |
| ENSDARG00000103962 | add1 | adducin 1 (alpha) | 555511 |
| ENSDARG00000074581 | add2 | adducin 2 (beta) | 570147 |
| ENSDARG00000040874 | add3a | adducin 3 (gamma) a | 556762 |
| ENSDARG00000056250 | add3b | adducin 3 (gamma) b | 553243 |
| ENSDARG00000075133 | adgrb1a | adhesion G protein-coupled receptor B1a | 563527 |
| ENSDARG00000078529 | adgrb1b | adhesion G protein-coupled receptor B1b | 0 |
| ENSDARG00000059832 | adgrb3 | adhesion G protein-coupled receptor B3 | 0 |
| ENSDARG00000089292 | adgrl1a | adhesion G protein-coupled receptor L1a | 0 |
| ENSDARG00000069356 | adgrl2a | adhesion G protein-coupled receptor L2a | 57987 |
| ENSDARG00000075899 | adgrl2b.1 | adhesion G protein-coupled receptor L2b, tandem duplicate 1 | 101882597 |
| ENSDARG00000061121 | adgrl3.1 | adhesion G protein-coupled receptor L3.1 | 560631 |
| ENSDARG00000090624 | ADGRL3 (2 of 2) | adhesion G protein-coupled receptor L3 | 0 |
| ENSDARG00000080010 | adh5 | alcohol dehydrogenase 5 | 116517 |
| ENSDARG00000063525 | adpgk | ADP-dependent glucokinase | 557919 |
| ENSDARG00000104094 | adrbk2 | adrenergic, beta, receptor kinase 2 | 569403 |
| ENSDARG00000017049 | adsl | adenylosuccinate lyase | 334431 |
| ENSDARG00000062272 | afg3l2 | AFG3-like AAA ATPase 2 | 569168 |
| ENSDARG00000062123 | agap1 | ArfGAP with GTPase domain, ankyrin repeat and PH domain 1 | 100148154 |
| ENSDARG00000099874 | agap2 | ArfGAP with GTPase domain, ankyrin repeat and PH domain 2 | 565109 |
| ENSDARG00000087822 | AGAP3 | ArfGAP with GTPase domain, ankyrin repeat and PH domain 3 | 0 |
| ENSDARG00000019526 | agfg1b | ArfGAP with FG repeats 1b | 393560 |
| ENSDARG00000009837 | agk | acylglycerol kinase | 334270 |
| ENSDARG00000103811 | agla | amylo-alpha-1, 6-glucosidase, 4-alpha-glucanotransferase a | 567798 |
| ENSDARG00000092644 | ago1 | argonaute RISC catalytic component 1 | 570775 |
| ENSDARG00000061268 | ago2 | argonaute RISC catalytic component 2 | 570630 |
| ENSDARG00000059888 | AGO3 (1 of 2) | argonaute RISC catalytic component 3 | 567622 |
| ENSDARG00000063187 | agpat5 | 1-acylglycerol-3-phosphate O-acyltransferase 5 (lysophosphatidic acid acyltransferase, epsilon) | 767778 |
| ENSDARG00000042821 | agps | alkylglycerone phosphate synthase | 386801 |
| ENSDARG00000079388 | agrn | agrin | 565373 |
| ENSDARG00000005191 | ahcy | adenosylhomocysteinase | 387530 |
| ENSDARG00000056331 | ahcyl1 | adenosylhomocysteinase-like 1 | 798918 |
| ENSDARG00000039343 | ahcyl2 | adenosylhomocysteinase-like 2 | 394240 |
| ENSDARG00000061764 | ahnak | AHNAK nucleoprotein | 559276 |
| ENSDARG00000069293 | ahsg2 | alpha-2-HS-glycoprotein 2 | 567406 |
| ENSDARG00000014532 | aida | axin interactor, dorsalization associated | 393763 |
| ENSDARG00000058088 | aifm1 | apoptosis-inducing factor, mitochondrion-associated 1 | 373125 |
| ENSDARG00000077549 | AIFM2 | apoptosis-inducing factor, mitochondrion-associated, 2 | 557507 |
| ENSDARG00000062780 | AIFM3 | apoptosis-inducing factor, mitochondrion-associated, 3 | 100150876 |
| ENSDARG00000060036 | aimp1 | aminoacyl tRNA synthetase complex-interacting multifunctional protein 1 | 562232 |
| ENSDARG00000018903 | aimp2 | aminoacyl tRNA synthetase complex-interacting multifunctional protein 2 | 406647 |
| ENSDARG00000038655 | ajap1 | adherens junctions associated protein 1 | 0 |
| ENSDARG00000001950 | ak1 | adenylate kinase 1 | 445486 |
| ENSDARG00000005926 | ak2 | adenylate kinase 2 | 321793 |
| ENSDARG00000006546 | ak4 | adenylate kinase 4 | 406590 |
| ENSDARG00000012555 | ak5 | adenylate kinase 5 | 100003595 |
| ENSDARG00000055678 | akap12b | A kinase (PRKA) anchor protein 12b | 567365 |
| ENSDARG00000006062 | akap1b | A kinase (PRKA) anchor protein 1b | 556208 |
| ENSDARG00000043843 | akap7 | A kinase (PRKA) anchor protein 7 | 0 |
| ENSDARG00000079610 | akap9 | A kinase (PRKA) anchor protein 9 | 0 |
| ENSDARG00000052030 | akr1a1b | aldo-keto reductase family 1, member A1b (aldehyde reductase) | 799805 |
| ENSDARG00000027394 | AL929252.1 | Uncharacterized protein | 559304 |
| ENSDARG00000100372 | alad | aminolevulinate dehydratase | 550338 |
| ENSDARG00000026531 | alcama | activated leukocyte cell adhesion molecule a | 30194 |
| ENSDARG00000037935 | aldh16a1 | aldehyde dehydrogenase 16 family, member A1 | 492710 |
| ENSDARG00000099579 | aldh18a1 | aldehyde dehydrogenase 18 family, member A1 | 557186 |
| ENSDARG00000053493 | aldh1a2 | aldehyde dehydrogenase 1 family, member A2 | 116713 |
| ENSDARG00000077004 | aldh1l1 | aldehyde dehydrogenase 1 family, member L1 | 100526683 |
| ENSDARG00000070230 | aldh1l2 | aldehyde dehydrogenase 1 family, member L2 | 100333269 |
| ENSDARG00000089924 | aldh2.1 | aldehyde dehydrogenase 2 family (mitochondrial), tandem duplicate 1 | 393462 |
| ENSDARG00000028087 | aldh2.2 | aldehyde dehydrogenase 2 family (mitochondrial), tandem duplicate 2 | 368239 |
| ENSDARG00000074791 | aldh3a1 | aldehyde dehydrogenase 3 family, member A1 | 100000026 |
| ENSDARG00000028259 | aldh3a2a | aldehyde dehydrogenase 3 family, member A2a | 323653 |
| ENSDARG00000029381 | aldh3a2b | aldehyde dehydrogenase 3 family, member A2b | 0 |
| ENSDARG00000038207 | aldh4a1 | aldehyde dehydrogenase 4 family, member A1 | 394133 |
| ENSDARG00000076544 | aldh5a1 | aldehyde dehydrogenase 5 family, member A1 (succinate-semialdehyde dehydrogenase) | 565235 |
| ENSDARG00000053485 | aldh6a1 | aldehyde dehydrogenase 6 family, member A1 | 436647 |
| ENSDARG00000018426 | aldh7a1 | aldehyde dehydrogenase 7 family, member A1 | 334197 |
| ENSDARG00000069100 | aldh9a1a.1 | aldehyde dehydrogenase 9 family, member A1a, tandem duplicate 1 | 100005587 |
| ENSDARG00000011665 | aldoaa | aldolase a, fructose-bisphosphate, a | 336425 |
| ENSDARG00000034470 | aldoab | aldolase a, fructose-bisphosphate, b | 406496 |
| ENSDARG00000057661 | aldoca | aldolase C, fructose-bisphosphate, a | 792692 |
| ENSDARG00000019702 | aldocb | aldolase C, fructose-bisphosphate, b | 369193 |
| ENSDARG00000070399 | alg2 | asparagine-linked glycosylation 2 (alpha-1,3-mannosyltransferase) | 0 |
| ENSDARG00000012840 | alg9 | ALG9, alpha-1,2-mannosyltransferase | 393598 |
| ENSDARG00000015546 | alpl | alkaline phosphatase, liver/bone/kidney | 393982 |
| ENSDARG00000029952 | ampd2 | adenosine monophosphate deaminase 2 (isoform L) | 571355 |
| ENSDARG00000007663 | amph | amphiphysin | 393804 |
| ENSDARG00000059093 | ANK1 (1 of 2) | ankyrin 1, erythrocytic | 0 |
| ENSDARG00000092143 | ank1a | ankyrin 1, erythrocytic a | 449796 |
| ENSDARG00000074777 | ank1b | ankyrin 1, erythrocytic b | 0 |
| ENSDARG00000009026 | ANK2 (1 of 2) | ankyrin 2, neuronal | 568926 |
| ENSDARG00000043313 | ank2b | ankyrin 2b, neuronal | 450043 |
| ENSDARG00000061736 | ank3a | ankyrin 3a | 103909035 |
| ENSDARG00000077582 | ank3b | ankyrin 3b | 100126016 |
| ENSDARG00000061013 | ankfy1 | ankyrin repeat and FYVE domain containing 1 | 568480 |
| ENSDARG00000076829 | ankib1b | ankyrin repeat and IBR domain containing 1b | 561277 |
| ENSDARG00000098140 | ankrd13d | ankyrin repeat domain 13 family, member D | 562931 |
| ENSDARG00000078901 | anks1ab | ankyrin repeat and sterile alpha motif domain containing 1Ab | 100003659 |
| ENSDARG00000003512 | anks1b | ankyrin repeat and sterile alpha motif domain containing 1B | 565408 |
| ENSDARG00000077327 | ano11 | anoctamin 11 | 0 |
| ENSDARG00000102335 | ano1 | anoctamin 1, calcium activated chloride channel | 407698 |
| ENSDARG00000077229 | ano8b | anoctamin 8b | 562782 |
| ENSDARG00000036809 | ANPEP (1 of 5) | alanyl (membrane) aminopeptidase | 555478 |
| ENSDARG00000077383 | anxa11a | annexin A11a | 368215 |
| ENSDARG00000002632 | anxa11b | annexin A11b | 103908630 |
| ENSDARG00000013976 | anxa13 | annexin A13 | 81880 |
| ENSDARG00000013613 | anxa13l | annexin A13, like | 554118 |
| ENSDARG00000026726 | anxa1a | annexin A1a | 334724 |
| ENSDARG00000003216 | anxa2a | annexin A2a | 325557 |
| ENSDARG00000044254 | anxa3b | annexin A3b | 492336 |
| ENSDARG00000036456 | anxa4 | annexin A4 | 353362 |
| ENSDARG00000016470 | anxa5b | annexin A5b | 337132 |
| ENSDARG00000013335 | anxa6 | annexin A6 | 0 |
| ENSDARG00000014646 | aoc2 | amine oxidase, copper containing 2 | 541429 |
| ENSDARG00000020054 | aox1 | aldehyde oxidase 1 | 570457 |
| ENSDARG00000020621 | ap1b1 | adaptor-related protein complex 1, beta 1 subunit | 562166 |
| ENSDARG00000102394 | ap1g1 | adaptor-related protein complex 1, gamma 1 subunit | 324466 |
| ENSDARG00000054337 | ap1g2 | adaptor-related protein complex 1, gamma 2 subunit | 767712 |
| ENSDARG00000096249 | ap1m1 | adaptor-related protein complex 1, mu 1 subunit | 100330411 |
| ENSDARG00000096454 | ap1m2 | adaptor-related protein complex 1, mu 2 subunit | 403021 |
| ENSDARG00000056803 | ap1s1 | adaptor-related protein complex 1, sigma 1 subunit | 393279 |
| ENSDARG00000058504 | ap1s2 | adaptor-related protein complex 1, sigma 2 subunit | 327237 |
| ENSDARG00000054320 | ap2a1 | adaptor-related protein complex 2, alpha 1 subunit | 558153 |
| ENSDARG00000035152 | ap2b1 | adaptor-related protein complex 2, beta 1 subunit | 334632 |
| ENSDARG00000002790 | ap2m1a | adaptor-related protein complex 2, mu 1 subunit, a | 321051 |
| ENSDARG00000033899 | ap2m1b | adaptor-related protein complex 2, mu 1 subunit, b | 394001 |
| ENSDARG00000026967 | ap2s1 | adaptor-related protein complex 2, sigma 1 subunit | 100148085 |
| ENSDARG00000105296 | AP3B2 | adaptor-related protein complex 3, beta 2 subunit | 100000165 |
| ENSDARG00000071424 | ap3d1 | adaptor-related protein complex 3, delta 1 subunit | 563359 |
| ENSDARG00000016128 | ap3m2 | adaptor-related protein complex 3, mu 2 subunit | 415244 |
| ENSDARG00000039882 | ap3s2 | adaptor-related protein complex 3, sigma 2 subunit | 436812 |
| ENSDARG00000074328 | apba1b | amyloid beta (A4) precursor protein-binding, family A, member 1b | 565507 |
| ENSDARG00000099995 | apbb2b | amyloid beta (A4) precursor protein-binding, family B, member 2b | 564990 |
| ENSDARG00000058868 | apc | adenomatous polyposis coli | 386762 |
| ENSDARG00000045843 | apex1 | APEX nuclease (multifunctional DNA repair enzyme) 1 | 406730 |
| ENSDARG00000033597 | api5 | apoptosis inhibitor 5 | 321676 |
| ENSDARG00000098368 | aplp1 | amyloid beta (A4) precursor-like protein 1 | 404233 |
| ENSDARG00000054864 | aplp2 | amyloid beta (A4) precursor-like protein 2 | 796801 |
| ENSDARG00000017422 | apmap | adipocyte plasma membrane associated protein | 266637 |
| ENSDARG00000012076 | apoa1a | apolipoprotein A-Ia | 30355 |
| ENSDARG00000101324 | apoa1b | apolipoprotein A-Ib | 100101640 |
| ENSDARG00000015866 | apoa2 | apolipoprotein A-II | 322327 |
| ENSDARG00000040298 | apoa4b.1 | apolipoprotein A-IV b, tandem duplicate 1 | 322543 |
| ENSDARG00000020866 | apoa4b.2 | apolipoprotein A-IV b, tandem duplicate 2 | 570354 |
| ENSDARG00000042780 | apoba | apolipoprotein Ba | 566465 |
| ENSDARG00000022767 | apobb.1 | apolipoprotein Bb, tandem duplicate 1 | 321166 |
| ENSDARG00000075016 | apobb.2 | apolipoprotein Bb, tandem duplicate 2 | 100330435 |
| ENSDARG00000092170 | apoc1l | apolipoprotein C-I like | 570638 |
| ENSDARG00000092155 | apoc2 | apolipoprotein C-II | 568972 |
| ENSDARG00000040295 | apoeb | apolipoprotein Eb | 30314 |
| ENSDARG00000104696 | apooa | apolipoprotein O, a | 641319 |
| ENSDARG00000026444 | apoob | apolipoprotein O, b | 449837 |
| ENSDARG00000039374 | apool | apolipoprotein O-like | 541537 |
| ENSDARG00000104279 | appa | amyloid beta (A4) precursor protein a | 58083 |
| ENSDARG00000055543 | appb | amyloid beta (A4) precursor protein b | 170846 |
| ENSDARG00000060734 | appl1 | adaptor protein, phosphotyrosine interaction, PH domain and leucine zipper containing 1 | 571540 |
| ENSDARG00000078139 | appl2 | adaptor protein, phosphotyrosine interaction, PH domain and leucine zipper containing 2 | 100000135 |
| ENSDARG00000003519 | aprt | adenine phosphoribosyltransferase | 393641 |
| ENSDARG00000023713 | aqp1a.1 | aquaporin 1a (Colton blood group), tandem duplicate 1 | 335821 |
| ENSDARG00000031214 | arcn1b | archain 1b | 399661 |
| ENSDARG00000036998 | arf3b | ADP-ribosylation factor 3b | 368822 |
| ENSDARG00000052928 | arf6b | ADP-ribosylation factor 6b | 336063 |
| ENSDARG00000077656 | arfgef3 | ARFGEF family member 3 | 562652 |
| ENSDARG00000056664 | arfip2b | ADP-ribosylation factor interacting protein 2b | 550342 |
| ENSDARG00000057857 | arfrp1 | ADP-ribosylation factor related protein 1 | 503602 |
| ENSDARG00000024324 | arhgap1 | Rho GTPase activating protein 1 | 550478 |
| ENSDARG00000075673 | arhgap21b | Rho GTPase activating protein 21b | 326745 |
| ENSDARG00000052950 | arhgap23a | Rho GTPase activating protein 23a | 100006233 |
| ENSDARG00000059472 | arhgap31 | Rho GTPase activating protein 31 | 101885883 |
| ENSDARG00000062577 | arhgap35a | Rho GTPase activating protein 35a | 100004299 |
| ENSDARG00000104086 | arhgap39 | Rho GTPase activating protein 39 | 101885231 |
| ENSDARG00000102424 | ARHGAP44 (1 of 2) | Rho GTPase activating protein 44 | 567298 |
| ENSDARG00000104549 | arhgap44 | Rho GTPase activating protein 44 | 0 |
| ENSDARG00000015003 | arhgap4b | Rho GTPase activating protein 4b | 561231 |
| ENSDARG00000043795 | arhgdia | Rho GDP dissociation inhibitor (GDI) alpha | 406770 |
| ENSDARG00000040380 | arhgef1a | Rho guanine nucleotide exchange factor (GEF) 1a | 368266 |
| ENSDARG00000006299 | arhgef7a | Rho guanine nucleotide exchange factor (GEF) 7a | 553493 |
| ENSDARG00000073848 | arhgef7b | Rho guanine nucleotide exchange factor (GEF) 7b | 494081 |
| ENSDARG00000061746 | arhgef9a | Cdc42 guanine nucleotide exchange factor (GEF) 9a | 559868 |
| ENSDARG00000078624 | arhgef9b | Cdc42 guanine nucleotide exchange factor (GEF) 9b | 560923 |
| ENSDARG00000057995 | arl15b | ADP-ribosylation factor-like 15b | 560030 |
| ENSDARG00000069496 | arl2 | ADP-ribosylation factor-like 2 | 449774 |
| ENSDARG00000071409 | arl3 | ADP-ribosylation factor-like 3 | 797451 |
| ENSDARG00000054578 | arl6ip1 | ADP-ribosylation factor-like 6 interacting protein 1 | 394087 |
| ENSDARG00000016238 | arl6ip5b | ADP-ribosylation factor-like 6 interacting protein 5b | 436790 |
| ENSDARG00000078686 | arl8a | ADP-ribosylation factor-like 8A | 567222 |
| ENSDARG00000006915 | arl8ba | ADP-ribosylation factor-like 8Ba | 337424 |
| ENSDARG00000071881 | arl9 | ADP-ribosylation factor-like 9 | 751670 |
| ENSDARG00000013861 | armc1 | armadillo repeat containing 1 | 494066 |
| ENSDARG00000103770 | ARMC6 | armadillo repeat containing 6 | 555425 |
| ENSDARG00000008383 | arpc1a | actin related protein 2/3 complex, subunit 1A | 394243 |
| ENSDARG00000075989 | arpc2 | actin related protein 2/3 complex, subunit 2 | 327068 |
| ENSDARG00000057882 | arpc3 | actin related protein 2/3 complex, subunit 3 | 415204 |
| ENSDARG00000058225 | arpc4l | actin related protein 2/3 complex, subunit 4, like | 114456 |
| ENSDARG00000039142 | arpc5a | actin related protein 2/3 complex, subunit 5A | 336504 |
| ENSDARG00000019062 | arpc5b | actin related protein 2/3 complex, subunit 5B | 399482 |
| ENSDARG00000041720 | arpc5la | actin related protein 2/3 complex, subunit 5-like, a | 393550 |
| ENSDARG00000035433 | arpc5lb | actin related protein 2/3 complex, subunit 5-like, b | 556104 |
| ENSDARG00000043241 | arrb1 | arrestin, beta 1 | 553266 |
| ENSDARG00000074329 | arvcfa | armadillo repeat gene deleted in velocardiofacial syndrome a | 572216 |
| ENSDARG00000061688 | arvcfb | armadillo repeat gene deleted in velocardiofacial syndrome b | 564669 |
| ENSDARG00000029463 | asap1a | ArfGAP with SH3 domain, ankyrin repeat and PH domain 1a | 100170829 |
| ENSDARG00000039729 | asap1b | ArfGAP with SH3 domain, ankyrin repeat and PH domain 1b | 565859 |
| ENSDARG00000105115 | ASIC1 (2 of 2) | acid sensing (proton gated) ion channel 1 | 0 |
| ENSDARG00000008329 | asic1a | acid-sensing (proton-gated) ion channel 1a | 791696 |
| ENSDARG00000006849 | asic2 | acid-sensing (proton-gated) ion channel 2 | 407669 |
| ENSDARG00000018190 | asna1 | arsA arsenite transporter, ATP-binding, homolog 1 (bacterial) | 325704 |
| ENSDARG00000078703 | ASPH | aspartate beta-hydroxylase | 559236 |
| ENSDARG00000059629 | asphd2 | aspartate beta-hydroxylase domain containing 2 | 569819 |
| ENSDARG00000021681 | asrgl1 | asparaginase like 1 | 541402 |
| ENSDARG00000068323 | astn1 | astrotactin 1 | 791157 |
| ENSDARG00000023267 | atad1a | ATPase family, AAA domain containing 1a | 368672 |
| ENSDARG00000056609 | atad1b | ATPase family, AAA domain containing 1b | 368412 |
| ENSDARG00000086848 | atad3b | ATPase family, AAA domain containing 3B | 0 |
| ENSDARG00000009350 | atg3 | autophagy related 3 | 336632 |
| ENSDARG00000044551 | atg9a | ATG9 autophagy related 9 homolog A (S. cerevisiae) | 100009663 |
| ENSDARG00000016706 | atic | 5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase/IMP cyclohydrolase | 140622 |
| ENSDARG00000060481 | atl1 | atlastin GTPase 1 | 571909 |
| ENSDARG00000029931 | atp13a1 | ATPase type 13A1 | 334466 |
| ENSDARG00000002791 | atp1a1a.1 | ATPase, Na+/K+ transporting, alpha 1a polypeptide, tandem duplicate 1 | 64612 |
| ENSDARG00000001870 | atp1a1a.4 | ATPase, Na+/K+ transporting, alpha 1a polypeptide, tandem duplicate 4 | 64615 |
| ENSDARG00000019856 | atp1a1b | ATPase, Na+/K+ transporting, alpha 1b polypeptide | 64616 |
| ENSDARG00000010472 | atp1a2a | ATPase, Na+/K+ transporting, alpha 2a polypeptide | 64609 |
| ENSDARG00000018259 | atp1a3a | ATPase, Na+/K+ transporting, alpha 3a polypeptide | 64610 |
| ENSDARG00000104139 | atp1a3b | ATPase, Na+/K+ transporting, alpha 3b polypeptide | 64611 |
| ENSDARG00000013144 | atp1b1a | ATPase, Na+/K+ transporting, beta 1a polypeptide | 64267 |
| ENSDARG00000076833 | atp1b1b | ATPase, Na+/K+ transporting, beta 1b polypeptide | 64273 |
| ENSDARG00000099203 | atp1b2a | ATPase, Na+/K+ transporting, beta 2a polypeptide | 64269 |
| ENSDARG00000034424 | atp1b2b | ATPase, Na+/K+ transporting, beta 2b polypeptide | 114457 |
| ENSDARG00000015790 | atp1b3a | ATPase, Na+/K+ transporting, beta 3a polypeptide | 30468 |
| ENSDARG00000042837 | atp1b3b | ATPase, Na+/K+ transporting, beta 3b polypeptide | 64272 |
| ENSDARG00000053262 | atp1b4 | ATPase, Na+/K+ transporting, beta 4 polypeptide | 100037383 |
| ENSDARG00000035458 | atp2a1l | ATPase, Ca++ transporting, cardiac muscle, fast twitch 1 like | 494489 |
| ENSDARG00000005122 | atp2a2b | ATPase, Ca++ transporting, cardiac muscle, slow twitch 2b | 568265 |
| ENSDARG00000060978 | atp2a3 | ATPase, Ca++ transporting, ubiquitous | 568671 |
| ENSDARG00000012684 | atp2b1a | ATPase, Ca++ transporting, plasma membrane 1a | 378746 |
| ENSDARG00000063433 | atp2b2 | ATPase, Ca++ transporting, plasma membrane 2 | 557430 |
| ENSDARG00000043474 | atp2b3a | ATPase, Ca++ transporting, plasma membrane 3a | 436745 |
| ENSDARG00000023445 | atp2b3b | ATPase, Ca++ transporting, plasma membrane 3b | 558525 |
| ENSDARG00000044902 | atp2b4 | ATPase, Ca++ transporting, plasma membrane 4 | 0 |
| ENSDARG00000010149 | atp5a1 | ATP synthase, H+ transporting, mitochondrial F1 complex, alpha subunit 1, cardiac muscle | 553755 |
| ENSDARG00000070083 | atp5b | ATP synthase, H+ transporting, mitochondrial F1 complex, beta polypeptide | 554135 |
| ENSDARG00000045514 | atp5c1 | ATP synthase, H+ transporting, mitochondrial F1 complex, gamma polypeptide 1 | 336957 |
| ENSDARG00000019404 | atp5d | ATP synthase, H+ transporting, mitochondrial F1 complex, delta subunit | 335709 |
| ENSDARG00000095897 | atp5e | ATP synthase, H+ transporting, mitochondrial F1 complex, epsilon subunit | 798291 |
| ENSDARG00000011553 | atp5f1 | ATP synthase, H+ transporting, mitochondrial Fo complex, subunit B1 | 449787 |
| ENSDARG00000098355 | atp5h | ATP synthase, H+ transporting, mitochondrial Fo complex, subunit d | 393675 |
| ENSDARG00000078113 | atp5ia | ATP synthase, H+ transporting, mitochondrial Fo complex, subunit Ea | 559659 |
| ENSDARG00000068940 | atp5ib | ATP synthase, H+ transporting, mitochondrial Fo complex, subunit Eb | 767750 |
| ENSDARG00000077967 | ATP5J (2 of 3) | ATP synthase, H+ transporting, mitochondrial Fo complex, subunit F6 | 570370 |
| ENSDARG00000014313 | atp5j | ATP synthase, H+ transporting, mitochondrial Fo complex, subunit F6 | 406599 |
| ENSDARG00000011841 | atp5l | ATP synthase, H+ transporting, mitochondrial F0 complex, subunit g | 326977 |
| ENSDARG00000001788 | atp5o | ATP synthase, H+ transporting, mitochondrial F1 complex, O subunit | 335191 |
| ENSDARG00000012072 | atp5s | ATP synthase, H+ transporting, mitochondrial F0 complex, subunit s | 0 |
| ENSDARG00000041417 | atp6ap1a | ATPase, H+ transporting, lysosomal accessory protein 1a | 100004566 |
| ENSDARG00000037153 | atp6ap1b | ATPase, H+ transporting, lysosomal accessory protein 1b | 195824 |
| ENSDARG00000008735 | atp6ap2 | ATPase, H+ transporting, lysosomal accessory protein 2 | 406296 |
| ENSDARG00000015174 | atp6v0a1b | ATPase, H+ transporting, lysosomal V0 subunit a1b | 553691 |
| ENSDARG00000035538 | atp6v0a2a | ATPase, H+ transporting, lysosomal V0 subunit a2a | 561469 |
| ENSDARG00000057853 | atp6v0ca | ATPase, H+ transporting, lysosomal, V0 subunit ca | 192336 |
| ENSDARG00000036577 | atp6v0cb | ATPase, H+ transporting, lysosomal, V0 subunit cb | 325402 |
| ENSDARG00000069090 | atp6v0d1 | ATPase, H+ transporting, lysosomal V0 subunit d1 | 322811 |
| ENSDARG00000034534 | atp6v1aa | ATPase, H+ transporting, lysosomal, V1 subunit Aa | 394110 |
| ENSDARG00000076318 | atp6v1ab | ATPase, H+ transporting, lysosomal, V1 subunit Ab | 337583 |
| ENSDARG00000043465 | atp6v1b2 | ATPase, H+ transporting, lysosomal, V1 subunit B2 | 359840 |
| ENSDARG00000013443 | atp6v1ba | ATPase, H+ transporting, lysosomal, V1 subunit B, member a | 359839 |
| ENSDARG00000023967 | atp6v1c1a | ATPase, H+ transporting, lysosomal, V1 subunit C1a | 368907 |
| ENSDARG00000035880 | atp6v1c1b | ATPase, H+ transporting, lysosomal, V1 subunit C1b | 449655 |
| ENSDARG00000011175 | atp6v1d | ATPase, H+ transporting, lysosomal V1 subunit D | 393935 |
| ENSDARG00000030694 | atp6v1e1b | ATPase, H+ transporting, lysosomal, V1 subunit E1b | 192335 |
| ENSDARG00000045543 | atp6v1f | ATPase, H+ transporting, lysosomal, V1 subunit F | 436799 |
| ENSDARG00000022315 | atp6v1g1 | ATPase, H+ transporting, lysosomal, V1 subunit G1 | 335025 |
| ENSDARG00000006370 | atp6v1h | ATPase, H+ transporting, lysosomal V1 subunit H | 286779 |
| ENSDARG00000063001 | atp8a1 | ATPase, aminophospholipid transporter (APLT), class I, type 8A, member 1 | 0 |
| ENSDARG00000077492 | atp8a2 | ATPase, aminophospholipid transporter, class I, type 8A, member 2 | 100330943 |
| ENSDARG00000062521 | atp9b | ATPase, class II, type 9B | 568160 |
| ENSDARG00000044092 | atpif1b | ATPase inhibitory factor 1b | 558368 |
| ENSDARG00000042975 | auh | AU RNA binding protein/enoyl-CoA hydratase | 445182 |
| ENSDARG00000038414 | b3galt6 | UDP-Gal:betaGal beta 1,3-galactosyltransferase polypeptide 6 | 572324 |
| ENSDARG00000077740 | b3galtlb | beta 1,3-galactosyltransferase-like b | 564156 |
| ENSDARG00000001939 | b3gat3 | beta-1,3-glucuronyltransferase 3 (glucuronosyltransferase I) | 192334 |
| ENSDARG00000076701 | b4galnt4b | beta-1,4-N-acetyl-galactosaminyl transferase 4b | 559305 |
| ENSDARG00000097238 | ba1l | ba1 globin, like | 30217 |
| ENSDARG00000020895 | bag1 | BCL2-associated athanogene 1 | 558108 |
| ENSDARG00000017316 | bag5 | BCL2-associated athanogene 5 | 569580 |
| ENSDARG00000075892 | bag6 | BCL2-associated athanogene 6 | 83910 |
| ENSDARG00000062799 | baiap2a | BAI1-associated protein 2a | 678518 |
| ENSDARG00000079264 | baiap3 | BAI1-associated protein 3 | 564728 |
| ENSDARG00000037009 | banf1 | barrier to autointegration factor 1 | 334717 |
| ENSDARG00000053358 | basp1 | brain abundant, membrane attached signal protein 1 | 569631 |
| ENSDARG00000090190 | bcam | basal cell adhesion molecule (Lutheran blood group) | 767689 |
| ENSDARG00000099412 | bcan | brevican | 334113 |
| ENSDARG00000016231 | bcap29 | B-cell receptor-associated protein 29 | 554151 |
| ENSDARG00000044972 | bcap31 | B-cell receptor-associated protein 31 | 368488 |
| ENSDARG00000045568 | bcat1 | branched chain amino-acid transaminase 1, cytosolic | 337412 |
| ENSDARG00000054849 | bcat2 | branched chain amino-acid transaminase 2, mitochondrial | 436949 |
| ENSDARG00000040555 | bckdha | branched chain keto acid dehydrogenase E1, alpha polypeptide | 554124 |
| ENSDARG00000016904 | bckdk | branched chain ketoacid dehydrogenase kinase | 282677 |
| ENSDARG00000094704 | bcl2a | B-cell CLL/lymphoma 2a | 570772 |
| ENSDARG00000062370 | bcl2l13 | BCL2-like 13 (apoptosis facilitator) | 559355 |
| ENSDARG00000008434 | bcl2l1 | bcl2-like 1 | 114401 |
| ENSDARG00000012295 | bcs1l | BC1 (ubiquinol-cytochrome c reductase) synthesis-like | 394157 |
| ENSDARG00000059388 | bdh1 | 3-hydroxybutyrate dehydrogenase, type 1 | 100037356 |
| ENSDARG00000018817 | bdnf | brain-derived neurotrophic factor | 58118 |
| ENSDARG00000043673 | BEGAIN | brain-enriched guanylate kinase-associated | 563117 |
| ENSDARG00000034885 | bet1 | Bet1 golgi vesicular membrane trafficking protein | 415135 |
| ENSDARG00000013430 | bhmt | betaine-homocysteine methyltransferase | 322228 |
| ENSDARG00000042114 | bin1a | bridging integrator 1a | 0 |
| ENSDARG00000059857 | blvra | biliverdin reductase A | 767661 |
| ENSDARG00000096829 | blvrb | biliverdin reductase B | 436959 |
| ENSDARG00000020057 | bmpr2b | bone morphogenetic protein receptor, type II b (serine/threonine kinase) | 555655 |
| ENSDARG00000099961 | bnip3 | BCL2/adenovirus E1B interacting protein 3 | 336116 |
| ENSDARG00000039915 | bola1 | bolA family member 1 | 0 |
| ENSDARG00000102877 | bphl | biphenyl hydrolase-like (serine hydrolase) | 402804 |
| ENSDARG00000017661 | braf | B-Raf proto-oncogene, serine/threonine kinase | 403065 |
| ENSDARG00000010108 | bri3bp | bri3 binding protein | 327046 |
| ENSDARG00000078302 | BRINP1 | bone morphogenetic protein/retinoic acid inducible neural-specific 1 | 100334375 |
| ENSDARG00000014302 | brinp2 | bone morphogenetic protein/retinoic acid inducible neural-specific 2 | 561009 |
| ENSDARG00000061051 | BRINP3 (1 of 3) | bone morphogenetic protein/retinoic acid inducible neural-specific 3 | 100001603 |
| ENSDARG00000041974 | brk1 | BRICK1, SCAR/WAVE actin-nucleating complex subunit | 415187 |
| ENSDARG00000013659 | brox | BRO1 domain and CAAX motif containing | 492786 |
| ENSDARG00000061421 | BRSK2 (1 of 2) | BR serine/threonine kinase 2 | 0 |
| ENSDARG00000037008 | bscl2 | Berardinelli-Seip congenital lipodystrophy 2 (seipin) | 567403 |
| ENSDARG00000019881 | bsg | basigin | 337692 |
| ENSDARG00000079161 | bsnb | bassoon (presynaptic cytomatrix protein) b | 100329660 |
| ENSDARG00000063255 | btbd11a | BTB (POZ) domain containing 11a | 562893 |
| ENSDARG00000099321 | btbd17a | BTB (POZ) domain containing 17a | 563263 |
| ENSDARG00000102472 | btbd17b | BTB (POZ) domain containing 17b | 563672 |
| ENSDARG00000035400 | btf3 | basic transcription factor 3 | 556625 |
| ENSDARG00000089681 | btf3l4 | basic transcription factor 3-like 4 | 393667 |
| ENSDARG00000099026 | BX004828.2 | 0 | 101885702 |
| ENSDARG00000090615 | BX957297.1 | Uncharacterized protein | 567192 |
| ENSDARG00000089562 | BX957322.1 | Uncharacterized protein | 100006797 |
| ENSDARG00000069977 | C11H12orf10 | chromosome 12 open reading frame 10 | 664752 |
| ENSDARG00000040582 | C18H3orf33 | chromosome 3 open reading frame 33 | 565816 |
| ENSDARG00000056165 | C19H6orf136 | chromosome 6 open reading frame 136 | 565228 |
| ENSDARG00000036335 | c1galt1c1 | C1GALT1-specific chaperone 1 | 324052 |
| ENSDARG00000039887 | c1qbp | complement component 1, q subcomponent binding protein | 334619 |
| ENSDARG00000030038 | C20H12orf57 | chromosome 12 open reading frame 57 | 323559 |
| ENSDARG00000043842 | C20H1orf198 | chromosome 1 open reading frame 198 | 568408 |
| ENSDARG00000041915 | C20H1orf95 | chromosome 1 open reading frame 95 | 562918 |
| ENSDARG00000095751 | C20H6orf58 (2 of 2) | chromosome 6 open reading frame 58 | 406484 |
| ENSDARG00000057556 | C21H18orf32 | chromosome 18 open reading frame 32 | 336641 |
| ENSDARG00000071378 | C22H19orf70 | chromosome 19 open reading frame 70 | 558738 |
| ENSDARG00000019332 | C24H11orf65 | chromosome 11 open reading frame 65 | 406306 |
| ENSDARG00000029307 | C24H18orf8 | chromosome 18 open reading frame 8 | 0 |
| ENSDARG00000100152 | C25H12orf75 | chromosome 12 open reading frame 75 | 100535414 |
| ENSDARG00000101297 | C25H15orf27 | chromosome 15 open reading frame 27 | 0 |
| ENSDARG00000036888 | C25HXorf38 (1 of 2) | chromosome X open reading frame 38 | 777611 |
| ENSDARG00000061066 | c2cd2l | c2cd2-like | 566488 |
| ENSDARG00000094590 | c2cd5 | C2 calcium-dependent domain containing 5 | 100333813 |
| ENSDARG00000012694 | c3a.1 | complement component c3a, duplicate 1 | 321046 |
| ENSDARG00000052207 | c3a.3 | complement component c3a, duplicate 3 | 30491 |
| ENSDARG00000061257 | C3H17orf75 | chromosome 17 open reading frame 75 | 751673 |
| ENSDARG00000037909 | C3H19orf52 | chromosome 19 open reading frame 52 | 553583 |
| ENSDARG00000076268 | C5H9orf172 (1 of 2) | chromosome 9 open reading frame 172 | 568961 |
| ENSDARG00000078891 | C7H7orf43 | chromosome 7 open reading frame 43 | 100003958 |
| ENSDARG00000076873 | C8H9orf172 (2 of 2) | chromosome 9 open reading frame 172 | 799850 |
| ENSDARG00000016319 | c9 | complement component 9 | 554141 |
| ENSDARG00000020618 | C9H21orf33 | chromosome 21 open reading frame 33 | 550355 |
| ENSDARG00000069671 | C9H2orf47 | chromosome 2 open reading frame 47 | 552941 |
| ENSDARG00000052644 | ca10a | carbonic anhydrase Xa | 641327 |
| ENSDARG00000060123 | ca16b | carbonic anhydrase XVI b | 569183 |
| ENSDARG00000043589 | ca4a | carbonic anhydrase IV a | 555196 |
| ENSDARG00000043451 | cab39 | calcium binding protein 39 | 415235 |
| ENSDARG00000036658 | cab39l | calcium binding protein 39-like | 492361 |
| ENSDARG00000086931 | CABZ01029822.1 | Uncharacterized protein | 0 |
| ENSDARG00000099511 | CABZ01034698.4 | 0 | 0 |
| ENSDARG00000073822 | CABZ01046283.1 | Uncharacterized protein | 0 |
| ENSDARG00000061660 | CABZ01055347.1 | Uncharacterized protein | 0 |
| ENSDARG00000025074 | CABZ01059390.1 | Uncharacterized protein | 0 |
| ENSDARG00000074844 | CABZ01061495.1 | Uncharacterized protein | 100334801 |
| ENSDARG00000100177 | CABZ01066957.1 | Uncharacterized protein | 0 |
| ENSDARG00000102813 | CABZ01069436.1 | 0 | 101883099 |
| ENSDARG00000104674 | CABZ01075268.1 | 0 | 0 |
| ENSDARG00000089720 | CABZ01077218.1 | Uncharacterized protein | 100334619 |
| ENSDARG00000091618 | CABZ01080009.1 | Uncharacterized protein | 0 |
| ENSDARG00000104937 | CABZ01084323.1 | 0 | 100330629 |
| ENSDARG00000104106 | CABZ01087953.1 | 0 | 101883442 |
| ENSDARG00000039322 | CABZ01088149.1 | Uncharacterized protein | 402879 |
| ENSDARG00000059349 | CABZ01090374.1 | Uncharacterized protein | 0 |
| ENSDARG00000069046 | CABZ01118678.1 | Uncharacterized protein | 0 |
| ENSDARG00000037905 | cacna1aa | calcium channel, voltage-dependent, P/Q type, alpha 1A subunit, a | 562059 |
| ENSDARG00000006923 | cacna1ab | calcium channel, voltage-dependent, P/Q type, alpha 1A subunit, b | 569528 |
| ENSDARG00000079295 | cacna1bb | calcium channel, voltage-dependent, N type, alpha 1B subunit, b | 562796 |
| ENSDARG00000008398 | cacna1c | calcium channel, voltage-dependent, L type, alpha 1C subunit | 170581 |
| ENSDARG00000089913 | cacna1g | calcium channel, voltage-dependent, T type, alpha 1G subunit | 0 |
| ENSDARG00000014804 | cacna2d1a | calcium channel, voltage-dependent, alpha 2/delta subunit 1a | 561434 |
| ENSDARG00000039182 | cacna2d2a | calcium channel, voltage-dependent, alpha 2/delta subunit 2a | 541376 |
| ENSDARG00000078169 | cacna2d2b | calcium channel, voltage-dependent, alpha 2/delta subunit 2b | 568759 |
| ENSDARG00000078760 | cacna2d3 | calcium channel, voltage dependent, alpha2/delta subunit 3 | 100148645 |
| ENSDARG00000002167 | cacnb1 | calcium channel, voltage-dependent, beta 1 subunit | 436925 |
| ENSDARG00000055565 | cacnb2b | calcium channel, voltage-dependent, beta 2b | 0 |
| ENSDARG00000001881 | cacnb3a | calcium channel, voltage-dependent, beta 3a | 565158 |
| ENSDARG00000076030 | cacnb3b | calcium channel, voltage-dependent, beta 3b | 559015 |
| ENSDARG00000009738 | cacnb4a | calcium channel, voltage-dependent, beta 4a subunit | 562422 |
| ENSDARG00000062174 | cacnb4b | calcium channel, voltage-dependent, beta 4b subunit | 678560 |
| ENSDARG00000032565 | cacng2a | calcium channel, voltage-dependent, gamma subunit 2a | 393614 |
| ENSDARG00000102376 | cacng2b | calcium channel, voltage-dependent, gamma subunit 2b | 569855 |
| ENSDARG00000076401 | cacng3b | calcium channel, voltage-dependent, gamma subunit 3b | 564883 |
| ENSDARG00000020450 | cacng8a | calcium channel, voltage-dependent, gamma subunit 8a | 550428 |
| ENSDARG00000070626 | cacng8b | calcium channel, voltage-dependent, gamma subunit 8b | 100003329 |
| ENSDARG00000041895 | cad | carbamoyl-phosphate synthetase 2, aspartate transcarbamylase, and dihydroorotase | 266992 |
| ENSDARG00000031075 | cadm1a | cell adhesion molecule 1a | 569931 |
| ENSDARG00000042677 | cadm1b | cell adhesion molecule 1b | 562183 |
| ENSDARG00000009930 | cadm2a | cell adhesion molecule 2a | 793954 |
| ENSDARG00000062633 | cadm2b | cell adhesion molecule 2b | 571698 |
| ENSDARG00000057013 | cadm3 | cell adhesion molecule 3 | 692273 |
| ENSDARG00000040291 | cadm4 | cell adhesion molecule 4 | 492787 |
| ENSDARG00000043661 | cadpsa | Ca2+-dependent activator protein for secretion a | 100333933 |
| ENSDARG00000070567 | cadpsb | Ca2+-dependent activator protein for secretion b | 100001129 |
| ENSDARG00000011166 | cahz | carbonic anhydrase | 30331 |
| ENSDARG00000041062 | calb2a | calbindin 2a | 393691 |
| ENSDARG00000036344 | calb2b | calbindin 2b | 393684 |
| ENSDARG00000015050 | calm2b | calmodulin 2b, (phosphorylase kinase, delta) | 192322 |
| ENSDARG00000103979 | calr3a | calreticulin 3a | 30248 |
| ENSDARG00000102808 | calr3b | calreticulin 3b | 321315 |
| ENSDARG00000076290 | calr | calreticulin | 325317 |
| ENSDARG00000060116 | camk1a | calcium/calmodulin-dependent protein kinase Ia | 555945 |
| ENSDARG00000053617 | camk2a | calcium/calmodulin-dependent protein kinase II alpha | 550436 |
| ENSDARG00000011065 | camk2b1 | calcium/calmodulin-dependent protein kinase (CaM kinase) II beta 1 | 562064 |
| ENSDARG00000100089 | camk2b2 | calcium/calmodulin-dependent protein kinase (CaM kinase) II beta 2 | 0 |
| ENSDARG00000043010 | camk2d1 | calcium/calmodulin-dependent protein kinase (CaM kinase) II delta 1 | 445208 |
| ENSDARG00000014273 | camk2d2 | calcium/calmodulin-dependent protein kinase (CaM kinase) II delta 2 | 436815 |
| ENSDARG00000071395 | camk2g1 | calcium/calmodulin-dependent protein kinase (CaM kinase) II gamma 1 | 791855 |
| ENSDARG00000056206 | camk2g2 | calcium/calmodulin-dependent protein kinase (CaM kinase) II gamma 2 | 791855 |
| ENSDARG00000005372 | camk4 | calcium/calmodulin-dependent protein kinase IV | 550270 |
| ENSDARG00000078645 | CAMKK2 | calcium/calmodulin-dependent protein kinase kinase 2, beta | 0 |
| ENSDARG00000027963 | camkva | CaM kinase-like vesicle-associated a | 567243 |
| ENSDARG00000005141 | camkvb | CaM kinase-like vesicle-associated b | 393422 |
| ENSDARG00000057000 | camkvl | CaM kinase-like vesicle-associated, like | 553431 |
| ENSDARG00000100409 | camlg | calcium modulating ligand | 404622 |
| ENSDARG00000062173 | camsap2a | calmodulin regulated spectrin-associated protein family, member 2a | 562741 |
| ENSDARG00000017841 | cand1 | cullin-associated and neddylation-dissociated 1 | 406806 |
| ENSDARG00000005749 | cand2 | cullin-associated and neddylation-dissociated 2 (putative) | 555955 |
| ENSDARG00000037488 | canx | calnexin | 406757 |
| ENSDARG00000016350 | cap1 | CAP, adenylate cyclase-associated protein 1 (yeast) | 334527 |
| ENSDARG00000052702 | capn1a | calpain 1, (mu/I) large subunit a | 393417 |
| ENSDARG00000052748 | capn1b | calpain 1, (mu/I) large subunit b | 565106 |
| ENSDARG00000069748 | capn5b | calpain 5b | 100006332 |
| ENSDARG00000035329 | capns1a | calpain, small subunit 1 a | 550598 |
| ENSDARG00000013804 | capns1b | calpain, small subunit 1 b | 560033 |
| ENSDARG00000034240 | capza1a | capping protein (actin filament) muscle Z-line, alpha 1a | 327029 |
| ENSDARG00000056090 | capza1b | capping protein (actin filament) muscle Z-line, alpha 1b | 474327 |
| ENSDARG00000100252 | capzb | capping protein (actin filament) muscle Z-line, beta | 335083 |
| ENSDARG00000060829 | car15 | carbonic anhydrase 15 | 568143 |
| ENSDARG00000053129 | carhsp1 | calcium regulated heat stable protein 1 | 393534 |
| ENSDARG00000036164 | cars | cysteinyl-tRNA synthetase | 368284 |
| ENSDARG00000045832 | cart2 | cocaine- and amphetamine-regulated transcript 2 | 550232 |
| ENSDARG00000074490 | casc4 | cancer susceptibility candidate 4 | 555654 |
| ENSDARG00000059809 | caska | calcium/calmodulin-dependent serine protein kinase a | 259195 |
| ENSDARG00000100119 | caskb | calcium/calmodulin-dependent serine protein kinase b | 0 |
| ENSDARG00000046107 | CASKIN1 (1 of 2) | CASK interacting protein 1 | 563678 |
| ENSDARG00000008982 | casq2 | calsequestrin 2 | 114410 |
| ENSDARG00000058693 | cast | calpastatin | 563084 |
| ENSDARG00000104702 | cat | catalase | 30068 |
| ENSDARG00000063376 | CBLN3 (1 of 2) | cerebellin 3 precursor | 560359 |
| ENSDARG00000036587 | cbr1 | carbonyl reductase 1 | 373866 |
| ENSDARG00000021149 | cbr1l | carbonyl reductase 1-like | 337696 |
| ENSDARG00000036281 | CC2D1A | coiled-coil and C2 domain containing 1A | 0 |
| ENSDARG00000006863 | CCDC108 | coiled-coil domain containing 108 | 100333283 |
| ENSDARG00000099190 | ccdc127a | coiled-coil domain containing 127a | 751722 |
| ENSDARG00000002880 | ccdc132 | coiled-coil domain containing 132 | 101883616 |
| ENSDARG00000030508 | CCDC134 (1 of 2) | coiled-coil domain containing 134 | 0 |
| ENSDARG00000057921 | ccdc136b | coiled-coil domain containing 136b | 100330186 |
| ENSDARG00000052379 | ccdc22 | coiled-coil domain containing 22 | 556259 |
| ENSDARG00000022983 | ccdc28a | coiled-coil domain containing 28A | 326808 |
| ENSDARG00000054362 | ccdc47 | coiled-coil domain containing 47 | 447812 |
| ENSDARG00000058828 | ccdc51 | coiled-coil domain containing 51 | 751752 |
| ENSDARG00000045351 | ccdc58 | coiled-coil domain containing 58 | 393780 |
| ENSDARG00000044626 | ccdc90b | coiled-coil domain containing 90B | 100149897 |
| ENSDARG00000100052 | cckb | cholecystokinin b | 100000095 |
| ENSDARG00000063677 | ccny | cyclin Y | 557738 |
| ENSDARG00000099692 | ccnyl1 | cyclin Y-like 1 | 767752 |
| ENSDARG00000087206 | CCT2 | chaperonin containing TCP1, subunit 2 (beta) | 0 |
| ENSDARG00000016173 | cct3 | chaperonin containing TCP1, subunit 3 (gamma) | 192327 |
| ENSDARG00000013475 | cct4 | chaperonin containing TCP1, subunit 4 (delta) | 393555 |
| ENSDARG00000045399 | cct5 | chaperonin containing TCP1, subunit 5 (epsilon) | 322258 |
| ENSDARG00000021252 | cct6a | chaperonin containing TCP1, subunit 6A (zeta 1) | 116994 |
| ENSDARG00000007385 | cct7 | chaperonin containing TCP1, subunit 7 (eta) | 192324 |
| ENSDARG00000008243 | cct8 | chaperonin containing TCP1, subunit 8 (theta) | 394037 |
| ENSDARG00000056559 | ccz1 | CCZ1 vacuolar protein trafficking and biogenesis associated homolog (S. cerevisiae) | 393103 |
| ENSDARG00000079191 | CD200 (2 of 3) | CD200 molecule | 571844 |
| ENSDARG00000055504 | CD68 | CD68 molecule | 100000085 |
| ENSDARG00000009087 | cd74a | CD74 molecule, major histocompatibility complex, class II invariant chain a | 58113 |
| ENSDARG00000036080 | cd81a | CD81 molecule a | 58031 |
| ENSDARG00000022437 | cd81b | CD81 molecule b | 445280 |
| ENSDARG00000019098 | cd82a | CD82 molecule a | 324098 |
| ENSDARG00000051975 | cd99 | CD99 molecule | 559896 |
| ENSDARG00000056722 | cd99l2 | CD99 molecule-like 2 | 323266 |
| ENSDARG00000099301 | CDC37 | cell division cycle 37 | 0 |
| ENSDARG00000016464 | CDC42BPA (1 of 2) | CDC42 binding protein kinase alpha (DMPK-like) | 566003 |
| ENSDARG00000104283 | cdc42bpaa | CDC42 binding protein kinase alpha (DMPK-like) a | 0 |
| ENSDARG00000019383 | cdc42bpb | CDC42 binding protein kinase beta (DMPK-like) | 567039 |
| ENSDARG00000044573 | cdc42 | cell division cycle 42 | 336839 |
| ENSDARG00000045036 | cdc42ep4b | CDC42 effector protein (Rho GTPase binding) 4b | 100002938 |
| ENSDARG00000040158 | cdc42l | cell division cycle 42, like | 793158 |
| ENSDARG00000094577 | cdc42se2 | CDC42 small effector 2 | 794234 |
| ENSDARG00000055843 | cdh10a | cadherin 10, type 2a (T2-cadherin) | 568370 |
| ENSDARG00000021442 | cdh11 | cadherin 11, type 2, OB-cadherin (osteoblast) | 30461 |
| ENSDARG00000014215 | cdh13 | cadherin 13, H-cadherin (heart) | 558732 |
| ENSDARG00000077996 | cdh24b | cadherin 24, type 2b | 0 |
| ENSDARG00000018693 | cdh2 | cadherin 2, type 1, N-cadherin (neuronal) | 30291 |
| ENSDARG00000015002 | cdh4 | cadherin 4, type 1, R-cadherin (retinal) | 30297 |
| ENSDARG00000014522 | cdh6 | cadherin 6 | 541393 |
| ENSDARG00000023542 | cdh7 | cadherin 7, type 2 | 567704 |
| ENSDARG00000037478 | cdh8 | cadherin 8 | 0 |
| ENSDARG00000070686 | cdipt | CDP-diacylglycerol--inositol 3-phosphatidyltransferase (phosphatidylinositol synthase) | 404620 |
| ENSDARG00000012204 | CDK18 | cyclin-dependent kinase 18 | 100150007 |
| ENSDARG00000056683 | cdk5 | cyclin-dependent kinase 5 | 65234 |
| ENSDARG00000045087 | cdk5r1b | cyclin-dependent kinase 5, regulatory subunit 1b (p35) | 436788 |
| ENSDARG00000015240 | cdkl5 | cyclin-dependent kinase-like 5 | 559341 |
| ENSDARG00000053395 | cdkn2aipnl | CDKN2A interacting protein N-terminal like | 450034 |
| ENSDARG00000019549 | cds1 | CDP-diacylglycerol synthase (phosphatidate cytidylyltransferase) 1 | 100002015 |
| ENSDARG00000035577 | cds2 | CDP-diacylglycerol synthase (phosphatidate cytidylyltransferase) 2 | 394161 |
| ENSDARG00000055825 | celsr3 | cadherin, EGF LAG seven-pass G-type receptor 3 | 0 |
| ENSDARG00000055133 | cenpf | centromere protein F | 497403 |
| ENSDARG00000006128 | cep170aa | centrosomal protein 170Aa | 557463 |
| ENSDARG00000058892 | cep170ab | centrosomal protein 170Ab | 566358 |
| ENSDARG00000041595 | ces3 | carboxylesterase 3 | 560651 |
| ENSDARG00000101510 | cetn2 | centrin, EF-hand protein, 2 | 100006257 |
| ENSDARG00000021124 | cfl1 | cofilin 1 | 321496 |
| ENSDARG00000012972 | cfl1l | cofilin 1 (non-muscle), like | 406738 |
| ENSDARG00000055607 | cgna | cingulin a | 561423 |
| ENSDARG00000015854 | chata | choline O-acetyltransferase a | 100170938 |
| ENSDARG00000059304 | chchd2 | coiled-coil-helix-coiled-coil-helix domain containing 2 | 393740 |
| ENSDARG00000032919 | chchd3a | coiled-coil-helix-coiled-coil-helix domain containing 3a | 407730 |
| ENSDARG00000045978 | chchd6b | coiled-coil-helix-coiled-coil-helix domain containing 6b | 492522 |
| ENSDARG00000068156 | chchd7 | coiled-coil-helix-coiled-coil-helix domain containing 7 | 561510 |
| ENSDARG00000100304 | chdh | choline dehydrogenase | 100331020 |
| ENSDARG00000069673 | chid1 | chitinase domain containing 1 | 337225 |
| ENSDARG00000103109 | chl1b | cell adhesion molecule L1-like b | 0 |
| ENSDARG00000099624 | chmp1b | chromatin modifying protein 1B | 336426 |
| ENSDARG00000016255 | chmp4ba | charged multivesicular body protein 4Ba | 393164 |
| ENSDARG00000007323 | chmp4bb | charged multivesicular body protein 4Bb | 406766 |
| ENSDARG00000102024 | chmp6b | charged multivesicular body protein 6b | 445397 |
| ENSDARG00000052859 | chp1 | calcineurin-like EF-hand protein 1 | 325361 |
| ENSDARG00000098612 | chrm2a | cholinergic receptor, muscarinic 2a | 352938 |
| ENSDARG00000069254 | chrm4a | cholinergic receptor, muscarinic 4a | 100150701 |
| ENSDARG00000044341 | chst7 | carbohydrate (N-acetylglucosamine 6-O) sulfotransferase 7 | 563748 |
| ENSDARG00000103987 | cib1 | calcium and integrin binding 1 (calmyrin) | 100536377 |
| ENSDARG00000103672 | cirbpa | cold inducible RNA binding protein a | 550495 |
| ENSDARG00000013351 | cirbpb | cold inducible RNA binding protein b | 678563 |
| ENSDARG00000079516 | cisd1 | CDGSH iron sulfur domain 1 | 393577 |
| ENSDARG00000052703 | cisd2 | CDGSH iron sulfur domain 2 | 393354 |
| ENSDARG00000088825 | CIT (1 of 2) | citron rho-interacting serine/threonine kinase | 0 |
| ENSDARG00000089856 | CIT (2 of 2) | citron rho-interacting serine/threonine kinase | 0 |
| ENSDARG00000032405 | CKAP4 | cytoskeleton-associated protein 4 | 406549 |
| ENSDARG00000102674 | ckap5 | cytoskeleton associated protein 5 | 492614 |
| ENSDARG00000043257 | ckbb | creatine kinase, brain b | 140744 |
| ENSDARG00000040565 | ckmb | creatine kinase, muscle b | 794752 |
| ENSDARG00000016598 | ckmt1 | creatine kinase, mitochondrial 1 | 321892 |
| ENSDARG00000069615 | ckmt2a | creatine kinase, mitochondrial 2a (sarcomeric) | 327506 |
| ENSDARG00000039929 | ckmt2b | creatine kinase, mitochondrial 2b (sarcomeric) | 799492 |
| ENSDARG00000010280 | clasp1a | cytoplasmic linker associated protein 1a | 562077 |
| ENSDARG00000020345 | clasp2 | cytoplasmic linker associated protein 2 | 404604 |
| ENSDARG00000035808 | clcn4 | chloride channel, voltage-sensitive 4 | 768175 |
| ENSDARG00000032577 | clcn6 | chloride channel 6 | 568122 |
| ENSDARG00000019556 | clcn7 | chloride channel 7 | 553351 |
| ENSDARG00000042357 | cldnk | claudin k | 445070 |
| ENSDARG00000009315 | clgn | calmegin | 556495 |
| ENSDARG00000059596 | clip2 | CAP-GLY domain containing linker protein 2 | 556406 |
| ENSDARG00000054456 | clip3 | CAP-GLY domain containing linker protein 3 | 562450 |
| ENSDARG00000096867 | clmn | calmin | 100330495 |
| ENSDARG00000020679 | clpp | caseinolytic mitochondrial matrix peptidase proteolytic subunit | 553713 |
| ENSDARG00000018973 | clptm1 | cleft lip and palate associated transmembrane protein 1 | 795480 |
| ENSDARG00000021048 | clptm1l | CLPTM1-like | 436653 |
| ENSDARG00000031720 | clstn1 | calsyntenin 1 | 777737 |
| ENSDARG00000073883 | clstn3 | calsyntenin 3 | 555627 |
| ENSDARG00000045618 | clta | clathrin, light chain A | 406318 |
| ENSDARG00000043493 | cltca | clathrin, heavy chain a (Hc) | 323579 |
| ENSDARG00000090716 | cltcb | clathrin, heavy chain b (Hc) | 503600 |
| ENSDARG00000053122 | clvs2 | clavesin 2 | 566769 |
| ENSDARG00000029729 | clybl | citrate lyase beta like | 641475 |
| ENSDARG00000103670 | cmc1 | C-x(9)-C motif containing 1 | 0 |
| ENSDARG00000019924 | cmpk | cytidylate kinase | 406383 |
| ENSDARG00000052020 | cmtm4 | CKLF-like MARVEL transmembrane domain containing 4 | 436765 |
| ENSDARG00000099631 | cmtm6 | CKLF-like MARVEL transmembrane domain containing 6 | 368348 |
| ENSDARG00000069583 | CNDP1 | carnosine dipeptidase 1 (metallopeptidase M20 family) | 619255 |
| ENSDARG00000003931 | cndp2 | CNDP dipeptidase 2 (metallopeptidase M20 family) | 327288 |
| ENSDARG00000074480 | cnksr2a | connector enhancer of kinase suppressor of Ras 2a | 561623 |
| ENSDARG00000073707 | cnksr2b | connector enhancer of kinase suppressor of Ras 2b | 558834 |
| ENSDARG00000099568 | CNNM3 | cyclin and CBS domain divalent metal cation transport mediator 3 | 100329721 |
| ENSDARG00000074309 | cnnm4b | cyclin and CBS domain divalent metal cation transport mediator 4b | 100150651 |
| ENSDARG00000004174 | cnot1 | CCR4-NOT transcription complex, subunit 1 | 448949 |
| ENSDARG00000070822 | cnp | 2',3'-cyclic nucleotide 3' phosphodiesterase | 325896 |
| ENSDARG00000009020 | cnr1 | cannabinoid receptor 1 | 404209 |
| ENSDARG00000091683 | cnrip1a | cannabinoid receptor interacting protein 1a | 445213 |
| ENSDARG00000028661 | cntfr | ciliary neurotrophic factor receptor | 368438 |
| ENSDARG00000045685 | cntn1b | contactin 1b | 541474 |
| ENSDARG00000000472 | cntn2 | contactin 2 | 0 |
| ENSDARG00000013647 | cntn3a.2 | contactin 3a, tandem duplicate 2 | 100000688 |
| ENSDARG00000087843 | CNTN4 (2 of 3) | contactin 4 | 0 |
| ENSDARG00000098161 | cntn4 | contactin 4 | 571349 |
| ENSDARG00000021584 | cntn5 | contactin 5 | 327429 |
| ENSDARG00000074524 | cntnap1 | contactin associated protein 1 | 566220 |
| ENSDARG00000058969 | cntnap2a | contactin associated protein-like 2a | 559157 |
| ENSDARG00000074558 | cntnap2b | contactin associated protein-like 2b | 563345 |
| ENSDARG00000075189 | cntnap5b | contactin associated protein-like 5b | 571891 |
| ENSDARG00000100585 | coa3 | cytochrome C oxidase assembly factor 3 | 100141351 |
| ENSDARG00000092677 | coa6 | cytochrome c oxidase assembly factor 6 | 100150991 |
| ENSDARG00000019355 | coa7 | cytochrome c oxidase assembly factor 7 | 555268 |
| ENSDARG00000078322 | col12a1a | collagen, type XII, alpha 1a | 100149543 |
| ENSDARG00000094258 | commd9 | COMM domain containing 9 | 562660 |
| ENSDARG00000015337 | comta | catechol-O-methyltransferase a | 561372 |
| ENSDARG00000025679 | comtb | catechol-O-methyltransferase b | 565370 |
| ENSDARG00000004173 | copa | coatomer protein complex, subunit alpha | 260439 |
| ENSDARG00000056557 | copb1 | coatomer protein complex, subunit beta 1 | 338138 |
| ENSDARG00000018562 | cope | coatomer protein complex, subunit epsilon | 492492 |
| ENSDARG00000034823 | copg2 | coatomer protein complex, subunit gamma 2 | 30187 |
| ENSDARG00000008623 | cops3 | COP9 signalosome subunit 3 | 0 |
| ENSDARG00000043732 | cops4 | COP9 constitutive photomorphogenic homolog subunit 4 (Arabidopsis) | 325592 |
| ENSDARG00000057624 | cops5 | COP9 signalosome subunit 5 | 393698 |
| ENSDARG00000022788 | cops7a | COP9 constitutive photomorphogenic homolog subunit 7A | 336736 |
| ENSDARG00000024846 | cops8 | COP9 signalosome subunit 8 | 393198 |
| ENSDARG00000017844 | copz1 | coatomer protein complex, subunit zeta 1 | 0 |
| ENSDARG00000035618 | coq5 | coenzyme Q5 homolog, methyltransferase (S. cerevisiae) | 447802 |
| ENSDARG00000060380 | coq6 | coenzyme Q6 monooxygenase | 751691 |
| ENSDARG00000062594 | coq7 | coenzyme Q7 homolog, ubiquinone (yeast) | 559504 |
| ENSDARG00000023583 | coq9 | coenzyme Q9 homolog (S. cerevisiae) | 567716 |
| ENSDARG00000054610 | coro1a | coronin, actin binding protein, 1A | 100002113 |
| ENSDARG00000008660 | coro1b | coronin, actin binding protein, 1B | 560168 |
| ENSDARG00000035598 | coro1ca | coronin, actin binding protein, 1Ca | 317741 |
| ENSDARG00000021193 | coro1cb | coronin, actin binding protein, 1Cb | 562849 |
| ENSDARG00000005210 | coro2a | coronin, actin binding protein, 2A | 323538 |
| ENSDARG00000079440 | coro2ba | coronin, actin binding protein, 2Ba | 560235 |
| ENSDARG00000026829 | cotl1 | coactosin-like F-actin binding protein 1 | 336381 |
| ENSDARG00000092124 | cox14 | COX14 cytochrome c oxidase assembly factor | 0 |
| ENSDARG00000075933 | cox15 | cytochrome c oxidase assembly homolog 15 (yeast) | 394181 |
| ENSDARG00000039136 | cox16 | COX16 cytochrome c oxidase assembly homolog (S. cerevisiae) | 550257 |
| ENSDARG00000063882 | cox19 | COX19 cytochrome c oxidase assembly factor | 100004173 |
| ENSDARG00000032970 | cox4i1 | cytochrome c oxidase subunit IV isoform 1 | 326975 |
| ENSDARG00000012388 | cox4i1l | cytochrome c oxidase subunit IV isoform 1, like | 402880 |
| ENSDARG00000022509 | cox4i2 | cytochrome c oxidase subunit IV isoform 2 | 393776 |
| ENSDARG00000088383 | cox5aa | cytochrome c oxidase subunit Vaa | 554101 |
| ENSDARG00000099663 | cox5ab | cytochrome c oxidase subunit Vab | 326962 |
| ENSDARG00000011208 | COX5B (1 of 3) | cytochrome c oxidase subunit Vb | 100002384 |
| ENSDARG00000068738 | cox5b2 | cytochrome c oxidase subunit Vb 2 | 751638 |
| ENSDARG00000015978 | COX5B (2 of 3) | cytochrome c oxidase subunit Vb | 415253 |
| ENSDARG00000022438 | cox6a1 | cytochrome c oxidase subunit VIa polypeptide 1 | 335774 |
| ENSDARG00000045230 | cox6b1 | cytochrome c oxidase subunit VIb polypeptide 1 | 436968 |
| ENSDARG00000037860 | cox6b2 | cytochrome c oxidase subunit VIb polypeptide 2 | 393478 |
| ENSDARG00000038577 | cox6c | cytochrome c oxidase subunit VIc | 494577 |
| ENSDARG00000053217 | cox7a2a | cytochrome c oxidase, subunit VIIa 2a | 554103 |
| ENSDARG00000054907 | cox7a3 | cytochrome c oxidase subunit VIIa polypeptide 3 | 335751 |
| ENSDARG00000104537 | cox7c | cytochrome c oxidase, subunit VIIc | 336118 |
| ENSDARG00000010312 | cp | ceruloplasmin | 84702 |
| ENSDARG00000055648 | cpda | carboxypeptidase D, a | 563008 |
| ENSDARG00000055874 | cpe | carboxypeptidase E | 407979 |
| ENSDARG00000077736 | CPLX1 (2 of 2) | complexin 1 | 563082 |
| ENSDARG00000018997 | cplx2l | complexin 2, like | 436732 |
| ENSDARG00000089486 | cplx3b | complexin 3b | 794623 |
| ENSDARG00000075536 | cpne1 | copine I | 324841 |
| ENSDARG00000007753 | cpne2 | copine II | 565225 |
| ENSDARG00000061466 | CPNE5 (1 of 2) | copine V | 568003 |
| ENSDARG00000070919 | cpne5 | copine V | 394003 |
| ENSDARG00000102584 | cpne7 | copine VII | 564556 |
| ENSDARG00000102191 | CPNE8 (2 of 2) | copine VIII | 101886030 |
| ENSDARG00000062025 | cpox | coproporphyrinogen oxidase | 321294 |
| ENSDARG00000018618 | cpsf6 | cleavage and polyadenylation specific factor 6 | 327069 |
| ENSDARG00000062054 | cpt1ab | carnitine palmitoyltransferase 1Ab (liver) | 560000 |
| ENSDARG00000038618 | cpt2 | carnitine palmitoyltransferase 2 | 100005717 |
| ENSDARG00000074235 | CR376783.1 | Uncharacterized protein | 797620 |
| ENSDARG00000044212 | CR735126.1 | Uncharacterized protein | 0 |
| ENSDARG00000086969 | CR774179.1 | Uncharacterized protein | 0 |
| ENSDARG00000045926 | crabp1a | cellular retinoic acid binding protein 1a | 171479 |
| ENSDARG00000016545 | CRAT (1 of 2) | carnitine O-acetyltransferase | 562075 |
| ENSDARG00000040248 | crata | carnitine O-acetyltransferase a | 449545 |
| ENSDARG00000098782 | cratb | carnitine O-acetyltransferase b | 447855 |
| ENSDARG00000055635 | crk | v-crk avian sarcoma virus CT10 oncogene homolog | 445234 |
| ENSDARG00000056742 | crmp1 | collapsin response mediator protein 1 | 553756 |
| ENSDARG00000040352 | crot | carnitine O-octanoyltransferase | 553204 |
| ENSDARG00000071454 | crp1 | C-reactive protein 1 | 570508 |
| ENSDARG00000059826 | crtac1a | cartilage acidic protein 1a | 559626 |
| ENSDARG00000003675 | cryl1 | crystallin, lambda 1 | 751596 |
| ENSDARG00000016074 | cryz | crystallin, zeta (quinone reductase) | 407640 |
| ENSDARG00000026902 | cryzl1 | crystallin, zeta (quinone reductase)-like 1 | 436906 |
| ENSDARG00000103364 | cs | citrate synthase | 322339 |
| ENSDARG00000006963 | cse1l | CSE1 chromosome segregation 1-like (yeast) | 30707 |
| ENSDARG00000001559 | csmd2 | CUB and Sushi multiple domains 2 | 565460 |
| ENSDARG00000077920 | CSMD3 | CUB and Sushi multiple domains 3 | 0 |
| ENSDARG00000052674 | csnk1a1 | casein kinase 1, alpha 1 | 192307 |
| ENSDARG00000008370 | csnk1da | casein kinase 1, delta a | 322106 |
| ENSDARG00000006125 | csnk1db | casein kinase 1, delta b | 406533 |
| ENSDARG00000045150 | csnk1e | casein kinase 1, epsilon | 100006858 |
| ENSDARG00000104342 | csnk1g1 | casein kinase 1, gamma 1 | 494092 |
| ENSDARG00000034056 | csnk1g2b | casein kinase 1, gamma 2b | 561724 |
| ENSDARG00000002082 | csnk2a1 | casein kinase 2, alpha 1 polypeptide | 30502 |
| ENSDARG00000012818 | csnk2a2a | casein kinase 2, alpha prime polypeptide a | 30488 |
| ENSDARG00000013582 | csnk2a2b | casein kinase 2, alpha prime polypeptide b | 556379 |
| ENSDARG00000077776 | csnk2b | casein kinase 2, beta polypeptide | 30428 |
| ENSDARG00000078227 | cspg4 | chondroitin sulfate proteoglycan 4 | 560393 |
| ENSDARG00000069981 | cspg5a | chondroitin sulfate proteoglycan 5a | 558206 |
| ENSDARG00000099793 | cspg5b | chondroitin sulfate proteoglycan 5b | 559358 |
| ENSDARG00000045352 | cst14a.2 | cystatin 14a, tandem duplicate 2 | 792225 |
| ENSDARG00000077343 | CT573264.1 | Uncharacterized protein | 100536351 |
| ENSDARG00000074376 | CT737192.1 | Uncharacterized protein | 0 |
| ENSDARG00000057007 | ctbp1 | C-terminal binding protein 1 | 65229 |
| ENSDARG00000044062 | ctbp2a | C-terminal binding protein 2a | 65230 |
| ENSDARG00000040674 | ctdsp1 | CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 1 | 553744 |
| ENSDARG00000032206 | cthl | cystathionase (cystathionine gamma-lyase), like | 445818 |
| ENSDARG00000102441 | ctnna1 | catenin (cadherin-associated protein), alpha 1 | 30741 |
| ENSDARG00000024785 | ctnna2 | catenin (cadherin-associated protein), alpha 2 | 567500 |
| ENSDARG00000018162 | ctnnal1 | catenin (cadherin-associated protein), alpha-like 1 | 100034581 |
| ENSDARG00000014571 | ctnnb1 | catenin (cadherin-associated protein), beta 1 | 30265 |
| ENSDARG00000023472 | ctnnb2 | catenin, beta 2 | 0 |
| ENSDARG00000078233 | ctnnd1 | catenin (cadherin-associated protein), delta 1 | 556726 |
| ENSDARG00000062415 | ctnnd2a | catenin (cadherin-associated protein), delta 2a | 368485 |
| ENSDARG00000003779 | ctnnd2b | catenin (cadherin-associated protein), delta 2b | 558069 |
| ENSDARG00000055120 | ctsba | cathepsin Ba | 406645 |
| ENSDARG00000101334 | ctsc | cathepsin C | 368704 |
| ENSDARG00000057698 | ctsd | cathepsin D | 65225 |
| ENSDARG00000040991 | cttnbp2 | cortactin binding protein 2 | 555837 |
| ENSDARG00000011777 | cttn | cortactin | 0 |
| ENSDARG00000019239 | cul1a | cullin 1a | 323883 |
| ENSDARG00000038967 | cul3a | cullin 3a | 324655 |
| ENSDARG00000069742 | cul5a | cullin 5a | 327215 |
| ENSDARG00000043134 | cux1b | cut-like homeobox 1b | 445120 |
| ENSDARG00000041799 | cx43 | connexin 43 | 30236 |
| ENSDARG00000073896 | cx47.1 | connexin 47.1 | 447835 |
| ENSDARG00000098589 | cyb5a | cytochrome b5 type A (microsomal) | 406409 |
| ENSDARG00000099774 | cyb5b | cytochrome b5 type B | 405812 |
| ENSDARG00000018966 | cyb5r1 | cytochrome b5 reductase 1 | 336553 |
| ENSDARG00000005891 | cyb5r3 | cytochrome b5 reductase 3 | 324560 |
| ENSDARG00000095577 | cybrd1 | cytochrome b reductase 1 | 552960 |
| ENSDARG00000038075 | cyc1 | cytochrome c-1 | 564379 |
| ENSDARG00000044562 | cycsb | cytochrome c, somatic b | 415158 |
| ENSDARG00000044345 | cyfip1 | cytoplasmic FMR1 interacting protein 1 | 336613 |
| ENSDARG00000036375 | cyfip2 | cytoplasmic FMR1 interacting protein 2 | 100002872 |
| ENSDARG00000098360 | cyp19a1b | cytochrome P450, family 19, subfamily A, polypeptide 1b | 0 |
| ENSDARG00000006040 | cyp20a1 | cytochrome P450, family 20, subfamily A, polypeptide 1 | 406641 |
| ENSDARG00000077121 | cyp26b1 | cytochrome P450, family 26, subfamily b, polypeptide 1 | 324188 |
| ENSDARG00000088013 | CYP27A1 (4 of 4) | cytochrome P450, family 27, subfamily A, polypeptide 1 | 723999 |
| ENSDARG00000033802 | cyp27a7 | cytochrome P450, family 27, subfamily A, polypeptide 7 | 402831 |
| ENSDARG00000042978 | cyp2p6 | cytochrome P450, family 2, subfamily P, polypeptide 6 | 393108 |
| ENSDARG00000004964 | cyp4t8 | cytochrome P450, family 4, subfamily T, polypeptide 8 | 387527 |
| ENSDARG00000042641 | cyp51 | cytochrome P450, family 51 | 414331 |
| ENSDARG00000028800 | CYTH2 | cytohesin 2 | 569358 |
| ENSDARG00000005159 | cyth3b | cytohesin 3b | 0 |
| ENSDARG00000015059 | daam1a | dishevelled associated activator of morphogenesis 1a | 557451 |
| ENSDARG00000009689 | daam1b | dishevelled associated activator of morphogenesis 1b | 560060 |
| ENSDARG00000028393 | daam2 | dishevelled associated activator of morphogenesis 2 | 0 |
| ENSDARG00000059939 | dab1a | Dab, reelin signal transducer, homolog 1a (Drosophila) | 692351 |
| ENSDARG00000069484 | dab2ipa | DAB2 interacting protein a | 100002454 |
| ENSDARG00000075110 | dab2ipb | DAB2 interacting protein b | 558237 |
| ENSDARG00000016153 | dag1 | dystroglycan 1 | 286829 |
| ENSDARG00000062956 | dagla | diacylglycerol lipase, alpha | 569398 |
| ENSDARG00000035603 | dao.2 | D-amino-acid oxidase, tandem duplicate 2 | 405800 |
| ENSDARG00000060093 | dapk1 | death-associated protein kinase 1 | 558314 |
| ENSDARG00000070043 | dars | aspartyl-tRNA synthetase | 324025 |
| ENSDARG00000069446 | dbh | dopamine beta-hydroxylase (dopamine beta-monooxygenase) | 30505 |
| ENSDARG00000026369 | dbi | diazepam binding inhibitor (GABA receptor modulator, acyl-CoA binding protein) | 393831 |
| ENSDARG00000069113 | dbn1 | drebrin 1 | 561153 |
| ENSDARG00000016016 | dbnlb | drebrin-like b | 553729 |
| ENSDARG00000101561 | dbt | dihydrolipoamide branched chain transacylase E2 | 541388 |
| ENSDARG00000054355 | dcaf7 | ddb1 and cul4 associated factor 7 | 337565 |
| ENSDARG00000061638 | dcakd | dephospho-CoA kinase domain containing | 751733 |
| ENSDARG00000104282 | DCC | DCC netrin 1 receptor | 0 |
| ENSDARG00000079850 | dchs1b | dachsous cadherin-related 1b | 0 |
| ENSDARG00000104664 | dckl1b | doublecortin-like kinase 1b | 100151759 |
| ENSDARG00000034093 | dclk2 | doublecortin-like kinase 2 | 572548 |
| ENSDARG00000019743 | dctn1a | dynactin 1a | 407638 |
| ENSDARG00000056753 | dctn1b | dynactin 1b | 100003026 |
| ENSDARG00000031388 | dctn2 | dynactin 2 (p50) | 394141 |
| ENSDARG00000041363 | dctn3 | dynactin 3 (p22) | 431767 |
| ENSDARG00000023988 | dctn4 | dynactin 4 | 550479 |
| ENSDARG00000105009 | ddah2 | dimethylarginine dimethylaminohydrolase 2 | 799954 |
| ENSDARG00000103083 | DDB1 (2 of 2) | damage-specific DNA binding protein 1, 127kDa | 0 |
| ENSDARG00000074431 | ddb1 | damage-specific DNA binding protein 1 | 0 |
| ENSDARG00000037318 | ddost | dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit (non-catalytic) | 406408 |
| ENSDARG00000037172 | ddrgk1 | DDRGK domain containing 1 | 0 |
| ENSDARG00000044751 | ddt | D-dopachrome tautomerase | 415237 |
| ENSDARG00000061865 | ddx10 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 10 | 569118 |
| ENSDARG00000005699 | ddx19 | DEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide 19 (DBP5 homolog, yeast) | 192339 |
| ENSDARG00000032117 | ddx1 | DEAD (Asp-Glu-Ala-Asp) box helicase 1 | 556790 |
| ENSDARG00000006225 | ddx39aa | DEAD (Asp-Glu-Ala-Asp) box polypeptide 39Aa | 325550 |
| ENSDARG00000020573 | ddx3 | DEAD (Asp-Glu-Ala-Asp) box helicase 3 | 566947 |
| ENSDARG00000038068 | ddx5 | DEAD (Asp-Glu-Ala-Asp) box helicase 5 | 322206 |
| ENSDARG00000061338 | ddx6 | DEAD (Asp-Glu-Ala-Asp) box helicase 6 | 564633 |
| ENSDARG00000003869 | decr1 | 2,4-dienoyl CoA reductase 1, mitochondrial | 436717 |
| ENSDARG00000037229 | denr | density-regulated protein | 436970 |
| ENSDARG00000005897 | dera | deoxyribose-phosphate aldolase (putative) | 450077 |
| ENSDARG00000103503 | dgat1a | diacylglycerol O-acyltransferase 1a | 325875 |
| ENSDARG00000060626 | dgkaa | diacylglycerol kinase, alpha a | 724010 |
| ENSDARG00000104793 | dgke | diacylglycerol kinase, epsilon | 100150463 |
| ENSDARG00000062696 | DGKG | diacylglycerol kinase, gamma 90kDa | 0 |
| ENSDARG00000018716 | dgkh | diacylglycerol kinase, eta | 100137124 |
| ENSDARG00000063578 | DGKI | diacylglycerol kinase, iota | 100331649 |
| ENSDARG00000074183 | DGKK | diacylglycerol kinase, kappa | 565122 |
| ENSDARG00000013236 | dhcr24 | 24-dehydrocholesterol reductase | 494102 |
| ENSDARG00000051889 | dhodh | dihydroorotate dehydrogenase | 494065 |
| ENSDARG00000011770 | dhrs12 | dehydrogenase/reductase (SDR family) member 12 | 556393 |
| ENSDARG00000026322 | dhrs13a.1 | dehydrogenase/reductase (SDR family) member 13a, tandem duplicate 1 | 492491 |
| ENSDARG00000041137 | dhrs13a.3 | dehydrogenase/reductase (SDR family) member 13a, duplicate 3 | 492783 |
| ENSDARG00000054300 | dhrs1 | dehydrogenase/reductase (SDR family) member 1 | 368670 |
| ENSDARG00000021135 | dhrs2 | dehydrogenase/reductase (SDR family) member 2 | 393539 |
| ENSDARG00000075726 | dhrs7b | dehydrogenase/reductase (SDR family) member 7B | 550454 |
| ENSDARG00000003444 | dhrs7 | dehydrogenase/reductase (SDR family) member 7 | 751626 |
| ENSDARG00000019260 | dhrs9 | dehydrogenase/reductase (SDR family) member 9 | 322529 |
| ENSDARG00000016415 | dhtkd1 | dehydrogenase E1 and transketolase domain containing 1 | 494076 |
| ENSDARG00000015392 | dhx15 | DEAH (Asp-Glu-Ala-His) box helicase 15 | 321931 |
| ENSDARG00000063419 | DHX35 | DEAH (Asp-Glu-Ala-His) box polypeptide 35 | 0 |
| ENSDARG00000062612 | dhx37 | DEAH (Asp-Glu-Ala-His) box polypeptide 37 | 100009635 |
| ENSDARG00000079725 | dhx9 | DEAH (Asp-Glu-Ala-His) box helicase 9 | 568043 |
| ENSDARG00000104172 | diabloa | diablo, IAP-binding mitochondrial protein a | 393317 |
| ENSDARG00000014956 | diablob | diablo, IAP-binding mitochondrial protein b | 570425 |
| ENSDARG00000005350 | dip2ba | DIP2 disco-interacting protein 2 homolog Ba (Drosophila) | 405899 |
| ENSDARG00000016348 | dip2bb | DIP2 disco-interacting protein 2 homolog Bb (Drosophila) | 568954 |
| ENSDARG00000028066 | diras1a | DIRAS family, GTP-binding RAS-like 1a | 327577 |
| ENSDARG00000009372 | diras1b | DIRAS family, GTP-binding RAS-like 1b | 565395 |
| ENSDARG00000033259 | dis3l2 | DIS3 like 3'-5' exoribonuclease 2 | 566106 |
| ENSDARG00000025302 | dixdc1a | DIX domain containing 1a | 360137 |
| ENSDARG00000015918 | dlat | dihydrolipoamide S-acetyltransferase (E2 component of pyruvate dehydrogenase complex) | 324201 |
| ENSDARG00000008785 | dldh | dihydrolipoamide dehydrogenase | 399479 |
| ENSDARG00000009677 | dlg1 | discs, large homolog 1 (Drosophila) | 114446 |
| ENSDARG00000102216 | dlg1l | discs, large (Drosophila) homolog 1, like | 497648 |
| ENSDARG00000099323 | dlg2 | discs, large homolog 2 (Drosophila) | 497638 |
| ENSDARG00000076796 | dlg3 | discs, large homolog 3 (Drosophila) | 564081 |
| ENSDARG00000041926 | DLG4 (2 of 2) | discs, large homolog 4 (Drosophila) | 0 |
| ENSDARG00000014280 | dlgap1a | discs, large (Drosophila) homolog-associated protein 1a | 569898 |
| ENSDARG00000076070 | DLGAP2 (2 of 3) | discs, large (Drosophila) homolog-associated protein 2 | 0 |
| ENSDARG00000059340 | dlgap2a | discs, large (Drosophila) homolog-associated protein 2a | 557621 |
| ENSDARG00000092376 | dlgap2b | discs, large (Drosophila) homolog-associated protein 2b | 0 |
| ENSDARG00000055459 | dlgap3 | discs, large (Drosophila) homolog-associated protein 3 | 557556 |
| ENSDARG00000012823 | dlgap4b | discs, large (Drosophila) homolog-associated protein 4b | 0 |
| ENSDARG00000014230 | dlst | dihydrolipoamide S-succinyltransferase | 368262 |
| ENSDARG00000006585 | DMWD | dystrophia myotonica, WD repeat containing | 571996 |
| ENSDARG00000098599 | DMXL1 | Dmx-like 1 | 100333625 |
| ENSDARG00000091293 | dmxl2 | Dmx-like 2 | 378958 |
| ENSDARG00000100115 | dnah1 | dynein, axonemal, heavy chain 1 | 100317899 |
| ENSDARG00000104066 | dnaja2 | DnaJ (Hsp40) homolog, subfamily A, member 2 | 406814 |
| ENSDARG00000010745 | dnaja2l | DnaJ (Hsp40) homolog, subfamily A, member 2, like | 324164 |
| ENSDARG00000058494 | dnaja3a | DnaJ (Hsp40) homolog, subfamily A, member 3A | 335894 |
| ENSDARG00000102295 | dnaja3b | DnaJ (Hsp40) homolog, subfamily A, member 3B | 394242 |
| ENSDARG00000015088 | dnajb11 | DnaJ (Hsp40) homolog, subfamily B, member 11 | 386709 |
| ENSDARG00000039363 | dnajb12a | DnaJ (Hsp40) homolog, subfamily B, member 12a | 324005 |
| ENSDARG00000058644 | dnajb2 | DnaJ (Hsp40) homolog, subfamily B, member 2 | 561686 |
| ENSDARG00000038978 | dnajb4 | DnaJ (Hsp40) homolog, subfamily B, member 4 | 445061 |
| ENSDARG00000004680 | dnajb6a | DnaJ (Hsp40) homolog, subfamily B, member 6a | 436626 |
| ENSDARG00000074727 | dnajc10 | DnaJ (Hsp40) homolog, subfamily C, member 10 | 557858 |
| ENSDARG00000102696 | DNAJC11 (2 of 2) | DnaJ (Hsp40) homolog, subfamily C, member 11 | 100329402 |
| ENSDARG00000011196 | dnajc11 | DnaJ (Hsp40) homolog, subfamily C, member 11 | 322953 |
| ENSDARG00000044420 | dnajc19 | DnaJ (Hsp40) homolog, subfamily C, member 19 | 393734 |
| ENSDARG00000042948 | dnajc5aa | DnaJ (Hsp40) homolog, subfamily C, member 5aa | 386768 |
| ENSDARG00000004836 | dnajc5ab | DnaJ (Hsp40) homolog, subfamily C, member 5ab | 797907 |
| ENSDARG00000041896 | dnajc5ga | DnaJ (Hsp40) homolog, subfamily C, member 5 gamma a | 322863 |
| ENSDARG00000079891 | dnajc6 | DnaJ (Hsp40) homolog, subfamily C, member 6 | 796345 |
| ENSDARG00000061031 | dner | delta/notch-like EGF repeat containing | 100332110 |
| ENSDARG00000010042 | dnm1a | dynamin 1a | 100307098 |
| ENSDARG00000009281 | dnm1b | dynamin 1b | 100333543 |
| ENSDARG00000015006 | dnm1l | dynamin 1-like | 393896 |
| ENSDARG00000100145 | DNM3 (2 of 2) | dynamin 3 | 0 |
| ENSDARG00000032238 | dnm3a | dynamin 3a | 557996 |
| ENSDARG00000022599 | dnpep | aspartyl aminopeptidase | 393122 |
| ENSDARG00000088293 | doc2b | double C2-like domains, beta | 0 |
| ENSDARG00000075327 | dock10 | dedicator of cytokinesis 10 | 100000949 |
| ENSDARG00000063180 | dock3 | dedicator of cytokinesis 3 | 561387 |
| ENSDARG00000076213 | DOCK4 (2 of 2) | dedicator of cytokinesis 4 | 566115 |
| ENSDARG00000078675 | dock7 | dedicator of cytokinesis 7 | 568817 |
| ENSDARG00000055225 | dock8 | dedicator of cytokinesis 8 | 403040 |
| ENSDARG00000055857 | dopey2 | dopey family member 2 | 0 |
| ENSDARG00000057011 | dpm1 | dolichyl-phosphate mannosyltransferase polypeptide 1, catalytic subunit | 445202 |
| ENSDARG00000104767 | DPP10 | dipeptidyl-peptidase 10 (non-functional) | 0 |
| ENSDARG00000020676 | dpp3 | dipeptidyl-peptidase 3 | 322527 |
| ENSDARG00000076826 | dpp6a | dipeptidyl-peptidase 6a | 100007104 |
| ENSDARG00000024744 | dpp6b | dipeptidyl-peptidase 6b | 566832 |
| ENSDARG00000052606 | dpp9 | dipeptidyl-peptidase 9 | 768170 |
| ENSDARG00000032083 | dpysl2b | dihydropyrimidinase-like 2b | 553412 |
| ENSDARG00000002587 | dpysl3 | dihydropyrimidinase-like 3 | 553166 |
| ENSDARG00000103490 | dpysl4 | dihydropyrimidinase-like 4 | 553411 |
| ENSDARG00000011141 | dpysl5a | dihydropyrimidinase-like 5a | 324416 |
| ENSDARG00000059311 | dpysl5b | dihydropyrimidinase-like 5b | 553410 |
| ENSDARG00000044241 | dram2b | DNA-damage regulated autophagy modulator 2b | 415225 |
| ENSDARG00000039345 | drg1 | developmentally regulated GTP binding protein 1 | 336823 |
| ENSDARG00000006642 | drg2 | developmentally regulated GTP binding protein 2 | 323355 |
| ENSDARG00000098057 | dscaml1 | Down syndrome cell adhesion molecule like 1 | 0 |
| ENSDARG00000101858 | DST (3 of 4) | dystonin | 0 |
| ENSDARG00000044628 | dtd1 | D-tyrosyl-tRNA deacylase 1 | 445046 |
| ENSDARG00000013020 | dtnbb | dystrobrevin, beta b | 559786 |
| ENSDARG00000076302 | dtx1 | deltex1 | 570481 |
| ENSDARG00000078016 | dtx4 | deltex 4, E3 ubiquitin ligase | 100037355 |
| ENSDARG00000045200 | dusp19b | dual specificity phosphatase 19b | 406252 |
| ENSDARG00000061255 | dusp3a | dual specificity phosphatase 3a | 100000665 |
| ENSDARG00000078386 | dync1i2a | dynein, cytoplasmic 1, intermediate chain 2a | 767729 |
| ENSDARG00000098317 | dync1li1 | dynein, cytoplasmic 1, light intermediate chain 1 | 556103 |
| ENSDARG00000021582 | dynlrb1 | dynein, light chain, roadblock-type 1 | 394163 |
| ENSDARG00000007370 | dysf | dysferlin, limb girdle muscular dystrophy 2B (autosomal recessive) | 560924 |
| ENSDARG00000040021 | ebag9 | estrogen receptor binding site associated, antigen, 9 | 394069 |
| ENSDARG00000102142 | ece2b | endothelin converting enzyme 2b | 793465 |
| ENSDARG00000012915 | ech1 | enoyl CoA hydratase 1, peroxisomal | 450048 |
| ENSDARG00000001578 | echs1 | enoyl CoA hydratase, short chain, 1, mitochondrial | 368912 |
| ENSDARG00000018002 | eci1 | enoyl-CoA delta isomerase 1 | 334101 |
| ENSDARG00000102412 | eci2 | enoyl-CoA delta isomerase 2 | 436918 |
| ENSDARG00000062834 | ecsit | ECSIT signalling integrator | 569243 |
| ENSDARG00000093413 | edil3 | EGF-like repeats and discoidin I-like domains 3 | 793444 |
| ENSDARG00000062868 | eea1 | early endosome antigen 1 | 562943 |
| ENSDARG00000039502 | eef1a1a | eukaryotic translation elongation factor 1 alpha 1a | 336334 |
| ENSDARG00000020850 | eef1a1l1 | eukaryotic translation elongation factor 1 alpha 1, like 1 | 30516 |
| ENSDARG00000044521 | eef1b2 | eukaryotic translation elongation factor 1 beta 2 | 335370 |
| ENSDARG00000030053 | eef1db | eukaryotic translation elongation factor 1 delta b (guanine nucleotide exchange protein) | 560745 |
| ENSDARG00000071774 | eef1e1 | eukaryotic translation elongation factor 1 epsilon 1 | 565440 |
| ENSDARG00000056119 | eef1g | eukaryotic translation elongation factor 1 gamma | 195822 |
| ENSDARG00000100371 | eef2b | eukaryotic translation elongation factor 2b | 326929 |
| ENSDARG00000035256 | eef2l2 | eukaryotic translation elongation factor 2, like 2 | 336168 |
| ENSDARG00000055759 | efhd2 | EF-hand domain family, member D2 | 571114 |
| ENSDARG00000007723 | efnb1 | ephrin-B1 | 114377 |
| ENSDARG00000020164 | efnb2a | ephrin-B2a | 30219 |
| ENSDARG00000042277 | efnb3b | ephrin-B3b | 114378 |
| ENSDARG00000033516 | efr3ba | EFR3 homolog Ba (S. cerevisiae) | 560858 |
| ENSDARG00000069318 | efr3bb | EFR3 homolog Bb (S. cerevisiae) | 561353 |
| ENSDARG00000014793 | ehd1b | EH-domain containing 1b | 447839 |
| ENSDARG00000007869 | ehd3 | EH-domain containing 3 | 562949 |
| ENSDARG00000070029 | ehhadh | enoyl-CoA, hydratase/3-hydroxyacyl CoA dehydrogenase | 100000859 |
| ENSDARG00000057912 | eif1axb | eukaryotic translation initiation factor 1A, X-linked, b | 322599 |
| ENSDARG00000052178 | eif2s1b | eukaryotic translation initiation factor 2, subunit 1 alpha b | 321564 |
| ENSDARG00000021257 | eif3d | eukaryotic translation initiation factor 3, subunit D | 336498 |
| ENSDARG00000077533 | eif3f | eukaryotic translation initiation factor 3, subunit F | 557270 |
| ENSDARG00000016889 | eif3g | eukaryotic translation initiation factor 3, subunit G | 393974 |
| ENSDARG00000068289 | eif3k | eukaryotic translation initiation factor 3, subunit K | 550245 |
| ENSDARG00000013931 | eif3m | eukaryotic translation initiation factor 3, subunit M | 334626 |
| ENSDARG00000076815 | eif3s10 | eukaryotic translation initiation factor 3, subunit 10 (theta) | 327515 |
| ENSDARG00000101082 | eif3s6ip | eukaryotic translation initiation factor 3, subunit 6 interacting protein | 406402 |
| ENSDARG00000003032 | eif4a1b | eukaryotic translation initiation factor 4A1B | 399484 |
| ENSDARG00000010194 | eif4bb | eukaryotic translation initiation factor 4Bb | 336619 |
| ENSDARG00000042252 | eif4h | eukaryotic translation initiation factor 4h | 402996 |
| ENSDARG00000017235 | eif5a | eukaryotic translation initiation factor 5A | 406464 |
| ENSDARG00000038695 | elavl1 | ELAV like RNA binding protein 1 | 30736 |
| ENSDARG00000014420 | elavl3 | ELAV like neuron-specific RNA binding protein 3 | 30732 |
| ENSDARG00000045639 | elavl4 | ELAV like neuron-specific RNA binding protein 4 | 30737 |
| ENSDARG00000078141 | ELFN1 (2 of 2) | extracellular leucine-rich repeat and fibronectin type III domain containing 1 | 562570 |
| ENSDARG00000074372 | elfn1b | extracellular leucine-rich repeat and fibronectin type III domain containing 1b | 100144401 |
| ENSDARG00000077609 | elfn2a | extracellular leucine-rich repeat and fibronectin type III domain containing 2a | 0 |
| ENSDARG00000098753 | elmo1 | engulfment and cell motility 1 (ced-12 homolog, C. elegans) | 406364 |
| ENSDARG00000063527 | elmo2 | engulfment and cell motility 2 | 100006857 |
| ENSDARG00000101669 | elmod2 | ELMO/CED-12 domain containing 2 | 553267 |
| ENSDARG00000059485 | EMB | embigin | 100141497 |
| ENSDARG00000054793 | emc10 | ER membrane protein complex subunit 10 | 565829 |
| ENSDARG00000057255 | emc1 | ER membrane protein complex subunit 1 | 565399 |
| ENSDARG00000003308 | emc2 | ER membrane protein complex subunit 2 | 407073 |
| ENSDARG00000020607 | emc3 | ER membrane protein complex subunit 3 | 406826 |
| ENSDARG00000039268 | emc4 | ER membrane protein complex subunit 4 | 0 |
| ENSDARG00000035282 | emc6 | ER membrane protein complex subunit 6 | 337220 |
| ENSDARG00000012144 | emc7 | ER membrane protein complex subunit 7 | 561549 |
| ENSDARG00000008808 | eml2 | echinoderm microtubule associated protein like 2 | 569375 |
| ENSDARG00000032049 | enah | enabled homolog (Drosophila) | 541394 |
| ENSDARG00000071224 | ENDOD1 (7 of 10) | endonuclease domain containing 1 | 559260 |
| ENSDARG00000022456 | eno1a | enolase 1a, (alpha) | 334116 |
| ENSDARG00000013750 | eno1b | enolase 1b, (alpha) | 393668 |
| ENSDARG00000014287 | eno2 | enolase 2 | 402874 |
| ENSDARG00000039007 | eno3 | enolase 3, (beta, muscle) | 378963 |
| ENSDARG00000040469 | enpp6 | ectonucleotide pyrophosphatase/phosphodiesterase 6 | 791915 |
| ENSDARG00000061644 | ensab | endosulfine alpha b | 100317933 |
| ENSDARG00000045066 | entpd1 | ectonucleoside triphosphate diphosphohydrolase 1 | 445151 |
| ENSDARG00000029019 | epb41b | erythrocyte membrane protein band 4.1b (elliptocytosis 1, RH-linked) | 326287 |
| ENSDARG00000076364 | EPB41L1 | erythrocyte membrane protein band 4.1-like 1 | 0 |
| ENSDARG00000093374 | EPB41L2 | erythrocyte membrane protein band 4.1-like 2 | 0 |
| ENSDARG00000002255 | epb41l3a | erythrocyte membrane protein band 4.1-like 3a | 445478 |
| ENSDARG00000040087 | epb41l4a | erythrocyte membrane protein band 4.1 like 4A | 30470 |
| ENSDARG00000061398 | epb41l4b | erythrocyte membrane protein band 4.1 like 4B | 100332627 |
| ENSDARG00000103498 | epd | ependymin | 30199 |
| ENSDARG00000045420 | epdr1 | ependymin related 1 | 436689 |
| ENSDARG00000017354 | epha2a | eph receptor A2 a | 30689 |
| ENSDARG00000054454 | epha4a | eph receptor A4a | 64271 |
| ENSDARG00000010767 | epha4l | eph receptor A4, like | 30687 |
| ENSDARG00000022971 | epha6 | eph receptor A6 | 100005538 |
| ENSDARG00000004635 | epha7 | eph receptor A7 | 562195 |
| ENSDARG00000076757 | ephb1 | EPH receptor B1 | 0 |
| ENSDARG00000037373 | ephb2a | eph receptor B2a | 562176 |
| ENSDARG00000021539 | ephb2b | eph receptor B2b | 554232 |
| ENSDARG00000031548 | ephb3a | eph receptor B3a | 30313 |
| ENSDARG00000100725 | ephb4a | eph receptor B4a | 30688 |
| ENSDARG00000042854 | ephx1 | epoxide hydrolase 1, microsomal (xenobiotic) | 394043 |
| ENSDARG00000040255 | ephx2 | epoxide hydrolase 2, cytoplasmic | 494099 |
| ENSDARG00000103298 | ephx5 | epoxide hydrolase 5 | 322331 |
| ENSDARG00000010411 | epn1 | epsin 1 | 406671 |
| ENSDARG00000060494 | eprs | glutamyl-prolyl-tRNA synthetase | 562037 |
| ENSDARG00000078216 | eps15 | epidermal growth factor receptor pathway substrate 15 | 795505 |
| ENSDARG00000042670 | eps15l1a | epidermal growth factor receptor pathway substrate 15-like 1a | 568170 |
| ENSDARG00000060484 | EPT1 | ethanolaminephosphotransferase 1 | 557632 |
| ENSDARG00000021859 | erap1b | endoplasmic reticulum aminopeptidase 1b | 393175 |
| ENSDARG00000013161 | ERAS | ES cell expressed Ras | 393228 |
| ENSDARG00000044281 | erbb2ip | erbb2 interacting protein | 566811 |
| ENSDARG00000063207 | erbb4a | erb-b2 receptor tyrosine kinase 4a | 100007677 |
| ENSDARG00000076014 | ERC1 (3 of 3) | ELKS/RAB6-interacting/CAST family member 1 | 560003 |
| ENSDARG00000009941 | erc1a | ELKS/RAB6-interacting/CAST family member 1a | 548606 |
| ENSDARG00000021991 | erlin1 | ER lipid raft associated 1 | 541490 |
| ENSDARG00000086523 | erlin2 | ER lipid raft associated 2 (erlin2), mRNA | 100151163 |
| ENSDARG00000019008 | erp44 | endoplasmic reticulum protein 44 | 393866 |
| ENSDARG00000019496 | esd | esterase D/formylglutathione hydrolase | 550494 |
| ENSDARG00000014239 | esyt1b | extended synaptotagmin-like protein 1b | 571083 |
| ENSDARG00000043976 | etf1b | eukaryotic translation termination factor 1b | 266989 |
| ENSDARG00000101631 | ETFA | electron-transfer-flavoprotein, alpha polypeptide | 0 |
| ENSDARG00000009250 | etfb | electron-transfer-flavoprotein, beta polypeptide | 406271 |
| ENSDARG00000038834 | etfdh | electron-transferring-flavoprotein dehydrogenase | 447859 |
| ENSDARG00000035650 | evla | Enah/Vasp-like a | 58003 |
| ENSDARG00000099720 | evlb | Enah/Vasp-like b | 0 |
| ENSDARG00000043019 | exoc1 | exocyst complex component 1 | 322338 |
| ENSDARG00000055610 | exoc2 | exocyst complex component 2 | 100004870 |
| ENSDARG00000014582 | exoc3 | exocyst complex component 3 | 334015 |
| ENSDARG00000004336 | exoc4 | exocyst complex component 4 | 559866 |
| ENSDARG00000060323 | exoc5 | exocyst complex component 5 | 559010 |
| ENSDARG00000098874 | exoc6b | exocyst complex component 6B | 767763 |
| ENSDARG00000020521 | exoc6 | exocyst complex component 6 | 406591 |
| ENSDARG00000087229 | exoc7 | exocyst complex component 7 | 406723 |
| ENSDARG00000063211 | exoc8 | exocyst complex component 8 | 568029 |
| ENSDARG00000056648 | ext2 | exostosin glycosyltransferase 2 | 493780 |
| ENSDARG00000026811 | extl3 | exostosin-like glycosyltransferase 3 | 493783 |
| ENSDARG00000025091 | ezrb | ezrin b | 561589 |
| ENSDARG00000099124 | f3a | coagulation factor IIIa | 567257 |
| ENSDARG00000008457 | faah2a | fatty acid amide hydrolase 2a | 436973 |
| ENSDARG00000077764 | FAAH | fatty acid amide hydrolase | 569067 |
| ENSDARG00000017299 | fabp11a | fatty acid binding protein 11a | 447944 |
| ENSDARG00000023290 | fabp3 | fatty acid binding protein 3, muscle and heart | 171478 |
| ENSDARG00000007697 | fabp7a | fatty acid binding protein 7, brain, a | 58128 |
| ENSDARG00000034650 | fabp7b | fatty acid binding protein 7, brain, b | 407736 |
| ENSDARG00000019532 | fads2 | fatty acid desaturase 2 | 140615 |
| ENSDARG00000052374 | faf2 | Fas associated factor family member 2 | 497596 |
| ENSDARG00000086300 | FAM107A | family with sequence similarity 107, member A | 794256 |
| ENSDARG00000019205 | fam120c | family with sequence similarity 120C | 557842 |
| ENSDARG00000026762 | fam126a | family with sequence similarity 126, member A | 402926 |
| ENSDARG00000100400 | FAM126B (1 of 2) | family with sequence similarity 126, member B | 0 |
| ENSDARG00000008026 | fam129bb | family with sequence similarity 129, member Bb | 556540 |
| ENSDARG00000070952 | fam131ba | family with sequence similarity 131, member Ba | 565996 |
| ENSDARG00000070575 | fam131bb | family with sequence similarity 131, member Bb | 556725 |
| ENSDARG00000098401 | fam136a | family with sequence similarity 136, member A | 0 |
| ENSDARG00000063344 | fam162a | family with sequence similarity 162, member A | 336363 |
| ENSDARG00000079656 | FAM171A2 (2 of 2) | family with sequence similarity 171, member A2 | 797104 |
| ENSDARG00000075249 | fam171a2 | family with sequence similarity 171, member A2 | 556398 |
| ENSDARG00000014081 | fam184a | family with sequence similarity 184, member A | 555640 |
| ENSDARG00000008573 | fam20b | family with sequence similarity 20, member B (H. sapiens) | 557060 |
| ENSDARG00000035985 | FAM210A (1 of 2) | family with sequence similarity 210, member A | 562734 |
| ENSDARG00000052638 | fam210b | family with sequence similarity 210, member B | 619270 |
| ENSDARG00000057378 | fam213aa | family with sequence similarity 213, member Aa | 323739 |
| ENSDARG00000016396 | fam219aa | family with sequence similarity 219, member Aa | 445197 |
| ENSDARG00000070675 | fam3a | family with sequence similarity 3, member A | 393773 |
| ENSDARG00000035907 | fam49a | family with sequence similarity 49, member A | 445212 |
| ENSDARG00000045417 | fam49bb | family with sequence similarity 49, member Bb | 404610 |
| ENSDARG00000098860 | FAM57A | family with sequence similarity 57, member A | 0 |
| ENSDARG00000017431 | fam63b | family with sequence similarity 63, member B | 570359 |
| ENSDARG00000003320 | fam91a1 | family with sequence similarity 91, member A1 | 393413 |
| ENSDARG00000052010 | fam96b | family with sequence similarity 96, member B | 436722 |
| ENSDARG00000074381 | farp1 | FERM, RhoGEF (ARHGEF) and pleckstrin domain protein 1 (chondrocyte-derived) | 0 |
| ENSDARG00000016429 | farp2 | FERM, RhoGEF and pleckstrin domain protein 2 | 567490 |
| ENSDARG00000098825 | farsa | phenylalanyl-tRNA synthetase, alpha subunit | 692329 |
| ENSDARG00000016864 | farsb | phenylalanyl-tRNA synthetase, beta subunit | 493608 |
| ENSDARG00000087657 | fasn | fatty acid synthase | 559001 |
| ENSDARG00000017591 | fat1a | FAT atypical cadherin 1a | 406172 |
| ENSDARG00000018923 | fat2 | FAT atypical cadherin 2 | 568468 |
| ENSDARG00000033840 | fat3a | FAT atypical cadherin 3a | 0 |
| ENSDARG00000099022 | faua | Finkel-Biskis-Reilly murine sarcoma virus (FBR-MuSV) ubiquitously expressed a | 393710 |
| ENSDARG00000099306 | FBXO2 | F-box protein 2 | 559776 |
| ENSDARG00000076310 | FBXO41 | F-box protein 41 | 101883316 |
| ENSDARG00000078908 | fbxo41 | F-box protein 41 | 567804 |
| ENSDARG00000040890 | fdps | farnesyl diphosphate synthase (farnesyl pyrophosphate synthetase, dimethylallyltranstransferase, geranyltranstransferase) | 552997 |
| ENSDARG00000003462 | fech | ferrochelatase | 58215 |
| ENSDARG00000053973 | fetub | fetuin B | 569489 |
| ENSDARG00000008969 | fgb | fibrinogen beta chain | 337315 |
| ENSDARG00000062542 | fgf12b | fibroblast growth factor 12b | 751749 |
| ENSDARG00000040162 | FGFBP3 | fibroblast growth factor binding protein 3 | 497419 |
| ENSDARG00000058115 | fgfr2 | fibroblast growth factor receptor 2 | 352940 |
| ENSDARG00000037281 | fgg | fibrinogen gamma chain | 406327 |
| ENSDARG00000075132 | fh | fumarate hydratase | 393938 |
| ENSDARG00000060393 | fibcd1 | fibrinogen C domain containing 1 | 567525 |
| ENSDARG00000016058 | fibpa | fibroblast growth factor (acidic) intracellular binding protein a | 393381 |
| ENSDARG00000095579 | FIS1 | fission 1 (mitochondrial outer membrane) homolog (S. cerevisiae) | 797988 |
| ENSDARG00000037000 | fkbp11 | FK506 binding protein 11 | 368823 |
| ENSDARG00000022684 | fkbp1aa | FK506 binding protein 1Aa | 335335 |
| ENSDARG00000033567 | fkbp1ab | FK506 binding protein 1Ab | 449552 |
| ENSDARG00000044540 | fkbp2 | FK506 binding protein 2 | 447939 |
| ENSDARG00000008447 | fkbp4 | FK506 binding protein 4 | 321795 |
| ENSDARG00000059701 | flii | flightless I homolog (Drosophila) | 560281 |
| ENSDARG00000010764 | flj13639 | flj13639 | 677747 |
| ENSDARG00000074201 | flna | filamin A, alpha (actin binding protein 280) | 0 |
| ENSDARG00000098374 | FLNB | filamin B, beta | 0 |
| ENSDARG00000097351 | flnbl | filamin B, like | 100534866 |
| ENSDARG00000001710 | flot1a | flotillin 1a | 561345 |
| ENSDARG00000037998 | flot1b | flotillin 1b | 245697 |
| ENSDARG00000004830 | flot2a | flotillin 2a | 245698 |
| ENSDARG00000069774 | flot2b | flotillin 2b | 393612 |
| ENSDARG00000019371 | flt1 | fms-related tyrosine kinase 1 (vascular endothelial growth factor/vascular permeability factor receptor) | 544667 |
| ENSDARG00000031506 | flvcr2b | feline leukemia virus subgroup C cellular receptor family, member 2b | 569329 |
| ENSDARG00000061778 | fmn2b | formin 2b | 566432 |
| ENSDARG00000012586 | fmnl2a | formin-like 2a | 100006456 |
| ENSDARG00000075041 | fmnl2b | formin-like 2b | 492608 |
| ENSDARG00000016357 | fmo5 | flavin containing monooxygenase 5 | 794311 |
| ENSDARG00000037433 | fmr1 | fragile X mental retardation 1 | 259191 |
| ENSDARG00000099143 | fnta | farnesyltransferase, CAAX box, alpha | 555882 |
| ENSDARG00000078399 | FO704819.2 | Uncharacterized protein | 100332846 |
| ENSDARG00000098751 | FQ323163.2 | Uncharacterized protein | 795227 |
| ENSDARG00000042249 | FRMD4A | FERM domain containing 4A | 564713 |
| ENSDARG00000074865 | frmpd3 | FERM and PDZ domain containing 3 | 798302 |
| ENSDARG00000078688 | frs3 | fibroblast growth factor receptor substrate 3 | 100330147 |
| ENSDARG00000056001 | fryb | furry homolog b (Drosophila) | 792949 |
| ENSDARG00000040001 | fryl | furry homolog, like | 558173 |
| ENSDARG00000059680 | fscn1a | fascin actin-bundling protein 1a | 558271 |
| ENSDARG00000037747 | fscn1b | fascin actin-bundling protein 1b | 570314 |
| ENSDARG00000015551 | fth1a | ferritin, heavy polypeptide 1a | 58108 |
| ENSDARG00000044216 | fto | fat mass and obesity associated | 553363 |
| ENSDARG00000029248 | fubp1 | far upstream element (FUSE) binding protein 1 | 406418 |
| ENSDARG00000040822 | fundc1 | FUN14 domain containing 1 | 436984 |
| ENSDARG00000103740 | fundc2 | fun14 domain containing 2 | 792181 |
| ENSDARG00000088117 | FUT8 (2 of 2) | fucosyltransferase 8 (alpha (1,6) fucosyltransferase) | 100331714 |
| ENSDARG00000015449 | fut8a | fucosyltransferase 8a (alpha (1,6) fucosyltransferase) | 797512 |
| ENSDARG00000100971 | fxyd6 | FXYD domain containing ion transport regulator 6 | 333987 |
| ENSDARG00000011370 | fyna | FYN proto-oncogene, Src family tyrosine kinase a | 373872 |
| ENSDARG00000025319 | fynb | FYN proto-oncogene, Src family tyrosine kinase b | 574422 |
| ENSDARG00000017741 | g3bp1 | GTPase activating protein (SH3 domain) binding protein 1 | 335512 |
| ENSDARG00000071065 | g6pd | glucose-6-phosphate dehydrogenase | 570579 |
| ENSDARG00000027200 | gabarapl2 | GABA(A) receptor-associated protein-like 2 | 403036 |
| ENSDARG00000018967 | gabbr1a | gamma-aminobutyric acid (GABA) B receptor, 1a | 373873 |
| ENSDARG00000016667 | gabbr1b | gamma-aminobutyric acid (GABA) B receptor, 1b | 558708 |
| ENSDARG00000061042 | gabbr2 | gamma-aminobutyric acid (GABA) B receptor, 2 | 560267 |
| ENSDARG00000068989 | gabra1 | gamma-aminobutyric acid (GABA) A receptor, alpha 1 | 768183 |
| ENSDARG00000034011 | GABRA2 (1 of 2) | gamma-aminobutyric acid (GABA) A receptor, alpha 2 | 0 |
| ENSDARG00000058736 | gabra6b | gamma-aminobutyric acid (GABA) A receptor, alpha 6b | 559693 |
| ENSDARG00000079586 | gabrb2 | gamma-aminobutyric acid (GABA) A receptor, beta 2 | 0 |
| ENSDARG00000023771 | gabrb3 | gamma-aminobutyric acid (GABA) A receptor, beta 3 | 566922 |
| ENSDARG00000099096 | gabrb4 | gamma-aminobutyric acid (GABA) A receptor, beta 4 | 566514 |
| ENSDARG00000059763 | gabrd | gamma-aminobutyric acid (GABA) A receptor, delta | 571422 |
| ENSDARG00000053665 | gabrg2 | gamma-aminobutyric acid (GABA) A receptor, gamma 2 | 553402 |
| ENSDARG00000094512 | gabrz | gamma-aminobutyric acid (GABA) A receptor, zeta | 561738 |
| ENSDARG00000027419 | gad1b | glutamate decarboxylase 1b | 378441 |
| ENSDARG00000015537 | gad2 | glutamate decarboxylase 2 | 550403 |
| ENSDARG00000090654 | gak | cyclin G associated kinase | 100151158 |
| ENSDARG00000061335 | galnt1 | UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 1 | 559073 |
| ENSDARG00000003829 | galnt2 | UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 2 | 570248 |
| ENSDARG00000057303 | galnt7 | UDP-N-acetyl-alpha-D-galactosamine: polypeptide N-acetylgalactosaminyltransferase 7 | 553668 |
| ENSDARG00000006832 | galnt9 | polypeptide N-acetylgalactosaminyltransferase 9 | 797549 |
| ENSDARG00000076888 | GANAB (2 of 2) | glucosidase, alpha; neutral AB | 100126019 |
| ENSDARG00000099744 | gap43 | growth associated protein 43 | 30608 |
| ENSDARG00000043457 | gapdh | glyceraldehyde-3-phosphate dehydrogenase | 317743 |
| ENSDARG00000039914 | gapdhs | glyceraldehyde-3-phosphate dehydrogenase, spermatogenic | 406367 |
| ENSDARG00000090183 | gapvd1 | GTPase activating protein and VPS9 domains 1 | 436693 |
| ENSDARG00000059070 | gars | glycyl-tRNA synthetase | 337230 |
| ENSDARG00000074443 | gas7a | growth arrest-specific 7a | 793701 |
| ENSDARG00000076058 | gba | glucosidase, beta, acid | 559072 |
| ENSDARG00000007264 | gbas | glioblastoma amplified sequence | 30232 |
| ENSDARG00000099524 | gbe1b | glucan (1,4-alpha-), branching enzyme 1b | 559212 |
| ENSDARG00000005643 | gcat | glycine C-acetyltransferase | 402822 |
| ENSDARG00000037057 | gcdha | glutaryl-CoA dehydrogenase a | 393860 |
| ENSDARG00000098831 | gcdhb | glutaryl-CoA dehydrogenase b | 450003 |
| ENSDARG00000058419 | gcn1l1 | GCN1 general control of amino-acid synthesis 1-like 1 (yeast) | 0 |
| ENSDARG00000091655 | gcsha | glycine cleavage system protein H (aminomethyl carrier), a | 541329 |
| ENSDARG00000058601 | gdap1 | ganglioside induced differentiation associated protein 1 | 553702 |
| ENSDARG00000028628 | gdap1l1 | ganglioside induced differentiation associated protein 1-like 1 | 562163 |
| ENSDARG00000055108 | gde1 | glycerophosphodiester phosphodiesterase 1 | 393213 |
| ENSDARG00000056122 | gdi1 | GDP dissociation inhibitor 1 | 554985 |
| ENSDARG00000005451 | gdi2 | GDP dissociation inhibitor 2 | 323765 |
| ENSDARG00000017261 | gdpd1 | glycerophosphodiester phosphodiesterase domain containing 1 | 445504 |
| ENSDARG00000076962 | gdpd5b | glycerophosphodiester phosphodiesterase domain containing 5b | 560645 |
| ENSDARG00000025301 | gfap | glial fibrillary acidic protein | 30646 |
| ENSDARG00000063624 | gfm1 | G elongation factor, mitochondrial 1 | 561840 |
| ENSDARG00000086866 | GFPT1 (2 of 2) | glutamine--fructose-6-phosphate transaminase 1 | 0 |
| ENSDARG00000057465 | gfpt1 | glutamine--fructose-6-phosphate transaminase 1 | 0 |
| ENSDARG00000042723 | gfra2a | GDNF family receptor alpha 2a | 405794 |
| ENSDARG00000044015 | gfra2b | GDNF family receptor alpha 2b | 564916 |
| ENSDARG00000056651 | gfra4a | GDNF family receptor alpha 4a | 100144564 |
| ENSDARG00000005085 | ggctb | gamma-glutamylcyclotransferase b | 406278 |
| ENSDARG00000078258 | GGT5 (2 of 2) | gamma-glutamyltransferase 5 | 100333757 |
| ENSDARG00000055569 | ghdc | GH3 domain containing | 497449 |
| ENSDARG00000098983 | GHITM | growth hormone inducible transmembrane protein | 0 |
| ENSDARG00000104483 | gipc1 | GIPC PDZ domain containing family, member 1 | 445568 |
| ENSDARG00000039489 | git1 | G protein-coupled receptor kinase interacting ArfGAP 1 | 568935 |
| ENSDARG00000053456 | GK2 | glycerol kinase 2 | 100136866 |
| ENSDARG00000068981 | glceb | glucuronic acid epimerase b | 100007670 |
| ENSDARG00000061282 | glg1a | golgi glycoprotein 1a | 792865 |
| ENSDARG00000101210 | glg1b | golgi glycoprotein 1b | 751680 |
| ENSDARG00000016837 | glipr2l | GLI pathogenesis-related 2, like | 449805 |
| ENSDARG00000068978 | glo1 | glyoxalase 1 | 368213 |
| ENSDARG00000042509 | glod4 | glyoxalase domain containing 4 | 447874 |
| ENSDARG00000012019 | glra1 | glycine receptor, alpha 1 | 30676 |
| ENSDARG00000100944 | glrx2 | glutaredoxin 2 | 436677 |
| ENSDARG00000098785 | glrx3 | glutaredoxin 3 | 449777 |
| ENSDARG00000043665 | glrx5 | glutaredoxin 5 homolog (S. cerevisiae) | 406294 |
| ENSDARG00000062812 | glsa | glutaminase a | 564147 |
| ENSDARG00000040705 | glsb | glutaminase b | 564746 |
| ENSDARG00000008816 | glud1a | glutamate dehydrogenase 1a | 317737 |
| ENSDARG00000101074 | glud1b | glutamate dehydrogenase 1b | 373092 |
| ENSDARG00000099776 | glula | glutamate-ammonia ligase (glutamine synthase) a | 100000775 |
| ENSDARG00000100003 | glulb | glutamate-ammonia ligase (glutamine synthase) b | 336473 |
| ENSDARG00000088439 | gm2a | GM2 ganglioside activator | 550529 |
| ENSDARG00000026629 | gmds | GDP-mannose 4,6-dehydratase | 393461 |
| ENSDARG00000028327 | gmfb | glia maturation factor, beta | 406384 |
| ENSDARG00000017658 | gmppb | GDP-mannose pyrophosphorylase B | 445097 |
| ENSDARG00000061301 | gmpr2 | guanosine monophosphate reductase 2 | 678545 |
| ENSDARG00000010002 | gna11a | guanine nucleotide binding protein (G protein), alpha 11a (Gq class) | 563953 |
| ENSDARG00000053326 | gna11b | guanine nucleotide binding protein (G protein), alpha 11b (Gq class) | 493613 |
| ENSDARG00000073765 | gna13a | guanine nucleotide binding protein (G protein), alpha 13a | 326825 |
| ENSDARG00000037924 | gna13b | guanine nucleotide binding protein (G protein), alpha 13b | 336333 |
| ENSDARG00000092948 | gna15.3 | guanine nucleotide binding protein (G protein), alpha 15 (Gq class), tandem duplicate 3 | 0 |
| ENSDARG00000021647 | gnai1 | guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 1 | 393946 |
| ENSDARG00000018174 | gnai2a | guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 2a | 336421 |
| ENSDARG00000030644 | gnai3 | guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 3 | 100006888 |
| ENSDARG00000044760 | gnaia | guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide a | 323509 |
| ENSDARG00000045415 | gnal | guanine nucleotide binding protein (G protein), alpha activating activity polypeptide, olfactory type | 492467 |
| ENSDARG00000016676 | gnao1a | guanine nucleotide binding protein (G protein), alpha activating activity polypeptide O, a | 393760 |
| ENSDARG00000036058 | gnao1b | guanine nucleotide binding protein (G protein), alpha activating activity polypeptide O, b | 393801 |
| ENSDARG00000011487 | gnaq | guanine nucleotide binding protein (G protein), q polypeptide | 570108 |
| ENSDARG00000043094 | gnas | GNAS complex locus | 557353 |
| ENSDARG00000099844 | GNAZ | guanine nucleotide binding protein (G protein), alpha z polypeptide | 0 |
| ENSDARG00000035357 | gnb2 | guanine nucleotide binding protein (G protein), beta polypeptide 2 | 541370 |
| ENSDARG00000041619 | gnb2l1 | guanine nucleotide binding protein (G protein), beta polypeptide 2-like 1 | 30722 |
| ENSDARG00000019278 | GNB4 | guanine nucleotide binding protein (G protein), beta polypeptide 4 | 0 |
| ENSDARG00000099685 | gnb5a | guanine nucleotide binding protein (G protein), beta 5a | 562813 |
| ENSDARG00000055377 | gnb5b | guanine nucleotide binding protein (G protein), beta 5b | 393719 |
| ENSDARG00000003375 | gng12a | guanine nucleotide binding protein (G protein), gamma 12a | 406604 |
| ENSDARG00000037921 | gng13b | guanine nucleotide binding protein (G protein), gamma 13b | 436673 |
| ENSDARG00000056831 | gng2 | guanine nucleotide binding protein (G protein), gamma 2 | 550256 |
| ENSDARG00000009553 | gng3 | guanine nucleotide binding protein (G protein), gamma 3 | 114462 |
| ENSDARG00000039830 | gng5 | guanine nucleotide binding protein (G protein), gamma 5 | 337838 |
| ENSDARG00000038693 | gng7 | guanine nucleotide binding protein (G protein), gamma 7 | 436670 |
| ENSDARG00000024443 | gnpda2 | glucosamine-6-phosphate deaminase 2 | 556291 |
| ENSDARG00000063197 | golga2 | golgin A2 | 565431 |
| ENSDARG00000062511 | golga3 | golgin A3 | 570879 |
| ENSDARG00000039279 | golga5 | golgin A5 | 406720 |
| ENSDARG00000036295 | golga7ba | golgin A7 family, member Ba | 550228 |
| ENSDARG00000105099 | golga7bb | golgin A7 family, member Bb | 0 |
| ENSDARG00000057676 | golga7 | golgin A7 | 798622 |
| ENSDARG00000061951 | golgb1 | golgin B1 | 566353 |
| ENSDARG00000025858 | GOLM1 | golgi membrane protein 1 | 393249 |
| ENSDARG00000015126 | gorasp2 | golgi reassembly stacking protein 2 | 393676 |
| ENSDARG00000026654 | gosr1 | golgi SNAP receptor complex member 1 | 100001316 |
| ENSDARG00000053070 | gosr2 | golgi SNAP receptor complex member 2 | 324569 |
| ENSDARG00000039093 | got1 | glutamic-oxaloacetic transaminase 1, soluble | 406330 |
| ENSDARG00000041068 | got2a | glutamic-oxaloacetic transaminase 2a, mitochondrial | 406688 |
| ENSDARG00000005840 | got2b | glutamic-oxaloacetic transaminase 2b, mitochondrial | 335974 |
| ENSDARG00000074169 | gpam | glycerol-3-phosphate acyltransferase, mitochondrial | 100332911 |
| ENSDARG00000019341 | gpc1a | glypican 1a | 553367 |
| ENSDARG00000090585 | gpc1b | glypican 1b | 393995 |
| ENSDARG00000032199 | gpc3 | glypican 3 | 0 |
| ENSDARG00000043180 | gpd1b | glycerol-3-phosphate dehydrogenase 1b | 325181 |
| ENSDARG00000040024 | gpd1l | glycerol-3-phosphate dehydrogenase 1-like | 449663 |
| ENSDARG00000062430 | gpd2 | glycerol-3-phosphate dehydrogenase 2 (mitochondrial) | 751628 |
| ENSDARG00000089076 | gphna | gephyrin a | 555470 |
| ENSDARG00000100851 | gphnb | gephyrin b | 0 |
| ENSDARG00000012987 | gpia | glucose-6-phosphate isomerase a | 246094 |
| ENSDARG00000055455 | gpm6aa | glycoprotein M6Aa | 368223 |
| ENSDARG00000004621 | gpm6ab | glycoprotein M6Ab | 407734 |
| ENSDARG00000005739 | gpm6ba | glycoprotein M6Ba | 368225 |
| ENSDARG00000001676 | gpm6bb | glycoprotein M6Bb | 503756 |
| ENSDARG00000103200 | gpr107 | G protein-coupled receptor 107 | 570589 |
| ENSDARG00000096651 | gpr108 | G protein-coupled receptor 108 | 0 |
| ENSDARG00000077134 | gpr158a | G protein-coupled receptor 158a | 570001 |
| ENSDARG00000079665 | gpr158b | G protein-coupled receptor 158b | 568637 |
| ENSDARG00000056774 | gpr37l1b | G protein-coupled receptor 37 like 1b | 567796 |
| ENSDARG00000077080 | gprc5ba | G protein-coupled receptor, class C, group 5, member Ba | 570207 |
| ENSDARG00000075141 | gprc5bb | G protein-coupled receptor, class C, group 5, member Bb | 100002676 |
| ENSDARG00000075043 | GPRIN3 | GPRIN family member 3 | 100536832 |
| ENSDARG00000040650 | gps1 | G protein pathway suppressor 1 | 777711 |
| ENSDARG00000018146 | gpx1a | glutathione peroxidase 1a | 0 |
| ENSDARG00000076836 | gpx4b | glutathione peroxidase 4b | 0 |
| ENSDARG00000038059 | grb2b | growth factor receptor-bound protein 2b | 406308 |
| ENSDARG00000068264 | grhpra | glyoxylate reductase/hydroxypyruvate reductase a | 497183 |
| ENSDARG00000021352 | gria1a | glutamate receptor, ionotropic, AMPA 1a | 798689 |
| ENSDARG00000032714 | gria1b | glutamate receptor, ionotropic, AMPA 1b | 403044 |
| ENSDARG00000070173 | gria2a | glutamate receptor, ionotropic, AMPA 2a | 170450 |
| ENSDARG00000052765 | gria2b | glutamate receptor, ionotropic, AMPA 2b | 170451 |
| ENSDARG00000032737 | gria3a | glutamate receptor, ionotropic, AMPA 3a | 170452 |
| ENSDARG00000037498 | gria3b | glutamate receptor, ionotropic, AMPA 3b | 368416 |
| ENSDARG00000037496 | gria4a | glutamate receptor, ionotropic, AMPA 4a | 407735 |
| ENSDARG00000059368 | gria4b | glutamate receptor, ionotropic, AMPA 4b | 336069 |
| ENSDARG00000074583 | grid1a | glutamate receptor, ionotropic, delta 1a | 793756 |
| ENSDARG00000044161 | grid1b | glutamate receptor, ionotropic, delta 1b | 557511 |
| ENSDARG00000055302 | grid2 | glutamate receptor, ionotropic, delta 2 | 448841 |
| ENSDARG00000076103 | grid2ipa | glutamate receptor, ionotropic, delta 2 (Grid2) interacting protein, a | 561017 |
| ENSDARG00000095603 | grid2ipb | glutamate receptor, ionotropic, delta 2 (Grid2) interacting protein, b | 559717 |
| ENSDARG00000077715 | GRIK3 | glutamate receptor, ionotropic, kainate 3 | 0 |
| ENSDARG00000027828 | grin1a | glutamate receptor, ionotropic, N-methyl D-aspartate 1a | 767745 |
| ENSDARG00000025728 | grin1b | glutamate receptor, ionotropic, N-methyl D-aspartate 1b | 100005675 |
| ENSDARG00000034493 | grin2aa | glutamate receptor, ionotropic, N-methyl D-aspartate 2A, a | 563297 |
| ENSDARG00000070543 | grin2ab | glutamate receptor, ionotropic, N-methyl D-aspartate 2A, b | 570493 |
| ENSDARG00000079348 | GRIN2B (2 of 2) | glutamate receptor, ionotropic, N-methyl D-aspartate 2B | 0 |
| ENSDARG00000030376 | grin2bb | glutamate receptor, ionotropic, N-methyl D-aspartate 2B, genome duplicate b | 559976 |
| ENSDARG00000070620 | grin2db | glutamate receptor, ionotropic, N-methyl D-aspartate 2D, b | 0 |
| ENSDARG00000101317 | grinaa | glutamate receptor, ionotropic, N-methyl D-aspartate-associated protein 1a (glutamate binding) | 724005 |
| ENSDARG00000015053 | grip1 | glutamate receptor interacting protein 1 | 558006 |
| ENSDARG00000027618 | grip2b | glutamate receptor interacting protein 2b | 562290 |
| ENSDARG00000063069 | gripap1 | GRIP1 associated protein 1 | 791150 |
| ENSDARG00000070448 | grk4 | G protein-coupled receptor kinase 4 | 560761 |
| ENSDARG00000078815 | grk5l | G protein-coupled receptor kinase 5 like | 567452 |
| ENSDARG00000032157 | grk6 | G protein-coupled receptor kinase 6 | 561288 |
| ENSDARG00000026796 | grm1a | glutamate receptor, metabotropic 1a | 555576 |
| ENSDARG00000098320 | grm1b | glutamate receptor, metabotropic 1b | 100150246 |
| ENSDARG00000007195 | GRM2 (2 of 2) | glutamate receptor, metabotropic 2 | 564461 |
| ENSDARG00000031712 | grm3 | glutamate receptor, metabotropic 3 | 565256 |
| ENSDARG00000040156 | grm4 | glutamate receptor, metabotropic 4 | 567181 |
| ENSDARG00000004445 | GRM5 (1 of 2) | glutamate receptor, metabotropic 5 | 0 |
| ENSDARG00000102067 | GRM5 (2 of 2) | glutamate receptor, metabotropic 5 | 100332913 |
| ENSDARG00000017742 | grm6a | glutamate receptor, metabotropic 6a | 568484 |
| ENSDARG00000100267 | GRM7 | glutamate receptor, metabotropic 7 | 0 |
| ENSDARG00000077654 | grm8a | glutamate receptor, metabotropic 8a | 792371 |
| ENSDARG00000076508 | grm8b | glutamate receptor, metabotropic 8b | 569768 |
| ENSDARG00000010021 | grpel1 | GrpE-like 1, mitochondrial | 559037 |
| ENSDARG00000029501 | gsk3aa | glycogen synthase kinase 3 alpha a | 560194 |
| ENSDARG00000015681 | gsk3ab | glycogen synthase kinase 3 alpha b | 30664 |
| ENSDARG00000017803 | gsk3b | glycogen synthase kinase 3 beta | 30654 |
| ENSDARG00000011459 | gsna | gelsolin a | 321099 |
| ENSDARG00000090228 | gstal | glutathione S-transferase, alpha-like | 406703 |
| ENSDARG00000056510 | gstk1 | glutathione S-transferase kappa 1 | 436833 |
| ENSDARG00000042533 | gstm | glutathione S-transferase M | 324366 |
| ENSDARG00000022183 | gsto1 | glutathione S-transferase omega 1 | 436894 |
| ENSDARG00000104068 | gstp1 | glutathione S-transferase pi 1 | 79381 |
| ENSDARG00000017388 | gstt1b | glutathione S-transferase theta 1b | 393556 |
| ENSDARG00000027984 | gstz1 | glutathione S-transferase zeta 1 | 436754 |
| ENSDARG00000053467 | gtpbp1 | GTP binding protein 1 | 378721 |
| ENSDARG00000093185 | GUCY1A2 | guanylate cyclase 1, soluble, alpha 2 | 0 |
| ENSDARG00000086790 | gucy1b3 | guanylate cyclase 1, soluble, beta 3 | 100150304 |
| ENSDARG00000001733 | gulp1a | GULP, engulfment adaptor PTB domain containing 1a | 368404 |
| ENSDARG00000011934 | gyg1a | glycogenin 1a | 406831 |
| ENSDARG00000099214 | h2afvb | H2A histone family, member Vb | 403077 |
| ENSDARG00000014806 | hacd2 | 3-hydroxyacyl-CoA dehydratase 2 | 334121 |
| ENSDARG00000020529 | hacl1 | 2-hydroxyacyl-CoA lyase 1 | 406358 |
| ENSDARG00000057128 | hadhaa | hydroxyacyl-CoA dehydrogenase/3-ketoacyl-CoA thiolase/enoyl-CoA hydratase (trifunctional protein), alpha subunit a | 553401 |
| ENSDARG00000060594 | hadhab | hydroxyacyl-CoA dehydrogenase/3-ketoacyl-CoA thiolase/enoyl-CoA hydratase (trifunctional protein), alpha subunit b | 793834 |
| ENSDARG00000102055 | hadhb | hydroxyacyl-CoA dehydrogenase/3-ketoacyl-CoA thiolase/enoyl-CoA hydratase (trifunctional protein), beta subunit | 0 |
| ENSDARG00000030765 | hadh | hydroxyacyl-CoA dehydrogenase | 445121 |
| ENSDARG00000025338 | hagh | hydroxyacylglutathione hydrolase | 336977 |
| ENSDARG00000068516 | hapln1b | hyaluronan and proteoglycan link protein 1b | 569074 |
| ENSDARG00000018542 | hapln4 | hyaluronan and proteoglycan link protein 4 | 559818 |
| ENSDARG00000003693 | hars | histidyl-tRNA synthetase | 447847 |
| ENSDARG00000097011 | hbaa1 | hemoglobin, alpha adult 1 | 30507 |
| ENSDARG00000012519 | hcfc1b | host cell factor C1b | 565206 |
| ENSDARG00000104480 | hcn1 | hyperpolarization activated cyclic nucleotide-gated potassium channel 1 | 0 |
| ENSDARG00000027192 | hcn3 | hyperpolarization activated cyclic nucleotide-gated potassium channel 3 | 560709 |
| ENSDARG00000074419 | hcn4l | hyperpolarization activated cyclic nucleotide-gated potassium channel 4l | 556759 |
| ENSDARG00000087573 | hdac11 | histone deacetylase 11 | 431718 |
| ENSDARG00000009830 | hdlbpa | high density lipoprotein binding protein a | 794178 |
| ENSDARG00000042630 | hebp2 | heme binding protein 2 | 393167 |
| ENSDARG00000101079 | HECTD4 | HECT domain containing E3 ubiquitin protein ligase 4 | 101885095 |
| ENSDARG00000062447 | hecw2a | HECT, C2 and WW domain containing E3 ubiquitin protein ligase 2a | 0 |
| ENSDARG00000063253 | hecw2b | HECT, C2 and WW domain containing E3 ubiquitin protein ligase 2b | 100331355 |
| ENSDARG00000077945 | HEPACAM (2 of 2) | hepatic and glial cell adhesion molecule | 0 |
| ENSDARG00000056934 | hepacama | hepatic and glial cell adhesion molecule a | 100005952 |
| ENSDARG00000059231 | hephl1a | hephaestin-like 1a | 561607 |
| ENSDARG00000034368 | hexb | hexosaminidase B (beta polypeptide) | 323613 |
| ENSDARG00000020960 | hgs | hepatocyte growth factor-regulated tyrosine kinase substrate | 334175 |
| ENSDARG00000007959 | hibadhb | 3-hydroxyisobutyrate dehydrogenase b | 394135 |
| ENSDARG00000054867 | hibch | 3-hydroxyisobutyryl-CoA hydrolase | 798364 |
| ENSDARG00000022303 | higd1a | HIG1 hypoxia inducible domain family, member 1A | 373084 |
| ENSDARG00000052371 | higd2a | HIG1 hypoxia inducible domain family, member 2A | 0 |
| ENSDARG00000103824 | hint1 | histidine triad nucleotide binding protein 1 | 449551 |
| ENSDARG00000062972 | hip1 | huntingtin interacting protein 1 | 793823 |
| ENSDARG00000060627 | HIP1R (1 of 2) | huntingtin interacting protein 1 related | 103910581 |
| ENSDARG00000102458 | hip1rb | huntingtin interacting protein 1 related b | 562138 |
| ENSDARG00000035519 | histh1l | histone H1 like | 550353 |
| ENSDARG00000039452 | hk1 | hexokinase 1 | 406791 |
| ENSDARG00000038703 | hkdc1 | hexokinase domain containing 1 | 321224 |
| ENSDARG00000037846 | hm13 | histocompatibility (minor) 13 | 259190 |
| ENSDARG00000099175 | hmgb1a | high mobility group box 1a | 321622 |
| ENSDARG00000030479 | hmgb1b | high mobility group box 1b | 795095 |
| ENSDARG00000029722 | hmgb2a | high mobility group box 2a | 641484 |
| ENSDARG00000056725 | hmgb3a | high mobility group box 3a | 561025 |
| ENSDARG00000021220 | hmgcl | 3-hydroxymethyl-3-methylglutaryl-CoA lyase | 394190 |
| ENSDARG00000055101 | hmox2a | heme oxygenase 2a | 100002875 |
| ENSDARG00000078452 | hmox2b | heme oxygenase 2b | 100329531 |
| ENSDARG00000102975 | hmp19 | HMP19 protein | 324971 |
| ENSDARG00000099312 | hn1b | hematological and neurological expressed 1b | 402906 |
| ENSDARG00000007960 | hnrnpaba | heterogeneous nuclear ribonucleoprotein A/Ba | 321466 |
| ENSDARG00000059246 | hnrnpd | heterogeneous nuclear ribonucleoprotein D | 560522 |
| ENSDARG00000079780 | HNRNPUL1 (2 of 2) | heterogeneous nuclear ribonucleoprotein U-like 1 | 0 |
| ENSDARG00000007034 | hnrpkl | heterogeneous nuclear ribonucleoprotein K, like | 406264 |
| ENSDARG00000101759 | homer1b | homer scaffolding protein 1b | 436769 |
| ENSDARG00000102532 | HOMER3 (2 of 2) | homer scaffolding protein 3 | 100330028 |
| ENSDARG00000010789 | homer3 | homer scaffolding protein 3 | 394088 |
| ENSDARG00000044669 | hp1bp3 | heterochromatin protein 1, binding protein 3 | 558102 |
| ENSDARG00000018397 | hpca | hippocalcin | 393696 |
| ENSDARG00000022763 | hpcal1 | hippocalcin-like 1 | 394139 |
| ENSDARG00000070491 | hpcal4 | hippocalcin like 4 | 564909 |
| ENSDARG00000008884 | hprt1 | hypoxanthine phosphoribosyltransferase 1 | 406259 |
| ENSDARG00000012609 | hpx | hemopexin | 327588 |
| ENSDARG00000098497 | hrasa | v-Ha-ras Harvey rat sarcoma viral oncogene homolog a | 550286 |
| ENSDARG00000035882 | hrsp12 | heat-responsive protein 12 | 436849 |
| ENSDARG00000062008 | hs2st1b | heparan sulfate 2-O-sulfotransferase 1b | 100151174 |
| ENSDARG00000012381 | hsc70 | heat shock cognate 70 | 393586 |
| ENSDARG00000017781 | hsd17b10 | hydroxysteroid (17-beta) dehydrogenase 10 | 450078 |
| ENSDARG00000035872 | hsd17b12b | hydroxysteroid (17-beta) dehydrogenase 12b | 322626 |
| ENSDARG00000101239 | hsd17b4 | hydroxysteroid (17-beta) dehydrogenase 4 | 393105 |
| ENSDARG00000041736 | hsdl1 | hydroxysteroid dehydrogenase like 1 | 494064 |
| ENSDARG00000002523 | hsdl2 | hydroxysteroid dehydrogenase like 2 | 322347 |
| ENSDARG00000024746 | hsp90aa1.2 | heat shock protein 90, alpha (cytosolic), class A member 1, tandem duplicate 2 | 565155 |
| ENSDARG00000029150 | hsp90ab1 | heat shock protein 90, alpha (cytosolic), class B member 1 | 30573 |
| ENSDARG00000003570 | hsp90b1 | heat shock protein 90, beta (grp94), member 1 | 386590 |
| ENSDARG00000070603 | hspa12a | heat shock protein 12A | 751725 |
| ENSDARG00000040984 | hspa13 | heat shock protein 70 family, member 13 | 560481 |
| ENSDARG00000004754 | hspa4a | heat shock protein 4a | 335865 |
| ENSDARG00000018989 | hspa4b | heat shock protein 4b | 334086 |
| ENSDARG00000053544 | hspa4l | heat shock protein 4 like | 562019 |
| ENSDARG00000103846 | hspa5 | heat shock protein 5 | 378848 |
| ENSDARG00000037403 | HSPA8 (1 of 2) | heat shock 70kDa protein 8 | 562935 |
| ENSDARG00000068992 | hspa8 | heat shock protein 8 | 573376 |
| ENSDARG00000003035 | hspa9 | heat shock protein 9 | 373085 |
| ENSDARG00000041065 | hspb1 | heat shock protein, alpha-crystallin-related, 1 | 368243 |
| ENSDARG00000056160 | hspd1 | heat shock 60 protein 1 | 282676 |
| ENSDARG00000056167 | hspe1 | heat shock 10 protein 1 | 58041 |
| ENSDARG00000028386 | htatip2 | HIV-1 Tat interactive protein 2 | 64605 |
| ENSDARG00000052866 | htt | huntingtin | 30214 |
| ENSDARG00000016782 | huwe1 | HECT, UBA and WWE domain containing 1 | 0 |
| ENSDARG00000013670 | hyou1 | hypoxia up-regulated 1 | 327133 |
| ENSDARG00000044134 | iars2 | isoleucyl-tRNA synthetase 2, mitochondrial | 100001219 |
| ENSDARG00000007955 | iars | isoleucyl-tRNA synthetase | 334393 |
| ENSDARG00000063446 | iba57 | IBA57, iron-sulfur cluster assembly homolog (S. cerevisiae) | 100330979 |
| ENSDARG00000009978 | icn | ictacalcin | 336655 |
| ENSDARG00000025375 | idh1 | isocitrate dehydrogenase 1 (NADP+), soluble | 100006589 |
| ENSDARG00000003795 | IDH2 | isocitrate dehydrogenase 2 (NADP+), mitochondrial | 0 |
| ENSDARG00000030278 | idh3a | isocitrate dehydrogenase 3 (NAD+) alpha | 393926 |
| ENSDARG00000044753 | idh3b | isocitrate dehydrogenase 3 (NAD+) beta | 415247 |
| ENSDARG00000023648 | idh3g | isocitrate dehydrogenase 3 (NAD+) gamma | 550579 |
| ENSDARG00000031886 | ift140 | intraflagellar transport 140 homolog (Chlamydomonas) | 553213 |
| ENSDARG00000002184 | igbp1 | immunoglobulin (CD79A) binding protein 1 | 406843 |
| ENSDARG00000034434 | igf1rb | insulin-like growth factor 1b receptor | 245702 |
| ENSDARG00000006094 | igf2r | insulin-like growth factor 2 receptor | 557061 |
| ENSDARG00000096355 | ighv1-4 | immunoglobulin heavy variable 1-4 | 0 |
| ENSDARG00000022176 | IGLON5 | IgLON family member 5 | 550472 |
| ENSDARG00000017217 | igsf11 | immunoglobulin superfamily member 11 | 100004469 |
| ENSDARG00000031049 | igsf21a | immunoglobin superfamily, member 21a | 569872 |
| ENSDARG00000077002 | igsf3 | immunoglobulin superfamily, member 3 | 557878 |
| ENSDARG00000038467 | igsf8 | immunoglobulin superfamily, member 8 | 0 |
| ENSDARG00000033845 | igsf9ba | immunoglobulin superfamily, member 9Ba | 567389 |
| ENSDARG00000069467 | igsf9bb | immunoglobulin superfamily, member 9Bb | 569554 |
| ENSDARG00000062045 | il1rapl1a | interleukin 1 receptor accessory protein-like 1a | 568868 |
| ENSDARG00000014591 | ilf2 | interleukin enhancer binding factor 2 | 406517 |
| ENSDARG00000017126 | ilvbl | ilvB (bacterial acetolactate synthase)-like | 393639 |
| ENSDARG00000021611 | im:7152756 | im:7152756 | 0 |
| ENSDARG00000102874 | immt | inner membrane protein, mitochondrial (mitofilin) | 327206 |
| ENSDARG00000003517 | impa1 | inositol(myo)-1(or 4)-monophosphatase 1 | 437018 |
| ENSDARG00000058114 | impad1 | inositol monophosphatase domain containing 1 | 641570 |
| ENSDARG00000006900 | impdh2 | IMP (inosine 5'-monophosphate) dehydrogenase 2 | 317745 |
| ENSDARG00000011862 | inaa | internexin neuronal intermediate filament protein, alpha a | 555251 |
| ENSDARG00000053248 | inab | internexin neuronal intermediate filament protein, alpha b | 30230 |
| ENSDARG00000063352 | inpp4aa | inositol polyphosphate-4-phosphatase, type Ia | 664741 |
| ENSDARG00000075201 | inpp4b | inositol polyphosphate-4-phosphatase, type II | 560006 |
| ENSDARG00000093359 | inpp5jb | inositol polyphosphate-5-phosphatase Jb | 0 |
| ENSDARG00000011948 | insra | insulin receptor a | 245699 |
| ENSDARG00000071524 | insrb | insulin receptor b | 245700 |
| ENSDARG00000035751 | ipo7 | importin 7 | 321739 |
| ENSDARG00000016551 | iqsec1b | IQ motif and Sec7 domain 1b | 100148736 |
| ENSDARG00000102125 | IQSEC2 (2 of 2) | IQ motif and Sec7 domain 2 | 560461 |
| ENSDARG00000077709 | iqsec2 | IQ motif and Sec7 domain 2 | 797187 |
| ENSDARG00000099850 | iqsec3a | IQ motif and Sec7 domain 3a | 797355 |
| ENSDARG00000093091 | iqsec3b | IQ motif and Sec7 domain 3b | 555340 |
| ENSDARG00000068657 | irgq2 | immunity-related GTPase family, q2 | 556650 |
| ENSDARG00000051875 | islr2 | immunoglobulin superfamily containing leucine-rich repeat 2 | 541396 |
| ENSDARG00000060069 | isoc1 | isochorismatase domain containing 1 | 559897 |
| ENSDARG00000013371 | isoc2 | isochorismatase domain containing 2 | 492617 |
| ENSDARG00000051888 | ist1 | increased sodium tolerance 1 homolog (yeast) | 321460 |
| ENSDARG00000099199 | itchb | itchy E3 ubiquitin protein ligase b | 100330031 |
| ENSDARG00000075584 | itfg1 | integrin alpha FG-GAP repeat containing 1 | 100331534 |
| ENSDARG00000006314 | itgav | integrin, alpha V | 100151255 |
| ENSDARG00000016939 | itgb2 | integrin, beta 2 | 557797 |
| ENSDARG00000012942 | itgb5 | integrin, beta 5 | 563556 |
| ENSDARG00000007098 | itm2ba | integral membrane protein 2Ba | 323632 |
| ENSDARG00000074149 | itpr1b | inositol 1,4,5-trisphosphate receptor, type 1b | 100149388 |
| ENSDARG00000011909 | itpr2 | inositol 1,4,5-trisphosphate receptor, type 2 | 562585 |
| ENSDARG00000061741 | itpr3 | inositol 1,4,5-trisphosphate receptor, type 3 | 794666 |
| ENSDARG00000000086 | itsn1 | intersectin 1 (SH3 domain protein) | 368504 |
| ENSDARG00000042853 | ivd | isovaleryl-CoA dehydrogenase | 368775 |
| ENSDARG00000053547 | jakmip2 | janus kinase and microtubule interacting protein 2 | 559839 |
| ENSDARG00000086037 | jam3a | junctional adhesion molecule 3a | 797651 |
| ENSDARG00000070787 | jupa | junction plakoglobin a | 30415 |
| ENSDARG00000059067 | jupb | junction plakoglobin b | 100330886 |
| ENSDARG00000059473 | kank4 | KN motif and ankyrin repeat domains 4 | 101884737 |
| ENSDARG00000103799 | kars | lysyl-tRNA synthetase | 280647 |
| ENSDARG00000012275 | katnal2 | katanin p60 subunit A-like 2 | 641431 |
| ENSDARG00000076923 | kazna | kazrin, periplakin interacting protein a | 100003663 |
| ENSDARG00000104958 | KBTBD11 | kelch repeat and BTB (POZ) domain containing 11 | 557467 |
| ENSDARG00000062942 | kcna1a | potassium voltage-gated channel, shaker-related subfamily, member 1a | 795942 |
| ENSDARG00000102064 | kcna2b | potassium voltage-gated channel, shaker-related subfamily, member 2b | 562853 |
| ENSDARG00000079491 | KCNA3 | potassium channel, voltage gated shaker related subfamily A, member 3 | 100150068 |
| ENSDARG00000087247 | KCNAB2 (2 of 2) | potassium channel, voltage gated subfamily A regulatory beta subunit 2 | 797337 |
| ENSDARG00000062134 | kcnab2b | potassium voltage-gated channel, shaker-related subfamily, beta member 2 b | 565014 |
| ENSDARG00000074320 | KCNAB3 | potassium channel, voltage gated subfamily A regulatory beta subunit 3 | 0 |
| ENSDARG00000051852 | kcnc1a | potassium voltage-gated channel, Shaw-related subfamily, member 1a | 565419 |
| ENSDARG00000055855 | kcnc3a | potassium voltage-gated channel, Shaw-related subfamily, member 3a | 559096 |
| ENSDARG00000074746 | kcnd1 | potassium voltage-gated channel, Shal-related subfamily, member 1 | 100001681 |
| ENSDARG00000032799 | kcnd2 | potassium voltage-gated channel, Shal-related subfamily, member 2 | 570188 |
| ENSDARG00000056101 | kcnd3 | potassium voltage-gated channel, Shal-related subfamily, member 3 | 327415 |
| ENSDARG00000069560 | kcnh3 | potassium voltage-gated channel, subfamily H (eag-related), member 3 | 0 |
| ENSDARG00000088100 | KCNIP4 (2 of 2) | Kv channel interacting protein 4 | 100536963 |
| ENSDARG00000090815 | KCNJ10 (1 of 3) | potassium channel, inwardly rectifying subfamily J, member 10 | 100538267 |
| ENSDARG00000103332 | KCNJ10 (2 of 3) | potassium channel, inwardly rectifying subfamily J, member 10 | 0 |
| ENSDARG00000031438 | kcnj11l | potassium inwardly-rectifying channel, subfamily J, member 11, like | 0 |
| ENSDARG00000062217 | kcnj3b | potassium inwardly-rectifying channel, subfamily J, member 3b | 557095 |
| ENSDARG00000079840 | kcnma1a | potassium large conductance calcium-activated channel, subfamily M, alpha member 1a | 568554 |
| ENSDARG00000091130 | kcnq2b | potassium voltage-gated channel, KQT-like subfamily, member 2b | 0 |
| ENSDARG00000079484 | kcnt1 | potassium channel, subfamily T, member 1 | 100004419 |
| ENSDARG00000069843 | kctd12.1 | potassium channel tetramerisation domain containing 12.1 | 373865 |
| ENSDARG00000053542 | kctd12.2 | potassium channel tetramerisation domain containing 12.2 | 553403 |
| ENSDARG00000004648 | kctd16a | potassium channel tetramerization domain containing 16a | 567207 |
| ENSDARG00000020102 | kctd16b | potassium channel tetramerization domain containing 16b | 569756 |
| ENSDARG00000060854 | kctd3 | potassium channel tetramerization domain containing 3 | 569113 |
| ENSDARG00000068691 | kctd4 | potassium channel tetramerization domain containing 4 | 792402 |
| ENSDARG00000067507 | kctd8 | potassium channel tetramerization domain containing 8 | 568933 |
| ENSDARG00000012021 | kdsr | 3-ketodihydrosphingosine reductase | 394114 |
| ENSDARG00000052856 | khdrbs1a | KH domain containing, RNA binding, signal transduction associated 1a | 30082 |
| ENSDARG00000062477 | kiaa1549la | KIAA1549-like a | 566592 |
| ENSDARG00000062086 | kiaa1549lb | KIAA1549-like b | 0 |
| ENSDARG00000017338 | kidins220b | kinase D-interacting substrate 220b | 335881 |
| ENSDARG00000061817 | kif1aa | kinesin family member 1Aa | 100330443 |
| ENSDARG00000061131 | kif21a | kinesin family member 21A | 566481 |
| ENSDARG00000043571 | KIF2A | kinesin heavy chain member 2A | 447926 |
| ENSDARG00000017162 | KIF3C (1 of 2) | kinesin family member 3C | 100331925 |
| ENSDARG00000098936 | kif5aa | kinesin family member 5A, a | 566086 |
| ENSDARG00000074131 | kif5ba | kinesin family member 5B, a | 563459 |
| ENSDARG00000103394 | kif5bb | kinesin family member 5B, b | 334108 |
| ENSDARG00000076027 | kif5c | kinesin family member 5C | 565563 |
| ENSDARG00000075806 | kirrel3a | kin of IRRE like 3 a | 571887 |
| ENSDARG00000104665 | kirrel3l | kin of IRRE like 3 like | 557315 |
| ENSDARG00000056998 | kirrelb | kin of IRRE like b | 100170816 |
| ENSDARG00000099740 | klc2 | kinesin light chain 2 | 562365 |
| ENSDARG00000023190 | kpna4 | karyopherin alpha 4 (importin alpha 3) | 327373 |
| ENSDARG00000104889 | kpnb1 | karyopherin (importin) beta 1 | 570700 |
| ENSDARG00000040245 | kpnb3 | karyopherin (importin) beta 3 | 569455 |
| ENSDARG00000010844 | kras | v-Ki-ras2 Kirsten rat sarcoma viral oncogene homolog | 445289 |
| ENSDARG00000028618 | KRT18 (2 of 3) | keratin 18, type I | 393540 |
| ENSDARG00000018404 | krt18 | keratin 18 | 352912 |
| ENSDARG00000058371 | krt5 | keratin 5 | 0 |
| ENSDARG00000058358 | krt8 | keratin 8 | 797433 |
| ENSDARG00000044975 | krt94 | keratin 94 | 768289 |
| ENSDARG00000094526 | ksr2 | kinase suppressor of ras 2 | 569127 |
| ENSDARG00000032802 | ktn1 | kinectin 1 | 368207 |
| ENSDARG00000040803 | lactb | lactamase, beta | 553619 |
| ENSDARG00000092610 | lamp1 | lysosomal-associated membrane protein 1 | 563328 |
| ENSDARG00000076464 | lamtor1 | late endosomal/lysosomal adaptor, MAPK and MTOR activator 1 | 100329656 |
| ENSDARG00000039872 | lamtor2 | late endosomal/lysosomal adaptor, MAPK and MTOR activator 2 | 445155 |
| ENSDARG00000057075 | lamtor3 | late endosomal/lysosomal adaptor, MAPK and MTOR activator 3 | 553597 |
| ENSDARG00000045542 | LAMTOR4 | late endosomal/lysosomal adaptor, MAPK and MTOR activator 4 | 561019 |
| ENSDARG00000090194 | lamtor5 | late endosomal/lysosomal adaptor, MAPK and MTOR activator 5 | 100333504 |
| ENSDARG00000013741 | lancl1 | LanC antibiotic synthetase component C-like 1 (bacterial) | 494154 |
| ENSDARG00000045465 | lancl2 | LanC lantibiotic synthetase component C-like 2 (bacterial) | 556310 |
| ENSDARG00000061981 | lap3 | leucine aminopeptidase 3 | 562854 |
| ENSDARG00000090988 | larsa | leucyl-tRNA synthetase a | 0 |
| ENSDARG00000019280 | larsb | leucyl-tRNA synthetase b | 569778 |
| ENSDARG00000057867 | lasp1 | LIM and SH3 protein 1 | 323216 |
| ENSDARG00000045140 | lcmt1 | leucine carboxyl methyltransferase 1 | 447907 |
| ENSDARG00000101251 | ldha | lactate dehydrogenase A4 | 30496 |
| ENSDARG00000019644 | ldhba | lactate dehydrogenase Ba | 30497 |
| ENSDARG00000071076 | ldhbb | lactate dehydrogenase Bb | 436747 |
| ENSDARG00000038845 | ldhd | lactate dehydrogenase D | 334208 |
| ENSDARG00000056978 | letm1 | leucine zipper-EF-hand containing transmembrane protein 1 | 570745 |
| ENSDARG00000077515 | letm2 | leucine zipper-EF-hand containing transmembrane protein 2 | 100006201 |
| ENSDARG00000030641 | letmd1 | LETM1 domain containing 1 | 567014 |
| ENSDARG00000054942 | lgals2a | lectin, galactoside-binding, soluble, 2a | 405830 |
| ENSDARG00000040528 | lgals3bpb | lectin, galactoside-binding, soluble, 3 binding protein b | 405809 |
| ENSDARG00000025903 | lgals9l1 | lectin, galactoside-binding, soluble, 9 (galectin 9)-like 1 | 337597 |
| ENSDARG00000055525 | lgalslb | lectin, galactoside-binding-like b | 368889 |
| ENSDARG00000020493 | lgi1a | leucine-rich, glioma inactivated 1a | 323011 |
| ENSDARG00000058421 | lgi1b | leucine-rich, glioma inactivated 1b | 654829 |
| ENSDARG00000089957 | lgi2a | leucine-rich repeat LGI family, member 2a | 654827 |
| ENSDARG00000041358 | lgi3 | leucine-rich repeat LGI family, member 3 | 654830 |
| ENSDARG00000021595 | lhfpl3 | lipoma HMGIC fusion partner-like 3 | 445050 |
| ENSDARG00000045023 | lhfpl5a | lipoma HMGIC fusion partner-like 5a | 558325 |
| ENSDARG00000060196 | lhpp | phospholysine phosphohistidine inorganic pyrophosphate phosphatase | 100073345 |
| ENSDARG00000103774 | limch1b | LIM and calponin homology domains 1b | 557200 |
| ENSDARG00000013414 | lin7a | lin-7 homolog A (C. elegans) | 393682 |
| ENSDARG00000037932 | lin7b | lin-7 homolog B (C. elegans) | 553768 |
| ENSDARG00000027322 | lin7c | lin-7 homolog C (C. elegans) | 447934 |
| ENSDARG00000034165 | lingo1a | leucine rich repeat and Ig domain containing 1a | 564943 |
| ENSDARG00000035899 | lingo1b | leucine rich repeat and Ig domain containing 1b | 447837 |
| ENSDARG00000074535 | lingo2a | leucine rich repeat and Ig domain containing 2a | 566713 |
| ENSDARG00000069970 | lingo2b | leucine rich repeat and Ig domain containing 2b | 564052 |
| ENSDARG00000067815 | LINGO3 (2 of 2) | leucine rich repeat and Ig domain containing 3 | 100151059 |
| ENSDARG00000009693 | llgl1 | lethal giant larvae homolog 1 (Drosophila) | 751760 |
| ENSDARG00000023920 | llgl2 | lethal giant larvae homolog 2 (Drosophila) | 795670 |
| ENSDARG00000069980 | lman1 | lectin, mannose-binding, 1 | 559775 |
| ENSDARG00000088663 | lman2lb | lectin, mannose-binding 2-like b | 767766 |
| ENSDARG00000061854 | lman2 | lectin, mannose-binding 2 | 557960 |
| ENSDARG00000017312 | lmbrd2b | LMBR1 domain containing 2b | 335257 |
| ENSDARG00000078094 | lmf2a | lipase maturation factor 2a | 571852 |
| ENSDARG00000025859 | lmf2b | lipase maturation factor 2b | 407708 |
| ENSDARG00000044299 | lmnb1 | lamin B1 | 195816 |
| ENSDARG00000101624 | lmnb2 | lamin B2 | 30196 |
| ENSDARG00000004930 | lmo7a | LIM domain 7a | 553341 |
| ENSDARG00000076324 | lmtk2 | lemur tyrosine kinase 2 | 564666 |
| ENSDARG00000029114 | LMTK3 | lemur tyrosine kinase 3 | 562728 |
| ENSDARG00000006639 | lnpb | limb and neural patterns b | 337509 |
| ENSDARG00000100394 | lnpep | leucyl/cystinyl aminopeptidase | 322814 |
| ENSDARG00000102765 | lonp1 | lon peptidase 1, mitochondrial | 563257 |
| ENSDARG00000013542 | lpgat1 | lysophosphatidylglycerol acyltransferase 1 | 561832 |
| ENSDARG00000010144 | lppr3a | lipid phosphate phosphatase-related protein type 3a | 553336 |
| ENSDARG00000028552 | lppr3b | lipid phosphate phosphatase-related protein type 3b | 570173 |
| ENSDARG00000079671 | lppr4a | lipid phosphate phosphatase-related protein type 4a | 559888 |
| ENSDARG00000075580 | lrch2 | leucine-rich repeats and calponin homology (CH) domain containing 2 | 100150488 |
| ENSDARG00000100679 | lrch3 | leucine-rich repeats and calponin homology (CH) domain containing 3 | 100126105 |
| ENSDARG00000027602 | lrfn1 | leucine rich repeat and fibronectin type III domain containing 1 | 393587 |
| ENSDARG00000039500 | LRFN3 (1 of 2) | leucine rich repeat and fibronectin type III domain containing 3 | 100333421 |
| ENSDARG00000071230 | lrfn5a | leucine rich repeat and fibronectin type III domain containing 5a | 100144400 |
| ENSDARG00000079396 | lrfn5b | leucine rich repeat and fibronectin type III domain containing 5b | 555839 |
| ENSDARG00000075625 | lrig1 | leucine-rich repeats and immunoglobulin-like domains 1 | 560325 |
| ENSDARG00000097827 | LRP1 (2 of 2) | low density lipoprotein receptor-related protein 1 | 0 |
| ENSDARG00000029146 | lrp1ab | low density lipoprotein receptor-related protein 1Ab | 565797 |
| ENSDARG00000061517 | lrp1bb | low density lipoprotein receptor-related protein 1Bb | 559293 |
| ENSDARG00000070074 | lrp8 | low density lipoprotein receptor-related protein 8, apolipoprotein e receptor | 557734 |
| ENSDARG00000033604 | lrpap1 | low density lipoprotein receptor-related protein associated protein 1 | 333939 |
| ENSDARG00000043970 | lrpprc | leucine-rich pentatricopeptide repeat containing | 557195 |
| ENSDARG00000086990 | lrrc16b | leucine rich repeat containing 16B | 0 |
| ENSDARG00000102684 | LRRC1 | leucine rich repeat containing 1 | 0 |
| ENSDARG00000069402 | lrrc4.1 | leucine rich repeat containing 4.1 | 565021 |
| ENSDARG00000003020 | lrrc4.2 | leucine rich repeat containing 4.2 | 566572 |
| ENSDARG00000053138 | lrrc47 | leucine rich repeat containing 47 | 553404 |
| ENSDARG00000004597 | lrrc4ba | leucine rich repeat containing 4Ba | 556747 |
| ENSDARG00000014792 | lrrc4bb | leucine rich repeat containing 4Bb | 100535350 |
| ENSDARG00000016739 | LRRC4C | leucine rich repeat containing 4C | 564462 |
| ENSDARG00000043938 | lrrc57 | leucine rich repeat containing 57 | 436900 |
| ENSDARG00000071426 | lrrc59 | leucine rich repeat containing 59 | 405868 |
| ENSDARG00000063411 | lrrc73 | leucine rich repeat containing 73 | 570766 |
| ENSDARG00000074308 | lrrc75ba | leucine rich repeat containing 75Ba | 101885211 |
| ENSDARG00000079670 | lrrc7 | leucine rich repeat containing 7 | 566023 |
| ENSDARG00000032188 | lrrc8a | leucine rich repeat containing 8 family, member A | 327642 |
| ENSDARG00000052713 | lrrtm1 | leucine rich repeat transmembrane neuronal 1 | 570385 |
| ENSDARG00000071374 | lrrtm2 | leucine rich repeat transmembrane neuronal 2 | 100538061 |
| ENSDARG00000078839 | LRRTM4 (1 of 3) | leucine rich repeat transmembrane neuronal 4 | 563710 |
| ENSDARG00000060683 | lrsam1 | leucine rich repeat and sterile alpha motif containing 1 | 562066 |
| ENSDARG00000103069 | LSAMP | limbic system-associated membrane protein | 0 |
| ENSDARG00000071573 | lsm5 | LSM5 homolog, U6 small nuclear RNA associated (S. cerevisiae) | 563948 |
| ENSDARG00000006029 | lta4h | leukotriene A4 hydrolase | 406575 |
| ENSDARG00000053113 | ly75 | lymphocyte antigen 75 | 504036 |
| ENSDARG00000056874 | lygl1 | lysozyme g-like 1 | 436979 |
| ENSDARG00000053656 | lypla2 | lysophospholipase II | 393722 |
| ENSDARG00000055899 | lzic | leucine zipper and CTNNBIP1 domain containing | 436871 |
| ENSDARG00000058558 | lzts3b | leucine zipper, putative tumor suppressor family member 3b | 560221 |
| ENSDARG00000018432 | m6pr | mannose-6-phosphate receptor (cation dependent) | 406486 |
| ENSDARG00000028533 | macf1a | microtubule-actin crosslinking factor 1a | 562190 |
| ENSDARG00000029609 | macrod1 | MACRO domain containing 1 | 447834 |
| ENSDARG00000003495 | madd | MAP-kinase activating death domain | 100150917 |
| ENSDARG00000070145 | magi1a | membrane associated guanylate kinase, WW and PDZ domain containing 1a | 100319849 |
| ENSDARG00000003169 | magi1b | membrane associated guanylate kinase, WW and PDZ domain containing 1b | 474349 |
| ENSDARG00000021590 | MAGI2 (1 of 2) | membrane associated guanylate kinase, WW and PDZ domain containing 2 | 564112 |
| ENSDARG00000073769 | MAGI2 (2 of 2) | membrane associated guanylate kinase, WW and PDZ domain containing 2 | 0 |
| ENSDARG00000101869 | magi3a | membrane associated guanylate kinase, WW and PDZ domain containing 3a | 561689 |
| ENSDARG00000025974 | magi3b | membrane associated guanylate kinase, WW and PDZ domain containing 3b | 0 |
| ENSDARG00000104023 | mag | myelin associated glycoprotein | 474346 |
| ENSDARG00000024017 | mamdc1 | MAM domain containing 1 | 368826 |
| ENSDARG00000041728 | man1a2 | mannosidase, alpha, class 1A, member 2 | 335538 |
| ENSDARG00000073792 | man1b1a | mannosidase, alpha, class 1B, member 1a | 562592 |
| ENSDARG00000102200 | man2a1 | mannosidase, alpha, class 2A, member 1 | 0 |
| ENSDARG00000063101 | man2a2 | mannosidase, alpha, class 2A, member 2 | 100333987 |
| ENSDARG00000059853 | manbal | mannosidase, beta A, lysosomal-like | 795041 |
| ENSDARG00000023712 | mao | monoamine oxidase | 404730 |
| ENSDARG00000059601 | map1aa | microtubule-associated protein 1Aa | 100151220 |
| ENSDARG00000022045 | map1ab | microtubule-associated protein 1Ab | 797545 |
| ENSDARG00000060434 | map1b | Hypothetical microtubule-associated protein 1b; Uncharacterized protein | 559032 |
| ENSDARG00000033609 | map1lc3a | microtubule-associated protein 1 light chain 3 alpha | 406466 |
| ENSDARG00000101127 | map1lc3b | microtubule-associated protein 1 light chain 3 beta | 322425 |
| ENSDARG00000060805 | map1sa | microtubule-associated protein 1Sa | 562808 |
| ENSDARG00000060326 | map1sb | microtubule-associated protein 1Sb | 565533 |
| ENSDARG00000007825 | map2k1 | mitogen-activated protein kinase kinase 1 | 406728 |
| ENSDARG00000006609 | map2k2a | mitogen-activated protein kinase kinase 2a | 563181 |
| ENSDARG00000077177 | map2k4b | mitogen-activated protein kinase kinase 4b | 568951 |
| ENSDARG00000099184 | map2k6 | mitogen-activated protein kinase kinase 6 | 0 |
| ENSDARG00000055052 | map2 | microtubule-associated protein 2 | 0 |
| ENSDARG00000068745 | MAP4 (1 of 3) | microtubule-associated protein 4 | 100537884 |
| ENSDARG00000101687 | MAP4 (3 of 3) | microtubule-associated protein 4 | 100001114 |
| ENSDARG00000071357 | map4k3b | mitogen-activated protein kinase kinase kinase kinase 3b | 100005183 |
| ENSDARG00000074521 | map6a | microtubule-associated protein 6a | 100000844 |
| ENSDARG00000074073 | map6b | microtubule-associated protein 6b | 100334879 |
| ENSDARG00000079777 | map6d1 | MAP6 domain containing 1 | 0 |
| ENSDARG00000104701 | map7d1b | MAP7 domain containing 1b | 796270 |
| ENSDARG00000068480 | map7d2a | MAP7 domain containing 2a | 100005195 |
| ENSDARG00000102730 | mapk10 | mitogen-activated protein kinase 10 | 569698 |
| ENSDARG00000027552 | mapk1 | mitogen-activated protein kinase 1 | 360144 |
| ENSDARG00000070573 | mapk3 | mitogen-activated protein kinase 3 | 399480 |
| ENSDARG00000062531 | mapk8ip3 | mitogen-activated protein kinase 8 interacting protein 3 | 100318355 |
| ENSDARG00000091777 | mapkap1 | mitogen-activated protein kinase associated protein 1 | 100537196 |
| ENSDARG00000002659 | mapre1b | microtubule-associated protein, RP/EB family, member 1b | 334728 |
| ENSDARG00000099943 | mapre2 | microtubule-associated protein, RP/EB family, member 2 | 557679 |
| ENSDARG00000020231 | mapre3a | microtubule-associated protein, RP/EB family, member 3a | 559217 |
| ENSDARG00000102878 | mapre3b | microtubule-associated protein, RP/EB family, member 3b | 431717 |
| ENSDARG00000089314 | mapta | microtubule-associated protein tau a | 0 |
| ENSDARG00000087616 | maptb | microtubule-associated protein tau b | 567833 |
| ENSDARG00000070487 | MARC2 | mitochondrial amidoxime reducing component 2 | 378742 |
| ENSDARG00000032552 | march5 | membrane-associated ring finger (C3HC4) 5 | 561952 |
| ENSDARG00000096271 | march7 | membrane-associated ring finger (C3HC4) 7 | 561242 |
| ENSDARG00000008803 | marcksb | myristoylated alanine-rich protein kinase C substrate b | 323201 |
| ENSDARG00000039034 | marcksl1a | MARCKS-like 1a | 406407 |
| ENSDARG00000035715 | marcksl1b | MARCKS-like 1b | 406504 |
| ENSDARG00000104014 | MARK1 | MAP/microtubule affinity-regulating kinase 1 | 0 |
| ENSDARG00000032458 | mark2b | MAP/microtubule affinity-regulating kinase 2b | 100005996 |
| ENSDARG00000019345 | mark3a | MAP/microtubule affinity-regulating kinase 3a | 334300 |
| ENSDARG00000026630 | mark3b | MAP/microtubule affinity-regulating kinase 3b | 767634 |
| ENSDARG00000023914 | mark4a | MAP/microtubule affinity-regulating kinase 4a | 569346 |
| ENSDARG00000034396 | mars | methionyl-tRNA synthetase | 338183 |
| ENSDARG00000040334 | mat2aa | methionine adenosyltransferase II, alpha a | 0 |
| ENSDARG00000012932 | mat2b | methionine adenosyltransferase II, beta | 541347 |
| ENSDARG00000036186 | mbpa | myelin basic protein a | 326281 |
| ENSDARG00000089413 | mbpb | myelin basic protein b | 368319 |
| ENSDARG00000089643 | mcama | melanoma cell adhesion molecule a | 566921 |
| ENSDARG00000102538 | mccc1 | methylcrotonoyl-CoA carboxylase 1 (alpha) | 777632 |
| ENSDARG00000017571 | mccc2 | methylcrotonoyl-CoA carboxylase 2 (beta) | 405863 |
| ENSDARG00000056603 | mcf2b | MCF.2 cell line derived transforming sequence b | 559907 |
| ENSDARG00000058848 | mcoln1b | mucolipin 1b | 795916 |
| ENSDARG00000025972 | mcts1 | malignant T cell amplified sequence 1 | 394046 |
| ENSDARG00000101175 | mcu | mitochondrial calcium uniporter | 768182 |
| ENSDARG00000017772 | mdh1aa | malate dehydrogenase 1Aa, NAD (soluble) | 399662 |
| ENSDARG00000043371 | mdh2 | malate dehydrogenase 2, NAD (mitochondrial) | 406405 |
| ENSDARG00000100718 | me2 | malic enzyme 2, NAD(+)-dependent, mitochondrial | 445233 |
| ENSDARG00000002305 | me3 | malic enzyme 3, NADP(+)-dependent, mitochondrial | 492719 |
| ENSDARG00000032326 | mecr | mitochondrial trans-2-enoyl-CoA reductase | 550550 |
| ENSDARG00000056726 | mettl7a | methyltransferase like 7A | 569131 |
| ENSDARG00000039203 | MFF (1 of 2) | mitochondrial fission factor | 553588 |
| ENSDARG00000053753 | mff | mitochondrial fission factor | 393508 |
| ENSDARG00000075159 | mfi2 | antigen p97 (melanoma associated) identified by monoclonal antibodies 133.2 and 96.5 | 565942 |
| ENSDARG00000079219 | MFN1 (2 of 3) | mitofusin 1 | 0 |
| ENSDARG00000101793 | mfn1b | mitofusin 1b | 0 |
| ENSDARG00000079504 | mfn2 | mitofusin 2 | 567448 |
| ENSDARG00000036820 | mgll | monoglyceride lipase | 378960 |
| ENSDARG00000004796 | mgrn1b | mahogunin, ring finger 1b | 553327 |
| ENSDARG00000032618 | mgst1.1 | microsomal glutathione S-transferase 1.1 | 449784 |
| ENSDARG00000022165 | mgst1.2 | microsomal glutathione S-transferase 1.2 | 431762 |
| ENSDARG00000102744 | mgst3 | microsomal glutathione S-transferase 3 | 406736 |
| ENSDARG00000075963 | mhc1uba | major histocompatibility complex class I UBA | 30757 |
| ENSDARG00000063358 | micu1 | mitochondrial calcium uptake 1 | 561210 |
| ENSDARG00000074255 | MICU3 (1 of 2) | mitochondrial calcium uptake family, member 3 | 100536173 |
| ENSDARG00000078599 | micu3a | mitochondrial calcium uptake family, member 3a | 559552 |
| ENSDARG00000071336 | mif | macrophage migration inhibitory factor | 751093 |
| ENSDARG00000035360 | mink1 | misshapen-like kinase 1 | 100038799 |
| ENSDARG00000101619 | minos1 | mitochondrial inner membrane organizing system 1 | 751699 |
| ENSDARG00000063026 | mlc1 | megalencephalic leukoencephalopathy with subcortical cysts 1 | 559990 |
| ENSDARG00000059630 | mlec | malectin | 569613 |
| ENSDARG00000034616 | mlf2 | myeloid leukemia factor 2 | 561675 |
| ENSDARG00000029472 | mllt4a | myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 4a | 558359 |
| ENSDARG00000104732 | mlycd | malonyl-CoA decarboxylase | 100037349 |
| ENSDARG00000060383 | mmaa | methylmalonic aciduria (cobalamin deficiency) cblA type | 564760 |
| ENSDARG00000010553 | mmgt1 | membrane magnesium transporter 1 | 402891 |
| ENSDARG00000079067 | mms19 | MMS19 nucleotide excision repair homolog (S. cerevisiae) | 563643 |
| ENSDARG00000012953 | mob1bb | MOB kinase activator 1Bb | 407654 |
| ENSDARG00000056085 | mob4 | MOB family member 4, phocein | 445045 |
| ENSDARG00000060252 | mogs | mannosyl-oligosaccharide glucosidase | 567913 |
| ENSDARG00000093448 | mpc1 | mitochondrial pyruvate carrier 1 | 436671 |
| ENSDARG00000037679 | MPC2 (2 of 2) | mitochondrial pyruvate carrier 2 | 0 |
| ENSDARG00000024478 | mpc2 | mitochondrial pyruvate carrier 2 | 321611 |
| ENSDARG00000030786 | mpi | mannose phosphate isomerase | 613246 |
| ENSDARG00000053453 | mpp2a | membrane protein, palmitoylated 2a (MAGUK p55 subfamily member 2) | 562207 |
| ENSDARG00000010957 | mpp2b | membrane protein, palmitoylated 2b (MAGUK p55 subfamily member 2) | 431770 |
| ENSDARG00000015184 | mpp3a | membrane protein, palmitoylated 3a (MAGUK p55 subfamily member 3) | 572079 |
| ENSDARG00000062667 | mpp3b | membrane protein, palmitoylated 3b (MAGUK p55 subfamily member 3) | 799884 |
| ENSDARG00000056892 | mpp6a | membrane protein, palmitoylated 6a (MAGUK p55 subfamily member 6) | 402878 |
| ENSDARG00000105209 | MPRIP (3 of 3) | myosin phosphatase Rho interacting protein | 562530 |
| ENSDARG00000102081 | mprip | myosin phosphatase Rho interacting protein | 558831 |
| ENSDARG00000032431 | mpv17 | MpV17 mitochondrial inner membrane protein | 394140 |
| ENSDARG00000059048 | mpzl1l | myelin protein zero-like 1 like | 555163 |
| ENSDARG00000038609 | mpz | myelin protein zero | 114417 |
| ENSDARG00000075285 | MRAS (2 of 2) | muscle RAS oncogene homolog | 100536317 |
| ENSDARG00000102100 | mrc1a | mannose receptor, C type 1a | 100286774 |
| ENSDARG00000075754 | mri1 | methylthioribose-1-phosphate isomerase 1 | 100002302 |
| ENSDARG00000013075 | mrpl11 | mitochondrial ribosomal protein L11 | 436829 |
| ENSDARG00000038768 | mrpl12 | mitochondrial ribosomal protein L12 | 550391 |
| ENSDARG00000068886 | mrpl14 | mitochondrial ribosomal protein L14 | 394122 |
| ENSDARG00000075677 | mrpl17 | mitochondrial ribosomal protein L17 | 447945 |
| ENSDARG00000090462 | mrpl20 | mitochondrial ribosomal protein L20 | 751640 |
| ENSDARG00000045696 | mrpl23 | mitochondrial ribosomal protein L23 | 415142 |
| ENSDARG00000043554 | mrpl40 | mitochondrial ribosomal protein L40 | 447873 |
| ENSDARG00000076334 | mrpl43 | mitochondrial ribosomal protein L43 | 436701 |
| ENSDARG00000016307 | mrpl46 | mitochondrial ribosomal protein L46 | 436791 |
| ENSDARG00000035167 | mrpl48 | mitochondrial ribosomal protein L48 | 503593 |
| ENSDARG00000068305 | mrps14 | mitochondrial ribosomal protein S14 | 751639 |
| ENSDARG00000059234 | mrps27 | mitochondrial ribosomal protein S27 | 445321 |
| ENSDARG00000104111 | mrps30 | mitochondrial ribosomal protein S30 | 555760 |
| ENSDARG00000057855 | mrps31 | mitochondrial ribosomal protein S31 | 407642 |
| ENSDARG00000079823 | mrps36 | mitochondrial ribosomal protein S36 | 567309 |
| ENSDARG00000058128 | msna | moesin a | 286739 |
| ENSDARG00000037261 | mtap | methylthioadenosine phosphorylase | 393526 |
| ENSDARG00000063911 | mt-atp6 | ATP synthase 6, mitochondrial | 140519 |
| ENSDARG00000063910 | mt-atp8 | ATP synthase 8, mitochondrial | 140520 |
| ENSDARG00000019732 | mtch2 | mitochondrial carrier homolog 2 | 30655 |
| ENSDARG00000063908 | mt-co2 | cytochrome c oxidase II, mitochondrial | 140540 |
| ENSDARG00000063924 | mt-cyb | cytochrome b, mitochondrial | 140512 |
| ENSDARG00000014159 | mtdha | metadherin a | 368910 |
| ENSDARG00000004939 | mtdhb | metadherin b | 553275 |
| ENSDARG00000099908 | MTFP1 | mitochondrial fission process 1 | 0 |
| ENSDARG00000003940 | mtfr1l | mitochondrial fission regulator 1-like | 334296 |
| ENSDARG00000040492 | mthfd1b | methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1b | 260441 |
| ENSDARG00000010601 | mtmr10 | myotubularin related protein 10 | 555239 |
| ENSDARG00000056690 | mtmr1a | myotubularin related protein 1a | 553750 |
| ENSDARG00000063895 | mt-nd1 | NADH dehydrogenase 1, mitochondrial | 140531 |
| ENSDARG00000063899 | mt-nd2 | NADH dehydrogenase 2, mitochondrial | 140532 |
| ENSDARG00000063917 | mt-nd4 | NADH dehydrogenase 4, mitochondrial | 140534 |
| ENSDARG00000063921 | mt-nd5 | NADH dehydrogenase 5, mitochondrial | 140535 |
| ENSDARG00000053196 | mtor | mechanistic target of rapamycin (serine/threonine kinase) | 324254 |
| ENSDARG00000076155 | mtpap | mitochondrial poly(A) polymerase | 563801 |
| ENSDARG00000104018 | mtpn | myotrophin | 406240 |
| ENSDARG00000087260 | MTSS1L (2 of 2) | metastasis suppressor 1-like | 0 |
| ENSDARG00000025500 | mtx1a | metaxin 1a | 556917 |
| ENSDARG00000036109 | mtx1b | metaxin 1b | 286740 |
| ENSDARG00000029415 | mtx2 | metaxin 2 | 286741 |
| ENSDARG00000090195 | mtx3 | metaxin 3 | 402916 |
| ENSDARG00000060554 | mut | methylmalonyl CoA mutase | 569581 |
| ENSDARG00000021242 | mvp | major vault protein | 373081 |
| ENSDARG00000001220 | mycbp2 | MYC binding protein 2 | 368439 |
| ENSDARG00000071679 | mydgf | myeloid-derived growth factor | 436753 |
| ENSDARG00000000103 | myh10 | myosin, heavy chain 10, non-muscle | 555454 |
| ENSDARG00000009782 | myh11a | myosin, heavy chain 11a, smooth muscle | 554168 |
| ENSDARG00000073732 | myh14 | myosin, heavy chain 14, non-muscle | 563011 |
| ENSDARG00000063295 | myh9a | myosin, heavy chain 9a, non-muscle | 333938 |
| ENSDARG00000001014 | myh9b | myosin, heavy chain 9b, non-muscle | 561431 |
| ENSDARG00000099766 | myl12.1 | myosin, light chain 12, genome duplicate 1 | 326719 |
| ENSDARG00000008494 | myl6 | myosin, light chain 6, alkali, smooth muscle and non-muscle | 798632 |
| ENSDARG00000008030 | myl9b | myosin, light chain 9b, regulatory | 406493 |
| ENSDARG00000017441 | mylz3 | myosin, light polypeptide 3, skeletal muscle | 58143 |
| ENSDARG00000074723 | myo10l1 | myosin X-like 1 | 566674 |
| ENSDARG00000075752 | myo18aa | myosin XVIIIAa | 566880 |
| ENSDARG00000061862 | myo18ab | myosin XVIIIAb | 0 |
| ENSDARG00000024694 | myo1b | myosin IB | 541515 |
| ENSDARG00000061579 | myo1c | myosin IC | 566459 |
| ENSDARG00000036863 | MYO1D | myosin ID | 559562 |
| ENSDARG00000078734 | myo1f | myosin IF | 565345 |
| ENSDARG00000078603 | myo1hb | myosin IHb | 0 |
| ENSDARG00000061635 | myo5aa | myosin VAa | 562188 |
| ENSDARG00000044016 | myo6a | myosin VIa | 445473 |
| ENSDARG00000042141 | myo6b | myosin VIb | 445472 |
| ENSDARG00000057928 | mzt2b | mitotic spindle organizing protein 2B | 445174 |
| ENSDARG00000005513 | naca | nascent polypeptide-associated complex alpha subunit | 195823 |
| ENSDARG00000015025 | nadl1.1 | neural adhesion molecule L1.1 | 30656 |
| ENSDARG00000007149 | nadl1.2 | neural adhesion molecule L1.2 | 30634 |
| ENSDARG00000011301 | nae1 | nedd8 activating enzyme E1 subunit 1 | 573336 |
| ENSDARG00000099116 | nagk | N-acetylglucosamine kinase | 100126013 |
| ENSDARG00000030598 | nampta | nicotinamide phosphoribosyltransferase a | 100330160 |
| ENSDARG00000045620 | nansa | N-acetylneuraminic acid synthase a | 322780 |
| ENSDARG00000070560 | nap1l4a | nucleosome assembly protein 1-like 4a | 337155 |
| ENSDARG00000068868 | nap1l4b | nucleosome assembly protein 1-like 4b | 492812 |
| ENSDARG00000067605 | napaa | N-ethylmaleimide-sensitive factor attachment protein, alpha a | 567306 |
| ENSDARG00000020405 | napab | N-ethylmaleimide-sensitive factor attachment protein, alpha b | 327082 |
| ENSDARG00000069101 | NAPB (2 of 2) | N-ethylmaleimide-sensitive factor attachment protein, beta | 568195 |
| ENSDARG00000013669 | napba | N-ethylmaleimide-sensitive factor attachment protein, beta a | 553682 |
| ENSDARG00000009252 | napepld | N-acyl phosphatidylethanolamine phospholipase D | 568061 |
| ENSDARG00000006617 | napga | N-ethylmaleimide-sensitive factor attachment protein, gamma a | 541353 |
| ENSDARG00000043012 | napgb | N-ethylmaleimide-sensitive factor attachment protein, gamma b | 436742 |
| ENSDARG00000061100 | nars | asparaginyl-tRNA synthetase | 568332 |
| ENSDARG00000090962 | nat14 | N-acetyltransferase 14 (GCN5-related, putative) | 751624 |
| ENSDARG00000008593 | nbas | neuroblastoma amplified sequence | 556592 |
| ENSDARG00000070080 | nbeaa | neurobeachin a | 541373 |
| ENSDARG00000070688 | ncalda | neurocalcin delta a | 556178 |
| ENSDARG00000011334 | ncaldb | neurocalcin delta b | 445319 |
| ENSDARG00000056181 | ncam1a | neural cell adhesion molecule 1a | 30447 |
| ENSDARG00000007220 | ncam1b | neural cell adhesion molecule 1b | 114442 |
| ENSDARG00000098523 | NCAM2 (2 of 2) | neural cell adhesion molecule 2 | 767777 |
| ENSDARG00000017466 | ncam2 | neural cell adhesion molecule 2 | 114441 |
| ENSDARG00000062087 | nceh1b | neutral cholesterol ester hydrolase 1b | 777713 |
| ENSDARG00000060853 | nckap1 | NCK-associated protein 1 | 561894 |
| ENSDARG00000079475 | nckipsd | NCK interacting protein with SH3 domain | 100003986 |
| ENSDARG00000058861 | ncl1 | nicalin | 336844 |
| ENSDARG00000002710 | ncl | nucleolin | 767714 |
| ENSDARG00000054186 | ncs1b | neuronal calcium sensor 1b | 553174 |
| ENSDARG00000103284 | NCSTN | nicastrin | 0 |
| ENSDARG00000103902 | nde1 | nudE neurodevelopment protein 1 | 566146 |
| ENSDARG00000010953 | ndel1a | nudE neurodevelopment protein 1-like 1a | 394244 |
| ENSDARG00000104225 | ndel1b | nudE neurodevelopment protein 1-like 1b | 333957 |
| ENSDARG00000032849 | ndrg1a | N-myc downstream regulated 1a | 792085 |
| ENSDARG00000011170 | ndrg2 | NDRG family member 2 | 494050 |
| ENSDARG00000013087 | ndrg3a | ndrg family member 3a | 30286 |
| ENSDARG00000010052 | ndrg3b | ndrg family member 3b | 327395 |
| ENSDARG00000103937 | ndrg4 | NDRG family member 4 | 692305 |
| ENSDARG00000013333 | ndufa10 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 10 | 0 |
| ENSDARG00000042777 | ndufa11 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 11 | 393497 |
| ENSDARG00000042469 | ndufa12 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 12 | 556538 |
| ENSDARG00000098584 | ndufa13 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 13 | 0 |
| ENSDARG00000036329 | ndufa1 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 1 | 415243 |
| ENSDARG00000021984 | ndufa2 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 2 | 554125 |
| ENSDARG00000056108 | ndufa4 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 4 | 406298 |
| ENSDARG00000039346 | ndufa5 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 5 | 541328 |
| ENSDARG00000038028 | ndufa6 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 6 | 393943 |
| ENSDARG00000036684 | ndufa7 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 7 | 445042 |
| ENSDARG00000058041 | ndufa8 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 8 | 393700 |
| ENSDARG00000070723 | ndufa9a | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 9a | 541331 |
| ENSDARG00000058463 | ndufab1a | NADH dehydrogenase (ubiquinone) 1, alpha/beta subcomplex, 1a | 573619 |
| ENSDARG00000014915 | ndufab1b | NADH dehydrogenase (ubiquinone) 1, alpha/beta subcomplex, 1b | 325712 |
| ENSDARG00000069286 | ndufaf2 | NADH dehydrogenase (ubiquinone) complex I, assembly factor 2 | 100003850 |
| ENSDARG00000077859 | ndufaf4 | NADH dehydrogenase (ubiquinone) complex I, assembly factor 4 | 794169 |
| ENSDARG00000053652 | ndufaf6 | NADH dehydrogenase (ubiquinone) complex I, assembly factor 6 | 569692 |
| ENSDARG00000028889 | ndufb10 | NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 10 | 393703 |
| ENSDARG00000043467 | ndufb11 | NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 11 | 562219 |
| ENSDARG00000087456 | NDUFB1 | NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 1, 7kDa | 100535765 |
| ENSDARG00000075709 | ndufb3 | NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 3 | 100332034 |
| ENSDARG00000070824 | ndufb5 | NADH dehydrogenase (ubiquinone) 1 beta subcomplex 5 | 797063 |
| ENSDARG00000037259 | ndufb6 | NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 6 | 393781 |
| ENSDARG00000033789 | ndufb7 | NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 7 | 393821 |
| ENSDARG00000010113 | ndufb8 | NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 8 | 393829 |
| ENSDARG00000041314 | ndufb9 | NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 9 | 573021 |
| ENSDARG00000102115 | ndufc2 | NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 2 | 541390 |
| ENSDARG00000028546 | ndufs1 | NADH dehydrogenase (ubiquinone) Fe-S protein 1 | 493605 |
| ENSDARG00000007526 | ndufs2 | NADH dehydrogenase (ubiquinone) Fe-S protein 2 | 553672 |
| ENSDARG00000015385 | ndufs3 | NADH dehydrogenase (ubiquinone) Fe-S protein 3, (NADH-coenzyme Q reductase) | 550451 |
| ENSDARG00000052840 | ndufs4 | NADH dehydrogenase (ubiquinone) Fe-S protein 4, (NADH-coenzyme Q reductase) | 573689 |
| ENSDARG00000006290 | ndufs5 | NADH dehydrogenase (ubiquinone) Fe-S protein 5 | 553660 |
| ENSDARG00000056583 | ndufs6 | NADH dehydrogenase (ubiquinone) Fe-S protein 6 | 447913 |
| ENSDARG00000074552 | ndufs7 | NADH dehydrogenase (ubiquinone) Fe-S protein 7, (NADH-coenzyme Q reductase) | 796997 |
| ENSDARG00000051986 | ndufs8a | NADH dehydrogenase (ubiquinone) Fe-S protein 8a | 406413 |
| ENSDARG00000036438 | ndufv1 | NADH dehydrogenase (ubiquinone) flavoprotein 1 | 445291 |
| ENSDARG00000013044 | ndufv2 | NADH dehydrogenase (ubiquinone) flavoprotein 2 | 393720 |
| ENSDARG00000090389 | ndufv3 | NADH dehydrogenase (ubiquinone) flavoprotein 3 | 0 |
| ENSDARG00000021200 | nebl | nebulette | 541446 |
| ENSDARG00000056566 | necab1 | N-terminal EF-hand calcium binding protein 1 | 550546 |
| ENSDARG00000056745 | necab2 | N-terminal EF-hand calcium binding protein 2 | 565258 |
| ENSDARG00000020798 | necap1 | NECAP endocytosis associated 1 | 393695 |
| ENSDARG00000060006 | nedd4l | neural precursor cell expressed, developmentally down-regulated 4-like | 559634 |
| ENSDARG00000007989 | nedd8 | neural precursor cell expressed, developmentally down-regulated 8 | 368667 |
| ENSDARG00000057568 | nefla | neurofilament, light polypeptide a | 0 |
| ENSDARG00000012426 | neflb | neurofilament, light polypeptide b | 664698 |
| ENSDARG00000021351 | nefma | neurofilament, medium polypeptide a | 553718 |
| ENSDARG00000043697 | nefmb | neurofilament, medium polypeptide b | 100033387 |
| ENSDARG00000021607 | negr1 | neuronal growth regulator 1 | 445374 |
| ENSDARG00000102855 | neo1a | neogenin 1a | 266983 |
| ENSDARG00000075100 | neo1b | neogenin 1b | 100332721 |
| ENSDARG00000012982 | nf1a | neurofibromin 1a | 326708 |
| ENSDARG00000061099 | nfasca | neurofascin homolog (chicken) a | 100141490 |
| ENSDARG00000062237 | nfs1 | NFS1 cysteine desulfurase | 562714 |
| ENSDARG00000068125 | nfu1 | NFU1 iron-sulfur cluster scaffold homolog (S. cerevisiae) | 403053 |
| ENSDARG00000076179 | ngef | neuronal guanine nucleotide exchange factor | 569090 |
| ENSDARG00000069878 | nhp2l1a | NHP2 non-histone chromosome protein 2-like 1a (S. cerevisiae) | 393282 |
| ENSDARG00000023299 | nhp2l1b | NHP2 non-histone chromosome protein 2-like 1b (S. cerevisiae) | 321114 |
| ENSDARG00000089066 | NHSL2 | NHS-like 2 | 794153 |
| ENSDARG00000045249 | nif3l1 | NIF3 NGG1 interacting factor 3-like 1 (S. cerevisiae) | 574421 |
| ENSDARG00000079824 | ninj2 | ninjurin 2 | 799267 |
| ENSDARG00000020387 | nipsnap3a | nipsnap homolog 3A (C. elegans) | 393234 |
| ENSDARG00000017590 | nit2 | nitrilase family, member 2 | 402904 |
| ENSDARG00000077710 | nlgn1 | neuroligin 1 | 0 |
| ENSDARG00000077329 | nlgn2a | neuroligin 2a | 556158 |
| ENSDARG00000079251 | nlgn2b | neuroligin 2b | 100002407 |
| ENSDARG00000104786 | nlgn3a | neuroligin 3a | 562702 |
| ENSDARG00000062376 | nlgn3b | neuroligin 3b | 567415 |
| ENSDARG00000079455 | nlgn4a | neuroligin 4a | 561122 |
| ENSDARG00000073724 | nlrx1 | NLR family member X1 | 557335 |
| ENSDARG00000103791 | nme2b.1 | NME/NM23 nucleoside diphosphate kinase 2b, tandem duplicate 1 | 30083 |
| ENSDARG00000100990 | nme3 | NME/NM23 nucleoside diphosphate kinase 3 | 30085 |
| ENSDARG00000021808 | nmt2 | N-myristoyltransferase 2 | 556483 |
| ENSDARG00000023536 | nnt | nicotinamide nucleotide transhydrogenase | 406619 |
| ENSDARG00000078592 | nomo | nodal modulator | 446115 |
| ENSDARG00000020482 | nono | non-POU domain containing, octamer-binding | 321994 |
| ENSDARG00000068910 | nos1 | nitric oxide synthase 1 (neuronal) | 60658 |
| ENSDARG00000017673 | nova2 | neuro-oncological ventral antigen 2 | 553201 |
| ENSDARG00000014717 | novel gene | 0 | 0 |
| ENSDARG00000067794 | novel gene | 0 | 0 |
| ENSDARG00000101726 | novel gene | 0 | 0 |
| ENSDARG00000102845 | novel gene | 0 | 0 |
| ENSDARG00000104615 | novel gene | 0 | 0 |
| ENSDARG00000068624 | novel gene | 0 | 100003123 |
| ENSDARG00000079483 | novel gene | 0 | 100537790 |
| ENSDARG00000090164 | novel gene | 0 | 101884007 |
| ENSDARG00000078770 | novel gene | 0 | 336430 |
| ENSDARG00000033201 | novel gene | 0 | 449795 |
| ENSDARG00000053454 | novel gene | 0 | 556251 |
| ENSDARG00000077545 | novel gene | 0 | 558390 |
| ENSDARG00000067824 | novel gene | 0 | 564195 |
| ENSDARG00000077124 | novel gene | 0 | 564701 |
| ENSDARG00000052263 | novel gene | 0 | 566914 |
| ENSDARG00000087599 | novel gene | 0 | 567508 |
| ENSDARG00000104264 | novel gene | 0 | 777704 |
| ENSDARG00000017180 | npc1 | Niemann-Pick disease, type C1 | 553330 |
| ENSDARG00000012871 | npepl1 | aminopeptidase-like 1 | 550383 |
| ENSDARG00000044943 | npepps | aminopeptidase puromycin sensitive | 407726 |
| ENSDARG00000043864 | nptnb | neuroplastin b | 403006 |
| ENSDARG00000074671 | nptx1l | neuronal pentraxin 1 like | 405872 |
| ENSDARG00000037794 | nptx2a | neuronal pentraxin 2a | 553230 |
| ENSDARG00000040430 | nptxra | neuronal pentraxin receptor a | 101884706 |
| ENSDARG00000010250 | nqo1 | NAD(P)H dehydrogenase, quinone 1 | 322506 |
| ENSDARG00000038225 | nras | neuroblastoma RAS viral (v-ras) oncogene homolog | 30380 |
| ENSDARG00000056089 | nrbp1 | nuclear receptor binding protein 1 | 569806 |
| ENSDARG00000006396 | nrcama | neuronal cell adhesion molecule a | 556537 |
| ENSDARG00000019596 | nrd1a | nardilysin a (N-arginine dibasic convertase) | 565850 |
| ENSDARG00000079985 | nrip2 | nuclear receptor interacting protein 2 | 565865 |
| ENSDARG00000102153 | nrp1a | neuropilin 1a | 353246 |
| ENSDARG00000027290 | nrp1b | neuropilin 1b | 402973 |
| ENSDARG00000096546 | nrp2a | neuropilin 2a | 405901 |
| ENSDARG00000038446 | nrp2b | neuropilin 2b | 405902 |
| ENSDARG00000044719 | nrsn1 | neurensin 1 | 335551 |
| ENSDARG00000061647 | nrxn1a | neurexin 1a | 565531 |
| ENSDARG00000061454 | nrxn2a | neurexin 2a | 558326 |
| ENSDARG00000063150 | nrxn2b | neurexin 2b | 0 |
| ENSDARG00000043746 | nrxn3a | neurexin 3a | 0 |
| ENSDARG00000062693 | nrxn3b | neurexin 3b | 570698 |
| ENSDARG00000007654 | nsfa | N-ethylmaleimide-sensitive factor a | 368483 |
| ENSDARG00000038991 | nsfb | N-ethylmaleimide-sensitive factor b | 554200 |
| ENSDARG00000087640 | nsfl1c | NSFL1 (p97) cofactor (p47) | 492805 |
| ENSDARG00000039399 | nt5c2a | 5'-nucleotidase, cytosolic IIa | 324845 |
| ENSDARG00000006797 | nt5dc1 | 5'-nucleotidase domain containing 1 | 568757 |
| ENSDARG00000062949 | nt5dc3 | 5'-nucleotidase domain containing 3 | 558469 |
| ENSDARG00000018065 | ntm | neurotrimin | 678534 |
| ENSDARG00000022531 | ntn1b | netrin 1b | 30192 |
| ENSDARG00000014973 | ntng1a | netrin g1a | 0 |
| ENSDARG00000077367 | ntng2a | netrin g2a | 560014 |
| ENSDARG00000059897 | ntrk2a | neurotrophic tyrosine kinase, receptor, type 2a | 65090 |
| ENSDARG00000077228 | ntrk3a | neurotrophic tyrosine kinase, receptor, type 3a | 568668 |
| ENSDARG00000054833 | nucb1 | nucleobindin 1 | 751753 |
| ENSDARG00000036299 | nucb2a | nucleobindin 2a | 373078 |
| ENSDARG00000062335 | nudt14 | nudix (nucleoside diphosphate linked moiety X)-type motif 14 | 559149 |
| ENSDARG00000078073 | nudt5 | nudix (nucleoside diphosphate linked moiety X)-type motif 5 | 415176 |
| ENSDARG00000101949 | numbl | numb homolog (Drosophila)-like | 497616 |
| ENSDARG00000027279 | numb | numb homolog (Drosophila) | 692064 |
| ENSDARG00000010078 | nup133 | nucleoporin 133 | 406852 |
| ENSDARG00000033978 | nxn | nucleoredoxin | 553621 |
| ENSDARG00000078425 | oat | ornithine aminotransferase | 572518 |
| ENSDARG00000019713 | oatx | organic anion transporter X | 0 |
| ENSDARG00000052334 | ociad1 | OCIA domain containing 1 | 393272 |
| ENSDARG00000034270 | ogdha | oxoglutarate (alpha-ketoglutarate) dehydrogenase a (lipoamide) | 564552 |
| ENSDARG00000103428 | ogdhb | oxoglutarate (alpha-ketoglutarate) dehydrogenase b (lipoamide) | 797715 |
| ENSDARG00000079249 | OGDHL | oxoglutarate dehydrogenase-like | 559207 |
| ENSDARG00000089444 | ogmb | oligodendrocyte myelin glycoprotein b | 101882436 |
| ENSDARG00000105243 | ogt.1 | O-linked N-acetylglucosamine (GlcNAc) transferase, tandem duplicate 1 | 337685 |
| ENSDARG00000044565 | ola1 | Obg-like ATPase 1 | 326864 |
| ENSDARG00000018270 | olfm1a | olfactomedin 1a | 436764 |
| ENSDARG00000014053 | olfm1b | olfactomedin 1b | 445092 |
| ENSDARG00000077847 | olfm2a | olfactomedin 2a | 334035 |
| ENSDARG00000102825 | olfm2b | olfactomedin 2b | 406544 |
| ENSDARG00000071493 | olfm3a | olfactomedin 3a | 563182 |
| ENSDARG00000086573 | omga | oligodendrocyte myelin glycoprotein a | 0 |
| ENSDARG00000070801 | opa1 | optic atrophy 1 (autosomal dominant) | 492332 |
| ENSDARG00000061761 | opa3 | optic atrophy 3 | 497278 |
| ENSDARG00000013005 | opcml | opioid binding protein/cell adhesion molecule-like | 449538 |
| ENSDARG00000058022 | ormdl2 | ORMDL sphingolipid biosynthesis regulator 2 | 677753 |
| ENSDARG00000025555 | ormdl3 | ORMDL sphingolipid biosynthesis regulator 3 | 450067 |
| ENSDARG00000039724 | OSBPL8 | oxysterol binding protein-like 8 | 101883298 |
| ENSDARG00000004634 | osbp | oxysterol binding protein | 564372 |
| ENSDARG00000030832 | otofa | otoferlin a | 557476 |
| ENSDARG00000011462 | otub1b | OTU deubiquitinase, ubiquitin aldehyde binding 1b | 0 |
| ENSDARG00000073931 | otulina | OTU deubiquitinase with linear linkage specificity a | 100001665 |
| ENSDARG00000069313 | oxa1l | oxidase (cytochrome c) assembly 1-like | 562963 |
| ENSDARG00000013063 | oxct1a | 3-oxoacid CoA transferase 1a | 442923 |
| ENSDARG00000054319 | oxct1b | 3-oxoacid CoA transferase 1b | 560273 |
| ENSDARG00000063310 | oxr1b | oxidation resistance 1b | 570476 |
| ENSDARG00000034189 | oxsr1a | oxidative stress responsive 1a | 568602 |
| ENSDARG00000105116 | p4hb | prolyl 4-hydroxylase, beta polypeptide | 406673 |
| ENSDARG00000039578 | pa2g4a | proliferation-associated 2G4, a | 368737 |
| ENSDARG00000070657 | pa2g4b | proliferation-associated 2G4, b | 323462 |
| ENSDARG00000017219 | pabpc1a | poly(A) binding protein, cytoplasmic 1a | 606498 |
| ENSDARG00000078185 | pacs2 | phosphofurin acidic cluster sorting protein 2 | 794778 |
| ENSDARG00000032865 | pacsin1a | protein kinase C and casein kinase substrate in neurons 1a | 553559 |
| ENSDARG00000042128 | pacsin1b | protein kinase C and casein kinase substrate in neurons 1b | 619246 |
| ENSDARG00000099339 | pacsin3 | protein kinase C and casein kinase substrate in neurons 3 | 393964 |
| ENSDARG00000044167 | padi2 | peptidyl arginine deiminase, type II | 386792 |
| ENSDARG00000032013 | pafah1b1a | platelet-activating factor acetylhydrolase 1b, regulatory subunit 1a | 394246 |
| ENSDARG00000026595 | pafah1b1b | platelet-activating factor acetylhydrolase 1b, regulatory subunit 1b | 394247 |
| ENSDARG00000035352 | pafah1b2 | platelet-activating factor acetylhydrolase 1b, catalytic subunit 2 | 567170 |
| ENSDARG00000058047 | pafah1b3 | platelet-activating factor acetylhydrolase, isoform Ib, gamma subunit | 394033 |
| ENSDARG00000055875 | pag1 | phosphoprotein membrane anchor with glycosphingolipid microdomains 1 | 100034498 |
| ENSDARG00000033539 | paics | phosphoribosylaminoimidazole carboxylase, phosphoribosylaminoimidazole succinocarboxamide synthetase | 321193 |
| ENSDARG00000103959 | pak1 | p21 protein (Cdc42/Rac)-activated kinase 1 | 373103 |
| ENSDARG00000068177 | pak2a | p21 protein (Cdc42/Rac)-activated kinase 2a | 266749 |
| ENSDARG00000030154 | pak7 | p21 protein (Cdc42/Rac)-activated kinase 7 | 0 |
| ENSDARG00000026882 | palm1a | paralemmin 1a | 550557 |
| ENSDARG00000007818 | palm1b | paralemmin 1b | 393573 |
| ENSDARG00000069608 | palm2 | paralemmin 2 | 100002543 |
| ENSDARG00000102822 | pam16 | presequence translocase-associated motor 16 homolog (S. cerevisiae) | 393777 |
| ENSDARG00000100170 | pam | peptidylglycine alpha-amidating monooxygenase | 570822 |
| ENSDARG00000003380 | PAOX | polyamine oxidase (exo-N4-amino) | 562105 |
| ENSDARG00000101719 | pargl | poly (ADP-ribose) glycohydrolase, like | 797792 |
| ENSDARG00000034826 | park7 | parkinson protein 7 | 449674 |
| ENSDARG00000019529 | parp1 | poly (ADP-ribose) polymerase 1 | 560788 |
| ENSDARG00000003961 | parp3 | poly (ADP-ribose) polymerase family, member 3 | 335495 |
| ENSDARG00000071015 | pbxip1a | pre-B-cell leukemia homeobox interacting protein 1a | 563463 |
| ENSDARG00000011824 | pbxip1b | pre-B-cell leukemia homeobox interacting protein 1b | 0 |
| ENSDARG00000031981 | pcbd1 | pterin-4 alpha-carbinolamine dehydratase/dimerization cofactor of hepatocyte nuclear factor 1 alpha | 393787 |
| ENSDARG00000099039 | pcbp2 | poly(rC) binding protein 2 | 321844 |
| ENSDARG00000028982 | pcca | propionyl CoA carboxylase, alpha polypeptide | 437019 |
| ENSDARG00000038910 | pccb | propionyl CoA carboxylase, beta polypeptide | 405861 |
| ENSDARG00000099729 | pcdh10a | protocadherin 10a | 0 |
| ENSDARG00000102824 | pcdh10b | protocadherin 10b | 360138 |
| ENSDARG00000027041 | pcdh17 | protocadherin 17 | 561336 |
| ENSDARG00000034344 | pcdh19 | protocadherin 19 | 0 |
| ENSDARG00000097170 | pcdh1a4 | protocadherin 1 alpha 4 | 497127 |
| ENSDARG00000062720 | pcdh1a | protocadherin 1a | 100006496 |
| ENSDARG00000036175 | pcdh1b | protocadherin 1b | 572016 |
| ENSDARG00000101232 | pcdh1g2 | protocadherin 1 gamma 2 | 553986 |
| ENSDARG00000100829 | pcdh1g32 | protocadherin 1 gamma 32 | 554016 |
| ENSDARG00000104826 | pcdh1gc5 | protocadherin 1 gamma c 5 | 503571 |
| ENSDARG00000088136 | pcdh2aa15 | protocadherin 2 alpha a 15 | 493601 |
| ENSDARG00000090348 | pcdh2ab5 | protocadherin 2 alpha b 5 | 493591 |
| ENSDARG00000099783 | pcdh2ac | protocadherin 2 alpha c | 493587 |
| ENSDARG00000098788 | pcdh2g16 | protocadherin 2 gamma 16 | 0 |
| ENSDARG00000103256 | pcdh2g1 | protocadherin 2 gamma 1 | 103912055 |
| ENSDARG00000078898 | pcdh7a | protocadherin 7a | 568198 |
| ENSDARG00000060610 | pcdh7b | protocadherin 7b | 555504 |
| ENSDARG00000063264 | PCDH9 | protocadherin 9 | 0 |
| ENSDARG00000077023 | pcdhb | protocadherin b | 100333352 |
| ENSDARG00000063299 | pcloa | piccolo presynaptic cytomatrix protein a | 100148002 |
| ENSDARG00000098880 | pclob | piccolo presynaptic cytomatrix protein b | 0 |
| ENSDARG00000042848 | pcmtl | l-isoaspartyl protein carboxyl methyltransferase, like | 393741 |
| ENSDARG00000015201 | pcmt | protein-L-isoaspartate (D-aspartate) O-methyltransferase | 30751 |
| ENSDARG00000033012 | pcnt | pericentrin | 325536 |
| ENSDARG00000053130 | pcp4a | Purkinje cell protein 4a | 386848 |
| ENSDARG00000023181 | pcp4l1 | Purkinje cell protein 4 like 1 | 100008021 |
| ENSDARG00000076170 | pcsk1nl | proprotein convertase subtilisin/kexin type 1 inhibitor, like | 570305 |
| ENSDARG00000002600 | pcsk1 | proprotein convertase subtilisin/kexin type 1 | 100005716 |
| ENSDARG00000019451 | pcsk2 | proprotein convertase subtilisin/kexin type 2 | 100004460 |
| ENSDARG00000098766 | pcxa | pyruvate carboxylase a | 0 |
| ENSDARG00000069003 | PCYOX1L | prenylcysteine oxidase 1 like | 0 |
| ENSDARG00000103052 | pcyox1 | prenylcysteine oxidase 1 | 550289 |
| ENSDARG00000058162 | pcyt1ba | phosphate cytidylyltransferase 1, choline, beta a | 100001552 |
| ENSDARG00000035054 | pdcd10a | programmed cell death 10a | 393527 |
| ENSDARG00000026072 | pdcd5 | programmed cell death 5 | 394152 |
| ENSDARG00000025269 | pdcd6ip | programmed cell death 6 interacting protein | 406669 |
| ENSDARG00000005220 | pdcd6 | programmed cell death 6 | 393925 |
| ENSDARG00000053609 | pddc1 | Parkinson disease 7 domain containing 1 | 100148360 |
| ENSDARG00000003449 | pde10a | phosphodiesterase 10A | 394077 |
| ENSDARG00000013950 | peptide deformylase (mitochondrial) | 619248 | |
| ENSDARG00000100897 | PDGFRB | platelet-derived growth factor receptor, beta polypeptide | 0 |
| ENSDARG00000012387 | pdha1a | pyruvate dehydrogenase (lipoamide) alpha 1a | 406702 |
| ENSDARG00000021346 | pdhb | pyruvate dehydrogenase (lipoamide) beta | 406428 |
| ENSDARG00000051756 | pdhx | pyruvate dehydrogenase complex, component X | 393532 |
| ENSDARG00000102640 | pdia3 | protein disulfide isomerase family A, member 3 | 378851 |
| ENSDARG00000018491 | pdia4 | protein disulfide isomerase family A, member 4 | 554998 |
| ENSDARG00000069242 | pdia5 | protein disulfide isomerase family A, member 5 | 556162 |
| ENSDARG00000009001 | pdia6 | protein disulfide isomerase family A, member 6 | 322160 |
| ENSDARG00000013128 | pdk1 | pyruvate dehydrogenase kinase, isozyme 1 | 555840 |
| ENSDARG00000014527 | pdk3a | pyruvate dehydrogenase kinase, isozyme 3a | 0 |
| ENSDARG00000018285 | pdpk1b | 3-phosphoinositide dependent protein kinase 1b | 403000 |
| ENSDARG00000103288 | PDPR | pyruvate dehydrogenase phosphatase regulatory subunit | 100330789 |
| ENSDARG00000088959 | pdxka | pyridoxal (pyridoxine, vitamin B6) kinase a | 791775 |
| ENSDARG00000036546 | pdxkb | pyridoxal (pyridoxine, vitamin B6) kinase b | 564697 |
| ENSDARG00000038315 | pdxp | pyridoxal (pyridoxine, vitamin B6) phosphatase | 561030 |
| ENSDARG00000053194 | pdzd11 | PDZ domain containing 11 | 405893 |
| ENSDARG00000088699 | pdzd8 | PDZ domain containing 8 | 561279 |
| ENSDARG00000079771 | PEA15 | phosphoprotein enriched in astrocytes 15 | 557745 |
| ENSDARG00000042069 | pebp1 | phosphatidylethanolamine binding protein 1 | 406627 |
| ENSDARG00000055976 | pecr | peroxisomal trans-2-enoyl-CoA reductase | 550422 |
| ENSDARG00000102249 | PEPD | peptidase D | 0 |
| ENSDARG00000025559 | pex11g | peroxisomal biogenesis factor 11 gamma | 563203 |
| ENSDARG00000028322 | pex14 | peroxisomal biogenesis factor 14 | 559933 |
| ENSDARG00000058202 | pex16 | peroxisomal biogenesis factor 16 | 573990 |
| ENSDARG00000014536 | pfdn1 | prefoldin subunit 1 | 503766 |
| ENSDARG00000025391 | pfdn2 | prefoldin subunit 2 | 323012 |
| ENSDARG00000035043 | pfdn5 | prefoldin 5 | 568182 |
| ENSDARG00000060504 | pfkla | phosphofructokinase, liver a | 570106 |
| ENSDARG00000099755 | pfklb | phosphofructokinase, liver b | 550259 |
| ENSDARG00000060797 | pfkmb | phosphofructokinase, muscle b | 568001 |
| ENSDARG00000028000 | pfkpa | phosphofructokinase, platelet a | 560944 |
| ENSDARG00000012801 | pfkpb | phosphofructokinase, platelet b | 561416 |
| ENSDARG00000088091 | pfn1 | profilin 1 | 799355 |
| ENSDARG00000012682 | pfn2l | profilin 2 like | 321383 |
| ENSDARG00000003952 | pfn2 | profilin 2 | 394235 |
| ENSDARG00000005423 | pgam1a | phosphoglycerate mutase 1a | 323107 |
| ENSDARG00000014068 | pgam1b | phosphoglycerate mutase 1b | 327165 |
| ENSDARG00000035608 | pgam5 | phosphoglycerate mutase family member 5 | 492357 |
| ENSDARG00000015343 | pgd | phosphogluconate dehydrogenase | 406762 |
| ENSDARG00000054191 | pgk1 | phosphoglycerate kinase 1 | 406696 |
| ENSDARG00000013561 | pgm1 | phosphoglucomutase 1 | 394000 |
| ENSDARG00000054172 | pgrmc1 | progesterone receptor membrane component 1 | 492520 |
| ENSDARG00000030237 | pgrmc2 | progesterone receptor membrane component 2 | 406378 |
| ENSDARG00000075857 | PGS1 | phosphatidylglycerophosphate synthase 1 | 0 |
| ENSDARG00000045297 | phb2a | prohibitin 2a | 324421 |
| ENSDARG00000017728 | phb2b | prohibitin 2b | 436954 |
| ENSDARG00000057414 | phb | prohibitin | 321346 |
| ENSDARG00000077596 | PHF24 | PHD finger protein 24 | 558943 |
| ENSDARG00000001873 | phgdh | phosphoglycerate dehydrogenase | 321928 |
| ENSDARG00000105159 | phka1 | phosphorylase kinase, alpha 1 (muscle) | 572183 |
| ENSDARG00000078284 | phkb | phosphorylase kinase, beta | 569706 |
| ENSDARG00000054208 | phkg2 | phosphorylase kinase, gamma 2 (testis) | 393937 |
| ENSDARG00000045567 | phpt1 | phosphohistidine phosphatase 1 | 445172 |
| ENSDARG00000003998 | phyhipla | phytanoyl-CoA 2-hydroxylase interacting protein-like a | 790943 |
| ENSDARG00000070498 | phyhiplb | phytanoyl-CoA 2-hydroxylase interacting protein-like b | 100004913 |
| ENSDARG00000086740 | phyh | phytanoyl-CoA 2-hydroxylase | 550521 |
| ENSDARG00000033666 | pi4k2a | phosphatidylinositol 4-kinase type 2 alpha | 406667 |
| ENSDARG00000076724 | pi4kaa | phosphatidylinositol 4-kinase, catalytic, alpha a | 556956 |
| ENSDARG00000062823 | pi4kab | phosphatidylinositol 4-kinase, catalytic, alpha b | 793349 |
| ENSDARG00000062445 | pias1b | protein inhibitor of activated STAT, 1b | 562982 |
| ENSDARG00000012866 | picalma | phosphatidylinositol binding clathrin assembly protein a | 393901 |
| ENSDARG00000014137 | picalmb | phosphatidylinositol binding clathrin assembly protein b | 445286 |
| ENSDARG00000038270 | PIGG | phosphatidylinositol glycan anchor biosynthesis, class G | 100538291 |
| ENSDARG00000024479 | pigk | phosphatidylinositol glycan anchor biosynthesis, class K | 415239 |
| ENSDARG00000017813 | pigs | phosphatidylinositol glycan anchor biosynthesis, class S | 767675 |
| ENSDARG00000060469 | pik3r4 | phosphoinositide-3-kinase, regulatory subunit 4 | 564088 |
| ENSDARG00000056112 | pikfyve | phosphoinositide kinase, FYVE finger containing | 337690 |
| ENSDARG00000101915 | PIN1 | peptidylprolyl cis/trans isomerase, NIMA-interacting 1 | 0 |
| ENSDARG00000004527 | pin4 | protein (peptidylprolyl cis/trans isomerase) NIMA-interacting, 4 (parvulin) | 553574 |
| ENSDARG00000003776 | pip4k2aa | phosphatidylinositol-5-phosphate 4-kinase, type II, alpha a | 560360 |
| ENSDARG00000063544 | pip4k2ab | phosphatidylinositol-5-phosphate 4-kinase, type II, alpha b | 555920 |
| ENSDARG00000024642 | pip5k1ab | phosphatidylinositol-4-phosphate 5-kinase, type I, alpha, b | 492549 |
| ENSDARG00000076001 | pip5k1ca | phosphatidylinositol-4-phosphate 5-kinase, type I, gamma a | 798876 |
| ENSDARG00000100313 | pip5k1cb | phosphatidylinositol-4-phosphate 5-kinase, type I, gamma b | 100005033 |
| ENSDARG00000031020 | pip5k2 | phosphatidylinositol-4-phosphate 5-kinase, type II | 373124 |
| ENSDARG00000055591 | pipox | pipecolic acid oxidase | 558601 |
| ENSDARG00000044091 | pitpnab | phosphatidylinositol transfer protein, alpha b | 100333060 |
| ENSDARG00000055255 | pitpnm3 | PITPNM family member 3 | 337272 |
| ENSDARG00000038563 | pitrm1 | pitrilysin metallopeptidase 1 | 406808 |
| ENSDARG00000099730 | pkma | pyruvate kinase, muscle, a | 335817 |
| ENSDARG00000079438 | PKP3 (2 of 2) | plakophilin 3 | 100330353 |
| ENSDARG00000045331 | pkp4 | plakophilin 4 | 561566 |
| ENSDARG00000005774 | pl10 | pl10 | 30116 |
| ENSDARG00000068246 | plcb3 | phospholipase C, beta 3 (phosphatidylinositol-specific) | 100001532 |
| ENSDARG00000100786 | PLCB4 | phospholipase C, beta 4 | 100537538 |
| ENSDARG00000052957 | plcd3a | phospholipase C, delta 3a | 569040 |
| ENSDARG00000079572 | plcd3b | phospholipase C, delta 3b | 562569 |
| ENSDARG00000059960 | plch2a | phospholipase C, eta 2a | 100334231 |
| ENSDARG00000067676 | plcl1 | phospholipase C-like 1 | 563105 |
| ENSDARG00000062590 | pleca | plectin a | 567639 |
| ENSDARG00000021987 | plecb | plectin b | 100321011 |
| ENSDARG00000045870 | plekha5 | pleckstrin homology domain containing, family A member 5 | 567300 |
| ENSDARG00000020328 | plekha6 | pleckstrin homology domain containing, family A member 6 | 568764 |
| ENSDARG00000062220 | plekha7b | pleckstrin homology domain containing, family A member 7b | 100005106 |
| ENSDARG00000100789 | plgrkt | plasminogen receptor, C-terminal lysine transmembrane protein | 606662 |
| ENSDARG00000011929 | plp1b | proteolipid protein 1b | 368234 |
| ENSDARG00000104668 | plxna1b | plexin A1b | 0 |
| ENSDARG00000060372 | plxna2 | plexin A2 | 561285 |
| ENSDARG00000007172 | plxna3 | plexin A3 | 567422 |
| ENSDARG00000019328 | plxna4 | plexin A4 | 325938 |
| ENSDARG00000017211 | plxnb1b | plexin b1b | 570222 |
| ENSDARG00000079892 | PLXNB2 (4 of 5) | plexin B2 | 561237 |
| ENSDARG00000003811 | plxnb2a | plexin b2a | 561433 |
| ENSDARG00000036985 | plxnb2b | plexin b2b | 0 |
| ENSDARG00000032990 | plxnb3 | plexin B3 | 566546 |
| ENSDARG00000062096 | pm20d1.2 | peptidase M20 domain containing 1, tandem duplicate 2 | 768158 |
| ENSDARG00000102607 | pmpca | peptidase (mitochondrial processing) alpha | 492801 |
| ENSDARG00000099802 | pnp5b | purine nucleoside phosphorylase 5b | 447889 |
| ENSDARG00000040942 | pnp6 | purine nucleoside phosphorylase 6 | 402953 |
| ENSDARG00000010773 | pnpla6 | patatin-like phospholipase domain containing 6 | 560986 |
| ENSDARG00000040002 | pnpla7b | patatin-like phospholipase domain containing 7b | 503936 |
| ENSDARG00000078745 | pnpla8 | patatin-like phospholipase domain containing 8 | 100126812 |
| ENSDARG00000017612 | pnpo | pyridoxamine 5'-phosphate oxidase | 0 |
| ENSDARG00000010941 | pomgnt2 | protein O-linked mannose N-acetylglucosaminyltransferase 2 (beta 1,4-) | 497644 |
| ENSDARG00000016856 | pon2 | paraoxonase 2 | 335176 |
| ENSDARG00000090444 | ponzr1 | plac8 onzin related protein 1 | 567858 |
| ENSDARG00000055092 | pora | P450 (cytochrome) oxidoreductase a | 568202 |
| ENSDARG00000059035 | porb | P450 (cytochrome) oxidoreductase b | 327556 |
| ENSDARG00000030441 | ppa1b | pyrophosphatase (inorganic) 1b | 550531 |
| ENSDARG00000009685 | ppa2 | pyrophosphatase (inorganic) 2 | 402961 |
| ENSDARG00000059933 | ppap2b | phosphatidic acid phosphatase type 2B | 557680 |
| ENSDARG00000069940 | ppap2d | phosphatidic acid phosphatase type 2D | 559124 |
| ENSDARG00000043527 | ppapdc2 | phosphatidic acid phosphatase type 2 domain containing 2 | 553687 |
| ENSDARG00000031317 | ppdpfb | pancreatic progenitor cell differentiation and proliferation factor b | 336303 |
| ENSDARG00000013000 | ppfia2 | protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2 | 566597 |
| ENSDARG00000077053 | ppfia3 | protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 3 | 567230 |
| ENSDARG00000053205 | ppfia4 | protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 4 | 100005543 |
| ENSDARG00000009212 | ppiaa | peptidylprolyl isomerase Aa (cyclophilin A) | 336612 |
| ENSDARG00000103994 | ppiab | peptidylprolyl isomerase Ab (cyclophilin A) | 335519 |
| ENSDARG00000092798 | ppib | peptidylprolyl isomerase B (cyclophilin B) | 406292 |
| ENSDARG00000104343 | PPIC | peptidylprolyl isomerase C (cyclophilin C) | 100333141 |
| ENSDARG00000038835 | ppid | peptidylprolyl isomerase D | 415155 |
| ENSDARG00000032155 | ppm1aa | protein phosphatase, Mg2+/Mn2+ dependent, 1Aa | 30704 |
| ENSDARG00000043250 | ppm1ab | protein phosphatase, Mg2+/Mn2+ dependent, 1Ab | 30703 |
| ENSDARG00000026499 | ppm1e | protein phosphatase, Mg2+/Mn2+ dependent, 1E | 553417 |
| ENSDARG00000063684 | ppm1h | protein phosphatase, Mg2+/Mn2+ dependent, 1H | 768291 |
| ENSDARG00000074690 | ppm1j | protein phosphatase, Mg2+/Mn2+ dependent, 1J | 556950 |
| ENSDARG00000009771 | ppme1 | protein phosphatase methylesterase 1 | 335124 |
| ENSDARG00000003486 | ppp1caa | protein phosphatase 1, catalytic subunit, alpha isozyme a | 407980 |
| ENSDARG00000071566 | ppp1cab | protein phosphatase 1, catalytic subunit, alpha isozyme b | 327301 |
| ENSDARG00000044153 | ppp1cb | protein phosphatase 1, catalytic subunit, beta isozyme | 100003223 |
| ENSDARG00000099226 | ppp1cc | protein phosphatase 1, catalytic subunit, gamma isozyme | 571753 |
| ENSDARG00000010784 | ppp1r12a | protein phosphatase 1, regulatory subunit 12A | 445393 |
| ENSDARG00000103144 | ppp1r21 | protein phosphatase 1, regulatory subunit 21 | 492333 |
| ENSDARG00000054007 | ppp1r2 | protein phosphatase 1, regulatory (inhibitor) subunit 2 | 402967 |
| ENSDARG00000009740 | ppp1r7 | protein phosphatase 1, regulatory (inhibitor) subunit 7 | 560323 |
| ENSDARG00000061304 | ppp1r9a | protein phosphatase 1, regulatory subunit 9A | 560862 |
| ENSDARG00000079366 | ppp1r9ba | protein phosphatase 1, regulatory subunit 9Ba | 558232 |
| ENSDARG00000071709 | ppp1r9bb | protein phosphatase 1, regulatory subunit 9Bb | 100331262 |
| ENSDARG00000099241 | ppp2cb | protein phosphatase 2, catalytic subunit, beta isozyme | 406582 |
| ENSDARG00000007791 | ppp2r1a | protein phosphatase 2, regulatory subunit A, alpha | 406685 |
| ENSDARG00000032430 | ppp2r1b | protein phosphatase 2, regulatory subunit A, beta | 449548 |
| ENSDARG00000021996 | ppp2r2aa | protein phosphatase 2, regulatory subunit B, alpha a | 558752 |
| ENSDARG00000102009 | ppp2r2d | protein phosphatase 2, regulatory subunit B, delta | 0 |
| ENSDARG00000031985 | ppp2r4 | protein phosphatase 2A activator, regulatory subunit 4 | 445031 |
| ENSDARG00000054931 | PPP2R5B | protein phosphatase 2, regulatory subunit B', beta | 562282 |
| ENSDARG00000031200 | ppp2r5cb | protein phosphatase 2, regulatory subunit B', gamma b | 557208 |
| ENSDARG00000015474 | ppp2r5ea | protein phosphatase 2, regulatory subunit B', epsilon isoform a | 373881 |
| ENSDARG00000069118 | ppp2r5eb | protein phosphatase 2, regulatory subunit B', epsilon isoform b | 373878 |
| ENSDARG00000004988 | ppp3ca | protein phosphatase 3, catalytic subunit, alpha isozyme | 794574 |
| ENSDARG00000025106 | ppp3cb | protein phosphatase 3, catalytic subunit, beta isozyme | 550554 |
| ENSDARG00000014962 | ppp3cca | protein phosphatase 3, catalytic subunit, gamma isozyme, a | 557926 |
| ENSDARG00000057456 | ppp3ccb | protein phosphatase 3, catalytic subunit, gamma isozyme, b | 565241 |
| ENSDARG00000069360 | ppp3r1b | protein phosphatase 3 (formerly 2B), regulatory s1ubunit B, alpha isoform, b | 447814 |
| ENSDARG00000034313 | ppp5c | protein phosphatase 5, catalytic subunit | 541536 |
| ENSDARG00000002949 | ppp6c | protein phosphatase 6, catalytic subunit | 393980 |
| ENSDARG00000045540 | ppp6r2a | protein phosphatase 6, regulatory subunit 2a | 560729 |
| ENSDARG00000069654 | ppp6r2b | protein phosphatase 6, regulatory subunit 2b | 570360 |
| ENSDARG00000039980 | ppt1 | palmitoyl-protein thioesterase 1 (ceroid-lipofuscinosis, neuronal 1, infantile) | 406648 |
| ENSDARG00000032535 | praf2 | PRA1 domain family, member 2 | 550393 |
| ENSDARG00000058734 | prdx1 | peroxiredoxin 1 | 541344 |
| ENSDARG00000025350 | prdx2 | peroxiredoxin 2 | 791455 |
| ENSDARG00000032102 | prdx3 | peroxiredoxin 3 | 0 |
| ENSDARG00000069013 | prdx4 | peroxiredoxin 4 | 570477 |
| ENSDARG00000055064 | prdx5 | peroxiredoxin 5 | 0 |
| ENSDARG00000043511 | prdx6 | peroxiredoxin 6 | 393778 |
| ENSDARG00000057151 | prep | prolyl endopeptidase | 553791 |
| ENSDARG00000075793 | prex1 | phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 1 | 570984 |
| ENSDARG00000020982 | prickle2a | prickle homolog 2a (Drosophila) | 777736 |
| ENSDARG00000060596 | prkaa1 | protein kinase, AMP-activated, alpha 1 catalytic subunit | 100001198 |
| ENSDARG00000100349 | prkacaa | protein kinase, cAMP-dependent, catalytic, alpha, genome duplicate a | 564064 |
| ENSDARG00000016809 | prkacab | protein kinase, cAMP-dependent, catalytic, alpha, genome duplicate b | 445076 |
| ENSDARG00000059125 | prkacbb | protein kinase, cAMP-dependent, catalytic, beta b | 563147 |
| ENSDARG00000076128 | prkar1aa | protein kinase, cAMP-dependent, regulatory, type I, alpha (tissue specific extinguisher 1) a | 494533 |
| ENSDARG00000101755 | prkar1ab | protein kinase, cAMP-dependent, regulatory, type I, alpha (tissue specific extinguisher 1) b | 550427 |
| ENSDARG00000103800 | prkar1b | protein kinase, cAMP-dependent, regulatory, type I, beta | 0 |
| ENSDARG00000033184 | prkar2aa | protein kinase, cAMP-dependent, regulatory, type II, alpha A | 556042 |
| ENSDARG00000036446 | PRKAR2B | protein kinase, cAMP-dependent, regulatory, type II, beta | 0 |
| ENSDARG00000039241 | prkca | protein kinase C, alpha | 497384 |
| ENSDARG00000008723 | prkcba | protein kinase C, beta a | 100006675 |
| ENSDARG00000022254 | prkcbb | protein kinase C, beta b | 393953 |
| ENSDARG00000009208 | prkcda | protein kinase C, delta a | 334571 |
| ENSDARG00000003008 | prkcea | protein kinase C, epsilon a | 100005075 |
| ENSDARG00000004561 | prkcg | protein kinase C, gamma | 100003514 |
| ENSDARG00000004470 | prkcsh | protein kinase C substrate 80K-H | 394028 |
| ENSDARG00000075949 | prkd1 | protein kinase D1 | 557123 |
| ENSDARG00000075083 | prkdc | protein kinase, DNA-activated, catalytic polypeptide | 562283 |
| ENSDARG00000076718 | prkra | protein kinase, interferon-inducible double stranded RNA dependent activator | 557370 |
| ENSDARG00000037946 | prl | prolactin | 791526 |
| ENSDARG00000010246 | prmt1 | protein arginine methyltransferase 1 | 321974 |
| ENSDARG00000003705 | prnprs3 | prion protein, related sequence 3 | 503702 |
| ENSDARG00000039966 | prom1a | prominin 1a | 322857 |
| ENSDARG00000034007 | prom1b | prominin 1 b | 378834 |
| ENSDARG00000060288 | prosc | proline synthetase co-transcribed homolog (bacterial) | 563548 |
| ENSDARG00000037783 | proza | protein Z, vitamin K-dependent plasma glycoprotein a | 768176 |
| ENSDARG00000015239 | prp19 | PRP19/PSO4 homolog (S. cerevisiae) | 321544 |
| ENSDARG00000028306 | prph | peripherin | 30259 |
| ENSDARG00000015524 | prps1a | phosphoribosyl pyrophosphate synthetase 1A | 140620 |
| ENSDARG00000037506 | prps1b | phosphoribosyl pyrophosphate synthetase 1B | 560827 |
| ENSDARG00000019326 | prpsap2 | phosphoribosyl pyrophosphate synthetase-associated protein 2 | 406369 |
| ENSDARG00000062208 | prrt1 | proline-rich transmembrane protein 1 | 559734 |
| ENSDARG00000076602 | prss16 | protease, serine, 16 (thymus) | 556307 |
| ENSDARG00000016733 | psat1 | phosphoserine aminotransferase 1 | 327512 |
| ENSDARG00000076702 | PSD (1 of 2) | pleckstrin and Sec7 domain containing | 101886286 |
| ENSDARG00000063036 | psd2 | pleckstrin and Sec7 domain containing 2 | 557199 |
| ENSDARG00000104820 | psd3l | pleckstrin and Sec7 domain containing 3, like | 0 |
| ENSDARG00000004870 | psen1 | presenilin 1 | 30221 |
| ENSDARG00000101560 | psma1 | proteasome (prosome, macropain) subunit, alpha type, 1 | 445033 |
| ENSDARG00000086618 | psma3 | proteasome (prosome, macropain) subunit, alpha type, 3 | 564370 |
| ENSDARG00000045928 | psma4 | proteasome (prosome, macropain) subunit, alpha type, 4 | 326687 |
| ENSDARG00000003526 | psma5 | proteasome (prosome, macropain) subunit, alpha type, 5 | 403011 |
| ENSDARG00000019398 | psma6a | proteasome (prosome, macropain) subunit, alpha type, 6a | 171585 |
| ENSDARG00000013966 | psma6b | proteasome (prosome, macropain) subunit, alpha type, 6b | 436862 |
| ENSDARG00000010965 | psma8 | proteasome (prosome, macropain) subunit, alpha type, 8 | 406445 |
| ENSDARG00000009640 | psmb1 | proteasome (prosome, macropain) subunit, beta type, 1 | 445413 |
| ENSDARG00000031511 | psmb2 | proteasome (prosome, macropain) subunit, beta type, 2 | 436882 |
| ENSDARG00000013938 | psmb3 | proteasome (prosome, macropain) subunit, beta type, 3 | 573095 |
| ENSDARG00000054696 | psmb4 | proteasome (prosome, macropain) subunit, beta type, 4 | 550499 |
| ENSDARG00000075445 | psmb5 | proteasome (prosome, macropain) subunit, beta type, 5 | 30387 |
| ENSDARG00000002240 | psmb6 | proteasome (prosome, macropain) subunit, beta type, 6 | 30388 |
| ENSDARG00000037962 | psmb7 | proteasome (prosome, macropain) subunit, beta type, 7 | 64278 |
| ENSDARG00000043561 | psmc1b | proteasome (prosome, macropain) 26S subunit, ATPase, 1b | 415181 |
| ENSDARG00000020101 | psmc2 | proteasome (prosome, macropain) 26S subunit, ATPase 2 | 393941 |
| ENSDARG00000007141 | psmc3 | proteasome (prosome, macropain) 26S subunit, ATPase, 3 | 321947 |
| ENSDARG00000027099 | psmc4 | proteasome (prosome, macropain) 26S subunit, ATPase, 4 | 326884 |
| ENSDARG00000015315 | psmc5 | proteasome (prosome, macropain) 26S subunit, ATPase, 5 | 445285 |
| ENSDARG00000037038 | psmc6 | proteasome (prosome, macropain) 26S subunit, ATPase, 6 | 321585 |
| ENSDARG00000060150 | psmd11a | proteasome (prosome, macropain) 26S subunit, non-ATPase, 11a | 503934 |
| ENSDARG00000005134 | psmd11b | proteasome (prosome, macropain) 26S subunit, non-ATPase, 11b | 322265 |
| ENSDARG00000052377 | psmd12 | proteasome (prosome, macropain) 26S subunit, non-ATPase, 12 | 321898 |
| ENSDARG00000016239 | psmd13 | proteasome (prosome, macropain) 26S subunit, non-ATPase, 13 | 393922 |
| ENSDARG00000063100 | psmd14 | proteasome (prosome, macropain) 26S subunit, non-ATPase, 14 | 799058 |
| ENSDARG00000003189 | psmd1 | proteasome (prosome, macropain) 26S subunit, non-ATPase, 1 | 554967 |
| ENSDARG00000103190 | PSMD2 (2 of 2) | proteasome (prosome, macropain) 26S subunit, non-ATPase, 2 | 0 |
| ENSDARG00000099653 | psmd2 | proteasome (prosome, macropain) 26S subunit, non-ATPase, 2 | 0 |
| ENSDARG00000018124 | psmd3 | proteasome (prosome, macropain) 26S subunit, non-ATPase, 3 | 393544 |
| ENSDARG00000023279 | psmd4b | proteasome (prosome, macropain) 26S subunit, non-ATPase, 4b | 550287 |
| ENSDARG00000070674 | psmd6 | proteasome (prosome, macropain) 26S subunit, non-ATPase, 6 | 393261 |
| ENSDARG00000102417 | psmd7 | proteasome (prosome, macropain) 26S subunit, non-ATPase, 7 (Mov34 homolog) | 327330 |
| ENSDARG00000020454 | psmd8 | proteasome (prosome, macropain) 26S subunit, non-ATPase, 8 | 415221 |
| ENSDARG00000012588 | ptdss1a | phosphatidylserine synthase 1a | 393931 |
| ENSDARG00000063375 | pter | phosphotriesterase related | 751664 |
| ENSDARG00000027088 | ptgdsb | prostaglandin D2 synthase b | 336492 |
| ENSDARG00000068415 | ptgesl | prostaglandin E synthase 2-like | 799964 |
| ENSDARG00000075505 | PTGFRN (1 of 2) | prostaglandin F2 receptor inhibitor | 101884699 |
| ENSDARG00000078172 | PTGFRN (2 of 2) | prostaglandin F2 receptor inhibitor | 101883373 |
| ENSDARG00000024877 | ptgr1 | prostaglandin reductase 1 | 494108 |
| ENSDARG00000022951 | pth2 | parathyroid hormone 2 | 402812 |
| ENSDARG00000011863 | ptk7a | protein tyrosine kinase 7a | 553690 |
| ENSDARG00000101766 | ptmab | prothymosin, alpha b | 559194 |
| ENSDARG00000035676 | ptp4a2b | protein tyrosine phosphatase type IVA, member 2b | 334483 |
| ENSDARG00000013860 | ptpmt1 | protein tyrosine phosphatase, mitochondrial 1 | 567019 |
| ENSDARG00000070124 | ptpn23a | protein tyrosine phosphatase, non-receptor type 23, a | 100007322 |
| ENSDARG00000000183 | ptpn4b | protein tyrosine phosphatase, non-receptor type 4b | 407634 |
| ENSDARG00000074866 | ptpn5 | protein tyrosine phosphatase, non-receptor type 5 | 559524 |
| ENSDARG00000073902 | ptpn9b | protein tyrosine phosphatase, non-receptor type 9, b | 562170 |
| ENSDARG00000001769 | ptpra | protein tyrosine phosphatase, receptor type, A | 140818 |
| ENSDARG00000071437 | ptprc | protein tyrosine phosphatase, receptor type, C | 0 |
| ENSDARG00000019945 | ptprdb | protein tyrosine phosphatase, receptor type, D, b | 100073332 |
| ENSDARG00000015891 | ptprea | protein tyrosine phosphatase, receptor type, E, a | 100499203 |
| ENSDARG00000103479 | ptprfa | protein tyrosine phosphatase, receptor type, f, a | 0 |
| ENSDARG00000005754 | ptprfb | protein tyrosine phosphatase, receptor type, f, b | 799867 |
| ENSDARG00000045006 | ptprga | protein tyrosine phosphatase, receptor type, g a | 561197 |
| ENSDARG00000063416 | ptprk | protein tyrosine phosphatase, receptor type, K | 0 |
| ENSDARG00000035970 | ptprn2 | protein tyrosine phosphatase, receptor type, N polypeptide 2 | 325033 |
| ENSDARG00000058646 | ptprna | protein tyrosine phosphatase, receptor type, Na | 324790 |
| ENSDARG00000077047 | ptprnb | protein tyrosine phosphatase, receptor type, Nb | 564351 |
| ENSDARG00000100422 | ptpro | protein tyrosine phosphatase, receptor type, O | 407676 |
| ENSDARG00000059738 | ptprsa | protein tyrosine phosphatase, receptor type, s, a | 140820 |
| ENSDARG00000097572 | ptprt | protein tyrosine phosphatase, receptor type, t | 0 |
| ENSDARG00000051814 | ptprz1a | protein tyrosine phosphatase, receptor-type, Z polypeptide 1a | 0 |
| ENSDARG00000020871 | ptprz1b | protein tyrosine phosphatase, receptor-type, Z polypeptide 1b | 553019 |
| ENSDARG00000077339 | ptrh2 | peptidyl-tRNA hydrolase 2 | 791795 |
| ENSDARG00000001241 | puf60b | poly-U binding splicing factor b | 415211 |
| ENSDARG00000067591 | pura | purine-rich element binding protein A | 415106 |
| ENSDARG00000068822 | purba | purine-rich element binding protein Ba | 564840 |
| ENSDARG00000002768 | pvalb2 | parvalbumin 2 | 58028 |
| ENSDARG00000024433 | pvalb4 | parvalbumin 4 | 337731 |
| ENSDARG00000009311 | pvalb6 | parvalbumin 6 | 402806 |
| ENSDARG00000034705 | pvalb7 | parvalbumin 7 | 402807 |
| ENSDARG00000086034 | pvrl1b | poliovirus receptor-related 1b | 0 |
| ENSDARG00000063390 | pvrl2l | poliovirus receptor-related 2 like | 560933 |
| ENSDARG00000040309 | pxdc1b | PX domain containing 1b | 566349 |
| ENSDARG00000102254 | pycr1a | pyrroline-5-carboxylate reductase 1a | 393799 |
| ENSDARG00000098639 | pycr1b | pyrroline-5-carboxylate reductase 1b | 100149838 |
| ENSDARG00000015236 | pycrl | pyrroline-5-carboxylate reductase-like | 560682 |
| ENSDARG00000002021 | pygb | phosphorylase, glycogen; brain | 403051 |
| ENSDARG00000013317 | pygmb | phosphorylase, glycogen, muscle b | 393444 |
| ENSDARG00000010316 | qars | glutaminyl-tRNA synthetase | 394188 |
| ENSDARG00000040190 | qdpra | quinoid dihydropteridine reductase a | 566323 |
| ENSDARG00000002026 | qkib | QKI, KH domain containing, RNA binding b | 100462714 |
| ENSDARG00000077174 | QSOX2 | quiescin Q6 sulfhydryl oxidase 2 | 559603 |
| ENSDARG00000046106 | rab10 | RAB10, member RAS oncogene family | 436839 |
| ENSDARG00000041450 | rab11a | RAB11a, member RAS oncogene family | 492487 |
| ENSDARG00000090086 | rab11bb | RAB11B, member RAS oncogene family, b | 436828 |
| ENSDARG00000104179 | rab11fip5a | RAB11 family interacting protein 5a (class I) | 562719 |
| ENSDARG00000035188 | rab14l | RAB14, member RAS oncogene family, like | 393656 |
| ENSDARG00000074246 | rab14 | RAB14, member RAS oncogene family | 373104 |
| ENSDARG00000026484 | rab15 | RAB15, member RAS oncogene family | 436590 |
| ENSDARG00000055291 | rab18a | RAB18A, member RAS oncogene family | 325255 |
| ENSDARG00000098344 | rab18b | RAB18B, member RAS oncogene family | 445055 |
| ENSDARG00000058044 | rab1ba | zRAB1B, member RAS oncogene family a | 394117 |
| ENSDARG00000056479 | rab1bb | RAB1B, member RAS oncogene family b | 415219 |
| ENSDARG00000015807 | rab22a | RAB22A, member RAS oncogene family | 100001531 |
| ENSDARG00000004151 | rab23 | RAB23, member RAS oncogene family | 553778 |
| ENSDARG00000054032 | rab24 | RAB24, member RAS oncogene family | 574423 |
| ENSDARG00000087762 | RAB27B | RAB27B, member RAS oncogene family | 561717 |
| ENSDARG00000020261 | rab2a | RAB2A, member RAS oncogene family | 140617 |
| ENSDARG00000037884 | rab30 | RAB30, member RAS oncogene family | 437023 |
| ENSDARG00000020128 | RAB31 | RAB31, member RAS oncogene family | 568800 |
| ENSDARG00000057394 | rab33a | RAB33A, member RAS oncogene family | 751718 |
| ENSDARG00000045628 | rab34a | RAB34, member RAS oncogene family a | 450074 |
| ENSDARG00000102078 | RAB35 (2 of 2) | RAB35, member RAS oncogene family | 796454 |
| ENSDARG00000058425 | rab35b | RAB35, member RAS oncogene family b | 445154 |
| ENSDARG00000036501 | rab39bb | RAB39B, member RAS oncogene family b | 566010 |
| ENSDARG00000056347 | rab3aa | RAB3A, member RAS oncogene family, a | 445024 |
| ENSDARG00000043835 | rab3ab | RAB3A, member RAS oncogene family, b | 550457 |
| ENSDARG00000042803 | rab3b | RAB3B, member RAS oncogene family | 557482 |
| ENSDARG00000014462 | rab3c | RAB3C, member RAS oncogene family | 436803 |
| ENSDARG00000029885 | rab41 | RAB41, member RAS oncogene family | 406674 |
| ENSDARG00000034215 | rab42a | RAB42, member RAS oncogene family a | 406315 |
| ENSDARG00000019144 | rab4a | RAB4a, member RAS oncogene family | 403031 |
| ENSDARG00000012177 | rab4b | RAB4B, member RAS oncogene family | 445498 |
| ENSDARG00000018602 | rab5aa | RAB5A, member RAS oncogene family, a | 337737 |
| ENSDARG00000007257 | rab5ab | RAB5A, member RAS oncogene family, b | 393945 |
| ENSDARG00000016059 | rab5b | RAB5B, member RAS oncogene family | 405821 |
| ENSDARG00000026712 | rab5c | RAB5C, member RAS oncogene family | 386807 |
| ENSDARG00000103714 | rab6a | RAB6A, member RAS oncogene family | 793785 |
| ENSDARG00000034522 | rab6ba | RAB6B, member RAS oncogene family a | 558900 |
| ENSDARG00000031343 | rab6bb | RAB6B, member RAS oncogene family b | 541339 |
| ENSDARG00000020497 | rab7 | RAB7, member RAS oncogene family | 393902 |
| ENSDARG00000067920 | rab8a | RAB8A, member RAS oncogene family | 571897 |
| ENSDARG00000068628 | rab8b | RAB8B, member RAS oncogene family | 100093706 |
| ENSDARG00000045027 | rab9a | RAB9A, member RAS oncogene family | 751756 |
| ENSDARG00000103391 | rab9b | RAB9B, member RAS oncogene family | 100003363 |
| ENSDARG00000059600 | rabep1 | rabaptin, RAB GTPase binding effector protein 1 | 568064 |
| ENSDARG00000098967 | RABGAP1 | RAB GTPase activating protein 1 | 559632 |
| ENSDARG00000002690 | rabif | RAB interacting factor | 436784 |
| ENSDARG00000038010 | rac2 | ras-related C3 botulinum toxin substrate 2 (rho family, small GTP binding protein Rac2) | 415151 |
| ENSDARG00000090062 | rac3a | ras-related C3 botulinum toxin substrate 3a (rho family, small GTP binding protein Rac3) | 437027 |
| ENSDARG00000020795 | rac3b | ras-related C3 botulinum toxin substrate 3b (rho family, small GTP binding protein Rac3) | 795167 |
| ENSDARG00000058984 | rad23ab | RAD23 homolog Ab (S. cerevisiae) | 562839 |
| ENSDARG00000039974 | rai14 | retinoic acid induced 14 | 794796 |
| ENSDARG00000045463 | ralaa | v-ral simian leukemia viral oncogene homolog Aa (ras related) | 393993 |
| ENSDARG00000016253 | ralab | v-ral simian leukemia viral oncogene homolog Ab (ras related) | 492355 |
| ENSDARG00000040778 | ralba | v-ral simian leukemia viral oncogene homolog Ba (ras related) | 415213 |
| ENSDARG00000076889 | ralgapa1 | Ral GTPase activating protein, alpha subunit 1 (catalytic) | 556427 |
| ENSDARG00000088899 | ralgapb | Ral GTPase activating protein, beta subunit (non-catalytic) | 559303 |
| ENSDARG00000057026 | ran | RAN, member RAS oncogene family | 30572 |
| ENSDARG00000087346 | rap1ab | RAP1A, member of RAS oncogene family b | 406714 |
| ENSDARG00000008867 | rap1b | RAP1B, member of RAS oncogene family | 321107 |
| ENSDARG00000061551 | rap1gap2a | RAP1 GTPase activating protein 2a | 0 |
| ENSDARG00000073741 | RAP1GDS1 | RAP1, GTP-GDP dissociation stimulator 1 | 100334870 |
| ENSDARG00000077553 | RAP2A (2 of 2) | RAP2A, member of RAS oncogene family | 100332969 |
| ENSDARG00000040246 | rap2ab | RAP2A, member of RAS oncogene family b | 100003903 |
| ENSDARG00000103032 | rap2b | RAP2B, member of RAS oncogene family | 414329 |
| ENSDARG00000015649 | rap2c | RAP2C, member of RAS oncogene family | 474323 |
| ENSDARG00000005482 | rapgef2 | Rap guanine nucleotide exchange factor (GEF) 2 | 557693 |
| ENSDARG00000075924 | RAPGEF4 (1 of 2) | Rap guanine nucleotide exchange factor (GEF) 4 | 100141331 |
| ENSDARG00000079872 | rapgef4 | Rap guanine nucleotide exchange factor (GEF) 4 | 0 |
| ENSDARG00000054530 | rars | arginyl-tRNA synthetase | 337070 |
| ENSDARG00000073824 | RASGRF1 | Ras protein-specific guanine nucleotide-releasing factor 1 | 568885 |
| ENSDARG00000002816 | rasgrf2b | Ras protein-specific guanine nucleotide-releasing factor 2b | 553520 |
| ENSDARG00000022129 | rbm4.3 | RNA binding motif protein 4.3 | 406277 |
| ENSDARG00000101199 | rbp4 | retinol binding protein 4, plasma | 30077 |
| ENSDARG00000078215 | rbsn | rabenosyn, RAB effector | 569240 |
| ENSDARG00000038030 | rbx1 | ring-box 1, E3 ubiquitin protein ligase | 449846 |
| ENSDARG00000056075 | rca2.1 | regulator of complement activation group 2 gene 1 | 541367 |
| ENSDARG00000039378 | rcn2 | reticulocalbin 2 | 571340 |
| ENSDARG00000045277 | rdh12l | retinol dehydrogenase 12, like | 494176 |
| ENSDARG00000018069 | rdh12 | retinol dehydrogenase 12 (all-trans/9-cis/11-cis) | 436597 |
| ENSDARG00000008491 | RDH13 (1 of 3) | retinol dehydrogenase 13 (all-trans/9-cis) | 407663 |
| ENSDARG00000002467 | rdh14b | retinol dehydrogenase 14b (all-trans/9-cis/11-cis) | 334665 |
| ENSDARG00000028048 | rdh8a | retinol dehydrogenase 8a | 280648 |
| ENSDARG00000100742 | reep5 | receptor accessory protein 5 | 337237 |
| ENSDARG00000098969 | RELN | reelin | 0 |
| ENSDARG00000074064 | rem2 | RAS (RAD and GEM)-like GTP binding 2 | 798285 |
| ENSDARG00000002877 | reps1 | RALBP1 associated Eps domain containing 1 | 564535 |
| ENSDARG00000070140 | RETSAT (2 of 2) | retinol saturase (all-trans-retinol 13,14-reductase) | 777751 |
| ENSDARG00000018600 | retsat | retinol saturase (all-trans-retinol 13,14-reductase) | 325922 |
| ENSDARG00000098654 | RFTN1 (2 of 2) | raftlin, lipid raft linker 1 | 101885083 |
| ENSDARG00000056078 | rftn2 | raftlin family member 2 | 406729 |
| ENSDARG00000012248 | rgma | repulsive guidance molecule family member a | 414272 |
| ENSDARG00000098645 | rgn | regucalcin | 403070 |
| ENSDARG00000054890 | rgra | retinal G protein coupled receptor a | 550575 |
| ENSDARG00000036774 | rgs12b | regulator of G-protein signaling 12b | 378970 |
| ENSDARG00000039435 | rgs17 | regulator of G-protein signaling 17 | 436796 |
| ENSDARG00000077385 | rgs19 | regulator of G-protein signaling 19 | 100170801 |
| ENSDARG00000038859 | rgs20 | regulator of G-protein signaling 20 | 436704 |
| ENSDARG00000015627 | rgs6 | regulator of G-protein signaling 6 | 562667 |
| ENSDARG00000016584 | rgs7a | regulator of G-protein signaling 7a | 436814 |
| ENSDARG00000096687 | rgs7b | regulator of G-protein signaling 7b | 100535325 |
| ENSDARG00000086756 | RGS9BP | regulator of G protein signaling 9 binding protein | 559232 |
| ENSDARG00000045156 | rgs9b | regulator of G-protein signaling 9b | 793087 |
| ENSDARG00000009018 | rhbg | Rh family, B glycoprotein (gene/pseudogene) | 337596 |
| ENSDARG00000090213 | rheb | Ras homolog enriched in brain | 0 |
| ENSDARG00000026845 | rhoaa | ras homolog gene family, member Aa | 406411 |
| ENSDARG00000090036 | RHOBTB3 | Rho-related BTB domain containing 3 | 562804 |
| ENSDARG00000068653 | rhoga | ras homolog family member Ga | 556424 |
| ENSDARG00000004301 | rhogb | ras homolog family member Gb | 336933 |
| ENSDARG00000028401 | rhogc | ras homolog gene family, member Gc | 324681 |
| ENSDARG00000025953 | rhoq | ras homolog family member Q | 327510 |
| ENSDARG00000099996 | rhot1a | ras homolog family member T1a | 327143 |
| ENSDARG00000103313 | rhot1b | ras homolog family member T1 | 561933 |
| ENSDARG00000056328 | rhot2 | ras homolog family member T2 | 557574 |
| ENSDARG00000007247 | ric8a | RIC8 guanine nucleotide exchange factor A | 449546 |
| ENSDARG00000104101 | RIMBP2 (2 of 2) | RIMS binding protein 2 | 100535071 |
| ENSDARG00000001154 | rimbp2 | RIMS binding protein 2 | 0 |
| ENSDARG00000074680 | rims1a | regulating synaptic membrane exocytosis 1a | 0 |
| ENSDARG00000078902 | rims1b | regulating synaptic membrane exocytosis 1b | 568486 |
| ENSDARG00000101606 | rims2a | regulating synaptic membrane exocytosis 2a | 393672 |
| ENSDARG00000062305 | rims3 | regulating synaptic membrane exocytosis 3 | 563414 |
| ENSDARG00000070100 | RIN1 (1 of 2) | Ras and Rab interactor 1 | 101882011 |
| ENSDARG00000005547 | rint1 | RAD50 interactor 1 | 562894 |
| ENSDARG00000069150 | rit1 | Ras-like without CAAX 1 | 793942 |
| ENSDARG00000070780 | rln3a | relaxin 3a | 503864 |
| ENSDARG00000056639 | rltpr | RGD motif, leucine rich repeats, tropomodulin domain and proline-rich containing | 567758 |
| ENSDARG00000074507 | rmdn1 | regulator of microtubule dynamics 1 | 558162 |
| ENSDARG00000078761 | RMDN2 | regulator of microtubule dynamics 2 | 0 |
| ENSDARG00000041407 | rmdn3 | regulator of microtubule dynamics 3 | 100034410 |
| ENSDARG00000104458 | rnasekb | ribonuclease, RNase K b | 336290 |
| ENSDARG00000010442 | rnf11a | ring finger protein 11a | 393996 |
| ENSDARG00000103551 | rnf213b | ring finger protein 213b | 563001 |
| ENSDARG00000026784 | robo1 | roundabout, axon guidance receptor, homolog 1 (Drosophila) | 30769 |
| ENSDARG00000014891 | robo2 | roundabout, axon guidance receptor, homolog 2 (Drosophila) | 60309 |
| ENSDARG00000005645 | robo3 | roundabout, axon guidance receptor, homolog 3 (Drosophila) | 30770 |
| ENSDARG00000104413 | rogdi | rogdi homolog (Drosophila) | 335642 |
| ENSDARG00000044339 | rp2 | retinitis pigmentosa 2 (X-linked recessive) | 406755 |
| ENSDARG00000007480 | rpe65a | retinal pigment epithelium-specific protein 65a | 393724 |
| ENSDARG00000054420 | rpe65c | retinal pigment epithelium-specific protein 65c | 100004076 |
| ENSDARG00000005251 | rpe | ribulose-5-phosphate-3-epimerase | 334897 |
| ENSDARG00000063436 | RPH3A | rabphilin 3A | 101886567 |
| ENSDARG00000042905 | rpl10a | ribosomal protein L10a | 323305 |
| ENSDARG00000025581 | rpl10 | ribosomal protein L10 | 336712 |
| ENSDARG00000043509 | rpl11 | ribosomal protein L11 | 415229 |
| ENSDARG00000006691 | rpl12 | ribosomal protein L12 | 327089 |
| ENSDARG00000044093 | rpl13a | ribosomal protein L13a | 560828 |
| ENSDARG00000099380 | rpl13 | ribosomal protein L13 | 378961 |
| ENSDARG00000103433 | rpl14 | ribosomal protein L14 | 323365 |
| ENSDARG00000009285 | rpl15 | ribosomal protein L15 | 445053 |
| ENSDARG00000025073 | rpl18a | ribosomal protein L18a | 394035 |
| ENSDARG00000029533 | rpl18 | ribosomal protein L18 | 445038 |
| ENSDARG00000013307 | rpl19 | ribosomal protein L19 | 406489 |
| ENSDARG00000010516 | rpl21 | ribosomal protein L21 | 336568 |
| ENSDARG00000070437 | rpl22 | ribosomal protein L22 | 557397 |
| ENSDARG00000006316 | rpl23a | ribosomal protein L23a | 335539 |
| ENSDARG00000053457 | rpl23 | ribosomal protein L23 | 336812 |
| ENSDARG00000099104 | rpl24 | ribosomal protein L24 | 192301 |
| ENSDARG00000102317 | rpl26 | ribosomal protein L26 | 406387 |
| ENSDARG00000015128 | rpl27 | ribosomal protein L27 | 325618 |
| ENSDARG00000005791 | rpl28 | ribosomal protein L28 | 394036 |
| ENSDARG00000035871 | rpl30 | ribosomal protein L30 | 336713 |
| ENSDARG00000053365 | rpl31 | ribosomal protein L31 | 677758 |
| ENSDARG00000054818 | rpl32 | ribosomal protein L32 | 556556 |
| ENSDARG00000029500 | rpl34 | ribosomal protein L34 | 394097 |
| ENSDARG00000088030 | rpl35a | ribosomal protein L35a | 436760 |
| ENSDARG00000018334 | rpl35 | ribosomal protein L35 | 192299 |
| ENSDARG00000100588 | rpl36 | ribosomal protein L36 | 405888 |
| ENSDARG00000076360 | rpl37 | ribosomal protein L37 | 0 |
| ENSDARG00000006413 | rpl38 | ribosomal protein L38 | 436759 |
| ENSDARG00000003599 | rpl3 | ribosomal protein L3 | 322571 |
| ENSDARG00000041182 | rpl4 | ribosomal protein L4 | 337090 |
| ENSDARG00000015862 | rpl5b | ribosomal protein L5b | 415196 |
| ENSDARG00000058451 | rpl6 | ribosomal protein L6 | 336727 |
| ENSDARG00000019230 | rpl7a | ribosomal protein L7a | 337010 |
| ENSDARG00000007320 | rpl7 | ribosomal protein L7 | 336710 |
| ENSDARG00000014867 | rpl8 | ribosomal protein L8 | 393686 |
| ENSDARG00000037350 | rpl9 | ribosomal protein L9 | 336702 |
| ENSDARG00000051783 | rplp0 | ribosomal protein, large, P0 | 58101 |
| ENSDARG00000021864 | rplp1 | ribosomal protein, large, P1 | 336719 |
| ENSDARG00000011201 | rplp2l | ribosomal protein, large P2, like | 558312 |
| ENSDARG00000101406 | rplp2 | ribosomal protein, large P2 | 335629 |
| ENSDARG00000059323 | rpn1 | ribophorin I | 325561 |
| ENSDARG00000009724 | rpn2 | ribophorin II | 335985 |
| ENSDARG00000034897 | rps10 | ribosomal protein S10 | 394121 |
| ENSDARG00000053058 | rps11 | ribosomal protein S11 | 406686 |
| ENSDARG00000036875 | rps12 | ribosomal protein S12 | 337007 |
| ENSDARG00000036298 | rps13 | ribosomal protein S13 | 415169 |
| ENSDARG00000036629 | rps14 | ribosomal protein S14 | 336687 |
| ENSDARG00000010160 | rps15a | ribosomal protein S15a | 336764 |
| ENSDARG00000070849 | rps15 | ribosomal protein S15 | 337148 |
| ENSDARG00000045487 | rps16 | ribosomal protein S16 | 767765 |
| ENSDARG00000046157 | RPS17 | ribosomal protein S17 | 541327 |
| ENSDARG00000100392 | rps18 | ribosomal protein S18 | 192300 |
| ENSDARG00000030602 | rps19 | ribosomal protein S19 | 393723 |
| ENSDARG00000025850 | rps21 | ribosomal protein S21 | 394166 |
| ENSDARG00000021838 | rps23 | ribosomal protein S23 | 100000341 |
| ENSDARG00000039347 | rps24 | ribosomal protein S24 | 394194 |
| ENSDARG00000041811 | rps25 | ribosomal protein S25 | 393788 |
| ENSDARG00000037071 | rps26 | ribosomal protein S26 | 336662 |
| ENSDARG00000023298 | rps27.1 | ribosomal protein S27, isoform 1 | 554155 |
| ENSDARG00000100513 | rps27.2 | ribosomal protein S27, isoform 2 | 677743 |
| ENSDARG00000032725 | rps27a | ribosomal protein S27a | 337810 |
| ENSDARG00000035860 | rps28 | ribosomal protein S28 | 406307 |
| ENSDARG00000041232 | rps29 | ribosomal protein S29 | 405889 |
| ENSDARG00000077291 | rps2 | ribosomal protein S2 | 406565 |
| ENSDARG00000035692 | rps3a | ribosomal protein S3A | 337240 |
| ENSDARG00000103007 | rps3 | ribosomal protein S3 | 336550 |
| ENSDARG00000014690 | rps4x | ribosomal protein S4, X-linked | 449547 |
| ENSDARG00000043453 | rps5 | ribosomal protein S5 | 192297 |
| ENSDARG00000033437 | rps6ka1 | ribosomal protein S6 kinase a, polypeptide 1 | 0 |
| ENSDARG00000019778 | rps6 | ribosomal protein S6 | 445028 |
| ENSDARG00000042566 | rps7 | ribosomal protein S7 | 393725 |
| ENSDARG00000055996 | rps8a | ribosomal protein S8a | 407690 |
| ENSDARG00000057368 | rps8b | ribosomal protein S8b | 100000836 |
| ENSDARG00000011405 | rps9 | ribosomal protein S9 | 114420 |
| ENSDARG00000019181 | rpsa | ribosomal protein SA | 394027 |
| ENSDARG00000069728 | rpz3 | rapunzel 3 | 100001029 |
| ENSDARG00000039871 | rpz4 | rapunzel 4 | 550609 |
| ENSDARG00000075718 | rpz5 | rapunzel 5 | 100003142 |
| ENSDARG00000091161 | rpz | rapunzel | 100002737 |
| ENSDARG00000036252 | rras2 | related RAS viral (r-ras) oncogene homolog 2 | 550513 |
| ENSDARG00000006553 | rras | related RAS viral (r-ras) oncogene homolog | 449660 |
| ENSDARG00000013763 | RRBP1 (1 of 2) | ribosome binding protein 1 | 553448 |
| ENSDARG00000041703 | rrbp1b | ribosome binding protein 1 homolog b (dog) | 0 |
| ENSDARG00000027236 | rs1a | retinoschisin 1a | 445044 |
| ENSDARG00000101047 | rtcb | RNA 2',3'-cyclic phosphate and 5'-OH ligase | 406376 |
| ENSDARG00000006497 | rtn1a | reticulon 1a | 323706 |
| ENSDARG00000021143 | rtn1b | reticulon 1b | 327065 |
| ENSDARG00000091348 | rtn3 | reticulon 3 | 394047 |
| ENSDARG00000044601 | rtn4a | reticulon 4a | 447816 |
| ENSDARG00000031366 | rtn4ip1 | reticulon 4 interacting protein 1 | 393323 |
| ENSDARG00000029692 | rufy3 | RUN and FYVE domain containing 3 | 541522 |
| ENSDARG00000078125 | rusc1 | RUN and SH3 domain containing 1 | 558727 |
| ENSDARG00000002591 | ruvbl1 | RuvB-like AAA ATPase 1 | 317679 |
| ENSDARG00000055639 | ruvbl2 | RuvB-like AAA ATPase 2 | 317678 |
| ENSDARG00000023797 | ryr1b | ryanodine receptor 1b (skeletal) | 570245 |
| ENSDARG00000071331 | ryr3 | ryanodine receptor 3 | 561350 |
| ENSDARG00000025254 | s100a10b | S100 calcium binding protein A10b | 406276 |
| ENSDARG00000057598 | s100b | S100 calcium binding protein, beta (neural) | 436825 |
| ENSDARG00000071014 | s100u | S100 calcium binding protein U | 563060 |
| ENSDARG00000070702 | s100v2 | S100 calcium binding protein V2 | 336965 |
| ENSDARG00000042690 | s1pr1 | sphingosine-1-phosphate receptor 1 | 64617 |
| ENSDARG00000059806 | sacm1lb | SAC1 suppressor of actin mutations 1-like b (yeast) | 791142 |
| ENSDARG00000091042 | sacs | sacsin molecular chaperone | 558150 |
| ENSDARG00000039681 | samm50l | sorting and assembly machinery component 50 homolog, like | 492471 |
| ENSDARG00000045814 | samm50 | SAMM50 sorting and assembly machinery component | 393111 |
| ENSDARG00000103403 | sar1b | secretion associated, Ras related GTPase 1B | 554177 |
| ENSDARG00000058102 | sardh | sarcosine dehydrogenase | 394103 |
| ENSDARG00000010610 | sarm1 | sterile alpha and TIR motif containing 1 | 403143 |
| ENSDARG00000008237 | sars | seryl-tRNA synthetase | 445405 |
| ENSDARG00000062968 | sbf1 | SET binding factor 1 | 568277 |
| ENSDARG00000060865 | scai | suppressor of cancer cell invasion | 556383 |
| ENSDARG00000036911 | scamp1 | secretory carrier membrane protein 1 | 402992 |
| ENSDARG00000010279 | scamp2 | secretory carrier membrane protein 2 | 406687 |
| ENSDARG00000016765 | scamp3 | secretory carrier membrane protein 3 | 492473 |
| ENSDARG00000010531 | scamp4 | secretory carrier membrane protein 4 | 562881 |
| ENSDARG00000018743 | scamp5a | secretory carrier membrane protein 5a | 324467 |
| ENSDARG00000098312 | scarb2 | scavenger receptor class B, member 2 | 192340 |
| ENSDARG00000024564 | sccpdhb | saccharopine dehydrogenase b | 407665 |
| ENSDARG00000079645 | sc:d217 | sc:d217 | 0 |
| ENSDARG00000000779 | scfd1 | sec1 family domain containing 1 | 317732 |
| ENSDARG00000038574 | scg2b | secretogranin II (chromogranin C), b | 777641 |
| ENSDARG00000077045 | scg5 | secretogranin V | 393699 |
| ENSDARG00000102415 | scinla | scinderin like a | 323285 |
| ENSDARG00000058348 | scinlb | scinderin like b | 406363 |
| ENSDARG00000010728 | scin | scinderin | 566764 |
| ENSDARG00000105003 | scn1bb | sodium channel, voltage-gated, type I, beta b | 100005325 |
| ENSDARG00000062744 | scn1lab | sodium channel, voltage-gated, type I like, alpha b | 559447 |
| ENSDARG00000101713 | scn2b | sodium channel, voltage-gated, type II, beta | 777666 |
| ENSDARG00000099031 | scn4ba | sodium channel, voltage-gated, type IV, beta a | 566069 |
| ENSDARG00000005775 | scn8aa | sodium channel, voltage gated, type VIII, alpha subunit a | 58152 |
| ENSDARG00000018032 | scn8ab | sodium channel, voltage gated, type VIII, alpha subunit b | 793996 |
| ENSDARG00000067593 | SCO1 | SCO1 cytochrome c oxidase assembly protein | 796572 |
| ENSDARG00000045555 | sco2 | SCO2 cytochrome c oxidase assembly protein | 606683 |
| ENSDARG00000012194 | scp2a | sterol carrier protein 2a | 393839 |
| ENSDARG00000102356 | scp2b | sterol carrier protein 2b | 450073 |
| ENSDARG00000000861 | scrib | scribbled planar cell polarity protein | 368473 |
| ENSDARG00000043497 | scrn2 | secernin 2 | 558306 |
| ENSDARG00000016611 | scrn3 | secernin 3 | 326058 |
| ENSDARG00000100562 | sdc3 | syndecan 3 | 337077 |
| ENSDARG00000012513 | sdcbp2 | syndecan binding protein (syntenin) 2 | 325004 |
| ENSDARG00000030097 | sdcbp | syndecan binding protein (syntenin) | 407719 |
| ENSDARG00000035631 | sdf2l1 | stromal cell-derived factor 2-like 1 | 445275 |
| ENSDARG00000070682 | sdf4 | stromal cell derived factor 4 | 406360 |
| ENSDARG00000016721 | sdha | succinate dehydrogenase complex, subunit A, flavoprotein (Fp) | 393884 |
| ENSDARG00000075768 | sdhb | succinate dehydrogenase complex, subunit B, iron sulfur (Ip) | 562149 |
| ENSDARG00000038608 | sdhc | succinate dehydrogenase complex, subunit C, integral membrane protein | 445129 |
| ENSDARG00000078866 | sdk1a | sidekick cell adhesion molecule 1a | 558391 |
| ENSDARG00000062854 | sdk1b | sidekick cell adhesion molecule 1b | 555553 |
| ENSDARG00000060452 | sdk2b | sidekick cell adhesion molecule 2b | 564922 |
| ENSDARG00000099617 | sdr39u1 | short chain dehydrogenase/reductase family 39U, member 1 | 767637 |
| ENSDARG00000008936 | sec11a | SEC11 homolog A (S. cerevisiae) | 436794 |
| ENSDARG00000078328 | sec22a | SEC22 vesicle trafficking protein homolog A (S. cerevisiae) | 324832 |
| ENSDARG00000030108 | sec22ba | SEC22 vesicle trafficking protein homolog B (S. cerevisiae), a | 114464 |
| ENSDARG00000100435 | sec22bb | SEC22 vesicle trafficking protein homolog B (S. cerevisiae), b | 327277 |
| ENSDARG00000104230 | sec23a | Sec23 homolog A (S. cerevisiae) | 406774 |
| ENSDARG00000061413 | sec23ip | SEC23 interacting protein | 560750 |
| ENSDARG00000104375 | sec24a | SEC24 family, member A (S. cerevisiae) | 100000526 |
| ENSDARG00000071906 | sec24b | SEC24 family member B | 334633 |
| ENSDARG00000103516 | sec24c | SEC24 family, member C (S. cerevisiae) | 571870 |
| ENSDARG00000021669 | sec61a1 | Sec61 alpha 1 subunit (S. cerevisiae) | 192298 |
| ENSDARG00000076568 | sec61b | Sec61 beta subunit | 791986 |
| ENSDARG00000018637 | sec61g | Sec61 gamma subunit | 436772 |
| ENSDARG00000019951 | sec62 | SEC62 homolog (S. cerevisiae) | 563660 |
| ENSDARG00000017740 | sec63 | SEC63 homolog (S. cerevisiae) | 436861 |
| ENSDARG00000004581 | sel1l | sel-1 suppressor of lin-12-like (C. elegans) | 568615 |
| ENSDARG00000024717 | selenbp1 | selenium binding protein 1 | 393542 |
| ENSDARG00000100823 | selk | selenoprotein K | 791738 |
| ENSDARG00000058354 | selt1a | selenoprotein T, 1a | 352919 |
| ENSDARG00000023220 | selt2 | selenoprotein T, 2 | 0 |
| ENSDARG00000079611 | sema4c | sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4C | 561895 |
| ENSDARG00000036626 | sema6ba | sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6Ba | 0 |
| ENSDARG00000099664 | sep15 | selenoprotein 15 | 352923 |
| ENSDARG00000099236 | sept10 | septin 10 | 550129 |
| ENSDARG00000019191 | sept12 | septin 12 | 564600 |
| ENSDARG00000102889 | sept15 | septin 15 | 792259 |
| ENSDARG00000027590 | sept2 | septin 2 | 323457 |
| ENSDARG00000030656 | sept3 | septin 3 | 554123 |
| ENSDARG00000013843 | sept5a | septin 5a | 0 |
| ENSDARG00000036031 | sept5b | septin 5b | 445325 |
| ENSDARG00000010721 | sept6 | septin 6 | 322694 |
| ENSDARG00000087647 | sept7a | septin 7a | 0 |
| ENSDARG00000052673 | sept7b | septin 7b | 100000597 |
| ENSDARG00000032606 | sept8a | septin 8a | 321590 |
| ENSDARG00000014233 | sept8b | septin 8b | 571702 |
| ENSDARG00000103900 | sept9b | septin 9b | 541494 |
| ENSDARG00000038228 | sepw2a | selenoprotein W, 2a | 378437 |
| ENSDARG00000074242 | serbp1a | SERPINE1 mRNA binding protein 1a | 335449 |
| ENSDARG00000032340 | serhl | serine hydrolase-like | 322648 |
| ENSDARG00000005924 | serpina10a | serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 10a | 565064 |
| ENSDARG00000090850 | serpina1l | serine (or cysteine) proteinase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1, like | 321195 |
| ENSDARG00000090286 | serpina1 | serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1 | 322701 |
| ENSDARG00000091801 | serpinb14 | serpin peptidase inhibitor, clade B (ovalbumin), member 14 | 569051 |
| ENSDARG00000040614 | sestd1 | SEC14 and spectrin domains 1 | 360133 |
| ENSDARG00000031495 | seta | SET translocation (myeloid leukemia-associated) A | 323501 |
| ENSDARG00000076052 | sez6l2 | seizure related 6 homolog (mouse)-like 2 | 557847 |
| ENSDARG00000100594 | sez6l | seizure related 6 homolog (mouse)-like | 0 |
| ENSDARG00000011564 | sfpq | splicing factor proline/glutamine-rich | 406564 |
| ENSDARG00000019963 | sfxn1 | sideroflexin 1 | 445143 |
| ENSDARG00000057374 | sfxn3 | sideroflexin 3 | 791182 |
| ENSDARG00000069832 | sfxn4 | sideroflexin 4 | 556121 |
| ENSDARG00000026137 | sfxn5b | sideroflexin 5b | 561995 |
| ENSDARG00000097897 | sgip1a | SH3-domain GRB2-like (endophilin) interacting protein 1a | 566542 |
| ENSDARG00000075376 | sgsm1b | small G protein signaling modulator 1b | 100535556 |
| ENSDARG00000036878 | sh3bgrl2 | SH3 domain binding glutamate-rich protein like 2 | 431744 |
| ENSDARG00000098075 | sh3bgrl3 | SH3 domain binding glutamate-rich protein like 3 | 393248 |
| ENSDARG00000058302 | sh3bgrl | SH3 domain binding glutamate-rich protein like | 394184 |
| ENSDARG00000007136 | sh3bp5lb | SH3-binding domain protein 5-like, b | 558670 |
| ENSDARG00000002642 | sh3gl1b | SH3-domain GRB2-like 1b | 378838 |
| ENSDARG00000039901 | SH3GL2 (2 of 2) | SH3-domain GRB2-like 2 | 0 |
| ENSDARG00000023600 | sh3gl2 | SH3-domain GRB2-like 2 | 394091 |
| ENSDARG00000013360 | sh3gl3a | SH3-domain GRB2-like 3a | 571626 |
| ENSDARG00000045294 | sh3glb1a | SH3-domain GRB2-like endophilin B1a | 792229 |
| ENSDARG00000008983 | sh3glb2a | SH3-domain GRB2-like endophilin B2a | 402885 |
| ENSDARG00000035470 | sh3glb2b | SH3-domain GRB2-like endophilin B2b | 394094 |
| ENSDARG00000075853 | sh3kbp1 | SH3-domain kinase binding protein 1 | 793623 |
| ENSDARG00000060539 | shank1 | SH3 and multiple ankyrin repeat domains 1 | 568908 |
| ENSDARG00000062325 | shank2 | SH3 and multiple ankyrin repeat domains 2 | 567595 |
| ENSDARG00000063332 | shank3a | SH3 and multiple ankyrin repeat domains 3a | 0 |
| ENSDARG00000062462 | shisa7a | shisa family member 7a | 0 |
| ENSDARG00000063144 | shisa7b | shisa family member 7 | 100034520 |
| ENSDARG00000052642 | shisa9b | shisa family member 9b | 566015 |
| ENSDARG00000104414 | shmt2 | serine hydroxymethyltransferase 2 (mitochondrial) | 100144628 |
| ENSDARG00000040853 | shoc2 | soc-2 suppressor of clear homolog (C. elegans) | 555476 |
| ENSDARG00000101804 | si:ch1073-140o9.2 | si:ch1073-140o9.2 | 0 |
| ENSDARG00000079651 | si:ch1073-174d20.2 | si:ch1073-174d20.2 | 570430 |
| ENSDARG00000088247 | si:ch1073-303k11.2 | si:ch1073-303k11.2 | 100333296 |
| ENSDARG00000102780 | si:ch1073-370j18.1 | si:ch1073-370j18.1 | 0 |
| ENSDARG00000070442 | si:ch211-113g11.6 | si:ch211-113g11.6 | 559008 |
| ENSDARG00000073764 | si:ch211-113j14.1 | si:ch211-113j14.1 | 558239 |
| ENSDARG00000092970 | si:ch211-11c15.3 | si:ch211-11c15.3 | 566329 |
| ENSDARG00000098644 | si:ch211-126j24.1 | si:ch211-126j24.1 | 571137 |
| ENSDARG00000092976 | si:ch211-127i16.2 | si:ch211-127i16.2 | 558050 |
| ENSDARG00000089477 | si:ch211-132g1.3 | si:ch211-132g1.3 | 103911757 |
| ENSDARG00000078748 | si:ch211-137a8.4 | si:ch211-137a8.4 | 560240 |
| ENSDARG00000088882 | si:ch211-149b19.2 | si:ch211-149b19.2 | 101884675 |
| ENSDARG00000092746 | si:ch211-150o23.2 | si:ch211-150o23.2 | 0 |
| ENSDARG00000097187 | si:ch211-153l6.6 | si:ch211-153l6.6 | 101884986 |
| ENSDARG00000056379 | si:ch211-154o6.6 | si:ch211-154o6.6 | 564061 |
| ENSDARG00000076970 | si:ch211-165b10.3 | si:ch211-165b10.3 | 100334077 |
| ENSDARG00000087857 | si:ch211-180a12.2 | si:ch211-180a12.2 | 100150385 |
| ENSDARG00000070757 | si:ch211-182e10.4 | si:ch211-182e10.4 | 553308 |
| ENSDARG00000078088 | si:ch211-186j3.6 | si:ch211-186j3.6 | 556807 |
| ENSDARG00000088276 | si:ch211-190p8.2 | si:ch211-190p8.2 | 436997 |
| ENSDARG00000095459 | si:ch211-191j22.3 | si:ch211-191j22.3 | 0 |
| ENSDARG00000035660 | si:ch211-193k19.1 | si:ch211-193k19.1 | 100034533 |
| ENSDARG00000100894 | si:ch211-193k8.5 | si:ch211-193k8.5 | 0 |
| ENSDARG00000037867 | si:ch211-194i10.5 | si:ch211-194i10.5 | 402883 |
| ENSDARG00000077087 | si:ch211-196g2.4 | si:ch211-196g2.4 | 0 |
| ENSDARG00000054400 | si:ch211-198n5.11 | si:ch211-198n5.11 | 100004018 |
| ENSDARG00000063361 | si:ch211-1e14.1 | si:ch211-1e14.1 | 556850 |
| ENSDARG00000029170 | si:ch211-200p22.4 | si:ch211-200p22.4 | 565799 |
| ENSDARG00000092856 | si:ch211-210c8.7 | si:ch211-210c8.7 | 0 |
| ENSDARG00000069595 | si:ch211-214c7.4 | si:ch211-214c7.4 | 560699 |
| ENSDARG00000007976 | si:ch211-220f16.2 | si:ch211-220f16.2 | 797650 |
| ENSDARG00000062330 | si:ch211-233a24.2 | si:ch211-233a24.2 | 564367 |
| ENSDARG00000071353 | si:ch211-235e9.8 | si:ch211-235e9.8 | 100002196 |
| ENSDARG00000098585 | si:ch211-237a4.2 | si:ch211-237a4.2 | 0 |
| ENSDARG00000007275 | si:ch211-251b21.1 | si:ch211-251b21.1 | 571720 |
| ENSDARG00000030945 | si:ch211-259g3.4 | si:ch211-259g3.4 | 0 |
| ENSDARG00000059333 | si:ch211-285f17.1 | si:ch211-285f17.1 | 562465 |
| ENSDARG00000096661 | si:ch211-286b5.5 | si:ch211-286b5.5 | 100003596 |
| ENSDARG00000089255 | si:ch211-286c4.6 | si:ch211-286c4.6 | 100003678 |
| ENSDARG00000097855 | si:ch211-3o3.9 | si:ch211-3o3.9 | 101886596 |
| ENSDARG00000069735 | si:ch211-5k11.6 | si:ch211-5k11.6 | 497166 |
| ENSDARG00000078824 | si:ch211-66e2.3 | si:ch211-66e2.3 | 100141487 |
| ENSDARG00000076144 | si:ch211-74m13.3 | si:ch211-74m13.3 | 100334824 |
| ENSDARG00000096346 | si:ch211-85n16.3 | si:ch211-85n16.3 | 0 |
| ENSDARG00000074282 | si:ch73-12o23.1 | si:ch73-12o23.1 | 100332784 |
| ENSDARG00000087364 | si:ch73-140j24.4 | si:ch73-140j24.4 | 101885000 |
| ENSDARG00000077468 | si:ch73-206p6.1 | si:ch73-206p6.1 | 571998 |
| ENSDARG00000086746 | si:ch73-209e20.3 | si:ch73-209e20.3 | 100333321 |
| ENSDARG00000062831 | si:ch73-22o12.1 | si:ch73-22o12.1 | 560816 |
| ENSDARG00000091269 | si:ch73-27e22.8 | si:ch73-27e22.8 | 553357 |
| ENSDARG00000036383 | si:ch73-335m24.5 | si:ch73-335m24.5 | 561209 |
| ENSDARG00000023940 | si:ch73-362m14.4 | si:ch73-362m14.4 | 555437 |
| ENSDARG00000100480 | si:ch73-364h19.2 | si:ch73-364h19.2 | 100537438 |
| ENSDARG00000098465 | si:ch73-366l1.5 | si:ch73-366l1.5 | 100003714 |
| ENSDARG00000057826 | si:ch73-61d6.3 | si:ch73-61d6.3 | 100001136 |
| ENSDARG00000090149 | si:dkey-108k21.18 | si:dkey-108k21.18 | 560380 |
| ENSDARG00000099799 | si:dkey-11f4.16 | si:dkey-11f4.16 | 0 |
| ENSDARG00000004386 | si:dkey-153k10.9 | si:dkey-153k10.9 | 566596 |
| ENSDARG00000037967 | si:dkey-16l2.16 | si:dkey-16l2.16 | 100003902 |
| ENSDARG00000080000 | si:dkey-178e17.1 | si:dkey-178e17.1 | 793282 |
| ENSDARG00000052680 | si:dkey-182g1.2 | si:dkey-182g1.2 | 100005482 |
| ENSDARG00000033413 | si:dkey-183c16.7 | si:dkey-183c16.7 | 556673 |
| ENSDARG00000070511 | si:dkey-183j2.10 | si:dkey-183j2.10 | 100151589 |
| ENSDARG00000040463 | si:dkey-185e18.6 | si:dkey-185e18.6 | 556422 |
| ENSDARG00000056080 | si:dkey-191g9.5 | si:dkey-191g9.5 | 100148873 |
| ENSDARG00000089886 | si:dkey-19e4.5 | si:dkey-19e4.5 | 570260 |
| ENSDARG00000104065 | si:dkey-1b17.11 | si:dkey-1b17.11 | 100006734 |
| ENSDARG00000094695 | si:dkey-204f11.64 | si:dkey-204f11.64 | 100537752 |
| ENSDARG00000096701 | si:dkey-21e13.3 | si:dkey-21e13.3 | 100150043 |
| ENSDARG00000095005 | si:dkey-21e2.10 | si:dkey-21e2.10 | 556860 |
| ENSDARG00000074894 | si:dkey-228b2.5 | si:dkey-228b2.5 | 100537539 |
| ENSDARG00000069537 | si:dkey-238c7.16 | si:dkey-238c7.16 | 559078 |
| ENSDARG00000078847 | si:dkey-238o13.4 | si:dkey-238o13.4 | 560780 |
| ENSDARG00000043518 | si:dkey-239i20.2 | si:dkey-239i20.2 | 557525 |
| ENSDARG00000043514 | si:dkey-239i20.4 | si:dkey-239i20.4 | 557628 |
| ENSDARG00000095848 | si:dkey-239n17.6 | si:dkey-239n17.6 | 0 |
| ENSDARG00000061719 | si:dkey-23p11.2 | si:dkey-23p11.2 | 565752 |
| ENSDARG00000034210 | si:dkey-240h12.4 | si:dkey-240h12.4 | 555536 |
| ENSDARG00000092161 | si:dkey-261e22.4 | si:dkey-261e22.4 | 0 |
| ENSDARG00000093195 | si:dkey-261m9.6 | si:dkey-261m9.6 | 563569 |
| ENSDARG00000073752 | si:dkey-266j7.2 | si:dkey-266j7.2 | 0 |
| ENSDARG00000090872 | si:dkey-276j7.1 | si:dkey-276j7.1 | 561769 |
| ENSDARG00000003146 | si:dkey-27m7.4 | si:dkey-27m7.4 | 0 |
| ENSDARG00000002840 | si:dkey-28b4.8 | si:dkey-28b4.8 | 562640 |
| ENSDARG00000099614 | si:dkey-29k15.2 | si:dkey-29k15.2 | 557266 |
| ENSDARG00000063159 | si:dkey-32e23.4 | si:dkey-32e23.4 | 566871 |
| ENSDARG00000002956 | si:dkey-32n7.4 | si:dkey-32n7.4 | 101882585 |
| ENSDARG00000057881 | si:dkey-33c12.3 | si:dkey-33c12.3 | 335380 |
| ENSDARG00000097110 | si:dkey-56f14.4 | si:dkey-56f14.4 | 100005476 |
| ENSDARG00000092039 | si:dkey-70p6.1 | si:dkey-70p6.1 | 570575 |
| ENSDARG00000097528 | si:dkey-7j14.5 | si:dkey-7j14.5 | 556563 |
| ENSDARG00000057262 | si:dkey-91i10.3 | si:dkey-91i10.3 | 565876 |
| ENSDARG00000096622 | si:dkey-9c18.4 | si:dkey-9c18.4 | 0 |
| ENSDARG00000011602 | si:dkeyp-117h8.2 | si:dkeyp-117h8.2 | 570321 |
| ENSDARG00000063008 | si:dkeyp-27e10.3 | si:dkeyp-27e10.3 | 0 |
| ENSDARG00000070516 | si:dkeyp-52c3.2 | si:dkeyp-52c3.2 | 436616 |
| ENSDARG00000087165 | si:dkeyp-69c1.7 | si:dkeyp-69c1.7 | 566978 |
| ENSDARG00000055731 | si:dkeyp-69e1.8 | si:dkeyp-69e1.8 | 100001340 |
| ENSDARG00000058248 | si:dkeyp-77h1.4 | si:dkeyp-77h1.4 | 497302 |
| ENSDARG00000011418 | sigmar1 | sigma non-opioid intracellular receptor 1 | 393952 |
| ENSDARG00000020134 | sipa1l1 | signal-induced proliferation-associated 1 like 1 | 562521 |
| ENSDARG00000076697 | SIPA1 | signal-induced proliferation-associated 1 | 555294 |
| ENSDARG00000071662 | si:rp71-36a1.3 | si:rp71-36a1.3 | 100034563 |
| ENSDARG00000035679 | si:rp71-45k5.4 | si:rp71-45k5.4 | 403015 |
| ENSDARG00000011488 | sirt2 | sirtuin 2 (silent mating type information regulation 2, homolog) 2 (S. cerevisiae) | 322309 |
| ENSDARG00000079078 | si:xx-by187g17.1 | si:xx-by187g17.1 | 497162 |
| ENSDARG00000003151 | skp1 | S-phase kinase-associated protein 1 | 393716 |
| ENSDARG00000086870 | slc12a2 | solute carrier family 12 (sodium/potassium/chloride transporter), member 2 | 0 |
| ENSDARG00000014378 | slc12a4 | solute carrier family 12 (potassium/chloride transporter), member 4 | 562826 |
| ENSDARG00000075815 | SLC12A5 (1 of 2) | solute carrier family 12 (potassium/chloride transporter), member 5 | 572215 |
| ENSDARG00000078187 | slc12a5b | solute carrier family 12 (potassium/chloride transporter), member 5b | 797331 |
| ENSDARG00000053853 | slc13a2 | solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 2 | 406537 |
| ENSDARG00000102793 | slc16a6a | solute carrier family 16, member 6a | 564872 |
| ENSDARG00000001127 | slc17a6a | solute carrier family 17 (vesicular glutamate transporter), member 6a | 494492 |
| ENSDARG00000041150 | slc17a6b | solute carrier family 17 (vesicular glutamate transporter), member 6b | 100149756 |
| ENSDARG00000016480 | slc17a7a | solute carrier family 17 (vesicular glutamate transporter), member 7a | 795293 |
| ENSDARG00000090106 | slc17a7b | solute carrier family 17 (vesicular glutamate transporter), member 7b | 100331980 |
| ENSDARG00000006356 | slc18a3a | solute carrier family 18 (vesicular acetylcholine transporter), member 3a | 559347 |
| ENSDARG00000033993 | SLC19A1 | solute carrier family 19 (folate transporter), member 1 | 497434 |
| ENSDARG00000102453 | slc1a2b | solute carrier family 1 (glial high affinity glutamate transporter), member 2b | 335836 |
| ENSDARG00000104431 | slc1a3a | solute carrier family 1 (glial high affinity glutamate transporter), member 3a | 323439 |
| ENSDARG00000043148 | slc1a3b | solute carrier family 1 (glial high affinity glutamate transporter), member 3b | 556181 |
| ENSDARG00000000551 | slc1a4 | solute carrier family 1 (glutamate/neutral amino acid transporter), member 4 | 368885 |
| ENSDARG00000010096 | slc1a6 | solute carrier family 1 (high affinity aspartate/glutamate transporter), member 6 | 559270 |
| ENSDARG00000060796 | slc20a2 | solute carrier family 20 (phosphate transporter), member 2 | 558346 |
| ENSDARG00000059166 | SLC22A13 | solute carrier family 22 (organic anion/urate transporter), member 13 | 556601 |
| ENSDARG00000078856 | slc22a23 | solute carrier family 22, member 23 | 555979 |
| ENSDARG00000056643 | slc22a7b.1 | solute carrier family 22 (organic anion transporter), member 7b, tandem duplicate 1 | 393320 |
| ENSDARG00000063158 | SLC24A2 (2 of 2) | solute carrier family 24 (sodium/potassium/calcium exchanger), member 2 | 0 |
| ENSDARG00000015425 | slc24a4a | solute carrier family 24 (sodium/potassium/calcium exchanger), member 4a | 550467 |
| ENSDARG00000067509 | slc24a4b | solute carrier family 24 (sodium/potassium/calcium exchanger), member 4b | 100334346 |
| ENSDARG00000035741 | slc25a11 | solute carrier family 25 (mitochondrial carrier; oxoglutarate carrier), member 11 | 415189 |
| ENSDARG00000102362 | slc25a12 | solute carrier family 25 (aspartate/glutamate carrier), member 12 | 337675 |
| ENSDARG00000099353 | slc25a13 | solute carrier family 25 (aspartate/glutamate carrier), member 13 | 0 |
| ENSDARG00000026680 | slc25a14 | solute carrier family 25 (mitochondrial carrier, brain), member 14 | 393133 |
| ENSDARG00000057287 | slc25a16 | solute carrier family 25 (mitochondrial carrier; Graves disease autoantigen), member 16 | 324578 |
| ENSDARG00000070717 | slc25a18 | solute carrier family 25 (glutamate carrier), member 18 | 568740 |
| ENSDARG00000076381 | slc25a1b | slc25a1 solute carrier family 25 (mitochondrial carrier; citrate transporter), member 1b | 795332 |
| ENSDARG00000040401 | slc25a20 | solute carrier family 25 (carnitine/acylcarnitine translocase), member 20 | 393833 |
| ENSDARG00000020718 | SLC25A22 (1 of 2) | solute carrier family 25 (mitochondrial carrier: glutamate), member 22 | 571044 |
| ENSDARG00000005690 | slc25a23a | solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 23a | 561939 |
| ENSDARG00000008568 | slc25a24 | solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 24 | 447867 |
| ENSDARG00000035468 | slc25a25b | solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25b | 569227 |
| ENSDARG00000058208 | slc25a26 | solute carrier family 25 (S-adenosylmethionine carrier), member 26 | 560478 |
| ENSDARG00000026835 | slc25a32b | solute carrier family 25 (mitochondrial folate carrier), member 32b | 503758 |
| ENSDARG00000038731 | slc25a36a | solute carrier family 25 (pyrimidine nucleotide carrier ), member 36a | 436940 |
| ENSDARG00000074533 | slc25a38b | solute carrier family 25, member 38b | 571961 |
| ENSDARG00000025566 | slc25a3b | solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 3b | 322362 |
| ENSDARG00000015856 | slc25a40 | solute carrier family 25, member 40 | 436633 |
| ENSDARG00000099368 | slc25a42 | solute carrier family 25, member 42 | 751743 |
| ENSDARG00000035181 | slc25a46 | solute carrier family 25, member 46 | 436831 |
| ENSDARG00000027355 | slc25a4 | solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 4 | 327067 |
| ENSDARG00000056163 | slc25a51a | solute carrier family 25, member 51a | 393728 |
| ENSDARG00000092553 | slc25a5 | solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 5 | 192321 |
| ENSDARG00000005853 | slc25a6 | solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 6 | 566370 |
| ENSDARG00000006240 | slc27a1a | solute carrier family 27 (fatty acid transporter), member 1a | 541410 |
| ENSDARG00000038352 | slc27a1b | solute carrier family 27 (fatty acid transporter), member 1b | 0 |
| ENSDARG00000100731 | slc27a2b | solute carrier family 27 (fatty acid transporter), member 2b | 100004228 |
| ENSDARG00000046054 | SLC27A3 | solute carrier family 27 (fatty acid transporter), member 3 | 568883 |
| ENSDARG00000017047 | slc27a4 | solute carrier family 27 (fatty acid transporter), member 4 | 550432 |
| ENSDARG00000101289 | slc29a1a | solute carrier family 29 (equilibrative nucleoside transporter), member 1a | 563580 |
| ENSDARG00000001437 | slc2a1a | solute carrier family 2 (facilitated glucose transporter), member 1a | 555778 |
| ENSDARG00000007412 | slc2a1b | solute carrier family 2 (facilitated glucose transporter), member 1b | 100321338 |
| ENSDARG00000013295 | slc2a3a | solute carrier family 2 (facilitated glucose transporter), member 3a | 436916 |
| ENSDARG00000057272 | slc30a9 | solute carrier family 30 (zinc transporter), member 9 | 497184 |
| ENSDARG00000013961 | slc31a1 | solute carrier family 31 (copper transporter), member 1 | 403028 |
| ENSDARG00000059775 | slc32a1 | solute carrier family 32 (GABA vesicular transporter), member 1 | 798575 |
| ENSDARG00000011945 | slc35e1 | solute carrier family 35, member E1 | 406347 |
| ENSDARG00000053003 | slc35f1 | solute carrier family 35, member F1 | 555352 |
| ENSDARG00000016745 | slc35f6 | solute carrier family 35, member F6 | 436832 |
| ENSDARG00000056262 | slc35g2a | solute carrier family 35, member G2a | 492818 |
| ENSDARG00000045447 | slc35g2b | solute carrier family 35, member G2b | 541438 |
| ENSDARG00000027065 | slc38a3a | solute carrier family 38, member 3a | 100073328 |
| ENSDARG00000102387 | SLC39A14 | solute carrier family 39 (zinc transporter), member 14 | 799782 |
| ENSDARG00000055791 | SLC3A2 (3 of 3) | solute carrier family 3 (amino acid transporter heavy chain), member 2 | 790938 |
| ENSDARG00000036427 | slc3a2a | solute carrier family 3 (amino acid transporter heavy chain), member 2a | 796322 |
| ENSDARG00000037012 | slc3a2b | solute carrier family 3 (amino acid transporter heavy chain), member 2b | 399488 |
| ENSDARG00000090982 | slc44a1b | solute carrier family 44 (choline transporter), member 1b | 0 |
| ENSDARG00000037059 | slc44a2 | solute carrier family 44 (choline transporter), member 2 | 324345 |
| ENSDARG00000063133 | slc4a10a | solute carrier family 4, sodium bicarbonate transporter, member 10a | 798983 |
| ENSDARG00000060303 | slc4a10b | solute carrier family 4, sodium bicarbonate transporter, member 10b | 557642 |
| ENSDARG00000012881 | slc4a1a | solute carrier family 4 (anion exchanger), member 1a (Diego blood group) | 84703 |
| ENSDARG00000013730 | slc4a4a | solute carrier family 4 (sodium bicarbonate cotransporter), member 4a | 568631 |
| ENSDARG00000015531 | slc4a8 | solute carrier family 4, sodium bicarbonate cotransporter, member 8 | 557494 |
| ENSDARG00000087981 | SLC6A11 | solute carrier family 6 (neurotransmitter transporter), member 11 | 0 |
| ENSDARG00000067567 | SLC6A13 | solute carrier family 6 (neurotransmitter transporter), member 13 | 100332592 |
| ENSDARG00000000730 | slc6a13 | solute carrier family 6 (neurotransmitter transporter), member 13 | 368923 |
| ENSDARG00000068787 | slc6a17 | solute carrier family 6 (neutral amino acid transporter), member 17 | 798635 |
| ENSDARG00000045944 | slc6a1a | solute carrier family 6 (neurotransmitter transporter), member 1a | 692318 |
| ENSDARG00000039647 | slc6a1b | solute carrier family 6 (neurotransmitter transporter), member 1b | 492490 |
| ENSDARG00000016141 | slc6a2 | solute carrier family 6 (neurotransmitter transporter), member 2 | 565776 |
| ENSDARG00000061165 | slc6a4a | solute carrier family 6 (neurotransmitter transporter), member 4a | 664719 |
| ENSDARG00000067964 | slc6a5 | solute carrier family 6 (neurotransmitter transporter), member 5 | 494450 |
| ENSDARG00000012534 | slc6a6a | solute carrier family 6 (neurotransmitter transporter), member 6a | 556480 |
| ENSDARG00000008100 | slc7a10a | solute carrier family 7 (neutral amino acid transporter light chain, asc system), member 10a | 567420 |
| ENSDARG00000010816 | slc7a14a | solute carrier family 7, member 14a | 566270 |
| ENSDARG00000020645 | slc7a3a | solute carrier family 7 (cationic amino acid transporter, y+ system), member 3a | 492363 |
| ENSDARG00000075831 | slc7a8b | solute carrier family 7 (amino acid transporter light chain, L system), member 8b | 100007704 |
| ENSDARG00000043406 | slc8a1b | solute carrier family 8 (sodium/calcium exchanger), member 1b | 797199 |
| ENSDARG00000100592 | slc8a2b | solute carrier family 8 (sodium/calcium exchanger), member 2b | 567212 |
| ENSDARG00000004931 | slc8a3 | solute carrier family 8 (sodium/calcium exchanger), member 3 | 560561 |
| ENSDARG00000055154 | slc8a4a | solute carrier family 8 (sodium/calcium exchanger), member 4a | 568512 |
| ENSDARG00000037145 | slc8a4b | solute carrier family 8 (sodium/calcium exchanger), member 4b | 557499 |
| ENSDARG00000067784 | SLC9A1 | solute carrier family 9, subfamily A (NHE1, cation proton antiporter 1), member 1 | 0 |
| ENSDARG00000000068 | slc9a3r1 | solute carrier family 9, subfamily A (NHE3, cation proton antiporter 3), member 3 regulator 1 | 327272 |
| ENSDARG00000009209 | slc9a6a | solute carrier family 9, subfamily A (NHE6, cation proton antiporter 6), member 6a | 0 |
| ENSDARG00000075382 | slc9a6b | solute carrier family 9, subfamily A (NHE6, cation proton antiporter 6), member 6b | 556638 |
| ENSDARG00000076754 | slc9a7 | solute carrier family 9, subfamily A (NHE7, cation proton antiporter 7), member 7 | 554532 |
| ENSDARG00000086104 | SLCO5A1 (2 of 2) | solute carrier organic anion transporter family, member 5A1 | 570033 |
| ENSDARG00000079781 | slitrk4 | SLIT and NTRK-like family, member 4 | 558123 |
| ENSDARG00000103359 | SLITRK5 (2 of 2) | SLIT and NTRK-like family, member 5 | 101882841 |
| ENSDARG00000061525 | slka | STE20-like kinase a | 568838 |
| ENSDARG00000054458 | slmapa | sarcolemma associated protein a | 572011 |
| ENSDARG00000020764 | slmapb | sarcolemma associated protein b | 393146 |
| ENSDARG00000055288 | slmo1 | slowmo homolog 1 (Drosophila) | 563270 |
| ENSDARG00000031302 | smap1 | small ArfGAP 1 | 777731 |
| ENSDARG00000077946 | smarcc2 | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 2 | 567476 |
| ENSDARG00000058203 | smc1al | structural maintenance of chromosomes 1A, like | 403060 |
| ENSDARG00000019000 | smc3 | structural maintenance of chromosomes 3 | 324475 |
| ENSDARG00000098226 | smpd3 | sphingomyelin phosphodiesterase 3, neutral | 568381 |
| ENSDARG00000008155 | sms | spermine synthase | 80872 |
| ENSDARG00000051970 | smu1b | smu-1 suppressor of mec-8 and unc-52 homolog b (C. elegans) | 570074 |
| ENSDARG00000038067 | smurf2 | SMAD specific E3 ubiquitin protein ligase 2 | 563633 |
| ENSDARG00000012874 | snap23.1 | synaptosomal-associated protein 23.1 | 393159 |
| ENSDARG00000020609 | snap25a | synaptosomal-associated protein, 25a | 30712 |
| ENSDARG00000058117 | snap25b | synaptosomal-associated protein, 25b | 30711 |
| ENSDARG00000038518 | snap29 | synaptosomal-associated protein 29 | 553460 |
| ENSDARG00000015931 | SNAP91 (1 of 2) | synaptosomal-associated protein, 91kDa | 559029 |
| ENSDARG00000098809 | snap91 | synaptosomal-associated protein 91 | 565384 |
| ENSDARG00000104945 | sncb | synuclein, beta | 393944 |
| ENSDARG00000034423 | sncga | synuclein, gamma a | 550229 |
| ENSDARG00000006766 | snd1 | staphylococcal nuclease and tudor domain containing 1 | 324404 |
| ENSDARG00000078485 | snpha | syntaphilin a | 0 |
| ENSDARG00000075539 | snphb | syntaphilin b | 569963 |
| ENSDARG00000019060 | snx12 | sorting nexin 12 | 394098 |
| ENSDARG00000071031 | snx1a | sorting nexin 1a | 337386 |
| ENSDARG00000103529 | snx1b | sorting nexin 1b | 619253 |
| ENSDARG00000053527 | snx24 | sorting nexin 24 | 619249 |
| ENSDARG00000033804 | snx27a | sorting nexin family member 27a | 566205 |
| ENSDARG00000016977 | snx27b | sorting nexin family member 27b | 393512 |
| ENSDARG00000008678 | snx3 | sorting nexin 3 | 563511 |
| ENSDARG00000022659 | snx4 | sorting nexin 4 | 323203 |
| ENSDARG00000058444 | snx6 | sorting nexin 6 | 567182 |
| ENSDARG00000043848 | sod1 | superoxide dismutase 1, soluble | 30553 |
| ENSDARG00000042644 | sod2 | superoxide dismutase 2, mitochondrial | 335799 |
| ENSDARG00000097245 | soga3a | SOGA family member 3a | 100332515 |
| ENSDARG00000075455 | soga3b | SOGA family member 3b | 563485 |
| ENSDARG00000103435 | sorbs1 | sorbin and SH3 domain containing 1 | 0 |
| ENSDARG00000061603 | sorbs2b | sorbin and SH3 domain containing 2b | 0 |
| ENSDARG00000077465 | SORCS2 | sortilin-related VPS10 domain containing receptor 2 | 100330371 |
| ENSDARG00000077349 | SORCS3 (1 of 2) | sortilin-related VPS10 domain containing receptor 3 | 0 |
| ENSDARG00000013892 | sorl1 | sortilin-related receptor, L(DLR class) A repeats containing | 497306 |
| ENSDARG00000056252 | sort1b | sortilin 1b | 799153 |
| ENSDARG00000004017 | spag1a | sperm associated antigen 1a | 0 |
| ENSDARG00000074531 | spag9a | sperm associated antigen 9a | 0 |
| ENSDARG00000059298 | spcs1 | signal peptidase complex subunit 1 homolog (S. cerevisiae) | 606666 |
| ENSDARG00000042823 | spcs2 | signal peptidase complex subunit 2 homolog (S. cerevisiae) | 541342 |
| ENSDARG00000089976 | spcs3 | signal peptidase complex subunit 3 homolog (S. cerevisiae) | 405884 |
| ENSDARG00000029480 | specc1 | sperm antigen with calponin homology and coiled-coil domains 1 | 562343 |
| ENSDARG00000103457 | spg20b | spastic paraplegia 20b (Troyer syndrome) | 751741 |
| ENSDARG00000068187 | spg7 | spastic paraplegia 7 | 794740 |
| ENSDARG00000035868 | spire1a | spire-type actin nucleation factor 1a | 557962 |
| ENSDARG00000019231 | spna2 | spectrin alpha 2 | 553383 |
| ENSDARG00000004406 | spra | sepiapterin reductase a | 554136 |
| ENSDARG00000057665 | sprn2 | shadow of prion protein 2 | 100006310 |
| ENSDARG00000023309 | spryd4 | SPRY domain containing 4 | 100005939 |
| ENSDARG00000102883 | SPTBN1 | spectrin, beta, non-erythrocytic 1 | 553451 |
| ENSDARG00000053956 | sptbn2 | spectrin, beta, non-erythrocytic 2 | 558044 |
| ENSDARG00000030490 | sptb | spectrin, beta, erythrocytic | 58040 |
| ENSDARG00000017034 | sqrdl | sulfide quinone reductase-like (yeast) | 550580 |
| ENSDARG00000059773 | SRCIN1 (1 of 2) | SRC kinase signaling inhibitor 1 | 0 |
| ENSDARG00000101812 | SRCIN1 (2 of 2) | SRC kinase signaling inhibitor 1 | 0 |
| ENSDARG00000008107 | src | v-src avian sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog | 325084 |
| ENSDARG00000032161 | srgap2a | SLIT-ROBO Rho GTPase activating protein 2a | 556198 |
| ENSDARG00000060309 | srgap3 | SLIT-ROBO Rho GTPase activating protein 3 | 561567 |
| ENSDARG00000035699 | srp19 | signal recognition particle 19 | 406652 |
| ENSDARG00000098367 | srp54 | signal recognition particle 54 | 393963 |
| ENSDARG00000014139 | srp72 | signal recognition particle 72 | 327335 |
| ENSDARG00000031560 | srpk1b | SRSF protein kinase 1b | 791187 |
| ENSDARG00000030949 | srprb | signal recognition particle receptor, B subunit | 436845 |
| ENSDARG00000027734 | srsf5b | serine/arginine-rich splicing factor 5b | 436883 |
| ENSDARG00000039350 | ssbp1 | single-stranded DNA binding protein 1 | 550504 |
| ENSDARG00000029252 | ssb | Sjogren syndrome antigen B (autoantigen La) | 321504 |
| ENSDARG00000101627 | ssr1 | signal sequence receptor, alpha | 373100 |
| ENSDARG00000014165 | ssr3 | signal sequence receptor, gamma | 337190 |
| ENSDARG00000019444 | ssr4 | signal sequence receptor, delta | 326701 |
| ENSDARG00000021973 | st13 | suppression of tumorigenicity 13 (colon carcinoma) (Hsp70 interacting protein) | 564225 |
| ENSDARG00000044514 | st6gal1 | ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 | 445376 |
| ENSDARG00000045748 | stab2 | stabilin 2 | 565194 |
| ENSDARG00000002127 | stam | signal transducing adaptor molecule (SH3 domain and ITAM motif) 1 | 386967 |
| ENSDARG00000045421 | stard3nl | STARD3 N-terminal like | 436592 |
| ENSDARG00000076117 | STARD5 | StAR-related lipid transfer (START) domain containing 5 | 0 |
| ENSDARG00000006266 | stat1a | signal transducer and activator of transcription 1, 91kDa | 30768 |
| ENSDARG00000026801 | stau2 | staufen double-stranded RNA binding protein 2 | 393899 |
| ENSDARG00000055901 | steap4 | STEAP family member 4 | 550429 |
| ENSDARG00000060723 | stim1a | stromal interaction molecule 1a | 556360 |
| ENSDARG00000004906 | stip1 | stress-induced phosphoprotein 1 | 493606 |
| ENSDARG00000039022 | stk25b | serine/threonine kinase 25b | 406600 |
| ENSDARG00000011312 | stk3 | serine/threonine kinase 3 (STE20 homolog, yeast) | 324125 |
| ENSDARG00000033655 | stmn1b | stathmin 1b | 550548 |
| ENSDARG00000103135 | stoml2 | stomatin (EPB72)-like 2 | 394006 |
| ENSDARG00000003835 | stom | stomatin | 81534 |
| ENSDARG00000051874 | stra6 | stimulated by retinoic acid 6 | 724007 |
| ENSDARG00000074084 | strip1 | striatin interacting protein 1 | 568805 |
| ENSDARG00000001729 | strn3 | striatin, calmodulin binding protein 3 | 563124 |
| ENSDARG00000104953 | stt3a | STT3A, subunit of the oligosaccharyltransferase complex (catalytic) | 266797 |
| ENSDARG00000001890 | STT3B | STT3B, subunit of the oligosaccharyltransferase complex (catalytic) | 0 |
| ENSDARG00000045228 | stub1 | STIP1 homology and U-Box containing protein 1 | 324243 |
| ENSDARG00000044605 | stx12l | syntaxin 12, like | 569124 |
| ENSDARG00000098813 | stx12 | syntaxin 12 | 415141 |
| ENSDARG00000003307 | stx16 | syntaxin 16 | 562857 |
| ENSDARG00000035763 | stx18 | syntaxin 18 | 554130 |
| ENSDARG00000000503 | stx1b | syntaxin 1B | 58038 |
| ENSDARG00000052518 | stx4 | syntaxin 4 | 323735 |
| ENSDARG00000103173 | stx8 | syntaxin 8 | 393346 |
| ENSDARG00000001994 | stxbp1a | syntaxin binding protein 1a | 574003 |
| ENSDARG00000056036 | stxbp1b | syntaxin binding protein 1b | 557717 |
| ENSDARG00000008142 | stxbp3 | syntaxin binding protein 3 | 406780 |
| ENSDARG00000002656 | stxbp5a | syntaxin binding protein 5a (tomosyn) | 556789 |
| ENSDARG00000029234 | stxbp5b | syntaxin binding protein 5b (tomosyn) | 571925 |
| ENSDARG00000006383 | stxbp5l | syntaxin binding protein 5-like | 567605 |
| ENSDARG00000088862 | STXBP6 (2 of 2) | syntaxin binding protein 6 (amisyn) | 0 |
| ENSDARG00000028354 | stxbp6l | syntaxin binding protein 6 (amisyn), like | 100002110 |
| ENSDARG00000053446 | sub1a | SUB1 homolog a (S. cerevisiae) | 553561 |
| ENSDARG00000007720 | sub1b | SUB1 homolog b (S. cerevisiae) | 436778 |
| ENSDARG00000005359 | sucla2 | succinate-CoA ligase, ADP-forming, beta subunit | 406299 |
| ENSDARG00000052712 | suclg1 | succinate-CoA ligase, alpha subunit | 436850 |
| ENSDARG00000044914 | suclg2 | succinate-CoA ligase, GDP-forming, beta subunit | 317746 |
| ENSDARG00000028275 | sult1st1 | sulfotransferase family 1, cytosolic sulfotransferase 1 | 323424 |
| ENSDARG00000035993 | sumo3b | small ubiquitin-like modifier 3b | 436950 |
| ENSDARG00000079949 | supt16h | suppressor of Ty 16 homolog (S. cerevisiae) | 562348 |
| ENSDARG00000003592 | surf1 | surfeit 1 | 557970 |
| ENSDARG00000035131 | surf4l | surfeit gene 4, like | 436743 |
| ENSDARG00000088277 | susd5 | sushi domain containing 5 | 101886789 |
| ENSDARG00000059945 | SV2A | synaptic vesicle glycoprotein 2A | 568033 |
| ENSDARG00000060711 | sv2bb | synaptic vesicle glycoprotein 2Bb | 100006427 |
| ENSDARG00000059997 | sv2c | synaptic vesicle glycoprotein 2C | 562345 |
| ENSDARG00000055075 | svila | supervillin a | 30112 |
| ENSDARG00000014651 | svilb | supervillin b | 0 |
| ENSDARG00000056833 | svopa | SV2 related protein homolog a (rat) | 559162 |
| ENSDARG00000057286 | swap70b | SWAP switching B-cell complex 70b subunit 70b | 562490 |
| ENSDARG00000030775 | sybl1 | synaptobrevin-like 1 | 393236 |
| ENSDARG00000060368 | syn1 | synapsin I | 570672 |
| ENSDARG00000045945 | syn2a | synapsin IIa | 436870 |
| ENSDARG00000098417 | syn3 | synapsin III | 0 |
| ENSDARG00000009499 | syne1a | spectrin repeat containing, nuclear envelope 1a | 0 |
| ENSDARG00000063068 | syne1b | spectrin repeat containing, nuclear envelope 1b | 0 |
| ENSDARG00000092624 | SYNE1 | spectrin repeat containing, nuclear envelope 1 | 0 |
| ENSDARG00000063713 | SYNGAP1 (1 of 2) | synaptic Ras GTPase activating protein 1 | 100034485 |
| ENSDARG00000069765 | SYNGAP1 (2 of 2) | synaptic Ras GTPase activating protein 1 | 0 |
| ENSDARG00000002564 | syngr1a | synaptogyrin 1a | 450047 |
| ENSDARG00000014871 | syngr3a | synaptogyrin 3a | 553617 |
| ENSDARG00000025034 | syngr3b | synaptogyrin 3b | 570094 |
| ENSDARG00000025011 | synj1 | synaptojanin 1 | 337236 |
| ENSDARG00000028321 | synj2bp | synaptojanin 2 binding protein | 334599 |
| ENSDARG00000062350 | synm | synemin, intermediate filament protein | 558800 |
| ENSDARG00000042974 | sypa | synaptophysin a | 559698 |
| ENSDARG00000002230 | sypb | synaptophysin b | 436774 |
| ENSDARG00000101776 | syt12 | synaptotagmin XII | 566397 |
| ENSDARG00000030614 | syt1a | synaptotagmin Ia | 436736 |
| ENSDARG00000014169 | SYT2 (1 of 2) | synaptotagmin II | 0 |
| ENSDARG00000025206 | syt2a | synaptotagmin IIa | 567877 |
| ENSDARG00000075830 | syt3 | synaptotagmin III | 100000615 |
| ENSDARG00000011640 | syt5b | synaptotagmin Vb | 553567 |
| ENSDARG00000063568 | syt7a | synaptotagmin VIIa | 561429 |
| ENSDARG00000078060 | syt7b | synaptotagmin VIIb | 100334336 |
| ENSDARG00000003994 | syt9a | synaptotagmin IXa | 798433 |
| ENSDARG00000017842 | syvn1 | synovial apoptosis inhibitor 1, synoviolin | 335226 |
| ENSDARG00000079805 | tagln3a | transgelin 3a | 563096 |
| ENSDARG00000058394 | tagln3b | transgelin 3b | 797861 |
| ENSDARG00000103369 | TALDO1 | transaldolase 1 | 0 |
| ENSDARG00000100157 | TAMM41 | TAM41, mitochondrial translocator assembly and maintenance protein, homolog (S. cerevisiae) | 0 |
| ENSDARG00000079004 | TANC2 (1 of 2) | tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2 | 0 |
| ENSDARG00000079097 | tanc2a | tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2a | 568810 |
| ENSDARG00000069603 | taok1a | TAO kinase 1a | 100004576 |
| ENSDARG00000074899 | taok2a | TAO kinase 2a | 556172 |
| ENSDARG00000100620 | taok2b | TAO kinase 2b | 100151551 |
| ENSDARG00000094052 | taok3b | TAO kinase 3b | 555650 |
| ENSDARG00000061143 | tapt1b | transmembrane anterior posterior transformation 1b | 559710 |
| ENSDARG00000013250 | tars | threonyl-tRNA synthetase | 449661 |
| ENSDARG00000078604 | tbc1d10b | TBC1 domain family, member 10b | 0 |
| ENSDARG00000069339 | tbc1d24 | TBC1 domain family, member 24 | 100536073 |
| ENSDARG00000016754 | tbca | tubulin cofactor a | 394029 |
| ENSDARG00000068404 | tbcb | tubulin folding cofactor B | 337350 |
| ENSDARG00000098749 | tbcd | tubulin folding cofactor D | 553265 |
| ENSDARG00000103095 | tbk1 | TANK-binding kinase 1 | 692289 |
| ENSDARG00000061473 | tbkbp1 | TBK1 binding protein 1 | 559568 |
| ENSDARG00000043357 | tbl2 | transducin (beta)-like 2 | 337068 |
| ENSDARG00000002249 | tbxas1 | thromboxane A synthase 1 (platelet) | 792041 |
| ENSDARG00000079881 | TCAIM | T cell activation inhibitor, mitochondrial | 559258 |
| ENSDARG00000012927 | tcea2 | transcription elongation factor A (SII), 2 | 393961 |
| ENSDARG00000045355 | tceb1a | transcription elongation factor B (SIII), polypeptide 1a | 447935 |
| ENSDARG00000037980 | tceb2 | transcription elongation factor B (SIII), polypeptide 2 | 192341 |
| ENSDARG00000105142 | tcirg1b | T-cell, immune regulator 1, ATPase, H+ transporting, lysosomal V0 subunit A3b | 406342 |
| ENSDARG00000031890 | tcp11l1 | t-complex 11, testis-specific-like 1 | 541488 |
| ENSDARG00000017891 | tcp1 | t-complex 1 | 30477 |
| ENSDARG00000069941 | TDP2 (2 of 2) | tyrosyl-DNA phosphodiesterase 2 | 101887157 |
| ENSDARG00000053474 | tdrkh | tudor and KH domain containing | 541541 |
| ENSDARG00000025904 | tecrb | trans-2,3-enoyl-CoA reductase b | 326954 |
| ENSDARG00000003403 | tenm1 | teneurin transmembrane protein 1 | 563094 |
| ENSDARG00000011171 | TENM2 (1 of 2) | teneurin transmembrane protein 2 | 568845 |
| ENSDARG00000037122 | TENM2 (2 of 2) | teneurin transmembrane protein 2 | 563197 |
| ENSDARG00000005479 | tenm3 | teneurin transmembrane protein 3 | 30155 |
| ENSDARG00000105088 | tenm4 | teneurin transmembrane protein 4 | 30156 |
| ENSDARG00000092154 | tex264a | testis expressed 264a | 402960 |
| ENSDARG00000063145 | tfam | transcription factor A, mitochondrial | 571106 |
| ENSDARG00000016771 | tfa | transferrin-a | 0 |
| ENSDARG00000060306 | tfcp2 | transcription factor CP2 | 100001906 |
| ENSDARG00000003641 | tfg | trk-fused gene | 336466 |
| ENSDARG00000101322 | tfr1a | transferrin receptor 1a | 494477 |
| ENSDARG00000077372 | tfr1b | transferrin receptor 1b | 494478 |
| ENSDARG00000092470 | THBD | thrombomodulin | 0 |
| ENSDARG00000103914 | THEM6 (2 of 2) | thioesterase superfamily member 6 | 100142654 |
| ENSDARG00000013776 | thop1 | thimet oligopeptidase 1 | 436628 |
| ENSDARG00000061479 | thsd7aa | thrombospondin, type I, domain containing 7Aa | 557991 |
| ENSDARG00000035018 | thy1 | Thy-1 cell surface antigen | 336617 |
| ENSDARG00000069116 | timm10 | translocase of inner mitochondrial membrane 10 homolog (yeast) | 436951 |
| ENSDARG00000058297 | timm13 | translocase of inner mitochondrial membrane 13 homolog (yeast) | 436738 |
| ENSDARG00000029510 | timm17a | translocase of inner mitochondrial membrane 17 homolog A (yeast) | 0 |
| ENSDARG00000060124 | timm17b | translocase of inner mitochondrial membrane 17 homolog B (yeast) | 562144 |
| ENSDARG00000071594 | TIMM21 | translocase of inner mitochondrial membrane 21 homolog (yeast) | 560369 |
| ENSDARG00000071396 | timm23 | translocase of inner mitochondrial membrane 23 homolog (yeast) | 798429 |
| ENSDARG00000104771 | timm44 | translocase of inner mitochondrial membrane 44 homolog (yeast) | 541380 |
| ENSDARG00000031098 | timm50 | translocase of inner mitochondrial membrane 50 homolog (S. cerevisiae) | 393638 |
| ENSDARG00000023672 | timm8a | translocase of inner mitochondrial membrane 8 homolog A (yeast) | 445243 |
| ENSDARG00000055708 | timm8b | translocase of inner mitochondrial membrane 8 homolog B (yeast) | 793663 |
| ENSDARG00000043865 | timm9 | translocase of inner mitochondrial membrane 9 homolog | 282678 |
| ENSDARG00000077506 | tjp1a | tight junction protein 1a | 378845 |
| ENSDARG00000079374 | tjp1b | tight junction protein 1b | 565459 |
| ENSDARG00000029689 | tkta | transketolase a | 557518 |
| ENSDARG00000006019 | tktb | transketolase b | 378713 |
| ENSDARG00000062951 | tldc1 | TBC/LysM-associated domain containing 1 | 561678 |
| ENSDARG00000100729 | tln1 | talin 1 | 494453 |
| ENSDARG00000017901 | tln2a | talin 2a | 556842 |
| ENSDARG00000104934 | tln2b | talin 2b | 0 |
| ENSDARG00000003866 | tm9sf2 | transmembrane 9 superfamily member 2 | 334370 |
| ENSDARG00000028748 | tm9sf3 | transmembrane 9 superfamily member 3 | 406698 |
| ENSDARG00000002536 | tm9sf4 | transmembrane 9 superfamily protein member 4 | 393482 |
| ENSDARG00000069099 | tmco1 | transmembrane and coiled-coil domains 1 | 378980 |
| ENSDARG00000041391 | tmed10 | transmembrane emp24-like trafficking protein 10 (yeast) | 326716 |
| ENSDARG00000026908 | tmed2 | transmembrane emp24 domain trafficking protein 2 | 321550 |
| ENSDARG00000024954 | tmed4 | transmembrane emp24 protein transport domain containing 4 | 415224 |
| ENSDARG00000008765 | tmed5 | transmembrane emp24 protein transport domain containing 5 | 393374 |
| ENSDARG00000046022 | tmed7 | transmembrane emp24 protein transport domain containing 7 | 405848 |
| ENSDARG00000052390 | tmed8 | transmembrane emp24 protein transport domain containing 8 | 553196 |
| ENSDARG00000004261 | tmed9 | transmembrane emp24 protein transport domain containing 9 | 613145 |
| ENSDARG00000051824 | tmeff2a | transmembrane protein with EGF-like and two follistatin-like domains 2a | 100004613 |
| ENSDARG00000035949 | tmem106bb | transmembrane protein 106Bb | 566699 |
| ENSDARG00000070866 | tmem11 | transmembrane protein 11 | 407695 |
| ENSDARG00000075180 | tmem14ca | transmembrane protein 14Ca | 751728 |
| ENSDARG00000069649 | tmem160 | transmembrane protein 160 | 559491 |
| ENSDARG00000079858 | tmem163a | transmembrane protein 163a | 564964 |
| ENSDARG00000088227 | tmem163b | transmembrane protein 163b | 100333673 |
| ENSDARG00000034600 | tmem165 | transmembrane protein 165 | 324501 |
| ENSDARG00000076376 | tmem175 | transmembrane protein 175 | 100002086 |
| ENSDARG00000037484 | tmem192 | transmembrane protein 192 | 797335 |
| ENSDARG00000016968 | tmem214 | transmembrane protein 214 | 402840 |
| ENSDARG00000086661 | TMEM221 | transmembrane protein 221 | 100537506 |
| ENSDARG00000069494 | tmem223 | transmembrane protein 223 | 445200 |
| ENSDARG00000087916 | tmem240a | transmembrane protein 240a | 568378 |
| ENSDARG00000090145 | tmem240b | transmembrane protein 240b | 0 |
| ENSDARG00000038901 | tmem256 | transmembrane protein 256 | 550283 |
| ENSDARG00000092787 | TMEM261 | transmembrane protein 261 | 558861 |
| ENSDARG00000030236 | tmem30aa | transmembrane protein 30Aa | 337397 |
| ENSDARG00000043555 | tmem30ab | transmembrane protein 30Ab | 333952 |
| ENSDARG00000041332 | tmem33 | transmembrane protein 33 | 327214 |
| ENSDARG00000015757 | tmem50a | transmembrane protein 50A | 406850 |
| ENSDARG00000038615 | tmem55ba | transmembrane protein 55Ba | 556136 |
| ENSDARG00000003655 | tmem59l | transmembrane protein 59-like | 406646 |
| ENSDARG00000063414 | tmem5 | transmembrane protein 5 | 557187 |
| ENSDARG00000076393 | tmem65 | transmembrane protein 65 | 100005403 |
| ENSDARG00000025693 | tmem9b | TMEM9 domain family, member B | 325931 |
| ENSDARG00000002571 | tmod2 | tropomodulin 2 | 447869 |
| ENSDARG00000020890 | tmod4 | tropomodulin 4 (muscle) | 30441 |
| ENSDARG00000022978 | tmpob | thymopoietin b | 553624 |
| ENSDARG00000077777 | tmsb4x | thymosin, beta 4 x | 793949 |
| ENSDARG00000063149 | tmtc1 | transmembrane and tetratricopeptide repeat containing 1 | 555449 |
| ENSDARG00000062846 | tmtc3 | transmembrane and tetratricopeptide repeat containing 3 | 100150181 |
| ENSDARG00000007786 | tmx2b | thioredoxin-related transmembrane protein 2b | 415203 |
| ENSDARG00000088466 | TMX3 (2 of 2) | thioredoxin-related transmembrane protein 3 | 0 |
| ENSDARG00000101512 | tmx4 | thioredoxin-related transmembrane protein 4 | 792311 |
| ENSDARG00000078362 | TNC (2 of 2) | tenascin C | 100149028 |
| ENSDARG00000021948 | tnc | tenascin C | 30037 |
| ENSDARG00000043416 | tnfaip2a | tumor necrosis factor, alpha-induced protein 2a | 567159 |
| ENSDARG00000056218 | tnika | TRAF2 and NCK interacting kinase a | 325042 |
| ENSDARG00000009031 | tnikb | TRAF2 and NCK interacting kinase b | 556959 |
| ENSDARG00000020771 | tnr | tenascin R (restrictin, janusin) | 369191 |
| ENSDARG00000078842 | tns1a | tensin 1a | 565181 |
| ENSDARG00000098486 | tollip | toll interacting protein | 336876 |
| ENSDARG00000056394 | TOM1L2 (2 of 2) | target of myb1-like 2 (chicken) | 100535615 |
| ENSDARG00000009217 | tom1l2 | target of myb1-like 2 (chicken) | 560327 |
| ENSDARG00000104581 | tom1 | target of myb1 (chicken) | 0 |
| ENSDARG00000045146 | tomm22 | translocase of outer mitochondrial membrane 22 homolog (yeast) | 321232 |
| ENSDARG00000001557 | tomm34 | translocase of outer mitochondrial membrane 34 | 323361 |
| ENSDARG00000036721 | tomm40l | translocase of outer mitochondrial membrane 40 homolog, like | 405895 |
| ENSDARG00000092112 | tomm40 | translocase of outer mitochondrial membrane 40 homolog (yeast) | 322645 |
| ENSDARG00000097797 | tomm6 | translocase of outer mitochondrial membrane 6 homolog (yeast) | 100001229 |
| ENSDARG00000029639 | tomm70a | translocase of outer mitochondrial membrane 70 homolog A (S. cerevisiae) | 572969 |
| ENSDARG00000067717 | tomm7 | translocase of outer mitochondrial membrane 7 homolog (yeast) | 751649 |
| ENSDARG00000079308 | tor3a | torsin family 3, member A | 792835 |
| ENSDARG00000054858 | tp53bp2b | tumor protein p53 binding protein, 2b | 568138 |
| ENSDARG00000069430 | tp53i11a | tumor protein p53 inducible protein 11a | 503774 |
| ENSDARG00000006025 | tp53i11b | tumor protein p53 inducible protein 11b | 556618 |
| ENSDARG00000013655 | tpd52l2b | tumor protein D52-like 2b | 322103 |
| ENSDARG00000061713 | tpd52 | tumor protein D52 | 559992 |
| ENSDARG00000057239 | tph2 | tryptophan hydroxylase 2 (tryptophan 5-monooxygenase) | 0 |
| ENSDARG00000025012 | tpi1a | triosephosphate isomerase 1a | 192309 |
| ENSDARG00000040988 | tpi1b | triosephosphate isomerase 1b | 560753 |
| ENSDARG00000087402 | TPM1 | tropomyosin 1 (alpha) | 393908 |
| ENSDARG00000005162 | tpm3 | tropomyosin 3 | 373076 |
| ENSDARG00000033683 | tpma | alpha-tropomyosin | 30324 |
| ENSDARG00000078751 | TPP2 | tripeptidyl peptidase II | 100331669 |
| ENSDARG00000035338 | tppp3l | tubulin polymerization promoting protein 3, like | 559490 |
| ENSDARG00000105029 | TPPP | tubulin polymerization promoting protein | 100333482 |
| ENSDARG00000068149 | tprg1l | tumor protein p63 regulated 1-like | 568867 |
| ENSDARG00000014941 | tprkb | Tp53rk binding protein | 492501 |
| ENSDARG00000073872 | tpst1 | tyrosylprotein sulfotransferase 1 | 30677 |
| ENSDARG00000092693 | tpt1 | tumor protein, translationally-controlled 1 | 30723 |
| ENSDARG00000010445 | trabd | TraB domain containing | 322581 |
| ENSDARG00000003963 | traf4a | tnf receptor-associated factor 4a | 404045 |
| ENSDARG00000024317 | trap1 | TNF receptor-associated protein 1 | 571959 |
| ENSDARG00000060101 | trappc12 | trafficking protein particle complex 12 | 791162 |
| ENSDARG00000039005 | trappc1 | trafficking protein particle complex 1 | 437015 |
| ENSDARG00000097660 | trappc2 | trafficking protein particle complex 2 | 767808 |
| ENSDARG00000045364 | trappc3 | trafficking protein particle complex 3 | 445207 |
| ENSDARG00000027418 | trappc4 | trafficking protein particle complex 4 | 393737 |
| ENSDARG00000026068 | trappc5 | trafficking protein particle complex 5 | 436755 |
| ENSDARG00000016463 | trappc6b | trafficking protein particle complex 6b | 450008 |
| ENSDARG00000038866 | TRAPPC8 | trafficking protein particle complex 8 | 570167 |
| ENSDARG00000099118 | TRAPPC9 (1 of 2) | trafficking protein particle complex 9 | 100002630 |
| ENSDARG00000062013 | TRHDE (1 of 2) | thyrotropin-releasing hormone degrading enzyme | 562409 |
| ENSDARG00000069420 | trim23 | tripartite motif containing 23 | 767706 |
| ENSDARG00000104396 | trim25 | tripartite motif containing 25 | 393144 |
| ENSDARG00000076174 | trim2b | tripartite motif containing 2b | 558046 |
| ENSDARG00000063711 | trim3a | tripartite motif containing 3a | 562489 |
| ENSDARG00000005397 | trim3b | tripartite motif containing 3b | 555391 |
| ENSDARG00000058649 | trim46b | tripartite motif containing 46b | 100034631 |
| ENSDARG00000019426 | trioa | trio Rho guanine nucleotide exchange factor a | 557667 |
| ENSDARG00000000370 | triob | trio Rho guanine nucleotide exchange factor b | 368847 |
| ENSDARG00000078381 | trip11 | thyroid hormone receptor interactor 11 | 100537975 |
| ENSDARG00000061397 | trip12 | thyroid hormone receptor interactor 12 | 564866 |
| ENSDARG00000011897 | TSG101 (1 of 3) | tumor susceptibility 101 | 100333057 |
| ENSDARG00000040854 | tsg101a | tumor susceptibility 101a | 415179 |
| ENSDARG00000041830 | tsn | translin | 334107 |
| ENSDARG00000003754 | tspan2a | tetraspanin 2a | 378854 |
| ENSDARG00000008407 | tspan7b | tetraspanin 7b | 449539 |
| ENSDARG00000025299 | tspan9a | tetraspanin 9a | 555341 |
| ENSDARG00000045706 | tspan9b | tetraspanin 9b | 431733 |
| ENSDARG00000039666 | tsta3 | tissue specific transplantation antigen P35B | 494077 |
| ENSDARG00000043026 | TTBK2 (2 of 2) | tau tubulin kinase 2 | 569881 |
| ENSDARG00000074435 | ttc19 | tetratricopeptide repeat domain 19 | 0 |
| ENSDARG00000061207 | ttc7b | tetratricopeptide repeat domain 7B | 568039 |
| ENSDARG00000068421 | ttc9b | tetratricopeptide repeat domain 9B | 559732 |
| ENSDARG00000007025 | ttc9c | tetratricopeptide repeat domain 9C | 393235 |
| ENSDARG00000028213 | ttna | titin a | 317731 |
| ENSDARG00000000563 | ttnb | titin b | 0 |
| ENSDARG00000076804 | ttyh1 | tweety family member 1 | 100537642 |
| ENSDARG00000034473 | ttyh3a | tweety family member 3a | 556314 |
| ENSDARG00000007678 | ttyh3b | tweety family member 3b | 798421 |
| ENSDARG00000045367 | tuba1b | tubulin, alpha 1b | 373080 |
| ENSDARG00000045014 | tuba2 | tubulin, alpha 2 | 406303 |
| ENSDARG00000074289 | tuba4l | tubulin, alpha 4 like | 327377 |
| ENSDARG00000042708 | tuba8l | tubulin, alpha 8 like | 337223 |
| ENSDARG00000053066 | tubb1 | tubulin, beta 1 class VI | 558552 |
| ENSDARG00000039522 | tubb2 | tubulin, beta 2A class IIa | 554127 |
| ENSDARG00000098591 | tubb4b | tubulin, beta 4B class IVb | 641421 |
| ENSDARG00000037997 | tubb5 | tubulin, beta 5 | 386701 |
| ENSDARG00000104801 | tubb6 | tubulin, beta 6 class V | 550458 |
| ENSDARG00000041723 | TUBB8P7 (2 of 3) | tubulin, beta 8 class VIII pseudogene 7 | 406811 |
| ENSDARG00000015610 | tubg1 | tubulin, gamma 1 | 393882 |
| ENSDARG00000013079 | tubgcp2 | tubulin, gamma complex associated protein 2 | 386975 |
| ENSDARG00000029133 | tubgcp3 | tubulin, gamma complex associated protein 3 | 563517 |
| ENSDARG00000104173 | tufm | Tu translation elongation factor, mitochondrial | 541378 |
| ENSDARG00000078854 | tusc3 | tumor suppressor candidate 3 | 553582 |
| ENSDARG00000090481 | tusc5a | tumor suppressor candidate 5a | 101886018 |
| ENSDARG00000011661 | twf1b | twinfilin actin-binding protein 1b | 432375 |
| ENSDARG00000034777 | txn2 | thioredoxin 2 | 402938 |
| ENSDARG00000038980 | txndc12 | thioredoxin domain containing 12 (endoplasmic reticulum) | 324900 |
| ENSDARG00000009342 | txndc5 | thioredoxin domain containing 5 | 406289 |
| ENSDARG00000011921 | txnl1 | thioredoxin-like 1 | 394113 |
| ENSDARG00000100374 | txnrd1 | thioredoxin reductase 1 | 352924 |
| ENSDARG00000044125 | txn | thioredoxin | 436734 |
| ENSDARG00000037559 | uba1 | ubiquitin-like modifier activating enzyme 1 | 406335 |
| ENSDARG00000063588 | uba5 | ubiquitin-like modifier activating enzyme 5 | 0 |
| ENSDARG00000027141 | ube2l3b | ubiquitin-conjugating enzyme E2L 3b | 321943 |
| ENSDARG00000008748 | ube2na | ubiquitin-conjugating enzyme E2Na | 406807 |
| ENSDARG00000041875 | ube2v1 | ubiquitin-conjugating enzyme E2 variant 1 | 0 |
| ENSDARG00000028198 | ube2v2 | ubiquitin-conjugating enzyme E2 variant 2 | 406836 |
| ENSDARG00000037017 | ube4b | ubiquitination factor E4B, UFD2 homolog (S. cerevisiae) | 170784 |
| ENSDARG00000021556 | ubl3b | ubiquitin-like 3b | 405782 |
| ENSDARG00000007359 | UBL4A | ubiquitin-like 4A | 393270 |
| ENSDARG00000009549 | ubr4 | ubiquitin protein ligase E3 component n-recognin 4 | 407625 |
| ENSDARG00000035066 | ubtf | upstream binding transcription factor, RNA polymerase I | 368509 |
| ENSDARG00000003087 | ubxn6 | UBX domain protein 6 | 554960 |
| ENSDARG00000026871 | uchl1 | ubiquitin carboxyl-terminal esterase L1 (ubiquitin thiolesterase) | 325119 |
| ENSDARG00000030177 | uchl3 | ubiquitin carboxyl-terminal esterase L3 (ubiquitin thiolesterase) | 554104 |
| ENSDARG00000103404 | uchl5 | ubiquitin carboxyl-terminal hydrolase L5 | 406357 |
| ENSDARG00000023151 | ucp1 | uncoupling protein 1 | 83908 |
| ENSDARG00000007496 | ufl1 | UFM1-specific ligase 1 | 393578 |
| ENSDARG00000043857 | ufm1 | ubiquitin-fold modifier 1 | 322756 |
| ENSDARG00000058221 | ugcg | UDP-glucose ceramide glucosyltransferase | 0 |
| ENSDARG00000054746 | uggt1 | UDP-glucose glycoprotein glucosyltransferase 1 | 553288 |
| ENSDARG00000062089 | uggt2 | UDP-glucose glycoprotein glucosyltransferase 2 | 497542 |
| ENSDARG00000008200 | ugp2b | UDP-glucose pyrophosphorylase 2b | 334420 |
| ENSDARG00000097491 | ugt1b1 | UDP glucuronosyltransferase 1 family, polypeptide B1 | 558508 |
| ENSDARG00000032862 | ugt5g1 | UDP glucuronosyltransferase 5 family, polypeptide G1 | 570913 |
| ENSDARG00000037455 | ugt8 | UDP glycosyltransferase 8 | 553508 |
| ENSDARG00000004055 | uhrf1bp1l | UHRF1 binding protein 1-like | 562208 |
| ENSDARG00000061829 | UNC13A | unc-13 homolog A (C. elegans) | 568618 |
| ENSDARG00000101233 | unc13c | unc-13 homolog C (C. elegans) | 0 |
| ENSDARG00000040455 | unc50 | unc-50 homolog (C. elegans) | 393217 |
| ENSDARG00000016302 | upf1 | upf1 regulator of nonsense transcripts homolog (yeast) | 406783 |
| ENSDARG00000068176 | uqcc1 | ubiquinol-cytochrome c reductase complex assembly factor 1 | 767788 |
| ENSDARG00000011272 | uqcc2 | ubiquinol-cytochrome c reductase complex assembly factor 2 | 393731 |
| ENSDARG00000093382 | uqcc3 | ubiquinol-cytochrome c reductase complex assembly factor 3 | 558925 |
| ENSDARG00000104517 | UQCR10 | ubiquinol-cytochrome c reductase, complex III subunit X | 100333064 |
| ENSDARG00000011146 | uqcrb | ubiquinol-cytochrome c reductase binding protein | 554154 |
| ENSDARG00000052304 | uqcrc1 | ubiquinol-cytochrome c reductase core protein I | 393793 |
| ENSDARG00000014794 | uqcrc2a | ubiquinol-cytochrome c reductase core protein IIa | 436930 |
| ENSDARG00000071691 | uqcrc2b | ubiquinol-cytochrome c reductase core protein IIb | 322549 |
| ENSDARG00000007745 | uqcrfs1 | ubiquinol-cytochrome c reductase, Rieske iron-sulfur polypeptide 1 | 406359 |
| ENSDARG00000059128 | uqcrh | ubiquinol-cytochrome c reductase hinge protein | 566723 |
| ENSDARG00000029064 | uqcrq | ubiquinol-cytochrome c reductase, complex III subunit VII | 436768 |
| ENSDARG00000007250 | use1 | unconventional SNARE in the ER 1 homolog (S. cerevisiae) | 393294 |
| ENSDARG00000096003 | USMG5 | up-regulated during skeletal muscle growth 5 homolog (mouse) | 794625 |
| ENSDARG00000019581 | uso1 | USO1 vesicle transport factor | 393124 |
| ENSDARG00000078109 | usp12a | ubiquitin specific peptidase 12a | 565736 |
| ENSDARG00000025889 | usp14 | ubiquitin specific peptidase 14 (tRNA-guanine transglycosylase) | 335736 |
| ENSDARG00000014517 | usp5 | ubiquitin specific peptidase 5 (isopeptidase T) | 406618 |
| ENSDARG00000073710 | usp7 | ubiquitin specific peptidase 7 (herpes virus-associated) | 553758 |
| ENSDARG00000063719 | usp8 | ubiquitin specific peptidase 8 | 565434 |
| ENSDARG00000013708 | usp9 | ubiquitin specific peptidase 9 | 568683 |
| ENSDARG00000056102 | uxs1 | UDP-glucuronate decarboxylase 1 | 192315 |
| ENSDARG00000014303 | vac14 | vac14 homolog (S. cerevisiae) | 447912 |
| ENSDARG00000097576 | VAMP1 (2 of 2) | vesicle-associated membrane protein 1 (synaptobrevin 1) | 436805 |
| ENSDARG00000031283 | vamp1 | vesicle-associated membrane protein 1 | 406707 |
| ENSDARG00000056877 | vamp2 | vesicle-associated membrane protein 2 | 336281 |
| ENSDARG00000043510 | vamp4 | vesicle-associated membrane protein 4 | 393708 |
| ENSDARG00000068262 | vamp5 | vesicle-associated membrane protein 5 | 325484 |
| ENSDARG00000024116 | vamp8 | vesicle-associated membrane protein 8 (endobrevin) | 327007 |
| ENSDARG00000004305 | vangl1 | VANGL planar cell polarity protein 1 | 403076 |
| ENSDARG00000027397 | vangl2 | VANGL planar cell polarity protein 2 | 245949 |
| ENSDARG00000046048 | vapal | VAMP (vesicle-associated membrane protein)-associated protein A, like | 436819 |
| ENSDARG00000070435 | vapb | VAMP (vesicle-associated membrane protein)-associated protein B and C | 323628 |
| ENSDARG00000099690 | vaspa | vasodilator-stimulated phosphoprotein a | 555805 |
| ENSDARG00000017105 | vaspb | vasodilator-stimulated phosphoprotein b | 550487 |
| ENSDARG00000056481 | vat1 | vesicle amine transport 1 | 368752 |
| ENSDARG00000073713 | vav3b | vav 3 guanine nucleotide exchange factor b | 0 |
| ENSDARG00000030241 | vbp1 | von Hippel-Lindau binding protein 1 | 553651 |
| ENSDARG00000009401 | vcanb | versican b | 323465 |
| ENSDARG00000044968 | vcla | vinculin a | 552980 |
| ENSDARG00000020008 | vcp | valosin containing protein | 327197 |
| ENSDARG00000045132 | vdac1 | voltage-dependent anion channel 1 | 334582 |
| ENSDARG00000013623 | vdac2 | voltage-dependent anion channel 2 | 322126 |
| ENSDARG00000021564 | VDAC3 (2 of 2) | voltage-dependent anion channel 3 | 393186 |
| ENSDARG00000003695 | vdac3 | voltage-dependent anion channel 3 | 406529 |
| ENSDARG00000044501 | viml | vimentin like | 393746 |
| ENSDARG00000070097 | vimp | VCP-interacting membrane protein | 735249 |
| ENSDARG00000010008 | vim | vimentin | 140599 |
| ENSDARG00000006257 | vldlr | very low density lipoprotein receptor | 393897 |
| ENSDARG00000093945 | VMA21 | VMA21 vacuolar H+-ATPase homolog (S. cerevisiae) | 100307085 |
| ENSDARG00000036338 | vps11 | vacuolar protein sorting 11 | 565852 |
| ENSDARG00000039225 | vps13a | vacuolar protein sorting 13 homolog A (S. cerevisiae) | 378837 |
| ENSDARG00000074465 | VPS13C | vacuolar protein sorting 13 homolog C (S. cerevisiae) | 798746 |
| ENSDARG00000017986 | vps13d | vacuolar protein sorting 13 homolog D (S. cerevisiae) | 567879 |
| ENSDARG00000059902 | vps16 | vacuolar protein sorting protein 16 | 798985 |
| ENSDARG00000070433 | vps18 | vacuolar protein sorting 18 homolog (S. cerevisiae) | 100005887 |
| ENSDARG00000015823 | vps26b | vacuolar protein sorting 26 homolog B | 406684 |
| ENSDARG00000103203 | vps28 | vacuolar protein sorting 28 (yeast) | 393562 |
| ENSDARG00000069521 | vps29 | vacuolar protein sorting 29 homolog (S. cerevisiae) | 573437 |
| ENSDARG00000008224 | vps35 | vacuolar protein sorting 35 homolog (S. cerevisiae) | 561697 |
| ENSDARG00000061180 | vps45 | vacuolar protein sorting 45 homolog (S. cerevisiae) | 561653 |
| ENSDARG00000062016 | vps51 | vacuolar protein sorting 51 homolog (S. cerevisiae) | 559339 |
| ENSDARG00000099258 | vps52 | vacuolar protein sorting 52 homolog (S. cerevisiae) | 0 |
| ENSDARG00000069324 | vps53 | vacuolar protein sorting 53 homolog (S. cerevisiae) | 492817 |
| ENSDARG00000060477 | vps8 | vacuolar protein sorting 8 homolog (S. cerevisiae) | 791134 |
| ENSDARG00000036056 | vrk3 | vaccinia related kinase 3 | 0 |
| ENSDARG00000023228 | vsnl1a | visinin-like 1a | 492494 |
| ENSDARG00000044053 | vsnl1b | visinin-like 1b | 561162 |
| ENSDARG00000077940 | VSTM2B (1 of 2) | V-set and transmembrane domain containing 2B | 793103 |
| ENSDARG00000013732 | vta1 | vesicle (multivesicular body) trafficking 1 | 406414 |
| ENSDARG00000092233 | vtg1 | vitellogenin 1 | 559475 |
| ENSDARG00000055809 | vtg2 | vitellogenin 2 | 559931 |
| ENSDARG00000092126 | vtg5 | vitellogenin 5 | 64260 |
| ENSDARG00000092419 | vtg7 | vitellogenin 7 | 559856 |
| ENSDARG00000045065 | vti1a | vesicle transport through interaction with t-SNAREs 1A | 567385 |
| ENSDARG00000039270 | vti1b | vesicle transport through interaction with t-SNAREs 1B | 394011 |
| ENSDARG00000073905 | VWA5A (2 of 4) | von Willebrand factor A domain containing 5A | 0 |
| ENSDARG00000078593 | vwa8 | von Willebrand factor A domain containing 8 | 561317 |
| ENSDARG00000091457 | WASF1 (2 of 2) | WAS protein family, member 1 | 101885310 |
| ENSDARG00000060349 | wasf1 | WAS protein family, member 1 | 100001221 |
| ENSDARG00000062948 | wasf3b | WAS protein family, member 3b | 563632 |
| ENSDARG00000014113 | wasla | Wiskott-Aldrich syndrome-like a | 100009637 |
| ENSDARG00000042418 | wbscr17 | Williams-Beuren syndrome chromosome region 17 | 567794 |
| ENSDARG00000005425 | wdfy1 | WD repeat and FYVE domain containing 1 | 474326 |
| ENSDARG00000078890 | wdfy3 | WD repeat and FYVE domain containing 3 | 568432 |
| ENSDARG00000075245 | wdr11 | WD repeat domain 11 | 558865 |
| ENSDARG00000075098 | wdr17 | WD repeat domain 17 | 561804 |
| ENSDARG00000037406 | wdr19 | WD repeat domain 19 | 556414 |
| ENSDARG00000062310 | WDR26 (2 of 2) | WD repeat domain 26 | 559461 |
| ENSDARG00000074611 | wdr37 | WD repeat domain 37 | 558075 |
| ENSDARG00000078136 | wdr47a | WD repeat domain 47a | 558528 |
| ENSDARG00000099757 | wdr47b | WD repeat domain 47b | 567031 |
| ENSDARG00000038543 | wdr48 | WD repeat domain 48 | 334682 |
| ENSDARG00000060312 | wdr62 | WD repeat domain 62 | 570949 |
| ENSDARG00000100274 | WDR7 | WD repeat domain 7 | 0 |
| ENSDARG00000074617 | wfs1b | Wolfram syndrome 1b (wolframin) | 556580 |
| ENSDARG00000006712 | wipf2b | WAS/WASL interacting protein family, member 2b | 415255 |
| ENSDARG00000039392 | wnk1b | WNK lysine deficient protein kinase 1b | 561159 |
| ENSDARG00000074271 | wrb | tryptophan rich basic protein | 335756 |
| ENSDARG00000088195 | wu:fb52c12 | wu:fb52c12 | 322178 |
| ENSDARG00000094133 | wu:fc21g02 | wu:fc21g02 | 324205 |
| ENSDARG00000043460 | wu:fj39g12 | wu:fj39g12 | 563937 |
| ENSDARG00000020443 | xkr6a | XK, Kell blood group complex subunit-related family, member 6a | 0 |
| ENSDARG00000043410 | xkr6b | XK, Kell blood group complex subunit-related family, member 6b | 792104 |
| ENSDARG00000026333 | xkr7 | XK, Kell blood group complex subunit-related family, member 7 | 497073 |
| ENSDARG00000029011 | xpnpep1 | X-prolyl aminopeptidase (aminopeptidase P) 1, soluble | 406253 |
| ENSDARG00000071551 | xrcc6 | X-ray repair complementing defective repair in Chinese hamster cells 6 | 334488 |
| ENSDARG00000035913 | yars | tyrosyl-tRNA synthetase | 368235 |
| ENSDARG00000005941 | yes1 | YES proto-oncogene 1, Src family tyrosine kinase | 407620 |
| ENSDARG00000020449 | yipf3 | Yip1 domain family, member 3 | 334511 |
| ENSDARG00000061692 | yjefn3 | YjeF N-terminal domain containing 3 | 557840 |
| ENSDARG00000038308 | ykt6 | YKT6 v-SNARE homolog (S. cerevisiae) | 394067 |
| ENSDARG00000075192 | yme1l1a | YME1-like 1a | 793098 |
| ENSDARG00000104401 | yme1l1b | YME1-like 1b | 557907 |
| ENSDARG00000055510 | ypel3 | yippee-like 3 | 368214 |
| ENSDARG00000013078 | ywhaba | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, beta polypeptide a | 323055 |
| ENSDARG00000075758 | ywhabb | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, beta polypeptide b | 406419 |
| ENSDARG00000040287 | ywhabl | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, beta polypeptide like | 321729 |
| ENSDARG00000006399 | ywhae1 | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, epsilon polypeptide 1 | 322060 |
| ENSDARG00000017014 | ywhae2 | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, epsilon polypeptide 2 | 503763 |
| ENSDARG00000067626 | ywhag1 | 3-monooxygenase/tryptophan 5-monooxygenase activation protein, gamma polypeptide 1 | 117604 |
| ENSDARG00000071658 | ywhag2 | 3-monooxygenase/tryptophan 5-monooxygenase activation protein, gamma polypeptide 2 | 560560 |
| ENSDARG00000005560 | ywhah | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, eta polypeptide | 406443 |
| ENSDARG00000042539 | ywhaqa | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta polypeptide a | 399487 |
| ENSDARG00000023323 | ywhaqb | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta polypeptide b | 335195 |
| ENSDARG00000032575 | ywhaz | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide | 336610 |
| ENSDARG00000027915 | zc2hc1a | zinc finger, C2HC-type containing 1A | 324412 |
| ENSDARG00000070441 | zdhhc17 | zinc finger, DHHC-type containing 17 | 100148543 |
| ENSDARG00000058178 | zdhhc20b | zinc finger, DHHC-type containing 20b | 100001188 |
| ENSDARG00000016263 | zdhhc5a | zinc finger, DHHC-type containing 5a | 571795 |
| ENSDARG00000042872 | zdhhc8a | zinc finger, DHHC-type containing 8a | 555918 |
| ENSDARG00000103453 | zfpl1 | zinc finger protein-like 1 | 393646 |
| ENSDARG00000027887 | zfyve21 | zinc finger, FYVE domain containing 21 | 100004144 |
| ENSDARG00000033599 | zgc:100906 | zgc:100906 | 445123 |
| ENSDARG00000087243 | zgc:100918 | zgc:100918 | 449549 |
| ENSDARG00000086342 | zgc:101566 | zgc:101566 | 449823 |
| ENSDARG00000038876 | zgc:101569 | zgc:101569 | 449822 |
| ENSDARG00000040965 | zgc:101731 | zgc:101731 | 447892 |
| ENSDARG00000038694 | zgc:101744 | zgc:101744 | 541409 |
| ENSDARG00000069600 | zgc:109889 | zgc:109889 | 553542 |
| ENSDARG00000021404 | zgc:110319 | zgc:110319 | 796111 |
| ENSDARG00000046030 | zgc:110339 | zgc:110339 | 550490 |
| ENSDARG00000073845 | zgc:110843 | zgc:110843 | 541492 |
| ENSDARG00000058966 | zgc:112332 | zgc:112332 | 553712 |
| ENSDARG00000003127 | zgc:123105 | zgc:123105 | 664766 |
| ENSDARG00000079932 | zgc:152830 | zgc:152830 | 767682 |
| ENSDARG00000071626 | zgc:153154 | zgc:153154 | 751701 |
| ENSDARG00000096992 | zgc:153405 | zgc:153405 | 751661 |
| ENSDARG00000045979 | zgc:153704 | zgc:153704 | 767759 |
| ENSDARG00000033738 | zgc:153867 | zgc:153867 | 337226 |
| ENSDARG00000069529 | zgc:153981 | zgc:153981 | 563986 |
| ENSDARG00000017785 | zgc:158689 | zgc:158689 | 791177 |
| ENSDARG00000019651 | zgc:162239 | zgc:162239 | 0 |
| ENSDARG00000069846 | zgc:162944 | zgc:162944 | 569993 |
| ENSDARG00000091525 | zgc:173552 | zgc:173552 | 794406 |
| ENSDARG00000029473 | zgc:173994 | zgc:173994 | 571365 |
| ENSDARG00000091136 | zgc:174259 | zgc:174259 | 791587 |
| ENSDARG00000092873 | zgc:193541 | zgc:193541 | 557217 |
| ENSDARG00000039356 | zgc:194209 | zgc:194209 | 570908 |
| ENSDARG00000079847 | zgc:194578 | zgc:194578 | 792915 |
| ENSDARG00000031435 | zgc:56493 | zgc:56493 | 336637 |
| ENSDARG00000017140 | zgc:63587 | zgc:63587 | 393431 |
| ENSDARG00000012281 | zgc:65851 | zgc:65851 | 321113 |
| ENSDARG00000016301 | zgc:65894 | zgc:65894 | 335798 |
| ENSDARG00000101443 | zgc:66014 | zgc:66014 | 393817 |
| ENSDARG00000095464 | zgc:66350 | zgc:66350 | 393493 |
| ENSDARG00000014015 | zgc:77086 | zgc:77086 | 405841 |
| ENSDARG00000014222 | zgc:86598 | zgc:86598 | 415254 |
| ENSDARG00000021287 | zgc:91909 | zgc:91909 | 431725 |
| ENSDARG00000044874 | zgc:92107 | zgc:92107 | 492329 |
| ENSDARG00000041060 | zgc:92326 | zgc:92326 | 327284 |
| ENSDARG00000069734 | zgc:92880 | zgc:92880 | 445037 |
| ENSDARG00000044090 | zmpste24 | zinc metallopeptidase, STE24 homolog | 334380 |
| ENSDARG00000061718 | znf704 | zinc finger protein 704 | 560056 |
| ENSDARG00000018738 | zw10 | zw10 kinetochore protein | 393110 |
| ENSDARG00000075733 | zyx | zyxin | 564246 |
Supplementary Table 2b. Postsynaptic density proteins identified in zebrafish.
Proteins identified in zebrafish whole brain synaptosomal preparations. For each protein the following database identifiers are given: Ensembl Gene, Associated Gene Name, Description and MGI ID.
| ENSEMBL ID | Associated Gene Name | Description | EntrezGene ID |
|---|---|---|---|
| ENSDARG00000011855 | aak1a | AP2 associated kinase 1a | 568999 |
| ENSDARG00000074876 | abcd1 | ATP-binding cassette, sub-family D (ALD), member 1 | 566367 |
| ENSDARG00000059409 | ABCD2 (1 of 2) | ATP-binding cassette, sub-family D (ALD), member 2 | 100537809 |
| ENSDARG00000104085 | abcd3a | ATP-binding cassette, sub-family D (ALD), member 3a | 406803 |
| ENSDARG00000007216 | abce1 | ATP-binding cassette, sub-family E (OABP), member 1 | 406324 |
| ENSDARG00000010155 | abi1a | abl-interactor 1a | 393711 |
| ENSDARG00000062991 | abi1b | abl-interactor 1b | 767738 |
| ENSDARG00000058207 | abi2a | abl-interactor 2a | 767646 |
| ENSDARG00000014875 | abi2b | abl-interactor 2b | 692331 |
| ENSDARG00000060072 | abi3a | ABI family, member 3a | 767807 |
| ENSDARG00000063283 | abi3b | ABI family, member 3b | 566059 |
| ENSDARG00000013841 | abl2 | c-abl oncogene 2, non-receptor tyrosine kinase | 570636 |
| ENSDARG00000060149 | ablim1a | actin binding LIM protein 1a | 100003558 |
| ENSDARG00000045064 | ablim1b | actin binding LIM protein 1b | 541550 |
| ENSDARG00000055223 | ablim2 | actin binding LIM protein family, member 2 | 568454 |
| ENSDARG00000088168 | ablim3 | actin binding LIM protein family, member 3 | 566821 |
| ENSDARG00000045411 | acad11 | acyl-CoA dehydrogenase family, member 11 | 393147 |
| ENSDARG00000045799 | acanb | aggrecan b | 559593 |
| ENSDARG00000034883 | acbd5a | acyl-CoA binding domain containing 5a | 553674 |
| ENSDARG00000031796 | ache | acetylcholinesterase | 114549 |
| ENSDARG00000058352 | acot9.1 | acyl-CoA thioesterase 9, tandem duplicate 1 | 798073 |
| ENSDARG00000020149 | acoxl | acyl-CoA oxidase-like | 569104 |
| ENSDARG00000037746 | actb1 | actin, beta 1 | 57934 |
| ENSDARG00000037870 | actb2 | actin, beta 2 | 57935 |
| ENSDARG00000099786 | ACTN1 (2 of 3) | actinin, alpha 1 | 0 |
| ENSDARG00000007219 | actn1 | actinin, alpha 1 | 560400 |
| ENSDARG00000011611 | actr1 | ARP1 actin-related protein 1, centractin (yeast) | 406681 |
| ENSDARG00000052438 | actr2a | ARP2 actin-related protein 2a homolog (yeast) | 406820 |
| ENSDARG00000091138 | ACTR3 (1 of 2) | ARP3 actin-related protein 3 homolog (yeast) | 0 |
| ENSDARG00000100510 | ACTR3 (2 of 2) | ARP3 actin-related protein 3 homolog (yeast) | 0 |
| ENSDARG00000008790 | actr3b | ARP3 actin-related protein 3 homolog B (yeast) | 100038776 |
| ENSDARG00000079204 | adam11 | ADAM metallopeptidase domain 11 | 567145 |
| ENSDARG00000060532 | adam22 | ADAM metallopeptidase domain 22 | 100126805 |
| ENSDARG00000062323 | adam23a | ADAM metallopeptidase domain 23a | 794175 |
| ENSDARG00000003544 | adarb1b | adenosine deaminase, RNA-specific, B1b | 558759 |
| ENSDARG00000062561 | adck1 | aarF domain containing kinase 1 | 100144553 |
| ENSDARG00000069026 | adcy1a | adenylate cyclase 1a | 557026 |
| ENSDARG00000088634 | adcy1b | adenylate cyclase 1b | 0 |
| ENSDARG00000077145 | ADCY3 | adenylate cyclase 3 | 571825 |
| ENSDARG00000027740 | adcyap1b | adenylate cyclase activating polypeptide 1b | 335625 |
| ENSDARG00000103962 | add1 | adducin 1 (alpha) | 555511 |
| ENSDARG00000074581 | add2 | adducin 2 (beta) | 570147 |
| ENSDARG00000040874 | add3a | adducin 3 (gamma) a | 556762 |
| ENSDARG00000056250 | add3b | adducin 3 (gamma) b | 553243 |
| ENSDARG00000054177 | adgra1 | adhesion G protein-coupled receptor A1 | 560847 |
| ENSDARG00000075133 | adgrb1a | adhesion G protein-coupled receptor B1a | 563527 |
| ENSDARG00000078529 | adgrb1b | adhesion G protein-coupled receptor B1b | 0 |
| ENSDARG00000059832 | adgrb3 | adhesion G protein-coupled receptor B3 | 0 |
| ENSDARG00000089292 | adgrl1a | adhesion G protein-coupled receptor L1a | 0 |
| ENSDARG00000075899 | adgrl2b.1 | adhesion G protein-coupled receptor L2b, tandem duplicate 1 | 101882597 |
| ENSDARG00000062123 | agap1 | ArfGAP with GTPase domain, ankyrin repeat and PH domain 1 | 100148154 |
| ENSDARG00000099874 | agap2 | ArfGAP with GTPase domain, ankyrin repeat and PH domain 2 | 565109 |
| ENSDARG00000087822 | AGAP3 | ArfGAP with GTPase domain, ankyrin repeat and PH domain 3 | 0 |
| ENSDARG00000069334 | AGFG2 | ArfGAP with FG repeats 2 | 0 |
| ENSDARG00000092644 | ago1 | argonaute RISC catalytic component 1 | 570775 |
| ENSDARG00000063079 | ago3b | argonaute RISC catalytic component 3b | 568159 |
| ENSDARG00000101139 | agpat2 | 1-acylglycerol-3-phosphate O-acyltransferase 2 (lysophosphatidic acid acyltransferase, beta) | 777624 |
| ENSDARG00000042821 | agps | alkylglycerone phosphate synthase | 386801 |
| ENSDARG00000079388 | agrn | agrin | 565373 |
| ENSDARG00000061764 | ahnak | AHNAK nucleoprotein | 559276 |
| ENSDARG00000062780 | AIFM3 | apoptosis-inducing factor, mitochondrion-associated, 3 | 100150876 |
| ENSDARG00000060036 | aimp1 | aminoacyl tRNA synthetase complex-interacting multifunctional protein 1 | 562232 |
| ENSDARG00000018903 | aimp2 | aminoacyl tRNA synthetase complex-interacting multifunctional protein 2 | 406647 |
| ENSDARG00000038655 | ajap1 | adherens junctions associated protein 1 | 0 |
| ENSDARG00000006546 | ak4 | adenylate kinase 4 | 406590 |
| ENSDARG00000012555 | ak5 | adenylate kinase 5 | 100003595 |
| ENSDARG00000079610 | akap9 | A kinase (PRKA) anchor protein 9 | 0 |
| ENSDARG00000037935 | aldh16a1 | aldehyde dehydrogenase 16 family, member A1 | 492710 |
| ENSDARG00000099579 | aldh18a1 | aldehyde dehydrogenase 18 family, member A1 | 557186 |
| ENSDARG00000074791 | aldh3a1 | aldehyde dehydrogenase 3 family, member A1 | 100000026 |
| ENSDARG00000028259 | aldh3a2a | aldehyde dehydrogenase 3 family, member A2a | 323653 |
| ENSDARG00000011665 | aldoaa | aldolase a, fructose-bisphosphate, a | 336425 |
| ENSDARG00000034470 | aldoab | aldolase a, fructose-bisphosphate, b | 406496 |
| ENSDARG00000057661 | aldoca | aldolase C, fructose-bisphosphate, a | 792692 |
| ENSDARG00000015546 | alpl | alkaline phosphatase, liver/bone/kidney | 393982 |
| ENSDARG00000077732 | alyref | Aly/REF export factor | 560127 |
| ENSDARG00000009026 | ANK2 (1 of 2) | ankyrin 2, neuronal | 568926 |
| ENSDARG00000043313 | ank2b | ankyrin 2b, neuronal | 450043 |
| ENSDARG00000061736 | ank3a | ankyrin 3a | 103909035 |
| ENSDARG00000077582 | ank3b | ankyrin 3b | 100126016 |
| ENSDARG00000076829 | ankib1b | ankyrin repeat and IBR domain containing 1b | 561277 |
| ENSDARG00000062103 | ankrd24 | ankyrin repeat domain 24 | 565326 |
| ENSDARG00000057159 | ankrd29 | ankyrin repeat domain 29 | 553798 |
| ENSDARG00000068274 | ANKRD65 | ankyrin repeat domain 65 | 566073 |
| ENSDARG00000062396 | anks1aa | ankyrin repeat and sterile alpha motif domain containing 1Aa | 0 |
| ENSDARG00000078901 | anks1ab | ankyrin repeat and sterile alpha motif domain containing 1Ab | 100003659 |
| ENSDARG00000003512 | anks1b | ankyrin repeat and sterile alpha motif domain containing 1B | 565408 |
| ENSDARG00000020621 | ap1b1 | adaptor-related protein complex 1, beta 1 subunit | 562166 |
| ENSDARG00000102394 | ap1g1 | adaptor-related protein complex 1, gamma 1 subunit | 324466 |
| ENSDARG00000054337 | ap1g2 | adaptor-related protein complex 1, gamma 2 subunit | 767712 |
| ENSDARG00000096249 | ap1m1 | adaptor-related protein complex 1, mu 1 subunit | 100330411 |
| ENSDARG00000096454 | ap1m2 | adaptor-related protein complex 1, mu 2 subunit | 403021 |
| ENSDARG00000056803 | ap1s1 | adaptor-related protein complex 1, sigma 1 subunit | 393279 |
| ENSDARG00000058504 | ap1s2 | adaptor-related protein complex 1, sigma 2 subunit | 327237 |
| ENSDARG00000054320 | ap2a1 | adaptor-related protein complex 2, alpha 1 subunit | 558153 |
| ENSDARG00000035152 | ap2b1 | adaptor-related protein complex 2, beta 1 subunit | 334632 |
| ENSDARG00000002790 | ap2m1a | adaptor-related protein complex 2, mu 1 subunit, a | 321051 |
| ENSDARG00000033899 | ap2m1b | adaptor-related protein complex 2, mu 1 subunit, b | 394001 |
| ENSDARG00000026967 | ap2s1 | adaptor-related protein complex 2, sigma 1 subunit | 100148085 |
| ENSDARG00000016128 | ap3m2 | adaptor-related protein complex 3, mu 2 subunit | 415244 |
| ENSDARG00000058868 | apc | adenomatous polyposis coli | 386762 |
| ENSDARG00000060734 | appl1 | adaptor protein, phosphotyrosine interaction, PH domain and leucine zipper containing 1 | 571540 |
| ENSDARG00000078139 | appl2 | adaptor protein, phosphotyrosine interaction, PH domain and leucine zipper containing 2 | 100000135 |
| ENSDARG00000023713 | aqp1a.1 | aquaporin 1a (Colton blood group), tandem duplicate 1 | 335821 |
| ENSDARG00000031214 | arcn1b | archain 1b | 399661 |
| ENSDARG00000075673 | arhgap21b | Rho GTPase activating protein 21b | 326745 |
| ENSDARG00000052950 | arhgap23a | Rho GTPase activating protein 23a | 100006233 |
| ENSDARG00000059472 | arhgap31 | Rho GTPase activating protein 31 | 101885883 |
| ENSDARG00000011333 | arhgap33 | Rho GTPase activating protein 33 | 569720 |
| ENSDARG00000104086 | arhgap39 | Rho GTPase activating protein 39 | 101885231 |
| ENSDARG00000104549 | arhgap44 | Rho GTPase activating protein 44 | 0 |
| ENSDARG00000061294 | arhgap5 | Rho GTPase activating protein 5 | 795752 |
| ENSDARG00000040380 | arhgef1a | Rho guanine nucleotide exchange factor (GEF) 1a | 368266 |
| ENSDARG00000006299 | arhgef7a | Rho guanine nucleotide exchange factor (GEF) 7a | 553493 |
| ENSDARG00000061746 | arhgef9a | Cdc42 guanine nucleotide exchange factor (GEF) 9a | 559868 |
| ENSDARG00000078624 | arhgef9b | Cdc42 guanine nucleotide exchange factor (GEF) 9b | 560923 |
| ENSDARG00000008383 | arpc1a | actin related protein 2/3 complex, subunit 1A | 394243 |
| ENSDARG00000075989 | arpc2 | actin related protein 2/3 complex, subunit 2 | 327068 |
| ENSDARG00000057882 | arpc3 | actin related protein 2/3 complex, subunit 3 | 415204 |
| ENSDARG00000058225 | arpc4l | actin related protein 2/3 complex, subunit 4, like | 114456 |
| ENSDARG00000039142 | arpc5a | actin related protein 2/3 complex, subunit 5A | 336504 |
| ENSDARG00000019062 | arpc5b | actin related protein 2/3 complex, subunit 5B | 399482 |
| ENSDARG00000041720 | arpc5la | actin related protein 2/3 complex, subunit 5-like, a | 393550 |
| ENSDARG00000035433 | arpc5lb | actin related protein 2/3 complex, subunit 5-like, b | 556104 |
| ENSDARG00000074329 | arvcfa | armadillo repeat gene deleted in velocardiofacial syndrome a | 572216 |
| ENSDARG00000061688 | arvcfb | armadillo repeat gene deleted in velocardiofacial syndrome b | 564669 |
| ENSDARG00000105115 | ASIC1 (2 of 2) | acid sensing (proton gated) ion channel 1 | 0 |
| ENSDARG00000008329 | asic1a | acid-sensing (proton-gated) ion channel 1a | 791696 |
| ENSDARG00000078703 | ASPH | aspartate beta-hydroxylase | 559236 |
| ENSDARG00000059629 | asphd2 | aspartate beta-hydroxylase domain containing 2 | 569819 |
| ENSDARG00000068323 | astn1 | astrotactin 1 | 791157 |
| ENSDARG00000023267 | atad1a | ATPase family, AAA domain containing 1a | 368672 |
| ENSDARG00000086848 | atad3b | ATPase family, AAA domain containing 3B | 0 |
| ENSDARG00000029931 | atp13a1 | ATPase type 13A1 | 334466 |
| ENSDARG00000104139 | atp1a3b | ATPase, Na+/K+ transporting, alpha 3b polypeptide | 64611 |
| ENSDARG00000041417 | atp6ap1a | ATPase, H+ transporting, lysosomal accessory protein 1a | 100004566 |
| ENSDARG00000037153 | atp6ap1b | ATPase, H+ transporting, lysosomal accessory protein 1b | 195824 |
| ENSDARG00000008735 | atp6ap2 | ATPase, H+ transporting, lysosomal accessory protein 2 | 406296 |
| ENSDARG00000015174 | atp6v0a1b | ATPase, H+ transporting, lysosomal V0 subunit a1b | 553691 |
| ENSDARG00000035538 | atp6v0a2a | ATPase, H+ transporting, lysosomal V0 subunit a2a | 561469 |
| ENSDARG00000057853 | atp6v0ca | ATPase, H+ transporting, lysosomal, V0 subunit ca | 192336 |
| ENSDARG00000036577 | atp6v0cb | ATPase, H+ transporting, lysosomal, V0 subunit cb | 325402 |
| ENSDARG00000069090 | atp6v0d1 | ATPase, H+ transporting, lysosomal V0 subunit d1 | 322811 |
| ENSDARG00000043465 | atp6v1b2 | ATPase, H+ transporting, lysosomal, V1 subunit B2 | 359840 |
| ENSDARG00000013443 | atp6v1ba | ATPase, H+ transporting, lysosomal, V1 subunit B, member a | 359839 |
| ENSDARG00000023967 | atp6v1c1a | ATPase, H+ transporting, lysosomal, V1 subunit C1a | 368907 |
| ENSDARG00000011175 | atp6v1d | ATPase, H+ transporting, lysosomal V1 subunit D | 393935 |
| ENSDARG00000030694 | atp6v1e1b | ATPase, H+ transporting, lysosomal, V1 subunit E1b | 192335 |
| ENSDARG00000045543 | atp6v1f | ATPase, H+ transporting, lysosomal, V1 subunit F | 436799 |
| ENSDARG00000022315 | atp6v1g1 | ATPase, H+ transporting, lysosomal, V1 subunit G1 | 335025 |
| ENSDARG00000006370 | atp6v1h | ATPase, H+ transporting, lysosomal V1 subunit H | 286779 |
| ENSDARG00000062521 | atp9b | ATPase, class II, type 9B | 568160 |
| ENSDARG00000001939 | b3gat3 | beta-1,3-glucuronyltransferase 3 (glucuronosyltransferase I) | 192334 |
| ENSDARG00000076701 | b4galnt4b | beta-1,4-N-acetyl-galactosaminyl transferase 4b | 559305 |
| ENSDARG00000039486 | bag3 | BCL2-associated athanogene 3 | 445139 |
| ENSDARG00000017316 | bag5 | BCL2-associated athanogene 5 | 569580 |
| ENSDARG00000062799 | baiap2a | BAI1-associated protein 2a | 678518 |
| ENSDARG00000103333 | baiap2b | BAI1-associated protein 2b | 100537012 |
| ENSDARG00000031119 | baiap2l1b | BAI1-associated protein 2-like 1b | 550528 |
| ENSDARG00000037009 | banf1 | barrier to autointegration factor 1 | 334717 |
| ENSDARG00000053358 | basp1 | brain abundant, membrane attached signal protein 1 | 569631 |
| ENSDARG00000053979 | bc2 | putative breast adenocarcinoma marker | 798901 |
| ENSDARG00000044972 | bcap31 | B-cell receptor-associated protein 31 | 368488 |
| ENSDARG00000016904 | bckdk | branched chain ketoacid dehydrogenase kinase | 282677 |
| ENSDARG00000043673 | BEGAIN | brain-enriched guanylate kinase-associated | 563117 |
| ENSDARG00000010108 | bri3bp | bri3 binding protein | 327046 |
| ENSDARG00000078302 | BRINP1 | bone morphogenetic protein/retinoic acid inducible neural-specific 1 | 100334375 |
| ENSDARG00000014302 | brinp2 | bone morphogenetic protein/retinoic acid inducible neural-specific 2 | 561009 |
| ENSDARG00000041974 | brk1 | BRICK1, SCAR/WAVE actin-nucleating complex subunit | 415187 |
| ENSDARG00000061421 | BRSK2 (1 of 2) | BR serine/threonine kinase 2 | 0 |
| ENSDARG00000079161 | bsnb | bassoon (presynaptic cytomatrix protein) b | 100329660 |
| ENSDARG00000063255 | btbd11a | BTB (POZ) domain containing 11a | 562893 |
| ENSDARG00000063040 | btbd11b | BTB (POZ) domain containing 11b | 100034519 |
| ENSDARG00000099321 | btbd17a | BTB (POZ) domain containing 17a | 563263 |
| ENSDARG00000102472 | btbd17b | BTB (POZ) domain containing 17b | 563672 |
| ENSDARG00000090615 | BX957297.1 | Uncharacterized protein | 567192 |
| ENSDARG00000089562 | BX957322.1 | Uncharacterized protein | 100006797 |
| ENSDARG00000040582 | C18H3orf33 | chromosome 3 open reading frame 33 | 565816 |
| ENSDARG00000039887 | c1qbp | complement component 1, q subcomponent binding protein | 334619 |
| ENSDARG00000030038 | C20H12orf57 | chromosome 12 open reading frame 57 | 323559 |
| ENSDARG00000043842 | C20H1orf198 | chromosome 1 open reading frame 198 | 568408 |
| ENSDARG00000057556 | C21H18orf32 | chromosome 18 open reading frame 32 | 336641 |
| ENSDARG00000029307 | C24H18orf8 | chromosome 18 open reading frame 8 | 0 |
| ENSDARG00000095034 | C24H8orf34 | chromosome 8 open reading frame 34 | 557144 |
| ENSDARG00000100152 | C25H12orf75 | chromosome 12 open reading frame 75 | 100535414 |
| ENSDARG00000036888 | C25HXorf38 (1 of 2) | chromosome X open reading frame 38 | 777611 |
| ENSDARG00000061066 | c2cd2l | c2cd2-like | 566488 |
| ENSDARG00000079556 | C2CD4C (1 of 2) | C2 calcium-dependent domain containing 4C | 568754 |
| ENSDARG00000094590 | c2cd5 | C2 calcium-dependent domain containing 5 | 100333813 |
| ENSDARG00000061257 | C3H17orf75 | chromosome 17 open reading frame 75 | 751673 |
| ENSDARG00000015065 | c4 | complement component 4 | 566261 |
| ENSDARG00000076268 | C5H9orf172 (1 of 2) | chromosome 9 open reading frame 172 | 568961 |
| ENSDARG00000078891 | C7H7orf43 | chromosome 7 open reading frame 43 | 100003958 |
| ENSDARG00000076873 | C8H9orf172 (2 of 2) | chromosome 9 open reading frame 172 | 799850 |
| ENSDARG00000052644 | ca10a | carbonic anhydrase Xa | 641327 |
| ENSDARG00000043589 | ca4a | carbonic anhydrase IV a | 555196 |
| ENSDARG00000073822 | CABZ01046283.1 | Uncharacterized protein | 0 |
| ENSDARG00000027564 | CABZ01052588.1 | Non-specific serine/threonine protein kinase | 0 |
| ENSDARG00000061660 | CABZ01055347.1 | Uncharacterized protein | 0 |
| ENSDARG00000025074 | CABZ01059390.1 | Uncharacterized protein | 0 |
| ENSDARG00000102813 | CABZ01069436.1 | 0 | 101883099 |
| ENSDARG00000104937 | CABZ01084323.1 | 0 | 100330629 |
| ENSDARG00000059349 | CABZ01090374.1 | Uncharacterized protein | 0 |
| ENSDARG00000069046 | CABZ01118678.1 | Uncharacterized protein | 0 |
| ENSDARG00000037905 | cacna1aa | calcium channel, voltage-dependent, P/Q type, alpha 1A subunit, a | 562059 |
| ENSDARG00000006923 | cacna1ab | calcium channel, voltage-dependent, P/Q type, alpha 1A subunit, b | 569528 |
| ENSDARG00000079295 | cacna1bb | calcium channel, voltage-dependent, N type, alpha 1B subunit, b | 562796 |
| ENSDARG00000014804 | cacna2d1a | calcium channel, voltage-dependent, alpha 2/delta subunit 1a | 561434 |
| ENSDARG00000039182 | cacna2d2a | calcium channel, voltage-dependent, alpha 2/delta subunit 2a | 541376 |
| ENSDARG00000078169 | cacna2d2b | calcium channel, voltage-dependent, alpha 2/delta subunit 2b | 568759 |
| ENSDARG00000078760 | cacna2d3 | calcium channel, voltage dependent, alpha2/delta subunit 3 | 100148645 |
| ENSDARG00000002167 | cacnb1 | calcium channel, voltage-dependent, beta 1 subunit | 436925 |
| ENSDARG00000055565 | cacnb2b | calcium channel, voltage-dependent, beta 2b | 0 |
| ENSDARG00000001881 | cacnb3a | calcium channel, voltage-dependent, beta 3a | 565158 |
| ENSDARG00000076030 | cacnb3b | calcium channel, voltage-dependent, beta 3b | 559015 |
| ENSDARG00000009738 | cacnb4a | calcium channel, voltage-dependent, beta 4a subunit | 562422 |
| ENSDARG00000062174 | cacnb4b | calcium channel, voltage-dependent, beta 4b subunit | 678560 |
| ENSDARG00000032565 | cacng2a | calcium channel, voltage-dependent, gamma subunit 2a | 393614 |
| ENSDARG00000102376 | cacng2b | calcium channel, voltage-dependent, gamma subunit 2b | 569855 |
| ENSDARG00000076401 | cacng3b | calcium channel, voltage-dependent, gamma subunit 3b | 564883 |
| ENSDARG00000020450 | cacng8a | calcium channel, voltage-dependent, gamma subunit 8a | 550428 |
| ENSDARG00000070626 | cacng8b | calcium channel, voltage-dependent, gamma subunit 8b | 100003329 |
| ENSDARG00000015050 | calm2b | calmodulin 2b, (phosphorylase kinase, delta) | 192322 |
| ENSDARG00000053617 | camk2a | calcium/calmodulin-dependent protein kinase II alpha | 550436 |
| ENSDARG00000011065 | camk2b1 | calcium/calmodulin-dependent protein kinase (CaM kinase) II beta 1 | 562064 |
| ENSDARG00000100089 | camk2b2 | calcium/calmodulin-dependent protein kinase (CaM kinase) II beta 2 | 0 |
| ENSDARG00000043010 | camk2d1 | calcium/calmodulin-dependent protein kinase (CaM kinase) II delta 1 | 445208 |
| ENSDARG00000014273 | camk2d2 | calcium/calmodulin-dependent protein kinase (CaM kinase) II delta 2 | 436815 |
| ENSDARG00000071395 | camk2g1 | calcium/calmodulin-dependent protein kinase (CaM kinase) II gamma 1 | 791855 |
| ENSDARG00000056206 | camk2g2 | calcium/calmodulin-dependent protein kinase (CaM kinase) II gamma 2 | 791855 |
| ENSDARG00000027963 | camkva | CaM kinase-like vesicle-associated a | 567243 |
| ENSDARG00000057000 | camkvl | CaM kinase-like vesicle-associated, like | 553431 |
| ENSDARG00000062173 | camsap2a | calmodulin regulated spectrin-associated protein family, member 2a | 562741 |
| ENSDARG00000103521 | capsla | calcyphosine-like a | 550371 |
| ENSDARG00000034240 | capza1a | capping protein (actin filament) muscle Z-line, alpha 1a | 327029 |
| ENSDARG00000056090 | capza1b | capping protein (actin filament) muscle Z-line, alpha 1b | 474327 |
| ENSDARG00000100252 | capzb | capping protein (actin filament) muscle Z-line, beta | 335083 |
| ENSDARG00000060829 | car15 | carbonic anhydrase 15 | 568143 |
| ENSDARG00000036164 | cars | cysteinyl-tRNA synthetase | 368284 |
| ENSDARG00000045832 | cart2 | cocaine- and amphetamine-regulated transcript 2 | 550232 |
| ENSDARG00000074490 | casc4 | cancer susceptibility candidate 4 | 555654 |
| ENSDARG00000059809 | caska | calcium/calmodulin-dependent serine protein kinase a | 259195 |
| ENSDARG00000100119 | caskb | calcium/calmodulin-dependent serine protein kinase b | 0 |
| ENSDARG00000046107 | CASKIN1 (1 of 2) | CASK interacting protein 1 | 563678 |
| ENSDARG00000008982 | casq2 | calsequestrin 2 | 114410 |
| ENSDARG00000036281 | CC2D1A | coiled-coil and C2 domain containing 1A | 0 |
| ENSDARG00000052379 | ccdc22 | coiled-coil domain containing 22 | 556259 |
| ENSDARG00000100052 | cckb | cholecystokinin b | 100000095 |
| ENSDARG00000063677 | ccny | cyclin Y | 557738 |
| ENSDARG00000056559 | ccz1 | CCZ1 vacuolar protein trafficking and biogenesis associated homolog (S. cerevisiae) | 393103 |
| ENSDARG00000079191 | CD200 (2 of 3) | CD200 molecule | 571844 |
| ENSDARG00000016464 | CDC42BPA (1 of 2) | CDC42 binding protein kinase alpha (DMPK-like) | 566003 |
| ENSDARG00000019383 | cdc42bpb | CDC42 binding protein kinase beta (DMPK-like) | 567039 |
| ENSDARG00000055843 | cdh10a | cadherin 10, type 2a (T2-cadherin) | 568370 |
| ENSDARG00000021442 | cdh11 | cadherin 11, type 2, OB-cadherin (osteoblast) | 30461 |
| ENSDARG00000014215 | cdh13 | cadherin 13, H-cadherin (heart) | 558732 |
| ENSDARG00000077996 | cdh24b | cadherin 24, type 2b | 0 |
| ENSDARG00000018693 | cdh2 | cadherin 2, type 1, N-cadherin (neuronal) | 30291 |
| ENSDARG00000015002 | cdh4 | cadherin 4, type 1, R-cadherin (retinal) | 30297 |
| ENSDARG00000014522 | cdh6 | cadherin 6 | 541393 |
| ENSDARG00000023542 | cdh7 | cadherin 7, type 2 | 567704 |
| ENSDARG00000037478 | cdh8 | cadherin 8 | 0 |
| ENSDARG00000012204 | CDK18 | cyclin-dependent kinase 18 | 100150007 |
| ENSDARG00000056683 | cdk5 | cyclin-dependent kinase 5 | 65234 |
| ENSDARG00000045087 | cdk5r1b | cyclin-dependent kinase 5, regulatory subunit 1b (p35) | 436788 |
| ENSDARG00000015240 | cdkl5 | cyclin-dependent kinase-like 5 | 559341 |
| ENSDARG00000055133 | cenpf | centromere protein F | 497403 |
| ENSDARG00000002991 | cep135 | centrosomal protein 135 | 553303 |
| ENSDARG00000006128 | cep170aa | centrosomal protein 170Aa | 557463 |
| ENSDARG00000058892 | cep170ab | centrosomal protein 170Ab | 566358 |
| ENSDARG00000078327 | CEP170B (2 of 2) | centrosomal protein 170B | 100318353 |
| ENSDARG00000020834 | CEP250 | centrosomal protein 250kDa | 564101 |
| ENSDARG00000101510 | cetn2 | centrin, EF-hand protein, 2 | 100006257 |
| ENSDARG00000055607 | cgna | cingulin a | 561423 |
| ENSDARG00000032919 | chchd3a | coiled-coil-helix-coiled-coil-helix domain containing 3a | 407730 |
| ENSDARG00000102643 | chmp1a | charged multivesicular body protein 1A | 393535 |
| ENSDARG00000099624 | chmp1b | chromatin modifying protein 1B | 336426 |
| ENSDARG00000016255 | chmp4ba | charged multivesicular body protein 4Ba | 393164 |
| ENSDARG00000102024 | chmp6b | charged multivesicular body protein 6b | 445397 |
| ENSDARG00000075007 | chst2a | carbohydrate (N-acetylglucosamine-6-O) sulfotransferase 2a | 561896 |
| ENSDARG00000044341 | chst7 | carbohydrate (N-acetylglucosamine 6-O) sulfotransferase 7 | 563748 |
| ENSDARG00000103987 | cib1 | calcium and integrin binding 1 (calmyrin) | 100536377 |
| ENSDARG00000013351 | cirbpb | cold inducible RNA binding protein b | 678563 |
| ENSDARG00000052703 | cisd2 | CDGSH iron sulfur domain 2 | 393354 |
| ENSDARG00000088825 | CIT (1 of 2) | citron rho-interacting serine/threonine kinase | 0 |
| ENSDARG00000089856 | CIT (2 of 2) | citron rho-interacting serine/threonine kinase | 0 |
| ENSDARG00000102674 | ckap5 | cytoskeleton associated protein 5 | 492614 |
| ENSDARG00000016598 | ckmt1 | creatine kinase, mitochondrial 1 | 321892 |
| ENSDARG00000069615 | ckmt2a | creatine kinase, mitochondrial 2a (sarcomeric) | 327506 |
| ENSDARG00000039929 | ckmt2b | creatine kinase, mitochondrial 2b (sarcomeric) | 799492 |
| ENSDARG00000104358 | CLASP1 (2 of 2) | cytoplasmic linker associated protein 1 | 0 |
| ENSDARG00000010280 | clasp1a | cytoplasmic linker associated protein 1a | 562077 |
| ENSDARG00000020345 | clasp2 | cytoplasmic linker associated protein 2 | 404604 |
| ENSDARG00000032577 | clcn6 | chloride channel 6 | 568122 |
| ENSDARG00000104439 | cldnd1a | claudin domain containing 1a | 335628 |
| ENSDARG00000042357 | cldnk | claudin k | 445070 |
| ENSDARG00000059596 | clip2 | CAP-GLY domain containing linker protein 2 | 556406 |
| ENSDARG00000054456 | clip3 | CAP-GLY domain containing linker protein 3 | 562450 |
| ENSDARG00000096867 | clmn | calmin | 100330495 |
| ENSDARG00000021048 | clptm1l | CLPTM1-like | 436653 |
| ENSDARG00000052020 | cmtm4 | CKLF-like MARVEL transmembrane domain containing 4 | 436765 |
| ENSDARG00000074480 | cnksr2a | connector enhancer of kinase suppressor of Ras 2a | 561623 |
| ENSDARG00000073707 | cnksr2b | connector enhancer of kinase suppressor of Ras 2b | 558834 |
| ENSDARG00000004174 | cnot1 | CCR4-NOT transcription complex, subunit 1 | 448949 |
| ENSDARG00000045685 | cntn1b | contactin 1b | 541474 |
| ENSDARG00000000472 | cntn2 | contactin 2 | 0 |
| ENSDARG00000013647 | cntn3a.2 | contactin 3a, tandem duplicate 2 | 100000688 |
| ENSDARG00000087843 | CNTN4 (2 of 3) | contactin 4 | 0 |
| ENSDARG00000098161 | cntn4 | contactin 4 | 571349 |
| ENSDARG00000021584 | cntn5 | contactin 5 | 327429 |
| ENSDARG00000074524 | cntnap1 | contactin associated protein 1 | 566220 |
| ENSDARG00000074558 | cntnap2b | contactin associated protein-like 2b | 563345 |
| ENSDARG00000078322 | col12a1a | collagen, type XII, alpha 1a | 100149543 |
| ENSDARG00000020007 | col1a2 | collagen, type I, alpha 2 | 336471 |
| ENSDARG00000070393 | COQ10A | coenzyme Q10 homolog A (S. cerevisiae) | 100002536 |
| ENSDARG00000054610 | coro1a | coronin, actin binding protein, 1A | 100002113 |
| ENSDARG00000008660 | coro1b | coronin, actin binding protein, 1B | 560168 |
| ENSDARG00000035598 | coro1ca | coronin, actin binding protein, 1Ca | 317741 |
| ENSDARG00000021193 | coro1cb | coronin, actin binding protein, 1Cb | 562849 |
| ENSDARG00000005210 | coro2a | coronin, actin binding protein, 2A | 323538 |
| ENSDARG00000079440 | coro2ba | coronin, actin binding protein, 2Ba | 560235 |
| ENSDARG00000039136 | cox16 | COX16 cytochrome c oxidase assembly homolog (S. cerevisiae) | 550257 |
| ENSDARG00000032970 | cox4i1 | cytochrome c oxidase subunit IV isoform 1 | 326975 |
| ENSDARG00000012388 | cox4i1l | cytochrome c oxidase subunit IV isoform 1, like | 402880 |
| ENSDARG00000022509 | cox4i2 | cytochrome c oxidase subunit IV isoform 2 | 393776 |
| ENSDARG00000088383 | cox5aa | cytochrome c oxidase subunit Vaa | 554101 |
| ENSDARG00000022438 | cox6a1 | cytochrome c oxidase subunit VIa polypeptide 1 | 335774 |
| ENSDARG00000038577 | cox6c | cytochrome c oxidase subunit VIc | 494577 |
| ENSDARG00000053217 | cox7a2a | cytochrome c oxidase, subunit VIIa 2a | 554103 |
| ENSDARG00000054907 | cox7a3 | cytochrome c oxidase subunit VIIa polypeptide 3 | 335751 |
| ENSDARG00000052604 | CPEB2 | cytoplasmic polyadenylation element binding protein 2 | 100330483 |
| ENSDARG00000007753 | cpne2 | copine II | 565225 |
| ENSDARG00000061466 | CPNE5 (1 of 2) | copine V | 568003 |
| ENSDARG00000070919 | cpne5 | copine V | 394003 |
| ENSDARG00000102584 | cpne7 | copine VII | 564556 |
| ENSDARG00000102191 | CPNE8 (2 of 2) | copine VIII | 101886030 |
| ENSDARG00000025189 | cpne8 | copine VIII | 100332028 |
| ENSDARG00000018618 | cpsf6 | cleavage and polyadenylation specific factor 6 | 327069 |
| ENSDARG00000059770 | cpt1aa | carnitine palmitoyltransferase 1Aa (liver) | 558088 |
| ENSDARG00000062054 | cpt1ab | carnitine palmitoyltransferase 1Ab (liver) | 560000 |
| ENSDARG00000074367 | CR381676.1 | Ubiquitin carboxyl-terminal hydrolase | 559878 |
| ENSDARG00000086969 | CR774179.1 | Uncharacterized protein | 0 |
| ENSDARG00000016545 | CRAT (1 of 2) | carnitine O-acetyltransferase | 562075 |
| ENSDARG00000098782 | cratb | carnitine O-acetyltransferase b | 447855 |
| ENSDARG00000059826 | crtac1a | cartilage acidic protein 1a | 559626 |
| ENSDARG00000052114 | crtc3 | CREB regulated transcription coactivator 3 | 0 |
| ENSDARG00000101380 | cryabb | crystallin, alpha B, b | 436943 |
| ENSDARG00000052674 | csnk1a1 | casein kinase 1, alpha 1 | 192307 |
| ENSDARG00000008370 | csnk1da | casein kinase 1, delta a | 322106 |
| ENSDARG00000045150 | csnk1e | casein kinase 1, epsilon | 100006858 |
| ENSDARG00000104342 | csnk1g1 | casein kinase 1, gamma 1 | 494092 |
| ENSDARG00000034056 | csnk1g2b | casein kinase 1, gamma 2b | 561724 |
| ENSDARG00000002082 | csnk2a1 | casein kinase 2, alpha 1 polypeptide | 30502 |
| ENSDARG00000012818 | csnk2a2a | casein kinase 2, alpha prime polypeptide a | 30488 |
| ENSDARG00000013582 | csnk2a2b | casein kinase 2, alpha prime polypeptide b | 556379 |
| ENSDARG00000077776 | csnk2b | casein kinase 2, beta polypeptide | 30428 |
| ENSDARG00000077343 | CT573264.1 | Uncharacterized protein | 100536351 |
| ENSDARG00000074376 | CT737192.1 | Uncharacterized protein | 0 |
| ENSDARG00000057007 | ctbp1 | C-terminal binding protein 1 | 65229 |
| ENSDARG00000044062 | ctbp2a | C-terminal binding protein 2a | 65230 |
| ENSDARG00000102441 | ctnna1 | catenin (cadherin-associated protein), alpha 1 | 30741 |
| ENSDARG00000024785 | ctnna2 | catenin (cadherin-associated protein), alpha 2 | 567500 |
| ENSDARG00000018162 | ctnnal1 | catenin (cadherin-associated protein), alpha-like 1 | 100034581 |
| ENSDARG00000014571 | ctnnb1 | catenin (cadherin-associated protein), beta 1 | 30265 |
| ENSDARG00000023472 | ctnnb2 | catenin, beta 2 | 0 |
| ENSDARG00000078233 | ctnnd1 | catenin (cadherin-associated protein), delta 1 | 556726 |
| ENSDARG00000062415 | ctnnd2a | catenin (cadherin-associated protein), delta 2a | 368485 |
| ENSDARG00000003779 | ctnnd2b | catenin (cadherin-associated protein), delta 2b | 558069 |
| ENSDARG00000040991 | cttnbp2 | cortactin binding protein 2 | 555837 |
| ENSDARG00000011777 | cttn | cortactin | 0 |
| ENSDARG00000054748 | cuedc1b | CUE domain containing 1b | 556847 |
| ENSDARG00000019239 | cul1a | cullin 1a | 323883 |
| ENSDARG00000038967 | cul3a | cullin 3a | 324655 |
| ENSDARG00000069742 | cul5a | cullin 5a | 327215 |
| ENSDARG00000043134 | cux1b | cut-like homeobox 1b | 445120 |
| ENSDARG00000071192 | cx40.8 | connexin 40.8 | 565638 |
| ENSDARG00000041799 | cx43 | connexin 43 | 30236 |
| ENSDARG00000077765 | cx44.2 | connexin 44.2 | 114404 |
| ENSDARG00000073896 | cx47.1 | connexin 47.1 | 447835 |
| ENSDARG00000038075 | cyc1 | cytochrome c-1 | 564379 |
| ENSDARG00000044345 | cyfip1 | cytoplasmic FMR1 interacting protein 1 | 336613 |
| ENSDARG00000036375 | cyfip2 | cytoplasmic FMR1 interacting protein 2 | 100002872 |
| ENSDARG00000076742 | cyth1a | cytohesin 1a | 321938 |
| ENSDARG00000028800 | CYTH2 | cytohesin 2 | 569358 |
| ENSDARG00000005159 | cyth3b | cytohesin 3b | 0 |
| ENSDARG00000015059 | daam1a | dishevelled associated activator of morphogenesis 1a | 557451 |
| ENSDARG00000009689 | daam1b | dishevelled associated activator of morphogenesis 1b | 560060 |
| ENSDARG00000028393 | daam2 | dishevelled associated activator of morphogenesis 2 | 0 |
| ENSDARG00000069484 | dab2ipa | DAB2 interacting protein a | 100002454 |
| ENSDARG00000075110 | dab2ipb | DAB2 interacting protein b | 558237 |
| ENSDARG00000062956 | dagla | diacylglycerol lipase, alpha | 569398 |
| ENSDARG00000060093 | dapk1 | death-associated protein kinase 1 | 558314 |
| ENSDARG00000070043 | dars | aspartyl-tRNA synthetase | 324025 |
| ENSDARG00000069113 | dbn1 | drebrin 1 | 561153 |
| ENSDARG00000016016 | dbnlb | drebrin-like b | 553729 |
| ENSDARG00000101561 | dbt | dihydrolipoamide branched chain transacylase E2 | 541388 |
| ENSDARG00000054355 | dcaf7 | ddb1 and cul4 associated factor 7 | 337565 |
| ENSDARG00000061638 | dcakd | dephospho-CoA kinase domain containing | 751733 |
| ENSDARG00000104282 | DCC | DCC netrin 1 receptor | 0 |
| ENSDARG00000104664 | dckl1b | doublecortin-like kinase 1b | 100151759 |
| ENSDARG00000034093 | dclk2 | doublecortin-like kinase 2 | 572548 |
| ENSDARG00000019743 | dctn1a | dynactin 1a | 407638 |
| ENSDARG00000031388 | dctn2 | dynactin 2 (p50) | 394141 |
| ENSDARG00000041363 | dctn3 | dynactin 3 (p22) | 431767 |
| ENSDARG00000103083 | DDB1 (2 of 2) | damage-specific DNA binding protein 1, 127kDa | 0 |
| ENSDARG00000037318 | ddost | dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit (non-catalytic) | 406408 |
| ENSDARG00000061865 | ddx10 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 10 | 569118 |
| ENSDARG00000032117 | ddx1 | DEAD (Asp-Glu-Ala-Asp) box helicase 1 | 556790 |
| ENSDARG00000006225 | ddx39aa | DEAD (Asp-Glu-Ala-Asp) box polypeptide 39Aa | 325550 |
| ENSDARG00000020573 | ddx3 | DEAD (Asp-Glu-Ala-Asp) box helicase 3 | 566947 |
| ENSDARG00000038068 | ddx5 | DEAD (Asp-Glu-Ala-Asp) box helicase 5 | 322206 |
| ENSDARG00000061338 | ddx6 | DEAD (Asp-Glu-Ala-Asp) box helicase 6 | 564633 |
| ENSDARG00000003869 | decr1 | 2,4-dienoyl CoA reductase 1, mitochondrial | 436717 |
| ENSDARG00000005897 | dera | deoxyribose-phosphate aldolase (putative) | 450077 |
| ENSDARG00000075362 | dfnb31a | deafness, autosomal recessive 31a | 100334777 |
| ENSDARG00000068166 | dfnb31b | deafness, autosomal recessive 31b | 100534775 |
| ENSDARG00000062696 | DGKG | diacylglycerol kinase, gamma 90kDa | 0 |
| ENSDARG00000063578 | DGKI | diacylglycerol kinase, iota | 100331649 |
| ENSDARG00000013236 | dhcr24 | 24-dehydrocholesterol reductase | 494102 |
| ENSDARG00000021135 | dhrs2 | dehydrogenase/reductase (SDR family) member 2 | 393539 |
| ENSDARG00000015392 | dhx15 | DEAH (Asp-Glu-Ala-His) box helicase 15 | 321931 |
| ENSDARG00000063419 | DHX35 | DEAH (Asp-Glu-Ala-His) box polypeptide 35 | 0 |
| ENSDARG00000062612 | dhx37 | DEAH (Asp-Glu-Ala-His) box polypeptide 37 | 100009635 |
| ENSDARG00000079725 | dhx9 | DEAH (Asp-Glu-Ala-His) box helicase 9 | 568043 |
| ENSDARG00000005350 | dip2ba | DIP2 disco-interacting protein 2 homolog Ba (Drosophila) | 405899 |
| ENSDARG00000016348 | dip2bb | DIP2 disco-interacting protein 2 homolog Bb (Drosophila) | 568954 |
| ENSDARG00000015918 | dlat | dihydrolipoamide S-acetyltransferase (E2 component of pyruvate dehydrogenase complex) | 324201 |
| ENSDARG00000008785 | dldh | dihydrolipoamide dehydrogenase | 399479 |
| ENSDARG00000009677 | dlg1 | discs, large homolog 1 (Drosophila) | 114446 |
| ENSDARG00000102216 | dlg1l | discs, large (Drosophila) homolog 1, like | 497648 |
| ENSDARG00000099323 | dlg2 | discs, large homolog 2 (Drosophila) | 497638 |
| ENSDARG00000076796 | dlg3 | discs, large homolog 3 (Drosophila) | 564081 |
| ENSDARG00000041926 | DLG4 (2 of 2) | discs, large homolog 4 (Drosophila) | 0 |
| ENSDARG00000074059 | dlg5a | discs, large homolog 5a (Drosophila) | 569065 |
| ENSDARG00000014280 | dlgap1a | discs, large (Drosophila) homolog-associated protein 1a | 569898 |
| ENSDARG00000076070 | DLGAP2 (2 of 3) | discs, large (Drosophila) homolog-associated protein 2 | 0 |
| ENSDARG00000059340 | dlgap2a | discs, large (Drosophila) homolog-associated protein 2a | 557621 |
| ENSDARG00000092376 | dlgap2b | discs, large (Drosophila) homolog-associated protein 2b | 0 |
| ENSDARG00000055459 | dlgap3 | discs, large (Drosophila) homolog-associated protein 3 | 557556 |
| ENSDARG00000012823 | dlgap4b | discs, large (Drosophila) homolog-associated protein 4b | 0 |
| ENSDARG00000014230 | dlst | dihydrolipoamide S-succinyltransferase | 368262 |
| ENSDARG00000008487 | dmd | dystrophin | 83773 |
| ENSDARG00000013110 | dmtn | dematin actin binding protein | 565367 |
| ENSDARG00000006585 | DMWD | dystrophia myotonica, WD repeat containing | 571996 |
| ENSDARG00000098599 | DMXL1 | Dmx-like 1 | 100333625 |
| ENSDARG00000091293 | dmxl2 | Dmx-like 2 | 378958 |
| ENSDARG00000078759 | dna2 | DNA replication helicase/nuclease 2 | 559535 |
| ENSDARG00000100115 | dnah1 | dynein, axonemal, heavy chain 1 | 100317899 |
| ENSDARG00000104066 | dnaja2 | DnaJ (Hsp40) homolog, subfamily A, member 2 | 406814 |
| ENSDARG00000010745 | dnaja2l | DnaJ (Hsp40) homolog, subfamily A, member 2, like | 324164 |
| ENSDARG00000058494 | dnaja3a | DnaJ (Hsp40) homolog, subfamily A, member 3A | 335894 |
| ENSDARG00000102295 | dnaja3b | DnaJ (Hsp40) homolog, subfamily A, member 3B | 394242 |
| ENSDARG00000039363 | dnajb12a | DnaJ (Hsp40) homolog, subfamily B, member 12a | 324005 |
| ENSDARG00000058644 | dnajb2 | DnaJ (Hsp40) homolog, subfamily B, member 2 | 561686 |
| ENSDARG00000038978 | dnajb4 | DnaJ (Hsp40) homolog, subfamily B, member 4 | 445061 |
| ENSDARG00000004680 | dnajb6a | DnaJ (Hsp40) homolog, subfamily B, member 6a | 436626 |
| ENSDARG00000020953 | dnajb6b | DnaJ (Hsp40) homolog, subfamily B, member 6b | 393275 |
| ENSDARG00000074727 | dnajc10 | DnaJ (Hsp40) homolog, subfamily C, member 10 | 557858 |
| ENSDARG00000011196 | dnajc11 | DnaJ (Hsp40) homolog, subfamily C, member 11 | 322953 |
| ENSDARG00000060725 | dnajc16l | DnaJ (Hsp40) homolog, subfamily C, member 16 like | 556590 |
| ENSDARG00000032238 | dnm3a | dynamin 3a | 557996 |
| ENSDARG00000088293 | doc2b | double C2-like domains, beta | 0 |
| ENSDARG00000075327 | dock10 | dedicator of cytokinesis 10 | 100000949 |
| ENSDARG00000063180 | dock3 | dedicator of cytokinesis 3 | 561387 |
| ENSDARG00000076213 | DOCK4 (2 of 2) | dedicator of cytokinesis 4 | 566115 |
| ENSDARG00000024874 | dock4b | dedicator of cytokinesis 4b | 654774 |
| ENSDARG00000078675 | dock7 | dedicator of cytokinesis 7 | 568817 |
| ENSDARG00000055225 | dock8 | dedicator of cytokinesis 8 | 403040 |
| ENSDARG00000055857 | dopey2 | dopey family member 2 | 0 |
| ENSDARG00000057011 | dpm1 | dolichyl-phosphate mannosyltransferase polypeptide 1, catalytic subunit | 445202 |
| ENSDARG00000098057 | dscaml1 | Down syndrome cell adhesion molecule like 1 | 0 |
| ENSDARG00000076673 | DSP (2 of 2) | desmoplakin | 327364 |
| ENSDARG00000101858 | DST (3 of 4) | dystonin | 0 |
| ENSDARG00000013020 | dtnbb | dystrobrevin, beta b | 559786 |
| ENSDARG00000076302 | dtx1 | deltex1 | 570481 |
| ENSDARG00000078016 | dtx4 | deltex 4, E3 ubiquitin ligase | 100037355 |
| ENSDARG00000063570 | dyrk1aa | dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1A, a | 100005019 |
| ENSDARG00000001578 | echs1 | enoyl CoA hydratase, short chain, 1, mitochondrial | 368912 |
| ENSDARG00000093413 | edil3 | EGF-like repeats and discoidin I-like domains 3 | 793444 |
| ENSDARG00000039502 | eef1a1a | eukaryotic translation elongation factor 1 alpha 1a | 336334 |
| ENSDARG00000069951 | eef1a1b | eukaryotic translation elongation factor 1 alpha 1b | 100004503 |
| ENSDARG00000020850 | eef1a1l1 | eukaryotic translation elongation factor 1 alpha 1, like 1 | 30516 |
| ENSDARG00000071774 | eef1e1 | eukaryotic translation elongation factor 1 epsilon 1 | 565440 |
| ENSDARG00000056119 | eef1g | eukaryotic translation elongation factor 1 gamma | 195822 |
| ENSDARG00000035256 | eef2l2 | eukaryotic translation elongation factor 2, like 2 | 336168 |
| ENSDARG00000007723 | efnb1 | ephrin-B1 | 114377 |
| ENSDARG00000005163 | efr3a | EFR3 homolog A (S. cerevisiae) | 393800 |
| ENSDARG00000033516 | efr3ba | EFR3 homolog Ba (S. cerevisiae) | 560858 |
| ENSDARG00000069318 | efr3bb | EFR3 homolog Bb (S. cerevisiae) | 561353 |
| ENSDARG00000014793 | ehd1b | EH-domain containing 1b | 447839 |
| ENSDARG00000070029 | ehhadh | enoyl-CoA, hydratase/3-hydroxyacyl CoA dehydrogenase | 100000859 |
| ENSDARG00000068289 | eif3k | eukaryotic translation initiation factor 3, subunit K | 550245 |
| ENSDARG00000076815 | eif3s10 | eukaryotic translation initiation factor 3, subunit 10 (theta) | 327515 |
| ENSDARG00000101082 | eif3s6ip | eukaryotic translation initiation factor 3, subunit 6 interacting protein | 406402 |
| ENSDARG00000038695 | elavl1 | ELAV like RNA binding protein 1 | 30736 |
| ENSDARG00000045639 | elavl4 | ELAV like neuron-specific RNA binding protein 4 | 30737 |
| ENSDARG00000078141 | ELFN1 (2 of 2) | extracellular leucine-rich repeat and fibronectin type III domain containing 1 | 562570 |
| ENSDARG00000074372 | elfn1b | extracellular leucine-rich repeat and fibronectin type III domain containing 1b | 100144401 |
| ENSDARG00000077609 | elfn2a | extracellular leucine-rich repeat and fibronectin type III domain containing 2a | 0 |
| ENSDARG00000098753 | elmo1 | engulfment and cell motility 1 (ced-12 homolog, C. elegans) | 406364 |
| ENSDARG00000063527 | elmo2 | engulfment and cell motility 2 | 100006857 |
| ENSDARG00000042126 | elmo3 | engulfment and cell motility 3 | 553310 |
| ENSDARG00000003308 | emc2 | ER membrane protein complex subunit 2 | 407073 |
| ENSDARG00000039268 | emc4 | ER membrane protein complex subunit 4 | 0 |
| ENSDARG00000035282 | emc6 | ER membrane protein complex subunit 6 | 337220 |
| ENSDARG00000012144 | emc7 | ER membrane protein complex subunit 7 | 561549 |
| ENSDARG00000053517 | EML5 | echinoderm microtubule associated protein like 5 | 570011 |
| ENSDARG00000102755 | EML6 | echinoderm microtubule associated protein like 6 | 0 |
| ENSDARG00000032049 | enah | enabled homolog (Drosophila) | 541394 |
| ENSDARG00000071224 | ENDOD1 (7 of 10) | endonuclease domain containing 1 | 559260 |
| ENSDARG00000040469 | enpp6 | ectonucleotide pyrophosphatase/phosphodiesterase 6 | 791915 |
| ENSDARG00000029019 | epb41b | erythrocyte membrane protein band 4.1b (elliptocytosis 1, RH-linked) | 326287 |
| ENSDARG00000040087 | epb41l4a | erythrocyte membrane protein band 4.1 like 4A | 30470 |
| ENSDARG00000061398 | epb41l4b | erythrocyte membrane protein band 4.1 like 4B | 100332627 |
| ENSDARG00000060494 | eprs | glutamyl-prolyl-tRNA synthetase | 562037 |
| ENSDARG00000078216 | eps15 | epidermal growth factor receptor pathway substrate 15 | 795505 |
| ENSDARG00000042670 | eps15l1a | epidermal growth factor receptor pathway substrate 15-like 1a | 568170 |
| ENSDARG00000013161 | ERAS | ES cell expressed Ras | 393228 |
| ENSDARG00000044281 | erbb2ip | erbb2 interacting protein | 566811 |
| ENSDARG00000063207 | erbb4a | erb-b2 receptor tyrosine kinase 4a | 100007677 |
| ENSDARG00000076014 | ERC1 (3 of 3) | ELKS/RAB6-interacting/CAST family member 1 | 560003 |
| ENSDARG00000009941 | erc1a | ELKS/RAB6-interacting/CAST family member 1a | 548606 |
| ENSDARG00000032866 | erh | enhancer of rudimentary homolog (Drosophila) | 30475 |
| ENSDARG00000043976 | etf1b | eukaryotic translation termination factor 1b | 266989 |
| ENSDARG00000035650 | evla | Enah/Vasp-like a | 58003 |
| ENSDARG00000099720 | evlb | Enah/Vasp-like b | 0 |
| ENSDARG00000043019 | exoc1 | exocyst complex component 1 | 322338 |
| ENSDARG00000055610 | exoc2 | exocyst complex component 2 | 100004870 |
| ENSDARG00000014582 | exoc3 | exocyst complex component 3 | 334015 |
| ENSDARG00000004336 | exoc4 | exocyst complex component 4 | 559866 |
| ENSDARG00000060323 | exoc5 | exocyst complex component 5 | 559010 |
| ENSDARG00000098874 | exoc6b | exocyst complex component 6B | 767763 |
| ENSDARG00000020521 | exoc6 | exocyst complex component 6 | 406591 |
| ENSDARG00000087229 | exoc7 | exocyst complex component 7 | 406723 |
| ENSDARG00000063211 | exoc8 | exocyst complex component 8 | 568029 |
| ENSDARG00000056648 | ext2 | exostosin glycosyltransferase 2 | 493780 |
| ENSDARG00000026811 | extl3 | exostosin-like glycosyltransferase 3 | 493783 |
| ENSDARG00000086300 | FAM107A | family with sequence similarity 107, member A | 794256 |
| ENSDARG00000088073 | fam110b | family with sequence similarity 110, member B | 573995 |
| ENSDARG00000019205 | fam120c | family with sequence similarity 120C | 557842 |
| ENSDARG00000026762 | fam126a | family with sequence similarity 126, member A | 402926 |
| ENSDARG00000100400 | FAM126B (1 of 2) | family with sequence similarity 126, member B | 0 |
| ENSDARG00000070952 | fam131ba | family with sequence similarity 131, member Ba | 565996 |
| ENSDARG00000070575 | fam131bb | family with sequence similarity 131, member Bb | 556725 |
| ENSDARG00000008573 | fam20b | family with sequence similarity 20, member B (H. sapiens) | 557060 |
| ENSDARG00000035985 | FAM210A (1 of 2) | family with sequence similarity 210, member A | 562734 |
| ENSDARG00000098860 | FAM57A | family with sequence similarity 57, member A | 0 |
| ENSDARG00000003320 | fam91a1 | family with sequence similarity 91, member A1 | 393413 |
| ENSDARG00000074381 | farp1 | FERM, RhoGEF (ARHGEF) and pleckstrin domain protein 1 (chondrocyte-derived) | 0 |
| ENSDARG00000099022 | faua | Finkel-Biskis-Reilly murine sarcoma virus (FBR-MuSV) ubiquitously expressed a | 393710 |
| ENSDARG00000021366 | fbp1a | fructose-1,6-bisphosphatase 1a | 335231 |
| ENSDARG00000099306 | FBXO2 | F-box protein 2 | 559776 |
| ENSDARG00000076310 | FBXO41 | F-box protein 41 | 101883316 |
| ENSDARG00000078908 | fbxo41 | F-box protein 41 | 567804 |
| ENSDARG00000020741 | fga | fibrinogen alpha chain | 378986 |
| ENSDARG00000008969 | fgb | fibrinogen beta chain | 337315 |
| ENSDARG00000003058 | fgfr1op | FGFR1 oncogene partner | 415215 |
| ENSDARG00000037281 | fgg | fibrinogen gamma chain | 406327 |
| ENSDARG00000052625 | fkbp1b | FK506 binding protein 1b | 393785 |
| ENSDARG00000059701 | flii | flightless I homolog (Drosophila) | 560281 |
| ENSDARG00000010764 | flj13639 | flj13639 | 677747 |
| ENSDARG00000098374 | FLNB | filamin B, beta | 0 |
| ENSDARG00000097351 | flnbl | filamin B, like | 100534866 |
| ENSDARG00000018820 | flncb | filamin C, gamma b (actin binding protein 280) | 570288 |
| ENSDARG00000001710 | flot1a | flotillin 1a | 561345 |
| ENSDARG00000037998 | flot1b | flotillin 1b | 245697 |
| ENSDARG00000004830 | flot2a | flotillin 2a | 245698 |
| ENSDARG00000069774 | flot2b | flotillin 2b | 393612 |
| ENSDARG00000019371 | flt1 | fms-related tyrosine kinase 1 (vascular endothelial growth factor/vascular permeability factor receptor) | 544667 |
| ENSDARG00000061778 | fmn2b | formin 2b | 566432 |
| ENSDARG00000012586 | fmnl2a | formin-like 2a | 100006456 |
| ENSDARG00000075041 | fmnl2b | formin-like 2b | 492608 |
| ENSDARG00000037433 | fmr1 | fragile X mental retardation 1 | 259191 |
| ENSDARG00000098751 | FQ323163.2 | Uncharacterized protein | 795227 |
| ENSDARG00000102626 | frem2b | Fras1 related extracellular matrix protein 2b | 566673 |
| ENSDARG00000042249 | FRMD4A | FERM domain containing 4A | 564713 |
| ENSDARG00000074865 | frmpd3 | FERM and PDZ domain containing 3 | 798302 |
| ENSDARG00000078688 | frs3 | fibroblast growth factor receptor substrate 3 | 100330147 |
| ENSDARG00000056001 | fryb | furry homolog b (Drosophila) | 792949 |
| ENSDARG00000040001 | fryl | furry homolog, like | 558173 |
| ENSDARG00000040822 | fundc1 | FUN14 domain containing 1 | 436984 |
| ENSDARG00000088117 | FUT8 (2 of 2) | fucosyltransferase 8 (alpha (1,6) fucosyltransferase) | 100331714 |
| ENSDARG00000015449 | fut8a | fucosyltransferase 8a (alpha (1,6) fucosyltransferase) | 797512 |
| ENSDARG00000011370 | fyna | FYN proto-oncogene, Src family tyrosine kinase a | 373872 |
| ENSDARG00000025319 | fynb | FYN proto-oncogene, Src family tyrosine kinase b | 574422 |
| ENSDARG00000027200 | gabarapl2 | GABA(A) receptor-associated protein-like 2 | 403036 |
| ENSDARG00000018967 | gabbr1a | gamma-aminobutyric acid (GABA) B receptor, 1a | 373873 |
| ENSDARG00000016667 | gabbr1b | gamma-aminobutyric acid (GABA) B receptor, 1b | 558708 |
| ENSDARG00000061042 | gabbr2 | gamma-aminobutyric acid (GABA) B receptor, 2 | 560267 |
| ENSDARG00000068989 | gabra1 | gamma-aminobutyric acid (GABA) A receptor, alpha 1 | 768183 |
| ENSDARG00000090883 | gabra3 | gamma-aminobutyric acid (GABA) A receptor, alpha 3 | 0 |
| ENSDARG00000079586 | gabrb2 | gamma-aminobutyric acid (GABA) A receptor, beta 2 | 0 |
| ENSDARG00000023771 | gabrb3 | gamma-aminobutyric acid (GABA) A receptor, beta 3 | 566922 |
| ENSDARG00000099096 | gabrb4 | gamma-aminobutyric acid (GABA) A receptor, beta 4 | 566514 |
| ENSDARG00000059763 | gabrd | gamma-aminobutyric acid (GABA) A receptor, delta | 571422 |
| ENSDARG00000053665 | gabrg2 | gamma-aminobutyric acid (GABA) A receptor, gamma 2 | 553402 |
| ENSDARG00000090654 | gak | cyclin G associated kinase | 100151158 |
| ENSDARG00000003829 | galnt2 | UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 2 | 570248 |
| ENSDARG00000006832 | galnt9 | polypeptide N-acetylgalactosaminyltransferase 9 | 797549 |
| ENSDARG00000099744 | gap43 | growth associated protein 43 | 30608 |
| ENSDARG00000007264 | gbas | glioblastoma amplified sequence | 30232 |
| ENSDARG00000037057 | gcdha | glutaryl-CoA dehydrogenase a | 393860 |
| ENSDARG00000098831 | gcdhb | glutaryl-CoA dehydrogenase b | 450003 |
| ENSDARG00000025301 | gfap | glial fibrillary acidic protein | 30646 |
| ENSDARG00000042723 | gfra2a | GDNF family receptor alpha 2a | 405794 |
| ENSDARG00000044015 | gfra2b | GDNF family receptor alpha 2b | 564916 |
| ENSDARG00000087704 | gfra3 | GDNF family receptor alpha 3 | 556647 |
| ENSDARG00000055569 | ghdc | GH3 domain containing | 497449 |
| ENSDARG00000037589 | gid8b | GID complex subunit 8 homolog b (S. cerevisiae) | 566067 |
| ENSDARG00000104483 | gipc1 | GIPC PDZ domain containing family, member 1 | 445568 |
| ENSDARG00000039489 | git1 | G protein-coupled receptor kinase interacting ArfGAP 1 | 568935 |
| ENSDARG00000068981 | glceb | glucuronic acid epimerase b | 100007670 |
| ENSDARG00000016837 | glipr2l | GLI pathogenesis-related 2, like | 449805 |
| ENSDARG00000012019 | glra1 | glycine receptor, alpha 1 | 30676 |
| ENSDARG00000052782 | glrba | glycine receptor, beta a | 83412 |
| ENSDARG00000008816 | glud1a | glutamate dehydrogenase 1a | 317737 |
| ENSDARG00000101074 | glud1b | glutamate dehydrogenase 1b | 373092 |
| ENSDARG00000010002 | gna11a | guanine nucleotide binding protein (G protein), alpha 11a (Gq class) | 563953 |
| ENSDARG00000092948 | gna15.3 | guanine nucleotide binding protein (G protein), alpha 15 (Gq class), tandem duplicate 3 | 0 |
| ENSDARG00000018174 | gnai2a | guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 2a | 336421 |
| ENSDARG00000030644 | gnai3 | guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 3 | 100006888 |
| ENSDARG00000044760 | gnaia | guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide a | 323509 |
| ENSDARG00000045415 | gnal | guanine nucleotide binding protein (G protein), alpha activating activity polypeptide, olfactory type | 492467 |
| ENSDARG00000016676 | gnao1a | guanine nucleotide binding protein (G protein), alpha activating activity polypeptide O, a | 393760 |
| ENSDARG00000036058 | gnao1b | guanine nucleotide binding protein (G protein), alpha activating activity polypeptide O, b | 393801 |
| ENSDARG00000043094 | gnas | GNAS complex locus | 557353 |
| ENSDARG00000099844 | GNAZ | guanine nucleotide binding protein (G protein), alpha z polypeptide | 0 |
| ENSDARG00000035357 | gnb2 | guanine nucleotide binding protein (G protein), beta polypeptide 2 | 541370 |
| ENSDARG00000099685 | gnb5a | guanine nucleotide binding protein (G protein), beta 5a | 562813 |
| ENSDARG00000055377 | gnb5b | guanine nucleotide binding protein (G protein), beta 5b | 393719 |
| ENSDARG00000003375 | gng12a | guanine nucleotide binding protein (G protein), gamma 12a | 406604 |
| ENSDARG00000037921 | gng13b | guanine nucleotide binding protein (G protein), gamma 13b | 436673 |
| ENSDARG00000056831 | gng2 | guanine nucleotide binding protein (G protein), gamma 2 | 550256 |
| ENSDARG00000009553 | gng3 | guanine nucleotide binding protein (G protein), gamma 3 | 114462 |
| ENSDARG00000038693 | gng7 | guanine nucleotide binding protein (G protein), gamma 7 | 436670 |
| ENSDARG00000062511 | golga3 | golgin A3 | 570879 |
| ENSDARG00000036295 | golga7ba | golgin A7 family, member Ba | 550228 |
| ENSDARG00000105099 | golga7bb | golgin A7 family, member Bb | 0 |
| ENSDARG00000057676 | golga7 | golgin A7 | 798622 |
| ENSDARG00000053070 | gosr2 | golgi SNAP receptor complex member 2 | 324569 |
| ENSDARG00000019341 | gpc1a | glypican 1a | 553367 |
| ENSDARG00000090585 | gpc1b | glypican 1b | 393995 |
| ENSDARG00000032199 | gpc3 | glypican 3 | 0 |
| ENSDARG00000089076 | gphna | gephyrin a | 555470 |
| ENSDARG00000100851 | gphnb | gephyrin b | 0 |
| ENSDARG00000077134 | gpr158a | G protein-coupled receptor 158a | 570001 |
| ENSDARG00000079665 | gpr158b | G protein-coupled receptor 158b | 568637 |
| ENSDARG00000077080 | gprc5ba | G protein-coupled receptor, class C, group 5, member Ba | 570207 |
| ENSDARG00000075141 | gprc5bb | G protein-coupled receptor, class C, group 5, member Bb | 100002676 |
| ENSDARG00000075043 | GPRIN3 | GPRIN family member 3 | 100536832 |
| ENSDARG00000092517 | gpsm1a | G-protein signaling modulator 1a | 100004265 |
| ENSDARG00000018146 | gpx1a | glutathione peroxidase 1a | 0 |
| ENSDARG00000022668 | grapb | GRB2-related adaptor protein b | 555184 |
| ENSDARG00000038059 | grb2b | growth factor receptor-bound protein 2b | 406308 |
| ENSDARG00000021352 | gria1a | glutamate receptor, ionotropic, AMPA 1a | 798689 |
| ENSDARG00000032714 | gria1b | glutamate receptor, ionotropic, AMPA 1b | 403044 |
| ENSDARG00000070173 | gria2a | glutamate receptor, ionotropic, AMPA 2a | 170450 |
| ENSDARG00000052765 | gria2b | glutamate receptor, ionotropic, AMPA 2b | 170451 |
| ENSDARG00000032737 | gria3a | glutamate receptor, ionotropic, AMPA 3a | 170452 |
| ENSDARG00000037498 | gria3b | glutamate receptor, ionotropic, AMPA 3b | 368416 |
| ENSDARG00000037496 | gria4a | glutamate receptor, ionotropic, AMPA 4a | 407735 |
| ENSDARG00000059368 | gria4b | glutamate receptor, ionotropic, AMPA 4b | 336069 |
| ENSDARG00000055302 | grid2 | glutamate receptor, ionotropic, delta 2 | 448841 |
| ENSDARG00000076103 | grid2ipa | glutamate receptor, ionotropic, delta 2 (Grid2) interacting protein, a | 561017 |
| ENSDARG00000095603 | grid2ipb | glutamate receptor, ionotropic, delta 2 (Grid2) interacting protein, b | 559717 |
| ENSDARG00000027828 | grin1a | glutamate receptor, ionotropic, N-methyl D-aspartate 1a | 767745 |
| ENSDARG00000025728 | grin1b | glutamate receptor, ionotropic, N-methyl D-aspartate 1b | 100005675 |
| ENSDARG00000034493 | grin2aa | glutamate receptor, ionotropic, N-methyl D-aspartate 2A, a | 563297 |
| ENSDARG00000070543 | grin2ab | glutamate receptor, ionotropic, N-methyl D-aspartate 2A, b | 570493 |
| ENSDARG00000079348 | GRIN2B (2 of 2) | glutamate receptor, ionotropic, N-methyl D-aspartate 2B | 0 |
| ENSDARG00000030376 | grin2bb | glutamate receptor, ionotropic, N-methyl D-aspartate 2B, genome duplicate b | 559976 |
| ENSDARG00000078149 | grin2ca | glutamate receptor, ionotropic, N-methyl D-aspartate 2Ca | 100329743 |
| ENSDARG00000077560 | grin2cb | glutamate receptor, ionotropic, N-methyl D-aspartate 2Cb | 100333648 |
| ENSDARG00000086207 | grin2da | glutamate receptor, ionotropic, N-methyl D-aspartate 2D, a | 449864 |
| ENSDARG00000070620 | grin2db | glutamate receptor, ionotropic, N-methyl D-aspartate 2D, b | 0 |
| ENSDARG00000015053 | grip1 | glutamate receptor interacting protein 1 | 558006 |
| ENSDARG00000027618 | grip2b | glutamate receptor interacting protein 2b | 562290 |
| ENSDARG00000070448 | grk4 | G protein-coupled receptor kinase 4 | 560761 |
| ENSDARG00000007195 | GRM2 (2 of 2) | glutamate receptor, metabotropic 2 | 564461 |
| ENSDARG00000040156 | grm4 | glutamate receptor, metabotropic 4 | 567181 |
| ENSDARG00000004445 | GRM5 (1 of 2) | glutamate receptor, metabotropic 5 | 0 |
| ENSDARG00000102067 | GRM5 (2 of 2) | glutamate receptor, metabotropic 5 | 100332913 |
| ENSDARG00000017742 | grm6a | glutamate receptor, metabotropic 6a | 568484 |
| ENSDARG00000100267 | GRM7 | glutamate receptor, metabotropic 7 | 0 |
| ENSDARG00000077654 | grm8a | glutamate receptor, metabotropic 8a | 792371 |
| ENSDARG00000076508 | grm8b | glutamate receptor, metabotropic 8b | 569768 |
| ENSDARG00000015681 | gsk3ab | glycogen synthase kinase 3 alpha b | 30664 |
| ENSDARG00000017803 | gsk3b | glycogen synthase kinase 3 beta | 30654 |
| ENSDARG00000011459 | gsna | gelsolin a | 321099 |
| ENSDARG00000053467 | gtpbp1 | GTP binding protein 1 | 378721 |
| ENSDARG00000086790 | gucy1b3 | guanylate cyclase 1, soluble, beta 3 | 100150304 |
| ENSDARG00000001733 | gulp1a | GULP, engulfment adaptor PTB domain containing 1a | 368404 |
| ENSDARG00000099214 | h2afvb | H2A histone family, member Vb | 403077 |
| ENSDARG00000087897 | h2afy2 | H2A histone family, member Y2 | 558711 |
| ENSDARG00000057128 | hadhaa | hydroxyacyl-CoA dehydrogenase/3-ketoacyl-CoA thiolase/enoyl-CoA hydratase (trifunctional protein), alpha subunit a | 553401 |
| ENSDARG00000060594 | hadhab | hydroxyacyl-CoA dehydrogenase/3-ketoacyl-CoA thiolase/enoyl-CoA hydratase (trifunctional protein), alpha subunit b | 793834 |
| ENSDARG00000102055 | hadhb | hydroxyacyl-CoA dehydrogenase/3-ketoacyl-CoA thiolase/enoyl-CoA hydratase (trifunctional protein), beta subunit | 0 |
| ENSDARG00000068516 | hapln1b | hyaluronan and proteoglycan link protein 1b | 569074 |
| ENSDARG00000018542 | hapln4 | hyaluronan and proteoglycan link protein 4 | 559818 |
| ENSDARG00000012519 | hcfc1b | host cell factor C1b | 565206 |
| ENSDARG00000087573 | hdac11 | histone deacetylase 11 | 431718 |
| ENSDARG00000101079 | HECTD4 | HECT domain containing E3 ubiquitin protein ligase 4 | 101885095 |
| ENSDARG00000062447 | hecw2a | HECT, C2 and WW domain containing E3 ubiquitin protein ligase 2a | 0 |
| ENSDARG00000063253 | hecw2b | HECT, C2 and WW domain containing E3 ubiquitin protein ligase 2b | 100331355 |
| ENSDARG00000056934 | hepacama | hepatic and glial cell adhesion molecule a | 100005952 |
| ENSDARG00000059231 | hephl1a | hephaestin-like 1a | 561607 |
| ENSDARG00000073841 | herc2 | HECT and RLD domain containing E3 ubiquitin protein ligase 2 | 0 |
| ENSDARG00000052371 | higd2a | HIG1 hypoxia inducible domain family, member 2A | 0 |
| ENSDARG00000035519 | histh1l | histone H1 like | 550353 |
| ENSDARG00000039452 | hk1 | hexokinase 1 | 406791 |
| ENSDARG00000038703 | hkdc1 | hexokinase domain containing 1 | 321224 |
| ENSDARG00000099175 | hmgb1a | high mobility group box 1a | 321622 |
| ENSDARG00000030479 | hmgb1b | high mobility group box 1b | 795095 |
| ENSDARG00000056725 | hmgb3a | high mobility group box 3a | 561025 |
| ENSDARG00000040881 | hnrnph1 | heterogeneous nuclear ribonucleoprotein H1 | 402984 |
| ENSDARG00000079780 | HNRNPUL1 (2 of 2) | heterogeneous nuclear ribonucleoprotein U-like 1 | 0 |
| ENSDARG00000007034 | hnrpkl | heterogeneous nuclear ribonucleoprotein K, like | 406264 |
| ENSDARG00000101759 | homer1b | homer scaffolding protein 1b | 436769 |
| ENSDARG00000102532 | HOMER3 (2 of 2) | homer scaffolding protein 3 | 100330028 |
| ENSDARG00000010789 | homer3 | homer scaffolding protein 3 | 394088 |
| ENSDARG00000044669 | hp1bp3 | heterochromatin protein 1, binding protein 3 | 558102 |
| ENSDARG00000099478 | hs2st1a | heparan sulfate 2-O-sulfotransferase 1a | 791188 |
| ENSDARG00000062008 | hs2st1b | heparan sulfate 2-O-sulfotransferase 1b | 100151174 |
| ENSDARG00000041921 | hsbp1b | heat shock factor binding protein 1b | 393727 |
| ENSDARG00000012381 | hsc70 | heat shock cognate 70 | 393586 |
| ENSDARG00000017781 | hsd17b10 | hydroxysteroid (17-beta) dehydrogenase 10 | 450078 |
| ENSDARG00000035872 | hsd17b12b | hydroxysteroid (17-beta) dehydrogenase 12b | 322626 |
| ENSDARG00000070603 | hspa12a | heat shock protein 12A | 751725 |
| ENSDARG00000040984 | hspa13 | heat shock protein 70 family, member 13 | 560481 |
| ENSDARG00000037403 | HSPA8 (1 of 2) | heat shock 70kDa protein 8 | 562935 |
| ENSDARG00000068992 | hspa8 | heat shock protein 8 | 573376 |
| ENSDARG00000062997 | hyal2b | hyaluronoglucosaminidase 2b | 559228 |
| ENSDARG00000007955 | iars | isoleucyl-tRNA synthetase | 334393 |
| ENSDARG00000009978 | icn | ictacalcin | 336655 |
| ENSDARG00000103563 | IFT122 | intraflagellar transport 122 | 0 |
| ENSDARG00000096355 | ighv1-4 | immunoglobulin heavy variable 1-4 | 0 |
| ENSDARG00000022176 | IGLON5 | IgLON family member 5 | 550472 |
| ENSDARG00000031049 | igsf21a | immunoglobin superfamily, member 21a | 569872 |
| ENSDARG00000033845 | igsf9ba | immunoglobulin superfamily, member 9Ba | 567389 |
| ENSDARG00000069467 | igsf9bb | immunoglobulin superfamily, member 9Bb | 569554 |
| ENSDARG00000062045 | il1rapl1a | interleukin 1 receptor accessory protein-like 1a | 568868 |
| ENSDARG00000037553 | il1rapl2 | interleukin 1 receptor accessory protein-like 2 | 100149452 |
| ENSDARG00000014591 | ilf2 | interleukin enhancer binding factor 2 | 406517 |
| ENSDARG00000017126 | ilvbl | ilvB (bacterial acetolactate synthase)-like | 393639 |
| ENSDARG00000006900 | impdh2 | IMP (inosine 5'-monophosphate) dehydrogenase 2 | 317745 |
| ENSDARG00000011862 | inaa | internexin neuronal intermediate filament protein, alpha a | 555251 |
| ENSDARG00000053248 | inab | internexin neuronal intermediate filament protein, alpha b | 30230 |
| ENSDARG00000063352 | inpp4aa | inositol polyphosphate-4-phosphatase, type Ia | 664741 |
| ENSDARG00000016551 | iqsec1b | IQ motif and Sec7 domain 1b | 100148736 |
| ENSDARG00000102125 | IQSEC2 (2 of 2) | IQ motif and Sec7 domain 2 | 560461 |
| ENSDARG00000077709 | iqsec2 | IQ motif and Sec7 domain 2 | 797187 |
| ENSDARG00000099850 | iqsec3a | IQ motif and Sec7 domain 3a | 797355 |
| ENSDARG00000093091 | iqsec3b | IQ motif and Sec7 domain 3b | 555340 |
| ENSDARG00000051888 | ist1 | increased sodium tolerance 1 homolog (yeast) | 321460 |
| ENSDARG00000055053 | itih1 | inter-alpha-trypsin inhibitor heavy chain 1 | 564761 |
| ENSDARG00000007098 | itm2ba | integral membrane protein 2Ba | 323632 |
| ENSDARG00000074149 | itpr1b | inositol 1,4,5-trisphosphate receptor, type 1b | 100149388 |
| ENSDARG00000011909 | itpr2 | inositol 1,4,5-trisphosphate receptor, type 2 | 562585 |
| ENSDARG00000061741 | itpr3 | inositol 1,4,5-trisphosphate receptor, type 3 | 794666 |
| ENSDARG00000053547 | jakmip2 | janus kinase and microtubule interacting protein 2 | 559839 |
| ENSDARG00000070787 | jupa | junction plakoglobin a | 30415 |
| ENSDARG00000059067 | jupb | junction plakoglobin b | 100330886 |
| ENSDARG00000104119 | kalrna | kalirin, RhoGEF kinase a | 0 |
| ENSDARG00000103799 | kars | lysyl-tRNA synthetase | 280647 |
| ENSDARG00000076923 | kazna | kazrin, periplakin interacting protein a | 100003663 |
| ENSDARG00000006601 | kaznb | kazrin, periplakin interacting protein b | 790937 |
| ENSDARG00000104958 | KBTBD11 | kelch repeat and BTB (POZ) domain containing 11 | 557467 |
| ENSDARG00000079491 | KCNA3 | potassium channel, voltage gated shaker related subfamily A, member 3 | 100150068 |
| ENSDARG00000087247 | KCNAB2 (2 of 2) | potassium channel, voltage gated subfamily A regulatory beta subunit 2 | 797337 |
| ENSDARG00000074320 | KCNAB3 | potassium channel, voltage gated subfamily A regulatory beta subunit 3 | 0 |
| ENSDARG00000074746 | kcnd1 | potassium voltage-gated channel, Shal-related subfamily, member 1 | 100001681 |
| ENSDARG00000032799 | kcnd2 | potassium voltage-gated channel, Shal-related subfamily, member 2 | 570188 |
| ENSDARG00000056101 | kcnd3 | potassium voltage-gated channel, Shal-related subfamily, member 3 | 327415 |
| ENSDARG00000088100 | KCNIP4 (2 of 2) | Kv channel interacting protein 4 | 100536963 |
| ENSDARG00000090815 | KCNJ10 (1 of 3) | potassium channel, inwardly rectifying subfamily J, member 10 | 100538267 |
| ENSDARG00000103332 | KCNJ10 (2 of 3) | potassium channel, inwardly rectifying subfamily J, member 10 | 0 |
| ENSDARG00000079840 | kcnma1a | potassium large conductance calcium-activated channel, subfamily M, alpha member 1a | 568554 |
| ENSDARG00000069843 | kctd12.1 | potassium channel tetramerisation domain containing 12.1 | 373865 |
| ENSDARG00000045893 | kctd15a | potassium channel tetramerization domain containing 15a | 445133 |
| ENSDARG00000004648 | kctd16a | potassium channel tetramerization domain containing 16a | 567207 |
| ENSDARG00000020102 | kctd16b | potassium channel tetramerization domain containing 16b | 569756 |
| ENSDARG00000060854 | kctd3 | potassium channel tetramerization domain containing 3 | 569113 |
| ENSDARG00000067507 | kctd8 | potassium channel tetramerization domain containing 8 | 568933 |
| ENSDARG00000052856 | khdrbs1a | KH domain containing, RNA binding, signal transduction associated 1a | 30082 |
| ENSDARG00000062477 | kiaa1549la | KIAA1549-like a | 566592 |
| ENSDARG00000062086 | kiaa1549lb | KIAA1549-like b | 0 |
| ENSDARG00000043571 | KIF2A | kinesin heavy chain member 2A | 447926 |
| ENSDARG00000087538 | kif3a | kinesin family member 3A | 550267 |
| ENSDARG00000017162 | KIF3C (1 of 2) | kinesin family member 3C | 100331925 |
| ENSDARG00000075806 | kirrel3a | kin of IRRE like 3 a | 571887 |
| ENSDARG00000010844 | kras | v-Ki-ras2 Kirsten rat sarcoma viral oncogene homolog | 445289 |
| ENSDARG00000036840 | krt15 | keratin 15 | 406844 |
| ENSDARG00000028618 | KRT18 (2 of 3) | keratin 18, type I | 393540 |
| ENSDARG00000018404 | krt18 | keratin 18 | 352912 |
| ENSDARG00000017624 | krt4 | keratin 4 | 794486 |
| ENSDARG00000058371 | krt5 | keratin 5 | 0 |
| ENSDARG00000058358 | krt8 | keratin 8 | 797433 |
| ENSDARG00000036830 | krt91 | keratin 91 | 445051 |
| ENSDARG00000044975 | krt94 | keratin 94 | 768289 |
| ENSDARG00000094526 | ksr2 | kinase suppressor of ras 2 | 569127 |
| ENSDARG00000040803 | lactb | lactamase, beta | 553619 |
| ENSDARG00000013741 | lancl1 | LanC antibiotic synthetase component C-like 1 (bacterial) | 494154 |
| ENSDARG00000045465 | lancl2 | LanC lantibiotic synthetase component C-like 2 (bacterial) | 556310 |
| ENSDARG00000057867 | lasp1 | LIM and SH3 protein 1 | 323216 |
| ENSDARG00000014013 | lbr | lamin B receptor | 368360 |
| ENSDARG00000077515 | letm2 | leucine zipper-EF-hand containing transmembrane protein 2 | 100006201 |
| ENSDARG00000030641 | letmd1 | LETM1 domain containing 1 | 567014 |
| ENSDARG00000040528 | lgals3bpb | lectin, galactoside-binding, soluble, 3 binding protein b | 405809 |
| ENSDARG00000025903 | lgals9l1 | lectin, galactoside-binding, soluble, 9 (galectin 9)-like 1 | 337597 |
| ENSDARG00000020493 | lgi1a | leucine-rich, glioma inactivated 1a | 323011 |
| ENSDARG00000058421 | lgi1b | leucine-rich, glioma inactivated 1b | 654829 |
| ENSDARG00000089957 | lgi2a | leucine-rich repeat LGI family, member 2a | 654827 |
| ENSDARG00000041358 | lgi3 | leucine-rich repeat LGI family, member 3 | 654830 |
| ENSDARG00000021595 | lhfpl3 | lipoma HMGIC fusion partner-like 3 | 445050 |
| ENSDARG00000078998 | lhfpl4a | lipoma HMGIC fusion partner-like 4a | 100151593 |
| ENSDARG00000045023 | lhfpl5a | lipoma HMGIC fusion partner-like 5a | 558325 |
| ENSDARG00000074275 | limch1a | LIM and calponin homology domains 1a | 561940 |
| ENSDARG00000103774 | limch1b | LIM and calponin homology domains 1b | 557200 |
| ENSDARG00000013414 | lin7a | lin-7 homolog A (C. elegans) | 393682 |
| ENSDARG00000037932 | lin7b | lin-7 homolog B (C. elegans) | 553768 |
| ENSDARG00000027322 | lin7c | lin-7 homolog C (C. elegans) | 447934 |
| ENSDARG00000034165 | lingo1a | leucine rich repeat and Ig domain containing 1a | 564943 |
| ENSDARG00000069970 | lingo2b | leucine rich repeat and Ig domain containing 2b | 564052 |
| ENSDARG00000067815 | LINGO3 (2 of 2) | leucine rich repeat and Ig domain containing 3 | 100151059 |
| ENSDARG00000044299 | lmnb1 | lamin B1 | 195816 |
| ENSDARG00000101624 | lmnb2 | lamin B2 | 30196 |
| ENSDARG00000029114 | LMTK3 | lemur tyrosine kinase 3 | 562728 |
| ENSDARG00000013542 | lpgat1 | lysophosphatidylglycerol acyltransferase 1 | 561832 |
| ENSDARG00000010144 | lppr3a | lipid phosphate phosphatase-related protein type 3a | 553336 |
| ENSDARG00000028552 | lppr3b | lipid phosphate phosphatase-related protein type 3b | 570173 |
| ENSDARG00000079671 | lppr4a | lipid phosphate phosphatase-related protein type 4a | 559888 |
| ENSDARG00000075580 | lrch2 | leucine-rich repeats and calponin homology (CH) domain containing 2 | 100150488 |
| ENSDARG00000100679 | lrch3 | leucine-rich repeats and calponin homology (CH) domain containing 3 | 100126105 |
| ENSDARG00000027602 | lrfn1 | leucine rich repeat and fibronectin type III domain containing 1 | 393587 |
| ENSDARG00000071230 | lrfn5a | leucine rich repeat and fibronectin type III domain containing 5a | 100144400 |
| ENSDARG00000079396 | lrfn5b | leucine rich repeat and fibronectin type III domain containing 5b | 555839 |
| ENSDARG00000086990 | lrrc16b | leucine rich repeat containing 16B | 0 |
| ENSDARG00000102684 | LRRC1 | leucine rich repeat containing 1 | 0 |
| ENSDARG00000069402 | lrrc4.1 | leucine rich repeat containing 4.1 | 565021 |
| ENSDARG00000003020 | lrrc4.2 | leucine rich repeat containing 4.2 | 566572 |
| ENSDARG00000043938 | lrrc57 | leucine rich repeat containing 57 | 436900 |
| ENSDARG00000063411 | lrrc73 | leucine rich repeat containing 73 | 570766 |
| ENSDARG00000074308 | lrrc75ba | leucine rich repeat containing 75Ba | 101885211 |
| ENSDARG00000079670 | lrrc7 | leucine rich repeat containing 7 | 566023 |
| ENSDARG00000032188 | lrrc8a | leucine rich repeat containing 8 family, member A | 327642 |
| ENSDARG00000095170 | lrrfip1b | leucine rich repeat (in FLII) interacting protein 1b | 100321881 |
| ENSDARG00000052713 | lrrtm1 | leucine rich repeat transmembrane neuronal 1 | 570385 |
| ENSDARG00000078839 | LRRTM4 (1 of 3) | leucine rich repeat transmembrane neuronal 4 | 563710 |
| ENSDARG00000099386 | lrrtm4l2 | leucine rich repeat transmembrane neuronal 4 like 2 | 0 |
| ENSDARG00000103069 | LSAMP | limbic system-associated membrane protein | 0 |
| ENSDARG00000071573 | lsm5 | LSM5 homolog, U6 small nuclear RNA associated (S. cerevisiae) | 563948 |
| ENSDARG00000075556 | luzp1 | leucine zipper protein 1 | 558606 |
| ENSDARG00000058558 | lzts3b | leucine zipper, putative tumor suppressor family member 3b | 560221 |
| ENSDARG00000028533 | macf1a | microtubule-actin crosslinking factor 1a | 562190 |
| ENSDARG00000070145 | magi1a | membrane associated guanylate kinase, WW and PDZ domain containing 1a | 100319849 |
| ENSDARG00000003169 | magi1b | membrane associated guanylate kinase, WW and PDZ domain containing 1b | 474349 |
| ENSDARG00000021590 | MAGI2 (1 of 2) | membrane associated guanylate kinase, WW and PDZ domain containing 2 | 564112 |
| ENSDARG00000073769 | MAGI2 (2 of 2) | membrane associated guanylate kinase, WW and PDZ domain containing 2 | 0 |
| ENSDARG00000101869 | magi3a | membrane associated guanylate kinase, WW and PDZ domain containing 3a | 561689 |
| ENSDARG00000025974 | magi3b | membrane associated guanylate kinase, WW and PDZ domain containing 3b | 0 |
| ENSDARG00000025108 | magixa | MAGI family member, X-linked a | 796857 |
| ENSDARG00000077633 | magixb | MAGI family member, X-linked b | 0 |
| ENSDARG00000104023 | mag | myelin associated glycoprotein | 474346 |
| ENSDARG00000024017 | mamdc1 | MAM domain containing 1 | 368826 |
| ENSDARG00000041728 | man1a2 | mannosidase, alpha, class 1A, member 2 | 335538 |
| ENSDARG00000073792 | man1b1a | mannosidase, alpha, class 1B, member 1a | 562592 |
| ENSDARG00000063101 | man2a2 | mannosidase, alpha, class 2A, member 2 | 100333987 |
| ENSDARG00000059853 | manbal | mannosidase, beta A, lysosomal-like | 795041 |
| ENSDARG00000059601 | map1aa | microtubule-associated protein 1Aa | 100151220 |
| ENSDARG00000022045 | map1ab | microtubule-associated protein 1Ab | 797545 |
| ENSDARG00000060434 | map1b | Hypothetical microtubule-associated protein 1b; Uncharacterized protein | 559032 |
| ENSDARG00000033609 | map1lc3a | microtubule-associated protein 1 light chain 3 alpha | 406466 |
| ENSDARG00000101127 | map1lc3b | microtubule-associated protein 1 light chain 3 beta | 322425 |
| ENSDARG00000060805 | map1sa | microtubule-associated protein 1Sa | 562808 |
| ENSDARG00000060326 | map1sb | microtubule-associated protein 1Sb | 565533 |
| ENSDARG00000007825 | map2k1 | mitogen-activated protein kinase kinase 1 | 406728 |
| ENSDARG00000006609 | map2k2a | mitogen-activated protein kinase kinase 2a | 563181 |
| ENSDARG00000099184 | map2k6 | mitogen-activated protein kinase kinase 6 | 0 |
| ENSDARG00000055052 | map2 | microtubule-associated protein 2 | 0 |
| ENSDARG00000068745 | MAP4 (1 of 3) | microtubule-associated protein 4 | 100537884 |
| ENSDARG00000101687 | MAP4 (3 of 3) | microtubule-associated protein 4 | 100001114 |
| ENSDARG00000071357 | map4k3b | mitogen-activated protein kinase kinase kinase kinase 3b | 100005183 |
| ENSDARG00000074521 | map6a | microtubule-associated protein 6a | 100000844 |
| ENSDARG00000074073 | map6b | microtubule-associated protein 6b | 100334879 |
| ENSDARG00000079777 | map6d1 | MAP6 domain containing 1 | 0 |
| ENSDARG00000104701 | map7d1b | MAP7 domain containing 1b | 796270 |
| ENSDARG00000068480 | map7d2a | MAP7 domain containing 2a | 100005195 |
| ENSDARG00000102730 | mapk10 | mitogen-activated protein kinase 10 | 569698 |
| ENSDARG00000091777 | mapkap1 | mitogen-activated protein kinase associated protein 1 | 100537196 |
| ENSDARG00000089314 | mapta | microtubule-associated protein tau a | 0 |
| ENSDARG00000087616 | maptb | microtubule-associated protein tau b | 567833 |
| ENSDARG00000104014 | MARK1 | MAP/microtubule affinity-regulating kinase 1 | 0 |
| ENSDARG00000032458 | mark2b | MAP/microtubule affinity-regulating kinase 2b | 100005996 |
| ENSDARG00000019345 | mark3a | MAP/microtubule affinity-regulating kinase 3a | 334300 |
| ENSDARG00000026630 | mark3b | MAP/microtubule affinity-regulating kinase 3b | 767634 |
| ENSDARG00000024966 | MARK4 (2 of 2) | MAP/microtubule affinity-regulating kinase 4 | 571279 |
| ENSDARG00000023914 | mark4a | MAP/microtubule affinity-regulating kinase 4a | 569346 |
| ENSDARG00000034396 | mars | methionyl-tRNA synthetase | 338183 |
| ENSDARG00000015947 | matn4 | matrilin 4 | 497348 |
| ENSDARG00000036186 | mbpa | myelin basic protein a | 326281 |
| ENSDARG00000089413 | mbpb | myelin basic protein b | 368319 |
| ENSDARG00000014634 | mbtps1 | membrane-bound transcription factor peptidase, site 1 | 326710 |
| ENSDARG00000056603 | mcf2b | MCF.2 cell line derived transforming sequence b | 559907 |
| ENSDARG00000075159 | mfi2 | antigen p97 (melanoma associated) identified by monoclonal antibodies 133.2 and 96.5 | 565942 |
| ENSDARG00000102744 | mgst3 | microsomal glutathione S-transferase 3 | 406736 |
| ENSDARG00000102184 | mib1 | mindbomb E3 ubiquitin protein ligase 1 | 352910 |
| ENSDARG00000063358 | micu1 | mitochondrial calcium uptake 1 | 561210 |
| ENSDARG00000074255 | MICU3 (1 of 2) | mitochondrial calcium uptake family, member 3 | 100536173 |
| ENSDARG00000078599 | micu3a | mitochondrial calcium uptake family, member 3a | 559552 |
| ENSDARG00000035360 | mink1 | misshapen-like kinase 1 | 100038799 |
| ENSDARG00000102587 | MIOS | missing oocyte, meiosis regulator, homolog (Drosophila) | 0 |
| ENSDARG00000063026 | mlc1 | megalencephalic leukoencephalopathy with subcortical cysts 1 | 559990 |
| ENSDARG00000034616 | mlf2 | myeloid leukemia factor 2 | 561675 |
| ENSDARG00000029472 | mllt4a | myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 4a | 558359 |
| ENSDARG00000060383 | mmaa | methylmalonic aciduria (cobalamin deficiency) cblA type | 564760 |
| ENSDARG00000010553 | mmgt1 | membrane magnesium transporter 1 | 402891 |
| ENSDARG00000056085 | mob4 | MOB family member 4, phocein | 445045 |
| ENSDARG00000060252 | mogs | mannosyl-oligosaccharide glucosidase | 567913 |
| ENSDARG00000031136 | moxd1 | monooxygenase, DBH-like 1 | 570584 |
| ENSDARG00000037679 | MPC2 (2 of 2) | mitochondrial pyruvate carrier 2 | 0 |
| ENSDARG00000024478 | mpc2 | mitochondrial pyruvate carrier 2 | 321611 |
| ENSDARG00000053453 | mpp2a | membrane protein, palmitoylated 2a (MAGUK p55 subfamily member 2) | 562207 |
| ENSDARG00000015184 | mpp3a | membrane protein, palmitoylated 3a (MAGUK p55 subfamily member 3) | 572079 |
| ENSDARG00000062667 | mpp3b | membrane protein, palmitoylated 3b (MAGUK p55 subfamily member 3) | 799884 |
| ENSDARG00000056892 | mpp6a | membrane protein, palmitoylated 6a (MAGUK p55 subfamily member 6) | 402878 |
| ENSDARG00000105209 | MPRIP (3 of 3) | myosin phosphatase Rho interacting protein | 562530 |
| ENSDARG00000102081 | mprip | myosin phosphatase Rho interacting protein | 558831 |
| ENSDARG00000038609 | mpz | myelin protein zero | 114417 |
| ENSDARG00000038768 | mrpl12 | mitochondrial ribosomal protein L12 | 550391 |
| ENSDARG00000075677 | mrpl17 | mitochondrial ribosomal protein L17 | 447945 |
| ENSDARG00000090462 | mrpl20 | mitochondrial ribosomal protein L20 | 751640 |
| ENSDARG00000045696 | mrpl23 | mitochondrial ribosomal protein L23 | 415142 |
| ENSDARG00000043554 | mrpl40 | mitochondrial ribosomal protein L40 | 447873 |
| ENSDARG00000076334 | mrpl43 | mitochondrial ribosomal protein L43 | 436701 |
| ENSDARG00000016307 | mrpl46 | mitochondrial ribosomal protein L46 | 436791 |
| ENSDARG00000068305 | mrps14 | mitochondrial ribosomal protein S14 | 751639 |
| ENSDARG00000104111 | mrps30 | mitochondrial ribosomal protein S30 | 555760 |
| ENSDARG00000057855 | mrps31 | mitochondrial ribosomal protein S31 | 407642 |
| ENSDARG00000079823 | mrps36 | mitochondrial ribosomal protein S36 | 567309 |
| ENSDARG00000063911 | mt-atp6 | ATP synthase 6, mitochondrial | 140519 |
| ENSDARG00000014159 | mtdha | metadherin a | 368910 |
| ENSDARG00000003940 | mtfr1l | mitochondrial fission regulator 1-like | 334296 |
| ENSDARG00000040492 | mthfd1b | methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1b | 260441 |
| ENSDARG00000063899 | mt-nd2 | NADH dehydrogenase 2, mitochondrial | 140532 |
| ENSDARG00000053196 | mtor | mechanistic target of rapamycin (serine/threonine kinase) | 324254 |
| ENSDARG00000076155 | mtpap | mitochondrial poly(A) polymerase | 563801 |
| ENSDARG00000062347 | mtus2 | microtubule associated tumor suppressor candidate 2 | 556195 |
| ENSDARG00000001220 | mycbp2 | MYC binding protein 2 | 368439 |
| ENSDARG00000000103 | myh10 | myosin, heavy chain 10, non-muscle | 555454 |
| ENSDARG00000009782 | myh11a | myosin, heavy chain 11a, smooth muscle | 554168 |
| ENSDARG00000100972 | myh11b | myosin, heavy chain 11b, smooth muscle | 566080 |
| ENSDARG00000073732 | myh14 | myosin, heavy chain 14, non-muscle | 563011 |
| ENSDARG00000063295 | myh9a | myosin, heavy chain 9a, non-muscle | 333938 |
| ENSDARG00000001014 | myh9b | myosin, heavy chain 9b, non-muscle | 561431 |
| ENSDARG00000012944 | myhz2 | myosin, heavy polypeptide 2, fast muscle specific | 246275 |
| ENSDARG00000099766 | myl12.1 | myosin, light chain 12, genome duplicate 1 | 326719 |
| ENSDARG00000008494 | myl6 | myosin, light chain 6, alkali, smooth muscle and non-muscle | 798632 |
| ENSDARG00000038123 | myl9a | myosin, light chain 9a, regulatory | 450006 |
| ENSDARG00000008030 | myl9b | myosin, light chain 9b, regulatory | 406493 |
| ENSDARG00000017441 | mylz3 | myosin, light polypeptide 3, skeletal muscle | 58143 |
| ENSDARG00000074723 | myo10l1 | myosin X-like 1 | 566674 |
| ENSDARG00000075752 | myo18aa | myosin XVIIIAa | 566880 |
| ENSDARG00000061862 | myo18ab | myosin XVIIIAb | 0 |
| ENSDARG00000024694 | myo1b | myosin IB | 541515 |
| ENSDARG00000061579 | myo1c | myosin IC | 566459 |
| ENSDARG00000036863 | MYO1D | myosin ID | 559562 |
| ENSDARG00000036179 | myo1ea | myosin IE, a | 555285 |
| ENSDARG00000078734 | myo1f | myosin IF | 565345 |
| ENSDARG00000036104 | MYO1G | myosin IG | 571487 |
| ENSDARG00000078603 | myo1hb | myosin IHb | 0 |
| ENSDARG00000061635 | myo5aa | myosin VAa | 562188 |
| ENSDARG00000062003 | myo5b | myosin VB | 567402 |
| ENSDARG00000104235 | myo5c | myosin VC | 562676 |
| ENSDARG00000044016 | myo6a | myosin VIa | 445473 |
| ENSDARG00000042141 | myo6b | myosin VIb | 445472 |
| ENSDARG00000073843 | myo9ab | myosin IXAb | 0 |
| ENSDARG00000012378 | naa35 | N(alpha)-acetyltransferase 35, NatC auxiliary subunit | 321587 |
| ENSDARG00000001835 | nalcn | sodium leak channel, non-selective | 548345 |
| ENSDARG00000060853 | nckap1 | NCK-associated protein 1 | 561894 |
| ENSDARG00000079475 | nckipsd | NCK interacting protein with SH3 domain | 100003986 |
| ENSDARG00000002710 | ncl | nucleolin | 767714 |
| ENSDARG00000103902 | nde1 | nudE neurodevelopment protein 1 | 566146 |
| ENSDARG00000010953 | ndel1a | nudE neurodevelopment protein 1-like 1a | 394244 |
| ENSDARG00000104225 | ndel1b | nudE neurodevelopment protein 1-like 1b | 333957 |
| ENSDARG00000103937 | ndrg4 | NDRG family member 4 | 692305 |
| ENSDARG00000042777 | ndufa11 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 11 | 393497 |
| ENSDARG00000042469 | ndufa12 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 12 | 556538 |
| ENSDARG00000098584 | ndufa13 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 13 | 0 |
| ENSDARG00000036329 | ndufa1 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 1 | 415243 |
| ENSDARG00000021984 | ndufa2 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 2 | 554125 |
| ENSDARG00000039346 | ndufa5 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 5 | 541328 |
| ENSDARG00000038028 | ndufa6 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 6 | 393943 |
| ENSDARG00000036684 | ndufa7 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 7 | 445042 |
| ENSDARG00000058463 | ndufab1a | NADH dehydrogenase (ubiquinone) 1, alpha/beta subcomplex, 1a | 573619 |
| ENSDARG00000014915 | ndufab1b | NADH dehydrogenase (ubiquinone) 1, alpha/beta subcomplex, 1b | 325712 |
| ENSDARG00000053652 | ndufaf6 | NADH dehydrogenase (ubiquinone) complex I, assembly factor 6 | 569692 |
| ENSDARG00000043467 | ndufb11 | NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 11 | 562219 |
| ENSDARG00000087456 | NDUFB1 | NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 1, 7kDa | 100535765 |
| ENSDARG00000075709 | ndufb3 | NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 3 | 100332034 |
| ENSDARG00000070824 | ndufb5 | NADH dehydrogenase (ubiquinone) 1 beta subcomplex 5 | 797063 |
| ENSDARG00000037259 | ndufb6 | NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 6 | 393781 |
| ENSDARG00000010113 | ndufb8 | NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 8 | 393829 |
| ENSDARG00000041314 | ndufb9 | NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 9 | 573021 |
| ENSDARG00000102115 | ndufc2 | NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 2 | 541390 |
| ENSDARG00000007526 | ndufs2 | NADH dehydrogenase (ubiquinone) Fe-S protein 2 | 553672 |
| ENSDARG00000015385 | ndufs3 | NADH dehydrogenase (ubiquinone) Fe-S protein 3, (NADH-coenzyme Q reductase) | 550451 |
| ENSDARG00000052840 | ndufs4 | NADH dehydrogenase (ubiquinone) Fe-S protein 4, (NADH-coenzyme Q reductase) | 573689 |
| ENSDARG00000006290 | ndufs5 | NADH dehydrogenase (ubiquinone) Fe-S protein 5 | 553660 |
| ENSDARG00000056583 | ndufs6 | NADH dehydrogenase (ubiquinone) Fe-S protein 6 | 447913 |
| ENSDARG00000013044 | ndufv2 | NADH dehydrogenase (ubiquinone) flavoprotein 2 | 393720 |
| ENSDARG00000021200 | nebl | nebulette | 541446 |
| ENSDARG00000056745 | necab2 | N-terminal EF-hand calcium binding protein 2 | 565258 |
| ENSDARG00000060006 | nedd4l | neural precursor cell expressed, developmentally down-regulated 4-like | 559634 |
| ENSDARG00000007989 | nedd8 | neural precursor cell expressed, developmentally down-regulated 8 | 368667 |
| ENSDARG00000057568 | nefla | neurofilament, light polypeptide a | 0 |
| ENSDARG00000012426 | neflb | neurofilament, light polypeptide b | 664698 |
| ENSDARG00000021351 | nefma | neurofilament, medium polypeptide a | 553718 |
| ENSDARG00000043697 | nefmb | neurofilament, medium polypeptide b | 100033387 |
| ENSDARG00000021607 | negr1 | neuronal growth regulator 1 | 445374 |
| ENSDARG00000102855 | neo1a | neogenin 1a | 266983 |
| ENSDARG00000075100 | neo1b | neogenin 1b | 100332721 |
| ENSDARG00000096368 | neu3.5 | sialidase 3 (membrane sialidase), tandem duplicate 5 | 100000684 |
| ENSDARG00000012982 | nf1a | neurofibromin 1a | 326708 |
| ENSDARG00000056910 | NFASC (1 of 2) | neurofascin | 559590 |
| ENSDARG00000062237 | nfs1 | NFS1 cysteine desulfurase | 562714 |
| ENSDARG00000069878 | nhp2l1a | NHP2 non-histone chromosome protein 2-like 1a (S. cerevisiae) | 393282 |
| ENSDARG00000023299 | nhp2l1b | NHP2 non-histone chromosome protein 2-like 1b (S. cerevisiae) | 321114 |
| ENSDARG00000089066 | NHSL2 | NHS-like 2 | 794153 |
| ENSDARG00000079824 | ninj2 | ninjurin 2 | 799267 |
| ENSDARG00000069427 | nkain2 | Na+/K+ transporting ATPase interacting 2 | 101883714 |
| ENSDARG00000077329 | nlgn2a | neuroligin 2a | 556158 |
| ENSDARG00000079251 | nlgn2b | neuroligin 2b | 100002407 |
| ENSDARG00000062376 | nlgn3b | neuroligin 3b | 567415 |
| ENSDARG00000073724 | nlrx1 | NLR family member X1 | 557335 |
| ENSDARG00000103791 | nme2b.1 | NME/NM23 nucleoside diphosphate kinase 2b, tandem duplicate 1 | 30083 |
| ENSDARG00000020482 | nono | non-POU domain containing, octamer-binding | 321994 |
| ENSDARG00000102845 | novel gene | 0 | 0 |
| ENSDARG00000104615 | novel gene | 0 | 0 |
| ENSDARG00000068624 | novel gene | 0 | 100003123 |
| ENSDARG00000059083 | novel gene | 0 | 100005981 |
| ENSDARG00000090164 | novel gene | 0 | 101884007 |
| ENSDARG00000078770 | novel gene | 0 | 336430 |
| ENSDARG00000033201 | novel gene | 0 | 449795 |
| ENSDARG00000053454 | novel gene | 0 | 556251 |
| ENSDARG00000077545 | novel gene | 0 | 558390 |
| ENSDARG00000077124 | novel gene | 0 | 564701 |
| ENSDARG00000104264 | novel gene | 0 | 777704 |
| ENSDARG00000043864 | nptnb | neuroplastin b | 403006 |
| ENSDARG00000037794 | nptx2a | neuronal pentraxin 2a | 553230 |
| ENSDARG00000040926 | nr2f2 | nuclear receptor subfamily 2, group F, member 2 | 30424 |
| ENSDARG00000069368 | nrn1la | neuritin 1-like a | 100004307 |
| ENSDARG00000061647 | nrxn1a | neurexin 1a | 565531 |
| ENSDARG00000063150 | nrxn2b | neurexin 2b | 0 |
| ENSDARG00000043746 | nrxn3a | neurexin 3a | 0 |
| ENSDARG00000062693 | nrxn3b | neurexin 3b | 570698 |
| ENSDARG00000071017 | nt5e | 5'-nucleotidase, ecto (CD73) | 393906 |
| ENSDARG00000018065 | ntm | neurotrimin | 678534 |
| ENSDARG00000014973 | ntng1a | netrin g1a | 0 |
| ENSDARG00000077367 | ntng2a | netrin g2a | 560014 |
| ENSDARG00000077228 | ntrk3a | neurotrophic tyrosine kinase, receptor, type 3a | 568668 |
| ENSDARG00000027279 | numb | numb homolog (Drosophila) | 692064 |
| ENSDARG00000010078 | nup133 | nucleoporin 133 | 406852 |
| ENSDARG00000103123 | NYAP1 | neuronal tyrosine-phosphorylated phosphoinositide-3-kinase adaptor 1 | 101885998 |
| ENSDARG00000028900 | odf2a | outer dense fiber of sperm tails 2a | 792921 |
| ENSDARG00000034270 | ogdha | oxoglutarate (alpha-ketoglutarate) dehydrogenase a (lipoamide) | 564552 |
| ENSDARG00000103428 | ogdhb | oxoglutarate (alpha-ketoglutarate) dehydrogenase b (lipoamide) | 797715 |
| ENSDARG00000079249 | OGDHL | oxoglutarate dehydrogenase-like | 559207 |
| ENSDARG00000089444 | ogmb | oligodendrocyte myelin glycoprotein b | 101882436 |
| ENSDARG00000105243 | ogt.1 | O-linked N-acetylglucosamine (GlcNAc) transferase, tandem duplicate 1 | 337685 |
| ENSDARG00000018270 | olfm1a | olfactomedin 1a | 436764 |
| ENSDARG00000014053 | olfm1b | olfactomedin 1b | 445092 |
| ENSDARG00000077847 | olfm2a | olfactomedin 2a | 334035 |
| ENSDARG00000102825 | olfm2b | olfactomedin 2b | 406544 |
| ENSDARG00000071493 | olfm3a | olfactomedin 3a | 563182 |
| ENSDARG00000086573 | omga | oligodendrocyte myelin glycoprotein a | 0 |
| ENSDARG00000061761 | opa3 | optic atrophy 3 | 497278 |
| ENSDARG00000013005 | opcml | opioid binding protein/cell adhesion molecule-like | 449538 |
| ENSDARG00000039724 | OSBPL8 | oxysterol binding protein-like 8 | 101883298 |
| ENSDARG00000069313 | oxa1l | oxidase (cytochrome c) assembly 1-like | 562963 |
| ENSDARG00000039578 | pa2g4a | proliferation-associated 2G4, a | 368737 |
| ENSDARG00000017219 | pabpc1a | poly(A) binding protein, cytoplasmic 1a | 606498 |
| ENSDARG00000099339 | pacsin3 | protein kinase C and casein kinase substrate in neurons 3 | 393964 |
| ENSDARG00000026595 | pafah1b1b | platelet-activating factor acetylhydrolase 1b, regulatory subunit 1b | 394247 |
| ENSDARG00000055875 | pag1 | phosphoprotein membrane anchor with glycosphingolipid microdomains 1 | 100034498 |
| ENSDARG00000018110 | pak4 | p21 protein (Cdc42/Rac)-activated kinase 4 | 431769 |
| ENSDARG00000030154 | pak7 | p21 protein (Cdc42/Rac)-activated kinase 7 | 0 |
| ENSDARG00000026882 | palm1a | paralemmin 1a | 550557 |
| ENSDARG00000069608 | palm2 | paralemmin 2 | 100002543 |
| ENSDARG00000102822 | pam16 | presequence translocase-associated motor 16 homolog (S. cerevisiae) | 393777 |
| ENSDARG00000100170 | pam | peptidylglycine alpha-amidating monooxygenase | 570822 |
| ENSDARG00000101719 | pargl | poly (ADP-ribose) glycohydrolase, like | 797792 |
| ENSDARG00000019529 | parp1 | poly (ADP-ribose) polymerase 1 | 560788 |
| ENSDARG00000079202 | parp2 | poly (ADP-ribose) polymerase 2 | 100007906 |
| ENSDARG00000003961 | parp3 | poly (ADP-ribose) polymerase family, member 3 | 335495 |
| ENSDARG00000011824 | pbxip1b | pre-B-cell leukemia homeobox interacting protein 1b | 0 |
| ENSDARG00000099729 | pcdh10a | protocadherin 10a | 0 |
| ENSDARG00000102824 | pcdh10b | protocadherin 10b | 360138 |
| ENSDARG00000034344 | pcdh19 | protocadherin 19 | 0 |
| ENSDARG00000097170 | pcdh1a4 | protocadherin 1 alpha 4 | 497127 |
| ENSDARG00000036175 | pcdh1b | protocadherin 1b | 572016 |
| ENSDARG00000100829 | pcdh1g32 | protocadherin 1 gamma 32 | 554016 |
| ENSDARG00000088136 | pcdh2aa15 | protocadherin 2 alpha a 15 | 493601 |
| ENSDARG00000099783 | pcdh2ac | protocadherin 2 alpha c | 493587 |
| ENSDARG00000103256 | pcdh2g1 | protocadherin 2 gamma 1 | 103912055 |
| ENSDARG00000063264 | PCDH9 | protocadherin 9 | 0 |
| ENSDARG00000077023 | pcdhb | protocadherin b | 100333352 |
| ENSDARG00000063299 | pcloa | piccolo presynaptic cytomatrix protein a | 100148002 |
| ENSDARG00000098880 | pclob | piccolo presynaptic cytomatrix protein b | 0 |
| ENSDARG00000058162 | pcyt1ba | phosphate cytidylyltransferase 1, choline, beta a | 100001552 |
| ENSDARG00000005220 | pdcd6 | programmed cell death 6 | 393925 |
| ENSDARG00000053609 | pddc1 | Parkinson disease 7 domain containing 1 | 100148360 |
| ENSDARG00000012387 | pdha1a | pyruvate dehydrogenase (lipoamide) alpha 1a | 406702 |
| ENSDARG00000051756 | pdhx | pyruvate dehydrogenase complex, component X | 393532 |
| ENSDARG00000013128 | pdk1 | pyruvate dehydrogenase kinase, isozyme 1 | 555840 |
| ENSDARG00000014527 | pdk3a | pyruvate dehydrogenase kinase, isozyme 3a | 0 |
| ENSDARG00000018285 | pdpk1b | 3-phosphoinositide dependent protein kinase 1b | 403000 |
| ENSDARG00000038315 | pdxp | pyridoxal (pyridoxine, vitamin B6) phosphatase | 561030 |
| ENSDARG00000053194 | pdzd11 | PDZ domain containing 11 | 405893 |
| ENSDARG00000055976 | pecr | peroxisomal trans-2-enoyl-CoA reductase | 550422 |
| ENSDARG00000058202 | pex16 | peroxisomal biogenesis factor 16 | 573990 |
| ENSDARG00000060504 | pfkla | phosphofructokinase, liver a | 570106 |
| ENSDARG00000099755 | pfklb | phosphofructokinase, liver b | 550259 |
| ENSDARG00000060797 | pfkmb | phosphofructokinase, muscle b | 568001 |
| ENSDARG00000028000 | pfkpa | phosphofructokinase, platelet a | 560944 |
| ENSDARG00000012801 | pfkpb | phosphofructokinase, platelet b | 561416 |
| ENSDARG00000035608 | pgam5 | phosphoglycerate mutase family member 5 | 492357 |
| ENSDARG00000045297 | phb2a | prohibitin 2a | 324421 |
| ENSDARG00000017728 | phb2b | prohibitin 2b | 436954 |
| ENSDARG00000057414 | phb | prohibitin | 321346 |
| ENSDARG00000030887 | phf23a | PHD finger protein 23a | 541372 |
| ENSDARG00000001873 | phgdh | phosphoglycerate dehydrogenase | 321928 |
| ENSDARG00000105159 | phka1 | phosphorylase kinase, alpha 1 (muscle) | 572183 |
| ENSDARG00000054208 | phkg2 | phosphorylase kinase, gamma 2 (testis) | 393937 |
| ENSDARG00000061093 | phldb1a | pleckstrin homology-like domain, family B, member 1a | 566287 |
| ENSDARG00000079378 | phldb1b | pleckstrin homology-like domain, family B, member 1b | 570275 |
| ENSDARG00000076724 | pi4kaa | phosphatidylinositol 4-kinase, catalytic, alpha a | 556956 |
| ENSDARG00000062823 | pi4kab | phosphatidylinositol 4-kinase, catalytic, alpha b | 793349 |
| ENSDARG00000024642 | pip5k1ab | phosphatidylinositol-4-phosphate 5-kinase, type I, alpha, b | 492549 |
| ENSDARG00000031020 | pip5k2 | phosphatidylinositol-4-phosphate 5-kinase, type II | 373124 |
| ENSDARG00000052462 | pisd | phosphatidylserine decarboxylase | 553433 |
| ENSDARG00000099730 | pkma | pyruvate kinase, muscle, a | 335817 |
| ENSDARG00000079438 | PKP3 (2 of 2) | plakophilin 3 | 100330353 |
| ENSDARG00000045331 | pkp4 | plakophilin 4 | 561566 |
| ENSDARG00000005774 | pl10 | pl10 | 30116 |
| ENSDARG00000052957 | plcd3a | phospholipase C, delta 3a | 569040 |
| ENSDARG00000057125 | pld1b | phospholipase D1b | 572492 |
| ENSDARG00000062590 | pleca | plectin a | 567639 |
| ENSDARG00000021987 | plecb | plectin b | 100321011 |
| ENSDARG00000045870 | plekha5 | pleckstrin homology domain containing, family A member 5 | 567300 |
| ENSDARG00000020328 | plekha6 | pleckstrin homology domain containing, family A member 6 | 568764 |
| ENSDARG00000060813 | plekha7a | pleckstrin homology domain containing, family A member 7a | 571486 |
| ENSDARG00000062220 | plekha7b | pleckstrin homology domain containing, family A member 7b | 100005106 |
| ENSDARG00000100789 | plgrkt | plasminogen receptor, C-terminal lysine transmembrane protein | 606662 |
| ENSDARG00000011929 | plp1b | proteolipid protein 1b | 368234 |
| ENSDARG00000039272 | pls1 | plastin 1 (I isoform) | 334273 |
| ENSDARG00000104668 | plxna1b | plexin A1b | 0 |
| ENSDARG00000040002 | pnpla7b | patatin-like phospholipase domain containing 7b | 503936 |
| ENSDARG00000078745 | pnpla8 | patatin-like phospholipase domain containing 8 | 100126812 |
| ENSDARG00000102790 | PNPT1 | polyribonucleotide nucleotidyltransferase 1 | 0 |
| ENSDARG00000010941 | pomgnt2 | protein O-linked mannose N-acetylglucosaminyltransferase 2 (beta 1,4-) | 497644 |
| ENSDARG00000104267 | postnb | periostin, osteoblast specific factor b | 337176 |
| ENSDARG00000043527 | ppapdc2 | phosphatidic acid phosphatase type 2 domain containing 2 | 553687 |
| ENSDARG00000013000 | ppfia2 | protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2 | 566597 |
| ENSDARG00000077053 | ppfia3 | protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 3 | 567230 |
| ENSDARG00000053205 | ppfia4 | protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 4 | 100005543 |
| ENSDARG00000045749 | PPFIBP1 (1 of 2) | PTPRF interacting protein, binding protein 1 (liprin beta 1) | 565020 |
| ENSDARG00000003486 | ppp1caa | protein phosphatase 1, catalytic subunit, alpha isozyme a | 407980 |
| ENSDARG00000071566 | ppp1cab | protein phosphatase 1, catalytic subunit, alpha isozyme b | 327301 |
| ENSDARG00000044153 | ppp1cb | protein phosphatase 1, catalytic subunit, beta isozyme | 100003223 |
| ENSDARG00000099226 | ppp1cc | protein phosphatase 1, catalytic subunit, gamma isozyme | 571753 |
| ENSDARG00000010784 | ppp1r12a | protein phosphatase 1, regulatory subunit 12A | 445393 |
| ENSDARG00000103144 | ppp1r21 | protein phosphatase 1, regulatory subunit 21 | 492333 |
| ENSDARG00000061304 | ppp1r9a | protein phosphatase 1, regulatory subunit 9A | 560862 |
| ENSDARG00000079366 | ppp1r9ba | protein phosphatase 1, regulatory subunit 9Ba | 558232 |
| ENSDARG00000071709 | ppp1r9bb | protein phosphatase 1, regulatory subunit 9Bb | 100331262 |
| ENSDARG00000007791 | ppp2r1a | protein phosphatase 2, regulatory subunit A, alpha | 406685 |
| ENSDARG00000032430 | ppp2r1b | protein phosphatase 2, regulatory subunit A, beta | 449548 |
| ENSDARG00000056797 | ppp2r2ca | protein phosphatase 2, regulatory subunit B, gamma a | 100002802 |
| ENSDARG00000054931 | PPP2R5B | protein phosphatase 2, regulatory subunit B', beta | 562282 |
| ENSDARG00000031200 | ppp2r5cb | protein phosphatase 2, regulatory subunit B', gamma b | 557208 |
| ENSDARG00000069118 | ppp2r5eb | protein phosphatase 2, regulatory subunit B', epsilon isoform b | 373878 |
| ENSDARG00000069360 | ppp3r1b | protein phosphatase 3 (formerly 2B), regulatory s1ubunit B, alpha isoform, b | 447814 |
| ENSDARG00000045540 | ppp6r2a | protein phosphatase 6, regulatory subunit 2a | 560729 |
| ENSDARG00000058734 | prdx1 | peroxiredoxin 1 | 541344 |
| ENSDARG00000025350 | prdx2 | peroxiredoxin 2 | 791455 |
| ENSDARG00000032102 | prdx3 | peroxiredoxin 3 | 0 |
| ENSDARG00000055064 | prdx5 | peroxiredoxin 5 | 0 |
| ENSDARG00000075793 | prex1 | phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 1 | 570984 |
| ENSDARG00000020982 | prickle2a | prickle homolog 2a (Drosophila) | 777736 |
| ENSDARG00000037593 | prickle2b | prickle homolog 2 (Drosophila) | 368250 |
| ENSDARG00000076128 | prkar1aa | protein kinase, cAMP-dependent, regulatory, type I, alpha (tissue specific extinguisher 1) a | 494533 |
| ENSDARG00000101755 | prkar1ab | protein kinase, cAMP-dependent, regulatory, type I, alpha (tissue specific extinguisher 1) b | 550427 |
| ENSDARG00000103800 | prkar1b | protein kinase, cAMP-dependent, regulatory, type I, beta | 0 |
| ENSDARG00000009208 | prkcda | protein kinase C, delta a | 334571 |
| ENSDARG00000075949 | prkd1 | protein kinase D1 | 557123 |
| ENSDARG00000075083 | prkdc | protein kinase, DNA-activated, catalytic polypeptide | 562283 |
| ENSDARG00000037946 | prl | prolactin | 791526 |
| ENSDARG00000003705 | prnprs3 | prion protein, related sequence 3 | 503702 |
| ENSDARG00000037783 | proza | protein Z, vitamin K-dependent plasma glycoprotein a | 768176 |
| ENSDARG00000015239 | prp19 | PRP19/PSO4 homolog (S. cerevisiae) | 321544 |
| ENSDARG00000028306 | prph | peripherin | 30259 |
| ENSDARG00000076702 | PSD (1 of 2) | pleckstrin and Sec7 domain containing | 101886286 |
| ENSDARG00000078102 | PSD (2 of 2) | pleckstrin and Sec7 domain containing | 0 |
| ENSDARG00000063036 | psd2 | pleckstrin and Sec7 domain containing 2 | 557199 |
| ENSDARG00000104820 | psd3l | pleckstrin and Sec7 domain containing 3, like | 0 |
| ENSDARG00000037038 | psmc6 | proteasome (prosome, macropain) 26S subunit, ATPase, 6 | 321585 |
| ENSDARG00000008249 | ptchd4 | patched domain containing 4 | 564097 |
| ENSDARG00000022951 | pth2 | parathyroid hormone 2 | 402812 |
| ENSDARG00000019945 | ptprdb | protein tyrosine phosphatase, receptor type, D, b | 100073332 |
| ENSDARG00000103479 | ptprfa | protein tyrosine phosphatase, receptor type, f, a | 0 |
| ENSDARG00000045006 | ptprga | protein tyrosine phosphatase, receptor type, g a | 561197 |
| ENSDARG00000035970 | ptprn2 | protein tyrosine phosphatase, receptor type, N polypeptide 2 | 325033 |
| ENSDARG00000058646 | ptprna | protein tyrosine phosphatase, receptor type, Na | 324790 |
| ENSDARG00000077047 | ptprnb | protein tyrosine phosphatase, receptor type, Nb | 564351 |
| ENSDARG00000059738 | ptprsa | protein tyrosine phosphatase, receptor type, s, a | 140820 |
| ENSDARG00000097572 | ptprt | protein tyrosine phosphatase, receptor type, t | 0 |
| ENSDARG00000051814 | ptprz1a | protein tyrosine phosphatase, receptor-type, Z polypeptide 1a | 0 |
| ENSDARG00000077339 | ptrh2 | peptidyl-tRNA hydrolase 2 | 791795 |
| ENSDARG00000001241 | puf60b | poly-U binding splicing factor b | 415211 |
| ENSDARG00000067591 | pura | purine-rich element binding protein A | 415106 |
| ENSDARG00000068822 | purba | purine-rich element binding protein Ba | 564840 |
| ENSDARG00000015921 | pwp1 | PWP1 homolog (S. cerevisiae) | 393262 |
| ENSDARG00000102254 | pycr1a | pyrroline-5-carboxylate reductase 1a | 393799 |
| ENSDARG00000098639 | pycr1b | pyrroline-5-carboxylate reductase 1b | 100149838 |
| ENSDARG00000015236 | pycrl | pyrroline-5-carboxylate reductase-like | 560682 |
| ENSDARG00000010316 | qars | glutaminyl-tRNA synthetase | 394188 |
| ENSDARG00000002026 | qkib | QKI, KH domain containing, RNA binding b | 100462714 |
| ENSDARG00000077174 | QSOX2 | quiescin Q6 sulfhydryl oxidase 2 | 559603 |
| ENSDARG00000026484 | rab15 | RAB15, member RAS oncogene family | 436590 |
| ENSDARG00000102078 | RAB35 (2 of 2) | RAB35, member RAS oncogene family | 796454 |
| ENSDARG00000058425 | rab35b | RAB35, member RAS oncogene family b | 445154 |
| ENSDARG00000056347 | rab3aa | RAB3A, member RAS oncogene family, a | 445024 |
| ENSDARG00000042803 | rab3b | RAB3B, member RAS oncogene family | 557482 |
| ENSDARG00000038010 | rac2 | ras-related C3 botulinum toxin substrate 2 (rho family, small GTP binding protein Rac2) | 415151 |
| ENSDARG00000090062 | rac3a | ras-related C3 botulinum toxin substrate 3a (rho family, small GTP binding protein Rac3) | 437027 |
| ENSDARG00000020795 | rac3b | ras-related C3 botulinum toxin substrate 3b (rho family, small GTP binding protein Rac3) | 795167 |
| ENSDARG00000007196 | rae1 | ribonucleic acid export 1 | 393973 |
| ENSDARG00000039974 | rai14 | retinoic acid induced 14 | 794796 |
| ENSDARG00000045463 | ralaa | v-ral simian leukemia viral oncogene homolog Aa (ras related) | 393993 |
| ENSDARG00000040778 | ralba | v-ral simian leukemia viral oncogene homolog Ba (ras related) | 415213 |
| ENSDARG00000076889 | ralgapa1 | Ral GTPase activating protein, alpha subunit 1 (catalytic) | 556427 |
| ENSDARG00000005861 | ralgapa2 | Ral GTPase activating protein, alpha subunit 2 (catalytic) | 568210 |
| ENSDARG00000088899 | ralgapb | Ral GTPase activating protein, beta subunit (non-catalytic) | 559303 |
| ENSDARG00000057026 | ran | RAN, member RAS oncogene family | 30572 |
| ENSDARG00000087346 | rap1ab | RAP1A, member of RAS oncogene family b | 406714 |
| ENSDARG00000103032 | rap2b | RAP2B, member of RAS oncogene family | 414329 |
| ENSDARG00000015649 | rap2c | RAP2C, member of RAS oncogene family | 474323 |
| ENSDARG00000075924 | RAPGEF4 (1 of 2) | Rap guanine nucleotide exchange factor (GEF) 4 | 100141331 |
| ENSDARG00000079872 | rapgef4 | Rap guanine nucleotide exchange factor (GEF) 4 | 0 |
| ENSDARG00000054530 | rars | arginyl-tRNA synthetase | 337070 |
| ENSDARG00000073824 | RASGRF1 | Ras protein-specific guanine nucleotide-releasing factor 1 | 568885 |
| ENSDARG00000002816 | rasgrf2b | Ras protein-specific guanine nucleotide-releasing factor 2b | 553520 |
| ENSDARG00000055080 | rbm4.2 | RNA binding motif protein 4.2 | 557528 |
| ENSDARG00000022129 | rbm4.3 | RNA binding motif protein 4.3 | 406277 |
| ENSDARG00000077060 | rbm6 | RNA binding motif protein 6 | 567058 |
| ENSDARG00000038030 | rbx1 | ring-box 1, E3 ubiquitin protein ligase | 449846 |
| ENSDARG00000056075 | rca2.1 | regulator of complement activation group 2 gene 1 | 541367 |
| ENSDARG00000008491 | RDH13 (1 of 3) | retinol dehydrogenase 13 (all-trans/9-cis) | 407663 |
| ENSDARG00000074064 | rem2 | RAS (RAD and GEM)-like GTP binding 2 | 798285 |
| ENSDARG00000098654 | RFTN1 (2 of 2) | raftlin, lipid raft linker 1 | 101885083 |
| ENSDARG00000056078 | rftn2 | raftlin family member 2 | 406729 |
| ENSDARG00000012248 | rgma | repulsive guidance molecule family member a | 414272 |
| ENSDARG00000036774 | rgs12b | regulator of G-protein signaling 12b | 378970 |
| ENSDARG00000039435 | rgs17 | regulator of G-protein signaling 17 | 436796 |
| ENSDARG00000077385 | rgs19 | regulator of G-protein signaling 19 | 100170801 |
| ENSDARG00000038859 | rgs20 | regulator of G-protein signaling 20 | 436704 |
| ENSDARG00000016584 | rgs7a | regulator of G-protein signaling 7a | 436814 |
| ENSDARG00000096687 | rgs7b | regulator of G-protein signaling 7b | 100535325 |
| ENSDARG00000086756 | RGS9BP | regulator of G protein signaling 9 binding protein | 559232 |
| ENSDARG00000045156 | rgs9b | regulator of G-protein signaling 9b | 793087 |
| ENSDARG00000009018 | rhbg | Rh family, B glycoprotein (gene/pseudogene) | 337596 |
| ENSDARG00000026845 | rhoaa | ras homolog gene family, member Aa | 406411 |
| ENSDARG00000021309 | rhoae | ras homolog gene family, member Ae | 394125 |
| ENSDARG00000090036 | RHOBTB3 | Rho-related BTB domain containing 3 | 562804 |
| ENSDARG00000004301 | rhogb | ras homolog family member Gb | 336933 |
| ENSDARG00000028401 | rhogc | ras homolog gene family, member Gc | 324681 |
| ENSDARG00000025953 | rhoq | ras homolog family member Q | 327510 |
| ENSDARG00000099996 | rhot1a | ras homolog family member T1a | 327143 |
| ENSDARG00000104101 | RIMBP2 (2 of 2) | RIMS binding protein 2 | 100535071 |
| ENSDARG00000001154 | rimbp2 | RIMS binding protein 2 | 0 |
| ENSDARG00000074680 | rims1a | regulating synaptic membrane exocytosis 1a | 0 |
| ENSDARG00000078902 | rims1b | regulating synaptic membrane exocytosis 1b | 568486 |
| ENSDARG00000101606 | rims2a | regulating synaptic membrane exocytosis 2a | 393672 |
| ENSDARG00000102690 | rims2b | regulating synaptic membrane exocytosis 2b | 563393 |
| ENSDARG00000062305 | rims3 | regulating synaptic membrane exocytosis 3 | 563414 |
| ENSDARG00000070100 | RIN1 (1 of 2) | Ras and Rab interactor 1 | 101882011 |
| ENSDARG00000069150 | rit1 | Ras-like without CAAX 1 | 793942 |
| ENSDARG00000104458 | rnasekb | ribonuclease, RNase K b | 336290 |
| ENSDARG00000014891 | robo2 | roundabout, axon guidance receptor, homolog 2 (Drosophila) | 60309 |
| ENSDARG00000104413 | rogdi | rogdi homolog (Drosophila) | 335642 |
| ENSDARG00000044339 | rp2 | retinitis pigmentosa 2 (X-linked recessive) | 406755 |
| ENSDARG00000042905 | rpl10a | ribosomal protein L10a | 323305 |
| ENSDARG00000025581 | rpl10 | ribosomal protein L10 | 336712 |
| ENSDARG00000043509 | rpl11 | ribosomal protein L11 | 415229 |
| ENSDARG00000006691 | rpl12 | ribosomal protein L12 | 327089 |
| ENSDARG00000044093 | rpl13a | ribosomal protein L13a | 560828 |
| ENSDARG00000099380 | rpl13 | ribosomal protein L13 | 378961 |
| ENSDARG00000103433 | rpl14 | ribosomal protein L14 | 323365 |
| ENSDARG00000009285 | rpl15 | ribosomal protein L15 | 445053 |
| ENSDARG00000025073 | rpl18a | ribosomal protein L18a | 394035 |
| ENSDARG00000029533 | rpl18 | ribosomal protein L18 | 445038 |
| ENSDARG00000013307 | rpl19 | ribosomal protein L19 | 406489 |
| ENSDARG00000010516 | rpl21 | ribosomal protein L21 | 336568 |
| ENSDARG00000070437 | rpl22 | ribosomal protein L22 | 557397 |
| ENSDARG00000006316 | rpl23a | ribosomal protein L23a | 335539 |
| ENSDARG00000053457 | rpl23 | ribosomal protein L23 | 336812 |
| ENSDARG00000099104 | rpl24 | ribosomal protein L24 | 192301 |
| ENSDARG00000102317 | rpl26 | ribosomal protein L26 | 406387 |
| ENSDARG00000015128 | rpl27 | ribosomal protein L27 | 325618 |
| ENSDARG00000005791 | rpl28 | ribosomal protein L28 | 394036 |
| ENSDARG00000035871 | rpl30 | ribosomal protein L30 | 336713 |
| ENSDARG00000053365 | rpl31 | ribosomal protein L31 | 677758 |
| ENSDARG00000054818 | rpl32 | ribosomal protein L32 | 556556 |
| ENSDARG00000029500 | rpl34 | ribosomal protein L34 | 394097 |
| ENSDARG00000088030 | rpl35a | ribosomal protein L35a | 436760 |
| ENSDARG00000018334 | rpl35 | ribosomal protein L35 | 192299 |
| ENSDARG00000100588 | rpl36 | ribosomal protein L36 | 405888 |
| ENSDARG00000098458 | RPL37A | ribosomal protein L37a | 572109 |
| ENSDARG00000076360 | rpl37 | ribosomal protein L37 | 0 |
| ENSDARG00000006413 | rpl38 | ribosomal protein L38 | 436759 |
| ENSDARG00000003599 | rpl3 | ribosomal protein L3 | 322571 |
| ENSDARG00000041182 | rpl4 | ribosomal protein L4 | 337090 |
| ENSDARG00000015862 | rpl5b | ribosomal protein L5b | 415196 |
| ENSDARG00000058451 | rpl6 | ribosomal protein L6 | 336727 |
| ENSDARG00000019230 | rpl7a | ribosomal protein L7a | 337010 |
| ENSDARG00000007320 | rpl7 | ribosomal protein L7 | 336710 |
| ENSDARG00000014867 | rpl8 | ribosomal protein L8 | 393686 |
| ENSDARG00000037350 | rpl9 | ribosomal protein L9 | 336702 |
| ENSDARG00000051783 | rplp0 | ribosomal protein, large, P0 | 58101 |
| ENSDARG00000021864 | rplp1 | ribosomal protein, large, P1 | 336719 |
| ENSDARG00000011201 | rplp2l | ribosomal protein, large P2, like | 558312 |
| ENSDARG00000101406 | rplp2 | ribosomal protein, large P2 | 335629 |
| ENSDARG00000034897 | rps10 | ribosomal protein S10 | 394121 |
| ENSDARG00000053058 | rps11 | ribosomal protein S11 | 406686 |
| ENSDARG00000036875 | rps12 | ribosomal protein S12 | 337007 |
| ENSDARG00000036298 | rps13 | ribosomal protein S13 | 415169 |
| ENSDARG00000036629 | rps14 | ribosomal protein S14 | 336687 |
| ENSDARG00000010160 | rps15a | ribosomal protein S15a | 336764 |
| ENSDARG00000070849 | rps15 | ribosomal protein S15 | 337148 |
| ENSDARG00000045487 | rps16 | ribosomal protein S16 | 767765 |
| ENSDARG00000046157 | RPS17 | ribosomal protein S17 | 541327 |
| ENSDARG00000100392 | rps18 | ribosomal protein S18 | 192300 |
| ENSDARG00000030602 | rps19 | ribosomal protein S19 | 393723 |
| ENSDARG00000025850 | rps21 | ribosomal protein S21 | 394166 |
| ENSDARG00000021838 | rps23 | ribosomal protein S23 | 100000341 |
| ENSDARG00000039347 | rps24 | ribosomal protein S24 | 394194 |
| ENSDARG00000041811 | rps25 | ribosomal protein S25 | 393788 |
| ENSDARG00000037071 | rps26 | ribosomal protein S26 | 336662 |
| ENSDARG00000023298 | rps27.1 | ribosomal protein S27, isoform 1 | 554155 |
| ENSDARG00000100513 | rps27.2 | ribosomal protein S27, isoform 2 | 677743 |
| ENSDARG00000032725 | rps27a | ribosomal protein S27a | 337810 |
| ENSDARG00000035860 | rps28 | ribosomal protein S28 | 406307 |
| ENSDARG00000041232 | rps29 | ribosomal protein S29 | 405889 |
| ENSDARG00000077291 | rps2 | ribosomal protein S2 | 406565 |
| ENSDARG00000035692 | rps3a | ribosomal protein S3A | 337240 |
| ENSDARG00000103007 | rps3 | ribosomal protein S3 | 336550 |
| ENSDARG00000014690 | rps4x | ribosomal protein S4, X-linked | 449547 |
| ENSDARG00000043453 | rps5 | ribosomal protein S5 | 192297 |
| ENSDARG00000019778 | rps6 | ribosomal protein S6 | 445028 |
| ENSDARG00000042566 | rps7 | ribosomal protein S7 | 393725 |
| ENSDARG00000055996 | rps8a | ribosomal protein S8a | 407690 |
| ENSDARG00000057368 | rps8b | ribosomal protein S8b | 100000836 |
| ENSDARG00000011405 | rps9 | ribosomal protein S9 | 114420 |
| ENSDARG00000069728 | rpz3 | rapunzel 3 | 100001029 |
| ENSDARG00000039871 | rpz4 | rapunzel 4 | 550609 |
| ENSDARG00000075718 | rpz5 | rapunzel 5 | 100003142 |
| ENSDARG00000091161 | rpz | rapunzel | 100002737 |
| ENSDARG00000013763 | RRBP1 (1 of 2) | ribosome binding protein 1 | 553448 |
| ENSDARG00000041703 | rrbp1b | ribosome binding protein 1 homolog b (dog) | 0 |
| ENSDARG00000062558 | rspry1 | ring finger and SPRY domain containing 1 | 565154 |
| ENSDARG00000101047 | rtcb | RNA 2',3'-cyclic phosphate and 5'-OH ligase | 406376 |
| ENSDARG00000090035 | rtn4r | reticulon 4 receptor | 403306 |
| ENSDARG00000029692 | rufy3 | RUN and FYVE domain containing 3 | 541522 |
| ENSDARG00000078125 | rusc1 | RUN and SH3 domain containing 1 | 558727 |
| ENSDARG00000002591 | ruvbl1 | RuvB-like AAA ATPase 1 | 317679 |
| ENSDARG00000055639 | ruvbl2 | RuvB-like AAA ATPase 2 | 317678 |
| ENSDARG00000023797 | ryr1b | ryanodine receptor 1b (skeletal) | 570245 |
| ENSDARG00000071331 | ryr3 | ryanodine receptor 3 | 561350 |
| ENSDARG00000071014 | s100u | S100 calcium binding protein U | 563060 |
| ENSDARG00000042690 | s1pr1 | sphingosine-1-phosphate receptor 1 | 64617 |
| ENSDARG00000059806 | sacm1lb | SAC1 suppressor of actin mutations 1-like b (yeast) | 791142 |
| ENSDARG00000091042 | sacs | sacsin molecular chaperone | 558150 |
| ENSDARG00000058102 | sardh | sarcosine dehydrogenase | 394103 |
| ENSDARG00000010610 | sarm1 | sterile alpha and TIR motif containing 1 | 403143 |
| ENSDARG00000060865 | scai | suppressor of cancer cell invasion | 556383 |
| ENSDARG00000079645 | sc:d217 | sc:d217 | 0 |
| ENSDARG00000103993 | scg2a | secretogranin II (chromogranin C) a | 793796 |
| ENSDARG00000038574 | scg2b | secretogranin II (chromogranin C), b | 777641 |
| ENSDARG00000058348 | scinlb | scinderin like b | 406363 |
| ENSDARG00000010728 | scin | scinderin | 566764 |
| ENSDARG00000000861 | scrib | scribbled planar cell polarity protein | 368473 |
| ENSDARG00000100562 | sdc3 | syndecan 3 | 337077 |
| ENSDARG00000012513 | sdcbp2 | syndecan binding protein (syntenin) 2 | 325004 |
| ENSDARG00000030097 | sdcbp | syndecan binding protein (syntenin) | 407719 |
| ENSDARG00000062854 | sdk1b | sidekick cell adhesion molecule 1b | 555553 |
| ENSDARG00000100435 | sec22bb | SEC22 vesicle trafficking protein homolog B (S. cerevisiae), b | 327277 |
| ENSDARG00000104230 | sec23a | Sec23 homolog A (S. cerevisiae) | 406774 |
| ENSDARG00000104375 | sec24a | SEC24 family, member A (S. cerevisiae) | 100000526 |
| ENSDARG00000071906 | sec24b | SEC24 family member B | 334633 |
| ENSDARG00000079611 | sema4c | sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4C | 561895 |
| ENSDARG00000036626 | sema6ba | sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6Ba | 0 |
| ENSDARG00000000189 | sema6e | sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6E | 556743 |
| ENSDARG00000099664 | sep15 | selenoprotein 15 | 352923 |
| ENSDARG00000099236 | sept10 | septin 10 | 550129 |
| ENSDARG00000019191 | sept12 | septin 12 | 564600 |
| ENSDARG00000102889 | sept15 | septin 15 | 792259 |
| ENSDARG00000027590 | sept2 | septin 2 | 323457 |
| ENSDARG00000030656 | sept3 | septin 3 | 554123 |
| ENSDARG00000013843 | sept5a | septin 5a | 0 |
| ENSDARG00000036031 | sept5b | septin 5b | 445325 |
| ENSDARG00000010721 | sept6 | septin 6 | 322694 |
| ENSDARG00000087647 | sept7a | septin 7a | 0 |
| ENSDARG00000052673 | sept7b | septin 7b | 100000597 |
| ENSDARG00000032606 | sept8a | septin 8a | 321590 |
| ENSDARG00000014233 | sept8b | septin 8b | 571702 |
| ENSDARG00000103900 | sept9b | septin 9b | 541494 |
| ENSDARG00000056121 | serac1 | serine active site containing 1 | 568022 |
| ENSDARG00000074242 | serbp1a | SERPINE1 mRNA binding protein 1a | 335449 |
| ENSDARG00000005924 | serpina10a | serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 10a | 565064 |
| ENSDARG00000040614 | sestd1 | SEC14 and spectrin domains 1 | 360133 |
| ENSDARG00000011564 | sfpq | splicing factor proline/glutamine-rich | 406564 |
| ENSDARG00000097897 | sgip1a | SH3-domain GRB2-like (endophilin) interacting protein 1a | 566542 |
| ENSDARG00000036878 | sh3bgrl2 | SH3 domain binding glutamate-rich protein like 2 | 431744 |
| ENSDARG00000060539 | shank1 | SH3 and multiple ankyrin repeat domains 1 | 568908 |
| ENSDARG00000062325 | shank2 | SH3 and multiple ankyrin repeat domains 2 | 567595 |
| ENSDARG00000063332 | shank3a | SH3 and multiple ankyrin repeat domains 3a | 0 |
| ENSDARG00000063054 | shank3b | SH3 and multiple ankyrin repeat domains 3b | 566152 |
| ENSDARG00000062462 | shisa7a | shisa family member 7a | 0 |
| ENSDARG00000063144 | shisa7b | shisa family member 7 | 100034520 |
| ENSDARG00000045145 | shisa9a | shisa family member 9a | 541382 |
| ENSDARG00000052642 | shisa9b | shisa family member 9b | 566015 |
| ENSDARG00000104414 | shmt2 | serine hydroxymethyltransferase 2 (mitochondrial) | 100144628 |
| ENSDARG00000040853 | shoc2 | soc-2 suppressor of clear homolog (C. elegans) | 555476 |
| ENSDARG00000102780 | si:ch1073-370j18.1 | si:ch1073-370j18.1 | 0 |
| ENSDARG00000070442 | si:ch211-113g11.6 | si:ch211-113g11.6 | 559008 |
| ENSDARG00000088882 | si:ch211-149b19.2 | si:ch211-149b19.2 | 101884675 |
| ENSDARG00000092746 | si:ch211-150o23.2 | si:ch211-150o23.2 | 0 |
| ENSDARG00000097187 | si:ch211-153l6.6 | si:ch211-153l6.6 | 101884986 |
| ENSDARG00000076970 | si:ch211-165b10.3 | si:ch211-165b10.3 | 100334077 |
| ENSDARG00000088276 | si:ch211-190p8.2 | si:ch211-190p8.2 | 436997 |
| ENSDARG00000095459 | si:ch211-191j22.3 | si:ch211-191j22.3 | 0 |
| ENSDARG00000063361 | si:ch211-1e14.1 | si:ch211-1e14.1 | 556850 |
| ENSDARG00000104157 | si:ch211-232h12.2 | si:ch211-232h12.2 | 0 |
| ENSDARG00000071353 | si:ch211-235e9.8 | si:ch211-235e9.8 | 100002196 |
| ENSDARG00000075491 | si:ch211-26b3.4 | si:ch211-26b3.4 | 568768 |
| ENSDARG00000059333 | si:ch211-285f17.1 | si:ch211-285f17.1 | 562465 |
| ENSDARG00000089255 | si:ch211-286c4.6 | si:ch211-286c4.6 | 100003678 |
| ENSDARG00000097855 | si:ch211-3o3.9 | si:ch211-3o3.9 | 101886596 |
| ENSDARG00000087364 | si:ch73-140j24.4 | si:ch73-140j24.4 | 101885000 |
| ENSDARG00000023940 | si:ch73-362m14.4 | si:ch73-362m14.4 | 555437 |
| ENSDARG00000057826 | si:ch73-61d6.3 | si:ch73-61d6.3 | 100001136 |
| ENSDARG00000090149 | si:dkey-108k21.18 | si:dkey-108k21.18 | 560380 |
| ENSDARG00000089418 | si:dkey-164f24.2 | si:dkey-164f24.2 | 566607 |
| ENSDARG00000078317 | si:dkey-175m17.7 | si:dkey-175m17.7 | 100007304 |
| ENSDARG00000080000 | si:dkey-178e17.1 | si:dkey-178e17.1 | 793282 |
| ENSDARG00000094695 | si:dkey-204f11.64 | si:dkey-204f11.64 | 100537752 |
| ENSDARG00000096701 | si:dkey-21e13.3 | si:dkey-21e13.3 | 100150043 |
| ENSDARG00000095005 | si:dkey-21e2.10 | si:dkey-21e2.10 | 556860 |
| ENSDARG00000061719 | si:dkey-23p11.2 | si:dkey-23p11.2 | 565752 |
| ENSDARG00000093195 | si:dkey-261m9.6 | si:dkey-261m9.6 | 563569 |
| ENSDARG00000073752 | si:dkey-266j7.2 | si:dkey-266j7.2 | 0 |
| ENSDARG00000003146 | si:dkey-27m7.4 | si:dkey-27m7.4 | 0 |
| ENSDARG00000099614 | si:dkey-29k15.2 | si:dkey-29k15.2 | 557266 |
| ENSDARG00000057881 | si:dkey-33c12.3 | si:dkey-33c12.3 | 335380 |
| ENSDARG00000032482 | si:dkey-40c11.2 | si:dkey-40c11.2 | 798260 |
| ENSDARG00000092039 | si:dkey-70p6.1 | si:dkey-70p6.1 | 570575 |
| ENSDARG00000097528 | si:dkey-7j14.5 | si:dkey-7j14.5 | 556563 |
| ENSDARG00000094074 | si:dkey-81h8.1 | si:dkey-81h8.1 | 100034657 |
| ENSDARG00000011602 | si:dkeyp-117h8.2 | si:dkeyp-117h8.2 | 570321 |
| ENSDARG00000063008 | si:dkeyp-27e10.3 | si:dkeyp-27e10.3 | 0 |
| ENSDARG00000045976 | sidt2 | SID1 transmembrane family, member 2 | 796833 |
| ENSDARG00000020134 | sipa1l1 | signal-induced proliferation-associated 1 like 1 | 562521 |
| ENSDARG00000061699 | SIPA1L3 | signal-induced proliferation-associated 1 like 3 | 561005 |
| ENSDARG00000076697 | SIPA1 | signal-induced proliferation-associated 1 | 555294 |
| ENSDARG00000003151 | skp1 | S-phase kinase-associated protein 1 | 393716 |
| ENSDARG00000035741 | slc25a11 | solute carrier family 25 (mitochondrial carrier; oxoglutarate carrier), member 11 | 415189 |
| ENSDARG00000026680 | slc25a14 | solute carrier family 25 (mitochondrial carrier, brain), member 14 | 393133 |
| ENSDARG00000057287 | slc25a16 | solute carrier family 25 (mitochondrial carrier; Graves disease autoantigen), member 16 | 324578 |
| ENSDARG00000070717 | slc25a18 | solute carrier family 25 (glutamate carrier), member 18 | 568740 |
| ENSDARG00000076381 | slc25a1b | slc25a1 solute carrier family 25 (mitochondrial carrier; citrate transporter), member 1b | 795332 |
| ENSDARG00000020718 | SLC25A22 (1 of 2) | solute carrier family 25 (mitochondrial carrier: glutamate), member 22 | 571044 |
| ENSDARG00000074533 | slc25a38b | solute carrier family 25, member 38b | 571961 |
| ENSDARG00000099368 | slc25a42 | solute carrier family 25, member 42 | 751743 |
| ENSDARG00000027355 | slc25a4 | solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 4 | 327067 |
| ENSDARG00000092553 | slc25a5 | solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 5 | 192321 |
| ENSDARG00000005853 | slc25a6 | solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 6 | 566370 |
| ENSDARG00000006240 | slc27a1a | solute carrier family 27 (fatty acid transporter), member 1a | 541410 |
| ENSDARG00000077368 | slc30a6 | solute carrier family 30 (zinc transporter), member 6 | 0 |
| ENSDARG00000102381 | SLC44A4 | solute carrier family 44, member 4 | 0 |
| ENSDARG00000012881 | slc4a1a | solute carrier family 4 (anion exchanger), member 1a (Diego blood group) | 84703 |
| ENSDARG00000086104 | SLCO5A1 (2 of 2) | solute carrier organic anion transporter family, member 5A1 | 570033 |
| ENSDARG00000079781 | slitrk4 | SLIT and NTRK-like family, member 4 | 558123 |
| ENSDARG00000103359 | SLITRK5 (2 of 2) | SLIT and NTRK-like family, member 5 | 101882841 |
| ENSDARG00000074153 | slitrk5b | SLIT and NTRK-like family, member 5b | 100331252 |
| ENSDARG00000077946 | smarcc2 | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 2 | 567476 |
| ENSDARG00000058203 | smc1al | structural maintenance of chromosomes 1A, like | 403060 |
| ENSDARG00000019000 | smc3 | structural maintenance of chromosomes 3 | 324475 |
| ENSDARG00000010130 | smpdl3b | sphingomyelin phosphodiesterase, acid-like 3B | 564392 |
| ENSDARG00000002538 | smu1a | smu-1 suppressor of mec-8 and unc-52 homolog a (C. elegans) | 393168 |
| ENSDARG00000051970 | smu1b | smu-1 suppressor of mec-8 and unc-52 homolog b (C. elegans) | 570074 |
| ENSDARG00000020609 | snap25a | synaptosomal-associated protein, 25a | 30712 |
| ENSDARG00000058117 | snap25b | synaptosomal-associated protein, 25b | 30711 |
| ENSDARG00000078485 | snpha | syntaphilin a | 0 |
| ENSDARG00000075539 | snphb | syntaphilin b | 569963 |
| ENSDARG00000071031 | snx1a | sorting nexin 1a | 337386 |
| ENSDARG00000053527 | snx24 | sorting nexin 24 | 619249 |
| ENSDARG00000097245 | soga3a | SOGA family member 3a | 100332515 |
| ENSDARG00000075455 | soga3b | SOGA family member 3b | 563485 |
| ENSDARG00000103435 | sorbs1 | sorbin and SH3 domain containing 1 | 0 |
| ENSDARG00000003046 | sorbs2a | sorbin and SH3 domain containing 2a | 571008 |
| ENSDARG00000061603 | sorbs2b | sorbin and SH3 domain containing 2b | 0 |
| ENSDARG00000077349 | SORCS3 (1 of 2) | sortilin-related VPS10 domain containing receptor 3 | 0 |
| ENSDARG00000004017 | spag1a | sperm associated antigen 1a | 0 |
| ENSDARG00000029480 | specc1 | sperm antigen with calponin homology and coiled-coil domains 1 | 562343 |
| ENSDARG00000009567 | SPEG (1 of 2) | SPEG complex locus | 0 |
| ENSDARG00000038445 | SPIRE1 (2 of 2) | spire-type actin nucleation factor 1 | 0 |
| ENSDARG00000035868 | spire1a | spire-type actin nucleation factor 1a | 557962 |
| ENSDARG00000019231 | spna2 | spectrin alpha 2 | 553383 |
| ENSDARG00000057665 | sprn2 | shadow of prion protein 2 | 100006310 |
| ENSDARG00000102883 | SPTBN1 | spectrin, beta, non-erythrocytic 1 | 553451 |
| ENSDARG00000053956 | sptbn2 | spectrin, beta, non-erythrocytic 2 | 558044 |
| ENSDARG00000030490 | sptb | spectrin, beta, erythrocytic | 58040 |
| ENSDARG00000017034 | sqrdl | sulfide quinone reductase-like (yeast) | 550580 |
| ENSDARG00000059773 | SRCIN1 (1 of 2) | SRC kinase signaling inhibitor 1 | 0 |
| ENSDARG00000101812 | SRCIN1 (2 of 2) | SRC kinase signaling inhibitor 1 | 0 |
| ENSDARG00000008107 | src | v-src avian sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog | 325084 |
| ENSDARG00000032161 | srgap2a | SLIT-ROBO Rho GTPase activating protein 2a | 556198 |
| ENSDARG00000035699 | srp19 | signal recognition particle 19 | 406652 |
| ENSDARG00000098367 | srp54 | signal recognition particle 54 | 393963 |
| ENSDARG00000014139 | srp72 | signal recognition particle 72 | 327335 |
| ENSDARG00000031560 | srpk1b | SRSF protein kinase 1b | 791187 |
| ENSDARG00000027734 | srsf5b | serine/arginine-rich splicing factor 5b | 436883 |
| ENSDARG00000039350 | ssbp1 | single-stranded DNA binding protein 1 | 550504 |
| ENSDARG00000029252 | ssb | Sjogren syndrome antigen B (autoantigen La) | 321504 |
| ENSDARG00000014165 | ssr3 | signal sequence receptor, gamma | 337190 |
| ENSDARG00000053668 | stag2b | stromal antigen 2b | 560751 |
| ENSDARG00000044182 | stau1 | staufen double-stranded RNA binding protein 1 | 334440 |
| ENSDARG00000026801 | stau2 | staufen double-stranded RNA binding protein 2 | 393899 |
| ENSDARG00000059003 | strip2 | striatin interacting protein 2 | 556392 |
| ENSDARG00000045228 | stub1 | STIP1 homology and U-Box containing protein 1 | 324243 |
| ENSDARG00000035763 | stx18 | syntaxin 18 | 554130 |
| ENSDARG00000029234 | stxbp5b | syntaxin binding protein 5b (tomosyn) | 571925 |
| ENSDARG00000028354 | stxbp6l | syntaxin binding protein 6 (amisyn), like | 100002110 |
| ENSDARG00000079949 | supt16h | suppressor of Ty 16 homolog (S. cerevisiae) | 562348 |
| ENSDARG00000055075 | svila | supervillin a | 30112 |
| ENSDARG00000014651 | svilb | supervillin b | 0 |
| ENSDARG00000060112 | sybu | syntabulin (syntaxin-interacting) | 568207 |
| ENSDARG00000060368 | syn1 | synapsin I | 570672 |
| ENSDARG00000045945 | syn2a | synapsin IIa | 436870 |
| ENSDARG00000098417 | syn3 | synapsin III | 0 |
| ENSDARG00000009499 | syne1a | spectrin repeat containing, nuclear envelope 1a | 0 |
| ENSDARG00000063068 | syne1b | spectrin repeat containing, nuclear envelope 1b | 0 |
| ENSDARG00000092624 | SYNE1 | spectrin repeat containing, nuclear envelope 1 | 0 |
| ENSDARG00000063713 | SYNGAP1 (1 of 2) | synaptic Ras GTPase activating protein 1 | 100034485 |
| ENSDARG00000069765 | SYNGAP1 (2 of 2) | synaptic Ras GTPase activating protein 1 | 0 |
| ENSDARG00000062350 | synm | synemin, intermediate filament protein | 558800 |
| ENSDARG00000075830 | syt3 | synaptotagmin III | 100000615 |
| ENSDARG00000063568 | syt7a | synaptotagmin VIIa | 561429 |
| ENSDARG00000078060 | syt7b | synaptotagmin VIIb | 100334336 |
| ENSDARG00000003994 | syt9a | synaptotagmin IXa | 798433 |
| ENSDARG00000029239 | syt9b | synaptotagmin IXb | 445476 |
| ENSDARG00000079805 | tagln3a | transgelin 3a | 563096 |
| ENSDARG00000089190 | TANC1 (2 of 2) | tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1 | 100334650 |
| ENSDARG00000079004 | TANC2 (1 of 2) | tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2 | 0 |
| ENSDARG00000079097 | tanc2a | tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2a | 568810 |
| ENSDARG00000069603 | taok1a | TAO kinase 1a | 100004576 |
| ENSDARG00000074899 | taok2a | TAO kinase 2a | 556172 |
| ENSDARG00000100620 | taok2b | TAO kinase 2b | 100151551 |
| ENSDARG00000013250 | tars | threonyl-tRNA synthetase | 449661 |
| ENSDARG00000103095 | tbk1 | TANK-binding kinase 1 | 692289 |
| ENSDARG00000061473 | tbkbp1 | TBK1 binding protein 1 | 559568 |
| ENSDARG00000043357 | tbl2 | transducin (beta)-like 2 | 337068 |
| ENSDARG00000079881 | TCAIM | T cell activation inhibitor, mitochondrial | 559258 |
| ENSDARG00000045355 | tceb1a | transcription elongation factor B (SIII), polypeptide 1a | 447935 |
| ENSDARG00000037980 | tceb2 | transcription elongation factor B (SIII), polypeptide 2 | 192341 |
| ENSDARG00000102365 | tceb3 | transcription elongation factor B (SIII), polypeptide 3 | 0 |
| ENSDARG00000035605 | tchp | trichoplein, keratin filament binding | 678595 |
| ENSDARG00000105142 | tcirg1b | T-cell, immune regulator 1, ATPase, H+ transporting, lysosomal V0 subunit A3b | 406342 |
| ENSDARG00000031890 | tcp11l1 | t-complex 11, testis-specific-like 1 | 541488 |
| ENSDARG00000025904 | tecrb | trans-2,3-enoyl-CoA reductase b | 326954 |
| ENSDARG00000011171 | TENM2 (1 of 2) | teneurin transmembrane protein 2 | 568845 |
| ENSDARG00000037122 | TENM2 (2 of 2) | teneurin transmembrane protein 2 | 563197 |
| ENSDARG00000063145 | tfam | transcription factor A, mitochondrial | 571106 |
| ENSDARG00000060306 | tfcp2 | transcription factor CP2 | 100001906 |
| ENSDARG00000035018 | thy1 | Thy-1 cell surface antigen | 336617 |
| ENSDARG00000058297 | timm13 | translocase of inner mitochondrial membrane 13 homolog (yeast) | 436738 |
| ENSDARG00000071594 | TIMM21 | translocase of inner mitochondrial membrane 21 homolog (yeast) | 560369 |
| ENSDARG00000023672 | timm8a | translocase of inner mitochondrial membrane 8 homolog A (yeast) | 445243 |
| ENSDARG00000077506 | tjp1a | tight junction protein 1a | 378845 |
| ENSDARG00000079374 | tjp1b | tight junction protein 1b | 565459 |
| ENSDARG00000023443 | tjp2b | tight junction protein 2b (zona occludens 2) | 436639 |
| ENSDARG00000029689 | tkta | transketolase a | 557518 |
| ENSDARG00000062951 | tldc1 | TBC/LysM-associated domain containing 1 | 561678 |
| ENSDARG00000100729 | tln1 | talin 1 | 494453 |
| ENSDARG00000028748 | tm9sf3 | transmembrane 9 superfamily member 3 | 406698 |
| ENSDARG00000069099 | tmco1 | transmembrane and coiled-coil domains 1 | 378980 |
| ENSDARG00000041391 | tmed10 | transmembrane emp24-like trafficking protein 10 (yeast) | 326716 |
| ENSDARG00000024954 | tmed4 | transmembrane emp24 protein transport domain containing 4 | 415224 |
| ENSDARG00000008765 | tmed5 | transmembrane emp24 protein transport domain containing 5 | 393374 |
| ENSDARG00000046022 | tmed7 | transmembrane emp24 protein transport domain containing 7 | 405848 |
| ENSDARG00000051824 | tmeff2a | transmembrane protein with EGF-like and two follistatin-like domains 2a | 100004613 |
| ENSDARG00000069649 | tmem160 | transmembrane protein 160 | 559491 |
| ENSDARG00000016968 | tmem214 | transmembrane protein 214 | 402840 |
| ENSDARG00000086661 | TMEM221 | transmembrane protein 221 | 100537506 |
| ENSDARG00000069494 | tmem223 | transmembrane protein 223 | 445200 |
| ENSDARG00000087916 | tmem240a | transmembrane protein 240a | 568378 |
| ENSDARG00000090145 | tmem240b | transmembrane protein 240b | 0 |
| ENSDARG00000063414 | tmem5 | transmembrane protein 5 | 557187 |
| ENSDARG00000076393 | tmem65 | transmembrane protein 65 | 100005403 |
| ENSDARG00000002571 | tmod2 | tropomodulin 2 | 447869 |
| ENSDARG00000020890 | tmod4 | tropomodulin 4 (muscle) | 30441 |
| ENSDARG00000022978 | tmpob | thymopoietin b | 553624 |
| ENSDARG00000063149 | tmtc1 | transmembrane and tetratricopeptide repeat containing 1 | 555449 |
| ENSDARG00000078362 | TNC (2 of 2) | tenascin C | 100149028 |
| ENSDARG00000021948 | tnc | tenascin C | 30037 |
| ENSDARG00000056218 | tnika | TRAF2 and NCK interacting kinase a | 325042 |
| ENSDARG00000009031 | tnikb | TRAF2 and NCK interacting kinase b | 556959 |
| ENSDARG00000020771 | tnr | tenascin R (restrictin, janusin) | 369191 |
| ENSDARG00000098486 | tollip | toll interacting protein | 336876 |
| ENSDARG00000056394 | TOM1L2 (2 of 2) | target of myb1-like 2 (chicken) | 100535615 |
| ENSDARG00000104581 | tom1 | target of myb1 (chicken) | 0 |
| ENSDARG00000034195 | top2b | topoisomerase (DNA) II beta | 569877 |
| ENSDARG00000057239 | tph2 | tryptophan hydroxylase 2 (tryptophan 5-monooxygenase) | 0 |
| ENSDARG00000087402 | TPM1 | tropomyosin 1 (alpha) | 393908 |
| ENSDARG00000005162 | tpm3 | tropomyosin 3 | 373076 |
| ENSDARG00000033683 | tpma | alpha-tropomyosin | 30324 |
| ENSDARG00000073872 | tpst1 | tyrosylprotein sulfotransferase 1 | 30677 |
| ENSDARG00000003963 | traf4a | tnf receptor-associated factor 4a | 404045 |
| ENSDARG00000098118 | TRAPPC10 | trafficking protein particle complex 10 | 0 |
| ENSDARG00000039005 | trappc1 | trafficking protein particle complex 1 | 437015 |
| ENSDARG00000097660 | trappc2 | trafficking protein particle complex 2 | 767808 |
| ENSDARG00000045364 | trappc3 | trafficking protein particle complex 3 | 445207 |
| ENSDARG00000026068 | trappc5 | trafficking protein particle complex 5 | 436755 |
| ENSDARG00000016463 | trappc6b | trafficking protein particle complex 6b | 450008 |
| ENSDARG00000099118 | TRAPPC9 (1 of 2) | trafficking protein particle complex 9 | 100002630 |
| ENSDARG00000069420 | trim23 | tripartite motif containing 23 | 767706 |
| ENSDARG00000104396 | trim25 | tripartite motif containing 25 | 393144 |
| ENSDARG00000076174 | trim2b | tripartite motif containing 2b | 558046 |
| ENSDARG00000062794 | trim36 | tripartite motif containing 36 | 406849 |
| ENSDARG00000063711 | trim3a | tripartite motif containing 3a | 562489 |
| ENSDARG00000005397 | trim3b | tripartite motif containing 3b | 555391 |
| ENSDARG00000012367 | trim46a | tripartite motif containing 46a | 569905 |
| ENSDARG00000058649 | trim46b | tripartite motif containing 46b | 100034631 |
| ENSDARG00000019426 | trioa | trio Rho guanine nucleotide exchange factor a | 557667 |
| ENSDARG00000000370 | triob | trio Rho guanine nucleotide exchange factor b | 368847 |
| ENSDARG00000061397 | trip12 | thyroid hormone receptor interactor 12 | 564866 |
| ENSDARG00000003754 | tspan2a | tetraspanin 2a | 378854 |
| ENSDARG00000043026 | TTBK2 (2 of 2) | tau tubulin kinase 2 | 569881 |
| ENSDARG00000061207 | ttc7b | tetratricopeptide repeat domain 7B | 568039 |
| ENSDARG00000045367 | tuba1b | tubulin, alpha 1b | 373080 |
| ENSDARG00000045014 | tuba2 | tubulin, alpha 2 | 406303 |
| ENSDARG00000074289 | tuba4l | tubulin, alpha 4 like | 327377 |
| ENSDARG00000031164 | tuba8l2 | tubulin, alpha 8 like 2 | 393664 |
| ENSDARG00000006260 | tuba8l4 | tubulin, alpha 8 like 4 | 393154 |
| ENSDARG00000042708 | tuba8l | tubulin, alpha 8 like | 337223 |
| ENSDARG00000053066 | tubb1 | tubulin, beta 1 class VI | 558552 |
| ENSDARG00000039522 | tubb2 | tubulin, beta 2A class IIa | 554127 |
| ENSDARG00000098591 | tubb4b | tubulin, beta 4B class IVb | 641421 |
| ENSDARG00000037997 | tubb5 | tubulin, beta 5 | 386701 |
| ENSDARG00000041723 | TUBB8P7 (2 of 3) | tubulin, beta 8 class VIII pseudogene 7 | 406811 |
| ENSDARG00000015610 | tubg1 | tubulin, gamma 1 | 393882 |
| ENSDARG00000013079 | tubgcp2 | tubulin, gamma complex associated protein 2 | 386975 |
| ENSDARG00000029133 | tubgcp3 | tubulin, gamma complex associated protein 3 | 563517 |
| ENSDARG00000104173 | tufm | Tu translation elongation factor, mitochondrial | 541378 |
| ENSDARG00000078854 | tusc3 | tumor suppressor candidate 3 | 553582 |
| ENSDARG00000011661 | twf1b | twinfilin actin-binding protein 1b | 432375 |
| ENSDARG00000060238 | uacab | uveal autoantigen with coiled-coil domains and ankyrin repeats b | 564040 |
| ENSDARG00000045877 | ube2nb | ubiquitin-conjugating enzyme E2Nb | 393313 |
| ENSDARG00000018000 | ubp1 | upstream binding protein 1 (LBP-1a) | 100136855 |
| ENSDARG00000035066 | ubtf | upstream binding transcription factor, RNA polymerase I | 368509 |
| ENSDARG00000062089 | uggt2 | UDP-glucose glycoprotein glucosyltransferase 2 | 497542 |
| ENSDARG00000061829 | UNC13A | unc-13 homolog A (C. elegans) | 568618 |
| ENSDARG00000101233 | unc13c | unc-13 homolog C (C. elegans) | 0 |
| ENSDARG00000040455 | unc50 | unc-50 homolog (C. elegans) | 393217 |
| ENSDARG00000098290 | UNC80 | unc-80 homolog (C. elegans) | 0 |
| ENSDARG00000034321 | uo:ion006 | uo:ion006 | 100000814 |
| ENSDARG00000016302 | upf1 | upf1 regulator of nonsense transcripts homolog (yeast) | 406783 |
| ENSDARG00000093382 | uqcc3 | ubiquinol-cytochrome c reductase complex assembly factor 3 | 558925 |
| ENSDARG00000104517 | UQCR10 | ubiquinol-cytochrome c reductase, complex III subunit X | 100333064 |
| ENSDARG00000007745 | uqcrfs1 | ubiquinol-cytochrome c reductase, Rieske iron-sulfur polypeptide 1 | 406359 |
| ENSDARG00000029064 | uqcrq | ubiquinol-cytochrome c reductase, complex III subunit VII | 436768 |
| ENSDARG00000004305 | vangl1 | VANGL planar cell polarity protein 1 | 403076 |
| ENSDARG00000027397 | vangl2 | VANGL planar cell polarity protein 2 | 245949 |
| ENSDARG00000046048 | vapal | VAMP (vesicle-associated membrane protein)-associated protein A, like | 436819 |
| ENSDARG00000070435 | vapb | VAMP (vesicle-associated membrane protein)-associated protein B and C | 323628 |
| ENSDARG00000044575 | vars | valyl-tRNA synthetase | 114427 |
| ENSDARG00000099690 | vaspa | vasodilator-stimulated phosphoprotein a | 555805 |
| ENSDARG00000017105 | vaspb | vasodilator-stimulated phosphoprotein b | 550487 |
| ENSDARG00000073713 | vav3b | vav 3 guanine nucleotide exchange factor b | 0 |
| ENSDARG00000009401 | vcanb | versican b | 323465 |
| ENSDARG00000044968 | vcla | vinculin a | 552980 |
| ENSDARG00000086665 | VGF | VGF nerve growth factor inducible | 0 |
| ENSDARG00000044501 | viml | vimentin like | 393746 |
| ENSDARG00000010008 | vim | vimentin | 140599 |
| ENSDARG00000078247 | vip | vasoactive intestinal peptide | 795653 |
| ENSDARG00000036338 | vps11 | vacuolar protein sorting 11 | 565852 |
| ENSDARG00000017986 | vps13d | vacuolar protein sorting 13 homolog D (S. cerevisiae) | 567879 |
| ENSDARG00000103203 | vps28 | vacuolar protein sorting 28 (yeast) | 393562 |
| ENSDARG00000062016 | vps51 | vacuolar protein sorting 51 homolog (S. cerevisiae) | 559339 |
| ENSDARG00000069324 | vps53 | vacuolar protein sorting 53 homolog (S. cerevisiae) | 492817 |
| ENSDARG00000036056 | vrk3 | vaccinia related kinase 3 | 0 |
| ENSDARG00000092126 | vtg5 | vitellogenin 5 | 64260 |
| ENSDARG00000016825 | vtg6 | vitellogenin 6 | 559229 |
| ENSDARG00000039270 | vti1b | vesicle transport through interaction with t-SNAREs 1B | 394011 |
| ENSDARG00000091457 | WASF1 (2 of 2) | WAS protein family, member 1 | 101885310 |
| ENSDARG00000060349 | wasf1 | WAS protein family, member 1 | 100001221 |
| ENSDARG00000062948 | wasf3b | WAS protein family, member 3b | 563632 |
| ENSDARG00000014113 | wasla | Wiskott-Aldrich syndrome-like a | 100009637 |
| ENSDARG00000042418 | wbscr17 | Williams-Beuren syndrome chromosome region 17 | 567794 |
| ENSDARG00000075245 | wdr11 | WD repeat domain 11 | 558865 |
| ENSDARG00000062310 | WDR26 (2 of 2) | WD repeat domain 26 | 559461 |
| ENSDARG00000069269 | wdr35 | WD repeat domain 35 | 565515 |
| ENSDARG00000074611 | wdr37 | WD repeat domain 37 | 558075 |
| ENSDARG00000078136 | wdr47a | WD repeat domain 47a | 558528 |
| ENSDARG00000038543 | wdr48 | WD repeat domain 48 | 334682 |
| ENSDARG00000060312 | wdr62 | WD repeat domain 62 | 570949 |
| ENSDARG00000100274 | WDR7 | WD repeat domain 7 | 0 |
| ENSDARG00000006712 | wipf2b | WAS/WASL interacting protein family, member 2b | 415255 |
| ENSDARG00000039392 | wnk1b | WNK lysine deficient protein kinase 1b | 561159 |
| ENSDARG00000088195 | wu:fb52c12 | wu:fb52c12 | 322178 |
| ENSDARG00000020443 | xkr6a | XK, Kell blood group complex subunit-related family, member 6a | 0 |
| ENSDARG00000043410 | xkr6b | XK, Kell blood group complex subunit-related family, member 6b | 792104 |
| ENSDARG00000026333 | xkr7 | XK, Kell blood group complex subunit-related family, member 7 | 497073 |
| ENSDARG00000007916 | xpnpep3 | X-prolyl aminopeptidase 3, mitochondrial | 573360 |
| ENSDARG00000071551 | xrcc6 | X-ray repair complementing defective repair in Chinese hamster cells 6 | 334488 |
| ENSDARG00000005941 | yes1 | YES proto-oncogene 1, Src family tyrosine kinase | 407620 |
| ENSDARG00000061692 | yjefn3 | YjeF N-terminal domain containing 3 | 557840 |
| ENSDARG00000038308 | ykt6 | YKT6 v-SNARE homolog (S. cerevisiae) | 394067 |
| ENSDARG00000075192 | yme1l1a | YME1-like 1a | 793098 |
| ENSDARG00000104401 | yme1l1b | YME1-like 1b | 557907 |
| ENSDARG00000055510 | ypel3 | yippee-like 3 | 368214 |
| ENSDARG00000013078 | ywhaba | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, beta polypeptide a | 323055 |
| ENSDARG00000075758 | ywhabb | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, beta polypeptide b | 406419 |
| ENSDARG00000040287 | ywhabl | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, beta polypeptide like | 321729 |
| ENSDARG00000005560 | ywhah | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, eta polypeptide | 406443 |
| ENSDARG00000032575 | ywhaz | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide | 336610 |
| ENSDARG00000027915 | zc2hc1a | zinc finger, C2HC-type containing 1A | 324412 |
| ENSDARG00000070441 | zdhhc17 | zinc finger, DHHC-type containing 17 | 100148543 |
| ENSDARG00000016263 | zdhhc5a | zinc finger, DHHC-type containing 5a | 571795 |
| ENSDARG00000086342 | zgc:101566 | zgc:101566 | 449823 |
| ENSDARG00000069600 | zgc:109889 | zgc:109889 | 553542 |
| ENSDARG00000071626 | zgc:153154 | zgc:153154 | 751701 |
| ENSDARG00000096992 | zgc:153405 | zgc:153405 | 751661 |
| ENSDARG00000033738 | zgc:153867 | zgc:153867 | 337226 |
| ENSDARG00000069529 | zgc:153981 | zgc:153981 | 563986 |
| ENSDARG00000017785 | zgc:158689 | zgc:158689 | 791177 |
| ENSDARG00000019651 | zgc:162239 | zgc:162239 | 0 |
| ENSDARG00000091525 | zgc:173552 | zgc:173552 | 794406 |
| ENSDARG00000092873 | zgc:193541 | zgc:193541 | 557217 |
| ENSDARG00000035340 | zgc:194261 | zgc:194261 | 572372 |
| ENSDARG00000017140 | zgc:63587 | zgc:63587 | 393431 |
| ENSDARG00000012281 | zgc:65851 | zgc:65851 | 321113 |
| ENSDARG00000016301 | zgc:65894 | zgc:65894 | 335798 |
| ENSDARG00000014222 | zgc:86598 | zgc:86598 | 415254 |
| ENSDARG00000044874 | zgc:92107 | zgc:92107 | 492329 |
| ENSDARG00000041060 | zgc:92326 | zgc:92326 | 327284 |
| ENSDARG00000061718 | znf704 | zinc finger protein 704 | 560056 |
| ENSDARG00000075733 | zyx | zyxin | 564246 |
Supplementary Table 3a. IPA cell location and molecular function categories in mouse SYN and PSD proteins.
Cell location and molecular function of mouse SYN and PSD proteins as described by the Ingenuity knowledgebase (www.qiagen.com/ingenuity). Proteins found in synaptosomes (SYN) and/or PSDs are indicated.
| ENSEBLE ID | Associated Gene Name | Description | MGI ID | SYN ONLY | PSD ONLY | EQUAL ABUNDANCE | PSD Depleted | PSD enriched | PSD component | SYN Component | Cell Location | Protein Type |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ENSMUSG00000037072 | Sep15 | selenoprotein | MGI:1927947 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000050705 | 2310061I04Rik | RIKEN cDNA 2310061I04 gene | MGI:1916912 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000040412 | 5330417C22Rik | RIKEN cDNA 5330417C22 gene | MGI:1923930 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000026944 | Abca2 | ATP-binding cassette, sub-family A (ABC1), member 2 | MGI:99606 | YES | YES | Plasma Membrane | transporter | |||||
| ENSMUSG00000041797 | Abca9 | ATP-binding cassette, sub-family A (ABC1), member 9 | MGI:2386796 | YES | YES | Cytoplasm | transporter | |||||
| ENSMUSG00000047368 | Abhd17b | abhydrolase domain containing 17B | MGI:1917816 | YES | YES | Other | peptidase | |||||
| ENSMUSG00000002475 | Abhd3 | abhydrolase domain containing 3 | MGI:2147183 | YES | YES | Other | enzyme | |||||
| ENSMUSG00000023832 | Acat2 | acetyl-Coenzyme A acetyltransferase 2 | MGI:87871 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000023328 | Ache | acetylcholinesterase | MGI:87876 | YES | YES | Plasma Membrane | enzyme | |||||
| ENSMUSG00000028405 | Aco1 | aconitase 1 | MGI:87879 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000002103 | Acp2 | acid phosphatase 2, lysosomal | MGI:87882 | YES | YES | Cytoplasm | phosphatase | |||||
| ENSMUSG00000028093 | Acp6 | acid phosphatase 6, lysophosphatidic | MGI:1931010 | YES | YES | Cytoplasm | phosphatase | |||||
| ENSMUSG00000020333 | Acsl6 | acyl-CoA synthetase long-chain family member 6 | MGI:894291 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000060923 | Acyp2 | acylphosphatase 2, muscle type | MGI:1922822 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000022376 | Adcy8 | adenylate cyclase 8 | MGI:1341110 | YES | YES | Plasma Membrane | enzyme | |||||
| ENSMUSG00000029778 | Adcyap1r1 | adenylate cyclase activating polypeptide 1 receptor 1 | MGI:108449 | YES | YES | Plasma Membrane | G-protein coupled receptor | |||||
| ENSMUSG00000023827 | Agpat4 | 1-acylglycerol-3-phosphate O-acyltransferase 4 (lysophosphatidic acid acyltransferase, delta) | MGI:1915512 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000042410 | Agps | alkylglycerone phosphate synthase | MGI:2443065 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000048087 | Gm4737 | predicted gene 4737 | MGI:3643647 | YES | YES | Other | other | |||||
| ENSMUSG00000029772 | Ahcyl2 | S-adenosylhomocysteine hydrolase-like 2 | MGI:1921590 | YES | YES | Other | enzyme | |||||
| ENSMUSG00000028792 | Ak2 | adenylate kinase 2 | MGI:87978 | YES | YES | Cytoplasm | kinase | |||||
| ENSMUSG00000028692 | Akr1a1 | aldo-keto reductase family 1, member A1 (aldehyde reductase) | MGI:1929955 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000001642 | Akr1b3 | aldo-keto reductase family 1, member B3 (aldose reductase) | MGI:1353494 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000025007 | Aldh18a1 | aldehyde dehydrogenase 18 family, member A1 | MGI:1888908 | YES | YES | Cytoplasm | kinase | |||||
| ENSMUSG00000053279 | Aldh1a1 | aldehyde dehydrogenase family 1, subfamily A1 | MGI:1353450 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000035936 | Aldh5a1 | aldhehyde dehydrogenase family 5, subfamily A1 | MGI:2441982 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000053644 | Aldh7a1 | aldehyde dehydrogenase family 7, member A1 | MGI:108186 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000031751 | Amfr | autocrine motility factor receptor | MGI:1345634 | YES | YES | Plasma Membrane | transmembrane receptor | |||||
| ENSMUSG00000037949 | Ano10 | anoctamin 10 | MGI:2143103 | YES | YES | Plasma Membrane | ion channel | |||||
| ENSMUSG00000027712 | Anxa5 | annexin A5 | MGI:106008 | YES | YES | Plasma Membrane | transporter | |||||
| ENSMUSG00000031996 | Aplp2 | amyloid beta (A4) precursor-like protein 2 | MGI:88047 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000032083 | Apoa1 | apolipoprotein A-I | MGI:88049 | YES | YES | Extracellular Space | transporter | |||||
| ENSMUSG00000028070 | Apoa1bp | apolipoprotein A-I binding protein | MGI:2180167 | YES | YES | Extracellular Space | enzyme | |||||
| ENSMUSG00000021877 | Arf4 | ADP-ribosylation factor 4 | MGI:99433 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000030881 | Arfip2 | ADP-ribosylation factor interacting protein 2 | MGI:1924182 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000042348 | Arl15 | ADP-ribosylation factor-like 15 | MGI:2442308 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000035199 | Arl6ip5 | ADP-ribosylation factor-like 6 interacting protein 5 | MGI:1929501 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000033124 | Atg9a | autophagy related 9A | MGI:2138446 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000007107 | Atp1a4 | ATPase, Na+/K+ transporting, alpha 4 polypeptide | MGI:1351335 | YES | YES | Plasma Membrane | transporter | |||||
| ENSMUSG00000032412 | Atp1b3 | ATPase, Na+/K+ transporting, beta 3 polypeptide | MGI:107788 | YES | YES | Plasma Membrane | transporter | |||||
| ENSMUSG00000020788 | Atp2a3 | ATPase, Ca++ transporting, ubiquitous | MGI:1194503 | YES | YES | Cytoplasm | transporter | |||||
| ENSMUSG00000016252 | Atp5e | ATP synthase, H+ transporting, mitochondrial F1 complex, epsilon subunit | MGI:1855697 | YES | YES | Cytoplasm | transporter | |||||
| ENSMUSG00000022890 | Atp5j | ATP synthase, H+ transporting, mitochondrial F0 complex, subunit F | MGI:107777 | YES | YES | Cytoplasm | transporter | |||||
| ENSMUSG00000028830 | AU040320 | expressed sequence AU040320 | MGI:2140475 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000006010 | BC003331 | cDNA sequence BC003331 | MGI:2385108 | YES | YES | Other | other | |||||
| ENSMUSG00000030268 | Bcat1 | branched chain aminotransferase 1, cytosolic | MGI:104861 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000026172 | Bcs1l | BCS1-like (yeast) | MGI:1914071 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000020840 | Blmh | bleomycin hydrolase | MGI:1345186 | YES | YES | Cytoplasm | peptidase | |||||
| ENSMUSG00000024191 | Bnip1 | BCL2/adenovirus E1B interacting protein 1 | MGI:109328 | YES | YES | Cytoplasm | transporter | |||||
| ENSMUSG00000026617 | Bpnt1 | bisphosphate 3'-nucleotidase 1 | MGI:1338800 | YES | YES | Nucleus | phosphatase | |||||
| ENSMUSG00000038526 | Car14 | carbonic anhydrase 14 | MGI:1344341 | YES | YES | Plasma Membrane | enzyme | |||||
| ENSMUSG00000014226 | Cacybp | calcyclin binding protein | MGI:1270839 | YES | YES | Nucleus | other | |||||
| ENSMUSG00000032076 | Cadm1 | cell adhesion molecule 1 | MGI:1889272 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000028222 | Calb1 | calbindin 1 | MGI:88248 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000003657 | Calb2 | calbindin 2 | MGI:101914 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000027187 | Cat | catalase | MGI:88271 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000031641 | Cbr4 | carbonyl reductase 4 | MGI:2384567 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000030613 | Ccdc90b | coiled-coil domain containing 90B | MGI:1913615 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000029084 | Cd38 | CD38 antigen | MGI:107474 | YES | YES | Plasma Membrane | enzyme | |||||
| ENSMUSG00000027215 | Cd82 | CD82 antigen | MGI:104651 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000035776 | Cd99l2 | CD99 antigen-like 2 | MGI:2177151 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000004071 | Cdip1 | cell death inducing Trp53 target 1 | MGI:1913876 | YES | YES | Nucleus | other | |||||
| ENSMUSG00000029330 | Cds1 | CDP-diacylglycerol synthase 1 | MGI:1921846 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000058793 | Cds2 | CDP-diacylglycerol synthase (phosphatidate cytidylyltransferase) 2 | MGI:1332236 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000032803 | Cdv3 | carnitine deficiency-associated gene expressed in ventricle 3 | MGI:2448759 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000087408 | Cers1 | ceramide synthase 1 | MGI:2136690 | YES | YES | #N/A | #N/A | |||||
| ENSMUSG00000025512 | Chid1 | chitinase domain containing 1 | MGI:1915288 | YES | YES | Extracellular Space | other | |||||
| ENSMUSG00000004319 | Clcn3 | chloride channel 3 | MGI:103555 | YES | YES | Plasma Membrane | ion channel | |||||
| ENSMUSG00000036636 | Clcn7 | chloride channel 7 | MGI:1347048 | YES | YES | Plasma Membrane | ion channel | |||||
| ENSMUSG00000079598 | Clec2l | C-type lectin domain family 2, member L | MGI:2141402 | YES | YES | Other | other | |||||
| ENSMUSG00000037242 | Clic4 | chloride intracellular channel 4 (mitochondrial) | MGI:1352754 | YES | YES | Plasma Membrane | ion channel | |||||
| ENSMUSG00000002660 | Clpp | caseinolytic mitochondrial matrix peptidase proteolytic subunit | MGI:1858213 | YES | YES | Cytoplasm | peptidase | |||||
| ENSMUSG00000025545 | Clybl | citrate lyase beta like | MGI:1916884 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000024644 | Cndp2 | CNDP dipeptidase 2 (metallopeptidase M20 family) | MGI:1913304 | YES | YES | Cytoplasm | peptidase | |||||
| ENSMUSG00000025189 | Cnnm1 | cyclin M1 | MGI:1891366 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000064105 | Cnnm2 | cyclin M2 | MGI:2151054 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000001138 | Cnnm3 | cyclin M3 | MGI:2151055 | YES | YES | Other | other | |||||
| ENSMUSG00000025381 | Cnpy2 | canopy 2 homolog (zebrafish) | MGI:1928477 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000031772 | Cntnap4 | contactin associated protein-like 4 | MGI:2183572 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000070695 | Cntnap5a | contactin associated protein-like 5A | MGI:3643623 | YES | YES | Other | other | |||||
| ENSMUSG00000048351 | Coa7 | cytochrome c oxidase assembly factor 7 | MGI:1917143 | YES | YES | Other | other | |||||
| ENSMUSG00000000326 | Comt | catechol-O-methyltransferase | MGI:88470 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000020841 | Cpd | carboxypeptidase D | MGI:107265 | YES | YES | Extracellular Space | peptidase | |||||
| ENSMUSG00000028607 | Cpt2 | carnitine palmitoyltransferase 2 | MGI:109176 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000016481 | Cr1l | complement component (3b/4b) receptor 1-like | MGI:88513 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000026853 | Crat | carnitine acetyltransferase | MGI:109501 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000030284 | Creld1 | cysteine-rich with EGF-like domains 1 | MGI:2152539 | YES | YES | Other | other | |||||
| ENSMUSG00000003623 | Crot | carnitine O-octanoyltransferase | MGI:1921364 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000028804 | Csmd2 | CUB and Sushi multiple domains 2 | MGI:2386401 | YES | YES | Other | other | |||||
| ENSMUSG00000031778 | Cx3cl1 | chemokine (C-X3-C motif) ligand 1 | MGI:1097153 | YES | YES | Extracellular Space | cytokine | |||||
| ENSMUSG00000022865 | Cxadr | coxsackie virus and adenovirus receptor | MGI:1201679 | YES | YES | Plasma Membrane | transmembrane receptor | |||||
| ENSMUSG00000026456 | Cyb5r1 | cytochrome b5 reductase 1 | MGI:1919267 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000061740 | Cyp2d22 | cytochrome P450, family 2, subfamily d, polypeptide 22 | MGI:1929474 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000015224 | Cyp2j9 | cytochrome P450, family 2, subfamily j, polypeptide 9 | MGI:1921769 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000001467 | Cyp51 | cytochrome P450, family 51 | MGI:106040 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000022174 | Dad1 | defender against cell death 1 | MGI:101912 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000039952 | Dag1 | dystroglycan 1 | MGI:101864 | YES | YES | Plasma Membrane | transmembrane receptor | |||||
| ENSMUSG00000007039 | Ddah2 | dimethylarginine dimethylaminohydrolase 2 | MGI:1859016 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000001666 | Ddt | D-dopachrome tautomerase | MGI:1298381 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000031730 | Dhodh | dihydroorotate dehydrogenase | MGI:1928378 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000026209 | Dnpep | aspartyl aminopeptidase | MGI:1278328 | YES | YES | Cytoplasm | peptidase | |||||
| ENSMUSG00000035640 | Dos | downstream of Stk11 | MGI:1354170 | YES | YES | Other | other | |||||
| ENSMUSG00000063904 | Dpp3 | dipeptidylpeptidase 3 | MGI:1922471 | YES | YES | Cytoplasm | peptidase | |||||
| ENSMUSG00000027430 | Dtd1 | D-tyrosyl-tRNA deacylase 1 | MGI:1913294 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000047459 | Dynlrb1 | dynein light chain roadblock-type 1 | MGI:1914318 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000031168 | Ebp | phenylalkylamine Ca2+ antagonist (emopamil) binding protein | MGI:107822 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000024132 | Eci1 | enoyl-Coenzyme A delta isomerase 1 | MGI:94871 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000031217 | Efnb1 | ephrin B1 | MGI:102708 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000042302 | Ehbp1 | EH domain binding protein 1 | MGI:2667252 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000078812 | Eif5a | eukaryotic translation initiation factor 5A | MGI:106248 | YES | YES | Cytoplasm | translation regulator | |||||
| ENSMUSG00000038619 | Ensa | endosulfine alpha | MGI:1891189 | YES | YES | Cytoplasm | transporter | |||||
| ENSMUSG00000028664 | Ephb2 | Eph receptor B2 | MGI:99611 | YES | YES | Plasma Membrane | kinase | |||||
| ENSMUSG00000005958 | Ephb3 | Eph receptor B3 | MGI:104770 | YES | YES | Plasma Membrane | kinase | |||||
| ENSMUSG00000038776 | Ephx1 | epoxide hydrolase 1, microsomal | MGI:95405 | YES | YES | Cytoplasm | peptidase | |||||
| ENSMUSG00000035203 | Epn1 | epsin 1 | MGI:1333763 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000001576 | Ergic1 | endoplasmic reticulum-golgi intermediate compartment (ERGIC) 1 | MGI:1914708 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000046324 | Ermp1 | endoplasmic reticulum metallopeptidase 1 | MGI:106250 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000021831 | Ero1l | ERO1-like (S. cerevisiae) | MGI:1354385 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000032314 | Etfa | electron transferring flavoprotein, alpha polypeptide | MGI:106092 | YES | YES | Cytoplasm | transporter | |||||
| ENSMUSG00000004610 | Etfb | electron transferring flavoprotein, beta polypeptide | MGI:106098 | YES | YES | Cytoplasm | transporter | |||||
| ENSMUSG00000027809 | Etfdh | electron transferring flavoprotein, dehydrogenase | MGI:106100 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000028773 | Fabp3 | fatty acid binding protein 3, muscle and heart | MGI:95476 | YES | YES | Cytoplasm | transporter | |||||
| ENSMUSG00000019874 | Fabp7 | fatty acid binding protein 7, brain | MGI:101916 | YES | YES | Cytoplasm | transporter | |||||
| ENSMUSG00000010663 | Fads1 | fatty acid desaturase 1 | MGI:1923517 | YES | YES | Plasma Membrane | enzyme | |||||
| ENSMUSG00000030630 | Fah | fumarylacetoacetate hydrolase | MGI:95482 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000027371 | Fahd2a | fumarylacetoacetate hydrolase domain containing 2A | MGI:1915376 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000057497 | Fam136a | family with sequence similarity 136, member A | MGI:1913738 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000094103 | 1700047I17Rik2 | RIKEN cDNA 1700047I17 gene 2 | MGI:3714351 | YES | YES | Other | other | |||||
| ENSMUSG00000020883 | Fbxl20 | F-box and leucine-rich repeat protein 20 | MGI:1919444 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000030357 | Fkbp4 | FK506 binding protein 4 | MGI:95543 | YES | YES | Nucleus | enzyme | |||||
| ENSMUSG00000075415 | Fnbp1 | formin binding protein 1 | MGI:109606 | YES | YES | Nucleus | enzyme | |||||
| ENSMUSG00000001773 | Folh1 | folate hydrolase 1 | MGI:1858193 | YES | YES | Plasma Membrane | peptidase | |||||
| ENSMUSG00000062382 | Gm10116 | predicted pseudogene 10116 | MGI:3779109 | YES | YES | #N/A | #N/A | |||||
| ENSMUSG00000031400 | G6pdx | glucose-6-phosphate dehydrogenase X-linked | MGI:105979 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000033917 | Gde1 | glycerophosphodiester phosphodiesterase 1 | MGI:1891827 | YES | YES | Plasma Membrane | enzyme | |||||
| ENSMUSG00000021218 | Gdi2 | guanosine diphosphate (GDP) dissociation inhibitor 2 | MGI:99845 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000061666 | Gdpd1 | glycerophosphodiester phosphodiesterase domain containing 1 | MGI:1913819 | YES | YES | Other | enzyme | |||||
| ENSMUSG00000040888 | Gfer | growth factor, erv1 (S. cerevisiae)-like (augmenter of liver regeneration) | MGI:107757 | YES | YES | Nucleus | enzyme | |||||
| ENSMUSG00000017286 | Glod4 | glyoxalase domain containing 4 | MGI:1914451 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000000253 | Gmpr | guanosine monophosphate reductase | MGI:1913605 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000020372 | Gnb2l1 | guanine nucleotide binding protein (G protein), beta polypeptide 2 like 1 | MGI:101849 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000024429 | Gnl1 | guanine nucleotide binding protein-like 1 | MGI:95764 | YES | YES | Nucleus | other | |||||
| ENSMUSG00000021192 | Golga5 | golgi autoantigen, golgin subfamily a, 5 | MGI:1351475 | YES | YES | Cytoplasm | kinase | |||||
| ENSMUSG00000030245 | Golt1b | golgi transport 1 homolog B (S. cerevisiae) | MGI:1914214 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000020946 | Gosr2 | golgi SNAP receptor complex member 2 | MGI:1927204 | YES | YES | Cytoplasm | transporter | |||||
| ENSMUSG00000050627 | Gpd1l | glycerol-3-phosphate dehydrogenase 1-like | MGI:1289257 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000059923 | Grb2 | growth factor receptor bound protein 2 | MGI:95805 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000035637 | Grhpr | glyoxylate reductase/hydroxypyruvate reductase | MGI:1923488 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000029198 | Grpel1 | GrpE-like 1, mitochondrial | MGI:1334417 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000031584 | Gsr | glutathione reductase | MGI:95804 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000021033 | Gstz1 | glutathione transferase zeta 1 (maleylacetoacetate isomerase) | MGI:1341859 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000019188 | H13 | histocompatibility 13 | MGI:95886 | YES | YES | Cytoplasm | peptidase | |||||
| ENSMUSG00000021884 | Hacl1 | 2-hydroxyacyl-CoA lyase 1 | MGI:1929657 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000027984 | Hadh | hydroxyacyl-Coenzyme A dehydrogenase | MGI:96009 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000024158 | Hagh | hydroxyacyl glutathione hydrolase | MGI:95745 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000025421 | Hdhd2 | haloacid dehalogenase-like hydrolase domain containing 2 | MGI:1924237 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000042770 | Hebp1 | heme binding protein 1 | MGI:1333880 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000029776 | Hibadh | 3-hydroxyisobutyrate dehydrogenase | MGI:1889802 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000028672 | Hmgcl | 3-hydroxy-3-methylglutaryl-Coenzyme A lyase | MGI:96158 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000025630 | Hprt | hypoxanthine guanine phosphoribosyl transferase | MGI:96217 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000022323 | Hrsp12 | heat-responsive protein 12 | MGI:1095401 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000029311 | Hsd17b11 | hydroxysteroid (17-beta) dehydrogenase 11 | MGI:2149821 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000068329 | Htra2 | HtrA serine peptidase 2 | MGI:1928676 | YES | YES | Cytoplasm | peptidase | |||||
| ENSMUSG00000026618 | Iars2 | isoleucine-tRNA synthetase 2, mitochondrial | MGI:1919586 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000025950 | Idh1 | isocitrate dehydrogenase 1 (NADP+), soluble | MGI:96413 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000023830 | Igf2r | insulin-like growth factor 2 receptor | MGI:96435 | YES | YES | Plasma Membrane | transmembrane receptor | |||||
| ENSMUSG00000000159 | Igsf5 | immunoglobulin superfamily, member 5 | MGI:1919308 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000021756 | Il6st | interleukin 6 signal transducer | MGI:96560 | YES | YES | Plasma Membrane | transmembrane receptor | |||||
| ENSMUSG00000027531 | Impa1 | inositol (myo)-1(or 4)-monophosphatase 1 | MGI:1933158 | YES | YES | Cytoplasm | phosphatase | |||||
| ENSMUSG00000066324 | Impad1 | inositol monophosphatase domain containing 1 | MGI:1915720 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000026102 | Inpp1 | inositol polyphosphate-1-phosphatase | MGI:104848 | YES | YES | Cytoplasm | phosphatase | |||||
| ENSMUSG00000051243 | Islr2 | immunoglobulin superfamily containing leucine-rich repeat 2 | MGI:2444277 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000086784 | Isoc2a | isochorismatase domain containing 2a | MGI:3609243 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000027111 | Itga6 | integrin alpha 6 | MGI:96605 | YES | YES | Plasma Membrane | transmembrane receptor | |||||
| ENSMUSG00000030786 | Itgam | integrin alpha M | MGI:96607 | YES | YES | Plasma Membrane | transmembrane receptor | |||||
| ENSMUSG00000027087 | Itgav | integrin alpha V | MGI:96608 | YES | YES | Plasma Membrane | ion channel | |||||
| ENSMUSG00000025809 | Itgb1 | integrin beta 1 (fibronectin receptor beta) | MGI:96610 | YES | YES | Plasma Membrane | transmembrane receptor | |||||
| ENSMUSG00000000290 | Itgb2 | integrin beta 2 | MGI:96611 | YES | YES | Plasma Membrane | transmembrane receptor | |||||
| ENSMUSG00000025321 | Itgb8 | integrin beta 8 | MGI:1338035 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000022629 | Kif21a | kinesin family member 21A | MGI:109188 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000020988 | L2hgdh | L-2-hydroxyglutarate dehydrogenase | MGI:2384968 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000016534 | Lamp2 | lysosomal-associated membrane protein 2 | MGI:96748 | YES | YES | Plasma Membrane | enzyme | |||||
| ENSMUSG00000027270 | Lamp5 | lysosomal-associated membrane protein family, member 5 | MGI:1923411 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000035202 | Lars2 | leucyl-tRNA synthetase, mitochondrial | MGI:2142973 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000063229 | Ldha | lactate dehydrogenase A | MGI:96759 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000045083 | Lingo2 | leucine rich repeat and Ig domain containing 2 | MGI:2442298 | YES | YES | Extracellular Space | other | |||||
| ENSMUSG00000021484 | Lman2 | lectin, mannose-binding 2 | MGI:1914140 | YES | YES | Cytoplasm | transporter | |||||
| ENSMUSG00000073725 | Lmbrd1 | LMBR1 domain containing 1 | MGI:1915671 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000039704 | Lmbrd2 | LMBR1 domain containing 2 | MGI:2444173 | YES | YES | Other | other | |||||
| ENSMUSG00000023845 | Lnpep | leucyl/cystinyl aminopeptidase | MGI:2387123 | YES | YES | Cytoplasm | peptidase | |||||
| ENSMUSG00000042992 | Loh12cr1 | loss of heterozygosity, 12, chromosomal region 1 homolog (human) | MGI:1915024 | YES | YES | Other | other | |||||
| ENSMUSG00000041168 | Lonp1 | lon peptidase 1, mitochondrial | MGI:1921392 | YES | YES | Cytoplasm | peptidase | |||||
| ENSMUSG00000035653 | Lrfn5 | leucine rich repeat and fibronectin type III domain containing 5 | MGI:2144814 | YES | YES | Nucleus | other | |||||
| ENSMUSG00000034648 | Lrrn1 | leucine rich repeat protein 1, neuronal | MGI:106038 | YES | YES | Other | other | |||||
| ENSMUSG00000015889 | Lta4h | leukotriene A4 hydrolase | MGI:96836 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000047557 | Lxn | latexin | MGI:107633 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000046573 | Lyrm4 | LYR motif containing 4 | MGI:2683538 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000007458 | M6pr | mannose-6-phosphate receptor, cation dependent | MGI:96904 | YES | YES | Cytoplasm | transporter | |||||
| ENSMUSG00000036278 | Macrod1 | MACRO domain containing 1 | MGI:2147583 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000032575 | Manf | mesencephalic astrocyte-derived neurotrophic factor | MGI:1922090 | YES | YES | Extracellular Space | other | |||||
| ENSMUSG00000047945 | Marcksl1 | MARCKS-like 1 | MGI:97143 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000051098 | Mblac2 | metallo-beta-lactamase domain containing 2 | MGI:1920102 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000035596 | Mboat7 | membrane bound O-acyltransferase domain containing 7 | MGI:1924832 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000021371 | Mcur1 | mitochondrial calcium uniporter regulator 1 | MGI:1923387 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000032418 | Me1 | malic enzyme 1, NADP(+)-dependent, cytosolic | MGI:97043 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000024556 | Me2 | malic enzyme 2, NAD(+)-dependent, mitochondrial | MGI:2147351 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000014361 | Mertk | c-mer proto-oncogene tyrosine kinase | MGI:96965 | YES | YES | Plasma Membrane | kinase | |||||
| ENSMUSG00000038503 | Mesdc2 | mesoderm development candidate 2 | MGI:1891421 | YES | YES | Extracellular Space | other | |||||
| ENSMUSG00000054619 | Mettl7a1 | methyltransferase like 7A1 | MGI:1916523 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000027668 | Mfn1 | mitofusin 1 | MGI:1914664 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000041439 | Mfsd6 | major facilitator superfamily domain containing 6 | MGI:1922925 | YES | YES | Other | other | |||||
| ENSMUSG00000033174 | Mgll | monoglyceride lipase | MGI:1346042 | YES | YES | Plasma Membrane | enzyme | |||||
| ENSMUSG00000008540 | Mgst1 | microsomal glutathione S-transferase 1 | MGI:1913850 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000040675 | Mthfd1l | methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1-like | MGI:1924836 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000023921 | Mut | methylmalonyl-Coenzyme A mutase | MGI:97239 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000019579 | Mydgf | myeloid derived growth factor | MGI:2156020 | YES | YES | Extracellular Space | cytokine | |||||
| ENSMUSG00000024587 | Nars | asparaginyl-tRNA synthetase | MGI:1917473 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000020576 | Nbas | neuroblastoma amplified sequence | MGI:1918419 | YES | YES | Nucleus | other | |||||
| ENSMUSG00000003458 | Ncstn | nicastrin | MGI:1891700 | YES | YES | Plasma Membrane | peptidase | |||||
| ENSMUSG00000035674 | Ndufa3 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 3 | MGI:1913341 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000068184 | Ndufaf2 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, assembly factor 2 | MGI:1922847 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000022454 | Nell2 | NEL-like 2 | MGI:1858510 | YES | YES | Extracellular Space | other | |||||
| ENSMUSG00000015247 | Nipsnap3b | nipsnap homolog 3B (C. elegans) | MGI:1913786 | YES | YES | Nucleus | other | |||||
| ENSMUSG00000021710 | Nln | neurolysin (metallopeptidase M3 family) | MGI:1923055 | YES | YES | Cytoplasm | peptidase | |||||
| ENSMUSG00000037601 | Nme1 | NME/NM23 nucleoside diphosphate kinase 1 | MGI:97355 | YES | YES | Cytoplasm | kinase | |||||
| ENSMUSG00000024413 | Npc1 | Niemann-Pick type C1 | MGI:1097712 | YES | YES | Cytoplasm | transporter | |||||
| ENSMUSG00000025969 | Nrp2 | neuropilin 2 | MGI:1100492 | YES | YES | Plasma Membrane | kinase | |||||
| ENSMUSG00000031349 | Nsdhl | NAD(P) dependent steroid dehydrogenase-like | MGI:1099438 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000059146 | Ntrk3 | neurotrophic tyrosine kinase, receptor, type 3 | MGI:97385 | YES | YES | Plasma Membrane | kinase | |||||
| ENSMUSG00000030824 | Nucb1 | nucleobindin 1 | MGI:97388 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000030659 | Nucb2 | nucleobindin 2 | MGI:1858179 | YES | YES | Nucleus | other | |||||
| ENSMUSG00000024213 | Nudt3 | nudix (nucleotide diphosphate linked moiety X)-type motif 3 | MGI:1928484 | YES | YES | Cytoplasm | phosphatase | |||||
| ENSMUSG00000030934 | Oat | ornithine aminotransferase | MGI:97394 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000038150 | Ormdl3 | ORM1-like 3 (S. cerevisiae) | MGI:1913862 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000036353 | P2ry12 | purinergic receptor P2Y, G-protein coupled 12 | MGI:1918089 | YES | YES | Plasma Membrane | G-protein coupled receptor | |||||
| ENSMUSG00000016664 | Pacsin2 | protein kinase C and casein kinase substrate in neurons 2 | MGI:1345153 | YES | YES | Cytoplasm | transporter | |||||
| ENSMUSG00000028964 | Park7 | Parkinson disease (autosomal recessive, early onset) 7 | MGI:2135637 | YES | YES | Nucleus | enzyme | |||||
| ENSMUSG00000041650 | Pcca | propionyl-Coenzyme A carboxylase, alpha polypeptide | MGI:97499 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000032527 | Pccb | propionyl Coenzyme A carboxylase, beta polypeptide | MGI:1914154 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000051375 | Pcdh1 | protocadherin 1 | MGI:104692 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000040618 | Pck2 | phosphoenolpyruvate carboxykinase 2 (mitochondrial) | MGI:1860456 | YES | YES | Cytoplasm | kinase | |||||
| ENSMUSG00000029998 | Pcyox1 | prenylcysteine oxidase 1 | MGI:1914131 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000024579 | Pcyox1l | prenylcysteine oxidase 1 like | MGI:3606062 | YES | YES | Other | other | |||||
| ENSMUSG00000035246 | Pcyt1b | phosphate cytidylyltransferase 1, choline, beta isoform | MGI:2147987 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000022436 | Sh3bp1 | SH3-domain binding protein 1 | MGI:104603 | YES | YES | #N/A | #N/A | |||||
| ENSMUSG00000028961 | Pgd | phosphogluconate dehydrogenase | MGI:97553 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000043445 | Pgp | phosphoglycolate phosphatase | MGI:1914328 | YES | YES | Other | enzyme | |||||
| ENSMUSG00000037747 | Phyhipl | phytanoyl-CoA hydroxylase interacting protein-like | MGI:1918161 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000032462 | Pik3cb | phosphatidylinositol 3-kinase, catalytic, beta polypeptide | MGI:1922019 | YES | YES | Cytoplasm | kinase | |||||
| ENSMUSG00000026640 | Plxna2 | plexin A2 | MGI:107684 | YES | YES | Plasma Membrane | transmembrane receptor | |||||
| ENSMUSG00000036606 | Plxnb2 | plexin B2 | MGI:2154239 | YES | YES | Plasma Membrane | transmembrane receptor | |||||
| ENSMUSG00000030123 | Plxnd1 | plexin D1 | MGI:2154244 | YES | YES | Plasma Membrane | transmembrane receptor | |||||
| ENSMUSG00000032667 | Pon2 | paraoxonase 2 | MGI:106687 | YES | YES | Plasma Membrane | enzyme | |||||
| ENSMUSG00000020089 | Ppa1 | pyrophosphatase (inorganic) 1 | MGI:97831 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000021868 | Ppif | peptidylprolyl isomerase F (cyclophilin F) | MGI:2145814 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000061718 | Ppp1r1b | protein phosphatase 1, regulatory (inhibitor) subunit 1B | MGI:94860 | YES | YES | Cytoplasm | phosphatase | |||||
| ENSMUSG00000003099 | Ppp5c | protein phosphatase 5, catalytic subunit | MGI:102666 | YES | YES | Nucleus | phosphatase | |||||
| ENSMUSG00000021948 | Prkcd | protein kinase C, delta | MGI:97598 | YES | YES | Cytoplasm | kinase | |||||
| ENSMUSG00000003402 | Prkcsh | protein kinase C substrate 80K-H | MGI:107877 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000029086 | Prom1 | prominin 1 | MGI:1100886 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000004207 | Psap | prosaposin | MGI:97783 | YES | YES | Extracellular Space | other | |||||
| ENSMUSG00000032301 | Psma4 | proteasome (prosome, macropain) subunit, alpha type 4 | MGI:1347060 | YES | YES | Cytoplasm | peptidase | |||||
| ENSMUSG00000027864 | Ptgfrn | prostaglandin F2 receptor negative regulator | MGI:1277114 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000026238 | Ptma | prothymosin alpha | MGI:97803 | YES | YES | Nucleus | other | |||||
| ENSMUSG00000028788 | Ptp4a2 | protein tyrosine phosphatase 4a2 | MGI:1277117 | YES | YES | Cytoplasm | phosphatase | |||||
| ENSMUSG00000030854 | Ptpn5 | protein tyrosine phosphatase, non-receptor type 5 | MGI:97807 | YES | YES | Plasma Membrane | phosphatase | |||||
| ENSMUSG00000041836 | Ptpre | protein tyrosine phosphatase, receptor type, E | MGI:97813 | YES | YES | Plasma Membrane | phosphatase | |||||
| ENSMUSG00000025314 | Ptprj | protein tyrosine phosphatase, receptor type, J | MGI:104574 | YES | YES | Plasma Membrane | phosphatase | |||||
| ENSMUSG00000026204 | Ptprn | protein tyrosine phosphatase, receptor type, N | MGI:102765 | YES | YES | Plasma Membrane | phosphatase | |||||
| ENSMUSG00000005716 | Pvalb | parvalbumin | MGI:97821 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000032012 | Pvrl1 | poliovirus receptor-related 1 | MGI:1926483 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000032648 | Pygm | muscle glycogen phosphorylase | MGI:97830 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000004768 | Rab23 | RAB23, member RAS oncogene family | MGI:99833 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000030643 | Rab30 | RAB30, member RAS oncogene family | MGI:1923235 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000039318 | Rab3gap2 | RAB3 GTPase activating protein subunit 2 | MGI:1916043 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000079316 | Rab9 | RAB9, member RAS oncogene family | MGI:1890695 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000050029 | Rap2c | RAP2C, member of RAS oncogene family | MGI:1919315 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000018848 | Rars | arginyl-tRNA synthetase | MGI:1914297 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000066441 | Rdh11 | retinol dehydrogenase 11 | MGI:102581 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000032026 | Rexo2 | REX2, RNA exonuclease 2 homolog (S. cerevisiae) | MGI:1888981 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000052087 | Rgs14 | regulator of G-protein signaling 14 | MGI:1859709 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000028229 | Rmdn1 | regulator of microtubule dynamics 1 | MGI:1913552 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000013878 | Rnf170 | ring finger protein 170 | MGI:1924983 | YES | YES | Other | other | |||||
| ENSMUSG00000032518 | Rpsa | ribosomal protein SA | MGI:105381 | YES | YES | Cytoplasm | translation regulator | |||||
| ENSMUSG00000033208 | S100b | S100 protein, beta polypeptide, neural | MGI:98217 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000078441 | Scamp4 | secretory carrier membrane protein 4 | MGI:1928947 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000029426 | Scarb2 | scavenger receptor class B, member 2 | MGI:1196458 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000049281 | Scn3b | sodium channel, voltage-gated, type III, beta | MGI:1918882 | YES | YES | Plasma Membrane | ion channel | |||||
| ENSMUSG00000028603 | Scp2 | sterol carrier protein 2, liver | MGI:98254 | YES | YES | Cytoplasm | transporter | |||||
| ENSMUSG00000069539 | Scyl2 | SCY1-like 2 (S. cerevisiae) | MGI:1289172 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000022223 | Sdr39u1 | short chain dehydrogenase/reductase family 39U, member 1 | MGI:1916876 | YES | YES | Nucleus | other | |||||
| ENSMUSG00000075702 | Selm | selenoprotein M | MGI:2149786 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000075700 | Selt | selenoprotein T | MGI:1916477 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000004937 | Sgta | small glutamine-rich tetratricopeptide repeat (TPR)-containing, alpha | MGI:1098703 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000037062 | Sh3glb1 | SH3-domain GRB2-like B1 (endophilin) | MGI:1859730 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000027130 | Slc12a6 | solute carrier family 12, member 6 | MGI:2135960 | YES | YES | Plasma Membrane | transporter | |||||
| ENSMUSG00000037344 | Slc12a9 | solute carrier family 12 (potassium/chloride transporters), member 9 | MGI:1933532 | YES | YES | Plasma Membrane | transporter | |||||
| ENSMUSG00000025094 | Slc18a2 | solute carrier family 18 (vesicular monoamine), member 2 | MGI:106677 | YES | YES | Plasma Membrane | transporter | |||||
| ENSMUSG00000020142 | Slc1a4 | solute carrier family 1 (glutamate/neutral amino acid transporter), member 4 | MGI:2135601 | YES | YES | Plasma Membrane | transporter | |||||
| ENSMUSG00000037656 | Slc20a2 | solute carrier family 20, member 2 | MGI:97851 | YES | YES | Plasma Membrane | transporter | |||||
| ENSMUSG00000020334 | Slc22a4 | solute carrier family 22 (organic cation transporter), member 4 | MGI:1353479 | YES | YES | Plasma Membrane | transporter | |||||
| ENSMUSG00000027340 | Slc23a2 | solute carrier family 23 (nucleobase transporters), member 2 | MGI:1859682 | YES | YES | Plasma Membrane | transporter | |||||
| ENSMUSG00000015112 | Slc25a13 | solute carrier family 25 (mitochondrial carrier, adenine nucleotide translocator), member 13 | MGI:1354721 | YES | YES | Cytoplasm | transporter | |||||
| ENSMUSG00000037434 | Slc30a1 | solute carrier family 30 (zinc transporter), member 1 | MGI:1345281 | YES | YES | Plasma Membrane | transporter | |||||
| ENSMUSG00000029221 | Slc30a9 | solute carrier family 30 (zinc transporter), member 9 | MGI:1923690 | YES | YES | Nucleus | transporter | |||||
| ENSMUSG00000038178 | Slc43a2 | solute carrier family 43, member 2 | MGI:2442746 | YES | YES | Plasma Membrane | transporter | |||||
| ENSMUSG00000057193 | Slc44a2 | solute carrier family 44, member 2 | MGI:1915932 | YES | YES | Extracellular Space | transporter | |||||
| ENSMUSG00000006576 | Slc4a3 | solute carrier family 4 (anion exchanger), member 3 | MGI:109350 | YES | YES | Plasma Membrane | transporter | |||||
| ENSMUSG00000021733 | Slc4a7 | solute carrier family 4, sodium bicarbonate cotransporter, member 7 | MGI:2443878 | YES | YES | Plasma Membrane | transporter | |||||
| ENSMUSG00000021609 | Slc6a3 | solute carrier family 6 (neurotransmitter transporter, dopamine), member 3 | MGI:94862 | YES | YES | Plasma Membrane | transporter | |||||
| ENSMUSG00000039728 | Slc6a5 | solute carrier family 6 (neurotransmitter transporter, glycine), member 5 | MGI:105090 | YES | YES | Plasma Membrane | transporter | |||||
| ENSMUSG00000028542 | Slc6a9 | solute carrier family 6 (neurotransmitter transporter, glycine), member 9 | MGI:95760 | YES | YES | Plasma Membrane | transporter | |||||
| ENSMUSG00000027737 | Slc7a11 | solute carrier family 7 (cationic amino acid transporter, y+ system), member 11 | MGI:1347355 | YES | YES | Plasma Membrane | transporter | |||||
| ENSMUSG00000040010 | Slc7a5 | solute carrier family 7 (cationic amino acid transporter, y+ system), member 5 | MGI:1298205 | YES | YES | Plasma Membrane | transporter | |||||
| ENSMUSG00000020733 | Slc9a3r1 | solute carrier family 9 (sodium/hydrogen exchanger), member 3 regulator 1 | MGI:1349482 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000060681 | Slc9a6 | solute carrier family 9 (sodium/hydrogen exchanger), member 6 | MGI:2443511 | YES | YES | Plasma Membrane | transporter | |||||
| ENSMUSG00000037341 | Slc9a7 | solute carrier family 9 (sodium/hydrogen exchanger), member 7 | MGI:2444530 | YES | YES | Cytoplasm | transporter | |||||
| ENSMUSG00000025060 | Slk | STE20-like kinase | MGI:103241 | YES | YES | Nucleus | kinase | |||||
| ENSMUSG00000032382 | Snx1 | sorting nexin 1 | MGI:1928395 | YES | YES | Cytoplasm | transporter | |||||
| ENSMUSG00000027534 | Snx16 | sorting nexin 16 | MGI:1921968 | YES | YES | Other | transporter | |||||
| ENSMUSG00000034484 | Snx2 | sorting nexin 2 | MGI:1915054 | YES | YES | Cytoplasm | transporter | |||||
| ENSMUSG00000022808 | Snx4 | sorting nexin 4 | MGI:1916400 | YES | YES | Cytoplasm | transporter | |||||
| ENSMUSG00000005656 | Snx6 | sorting nexin 6 | MGI:1919433 | YES | YES | Cytoplasm | transporter | |||||
| ENSMUSG00000003161 | Sri | sorcin | MGI:98419 | YES | YES | Cytoplasm | transporter | |||||
| ENSMUSG00000026718 | Stam | signal transducing adaptor molecule (SH3 domain and ITAM motif) 1 | MGI:1329014 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000028832 | Stmn1 | stathmin 1 | MGI:96739 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000030805 | Stx4a | syntaxin 4A (placental) | MGI:893577 | YES | YES | Plasma Membrane | transporter | |||||
| ENSMUSG00000027882 | Stxbp3 | syntaxin binding protein 3 | MGI:107362 | YES | YES | Plasma Membrane | transporter | |||||
| ENSMUSG00000014867 | Surf4 | surfeit gene 4 | MGI:98445 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000051111 | Sv2c | synaptic vesicle glycoprotein 2c | MGI:1922459 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000042078 | Svop | SV2 related protein | MGI:1915916 | YES | YES | Cytoplasm | transporter | |||||
| ENSMUSG00000025503 | Taldo1 | transaldolase 1 | MGI:1274789 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000046985 | Tapt1 | transmembrane anterior posterior transformation 1 | MGI:2683537 | YES | YES | Plasma Membrane | G-protein coupled receptor | |||||
| ENSMUSG00000022241 | Tars | threonyl-tRNA synthetase | MGI:106314 | YES | YES | Nucleus | enzyme | |||||
| ENSMUSG00000029359 | Tesc | tescalcin | MGI:1930803 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000040813 | Tex264 | testis expressed gene 264 | MGI:1096570 | YES | YES | Extracellular Space | other | |||||
| ENSMUSG00000028145 | Them4 | thioesterase superfamily member 4 | MGI:1923028 | YES | YES | Plasma Membrane | enzyme | |||||
| ENSMUSG00000048550 | Thnsl1 | threonine synthase-like 1 (bacterial) | MGI:2139347 | YES | YES | Nucleus | kinase | |||||
| ENSMUSG00000004929 | Thop1 | thimet oligopeptidase 1 | MGI:1354165 | YES | YES | Cytoplasm | peptidase | |||||
| ENSMUSG00000013701 | Timm23 | translocase of inner mitochondrial membrane 23 | MGI:1858317 | YES | YES | Cytoplasm | transporter | |||||
| ENSMUSG00000025544 | Tm9sf2 | transmembrane 9 superfamily member 2 | MGI:1915309 | YES | YES | Plasma Membrane | transporter | |||||
| ENSMUSG00000025016 | Tm9sf3 | transmembrane 9 superfamily member 3 | MGI:1914262 | YES | YES | Cytoplasm | transporter | |||||
| ENSMUSG00000068040 | Tm9sf4 | transmembrane 9 superfamily protein member 4 | MGI:2139220 | YES | YES | Cytoplasm | transporter | |||||
| ENSMUSG00000033184 | Tmed7 | transmembrane emp24 protein transport domain containing 7 | MGI:1913926 | YES | YES | Cytoplasm | transporter | |||||
| ENSMUSG00000013495 | Tmem175 | transmembrane protein 175 | MGI:1919642 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000028857 | Tmem222 | transmembrane protein 222 | MGI:1098568 | YES | YES | Other | other | |||||
| ENSMUSG00000048022 | Tmem229a | transmembrane protein 229A | MGI:2442812 | YES | YES | Other | other | |||||
| ENSMUSG00000030095 | Tmem43 | transmembrane protein 43 | MGI:1921372 | YES | YES | Nucleus | other | |||||
| ENSMUSG00000026411 | Tmem9 | transmembrane protein 9 | MGI:1913491 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000021072 | Tmx1 | thioredoxin-related transmembrane protein 1 | MGI:1919986 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000035274 | Tpbg | trophoblast glycoprotein | MGI:1341264 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000029030 | Tprgl | transformation related protein 63 regulated like | MGI:1915058 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000005981 | Trap1 | TNF receptor-associated protein 1 | MGI:1915265 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000032112 | Trappc4 | trafficking protein particle complex 4 | MGI:1926211 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000058254 | Tspan7 | tetraspanin 7 | MGI:1298407 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000044986 | Tst | thiosulfate sulfurtransferase, mitochondrial | MGI:98852 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000020250 | Txnrd1 | thioredoxin reductase 1 | MGI:1354175 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000032557 | Uba5 | ubiquitin-like modifier activating enzyme 5 | MGI:1913913 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000038965 | Ube2l3 | ubiquitin-conjugating enzyme E2L 3 | MGI:109240 | YES | YES | Nucleus | enzyme | |||||
| ENSMUSG00000020802 | Ube2o | ubiquitin-conjugating enzyme E2O | MGI:2444266 | YES | YES | Nucleus | enzyme | |||||
| ENSMUSG00000005312 | Ubqln1 | ubiquilin 1 | MGI:1860276 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000050148 | Ubqln2 | ubiquilin 2 | MGI:1860283 | YES | YES | Nucleus | other | |||||
| ENSMUSG00000063882 | Uqcrh | ubiquinol-cytochrome c reductase hinge protein | MGI:1913826 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000002395 | Use1 | unconventional SNARE in the ER 1 homolog (S. cerevisiae) | MGI:1914273 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000050732 | Vamp8 | vesicle-associated membrane protein 8 | MGI:1336882 | YES | YES | Plasma Membrane | transporter | |||||
| ENSMUSG00000046844 | Vat1l | vesicle amine transport protein 1 homolog-like (T. californica) | MGI:2142534 | YES | YES | Other | enzyme | |||||
| ENSMUSG00000027962 | Vcam1 | vascular cell adhesion molecule 1 | MGI:98926 | YES | YES | Plasma Membrane | transmembrane receptor | |||||
| ENSMUSG00000018171 | Vmp1 | vacuole membrane protein 1 | MGI:1923159 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000035284 | Vps13c | vacuolar protein sorting 13C (yeast) | MGI:2444207 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000020220 | Vps13d | vacuolar protein sorting 13 D (yeast) | MGI:2448530 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000020078 | Vps26a | vacuolar protein sorting 26 homolog A (yeast) | MGI:1353654 | YES | YES | Cytoplasm | transporter | |||||
| ENSMUSG00000073643 | Wdfy1 | WD repeat and FYVE domain containing 1 | MGI:1916618 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000032313 | AI118078 | expressed sequence AI118078 | MGI:2142980 | YES | YES | Other | other | |||||
| ENSMUSG00000054091 | 1810037I17Rik | RIKEN cDNA 1810037I17 gene | MGI:1914954 | YES | YES | Other | other | |||||
| ENSMUSG00000050875 | A730017C20Rik | RIKEN cDNA A730017C20 gene | MGI:2442934 | YES | YES | Other | other | |||||
| ENSMUSG00000027942 | 4933434E20Rik | RIKEN cDNA 4933434E20 gene | MGI:1914027 | YES | YES | #N/A | #N/A | |||||
| ENSMUSG00000023938 | Aars2 | alanyl-tRNA synthetase 2, mitochondrial (putative) | MGI:2681839 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000025375 | Aatk | apoptosis-associated tyrosine kinase | MGI:1197518 | YES | YES | Cytoplasm | kinase | |||||
| ENSMUSG00000015243 | Abca1 | ATP-binding cassette, sub-family A (ABC1), member 1 | MGI:99607 | YES | YES | Plasma Membrane | transporter | |||||
| ENSMUSG00000031974 | Abcb10 | ATP-binding cassette, sub-family B (MDR/TAP), member 10 | MGI:1860508 | YES | YES | Cytoplasm | transporter | |||||
| ENSMUSG00000032842 | Abcc10 | ATP-binding cassette, sub-family C (CFTR/MRP), member 10 | MGI:2386976 | YES | YES | Plasma Membrane | transporter | |||||
| ENSMUSG00000020681 | Ace | angiotensin I converting enzyme (peptidyl-dipeptidase A) 1 | MGI:87874 | YES | YES | Plasma Membrane | peptidase | |||||
| ENSMUSG00000076435 | Acsf2 | acyl-CoA synthetase family member 2 | MGI:2388287 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000023262 | Acy1 | aminoacylase 1 | MGI:87913 | YES | YES | Cytoplasm | peptidase | |||||
| ENSMUSG00000008822 | Acyp1 | acylphosphatase 1, erythrocyte (common) type | MGI:1913454 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000031555 | Adam9 | a disintegrin and metallopeptidase domain 9 (meltrin gamma) | MGI:105376 | YES | YES | Plasma Membrane | peptidase | |||||
| ENSMUSG00000028138 | Adh5 | alcohol dehydrogenase 5 (class III), chi polypeptide | MGI:87929 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000025236 | Adpgk | ADP-dependent glucokinase | MGI:1919391 | YES | YES | Plasma Membrane | kinase | |||||
| ENSMUSG00000039313 | AF529169 | cDNA sequence AF529169 | MGI:2667167 | YES | YES | Other | other | |||||
| ENSMUSG00000041936 | Agrn | agrin | MGI:87961 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000047804 | Akap10 | A kinase (PRKA) anchor protein 10 | MGI:1890218 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000028393 | Alad | aminolevulinate, delta-, dehydratase | MGI:96853 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000028737 | Aldh4a1 | aldehyde dehydrogenase 4 family, member A1 | MGI:2443883 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000026024 | Als2 | amyotrophic lateral sclerosis 2 (juvenile) | MGI:1921268 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000050947 | Amigo1 | adhesion molecule with Ig like domain 1 | MGI:2653612 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000018340 | Anxa6 | annexin A6 | MGI:88255 | YES | YES | Plasma Membrane | ion channel | |||||
| ENSMUSG00000035960 | Apex1 | apurinic/apyrimidinic endonuclease 1 | MGI:88042 | YES | YES | Nucleus | enzyme | |||||
| ENSMUSG00000022548 | Apod | apolipoprotein D | MGI:88056 | YES | YES | Extracellular Space | transporter | |||||
| ENSMUSG00000035133 | Arhgap5 | Rho GTPase activating protein 5 | MGI:1332637 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000060904 | Arl1 | ADP-ribosylation factor-like 1 | MGI:99436 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000022620 | Arsa | arylsulfatase A | MGI:88077 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000024887 | Asah2 | N-acylsphingosine amidohydrolase 2 | MGI:1859310 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000022663 | Atg3 | autophagy related 3 | MGI:1915091 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000062949 | Atp11c | ATPase, class VI, type 11C | MGI:1859661 | YES | YES | Plasma Membrane | transporter | |||||
| ENSMUSG00000036622 | Atp13a2 | ATPase type 13A2 | MGI:1922022 | YES | YES | Cytoplasm | transporter | |||||
| ENSMUSG00000038094 | Atp13a4 | ATPase type 13A4 | MGI:1924456 | YES | YES | Cytoplasm | transporter | |||||
| ENSMUSG00000024566 | Atp9b | ATPase, class II, type 9B | MGI:1354757 | YES | YES | Cytoplasm | transporter | |||||
| ENSMUSG00000047379 | B4gat1 | beta-1,4-glucuronyltransferase 1 | MGI:1919680 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000007659 | Bcl2l1 | BCL2-like 1 | MGI:88139 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000032757 | Bet1 | blocked early in transport 1 homolog (S. cerevisiae) | MGI:1343104 | YES | YES | Cytoplasm | transporter | |||||
| ENSMUSG00000098112 | Bin2 | bridging integrator 2 | MGI:3611448 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000040466 | Blvrb | biliverdin reductase B (flavin reductase (NADPH)) | MGI:2385271 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000038286 | Bphl | biphenyl hydrolase-like (serine hydrolase, breast epithelial mucin-associated antigen) | MGI:1915271 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000015488 | Cacfd1 | calcium channel flower domain containing 1 | MGI:1924317 | YES | YES | Plasma Membrane | ion channel | |||||
| ENSMUSG00000038498 | Catsper1 | cation channel, sperm associated 1 | MGI:2179947 | YES | YES | Plasma Membrane | ion channel | |||||
| ENSMUSG00000001376 | Ccdc132 | coiled-coil domain containing 132 | MGI:1920538 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000031143 | Ccdc22 | coiled-coil domain containing 22 | MGI:1859608 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000070871 | Ccnyl1 | cyclin Y-like 1 | MGI:2138614 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000067722 | BC003965 | cDNA sequence BC003965 | MGI:2670966 | YES | YES | Other | other | |||||
| ENSMUSG00000030342 | Cd9 | CD9 antigen | MGI:88348 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000026605 | Cenpf | centromere protein F | MGI:1313302 | YES | YES | Nucleus | other | |||||
| ENSMUSG00000015714 | Cers2 | ceramide synthase 2 | MGI:1924143 | YES | YES | Nucleus | transcription regulator | |||||
| ENSMUSG00000027884 | Clcc1 | chloride channel CLIC-like 1 | MGI:2385186 | YES | YES | Cytoplasm | ion channel | |||||
| ENSMUSG00000032024 | Clmp | CXADR-like membrane protein | MGI:1918816 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000021610 | Clptm1l | CLPTM1-like | MGI:2442892 | YES | YES | Other | other | |||||
| ENSMUSG00000040759 | Cmtm5 | CKLF-like MARVEL transmembrane domain containing 5 | MGI:2447164 | YES | YES | Other | cytokine | |||||
| ENSMUSG00000003135 | Cnot11 | CCR4-NOT transcription complex, subunit 11 | MGI:106580 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000032458 | Copb2 | coatomer protein complex, subunit beta 2 (beta prime) | MGI:1354962 | YES | YES | Cytoplasm | transporter | |||||
| ENSMUSG00000027206 | Cops2 | COP9 (constitutive photomorphogenic) homolog, subunit 2 (Arabidopsis thaliana) | MGI:1330276 | YES | YES | Nucleus | transcription regulator | |||||
| ENSMUSG00000019494 | Cops6 | COP9 (constitutive photomorphogenic) homolog, subunit 6 (Arabidopsis thaliana) | MGI:1349439 | YES | YES | Nucleus | other | |||||
| ENSMUSG00000028247 | Coq3 | coenzyme Q3 homolog, methyltransferase (yeast) | MGI:101813 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000030652 | Coq7 | demethyl-Q 7 | MGI:107207 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000031782 | Coq9 | coenzyme Q9 homolog (yeast) | MGI:1915164 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000060924 | Csmd1 | CUB and Sushi multiple domains 1 | MGI:2137383 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000026176 | Ctdsp1 | CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 1 | MGI:2654470 | YES | YES | Nucleus | phosphatase | |||||
| ENSMUSG00000031095 | Cul4b | cullin 4B | MGI:1919834 | YES | YES | Nucleus | other | |||||
| ENSMUSG00000032040 | Dcps | decapping enzyme, scavenger | MGI:1916555 | YES | YES | Nucleus | enzyme | |||||
| ENSMUSG00000038633 | Degs1 | degenerative spermatocyte homolog 1 (Drosophila) | MGI:1097711 | YES | YES | Plasma Membrane | enzyme | |||||
| ENSMUSG00000042426 | Dhx29 | DEAH (Asp-Glu-Ala-His) box polypeptide 29 | MGI:2145374 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000029433 | Diablo | diablo homolog (Drosophila) | MGI:1913843 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000022848 | Dirc2 | disrupted in renal carcinoma 2 (human) | MGI:2387188 | YES | YES | Other | other | |||||
| ENSMUSG00000033987 | Dnah17 | dynein, axonemal, heavy chain 17 | MGI:1917176 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000032285 | Dnaja4 | DnaJ (Hsp40) homolog, subfamily A, member 4 | MGI:1927638 | YES | YES | Nucleus | other | |||||
| ENSMUSG00000041035 | Dpcd | deleted in primary ciliary dyskinesia | MGI:1924407 | YES | YES | Nucleus | other | |||||
| ENSMUSG00000061576 | Dpp6 | dipeptidylpeptidase 6 | MGI:94921 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000053898 | Ech1 | enoyl coenzyme A hydratase 1, peroxisomal | MGI:1858208 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000025967 | Eef1b2 | eukaryotic translation elongation factor 1 beta 2 | MGI:1929520 | YES | YES | Cytoplasm | translation regulator | |||||
| ENSMUSG00000003934 | Efnb3 | ephrin B3 | MGI:109196 | YES | YES | Plasma Membrane | kinase | |||||
| ENSMUSG00000020122 | Egfr | epidermal growth factor receptor | MGI:95294 | YES | YES | Plasma Membrane | kinase | |||||
| ENSMUSG00000062762 | Ei24 | etoposide induced 2.4 mRNA | MGI:108090 | YES | YES | Other | other | |||||
| ENSMUSG00000067194 | Eif1ax | eukaryotic translation initiation factor 1A, X-linked | MGI:1913485 | YES | YES | Other | translation regulator | |||||
| ENSMUSG00000040028 | Elavl1 | ELAV (embryonic lethal, abnormal vision)-like 1 (Hu antigen R) | MGI:1100851 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000006390 | Elovl1 | elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 1 | MGI:1858959 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000032349 | Elovl5 | ELOVL family member 5, elongation of long chain fatty acids (yeast) | MGI:1916051 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000047260 | Emc6 | ER membrane protein complex subunit 6 | MGI:1913298 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000058070 | Eml1 | echinoderm microtubule associated protein like 1 | MGI:1915769 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000048120 | Entpd1 | ectonucleoside triphosphate diphosphohydrolase 1 | MGI:102805 | YES | YES | Plasma Membrane | enzyme | |||||
| ENSMUSG00000041608 | Entpd3 | ectonucleoside triphosphate diphosphohydrolase 3 | MGI:1321386 | YES | YES | Plasma Membrane | enzyme | |||||
| ENSMUSG00000075703 | Ept1 | ethanolaminephosphotransferase 1 (CDP-ethanolamine-specific) | MGI:107898 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000021583 | Erap1 | endoplasmic reticulum aminopeptidase 1 | MGI:1933403 | YES | YES | Extracellular Space | peptidase | |||||
| ENSMUSG00000020311 | Erlec1 | endoplasmic reticulum lectin 1 | MGI:1914003 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000019232 | Etnppl | ethanolamine phosphate phospholyase | MGI:1919010 | YES | YES | Other | enzyme | |||||
| ENSMUSG00000049339 | Fam134a | family with sequence similarity 134, member A | MGI:2388278 | YES | YES | Other | other | |||||
| ENSMUSG00000017802 | Fam134c | family with sequence similarity 134, member C | MGI:1915248 | YES | YES | Other | other | |||||
| ENSMUSG00000038121 | Fam210a | family with sequence similarity 210, member A | MGI:1914000 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000028439 | Fam219a | family with sequence similarity 219, member A | MGI:1919151 | YES | YES | Other | other | |||||
| ENSMUSG00000020589 | Fam49a | family with sequence similarity 49, member A | MGI:1261783 | YES | YES | Other | other | |||||
| ENSMUSG00000042444 | Fam63b | family with sequence similarity 63, member B | MGI:2443086 | YES | YES | Nucleus | transporter | |||||
| ENSMUSG00000025738 | Fbxl16 | F-box and leucine-rich repeat protein 16 | MGI:2448488 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000054252 | Fgfr3 | fibroblast growth factor receptor 3 | MGI:95524 | YES | YES | Plasma Membrane | kinase | |||||
| ENSMUSG00000039735 | Fnbp1l | formin binding protein 1-like | MGI:1925642 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000028048 | Gba | glucosidase, beta, acid | MGI:95665 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000022707 | Gbe1 | glucan (1,4-alpha-), branching enzyme 1 | MGI:1921435 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000027774 | Gfm1 | G elongation factor, mitochondrial 1 | MGI:107339 | YES | YES | Cytoplasm | translation regulator | |||||
| ENSMUSG00000041028 | Ghitm | growth hormone inducible transmembrane protein | MGI:1913342 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000018196 | Glrx2 | glutaredoxin 2 (thioltransferase) | MGI:1916617 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000100679 | Gm28778 | predicted gene 28778 | MGI:5579484 | YES | YES | #N/A | #N/A | |||||
| ENSMUSG00000042165 | Gm9774 | predicted pseudogene 9774 | MGI:3642386 | YES | YES | Other | other | |||||
| ENSMUSG00000002546 | Golga2 | golgi autoantigen, golgin subfamily a, 2 | MGI:2139395 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000029502 | Golga3 | golgi autoantigen, golgin subfamily a, 3 | MGI:96958 | YES | YES | Cytoplasm | transporter | |||||
| ENSMUSG00000036357 | Gpr101 | G protein-coupled receptor 101 | MGI:2685211 | YES | YES | Plasma Membrane | G-protein coupled receptor | |||||
| ENSMUSG00000028051 | Hcn3 | hyperpolarization-activated, cyclic nucleotide-gated K+ 3 | MGI:1298211 | YES | YES | Plasma Membrane | ion channel | |||||
| ENSMUSG00000030532 | Hddc3 | HD domain containing 3 | MGI:1915945 | YES | YES | Other | other | |||||
| ENSMUSG00000034088 | Hdlbp | high density lipoprotein (HDL) binding protein | MGI:99256 | YES | YES | Nucleus | transporter | |||||
| ENSMUSG00000021301 | Hecw1 | HECT, C2 and WW domain containing E3 ubiquitin protein ligase 1 | MGI:2444115 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000032523 | Hhatl | hedgehog acyltransferase-like | MGI:1922020 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000021670 | Hmgcr | 3-hydroxy-3-methylglutaryl-Coenzyme A reductase | MGI:96159 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000093930 | Hmgcs1 | 3-hydroxy-3-methylglutaryl-Coenzyme A synthase 1 | MGI:107592 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000032932 | Hspa13 | heat shock protein 70 family, member 13 | MGI:1309463 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000049287 | Iba57 | IBA57, iron-sulfur cluster assembly homolog (S. cerevisiae) | MGI:3041174 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000032816 | Igdcc4 | immunoglobulin superfamily, DCC subclass, member 4 | MGI:1858497 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000042035 | Igsf3 | immunoglobulin superfamily, member 3 | MGI:1926158 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000022514 | Il1rap | interleukin 1 receptor accessory protein | MGI:104975 | YES | YES | Plasma Membrane | transmembrane receptor | |||||
| ENSMUSG00000024423 | Impact | impact, RWD domain protein | MGI:1098233 | YES | YES | Other | other | |||||
| ENSMUSG00000021241 | Isca2 | iron-sulfur cluster assembly 2 homolog (S. cerevisiae) | MGI:1921566 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000020689 | Itgb3 | integrin beta 3 | MGI:96612 | YES | YES | Plasma Membrane | transmembrane receptor | |||||
| ENSMUSG00000022817 | Itgb5 | integrin beta 5 | MGI:96614 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000026971 | Itgb6 | integrin beta 6 | MGI:96615 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000074797 | Itpa | inosine triphosphatase (nucleoside triphosphate pyrophosphatase) | MGI:96622 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000042644 | Itpr3 | inositol 1,4,5-triphosphate receptor 3 | MGI:96624 | YES | YES | Cytoplasm | ion channel | |||||
| ENSMUSG00000053062 | Jam2 | junction adhesion molecule 2 | MGI:1933820 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000018470 | Kcnab3 | potassium voltage-gated channel, shaker-related subfamily, beta member 3 | MGI:1336208 | YES | YES | Plasma Membrane | ion channel | |||||
| ENSMUSG00000028033 | Kcnq5 | potassium voltage-gated channel, subfamily Q, member 5 | MGI:1924937 | YES | YES | Plasma Membrane | ion channel | |||||
| ENSMUSG00000010277 | 2610507B11Rik | RIKEN cDNA 2610507B11 gene | MGI:1919753 | YES | YES | Extracellular Space | other | |||||
| ENSMUSG00000060012 | Kif13b | kinesin family member 13B | MGI:1098265 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000074657 | Kif5a | kinesin family member 5A | MGI:109564 | YES | YES | Cytoplasm | transporter | |||||
| ENSMUSG00000021929 | Kpna3 | karyopherin (importin) alpha 3 | MGI:1100863 | YES | YES | Nucleus | transporter | |||||
| ENSMUSG00000004383 | Large | like-glycosyltransferase | MGI:1342270 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000037172 | E330009J07Rik | RIKEN cDNA E330009J07 gene | MGI:2444256 | YES | YES | Other | other | |||||
| ENSMUSG00000037669 | Ldah | lipid droplet associated hydrolase | MGI:1916082 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000042363 | Lgalsl | lectin, galactoside binding-like | MGI:1916114 | YES | YES | Other | other | |||||
| ENSMUSG00000001143 | Lman2l | lectin, mannose-binding 2-like | MGI:2443010 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000021608 | Lpcat1 | lysophosphatidylcholine acyltransferase 1 | MGI:2384812 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000063446 | Lppr1 | lipid phosphate phosphatase-related protein type 1 | MGI:2445015 | YES | YES | Other | other | |||||
| ENSMUSG00000040490 | Lrfn2 | leucine rich repeat and fibronectin type III domain containing 2 | MGI:1917780 | YES | YES | Other | other | |||||
| ENSMUSG00000075605 | 2300005B03Rik | RIKEN cDNA 2300005B03 gene | MGI:1916712 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000025903 | Lypla1 | lysophospholipase 1 | MGI:1344588 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000053907 | Mat2a | methionine adenosyltransferase II, alpha | MGI:2443731 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000051065 | Mb21d2 | Mab-21 domain containing 2 | MGI:1917028 | YES | YES | Other | other | |||||
| ENSMUSG00000048755 | Mcat | malonyl CoA:ACP acyltransferase (mitochondrial) | MGI:2388651 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000004567 | Mcoln1 | mucolipin 1 | MGI:1890498 | YES | YES | Cytoplasm | ion channel | |||||
| ENSMUSG00000029575 | Mmab | methylmalonic aciduria (cobalamin deficiency) type B homolog (human) | MGI:1924947 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000107283 | Mpv17 | MpV17 mitochondrial inner membrane protein | MGI:97138 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000028622 | Mrpl37 | mitochondrial ribosomal protein L37 | MGI:1926268 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000022889 | Mrpl39 | mitochondrial ribosomal protein L39 | MGI:1351620 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000030612 | Mrpl46 | mitochondrial ribosomal protein L46 | MGI:1914558 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000030706 | Mrpl48 | mitochondrial ribosomal protein L48 | MGI:1289321 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000007338 | Mrpl49 | mitochondrial ribosomal protein L49 | MGI:108180 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000054733 | Msra | methionine sulfoxide reductase A | MGI:106916 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000064351 | mt-Co1 | mitochondrially encoded cytochrome c oxidase I | MGI:102504 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000033196 | Myh2 | myosin, heavy polypeptide 2, skeletal muscle, adult | MGI:1339710 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000041794 | Myrip | myosin VIIA and Rab interacting protein | MGI:2384407 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000022698 | Naa50 | N(alpha)-acetyltransferase 50, NatE catalytic subunit | MGI:1919367 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000041073 | Nacad | NAC alpha domain containing | MGI:3603030 | YES | YES | Other | other | |||||
| ENSMUSG00000022253 | Nadk2 | NAD kinase 2, mitochondrial | MGI:1915896 | YES | YES | Cytoplasm | kinase | |||||
| ENSMUSG00000028334 | Nans | N-acetylneuraminic acid synthase (sialic acid synthase) | MGI:2149820 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000028614 | Ndc1 | NDC1 transmembrane nucleoporin | MGI:1920037 | YES | YES | Nucleus | other | |||||
| ENSMUSG00000053253 | Ndfip2 | Nedd4 family interacting protein 2 | MGI:1923523 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000037499 | Nenf | neuron derived neurotrophic factor | MGI:1913458 | YES | YES | Extracellular Space | growth factor | |||||
| ENSMUSG00000026036 | Nif3l1 | Ngg1 interacting factor 3-like 1 (S. pombe) | MGI:1929485 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000029112 | Nkx1-1 | NK1 transcription factor related, locus 1 (Drosophila) | MGI:109346 | YES | YES | Other | other | |||||
| ENSMUSG00000063887 | Nlgn1 | neuroligin 1 | MGI:2179435 | YES | YES | Plasma Membrane | enzyme | |||||
| ENSMUSG00000015094 | Npdc1 | neural proliferation, differentiation and control 1 | MGI:1099802 | YES | YES | Extracellular Space | other | |||||
| ENSMUSG00000032558 | Nphp3 | nephronophthisis 3 (adolescent) | MGI:1921275 | YES | YES | Extracellular Space | other | |||||
| ENSMUSG00000027852 | Nras | neuroblastoma ras oncogene | MGI:97376 | YES | YES | Plasma Membrane | enzyme | |||||
| ENSMUSG00000032615 | Nt5m | 5',3'-nucleotidase, mitochondrial | MGI:1917127 | YES | YES | Cytoplasm | phosphatase | |||||
| ENSMUSG00000028443 | Nudt2 | nudix (nucleoside diphosphate linked moiety X)-type motif 2 | MGI:1913651 | YES | YES | Plasma Membrane | phosphatase | |||||
| ENSMUSG00000001173 | Ocrl | oculocerebrorenal syndrome of Lowe | MGI:109589 | YES | YES | Cytoplasm | phosphatase | |||||
| ENSMUSG00000021252 | 0610007P14Rik | RIKEN cDNA 0610007P14 gene | MGI:1915571 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000028927 | Padi2 | peptidyl arginine deiminase, type II | MGI:1338892 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000005447 | Pafah1b3 | platelet-activating factor acetylhydrolase, isoform 1b, subunit 3 | MGI:108414 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000037514 | Pank2 | pantothenate kinase 2 | MGI:1921700 | YES | YES | Cytoplasm | kinase | |||||
| ENSMUSG00000034981 | Parm1 | prostate androgen-regulated mucin-like protein 1 | MGI:2443349 | YES | YES | Extracellular Space | other | |||||
| ENSMUSG00000043572 | Pars2 | prolyl-tRNA synthetase (mitochondrial)(putative) | MGI:2386296 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000051323 | Pcdh19 | protocadherin 19 | MGI:2685563 | YES | YES | Extracellular Space | other | |||||
| ENSMUSG00000036422 | Pcdh8 | protocadherin 8 | MGI:1306800 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000102918 | Pcdhgc3 | protocadherin gamma subfamily C, 3 | MGI:1935201 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000038370 | Pcp4l1 | Purkinje cell protein 4-like 1 | MGI:1913675 | YES | YES | Other | other | |||||
| ENSMUSG00000059173 | Pde1a | phosphodiesterase 1A, calmodulin-dependent | MGI:1201792 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000029231 | Pdgfra | platelet derived growth factor receptor, alpha polypeptide | MGI:97530 | YES | YES | Plasma Membrane | kinase | |||||
| ENSMUSG00000024620 | Pdgfrb | platelet derived growth factor receptor, beta polypeptide | MGI:97531 | YES | YES | Plasma Membrane | kinase | |||||
| ENSMUSG00000006494 | Pdk1 | pyruvate dehydrogenase kinase, isoenzyme 1 | MGI:1926119 | YES | YES | Cytoplasm | kinase | |||||
| ENSMUSG00000038967 | Pdk2 | pyruvate dehydrogenase kinase, isoenzyme 2 | MGI:1343087 | YES | YES | Cytoplasm | kinase | |||||
| ENSMUSG00000028273 | Pdlim5 | PDZ and LIM domain 5 | MGI:1927489 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000006412 | Pfdn2 | prefoldin 2 | MGI:1276111 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000052033 | Pfdn4 | prefoldin 4 | MGI:1923512 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000024309 | Pfdn6 | prefoldin subunit 6 | MGI:95908 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000039047 | Pigk | phosphatidylinositol glycan anchor biosynthesis, class K | MGI:1913863 | YES | YES | Cytoplasm | peptidase | |||||
| ENSMUSG00000033628 | Pik3c3 | phosphoinositide-3-kinase, class 3 | MGI:2445019 | YES | YES | Cytoplasm | kinase | |||||
| ENSMUSG00000079480 | Pin4 | protein (peptidyl-prolyl cis/trans isomerase) NIMA-interacting, 4 (parvulin) | MGI:1916963 | YES | YES | Nucleus | enzyme | |||||
| ENSMUSG00000040430 | Pitpnc1 | phosphatidylinositol transfer protein, cytoplasmic 1 | MGI:1919045 | YES | YES | Cytoplasm | transporter | |||||
| ENSMUSG00000027499 | Pkia | protein kinase inhibitor, alpha | MGI:104747 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000060675 | Pla2g16 | phospholipase A2, group XVI | MGI:2179715 | YES | YES | Nucleus | enzyme | |||||
| ENSMUSG00000020828 | Pld2 | phospholipase D2 | MGI:892877 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000031146 | Plp2 | proteolipid protein 2 | MGI:1298382 | YES | YES | #N/A | #N/A | |||||
| ENSMUSG00000026626 | Ppp2r5a | protein phosphatase 2, regulatory subunit B', alpha | MGI:2388479 | YES | YES | Cytoplasm | phosphatase | |||||
| ENSMUSG00000019849 | Prep | prolyl endopeptidase | MGI:1270863 | YES | YES | Cytoplasm | peptidase | |||||
| ENSMUSG00000067713 | Prkag1 | protein kinase, AMP-activated, gamma 1 non-catalytic subunit | MGI:108411 | YES | YES | Nucleus | kinase | |||||
| ENSMUSG00000028944 | Prkag2 | protein kinase, AMP-activated, gamma 2 non-catalytic subunit | MGI:1336153 | YES | YES | Cytoplasm | kinase | |||||
| ENSMUSG00000023110 | Prmt5 | protein arginine N-methyltransferase 5 | MGI:1351645 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000024347 | Psd2 | pleckstrin and Sec7 domain containing 2 | MGI:1921252 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000005625 | Psmd4 | proteasome (prosome, macropain) 26S subunit, non-ATPase, 4 | MGI:1201670 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000015090 | Ptgds | prostaglandin D2 synthase (brain) | MGI:99261 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000023972 | Ptk7 | PTK7 protein tyrosine kinase 7 | MGI:1918711 | YES | YES | Plasma Membrane | kinase | |||||
| ENSMUSG00000030122 | Ptms | parathymosin | MGI:1916452 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000026395 | Ptprc | protein tyrosine phosphatase, receptor type, C | MGI:97810 | YES | YES | Plasma Membrane | phosphatase | |||||
| ENSMUSG00000022656 | Pvrl3 | poliovirus receptor-related 3 | MGI:1930171 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000020149 | Rab1 | RAB1, member RAS oncogene family | MGI:97842 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000022159 | Rab2b | RAB2B, member RAS oncogene family | MGI:1923588 | YES | YES | Plasma Membrane | enzyme | |||||
| ENSMUSG00000020732 | Rab37 | RAB37, member RAS oncogene family | MGI:1929945 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000036104 | Rab3gap1 | RAB3 GTPase activating protein subunit 1 | MGI:2445001 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000025907 | Rb1cc1 | RB1-inducible coiled-coil 1 | MGI:1341850 | YES | YES | Nucleus | other | |||||
| ENSMUSG00000022400 | Rbx1 | ring-box 1 | MGI:1891829 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000017144 | Rnd3 | Rho family GTPase 3 | MGI:1921444 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000035890 | Rnf126 | ring finger protein 126 | MGI:1917544 | YES | YES | Other | other | |||||
| ENSMUSG00000041926 | Rnpep | arginyl aminopeptidase (aminopeptidase B) | MGI:2384902 | YES | YES | Cytoplasm | peptidase | |||||
| ENSMUSG00000019864 | Rtn4ip1 | reticulon 4 interacting protein 1 | MGI:2178759 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000044080 | S100a1 | S100 calcium binding protein A1 | MGI:1338917 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000040188 | Scamp2 | secretory carrier membrane protein 2 | MGI:1346518 | YES | YES | Cytoplasm | transporter | |||||
| ENSMUSG00000057182 | Scn3a | sodium channel, voltage-gated, type III, alpha | MGI:98249 | YES | YES | Plasma Membrane | ion channel | |||||
| ENSMUSG00000074211 | Sdhaf1 | succinate dehydrogenase complex assembly factor 1 | MGI:1915582 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000058076 | Sdhc | succinate dehydrogenase complex, subunit C, integral membrane protein | MGI:1913302 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000000171 | Sdhd | succinate dehydrogenase complex, subunit D, integral membrane protein | MGI:1914175 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000035325 | Sec31a | Sec31 homolog A (S. cerevisiae) | MGI:1916412 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000078974 | Sec61g | SEC61, gamma subunit | MGI:1202066 | YES | YES | Plasma Membrane | transporter | |||||
| ENSMUSG00000030539 | Sema4b | sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4B | MGI:107559 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000001227 | Sema6b | sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6B | MGI:1202889 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000060147 | Serpinb6a | serine (or cysteine) peptidase inhibitor, clade B, member 6a | MGI:103123 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000041889 | Shisa4 | shisa family member 4 | MGI:1924802 | YES | YES | Other | other | |||||
| ENSMUSG00000018459 | Slc13a3 | solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 3 | MGI:2149635 | YES | YES | Plasma Membrane | transporter | |||||
| ENSMUSG00000020805 | Slc13a5 | solute carrier family 13 (sodium-dependent citrate transporter), member 5 | MGI:3037150 | YES | YES | Plasma Membrane | transporter | |||||
| ENSMUSG00000024935 | Slc1a1 | solute carrier family 1 (neuronal/epithelial high affinity glutamate transporter, system Xag), member 1 | MGI:105083 | YES | YES | Plasma Membrane | transporter | |||||
| ENSMUSG00000027397 | Slc20a1 | solute carrier family 20, member 1 | MGI:108392 | YES | YES | Plasma Membrane | transporter | |||||
| ENSMUSG00000041771 | Slc24a4 | solute carrier family 24 (sodium/potassium/calcium exchanger), member 4 | MGI:2447362 | YES | YES | Plasma Membrane | transporter | |||||
| ENSMUSG00000018740 | Slc25a35 | solute carrier family 25, member 35 | MGI:1919248 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000024891 | Slc29a2 | solute carrier family 29 (nucleoside transporters), member 2 | MGI:1345278 | YES | YES | Plasma Membrane | transporter | |||||
| ENSMUSG00000089774 | Slc5a3 | solute carrier family 5 (inositol transporters), member 3 | MGI:1858226 | YES | YES | Plasma Membrane | transporter | |||||
| ENSMUSG00000041313 | Slc7a1 | solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 | MGI:88117 | YES | YES | Plasma Membrane | transporter | |||||
| ENSMUSG00000022180 | Slc7a8 | solute carrier family 7 (cationic amino acid transporter, y+ system), member 8 | MGI:1355323 | YES | YES | Plasma Membrane | transporter | |||||
| ENSMUSG00000037972 | Snn | stannin | MGI:1276549 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000020018 | Snrpf | small nuclear ribonucleoprotein polypeptide F | MGI:1917128 | YES | YES | Nucleus | other | |||||
| ENSMUSG00000060429 | Sntb1 | syntrophin, basic 1 | MGI:101781 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000038291 | Snx25 | sorting nexin 25 | MGI:2142610 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000028385 | Snx30 | sorting nexin family member 30 | MGI:2443882 | YES | YES | Other | other | |||||
| ENSMUSG00000027423 | Snx5 | sorting nexin 5 | MGI:1916428 | YES | YES | Cytoplasm | transporter | |||||
| ENSMUSG00000020859 | Spag9 | sperm associated antigen 9 | MGI:1918084 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000056222 | Spock1 | sparc/osteonectin, cwcv and kazal-like domains proteoglycan 1 | MGI:105371 | YES | YES | Extracellular Space | other | |||||
| ENSMUSG00000021468 | Sptlc1 | serine palmitoyltransferase, long chain base subunit 1 | MGI:1099431 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000032042 | Srpr | signal recognition particle receptor ('docking protein') | MGI:1914648 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000021427 | Ssr1 | signal sequence receptor, alpha | MGI:105082 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000003062 | Stard3nl | STARD3 N-terminal like | MGI:1923455 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000029428 | Stx2 | syntaxin 2 | MGI:108059 | YES | YES | Cytoplasm | transporter | |||||
| ENSMUSG00000020265 | Sumo3 | SMT3 suppressor of mif two 3 homolog 3 (yeast) | MGI:1336201 | YES | YES | Nucleus | other | |||||
| ENSMUSG00000049858 | Suox | sulfite oxidase | MGI:2446117 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000020570 | Sypl | synaptophysin-like protein | MGI:108081 | YES | YES | Plasma Membrane | transporter | |||||
| ENSMUSG00000024261 | Syt4 | synaptotagmin IV | MGI:101759 | YES | YES | Cytoplasm | transporter | |||||
| ENSMUSG00000030515 | Tarsl2 | threonyl-tRNA synthetase-like 2 | MGI:2444486 | YES | YES | Other | enzyme | |||||
| ENSMUSG00000038520 | Tbc1d17 | TBC1 domain family, member 17 | MGI:2449973 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000040548 | Tex2 | testis expressed gene 2 | MGI:102465 | YES | YES | Other | other | |||||
| ENSMUSG00000062580 | Timm17a | translocase of inner mitochondrial membrane 17a | MGI:1343131 | YES | YES | Cytoplasm | transporter | |||||
| ENSMUSG00000031397 | Tktl1 | transketolase-like 1 | MGI:1933244 | YES | YES | Other | enzyme | |||||
| ENSMUSG00000039428 | Tmem135 | transmembrane protein 135 | MGI:1920009 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000043843 | Tmem145 | transmembrane protein 145 | MGI:3607779 | YES | YES | Other | other | |||||
| ENSMUSG00000025521 | Tmem192 | transmembrane protein 192 | MGI:1920317 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000040883 | Tmem205 | transmembrane protein 205 | MGI:3045495 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000031791 | Tmem38a | transmembrane protein 38A | MGI:1921416 | YES | YES | Cytoplasm | ion channel | |||||
| ENSMUSG00000035953 | Tmem55b | transmembrane protein 55b | MGI:2448501 | YES | YES | Cytoplasm | phosphatase | |||||
| ENSMUSG00000033808 | Tmem87a | transmembrane protein 87A | MGI:2441844 | YES | YES | Other | other | |||||
| ENSMUSG00000028958 | Tmub1 | transmembrane and ubiquitin-like domain containing 1 | MGI:1923764 | YES | YES | Extracellular Space | other | |||||
| ENSMUSG00000018322 | Tomm34 | translocase of outer mitochondrial membrane 34 | MGI:1914395 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000015363 | Trabd | TraB domain containing | MGI:1915226 | YES | YES | Extracellular Space | other | |||||
| ENSMUSG00000020628 | Trappc12 | trafficking protein particle complex 12 | MGI:2445089 | YES | YES | Other | other | |||||
| ENSMUSG00000051675 | Trim32 | tripartite motif-containing 32 | MGI:1917057 | YES | YES | Nucleus | transcription regulator | |||||
| ENSMUSG00000038812 | Trmt112 | tRNA methyltransferase 11-2 | MGI:1914924 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000040521 | Tsfm | Ts translation elongation factor, mitochondrial | MGI:1913649 | YES | YES | Cytoplasm | translation regulator | |||||
| ENSMUSG00000026374 | Tsn | translin | MGI:109263 | YES | YES | Nucleus | other | |||||
| ENSMUSG00000041278 | Ttc1 | tetratricopeptide repeat domain 1 | MGI:1914077 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000025474 | Tubgcp2 | tubulin, gamma complex associated protein 2 | MGI:1921487 | YES | YES | Cytoplasm | peptidase | |||||
| ENSMUSG00000075704 | Txnrd2 | thioredoxin reductase 2 | MGI:1347023 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000042345 | Ubash3a | ubiquitin associated and SH3 domain containing, A | MGI:1926074 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000032020 | Ubash3b | ubiquitin associated and SH3 domain containing, B | MGI:1920078 | YES | YES | Other | enzyme | |||||
| ENSMUSG00000028277 | Ube2j1 | ubiquitin-conjugating enzyme E2J 1 | MGI:1926245 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000005575 | Ube2m | ubiquitin-conjugating enzyme E2M | MGI:108278 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000059890 | Ube4a | ubiquitination factor E4A | MGI:2154580 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000018189 | Uchl5 | ubiquitin carboxyl-terminal esterase L5 | MGI:1914848 | YES | YES | Cytoplasm | peptidase | |||||
| ENSMUSG00000027746 | Ufm1 | ubiquitin-fold modifier 1 | MGI:1915140 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000005882 | Uqcc1 | ubiquinol-cytochrome c reductase complex assembly factor 1 | MGI:1929472 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000071654 | Uqcc3 | ubiquinol-cytochrome c reductase complex assembly factor 3 | MGI:2147553 | YES | YES | Extracellular Space | other | |||||
| ENSMUSG00000028684 | Urod | uroporphyrinogen decarboxylase | MGI:98916 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000031066 | Usp11 | ubiquitin specific peptidase 11 | MGI:2384312 | YES | YES | Nucleus | peptidase | |||||
| ENSMUSG00000047879 | Usp14 | ubiquitin specific peptidase 14 | MGI:1928898 | YES | YES | Cytoplasm | peptidase | |||||
| ENSMUSG00000073131 | Vma21 | VMA21 vacuolar H+-ATPase homolog (S. cerevisiae) | MGI:1914298 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000028753 | Vwa5b1 | von Willebrand factor A domain containing 5B1 | MGI:1922968 | YES | YES | Extracellular Space | other | |||||
| ENSMUSG00000025173 | Wdr45b | WD repeat domain 45B | MGI:1914090 | YES | YES | Other | other | |||||
| ENSMUSG00000041245 | Wnk3 | WNK lysine deficient protein kinase 3 | MGI:2652875 | YES | YES | Plasma Membrane | kinase | |||||
| ENSMUSG00000022100 | Xpo7 | exportin 7 | MGI:1929705 | YES | YES | Nucleus | transporter | |||||
| ENSMUSG00000071074 | Yipf3 | Yip1 domain family, member 3 | MGI:106280 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000024487 | Yipf5 | Yip1 domain family, member 5 | MGI:1914430 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000035798 | Zdhhc17 | zinc finger, DHHC domain containing 17 | MGI:2445110 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000021969 | Zdhhc20 | zinc finger, DHHC domain containing 20 | MGI:1923215 | YES | YES | Plasma Membrane | enzyme | |||||
| ENSMUSG00000025971 | 9430016H08Rik | RIKEN cDNA 9430016H08 gene | MGI:1915365 | YES | YES | Other | other | |||||
| ENSMUSG00000037977 | 6430571L13Rik | RIKEN cDNA 6430571L13 gene | MGI:2445137 | YES | YES | Other | other | |||||
| ENSMUSG00000045176 | 2310047M10Rik | RIKEN cDNA 2310047M10 gene | MGI:1919173 | YES | YES | Other | other | |||||
| ENSMUSG00000062376 | 2010012O05Rik | RIKEN cDNA 2010012O05 gene | MGI:1913689 | YES | YES | Other | other | |||||
| ENSMUSG00000025731 | 0610011F06Rik | RIKEN cDNA 0610011F06 gene | MGI:1915597 | YES | YES | Other | other | |||||
| ENSMUSG00000035722 | Abca7 | ATP-binding cassette, sub-family A (ABC1), member 7 | MGI:1351646 | YES | YES | Plasma Membrane | transporter | |||||
| ENSMUSG00000022822 | Abcc5 | ATP-binding cassette, sub-family C (CFTR/MRP), member 5 | MGI:1351644 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000020532 | Acaca | acetyl-Coenzyme A carboxylase alpha | MGI:108451 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000026003 | Acadl | acyl-Coenzyme A dehydrogenase, long-chain | MGI:87866 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000029033 | Acap3 | ArfGAP with coiled-coil, ankyrin repeat and PH domains 3 | MGI:2153589 | YES | YES | Nucleus | transcription regulator | |||||
| ENSMUSG00000029098 | Acox3 | acyl-Coenzyme A oxidase 3, pristanoyl | MGI:1933156 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000015016 | Acsf3 | acyl-CoA synthetase family member 3 | MGI:2182591 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000024981 | Acsl5 | acyl-CoA synthetase long-chain family member 5 | MGI:1919129 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000020654 | Adcy3 | adenylate cyclase 3 | MGI:99675 | YES | YES | Plasma Membrane | enzyme | |||||
| ENSMUSG00000026457 | Adipor1 | adiponectin receptor 1 | MGI:1919924 | YES | YES | Plasma Membrane | transmembrane receptor | |||||
| ENSMUSG00000022407 | Adsl | adenylosuccinate lyase | MGI:103202 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000033400 | Agl | amylo-1,6-glucosidase, 4-alpha-glucanotransferase | MGI:1924809 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000001211 | Agpat3 | 1-acylglycerol-3-phosphate O-acyltransferase 3 | MGI:1336186 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000042901 | Aida | axin interactor, dorsalization associated | MGI:1919737 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000028743 | Akr7a5 | aldo-keto reductase family 7, member A5 (aflatoxin aldehyde reductase) | MGI:107796 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000026687 | Aldh9a1 | aldehyde dehydrogenase 9, subfamily A1 | MGI:1861622 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000034863 | Ano8 | anoctamin 8 | MGI:2687327 | YES | YES | Plasma Membrane | ion channel | |||||
| ENSMUSG00000032590 | Apeh | acylpeptide hydrolase | MGI:88041 | YES | YES | Cytoplasm | peptidase | |||||
| ENSMUSG00000040760 | Appl1 | adaptor protein, phosphotyrosine interaction, PH domain and leucine zipper containing 1 | MGI:1920243 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000027247 | Arhgap1 | Rho GTPase activating protein 1 | MGI:2445003 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000032468 | Armc8 | armadillo repeat containing 8 | MGI:1921375 | YES | YES | Nucleus | other | |||||
| ENSMUSG00000049047 | Armcx3 | armadillo repeat containing, X-linked 3 | MGI:1918953 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000018909 | Arrb1 | arrestin, beta 1 | MGI:99473 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000024759 | Atl3 | atlastin GTPase 3 | MGI:1924270 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000027312 | Atrn | attractin | MGI:1341628 | YES | YES | Extracellular Space | other | |||||
| ENSMUSG00000002980 | Bcam | basal cell adhesion molecule | MGI:1929940 | YES | YES | Plasma Membrane | transmembrane receptor | |||||
| ENSMUSG00000003452 | Bicd1 | bicaudal D homolog 1 (Drosophila) | MGI:1101760 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000021796 | Bmpr1a | bone morphogenetic protein receptor, type 1A | MGI:1338938 | YES | YES | Plasma Membrane | kinase | |||||
| ENSMUSG00000021501 | Caml | calcium modulating ligand | MGI:104728 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000075229 | Ccdc58 | coiled-coil domain containing 58 | MGI:2146423 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000016494 | Cd34 | CD34 antigen | MGI:88329 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000060002 | Chpt1 | choline phosphotransferase 1 | MGI:2384841 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000000605 | Clcn4-2 | chloride channel 4-2 | MGI:104571 | YES | YES | Plasma Membrane | ion channel | |||||
| ENSMUSG00000096188 | Cmtm4 | CKLF-like MARVEL transmembrane domain containing 4 | MGI:2142888 | YES | YES | Extracellular Space | cytokine | |||||
| ENSMUSG00000023973 | Cnpy3 | canopy 3 homolog (zebrafish) | MGI:1919279 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000038949 | Cnst | consortin, connexin sorting protein | MGI:2445141 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000001755 | Coasy | Coenzyme A synthase | MGI:1918993 | YES | YES | Cytoplasm | kinase | |||||
| ENSMUSG00000030754 | Copb1 | coatomer protein complex, subunit beta 1 | MGI:1917599 | YES | YES | Cytoplasm | transporter | |||||
| ENSMUSG00000031827 | Cotl1 | coactosin-like 1 (Dictyostelium) | MGI:1919292 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000017778 | Cox7c | cytochrome c oxidase subunit VIIc | MGI:103226 | YES | YES | #N/A | #N/A | |||||
| ENSMUSG00000021477 | Ctsl | cathepsin L | MGI:88564 | YES | YES | Cytoplasm | peptidase | |||||
| ENSMUSG00000060534 | Dcc | deleted in colorectal carcinoma | MGI:94869 | YES | YES | Plasma Membrane | transmembrane receptor | |||||
| ENSMUSG00000048787 | Dcun1d3 | DCN1, defective in cullin neddylation 1, domain containing 3 (S. cerevisiae) | MGI:2679003 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000042404 | Dennd4b | DENN/MADD domain containing 4B | MGI:2446201 | YES | YES | Extracellular Space | other | |||||
| ENSMUSG00000022365 | Derl1 | Der1-like domain family, member 1 | MGI:1915069 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000024991 | Eif3a | eukaryotic translation initiation factor 3, subunit A | MGI:95301 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000028760 | Eif4g3 | eukaryotic translation initiation factor 4 gamma, 3 | MGI:1923935 | YES | YES | Cytoplasm | translation regulator | |||||
| ENSMUSG00000021282 | Eif5 | eukaryotic translation initiation factor 5 | MGI:95309 | YES | YES | Cytoplasm | translation regulator | |||||
| ENSMUSG00000062209 | Erbb4 | v-erb-a erythroblastic leukemia viral oncogene homolog 4 (avian) | MGI:104771 | YES | YES | Plasma Membrane | kinase | |||||
| ENSMUSG00000026858 | Fam73b | family with sequence similarity 73, member B | MGI:1922035 | YES | YES | Other | other | |||||
| ENSMUSG00000059743 | Fdps | farnesyl diphosphate synthetase | MGI:104888 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000079677 | Fdx1l | ferredoxin 1-like | MGI:1915415 | YES | YES | Cytoplasm | transporter | |||||
| ENSMUSG00000018861 | Fdxr | ferredoxin reductase | MGI:104724 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000066151 | Fkbp15 | FK506 binding protein 15 | MGI:2444782 | YES | YES | Plasma Membrane | enzyme | |||||
| ENSMUSG00000025466 | Fuom | fucose mutarotase | MGI:1916314 | YES | YES | Other | enzyme | |||||
| ENSMUSG00000029638 | Glcci1 | glucocorticoid induced transcript 1 | MGI:2179717 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000021102 | Glrx5 | glutaredoxin 5 homolog (S. cerevisiae) | MGI:1920296 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000043192 | Gm1840 | predicted gene 1840 | MGI:3037698 | YES | YES | Other | other | |||||
| ENSMUSG00000000594 | Gm2a | GM2 ganglioside activator protein | MGI:95762 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000022561 | Gpaa1 | GPI anchor attachment protein 1 | MGI:1202392 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000040111 | Gramd1b | GRAM domain containing 1B | MGI:1925037 | YES | YES | Other | other | |||||
| ENSMUSG00000038422 | Hdhd3 | haloacid dehalogenase-like hydrolase domain containing 3 | MGI:1919998 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000039414 | Heatr5b | HEAT repeat containing 5B | MGI:2444098 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000049511 | Htr1b | 5-hydroxytryptamine (serotonin) receptor 1B | MGI:96274 | YES | YES | Plasma Membrane | G-protein coupled receptor | |||||
| ENSMUSG00000025261 | Huwe1 | HECT, UBA and WWE domain containing 1 | MGI:1926884 | YES | YES | Nucleus | transcription regulator | |||||
| ENSMUSG00000005533 | Igf1r | insulin-like growth factor I receptor | MGI:96433 | YES | YES | Plasma Membrane | transmembrane receptor | |||||
| ENSMUSG00000037679 | Inf2 | inverted formin, FH2 and WH2 domain containing | MGI:1917685 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000031948 | Kars | lysyl-tRNA synthetase | MGI:1934754 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000028931 | Kcnab2 | potassium voltage-gated channel, shaker-related subfamily, beta member 2 | MGI:109239 | YES | YES | Plasma Membrane | ion channel | |||||
| ENSMUSG00000059742 | Kcnh7 | potassium voltage-gated channel, subfamily H (eag-related), member 7 | MGI:2159566 | YES | YES | Plasma Membrane | ion channel | |||||
| ENSMUSG00000043301 | Kcnj6 | potassium inwardly-rectifying channel, subfamily J, member 6 | MGI:104781 | YES | YES | Plasma Membrane | ion channel | |||||
| ENSMUSG00000033998 | Kcnk1 | potassium channel, subfamily K, member 1 | MGI:109322 | YES | YES | Plasma Membrane | ion channel | |||||
| ENSMUSG00000003731 | Kpna6 | karyopherin (importin) alpha 6 | MGI:1100836 | YES | YES | Nucleus | transporter | |||||
| ENSMUSG00000054469 | Lclat1 | lysocardiolipin acyltransferase 1 | MGI:2684937 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000030029 | Lrig1 | leucine-rich repeats and immunoglobulin-like domains 1 | MGI:107935 | YES | YES | Extracellular Space | other | |||||
| ENSMUSG00000021596 | Mctp1 | multiple C2 domains, transmembrane 1 | MGI:1926021 | YES | YES | Other | other | |||||
| ENSMUSG00000034602 | Mon2 | MON2 homolog (yeast) | MGI:1914324 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000040112 | Mrps35 | mitochondrial ribosomal protein S35 | MGI:2385255 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000064345 | mt-Nd2 | mitochondrially encoded NADH dehydrogenase 2 | MGI:102500 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000040752 | Myh6 | myosin, heavy polypeptide 6, cardiac muscle, alpha | MGI:97255 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000030869 | Ndufab1 | NADH dehydrogenase (ubiquinone) 1, alpha/beta subcomplex, 1 | MGI:1917566 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000032216 | Nedd4 | neural precursor cell expressed, developmentally down-regulated 4 | MGI:97297 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000029361 | Nos1 | nitric oxide synthase 1, neuronal | MGI:97360 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000028851 | Nudc | nuclear distribution gene C homolog (Aspergillus) | MGI:106014 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000040462 | Os9 | amplified in osteosarcoma | MGI:1924301 | YES | YES | Nucleus | other | |||||
| ENSMUSG00000024687 | Osbp | oxysterol binding protein | MGI:97447 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000040875 | Osbpl10 | oxysterol binding protein-like 10 | MGI:1921736 | YES | YES | Other | other | |||||
| ENSMUSG00000027419 | Pcsk2 | proprotein convertase subtilisin/kexin type 2 | MGI:97512 | YES | YES | Extracellular Space | peptidase | |||||
| ENSMUSG00000049225 | Pdp1 | pyruvate dehyrogenase phosphatase catalytic subunit 1 | MGI:2685870 | YES | YES | Cytoplasm | phosphatase | |||||
| ENSMUSG00000033624 | Pdpr | pyruvate dehydrogenase phosphatase regulatory subunit | MGI:2442188 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000074746 | Pdzd8 | PDZ domain containing 8 | MGI:2677270 | YES | YES | Extracellular Space | other | |||||
| ENSMUSG00000001289 | Pfdn5 | prefoldin 5 | MGI:1928753 | YES | YES | Nucleus | transcription regulator | |||||
| ENSMUSG00000031807 | Pgls | 6-phosphogluconolactonase | MGI:1913421 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000021193 | Pitrm1 | pitrilysin metallepetidase 1 | MGI:1916867 | YES | YES | Cytoplasm | peptidase | |||||
| ENSMUSG00000010660 | Plcd1 | phospholipase C, delta 1 | MGI:97614 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000042251 | Pm20d1 | peptidase M20 domain containing 1 | MGI:2442939 | YES | YES | Cytoplasm | peptidase | |||||
| ENSMUSG00000026926 | Pmpca | peptidase (mitochondrial processing) alpha | MGI:1918568 | YES | YES | Cytoplasm | peptidase | |||||
| ENSMUSG00000034064 | Poglut1 | protein O-glucosyltransferase 1 | MGI:2444232 | YES | YES | Extracellular Space | enzyme | |||||
| ENSMUSG00000021018 | Polr2h | polymerase (RNA) II (DNA directed) polypeptide H | MGI:2384309 | YES | YES | Nucleus | enzyme | |||||
| ENSMUSG00000028013 | Ppa2 | pyrophosphatase (inorganic) 2 | MGI:1922026 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000021096 | Ppm1a | protein phosphatase 1A, magnesium dependent, alpha isoform | MGI:99878 | YES | YES | Cytoplasm | phosphatase | |||||
| ENSMUSG00000033819 | Ppp1r16a | protein phosphatase 1, regulatory (inhibitor) subunit 16A | MGI:1920312 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000059409 | Ppp2r5d | protein phosphatase 2, regulatory subunit B', delta | MGI:2388481 | YES | YES | Nucleus | phosphatase | |||||
| ENSMUSG00000050697 | Prkaa1 | protein kinase, AMP-activated, alpha 1 catalytic subunit | MGI:2145955 | YES | YES | Cytoplasm | kinase | |||||
| ENSMUSG00000021178 | Psmc1 | protease (prosome, macropain) 26S subunit, ATPase 1 | MGI:106054 | YES | YES | Nucleus | peptidase | |||||
| ENSMUSG00000047250 | Ptgs1 | prostaglandin-endoperoxide synthase 1 | MGI:97797 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000026064 | Ptp4a1 | protein tyrosine phosphatase 4a1 | MGI:1277096 | YES | YES | Cytoplasm | phosphatase | |||||
| ENSMUSG00000027540 | Ptpn1 | protein tyrosine phosphatase, non-receptor type 1 | MGI:97805 | YES | YES | Cytoplasm | phosphatase | |||||
| ENSMUSG00000032290 | Ptpn9 | protein tyrosine phosphatase, non-receptor type 9 | MGI:1928376 | YES | YES | Cytoplasm | phosphatase | |||||
| ENSMUSG00000015087 | Rabl6 | RAB, member RAS oncogene family-like 6 | MGI:2442633 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000044024 | Rell2 | RELT-like 2 | MGI:1918044 | YES | YES | Extracellular Space | other | |||||
| ENSMUSG00000029048 | Rer1 | RER1 retention in endoplasmic reticulum 1 homolog (S. cerevisiae) | MGI:1915080 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000038650 | Rnh1 | ribonuclease/angiogenin inhibitor 1 | MGI:1195456 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000046480 | Scn4b | sodium channel, type IV, beta | MGI:2687406 | YES | YES | Plasma Membrane | ion channel | |||||
| ENSMUSG00000069844 | Sco1 | SCO cytochrome oxidase deficient homolog 1 (yeast) | MGI:106362 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000019802 | Sec63 | SEC63-like (S. cerevisiae) | MGI:2155302 | YES | YES | Cytoplasm | transporter | |||||
| ENSMUSG00000028064 | Sema4a | sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A | MGI:107560 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000058207 | Serpina3k | serine (or cysteine) peptidase inhibitor, clade A, member 3K | MGI:98377 | YES | YES | Extracellular Space | other | |||||
| ENSMUSG00000022494 | Shisa9 | shisa family member 9 | MGI:1919805 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000029219 | Slc10a4 | solute carrier family 10 (sodium/bile acid cotransporter family), member 4 | MGI:3606480 | YES | YES | Plasma Membrane | transporter | |||||
| ENSMUSG00000022199 | Slc22a17 | solute carrier family 22 (organic cation transporter), member 17 | MGI:1926225 | YES | YES | Plasma Membrane | transporter | |||||
| ENSMUSG00000031105 | Slc25a14 | solute carrier family 25 (mitochondrial carrier, brain), member 14 | MGI:1330823 | YES | YES | Cytoplasm | transporter | |||||
| ENSMUSG00000023942 | Slc29a1 | solute carrier family 29 (nucleoside transporters), member 1 | MGI:1927073 | YES | YES | Plasma Membrane | transporter | |||||
| ENSMUSG00000033272 | Slc35a4 | solute carrier family 35, member A4 | MGI:1915093 | YES | YES | Other | transporter | |||||
| ENSMUSG00000038602 | Slc35f1 | solute carrier family 35, member F1 | MGI:2139810 | YES | YES | Other | other | |||||
| ENSMUSG00000070287 | Slc35g2 | solute carrier family 35, member G2 | MGI:2685365 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000039838 | Slc45a1 | solute carrier family 45, member 1 | MGI:2653235 | YES | YES | Other | transporter | |||||
| ENSMUSG00000020838 | Slc6a4 | solute carrier family 6 (neurotransmitter transporter, serotonin), member 4 | MGI:96285 | YES | YES | Plasma Membrane | transporter | |||||
| ENSMUSG00000031596 | Slc7a2 | solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 | MGI:99828 | YES | YES | Plasma Membrane | transporter | |||||
| ENSMUSG00000031904 | Slc7a6 | solute carrier family 7 (cationic amino acid transporter, y+ system), member 6 | MGI:2142598 | YES | YES | Plasma Membrane | transporter | |||||
| ENSMUSG00000079055 | Slc8a3 | solute carrier family 8 (sodium/calcium exchanger), member 3 | MGI:107976 | YES | YES | Plasma Membrane | transporter | |||||
| ENSMUSG00000025020 | Slit1 | slit homolog 1 (Drosophila) | MGI:1315203 | YES | YES | Extracellular Space | other | |||||
| ENSMUSG00000075478 | Slitrk1 | SLIT and NTRK-like family, member 1 | MGI:2679446 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000046699 | Slitrk4 | SLIT and NTRK-like family, member 4 | MGI:2442509 | YES | YES | Extracellular Space | other | |||||
| ENSMUSG00000001018 | Snapin | SNAP-associated protein | MGI:1333745 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000023064 | Sncg | synuclein, gamma | MGI:1298397 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000000738 | Spg7 | spastic paraplegia 7 homolog (human) | MGI:2385906 | YES | YES | Cytoplasm | peptidase | |||||
| ENSMUSG00000027500 | Stmn2 | stathmin-like 2 | MGI:98241 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000014601 | Strip1 | striatin interacting protein 1 | MGI:2443884 | YES | YES | Nucleus | other | |||||
| ENSMUSG00000030126 | Tmcc1 | transmembrane and coiled coil domains 1 | MGI:2442368 | YES | YES | Other | other | |||||
| ENSMUSG00000004394 | Tmed4 | transmembrane emp24 protein transport domain containing 4 | MGI:1915070 | YES | YES | Cytoplasm | transporter | |||||
| ENSMUSG00000020701 | Tmem132e | transmembrane protein 132E | MGI:2685490 | YES | YES | Other | other | |||||
| ENSMUSG00000029234 | Tmem165 | transmembrane protein 165 | MGI:894407 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000013858 | Tmem259 | transmembrane protein 259 | MGI:2177957 | YES | YES | Other | other | |||||
| ENSMUSG00000034145 | Tmem63c | transmembrane protein 63c | MGI:2444386 | YES | YES | Other | ion channel | |||||
| ENSMUSG00000041763 | Tpp2 | tripeptidyl peptidase II | MGI:102724 | YES | YES | Cytoplasm | peptidase | |||||
| ENSMUSG00000061808 | Ttr | transthyretin | MGI:98865 | YES | YES | Extracellular Space | transporter | |||||
| ENSMUSG00000028367 | Txn1 | thioredoxin 1 | MGI:98874 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000025326 | Ube3a | ubiquitin protein ligase E3A | MGI:105098 | YES | YES | Nucleus | enzyme | |||||
| ENSMUSG00000019951 | Uhrf1bp1l | UHRF1 (ICBP90) binding protein 1-like | MGI:2442888 | YES | YES | Other | other | |||||
| ENSMUSG00000029407 | Uso1 | USO1 vesicle docking factor | MGI:1929095 | YES | YES | Cytoplasm | transporter | |||||
| ENSMUSG00000022710 | Usp7 | ubiquitin specific peptidase 7 | MGI:2182061 | YES | YES | Nucleus | peptidase | |||||
| ENSMUSG00000045210 | Vcpip1 | valosin containing protein (p97)/p47 complex interacting protein 1 | MGI:1917925 | YES | YES | Cytoplasm | peptidase | |||||
| ENSMUSG00000024924 | Vldlr | very low density lipoprotein receptor | MGI:98935 | YES | YES | Plasma Membrane | transporter | |||||
| ENSMUSG00000034341 | Wbp2 | WW domain binding protein 2 | MGI:104709 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000045962 | Wnk1 | WNK lysine deficient protein kinase 1 | MGI:2442092 | YES | YES | Cytoplasm | kinase | |||||
| ENSMUSG00000025027 | Xpnpep1 | X-prolyl aminopeptidase (aminopeptidase P) 1, soluble | MGI:2180003 | YES | YES | Cytoplasm | peptidase | |||||
| ENSMUSG00000020290 | Xpo1 | exportin 1, CRM1 homolog (yeast) | MGI:2144013 | YES | YES | Nucleus | transporter | |||||
| ENSMUSG00000026469 | Xpr1 | xenotropic and polytropic retrovirus receptor 1 | MGI:97932 | YES | YES | Plasma Membrane | G-protein coupled receptor | |||||
| ENSMUSG00000056004 | 9330182L06Rik | RIKEN cDNA 9330182L06 gene | MGI:2443264 | YES | YES | Other | other | |||||
| ENSMUSG00000029095 | Ablim2 | actin-binding LIM protein 2 | MGI:2385758 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000025475 | Adgra1 | adhesion G protein-coupled receptor A1 | MGI:1277167 | YES | YES | Plasma Membrane | G-protein coupled receptor | |||||
| ENSMUSG00000039546 | Ajap1 | adherens junction associated protein 1 | MGI:2685419 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000022602 | Arc | activity regulated cytoskeletal-associated protein | MGI:88067 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000019467 | Arhgef25 | Rho guanine nucleotide exchange factor (GEF) 25 | MGI:1277173 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000025656 | Arhgef9 | CDC42 guanine nucleotide exchange factor (GEF) 9 | MGI:2442233 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000057897 | Camk2b | calcium/calmodulin-dependent protein kinase II, beta | MGI:88257 | YES | YES | Cytoplasm | kinase | |||||
| ENSMUSG00000037979 | Ccdc92 | coiled-coil domain containing 92 | MGI:106485 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000031971 | Ccsap | centriole, cilia and spindle associated protein | MGI:1920670 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000000305 | Cdh4 | cadherin 4 | MGI:99218 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000078695 | Cisd3 | CDGSH iron sulfur domain 3 | MGI:101788 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000039652 | Cpeb3 | cytoplasmic polyadenylation element binding protein 3 | MGI:2443075 | YES | YES | Cytoplasm | translation regulator | |||||
| ENSMUSG00000030270 | Cpne9 | copine family member IX | MGI:2443052 | YES | YES | Extracellular Space | other | |||||
| ENSMUSG00000060843 | Ctnna3 | catenin (cadherin associated protein), alpha 3 | MGI:2661445 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000036712 | Cyld | cylindromatosis (turban tumor syndrome) | MGI:1921506 | YES | YES | Nucleus | transcription regulator | |||||
| ENSMUSG00000017132 | Cyth1 | cytohesin 1 | MGI:1334257 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000027797 | Dclk1 | doublecortin-like kinase 1 | MGI:1330861 | YES | YES | Other | kinase | |||||
| ENSMUSG00000000881 | Dlg3 | discs, large homolog 3 (Drosophila) | MGI:1888986 | YES | YES | Plasma Membrane | kinase | |||||
| ENSMUSG00000036052 | Dnajb5 | DnaJ (Hsp40) homolog, subfamily B, member 5 | MGI:1930018 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000021131 | Erh | enhancer of rudimentary homolog (Drosophila) | MGI:108089 | YES | YES | Nucleus | other | |||||
| ENSMUSG00000021750 | Fam107a | family with sequence similarity 107, member A | MGI:3041256 | YES | YES | Nucleus | other | |||||
| ENSMUSG00000049119 | Fam110b | family with sequence similarity 110, member B | MGI:1916593 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000073805 | Fam196a | family with sequence similarity 196, member A | MGI:3605068 | YES | YES | Other | other | |||||
| ENSMUSG00000000531 | Grasp | GRP1 (general receptor for phosphoinositides 1)-associated scaffold protein | MGI:1860303 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000010825 | Grid2ip | glutamate receptor, ionotropic, delta 2 (Grid2) interacting protein 1 | MGI:2176213 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000030098 | Grip2 | glutamate receptor interacting protein 2 | MGI:2681173 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000078249 | Hmga1-rs1 | high mobility group AT-hook I, related sequence 1 | MGI:96161 | YES | YES | Nucleus | transcription regulator | |||||
| ENSMUSG00000034275 | Igsf9b | immunoglobulin superfamily, member 9B | MGI:2685354 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000001016 | Ilf2 | interleukin enhancer binding factor 2 | MGI:1915031 | YES | YES | Nucleus | transcription regulator | |||||
| ENSMUSG00000033960 | 9430020K01Rik | RIKEN cDNA 9430020K01 gene | MGI:2685174 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000044216 | Kcnj4 | potassium inwardly-rectifying channel, subfamily J, member 4 | MGI:104743 | YES | YES | Plasma Membrane | ion channel | |||||
| ENSMUSG00000046417 | Lrrc75a | leucine rich repeat containing 75A | MGI:2682293 | YES | YES | Other | other | |||||
| ENSMUSG00000052581 | Lrrtm4 | leucine rich repeat transmembrane neuronal 4 | MGI:2389180 | YES | YES | Extracellular Space | other | |||||
| ENSMUSG00000018819 | Lsp1 | lymphocyte specific 1 | MGI:96832 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000001089 | Luzp1 | leucine zipper protein 1 | MGI:107629 | YES | YES | Nucleus | other | |||||
| ENSMUSG00000033900 | Map9 | microtubule-associated protein 9 | MGI:2442208 | YES | YES | Other | other | |||||
| ENSMUSG00000053693 | Mast1 | microtubule associated serine/threonine kinase 1 | MGI:1861901 | YES | YES | Cytoplasm | kinase | |||||
| ENSMUSG00000043557 | Mdga1 | MAM domain containing glycosylphosphatidylinositol anchor 1 | MGI:1922012 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000029436 | Mmp17 | matrix metallopeptidase 17 | MGI:1346076 | YES | YES | Extracellular Space | peptidase | |||||
| ENSMUSG00000021339 | Mrs2 | MRS2 magnesium homeostasis factor homolog (S. cerevisiae) | MGI:2685748 | YES | YES | Cytoplasm | transporter | |||||
| ENSMUSG00000061086 | Myl4 | myosin, light polypeptide 4 | MGI:97267 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000024300 | Myo1f | myosin IF | MGI:107711 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000027233 | Patl2 | protein associated with topoisomerase II homolog 2 (yeast) | MGI:1914828 | YES | YES | Cytoplasm | translation regulator | |||||
| ENSMUSG00000035383 | Pmch | pro-melanin-concentrating hormone | MGI:97629 | YES | YES | Extracellular Space | other | |||||
| ENSMUSG00000040105 | Ppapdc2 | phosphatidic acid phosphatase type 2 domain containing 2 | MGI:1921661 | YES | YES | Other | phosphatase | |||||
| ENSMUSG00000002731 | Prkra | protein kinase, interferon inducible double stranded RNA dependent activator | MGI:1344375 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000023484 | Prph | peripherin | MGI:97774 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000034686 | Prr7 | proline rich 7 (synaptic) | MGI:3487246 | YES | YES | Other | other | |||||
| ENSMUSG00000006456 | Rbm14 | RNA binding motif protein 14 | MGI:1929092 | YES | YES | Nucleus | transcription regulator | |||||
| ENSMUSG00000031134 | Rbmx | RNA binding motif protein, X chromosome | MGI:1343044 | YES | YES | Nucleus | other | |||||
| ENSMUSG00000048758 | Rpl29 | ribosomal protein L29 | MGI:99687 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000049517 | Rps23 | ribosomal protein S23 | MGI:1913725 | YES | YES | Cytoplasm | translation regulator | |||||
| ENSMUSG00000036781 | Rps27l | ribosomal protein S27-like | MGI:1915191 | YES | YES | Cytoplasm | translation regulator | |||||
| ENSMUSG00000036371 | Serbp1 | serpine1 mRNA binding protein 1 | MGI:1914120 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000022623 | Shank3 | SH3/ankyrin domain gene 3 | MGI:1930016 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000024068 | Spast | spastin | MGI:1858896 | YES | YES | Nucleus | enzyme | |||||
| ENSMUSG00000004366 | Sst | somatostatin | MGI:98326 | YES | YES | Extracellular Space | other | |||||
| ENSMUSG00000020115 | Tbk1 | TANK-binding kinase 1 | MGI:1929658 | YES | YES | Cytoplasm | kinase | |||||
| ENSMUSG00000038517 | Tbkbp1 | TBK1 binding protein 1 | MGI:1920424 | YES | YES | Other | other | |||||
| ENSMUSG00000084845 | Tmem240 | transmembrane protein 240 | MGI:3648074 | YES | YES | Other | other | |||||
| ENSMUSG00000019961 | Tmpo | thymopoietin | MGI:106920 | YES | YES | Nucleus | other | |||||
| ENSMUSG00000000296 | Tpd52l1 | tumor protein D52-like 1 | MGI:1298386 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000026556 | Vangl2 | vang-like 2 (van gogh, Drosophila) | MGI:2135272 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000031913 | Vps4a | vacuolar protein sorting 4a (yeast) | MGI:1890520 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000031930 | Wwp2 | WW domain containing E3 ubiquitin protein ligase 2 | MGI:1914144 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000022401 | Xpnpep3 | X-prolyl aminopeptidase (aminopeptidase P) 3, putative | MGI:2445217 | YES | YES | Cytoplasm | peptidase | |||||
| ENSMUSG00000028639 | Ybx1 | Y box protein 1 | MGI:99146 | YES | YES | Nucleus | transcription regulator | |||||
| ENSMUSG00000060166 | Zdhhc8 | zinc finger, DHHC domain containing 8 | MGI:1338012 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000041530 | Ago1 | argonaute RISC catalytic subunit 1 | MGI:2446630 | YES | YES | Cytoplasm | translation regulator | |||||
| ENSMUSG00000045411 | 2410002F23Rik | RIKEN cDNA 2410002F23 gene | MGI:1914226 | YES | YES | Other | other | |||||
| ENSMUSG00000036046 | 5031439G07Rik | RIKEN cDNA 5031439G07 gene | MGI:2444899 | YES | YES | Other | other | |||||
| ENSMUSG00000029106 | Add1 | adducin 1 (alpha) | MGI:87918 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000078139 | AK157302 | cDNA sequence AK157302 | MGI:3574096 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000021866 | Anxa11 | annexin A11 | MGI:108481 | YES | YES | Nucleus | other | |||||
| ENSMUSG00000021824 | Ap3m1 | adaptor-related protein complex 3, mu 1 subunit | MGI:1929212 | YES | YES | Cytoplasm | transporter | |||||
| ENSMUSG00000063801 | Ap3s2 | adaptor-related protein complex 3, sigma 2 subunit | MGI:1337060 | YES | YES | Cytoplasm | transporter | |||||
| ENSMUSG00000032081 | Apoc3 | apolipoprotein C-III | MGI:88055 | YES | YES | Extracellular Space | transporter | |||||
| ENSMUSG00000001127 | Araf | v-raf murine sarcoma 3611 viral oncogene homolog | MGI:88065 | YES | YES | Cytoplasm | kinase | |||||
| ENSMUSG00000054277 | Arfgap3 | ADP-ribosylation factor GTPase activating protein 3 | MGI:1913501 | YES | YES | Cytoplasm | transporter | |||||
| ENSMUSG00000022799 | Arhgap31 | Rho GTPase activating protein 31 | MGI:1333857 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000023017 | Asic1 | acid-sensing (proton-gated) ion channel 1 | MGI:1194915 | YES | YES | Plasma Membrane | ion channel | |||||
| ENSMUSG00000047507 | Baiap3 | BAI1-associated protein 3 | MGI:2685783 | YES | YES | Extracellular Space | other | |||||
| ENSMUSG00000006464 | Bbs1 | Bardet-Biedl syndrome 1 (human) | MGI:1277215 | YES | YES | Extracellular Space | other | |||||
| ENSMUSG00000036896 | C1qc | complement component 1, q subcomponent, C chain | MGI:88225 | YES | YES | Extracellular Space | other | |||||
| ENSMUSG00000036377 | C530008M17Rik | RIKEN cDNA C530008M17 gene | MGI:2444817 | YES | YES | Other | other | |||||
| ENSMUSG00000053819 | Camk2d | calcium/calmodulin-dependent protein kinase II, delta | MGI:1341265 | YES | YES | Cytoplasm | kinase | |||||
| ENSMUSG00000029471 | Camkk2 | calcium/calmodulin-dependent protein kinase kinase 2, beta | MGI:2444812 | YES | YES | Cytoplasm | kinase | |||||
| ENSMUSG00000034471 | Caskin2 | CASK-interacting protein 2 | MGI:2157062 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000040380 | Cbln3 | cerebellin 3 precursor protein | MGI:1889286 | YES | YES | Extracellular Space | other | |||||
| ENSMUSG00000022428 | Cby1 | chibby homolog 1 (Drosophila) | MGI:1920989 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000031150 | Ccdc120 | coiled-coil domain containing 120 | MGI:1859619 | YES | YES | Other | other | |||||
| ENSMUSG00000041609 | Ccdc64 | coiled-coil domain containing 64 | MGI:1922915 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000032878 | Ccdc85a | coiled-coil domain containing 85A | MGI:2445069 | YES | YES | Other | other | |||||
| ENSMUSG00000000378 | Ccm2 | cerebral cavernous malformation 2 | MGI:2384924 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000023932 | Cdc5l | cell division cycle 5-like (S. pombe) | MGI:1918952 | YES | YES | Nucleus | other | |||||
| ENSMUSG00000050840 | Cdh20 | cadherin 20 | MGI:1346069 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000027350 | Chgb | chromogranin B | MGI:88395 | YES | YES | Extracellular Space | other | |||||
| ENSMUSG00000036550 | Cnot1 | CCR4-NOT transcription complex, subunit 1 | MGI:2442402 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000036103 | Colec12 | collectin sub-family member 12 | MGI:2152907 | YES | YES | Plasma Membrane | transmembrane receptor | |||||
| ENSMUSG00000034807 | Colgalt1 | collagen beta(1-O)galactosyltransferase 1 | MGI:1924348 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000023020 | Cox14 | cytochrome c oxidase assembly protein 14 | MGI:1913629 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000032564 | Cpne4 | copine IV | MGI:1921270 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000034820 | Cpsf7 | cleavage and polyadenylation specific factor 7 | MGI:1917826 | YES | YES | Nucleus | other | |||||
| ENSMUSG00000062127 | Cttnbp2nl | CTTNBP2 N-terminal like | MGI:1933137 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000022150 | Dab2 | disabled 2, mitogen-responsive phosphoprotein | MGI:109175 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000034974 | Dapk3 | death-associated protein kinase 3 | MGI:1203520 | YES | YES | Cytoplasm | kinase | |||||
| ENSMUSG00000053289 | Ddx10 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 10 | MGI:1924841 | YES | YES | Nucleus | enzyme | |||||
| ENSMUSG00000040620 | Dhx33 | DEAH (Asp-Glu-Ala-His) box polypeptide 33 | MGI:2445102 | YES | YES | Nucleus | enzyme | |||||
| ENSMUSG00000044447 | Dock5 | dedicator of cytokinesis 5 | MGI:2652871 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000005610 | Eif4g2 | eukaryotic translation initiation factor 4, gamma 2 | MGI:109207 | YES | YES | Cytoplasm | translation regulator | |||||
| ENSMUSG00000003410 | Elavl3 | ELAV (embryonic lethal, abnormal vision, Drosophila)-like 3 (Hu antigen C) | MGI:109157 | YES | YES | Nucleus | other | |||||
| ENSMUSG00000023960 | Enpp5 | ectonucleotide pyrophosphatase/phosphodiesterase 5 | MGI:1933830 | YES | YES | Extracellular Space | enzyme | |||||
| ENSMUSG00000009079 | Ewsr1 | Ewing sarcoma breakpoint region 1 | MGI:99960 | YES | YES | Nucleus | other | |||||
| ENSMUSG00000027963 | Extl2 | exostoses (multiple)-like 2 | MGI:1889574 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000041685 | Fcho2 | FCH domain only 2 | MGI:3505790 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000028001 | Fga | fibrinogen alpha chain | MGI:1316726 | YES | YES | Extracellular Space | other | |||||
| ENSMUSG00000033860 | Fgg | fibrinogen gamma chain | MGI:95526 | YES | YES | Extracellular Space | other | |||||
| ENSMUSG00000051379 | Flrt3 | fibronectin leucine rich transmembrane protein 3 | MGI:1918686 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000030795 | Fus | fused in sarcoma | MGI:1353633 | YES | YES | Nucleus | transcription regulator | |||||
| ENSMUSG00000018583 | G3bp1 | GTPase activating protein (SH3 domain) binding protein 1 | MGI:1351465 | YES | YES | Nucleus | enzyme | |||||
| ENSMUSG00000030161 | Gabarapl1 | gamma-aminobutyric acid (GABA) A receptor-associated protein-like 1 | MGI:1914980 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000033751 | Gadd45gip1 | growth arrest and DNA-damage-inducible, gamma interacting protein 1 | MGI:1914947 | YES | YES | Nucleus | other | |||||
| ENSMUSG00000025858 | Get4 | golgi to ER traffic protein 4 homolog (S. cerevisiae) | MGI:1914854 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000027573 | Gid8 | GID complex subunit 8 homolog (S. cerevisiae) | MGI:1923675 | YES | YES | Nucleus | other | |||||
| ENSMUSG00000021552 | Gkap1 | G kinase anchoring protein 1 | MGI:1891694 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000050490 | Gm8394 | predicted gene 8394 | MGI:3647964 | YES | YES | Other | other | |||||
| ENSMUSG00000003378 | Grik5 | glutamate receptor, ionotropic, kainate 5 (gamma 2) | MGI:95818 | YES | YES | Plasma Membrane | ion channel | |||||
| ENSMUSG00000026959 | Grin1 | glutamate receptor, ionotropic, NMDA1 (zeta 1) | MGI:95819 | YES | YES | Plasma Membrane | ion channel | |||||
| ENSMUSG00000046182 | Gsg1l | GSG1-like | MGI:2685483 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000006930 | Hap1 | huntingtin-associated protein 1 | MGI:1261831 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000060373 | Hnrnpc | heterogeneous nuclear ribonucleoprotein C | MGI:107795 | YES | YES | Nucleus | other | |||||
| ENSMUSG00000028759 | Hp1bp3 | heterochromatin protein 1, binding protein 3 | MGI:109369 | YES | YES | Nucleus | other | |||||
| ENSMUSG00000020279 | Il9r | interleukin 9 receptor | MGI:96564 | YES | YES | Plasma Membrane | transmembrane receptor | |||||
| ENSMUSG00000024187 | Itfg3 | integrin alpha FG-GAP repeat containing 3 | MGI:2146854 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000053519 | Kcnip1 | Kv channel-interacting protein 1 | MGI:1917607 | YES | YES | Plasma Membrane | ion channel | |||||
| ENSMUSG00000003779 | Kif20a | kinesin family member 20A | MGI:1201682 | YES | YES | Cytoplasm | transporter | |||||
| ENSMUSG00000000600 | Krit1 | KRIT1, ankyrin repeat containing | MGI:1930618 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000020585 | Laptm4a | lysosomal-associated protein transmembrane 4A | MGI:108017 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000037353 | Letmd1 | LETM1 domain containing 1 | MGI:1915864 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000042873 | Lhfpl4 | lipoma HMGIC fusion partner-like protein 4 | MGI:3057108 | YES | YES | Other | other | |||||
| ENSMUSG00000062075 | Lmnb2 | lamin B2 | MGI:96796 | YES | YES | Nucleus | other | |||||
| ENSMUSG00000040563 | Lppr2 | lipid phosphate phosphatase-related protein type 2 | MGI:2384575 | YES | YES | Other | phosphatase | |||||
| ENSMUSG00000022801 | Lrch3 | leucine-rich repeats and calponin homology (CH) domain containing 3 | MGI:1917394 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000042846 | Lrrtm3 | leucine rich repeat transmembrane neuronal 3 | MGI:2389177 | YES | YES | Other | other | |||||
| ENSMUSG00000055003 | Lrtm2 | leucine-rich repeats and transmembrane domains 2 | MGI:2141485 | YES | YES | Other | other | |||||
| ENSMUSG00000043807 | Ly6g5b | lymphocyte antigen 6 complex, locus G5B | MGI:2385809 | YES | YES | Other | other | |||||
| ENSMUSG00000034751 | Mast4 | microtubule associated serine/threonine kinase family member 4 | MGI:1918885 | YES | YES | Other | kinase | |||||
| ENSMUSG00000018076 | Med13l | mediator complex subunit 13-like | MGI:2670178 | YES | YES | Nucleus | other | |||||
| ENSMUSG00000070462 | Mesdc1 | mesoderm development candidate 1 | MGI:1891420 | YES | YES | Other | other | |||||
| ENSMUSG00000032517 | Mobp | myelin-associated oligodendrocytic basic protein | MGI:108511 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000044018 | Mrpl50 | mitochondrial ribosomal protein L50 | MGI:107329 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000049230 | Gm9833 | predicted gene 9833 | MGI:3641855 | YES | YES | Other | other | |||||
| ENSMUSG00000035285 | Nat14 | N-acetyltransferase 14 | MGI:3039561 | YES | YES | Extracellular Space | other | |||||
| ENSMUSG00000050321 | Neto1 | neuropilin (NRP) and tolloid (TLL)-like 1 | MGI:2180216 | YES | YES | Extracellular Space | other | |||||
| ENSMUSG00000036902 | Neto2 | neuropilin (NRP) and tolloid (TLL)-like 2 | MGI:1921763 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000009073 | Nf2 | neurofibromatosis 2 | MGI:97307 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000032606 | Nicn1 | nicolin 1 | MGI:1913507 | YES | YES | Nucleus | other | |||||
| ENSMUSG00000078532 | Nkain1 | Na+/K+ transporting ATPase interacting 1 | MGI:1914399 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000056209 | Npm3 | nucleoplasmin 3 | MGI:894653 | YES | YES | Nucleus | other | |||||
| ENSMUSG00000044287 | Nrn1l | neuritin 1-like | MGI:2443642 | YES | YES | Other | other | |||||
| ENSMUSG00000036992 | Nxt1 | NTF2-related export protein 1 | MGI:1929619 | YES | YES | Nucleus | transporter | |||||
| ENSMUSG00000045348 | Nyap1 | neuronal tyrosine-phosphorylated phosphoinositide 3-kinase adaptor 1 | MGI:2443880 | YES | YES | Other | other | |||||
| ENSMUSG00000031592 | Pcm1 | pericentriolar material 1 | MGI:1277958 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000021140 | Pcnx | pecanex homolog (Drosophila) | MGI:1891924 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000054874 | Pcnxl3 | pecanex-like 3 (Drosophila) | MGI:1861733 | YES | YES | Other | other | |||||
| ENSMUSG00000025137 | Pcyt2 | phosphate cytidylyltransferase 2, ethanolamine | MGI:1915921 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000002006 | Pdzd4 | PDZ domain containing 4 | MGI:2443483 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000074305 | Peak1 | pseudopodium-enriched atypical kinase 1 | MGI:2442366 | YES | YES | Plasma Membrane | kinase | |||||
| ENSMUSG00000042275 | Pelo | pelota homolog (Drosophila) | MGI:2145154 | YES | YES | Nucleus | other | |||||
| ENSMUSG00000035395 | Pet2 | plasmacytoma expressed transcript 2 | MGI:101758 | YES | YES | Other | other | |||||
| ENSMUSG00000041417 | Pik3r1 | phosphatidylinositol 3-kinase, regulatory subunit, polypeptide 1 (p85 alpha) | MGI:97583 | YES | YES | Cytoplasm | kinase | |||||
| ENSMUSG00000028126 | Pip5k1a | phosphatidylinositol-4-phosphate 5-kinase, type 1 alpha | MGI:107929 | YES | YES | Cytoplasm | kinase | |||||
| ENSMUSG00000026179 | Pnkd | paroxysmal nonkinesiogenic dyskinesia | MGI:1930773 | YES | YES | Nucleus | other | |||||
| ENSMUSG00000045731 | Pnoc | prepronociceptin | MGI:105308 | YES | YES | Extracellular Space | other | |||||
| ENSMUSG00000026181 | Ppm1f | protein phosphatase 1F (PP2C domain containing) | MGI:1918464 | YES | YES | Cytoplasm | phosphatase | |||||
| ENSMUSG00000073557 | Ppp1r12b | protein phosphatase 1, regulatory (inhibitor) subunit 12B | MGI:1916417 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000040472 | Rabggta | Rab geranylgeranyl transferase, a subunit | MGI:1860443 | YES | YES | Extracellular Space | enzyme | |||||
| ENSMUSG00000022246 | Rai14 | retinoic acid induced 14 | MGI:1922896 | YES | YES | Nucleus | transcription regulator | |||||
| ENSMUSG00000029602 | Rasal1 | RAS protein activator like 1 (GAP1 like) | MGI:1330842 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000030134 | Rasgef1a | RasGEF domain family, member 1A | MGI:1917977 | YES | YES | Other | other | |||||
| ENSMUSG00000027347 | Rasgrp1 | RAS guanyl releasing protein 1 | MGI:1314635 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000031167 | Rbm3 | RNA binding motif protein 3 | MGI:1099460 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000022176 | Rem2 | rad and gem related GTP binding protein 2 | MGI:2155260 | YES | YES | Plasma Membrane | enzyme | |||||
| ENSMUSG00000002458 | Rgs19 | regulator of G-protein signaling 19 | MGI:1915153 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000035226 | Rims4 | regulating synaptic membrane exocytosis 4 | MGI:2674366 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000050357 | Rltpr | RGD motif, leucine rich repeats, tropomodulin domain and proline-rich containing | MGI:2685431 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000010086 | Rnf112 | ring finger protein 112 | MGI:106611 | YES | YES | Nucleus | transcription regulator | |||||
| ENSMUSG00000042790 | Rnf214 | ring finger protein 214 | MGI:2444451 | YES | YES | Other | other | |||||
| ENSMUSG00000059291 | Rpl11 | ribosomal protein L11 | MGI:1914275 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000060636 | Rpl35a | ribosomal protein L35A | MGI:1928894 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000003644 | Rps6ka1 | ribosomal protein S6 kinase polypeptide 1 | MGI:104558 | YES | YES | Cytoplasm | kinase | |||||
| ENSMUSG00000027429 | Sec23b | SEC23B (S. cerevisiae) | MGI:1350925 | YES | YES | Extracellular Space | transporter | |||||
| ENSMUSG00000004631 | Sgce | sarcoglycan, epsilon | MGI:1329042 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000001995 | Sipa1l2 | signal-induced proliferation-associated 1 like 2 | MGI:2676970 | YES | YES | Other | other | |||||
| ENSMUSG00000041133 | Smc1a | structural maintenance of chromosomes 1A | MGI:1344345 | YES | YES | Nucleus | transporter | |||||
| ENSMUSG00000021039 | Snw1 | SNW domain containing 1 | MGI:1913604 | YES | YES | Nucleus | transcription regulator | |||||
| ENSMUSG00000031626 | Sorbs2 | sorbin and SH3 domain containing 2 | MGI:1924574 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000044352 | Sowaha | sosondowah ankyrin repeat domain family member A | MGI:2687280 | YES | YES | Other | other | |||||
| ENSMUSG00000033594 | Spata2l | spermatogenesis associated 2-like | MGI:1926029 | YES | YES | Other | other | |||||
| ENSMUSG00000021917 | Spcs1 | signal peptidase complex subunit 1 homolog (S. cerevisiae) | MGI:1916269 | YES | YES | Cytoplasm | peptidase | |||||
| ENSMUSG00000079108 | Srp54b | signal recognition particle 54B | MGI:3714357 | YES | YES | #N/A | #N/A | |||||
| ENSMUSG00000054453 | Sytl5 | synaptotagmin-like 5 | MGI:2668451 | YES | YES | Other | other | |||||
| ENSMUSG00000034412 | Tbc1d10a | TBC1 domain family, member 10a | MGI:2144164 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000038079 | Tmem237 | transmembrane protein 237 | MGI:2138365 | YES | YES | Other | other | |||||
| ENSMUSG00000028826 | Tmem57 | transmembrane protein 57 | MGI:1913396 | YES | YES | Nucleus | other | |||||
| ENSMUSG00000014353 | Tmem87b | transmembrane protein 87B | MGI:1919727 | YES | YES | Other | other | |||||
| ENSMUSG00000049775 | Tmsb4x | thymosin, beta 4, X chromosome | MGI:99510 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000005674 | Tomm40l | translocase of outer mitochondrial membrane 40 homolog-like (yeast) | MGI:3589112 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000078713 | Tomm5 | translocase of outer mitochondrial membrane 5 homolog (yeast) | MGI:1915762 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000033475 | Tomm6 | translocase of outer mitochondrial membrane 6 homolog (yeast) | MGI:1913369 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000048707 | Tprn | taperin | MGI:2139535 | YES | YES | Extracellular Space | other | |||||
| ENSMUSG00000021188 | Trip11 | thyroid hormone receptor interactor 11 | MGI:1924393 | YES | YES | Cytoplasm | transcription regulator | |||||
| ENSMUSG00000026510 | Trp53bp2 | transformation related protein 53 binding protein 2 | MGI:2138319 | YES | YES | Nucleus | other | |||||
| ENSMUSG00000042734 | Ttc9 | tetratricopeptide repeat domain 9 | MGI:1916730 | YES | YES | Extracellular Space | other | |||||
| ENSMUSG00000043091 | Tuba1c | tubulin, alpha 1C | MGI:1095409 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000030137 | Tuba8 | tubulin, alpha 8 | MGI:1858225 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000000759 | Tubgcp3 | tubulin, gamma complex associated protein 3 | MGI:2183752 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000034485 | Uaca | uveal autoantigen with coiled-coil domains and ankyrin repeats | MGI:1919815 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000052981 | Ube2ql1 | ubiquitin-conjugating enzyme E2Q family-like 1 | MGI:1924230 | YES | YES | Nucleus | other | |||||
| ENSMUSG00000009741 | Ubp1 | upstream binding protein 1 | MGI:104889 | YES | YES | Cytoplasm | transcription regulator | |||||
| ENSMUSG00000020923 | Ubtf | upstream binding transcription factor, RNA polymerase I | MGI:98512 | YES | YES | Nucleus | transcription regulator | |||||
| ENSMUSG00000028456 | Unc13b | unc-13 homolog B (C. elegans) | MGI:1342278 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000021198 | Unc79 | unc-79 homolog (C. elegans) | MGI:2684729 | YES | YES | Other | other | |||||
| ENSMUSG00000039929 | Urb1 | URB1 ribosome biogenesis 1 homolog (S. cerevisiae) | MGI:2146468 | YES | YES | Nucleus | other | |||||
| ENSMUSG00000039701 | Usp53 | ubiquitin specific peptidase 53 | MGI:2139607 | YES | YES | Extracellular Space | enzyme | |||||
| ENSMUSG00000027363 | Usp8 | ubiquitin specific peptidase 8 | MGI:1934029 | YES | YES | Cytoplasm | peptidase | |||||
| ENSMUSG00000030403 | Vasp | vasodilator-stimulated phosphoprotein | MGI:109268 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000066278 | Vps37b | vacuolar protein sorting 37B (yeast) | MGI:1916724 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000021994 | Wnt5a | wingless-type MMTV integration site family, member 5A | MGI:98958 | YES | YES | Extracellular Space | cytokine | |||||
| ENSMUSG00000022390 | Zc3h7b | zinc finger CCCH type containing 7B | MGI:1328310 | YES | YES | Nucleus | other | |||||
| ENSMUSG00000043629 | 1700019D03Rik | RIKEN cDNA 1700019D03 gene | MGI:1914330 | YES | YES | Other | other | |||||
| ENSMUSG00000087260 | Lamtor5 | late endosomal/lysosomal adaptor, MAPK and MTOR activator 5 | MGI:1915826 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000028953 | Abcf2 | ATP-binding cassette, sub-family F (GCN20), member 2 | MGI:1351657 | YES | YES | Cytoplasm | transporter | |||||
| ENSMUSG00000069833 | Ahnak | AHNAK nucleoprotein (desmoyokin) | MGI:1316648 | YES | YES | Nucleus | other | |||||
| ENSMUSG00000072812 | Ahnak2 | AHNAK nucleoprotein 2 | MGI:2144831 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000036591 | Arhgap21 | Rho GTPase activating protein 21 | MGI:1918685 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000049804 | Armcx4 | armadillo repeat containing, X-linked 4 | MGI:2147887 | YES | YES | Other | other | |||||
| ENSMUSG00000042605 | Atxn2 | ataxin 2 | MGI:1277223 | YES | YES | Nucleus | other | |||||
| ENSMUSG00000013523 | Bcas1 | breast carcinoma amplified sequence 1 | MGI:1924210 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000030279 | C2cd5 | C2 calcium-dependent domain containing 5 | MGI:1921991 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000057914 | Cacnb2 | calcium channel, voltage-dependent, beta 2 subunit | MGI:894644 | YES | YES | Plasma Membrane | ion channel | |||||
| ENSMUSG00000029761 | Cald1 | caldesmon 1 | MGI:88250 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000031012 | Cask | calcium/calmodulin-dependent serine protein kinase (MAGUK family) | MGI:1309489 | YES | YES | Plasma Membrane | kinase | |||||
| ENSMUSG00000026312 | Cdh7 | cadherin 7, type 2 | MGI:2442792 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000025370 | Cdh9 | cadherin 9 | MGI:107433 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000046691 | Chtf8 | CTF8, chromosome transmission fidelity factor 8 | MGI:2443370 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000058897 | Col25a1 | collagen, type XXV, alpha 1 | MGI:1924268 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000034361 | Cpne2 | copine II | MGI:2387578 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000018001 | Cyth3 | cytohesin 3 | MGI:1335107 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000078794 | Dact3 | dapper homolog 3, antagonist of beta-catenin (xenopus) | MGI:3654828 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000021559 | Dapk1 | death associated protein kinase 1 | MGI:1916885 | YES | YES | Cytoplasm | kinase | |||||
| ENSMUSG00000049354 | Dcaf7 | DDB1 and CUL4 associated factor 7 | MGI:1919083 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000059213 | Ddn | dendrin | MGI:108101 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000038665 | Dgki | diacylglycerol kinase, iota | MGI:2443430 | YES | YES | Cytoplasm | kinase | |||||
| ENSMUSG00000029169 | Dhx15 | DEAH (Asp-Glu-Ala-His) box polypeptide 15 | MGI:1099786 | YES | YES | Nucleus | enzyme | |||||
| ENSMUSG00000042699 | Dhx9 | DEAH (Asp-Glu-Ala-His) box polypeptide 9 | MGI:108177 | YES | YES | Nucleus | enzyme | |||||
| ENSMUSG00000028035 | Dnajb4 | DnaJ (Hsp40) homolog, subfamily B, member 4 | MGI:1914285 | YES | YES | Nucleus | other | |||||
| ENSMUSG00000024350 | Dnajc18 | DnaJ (Hsp40) homolog, subfamily C, member 18 | MGI:1923844 | YES | YES | Other | enzyme | |||||
| ENSMUSG00000038608 | Dock10 | dedicator of cytokinesis 10 | MGI:2146320 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000001229 | Dpp9 | dipeptidylpeptidase 9 | MGI:2443967 | YES | YES | Cytoplasm | peptidase | |||||
| ENSMUSG00000022897 | Dyrk1a | dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1a | MGI:1330299 | YES | YES | Nucleus | kinase | |||||
| ENSMUSG00000027810 | Eif2a | eukaryotic translation initiation factor 2A | MGI:1098684 | YES | YES | Cytoplasm | translation regulator | |||||
| ENSMUSG00000031029 | Eif3f | eukaryotic translation initiation factor 3, subunit F | MGI:1913335 | YES | YES | Cytoplasm | translation regulator | |||||
| ENSMUSG00000058655 | Eif4b | eukaryotic translation initiation factor 4B | MGI:95304 | YES | YES | Cytoplasm | translation regulator | |||||
| ENSMUSG00000015337 | Endog | endonuclease G | MGI:1261433 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000028434 | Epb4.1l4b | erythrocyte protein band 4.1 like 4b | MGI:1859149 | YES | YES | Cytoplasm | transporter | |||||
| ENSMUSG00000053799 | Exoc6 | exocyst complex component 6 | MGI:1351611 | YES | YES | Plasma Membrane | transporter | |||||
| ENSMUSG00000002017 | Fam98a | family with sequence similarity 98, member A | MGI:1919972 | YES | YES | Other | other | |||||
| ENSMUSG00000030759 | Far1 | fatty acyl CoA reductase 1 | MGI:1914670 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000036585 | Fgf1 | fibroblast growth factor 1 | MGI:95515 | YES | YES | Extracellular Space | growth factor | |||||
| ENSMUSG00000026841 | Fibcd1 | fibrinogen C domain containing 1 | MGI:2138953 | YES | YES | Other | other | |||||
| ENSMUSG00000026657 | Frmd4a | FERM domain containing 4A | MGI:1919850 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000042425 | Frmpd3 | FERM and PDZ domain containing 3 | MGI:3646547 | YES | YES | Other | other | |||||
| ENSMUSG00000021065 | Fut8 | fucosyltransferase 8 | MGI:1858901 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000025241 | Fyco1 | FYVE and coiled-coil domain containing 1 | MGI:107277 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000034201 | Gas2l1 | growth arrest-specific 2 like 1 | MGI:1926176 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000022103 | Gfra2 | glial cell line derived neurotrophic factor family receptor alpha 2 | MGI:1195462 | YES | YES | Plasma Membrane | transmembrane receptor | |||||
| ENSMUSG00000028020 | Glrb | glycine receptor, beta subunit | MGI:95751 | YES | YES | Plasma Membrane | ion channel | |||||
| ENSMUSG00000043384 | Gprasp1 | G protein-coupled receptor associated sorting protein 1 | MGI:1917418 | YES | YES | Cytoplasm | transporter | |||||
| ENSMUSG00000071531 | Gprin2 | G protein regulated inducer of neurite outgrowth 2 | MGI:2444560 | YES | YES | Other | other | |||||
| ENSMUSG00000020734 | Grin2c | glutamate receptor, ionotropic, NMDA2C (epsilon 3) | MGI:95822 | YES | YES | Plasma Membrane | ion channel | |||||
| ENSMUSG00000034813 | Grip1 | glutamate receptor interacting protein 1 | MGI:1921303 | YES | YES | Plasma Membrane | transcription regulator | |||||
| ENSMUSG00000035376 | Hacd2 | 3-hydroxyacyl-CoA dehydratase 2 | MGI:1918007 | YES | YES | Cytoplasm | phosphatase | |||||
| ENSMUSG00000004894 | Hapln2 | hyaluronan and proteoglycan link protein 2 | MGI:2137300 | YES | YES | Extracellular Space | other | |||||
| ENSMUSG00000015165 | Hnrnpl | heterogeneous nuclear ribonucleoprotein L | MGI:104816 | YES | YES | Nucleus | other | |||||
| ENSMUSG00000026925 | Inpp5e | inositol polyphosphate-5-phosphatase E | MGI:1927753 | YES | YES | Cytoplasm | phosphatase | |||||
| ENSMUSG00000022208 | Jph4 | junctophilin 4 | MGI:2443113 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000061751 | Kalrn | kalirin, RhoGEF kinase | MGI:2685385 | YES | YES | Cytoplasm | kinase | |||||
| ENSMUSG00000028790 | Khdrbs1 | KH domain containing, RNA binding, signal transduction associated 1 | MGI:893579 | YES | YES | Nucleus | transcription regulator | |||||
| ENSMUSG00000046312 | AI464131 | expressed sequence AI464131 | MGI:2140300 | YES | YES | Nucleus | other | |||||
| ENSMUSG00000050390 | C77080 | expressed sequence C77080 | MGI:2140651 | YES | YES | Other | other | |||||
| ENSMUSG00000020668 | Kif3c | kinesin family member 3C | MGI:107979 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000066129 | Kndc1 | kinase non-catalytic C-lobe domain (KIND) containing 1 | MGI:1923734 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000021843 | Ktn1 | kinectin 1 | MGI:109153 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000036560 | Lgi4 | leucine-rich repeat LGI family, member 4 | MGI:2180197 | YES | YES | Extracellular Space | other | |||||
| ENSMUSG00000051067 | Lingo3 | leucine rich repeat and Ig domain containing 3 | MGI:3609246 | YES | YES | Other | other | |||||
| ENSMUSG00000022802 | Lmln | leishmanolysin-like (metallopeptidase M8 family) | MGI:2444736 | YES | YES | Plasma Membrane | peptidase | |||||
| ENSMUSG00000068015 | Lrch1 | leucine-rich repeats and calponin homology (CH) domain containing 1 | MGI:2443390 | YES | YES | Other | other | |||||
| ENSMUSG00000063297 | Luzp2 | leucine zipper protein 2 | MGI:1889615 | YES | YES | Other | other | |||||
| ENSMUSG00000025151 | Maged1 | melanoma antigen, family D, 1 | MGI:1930187 | YES | YES | Plasma Membrane | transcription regulator | |||||
| ENSMUSG00000052539 | Magi3 | membrane associated guanylate kinase, WW and PDZ domain containing 3 | MGI:1923484 | YES | YES | Cytoplasm | kinase | |||||
| ENSMUSG00000002948 | Map2k7 | mitogen-activated protein kinase kinase 7 | MGI:1346871 | YES | YES | Cytoplasm | kinase | |||||
| ENSMUSG00000019996 | Map7 | microtubule-associated protein 7 | MGI:1328328 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000031833 | Mast3 | microtubule associated serine/threonine kinase 3 | MGI:2683541 | YES | YES | Other | kinase | |||||
| ENSMUSG00000016995 | Matn4 | matrilin 4 | MGI:1328314 | YES | YES | Extracellular Space | other | |||||
| ENSMUSG00000021112 | Mpp5 | membrane protein, palmitoylated 5 (MAGUK p55 subfamily member 5) | MGI:1927339 | YES | YES | Plasma Membrane | kinase | |||||
| ENSMUSG00000023939 | Mrpl14 | mitochondrial ribosomal protein L14 | MGI:1333864 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000035772 | Mrps2 | mitochondrial ribosomal protein S2 | MGI:2153089 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000021731 | Mrps30 | mitochondrial ribosomal protein S30 | MGI:1926237 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000069769 | Msi2 | musashi RNA-binding protein 2 | MGI:1923876 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000032220 | Myo1e | myosin IE | MGI:106621 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000033577 | Myo6 | myosin VI | MGI:104785 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000039585 | Myo9a | myosin IXa | MGI:107735 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000034000 | Neu4 | sialidase 4 | MGI:2661364 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000039103 | Nexn | nexilin | MGI:1916060 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000060275 | Nrg2 | neuregulin 2 | MGI:1098246 | YES | YES | Extracellular Space | growth factor | |||||
| ENSMUSG00000035513 | Ntng2 | netrin G2 | MGI:2159341 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000031754 | Nudt21 | nudix (nucleoside diphosphate linked moiety X)-type motif 21 | MGI:1915469 | YES | YES | Nucleus | other | |||||
| ENSMUSG00000044252 | Osbpl1a | oxysterol binding protein-like 1A | MGI:1927551 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000074923 | Pak6 | p21 protein (Cdc42/Rac)-activated kinase 6 | MGI:2679420 | YES | YES | Cytoplasm | kinase | |||||
| ENSMUSG00000051242 | Pcdhb9 | protocadherin beta 9 | MGI:2136744 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000021699 | Pde4d | phosphodiesterase 4D, cAMP specific | MGI:99555 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000021684 | Pde8b | phosphodiesterase 8B | MGI:2443999 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000041119 | Pde9a | phosphodiesterase 9A | MGI:1277179 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000024960 | Plcb3 | phospholipase C, beta 3 | MGI:104778 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000027695 | Pld1 | phospholipase D1 | MGI:109585 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000045659 | Plekha7 | pleckstrin homology domain containing, family A member 7 | MGI:2445094 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000017843 | Ppp2r5c | protein phosphatase 2, regulatory subunit B', gamma | MGI:1349475 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000079104 | Prps1l3 | phosphoribosyl pyrophosphate synthetase 1-like 3 | MGI:3779453 | YES | YES | Other | kinase | |||||
| ENSMUSG00000006498 | Ptbp1 | polypyrimidine tract binding protein 1 | MGI:97791 | YES | YES | Nucleus | enzyme | |||||
| ENSMUSG00000028909 | Ptpru | protein tyrosine phosphatase, receptor type, U | MGI:1321151 | YES | YES | Plasma Membrane | phosphatase | |||||
| ENSMUSG00000024096 | Ralbp1 | ralA binding protein 1 | MGI:108466 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000028292 | Rars2 | arginyl-tRNA synthetase 2, mitochondrial | MGI:1923596 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000032946 | Rasgrp2 | RAS, guanyl releasing protein 2 | MGI:1333849 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000035469 | Rcbtb1 | regulator of chromosome condensation (RCC1) and BTB (POZ) domain containing protein 1 | MGI:1918580 | YES | YES | Nucleus | other | |||||
| ENSMUSG00000031320 | Rps4x | ribosomal protein S4, X-linked | MGI:98158 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000050079 | Rspry1 | ring finger and SPRY domain containing 1 | MGI:1914860 | YES | YES | Other | other | |||||
| ENSMUSG00000000339 | Rtca | RNA 3'-terminal phosphate cyclase | MGI:1913618 | YES | YES | Nucleus | enzyme | |||||
| ENSMUSG00000043811 | Rtn4r | reticulon 4 receptor | MGI:2136886 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000057378 | Ryr3 | ryanodine receptor 3 | MGI:99684 | YES | YES | Plasma Membrane | ion channel | |||||
| ENSMUSG00000030602 | Pak4 | p21 protein (Cdc42/Rac)-activated kinase 4 | MGI:1917834 | YES | YES | #N/A | #N/A | |||||
| ENSMUSG00000025816 | Sec61a2 | Sec61, alpha subunit 2 (S. cerevisiae) | MGI:1931071 | YES | YES | Cytoplasm | transporter | |||||
| ENSMUSG00000053617 | Sh3pxd2a | SH3 and PX domains 2A | MGI:1298393 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000030583 | Sipa1l3 | signal-induced proliferation-associated 1 like 3 | MGI:1921456 | YES | YES | Extracellular Space | other | |||||
| ENSMUSG00000027075 | Slc43a1 | solute carrier family 43, member 1 | MGI:1931352 | YES | YES | Plasma Membrane | transporter | |||||
| ENSMUSG00000024974 | Smc3 | structural maintenance of chromosomes 3 | MGI:1339795 | YES | YES | Nucleus | other | |||||
| ENSMUSG00000026207 | Speg | SPEG complex locus | MGI:109282 | YES | YES | Nucleus | kinase | |||||
| ENSMUSG00000039536 | Stau1 | staufen (RNA binding protein) homolog 1 (Drosophila) | MGI:1338864 | YES | YES | Cytoplasm | transporter | |||||
| ENSMUSG00000027220 | Syt13 | synaptotagmin XIII | MGI:1933945 | YES | YES | Plasma Membrane | transporter | |||||
| ENSMUSG00000022019 | Tdrd3 | tudor domain containing 3 | MGI:2444023 | YES | YES | Nucleus | transcription regulator | |||||
| ENSMUSG00000038828 | Tmem214 | transmembrane protein 214 | MGI:1916046 | YES | YES | Extracellular Space | other | |||||
| ENSMUSG00000000827 | Tpd52l2 | tumor protein D52-like 2 | MGI:1913564 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000020308 | Tpgs1 | tubulin polyglutamylase complex subunit 1 | MGI:106618 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000034799 | Unc13a | unc-13 homolog A (C. elegans) | MGI:3051532 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000038013 | Wipf2 | WAS/WASL interacting protein family, member 2 | MGI:1924462 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000045414 | 1190002N15Rik | RIKEN cDNA 1190002N15 gene | MGI:1916111 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000042647 | Acad12 | acyl-Coenzyme A dehydrogenase family, member 12 | MGI:2443320 | YES | YES | YES | #N/A | #N/A | ||||
| ENSMUSG00000030861 | Acadsb | acyl-Coenzyme A dehydrogenase, short/branched chain | MGI:1914135 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000029610 | Aimp2 | aminoacyl tRNA synthetase complex-interacting multifunctional protein 2 | MGI:2385237 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000039058 | Ak5 | adenylate kinase 5 | MGI:2677491 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSMUSG00000061758 | Akr1b10 | aldo-keto reductase family 1, member B10 (aldose reductase) | MGI:1915111 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000032826 | Ank2 | ankyrin 2, brain | MGI:88025 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000069601 | Ank3 | ankyrin 3, epithelial | MGI:88026 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000040351 | Ankib1 | ankyrin repeat and IBR domain containing 1 | MGI:1918047 | YES | YES | YES | Nucleus | transcription regulator | ||||
| ENSMUSG00000029994 | Anxa4 | annexin A4 | MGI:88030 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000031367 | Ap1s2 | adaptor-related protein complex 1, sigma 2 subunit | MGI:1889383 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSMUSG00000033436 | Armcx2 | armadillo repeat containing, X-linked 2 | MGI:1914666 | YES | YES | YES | Other | other | ||||
| ENSMUSG00000015943 | Bola1 | bolA-like 1 (E. coli) | MGI:1916418 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000020042 | Btbd11 | BTB (POZ) domain containing 11 | MGI:1921257 | YES | YES | YES | Other | transcription regulator | ||||
| ENSMUSG00000036707 | Cab39 | calcium binding protein 39 | MGI:107438 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000090071 | Cdk5r2 | cyclin-dependent kinase 5, regulatory subunit 2 (p39) | MGI:1330828 | YES | YES | YES | Nucleus | other | ||||
| ENSMUSG00000022604 | Cep97 | centrosomal protein 97 | MGI:1921451 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000033916 | Chmp2a | charged multivesicular body protein 2A | MGI:1916203 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000053119 | Chmp3 | charged multivesicular body protein 3 | MGI:1913950 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000040495 | Chrm4 | cholinergic receptor, muscarinic 4 | MGI:88399 | YES | YES | YES | Plasma Membrane | G-protein coupled receptor | ||||
| ENSMUSG00000022132 | Cldn10 | claudin 10 | MGI:1913101 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000030282 | Cmas | cytidine monophospho-N-acetylneuraminic acid synthetase | MGI:1337124 | YES | YES | YES | Nucleus | enzyme | ||||
| ENSMUSG00000034390 | Cmip | c-Maf inducing protein | MGI:1921690 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000053931 | Cnn3 | calponin 3, acidic | MGI:1919244 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000020300 | Cpeb4 | cytoplasmic polyadenylation element binding protein 4 | MGI:1914829 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000032060 | Cryab | crystallin, alpha B | MGI:88516 | YES | YES | YES | Nucleus | other | ||||
| ENSMUSG00000031446 | Cul4a | cullin 4A | MGI:1914487 | YES | YES | YES | Nucleus | other | ||||
| ENSMUSG00000055065 | Ddx17 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 17 | MGI:1914290 | YES | YES | YES | Nucleus | enzyme | ||||
| ENSMUSG00000040479 | Dgkz | diacylglycerol kinase zeta | MGI:1278339 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSMUSG00000042662 | Dusp15 | dual specificity phosphatase-like 15 | MGI:1934928 | YES | YES | YES | Cytoplasm | phosphatase | ||||
| ENSMUSG00000021791 | Dydc2 | DPY30 domain containing 2 | MGI:1918450 | YES | YES | YES | Extracellular Space | other | ||||
| ENSMUSG00000026247 | Ecel1 | endothelin converting enzyme-like 1 | MGI:1343461 | YES | YES | YES | Plasma Membrane | peptidase | ||||
| ENSMUSG00000033047 | Eif3l | eukaryotic translation initiation factor 3, subunit L | MGI:2386251 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000025580 | Eif4a3 | eukaryotic translation initiation factor 4A3 | MGI:1923731 | YES | YES | YES | Nucleus | enzyme | ||||
| ENSMUSG00000028546 | Elavl4 | ELAV (embryonic lethal, abnormal vision, Drosophila)-like 4 (Hu antigen D) | MGI:107427 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000033068 | Entpd6 | ectonucleoside triphosphate diphosphohydrolase 6 | MGI:1202295 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000027624 | Epb4.1l1 | erythrocyte protein band 4.1 like 1 | MGI:103010 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000010080 | Epn3 | epsin 3 | MGI:1919139 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000025198 | Erlin1 | ER lipid raft associated 1 | MGI:2387613 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000028995 | Fam126a | family with sequence similarity 126, member A | MGI:2149839 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000054942 | Fam73a | family with sequence similarity 73, member A | MGI:1924567 | YES | YES | YES | Other | other | ||||
| ENSMUSG00000058715 | Fcer1g | Fc receptor, IgE, high affinity I, gamma polypeptide | MGI:95496 | YES | YES | YES | Plasma Membrane | transmembrane receptor | ||||
| ENSMUSG00000020170 | Frs2 | fibroblast growth factor receptor substrate 2 | MGI:1100860 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000036570 | Fxyd1 | FXYD domain-containing ion transport regulator 1 | MGI:1889273 | YES | YES | YES | Plasma Membrane | ion channel | ||||
| ENSMUSG00000029714 | Gigyf1 | GRB10 interacting GYF protein 1 | MGI:1888677 | YES | YES | YES | Extracellular Space | other | ||||
| ENSMUSG00000048000 | Gigyf2 | GRB10 interacting GYF protein 2 | MGI:2138584 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000000263 | Glra1 | glycine receptor, alpha 1 subunit | MGI:95747 | YES | YES | YES | Plasma Membrane | ion channel | ||||
| ENSMUSG00000021591 | Glrx | glutaredoxin | MGI:2135625 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000031068 | Glrx3 | glutaredoxin 3 | MGI:1353653 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000036568 | Gltscr1l | GLTSCR1-like | MGI:2673855 | YES | YES | YES | Other | other | ||||
| ENSMUSG00000066487 | Gm5786 | predicted pseudogene 5786 | MGI:3645003 | YES | YES | YES | Other | other | ||||
| ENSMUSG00000023439 | Gnb3 | guanine nucleotide binding protein (G protein), beta 3 | MGI:95785 | YES | YES | YES | Plasma Membrane | enzyme | ||||
| ENSMUSG00000022112 | Gpc5 | glypican 5 | MGI:1194894 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000035900 | Gramd4 | GRAM domain containing 4 | MGI:2676308 | YES | YES | YES | Other | other | ||||
| ENSMUSG00000033981 | Gria2 | glutamate receptor, ionotropic, AMPA2 (alpha 2) | MGI:95809 | YES | YES | YES | Plasma Membrane | ion channel | ||||
| ENSMUSG00000025892 | Gria4 | glutamate receptor, ionotropic, AMPA4 (alpha 4) | MGI:95811 | YES | YES | YES | Plasma Membrane | ion channel | ||||
| ENSMUSG00000042535 | Gtpbp1 | GTP binding protein 1 | MGI:109443 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000000628 | Hk2 | hexokinase 2 | MGI:1315197 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSMUSG00000031839 | Hsbp1 | heat shock factor binding protein 1 | MGI:1915446 | YES | YES | YES | Nucleus | transcription regulator | ||||
| ENSMUSG00000004951 | Hspb1 | heat shock protein 1 | MGI:96240 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000037851 | Iars | isoleucine-tRNA synthetase | MGI:2145219 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000018858 | Ict1 | immature colon carcinoma transcript 1 | MGI:1915822 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000066232 | Ipo7 | importin 7 | MGI:2152414 | YES | YES | YES | Nucleus | transporter | ||||
| ENSMUSG00000046879 | Irgm1 | immunity-related GTPase family M member 1 | MGI:107567 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000009731 | Kcnd1 | potassium voltage-gated channel, Shal-related family, member 1 | MGI:96671 | YES | YES | YES | Plasma Membrane | ion channel | ||||
| ENSMUSG00000075402 | Krt76 | keratin 76 | MGI:1924305 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000106379 | Lhfpl3 | lipoma HMGIC fusion partner-like 3 | MGI:1925076 | YES | YES | YES | Other | other | ||||
| ENSMUSG00000036957 | Lrfn3 | leucine rich repeat and fibronectin type III domain containing 3 | MGI:2442512 | YES | YES | YES | Other | other | ||||
| ENSMUSG00000026344 | Lypd1 | Ly6/Plaur domain containing 1 | MGI:1919835 | YES | YES | YES | Plasma Membrane | G-protein coupled receptor | ||||
| ENSMUSG00000032295 | Man2c1 | mannosidase, alpha, class 2C, member 1 | MGI:1920994 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000004933 | Matk | megakaryocyte-associated tyrosine kinase | MGI:99259 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSMUSG00000041607 | Mbp | myelin basic protein | MGI:96925 | YES | YES | YES | Extracellular Space | other | ||||
| ENSMUSG00000031442 | Mcf2l | mcf.2 transforming sequence-like | MGI:103263 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000021290 | 2010107E04Rik | RIKEN cDNA 2010107E04 gene | MGI:1917507 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000028402 | Mpdz | multiple PDZ domain protein | MGI:1343489 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000024902 | Mrpl11 | mitochondrial ribosomal protein L11 | MGI:2137215 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000022706 | Mrpl40 | mitochondrial ribosomal protein L40 | MGI:1332635 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000030037 | Mrpl53 | mitochondrial ribosomal protein L53 | MGI:1915749 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000034729 | Mrps10 | mitochondrial ribosomal protein S10 | MGI:1928139 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000054312 | Mrps21 | mitochondrial ribosomal protein S21 | MGI:1913542 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000032459 | Mrps22 | mitochondrial ribosomal protein S22 | MGI:1928137 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000029840 | Mtpn | myotrophin | MGI:99445 | YES | YES | YES | Nucleus | transcription regulator | ||||
| ENSMUSG00000016427 | Ndufa1 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 1 | MGI:1929511 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000024038 | Ndufv3 | NADH dehydrogenase (ubiquinone) flavoprotein 3 | MGI:1890894 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000017837 | Nkiras2 | NFKB inhibitor interacting Ras-like protein 2 | MGI:1919216 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000006476 | Nsmf | NMDA receptor synaptonuclear signaling and neuronal migration factor | MGI:1861755 | YES | YES | YES | Extracellular Space | other | ||||
| ENSMUSG00000045211 | Nudt18 | nudix (nucleoside diphosphate linked moiety X)-type motif 18 | MGI:2385853 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000047986 | Palm3 | paralemmin 3 | MGI:1921587 | YES | YES | YES | Other | other | ||||
| ENSMUSG00000029623 | Pdap1 | PDGFA associated protein 1 | MGI:2448536 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000050890 | Pdik1l | PDLIM1 interacting kinase 1 like | MGI:2385213 | YES | YES | YES | Nucleus | kinase | ||||
| ENSMUSG00000018733 | Pex12 | peroxisomal biogenesis factor 12 | MGI:2144177 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000019809 | Pex3 | peroxisomal biogenesis factor 3 | MGI:1929646 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000003469 | Phyhip | phytanoyl-CoA hydroxylase interacting protein | MGI:1860417 | YES | YES | YES | Other | other | ||||
| ENSMUSG00000023452 | Pisd | phosphatidylserine decarboxylase | MGI:2445114 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000029055 | Plch2 | phospholipase C, eta 2 | MGI:2443078 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000016495 | Plgrkt | plasminogen receptor, C-terminal lysine transmembrane protein | MGI:1915009 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000039254 | Pomt1 | protein-O-mannosyltransferase 1 | MGI:2138994 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000037519 | Ppfia1 | protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 1 | MGI:1924750 | YES | YES | YES | Plasma Membrane | phosphatase | ||||
| ENSMUSG00000039457 | Ppl | periplakin | MGI:1194898 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000015869 | Prpsap1 | phosphoribosyl pyrophosphate synthetase-associated protein 1 | MGI:1915013 | YES | YES | YES | Extracellular Space | other | ||||
| ENSMUSG00000071519 | Prss3 | protease, serine 3 | MGI:102758 | YES | YES | YES | Extracellular Space | peptidase | ||||
| ENSMUSG00000028484 | Psip1 | PC4 and SFRS1 interacting protein 1 | MGI:2142116 | YES | YES | YES | Nucleus | other | ||||
| ENSMUSG00000026914 | Psmd14 | proteasome (prosome, macropain) 26S subunit, non-ATPase, 14 | MGI:1913284 | YES | YES | YES | Cytoplasm | peptidase | ||||
| ENSMUSG00000030359 | Pzp | pregnancy zone protein | MGI:87854 | YES | YES | YES | Extracellular Space | other | ||||
| ENSMUSG00000027935 | Rab13 | RAB13, member RAS oncogene family | MGI:1927232 | YES | YES | YES | Plasma Membrane | enzyme | ||||
| ENSMUSG00000041351 | Rap1gap | Rap1 GTPase-activating protein | MGI:109338 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000070509 | Rgma | repulsive guidance molecule family member A | MGI:2679262 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000025257 | Ribc1 | RIB43A domain with coiled-coils 1 | MGI:1913861 | YES | YES | YES | Other | other | ||||
| ENSMUSG00000024883 | Rin1 | Ras and Rab interactor 1 | MGI:2385695 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000019763 | Rmnd1 | required for meiotic nuclear division 1 homolog (S. cerevisiae) | MGI:1913334 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000009535 | Rnmt | RNA (guanine-7-) methyltransferase | MGI:1915147 | YES | YES | YES | Nucleus | enzyme | ||||
| ENSMUSG00000046330 | Rpl37a | ribosomal protein L37a | MGI:98068 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000050896 | Rtn4rl2 | reticulon 4 receptor-like 2 | MGI:2669796 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000074457 | S100a16 | S100 calcium binding protein A16 | MGI:1915110 | YES | YES | YES | Nucleus | other | ||||
| ENSMUSG00000042682 | Selk | selenoprotein K | MGI:1931466 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000059336 | Slc14a1 | solute carrier family 14 (urea transporter), member 1 | MGI:1351654 | YES | YES | YES | Plasma Membrane | transporter | ||||
| ENSMUSG00000054099 | Slc25a40 | solute carrier family 25, member 40 | MGI:2442486 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000024270 | Slc39a6 | solute carrier family 39 (metal ion transporter), member 6 | MGI:2147279 | YES | YES | YES | Plasma Membrane | transporter | ||||
| ENSMUSG00000078350 | Smim1 | small integral membrane protein 1 | MGI:1916109 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000022114 | Spry2 | sprouty homolog 2 (Drosophila) | MGI:1345138 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000015837 | Sqstm1 | sequestosome 1 | MGI:107931 | YES | YES | YES | Cytoplasm | transcription regulator | ||||
| ENSMUSG00000056812 | St8sia3 | ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 3 | MGI:106019 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000061455 | Stx17 | syntaxin 17 | MGI:1914977 | YES | YES | YES | Plasma Membrane | transporter | ||||
| ENSMUSG00000029125 | Stx18 | syntaxin 18 | MGI:1918366 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSMUSG00000074736 | Syndig1 | synapse differentiation inducing 1 | MGI:3702158 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000009670 | Tex11 | testis expressed gene 11 | MGI:1933237 | YES | YES | YES | Nucleus | other | ||||
| ENSMUSG00000038028 | Tigar | Trp53 induced glycolysis repulatory phosphatase | MGI:2442752 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000034659 | Tmem109 | transmembrane protein 109 | MGI:1915789 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000057716 | Tmem178b | transmembrane protein 178B | MGI:3647581 | YES | YES | YES | Other | other | ||||
| ENSMUSG00000036372 | Tmem258 | transmembrane protein 258 | MGI:1916288 | YES | YES | YES | Other | other | ||||
| ENSMUSG00000025666 | Tmem47 | transmembrane protein 47 | MGI:2177570 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000028232 | Tmem68 | transmembrane protein 68 | MGI:1919348 | YES | YES | YES | Other | other | ||||
| ENSMUSG00000025139 | Tollip | toll interacting protein | MGI:1891808 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000071660 | Ttc9c | tetratricopeptide repeat domain 9C | MGI:1917637 | YES | YES | YES | Other | other | ||||
| ENSMUSG00000001473 | Tubb6 | tubulin, beta 6 class V | MGI:1915201 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000062963 | Ufc1 | ubiquitin-fold modifier conjugating enzyme 1 | MGI:1913405 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000024208 | Uqcc2 | ubiquinol-cytochrome c reductase complex assembly factor 2 | MGI:1914517 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000073002 | Vamp5 | vesicle-associated membrane protein 5 | MGI:1858622 | YES | YES | YES | Plasma Membrane | transporter | ||||
| ENSMUSG00000078656 | Vps25 | vacuolar protein sorting 25 (yeast) | MGI:106354 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000028868 | Wasf2 | WAS protein family, member 2 | MGI:1098641 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000024101 | Wash1 | WAS protein family homolog 1 | MGI:1916017 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000064030 | Wibg | within bgcn homolog (Drosophila) | MGI:1925678 | YES | YES | YES | Nucleus | other | ||||
| ENSMUSG00000022792 | Yars2 | tyrosyl-tRNA synthetase 2 (mitochondrial) | MGI:1917370 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000032264 | Zw10 | zw10 kinetochore protein | MGI:1349478 | YES | YES | YES | Nucleus | other | ||||
| ENSMUSG00000036698 | Ago2 | argonaute RISC catalytic subunit 2 | MGI:2446632 | YES | YES | YES | Cytoplasm | translation regulator | ||||
| ENSMUSG00000001833 | Sept7 | septin 7 | MGI:1335094 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000018398 | Sept8 | septin 8 | MGI:894310 | YES | YES | YES | Extracellular Space | other | ||||
| ENSMUSG00000026090 | 2010300C02Rik | RIKEN cDNA 2010300C02 gene | MGI:1919347 | YES | YES | YES | Other | other | ||||
| ENSMUSG00000026319 | 2310035C23Rik | RIKEN cDNA 2310035C23 gene | MGI:1922832 | YES | YES | YES | Other | other | ||||
| ENSMUSG00000053963 | 6330403A02Rik | RIKEN cDNA 6330403A02 gene | MGI:2138735 | YES | YES | YES | Other | other | ||||
| ENSMUSG00000044060 | A830010M20Rik | RIKEN cDNA A830010M20 gene | MGI:2445097 | YES | YES | YES | Other | other | ||||
| ENSMUSG00000057230 | Aak1 | AP2 associated kinase 1 | MGI:1098687 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSMUSG00000040584 | Abcb1a | ATP-binding cassette, sub-family B (MDR/TAP), member 1A | MGI:97570 | YES | YES | YES | Plasma Membrane | transporter | ||||
| ENSMUSG00000031333 | Abcb7 | ATP-binding cassette, sub-family B (MDR/TAP), member 7 | MGI:109533 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSMUSG00000028973 | Abcb8 | ATP-binding cassette, sub-family B (MDR/TAP), member 8 | MGI:1351667 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSMUSG00000032046 | Abhd12 | abhydrolase domain containing 12 | MGI:1923442 | YES | YES | YES | Other | enzyme | ||||
| ENSMUSG00000007036 | Abhd16a | abhydrolase domain containing 16A | MGI:99476 | YES | YES | YES | Other | other | ||||
| ENSMUSG00000025277 | Abhd6 | abhydrolase domain containing 6 | MGI:1913332 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000026782 | Abi2 | abl-interactor 2 | MGI:106913 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000026596 | Abl2 | v-abl Abelson murine leukemia viral oncogene 2 (arg, Abelson-related gene) | MGI:87860 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSMUSG00000036880 | Acaa2 | acetyl-Coenzyme A acyltransferase 2 (mitochondrial 3-oxoacyl-Coenzyme A thiolase) | MGI:1098623 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000031969 | Acad8 | acyl-Coenzyme A dehydrogenase family, member 8 | MGI:1914198 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000030607 | Acan | aggrecan | MGI:99602 | YES | YES | YES | Extracellular Space | other | ||||
| ENSMUSG00000049076 | Acap2 | ArfGAP with coiled-coil, ankyrin repeat and PH domains 2 | MGI:1925868 | YES | YES | YES | Nucleus | other | ||||
| ENSMUSG00000032047 | Acat1 | acetyl-Coenzyme A acetyltransferase 1 | MGI:87870 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000026499 | Acbd3 | acyl-Coenzyme A binding domain containing 3 | MGI:2181074 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000025287 | Acot9 | acyl-CoA thioesterase 9 | MGI:1928939 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000032883 | Acsl3 | acyl-CoA synthetase long-chain family member 3 | MGI:1921455 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000031278 | Acsl4 | acyl-CoA synthetase long-chain family member 4 | MGI:1354713 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000054808 | Actn4 | actinin alpha 4 | MGI:1890773 | YES | YES | YES | Cytoplasm | transcription regulator | ||||
| ENSMUSG00000021076 | Actr10 | ARP10 actin-related protein 10 | MGI:1891654 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000025228 | Actr1a | ARP1 actin-related protein 1A, centractin alpha | MGI:1858964 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000037351 | Actr1b | ARP1 actin-related protein 1B, centractin beta | MGI:1917446 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000020152 | Actr2 | ARP2 actin-related protein 2 | MGI:1913963 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000026341 | Actr3 | ARP3 actin-related protein 3 | MGI:1921367 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000056367 | Actr3b | ARP3 actin-related protein 3B | MGI:2661120 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000020926 | Adam11 | a disintegrin and metallopeptidase domain 11 | MGI:1098667 | YES | YES | YES | Plasma Membrane | peptidase | ||||
| ENSMUSG00000025964 | Adam23 | a disintegrin and metallopeptidase domain 23 | MGI:1345162 | YES | YES | YES | Plasma Membrane | peptidase | ||||
| ENSMUSG00000056413 | Adap1 | ArfGAP with dual PH domains 1 | MGI:2442201 | YES | YES | YES | Nucleus | other | ||||
| ENSMUSG00000020431 | Adcy1 | adenylate cyclase 1 | MGI:99677 | YES | YES | YES | Plasma Membrane | enzyme | ||||
| ENSMUSG00000021536 | Adcy2 | adenylate cyclase 2 | MGI:99676 | YES | YES | YES | Plasma Membrane | enzyme | ||||
| ENSMUSG00000022994 | Adcy6 | adenylate cyclase 6 | MGI:87917 | YES | YES | YES | Plasma Membrane | enzyme | ||||
| ENSMUSG00000030000 | Add2 | adducin 2 (beta) | MGI:87919 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000025026 | Add3 | adducin 3 (gamma) | MGI:1351615 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000028782 | Adgrb2 | adhesion G protein-coupled receptor B2 | MGI:2451244 | YES | YES | YES | Plasma Membrane | G-protein coupled receptor | ||||
| ENSMUSG00000028184 | Adgrl2 | adhesion G protein-coupled receptor L2 | MGI:2139714 | YES | YES | YES | Plasma Membrane | G-protein coupled receptor | ||||
| ENSMUSG00000033717 | Adra2a | adrenergic receptor, alpha 2a | MGI:87934 | YES | YES | YES | Plasma Membrane | G-protein coupled receptor | ||||
| ENSMUSG00000024858 | Adrbk1 | adrenergic receptor kinase, beta 1 | MGI:87940 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSMUSG00000024527 | Afg3l2 | AFG3-like AAA ATPase 2 | MGI:1916847 | YES | YES | YES | Cytoplasm | peptidase | ||||
| ENSMUSG00000055013 | Agap1 | ArfGAP with GTPase domain, ankyrin repeat and PH domain 1 | MGI:2653690 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000023353 | Agap3 | ArfGAP with GTPase domain, ankyrin repeat and PH domain 3 | MGI:2183446 | YES | YES | YES | Nucleus | transcription regulator | ||||
| ENSMUSG00000026159 | Agfg1 | ArfGAP with FG repeats 1 | MGI:1333754 | YES | YES | YES | Nucleus | other | ||||
| ENSMUSG00000029916 | Agk | acylglycerol kinase | MGI:1917173 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSMUSG00000034254 | Agpat1 | 1-acylglycerol-3-phosphate O-acyltransferase 1 (lysophosphatidic acid acyltransferase, alpha) | MGI:1932075 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000021037 | Ahsa1 | AHA1, activator of heat shock protein ATPase 1 | MGI:2387603 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000036932 | Aifm1 | apoptosis-inducing factor, mitochondrion-associated 1 | MGI:1349419 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000028029 | Aimp1 | aminoacyl tRNA synthetase complex-interacting multifunctional protein 1 | MGI:102774 | YES | YES | YES | Extracellular Space | cytokine | ||||
| ENSMUSG00000026817 | Ak1 | adenylate kinase 1 | MGI:87977 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSMUSG00000024782 | Ak3 | adenylate kinase 3 | MGI:1860835 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSMUSG00000028527 | Ak4 | adenylate kinase 4 | MGI:87979 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSMUSG00000021057 | Akap5 | A kinase (PRKA) anchor protein 5 | MGI:2685104 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000010025 | Aldh3a2 | aldehyde dehydrogenase family 3, subfamily A2 | MGI:1353452 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000030695 | Aldoa | aldolase A, fructose-bisphosphate | MGI:87994 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000039740 | Alg2 | asparagine-linked glycosylation 2 (alpha-1,3-mannosyltransferase) | MGI:1914731 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000028766 | Alpl | alkaline phosphatase, liver/bone/kidney | MGI:87983 | YES | YES | YES | Plasma Membrane | phosphatase | ||||
| ENSMUSG00000027889 | Ampd2 | adenosine monophosphate deaminase 2 | MGI:88016 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000005686 | Ampd3 | adenosine monophosphate deaminase 3 | MGI:1096344 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000022265 | Ank | progressive ankylosis | MGI:3045421 | YES | YES | YES | Plasma Membrane | transporter | ||||
| ENSMUSG00000039062 | Anpep | alanyl (membrane) aminopeptidase | MGI:5000466 | YES | YES | YES | Plasma Membrane | peptidase | ||||
| ENSMUSG00000032231 | Anxa2 | annexin A2 | MGI:88246 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000021814 | Anxa7 | annexin A7 | MGI:88031 | YES | YES | YES | Plasma Membrane | ion channel | ||||
| ENSMUSG00000009090 | Ap1b1 | adaptor protein complex AP-1, beta 1 subunit | MGI:1096368 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSMUSG00000003033 | Ap1m1 | adaptor-related protein complex AP-1, mu subunit 1 | MGI:102776 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSMUSG00000060279 | Ap2a1 | adaptor-related protein complex 2, alpha 1 subunit | MGI:101921 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSMUSG00000002957 | Ap2a2 | adaptor-related protein complex 2, alpha 2 subunit | MGI:101920 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSMUSG00000035152 | Ap2b1 | adaptor-related protein complex 2, beta 1 subunit | MGI:1919020 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000008036 | Ap2s1 | adaptor-related protein complex 2, sigma 1 subunit | MGI:2141861 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSMUSG00000062444 | Ap3b2 | adaptor-related protein complex 3, beta 2 subunit | MGI:1100869 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSMUSG00000020198 | Ap3d1 | adaptor-related protein complex 3, delta 1 subunit | MGI:107734 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSMUSG00000031539 | Ap3m2 | adaptor-related protein complex 3, mu 2 subunit | MGI:1929214 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSMUSG00000024480 | Ap3s1 | adaptor-related protein complex 3, sigma 1 subunit | MGI:1337062 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSMUSG00000005871 | Apc | adenomatosis polyposis coli | MGI:88039 | YES | YES | YES | Nucleus | enzyme | ||||
| ENSMUSG00000025525 | Apool | apolipoprotein O-like | MGI:1915367 | YES | YES | YES | Extracellular Space | other | ||||
| ENSMUSG00000020263 | Appl2 | adaptor protein, phosphotyrosine interaction, PH domain and leucine zipper containing 2 | MGI:2384914 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000032096 | Arcn1 | archain 1 | MGI:2387591 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000048076 | Arf1 | ADP-ribosylation factor 1 | MGI:99431 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000020440 | Arf5 | ADP-ribosylation factor 5 | MGI:99434 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000044147 | Arf6 | ADP-ribosylation factor 6 | MGI:99435 | YES | YES | YES | Plasma Membrane | transporter | ||||
| ENSMUSG00000027575 | Arfgap1 | ADP-ribosylation factor GTPase activating protein 1 | MGI:2183559 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSMUSG00000027255 | Arfgap2 | ADP-ribosylation factor GTPase activating protein 2 | MGI:1924288 | YES | YES | YES | Nucleus | other | ||||
| ENSMUSG00000049807 | Arhgap23 | Rho GTPase activating protein 23 | MGI:3697726 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000036882 | Arhgap33 | Rho GTPase activating protein 33 | MGI:2673998 | YES | YES | YES | Plasma Membrane | transporter | ||||
| ENSMUSG00000033389 | Arhgap44 | Rho GTPase activating protein 44 | MGI:2144423 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000025132 | Arhgdia | Rho GDP dissociation inhibitor (GDI) alpha | MGI:2178103 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000041977 | Arhgef11 | Rho guanine nucleotide exchange factor (GEF) 11 | MGI:2441869 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000028059 | Arhgef2 | rho/rac guanine nucleotide exchange factor (GEF) 2 | MGI:103264 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000054901 | Arhgef33 | Rho guanine nucleotide exchange factor (GEF) 33 | MGI:2685787 | YES | YES | YES | Other | other | ||||
| ENSMUSG00000031511 | Arhgef7 | Rho guanine nucleotide exchange factor (GEF7) | MGI:1860493 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000026426 | Arl8a | ADP-ribosylation factor-like 8A | MGI:1915974 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000027599 | Armc1 | armadillo repeat containing 1 | MGI:1921502 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000038525 | Armc10 | armadillo repeat containing 10 | MGI:1914461 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000029621 | Arpc1a | actin related protein 2/3 complex, subunit 1A | MGI:1928896 | YES | YES | YES | Extracellular Space | other | ||||
| ENSMUSG00000006304 | Arpc2 | actin related protein 2/3 complex, subunit 2 | MGI:1923959 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000029465 | Arpc3 | actin related protein 2/3 complex, subunit 3 | MGI:1928375 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000079426 | Arpc4 | actin related protein 2/3 complex, subunit 4 | MGI:1915339 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000026755 | Arpc5l | actin related protein 2/3 complex, subunit 5-like | MGI:1921442 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000000325 | Arvcf | armadillo repeat gene deleted in velo-cardio-facial syndrome | MGI:109620 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000031591 | Asah1 | N-acylsphingosine amidohydrolase 1 | MGI:1277124 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000028207 | Asph | aspartate-beta-hydroxylase | MGI:1914186 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000026587 | Astn1 | astrotactin 1 | MGI:1098567 | YES | YES | YES | Other | other | ||||
| ENSMUSG00000028373 | Astn2 | astrotactin 2 | MGI:1889277 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000013662 | Atad1 | ATPase family, AAA domain containing 1 | MGI:1915229 | YES | YES | YES | Other | enzyme | ||||
| ENSMUSG00000024426 | Atat1 | alpha tubulin acetyltransferase 1 | MGI:1913869 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000034958 | Atcay | ataxia, cerebellar, Cayman type homolog (human) | MGI:2448730 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000034566 | Atp5h | ATP synthase, H+ transporting, mitochondrial F0 complex, subunit D | MGI:1918929 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000050856 | Atp5k | ATP synthase, H+ transporting, mitochondrial F1F0 complex, subunit E | MGI:106636 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000022956 | Atp5o | ATP synthase, H+ transporting, mitochondrial F1 complex, O subunit | MGI:106341 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSMUSG00000019087 | Atp6ap1 | ATPase, H+ transporting, lysosomal accessory protein 1 | MGI:109629 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSMUSG00000031007 | Atp6ap2 | ATPase, H+ transporting, lysosomal accessory protein 2 | MGI:1917745 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSMUSG00000019302 | Atp6v0a1 | ATPase, H+ transporting, lysosomal V0 subunit A1 | MGI:103286 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSMUSG00000024121 | Atp6v0c | ATPase, H+ transporting, lysosomal V0 subunit C | MGI:88116 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSMUSG00000013160 | Atp6v0d1 | ATPase, H+ transporting, lysosomal V0 subunit D1 | MGI:1201778 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSMUSG00000052459 | Atp6v1a | ATPase, H+ transporting, lysosomal V1 subunit A | MGI:1201780 | YES | YES | YES | Plasma Membrane | transporter | ||||
| ENSMUSG00000006273 | Atp6v1b2 | ATPase, H+ transporting, lysosomal V1 subunit B2 | MGI:109618 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSMUSG00000022295 | Atp6v1c1 | ATPase, H+ transporting, lysosomal V1 subunit C1 | MGI:1913585 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSMUSG00000021114 | Atp6v1d | ATPase, H+ transporting, lysosomal V1 subunit D | MGI:1921084 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSMUSG00000019210 | Atp6v1e1 | ATPase, H+ transporting, lysosomal V1 subunit E1 | MGI:894326 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSMUSG00000004285 | Atp6v1f | ATPase, H+ transporting, lysosomal V1 subunit F | MGI:1913394 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000039105 | Atp6v1g1 | ATPase, H+ transporting, lysosomal V1 subunit G1 | MGI:1913540 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSMUSG00000024403 | Atp6v1g2 | ATPase, H+ transporting, lysosomal V1 subunit G2 | MGI:1913487 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSMUSG00000033793 | Atp6v1h | ATPase, H+ transporting, lysosomal V1 subunit H | MGI:1914864 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSMUSG00000027546 | Atp9a | ATPase, class II, type 9A | MGI:1330826 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSMUSG00000016541 | Atxn10 | ataxin 10 | MGI:1859293 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000021460 | Auh | AU RNA binding protein/enoyl-coenzyme A hydratase | MGI:1338011 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000071649 | B3gat3 | beta-1,3-glucuronyltransferase 3 (glucuronosyltransferase I) | MGI:1919977 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000006731 | B4galnt1 | beta-1,4-N-acetyl-galactosaminyl transferase 1 | MGI:1342057 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000049792 | Bag5 | BCL2-associated athanogene 5 | MGI:1917619 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000034730 | Adgrb1 | adhesion G protein-coupled receptor B1 | MGI:1933736 | YES | YES | YES | Plasma Membrane | G-protein coupled receptor | ||||
| ENSMUSG00000033569 | Adgrb3 | adhesion G protein-coupled receptor B3 | MGI:2441837 | YES | YES | YES | Plasma Membrane | G-protein coupled receptor | ||||
| ENSMUSG00000045763 | Basp1 | brain abundant, membrane attached signal protein 1 | MGI:1917600 | YES | YES | YES | Nucleus | transcription regulator | ||||
| ENSMUSG00000004892 | Bcan | brevican | MGI:1096385 | YES | YES | YES | Extracellular Space | other | ||||
| ENSMUSG00000002015 | Bcap31 | B cell receptor associated protein 31 | MGI:1350933 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSMUSG00000059439 | Bcas3 | breast carcinoma amplified sequence 3 | MGI:2385848 | YES | YES | YES | Nucleus | other | ||||
| ENSMUSG00000009681 | Bcr | breakpoint cluster region | MGI:88141 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSMUSG00000037905 | Bri3bp | Bri3 binding protein | MGI:1924059 | YES | YES | YES | Extracellular Space | other | ||||
| ENSMUSG00000028351 | Brinp1 | bone morphogenic protein/retinoic acid inducible neural specific 1 | MGI:1928478 | YES | YES | YES | Nucleus | peptidase | ||||
| ENSMUSG00000004031 | Brinp2 | bone morphogenic protein/retinoic acid inducible neural-specific 2 | MGI:2443333 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000035390 | Brsk1 | BR serine/threonine kinase 1 | MGI:2685946 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSMUSG00000053046 | Brsk2 | BR serine/threonine kinase 2 | MGI:1923020 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSMUSG00000032589 | Bsn | bassoon | MGI:1277955 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000036905 | C1qb | complement component 1, q subcomponent, beta polypeptide | MGI:88224 | YES | YES | YES | Extracellular Space | other | ||||
| ENSMUSG00000018446 | C1qbp | complement component 1, q subcomponent binding protein | MGI:1194505 | YES | YES | YES | Cytoplasm | transcription regulator | ||||
| ENSMUSG00000040794 | C1qtnf4 | C1q and tumor necrosis factor related protein 4 | MGI:1914695 | YES | YES | YES | Extracellular Space | other | ||||
| ENSMUSG00000045912 | C2cd4c | C2 calcium-dependent domain containing 4C | MGI:2685084 | YES | YES | YES | Extracellular Space | other | ||||
| ENSMUSG00000000805 | Car4 | carbonic anhydrase 4 | MGI:1096574 | YES | YES | YES | Plasma Membrane | enzyme | ||||
| ENSMUSG00000034656 | Cacna1a | calcium channel, voltage-dependent, P/Q type, alpha 1A subunit | MGI:109482 | YES | YES | YES | Plasma Membrane | ion channel | ||||
| ENSMUSG00000004113 | Cacna1b | calcium channel, voltage-dependent, N type, alpha 1B subunit | MGI:88296 | YES | YES | YES | Plasma Membrane | ion channel | ||||
| ENSMUSG00000040118 | Cacna2d1 | calcium channel, voltage-dependent, alpha2/delta subunit 1 | MGI:88295 | YES | YES | YES | Plasma Membrane | ion channel | ||||
| ENSMUSG00000010066 | Cacna2d2 | calcium channel, voltage-dependent, alpha 2/delta subunit 2 | MGI:1929813 | YES | YES | YES | Plasma Membrane | ion channel | ||||
| ENSMUSG00000021991 | Cacna2d3 | calcium channel, voltage-dependent, alpha2/delta subunit 3 | MGI:1338890 | YES | YES | YES | Plasma Membrane | ion channel | ||||
| ENSMUSG00000020882 | Cacnb1 | calcium channel, voltage-dependent, beta 1 subunit | MGI:102522 | YES | YES | YES | Other | ion channel | ||||
| ENSMUSG00000003352 | Cacnb3 | calcium channel, voltage-dependent, beta 3 subunit | MGI:103307 | YES | YES | YES | Plasma Membrane | ion channel | ||||
| ENSMUSG00000017412 | Cacnb4 | calcium channel, voltage-dependent, beta 4 subunit | MGI:103301 | YES | YES | YES | Plasma Membrane | ion channel | ||||
| ENSMUSG00000032936 | Camkv | CaM kinase-like vesicle-associated | MGI:2384296 | YES | YES | YES | Other | kinase | ||||
| ENSMUSG00000044433 | Camsap3 | calmodulin regulated spectrin-associated protein family, member 3 | MGI:1916947 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000021373 | Cap2 | CAP, adenylate cyclase-associated protein, 2 (yeast) | MGI:1914502 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000035547 | Capn5 | calpain 5 | MGI:1100859 | YES | YES | YES | Cytoplasm | peptidase | ||||
| ENSMUSG00000001794 | Capns1 | calpain, small subunit 1 | MGI:88266 | YES | YES | YES | Cytoplasm | peptidase | ||||
| ENSMUSG00000027184 | Caprin1 | cell cycle associated protein 1 | MGI:1858234 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000060227 | Casc4 | cancer susceptibility candidate 4 | MGI:2443129 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000033597 | Caskin1 | CASK interacting protein 1 | MGI:2442952 | YES | YES | YES | Nucleus | transcription regulator | ||||
| ENSMUSG00000051483 | Cbr1 | carbonyl reductase 1 | MGI:88284 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000029769 | Ccdc136 | coiled-coil domain containing 136 | MGI:1918128 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000059554 | Ccdc28a | coiled-coil domain containing 28A | MGI:2443508 | YES | YES | YES | Other | other | ||||
| ENSMUSG00000078622 | Ccdc47 | coiled-coil domain containing 47 | MGI:1914413 | YES | YES | YES | Extracellular Space | other | ||||
| ENSMUSG00000025645 | Ccdc51 | coiled-coil domain containing 51 | MGI:1913908 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000032740 | Ccdc88a | coiled coil domain containing 88A | MGI:1925177 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000034024 | Cct2 | chaperonin containing Tcp1, subunit 2 (beta) | MGI:107186 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSMUSG00000001416 | Cct3 | chaperonin containing Tcp1, subunit 3 (gamma) | MGI:104708 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000007739 | Cct4 | chaperonin containing Tcp1, subunit 4 (delta) | MGI:104689 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000022234 | Cct5 | chaperonin containing Tcp1, subunit 5 (epsilon) | MGI:107185 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000029447 | Cct6a | chaperonin containing Tcp1, subunit 6a (zeta) | MGI:107943 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000030007 | Cct7 | chaperonin containing Tcp1, subunit 7 (eta) | MGI:107184 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000025613 | Cct8 | chaperonin containing Tcp1, subunit 8 (theta) | MGI:107183 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000029617 | Ccz1 | CCZ1 vacuolar protein trafficking and biogenesis associated | MGI:2141070 | YES | YES | YES | Other | other | ||||
| ENSMUSG00000055447 | Cd47 | CD47 antigen (Rh-related antigen, integrin-associated signal transducer) | MGI:96617 | YES | YES | YES | Plasma Membrane | transmembrane receptor | ||||
| ENSMUSG00000037706 | Cd81 | CD81 antigen | MGI:1096398 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000026490 | Cdc42bpa | CDC42 binding protein kinase alpha | MGI:2441841 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSMUSG00000021279 | Cdc42bpb | CDC42 binding protein kinase beta | MGI:2136459 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSMUSG00000049521 | Cdc42ep1 | CDC42 effector protein (Rho GTPase binding) 1 | MGI:1929763 | YES | YES | YES | Extracellular Space | other | ||||
| ENSMUSG00000041598 | Cdc42ep4 | CDC42 effector protein (Rho GTPase binding) 4 | MGI:1929760 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000052298 | Cdc42se2 | CDC42 small effector 2 | MGI:1919979 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000022321 | Cdh10 | cadherin 10 | MGI:107436 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000031673 | Cdh11 | cadherin 11 | MGI:99217 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000031841 | Cdh13 | cadherin 13 | MGI:99551 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000040420 | Cdh18 | cadherin 18 | MGI:1344366 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000024304 | Cdh2 | cadherin 2 | MGI:88355 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000039385 | Cdh6 | cadherin 6 | MGI:107435 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000030682 | Cdipt | CDP-diacylglycerol--inositol 3-phosphatidyltransferase (phosphatidylinositol synthase) | MGI:105491 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000020015 | Cdk17 | cyclin-dependent kinase 17 | MGI:97517 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSMUSG00000028969 | Cdk5 | cyclin-dependent kinase 5 | MGI:101765 | YES | YES | YES | Nucleus | kinase | ||||
| ENSMUSG00000018669 | Cdk5rap3 | CDK5 regulatory subunit associated protein 3 | MGI:1933126 | YES | YES | YES | Nucleus | other | ||||
| ENSMUSG00000002107 | Celf2 | CUGBP, Elav-like family member 2 | MGI:1338822 | YES | YES | YES | Nucleus | other | ||||
| ENSMUSG00000060240 | Cend1 | cell cycle exit and neuronal differentiation 1 | MGI:1929898 | YES | YES | YES | Other | other | ||||
| ENSMUSG00000057335 | Cep170 | centrosomal protein 170 | MGI:1918348 | YES | YES | YES | Nucleus | other | ||||
| ENSMUSG00000072825 | Cep170b | centrosomal protein 170B | MGI:2145043 | YES | YES | YES | Other | other | ||||
| ENSMUSG00000031347 | Cetn2 | centrin 2 | MGI:1347085 | YES | YES | YES | Nucleus | enzyme | ||||
| ENSMUSG00000056201 | Cfl1 | cofilin 1, non-muscle | MGI:101757 | YES | YES | YES | Nucleus | other | ||||
| ENSMUSG00000062929 | Cfl2 | cofilin 2, muscle | MGI:101763 | YES | YES | YES | Extracellular Space | other | ||||
| ENSMUSG00000030086 | Chchd6 | coiled-coil-helix-coiled-coil-helix domain containing 6 | MGI:1913348 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000004843 | Chmp2b | charged multivesicular body protein 2B | MGI:1916192 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000014077 | Chp1 | calcineurin-like EF hand protein 1 | MGI:1927185 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSMUSG00000032773 | Chrm1 | cholinergic receptor, muscarinic 1, CNS | MGI:88396 | YES | YES | YES | Plasma Membrane | G-protein coupled receptor | ||||
| ENSMUSG00000046159 | Chrm3 | cholinergic receptor, muscarinic 3, cardiac | MGI:88398 | YES | YES | YES | Plasma Membrane | G-protein coupled receptor | ||||
| ENSMUSG00000045193 | Cirbp | cold inducible RNA binding protein | MGI:893588 | YES | YES | YES | Nucleus | translation regulator | ||||
| ENSMUSG00000037710 | Cisd1 | CDGSH iron sulfur domain 1 | MGI:1261855 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000046841 | Ckap4 | cytoskeleton-associated protein 4 | MGI:2444926 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000040549 | Ckap5 | cytoskeleton associated protein 5 | MGI:1923036 | YES | YES | YES | Nucleus | transcription regulator | ||||
| ENSMUSG00000000308 | Ckmt1 | creatine kinase, mitochondrial 1, ubiquitous | MGI:99441 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSMUSG00000064302 | Clasp1 | CLIP associating protein 1 | MGI:1923957 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000033392 | Clasp2 | CLIP associating protein 2 | MGI:1923749 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000022843 | Clcn2 | chloride channel 2 | MGI:105061 | YES | YES | YES | Plasma Membrane | ion channel | ||||
| ENSMUSG00000029016 | Clcn6 | chloride channel 6 | MGI:1347049 | YES | YES | YES | Plasma Membrane | ion channel | ||||
| ENSMUSG00000037625 | Cldn11 | claudin 11 | MGI:106925 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000006169 | Clint1 | clathrin interactor 1 | MGI:2144243 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000049550 | Clip1 | CAP-GLY domain containing linker protein 1 | MGI:1928401 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000063146 | Clip2 | CAP-GLY domain containing linker protein 2 | MGI:1313136 | YES | YES | YES | Cytoplasm | transcription regulator | ||||
| ENSMUSG00000013921 | Clip3 | CAP-GLY domain containing linker protein 3 | MGI:1923936 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000001829 | Clpb | ClpB caseinolytic peptidase B | MGI:1100517 | YES | YES | YES | Nucleus | transcription regulator | ||||
| ENSMUSG00000028478 | Clta | clathrin, light polypeptide (Lca) | MGI:894297 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000047547 | Cltb | clathrin, light polypeptide (Lcb) | MGI:1921575 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000022037 | Clu | clusterin | MGI:88423 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000028719 | Cmpk1 | cytidine monophosphate (UMP-CMP) kinase 1 | MGI:1913838 | YES | YES | YES | Nucleus | kinase | ||||
| ENSMUSG00000025658 | Cnksr2 | connector enhancer of kinase suppressor of Ras 2 | MGI:2661175 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000028444 | Cntfr | ciliary neurotrophic factor receptor | MGI:99605 | YES | YES | YES | Plasma Membrane | transmembrane receptor | ||||
| ENSMUSG00000055022 | Cntn1 | contactin 1 | MGI:105980 | YES | YES | YES | Plasma Membrane | enzyme | ||||
| ENSMUSG00000053024 | Cntn2 | contactin 2 | MGI:104518 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000030075 | Cntn3 | contactin 3 | MGI:99534 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000064293 | Cntn4 | contactin 4 | MGI:1095737 | YES | YES | YES | Plasma Membrane | enzyme | ||||
| ENSMUSG00000039488 | Cntn5 | contactin 5 | MGI:3042287 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000030092 | Cntn6 | contactin 6 | MGI:1858223 | YES | YES | YES | Plasma Membrane | enzyme | ||||
| ENSMUSG00000017167 | Cntnap1 | contactin associated protein-like 1 | MGI:1858201 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000017188 | Coa3 | cytochrome C oxidase assembly factor 3 | MGI:1098757 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000051355 | Commd1 | COMM domain containing 1 | MGI:109474 | YES | YES | YES | Nucleus | transporter | ||||
| ENSMUSG00000042705 | Commd10 | COMM domain containing 10 | MGI:1916706 | YES | YES | YES | Other | other | ||||
| ENSMUSG00000027163 | Commd9 | COMM domain containing 9 | MGI:1923751 | YES | YES | YES | Other | other | ||||
| ENSMUSG00000021773 | Comtd1 | catechol-O-methyltransferase domain containing 1 | MGI:1916406 | YES | YES | YES | Plasma Membrane | enzyme | ||||
| ENSMUSG00000026553 | Copa | coatomer protein complex subunit alpha | MGI:1334462 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSMUSG00000030058 | Copg1 | coatomer protein complex, subunit gamma 1 | MGI:1858696 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSMUSG00000019373 | Cops3 | COP9 (constitutive photomorphogenic) homolog, subunit 3 (Arabidopsis thaliana) | MGI:1349409 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000035297 | Cops4 | COP9 (constitutive photomorphogenic) homolog, subunit 4 (Arabidopsis thaliana) | MGI:1349414 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000030127 | Cops7a | COP9 (constitutive photomorphogenic) homolog, subunit 7a (Arabidopsis thaliana) | MGI:1349400 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000034432 | Cops8 | COP9 (constitutive photomorphogenic) homolog, subunit 8 (Arabidopsis thaliana) | MGI:1915363 | YES | YES | YES | Nucleus | other | ||||
| ENSMUSG00000060992 | Copz1 | coatomer protein complex, subunit zeta 1 | MGI:1929063 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSMUSG00000024835 | Coro1b | coronin, actin binding protein 1B | MGI:1345963 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000031818 | Cox4i1 | cytochrome c oxidase subunit IV isoform 1 | MGI:88473 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000041697 | Cox6a1 | cytochrome c oxidase subunit VIa polypeptide 1 | MGI:103099 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000024248 | Cox7a2l | cytochrome c oxidase subunit VIIa polypeptide 2-like | MGI:106015 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000024008 | Cpne5 | copine V | MGI:2385908 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000022212 | Cpne6 | copine VI | MGI:1334445 | YES | YES | YES | Plasma Membrane | transporter | ||||
| ENSMUSG00000055531 | Cpsf6 | cleavage and polyadenylation specific factor 6 | MGI:1913948 | YES | YES | YES | Nucleus | other | ||||
| ENSMUSG00000024900 | Cpt1a | carnitine palmitoyltransferase 1a, liver | MGI:1098296 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000017776 | Crk | v-crk sarcoma virus CT10 oncogene homolog (avian) | MGI:88508 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000040860 | Crocc | ciliary rootlet coiled-coil, rootletin | MGI:3529431 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000042401 | Crtac1 | cartilage acidic protein 1 | MGI:1920082 | YES | YES | YES | Extracellular Space | other | ||||
| ENSMUSG00000068823 | Csde1 | cold shock domain containing E1, RNA binding | MGI:92356 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000024576 | Csnk1a1 | casein kinase 1, alpha 1 | MGI:1934950 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSMUSG00000032384 | Csnk1g1 | casein kinase 1, gamma 1 | MGI:2660884 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSMUSG00000003345 | Csnk1g2 | casein kinase 1, gamma 2 | MGI:1920014 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSMUSG00000073563 | Csnk1g3 | casein kinase 1, gamma 3 | MGI:1917675 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSMUSG00000024387 | Csnk2b | casein kinase 2, beta polypeptide | MGI:88548 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSMUSG00000026421 | Csrp1 | cysteine and glycine-rich protein 1 | MGI:88549 | YES | YES | YES | Nucleus | other | ||||
| ENSMUSG00000027447 | Cst3 | cystatin C | MGI:102519 | YES | YES | YES | Extracellular Space | other | ||||
| ENSMUSG00000037373 | Ctbp1 | C-terminal binding protein 1 | MGI:1201685 | YES | YES | YES | Nucleus | enzyme | ||||
| ENSMUSG00000037815 | Ctnna1 | catenin (cadherin associated protein), alpha 1 | MGI:88274 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000063063 | Ctnna2 | catenin (cadherin associated protein), alpha 2 | MGI:88275 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000006932 | Ctnnb1 | catenin (cadherin associated protein), beta 1 | MGI:88276 | YES | YES | YES | Nucleus | transcription regulator | ||||
| ENSMUSG00000022240 | Ctnnd2 | catenin (cadherin associated protein), delta 2 | MGI:1195966 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000031078 | Cttn | cortactin | MGI:99695 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000029686 | Cul1 | cullin 1 | MGI:1349658 | YES | YES | YES | Nucleus | enzyme | ||||
| ENSMUSG00000004364 | Cul3 | cullin 3 | MGI:1347360 | YES | YES | YES | Nucleus | enzyme | ||||
| ENSMUSG00000032030 | Cul5 | cullin 5 | MGI:1922967 | YES | YES | YES | Nucleus | ion channel | ||||
| ENSMUSG00000031924 | Cyb5b | cytochrome b5 type B | MGI:1913677 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000027015 | Cybrd1 | cytochrome b reductase 1 | MGI:2654575 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000022551 | Cyc1 | cytochrome c-1 | MGI:1913695 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000030447 | Cyfip1 | cytoplasmic FMR1 interacting protein 1 | MGI:1338801 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000020340 | Cyfip2 | cytoplasmic FMR1 interacting protein 2 | MGI:1924134 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000021259 | Cyp46a1 | cytochrome P450, family 46, subfamily a, polypeptide 1 | MGI:1341877 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000053329 | D10Jhu81e | DNA segment, Chr 10, Johns Hopkins University 81 expressed | MGI:1351861 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000034574 | Daam1 | dishevelled associated activator of morphogenesis 1 | MGI:1914596 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000040260 | Daam2 | dishevelled associated activator of morphogenesis 2 | MGI:1923691 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000028519 | Dab1 | disabled 1 | MGI:108554 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000035735 | Dagla | diacylglycerol lipase, alpha | MGI:2677061 | YES | YES | YES | Plasma Membrane | enzyme | ||||
| ENSMUSG00000042096 | Dao | D-amino acid oxidase | MGI:94859 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000026356 | Dars | aspartyl-tRNA synthetase | MGI:2442544 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000034675 | Dbn1 | drebrin 1 | MGI:1931838 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000020476 | Dbnl | drebrin-like | MGI:700006 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000000340 | Dbt | dihydrolipoamide branched chain transacylase E2 | MGI:105386 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000020935 | Dcakd | dephospho-CoA kinase domain containing | MGI:1915337 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000025410 | Dctn2 | dynactin 2 | MGI:107733 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000028447 | Dctn3 | dynactin 3 | MGI:1859251 | YES | YES | YES | Nucleus | other | ||||
| ENSMUSG00000024603 | Dctn4 | dynactin 4 | MGI:1914915 | YES | YES | YES | Nucleus | other | ||||
| ENSMUSG00000030868 | Dctn5 | dynactin 5 | MGI:1891689 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000024740 | Ddb1 | damage specific DNA binding protein 1 | MGI:1202384 | YES | YES | YES | Nucleus | other | ||||
| ENSMUSG00000028757 | Ddost | dolichyl-di-phosphooligosaccharide-protein glycotransferase | MGI:1194508 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000068290 | Ddrgk1 | DDRGK domain containing 1 | MGI:1924256 | YES | YES | YES | Extracellular Space | other | ||||
| ENSMUSG00000037149 | Ddx1 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 1 | MGI:2144727 | YES | YES | YES | Nucleus | enzyme | ||||
| ENSMUSG00000000787 | Ddx3x | DEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide 3, X-linked | MGI:103064 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000069045 | Ddx3y | DEAD (Asp-Glu-Ala-Asp) box polypeptide 3, Y-linked | MGI:1349406 | YES | YES | YES | Other | enzyme | ||||
| ENSMUSG00000032097 | Ddx6 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 6 | MGI:104976 | YES | YES | YES | Nucleus | enzyme | ||||
| ENSMUSG00000028223 | Decr1 | 2,4-dienoyl CoA reductase 1, mitochondrial | MGI:1914710 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000035392 | Dennd1a | DENN/MADD domain containing 1A | MGI:2442794 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000000276 | Dgke | diacylglycerol kinase, epsilon | MGI:1889276 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSMUSG00000022861 | Dgkg | diacylglycerol kinase, gamma | MGI:105060 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSMUSG00000020834 | Dhrs13 | dehydrogenase/reductase (SDR family) member 13 | MGI:1917701 | YES | YES | YES | Other | enzyme | ||||
| ENSMUSG00000023026 | Dip2b | DIP2 disco-interacting protein 2 homolog B (Drosophila) | MGI:2145977 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000043670 | Diras1 | DIRAS family, GTP-binding RAS-like 1 | MGI:2183442 | YES | YES | YES | Plasma Membrane | enzyme | ||||
| ENSMUSG00000047842 | Diras2 | DIRAS family, GTP-binding RAS-like 2 | MGI:1915453 | YES | YES | YES | Plasma Membrane | enzyme | ||||
| ENSMUSG00000020664 | Dld | dihydrolipoamide dehydrogenase | MGI:107450 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000022770 | Dlg1 | discs, large homolog 1 (Drosophila) | MGI:107231 | YES | YES | YES | Plasma Membrane | kinase | ||||
| ENSMUSG00000004789 | Dlst | dihydrolipoamide S-succinyltransferase (E2 component of 2-oxo-glutarate complex) | MGI:1926170 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000030410 | Dmwd | dystrophia myotonica-containing WD repeat motif | MGI:94907 | YES | YES | YES | Nucleus | other | ||||
| ENSMUSG00000005483 | Dnajb1 | DnaJ (Hsp40) homolog, subfamily B, member 1 | MGI:1931874 | YES | YES | YES | Nucleus | other | ||||
| ENSMUSG00000004460 | Dnajb11 | DnaJ (Hsp40) homolog, subfamily B, member 11 | MGI:1915088 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000020109 | Dnajb12 | DnaJ (Hsp40) homolog, subfamily B, member 12 | MGI:1931881 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000026203 | Dnajb2 | DnaJ (Hsp40) homolog, subfamily B, member 2 | MGI:1928739 | YES | YES | YES | Nucleus | other | ||||
| ENSMUSG00000027006 | Dnajc10 | DnaJ (Hsp40) homolog, subfamily C, member 10 | MGI:1914111 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000040697 | Dnajc16 | DnaJ (Hsp40) homolog, subfamily C, member 16 | MGI:2442146 | YES | YES | YES | Other | other | ||||
| ENSMUSG00000027679 | Dnajc19 | DnaJ (Hsp40) homolog, subfamily C, member 19 | MGI:1914963 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000026825 | Dnm1 | dynamin 1 | MGI:107384 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000022789 | Dnm1l | dynamin 1-like | MGI:1921256 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000033335 | Dnm2 | dynamin 2 | MGI:109547 | YES | YES | YES | Plasma Membrane | enzyme | ||||
| ENSMUSG00000040265 | Dnm3 | dynamin 3 | MGI:1341299 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000058325 | Dock1 | dedicator of cytokinesis 1 | MGI:2429765 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000039716 | Dock3 | dedicator of cyto-kinesis 3 | MGI:2429763 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000035954 | Dock4 | dedicator of cytokinesis 4 | MGI:1918006 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000028556 | Dock7 | dedicator of cytokinesis 7 | MGI:1914549 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000025558 | Dock9 | dedicator of cytokinesis 9 | MGI:106321 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000020537 | Drg2 | developmentally regulated GTP binding protein 2 | MGI:1342307 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000103888 | Dst | dystonin | MGI:104627 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000003518 | Dusp3 | dual specificity phosphatase 3 (vaccinia virus phosphatase VH1-related) | MGI:1919599 | YES | YES | YES | Cytoplasm | phosphatase | ||||
| ENSMUSG00000029757 | Dync1i1 | dynein cytoplasmic 1 intermediate chain 1 | MGI:107743 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000027012 | Dync1i2 | dynein cytoplasmic 1 intermediate chain 2 | MGI:107750 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000032435 | Dync1li1 | dynein cytoplasmic 1 light intermediate chain 1 | MGI:2135610 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000035770 | Dync1li2 | dynein, cytoplasmic 1 light intermediate chain 2 | MGI:107738 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000009013 | Dynll1 | dynein light chain LC8-type 1 | MGI:1861457 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000020483 | Dynll2 | dynein light chain LC8-type 2 | MGI:1915347 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000025465 | Echs1 | enoyl Coenzyme A hydratase, short chain, 1, mitochondrial | MGI:2136460 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000066839 | Ecsit | ECSIT homolog (Drosophila) | MGI:1349469 | YES | YES | YES | Nucleus | transcription regulator | ||||
| ENSMUSG00000015092 | Edf1 | endothelial differentiation-related factor 1 | MGI:1891227 | YES | YES | YES | Nucleus | transcription regulator | ||||
| ENSMUSG00000034488 | Edil3 | EGF-like repeats and discoidin I-like domains 3 | MGI:1329025 | YES | YES | YES | Extracellular Space | other | ||||
| ENSMUSG00000037742 | Eef1a1 | eukaryotic translation elongation factor 1 alpha 1 | MGI:1096881 | YES | YES | YES | Cytoplasm | translation regulator | ||||
| ENSMUSG00000016349 | Eef1a2 | eukaryotic translation elongation factor 1 alpha 2 | MGI:1096317 | YES | YES | YES | Cytoplasm | translation regulator | ||||
| ENSMUSG00000055762 | Eef1d | eukaryotic translation elongation factor 1 delta (guanine nucleotide exchange protein) | MGI:1913906 | YES | YES | YES | Cytoplasm | translation regulator | ||||
| ENSMUSG00000001707 | Eef1e1 | eukaryotic translation elongation factor 1 epsilon 1 | MGI:1913393 | YES | YES | YES | Cytoplasm | translation regulator | ||||
| ENSMUSG00000071644 | Eef1g | eukaryotic translation elongation factor 1 gamma | MGI:1914410 | YES | YES | YES | Cytoplasm | translation regulator | ||||
| ENSMUSG00000036611 | Eepd1 | endonuclease/exonuclease/phosphatase family domain containing 1 | MGI:1914734 | YES | YES | YES | Other | other | ||||
| ENSMUSG00000020658 | Efr3b | EFR3 homolog B (S. cerevisiae) | MGI:2444851 | YES | YES | YES | Other | other | ||||
| ENSMUSG00000024772 | Ehd1 | EH-domain containing 1 | MGI:1341878 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000024065 | Ehd3 | EH-domain containing 3 | MGI:1928900 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000035150 | Eif2s3x | eukaryotic translation initiation factor 2, subunit 3, structural gene X-linked | MGI:1349431 | YES | YES | YES | Cytoplasm | translation regulator | ||||
| ENSMUSG00000022884 | Eif4a2 | eukaryotic translation initiation factor 4A2 | MGI:106906 | YES | YES | YES | Cytoplasm | translation regulator | ||||
| ENSMUSG00000045983 | Eif4g1 | eukaryotic translation initiation factor 4, gamma 1 | MGI:2384784 | YES | YES | YES | Cytoplasm | translation regulator | ||||
| ENSMUSG00000048988 | Elfn1 | leucine rich repeat and fibronectin type III, extracellular 1 | MGI:2442479 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000043460 | Elfn2 | leucine rich repeat and fibronectin type III, extracellular 2 | MGI:3608416 | YES | YES | YES | Extracellular Space | other | ||||
| ENSMUSG00000041986 | Elmod1 | ELMO/CED-12 domain containing 1 | MGI:3583900 | YES | YES | YES | Other | other | ||||
| ENSMUSG00000078517 | Emc1 | ER membrane protein complex subunit 1 | MGI:2443696 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000022337 | Emc2 | ER membrane protein complex subunit 2 | MGI:1913986 | YES | YES | YES | Nucleus | other | ||||
| ENSMUSG00000030286 | Emc3 | ER membrane protein complex subunit 3 | MGI:1913337 | YES | YES | YES | Nucleus | other | ||||
| ENSMUSG00000055943 | Emc7 | ER membrane protein complex subunit 7 | MGI:1920274 | YES | YES | YES | Other | other | ||||
| ENSMUSG00000001964 | Emd | emerin | MGI:108117 | YES | YES | YES | Nucleus | other | ||||
| ENSMUSG00000024044 | Epb4.1l3 | erythrocyte protein band 4.1 like 3 | MGI:103008 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000002808 | Epdr1 | ependymin related protein 1 (zebrafish) | MGI:2145369 | YES | YES | YES | Nucleus | other | ||||
| ENSMUSG00000026235 | Epha4 | Eph receptor A4 | MGI:98277 | YES | YES | YES | Plasma Membrane | kinase | ||||
| ENSMUSG00000021709 | Erbb2ip | Erbb2 interacting protein | MGI:1890169 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000031483 | Erlin2 | ER lipid raft associated 2 | MGI:2387215 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000026830 | Ermn | ermin, ERM-like protein | MGI:1925017 | YES | YES | YES | Extracellular Space | other | ||||
| ENSMUSG00000021171 | Esyt2 | extended synaptotagmin-like protein 2 | MGI:1261845 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000024360 | Etf1 | eukaryotic translation termination factor 1 | MGI:2385071 | YES | YES | YES | Cytoplasm | translation regulator | ||||
| ENSMUSG00000011832 | Evi5l | ecotropic viral integration site 5 like | MGI:2442167 | YES | YES | YES | Other | other | ||||
| ENSMUSG00000032705 | Exd2 | exonuclease 3'-5' domain containing 2 | MGI:1922485 | YES | YES | YES | Other | other | ||||
| ENSMUSG00000036435 | Exoc1 | exocyst complex component 1 | MGI:2445020 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSMUSG00000021357 | Exoc2 | exocyst complex component 2 | MGI:1913732 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSMUSG00000029763 | Exoc4 | exocyst complex component 4 | MGI:1096376 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSMUSG00000061244 | Exoc5 | exocyst complex component 5 | MGI:2145645 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000033769 | Exoc6b | exocyst complex component 6B | MGI:1923164 | YES | YES | YES | Other | other | ||||
| ENSMUSG00000074030 | Exoc8 | exocyst complex component 8 | MGI:2142527 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000028128 | F3 | coagulation factor III | MGI:88381 | YES | YES | YES | Plasma Membrane | transmembrane receptor | ||||
| ENSMUSG00000034171 | Faah | fatty acid amide hydrolase | MGI:109609 | YES | YES | YES | Plasma Membrane | enzyme | ||||
| ENSMUSG00000045316 | Fahd1 | fumarylacetoacetate hydrolase domain containing 1 | MGI:1915886 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000023011 | Faim2 | Fas apoptotic inhibitory molecule 2 | MGI:1919643 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000038014 | Fam120a | family with sequence similarity 120, member A | MGI:2446163 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000025262 | Fam120c | family with sequence similarity 120, member C | MGI:2387687 | YES | YES | YES | Other | other | ||||
| ENSMUSG00000038174 | Fam126b | family with sequence similarity 126, member B | MGI:1098784 | YES | YES | YES | Other | other | ||||
| ENSMUSG00000029861 | Fam131b | family with sequence similarity 131, member B | MGI:1923406 | YES | YES | YES | Other | other | ||||
| ENSMUSG00000003955 | Fam162a | family with sequence similarity 162, member A | MGI:1917436 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000050530 | Fam171a1 | family with sequence similarity 171, member A1 | MGI:2442917 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000034685 | Fam171a2 | family with sequence similarity 171, member A2 | MGI:2448496 | YES | YES | YES | Other | other | ||||
| ENSMUSG00000048388 | Fam171b | family with sequence similarity 171, member B | MGI:2444579 | YES | YES | YES | Other | other | ||||
| ENSMUSG00000021792 | Fam213a | family with sequence similarity 213, member A | MGI:1917814 | YES | YES | YES | Extracellular Space | other | ||||
| ENSMUSG00000029059 | Fam213b | family with sequence similarity 213, member B | MGI:1913719 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000058966 | Fam57b | family with sequence similarity 57, member B | MGI:1916202 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000025555 | Farp1 | FERM, RhoGEF (Arhgef) and pleckstrin domain protein 1 (chondrocyte-derived) | MGI:2446173 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000026245 | Farsb | phenylalanyl-tRNA synthetase, beta subunit | MGI:1346035 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000070047 | Fat1 | FAT tumor suppressor homolog 1 (Drosophila) | MGI:109168 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000055333 | Fat2 | FAT tumor suppressor homolog 2 (Drosophila) | MGI:2685369 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000074505 | Fat3 | FAT tumor suppressor homolog 3 (Drosophila) | MGI:2444314 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000041556 | Fbxo2 | F-box protein 2 | MGI:2446216 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000047013 | Fbxo41 | F-box protein 41 | MGI:1261912 | YES | YES | YES | Other | other | ||||
| ENSMUSG00000024588 | Fech | ferrochelatase | MGI:95513 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000022523 | Fgf12 | fibroblast growth factor 12 | MGI:109183 | YES | YES | YES | Extracellular Space | other | ||||
| ENSMUSG00000040242 | Fgfr1op2 | FGFR1 oncogene partner 2 | MGI:1914779 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000024911 | Fibp | fibroblast growth factor (acidic) intracellular binding protein | MGI:1926233 | YES | YES | YES | Nucleus | other | ||||
| ENSMUSG00000020635 | Fkbp1b | FK506 binding protein 1b | MGI:1336205 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000019428 | Fkbp8 | FK506 binding protein 8 | MGI:1341070 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000002812 | Flii | flightless I homolog (Drosophila) | MGI:1342286 | YES | YES | YES | Nucleus | other | ||||
| ENSMUSG00000059714 | Flot1 | flotillin 1 | MGI:1100500 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000047414 | Flrt2 | fibronectin leucine rich transmembrane protein 2 | MGI:3603594 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000028354 | Fmn2 | formin 2 | MGI:1859252 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000055805 | Fmnl1 | formin-like 1 | MGI:1888994 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000036053 | Fmnl2 | formin-like 2 | MGI:1918659 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000000838 | Fmr1 | fragile X mental retardation syndrome 1 | MGI:95564 | YES | YES | YES | Nucleus | other | ||||
| ENSMUSG00000045589 | Frrs1l | ferric-chelate reductase 1 like | MGI:2442704 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSMUSG00000023266 | Frs3 | fibroblast growth factor receptor substrate 3 | MGI:2135965 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000056602 | Fry | furry homolog (Drosophila) | MGI:2443895 | YES | YES | YES | Extracellular Space | other | ||||
| ENSMUSG00000011589 | Fsd1 | fibronectin type 3 and SPRY domain-containing protein | MGI:1934858 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000024661 | Fth1 | ferritin heavy chain 1 | MGI:95588 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000025040 | Fundc1 | FUN14 domain containing 1 | MGI:1919268 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000031198 | Fundc2 | FUN14 domain containing 2 | MGI:1914641 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000031950 | Gabarapl2 | gamma-aminobutyric acid (GABA) A receptor-associated protein-like 2 | MGI:1890602 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000039809 | Gabbr2 | gamma-aminobutyric acid (GABA) B receptor, 2 | MGI:2386030 | YES | YES | YES | Plasma Membrane | G-protein coupled receptor | ||||
| ENSMUSG00000010803 | Gabra1 | gamma-aminobutyric acid (GABA) A receptor, subunit alpha 1 | MGI:95613 | YES | YES | YES | Plasma Membrane | ion channel | ||||
| ENSMUSG00000000560 | Gabra2 | gamma-aminobutyric acid (GABA) A receptor, subunit alpha 2 | MGI:95614 | YES | YES | YES | Plasma Membrane | ion channel | ||||
| ENSMUSG00000031343 | Gabra3 | gamma-aminobutyric acid (GABA) A receptor, subunit alpha 3 | MGI:95615 | YES | YES | YES | Plasma Membrane | ion channel | ||||
| ENSMUSG00000029211 | Gabra4 | gamma-aminobutyric acid (GABA) A receptor, subunit alpha 4 | MGI:95616 | YES | YES | YES | Plasma Membrane | ion channel | ||||
| ENSMUSG00000020428 | Gabra6 | gamma-aminobutyric acid (GABA) A receptor, subunit alpha 6 | MGI:95618 | YES | YES | YES | Plasma Membrane | ion channel | ||||
| ENSMUSG00000029212 | Gabrb1 | gamma-aminobutyric acid (GABA) A receptor, subunit beta 1 | MGI:95619 | YES | YES | YES | Plasma Membrane | ion channel | ||||
| ENSMUSG00000007653 | Gabrb2 | gamma-aminobutyric acid (GABA) A receptor, subunit beta 2 | MGI:95620 | YES | YES | YES | Plasma Membrane | ion channel | ||||
| ENSMUSG00000033676 | Gabrb3 | gamma-aminobutyric acid (GABA) A receptor, subunit beta 3 | MGI:95621 | YES | YES | YES | Plasma Membrane | ion channel | ||||
| ENSMUSG00000020436 | Gabrg2 | gamma-aminobutyric acid (GABA) A receptor, subunit gamma 2 | MGI:95623 | YES | YES | YES | Plasma Membrane | ion channel | ||||
| ENSMUSG00000062234 | Gak | cyclin G associated kinase | MGI:2442153 | YES | YES | YES | Nucleus | kinase | ||||
| ENSMUSG00000057666 | Gapdh | glyceraldehyde-3-phosphate dehydrogenase | MGI:95640 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000033066 | Gas7 | growth arrest specific 7 | MGI:1202388 | YES | YES | YES | Cytoplasm | transcription regulator | ||||
| ENSMUSG00000003809 | Gcdh | glutaryl-Coenzyme A dehydrogenase | MGI:104541 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000041638 | Gcn1l1 | GCN1 general control of amino-acid synthesis 1-like 1 (yeast) | MGI:2444248 | YES | YES | YES | Cytoplasm | translation regulator | ||||
| ENSMUSG00000025777 | Gdap1 | ganglioside-induced differentiation-associated-protein 1 | MGI:1338002 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000017943 | Gdap1l1 | ganglioside-induced differentiation-associated protein 1-like 1 | MGI:2385163 | YES | YES | YES | Other | other | ||||
| ENSMUSG00000019433 | Gipc1 | GIPC PDZ domain containing family, member 1 | MGI:1926252 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000011877 | Git1 | G protein-coupled receptor kinase-interactor 1 | MGI:1927140 | YES | YES | YES | Nucleus | kinase | ||||
| ENSMUSG00000011884 | Gltp | glycolipid transfer protein | MGI:1929253 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSMUSG00000021794 | Glud1 | glutamate dehydrogenase 1 | MGI:95753 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000057262 | Gm10020 | predicted pseudogene 10020 | MGI:3642192 | YES | YES | YES | Other | other | ||||
| ENSMUSG00000042744 | Gm15800 | predicted gene 15800 | MGI:3647820 | YES | YES | YES | Nucleus | other | ||||
| ENSMUSG00000091228 | Gm20390 | predicted gene 20390 | MGI:5141855 | YES | YES | YES | #N/A | #N/A | ||||
| ENSMUSG00000090639 | Gm20425 | predicted gene 20425 | MGI:5141890 | YES | YES | YES | #N/A | #N/A | ||||
| ENSMUSG00000100725 | Gm28062 | predicted gene 28062 | MGI:5578768 | YES | YES | YES | Nucleus | transporter | ||||
| ENSMUSG00000063696 | Gm8730 | predicted pseudogene 8730 | MGI:3644565 | YES | YES | YES | Other | other | ||||
| ENSMUSG00000044424 | Gm9493 | predicted gene 9493 | MGI:3779903 | YES | YES | YES | Other | other | ||||
| ENSMUSG00000062014 | Gmfb | glia maturation factor, beta | MGI:1927133 | YES | YES | YES | Cytoplasm | growth factor | ||||
| ENSMUSG00000000149 | Gna12 | guanine nucleotide binding protein, alpha 12 | MGI:95767 | YES | YES | YES | Plasma Membrane | enzyme | ||||
| ENSMUSG00000057614 | Gnai1 | guanine nucleotide binding protein (G protein), alpha inhibiting 1 | MGI:95771 | YES | YES | YES | Plasma Membrane | enzyme | ||||
| ENSMUSG00000032562 | Gnai2 | guanine nucleotide binding protein (G protein), alpha inhibiting 2 | MGI:95772 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000000001 | Gnai3 | guanine nucleotide binding protein (G protein), alpha inhibiting 3 | MGI:95773 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000024524 | Gnal | guanine nucleotide binding protein, alpha stimulating, olfactory type | MGI:95774 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000031748 | Gnao1 | guanine nucleotide binding protein, alpha O | MGI:95775 | YES | YES | YES | Plasma Membrane | enzyme | ||||
| ENSMUSG00000024639 | Gnaq | guanine nucleotide binding protein, alpha q polypeptide | MGI:95776 | YES | YES | YES | Plasma Membrane | enzyme | ||||
| ENSMUSG00000029713 | Gnb2 | guanine nucleotide binding protein (G protein), beta 2 | MGI:95784 | YES | YES | YES | Plasma Membrane | enzyme | ||||
| ENSMUSG00000027669 | Gnb4 | guanine nucleotide binding protein (G protein), beta 4 | MGI:104581 | YES | YES | YES | Plasma Membrane | enzyme | ||||
| ENSMUSG00000032192 | Gnb5 | guanine nucleotide binding protein (G protein), beta 5 | MGI:101848 | YES | YES | YES | Plasma Membrane | enzyme | ||||
| ENSMUSG00000025739 | Gng13 | guanine nucleotide binding protein (G protein), gamma 13 | MGI:1925616 | YES | YES | YES | Plasma Membrane | enzyme | ||||
| ENSMUSG00000043004 | Gng2 | guanine nucleotide binding protein (G protein), gamma 2 | MGI:102705 | YES | YES | YES | Plasma Membrane | enzyme | ||||
| ENSMUSG00000021303 | Gng4 | guanine nucleotide binding protein (G protein), gamma 4 | MGI:102703 | YES | YES | YES | Plasma Membrane | enzyme | ||||
| ENSMUSG00000031985 | Gnpat | glyceronephosphate O-acyltransferase | MGI:1343460 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000015341 | Golga7 | golgi autoantigen, golgin subfamily a, 7 | MGI:1931029 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000034243 | Golgb1 | golgi autoantigen, golgin subfamily b, macrogolgin 1 | MGI:1099447 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000010392 | Gosr1 | golgi SNAP receptor complex member 1 | MGI:1858260 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSMUSG00000034220 | Gpc1 | glypican 1 | MGI:1194891 | YES | YES | YES | Plasma Membrane | transmembrane receptor | ||||
| ENSMUSG00000045967 | Gpr158 | G protein-coupled receptor 158 | MGI:2441697 | YES | YES | YES | Plasma Membrane | G-protein coupled receptor | ||||
| ENSMUSG00000038390 | Gpr162 | G protein-coupled receptor 162 | MGI:1315214 | YES | YES | YES | Plasma Membrane | G-protein coupled receptor | ||||
| ENSMUSG00000045441 | Gprin3 | GPRIN family member 3 | MGI:1924785 | YES | YES | YES | Other | other | ||||
| ENSMUSG00000034786 | Gpsm3 | G-protein signalling modulator 3 (AGS3-like, C. elegans) | MGI:2146785 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000075706 | Gpx4 | glutathione peroxidase 4 | MGI:104767 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000020524 | Gria1 | glutamate receptor, ionotropic, AMPA1 (alpha 1) | MGI:95808 | YES | YES | YES | Plasma Membrane | ion channel | ||||
| ENSMUSG00000071424 | Grid2 | glutamate receptor, ionotropic, delta 2 | MGI:95813 | YES | YES | YES | Plasma Membrane | ion channel | ||||
| ENSMUSG00000056073 | Grik2 | glutamate receptor, ionotropic, kainate 2 (beta 2) | MGI:95815 | YES | YES | YES | Plasma Membrane | ion channel | ||||
| ENSMUSG00000001985 | Grik3 | glutamate receptor, ionotropic, kainate 3 | MGI:95816 | YES | YES | YES | Plasma Membrane | ion channel | ||||
| ENSMUSG00000030209 | Grin2b | glutamate receptor, ionotropic, NMDA2B (epsilon 2) | MGI:95821 | YES | YES | YES | Plasma Membrane | ion channel | ||||
| ENSMUSG00000031153 | Gripap1 | GRIP1 associated protein 1 | MGI:1859616 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000074886 | Grk6 | G protein-coupled receptor kinase 6 | MGI:1347078 | YES | YES | YES | Plasma Membrane | kinase | ||||
| ENSMUSG00000023192 | Grm2 | glutamate receptor, metabotropic 2 | MGI:1351339 | YES | YES | YES | Plasma Membrane | G-protein coupled receptor | ||||
| ENSMUSG00000003974 | Grm3 | glutamate receptor, metabotropic 3 | MGI:1351340 | YES | YES | YES | Plasma Membrane | G-protein coupled receptor | ||||
| ENSMUSG00000063239 | Grm4 | glutamate receptor, metabotropic 4 | MGI:1351341 | YES | YES | YES | Plasma Membrane | G-protein coupled receptor | ||||
| ENSMUSG00000049583 | Grm5 | glutamate receptor, metabotropic 5 | MGI:1351342 | YES | YES | YES | Plasma Membrane | G-protein coupled receptor | ||||
| ENSMUSG00000056755 | Grm7 | glutamate receptor, metabotropic 7 | MGI:1351344 | YES | YES | YES | Plasma Membrane | G-protein coupled receptor | ||||
| ENSMUSG00000024211 | Grm8 | glutamate receptor, metabotropic 8 | MGI:1351345 | YES | YES | YES | Plasma Membrane | G-protein coupled receptor | ||||
| ENSMUSG00000022812 | Gsk3b | glycogen synthase kinase 3 beta | MGI:1861437 | YES | YES | YES | Nucleus | kinase | ||||
| ENSMUSG00000026879 | Gsn | gelsolin | MGI:95851 | YES | YES | YES | Extracellular Space | other | ||||
| ENSMUSG00000032348 | Gsta4 | glutathione S-transferase, alpha 4 | MGI:1309515 | YES | YES | YES | Other | enzyme | ||||
| ENSMUSG00000060261 | Gtf2i | general transcription factor II I | MGI:1202722 | YES | YES | YES | Nucleus | transcription regulator | ||||
| ENSMUSG00000041624 | Gucy1a2 | guanylate cyclase 1, soluble, alpha 2 | MGI:2660877 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000028005 | Gucy1b3 | guanylate cyclase 1, soluble, beta 3 | MGI:1860604 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000020444 | Guk1 | guanylate kinase 1 | MGI:95871 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSMUSG00000025059 | Gyk | glycerol kinase | MGI:106594 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSMUSG00000051839 | Gypa | glycophorin A | MGI:95880 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000060743 | H3f3a | H3 histone, family 3A | MGI:1097686 | YES | YES | YES | Nucleus | other | ||||
| ENSMUSG00000033629 | Hacd3 | 3-hydroxyacyl-CoA dehydratase 3 | MGI:1889341 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000025745 | Hadha | hydroxyacyl-Coenzyme A dehydrogenase/3-ketoacyl-Coenzyme A thiolase/enoyl-Coenzyme A hydratase (trifunctional protein), alpha subunit | MGI:2135593 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000059447 | Hadhb | hydroxyacyl-Coenzyme A dehydrogenase/3-ketoacyl-Coenzyme A thiolase/enoyl-Coenzyme A hydratase (trifunctional protein), beta subunit | MGI:2136381 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000031386 | Hcfc1 | host cell factor C1 | MGI:105942 | YES | YES | YES | Nucleus | transcription regulator | ||||
| ENSMUSG00000042807 | Hecw2 | HECT, C2 and WW domain containing E3 ubiquitin protein ligase 2 | MGI:2685817 | YES | YES | YES | Extracellular Space | enzyme | ||||
| ENSMUSG00000046240 | Hepacam | hepatocyte cell adhesion molecule | MGI:1920177 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000021665 | Hexb | hexosaminidase B | MGI:96074 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000020267 | Hint1 | histidine triad nucleotide binding protein 1 | MGI:1321133 | YES | YES | YES | Nucleus | enzyme | ||||
| ENSMUSG00000039959 | Hip1 | huntingtin interacting protein 1 | MGI:1099804 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000000915 | Hip1r | huntingtin interacting protein 1 related | MGI:1352504 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000060081 | Hist1h2aa | histone cluster 1, H2aa | MGI:2448285 | YES | YES | YES | Nucleus | other | ||||
| ENSMUSG00000037012 | Hk1 | hexokinase 1 | MGI:96103 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSMUSG00000004980 | Hnrnpa2b1 | heterogeneous nuclear ribonucleoprotein A2/B1 | MGI:104819 | YES | YES | YES | #N/A | #N/A | ||||
| ENSMUSG00000020358 | Hnrnpab | heterogeneous nuclear ribonucleoprotein A/B | MGI:1330294 | YES | YES | YES | Nucleus | enzyme | ||||
| ENSMUSG00000000568 | Hnrnpd | heterogeneous nuclear ribonucleoprotein D | MGI:101947 | YES | YES | YES | Nucleus | transcription regulator | ||||
| ENSMUSG00000021546 | Hnrnpk | heterogeneous nuclear ribonucleoprotein K | MGI:99894 | YES | YES | YES | Nucleus | other | ||||
| ENSMUSG00000037234 | Hook3 | hook homolog 3 (Drosophila) | MGI:2443554 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000028785 | Hpca | hippocalcin | MGI:1336200 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000071379 | Hpcal1 | hippocalcin-like 1 | MGI:1855689 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000046093 | Hpcal4 | hippocalcin-like 4 | MGI:2157521 | YES | YES | YES | Other | transporter | ||||
| ENSMUSG00000025499 | Hras | Harvey rat sarcoma virus oncogene | MGI:96224 | YES | YES | YES | Plasma Membrane | enzyme | ||||
| ENSMUSG00000025260 | Hsd17b10 | hydroxysteroid (17-beta) dehydrogenase 10 | MGI:1333871 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000027195 | Hsd17b12 | hydroxysteroid (17-beta) dehydrogenase 12 | MGI:1926967 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000024507 | Hsd17b4 | hydroxysteroid (17-beta) dehydrogenase 4 | MGI:105089 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000034189 | Hsdl1 | hydroxysteroid dehydrogenase like 1 | MGI:1919802 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000028383 | Hsdl2 | hydroxysteroid dehydrogenase like 2 | MGI:1919729 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000025092 | Hspa12a | heat shock protein 12A | MGI:1920692 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000074793 | Hspa12b | heat shock protein 12B | MGI:1919880 | YES | YES | YES | Other | other | ||||
| ENSMUSG00000091971 | Hspa1a | heat shock protein 1A | MGI:96244 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000059970 | Hspa2 | heat shock protein 2 | MGI:96243 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000015656 | Hspa8 | heat shock protein 8 | MGI:105384 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000002010 | Idh3g | isocitrate dehydrogenase 3 (NAD+), gamma | MGI:1099463 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000076617 | Ighm | immunoglobulin heavy constant mu | MGI:96448 | YES | YES | YES | Plasma Membrane | transmembrane receptor | ||||
| ENSMUSG00000038034 | Igsf8 | immunoglobulin superfamily, member 8 | MGI:2154090 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000052372 | Il1rapl1 | interleukin 1 receptor accessory protein-like 1 | MGI:2687319 | YES | YES | YES | Plasma Membrane | transmembrane receptor | ||||
| ENSMUSG00000032763 | Ilvbl | ilvB (bacterial acetolactate synthase)-like | MGI:1351911 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000052337 | Immt | inner membrane protein, mitochondrial | MGI:1923864 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000026113 | Inpp4a | inositol polyphosphate-4-phosphatase, type I | MGI:1931123 | YES | YES | YES | Cytoplasm | phosphatase | ||||
| ENSMUSG00000025477 | Inpp5a | inositol polyphosphate-5-phosphatase A | MGI:2686961 | YES | YES | YES | Plasma Membrane | phosphatase | ||||
| ENSMUSG00000040797 | Iqsec3 | IQ motif and Sec7 domain 3 | MGI:2677208 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000031729 | Ist1 | increased sodium tolerance 1 homolog (yeast) | MGI:1919205 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000031703 | Itfg1 | integrin alpha FG-GAP repeat containing 1 | MGI:106419 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000006522 | Itih3 | inter-alpha trypsin inhibitor, heavy chain 3 | MGI:96620 | YES | YES | YES | Extracellular Space | other | ||||
| ENSMUSG00000022108 | Itm2b | integral membrane protein 2B | MGI:1309517 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000027296 | Itpka | inositol 1,4,5-trisphosphate 3-kinase A | MGI:1333822 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSMUSG00000030102 | Itpr1 | inositol 1,4,5-trisphosphate receptor 1 | MGI:96623 | YES | YES | YES | Cytoplasm | ion channel | ||||
| ENSMUSG00000030287 | Itpr2 | inositol 1,4,5-triphosphate receptor 2 | MGI:99418 | YES | YES | YES | Cytoplasm | ion channel | ||||
| ENSMUSG00000028530 | Jak1 | Janus kinase 1 | MGI:96628 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSMUSG00000063646 | Jakmip1 | janus kinase and microtubule interacting protein 1 | MGI:1923321 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000024502 | Jakmip2 | janus kinase and microtubule interacting protein 2 | MGI:1923467 | YES | YES | YES | Extracellular Space | other | ||||
| ENSMUSG00000056856 | Jakmip3 | janus kinase and microtubule interacting protein 3 | MGI:1921254 | YES | YES | YES | Other | other | ||||
| ENSMUSG00000031990 | Jam3 | junction adhesion molecule 3 | MGI:1933825 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000042686 | Jph1 | junctophilin 1 | MGI:1891495 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000001552 | Jup | junction plakoglobin | MGI:96650 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000047976 | Kcna1 | potassium voltage-gated channel, shaker-related subfamily, member 1 | MGI:96654 | YES | YES | YES | Plasma Membrane | ion channel | ||||
| ENSMUSG00000040724 | Kcna2 | potassium voltage-gated channel, shaker-related subfamily, member 2 | MGI:96659 | YES | YES | YES | Plasma Membrane | ion channel | ||||
| ENSMUSG00000047959 | Kcna3 | potassium voltage-gated channel, shaker-related subfamily, member 3 | MGI:96660 | YES | YES | YES | Plasma Membrane | ion channel | ||||
| ENSMUSG00000042604 | Kcna4 | potassium voltage-gated channel, shaker-related subfamily, member 4 | MGI:96661 | YES | YES | YES | Plasma Membrane | ion channel | ||||
| ENSMUSG00000038077 | Kcna6 | potassium voltage-gated channel, shaker-related, subfamily, member 6 | MGI:96663 | YES | YES | YES | Plasma Membrane | ion channel | ||||
| ENSMUSG00000027827 | Kcnab1 | potassium voltage-gated channel, shaker-related subfamily, beta member 1 | MGI:109155 | YES | YES | YES | Plasma Membrane | ion channel | ||||
| ENSMUSG00000050556 | Kcnb1 | potassium voltage gated channel, Shab-related subfamily, member 1 | MGI:96666 | YES | YES | YES | Plasma Membrane | ion channel | ||||
| ENSMUSG00000062785 | Kcnc3 | potassium voltage gated channel, Shaw-related subfamily, member 3 | MGI:96669 | YES | YES | YES | Plasma Membrane | ion channel | ||||
| ENSMUSG00000060882 | Kcnd2 | potassium voltage-gated channel, Shal-related family, member 2 | MGI:102663 | YES | YES | YES | Plasma Membrane | ion channel | ||||
| ENSMUSG00000040896 | Kcnd3 | potassium voltage-gated channel, Shal-related family, member 3 | MGI:1928743 | YES | YES | YES | Plasma Membrane | ion channel | ||||
| ENSMUSG00000054477 | Kcnn2 | potassium intermediate/small conductance calcium-activated channel, subfamily N, member 2 | MGI:2153182 | YES | YES | YES | Plasma Membrane | ion channel | ||||
| ENSMUSG00000016346 | Kcnq2 | potassium voltage-gated channel, subfamily Q, member 2 | MGI:1309503 | YES | YES | YES | Plasma Membrane | ion channel | ||||
| ENSMUSG00000022350 | E430025E21Rik | RIKEN cDNA E430025E21 gene | MGI:2146110 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000034560 | A230046K03Rik | RIKEN cDNA A230046K03 gene | MGI:2441787 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000021693 | Kif2a | kinesin family member 2A | MGI:108390 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000006740 | Kif5b | kinesin family member 5B | MGI:1098268 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000026764 | Kif5c | kinesin family member 5C | MGI:1098269 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000026585 | Kifap3 | kinesin-associated protein 3 | MGI:107566 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000032036 | Kirrel3 | kin of IRRE like 3 (Drosophila) | MGI:1914953 | YES | YES | YES | Extracellular Space | other | ||||
| ENSMUSG00000021288 | Klc1 | kinesin light chain 1 | MGI:107978 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000024862 | Klc2 | kinesin light chain 2 | MGI:107953 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000030265 | Kras | v-Ki-ras2 Kirsten rat sarcoma viral oncogene homolog | MGI:96680 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000031391 | L1cam | L1 cell adhesion molecule | MGI:96721 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000032370 | Lactb | lactamase, beta | MGI:1933395 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000030842 | Lamtor1 | late endosomal/lysosomal adaptor, MAPK and MTOR activator 1 | MGI:1913758 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000094685 | Gm5900 | predicted pseudogene 5900 | MGI:3643907 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000026000 | Lancl1 | LanC (bacterial lantibiotic synthetase component C)-like 1 | MGI:1336997 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000062190 | Lancl2 | LanC (bacterial lantibiotic synthetase component C)-like 2 | MGI:1919085 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000037331 | Larp1 | La ribonucleoprotein domain family, member 1 | MGI:1890165 | YES | YES | YES | Cytoplasm | translation regulator | ||||
| ENSMUSG00000024493 | Lars | leucyl-tRNA synthetase | MGI:1913808 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000038366 | Lasp1 | LIM and SH3 protein 1 | MGI:109656 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSMUSG00000067242 | Lgi1 | leucine-rich repeat LGI family, member 1 | MGI:1861691 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000039252 | Lgi2 | leucine-rich repeat LGI family, member 2 | MGI:2180196 | YES | YES | YES | Extracellular Space | other | ||||
| ENSMUSG00000023022 | Lima1 | LIM domain and actin binding 1 | MGI:1920992 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000037736 | Limch1 | LIM and calponin homology domains 1 | MGI:1924819 | YES | YES | YES | Other | other | ||||
| ENSMUSG00000040699 | Limd2 | LIM domain containing 2 | MGI:1915053 | YES | YES | YES | Other | other | ||||
| ENSMUSG00000019906 | Lin7a | lin-7 homolog A (C. elegans) | MGI:2135609 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000027162 | Lin7c | lin-7 homolog C (C. elegans) | MGI:1330839 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000028063 | Lmna | lamin A | MGI:96794 | YES | YES | YES | Nucleus | other | ||||
| ENSMUSG00000024590 | Lmnb1 | lamin B1 | MGI:96795 | YES | YES | YES | Nucleus | other | ||||
| ENSMUSG00000038970 | Lmtk2 | lemur tyrosine kinase 2 | MGI:3036247 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSMUSG00000062044 | Lmtk3 | lemur tyrosine kinase 3 | MGI:3039582 | YES | YES | YES | Other | kinase | ||||
| ENSMUSG00000047866 | Lonp2 | lon peptidase 2, peroxisomal | MGI:1914137 | YES | YES | YES | Cytoplasm | peptidase | ||||
| ENSMUSG00000026623 | Lpgat1 | lysophosphatidylglycerol acyltransferase 1 | MGI:2446186 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000013033 | Adgrl1 | adhesion G protein-coupled receptor L1 | MGI:1929461 | YES | YES | YES | Plasma Membrane | G-protein coupled receptor | ||||
| ENSMUSG00000037605 | Adgrl3 | adhesion G protein-coupled receptor L3 | MGI:2441950 | YES | YES | YES | Plasma Membrane | G-protein coupled receptor | ||||
| ENSMUSG00000035835 | Lppr3 | lipid phosphate phosphatase-related protein type 3 | MGI:2388640 | YES | YES | YES | Other | phosphatase | ||||
| ENSMUSG00000044667 | Lppr4 | lipid phosphate phosphatase-related protein type 4 | MGI:106530 | YES | YES | YES | Plasma Membrane | phosphatase | ||||
| ENSMUSG00000030600 | Lrfn1 | leucine rich repeat and fibronectin type III domain containing 1 | MGI:2136810 | YES | YES | YES | Nucleus | other | ||||
| ENSMUSG00000045045 | Lrfn4 | leucine rich repeat and fibronectin type III domain containing 4 | MGI:2385612 | YES | YES | YES | Other | other | ||||
| ENSMUSG00000063052 | Lrrc40 | leucine rich repeat containing 40 | MGI:1914394 | YES | YES | YES | Nucleus | other | ||||
| ENSMUSG00000050587 | Lrrc4c | leucine rich repeat containing 4C | MGI:2442636 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000027286 | Lrrc57 | leucine rich repeat containing 57 | MGI:1913856 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000020869 | Lrrc59 | leucine rich repeat containing 59 | MGI:2138133 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000007476 | Lrrc8a | leucine rich repeat containing 8A | MGI:2652847 | YES | YES | YES | Extracellular Space | other | ||||
| ENSMUSG00000070639 | Lrrc8b | leucine rich repeat containing 8 family, member B | MGI:2141353 | YES | YES | YES | Other | other | ||||
| ENSMUSG00000054720 | Lrrc8c | leucine rich repeat containing 8 family, member C | MGI:2140839 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000046079 | Lrrc8d | leucine rich repeat containing 8D | MGI:1922368 | YES | YES | YES | Plasma Membrane | G-protein coupled receptor | ||||
| ENSMUSG00000061080 | Lsamp | limbic system-associated membrane protein | MGI:1261760 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000042228 | Lyn | Yamaguchi sarcoma viral (v-yes-1) oncogene homolog | MGI:96892 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSMUSG00000045095 | Magi1 | membrane associated guanylate kinase, WW and PDZ domain containing 1 | MGI:1203522 | YES | YES | YES | Plasma Membrane | kinase | ||||
| ENSMUSG00000040003 | Magi2 | membrane associated guanylate kinase, WW and PDZ domain containing 2 | MGI:1354953 | YES | YES | YES | Plasma Membrane | kinase | ||||
| ENSMUSG00000027254 | Map1a | microtubule-associated protein 1 A | MGI:1306776 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000052727 | Map1b | microtubule-associated protein 1B | MGI:1306778 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000027602 | Map1lc3a | microtubule-associated protein 1 light chain 3 alpha | MGI:1915661 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000019261 | Map1s | microtubule-associated protein 1S | MGI:2443304 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000004936 | Map2k1 | mitogen-activated protein kinase kinase 1 | MGI:1346866 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSMUSG00000033352 | Map2k4 | mitogen-activated protein kinase kinase 4 | MGI:1346869 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSMUSG00000026074 | Map4k4 | mitogen-activated protein kinase kinase kinase kinase 4 | MGI:1349394 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSMUSG00000055407 | Map6 | microtubule-associated protein 6 | MGI:1201690 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000028849 | Map7d1 | MAP7 domain containing 1 | MGI:2384297 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000041020 | Map7d2 | MAP7 domain containing 2 | MGI:1917474 | YES | YES | YES | Other | other | ||||
| ENSMUSG00000063358 | Mapk1 | mitogen-activated protein kinase 1 | MGI:1346858 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSMUSG00000046709 | Mapk10 | mitogen-activated protein kinase 10 | MGI:1346863 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSMUSG00000063065 | Mapk3 | mitogen-activated protein kinase 3 | MGI:1346859 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSMUSG00000024163 | Mapk8ip3 | mitogen-activated protein kinase 8 interacting protein 3 | MGI:1353598 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000027479 | Mapre1 | microtubule-associated protein, RP/EB family, member 1 | MGI:891995 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000024277 | Mapre2 | microtubule-associated protein, RP/EB family, member 2 | MGI:106271 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000029166 | Mapre3 | microtubule-associated protein, RP/EB family, member 3 | MGI:2140967 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000018411 | Mapt | microtubule-associated protein tau | MGI:97180 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000073481 | Marc2 | mitochondrial amidoxime reducing component 2 | MGI:1914497 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000023307 | March5 | membrane-associated ring finger (C3HC4) 5 | MGI:1915207 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000037236 | Matr3 | matrin 3 | MGI:1298379 | YES | YES | YES | Nucleus | other | ||||
| ENSMUSG00000027709 | Mccc1 | methylcrotonoyl-Coenzyme A carboxylase 1 (alpha) | MGI:1919289 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000021646 | Mccc2 | methylcrotonoyl-Coenzyme A carboxylase 2 (beta) | MGI:1925288 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000009647 | Mcu | mitochondrial calcium uniporter | MGI:3026965 | YES | YES | YES | Cytoplasm | ion channel | ||||
| ENSMUSG00000034912 | Mdga2 | MAM domain containing glycosylphosphatidylinositol anchor 2 | MGI:2444706 | YES | YES | YES | Other | other | ||||
| ENSMUSG00000026150 | Mff | mitochondrial fission factor | MGI:1922984 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000026688 | Mgst3 | microsomal glutathione S-transferase 3 | MGI:1913697 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000024410 | 3110002H16Rik | RIKEN cDNA 3110002H16 gene | MGI:1916528 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000049760 | 2410015M20Rik | RIKEN cDNA 2410015M20 gene | MGI:2442174 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000003178 | Mical3 | microtubule associated monooxygenase, calponin and LIM domain containing 3 | MGI:2442733 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000020111 | Micu1 | mitochondrial calcium uptake 1 | MGI:2384909 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000039478 | Micu3 | mitochondrial calcium uptake family, member 3 | MGI:1925756 | YES | YES | YES | Other | other | ||||
| ENSMUSG00000020827 | Mink1 | misshapen-like kinase 1 (zebrafish) | MGI:1355329 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSMUSG00000042447 | Mios | missing oocyte, meiosis regulator, homolog (Drosophila) | MGI:2182066 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000035805 | Mlc1 | megalencephalic leukoencephalopathy with subcortical cysts 1 homolog (human) | MGI:2157910 | YES | YES | YES | Plasma Membrane | transporter | ||||
| ENSMUSG00000048578 | Mlec | malectin | MGI:1924015 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000068036 | Mllt4 | myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila) translocated to, 4 | MGI:1314653 | YES | YES | YES | Nucleus | other | ||||
| ENSMUSG00000074064 | Mlycd | malonyl-CoA decarboxylase | MGI:1928485 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000076439 | Mog | myelin oligodendrocyte glycoprotein | MGI:97435 | YES | YES | YES | Extracellular Space | other | ||||
| ENSMUSG00000032583 | Mon1a | MON1 homolog A (yeast) | MGI:1920075 | YES | YES | YES | Other | other | ||||
| ENSMUSG00000078908 | Mon1b | MON1 homolog b (yeast) | MGI:1923231 | YES | YES | YES | Other | other | ||||
| ENSMUSG00000026568 | Mpc2 | mitochondrial pyruvate carrier 2 | MGI:1917706 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000017314 | Mpp2 | membrane protein, palmitoylated 2 (MAGUK p55 subfamily member 2) | MGI:1858257 | YES | YES | YES | Plasma Membrane | kinase | ||||
| ENSMUSG00000052373 | Mpp3 | membrane protein, palmitoylated 3 (MAGUK p55 subfamily member 3) | MGI:1328354 | YES | YES | YES | Plasma Membrane | kinase | ||||
| ENSMUSG00000032470 | Mras | muscle and microspikes RAS | MGI:1100856 | YES | YES | YES | Plasma Membrane | enzyme | ||||
| ENSMUSG00000039640 | Mrpl12 | mitochondrial ribosomal protein L12 | MGI:1926273 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000020514 | Mrpl22 | mitochondrial ribosomal protein L22 | MGI:1333794 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000037772 | Mrpl23 | mitochondrial ribosomal protein L23 | MGI:1196612 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000025208 | Mrpl43 | mitochondrial ribosomal protein L43 | MGI:2137229 | YES | YES | YES | Cytoplasm | translation regulator | ||||
| ENSMUSG00000041632 | Mrps27 | mitochondrial ribosomal protein S27 | MGI:1919064 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000026887 | Mrrf | mitochondrial ribosome recycling factor | MGI:1915121 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000064356 | mt-Atp8 | mitochondrially encoded ATP synthase 8 | MGI:99926 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000027282 | Mtch2 | mitochondrial carrier homolog 2 (C. elegans) | MGI:1929260 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000064354 | mt-Co2 | mitochondrially encoded cytochrome c oxidase II | MGI:102503 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000046671 | Mtfr1l | mitochondrial fission regulator 1-like | MGI:1924074 | YES | YES | YES | Other | other | ||||
| ENSMUSG00000021048 | Mthfd1 | methylenetetrahydrofolate dehydrogenase (NADP+ dependent), methenyltetrahydrofolate cyclohydrolase, formyltetrahydrofolate synthase | MGI:1342005 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000015214 | Mtmr1 | myotubularin related protein 1 | MGI:1858271 | YES | YES | YES | Cytoplasm | phosphatase | ||||
| ENSMUSG00000031918 | Mtmr2 | myotubularin related protein 2 | MGI:1924366 | YES | YES | YES | Cytoplasm | phosphatase | ||||
| ENSMUSG00000028991 | Mtor | mechanistic target of rapamycin (serine/threonine kinase) | MGI:1928394 | YES | YES | YES | Nucleus | kinase | ||||
| ENSMUSG00000024234 | Mtpap | mitochondrial poly(A) polymerase | MGI:1914690 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000033763 | Mtss1l | metastasis suppressor 1-like | MGI:3039591 | YES | YES | YES | Other | other | ||||
| ENSMUSG00000029651 | Mtus2 | microtubule associated tumor suppressor candidate 2 | MGI:1915388 | YES | YES | YES | Nucleus | other | ||||
| ENSMUSG00000064068 | Mtx1 | metaxin 1 | MGI:103025 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSMUSG00000027099 | Mtx2 | metaxin 2 | MGI:1859652 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSMUSG00000021704 | Mtx3 | metaxin 3 | MGI:2686040 | YES | YES | YES | Other | other | ||||
| ENSMUSG00000038740 | Mvb12b | multivesicular body subunit 12B | MGI:1919793 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000068566 | Myadm | myeloid-associated differentiation marker | MGI:1355332 | YES | YES | YES | Nucleus | other | ||||
| ENSMUSG00000033004 | Mycbp2 | MYC binding protein 2 | MGI:2179432 | YES | YES | YES | Nucleus | enzyme | ||||
| ENSMUSG00000020900 | Myh10 | myosin, heavy polypeptide 10, non-muscle | MGI:1930780 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000030739 | Myh14 | myosin, heavy polypeptide 14 | MGI:1919210 | YES | YES | YES | Extracellular Space | other | ||||
| ENSMUSG00000022443 | Myh9 | myosin, heavy polypeptide 9, non-muscle | MGI:107717 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000000631 | Myo18a | myosin XVIIIA | MGI:2667185 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000035441 | Myo1d | myosin ID | MGI:107728 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000034593 | Myo5a | myosin VA | MGI:105976 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000058799 | Nap1l1 | nucleosome assembly protein 1-like 1 | MGI:1855693 | YES | YES | YES | Nucleus | other | ||||
| ENSMUSG00000059119 | Nap1l4 | nucleosome assembly protein 1-like 4 | MGI:1316687 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000002341 | Ncan | neurocan | MGI:104694 | YES | YES | YES | Extracellular Space | other | ||||
| ENSMUSG00000027002 | Nckap1 | NCK-associated protein 1 | MGI:1355333 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000032598 | Nckipsd | NCK interacting protein with SH3 domain | MGI:1931834 | YES | YES | YES | Other | other | ||||
| ENSMUSG00000026234 | Ncl | nucleolin | MGI:97286 | YES | YES | YES | Nucleus | other | ||||
| ENSMUSG00000020238 | Ncln | nicalin homolog (zebrafish) | MGI:1926081 | YES | YES | YES | Cytoplasm | peptidase | ||||
| ENSMUSG00000062661 | Ncs1 | neuronal calcium sensor 1 | MGI:109166 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000020022 | Ndufa12 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 12 | MGI:1913664 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000036199 | Ndufa13 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 13 | MGI:1914434 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000014294 | Ndufa2 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 2 | MGI:1343103 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000029632 | Ndufa4 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 4 | MGI:107686 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000022450 | Ndufa6 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 6 (B14) | MGI:1914380 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000041881 | Ndufa7 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 7 (B14.5a) | MGI:1913666 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000026895 | Ndufa8 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 8 | MGI:1915625 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000000399 | Ndufa9 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 9 | MGI:1913358 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000070283 | Ndufaf3 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, assembly factor 3 | MGI:1913956 | YES | YES | YES | Nucleus | other | ||||
| ENSMUSG00000028261 | Ndufaf4 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, assembly factor 4 | MGI:1915743 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000050323 | Ndufaf6 | NADH dehydrogenase (ubiquinone) complex I, assembly factor 6 | MGI:1924197 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000040048 | Ndufb10 | NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 10 | MGI:1915592 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000031059 | Ndufb11 | NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 11 | MGI:1349919 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000002416 | Ndufb2 | NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 2 | MGI:1915448 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000026032 | Ndufb3 | NADH dehydrogenase (ubiquinone) 1 beta subcomplex 3 | MGI:1913745 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000022820 | Ndufb4 | NADH dehydrogenase (ubiquinone) 1 beta subcomplex 4 | MGI:1915444 | YES | YES | YES | #N/A | #N/A | ||||
| ENSMUSG00000027673 | Ndufb5 | NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 5 | MGI:1913296 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000071014 | Ndufb6 | NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 6 | MGI:2684983 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000033938 | Ndufb7 | NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 7 | MGI:1914166 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000025204 | Ndufb8 | NADH dehydrogenase (ubiquinone) 1 beta subcomplex 8 | MGI:1914514 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000022354 | Ndufb9 | NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 9 | MGI:1913468 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000030647 | Ndufc2 | NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 2 | MGI:1344370 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000005510 | Ndufs3 | NADH dehydrogenase (ubiquinone) Fe-S protein 3 | MGI:1915599 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000021606 | Ndufs6 | NADH dehydrogenase (ubiquinone) Fe-S protein 6 | MGI:107932 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000020153 | Ndufs7 | NADH dehydrogenase (ubiquinone) Fe-S protein 7 | MGI:1922656 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000024099 | Ndufv2 | NADH dehydrogenase (ubiquinone) flavoprotein 2 | MGI:1920150 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000053702 | Nebl | nebulette | MGI:1921353 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000031837 | Necab2 | N-terminal EF-hand calcium binding protein 2 | MGI:2152211 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000030327 | Necap1 | NECAP endocytosis associated 1 | MGI:1914852 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000024589 | Nedd4l | neural precursor cell expressed, developmentally down-regulated gene 4-like | MGI:1933754 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000010376 | Nedd8 | neural precursor cell expressed, developmentally down-regulated gene 8 | MGI:97301 | YES | YES | YES | Nucleus | enzyme | ||||
| ENSMUSG00000020396 | Nefh | neurofilament, heavy polypeptide | MGI:97309 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000040037 | Negr1 | neuronal growth regulator 1 | MGI:2444846 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000020716 | Nf1 | neurofibromatosis 1 | MGI:97306 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000027618 | Nfs1 | nitrogen fixation gene 1 (S. cerevisiae) | MGI:1316706 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000026259 | Ngef | neuronal guanine nucleotide exchange factor | MGI:1858414 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000047037 | Nipa1 | non imprinted in Prader-Willi/Angelman syndrome 1 homolog (human) | MGI:2442058 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000021910 | Nisch | nischarin | MGI:1928323 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000051790 | Nlgn2 | neuroligin 2 | MGI:2681835 | YES | YES | YES | Plasma Membrane | enzyme | ||||
| ENSMUSG00000031302 | Nlgn3 | neuroligin 3 | MGI:2444609 | YES | YES | YES | Plasma Membrane | enzyme | ||||
| ENSMUSG00000073435 | Nme3 | NME/NM23 nucleoside diphosphate kinase 3 | MGI:1930182 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSMUSG00000021047 | Nova1 | neuro-oncological ventral antigen 1 | MGI:104297 | YES | YES | YES | Nucleus | other | ||||
| ENSMUSG00000025582 | Nptx1 | neuronal pentraxin 1 | MGI:107811 | YES | YES | YES | Extracellular Space | other | ||||
| ENSMUSG00000039114 | Nrn1 | neuritin 1 | MGI:1915654 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000066392 | Nrxn3 | neurexin III | MGI:1096389 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000029126 | Nsg1 | neuron specific gene family member 1 | MGI:109149 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000020297 | Nsg2 | neuron specific gene family member 2 | MGI:1202070 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000059974 | Ntm | neurotrimin | MGI:2446259 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000059857 | Ntng1 | netrin G1 | MGI:1934028 | YES | YES | YES | Extracellular Space | other | ||||
| ENSMUSG00000021224 | Numb | numb gene homolog (Drosophila) | MGI:107423 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000029152 | Ociad1 | OCIA domain containing 1 | MGI:1915345 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000020456 | Ogdh | oxoglutarate (alpha-ketoglutarate) dehydrogenase (lipoamide) | MGI:1098267 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000021913 | Ogdhl | oxoglutarate dehydrogenase-like | MGI:3616088 | YES | YES | YES | Other | enzyme | ||||
| ENSMUSG00000034160 | Ogt | O-linked N-acetylglucosamine (GlcNAc) transferase (UDP-N-acetylglucosamine:polypeptide-N-acetylglucosaminyl transferase) | MGI:1339639 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000049612 | Omg | oligodendrocyte myelin glycoprotein | MGI:106586 | YES | YES | YES | Plasma Membrane | G-protein coupled receptor | ||||
| ENSMUSG00000052214 | Opa3 | optic atrophy 3 | MGI:2686271 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000062257 | Opcml | opioid binding protein/cell adhesion molecule-like | MGI:97397 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000024767 | Otub1 | OTU domain, ubiquitin aldehyde binding 1 | MGI:2147616 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000025364 | Pa2g4 | proliferation-associated 2G4 | MGI:894684 | YES | YES | YES | Nucleus | transcription regulator | ||||
| ENSMUSG00000022283 | Pabpc1 | poly(A) binding protein, cytoplasmic 1 | MGI:1349722 | YES | YES | YES | Cytoplasm | translation regulator | ||||
| ENSMUSG00000024855 | Pacs1 | phosphofurin acidic cluster sorting protein 1 | MGI:1277113 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000020745 | Pafah1b1 | platelet-activating factor acetylhydrolase, isoform 1b, subunit 1 | MGI:109520 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000003131 | Pafah1b2 | platelet-activating factor acetylhydrolase, isoform 1b, subunit 2 | MGI:108415 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000027508 | Pag1 | phosphoprotein associated with glycosphingolipid microdomains 1 | MGI:2443160 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000029247 | Paics | phosphoribosylaminoimidazole carboxylase, phosphoribosylaminoribosylaminoimidazole, succinocarboxamide synthetase | MGI:1914304 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000030774 | Pak1 | p21 protein (Cdc42/Rac)-activated kinase 1 | MGI:1339975 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSMUSG00000035863 | Palm | paralemmin | MGI:1261814 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000090053 | Palm2 | paralemmin 2 | MGI:1934601 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000026335 | Pam | peptidylglycine alpha-amidating monooxygenase | MGI:97475 | YES | YES | YES | Plasma Membrane | enzyme | ||||
| ENSMUSG00000014301 | Pam16 | presequence translocase-asssociated motor 16 homolog (S. cerevisiae) | MGI:1913699 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000051695 | Pcbp1 | poly(rC) binding protein 1 | MGI:1345635 | YES | YES | YES | Nucleus | translation regulator | ||||
| ENSMUSG00000056851 | Pcbp2 | poly(rC) binding protein 2 | MGI:108202 | YES | YES | YES | Nucleus | other | ||||
| ENSMUSG00000001120 | Pcbp3 | poly(rC) binding protein 3 | MGI:1890470 | YES | YES | YES | Nucleus | other | ||||
| ENSMUSG00000049100 | Pcdh10 | protocadherin 10 | MGI:1338042 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000102543 | Pcdhgc5 | protocadherin gamma subfamily C, 5 | MGI:1935205 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000061601 | Pclo | piccolo (presynaptic cytomatrix protein) | MGI:1349390 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSMUSG00000019795 | Pcmt1 | protein-L-isoaspartate (D-aspartate) O-methyltransferase 1 | MGI:97502 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000027835 | Pdcd10 | programmed cell death 10 | MGI:1928396 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000021576 | Pdcd6 | programmed cell death 6 | MGI:109283 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000022489 | Pde1b | phosphodiesterase 1B, Ca2+-calmodulin dependent | MGI:97523 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000030653 | Pde2a | phosphodiesterase 2A, cGMP-stimulated | MGI:2446107 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000025584 | Pde8a | phosphodiesterase 8A | MGI:1277116 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000031299 | Pdha1 | pyruvate dehydrogenase E1 alpha 1 | MGI:97532 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000021748 | Pdhb | pyruvate dehydrogenase (lipoamide) beta | MGI:1915513 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000010914 | Pdhx | pyruvate dehydrogenase complex, component X | MGI:1351627 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000035232 | Pdk3 | pyruvate dehydrogenase kinase, isoenzyme 3 | MGI:2384308 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSMUSG00000024122 | Pdpk1 | 3-phosphoinositide dependent protein kinase 1 | MGI:1338068 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSMUSG00000013698 | Pea15a | phosphoprotein enriched in astrocytes 15A | MGI:104799 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSMUSG00000028102 | Pex11b | peroxisomal biogenesis factor 11 beta | MGI:1338882 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000028975 | Pex14 | peroxisomal biogenesis factor 14 | MGI:1927868 | YES | YES | YES | Cytoplasm | transcription regulator | ||||
| ENSMUSG00000005069 | Pex5 | peroxisomal biogenesis factor 5 | MGI:1098808 | YES | YES | YES | Cytoplasm | transmembrane receptor | ||||
| ENSMUSG00000020277 | Pfkl | phosphofructokinase, liver, B-type | MGI:97547 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSMUSG00000033065 | Pfkm | phosphofructokinase, muscle | MGI:97548 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSMUSG00000021196 | Pfkp | phosphofructokinase, platelet | MGI:1891833 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSMUSG00000011752 | Pgam1 | phosphoglycerate mutase 1 | MGI:97552 | YES | YES | YES | Cytoplasm | phosphatase | ||||
| ENSMUSG00000017715 | Pgs1 | phosphatidylglycerophosphate synthase 1 | MGI:1921701 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000038845 | Phb | prohibitin | MGI:97572 | YES | YES | YES | Nucleus | transcription regulator | ||||
| ENSMUSG00000004264 | Phb2 | prohibitin 2 | MGI:102520 | YES | YES | YES | Cytoplasm | transcription regulator | ||||
| ENSMUSG00000048537 | Phldb1 | pleckstrin homology-like domain, family B, member 1 | MGI:2143230 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000041720 | Pi4ka | phosphatidylinositol 4-kinase, catalytic, alpha polypeptide | MGI:2448506 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSMUSG00000041958 | Pigs | phosphatidylinositol glycan anchor biosynthesis, class S | MGI:2687325 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000017721 | Pigt | phosphatidylinositol glycan anchor biosynthesis, class T | MGI:1926178 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000032571 | Pik3r4 | phosphatidylinositol 3 kinase, regulatory subunit, polypeptide 4, p150 | MGI:1922919 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSMUSG00000032171 | Pin1 | protein (peptidyl-prolyl cis/trans isomerase) NIMA-interacting 1 | MGI:1346036 | YES | YES | YES | Nucleus | enzyme | ||||
| ENSMUSG00000026737 | Pip4k2a | phosphatidylinositol-5-phosphate 4-kinase, type II, alpha | MGI:1298206 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSMUSG00000018547 | Pip4k2b | phosphatidylinositol-5-phosphate 4-kinase, type II, beta | MGI:1934234 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSMUSG00000034902 | Pip5k1c | phosphatidylinositol-4-phosphate 5-kinase, type 1 gamma | MGI:1298224 | YES | YES | YES | Plasma Membrane | kinase | ||||
| ENSMUSG00000017781 | Pitpna | phosphatidylinositol transfer protein, alpha | MGI:99887 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSMUSG00000029406 | Pitpnm2 | phosphatidylinositol transfer protein, membrane-associated 2 | MGI:1336192 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000050211 | Pla2g4e | phospholipase A2, group IVE | MGI:1919144 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000039943 | Plcb4 | phospholipase C, beta 4 | MGI:107464 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000038910 | Plcl2 | phospholipase C-like 2 | MGI:1352756 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000003363 | Pld3 | phospholipase D family, member 3 | MGI:1333782 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000022565 | Plec | plectin | MGI:1277961 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000030231 | Plekha5 | pleckstrin homology domain containing, family A member 5 | MGI:1923802 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000030701 | Plekhb1 | pleckstrin homology domain containing, family B (evectins) member 1 | MGI:1351469 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000040624 | Plekhg1 | pleckstrin homology domain containing, family G (with RhoGef domain) member 1 | MGI:2676551 | YES | YES | YES | Extracellular Space | other | ||||
| ENSMUSG00000031775 | Pllp | plasma membrane proteolipid | MGI:1915051 | YES | YES | YES | Plasma Membrane | transporter | ||||
| ENSMUSG00000004565 | Pnpla6 | patatin-like phospholipase domain containing 6 | MGI:1354723 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000066235 | Pomgnt2 | protein O-linked mannose beta 1,4-N-acetylglucosaminyltransferase 2 | MGI:2143424 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000028517 | Ppap2b | phosphatidic acid phosphatase type 2B | MGI:1915166 | YES | YES | YES | Plasma Membrane | phosphatase | ||||
| ENSMUSG00000053825 | Ppfia2 | protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2 | MGI:2443834 | YES | YES | YES | Plasma Membrane | phosphatase | ||||
| ENSMUSG00000003863 | Ppfia3 | protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 3 | MGI:1924037 | YES | YES | YES | Plasma Membrane | phosphatase | ||||
| ENSMUSG00000032383 | Ppib | peptidylprolyl isomerase B | MGI:97750 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000027804 | Ppid | peptidylprolyl isomerase D (cyclophilin D) | MGI:1914988 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000034613 | Ppm1h | protein phosphatase 1H (PP2C domain containing) | MGI:2442087 | YES | YES | YES | Other | phosphatase | ||||
| ENSMUSG00000030718 | Ppme1 | protein phosphatase methylesterase 1 | MGI:1919840 | YES | YES | YES | Other | enzyme | ||||
| ENSMUSG00000040385 | Ppp1ca | protein phosphatase 1, catalytic subunit, alpha isoform | MGI:103016 | YES | YES | YES | Cytoplasm | phosphatase | ||||
| ENSMUSG00000014956 | Ppp1cb | protein phosphatase 1, catalytic subunit, beta isoform | MGI:104871 | YES | YES | YES | Cytoplasm | phosphatase | ||||
| ENSMUSG00000019907 | Ppp1r12a | protein phosphatase 1, regulatory (inhibitor) subunit 12A | MGI:1309528 | YES | YES | YES | Cytoplasm | phosphatase | ||||
| ENSMUSG00000020349 | Ppp2ca | protein phosphatase 2 (formerly 2A), catalytic subunit, alpha isoform | MGI:1321159 | YES | YES | YES | Cytoplasm | phosphatase | ||||
| ENSMUSG00000007564 | Ppp2r1a | protein phosphatase 2, regulatory subunit A, alpha | MGI:1926334 | YES | YES | YES | Cytoplasm | phosphatase | ||||
| ENSMUSG00000022052 | Ppp2r2a | protein phosphatase 2, regulatory subunit B, alpha | MGI:1919228 | YES | YES | YES | Cytoplasm | phosphatase | ||||
| ENSMUSG00000039515 | Ppp2r4 | protein phosphatase 2A activator, regulatory subunit B | MGI:1346006 | YES | YES | YES | Cytoplasm | phosphatase | ||||
| ENSMUSG00000024777 | Ppp2r5b | protein phosphatase 2, regulatory subunit B', beta | MGI:2388480 | YES | YES | YES | Cytoplasm | phosphatase | ||||
| ENSMUSG00000021051 | Ppp2r5e | protein phosphatase 2, regulatory subunit B', epsilon | MGI:1349473 | YES | YES | YES | Cytoplasm | phosphatase | ||||
| ENSMUSG00000028657 | Ppt1 | palmitoyl-protein thioesterase 1 | MGI:1298204 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000028691 | Prdx1 | peroxiredoxin 1 | MGI:99523 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000005161 | Prdx2 | peroxiredoxin 2 | MGI:109486 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000025289 | Prdx4 | peroxiredoxin 4 | MGI:1859815 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000045302 | Preb | prolactin regulatory element binding | MGI:1355326 | YES | YES | YES | Nucleus | transcription regulator | ||||
| ENSMUSG00000005469 | Prkaca | protein kinase, cAMP dependent, catalytic, alpha | MGI:97592 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSMUSG00000005034 | Prkacb | protein kinase, cAMP dependent, catalytic, beta | MGI:97594 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSMUSG00000020612 | Prkar1a | protein kinase, cAMP dependent regulatory, type I, alpha | MGI:104878 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSMUSG00000032601 | Prkar2a | protein kinase, cAMP dependent regulatory, type II alpha | MGI:108025 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSMUSG00000002997 | Prkar2b | protein kinase, cAMP dependent regulatory, type II beta | MGI:97760 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSMUSG00000050965 | Prkca | protein kinase C, alpha | MGI:97595 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSMUSG00000052889 | Prkcb | protein kinase C, beta | MGI:97596 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSMUSG00000078816 | Prkcg | protein kinase C, gamma | MGI:97597 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSMUSG00000030350 | Prmt8 | protein arginine N-methyltransferase 8 | MGI:3043083 | YES | YES | YES | Nucleus | enzyme | ||||
| ENSMUSG00000092305 | Prps1l1 | phosphoribosyl pyrophosphate synthetase 1-like 1 | MGI:1922706 | YES | YES | YES | Other | kinase | ||||
| ENSMUSG00000020528 | Prpsap2 | phosphoribosyl pyrophosphate synthetase-associated protein 2 | MGI:2384838 | YES | YES | YES | Other | other | ||||
| ENSMUSG00000064125 | Prr36 | proline rich 36 | MGI:3605626 | YES | YES | YES | Other | other | ||||
| ENSMUSG00000024393 | Prrc2a | proline-rich coiled-coil 2A | MGI:1915467 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000040225 | Prrc2c | proline-rich coiled-coil 2C | MGI:1913754 | YES | YES | YES | Other | other | ||||
| ENSMUSG00000015476 | Prrt1 | proline-rich transmembrane protein 1 | MGI:1932118 | YES | YES | YES | Other | other | ||||
| ENSMUSG00000037126 | Psd | pleckstrin and Sec7 domain containing | MGI:1920978 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000030465 | Psd3 | pleckstrin and Sec7 domain containing 3 | MGI:1918215 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000019969 | Psen1 | presenilin 1 | MGI:1202717 | YES | YES | YES | Plasma Membrane | peptidase | ||||
| ENSMUSG00000060073 | Psma3 | proteasome (prosome, macropain) subunit, alpha type 3 | MGI:104883 | YES | YES | YES | Cytoplasm | peptidase | ||||
| ENSMUSG00000068749 | Psma5 | proteasome (prosome, macropain) subunit, alpha type 5 | MGI:1347009 | YES | YES | YES | Cytoplasm | peptidase | ||||
| ENSMUSG00000021024 | Psma6 | proteasome (prosome, macropain) subunit, alpha type 6 | MGI:1347006 | YES | YES | YES | Cytoplasm | peptidase | ||||
| ENSMUSG00000027566 | Psma7 | proteasome (prosome, macropain) subunit, alpha type 7 | MGI:1347070 | YES | YES | YES | Cytoplasm | peptidase | ||||
| ENSMUSG00000014769 | Psmb1 | proteasome (prosome, macropain) subunit, beta type 1 | MGI:104884 | YES | YES | YES | Cytoplasm | peptidase | ||||
| ENSMUSG00000005779 | Psmb4 | proteasome (prosome, macropain) subunit, beta type 4 | MGI:1098257 | YES | YES | YES | Cytoplasm | peptidase | ||||
| ENSMUSG00000018286 | Psmb6 | proteasome (prosome, macropain) subunit, beta type 6 | MGI:104880 | YES | YES | YES | Nucleus | peptidase | ||||
| ENSMUSG00000028932 | Psmc2 | proteasome (prosome, macropain) 26S subunit, ATPase 2 | MGI:109555 | YES | YES | YES | Nucleus | peptidase | ||||
| ENSMUSG00000002102 | Psmc3 | proteasome (prosome, macropain) 26S subunit, ATPase 3 | MGI:1098754 | YES | YES | YES | Nucleus | transcription regulator | ||||
| ENSMUSG00000030603 | Psmc4 | proteasome (prosome, macropain) 26S subunit, ATPase, 4 | MGI:1346093 | YES | YES | YES | Nucleus | peptidase | ||||
| ENSMUSG00000020708 | Psmc5 | protease (prosome, macropain) 26S subunit, ATPase 5 | MGI:105047 | YES | YES | YES | Nucleus | transcription regulator | ||||
| ENSMUSG00000021832 | Psmc6 | proteasome (prosome, macropain) 26S subunit, ATPase, 6 | MGI:1914339 | YES | YES | YES | Nucleus | peptidase | ||||
| ENSMUSG00000017428 | Psmd11 | proteasome (prosome, macropain) 26S subunit, non-ATPase, 11 | MGI:1916327 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000020720 | Psmd12 | proteasome (prosome, macropain) 26S subunit, non-ATPase, 12 | MGI:1914247 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000025487 | Psmd13 | proteasome (prosome, macropain) 26S subunit, non-ATPase, 13 | MGI:1345192 | YES | YES | YES | Cytoplasm | peptidase | ||||
| ENSMUSG00000017221 | Psmd3 | proteasome (prosome, macropain) 26S subunit, non-ATPase, 3 | MGI:98858 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000021737 | Psmd6 | proteasome (prosome, macropain) 26S subunit, non-ATPase, 6 | MGI:1913663 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000039067 | Psmd7 | proteasome (prosome, macropain) 26S subunit, non-ATPase, 7 | MGI:1351511 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000021938 | Pspc1 | paraspeckle protein 1 | MGI:1913895 | YES | YES | YES | Nucleus | other | ||||
| ENSMUSG00000021518 | Ptdss1 | phosphatidylserine synthase 1 | MGI:1276575 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000063235 | Ptpmt1 | protein tyrosine phosphatase, mitochondrial 1 | MGI:1913711 | YES | YES | YES | Cytoplasm | phosphatase | ||||
| ENSMUSG00000028399 | Ptprd | protein tyrosine phosphatase, receptor type, D | MGI:97812 | YES | YES | YES | Plasma Membrane | phosphatase | ||||
| ENSMUSG00000056553 | Ptprn2 | protein tyrosine phosphatase, receptor type, N polypeptide 2 | MGI:107418 | YES | YES | YES | Plasma Membrane | phosphatase | ||||
| ENSMUSG00000013236 | Ptprs | protein tyrosine phosphatase, receptor type, S | MGI:97815 | YES | YES | YES | Plasma Membrane | phosphatase | ||||
| ENSMUSG00000053141 | Ptprt | protein tyrosine phosphatase, receptor type, T | MGI:1321152 | YES | YES | YES | Plasma Membrane | phosphatase | ||||
| ENSMUSG00000068748 | Ptprz1 | protein tyrosine phosphatase, receptor type Z, polypeptide 1 | MGI:97816 | YES | YES | YES | Plasma Membrane | phosphatase | ||||
| ENSMUSG00000072582 | Ptrh2 | peptidyl-tRNA hydrolase 2 | MGI:2444848 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000022571 | Pycrl | pyrroline-5-carboxylate reductase-like | MGI:1913444 | YES | YES | YES | Other | enzyme | ||||
| ENSMUSG00000032604 | Qars | glutaminyl-tRNA synthetase | MGI:1915851 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000062078 | Qk | quaking | MGI:97837 | YES | YES | YES | Nucleus | other | ||||
| ENSMUSG00000023460 | Rab12 | RAB12, member RAS oncogene family | MGI:894284 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000021062 | Rab15 | RAB15, member RAS oncogene family | MGI:1916865 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000020132 | Rab21 | RAB21, member RAS oncogene family | MGI:894308 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000027519 | Rab22a | RAB22A, member RAS oncogene family | MGI:105072 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000027739 | Rab33b | RAB33B, member RAS oncogene family | MGI:1330805 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000019066 | Rab3d | RAB3D, member RAS oncogene family | MGI:97844 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000003037 | Rab8a | RAB8A, member RAS oncogene family | MGI:96960 | YES | YES | YES | Plasma Membrane | enzyme | ||||
| ENSMUSG00000036943 | Rab8b | RAB8B, member RAS oncogene family | MGI:2442982 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000020817 | Rabep1 | rabaptin, RAB GTPase binding effector protein 1 | MGI:1860236 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSMUSG00000021027 | Ralgapa1 | Ral GTPase activating protein, alpha subunit 1 | MGI:1931050 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000027652 | Ralgapb | Ral GTPase activating protein, beta subunit (non-catalytic) | MGI:2444531 | YES | YES | YES | Other | other | ||||
| ENSMUSG00000029430 | Ran | RAN, member RAS oncogene family | MGI:1333112 | YES | YES | YES | Nucleus | enzyme | ||||
| ENSMUSG00000068798 | Rap1a | RAS-related protein-1a | MGI:97852 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000052681 | Rap1b | RAS related protein 1b | MGI:894315 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000062232 | Rapgef2 | Rap guanine nucleotide exchange factor (GEF) 2 | MGI:2659071 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000031453 | Rasa3 | RAS p21 protein activator 3 | MGI:1197013 | YES | YES | YES | Plasma Membrane | ion channel | ||||
| ENSMUSG00000070565 | Rasal2 | RAS protein activator like 2 | MGI:2443881 | YES | YES | YES | Extracellular Space | other | ||||
| ENSMUSG00000032356 | Rasgrf1 | RAS protein-specific guanine nucleotide-releasing factor 1 | MGI:99694 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000032320 | Rcn2 | reticulocalbin 2 | MGI:1349765 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000008435 | Rdh13 | retinol dehydrogenase 13 (all-trans and 9-cis) | MGI:1918732 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000021219 | Rgs6 | regulator of G-protein signaling 6 | MGI:1354730 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000026527 | Rgs7 | regulator of G protein signaling 7 | MGI:1346089 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000028945 | Rheb | Ras homolog enriched in brain | MGI:97912 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000029449 | Rhof | ras homolog gene family, member f | MGI:1345629 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000024143 | Rhoq | ras homolog gene family, member Q | MGI:1931553 | YES | YES | YES | Plasma Membrane | enzyme | ||||
| ENSMUSG00000017686 | Rhot1 | ras homolog gene family, member T1 | MGI:1926078 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000025485 | Ric8 | resistance to inhibitors of cholinesterase 8 homolog (C. elegans) | MGI:2141866 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000032890 | Rims3 | regulating synaptic membrane exocytosis 3 | MGI:2443331 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000070730 | Rmdn3 | regulator of microtubule dynamics 3 | MGI:1915059 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000022883 | Robo1 | roundabout homolog 1 (Drosophila) | MGI:1274781 | YES | YES | YES | Plasma Membrane | transmembrane receptor | ||||
| ENSMUSG00000052516 | Robo2 | roundabout homolog 2 (Drosophila) | MGI:1890110 | YES | YES | YES | Plasma Membrane | transmembrane receptor | ||||
| ENSMUSG00000022540 | Rogdi | rogdi homolog (Drosophila) | MGI:1913299 | YES | YES | YES | Nucleus | other | ||||
| ENSMUSG00000038510 | Rpf2 | ribosome production factor 2 homolog (S. cerevisiae) | MGI:1914489 | YES | YES | YES | Nucleus | other | ||||
| ENSMUSG00000029608 | Rph3a | rabphilin 3A | MGI:102788 | YES | YES | YES | Plasma Membrane | transporter | ||||
| ENSMUSG00000045128 | Rpl18a | ribosomal protein L18A | MGI:1924058 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000028936 | Rpl22 | ribosomal protein L22 | MGI:99262 | YES | YES | YES | Nucleus | other | ||||
| ENSMUSG00000071415 | Rpl23 | ribosomal protein L23 | MGI:1929455 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000058600 | Rpl30 | ribosomal protein L30 | MGI:98037 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000058558 | Rpl5 | ribosomal protein L5 | MGI:102854 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000067274 | Rplp0 | ribosomal protein, large, P0 | MGI:1927636 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000007892 | Rplp1 | ribosomal protein, large, P1 | MGI:1927099 | YES | YES | YES | Nucleus | other | ||||
| ENSMUSG00000030062 | Rpn1 | ribophorin I | MGI:98084 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000027642 | Rpn2 | ribophorin II | MGI:98085 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000067038 | Rps12-ps3 | ribosomal protein S12, pseudogene 3 | MGI:3704503 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000008683 | Rps15a | ribosomal protein S15A | MGI:2389091 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000009927 | Rps25 | ribosomal protein S25 | MGI:1922867 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000020460 | Rps27a | ribosomal protein S27A | MGI:1925544 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000030744 | Rps3 | ribosomal protein S3 | MGI:1350917 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000012848 | Rps5 | ribosomal protein S5 | MGI:1097682 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000038387 | Rras | Harvey rat sarcoma oncogene, subgroup R | MGI:98179 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000055723 | Rras2 | related RAS viral (r-ras) oncogene homolog 2 | MGI:1914172 | YES | YES | YES | Plasma Membrane | enzyme | ||||
| ENSMUSG00000027422 | Rrbp1 | ribosome binding protein 1 | MGI:1932395 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSMUSG00000029291 | Rufy3 | RUN and FYVE domain containing 3 | MGI:106484 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000006575 | Rundc3a | RUN domain containing 3A | MGI:1858752 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000021313 | Ryr2 | ryanodine receptor 2, cardiac | MGI:99685 | YES | YES | YES | Plasma Membrane | ion channel | ||||
| ENSMUSG00000045092 | S1pr1 | sphingosine-1-phosphate receptor 1 | MGI:1096355 | YES | YES | YES | Plasma Membrane | G-protein coupled receptor | ||||
| ENSMUSG00000025240 | Sacm1l | SAC1 (suppressor of actin mutations 1, homolog)-like (S. cerevisiae) | MGI:1933169 | YES | YES | YES | Cytoplasm | phosphatase | ||||
| ENSMUSG00000048279 | Sacs | sacsin | MGI:1354724 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000022437 | Samm50 | sorting and assembly machinery component 50 homolog (S. cerevisiae) | MGI:1915903 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000020088 | Sar1a | SAR1 gene homolog A (S. cerevisiae) | MGI:98230 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000050132 | Sarm1 | sterile alpha and HEAT/Armadillo motif containing 1 | MGI:2136419 | YES | YES | YES | Plasma Membrane | transmembrane receptor | ||||
| ENSMUSG00000068739 | Sars | seryl-aminoacyl-tRNA synthetase | MGI:102809 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000040722 | Scamp5 | secretory carrier membrane protein 5 | MGI:1928948 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000038936 | Sccpdh | saccharopine dehydrogenase (putative) | MGI:1924486 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000062110 | Scfd2 | Sec1 family domain containing 2 | MGI:2443446 | YES | YES | YES | Other | transporter | ||||
| ENSMUSG00000050711 | Scg2 | secretogranin II | MGI:103033 | YES | YES | YES | Extracellular Space | cytokine | ||||
| ENSMUSG00000091780 | Sco2 | SCO cytochrome oxidase deficient homolog 2 (yeast) | MGI:3818630 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000022568 | Scrib | scribbled homolog (Drosophila) | MGI:2145950 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000019124 | Scrn1 | secernin 1 | MGI:1917188 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000025743 | Sdc3 | syndecan 3 | MGI:1349163 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000028249 | Sdcbp | syndecan binding protein | MGI:1337026 | YES | YES | YES | Plasma Membrane | enzyme | ||||
| ENSMUSG00000002064 | Sdf2 | stromal cell derived factor 2 | MGI:108019 | YES | YES | YES | Extracellular Space | enzyme | ||||
| ENSMUSG00000025724 | Sec11a | SEC11 homolog A (S. cerevisiae) | MGI:1929464 | YES | YES | YES | Cytoplasm | peptidase | ||||
| ENSMUSG00000024516 | Sec11c | SEC11 homolog C (S. cerevisiae) | MGI:1913536 | YES | YES | YES | Cytoplasm | peptidase | ||||
| ENSMUSG00000027879 | Sec22b | SEC22 vesicle trafficking protein homolog B (S. cerevisiae) | MGI:1338759 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000020986 | Sec23a | SEC23A (S. cerevisiae) | MGI:1349635 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSMUSG00000001052 | Sec24b | Sec24 related gene family, member B (S. cerevisiae) | MGI:2139764 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSMUSG00000039367 | Sec24c | Sec24 related gene family, member C (S. cerevisiae) | MGI:1919746 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSMUSG00000053317 | Sec61b | Sec61 beta subunit | MGI:1913462 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSMUSG00000021451 | Sema4d | sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4D | MGI:109244 | YES | YES | YES | Plasma Membrane | transmembrane receptor | ||||
| ENSMUSG00000000627 | Sema4f | sema domain, immunoglobulin domain (Ig), TM domain, and short cytoplasmic domain | MGI:1340055 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000038264 | Sema7a | sema domain, immunoglobulin domain (Ig), and GPI membrane anchor, (semaphorin) 7A | MGI:1306826 | YES | YES | YES | Plasma Membrane | transmembrane receptor | ||||
| ENSMUSG00000026249 | Serpine2 | serine (or cysteine) peptidase inhibitor, clade E, member 2 | MGI:101780 | YES | YES | YES | Extracellular Space | other | ||||
| ENSMUSG00000070436 | Serpinh1 | serine (or cysteine) peptidase inhibitor, clade H, member 1 | MGI:88283 | YES | YES | YES | Extracellular Space | other | ||||
| ENSMUSG00000042272 | Sestd1 | SEC14 and spectrin domains 1 | MGI:1916262 | YES | YES | YES | Extracellular Space | other | ||||
| ENSMUSG00000006527 | Sfmbt1 | Scm-like with four mbt domains 1 | MGI:1859609 | YES | YES | YES | Nucleus | transcription regulator | ||||
| ENSMUSG00000028820 | Sfpq | splicing factor proline/glutamine rich (polypyrimidine tract binding protein associated) | MGI:1918764 | YES | YES | YES | Nucleus | other | ||||
| ENSMUSG00000025212 | Sfxn3 | sideroflexin 3 | MGI:2137679 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSMUSG00000028524 | Sgip1 | SH3-domain GRB2-like (endophilin) interacting protein 1 | MGI:1920344 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000020097 | Sgpl1 | sphingosine phosphate lyase 1 | MGI:1261415 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000025403 | Shmt2 | serine hydroxymethyltransferase 2 (mitochondrial) | MGI:1277989 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000024976 | Shoc2 | soc-2 (suppressor of clear) homolog (C. elegans) | MGI:1927197 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000034135 | Sik3 | SIK family kinase 3 | MGI:2446296 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSMUSG00000036087 | Slain2 | SLAIN motif family, member 2 | MGI:1923241 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000024597 | Slc12a2 | solute carrier family 12, member 2 | MGI:101924 | YES | YES | YES | Plasma Membrane | transporter | ||||
| ENSMUSG00000005360 | Slc1a3 | solute carrier family 1 (glial high affinity glutamate transporter), member 3 | MGI:99917 | YES | YES | YES | Plasma Membrane | transporter | ||||
| ENSMUSG00000003528 | Slc25a1 | solute carrier family 25 (mitochondrial carrier, citrate transporter), member 1 | MGI:1345283 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSMUSG00000025792 | Slc25a10 | solute carrier family 25 (mitochondrial carrier, dicarboxylate transporter), member 10 | MGI:1353497 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSMUSG00000014606 | Slc25a11 | solute carrier family 25 (mitochondrial carrier oxoglutarate carrier), member 11 | MGI:1915113 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSMUSG00000027010 | Slc25a12 | solute carrier family 25 (mitochondrial carrier, Aralar), member 12 | MGI:1926080 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSMUSG00000004902 | Slc25a18 | solute carrier family 25 (mitochondrial carrier), member 18 | MGI:1919053 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSMUSG00000019082 | Slc25a22 | solute carrier family 25 (mitochondrial carrier, glutamate), member 22 | MGI:1915517 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSMUSG00000023912 | Slc25a27 | solute carrier family 25, member 27 | MGI:1921261 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSMUSG00000031633 | Slc25a4 | solute carrier family 25 (mitochondrial carrier, adenine nucleotide translocator), member 4 | MGI:1353495 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSMUSG00000002346 | Slc25a42 | solute carrier family 25, member 42 | MGI:1920345 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSMUSG00000024259 | Slc25a46 | solute carrier family 25, member 46 | MGI:1914703 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000016319 | Slc25a5 | solute carrier family 25 (mitochondrial carrier, adenine nucleotide translocator), member 5 | MGI:1353496 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSMUSG00000031808 | Slc27a1 | solute carrier family 27 (fatty acid transporter), member 1 | MGI:1347098 | YES | YES | YES | Plasma Membrane | transporter | ||||
| ENSMUSG00000059316 | Slc27a4 | solute carrier family 27 (fatty acid transporter), member 4 | MGI:1347347 | YES | YES | YES | Plasma Membrane | transporter | ||||
| ENSMUSG00000028645 | Slc2a1 | solute carrier family 2 (facilitated glucose transporter), member 1 | MGI:95755 | YES | YES | YES | Plasma Membrane | transporter | ||||
| ENSMUSG00000024327 | Slc39a7 | solute carrier family 39 (zinc transporter), member 7 | MGI:95909 | YES | YES | YES | Plasma Membrane | transporter | ||||
| ENSMUSG00000006574 | Slc4a1 | solute carrier family 4 (anion exchanger), member 1 | MGI:109393 | YES | YES | YES | Plasma Membrane | transporter | ||||
| ENSMUSG00000002504 | Slc9a3r2 | solute carrier family 9 (sodium/hydrogen exchanger), member 3 regulator 2 | MGI:1890662 | YES | YES | YES | Plasma Membrane | transporter | ||||
| ENSMUSG00000048304 | Slitrk3 | SLIT and NTRK-like family, member 3 | MGI:2679447 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000021870 | Slmap | sarcolemma associated protein | MGI:1933549 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000026155 | Smap1 | small ArfGAP 1 | MGI:2138261 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000061461 | Smim20 | small integral membrane protein 20 | MGI:1913528 | YES | YES | YES | Other | other | ||||
| ENSMUSG00000027273 | Snap25 | synaptosomal-associated protein 25 | MGI:98331 | YES | YES | YES | Plasma Membrane | transporter | ||||
| ENSMUSG00000022765 | Snap29 | synaptosomal-associated protein 29 | MGI:1914724 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSMUSG00000009894 | Snap47 | synaptosomal-associated protein, 47 | MGI:1915076 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000001424 | Snd1 | staphylococcal nuclease and tudor domain containing 1 | MGI:1929266 | YES | YES | YES | Nucleus | enzyme | ||||
| ENSMUSG00000028136 | Snx27 | sorting nexin family member 27 | MGI:1923992 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000038916 | Soga3 | SOGA family member 3 | MGI:1914662 | YES | YES | YES | Other | other | ||||
| ENSMUSG00000029093 | Sorcs2 | sortilin-related VPS10 domain containing receptor 2 | MGI:1932289 | YES | YES | YES | Plasma Membrane | transporter | ||||
| ENSMUSG00000018593 | Sparc | secreted acidic cysteine rich glycoprotein | MGI:98373 | YES | YES | YES | Extracellular Space | other | ||||
| ENSMUSG00000035227 | Spcs2 | signal peptidase complex subunit 2 homolog (S. cerevisiae) | MGI:1913874 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000026163 | Sphkap | SPHK1 interactor, AKAP domain containing | MGI:1924879 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000027351 | Spred1 | sprouty protein with EVH-1 domain 1, related sequence | MGI:2150016 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000021930 | Spryd7 | SPRY domain containing 7 | MGI:1913924 | YES | YES | YES | Other | other | ||||
| ENSMUSG00000057738 | Sptan1 | spectrin alpha, non-erythrocytic 1 | MGI:98386 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000020315 | Sptbn1 | spectrin beta, non-erythrocytic 1 | MGI:98388 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000067889 | Sptbn2 | spectrin beta, non-erythrocytic 2 | MGI:1313261 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000011751 | Sptbn4 | spectrin beta, non-erythrocytic 4 | MGI:1890574 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000005803 | Sqrdl | sulfide quinone reductase-like (yeast) | MGI:1929899 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000027646 | Src | Rous sarcoma oncogene | MGI:98397 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSMUSG00000026425 | Srgap2 | SLIT-ROBO Rho GTPase activating protein 2 | MGI:109605 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000030257 | Srgap3 | SLIT-ROBO Rho GTPase activating protein 3 | MGI:2152938 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000009549 | Srp14 | signal recognition particle 14 | MGI:107169 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000020780 | Srp68 | signal recognition particle 68 | MGI:1917447 | YES | YES | YES | Nucleus | other | ||||
| ENSMUSG00000062604 | Srpk2 | serine/arginine-rich protein specific kinase 2 | MGI:1201408 | YES | YES | YES | Nucleus | kinase | ||||
| ENSMUSG00000001323 | Srr | serine racemase | MGI:1351636 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000029911 | Ssbp1 | single-stranded DNA binding protein 1 | MGI:1920040 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000002014 | Ssr4 | signal sequence receptor, delta | MGI:1099464 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000039156 | Stim2 | stromal interaction molecule 2 | MGI:2151156 | YES | YES | YES | Plasma Membrane | transporter | ||||
| ENSMUSG00000063410 | Stk24 | serine/threonine kinase 24 | MGI:2385007 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSMUSG00000027581 | Stmn3 | stathmin-like 3 | MGI:1277137 | YES | YES | YES | Nucleus | other | ||||
| ENSMUSG00000028455 | Stoml2 | stomatin (Epb7.2)-like 2 | MGI:1913842 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000030224 | Strap | serine/threonine kinase receptor associated protein | MGI:1329037 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000024077 | Strn | striatin, calmodulin binding protein | MGI:1333757 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000020954 | Strn3 | striatin, calmodulin binding protein 3 | MGI:2151064 | YES | YES | YES | Nucleus | transcription regulator | ||||
| ENSMUSG00000030374 | Strn4 | striatin, calmodulin binding protein 4 | MGI:2142346 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000032116 | Stt3a | STT3, subunit of the oligosaccharyltransferase complex, homolog A (S. cerevisiae) | MGI:105124 | YES | YES | YES | Plasma Membrane | enzyme | ||||
| ENSMUSG00000032437 | Stt3b | STT3, subunit of the oligosaccharyltransferase complex, homolog B (S. cerevisiae) | MGI:1915542 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000039615 | Stub1 | STIP1 homology and U-Box containing protein 1 | MGI:1891731 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000028879 | Stx12 | syntaxin 12 | MGI:1931027 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000027522 | Stx16 | syntaxin 16 | MGI:1923396 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSMUSG00000010110 | Stx5a | syntaxin 5A | MGI:1928483 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSMUSG00000026470 | Stx6 | syntaxin 6 | MGI:1926235 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSMUSG00000019998 | Stx7 | syntaxin 7 | MGI:1858210 | YES | YES | YES | Plasma Membrane | transporter | ||||
| ENSMUSG00000020903 | Stx8 | syntaxin 8 | MGI:1890156 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000046314 | Stxbp6 | syntaxin binding protein 6 (amisyn) | MGI:2384963 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000052738 | Suclg1 | succinate-CoA ligase, GDP-forming, alpha subunit | MGI:1927234 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000022024 | Sugt1 | SGT1, suppressor of G2 allele of SKP1 (S. cerevisiae) | MGI:1915205 | YES | YES | YES | Nucleus | other | ||||
| ENSMUSG00000022340 | Sybu | syntabulin (syntaxin-interacting) | MGI:2442392 | YES | YES | YES | Other | other | ||||
| ENSMUSG00000009394 | Syn2 | synapsin II | MGI:103020 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000032423 | Syncrip | synaptotagmin binding, cytoplasmic RNA interacting protein | MGI:1891690 | YES | YES | YES | Nucleus | other | ||||
| ENSMUSG00000007021 | Syngr3 | synaptogyrin 3 | MGI:1341881 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000021139 | Gm20498 | predicted gene 20498 | MGI:5141963 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000035864 | Syt1 | synaptotagmin I | MGI:99667 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSMUSG00000058420 | Syt17 | synaptotagmin XVII | MGI:104966 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000030731 | Syt3 | synaptotagmin III | MGI:99665 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSMUSG00000065954 | Tacc1 | transforming, acidic coiled-coil containing protein 1 | MGI:2443510 | YES | YES | YES | Nucleus | other | ||||
| ENSMUSG00000022658 | Tagln3 | transgelin 3 | MGI:1926784 | YES | YES | YES | Extracellular Space | other | ||||
| ENSMUSG00000030316 | Tamm41 | TAM41, mitochondrial translocator assembly and maintenance protein, homolog (S. cerevisiae) | MGI:1916221 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000035168 | Tanc1 | tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1 | MGI:1914110 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000053580 | Tanc2 | tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2 | MGI:2444121 | YES | YES | YES | Nucleus | transcription regulator | ||||
| ENSMUSG00000017291 | Taok1 | TAO kinase 1 | MGI:1914490 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSMUSG00000041459 | Tardbp | TAR DNA binding protein | MGI:2387629 | YES | YES | YES | Nucleus | transcription regulator | ||||
| ENSMUSG00000036473 | Tbc1d24 | TBC1 domain family, member 24 | MGI:2443456 | YES | YES | YES | #N/A | #N/A | ||||
| ENSMUSG00000079658 | Tceb1 | transcription elongation factor B (SIII), polypeptide 1 | MGI:1915173 | YES | YES | YES | Nucleus | transcription regulator | ||||
| ENSMUSG00000055839 | Tceb2 | transcription elongation factor B (SIII), polypeptide 2 | MGI:1914923 | YES | YES | YES | Nucleus | transcription regulator | ||||
| ENSMUSG00000068039 | Tcp1 | t-complex protein 1 | MGI:98535 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000041912 | Tdrkh | tudor and KH domain containing protein | MGI:1919884 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000031708 | Tecr | trans-2,3-enoyl-CoA reductase | MGI:1915408 | YES | YES | YES | Plasma Membrane | enzyme | ||||
| ENSMUSG00000049336 | Tenm2 | teneurin transmembrane protein 2 | MGI:1345184 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000031561 | Tenm3 | teneurin transmembrane protein 3 | MGI:1345183 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000020219 | Timm13 | translocase of inner mitochondrial membrane 13 | MGI:1353432 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSMUSG00000031158 | Timm17b | translocase of inner mitochondrial membrane 17b | MGI:1343176 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSMUSG00000020843 | Timm22 | translocase of inner mitochondrial membrane 22 | MGI:1929742 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSMUSG00000002949 | Timm44 | translocase of inner mitochondrial membrane 44 | MGI:1343262 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSMUSG00000045455 | Gm9797 | predicted pseudogene 9797 | MGI:3704349 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSMUSG00000030516 | Tjp1 | tight junction protein 1 | MGI:98759 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000024812 | Tjp2 | tight junction protein 2 | MGI:1341872 | YES | YES | YES | Plasma Membrane | kinase | ||||
| ENSMUSG00000042066 | Tmcc2 | transmembrane and coiled-coil domains 2 | MGI:1916125 | YES | YES | YES | Extracellular Space | other | ||||
| ENSMUSG00000052428 | Tmco1 | transmembrane and coiled-coil domains 1 | MGI:1921173 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000021248 | Tmed10 | transmembrane emp24-like trafficking protein 10 (yeast) | MGI:1915831 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSMUSG00000058569 | Tmed9 | transmembrane emp24 protein transport domain containing 9 | MGI:1914761 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSMUSG00000028347 | Tmeff1 | transmembrane protein with EGF-like and two follistatin-like domains 1 | MGI:1926810 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000026109 | Tmeff2 | transmembrane protein with EGF-like and two follistatin-like domains 2 | MGI:1861735 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000043284 | Tmem11 | transmembrane protein 11 | MGI:2144726 | YES | YES | YES | Plasma Membrane | G-protein coupled receptor | ||||
| ENSMUSG00000030615 | Tmem126a | transmembrane protein 126A | MGI:1913521 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000026347 | Tmem163 | transmembrane protein 163 | MGI:1919410 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000070394 | Tmem256 | transmembrane protein 256 | MGI:1916436 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000032328 | Tmem30a | transmembrane protein 30A | MGI:106402 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000037720 | Tmem33 | transmembrane protein 33 | MGI:1915128 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000028221 | Tmem55a | transmembrane protein 55A | MGI:1919769 | YES | YES | YES | Cytoplasm | phosphatase | ||||
| ENSMUSG00000036026 | Tmem63b | transmembrane protein 63b | MGI:2387609 | YES | YES | YES | Other | other | ||||
| ENSMUSG00000031021 | Tmem9b | TMEM9 domain family, member B | MGI:1915254 | YES | YES | YES | Other | other | ||||
| ENSMUSG00000027692 | Tnik | TRAF2 and NCK interacting kinase | MGI:1916264 | YES | YES | YES | Plasma Membrane | kinase | ||||
| ENSMUSG00000015829 | Tnr | tenascin R | MGI:99516 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000042870 | Tom1 | target of myb1 homolog (chicken) | MGI:1338026 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSMUSG00000000538 | Tom1l2 | target of myb1-like 2 (chicken) | MGI:2443306 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSMUSG00000002984 | Tomm40 | translocase of outer mitochondrial membrane 40 homolog (yeast) | MGI:1858259 | YES | YES | YES | Cytoplasm | ion channel | ||||
| ENSMUSG00000022752 | Tomm70a | translocase of outer mitochondrial membrane 70 homolog A (yeast) | MGI:106295 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSMUSG00000027506 | Tpd52 | tumor protein D52 | MGI:107749 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000023456 | Tpi1 | triosephosphate isomerase 1 | MGI:98797 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000032366 | Tpm1 | tropomyosin 1, alpha | MGI:98809 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000027940 | Tpm3 | tropomyosin 3, gamma | MGI:1890149 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000021573 | Tppp | tubulin polymerization promoting protein | MGI:1920198 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000000374 | Trappc10 | trafficking protein particle complex 10 | MGI:1336209 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSMUSG00000028847 | Trappc3 | trafficking protein particle complex 3 | MGI:1351486 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000040236 | Trappc5 | trafficking protein particle complex 5 | MGI:1913932 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000020993 | Trappc6b | trafficking protein particle complex 6B | MGI:1925482 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000033382 | Trappc8 | trafficking protein particle complex 8 | MGI:2443008 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSMUSG00000047921 | Trappc9 | trafficking protein particle complex 9 | MGI:1923760 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000050663 | Trhde | TRH-degrading enzyme | MGI:2384311 | YES | YES | YES | Plasma Membrane | peptidase | ||||
| ENSMUSG00000027993 | Trim2 | tripartite motif-containing 2 | MGI:1933163 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000021712 | Trim23 | tripartite motif-containing 23 | MGI:1933161 | YES | YES | YES | Nucleus | enzyme | ||||
| ENSMUSG00000036989 | Trim3 | tripartite motif-containing 3 | MGI:1860040 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000021071 | Trim9 | tripartite motif-containing 9 | MGI:2137354 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000022263 | Trio | triple functional domain (PTPRF interacting) | MGI:1927230 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSMUSG00000055963 | Triqk | triple QxxK/R motif containing | MGI:3650048 | YES | YES | YES | Other | other | ||||
| ENSMUSG00000018507 | Trpv2 | transient receptor potential cation channel, subfamily V, member 2 | MGI:1341836 | YES | YES | YES | Plasma Membrane | ion channel | ||||
| ENSMUSG00000002496 | Tsc2 | tuberous sclerosis 2 | MGI:102548 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000014402 | Tsg101 | tumor susceptibility gene 101 | MGI:106581 | YES | YES | YES | Cytoplasm | transcription regulator | ||||
| ENSMUSG00000027858 | Tspan2 | tetraspanin 2 | MGI:1917997 | YES | YES | YES | Extracellular Space | other | ||||
| ENSMUSG00000033530 | Ttc7b | tetratricopeptide repeat domain 7B | MGI:2144724 | YES | YES | YES | Other | other | ||||
| ENSMUSG00000007944 | Ttc9b | tetratricopeptide repeat domain 9B | MGI:1920282 | YES | YES | YES | Other | other | ||||
| ENSMUSG00000026202 | Tuba4a | tubulin, alpha 4A | MGI:1095410 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000058672 | Tubb2a | tubulin, beta 2A class IIA | MGI:107861 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000045136 | Tubb2b | tubulin, beta 2B class IIB | MGI:1920960 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000062380 | Tubb3 | tubulin, beta 3 class III | MGI:107813 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000062591 | Tubb4a | tubulin, beta 4A class IVA | MGI:107848 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000036752 | Tubb4b | tubulin, beta 4B class IVB | MGI:1915472 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000001525 | Tubb5 | tubulin, beta 5 class I | MGI:107812 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000045007 | Tubg2 | tubulin, gamma 2 | MGI:2144208 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000073838 | Tufm | Tu translation elongation factor, mitochondrial | MGI:1923686 | YES | YES | YES | Cytoplasm | translation regulator | ||||
| ENSMUSG00000039530 | Tusc3 | tumor suppressor candidate 3 | MGI:1933134 | YES | YES | YES | Plasma Membrane | transporter | ||||
| ENSMUSG00000022451 | Twf1 | twinfilin, actin-binding protein, homolog 1 (Drosophila) | MGI:1100520 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSMUSG00000023277 | Twf2 | twinfilin, actin-binding protein, homolog 2 (Drosophila) | MGI:1346078 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSMUSG00000028567 | Txndc12 | thioredoxin domain containing 12 (endoplasmic reticulum) | MGI:1913323 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000024583 | Txnl1 | thioredoxin-like 1 | MGI:1860078 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000042520 | Ubap2l | ubiquitin-associated protein 2-like | MGI:1921633 | YES | YES | YES | Other | other | ||||
| ENSMUSG00000074781 | Ube2n | ubiquitin-conjugating enzyme E2N | MGI:1934835 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000078923 | Ube2v1 | ubiquitin-conjugating enzyme E2 variant 1 | MGI:1913839 | YES | YES | YES | Nucleus | transcription regulator | ||||
| ENSMUSG00000022674 | Ube2v2 | ubiquitin-conjugating enzyme E2 variant 2 | MGI:1917870 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000001687 | Ubl3 | ubiquitin-like 3 | MGI:1344373 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000015290 | Ubl4 | ubiquitin-like 4 | MGI:95049 | YES | YES | YES | #N/A | #N/A | ||||
| ENSMUSG00000066036 | Ubr4 | ubiquitin protein ligase E3 component n-recognin 4 | MGI:1916366 | YES | YES | YES | Nucleus | enzyme | ||||
| ENSMUSG00000026353 | Ubxn4 | UBX domain protein 4 | MGI:1915062 | YES | YES | YES | Extracellular Space | other | ||||
| ENSMUSG00000019578 | Ubxn6 | UBX domain protein 6 | MGI:1913780 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000040359 | Ufl1 | UFM1 specific ligase 1 | MGI:1914740 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000032854 | Ugt8a | UDP galactosyltransferase 8A | MGI:109522 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000062151 | Unc13c | unc-13 homolog C (C. elegans) | MGI:2149021 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000055567 | Unc80 | unc-80 homolog (C. elegans) | MGI:2652882 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000058301 | Upf1 | UPF1 regulator of nonsense transcripts homolog (yeast) | MGI:107995 | YES | YES | YES | Nucleus | enzyme | ||||
| ENSMUSG00000044894 | Uqcrq | ubiquinol-cytochrome c reductase, complex III subunit VII | MGI:107807 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000071528 | Usmg5 | upregulated during skeletal muscle growth 5 | MGI:1891435 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000063317 | Usp31 | ubiquitin specific peptidase 31 | MGI:1923429 | YES | YES | YES | Nucleus | peptidase | ||||
| ENSMUSG00000054814 | Usp46 | ubiquitin specific peptidase 46 | MGI:1916977 | YES | YES | YES | Other | peptidase | ||||
| ENSMUSG00000019820 | Utrn | utrophin | MGI:104631 | YES | YES | YES | Plasma Membrane | transmembrane receptor | ||||
| ENSMUSG00000010936 | Vac14 | Vac14 homolog (S. cerevisiae) | MGI:2157980 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000030337 | Vamp1 | vesicle-associated membrane protein 1 | MGI:1313276 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSMUSG00000020894 | Vamp2 | vesicle-associated membrane protein 2 | MGI:1313277 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000028955 | Vamp3 | vesicle-associated membrane protein 3 | MGI:1321389 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000026696 | Vamp4 | vesicle-associated membrane protein 4 | MGI:1858730 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000051412 | Vamp7 | vesicle-associated membrane protein 7 | MGI:1096399 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSMUSG00000024091 | Vapa | vesicle-associated membrane protein, associated protein A | MGI:1353561 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000054455 | Vapb | vesicle-associated membrane protein, associated protein B and C | MGI:1928744 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000007029 | Vars | valyl-tRNA synthetase | MGI:90675 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000021614 | Vcan | versican | MGI:102889 | YES | YES | YES | Extracellular Space | other | ||||
| ENSMUSG00000021823 | Vcl | vinculin | MGI:98927 | YES | YES | YES | Plasma Membrane | enzyme | ||||
| ENSMUSG00000020402 | Vdac1 | voltage-dependent anion channel 1 | MGI:106919 | YES | YES | YES | Cytoplasm | ion channel | ||||
| ENSMUSG00000021771 | Vdac2 | voltage-dependent anion channel 2 | MGI:106915 | YES | YES | YES | Cytoplasm | ion channel | ||||
| ENSMUSG00000008892 | Vdac3 | voltage-dependent anion channel 3 | MGI:106922 | YES | YES | YES | Cytoplasm | ion channel | ||||
| ENSMUSG00000037428 | Vgf | VGF nerve growth factor inducible | MGI:1343180 | YES | YES | YES | Extracellular Space | growth factor | ||||
| ENSMUSG00000075701 | Vimp | VCP-interacting membrane protein | MGI:95994 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000032127 | Vps11 | vacuolar protein sorting 11 (yeast) | MGI:1918982 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSMUSG00000027411 | Vps16 | vacuolar protein sorting 16 (yeast) | MGI:2136772 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSMUSG00000034216 | Vps18 | vacuolar protein sorting 18 (yeast) | MGI:2443626 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSMUSG00000031988 | Vps26b | vacuolar protein sorting 26 homolog B (yeast) | MGI:1917656 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSMUSG00000059323 | Tonsl | tonsoku-like, DNA repair protein | MGI:1919999 | YES | YES | YES | #N/A | #N/A | ||||
| ENSMUSG00000029462 | Vps29 | vacuolar protein sorting 29 (S. pombe) | MGI:1928344 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSMUSG00000029434 | Vps33a | vacuolar protein sorting 33A (yeast) | MGI:1924823 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSMUSG00000048832 | Vps37c | vacuolar protein sorting 37C (yeast) | MGI:2147661 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000027291 | Vps39 | vacuolar protein sorting 39 (yeast) | MGI:2443189 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSMUSG00000041236 | Vps41 | vacuolar protein sorting 41 (yeast) | MGI:1929215 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSMUSG00000015747 | Vps45 | vacuolar protein sorting 45 (yeast) | MGI:891965 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSMUSG00000024797 | Vps51 | vacuolar protein sorting 51 homolog (S. cerevisiae) | MGI:1915755 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000024319 | Vps52 | vacuolar protein sorting 52 (yeast) | MGI:1330304 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000017288 | Vps53 | vacuolar protein sorting 53 (yeast) | MGI:1915549 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000001062 | Vps9d1 | VPS9 domain containing 1 | MGI:1914143 | YES | YES | YES | Other | transporter | ||||
| ENSMUSG00000054459 | Vsnl1 | visinin-like 1 | MGI:1349453 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000019868 | Vta1 | Vps20-associated 1 homolog (S. cerevisiae) | MGI:1913451 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000024983 | Vti1a | vesicle transport through interaction with t-SNAREs 1A | MGI:1855699 | YES | YES | YES | Plasma Membrane | transporter | ||||
| ENSMUSG00000021124 | Vti1b | vesicle transport through interaction with t-SNAREs 1B | MGI:1855688 | YES | YES | YES | Plasma Membrane | transporter | ||||
| ENSMUSG00000058997 | Vwa8 | von Willebrand factor A domain containing 8 | MGI:1919008 | YES | YES | YES | Other | other | ||||
| ENSMUSG00000019831 | Wasf1 | WAS protein family, member 1 | MGI:1890563 | YES | YES | YES | Nucleus | other | ||||
| ENSMUSG00000029636 | Wasf3 | WAS protein family, member 3 | MGI:2658986 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000029684 | Wasl | Wiskott-Aldrich syndrome-like (human) | MGI:1920428 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000034040 | Wbscr17 | Williams-Beuren syndrome chromosome region 17 homolog (human) | MGI:2137594 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000021147 | Wdr37 | WD repeat domain 37 | MGI:1920393 | YES | YES | YES | Other | other | ||||
| ENSMUSG00000040389 | Wdr47 | WD repeat domain 47 | MGI:2139593 | YES | YES | YES | Other | other | ||||
| ENSMUSG00000032512 | Wdr48 | WD repeat domain 48 | MGI:1914811 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000040560 | Wdr7 | WD repeat domain 7 | MGI:1860197 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000039474 | Wfs1 | Wolfram syndrome 1 homolog (human) | MGI:1328355 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000086040 | Wipf3 | WAS/WASL interacting protein family, member 3 | MGI:3044681 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000037989 | Wnk2 | WNK lysine deficient protein kinase 2 | MGI:1922857 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSMUSG00000004637 | Wwox | WW domain-containing oxidoreductase | MGI:1931237 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000027022 | Xirp2 | xin actin-binding repeat containing 2 | MGI:2685198 | YES | YES | YES | Other | other | ||||
| ENSMUSG00000051951 | Xkr4 | X Kell blood group precursor related family member 4 | MGI:3528744 | YES | YES | YES | Other | other | ||||
| ENSMUSG00000028811 | Yars | tyrosyl-tRNA synthetase | MGI:2147627 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000002741 | Ykt6 | YKT6 homolog (S. Cerevisiae) | MGI:1927550 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000026775 | Yme1l1 | YME1-like 1 (S. cerevisiae) | MGI:1351651 | YES | YES | YES | Cytoplasm | peptidase | ||||
| ENSMUSG00000018326 | Ywhab | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, beta polypeptide | MGI:1891917 | YES | YES | YES | Cytoplasm | transcription regulator | ||||
| ENSMUSG00000020849 | Ywhae | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, epsilon polypeptide | MGI:894689 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000051391 | Ywhag | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, gamma polypeptide | MGI:108109 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000018965 | Ywhah | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, eta polypeptide | MGI:109194 | YES | YES | YES | Cytoplasm | transcription regulator | ||||
| ENSMUSG00000076432 | Ywhaq | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta polypeptide | MGI:891963 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000022285 | Ywhaz | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide | MGI:109484 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000034173 | Zbed5 | zinc finger, BED type containing 5 | MGI:1919220 | YES | YES | YES | Nucleus | other | ||||
| ENSMUSG00000020083 | 2010107G23Rik | RIKEN cDNA 2010107G23 gene | MGI:1917144 | YES | YES | Other | other | |||||
| ENSMUSG00000031824 | 6430548M08Rik | RIKEN cDNA 6430548M08 gene | MGI:2443793 | YES | YES | Other | other | |||||
| ENSMUSG00000031960 | Aars | alanyl-tRNA synthetase | MGI:2384560 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000057880 | Abat | 4-aminobutyrate aminotransferase | MGI:2443582 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000024130 | Abca3 | ATP-binding cassette, sub-family A (ABC1), member 3 | MGI:1351617 | YES | YES | Plasma Membrane | transporter | |||||
| ENSMUSG00000018800 | Abca5 | ATP-binding cassette, sub-family A (ABC1), member 5 | MGI:2386607 | YES | YES | Plasma Membrane | transporter | |||||
| ENSMUSG00000026198 | Abcb6 | ATP-binding cassette, sub-family B (MDR/TAP), member 6 | MGI:1921354 | YES | YES | Cytoplasm | transporter | |||||
| ENSMUSG00000029408 | Abcb9 | ATP-binding cassette, sub-family B (MDR/TAP), member 9 | MGI:1861729 | YES | YES | Cytoplasm | transporter | |||||
| ENSMUSG00000029802 | Abcg2 | ATP-binding cassette, sub-family G (WHITE), member 2 | MGI:1347061 | YES | YES | Plasma Membrane | transporter | |||||
| ENSMUSG00000017631 | Abr | active BCR-related gene | MGI:107771 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000036138 | Acaa1a | acetyl-Coenzyme A acyltransferase 1A | MGI:2148491 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000027710 | Acad9 | acyl-Coenzyme A dehydrogenase family, member 9 | MGI:1914272 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000062908 | Acadm | acyl-Coenzyme A dehydrogenase, medium chain | MGI:87867 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000018574 | Acadvl | acyl-Coenzyme A dehydrogenase, very long chain | MGI:895149 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000020917 | Acly | ATP citrate lyase | MGI:103251 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000022477 | Aco2 | aconitase 2, mitochondrial | MGI:87880 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000034853 | Acot11 | acyl-CoA thioesterase 11 | MGI:1913736 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000006717 | Acot13 | acyl-CoA thioesterase 13 | MGI:1914084 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000028937 | Acot7 | acyl-CoA thioesterase 7 | MGI:1917275 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000020777 | Acox1 | acyl-Coenzyme A oxidase 1, palmitoyl | MGI:1330812 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000032281 | Acsbg1 | acyl-CoA synthetase bubblegum family member 1 | MGI:2385656 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000018796 | Acsl1 | acyl-CoA synthetase long-chain family member 1 | MGI:102797 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000054693 | Adam10 | a disintegrin and metallopeptidase domain 10 | MGI:109548 | YES | YES | Plasma Membrane | peptidase | |||||
| ENSMUSG00000021044 | Adck1 | aarF domain containing kinase 1 | MGI:1919363 | YES | YES | Cytoplasm | kinase | |||||
| ENSMUSG00000022840 | Adcy5 | adenylate cyclase 5 | MGI:99673 | YES | YES | Plasma Membrane | enzyme | |||||
| ENSMUSG00000005580 | Adcy9 | adenylate cyclase 9 | MGI:108450 | YES | YES | Plasma Membrane | enzyme | |||||
| ENSMUSG00000027893 | Ahcyl1 | S-adenosylhomocysteine hydrolase-like 1 | MGI:2385184 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000038587 | Akap12 | A kinase (PRKA) anchor protein (gravin) 12 | MGI:1932576 | YES | YES | Cytoplasm | transporter | |||||
| ENSMUSG00000039166 | Akap7 | A kinase (PRKA) anchor protein 7 | MGI:1859150 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000029368 | Alb | albumin | MGI:87991 | YES | YES | Extracellular Space | transporter | |||||
| ENSMUSG00000022636 | Alcam | activated leukocyte cell adhesion molecule | MGI:1313266 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000035561 | Aldh1b1 | aldehyde dehydrogenase 1 family, member B1 | MGI:1919785 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000030088 | Aldh1l1 | aldehyde dehydrogenase 1 family, member L1 | MGI:1340024 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000029455 | Aldh2 | aldehyde dehydrogenase 2, mitochondrial | MGI:99600 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000021238 | Aldh6a1 | aldehyde dehydrogenase family 6, subfamily A1 | MGI:1915077 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000017390 | Aldoc | aldolase C, fructose-bisphosphate | MGI:101863 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000021314 | Amph | amphiphysin | MGI:103574 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000048307 | Ankrd46 | ankyrin repeat domain 46 | MGI:1916089 | YES | YES | Nucleus | transcription regulator | |||||
| ENSMUSG00000031731 | Ap1g1 | adaptor protein complex AP-1, gamma 1 subunit | MGI:101919 | YES | YES | Cytoplasm | transporter | |||||
| ENSMUSG00000006651 | Aplp1 | amyloid beta (A4) precursor-like protein 1 | MGI:88046 | YES | YES | Extracellular Space | other | |||||
| ENSMUSG00000033096 | Apmap | adipocyte plasma membrane associated protein | MGI:1919131 | YES | YES | Plasma Membrane | enzyme | |||||
| ENSMUSG00000002985 | Apoe | apolipoprotein E | MGI:88057 | YES | YES | Extracellular Space | transporter | |||||
| ENSMUSG00000079508 | Apoo | apolipoprotein O | MGI:1915566 | YES | YES | Extracellular Space | other | |||||
| ENSMUSG00000022892 | App | amyloid beta (A4) precursor protein | MGI:88059 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000024411 | Aqp4 | aquaporin 4 | MGI:107387 | YES | YES | Plasma Membrane | transporter | |||||
| ENSMUSG00000059495 | Arhgef12 | Rho guanine nucleotide exchange factor (GEF) 12 | MGI:1916882 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000030105 | Arl8b | ADP-ribosylation factor-like 8B | MGI:1914416 | YES | YES | Plasma Membrane | enzyme | |||||
| ENSMUSG00000008475 | Arpc5 | actin related protein 2/3 complex, subunit 5 | MGI:1915021 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000052456 | Asna1 | arsA arsenite transporter, ATP-binding, homolog 1 (bacterial) | MGI:1928379 | YES | YES | Nucleus | transporter | |||||
| ENSMUSG00000024654 | Asrgl1 | asparaginase like 1 | MGI:1913764 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000026192 | Atic | 5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase/IMP cyclohydrolase | MGI:1351352 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000021066 | Atl1 | atlastin GTPase 1 | MGI:1921241 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000059811 | Atl2 | atlastin GTPase 2 | MGI:1929492 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000031862 | Atp13a1 | ATPase type 13A1 | MGI:2180801 | YES | YES | Extracellular Space | transporter | |||||
| ENSMUSG00000033161 | Atp1a1 | ATPase, Na+/K+ transporting, alpha 1 polypeptide | MGI:88105 | YES | YES | Plasma Membrane | transporter | |||||
| ENSMUSG00000007097 | Atp1a2 | ATPase, Na+/K+ transporting, alpha 2 polypeptide | MGI:88106 | YES | YES | Plasma Membrane | transporter | |||||
| ENSMUSG00000040907 | Atp1a3 | ATPase, Na+/K+ transporting, alpha 3 polypeptide | MGI:88107 | YES | YES | Plasma Membrane | transporter | |||||
| ENSMUSG00000026576 | Atp1b1 | ATPase, Na+/K+ transporting, beta 1 polypeptide | MGI:88108 | YES | YES | Plasma Membrane | transporter | |||||
| ENSMUSG00000041329 | Atp1b2 | ATPase, Na+/K+ transporting, beta 2 polypeptide | MGI:88109 | YES | YES | Plasma Membrane | transporter | |||||
| ENSMUSG00000029467 | Atp2a2 | ATPase, Ca++ transporting, cardiac muscle, slow twitch 2 | MGI:88110 | YES | YES | Cytoplasm | transporter | |||||
| ENSMUSG00000019943 | Atp2b1 | ATPase, Ca++ transporting, plasma membrane 1 | MGI:104653 | YES | YES | Plasma Membrane | transporter | |||||
| ENSMUSG00000030302 | Atp2b2 | ATPase, Ca++ transporting, plasma membrane 2 | MGI:105368 | YES | YES | Plasma Membrane | transporter | |||||
| ENSMUSG00000031376 | Atp2b3 | ATPase, Ca++ transporting, plasma membrane 3 | MGI:1347353 | YES | YES | Plasma Membrane | transporter | |||||
| ENSMUSG00000026463 | Atp2b4 | ATPase, Ca++ transporting, plasma membrane 4 | MGI:88111 | YES | YES | Plasma Membrane | transporter | |||||
| ENSMUSG00000032570 | Atp2c1 | ATPase, Ca++-sequestering | MGI:1889008 | YES | YES | Cytoplasm | transporter | |||||
| ENSMUSG00000025428 | Atp5a1 | ATP synthase, H+ transporting, mitochondrial F1 complex, alpha subunit 1 | MGI:88115 | YES | YES | Cytoplasm | transporter | |||||
| ENSMUSG00000025393 | Atp5b | ATP synthase, H+ transporting mitochondrial F1 complex, beta subunit | MGI:107801 | YES | YES | Cytoplasm | transporter | |||||
| ENSMUSG00000025781 | Atp5c1 | ATP synthase, H+ transporting, mitochondrial F1 complex, gamma polypeptide 1 | MGI:1261437 | YES | YES | Cytoplasm | transporter | |||||
| ENSMUSG00000003072 | Atp5d | ATP synthase, H+ transporting, mitochondrial F1 complex, delta subunit | MGI:1913293 | YES | YES | Cytoplasm | transporter | |||||
| ENSMUSG00000000563 | Atp5f1 | ATP synthase, H+ transporting, mitochondrial F0 complex, subunit B1 | MGI:1100495 | YES | YES | Cytoplasm | transporter | |||||
| ENSMUSG00000038690 | Atp5j2 | ATP synthase, H+ transporting, mitochondrial F0 complex, subunit F2 | MGI:1927558 | YES | YES | Cytoplasm | transporter | |||||
| ENSMUSG00000038717 | Atp5l | ATP synthase, H+ transporting, mitochondrial F0 complex, subunit G | MGI:1351597 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000037685 | Atp8a1 | ATPase, aminophospholipid transporter (APLT), class I, type 8A, member 1 | MGI:1330848 | YES | YES | Cytoplasm | transporter | |||||
| ENSMUSG00000021983 | Atp8a2 | ATPase, aminophospholipid transporter-like, class I, type 8A, member 2 | MGI:1354710 | YES | YES | Plasma Membrane | transporter | |||||
| ENSMUSG00000024392 | Bag6 | BCL2-associated athanogene 6 | MGI:1919439 | YES | YES | Nucleus | enzyme | |||||
| ENSMUSG00000009112 | Bcl2l13 | BCL2-like 13 (apoptosis facilitator) | MGI:2136959 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000046598 | Bdh1 | 3-hydroxybutyrate dehydrogenase, type 1 | MGI:1919161 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000024381 | Bin1 | bridging integrator 1 | MGI:108092 | YES | YES | Nucleus | other | |||||
| ENSMUSG00000067336 | Bmpr2 | bone morphogenetic protein receptor, type II (serine/threonine kinase) | MGI:1095407 | YES | YES | Plasma Membrane | kinase | |||||
| ENSMUSG00000023175 | Bsg | basigin | MGI:88208 | YES | YES | Plasma Membrane | transporter | |||||
| ENSMUSG00000032120 | C2cd2l | C2 calcium-dependent domain containing 2-like | MGI:1919014 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000027562 | Car2 | carbonic anhydrase 2 | MGI:88269 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000051331 | Cacna1c | calcium channel, voltage-dependent, L type, alpha 1C subunit | MGI:103013 | YES | YES | Plasma Membrane | ion channel | |||||
| ENSMUSG00000004110 | Cacna1e | calcium channel, voltage-dependent, R type, alpha 1E subunit | MGI:106217 | YES | YES | Plasma Membrane | ion channel | |||||
| ENSMUSG00000022416 | Cacna1i | calcium channel, voltage-dependent, alpha 1I subunit | MGI:2178051 | YES | YES | Plasma Membrane | ion channel | |||||
| ENSMUSG00000013629 | Cad | carbamoyl-phosphate synthetase 2, aspartate transcarbamylase, and dihydroorotase | MGI:1916969 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000064115 | Cadm2 | cell adhesion molecule 2 | MGI:2442722 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000005338 | Cadm3 | cell adhesion molecule 3 | MGI:2137858 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000054793 | Cadm4 | cell adhesion molecule 4 | MGI:2449088 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000054423 | Cadps | Ca2+-dependent secretion activator | MGI:1350922 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000003814 | Calr | calreticulin | MGI:88252 | YES | YES | Cytoplasm | transcription regulator | |||||
| ENSMUSG00000020114 | Cand1 | cullin associated and neddylation disassociated 1 | MGI:1261820 | YES | YES | Cytoplasm | transcription regulator | |||||
| ENSMUSG00000020368 | Canx | calnexin | MGI:88261 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000028656 | Cap1 | CAP, adenylate cyclase-associated protein 1 (yeast) | MGI:88262 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000031505 | Carkd | carbohydrate kinase domain containing | MGI:1913353 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000022661 | Cd200 | CD200 antigen | MGI:1196990 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000002944 | Cd36 | CD36 antigen | MGI:107899 | YES | YES | Plasma Membrane | transmembrane receptor | |||||
| ENSMUSG00000019471 | Cdc37 | cell division cycle 37 | MGI:109531 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000006699 | Cdc42 | cell division cycle 42 | MGI:106211 | YES | YES | Plasma Membrane | enzyme | |||||
| ENSMUSG00000068740 | Celsr2 | cadherin, EGF LAG seven-pass G-type receptor 2 (flamingo homolog, Drosophila) | MGI:1858235 | YES | YES | Plasma Membrane | G-protein coupled receptor | |||||
| ENSMUSG00000030077 | Chl1 | cell adhesion molecule with homology to L1CAM | MGI:1098266 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000025371 | Chmp6 | charged multivesicular body protein 6 | MGI:3583942 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000028165 | Cisd2 | CDGSH iron sulfur domain 2 | MGI:1914256 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000001270 | Ckb | creatine kinase, brain | MGI:88407 | YES | YES | Cytoplasm | kinase | |||||
| ENSMUSG00000002981 | Clptm1 | cleft lip and palate associated transmembrane protein 1 | MGI:1927155 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000039953 | Clstn1 | calsyntenin 1 | MGI:1929895 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000008153 | Clstn3 | calsyntenin 3 | MGI:2178323 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000047126 | Cltc | clathrin, heavy polypeptide (Hc) | MGI:2388633 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000039163 | Cmc1 | COX assembly mitochondrial protein 1 | MGI:1915149 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000044288 | Cnr1 | cannabinoid receptor 1 (brain) | MGI:104615 | YES | YES | Plasma Membrane | G-protein coupled receptor | |||||
| ENSMUSG00000044629 | Cnrip1 | cannabinoid receptor interacting protein 1 | MGI:1917505 | YES | YES | Extracellular Space | other | |||||
| ENSMUSG00000039419 | Cntnap2 | contactin associated protein-like 2 | MGI:1914047 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000036513 | Commd2 | COMM domain containing 2 | MGI:1098806 | YES | YES | Other | other | |||||
| ENSMUSG00000030707 | Coro1a | coronin, actin binding protein 1A | MGI:1345961 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000000088 | Cox5a | cytochrome c oxidase subunit Va | MGI:88474 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000061518 | Cox5b | cytochrome c oxidase subunit Vb | MGI:88475 | YES | YES | #N/A | #N/A | |||||
| ENSMUSG00000036751 | Cox6b1 | cytochrome c oxidase, subunit VIb polypeptide 1 | MGI:107460 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000014313 | Cox6c | cytochrome c oxidase subunit VIc | MGI:104614 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000032330 | Cox7a2 | cytochrome c oxidase subunit VIIa 2 | MGI:1316715 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000037852 | Cpe | carboxypeptidase E | MGI:101932 | YES | YES | Cytoplasm | peptidase | |||||
| ENSMUSG00000033615 | Cplx1 | complexin 1 | MGI:104727 | YES | YES | Plasma Membrane | transporter | |||||
| ENSMUSG00000007783 | Cpt1c | carnitine palmitoyltransferase 1c | MGI:2446526 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000029121 | Crmp1 | collapsin response mediator protein 1 | MGI:107793 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000030905 | Crym | crystallin, mu | MGI:102675 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000005683 | Cs | citrate synthase | MGI:88529 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000002718 | Cse1l | chromosome segregation 1-like (S. cerevisiae) | MGI:1339951 | YES | YES | Nucleus | transporter | |||||
| ENSMUSG00000032911 | Cspg4 | chondroitin sulfate proteoglycan 4 | MGI:2153093 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000032482 | Cspg5 | chondroitin sulfate proteoglycan 5 | MGI:1352747 | YES | YES | Extracellular Space | growth factor | |||||
| ENSMUSG00000021000 | Ctage5 | CTAGE family, member 5 | MGI:1346056 | YES | YES | Other | enzyme | |||||
| ENSMUSG00000021939 | Ctsb | cathepsin B | MGI:88561 | YES | YES | Cytoplasm | peptidase | |||||
| ENSMUSG00000007891 | Ctsd | cathepsin D | MGI:88562 | YES | YES | Cytoplasm | peptidase | |||||
| ENSMUSG00000024646 | Cyb5a | cytochrome b5 type A (microsomal) | MGI:1926952 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000018042 | Cyb5r3 | cytochrome b5 reductase 3 | MGI:94893 | YES | YES | Other | enzyme | |||||
| ENSMUSG00000026385 | Dbi | diazepam binding inhibitor | MGI:94865 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000031865 | Dctn1 | dynactin 1 | MGI:107745 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000028194 | Ddah1 | dimethylarginine dimethylaminohydrolase 1 | MGI:1916469 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000002332 | Dhrs1 | dehydrogenase/reductase (SDR family) member 1 | MGI:1196314 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000040035 | Disp2 | dispatched homolog 2 (Drosophila) | MGI:2388733 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000000168 | Dlat | dihydrolipoamide S-acetyltransferase (E2 component of pyruvate dehydrogenase complex) | MGI:2385311 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000045103 | Dmd | dystrophin, muscular dystrophy | MGI:94909 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000032560 | Dnajc13 | DnaJ (Hsp40) homolog, subfamily C, member 13 | MGI:2676368 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000022136 | Dnajc3 | DnaJ (Hsp40) homolog, subfamily C, member 3 | MGI:107373 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000000826 | Dnajc5 | DnaJ (Hsp40) homolog, subfamily C, member 5 | MGI:892995 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000028528 | Dnajc6 | DnaJ (Hsp40) homolog, subfamily C, member 6 | MGI:1919935 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000036766 | Dner | delta/notch-like EGF-related receptor | MGI:2152889 | YES | YES | Plasma Membrane | transmembrane receptor | |||||
| ENSMUSG00000022048 | Dpysl2 | dihydropyrimidinase-like 2 | MGI:1349763 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000024501 | Dpysl3 | dihydropyrimidinase-like 3 | MGI:1349762 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000025478 | Dpysl4 | dihydropyrimidinase-like 4 | MGI:1349764 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000029168 | Dpysl5 | dihydropyrimidinase-like 5 | MGI:1929772 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000015932 | Dstn | destrin | MGI:1929270 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000024302 | Dtna | dystrobrevin alpha | MGI:106039 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000018707 | Dync1h1 | dynein cytoplasmic 1 heavy chain 1 | MGI:103147 | YES | YES | Cytoplasm | peptidase | |||||
| ENSMUSG00000021417 | Eci2 | enoyl-Coenzyme A delta isomerase 2 | MGI:1346064 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000050812 | AI314180 | expressed sequence AI314180 | MGI:2140220 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000036499 | Eea1 | early endosome antigen 1 | MGI:2442192 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000034994 | Eef2 | eukaryotic translation elongation factor 2 | MGI:95288 | YES | YES | Cytoplasm | translation regulator | |||||
| ENSMUSG00000040659 | Efhd2 | EF hand domain containing 2 | MGI:106504 | YES | YES | Other | other | |||||
| ENSMUSG00000027293 | Ehd4 | EH-domain containing 4 | MGI:1919619 | YES | YES | Plasma Membrane | enzyme | |||||
| ENSMUSG00000021116 | Eif2s1 | eukaryotic translation initiation factor 2, subunit 1 alpha | MGI:95299 | YES | YES | Cytoplasm | translation regulator | |||||
| ENSMUSG00000040731 | Eif4h | eukaryotic translation initiation factor 4H | MGI:1341822 | YES | YES | Cytoplasm | translation regulator | |||||
| ENSMUSG00000021728 | Emb | embigin | MGI:95321 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000037419 | Endod1 | endonuclease domain containing 1 | MGI:1919196 | YES | YES | Extracellular Space | enzyme | |||||
| ENSMUSG00000059040 | Eno1b | enolase 1B, retrotransposed | MGI:3648653 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000004267 | Eno2 | enolase 2, gamma neuronal | MGI:95394 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000015085 | Entpd2 | ectonucleoside triphosphate diphosphohydrolase 2 | MGI:1096863 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000019978 | Epb4.1l2 | erythrocyte protein band 4.1 like 2 | MGI:103009 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000026615 | Eprs | glutamyl-prolyl-tRNA synthetase | MGI:97838 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000028552 | Eps15 | epidermal growth factor receptor pathway substrate 15 | MGI:104583 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000029616 | Erp29 | endoplasmic reticulum protein 29 | MGI:1914647 | YES | YES | Cytoplasm | transporter | |||||
| ENSMUSG00000028343 | Erp44 | endoplasmic reticulum protein 44 | MGI:1923549 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000025366 | Esyt1 | extended synaptotagmin-like protein 1 | MGI:1344426 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000052397 | Ezr | ezrin | MGI:98931 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000027533 | Fabp5 | fatty acid binding protein 5, epidermal | MGI:101790 | YES | YES | Cytoplasm | transporter | |||||
| ENSMUSG00000025873 | Faf2 | Fas associated factor family member 2 | MGI:1923827 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000022378 | Fam49b | family with sequence similarity 49, member B | MGI:1923520 | YES | YES | Extracellular Space | other | |||||
| ENSMUSG00000025153 | Fasn | fatty acid synthase | MGI:95485 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000026526 | Fh1 | fumarate hydratase 1 | MGI:95530 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000019054 | Fis1 | fission 1 (mitochondrial outer membrane) homolog (yeast) | MGI:1913687 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000032966 | Fkbp1a | FK506 binding protein 1a | MGI:95541 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000056629 | Fkbp2 | FK506 binding protein 2 | MGI:95542 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000020949 | Fkbp3 | FK506 binding protein 3 | MGI:1353460 | YES | YES | Nucleus | enzyme | |||||
| ENSMUSG00000029581 | Fscn1 | fascin homolog 1, actin bundling protein (Strongylocentrotus purpuratus) | MGI:1352745 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000066705 | Fxyd6 | FXYD domain-containing ion transport regulator 6 | MGI:1890226 | YES | YES | Plasma Membrane | ion channel | |||||
| ENSMUSG00000036578 | Fxyd7 | FXYD domain-containing ion transport regulator 7 | MGI:1889006 | YES | YES | Plasma Membrane | ion channel | |||||
| ENSMUSG00000025579 | Gaa | glucosidase, alpha, acid | MGI:95609 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000070880 | Gad1 | glutamate decarboxylase 1 | MGI:95632 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000026787 | Gad2 | glutamic acid decarboxylase 2 | MGI:95634 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000071650 | Ganab | alpha glucosidase 2 alpha neutral subunit | MGI:1097667 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000029777 | Gars | glycyl-tRNA synthetase | MGI:2449057 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000058624 | Gda | guanine deaminase | MGI:95678 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000015291 | Gdi1 | guanosine diphosphate (GDP) dissociation inhibitor 1 | MGI:99846 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000027603 | Ggt7 | gamma-glutamyltransferase 7 | MGI:1913385 | YES | YES | Plasma Membrane | enzyme | |||||
| ENSMUSG00000056966 | Gjc3 | gap junction protein, gamma 3 | MGI:2153041 | YES | YES | Extracellular Space | transporter | |||||
| ENSMUSG00000003316 | Glg1 | golgi apparatus protein 1 | MGI:104967 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000024026 | Glo1 | glyoxalase 1 | MGI:95742 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000026103 | Gls | glutaminase | MGI:95752 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000026473 | Glul | glutamate-ammonia ligase (glutamine synthetase) | MGI:95739 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000058927 | Gm10053 | predicted gene 10053 | MGI:3704493 | YES | YES | Cytoplasm | transporter | |||||
| ENSMUSG00000034781 | Gna11 | guanine nucleotide binding protein, alpha 11 | MGI:95766 | YES | YES | Plasma Membrane | enzyme | |||||
| ENSMUSG00000020611 | Gna13 | guanine nucleotide binding protein, alpha 13 | MGI:95768 | YES | YES | Plasma Membrane | enzyme | |||||
| ENSMUSG00000029064 | Gnb1 | guanine nucleotide binding protein (G protein), beta 1 | MGI:95781 | YES | YES | Plasma Membrane | enzyme | |||||
| ENSMUSG00000071658 | Gng3 | guanine nucleotide binding protein (G protein), gamma 3 | MGI:102704 | YES | YES | Plasma Membrane | enzyme | |||||
| ENSMUSG00000025190 | Got1 | glutamic-oxaloacetic transaminase 1, soluble | MGI:95791 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000031672 | Got2 | glutamatic-oxaloacetic transaminase 2, mitochondrial | MGI:95792 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000031119 | Gpc4 | glypican 4 | MGI:104902 | YES | YES | Plasma Membrane | transmembrane receptor | |||||
| ENSMUSG00000023019 | Gpd1 | glycerol-3-phosphate dehydrogenase 1 (soluble) | MGI:95679 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000026827 | Gpd2 | glycerol phosphate dehydrogenase 2, mitochondrial | MGI:99778 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000036427 | Gpi1 | glucose phosphate isomerase 1 | MGI:95797 | YES | YES | Extracellular Space | enzyme | |||||
| ENSMUSG00000031517 | Gpm6a | glycoprotein m6a | MGI:107671 | YES | YES | Plasma Membrane | ion channel | |||||
| ENSMUSG00000031342 | Gpm6b | glycoprotein m6b | MGI:107672 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000031785 | Adgrg1 | adhesion G protein-coupled receptor G1 | MGI:1340051 | YES | YES | Plasma Membrane | G-protein coupled receptor | |||||
| ENSMUSG00000025156 | Gps1 | G protein pathway suppressor 1 | MGI:2384801 | YES | YES | Other | other | |||||
| ENSMUSG00000041078 | Grid1 | glutamate receptor, ionotropic, delta 1 | MGI:95812 | YES | YES | Plasma Membrane | ion channel | |||||
| ENSMUSG00000019828 | Grm1 | glutamate receptor, metabotropic 1 | MGI:1351338 | YES | YES | Plasma Membrane | G-protein coupled receptor | |||||
| ENSMUSG00000057177 | Gsk3a | glycogen synthase kinase 3 alpha | MGI:2152453 | YES | YES | Nucleus | kinase | |||||
| ENSMUSG00000029864 | Gstk1 | glutathione S-transferase kappa 1 | MGI:1923513 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000058135 | Gstm1 | glutathione S-transferase, mu 1 | MGI:95860 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000004032 | Gstm5 | glutathione S-transferase, mu 5 | MGI:1309466 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000060803 | Gstp1 | glutathione S-transferase, pi 1 | MGI:95865 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000069919 | Hba-a1 | hemoglobin alpha, adult chain 1 | MGI:96015 | YES | YES | Extracellular Space | transporter | |||||
| ENSMUSG00000073940 | Hbb-bt | hemoglobin, beta adult t chain | MGI:5474850 | YES | YES | Cytoplasm | transporter | |||||
| ENSMUSG00000021730 | Hcn1 | hyperpolarization-activated, cyclic nucleotide-gated K+ 1 | MGI:1096392 | YES | YES | Plasma Membrane | ion channel | |||||
| ENSMUSG00000020331 | Hcn2 | hyperpolarization-activated, cyclic nucleotide-gated K+ 2 | MGI:1298210 | YES | YES | Plasma Membrane | ion channel | |||||
| ENSMUSG00000032338 | Hcn4 | hyperpolarization-activated, cyclic nucleotide-gated K+ 4 | MGI:1298209 | YES | YES | Plasma Membrane | ion channel | |||||
| ENSMUSG00000025232 | Hexa | hexosaminidase A | MGI:96073 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000025793 | Hgs | HGF-regulated tyrosine kinase substrate | MGI:104681 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000041426 | Hibch | 3-hydroxyisobutyryl-Coenzyme A hydrolase | MGI:1923792 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000038412 | Higd1a | HIG1 domain family, member 1A | MGI:1930666 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000028470 | Hint2 | histidine triad nucleotide binding protein 2 | MGI:1916167 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000019791 | Hint3 | histidine triad nucleotide binding protein 3 | MGI:1914097 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000004070 | Hmox2 | heme oxygenase 2 | MGI:109373 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000073422 | H2-Ke6 | H2-K region expressed gene 6 | MGI:95911 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000021270 | Hsp90aa1 | heat shock protein 90, alpha (cytosolic), class A member 1 | MGI:96250 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000023944 | Hsp90ab1 | heat shock protein 90 alpha (cytosolic), class B member 1 | MGI:96247 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000020048 | Hsp90b1 | heat shock protein 90, beta (Grp94), member 1 | MGI:98817 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000020361 | Hspa4 | heat shock protein 4 | MGI:1342292 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000025757 | Hspa4l | heat shock protein 4 like | MGI:107422 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000026864 | Hspa5 | heat shock protein 5 | MGI:95835 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000024359 | Hspa9 | heat shock protein 9 | MGI:96245 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000025980 | Hspd1 | heat shock protein 1 (chaperonin) | MGI:96242 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000073676 | Hspe1 | heat shock protein 1 (chaperonin 10) | MGI:104680 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000029657 | Hsph1 | heat shock 105kDa/110kDa protein 1 | MGI:105053 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000029104 | Htt | huntingtin | MGI:96067 | YES | YES | Cytoplasm | transcription regulator | |||||
| ENSMUSG00000032115 | Hyou1 | hypoxia up-regulated 1 | MGI:108030 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000032174 | Icam5 | intercellular adhesion molecule 5, telencephalin | MGI:109430 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000030541 | Idh2 | isocitrate dehydrogenase 2 (NADP+), mitochondrial | MGI:96414 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000032279 | Idh3a | isocitrate dehydrogenase 3 (NAD+) alpha | MGI:1915084 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000027406 | Idh3b | isocitrate dehydrogenase 3 (NAD+) beta | MGI:2158650 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000030662 | Ipo5 | importin 5 | MGI:1917822 | YES | YES | Nucleus | transporter | |||||
| ENSMUSG00000058975 | Kcnc1 | potassium voltage gated channel, Shaw-related subfamily, member 1 | MGI:96667 | YES | YES | Plasma Membrane | ion channel | |||||
| ENSMUSG00000044708 | Kcnj10 | potassium inwardly-rectifying channel, subfamily J, member 10 | MGI:1194504 | YES | YES | Plasma Membrane | ion channel | |||||
| ENSMUSG00000063142 | Kcnma1 | potassium large conductance calcium-activated channel, subfamily M, alpha member 1 | MGI:99923 | YES | YES | Plasma Membrane | ion channel | |||||
| ENSMUSG00000036062 | N28178 | expressed sequence N28178 | MGI:2140712 | YES | YES | Other | other | |||||
| ENSMUSG00000037270 | 4932438A13Rik | RIKEN cDNA 4932438A13 gene | MGI:2444631 | YES | YES | Other | other | |||||
| ENSMUSG00000030207 | 8430419L09Rik | RIKEN cDNA 8430419L09 gene | MGI:1921775 | YES | YES | Other | other | |||||
| ENSMUSG00000036333 | Kidins220 | kinase D-interacting substrate 220 | MGI:1924730 | YES | YES | Nucleus | transcription regulator | |||||
| ENSMUSG00000014602 | Kif1a | kinesin family member 1A | MGI:108391 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000005672 | Kit | kit oncogene | MGI:96677 | YES | YES | Plasma Membrane | transmembrane receptor | |||||
| ENSMUSG00000001440 | Kpnb1 | karyopherin (importin) beta 1 | MGI:107532 | YES | YES | Nucleus | transporter | |||||
| ENSMUSG00000032796 | Lama1 | laminin, alpha 1 | MGI:99892 | YES | YES | Extracellular Space | other | |||||
| ENSMUSG00000031447 | Lamp1 | lysosomal-associated membrane protein 1 | MGI:96745 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000028062 | Lamtor2 | late endosomal/lysosomal adaptor, MAPK and MTOR activator 2 | MGI:1932697 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000050552 | Lamtor4 | late endosomal/lysosomal adaptor, MAPK and MTOR activator 4 | MGI:1913346 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000039682 | Lap3 | leucine aminopeptidase 3 | MGI:1914238 | YES | YES | Cytoplasm | peptidase | |||||
| ENSMUSG00000030246 | Ldhb | lactate dehydrogenase B | MGI:96763 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000005299 | Letm1 | leucine zipper-EF-hand containing transmembrane protein 1 | MGI:1932557 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000049556 | Lingo1 | leucine rich repeat and Ig domain containing 1 | MGI:1915522 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000020536 | Llgl1 | lethal giant larvae homolog 1 (Drosophila) | MGI:102682 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000009207 | Lnp | limb and neural patterns | MGI:1918115 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000027134 | Lpcat4 | lysophosphatidylcholine acyltransferase 4 | MGI:2138993 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000040249 | Lrp1 | low density lipoprotein receptor-related protein 1 | MGI:96828 | YES | YES | Plasma Membrane | transmembrane receptor | |||||
| ENSMUSG00000049252 | Lrp1b | low density lipoprotein-related protein 1B (deleted in tumors) | MGI:2151136 | YES | YES | Plasma Membrane | transmembrane receptor | |||||
| ENSMUSG00000029103 | Lrpap1 | low density lipoprotein receptor-related protein associated protein 1 | MGI:96829 | YES | YES | Plasma Membrane | transmembrane receptor | |||||
| ENSMUSG00000024120 | Lrpprc | leucine-rich PPR-motif containing | MGI:1919666 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000021338 | Lrrc16a | leucine rich repeat containing 16A | MGI:1915982 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000029028 | Lrrc47 | leucine rich repeat containing 47 | MGI:1920196 | YES | YES | Other | other | |||||
| ENSMUSG00000047085 | Lrrc4b | leucine rich repeat containing 4B | MGI:3027390 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000020922 | Lsm12 | LSM12 homolog (S. cerevisiae) | MGI:1919592 | YES | YES | Other | other | |||||
| ENSMUSG00000022577 | Ly6h | lymphocyte antigen 6 complex, locus H | MGI:1346030 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000040687 | Madd | MAP-kinase activating death domain | MGI:2444672 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000036634 | Mag | myelin-associated glycoprotein | MGI:96912 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000025037 | Maoa | monoamine oxidase A | MGI:96915 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000040147 | Maob | monoamine oxidase B | MGI:96916 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000015222 | Map2 | microtubule-associated protein 2 | MGI:97175 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000032479 | Map4 | microtubule-associated protein 4 | MGI:97178 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000069662 | Marcks | myristoylated alanine rich protein kinase C substrate | MGI:96907 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000040354 | Mars | methionine-tRNA synthetase | MGI:1345633 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000037523 | Mavs | mitochondrial antiviral signaling protein | MGI:2444773 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000032135 | Mcam | melanoma cell adhesion molecule | MGI:1933966 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000020321 | Mdh1 | malate dehydrogenase 1, NAD (soluble) | MGI:97051 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000019179 | Mdh2 | malate dehydrogenase 2, NAD (mitochondrial) | MGI:97050 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000030621 | Me3 | malic enzyme 3, NADP(+)-dependent, mitochondrial | MGI:1916679 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000028910 | Mecr | mitochondrial trans-2-enoyl-CoA reductase | MGI:1349441 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000029020 | Mfn2 | mitofusin 2 | MGI:2442230 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000056050 | Mia3 | melanoma inhibitory activity 3 | MGI:2443183 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000033307 | Mif | macrophage migration inhibitory factor | MGI:96982 | YES | YES | Extracellular Space | cytokine | |||||
| ENSMUSG00000023861 | Mpc1 | mitochondrial pyruvate carrier 1 | MGI:1915240 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000038388 | Mpp6 | membrane protein, palmitoylated 6 (MAGUK p55 subfamily member 6) | MGI:1927340 | YES | YES | Plasma Membrane | kinase | |||||
| ENSMUSG00000071711 | Mpst | mercaptopyruvate sulfurtransferase | MGI:2179733 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000037740 | Mrps26 | mitochondrial ribosomal protein S26 | MGI:1333830 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000031207 | Msn | moesin | MGI:97167 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000064358 | mt-Co3 | mitochondrially encoded cytochrome c oxidase III | MGI:102502 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000004748 | Mtfp1 | mitochondrial fission process 1 | MGI:1916686 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000064341 | mt-Nd1 | mitochondrially encoded NADH dehydrogenase 1 | MGI:101787 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000064363 | mt-Nd4 | mitochondrially encoded NADH dehydrogenase 4 | MGI:102498 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000064367 | mt-Nd5 | mitochondrially encoded NADH dehydrogenase 5 | MGI:102496 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000006024 | Napa | N-ethylmaleimide sensitive fusion protein attachment protein alpha | MGI:104563 | YES | YES | Cytoplasm | transporter | |||||
| ENSMUSG00000027438 | Napb | N-ethylmaleimide sensitive fusion protein attachment protein beta | MGI:104562 | YES | YES | Cytoplasm | transporter | |||||
| ENSMUSG00000024581 | Napg | N-ethylmaleimide sensitive fusion protein attachment protein gamma | MGI:104561 | YES | YES | Cytoplasm | transporter | |||||
| ENSMUSG00000027799 | Nbea | neurobeachin | MGI:1347075 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000051359 | Ncald | neurocalcin delta | MGI:1196326 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000039542 | Ncam1 | neural cell adhesion molecule 1 | MGI:97281 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000022762 | Ncam2 | neural cell adhesion molecule 2 | MGI:97282 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000028833 | Ncdn | neurochondrin | MGI:1347351 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000027698 | Nceh1 | neutral cholesterol ester hydrolase 1 | MGI:2443191 | YES | YES | Plasma Membrane | enzyme | |||||
| ENSMUSG00000005125 | Ndrg1 | N-myc downstream regulated gene 1 | MGI:1341799 | YES | YES | Nucleus | kinase | |||||
| ENSMUSG00000004558 | Ndrg2 | N-myc downstream regulated gene 2 | MGI:1352498 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000027634 | Ndrg3 | N-myc downstream regulated gene 3 | MGI:1352499 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000036564 | Ndrg4 | N-myc downstream regulated gene 4 | MGI:2384590 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000026260 | Ndufa10 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex 10 | MGI:1914523 | YES | YES | Cytoplasm | transporter | |||||
| ENSMUSG00000002379 | Ndufa11 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex 11 | MGI:1917125 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000023089 | Ndufa5 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 5 | MGI:1915452 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000025968 | Ndufs1 | NADH dehydrogenase (ubiquinone) Fe-S protein 1 | MGI:2443241 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000013593 | Ndufs2 | NADH dehydrogenase (ubiquinone) Fe-S protein 2 | MGI:2385112 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000021764 | Ndufs4 | NADH dehydrogenase (ubiquinone) Fe-S protein 4 | MGI:1343135 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000028648 | Ndufs5 | NADH dehydrogenase (ubiquinone) Fe-S protein 5 | MGI:1890889 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000059734 | Ndufs8 | NADH dehydrogenase (ubiquinone) Fe-S protein 8 | MGI:2385079 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000037916 | Ndufv1 | NADH dehydrogenase (ubiquinone) flavoprotein 1 | MGI:107851 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000032340 | Neo1 | neogenin | MGI:1097159 | YES | YES | Plasma Membrane | transcription regulator | |||||
| ENSMUSG00000026442 | Nfasc | neurofascin | MGI:104753 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000032109 | Nlrx1 | NLR family member X1 | MGI:2429611 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000025453 | Nnt | nicotinamide nucleotide transhydrogenase | MGI:109279 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000030835 | Nomo1 | nodal modulator 1 | MGI:2385850 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000001441 | Npepps | aminopeptidase puromycin sensitive | MGI:1101358 | YES | YES | Cytoplasm | peptidase | |||||
| ENSMUSG00000032336 | Nptn | neuroplastin | MGI:108077 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000022421 | Nptxr | neuronal pentraxin receptor | MGI:1920590 | YES | YES | Plasma Membrane | transmembrane receptor | |||||
| ENSMUSG00000020598 | Nrcam | neuronal cell adhesion molecule | MGI:104750 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000025810 | Nrp1 | neuropilin 1 | MGI:106206 | YES | YES | Plasma Membrane | transmembrane receptor | |||||
| ENSMUSG00000024109 | Nrxn1 | neurexin I | MGI:1096391 | YES | YES | Plasma Membrane | transporter | |||||
| ENSMUSG00000033768 | Nrxn2 | neurexin II | MGI:1096362 | YES | YES | Plasma Membrane | transporter | |||||
| ENSMUSG00000034187 | Nsf | N-ethylmaleimide sensitive fusion protein | MGI:104560 | YES | YES | Cytoplasm | transporter | |||||
| ENSMUSG00000027455 | Nsfl1c | NSFL1 (p97) cofactor (p47) | MGI:3042273 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000020736 | Nt5c | 5',3'-nucleotidase, cytosolic | MGI:1354954 | YES | YES | Cytoplasm | phosphatase | |||||
| ENSMUSG00000054027 | Nt5dc3 | 5'-nucleotidase domain containing 3 | MGI:3513266 | YES | YES | Other | other | |||||
| ENSMUSG00000055254 | Ntrk2 | neurotrophic tyrosine kinase, receptor, type 2 | MGI:97384 | YES | YES | Plasma Membrane | kinase | |||||
| ENSMUSG00000029153 | Ociad2 | OCIA domain containing 2 | MGI:1916377 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000027108 | Ola1 | Obg-like ATPase 1 | MGI:1914309 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000038084 | Opa1 | optic atrophy 1 | MGI:1921393 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000050121 | Opalin | oligodendrocytic myelin paranodal and inner loop protein | MGI:2657025 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000020189 | Osbpl8 | oxysterol binding protein-like 8 | MGI:2443807 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000022186 | Oxct1 | 3-oxoacid CoA transferase 1 | MGI:1914291 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000022307 | Oxr1 | oxidation resistance 1 | MGI:2179326 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000025130 | P4hb | prolyl 4-hydroxylase, beta polypeptide | MGI:97464 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000040276 | Pacsin1 | protein kinase C and casein kinase substrate in neurons 1 | MGI:1345181 | YES | YES | Cytoplasm | kinase | |||||
| ENSMUSG00000035566 | Pcdh17 | protocadherin 17 | MGI:2684924 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000029108 | Pcdh7 | protocadherin 7 | MGI:1860487 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000055421 | Pcdh9 | protocadherin 9 | MGI:1306801 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000039278 | Pcsk1n | proprotein convertase subtilisin/kexin type 1 inhibitor | MGI:1353431 | YES | YES | Extracellular Space | other | |||||
| ENSMUSG00000024892 | Pcx | pyruvate carboxylase | MGI:97520 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000032504 | Pdcd6ip | programmed cell death 6 interacting protein | MGI:1333753 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000051007 | Pddc1 | Parkinson disease 7 domain containing 1 | MGI:2387178 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000023868 | Pde10a | phosphodiesterase 10A | MGI:1345143 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000027248 | Pdia3 | protein disulfide isomerase associated 3 | MGI:95834 | YES | YES | Cytoplasm | peptidase | |||||
| ENSMUSG00000025823 | Pdia4 | protein disulfide isomerase associated 4 | MGI:104864 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000020571 | Pdia6 | protein disulfide isomerase associated 6 | MGI:1919103 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000032788 | Pdxk | pyridoxal (pyridoxine, vitamin B6) kinase | MGI:1351869 | YES | YES | Cytoplasm | kinase | |||||
| ENSMUSG00000032959 | Pebp1 | phosphatidylethanolamine binding protein 1 | MGI:1344408 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000027674 | Pex5l | peroxisomal biogenesis factor 5-like | MGI:1916672 | YES | YES | Cytoplasm | ion channel | |||||
| ENSMUSG00000027805 | Pfn2 | profilin 2 | MGI:97550 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000062070 | Pgk1 | phosphoglycerate kinase 1 | MGI:97555 | YES | YES | Cytoplasm | kinase | |||||
| ENSMUSG00000025791 | Pgm2 | phosphoglucomutase 2 | MGI:97565 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000030729 | Pgm2l1 | phosphoglucomutase 2-like 1 | MGI:1918224 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000006373 | Pgrmc1 | progesterone receptor membrane component 1 | MGI:1858305 | YES | YES | Plasma Membrane | transmembrane receptor | |||||
| ENSMUSG00000049940 | Pgrmc2 | progesterone receptor membrane component 2 | MGI:1918054 | YES | YES | Nucleus | other | |||||
| ENSMUSG00000053398 | Phgdh | 3-phosphoglycerate dehydrogenase | MGI:1355330 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000010760 | Phlda2 | pleckstrin homology-like domain, family A, member 2 | MGI:1202307 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000036504 | Phpt1 | phosphohistidine phosphatase 1 | MGI:1922704 | YES | YES | Cytoplasm | phosphatase | |||||
| ENSMUSG00000025178 | Pi4k2a | phosphatidylinositol 4-kinase type 2 alpha | MGI:1934031 | YES | YES | Cytoplasm | kinase | |||||
| ENSMUSG00000039361 | Picalm | phosphatidylinositol binding clathrin assembly protein | MGI:2385902 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000024851 | Pitpnm1 | phosphatidylinositol transfer protein, membrane-associated 1 | MGI:1197524 | YES | YES | Cytoplasm | transporter | |||||
| ENSMUSG00000032294 | Pkm | pyruvate kinase, muscle | MGI:97591 | YES | YES | Cytoplasm | kinase | |||||
| ENSMUSG00000051177 | Plcb1 | phospholipase C, beta 1 | MGI:97613 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000031425 | Plp1 | proteolipid protein (myelin) 1 | MGI:97623 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000030084 | Plxna1 | plexin A1 | MGI:107685 | YES | YES | Plasma Membrane | transmembrane receptor | |||||
| ENSMUSG00000029765 | Plxna4 | plexin A4 | MGI:2179061 | YES | YES | Plasma Membrane | transmembrane receptor | |||||
| ENSMUSG00000053646 | Plxnb1 | plexin B1 | MGI:2154238 | YES | YES | Plasma Membrane | transmembrane receptor | |||||
| ENSMUSG00000074785 | Plxnc1 | plexin C1 | MGI:1890127 | YES | YES | Plasma Membrane | transmembrane receptor | |||||
| ENSMUSG00000036257 | Pnpla8 | patatin-like phospholipase domain containing 8 | MGI:1914702 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000033152 | Podxl2 | podocalyxin-like 2 | MGI:2442488 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000005514 | Por | P450 (cytochrome) oxidoreductase | MGI:97744 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000071866 | Ppia | peptidylprolyl isomerase A | MGI:97749 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000034709 | Ppp1r21 | protein phosphatase 1, regulatory subunit 21 | MGI:1921075 | YES | YES | Other | other | |||||
| ENSMUSG00000026275 | Ppp1r7 | protein phosphatase 1, regulatory (inhibitor) subunit 7 | MGI:1913635 | YES | YES | Nucleus | phosphatase | |||||
| ENSMUSG00000028161 | Ppp3ca | protein phosphatase 3, catalytic subunit, alpha isoform | MGI:107164 | YES | YES | Cytoplasm | phosphatase | |||||
| ENSMUSG00000021816 | Ppp3cb | protein phosphatase 3, catalytic subunit, beta isoform | MGI:107163 | YES | YES | Plasma Membrane | phosphatase | |||||
| ENSMUSG00000033953 | Ppp3r1 | protein phosphatase 3, regulatory subunit B, alpha isoform (calcineurin B, type I) | MGI:107172 | YES | YES | Cytoplasm | phosphatase | |||||
| ENSMUSG00000031149 | Praf2 | PRA1 domain family 2 | MGI:1859607 | YES | YES | Other | other | |||||
| ENSMUSG00000024997 | Prdx3 | peroxiredoxin 3 | MGI:88034 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000024953 | Prdx5 | peroxiredoxin 5 | MGI:1859821 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000026701 | Prdx6 | peroxiredoxin 6 | MGI:894320 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000045038 | Prkce | protein kinase C, epsilon | MGI:97599 | YES | YES | Cytoplasm | kinase | |||||
| ENSMUSG00000079037 | Prnp | prion protein | MGI:97769 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000003526 | Prodh | proline dehydrogenase | MGI:97770 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000045114 | Prrt2 | proline-rich transmembrane protein 2 | MGI:1916267 | YES | YES | Other | other | |||||
| ENSMUSG00000045009 | Prrt3 | proline-rich transmembrane protein 3 | MGI:2444810 | YES | YES | Other | other | |||||
| ENSMUSG00000024640 | Psat1 | phosphoserine aminotransferase 1 | MGI:2183441 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000030751 | Psma1 | proteasome (prosome, macropain) subunit, alpha type 1 | MGI:1347005 | YES | YES | Cytoplasm | peptidase | |||||
| ENSMUSG00000015671 | Psma2 | proteasome (prosome, macropain) subunit, alpha type 2 | MGI:104885 | YES | YES | Cytoplasm | peptidase | |||||
| ENSMUSG00000028837 | Psmb2 | proteasome (prosome, macropain) subunit, beta type 2 | MGI:1347045 | YES | YES | Cytoplasm | peptidase | |||||
| ENSMUSG00000069744 | Psmb3 | proteasome (prosome, macropain) subunit, beta type 3 | MGI:1347014 | YES | YES | Cytoplasm | peptidase | |||||
| ENSMUSG00000022193 | Psmb5 | proteasome (prosome, macropain) subunit, beta type 5 | MGI:1194513 | YES | YES | Cytoplasm | peptidase | |||||
| ENSMUSG00000026229 | Psmd1 | proteasome (prosome, macropain) 26S subunit, non-ATPase, 1 | MGI:1917497 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000006998 | Psmd2 | proteasome (prosome, macropain) 26S subunit, non-ATPase, 2 | MGI:1096584 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000026869 | Psmd5 | proteasome (prosome, macropain) 26S subunit, non-ATPase, 5 | MGI:1914248 | YES | YES | Other | other | |||||
| ENSMUSG00000026820 | Ptges2 | prostaglandin E synthase 2 | MGI:1917592 | YES | YES | Cytoplasm | transcription regulator | |||||
| ENSMUSG00000027303 | Ptpra | protein tyrosine phosphatase, receptor type, A | MGI:97808 | YES | YES | Plasma Membrane | phosphatase | |||||
| ENSMUSG00000033295 | Ptprf | protein tyrosine phosphatase, receptor type, F | MGI:102695 | YES | YES | Plasma Membrane | phosphatase | |||||
| ENSMUSG00000033059 | Pygb | brain glycogen phosphorylase | MGI:97828 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000015806 | Qdpr | quinoid dihydropteridine reductase | MGI:97836 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000020671 | Rab10 | RAB10, member RAS oncogene family | MGI:105066 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000077450 | Rab11b | RAB11B, member RAS oncogene family | MGI:99425 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000026878 | Rab14 | RAB14, member RAS oncogene family | MGI:1915615 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000073639 | Rab18 | RAB18, member RAS oncogene family | MGI:102790 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000024870 | Rab1b | RAB1B, member RAS oncogene family | MGI:1923558 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000034789 | Rab24 | RAB24, member RAS oncogene family | MGI:105065 | YES | YES | Other | other | |||||
| ENSMUSG00000024511 | Rab27b | RAB27B, member RAS oncogene family | MGI:1931295 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000047187 | Rab2a | RAB2A, member RAS oncogene family | MGI:1928750 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000056515 | Rab31 | RAB31, member RAS oncogene family | MGI:1914603 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000055069 | Rab39 | RAB39, member RAS oncogene family | MGI:2442855 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000031202 | Rab39b | RAB39B, member RAS oncogene family | MGI:1915040 | YES | YES | Plasma Membrane | enzyme | |||||
| ENSMUSG00000031840 | Rab3a | RAB3A, member RAS oncogene family | MGI:97843 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000003411 | Rab3b | RAB3B, member RAS oncogene family | MGI:1917158 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000021700 | Rab3c | RAB3C, member RAS oncogene family | MGI:1914545 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000019478 | Rab4a | RAB4A, member RAS oncogene family | MGI:105069 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000053291 | Rab4b | RAB4B, member RAS oncogene family | MGI:105071 | YES | YES | Plasma Membrane | enzyme | |||||
| ENSMUSG00000017831 | Rab5a | RAB5A, member RAS oncogene family | MGI:105926 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000000711 | Rab5b | RAB5B, member RAS oncogene family | MGI:105938 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000019173 | Rab5c | RAB5C, member RAS oncogene family | MGI:105306 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000030704 | Rab6a | RAB6A, member RAS oncogene family | MGI:894313 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000032549 | Rab6b | RAB6B, member RAS oncogene family | MGI:107283 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000079477 | Rab7 | RAB7, member RAS oncogene family | MGI:105068 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000043463 | Rab9b | RAB9B, member RAS oncogene family | MGI:2442454 | YES | YES | Plasma Membrane | enzyme | |||||
| ENSMUSG00000028149 | Rap1gds1 | RAP1, GTP-GDP dissociation stimulator 1 | MGI:2385189 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000051615 | Rap2a | RAS related protein 2a | MGI:97855 | YES | YES | Plasma Membrane | enzyme | |||||
| ENSMUSG00000036894 | Rap2b | RAP2B, member of RAS oncogene family | MGI:1921262 | YES | YES | Plasma Membrane | enzyme | |||||
| ENSMUSG00000020621 | Rdh14 | retinol dehydrogenase 14 (all-trans and 9-cis) | MGI:1920402 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000032050 | Rdx | radixin | MGI:97887 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000052852 | Reep1 | receptor accessory protein 1 | MGI:1098827 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000005873 | Reep5 | receptor accessory protein 5 | MGI:1270152 | YES | YES | Extracellular Space | transporter | |||||
| ENSMUSG00000025978 | Rftn2 | raftlin family member 2 | MGI:1921263 | YES | YES | Other | other | |||||
| ENSMUSG00000029101 | Rgs12 | regulator of G-protein signaling 12 | MGI:1918979 | YES | YES | Nucleus | other | |||||
| ENSMUSG00000007815 | Rhoa | ras homolog gene family, member A | MGI:1096342 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000054364 | Rhob | ras homolog gene family, member B | MGI:107949 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000025733 | Rhot2 | ras homolog gene family, member T2 | MGI:2384892 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000020580 | Rock2 | Rho-associated coiled-coil containing protein kinase 2 | MGI:107926 | YES | YES | Cytoplasm | kinase | |||||
| ENSMUSG00000021087 | Rtn1 | reticulon 1 | MGI:1933947 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000024758 | Rtn3 | reticulon 3 | MGI:1339970 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000020458 | Rtn4 | reticulon 4 | MGI:1915835 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000003868 | Ruvbl2 | RuvB-like protein 2 | MGI:1342299 | YES | YES | Nucleus | transcription regulator | |||||
| ENSMUSG00000036529 | Sbf1 | SET binding factor 1 | MGI:1925230 | YES | YES | Plasma Membrane | phosphatase | |||||
| ENSMUSG00000021687 | Scamp1 | secretory carrier membrane protein 1 | MGI:1349480 | YES | YES | Cytoplasm | transporter | |||||
| ENSMUSG00000028049 | Scamp3 | secretory carrier membrane protein 3 | MGI:1346346 | YES | YES | Cytoplasm | transporter | |||||
| ENSMUSG00000020952 | Scfd1 | Sec1 family domain containing 1 | MGI:1924233 | YES | YES | Cytoplasm | transporter | |||||
| ENSMUSG00000064329 | Scn1a | sodium channel, voltage-gated, type I, alpha | MGI:98246 | YES | YES | Plasma Membrane | ion channel | |||||
| ENSMUSG00000019194 | Scn1b | sodium channel, voltage-gated, type I, beta | MGI:98247 | YES | YES | Plasma Membrane | ion channel | |||||
| ENSMUSG00000075318 | Scn2a1 | sodium channel, voltage-gated, type II, alpha 1 | MGI:98248 | YES | YES | Plasma Membrane | ion channel | |||||
| ENSMUSG00000070304 | Scn2b | sodium channel, voltage-gated, type II, beta | MGI:106921 | YES | YES | Plasma Membrane | ion channel | |||||
| ENSMUSG00000023033 | Scn8a | sodium channel, voltage-gated, type VIII, alpha | MGI:103169 | YES | YES | Plasma Membrane | ion channel | |||||
| ENSMUSG00000017009 | Sdc4 | syndecan 4 | MGI:1349164 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000022769 | Sdf2l1 | stromal cell-derived factor 2-like 1 | MGI:2149842 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000021577 | Sdha | succinate dehydrogenase complex, subunit A, flavoprotein (Fp) | MGI:1914195 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000009863 | Sdhb | succinate dehydrogenase complex, subunit B, iron sulfur (Ip) | MGI:1914930 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000041592 | Sdk2 | sidekick homolog 2 (chicken) | MGI:2443847 | YES | YES | Other | other | |||||
| ENSMUSG00000020964 | Sel1l | sel-1 suppressor of lin-12-like (C. elegans) | MGI:1329016 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000019877 | Serinc1 | serine incorporator 1 | MGI:1926228 | YES | YES | Plasma Membrane | transporter | |||||
| ENSMUSG00000044734 | Serpinb1a | serine (or cysteine) peptidase inhibitor, clade B, member 1a | MGI:1913472 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000058153 | Sez6l | seizure related 6 homolog like | MGI:1935121 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000030683 | Sez6l2 | seizure related 6 homolog like 2 | MGI:2385295 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000021474 | Sfxn1 | sideroflexin 1 | MGI:2137677 | YES | YES | Cytoplasm | transporter | |||||
| ENSMUSG00000033720 | Sfxn5 | sideroflexin 5 | MGI:2137681 | YES | YES | Cytoplasm | transporter | |||||
| ENSMUSG00000028843 | Sh3bgrl3 | SH3 domain binding glutamic acid-rich protein-like 3 | MGI:1920973 | YES | YES | Nucleus | other | |||||
| ENSMUSG00000003200 | Sh3gl1 | SH3-domain GRB2-like 1 | MGI:700010 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000028488 | Sh3gl2 | SH3-domain GRB2-like 2 | MGI:700009 | YES | YES | Plasma Membrane | enzyme | |||||
| ENSMUSG00000026860 | Sh3glb2 | SH3-domain GRB2-like endophilin B2 | MGI:2385131 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000037902 | Sirpa | signal-regulatory protein alpha | MGI:108563 | YES | YES | Plasma Membrane | phosphatase | |||||
| ENSMUSG00000015149 | Sirt2 | sirtuin 2 | MGI:1927664 | YES | YES | Nucleus | transcription regulator | |||||
| ENSMUSG00000017740 | Slc12a5 | solute carrier family 12, member 5 | MGI:1862037 | YES | YES | Plasma Membrane | transporter | |||||
| ENSMUSG00000032902 | Slc16a1 | solute carrier family 16 (monocarboxylic acid transporters), member 1 | MGI:106013 | YES | YES | Plasma Membrane | transporter | |||||
| ENSMUSG00000030500 | Slc17a6 | solute carrier family 17 (sodium-dependent inorganic phosphate cotransporter), member 6 | MGI:2156052 | YES | YES | Plasma Membrane | transporter | |||||
| ENSMUSG00000070570 | Slc17a7 | solute carrier family 17 (sodium-dependent inorganic phosphate cotransporter), member 7 | MGI:1920211 | YES | YES | Plasma Membrane | transporter | |||||
| ENSMUSG00000005089 | Slc1a2 | solute carrier family 1 (glial high affinity glutamate transporter), member 2 | MGI:101931 | YES | YES | Plasma Membrane | transporter | |||||
| ENSMUSG00000005357 | Slc1a6 | solute carrier family 1 (high affinity aspartate/glutamate transporter), member 6 | MGI:1096331 | YES | YES | Plasma Membrane | transporter | |||||
| ENSMUSG00000038267 | Slc22a23 | solute carrier family 22, member 23 | MGI:1920352 | YES | YES | Other | transporter | |||||
| ENSMUSG00000037996 | Slc24a2 | solute carrier family 24 (sodium/potassium/calcium exchanger), member 2 | MGI:1923626 | YES | YES | Plasma Membrane | transporter | |||||
| ENSMUSG00000032602 | Slc25a20 | solute carrier family 25 (mitochondrial carnitine/acylcarnitine translocase), member 20 | MGI:1928738 | YES | YES | Cytoplasm | transporter | |||||
| ENSMUSG00000046329 | Slc25a23 | solute carrier family 25 (mitochondrial carrier phosphate carrier), member 23 | MGI:1914222 | YES | YES | Cytoplasm | transporter | |||||
| ENSMUSG00000026819 | Slc25a25 | solute carrier family 25 (mitochondrial carrier, phosphate carrier), member 25 | MGI:1915913 | YES | YES | Cytoplasm | transporter | |||||
| ENSMUSG00000061904 | Slc25a3 | solute carrier family 25 (mitochondrial carrier, phosphate carrier), member 3 | MGI:1353498 | YES | YES | Cytoplasm | transporter | |||||
| ENSMUSG00000045973 | Slc25a51 | solute carrier family 25, member 51 | MGI:2684984 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000036298 | Slc2a13 | solute carrier family 2 (facilitated glucose transporter), member 13 | MGI:2146030 | YES | YES | Plasma Membrane | transporter | |||||
| ENSMUSG00000003153 | Slc2a3 | solute carrier family 2 (facilitated glucose transporter), member 3 | MGI:95757 | YES | YES | Plasma Membrane | transporter | |||||
| ENSMUSG00000029151 | Slc30a3 | solute carrier family 30 (zinc transporter), member 3 | MGI:1345280 | YES | YES | Plasma Membrane | transporter | |||||
| ENSMUSG00000037771 | Slc32a1 | solute carrier family 32 (GABA vesicular transporter), member 1 | MGI:1194488 | YES | YES | Plasma Membrane | transporter | |||||
| ENSMUSG00000010064 | Slc38a3 | solute carrier family 38, member 3 | MGI:1923507 | YES | YES | Plasma Membrane | transporter | |||||
| ENSMUSG00000025986 | Slc39a10 | solute carrier family 39 (zinc transporter), member 10 | MGI:1914515 | YES | YES | Extracellular Space | transporter | |||||
| ENSMUSG00000010095 | Slc3a2 | solute carrier family 3 (activators of dibasic and neutral amino acid transport), member 2 | MGI:96955 | YES | YES | Plasma Membrane | transporter | |||||
| ENSMUSG00000028412 | Slc44a1 | solute carrier family 44, member 1 | MGI:2140592 | YES | YES | Plasma Membrane | transporter | |||||
| ENSMUSG00000026904 | Slc4a10 | solute carrier family 4, sodium bicarbonate cotransporter-like, member 10 | MGI:2150150 | YES | YES | Plasma Membrane | transporter | |||||
| ENSMUSG00000060961 | Slc4a4 | solute carrier family 4 (anion exchanger), member 4 | MGI:1927555 | YES | YES | Plasma Membrane | transporter | |||||
| ENSMUSG00000023032 | Slc4a8 | solute carrier family 4 (anion exchanger), member 8 | MGI:1928745 | YES | YES | Plasma Membrane | transporter | |||||
| ENSMUSG00000030310 | Slc6a1 | solute carrier family 6 (neurotransmitter transporter, GABA), member 1 | MGI:95627 | YES | YES | Plasma Membrane | transporter | |||||
| ENSMUSG00000030307 | Slc6a11 | solute carrier family 6 (neurotransmitter transporter, GABA), member 11 | MGI:95630 | YES | YES | Plasma Membrane | transporter | |||||
| ENSMUSG00000027894 | Slc6a17 | solute carrier family 6 (neurotransmitter transporter), member 17 | MGI:2442535 | YES | YES | Cytoplasm | transporter | |||||
| ENSMUSG00000052026 | Slc6a7 | solute carrier family 6 (neurotransmitter transporter, L-proline), member 7 | MGI:2147363 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000069072 | Slc7a14 | solute carrier family 7 (cationic amino acid transporter, y+ system), member 14 | MGI:3040688 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000054640 | Slc8a1 | solute carrier family 8 (sodium/calcium exchanger), member 1 | MGI:107956 | YES | YES | Plasma Membrane | transporter | |||||
| ENSMUSG00000030376 | Slc8a2 | solute carrier family 8 (sodium/calcium exchanger), member 2 | MGI:107996 | YES | YES | Cytoplasm | transporter | |||||
| ENSMUSG00000028854 | Slc9a1 | solute carrier family 9 (sodium/hydrogen exchanger), member 1 | MGI:102462 | YES | YES | Plasma Membrane | ion channel | |||||
| ENSMUSG00000031906 | Smpd3 | sphingomyelin phosphodiesterase 3, neutral | MGI:1927578 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000033419 | Snap91 | synaptosomal-associated protein 91 | MGI:109132 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000025889 | Snca | synuclein, alpha | MGI:1277151 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000034891 | Sncb | synuclein, beta | MGI:1889011 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000027488 | Snta1 | syntrophin, acidic 1 | MGI:101772 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000046032 | Snx12 | sorting nexin 12 | MGI:1919331 | YES | YES | Cytoplasm | transporter | |||||
| ENSMUSG00000019804 | Snx3 | sorting nexin 3 | MGI:1860188 | YES | YES | Cytoplasm | transporter | |||||
| ENSMUSG00000022982 | Sod1 | superoxide dismutase 1, soluble | MGI:98351 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000006818 | Sod2 | superoxide dismutase 2, mitochondrial | MGI:98352 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000049313 | Sorl1 | sortilin-related receptor, LDLR class A repeats-containing | MGI:1202296 | YES | YES | Cytoplasm | transporter | |||||
| ENSMUSG00000068747 | Sort1 | sortilin 1 | MGI:1338015 | YES | YES | Plasma Membrane | G-protein coupled receptor | |||||
| ENSMUSG00000029309 | Sparcl1 | SPARC-like 1 | MGI:108110 | YES | YES | Extracellular Space | other | |||||
| ENSMUSG00000048040 | Arxes2 | adipocyte-related X-chromosome expressed sequence 2 | MGI:1924226 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000033735 | Spr | sepiapterin reductase | MGI:103078 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000022403 | St13 | suppression of tumorigenicity 13 | MGI:1917606 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000009588 | St6galnac1 | ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 1 | MGI:1341826 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000026389 | Steap3 | STEAP family member 3 | MGI:1915678 | YES | YES | Cytoplasm | transporter | |||||
| ENSMUSG00000030987 | Stim1 | stromal interaction molecule 1 | MGI:107476 | YES | YES | Plasma Membrane | ion channel | |||||
| ENSMUSG00000024966 | Stip1 | stress-induced phosphoprotein 1 | MGI:109130 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000026213 | Stk11ip | serine/threonine kinase 11 interacting protein | MGI:1918978 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000007207 | Stx1a | syntaxin 1A (brain) | MGI:109355 | YES | YES | Cytoplasm | transporter | |||||
| ENSMUSG00000030806 | Stx1b | syntaxin 1B | MGI:1930705 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000026797 | Stxbp1 | syntaxin binding protein 1 | MGI:107363 | YES | YES | Cytoplasm | transporter | |||||
| ENSMUSG00000019790 | Stxbp5 | syntaxin binding protein 5 (tomosyn) | MGI:1926058 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000022829 | Stxbp5l | syntaxin binding protein 5-like | MGI:2443815 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000022110 | Sucla2 | succinate-Coenzyme A ligase, ADP-forming, beta subunit | MGI:1306775 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000038486 | Sv2a | synaptic vesicle glycoprotein 2 a | MGI:1927139 | YES | YES | Cytoplasm | transporter | |||||
| ENSMUSG00000053025 | Sv2b | synaptic vesicle glycoprotein 2 b | MGI:1927338 | YES | YES | Plasma Membrane | transporter | |||||
| ENSMUSG00000074093 | Svip | small VCP/p97-interacting protein | MGI:1922994 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000022415 | Syngr1 | synaptogyrin 1 | MGI:1328323 | YES | YES | Plasma Membrane | transporter | |||||
| ENSMUSG00000022973 | Synj1 | synaptojanin 1 | MGI:1354961 | YES | YES | Cytoplasm | phosphatase | |||||
| ENSMUSG00000031144 | Syp | synaptophysin | MGI:98467 | YES | YES | Cytoplasm | transporter | |||||
| ENSMUSG00000049303 | Syt12 | synaptotagmin XII | MGI:2159601 | YES | YES | Plasma Membrane | transporter | |||||
| ENSMUSG00000026452 | Syt2 | synaptotagmin II | MGI:99666 | YES | YES | Cytoplasm | transporter | |||||
| ENSMUSG00000066621 | Tecpr1 | tectonin beta-propeller repeat containing 1 | MGI:1917631 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000016150 | Tenm1 | teneurin transmembrane protein 1 | MGI:1345185 | YES | YES | Plasma Membrane | peptidase | |||||
| ENSMUSG00000048078 | Tenm4 | teneurin transmembrane protein 4 | MGI:2447063 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000022797 | Tfrc | transferrin receptor | MGI:98822 | YES | YES | Plasma Membrane | transporter | |||||
| ENSMUSG00000056429 | Tgoln1 | trans-golgi network protein | MGI:105080 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000032625 | Thsd7a | thrombospondin, type I, domain containing 7A | MGI:2685683 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000027076 | Timm10 | translocase of inner mitochondrial membrane 10 | MGI:1353429 | YES | YES | Cytoplasm | transporter | |||||
| ENSMUSG00000089847 | Timm10b | translocase of inner mitochondrial membrane 10B | MGI:1315196 | YES | YES | Cytoplasm | transporter | |||||
| ENSMUSG00000003438 | Timm50 | translocase of inner mitochondrial membrane 50 | MGI:1913775 | YES | YES | Cytoplasm | phosphatase | |||||
| ENSMUSG00000039016 | Timm8b | translocase of inner mitochondrial membrane 8B | MGI:1353424 | YES | YES | Cytoplasm | transporter | |||||
| ENSMUSG00000021079 | Timm9 | translocase of inner mitochondrial membrane 9 | MGI:1353436 | YES | YES | Cytoplasm | transporter | |||||
| ENSMUSG00000021957 | Tkt | transketolase | MGI:105992 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000028465 | Tln1 | talin 1 | MGI:1099832 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000052698 | Tln2 | talin 2 | MGI:1917799 | YES | YES | Nucleus | other | |||||
| ENSMUSG00000029390 | Tmed2 | transmembrane emp24 domain trafficking protein 2 | MGI:1929269 | YES | YES | Cytoplasm | transporter | |||||
| ENSMUSG00000029571 | Tmem106b | transmembrane protein 106B | MGI:1919150 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000024736 | Tmem132a | transmembrane protein 132A | MGI:2147810 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000021361 | Tmem14c | transmembrane protein 14C | MGI:1913404 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000062373 | Tmem65 | transmembrane protein 65 | MGI:1922118 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000050043 | Tmx2 | thioredoxin-related transmembrane protein 2 | MGI:1914208 | YES | YES | Other | enzyme | |||||
| ENSMUSG00000024614 | Tmx3 | thioredoxin-related transmembrane protein 3 | MGI:2442418 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000034723 | Tmx4 | thioredoxin-related transmembrane protein 4 | MGI:106558 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000023915 | Tnfrsf21 | tumor necrosis factor receptor superfamily, member 21 | MGI:2151075 | YES | YES | Plasma Membrane | transmembrane receptor | |||||
| ENSMUSG00000014846 | Tppp3 | tubulin polymerization-promoting protein family member 3 | MGI:1915221 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000060126 | Tpt1 | tumor protein, translationally-controlled 1 | MGI:104890 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000052387 | Trpm3 | transient receptor potential cation channel, subfamily M, member 3 | MGI:2443101 | YES | YES | Plasma Membrane | ion channel | |||||
| ENSMUSG00000030352 | Tspan9 | tetraspanin 9 | MGI:1924558 | YES | YES | Plasma Membrane | other | |||||
| ENSMUSG00000030428 | Ttyh1 | tweety homolog 1 (Drosophila) | MGI:1889007 | YES | YES | Plasma Membrane | ion channel | |||||
| ENSMUSG00000036565 | Ttyh3 | tweety homolog 3 (Drosophila) | MGI:1925589 | YES | YES | Plasma Membrane | ion channel | |||||
| ENSMUSG00000072235 | Tuba1a | tubulin, alpha 1A | MGI:98869 | YES | YES | Cytoplasm | other | |||||
| ENSMUSG00000038991 | Txndc5 | thioredoxin domain containing 5 | MGI:2145316 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000001924 | Uba1 | ubiquitin-like modifier activating enzyme 1 | MGI:98890 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000029223 | Uchl1 | ubiquitin carboxy-terminal hydrolase L1 | MGI:103149 | YES | YES | Cytoplasm | peptidase | |||||
| ENSMUSG00000031634 | Ufsp2 | UFM1-specific peptidase 2 | MGI:1913679 | YES | YES | Other | enzyme | |||||
| ENSMUSG00000037470 | Uggt1 | UDP-glucose glycoprotein glucosyltransferase 1 | MGI:2443162 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000001891 | Ugp2 | UDP-glucose pyrophosphorylase 2 | MGI:2183447 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000059534 | Uqcr10 | ubiquinol-cytochrome c reductase, complex III subunit X | MGI:1913402 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000021520 | Uqcrb | ubiquinol-cytochrome c reductase binding protein | MGI:1914780 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000025651 | Uqcrc1 | ubiquinol-cytochrome c reductase core protein 1 | MGI:107876 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000030884 | Uqcrc2 | ubiquinol cytochrome c reductase core protein 2 | MGI:1914253 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000038462 | Uqcrfs1 | ubiquinol-cytochrome c reductase, Rieske iron-sulfur polypeptide 1 | MGI:1913944 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000038429 | Usp5 | ubiquitin specific peptidase 5 (isopeptidase T) | MGI:1347343 | YES | YES | Cytoplasm | peptidase | |||||
| ENSMUSG00000031010 | Usp9x | ubiquitin specific peptidase 9, X chromosome | MGI:894681 | YES | YES | Plasma Membrane | peptidase | |||||
| ENSMUSG00000034993 | Vat1 | vesicle amine transport protein 1 homolog (T californica) | MGI:1349450 | YES | YES | Plasma Membrane | transporter | |||||
| ENSMUSG00000028452 | Vcp | valosin containing protein | MGI:99919 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000046230 | Vps13a | vacuolar protein sorting 13A (yeast) | MGI:2444304 | YES | YES | Cytoplasm | transporter | |||||
| ENSMUSG00000031696 | Vps35 | vacuolar protein sorting 35 | MGI:1890467 | YES | YES | Cytoplasm | transporter | |||||
| ENSMUSG00000023186 | Vwa5a | von Willebrand factor A domain containing 5A | MGI:1915026 | YES | YES | Nucleus | other | |||||
| ENSMUSG00000021266 | Wars | tryptophanyl-tRNA synthetase | MGI:104630 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000043940 | Wdfy3 | WD repeat and FYVE domain containing 3 | MGI:1096875 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000005103 | Wdr1 | WD repeat domain 1 | MGI:1337100 | YES | YES | Extracellular Space | other | |||||
| ENSMUSG00000039828 | Wdr70 | WD repeat domain 70 | MGI:1921020 | YES | YES | Other | other | |||||
| ENSMUSG00000033488 | BC026585 | cDNA sequence BC026585 | MGI:2448516 | YES | YES | Cytoplasm | enzyme | |||||
| ENSMUSG00000048429 | 1810026J23Rik | RIKEN cDNA 1810026J23 gene | MGI:1917023 | YES | YES | Other | other | |||||
| ENSMUSG00000026276 | Sept2 | septin 2 | MGI:97298 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000022456 | Sept3 | septin 3 | MGI:1345148 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000020486 | Sept4 | septin 4 | MGI:1270156 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000072214 | Sept5 | septin 5 | MGI:1195461 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000050379 | Sept6 | septin 6 | MGI:1888939 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000059248 | Sept9 | septin 9 | MGI:1858222 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000058013 | Sept11 | septin 11 | MGI:1277214 | YES | YES | YES | Nucleus | other | ||||
| ENSMUSG00000055782 | Abcd2 | ATP-binding cassette, sub-family D (ALD), member 2 | MGI:1349467 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSMUSG00000028127 | Abcd3 | ATP-binding cassette, sub-family D (ALD), member 3 | MGI:1349216 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSMUSG00000058835 | Abi1 | abl-interactor 1 | MGI:104913 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000025085 | Ablim1 | actin-binding LIM protein 1 | MGI:1194500 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000032735 | Ablim3 | actin binding LIM protein family, member 3 | MGI:2442582 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000090150 | Acad11 | acyl-Coenzyme A dehydrogenase family, member 11 | MGI:2143169 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000017307 | Acot8 | acyl-CoA thioesterase 8 | MGI:2158201 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000031972 | Acta1 | actin, alpha 1, skeletal muscle | MGI:87902 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000029580 | Actb | actin, beta | MGI:87904 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000055194 | Actbl2 | actin, beta-like 2 | MGI:2444552 | YES | YES | YES | Nucleus | other | ||||
| ENSMUSG00000062825 | Actg1 | actin, gamma, cytoplasmic 1 | MGI:87906 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000052374 | Actn2 | actinin alpha 2 | MGI:109192 | YES | YES | YES | Nucleus | transcription regulator | ||||
| ENSMUSG00000025422 | Agap2 | ArfGAP with GTPase domain, ankyrin repeat and PH domain 2 | MGI:3580016 | YES | YES | YES | Nucleus | enzyme | ||||
| ENSMUSG00000031467 | Agpat5 | 1-acylglycerol-3-phosphate O-acyltransferase 5 (lysophosphatidic acid acyltransferase, epsilon) | MGI:1196345 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000019986 | Ahi1 | Abelson helper integration site 1 | MGI:87971 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000022763 | Aifm3 | apoptosis-inducing factor, mitochondrion-associated 3 | MGI:1919418 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000024885 | Aldh3b1 | aldehyde dehydrogenase 3 family, member B1 | MGI:1914939 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000021986 | Amer2 | APC membrane recruitment 2 | MGI:1919375 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000041688 | Amot | angiomotin | MGI:108440 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000078137 | Ankrd63 | ankyrin repeat domain 63 | MGI:2686183 | YES | YES | YES | Other | other | ||||
| ENSMUSG00000058589 | Anks1b | ankyrin repeat and sterile alpha motif domain containing 1B | MGI:1924781 | YES | YES | YES | Nucleus | other | ||||
| ENSMUSG00000004849 | Ap1s1 | adaptor protein complex AP-1, sigma 1 | MGI:1098244 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSMUSG00000022841 | Ap2m1 | adaptor-related protein complex 2, mu 1 subunit | MGI:1298405 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSMUSG00000004655 | Aqp1 | aquaporin 1 | MGI:103201 | YES | YES | YES | Plasma Membrane | transporter | ||||
| ENSMUSG00000036452 | Arhgap26 | Rho GTPase activating protein 26 | MGI:1918552 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000041444 | Arhgap32 | Rho GTPase activating protein 32 | MGI:2450166 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000033697 | Arhgap39 | Rho GTPase activating protein 39 | MGI:107858 | YES | YES | YES | Nucleus | other | ||||
| ENSMUSG00000029348 | Asphd2 | aspartate beta-hydroxylase domain containing 2 | MGI:1920148 | YES | YES | YES | Other | enzyme | ||||
| ENSMUSG00000029036 | Atad3a | ATPase family, AAA domain containing 3A | MGI:1919214 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000052364 | B630019K06Rik | RIKEN cDNA B630019K06 gene | MGI:2147918 | YES | YES | YES | Other | other | ||||
| ENSMUSG00000042215 | Bag2 | BCL2-associated athanogene 2 | MGI:1891254 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000025372 | Baiap2 | brain-specific angiogenesis inhibitor 1-associated protein 2 | MGI:2137336 | YES | YES | YES | Plasma Membrane | kinase | ||||
| ENSMUSG00000024844 | Banf1 | barrier to autointegration factor 1 | MGI:1346330 | YES | YES | YES | Nucleus | other | ||||
| ENSMUSG00000036948 | BC037034 | cDNA sequence BC037034 | MGI:2385896 | YES | YES | YES | Other | other | ||||
| ENSMUSG00000020650 | Bcap29 | B cell receptor associated protein 29 | MGI:101917 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSMUSG00000030802 | Bckdk | branched chain ketoacid dehydrogenase kinase | MGI:1276121 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSMUSG00000102226 | Begain | brain-enriched guanylate kinase-associated | MGI:3044626 | YES | YES | YES | Nucleus | other | ||||
| ENSMUSG00000033940 | Brk1 | BRICK1, SCAR/WAVE actin-nucleating complex subunit | MGI:1915406 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000000202 | Btbd17 | BTB (POZ) domain containing 17 | MGI:1919264 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000019146 | Cacng2 | calcium channel, voltage-dependent, gamma subunit 2 | MGI:1316660 | YES | YES | YES | Plasma Membrane | ion channel | ||||
| ENSMUSG00000066189 | Cacng3 | calcium channel, voltage-dependent, gamma subunit 3 | MGI:1859165 | YES | YES | YES | Plasma Membrane | ion channel | ||||
| ENSMUSG00000053395 | Cacng8 | calcium channel, voltage-dependent, gamma subunit 8 | MGI:1932376 | YES | YES | YES | Plasma Membrane | ion channel | ||||
| ENSMUSG00000023055 | Calcoco1 | calcium binding and coiled coil domain 1 | MGI:1914738 | YES | YES | YES | Nucleus | transcription regulator | ||||
| ENSMUSG00000019370 | Calm3 | calmodulin 3 | MGI:103249 | YES | YES | YES | Nucleus | other | ||||
| ENSMUSG00000024617 | Camk2a | calcium/calmodulin-dependent protein kinase II alpha | MGI:88256 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSMUSG00000021820 | Camk2g | calcium/calmodulin-dependent protein kinase II gamma | MGI:88259 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSMUSG00000041570 | Camsap2 | calmodulin regulated spectrin-associated protein family, member 2 | MGI:1922434 | YES | YES | YES | Other | other | ||||
| ENSMUSG00000070372 | Capza1 | capping protein (actin filament) muscle Z-line, alpha 1 | MGI:106227 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000015733 | Capza2 | capping protein (actin filament) muscle Z-line, alpha 2 | MGI:106222 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000028745 | Capzb | capping protein (actin filament) muscle Z-line, beta | MGI:104652 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000056158 | Car10 | carbonic anhydrase 10 | MGI:1919855 | YES | YES | YES | Other | enzyme | ||||
| ENSMUSG00000031654 | Cbln1 | cerebellin 1 precursor protein | MGI:88281 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000036686 | Cc2d1a | coiled-coil and C2 domain containing 1A | MGI:2384831 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000021578 | Ccdc127 | coiled-coil domain containing 127 | MGI:1914683 | YES | YES | YES | Other | other | ||||
| ENSMUSG00000062961 | Ccdc177 | coiled-coil domain containing 177 | MGI:2686414 | YES | YES | YES | Other | other | ||||
| ENSMUSG00000024286 | Ccny | cyclin Y | MGI:1915224 | YES | YES | YES | Nucleus | other | ||||
| ENSMUSG00000026437 | Cdk18 | cyclin-dependent kinase 18 | MGI:97518 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSMUSG00000031292 | Cdkl5 | cyclin-dependent kinase-like 5 | MGI:1278336 | YES | YES | YES | Nucleus | kinase | ||||
| ENSMUSG00000094626 | Cecr6 | cat eye syndrome chromosome region, candidate 6 | MGI:2136977 | YES | YES | YES | Nucleus | transcription regulator | ||||
| ENSMUSG00000053768 | Chchd3 | coiled-coil-helix-coiled-coil-helix domain containing 3 | MGI:1913325 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000000743 | Chmp1a | charged multivesicular body protein 1A | MGI:1920159 | YES | YES | YES | Extracellular Space | peptidase | ||||
| ENSMUSG00000038467 | Chmp4b | charged multivesicular body protein 4B | MGI:1922858 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000029516 | Cit | citron | MGI:105313 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSMUSG00000046798 | Cldn12 | claudin 12 | MGI:1929288 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000021097 | Clmn | calmin | MGI:2136957 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000006782 | Cnp | 2',3'-cyclic nucleotide 3' phosphodiesterase | MGI:88437 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000051154 | Commd3 | COMM domain containing 3 | MGI:88218 | YES | YES | YES | Other | other | ||||
| ENSMUSG00000004530 | Coro1c | coronin, actin binding protein 1C | MGI:1345964 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000028337 | Coro2a | coronin, actin binding protein 2A | MGI:1345966 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000041729 | Coro2b | coronin, actin binding protein, 2B | MGI:2444283 | YES | YES | YES | Other | other | ||||
| ENSMUSG00000020183 | Cpm | carboxypeptidase M | MGI:1917824 | YES | YES | YES | Plasma Membrane | peptidase | ||||
| ENSMUSG00000006356 | Crip2 | cysteine rich protein 2 | MGI:1915587 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000025162 | Csnk1d | casein kinase 1, delta | MGI:1355272 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSMUSG00000022433 | Csnk1e | casein kinase 1, epsilon | MGI:1351660 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSMUSG00000074698 | Csnk2a1 | casein kinase 2, alpha 1 polypeptide | MGI:88543 | YES | YES | YES | Nucleus | kinase | ||||
| ENSMUSG00000046707 | Csnk2a2 | casein kinase 2, alpha prime polypeptide | MGI:88547 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSMUSG00000000416 | Cttnbp2 | cortactin binding protein 2 | MGI:1353467 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000003269 | Cyth2 | cytohesin 2 | MGI:1334255 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000068373 | D430041D05Rik | RIKEN cDNA D430041D05 gene | MGI:2181743 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000063455 | D630045J12Rik | RIKEN cDNA D630045J12 gene | MGI:2669829 | YES | YES | YES | Other | other | ||||
| ENSMUSG00000026883 | Dab2ip | disabled 2 interacting protein | MGI:1916851 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000028078 | Dclk2 | doublecortin-like kinase 2 | MGI:1918012 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSMUSG00000020719 | Ddx5 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 5 | MGI:105037 | YES | YES | YES | Nucleus | enzyme | ||||
| ENSMUSG00000036095 | Dgkb | diacylglycerol kinase, beta | MGI:2442474 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSMUSG00000022210 | Dhrs4 | dehydrogenase/reductase (SDR family) member 4 | MGI:90169 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000042569 | Dhrs7b | dehydrogenase/reductase (SDR family) member 7B | MGI:2384931 | YES | YES | YES | Other | other | ||||
| ENSMUSG00000032480 | Dhx30 | DEAH (Asp-Glu-Ala-His) box polypeptide 30 | MGI:1920081 | YES | YES | YES | Nucleus | enzyme | ||||
| ENSMUSG00000052572 | Dlg2 | discs, large homolog 2 (Drosophila) | MGI:1344351 | YES | YES | YES | Plasma Membrane | kinase | ||||
| ENSMUSG00000020886 | Dlg4 | discs, large homolog 4 (Drosophila) | MGI:1277959 | YES | YES | YES | Plasma Membrane | kinase | ||||
| ENSMUSG00000003279 | Dlgap1 | discs, large (Drosophila) homolog-associated protein 1 | MGI:1346065 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000047495 | Dlgap2 | discs, large (Drosophila) homolog-associated protein 2 | MGI:2443181 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000042388 | Dlgap3 | discs, large (Drosophila) homolog-associated protein 3 | MGI:3039563 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000061689 | Dlgap4 | discs, large homolog-associated protein 4 (Drosophila) | MGI:2138865 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000022099 | Dmtn | dematin actin binding protein | MGI:99670 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000028410 | Dnaja1 | DnaJ (Hsp40) homolog, subfamily A, member 1 | MGI:1270129 | YES | YES | YES | Nucleus | other | ||||
| ENSMUSG00000031701 | Dnaja2 | DnaJ (Hsp40) homolog, subfamily A, member 2 | MGI:1931882 | YES | YES | YES | Nucleus | enzyme | ||||
| ENSMUSG00000004069 | Dnaja3 | DnaJ (Hsp40) homolog, subfamily A, member 3 | MGI:1933786 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000074212 | Dnajb14 | DnaJ (Hsp40) homolog, subfamily B, member 14 | MGI:1917854 | YES | YES | YES | Other | enzyme | ||||
| ENSMUSG00000029131 | Dnajb6 | DnaJ (Hsp40) homolog, subfamily B, member 6 | MGI:1344381 | YES | YES | YES | Nucleus | transcription regulator | ||||
| ENSMUSG00000039768 | Dnajc11 | DnaJ (Hsp40) homolog, subfamily C, member 11 | MGI:2443386 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000078919 | Dpm1 | dolichol-phosphate (beta-D) mannosyltransferase 1 | MGI:1330239 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000037259 | Dzank1 | double zinc ribbon and ankyrin repeat domains 1 | MGI:2139080 | YES | YES | YES | Other | other | ||||
| ENSMUSG00000028039 | Efna3 | ephrin A3 | MGI:106644 | YES | YES | YES | Plasma Membrane | kinase | ||||
| ENSMUSG00000015002 | Efr3a | EFR3 homolog A (S. cerevisiae) | MGI:1923990 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000008489 | Elavl2 | ELAV (embryonic lethal, abnormal vision, Drosophila)-like 2 (Hu antigen B) | MGI:1100887 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000041112 | Elmo1 | engulfment and cell motility 1 | MGI:2153044 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000017670 | Elmo2 | engulfment and cell motility 2 | MGI:2153045 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000008140 | Emc10 | ER membrane protein complex subunit 10 | MGI:1916933 | YES | YES | YES | Other | other | ||||
| ENSMUSG00000027131 | Emc4 | ER membrane protein complex subunit 4 | MGI:1915282 | YES | YES | YES | Other | other | ||||
| ENSMUSG00000031819 | Emc8 | ER membrane protein complex subunit 8 | MGI:1343095 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000022995 | Enah | enabled homolog (Drosophila) | MGI:108360 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000038173 | Enpp6 | ectonucleotide pyrophosphatase/phosphodiesterase 6 | MGI:2445171 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000023216 | Epb4.2 | erythrocyte protein band 4.2 | MGI:95402 | YES | YES | YES | Plasma Membrane | transporter | ||||
| ENSMUSG00000006276 | Eps15l1 | epidermal growth factor receptor pathway substrate 15-like 1 | MGI:104582 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000015766 | Eps8 | epidermal growth factor receptor pathway substrate 8 | MGI:104684 | YES | YES | YES | Plasma Membrane | peptidase | ||||
| ENSMUSG00000030172 | Erc1 | ELKS/RAB6-interacting/CAST family member 1 | MGI:2151013 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000040640 | Erc2 | ELKS/RAB6-interacting/CAST family member 2 | MGI:1098749 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000036617 | Etl4 | enhancer trap locus 4 | MGI:95454 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000035104 | Eva1a | eva-1 homolog A (C. elegans) | MGI:2385247 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000021262 | Evl | Ena-vasodilator stimulated phosphoprotein | MGI:1194884 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000034152 | Exoc3 | exocyst complex component 3 | MGI:2443972 | YES | YES | YES | Plasma Membrane | transporter | ||||
| ENSMUSG00000020792 | Exoc7 | exocyst complex component 7 | MGI:1859270 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSMUSG00000042787 | Exog | endo/exonuclease (5'-3'), endonuclease G-like | MGI:2143333 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000009216 | Fam163b | family with sequence similarity 163, member B | MGI:1926106 | YES | YES | YES | Other | other | ||||
| ENSMUSG00000061111 | Fam195b | family with sequence similarity 195, member B | MGI:2384752 | YES | YES | YES | Other | other | ||||
| ENSMUSG00000032224 | Fam81a | family with sequence similarity 81, member A | MGI:1924136 | YES | YES | YES | Other | other | ||||
| ENSMUSG00000027349 | Fam98b | family with sequence similarity 98, member B | MGI:1915465 | YES | YES | YES | Other | other | ||||
| ENSMUSG00000003808 | Farsa | phenylalanyl-tRNA synthetase, alpha subunit | MGI:1913840 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000038274 | Fau | Finkel-Biskis-Reilly murine sarcoma virus (FBR-MuSV) ubiquitously expressed (fox derived) | MGI:102547 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000061981 | Flot2 | flotillin 2 | MGI:103309 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000027680 | Fxr1 | fragile X mental retardation gene 1, autosomal homolog | MGI:104860 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000018765 | Fxr2 | fragile X mental retardation, autosomal homolog 2 | MGI:1346074 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000019843 | Fyn | Fyn proto-oncogene | MGI:95602 | YES | YES | YES | Plasma Membrane | kinase | ||||
| ENSMUSG00000029405 | G3bp2 | GTPase activating protein (SH3 domain) binding protein 2 | MGI:2442040 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000024462 | Gabbr1 | gamma-aminobutyric acid (GABA) B receptor, 1 | MGI:1860139 | YES | YES | YES | Plasma Membrane | G-protein coupled receptor | ||||
| ENSMUSG00000021003 | Galc | galactosylceramidase | MGI:95636 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000047261 | Gap43 | growth associated protein 43 | MGI:95639 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000029432 | Gbas | glioblastoma amplified sequence | MGI:1278343 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000020932 | Gfap | glial fibrillary acidic protein | MGI:95697 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000050953 | Gja1 | gap junction protein, alpha 1 | MGI:95713 | YES | YES | YES | Plasma Membrane | transporter | ||||
| ENSMUSG00000040055 | Gjb6 | gap junction protein, beta 6 | MGI:107588 | YES | YES | YES | Plasma Membrane | transporter | ||||
| ENSMUSG00000043448 | Gjc2 | gap junction protein, gamma 2 | MGI:2153060 | YES | YES | YES | Plasma Membrane | transporter | ||||
| ENSMUSG00000028480 | Glipr2 | GLI pathogenesis-related 2 | MGI:1917770 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000058064 | Gm10036 | predicted gene 10036 | MGI:3642334 | YES | YES | YES | Other | other | ||||
| ENSMUSG00000069117 | Gm10260 | predicted gene 10260 | MGI:3642298 | YES | YES | YES | Other | other | ||||
| ENSMUSG00000029419 | Gm996 | predicted gene 996 | MGI:2685842 | YES | YES | YES | Other | other | ||||
| ENSMUSG00000027523 | Gnas | GNAS (guanine nucleotide binding protein, alpha stimulating) complex locus | MGI:95777 | YES | YES | YES | Plasma Membrane | enzyme | ||||
| ENSMUSG00000040009 | Gnaz | guanine nucleotide binding protein, alpha z subunit | MGI:95780 | YES | YES | YES | Plasma Membrane | enzyme | ||||
| ENSMUSG00000036402 | Gng12 | guanine nucleotide binding protein (G protein), gamma 12 | MGI:1336171 | YES | YES | YES | Plasma Membrane | enzyme | ||||
| ENSMUSG00000042532 | Golga7b | golgi autoantigen, golgin subfamily a, 7B | MGI:1918396 | YES | YES | YES | Other | other | ||||
| ENSMUSG00000047454 | Gphn | gephyrin | MGI:109602 | YES | YES | YES | Plasma Membrane | enzyme | ||||
| ENSMUSG00000008734 | Gprc5b | G protein-coupled receptor, family C, group 5, member B | MGI:1927596 | YES | YES | YES | Plasma Membrane | G-protein coupled receptor | ||||
| ENSMUSG00000069227 | Gprin1 | G protein-regulated inducer of neurite outgrowth 1 | MGI:1349455 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000026930 | Gpsm1 | G-protein signalling modulator 1 (AGS3-like, C. elegans) | MGI:1915089 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000063856 | Gpx1 | glutathione peroxidase 1 | MGI:104887 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000072772 | Grcc10 | gene rich cluster, C10 gene | MGI:1315201 | YES | YES | YES | Other | other | ||||
| ENSMUSG00000001986 | Gria3 | glutamate receptor, ionotropic, AMPA3 (alpha 3) | MGI:95810 | YES | YES | YES | Plasma Membrane | ion channel | ||||
| ENSMUSG00000059003 | Grin2a | glutamate receptor, ionotropic, NMDA2A (epsilon 1) | MGI:95820 | YES | YES | YES | Plasma Membrane | ion channel | ||||
| ENSMUSG00000002771 | Grin2d | glutamate receptor, ionotropic, NMDA2D (epsilon 4) | MGI:95823 | YES | YES | YES | Plasma Membrane | ion channel | ||||
| ENSMUSG00000090523 | Gypc | glycophorin C | MGI:1098566 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000021613 | Hapln1 | hyaluronan and proteoglycan link protein 1 | MGI:1337006 | YES | YES | YES | Extracellular Space | other | ||||
| ENSMUSG00000007594 | Hapln4 | hyaluronan and proteoglycan link protein 4 | MGI:2679531 | YES | YES | YES | Extracellular Space | other | ||||
| ENSMUSG00000034245 | Hdac11 | histone deacetylase 11 | MGI:2385252 | YES | YES | YES | Nucleus | transcription regulator | ||||
| ENSMUSG00000069308 | Hist1h2bp | histone cluster 1, H2bp | MGI:2448409 | YES | YES | YES | Nucleus | other | ||||
| ENSMUSG00000069266 | Hist1h4b | histone cluster 1, H4b | MGI:2448420 | YES | YES | YES | #N/A | #N/A | ||||
| ENSMUSG00000046434 | Hnrnpa1 | heterogeneous nuclear ribonucleoprotein A1 | MGI:104820 | YES | YES | YES | Nucleus | other | ||||
| ENSMUSG00000059005 | Hnrnpa3 | heterogeneous nuclear ribonucleoprotein A3 | MGI:1917171 | YES | YES | YES | Nucleus | other | ||||
| ENSMUSG00000059208 | Hnrnpm | heterogeneous nuclear ribonucleoprotein M | MGI:1926465 | YES | YES | YES | Nucleus | other | ||||
| ENSMUSG00000007617 | Homer1 | homer homolog 1 (Drosophila) | MGI:1347345 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000025813 | Homer2 | homer homolog 2 (Drosophila) | MGI:1347354 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000003573 | Homer3 | homer homolog 3 (Drosophila) | MGI:1347359 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000006205 | Htra1 | HtrA serine peptidase 1 | MGI:1929076 | YES | YES | YES | Extracellular Space | peptidase | ||||
| ENSMUSG00000013367 | Iglon5 | IgLON family member 5 | MGI:2686277 | YES | YES | YES | Other | other | ||||
| ENSMUSG00000040972 | Igsf21 | immunoglobulin superfamily, member 21 | MGI:2681842 | YES | YES | YES | Extracellular Space | other | ||||
| ENSMUSG00000040612 | Ildr2 | immunoglobulin-like domain containing receptor 2 | MGI:1196370 | YES | YES | YES | Other | other | ||||
| ENSMUSG00000034336 | Ina | internexin neuronal intermediate filament protein, alpha | MGI:96568 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000034312 | Iqsec1 | IQ motif and Sec7 domain 1 | MGI:1196356 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000041115 | Iqsec2 | IQ motif and Sec7 domain 2 | MGI:3528396 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000026223 | Itm2c | integral membrane protein 2C | MGI:1927594 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000020640 | Itsn2 | intersectin 2 | MGI:1338049 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000025318 | Jph3 | junctophilin 3 | MGI:1891497 | YES | YES | YES | Plasma Membrane | ion channel | ||||
| ENSMUSG00000040606 | Kazn | kazrin, periplakin interacting protein | MGI:1918779 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000029088 | Kcnip4 | Kv channel interacting protein 4 | MGI:1933131 | YES | YES | YES | Plasma Membrane | ion channel | ||||
| ENSMUSG00000098557 | Kctd12 | potassium channel tetramerisation domain containing 12 | MGI:2145823 | YES | YES | YES | Plasma Membrane | ion channel | ||||
| ENSMUSG00000051401 | Kctd16 | potassium channel tetramerisation domain containing 16 | MGI:1914659 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000037653 | Kctd8 | potassium channel tetramerisation domain containing 8 | MGI:2443804 | YES | YES | YES | Other | other | ||||
| ENSMUSG00000033595 | Lgi3 | leucine-rich repeat LGI family, member 3 | MGI:2182619 | YES | YES | YES | Extracellular Space | other | ||||
| ENSMUSG00000003872 | Lin7b | lin-7 homolog B (C. elegans) | MGI:1330858 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000033060 | Lmo7 | LIM domain only 7 | MGI:1353586 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000028176 | Lrrc7 | leucine rich repeat containing 7 | MGI:2676665 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000071073 | Lrrc73 | leucine rich repeat containing 73 | MGI:2684934 | YES | YES | YES | Other | other | ||||
| ENSMUSG00000060780 | Lrrtm1 | leucine rich repeat transmembrane neuronal 1 | MGI:2389173 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000071862 | Lrrtm2 | leucine rich repeat transmembrane neuronal 2 | MGI:2389174 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000044847 | Lsm11 | U7 snRNP-specific Sm-like protein LSM11 | MGI:1919540 | YES | YES | YES | Nucleus | other | ||||
| ENSMUSG00000036306 | Lzts1 | leucine zipper, putative tumor suppressor 1 | MGI:2684762 | YES | YES | YES | Nucleus | transcription regulator | ||||
| ENSMUSG00000037703 | Lzts3 | leucine zipper, putative tumor suppressor family member 3 | MGI:2656976 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000024479 | Mal2 | mal, T cell differentiation protein 2 | MGI:2146021 | YES | YES | YES | Plasma Membrane | transporter | ||||
| ENSMUSG00000031812 | Map1lc3b | microtubule-associated protein 1 light chain 3 beta | MGI:1914693 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000041205 | Map6d1 | MAP6 domain containing 1 | MGI:3607784 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000026620 | Mark1 | MAP/microtubule affinity regulating kinase 1 | MGI:2664902 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSMUSG00000024969 | Mark2 | MAP/microtubule affinity regulating kinase 2 | MGI:99638 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSMUSG00000007411 | Mark3 | MAP/microtubule affinity regulating kinase 3 | MGI:1341865 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSMUSG00000030397 | Mark4 | MAP/microtubule affinity regulating kinase 4 | MGI:1920955 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSMUSG00000030605 | Mfge8 | milk fat globule-EGF factor 8 protein | MGI:102768 | YES | YES | YES | Extracellular Space | other | ||||
| ENSMUSG00000030120 | Mlf2 | myeloid leukemia factor 2 | MGI:1353554 | YES | YES | YES | Nucleus | other | ||||
| ENSMUSG00000032355 | Mlip | muscular LMNA-interacting protein | MGI:1916892 | YES | YES | YES | Nucleus | other | ||||
| ENSMUSG00000037022 | Mmaa | methylmalonic aciduria (cobalamin deficiency) type A | MGI:1923805 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000061273 | Mmgt1 | membrane magnesium transporter 1 | MGI:2384305 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSMUSG00000025979 | Mob4 | MOB family member 4, phocein | MGI:104899 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000030036 | Mogs | mannosyl-oligosaccharide glucosidase | MGI:1929872 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000031402 | Mpp1 | membrane protein, palmitoylated | MGI:105941 | YES | YES | YES | Plasma Membrane | kinase | ||||
| ENSMUSG00000005417 | Mprip | myosin phosphatase Rho interacting protein | MGI:1349438 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000023723 | Mrps23 | mitochondrial ribosomal protein S23 | MGI:1928138 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000040269 | Mrps28 | mitochondrial ribosomal protein S28 | MGI:1913480 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000061474 | Mrps36 | mitochondrial ribosomal protein S36 | MGI:1913378 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000024012 | Mtch1 | mitochondrial carrier homolog 1 (C. elegans) | MGI:1929261 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000022255 | Mtdh | metadherin | MGI:1914404 | YES | YES | YES | Cytoplasm | transcription regulator | ||||
| ENSMUSG00000018830 | Myh11 | myosin, heavy polypeptide 11, smooth muscle | MGI:102643 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000061816 | Myl1 | myosin, light polypeptide 1 | MGI:97269 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000034868 | Myl12b | myosin, light chain 12B, regulatory | MGI:107494 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000090841 | Myl6 | myosin, light polypeptide 6, alkali, smooth muscle and non-muscle | MGI:109318 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000039824 | Myl6b | myosin, light polypeptide 6B | MGI:1917789 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000022836 | Mylk | myosin, light polypeptide kinase | MGI:894806 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSMUSG00000018417 | Myo1b | myosin IB | MGI:107752 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000017774 | Myo1c | myosin IC | MGI:106612 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000022671 | Mzt2 | mitotic spindle organizing protein 2 | MGI:1922845 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000009418 | Nav1 | neuron navigator 1 | MGI:2183683 | YES | YES | YES | Other | enzyme | ||||
| ENSMUSG00000018736 | Ndel1 | nuclear distribution gene E-like homolog 1 (A. nidulans) | MGI:1932915 | YES | YES | YES | Nucleus | other | ||||
| ENSMUSG00000022055 | Nefl | neurofilament, light polypeptide | MGI:97313 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000022054 | Nefm | neurofilament, medium polypeptide | MGI:97314 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000079481 | Nhsl2 | NHS-like 2 | MGI:3645090 | YES | YES | YES | Other | other | ||||
| ENSMUSG00000034285 | Nipsnap1 | 4-nitrophenylphosphatase domain and non-neuronal SNAP25-like protein homolog 1 (C. elegans) | MGI:1278344 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000021772 | Nkiras1 | NFKB inhibitor interacting Ras-like protein 1 | MGI:1916971 | YES | YES | YES | Other | enzyme | ||||
| ENSMUSG00000031311 | Nono | non-POU-domain-containing, octamer binding protein | MGI:1855692 | YES | YES | YES | Nucleus | other | ||||
| ENSMUSG00000057113 | Npm1 | nucleophosmin 1 | MGI:106184 | YES | YES | YES | Nucleus | transcription regulator | ||||
| ENSMUSG00000032420 | Nt5e | 5' nucleotidase, ecto | MGI:99782 | YES | YES | YES | Plasma Membrane | phosphatase | ||||
| ENSMUSG00000063160 | Numbl | numb-like | MGI:894702 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000054976 | Nyap2 | neuronal tyrosine-phophorylated phosphoinositide 3-kinase adaptor 2 | MGI:2443135 | YES | YES | YES | Other | other | ||||
| ENSMUSG00000026833 | Olfm1 | olfactomedin 1 | MGI:1860437 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000032172 | Olfm2 | olfactomedin 2 | MGI:3045350 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000027965 | Olfm3 | olfactomedin 3 | MGI:2387329 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000027257 | Pacsin3 | protein kinase C and casein kinase substrate in neurons 3 | MGI:1891410 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000039913 | Pak7 | p21 protein (Cdc42/Rac)-activated kinase 7 | MGI:1920334 | YES | YES | YES | Nucleus | kinase | ||||
| ENSMUSG00000089945 | Pakap | paralemmin A kinase anchor protein | MGI:5141924 | YES | YES | YES | Other | other | ||||
| ENSMUSG00000058441 | Panx2 | pannexin 2 | MGI:1890615 | YES | YES | YES | Plasma Membrane | transporter | ||||
| ENSMUSG00000102697 | Pcdhac2 | protocadherin alpha subfamily C, 2 | MGI:1891443 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000028525 | Pde4b | phosphodiesterase 4B, cAMP specific | MGI:99557 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000015668 | Pdzd11 | PDZ domain containing 11 | MGI:1919871 | YES | YES | YES | Extracellular Space | other | ||||
| ENSMUSG00000029500 | Pgam5 | phosphoglycerate mutase family member 5 | MGI:1919792 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000073678 | Pgap1 | post-GPI attachment to proteins 1 | MGI:2443342 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000054728 | Phactr1 | phosphatase and actin regulator 1 | MGI:2659021 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000041957 | Pkp2 | plakophilin 2 | MGI:1914701 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000026991 | Pkp4 | plakophilin 4 | MGI:109281 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000041757 | Plekha6 | pleckstrin homology domain containing, family A member 6 | MGI:2388662 | YES | YES | YES | Other | other | ||||
| ENSMUSG00000066438 | Plekhd1 | pleckstrin homology domain containing, family D (with coiled-coil domains) member 1 | MGI:3036228 | YES | YES | YES | Other | other | ||||
| ENSMUSG00000026458 | Ppfia4 | protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 4 | MGI:1915757 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000004455 | Ppp1cc | protein phosphatase 1, catalytic subunit, gamma isoform | MGI:104872 | YES | YES | YES | Cytoplasm | phosphatase | ||||
| ENSMUSG00000037754 | Ppp1r16b | protein phosphatase 1, regulatory (inhibitor) subunit 16B | MGI:2151841 | YES | YES | YES | Plasma Membrane | phosphatase | ||||
| ENSMUSG00000032827 | Ppp1r9a | protein phosphatase 1, regulatory (inhibitor) subunit 9A | MGI:2442401 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000038976 | Ppp1r9b | protein phosphatase 1, regulatory subunit 9B | MGI:2387581 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000029120 | Ppp2r2c | protein phosphatase 2, regulatory subunit B, gamma | MGI:2442660 | YES | YES | YES | Other | phosphatase | ||||
| ENSMUSG00000030020 | Prickle2 | prickle homolog 2 (Drosophila) | MGI:1925144 | YES | YES | YES | Nucleus | other | ||||
| ENSMUSG00000025855 | Prkar1b | protein kinase, cAMP dependent regulatory, type I beta | MGI:97759 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSMUSG00000029334 | Prkg2 | protein kinase, cGMP-dependent, type II | MGI:108173 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSMUSG00000052429 | Prmt1 | protein arginine N-methyltransferase 1 | MGI:107846 | YES | YES | YES | Nucleus | enzyme | ||||
| ENSMUSG00000028134 | Ptbp2 | polypyrimidine tract binding protein 2 | MGI:1860489 | YES | YES | YES | Nucleus | other | ||||
| ENSMUSG00000021745 | Ptprg | protein tyrosine phosphatase, receptor type, G | MGI:97814 | YES | YES | YES | Plasma Membrane | phosphatase | ||||
| ENSMUSG00000043991 | Pura | purine rich element binding protein A | MGI:103079 | YES | YES | YES | Nucleus | transcription regulator | ||||
| ENSMUSG00000094483 | Purb | purine rich element binding protein B | MGI:1338779 | YES | YES | YES | Nucleus | transcription regulator | ||||
| ENSMUSG00000049184 | Purg | purine-rich element binding protein G | MGI:1922279 | YES | YES | YES | Other | other | ||||
| ENSMUSG00000026520 | Pycr2 | pyrroline-5-carboxylate reductase family, member 2 | MGI:1277956 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000040022 | Rab11fip2 | RAB11 family interacting protein 2 (class I) | MGI:1922248 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000051343 | Rab11fip5 | RAB11 family interacting protein 5 (class I) | MGI:1098586 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000029518 | Rab35 | RAB35, member RAS oncogene family | MGI:1924657 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000001847 | Rac1 | RAS-related C3 botulinum substrate 1 | MGI:97845 | YES | YES | YES | Plasma Membrane | enzyme | ||||
| ENSMUSG00000018012 | Rac3 | RAS-related C3 botulinum substrate 3 | MGI:2180784 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000008859 | Rala | v-ral simian leukemia viral oncogene homolog A (ras related) | MGI:1927243 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000004451 | Ralb | v-ral simian leukemia viral oncogene homolog B (ras related) | MGI:1927244 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000049044 | Rapgef4 | Rap guanine nucleotide exchange factor (GEF) 4 | MGI:1917723 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000026014 | Raph1 | Ras association (RalGDS/AF-6) and pleckstrin homology domains 1 | MGI:1924550 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000021708 | Rasgrf2 | RAS protein-specific guanine nucleotide-releasing factor 2 | MGI:109137 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000038555 | Reep2 | receptor accessory protein 2 | MGI:2385070 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000039316 | Rftn1 | raftlin lipid raft linker 1 | MGI:1923688 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000019775 | Rgs17 | regulator of G-protein signaling 17 | MGI:1927469 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000002459 | Rgs20 | regulator of G-protein signaling 20 | MGI:1929866 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000021719 | Rgs7bp | regulator of G-protein signalling 7 binding protein | MGI:106334 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000042671 | Rgs8 | regulator of G-protein signaling 8 | MGI:108408 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000020599 | Rgs9 | regulator of G-protein signaling 9 | MGI:1338824 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000073982 | Rhog | ras homolog gene family, member G | MGI:1928370 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000029420 | Rimbp2 | RIMS binding protein 2 | MGI:2443235 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000041670 | Rims1 | regulating synaptic membrane exocytosis 1 | MGI:2152971 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000037386 | Rims2 | regulating synaptic membrane exocytosis 2 | MGI:2152972 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000060090 | Rp2h | retinitis pigmentosa 2 homolog (human) | MGI:1277953 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000058443 | Rpl10-ps3 | ribosomal protein L10, pseudogene 3 | MGI:3704336 | YES | YES | YES | Other | other | ||||
| ENSMUSG00000037805 | Rpl10a | ribosomal protein L10A | MGI:1343877 | YES | YES | YES | Nucleus | other | ||||
| ENSMUSG00000038900 | Rpl12 | ribosomal protein L12 | MGI:98002 | YES | YES | YES | Nucleus | other | ||||
| ENSMUSG00000000740 | Rpl13 | ribosomal protein L13 | MGI:105922 | YES | YES | YES | Nucleus | other | ||||
| ENSMUSG00000074129 | Rpl13a | ribosomal protein L13A | MGI:1351455 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000025794 | Rpl14 | ribosomal protein L14 | MGI:1914365 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000062328 | Rpl17 | ribosomal protein L17 | MGI:2448270 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000059070 | Rpl18 | ribosomal protein L18 | MGI:98003 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000017404 | Rpl19 | ribosomal protein L19 | MGI:98020 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000041453 | Rpl21 | ribosomal protein L21 | MGI:1278340 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000039221 | Rpl22l1 | ribosomal protein L22 like 1 | MGI:1915278 | YES | YES | YES | Other | other | ||||
| ENSMUSG00000058546 | Rpl23a | ribosomal protein L23A | MGI:3040672 | YES | YES | YES | Nucleus | other | ||||
| ENSMUSG00000098274 | Rpl24 | ribosomal protein L24 | MGI:1915443 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000060938 | Rpl26 | ribosomal protein L26 | MGI:106022 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000073640 | Rpl27-ps3 | ribosomal protein L27, pseudogene 3 | MGI:3646174 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000046364 | Rpl27a | ribosomal protein L27A | MGI:1347076 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000030432 | Rpl28 | ribosomal protein L28 | MGI:101839 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000060036 | Rpl3 | ribosomal protein L3 | MGI:1351605 | YES | YES | YES | Nucleus | other | ||||
| ENSMUSG00000073702 | Rpl31 | ribosomal protein L31 | MGI:2149632 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000057841 | Rpl32 | ribosomal protein L32 | MGI:98038 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000062006 | Rpl34 | ribosomal protein L34 | MGI:1915686 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000078193 | Gm2000 | predicted gene 2000 | MGI:3780170 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000057863 | Rpl36 | ribosomal protein L36 | MGI:1860603 | YES | YES | YES | Nucleus | other | ||||
| ENSMUSG00000057322 | Rpl38 | ribosomal protein L38 | MGI:1914921 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000032399 | Rpl4 | ribosomal protein L4 | MGI:1915141 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000029614 | Rpl6 | ribosomal protein L6 | MGI:108057 | YES | YES | YES | Nucleus | other | ||||
| ENSMUSG00000043716 | Rpl7 | ribosomal protein L7 | MGI:98073 | YES | YES | YES | Nucleus | transcription regulator | ||||
| ENSMUSG00000062647 | Rpl7a | ribosomal protein L7A | MGI:1353472 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000003970 | Rpl8 | ribosomal protein L8 | MGI:1350927 | YES | YES | YES | Other | other | ||||
| ENSMUSG00000047215 | Rpl9 | ribosomal protein L9 | MGI:1298373 | YES | YES | YES | Nucleus | other | ||||
| ENSMUSG00000025508 | Rplp2 | ribosomal protein, large P2 | MGI:1914436 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000052146 | Rps10 | ribosomal protein S10 | MGI:1914347 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000003429 | Rps11 | ribosomal protein S11 | MGI:1351329 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000090862 | Rps13 | ribosomal protein S13 | MGI:1915302 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000024608 | Rps14 | ribosomal protein S14 | MGI:98107 | YES | YES | YES | Cytoplasm | translation regulator | ||||
| ENSMUSG00000063457 | Rps15 | ribosomal protein S15 | MGI:98117 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000037563 | Rps16 | ribosomal protein S16 | MGI:98118 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000061787 | Rps17 | ribosomal protein S17 | MGI:1309526 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000040952 | Rps19 | ribosomal protein S19 | MGI:1333780 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000028234 | Rps20 | ribosomal protein S20 | MGI:1914677 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000025290 | Rps24 | ribosomal protein S24 | MGI:98147 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000059775 | Rps26-ps1 | ribosomal protein S26, pseudogene 1 | MGI:3704322 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000050621 | Rps27rt | ribosomal protein S27, retrogene | MGI:3704345 | YES | YES | YES | Other | other | ||||
| ENSMUSG00000067288 | Rps28 | ribosomal protein S28 | MGI:1859516 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000034892 | Rps29 | ribosomal protein S29 | MGI:107681 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000028081 | Rps3a1 | ribosomal protein S3A1 | MGI:1202063 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000028495 | Rps6 | ribosomal protein S6 | MGI:98159 | YES | YES | YES | #N/A | #N/A | ||||
| ENSMUSG00000047675 | Rps8 | ribosomal protein S8 | MGI:98166 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000006333 | Rps9 | ribosomal protein S9 | MGI:1924096 | YES | YES | YES | Cytoplasm | translation regulator | ||||
| ENSMUSG00000001783 | Rtcb | RNA 2',3'-cyclic phosphate and 5'-OH ligase | MGI:106379 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000035236 | Scai | suppressor of cancer cell invasion | MGI:2443716 | YES | YES | YES | Nucleus | transcription regulator | ||||
| ENSMUSG00000032261 | Sh3bgrl2 | SH3 domain binding glutamic acid-rich protein like 2 | MGI:1915350 | YES | YES | YES | Nucleus | other | ||||
| ENSMUSG00000040990 | Sh3kbp1 | SH3-domain kinase binding protein 1 | MGI:1889583 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000053930 | Shisa6 | shisa family member 6 | MGI:2685725 | YES | YES | YES | Other | other | ||||
| ENSMUSG00000053550 | Shisa7 | shisa family member 7 | MGI:3605641 | YES | YES | YES | Other | other | ||||
| ENSMUSG00000042700 | Sipa1l1 | signal-induced proliferation-associated 1 like 1 | MGI:2443679 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000036309 | Skp1a | S-phase kinase-associated protein 1A | MGI:103575 | YES | YES | YES | Nucleus | transcription regulator | ||||
| ENSMUSG00000027287 | Snap23 | synaptosomal-associated protein 23 | MGI:109356 | YES | YES | YES | Plasma Membrane | transporter | ||||
| ENSMUSG00000027457 | Snph | syntaphilin | MGI:2139270 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000025006 | Sorbs1 | sorbin and SH3 domain containing 1 | MGI:700014 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000051934 | Spats2 | spermatogenesis associated, serine-rich 2 | MGI:1919822 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000042331 | Specc1 | sperm antigen with calponin homology and coiled-coil domains 1 | MGI:2442356 | YES | YES | YES | Nucleus | other | ||||
| ENSMUSG00000024533 | Spire1 | spire homolog 1 (Drosophila) | MGI:1915416 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000045671 | Spred2 | sprouty-related, EVH1 domain containing 2 | MGI:2150019 | YES | YES | YES | Extracellular Space | cytokine | ||||
| ENSMUSG00000026532 | Spta1 | spectrin alpha, erythrocytic 1 | MGI:98385 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000021061 | Sptb | spectrin beta, erythrocytic | MGI:98387 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000038453 | Srcin1 | SRC kinase signaling inhibitor 1 | MGI:1933179 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000071172 | Srsf3 | serine/arginine-rich splicing factor 3 | MGI:98285 | YES | YES | YES | Nucleus | other | ||||
| ENSMUSG00000027828 | Ssr3 | signal sequence receptor, gamma | MGI:1914687 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000026880 | Stom | stomatin | MGI:95403 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000037217 | Syn1 | synapsin I | MGI:98460 | YES | YES | YES | Plasma Membrane | transporter | ||||
| ENSMUSG00000059602 | Syn3 | synapsin III | MGI:1351334 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000096054 | Syne1 | spectrin repeat containing, nuclear envelope 1 | MGI:1927152 | YES | YES | YES | Nucleus | other | ||||
| ENSMUSG00000067629 | Syngap1 | synaptic Ras GTPase activating protein 1 homolog (rat) | MGI:3039785 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000043079 | Synpo | synaptopodin | MGI:1099446 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000068923 | Syt11 | synaptotagmin XI | MGI:1859547 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSMUSG00000004961 | Syt5 | synaptotagmin V | MGI:1926368 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSMUSG00000024743 | Syt7 | synaptotagmin VII | MGI:1859545 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSMUSG00000059981 | Taok2 | TAO kinase 2 | MGI:1915919 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSMUSG00000042492 | Tbc1d10b | TBC1 domain family, member 10b | MGI:1915699 | YES | YES | YES | Plasma Membrane | enzyme | ||||
| ENSMUSG00000005374 | Tbl2 | transducin (beta)-like 2 | MGI:1351652 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000003923 | Tfam | transcription factor A, mitochondrial | MGI:107810 | YES | YES | YES | Cytoplasm | transcription regulator | ||||
| ENSMUSG00000056665 | Them6 | thioesterase superfamily member 6 | MGI:1925301 | YES | YES | YES | Other | other | ||||
| ENSMUSG00000032011 | Thy1 | thymus cell antigen 1, theta | MGI:98747 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSMUSG00000070498 | Tmem132b | transmembrane protein 132B | MGI:3609245 | YES | YES | YES | Other | other | ||||
| ENSMUSG00000033578 | Tmem35 | transmembrane protein 35 | MGI:1914814 | YES | YES | YES | Other | other | ||||
| ENSMUSG00000028328 | Tmod1 | tropomodulin 1 | MGI:98775 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSMUSG00000032186 | Tmod2 | tropomodulin 2 | MGI:1355335 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000058587 | Tmod3 | tropomodulin 3 | MGI:1355315 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000028364 | Tnc | tenascin C | MGI:101922 | YES | YES | YES | Extracellular Space | other | ||||
| ENSMUSG00000093904 | Tomm20 | translocase of outer mitochondrial membrane 20 homolog (yeast) | MGI:1915202 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSMUSG00000022427 | Tomm22 | translocase of outer mitochondrial membrane 22 homolog (yeast) | MGI:2450248 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSMUSG00000021277 | Traf3 | TNF receptor-associated factor 3 | MGI:108041 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000042766 | Trim46 | tripartite motif-containing 46 | MGI:2673000 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000068735 | Trp53i11 | transformation related protein 53 inducible protein 11 | MGI:2670995 | YES | YES | YES | Other | other | ||||
| ENSMUSG00000029723 | Tsc22d4 | TSC22 domain family, member 4 | MGI:1926079 | YES | YES | YES | Nucleus | transcription regulator | ||||
| ENSMUSG00000051747 | Ttn | titin | MGI:98864 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSMUSG00000029592 | Usp30 | ubiquitin specific peptidase 30 | MGI:2140991 | YES | YES | YES | Other | other | ||||
| ENSMUSG00000026728 | Vim | vimentin | MGI:98932 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000037957 | Wdr20 | WD repeat domain 20 | MGI:1916891 | YES | YES | YES | Other | other | ||||
| ENSMUSG00000035067 | Xkr6 | X Kell blood group precursor related family member 6 homolog | MGI:2447765 | YES | YES | YES | Other | other | ||||
| ENSMUSG00000014932 | Yes1 | Yamaguchi sarcoma viral (v-yes) oncogene homolog 1 | MGI:99147 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSMUSG00000043542 | Zc2hc1a | zinc finger, C2HC-type containing 1A | MGI:1914556 | YES | YES | YES | Other | other | ||||
| ENSMUSG00000034075 | Zdhhc5 | zinc finger, DHHC domain containing 5 | MGI:1923573 | YES | YES | YES | Nucleus | enzyme | ||||
| ENSMUSG00000037855 | Zfp365 | zinc finger protein 365 | MGI:2143676 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000019923 | Zwint | ZW10 interactor | MGI:1289227 | YES | YES | YES | Nucleus | other | ||||
| ENSMUSG00000021807 | 2700060E02Rik | RIKEN cDNA 2700060E02 gene | MGI:1915295 | YES | YES | YES | Nucleus | other | ||||
| ENSMUSG00000031983 | 2310022B05Rik | RIKEN cDNA 2310022B05 gene | MGI:1916801 | YES | YES | YES | Cytoplasm | other | ||||
| ENSMUSG00000032666 | 1700025G04Rik | RIKEN cDNA 1700025G04 gene | MGI:1916649 | YES | YES | YES | Other | other | ||||
| ENSMUSG00000087651 | 1500009L16Rik | RIKEN cDNA 1500009L16 gene | MGI:1917034 | YES | YES | YES | Other | other |
Supplementary Table 3b. IPA cell location and molecular function categories in zebrafish SYN and PSD proteins.
Cell location and molecular function of mouse SYN and PSD proteins as described by the Ingenuity knowledgebase (www.qiagen.com/ingenuity). Proteins found in synaptosomes (SYN) and/or PSDs are indicated.
| ENSEMBL ID | Associated Gene Name | Description | EntrezGene ID | SYN ONLY | PSD ONLY | EQUAL ABUNDANCE | PSD Depeleted | PSD enriched | PSD component | SYN Components | Cell location | Protein type |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ENSDARG00000104264 | novel gene | 0 | 777704 | YES | YES | YES | Other | other | ||||
| ENSDARG00000075133 | adgrb1a | adhesion G protein-coupled receptor B1a [Source:ZFIN;Acc:ZDB-GENE-060607-12] | 563527 | YES | YES | YES | Plasma Membrane | G-protein coupled receptor | ||||
| ENSDARG00000075899 | adgrl2b.1 | adhesion G protein-coupled receptor L2b, tandem duplicate 1 [Source:ZFIN;Acc:ZDB-GENE-130116-4] | 101882597 | YES | YES | YES | Plasma Membrane | G-protein coupled receptor | ||||
| ENSDARG00000062780 | AIFM3 | apoptosis-inducing factor, mitochondrion-associated, 3 [Source:HGNC Symbol;Acc:HGNC:26398] | 100150876 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000018903 | aimp2 | aminoacyl tRNA synthetase complex-interacting multifunctional protein 2 [Source:ZFIN;Acc:ZDB-GENE-040426-2652] | 406647 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000079610 | akap9 | A kinase (PRKA) anchor protein 9 [Source:ZFIN;Acc:ZDB-GENE-030131-7276] | 0 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000076829 | ankib1b | ankyrin repeat and IBR domain containing 1b [Source:ZFIN;Acc:ZDB-GENE-100920-6] | 561277 | YES | YES | YES | Nucleus | transcription regulator | ||||
| ENSDARG00000096454 | ap1m2 | adaptor-related protein complex 1, mu 2 subunit [Source:ZFIN;Acc:ZDB-GENE-041114-20] | 403021 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSDARG00000056803 | ap1s1 | adaptor-related protein complex 1, sigma 1 subunit [Source:ZFIN;Acc:ZDB-GENE-040426-1119] | 393279 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSDARG00000059472 | arhgap31 | Rho GTPase activating protein 31 [Source:ZFIN;Acc:ZDB-GENE-100922-256] | 101885883 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000105115 | ASIC1 (2 of 2) | acid sensing (proton gated) ion channel 1 [Source:HGNC Symbol;Acc:HGNC:100] | 0 | YES | YES | YES | Plasma Membrane | ion channel | ||||
| ENSDARG00000029931 | atp13a1 | ATPase type 13A1 [Source:ZFIN;Acc:ZDB-GENE-040426-2804] | 334466 | YES | YES | YES | Extracellular Space | transporter | ||||
| ENSDARG00000104139 | atp1a3b | ATPase, Na+/K+ transporting, alpha 3b polypeptide [Source:ZFIN;Acc:ZDB-GENE-001212-8] | 64611 | YES | YES | YES | Plasma Membrane | transporter | ||||
| ENSDARG00000008735 | atp6ap2 | ATPase, H+ transporting, lysosomal accessory protein 2 [Source:ZFIN;Acc:ZDB-GENE-040426-1960] | 406296 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSDARG00000062521 | atp9b | ATPase, class II, type 9B [Source:ZFIN;Acc:ZDB-GENE-060503-583] | 568160 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSDARG00000076701 | b4galnt4b | beta-1,4-N-acetyl-galactosaminyl transferase 4b [Source:ZFIN;Acc:ZDB-GENE-100422-5] | 559305 | YES | YES | YES | Extracellular Space | enzyme | ||||
| ENSDARG00000061421 | BRSK2 (1 of 2) | BR serine/threonine kinase 2 [Source:HGNC Symbol;Acc:HGNC:11405] | 0 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSDARG00000029307 | C24H18orf8 | chromosome 18 open reading frame 8 [Source:HGNC Symbol;Acc:HGNC:24326] | 0 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000036888 | C25HXorf38 (1 of 2) | chromosome X open reading frame 38 [Source:HGNC Symbol;Acc:HGNC:28589] | 777611 | YES | YES | YES | Other | other | ||||
| ENSDARG00000094590 | c2cd5 | C2 calcium-dependent domain containing 5 [Source:ZFIN;Acc:ZDB-GENE-041114-193] | 100333813 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000061660 | CABZ01055347.1 | Uncharacterized protein [Source:UniProtKB/TrEMBL;Acc:E7F427] | 0 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000055565 | cacnb2b | calcium channel, voltage-dependent, beta 2b [Source:ZFIN;Acc:ZDB-GENE-050208-129] | 0 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000045832 | cart2 | cocaine- and amphetamine-regulated transcript 2 [Source:ZFIN;Acc:ZDB-GENE-050417-23] | 550232 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000052379 | ccdc22 | coiled-coil domain containing 22 [Source:ZFIN;Acc:ZDB-GENE-050913-93] | 556259 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000100052 | cckb | cholecystokinin b [Source:ZFIN;Acc:ZDB-GENE-060720-30] | 100000095 | YES | YES | YES | Extracellular Space | other | ||||
| ENSDARG00000056559 | ccz1 | CCZ1 vacuolar protein trafficking and biogenesis associated homolog (S. cerevisiae) [Source:ZFIN;Acc:ZDB-GENE-040426-698] | 393103 | YES | YES | YES | Other | other | ||||
| ENSDARG00000014522 | cdh6 | cadherin 6 [Source:ZFIN;Acc:ZDB-GENE-050320-92] | 541393 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000055133 | cenpf | centromere protein F [Source:ZFIN;Acc:ZDB-GENE-041111-205] | 497403 | YES | YES | YES | Nucleus | other | ||||
| ENSDARG00000006128 | cep170aa | centrosomal protein 170Aa [Source:ZFIN;Acc:ZDB-GENE-090312-109] | 557463 | YES | YES | YES | Nucleus | other | ||||
| ENSDARG00000055607 | cgna | cingulin a [Source:ZFIN;Acc:ZDB-GENE-100921-78] | 561423 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000099624 | chmp1b | chromatin modifying protein 1B [Source:ZFIN;Acc:ZDB-GENE-030131-8370] | 336426 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000044341 | chst7 | carbohydrate (N-acetylglucosamine 6-O) sulfotransferase 7 [Source:ZFIN;Acc:ZDB-GENE-131121-150] | 563748 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000039929 | ckmt2b | creatine kinase, mitochondrial 2b (sarcomeric) [Source:ZFIN;Acc:ZDB-GENE-040426-1654] | 799492 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSDARG00000020345 | clasp2 | cytoplasmic linker associated protein 2 [Source:ZFIN;Acc:ZDB-GENE-040426-2343] | 404604 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000013647 | cntn3a.2 | contactin 3a, tandem duplicate 2 [Source:ZFIN;Acc:ZDB-GENE-090312-57] | 100000688 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000074558 | cntnap2b | contactin associated protein-like 2b [Source:ZFIN;Acc:ZDB-GENE-070912-26] | 563345 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000077343 | CT573264.1 | Uncharacterized protein [Source:UniProtKB/TrEMBL;Acc:F1QPB9] | 100536351 | YES | YES | YES | Other | other | ||||
| ENSDARG00000044062 | ctbp2a | C-terminal binding protein 2a [Source:ZFIN;Acc:ZDB-GENE-010130-2] | 65230 | YES | YES | YES | Nucleus | transcription regulator | ||||
| ENSDARG00000024785 | ctnna2 | catenin (cadherin-associated protein), alpha 2 [Source:ZFIN;Acc:ZDB-GENE-060815-3] | 567500 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000061865 | ddx10 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 10 [Source:ZFIN;Acc:ZDB-GENE-071022-2] | 569118 | YES | YES | YES | Nucleus | enzyme | ||||
| ENSDARG00000006225 | ddx39aa | DEAD (Asp-Glu-Ala-Asp) box polypeptide 39Aa [Source:ZFIN;Acc:ZDB-GENE-030131-4275] | 325550 | YES | YES | YES | Nucleus | enzyme | ||||
| ENSDARG00000013236 | dhcr24 | 24-dehydrocholesterol reductase [Source:ZFIN;Acc:ZDB-GENE-041212-73] | 494102 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000005350 | dip2ba | DIP2 disco-interacting protein 2 homolog Ba (Drosophila) [Source:ZFIN;Acc:ZDB-GENE-040426-2305] | 405899 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000100115 | dnah1 | dynein, axonemal, heavy chain 1 [Source:ZFIN;Acc:ZDB-GENE-081015-3] | 100317899 | YES | YES | YES | Extracellular Space | other | ||||
| ENSDARG00000007723 | efnb1 | ephrin-B1 [Source:ZFIN;Acc:ZDB-GENE-010618-2] | 114377 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000014793 | ehd1b | EH-domain containing 1b [Source:ZFIN;Acc:ZDB-GENE-030131-2069] | 447839 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000068289 | eif3k | eukaryotic translation initiation factor 3, subunit K [Source:ZFIN;Acc:ZDB-GENE-050417-45] | 550245 | YES | YES | YES | Cytoplasm | translation regulator | ||||
| ENSDARG00000038695 | elavl1 | ELAV like RNA binding protein 1 [Source:ZFIN;Acc:ZDB-GENE-990415-245] | 30736 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000039268 | emc4 | ER membrane protein complex subunit 4 [Source:ZFIN;Acc:ZDB-GENE-040426-1891] | 0 | YES | YES | YES | Other | other | ||||
| ENSDARG00000035282 | emc6 | ER membrane protein complex subunit 6 [Source:ZFIN;Acc:ZDB-GENE-030131-9164] | 337220 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000088117 | FUT8 (2 of 2) | fucosyltransferase 8 (alpha (1,6) fucosyltransferase) [Source:HGNC Symbol;Acc:HGNC:4019] | 100331714 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000023771 | gabrb3 | gamma-aminobutyric acid (GABA) A receptor, beta 3 [Source:ZFIN;Acc:ZDB-GENE-101102-2] | 566922 | YES | YES | YES | Plasma Membrane | ion channel | ||||
| ENSDARG00000025301 | gfap | glial fibrillary acidic protein [Source:ZFIN;Acc:ZDB-GENE-990914-3] | 30646 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000044015 | gfra2b | GDNF family receptor alpha 2b [Source:ZFIN;Acc:ZDB-GENE-091204-26] | 564916 | YES | YES | YES | Plasma Membrane | transmembrane receptor | ||||
| ENSDARG00000068981 | glceb | glucuronic acid epimerase b [Source:ZFIN;Acc:ZDB-GENE-040630-8] | 100007670 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000092948 | gna15.3 | guanine nucleotide binding protein (G protein), alpha 15 (Gq class), tandem duplicate 3 [Source:ZFIN;Acc:ZDB-GENE-050208-700] | 0 | YES | YES | YES | Plasma Membrane | enzyme | ||||
| ENSDARG00000062511 | golga3 | golgin A3 [Source:ZFIN;Acc:ZDB-GENE-060526-252] | 570879 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSDARG00000036295 | golga7ba | golgin A7 family, member Ba [Source:ZFIN;Acc:ZDB-GENE-050417-14] | 550228 | YES | YES | YES | Other | other | ||||
| ENSDARG00000021352 | gria1a | glutamate receptor, ionotropic, AMPA 1a [Source:ZFIN;Acc:ZDB-GENE-020125-1] | 798689 | YES | YES | YES | Plasma Membrane | ion channel | ||||
| ENSDARG00000032714 | gria1b | glutamate receptor, ionotropic, AMPA 1b [Source:ZFIN;Acc:ZDB-GENE-020125-2] | 403044 | YES | YES | YES | Plasma Membrane | ion channel | ||||
| ENSDARG00000063253 | hecw2b | HECT, C2 and WW domain containing E3 ubiquitin protein ligase 2b [Source:ZFIN;Acc:ZDB-GENE-101001-3] | 100331355 | YES | YES | YES | Extracellular Space | enzyme | ||||
| ENSDARG00000056725 | hmgb3a | high mobility group box 3a [Source:ZFIN;Acc:ZDB-GENE-050428-1] | 561025 | YES | YES | YES | Nucleus | other | ||||
| ENSDARG00000009978 | icn | ictacalcin [Source:ZFIN;Acc:ZDB-GENE-030131-8599] | 336655 | YES | YES | YES | Other | transporter | ||||
| ENSDARG00000014591 | ilf2 | interleukin enhancer binding factor 2 [Source:ZFIN;Acc:ZDB-GENE-040426-2345] | 406517 | YES | YES | YES | Nucleus | transcription regulator | ||||
| ENSDARG00000076923 | kazna | kazrin, periplakin interacting protein a [Source:ZFIN;Acc:ZDB-GENE-081105-97] | 100003663 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000090815 | KCNJ10 (1 of 3) | potassium channel, inwardly rectifying subfamily J, member 10 [Source:HGNC Symbol;Acc:HGNC:6256] | 100538267 | YES | YES | YES | Plasma Membrane | ion channel | ||||
| ENSDARG00000060854 | kctd3 | potassium channel tetramerization domain containing 3 [Source:ZFIN;Acc:ZDB-GENE-060503-169] | 569113 | YES | YES | YES | Other | ion channel | ||||
| ENSDARG00000017162 | KIF3C (1 of 2) | kinesin family member 3C [Source:HGNC Symbol;Acc:HGNC:6321] | 100331925 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000094526 | ksr2 | kinase suppressor of ras 2 [Source:ZFIN;Acc:ZDB-GENE-030131-8047] | 569127 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSDARG00000045023 | lhfpl5a | lipoma HMGIC fusion partner-like 5a [Source:ZFIN;Acc:ZDB-GENE-110131-8] | 558325 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000071230 | lrfn5a | leucine rich repeat and fibronectin type III domain containing 5a [Source:ZFIN;Acc:ZDB-GENE-080327-17] | 100144400 | YES | YES | YES | Nucleus | other | ||||
| ENSDARG00000102684 | LRRC1 | leucine rich repeat containing 1 [Source:HGNC Symbol;Acc:HGNC:14307] | 0 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000003020 | lrrc4.2 | leucine rich repeat containing 4.2 [Source:ZFIN;Acc:ZDB-GENE-030131-7997] | 566572 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000074308 | lrrc75ba | leucine rich repeat containing 75Ba [Source:ZFIN;Acc:ZDB-GENE-081104-225] | 101885211 | YES | YES | YES | Other | other | ||||
| ENSDARG00000060805 | map1sa | microtubule-associated protein 1Sa [Source:ZFIN;Acc:ZDB-GENE-030131-3982] | 562808 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000099184 | map2k6 | mitogen-activated protein kinase kinase 6 [Source:ZFIN;Acc:ZDB-GENE-010202-3] | 0 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSDARG00000019345 | mark3a | MAP/microtubule affinity-regulating kinase 3a [Source:ZFIN;Acc:ZDB-GENE-030131-6232] | 334300 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSDARG00000056603 | mcf2b | MCF.2 cell line derived transforming sequence b [Source:ZFIN;Acc:ZDB-GENE-091118-4] | 559907 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000035360 | mink1 | misshapen-like kinase 1 [Source:ZFIN;Acc:ZDB-GENE-060526-45] | 100038799 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSDARG00000090462 | mrpl20 | mitochondrial ribosomal protein L20 [Source:ZFIN;Acc:ZDB-GENE-030131-5299] | 751640 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000068305 | mrps14 | mitochondrial ribosomal protein S14 [Source:ZFIN;Acc:ZDB-GENE-060825-75] | 751639 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000104111 | mrps30 | mitochondrial ribosomal protein S30 [Source:ZFIN;Acc:ZDB-GENE-050809-4] | 555760 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000057855 | mrps31 | mitochondrial ribosomal protein S31 [Source:ZFIN;Acc:ZDB-GENE-030131-3715] | 407642 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000063911 | mt-atp6 | ATP synthase 6, mitochondrial [Source:ZFIN;Acc:ZDB-GENE-011205-18] | 140519 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSDARG00000063899 | mt-nd2 | NADH dehydrogenase 2, mitochondrial [Source:ZFIN;Acc:ZDB-GENE-011205-8] | 140532 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000060853 | nckap1 | NCK-associated protein 1 [Source:ZFIN;Acc:ZDB-GENE-030131-4426] | 561894 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000010953 | ndel1a | nudE neurodevelopment protein 1-like 1a [Source:ZFIN;Acc:ZDB-GENE-040115-2] | 394244 | YES | YES | YES | Nucleus | other | ||||
| ENSDARG00000103937 | ndrg4 | NDRG family member 4 [Source:ZFIN;Acc:ZDB-GENE-060512-226] | 692305 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000087456 | NDUFB1 | NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 1, 7kDa [Source:HGNC Symbol;Acc:HGNC:7695] | 100535765 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000043467 | ndufb11 | NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 11 [Source:ZFIN;Acc:ZDB-GENE-050309-25] | 562219 | YES | YES | YES | Extracellular Space | other | ||||
| ENSDARG00000069878 | nhp2l1a | NHP2 non-histone chromosome protein 2-like 1a (S. cerevisiae) [Source:ZFIN;Acc:ZDB-GENE-040426-879] | 393282 | YES | YES | YES | Other | other | ||||
| ENSDARG00000023299 | nhp2l1b | NHP2 non-histone chromosome protein 2-like 1b (S. cerevisiae) [Source:ZFIN;Acc:ZDB-GENE-030131-9670] | 321114 | YES | YES | YES | Other | other | ||||
| ENSDARG00000103791 | nme2b.1 | NME/NM23 nucleoside diphosphate kinase 2b, tandem duplicate 1 [Source:ZFIN;Acc:ZDB-GENE-000210-32] | 30083 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000014973 | ntng1a | netrin g1a [Source:ZFIN;Acc:ZDB-GENE-091118-13] | 0 | YES | YES | YES | Extracellular Space | other | ||||
| ENSDARG00000077228 | ntrk3a | neurotrophic tyrosine kinase, receptor, type 3a [Source:ZFIN;Acc:ZDB-GENE-010126-3] | 568668 | YES | YES | YES | Plasma Membrane | kinase | ||||
| ENSDARG00000010078 | nup133 | nucleoporin 133 [Source:ZFIN;Acc:ZDB-GENE-040426-2941] | 406852 | YES | YES | YES | Nucleus | transporter | ||||
| ENSDARG00000030154 | pak7 | p21 protein (Cdc42/Rac)-activated kinase 7 [Source:ZFIN;Acc:ZDB-GENE-040426-2085] | 0 | YES | YES | YES | Nucleus | kinase | ||||
| ENSDARG00000003961 | parp3 | poly (ADP-ribose) polymerase family, member 3 [Source:ZFIN;Acc:ZDB-GENE-030131-7435] | 335495 | YES | YES | YES | Nucleus | enzyme | ||||
| ENSDARG00000098880 | pclob | piccolo presynaptic cytomatrix protein b [Source:ZFIN;Acc:ZDB-GENE-030131-8100] | 0 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSDARG00000038315 | pdxp | pyridoxal (pyridoxine, vitamin B6) phosphatase [Source:ZFIN;Acc:ZDB-GENE-080723-69] | 561030 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000105159 | phka1 | phosphorylase kinase, alpha 1 (muscle) [Source:ZFIN;Acc:ZDB-GENE-031118-56] | 572183 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSDARG00000099730 | pkma | pyruvate kinase, muscle, a [Source:ZFIN;Acc:ZDB-GENE-031201-4] | 335817 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSDARG00000100789 | plgrkt | plasminogen receptor, C-terminal lysine transmembrane protein [Source:ZFIN;Acc:ZDB-GENE-050809-125] | 606662 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000043527 | ppapdc2 | phosphatidic acid phosphatase type 2 domain containing 2 [Source:ZFIN;Acc:ZDB-GENE-040724-247] | 553687 | YES | YES | YES | Other | phosphatase | ||||
| ENSDARG00000075793 | prex1 | phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 1 [Source:ZFIN;Acc:ZDB-GENE-030131-1428] | 570984 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000015236 | pycrl | pyrroline-5-carboxylate reductase-like [Source:ZFIN;Acc:ZDB-GENE-041014-244] | 560682 | YES | YES | YES | Other | enzyme | ||||
| ENSDARG00000077174 | QSOX2 | quiescin Q6 sulfhydryl oxidase 2 [Source:HGNC Symbol;Acc:HGNC:30249] | 559603 | YES | YES | YES | Other | enzyme | ||||
| ENSDARG00000022129 | rbm4.3 | RNA binding motif protein 4.3 [Source:ZFIN;Acc:ZDB-GENE-040426-1938] | 406277 | YES | YES | YES | Nucleus | other | ||||
| ENSDARG00000038859 | rgs20 | regulator of G-protein signaling 20 [Source:ZFIN;Acc:ZDB-GENE-040718-128] | 436704 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000045156 | rgs9b | regulator of G-protein signaling 9b [Source:ZFIN;Acc:ZDB-GENE-040426-1708] | 793087 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000026845 | rhoaa | ras homolog gene family, member Aa [Source:ZFIN;Acc:ZDB-GENE-040426-2150] | 406411 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000025953 | rhoq | ras homolog family member Q [Source:ZFIN;Acc:ZDB-GENE-030131-5721] | 327510 | YES | YES | YES | Plasma Membrane | enzyme | ||||
| ENSDARG00000104101 | RIMBP2 (2 of 2) | RIMS binding protein 2 [Source:HGNC Symbol;Acc:HGNC:30339] | 100535071 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000025850 | rps21 | ribosomal protein S21 [Source:ZFIN;Acc:ZDB-GENE-040426-1102] | 394166 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000002591 | ruvbl1 | RuvB-like AAA ATPase 1 [Source:ZFIN;Acc:ZDB-GENE-030109-2] | 317679 | YES | YES | YES | Nucleus | transcription regulator | ||||
| ENSDARG00000055639 | ruvbl2 | RuvB-like AAA ATPase 2 [Source:ZFIN;Acc:ZDB-GENE-030109-1] | 317678 | YES | YES | YES | Nucleus | transcription regulator | ||||
| ENSDARG00000058102 | sardh | sarcosine dehydrogenase [Source:ZFIN;Acc:ZDB-GENE-040426-996] | 394103 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000104375 | sec24a | SEC24 family, member A (S. cerevisiae) [Source:ZFIN;Acc:ZDB-GENE-120926-1] | 100000526 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSDARG00000060539 | shank1 | SH3 and multiple ankyrin repeat domains 1 [Source:ZFIN;Acc:ZDB-GENE-130530-691] | 568908 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000062462 | shisa7a | shisa family member 7a [Source:ZFIN;Acc:ZDB-GENE-100922-37] | 0 | YES | YES | YES | Other | other | ||||
| ENSDARG00000088276 | si:ch211-190p8.2 | si:ch211-190p8.2 [Source:ZFIN;Acc:ZDB-GENE-070424-145] | 436997 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000095005 | si:dkey-21e2.10 | si:dkey-21e2.10 [Source:ZFIN;Acc:ZDB-GENE-050208-778] | 556860 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000061719 | si:dkey-23p11.2 | si:dkey-23p11.2 [Source:ZFIN;Acc:ZDB-GENE-100427-4] | 565752 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000076381 | slc25a1b | slc25a1 solute carrier family 25 (mitochondrial carrier; citrate transporter), member 1b [Source:ZFIN;Acc:ZDB-GENE-130114-1] | 795332 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000074533 | slc25a38b | solute carrier family 25, member 38b [Source:ZFIN;Acc:ZDB-GENE-110214-1] | 571961 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000077946 | smarcc2 | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 2 [Source:ZFIN;Acc:ZDB-GENE-100728-1] | 567476 | YES | YES | YES | Nucleus | transcription regulator | ||||
| ENSDARG00000078485 | snpha | syntaphilin a [Source:ZFIN;Acc:ZDB-GENE-081104-227] | 0 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000077349 | SORCS3 (1 of 2) | sortilin-related VPS10 domain containing receptor 3 [Source:HGNC Symbol;Acc:HGNC:16699] | 0 | YES | YES | YES | Nucleus | transporter | ||||
| ENSDARG00000035699 | srp19 | signal recognition particle 19 [Source:ZFIN;Acc:ZDB-GENE-040426-2657] | 406652 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000027734 | srsf5b | serine/arginine-rich splicing factor 5b [Source:ZFIN;Acc:ZDB-GENE-040718-354] | 436883 | YES | YES | YES | Nucleus | other | ||||
| ENSDARG00000079949 | supt16h | suppressor of Ty 16 homolog (S. cerevisiae) [Source:ZFIN;Acc:ZDB-GENE-031118-96] | 562348 | YES | YES | YES | Nucleus | transcription regulator | ||||
| ENSDARG00000031890 | tcp11l1 | t-complex 11, testis-specific-like 1 [Source:ZFIN;Acc:ZDB-GENE-050327-11] | 541488 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000071594 | TIMM21 | translocase of inner mitochondrial membrane 21 homolog (yeast) [Source:HGNC Symbol;Acc:HGNC:25010] | 560369 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000008765 | tmed5 | transmembrane emp24 protein transport domain containing 5 [Source:ZFIN;Acc:ZDB-GENE-040426-1302] | 393374 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000069494 | tmem223 | transmembrane protein 223 [Source:ZFIN;Acc:ZDB-GENE-040801-113] | 445200 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000063149 | tmtc1 | transmembrane and tetratricopeptide repeat containing 1 [Source:ZFIN;Acc:ZDB-GENE-070705-219] | 555449 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000039005 | trappc1 | trafficking protein particle complex 1 [Source:ZFIN;Acc:ZDB-GENE-040718-242] | 437015 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000097660 | trappc2 | trafficking protein particle complex 2 [Source:ZFIN;Acc:ZDB-GENE-060929-1266] | 767808 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000026068 | trappc5 | trafficking protein particle complex 5 [Source:ZFIN;Acc:ZDB-GENE-040718-185] | 436755 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000069420 | trim23 | tripartite motif containing 23 [Source:ZFIN;Acc:ZDB-GENE-060929-804] | 767706 | YES | YES | YES | Nucleus | enzyme | ||||
| ENSDARG00000003754 | tspan2a | tetraspanin 2a [Source:ZFIN;Acc:ZDB-GENE-050522-511] | 378854 | YES | YES | YES | Extracellular Space | other | ||||
| ENSDARG00000043026 | TTBK2 (2 of 2) | tau tubulin kinase 2 [Source:HGNC Symbol;Acc:HGNC:19141] | 569881 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSDARG00000042708 | tuba8l | tubulin, alpha 8 like [Source:ZFIN;Acc:ZDB-GENE-030131-9167] | 337223 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000013079 | tubgcp2 | tubulin, gamma complex associated protein 2 [Source:ZFIN;Acc:ZDB-GENE-031118-150] | 386975 | YES | YES | YES | Cytoplasm | peptidase | ||||
| ENSDARG00000040455 | unc50 | unc-50 homolog (C. elegans) [Source:ZFIN;Acc:ZDB-GENE-040426-907] | 393217 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000042418 | wbscr17 | Williams-Beuren syndrome chromosome region 17 [Source:ZFIN;Acc:ZDB-GENE-110419-5] | 567794 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000062310 | WDR26 (2 of 2) | WD repeat domain 26 [Source:HGNC Symbol;Acc:HGNC:21208] | 559461 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000060312 | wdr62 | WD repeat domain 62 [Source:ZFIN;Acc:ZDB-GENE-060503-291] | 570949 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000039392 | wnk1b | WNK lysine deficient protein kinase 1b [Source:ZFIN;Acc:ZDB-GENE-030131-2656] | 561159 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSDARG00000026333 | xkr7 | XK, Kell blood group complex subunit-related family, member 7 [Source:ZFIN;Acc:ZDB-GENE-051113-336] | 497073 | YES | YES | YES | Other | other | ||||
| ENSDARG00000038308 | ykt6 | YKT6 v-SNARE homolog (S. cerevisiae) [Source:ZFIN;Acc:ZDB-GENE-040426-733] | 394067 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000075758 | ywhabb | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, beta polypeptide b [Source:ZFIN;Acc:ZDB-GENE-040426-2160] | 406419 | YES | YES | YES | Cytoplasm | transcription regulator | ||||
| ENSDARG00000069529 | zgc:153981 | zgc:153981 [Source:ZFIN;Acc:ZDB-GENE-061103-367] | 563986 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000092873 | zgc:193541 | zgc:193541 [Source:ZFIN;Acc:ZDB-GENE-081022-15] | 557217 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000075733 | zyx | zyxin [Source:ZFIN;Acc:ZDB-GENE-070928-18] | 564246 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000033201 | novel gene | 0 | 449795 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000053454 | novel gene | 0 | 556251 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000068624 | novel gene | 0 | 100003123 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000077124 | novel gene | 0 | 564701 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSDARG00000078770 | novel gene | 0 | 336430 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000102845 | novel gene | 0 | 0 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSDARG00000104615 | novel gene | 0 | 0 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSDARG00000092644 | ago1 | argonaute RISC catalytic component 1 [Source:ZFIN;Acc:ZDB-GENE-110606-3] | 570775 | YES | YES | YES | Cytoplasm | translation regulator | ||||
| ENSDARG00000027590 | sept2 | septin 2 [Source:ZFIN;Acc:ZDB-GENE-030131-911] | 323457 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000099236 | sept10 | septin 10 [Source:ZFIN;Acc:ZDB-GENE-050417-3] | 550129 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000019191 | sept12 | septin 12 [Source:ZFIN;Acc:ZDB-GENE-120215-68] | 564600 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000099664 | sep15 | selenoprotein 15 [Source:ZFIN;Acc:ZDB-GENE-030327-1] | 352923 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000102889 | sept15 | septin 15 [Source:ZFIN;Acc:ZDB-GENE-061103-265] | 792259 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000011855 | aak1a | AP2 associated kinase 1a [Source:ZFIN;Acc:ZDB-GENE-081105-101] | 568999 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSDARG00000074876 | abcd1 | ATP-binding cassette, sub-family D (ALD), member 1 [Source:ZFIN;Acc:ZDB-GENE-050517-27] | 566367 | YES | YES | YES | Plasma Membrane | transporter | ||||
| ENSDARG00000059409 | ABCD2 (1 of 2) | ATP-binding cassette, sub-family D (ALD), member 2 [Source:HGNC Symbol;Acc:HGNC:66] | 100537809 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSDARG00000007216 | abce1 | ATP-binding cassette, sub-family E (OABP), member 1 [Source:ZFIN;Acc:ZDB-GENE-040426-1995] | 406324 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSDARG00000014875 | abi2b | abl-interactor 2b [Source:ZFIN;Acc:ZDB-GENE-060519-5] | 692331 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000060072 | abi3a | ABI family, member 3a [Source:ZFIN;Acc:ZDB-GENE-060929-958] | 767807 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000063283 | abi3b | ABI family, member 3b [Source:ZFIN;Acc:ZDB-GENE-121214-259] | 566059 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000013841 | abl2 | c-abl oncogene 2, non-receptor tyrosine kinase [Source:ZFIN;Acc:ZDB-GENE-020809-2] | 570636 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSDARG00000045799 | acanb | aggrecan b [Source:ZFIN;Acc:ZDB-GENE-100422-16] | 559593 | YES | YES | YES | Extracellular Space | other | ||||
| ENSDARG00000034883 | acbd5a | acyl-CoA binding domain containing 5a [Source:ZFIN;Acc:ZDB-GENE-050522-268] | 553674 | YES | YES | YES | Nucleus | other | ||||
| ENSDARG00000031796 | ache | acetylcholinesterase [Source:ZFIN;Acc:ZDB-GENE-010906-1] | 114549 | YES | YES | YES | Plasma Membrane | enzyme | ||||
| ENSDARG00000058352 | acot9.1 | acyl-CoA thioesterase 9, tandem duplicate 1 [Source:ZFIN;Acc:ZDB-GENE-040724-139] | 798073 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000020149 | acoxl | acyl-CoA oxidase-like [Source:ZFIN;Acc:ZDB-GENE-081107-18] | 569104 | YES | YES | YES | Other | enzyme | ||||
| ENSDARG00000007219 | actn1 | actinin, alpha 1 [Source:ZFIN;Acc:ZDB-GENE-081113-4] | 560400 | YES | YES | YES | Cytoplasm | transcription regulator | ||||
| ENSDARG00000099786 | ACTN1 (2 of 3) | actinin, alpha 1 [Source:HGNC Symbol;Acc:HGNC:163] | 0 | YES | YES | YES | Cytoplasm | transcription regulator | ||||
| ENSDARG00000011611 | actr1 | ARP1 actin-related protein 1, centractin (yeast) [Source:ZFIN;Acc:ZDB-GENE-040426-2695] | 406681 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000079204 | adam11 | ADAM metallopeptidase domain 11 [Source:ZFIN;Acc:ZDB-GENE-070808-3] | 567145 | YES | YES | YES | Plasma Membrane | peptidase | ||||
| ENSDARG00000060532 | adam22 | ADAM metallopeptidase domain 22 [Source:ZFIN;Acc:ZDB-GENE-070810-1] | 100126805 | YES | YES | YES | Plasma Membrane | peptidase | ||||
| ENSDARG00000062323 | adam23a | ADAM metallopeptidase domain 23a [Source:ZFIN;Acc:ZDB-GENE-070810-2] | 794175 | YES | YES | YES | Plasma Membrane | peptidase | ||||
| ENSDARG00000003544 | adarb1b | adenosine deaminase, RNA-specific, B1b [Source:ZFIN;Acc:ZDB-GENE-030219-15] | 558759 | YES | YES | YES | Nucleus | enzyme | ||||
| ENSDARG00000062561 | adck1 | aarF domain containing kinase 1 [Source:ZFIN;Acc:ZDB-GENE-080401-5] | 100144553 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSDARG00000069026 | adcy1a | adenylate cyclase 1a [Source:ZFIN;Acc:ZDB-GENE-070705-302] | 557026 | YES | YES | YES | Plasma Membrane | enzyme | ||||
| ENSDARG00000088634 | adcy1b | adenylate cyclase 1b [Source:ZFIN;Acc:ZDB-GENE-100805-1] | 0 | YES | YES | YES | Plasma Membrane | enzyme | ||||
| ENSDARG00000077145 | ADCY3 | adenylate cyclase 3 [Source:HGNC Symbol;Acc:HGNC:234] | 571825 | YES | YES | YES | Plasma Membrane | enzyme | ||||
| ENSDARG00000103962 | add1 | adducin 1 (alpha) [Source:ZFIN;Acc:ZDB-GENE-030909-2] | 555511 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000074581 | add2 | adducin 2 (beta) [Source:ZFIN;Acc:ZDB-GENE-080718-4] | 570147 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000040874 | add3a | adducin 3 (gamma) a [Source:ZFIN;Acc:ZDB-GENE-030131-2721] | 556762 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000056250 | add3b | adducin 3 (gamma) b [Source:ZFIN;Acc:ZDB-GENE-080718-6] | 553243 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000078529 | adgrb1b | adhesion G protein-coupled receptor B1b [Source:ZFIN;Acc:ZDB-GENE-081031-45] | 0 | YES | YES | YES | Plasma Membrane | G-protein coupled receptor | ||||
| ENSDARG00000059832 | adgrb3 | adhesion G protein-coupled receptor B3 [Source:ZFIN;Acc:ZDB-GENE-070424-99] | 0 | YES | YES | YES | Plasma Membrane | G-protein coupled receptor | ||||
| ENSDARG00000089292 | adgrl1a | adhesion G protein-coupled receptor L1a [Source:ZFIN;Acc:ZDB-GENE-131127-267] | 0 | YES | YES | YES | Plasma Membrane | G-protein coupled receptor | ||||
| ENSDARG00000099874 | agap2 | ArfGAP with GTPase domain, ankyrin repeat and PH domain 2 [Source:ZFIN;Acc:ZDB-GENE-061103-343] | 565109 | YES | YES | YES | Nucleus | enzyme | ||||
| ENSDARG00000042821 | agps | alkylglycerone phosphate synthase [Source:ZFIN;Acc:ZDB-GENE-031118-14] | 386801 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000079388 | agrn | agrin [Source:ZFIN;Acc:ZDB-GENE-030131-1033] | 565373 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000061764 | ahnak | AHNAK nucleoprotein [Source:ZFIN;Acc:ZDB-GENE-030131-8719] | 559276 | YES | YES | YES | Nucleus | other | ||||
| ENSDARG00000060036 | aimp1 | aminoacyl tRNA synthetase complex-interacting multifunctional protein 1 [Source:ZFIN;Acc:ZDB-GENE-060825-144] | 562232 | YES | YES | YES | Extracellular Space | cytokine | ||||
| ENSDARG00000006546 | ak4 | adenylate kinase 4 [Source:ZFIN;Acc:ZDB-GENE-040426-2505] | 406590 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSDARG00000012555 | ak5 | adenylate kinase 5 [Source:ZFIN;Acc:ZDB-GENE-030131-8256] | 100003595 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSDARG00000037935 | aldh16a1 | aldehyde dehydrogenase 16 family, member A1 [Source:ZFIN;Acc:ZDB-GENE-070112-2062] | 492710 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000099579 | aldh18a1 | aldehyde dehydrogenase 18 family, member A1 [Source:ZFIN;Acc:ZDB-GENE-030131-5602] | 557186 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSDARG00000028259 | aldh3a2a | aldehyde dehydrogenase 3 family, member A2a [Source:ZFIN;Acc:ZDB-GENE-040718-74] | 323653 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000011665 | aldoaa | aldolase a, fructose-bisphosphate, a [Source:ZFIN;Acc:ZDB-GENE-030131-8369] | 336425 | YES | YES | YES | Other | other | ||||
| ENSDARG00000034470 | aldoab | aldolase a, fructose-bisphosphate, b [Source:ZFIN;Acc:ZDB-GENE-040426-2299] | 406496 | YES | YES | YES | Other | other | ||||
| ENSDARG00000057661 | aldoca | aldolase C, fructose-bisphosphate, a [Source:ZFIN;Acc:ZDB-GENE-050706-128] | 792692 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000009026 | ANK2 (1 of 2) | ankyrin 2, neuronal [Source:HGNC Symbol;Acc:HGNC:493] | 568926 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000043313 | ank2b | ankyrin 2b, neuronal [Source:ZFIN;Acc:ZDB-GENE-041010-165] | 450043 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000061736 | ank3a | ankyrin 3a [Source:ZFIN;Acc:ZDB-GENE-060621-1] | 103909035 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000077582 | ank3b | ankyrin 3b [Source:ZFIN;Acc:ZDB-GENE-060621-2] | 100126016 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000020621 | ap1b1 | adaptor-related protein complex 1, beta 1 subunit [Source:ZFIN;Acc:ZDB-GENE-061025-1] | 562166 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSDARG00000102394 | ap1g1 | adaptor-related protein complex 1, gamma 1 subunit [Source:ZFIN;Acc:ZDB-GENE-030131-3187] | 324466 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSDARG00000054337 | ap1g2 | adaptor-related protein complex 1, gamma 2 subunit [Source:ZFIN;Acc:ZDB-GENE-030616-587] | 767712 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSDARG00000096249 | ap1m1 | adaptor-related protein complex 1, mu 1 subunit [Source:ZFIN;Acc:ZDB-GENE-110126-1] | 100330411 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSDARG00000054320 | ap2a1 | adaptor-related protein complex 2, alpha 1 subunit [Source:ZFIN;Acc:ZDB-GENE-090302-2] | 558153 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSDARG00000035152 | ap2b1 | adaptor-related protein complex 2, beta 1 subunit [Source:ZFIN;Acc:ZDB-GENE-030131-6564] | 334632 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000033899 | ap2m1b | adaptor-related protein complex 2, mu 1 subunit, b [Source:ZFIN;Acc:ZDB-GENE-040426-1103] | 394001 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSDARG00000016128 | ap3m2 | adaptor-related protein complex 3, mu 2 subunit [Source:ZFIN;Acc:ZDB-GENE-021022-2] | 415244 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSDARG00000058868 | apc | adenomatous polyposis coli [Source:ZFIN;Acc:ZDB-GENE-031112-7] | 386762 | YES | YES | YES | Nucleus | enzyme | ||||
| ENSDARG00000060734 | appl1 | adaptor protein, phosphotyrosine interaction, PH domain and leucine zipper containing 1 [Source:ZFIN;Acc:ZDB-GENE-081016-1] | 571540 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000078139 | appl2 | adaptor protein, phosphotyrosine interaction, PH domain and leucine zipper containing 2 [Source:ZFIN;Acc:ZDB-GENE-081016-2] | 100000135 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000031214 | arcn1b | archain 1b [Source:ZFIN;Acc:ZDB-GENE-040121-7] | 399661 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000075673 | arhgap21b | Rho GTPase activating protein 21b [Source:ZFIN;Acc:ZDB-GENE-030131-4944] | 326745 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000052950 | arhgap23a | Rho GTPase activating protein 23a [Source:ZFIN;Acc:ZDB-GENE-121019-1] | 100006233 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000104549 | arhgap44 | Rho GTPase activating protein 44 [Source:ZFIN;Acc:ZDB-GENE-120206-2] | 0 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000040380 | arhgef1a | Rho guanine nucleotide exchange factor (GEF) 1a [Source:ZFIN;Acc:ZDB-GENE-030722-5] | 368266 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000006299 | arhgef7a | Rho guanine nucleotide exchange factor (GEF) 7a [Source:ZFIN;Acc:ZDB-GENE-060322-11] | 553493 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000008383 | arpc1a | actin related protein 2/3 complex, subunit 1A [Source:ZFIN;Acc:ZDB-GENE-040116-1] | 394243 | YES | YES | YES | Extracellular Space | other | ||||
| ENSDARG00000075989 | arpc2 | actin related protein 2/3 complex, subunit 2 [Source:ZFIN;Acc:ZDB-GENE-030131-5276] | 327068 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000039142 | arpc5a | actin related protein 2/3 complex, subunit 5A [Source:ZFIN;Acc:ZDB-GENE-030131-8448] | 336504 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000019062 | arpc5b | actin related protein 2/3 complex, subunit 5B [Source:ZFIN;Acc:ZDB-GENE-040120-7] | 399482 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000035433 | arpc5lb | actin related protein 2/3 complex, subunit 5-like, b [Source:ZFIN;Acc:ZDB-GENE-060531-161] | 556104 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000061688 | arvcfb | armadillo repeat gene deleted in velocardiofacial syndrome b [Source:ZFIN;Acc:ZDB-GENE-060526-60] | 564669 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000008329 | asic1a | acid-sensing (proton-gated) ion channel 1a [Source:ZFIN;Acc:ZDB-GENE-040513-2] | 791696 | YES | YES | YES | Plasma Membrane | ion channel | ||||
| ENSDARG00000078703 | ASPH | aspartate beta-hydroxylase [Source:HGNC Symbol;Acc:HGNC:757] | 559236 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000059629 | asphd2 | aspartate beta-hydroxylase domain containing 2 [Source:ZFIN;Acc:ZDB-GENE-061103-142] | 569819 | YES | YES | YES | Other | enzyme | ||||
| ENSDARG00000068323 | astn1 | astrotactin 1 [Source:ZFIN;Acc:ZDB-GENE-050320-140] | 791157 | YES | YES | YES | Other | other | ||||
| ENSDARG00000023267 | atad1a | ATPase family, AAA domain containing 1a [Source:ZFIN;Acc:ZDB-GENE-030616-593] | 368672 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000041417 | atp6ap1a | ATPase, H+ transporting, lysosomal accessory protein 1a [Source:ZFIN;Acc:ZDB-GENE-090312-136] | 100004566 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000037153 | atp6ap1b | ATPase, H+ transporting, lysosomal accessory protein 1b [Source:ZFIN;Acc:ZDB-GENE-020423-2] | 195824 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000015174 | atp6v0a1b | ATPase, H+ transporting, lysosomal V0 subunit a1b [Source:ZFIN;Acc:ZDB-GENE-050522-215] | 553691 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSDARG00000035538 | atp6v0a2a | ATPase, H+ transporting, lysosomal V0 subunit a2a [Source:ZFIN;Acc:ZDB-GENE-091113-4] | 561469 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSDARG00000057853 | atp6v0ca | ATPase, H+ transporting, lysosomal, V0 subunit ca [Source:ZFIN;Acc:ZDB-GENE-020419-23] | 192336 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSDARG00000069090 | atp6v0d1 | ATPase, H+ transporting, lysosomal V0 subunit d1 [Source:ZFIN;Acc:ZDB-GENE-030131-1531] | 322811 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSDARG00000043465 | atp6v1b2 | ATPase, H+ transporting, lysosomal, V1 subunit B2 [Source:ZFIN;Acc:ZDB-GENE-030711-4] | 359840 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSDARG00000013443 | atp6v1ba | ATPase, H+ transporting, lysosomal, V1 subunit B, member a [Source:ZFIN;Acc:ZDB-GENE-030711-3] | 359839 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000023967 | atp6v1c1a | ATPase, H+ transporting, lysosomal, V1 subunit C1a [Source:ZFIN;Acc:ZDB-GENE-030616-612] | 368907 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSDARG00000011175 | atp6v1d | ATPase, H+ transporting, lysosomal V1 subunit D [Source:ZFIN;Acc:ZDB-GENE-040426-727] | 393935 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSDARG00000030694 | atp6v1e1b | ATPase, H+ transporting, lysosomal, V1 subunit E1b [Source:ZFIN;Acc:ZDB-GENE-020419-11] | 192335 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSDARG00000045543 | atp6v1f | ATPase, H+ transporting, lysosomal, V1 subunit F [Source:ZFIN;Acc:ZDB-GENE-040718-259] | 436799 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000022315 | atp6v1g1 | ATPase, H+ transporting, lysosomal, V1 subunit G1 [Source:ZFIN;Acc:ZDB-GENE-030131-6965] | 335025 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSDARG00000006370 | atp6v1h | ATPase, H+ transporting, lysosomal V1 subunit H [Source:ZFIN;Acc:ZDB-GENE-021219-2] | 286779 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSDARG00000001939 | b3gat3 | beta-1,3-glucuronyltransferase 3 (glucuronosyltransferase I) [Source:ZFIN;Acc:ZDB-GENE-020419-3] | 192334 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000017316 | bag5 | BCL2-associated athanogene 5 [Source:ZFIN;Acc:ZDB-GENE-050913-58] | 569580 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000053358 | basp1 | brain abundant, membrane attached signal protein 1 [Source:ZFIN;Acc:ZDB-GENE-070424-158] | 569631 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000044972 | bcap31 | B-cell receptor-associated protein 31 [Source:ZFIN;Acc:ZDB-GENE-030616-63] | 368488 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSDARG00000010108 | bri3bp | bri3 binding protein [Source:ZFIN;Acc:ZDB-GENE-030131-5254] | 327046 | YES | YES | YES | Extracellular Space | other | ||||
| ENSDARG00000078302 | BRINP1 | bone morphogenetic protein/retinoic acid inducible neural-specific 1 [Source:HGNC Symbol;Acc:HGNC:2687] | 100334375 | YES | YES | YES | Nucleus | peptidase | ||||
| ENSDARG00000014302 | brinp2 | bone morphogenetic protein/retinoic acid inducible neural-specific 2 [Source:ZFIN;Acc:ZDB-GENE-070912-621] | 561009 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000090615 | BX957297.1 | Uncharacterized protein [Source:UniProtKB/TrEMBL;Acc:E7F2M5] | 567192 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000089562 | BX957322.1 | Uncharacterized protein [Source:UniProtKB/TrEMBL;Acc:E7EYF4] | 100006797 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000040582 | C18H3orf33 | chromosome 3 open reading frame 33 [Source:HGNC Symbol;Acc:HGNC:26434] | 565816 | YES | YES | YES | Extracellular Space | other | ||||
| ENSDARG00000039887 | c1qbp | complement component 1, q subcomponent binding protein [Source:ZFIN;Acc:ZDB-GENE-050417-408] | 334619 | YES | YES | YES | Cytoplasm | transcription regulator | ||||
| ENSDARG00000061066 | c2cd2l | c2cd2-like [Source:ZFIN;Acc:ZDB-GENE-041019-1] | 566488 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000061257 | C3H17orf75 | chromosome 17 open reading frame 75 [Source:HGNC Symbol;Acc:HGNC:30173] | 751673 | YES | YES | YES | Extracellular Space | other | ||||
| ENSDARG00000043589 | ca4a | carbonic anhydrase IV a [Source:ZFIN;Acc:ZDB-GENE-080204-85] | 555196 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000073822 | CABZ01046283.1 | Uncharacterized protein [Source:UniProtKB/TrEMBL;Acc:F1QCJ4] | 0 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000025074 | CABZ01059390.1 | Uncharacterized protein [Source:UniProtKB/TrEMBL;Acc:E7F238] | 0 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSDARG00000102813 | CABZ01069436.1 | 0 | 101883099 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000104937 | CABZ01084323.1 | 0 | 100330629 | YES | YES | YES | Plasma Membrane | phosphatase | ||||
| ENSDARG00000079295 | cacna1bb | calcium channel, voltage-dependent, N type, alpha 1B subunit, b [Source:ZFIN;Acc:ZDB-GENE-090514-4] | 562796 | YES | YES | YES | Plasma Membrane | ion channel | ||||
| ENSDARG00000014804 | cacna2d1a | calcium channel, voltage-dependent, alpha 2/delta subunit 1a [Source:ZFIN;Acc:ZDB-GENE-041210-215] | 561434 | YES | YES | YES | Plasma Membrane | ion channel | ||||
| ENSDARG00000039182 | cacna2d2a | calcium channel, voltage-dependent, alpha 2/delta subunit 2a [Source:ZFIN;Acc:ZDB-GENE-050320-71] | 541376 | YES | YES | YES | Plasma Membrane | ion channel | ||||
| ENSDARG00000078169 | cacna2d2b | calcium channel, voltage-dependent, alpha 2/delta subunit 2b [Source:ZFIN;Acc:ZDB-GENE-060503-99] | 568759 | YES | YES | YES | Plasma Membrane | ion channel | ||||
| ENSDARG00000078760 | cacna2d3 | calcium channel, voltage dependent, alpha2/delta subunit 3 [Source:ZFIN;Acc:ZDB-GENE-030616-133] | 100148645 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000002167 | cacnb1 | calcium channel, voltage-dependent, beta 1 subunit [Source:ZFIN;Acc:ZDB-GENE-040718-399] | 436925 | YES | YES | YES | Other | ion channel | ||||
| ENSDARG00000076401 | cacng3b | calcium channel, voltage-dependent, gamma subunit 3b [Source:ZFIN;Acc:ZDB-GENE-090828-1] | 564883 | YES | YES | YES | Plasma Membrane | ion channel | ||||
| ENSDARG00000015050 | calm2b | calmodulin 2b, (phosphorylase kinase, delta) [Source:ZFIN;Acc:ZDB-GENE-020415-2] | 192322 | YES | YES | YES | Nucleus | other | ||||
| ENSDARG00000053617 | camk2a | calcium/calmodulin-dependent protein kinase II alpha [Source:ZFIN;Acc:ZDB-GENE-051113-72] | 550436 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSDARG00000011065 | camk2b1 | calcium/calmodulin-dependent protein kinase (CaM kinase) II beta 1 [Source:ZFIN;Acc:ZDB-GENE-030131-9588] | 562064 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSDARG00000043010 | camk2d1 | calcium/calmodulin-dependent protein kinase (CaM kinase) II delta 1 [Source:ZFIN;Acc:ZDB-GENE-040801-121] | 445208 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000027963 | camkva | CaM kinase-like vesicle-associated a [Source:ZFIN;Acc:ZDB-GENE-100819-2] | 567243 | YES | YES | YES | Other | kinase | ||||
| ENSDARG00000057000 | camkvl | CaM kinase-like vesicle-associated, like [Source:ZFIN;Acc:ZDB-GENE-090311-58] | 553431 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000062173 | camsap2a | calmodulin regulated spectrin-associated protein family, member 2a [Source:ZFIN;Acc:ZDB-GENE-030131-3016] | 562741 | YES | YES | YES | Other | other | ||||
| ENSDARG00000034240 | capza1a | capping protein (actin filament) muscle Z-line, alpha 1a [Source:ZFIN;Acc:ZDB-GENE-030131-5237] | 327029 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000060829 | car15 | carbonic anhydrase 15 [Source:ZFIN;Acc:ZDB-GENE-091204-152] | 568143 | YES | YES | YES | Extracellular Space | enzyme | ||||
| ENSDARG00000036164 | cars | cysteinyl-tRNA synthetase [Source:ZFIN;Acc:ZDB-GENE-030328-29] | 368284 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000074490 | casc4 | cancer susceptibility candidate 4 [Source:ZFIN;Acc:ZDB-GENE-091130-1] | 555654 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000059809 | caska | calcium/calmodulin-dependent serine protein kinase a [Source:ZFIN;Acc:ZDB-GENE-020802-4] | 259195 | YES | YES | YES | Plasma Membrane | kinase | ||||
| ENSDARG00000100119 | caskb | calcium/calmodulin-dependent serine protein kinase b [Source:ZFIN;Acc:ZDB-GENE-030131-4473] | 0 | YES | YES | YES | Plasma Membrane | kinase | ||||
| ENSDARG00000046107 | CASKIN1 (1 of 2) | CASK interacting protein 1 [Source:HGNC Symbol;Acc:HGNC:20879] | 563678 | YES | YES | YES | Nucleus | transcription regulator | ||||
| ENSDARG00000008982 | casq2 | calsequestrin 2 [Source:ZFIN;Acc:ZDB-GENE-010724-12] | 114410 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000063677 | ccny | cyclin Y [Source:ZFIN;Acc:ZDB-GENE-091117-45] | 557738 | YES | YES | YES | Nucleus | other | ||||
| ENSDARG00000016464 | CDC42BPA (1 of 2) | CDC42 binding protein kinase alpha (DMPK-like) [Source:HGNC Symbol;Acc:HGNC:1737] | 566003 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSDARG00000019383 | cdc42bpb | CDC42 binding protein kinase beta (DMPK-like) [Source:ZFIN;Acc:ZDB-GENE-030131-3647] | 567039 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSDARG00000055843 | cdh10a | cadherin 10, type 2a (T2-cadherin) [Source:ZFIN;Acc:ZDB-GENE-060614-1] | 568370 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000021442 | cdh11 | cadherin 11, type 2, OB-cadherin (osteoblast) [Source:ZFIN;Acc:ZDB-GENE-980526-170] | 30461 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000018693 | cdh2 | cadherin 2, type 1, N-cadherin (neuronal) [Source:ZFIN;Acc:ZDB-GENE-990415-171] | 30291 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000077996 | cdh24b | cadherin 24, type 2b [Source:ZFIN;Acc:ZDB-GENE-081104-50] | 0 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000015002 | cdh4 | cadherin 4, type 1, R-cadherin (retinal) [Source:ZFIN;Acc:ZDB-GENE-991207-1] | 30297 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000023542 | cdh7 | cadherin 7, type 2 [Source:ZFIN;Acc:ZDB-GENE-061019-3] | 567704 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000037478 | cdh8 | cadherin 8 [Source:ZFIN;Acc:ZDB-GENE-130530-597] | 0 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000056683 | cdk5 | cyclin-dependent kinase 5 [Source:ZFIN;Acc:ZDB-GENE-010131-2] | 65234 | YES | YES | YES | Nucleus | kinase | ||||
| ENSDARG00000045087 | cdk5r1b | cyclin-dependent kinase 5, regulatory subunit 1b (p35) [Source:ZFIN;Acc:ZDB-GENE-040718-220] | 436788 | YES | YES | YES | Nucleus | kinase | ||||
| ENSDARG00000058892 | cep170ab | centrosomal protein 170Ab [Source:ZFIN;Acc:ZDB-GENE-050309-106] | 566358 | YES | YES | YES | Nucleus | other | ||||
| ENSDARG00000101510 | cetn2 | centrin, EF-hand protein, 2 [Source:ZFIN;Acc:ZDB-GENE-091118-94] | 100006257 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000032919 | chchd3a | coiled-coil-helix-coiled-coil-helix domain containing 3a [Source:ZFIN;Acc:ZDB-GENE-030131-5005] | 407730 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000016255 | chmp4ba | charged multivesicular body protein 4Ba [Source:ZFIN;Acc:ZDB-GENE-040426-906] | 393164 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000102024 | chmp6b | charged multivesicular body protein 6b [Source:ZFIN;Acc:ZDB-GENE-040924-2] | 445397 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000103987 | cib1 | calcium and integrin binding 1 (calmyrin) [Source:ZFIN;Acc:ZDB-GENE-050417-170] | 100536377 | YES | YES | YES | Nucleus | other | ||||
| ENSDARG00000013351 | cirbpb | cold inducible RNA binding protein b [Source:ZFIN;Acc:ZDB-GENE-030131-5841] | 678563 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000052703 | cisd2 | CDGSH iron sulfur domain 2 [Source:ZFIN;Acc:ZDB-GENE-040426-1381] | 393354 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000102674 | ckap5 | cytoskeleton associated protein 5 [Source:ZFIN;Acc:ZDB-GENE-051120-174] | 492614 | YES | YES | YES | Nucleus | transcription regulator | ||||
| ENSDARG00000016598 | ckmt1 | creatine kinase, mitochondrial 1 [Source:ZFIN;Acc:ZDB-GENE-030131-611] | 321892 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSDARG00000010280 | clasp1a | cytoplasmic linker associated protein 1a [Source:ZFIN;Acc:ZDB-GENE-081104-520] | 562077 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000032577 | clcn6 | chloride channel 6 [Source:ZFIN;Acc:ZDB-GENE-030131-2056] | 568122 | YES | YES | YES | Plasma Membrane | ion channel | ||||
| ENSDARG00000042357 | cldnk | claudin k [Source:ZFIN;Acc:ZDB-GENE-040801-201] | 445070 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000059596 | clip2 | CAP-GLY domain containing linker protein 2 [Source:ZFIN;Acc:ZDB-GENE-100727-3] | 556406 | YES | YES | YES | Cytoplasm | transcription regulator | ||||
| ENSDARG00000054456 | clip3 | CAP-GLY domain containing linker protein 3 [Source:ZFIN;Acc:ZDB-GENE-131127-193] | 562450 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000096867 | clmn | calmin [Source:ZFIN;Acc:ZDB-GENE-131127-546] | 100330495 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000021048 | clptm1l | CLPTM1-like [Source:ZFIN;Acc:ZDB-GENE-040718-75] | 436653 | YES | YES | YES | Other | other | ||||
| ENSDARG00000052020 | cmtm4 | CKLF-like MARVEL transmembrane domain containing 4 [Source:ZFIN;Acc:ZDB-GENE-040718-195] | 436765 | YES | YES | YES | Extracellular Space | cytokine | ||||
| ENSDARG00000004174 | cnot1 | CCR4-NOT transcription complex, subunit 1 [Source:ZFIN;Acc:ZDB-GENE-040915-1] | 448949 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000045685 | cntn1b | contactin 1b [Source:ZFIN;Acc:ZDB-GENE-041210-236] | 541474 | YES | YES | YES | Plasma Membrane | enzyme | ||||
| ENSDARG00000000472 | cntn2 | contactin 2 [Source:ZFIN;Acc:ZDB-GENE-990630-12] | 0 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000098161 | cntn4 | contactin 4 [Source:ZFIN;Acc:ZDB-GENE-060929-776] | 571349 | YES | YES | YES | Plasma Membrane | enzyme | ||||
| ENSDARG00000087843 | CNTN4 (2 of 3) | contactin 4 [Source:HGNC Symbol;Acc:HGNC:2174] | 0 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000021584 | cntn5 | contactin 5 [Source:ZFIN;Acc:ZDB-GENE-030131-5640] | 327429 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000074524 | cntnap1 | contactin associated protein 1 [Source:ZFIN;Acc:ZDB-GENE-030916-2] | 566220 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000078322 | col12a1a | collagen, type XII, alpha 1a [Source:ZFIN;Acc:ZDB-GENE-090728-1] | 100149543 | YES | YES | YES | Extracellular Space | other | ||||
| ENSDARG00000054610 | coro1a | coronin, actin binding protein, 1A [Source:ZFIN;Acc:ZDB-GENE-030131-9512] | 100002113 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000008660 | coro1b | coronin, actin binding protein, 1B [Source:ZFIN;Acc:ZDB-GENE-040123-3] | 560168 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000035598 | coro1ca | coronin, actin binding protein, 1Ca [Source:ZFIN;Acc:ZDB-GENE-030114-6] | 317741 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000021193 | coro1cb | coronin, actin binding protein, 1Cb [Source:ZFIN;Acc:ZDB-GENE-030131-418] | 562849 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000039136 | cox16 | COX16 cytochrome c oxidase assembly homolog (S. cerevisiae) [Source:ZFIN;Acc:ZDB-GENE-050417-60] | 550257 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000032970 | cox4i1 | cytochrome c oxidase subunit IV isoform 1 [Source:ZFIN;Acc:ZDB-GENE-030131-5175] | 326975 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000012388 | cox4i1l | cytochrome c oxidase subunit IV isoform 1, like [Source:ZFIN;Acc:ZDB-GENE-130814-2] | 402880 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000022509 | cox4i2 | cytochrome c oxidase subunit IV isoform 2 [Source:ZFIN;Acc:ZDB-GENE-040426-1775] | 393776 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000088383 | cox5aa | cytochrome c oxidase subunit Vaa [Source:ZFIN;Acc:ZDB-GENE-050522-133] | 554101 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000022438 | cox6a1 | cytochrome c oxidase subunit VIa polypeptide 1 [Source:ZFIN;Acc:ZDB-GENE-030131-7715] | 335774 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000038577 | cox6c | cytochrome c oxidase subunit VIc [Source:ZFIN;Acc:ZDB-GENE-040724-95] | 494577 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000053217 | cox7a2a | cytochrome c oxidase, subunit VIIa 2a [Source:ZFIN;Acc:ZDB-GENE-050522-153] | 554103 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000054907 | cox7a3 | cytochrome c oxidase subunit VIIa polypeptide 3 [Source:ZFIN;Acc:ZDB-GENE-030131-7691] | 335751 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000007753 | cpne2 | copine II [Source:ZFIN;Acc:ZDB-GENE-030131-7896] | 565225 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000102584 | cpne7 | copine VII [Source:ZFIN;Acc:ZDB-GENE-131125-70] | 564556 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSDARG00000018618 | cpsf6 | cleavage and polyadenylation specific factor 6 [Source:ZFIN;Acc:ZDB-GENE-030131-5277] | 327069 | YES | YES | YES | Nucleus | other | ||||
| ENSDARG00000062054 | cpt1ab | carnitine palmitoyltransferase 1Ab (liver) [Source:ZFIN;Acc:ZDB-GENE-030131-3250] | 560000 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000086969 | CR774179.1 | Uncharacterized protein [Source:UniProtKB/TrEMBL;Acc:E7F3Q3] | 0 | YES | YES | YES | Other | other | ||||
| ENSDARG00000059826 | crtac1a | cartilage acidic protein 1a [Source:ZFIN;Acc:ZDB-GENE-030131-9704] | 559626 | YES | YES | YES | Extracellular Space | other | ||||
| ENSDARG00000052674 | csnk1a1 | casein kinase 1, alpha 1 [Source:ZFIN;Acc:ZDB-GENE-020419-19] | 192307 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSDARG00000012818 | csnk2a2a | casein kinase 2, alpha prime polypeptide a [Source:ZFIN;Acc:ZDB-GENE-990415-28] | 30488 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSDARG00000074376 | CT737192.1 | Uncharacterized protein [Source:UniProtKB/TrEMBL;Acc:E7FFL5] | 0 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000057007 | ctbp1 | C-terminal binding protein 1 [Source:ZFIN;Acc:ZDB-GENE-010130-1] | 65229 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000102441 | ctnna1 | catenin (cadherin-associated protein), alpha 1 [Source:ZFIN;Acc:ZDB-GENE-991207-24] | 30741 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000014571 | ctnnb1 | catenin (cadherin-associated protein), beta 1 [Source:ZFIN;Acc:ZDB-GENE-980526-362] | 30265 | YES | YES | YES | Nucleus | transcription regulator | ||||
| ENSDARG00000023472 | ctnnb2 | catenin, beta 2 [Source:ZFIN;Acc:ZDB-GENE-040426-2575] | 0 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000078233 | ctnnd1 | catenin (cadherin-associated protein), delta 1 [Source:ZFIN;Acc:ZDB-GENE-110208-9] | 556726 | YES | YES | YES | Nucleus | other | ||||
| ENSDARG00000062415 | ctnnd2a | catenin (cadherin-associated protein), delta 2a [Source:ZFIN;Acc:ZDB-GENE-030616-60] | 368485 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000003779 | ctnnd2b | catenin (cadherin-associated protein), delta 2b [Source:ZFIN;Acc:ZDB-GENE-070912-532] | 558069 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000011777 | cttn | cortactin [Source:ZFIN;Acc:ZDB-GENE-040715-3] | 0 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000040991 | cttnbp2 | cortactin binding protein 2 [Source:ZFIN;Acc:ZDB-GENE-030131-8134] | 555837 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000019239 | cul1a | cullin 1a [Source:ZFIN;Acc:ZDB-GENE-030131-2603] | 323883 | YES | YES | YES | Nucleus | enzyme | ||||
| ENSDARG00000038967 | cul3a | cullin 3a [Source:ZFIN;Acc:ZDB-GENE-030131-3376] | 324655 | YES | YES | YES | Nucleus | enzyme | ||||
| ENSDARG00000069742 | cul5a | cullin 5a [Source:ZFIN;Acc:ZDB-GENE-030131-5426] | 327215 | YES | YES | YES | Nucleus | ion channel | ||||
| ENSDARG00000043134 | cux1b | cut-like homeobox 1b [Source:ZFIN;Acc:ZDB-GENE-040801-260] | 445120 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000038075 | cyc1 | cytochrome c-1 [Source:ZFIN;Acc:ZDB-GENE-031105-2] | 564379 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000044345 | cyfip1 | cytoplasmic FMR1 interacting protein 1 [Source:ZFIN;Acc:ZDB-GENE-030131-8557] | 336613 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000036375 | cyfip2 | cytoplasmic FMR1 interacting protein 2 [Source:ZFIN;Acc:ZDB-GENE-080724-2] | 100002872 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000015059 | daam1a | dishevelled associated activator of morphogenesis 1a [Source:ZFIN;Acc:ZDB-GENE-030911-6] | 557451 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000009689 | daam1b | dishevelled associated activator of morphogenesis 1b [Source:ZFIN;Acc:ZDB-GENE-030131-4212] | 560060 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000028393 | daam2 | dishevelled associated activator of morphogenesis 2 [Source:ZFIN;Acc:ZDB-GENE-070709-1] | 0 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000069484 | dab2ipa | DAB2 interacting protein a [Source:ZFIN;Acc:ZDB-GENE-081104-516] | 100002454 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000075110 | dab2ipb | DAB2 interacting protein b [Source:ZFIN;Acc:ZDB-GENE-030131-2449] | 558237 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000062956 | dagla | diacylglycerol lipase, alpha [Source:ZFIN;Acc:ZDB-GENE-070619-1] | 569398 | YES | YES | YES | Plasma Membrane | enzyme | ||||
| ENSDARG00000070043 | dars | aspartyl-tRNA synthetase [Source:ZFIN;Acc:ZDB-GENE-061110-135] | 324025 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000069113 | dbn1 | drebrin 1 [Source:ZFIN;Acc:ZDB-GENE-091204-284] | 561153 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000016016 | dbnlb | drebrin-like b [Source:ZFIN;Acc:ZDB-GENE-050522-293] | 553729 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000101561 | dbt | dihydrolipoamide branched chain transacylase E2 [Source:ZFIN;Acc:ZDB-GENE-050320-85] | 541388 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000054355 | dcaf7 | ddb1 and cul4 associated factor 7 [Source:ZFIN;Acc:ZDB-GENE-030131-9511] | 337565 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000061638 | dcakd | dephospho-CoA kinase domain containing [Source:ZFIN;Acc:ZDB-GENE-060825-226] | 751733 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000104282 | DCC | DCC netrin 1 receptor [Source:HGNC Symbol;Acc:HGNC:2701] | 0 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000104664 | dckl1b | doublecortin-like kinase 1b [Source:ZFIN;Acc:ZDB-GENE-080516-7] | 100151759 | YES | YES | YES | Other | kinase | ||||
| ENSDARG00000019743 | dctn1a | dynactin 1a [Source:ZFIN;Acc:ZDB-GENE-070117-2205] | 407638 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000031388 | dctn2 | dynactin 2 (p50) [Source:ZFIN;Acc:ZDB-GENE-040426-1279] | 394141 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000041363 | dctn3 | dynactin 3 (p22) [Source:ZFIN;Acc:ZDB-GENE-040704-65] | 431767 | YES | YES | YES | Nucleus | other | ||||
| ENSDARG00000103083 | DDB1 (2 of 2) | damage-specific DNA binding protein 1, 127kDa [Source:HGNC Symbol;Acc:HGNC:2717] | 0 | YES | YES | YES | Nucleus | other | ||||
| ENSDARG00000037318 | ddost | dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit (non-catalytic) [Source:ZFIN;Acc:ZDB-GENE-040426-2147] | 406408 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000032117 | ddx1 | DEAD (Asp-Glu-Ala-Asp) box helicase 1 [Source:ZFIN;Acc:ZDB-GENE-041001-215] | 556790 | YES | YES | YES | Nucleus | enzyme | ||||
| ENSDARG00000020573 | ddx3 | DEAD (Asp-Glu-Ala-Asp) box helicase 3 [Source:ZFIN;Acc:ZDB-GENE-030131-1565] | 566947 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000061338 | ddx6 | DEAD (Asp-Glu-Ala-Asp) box helicase 6 [Source:ZFIN;Acc:ZDB-GENE-070912-83] | 564633 | YES | YES | YES | Nucleus | enzyme | ||||
| ENSDARG00000005897 | dera | deoxyribose-phosphate aldolase (putative) [Source:ZFIN;Acc:ZDB-GENE-041010-200] | 450077 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000062696 | DGKG | diacylglycerol kinase, gamma 90kDa [Source:HGNC Symbol;Acc:HGNC:2853] | 0 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSDARG00000063578 | DGKI | diacylglycerol kinase, iota [Source:HGNC Symbol;Acc:HGNC:2855] | 100331649 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSDARG00000079725 | dhx9 | DEAH (Asp-Glu-Ala-His) box helicase 9 [Source:ZFIN;Acc:ZDB-GENE-070912-171] | 568043 | YES | YES | YES | Nucleus | enzyme | ||||
| ENSDARG00000016348 | dip2bb | DIP2 disco-interacting protein 2 homolog Bb (Drosophila) [Source:ZFIN;Acc:ZDB-GENE-100729-4] | 568954 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000015918 | dlat | dihydrolipoamide S-acetyltransferase (E2 component of pyruvate dehydrogenase complex) [Source:ZFIN;Acc:ZDB-GENE-030131-2921] | 324201 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000008785 | dldh | dihydrolipoamide dehydrogenase [Source:ZFIN;Acc:ZDB-GENE-040120-4] | 399479 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000009677 | dlg1 | discs, large homolog 1 (Drosophila) [Source:ZFIN;Acc:ZDB-GENE-010724-8] | 114446 | YES | YES | YES | Plasma Membrane | kinase | ||||
| ENSDARG00000102216 | dlg1l | discs, large (Drosophila) homolog 1, like [Source:ZFIN;Acc:ZDB-GENE-050222-3] | 497648 | YES | YES | YES | Plasma Membrane | kinase | ||||
| ENSDARG00000014230 | dlst | dihydrolipoamide S-succinyltransferase [Source:ZFIN;Acc:ZDB-GENE-030326-1] | 368262 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000006585 | DMWD | dystrophia myotonica, WD repeat containing [Source:HGNC Symbol;Acc:HGNC:2936] | 571996 | YES | YES | YES | Nucleus | other | ||||
| ENSDARG00000098599 | DMXL1 | Dmx-like 1 [Source:HGNC Symbol;Acc:HGNC:2937] | 100333625 | YES | YES | YES | Extracellular Space | other | ||||
| ENSDARG00000091293 | dmxl2 | Dmx-like 2 [Source:ZFIN;Acc:ZDB-GENE-031007-6] | 378958 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000010745 | dnaja2l | DnaJ (Hsp40) homolog, subfamily A, member 2, like [Source:ZFIN;Acc:ZDB-GENE-030131-2884] | 324164 | YES | YES | YES | Nucleus | enzyme | ||||
| ENSDARG00000102295 | dnaja3b | DnaJ (Hsp40) homolog, subfamily A, member 3B [Source:ZFIN;Acc:ZDB-GENE-040115-3] | 394242 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000058644 | dnajb2 | DnaJ (Hsp40) homolog, subfamily B, member 2 [Source:ZFIN;Acc:ZDB-GENE-061013-537] | 561686 | YES | YES | YES | Nucleus | other | ||||
| ENSDARG00000038978 | dnajb4 | DnaJ (Hsp40) homolog, subfamily B, member 4 [Source:ZFIN;Acc:ZDB-GENE-040801-192] | 445061 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000074727 | dnajc10 | DnaJ (Hsp40) homolog, subfamily C, member 10 [Source:ZFIN;Acc:ZDB-GENE-070327-1] | 557858 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000011196 | dnajc11 | DnaJ (Hsp40) homolog, subfamily C, member 11 [Source:ZFIN;Acc:ZDB-GENE-030131-1673] | 322953 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000032238 | dnm3a | dynamin 3a [Source:ZFIN;Acc:ZDB-GENE-040724-76] | 557996 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000075327 | dock10 | dedicator of cytokinesis 10 [Source:ZFIN;Acc:ZDB-GENE-130530-773] | 100000949 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000063180 | dock3 | dedicator of cytokinesis 3 [Source:ZFIN;Acc:ZDB-GENE-100624-1] | 561387 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000076213 | DOCK4 (2 of 2) | dedicator of cytokinesis 4 [Source:HGNC Symbol;Acc:HGNC:19192] | 566115 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000078675 | dock7 | dedicator of cytokinesis 7 [Source:ZFIN;Acc:ZDB-GENE-050302-15] | 568817 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000055225 | dock8 | dedicator of cytokinesis 8 [Source:ZFIN;Acc:ZDB-GENE-060503-743] | 403040 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000055857 | dopey2 | dopey family member 2 [Source:ZFIN;Acc:ZDB-GENE-040426-1195] | 0 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000057011 | dpm1 | dolichyl-phosphate mannosyltransferase polypeptide 1, catalytic subunit [Source:ZFIN;Acc:ZDB-GENE-040801-115] | 445202 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000098057 | dscaml1 | Down syndrome cell adhesion molecule like 1 [Source:ZFIN;Acc:ZDB-GENE-110601-1] | 0 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000101858 | DST (3 of 4) | dystonin [Source:HGNC Symbol;Acc:HGNC:1090] | 0 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000013020 | dtnbb | dystrobrevin, beta b [Source:ZFIN;Acc:ZDB-GENE-070117-3] | 559786 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000078016 | dtx4 | deltex 4, E3 ubiquitin ligase [Source:ZFIN;Acc:ZDB-GENE-070410-129] | 100037355 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000001578 | echs1 | enoyl CoA hydratase, short chain, 1, mitochondrial [Source:ZFIN;Acc:ZDB-GENE-030616-617] | 368912 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000093413 | edil3 | EGF-like repeats and discoidin I-like domains 3 [Source:ZFIN;Acc:ZDB-GENE-060503-366] | 793444 | YES | YES | YES | Extracellular Space | other | ||||
| ENSDARG00000039502 | eef1a1a | eukaryotic translation elongation factor 1 alpha 1a [Source:ZFIN;Acc:ZDB-GENE-030131-8278] | 336334 | YES | YES | YES | Cytoplasm | translation regulator | ||||
| ENSDARG00000020850 | eef1a1l1 | eukaryotic translation elongation factor 1 alpha 1, like 1 [Source:ZFIN;Acc:ZDB-GENE-990415-52] | 30516 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000071774 | eef1e1 | eukaryotic translation elongation factor 1 epsilon 1 [Source:ZFIN;Acc:ZDB-GENE-030131-4949] | 565440 | YES | YES | YES | Cytoplasm | translation regulator | ||||
| ENSDARG00000056119 | eef1g | eukaryotic translation elongation factor 1 gamma [Source:ZFIN;Acc:ZDB-GENE-020423-3] | 195822 | YES | YES | YES | Cytoplasm | translation regulator | ||||
| ENSDARG00000035256 | eef2l2 | eukaryotic translation elongation factor 2, like 2 [Source:ZFIN;Acc:ZDB-GENE-030131-8112] | 336168 | YES | YES | YES | Cytoplasm | translation regulator | ||||
| ENSDARG00000033516 | efr3ba | EFR3 homolog Ba (S. cerevisiae) [Source:ZFIN;Acc:ZDB-GENE-030616-64] | 560858 | YES | YES | YES | Other | other | ||||
| ENSDARG00000069318 | efr3bb | EFR3 homolog Bb (S. cerevisiae) [Source:ZFIN;Acc:ZDB-GENE-041014-293] | 561353 | YES | YES | YES | Other | other | ||||
| ENSDARG00000070029 | ehhadh | enoyl-CoA, hydratase/3-hydroxyacyl CoA dehydrogenase [Source:ZFIN;Acc:ZDB-GENE-040426-2581] | 100000859 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000076815 | eif3s10 | eukaryotic translation initiation factor 3, subunit 10 (theta) [Source:ZFIN;Acc:ZDB-GENE-030131-5726] | 327515 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000101082 | eif3s6ip | eukaryotic translation initiation factor 3, subunit 6 interacting protein [Source:ZFIN;Acc:ZDB-GENE-040426-2138] | 406402 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000045639 | elavl4 | ELAV like neuron-specific RNA binding protein 4 [Source:ZFIN;Acc:ZDB-GENE-990415-246] | 30737 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000078141 | ELFN1 (2 of 2) | extracellular leucine-rich repeat and fibronectin type III domain containing 1 [Source:HGNC Symbol;Acc:HGNC:33154] | 562570 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000077609 | elfn2a | extracellular leucine-rich repeat and fibronectin type III domain containing 2a [Source:ZFIN;Acc:ZDB-GENE-080327-26] | 0 | YES | YES | YES | Extracellular Space | other | ||||
| ENSDARG00000098753 | elmo1 | engulfment and cell motility 1 (ced-12 homolog, C. elegans) [Source:ZFIN;Acc:ZDB-GENE-040426-2069] | 406364 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000063527 | elmo2 | engulfment and cell motility 2 [Source:EntrezGene;Acc:100006857] | 100006857 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000003308 | emc2 | ER membrane protein complex subunit 2 [Source:ZFIN;Acc:ZDB-GENE-040625-82] | 407073 | YES | YES | YES | Nucleus | other | ||||
| ENSDARG00000012144 | emc7 | ER membrane protein complex subunit 7 [Source:ZFIN;Acc:ZDB-GENE-041001-170] | 561549 | YES | YES | YES | Other | other | ||||
| ENSDARG00000040087 | epb41l4a | erythrocyte membrane protein band 4.1 like 4A [Source:ZFIN;Acc:ZDB-GENE-990415-20] | 30470 | YES | YES | YES | Extracellular Space | other | ||||
| ENSDARG00000060494 | eprs | glutamyl-prolyl-tRNA synthetase [Source:ZFIN;Acc:ZDB-GENE-030131-638] | 562037 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000078216 | eps15 | epidermal growth factor receptor pathway substrate 15 [Source:ZFIN;Acc:ZDB-GENE-081104-264] | 795505 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000042670 | eps15l1a | epidermal growth factor receptor pathway substrate 15-like 1a [Source:ZFIN;Acc:ZDB-GENE-060503-274] | 568170 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000013161 | ERAS | ES cell expressed Ras [Source:HGNC Symbol;Acc:HGNC:5174] | 393228 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000044281 | erbb2ip | erbb2 interacting protein [Source:ZFIN;Acc:ZDB-GENE-061103-403] | 566811 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000063207 | erbb4a | erb-b2 receptor tyrosine kinase 4a [Source:ZFIN;Acc:ZDB-GENE-030918-4] | 100007677 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000043976 | etf1b | eukaryotic translation termination factor 1b [Source:ZFIN;Acc:ZDB-GENE-021029-1] | 266989 | YES | YES | YES | Cytoplasm | translation regulator | ||||
| ENSDARG00000043019 | exoc1 | exocyst complex component 1 [Source:ZFIN;Acc:ZDB-GENE-030131-1057] | 322338 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSDARG00000055610 | exoc2 | exocyst complex component 2 [Source:ZFIN;Acc:ZDB-GENE-040426-1160] | 100004870 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSDARG00000014582 | exoc3 | exocyst complex component 3 [Source:ZFIN;Acc:ZDB-GENE-030131-5947] | 334015 | YES | YES | YES | Plasma Membrane | transporter | ||||
| ENSDARG00000004336 | exoc4 | exocyst complex component 4 [Source:ZFIN;Acc:ZDB-GENE-041210-112] | 559866 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSDARG00000060323 | exoc5 | exocyst complex component 5 [Source:ZFIN;Acc:ZDB-GENE-080204-126] | 559010 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000020521 | exoc6 | exocyst complex component 6 [Source:ZFIN;Acc:ZDB-GENE-040426-2511] | 406591 | YES | YES | YES | Plasma Membrane | transporter | ||||
| ENSDARG00000098874 | exoc6b | exocyst complex component 6B [Source:ZFIN;Acc:ZDB-GENE-060929-132] | 767763 | YES | YES | YES | Other | other | ||||
| ENSDARG00000087229 | exoc7 | exocyst complex component 7 [Source:ZFIN;Acc:ZDB-GENE-040426-2752] | 406723 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSDARG00000063211 | exoc8 | exocyst complex component 8 [Source:ZFIN;Acc:ZDB-GENE-070410-60] | 568029 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000056648 | ext2 | exostosin glycosyltransferase 2 [Source:ZFIN;Acc:ZDB-GENE-041124-3] | 493780 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000026811 | extl3 | exostosin-like glycosyltransferase 3 [Source:ZFIN;Acc:ZDB-GENE-041124-2] | 493783 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000086300 | FAM107A | family with sequence similarity 107, member A [Source:HGNC Symbol;Acc:HGNC:30827] | 794256 | YES | YES | YES | Nucleus | other | ||||
| ENSDARG00000019205 | fam120c | family with sequence similarity 120C [Source:ZFIN;Acc:ZDB-GENE-090313-228] | 557842 | YES | YES | YES | Other | other | ||||
| ENSDARG00000100400 | FAM126B (1 of 2) | family with sequence similarity 126, member B [Source:HGNC Symbol;Acc:HGNC:28593] | 0 | YES | YES | YES | Other | other | ||||
| ENSDARG00000070952 | fam131ba | family with sequence similarity 131, member Ba [Source:ZFIN;Acc:ZDB-GENE-060503-616] | 565996 | YES | YES | YES | Other | other | ||||
| ENSDARG00000070575 | fam131bb | family with sequence similarity 131, member Bb [Source:ZFIN;Acc:ZDB-GENE-100922-12] | 556725 | YES | YES | YES | Other | other | ||||
| ENSDARG00000008573 | fam20b | family with sequence similarity 20, member B (H. sapiens) [Source:ZFIN;Acc:ZDB-GENE-040724-125] | 557060 | YES | YES | YES | Extracellular Space | enzyme | ||||
| ENSDARG00000035985 | FAM210A (1 of 2) | family with sequence similarity 210, member A [Source:HGNC Symbol;Acc:HGNC:28346] | 562734 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000098860 | FAM57A | family with sequence similarity 57, member A [Source:HGNC Symbol;Acc:HGNC:29646] | 0 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000003320 | fam91a1 | family with sequence similarity 91, member A1 [Source:ZFIN;Acc:ZDB-GENE-040426-1187] | 393413 | YES | YES | YES | Other | other | ||||
| ENSDARG00000074381 | farp1 | FERM, RhoGEF (ARHGEF) and pleckstrin domain protein 1 (chondrocyte-derived) [Source:ZFIN;Acc:ZDB-GENE-070424-163] | 0 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000099306 | FBXO2 | F-box protein 2 [Source:HGNC Symbol;Acc:HGNC:13581] | 559776 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000076310 | FBXO41 | F-box protein 41 [Source:HGNC Symbol;Acc:HGNC:29409] | 101883316 | YES | YES | YES | Other | other | ||||
| ENSDARG00000059701 | flii | flightless I homolog (Drosophila) [Source:ZFIN;Acc:ZDB-GENE-071212-3] | 560281 | YES | YES | YES | Nucleus | other | ||||
| ENSDARG00000010764 | flj13639 | flj13639 [Source:ZFIN;Acc:ZDB-GENE-030131-8104] | 677747 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000098374 | FLNB | filamin B, beta [Source:HGNC Symbol;Acc:HGNC:3755] | 0 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000097351 | flnbl | filamin B, like [Source:ZFIN;Acc:ZDB-GENE-031112-6] | 100534866 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000001710 | flot1a | flotillin 1a [Source:ZFIN;Acc:ZDB-GENE-020430-1] | 561345 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000004830 | flot2a | flotillin 2a [Source:ZFIN;Acc:ZDB-GENE-020430-3] | 245698 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000069774 | flot2b | flotillin 2b [Source:ZFIN;Acc:ZDB-GENE-040426-1368] | 393612 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000019371 | flt1 | fms-related tyrosine kinase 1 (vascular endothelial growth factor/vascular permeability factor receptor) [Source:ZFIN;Acc:ZDB-GENE-050407-1] | 544667 | YES | YES | YES | Plasma Membrane | kinase | ||||
| ENSDARG00000061778 | fmn2b | formin 2b [Source:ZFIN;Acc:ZDB-GENE-080225-35] | 566432 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000012586 | fmnl2a | formin-like 2a [Source:ZFIN;Acc:ZDB-GENE-081105-70] | 100006456 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000075041 | fmnl2b | formin-like 2b [Source:ZFIN;Acc:ZDB-GENE-041111-182] | 492608 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000037433 | fmr1 | fragile X mental retardation 1 [Source:ZFIN;Acc:ZDB-GENE-020731-6] | 259191 | YES | YES | YES | Nucleus | other | ||||
| ENSDARG00000042249 | FRMD4A | FERM domain containing 4A [Source:HGNC Symbol;Acc:HGNC:25491] | 564713 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000056001 | fryb | furry homolog b (Drosophila) [Source:ZFIN;Acc:ZDB-GENE-080215-5] | 792949 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000040001 | fryl | furry homolog, like [Source:ZFIN;Acc:ZDB-GENE-040914-58] | 558173 | YES | YES | YES | Other | other | ||||
| ENSDARG00000040822 | fundc1 | FUN14 domain containing 1 [Source:ZFIN;Acc:ZDB-GENE-040718-466] | 436984 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000015449 | fut8a | fucosyltransferase 8a (alpha (1,6) fucosyltransferase) [Source:ZFIN;Acc:ZDB-GENE-031118-20] | 797512 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000011370 | fyna | FYN proto-oncogene, Src family tyrosine kinase a [Source:ZFIN;Acc:ZDB-GENE-030903-5] | 373872 | YES | YES | YES | Plasma Membrane | kinase | ||||
| ENSDARG00000025319 | fynb | FYN proto-oncogene, Src family tyrosine kinase b [Source:ZFIN;Acc:ZDB-GENE-050706-89] | 574422 | YES | YES | YES | Plasma Membrane | kinase | ||||
| ENSDARG00000061042 | gabbr2 | gamma-aminobutyric acid (GABA) B receptor, 2 [Source:ZFIN;Acc:ZDB-GENE-060503-620] | 560267 | YES | YES | YES | Plasma Membrane | G-protein coupled receptor | ||||
| ENSDARG00000068989 | gabra1 | gamma-aminobutyric acid (GABA) A receptor, alpha 1 [Source:ZFIN;Acc:ZDB-GENE-061013-194] | 768183 | YES | YES | YES | Plasma Membrane | ion channel | ||||
| ENSDARG00000099096 | gabrb4 | gamma-aminobutyric acid (GABA) A receptor, beta 4 [Source:ZFIN;Acc:ZDB-GENE-070424-211] | 566514 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000059763 | gabrd | gamma-aminobutyric acid (GABA) A receptor, delta [Source:ZFIN;Acc:ZDB-GENE-081105-170] | 571422 | YES | YES | YES | Plasma Membrane | ion channel | ||||
| ENSDARG00000090654 | gak | cyclin G associated kinase [Source:ZFIN;Acc:ZDB-GENE-041210-358] | 100151158 | YES | YES | YES | Nucleus | kinase | ||||
| ENSDARG00000003829 | galnt2 | UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 2 [Source:ZFIN;Acc:ZDB-GENE-041111-110] | 570248 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000006832 | galnt9 | polypeptide N-acetylgalactosaminyltransferase 9 [Source:ZFIN;Acc:ZDB-GENE-060526-21] | 797549 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000099744 | gap43 | growth associated protein 43 [Source:ZFIN;Acc:ZDB-GENE-990415-87] | 30608 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000037057 | gcdha | glutaryl-CoA dehydrogenase a [Source:ZFIN;Acc:ZDB-GENE-040426-1855] | 393860 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000042723 | gfra2a | GDNF family receptor alpha 2a [Source:ZFIN;Acc:ZDB-GENE-040630-1] | 405794 | YES | YES | YES | Plasma Membrane | transmembrane receptor | ||||
| ENSDARG00000055569 | ghdc | GH3 domain containing [Source:ZFIN;Acc:ZDB-GENE-050208-165] | 497449 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000104483 | gipc1 | GIPC PDZ domain containing family, member 1 [Source:ZFIN;Acc:ZDB-GENE-060726-1] | 445568 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000039489 | git1 | G protein-coupled receptor kinase interacting ArfGAP 1 [Source:ZFIN;Acc:ZDB-GENE-100408-5] | 568935 | YES | YES | YES | Nucleus | kinase | ||||
| ENSDARG00000010002 | gna11a | guanine nucleotide binding protein (G protein), alpha 11a (Gq class) [Source:ZFIN;Acc:ZDB-GENE-050208-597] | 563953 | YES | YES | YES | Plasma Membrane | enzyme | ||||
| ENSDARG00000018174 | gnai2a | guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 2a [Source:ZFIN;Acc:ZDB-GENE-030131-8365] | 336421 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000030644 | gnai3 | guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 3 [Source:ZFIN;Acc:ZDB-GENE-070713-6] | 100006888 | YES | YES | YES | Plasma Membrane | enzyme | ||||
| ENSDARG00000044760 | gnaia | guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide a [Source:ZFIN;Acc:ZDB-GENE-030131-2229] | 323509 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000045415 | gnal | guanine nucleotide binding protein (G protein), alpha activating activity polypeptide, olfactory type [Source:ZFIN;Acc:ZDB-GENE-041114-26] | 492467 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000016676 | gnao1a | guanine nucleotide binding protein (G protein), alpha activating activity polypeptide O, a [Source:ZFIN;Acc:ZDB-GENE-040426-1757] | 393760 | YES | YES | YES | Plasma Membrane | enzyme | ||||
| ENSDARG00000036058 | gnao1b | guanine nucleotide binding protein (G protein), alpha activating activity polypeptide O, b [Source:ZFIN;Acc:ZDB-GENE-040426-1693] | 393801 | YES | YES | YES | Plasma Membrane | enzyme | ||||
| ENSDARG00000043094 | gnas | GNAS complex locus [Source:ZFIN;Acc:ZDB-GENE-090417-2] | 557353 | YES | YES | YES | Plasma Membrane | enzyme | ||||
| ENSDARG00000099844 | GNAZ | guanine nucleotide binding protein (G protein), alpha z polypeptide [Source:HGNC Symbol;Acc:HGNC:4395] | 0 | YES | YES | YES | Plasma Membrane | enzyme | ||||
| ENSDARG00000035357 | gnb2 | guanine nucleotide binding protein (G protein), beta polypeptide 2 [Source:ZFIN;Acc:ZDB-GENE-050320-65] | 541370 | YES | YES | YES | Plasma Membrane | enzyme | ||||
| ENSDARG00000099685 | gnb5a | guanine nucleotide binding protein (G protein), beta 5a [Source:ZFIN;Acc:ZDB-GENE-070112-342] | 562813 | YES | YES | YES | Plasma Membrane | enzyme | ||||
| ENSDARG00000055377 | gnb5b | guanine nucleotide binding protein (G protein), beta 5b [Source:ZFIN;Acc:ZDB-GENE-040426-1712] | 393719 | YES | YES | YES | Plasma Membrane | enzyme | ||||
| ENSDARG00000003375 | gng12a | guanine nucleotide binding protein (G protein), gamma 12a [Source:ZFIN;Acc:ZDB-GENE-040426-2555] | 406604 | YES | YES | YES | Plasma Membrane | enzyme | ||||
| ENSDARG00000037921 | gng13b | guanine nucleotide binding protein (G protein), gamma 13b [Source:ZFIN;Acc:ZDB-GENE-040718-97] | 436673 | YES | YES | YES | Plasma Membrane | enzyme | ||||
| ENSDARG00000056831 | gng2 | guanine nucleotide binding protein (G protein), gamma 2 [Source:ZFIN;Acc:ZDB-GENE-050417-59] | 550256 | YES | YES | YES | Plasma Membrane | enzyme | ||||
| ENSDARG00000009553 | gng3 | guanine nucleotide binding protein (G protein), gamma 3 [Source:ZFIN;Acc:ZDB-GENE-010705-1] | 114462 | YES | YES | YES | Plasma Membrane | enzyme | ||||
| ENSDARG00000057676 | golga7 | golgin A7 [Source:ZFIN;Acc:ZDB-GENE-040625-142] | 798622 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000105099 | golga7bb | golgin A7 family, member Bb [Source:ZFIN;Acc:ZDB-GENE-090312-173] | 0 | YES | YES | YES | Other | other | ||||
| ENSDARG00000053070 | gosr2 | golgi SNAP receptor complex member 2 [Source:ZFIN;Acc:ZDB-GENE-030131-3290] | 324569 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSDARG00000019341 | gpc1a | glypican 1a [Source:ZFIN;Acc:ZDB-GENE-051120-147] | 553367 | YES | YES | YES | Plasma Membrane | transmembrane receptor | ||||
| ENSDARG00000090585 | gpc1b | glypican 1b [Source:ZFIN;Acc:ZDB-GENE-040426-1306] | 393995 | YES | YES | YES | Plasma Membrane | transmembrane receptor | ||||
| ENSDARG00000032199 | gpc3 | glypican 3 [Source:ZFIN;Acc:ZDB-GENE-031212-1] | 0 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000077134 | gpr158a | G protein-coupled receptor 158a [Source:ZFIN;Acc:ZDB-GENE-060130-18] | 570001 | YES | YES | YES | Plasma Membrane | G-protein coupled receptor | ||||
| ENSDARG00000079665 | gpr158b | G protein-coupled receptor 158b [Source:ZFIN;Acc:ZDB-GENE-130530-875] | 568637 | YES | YES | YES | Plasma Membrane | G-protein coupled receptor | ||||
| ENSDARG00000018146 | gpx1a | glutathione peroxidase 1a [Source:ZFIN;Acc:ZDB-GENE-030410-1] | 0 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000055302 | grid2 | glutamate receptor, ionotropic, delta 2 [Source:ZFIN;Acc:ZDB-GENE-040913-1] | 448841 | YES | YES | YES | Plasma Membrane | ion channel | ||||
| ENSDARG00000070448 | grk4 | G protein-coupled receptor kinase 4 [Source:ZFIN;Acc:ZDB-GENE-060929-1198] | 560761 | YES | YES | YES | Plasma Membrane | kinase | ||||
| ENSDARG00000007195 | GRM2 (2 of 2) | glutamate receptor, metabotropic 2 [Source:HGNC Symbol;Acc:HGNC:4594] | 564461 | YES | YES | YES | Plasma Membrane | G-protein coupled receptor | ||||
| ENSDARG00000004445 | GRM5 (1 of 2) | glutamate receptor, metabotropic 5 [Source:HGNC Symbol;Acc:HGNC:4597] | 0 | YES | YES | YES | Plasma Membrane | G-protein coupled receptor | ||||
| ENSDARG00000102067 | GRM5 (2 of 2) | glutamate receptor, metabotropic 5 [Source:HGNC Symbol;Acc:HGNC:4597] | 100332913 | YES | YES | YES | Plasma Membrane | G-protein coupled receptor | ||||
| ENSDARG00000017742 | grm6a | glutamate receptor, metabotropic 6a [Source:ZFIN;Acc:ZDB-GENE-060208-1] | 568484 | YES | YES | YES | Plasma Membrane | G-protein coupled receptor | ||||
| ENSDARG00000100267 | GRM7 | glutamate receptor, metabotropic 7 [Source:HGNC Symbol;Acc:HGNC:4599] | 0 | YES | YES | YES | Plasma Membrane | G-protein coupled receptor | ||||
| ENSDARG00000077654 | grm8a | glutamate receptor, metabotropic 8a [Source:ZFIN;Acc:ZDB-GENE-110421-2] | 792371 | YES | YES | YES | Plasma Membrane | G-protein coupled receptor | ||||
| ENSDARG00000076508 | grm8b | glutamate receptor, metabotropic 8b [Source:ZFIN;Acc:ZDB-GENE-110421-3] | 569768 | YES | YES | YES | Plasma Membrane | G-protein coupled receptor | ||||
| ENSDARG00000015681 | gsk3ab | glycogen synthase kinase 3 alpha b [Source:ZFIN;Acc:ZDB-GENE-990714-3] | 30664 | YES | YES | YES | Nucleus | kinase | ||||
| ENSDARG00000017803 | gsk3b | glycogen synthase kinase 3 beta [Source:ZFIN;Acc:ZDB-GENE-990714-4] | 30654 | YES | YES | YES | Nucleus | kinase | ||||
| ENSDARG00000053467 | gtpbp1 | GTP binding protein 1 [Source:ZFIN;Acc:ZDB-GENE-030909-12] | 378721 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000086790 | gucy1b3 | guanylate cyclase 1, soluble, beta 3 [Source:ZFIN;Acc:ZDB-GENE-090313-160] | 100150304 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000001733 | gulp1a | GULP, engulfment adaptor PTB domain containing 1a [Source:ZFIN;Acc:ZDB-GENE-030616-21] | 368404 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000057128 | hadhaa | hydroxyacyl-CoA dehydrogenase/3-ketoacyl-CoA thiolase/enoyl-CoA hydratase (trifunctional protein), alpha subunit a [Source:ZFIN;Acc:ZDB-GENE-031222-5] | 553401 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000060594 | hadhab | hydroxyacyl-CoA dehydrogenase/3-ketoacyl-CoA thiolase/enoyl-CoA hydratase (trifunctional protein), alpha subunit b [Source:ZFIN;Acc:ZDB-GENE-041111-204] | 793834 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000102055 | hadhb | hydroxyacyl-CoA dehydrogenase/3-ketoacyl-CoA thiolase/enoyl-CoA hydratase (trifunctional protein), beta subunit [Source:ZFIN;Acc:ZDB-GENE-030131-8550] | 0 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000018542 | hapln4 | hyaluronan and proteoglycan link protein 4 [Source:ZFIN;Acc:ZDB-GENE-060503-243] | 559818 | YES | YES | YES | Extracellular Space | other | ||||
| ENSDARG00000012519 | hcfc1b | host cell factor C1b [Source:ZFIN;Acc:ZDB-GENE-030131-2411] | 565206 | YES | YES | YES | Nucleus | transcription regulator | ||||
| ENSDARG00000087573 | hdac11 | histone deacetylase 11 [Source:ZFIN;Acc:ZDB-GENE-040704-7] | 431718 | YES | YES | YES | Nucleus | transcription regulator | ||||
| ENSDARG00000101079 | HECTD4 | HECT domain containing E3 ubiquitin protein ligase 4 [Source:HGNC Symbol;Acc:HGNC:26611] | 101885095 | YES | YES | YES | Nucleus | other | ||||
| ENSDARG00000062447 | hecw2a | HECT, C2 and WW domain containing E3 ubiquitin protein ligase 2a [Source:ZFIN;Acc:ZDB-GENE-090311-22] | 0 | YES | YES | YES | Extracellular Space | enzyme | ||||
| ENSDARG00000056934 | hepacama | hepatic and glial cell adhesion molecule a [Source:ZFIN;Acc:ZDB-GENE-050522-198] | 100005952 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000059231 | hephl1a | hephaestin-like 1a [Source:ZFIN;Acc:ZDB-GENE-070801-4] | 561607 | YES | YES | YES | Other | enzyme | ||||
| ENSDARG00000052371 | higd2a | HIG1 hypoxia inducible domain family, member 2A [Source:ZFIN;Acc:ZDB-GENE-050417-119] | 0 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000039452 | hk1 | hexokinase 1 [Source:ZFIN;Acc:ZDB-GENE-040426-2848] | 406791 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSDARG00000038703 | hkdc1 | hexokinase domain containing 1 [Source:ZFIN;Acc:ZDB-GENE-030131-9801] | 321224 | YES | YES | YES | Other | kinase | ||||
| ENSDARG00000099175 | hmgb1a | high mobility group box 1a [Source:ZFIN;Acc:ZDB-GENE-030131-341] | 321622 | YES | YES | YES | Nucleus | transcription regulator | ||||
| ENSDARG00000030479 | hmgb1b | high mobility group box 1b [Source:ZFIN;Acc:ZDB-GENE-030131-8480] | 795095 | YES | YES | YES | Nucleus | transcription regulator | ||||
| ENSDARG00000079780 | HNRNPUL1 (2 of 2) | heterogeneous nuclear ribonucleoprotein U-like 1 [Source:HGNC Symbol;Acc:HGNC:17011] | 0 | YES | YES | YES | Nucleus | other | ||||
| ENSDARG00000007034 | hnrpkl | heterogeneous nuclear ribonucleoprotein K, like [Source:ZFIN;Acc:ZDB-GENE-040426-1923] | 406264 | YES | YES | YES | Nucleus | other | ||||
| ENSDARG00000012381 | hsc70 | heat shock cognate 70 [Source:ZFIN;Acc:ZDB-GENE-040426-1221] | 393586 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000017781 | hsd17b10 | hydroxysteroid (17-beta) dehydrogenase 10 [Source:ZFIN;Acc:ZDB-GENE-041010-201] | 450078 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000035872 | hsd17b12b | hydroxysteroid (17-beta) dehydrogenase 12b [Source:ZFIN;Acc:ZDB-GENE-030131-1346] | 322626 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000040984 | hspa13 | heat shock protein 70 family, member 13 [Source:ZFIN;Acc:ZDB-GENE-070410-58] | 560481 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000068992 | hspa8 | heat shock protein 8 [Source:ZFIN;Acc:ZDB-GENE-990415-92] | 573376 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000037403 | HSPA8 (1 of 2) | heat shock 70kDa protein 8 [Source:HGNC Symbol;Acc:HGNC:5241] | 562935 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000007955 | iars | isoleucyl-tRNA synthetase [Source:ZFIN;Acc:ZDB-GENE-030131-6325] | 334393 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000096355 | ighv1-4 | immunoglobulin heavy variable 1-4 [Source:ZFIN;Acc:ZDB-GENE-040514-1] | 0 | YES | YES | YES | Other | other | ||||
| ENSDARG00000062045 | il1rapl1a | interleukin 1 receptor accessory protein-like 1a [Source:ZFIN;Acc:ZDB-GENE-061013-249] | 568868 | YES | YES | YES | Plasma Membrane | transmembrane receptor | ||||
| ENSDARG00000017126 | ilvbl | ilvB (bacterial acetolactate synthase)-like [Source:ZFIN;Acc:ZDB-GENE-040426-1623] | 393639 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000006900 | impdh2 | IMP (inosine 5'-monophosphate) dehydrogenase 2 [Source:ZFIN;Acc:ZDB-GENE-030114-5] | 317745 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000063352 | inpp4aa | inositol polyphosphate-4-phosphatase, type Ia [Source:ZFIN;Acc:ZDB-GENE-060312-5] | 664741 | YES | YES | YES | Cytoplasm | phosphatase | ||||
| ENSDARG00000051888 | ist1 | increased sodium tolerance 1 homolog (yeast) [Source:ZFIN;Acc:ZDB-GENE-030131-179] | 321460 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000007098 | itm2ba | integral membrane protein 2Ba [Source:ZFIN;Acc:ZDB-GENE-030131-2352] | 323632 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000074149 | itpr1b | inositol 1,4,5-trisphosphate receptor, type 1b [Source:ZFIN;Acc:ZDB-GENE-070604-2] | 100149388 | YES | YES | YES | Cytoplasm | ion channel | ||||
| ENSDARG00000011909 | itpr2 | inositol 1,4,5-trisphosphate receptor, type 2 [Source:ZFIN;Acc:ZDB-GENE-050419-192] | 562585 | YES | YES | YES | Cytoplasm | ion channel | ||||
| ENSDARG00000061741 | itpr3 | inositol 1,4,5-trisphosphate receptor, type 3 [Source:ZFIN;Acc:ZDB-GENE-070605-1] | 794666 | YES | YES | YES | Cytoplasm | ion channel | ||||
| ENSDARG00000070787 | jupa | junction plakoglobin a [Source:ZFIN;Acc:ZDB-GENE-991207-22] | 30415 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000059067 | jupb | junction plakoglobin b [Source:ZFIN;Acc:ZDB-GENE-110407-10] | 100330886 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000103799 | kars | lysyl-tRNA synthetase [Source:ZFIN;Acc:ZDB-GENE-021115-8] | 280647 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000104958 | KBTBD11 | kelch repeat and BTB (POZ) domain containing 11 [Source:HGNC Symbol;Acc:HGNC:29104] | 557467 | YES | YES | YES | Other | other | ||||
| ENSDARG00000079491 | KCNA3 | potassium channel, voltage gated shaker related subfamily A, member 3 [Source:HGNC Symbol;Acc:HGNC:6221] | 100150068 | YES | YES | YES | Plasma Membrane | ion channel | ||||
| ENSDARG00000087247 | KCNAB2 (2 of 2) | potassium channel, voltage gated subfamily A regulatory beta subunit 2 [Source:HGNC Symbol;Acc:HGNC:6229] | 797337 | YES | YES | YES | Plasma Membrane | ion channel | ||||
| ENSDARG00000074320 | KCNAB3 | potassium channel, voltage gated subfamily A regulatory beta subunit 3 [Source:HGNC Symbol;Acc:HGNC:6230] | 0 | YES | YES | YES | Plasma Membrane | ion channel | ||||
| ENSDARG00000074746 | kcnd1 | potassium voltage-gated channel, Shal-related subfamily, member 1 [Source:ZFIN;Acc:ZDB-GENE-081105-40] | 100001681 | YES | YES | YES | Plasma Membrane | ion channel | ||||
| ENSDARG00000032799 | kcnd2 | potassium voltage-gated channel, Shal-related subfamily, member 2 [Source:ZFIN;Acc:ZDB-GENE-041210-70] | 570188 | YES | YES | YES | Plasma Membrane | ion channel | ||||
| ENSDARG00000056101 | kcnd3 | potassium voltage-gated channel, Shal-related subfamily, member 3 [Source:ZFIN;Acc:ZDB-GENE-030131-5626] | 327415 | YES | YES | YES | Plasma Membrane | ion channel | ||||
| ENSDARG00000088100 | KCNIP4 (2 of 2) | Kv channel interacting protein 4 [Source:HGNC Symbol;Acc:HGNC:30083] | 100536963 | YES | YES | YES | Plasma Membrane | ion channel | ||||
| ENSDARG00000103332 | KCNJ10 (2 of 3) | potassium channel, inwardly rectifying subfamily J, member 10 [Source:HGNC Symbol;Acc:HGNC:6256] | 0 | YES | YES | YES | Plasma Membrane | ion channel | ||||
| ENSDARG00000079840 | kcnma1a | potassium large conductance calcium-activated channel, subfamily M, alpha member 1a [Source:ZFIN;Acc:ZDB-GENE-070202-9] | 568554 | YES | YES | YES | Plasma Membrane | ion channel | ||||
| ENSDARG00000062477 | kiaa1549la | KIAA1549-like a [Source:ZFIN;Acc:ZDB-GENE-061207-55] | 566592 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000043571 | KIF2A | kinesin heavy chain member 2A [Source:HGNC Symbol;Acc:HGNC:6318] | 447926 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000075806 | kirrel3a | kin of IRRE like 3 a [Source:ZFIN;Acc:ZDB-GENE-091117-44] | 571887 | YES | YES | YES | Extracellular Space | other | ||||
| ENSDARG00000013741 | lancl1 | LanC antibiotic synthetase component C-like 1 (bacterial) [Source:ZFIN;Acc:ZDB-GENE-040924-1] | 494154 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000045465 | lancl2 | LanC lantibiotic synthetase component C-like 2 (bacterial) [Source:ZFIN;Acc:ZDB-GENE-070209-191] | 556310 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000057867 | lasp1 | LIM and SH3 protein 1 [Source:ZFIN;Acc:ZDB-GENE-030131-1936] | 323216 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSDARG00000077515 | letm2 | leucine zipper-EF-hand containing transmembrane protein 2 [Source:ZFIN;Acc:ZDB-GENE-090521-4] | 100006201 | YES | YES | YES | Other | other | ||||
| ENSDARG00000030641 | letmd1 | LETM1 domain containing 1 [Source:ZFIN;Acc:ZDB-GENE-080723-8] | 567014 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000040528 | lgals3bpb | lectin, galactoside-binding, soluble, 3 binding protein b [Source:ZFIN;Acc:ZDB-GENE-040426-2262] | 405809 | YES | YES | YES | Plasma Membrane | transmembrane receptor | ||||
| ENSDARG00000025903 | lgals9l1 | lectin, galactoside-binding, soluble, 9 (galectin 9)-like 1 [Source:ZFIN;Acc:ZDB-GENE-030131-9543] | 337597 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000020493 | lgi1a | leucine-rich, glioma inactivated 1a [Source:ZFIN;Acc:ZDB-GENE-030131-1731] | 323011 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000058421 | lgi1b | leucine-rich, glioma inactivated 1b [Source:ZFIN;Acc:ZDB-GENE-060217-1] | 654829 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000089957 | lgi2a | leucine-rich repeat LGI family, member 2a [Source:ZFIN;Acc:ZDB-GENE-060217-2] | 654827 | YES | YES | YES | Extracellular Space | other | ||||
| ENSDARG00000034165 | lingo1a | leucine rich repeat and Ig domain containing 1a [Source:ZFIN;Acc:ZDB-GENE-080327-16] | 564943 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000069970 | lingo2b | leucine rich repeat and Ig domain containing 2b [Source:ZFIN;Acc:ZDB-GENE-080723-57] | 564052 | YES | YES | YES | Extracellular Space | other | ||||
| ENSDARG00000067815 | LINGO3 (2 of 2) | leucine rich repeat and Ig domain containing 3 [Source:HGNC Symbol;Acc:HGNC:21206] | 100151059 | YES | YES | YES | Other | other | ||||
| ENSDARG00000029114 | LMTK3 | lemur tyrosine kinase 3 [Source:HGNC Symbol;Acc:HGNC:19295] | 562728 | YES | YES | YES | Other | kinase | ||||
| ENSDARG00000013542 | lpgat1 | lysophosphatidylglycerol acyltransferase 1 [Source:ZFIN;Acc:ZDB-GENE-030131-2906] | 561832 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000010144 | lppr3a | lipid phosphate phosphatase-related protein type 3a [Source:ZFIN;Acc:ZDB-GENE-070912-555] | 553336 | YES | YES | YES | Other | phosphatase | ||||
| ENSDARG00000028552 | lppr3b | lipid phosphate phosphatase-related protein type 3b [Source:ZFIN;Acc:ZDB-GENE-120514-1] | 570173 | YES | YES | YES | Other | phosphatase | ||||
| ENSDARG00000079671 | lppr4a | lipid phosphate phosphatase-related protein type 4a [Source:ZFIN;Acc:ZDB-GENE-070705-280] | 559888 | YES | YES | YES | Plasma Membrane | phosphatase | ||||
| ENSDARG00000075580 | lrch2 | leucine-rich repeats and calponin homology (CH) domain containing 2 [Source:ZFIN;Acc:ZDB-GENE-080114-1] | 100150488 | YES | YES | YES | Other | other | ||||
| ENSDARG00000100679 | lrch3 | leucine-rich repeats and calponin homology (CH) domain containing 3 [Source:ZFIN;Acc:ZDB-GENE-071004-44] | 100126105 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000027602 | lrfn1 | leucine rich repeat and fibronectin type III domain containing 1 [Source:ZFIN;Acc:ZDB-GENE-040426-1227] | 393587 | YES | YES | YES | Nucleus | other | ||||
| ENSDARG00000079396 | lrfn5b | leucine rich repeat and fibronectin type III domain containing 5b [Source:ZFIN;Acc:ZDB-GENE-080327-20] | 555839 | YES | YES | YES | Nucleus | other | ||||
| ENSDARG00000086990 | lrrc16b | leucine rich repeat containing 16B [Source:ZFIN;Acc:ZDB-GENE-070912-183] | 0 | YES | YES | YES | Other | other | ||||
| ENSDARG00000069402 | lrrc4.1 | leucine rich repeat containing 4.1 [Source:ZFIN;Acc:ZDB-GENE-041210-129] | 565021 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000043938 | lrrc57 | leucine rich repeat containing 57 [Source:ZFIN;Acc:ZDB-GENE-040718-372] | 436900 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000052713 | lrrtm1 | leucine rich repeat transmembrane neuronal 1 [Source:ZFIN;Acc:ZDB-GENE-050506-80] | 570385 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000078839 | LRRTM4 (1 of 3) | leucine rich repeat transmembrane neuronal 4 [Source:HGNC Symbol;Acc:HGNC:19411] | 563710 | YES | YES | YES | Extracellular Space | other | ||||
| ENSDARG00000103069 | LSAMP | limbic system-associated membrane protein [Source:HGNC Symbol;Acc:HGNC:6705] | 0 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000071573 | lsm5 | LSM5 homolog, U6 small nuclear RNA associated (S. cerevisiae) [Source:ZFIN;Acc:ZDB-GENE-070822-23] | 563948 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000028533 | macf1a | microtubule-actin crosslinking factor 1a [Source:ZFIN;Acc:ZDB-GENE-030131-3606] | 562190 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000104023 | mag | myelin associated glycoprotein [Source:ZFIN;Acc:ZDB-GENE-041217-24] | 474346 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000070145 | magi1a | membrane associated guanylate kinase, WW and PDZ domain containing 1a [Source:ZFIN;Acc:ZDB-GENE-081105-13] | 100319849 | YES | YES | YES | Plasma Membrane | kinase | ||||
| ENSDARG00000003169 | magi1b | membrane associated guanylate kinase, WW and PDZ domain containing 1b [Source:ZFIN;Acc:ZDB-GENE-030131-5110] | 474349 | YES | YES | YES | Plasma Membrane | kinase | ||||
| ENSDARG00000021590 | MAGI2 (1 of 2) | membrane associated guanylate kinase, WW and PDZ domain containing 2 [Source:HGNC Symbol;Acc:HGNC:18957] | 564112 | YES | YES | YES | Plasma Membrane | kinase | ||||
| ENSDARG00000073769 | MAGI2 (2 of 2) | membrane associated guanylate kinase, WW and PDZ domain containing 2 [Source:HGNC Symbol;Acc:HGNC:18957] | 0 | YES | YES | YES | Plasma Membrane | kinase | ||||
| ENSDARG00000101869 | magi3a | membrane associated guanylate kinase, WW and PDZ domain containing 3a [Source:ZFIN;Acc:ZDB-GENE-090312-48] | 561689 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSDARG00000041728 | man1a2 | mannosidase, alpha, class 1A, member 2 [Source:ZFIN;Acc:ZDB-GENE-070912-390] | 335538 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000073792 | man1b1a | mannosidase, alpha, class 1B, member 1a [Source:ZFIN;Acc:ZDB-GENE-091116-34] | 562592 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000063101 | man2a2 | mannosidase, alpha, class 2A, member 2 [Source:ZFIN;Acc:ZDB-GENE-110331-1] | 100333987 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000059853 | manbal | mannosidase, beta A, lysosomal-like [Source:EntrezGene;Acc:795041] | 795041 | YES | YES | YES | Other | other | ||||
| ENSDARG00000059601 | map1aa | microtubule-associated protein 1Aa [Source:ZFIN;Acc:ZDB-GENE-100426-2] | 100151220 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000022045 | map1ab | microtubule-associated protein 1Ab [Source:ZFIN;Acc:ZDB-GENE-100426-3] | 797545 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000060434 | map1b | Hypothetical microtubule-associated protein 1b; Uncharacterized protein [Source:UniProtKB/TrEMBL;Acc:B2ZZ14] | 559032 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000101127 | map1lc3b | microtubule-associated protein 1 light chain 3 beta [Source:ZFIN;Acc:ZDB-GENE-030131-1145] | 322425 | YES | YES | YES | Other | other | ||||
| ENSDARG00000060326 | map1sb | microtubule-associated protein 1Sb [Source:ZFIN;Acc:ZDB-GENE-070912-412] | 565533 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000055052 | map2 | microtubule-associated protein 2 [Source:ZFIN;Acc:ZDB-GENE-041010-118] | 0 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000007825 | map2k1 | mitogen-activated protein kinase kinase 1 [Source:ZFIN;Acc:ZDB-GENE-040426-2759] | 406728 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSDARG00000006609 | map2k2a | mitogen-activated protein kinase kinase 2a [Source:ZFIN;Acc:ZDB-GENE-041027-1] | 563181 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSDARG00000068745 | MAP4 (1 of 3) | microtubule-associated protein 4 [Source:HGNC Symbol;Acc:HGNC:6862] | 100537884 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000101687 | MAP4 (3 of 3) | microtubule-associated protein 4 [Source:HGNC Symbol;Acc:HGNC:6862] | 100001114 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000071357 | map4k3b | mitogen-activated protein kinase kinase kinase kinase 3b [Source:ZFIN;Acc:ZDB-GENE-090428-1] | 100005183 | YES | YES | YES | Other | kinase | ||||
| ENSDARG00000104701 | map7d1b | MAP7 domain containing 1b [Source:ZFIN;Acc:ZDB-GENE-080215-23] | 796270 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000068480 | map7d2a | MAP7 domain containing 2a [Source:ZFIN;Acc:ZDB-GENE-060526-150] | 100005195 | YES | YES | YES | Other | other | ||||
| ENSDARG00000102730 | mapk10 | mitogen-activated protein kinase 10 [Source:ZFIN;Acc:ZDB-GENE-051120-117] | 569698 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSDARG00000091777 | mapkap1 | mitogen-activated protein kinase associated protein 1 [Source:ZFIN;Acc:ZDB-GENE-041111-178] | 100537196 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000089314 | mapta | microtubule-associated protein tau a [Source:ZFIN;Acc:ZDB-GENE-081027-1] | 0 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000087616 | maptb | microtubule-associated protein tau b [Source:ZFIN;Acc:ZDB-GENE-081027-2] | 567833 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000034396 | mars | methionyl-tRNA synthetase [Source:ZFIN;Acc:ZDB-GENE-030219-83] | 338183 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000075159 | mfi2 | antigen p97 (melanoma associated) identified by monoclonal antibodies 133.2 and 96.5 [Source:ZFIN;Acc:ZDB-GENE-050309-40] | 565942 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000063358 | micu1 | mitochondrial calcium uptake 1 [Source:ZFIN;Acc:ZDB-GENE-070410-22] | 561210 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000074255 | MICU3 (1 of 2) | mitochondrial calcium uptake family, member 3 [Source:HGNC Symbol;Acc:HGNC:27820] | 100536173 | YES | YES | YES | Other | other | ||||
| ENSDARG00000078599 | micu3a | mitochondrial calcium uptake family, member 3a [Source:ZFIN;Acc:ZDB-GENE-070713-1] | 559552 | YES | YES | YES | Other | other | ||||
| ENSDARG00000063026 | mlc1 | megalencephalic leukoencephalopathy with subcortical cysts 1 [Source:ZFIN;Acc:ZDB-GENE-050419-145] | 559990 | YES | YES | YES | Plasma Membrane | transporter | ||||
| ENSDARG00000029472 | mllt4a | myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 4a [Source:ZFIN;Acc:ZDB-GENE-050419-52] | 558359 | YES | YES | YES | Nucleus | other | ||||
| ENSDARG00000010553 | mmgt1 | membrane magnesium transporter 1 [Source:ZFIN;Acc:ZDB-GENE-040801-65] | 402891 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSDARG00000060252 | mogs | mannosyl-oligosaccharide glucosidase [Source:ZFIN;Acc:ZDB-GENE-070112-412] | 567913 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000024478 | mpc2 | mitochondrial pyruvate carrier 2 [Source:ZFIN;Acc:ZDB-GENE-030131-330] | 321611 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000053453 | mpp2a | membrane protein, palmitoylated 2a (MAGUK p55 subfamily member 2) [Source:ZFIN;Acc:ZDB-GENE-071004-82] | 562207 | YES | YES | YES | Plasma Membrane | kinase | ||||
| ENSDARG00000015184 | mpp3a | membrane protein, palmitoylated 3a (MAGUK p55 subfamily member 3) [Source:ZFIN;Acc:ZDB-GENE-130530-555] | 572079 | YES | YES | YES | Plasma Membrane | kinase | ||||
| ENSDARG00000062667 | mpp3b | membrane protein, palmitoylated 3b (MAGUK p55 subfamily member 3) [Source:ZFIN;Acc:ZDB-GENE-121214-128] | 799884 | YES | YES | YES | Plasma Membrane | kinase | ||||
| ENSDARG00000056892 | mpp6a | membrane protein, palmitoylated 6a (MAGUK p55 subfamily member 6) [Source:ZFIN;Acc:ZDB-GENE-050506-35] | 402878 | YES | YES | YES | Plasma Membrane | kinase | ||||
| ENSDARG00000038768 | mrpl12 | mitochondrial ribosomal protein L12 [Source:ZFIN;Acc:ZDB-GENE-050417-187] | 550391 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000075677 | mrpl17 | mitochondrial ribosomal protein L17 [Source:ZFIN;Acc:ZDB-GENE-040912-133] | 447945 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000045696 | mrpl23 | mitochondrial ribosomal protein L23 [Source:ZFIN;Acc:ZDB-GENE-040625-12] | 415142 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000016307 | mrpl46 | mitochondrial ribosomal protein L46 [Source:ZFIN;Acc:ZDB-GENE-040718-224] | 436791 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000014159 | mtdha | metadherin a [Source:ZFIN;Acc:ZDB-GENE-030616-615] | 368910 | YES | YES | YES | Cytoplasm | transcription regulator | ||||
| ENSDARG00000003940 | mtfr1l | mitochondrial fission regulator 1-like [Source:ZFIN;Acc:ZDB-GENE-030131-6228] | 334296 | YES | YES | YES | Other | other | ||||
| ENSDARG00000040492 | mthfd1b | methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1b [Source:ZFIN;Acc:ZDB-GENE-020905-4] | 260441 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000053196 | mtor | mechanistic target of rapamycin (serine/threonine kinase) [Source:ZFIN;Acc:ZDB-GENE-030131-2974] | 324254 | YES | YES | YES | Nucleus | kinase | ||||
| ENSDARG00000001220 | mycbp2 | MYC binding protein 2 [Source:ZFIN;Acc:ZDB-GENE-030616-132] | 368439 | YES | YES | YES | Nucleus | enzyme | ||||
| ENSDARG00000063295 | myh9a | myosin, heavy chain 9a, non-muscle [Source:ZFIN;Acc:ZDB-GENE-030131-5870] | 333938 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000074723 | myo10l1 | myosin X-like 1 [Source:ZFIN;Acc:ZDB-GENE-080425-1] | 566674 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000075752 | myo18aa | myosin XVIIIAa [Source:ZFIN;Acc:ZDB-GENE-080425-5] | 566880 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000061862 | myo18ab | myosin XVIIIAb [Source:ZFIN;Acc:ZDB-GENE-080423-1] | 0 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000024694 | myo1b | myosin IB [Source:ZFIN;Acc:ZDB-GENE-030131-695] | 541515 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000078603 | myo1hb | myosin IHb [Source:ZFIN;Acc:ZDB-GENE-070705-355] | 0 | YES | YES | YES | Other | other | ||||
| ENSDARG00000042141 | myo6b | myosin VIb [Source:ZFIN;Acc:ZDB-GENE-030318-3] | 445472 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000079475 | nckipsd | NCK interacting protein with SH3 domain [Source:ZFIN;Acc:ZDB-GENE-130326-2] | 100003986 | YES | YES | YES | Other | other | ||||
| ENSDARG00000002710 | ncl | nucleolin [Source:ZFIN;Acc:ZDB-GENE-030131-6986] | 767714 | YES | YES | YES | Nucleus | other | ||||
| ENSDARG00000103902 | nde1 | nudE neurodevelopment protein 1 [Source:ZFIN;Acc:ZDB-GENE-050913-61] | 566146 | YES | YES | YES | Nucleus | other | ||||
| ENSDARG00000104225 | ndel1b | nudE neurodevelopment protein 1-like 1b [Source:ZFIN;Acc:ZDB-GENE-030131-5889] | 333957 | YES | YES | YES | Nucleus | other | ||||
| ENSDARG00000036329 | ndufa1 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 1 [Source:ZFIN;Acc:ZDB-GENE-040625-168] | 415243 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000042777 | ndufa11 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 11 [Source:ZFIN;Acc:ZDB-GENE-040426-1631] | 393497 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000042469 | ndufa12 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 12 [Source:ZFIN;Acc:ZDB-GENE-050828-1] | 556538 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000098584 | ndufa13 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 13 [Source:ZFIN;Acc:ZDB-GENE-040426-1672] | 0 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000021984 | ndufa2 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 2 [Source:ZFIN;Acc:ZDB-GENE-050522-377] | 554125 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000039346 | ndufa5 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 5 [Source:ZFIN;Acc:ZDB-GENE-050320-17] | 541328 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000038028 | ndufa6 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 6 [Source:ZFIN;Acc:ZDB-GENE-040426-1124] | 393943 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000036684 | ndufa7 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 7 [Source:ZFIN;Acc:ZDB-GENE-040801-169] | 445042 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000058463 | ndufab1a | NADH dehydrogenase (ubiquinone) 1, alpha/beta subcomplex, 1a [Source:ZFIN;Acc:ZDB-GENE-040801-1] | 573619 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000014915 | ndufab1b | NADH dehydrogenase (ubiquinone) 1, alpha/beta subcomplex, 1b [Source:ZFIN;Acc:ZDB-GENE-030131-4437] | 325712 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000075709 | ndufb3 | NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 3 [Source:RefSeq peptide;Acc:NP_001289399] | 100332034 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000070824 | ndufb5 | NADH dehydrogenase (ubiquinone) 1 beta subcomplex 5 [Source:ZFIN;Acc:ZDB-GENE-011010-1] | 797063 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000037259 | ndufb6 | NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 6 [Source:ZFIN;Acc:ZDB-GENE-040426-1781] | 393781 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000010113 | ndufb8 | NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 8 [Source:ZFIN;Acc:ZDB-GENE-040426-1858] | 393829 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000041314 | ndufb9 | NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 9 [Source:ZFIN;Acc:ZDB-GENE-030131-8364] | 573021 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000102115 | ndufc2 | NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 2 [Source:ZFIN;Acc:ZDB-GENE-050320-87] | 541390 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000007526 | ndufs2 | NADH dehydrogenase (ubiquinone) Fe-S protein 2 [Source:ZFIN;Acc:ZDB-GENE-050522-273] | 553672 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000015385 | ndufs3 | NADH dehydrogenase (ubiquinone) Fe-S protein 3, (NADH-coenzyme Q reductase) [Source:ZFIN;Acc:ZDB-GENE-050417-274] | 550451 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000052840 | ndufs4 | NADH dehydrogenase (ubiquinone) Fe-S protein 4, (NADH-coenzyme Q reductase) [Source:ZFIN;Acc:ZDB-GENE-050522-421] | 573689 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000006290 | ndufs5 | NADH dehydrogenase (ubiquinone) Fe-S protein 5 [Source:ZFIN;Acc:ZDB-GENE-050522-437] | 553660 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000056583 | ndufs6 | NADH dehydrogenase (ubiquinone) Fe-S protein 6 [Source:ZFIN;Acc:ZDB-GENE-040912-86] | 447913 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000013044 | ndufv2 | NADH dehydrogenase (ubiquinone) flavoprotein 2 [Source:ZFIN;Acc:ZDB-GENE-040426-1713] | 393720 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000056745 | necab2 | N-terminal EF-hand calcium binding protein 2 [Source:ZFIN;Acc:ZDB-GENE-050706-167] | 565258 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000060006 | nedd4l | neural precursor cell expressed, developmentally down-regulated 4-like [Source:ZFIN;Acc:ZDB-GENE-051118-2] | 559634 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000007989 | nedd8 | neural precursor cell expressed, developmentally down-regulated 8 [Source:ZFIN;Acc:ZDB-GENE-030616-588] | 368667 | YES | YES | YES | Nucleus | enzyme | ||||
| ENSDARG00000102855 | neo1a | neogenin 1a [Source:ZFIN;Acc:ZDB-GENE-021031-1] | 266983 | YES | YES | YES | Plasma Membrane | transcription regulator | ||||
| ENSDARG00000075100 | neo1b | neogenin 1b [Source:ZFIN;Acc:ZDB-GENE-131121-320] | 100332721 | YES | YES | YES | Plasma Membrane | transcription regulator | ||||
| ENSDARG00000012982 | nf1a | neurofibromin 1a [Source:ZFIN;Acc:ZDB-GENE-030131-4907] | 326708 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000062237 | nfs1 | NFS1 cysteine desulfurase [Source:ZFIN;Acc:ZDB-GENE-060405-1] | 562714 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000077329 | nlgn2a | neuroligin 2a [Source:ZFIN;Acc:ZDB-GENE-090918-2] | 556158 | YES | YES | YES | Plasma Membrane | enzyme | ||||
| ENSDARG00000079251 | nlgn2b | neuroligin 2b [Source:ZFIN;Acc:ZDB-GENE-090918-3] | 100002407 | YES | YES | YES | Plasma Membrane | enzyme | ||||
| ENSDARG00000062376 | nlgn3b | neuroligin 3b [Source:ZFIN;Acc:ZDB-GENE-060526-197] | 567415 | YES | YES | YES | Plasma Membrane | enzyme | ||||
| ENSDARG00000073724 | nlrx1 | NLR family member X1 [Source:ZFIN;Acc:ZDB-GENE-071119-3] | 557335 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000020482 | nono | non-POU domain containing, octamer-binding [Source:ZFIN;Acc:ZDB-GENE-030131-713] | 321994 | YES | YES | YES | Nucleus | other | ||||
| ENSDARG00000043864 | nptnb | neuroplastin b [Source:ZFIN;Acc:ZDB-GENE-040426-1821] | 403006 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000037794 | nptx2a | neuronal pentraxin 2a [Source:ZFIN;Acc:ZDB-GENE-060707-1] | 553230 | YES | YES | YES | Extracellular Space | other | ||||
| ENSDARG00000061647 | nrxn1a | neurexin 1a [Source:ZFIN;Acc:ZDB-GENE-070206-1] | 565531 | YES | YES | YES | Plasma Membrane | transporter | ||||
| ENSDARG00000063150 | nrxn2b | neurexin 2b [Source:ZFIN;Acc:ZDB-GENE-070206-7] | 0 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000043746 | nrxn3a | neurexin 3a [Source:ZFIN;Acc:ZDB-GENE-070206-9] | 0 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000062693 | nrxn3b | neurexin 3b [Source:ZFIN;Acc:ZDB-GENE-070206-10] | 570698 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000018065 | ntm | neurotrimin [Source:ZFIN;Acc:ZDB-GENE-060421-6020] | 678534 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000027279 | numb | numb homolog (Drosophila) [Source:ZFIN;Acc:ZDB-GENE-060422-1] | 692064 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000034270 | ogdha | oxoglutarate (alpha-ketoglutarate) dehydrogenase a (lipoamide) [Source:ZFIN;Acc:ZDB-GENE-030131-2143] | 564552 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000103428 | ogdhb | oxoglutarate (alpha-ketoglutarate) dehydrogenase b (lipoamide) [Source:ZFIN;Acc:ZDB-GENE-090311-8] | 797715 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000079249 | OGDHL | oxoglutarate dehydrogenase-like [Source:HGNC Symbol;Acc:HGNC:25590] | 559207 | YES | YES | YES | Other | enzyme | ||||
| ENSDARG00000105243 | ogt.1 | O-linked N-acetylglucosamine (GlcNAc) transferase, tandem duplicate 1 [Source:ZFIN;Acc:ZDB-GENE-030131-9631] | 337685 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000039724 | OSBPL8 | oxysterol binding protein-like 8 [Source:HGNC Symbol;Acc:HGNC:16396] | 101883298 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000069313 | oxa1l | oxidase (cytochrome c) assembly 1-like [Source:ZFIN;Acc:ZDB-GENE-071004-49] | 562963 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000039578 | pa2g4a | proliferation-associated 2G4, a [Source:ZFIN;Acc:ZDB-GENE-030616-161] | 368737 | YES | YES | YES | Nucleus | transcription regulator | ||||
| ENSDARG00000017219 | pabpc1a | poly(A) binding protein, cytoplasmic 1a [Source:ZFIN;Acc:ZDB-GENE-030131-3238] | 606498 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000099339 | pacsin3 | protein kinase C and casein kinase substrate in neurons 3 [Source:ZFIN;Acc:ZDB-GENE-040426-984] | 393964 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000026595 | pafah1b1b | platelet-activating factor acetylhydrolase 1b, regulatory subunit 1b [Source:ZFIN;Acc:ZDB-GENE-040116-3] | 394247 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000026882 | palm1a | paralemmin 1a [Source:ZFIN;Acc:ZDB-GENE-050417-409] | 550557 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000069608 | palm2 | paralemmin 2 [Source:ZFIN;Acc:ZDB-GENE-050208-62] | 100002543 | YES | YES | YES | Other | other | ||||
| ENSDARG00000100170 | pam | peptidylglycine alpha-amidating monooxygenase [Source:ZFIN;Acc:ZDB-GENE-090313-384] | 570822 | YES | YES | YES | Plasma Membrane | enzyme | ||||
| ENSDARG00000102822 | pam16 | presequence translocase-associated motor 16 homolog (S. cerevisiae) [Source:ZFIN;Acc:ZDB-GENE-040426-1776] | 393777 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000101719 | pargl | poly (ADP-ribose) glycohydrolase, like [Source:ZFIN;Acc:ZDB-GENE-090313-286] | 797792 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000019529 | parp1 | poly (ADP-ribose) polymerase 1 [Source:ZFIN;Acc:ZDB-GENE-030131-3955] | 560788 | YES | YES | YES | Nucleus | enzyme | ||||
| ENSDARG00000011824 | pbxip1b | pre-B-cell leukemia homeobox interacting protein 1b [Source:ZFIN;Acc:ZDB-GENE-070112-2032] | 0 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000099729 | pcdh10a | protocadherin 10a [Source:ZFIN;Acc:ZDB-GENE-020731-1] | 0 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000102824 | pcdh10b | protocadherin 10b [Source:ZFIN;Acc:ZDB-GENE-030721-4] | 360138 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000034344 | pcdh19 | protocadherin 19 [Source:ZFIN;Acc:ZDB-GENE-030131-4218] | 0 | YES | YES | YES | Extracellular Space | other | ||||
| ENSDARG00000097170 | pcdh1a4 | protocadherin 1 alpha 4 [Source:ZFIN;Acc:ZDB-GENE-050202-4] | 497127 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000036175 | pcdh1b | protocadherin 1b [Source:ZFIN;Acc:ZDB-GENE-091015-2] | 572016 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000100829 | pcdh1g32 | protocadherin 1 gamma 32 [Source:ZFIN;Acc:ZDB-GENE-050609-25] | 554016 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000088136 | pcdh2aa15 | protocadherin 2 alpha a 15 [Source:ZFIN;Acc:ZDB-GENE-041118-15] | 493601 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000099783 | pcdh2ac | protocadherin 2 alpha c [Source:ZFIN;Acc:ZDB-GENE-041118-12] | 493587 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000103256 | pcdh2g1 | protocadherin 2 gamma 1 [Source:ZFIN;Acc:ZDB-GENE-050610-10] | 103912055 | YES | YES | YES | Other | other | ||||
| ENSDARG00000063264 | PCDH9 | protocadherin 9 [Source:HGNC Symbol;Acc:HGNC:8661] | 0 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000077023 | pcdhb | protocadherin b [Source:ZFIN;Acc:ZDB-GENE-020731-3] | 100333352 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000058162 | pcyt1ba | phosphate cytidylyltransferase 1, choline, beta a [Source:ZFIN;Acc:ZDB-GENE-061220-4] | 100001552 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000053609 | pddc1 | Parkinson disease 7 domain containing 1 [Source:ZFIN;Acc:ZDB-GENE-051030-96] | 100148360 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000012387 | pdha1a | pyruvate dehydrogenase (lipoamide) alpha 1a [Source:ZFIN;Acc:ZDB-GENE-040426-2719] | 406702 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000051756 | pdhx | pyruvate dehydrogenase complex, component X [Source:ZFIN;Acc:ZDB-GENE-040426-1539] | 393532 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000013128 | pdk1 | pyruvate dehydrogenase kinase, isozyme 1 [Source:ZFIN;Acc:ZDB-GENE-091204-402] | 555840 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSDARG00000014527 | pdk3a | pyruvate dehydrogenase kinase, isozyme 3a [Source:ZFIN;Acc:ZDB-GENE-061220-3] | 0 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSDARG00000018285 | pdpk1b | 3-phosphoinositide dependent protein kinase 1b [Source:ZFIN;Acc:ZDB-GENE-040426-1820] | 403000 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSDARG00000055976 | pecr | peroxisomal trans-2-enoyl-CoA reductase [Source:ZFIN;Acc:ZDB-GENE-050417-232] | 550422 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000058202 | pex16 | peroxisomal biogenesis factor 16 [Source:ZFIN;Acc:ZDB-GENE-050626-49] | 573990 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000060504 | pfkla | phosphofructokinase, liver a [Source:ZFIN;Acc:ZDB-GENE-121207-1] | 570106 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSDARG00000099755 | pfklb | phosphofructokinase, liver b [Source:ZFIN;Acc:ZDB-GENE-050417-62] | 550259 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSDARG00000060797 | pfkmb | phosphofructokinase, muscle b [Source:ZFIN;Acc:ZDB-GENE-081114-1] | 568001 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSDARG00000028000 | pfkpa | phosphofructokinase, platelet a [Source:ZFIN;Acc:ZDB-GENE-091112-22] | 560944 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSDARG00000012801 | pfkpb | phosphofructokinase, platelet b [Source:ZFIN;Acc:ZDB-GENE-070912-634] | 561416 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSDARG00000057414 | phb | prohibitin [Source:ZFIN;Acc:ZDB-GENE-030131-6577] | 321346 | YES | YES | YES | Nucleus | transcription regulator | ||||
| ENSDARG00000045297 | phb2a | prohibitin 2a [Source:ZFIN;Acc:ZDB-GENE-030131-3141] | 324421 | YES | YES | YES | Cytoplasm | transcription regulator | ||||
| ENSDARG00000017728 | phb2b | prohibitin 2b [Source:ZFIN;Acc:ZDB-GENE-040718-430] | 436954 | YES | YES | YES | Cytoplasm | transcription regulator | ||||
| ENSDARG00000054208 | phkg2 | phosphorylase kinase, gamma 2 (testis) [Source:ZFIN;Acc:ZDB-GENE-040426-825] | 393937 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSDARG00000076724 | pi4kaa | phosphatidylinositol 4-kinase, catalytic, alpha a [Source:ZFIN;Acc:ZDB-GENE-100204-2] | 556956 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSDARG00000062823 | pi4kab | phosphatidylinositol 4-kinase, catalytic, alpha b [Source:ZFIN;Acc:ZDB-GENE-080220-34] | 793349 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSDARG00000024642 | pip5k1ab | phosphatidylinositol-4-phosphate 5-kinase, type I, alpha, b [Source:ZFIN;Acc:ZDB-GENE-041111-123] | 492549 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSDARG00000031020 | pip5k2 | phosphatidylinositol-4-phosphate 5-kinase, type II [Source:ZFIN;Acc:ZDB-GENE-030828-9] | 373124 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSDARG00000052957 | plcd3a | phospholipase C, delta 3a [Source:ZFIN;Acc:ZDB-GENE-070620-1] | 569040 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000045870 | plekha5 | pleckstrin homology domain containing, family A member 5 [Source:ZFIN;Acc:ZDB-GENE-041210-25] | 567300 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000104668 | plxna1b | plexin A1b [Source:ZFIN;Acc:ZDB-GENE-061007-1] | 0 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000040002 | pnpla7b | patatin-like phospholipase domain containing 7b [Source:ZFIN;Acc:ZDB-GENE-050309-66] | 503936 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000010941 | pomgnt2 | protein O-linked mannose N-acetylglucosaminyltransferase 2 (beta 1,4-) [Source:ZFIN;Acc:ZDB-GENE-050522-242] | 497644 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000077053 | ppfia3 | protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 3 [Source:ZFIN;Acc:ZDB-GENE-100311-3] | 567230 | YES | YES | YES | Plasma Membrane | phosphatase | ||||
| ENSDARG00000044153 | ppp1cb | protein phosphatase 1, catalytic subunit, beta isozyme [Source:ZFIN;Acc:ZDB-GENE-030616-609] | 100003223 | YES | YES | YES | Cytoplasm | phosphatase | ||||
| ENSDARG00000099226 | ppp1cc | protein phosphatase 1, catalytic subunit, gamma isozyme [Source:ZFIN;Acc:ZDB-GENE-030131-5877] | 571753 | YES | YES | YES | Cytoplasm | phosphatase | ||||
| ENSDARG00000010784 | ppp1r12a | protein phosphatase 1, regulatory subunit 12A [Source:ZFIN;Acc:ZDB-GENE-041011-3] | 445393 | YES | YES | YES | Cytoplasm | phosphatase | ||||
| ENSDARG00000103144 | ppp1r21 | protein phosphatase 1, regulatory subunit 21 [Source:ZFIN;Acc:ZDB-GENE-041114-8] | 492333 | YES | YES | YES | Other | other | ||||
| ENSDARG00000007791 | ppp2r1a | protein phosphatase 2, regulatory subunit A, alpha [Source:ZFIN;Acc:ZDB-GENE-040426-2700] | 406685 | YES | YES | YES | Plasma Membrane | phosphatase | ||||
| ENSDARG00000032430 | ppp2r1b | protein phosphatase 2, regulatory subunit A, beta [Source:ZFIN;Acc:ZDB-GENE-040927-20] | 449548 | YES | YES | YES | Plasma Membrane | phosphatase | ||||
| ENSDARG00000054931 | PPP2R5B | protein phosphatase 2, regulatory subunit B', beta [Source:HGNC Symbol;Acc:HGNC:9310] | 562282 | YES | YES | YES | Cytoplasm | phosphatase | ||||
| ENSDARG00000069118 | ppp2r5eb | protein phosphatase 2, regulatory subunit B', epsilon isoform b [Source:ZFIN;Acc:ZDB-GENE-030904-6] | 373878 | YES | YES | YES | Cytoplasm | phosphatase | ||||
| ENSDARG00000069360 | ppp3r1b | protein phosphatase 3 (formerly 2B), regulatory s1ubunit B, alpha isoform, b [Source:ZFIN;Acc:ZDB-GENE-040912-18] | 447814 | YES | YES | YES | Other | other | ||||
| ENSDARG00000045540 | ppp6r2a | protein phosphatase 6, regulatory subunit 2a [Source:ZFIN;Acc:ZDB-GENE-030131-5428] | 560729 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000058734 | prdx1 | peroxiredoxin 1 [Source:ZFIN;Acc:ZDB-GENE-050320-35] | 541344 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000025350 | prdx2 | peroxiredoxin 2 [Source:ZFIN;Acc:ZDB-GENE-030326-2] | 791455 | YES | YES | YES | Other | other | ||||
| ENSDARG00000032102 | prdx3 | peroxiredoxin 3 [Source:ZFIN;Acc:ZDB-GENE-030826-18] | 0 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000055064 | prdx5 | peroxiredoxin 5 [Source:ZFIN;Acc:ZDB-GENE-050522-159] | 0 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000076128 | prkar1aa | protein kinase, cAMP-dependent, regulatory, type I, alpha (tissue specific extinguisher 1) a [Source:ZFIN;Acc:ZDB-GENE-050116-2] | 494533 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSDARG00000101755 | prkar1ab | protein kinase, cAMP-dependent, regulatory, type I, alpha (tissue specific extinguisher 1) b [Source:ZFIN;Acc:ZDB-GENE-050417-238] | 550427 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSDARG00000103800 | prkar1b | protein kinase, cAMP-dependent, regulatory, type I, beta [Source:ZFIN;Acc:ZDB-GENE-060929-580] | 0 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000009208 | prkcda | protein kinase C, delta a [Source:ZFIN;Acc:ZDB-GENE-030131-6503] | 334571 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSDARG00000075949 | prkd1 | protein kinase D1 [Source:ZFIN;Acc:ZDB-GENE-080131-1] | 557123 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSDARG00000075083 | prkdc | protein kinase, DNA-activated, catalytic polypeptide [Source:ZFIN;Acc:ZDB-GENE-030131-9008] | 562283 | YES | YES | YES | Nucleus | kinase | ||||
| ENSDARG00000015239 | prp19 | PRP19/PSO4 homolog (S. cerevisiae) [Source:ZFIN;Acc:ZDB-GENE-030131-263] | 321544 | YES | YES | YES | Nucleus | other | ||||
| ENSDARG00000076702 | PSD (1 of 2) | pleckstrin and Sec7 domain containing [Source:HGNC Symbol;Acc:HGNC:9507] | 101886286 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000063036 | psd2 | pleckstrin and Sec7 domain containing 2 [Source:ZFIN;Acc:ZDB-GENE-030131-3033] | 557199 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000104820 | psd3l | pleckstrin and Sec7 domain containing 3, like [Source:ZFIN;Acc:ZDB-GENE-030131-8195] | 0 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000037038 | psmc6 | proteasome (prosome, macropain) 26S subunit, ATPase, 6 [Source:ZFIN;Acc:ZDB-GENE-030131-304] | 321585 | YES | YES | YES | Nucleus | peptidase | ||||
| ENSDARG00000019945 | ptprdb | protein tyrosine phosphatase, receptor type, D, b [Source:ZFIN;Acc:ZDB-GENE-070615-38] | 100073332 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000103479 | ptprfa | protein tyrosine phosphatase, receptor type, f, a [Source:ZFIN;Acc:ZDB-GENE-020107-2] | 0 | YES | YES | YES | Plasma Membrane | phosphatase | ||||
| ENSDARG00000045006 | ptprga | protein tyrosine phosphatase, receptor type, g a [Source:ZFIN;Acc:ZDB-GENE-101101-4] | 561197 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000058646 | ptprna | protein tyrosine phosphatase, receptor type, Na [Source:ZFIN;Acc:ZDB-GENE-030131-3511] | 324790 | YES | YES | YES | Plasma Membrane | phosphatase | ||||
| ENSDARG00000077047 | ptprnb | protein tyrosine phosphatase, receptor type, Nb [Source:ZFIN;Acc:ZDB-GENE-100505-1] | 564351 | YES | YES | YES | Plasma Membrane | phosphatase | ||||
| ENSDARG00000059738 | ptprsa | protein tyrosine phosphatase, receptor type, s, a [Source:ZFIN;Acc:ZDB-GENE-020107-3] | 140820 | YES | YES | YES | Plasma Membrane | phosphatase | ||||
| ENSDARG00000097572 | ptprt | protein tyrosine phosphatase, receptor type, t [Source:ZFIN;Acc:ZDB-GENE-101028-4] | 0 | YES | YES | YES | Plasma Membrane | phosphatase | ||||
| ENSDARG00000051814 | ptprz1a | protein tyrosine phosphatase, receptor-type, Z polypeptide 1a [Source:ZFIN;Acc:ZDB-GENE-090406-1] | 0 | YES | YES | YES | Plasma Membrane | phosphatase | ||||
| ENSDARG00000077339 | ptrh2 | peptidyl-tRNA hydrolase 2 [Source:ZFIN;Acc:ZDB-GENE-050522-163] | 791795 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000068822 | purba | purine-rich element binding protein Ba [Source:ZFIN;Acc:ZDB-GENE-060531-100] | 564840 | YES | YES | YES | Nucleus | transcription regulator | ||||
| ENSDARG00000010316 | qars | glutaminyl-tRNA synthetase [Source:ZFIN;Acc:ZDB-GENE-040426-1011] | 394188 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000002026 | qkib | QKI, KH domain containing, RNA binding b [Source:ZFIN;Acc:ZDB-GENE-081028-54] | 100462714 | YES | YES | YES | Nucleus | other | ||||
| ENSDARG00000026484 | rab15 | RAB15, member RAS oncogene family [Source:ZFIN;Acc:ZDB-GENE-040718-2] | 436590 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000056347 | rab3aa | RAB3A, member RAS oncogene family, a [Source:ZFIN;Acc:ZDB-GENE-040801-2] | 445024 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000042803 | rab3b | RAB3B, member RAS oncogene family [Source:ZFIN;Acc:ZDB-GENE-050208-660] | 557482 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000038010 | rac2 | ras-related C3 botulinum toxin substrate 2 (rho family, small GTP binding protein Rac2) [Source:ZFIN;Acc:ZDB-GENE-040625-27] | 415151 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000090062 | rac3a | ras-related C3 botulinum toxin substrate 3a (rho family, small GTP binding protein Rac3) [Source:ZFIN;Acc:ZDB-GENE-040718-256] | 437027 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000045463 | ralaa | v-ral simian leukemia viral oncogene homolog Aa (ras related) [Source:ZFIN;Acc:ZDB-GENE-040426-1344] | 393993 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000040778 | ralba | v-ral simian leukemia viral oncogene homolog Ba (ras related) [Source:ZFIN;Acc:ZDB-GENE-040801-267] | 415213 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000076889 | ralgapa1 | Ral GTPase activating protein, alpha subunit 1 (catalytic) [Source:ZFIN;Acc:ZDB-GENE-080806-1] | 556427 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000088899 | ralgapb | Ral GTPase activating protein, beta subunit (non-catalytic) [Source:ZFIN;Acc:ZDB-GENE-130530-945] | 559303 | YES | YES | YES | Other | other | ||||
| ENSDARG00000057026 | ran | RAN, member RAS oncogene family [Source:ZFIN;Acc:ZDB-GENE-990415-88] | 30572 | YES | YES | YES | Nucleus | enzyme | ||||
| ENSDARG00000087346 | rap1ab | RAP1A, member of RAS oncogene family b [Source:ZFIN;Acc:ZDB-GENE-040426-2738] | 406714 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000103032 | rap2b | RAP2B, member of RAS oncogene family [Source:ZFIN;Acc:ZDB-GENE-040426-2818] | 414329 | YES | YES | YES | Plasma Membrane | enzyme | ||||
| ENSDARG00000015649 | rap2c | RAP2C, member of RAS oncogene family [Source:ZFIN;Acc:ZDB-GENE-041024-1] | 474323 | YES | YES | YES | Other | other | ||||
| ENSDARG00000054530 | rars | arginyl-tRNA synthetase [Source:ZFIN;Acc:ZDB-GENE-030131-9014] | 337070 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000073824 | RASGRF1 | Ras protein-specific guanine nucleotide-releasing factor 1 [Source:HGNC Symbol;Acc:HGNC:9875] | 568885 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000002816 | rasgrf2b | Ras protein-specific guanine nucleotide-releasing factor 2b [Source:ZFIN;Acc:ZDB-GENE-060504-1] | 553520 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000038030 | rbx1 | ring-box 1, E3 ubiquitin protein ligase [Source:ZFIN;Acc:ZDB-GENE-041008-106] | 449846 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000008491 | RDH13 (1 of 3) | retinol dehydrogenase 13 (all-trans/9-cis) [Source:HGNC Symbol;Acc:HGNC:19978] | 407663 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000098654 | RFTN1 (2 of 2) | raftlin, lipid raft linker 1 [Source:HGNC Symbol;Acc:HGNC:30278] | 101885083 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000056078 | rftn2 | raftlin family member 2 [Source:ZFIN;Acc:ZDB-GENE-040426-2760] | 406729 | YES | YES | YES | Other | other | ||||
| ENSDARG00000012248 | rgma | repulsive guidance molecule family member a [Source:ZFIN;Acc:ZDB-GENE-040527-1] | 414272 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000036774 | rgs12b | regulator of G-protein signaling 12b [Source:ZFIN;Acc:ZDB-GENE-031006-13] | 378970 | YES | YES | YES | Nucleus | other | ||||
| ENSDARG00000016584 | rgs7a | regulator of G-protein signaling 7a [Source:ZFIN;Acc:ZDB-GENE-040718-276] | 436814 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000096687 | rgs7b | regulator of G-protein signaling 7b [Source:ZFIN;Acc:ZDB-GENE-130531-78] | 100535325 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000086756 | RGS9BP | regulator of G protein signaling 9 binding protein [Source:HGNC Symbol;Acc:HGNC:30304] | 559232 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000004301 | rhogb | ras homolog family member Gb [Source:ZFIN;Acc:ZDB-GENE-030131-8877] | 336933 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000028401 | rhogc | ras homolog gene family, member Gc [Source:ZFIN;Acc:ZDB-GENE-030131-3402] | 324681 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000099996 | rhot1a | ras homolog family member T1a [Source:ZFIN;Acc:ZDB-GENE-030131-5354] | 327143 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000062305 | rims3 | regulating synaptic membrane exocytosis 3 [Source:ZFIN;Acc:ZDB-GENE-060503-788] | 563414 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000070100 | RIN1 (1 of 2) | Ras and Rab interactor 1 [Source:HGNC Symbol;Acc:HGNC:18749] | 101882011 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000069150 | rit1 | Ras-like without CAAX 1 [Source:ZFIN;Acc:ZDB-GENE-070912-329] | 793942 | YES | YES | YES | Plasma Membrane | enzyme | ||||
| ENSDARG00000014891 | robo2 | roundabout, axon guidance receptor, homolog 2 (Drosophila) [Source:ZFIN;Acc:ZDB-GENE-001019-1] | 60309 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000104413 | rogdi | rogdi homolog (Drosophila) [Source:ZFIN;Acc:ZDB-GENE-030131-7582] | 335642 | YES | YES | YES | Nucleus | other | ||||
| ENSDARG00000044339 | rp2 | retinitis pigmentosa 2 (X-linked recessive) [Source:ZFIN;Acc:ZDB-GENE-040426-2795] | 406755 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000025073 | rpl18a | ribosomal protein L18a [Source:ZFIN;Acc:ZDB-GENE-040426-1071] | 394035 | YES | YES | YES | Other | other | ||||
| ENSDARG00000076360 | rpl37 | ribosomal protein L37 [Source:ZFIN;Acc:ZDB-GENE-040625-39] | 0 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000051783 | rplp0 | ribosomal protein, large, P0 [Source:ZFIN;Acc:ZDB-GENE-000629-1] | 58101 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000070849 | rps15 | ribosomal protein S15 [Source:ZFIN;Acc:ZDB-GENE-030131-9092] | 337148 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000032725 | rps27a | ribosomal protein S27a [Source:ZFIN;Acc:ZDB-GENE-030131-10018] | 337810 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000043453 | rps5 | ribosomal protein S5 [Source:ZFIN;Acc:ZDB-GENE-020419-12] | 192297 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000042566 | rps7 | ribosomal protein S7 [Source:ZFIN;Acc:ZDB-GENE-040426-1718] | 393725 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000075718 | rpz5 | rapunzel 5 [Source:ZFIN;Acc:ZDB-GENE-030131-4678] | 100003142 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000013763 | RRBP1 (1 of 2) | ribosome binding protein 1 [Source:HGNC Symbol;Acc:HGNC:10448] | 553448 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSDARG00000041703 | rrbp1b | ribosome binding protein 1 homolog b (dog) [Source:ZFIN;Acc:ZDB-GENE-030429-36] | 0 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSDARG00000029692 | rufy3 | RUN and FYVE domain containing 3 [Source:ZFIN;Acc:ZDB-GENE-050327-58] | 541522 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000078125 | rusc1 | RUN and SH3 domain containing 1 [Source:ZFIN;Acc:ZDB-GENE-100922-274] | 558727 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000023797 | ryr1b | ryanodine receptor 1b (skeletal) [Source:ZFIN;Acc:ZDB-GENE-070705-417] | 570245 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000071331 | ryr3 | ryanodine receptor 3 [Source:ZFIN;Acc:ZDB-GENE-041001-165] | 561350 | YES | YES | YES | Plasma Membrane | ion channel | ||||
| ENSDARG00000071014 | s100u | S100 calcium binding protein U [Source:ZFIN;Acc:ZDB-GENE-030131-3121] | 563060 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000042690 | s1pr1 | sphingosine-1-phosphate receptor 1 [Source:ZFIN;Acc:ZDB-GENE-001228-2] | 64617 | YES | YES | YES | Plasma Membrane | G-protein coupled receptor | ||||
| ENSDARG00000059806 | sacm1lb | SAC1 suppressor of actin mutations 1-like b (yeast) [Source:ZFIN;Acc:ZDB-GENE-070112-542] | 791142 | YES | YES | YES | Cytoplasm | phosphatase | ||||
| ENSDARG00000091042 | sacs | sacsin molecular chaperone [Source:ZFIN;Acc:ZDB-GENE-110411-146] | 558150 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000010610 | sarm1 | sterile alpha and TIR motif containing 1 [Source:ZFIN;Acc:ZDB-GENE-040219-1] | 403143 | YES | YES | YES | Plasma Membrane | transmembrane receptor | ||||
| ENSDARG00000079645 | sc:d217 | sc:d217 [Source:ZFIN;Acc:ZDB-GENE-080303-15] | 0 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000010728 | scin | scinderin [Source:ZFIN;Acc:ZDB-GENE-060503-6] | 566764 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000058348 | scinlb | scinderin like b [Source:ZFIN;Acc:ZDB-GENE-040426-2068] | 406363 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000000861 | scrib | scribbled planar cell polarity protein [Source:ZFIN;Acc:ZDB-GENE-030616-572] | 368473 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000100562 | sdc3 | syndecan 3 [Source:ZFIN;Acc:ZDB-GENE-030131-9021] | 337077 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000030097 | sdcbp | syndecan binding protein (syntenin) [Source:ZFIN;Acc:ZDB-GENE-081104-57] | 407719 | YES | YES | YES | Plasma Membrane | enzyme | ||||
| ENSDARG00000012513 | sdcbp2 | syndecan binding protein (syntenin) 2 [Source:ZFIN;Acc:ZDB-GENE-030131-3727] | 325004 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000062854 | sdk1b | sidekick cell adhesion molecule 1b [Source:ZFIN;Acc:ZDB-GENE-090311-17] | 555553 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000100435 | sec22bb | SEC22 vesicle trafficking protein homolog B (S. cerevisiae), b [Source:ZFIN;Acc:ZDB-GENE-030131-5488] | 327277 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000104230 | sec23a | Sec23 homolog A (S. cerevisiae) [Source:ZFIN;Acc:ZDB-GENE-040426-2823] | 406774 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSDARG00000071906 | sec24b | SEC24 family member B [Source:ZFIN;Acc:ZDB-GENE-030131-6565] | 334633 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSDARG00000079611 | sema4c | sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4C [Source:ZFIN;Acc:ZDB-GENE-080303-16] | 561895 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000036626 | sema6ba | sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6Ba [Source:ZFIN;Acc:ZDB-GENE-131024-1] | 0 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000014233 | sept8b | septin 8b [Source:ZFIN;Acc:ZDB-GENE-070424-3] | 571702 | YES | YES | YES | Extracellular Space | other | ||||
| ENSDARG00000103900 | sept9b | septin 9b [Source:ZFIN;Acc:ZDB-GENE-050327-17] | 541494 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000074242 | serbp1a | SERPINE1 mRNA binding protein 1a [Source:ZFIN;Acc:ZDB-GENE-030131-7389] | 335449 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000040614 | sestd1 | SEC14 and spectrin domains 1 [Source:ZFIN;Acc:ZDB-GENE-030801-1] | 360133 | YES | YES | YES | Extracellular Space | other | ||||
| ENSDARG00000097897 | sgip1a | SH3-domain GRB2-like (endophilin) interacting protein 1a [Source:ZFIN;Acc:ZDB-GENE-131121-25] | 566542 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000104414 | shmt2 | serine hydroxymethyltransferase 2 (mitochondrial) [Source:ZFIN;Acc:ZDB-GENE-071213-1] | 100144628 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000040853 | shoc2 | soc-2 suppressor of clear homolog (C. elegans) [Source:ZFIN;Acc:ZDB-GENE-050208-523] | 555476 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000102780 | si:ch1073-370j18.1 | si:ch1073-370j18.1 [Source:ZFIN;Acc:ZDB-GENE-131121-412] | 0 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000070442 | si:ch211-113g11.6 | si:ch211-113g11.6 [Source:ZFIN;Acc:ZDB-GENE-060503-473] | 559008 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000076970 | si:ch211-165b10.3 | si:ch211-165b10.3 [Source:ZFIN;Acc:ZDB-GENE-131121-6] | 100334077 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000095459 | si:ch211-191j22.3 | si:ch211-191j22.3 [Source:ZFIN;Acc:ZDB-GENE-030131-4242] | 0 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000063361 | si:ch211-1e14.1 | si:ch211-1e14.1 [Source:ZFIN;Acc:ZDB-GENE-050420-149] | 556850 | YES | YES | YES | Other | other | ||||
| ENSDARG00000071353 | si:ch211-235e9.8 | si:ch211-235e9.8 [Source:ZFIN;Acc:ZDB-GENE-141219-2] | 100002196 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000087364 | si:ch73-140j24.4 | si:ch73-140j24.4 [Source:ZFIN;Acc:ZDB-GENE-131120-193] | 101885000 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000023940 | si:ch73-362m14.4 | si:ch73-362m14.4 [Source:ZFIN;Acc:ZDB-GENE-120215-30] | 555437 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000080000 | si:dkey-178e17.1 | si:dkey-178e17.1 [Source:ZFIN;Acc:ZDB-GENE-081104-41] | 793282 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000094695 | si:dkey-204f11.64 | si:dkey-204f11.64 [Source:ZFIN;Acc:ZDB-GENE-030131-8332] | 100537752 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000096701 | si:dkey-21e13.3 | si:dkey-21e13.3 [Source:ZFIN;Acc:ZDB-GENE-130531-75] | 100150043 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000073752 | si:dkey-266j7.2 | si:dkey-266j7.2 [Source:ZFIN;Acc:ZDB-GENE-060503-162] | 0 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000011602 | si:dkeyp-117h8.2 | si:dkeyp-117h8.2 [Source:ZFIN;Acc:ZDB-GENE-041210-321] | 570321 | YES | YES | YES | Other | other | ||||
| ENSDARG00000035741 | slc25a11 | solute carrier family 25 (mitochondrial carrier; oxoglutarate carrier), member 11 [Source:ZFIN;Acc:ZDB-GENE-040625-79] | 415189 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSDARG00000026680 | slc25a14 | solute carrier family 25 (mitochondrial carrier, brain), member 14 [Source:ZFIN;Acc:ZDB-GENE-040426-749] | 393133 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSDARG00000057287 | slc25a16 | solute carrier family 25 (mitochondrial carrier; Graves disease autoantigen), member 16 [Source:ZFIN;Acc:ZDB-GENE-030131-3299] | 324578 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSDARG00000070717 | slc25a18 | solute carrier family 25 (glutamate carrier), member 18 [Source:ZFIN;Acc:ZDB-GENE-041111-192] | 568740 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSDARG00000020718 | SLC25A22 (1 of 2) | solute carrier family 25 (mitochondrial carrier: glutamate), member 22 [Source:HGNC Symbol;Acc:HGNC:19954] | 571044 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSDARG00000027355 | slc25a4 | solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 4 [Source:ZFIN;Acc:ZDB-GENE-030131-5275] | 327067 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSDARG00000099368 | slc25a42 | solute carrier family 25, member 42 [Source:ZFIN;Acc:ZDB-GENE-060825-313] | 751743 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSDARG00000092553 | slc25a5 | solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 5 [Source:ZFIN;Acc:ZDB-GENE-020419-9] | 192321 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000005853 | slc25a6 | solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 6 [Source:ZFIN;Acc:ZDB-GENE-070912-446] | 566370 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSDARG00000012881 | slc4a1a | solute carrier family 4 (anion exchanger), member 1a (Diego blood group) [Source:ZFIN;Acc:ZDB-GENE-010525-1] | 84703 | YES | YES | YES | Plasma Membrane | transporter | ||||
| ENSDARG00000086104 | SLCO5A1 (2 of 2) | solute carrier organic anion transporter family, member 5A1 [Source:HGNC Symbol;Acc:HGNC:19046] | 570033 | YES | YES | YES | Other | transporter | ||||
| ENSDARG00000079781 | slitrk4 | SLIT and NTRK-like family, member 4 [Source:ZFIN;Acc:ZDB-GENE-140303-1] | 558123 | YES | YES | YES | Extracellular Space | other | ||||
| ENSDARG00000103359 | SLITRK5 (2 of 2) | SLIT and NTRK-like family, member 5 [Source:HGNC Symbol;Acc:HGNC:20295] | 101882841 | YES | YES | YES | Other | other | ||||
| ENSDARG00000058203 | smc1al | structural maintenance of chromosomes 1A, like [Source:ZFIN;Acc:ZDB-GENE-040426-57] | 403060 | YES | YES | YES | Nucleus | transporter | ||||
| ENSDARG00000020609 | snap25a | synaptosomal-associated protein, 25a [Source:ZFIN;Acc:ZDB-GENE-980526-468] | 30712 | YES | YES | YES | Plasma Membrane | transporter | ||||
| ENSDARG00000058117 | snap25b | synaptosomal-associated protein, 25b [Source:ZFIN;Acc:ZDB-GENE-980526-392] | 30711 | YES | YES | YES | Plasma Membrane | transporter | ||||
| ENSDARG00000075539 | snphb | syntaphilin b [Source:ZFIN;Acc:ZDB-GENE-090313-92] | 569963 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000071031 | snx1a | sorting nexin 1a [Source:ZFIN;Acc:ZDB-GENE-060302-3] | 337386 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSDARG00000053527 | snx24 | sorting nexin 24 [Source:ZFIN;Acc:ZDB-GENE-050913-45] | 619249 | YES | YES | YES | Other | transporter | ||||
| ENSDARG00000097245 | soga3a | SOGA family member 3a [Source:ZFIN;Acc:ZDB-GENE-100921-84] | 100332515 | YES | YES | YES | Other | other | ||||
| ENSDARG00000075455 | soga3b | SOGA family member 3b [Source:ZFIN;Acc:ZDB-GENE-081022-198] | 563485 | YES | YES | YES | Other | other | ||||
| ENSDARG00000004017 | spag1a | sperm associated antigen 1a [Source:ZFIN;Acc:ZDB-GENE-030131-9443] | 0 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000029480 | specc1 | sperm antigen with calponin homology and coiled-coil domains 1 [Source:ZFIN;Acc:ZDB-GENE-060526-207] | 562343 | YES | YES | YES | Nucleus | other | ||||
| ENSDARG00000019231 | spna2 | spectrin alpha 2 [Source:ZFIN;Acc:ZDB-GENE-051113-60] | 553383 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000057665 | sprn2 | shadow of prion protein 2 [Source:ZFIN;Acc:ZDB-GENE-041221-1] | 100006310 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000102883 | SPTBN1 | spectrin, beta, non-erythrocytic 1 [Source:HGNC Symbol;Acc:HGNC:11275] | 553451 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000053956 | sptbn2 | spectrin, beta, non-erythrocytic 2 [Source:ZFIN;Acc:ZDB-GENE-030131-787] | 558044 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000017034 | sqrdl | sulfide quinone reductase-like (yeast) [Source:ZFIN;Acc:ZDB-GENE-050417-436] | 550580 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000008107 | src | v-src avian sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog [Source:ZFIN;Acc:ZDB-GENE-030131-3809] | 325084 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSDARG00000032161 | srgap2a | SLIT-ROBO Rho GTPase activating protein 2a [Source:ZFIN;Acc:ZDB-GENE-060915-2] | 556198 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000098367 | srp54 | signal recognition particle 54 [Source:ZFIN;Acc:ZDB-GENE-040426-818] | 393963 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000014139 | srp72 | signal recognition particle 72 [Source:ZFIN;Acc:ZDB-GENE-030131-5546] | 327335 | YES | YES | YES | Nucleus | kinase | ||||
| ENSDARG00000031560 | srpk1b | SRSF protein kinase 1b [Source:ZFIN;Acc:ZDB-GENE-070112-2292] | 791187 | YES | YES | YES | Nucleus | kinase | ||||
| ENSDARG00000029252 | ssb | Sjogren syndrome antigen B (autoantigen La) [Source:ZFIN;Acc:ZDB-GENE-030131-223] | 321504 | YES | YES | YES | Nucleus | enzyme | ||||
| ENSDARG00000014165 | ssr3 | signal sequence receptor, gamma [Source:ZFIN;Acc:ZDB-GENE-030131-9134] | 337190 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000045228 | stub1 | STIP1 homology and U-Box containing protein 1 [Source:ZFIN;Acc:ZDB-GENE-030131-2963] | 324243 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000035763 | stx18 | syntaxin 18 [Source:ZFIN;Acc:ZDB-GENE-050522-397] | 554130 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSDARG00000029234 | stxbp5b | syntaxin binding protein 5b (tomosyn) [Source:ZFIN;Acc:ZDB-GENE-090821-5] | 571925 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000055075 | svila | supervillin a [Source:ZFIN;Acc:ZDB-GENE-990706-5] | 30112 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000045945 | syn2a | synapsin IIa [Source:ZFIN;Acc:ZDB-GENE-040718-341] | 436870 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000063068 | syne1b | spectrin repeat containing, nuclear envelope 1b [Source:ZFIN;Acc:ZDB-GENE-071218-4] | 0 | YES | YES | YES | Nucleus | other | ||||
| ENSDARG00000075830 | syt3 | synaptotagmin III [Source:ZFIN;Acc:ZDB-GENE-090601-1] | 100000615 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSDARG00000003994 | syt9a | synaptotagmin IXa [Source:ZFIN;Acc:ZDB-GENE-031002-18] | 798433 | YES | YES | YES | Plasma Membrane | transporter | ||||
| ENSDARG00000079805 | tagln3a | transgelin 3a [Source:ZFIN;Acc:ZDB-GENE-060531-53] | 563096 | YES | YES | YES | Extracellular Space | other | ||||
| ENSDARG00000079004 | TANC2 (1 of 2) | tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2 [Source:HGNC Symbol;Acc:HGNC:30212] | 0 | YES | YES | YES | Nucleus | transcription regulator | ||||
| ENSDARG00000079097 | tanc2a | tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2a [Source:ZFIN;Acc:ZDB-GENE-060130-180] | 568810 | YES | YES | YES | Nucleus | transcription regulator | ||||
| ENSDARG00000069603 | taok1a | TAO kinase 1a [Source:ZFIN;Acc:ZDB-GENE-130103-9] | 100004576 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSDARG00000013250 | tars | threonyl-tRNA synthetase [Source:ZFIN;Acc:ZDB-GENE-041010-218] | 449661 | YES | YES | YES | Nucleus | enzyme | ||||
| ENSDARG00000061473 | tbkbp1 | TBK1 binding protein 1 [Source:ZFIN;Acc:ZDB-GENE-100203-1] | 559568 | YES | YES | YES | Other | other | ||||
| ENSDARG00000043357 | tbl2 | transducin (beta)-like 2 [Source:ZFIN;Acc:ZDB-GENE-030131-9012] | 337068 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000079881 | TCAIM | T cell activation inhibitor, mitochondrial [Source:HGNC Symbol;Acc:HGNC:25241] | 559258 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000045355 | tceb1a | transcription elongation factor B (SIII), polypeptide 1a [Source:ZFIN;Acc:ZDB-GENE-040912-120] | 447935 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000037980 | tceb2 | transcription elongation factor B (SIII), polypeptide 2 [Source:ZFIN;Acc:ZDB-GENE-020415-1] | 192341 | YES | YES | YES | Nucleus | transcription regulator | ||||
| ENSDARG00000105142 | tcirg1b | T-cell, immune regulator 1, ATPase, H+ transporting, lysosomal V0 subunit A3b [Source:ZFIN;Acc:ZDB-GENE-040426-2022] | 406342 | YES | YES | YES | Plasma Membrane | enzyme | ||||
| ENSDARG00000025904 | tecrb | trans-2,3-enoyl-CoA reductase b [Source:ZFIN;Acc:ZDB-GENE-030131-5154] | 326954 | YES | YES | YES | Plasma Membrane | enzyme | ||||
| ENSDARG00000011171 | TENM2 (1 of 2) | teneurin transmembrane protein 2 [Source:HGNC Symbol;Acc:HGNC:29943] | 568845 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000037122 | TENM2 (2 of 2) | teneurin transmembrane protein 2 [Source:HGNC Symbol;Acc:HGNC:29943] | 563197 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000058297 | timm13 | translocase of inner mitochondrial membrane 13 homolog (yeast) [Source:ZFIN;Acc:ZDB-GENE-040718-167] | 436738 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSDARG00000023672 | timm8a | translocase of inner mitochondrial membrane 8 homolog A (yeast) [Source:ZFIN;Acc:ZDB-GENE-040801-158] | 445243 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSDARG00000029689 | tkta | transketolase a [Source:ZFIN;Acc:ZDB-GENE-130530-578] | 557518 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000062951 | tldc1 | TBC/LysM-associated domain containing 1 [Source:ZFIN;Acc:ZDB-GENE-050419-140] | 561678 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000100729 | tln1 | talin 1 [Source:ZFIN;Acc:ZDB-GENE-031002-48] | 494453 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000028748 | tm9sf3 | transmembrane 9 superfamily member 3 [Source:ZFIN;Acc:ZDB-GENE-040426-2714] | 406698 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSDARG00000069099 | tmco1 | transmembrane and coiled-coil domains 1 [Source:ZFIN;Acc:ZDB-GENE-050417-344] | 378980 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000041391 | tmed10 | transmembrane emp24-like trafficking protein 10 (yeast) [Source:ZFIN;Acc:ZDB-GENE-030131-4915] | 326716 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSDARG00000024954 | tmed4 | transmembrane emp24 protein transport domain containing 4 [Source:ZFIN;Acc:ZDB-GENE-040625-140] | 415224 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSDARG00000046022 | tmed7 | transmembrane emp24 protein transport domain containing 7 [Source:ZFIN;Acc:ZDB-GENE-040426-2570] | 405848 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSDARG00000051824 | tmeff2a | transmembrane protein with EGF-like and two follistatin-like domains 2a [Source:ZFIN;Acc:ZDB-GENE-070912-622] | 100004613 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000016968 | tmem214 | transmembrane protein 214 [Source:ZFIN;Acc:ZDB-GENE-110307-2] | 402840 | YES | YES | YES | Extracellular Space | other | ||||
| ENSDARG00000063414 | tmem5 | transmembrane protein 5 [Source:ZFIN;Acc:ZDB-GENE-060929-1018] | 557187 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000076393 | tmem65 | transmembrane protein 65 [Source:ZFIN;Acc:ZDB-GENE-081022-99] | 100005403 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000022978 | tmpob | thymopoietin b [Source:ZFIN;Acc:ZDB-GENE-050522-8] | 553624 | YES | YES | YES | Nucleus | other | ||||
| ENSDARG00000078362 | TNC (2 of 2) | tenascin C [Source:HGNC Symbol;Acc:HGNC:5318] | 100149028 | YES | YES | YES | Extracellular Space | other | ||||
| ENSDARG00000056218 | tnika | TRAF2 and NCK interacting kinase a [Source:ZFIN;Acc:ZDB-GENE-030131-3767] | 325042 | YES | YES | YES | Plasma Membrane | kinase | ||||
| ENSDARG00000020771 | tnr | tenascin R (restrictin, janusin) [Source:ZFIN;Acc:ZDB-GENE-030804-1] | 369191 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000098486 | tollip | toll interacting protein [Source:ZFIN;Acc:ZDB-GENE-030131-8820] | 336876 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000056394 | TOM1L2 (2 of 2) | target of myb1-like 2 (chicken) [Source:HGNC Symbol;Acc:HGNC:11984] | 100535615 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSDARG00000057239 | tph2 | tryptophan hydroxylase 2 (tryptophan 5-monooxygenase) [Source:ZFIN;Acc:ZDB-GENE-040624-4] | 0 | YES | YES | YES | Plasma Membrane | enzyme | ||||
| ENSDARG00000087402 | TPM1 | tropomyosin 1 (alpha) [Source:HGNC Symbol;Acc:HGNC:12010] | 393908 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000005162 | tpm3 | tropomyosin 3 [Source:ZFIN;Acc:ZDB-GENE-030826-16] | 373076 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000033683 | tpma | alpha-tropomyosin [Source:ZFIN;Acc:ZDB-GENE-990415-269] | 30324 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000003963 | traf4a | tnf receptor-associated factor 4a [Source:ZFIN;Acc:ZDB-GENE-040308-1] | 404045 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000099118 | TRAPPC9 (1 of 2) | trafficking protein particle complex 9 [Source:HGNC Symbol;Acc:HGNC:30832] | 100002630 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000104396 | trim25 | tripartite motif containing 25 [Source:ZFIN;Acc:ZDB-GENE-040426-797] | 393144 | YES | YES | YES | Cytoplasm | transcription regulator | ||||
| ENSDARG00000076174 | trim2b | tripartite motif containing 2b [Source:ZFIN;Acc:ZDB-GENE-090312-70] | 558046 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000063711 | trim3a | tripartite motif containing 3a [Source:ZFIN;Acc:ZDB-GENE-080522-4] | 562489 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000005397 | trim3b | tripartite motif containing 3b [Source:ZFIN;Acc:ZDB-GENE-070112-1132] | 555391 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000019426 | trioa | trio Rho guanine nucleotide exchange factor a [Source:ZFIN;Acc:ZDB-GENE-060503-334] | 557667 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSDARG00000000370 | triob | trio Rho guanine nucleotide exchange factor b [Source:ZFIN;Acc:ZDB-GENE-030616-399] | 368847 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSDARG00000061397 | trip12 | thyroid hormone receptor interactor 12 [Source:ZFIN;Acc:ZDB-GENE-041111-262] | 564866 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000061207 | ttc7b | tetratricopeptide repeat domain 7B [Source:ZFIN;Acc:ZDB-GENE-070105-3] | 568039 | YES | YES | YES | Other | other | ||||
| ENSDARG00000045367 | tuba1b | tubulin, alpha 1b [Source:ZFIN;Acc:ZDB-GENE-030822-1] | 373080 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000045014 | tuba2 | tubulin, alpha 2 [Source:ZFIN;Acc:ZDB-GENE-040426-1970] | 406303 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000074289 | tuba4l | tubulin, alpha 4 like [Source:ZFIN;Acc:ZDB-GENE-030131-5588] | 327377 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000053066 | tubb1 | tubulin, beta 1 class VI [Source:ZFIN;Acc:ZDB-GENE-110408-5] | 558552 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000039522 | tubb2 | tubulin, beta 2A class IIa [Source:ZFIN;Acc:ZDB-GENE-050522-384] | 554127 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000098591 | tubb4b | tubulin, beta 4B class IVb [Source:ZFIN;Acc:ZDB-GENE-030131-8625] | 641421 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000037997 | tubb5 | tubulin, beta 5 [Source:ZFIN;Acc:ZDB-GENE-031110-4] | 386701 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000041723 | TUBB8P7 (2 of 3) | tubulin, beta 8 class VIII pseudogene 7 [Source:HGNC Symbol;Acc:HGNC:42345] | 406811 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000015610 | tubg1 | tubulin, gamma 1 [Source:ZFIN;Acc:ZDB-GENE-040426-836] | 393882 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000029133 | tubgcp3 | tubulin, gamma complex associated protein 3 [Source:ZFIN;Acc:ZDB-GENE-030616-538] | 563517 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000104173 | tufm | Tu translation elongation factor, mitochondrial [Source:ZFIN;Acc:ZDB-GENE-050320-73] | 541378 | YES | YES | YES | Other | other | ||||
| ENSDARG00000078854 | tusc3 | tumor suppressor candidate 3 [Source:ZFIN;Acc:ZDB-GENE-050522-381] | 553582 | YES | YES | YES | Plasma Membrane | transporter | ||||
| ENSDARG00000011661 | twf1b | twinfilin actin-binding protein 1b [Source:ZFIN;Acc:ZDB-GENE-040711-3] | 432375 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSDARG00000062089 | uggt2 | UDP-glucose glycoprotein glucosyltransferase 2 [Source:ZFIN;Acc:ZDB-GENE-050517-21] | 497542 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000016302 | upf1 | upf1 regulator of nonsense transcripts homolog (yeast) [Source:ZFIN;Acc:ZDB-GENE-040426-2836] | 406783 | YES | YES | YES | Nucleus | enzyme | ||||
| ENSDARG00000093382 | uqcc3 | ubiquinol-cytochrome c reductase complex assembly factor 3 [Source:ZFIN;Acc:ZDB-GENE-030131-8451] | 558925 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000104517 | UQCR10 | ubiquinol-cytochrome c reductase, complex III subunit X [Source:HGNC Symbol;Acc:HGNC:30863] | 100333064 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000007745 | uqcrfs1 | ubiquinol-cytochrome c reductase, Rieske iron-sulfur polypeptide 1 [Source:ZFIN;Acc:ZDB-GENE-040426-2060] | 406359 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000029064 | uqcrq | ubiquinol-cytochrome c reductase, complex III subunit VII [Source:ZFIN;Acc:ZDB-GENE-040718-199] | 436768 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000004305 | vangl1 | VANGL planar cell polarity protein 1 [Source:ZFIN;Acc:ZDB-GENE-040621-2] | 403076 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000027397 | vangl2 | VANGL planar cell polarity protein 2 [Source:ZFIN;Acc:ZDB-GENE-020507-3] | 245949 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000046048 | vapal | VAMP (vesicle-associated membrane protein)-associated protein A, like [Source:ZFIN;Acc:ZDB-GENE-040718-281] | 436819 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000070435 | vapb | VAMP (vesicle-associated membrane protein)-associated protein B and C [Source:ZFIN;Acc:ZDB-GENE-030131-2348] | 323628 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000099690 | vaspa | vasodilator-stimulated phosphoprotein a [Source:ZFIN;Acc:ZDB-GENE-070424-68] | 555805 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000044968 | vcla | vinculin a [Source:ZFIN;Acc:ZDB-GENE-050506-61] | 552980 | YES | YES | YES | Plasma Membrane | enzyme | ||||
| ENSDARG00000036338 | vps11 | vacuolar protein sorting 11 [Source:ZFIN;Acc:ZDB-GENE-050731-5] | 565852 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSDARG00000017986 | vps13d | vacuolar protein sorting 13 homolog D (S. cerevisiae) [Source:ZFIN;Acc:ZDB-GENE-050912-1] | 567879 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000103203 | vps28 | vacuolar protein sorting 28 (yeast) [Source:ZFIN;Acc:ZDB-GENE-040426-1527] | 393562 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000062016 | vps51 | vacuolar protein sorting 51 homolog (S. cerevisiae) [Source:ZFIN;Acc:ZDB-GENE-030131-6008] | 559339 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000069324 | vps53 | vacuolar protein sorting 53 homolog (S. cerevisiae) [Source:ZFIN;Acc:ZDB-GENE-041114-199] | 492817 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000092126 | vtg5 | vitellogenin 5 [Source:ZFIN;Acc:ZDB-GENE-001201-4] | 64260 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000039270 | vti1b | vesicle transport through interaction with t-SNAREs 1B [Source:ZFIN;Acc:ZDB-GENE-040426-1148] | 394011 | YES | YES | YES | Plasma Membrane | transporter | ||||
| ENSDARG00000060349 | wasf1 | WAS protein family, member 1 [Source:ZFIN;Acc:ZDB-GENE-070705-555] | 100001221 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000091457 | WASF1 (2 of 2) | WAS protein family, member 1 [Source:HGNC Symbol;Acc:HGNC:12732] | 101885310 | YES | YES | YES | Other | other | ||||
| ENSDARG00000014113 | wasla | Wiskott-Aldrich syndrome-like a [Source:ZFIN;Acc:ZDB-GENE-070209-220] | 100009637 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000075245 | wdr11 | WD repeat domain 11 [Source:ZFIN;Acc:ZDB-GENE-081107-28] | 558865 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000074611 | wdr37 | WD repeat domain 37 [Source:ZFIN;Acc:ZDB-GENE-030131-8101] | 558075 | YES | YES | YES | Other | other | ||||
| ENSDARG00000078136 | wdr47a | WD repeat domain 47a [Source:ZFIN;Acc:ZDB-GENE-070216-3] | 558528 | YES | YES | YES | Other | other | ||||
| ENSDARG00000038543 | wdr48 | WD repeat domain 48 [Source:ZFIN;Acc:ZDB-GENE-030131-6622] | 334682 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000100274 | WDR7 | WD repeat domain 7 [Source:HGNC Symbol;Acc:HGNC:13490] | 0 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000088195 | wu:fb52c12 | wu:fb52c12 [Source:ZFIN;Acc:ZDB-GENE-030131-897] | 322178 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000071551 | xrcc6 | X-ray repair complementing defective repair in Chinese hamster cells 6 [Source:ZFIN;Acc:ZDB-GENE-030131-6420] | 334488 | YES | YES | YES | Nucleus | enzyme | ||||
| ENSDARG00000005941 | yes1 | YES proto-oncogene 1, Src family tyrosine kinase [Source:ZFIN;Acc:ZDB-GENE-050126-1] | 407620 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSDARG00000061692 | yjefn3 | YjeF N-terminal domain containing 3 [Source:ZFIN;Acc:ZDB-GENE-050208-755] | 557840 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000075192 | yme1l1a | YME1-like 1a [Source:ZFIN;Acc:ZDB-GENE-091113-41] | 793098 | YES | YES | YES | Cytoplasm | peptidase | ||||
| ENSDARG00000104401 | yme1l1b | YME1-like 1b [Source:ZFIN;Acc:ZDB-GENE-070410-25] | 557907 | YES | YES | YES | Cytoplasm | peptidase | ||||
| ENSDARG00000055510 | ypel3 | yippee-like 3 [Source:ZFIN;Acc:ZDB-GENE-030516-4] | 368214 | YES | YES | YES | Other | other | ||||
| ENSDARG00000013078 | ywhaba | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, beta polypeptide a [Source:ZFIN;Acc:ZDB-GENE-030131-6583] | 323055 | YES | YES | YES | Cytoplasm | transcription regulator | ||||
| ENSDARG00000040287 | ywhabl | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, beta polypeptide like [Source:ZFIN;Acc:ZDB-GENE-030131-448] | 321729 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000005560 | ywhah | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, eta polypeptide [Source:ZFIN;Acc:ZDB-GENE-040426-2191] | 406443 | YES | YES | YES | Cytoplasm | transcription regulator | ||||
| ENSDARG00000032575 | ywhaz | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide [Source:ZFIN;Acc:ZDB-GENE-030131-8554] | 336610 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000027915 | zc2hc1a | zinc finger, C2HC-type containing 1A [Source:ZFIN;Acc:ZDB-GENE-030131-3132] | 324412 | YES | YES | YES | Other | other | ||||
| ENSDARG00000070441 | zdhhc17 | zinc finger, DHHC-type containing 17 [Source:ZFIN;Acc:ZDB-GENE-070424-194] | 100148543 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000016263 | zdhhc5a | zinc finger, DHHC-type containing 5a [Source:ZFIN;Acc:ZDB-GENE-090312-92] | 571795 | YES | YES | YES | Nucleus | enzyme | ||||
| ENSDARG00000086342 | zgc:101566 | zgc:101566 [Source:ZFIN;Acc:ZDB-GENE-041010-73] | 449823 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000071626 | zgc:153154 | zgc:153154 [Source:ZFIN;Acc:ZDB-GENE-060825-337] | 751701 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000019651 | zgc:162239 | zgc:162239 [Source:ZFIN;Acc:ZDB-GENE-070424-28] | 0 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000017140 | zgc:63587 | zgc:63587 [Source:ZFIN;Acc:ZDB-GENE-040426-1175] | 393431 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000016301 | zgc:65894 | zgc:65894 [Source:ZFIN;Acc:ZDB-GENE-030131-7741] | 335798 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000044874 | zgc:92107 | zgc:92107 [Source:ZFIN;Acc:ZDB-GENE-041114-4] | 492329 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000041060 | zgc:92326 | zgc:92326 [Source:ZFIN;Acc:ZDB-GENE-040718-159] | 327284 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000061718 | znf704 | zinc finger protein 704 [Source:ZFIN;Acc:ZDB-GENE-060503-775] | 560056 | YES | YES | YES | Other | other | ||||
| ENSDARG00000014717 | novel gene | 0 | 0 | YES | YES | Cytoplasm | peptidase | |||||
| ENSDARG00000087599 | novel gene | 0 | 567508 | YES | YES | NA | NA | |||||
| ENSDARG00000101726 | novel gene | 0 | 0 | YES | YES | Other | other | |||||
| ENSDARG00000077686 | aak1b | AP2 associated kinase 1b [Source:ZFIN;Acc:ZDB-GENE-091118-25] | 100534954 | YES | YES | Cytoplasm | kinase | |||||
| ENSDARG00000069142 | aars | alanyl-tRNA synthetase [Source:ZFIN;Acc:ZDB-GENE-030131-3663] | 324940 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000074041 | abca5 | ATP-binding cassette, sub-family A (ABC1), member 5 [Source:ZFIN;Acc:ZDB-GENE-050517-5] | 100151075 | YES | YES | Plasma Membrane | transporter | |||||
| ENSDARG00000061591 | abcb10 | ATP-binding cassette, sub-family B (MDR/TAP), member 10 [Source:ZFIN;Acc:ZDB-GENE-040525-2] | 100003744 | YES | YES | Cytoplasm | transporter | |||||
| ENSDARG00000071004 | abhd12 | abhydrolase domain containing 12 [Source:ZFIN;Acc:ZDB-GENE-060929-268] | 767657 | YES | YES | Other | enzyme | |||||
| ENSDARG00000058367 | abhd14a | abhydrolase domain containing 14A [Source:ZFIN;Acc:ZDB-GENE-040426-2428] | 404598 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000078929 | abhd16a | abhydrolase domain containing 16A [Source:ZFIN;Acc:ZDB-GENE-010601-2] | 564613 | YES | YES | Other | other | |||||
| ENSDARG00000004687 | acaa1 | acetyl-CoA acyltransferase 1 [Source:ZFIN;Acc:ZDB-GENE-040704-48] | 431754 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000038881 | acaa2 | acetyl-CoA acyltransferase 2 [Source:ZFIN;Acc:ZDB-GENE-040426-1996] | 406325 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000099056 | ACAA2 (2 of 2) | acetyl-CoA acyltransferase 2 [Source:HGNC Symbol;Acc:HGNC:83] | 0 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000078512 | acaca | acetyl-CoA carboxylase alpha [Source:ZFIN;Acc:ZDB-GENE-060526-74] | 559403 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000055620 | acad9 | acyl-CoA dehydrogenase family, member 9 [Source:ZFIN;Acc:ZDB-GENE-060616-196] | 724002 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000088357 | acadl | acyl-CoA dehydrogenase, long chain [Source:ZFIN;Acc:ZDB-GENE-040426-771] | 394156 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000038900 | acadm | acyl-CoA dehydrogenase, C-4 to C-12 straight chain [Source:ZFIN;Acc:ZDB-GENE-040426-1945] | 406283 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000076780 | acadsb | acyl-CoA dehydrogenase, short/branched chain [Source:ZFIN;Acc:ZDB-GENE-070410-109] | 0 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000016687 | acadvl | acyl-CoA dehydrogenase, very long chain [Source:ZFIN;Acc:ZDB-GENE-030131-899] | 573723 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000045888 | acat1 | acetyl-CoA acetyltransferase 1 [Source:ZFIN;Acc:ZDB-GENE-040808-68] | 445290 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000099079 | aclya | ATP citrate lyase a [Source:ZFIN;Acc:ZDB-GENE-031113-1] | 436922 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000007294 | aco2 | aconitase 2, mitochondrial [Source:ZFIN;Acc:ZDB-GENE-030131-1390] | 322670 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000014727 | acox1 | acyl-CoA oxidase 1, palmitoyl [Source:ZFIN;Acc:ZDB-GENE-041010-219] | 449662 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000014674 | acsl3b | acyl-CoA synthetase long-chain family member 3b [Source:ZFIN;Acc:ZDB-GENE-030131-492] | 559656 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000099648 | ACSL6 | acyl-CoA synthetase long-chain family member 6 [Source:HGNC Symbol;Acc:HGNC:16496] | 0 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000038432 | actr10 | actin-related protein 10 homolog (S. cerevisiae) [Source:ZFIN;Acc:ZDB-GENE-040426-768] | 393139 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000061445 | adcy6a | adenylate cyclase 6a [Source:ZFIN;Acc:ZDB-GENE-060221-1] | 0 | YES | YES | Plasma Membrane | enzyme | |||||
| ENSDARG00000069356 | adgrl2a | adhesion G protein-coupled receptor L2a [Source:ZFIN;Acc:ZDB-GENE-000607-49] | 57987 | YES | YES | Plasma Membrane | G-protein coupled receptor | |||||
| ENSDARG00000090624 | ADGRL3 (2 of 2) | adhesion G protein-coupled receptor L3 [Source:HGNC Symbol;Acc:HGNC:20974] | 0 | YES | YES | Plasma Membrane | G-protein coupled receptor | |||||
| ENSDARG00000061121 | adgrl3.1 | adhesion G protein-coupled receptor L3.1 [Source:ZFIN;Acc:ZDB-GENE-030131-7856] | 560631 | YES | YES | Plasma Membrane | G-protein coupled receptor | |||||
| ENSDARG00000104094 | adrbk2 | adrenergic, beta, receptor kinase 2 [Source:ZFIN;Acc:ZDB-GENE-030616-382] | 569403 | YES | YES | Cytoplasm | kinase | |||||
| ENSDARG00000062272 | afg3l2 | AFG3-like AAA ATPase 2 [Source:ZFIN;Acc:ZDB-GENE-070912-46] | 569168 | YES | YES | Cytoplasm | peptidase | |||||
| ENSDARG00000103811 | agla | amylo-alpha-1, 6-glucosidase, 4-alpha-glucanotransferase a [Source:ZFIN;Acc:ZDB-GENE-071004-6] | 567798 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000063187 | agpat5 | 1-acylglycerol-3-phosphate O-acyltransferase 5 (lysophosphatidic acid acyltransferase, epsilon) [Source:ZFIN;Acc:ZDB-GENE-060929-914] | 767778 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000058088 | aifm1 | apoptosis-inducing factor, mitochondrion-associated 1 [Source:ZFIN;Acc:ZDB-GENE-030826-11] | 373125 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000077549 | AIFM2 | apoptosis-inducing factor, mitochondrion-associated, 2 [Source:HGNC Symbol;Acc:HGNC:21411] | 557507 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000001950 | ak1 | adenylate kinase 1 [Source:ZFIN;Acc:ZDB-GENE-040822-37] | 445486 | YES | YES | Cytoplasm | kinase | |||||
| ENSDARG00000055678 | akap12b | A kinase (PRKA) anchor protein 12b [Source:ZFIN;Acc:ZDB-GENE-030131-9753] | 567365 | YES | YES | Cytoplasm | transporter | |||||
| ENSDARG00000006062 | akap1b | A kinase (PRKA) anchor protein 1b [Source:ZFIN;Acc:ZDB-GENE-030131-6844] | 556208 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000026531 | alcama | activated leukocyte cell adhesion molecule a [Source:ZFIN;Acc:ZDB-GENE-990415-30] | 30194 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000089924 | aldh2.1 | aldehyde dehydrogenase 2 family (mitochondrial), tandem duplicate 1 [Source:ZFIN;Acc:ZDB-GENE-040426-1262] | 393462 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000029381 | aldh3a2b | aldehyde dehydrogenase 3 family, member A2b [Source:ZFIN;Acc:ZDB-GENE-040912-103] | 0 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000038207 | aldh4a1 | aldehyde dehydrogenase 4 family, member A1 [Source:ZFIN;Acc:ZDB-GENE-040426-1179] | 394133 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000053485 | aldh6a1 | aldehyde dehydrogenase 6 family, member A1 [Source:ZFIN;Acc:ZDB-GENE-030131-9192] | 436647 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000019702 | aldocb | aldolase C, fructose-bisphosphate, b [Source:ZFIN;Acc:ZDB-GENE-030821-1] | 369193 | YES | YES | NA | NA | |||||
| ENSDARG00000007663 | amph | amphiphysin [Source:ZFIN;Acc:ZDB-GENE-040426-1711] | 393804 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000059093 | ANK1 (1 of 2) | ankyrin 1, erythrocytic [Source:HGNC Symbol;Acc:HGNC:492] | 0 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000074777 | ank1b | ankyrin 1, erythrocytic b [Source:ZFIN;Acc:ZDB-GENE-091113-6] | 0 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000036809 | ANPEP (1 of 5) | alanyl (membrane) aminopeptidase [Source:HGNC Symbol;Acc:HGNC:500] | 555478 | YES | YES | Plasma Membrane | peptidase | |||||
| ENSDARG00000077383 | anxa11a | annexin A11a [Source:ZFIN;Acc:ZDB-GENE-030707-4] | 368215 | YES | YES | Nucleus | other | |||||
| ENSDARG00000002632 | anxa11b | annexin A11b [Source:ZFIN;Acc:ZDB-GENE-030707-5] | 103908630 | YES | YES | Nucleus | other | |||||
| ENSDARG00000003216 | anxa2a | annexin A2a [Source:ZFIN;Acc:ZDB-GENE-030131-4282] | 325557 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000105296 | AP3B2 | adaptor-related protein complex 3, beta 2 subunit [Source:HGNC Symbol;Acc:HGNC:567] | 100000165 | YES | YES | Cytoplasm | transporter | |||||
| ENSDARG00000071424 | ap3d1 | adaptor-related protein complex 3, delta 1 subunit [Source:ZFIN;Acc:ZDB-GENE-050208-437] | 563359 | YES | YES | Cytoplasm | transporter | |||||
| ENSDARG00000017422 | apmap | adipocyte plasma membrane associated protein [Source:ZFIN;Acc:ZDB-GENE-020919-1] | 266637 | YES | YES | Plasma Membrane | enzyme | |||||
| ENSDARG00000012076 | apoa1a | apolipoprotein A-Ia [Source:ZFIN;Acc:ZDB-GENE-990415-14] | 30355 | YES | YES | Extracellular Space | transporter | |||||
| ENSDARG00000101324 | apoa1b | apolipoprotein A-Ib [Source:ZFIN;Acc:ZDB-GENE-050302-172] | 100101640 | YES | YES | Extracellular Space | transporter | |||||
| ENSDARG00000015866 | apoa2 | apolipoprotein A-II [Source:ZFIN;Acc:ZDB-GENE-030131-1046] | 322327 | YES | YES | NA | NA | |||||
| ENSDARG00000022767 | apobb.1 | apolipoprotein Bb, tandem duplicate 1 [Source:ZFIN;Acc:ZDB-GENE-030131-9732] | 321166 | YES | YES | Extracellular Space | transporter | |||||
| ENSDARG00000075016 | apobb.2 | apolipoprotein Bb, tandem duplicate 2 [Source:ZFIN;Acc:ZDB-GENE-070705-554] | 100330435 | YES | YES | Extracellular Space | transporter | |||||
| ENSDARG00000039374 | apool | apolipoprotein O-like [Source:ZFIN;Acc:ZDB-GENE-050327-76] | 541537 | YES | YES | Extracellular Space | other | |||||
| ENSDARG00000036998 | arf3b | ADP-ribosylation factor 3b [Source:ZFIN;Acc:ZDB-GENE-030616-356] | 368822 | YES | YES | NA | NA | |||||
| ENSDARG00000052928 | arf6b | ADP-ribosylation factor 6b [Source:ZFIN;Acc:ZDB-GENE-030131-9056] | 336063 | YES | YES | NA | NA | |||||
| ENSDARG00000024324 | arhgap1 | Rho GTPase activating protein 1 [Source:ZFIN;Acc:ZDB-GENE-050417-305] | 550478 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000102424 | ARHGAP44 (1 of 2) | Rho GTPase activating protein 44 [Source:HGNC Symbol;Acc:HGNC:29096] | 567298 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000015003 | arhgap4b | Rho GTPase activating protein 4b [Source:ZFIN;Acc:ZDB-GENE-090311-38] | 561231 | YES | YES | NA | NA | |||||
| ENSDARG00000057995 | arl15b | ADP-ribosylation factor-like 15b [Source:ZFIN;Acc:ZDB-GENE-080813-2] | 560030 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000078686 | arl8a | ADP-ribosylation factor-like 8A [Source:ZFIN;Acc:ZDB-GENE-030131-5025] | 567222 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000029463 | asap1a | ArfGAP with SH3 domain, ankyrin repeat and PH domain 1a [Source:ZFIN;Acc:ZDB-GENE-070912-129] | 100170829 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000056609 | atad1b | ATPase family, AAA domain containing 1b [Source:ZFIN;Acc:ZDB-GENE-030616-44] | 368412 | YES | YES | Other | enzyme | |||||
| ENSDARG00000044551 | atg9a | ATG9 autophagy related 9 homolog A (S. cerevisiae) [Source:ZFIN;Acc:ZDB-GENE-070209-65] | 100009663 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000060481 | atl1 | atlastin GTPase 1 [Source:ZFIN;Acc:ZDB-GENE-030131-8003] | 571909 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000002791 | atp1a1a.1 | ATPase, Na+/K+ transporting, alpha 1a polypeptide, tandem duplicate 1 [Source:ZFIN;Acc:ZDB-GENE-001212-1] | 64612 | YES | YES | Plasma Membrane | transporter | |||||
| ENSDARG00000019856 | atp1a1b | ATPase, Na+/K+ transporting, alpha 1b polypeptide [Source:ZFIN;Acc:ZDB-GENE-001212-5] | 64616 | YES | YES | Plasma Membrane | transporter | |||||
| ENSDARG00000018259 | atp1a3a | ATPase, Na+/K+ transporting, alpha 3a polypeptide [Source:ZFIN;Acc:ZDB-GENE-001212-7] | 64610 | YES | YES | Plasma Membrane | transporter | |||||
| ENSDARG00000076833 | atp1b1b | ATPase, Na+/K+ transporting, beta 1b polypeptide [Source:ZFIN;Acc:ZDB-GENE-001127-4] | 64273 | YES | YES | Plasma Membrane | transporter | |||||
| ENSDARG00000099203 | atp1b2a | ATPase, Na+/K+ transporting, beta 2a polypeptide [Source:ZFIN;Acc:ZDB-GENE-001127-2] | 64269 | YES | YES | Plasma Membrane | transporter | |||||
| ENSDARG00000053262 | atp1b4 | ATPase, Na+/K+ transporting, beta 4 polypeptide [Source:ZFIN;Acc:ZDB-GENE-070412-1] | 100037383 | YES | YES | Plasma Membrane | transporter | |||||
| ENSDARG00000005122 | atp2a2b | ATPase, Ca++ transporting, cardiac muscle, slow twitch 2b [Source:ZFIN;Acc:ZDB-GENE-030131-867] | 568265 | YES | YES | Cytoplasm | transporter | |||||
| ENSDARG00000012684 | atp2b1a | ATPase, Ca++ transporting, plasma membrane 1a [Source:ZFIN;Acc:ZDB-GENE-030925-29] | 378746 | YES | YES | Plasma Membrane | transporter | |||||
| ENSDARG00000063433 | atp2b2 | ATPase, Ca++ transporting, plasma membrane 2 [Source:ZFIN;Acc:ZDB-GENE-061016-1] | 557430 | YES | YES | Plasma Membrane | transporter | |||||
| ENSDARG00000043474 | atp2b3a | ATPase, Ca++ transporting, plasma membrane 3a [Source:ZFIN;Acc:ZDB-GENE-040718-174] | 436745 | YES | YES | Plasma Membrane | transporter | |||||
| ENSDARG00000023445 | atp2b3b | ATPase, Ca++ transporting, plasma membrane 3b [Source:ZFIN;Acc:ZDB-GENE-080409-2] | 558525 | YES | YES | Plasma Membrane | transporter | |||||
| ENSDARG00000044902 | atp2b4 | ATPase, Ca++ transporting, plasma membrane 4 [Source:ZFIN;Acc:ZDB-GENE-061027-60] | 0 | YES | YES | Plasma Membrane | transporter | |||||
| ENSDARG00000010149 | atp5a1 | ATP synthase, H+ transporting, mitochondrial F1 complex, alpha subunit 1, cardiac muscle [Source:ZFIN;Acc:ZDB-GENE-060201-1] | 553755 | YES | YES | Cytoplasm | transporter | |||||
| ENSDARG00000070083 | atp5b | ATP synthase, H+ transporting, mitochondrial F1 complex, beta polypeptide [Source:ZFIN;Acc:ZDB-GENE-030131-124] | 554135 | YES | YES | Cytoplasm | transporter | |||||
| ENSDARG00000045514 | atp5c1 | ATP synthase, H+ transporting, mitochondrial F1 complex, gamma polypeptide 1 [Source:ZFIN;Acc:ZDB-GENE-030131-8901] | 336957 | YES | YES | Cytoplasm | transporter | |||||
| ENSDARG00000019404 | atp5d | ATP synthase, H+ transporting, mitochondrial F1 complex, delta subunit [Source:ZFIN;Acc:ZDB-GENE-030131-7649] | 335709 | YES | YES | Cytoplasm | transporter | |||||
| ENSDARG00000095897 | atp5e | ATP synthase, H+ transporting, mitochondrial F1 complex, epsilon subunit [Source:ZFIN;Acc:ZDB-GENE-110411-22] | 798291 | YES | YES | Cytoplasm | transporter | |||||
| ENSDARG00000011553 | atp5f1 | ATP synthase, H+ transporting, mitochondrial Fo complex, subunit B1 [Source:ZFIN;Acc:ZDB-GENE-041010-33] | 449787 | YES | YES | Cytoplasm | transporter | |||||
| ENSDARG00000098355 | atp5h | ATP synthase, H+ transporting, mitochondrial Fo complex, subunit d [Source:ZFIN;Acc:ZDB-GENE-040426-1658] | 393675 | YES | YES | Other | other | |||||
| ENSDARG00000078113 | atp5ia | ATP synthase, H+ transporting, mitochondrial Fo complex, subunit Ea [Source:ZFIN;Acc:ZDB-GENE-070928-12] | 559659 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000068940 | atp5ib | ATP synthase, H+ transporting, mitochondrial Fo complex, subunit Eb [Source:ZFIN;Acc:ZDB-GENE-051130-1] | 767750 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000014313 | atp5j | ATP synthase, H+ transporting, mitochondrial Fo complex, subunit F6 [Source:ZFIN;Acc:ZDB-GENE-040426-2534] | 406599 | YES | YES | Cytoplasm | transporter | |||||
| ENSDARG00000011841 | atp5l | ATP synthase, H+ transporting, mitochondrial F0 complex, subunit g [Source:ZFIN;Acc:ZDB-GENE-030131-5177] | 326977 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000001788 | atp5o | ATP synthase, H+ transporting, mitochondrial F1 complex, O subunit [Source:ZFIN;Acc:ZDB-GENE-050522-147] | 335191 | YES | YES | Cytoplasm | transporter | |||||
| ENSDARG00000034534 | atp6v1aa | ATPase, H+ transporting, lysosomal, V1 subunit Aa [Source:ZFIN;Acc:ZDB-GENE-040426-1143] | 394110 | YES | YES | Plasma Membrane | transporter | |||||
| ENSDARG00000076318 | atp6v1ab | ATPase, H+ transporting, lysosomal, V1 subunit Ab [Source:ZFIN;Acc:ZDB-GENE-030131-9529] | 337583 | YES | YES | Plasma Membrane | transporter | |||||
| ENSDARG00000035880 | atp6v1c1b | ATPase, H+ transporting, lysosomal, V1 subunit C1b [Source:ZFIN;Acc:ZDB-GENE-041010-104] | 449655 | YES | YES | Cytoplasm | transporter | |||||
| ENSDARG00000077492 | atp8a2 | ATPase, aminophospholipid transporter, class I, type 8A, member 2 [Source:ZFIN;Acc:ZDB-GENE-100209-2] | 100330943 | YES | YES | Plasma Membrane | transporter | |||||
| ENSDARG00000044092 | atpif1b | ATPase inhibitory factor 1b [Source:ZFIN;Acc:ZDB-GENE-050506-113] | 558368 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000097238 | ba1l | ba1 globin, like [Source:ZFIN;Acc:ZDB-GENE-040901-4] | 30217 | YES | YES | Cytoplasm | transporter | |||||
| ENSDARG00000020895 | bag1 | BCL2-associated athanogene 1 [Source:ZFIN;Acc:ZDB-GENE-050309-89] | 558108 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000079264 | baiap3 | BAI1-associated protein 3 [Source:ZFIN;Acc:ZDB-GENE-130530-863] | 564728 | YES | YES | Extracellular Space | other | |||||
| ENSDARG00000090190 | bcam | basal cell adhesion molecule (Lutheran blood group) [Source:ZFIN;Acc:ZDB-GENE-060929-620] | 767689 | YES | YES | Plasma Membrane | transmembrane receptor | |||||
| ENSDARG00000099412 | bcan | brevican [Source:ZFIN;Acc:ZDB-GENE-030131-6045] | 334113 | YES | YES | Extracellular Space | other | |||||
| ENSDARG00000040555 | bckdha | branched chain keto acid dehydrogenase E1, alpha polypeptide [Source:ZFIN;Acc:ZDB-GENE-050522-376] | 554124 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000012295 | bcs1l | BC1 (ubiquinol-cytochrome c reductase) synthesis-like [Source:ZFIN;Acc:ZDB-GENE-040426-938] | 394157 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000042114 | bin1a | bridging integrator 1a [Source:ZFIN;Acc:ZDB-GENE-070912-715] | 0 | YES | YES | Nucleus | other | |||||
| ENSDARG00000056165 | C19H6orf136 | chromosome 6 open reading frame 136 [Source:HGNC Symbol;Acc:HGNC:21301] | 565228 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000041915 | C20H1orf95 | chromosome 1 open reading frame 95 [Source:HGNC Symbol;Acc:HGNC:30491] | 562918 | YES | YES | Other | other | |||||
| ENSDARG00000071378 | C22H19orf70 | chromosome 19 open reading frame 70 [Source:HGNC Symbol;Acc:HGNC:33702] | 558738 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000019332 | C24H11orf65 | chromosome 11 open reading frame 65 [Source:HGNC Symbol;Acc:HGNC:28519] | 406306 | YES | YES | Other | other | |||||
| ENSDARG00000020618 | C9H21orf33 | chromosome 21 open reading frame 33 [Source:HGNC Symbol;Acc:HGNC:1273] | 550355 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000060123 | ca16b | carbonic anhydrase XVI b [Source:ZFIN;Acc:ZDB-GENE-080818-1] | 569183 | YES | YES | Plasma Membrane | phosphatase | |||||
| ENSDARG00000086931 | CABZ01029822.1 | Uncharacterized protein [Source:UniProtKB/TrEMBL;Acc:F1QIC8] | 0 | YES | YES | NA | NA | |||||
| ENSDARG00000104674 | CABZ01075268.1 | 0 | 0 | YES | YES | NA | NA | |||||
| ENSDARG00000091618 | CABZ01080009.1 | Uncharacterized protein [Source:UniProtKB/TrEMBL;Acc:E7FBE5] | 0 | YES | YES | NA | NA | |||||
| ENSDARG00000008398 | cacna1c | calcium channel, voltage-dependent, L type, alpha 1C subunit [Source:ZFIN;Acc:ZDB-GENE-020129-1] | 170581 | YES | YES | Plasma Membrane | ion channel | |||||
| ENSDARG00000041895 | cad | carbamoyl-phosphate synthetase 2, aspartate transcarbamylase, and dihydroorotase [Source:ZFIN;Acc:ZDB-GENE-021030-4] | 266992 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000057013 | cadm3 | cell adhesion molecule 3 [Source:ZFIN;Acc:ZDB-GENE-060512-196] | 692273 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000040291 | cadm4 | cell adhesion molecule 4 [Source:ZFIN;Acc:ZDB-GENE-041114-138] | 492787 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000070567 | cadpsb | Ca2+-dependent activator protein for secretion b [Source:ZFIN;Acc:ZDB-GENE-110621-2] | 100001129 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000011166 | cahz | carbonic anhydrase [Source:ZFIN;Acc:ZDB-GENE-980526-39] | 30331 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000005372 | camk4 | calcium/calmodulin-dependent protein kinase IV [Source:ZFIN;Acc:ZDB-GENE-050417-76] | 550270 | YES | YES | Nucleus | kinase | |||||
| ENSDARG00000005141 | camkvb | CaM kinase-like vesicle-associated b [Source:ZFIN;Acc:ZDB-GENE-040426-1140] | 393422 | YES | YES | Other | kinase | |||||
| ENSDARG00000017841 | cand1 | cullin-associated and neddylation-dissociated 1 [Source:ZFIN;Acc:ZDB-GENE-040426-2872] | 406806 | YES | YES | Cytoplasm | transcription regulator | |||||
| ENSDARG00000016350 | cap1 | CAP, adenylate cyclase-associated protein 1 (yeast) [Source:ZFIN;Acc:ZDB-GENE-030131-6459] | 334527 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000104702 | cat | catalase [Source:ZFIN;Acc:ZDB-GENE-000210-20] | 30068 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000002880 | ccdc132 | coiled-coil domain containing 132 [Source:ZFIN;Acc:ZDB-GENE-060503-325] | 101883616 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000057921 | ccdc136b | coiled-coil domain containing 136b [Source:ZFIN;Acc:ZDB-GENE-081104-244] | 100330186 | YES | YES | NA | NA | |||||
| ENSDARG00000022983 | ccdc28a | coiled-coil domain containing 28A [Source:ZFIN;Acc:ZDB-GENE-030131-5007] | 326808 | YES | YES | Other | other | |||||
| ENSDARG00000054362 | ccdc47 | coiled-coil domain containing 47 [Source:ZFIN;Acc:ZDB-GENE-040912-15] | 447812 | YES | YES | Extracellular Space | other | |||||
| ENSDARG00000058828 | ccdc51 | coiled-coil domain containing 51 [Source:ZFIN;Acc:ZDB-GENE-060825-200] | 751752 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000099692 | ccnyl1 | cyclin Y-like 1 [Source:ZFIN;Acc:ZDB-GENE-060929-732] | 767752 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000087206 | CCT2 | chaperonin containing TCP1, subunit 2 (beta) [Source:HGNC Symbol;Acc:HGNC:1615] | 0 | YES | YES | Cytoplasm | kinase | |||||
| ENSDARG00000016173 | cct3 | chaperonin containing TCP1, subunit 3 (gamma) [Source:ZFIN;Acc:ZDB-GENE-020419-5] | 192327 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000013475 | cct4 | chaperonin containing TCP1, subunit 4 (delta) [Source:ZFIN;Acc:ZDB-GENE-040426-1421] | 393555 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000045399 | cct5 | chaperonin containing TCP1, subunit 5 (epsilon) [Source:ZFIN;Acc:ZDB-GENE-030131-977] | 322258 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000021252 | cct6a | chaperonin containing TCP1, subunit 6A (zeta 1) [Source:ZFIN;Acc:ZDB-GENE-011018-2] | 116994 | YES | YES | Cytoplasm | transporter | |||||
| ENSDARG00000007385 | cct7 | chaperonin containing TCP1, subunit 7 (eta) [Source:ZFIN;Acc:ZDB-GENE-020419-7] | 192324 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000008243 | cct8 | chaperonin containing TCP1, subunit 8 (theta) [Source:ZFIN;Acc:ZDB-GENE-040426-876] | 394037 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000044573 | cdc42 | cell division cycle 42 [Source:ZFIN;Acc:ZDB-GENE-030131-8783] | 336839 | YES | YES | Plasma Membrane | enzyme | |||||
| ENSDARG00000040158 | cdc42l | cell division cycle 42, like [Source:ZFIN;Acc:ZDB-GENE-030131-6069] | 793158 | YES | YES | NA | NA | |||||
| ENSDARG00000070686 | cdipt | CDP-diacylglycerol--inositol 3-phosphatidyltransferase (phosphatidylinositol synthase) [Source:ZFIN;Acc:ZDB-GENE-040426-2536] | 404620 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000021124 | cfl1 | cofilin 1 [Source:ZFIN;Acc:ZDB-GENE-030131-215] | 321496 | YES | YES | Nucleus | other | |||||
| ENSDARG00000059304 | chchd2 | coiled-coil-helix-coiled-coil-helix domain containing 2 [Source:ZFIN;Acc:ZDB-GENE-040426-1737] | 393740 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000045978 | chchd6b | coiled-coil-helix-coiled-coil-helix domain containing 6b [Source:ZFIN;Acc:ZDB-GENE-041114-95] | 492522 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000052859 | chp1 | calcineurin-like EF-hand protein 1 [Source:ZFIN;Acc:ZDB-GENE-030131-4086] | 325361 | YES | YES | Cytoplasm | transporter | |||||
| ENSDARG00000103672 | cirbpa | cold inducible RNA binding protein a [Source:ZFIN;Acc:ZDB-GENE-050417-329] | 550495 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000079516 | cisd1 | CDGSH iron sulfur domain 1 [Source:ZFIN;Acc:ZDB-GENE-040426-1162] | 393577 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000043257 | ckbb | creatine kinase, brain b [Source:ZFIN;Acc:ZDB-GENE-020103-2] | 140744 | YES | YES | Cytoplasm | kinase | |||||
| ENSDARG00000043493 | cltca | clathrin, heavy chain a (Hc) [Source:ZFIN;Acc:ZDB-GENE-030131-2299] | 323579 | YES | YES | NA | NA | |||||
| ENSDARG00000090716 | cltcb | clathrin, heavy chain b (Hc) [Source:ZFIN;Acc:ZDB-GENE-050227-12] | 503600 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000103670 | cmc1 | C-x(9)-C motif containing 1 [Source:ZFIN;Acc:ZDB-GENE-040426-2626] | 0 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000019924 | cmpk | cytidylate kinase [Source:ZFIN;Acc:ZDB-GENE-040426-2113] | 406383 | YES | YES | Nucleus | kinase | |||||
| ENSDARG00000099631 | cmtm6 | CKLF-like MARVEL transmembrane domain containing 6 [Source:ZFIN;Acc:ZDB-GENE-030729-15] | 368348 | YES | YES | Extracellular Space | cytokine | |||||
| ENSDARG00000099568 | CNNM3 | cyclin and CBS domain divalent metal cation transport mediator 3 [Source:HGNC Symbol;Acc:HGNC:104] | 100329721 | YES | YES | Other | other | |||||
| ENSDARG00000091683 | cnrip1a | cannabinoid receptor interacting protein 1a [Source:ZFIN;Acc:ZDB-GENE-040801-126] | 445213 | YES | YES | Extracellular Space | other | |||||
| ENSDARG00000028661 | cntfr | ciliary neurotrophic factor receptor [Source:ZFIN;Acc:ZDB-GENE-030616-82] | 368438 | YES | YES | Plasma Membrane | transmembrane receptor | |||||
| ENSDARG00000075189 | cntnap5b | contactin associated protein-like 5b [Source:ZFIN;Acc:ZDB-GENE-070705-209] | 571891 | YES | YES | Other | other | |||||
| ENSDARG00000100585 | coa3 | cytochrome C oxidase assembly factor 3 [Source:ZFIN;Acc:ZDB-GENE-060810-187] | 100141351 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000019355 | coa7 | cytochrome c oxidase assembly factor 7 [Source:ZFIN;Acc:ZDB-GENE-041014-16] | 555268 | YES | YES | Other | other | |||||
| ENSDARG00000025679 | comtb | catechol-O-methyltransferase b [Source:ZFIN;Acc:ZDB-GENE-040724-164] | 565370 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000004173 | copa | coatomer protein complex, subunit alpha [Source:ZFIN;Acc:ZDB-GENE-020905-2] | 260439 | YES | YES | Cytoplasm | transporter | |||||
| ENSDARG00000026829 | cotl1 | coactosin-like F-actin binding protein 1 [Source:ZFIN;Acc:ZDB-GENE-030131-8325] | 336381 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000099663 | cox5ab | cytochrome c oxidase subunit Vab [Source:ZFIN;Acc:ZDB-GENE-030131-5162] | 326962 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000011208 | COX5B (1 of 3) | cytochrome c oxidase subunit Vb [Source:HGNC Symbol;Acc:HGNC:2269] | 100002384 | YES | YES | Other | other | |||||
| ENSDARG00000015978 | COX5B (2 of 3) | cytochrome c oxidase subunit Vb [Source:HGNC Symbol;Acc:HGNC:2269] | 415253 | YES | YES | Other | other | |||||
| ENSDARG00000045230 | cox6b1 | cytochrome c oxidase subunit VIb polypeptide 1 [Source:ZFIN;Acc:ZDB-GENE-040718-448] | 436968 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000037860 | cox6b2 | cytochrome c oxidase subunit VIb polypeptide 2 [Source:ZFIN;Acc:ZDB-GENE-040426-1566] | 393478 | YES | YES | NA | NA | |||||
| ENSDARG00000104537 | cox7c | cytochrome c oxidase, subunit VIIc [Source:ZFIN;Acc:ZDB-GENE-030131-8062] | 336118 | YES | YES | NA | NA | |||||
| ENSDARG00000010312 | cp | ceruloplasmin [Source:ZFIN;Acc:ZDB-GENE-010522-1] | 84702 | YES | YES | Extracellular Space | enzyme | |||||
| ENSDARG00000055874 | cpe | carboxypeptidase E [Source:ZFIN;Acc:ZDB-GENE-090313-68] | 407979 | YES | YES | Cytoplasm | peptidase | |||||
| ENSDARG00000077736 | CPLX1 (2 of 2) | complexin 1 [Source:HGNC Symbol;Acc:HGNC:2309] | 563082 | YES | YES | Plasma Membrane | transporter | |||||
| ENSDARG00000075536 | cpne1 | copine I [Source:ZFIN;Acc:ZDB-GENE-030131-3562] | 324841 | YES | YES | Nucleus | transporter | |||||
| ENSDARG00000038618 | cpt2 | carnitine palmitoyltransferase 2 [Source:ZFIN;Acc:ZDB-GENE-030131-6719] | 100005717 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000045926 | crabp1a | cellular retinoic acid binding protein 1a [Source:ZFIN;Acc:ZDB-GENE-020320-3] | 171479 | YES | YES | Cytoplasm | transporter | |||||
| ENSDARG00000040248 | crata | carnitine O-acetyltransferase a [Source:ZFIN;Acc:ZDB-GENE-040927-17] | 449545 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000016074 | cryz | crystallin, zeta (quinone reductase) [Source:ZFIN;Acc:ZDB-GENE-050306-24] | 407640 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000103364 | cs | citrate synthase [Source:ZFIN;Acc:ZDB-GENE-030131-1058] | 322339 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000001559 | csmd2 | CUB and Sushi multiple domains 2 [Source:ZFIN;Acc:ZDB-GENE-030616-39] | 565460 | YES | YES | Other | other | |||||
| ENSDARG00000069981 | cspg5a | chondroitin sulfate proteoglycan 5a [Source:ZFIN;Acc:ZDB-GENE-080425-4] | 558206 | YES | YES | Extracellular Space | growth factor | |||||
| ENSDARG00000045352 | cst14a.2 | cystatin 14a, tandem duplicate 2 [Source:ZFIN;Acc:ZDB-GENE-040426-1065] | 792225 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000040674 | ctdsp1 | CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 1 [Source:ZFIN;Acc:ZDB-GENE-050522-523] | 553744 | YES | YES | Nucleus | phosphatase | |||||
| ENSDARG00000044562 | cycsb | cytochrome c, somatic b [Source:ZFIN;Acc:ZDB-GENE-040625-38] | 415158 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000098360 | cyp19a1b | cytochrome P450, family 19, subfamily A, polypeptide 1b [Source:ZFIN;Acc:ZDB-GENE-001103-4] | 0 | YES | YES | NA | NA | |||||
| ENSDARG00000033802 | cyp27a7 | cytochrome P450, family 27, subfamily A, polypeptide 7 [Source:ZFIN;Acc:ZDB-GENE-081104-519] | 402831 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000004964 | cyp4t8 | cytochrome P450, family 4, subfamily T, polypeptide 8 [Source:ZFIN;Acc:ZDB-GENE-031219-3] | 387527 | YES | YES | Other | other | |||||
| ENSDARG00000059939 | dab1a | Dab, reelin signal transducer, homolog 1a (Drosophila) [Source:ZFIN;Acc:ZDB-GENE-060528-1] | 692351 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000079850 | dchs1b | dachsous cadherin-related 1b [Source:ZFIN;Acc:ZDB-GENE-050208-41] | 0 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000056753 | dctn1b | dynactin 1b [Source:ZFIN;Acc:ZDB-GENE-050419-28] | 100003026 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000060626 | dgkaa | diacylglycerol kinase, alpha a [Source:ZFIN;Acc:ZDB-GENE-060616-305] | 724010 | YES | YES | Cytoplasm | kinase | |||||
| ENSDARG00000104793 | dgke | diacylglycerol kinase, epsilon [Source:ZFIN;Acc:ZDB-GENE-090512-7] | 100150463 | YES | YES | Cytoplasm | kinase | |||||
| ENSDARG00000018716 | dgkh | diacylglycerol kinase, eta [Source:ZFIN;Acc:ZDB-GENE-070912-328] | 100137124 | YES | YES | Cytoplasm | kinase | |||||
| ENSDARG00000051889 | dhodh | dihydroorotate dehydrogenase [Source:ZFIN;Acc:ZDB-GENE-030131-3157] | 494065 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000028066 | diras1a | DIRAS family, GTP-binding RAS-like 1a [Source:ZFIN;Acc:ZDB-GENE-030131-5788] | 327577 | YES | YES | Plasma Membrane | enzyme | |||||
| ENSDARG00000025302 | dixdc1a | DIX domain containing 1a [Source:ZFIN;Acc:ZDB-GENE-030721-2] | 360137 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000102696 | DNAJC11 (2 of 2) | DnaJ (Hsp40) homolog, subfamily C, member 11 [Source:HGNC Symbol;Acc:HGNC:25570] | 100329402 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000010042 | dnm1a | dynamin 1a [Source:ZFIN;Acc:ZDB-GENE-081104-27] | 100307098 | YES | YES | NA | NA | |||||
| ENSDARG00000009281 | dnm1b | dynamin 1b [Source:ZFIN;Acc:ZDB-GENE-100920-3] | 100333543 | YES | YES | NA | NA | |||||
| ENSDARG00000015006 | dnm1l | dynamin 1-like [Source:ZFIN;Acc:ZDB-GENE-040426-1556] | 393896 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000100145 | DNM3 (2 of 2) | dynamin 3 [Source:HGNC Symbol;Acc:HGNC:29125] | 0 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000104767 | DPP10 | dipeptidyl-peptidase 10 (non-functional) [Source:HGNC Symbol;Acc:HGNC:20823] | 0 | YES | YES | Extracellular Space | peptidase | |||||
| ENSDARG00000076826 | dpp6a | dipeptidyl-peptidase 6a [Source:ZFIN;Acc:ZDB-GENE-090918-4] | 100007104 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000024744 | dpp6b | dipeptidyl-peptidase 6b [Source:ZFIN;Acc:ZDB-GENE-070705-88] | 566832 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000032083 | dpysl2b | dihydropyrimidinase-like 2b [Source:ZFIN;Acc:ZDB-GENE-031105-1] | 553412 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000002587 | dpysl3 | dihydropyrimidinase-like 3 [Source:ZFIN;Acc:ZDB-GENE-050720-2] | 553166 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000103490 | dpysl4 | dihydropyrimidinase-like 4 [Source:ZFIN;Acc:ZDB-GENE-050720-3] | 553411 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000011141 | dpysl5a | dihydropyrimidinase-like 5a [Source:ZFIN;Acc:ZDB-GENE-030131-3136] | 324416 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000059311 | dpysl5b | dihydropyrimidinase-like 5b [Source:ZFIN;Acc:ZDB-GENE-050720-4] | 553410 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000078386 | dync1i2a | dynein, cytoplasmic 1, intermediate chain 2a [Source:ZFIN;Acc:ZDB-GENE-060929-1086] | 767729 | YES | YES | NA | NA | |||||
| ENSDARG00000098317 | dync1li1 | dynein, cytoplasmic 1, light intermediate chain 1 [Source:ZFIN;Acc:ZDB-GENE-030131-4108] | 556103 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000007370 | dysf | dysferlin, limb girdle muscular dystrophy 2B (autosomal recessive) [Source:ZFIN;Acc:ZDB-GENE-070501-1] | 560924 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000102412 | eci2 | enoyl-CoA delta isomerase 2 [Source:ZFIN;Acc:ZDB-GENE-040718-392] | 436918 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000044521 | eef1b2 | eukaryotic translation elongation factor 1 beta 2 [Source:ZFIN;Acc:ZDB-GENE-030131-7310] | 335370 | YES | YES | Cytoplasm | translation regulator | |||||
| ENSDARG00000100371 | eef2b | eukaryotic translation elongation factor 2b [Source:ZFIN;Acc:ZDB-GENE-030131-5128] | 326929 | YES | YES | NA | NA | |||||
| ENSDARG00000055759 | efhd2 | EF-hand domain family, member D2 [Source:ZFIN;Acc:ZDB-GENE-060531-152] | 571114 | YES | YES | Other | other | |||||
| ENSDARG00000007869 | ehd3 | EH-domain containing 3 [Source:ZFIN;Acc:ZDB-GENE-041014-352] | 562949 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000052178 | eif2s1b | eukaryotic translation initiation factor 2, subunit 1 alpha b [Source:ZFIN;Acc:ZDB-GENE-030131-283] | 321564 | YES | YES | Cytoplasm | translation regulator | |||||
| ENSDARG00000013931 | eif3m | eukaryotic translation initiation factor 3, subunit M [Source:ZFIN;Acc:ZDB-GENE-040426-2643] | 334626 | YES | YES | Other | other | |||||
| ENSDARG00000003032 | eif4a1b | eukaryotic translation initiation factor 4A1B [Source:ZFIN;Acc:ZDB-GENE-040120-6] | 399484 | YES | YES | Cytoplasm | translation regulator | |||||
| ENSDARG00000057255 | emc1 | ER membrane protein complex subunit 1 [Source:ZFIN;Acc:ZDB-GENE-060810-98] | 565399 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000054793 | emc10 | ER membrane protein complex subunit 10 [Source:ZFIN;Acc:ZDB-GENE-030131-9045] | 565829 | YES | YES | Other | other | |||||
| ENSDARG00000022456 | eno1a | enolase 1a, (alpha) [Source:ZFIN;Acc:ZDB-GENE-030131-6048] | 334116 | YES | YES | NA | NA | |||||
| ENSDARG00000014287 | eno2 | enolase 2 [Source:ZFIN;Acc:ZDB-GENE-040704-27] | 402874 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000045066 | entpd1 | ectonucleoside triphosphate diphosphohydrolase 1 [Source:ZFIN;Acc:ZDB-GENE-040801-58] | 445151 | YES | YES | Plasma Membrane | enzyme | |||||
| ENSDARG00000076364 | EPB41L1 | erythrocyte membrane protein band 4.1-like 1 [Source:HGNC Symbol;Acc:HGNC:3378] | 0 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000002255 | epb41l3a | erythrocyte membrane protein band 4.1-like 3a [Source:ZFIN;Acc:ZDB-GENE-040822-8] | 445478 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000103498 | epd | ependymin [Source:ZFIN;Acc:ZDB-GENE-980526-111] | 30199 | YES | YES | NA | NA | |||||
| ENSDARG00000054454 | epha4a | eph receptor A4a [Source:ZFIN;Acc:ZDB-GENE-001207-7] | 64271 | YES | YES | Plasma Membrane | kinase | |||||
| ENSDARG00000031548 | ephb3a | eph receptor B3a [Source:ZFIN;Acc:ZDB-GENE-990415-60] | 30313 | YES | YES | Plasma Membrane | kinase | |||||
| ENSDARG00000042854 | ephx1 | epoxide hydrolase 1, microsomal (xenobiotic) [Source:ZFIN;Acc:ZDB-GENE-040426-913] | 394043 | YES | YES | Cytoplasm | peptidase | |||||
| ENSDARG00000021991 | erlin1 | ER lipid raft associated 1 [Source:ZFIN;Acc:ZDB-GENE-050327-13] | 541490 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000086523 | erlin2 | ER lipid raft associated 2 (erlin2), mRNA [Source:RefSeq mRNA;Acc:NM_001128415] | 100151163 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000101631 | ETFA | electron-transfer-flavoprotein, alpha polypeptide [Source:HGNC Symbol;Acc:HGNC:3481] | 0 | YES | YES | Cytoplasm | transporter | |||||
| ENSDARG00000038834 | etfdh | electron-transferring-flavoprotein dehydrogenase [Source:ZFIN;Acc:ZDB-GENE-040912-168] | 447859 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000025091 | ezrb | ezrin b [Source:ZFIN;Acc:ZDB-GENE-050803-1] | 561589 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000023290 | fabp3 | fatty acid binding protein 3, muscle and heart [Source:ZFIN;Acc:ZDB-GENE-020318-2] | 171478 | YES | YES | NA | NA | |||||
| ENSDARG00000007697 | fabp7a | fatty acid binding protein 7, brain, a [Source:ZFIN;Acc:ZDB-GENE-000627-1] | 58128 | YES | YES | Cytoplasm | transporter | |||||
| ENSDARG00000063344 | fam162a | family with sequence similarity 162, member A [Source:ZFIN;Acc:ZDB-GENE-030131-8307] | 336363 | YES | YES | Other | other | |||||
| ENSDARG00000075249 | fam171a2 | family with sequence similarity 171, member A2 [Source:ZFIN;Acc:ZDB-GENE-110606-5] | 556398 | YES | YES | Other | other | |||||
| ENSDARG00000079656 | FAM171A2 (2 of 2) | family with sequence similarity 171, member A2 [Source:HGNC Symbol;Acc:HGNC:30480] | 797104 | YES | YES | Other | other | |||||
| ENSDARG00000057378 | fam213aa | family with sequence similarity 213, member Aa [Source:ZFIN;Acc:ZDB-GENE-030131-2459] | 323739 | YES | YES | Extracellular Space | other | |||||
| ENSDARG00000098825 | farsa | phenylalanyl-tRNA synthetase, alpha subunit [Source:ZFIN;Acc:ZDB-GENE-050512-2] | 692329 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000017591 | fat1a | FAT atypical cadherin 1a [Source:ZFIN;Acc:ZDB-GENE-050425-1] | 406172 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000018923 | fat2 | FAT atypical cadherin 2 [Source:ZFIN;Acc:ZDB-GENE-111031-1] | 568468 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000033840 | fat3a | FAT atypical cadherin 3a [Source:ZFIN;Acc:ZDB-GENE-060929-1254] | 0 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000040890 | fdps | farnesyl diphosphate synthase (farnesyl pyrophosphate synthetase, dimethylallyltranstransferase, geranyltranstransferase) [Source:ZFIN;Acc:ZDB-GENE-050506-78] | 552997 | YES | YES | NA | NA | |||||
| ENSDARG00000016058 | fibpa | fibroblast growth factor (acidic) intracellular binding protein a [Source:ZFIN;Acc:ZDB-GENE-040426-1363] | 393381 | YES | YES | Nucleus | other | |||||
| ENSDARG00000095579 | FIS1 | fission 1 (mitochondrial outer membrane) homolog (S. cerevisiae) [Source:HGNC Symbol;Acc:HGNC:21689] | 797988 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000074201 | flna | filamin A, alpha (actin binding protein 280) [Source:ZFIN;Acc:ZDB-GENE-030131-2145] | 0 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000016357 | fmo5 | flavin containing monooxygenase 5 [Source:ZFIN;Acc:ZDB-GENE-030131-6606] | 794311 | YES | YES | Other | other | |||||
| ENSDARG00000078399 | FO704819.2 | Uncharacterized protein [Source:UniProtKB/TrEMBL;Acc:E7F351] | 100332846 | YES | YES | NA | NA | |||||
| ENSDARG00000103740 | fundc2 | fun14 domain containing 2 [Source:ZFIN;Acc:ZDB-GENE-041010-26] | 792181 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000100971 | fxyd6 | FXYD domain containing ion transport regulator 6 [Source:ZFIN;Acc:ZDB-GENE-030131-5919] | 333987 | YES | YES | Plasma Membrane | ion channel | |||||
| ENSDARG00000027419 | gad1b | glutamate decarboxylase 1b [Source:ZFIN;Acc:ZDB-GENE-030909-3] | 378441 | YES | YES | NA | NA | |||||
| ENSDARG00000015537 | gad2 | glutamate decarboxylase 2 [Source:ZFIN;Acc:ZDB-GENE-030909-9] | 550403 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000039914 | gapdhs | glyceraldehyde-3-phosphate dehydrogenase, spermatogenic [Source:ZFIN;Acc:ZDB-GENE-020913-1] | 406367 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000074443 | gas7a | growth arrest-specific 7a [Source:ZFIN;Acc:ZDB-GENE-130128-1] | 793701 | YES | YES | Cytoplasm | transcription regulator | |||||
| ENSDARG00000058419 | gcn1l1 | GCN1 general control of amino-acid synthesis 1-like 1 (yeast) [Source:ZFIN;Acc:ZDB-GENE-040724-264] | 0 | YES | YES | Cytoplasm | translation regulator | |||||
| ENSDARG00000028628 | gdap1l1 | ganglioside induced differentiation associated protein 1-like 1 [Source:ZFIN;Acc:ZDB-GENE-080812-2] | 562163 | YES | YES | NA | NA | |||||
| ENSDARG00000005451 | gdi2 | GDP dissociation inhibitor 2 [Source:ZFIN;Acc:ZDB-GENE-030131-2485] | 323765 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000057465 | gfpt1 | glutamine--fructose-6-phosphate transaminase 1 [Source:ZFIN;Acc:ZDB-GENE-070423-1] | 0 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000078258 | GGT5 (2 of 2) | gamma-glutamyltransferase 5 [Source:HGNC Symbol;Acc:HGNC:4260] | 100333757 | YES | YES | Plasma Membrane | enzyme | |||||
| ENSDARG00000053456 | GK2 | glycerol kinase 2 [Source:HGNC Symbol;Acc:HGNC:4291] | 100136866 | YES | YES | Cytoplasm | kinase | |||||
| ENSDARG00000061282 | glg1a | golgi glycoprotein 1a [Source:ZFIN;Acc:ZDB-GENE-030131-6448] | 792865 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000101210 | glg1b | golgi glycoprotein 1b [Source:ZFIN;Acc:ZDB-GENE-060825-255] | 751680 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000062812 | glsa | glutaminase a [Source:ZFIN;Acc:ZDB-GENE-050204-3] | 564147 | YES | YES | NA | NA | |||||
| ENSDARG00000099776 | glula | glutamate-ammonia ligase (glutamine synthase) a [Source:ZFIN;Acc:ZDB-GENE-030131-688] | 100000775 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000061301 | gmpr2 | guanosine monophosphate reductase 2 [Source:ZFIN;Acc:ZDB-GENE-030131-1238] | 678545 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000053326 | gna11b | guanine nucleotide binding protein (G protein), alpha 11b (Gq class) [Source:ZFIN;Acc:ZDB-GENE-041121-9] | 493613 | YES | YES | Plasma Membrane | enzyme | |||||
| ENSDARG00000073765 | gna13a | guanine nucleotide binding protein (G protein), alpha 13a [Source:ZFIN;Acc:ZDB-GENE-030131-5024] | 326825 | YES | YES | Plasma Membrane | enzyme | |||||
| ENSDARG00000037924 | gna13b | guanine nucleotide binding protein (G protein), alpha 13b [Source:ZFIN;Acc:ZDB-GENE-030131-8277] | 336333 | YES | YES | Plasma Membrane | enzyme | |||||
| ENSDARG00000021647 | gnai1 | guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 1 [Source:ZFIN;Acc:ZDB-GENE-040426-1310] | 393946 | YES | YES | NA | NA | |||||
| ENSDARG00000039279 | golga5 | golgin A5 [Source:ZFIN;Acc:ZDB-GENE-040426-2749] | 406720 | YES | YES | Cytoplasm | kinase | |||||
| ENSDARG00000061951 | golgb1 | golgin B1 [Source:ZFIN;Acc:ZDB-GENE-030429-9] | 566353 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000041068 | got2a | glutamic-oxaloacetic transaminase 2a, mitochondrial [Source:ZFIN;Acc:ZDB-GENE-040426-2703] | 406688 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000005840 | got2b | glutamic-oxaloacetic transaminase 2b, mitochondrial [Source:ZFIN;Acc:ZDB-GENE-030131-7917] | 335974 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000074169 | gpam | glycerol-3-phosphate acyltransferase, mitochondrial [Source:ZFIN;Acc:ZDB-GENE-101026-1] | 100332911 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000012987 | gpia | glucose-6-phosphate isomerase a [Source:ZFIN;Acc:ZDB-GENE-020513-2] | 246094 | YES | YES | Other | other | |||||
| ENSDARG00000055455 | gpm6aa | glycoprotein M6Aa [Source:ZFIN;Acc:ZDB-GENE-030710-7] | 368223 | YES | YES | Plasma Membrane | ion channel | |||||
| ENSDARG00000004621 | gpm6ab | glycoprotein M6Ab [Source:ZFIN;Acc:ZDB-GENE-030710-8] | 407734 | YES | YES | Plasma Membrane | ion channel | |||||
| ENSDARG00000005739 | gpm6ba | glycoprotein M6Ba [Source:ZFIN;Acc:ZDB-GENE-030710-9] | 368225 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000001676 | gpm6bb | glycoprotein M6Bb [Source:ZFIN;Acc:ZDB-GENE-030710-10] | 503756 | YES | YES | NA | NA | |||||
| ENSDARG00000076836 | gpx4b | glutathione peroxidase 4b [Source:ZFIN;Acc:ZDB-GENE-030410-3] | 0 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000074583 | grid1a | glutamate receptor, ionotropic, delta 1a [Source:ZFIN;Acc:ZDB-GENE-120411-28] | 793756 | YES | YES | Plasma Membrane | ion channel | |||||
| ENSDARG00000044161 | grid1b | glutamate receptor, ionotropic, delta 1b [Source:ZFIN;Acc:ZDB-GENE-121214-84] | 557511 | YES | YES | Plasma Membrane | ion channel | |||||
| ENSDARG00000077715 | GRIK3 | glutamate receptor, ionotropic, kainate 3 [Source:HGNC Symbol;Acc:HGNC:4581] | 0 | YES | YES | Plasma Membrane | ion channel | |||||
| ENSDARG00000101317 | grinaa | glutamate receptor, ionotropic, N-methyl D-aspartate-associated protein 1a (glutamate binding) [Source:ZFIN;Acc:ZDB-GENE-060616-231] | 724005 | YES | YES | NA | NA | |||||
| ENSDARG00000026796 | grm1a | glutamate receptor, metabotropic 1a [Source:ZFIN;Acc:ZDB-GENE-030131-7893] | 555576 | YES | YES | Plasma Membrane | G-protein coupled receptor | |||||
| ENSDARG00000098320 | grm1b | glutamate receptor, metabotropic 1b [Source:ZFIN;Acc:ZDB-GENE-090821-3] | 100150246 | YES | YES | Plasma Membrane | G-protein coupled receptor | |||||
| ENSDARG00000031712 | grm3 | glutamate receptor, metabotropic 3 [Source:ZFIN;Acc:ZDB-GENE-061009-13] | 565256 | YES | YES | Plasma Membrane | G-protein coupled receptor | |||||
| ENSDARG00000056510 | gstk1 | glutathione S-transferase kappa 1 [Source:ZFIN;Acc:ZDB-GENE-040718-298] | 436833 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000104068 | gstp1 | glutathione S-transferase pi 1 [Source:ZFIN;Acc:ZDB-GENE-020806-4] | 79381 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000093185 | GUCY1A2 | guanylate cyclase 1, soluble, alpha 2 [Source:HGNC Symbol;Acc:HGNC:4684] | 0 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000014806 | hacd2 | 3-hydroxyacyl-CoA dehydratase 2 [Source:ZFIN;Acc:ZDB-GENE-030131-6053] | 334121 | YES | YES | Cytoplasm | phosphatase | |||||
| ENSDARG00000097011 | hbaa1 | hemoglobin, alpha adult 1 [Source:ZFIN;Acc:ZDB-GENE-980526-79] | 30507 | YES | YES | NA | NA | |||||
| ENSDARG00000104480 | hcn1 | hyperpolarization activated cyclic nucleotide-gated potassium channel 1 [Source:ZFIN;Acc:ZDB-GENE-110520-1] | 0 | YES | YES | Plasma Membrane | ion channel | |||||
| ENSDARG00000027192 | hcn3 | hyperpolarization activated cyclic nucleotide-gated potassium channel 3 [Source:ZFIN;Acc:ZDB-GENE-060503-193] | 560709 | YES | YES | Plasma Membrane | ion channel | |||||
| ENSDARG00000077945 | HEPACAM (2 of 2) | hepatic and glial cell adhesion molecule [Source:HGNC Symbol;Acc:HGNC:26361] | 0 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000022303 | higd1a | HIG1 hypoxia inducible domain family, member 1A [Source:ZFIN;Acc:ZDB-GENE-030826-15] | 373084 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000102975 | hmp19 | HMP19 protein [Source:ZFIN;Acc:ZDB-GENE-030131-3694] | 324971 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000018397 | hpca | hippocalcin [Source:ZFIN;Acc:ZDB-GENE-040426-1683] | 393696 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000002523 | hsdl2 | hydroxysteroid dehydrogenase like 2 [Source:ZFIN;Acc:ZDB-GENE-030131-1066] | 322347 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000024746 | hsp90aa1.2 | heat shock protein 90, alpha (cytosolic), class A member 1, tandem duplicate 2 [Source:ZFIN;Acc:ZDB-GENE-031001-3] | 565155 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000029150 | hsp90ab1 | heat shock protein 90, alpha (cytosolic), class B member 1 [Source:ZFIN;Acc:ZDB-GENE-990415-95] | 30573 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000003570 | hsp90b1 | heat shock protein 90, beta (grp94), member 1 [Source:ZFIN;Acc:ZDB-GENE-031002-1] | 386590 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000018989 | hspa4b | heat shock protein 4b [Source:ZFIN;Acc:ZDB-GENE-030131-6018] | 334086 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000053544 | hspa4l | heat shock protein 4 like [Source:ZFIN;Acc:ZDB-GENE-030131-2412] | 562019 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000103846 | hspa5 | heat shock protein 5 [Source:ZFIN;Acc:ZDB-GENE-031001-11] | 378848 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000003035 | hspa9 | heat shock protein 9 [Source:ZFIN;Acc:ZDB-GENE-030828-12] | 373085 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000041065 | hspb1 | heat shock protein, alpha-crystallin-related, 1 [Source:ZFIN;Acc:ZDB-GENE-030326-4] | 368243 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000056160 | hspd1 | heat shock 60 protein 1 [Source:ZFIN;Acc:ZDB-GENE-021206-1] | 282676 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000052866 | htt | huntingtin [Source:ZFIN;Acc:ZDB-GENE-990415-131] | 30214 | YES | YES | Cytoplasm | transcription regulator | |||||
| ENSDARG00000016782 | huwe1 | HECT, UBA and WWE domain containing 1 [Source:ZFIN;Acc:ZDB-GENE-081104-387] | 0 | YES | YES | Nucleus | transcription regulator | |||||
| ENSDARG00000025375 | idh1 | isocitrate dehydrogenase 1 (NADP+), soluble [Source:ZFIN;Acc:ZDB-GENE-031006-1] | 100006589 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000003795 | IDH2 | isocitrate dehydrogenase 2 (NADP+), mitochondrial [Source:HGNC Symbol;Acc:HGNC:5383] | 0 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000030278 | idh3a | isocitrate dehydrogenase 3 (NAD+) alpha [Source:ZFIN;Acc:ZDB-GENE-040426-1007] | 393926 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000044753 | idh3b | isocitrate dehydrogenase 3 (NAD+) beta [Source:ZFIN;Acc:ZDB-GENE-040625-174] | 415247 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000023648 | idh3g | isocitrate dehydrogenase 3 (NAD+) gamma [Source:ZFIN;Acc:ZDB-GENE-050417-435] | 550579 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000002184 | igbp1 | immunoglobulin (CD79A) binding protein 1 [Source:ZFIN;Acc:ZDB-GENE-040426-2928] | 406843 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000077002 | igsf3 | immunoglobulin superfamily, member 3 [Source:ZFIN;Acc:ZDB-GENE-070912-243] | 557878 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000038467 | igsf8 | immunoglobulin superfamily, member 8 [Source:ZFIN;Acc:ZDB-GENE-040426-2444] | 0 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000102874 | immt | inner membrane protein, mitochondrial (mitofilin) [Source:ZFIN;Acc:ZDB-GENE-030131-5417] | 327206 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000075201 | inpp4b | inositol polyphosphate-4-phosphatase, type II [Source:ZFIN;Acc:ZDB-GENE-090312-97] | 560006 | YES | YES | Cytoplasm | phosphatase | |||||
| ENSDARG00000013371 | isoc2 | isochorismatase domain containing 2 [Source:ZFIN;Acc:ZDB-GENE-070518-1] | 492617 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000075584 | itfg1 | integrin alpha FG-GAP repeat containing 1 [Source:ZFIN;Acc:ZDB-GENE-121003-3] | 100331534 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000000086 | itsn1 | intersectin 1 (SH3 domain protein) [Source:ZFIN;Acc:ZDB-GENE-030616-226] | 368504 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000059473 | kank4 | KN motif and ankyrin repeat domains 4 [Source:ZFIN;Acc:ZDB-GENE-120423-1] | 101884737 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000055855 | kcnc3a | potassium voltage-gated channel, Shaw-related subfamily, member 3a [Source:ZFIN;Acc:ZDB-GENE-100901-1] | 559096 | YES | YES | Plasma Membrane | ion channel | |||||
| ENSDARG00000053542 | kctd12.2 | potassium channel tetramerisation domain containing 12.2 [Source:ZFIN;Acc:ZDB-GENE-060117-4] | 553403 | YES | YES | NA | NA | |||||
| ENSDARG00000017338 | kidins220b | kinase D-interacting substrate 220b [Source:ZFIN;Acc:ZDB-GENE-030131-7824] | 335881 | YES | YES | Nucleus | transcription regulator | |||||
| ENSDARG00000103394 | kif5bb | kinesin family member 5B, b [Source:ZFIN;Acc:ZDB-GENE-070629-4] | 334108 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000076027 | kif5c | kinesin family member 5C [Source:ZFIN;Acc:ZDB-GENE-081105-14] | 565563 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000104889 | kpnb1 | karyopherin (importin) beta 1 [Source:ZFIN;Acc:ZDB-GENE-030131-2579] | 570700 | YES | YES | Nucleus | transporter | |||||
| ENSDARG00000032802 | ktn1 | kinectin 1 [Source:ZFIN;Acc:ZDB-GENE-030408-4] | 368207 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000057075 | lamtor3 | late endosomal/lysosomal adaptor, MAPK and MTOR activator 3 [Source:ZFIN;Acc:ZDB-GENE-050522-345] | 553597 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000019280 | larsb | leucyl-tRNA synthetase b [Source:ZFIN;Acc:ZDB-GENE-030114-7] | 569778 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000101251 | ldha | lactate dehydrogenase A4 [Source:ZFIN;Acc:ZDB-GENE-991026-5] | 30496 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000019644 | ldhba | lactate dehydrogenase Ba [Source:ZFIN;Acc:ZDB-GENE-991026-6] | 30497 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000056978 | letm1 | leucine zipper-EF-hand containing transmembrane protein 1 [Source:ZFIN;Acc:ZDB-GENE-050522-154] | 570745 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000054942 | lgals2a | lectin, galactoside-binding, soluble, 2a [Source:ZFIN;Acc:ZDB-GENE-050318-2] | 405830 | YES | YES | Extracellular Space | other | |||||
| ENSDARG00000035899 | lingo1b | leucine rich repeat and Ig domain containing 1b [Source:ZFIN;Acc:ZDB-GENE-040912-136] | 447837 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000075625 | lrig1 | leucine-rich repeats and immunoglobulin-like domains 1 [Source:ZFIN;Acc:ZDB-GENE-031113-23] | 560325 | YES | YES | Extracellular Space | other | |||||
| ENSDARG00000029146 | lrp1ab | low density lipoprotein receptor-related protein 1Ab [Source:ZFIN;Acc:ZDB-GENE-030131-7126] | 565797 | YES | YES | Plasma Membrane | transmembrane receptor | |||||
| ENSDARG00000043970 | lrpprc | leucine-rich pentatricopeptide repeat containing [Source:ZFIN;Acc:ZDB-GENE-030131-7887] | 557195 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000004597 | lrrc4ba | leucine rich repeat containing 4Ba [Source:ZFIN;Acc:ZDB-GENE-080327-25] | 556747 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000014792 | lrrc4bb | leucine rich repeat containing 4Bb [Source:ZFIN;Acc:ZDB-GENE-091116-44] | 100535350 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000016739 | LRRC4C | leucine rich repeat containing 4C [Source:HGNC Symbol;Acc:HGNC:29317] | 564462 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000053656 | lypla2 | lysophospholipase II [Source:ZFIN;Acc:ZDB-GENE-040426-1715] | 393722 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000029609 | macrod1 | MACRO domain containing 1 [Source:ZFIN;Acc:ZDB-GENE-040912-100] | 447834 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000003495 | madd | MAP-kinase activating death domain [Source:ZFIN;Acc:ZDB-GENE-030131-8076] | 100150917 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000023712 | mao | monoamine oxidase [Source:ZFIN;Acc:ZDB-GENE-040329-3] | 404730 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000027552 | mapk1 | mitogen-activated protein kinase 1 [Source:ZFIN;Acc:ZDB-GENE-030722-2] | 360144 | YES | YES | Cytoplasm | kinase | |||||
| ENSDARG00000020231 | mapre3a | microtubule-associated protein, RP/EB family, member 3a [Source:ZFIN;Acc:ZDB-GENE-050913-88] | 559217 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000102878 | mapre3b | microtubule-associated protein, RP/EB family, member 3b [Source:ZFIN;Acc:ZDB-GENE-040704-6] | 431717 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000032552 | march5 | membrane-associated ring finger (C3HC4) 5 [Source:ZFIN;Acc:ZDB-GENE-070424-47] | 561952 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000035715 | marcksl1b | MARCKS-like 1b [Source:ZFIN;Acc:ZDB-GENE-040426-2315] | 406504 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000102538 | mccc1 | methylcrotonoyl-CoA carboxylase 1 (alpha) [Source:ZFIN;Acc:ZDB-GENE-050208-450] | 777632 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000017571 | mccc2 | methylcrotonoyl-CoA carboxylase 2 (beta) [Source:ZFIN;Acc:ZDB-GENE-040426-2493] | 405863 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000101175 | mcu | mitochondrial calcium uniporter [Source:ZFIN;Acc:ZDB-GENE-061013-24] | 768182 | YES | YES | Cytoplasm | ion channel | |||||
| ENSDARG00000017772 | mdh1aa | malate dehydrogenase 1Aa, NAD (soluble) [Source:ZFIN;Acc:ZDB-GENE-040204-1] | 399662 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000043371 | mdh2 | malate dehydrogenase 2, NAD (mitochondrial) [Source:ZFIN;Acc:ZDB-GENE-040426-2143] | 406405 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000032326 | mecr | mitochondrial trans-2-enoyl-CoA reductase [Source:ZFIN;Acc:ZDB-GENE-050417-399] | 550550 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000053753 | mff | mitochondrial fission factor [Source:ZFIN;Acc:ZDB-GENE-040426-1510] | 393508 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000039203 | MFF (1 of 2) | mitochondrial fission factor [Source:HGNC Symbol;Acc:HGNC:24858] | 553588 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000079504 | mfn2 | mitofusin 2 [Source:ZFIN;Acc:ZDB-GENE-081105-44] | 567448 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000071336 | mif | macrophage migration inhibitory factor [Source:ZFIN;Acc:ZDB-GENE-080708-1] | 751093 | YES | YES | Extracellular Space | cytokine | |||||
| ENSDARG00000101619 | minos1 | mitochondrial inner membrane organizing system 1 [Source:ZFIN;Acc:ZDB-GENE-060825-325] | 751699 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000059630 | mlec | malectin [Source:ZFIN;Acc:ZDB-GENE-081105-187] | 569613 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000104732 | mlycd | malonyl-CoA decarboxylase [Source:ZFIN;Acc:ZDB-GENE-070410-120] | 100037349 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000012953 | mob1bb | MOB kinase activator 1Bb [Source:ZFIN;Acc:ZDB-GENE-050522-58] | 407654 | YES | YES | NA | NA | |||||
| ENSDARG00000010957 | mpp2b | membrane protein, palmitoylated 2b (MAGUK p55 subfamily member 2) [Source:ZFIN;Acc:ZDB-GENE-040704-70] | 431770 | YES | YES | Plasma Membrane | kinase | |||||
| ENSDARG00000013075 | mrpl11 | mitochondrial ribosomal protein L11 [Source:ZFIN;Acc:ZDB-GENE-040718-294] | 436829 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000058128 | msna | moesin a [Source:ZFIN;Acc:ZDB-GENE-021211-2] | 286739 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000063910 | mt-atp8 | ATP synthase 8, mitochondrial [Source:ZFIN;Acc:ZDB-GENE-011205-19] | 140520 | YES | YES | NA | NA | |||||
| ENSDARG00000063908 | mt-co2 | cytochrome c oxidase II, mitochondrial [Source:ZFIN;Acc:ZDB-GENE-011205-15] | 140540 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000063921 | mt-nd5 | NADH dehydrogenase 5, mitochondrial [Source:ZFIN;Acc:ZDB-GENE-011205-12] | 140535 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000019732 | mtch2 | mitochondrial carrier homolog 2 [Source:ZFIN;Acc:ZDB-GENE-991008-9] | 30655 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000004939 | mtdhb | metadherin b [Source:ZFIN;Acc:ZDB-GENE-090929-2] | 553275 | YES | YES | Cytoplasm | transcription regulator | |||||
| ENSDARG00000099908 | MTFP1 | mitochondrial fission process 1 [Source:HGNC Symbol;Acc:HGNC:26945] | 0 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000010601 | mtmr10 | myotubularin related protein 10 [Source:ZFIN;Acc:ZDB-GENE-061103-52] | 555239 | YES | YES | Other | other | |||||
| ENSDARG00000025500 | mtx1a | metaxin 1a [Source:ZFIN;Acc:ZDB-GENE-060606-2] | 556917 | YES | YES | Cytoplasm | transporter | |||||
| ENSDARG00000029415 | mtx2 | metaxin 2 [Source:ZFIN;Acc:ZDB-GENE-021210-2] | 286741 | YES | YES | Cytoplasm | transporter | |||||
| ENSDARG00000090195 | mtx3 | metaxin 3 [Source:ZFIN;Acc:ZDB-GENE-021210-3] | 402916 | YES | YES | Other | other | |||||
| ENSDARG00000060554 | mut | methylmalonyl CoA mutase [Source:ZFIN;Acc:ZDB-GENE-010430-3] | 569581 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000015025 | nadl1.1 | neural adhesion molecule L1.1 [Source:ZFIN;Acc:ZDB-GENE-980526-512] | 30656 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000007149 | nadl1.2 | neural adhesion molecule L1.2 [Source:ZFIN;Acc:ZDB-GENE-990415-10] | 30634 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000070560 | nap1l4a | nucleosome assembly protein 1-like 4a [Source:ZFIN;Acc:ZDB-GENE-030131-9099] | 337155 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000068868 | nap1l4b | nucleosome assembly protein 1-like 4b [Source:ZFIN;Acc:ZDB-GENE-041114-168] | 492812 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000013669 | napba | N-ethylmaleimide-sensitive factor attachment protein, beta a [Source:ZFIN;Acc:ZDB-GENE-050522-230] | 553682 | YES | YES | Cytoplasm | transporter | |||||
| ENSDARG00000043012 | napgb | N-ethylmaleimide-sensitive factor attachment protein, gamma b [Source:ZFIN;Acc:ZDB-GENE-040718-171] | 436742 | YES | YES | Cytoplasm | transporter | |||||
| ENSDARG00000061100 | nars | asparaginyl-tRNA synthetase [Source:ZFIN;Acc:ZDB-GENE-030131-1016] | 568332 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000070080 | nbeaa | neurobeachin a [Source:ZFIN;Acc:ZDB-GENE-050320-68] | 541373 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000056181 | ncam1a | neural cell adhesion molecule 1a [Source:ZFIN;Acc:ZDB-GENE-990415-31] | 30447 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000007220 | ncam1b | neural cell adhesion molecule 1b [Source:ZFIN;Acc:ZDB-GENE-010822-2] | 114442 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000017466 | ncam2 | neural cell adhesion molecule 2 [Source:ZFIN;Acc:ZDB-GENE-010822-1] | 114441 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000098523 | NCAM2 (2 of 2) | neural cell adhesion molecule 2 [Source:HGNC Symbol;Acc:HGNC:7657] | 767777 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000058861 | ncl1 | nicalin [Source:ZFIN;Acc:ZDB-GENE-030131-8788] | 336844 | YES | YES | Cytoplasm | peptidase | |||||
| ENSDARG00000010052 | ndrg3b | ndrg family member 3b [Source:ZFIN;Acc:ZDB-GENE-030131-5606] | 327395 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000013333 | ndufa10 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 10 [Source:ZFIN;Acc:ZDB-GENE-030131-670] | 0 | YES | YES | Cytoplasm | transporter | |||||
| ENSDARG00000056108 | ndufa4 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 4 [Source:ZFIN;Acc:ZDB-GENE-040426-1962] | 406298 | YES | YES | NA | NA | |||||
| ENSDARG00000058041 | ndufa8 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 8 [Source:ZFIN;Acc:ZDB-GENE-040426-1688] | 393700 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000070723 | ndufa9a | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 9a [Source:ZFIN;Acc:ZDB-GENE-050320-20] | 541331 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000077859 | ndufaf4 | NADH dehydrogenase (ubiquinone) complex I, assembly factor 4 [Source:ZFIN;Acc:ZDB-GENE-060810-114] | 794169 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000028889 | ndufb10 | NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 10 [Source:ZFIN;Acc:ZDB-GENE-040426-1691] | 393703 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000033789 | ndufb7 | NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 7 [Source:ZFIN;Acc:ZDB-GENE-040426-1886] | 393821 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000028546 | ndufs1 | NADH dehydrogenase (ubiquinone) Fe-S protein 1 [Source:ZFIN;Acc:ZDB-GENE-030131-478] | 493605 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000074552 | ndufs7 | NADH dehydrogenase (ubiquinone) Fe-S protein 7, (NADH-coenzyme Q reductase) [Source:ZFIN;Acc:ZDB-GENE-041111-261] | 796997 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000051986 | ndufs8a | NADH dehydrogenase (ubiquinone) Fe-S protein 8a [Source:ZFIN;Acc:ZDB-GENE-040426-2153] | 406413 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000036438 | ndufv1 | NADH dehydrogenase (ubiquinone) flavoprotein 1 [Source:ZFIN;Acc:ZDB-GENE-040808-73] | 445291 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000061099 | nfasca | neurofascin homolog (chicken) a [Source:ZFIN;Acc:ZDB-GENE-080229-6] | 100141490 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000076179 | ngef | neuronal guanine nucleotide exchange factor [Source:ZFIN;Acc:ZDB-GENE-130530-794] | 569090 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000020387 | nipsnap3a | nipsnap homolog 3A (C. elegans) [Source:ZFIN;Acc:ZDB-GENE-040426-1037] | 393234 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000104786 | nlgn3a | neuroligin 3a [Source:ZFIN;Acc:ZDB-GENE-071219-1] | 562702 | YES | YES | Plasma Membrane | enzyme | |||||
| ENSDARG00000079455 | nlgn4a | neuroligin 4a [Source:ZFIN;Acc:ZDB-GENE-100309-2] | 561122 | YES | YES | NA | NA | |||||
| ENSDARG00000100990 | nme3 | NME/NM23 nucleoside diphosphate kinase 3 [Source:ZFIN;Acc:ZDB-GENE-000210-34] | 30085 | YES | YES | Cytoplasm | kinase | |||||
| ENSDARG00000023536 | nnt | nicotinamide nucleotide transhydrogenase [Source:ZFIN;Acc:ZDB-GENE-040426-2592] | 406619 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000078592 | nomo | nodal modulator [Source:ZFIN;Acc:ZDB-GENE-040826-3] | 446115 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000012871 | npepl1 | aminopeptidase-like 1 [Source:ZFIN;Acc:ZDB-GENE-050417-177] | 550383 | YES | YES | Nucleus | peptidase | |||||
| ENSDARG00000044943 | npepps | aminopeptidase puromycin sensitive [Source:ZFIN;Acc:ZDB-GENE-060524-3] | 407726 | YES | YES | Cytoplasm | peptidase | |||||
| ENSDARG00000074671 | nptx1l | neuronal pentraxin 1 like [Source:ZFIN;Acc:ZDB-GENE-040426-2418] | 405872 | YES | YES | Extracellular Space | other | |||||
| ENSDARG00000006396 | nrcama | neuronal cell adhesion molecule a [Source:ZFIN;Acc:ZDB-GENE-041210-235] | 556537 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000102153 | nrp1a | neuropilin 1a [Source:ZFIN;Acc:ZDB-GENE-030519-2] | 353246 | YES | YES | Plasma Membrane | transmembrane receptor | |||||
| ENSDARG00000061454 | nrxn2a | neurexin 2a [Source:ZFIN;Acc:ZDB-GENE-070206-5] | 558326 | YES | YES | NA | NA | |||||
| ENSDARG00000007654 | nsfa | N-ethylmaleimide-sensitive factor a [Source:ZFIN;Acc:ZDB-GENE-030616-37] | 368483 | YES | YES | Cytoplasm | transporter | |||||
| ENSDARG00000038991 | nsfb | N-ethylmaleimide-sensitive factor b [Source:ZFIN;Acc:ZDB-GENE-050808-1] | 554200 | YES | YES | Cytoplasm | transporter | |||||
| ENSDARG00000062949 | nt5dc3 | 5'-nucleotidase domain containing 3 [Source:ZFIN;Acc:ZDB-GENE-080523-3] | 558469 | YES | YES | Other | other | |||||
| ENSDARG00000022531 | ntn1b | netrin 1b [Source:ZFIN;Acc:ZDB-GENE-990415-168] | 30192 | YES | YES | Extracellular Space | other | |||||
| ENSDARG00000052334 | ociad1 | OCIA domain containing 1 [Source:ZFIN;Acc:ZDB-GENE-040426-1100] | 393272 | YES | YES | NA | NA | |||||
| ENSDARG00000044565 | ola1 | Obg-like ATPase 1 [Source:ZFIN;Acc:ZDB-GENE-030131-5063] | 326864 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000070801 | opa1 | optic atrophy 1 (autosomal dominant) [Source:ZFIN;Acc:ZDB-GENE-041114-7] | 492332 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000013063 | oxct1a | 3-oxoacid CoA transferase 1a [Source:ZFIN;Acc:ZDB-GENE-040723-3] | 442923 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000105116 | p4hb | prolyl 4-hydroxylase, beta polypeptide [Source:ZFIN;Acc:ZDB-GENE-080610-1] | 406673 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000070657 | pa2g4b | proliferation-associated 2G4, b [Source:ZFIN;Acc:ZDB-GENE-030131-2182] | 323462 | YES | YES | Nucleus | transcription regulator | |||||
| ENSDARG00000032865 | pacsin1a | protein kinase C and casein kinase substrate in neurons 1a [Source:ZFIN;Acc:ZDB-GENE-050522-155] | 553559 | YES | YES | Cytoplasm | kinase | |||||
| ENSDARG00000007818 | palm1b | paralemmin 1b [Source:ZFIN;Acc:ZDB-GENE-040426-1147] | 393573 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000099039 | pcbp2 | poly(rC) binding protein 2 [Source:ZFIN;Acc:ZDB-GENE-030131-563] | 321844 | YES | YES | Nucleus | other | |||||
| ENSDARG00000038910 | pccb | propionyl CoA carboxylase, beta polypeptide [Source:ZFIN;Acc:ZDB-GENE-040426-2467] | 405861 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000027041 | pcdh17 | protocadherin 17 [Source:ZFIN;Acc:ZDB-GENE-090608-3] | 561336 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000104826 | pcdh1gc5 | protocadherin 1 gamma c 5 [Source:ZFIN;Acc:ZDB-GENE-041118-16] | 503571 | YES | YES | Extracellular Space | other | |||||
| ENSDARG00000078898 | pcdh7a | protocadherin 7a [Source:ZFIN;Acc:ZDB-GENE-120206-1] | 568198 | YES | YES | NA | NA | |||||
| ENSDARG00000060610 | pcdh7b | protocadherin 7b [Source:ZFIN;Acc:ZDB-GENE-081104-299] | 555504 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000015201 | pcmt | protein-L-isoaspartate (D-aspartate) O-methyltransferase [Source:ZFIN;Acc:ZDB-GENE-990415-134] | 30751 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000002600 | pcsk1 | proprotein convertase subtilisin/kexin type 1 [Source:ZFIN;Acc:ZDB-GENE-071009-1] | 100005716 | YES | YES | Cytoplasm | peptidase | |||||
| ENSDARG00000076170 | pcsk1nl | proprotein convertase subtilisin/kexin type 1 inhibitor, like [Source:ZFIN;Acc:ZDB-GENE-090427-2] | 570305 | YES | YES | NA | NA | |||||
| ENSDARG00000098766 | pcxa | pyruvate carboxylase a [Source:ZFIN;Acc:ZDB-GENE-090908-3] | 0 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000021346 | pdhb | pyruvate dehydrogenase (lipoamide) beta [Source:ZFIN;Acc:ZDB-GENE-040426-2173] | 406428 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000018491 | pdia4 | protein disulfide isomerase family A, member 4 [Source:ZFIN;Acc:ZDB-GENE-030131-5493] | 554998 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000054191 | pgk1 | phosphoglycerate kinase 1 [Source:ZFIN;Acc:ZDB-GENE-040426-2711] | 406696 | YES | YES | Cytoplasm | kinase | |||||
| ENSDARG00000054172 | pgrmc1 | progesterone receptor membrane component 1 [Source:ZFIN;Acc:ZDB-GENE-041114-91] | 492520 | YES | YES | Plasma Membrane | transmembrane receptor | |||||
| ENSDARG00000077596 | PHF24 | PHD finger protein 24 [Source:HGNC Symbol;Acc:HGNC:29180] | 558943 | YES | YES | Other | other | |||||
| ENSDARG00000078284 | phkb | phosphorylase kinase, beta [Source:ZFIN;Acc:ZDB-GENE-110411-149] | 569706 | YES | YES | Cytoplasm | kinase | |||||
| ENSDARG00000012866 | picalma | phosphatidylinositol binding clathrin assembly protein a [Source:ZFIN;Acc:ZDB-GENE-040426-761] | 393901 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000014137 | picalmb | phosphatidylinositol binding clathrin assembly protein b [Source:ZFIN;Acc:ZDB-GENE-030131-6795] | 445286 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000017813 | pigs | phosphatidylinositol glycan anchor biosynthesis, class S [Source:ZFIN;Acc:ZDB-GENE-060929-436] | 767675 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000060469 | pik3r4 | phosphoinositide-3-kinase, regulatory subunit 4 [Source:ZFIN;Acc:ZDB-GENE-050309-84] | 564088 | YES | YES | Cytoplasm | kinase | |||||
| ENSDARG00000101915 | PIN1 | peptidylprolyl cis/trans isomerase, NIMA-interacting 1 [Source:HGNC Symbol;Acc:HGNC:8988] | 0 | YES | YES | Other | other | |||||
| ENSDARG00000063544 | pip4k2ab | phosphatidylinositol-5-phosphate 4-kinase, type II, alpha b [Source:ZFIN;Acc:ZDB-GENE-070912-272] | 555920 | YES | YES | Cytoplasm | kinase | |||||
| ENSDARG00000076001 | pip5k1ca | phosphatidylinositol-4-phosphate 5-kinase, type I, gamma a [Source:ZFIN;Acc:ZDB-GENE-050208-358] | 798876 | YES | YES | Plasma Membrane | kinase | |||||
| ENSDARG00000068246 | plcb3 | phospholipase C, beta 3 (phosphatidylinositol-specific) [Source:ZFIN;Acc:ZDB-GENE-030616-594] | 100001532 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000100786 | PLCB4 | phospholipase C, beta 4 [Source:HGNC Symbol;Acc:HGNC:9059] | 100537538 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000007172 | plxna3 | plexin A3 [Source:ZFIN;Acc:ZDB-GENE-070613-1] | 567422 | YES | YES | Plasma Membrane | transmembrane receptor | |||||
| ENSDARG00000019328 | plxna4 | plexin A4 [Source:ZFIN;Acc:ZDB-GENE-030131-4663] | 325938 | YES | YES | Plasma Membrane | transmembrane receptor | |||||
| ENSDARG00000017211 | plxnb1b | plexin b1b [Source:ZFIN;Acc:ZDB-GENE-090812-4] | 570222 | YES | YES | Plasma Membrane | transmembrane receptor | |||||
| ENSDARG00000003811 | plxnb2a | plexin b2a [Source:ZFIN;Acc:ZDB-GENE-030131-8917] | 561433 | YES | YES | Plasma Membrane | transmembrane receptor | |||||
| ENSDARG00000010773 | pnpla6 | patatin-like phospholipase domain containing 6 [Source:ZFIN;Acc:ZDB-GENE-050107-4] | 560986 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000059933 | ppap2b | phosphatidic acid phosphatase type 2B [Source:ZFIN;Acc:ZDB-GENE-060526-241] | 557680 | YES | YES | Plasma Membrane | phosphatase | |||||
| ENSDARG00000009212 | ppiaa | peptidylprolyl isomerase Aa (cyclophilin A) [Source:ZFIN;Acc:ZDB-GENE-030131-8556] | 336612 | YES | YES | NA | NA | |||||
| ENSDARG00000103994 | ppiab | peptidylprolyl isomerase Ab (cyclophilin A) [Source:ZFIN;Acc:ZDB-GENE-030131-7459] | 335519 | YES | YES | NA | NA | |||||
| ENSDARG00000092798 | ppib | peptidylprolyl isomerase B (cyclophilin B) [Source:ZFIN;Acc:ZDB-GENE-040426-1955] | 406292 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000026499 | ppm1e | protein phosphatase, Mg2+/Mn2+ dependent, 1E [Source:ZFIN;Acc:ZDB-GENE-070326-2] | 553417 | YES | YES | Nucleus | phosphatase | |||||
| ENSDARG00000063684 | ppm1h | protein phosphatase, Mg2+/Mn2+ dependent, 1H [Source:ZFIN;Acc:ZDB-GENE-061027-190] | 768291 | YES | YES | Other | phosphatase | |||||
| ENSDARG00000009740 | ppp1r7 | protein phosphatase 1, regulatory (inhibitor) subunit 7 [Source:ZFIN;Acc:ZDB-GENE-051113-288] | 560323 | YES | YES | Nucleus | phosphatase | |||||
| ENSDARG00000102009 | ppp2r2d | protein phosphatase 2, regulatory subunit B, delta [Source:ZFIN;Acc:ZDB-GENE-040426-2086] | 0 | YES | YES | Nucleus | phosphatase | |||||
| ENSDARG00000015474 | ppp2r5ea | protein phosphatase 2, regulatory subunit B', epsilon isoform a [Source:ZFIN;Acc:ZDB-GENE-030904-7] | 373881 | YES | YES | Cytoplasm | phosphatase | |||||
| ENSDARG00000004988 | ppp3ca | protein phosphatase 3, catalytic subunit, alpha isozyme [Source:ZFIN;Acc:ZDB-GENE-091113-24] | 794574 | YES | YES | Cytoplasm | phosphatase | |||||
| ENSDARG00000025106 | ppp3cb | protein phosphatase 3, catalytic subunit, beta isozyme [Source:ZFIN;Acc:ZDB-GENE-050417-406] | 550554 | YES | YES | Cytoplasm | phosphatase | |||||
| ENSDARG00000014962 | ppp3cca | protein phosphatase 3, catalytic subunit, gamma isozyme, a [Source:ZFIN;Acc:ZDB-GENE-030829-36] | 557926 | YES | YES | Cytoplasm | phosphatase | |||||
| ENSDARG00000057456 | ppp3ccb | protein phosphatase 3, catalytic subunit, gamma isozyme, b [Source:ZFIN;Acc:ZDB-GENE-070112-1102] | 565241 | YES | YES | Cytoplasm | phosphatase | |||||
| ENSDARG00000100349 | prkacaa | protein kinase, cAMP-dependent, catalytic, alpha, genome duplicate a [Source:ZFIN;Acc:ZDB-GENE-050114-6] | 564064 | YES | YES | Cytoplasm | kinase | |||||
| ENSDARG00000059125 | prkacbb | protein kinase, cAMP-dependent, catalytic, beta b [Source:ZFIN;Acc:ZDB-GENE-050904-4] | 563147 | YES | YES | Cytoplasm | kinase | |||||
| ENSDARG00000033184 | prkar2aa | protein kinase, cAMP-dependent, regulatory, type II, alpha A [Source:ZFIN;Acc:ZDB-GENE-030131-1007] | 556042 | YES | YES | Cytoplasm | kinase | |||||
| ENSDARG00000039241 | prkca | protein kinase C, alpha [Source:ZFIN;Acc:ZDB-GENE-050208-100] | 497384 | YES | YES | Cytoplasm | kinase | |||||
| ENSDARG00000008723 | prkcba | protein kinase C, beta a [Source:ZFIN;Acc:ZDB-GENE-040426-1354] | 100006675 | YES | YES | Cytoplasm | kinase | |||||
| ENSDARG00000022254 | prkcbb | protein kinase C, beta b [Source:ZFIN;Acc:ZDB-GENE-040426-1178] | 393953 | YES | YES | Cytoplasm | kinase | |||||
| ENSDARG00000003008 | prkcea | protein kinase C, epsilon a [Source:ZFIN;Acc:ZDB-GENE-030131-8191] | 100005075 | YES | YES | Cytoplasm | kinase | |||||
| ENSDARG00000039966 | prom1a | prominin 1a [Source:ZFIN;Acc:ZDB-GENE-030131-1577] | 322857 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000019326 | prpsap2 | phosphoribosyl pyrophosphate synthetase-associated protein 2 [Source:ZFIN;Acc:ZDB-GENE-040426-2077] | 406369 | YES | YES | Other | other | |||||
| ENSDARG00000101560 | psma1 | proteasome (prosome, macropain) subunit, alpha type, 1 [Source:ZFIN;Acc:ZDB-GENE-040801-15] | 445033 | YES | YES | Cytoplasm | peptidase | |||||
| ENSDARG00000086618 | psma3 | proteasome (prosome, macropain) subunit, alpha type, 3 [Source:ZFIN;Acc:ZDB-GENE-050913-120] | 564370 | YES | YES | Cytoplasm | peptidase | |||||
| ENSDARG00000003526 | psma5 | proteasome (prosome, macropain) subunit, alpha type, 5 [Source:ZFIN;Acc:ZDB-GENE-040625-96] | 403011 | YES | YES | Cytoplasm | peptidase | |||||
| ENSDARG00000009640 | psmb1 | proteasome (prosome, macropain) subunit, beta type, 1 [Source:ZFIN;Acc:ZDB-GENE-040618-2] | 445413 | YES | YES | Cytoplasm | peptidase | |||||
| ENSDARG00000031511 | psmb2 | proteasome (prosome, macropain) subunit, beta type, 2 [Source:ZFIN;Acc:ZDB-GENE-040718-353] | 436882 | YES | YES | Cytoplasm | peptidase | |||||
| ENSDARG00000013938 | psmb3 | proteasome (prosome, macropain) subunit, beta type, 3 [Source:ZFIN;Acc:ZDB-GENE-040426-2682] | 573095 | YES | YES | Other | other | |||||
| ENSDARG00000075445 | psmb5 | proteasome (prosome, macropain) subunit, beta type, 5 [Source:ZFIN;Acc:ZDB-GENE-990415-215] | 30387 | YES | YES | Cytoplasm | peptidase | |||||
| ENSDARG00000002240 | psmb6 | proteasome (prosome, macropain) subunit, beta type, 6 [Source:ZFIN;Acc:ZDB-GENE-990415-216] | 30388 | YES | YES | Nucleus | peptidase | |||||
| ENSDARG00000020101 | psmc2 | proteasome (prosome, macropain) 26S subunit, ATPase 2 [Source:ZFIN;Acc:ZDB-GENE-040426-1327] | 393941 | YES | YES | Nucleus | peptidase | |||||
| ENSDARG00000007141 | psmc3 | proteasome (prosome, macropain) 26S subunit, ATPase, 3 [Source:ZFIN;Acc:ZDB-GENE-030131-666] | 321947 | YES | YES | Nucleus | transcription regulator | |||||
| ENSDARG00000027099 | psmc4 | proteasome (prosome, macropain) 26S subunit, ATPase, 4 [Source:ZFIN;Acc:ZDB-GENE-030131-5083] | 326884 | YES | YES | Nucleus | peptidase | |||||
| ENSDARG00000015315 | psmc5 | proteasome (prosome, macropain) 26S subunit, ATPase, 5 [Source:ZFIN;Acc:ZDB-GENE-030131-6547] | 445285 | YES | YES | Nucleus | transcription regulator | |||||
| ENSDARG00000060150 | psmd11a | proteasome (prosome, macropain) 26S subunit, non-ATPase, 11a [Source:ZFIN;Acc:ZDB-GENE-030131-2711] | 503934 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000016239 | psmd13 | proteasome (prosome, macropain) 26S subunit, non-ATPase, 13 [Source:ZFIN;Acc:ZDB-GENE-040426-1004] | 393922 | YES | YES | Cytoplasm | peptidase | |||||
| ENSDARG00000099653 | psmd2 | proteasome (prosome, macropain) 26S subunit, non-ATPase, 2 [Source:ZFIN;Acc:ZDB-GENE-040426-1480] | 0 | YES | YES | Other | other | |||||
| ENSDARG00000018124 | psmd3 | proteasome (prosome, macropain) 26S subunit, non-ATPase, 3 [Source:ZFIN;Acc:ZDB-GENE-040426-1444] | 393544 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000070674 | psmd6 | proteasome (prosome, macropain) 26S subunit, non-ATPase, 6 [Source:ZFIN;Acc:ZDB-GENE-040426-1038] | 393261 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000102417 | psmd7 | proteasome (prosome, macropain) 26S subunit, non-ATPase, 7 (Mov34 homolog) [Source:ZFIN;Acc:ZDB-GENE-030131-5541] | 327330 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000068415 | ptgesl | prostaglandin E synthase 2-like [Source:ZFIN;Acc:ZDB-GENE-040426-1063] | 799964 | YES | YES | Cytoplasm | transcription regulator | |||||
| ENSDARG00000075505 | PTGFRN (1 of 2) | prostaglandin F2 receptor inhibitor [Source:HGNC Symbol;Acc:HGNC:9601] | 101884699 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000001769 | ptpra | protein tyrosine phosphatase, receptor type, A [Source:ZFIN;Acc:ZDB-GENE-020107-1] | 140818 | YES | YES | Plasma Membrane | phosphatase | |||||
| ENSDARG00000005754 | ptprfb | protein tyrosine phosphatase, receptor type, f, b [Source:ZFIN;Acc:ZDB-GENE-060503-530] | 799867 | YES | YES | Plasma Membrane | phosphatase | |||||
| ENSDARG00000063416 | ptprk | protein tyrosine phosphatase, receptor type, K [Source:ZFIN;Acc:ZDB-GENE-030131-9834] | 0 | YES | YES | Plasma Membrane | phosphatase | |||||
| ENSDARG00000100422 | ptpro | protein tyrosine phosphatase, receptor type, O [Source:ZFIN;Acc:ZDB-GENE-070424-54] | 407676 | YES | YES | Plasma Membrane | phosphatase | |||||
| ENSDARG00000020871 | ptprz1b | protein tyrosine phosphatase, receptor-type, Z polypeptide 1b [Source:ZFIN;Acc:ZDB-GENE-050506-100] | 553019 | YES | YES | Plasma Membrane | phosphatase | |||||
| ENSDARG00000086034 | pvrl1b | poliovirus receptor-related 1b [Source:ZFIN;Acc:ZDB-GENE-090224-2] | 0 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000013317 | pygmb | phosphorylase, glycogen, muscle b [Source:ZFIN;Acc:ZDB-GENE-040426-1206] | 393444 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000046106 | rab10 | RAB10, member RAS oncogene family [Source:ZFIN;Acc:ZDB-GENE-040718-304] | 436839 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000074246 | rab14 | RAB14, member RAS oncogene family [Source:ZFIN;Acc:ZDB-GENE-030826-20] | 373104 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000035188 | rab14l | RAB14, member RAS oncogene family, like [Source:ZFIN;Acc:ZDB-GENE-040426-1207] | 393656 | YES | YES | NA | NA | |||||
| ENSDARG00000058044 | rab1ba | zRAB1B, member RAS oncogene family a [Source:ZFIN;Acc:ZDB-GENE-040426-873] | 394117 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000020261 | rab2a | RAB2A, member RAS oncogene family [Source:ZFIN;Acc:ZDB-GENE-011212-2] | 140617 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000036501 | rab39bb | RAB39B, member RAS oncogene family b [Source:ZFIN;Acc:ZDB-GENE-061215-104] | 566010 | YES | YES | Plasma Membrane | enzyme | |||||
| ENSDARG00000043835 | rab3ab | RAB3A, member RAS oncogene family, b [Source:ZFIN;Acc:ZDB-GENE-041210-268] | 550457 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000034215 | rab42a | RAB42, member RAS oncogene family a [Source:ZFIN;Acc:ZDB-GENE-040426-1983] | 406315 | YES | YES | Other | other | |||||
| ENSDARG00000018602 | rab5aa | RAB5A, member RAS oncogene family, a [Source:ZFIN;Acc:ZDB-GENE-030131-139] | 337737 | YES | YES | NA | NA | |||||
| ENSDARG00000007257 | rab5ab | RAB5A, member RAS oncogene family, b [Source:ZFIN;Acc:ZDB-GENE-040122-3] | 393945 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000016059 | rab5b | RAB5B, member RAS oncogene family [Source:ZFIN;Acc:ZDB-GENE-040426-2593] | 405821 | YES | YES | NA | NA | |||||
| ENSDARG00000026712 | rab5c | RAB5C, member RAS oncogene family [Source:ZFIN;Acc:ZDB-GENE-031118-30] | 386807 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000034522 | rab6ba | RAB6B, member RAS oncogene family a [Source:ZFIN;Acc:ZDB-GENE-050809-136] | 558900 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000067920 | rab8a | RAB8A, member RAS oncogene family [Source:ZFIN;Acc:ZDB-GENE-070424-36] | 571897 | YES | YES | Plasma Membrane | enzyme | |||||
| ENSDARG00000068628 | rab8b | RAB8B, member RAS oncogene family [Source:ZFIN;Acc:ZDB-GENE-070620-16] | 100093706 | YES | YES | NA | NA | |||||
| ENSDARG00000016253 | ralab | v-ral simian leukemia viral oncogene homolog Ab (ras related) [Source:ZFIN;Acc:ZDB-GENE-041114-195] | 492355 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000061551 | rap1gap2a | RAP1 GTPase activating protein 2a [Source:ZFIN;Acc:ZDB-GENE-061013-179] | 0 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000073741 | RAP1GDS1 | RAP1, GTP-GDP dissociation stimulator 1 [Source:HGNC Symbol;Acc:HGNC:9859] | 100334870 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000040246 | rap2ab | RAP2A, member of RAS oncogene family b [Source:ZFIN;Acc:ZDB-GENE-090312-116] | 100003903 | YES | YES | NA | NA | |||||
| ENSDARG00000005482 | rapgef2 | Rap guanine nucleotide exchange factor (GEF) 2 [Source:ZFIN;Acc:ZDB-GENE-030131-6959] | 557693 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000100742 | reep5 | receptor accessory protein 5 [Source:ZFIN;Acc:ZDB-GENE-030131-9181] | 337237 | YES | YES | Extracellular Space | transporter | |||||
| ENSDARG00000098969 | RELN | reelin [Source:HGNC Symbol;Acc:HGNC:9957] | 0 | YES | YES | Extracellular Space | peptidase | |||||
| ENSDARG00000018600 | retsat | retinol saturase (all-trans-retinol 13,14-reductase) [Source:ZFIN;Acc:ZDB-GENE-050320-11] | 325922 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000068653 | rhoga | ras homolog family member Ga [Source:ZFIN;Acc:ZDB-GENE-060503-820] | 556424 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000056328 | rhot2 | ras homolog family member T2 [Source:ZFIN;Acc:ZDB-GENE-051120-96] | 557574 | YES | YES | NA | NA | |||||
| ENSDARG00000007247 | ric8a | RIC8 guanine nucleotide exchange factor A [Source:ZFIN;Acc:ZDB-GENE-040927-18] | 449546 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000070780 | rln3a | relaxin 3a [Source:ZFIN;Acc:ZDB-GENE-050310-3] | 503864 | YES | YES | Extracellular Space | other | |||||
| ENSDARG00000078761 | RMDN2 | regulator of microtubule dynamics 2 [Source:HGNC Symbol;Acc:HGNC:26567] | 0 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000041407 | rmdn3 | regulator of microtubule dynamics 3 [Source:ZFIN;Acc:ZDB-GENE-041014-256] | 100034410 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000005645 | robo3 | roundabout, axon guidance receptor, homolog 3 (Drosophila) [Source:ZFIN;Acc:ZDB-GENE-000209-4] | 30770 | YES | YES | Plasma Membrane | transmembrane receptor | |||||
| ENSDARG00000059323 | rpn1 | ribophorin I [Source:ZFIN;Acc:ZDB-GENE-030131-4286] | 325561 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000009724 | rpn2 | ribophorin II [Source:ZFIN;Acc:ZDB-GENE-030131-7928] | 335985 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000033437 | rps6ka1 | ribosomal protein S6 kinase a, polypeptide 1 [Source:ZFIN;Acc:ZDB-GENE-060929-516] | 0 | YES | YES | Cytoplasm | kinase | |||||
| ENSDARG00000019181 | rpsa | ribosomal protein SA [Source:ZFIN;Acc:ZDB-GENE-040426-811] | 394027 | YES | YES | Cytoplasm | translation regulator | |||||
| ENSDARG00000036252 | rras2 | related RAS viral (r-ras) oncogene homolog 2 [Source:ZFIN;Acc:ZDB-GENE-050417-352] | 550513 | YES | YES | Plasma Membrane | enzyme | |||||
| ENSDARG00000006497 | rtn1a | reticulon 1a [Source:ZFIN;Acc:ZDB-GENE-030131-2426] | 323706 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000021143 | rtn1b | reticulon 1b [Source:ZFIN;Acc:ZDB-GENE-030710-3] | 327065 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000091348 | rtn3 | reticulon 3 [Source:ZFIN;Acc:ZDB-GENE-030710-5] | 394047 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000057598 | s100b | S100 calcium binding protein, beta (neural) [Source:ZFIN;Acc:ZDB-GENE-040718-290] | 436825 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000045814 | samm50 | SAMM50 sorting and assembly machinery component [Source:ZFIN;Acc:ZDB-GENE-040426-806] | 393111 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000039681 | samm50l | sorting and assembly machinery component 50 homolog, like [Source:ZFIN;Acc:ZDB-GENE-041114-30] | 492471 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000008237 | sars | seryl-tRNA synthetase [Source:ZFIN;Acc:ZDB-GENE-040831-1] | 445405 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000062968 | sbf1 | SET binding factor 1 [Source:ZFIN;Acc:ZDB-GENE-040718-139] | 568277 | YES | YES | Plasma Membrane | phosphatase | |||||
| ENSDARG00000036911 | scamp1 | secretory carrier membrane protein 1 [Source:ZFIN;Acc:ZDB-GENE-040426-1803] | 402992 | YES | YES | Cytoplasm | transporter | |||||
| ENSDARG00000024564 | sccpdhb | saccharopine dehydrogenase b [Source:ZFIN;Acc:ZDB-GENE-041010-211] | 407665 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000105003 | scn1bb | sodium channel, voltage-gated, type I, beta b [Source:ZFIN;Acc:ZDB-GENE-090408-1] | 100005325 | YES | YES | Plasma Membrane | ion channel | |||||
| ENSDARG00000062744 | scn1lab | sodium channel, voltage-gated, type I like, alpha b [Source:ZFIN;Acc:ZDB-GENE-060906-1] | 559447 | YES | YES | Plasma Membrane | ion channel | |||||
| ENSDARG00000005775 | scn8aa | sodium channel, voltage gated, type VIII, alpha subunit a [Source:ZFIN;Acc:ZDB-GENE-000828-1] | 58152 | YES | YES | Plasma Membrane | ion channel | |||||
| ENSDARG00000018032 | scn8ab | sodium channel, voltage gated, type VIII, alpha subunit b [Source:ZFIN;Acc:ZDB-GENE-060906-2] | 793996 | YES | YES | Plasma Membrane | ion channel | |||||
| ENSDARG00000067593 | SCO1 | SCO1 cytochrome c oxidase assembly protein [Source:HGNC Symbol;Acc:HGNC:10603] | 796572 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000102356 | scp2b | sterol carrier protein 2b [Source:ZFIN;Acc:ZDB-GENE-041010-196] | 450073 | YES | YES | NA | NA | |||||
| ENSDARG00000035631 | sdf2l1 | stromal cell-derived factor 2-like 1 [Source:ZFIN;Acc:ZDB-GENE-040808-46] | 445275 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000016721 | sdha | succinate dehydrogenase complex, subunit A, flavoprotein (Fp) [Source:ZFIN;Acc:ZDB-GENE-040426-874] | 393884 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000075768 | sdhb | succinate dehydrogenase complex, subunit B, iron sulfur (Ip) [Source:ZFIN;Acc:ZDB-GENE-030131-8005] | 562149 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000078866 | sdk1a | sidekick cell adhesion molecule 1a [Source:ZFIN;Acc:ZDB-GENE-081104-374] | 558391 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000060452 | sdk2b | sidekick cell adhesion molecule 2b [Source:ZFIN;Acc:ZDB-GENE-030131-8203] | 564922 | YES | YES | Other | other | |||||
| ENSDARG00000008936 | sec11a | SEC11 homolog A (S. cerevisiae) [Source:ZFIN;Acc:ZDB-GENE-040718-227] | 436794 | YES | YES | Cytoplasm | peptidase | |||||
| ENSDARG00000076568 | sec61b | Sec61 beta subunit [Source:ZFIN;Acc:ZDB-GENE-040718-260] | 791986 | YES | YES | Cytoplasm | transporter | |||||
| ENSDARG00000017740 | sec63 | SEC63 homolog (S. cerevisiae) [Source:ZFIN;Acc:ZDB-GENE-040718-328] | 436861 | YES | YES | Cytoplasm | transporter | |||||
| ENSDARG00000019963 | sfxn1 | sideroflexin 1 [Source:ZFIN;Acc:ZDB-GENE-040801-45] | 445143 | YES | YES | Cytoplasm | transporter | |||||
| ENSDARG00000057374 | sfxn3 | sideroflexin 3 [Source:ZFIN;Acc:ZDB-GENE-070112-2092] | 791182 | YES | YES | Cytoplasm | transporter | |||||
| ENSDARG00000069832 | sfxn4 | sideroflexin 4 [Source:ZFIN;Acc:ZDB-GENE-050309-187] | 556121 | YES | YES | Cytoplasm | transporter | |||||
| ENSDARG00000026137 | sfxn5b | sideroflexin 5b [Source:ZFIN;Acc:ZDB-GENE-050706-107] | 561995 | YES | YES | Cytoplasm | transporter | |||||
| ENSDARG00000023600 | sh3gl2 | SH3-domain GRB2-like 2 [Source:ZFIN;Acc:ZDB-GENE-040121-3] | 394091 | YES | YES | Plasma Membrane | enzyme | |||||
| ENSDARG00000039901 | SH3GL2 (2 of 2) | SH3-domain GRB2-like 2 [Source:HGNC Symbol;Acc:HGNC:10831] | 0 | YES | YES | Plasma Membrane | enzyme | |||||
| ENSDARG00000101804 | si:ch1073-140o9.2 | si:ch1073-140o9.2 [Source:ZFIN;Acc:ZDB-GENE-120709-104] | 0 | YES | YES | NA | NA | |||||
| ENSDARG00000079651 | si:ch1073-174d20.2 | si:ch1073-174d20.2 [Source:ZFIN;Acc:ZDB-GENE-121214-51] | 570430 | YES | YES | Cytoplasm | peptidase | |||||
| ENSDARG00000073764 | si:ch211-113j14.1 | si:ch211-113j14.1 [Source:ZFIN;Acc:ZDB-GENE-120215-110] | 558239 | YES | YES | NA | NA | |||||
| ENSDARG00000092976 | si:ch211-127i16.2 | si:ch211-127i16.2 [Source:ZFIN;Acc:ZDB-GENE-091204-54] | 558050 | YES | YES | NA | NA | |||||
| ENSDARG00000087857 | si:ch211-180a12.2 | si:ch211-180a12.2 [Source:ZFIN;Acc:ZDB-GENE-121214-236] | 100150385 | YES | YES | NA | NA | |||||
| ENSDARG00000078088 | si:ch211-186j3.6 | si:ch211-186j3.6 [Source:ZFIN;Acc:ZDB-GENE-130530-842] | 556807 | YES | YES | NA | NA | |||||
| ENSDARG00000035660 | si:ch211-193k19.1 | si:ch211-193k19.1 [Source:ZFIN;Acc:ZDB-GENE-060503-110] | 100034533 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000037867 | si:ch211-194i10.5 | si:ch211-194i10.5 [Source:ZFIN;Acc:ZDB-GENE-131120-154] | 402883 | YES | YES | NA | NA | |||||
| ENSDARG00000029170 | si:ch211-200p22.4 | si:ch211-200p22.4 [Source:ZFIN;Acc:ZDB-GENE-081104-61] | 565799 | YES | YES | NA | NA | |||||
| ENSDARG00000007976 | si:ch211-220f16.2 | si:ch211-220f16.2 [Source:ZFIN;Acc:ZDB-GENE-070912-199] | 797650 | YES | YES | NA | NA | |||||
| ENSDARG00000062330 | si:ch211-233a24.2 | si:ch211-233a24.2 [Source:ZFIN;Acc:ZDB-GENE-090313-98] | 564367 | YES | YES | Other | other | |||||
| ENSDARG00000007275 | si:ch211-251b21.1 | si:ch211-251b21.1 [Source:ZFIN;Acc:ZDB-GENE-060809-5] | 571720 | YES | YES | NA | NA | |||||
| ENSDARG00000096661 | si:ch211-286b5.5 | si:ch211-286b5.5 [Source:ZFIN;Acc:ZDB-GENE-121214-209] | 100003596 | YES | YES | NA | NA | |||||
| ENSDARG00000077468 | si:ch73-206p6.1 | si:ch73-206p6.1 [Source:ZFIN;Acc:ZDB-GENE-030131-5048] | 571998 | YES | YES | Plasma Membrane | enzyme | |||||
| ENSDARG00000062831 | si:ch73-22o12.1 | si:ch73-22o12.1 [Source:ZFIN;Acc:ZDB-GENE-110411-27] | 560816 | YES | YES | Plasma Membrane | transmembrane receptor | |||||
| ENSDARG00000070511 | si:dkey-183j2.10 | si:dkey-183j2.10 [Source:ZFIN;Acc:ZDB-GENE-130530-729] | 100151589 | YES | YES | NA | NA | |||||
| ENSDARG00000074894 | si:dkey-228b2.5 | si:dkey-228b2.5 [Source:ZFIN;Acc:ZDB-GENE-091113-62] | 100537539 | YES | YES | Nucleus | other | |||||
| ENSDARG00000043514 | si:dkey-239i20.4 | si:dkey-239i20.4 [Source:ZFIN;Acc:ZDB-GENE-030131-2644] | 557628 | YES | YES | Other | other | |||||
| ENSDARG00000002840 | si:dkey-28b4.8 | si:dkey-28b4.8 [Source:ZFIN;Acc:ZDB-GENE-090501-2] | 562640 | YES | YES | NA | NA | |||||
| ENSDARG00000063159 | si:dkey-32e23.4 | si:dkey-32e23.4 [Source:ZFIN;Acc:ZDB-GENE-081105-41] | 566871 | YES | YES | NA | NA | |||||
| ENSDARG00000058248 | si:dkeyp-77h1.4 | si:dkeyp-77h1.4 [Source:ZFIN;Acc:ZDB-GENE-050208-18] | 497302 | YES | YES | NA | NA | |||||
| ENSDARG00000035679 | si:rp71-45k5.4 | si:rp71-45k5.4 [Source:ZFIN;Acc:ZDB-GENE-060503-941] | 403015 | YES | YES | Cytoplasm | peptidase | |||||
| ENSDARG00000079078 | si:xx-by187g17.1 | si:xx-by187g17.1 [Source:ZFIN;Acc:ZDB-GENE-041001-43] | 497162 | YES | YES | NA | NA | |||||
| ENSDARG00000075815 | SLC12A5 (1 of 2) | solute carrier family 12 (potassium/chloride transporter), member 5 [Source:HGNC Symbol;Acc:HGNC:13818] | 572215 | YES | YES | Plasma Membrane | transporter | |||||
| ENSDARG00000078187 | slc12a5b | solute carrier family 12 (potassium/chloride transporter), member 5b [Source:ZFIN;Acc:ZDB-GENE-080707-1] | 797331 | YES | YES | Plasma Membrane | transporter | |||||
| ENSDARG00000001127 | slc17a6a | solute carrier family 17 (vesicular glutamate transporter), member 6a [Source:ZFIN;Acc:ZDB-GENE-050105-4] | 494492 | YES | YES | NA | NA | |||||
| ENSDARG00000041150 | slc17a6b | solute carrier family 17 (vesicular glutamate transporter), member 6b [Source:ZFIN;Acc:ZDB-GENE-030616-554] | 100149756 | YES | YES | NA | NA | |||||
| ENSDARG00000102453 | slc1a2b | solute carrier family 1 (glial high affinity glutamate transporter), member 2b [Source:ZFIN;Acc:ZDB-GENE-030131-7779] | 335836 | YES | YES | Plasma Membrane | transporter | |||||
| ENSDARG00000043148 | slc1a3b | solute carrier family 1 (glial high affinity glutamate transporter), member 3b [Source:ZFIN;Acc:ZDB-GENE-090708-3] | 556181 | YES | YES | NA | NA | |||||
| ENSDARG00000010096 | slc1a6 | solute carrier family 1 (high affinity aspartate/glutamate transporter), member 6 [Source:ZFIN;Acc:ZDB-GENE-071004-45] | 559270 | YES | YES | Plasma Membrane | transporter | |||||
| ENSDARG00000102362 | slc25a12 | solute carrier family 25 (aspartate/glutamate carrier), member 12 [Source:ZFIN;Acc:ZDB-GENE-031006-11] | 337675 | YES | YES | Cytoplasm | transporter | |||||
| ENSDARG00000099353 | slc25a13 | solute carrier family 25 (aspartate/glutamate carrier), member 13 [Source:ZFIN;Acc:ZDB-GENE-080319-2] | 0 | YES | YES | Cytoplasm | transporter | |||||
| ENSDARG00000040401 | slc25a20 | solute carrier family 25 (carnitine/acylcarnitine translocase), member 20 [Source:ZFIN;Acc:ZDB-GENE-040426-1869] | 393833 | YES | YES | Cytoplasm | transporter | |||||
| ENSDARG00000005690 | slc25a23a | solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 23a [Source:ZFIN;Acc:ZDB-GENE-121214-154] | 561939 | YES | YES | Cytoplasm | transporter | |||||
| ENSDARG00000008568 | slc25a24 | solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 24 [Source:ZFIN;Acc:ZDB-GENE-040912-183] | 447867 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000038731 | slc25a36a | solute carrier family 25 (pyrimidine nucleotide carrier ), member 36a [Source:ZFIN;Acc:ZDB-GENE-040718-415] | 436940 | YES | YES | Cytoplasm | transporter | |||||
| ENSDARG00000025566 | slc25a3b | solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 3b [Source:ZFIN;Acc:ZDB-GENE-040426-1916] | 322362 | YES | YES | Cytoplasm | transporter | |||||
| ENSDARG00000015856 | slc25a40 | solute carrier family 25, member 40 [Source:ZFIN;Acc:ZDB-GENE-040718-52] | 436633 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000035181 | slc25a46 | solute carrier family 25, member 46 [Source:ZFIN;Acc:ZDB-GENE-040718-296] | 436831 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000056163 | slc25a51a | solute carrier family 25, member 51a [Source:ZFIN;Acc:ZDB-GENE-040426-1722] | 393728 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000038352 | slc27a1b | solute carrier family 27 (fatty acid transporter), member 1b [Source:ZFIN;Acc:ZDB-GENE-061013-672] | 0 | YES | YES | NA | NA | |||||
| ENSDARG00000017047 | slc27a4 | solute carrier family 27 (fatty acid transporter), member 4 [Source:ZFIN;Acc:ZDB-GENE-050417-248] | 550432 | YES | YES | Plasma Membrane | transporter | |||||
| ENSDARG00000001437 | slc2a1a | solute carrier family 2 (facilitated glucose transporter), member 1a [Source:ZFIN;Acc:ZDB-GENE-030131-3158] | 555778 | YES | YES | Plasma Membrane | transporter | |||||
| ENSDARG00000007412 | slc2a1b | solute carrier family 2 (facilitated glucose transporter), member 1b [Source:ZFIN;Acc:ZDB-GENE-090915-1] | 100321338 | YES | YES | Plasma Membrane | transporter | |||||
| ENSDARG00000013295 | slc2a3a | solute carrier family 2 (facilitated glucose transporter), member 3a [Source:ZFIN;Acc:ZDB-GENE-040718-390] | 436916 | YES | YES | Plasma Membrane | transporter | |||||
| ENSDARG00000059775 | slc32a1 | solute carrier family 32 (GABA vesicular transporter), member 1 [Source:ZFIN;Acc:ZDB-GENE-061201-1] | 798575 | YES | YES | Plasma Membrane | transporter | |||||
| ENSDARG00000056262 | slc35g2a | solute carrier family 35, member G2a [Source:ZFIN;Acc:ZDB-GENE-041114-202] | 492818 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000036427 | slc3a2a | solute carrier family 3 (amino acid transporter heavy chain), member 2a [Source:ZFIN;Acc:ZDB-GENE-000831-3] | 796322 | YES | YES | Plasma Membrane | transporter | |||||
| ENSDARG00000037012 | slc3a2b | solute carrier family 3 (amino acid transporter heavy chain), member 2b [Source:ZFIN;Acc:ZDB-GENE-040122-2] | 399488 | YES | YES | Plasma Membrane | transporter | |||||
| ENSDARG00000063133 | slc4a10a | solute carrier family 4, sodium bicarbonate transporter, member 10a [Source:ZFIN;Acc:ZDB-GENE-100215-3] | 798983 | YES | YES | Plasma Membrane | transporter | |||||
| ENSDARG00000060303 | slc4a10b | solute carrier family 4, sodium bicarbonate transporter, member 10b [Source:ZFIN;Acc:ZDB-GENE-081105-62] | 557642 | YES | YES | Plasma Membrane | transporter | |||||
| ENSDARG00000013730 | slc4a4a | solute carrier family 4 (sodium bicarbonate cotransporter), member 4a [Source:ZFIN;Acc:ZDB-GENE-060526-274] | 568631 | YES | YES | Plasma Membrane | transporter | |||||
| ENSDARG00000015531 | slc4a8 | solute carrier family 4, sodium bicarbonate cotransporter, member 8 [Source:ZFIN;Acc:ZDB-GENE-131119-88] | 557494 | YES | YES | Plasma Membrane | transporter | |||||
| ENSDARG00000087981 | SLC6A11 | solute carrier family 6 (neurotransmitter transporter), member 11 [Source:HGNC Symbol;Acc:HGNC:11044] | 0 | YES | YES | Plasma Membrane | transporter | |||||
| ENSDARG00000067567 | SLC6A13 | solute carrier family 6 (neurotransmitter transporter), member 13 [Source:HGNC Symbol;Acc:HGNC:11046] | 100332592 | YES | YES | Plasma Membrane | transporter | |||||
| ENSDARG00000045944 | slc6a1a | solute carrier family 6 (neurotransmitter transporter), member 1a [Source:ZFIN;Acc:ZDB-GENE-060519-23] | 692318 | YES | YES | Plasma Membrane | transporter | |||||
| ENSDARG00000039647 | slc6a1b | solute carrier family 6 (neurotransmitter transporter), member 1b [Source:ZFIN;Acc:ZDB-GENE-041114-57] | 492490 | YES | YES | Plasma Membrane | transporter | |||||
| ENSDARG00000012534 | slc6a6a | solute carrier family 6 (neurotransmitter transporter), member 6a [Source:ZFIN;Acc:ZDB-GENE-081104-1] | 556480 | YES | YES | Plasma Membrane | transporter | |||||
| ENSDARG00000010816 | slc7a14a | solute carrier family 7, member 14a [Source:ZFIN;Acc:ZDB-GENE-070912-112] | 566270 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000100592 | slc8a2b | solute carrier family 8 (sodium/calcium exchanger), member 2b [Source:ZFIN;Acc:ZDB-GENE-050419-209] | 567212 | YES | YES | Cytoplasm | transporter | |||||
| ENSDARG00000004931 | slc8a3 | solute carrier family 8 (sodium/calcium exchanger), member 3 [Source:ZFIN;Acc:ZDB-GENE-050810-1] | 560561 | YES | YES | Plasma Membrane | transporter | |||||
| ENSDARG00000055154 | slc8a4a | solute carrier family 8 (sodium/calcium exchanger), member 4a [Source:ZFIN;Acc:ZDB-GENE-060110-2] | 568512 | YES | YES | NA | NA | |||||
| ENSDARG00000020764 | slmapb | sarcolemma associated protein b [Source:ZFIN;Acc:ZDB-GENE-040426-809] | 393146 | YES | YES | NA | NA | |||||
| ENSDARG00000012874 | snap23.1 | synaptosomal-associated protein 23.1 [Source:ZFIN;Acc:ZDB-GENE-040426-883] | 393159 | YES | YES | Plasma Membrane | transporter | |||||
| ENSDARG00000098809 | snap91 | synaptosomal-associated protein 91 [Source:ZFIN;Acc:ZDB-GENE-100422-6] | 565384 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000015931 | SNAP91 (1 of 2) | synaptosomal-associated protein, 91kDa [Source:HGNC Symbol;Acc:HGNC:14986] | 559029 | YES | YES | NA | NA | |||||
| ENSDARG00000104945 | sncb | synuclein, beta [Source:ZFIN;Acc:ZDB-GENE-040426-1615] | 393944 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000006766 | snd1 | staphylococcal nuclease and tudor domain containing 1 [Source:ZFIN;Acc:ZDB-GENE-030131-3124] | 324404 | YES | YES | Nucleus | enzyme | |||||
| ENSDARG00000019060 | snx12 | sorting nexin 12 [Source:ZFIN;Acc:ZDB-GENE-040426-787] | 394098 | YES | YES | Cytoplasm | transporter | |||||
| ENSDARG00000033804 | snx27a | sorting nexin family member 27a [Source:ZFIN;Acc:ZDB-GENE-060503-371] | 566205 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000016977 | snx27b | sorting nexin family member 27b [Source:ZFIN;Acc:ZDB-GENE-040426-1529] | 393512 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000077465 | SORCS2 | sortilin-related VPS10 domain containing receptor 2 [Source:HGNC Symbol;Acc:HGNC:16698] | 100330371 | YES | YES | Plasma Membrane | transporter | |||||
| ENSDARG00000013892 | sorl1 | sortilin-related receptor, L(DLR class) A repeats containing [Source:ZFIN;Acc:ZDB-GENE-050208-22] | 497306 | YES | YES | Cytoplasm | transporter | |||||
| ENSDARG00000103457 | spg20b | spastic paraplegia 20b (Troyer syndrome) [Source:ZFIN;Acc:ZDB-GENE-060825-146] | 751741 | YES | YES | Other | other | |||||
| ENSDARG00000068187 | spg7 | spastic paraplegia 7 [Source:ZFIN;Acc:ZDB-GENE-030131-5391] | 794740 | YES | YES | Cytoplasm | peptidase | |||||
| ENSDARG00000023309 | spryd4 | SPRY domain containing 4 [Source:ZFIN;Acc:ZDB-GENE-040718-180] | 100005939 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000060309 | srgap3 | SLIT-ROBO Rho GTPase activating protein 3 [Source:ZFIN;Acc:ZDB-GENE-060524-4] | 561567 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000101627 | ssr1 | signal sequence receptor, alpha [Source:ZFIN;Acc:ZDB-GENE-030826-28] | 373100 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000019444 | ssr4 | signal sequence receptor, delta [Source:ZFIN;Acc:ZDB-GENE-030131-4900] | 326701 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000039022 | stk25b | serine/threonine kinase 25b [Source:ZFIN;Acc:ZDB-GENE-040426-2537] | 406600 | YES | YES | Cytoplasm | kinase | |||||
| ENSDARG00000033655 | stmn1b | stathmin 1b [Source:ZFIN;Acc:ZDB-GENE-050417-397] | 550548 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000003835 | stom | stomatin [Source:ZFIN;Acc:ZDB-GENE-980526-486] | 81534 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000103135 | stoml2 | stomatin (EPB72)-like 2 [Source:ZFIN;Acc:ZDB-GENE-040426-1139] | 394006 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000044605 | stx12l | syntaxin 12, like [Source:ZFIN;Acc:ZDB-GENE-110715-1] | 569124 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000003307 | stx16 | syntaxin 16 [Source:ZFIN;Acc:ZDB-GENE-060810-113] | 562857 | YES | YES | Cytoplasm | transporter | |||||
| ENSDARG00000000503 | stx1b | syntaxin 1B [Source:ZFIN;Acc:ZDB-GENE-000330-4] | 58038 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000103173 | stx8 | syntaxin 8 [Source:ZFIN;Acc:ZDB-GENE-040426-1360] | 393346 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000001994 | stxbp1a | syntaxin binding protein 1a [Source:ZFIN;Acc:ZDB-GENE-050626-106] | 574003 | YES | YES | Cytoplasm | transporter | |||||
| ENSDARG00000056036 | stxbp1b | syntaxin binding protein 1b [Source:ZFIN;Acc:ZDB-GENE-060531-166] | 557717 | YES | YES | Cytoplasm | transporter | |||||
| ENSDARG00000002656 | stxbp5a | syntaxin binding protein 5a (tomosyn) [Source:ZFIN;Acc:ZDB-GENE-041001-161] | 556789 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000007720 | sub1b | SUB1 homolog b (S. cerevisiae) [Source:ZFIN;Acc:ZDB-GENE-040718-209] | 436778 | YES | YES | Nucleus | transcription regulator | |||||
| ENSDARG00000005359 | sucla2 | succinate-CoA ligase, ADP-forming, beta subunit [Source:ZFIN;Acc:ZDB-GENE-040426-1963] | 406299 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000088277 | susd5 | sushi domain containing 5 [Source:ZFIN;Acc:ZDB-GENE-120215-196] | 101886789 | YES | YES | Other | other | |||||
| ENSDARG00000059945 | SV2A | synaptic vesicle glycoprotein 2A [Source:HGNC Symbol;Acc:HGNC:20566] | 568033 | YES | YES | Cytoplasm | transporter | |||||
| ENSDARG00000014871 | syngr3a | synaptogyrin 3a [Source:ZFIN;Acc:ZDB-GENE-050522-541] | 553617 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000025011 | synj1 | synaptojanin 1 [Source:ZFIN;Acc:ZDB-GENE-030131-9180] | 337236 | YES | YES | Cytoplasm | phosphatase | |||||
| ENSDARG00000042974 | sypa | synaptophysin a [Source:ZFIN;Acc:ZDB-GENE-031104-2] | 559698 | YES | YES | Cytoplasm | transporter | |||||
| ENSDARG00000030614 | syt1a | synaptotagmin Ia [Source:ZFIN;Acc:ZDB-GENE-040718-165] | 436736 | YES | YES | Cytoplasm | transporter | |||||
| ENSDARG00000014169 | SYT2 (1 of 2) | synaptotagmin II [Source:HGNC Symbol;Acc:HGNC:11510] | 0 | YES | YES | Cytoplasm | transporter | |||||
| ENSDARG00000025206 | syt2a | synaptotagmin IIa [Source:ZFIN;Acc:ZDB-GENE-060503-315] | 567877 | YES | YES | Cytoplasm | transporter | |||||
| ENSDARG00000011640 | syt5b | synaptotagmin Vb [Source:ZFIN;Acc:ZDB-GENE-050522-134] | 553567 | YES | YES | NA | NA | |||||
| ENSDARG00000078604 | tbc1d10b | TBC1 domain family, member 10b [Source:ZFIN;Acc:ZDB-GENE-030131-7754] | 0 | YES | YES | Plasma Membrane | enzyme | |||||
| ENSDARG00000069339 | tbc1d24 | TBC1 domain family, member 24 [Source:ZFIN;Acc:ZDB-GENE-050809-65] | 100536073 | YES | YES | NA | NA | |||||
| ENSDARG00000016754 | tbca | tubulin cofactor a [Source:ZFIN;Acc:ZDB-GENE-040426-962] | 394029 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000017891 | tcp1 | t-complex 1 [Source:ZFIN;Acc:ZDB-GENE-990714-24] | 30477 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000069941 | TDP2 (2 of 2) | tyrosyl-DNA phosphodiesterase 2 [Source:HGNC Symbol;Acc:HGNC:17768] | 101887157 | YES | YES | Cytoplasm | transcription regulator | |||||
| ENSDARG00000053474 | tdrkh | tudor and KH domain containing [Source:ZFIN;Acc:ZDB-GENE-050327-81] | 541541 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000003403 | tenm1 | teneurin transmembrane protein 1 [Source:ZFIN;Acc:ZDB-GENE-060531-26] | 563094 | YES | YES | Plasma Membrane | peptidase | |||||
| ENSDARG00000005479 | tenm3 | teneurin transmembrane protein 3 [Source:ZFIN;Acc:ZDB-GENE-990714-19] | 30155 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000105088 | tenm4 | teneurin transmembrane protein 4 [Source:ZFIN;Acc:ZDB-GENE-990714-20] | 30156 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000016771 | tfa | transferrin-a [Source:ZFIN;Acc:ZDB-GENE-980526-35] | 0 | YES | YES | Extracellular Space | other | |||||
| ENSDARG00000069116 | timm10 | translocase of inner mitochondrial membrane 10 homolog (yeast) [Source:ZFIN;Acc:ZDB-GENE-040718-427] | 436951 | YES | YES | Cytoplasm | transporter | |||||
| ENSDARG00000071396 | timm23 | translocase of inner mitochondrial membrane 23 homolog (yeast) [Source:ZFIN;Acc:ZDB-GENE-020419-17] | 798429 | YES | YES | Cytoplasm | transporter | |||||
| ENSDARG00000055708 | timm8b | translocase of inner mitochondrial membrane 8 homolog B (yeast) [Source:ZFIN;Acc:ZDB-GENE-031019-1] | 793663 | YES | YES | Cytoplasm | transporter | |||||
| ENSDARG00000043865 | timm9 | translocase of inner mitochondrial membrane 9 homolog [Source:ZFIN;Acc:ZDB-GENE-021206-14] | 282678 | YES | YES | Cytoplasm | transporter | |||||
| ENSDARG00000017901 | tln2a | talin 2a [Source:ZFIN;Acc:ZDB-GENE-040724-263] | 556842 | YES | YES | Nucleus | other | |||||
| ENSDARG00000104934 | tln2b | talin 2b [Source:ZFIN;Acc:ZDB-GENE-130325-1] | 0 | YES | YES | NA | NA | |||||
| ENSDARG00000004261 | tmed9 | transmembrane emp24 protein transport domain containing 9 [Source:ZFIN;Acc:ZDB-GENE-050417-434] | 613145 | YES | YES | Cytoplasm | transporter | |||||
| ENSDARG00000070866 | tmem11 | transmembrane protein 11 [Source:ZFIN;Acc:ZDB-GENE-050417-37] | 407695 | YES | YES | Plasma Membrane | G-protein coupled receptor | |||||
| ENSDARG00000075180 | tmem14ca | transmembrane protein 14Ca [Source:ZFIN;Acc:ZDB-GENE-060825-156] | 751728 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000079858 | tmem163a | transmembrane protein 163a [Source:ZFIN;Acc:ZDB-GENE-081104-429] | 564964 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000088227 | tmem163b | transmembrane protein 163b [Source:ZFIN;Acc:ZDB-GENE-091204-410] | 100333673 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000038901 | tmem256 | transmembrane protein 256 [Source:ZFIN;Acc:ZDB-GENE-050417-92] | 550283 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000041332 | tmem33 | transmembrane protein 33 [Source:ZFIN;Acc:ZDB-GENE-030131-5425] | 327214 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000025693 | tmem9b | TMEM9 domain family, member B [Source:ZFIN;Acc:ZDB-GENE-030131-4656] | 325931 | YES | YES | Other | other | |||||
| ENSDARG00000101512 | tmx4 | thioredoxin-related transmembrane protein 4 [Source:ZFIN;Acc:ZDB-GENE-050706-155] | 792311 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000078842 | tns1a | tensin 1a [Source:ZFIN;Acc:ZDB-GENE-120215-32] | 565181 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000009217 | tom1l2 | target of myb1-like 2 (chicken) [Source:ZFIN;Acc:ZDB-GENE-061013-3] | 560327 | YES | YES | NA | NA | |||||
| ENSDARG00000045146 | tomm22 | translocase of outer mitochondrial membrane 22 homolog (yeast) [Source:ZFIN;Acc:ZDB-GENE-030131-9810] | 321232 | YES | YES | Cytoplasm | transporter | |||||
| ENSDARG00000001557 | tomm34 | translocase of outer mitochondrial membrane 34 [Source:ZFIN;Acc:ZDB-GENE-030131-2081] | 323361 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000029639 | tomm70a | translocase of outer mitochondrial membrane 70 homolog A (S. cerevisiae) [Source:ZFIN;Acc:ZDB-GENE-030131-8173] | 572969 | YES | YES | Cytoplasm | transporter | |||||
| ENSDARG00000006025 | tp53i11b | tumor protein p53 inducible protein 11b [Source:ZFIN;Acc:ZDB-GENE-030131-6328] | 556618 | YES | YES | Other | other | |||||
| ENSDARG00000061713 | tpd52 | tumor protein D52 [Source:ZFIN;Acc:ZDB-GENE-060503-680] | 559992 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000013655 | tpd52l2b | tumor protein D52-like 2b [Source:ZFIN;Acc:ZDB-GENE-030131-822] | 322103 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000105029 | TPPP | tubulin polymerization promoting protein [Source:HGNC Symbol;Acc:HGNC:24164] | 100333482 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000035338 | tppp3l | tubulin polymerization promoting protein 3, like [Source:ZFIN;Acc:ZDB-GENE-140106-24] | 559490 | YES | YES | Nucleus | other | |||||
| ENSDARG00000060101 | trappc12 | trafficking protein particle complex 12 [Source:ZFIN;Acc:ZDB-GENE-070112-1282] | 791162 | YES | YES | Other | other | |||||
| ENSDARG00000028213 | ttna | titin a [Source:ZFIN;Acc:ZDB-GENE-030113-2] | 317731 | YES | YES | NA | NA | |||||
| ENSDARG00000076804 | ttyh1 | tweety family member 1 [Source:ZFIN;Acc:ZDB-GENE-100922-257] | 100537642 | YES | YES | Plasma Membrane | ion channel | |||||
| ENSDARG00000034473 | ttyh3a | tweety family member 3a [Source:ZFIN;Acc:ZDB-GENE-120508-1] | 556314 | YES | YES | Plasma Membrane | ion channel | |||||
| ENSDARG00000037559 | uba1 | ubiquitin-like modifier activating enzyme 1 [Source:ZFIN;Acc:ZDB-GENE-040426-2009] | 406335 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000008748 | ube2na | ubiquitin-conjugating enzyme E2Na [Source:ZFIN;Acc:ZDB-GENE-040426-2873] | 406807 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000041875 | ube2v1 | ubiquitin-conjugating enzyme E2 variant 1 [Source:ZFIN;Acc:ZDB-GENE-051030-102] | 0 | YES | YES | NA | NA | |||||
| ENSDARG00000007359 | UBL4A | ubiquitin-like 4A [Source:HGNC Symbol;Acc:HGNC:12505] | 393270 | YES | YES | Other | other | |||||
| ENSDARG00000009549 | ubr4 | ubiquitin protein ligase E3 component n-recognin 4 [Source:ZFIN;Acc:ZDB-GENE-090313-341] | 407625 | YES | YES | Nucleus | enzyme | |||||
| ENSDARG00000026871 | uchl1 | ubiquitin carboxyl-terminal esterase L1 (ubiquitin thiolesterase) [Source:ZFIN;Acc:ZDB-GENE-030131-3844] | 325119 | YES | YES | Cytoplasm | peptidase | |||||
| ENSDARG00000054746 | uggt1 | UDP-glucose glycoprotein glucosyltransferase 1 [Source:ZFIN;Acc:ZDB-GENE-061103-619] | 553288 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000097491 | ugt1b1 | UDP glucuronosyltransferase 1 family, polypeptide B1 [Source:ZFIN;Acc:ZDB-GENE-080227-10] | 558508 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000011146 | uqcrb | ubiquinol-cytochrome c reductase binding protein [Source:ZFIN;Acc:ZDB-GENE-050522-542] | 554154 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000052304 | uqcrc1 | ubiquinol-cytochrome c reductase core protein I [Source:ZFIN;Acc:ZDB-GENE-040426-1792] | 393793 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000014794 | uqcrc2a | ubiquinol-cytochrome c reductase core protein IIa [Source:ZFIN;Acc:ZDB-GENE-040718-405] | 436930 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000071691 | uqcrc2b | ubiquinol-cytochrome c reductase core protein IIb [Source:ZFIN;Acc:ZDB-GENE-030131-1269] | 322549 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000059128 | uqcrh | ubiquinol-cytochrome c reductase hinge protein [Source:ZFIN;Acc:ZDB-GENE-051030-93] | 566723 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000096003 | USMG5 | up-regulated during skeletal muscle growth 5 homolog (mouse) [Source:HGNC Symbol;Acc:HGNC:30889] | 794625 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000025889 | usp14 | ubiquitin specific peptidase 14 (tRNA-guanine transglycosylase) [Source:ZFIN;Acc:ZDB-GENE-030131-7676] | 335736 | YES | YES | Cytoplasm | peptidase | |||||
| ENSDARG00000073710 | usp7 | ubiquitin specific peptidase 7 (herpes virus-associated) [Source:ZFIN;Acc:ZDB-GENE-030131-3656] | 553758 | YES | YES | Nucleus | peptidase | |||||
| ENSDARG00000014303 | vac14 | vac14 homolog (S. cerevisiae) [Source:ZFIN;Acc:ZDB-GENE-040912-82] | 447912 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000031283 | vamp1 | vesicle-associated membrane protein 1 [Source:ZFIN;Acc:ZDB-GENE-040426-2725] | 406707 | YES | YES | Cytoplasm | transporter | |||||
| ENSDARG00000097576 | VAMP1 (2 of 2) | vesicle-associated membrane protein 1 (synaptobrevin 1) [Source:HGNC Symbol;Acc:HGNC:12642] | 436805 | YES | YES | Cytoplasm | transporter | |||||
| ENSDARG00000056877 | vamp2 | vesicle-associated membrane protein 2 [Source:ZFIN;Acc:ZDB-GENE-030131-8225] | 336281 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000043510 | vamp4 | vesicle-associated membrane protein 4 [Source:ZFIN;Acc:ZDB-GENE-040625-92] | 393708 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000068262 | vamp5 | vesicle-associated membrane protein 5 [Source:ZFIN;Acc:ZDB-GENE-030131-4209] | 325484 | YES | YES | Plasma Membrane | transporter | |||||
| ENSDARG00000056481 | vat1 | vesicle amine transport 1 [Source:ZFIN;Acc:ZDB-GENE-030616-178] | 368752 | YES | YES | Plasma Membrane | transporter | |||||
| ENSDARG00000020008 | vcp | valosin containing protein [Source:ZFIN;Acc:ZDB-GENE-030131-5408] | 327197 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000045132 | vdac1 | voltage-dependent anion channel 1 [Source:ZFIN;Acc:ZDB-GENE-030131-6514] | 334582 | YES | YES | Cytoplasm | ion channel | |||||
| ENSDARG00000013623 | vdac2 | voltage-dependent anion channel 2 [Source:ZFIN;Acc:ZDB-GENE-030131-845] | 322126 | YES | YES | Cytoplasm | ion channel | |||||
| ENSDARG00000003695 | vdac3 | voltage-dependent anion channel 3 [Source:ZFIN;Acc:ZDB-GENE-040426-2380] | 406529 | YES | YES | NA | NA | |||||
| ENSDARG00000021564 | VDAC3 (2 of 2) | voltage-dependent anion channel 3 [Source:HGNC Symbol;Acc:HGNC:12674] | 393186 | YES | YES | NA | NA | |||||
| ENSDARG00000070097 | vimp | VCP-interacting membrane protein [Source:ZFIN;Acc:ZDB-GENE-060804-1] | 735249 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000039225 | vps13a | vacuolar protein sorting 13 homolog A (S. cerevisiae) [Source:ZFIN;Acc:ZDB-GENE-030926-1] | 378837 | YES | YES | Cytoplasm | transporter | |||||
| ENSDARG00000074465 | VPS13C | vacuolar protein sorting 13 homolog C (S. cerevisiae) [Source:HGNC Symbol;Acc:HGNC:23594] | 798746 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000008224 | vps35 | vacuolar protein sorting 35 homolog (S. cerevisiae) [Source:ZFIN;Acc:ZDB-GENE-030131-2042] | 561697 | YES | YES | Cytoplasm | transporter | |||||
| ENSDARG00000061180 | vps45 | vacuolar protein sorting 45 homolog (S. cerevisiae) [Source:ZFIN;Acc:ZDB-GENE-060503-526] | 561653 | YES | YES | Cytoplasm | transporter | |||||
| ENSDARG00000023228 | vsnl1a | visinin-like 1a [Source:ZFIN;Acc:ZDB-GENE-041001-211] | 492494 | YES | YES | NA | NA | |||||
| ENSDARG00000044053 | vsnl1b | visinin-like 1b [Source:ZFIN;Acc:ZDB-GENE-080225-39] | 561162 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000077940 | VSTM2B (1 of 2) | V-set and transmembrane domain containing 2B [Source:HGNC Symbol;Acc:HGNC:33595] | 793103 | YES | YES | Other | other | |||||
| ENSDARG00000092233 | vtg1 | vitellogenin 1 [Source:ZFIN;Acc:ZDB-GENE-001201-1] | 559475 | YES | YES | NA | NA | |||||
| ENSDARG00000055809 | vtg2 | vitellogenin 2 [Source:ZFIN;Acc:ZDB-GENE-001201-2] | 559931 | YES | YES | NA | NA | |||||
| ENSDARG00000092419 | vtg7 | vitellogenin 7 [Source:ZFIN;Acc:ZDB-GENE-001201-6] | 559856 | YES | YES | NA | NA | |||||
| ENSDARG00000045065 | vti1a | vesicle transport through interaction with t-SNAREs 1A [Source:ZFIN;Acc:ZDB-GENE-050809-135] | 567385 | YES | YES | Plasma Membrane | transporter | |||||
| ENSDARG00000073905 | VWA5A (2 of 4) | von Willebrand factor A domain containing 5A [Source:HGNC Symbol;Acc:HGNC:6658] | 0 | YES | YES | Other | other | |||||
| ENSDARG00000078890 | wdfy3 | WD repeat and FYVE domain containing 3 [Source:ZFIN;Acc:ZDB-GENE-070705-153] | 568432 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000006399 | ywhae1 | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, epsilon polypeptide 1 [Source:ZFIN;Acc:ZDB-GENE-030131-779] | 322060 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000017014 | ywhae2 | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, epsilon polypeptide 2 [Source:ZFIN;Acc:ZDB-GENE-050306-44] | 503763 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000042539 | ywhaqa | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta polypeptide a [Source:ZFIN;Acc:ZDB-GENE-040122-5] | 399487 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000023323 | ywhaqb | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta polypeptide b [Source:ZFIN;Acc:ZDB-GENE-030131-7135] | 335195 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000033599 | zgc:100906 | zgc:100906 [Source:ZFIN;Acc:ZDB-GENE-040801-20] | 445123 | YES | YES | NA | NA | |||||
| ENSDARG00000087243 | zgc:100918 | zgc:100918 [Source:ZFIN;Acc:ZDB-GENE-040927-2] | 449549 | YES | YES | NA | NA | |||||
| ENSDARG00000073845 | zgc:110843 | zgc:110843 [Source:ZFIN;Acc:ZDB-GENE-050327-15] | 541492 | YES | YES | NA | NA | |||||
| ENSDARG00000003127 | zgc:123105 | zgc:123105 [Source:ZFIN;Acc:ZDB-GENE-051113-272] | 664766 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000069846 | zgc:162944 | zgc:162944 [Source:ZFIN;Acc:ZDB-GENE-070424-75] | 569993 | YES | YES | NA | NA | |||||
| ENSDARG00000031435 | zgc:56493 | zgc:56493 [Source:ZFIN;Acc:ZDB-GENE-030131-8581] | 336637 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000069734 | zgc:92880 | zgc:92880 [Source:ZFIN;Acc:ZDB-GENE-040801-164] | 445037 | YES | YES | Cytoplasm | transporter | |||||
| ENSDARG00000077545 | novel gene | 0 | 558390 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSDARG00000090164 | novel gene | 0 | 101884007 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000030656 | sept3 | septin 3 [Source:ZFIN;Acc:ZDB-GENE-050522-375] | 554123 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000010721 | sept6 | septin 6 [Source:ZFIN;Acc:ZDB-GENE-030131-1414] | 322694 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000104085 | abcd3a | ATP-binding cassette, sub-family D (ALD), member 3a [Source:ZFIN;Acc:ZDB-GENE-040426-2868] | 406803 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSDARG00000010155 | abi1a | abl-interactor 1a [Source:ZFIN;Acc:ZDB-GENE-040426-1701] | 393711 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000062991 | abi1b | abl-interactor 1b [Source:ZFIN;Acc:ZDB-GENE-060929-1182] | 767738 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000058207 | abi2a | abl-interactor 2a [Source:ZFIN;Acc:ZDB-GENE-060929-192] | 767646 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000060149 | ablim1a | actin binding LIM protein 1a [Source:ZFIN;Acc:ZDB-GENE-080219-41] | 100003558 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000055223 | ablim2 | actin binding LIM protein family, member 2 [Source:ZFIN;Acc:ZDB-GENE-100316-5] | 568454 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000045411 | acad11 | acyl-CoA dehydrogenase family, member 11 [Source:ZFIN;Acc:ZDB-GENE-040426-814] | 393147 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000037746 | actb1 | actin, beta 1 [Source:ZFIN;Acc:ZDB-GENE-000329-1] | 57934 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000037870 | actb2 | actin, beta 2 [Source:ZFIN;Acc:ZDB-GENE-000329-3] | 57935 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000091138 | ACTR3 (1 of 2) | ARP3 actin-related protein 3 homolog (yeast) [Source:HGNC Symbol;Acc:HGNC:170] | 0 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000100510 | ACTR3 (2 of 2) | ARP3 actin-related protein 3 homolog (yeast) [Source:HGNC Symbol;Acc:HGNC:170] | 0 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000008790 | actr3b | ARP3 actin-related protein 3 homolog B (yeast) [Source:ZFIN;Acc:ZDB-GENE-070424-10] | 100038776 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000027740 | adcyap1b | adenylate cyclase activating polypeptide 1b [Source:ZFIN;Acc:ZDB-GENE-041010-89] | 335625 | YES | YES | YES | Extracellular Space | other | ||||
| ENSDARG00000062123 | agap1 | ArfGAP with GTPase domain, ankyrin repeat and PH domain 1 [Source:ZFIN;Acc:ZDB-GENE-130405-1] | 100148154 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000087822 | AGAP3 | ArfGAP with GTPase domain, ankyrin repeat and PH domain 3 [Source:HGNC Symbol;Acc:HGNC:16923] | 0 | YES | YES | YES | Nucleus | transcription regulator | ||||
| ENSDARG00000038655 | ajap1 | adherens junctions associated protein 1 [Source:ZFIN;Acc:ZDB-GENE-041210-353] | 0 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000074791 | aldh3a1 | aldehyde dehydrogenase 3 family, member A1 [Source:ZFIN;Acc:ZDB-GENE-120823-1] | 100000026 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000015546 | alpl | alkaline phosphatase, liver/bone/kidney [Source:ZFIN;Acc:ZDB-GENE-040420-1] | 393982 | YES | YES | YES | Plasma Membrane | phosphatase | ||||
| ENSDARG00000078901 | anks1ab | ankyrin repeat and sterile alpha motif domain containing 1Ab [Source:ZFIN;Acc:ZDB-GENE-110107-1] | 100003659 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000003512 | anks1b | ankyrin repeat and sterile alpha motif domain containing 1B [Source:ZFIN;Acc:ZDB-GENE-041210-349] | 565408 | YES | YES | YES | Nucleus | other | ||||
| ENSDARG00000058504 | ap1s2 | adaptor-related protein complex 1, sigma 2 subunit [Source:ZFIN;Acc:ZDB-GENE-030131-5448] | 327237 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSDARG00000002790 | ap2m1a | adaptor-related protein complex 2, mu 1 subunit, a [Source:ZFIN;Acc:ZDB-GENE-030131-9784] | 321051 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSDARG00000026967 | ap2s1 | adaptor-related protein complex 2, sigma 1 subunit [Source:ZFIN;Acc:ZDB-GENE-040426-2174] | 100148085 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSDARG00000023713 | aqp1a.1 | aquaporin 1a (Colton blood group), tandem duplicate 1 [Source:ZFIN;Acc:ZDB-GENE-030131-7764] | 335821 | YES | YES | YES | Plasma Membrane | transporter | ||||
| ENSDARG00000104086 | arhgap39 | Rho GTPase activating protein 39 [Source:ZFIN;Acc:ZDB-GENE-041014-247] | 101885231 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000061746 | arhgef9a | Cdc42 guanine nucleotide exchange factor (GEF) 9a [Source:ZFIN;Acc:ZDB-GENE-070705-271] | 559868 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000078624 | arhgef9b | Cdc42 guanine nucleotide exchange factor (GEF) 9b [Source:ZFIN;Acc:ZDB-GENE-030131-7745] | 560923 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000057882 | arpc3 | actin related protein 2/3 complex, subunit 3 [Source:ZFIN;Acc:ZDB-GENE-040625-107] | 415204 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000058225 | arpc4l | actin related protein 2/3 complex, subunit 4, like [Source:ZFIN;Acc:ZDB-GENE-040808-18] | 114456 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000041720 | arpc5la | actin related protein 2/3 complex, subunit 5-like, a [Source:ZFIN;Acc:ZDB-GENE-040426-1453] | 393550 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000074329 | arvcfa | armadillo repeat gene deleted in velocardiofacial syndrome a [Source:ZFIN;Acc:ZDB-GENE-091113-43] | 572216 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000086848 | atad3b | ATPase family, AAA domain containing 3B [Source:ZFIN;Acc:ZDB-GENE-040426-1826] | 0 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000036577 | atp6v0cb | ATPase, H+ transporting, lysosomal, V0 subunit cb [Source:ZFIN;Acc:ZDB-GENE-030131-4127] | 325402 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSDARG00000062799 | baiap2a | BAI1-associated protein 2a [Source:ZFIN;Acc:ZDB-GENE-060421-2756] | 678518 | YES | YES | YES | Plasma Membrane | kinase | ||||
| ENSDARG00000037009 | banf1 | barrier to autointegration factor 1 [Source:ZFIN;Acc:ZDB-GENE-030131-6657] | 334717 | YES | YES | YES | Nucleus | other | ||||
| ENSDARG00000016904 | bckdk | branched chain ketoacid dehydrogenase kinase [Source:ZFIN;Acc:ZDB-GENE-021204-1] | 282677 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSDARG00000043673 | BEGAIN | brain-enriched guanylate kinase-associated [Source:HGNC Symbol;Acc:HGNC:24163] | 563117 | YES | YES | YES | Nucleus | other | ||||
| ENSDARG00000041974 | brk1 | BRICK1, SCAR/WAVE actin-nucleating complex subunit [Source:ZFIN;Acc:ZDB-GENE-040625-77] | 415187 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000079161 | bsnb | bassoon (presynaptic cytomatrix protein) b [Source:ZFIN;Acc:ZDB-GENE-120628-1] | 100329660 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000063255 | btbd11a | BTB (POZ) domain containing 11a [Source:ZFIN;Acc:ZDB-GENE-050419-142] | 562893 | YES | YES | YES | Other | transcription regulator | ||||
| ENSDARG00000099321 | btbd17a | BTB (POZ) domain containing 17a [Source:ZFIN;Acc:ZDB-GENE-130531-68] | 563263 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000102472 | btbd17b | BTB (POZ) domain containing 17b [Source:ZFIN;Acc:ZDB-GENE-091204-214] | 563672 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000030038 | C20H12orf57 | chromosome 12 open reading frame 57 [Source:HGNC Symbol;Acc:HGNC:29521] | 323559 | YES | YES | YES | Other | other | ||||
| ENSDARG00000043842 | C20H1orf198 | chromosome 1 open reading frame 198 [Source:HGNC Symbol;Acc:HGNC:25900] | 568408 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000057556 | C21H18orf32 | chromosome 18 open reading frame 32 [Source:HGNC Symbol;Acc:HGNC:31690] | 336641 | YES | YES | YES | Other | other | ||||
| ENSDARG00000100152 | C25H12orf75 | chromosome 12 open reading frame 75 [Source:HGNC Symbol;Acc:HGNC:35164] | 100535414 | YES | YES | YES | Other | other | ||||
| ENSDARG00000076268 | C5H9orf172 (1 of 2) | chromosome 9 open reading frame 172 [Source:HGNC Symbol;Acc:HGNC:37284] | 568961 | YES | YES | YES | Other | other | ||||
| ENSDARG00000078891 | C7H7orf43 | chromosome 7 open reading frame 43 [Source:HGNC Symbol;Acc:HGNC:25604] | 100003958 | YES | YES | YES | Other | other | ||||
| ENSDARG00000076873 | C8H9orf172 (2 of 2) | chromosome 9 open reading frame 172 [Source:HGNC Symbol;Acc:HGNC:37284] | 799850 | YES | YES | YES | Other | other | ||||
| ENSDARG00000052644 | ca10a | carbonic anhydrase Xa [Source:ZFIN;Acc:ZDB-GENE-051030-123] | 641327 | YES | YES | YES | Other | enzyme | ||||
| ENSDARG00000059349 | CABZ01090374.1 | Uncharacterized protein [Source:UniProtKB/TrEMBL;Acc:F1QU32] | 0 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000069046 | CABZ01118678.1 | Uncharacterized protein [Source:UniProtKB/TrEMBL;Acc:E7FCW1] | 0 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000037905 | cacna1aa | calcium channel, voltage-dependent, P/Q type, alpha 1A subunit, a [Source:ZFIN;Acc:ZDB-GENE-040724-26] | 562059 | YES | YES | YES | Plasma Membrane | ion channel | ||||
| ENSDARG00000006923 | cacna1ab | calcium channel, voltage-dependent, P/Q type, alpha 1A subunit, b [Source:ZFIN;Acc:ZDB-GENE-090514-3] | 569528 | YES | YES | YES | Plasma Membrane | ion channel | ||||
| ENSDARG00000001881 | cacnb3a | calcium channel, voltage-dependent, beta 3a [Source:ZFIN;Acc:ZDB-GENE-060221-2] | 565158 | YES | YES | YES | Plasma Membrane | ion channel | ||||
| ENSDARG00000076030 | cacnb3b | calcium channel, voltage-dependent, beta 3b [Source:ZFIN;Acc:ZDB-GENE-090127-1] | 559015 | YES | YES | YES | Plasma Membrane | ion channel | ||||
| ENSDARG00000009738 | cacnb4a | calcium channel, voltage-dependent, beta 4a subunit [Source:ZFIN;Acc:ZDB-GENE-080320-2] | 562422 | YES | YES | YES | Plasma Membrane | ion channel | ||||
| ENSDARG00000062174 | cacnb4b | calcium channel, voltage-dependent, beta 4b subunit [Source:ZFIN;Acc:ZDB-GENE-060421-7907] | 678560 | YES | YES | YES | Plasma Membrane | ion channel | ||||
| ENSDARG00000032565 | cacng2a | calcium channel, voltage-dependent, gamma subunit 2a [Source:ZFIN;Acc:ZDB-GENE-040426-1401] | 393614 | YES | YES | YES | Plasma Membrane | ion channel | ||||
| ENSDARG00000102376 | cacng2b | calcium channel, voltage-dependent, gamma subunit 2b [Source:ZFIN;Acc:ZDB-GENE-090624-3] | 569855 | YES | YES | YES | Plasma Membrane | ion channel | ||||
| ENSDARG00000020450 | cacng8a | calcium channel, voltage-dependent, gamma subunit 8a [Source:ZFIN;Acc:ZDB-GENE-050417-239] | 550428 | YES | YES | YES | Plasma Membrane | ion channel | ||||
| ENSDARG00000070626 | cacng8b | calcium channel, voltage-dependent, gamma subunit 8b [Source:ZFIN;Acc:ZDB-GENE-100921-72] | 100003329 | YES | YES | YES | Plasma Membrane | ion channel | ||||
| ENSDARG00000100089 | camk2b2 | calcium/calmodulin-dependent protein kinase (CaM kinase) II beta 2 [Source:ZFIN;Acc:ZDB-GENE-090312-34] | 0 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSDARG00000014273 | camk2d2 | calcium/calmodulin-dependent protein kinase (CaM kinase) II delta 2 [Source:ZFIN;Acc:ZDB-GENE-040718-277] | 436815 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000071395 | camk2g1 | calcium/calmodulin-dependent protein kinase (CaM kinase) II gamma 1 [Source:ZFIN;Acc:ZDB-GENE-050913-146] | 791855 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSDARG00000056206 | camk2g2 | calcium/calmodulin-dependent protein kinase (CaM kinase) II gamma 2 [Source:ZFIN;Acc:ZDB-GENE-070323-2] | 791855 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSDARG00000056090 | capza1b | capping protein (actin filament) muscle Z-line, alpha 1b [Source:ZFIN;Acc:ZDB-GENE-031002-24] | 474327 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000100252 | capzb | capping protein (actin filament) muscle Z-line, beta [Source:ZFIN;Acc:ZDB-GENE-030131-7023] | 335083 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000036281 | CC2D1A | coiled-coil and C2 domain containing 1A [Source:HGNC Symbol;Acc:HGNC:30237] | 0 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000079191 | CD200 (2 of 3) | CD200 molecule [Source:HGNC Symbol;Acc:HGNC:7203] | 571844 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000014215 | cdh13 | cadherin 13, H-cadherin (heart) [Source:ZFIN;Acc:ZDB-GENE-060503-286] | 558732 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000012204 | CDK18 | cyclin-dependent kinase 18 [Source:HGNC Symbol;Acc:HGNC:8751] | 100150007 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSDARG00000015240 | cdkl5 | cyclin-dependent kinase-like 5 [Source:ZFIN;Acc:ZDB-GENE-081022-110] | 559341 | YES | YES | YES | Nucleus | kinase | ||||
| ENSDARG00000088825 | CIT (1 of 2) | citron rho-interacting serine/threonine kinase [Source:HGNC Symbol;Acc:HGNC:1985] | 0 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSDARG00000089856 | CIT (2 of 2) | citron rho-interacting serine/threonine kinase [Source:HGNC Symbol;Acc:HGNC:1985] | 0 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSDARG00000069615 | ckmt2a | creatine kinase, mitochondrial 2a (sarcomeric) [Source:ZFIN;Acc:ZDB-GENE-030131-5717] | 327506 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSDARG00000074480 | cnksr2a | connector enhancer of kinase suppressor of Ras 2a [Source:ZFIN;Acc:ZDB-GENE-060228-5] | 561623 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000073707 | cnksr2b | connector enhancer of kinase suppressor of Ras 2b [Source:ZFIN;Acc:ZDB-GENE-030131-8048] | 558834 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000005210 | coro2a | coronin, actin binding protein, 2A [Source:ZFIN;Acc:ZDB-GENE-030131-2258] | 323538 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000079440 | coro2ba | coronin, actin binding protein, 2Ba [Source:ZFIN;Acc:ZDB-GENE-050518-1] | 560235 | YES | YES | YES | Other | other | ||||
| ENSDARG00000070919 | cpne5 | copine V [Source:ZFIN;Acc:ZDB-GENE-040426-1398] | 394003 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000061466 | CPNE5 (1 of 2) | copine V [Source:HGNC Symbol;Acc:HGNC:2318] | 568003 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000102191 | CPNE8 (2 of 2) | copine VIII [Source:HGNC Symbol;Acc:HGNC:23498] | 101886030 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000016545 | CRAT (1 of 2) | carnitine O-acetyltransferase [Source:HGNC Symbol;Acc:HGNC:2342] | 562075 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000098782 | cratb | carnitine O-acetyltransferase b [Source:ZFIN;Acc:ZDB-GENE-040912-162] | 447855 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000008370 | csnk1da | casein kinase 1, delta a [Source:ZFIN;Acc:ZDB-GENE-030131-825] | 322106 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSDARG00000045150 | csnk1e | casein kinase 1, epsilon [Source:ZFIN;Acc:ZDB-GENE-030131-7873] | 100006858 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSDARG00000104342 | csnk1g1 | casein kinase 1, gamma 1 [Source:ZFIN;Acc:ZDB-GENE-041212-63] | 494092 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSDARG00000034056 | csnk1g2b | casein kinase 1, gamma 2b [Source:ZFIN;Acc:ZDB-GENE-060825-285] | 561724 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSDARG00000002082 | csnk2a1 | casein kinase 2, alpha 1 polypeptide [Source:ZFIN;Acc:ZDB-GENE-990415-27] | 30502 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000013582 | csnk2a2b | casein kinase 2, alpha prime polypeptide b [Source:ZFIN;Acc:ZDB-GENE-090406-4] | 556379 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSDARG00000077776 | csnk2b | casein kinase 2, beta polypeptide [Source:ZFIN;Acc:ZDB-GENE-990415-29] | 30428 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSDARG00000018162 | ctnnal1 | catenin (cadherin-associated protein), alpha-like 1 [Source:ZFIN;Acc:ZDB-GENE-060503-507] | 100034581 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000041799 | cx43 | connexin 43 [Source:ZFIN;Acc:ZDB-GENE-991105-4] | 30236 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000073896 | cx47.1 | connexin 47.1 [Source:ZFIN;Acc:ZDB-GENE-040912-134] | 447835 | YES | YES | YES | Plasma Membrane | transporter | ||||
| ENSDARG00000028800 | CYTH2 | cytohesin 2 [Source:HGNC Symbol;Acc:HGNC:9502] | 569358 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000005159 | cyth3b | cytohesin 3b [Source:ZFIN;Acc:ZDB-GENE-130530-529] | 0 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000060093 | dapk1 | death-associated protein kinase 1 [Source:ZFIN;Acc:ZDB-GENE-060526-177] | 558314 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSDARG00000034093 | dclk2 | doublecortin-like kinase 2 [Source:ZFIN;Acc:ZDB-GENE-050420-170] | 572548 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSDARG00000038068 | ddx5 | DEAD (Asp-Glu-Ala-Asp) box helicase 5 [Source:ZFIN;Acc:ZDB-GENE-030131-925] | 322206 | YES | YES | YES | Nucleus | enzyme | ||||
| ENSDARG00000003869 | decr1 | 2,4-dienoyl CoA reductase 1, mitochondrial [Source:ZFIN;Acc:ZDB-GENE-040718-142] | 436717 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000021135 | dhrs2 | dehydrogenase/reductase (SDR family) member 2 [Source:ZFIN;Acc:ZDB-GENE-040426-1498] | 393539 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000015392 | dhx15 | DEAH (Asp-Glu-Ala-His) box helicase 15 [Source:ZFIN;Acc:ZDB-GENE-030131-650] | 321931 | YES | YES | YES | Nucleus | enzyme | ||||
| ENSDARG00000063419 | DHX35 | DEAH (Asp-Glu-Ala-His) box polypeptide 35 [Source:HGNC Symbol;Acc:HGNC:15861] | 0 | YES | YES | YES | Other | enzyme | ||||
| ENSDARG00000062612 | dhx37 | DEAH (Asp-Glu-Ala-His) box polypeptide 37 [Source:ZFIN;Acc:ZDB-GENE-030131-4505] | 100009635 | YES | YES | YES | Other | enzyme | ||||
| ENSDARG00000099323 | dlg2 | discs, large homolog 2 (Drosophila) [Source:ZFIN;Acc:ZDB-GENE-050221-3] | 497638 | YES | YES | YES | Plasma Membrane | kinase | ||||
| ENSDARG00000076796 | dlg3 | discs, large homolog 3 (Drosophila) [Source:ZFIN;Acc:ZDB-GENE-050208-93] | 564081 | YES | YES | YES | Plasma Membrane | kinase | ||||
| ENSDARG00000041926 | DLG4 (2 of 2) | discs, large homolog 4 (Drosophila) [Source:HGNC Symbol;Acc:HGNC:2903] | 0 | YES | YES | YES | Plasma Membrane | kinase | ||||
| ENSDARG00000014280 | dlgap1a | discs, large (Drosophila) homolog-associated protein 1a [Source:ZFIN;Acc:ZDB-GENE-030616-580] | 569898 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000076070 | DLGAP2 (2 of 3) | discs, large (Drosophila) homolog-associated protein 2 [Source:HGNC Symbol;Acc:HGNC:2906] | 0 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000059340 | dlgap2a | discs, large (Drosophila) homolog-associated protein 2a [Source:ZFIN;Acc:ZDB-GENE-101215-1] | 557621 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000092376 | dlgap2b | discs, large (Drosophila) homolog-associated protein 2b [Source:ZFIN;Acc:ZDB-GENE-040724-191] | 0 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000055459 | dlgap3 | discs, large (Drosophila) homolog-associated protein 3 [Source:ZFIN;Acc:ZDB-GENE-060503-413] | 557556 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000012823 | dlgap4b | discs, large (Drosophila) homolog-associated protein 4b [Source:ZFIN;Acc:ZDB-GENE-101215-2] | 0 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000104066 | dnaja2 | DnaJ (Hsp40) homolog, subfamily A, member 2 [Source:ZFIN;Acc:ZDB-GENE-040426-2884] | 406814 | YES | YES | YES | Nucleus | enzyme | ||||
| ENSDARG00000058494 | dnaja3a | DnaJ (Hsp40) homolog, subfamily A, member 3A [Source:ZFIN;Acc:ZDB-GENE-030131-7837] | 335894 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000039363 | dnajb12a | DnaJ (Hsp40) homolog, subfamily B, member 12a [Source:ZFIN;Acc:ZDB-GENE-030131-2725] | 324005 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000004680 | dnajb6a | DnaJ (Hsp40) homolog, subfamily B, member 6a [Source:ZFIN;Acc:ZDB-GENE-040718-45] | 436626 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000088293 | doc2b | double C2-like domains, beta [Source:ZFIN;Acc:ZDB-GENE-090615-1] | 0 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSDARG00000076302 | dtx1 | deltex1 [Source:EntrezGene;Acc:570481] | 570481 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000074372 | elfn1b | extracellular leucine-rich repeat and fibronectin type III domain containing 1b [Source:ZFIN;Acc:ZDB-GENE-080327-21] | 100144401 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000032049 | enah | enabled homolog (Drosophila) [Source:ZFIN;Acc:ZDB-GENE-010323-11] | 541394 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000071224 | ENDOD1 (7 of 10) | endonuclease domain containing 1 [Source:HGNC Symbol;Acc:HGNC:29129] | 559260 | YES | YES | YES | Extracellular Space | enzyme | ||||
| ENSDARG00000040469 | enpp6 | ectonucleotide pyrophosphatase/phosphodiesterase 6 [Source:ZFIN;Acc:ZDB-GENE-031205-1] | 791915 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000029019 | epb41b | erythrocyte membrane protein band 4.1b (elliptocytosis 1, RH-linked) [Source:ZFIN;Acc:ZDB-GENE-030130-1] | 326287 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000061398 | epb41l4b | erythrocyte membrane protein band 4.1 like 4B [Source:ZFIN;Acc:ZDB-GENE-100921-92] | 100332627 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSDARG00000076014 | ERC1 (3 of 3) | ELKS/RAB6-interacting/CAST family member 1 [Source:HGNC Symbol;Acc:HGNC:17072] | 560003 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000009941 | erc1a | ELKS/RAB6-interacting/CAST family member 1a [Source:ZFIN;Acc:ZDB-GENE-091214-5] | 548606 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000035650 | evla | Enah/Vasp-like a [Source:ZFIN;Acc:ZDB-GENE-000607-65] | 58003 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000099720 | evlb | Enah/Vasp-like b [Source:ZFIN;Acc:ZDB-GENE-040426-1804] | 0 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000026762 | fam126a | family with sequence similarity 126, member A [Source:ZFIN;Acc:ZDB-GENE-040426-1806] | 402926 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000099022 | faua | Finkel-Biskis-Reilly murine sarcoma virus (FBR-MuSV) ubiquitously expressed a [Source:ZFIN;Acc:ZDB-GENE-040426-1700] | 393710 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000078908 | fbxo41 | F-box protein 41 [Source:ZFIN;Acc:ZDB-GENE-090313-350] | 567804 | YES | YES | YES | Other | other | ||||
| ENSDARG00000008969 | fgb | fibrinogen beta chain [Source:ZFIN;Acc:ZDB-GENE-030131-9261] | 337315 | YES | YES | YES | Extracellular Space | other | ||||
| ENSDARG00000037281 | fgg | fibrinogen gamma chain [Source:ZFIN;Acc:ZDB-GENE-040426-1998] | 406327 | YES | YES | YES | Extracellular Space | other | ||||
| ENSDARG00000037998 | flot1b | flotillin 1b [Source:ZFIN;Acc:ZDB-GENE-020430-2] | 245697 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000098751 | FQ323163.2 | Uncharacterized protein [Source:UniProtKB/TrEMBL;Acc:E7FDI0] | 795227 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000074865 | frmpd3 | FERM and PDZ domain containing 3 [Source:ZFIN;Acc:ZDB-GENE-100316-9] | 798302 | YES | YES | YES | Other | other | ||||
| ENSDARG00000078688 | frs3 | fibroblast growth factor receptor substrate 3 [Source:ZFIN;Acc:ZDB-GENE-131121-119] | 100330147 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000027200 | gabarapl2 | GABA(A) receptor-associated protein-like 2 [Source:ZFIN;Acc:ZDB-GENE-040718-336] | 403036 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000018967 | gabbr1a | gamma-aminobutyric acid (GABA) B receptor, 1a [Source:ZFIN;Acc:ZDB-GENE-030904-5] | 373873 | YES | YES | YES | Plasma Membrane | G-protein coupled receptor | ||||
| ENSDARG00000016667 | gabbr1b | gamma-aminobutyric acid (GABA) B receptor, 1b [Source:ZFIN;Acc:ZDB-GENE-060503-5] | 558708 | YES | YES | YES | Plasma Membrane | G-protein coupled receptor | ||||
| ENSDARG00000079586 | gabrb2 | gamma-aminobutyric acid (GABA) A receptor, beta 2 [Source:ZFIN;Acc:ZDB-GENE-030131-8196] | 0 | YES | YES | YES | Plasma Membrane | ion channel | ||||
| ENSDARG00000053665 | gabrg2 | gamma-aminobutyric acid (GABA) A receptor, gamma 2 [Source:ZFIN;Acc:ZDB-GENE-091118-65] | 553402 | YES | YES | YES | Plasma Membrane | ion channel | ||||
| ENSDARG00000007264 | gbas | glioblastoma amplified sequence [Source:ZFIN;Acc:ZDB-GENE-991008-18] | 30232 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000098831 | gcdhb | glutaryl-CoA dehydrogenase b [Source:ZFIN;Acc:ZDB-GENE-041010-117] | 450003 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000016837 | glipr2l | GLI pathogenesis-related 2, like [Source:ZFIN;Acc:ZDB-GENE-041010-53] | 449805 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000012019 | glra1 | glycine receptor, alpha 1 [Source:ZFIN;Acc:ZDB-GENE-991117-1] | 30676 | YES | YES | YES | Plasma Membrane | ion channel | ||||
| ENSDARG00000008816 | glud1a | glutamate dehydrogenase 1a [Source:ZFIN;Acc:ZDB-GENE-030114-2] | 317737 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000101074 | glud1b | glutamate dehydrogenase 1b [Source:ZFIN;Acc:ZDB-GENE-030828-1] | 373092 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000038693 | gng7 | guanine nucleotide binding protein (G protein), gamma 7 [Source:ZFIN;Acc:ZDB-GENE-040718-93] | 436670 | YES | YES | YES | Plasma Membrane | enzyme | ||||
| ENSDARG00000089076 | gphna | gephyrin a [Source:ZFIN;Acc:ZDB-GENE-110127-3] | 555470 | YES | YES | YES | Plasma Membrane | enzyme | ||||
| ENSDARG00000100851 | gphnb | gephyrin b [Source:ZFIN;Acc:ZDB-GENE-070705-4] | 0 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000077080 | gprc5ba | G protein-coupled receptor, class C, group 5, member Ba [Source:ZFIN;Acc:ZDB-GENE-030131-8162] | 570207 | YES | YES | YES | Plasma Membrane | G-protein coupled receptor | ||||
| ENSDARG00000075141 | gprc5bb | G protein-coupled receptor, class C, group 5, member Bb [Source:ZFIN;Acc:ZDB-GENE-090313-75] | 100002676 | YES | YES | YES | Plasma Membrane | G-protein coupled receptor | ||||
| ENSDARG00000075043 | GPRIN3 | GPRIN family member 3 [Source:HGNC Symbol;Acc:HGNC:27733] | 100536832 | YES | YES | YES | Other | other | ||||
| ENSDARG00000038059 | grb2b | growth factor receptor-bound protein 2b [Source:ZFIN;Acc:ZDB-GENE-040426-1975] | 406308 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000070173 | gria2a | glutamate receptor, ionotropic, AMPA 2a [Source:ZFIN;Acc:ZDB-GENE-020125-3] | 170450 | YES | YES | YES | Plasma Membrane | ion channel | ||||
| ENSDARG00000052765 | gria2b | glutamate receptor, ionotropic, AMPA 2b [Source:ZFIN;Acc:ZDB-GENE-020125-4] | 170451 | YES | YES | YES | Plasma Membrane | ion channel | ||||
| ENSDARG00000032737 | gria3a | glutamate receptor, ionotropic, AMPA 3a [Source:ZFIN;Acc:ZDB-GENE-020125-5] | 170452 | YES | YES | YES | Plasma Membrane | ion channel | ||||
| ENSDARG00000037498 | gria3b | glutamate receptor, ionotropic, AMPA 3b [Source:ZFIN;Acc:ZDB-GENE-030616-53] | 368416 | YES | YES | YES | Plasma Membrane | ion channel | ||||
| ENSDARG00000037496 | gria4a | glutamate receptor, ionotropic, AMPA 4a [Source:ZFIN;Acc:ZDB-GENE-020125-7] | 407735 | YES | YES | YES | Plasma Membrane | ion channel | ||||
| ENSDARG00000059368 | gria4b | glutamate receptor, ionotropic, AMPA 4b [Source:ZFIN;Acc:ZDB-GENE-030131-8013] | 336069 | YES | YES | YES | Plasma Membrane | ion channel | ||||
| ENSDARG00000076103 | grid2ipa | glutamate receptor, ionotropic, delta 2 (Grid2) interacting protein, a [Source:ZFIN;Acc:ZDB-GENE-091113-42] | 561017 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000095603 | grid2ipb | glutamate receptor, ionotropic, delta 2 (Grid2) interacting protein, b [Source:ZFIN;Acc:ZDB-GENE-040724-161] | 559717 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000027828 | grin1a | glutamate receptor, ionotropic, N-methyl D-aspartate 1a [Source:ZFIN;Acc:ZDB-GENE-051202-1] | 767745 | YES | YES | YES | Plasma Membrane | ion channel | ||||
| ENSDARG00000025728 | grin1b | glutamate receptor, ionotropic, N-methyl D-aspartate 1b [Source:ZFIN;Acc:ZDB-GENE-051202-2] | 100005675 | YES | YES | YES | Plasma Membrane | ion channel | ||||
| ENSDARG00000034493 | grin2aa | glutamate receptor, ionotropic, N-methyl D-aspartate 2A, a [Source:ZFIN;Acc:ZDB-GENE-070424-129] | 563297 | YES | YES | YES | Plasma Membrane | ion channel | ||||
| ENSDARG00000070543 | grin2ab | glutamate receptor, ionotropic, N-methyl D-aspartate 2A, b [Source:ZFIN;Acc:ZDB-GENE-070424-223] | 570493 | YES | YES | YES | Plasma Membrane | ion channel | ||||
| ENSDARG00000079348 | GRIN2B (2 of 2) | glutamate receptor, ionotropic, N-methyl D-aspartate 2B [Source:HGNC Symbol;Acc:HGNC:4586] | 0 | YES | YES | YES | Plasma Membrane | ion channel | ||||
| ENSDARG00000030376 | grin2bb | glutamate receptor, ionotropic, N-methyl D-aspartate 2B, genome duplicate b [Source:ZFIN;Acc:ZDB-GENE-061207-27] | 559976 | YES | YES | YES | Plasma Membrane | ion channel | ||||
| ENSDARG00000070620 | grin2db | glutamate receptor, ionotropic, N-methyl D-aspartate 2D, b [Source:ZFIN;Acc:ZDB-GENE-100920-7] | 0 | YES | YES | YES | Plasma Membrane | ion channel | ||||
| ENSDARG00000015053 | grip1 | glutamate receptor interacting protein 1 [Source:ZFIN;Acc:ZDB-GENE-041210-125] | 558006 | YES | YES | YES | Plasma Membrane | transcription regulator | ||||
| ENSDARG00000027618 | grip2b | glutamate receptor interacting protein 2b [Source:ZFIN;Acc:ZDB-GENE-070705-172] | 562290 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000040156 | grm4 | glutamate receptor, metabotropic 4 [Source:ZFIN;Acc:ZDB-GENE-030131-5781] | 567181 | YES | YES | YES | Plasma Membrane | G-protein coupled receptor | ||||
| ENSDARG00000011459 | gsna | gelsolin a [Source:ZFIN;Acc:ZDB-GENE-030131-9653] | 321099 | YES | YES | YES | Extracellular Space | other | ||||
| ENSDARG00000099214 | h2afvb | H2A histone family, member Vb [Source:ZFIN;Acc:ZDB-GENE-050506-24] | 403077 | YES | YES | YES | Nucleus | other | ||||
| ENSDARG00000068516 | hapln1b | hyaluronan and proteoglycan link protein 1b [Source:ZFIN;Acc:ZDB-GENE-050920-1] | 569074 | YES | YES | YES | Extracellular Space | other | ||||
| ENSDARG00000035519 | histh1l | histone H1 like [Source:ZFIN;Acc:ZDB-GENE-050417-145] | 550353 | YES | YES | YES | Nucleus | other | ||||
| ENSDARG00000101759 | homer1b | homer scaffolding protein 1b [Source:ZFIN;Acc:ZDB-GENE-040801-44] | 436769 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000010789 | homer3 | homer scaffolding protein 3 [Source:ZFIN;Acc:ZDB-GENE-040426-739] | 394088 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000102532 | HOMER3 (2 of 2) | homer scaffolding protein 3 [Source:HGNC Symbol;Acc:HGNC:17514] | 100330028 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000044669 | hp1bp3 | heterochromatin protein 1, binding protein 3 [Source:ZFIN;Acc:ZDB-GENE-060810-79] | 558102 | YES | YES | YES | Nucleus | other | ||||
| ENSDARG00000062008 | hs2st1b | heparan sulfate 2-O-sulfotransferase 1b [Source:ZFIN;Acc:ZDB-GENE-100304-1] | 100151174 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000070603 | hspa12a | heat shock protein 12A [Source:ZFIN;Acc:ZDB-GENE-031001-1] | 751725 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000022176 | IGLON5 | IgLON family member 5 [Source:HGNC Symbol;Acc:HGNC:34550] | 550472 | YES | YES | YES | Other | other | ||||
| ENSDARG00000031049 | igsf21a | immunoglobin superfamily, member 21a [Source:ZFIN;Acc:ZDB-GENE-050809-118] | 569872 | YES | YES | YES | Extracellular Space | other | ||||
| ENSDARG00000033845 | igsf9ba | immunoglobulin superfamily, member 9Ba [Source:ZFIN;Acc:ZDB-GENE-060503-729] | 567389 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000069467 | igsf9bb | immunoglobulin superfamily, member 9Bb [Source:ZFIN;Acc:ZDB-GENE-091112-15] | 569554 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000011862 | inaa | internexin neuronal intermediate filament protein, alpha a [Source:ZFIN;Acc:ZDB-GENE-060531-65] | 555251 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000053248 | inab | internexin neuronal intermediate filament protein, alpha b [Source:ZFIN;Acc:ZDB-GENE-990415-83] | 30230 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000016551 | iqsec1b | IQ motif and Sec7 domain 1b [Source:ZFIN;Acc:ZDB-GENE-120813-1] | 100148736 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000077709 | iqsec2 | IQ motif and Sec7 domain 2 [Source:ZFIN;Acc:ZDB-GENE-081105-73] | 797187 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000102125 | IQSEC2 (2 of 2) | IQ motif and Sec7 domain 2 [Source:HGNC Symbol;Acc:HGNC:29059] | 560461 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000099850 | iqsec3a | IQ motif and Sec7 domain 3a [Source:ZFIN;Acc:ZDB-GENE-041210-343] | 797355 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000093091 | iqsec3b | IQ motif and Sec7 domain 3b [Source:ZFIN;Acc:ZDB-GENE-050420-210] | 555340 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000053547 | jakmip2 | janus kinase and microtubule interacting protein 2 [Source:ZFIN;Acc:ZDB-GENE-091204-293] | 559839 | YES | YES | YES | Extracellular Space | other | ||||
| ENSDARG00000069843 | kctd12.1 | potassium channel tetramerisation domain containing 12.1 [Source:ZFIN;Acc:ZDB-GENE-030902-5] | 373865 | YES | YES | YES | Plasma Membrane | ion channel | ||||
| ENSDARG00000004648 | kctd16a | potassium channel tetramerization domain containing 16a [Source:ZFIN;Acc:ZDB-GENE-060117-1] | 567207 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000020102 | kctd16b | potassium channel tetramerization domain containing 16b [Source:ZFIN;Acc:ZDB-GENE-060117-2] | 569756 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000067507 | kctd8 | potassium channel tetramerization domain containing 8 [Source:ZFIN;Acc:ZDB-GENE-060117-3] | 568933 | YES | YES | YES | Other | other | ||||
| ENSDARG00000052856 | khdrbs1a | KH domain containing, RNA binding, signal transduction associated 1a [Source:ZFIN;Acc:ZDB-GENE-000210-25] | 30082 | YES | YES | YES | Nucleus | transcription regulator | ||||
| ENSDARG00000062086 | kiaa1549lb | KIAA1549-like b [Source:ZFIN;Acc:ZDB-GENE-050419-18] | 0 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000010844 | kras | v-Ki-ras2 Kirsten rat sarcoma viral oncogene homolog [Source:ZFIN;Acc:ZDB-GENE-040808-67] | 445289 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000018404 | krt18 | keratin 18 [Source:ZFIN;Acc:ZDB-GENE-030411-6] | 352912 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000028618 | KRT18 (2 of 3) | keratin 18, type I [Source:HGNC Symbol;Acc:HGNC:6430] | 393540 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000058371 | krt5 | keratin 5 [Source:ZFIN;Acc:ZDB-GENE-991110-23] | 0 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000058358 | krt8 | keratin 8 [Source:ZFIN;Acc:ZDB-GENE-030411-5] | 797433 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000044975 | krt94 | keratin 94 [Source:ZFIN;Acc:ZDB-GENE-061027-116] | 768289 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000040803 | lactb | lactamase, beta [Source:ZFIN;Acc:ZDB-GENE-020111-1] | 553619 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000041358 | lgi3 | leucine-rich repeat LGI family, member 3 [Source:ZFIN;Acc:ZDB-GENE-060217-4] | 654830 | YES | YES | YES | Extracellular Space | other | ||||
| ENSDARG00000021595 | lhfpl3 | lipoma HMGIC fusion partner-like 3 [Source:ZFIN;Acc:ZDB-GENE-040801-180] | 445050 | YES | YES | YES | Other | other | ||||
| ENSDARG00000103774 | limch1b | LIM and calponin homology domains 1b [Source:ZFIN;Acc:ZDB-GENE-030131-9210] | 557200 | YES | YES | YES | Other | other | ||||
| ENSDARG00000013414 | lin7a | lin-7 homolog A (C. elegans) [Source:ZFIN;Acc:ZDB-GENE-040426-1665] | 393682 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000037932 | lin7b | lin-7 homolog B (C. elegans) [Source:ZFIN;Acc:ZDB-GENE-041011-1] | 553768 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000027322 | lin7c | lin-7 homolog C (C. elegans) [Source:ZFIN;Acc:ZDB-GENE-041011-2] | 447934 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000044299 | lmnb1 | lamin B1 [Source:ZFIN;Acc:ZDB-GENE-020424-2] | 195816 | YES | YES | YES | Nucleus | other | ||||
| ENSDARG00000101624 | lmnb2 | lamin B2 [Source:ZFIN;Acc:ZDB-GENE-990630-13] | 30196 | YES | YES | YES | Nucleus | other | ||||
| ENSDARG00000079670 | lrrc7 | leucine rich repeat containing 7 [Source:ZFIN;Acc:ZDB-GENE-130530-876] | 566023 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000063411 | lrrc73 | leucine rich repeat containing 73 [Source:ZFIN;Acc:ZDB-GENE-111118-2] | 570766 | YES | YES | YES | Other | other | ||||
| ENSDARG00000032188 | lrrc8a | leucine rich repeat containing 8 family, member A [Source:ZFIN;Acc:ZDB-GENE-030131-5853] | 327642 | YES | YES | YES | Extracellular Space | other | ||||
| ENSDARG00000058558 | lzts3b | leucine zipper, putative tumor suppressor family member 3b [Source:ZFIN;Acc:ZDB-GENE-110411-249] | 560221 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000025974 | magi3b | membrane associated guanylate kinase, WW and PDZ domain containing 3b [Source:ZFIN;Acc:ZDB-GENE-060503-301] | 0 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSDARG00000024017 | mamdc1 | MAM domain containing 1 [Source:ZFIN;Acc:ZDB-GENE-030616-360] | 368826 | YES | YES | YES | Other | other | ||||
| ENSDARG00000033609 | map1lc3a | microtubule-associated protein 1 light chain 3 alpha [Source:ZFIN;Acc:ZDB-GENE-040426-2232] | 406466 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000074521 | map6a | microtubule-associated protein 6a [Source:ZFIN;Acc:ZDB-GENE-050208-42] | 100000844 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000074073 | map6b | microtubule-associated protein 6b [Source:ZFIN;Acc:ZDB-GENE-130530-741] | 100334879 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000079777 | map6d1 | MAP6 domain containing 1 [Source:ZFIN;Acc:ZDB-GENE-081104-293] | 0 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000104014 | MARK1 | MAP/microtubule affinity-regulating kinase 1 [Source:HGNC Symbol;Acc:HGNC:6896] | 0 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSDARG00000032458 | mark2b | MAP/microtubule affinity-regulating kinase 2b [Source:ZFIN;Acc:ZDB-GENE-080215-4] | 100005996 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSDARG00000026630 | mark3b | MAP/microtubule affinity-regulating kinase 3b [Source:ZFIN;Acc:ZDB-GENE-060929-80] | 767634 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSDARG00000023914 | mark4a | MAP/microtubule affinity-regulating kinase 4a [Source:ZFIN;Acc:ZDB-GENE-060531-156] | 569346 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSDARG00000036186 | mbpa | myelin basic protein a [Source:ZFIN;Acc:ZDB-GENE-030128-2] | 326281 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000089413 | mbpb | myelin basic protein b [Source:ZFIN;Acc:ZDB-GENE-030429-21] | 368319 | YES | YES | YES | Extracellular Space | other | ||||
| ENSDARG00000102744 | mgst3 | microsomal glutathione S-transferase 3 [Source:ZFIN;Acc:ZDB-GENE-040426-2767] | 406736 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000034616 | mlf2 | myeloid leukemia factor 2 [Source:ZFIN;Acc:ZDB-GENE-050913-67] | 561675 | YES | YES | YES | Nucleus | other | ||||
| ENSDARG00000060383 | mmaa | methylmalonic aciduria (cobalamin deficiency) cblA type [Source:ZFIN;Acc:ZDB-GENE-070928-15] | 564760 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000056085 | mob4 | MOB family member 4, phocein [Source:ZFIN;Acc:ZDB-GENE-040801-174] | 445045 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000037679 | MPC2 (2 of 2) | mitochondrial pyruvate carrier 2 [Source:HGNC Symbol;Acc:HGNC:24515] | 0 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000102081 | mprip | myosin phosphatase Rho interacting protein [Source:ZFIN;Acc:ZDB-GENE-030131-4471] | 558831 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000105209 | MPRIP (3 of 3) | myosin phosphatase Rho interacting protein [Source:HGNC Symbol;Acc:HGNC:30321] | 562530 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000038609 | mpz | myelin protein zero [Source:ZFIN;Acc:ZDB-GENE-010724-4] | 114417 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000043554 | mrpl40 | mitochondrial ribosomal protein L40 [Source:ZFIN;Acc:ZDB-GENE-040912-37] | 447873 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000076334 | mrpl43 | mitochondrial ribosomal protein L43 [Source:ZFIN;Acc:ZDB-GENE-040718-125] | 436701 | YES | YES | YES | Cytoplasm | translation regulator | ||||
| ENSDARG00000079823 | mrps36 | mitochondrial ribosomal protein S36 [Source:ZFIN;Acc:ZDB-GENE-081104-343] | 567309 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000076155 | mtpap | mitochondrial poly(A) polymerase [Source:ZFIN;Acc:ZDB-GENE-101208-3] | 563801 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000000103 | myh10 | myosin, heavy chain 10, non-muscle [Source:ZFIN;Acc:ZDB-GENE-030616-162] | 555454 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000009782 | myh11a | myosin, heavy chain 11a, smooth muscle [Source:ZFIN;Acc:ZDB-GENE-050531-1] | 554168 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000073732 | myh14 | myosin, heavy chain 14, non-muscle [Source:ZFIN;Acc:ZDB-GENE-100921-1] | 563011 | YES | YES | YES | Extracellular Space | other | ||||
| ENSDARG00000001014 | myh9b | myosin, heavy chain 9b, non-muscle [Source:ZFIN;Acc:ZDB-GENE-030131-998] | 561431 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000099766 | myl12.1 | myosin, light chain 12, genome duplicate 1 [Source:ZFIN;Acc:ZDB-GENE-030131-4918] | 326719 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000008494 | myl6 | myosin, light chain 6, alkali, smooth muscle and non-muscle [Source:ZFIN;Acc:ZDB-GENE-041010-28] | 798632 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000008030 | myl9b | myosin, light chain 9b, regulatory [Source:ZFIN;Acc:ZDB-GENE-040426-2296] | 406493 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000017441 | mylz3 | myosin, light polypeptide 3, skeletal muscle [Source:ZFIN;Acc:ZDB-GENE-000322-6] | 58143 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000061579 | myo1c | myosin IC [Source:ZFIN;Acc:ZDB-GENE-090429-3] | 566459 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000036863 | MYO1D | myosin ID [Source:HGNC Symbol;Acc:HGNC:7598] | 559562 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000078734 | myo1f | myosin IF [Source:ZFIN;Acc:ZDB-GENE-081104-506] | 565345 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000061635 | myo5aa | myosin VAa [Source:ZFIN;Acc:ZDB-GENE-041027-2] | 562188 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000044016 | myo6a | myosin VIa [Source:ZFIN;Acc:ZDB-GENE-040819-2] | 445473 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000053652 | ndufaf6 | NADH dehydrogenase (ubiquinone) complex I, assembly factor 6 [Source:ZFIN;Acc:ZDB-GENE-100112-1] | 569692 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000021200 | nebl | nebulette [Source:ZFIN;Acc:ZDB-GENE-050320-151] | 541446 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000057568 | nefla | neurofilament, light polypeptide a [Source:ZFIN;Acc:ZDB-GENE-091117-1] | 0 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000012426 | neflb | neurofilament, light polypeptide b [Source:ZFIN;Acc:ZDB-GENE-060312-44] | 664698 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000021351 | nefma | neurofilament, medium polypeptide a [Source:ZFIN;Acc:ZDB-GENE-050522-205] | 553718 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000043697 | nefmb | neurofilament, medium polypeptide b [Source:ZFIN;Acc:ZDB-GENE-070103-4] | 100033387 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000021607 | negr1 | neuronal growth regulator 1 [Source:ZFIN;Acc:ZDB-GENE-040822-27] | 445374 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000089066 | NHSL2 | NHS-like 2 [Source:HGNC Symbol;Acc:HGNC:33737] | 794153 | YES | YES | YES | Other | other | ||||
| ENSDARG00000079824 | ninj2 | ninjurin 2 [Source:ZFIN;Acc:ZDB-GENE-100422-19] | 799267 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000077367 | ntng2a | netrin g2a [Source:ZFIN;Acc:ZDB-GENE-081105-186] | 560014 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000089444 | ogmb | oligodendrocyte myelin glycoprotein b [Source:ZFIN;Acc:ZDB-GENE-110628-1] | 101882436 | YES | YES | YES | Plasma Membrane | G-protein coupled receptor | ||||
| ENSDARG00000018270 | olfm1a | olfactomedin 1a [Source:ZFIN;Acc:ZDB-GENE-040718-194] | 436764 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000014053 | olfm1b | olfactomedin 1b [Source:ZFIN;Acc:ZDB-GENE-040801-228] | 445092 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000077847 | olfm2a | olfactomedin 2a [Source:ZFIN;Acc:ZDB-GENE-040426-2882] | 334035 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000102825 | olfm2b | olfactomedin 2b [Source:ZFIN;Acc:ZDB-GENE-040426-2402] | 406544 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000071493 | olfm3a | olfactomedin 3a [Source:ZFIN;Acc:ZDB-GENE-080219-11] | 563182 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000086573 | omga | oligodendrocyte myelin glycoprotein a [Source:ZFIN;Acc:ZDB-GENE-091204-203] | 0 | YES | YES | YES | Plasma Membrane | G-protein coupled receptor | ||||
| ENSDARG00000061761 | opa3 | optic atrophy 3 [Source:ZFIN;Acc:ZDB-GENE-050210-1] | 497278 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000013005 | opcml | opioid binding protein/cell adhesion molecule-like [Source:ZFIN;Acc:ZDB-GENE-040927-3] | 449538 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000055875 | pag1 | phosphoprotein membrane anchor with glycosphingolipid microdomains 1 [Source:ZFIN;Acc:ZDB-GENE-060503-586] | 100034498 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000063299 | pcloa | piccolo presynaptic cytomatrix protein a [Source:ZFIN;Acc:ZDB-GENE-030131-8115] | 100148002 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSDARG00000005220 | pdcd6 | programmed cell death 6 [Source:ZFIN;Acc:ZDB-GENE-040426-1307] | 393925 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000053194 | pdzd11 | PDZ domain containing 11 [Source:ZFIN;Acc:ZDB-GENE-040426-2544] | 405893 | YES | YES | YES | Extracellular Space | other | ||||
| ENSDARG00000035608 | pgam5 | phosphoglycerate mutase family member 5 [Source:ZFIN;Acc:ZDB-GENE-030131-683] | 492357 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000001873 | phgdh | phosphoglycerate dehydrogenase [Source:ZFIN;Acc:ZDB-GENE-030131-647] | 321928 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000079438 | PKP3 (2 of 2) | plakophilin 3 [Source:HGNC Symbol;Acc:HGNC:9025] | 100330353 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000045331 | pkp4 | plakophilin 4 [Source:ZFIN;Acc:ZDB-GENE-120411-14] | 561566 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000005774 | pl10 | pl10 [Source:ZFIN;Acc:ZDB-GENE-980526-150] | 30116 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000062590 | pleca | plectin a [Source:ZFIN;Acc:ZDB-GENE-030131-8984] | 567639 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000021987 | plecb | plectin b [Source:ZFIN;Acc:ZDB-GENE-100917-2] | 100321011 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000020328 | plekha6 | pleckstrin homology domain containing, family A member 6 [Source:ZFIN;Acc:ZDB-GENE-140106-14] | 568764 | YES | YES | YES | Other | other | ||||
| ENSDARG00000062220 | plekha7b | pleckstrin homology domain containing, family A member 7b [Source:ZFIN;Acc:ZDB-GENE-091014-2] | 100005106 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000011929 | plp1b | proteolipid protein 1b [Source:ZFIN;Acc:ZDB-GENE-030710-6] | 368234 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000078745 | pnpla8 | patatin-like phospholipase domain containing 8 [Source:ZFIN;Acc:ZDB-GENE-070705-553] | 100126812 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000013000 | ppfia2 | protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2 [Source:ZFIN;Acc:ZDB-GENE-030131-7905] | 566597 | YES | YES | YES | Plasma Membrane | phosphatase | ||||
| ENSDARG00000053205 | ppfia4 | protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 4 [Source:ZFIN;Acc:ZDB-GENE-070720-14] | 100005543 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000003486 | ppp1caa | protein phosphatase 1, catalytic subunit, alpha isozyme a [Source:ZFIN;Acc:ZDB-GENE-040516-3] | 407980 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000071566 | ppp1cab | protein phosphatase 1, catalytic subunit, alpha isozyme b [Source:ZFIN;Acc:ZDB-GENE-030131-5512] | 327301 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000061304 | ppp1r9a | protein phosphatase 1, regulatory subunit 9A [Source:ZFIN;Acc:ZDB-GENE-060503-660] | 560862 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000079366 | ppp1r9ba | protein phosphatase 1, regulatory subunit 9Ba [Source:ZFIN;Acc:ZDB-GENE-091006-1] | 558232 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000071709 | ppp1r9bb | protein phosphatase 1, regulatory subunit 9Bb [Source:ZFIN;Acc:ZDB-GENE-121214-117] | 100331262 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000031200 | ppp2r5cb | protein phosphatase 2, regulatory subunit B', gamma b [Source:ZFIN;Acc:ZDB-GENE-041014-4] | 557208 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000020982 | prickle2a | prickle homolog 2a (Drosophila) [Source:ZFIN;Acc:ZDB-GENE-061110-112] | 777736 | YES | YES | YES | Nucleus | other | ||||
| ENSDARG00000037946 | prl | prolactin [Source:ZFIN;Acc:ZDB-GENE-030513-1] | 791526 | YES | YES | YES | Extracellular Space | other | ||||
| ENSDARG00000003705 | prnprs3 | prion protein, related sequence 3 [Source:ZFIN;Acc:ZDB-GENE-041221-3] | 503702 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000037783 | proza | protein Z, vitamin K-dependent plasma glycoprotein a [Source:ZFIN;Acc:ZDB-GENE-061013-403] | 768176 | YES | YES | YES | Extracellular Space | peptidase | ||||
| ENSDARG00000028306 | prph | peripherin [Source:ZFIN;Acc:ZDB-GENE-990415-207] | 30259 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000022951 | pth2 | parathyroid hormone 2 [Source:ZFIN;Acc:ZDB-GENE-041102-1] | 402812 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000035970 | ptprn2 | protein tyrosine phosphatase, receptor type, N polypeptide 2 [Source:ZFIN;Acc:ZDB-GENE-030131-3758] | 325033 | YES | YES | YES | Plasma Membrane | phosphatase | ||||
| ENSDARG00000001241 | puf60b | poly-U binding splicing factor b [Source:ZFIN;Acc:ZDB-GENE-040625-116] | 415211 | YES | YES | YES | Nucleus | other | ||||
| ENSDARG00000067591 | pura | purine-rich element binding protein A [Source:ZFIN;Acc:ZDB-GENE-040624-8] | 415106 | YES | YES | YES | Nucleus | transcription regulator | ||||
| ENSDARG00000102254 | pycr1a | pyrroline-5-carboxylate reductase 1a [Source:ZFIN;Acc:ZDB-GENE-040426-1675] | 393799 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000098639 | pycr1b | pyrroline-5-carboxylate reductase 1b [Source:ZFIN;Acc:ZDB-GENE-050522-26] | 100149838 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000102078 | RAB35 (2 of 2) | RAB35, member RAS oncogene family [Source:HGNC Symbol;Acc:HGNC:9774] | 796454 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000058425 | rab35b | RAB35, member RAS oncogene family b [Source:ZFIN;Acc:ZDB-GENE-040801-62] | 445154 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000020795 | rac3b | ras-related C3 botulinum toxin substrate 3b (rho family, small GTP binding protein Rac3) [Source:ZFIN;Acc:ZDB-GENE-080220-21] | 795167 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000039974 | rai14 | retinoic acid induced 14 [Source:ZFIN;Acc:ZDB-GENE-040718-235] | 794796 | YES | YES | YES | Nucleus | transcription regulator | ||||
| ENSDARG00000079872 | rapgef4 | Rap guanine nucleotide exchange factor (GEF) 4 [Source:ZFIN;Acc:ZDB-GENE-070912-385] | 0 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000075924 | RAPGEF4 (1 of 2) | Rap guanine nucleotide exchange factor (GEF) 4 [Source:HGNC Symbol;Acc:HGNC:16626] | 100141331 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000056075 | rca2.1 | regulator of complement activation group 2 gene 1 [Source:ZFIN;Acc:ZDB-GENE-050320-60] | 541367 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000074064 | rem2 | RAS (RAD and GEM)-like GTP binding 2 [Source:ZFIN;Acc:ZDB-GENE-081110-1] | 798285 | YES | YES | YES | Plasma Membrane | enzyme | ||||
| ENSDARG00000039435 | rgs17 | regulator of G-protein signaling 17 [Source:ZFIN;Acc:ZDB-GENE-040718-229] | 436796 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000077385 | rgs19 | regulator of G-protein signaling 19 [Source:ZFIN;Acc:ZDB-GENE-080723-72] | 100170801 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000009018 | rhbg | Rh family, B glycoprotein (gene/pseudogene) [Source:ZFIN;Acc:ZDB-GENE-030131-9542] | 337596 | YES | YES | YES | Plasma Membrane | transporter | ||||
| ENSDARG00000090036 | RHOBTB3 | Rho-related BTB domain containing 3 [Source:HGNC Symbol;Acc:HGNC:18757] | 562804 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000001154 | rimbp2 | RIMS binding protein 2 [Source:ZFIN;Acc:ZDB-GENE-040724-96] | 0 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000074680 | rims1a | regulating synaptic membrane exocytosis 1a [Source:ZFIN;Acc:ZDB-GENE-090312-135] | 0 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000078902 | rims1b | regulating synaptic membrane exocytosis 1b [Source:ZFIN;Acc:ZDB-GENE-101223-4] | 568486 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000101606 | rims2a | regulating synaptic membrane exocytosis 2a [Source:ZFIN;Acc:ZDB-GENE-040426-1656] | 393672 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000104458 | rnasekb | ribonuclease, RNase K b [Source:ZFIN;Acc:ZDB-GENE-060825-160] | 336290 | YES | YES | YES | Other | peptidase | ||||
| ENSDARG00000025581 | rpl10 | ribosomal protein L10 [Source:ZFIN;Acc:ZDB-GENE-030131-8656] | 336712 | YES | YES | YES | Other | other | ||||
| ENSDARG00000042905 | rpl10a | ribosomal protein L10a [Source:ZFIN;Acc:ZDB-GENE-030131-2025] | 323305 | YES | YES | YES | Nucleus | other | ||||
| ENSDARG00000043509 | rpl11 | ribosomal protein L11 [Source:ZFIN;Acc:ZDB-GENE-040625-147] | 415229 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000006691 | rpl12 | ribosomal protein L12 [Source:ZFIN;Acc:ZDB-GENE-030131-5297] | 327089 | YES | YES | YES | Other | other | ||||
| ENSDARG00000099380 | rpl13 | ribosomal protein L13 [Source:ZFIN;Acc:ZDB-GENE-031007-1] | 378961 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000044093 | rpl13a | ribosomal protein L13a [Source:ZFIN;Acc:ZDB-GENE-030131-168] | 560828 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000103433 | rpl14 | ribosomal protein L14 [Source:ZFIN;Acc:ZDB-GENE-030131-2085] | 323365 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000009285 | rpl15 | ribosomal protein L15 [Source:ZFIN;Acc:ZDB-GENE-040801-183] | 445053 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000029533 | rpl18 | ribosomal protein L18 [Source:ZFIN;Acc:ZDB-GENE-040801-165] | 445038 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000013307 | rpl19 | ribosomal protein L19 [Source:ZFIN;Acc:ZDB-GENE-040426-2290] | 406489 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000010516 | rpl21 | ribosomal protein L21 [Source:ZFIN;Acc:ZDB-GENE-030131-8512] | 336568 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000070437 | rpl22 | ribosomal protein L22 [Source:ZFIN;Acc:ZDB-GENE-051113-276] | 557397 | YES | YES | YES | Nucleus | other | ||||
| ENSDARG00000053457 | rpl23 | ribosomal protein L23 [Source:ZFIN;Acc:ZDB-GENE-030131-8756] | 336812 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000006316 | rpl23a | ribosomal protein L23a [Source:ZFIN;Acc:ZDB-GENE-030131-7479] | 335539 | YES | YES | YES | Other | other | ||||
| ENSDARG00000099104 | rpl24 | ribosomal protein L24 [Source:ZFIN;Acc:ZDB-GENE-020419-25] | 192301 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000102317 | rpl26 | ribosomal protein L26 [Source:ZFIN;Acc:ZDB-GENE-040426-2117] | 406387 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000015128 | rpl27 | ribosomal protein L27 [Source:ZFIN;Acc:ZDB-GENE-030131-4343] | 325618 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000005791 | rpl28 | ribosomal protein L28 [Source:ZFIN;Acc:ZDB-GENE-040930-10] | 394036 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000003599 | rpl3 | ribosomal protein L3 [Source:ZFIN;Acc:ZDB-GENE-030131-1291] | 322571 | YES | YES | YES | Nucleus | other | ||||
| ENSDARG00000035871 | rpl30 | ribosomal protein L30 [Source:ZFIN;Acc:ZDB-GENE-030131-8657] | 336713 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000053365 | rpl31 | ribosomal protein L31 [Source:ZFIN;Acc:ZDB-GENE-060331-121] | 677758 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000054818 | rpl32 | ribosomal protein L32 [Source:ZFIN;Acc:ZDB-GENE-060331-105] | 556556 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000029500 | rpl34 | ribosomal protein L34 [Source:ZFIN;Acc:ZDB-GENE-040426-1033] | 394097 | YES | YES | YES | Other | other | ||||
| ENSDARG00000018334 | rpl35 | ribosomal protein L35 [Source:ZFIN;Acc:ZDB-GENE-020419-2] | 192299 | YES | YES | YES | Other | other | ||||
| ENSDARG00000088030 | rpl35a | ribosomal protein L35a [Source:ZFIN;Acc:ZDB-GENE-040718-190] | 436760 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000100588 | rpl36 | ribosomal protein L36 [Source:ZFIN;Acc:ZDB-GENE-040622-2] | 405888 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000006413 | rpl38 | ribosomal protein L38 [Source:ZFIN;Acc:ZDB-GENE-030131-8752] | 436759 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000041182 | rpl4 | ribosomal protein L4 [Source:ZFIN;Acc:ZDB-GENE-030131-9034] | 337090 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000015862 | rpl5b | ribosomal protein L5b [Source:ZFIN;Acc:ZDB-GENE-040625-93] | 415196 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000058451 | rpl6 | ribosomal protein L6 [Source:ZFIN;Acc:ZDB-GENE-030131-8671] | 336727 | YES | YES | YES | Nucleus | other | ||||
| ENSDARG00000007320 | rpl7 | ribosomal protein L7 [Source:ZFIN;Acc:ZDB-GENE-030131-8654] | 336710 | YES | YES | YES | Nucleus | transcription regulator | ||||
| ENSDARG00000019230 | rpl7a | ribosomal protein L7a [Source:ZFIN;Acc:ZDB-GENE-031001-9] | 337010 | YES | YES | YES | Other | other | ||||
| ENSDARG00000014867 | rpl8 | ribosomal protein L8 [Source:ZFIN;Acc:ZDB-GENE-040426-1670] | 393686 | YES | YES | YES | Other | other | ||||
| ENSDARG00000037350 | rpl9 | ribosomal protein L9 [Source:ZFIN;Acc:ZDB-GENE-030131-8646] | 336702 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000021864 | rplp1 | ribosomal protein, large, P1 [Source:ZFIN;Acc:ZDB-GENE-030131-8663] | 336719 | YES | YES | YES | Other | other | ||||
| ENSDARG00000101406 | rplp2 | ribosomal protein, large P2 [Source:ZFIN;Acc:ZDB-GENE-031018-2] | 335629 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000011201 | rplp2l | ribosomal protein, large P2, like [Source:ZFIN;Acc:ZDB-GENE-070327-2] | 558312 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000034897 | rps10 | ribosomal protein S10 [Source:ZFIN;Acc:ZDB-GENE-040426-1481] | 394121 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000053058 | rps11 | ribosomal protein S11 [Source:ZFIN;Acc:ZDB-GENE-040426-2701] | 406686 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000036875 | rps12 | ribosomal protein S12 [Source:ZFIN;Acc:ZDB-GENE-030131-8951] | 337007 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000036298 | rps13 | ribosomal protein S13 [Source:ZFIN;Acc:ZDB-GENE-040625-52] | 415169 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000036629 | rps14 | ribosomal protein S14 [Source:ZFIN;Acc:ZDB-GENE-030131-8631] | 336687 | YES | YES | YES | Cytoplasm | translation regulator | ||||
| ENSDARG00000010160 | rps15a | ribosomal protein S15a [Source:ZFIN;Acc:ZDB-GENE-030131-8708] | 336764 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000045487 | rps16 | ribosomal protein S16 [Source:ZFIN;Acc:ZDB-GENE-050506-107] | 767765 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000046157 | RPS17 | ribosomal protein S17 [Source:HGNC Symbol;Acc:HGNC:10397] | 541327 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000100392 | rps18 | ribosomal protein S18 [Source:ZFIN;Acc:ZDB-GENE-020419-20] | 192300 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000030602 | rps19 | ribosomal protein S19 [Source:ZFIN;Acc:ZDB-GENE-040426-1716] | 393723 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000077291 | rps2 | ribosomal protein S2 [Source:ZFIN;Acc:ZDB-GENE-040426-2454] | 406565 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000021838 | rps23 | ribosomal protein S23 [Source:ZFIN;Acc:ZDB-GENE-080220-50] | 100000341 | YES | YES | YES | Cytoplasm | translation regulator | ||||
| ENSDARG00000039347 | rps24 | ribosomal protein S24 [Source:ZFIN;Acc:ZDB-GENE-040109-5] | 394194 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000041811 | rps25 | ribosomal protein S25 [Source:ZFIN;Acc:ZDB-GENE-040426-1788] | 393788 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000037071 | rps26 | ribosomal protein S26 [Source:ZFIN;Acc:ZDB-GENE-030131-8606] | 336662 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000023298 | rps27.1 | ribosomal protein S27, isoform 1 [Source:ZFIN;Acc:ZDB-GENE-050522-549] | 554155 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000100513 | rps27.2 | ribosomal protein S27, isoform 2 [Source:ZFIN;Acc:ZDB-GENE-040426-1735] | 677743 | YES | YES | YES | Cytoplasm | translation regulator | ||||
| ENSDARG00000035860 | rps28 | ribosomal protein S28 [Source:ZFIN;Acc:ZDB-GENE-030131-2022] | 406307 | YES | YES | YES | Other | other | ||||
| ENSDARG00000041232 | rps29 | ribosomal protein S29 [Source:ZFIN;Acc:ZDB-GENE-040622-5] | 405889 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000103007 | rps3 | ribosomal protein S3 [Source:ZFIN;Acc:ZDB-GENE-030131-8494] | 336550 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000035692 | rps3a | ribosomal protein S3A [Source:ZFIN;Acc:ZDB-GENE-030131-9184] | 337240 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000014690 | rps4x | ribosomal protein S4, X-linked [Source:ZFIN;Acc:ZDB-GENE-040927-19] | 449547 | YES | YES | YES | Other | other | ||||
| ENSDARG00000019778 | rps6 | ribosomal protein S6 [Source:ZFIN;Acc:ZDB-GENE-040801-8] | 445028 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000055996 | rps8a | ribosomal protein S8a [Source:ZFIN;Acc:ZDB-GENE-030131-8626] | 407690 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000057368 | rps8b | ribosomal protein S8b [Source:ZFIN;Acc:ZDB-GENE-050522-202] | 100000836 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000011405 | rps9 | ribosomal protein S9 [Source:ZFIN;Acc:ZDB-GENE-010724-15] | 114420 | YES | YES | YES | Cytoplasm | translation regulator | ||||
| ENSDARG00000091161 | rpz | rapunzel [Source:ZFIN;Acc:ZDB-GENE-070117-651] | 100002737 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000069728 | rpz3 | rapunzel 3 [Source:ZFIN;Acc:ZDB-GENE-030131-4784] | 100001029 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000039871 | rpz4 | rapunzel 4 [Source:ZFIN;Acc:ZDB-GENE-050417-472] | 550609 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000101047 | rtcb | RNA 2',3'-cyclic phosphate and 5'-OH ligase [Source:ZFIN;Acc:ZDB-GENE-040426-2096] | 406376 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000060865 | scai | suppressor of cancer cell invasion [Source:ZFIN;Acc:ZDB-GENE-060526-57] | 556383 | YES | YES | YES | Nucleus | transcription regulator | ||||
| ENSDARG00000038574 | scg2b | secretogranin II (chromogranin C), b [Source:ZFIN;Acc:ZDB-GENE-061103-160] | 777641 | YES | YES | YES | Extracellular Space | cytokine | ||||
| ENSDARG00000013843 | sept5a | septin 5a [Source:ZFIN;Acc:ZDB-GENE-030131-7868] | 0 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000036031 | sept5b | septin 5b [Source:ZFIN;Acc:ZDB-GENE-040808-43] | 445325 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000087647 | sept7a | septin 7a [Source:ZFIN;Acc:ZDB-GENE-040426-1008] | 0 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000052673 | sept7b | septin 7b [Source:ZFIN;Acc:ZDB-GENE-030131-37] | 100000597 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000032606 | sept8a | septin 8a [Source:ZFIN;Acc:ZDB-GENE-030131-309] | 321590 | YES | YES | YES | Extracellular Space | other | ||||
| ENSDARG00000005924 | serpina10a | serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 10a [Source:ZFIN;Acc:ZDB-GENE-041014-246] | 565064 | YES | YES | YES | Extracellular Space | other | ||||
| ENSDARG00000011564 | sfpq | splicing factor proline/glutamine-rich [Source:ZFIN;Acc:ZDB-GENE-040426-2452] | 406564 | YES | YES | YES | Nucleus | other | ||||
| ENSDARG00000036878 | sh3bgrl2 | SH3 domain binding glutamate-rich protein like 2 [Source:ZFIN;Acc:ZDB-GENE-040704-38] | 431744 | YES | YES | YES | Nucleus | other | ||||
| ENSDARG00000062325 | shank2 | SH3 and multiple ankyrin repeat domains 2 [Source:ZFIN;Acc:ZDB-GENE-061207-58] | 567595 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000063332 | shank3a | SH3 and multiple ankyrin repeat domains 3a [Source:ZFIN;Acc:ZDB-GENE-060503-369] | 0 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000063144 | shisa7b | shisa family member 7 [Source:ZFIN;Acc:ZDB-GENE-060503-31] | 100034520 | YES | YES | YES | Other | other | ||||
| ENSDARG00000052642 | shisa9b | shisa family member 9b [Source:ZFIN;Acc:ZDB-GENE-081022-105] | 566015 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000088882 | si:ch211-149b19.2 | si:ch211-149b19.2 [Source:ZFIN;Acc:ZDB-GENE-110411-60] | 101884675 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000092746 | si:ch211-150o23.2 | si:ch211-150o23.2 [Source:ZFIN;Acc:ZDB-GENE-060503-386] | 0 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000097187 | si:ch211-153l6.6 | si:ch211-153l6.6 [Source:ZFIN;Acc:ZDB-GENE-131121-98] | 101884986 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000059333 | si:ch211-285f17.1 | si:ch211-285f17.1 [Source:ZFIN;Acc:ZDB-GENE-000607-77] | 562465 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000089255 | si:ch211-286c4.6 | si:ch211-286c4.6 [Source:ZFIN;Acc:ZDB-GENE-131127-232] | 100003678 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000097855 | si:ch211-3o3.9 | si:ch211-3o3.9 [Source:ZFIN;Acc:ZDB-GENE-131127-12] | 101886596 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000057826 | si:ch73-61d6.3 | si:ch73-61d6.3 [Source:ZFIN;Acc:ZDB-GENE-091204-109] | 100001136 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000090149 | si:dkey-108k21.18 | si:dkey-108k21.18 [Source:ZFIN;Acc:ZDB-GENE-131121-139] | 560380 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000093195 | si:dkey-261m9.6 | si:dkey-261m9.6 [Source:ZFIN;Acc:ZDB-GENE-131127-103] | 563569 | YES | YES | YES | Nucleus | other | ||||
| ENSDARG00000003146 | si:dkey-27m7.4 | si:dkey-27m7.4 [Source:ZFIN;Acc:ZDB-GENE-090313-297] | 0 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000099614 | si:dkey-29k15.2 | si:dkey-29k15.2 [Source:ZFIN;Acc:ZDB-GENE-141216-78] | 557266 | YES | YES | YES | Nucleus | transcription regulator | ||||
| ENSDARG00000057881 | si:dkey-33c12.3 | si:dkey-33c12.3 [Source:ZFIN;Acc:ZDB-GENE-030131-7320] | 335380 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000092039 | si:dkey-70p6.1 | si:dkey-70p6.1 [Source:ZFIN;Acc:ZDB-GENE-040724-140] | 570575 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000097528 | si:dkey-7j14.5 | si:dkey-7j14.5 [Source:ZFIN;Acc:ZDB-GENE-030131-8110] | 556563 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000063008 | si:dkeyp-27e10.3 | si:dkeyp-27e10.3 [Source:ZFIN;Acc:ZDB-GENE-041210-96] | 0 | YES | YES | YES | Other | other | ||||
| ENSDARG00000076697 | SIPA1 | signal-induced proliferation-associated 1 [Source:HGNC Symbol;Acc:HGNC:10885] | 555294 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000020134 | sipa1l1 | signal-induced proliferation-associated 1 like 1 [Source:ZFIN;Acc:ZDB-GENE-030131-9909] | 562521 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000003151 | skp1 | S-phase kinase-associated protein 1 [Source:ZFIN;Acc:ZDB-GENE-040426-1707] | 393716 | YES | YES | YES | Nucleus | transcription regulator | ||||
| ENSDARG00000006240 | slc27a1a | solute carrier family 27 (fatty acid transporter), member 1a [Source:ZFIN;Acc:ZDB-GENE-050320-112] | 541410 | YES | YES | YES | Plasma Membrane | transporter | ||||
| ENSDARG00000019000 | smc3 | structural maintenance of chromosomes 3 [Source:ZFIN;Acc:ZDB-GENE-030131-3196] | 324475 | YES | YES | YES | Nucleus | other | ||||
| ENSDARG00000051970 | smu1b | smu-1 suppressor of mec-8 and unc-52 homolog b (C. elegans) [Source:ZFIN;Acc:ZDB-GENE-130530-635] | 570074 | YES | YES | YES | Nucleus | other | ||||
| ENSDARG00000103435 | sorbs1 | sorbin and SH3 domain containing 1 [Source:ZFIN;Acc:ZDB-GENE-090313-168] | 0 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000061603 | sorbs2b | sorbin and SH3 domain containing 2b [Source:ZFIN;Acc:ZDB-GENE-070308-3] | 0 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000035868 | spire1a | spire-type actin nucleation factor 1a [Source:ZFIN;Acc:ZDB-GENE-061013-119] | 557962 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000030490 | sptb | spectrin, beta, erythrocytic [Source:ZFIN;Acc:ZDB-GENE-000906-1] | 58040 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000059773 | SRCIN1 (1 of 2) | SRC kinase signaling inhibitor 1 [Source:HGNC Symbol;Acc:HGNC:29506] | 0 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000101812 | SRCIN1 (2 of 2) | SRC kinase signaling inhibitor 1 [Source:HGNC Symbol;Acc:HGNC:29506] | 0 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000039350 | ssbp1 | single-stranded DNA binding protein 1 [Source:ZFIN;Acc:ZDB-GENE-050417-340] | 550504 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000026801 | stau2 | staufen double-stranded RNA binding protein 2 [Source:ZFIN;Acc:ZDB-GENE-040426-687] | 393899 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000028354 | stxbp6l | syntaxin binding protein 6 (amisyn), like [Source:ZFIN;Acc:ZDB-GENE-040724-16] | 100002110 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000014651 | svilb | supervillin b [Source:ZFIN;Acc:ZDB-GENE-081104-411] | 0 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000060368 | syn1 | synapsin I [Source:ZFIN;Acc:ZDB-GENE-081105-93] | 570672 | YES | YES | YES | Plasma Membrane | transporter | ||||
| ENSDARG00000098417 | syn3 | synapsin III [Source:ZFIN;Acc:ZDB-GENE-141212-325] | 0 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000092624 | SYNE1 | spectrin repeat containing, nuclear envelope 1 [Source:HGNC Symbol;Acc:HGNC:17089] | 0 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000009499 | syne1a | spectrin repeat containing, nuclear envelope 1a [Source:ZFIN;Acc:ZDB-GENE-030131-4172] | 0 | YES | YES | YES | Nucleus | other | ||||
| ENSDARG00000063713 | SYNGAP1 (1 of 2) | synaptic Ras GTPase activating protein 1 [Source:HGNC Symbol;Acc:HGNC:11497] | 100034485 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000069765 | SYNGAP1 (2 of 2) | synaptic Ras GTPase activating protein 1 [Source:HGNC Symbol;Acc:HGNC:11497] | 0 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000062350 | synm | synemin, intermediate filament protein [Source:ZFIN;Acc:ZDB-GENE-050419-81] | 558800 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000063568 | syt7a | synaptotagmin VIIa [Source:ZFIN;Acc:ZDB-GENE-090601-5] | 561429 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSDARG00000078060 | syt7b | synaptotagmin VIIb [Source:ZFIN;Acc:ZDB-GENE-090601-6] | 100334336 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSDARG00000074899 | taok2a | TAO kinase 2a [Source:ZFIN;Acc:ZDB-GENE-030131-9416] | 556172 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSDARG00000100620 | taok2b | TAO kinase 2b [Source:ZFIN;Acc:ZDB-GENE-110603-3] | 100151551 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSDARG00000103095 | tbk1 | TANK-binding kinase 1 [Source:ZFIN;Acc:ZDB-GENE-060512-359] | 692289 | YES | YES | YES | Cytoplasm | kinase | ||||
| ENSDARG00000063145 | tfam | transcription factor A, mitochondrial [Source:ZFIN;Acc:ZDB-GENE-061013-552] | 571106 | YES | YES | YES | Cytoplasm | transcription regulator | ||||
| ENSDARG00000060306 | tfcp2 | transcription factor CP2 [Source:ZFIN;Acc:ZDB-GENE-030131-2523] | 100001906 | YES | YES | YES | Nucleus | transcription regulator | ||||
| ENSDARG00000035018 | thy1 | Thy-1 cell surface antigen [Source:ZFIN;Acc:ZDB-GENE-030131-8561] | 336617 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000077506 | tjp1a | tight junction protein 1a [Source:ZFIN;Acc:ZDB-GENE-031001-2] | 378845 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000079374 | tjp1b | tight junction protein 1b [Source:ZFIN;Acc:ZDB-GENE-070925-1] | 565459 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000069649 | tmem160 | transmembrane protein 160 [Source:ZFIN;Acc:ZDB-GENE-060503-851] | 559491 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000086661 | TMEM221 | transmembrane protein 221 [Source:HGNC Symbol;Acc:HGNC:21943] | 100537506 | YES | YES | YES | Other | other | ||||
| ENSDARG00000087916 | tmem240a | transmembrane protein 240a [Source:ZFIN;Acc:ZDB-GENE-090313-306] | 568378 | YES | YES | YES | Other | other | ||||
| ENSDARG00000090145 | tmem240b | transmembrane protein 240b [Source:ZFIN;Acc:ZDB-GENE-131127-474] | 0 | YES | YES | YES | Other | other | ||||
| ENSDARG00000002571 | tmod2 | tropomodulin 2 [Source:ZFIN;Acc:ZDB-GENE-040912-185] | 447869 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000020890 | tmod4 | tropomodulin 4 (muscle) [Source:ZFIN;Acc:ZDB-GENE-991019-9] | 30441 | YES | YES | YES | Other | other | ||||
| ENSDARG00000021948 | tnc | tenascin C [Source:ZFIN;Acc:ZDB-GENE-980526-104] | 30037 | YES | YES | YES | Extracellular Space | other | ||||
| ENSDARG00000009031 | tnikb | TRAF2 and NCK interacting kinase b [Source:ZFIN;Acc:ZDB-GENE-051113-320] | 556959 | YES | YES | YES | Plasma Membrane | kinase | ||||
| ENSDARG00000104581 | tom1 | target of myb1 (chicken) [Source:ZFIN;Acc:ZDB-GENE-060721-1] | 0 | YES | YES | YES | Cytoplasm | transporter | ||||
| ENSDARG00000073872 | tpst1 | tyrosylprotein sulfotransferase 1 [Source:ZFIN;Acc:ZDB-GENE-000210-10] | 30677 | YES | YES | YES | Cytoplasm | enzyme | ||||
| ENSDARG00000045364 | trappc3 | trafficking protein particle complex 3 [Source:ZFIN;Acc:ZDB-GENE-040801-120] | 445207 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000016463 | trappc6b | trafficking protein particle complex 6b [Source:ZFIN;Acc:ZDB-GENE-041010-122] | 450008 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000058649 | trim46b | tripartite motif containing 46b [Source:ZFIN;Acc:ZDB-GENE-060503-466] | 100034631 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000035066 | ubtf | upstream binding transcription factor, RNA polymerase I [Source:ZFIN;Acc:ZDB-GENE-030616-252] | 368509 | YES | YES | YES | Nucleus | transcription regulator | ||||
| ENSDARG00000061829 | UNC13A | unc-13 homolog A (C. elegans) [Source:HGNC Symbol;Acc:HGNC:23150] | 568618 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000101233 | unc13c | unc-13 homolog C (C. elegans) [Source:ZFIN;Acc:ZDB-GENE-060503-478] | 0 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000017105 | vaspb | vasodilator-stimulated phosphoprotein b [Source:ZFIN;Acc:ZDB-GENE-030131-7118] | 550487 | YES | YES | YES | Plasma Membrane | other | ||||
| ENSDARG00000073713 | vav3b | vav 3 guanine nucleotide exchange factor b [Source:ZFIN;Acc:ZDB-GENE-070912-251] | 0 | YES | YES | YES | Extracellular Space | other | ||||
| ENSDARG00000009401 | vcanb | versican b [Source:ZFIN;Acc:ZDB-GENE-030131-2185] | 323465 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000010008 | vim | vimentin [Source:ZFIN;Acc:ZDB-GENE-011212-3] | 140599 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000044501 | viml | vimentin like [Source:ZFIN;Acc:ZDB-GENE-040426-1743] | 393746 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000036056 | vrk3 | vaccinia related kinase 3 [Source:ZFIN;Acc:ZDB-GENE-030131-246] | 0 | YES | YES | YES | Nucleus | kinase | ||||
| ENSDARG00000062948 | wasf3b | WAS protein family, member 3b [Source:ZFIN;Acc:ZDB-GENE-070112-1512] | 563632 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000006712 | wipf2b | WAS/WASL interacting protein family, member 2b [Source:ZFIN;Acc:ZDB-GENE-040625-182] | 415255 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000020443 | xkr6a | XK, Kell blood group complex subunit-related family, member 6a [Source:ZFIN;Acc:ZDB-GENE-131121-106] | 0 | YES | YES | YES | Other | other | ||||
| ENSDARG00000043410 | xkr6b | XK, Kell blood group complex subunit-related family, member 6b [Source:ZFIN;Acc:ZDB-GENE-040724-153] | 792104 | YES | YES | YES | Other | other | ||||
| ENSDARG00000069600 | zgc:109889 | zgc:109889 [Source:ZFIN;Acc:ZDB-GENE-050522-547] | 553542 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000096992 | zgc:153405 | zgc:153405 [Source:ZFIN;Acc:ZDB-GENE-060825-170] | 751661 | YES | YES | YES | Nucleus | other | ||||
| ENSDARG00000033738 | zgc:153867 | zgc:153867 [Source:ZFIN;Acc:ZDB-GENE-030131-9170] | 337226 | YES | YES | YES | Cytoplasm | other | ||||
| ENSDARG00000017785 | zgc:158689 | zgc:158689 [Source:ZFIN;Acc:ZDB-GENE-070112-1902] | 791177 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000091525 | zgc:173552 | zgc:173552 [Source:ZFIN;Acc:ZDB-GENE-080220-24] | 794406 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000012281 | zgc:65851 | zgc:65851 [Source:ZFIN;Acc:ZDB-GENE-030131-9669] | 321113 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000014222 | zgc:86598 | zgc:86598 [Source:ZFIN;Acc:ZDB-GENE-040625-181] | 415254 | YES | YES | YES | NA | NA | ||||
| ENSDARG00000061268 | ago2 | argonaute RISC catalytic component 2 [Source:ZFIN;Acc:ZDB-GENE-110606-6] | 570630 | YES | YES | Cytoplasm | translation regulator | |||||
| ENSDARG00000041685 | A2ML1 (3 of 13) | alpha-2-macroglobulin-like 1 [Source:HGNC Symbol;Acc:HGNC:23336] | 100006782 | YES | YES | Extracellular Space | other | |||||
| ENSDARG00000057169 | abca4a | ATP-binding cassette, sub-family A (ABC1), member 4a [Source:ZFIN;Acc:ZDB-GENE-050517-3] | 798993 | YES | YES | Plasma Membrane | transporter | |||||
| ENSDARG00000031795 | abcf1 | ATP-binding cassette, sub-family F (GCN20), member 1 [Source:ZFIN;Acc:ZDB-GENE-050517-31] | 406467 | YES | YES | Cytoplasm | transporter | |||||
| ENSDARG00000100075 | abcg2a | ATP-binding cassette, sub-family G (WHITE), member 2a [Source:ZFIN;Acc:ZDB-GENE-050517-35] | 735312 | YES | YES | Plasma Membrane | transporter | |||||
| ENSDARG00000058574 | abcg2c | ATP-binding cassette, sub-family G (WHITE), member 2c [Source:ZFIN;Acc:ZDB-GENE-050517-37] | 569858 | YES | YES | NA | NA | |||||
| ENSDARG00000069501 | abhd11 | abhydrolase domain containing 11 [Source:ZFIN;Acc:ZDB-GENE-040909-1] | 446169 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000018653 | acot7 | acyl-CoA thioesterase 7 [Source:ZFIN;Acc:ZDB-GENE-040912-42] | 447878 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000076486 | acp1 | acid phosphatase 1, soluble [Source:ZFIN;Acc:ZDB-GENE-050327-12] | 541489 | YES | YES | Cytoplasm | phosphatase | |||||
| ENSDARG00000015502 | adam10b | ADAM metallopeptidase domain 10b [Source:ZFIN;Acc:ZDB-GENE-071115-1] | 0 | YES | YES | Plasma Membrane | peptidase | |||||
| ENSDARG00000010070 | adam9 | ADAM metallopeptidase domain 9 [Source:ZFIN;Acc:ZDB-GENE-040912-127] | 447940 | YES | YES | Plasma Membrane | peptidase | |||||
| ENSDARG00000019526 | agfg1b | ArfGAP with FG repeats 1b [Source:ZFIN;Acc:ZDB-GENE-040426-1520] | 393560 | YES | YES | NA | NA | |||||
| ENSDARG00000052030 | akr1a1b | aldo-keto reductase family 1, member A1b (aldehyde reductase) [Source:ZFIN;Acc:ZDB-GENE-050417-118] | 799805 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000027394 | AL929252.1 | Uncharacterized protein [Source:UniProtKB/TrEMBL;Acc:E7F7K7] | 559304 | YES | YES | NA | NA | |||||
| ENSDARG00000028087 | aldh2.2 | aldehyde dehydrogenase 2 family (mitochondrial), tandem duplicate 2 [Source:ZFIN;Acc:ZDB-GENE-030326-5] | 368239 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000070399 | alg2 | asparagine-linked glycosylation 2 (alpha-1,3-mannosyltransferase) [Source:ZFIN;Acc:ZDB-GENE-060502-2] | 0 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000012840 | alg9 | ALG9, alpha-1,2-mannosyltransferase [Source:ZFIN;Acc:ZDB-GENE-040426-1270] | 393598 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000061013 | ankfy1 | ankyrin repeat and FYVE domain containing 1 [Source:ZFIN;Acc:ZDB-GENE-041222-1] | 568480 | YES | YES | Cytoplasm | transcription regulator | |||||
| ENSDARG00000077327 | ano11 | anoctamin 11 [Source:ZFIN;Acc:ZDB-GENE-090313-245] | 0 | YES | YES | NA | NA | |||||
| ENSDARG00000077229 | ano8b | anoctamin 8b [Source:ZFIN;Acc:ZDB-GENE-121024-1] | 562782 | YES | YES | Plasma Membrane | ion channel | |||||
| ENSDARG00000039882 | ap3s2 | adaptor-related protein complex 3, sigma 2 subunit [Source:ZFIN;Acc:ZDB-GENE-040718-274] | 436812 | YES | YES | Cytoplasm | transporter | |||||
| ENSDARG00000099995 | apbb2b | amyloid beta (A4) precursor protein-binding, family B, member 2b [Source:ZFIN;Acc:ZDB-GENE-090313-73] | 564990 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000033597 | api5 | apoptosis inhibitor 5 [Source:ZFIN;Acc:ZDB-GENE-030131-395] | 321676 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000098368 | aplp1 | amyloid beta (A4) precursor-like protein 1 [Source:ZFIN;Acc:ZDB-GENE-040319-1] | 404233 | YES | YES | Extracellular Space | other | |||||
| ENSDARG00000054864 | aplp2 | amyloid beta (A4) precursor-like protein 2 [Source:ZFIN;Acc:ZDB-GENE-061009-28] | 796801 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000020866 | apoa4b.2 | apolipoprotein A-IV b, tandem duplicate 2 [Source:ZFIN;Acc:ZDB-GENE-030131-3143] | 570354 | YES | YES | Extracellular Space | transporter | |||||
| ENSDARG00000042780 | apoba | apolipoprotein Ba [Source:ZFIN;Acc:ZDB-GENE-070702-4] | 566465 | YES | YES | Extracellular Space | transporter | |||||
| ENSDARG00000092155 | apoc2 | apolipoprotein C-II [Source:ZFIN;Acc:ZDB-GENE-030131-2168] | 568972 | YES | YES | NA | NA | |||||
| ENSDARG00000003519 | aprt | adenine phosphoribosyltransferase [Source:ZFIN;Acc:ZDB-GENE-040426-1492] | 393641 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000077656 | arfgef3 | ARFGEF family member 3 [Source:ZFIN;Acc:ZDB-GENE-081107-37] | 562652 | YES | YES | Other | other | |||||
| ENSDARG00000073848 | arhgef7b | Rho guanine nucleotide exchange factor (GEF) 7b [Source:ZFIN;Acc:ZDB-GENE-041212-49] | 494081 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000069496 | arl2 | ADP-ribosylation factor-like 2 [Source:ZFIN;Acc:ZDB-GENE-041010-19] | 449774 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000054578 | arl6ip1 | ADP-ribosylation factor-like 6 interacting protein 1 [Source:ZFIN;Acc:ZDB-GENE-040426-1087] | 394087 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000006915 | arl8ba | ADP-ribosylation factor-like 8Ba [Source:ZFIN;Acc:ZDB-GENE-030131-9370] | 337424 | YES | YES | NA | NA | |||||
| ENSDARG00000103770 | ARMC6 | armadillo repeat containing 6 [Source:HGNC Symbol;Acc:HGNC:25049] | 555425 | YES | YES | Other | other | |||||
| ENSDARG00000039729 | asap1b | ArfGAP with SH3 domain, ankyrin repeat and PH domain 1b [Source:ZFIN;Acc:ZDB-GENE-091112-16] | 565859 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000006849 | asic2 | acid-sensing (proton-gated) ion channel 2 [Source:ZFIN;Acc:ZDB-GENE-040513-4] | 407669 | YES | YES | Plasma Membrane | ion channel | |||||
| ENSDARG00000010472 | atp1a2a | ATPase, Na+/K+ transporting, alpha 2a polypeptide [Source:ZFIN;Acc:ZDB-GENE-001212-6] | 64609 | YES | YES | Plasma Membrane | transporter | |||||
| ENSDARG00000042975 | auh | AU RNA binding protein/enoyl-CoA hydratase [Source:ZFIN;Acc:ZDB-GENE-040801-95] | 445182 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000077740 | b3galtlb | beta 1,3-galactosyltransferase-like b [Source:ZFIN;Acc:ZDB-GENE-091204-128] | 564156 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000075892 | bag6 | BCL2-associated athanogene 6 [Source:ZFIN;Acc:ZDB-GENE-010501-5] | 83910 | YES | YES | Nucleus | enzyme | |||||
| ENSDARG00000094704 | bcl2a | B-cell CLL/lymphoma 2a [Source:ZFIN;Acc:ZDB-GENE-051012-1] | 570772 | YES | YES | Cytoplasm | transporter | |||||
| ENSDARG00000059388 | bdh1 | 3-hydroxybutyrate dehydrogenase, type 1 [Source:ZFIN;Acc:ZDB-GENE-070410-130] | 100037356 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000018817 | bdnf | brain-derived neurotrophic factor [Source:ZFIN;Acc:ZDB-GENE-000412-1] | 58118 | YES | YES | Extracellular Space | growth factor | |||||
| ENSDARG00000099961 | bnip3 | BCL2/adenovirus E1B interacting protein 3 [Source:ZFIN;Acc:ZDB-GENE-030131-8060] | 336116 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000039915 | bola1 | bolA family member 1 [Source:ZFIN;Acc:ZDB-GENE-040801-76] | 0 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000061051 | BRINP3 (1 of 3) | bone morphogenetic protein/retinoic acid inducible neural-specific 3 [Source:HGNC Symbol;Acc:HGNC:22393] | 100001603 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000013659 | brox | BRO1 domain and CAAX motif containing [Source:ZFIN;Acc:ZDB-GENE-041114-137] | 492786 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000099026 | BX004828.2 | 0 | 101885702 | YES | YES | NA | NA | |||||
| ENSDARG00000036335 | c1galt1c1 | C1GALT1-specific chaperone 1 [Source:ZFIN;Acc:ZDB-GENE-030131-2772] | 324052 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000095751 | C20H6orf58 (2 of 2) | chromosome 6 open reading frame 58 [Source:HGNC Symbol;Acc:HGNC:20960] | 406484 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000101297 | C25H15orf27 | chromosome 15 open reading frame 27 [Source:HGNC Symbol;Acc:HGNC:26763] | 0 | YES | YES | Other | other | |||||
| ENSDARG00000043451 | cab39 | calcium binding protein 39 [Source:ZFIN;Acc:ZDB-GENE-040625-158] | 415235 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000089913 | cacna1g | calcium channel, voltage-dependent, T type, alpha 1G subunit [Source:ZFIN;Acc:ZDB-GENE-090514-2] | 0 | YES | YES | Plasma Membrane | ion channel | |||||
| ENSDARG00000043661 | cadpsa | Ca2+-dependent activator protein for secretion a [Source:ZFIN;Acc:ZDB-GENE-030616-525] | 100333933 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000005749 | cand2 | cullin-associated and neddylation-dissociated 2 (putative) [Source:ZFIN;Acc:ZDB-GENE-060503-645] | 555955 | YES | YES | Nucleus | transcription regulator | |||||
| ENSDARG00000053129 | carhsp1 | calcium regulated heat stable protein 1 [Source:ZFIN;Acc:ZDB-GENE-040426-1479] | 393534 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000006863 | CCDC108 | coiled-coil domain containing 108 [Source:HGNC Symbol;Acc:HGNC:25325] | 100333283 | YES | YES | Other | other | |||||
| ENSDARG00000099190 | ccdc127a | coiled-coil domain containing 127a [Source:ZFIN;Acc:ZDB-GENE-060825-61] | 751722 | YES | YES | Other | other | |||||
| ENSDARG00000044626 | ccdc90b | coiled-coil domain containing 90B [Source:ZFIN;Acc:ZDB-GENE-040724-162] | 100149897 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000055504 | CD68 | CD68 molecule [Source:HGNC Symbol;Acc:HGNC:1693] | 100000085 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000009087 | cd74a | CD74 molecule, major histocompatibility complex, class II invariant chain a [Source:ZFIN;Acc:ZDB-GENE-000901-1] | 58113 | YES | YES | Plasma Membrane | transmembrane receptor | |||||
| ENSDARG00000104283 | cdc42bpaa | CDC42 binding protein kinase alpha (DMPK-like) a [Source:ZFIN;Acc:ZDB-GENE-120727-12] | 0 | YES | YES | NA | NA | |||||
| ENSDARG00000045036 | cdc42ep4b | CDC42 effector protein (Rho GTPase binding) 4b [Source:ZFIN;Acc:ZDB-GENE-091008-2] | 100002938 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000094577 | cdc42se2 | CDC42 small effector 2 [Source:ZFIN;Acc:ZDB-GENE-080204-45] | 794234 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000053395 | cdkn2aipnl | CDKN2A interacting protein N-terminal like [Source:ZFIN;Acc:ZDB-GENE-041010-154] | 450034 | YES | YES | Other | other | |||||
| ENSDARG00000019549 | cds1 | CDP-diacylglycerol synthase (phosphatidate cytidylyltransferase) 1 [Source:ZFIN;Acc:ZDB-GENE-070705-78] | 100002015 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000015854 | chata | choline O-acetyltransferase a [Source:ZFIN;Acc:ZDB-GENE-080102-2] | 100170938 | YES | YES | Nucleus | enzyme | |||||
| ENSDARG00000068156 | chchd7 | coiled-coil-helix-coiled-coil-helix domain containing 7 [Source:ZFIN;Acc:ZDB-GENE-080204-55] | 561510 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000007323 | chmp4bb | charged multivesicular body protein 4Bb [Source:ZFIN;Acc:ZDB-GENE-040426-2812] | 406766 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000019556 | clcn7 | chloride channel 7 [Source:ZFIN;Acc:ZDB-GENE-061103-196] | 553351 | YES | YES | Plasma Membrane | ion channel | |||||
| ENSDARG00000020679 | clpp | caseinolytic mitochondrial matrix peptidase proteolytic subunit [Source:ZFIN;Acc:ZDB-GENE-030131-7860] | 553713 | YES | YES | Cytoplasm | peptidase | |||||
| ENSDARG00000073883 | clstn3 | calsyntenin 3 [Source:ZFIN;Acc:ZDB-GENE-100921-73] | 555627 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000053122 | clvs2 | clavesin 2 [Source:ZFIN;Acc:ZDB-GENE-041014-313] | 566769 | YES | YES | Cytoplasm | transporter | |||||
| ENSDARG00000070822 | cnp | 2',3'-cyclic nucleotide 3' phosphodiesterase [Source:ZFIN;Acc:ZDB-GENE-030521-29] | 325896 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000092677 | coa6 | cytochrome c oxidase assembly factor 6 [Source:ZFIN;Acc:ZDB-GENE-070705-152] | 100150991 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000094258 | commd9 | COMM domain containing 9 [Source:ZFIN;Acc:ZDB-GENE-070424-139] | 562660 | YES | YES | Other | other | |||||
| ENSDARG00000015337 | comta | catechol-O-methyltransferase a [Source:ZFIN;Acc:ZDB-GENE-050913-117] | 561372 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000018562 | cope | coatomer protein complex, subunit epsilon [Source:ZFIN;Acc:ZDB-GENE-041114-59] | 492492 | YES | YES | Cytoplasm | transporter | |||||
| ENSDARG00000057624 | cops5 | COP9 signalosome subunit 5 [Source:ZFIN;Acc:ZDB-GENE-040426-1686] | 393698 | YES | YES | Nucleus | transcription regulator | |||||
| ENSDARG00000035618 | coq5 | coenzyme Q5 homolog, methyltransferase (S. cerevisiae) [Source:ZFIN;Acc:ZDB-GENE-040912-5] | 447802 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000092124 | cox14 | COX14 cytochrome c oxidase assembly factor [Source:ZFIN;Acc:ZDB-GENE-060503-626] | 0 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000075933 | cox15 | cytochrome c oxidase assembly homolog 15 (yeast) [Source:ZFIN;Acc:ZDB-GENE-040426-955] | 394181 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000018997 | cplx2l | complexin 2, like [Source:ZFIN;Acc:ZDB-GENE-040718-160] | 436732 | YES | YES | Plasma Membrane | transporter | |||||
| ENSDARG00000044212 | CR735126.1 | Uncharacterized protein [Source:UniProtKB/TrEMBL;Acc:F1Q4P0] | 0 | YES | YES | NA | NA | |||||
| ENSDARG00000056742 | crmp1 | collapsin response mediator protein 1 [Source:ZFIN;Acc:ZDB-GENE-050720-1] | 553756 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000071454 | crp1 | C-reactive protein 1 [Source:ZFIN;Acc:ZDB-GENE-060503-314] | 570508 | YES | YES | NA | NA | |||||
| ENSDARG00000101334 | ctsc | cathepsin C [Source:ZFIN;Acc:ZDB-GENE-030619-9] | 368704 | YES | YES | Cytoplasm | peptidase | |||||
| ENSDARG00000095577 | cybrd1 | cytochrome b reductase 1 [Source:ZFIN;Acc:ZDB-GENE-050522-365] | 552960 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000006040 | cyp20a1 | cytochrome P450, family 20, subfamily A, polypeptide 1 [Source:ZFIN;Acc:ZDB-GENE-040426-2646] | 406641 | YES | YES | Other | enzyme | |||||
| ENSDARG00000077121 | cyp26b1 | cytochrome P450, family 26, subfamily b, polypeptide 1 [Source:ZFIN;Acc:ZDB-GENE-030131-2908] | 324188 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000105009 | ddah2 | dimethylarginine dimethylaminohydrolase 2 [Source:ZFIN;Acc:ZDB-GENE-030131-913] | 799954 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000037229 | denr | density-regulated protein [Source:ZFIN;Acc:ZDB-GENE-040718-450] | 436970 | YES | YES | Other | other | |||||
| ENSDARG00000103503 | dgat1a | diacylglycerol O-acyltransferase 1a [Source:ZFIN;Acc:ZDB-GENE-030131-4600] | 325875 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000003444 | dhrs7 | dehydrogenase/reductase (SDR family) member 7 [Source:ZFIN;Acc:ZDB-GENE-060825-21] | 751626 | YES | YES | Other | enzyme | |||||
| ENSDARG00000019260 | dhrs9 | dehydrogenase/reductase (SDR family) member 9 [Source:ZFIN;Acc:ZDB-GENE-030131-1249] | 322529 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000009372 | diras1b | DIRAS family, GTP-binding RAS-like 1b [Source:ZFIN;Acc:ZDB-GENE-070912-620] | 565395 | YES | YES | Plasma Membrane | enzyme | |||||
| ENSDARG00000033259 | dis3l2 | DIS3 like 3'-5' exoribonuclease 2 [Source:ZFIN;Acc:ZDB-GENE-070705-171] | 566106 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000015088 | dnajb11 | DnaJ (Hsp40) homolog, subfamily B, member 11 [Source:ZFIN;Acc:ZDB-GENE-031113-9] | 386709 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000044241 | dram2b | DNA-damage regulated autophagy modulator 2b [Source:ZFIN;Acc:ZDB-GENE-040625-141] | 415225 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000061255 | dusp3a | dual specificity phosphatase 3a [Source:ZFIN;Acc:ZDB-GENE-111207-3] | 100000665 | YES | YES | Cytoplasm | phosphatase | |||||
| ENSDARG00000040021 | ebag9 | estrogen receptor binding site associated, antigen, 9 [Source:ZFIN;Acc:ZDB-GENE-040426-1093] | 394069 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000012915 | ech1 | enoyl CoA hydratase 1, peroxisomal [Source:ZFIN;Acc:ZDB-GENE-041010-170] | 450048 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000021257 | eif3d | eukaryotic translation initiation factor 3, subunit D [Source:ZFIN;Acc:ZDB-GENE-030131-8442] | 336498 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000077533 | eif3f | eukaryotic translation initiation factor 3, subunit F [Source:ZFIN;Acc:ZDB-GENE-100215-2] | 557270 | YES | YES | Cytoplasm | translation regulator | |||||
| ENSDARG00000016889 | eif3g | eukaryotic translation initiation factor 3, subunit G [Source:ZFIN;Acc:ZDB-GENE-040426-1076] | 393974 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000010194 | eif4bb | eukaryotic translation initiation factor 4Bb [Source:ZFIN;Acc:ZDB-GENE-030131-8563] | 336619 | YES | YES | Cytoplasm | translation regulator | |||||
| ENSDARG00000014420 | elavl3 | ELAV like neuron-specific RNA binding protein 3 [Source:ZFIN;Acc:ZDB-GENE-980526-76] | 30732 | YES | YES | Nucleus | other | |||||
| ENSDARG00000059485 | EMB | embigin [Source:HGNC Symbol;Acc:HGNC:30465] | 100141497 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000061644 | ensab | endosulfine alpha b [Source:ZFIN;Acc:ZDB-GENE-060526-294] | 100317933 | YES | YES | Cytoplasm | transporter | |||||
| ENSDARG00000004635 | epha7 | eph receptor A7 [Source:ZFIN;Acc:ZDB-GENE-030131-3745] | 562195 | YES | YES | Plasma Membrane | kinase | |||||
| ENSDARG00000010411 | epn1 | epsin 1 [Source:ZFIN;Acc:ZDB-GENE-040426-2680] | 406671 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000060484 | EPT1 | ethanolaminephosphotransferase 1 [Source:HGNC Symbol;Acc:HGNC:29361] | 557632 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000021859 | erap1b | endoplasmic reticulum aminopeptidase 1b [Source:ZFIN;Acc:ZDB-GENE-040426-934] | 393175 | YES | YES | Extracellular Space | peptidase | |||||
| ENSDARG00000008026 | fam129bb | family with sequence similarity 129, member Bb [Source:ZFIN;Acc:ZDB-GENE-030131-6218] | 556540 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000016396 | fam219aa | family with sequence similarity 219, member Aa [Source:ZFIN;Acc:ZDB-GENE-040801-110] | 445197 | YES | YES | Other | other | |||||
| ENSDARG00000052010 | fam96b | family with sequence similarity 96, member B [Source:ZFIN;Acc:ZDB-GENE-040718-148] | 436722 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000016429 | farp2 | FERM, RhoGEF and pleckstrin domain protein 2 [Source:ZFIN;Acc:ZDB-GENE-030131-2993] | 567490 | YES | YES | NA | NA | |||||
| ENSDARG00000003462 | fech | ferrochelatase [Source:ZFIN;Acc:ZDB-GENE-000928-1] | 58215 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000053973 | fetub | fetuin B [Source:ZFIN;Acc:ZDB-GENE-050208-4] | 569489 | YES | YES | Extracellular Space | other | |||||
| ENSDARG00000040162 | FGFBP3 | fibroblast growth factor binding protein 3 [Source:HGNC Symbol;Acc:HGNC:23428] | 497419 | YES | YES | Extracellular Space | other | |||||
| ENSDARG00000058115 | fgfr2 | fibroblast growth factor receptor 2 [Source:ZFIN;Acc:ZDB-GENE-030323-1] | 352940 | YES | YES | Plasma Membrane | kinase | |||||
| ENSDARG00000037000 | fkbp11 | FK506 binding protein 11 [Source:ZFIN;Acc:ZDB-GENE-030616-357] | 368823 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000099143 | fnta | farnesyltransferase, CAAX box, alpha [Source:ZFIN;Acc:ZDB-GENE-030131-866] | 555882 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000044216 | fto | fat mass and obesity associated [Source:ZFIN;Acc:ZDB-GENE-061108-1] | 553363 | YES | YES | Nucleus | other | |||||
| ENSDARG00000029248 | fubp1 | far upstream element (FUSE) binding protein 1 [Source:ZFIN;Acc:ZDB-GENE-040426-2159] | 406418 | YES | YES | Nucleus | transcription regulator | |||||
| ENSDARG00000017741 | g3bp1 | GTPase activating protein (SH3 domain) binding protein 1 [Source:ZFIN;Acc:ZDB-GENE-030131-7452] | 335512 | YES | YES | Nucleus | enzyme | |||||
| ENSDARG00000058736 | gabra6b | gamma-aminobutyric acid (GABA) A receptor, alpha 6b [Source:ZFIN;Acc:ZDB-GENE-080815-1] | 559693 | YES | YES | Plasma Membrane | ion channel | |||||
| ENSDARG00000094512 | gabrz | gamma-aminobutyric acid (GABA) A receptor, zeta [Source:ZFIN;Acc:ZDB-GENE-080303-26] | 561738 | YES | YES | NA | NA | |||||
| ENSDARG00000061335 | galnt1 | UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 1 [Source:ZFIN;Acc:ZDB-GENE-030131-2360] | 559073 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000076058 | gba | glucosidase, beta, acid [Source:ZFIN;Acc:ZDB-GENE-100922-270] | 559072 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000005643 | gcat | glycine C-acetyltransferase [Source:ZFIN;Acc:ZDB-GENE-060518-3] | 402822 | YES | YES | NA | NA | |||||
| ENSDARG00000091655 | gcsha | glycine cleavage system protein H (aminomethyl carrier), a [Source:ZFIN;Acc:ZDB-GENE-050320-18] | 541329 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000076962 | gdpd5b | glycerophosphodiester phosphodiesterase domain containing 5b [Source:ZFIN;Acc:ZDB-GENE-030131-6028] | 560645 | YES | YES | Other | enzyme | |||||
| ENSDARG00000056651 | gfra4a | GDNF family receptor alpha 4a [Source:ZFIN;Acc:ZDB-GENE-080402-10] | 100144564 | YES | YES | Plasma Membrane | transmembrane receptor | |||||
| ENSDARG00000098983 | GHITM | growth hormone inducible transmembrane protein [Source:HGNC Symbol;Acc:HGNC:17281] | 0 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000068978 | glo1 | glyoxalase 1 [Source:ZFIN;Acc:ZDB-GENE-030722-9] | 368213 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000100944 | glrx2 | glutaredoxin 2 [Source:ZFIN;Acc:ZDB-GENE-040718-101] | 436677 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000088439 | gm2a | GM2 ganglioside activator [Source:ZFIN;Acc:ZDB-GENE-050417-373] | 550529 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000017658 | gmppb | GDP-mannose pyrophosphorylase B [Source:ZFIN;Acc:ZDB-GENE-040801-234] | 445097 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000019278 | GNB4 | guanine nucleotide binding protein (G protein), beta polypeptide 4 [Source:HGNC Symbol;Acc:HGNC:20731] | 0 | YES | YES | Plasma Membrane | enzyme | |||||
| ENSDARG00000039830 | gng5 | guanine nucleotide binding protein (G protein), gamma 5 [Source:ZFIN;Acc:ZDB-GENE-030131-9966] | 337838 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000103200 | gpr107 | G protein-coupled receptor 107 [Source:ZFIN;Acc:ZDB-GENE-061103-130] | 570589 | YES | YES | Plasma Membrane | G-protein coupled receptor | |||||
| ENSDARG00000096651 | gpr108 | G protein-coupled receptor 108 [Source:ZFIN;Acc:ZDB-GENE-121214-129] | 0 | YES | YES | Plasma Membrane | G-protein coupled receptor | |||||
| ENSDARG00000010021 | grpel1 | GrpE-like 1, mitochondrial [Source:ZFIN;Acc:ZDB-GENE-051120-111] | 559037 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000029501 | gsk3aa | glycogen synthase kinase 3 alpha a [Source:ZFIN;Acc:ZDB-GENE-060503-796] | 560194 | YES | YES | Nucleus | kinase | |||||
| ENSDARG00000017388 | gstt1b | glutathione S-transferase theta 1b [Source:ZFIN;Acc:ZDB-GENE-040426-1491] | 393556 | YES | YES | Other | other | |||||
| ENSDARG00000011934 | gyg1a | glycogenin 1a [Source:ZFIN;Acc:ZDB-GENE-040426-2910] | 406831 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000060627 | HIP1R (1 of 2) | huntingtin interacting protein 1 related [Source:HGNC Symbol;Acc:HGNC:18415] | 103910581 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000102458 | hip1rb | huntingtin interacting protein 1 related b [Source:ZFIN;Acc:ZDB-GENE-070209-59] | 562138 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000007960 | hnrnpaba | heterogeneous nuclear ribonucleoprotein A/Ba [Source:ZFIN;Acc:ZDB-GENE-030131-185] | 321466 | YES | YES | Nucleus | enzyme | |||||
| ENSDARG00000059246 | hnrnpd | heterogeneous nuclear ribonucleoprotein D [Source:ZFIN;Acc:ZDB-GENE-070424-97] | 560522 | YES | YES | Nucleus | transcription regulator | |||||
| ENSDARG00000022763 | hpcal1 | hippocalcin-like 1 [Source:ZFIN;Acc:ZDB-GENE-040426-1242] | 394139 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000063446 | iba57 | IBA57, iron-sulfur cluster assembly homolog (S. cerevisiae) [Source:ZFIN;Acc:ZDB-GENE-060929-712] | 100330979 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000031886 | ift140 | intraflagellar transport 140 homolog (Chlamydomonas) [Source:ZFIN;Acc:ZDB-GENE-040724-165] | 553213 | YES | YES | Extracellular Space | other | |||||
| ENSDARG00000011948 | insra | insulin receptor a [Source:ZFIN;Acc:ZDB-GENE-020503-3] | 245699 | YES | YES | Plasma Membrane | kinase | |||||
| ENSDARG00000071524 | insrb | insulin receptor b [Source:ZFIN;Acc:ZDB-GENE-020503-4] | 245700 | YES | YES | Plasma Membrane | kinase | |||||
| ENSDARG00000035751 | ipo7 | importin 7 [Source:ZFIN;Acc:ZDB-GENE-030131-458] | 321739 | YES | YES | Nucleus | transporter | |||||
| ENSDARG00000068657 | irgq2 | immunity-related GTPase family, q2 [Source:ZFIN;Acc:ZDB-GENE-051212-7] | 556650 | YES | YES | NA | NA | |||||
| ENSDARG00000099199 | itchb | itchy E3 ubiquitin protein ligase b [Source:ZFIN;Acc:ZDB-GENE-131024-2] | 100330031 | YES | YES | Nucleus | enzyme | |||||
| ENSDARG00000012275 | katnal2 | katanin p60 subunit A-like 2 [Source:ZFIN;Acc:ZDB-GENE-051113-156] | 641431 | YES | YES | Other | other | |||||
| ENSDARG00000102064 | kcna2b | potassium voltage-gated channel, shaker-related subfamily, member 2b [Source:ZFIN;Acc:ZDB-GENE-080204-89] | 562853 | YES | YES | Plasma Membrane | ion channel | |||||
| ENSDARG00000062134 | kcnab2b | potassium voltage-gated channel, shaker-related subfamily, beta member 2 b [Source:ZFIN;Acc:ZDB-GENE-080219-36] | 565014 | YES | YES | Plasma Membrane | ion channel | |||||
| ENSDARG00000069560 | kcnh3 | potassium voltage-gated channel, subfamily H (eag-related), member 3 [Source:ZFIN;Acc:ZDB-GENE-070912-23] | 0 | YES | YES | Plasma Membrane | ion channel | |||||
| ENSDARG00000091130 | kcnq2b | potassium voltage-gated channel, KQT-like subfamily, member 2b [Source:ZFIN;Acc:ZDB-GENE-120130-1] | 0 | YES | YES | Plasma Membrane | ion channel | |||||
| ENSDARG00000079484 | kcnt1 | potassium channel, subfamily T, member 1 [Source:ZFIN;Acc:ZDB-GENE-091231-1] | 100004419 | YES | YES | Plasma Membrane | ion channel | |||||
| ENSDARG00000068691 | kctd4 | potassium channel tetramerization domain containing 4 [Source:ZFIN;Acc:ZDB-GENE-040704-71] | 792402 | YES | YES | Other | ion channel | |||||
| ENSDARG00000098936 | kif5aa | kinesin family member 5A, a [Source:ZFIN;Acc:ZDB-GENE-070912-141] | 566086 | YES | YES | Cytoplasm | transporter | |||||
| ENSDARG00000056998 | kirrelb | kin of IRRE like b [Source:ZFIN;Acc:ZDB-GENE-070912-173] | 100170816 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000045140 | lcmt1 | leucine carboxyl methyltransferase 1 [Source:ZFIN;Acc:ZDB-GENE-040912-75] | 447907 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000038845 | ldhd | lactate dehydrogenase D [Source:ZFIN;Acc:ZDB-GENE-030131-6140] | 334208 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000055525 | lgalslb | lectin, galactoside-binding-like b [Source:ZFIN;Acc:ZDB-GENE-030616-570] | 368889 | YES | YES | Other | other | |||||
| ENSDARG00000088663 | lman2lb | lectin, mannose-binding 2-like b [Source:ZFIN;Acc:ZDB-GENE-060929-256] | 767766 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000078094 | lmf2a | lipase maturation factor 2a [Source:ZFIN;Acc:ZDB-GENE-080723-47] | 571852 | YES | YES | Other | other | |||||
| ENSDARG00000004930 | lmo7a | LIM domain 7a [Source:ZFIN;Acc:ZDB-GENE-030219-74] | 553341 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000061517 | lrp1bb | low density lipoprotein receptor-related protein 1Bb [Source:ZFIN;Acc:ZDB-GENE-070912-401] | 559293 | YES | YES | Plasma Membrane | transmembrane receptor | |||||
| ENSDARG00000070074 | lrp8 | low density lipoprotein receptor-related protein 8, apolipoprotein e receptor [Source:ZFIN;Acc:ZDB-GENE-050506-134] | 557734 | YES | YES | Plasma Membrane | transmembrane receptor | |||||
| ENSDARG00000060683 | lrsam1 | leucine rich repeat and sterile alpha motif containing 1 [Source:ZFIN;Acc:ZDB-GENE-060526-97] | 562066 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000006029 | lta4h | leukotriene A4 hydrolase [Source:ZFIN;Acc:ZDB-GENE-040426-2477] | 406575 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000056874 | lygl1 | lysozyme g-like 1 [Source:ZFIN;Acc:ZDB-GENE-040718-461] | 436979 | YES | YES | Extracellular Space | enzyme | |||||
| ENSDARG00000018432 | m6pr | mannose-6-phosphate receptor (cation dependent) [Source:ZFIN;Acc:ZDB-GENE-040426-2285] | 406486 | YES | YES | Cytoplasm | transporter | |||||
| ENSDARG00000102200 | man2a1 | mannosidase, alpha, class 2A, member 1 [Source:ZFIN;Acc:ZDB-GENE-060526-31] | 0 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000077177 | map2k4b | mitogen-activated protein kinase kinase 4b [Source:ZFIN;Acc:ZDB-GENE-080721-13] | 568951 | YES | YES | Cytoplasm | kinase | |||||
| ENSDARG00000099943 | mapre2 | microtubule-associated protein, RP/EB family, member 2 [Source:ZFIN;Acc:ZDB-GENE-071004-41] | 557679 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000058848 | mcoln1b | mucolipin 1b [Source:ZFIN;Acc:ZDB-GENE-130514-1] | 795916 | YES | YES | Cytoplasm | ion channel | |||||
| ENSDARG00000025972 | mcts1 | malignant T cell amplified sequence 1 [Source:ZFIN;Acc:ZDB-GENE-040426-957] | 394046 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000004796 | mgrn1b | mahogunin, ring finger 1b [Source:ZFIN;Acc:ZDB-GENE-110401-3] | 553327 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000032431 | mpv17 | MpV17 mitochondrial inner membrane protein [Source:ZFIN;Acc:ZDB-GENE-040426-1168] | 394140 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000068886 | mrpl14 | mitochondrial ribosomal protein L14 [Source:ZFIN;Acc:ZDB-GENE-040426-1066] | 394122 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000063924 | mt-cyb | cytochrome b, mitochondrial [Source:ZFIN;Acc:ZDB-GENE-011205-17] | 140512 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000063895 | mt-nd1 | NADH dehydrogenase 1, mitochondrial [Source:ZFIN;Acc:ZDB-GENE-011205-7] | 140531 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000056690 | mtmr1a | myotubularin related protein 1a [Source:ZFIN;Acc:ZDB-GENE-050522-518] | 553750 | YES | YES | Cytoplasm | phosphatase | |||||
| ENSDARG00000057928 | mzt2b | mitotic spindle organizing protein 2B [Source:ZFIN;Acc:ZDB-GENE-040801-87] | 445174 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000005513 | naca | nascent polypeptide-associated complex alpha subunit [Source:ZFIN;Acc:ZDB-GENE-020423-4] | 195823 | YES | YES | Cytoplasm | transcription regulator | |||||
| ENSDARG00000011301 | nae1 | nedd8 activating enzyme E1 subunit 1 [Source:ZFIN;Acc:ZDB-GENE-040426-1552] | 573336 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000099116 | nagk | N-acetylglucosamine kinase [Source:ZFIN;Acc:ZDB-GENE-070928-19] | 100126013 | YES | YES | Cytoplasm | kinase | |||||
| ENSDARG00000011334 | ncaldb | neurocalcin delta b [Source:ZFIN;Acc:ZDB-GENE-040808-37] | 445319 | YES | YES | NA | NA | |||||
| ENSDARG00000062087 | nceh1b | neutral cholesterol ester hydrolase 1b [Source:ZFIN;Acc:ZDB-GENE-061110-43] | 777713 | YES | YES | Plasma Membrane | enzyme | |||||
| ENSDARG00000090389 | ndufv3 | NADH dehydrogenase (ubiquinone) flavoprotein 3 [Source:ZFIN;Acc:ZDB-GENE-030131-6500] | 0 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000056566 | necab1 | N-terminal EF-hand calcium binding protein 1 [Source:ZFIN;Acc:ZDB-GENE-050417-395] | 550546 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000068125 | nfu1 | NFU1 iron-sulfur cluster scaffold homolog (S. cerevisiae) [Source:ZFIN;Acc:ZDB-GENE-050417-365] | 403053 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000077710 | nlgn1 | neuroligin 1 [Source:ZFIN;Acc:ZDB-GENE-090918-1] | 0 | YES | YES | Plasma Membrane | enzyme | |||||
| ENSDARG00000068910 | nos1 | nitric oxide synthase 1 (neuronal) [Source:ZFIN;Acc:ZDB-GENE-001101-1] | 60658 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000040430 | nptxra | neuronal pentraxin receptor a [Source:ZFIN;Acc:ZDB-GENE-050208-280] | 101884706 | YES | YES | Plasma Membrane | transmembrane receptor | |||||
| ENSDARG00000010250 | nqo1 | NAD(P)H dehydrogenase, quinone 1 [Source:ZFIN;Acc:ZDB-GENE-030131-1226] | 322506 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000019596 | nrd1a | nardilysin a (N-arginine dibasic convertase) [Source:ZFIN;Acc:ZDB-GENE-070912-417] | 565850 | YES | YES | Cytoplasm | peptidase | |||||
| ENSDARG00000062335 | nudt14 | nudix (nucleoside diphosphate linked moiety X)-type motif 14 [Source:ZFIN;Acc:ZDB-GENE-041014-254] | 559149 | YES | YES | Cytoplasm | phosphatase | |||||
| ENSDARG00000030832 | otofa | otoferlin a [Source:ZFIN;Acc:ZDB-GENE-030131-7778] | 557476 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000073931 | otulina | OTU deubiquitinase with linear linkage specificity a [Source:ZFIN;Acc:ZDB-GENE-040426-2157] | 100001665 | YES | YES | Cytoplasm | peptidase | |||||
| ENSDARG00000054319 | oxct1b | 3-oxoacid CoA transferase 1b [Source:ZFIN;Acc:ZDB-GENE-060929-212] | 560273 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000078185 | pacs2 | phosphofurin acidic cluster sorting protein 2 [Source:ZFIN;Acc:ZDB-GENE-081218-1] | 794778 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000058047 | pafah1b3 | platelet-activating factor acetylhydrolase, isoform Ib, gamma subunit [Source:ZFIN;Acc:ZDB-GENE-040426-1026] | 394033 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000068177 | pak2a | p21 protein (Cdc42/Rac)-activated kinase 2a [Source:ZFIN;Acc:ZDB-GENE-021011-2] | 266749 | YES | YES | Cytoplasm | kinase | |||||
| ENSDARG00000034826 | park7 | parkinson protein 7 [Source:ZFIN;Acc:ZDB-GENE-041010-5] | 449674 | YES | YES | Nucleus | enzyme | |||||
| ENSDARG00000101232 | pcdh1g2 | protocadherin 1 gamma 2 [Source:ZFIN;Acc:ZDB-GENE-050608-2] | 553986 | YES | YES | Other | other | |||||
| ENSDARG00000090348 | pcdh2ab5 | protocadherin 2 alpha b 5 [Source:ZFIN;Acc:ZDB-GENE-041118-4] | 493591 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000033012 | pcnt | pericentrin [Source:ZFIN;Acc:ZDB-GENE-030829-26] | 325536 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000053130 | pcp4a | Purkinje cell protein 4a [Source:ZFIN;Acc:ZDB-GENE-031118-105] | 386848 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000045567 | phpt1 | phosphohistidine phosphatase 1 [Source:ZFIN;Acc:ZDB-GENE-040801-85] | 445172 | YES | YES | Cytoplasm | phosphatase | |||||
| ENSDARG00000003998 | phyhipla | phytanoyl-CoA 2-hydroxylase interacting protein-like a [Source:ZFIN;Acc:ZDB-GENE-061215-64] | 790943 | YES | YES | NA | NA | |||||
| ENSDARG00000038270 | PIGG | phosphatidylinositol glycan anchor biosynthesis, class G [Source:HGNC Symbol;Acc:HGNC:25985] | 100538291 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000004527 | pin4 | protein (peptidylprolyl cis/trans isomerase) NIMA-interacting, 4 (parvulin) [Source:ZFIN;Acc:ZDB-GENE-050522-117] | 553574 | YES | YES | Nucleus | enzyme | |||||
| ENSDARG00000055255 | pitpnm3 | PITPNM family member 3 [Source:ZFIN;Acc:ZDB-GENE-030131-9216] | 337272 | YES | YES | Cytoplasm | transporter | |||||
| ENSDARG00000059960 | plch2a | phospholipase C, eta 2a [Source:ZFIN;Acc:ZDB-GENE-030131-6367] | 100334231 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000079892 | PLXNB2 (4 of 5) | plexin B2 [Source:HGNC Symbol;Acc:HGNC:9104] | 561237 | YES | YES | Plasma Membrane | transmembrane receptor | |||||
| ENSDARG00000102607 | pmpca | peptidase (mitochondrial processing) alpha [Source:ZFIN;Acc:ZDB-GENE-030131-5809] | 492801 | YES | YES | Cytoplasm | peptidase | |||||
| ENSDARG00000099802 | pnp5b | purine nucleoside phosphorylase 5b [Source:ZFIN;Acc:ZDB-GENE-040912-54] | 447889 | YES | YES | Nucleus | enzyme | |||||
| ENSDARG00000017612 | pnpo | pyridoxamine 5'-phosphate oxidase [Source:ZFIN;Acc:ZDB-GENE-060602-2] | 0 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000016856 | pon2 | paraoxonase 2 [Source:ZFIN;Acc:ZDB-GENE-030131-7116] | 335176 | YES | YES | Extracellular Space | enzyme | |||||
| ENSDARG00000031317 | ppdpfb | pancreatic progenitor cell differentiation and proliferation factor b [Source:ZFIN;Acc:ZDB-GENE-030131-8247] | 336303 | YES | YES | Other | other | |||||
| ENSDARG00000009771 | ppme1 | protein phosphatase methylesterase 1 [Source:ZFIN;Acc:ZDB-GENE-030131-7064] | 335124 | YES | YES | Other | enzyme | |||||
| ENSDARG00000054007 | ppp1r2 | protein phosphatase 1, regulatory (inhibitor) subunit 2 [Source:ZFIN;Acc:ZDB-GENE-040426-1814] | 402967 | YES | YES | Cytoplasm | phosphatase | |||||
| ENSDARG00000069654 | ppp6r2b | protein phosphatase 6, regulatory subunit 2b [Source:ZFIN;Acc:ZDB-GENE-070705-441] | 570360 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000060596 | prkaa1 | protein kinase, AMP-activated, alpha 1 catalytic subunit [Source:ZFIN;Acc:ZDB-GENE-030325-1] | 100001198 | YES | YES | Cytoplasm | kinase | |||||
| ENSDARG00000016809 | prkacab | protein kinase, cAMP-dependent, catalytic, alpha, genome duplicate b [Source:ZFIN;Acc:ZDB-GENE-040801-208] | 445076 | YES | YES | Cytoplasm | kinase | |||||
| ENSDARG00000004561 | prkcg | protein kinase C, gamma [Source:ZFIN;Acc:ZDB-GENE-090206-1] | 100003514 | YES | YES | Cytoplasm | kinase | |||||
| ENSDARG00000076718 | prkra | protein kinase, interferon-inducible double stranded RNA dependent activator [Source:ZFIN;Acc:ZDB-GENE-050309-206] | 557370 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000037506 | prps1b | phosphoribosyl pyrophosphate synthetase 1B [Source:ZFIN;Acc:ZDB-GENE-030131-4011] | 560827 | YES | YES | Other | kinase | |||||
| ENSDARG00000045928 | psma4 | proteasome (prosome, macropain) subunit, alpha type, 4 [Source:ZFIN;Acc:ZDB-GENE-040426-1932] | 326687 | YES | YES | Cytoplasm | peptidase | |||||
| ENSDARG00000019398 | psma6a | proteasome (prosome, macropain) subunit, alpha type, 6a [Source:ZFIN;Acc:ZDB-GENE-020326-1] | 171585 | YES | YES | Cytoplasm | peptidase | |||||
| ENSDARG00000027088 | ptgdsb | prostaglandin D2 synthase b [Source:ZFIN;Acc:ZDB-GENE-030131-8436] | 336492 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000073902 | ptpn9b | protein tyrosine phosphatase, non-receptor type 9, b [Source:ZFIN;Acc:ZDB-GENE-090313-52] | 562170 | YES | YES | NA | NA | |||||
| ENSDARG00000040309 | pxdc1b | PX domain containing 1b [Source:ZFIN;Acc:ZDB-GENE-091204-294] | 566349 | YES | YES | Other | other | |||||
| ENSDARG00000090086 | rab11bb | RAB11B, member RAS oncogene family, b [Source:ZFIN;Acc:ZDB-GENE-040718-293] | 436828 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000104179 | rab11fip5a | RAB11 family interacting protein 5a (class I) [Source:ZFIN;Acc:ZDB-GENE-090313-206] | 562719 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000045628 | rab34a | RAB34, member RAS oncogene family a [Source:ZFIN;Acc:ZDB-GENE-041010-197] | 450074 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000077553 | RAP2A (2 of 2) | RAP2A, member of RAS oncogene family [Source:HGNC Symbol;Acc:HGNC:9861] | 100332969 | YES | YES | NA | NA | |||||
| ENSDARG00000101199 | rbp4 | retinol binding protein 4, plasma [Source:ZFIN;Acc:ZDB-GENE-000210-19] | 30077 | YES | YES | Extracellular Space | transporter | |||||
| ENSDARG00000078215 | rbsn | rabenosyn, RAB effector [Source:ZFIN;Acc:ZDB-GENE-100525-1] | 569240 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000039378 | rcn2 | reticulocalbin 2 [Source:ZFIN;Acc:ZDB-GENE-050706-62] | 571340 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000045277 | rdh12l | retinol dehydrogenase 12, like [Source:ZFIN;Acc:ZDB-GENE-040801-48] | 494176 | YES | YES | NA | NA | |||||
| ENSDARG00000070140 | RETSAT (2 of 2) | retinol saturase (all-trans-retinol 13,14-reductase) [Source:HGNC Symbol;Acc:HGNC:25991] | 777751 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000054890 | rgra | retinal G protein coupled receptor a [Source:ZFIN;Acc:ZDB-GENE-040924-6] | 550575 | YES | YES | Plasma Membrane | G-protein coupled receptor | |||||
| ENSDARG00000103313 | rhot1b | ras homolog family member T1 [Source:ZFIN;Acc:ZDB-GENE-061009-52] | 561933 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000103551 | rnf213b | ring finger protein 213b [Source:ZFIN;Acc:ZDB-GENE-110822-1] | 563001 | YES | YES | NA | NA | |||||
| ENSDARG00000005251 | rpe | ribulose-5-phosphate-3-epimerase [Source:ZFIN;Acc:ZDB-GENE-030131-6837] | 334897 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000070702 | s100v2 | S100 calcium binding protein V2 [Source:ZFIN;Acc:ZDB-GENE-030131-8909] | 336965 | YES | YES | NA | NA | |||||
| ENSDARG00000102415 | scinla | scinderin like a [Source:ZFIN;Acc:ZDB-GENE-030131-2005] | 323285 | YES | YES | NA | NA | |||||
| ENSDARG00000099031 | scn4ba | sodium channel, voltage-gated, type IV, beta a [Source:ZFIN;Acc:ZDB-GENE-070615-18] | 566069 | YES | YES | Plasma Membrane | ion channel | |||||
| ENSDARG00000078328 | sec22a | SEC22 vesicle trafficking protein homolog A (S. cerevisiae) [Source:ZFIN;Acc:ZDB-GENE-030131-3553] | 324832 | YES | YES | Cytoplasm | transporter | |||||
| ENSDARG00000030108 | sec22ba | SEC22 vesicle trafficking protein homolog B (S. cerevisiae), a [Source:ZFIN;Acc:ZDB-GENE-010724-3] | 114464 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000061413 | sec23ip | SEC23 interacting protein [Source:ZFIN;Acc:ZDB-GENE-030131-5259] | 560750 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000018637 | sec61g | Sec61 gamma subunit [Source:ZFIN;Acc:ZDB-GENE-040718-203] | 436772 | YES | YES | Plasma Membrane | transporter | |||||
| ENSDARG00000019951 | sec62 | SEC62 homolog (S. cerevisiae) [Source:ZFIN;Acc:ZDB-GENE-050913-83] | 563660 | YES | YES | Cytoplasm | transporter | |||||
| ENSDARG00000038228 | sepw2a | selenoprotein W, 2a [Source:ZFIN;Acc:ZDB-GENE-030428-1] | 378437 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000032340 | serhl | serine hydrolase-like [Source:ZFIN;Acc:ZDB-GENE-030131-1368] | 322648 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000031495 | seta | SET translocation (myeloid leukemia-associated) A [Source:ZFIN;Acc:ZDB-GENE-030131-2221] | 323501 | YES | YES | Nucleus | phosphatase | |||||
| ENSDARG00000075376 | sgsm1b | small G protein signaling modulator 1b [Source:ZFIN;Acc:ZDB-GENE-091117-39] | 100535556 | YES | YES | Nucleus | other | |||||
| ENSDARG00000075853 | sh3kbp1 | SH3-domain kinase binding protein 1 [Source:ZFIN;Acc:ZDB-GENE-091204-186] | 793623 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000056379 | si:ch211-154o6.6 | si:ch211-154o6.6 [Source:ZFIN;Acc:ZDB-GENE-060503-576] | 564061 | YES | YES | NA | NA | |||||
| ENSDARG00000070757 | si:ch211-182e10.4 | si:ch211-182e10.4 [Source:ZFIN;Acc:ZDB-GENE-041014-76] | 553308 | YES | YES | NA | NA | |||||
| ENSDARG00000100894 | si:ch211-193k8.5 | si:ch211-193k8.5 [Source:ZFIN;Acc:ZDB-GENE-030131-5121] | 0 | YES | YES | NA | NA | |||||
| ENSDARG00000092856 | si:ch211-210c8.7 | si:ch211-210c8.7 [Source:ZFIN;Acc:ZDB-GENE-090313-81] | 0 | YES | YES | NA | NA | |||||
| ENSDARG00000098585 | si:ch211-237a4.2 | si:ch211-237a4.2 [Source:ZFIN;Acc:ZDB-GENE-060503-504] | 0 | YES | YES | NA | NA | |||||
| ENSDARG00000030945 | si:ch211-259g3.4 | si:ch211-259g3.4 [Source:ZFIN;Acc:ZDB-GENE-030131-8118] | 0 | YES | YES | Extracellular Space | other | |||||
| ENSDARG00000069735 | si:ch211-5k11.6 | si:ch211-5k11.6 [Source:ZFIN;Acc:ZDB-GENE-081104-38] | 497166 | YES | YES | NA | NA | |||||
| ENSDARG00000078824 | si:ch211-66e2.3 | si:ch211-66e2.3 [Source:ZFIN;Acc:ZDB-GENE-080226-7] | 100141487 | YES | YES | NA | NA | |||||
| ENSDARG00000096346 | si:ch211-85n16.3 | si:ch211-85n16.3 [Source:ZFIN;Acc:ZDB-GENE-120215-137] | 0 | YES | YES | NA | NA | |||||
| ENSDARG00000086746 | si:ch73-209e20.3 | si:ch73-209e20.3 [Source:ZFIN;Acc:ZDB-GENE-120215-75] | 100333321 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000052680 | si:dkey-182g1.2 | si:dkey-182g1.2 [Source:ZFIN;Acc:ZDB-GENE-060503-619] | 100005482 | YES | YES | NA | NA | |||||
| ENSDARG00000033413 | si:dkey-183c16.7 | si:dkey-183c16.7 [Source:ZFIN;Acc:ZDB-GENE-041210-254] | 556673 | YES | YES | NA | NA | |||||
| ENSDARG00000104065 | si:dkey-1b17.11 | si:dkey-1b17.11 [Source:ZFIN;Acc:ZDB-GENE-050208-786] | 100006734 | YES | YES | NA | NA | |||||
| ENSDARG00000043518 | si:dkey-239i20.2 | si:dkey-239i20.2 [Source:ZFIN;Acc:ZDB-GENE-041001-98] | 557525 | YES | YES | Other | other | |||||
| ENSDARG00000095848 | si:dkey-239n17.6 | si:dkey-239n17.6 [Source:ZFIN;Acc:ZDB-GENE-110411-211] | 0 | YES | YES | NA | NA | |||||
| ENSDARG00000034210 | si:dkey-240h12.4 | si:dkey-240h12.4 [Source:ZFIN;Acc:ZDB-GENE-070705-406] | 555536 | YES | YES | NA | NA | |||||
| ENSDARG00000092161 | si:dkey-261e22.4 | si:dkey-261e22.4 [Source:ZFIN;Acc:ZDB-GENE-041210-161] | 0 | YES | YES | NA | NA | |||||
| ENSDARG00000097110 | si:dkey-56f14.4 | si:dkey-56f14.4 [Source:ZFIN;Acc:ZDB-GENE-131121-272] | 100005476 | YES | YES | NA | NA | |||||
| ENSDARG00000096622 | si:dkey-9c18.4 | si:dkey-9c18.4 [Source:ZFIN;Acc:ZDB-GENE-121214-46] | 0 | YES | YES | Plasma Membrane | G-protein coupled receptor | |||||
| ENSDARG00000070516 | si:dkeyp-52c3.2 | si:dkeyp-52c3.2 [Source:ZFIN;Acc:ZDB-GENE-110411-46] | 436616 | YES | YES | Other | other | |||||
| ENSDARG00000086870 | slc12a2 | solute carrier family 12 (sodium/potassium/chloride transporter), member 2 [Source:ZFIN;Acc:ZDB-GENE-040625-53] | 0 | YES | YES | NA | NA | |||||
| ENSDARG00000053853 | slc13a2 | solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 2 [Source:ZFIN;Acc:ZDB-GENE-040426-2389] | 406537 | YES | YES | Plasma Membrane | transporter | |||||
| ENSDARG00000006356 | slc18a3a | solute carrier family 18 (vesicular acetylcholine transporter), member 3a [Source:ZFIN;Acc:ZDB-GENE-060929-990] | 559347 | YES | YES | Plasma Membrane | transporter | |||||
| ENSDARG00000059166 | SLC22A13 | solute carrier family 22 (organic anion/urate transporter), member 13 [Source:HGNC Symbol;Acc:HGNC:8494] | 556601 | YES | YES | Plasma Membrane | transporter | |||||
| ENSDARG00000056643 | slc22a7b.1 | solute carrier family 22 (organic anion transporter), member 7b, tandem duplicate 1 [Source:ZFIN;Acc:ZDB-GENE-040426-1311] | 393320 | YES | YES | Plasma Membrane | transporter | |||||
| ENSDARG00000063158 | SLC24A2 (2 of 2) | solute carrier family 24 (sodium/potassium/calcium exchanger), member 2 [Source:HGNC Symbol;Acc:HGNC:10976] | 0 | YES | YES | Plasma Membrane | transporter | |||||
| ENSDARG00000015425 | slc24a4a | solute carrier family 24 (sodium/potassium/calcium exchanger), member 4a [Source:ZFIN;Acc:ZDB-GENE-050417-291] | 550467 | YES | YES | Plasma Membrane | transporter | |||||
| ENSDARG00000058208 | slc25a26 | solute carrier family 25 (S-adenosylmethionine carrier), member 26 [Source:ZFIN;Acc:ZDB-GENE-050913-126] | 560478 | YES | YES | Cytoplasm | transporter | |||||
| ENSDARG00000026835 | slc25a32b | solute carrier family 25 (mitochondrial folate carrier), member 32b [Source:ZFIN;Acc:ZDB-GENE-050306-39] | 503758 | YES | YES | Cytoplasm | transporter | |||||
| ENSDARG00000046054 | SLC27A3 | solute carrier family 27 (fatty acid transporter), member 3 [Source:HGNC Symbol;Acc:HGNC:10997] | 568883 | YES | YES | Cytoplasm | transporter | |||||
| ENSDARG00000101289 | slc29a1a | solute carrier family 29 (equilibrative nucleoside transporter), member 1a [Source:ZFIN;Acc:ZDB-GENE-050913-138] | 563580 | YES | YES | Plasma Membrane | transporter | |||||
| ENSDARG00000016745 | slc35f6 | solute carrier family 35, member F6 [Source:ZFIN;Acc:ZDB-GENE-040718-297] | 436832 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000102387 | SLC39A14 | solute carrier family 39 (zinc transporter), member 14 [Source:HGNC Symbol;Acc:HGNC:20858] | 799782 | YES | YES | Plasma Membrane | transporter | |||||
| ENSDARG00000000730 | slc6a13 | solute carrier family 6 (neurotransmitter transporter), member 13 [Source:ZFIN;Acc:ZDB-GENE-030616-629] | 368923 | YES | YES | NA | NA | |||||
| ENSDARG00000016141 | slc6a2 | solute carrier family 6 (neurotransmitter transporter), member 2 [Source:ZFIN;Acc:ZDB-GENE-110408-4] | 565776 | YES | YES | Plasma Membrane | transporter | |||||
| ENSDARG00000008100 | slc7a10a | solute carrier family 7 (neutral amino acid transporter light chain, asc system), member 10a [Source:ZFIN;Acc:ZDB-GENE-080116-1] | 567420 | YES | YES | Plasma Membrane | transporter | |||||
| ENSDARG00000067784 | SLC9A1 | solute carrier family 9, subfamily A (NHE1, cation proton antiporter 1), member 1 [Source:HGNC Symbol;Acc:HGNC:11071] | 0 | YES | YES | Plasma Membrane | ion channel | |||||
| ENSDARG00000075382 | slc9a6b | solute carrier family 9, subfamily A (NHE6, cation proton antiporter 6), member 6b [Source:ZFIN;Acc:ZDB-GENE-080226-2] | 556638 | YES | YES | Plasma Membrane | transporter | |||||
| ENSDARG00000022659 | snx4 | sorting nexin 4 [Source:ZFIN;Acc:ZDB-GENE-030131-1923] | 323203 | YES | YES | Cytoplasm | transporter | |||||
| ENSDARG00000059298 | spcs1 | signal peptidase complex subunit 1 homolog (S. cerevisiae) [Source:ZFIN;Acc:ZDB-GENE-050809-130] | 606666 | YES | YES | Cytoplasm | peptidase | |||||
| ENSDARG00000044514 | st6gal1 | ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 [Source:ZFIN;Acc:ZDB-GENE-060322-3] | 445376 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000045421 | stard3nl | STARD3 N-terminal like [Source:ZFIN;Acc:ZDB-GENE-040718-4] | 436592 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000055901 | steap4 | STEAP family member 4 [Source:ZFIN;Acc:ZDB-GENE-070424-244] | 550429 | YES | YES | Plasma Membrane | enzyme | |||||
| ENSDARG00000011312 | stk3 | serine/threonine kinase 3 (STE20 homolog, yeast) [Source:ZFIN;Acc:ZDB-GENE-030131-2845] | 324125 | YES | YES | Cytoplasm | kinase | |||||
| ENSDARG00000074084 | strip1 | striatin interacting protein 1 [Source:ZFIN;Acc:ZDB-GENE-040426-2927] | 568805 | YES | YES | Nucleus | other | |||||
| ENSDARG00000104953 | stt3a | STT3A, subunit of the oligosaccharyltransferase complex (catalytic) [Source:ZFIN;Acc:ZDB-GENE-021015-3] | 266797 | YES | YES | Plasma Membrane | enzyme | |||||
| ENSDARG00000053446 | sub1a | SUB1 homolog a (S. cerevisiae) [Source:ZFIN;Acc:ZDB-GENE-050522-151] | 553561 | YES | YES | Nucleus | transcription regulator | |||||
| ENSDARG00000028275 | sult1st1 | sulfotransferase family 1, cytosolic sulfotransferase 1 [Source:ZFIN;Acc:ZDB-GENE-030131-2144] | 323424 | YES | YES | NA | NA | |||||
| ENSDARG00000060711 | sv2bb | synaptic vesicle glycoprotein 2Bb [Source:ZFIN;Acc:ZDB-GENE-030131-2789] | 100006427 | YES | YES | Plasma Membrane | transporter | |||||
| ENSDARG00000059997 | sv2c | synaptic vesicle glycoprotein 2C [Source:ZFIN;Acc:ZDB-GENE-060526-233] | 562345 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000017842 | syvn1 | synovial apoptosis inhibitor 1, synoviolin [Source:ZFIN;Acc:ZDB-GENE-030131-7166] | 335226 | YES | YES | Cytoplasm | transporter | |||||
| ENSDARG00000068404 | tbcb | tubulin folding cofactor B [Source:ZFIN;Acc:ZDB-GENE-030131-9296] | 337350 | YES | YES | Other | other | |||||
| ENSDARG00000002249 | tbxas1 | thromboxane A synthase 1 (platelet) [Source:ZFIN;Acc:ZDB-GENE-030131-8805] | 792041 | YES | YES | Plasma Membrane | enzyme | |||||
| ENSDARG00000012927 | tcea2 | transcription elongation factor A (SII), 2 [Source:ZFIN;Acc:ZDB-GENE-040426-985] | 393961 | YES | YES | Nucleus | transcription regulator | |||||
| ENSDARG00000092154 | tex264a | testis expressed 264a [Source:ZFIN;Acc:ZDB-GENE-040426-1885] | 402960 | YES | YES | Extracellular Space | other | |||||
| ENSDARG00000092470 | THBD | thrombomodulin [Source:HGNC Symbol;Acc:HGNC:11784] | 0 | YES | YES | Plasma Membrane | transmembrane receptor | |||||
| ENSDARG00000061479 | thsd7aa | thrombospondin, type I, domain containing 7Aa [Source:ZFIN;Acc:ZDB-GENE-060503-709] | 557991 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000060124 | timm17b | translocase of inner mitochondrial membrane 17 homolog B (yeast) [Source:ZFIN;Acc:ZDB-GENE-081104-144] | 562144 | YES | YES | Cytoplasm | transporter | |||||
| ENSDARG00000002536 | tm9sf4 | transmembrane 9 superfamily protein member 4 [Source:ZFIN;Acc:ZDB-GENE-040426-1575] | 393482 | YES | YES | Cytoplasm | transporter | |||||
| ENSDARG00000035949 | tmem106bb | transmembrane protein 106Bb [Source:ZFIN;Acc:ZDB-GENE-050913-37] | 566699 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000037484 | tmem192 | transmembrane protein 192 [Source:ZFIN;Acc:ZDB-GENE-040426-2254] | 797335 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000015757 | tmem50a | transmembrane protein 50A [Source:ZFIN;Acc:ZDB-GENE-040426-2937] | 406850 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000038615 | tmem55ba | transmembrane protein 55Ba [Source:ZFIN;Acc:ZDB-GENE-051113-312] | 556136 | YES | YES | Cytoplasm | phosphatase | |||||
| ENSDARG00000062846 | tmtc3 | transmembrane and tetratricopeptide repeat containing 3 [Source:ZFIN;Acc:ZDB-GENE-061221-2] | 100150181 | YES | YES | Other | other | |||||
| ENSDARG00000067717 | tomm7 | translocase of outer mitochondrial membrane 7 homolog (yeast) [Source:ZFIN;Acc:ZDB-GENE-060825-115] | 751649 | YES | YES | Cytoplasm | transporter | |||||
| ENSDARG00000054858 | tp53bp2b | tumor protein p53 binding protein, 2b [Source:ZFIN;Acc:ZDB-GENE-050208-453] | 568138 | YES | YES | Nucleus | other | |||||
| ENSDARG00000040988 | tpi1b | triosephosphate isomerase 1b [Source:ZFIN;Acc:ZDB-GENE-020416-4] | 560753 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000014941 | tprkb | Tp53rk binding protein [Source:ZFIN;Acc:ZDB-GENE-041114-68] | 492501 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000010445 | trabd | TraB domain containing [Source:ZFIN;Acc:ZDB-GENE-030131-1301] | 322581 | YES | YES | Extracellular Space | other | |||||
| ENSDARG00000025299 | tspan9a | tetraspanin 9a [Source:ZFIN;Acc:ZDB-GENE-060503-632] | 555341 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000045706 | tspan9b | tetraspanin 9b [Source:ZFIN;Acc:ZDB-GENE-040704-25] | 431733 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000039666 | tsta3 | tissue specific transplantation antigen P35B [Source:ZFIN;Acc:ZDB-GENE-040722-1] | 494077 | YES | YES | Plasma Membrane | enzyme | |||||
| ENSDARG00000007025 | ttc9c | tetratricopeptide repeat domain 9C [Source:ZFIN;Acc:ZDB-GENE-040426-1053] | 393235 | YES | YES | Other | other | |||||
| ENSDARG00000000563 | ttnb | titin b [Source:ZFIN;Acc:ZDB-GENE-030616-413] | 0 | YES | YES | Cytoplasm | kinase | |||||
| ENSDARG00000100374 | txnrd1 | thioredoxin reductase 1 [Source:ZFIN;Acc:ZDB-GENE-030327-2] | 352924 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000037017 | ube4b | ubiquitination factor E4B, UFD2 homolog (S. cerevisiae) [Source:ZFIN;Acc:ZDB-GENE-020205-1] | 170784 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000021556 | ubl3b | ubiquitin-like 3b [Source:ZFIN;Acc:ZDB-GENE-040630-4] | 405782 | YES | YES | NA | NA | |||||
| ENSDARG00000023151 | ucp1 | uncoupling protein 1 [Source:ZFIN;Acc:ZDB-GENE-010503-1] | 83908 | YES | YES | Cytoplasm | transporter | |||||
| ENSDARG00000043857 | ufm1 | ubiquitin-fold modifier 1 [Source:ZFIN;Acc:ZDB-GENE-040426-2930] | 322756 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000032862 | ugt5g1 | UDP glucuronosyltransferase 5 family, polypeptide G1 [Source:ZFIN;Acc:ZDB-GENE-080305-10] | 570913 | YES | YES | NA | NA | |||||
| ENSDARG00000004055 | uhrf1bp1l | UHRF1 binding protein 1-like [Source:ZFIN;Acc:ZDB-GENE-041210-177] | 562208 | YES | YES | Other | other | |||||
| ENSDARG00000011272 | uqcc2 | ubiquinol-cytochrome c reductase complex assembly factor 2 [Source:ZFIN;Acc:ZDB-GENE-040426-1726] | 393731 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000007250 | use1 | unconventional SNARE in the ER 1 homolog (S. cerevisiae) [Source:ZFIN;Acc:ZDB-GENE-040426-1015] | 393294 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000063719 | usp8 | ubiquitin specific peptidase 8 [Source:ZFIN;Acc:ZDB-GENE-030131-1949] | 565434 | YES | YES | Cytoplasm | peptidase | |||||
| ENSDARG00000056102 | uxs1 | UDP-glucuronate decarboxylase 1 [Source:ZFIN;Acc:ZDB-GENE-020419-37] | 192315 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000030241 | vbp1 | von Hippel-Lindau binding protein 1 [Source:ZFIN;Acc:ZDB-GENE-050522-494] | 553651 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000093945 | VMA21 | VMA21 vacuolar H+-ATPase homolog (S. cerevisiae) [Source:HGNC Symbol;Acc:HGNC:22082] | 100307085 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000078593 | vwa8 | von Willebrand factor A domain containing 8 [Source:ZFIN;Acc:ZDB-GENE-070912-407] | 561317 | YES | YES | Other | other | |||||
| ENSDARG00000037406 | wdr19 | WD repeat domain 19 [Source:ZFIN;Acc:ZDB-GENE-090313-277] | 556414 | YES | YES | Extracellular Space | other | |||||
| ENSDARG00000099757 | wdr47b | WD repeat domain 47b [Source:ZFIN;Acc:ZDB-GENE-121114-10] | 567031 | YES | YES | Other | other | |||||
| ENSDARG00000074271 | wrb | tryptophan rich basic protein [Source:ZFIN;Acc:ZDB-GENE-030131-7696] | 335756 | YES | YES | Extracellular Space | other | |||||
| ENSDARG00000094133 | wu:fc21g02 | wu:fc21g02 [Source:ZFIN;Acc:ZDB-GENE-030131-2925] | 324205 | YES | YES | NA | NA | |||||
| ENSDARG00000058178 | zdhhc20b | zinc finger, DHHC-type containing 20b [Source:ZFIN;Acc:ZDB-GENE-091117-30] | 100001188 | YES | YES | Plasma Membrane | enzyme | |||||
| ENSDARG00000042872 | zdhhc8a | zinc finger, DHHC-type containing 8a [Source:ZFIN;Acc:ZDB-GENE-091118-26] | 555918 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000027887 | zfyve21 | zinc finger, FYVE domain containing 21 [Source:ZFIN;Acc:ZDB-GENE-030131-1296] | 100004144 | YES | YES | Other | other | |||||
| ENSDARG00000040965 | zgc:101731 | zgc:101731 [Source:ZFIN;Acc:ZDB-GENE-040912-57] | 447892 | YES | YES | NA | NA | |||||
| ENSDARG00000046030 | zgc:110339 | zgc:110339 [Source:ZFIN;Acc:ZDB-GENE-050417-323] | 550490 | YES | YES | NA | NA | |||||
| ENSDARG00000029473 | zgc:173994 | zgc:173994 [Source:ZFIN;Acc:ZDB-GENE-080218-30] | 571365 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000095464 | zgc:66350 | zgc:66350 [Source:ZFIN;Acc:ZDB-GENE-040426-1617] | 393493 | YES | YES | NA | NA | |||||
| ENSDARG00000044090 | zmpste24 | zinc metallopeptidase, STE24 homolog [Source:ZFIN;Acc:ZDB-GENE-030131-6312] | 334380 | YES | YES | Nucleus | peptidase | |||||
| ENSDARG00000018738 | zw10 | zw10 kinetochore protein [Source:ZFIN;Acc:ZDB-GENE-040426-800] | 393110 | YES | YES | Nucleus | other | |||||
| ENSDARG00000052263 | novel gene | 0 | 566914 | YES | YES | NA | NA | |||||
| ENSDARG00000067794 | novel gene | 0 | 0 | YES | YES | NA | NA | |||||
| ENSDARG00000067824 | novel gene | 0 | 564195 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000079483 | novel gene | 0 | 100537790 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000056314 | a2ml | alpha-2-macroglobulin-like [Source:ZFIN;Acc:ZDB-GENE-090212-1] | 100006972 | YES | YES | Extracellular Space | other | |||||
| ENSDARG00000051816 | aass | aminoadipate-semialdehyde synthase [Source:ZFIN;Acc:ZDB-GENE-061220-8] | 556229 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000006031 | abat | 4-aminobutyrate aminotransferase [Source:ZFIN;Acc:ZDB-GENE-031006-4] | 378968 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000013500 | abca2 | ATP-binding cassette, sub-family A (ABC1), member 2 [Source:ZFIN;Acc:ZDB-GENE-050517-1] | 100006090 | YES | YES | Plasma Membrane | transporter | |||||
| ENSDARG00000010936 | abcb4 | ATP-binding cassette, sub-family B (MDR/TAP), member 4 [Source:ZFIN;Acc:ZDB-GENE-080204-52] | 100136865 | YES | YES | Plasma Membrane | transporter | |||||
| ENSDARG00000063297 | abcb6a | ATP-binding cassette, sub-family B (MDR/TAP), member 6a [Source:ZFIN;Acc:ZDB-GENE-050517-9] | 564067 | YES | YES | Cytoplasm | transporter | |||||
| ENSDARG00000062795 | abcb7 | ATP-binding cassette, sub-family B (MDR/TAP), member 7 [Source:ZFIN;Acc:ZDB-GENE-050517-10] | 566515 | YES | YES | Cytoplasm | transporter | |||||
| ENSDARG00000056672 | abcb8 | ATP-binding cassette, sub-family B (MDR/TAP), member 8 [Source:ZFIN;Acc:ZDB-GENE-050410-6] | 548340 | YES | YES | Cytoplasm | transporter | |||||
| ENSDARG00000014031 | abcc2 | ATP-binding cassette, sub-family C (CFTR/MRP), member 2 [Source:ZFIN;Acc:ZDB-GENE-040426-1523] | 393561 | YES | YES | Other | other | |||||
| ENSDARG00000051879 | abcc8 | ATP-binding cassette, sub-family C (CFTR/MRP), member 8 [Source:ZFIN;Acc:ZDB-GENE-050517-22] | 553281 | YES | YES | Plasma Membrane | transporter | |||||
| ENSDARG00000100531 | abhd14b | abhydrolase domain containing 14B [Source:ZFIN;Acc:ZDB-GENE-040426-1342] | 393338 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000091433 | abhd17aa | abhydrolase domain containing 17Aa [Source:ZFIN;Acc:ZDB-GENE-030131-840] | 322121 | YES | YES | Other | enzyme | |||||
| ENSDARG00000042658 | acad8 | acyl-CoA dehydrogenase family, member 8 [Source:ZFIN;Acc:ZDB-GENE-040426-828] | 394130 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000030781 | acads | acyl-CoA dehydrogenase, C-2 to C-3 short chain [Source:ZFIN;Acc:ZDB-GENE-040808-64] | 445288 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000007127 | acat2 | acetyl-CoA acetyltransferase 2 [Source:ZFIN;Acc:ZDB-GENE-990714-22] | 30643 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000094730 | acbd7 | acyl-CoA binding domain containing 7 [Source:ZFIN;Acc:ZDB-GENE-050913-108] | 619256 | YES | YES | Other | other | |||||
| ENSDARG00000079166 | ace | angiotensin I converting enzyme (peptidyl-dipeptidase A) 1 [Source:ZFIN;Acc:ZDB-GENE-030131-1826] | 565980 | YES | YES | Plasma Membrane | peptidase | |||||
| ENSDARG00000026376 | aco1 | aconitase 1, soluble [Source:ZFIN;Acc:ZDB-GENE-031118-76] | 568448 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000061728 | acot13 | acyl-CoA thioesterase 13 [Source:ZFIN;Acc:ZDB-GENE-060503-437] | 564010 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000038865 | acox3 | acyl-CoA oxidase 3, pristanoyl [Source:ZFIN;Acc:ZDB-GENE-040426-2163] | 406421 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000007244 | ACP2 | acid phosphatase 2, lysosomal [Source:HGNC Symbol;Acc:HGNC:123] | 0 | YES | YES | Cytoplasm | phosphatase | |||||
| ENSDARG00000040064 | acp6 | acid phosphatase 6, lysophosphatidic [Source:ZFIN;Acc:ZDB-GENE-050208-290] | 558758 | YES | YES | Cytoplasm | phosphatase | |||||
| ENSDARG00000030514 | acsl1a | acyl-CoA synthetase long-chain family member 1a [Source:ZFIN;Acc:ZDB-GENE-050809-115] | 555308 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000003854 | acsl1b | acyl-CoA synthetase long-chain family member 1b [Source:ZFIN;Acc:ZDB-GENE-040801-88] | 445175 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000004078 | acsl4a | acyl-CoA synthetase long-chain family member 4a [Source:ZFIN;Acc:ZDB-GENE-040426-1565] | 393622 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000044142 | acss1 | acyl-CoA synthetase short-chain family member 1 [Source:ZFIN;Acc:ZDB-GENE-050320-139] | 541435 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000060126 | acyp1 | acylphosphatase 1, erythrocyte (common) type [Source:ZFIN;Acc:ZDB-GENE-050309-125] | 0 | YES | YES | NA | NA | |||||
| ENSDARG00000053468 | adam10a | ADAM metallopeptidase domain 10a [Source:ZFIN;Acc:ZDB-GENE-040917-2] | 566044 | YES | YES | NA | NA | |||||
| ENSDARG00000073923 | adck2 | aarF domain containing kinase 2 [Source:ZFIN;Acc:ZDB-GENE-131121-369] | 572087 | YES | YES | NA | NA | |||||
| ENSDARG00000020123 | adck3 | aarF domain containing kinase 3 [Source:ZFIN;Acc:ZDB-GENE-040718-487] | 437001 | YES | YES | Cytoplasm | kinase | |||||
| ENSDARG00000101130 | adck4 | aarF domain containing kinase 4 [Source:ZFIN;Acc:ZDB-GENE-060503-803] | 799071 | YES | YES | Cytoplasm | kinase | |||||
| ENSDARG00000100869 | adcy9 | adenylate cyclase 9 [Source:ZFIN;Acc:ZDB-GENE-050208-455] | 100333702 | YES | YES | Plasma Membrane | enzyme | |||||
| ENSDARG00000103847 | ADCY9 (2 of 2) | adenylate cyclase 9 [Source:HGNC Symbol;Acc:HGNC:240] | 0 | YES | YES | Plasma Membrane | enzyme | |||||
| ENSDARG00000105201 | adcyap1r1a | adenylate cyclase activating polypeptide 1a (pituitary) receptor type I [Source:ZFIN;Acc:ZDB-GENE-050315-1] | 0 | YES | YES | Plasma Membrane | G-protein coupled receptor | |||||
| ENSDARG00000080010 | adh5 | alcohol dehydrogenase 5 [Source:ZFIN;Acc:ZDB-GENE-011003-1] | 116517 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000063525 | adpgk | ADP-dependent glucokinase [Source:ZFIN;Acc:ZDB-GENE-061103-421] | 557919 | YES | YES | Plasma Membrane | kinase | |||||
| ENSDARG00000017049 | adsl | adenylosuccinate lyase [Source:ZFIN;Acc:ZDB-GENE-030131-6363] | 334431 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000009837 | agk | acylglycerol kinase [Source:ZFIN;Acc:ZDB-GENE-030131-6202] | 334270 | YES | YES | Cytoplasm | kinase | |||||
| ENSDARG00000059888 | AGO3 (1 of 2) | argonaute RISC catalytic component 3 [Source:HGNC Symbol;Acc:HGNC:18421] | 567622 | YES | YES | Cytoplasm | translation regulator | |||||
| ENSDARG00000005191 | ahcy | adenosylhomocysteinase [Source:ZFIN;Acc:ZDB-GENE-031219-6] | 387530 | YES | YES | Other | other | |||||
| ENSDARG00000056331 | ahcyl1 | adenosylhomocysteinase-like 1 [Source:ZFIN;Acc:ZDB-GENE-021206-7] | 798918 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000039343 | ahcyl2 | adenosylhomocysteinase-like 2 [Source:ZFIN;Acc:ZDB-GENE-040115-5] | 394240 | YES | YES | Other | enzyme | |||||
| ENSDARG00000069293 | ahsg2 | alpha-2-HS-glycoprotein 2 [Source:ZFIN;Acc:ZDB-GENE-030131-1133] | 567406 | YES | YES | Extracellular Space | other | |||||
| ENSDARG00000014532 | aida | axin interactor, dorsalization associated [Source:ZFIN;Acc:ZDB-GENE-071126-2] | 393763 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000005926 | ak2 | adenylate kinase 2 [Source:ZFIN;Acc:ZDB-GENE-030131-512] | 321793 | YES | YES | Cytoplasm | kinase | |||||
| ENSDARG00000043843 | akap7 | A kinase (PRKA) anchor protein 7 [Source:ZFIN;Acc:ZDB-GENE-040724-128] | 0 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000100372 | alad | aminolevulinate dehydratase [Source:ZFIN;Acc:ZDB-GENE-050417-123] | 550338 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000053493 | aldh1a2 | aldehyde dehydrogenase 1 family, member A2 [Source:ZFIN;Acc:ZDB-GENE-011010-3] | 116713 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000077004 | aldh1l1 | aldehyde dehydrogenase 1 family, member L1 [Source:ZFIN;Acc:ZDB-GENE-100519-4] | 100526683 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000070230 | aldh1l2 | aldehyde dehydrogenase 1 family, member L2 [Source:ZFIN;Acc:ZDB-GENE-100426-6] | 100333269 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000076544 | aldh5a1 | aldehyde dehydrogenase 5 family, member A1 (succinate-semialdehyde dehydrogenase) [Source:ZFIN;Acc:ZDB-GENE-070228-2] | 565235 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000018426 | aldh7a1 | aldehyde dehydrogenase 7 family, member A1 [Source:ZFIN;Acc:ZDB-GENE-030131-6129] | 334197 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000069100 | aldh9a1a.1 | aldehyde dehydrogenase 9 family, member A1a, tandem duplicate 1 [Source:ZFIN;Acc:ZDB-GENE-030131-1257] | 100005587 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000029952 | ampd2 | adenosine monophosphate deaminase 2 (isoform L) [Source:ZFIN;Acc:ZDB-GENE-070713-5] | 571355 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000092143 | ank1a | ankyrin 1, erythrocytic a [Source:ZFIN;Acc:ZDB-GENE-041010-44] | 449796 | YES | YES | NA | NA | |||||
| ENSDARG00000098140 | ankrd13d | ankyrin repeat domain 13 family, member D [Source:ZFIN;Acc:ZDB-GENE-101007-3] | 562931 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000102335 | ano1 | anoctamin 1, calcium activated chloride channel [Source:ZFIN;Acc:ZDB-GENE-121127-3] | 407698 | YES | YES | Plasma Membrane | ion channel | |||||
| ENSDARG00000013976 | anxa13 | annexin A13 [Source:ZFIN;Acc:ZDB-GENE-010406-1] | 81880 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000013613 | anxa13l | annexin A13, like [Source:ZFIN;Acc:ZDB-GENE-050522-310] | 554118 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000026726 | anxa1a | annexin A1a [Source:ZFIN;Acc:ZDB-GENE-030131-6664] | 334724 | YES | YES | Plasma Membrane | enzyme | |||||
| ENSDARG00000044254 | anxa3b | annexin A3b [Source:ZFIN;Acc:ZDB-GENE-041114-128] | 492336 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000036456 | anxa4 | annexin A4 [Source:ZFIN;Acc:ZDB-GENE-030707-1] | 353362 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000016470 | anxa5b | annexin A5b [Source:ZFIN;Acc:ZDB-GENE-030131-9076] | 337132 | YES | YES | Plasma Membrane | transporter | |||||
| ENSDARG00000013335 | anxa6 | annexin A6 [Source:ZFIN;Acc:ZDB-GENE-030707-3] | 0 | YES | YES | Plasma Membrane | ion channel | |||||
| ENSDARG00000014646 | aoc2 | amine oxidase, copper containing 2 [Source:ZFIN;Acc:ZDB-GENE-050320-133] | 541429 | YES | YES | Plasma Membrane | enzyme | |||||
| ENSDARG00000020054 | aox1 | aldehyde oxidase 1 [Source:ZFIN;Acc:ZDB-GENE-050208-742] | 570457 | YES | YES | NA | NA | |||||
| ENSDARG00000074328 | apba1b | amyloid beta (A4) precursor protein-binding, family A, member 1b [Source:ZFIN;Acc:ZDB-GENE-070925-3] | 565507 | YES | YES | Cytoplasm | transporter | |||||
| ENSDARG00000045843 | apex1 | APEX nuclease (multifunctional DNA repair enzyme) 1 [Source:ZFIN;Acc:ZDB-GENE-040426-2761] | 406730 | YES | YES | Nucleus | enzyme | |||||
| ENSDARG00000040298 | apoa4b.1 | apolipoprotein A-IV b, tandem duplicate 1 [Source:ZFIN;Acc:ZDB-GENE-030131-1263] | 322543 | YES | YES | Extracellular Space | transporter | |||||
| ENSDARG00000092170 | apoc1l | apolipoprotein C-I like [Source:ZFIN;Acc:ZDB-GENE-030131-1074] | 570638 | YES | YES | NA | NA | |||||
| ENSDARG00000040295 | apoeb | apolipoprotein Eb [Source:ZFIN;Acc:ZDB-GENE-980526-368] | 30314 | YES | YES | Extracellular Space | transporter | |||||
| ENSDARG00000104696 | apooa | apolipoprotein O, a [Source:ZFIN;Acc:ZDB-GENE-031118-91] | 641319 | YES | YES | Extracellular Space | other | |||||
| ENSDARG00000026444 | apoob | apolipoprotein O, b [Source:ZFIN;Acc:ZDB-GENE-041001-40] | 449837 | YES | YES | Extracellular Space | other | |||||
| ENSDARG00000104279 | appa | amyloid beta (A4) precursor protein a [Source:ZFIN;Acc:ZDB-GENE-000616-13] | 58083 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000055543 | appb | amyloid beta (A4) precursor protein b [Source:ZFIN;Acc:ZDB-GENE-020220-1] | 170846 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000056664 | arfip2b | ADP-ribosylation factor interacting protein 2b [Source:ZFIN;Acc:ZDB-GENE-050417-131] | 550342 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000057857 | arfrp1 | ADP-ribosylation factor related protein 1 [Source:ZFIN;Acc:ZDB-GENE-050227-14] | 503602 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000062577 | arhgap35a | Rho GTPase activating protein 35a [Source:ZFIN;Acc:ZDB-GENE-120516-2] | 100004299 | YES | YES | Nucleus | transcription regulator | |||||
| ENSDARG00000043795 | arhgdia | Rho GDP dissociation inhibitor (GDI) alpha [Source:ZFIN;Acc:ZDB-GENE-040426-2816] | 406770 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000071409 | arl3 | ADP-ribosylation factor-like 3 [Source:ZFIN;Acc:ZDB-GENE-030131-7518] | 797451 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000016238 | arl6ip5b | ADP-ribosylation factor-like 6 interacting protein 5b [Source:ZFIN;Acc:ZDB-GENE-040718-223] | 436790 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000071881 | arl9 | ADP-ribosylation factor-like 9 [Source:ZFIN;Acc:ZDB-GENE-060825-210] | 751670 | YES | YES | Other | other | |||||
| ENSDARG00000013861 | armc1 | armadillo repeat containing 1 [Source:ZFIN;Acc:ZDB-GENE-041212-33] | 494066 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000043241 | arrb1 | arrestin, beta 1 [Source:ZFIN;Acc:ZDB-GENE-060824-1] | 553266 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000018190 | asna1 | arsA arsenite transporter, ATP-binding, homolog 1 (bacterial) [Source:ZFIN;Acc:ZDB-GENE-040625-120] | 325704 | YES | YES | Nucleus | transporter | |||||
| ENSDARG00000021681 | asrgl1 | asparaginase like 1 [Source:ZFIN;Acc:ZDB-GENE-050320-102] | 541402 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000009350 | atg3 | autophagy related 3 [Source:ZFIN;Acc:ZDB-GENE-030131-8576] | 336632 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000016706 | atic | 5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase/IMP cyclohydrolase [Source:ZFIN;Acc:ZDB-GENE-011212-4] | 140622 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000001870 | atp1a1a.4 | ATPase, Na+/K+ transporting, alpha 1a polypeptide, tandem duplicate 4 [Source:ZFIN;Acc:ZDB-GENE-001212-4] | 64615 | YES | YES | Plasma Membrane | transporter | |||||
| ENSDARG00000013144 | atp1b1a | ATPase, Na+/K+ transporting, beta 1a polypeptide [Source:ZFIN;Acc:ZDB-GENE-001127-3] | 64267 | YES | YES | Plasma Membrane | transporter | |||||
| ENSDARG00000034424 | atp1b2b | ATPase, Na+/K+ transporting, beta 2b polypeptide [Source:ZFIN;Acc:ZDB-GENE-010718-1] | 114457 | YES | YES | Plasma Membrane | transporter | |||||
| ENSDARG00000015790 | atp1b3a | ATPase, Na+/K+ transporting, beta 3a polypeptide [Source:ZFIN;Acc:ZDB-GENE-990415-167] | 30468 | YES | YES | Plasma Membrane | transporter | |||||
| ENSDARG00000042837 | atp1b3b | ATPase, Na+/K+ transporting, beta 3b polypeptide [Source:ZFIN;Acc:ZDB-GENE-001127-1] | 64272 | YES | YES | Plasma Membrane | transporter | |||||
| ENSDARG00000035458 | atp2a1l | ATPase, Ca++ transporting, cardiac muscle, fast twitch 1 like [Source:ZFIN;Acc:ZDB-GENE-041229-2] | 494489 | YES | YES | Cytoplasm | transporter | |||||
| ENSDARG00000060978 | atp2a3 | ATPase, Ca++ transporting, ubiquitous [Source:ZFIN;Acc:ZDB-GENE-060531-103] | 568671 | YES | YES | Cytoplasm | transporter | |||||
| ENSDARG00000077967 | ATP5J (2 of 3) | ATP synthase, H+ transporting, mitochondrial Fo complex, subunit F6 [Source:HGNC Symbol;Acc:HGNC:847] | 570370 | YES | YES | Cytoplasm | transporter | |||||
| ENSDARG00000012072 | atp5s | ATP synthase, H+ transporting, mitochondrial F0 complex, subunit s [Source:ZFIN;Acc:ZDB-GENE-040426-959] | 0 | YES | YES | Cytoplasm | transporter | |||||
| ENSDARG00000063001 | atp8a1 | ATPase, aminophospholipid transporter (APLT), class I, type 8A, member 1 [Source:ZFIN;Acc:ZDB-GENE-060929-36] | 0 | YES | YES | NA | NA | |||||
| ENSDARG00000038414 | b3galt6 | UDP-Gal:betaGal beta 1,3-galactosyltransferase polypeptide 6 [Source:ZFIN;Acc:ZDB-GENE-101104-13] | 572324 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000016231 | bcap29 | B-cell receptor-associated protein 29 [Source:ZFIN;Acc:ZDB-GENE-050522-468] | 554151 | YES | YES | Cytoplasm | transporter | |||||
| ENSDARG00000045568 | bcat1 | branched chain amino-acid transaminase 1, cytosolic [Source:ZFIN;Acc:ZDB-GENE-030131-9358] | 337412 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000054849 | bcat2 | branched chain amino-acid transaminase 2, mitochondrial [Source:ZFIN;Acc:ZDB-GENE-040718-425] | 436949 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000008434 | bcl2l1 | bcl2-like 1 [Source:ZFIN;Acc:ZDB-GENE-010730-1] | 114401 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000062370 | bcl2l13 | BCL2-like 13 (apoptosis facilitator) [Source:ZFIN;Acc:ZDB-GENE-050419-215] | 559355 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000034885 | bet1 | Bet1 golgi vesicular membrane trafficking protein [Source:ZFIN;Acc:ZDB-GENE-040625-3] | 415135 | YES | YES | Cytoplasm | transporter | |||||
| ENSDARG00000013430 | bhmt | betaine-homocysteine methyltransferase [Source:ZFIN;Acc:ZDB-GENE-030131-947] | 322228 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000059857 | blvra | biliverdin reductase A [Source:ZFIN;Acc:ZDB-GENE-060929-312] | 767661 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000096829 | blvrb | biliverdin reductase B [Source:ZFIN;Acc:ZDB-GENE-030131-1516] | 436959 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000020057 | bmpr2b | bone morphogenetic protein receptor, type II b (serine/threonine kinase) [Source:ZFIN;Acc:ZDB-GENE-070618-2] | 555655 | YES | YES | Plasma Membrane | kinase | |||||
| ENSDARG00000102877 | bphl | biphenyl hydrolase-like (serine hydrolase) [Source:EntrezGene;Acc:402804] | 402804 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000017661 | braf | B-Raf proto-oncogene, serine/threonine kinase [Source:ZFIN;Acc:ZDB-GENE-040805-1] | 403065 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000037008 | bscl2 | Berardinelli-Seip congenital lipodystrophy 2 (seipin) [Source:ZFIN;Acc:ZDB-GENE-051113-140] | 567403 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000019881 | bsg | basigin [Source:ZFIN;Acc:ZDB-GENE-030131-9638] | 337692 | YES | YES | Plasma Membrane | transporter | |||||
| ENSDARG00000035400 | btf3 | basic transcription factor 3 [Source:ZFIN;Acc:ZDB-GENE-030131-8731] | 556625 | YES | YES | NA | NA | |||||
| ENSDARG00000089681 | btf3l4 | basic transcription factor 3-like 4 [Source:ZFIN;Acc:ZDB-GENE-040426-1650] | 393667 | YES | YES | Other | other | |||||
| ENSDARG00000069977 | C11H12orf10 | chromosome 12 open reading frame 10 [Source:HGNC Symbol;Acc:HGNC:17590] | 664752 | YES | YES | Nucleus | other | |||||
| ENSDARG00000012694 | c3a.1 | complement component c3a, duplicate 1 [Source:ZFIN;Acc:ZDB-GENE-990415-35] | 321046 | YES | YES | Extracellular Space | peptidase | |||||
| ENSDARG00000052207 | c3a.3 | complement component c3a, duplicate 3 [Source:ZFIN;Acc:ZDB-GENE-990415-37] | 30491 | YES | YES | Extracellular Space | peptidase | |||||
| ENSDARG00000037909 | C3H19orf52 | chromosome 19 open reading frame 52 [Source:HGNC Symbol;Acc:HGNC:25152] | 553583 | YES | YES | Other | other | |||||
| ENSDARG00000016319 | c9 | complement component 9 [Source:ZFIN;Acc:ZDB-GENE-050522-442] | 554141 | YES | YES | Extracellular Space | other | |||||
| ENSDARG00000069671 | C9H2orf47 | chromosome 2 open reading frame 47 [Source:HGNC Symbol;Acc:HGNC:26198] | 552941 | YES | YES | Other | other | |||||
| ENSDARG00000036658 | cab39l | calcium binding protein 39-like [Source:ZFIN;Acc:ZDB-GENE-041114-204] | 492361 | YES | YES | Cytoplasm | kinase | |||||
| ENSDARG00000099511 | CABZ01034698.4 | 0 | 0 | YES | YES | NA | NA | |||||
| ENSDARG00000074844 | CABZ01061495.1 | Uncharacterized protein [Source:UniProtKB/TrEMBL;Acc:F1R882] | 100334801 | YES | YES | NA | NA | |||||
| ENSDARG00000100177 | CABZ01066957.1 | Uncharacterized protein [Source:UniProtKB/TrEMBL;Acc:H9GZ07] | 0 | YES | YES | NA | NA | |||||
| ENSDARG00000089720 | CABZ01077218.1 | Uncharacterized protein [Source:UniProtKB/TrEMBL;Acc:E7FAI0] | 100334619 | YES | YES | NA | NA | |||||
| ENSDARG00000104106 | CABZ01087953.1 | 0 | 101883442 | YES | YES | NA | NA | |||||
| ENSDARG00000039322 | CABZ01088149.1 | Uncharacterized protein [Source:UniProtKB/TrEMBL;Acc:F1Q8T3] | 402879 | YES | YES | NA | NA | |||||
| ENSDARG00000031075 | cadm1a | cell adhesion molecule 1a [Source:ZFIN;Acc:ZDB-GENE-080505-2] | 569931 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000042677 | cadm1b | cell adhesion molecule 1b [Source:ZFIN;Acc:ZDB-GENE-080327-34] | 562183 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000009930 | cadm2a | cell adhesion molecule 2a [Source:ZFIN;Acc:ZDB-GENE-040426-1614] | 793954 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000062633 | cadm2b | cell adhesion molecule 2b [Source:ZFIN;Acc:ZDB-GENE-080506-3] | 571698 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000041062 | calb2a | calbindin 2a [Source:ZFIN;Acc:ZDB-GENE-040426-1677] | 393691 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000036344 | calb2b | calbindin 2b [Source:ZFIN;Acc:ZDB-GENE-040426-1668] | 393684 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000076290 | calr | calreticulin [Source:ZFIN;Acc:ZDB-GENE-030131-4042] | 325317 | YES | YES | Cytoplasm | transcription regulator | |||||
| ENSDARG00000103979 | calr3a | calreticulin 3a [Source:ZFIN;Acc:ZDB-GENE-000208-17] | 30248 | YES | YES | NA | NA | |||||
| ENSDARG00000102808 | calr3b | calreticulin 3b [Source:ZFIN;Acc:ZDB-GENE-030131-9907] | 321315 | YES | YES | NA | NA | |||||
| ENSDARG00000060116 | camk1a | calcium/calmodulin-dependent protein kinase Ia [Source:ZFIN;Acc:ZDB-GENE-131120-147] | 555945 | YES | YES | Cytoplasm | kinase | |||||
| ENSDARG00000078645 | CAMKK2 | calcium/calmodulin-dependent protein kinase kinase 2, beta [Source:HGNC Symbol;Acc:HGNC:1470] | 0 | YES | YES | Cytoplasm | kinase | |||||
| ENSDARG00000100409 | camlg | calcium modulating ligand [Source:ZFIN;Acc:ZDB-GENE-040426-2407] | 404622 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000037488 | canx | calnexin [Source:ZFIN;Acc:ZDB-GENE-040426-2797] | 406757 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000052702 | capn1a | calpain 1, (mu/I) large subunit a [Source:ZFIN;Acc:ZDB-GENE-040426-1263] | 393417 | YES | YES | NA | NA | |||||
| ENSDARG00000052748 | capn1b | calpain 1, (mu/I) large subunit b [Source:ZFIN;Acc:ZDB-GENE-060503-491] | 565106 | YES | YES | NA | NA | |||||
| ENSDARG00000069748 | capn5b | calpain 5b [Source:ZFIN;Acc:ZDB-GENE-091116-29] | 100006332 | YES | YES | Cytoplasm | peptidase | |||||
| ENSDARG00000035329 | capns1a | calpain, small subunit 1 a [Source:ZFIN;Acc:ZDB-GENE-030113-3] | 550598 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000013804 | capns1b | calpain, small subunit 1 b [Source:ZFIN;Acc:ZDB-GENE-030131-5206] | 560033 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000058693 | cast | calpastatin [Source:ZFIN;Acc:ZDB-GENE-050810-2] | 563084 | YES | YES | Cytoplasm | peptidase | |||||
| ENSDARG00000063376 | CBLN3 (1 of 2) | cerebellin 3 precursor [Source:HGNC Symbol;Acc:HGNC:20146] | 560359 | YES | YES | Extracellular Space | other | |||||
| ENSDARG00000036587 | cbr1 | carbonyl reductase 1 [Source:ZFIN;Acc:ZDB-GENE-030902-2] | 373866 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000021149 | cbr1l | carbonyl reductase 1-like [Source:ZFIN;Acc:ZDB-GENE-030131-9642] | 337696 | YES | YES | NA | NA | |||||
| ENSDARG00000030508 | CCDC134 (1 of 2) | coiled-coil domain containing 134 [Source:HGNC Symbol;Acc:HGNC:26185] | 0 | YES | YES | Other | other | |||||
| ENSDARG00000045351 | ccdc58 | coiled-coil domain containing 58 [Source:ZFIN;Acc:ZDB-GENE-040426-1780] | 393780 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000036080 | cd81a | CD81 molecule a [Source:ZFIN;Acc:ZDB-GENE-000831-5] | 58031 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000022437 | cd81b | CD81 molecule b [Source:ZFIN;Acc:ZDB-GENE-040808-52] | 445280 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000019098 | cd82a | CD82 molecule a [Source:ZFIN;Acc:ZDB-GENE-030131-2818] | 324098 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000051975 | cd99 | CD99 molecule [Source:ZFIN;Acc:ZDB-GENE-061103-391] | 559896 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000056722 | cd99l2 | CD99 molecule-like 2 [Source:ZFIN;Acc:ZDB-GENE-030131-1986] | 323266 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000099301 | CDC37 | cell division cycle 37 [Source:HGNC Symbol;Acc:HGNC:1735] | 0 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000035577 | cds2 | CDP-diacylglycerol synthase (phosphatidate cytidylyltransferase) 2 [Source:ZFIN;Acc:ZDB-GENE-030717-4] | 394161 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000055825 | celsr3 | cadherin, EGF LAG seven-pass G-type receptor 3 [Source:ZFIN;Acc:ZDB-GENE-070122-3] | 0 | YES | YES | Plasma Membrane | G-protein coupled receptor | |||||
| ENSDARG00000041595 | ces3 | carboxylesterase 3 [Source:ZFIN;Acc:ZDB-GENE-030131-9563] | 560651 | YES | YES | Other | other | |||||
| ENSDARG00000012972 | cfl1l | cofilin 1 (non-muscle), like [Source:ZFIN;Acc:ZDB-GENE-040426-2770] | 406738 | YES | YES | NA | NA | |||||
| ENSDARG00000100304 | chdh | choline dehydrogenase [Source:ZFIN;Acc:ZDB-GENE-120111-1] | 100331020 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000069673 | chid1 | chitinase domain containing 1 [Source:ZFIN;Acc:ZDB-GENE-030131-9169] | 337225 | YES | YES | Extracellular Space | other | |||||
| ENSDARG00000103109 | chl1b | cell adhesion molecule L1-like b [Source:ZFIN;Acc:ZDB-GENE-091105-1] | 0 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000098612 | chrm2a | cholinergic receptor, muscarinic 2a [Source:ZFIN;Acc:ZDB-GENE-030314-1] | 352938 | YES | YES | Plasma Membrane | G-protein coupled receptor | |||||
| ENSDARG00000069254 | chrm4a | cholinergic receptor, muscarinic 4a [Source:ZFIN;Acc:ZDB-GENE-090410-6] | 100150701 | YES | YES | Plasma Membrane | G-protein coupled receptor | |||||
| ENSDARG00000032405 | CKAP4 | cytoskeleton-associated protein 4 [Source:HGNC Symbol;Acc:HGNC:16991] | 406549 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000040565 | ckmb | creatine kinase, muscle b [Source:ZFIN;Acc:ZDB-GENE-040426-2128] | 794752 | YES | YES | Cytoplasm | kinase | |||||
| ENSDARG00000035808 | clcn4 | chloride channel, voltage-sensitive 4 [Source:ZFIN;Acc:ZDB-GENE-061013-353] | 768175 | YES | YES | Plasma Membrane | ion channel | |||||
| ENSDARG00000009315 | clgn | calmegin [Source:ZFIN;Acc:ZDB-GENE-060929-708] | 556495 | YES | YES | Cytoplasm | peptidase | |||||
| ENSDARG00000018973 | clptm1 | cleft lip and palate associated transmembrane protein 1 [Source:ZFIN;Acc:ZDB-GENE-040801-265] | 795480 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000031720 | clstn1 | calsyntenin 1 [Source:ZFIN;Acc:ZDB-GENE-040426-1064] | 777737 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000045618 | clta | clathrin, light chain A [Source:ZFIN;Acc:ZDB-GENE-040426-1986] | 406318 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000029729 | clybl | citrate lyase beta like [Source:ZFIN;Acc:ZDB-GENE-030516-6] | 641475 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000069583 | CNDP1 | carnosine dipeptidase 1 (metallopeptidase M20 family) [Source:HGNC Symbol;Acc:HGNC:20675] | 619255 | YES | YES | Cytoplasm | peptidase | |||||
| ENSDARG00000003931 | cndp2 | CNDP dipeptidase 2 (metallopeptidase M20 family) [Source:ZFIN;Acc:ZDB-GENE-030131-5499] | 327288 | YES | YES | Cytoplasm | peptidase | |||||
| ENSDARG00000074309 | cnnm4b | cyclin and CBS domain divalent metal cation transport mediator 4b [Source:ZFIN;Acc:ZDB-GENE-070705-181] | 100150651 | YES | YES | Plasma Membrane | transporter | |||||
| ENSDARG00000009020 | cnr1 | cannabinoid receptor 1 [Source:ZFIN;Acc:ZDB-GENE-040312-3] | 404209 | YES | YES | Plasma Membrane | G-protein coupled receptor | |||||
| ENSDARG00000058969 | cntnap2a | contactin associated protein-like 2a [Source:ZFIN;Acc:ZDB-GENE-120328-3] | 559157 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000056557 | copb1 | coatomer protein complex, subunit beta 1 [Source:ZFIN;Acc:ZDB-GENE-030219-38] | 338138 | YES | YES | Cytoplasm | transporter | |||||
| ENSDARG00000034823 | copg2 | coatomer protein complex, subunit gamma 2 [Source:ZFIN;Acc:ZDB-GENE-000208-8] | 30187 | YES | YES | Cytoplasm | transporter | |||||
| ENSDARG00000008623 | cops3 | COP9 signalosome subunit 3 [Source:ZFIN;Acc:ZDB-GENE-030131-7660] | 0 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000043732 | cops4 | COP9 constitutive photomorphogenic homolog subunit 4 (Arabidopsis) [Source:ZFIN;Acc:ZDB-GENE-030131-4317] | 325592 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000022788 | cops7a | COP9 constitutive photomorphogenic homolog subunit 7A [Source:ZFIN;Acc:ZDB-GENE-030131-8680] | 336736 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000024846 | cops8 | COP9 signalosome subunit 8 [Source:ZFIN;Acc:ZDB-GENE-040426-982] | 393198 | YES | YES | Nucleus | other | |||||
| ENSDARG00000017844 | copz1 | coatomer protein complex, subunit zeta 1 [Source:ZFIN;Acc:ZDB-GENE-000406-6] | 0 | YES | YES | Cytoplasm | transporter | |||||
| ENSDARG00000060380 | coq6 | coenzyme Q6 monooxygenase [Source:ZFIN;Acc:ZDB-GENE-060825-297] | 751691 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000062594 | coq7 | coenzyme Q7 homolog, ubiquinone (yeast) [Source:ZFIN;Acc:ZDB-GENE-070209-262] | 559504 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000023583 | coq9 | coenzyme Q9 homolog (S. cerevisiae) [Source:ZFIN;Acc:ZDB-GENE-061207-37] | 567716 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000063882 | cox19 | COX19 cytochrome c oxidase assembly factor [Source:ZFIN;Acc:ZDB-GENE-070410-29] | 100004173 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000068738 | cox5b2 | cytochrome c oxidase subunit Vb 2 [Source:ZFIN;Acc:ZDB-GENE-060825-71] | 751638 | YES | YES | Other | other | |||||
| ENSDARG00000055648 | cpda | carboxypeptidase D, a [Source:ZFIN;Acc:ZDB-GENE-081112-2] | 563008 | YES | YES | Extracellular Space | peptidase | |||||
| ENSDARG00000089486 | cplx3b | complexin 3b [Source:ZFIN;Acc:ZDB-GENE-091204-403] | 794623 | YES | YES | Nucleus | transporter | |||||
| ENSDARG00000062025 | cpox | coproporphyrinogen oxidase [Source:ZFIN;Acc:ZDB-GENE-030131-9884] | 321294 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000074235 | CR376783.1 | Uncharacterized protein [Source:UniProtKB/TrEMBL;Acc:E7F5Q2] | 797620 | YES | YES | NA | NA | |||||
| ENSDARG00000055635 | crk | v-crk avian sarcoma virus CT10 oncogene homolog [Source:ZFIN;Acc:ZDB-GENE-040801-148] | 445234 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000040352 | crot | carnitine O-octanoyltransferase [Source:ZFIN;Acc:ZDB-GENE-050522-35] | 553204 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000003675 | cryl1 | crystallin, lambda 1 [Source:ZFIN;Acc:ZDB-GENE-060810-7] | 751596 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000026902 | cryzl1 | crystallin, zeta (quinone reductase)-like 1 [Source:ZFIN;Acc:ZDB-GENE-040718-378] | 436906 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000006963 | cse1l | CSE1 chromosome segregation 1-like (yeast) [Source:ZFIN;Acc:ZDB-GENE-990603-1] | 30707 | YES | YES | Nucleus | transporter | |||||
| ENSDARG00000077920 | CSMD3 | CUB and Sushi multiple domains 3 [Source:HGNC Symbol;Acc:HGNC:19291] | 0 | YES | YES | Other | enzyme | |||||
| ENSDARG00000006125 | csnk1db | casein kinase 1, delta b [Source:ZFIN;Acc:ZDB-GENE-040426-2385] | 406533 | YES | YES | Cytoplasm | kinase | |||||
| ENSDARG00000078227 | cspg4 | chondroitin sulfate proteoglycan 4 [Source:ZFIN;Acc:ZDB-GENE-050519-1] | 560393 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000099793 | cspg5b | chondroitin sulfate proteoglycan 5b [Source:ZFIN;Acc:ZDB-GENE-060503-368] | 559358 | YES | YES | Extracellular Space | growth factor | |||||
| ENSDARG00000032206 | cthl | cystathionase (cystathionine gamma-lyase), like [Source:ZFIN;Acc:ZDB-GENE-040905-3] | 445818 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000055120 | ctsba | cathepsin Ba [Source:ZFIN;Acc:ZDB-GENE-040426-2650] | 406645 | YES | YES | Cytoplasm | peptidase | |||||
| ENSDARG00000057698 | ctsd | cathepsin D [Source:ZFIN;Acc:ZDB-GENE-010131-8] | 65225 | YES | YES | Cytoplasm | peptidase | |||||
| ENSDARG00000098589 | cyb5a | cytochrome b5 type A (microsomal) [Source:ZFIN;Acc:ZDB-GENE-040426-2148] | 406409 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000099774 | cyb5b | cytochrome b5 type B [Source:ZFIN;Acc:ZDB-GENE-040426-2614] | 405812 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000018966 | cyb5r1 | cytochrome b5 reductase 1 [Source:ZFIN;Acc:ZDB-GENE-030131-8497] | 336553 | YES | YES | NA | NA | |||||
| ENSDARG00000005891 | cyb5r3 | cytochrome b5 reductase 3 [Source:ZFIN;Acc:ZDB-GENE-030131-3281] | 324560 | YES | YES | Other | enzyme | |||||
| ENSDARG00000088013 | CYP27A1 (4 of 4) | cytochrome P450, family 27, subfamily A, polypeptide 1 [Source:HGNC Symbol;Acc:HGNC:2605] | 723999 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000042978 | cyp2p6 | cytochrome P450, family 2, subfamily P, polypeptide 6 [Source:ZFIN;Acc:ZDB-GENE-040426-790] | 393108 | YES | YES | Other | other | |||||
| ENSDARG00000042641 | cyp51 | cytochrome P450, family 51 [Source:ZFIN;Acc:ZDB-GENE-040625-2] | 414331 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000016153 | dag1 | dystroglycan 1 [Source:ZFIN;Acc:ZDB-GENE-021223-1] | 286829 | YES | YES | Plasma Membrane | transmembrane receptor | |||||
| ENSDARG00000035603 | dao.2 | D-amino-acid oxidase, tandem duplicate 2 [Source:ZFIN;Acc:ZDB-GENE-040426-2634] | 405800 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000069446 | dbh | dopamine beta-hydroxylase (dopamine beta-monooxygenase) [Source:ZFIN;Acc:ZDB-GENE-990621-3] | 30505 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000026369 | dbi | diazepam binding inhibitor (GABA receptor modulator, acyl-CoA binding protein) [Source:ZFIN;Acc:ZDB-GENE-040426-1861] | 393831 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000023988 | dctn4 | dynactin 4 [Source:ZFIN;Acc:ZDB-GENE-050417-306] | 550479 | YES | YES | Nucleus | other | |||||
| ENSDARG00000074431 | ddb1 | damage-specific DNA binding protein 1 [Source:ZFIN;Acc:ZDB-GENE-040426-1272] | 0 | YES | YES | Nucleus | other | |||||
| ENSDARG00000037172 | ddrgk1 | DDRGK domain containing 1 [Source:ZFIN;Acc:ZDB-GENE-040426-1050] | 0 | YES | YES | Extracellular Space | other | |||||
| ENSDARG00000044751 | ddt | D-dopachrome tautomerase [Source:ZFIN;Acc:ZDB-GENE-040625-160] | 415237 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000005699 | ddx19 | DEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide 19 (DBP5 homolog, yeast) [Source:ZFIN;Acc:ZDB-GENE-020419-30] | 192339 | YES | YES | Nucleus | enzyme | |||||
| ENSDARG00000074183 | DGKK | diacylglycerol kinase, kappa [Source:HGNC Symbol;Acc:HGNC:32395] | 565122 | YES | YES | NA | NA | |||||
| ENSDARG00000054300 | dhrs1 | dehydrogenase/reductase (SDR family) member 1 [Source:ZFIN;Acc:ZDB-GENE-030616-591] | 368670 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000011770 | dhrs12 | dehydrogenase/reductase (SDR family) member 12 [Source:ZFIN;Acc:ZDB-GENE-060929-1134] | 556393 | YES | YES | NA | NA | |||||
| ENSDARG00000026322 | dhrs13a.1 | dehydrogenase/reductase (SDR family) member 13a, tandem duplicate 1 [Source:ZFIN;Acc:ZDB-GENE-041114-58] | 492491 | YES | YES | Other | enzyme | |||||
| ENSDARG00000041137 | dhrs13a.3 | dehydrogenase/reductase (SDR family) member 13a, duplicate 3 [Source:ZFIN;Acc:ZDB-GENE-041114-134] | 492783 | YES | YES | Other | enzyme | |||||
| ENSDARG00000075726 | dhrs7b | dehydrogenase/reductase (SDR family) member 7B [Source:ZFIN;Acc:ZDB-GENE-050417-277] | 550454 | YES | YES | Other | other | |||||
| ENSDARG00000016415 | dhtkd1 | dehydrogenase E1 and transketolase domain containing 1 [Source:ZFIN;Acc:ZDB-GENE-041212-44] | 494076 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000104172 | diabloa | diablo, IAP-binding mitochondrial protein a [Source:ZFIN;Acc:ZDB-GENE-040426-1303] | 393317 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000014956 | diablob | diablo, IAP-binding mitochondrial protein b [Source:ZFIN;Acc:ZDB-GENE-070112-202] | 570425 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000044420 | dnajc19 | DnaJ (Hsp40) homolog, subfamily C, member 19 [Source:ZFIN;Acc:ZDB-GENE-040426-1730] | 393734 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000042948 | dnajc5aa | DnaJ (Hsp40) homolog, subfamily C, member 5aa [Source:ZFIN;Acc:ZDB-GENE-031113-20] | 386768 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000004836 | dnajc5ab | DnaJ (Hsp40) homolog, subfamily C, member 5ab [Source:ZFIN;Acc:ZDB-GENE-081021-2] | 797907 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000041896 | dnajc5ga | DnaJ (Hsp40) homolog, subfamily C, member 5 gamma a [Source:ZFIN;Acc:ZDB-GENE-030131-1583] | 322863 | YES | YES | NA | NA | |||||
| ENSDARG00000079891 | dnajc6 | DnaJ (Hsp40) homolog, subfamily C, member 6 [Source:ZFIN;Acc:ZDB-GENE-080104-2] | 796345 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000061031 | dner | delta/notch-like EGF repeat containing [Source:ZFIN;Acc:ZDB-GENE-130502-1] | 100332110 | YES | YES | NA | NA | |||||
| ENSDARG00000022599 | dnpep | aspartyl aminopeptidase [Source:ZFIN;Acc:ZDB-GENE-040426-852] | 393122 | YES | YES | Cytoplasm | peptidase | |||||
| ENSDARG00000020676 | dpp3 | dipeptidyl-peptidase 3 [Source:ZFIN;Acc:ZDB-GENE-030131-1247] | 322527 | YES | YES | Cytoplasm | peptidase | |||||
| ENSDARG00000052606 | dpp9 | dipeptidyl-peptidase 9 [Source:ZFIN;Acc:ZDB-GENE-061013-777] | 768170 | YES | YES | Cytoplasm | peptidase | |||||
| ENSDARG00000039345 | drg1 | developmentally regulated GTP binding protein 1 [Source:ZFIN;Acc:ZDB-GENE-030131-8767] | 336823 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000006642 | drg2 | developmentally regulated GTP binding protein 2 [Source:ZFIN;Acc:ZDB-GENE-040808-29] | 323355 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000044628 | dtd1 | D-tyrosyl-tRNA deacylase 1 [Source:ZFIN;Acc:ZDB-GENE-040801-175] | 445046 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000045200 | dusp19b | dual specificity phosphatase 19b [Source:ZFIN;Acc:ZDB-GENE-040502-1] | 406252 | YES | YES | Cytoplasm | phosphatase | |||||
| ENSDARG00000021582 | dynlrb1 | dynein, light chain, roadblock-type 1 [Source:ZFIN;Acc:ZDB-GENE-040426-989] | 394163 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000102142 | ece2b | endothelin converting enzyme 2b [Source:ZFIN;Acc:ZDB-GENE-131120-140] | 793465 | YES | YES | Plasma Membrane | peptidase | |||||
| ENSDARG00000018002 | eci1 | enoyl-CoA delta isomerase 1 [Source:ZFIN;Acc:ZDB-GENE-030131-6033] | 334101 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000062834 | ecsit | ECSIT signalling integrator [Source:ZFIN;Acc:ZDB-GENE-030131-1332] | 569243 | YES | YES | Nucleus | transcription regulator | |||||
| ENSDARG00000062868 | eea1 | early endosome antigen 1 [Source:ZFIN;Acc:ZDB-GENE-041111-270] | 562943 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000030053 | eef1db | eukaryotic translation elongation factor 1 delta b (guanine nucleotide exchange protein) [Source:ZFIN;Acc:ZDB-GENE-030131-6544] | 560745 | YES | YES | Cytoplasm | translation regulator | |||||
| ENSDARG00000020164 | efnb2a | ephrin-B2a [Source:ZFIN;Acc:ZDB-GENE-990415-67] | 30219 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000042277 | efnb3b | ephrin-B3b [Source:ZFIN;Acc:ZDB-GENE-010618-3] | 114378 | YES | YES | Plasma Membrane | kinase | |||||
| ENSDARG00000057912 | eif1axb | eukaryotic translation initiation factor 1A, X-linked, b [Source:ZFIN;Acc:ZDB-GENE-030131-1319] | 322599 | YES | YES | Other | other | |||||
| ENSDARG00000042252 | eif4h | eukaryotic translation initiation factor 4h [Source:ZFIN;Acc:ZDB-GENE-010328-19] | 402996 | YES | YES | Cytoplasm | translation regulator | |||||
| ENSDARG00000017235 | eif5a | eukaryotic translation initiation factor 5A [Source:ZFIN;Acc:ZDB-GENE-040426-2229] | 406464 | YES | YES | Cytoplasm | translation regulator | |||||
| ENSDARG00000101669 | elmod2 | ELMO/CED-12 domain containing 2 [Source:ZFIN;Acc:ZDB-GENE-060824-2] | 553267 | YES | YES | Other | other | |||||
| ENSDARG00000020607 | emc3 | ER membrane protein complex subunit 3 [Source:ZFIN;Acc:ZDB-GENE-040426-2904] | 406826 | YES | YES | Nucleus | other | |||||
| ENSDARG00000008808 | eml2 | echinoderm microtubule associated protein like 2 [Source:ZFIN;Acc:ZDB-GENE-050706-71] | 569375 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000013750 | eno1b | enolase 1b, (alpha) [Source:ZFIN;Acc:ZDB-GENE-040426-1651] | 393668 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000039007 | eno3 | enolase 3, (beta, muscle) [Source:ZFIN;Acc:ZDB-GENE-031006-5] | 378963 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000093374 | EPB41L2 | erythrocyte membrane protein band 4.1-like 2 [Source:HGNC Symbol;Acc:HGNC:3379] | 0 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000045420 | epdr1 | ependymin related 1 [Source:ZFIN;Acc:ZDB-GENE-040718-113] | 436689 | YES | YES | Nucleus | other | |||||
| ENSDARG00000017354 | epha2a | eph receptor A2 a [Source:ZFIN;Acc:ZDB-GENE-990415-63] | 30689 | YES | YES | Plasma Membrane | kinase | |||||
| ENSDARG00000010767 | epha4l | eph receptor A4, like [Source:ZFIN;Acc:ZDB-GENE-990415-61] | 30687 | YES | YES | Plasma Membrane | kinase | |||||
| ENSDARG00000022971 | epha6 | eph receptor A6 [Source:ZFIN;Acc:ZDB-GENE-090311-43] | 100005538 | YES | YES | NA | NA | |||||
| ENSDARG00000076757 | ephb1 | EPH receptor B1 [Source:ZFIN;Acc:ZDB-GENE-070912-219] | 0 | YES | YES | Plasma Membrane | kinase | |||||
| ENSDARG00000037373 | ephb2a | eph receptor B2a [Source:ZFIN;Acc:ZDB-GENE-070713-2] | 562176 | YES | YES | Plasma Membrane | kinase | |||||
| ENSDARG00000021539 | ephb2b | eph receptor B2b [Source:ZFIN;Acc:ZDB-GENE-050522-415] | 554232 | YES | YES | Plasma Membrane | kinase | |||||
| ENSDARG00000100725 | ephb4a | eph receptor B4a [Source:ZFIN;Acc:ZDB-GENE-990415-62] | 30688 | YES | YES | Plasma Membrane | kinase | |||||
| ENSDARG00000040255 | ephx2 | epoxide hydrolase 2, cytoplasmic [Source:ZFIN;Acc:ZDB-GENE-041212-70] | 494099 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000103298 | ephx5 | epoxide hydrolase 5 [Source:ZFIN;Acc:ZDB-GENE-030131-1050] | 322331 | YES | YES | NA | NA | |||||
| ENSDARG00000019008 | erp44 | endoplasmic reticulum protein 44 [Source:ZFIN;Acc:ZDB-GENE-040426-1902] | 393866 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000019496 | esd | esterase D/formylglutathione hydrolase [Source:ZFIN;Acc:ZDB-GENE-050417-328] | 550494 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000014239 | esyt1b | extended synaptotagmin-like protein 1b [Source:ZFIN;Acc:ZDB-GENE-040724-209] | 571083 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000009250 | etfb | electron-transfer-flavoprotein, beta polypeptide [Source:ZFIN;Acc:ZDB-GENE-040426-1931] | 406271 | YES | YES | NA | NA | |||||
| ENSDARG00000099124 | f3a | coagulation factor IIIa [Source:ZFIN;Acc:ZDB-GENE-071119-5] | 567257 | YES | YES | Plasma Membrane | transmembrane receptor | |||||
| ENSDARG00000077764 | FAAH | fatty acid amide hydrolase [Source:HGNC Symbol;Acc:HGNC:3553] | 569067 | YES | YES | Plasma Membrane | enzyme | |||||
| ENSDARG00000008457 | faah2a | fatty acid amide hydrolase 2a [Source:ZFIN;Acc:ZDB-GENE-040718-453] | 436973 | YES | YES | NA | NA | |||||
| ENSDARG00000017299 | fabp11a | fatty acid binding protein 11a [Source:ZFIN;Acc:ZDB-GENE-040912-132] | 447944 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000034650 | fabp7b | fatty acid binding protein 7, brain, b [Source:ZFIN;Acc:ZDB-GENE-040614-4] | 407736 | YES | YES | Cytoplasm | transporter | |||||
| ENSDARG00000019532 | fads2 | fatty acid desaturase 2 [Source:ZFIN;Acc:ZDB-GENE-011212-1] | 140615 | YES | YES | Plasma Membrane | enzyme | |||||
| ENSDARG00000052374 | faf2 | Fas associated factor family member 2 [Source:ZFIN;Acc:ZDB-GENE-050208-312] | 497596 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000098401 | fam136a | family with sequence similarity 136, member A [Source:ZFIN;Acc:ZDB-GENE-040426-2729] | 0 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000014081 | fam184a | family with sequence similarity 184, member A [Source:ZFIN;Acc:ZDB-GENE-041014-214] | 555640 | YES | YES | Extracellular Space | other | |||||
| ENSDARG00000052638 | fam210b | family with sequence similarity 210, member B [Source:ZFIN;Acc:ZDB-GENE-050913-122] | 619270 | YES | YES | Extracellular Space | other | |||||
| ENSDARG00000070675 | fam3a | family with sequence similarity 3, member A [Source:ZFIN;Acc:ZDB-GENE-040426-1772] | 393773 | YES | YES | Extracellular Space | other | |||||
| ENSDARG00000035907 | fam49a | family with sequence similarity 49, member A [Source:ZFIN;Acc:ZDB-GENE-040801-125] | 445212 | YES | YES | NA | NA | |||||
| ENSDARG00000045417 | fam49bb | family with sequence similarity 49, member Bb [Source:ZFIN;Acc:ZDB-GENE-040426-2590] | 404610 | YES | YES | Extracellular Space | other | |||||
| ENSDARG00000017431 | fam63b | family with sequence similarity 63, member B [Source:ZFIN;Acc:ZDB-GENE-030616-328] | 570359 | YES | YES | Nucleus | transporter | |||||
| ENSDARG00000016864 | farsb | phenylalanyl-tRNA synthetase, beta subunit [Source:ZFIN;Acc:ZDB-GENE-021206-2] | 493608 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000087657 | fasn | fatty acid synthase [Source:ZFIN;Acc:ZDB-GENE-030131-7802] | 559001 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000062542 | fgf12b | fibroblast growth factor 12b [Source:ZFIN;Acc:ZDB-GENE-060825-134] | 751749 | YES | YES | Extracellular Space | other | |||||
| ENSDARG00000075132 | fh | fumarate hydratase [Source:ZFIN;Acc:ZDB-GENE-010724-6] | 393938 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000060393 | fibcd1 | fibrinogen C domain containing 1 [Source:ZFIN;Acc:ZDB-GENE-070424-245] | 567525 | YES | YES | Other | other | |||||
| ENSDARG00000022684 | fkbp1aa | FK506 binding protein 1Aa [Source:ZFIN;Acc:ZDB-GENE-030131-7275] | 335335 | YES | YES | NA | NA | |||||
| ENSDARG00000033567 | fkbp1ab | FK506 binding protein 1Ab [Source:ZFIN;Acc:ZDB-GENE-040927-9] | 449552 | YES | YES | NA | NA | |||||
| ENSDARG00000044540 | fkbp2 | FK506 binding protein 2 [Source:ZFIN;Acc:ZDB-GENE-040912-126] | 447939 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000008447 | fkbp4 | FK506 binding protein 4 [Source:ZFIN;Acc:ZDB-GENE-030131-514] | 321795 | YES | YES | Nucleus | enzyme | |||||
| ENSDARG00000031506 | flvcr2b | feline leukemia virus subgroup C cellular receptor family, member 2b [Source:ZFIN;Acc:ZDB-GENE-041210-312] | 569329 | YES | YES | Plasma Membrane | transporter | |||||
| ENSDARG00000059680 | fscn1a | fascin actin-bundling protein 1a [Source:ZFIN;Acc:ZDB-GENE-031009-1] | 558271 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000037747 | fscn1b | fascin actin-bundling protein 1b [Source:ZFIN;Acc:ZDB-GENE-120507-1] | 570314 | YES | YES | NA | NA | |||||
| ENSDARG00000015551 | fth1a | ferritin, heavy polypeptide 1a [Source:ZFIN;Acc:ZDB-GENE-000831-2] | 58108 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000071065 | g6pd | glucose-6-phosphate dehydrogenase [Source:ZFIN;Acc:ZDB-GENE-070508-4] | 570579 | YES | YES | Other | enzyme | |||||
| ENSDARG00000034011 | GABRA2 (1 of 2) | gamma-aminobutyric acid (GABA) A receptor, alpha 2 [Source:HGNC Symbol;Acc:HGNC:4076] | 0 | YES | YES | Plasma Membrane | ion channel | |||||
| ENSDARG00000057303 | galnt7 | UDP-N-acetyl-alpha-D-galactosamine: polypeptide N-acetylgalactosaminyltransferase 7 [Source:ZFIN;Acc:ZDB-GENE-050522-284] | 553668 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000076888 | GANAB (2 of 2) | glucosidase, alpha; neutral AB [Source:HGNC Symbol;Acc:HGNC:4138] | 100126019 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000043457 | gapdh | glyceraldehyde-3-phosphate dehydrogenase [Source:ZFIN;Acc:ZDB-GENE-030115-1] | 317743 | YES | YES | Other | other | |||||
| ENSDARG00000090183 | gapvd1 | GTPase activating protein and VPS9 domains 1 [Source:ZFIN;Acc:ZDB-GENE-040718-117] | 436693 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000059070 | gars | glycyl-tRNA synthetase [Source:ZFIN;Acc:ZDB-GENE-030131-9174] | 337230 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000099524 | gbe1b | glucan (1,4-alpha-), branching enzyme 1b [Source:ZFIN;Acc:ZDB-GENE-110914-16] | 559212 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000058601 | gdap1 | ganglioside induced differentiation associated protein 1 [Source:ZFIN;Acc:ZDB-GENE-050522-424] | 553702 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000055108 | gde1 | glycerophosphodiester phosphodiesterase 1 [Source:ZFIN;Acc:ZDB-GENE-040426-881] | 393213 | YES | YES | Plasma Membrane | enzyme | |||||
| ENSDARG00000056122 | gdi1 | GDP dissociation inhibitor 1 [Source:ZFIN;Acc:ZDB-GENE-050522-504] | 554985 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000017261 | gdpd1 | glycerophosphodiester phosphodiesterase domain containing 1 [Source:ZFIN;Acc:ZDB-GENE-040822-22] | 445504 | YES | YES | Other | enzyme | |||||
| ENSDARG00000063624 | gfm1 | G elongation factor, mitochondrial 1 [Source:ZFIN;Acc:ZDB-GENE-061013-79] | 561840 | YES | YES | Cytoplasm | translation regulator | |||||
| ENSDARG00000086866 | GFPT1 (2 of 2) | glutamine--fructose-6-phosphate transaminase 1 [Source:HGNC Symbol;Acc:HGNC:4241] | 0 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000005085 | ggctb | gamma-glutamylcyclotransferase b [Source:ZFIN;Acc:ZDB-GENE-040426-1939] | 406278 | YES | YES | NA | NA | |||||
| ENSDARG00000042509 | glod4 | glyoxalase domain containing 4 [Source:ZFIN;Acc:ZDB-GENE-040912-38] | 447874 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000098785 | glrx3 | glutaredoxin 3 [Source:ZFIN;Acc:ZDB-GENE-041010-22] | 449777 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000043665 | glrx5 | glutaredoxin 5 homolog (S. cerevisiae) [Source:ZFIN;Acc:ZDB-GENE-040426-1957] | 406294 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000040705 | glsb | glutaminase b [Source:ZFIN;Acc:ZDB-GENE-030616-550] | 564746 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000100003 | glulb | glutamate-ammonia ligase (glutamine synthase) b [Source:ZFIN;Acc:ZDB-GENE-030131-8417] | 336473 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000026629 | gmds | GDP-mannose 4,6-dehydratase [Source:ZFIN;Acc:ZDB-GENE-050419-45] | 393461 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000028327 | gmfb | glia maturation factor, beta [Source:ZFIN;Acc:ZDB-GENE-040426-2114] | 406384 | YES | YES | Cytoplasm | growth factor | |||||
| ENSDARG00000011487 | gnaq | guanine nucleotide binding protein (G protein), q polypeptide [Source:ZFIN;Acc:ZDB-GENE-081104-25] | 570108 | YES | YES | Plasma Membrane | enzyme | |||||
| ENSDARG00000041619 | gnb2l1 | guanine nucleotide binding protein (G protein), beta polypeptide 2-like 1 [Source:ZFIN;Acc:ZDB-GENE-990415-89] | 30722 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000024443 | gnpda2 | glucosamine-6-phosphate deaminase 2 [Source:ZFIN;Acc:ZDB-GENE-091117-41] | 556291 | YES | YES | Nucleus | enzyme | |||||
| ENSDARG00000063197 | golga2 | golgin A2 [Source:ZFIN;Acc:ZDB-GENE-081030-1] | 565431 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000025858 | GOLM1 | golgi membrane protein 1 [Source:HGNC Symbol;Acc:HGNC:15451] | 393249 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000015126 | gorasp2 | golgi reassembly stacking protein 2 [Source:ZFIN;Acc:ZDB-GENE-040426-1659] | 393676 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000026654 | gosr1 | golgi SNAP receptor complex member 1 [Source:ZFIN;Acc:ZDB-GENE-050417-133] | 100001316 | YES | YES | Cytoplasm | transporter | |||||
| ENSDARG00000039093 | got1 | glutamic-oxaloacetic transaminase 1, soluble [Source:ZFIN;Acc:ZDB-GENE-040426-2003] | 406330 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000043180 | gpd1b | glycerol-3-phosphate dehydrogenase 1b [Source:ZFIN;Acc:ZDB-GENE-030131-3906] | 325181 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000040024 | gpd1l | glycerol-3-phosphate dehydrogenase 1-like [Source:ZFIN;Acc:ZDB-GENE-041010-220] | 449663 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000062430 | gpd2 | glycerol-3-phosphate dehydrogenase 2 (mitochondrial) [Source:ZFIN;Acc:ZDB-GENE-030131-4869] | 751628 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000056774 | gpr37l1b | G protein-coupled receptor 37 like 1b [Source:ZFIN;Acc:ZDB-GENE-081104-324] | 567796 | YES | YES | Plasma Membrane | G-protein coupled receptor | |||||
| ENSDARG00000040650 | gps1 | G protein pathway suppressor 1 [Source:ZFIN;Acc:ZDB-GENE-041111-240] | 777711 | YES | YES | Other | other | |||||
| ENSDARG00000068264 | grhpra | glyoxylate reductase/hydroxypyruvate reductase a [Source:ZFIN;Acc:ZDB-GENE-040724-230] | 497183 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000063069 | gripap1 | GRIP1 associated protein 1 [Source:ZFIN;Acc:ZDB-GENE-070112-922] | 791150 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000078815 | grk5l | G protein-coupled receptor kinase 5 like [Source:ZFIN;Acc:ZDB-GENE-081105-95] | 567452 | YES | YES | NA | NA | |||||
| ENSDARG00000032157 | grk6 | G protein-coupled receptor kinase 6 [Source:ZFIN;Acc:ZDB-GENE-101006-5] | 561288 | YES | YES | Plasma Membrane | kinase | |||||
| ENSDARG00000090228 | gstal | glutathione S-transferase, alpha-like [Source:ZFIN;Acc:ZDB-GENE-040426-2720] | 406703 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000042533 | gstm | glutathione S-transferase M [Source:ZFIN;Acc:ZDB-GENE-030911-2] | 324366 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000022183 | gsto1 | glutathione S-transferase omega 1 [Source:ZFIN;Acc:ZDB-GENE-040718-365] | 436894 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000027984 | gstz1 | glutathione S-transferase zeta 1 [Source:ZFIN;Acc:ZDB-GENE-040718-184] | 436754 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000020529 | hacl1 | 2-hydroxyacyl-CoA lyase 1 [Source:ZFIN;Acc:ZDB-GENE-040426-2058] | 406358 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000030765 | hadh | hydroxyacyl-CoA dehydrogenase [Source:ZFIN;Acc:ZDB-GENE-040801-261] | 445121 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000025338 | hagh | hydroxyacylglutathione hydrolase [Source:ZFIN;Acc:ZDB-GENE-030131-8921] | 336977 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000003693 | hars | histidyl-tRNA synthetase [Source:ZFIN;Acc:ZDB-GENE-040912-152] | 447847 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000074419 | hcn4l | hyperpolarization activated cyclic nucleotide-gated potassium channel 4l [Source:ZFIN;Acc:ZDB-GENE-110411-3] | 556759 | YES | YES | Plasma Membrane | ion channel | |||||
| ENSDARG00000009830 | hdlbpa | high density lipoprotein binding protein a [Source:ZFIN;Acc:ZDB-GENE-030131-2032] | 794178 | YES | YES | Other | other | |||||
| ENSDARG00000042630 | hebp2 | heme binding protein 2 [Source:ZFIN;Acc:ZDB-GENE-040426-914] | 393167 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000034368 | hexb | hexosaminidase B (beta polypeptide) [Source:ZFIN;Acc:ZDB-GENE-030131-2333] | 323613 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000020960 | hgs | hepatocyte growth factor-regulated tyrosine kinase substrate [Source:ZFIN;Acc:ZDB-GENE-030131-6107] | 334175 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000007959 | hibadhb | 3-hydroxyisobutyrate dehydrogenase b [Source:ZFIN;Acc:ZDB-GENE-040426-1582] | 394135 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000054867 | hibch | 3-hydroxyisobutyryl-CoA hydrolase [Source:ZFIN;Acc:ZDB-GENE-050327-29] | 798364 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000103824 | hint1 | histidine triad nucleotide binding protein 1 [Source:ZFIN;Acc:ZDB-GENE-040927-8] | 449551 | YES | YES | Nucleus | enzyme | |||||
| ENSDARG00000062972 | hip1 | huntingtin interacting protein 1 [Source:ZFIN;Acc:ZDB-GENE-010328-15] | 793823 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000037846 | hm13 | histocompatibility (minor) 13 [Source:ZFIN;Acc:ZDB-GENE-020802-3] | 259190 | YES | YES | Cytoplasm | peptidase | |||||
| ENSDARG00000029722 | hmgb2a | high mobility group box 2a [Source:ZFIN;Acc:ZDB-GENE-051120-126] | 641484 | YES | YES | Nucleus | transcription regulator | |||||
| ENSDARG00000021220 | hmgcl | 3-hydroxymethyl-3-methylglutaryl-CoA lyase [Source:ZFIN;Acc:ZDB-GENE-040426-958] | 394190 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000055101 | hmox2a | heme oxygenase 2a [Source:ZFIN;Acc:ZDB-GENE-040914-13] | 100002875 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000078452 | hmox2b | heme oxygenase 2b [Source:ZFIN;Acc:ZDB-GENE-091118-52] | 100329531 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000099312 | hn1b | hematological and neurological expressed 1b [Source:ZFIN;Acc:ZDB-GENE-030131-2784] | 402906 | YES | YES | Nucleus | other | |||||
| ENSDARG00000070491 | hpcal4 | hippocalcin like 4 [Source:ZFIN;Acc:ZDB-GENE-060503-307] | 564909 | YES | YES | Other | transporter | |||||
| ENSDARG00000008884 | hprt1 | hypoxanthine phosphoribosyltransferase 1 [Source:ZFIN;Acc:ZDB-GENE-040426-1918] | 406259 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000012609 | hpx | hemopexin [Source:ZFIN;Acc:ZDB-GENE-030131-5773] | 327588 | YES | YES | Extracellular Space | transporter | |||||
| ENSDARG00000098497 | hrasa | v-Ha-ras Harvey rat sarcoma viral oncogene homolog a [Source:ZFIN;Acc:ZDB-GENE-050417-95] | 550286 | YES | YES | Plasma Membrane | enzyme | |||||
| ENSDARG00000035882 | hrsp12 | heat-responsive protein 12 [Source:ZFIN;Acc:ZDB-GENE-040718-315] | 436849 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000101239 | hsd17b4 | hydroxysteroid (17-beta) dehydrogenase 4 [Source:ZFIN;Acc:ZDB-GENE-040421-1] | 393105 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000041736 | hsdl1 | hydroxysteroid dehydrogenase like 1 [Source:ZFIN;Acc:ZDB-GENE-041212-31] | 494064 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000004754 | hspa4a | heat shock protein 4a [Source:ZFIN;Acc:ZDB-GENE-040426-2832] | 335865 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000056167 | hspe1 | heat shock 10 protein 1 [Source:ZFIN;Acc:ZDB-GENE-000906-2] | 58041 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000028386 | htatip2 | HIV-1 Tat interactive protein 2 [Source:ZFIN;Acc:ZDB-GENE-001219-1] | 64605 | YES | YES | Nucleus | transcription regulator | |||||
| ENSDARG00000013670 | hyou1 | hypoxia up-regulated 1 [Source:ZFIN;Acc:ZDB-GENE-030131-5344] | 327133 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000044134 | iars2 | isoleucyl-tRNA synthetase 2, mitochondrial [Source:ZFIN;Acc:ZDB-GENE-030616-608] | 100001219 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000034434 | igf1rb | insulin-like growth factor 1b receptor [Source:ZFIN;Acc:ZDB-GENE-020503-2] | 245702 | YES | YES | Plasma Membrane | transmembrane receptor | |||||
| ENSDARG00000006094 | igf2r | insulin-like growth factor 2 receptor [Source:ZFIN;Acc:ZDB-GENE-041014-300] | 557061 | YES | YES | Plasma Membrane | transmembrane receptor | |||||
| ENSDARG00000017217 | igsf11 | immunoglobulin superfamily member 11 [Source:ZFIN;Acc:ZDB-GENE-080303-21] | 100004469 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000021611 | im:7152756 | im:7152756 [Source:ZFIN;Acc:ZDB-GENE-050309-232] | 0 | YES | YES | NA | NA | |||||
| ENSDARG00000003517 | impa1 | inositol(myo)-1(or 4)-monophosphatase 1 [Source:ZFIN;Acc:ZDB-GENE-040718-245] | 437018 | YES | YES | Cytoplasm | phosphatase | |||||
| ENSDARG00000058114 | impad1 | inositol monophosphatase domain containing 1 [Source:ZFIN;Acc:ZDB-GENE-051127-23] | 641570 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000093359 | inpp5jb | inositol polyphosphate-5-phosphatase Jb [Source:ZFIN;Acc:ZDB-GENE-081104-46] | 0 | YES | YES | Plasma Membrane | phosphatase | |||||
| ENSDARG00000051875 | islr2 | immunoglobulin superfamily containing leucine-rich repeat 2 [Source:ZFIN;Acc:ZDB-GENE-050320-95] | 541396 | YES | YES | Extracellular Space | other | |||||
| ENSDARG00000060069 | isoc1 | isochorismatase domain containing 1 [Source:ZFIN;Acc:ZDB-GENE-061020-1] | 559897 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000006314 | itgav | integrin, alpha V [Source:ZFIN;Acc:ZDB-GENE-060616-382] | 100151255 | YES | YES | Plasma Membrane | ion channel | |||||
| ENSDARG00000016939 | itgb2 | integrin, beta 2 [Source:ZFIN;Acc:ZDB-GENE-110411-4] | 557797 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000012942 | itgb5 | integrin, beta 5 [Source:ZFIN;Acc:ZDB-GENE-030131-6022] | 563556 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000042853 | ivd | isovaleryl-CoA dehydrogenase [Source:ZFIN;Acc:ZDB-GENE-030616-262] | 368775 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000086037 | jam3a | junctional adhesion molecule 3a [Source:ZFIN;Acc:ZDB-GENE-070424-177] | 797651 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000062942 | kcna1a | potassium voltage-gated channel, shaker-related subfamily, member 1a [Source:ZFIN;Acc:ZDB-GENE-110408-15] | 795942 | YES | YES | Plasma Membrane | ion channel | |||||
| ENSDARG00000051852 | kcnc1a | potassium voltage-gated channel, Shaw-related subfamily, member 1a [Source:ZFIN;Acc:ZDB-GENE-061220-10] | 565419 | YES | YES | Plasma Membrane | ion channel | |||||
| ENSDARG00000031438 | kcnj11l | potassium inwardly-rectifying channel, subfamily J, member 11, like [Source:ZFIN;Acc:ZDB-GENE-050222-2] | 0 | YES | YES | NA | NA | |||||
| ENSDARG00000062217 | kcnj3b | potassium inwardly-rectifying channel, subfamily J, member 3b [Source:ZFIN;Acc:ZDB-GENE-120215-38] | 557095 | YES | YES | Plasma Membrane | ion channel | |||||
| ENSDARG00000012021 | kdsr | 3-ketodihydrosphingosine reductase [Source:ZFIN;Acc:ZDB-GENE-040426-853] | 394114 | YES | YES | Plasma Membrane | enzyme | |||||
| ENSDARG00000061817 | kif1aa | kinesin family member 1Aa [Source:ZFIN;Acc:ZDB-GENE-100913-3] | 100330443 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000061131 | kif21a | kinesin family member 21A [Source:ZFIN;Acc:ZDB-GENE-110411-237] | 566481 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000074131 | kif5ba | kinesin family member 5B, a [Source:ZFIN;Acc:ZDB-GENE-070629-2] | 563459 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000104665 | kirrel3l | kin of IRRE like 3 like [Source:ZFIN;Acc:ZDB-GENE-070831-1] | 557315 | YES | YES | NA | NA | |||||
| ENSDARG00000099740 | klc2 | kinesin light chain 2 [Source:ZFIN;Acc:ZDB-GENE-030131-2670] | 562365 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000023190 | kpna4 | karyopherin alpha 4 (importin alpha 3) [Source:ZFIN;Acc:ZDB-GENE-030131-5584] | 327373 | YES | YES | Nucleus | transporter | |||||
| ENSDARG00000040245 | kpnb3 | karyopherin (importin) beta 3 [Source:ZFIN;Acc:ZDB-GENE-030424-2] | 569455 | YES | YES | Nucleus | transporter | |||||
| ENSDARG00000092610 | lamp1 | lysosomal-associated membrane protein 1 [Source:ZFIN;Acc:ZDB-GENE-030131-9303] | 563328 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000076464 | lamtor1 | late endosomal/lysosomal adaptor, MAPK and MTOR activator 1 [Source:ZFIN;Acc:ZDB-GENE-130819-1] | 100329656 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000039872 | lamtor2 | late endosomal/lysosomal adaptor, MAPK and MTOR activator 2 [Source:ZFIN;Acc:ZDB-GENE-040801-63] | 445155 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000045542 | LAMTOR4 | late endosomal/lysosomal adaptor, MAPK and MTOR activator 4 [Source:HGNC Symbol;Acc:HGNC:33772] | 561019 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000090194 | lamtor5 | late endosomal/lysosomal adaptor, MAPK and MTOR activator 5 [Source:ZFIN;Acc:ZDB-GENE-110411-193] | 100333504 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000061981 | lap3 | leucine aminopeptidase 3 [Source:ZFIN;Acc:ZDB-GENE-060719-1] | 562854 | YES | YES | Cytoplasm | peptidase | |||||
| ENSDARG00000090988 | larsa | leucyl-tRNA synthetase a [Source:ZFIN;Acc:ZDB-GENE-110411-16] | 0 | YES | YES | NA | NA | |||||
| ENSDARG00000071076 | ldhbb | lactate dehydrogenase Bb [Source:ZFIN;Acc:ZDB-GENE-040718-176] | 436747 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000060196 | lhpp | phospholysine phosphohistidine inorganic pyrophosphate phosphatase [Source:ZFIN;Acc:ZDB-GENE-070615-43] | 100073345 | YES | YES | Cytoplasm | phosphatase | |||||
| ENSDARG00000074535 | lingo2a | leucine rich repeat and Ig domain containing 2a [Source:ZFIN;Acc:ZDB-GENE-080327-19] | 566713 | YES | YES | Extracellular Space | other | |||||
| ENSDARG00000009693 | llgl1 | lethal giant larvae homolog 1 (Drosophila) [Source:ZFIN;Acc:ZDB-GENE-060825-69] | 751760 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000023920 | llgl2 | lethal giant larvae homolog 2 (Drosophila) [Source:ZFIN;Acc:ZDB-GENE-030131-9877] | 795670 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000069980 | lman1 | lectin, mannose-binding, 1 [Source:ZFIN;Acc:ZDB-GENE-060201-4] | 559775 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000061854 | lman2 | lectin, mannose-binding 2 [Source:ZFIN;Acc:ZDB-GENE-070209-149] | 557960 | YES | YES | Cytoplasm | transporter | |||||
| ENSDARG00000017312 | lmbrd2b | LMBR1 domain containing 2b [Source:ZFIN;Acc:ZDB-GENE-030131-7197] | 335257 | YES | YES | NA | NA | |||||
| ENSDARG00000025859 | lmf2b | lipase maturation factor 2b [Source:ZFIN;Acc:ZDB-GENE-070206-11] | 407708 | YES | YES | NA | NA | |||||
| ENSDARG00000076324 | lmtk2 | lemur tyrosine kinase 2 [Source:ZFIN;Acc:ZDB-GENE-100812-11] | 564666 | YES | YES | Cytoplasm | kinase | |||||
| ENSDARG00000006639 | lnpb | limb and neural patterns b [Source:ZFIN;Acc:ZDB-GENE-030131-9455] | 337509 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000100394 | lnpep | leucyl/cystinyl aminopeptidase [Source:ZFIN;Acc:ZDB-GENE-030131-1534] | 322814 | YES | YES | Cytoplasm | peptidase | |||||
| ENSDARG00000102765 | lonp1 | lon peptidase 1, mitochondrial [Source:ZFIN;Acc:ZDB-GENE-030131-4006] | 563257 | YES | YES | Cytoplasm | peptidase | |||||
| ENSDARG00000039500 | LRFN3 (1 of 2) | leucine rich repeat and fibronectin type III domain containing 3 [Source:HGNC Symbol;Acc:HGNC:28370] | 100333421 | YES | YES | Other | other | |||||
| ENSDARG00000097827 | LRP1 (2 of 2) | low density lipoprotein receptor-related protein 1 [Source:HGNC Symbol;Acc:HGNC:6692] | 0 | YES | YES | Plasma Membrane | transmembrane receptor | |||||
| ENSDARG00000033604 | lrpap1 | low density lipoprotein receptor-related protein associated protein 1 [Source:ZFIN;Acc:ZDB-GENE-030131-5871] | 333939 | YES | YES | Plasma Membrane | transmembrane receptor | |||||
| ENSDARG00000053138 | lrrc47 | leucine rich repeat containing 47 [Source:ZFIN;Acc:ZDB-GENE-060503-289] | 553404 | YES | YES | Other | other | |||||
| ENSDARG00000071426 | lrrc59 | leucine rich repeat containing 59 [Source:ZFIN;Acc:ZDB-GENE-040426-2547] | 405868 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000071374 | lrrtm2 | leucine rich repeat transmembrane neuronal 2 [Source:ZFIN;Acc:ZDB-GENE-080327-8] | 100538061 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000053113 | ly75 | lymphocyte antigen 75 [Source:ZFIN;Acc:ZDB-GENE-050309-166] | 504036 | YES | YES | Plasma Membrane | transmembrane receptor | |||||
| ENSDARG00000055899 | lzic | leucine zipper and CTNNBIP1 domain containing [Source:ZFIN;Acc:ZDB-GENE-040718-342] | 436871 | YES | YES | Other | other | |||||
| ENSDARG00000070573 | mapk3 | mitogen-activated protein kinase 3 [Source:ZFIN;Acc:ZDB-GENE-040121-1] | 399480 | YES | YES | Cytoplasm | kinase | |||||
| ENSDARG00000062531 | mapk8ip3 | mitogen-activated protein kinase 8 interacting protein 3 [Source:ZFIN;Acc:ZDB-GENE-090303-6] | 100318355 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000002659 | mapre1b | microtubule-associated protein, RP/EB family, member 1b [Source:ZFIN;Acc:ZDB-GENE-040426-2256] | 334728 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000070487 | MARC2 | mitochondrial amidoxime reducing component 2 [Source:HGNC Symbol;Acc:HGNC:26064] | 378742 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000096271 | march7 | membrane-associated ring finger (C3HC4) 7 [Source:ZFIN;Acc:ZDB-GENE-080204-121] | 561242 | YES | YES | Other | other | |||||
| ENSDARG00000008803 | marcksb | myristoylated alanine-rich protein kinase C substrate b [Source:ZFIN;Acc:ZDB-GENE-030131-1921] | 323201 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000039034 | marcksl1a | MARCKS-like 1a [Source:ZFIN;Acc:ZDB-GENE-040426-2146] | 406407 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000040334 | mat2aa | methionine adenosyltransferase II, alpha a [Source:ZFIN;Acc:ZDB-GENE-030131-2049] | 0 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000012932 | mat2b | methionine adenosyltransferase II, beta [Source:ZFIN;Acc:ZDB-GENE-030131-786] | 541347 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000089643 | mcama | melanoma cell adhesion molecule a [Source:ZFIN;Acc:ZDB-GENE-090218-29] | 566921 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000100718 | me2 | malic enzyme 2, NAD(+)-dependent, mitochondrial [Source:ZFIN;Acc:ZDB-GENE-040801-147] | 445233 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000002305 | me3 | malic enzyme 3, NADP(+)-dependent, mitochondrial [Source:ZFIN;Acc:ZDB-GENE-041111-294] | 492719 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000056726 | mettl7a | methyltransferase like 7A [Source:ZFIN;Acc:ZDB-GENE-061027-239] | 569131 | YES | YES | Other | enzyme | |||||
| ENSDARG00000079219 | MFN1 (2 of 3) | mitofusin 1 [Source:HGNC Symbol;Acc:HGNC:18262] | 0 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000101793 | mfn1b | mitofusin 1b [Source:ZFIN;Acc:ZDB-GENE-040426-1553] | 0 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000036820 | mgll | monoglyceride lipase [Source:ZFIN;Acc:ZDB-GENE-031006-9] | 378960 | YES | YES | Plasma Membrane | enzyme | |||||
| ENSDARG00000032618 | mgst1.1 | microsomal glutathione S-transferase 1.1 [Source:ZFIN;Acc:ZDB-GENE-041010-30] | 449784 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000022165 | mgst1.2 | microsomal glutathione S-transferase 1.2 [Source:ZFIN;Acc:ZDB-GENE-040704-59] | 431762 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000075963 | mhc1uba | major histocompatibility complex class I UBA [Source:ZFIN;Acc:ZDB-GENE-990415-145] | 30757 | YES | YES | NA | NA | |||||
| ENSDARG00000079067 | mms19 | MMS19 nucleotide excision repair homolog (S. cerevisiae) [Source:ZFIN;Acc:ZDB-GENE-120316-2] | 563643 | YES | YES | Nucleus | transcription regulator | |||||
| ENSDARG00000093448 | mpc1 | mitochondrial pyruvate carrier 1 [Source:ZFIN;Acc:ZDB-GENE-040718-94] | 436671 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000030786 | mpi | mannose phosphate isomerase [Source:ZFIN;Acc:ZDB-GENE-050904-6] | 613246 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000059048 | mpzl1l | myelin protein zero-like 1 like [Source:ZFIN;Acc:ZDB-GENE-050417-455] | 555163 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000075285 | MRAS (2 of 2) | muscle RAS oncogene homolog [Source:HGNC Symbol;Acc:HGNC:7227] | 100536317 | YES | YES | Plasma Membrane | enzyme | |||||
| ENSDARG00000102100 | mrc1a | mannose receptor, C type 1a [Source:ZFIN;Acc:ZDB-GENE-090915-4] | 100286774 | YES | YES | Plasma Membrane | transmembrane receptor | |||||
| ENSDARG00000075754 | mri1 | methylthioribose-1-phosphate isomerase 1 [Source:ZFIN;Acc:ZDB-GENE-080204-109] | 100002302 | YES | YES | Cytoplasm | translation regulator | |||||
| ENSDARG00000035167 | mrpl48 | mitochondrial ribosomal protein L48 [Source:ZFIN;Acc:ZDB-GENE-041008-125] | 503593 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000059234 | mrps27 | mitochondrial ribosomal protein S27 [Source:ZFIN;Acc:ZDB-GENE-040808-39] | 445321 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000063917 | mt-nd4 | NADH dehydrogenase 4, mitochondrial [Source:ZFIN;Acc:ZDB-GENE-011205-10] | 140534 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000037261 | mtap | methylthioadenosine phosphorylase [Source:ZFIN;Acc:ZDB-GENE-040426-1505] | 393526 | YES | YES | Nucleus | enzyme | |||||
| ENSDARG00000104018 | mtpn | myotrophin [Source:ZFIN;Acc:ZDB-GENE-040426-2166] | 406240 | YES | YES | Nucleus | transcription regulator | |||||
| ENSDARG00000087260 | MTSS1L (2 of 2) | metastasis suppressor 1-like [Source:HGNC Symbol;Acc:HGNC:25094] | 0 | YES | YES | Other | other | |||||
| ENSDARG00000036109 | mtx1b | metaxin 1b [Source:ZFIN;Acc:ZDB-GENE-021210-1] | 286740 | YES | YES | Cytoplasm | transporter | |||||
| ENSDARG00000021242 | mvp | major vault protein [Source:ZFIN;Acc:ZDB-GENE-030826-33] | 373081 | YES | YES | Nucleus | other | |||||
| ENSDARG00000071679 | mydgf | myeloid-derived growth factor [Source:ZFIN;Acc:ZDB-GENE-040718-183] | 436753 | YES | YES | Extracellular Space | cytokine | |||||
| ENSDARG00000030598 | nampta | nicotinamide phosphoribosyltransferase a [Source:ZFIN;Acc:ZDB-GENE-110420-1] | 100330160 | YES | YES | NA | NA | |||||
| ENSDARG00000045620 | nansa | N-acetylneuraminic acid synthase a [Source:ZFIN;Acc:ZDB-GENE-030131-1500] | 322780 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000067605 | napaa | N-ethylmaleimide-sensitive factor attachment protein, alpha a [Source:ZFIN;Acc:ZDB-GENE-060531-160] | 567306 | YES | YES | Cytoplasm | transporter | |||||
| ENSDARG00000020405 | napab | N-ethylmaleimide-sensitive factor attachment protein, alpha b [Source:ZFIN;Acc:ZDB-GENE-030131-5290] | 327082 | YES | YES | Cytoplasm | transporter | |||||
| ENSDARG00000069101 | NAPB (2 of 2) | N-ethylmaleimide-sensitive factor attachment protein, beta [Source:HGNC Symbol;Acc:HGNC:15751] | 568195 | YES | YES | Cytoplasm | transporter | |||||
| ENSDARG00000009252 | napepld | N-acyl phosphatidylethanolamine phospholipase D [Source:ZFIN;Acc:ZDB-GENE-030131-3856] | 568061 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000006617 | napga | N-ethylmaleimide-sensitive factor attachment protein, gamma a [Source:ZFIN;Acc:ZDB-GENE-050320-46] | 541353 | YES | YES | Cytoplasm | transporter | |||||
| ENSDARG00000090962 | nat14 | N-acetyltransferase 14 (GCN5-related, putative) [Source:ZFIN;Acc:ZDB-GENE-060825-15] | 751624 | YES | YES | Extracellular Space | other | |||||
| ENSDARG00000008593 | nbas | neuroblastoma amplified sequence [Source:ZFIN;Acc:ZDB-GENE-041014-367] | 556592 | YES | YES | Nucleus | other | |||||
| ENSDARG00000070688 | ncalda | neurocalcin delta a [Source:ZFIN;Acc:ZDB-GENE-080220-28] | 556178 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000054186 | ncs1b | neuronal calcium sensor 1b [Source:ZFIN;Acc:ZDB-GENE-050930-1] | 553174 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000103284 | NCSTN | nicastrin [Source:HGNC Symbol;Acc:HGNC:17091] | 0 | YES | YES | Plasma Membrane | peptidase | |||||
| ENSDARG00000032849 | ndrg1a | N-myc downstream regulated 1a [Source:ZFIN;Acc:ZDB-GENE-030826-34] | 792085 | YES | YES | Nucleus | kinase | |||||
| ENSDARG00000011170 | ndrg2 | NDRG family member 2 [Source:ZFIN;Acc:ZDB-GENE-041212-15] | 494050 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000013087 | ndrg3a | ndrg family member 3a [Source:ZFIN;Acc:ZDB-GENE-980526-124] | 30286 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000069286 | ndufaf2 | NADH dehydrogenase (ubiquinone) complex I, assembly factor 2 [Source:ZFIN;Acc:ZDB-GENE-040808-28] | 100003850 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000020798 | necap1 | NECAP endocytosis associated 1 [Source:ZFIN;Acc:ZDB-GENE-040426-1682] | 393695 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000045249 | nif3l1 | NIF3 NGG1 interacting factor 3-like 1 (S. cerevisiae) [Source:ZFIN;Acc:ZDB-GENE-050706-80] | 574421 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000017590 | nit2 | nitrilase family, member 2 [Source:ZFIN;Acc:ZDB-GENE-050522-65] | 402904 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000021808 | nmt2 | N-myristoyltransferase 2 [Source:ZFIN;Acc:ZDB-GENE-030131-717] | 556483 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000017673 | nova2 | neuro-oncological ventral antigen 2 [Source:ZFIN;Acc:ZDB-GENE-080211-1] | 553201 | YES | YES | Nucleus | other | |||||
| ENSDARG00000017180 | npc1 | Niemann-Pick disease, type C1 [Source:ZFIN;Acc:ZDB-GENE-030131-3161] | 553330 | YES | YES | Cytoplasm | transporter | |||||
| ENSDARG00000038225 | nras | neuroblastoma RAS viral (v-ras) oncogene homolog [Source:ZFIN;Acc:ZDB-GENE-990415-166] | 30380 | YES | YES | Plasma Membrane | enzyme | |||||
| ENSDARG00000056089 | nrbp1 | nuclear receptor binding protein 1 [Source:ZFIN;Acc:ZDB-GENE-041001-146] | 569806 | YES | YES | Nucleus | kinase | |||||
| ENSDARG00000079985 | nrip2 | nuclear receptor interacting protein 2 [Source:ZFIN;Acc:ZDB-GENE-030131-9425] | 565865 | YES | YES | NA | NA | |||||
| ENSDARG00000027290 | nrp1b | neuropilin 1b [Source:ZFIN;Acc:ZDB-GENE-040611-1] | 402973 | YES | YES | Plasma Membrane | transmembrane receptor | |||||
| ENSDARG00000096546 | nrp2a | neuropilin 2a [Source:ZFIN;Acc:ZDB-GENE-040611-2] | 405901 | YES | YES | Plasma Membrane | kinase | |||||
| ENSDARG00000038446 | nrp2b | neuropilin 2b [Source:ZFIN;Acc:ZDB-GENE-040611-3] | 405902 | YES | YES | Plasma Membrane | kinase | |||||
| ENSDARG00000044719 | nrsn1 | neurensin 1 [Source:ZFIN;Acc:ZDB-GENE-030131-7491] | 335551 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000087640 | nsfl1c | NSFL1 (p97) cofactor (p47) [Source:ZFIN;Acc:ZDB-GENE-041114-160] | 492805 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000039399 | nt5c2a | 5'-nucleotidase, cytosolic IIa [Source:ZFIN;Acc:ZDB-GENE-030131-3566] | 324845 | YES | YES | Cytoplasm | phosphatase | |||||
| ENSDARG00000006797 | nt5dc1 | 5'-nucleotidase domain containing 1 [Source:ZFIN;Acc:ZDB-GENE-030131-8274] | 568757 | YES | YES | Other | other | |||||
| ENSDARG00000059897 | ntrk2a | neurotrophic tyrosine kinase, receptor, type 2a [Source:ZFIN;Acc:ZDB-GENE-010126-1] | 65090 | YES | YES | Plasma Membrane | kinase | |||||
| ENSDARG00000054833 | nucb1 | nucleobindin 1 [Source:ZFIN;Acc:ZDB-GENE-060825-222] | 751753 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000036299 | nucb2a | nucleobindin 2a [Source:ZFIN;Acc:ZDB-GENE-030826-14] | 373078 | YES | YES | Nucleus | other | |||||
| ENSDARG00000078073 | nudt5 | nudix (nucleoside diphosphate linked moiety X)-type motif 5 [Source:ZFIN;Acc:ZDB-GENE-040625-63] | 415176 | YES | YES | Cytoplasm | phosphatase | |||||
| ENSDARG00000101949 | numbl | numb homolog (Drosophila)-like [Source:ZFIN;Acc:ZDB-GENE-051113-340] | 497616 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000033978 | nxn | nucleoredoxin [Source:ZFIN;Acc:ZDB-GENE-050522-75] | 553621 | YES | YES | Nucleus | enzyme | |||||
| ENSDARG00000078425 | oat | ornithine aminotransferase [Source:ZFIN;Acc:ZDB-GENE-110411-148] | 572518 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000019713 | oatx | organic anion transporter X [Source:ZFIN;Acc:ZDB-GENE-070115-1] | 0 | YES | YES | NA | NA | |||||
| ENSDARG00000058022 | ormdl2 | ORMDL sphingolipid biosynthesis regulator 2 [Source:ZFIN;Acc:ZDB-GENE-060331-113] | 677753 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000025555 | ormdl3 | ORMDL sphingolipid biosynthesis regulator 3 [Source:ZFIN;Acc:ZDB-GENE-041010-190] | 450067 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000004634 | osbp | oxysterol binding protein [Source:ZFIN;Acc:ZDB-GENE-090312-18] | 564372 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000011462 | otub1b | OTU deubiquitinase, ubiquitin aldehyde binding 1b [Source:ZFIN;Acc:ZDB-GENE-040718-204] | 0 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000063310 | oxr1b | oxidation resistance 1b [Source:ZFIN;Acc:ZDB-GENE-030131-2438] | 570476 | YES | YES | NA | NA | |||||
| ENSDARG00000034189 | oxsr1a | oxidative stress responsive 1a [Source:ZFIN;Acc:ZDB-GENE-070620-25] | 568602 | YES | YES | Nucleus | kinase | |||||
| ENSDARG00000042128 | pacsin1b | protein kinase C and casein kinase substrate in neurons 1b [Source:ZFIN;Acc:ZDB-GENE-050913-35] | 619246 | YES | YES | Cytoplasm | kinase | |||||
| ENSDARG00000044167 | padi2 | peptidyl arginine deiminase, type II [Source:ZFIN;Acc:ZDB-GENE-031116-67] | 386792 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000032013 | pafah1b1a | platelet-activating factor acetylhydrolase 1b, regulatory subunit 1a [Source:ZFIN;Acc:ZDB-GENE-040116-2] | 394246 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000035352 | pafah1b2 | platelet-activating factor acetylhydrolase 1b, catalytic subunit 2 [Source:ZFIN;Acc:ZDB-GENE-060526-43] | 567170 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000033539 | paics | phosphoribosylaminoimidazole carboxylase, phosphoribosylaminoimidazole succinocarboxamide synthetase [Source:ZFIN;Acc:ZDB-GENE-030131-9762] | 321193 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000103959 | pak1 | p21 protein (Cdc42/Rac)-activated kinase 1 [Source:ZFIN;Acc:ZDB-GENE-030826-29] | 373103 | YES | YES | NA | NA | |||||
| ENSDARG00000003380 | PAOX | polyamine oxidase (exo-N4-amino) [Source:HGNC Symbol;Acc:HGNC:20837] | 562105 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000071015 | pbxip1a | pre-B-cell leukemia homeobox interacting protein 1a [Source:ZFIN;Acc:ZDB-GENE-060503-871] | 563463 | YES | YES | NA | NA | |||||
| ENSDARG00000031981 | pcbd1 | pterin-4 alpha-carbinolamine dehydratase/dimerization cofactor of hepatocyte nuclear factor 1 alpha [Source:ZFIN;Acc:ZDB-GENE-040426-1787] | 393787 | YES | YES | Nucleus | transcription regulator | |||||
| ENSDARG00000028982 | pcca | propionyl CoA carboxylase, alpha polypeptide [Source:ZFIN;Acc:ZDB-GENE-040718-246] | 437019 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000062720 | pcdh1a | protocadherin 1a [Source:ZFIN;Acc:ZDB-GENE-091116-32] | 100006496 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000098788 | pcdh2g16 | protocadherin 2 gamma 16 [Source:ZFIN;Acc:ZDB-GENE-050610-6] | 0 | YES | YES | NA | NA | |||||
| ENSDARG00000042848 | pcmtl | l-isoaspartyl protein carboxyl methyltransferase, like [Source:ZFIN;Acc:ZDB-GENE-040426-1738] | 393741 | YES | YES | NA | NA | |||||
| ENSDARG00000023181 | pcp4l1 | Purkinje cell protein 4 like 1 [Source:ZFIN;Acc:ZDB-GENE-040724-35] | 100008021 | YES | YES | Other | other | |||||
| ENSDARG00000019451 | pcsk2 | proprotein convertase subtilisin/kexin type 2 [Source:ZFIN;Acc:ZDB-GENE-090608-1] | 100004460 | YES | YES | Extracellular Space | peptidase | |||||
| ENSDARG00000103052 | pcyox1 | prenylcysteine oxidase 1 [Source:ZFIN;Acc:ZDB-GENE-050417-98] | 550289 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000069003 | PCYOX1L | prenylcysteine oxidase 1 like [Source:HGNC Symbol;Acc:HGNC:28477] | 0 | YES | YES | Other | other | |||||
| ENSDARG00000035054 | pdcd10a | programmed cell death 10a [Source:ZFIN;Acc:ZDB-GENE-040426-1432] | 393527 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000026072 | pdcd5 | programmed cell death 5 [Source:ZFIN;Acc:ZDB-GENE-040426-980] | 394152 | YES | YES | Nucleus | other | |||||
| ENSDARG00000025269 | pdcd6ip | programmed cell death 6 interacting protein [Source:ZFIN;Acc:ZDB-GENE-040426-2678] | 406669 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000003449 | pde10a | phosphodiesterase 10A [Source:ZFIN;Acc:ZDB-GENE-040426-1115] | 394077 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000013950 | peptide deformylase (mitochondrial) [Source:ZFIN;Acc:ZDB-GENE-050913-42] | 619248 | YES | YES | Cytoplasm | enzyme | ||||||
| ENSDARG00000100897 | PDGFRB | platelet-derived growth factor receptor, beta polypeptide [Source:HGNC Symbol;Acc:HGNC:8804] | 0 | YES | YES | Plasma Membrane | kinase | |||||
| ENSDARG00000102640 | pdia3 | protein disulfide isomerase family A, member 3 [Source:ZFIN;Acc:ZDB-GENE-031002-9] | 378851 | YES | YES | Cytoplasm | peptidase | |||||
| ENSDARG00000069242 | pdia5 | protein disulfide isomerase family A, member 5 [Source:ZFIN;Acc:ZDB-GENE-030521-5] | 556162 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000009001 | pdia6 | protein disulfide isomerase family A, member 6 [Source:ZFIN;Acc:ZDB-GENE-030131-879] | 322160 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000103288 | PDPR | pyruvate dehydrogenase phosphatase regulatory subunit [Source:HGNC Symbol;Acc:HGNC:30264] | 100330789 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000088959 | pdxka | pyridoxal (pyridoxine, vitamin B6) kinase a [Source:ZFIN;Acc:ZDB-GENE-030131-8376] | 791775 | YES | YES | Cytoplasm | kinase | |||||
| ENSDARG00000036546 | pdxkb | pyridoxal (pyridoxine, vitamin B6) kinase b [Source:ZFIN;Acc:ZDB-GENE-030616-521] | 564697 | YES | YES | Cytoplasm | kinase | |||||
| ENSDARG00000088699 | pdzd8 | PDZ domain containing 8 [Source:ZFIN;Acc:ZDB-GENE-110310-2] | 561279 | YES | YES | Extracellular Space | other | |||||
| ENSDARG00000079771 | PEA15 | phosphoprotein enriched in astrocytes 15 [Source:HGNC Symbol;Acc:HGNC:8822] | 557745 | YES | YES | Cytoplasm | transporter | |||||
| ENSDARG00000042069 | pebp1 | phosphatidylethanolamine binding protein 1 [Source:ZFIN;Acc:ZDB-GENE-040426-2621] | 406627 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000102249 | PEPD | peptidase D [Source:HGNC Symbol;Acc:HGNC:8840] | 0 | YES | YES | Cytoplasm | peptidase | |||||
| ENSDARG00000025559 | pex11g | peroxisomal biogenesis factor 11 gamma [Source:ZFIN;Acc:ZDB-GENE-050913-79] | 563203 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000028322 | pex14 | peroxisomal biogenesis factor 14 [Source:ZFIN;Acc:ZDB-GENE-060130-169] | 559933 | YES | YES | Cytoplasm | transcription regulator | |||||
| ENSDARG00000014536 | pfdn1 | prefoldin subunit 1 [Source:ZFIN;Acc:ZDB-GENE-050306-50] | 503766 | YES | YES | Cytoplasm | transcription regulator | |||||
| ENSDARG00000025391 | pfdn2 | prefoldin subunit 2 [Source:ZFIN;Acc:ZDB-GENE-060519-27] | 323012 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000035043 | pfdn5 | prefoldin 5 [Source:ZFIN;Acc:ZDB-GENE-030131-6858] | 568182 | YES | YES | Nucleus | transcription regulator | |||||
| ENSDARG00000088091 | pfn1 | profilin 1 [Source:ZFIN;Acc:ZDB-GENE-031002-33] | 799355 | YES | YES | NA | NA | |||||
| ENSDARG00000003952 | pfn2 | profilin 2 [Source:ZFIN;Acc:ZDB-GENE-040115-4] | 394235 | YES | YES | NA | NA | |||||
| ENSDARG00000012682 | pfn2l | profilin 2 like [Source:ZFIN;Acc:ZDB-GENE-030131-34] | 321383 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000005423 | pgam1a | phosphoglycerate mutase 1a [Source:ZFIN;Acc:ZDB-GENE-030131-1827] | 323107 | YES | YES | Cytoplasm | phosphatase | |||||
| ENSDARG00000014068 | pgam1b | phosphoglycerate mutase 1b [Source:ZFIN;Acc:ZDB-GENE-030131-5376] | 327165 | YES | YES | Cytoplasm | phosphatase | |||||
| ENSDARG00000015343 | pgd | phosphogluconate dehydrogenase [Source:ZFIN;Acc:ZDB-GENE-040426-2807] | 406762 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000013561 | pgm1 | phosphoglucomutase 1 [Source:ZFIN;Acc:ZDB-GENE-040426-1245] | 394000 | YES | YES | NA | NA | |||||
| ENSDARG00000030237 | pgrmc2 | progesterone receptor membrane component 2 [Source:ZFIN;Acc:ZDB-GENE-040426-2102] | 406378 | YES | YES | Nucleus | other | |||||
| ENSDARG00000075857 | PGS1 | phosphatidylglycerophosphate synthase 1 [Source:HGNC Symbol;Acc:HGNC:30029] | 0 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000086740 | phyh | phytanoyl-CoA 2-hydroxylase [Source:ZFIN;Acc:ZDB-GENE-050417-361] | 550521 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000070498 | phyhiplb | phytanoyl-CoA 2-hydroxylase interacting protein-like b [Source:ZFIN;Acc:ZDB-GENE-040927-27] | 100004913 | YES | YES | NA | NA | |||||
| ENSDARG00000033666 | pi4k2a | phosphatidylinositol 4-kinase type 2 alpha [Source:ZFIN;Acc:ZDB-GENE-040426-2675] | 406667 | YES | YES | Cytoplasm | kinase | |||||
| ENSDARG00000062445 | pias1b | protein inhibitor of activated STAT, 1b [Source:ZFIN;Acc:ZDB-GENE-050419-202] | 562982 | YES | YES | Nucleus | transcription regulator | |||||
| ENSDARG00000024479 | pigk | phosphatidylinositol glycan anchor biosynthesis, class K [Source:ZFIN;Acc:ZDB-GENE-040625-162] | 415239 | YES | YES | Cytoplasm | peptidase | |||||
| ENSDARG00000056112 | pikfyve | phosphoinositide kinase, FYVE finger containing [Source:ZFIN;Acc:ZDB-GENE-030131-9636] | 337690 | YES | YES | Cytoplasm | kinase | |||||
| ENSDARG00000003776 | pip4k2aa | phosphatidylinositol-5-phosphate 4-kinase, type II, alpha a [Source:ZFIN;Acc:ZDB-GENE-080721-18] | 560360 | YES | YES | Cytoplasm | kinase | |||||
| ENSDARG00000100313 | pip5k1cb | phosphatidylinositol-4-phosphate 5-kinase, type I, gamma b [Source:ZFIN;Acc:ZDB-GENE-110408-21] | 100005033 | YES | YES | Plasma Membrane | kinase | |||||
| ENSDARG00000055591 | pipox | pipecolic acid oxidase [Source:ZFIN;Acc:ZDB-GENE-110627-2] | 558601 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000044091 | pitpnab | phosphatidylinositol transfer protein, alpha b [Source:ZFIN;Acc:ZDB-GENE-110408-51] | 100333060 | YES | YES | Cytoplasm | transporter | |||||
| ENSDARG00000038563 | pitrm1 | pitrilysin metallopeptidase 1 [Source:ZFIN;Acc:ZDB-GENE-040426-2876] | 406808 | YES | YES | Cytoplasm | peptidase | |||||
| ENSDARG00000079572 | plcd3b | phospholipase C, delta 3b [Source:ZFIN;Acc:ZDB-GENE-081222-2] | 562569 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000067676 | plcl1 | phospholipase C-like 1 [Source:ZFIN;Acc:ZDB-GENE-090313-194] | 563105 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000060372 | plxna2 | plexin A2 [Source:ZFIN;Acc:ZDB-GENE-090311-6] | 561285 | YES | YES | Plasma Membrane | transmembrane receptor | |||||
| ENSDARG00000036985 | plxnb2b | plexin b2b [Source:ZFIN;Acc:ZDB-GENE-080902-1] | 0 | YES | YES | Plasma Membrane | transmembrane receptor | |||||
| ENSDARG00000032990 | plxnb3 | plexin B3 [Source:ZFIN;Acc:ZDB-GENE-081104-53] | 566546 | YES | YES | Plasma Membrane | transmembrane receptor | |||||
| ENSDARG00000062096 | pm20d1.2 | peptidase M20 domain containing 1, tandem duplicate 2 [Source:ZFIN;Acc:ZDB-GENE-061013-637] | 768158 | YES | YES | Cytoplasm | peptidase | |||||
| ENSDARG00000040942 | pnp6 | purine nucleoside phosphorylase 6 [Source:ZFIN;Acc:ZDB-GENE-040426-1800] | 402953 | YES | YES | NA | NA | |||||
| ENSDARG00000090444 | ponzr1 | plac8 onzin related protein 1 [Source:ZFIN;Acc:ZDB-GENE-070727-1] | 567858 | YES | YES | NA | NA | |||||
| ENSDARG00000055092 | pora | P450 (cytochrome) oxidoreductase a [Source:ZFIN;Acc:ZDB-GENE-050809-121] | 568202 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000059035 | porb | P450 (cytochrome) oxidoreductase b [Source:ZFIN;Acc:ZDB-GENE-030131-5767] | 327556 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000030441 | ppa1b | pyrophosphatase (inorganic) 1b [Source:ZFIN;Acc:ZDB-GENE-050417-375] | 550531 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000009685 | ppa2 | pyrophosphatase (inorganic) 2 [Source:ZFIN;Acc:ZDB-GENE-040426-1857] | 402961 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000069940 | ppap2d | phosphatidic acid phosphatase type 2D [Source:ZFIN;Acc:ZDB-GENE-061201-42] | 559124 | YES | YES | NA | NA | |||||
| ENSDARG00000104343 | PPIC | peptidylprolyl isomerase C (cyclophilin C) [Source:HGNC Symbol;Acc:HGNC:9256] | 100333141 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000038835 | ppid | peptidylprolyl isomerase D [Source:ZFIN;Acc:ZDB-GENE-040625-34] | 415155 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000032155 | ppm1aa | protein phosphatase, Mg2+/Mn2+ dependent, 1Aa [Source:ZFIN;Acc:ZDB-GENE-991102-15] | 30704 | YES | YES | Cytoplasm | phosphatase | |||||
| ENSDARG00000043250 | ppm1ab | protein phosphatase, Mg2+/Mn2+ dependent, 1Ab [Source:ZFIN;Acc:ZDB-GENE-991102-14] | 30703 | YES | YES | Cytoplasm | phosphatase | |||||
| ENSDARG00000074690 | ppm1j | protein phosphatase, Mg2+/Mn2+ dependent, 1J [Source:ZFIN;Acc:ZDB-GENE-091230-9] | 556950 | YES | YES | Other | phosphatase | |||||
| ENSDARG00000099241 | ppp2cb | protein phosphatase 2, catalytic subunit, beta isozyme [Source:ZFIN;Acc:ZDB-GENE-040426-2487] | 406582 | YES | YES | NA | NA | |||||
| ENSDARG00000021996 | ppp2r2aa | protein phosphatase 2, regulatory subunit B, alpha a [Source:ZFIN;Acc:ZDB-GENE-130530-565] | 558752 | YES | YES | Cytoplasm | phosphatase | |||||
| ENSDARG00000031985 | ppp2r4 | protein phosphatase 2A activator, regulatory subunit 4 [Source:ZFIN;Acc:ZDB-GENE-040801-11] | 445031 | YES | YES | Cytoplasm | phosphatase | |||||
| ENSDARG00000034313 | ppp5c | protein phosphatase 5, catalytic subunit [Source:ZFIN;Acc:ZDB-GENE-050327-75] | 541536 | YES | YES | Nucleus | phosphatase | |||||
| ENSDARG00000002949 | ppp6c | protein phosphatase 6, catalytic subunit [Source:ZFIN;Acc:ZDB-GENE-040426-949] | 393980 | YES | YES | Nucleus | phosphatase | |||||
| ENSDARG00000039980 | ppt1 | palmitoyl-protein thioesterase 1 (ceroid-lipofuscinosis, neuronal 1, infantile) [Source:ZFIN;Acc:ZDB-GENE-040426-2653] | 406648 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000032535 | praf2 | PRA1 domain family, member 2 [Source:ZFIN;Acc:ZDB-GENE-050417-190] | 550393 | YES | YES | Other | other | |||||
| ENSDARG00000069013 | prdx4 | peroxiredoxin 4 [Source:ZFIN;Acc:ZDB-GENE-030131-1096] | 570477 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000043511 | prdx6 | peroxiredoxin 6 [Source:ZFIN;Acc:ZDB-GENE-040426-1778] | 393778 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000057151 | prep | prolyl endopeptidase [Source:ZFIN;Acc:ZDB-GENE-050522-14] | 553791 | YES | YES | Cytoplasm | peptidase | |||||
| ENSDARG00000036446 | PRKAR2B | protein kinase, cAMP-dependent, regulatory, type II, beta [Source:HGNC Symbol;Acc:HGNC:9392] | 0 | YES | YES | Cytoplasm | kinase | |||||
| ENSDARG00000004470 | prkcsh | protein kinase C substrate 80K-H [Source:ZFIN;Acc:ZDB-GENE-040426-770] | 394028 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000010246 | prmt1 | protein arginine methyltransferase 1 [Source:ZFIN;Acc:ZDB-GENE-030131-693] | 321974 | YES | YES | Nucleus | enzyme | |||||
| ENSDARG00000034007 | prom1b | prominin 1 b [Source:ZFIN;Acc:ZDB-GENE-031003-1] | 378834 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000060288 | prosc | proline synthetase co-transcribed homolog (bacterial) [Source:ZFIN;Acc:ZDB-GENE-030131-1378] | 563548 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000015524 | prps1a | phosphoribosyl pyrophosphate synthetase 1A [Source:ZFIN;Acc:ZDB-GENE-011212-5] | 140620 | YES | YES | Other | kinase | |||||
| ENSDARG00000062208 | prrt1 | proline-rich transmembrane protein 1 [Source:ZFIN;Acc:ZDB-GENE-060503-686] | 559734 | YES | YES | Other | other | |||||
| ENSDARG00000076602 | prss16 | protease, serine, 16 (thymus) [Source:ZFIN;Acc:ZDB-GENE-070112-732] | 556307 | YES | YES | Extracellular Space | peptidase | |||||
| ENSDARG00000016733 | psat1 | phosphoserine aminotransferase 1 [Source:ZFIN;Acc:ZDB-GENE-030131-5723] | 327512 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000004870 | psen1 | presenilin 1 [Source:ZFIN;Acc:ZDB-GENE-991119-4] | 30221 | YES | YES | Plasma Membrane | peptidase | |||||
| ENSDARG00000013966 | psma6b | proteasome (prosome, macropain) subunit, alpha type, 6b [Source:ZFIN;Acc:ZDB-GENE-040718-329] | 436862 | YES | YES | Cytoplasm | peptidase | |||||
| ENSDARG00000010965 | psma8 | proteasome (prosome, macropain) subunit, alpha type, 8 [Source:ZFIN;Acc:ZDB-GENE-040426-2194] | 406445 | YES | YES | Nucleus | peptidase | |||||
| ENSDARG00000054696 | psmb4 | proteasome (prosome, macropain) subunit, beta type, 4 [Source:ZFIN;Acc:ZDB-GENE-050417-333] | 550499 | YES | YES | Cytoplasm | peptidase | |||||
| ENSDARG00000037962 | psmb7 | proteasome (prosome, macropain) subunit, beta type, 7 [Source:ZFIN;Acc:ZDB-GENE-001208-4] | 64278 | YES | YES | Cytoplasm | peptidase | |||||
| ENSDARG00000043561 | psmc1b | proteasome (prosome, macropain) 26S subunit, ATPase, 1b [Source:ZFIN;Acc:ZDB-GENE-040625-69] | 415181 | YES | YES | Nucleus | peptidase | |||||
| ENSDARG00000003189 | psmd1 | proteasome (prosome, macropain) 26S subunit, non-ATPase, 1 [Source:ZFIN;Acc:ZDB-GENE-040426-810] | 554967 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000005134 | psmd11b | proteasome (prosome, macropain) 26S subunit, non-ATPase, 11b [Source:ZFIN;Acc:ZDB-GENE-030131-984] | 322265 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000052377 | psmd12 | proteasome (prosome, macropain) 26S subunit, non-ATPase, 12 [Source:ZFIN;Acc:ZDB-GENE-030131-617] | 321898 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000063100 | psmd14 | proteasome (prosome, macropain) 26S subunit, non-ATPase, 14 [Source:ZFIN;Acc:ZDB-GENE-070410-56] | 799058 | YES | YES | Cytoplasm | peptidase | |||||
| ENSDARG00000103190 | PSMD2 (2 of 2) | proteasome (prosome, macropain) 26S subunit, non-ATPase, 2 [Source:HGNC Symbol;Acc:HGNC:9559] | 0 | YES | YES | Other | other | |||||
| ENSDARG00000023279 | psmd4b | proteasome (prosome, macropain) 26S subunit, non-ATPase, 4b [Source:ZFIN;Acc:ZDB-GENE-050417-96] | 550287 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000020454 | psmd8 | proteasome (prosome, macropain) 26S subunit, non-ATPase, 8 [Source:ZFIN;Acc:ZDB-GENE-040625-136] | 415221 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000012588 | ptdss1a | phosphatidylserine synthase 1a [Source:ZFIN;Acc:ZDB-GENE-040426-837] | 393931 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000063375 | pter | phosphotriesterase related [Source:ZFIN;Acc:ZDB-GENE-060825-190] | 751664 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000078172 | PTGFRN (2 of 2) | prostaglandin F2 receptor inhibitor [Source:HGNC Symbol;Acc:HGNC:9601] | 101883373 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000024877 | ptgr1 | prostaglandin reductase 1 [Source:ZFIN;Acc:ZDB-GENE-041212-80] | 494108 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000011863 | ptk7a | protein tyrosine kinase 7a [Source:ZFIN;Acc:ZDB-GENE-050522-216] | 553690 | YES | YES | Plasma Membrane | kinase | |||||
| ENSDARG00000101766 | ptmab | prothymosin, alpha b [Source:ZFIN;Acc:ZDB-GENE-030131-8681] | 559194 | YES | YES | NA | NA | |||||
| ENSDARG00000035676 | ptp4a2b | protein tyrosine phosphatase type IVA, member 2b [Source:ZFIN;Acc:ZDB-GENE-041114-111] | 334483 | YES | YES | Cytoplasm | phosphatase | |||||
| ENSDARG00000013860 | ptpmt1 | protein tyrosine phosphatase, mitochondrial 1 [Source:ZFIN;Acc:ZDB-GENE-070112-272] | 567019 | YES | YES | NA | NA | |||||
| ENSDARG00000070124 | ptpn23a | protein tyrosine phosphatase, non-receptor type 23, a [Source:ZFIN;Acc:ZDB-GENE-100921-24] | 100007322 | YES | YES | Cytoplasm | phosphatase | |||||
| ENSDARG00000000183 | ptpn4b | protein tyrosine phosphatase, non-receptor type 4b [Source:ZFIN;Acc:ZDB-GENE-030131-6259] | 407634 | YES | YES | Cytoplasm | phosphatase | |||||
| ENSDARG00000074866 | ptpn5 | protein tyrosine phosphatase, non-receptor type 5 [Source:ZFIN;Acc:ZDB-GENE-101028-3] | 559524 | YES | YES | Plasma Membrane | phosphatase | |||||
| ENSDARG00000071437 | ptprc | protein tyrosine phosphatase, receptor type, C [Source:ZFIN;Acc:ZDB-GENE-050208-585] | 0 | YES | YES | Plasma Membrane | phosphatase | |||||
| ENSDARG00000015891 | ptprea | protein tyrosine phosphatase, receptor type, E, a [Source:ZFIN;Acc:ZDB-GENE-100714-1] | 100499203 | YES | YES | Plasma Membrane | phosphatase | |||||
| ENSDARG00000002768 | pvalb2 | parvalbumin 2 [Source:ZFIN;Acc:ZDB-GENE-000322-4] | 58028 | YES | YES | NA | NA | |||||
| ENSDARG00000024433 | pvalb4 | parvalbumin 4 [Source:ZFIN;Acc:ZDB-GENE-040625-48] | 337731 | YES | YES | NA | NA | |||||
| ENSDARG00000009311 | pvalb6 | parvalbumin 6 [Source:ZFIN;Acc:ZDB-GENE-040912-95] | 402806 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000034705 | pvalb7 | parvalbumin 7 [Source:ZFIN;Acc:ZDB-GENE-040718-285] | 402807 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000063390 | pvrl2l | poliovirus receptor-related 2 like [Source:ZFIN;Acc:ZDB-GENE-060503-249] | 560933 | YES | YES | Plasma Membrane | transmembrane receptor | |||||
| ENSDARG00000002021 | pygb | phosphorylase, glycogen; brain [Source:ZFIN;Acc:ZDB-GENE-040928-2] | 403051 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000040190 | qdpra | quinoid dihydropteridine reductase a [Source:ZFIN;Acc:ZDB-GENE-070705-197] | 566323 | YES | YES | NA | NA | |||||
| ENSDARG00000041450 | rab11a | RAB11a, member RAS oncogene family [Source:ZFIN;Acc:ZDB-GENE-041114-53] | 492487 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000055291 | rab18a | RAB18A, member RAS oncogene family [Source:ZFIN;Acc:ZDB-GENE-030131-3980] | 325255 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000098344 | rab18b | RAB18B, member RAS oncogene family [Source:ZFIN;Acc:ZDB-GENE-040801-185] | 445055 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000056479 | rab1bb | RAB1B, member RAS oncogene family b [Source:ZFIN;Acc:ZDB-GENE-040625-133] | 415219 | YES | YES | NA | NA | |||||
| ENSDARG00000015807 | rab22a | RAB22A, member RAS oncogene family [Source:ZFIN;Acc:ZDB-GENE-041114-161] | 100001531 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000004151 | rab23 | RAB23, member RAS oncogene family [Source:ZFIN;Acc:ZDB-GENE-050522-328] | 553778 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000054032 | rab24 | RAB24, member RAS oncogene family [Source:ZFIN;Acc:ZDB-GENE-050706-110] | 574423 | YES | YES | Other | other | |||||
| ENSDARG00000087762 | RAB27B | RAB27B, member RAS oncogene family [Source:HGNC Symbol;Acc:HGNC:9767] | 561717 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000037884 | rab30 | RAB30, member RAS oncogene family [Source:ZFIN;Acc:ZDB-GENE-040718-250] | 437023 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000020128 | RAB31 | RAB31, member RAS oncogene family [Source:HGNC Symbol;Acc:HGNC:9771] | 568800 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000057394 | rab33a | RAB33A, member RAS oncogene family [Source:ZFIN;Acc:ZDB-GENE-060825-283] | 751718 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000014462 | rab3c | RAB3C, member RAS oncogene family [Source:ZFIN;Acc:ZDB-GENE-040718-263] | 436803 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000029885 | rab41 | RAB41, member RAS oncogene family [Source:ZFIN;Acc:ZDB-GENE-040426-2686] | 406674 | YES | YES | NA | NA | |||||
| ENSDARG00000019144 | rab4a | RAB4a, member RAS oncogene family [Source:ZFIN;Acc:ZDB-GENE-040525-1] | 403031 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000012177 | rab4b | RAB4B, member RAS oncogene family [Source:ZFIN;Acc:ZDB-GENE-040822-15] | 445498 | YES | YES | NA | NA | |||||
| ENSDARG00000103714 | rab6a | RAB6A, member RAS oncogene family [Source:ZFIN;Acc:ZDB-GENE-040426-2849] | 793785 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000031343 | rab6bb | RAB6B, member RAS oncogene family b [Source:ZFIN;Acc:ZDB-GENE-050320-28] | 541339 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000020497 | rab7 | RAB7, member RAS oncogene family [Source:ZFIN;Acc:ZDB-GENE-040426-1352] | 393902 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000045027 | rab9a | RAB9A, member RAS oncogene family [Source:ZFIN;Acc:ZDB-GENE-060825-293] | 751756 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000103391 | rab9b | RAB9B, member RAS oncogene family [Source:ZFIN;Acc:ZDB-GENE-091118-112] | 100003363 | YES | YES | Plasma Membrane | enzyme | |||||
| ENSDARG00000059600 | rabep1 | rabaptin, RAB GTPase binding effector protein 1 [Source:ZFIN;Acc:ZDB-GENE-060526-165] | 568064 | YES | YES | Cytoplasm | transporter | |||||
| ENSDARG00000098967 | RABGAP1 | RAB GTPase activating protein 1 [Source:HGNC Symbol;Acc:HGNC:17155] | 559632 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000002690 | rabif | RAB interacting factor [Source:ZFIN;Acc:ZDB-GENE-040718-216] | 436784 | YES | YES | Other | transporter | |||||
| ENSDARG00000058984 | rad23ab | RAD23 homolog Ab (S. cerevisiae) [Source:ZFIN;Acc:ZDB-GENE-051113-160] | 562839 | YES | YES | NA | NA | |||||
| ENSDARG00000008867 | rap1b | RAP1B, member of RAS oncogene family [Source:ZFIN;Acc:ZDB-GENE-030131-9662] | 321107 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000018069 | rdh12 | retinol dehydrogenase 12 (all-trans/9-cis/11-cis) [Source:ZFIN;Acc:ZDB-GENE-040718-9] | 436597 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000002467 | rdh14b | retinol dehydrogenase 14b (all-trans/9-cis/11-cis) [Source:ZFIN;Acc:ZDB-GENE-030131-6605] | 334665 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000028048 | rdh8a | retinol dehydrogenase 8a [Source:ZFIN;Acc:ZDB-GENE-021115-3] | 280648 | YES | YES | Plasma Membrane | enzyme | |||||
| ENSDARG00000002877 | reps1 | RALBP1 associated Eps domain containing 1 [Source:ZFIN;Acc:ZDB-GENE-030131-9536] | 564535 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000098645 | rgn | regucalcin [Source:ZFIN;Acc:ZDB-GENE-040718-68] | 403070 | YES | YES | Nucleus | enzyme | |||||
| ENSDARG00000015627 | rgs6 | regulator of G-protein signaling 6 [Source:ZFIN;Acc:ZDB-GENE-030131-31] | 562667 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000090213 | rheb | Ras homolog enriched in brain [Source:ZFIN;Acc:ZDB-GENE-040426-1690] | 0 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000005547 | rint1 | RAD50 interactor 1 [Source:ZFIN;Acc:ZDB-GENE-041210-174] | 562894 | YES | YES | Nucleus | other | |||||
| ENSDARG00000056639 | rltpr | RGD motif, leucine rich repeats, tropomodulin domain and proline-rich containing [Source:ZFIN;Acc:ZDB-GENE-050419-197] | 567758 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000074507 | rmdn1 | regulator of microtubule dynamics 1 [Source:ZFIN;Acc:ZDB-GENE-070928-17] | 558162 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000010442 | rnf11a | ring finger protein 11a [Source:ZFIN;Acc:ZDB-GENE-040426-1277] | 393996 | YES | YES | NA | NA | |||||
| ENSDARG00000026784 | robo1 | roundabout, axon guidance receptor, homolog 1 (Drosophila) [Source:ZFIN;Acc:ZDB-GENE-000209-3] | 30769 | YES | YES | Plasma Membrane | transmembrane receptor | |||||
| ENSDARG00000007480 | rpe65a | retinal pigment epithelium-specific protein 65a [Source:ZFIN;Acc:ZDB-GENE-040426-1717] | 393724 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000054420 | rpe65c | retinal pigment epithelium-specific protein 65c [Source:ZFIN;Acc:ZDB-GENE-081104-505] | 100004076 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000063436 | RPH3A | rabphilin 3A [Source:HGNC Symbol;Acc:HGNC:17056] | 101886567 | YES | YES | Plasma Membrane | transporter | |||||
| ENSDARG00000006553 | rras | related RAS viral (r-ras) oncogene homolog [Source:ZFIN;Acc:ZDB-GENE-041010-217] | 449660 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000027236 | rs1a | retinoschisin 1a [Source:ZFIN;Acc:ZDB-GENE-040801-171] | 445044 | YES | YES | Extracellular Space | other | |||||
| ENSDARG00000044601 | rtn4a | reticulon 4a [Source:ZFIN;Acc:ZDB-GENE-030710-1] | 447816 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000031366 | rtn4ip1 | reticulon 4 interacting protein 1 [Source:ZFIN;Acc:ZDB-GENE-040426-1314] | 393323 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000025254 | s100a10b | S100 calcium binding protein A10b [Source:ZFIN;Acc:ZDB-GENE-040426-1937] | 406276 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000103403 | sar1b | secretion associated, Ras related GTPase 1B [Source:ZFIN;Acc:ZDB-GENE-040426-1958] | 554177 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000010279 | scamp2 | secretory carrier membrane protein 2 [Source:ZFIN;Acc:ZDB-GENE-040426-2702] | 406687 | YES | YES | Cytoplasm | transporter | |||||
| ENSDARG00000016765 | scamp3 | secretory carrier membrane protein 3 [Source:ZFIN;Acc:ZDB-GENE-041114-33] | 492473 | YES | YES | Cytoplasm | transporter | |||||
| ENSDARG00000010531 | scamp4 | secretory carrier membrane protein 4 [Source:ZFIN;Acc:ZDB-GENE-050913-40] | 562881 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000018743 | scamp5a | secretory carrier membrane protein 5a [Source:ZFIN;Acc:ZDB-GENE-030131-3188] | 324467 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000098312 | scarb2 | scavenger receptor class B, member 2 [Source:ZFIN;Acc:ZDB-GENE-020419-29] | 192340 | YES | YES | NA | NA | |||||
| ENSDARG00000000779 | scfd1 | sec1 family domain containing 1 [Source:ZFIN;Acc:ZDB-GENE-030116-3] | 317732 | YES | YES | Cytoplasm | transporter | |||||
| ENSDARG00000077045 | scg5 | secretogranin V [Source:ZFIN;Acc:ZDB-GENE-040426-1687] | 393699 | YES | YES | Extracellular Space | other | |||||
| ENSDARG00000101713 | scn2b | sodium channel, voltage-gated, type II, beta [Source:ZFIN;Acc:ZDB-GENE-070920-1] | 777666 | YES | YES | Plasma Membrane | ion channel | |||||
| ENSDARG00000045555 | sco2 | SCO2 cytochrome c oxidase assembly protein [Source:ZFIN;Acc:ZDB-GENE-041210-173] | 606683 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000012194 | scp2a | sterol carrier protein 2a [Source:ZFIN;Acc:ZDB-GENE-040426-1846] | 393839 | YES | YES | Cytoplasm | transporter | |||||
| ENSDARG00000043497 | scrn2 | secernin 2 [Source:ZFIN;Acc:ZDB-GENE-040724-75] | 558306 | YES | YES | Extracellular Space | other | |||||
| ENSDARG00000016611 | scrn3 | secernin 3 [Source:ZFIN;Acc:ZDB-GENE-030131-4783] | 326058 | YES | YES | Other | other | |||||
| ENSDARG00000070682 | sdf4 | stromal cell derived factor 4 [Source:ZFIN;Acc:ZDB-GENE-040426-2061] | 406360 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000038608 | sdhc | succinate dehydrogenase complex, subunit C, integral membrane protein [Source:ZFIN;Acc:ZDB-GENE-040801-26] | 445129 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000099617 | sdr39u1 | short chain dehydrogenase/reductase family 39U, member 1 [Source:ZFIN;Acc:ZDB-GENE-060929-104] | 767637 | YES | YES | Nucleus | other | |||||
| ENSDARG00000103516 | sec24c | SEC24 family, member C (S. cerevisiae) [Source:ZFIN;Acc:ZDB-GENE-030131-4487] | 571870 | YES | YES | Cytoplasm | transporter | |||||
| ENSDARG00000021669 | sec61a1 | Sec61 alpha 1 subunit (S. cerevisiae) [Source:ZFIN;Acc:ZDB-GENE-020418-2] | 192298 | YES | YES | Cytoplasm | transporter | |||||
| ENSDARG00000004581 | sel1l | sel-1 suppressor of lin-12-like (C. elegans) [Source:ZFIN;Acc:ZDB-GENE-041001-122] | 568615 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000024717 | selenbp1 | selenium binding protein 1 [Source:ZFIN;Acc:ZDB-GENE-040426-1436] | 393542 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000100823 | selk | selenoprotein K [Source:ZFIN;Acc:ZDB-GENE-040912-131] | 791738 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000058354 | selt1a | selenoprotein T, 1a [Source:ZFIN;Acc:ZDB-GENE-030327-6] | 352919 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000023220 | selt2 | selenoprotein T, 2 [Source:ZFIN;Acc:ZDB-GENE-030411-4] | 0 | YES | YES | NA | NA | |||||
| ENSDARG00000090286 | serpina1 | serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1 [Source:ZFIN;Acc:ZDB-GENE-030131-1421] | 322701 | YES | YES | Other | other | |||||
| ENSDARG00000090850 | serpina1l | serine (or cysteine) proteinase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1, like [Source:ZFIN;Acc:ZDB-GENE-040721-3] | 321195 | YES | YES | Other | other | |||||
| ENSDARG00000091801 | serpinb14 | serpin peptidase inhibitor, clade B (ovalbumin), member 14 [Source:ZFIN;Acc:ZDB-GENE-050506-148] | 569051 | YES | YES | Other | other | |||||
| ENSDARG00000100594 | sez6l | seizure related 6 homolog (mouse)-like [Source:ZFIN;Acc:ZDB-GENE-091204-162] | 0 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000076052 | sez6l2 | seizure related 6 homolog (mouse)-like 2 [Source:ZFIN;Acc:ZDB-GENE-091204-189] | 557847 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000058302 | sh3bgrl | SH3 domain binding glutamate-rich protein like [Source:ZFIN;Acc:ZDB-GENE-040426-1376] | 394184 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000098075 | sh3bgrl3 | SH3 domain binding glutamate-rich protein like 3 [Source:ZFIN;Acc:ZDB-GENE-040426-1059] | 393248 | YES | YES | Nucleus | other | |||||
| ENSDARG00000007136 | sh3bp5lb | SH3-binding domain protein 5-like, b [Source:ZFIN;Acc:ZDB-GENE-060503-310] | 558670 | YES | YES | Other | other | |||||
| ENSDARG00000002642 | sh3gl1b | SH3-domain GRB2-like 1b [Source:ZFIN;Acc:ZDB-GENE-031001-6] | 378838 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000013360 | sh3gl3a | SH3-domain GRB2-like 3a [Source:ZFIN;Acc:ZDB-GENE-070112-242] | 571626 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000045294 | sh3glb1a | SH3-domain GRB2-like endophilin B1a [Source:ZFIN;Acc:ZDB-GENE-050417-89] | 792229 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000008983 | sh3glb2a | SH3-domain GRB2-like endophilin B2a [Source:ZFIN;Acc:ZDB-GENE-061201-8] | 402885 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000035470 | sh3glb2b | SH3-domain GRB2-like endophilin B2b [Source:ZFIN;Acc:ZDB-GENE-040426-833] | 394094 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000088247 | si:ch1073-303k11.2 | si:ch1073-303k11.2 [Source:ZFIN;Acc:ZDB-GENE-141216-99] | 100333296 | YES | YES | Other | other | |||||
| ENSDARG00000092970 | si:ch211-11c15.3 | si:ch211-11c15.3 [Source:ZFIN;Acc:ZDB-GENE-070126-5] | 566329 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000098644 | si:ch211-126j24.1 | si:ch211-126j24.1 [Source:ZFIN;Acc:ZDB-GENE-050208-133] | 571137 | YES | YES | NA | NA | |||||
| ENSDARG00000089477 | si:ch211-132g1.3 | si:ch211-132g1.3 [Source:ZFIN;Acc:ZDB-GENE-090312-148] | 103911757 | YES | YES | NA | NA | |||||
| ENSDARG00000078748 | si:ch211-137a8.4 | si:ch211-137a8.4 [Source:ZFIN;Acc:ZDB-GENE-030131-3742] | 560240 | YES | YES | NA | NA | |||||
| ENSDARG00000077087 | si:ch211-196g2.4 | si:ch211-196g2.4 [Source:ZFIN;Acc:ZDB-GENE-081104-160] | 0 | YES | YES | NA | NA | |||||
| ENSDARG00000054400 | si:ch211-198n5.11 | si:ch211-198n5.11 [Source:ZFIN;Acc:ZDB-GENE-050411-52] | 100004018 | YES | YES | NA | NA | |||||
| ENSDARG00000069595 | si:ch211-214c7.4 | si:ch211-214c7.4 [Source:ZFIN;Acc:ZDB-GENE-030131-6275] | 560699 | YES | YES | NA | NA | |||||
| ENSDARG00000076144 | si:ch211-74m13.3 | si:ch211-74m13.3 [Source:ZFIN;Acc:ZDB-GENE-060503-219] | 100334824 | YES | YES | NA | NA | |||||
| ENSDARG00000074282 | si:ch73-12o23.1 | si:ch73-12o23.1 [Source:ZFIN;Acc:ZDB-GENE-130530-749] | 100332784 | YES | YES | NA | NA | |||||
| ENSDARG00000091269 | si:ch73-27e22.8 | si:ch73-27e22.8 [Source:ZFIN;Acc:ZDB-GENE-100917-4] | 553357 | YES | YES | NA | NA | |||||
| ENSDARG00000036383 | si:ch73-335m24.5 | si:ch73-335m24.5 [Source:ZFIN;Acc:ZDB-GENE-100922-39] | 561209 | YES | YES | NA | NA | |||||
| ENSDARG00000100480 | si:ch73-364h19.2 | si:ch73-364h19.2 [Source:ZFIN;Acc:ZDB-GENE-131119-21] | 100537438 | YES | YES | Other | other | |||||
| ENSDARG00000098465 | si:ch73-366l1.5 | si:ch73-366l1.5 [Source:ZFIN;Acc:ZDB-GENE-131127-426] | 100003714 | YES | YES | NA | NA | |||||
| ENSDARG00000099799 | si:dkey-11f4.16 | si:dkey-11f4.16 [Source:ZFIN;Acc:ZDB-GENE-070912-357] | 0 | YES | YES | NA | NA | |||||
| ENSDARG00000004386 | si:dkey-153k10.9 | si:dkey-153k10.9 [Source:ZFIN;Acc:ZDB-GENE-030131-3893] | 566596 | YES | YES | Other | other | |||||
| ENSDARG00000037967 | si:dkey-16l2.16 | si:dkey-16l2.16 [Source:ZFIN;Acc:ZDB-GENE-141219-36] | 100003902 | YES | YES | NA | NA | |||||
| ENSDARG00000040463 | si:dkey-185e18.6 | si:dkey-185e18.6 [Source:ZFIN;Acc:ZDB-GENE-141216-55] | 556422 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000056080 | si:dkey-191g9.5 | si:dkey-191g9.5 [Source:ZFIN;Acc:ZDB-GENE-090313-217] | 100148873 | YES | YES | NA | NA | |||||
| ENSDARG00000089886 | si:dkey-19e4.5 | si:dkey-19e4.5 [Source:ZFIN;Acc:ZDB-GENE-030131-1852] | 570260 | YES | YES | NA | NA | |||||
| ENSDARG00000069537 | si:dkey-238c7.16 | si:dkey-238c7.16 [Source:ZFIN;Acc:ZDB-GENE-030131-4169] | 559078 | YES | YES | NA | NA | |||||
| ENSDARG00000078847 | si:dkey-238o13.4 | si:dkey-238o13.4 [Source:ZFIN;Acc:ZDB-GENE-070705-403] | 560780 | YES | YES | NA | NA | |||||
| ENSDARG00000090872 | si:dkey-276j7.1 | si:dkey-276j7.1 [Source:ZFIN;Acc:ZDB-GENE-060720-44] | 561769 | YES | YES | NA | NA | |||||
| ENSDARG00000002956 | si:dkey-32n7.4 | si:dkey-32n7.4 [Source:ZFIN;Acc:ZDB-GENE-110411-175] | 101882585 | YES | YES | Plasma Membrane | transmembrane receptor | |||||
| ENSDARG00000057262 | si:dkey-91i10.3 | si:dkey-91i10.3 [Source:ZFIN;Acc:ZDB-GENE-081105-124] | 565876 | YES | YES | NA | NA | |||||
| ENSDARG00000087165 | si:dkeyp-69c1.7 | si:dkeyp-69c1.7 [Source:ZFIN;Acc:ZDB-GENE-091204-69] | 566978 | YES | YES | NA | NA | |||||
| ENSDARG00000055731 | si:dkeyp-69e1.8 | si:dkeyp-69e1.8 [Source:ZFIN;Acc:ZDB-GENE-070912-676] | 100001340 | YES | YES | NA | NA | |||||
| ENSDARG00000071662 | si:rp71-36a1.3 | si:rp71-36a1.3 [Source:ZFIN;Acc:ZDB-GENE-050208-566] | 100034563 | YES | YES | NA | NA | |||||
| ENSDARG00000011418 | sigmar1 | sigma non-opioid intracellular receptor 1 [Source:ZFIN;Acc:ZDB-GENE-040426-1006] | 393952 | YES | YES | Plasma Membrane | transmembrane receptor | |||||
| ENSDARG00000011488 | sirt2 | sirtuin 2 (silent mating type information regulation 2, homolog) 2 (S. cerevisiae) [Source:ZFIN;Acc:ZDB-GENE-030131-1028] | 322309 | YES | YES | Nucleus | transcription regulator | |||||
| ENSDARG00000014378 | slc12a4 | solute carrier family 12 (potassium/chloride transporter), member 4 [Source:ZFIN;Acc:ZDB-GENE-090218-27] | 562826 | YES | YES | Plasma Membrane | transporter | |||||
| ENSDARG00000102793 | slc16a6a | solute carrier family 16, member 6a [Source:ZFIN;Acc:ZDB-GENE-110208-1] | 564872 | YES | YES | Plasma Membrane | transporter | |||||
| ENSDARG00000016480 | slc17a7a | solute carrier family 17 (vesicular glutamate transporter), member 7a [Source:ZFIN;Acc:ZDB-GENE-050105-5] | 795293 | YES | YES | Plasma Membrane | transporter | |||||
| ENSDARG00000090106 | slc17a7b | solute carrier family 17 (vesicular glutamate transporter), member 7b [Source:ZFIN;Acc:ZDB-GENE-131125-32] | 100331980 | YES | YES | Plasma Membrane | transporter | |||||
| ENSDARG00000033993 | SLC19A1 | solute carrier family 19 (folate transporter), member 1 [Source:HGNC Symbol;Acc:HGNC:10937] | 497434 | YES | YES | Plasma Membrane | transporter | |||||
| ENSDARG00000104431 | slc1a3a | solute carrier family 1 (glial high affinity glutamate transporter), member 3a [Source:ZFIN;Acc:ZDB-GENE-030131-2159] | 323439 | YES | YES | NA | NA | |||||
| ENSDARG00000000551 | slc1a4 | solute carrier family 1 (glutamate/neutral amino acid transporter), member 4 [Source:ZFIN;Acc:ZDB-GENE-030616-566] | 368885 | YES | YES | Plasma Membrane | transporter | |||||
| ENSDARG00000060796 | slc20a2 | solute carrier family 20 (phosphate transporter), member 2 [Source:ZFIN;Acc:ZDB-GENE-060929-828] | 558346 | YES | YES | Plasma Membrane | transporter | |||||
| ENSDARG00000078856 | slc22a23 | solute carrier family 22, member 23 [Source:ZFIN;Acc:ZDB-GENE-030131-3564] | 555979 | YES | YES | Other | transporter | |||||
| ENSDARG00000067509 | slc24a4b | solute carrier family 24 (sodium/potassium/calcium exchanger), member 4b [Source:ZFIN;Acc:ZDB-GENE-081031-75] | 100334346 | YES | YES | Plasma Membrane | transporter | |||||
| ENSDARG00000035468 | slc25a25b | solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25b [Source:ZFIN;Acc:ZDB-GENE-060526-340] | 569227 | YES | YES | Cytoplasm | transporter | |||||
| ENSDARG00000100731 | slc27a2b | solute carrier family 27 (fatty acid transporter), member 2b [Source:ZFIN;Acc:ZDB-GENE-081104-49] | 100004228 | YES | YES | Cytoplasm | transporter | |||||
| ENSDARG00000057272 | slc30a9 | solute carrier family 30 (zinc transporter), member 9 [Source:ZFIN;Acc:ZDB-GENE-040724-254] | 497184 | YES | YES | Nucleus | transporter | |||||
| ENSDARG00000013961 | slc31a1 | solute carrier family 31 (copper transporter), member 1 [Source:ZFIN;Acc:ZDB-GENE-040415-3] | 403028 | YES | YES | Plasma Membrane | transporter | |||||
| ENSDARG00000011945 | slc35e1 | solute carrier family 35, member E1 [Source:ZFIN;Acc:ZDB-GENE-040426-2033] | 406347 | YES | YES | Other | other | |||||
| ENSDARG00000053003 | slc35f1 | solute carrier family 35, member F1 [Source:ZFIN;Acc:ZDB-GENE-060503-463] | 555352 | YES | YES | Other | other | |||||
| ENSDARG00000045447 | slc35g2b | solute carrier family 35, member G2b [Source:ZFIN;Acc:ZDB-GENE-050320-142] | 541438 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000027065 | slc38a3a | solute carrier family 38, member 3a [Source:ZFIN;Acc:ZDB-GENE-070615-25] | 100073328 | YES | YES | Plasma Membrane | transporter | |||||
| ENSDARG00000055791 | SLC3A2 (3 of 3) | solute carrier family 3 (amino acid transporter heavy chain), member 2 [Source:HGNC Symbol;Acc:HGNC:11026] | 790938 | YES | YES | Plasma Membrane | transporter | |||||
| ENSDARG00000090982 | slc44a1b | solute carrier family 44 (choline transporter), member 1b [Source:ZFIN;Acc:ZDB-GENE-050309-63] | 0 | YES | YES | Plasma Membrane | transporter | |||||
| ENSDARG00000037059 | slc44a2 | solute carrier family 44 (choline transporter), member 2 [Source:ZFIN;Acc:ZDB-GENE-030131-3065] | 324345 | YES | YES | Extracellular Space | transporter | |||||
| ENSDARG00000068787 | slc6a17 | solute carrier family 6 (neutral amino acid transporter), member 17 [Source:ZFIN;Acc:ZDB-GENE-030131-8187] | 798635 | YES | YES | Cytoplasm | transporter | |||||
| ENSDARG00000061165 | slc6a4a | solute carrier family 6 (neurotransmitter transporter), member 4a [Source:ZFIN;Acc:ZDB-GENE-060314-1] | 664719 | YES | YES | Plasma Membrane | transporter | |||||
| ENSDARG00000067964 | slc6a5 | solute carrier family 6 (neurotransmitter transporter), member 5 [Source:ZFIN;Acc:ZDB-GENE-050105-2] | 494450 | YES | YES | Plasma Membrane | transporter | |||||
| ENSDARG00000020645 | slc7a3a | solute carrier family 7 (cationic amino acid transporter, y+ system), member 3a [Source:ZFIN;Acc:ZDB-GENE-041114-206] | 492363 | YES | YES | Plasma Membrane | transporter | |||||
| ENSDARG00000075831 | slc7a8b | solute carrier family 7 (amino acid transporter light chain, L system), member 8b [Source:ZFIN;Acc:ZDB-GENE-030616-586] | 100007704 | YES | YES | Plasma Membrane | transporter | |||||
| ENSDARG00000043406 | slc8a1b | solute carrier family 8 (sodium/calcium exchanger), member 1b [Source:ZFIN;Acc:ZDB-GENE-060110-3] | 797199 | YES | YES | Plasma Membrane | transporter | |||||
| ENSDARG00000037145 | slc8a4b | solute carrier family 8 (sodium/calcium exchanger), member 4b [Source:ZFIN;Acc:ZDB-GENE-050809-103] | 557499 | YES | YES | NA | NA | |||||
| ENSDARG00000000068 | slc9a3r1 | solute carrier family 9, subfamily A (NHE3, cation proton antiporter 3), member 3 regulator 1 [Source:ZFIN;Acc:ZDB-GENE-031006-7] | 327272 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000009209 | slc9a6a | solute carrier family 9, subfamily A (NHE6, cation proton antiporter 6), member 6a [Source:ZFIN;Acc:ZDB-GENE-070424-104] | 0 | YES | YES | Plasma Membrane | transporter | |||||
| ENSDARG00000076754 | slc9a7 | solute carrier family 9, subfamily A (NHE7, cation proton antiporter 7), member 7 [Source:ZFIN;Acc:ZDB-GENE-050913-8] | 554532 | YES | YES | Cytoplasm | transporter | |||||
| ENSDARG00000061525 | slka | STE20-like kinase a [Source:ZFIN;Acc:ZDB-GENE-050411-66] | 568838 | YES | YES | Nucleus | kinase | |||||
| ENSDARG00000054458 | slmapa | sarcolemma associated protein a [Source:ZFIN;Acc:ZDB-GENE-061013-184] | 572011 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000055288 | slmo1 | slowmo homolog 1 (Drosophila) [Source:ZFIN;Acc:ZDB-GENE-130530-654] | 563270 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000031302 | smap1 | small ArfGAP 1 [Source:ZFIN;Acc:ZDB-GENE-060920-2] | 777731 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000098226 | smpd3 | sphingomyelin phosphodiesterase 3, neutral [Source:ZFIN;Acc:ZDB-GENE-081104-280] | 568381 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000008155 | sms | spermine synthase [Source:ZFIN;Acc:ZDB-GENE-040625-150] | 80872 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000038067 | smurf2 | SMAD specific E3 ubiquitin protein ligase 2 [Source:ZFIN;Acc:ZDB-GENE-030131-1830] | 563633 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000038518 | snap29 | synaptosomal-associated protein 29 [Source:ZFIN;Acc:ZDB-GENE-041111-226] | 553460 | YES | YES | Cytoplasm | transporter | |||||
| ENSDARG00000034423 | sncga | synuclein, gamma a [Source:ZFIN;Acc:ZDB-GENE-050417-18] | 550229 | YES | YES | NA | NA | |||||
| ENSDARG00000103529 | snx1b | sorting nexin 1b [Source:ZFIN;Acc:ZDB-GENE-050913-82] | 619253 | YES | YES | Cytoplasm | transporter | |||||
| ENSDARG00000008678 | snx3 | sorting nexin 3 [Source:ZFIN;Acc:ZDB-GENE-030131-921] | 563511 | YES | YES | Cytoplasm | transporter | |||||
| ENSDARG00000058444 | snx6 | sorting nexin 6 [Source:ZFIN;Acc:ZDB-GENE-050913-152] | 567182 | YES | YES | Cytoplasm | transporter | |||||
| ENSDARG00000043848 | sod1 | superoxide dismutase 1, soluble [Source:ZFIN;Acc:ZDB-GENE-990415-258] | 30553 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000042644 | sod2 | superoxide dismutase 2, mitochondrial [Source:ZFIN;Acc:ZDB-GENE-030131-7742] | 335799 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000056252 | sort1b | sortilin 1b [Source:ZFIN;Acc:ZDB-GENE-030131-7447] | 799153 | YES | YES | Plasma Membrane | G-protein coupled receptor | |||||
| ENSDARG00000074531 | spag9a | sperm associated antigen 9a [Source:ZFIN;Acc:ZDB-GENE-070820-7] | 0 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000042823 | spcs2 | signal peptidase complex subunit 2 homolog (S. cerevisiae) [Source:ZFIN;Acc:ZDB-GENE-050320-32] | 541342 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000089976 | spcs3 | signal peptidase complex subunit 3 homolog (S. cerevisiae) [Source:ZFIN;Acc:ZDB-GENE-040426-2206] | 405884 | YES | YES | Cytoplasm | peptidase | |||||
| ENSDARG00000004406 | spra | sepiapterin reductase a [Source:ZFIN;Acc:ZDB-GENE-050522-412] | 554136 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000030949 | srprb | signal recognition particle receptor, B subunit [Source:ZFIN;Acc:ZDB-GENE-040718-311] | 436845 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000021973 | st13 | suppression of tumorigenicity 13 (colon carcinoma) (Hsp70 interacting protein) [Source:ZFIN;Acc:ZDB-GENE-030131-5395] | 564225 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000045748 | stab2 | stabilin 2 [Source:ZFIN;Acc:ZDB-GENE-041210-336] | 565194 | YES | YES | Plasma Membrane | transmembrane receptor | |||||
| ENSDARG00000002127 | stam | signal transducing adaptor molecule (SH3 domain and ITAM motif) 1 [Source:ZFIN;Acc:ZDB-GENE-031118-123] | 386967 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000076117 | STARD5 | StAR-related lipid transfer (START) domain containing 5 [Source:HGNC Symbol;Acc:HGNC:18065] | 0 | YES | YES | Cytoplasm | transporter | |||||
| ENSDARG00000006266 | stat1a | signal transducer and activator of transcription 1, 91kDa [Source:ZFIN;Acc:ZDB-GENE-980526-499] | 30768 | YES | YES | Nucleus | transcription regulator | |||||
| ENSDARG00000060723 | stim1a | stromal interaction molecule 1a [Source:ZFIN;Acc:ZDB-GENE-060503-914] | 556360 | YES | YES | Plasma Membrane | ion channel | |||||
| ENSDARG00000004906 | stip1 | stress-induced phosphoprotein 1 [Source:ZFIN;Acc:ZDB-GENE-041121-17] | 493606 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000051874 | stra6 | stimulated by retinoic acid 6 [Source:ZFIN;Acc:ZDB-GENE-060616-252] | 724007 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000001729 | strn3 | striatin, calmodulin binding protein 3 [Source:ZFIN;Acc:ZDB-GENE-030616-405] | 563124 | YES | YES | Nucleus | transcription regulator | |||||
| ENSDARG00000001890 | STT3B | STT3B, subunit of the oligosaccharyltransferase complex (catalytic) [Source:HGNC Symbol;Acc:HGNC:30611] | 0 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000098813 | stx12 | syntaxin 12 [Source:ZFIN;Acc:ZDB-GENE-040625-11] | 415141 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000052518 | stx4 | syntaxin 4 [Source:ZFIN;Acc:ZDB-GENE-030131-2455] | 323735 | YES | YES | Plasma Membrane | transporter | |||||
| ENSDARG00000008142 | stxbp3 | syntaxin binding protein 3 [Source:ZFIN;Acc:ZDB-GENE-040426-2833] | 406780 | YES | YES | Plasma Membrane | transporter | |||||
| ENSDARG00000006383 | stxbp5l | syntaxin binding protein 5-like [Source:ZFIN;Acc:ZDB-GENE-041210-220] | 567605 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000088862 | STXBP6 (2 of 2) | syntaxin binding protein 6 (amisyn) [Source:HGNC Symbol;Acc:HGNC:19666] | 0 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000052712 | suclg1 | succinate-CoA ligase, alpha subunit [Source:ZFIN;Acc:ZDB-GENE-040912-101] | 436850 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000044914 | suclg2 | succinate-CoA ligase, GDP-forming, beta subunit [Source:ZFIN;Acc:ZDB-GENE-030114-3] | 317746 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000035993 | sumo3b | small ubiquitin-like modifier 3b [Source:ZFIN;Acc:ZDB-GENE-040718-426] | 436950 | YES | YES | Nucleus | other | |||||
| ENSDARG00000003592 | surf1 | surfeit 1 [Source:ZFIN;Acc:ZDB-GENE-041108-2] | 557970 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000035131 | surf4l | surfeit gene 4, like [Source:ZFIN;Acc:ZDB-GENE-040718-172] | 436743 | YES | YES | NA | NA | |||||
| ENSDARG00000056833 | svopa | SV2 related protein homolog a (rat) [Source:ZFIN;Acc:ZDB-GENE-060526-336] | 559162 | YES | YES | Cytoplasm | transporter | |||||
| ENSDARG00000057286 | swap70b | SWAP switching B-cell complex 70b subunit 70b [Source:ZFIN;Acc:ZDB-GENE-030131-3587] | 562490 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000030775 | sybl1 | synaptobrevin-like 1 [Source:ZFIN;Acc:ZDB-GENE-040426-1055] | 393236 | YES | YES | Cytoplasm | transporter | |||||
| ENSDARG00000002564 | syngr1a | synaptogyrin 1a [Source:ZFIN;Acc:ZDB-GENE-041010-169] | 450047 | YES | YES | Plasma Membrane | transporter | |||||
| ENSDARG00000025034 | syngr3b | synaptogyrin 3b [Source:ZFIN;Acc:ZDB-GENE-070822-6] | 570094 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000028321 | synj2bp | synaptojanin 2 binding protein [Source:ZFIN;Acc:ZDB-GENE-030131-6531] | 334599 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000002230 | sypb | synaptophysin b [Source:ZFIN;Acc:ZDB-GENE-040718-205] | 436774 | YES | YES | Cytoplasm | transporter | |||||
| ENSDARG00000101776 | syt12 | synaptotagmin XII [Source:ZFIN;Acc:ZDB-GENE-081022-147] | 566397 | YES | YES | Plasma Membrane | transporter | |||||
| ENSDARG00000058394 | tagln3b | transgelin 3b [Source:ZFIN;Acc:ZDB-GENE-030131-4199] | 797861 | YES | YES | Extracellular Space | other | |||||
| ENSDARG00000103369 | TALDO1 | transaldolase 1 [Source:HGNC Symbol;Acc:HGNC:11559] | 0 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000100157 | TAMM41 | TAM41, mitochondrial translocator assembly and maintenance protein, homolog (S. cerevisiae) [Source:HGNC Symbol;Acc:HGNC:25187] | 0 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000094052 | taok3b | TAO kinase 3b [Source:ZFIN;Acc:ZDB-GENE-060526-59] | 555650 | YES | YES | Cytoplasm | kinase | |||||
| ENSDARG00000061143 | tapt1b | transmembrane anterior posterior transformation 1b [Source:ZFIN;Acc:ZDB-GENE-030131-4025] | 559710 | YES | YES | Plasma Membrane | G-protein coupled receptor | |||||
| ENSDARG00000098749 | tbcd | tubulin folding cofactor D [Source:ZFIN;Acc:ZDB-GENE-060823-1] | 553265 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000003641 | tfg | trk-fused gene [Source:ZFIN;Acc:ZDB-GENE-030131-8410] | 336466 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000101322 | tfr1a | transferrin receptor 1a [Source:ZFIN;Acc:ZDB-GENE-041220-1] | 494477 | YES | YES | Plasma Membrane | transporter | |||||
| ENSDARG00000077372 | tfr1b | transferrin receptor 1b [Source:ZFIN;Acc:ZDB-GENE-041220-2] | 494478 | YES | YES | Plasma Membrane | transporter | |||||
| ENSDARG00000103914 | THEM6 (2 of 2) | thioesterase superfamily member 6 [Source:HGNC Symbol;Acc:HGNC:29656] | 100142654 | YES | YES | Other | other | |||||
| ENSDARG00000013776 | thop1 | thimet oligopeptidase 1 [Source:ZFIN;Acc:ZDB-GENE-040718-47] | 436628 | YES | YES | Cytoplasm | peptidase | |||||
| ENSDARG00000029510 | timm17a | translocase of inner mitochondrial membrane 17 homolog A (yeast) [Source:ZFIN;Acc:ZDB-GENE-031030-6] | 0 | YES | YES | Cytoplasm | transporter | |||||
| ENSDARG00000104771 | timm44 | translocase of inner mitochondrial membrane 44 homolog (yeast) [Source:ZFIN;Acc:ZDB-GENE-030131-145] | 541380 | YES | YES | Cytoplasm | transporter | |||||
| ENSDARG00000031098 | timm50 | translocase of inner mitochondrial membrane 50 homolog (S. cerevisiae) [Source:ZFIN;Acc:ZDB-GENE-040426-1618] | 393638 | YES | YES | Cytoplasm | phosphatase | |||||
| ENSDARG00000006019 | tktb | transketolase b [Source:ZFIN;Acc:ZDB-GENE-030909-13] | 378713 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000003866 | tm9sf2 | transmembrane 9 superfamily member 2 [Source:ZFIN;Acc:ZDB-GENE-030131-6302] | 334370 | YES | YES | Plasma Membrane | transporter | |||||
| ENSDARG00000026908 | tmed2 | transmembrane emp24 domain trafficking protein 2 [Source:ZFIN;Acc:ZDB-GENE-030131-269] | 321550 | YES | YES | Cytoplasm | transporter | |||||
| ENSDARG00000052390 | tmed8 | transmembrane emp24 protein transport domain containing 8 [Source:ZFIN;Acc:ZDB-GENE-050522-283] | 553196 | YES | YES | Other | other | |||||
| ENSDARG00000034600 | tmem165 | transmembrane protein 165 [Source:ZFIN;Acc:ZDB-GENE-030131-3222] | 324501 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000076376 | tmem175 | transmembrane protein 175 [Source:ZFIN;Acc:ZDB-GENE-070705-79] | 100002086 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000092787 | TMEM261 | transmembrane protein 261 [Source:HGNC Symbol;Acc:HGNC:30536] | 558861 | YES | YES | Other | other | |||||
| ENSDARG00000030236 | tmem30aa | transmembrane protein 30Aa [Source:ZFIN;Acc:ZDB-GENE-040704-26] | 337397 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000043555 | tmem30ab | transmembrane protein 30Ab [Source:ZFIN;Acc:ZDB-GENE-030131-5884] | 333952 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000003655 | tmem59l | transmembrane protein 59-like [Source:ZFIN;Acc:ZDB-GENE-040426-2651] | 406646 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000077777 | tmsb4x | thymosin, beta 4 x [Source:ZFIN;Acc:ZDB-GENE-030131-8304] | 793949 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000007786 | tmx2b | thioredoxin-related transmembrane protein 2b [Source:ZFIN;Acc:ZDB-GENE-040625-105] | 415203 | YES | YES | Other | enzyme | |||||
| ENSDARG00000088466 | TMX3 (2 of 2) | thioredoxin-related transmembrane protein 3 [Source:HGNC Symbol;Acc:HGNC:24718] | 0 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000043416 | tnfaip2a | tumor necrosis factor, alpha-induced protein 2a [Source:ZFIN;Acc:ZDB-GENE-041210-301] | 567159 | YES | YES | Extracellular Space | other | |||||
| ENSDARG00000092112 | tomm40 | translocase of outer mitochondrial membrane 40 homolog (yeast) [Source:ZFIN;Acc:ZDB-GENE-030131-1365] | 322645 | YES | YES | Cytoplasm | ion channel | |||||
| ENSDARG00000036721 | tomm40l | translocase of outer mitochondrial membrane 40 homolog, like [Source:ZFIN;Acc:ZDB-GENE-040426-2319] | 405895 | YES | YES | Cytoplasm | ion channel | |||||
| ENSDARG00000097797 | tomm6 | translocase of outer mitochondrial membrane 6 homolog (yeast) [Source:ZFIN;Acc:ZDB-GENE-131121-445] | 100001229 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000079308 | tor3a | torsin family 3, member A [Source:ZFIN;Acc:ZDB-GENE-081104-165] | 792835 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000069430 | tp53i11a | tumor protein p53 inducible protein 11a [Source:ZFIN;Acc:ZDB-GENE-050306-58] | 503774 | YES | YES | Other | other | |||||
| ENSDARG00000025012 | tpi1a | triosephosphate isomerase 1a [Source:ZFIN;Acc:ZDB-GENE-020416-3] | 192309 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000078751 | TPP2 | tripeptidyl peptidase II [Source:HGNC Symbol;Acc:HGNC:12016] | 100331669 | YES | YES | Cytoplasm | peptidase | |||||
| ENSDARG00000068149 | tprg1l | tumor protein p63 regulated 1-like [Source:ZFIN;Acc:ZDB-GENE-070531-2] | 568867 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000092693 | tpt1 | tumor protein, translationally-controlled 1 [Source:ZFIN;Acc:ZDB-GENE-990603-10] | 30723 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000024317 | trap1 | TNF receptor-associated protein 1 [Source:ZFIN;Acc:ZDB-GENE-030131-4257] | 571959 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000027418 | trappc4 | trafficking protein particle complex 4 [Source:ZFIN;Acc:ZDB-GENE-040426-1734] | 393737 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000038866 | TRAPPC8 | trafficking protein particle complex 8 [Source:HGNC Symbol;Acc:HGNC:29169] | 570167 | YES | YES | Cytoplasm | transporter | |||||
| ENSDARG00000062013 | TRHDE (1 of 2) | thyrotropin-releasing hormone degrading enzyme [Source:HGNC Symbol;Acc:HGNC:30748] | 562409 | YES | YES | Plasma Membrane | peptidase | |||||
| ENSDARG00000078381 | trip11 | thyroid hormone receptor interactor 11 [Source:ZFIN;Acc:ZDB-GENE-030131-9833] | 100537975 | YES | YES | Cytoplasm | transcription regulator | |||||
| ENSDARG00000011897 | TSG101 (1 of 3) | tumor susceptibility 101 [Source:HGNC Symbol;Acc:HGNC:15971] | 100333057 | YES | YES | Cytoplasm | transcription regulator | |||||
| ENSDARG00000040854 | tsg101a | tumor susceptibility 101a [Source:ZFIN;Acc:ZDB-GENE-030217-1] | 415179 | YES | YES | Cytoplasm | transcription regulator | |||||
| ENSDARG00000041830 | tsn | translin [Source:ZFIN;Acc:ZDB-GENE-030131-6039] | 334107 | YES | YES | Nucleus | other | |||||
| ENSDARG00000008407 | tspan7b | tetraspanin 7b [Source:ZFIN;Acc:ZDB-GENE-040927-5] | 449539 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000074435 | ttc19 | tetratricopeptide repeat domain 19 [Source:ZFIN;Acc:ZDB-GENE-070912-490] | 0 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000068421 | ttc9b | tetratricopeptide repeat domain 9B [Source:ZFIN;Acc:ZDB-GENE-050419-67] | 559732 | YES | YES | Other | other | |||||
| ENSDARG00000007678 | ttyh3b | tweety family member 3b [Source:ZFIN;Acc:ZDB-GENE-051120-105] | 798421 | YES | YES | Plasma Membrane | ion channel | |||||
| ENSDARG00000104801 | tubb6 | tubulin, beta 6 class V [Source:ZFIN;Acc:ZDB-GENE-070424-195] | 550458 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000090481 | tusc5a | tumor suppressor candidate 5a [Source:ZFIN;Acc:ZDB-GENE-131121-306] | 101886018 | YES | YES | Other | other | |||||
| ENSDARG00000044125 | txn | thioredoxin [Source:ZFIN;Acc:ZDB-GENE-040718-162] | 436734 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000034777 | txn2 | thioredoxin 2 [Source:ZFIN;Acc:ZDB-GENE-040426-1795] | 402938 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000038980 | txndc12 | thioredoxin domain containing 12 (endoplasmic reticulum) [Source:ZFIN;Acc:ZDB-GENE-050522-497] | 324900 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000009342 | txndc5 | thioredoxin domain containing 5 [Source:ZFIN;Acc:ZDB-GENE-040426-1951] | 406289 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000011921 | txnl1 | thioredoxin-like 1 [Source:ZFIN;Acc:ZDB-GENE-040426-701] | 394113 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000063588 | uba5 | ubiquitin-like modifier activating enzyme 5 [Source:ZFIN;Acc:ZDB-GENE-031112-2] | 0 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000027141 | ube2l3b | ubiquitin-conjugating enzyme E2L 3b [Source:ZFIN;Acc:ZDB-GENE-030131-662] | 321943 | YES | YES | Nucleus | enzyme | |||||
| ENSDARG00000028198 | ube2v2 | ubiquitin-conjugating enzyme E2 variant 2 [Source:ZFIN;Acc:ZDB-GENE-040426-2919] | 406836 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000003087 | ubxn6 | UBX domain protein 6 [Source:ZFIN;Acc:ZDB-GENE-030131-5680] | 554960 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000030177 | uchl3 | ubiquitin carboxyl-terminal esterase L3 (ubiquitin thiolesterase) [Source:ZFIN;Acc:ZDB-GENE-050522-158] | 554104 | YES | YES | Cytoplasm | peptidase | |||||
| ENSDARG00000103404 | uchl5 | ubiquitin carboxyl-terminal hydrolase L5 [Source:ZFIN;Acc:ZDB-GENE-040426-2051] | 406357 | YES | YES | Cytoplasm | peptidase | |||||
| ENSDARG00000007496 | ufl1 | UFM1-specific ligase 1 [Source:ZFIN;Acc:ZDB-GENE-040426-1163] | 393578 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000058221 | ugcg | UDP-glucose ceramide glucosyltransferase [Source:ZFIN;Acc:ZDB-GENE-030131-9885] | 0 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000008200 | ugp2b | UDP-glucose pyrophosphorylase 2b [Source:ZFIN;Acc:ZDB-GENE-030131-6352] | 334420 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000037455 | ugt8 | UDP glycosyltransferase 8 [Source:ZFIN;Acc:ZDB-GENE-060616-129] | 553508 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000068176 | uqcc1 | ubiquinol-cytochrome c reductase complex assembly factor 1 [Source:ZFIN;Acc:ZDB-GENE-060929-1034] | 767788 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000019581 | uso1 | USO1 vesicle transport factor [Source:ZFIN;Acc:ZDB-GENE-040426-688] | 393124 | YES | YES | Cytoplasm | transporter | |||||
| ENSDARG00000078109 | usp12a | ubiquitin specific peptidase 12a [Source:ZFIN;Acc:ZDB-GENE-060228-3] | 565736 | YES | YES | Cytoplasm | peptidase | |||||
| ENSDARG00000014517 | usp5 | ubiquitin specific peptidase 5 (isopeptidase T) [Source:ZFIN;Acc:ZDB-GENE-040426-2584] | 406618 | YES | YES | Cytoplasm | peptidase | |||||
| ENSDARG00000013708 | usp9 | ubiquitin specific peptidase 9 [Source:ZFIN;Acc:ZDB-GENE-061019-1] | 568683 | YES | YES | Plasma Membrane | peptidase | |||||
| ENSDARG00000024116 | vamp8 | vesicle-associated membrane protein 8 (endobrevin) [Source:ZFIN;Acc:ZDB-GENE-030131-5215] | 327007 | YES | YES | Plasma Membrane | transporter | |||||
| ENSDARG00000006257 | vldlr | very low density lipoprotein receptor [Source:ZFIN;Acc:ZDB-GENE-040426-803] | 393897 | YES | YES | Plasma Membrane | transporter | |||||
| ENSDARG00000059902 | vps16 | vacuolar protein sorting protein 16 [Source:ZFIN;Acc:ZDB-GENE-050809-9] | 798985 | YES | YES | Cytoplasm | transporter | |||||
| ENSDARG00000070433 | vps18 | vacuolar protein sorting 18 homolog (S. cerevisiae) [Source:ZFIN;Acc:ZDB-GENE-020419-33] | 100005887 | YES | YES | Cytoplasm | transporter | |||||
| ENSDARG00000015823 | vps26b | vacuolar protein sorting 26 homolog B [Source:ZFIN;Acc:ZDB-GENE-040426-2699] | 406684 | YES | YES | Cytoplasm | transporter | |||||
| ENSDARG00000069521 | vps29 | vacuolar protein sorting 29 homolog (S. cerevisiae) [Source:ZFIN;Acc:ZDB-GENE-030131-8764] | 573437 | YES | YES | Cytoplasm | transporter | |||||
| ENSDARG00000099258 | vps52 | vacuolar protein sorting 52 homolog (S. cerevisiae) [Source:ZFIN;Acc:ZDB-GENE-010430-2] | 0 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000060477 | vps8 | vacuolar protein sorting 8 homolog (S. cerevisiae) [Source:ZFIN;Acc:ZDB-GENE-060503-348] | 791134 | YES | YES | Other | other | |||||
| ENSDARG00000013732 | vta1 | vesicle (multivesicular body) trafficking 1 [Source:ZFIN;Acc:ZDB-GENE-040426-2154] | 406414 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000005425 | wdfy1 | WD repeat and FYVE domain containing 1 [Source:ZFIN;Acc:ZDB-GENE-041024-4] | 474326 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000075098 | wdr17 | WD repeat domain 17 [Source:ZFIN;Acc:ZDB-GENE-090313-131] | 561804 | YES | YES | Other | other | |||||
| ENSDARG00000074617 | wfs1b | Wolfram syndrome 1b (wolframin) [Source:ZFIN;Acc:ZDB-GENE-050809-19] | 556580 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000043460 | wu:fj39g12 | wu:fj39g12 [Source:ZFIN;Acc:ZDB-GENE-030131-7901] | 563937 | YES | YES | NA | NA | |||||
| ENSDARG00000029011 | xpnpep1 | X-prolyl aminopeptidase (aminopeptidase P) 1, soluble [Source:ZFIN;Acc:ZDB-GENE-040426-999] | 406253 | YES | YES | Cytoplasm | peptidase | |||||
| ENSDARG00000035913 | yars | tyrosyl-tRNA synthetase [Source:ZFIN;Acc:ZDB-GENE-030425-2] | 368235 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000020449 | yipf3 | Yip1 domain family, member 3 [Source:ZFIN;Acc:ZDB-GENE-030131-6443] | 334511 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000067626 | ywhag1 | 3-monooxygenase/tryptophan 5-monooxygenase activation protein, gamma polypeptide 1 [Source:ZFIN;Acc:ZDB-GENE-011115-1] | 117604 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000071658 | ywhag2 | 3-monooxygenase/tryptophan 5-monooxygenase activation protein, gamma polypeptide 2 [Source:ZFIN;Acc:ZDB-GENE-061023-2] | 560560 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000103453 | zfpl1 | zinc finger protein-like 1 [Source:ZFIN;Acc:ZDB-GENE-040426-1255] | 393646 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000038876 | zgc:101569 | zgc:101569 [Source:ZFIN;Acc:ZDB-GENE-041010-72] | 449822 | YES | YES | NA | NA | |||||
| ENSDARG00000038694 | zgc:101744 | zgc:101744 [Source:ZFIN;Acc:ZDB-GENE-050320-111] | 541409 | YES | YES | NA | NA | |||||
| ENSDARG00000021404 | zgc:110319 | zgc:110319 [Source:ZFIN;Acc:ZDB-GENE-050417-345] | 796111 | YES | YES | NA | NA | |||||
| ENSDARG00000058966 | zgc:112332 | zgc:112332 [Source:ZFIN;Acc:ZDB-GENE-050522-387] | 553712 | YES | YES | NA | NA | |||||
| ENSDARG00000079932 | zgc:152830 | zgc:152830 [Source:ZFIN;Acc:ZDB-GENE-090312-55] | 767682 | YES | YES | NA | NA | |||||
| ENSDARG00000045979 | zgc:153704 | zgc:153704 [Source:ZFIN;Acc:ZDB-GENE-060929-1114] | 767759 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000091136 | zgc:174259 | zgc:174259 [Source:ZFIN;Acc:ZDB-GENE-080213-10] | 791587 | YES | YES | Other | other | |||||
| ENSDARG00000039356 | zgc:194209 | zgc:194209 [Source:ZFIN;Acc:ZDB-GENE-081022-75] | 570908 | YES | YES | NA | NA | |||||
| ENSDARG00000079847 | zgc:194578 | zgc:194578 [Source:ZFIN;Acc:ZDB-GENE-081022-126] | 792915 | YES | YES | NA | NA | |||||
| ENSDARG00000101443 | zgc:66014 | zgc:66014 [Source:ZFIN;Acc:ZDB-GENE-040426-1507] | 393817 | YES | YES | Other | other | |||||
| ENSDARG00000014015 | zgc:77086 | zgc:77086 [Source:ZFIN;Acc:ZDB-GENE-040426-2238] | 405841 | YES | YES | NA | NA | |||||
| ENSDARG00000021287 | zgc:91909 | zgc:91909 [Source:ZFIN;Acc:ZDB-GENE-040704-16] | 431725 | YES | YES | NA | NA | |||||
| ENSDARG00000059083 | novel gene | 0 | 100005981 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000069334 | AGFG2 | ArfGAP with FG repeats 2 [Source:HGNC Symbol;Acc:HGNC:5177] | 0 | YES | YES | Other | other | |||||
| ENSDARG00000063079 | ago3b | argonaute RISC catalytic component 3b [Source:ZFIN;Acc:ZDB-GENE-060503-452] | 568159 | YES | YES | Cytoplasm | translation regulator | |||||
| ENSDARG00000101139 | agpat2 | 1-acylglycerol-3-phosphate O-acyltransferase 2 (lysophosphatidic acid acyltransferase, beta) [Source:ZFIN;Acc:ZDB-GENE-061103-541] | 777624 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000057159 | ankrd29 | ankyrin repeat domain 29 [Source:ZFIN;Acc:ZDB-GENE-050208-655] | 553798 | YES | YES | Other | other | |||||
| ENSDARG00000062396 | anks1aa | ankyrin repeat and sterile alpha motif domain containing 1Aa [Source:ZFIN;Acc:ZDB-GENE-090312-152] | 0 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000011333 | arhgap33 | Rho GTPase activating protein 33 [Source:ZFIN;Acc:ZDB-GENE-121105-12] | 569720 | YES | YES | Plasma Membrane | transporter | |||||
| ENSDARG00000061294 | arhgap5 | Rho GTPase activating protein 5 [Source:ZFIN;Acc:ZDB-GENE-081014-1] | 795752 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000039486 | bag3 | BCL2-associated athanogene 3 [Source:ZFIN;Acc:ZDB-GENE-040801-40] | 445139 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000031119 | baiap2l1b | BAI1-associated protein 2-like 1b [Source:ZFIN;Acc:ZDB-GENE-050417-371] | 550528 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000053979 | bc2 | putative breast adenocarcinoma marker [Source:ZFIN;Acc:ZDB-GENE-040426-2922] | 798901 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000063040 | btbd11b | BTB (POZ) domain containing 11b [Source:ZFIN;Acc:ZDB-GENE-050419-99] | 100034519 | YES | YES | Other | transcription regulator | |||||
| ENSDARG00000015065 | c4 | complement component 4 [Source:ZFIN;Acc:ZDB-GENE-030131-5697] | 566261 | YES | YES | Extracellular Space | other | |||||
| ENSDARG00000103521 | capsla | calcyphosine-like a [Source:ZFIN;Acc:ZDB-GENE-030131-7291] | 550371 | YES | YES | NA | NA | |||||
| ENSDARG00000078327 | CEP170B (2 of 2) | centrosomal protein 170B [Source:HGNC Symbol;Acc:HGNC:20362] | 100318353 | YES | YES | Other | other | |||||
| ENSDARG00000102643 | chmp1a | charged multivesicular body protein 1A [Source:ZFIN;Acc:ZDB-GENE-040426-1474] | 393535 | YES | YES | Extracellular Space | peptidase | |||||
| ENSDARG00000104358 | CLASP1 (2 of 2) | cytoplasmic linker associated protein 1 [Source:HGNC Symbol;Acc:HGNC:17088] | 0 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000104439 | cldnd1a | claudin domain containing 1a [Source:ZFIN;Acc:ZDB-GENE-030131-7568] | 335628 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000020007 | col1a2 | collagen, type I, alpha 2 [Source:ZFIN;Acc:ZDB-GENE-030131-8415] | 336471 | YES | YES | Extracellular Space | other | |||||
| ENSDARG00000070393 | COQ10A | coenzyme Q10 homolog A (S. cerevisiae) [Source:HGNC Symbol;Acc:HGNC:26515] | 100002536 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000052604 | CPEB2 | cytoplasmic polyadenylation element binding protein 2 [Source:HGNC Symbol;Acc:HGNC:21745] | 100330483 | YES | YES | Cytoplasm | translation regulator | |||||
| ENSDARG00000025189 | cpne8 | copine VIII [Source:ZFIN;Acc:ZDB-GENE-100809-1] | 100332028 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000052114 | crtc3 | CREB regulated transcription coactivator 3 [Source:ZFIN;Acc:ZDB-GENE-070712-2] | 0 | YES | YES | Nucleus | other | |||||
| ENSDARG00000076742 | cyth1a | cytohesin 1a [Source:ZFIN;Acc:ZDB-GENE-030131-657] | 321938 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000075362 | dfnb31a | deafness, autosomal recessive 31a [Source:ZFIN;Acc:ZDB-GENE-091118-27] | 100334777 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000074059 | dlg5a | discs, large homolog 5a (Drosophila) [Source:ZFIN;Acc:ZDB-GENE-030131-3149] | 569065 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000008487 | dmd | dystrophin [Source:ZFIN;Acc:ZDB-GENE-010426-1] | 83773 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000078759 | dna2 | DNA replication helicase/nuclease 2 [Source:ZFIN;Acc:ZDB-GENE-090313-71] | 559535 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000060725 | dnajc16l | DnaJ (Hsp40) homolog, subfamily C, member 16 like [Source:ZFIN;Acc:ZDB-GENE-130530-696] | 556590 | YES | YES | Other | other | |||||
| ENSDARG00000076673 | DSP (2 of 2) | desmoplakin [Source:HGNC Symbol;Acc:HGNC:3052] | 327364 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000063570 | dyrk1aa | dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1A, a [Source:ZFIN;Acc:ZDB-GENE-050302-29] | 100005019 | YES | YES | NA | NA | |||||
| ENSDARG00000102755 | EML6 | echinoderm microtubule associated protein like 6 [Source:HGNC Symbol;Acc:HGNC:35412] | 0 | YES | YES | Other | other | |||||
| ENSDARG00000032866 | erh | enhancer of rudimentary homolog (Drosophila) [Source:ZFIN;Acc:ZDB-GENE-990415-57] | 30475 | YES | YES | Nucleus | other | |||||
| ENSDARG00000088073 | fam110b | family with sequence similarity 110, member B [Source:ZFIN;Acc:ZDB-GENE-050626-70] | 573995 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000021366 | fbp1a | fructose-1,6-bisphosphatase 1a [Source:ZFIN;Acc:ZDB-GENE-030131-7171] | 335231 | YES | YES | Cytoplasm | phosphatase | |||||
| ENSDARG00000003058 | fgfr1op | FGFR1 oncogene partner [Source:ZFIN;Acc:ZDB-GENE-090417-1] | 415215 | YES | YES | Cytoplasm | kinase | |||||
| ENSDARG00000102626 | frem2b | Fras1 related extracellular matrix protein 2b [Source:ZFIN;Acc:ZDB-GENE-081119-4] | 566673 | YES | YES | Extracellular Space | other | |||||
| ENSDARG00000090883 | gabra3 | gamma-aminobutyric acid (GABA) A receptor, alpha 3 [Source:ZFIN;Acc:ZDB-GENE-091204-365] | 0 | YES | YES | Plasma Membrane | ion channel | |||||
| ENSDARG00000087704 | gfra3 | GDNF family receptor alpha 3 [Source:ZFIN;Acc:ZDB-GENE-110512-1] | 556647 | YES | YES | Plasma Membrane | transmembrane receptor | |||||
| ENSDARG00000037589 | gid8b | GID complex subunit 8 homolog b (S. cerevisiae) [Source:ZFIN;Acc:ZDB-GENE-030515-4] | 566067 | YES | YES | Nucleus | other | |||||
| ENSDARG00000073841 | herc2 | HECT and RLD domain containing E3 ubiquitin protein ligase 2 [Source:ZFIN;Acc:ZDB-GENE-070718-6] | 0 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000040881 | hnrnph1 | heterogeneous nuclear ribonucleoprotein H1 [Source:ZFIN;Acc:ZDB-GENE-040426-1856] | 402984 | YES | YES | Nucleus | other | |||||
| ENSDARG00000062997 | hyal2b | hyaluronoglucosaminidase 2b [Source:ZFIN;Acc:ZDB-GENE-050208-414] | 559228 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000037553 | il1rapl2 | interleukin 1 receptor accessory protein-like 2 [Source:ZFIN;Acc:ZDB-GENE-040219-5] | 100149452 | YES | YES | Plasma Membrane | transmembrane receptor | |||||
| ENSDARG00000045893 | kctd15a | potassium channel tetramerization domain containing 15a [Source:ZFIN;Acc:ZDB-GENE-041010-105] | 445133 | YES | YES | NA | NA | |||||
| ENSDARG00000087538 | kif3a | kinesin family member 3A [Source:ZFIN;Acc:ZDB-GENE-050417-71] | 550267 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000036840 | krt15 | keratin 15 [Source:ZFIN;Acc:ZDB-GENE-040426-2931] | 406844 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000014013 | lbr | lamin B receptor [Source:ZFIN;Acc:ZDB-GENE-030804-11] | 368360 | YES | YES | Nucleus | enzyme | |||||
| ENSDARG00000078998 | lhfpl4a | lipoma HMGIC fusion partner-like 4a [Source:ZFIN;Acc:ZDB-GENE-111017-1] | 100151593 | YES | YES | Other | other | |||||
| ENSDARG00000095170 | lrrfip1b | leucine rich repeat (in FLII) interacting protein 1b [Source:ZFIN;Acc:ZDB-GENE-050208-324] | 100321881 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000024966 | MARK4 (2 of 2) | MAP/microtubule affinity-regulating kinase 4 [Source:HGNC Symbol;Acc:HGNC:13538] | 571279 | YES | YES | Cytoplasm | kinase | |||||
| ENSDARG00000015947 | matn4 | matrilin 4 [Source:ZFIN;Acc:ZDB-GENE-050208-64] | 497348 | YES | YES | Extracellular Space | other | |||||
| ENSDARG00000014634 | mbtps1 | membrane-bound transcription factor peptidase, site 1 [Source:ZFIN;Acc:ZDB-GENE-030131-4909] | 326710 | YES | YES | Cytoplasm | peptidase | |||||
| ENSDARG00000102184 | mib1 | mindbomb E3 ubiquitin protein ligase 1 [Source:ZFIN;Acc:ZDB-GENE-030404-2] | 352910 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000031136 | moxd1 | monooxygenase, DBH-like 1 [Source:ZFIN;Acc:ZDB-GENE-030131-9320] | 570584 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000062347 | mtus2 | microtubule associated tumor suppressor candidate 2 [Source:ZFIN;Acc:ZDB-GENE-091204-75] | 556195 | YES | YES | Nucleus | other | |||||
| ENSDARG00000036104 | MYO1G | myosin IG [Source:HGNC Symbol;Acc:HGNC:13880] | 571487 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000073843 | myo9ab | myosin IXAb [Source:ZFIN;Acc:ZDB-GENE-080424-6] | 0 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000012378 | naa35 | N(alpha)-acetyltransferase 35, NatC auxiliary subunit [Source:ZFIN;Acc:ZDB-GENE-030131-306] | 321587 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000001835 | nalcn | sodium leak channel, non-selective [Source:ZFIN;Acc:ZDB-GENE-050410-12] | 548345 | YES | YES | Plasma Membrane | ion channel | |||||
| ENSDARG00000096368 | neu3.5 | sialidase 3 (membrane sialidase), tandem duplicate 5 [Source:ZFIN;Acc:ZDB-GENE-071220-2] | 100000684 | YES | YES | Plasma Membrane | enzyme | |||||
| ENSDARG00000040926 | nr2f2 | nuclear receptor subfamily 2, group F, member 2 [Source:ZFIN;Acc:ZDB-GENE-990415-252] | 30424 | YES | YES | Nucleus | ligand-dependent nuclear receptor | |||||
| ENSDARG00000069368 | nrn1la | neuritin 1-like a [Source:ZFIN;Acc:ZDB-GENE-050419-106] | 100004307 | YES | YES | Other | other | |||||
| ENSDARG00000071017 | nt5e | 5'-nucleotidase, ecto (CD73) [Source:ZFIN;Acc:ZDB-GENE-040426-1261] | 393906 | YES | YES | Plasma Membrane | phosphatase | |||||
| ENSDARG00000018110 | pak4 | p21 protein (Cdc42/Rac)-activated kinase 4 [Source:ZFIN;Acc:ZDB-GENE-040704-69] | 431769 | YES | YES | NA | NA | |||||
| ENSDARG00000079202 | parp2 | poly (ADP-ribose) polymerase 2 [Source:ZFIN;Acc:ZDB-GENE-031113-6] | 100007906 | YES | YES | Nucleus | enzyme | |||||
| ENSDARG00000030887 | phf23a | PHD finger protein 23a [Source:ZFIN;Acc:ZDB-GENE-050320-67] | 541372 | YES | YES | Other | other | |||||
| ENSDARG00000061093 | phldb1a | pleckstrin homology-like domain, family B, member 1a [Source:ZFIN;Acc:ZDB-GENE-060526-272] | 566287 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000052462 | pisd | phosphatidylserine decarboxylase [Source:ZFIN;Acc:ZDB-GENE-061215-46] | 553433 | YES | YES | NA | NA | |||||
| ENSDARG00000057125 | pld1b | phospholipase D1b [Source:ZFIN;Acc:ZDB-GENE-070510-3] | 572492 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000039272 | pls1 | plastin 1 (I isoform) [Source:ZFIN;Acc:ZDB-GENE-030131-6205] | 334273 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000102790 | PNPT1 | polyribonucleotide nucleotidyltransferase 1 [Source:HGNC Symbol;Acc:HGNC:23166] | 0 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000045749 | PPFIBP1 (1 of 2) | PTPRF interacting protein, binding protein 1 (liprin beta 1) [Source:HGNC Symbol;Acc:HGNC:9249] | 565020 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000078102 | PSD (2 of 2) | pleckstrin and Sec7 domain containing [Source:HGNC Symbol;Acc:HGNC:9507] | 0 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000008249 | ptchd4 | patched domain containing 4 [Source:ZFIN;Acc:ZDB-GENE-041001-178] | 564097 | YES | YES | Other | other | |||||
| ENSDARG00000007196 | rae1 | ribonucleic acid export 1 [Source:ZFIN;Acc:ZDB-GENE-040426-1029] | 393973 | YES | YES | Nucleus | other | |||||
| ENSDARG00000005861 | ralgapa2 | Ral GTPase activating protein, alpha subunit 2 (catalytic) [Source:ZFIN;Acc:ZDB-GENE-131122-56] | 568210 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000077060 | rbm6 | RNA binding motif protein 6 [Source:ZFIN;Acc:ZDB-GENE-050309-79] | 567058 | YES | YES | Nucleus | other | |||||
| ENSDARG00000098458 | RPL37A | ribosomal protein L37a [Source:HGNC Symbol;Acc:HGNC:10348] | 572109 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000062558 | rspry1 | ring finger and SPRY domain containing 1 [Source:ZFIN;Acc:ZDB-GENE-061026-2] | 565154 | YES | YES | Other | other | |||||
| ENSDARG00000000189 | sema6e | sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6E [Source:ZFIN;Acc:ZDB-GENE-040724-119] | 556743 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000089418 | si:dkey-164f24.2 | si:dkey-164f24.2 [Source:ZFIN;Acc:ZDB-GENE-030131-7035] | 566607 | YES | YES | NA | NA | |||||
| ENSDARG00000094074 | si:dkey-81h8.1 | si:dkey-81h8.1 [Source:ZFIN;Acc:ZDB-GENE-060503-931] | 100034657 | YES | YES | NA | NA | |||||
| ENSDARG00000045976 | sidt2 | SID1 transmembrane family, member 2 [Source:ZFIN;Acc:ZDB-GENE-030131-7356] | 796833 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000102381 | SLC44A4 | solute carrier family 44, member 4 [Source:HGNC Symbol;Acc:HGNC:13941] | 0 | YES | YES | Plasma Membrane | transporter | |||||
| ENSDARG00000074153 | slitrk5b | SLIT and NTRK-like family, member 5b [Source:ZFIN;Acc:ZDB-GENE-110408-55] | 100331252 | YES | YES | Other | other | |||||
| ENSDARG00000002538 | smu1a | smu-1 suppressor of mec-8 and unc-52 homolog a (C. elegans) [Source:ZFIN;Acc:ZDB-GENE-040426-916] | 393168 | YES | YES | Nucleus | other | |||||
| ENSDARG00000009567 | SPEG (1 of 2) | SPEG complex locus [Source:HGNC Symbol;Acc:HGNC:16901] | 0 | YES | YES | Nucleus | kinase | |||||
| ENSDARG00000038445 | SPIRE1 (2 of 2) | spire-type actin nucleation factor 1 [Source:HGNC Symbol;Acc:HGNC:30622] | 0 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000053668 | stag2b | stromal antigen 2b [Source:ZFIN;Acc:ZDB-GENE-030131-2785] | 560751 | YES | YES | Nucleus | other | |||||
| ENSDARG00000059003 | strip2 | striatin interacting protein 2 [Source:ZFIN;Acc:ZDB-GENE-120328-1] | 556392 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000060112 | sybu | syntabulin (syntaxin-interacting) [Source:ZFIN;Acc:ZDB-GENE-060929-1202] | 568207 | YES | YES | Other | other | |||||
| ENSDARG00000089190 | TANC1 (2 of 2) | tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1 [Source:HGNC Symbol;Acc:HGNC:29364] | 100334650 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000102365 | tceb3 | transcription elongation factor B (SIII), polypeptide 3 [Source:ZFIN;Acc:ZDB-GENE-040426-2810] | 0 | YES | YES | Nucleus | transcription regulator | |||||
| ENSDARG00000035605 | tchp | trichoplein, keratin filament binding [Source:ZFIN;Acc:ZDB-GENE-060421-3368] | 678595 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000034195 | top2b | topoisomerase (DNA) II beta [Source:ZFIN;Acc:ZDB-GENE-041008-136] | 569877 | YES | YES | Nucleus | enzyme | |||||
| ENSDARG00000062794 | trim36 | tripartite motif containing 36 [Source:ZFIN;Acc:ZDB-GENE-040426-2936] | 406849 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000045877 | ube2nb | ubiquitin-conjugating enzyme E2Nb [Source:ZFIN;Acc:ZDB-GENE-040426-1291] | 393313 | YES | YES | NA | NA | |||||
| ENSDARG00000018000 | ubp1 | upstream binding protein 1 (LBP-1a) [Source:ZFIN;Acc:ZDB-GENE-060503-870] | 100136855 | YES | YES | Cytoplasm | transcription regulator | |||||
| ENSDARG00000098290 | UNC80 | unc-80 homolog (C. elegans) [Source:HGNC Symbol;Acc:HGNC:26582] | 0 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000086665 | VGF | VGF nerve growth factor inducible [Source:HGNC Symbol;Acc:HGNC:12684] | 0 | YES | YES | Extracellular Space | growth factor | |||||
| ENSDARG00000078247 | vip | vasoactive intestinal peptide [Source:ZFIN;Acc:ZDB-GENE-080204-3] | 795653 | YES | YES | Extracellular Space | other | |||||
| ENSDARG00000016825 | vtg6 | vitellogenin 6 [Source:ZFIN;Acc:ZDB-GENE-001201-5] | 559229 | YES | YES | NA | NA | |||||
| ENSDARG00000069269 | wdr35 | WD repeat domain 35 [Source:ZFIN;Acc:ZDB-GENE-060810-148] | 565515 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000045064 | ablim1b | actin binding LIM protein 1b [Source:ZFIN;Acc:ZDB-GENE-050327-91] | 541550 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000088168 | ablim3 | actin binding LIM protein family, member 3 [Source:ZFIN;Acc:ZDB-GENE-070112-1122] | 566821 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000052438 | actr2a | ARP2 actin-related protein 2a homolog (yeast) [Source:ZFIN;Acc:ZDB-GENE-040426-2894] | 406820 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000054177 | adgra1 | adhesion G protein-coupled receptor A1 [Source:ZFIN;Acc:ZDB-GENE-130603-101] | 560847 | YES | YES | Plasma Membrane | G-protein coupled receptor | |||||
| ENSDARG00000077732 | alyref | Aly/REF export factor [Source:ZFIN;Acc:ZDB-GENE-070928-29] | 560127 | YES | YES | Other | other | |||||
| ENSDARG00000062103 | ankrd24 | ankyrin repeat domain 24 [Source:ZFIN;Acc:ZDB-GENE-050208-747] | 565326 | YES | YES | Other | other | |||||
| ENSDARG00000068274 | ANKRD65 | ankyrin repeat domain 65 [Source:HGNC Symbol;Acc:HGNC:42950] | 566073 | YES | YES | Other | other | |||||
| ENSDARG00000103333 | baiap2b | BAI1-associated protein 2b [Source:ZFIN;Acc:ZDB-GENE-100812-7] | 100537012 | YES | YES | Plasma Membrane | kinase | |||||
| ENSDARG00000095034 | C24H8orf34 | chromosome 8 open reading frame 34 [Source:HGNC Symbol;Acc:HGNC:30905] | 557144 | YES | YES | Other | other | |||||
| ENSDARG00000079556 | C2CD4C (1 of 2) | C2 calcium-dependent domain containing 4C [Source:HGNC Symbol;Acc:HGNC:29417] | 568754 | YES | YES | Extracellular Space | other | |||||
| ENSDARG00000027564 | CABZ01052588.1 | Non-specific serine/threonine protein kinase [Source:UniProtKB/TrEMBL;Acc:E7FE80] | 0 | YES | YES | Cytoplasm | kinase | |||||
| ENSDARG00000002991 | cep135 | centrosomal protein 135 [Source:ZFIN;Acc:ZDB-GENE-041210-325] | 553303 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000020834 | CEP250 | centrosomal protein 250kDa [Source:HGNC Symbol;Acc:HGNC:1859] | 564101 | YES | YES | Nucleus | other | |||||
| ENSDARG00000075007 | chst2a | carbohydrate (N-acetylglucosamine-6-O) sulfotransferase 2a [Source:ZFIN;Acc:ZDB-GENE-060929-40] | 561896 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000059770 | cpt1aa | carnitine palmitoyltransferase 1Aa (liver) [Source:ZFIN;Acc:ZDB-GENE-060503-925] | 558088 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000074367 | CR381676.1 | Ubiquitin carboxyl-terminal hydrolase [Source:UniProtKB/TrEMBL;Acc:E7F4N9] | 559878 | YES | YES | NA | NA | |||||
| ENSDARG00000101380 | cryabb | crystallin, alpha B, b [Source:ZFIN;Acc:ZDB-GENE-040718-419] | 436943 | YES | YES | Nucleus | other | |||||
| ENSDARG00000054748 | cuedc1b | CUE domain containing 1b [Source:ZFIN;Acc:ZDB-GENE-070424-29] | 556847 | YES | YES | Other | other | |||||
| ENSDARG00000071192 | cx40.8 | connexin 40.8 [Source:ZFIN;Acc:ZDB-GENE-050616-10] | 565638 | YES | YES | Plasma Membrane | transporter | |||||
| ENSDARG00000077765 | cx44.2 | connexin 44.2 [Source:ZFIN;Acc:ZDB-GENE-010619-1] | 114404 | YES | YES | NA | NA | |||||
| ENSDARG00000068166 | dfnb31b | deafness, autosomal recessive 31b [Source:ZFIN;Acc:ZDB-GENE-060526-377] | 100534775 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000013110 | dmtn | dematin actin binding protein [Source:ZFIN;Acc:ZDB-GENE-030131-9438] | 565367 | YES | YES | NA | NA | |||||
| ENSDARG00000020953 | dnajb6b | DnaJ (Hsp40) homolog, subfamily B, member 6b [Source:ZFIN;Acc:ZDB-GENE-040426-1122] | 393275 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000024874 | dock4b | dedicator of cytokinesis 4b [Source:ZFIN;Acc:ZDB-GENE-060130-74] | 654774 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000069951 | eef1a1b | eukaryotic translation elongation factor 1 alpha 1b [Source:ZFIN;Acc:ZDB-GENE-050417-327] | 100004503 | YES | YES | Cytoplasm | translation regulator | |||||
| ENSDARG00000005163 | efr3a | EFR3 homolog A (S. cerevisiae) [Source:ZFIN;Acc:ZDB-GENE-040426-1681] | 393800 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000042126 | elmo3 | engulfment and cell motility 3 [Source:ZFIN;Acc:ZDB-GENE-050309-39] | 553310 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000053517 | EML5 | echinoderm microtubule associated protein like 5 [Source:HGNC Symbol;Acc:HGNC:18197] | 570011 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000020741 | fga | fibrinogen alpha chain [Source:ZFIN;Acc:ZDB-GENE-031010-21] | 378986 | YES | YES | Extracellular Space | other | |||||
| ENSDARG00000052625 | fkbp1b | FK506 binding protein 1b [Source:ZFIN;Acc:ZDB-GENE-040426-1785] | 393785 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000018820 | flncb | filamin C, gamma b (actin binding protein 280) [Source:ZFIN;Acc:ZDB-GENE-041008-175] | 570288 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000052782 | glrba | glycine receptor, beta a [Source:ZFIN;Acc:ZDB-GENE-010410-2] | 83412 | YES | YES | Plasma Membrane | ion channel | |||||
| ENSDARG00000092517 | gpsm1a | G-protein signaling modulator 1a [Source:ZFIN;Acc:ZDB-GENE-060526-302] | 100004265 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000022668 | grapb | GRB2-related adaptor protein b [Source:ZFIN;Acc:ZDB-GENE-050913-4] | 555184 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000078149 | grin2ca | glutamate receptor, ionotropic, N-methyl D-aspartate 2Ca [Source:ZFIN;Acc:ZDB-GENE-070822-3] | 100329743 | YES | YES | Plasma Membrane | ion channel | |||||
| ENSDARG00000077560 | grin2cb | glutamate receptor, ionotropic, N-methyl D-aspartate 2Cb [Source:ZFIN;Acc:ZDB-GENE-100308-2] | 100333648 | YES | YES | Plasma Membrane | ion channel | |||||
| ENSDARG00000086207 | grin2da | glutamate receptor, ionotropic, N-methyl D-aspartate 2D, a [Source:ZFIN;Acc:ZDB-GENE-041008-124] | 449864 | YES | YES | Plasma Membrane | ion channel | |||||
| ENSDARG00000087897 | h2afy2 | H2A histone family, member Y2 [Source:ZFIN;Acc:ZDB-GENE-050913-114] | 558711 | YES | YES | Nucleus | other | |||||
| ENSDARG00000099478 | hs2st1a | heparan sulfate 2-O-sulfotransferase 1a [Source:ZFIN;Acc:ZDB-GENE-070112-2312] | 791188 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000041921 | hsbp1b | heat shock factor binding protein 1b [Source:ZFIN;Acc:ZDB-GENE-040426-1721] | 393727 | YES | YES | Nucleus | transcription regulator | |||||
| ENSDARG00000103563 | IFT122 | intraflagellar transport 122 [Source:HGNC Symbol;Acc:HGNC:13556] | 0 | YES | YES | Nucleus | other | |||||
| ENSDARG00000055053 | itih1 | inter-alpha-trypsin inhibitor heavy chain 1 [Source:ZFIN;Acc:ZDB-GENE-130530-650] | 564761 | YES | YES | NA | NA | |||||
| ENSDARG00000104119 | kalrna | kalirin, RhoGEF kinase a [Source:ZFIN;Acc:ZDB-GENE-100921-4] | 0 | YES | YES | Cytoplasm | kinase | |||||
| ENSDARG00000006601 | kaznb | kazrin, periplakin interacting protein b [Source:ZFIN;Acc:ZDB-GENE-061215-31] | 790937 | YES | YES | NA | NA | |||||
| ENSDARG00000017624 | krt4 | keratin 4 [Source:ZFIN;Acc:ZDB-GENE-000607-83] | 794486 | YES | YES | NA | NA | |||||
| ENSDARG00000036830 | krt91 | keratin 91 [Source:ZFIN;Acc:ZDB-GENE-040801-181] | 445051 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000074275 | limch1a | LIM and calponin homology domains 1a [Source:ZFIN;Acc:ZDB-GENE-090312-153] | 561940 | YES | YES | Other | other | |||||
| ENSDARG00000099386 | lrrtm4l2 | leucine rich repeat transmembrane neuronal 4 like 2 [Source:ZFIN;Acc:ZDB-GENE-080327-14] | 0 | YES | YES | Extracellular Space | other | |||||
| ENSDARG00000075556 | luzp1 | leucine zipper protein 1 [Source:ZFIN;Acc:ZDB-GENE-060810-90] | 558606 | YES | YES | Nucleus | other | |||||
| ENSDARG00000025108 | magixa | MAGI family member, X-linked a [Source:ZFIN;Acc:ZDB-GENE-060503-115] | 796857 | YES | YES | NA | NA | |||||
| ENSDARG00000077633 | magixb | MAGI family member, X-linked b [Source:ZFIN;Acc:ZDB-GENE-081105-33] | 0 | YES | YES | NA | NA | |||||
| ENSDARG00000102587 | MIOS | missing oocyte, meiosis regulator, homolog (Drosophila) [Source:HGNC Symbol;Acc:HGNC:21905] | 0 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000100972 | myh11b | myosin, heavy chain 11b, smooth muscle [Source:ZFIN;Acc:ZDB-GENE-101124-2] | 566080 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000012944 | myhz2 | myosin, heavy polypeptide 2, fast muscle specific [Source:ZFIN;Acc:ZDB-GENE-020604-1] | 246275 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000038123 | myl9a | myosin, light chain 9a, regulatory [Source:ZFIN;Acc:ZDB-GENE-041010-120] | 450006 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000036179 | myo1ea | myosin IE, a [Source:ZFIN;Acc:ZDB-GENE-030131-5127] | 555285 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000062003 | myo5b | myosin VB [Source:ZFIN;Acc:ZDB-GENE-031219-7] | 567402 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000104235 | myo5c | myosin VC [Source:ZFIN;Acc:ZDB-GENE-131127-196] | 562676 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000056910 | NFASC (1 of 2) | neurofascin [Source:HGNC Symbol;Acc:HGNC:29866] | 559590 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000069427 | nkain2 | Na+/K+ transporting ATPase interacting 2 [Source:ZFIN;Acc:ZDB-GENE-041014-77] | 101883714 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000103123 | NYAP1 | neuronal tyrosine-phosphorylated phosphoinositide-3-kinase adaptor 1 [Source:HGNC Symbol;Acc:HGNC:22009] | 101885998 | YES | YES | Other | other | |||||
| ENSDARG00000028900 | odf2a | outer dense fiber of sperm tails 2a [Source:ZFIN;Acc:ZDB-GENE-091117-46] | 792921 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000079378 | phldb1b | pleckstrin homology-like domain, family B, member 1b [Source:ZFIN;Acc:ZDB-GENE-121114-8] | 570275 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000060813 | plekha7a | pleckstrin homology domain containing, family A member 7a [Source:ZFIN;Acc:ZDB-GENE-050419-75] | 571486 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000104267 | postnb | periostin, osteoblast specific factor b [Source:ZFIN;Acc:ZDB-GENE-030131-9120] | 337176 | YES | YES | Extracellular Space | other | |||||
| ENSDARG00000056797 | ppp2r2ca | protein phosphatase 2, regulatory subunit B, gamma a [Source:ZFIN;Acc:ZDB-GENE-051120-168] | 100002802 | YES | YES | Other | phosphatase | |||||
| ENSDARG00000037593 | prickle2b | prickle homolog 2 (Drosophila) [Source:ZFIN;Acc:ZDB-GENE-030724-6] | 368250 | YES | YES | Nucleus | other | |||||
| ENSDARG00000015921 | pwp1 | PWP1 homolog (S. cerevisiae) [Source:ZFIN;Acc:ZDB-GENE-040426-1049] | 393262 | YES | YES | Nucleus | other | |||||
| ENSDARG00000055080 | rbm4.2 | RNA binding motif protein 4.2 [Source:ZFIN;Acc:ZDB-GENE-030131-3019] | 557528 | YES | YES | Nucleus | other | |||||
| ENSDARG00000021309 | rhoae | ras homolog gene family, member Ae [Source:ZFIN;Acc:ZDB-GENE-040426-1337] | 394125 | YES | YES | Plasma Membrane | enzyme | |||||
| ENSDARG00000102690 | rims2b | regulating synaptic membrane exocytosis 2b [Source:ZFIN;Acc:ZDB-GENE-060503-893] | 563393 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000090035 | rtn4r | reticulon 4 receptor [Source:ZFIN;Acc:ZDB-GENE-040310-1] | 403306 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000103993 | scg2a | secretogranin II (chromogranin C) a [Source:ZFIN;Acc:ZDB-GENE-030131-8214] | 793796 | YES | YES | Extracellular Space | cytokine | |||||
| ENSDARG00000056121 | serac1 | serine active site containing 1 [Source:ZFIN;Acc:ZDB-GENE-040616-1] | 568022 | YES | YES | Extracellular Space | other | |||||
| ENSDARG00000063054 | shank3b | SH3 and multiple ankyrin repeat domains 3b [Source:ZFIN;Acc:ZDB-GENE-041210-74] | 566152 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000045145 | shisa9a | shisa family member 9a [Source:ZFIN;Acc:ZDB-GENE-050320-78] | 541382 | YES | YES | Plasma Membrane | other | |||||
| ENSDARG00000104157 | si:ch211-232h12.2 | si:ch211-232h12.2 [Source:ZFIN;Acc:ZDB-GENE-130613-4] | 0 | YES | YES | NA | NA | |||||
| ENSDARG00000075491 | si:ch211-26b3.4 | si:ch211-26b3.4 [Source:ZFIN;Acc:ZDB-GENE-081104-60] | 568768 | YES | YES | NA | NA | |||||
| ENSDARG00000078317 | si:dkey-175m17.7 | si:dkey-175m17.7 [Source:ZFIN;Acc:ZDB-GENE-091204-18] | 100007304 | YES | YES | NA | NA | |||||
| ENSDARG00000032482 | si:dkey-40c11.2 | si:dkey-40c11.2 [Source:ZFIN;Acc:ZDB-GENE-060526-300] | 798260 | YES | YES | NA | NA | |||||
| ENSDARG00000061699 | SIPA1L3 | signal-induced proliferation-associated 1 like 3 [Source:HGNC Symbol;Acc:HGNC:23801] | 561005 | YES | YES | Extracellular Space | other | |||||
| ENSDARG00000077368 | slc30a6 | solute carrier family 30 (zinc transporter), member 6 [Source:ZFIN;Acc:ZDB-GENE-040426-1838] | 0 | YES | YES | Cytoplasm | transporter | |||||
| ENSDARG00000010130 | smpdl3b | sphingomyelin phosphodiesterase, acid-like 3B [Source:ZFIN;Acc:ZDB-GENE-041008-132] | 564392 | YES | YES | Extracellular Space | enzyme | |||||
| ENSDARG00000003046 | sorbs2a | sorbin and SH3 domain containing 2a [Source:ZFIN;Acc:ZDB-GENE-070308-2] | 571008 | YES | YES | NA | NA | |||||
| ENSDARG00000044182 | stau1 | staufen double-stranded RNA binding protein 1 [Source:ZFIN;Acc:ZDB-GENE-030131-6372] | 334440 | YES | YES | Cytoplasm | transporter | |||||
| ENSDARG00000029239 | syt9b | synaptotagmin IXb [Source:ZFIN;Acc:ZDB-GENE-040822-6] | 445476 | YES | YES | Plasma Membrane | transporter | |||||
| ENSDARG00000023443 | tjp2b | tight junction protein 2b (zona occludens 2) [Source:ZFIN;Acc:ZDB-GENE-040718-58] | 436639 | YES | YES | Plasma Membrane | kinase | |||||
| ENSDARG00000098118 | TRAPPC10 | trafficking protein particle complex 10 [Source:HGNC Symbol;Acc:HGNC:11868] | 0 | YES | YES | Cytoplasm | transporter | |||||
| ENSDARG00000012367 | trim46a | tripartite motif containing 46a [Source:ZFIN;Acc:ZDB-GENE-110825-1] | 569905 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000031164 | tuba8l2 | tubulin, alpha 8 like 2 [Source:ZFIN;Acc:ZDB-GENE-040426-1646] | 393664 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000006260 | tuba8l4 | tubulin, alpha 8 like 4 [Source:ZFIN;Acc:ZDB-GENE-040426-860] | 393154 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000060238 | uacab | uveal autoantigen with coiled-coil domains and ankyrin repeats b [Source:ZFIN;Acc:ZDB-GENE-030521-32] | 564040 | YES | YES | Cytoplasm | other | |||||
| ENSDARG00000034321 | uo:ion006 | uo:ion006 [Source:ZFIN;Acc:ZDB-GENE-090713-4] | 100000814 | YES | YES | NA | NA | |||||
| ENSDARG00000044575 | vars | valyl-tRNA synthetase [Source:ZFIN;Acc:ZDB-GENE-010601-1] | 114427 | YES | YES | Cytoplasm | enzyme | |||||
| ENSDARG00000007916 | xpnpep3 | X-prolyl aminopeptidase 3, mitochondrial [Source:ZFIN;Acc:ZDB-GENE-040426-2076] | 573360 | YES | YES | Cytoplasm | peptidase | |||||
| ENSDARG00000035340 | zgc:194261 | zgc:194261 [Source:ZFIN;Acc:ZDB-GENE-080723-77] | 572372 | YES | YES | NA | NA |
Supplementary Table 4a. Ensemble Families from mouse synaptosomal proteins
For each mouse synaptosomal protein its corresponding Ensembl Family code and descriptor are given.
| Ensembl Gene Id | Ensembl Protein Family Id(S) | Ensembl Family Description |
|---|---|---|
| ENSMUSG00000000001 | ENSFM00410000138458 | Guanine Nucleotide Binding Subunit |
| ENSMUSG00000000088 | ENSFM00500000272199 | Cytochrome C Oxidase Subunit 5A Mitochondrial Precursor Cytochrome C Oxidase Polypeptide Va |
| ENSMUSG00000000149 | ENSFM00730001522052 | Guanine Nucleotide Binding Subunit Alpha G Alpha G Subunit Alpha |
| ENSMUSG00000000159 | ENSFM00500000282325 | Unknown |
| ENSMUSG00000000159 | ENSFM00570000893288 | Unknown |
| ENSMUSG00000000159 | ENSFM00500000273376 | Immunoglobulin Superfamily Member 5 IGSF5 Junctional Adhesion Molecule 4 Jam 4 |
| ENSMUSG00000000168 | ENSFM00810002040672 | Dihydrolipoyllysine Residue Acetyltransferase Component Of Pyruvate Dehydrogenase Complex Mitochondrial EC_2.3.1.12 Dihydrolipoamide Acetyltransferase Component Of Pyruvate Dehydrogenase Complex Pyruvate Dehydrogenase Complex Component E2 Pdc E2 PDCE2 |
| ENSMUSG00000000171 | ENSFM00670001236122 | Succinate Dehydrogenase Cytochrome B Small Subunit Mitochondrial Precursor Cybs Succinate Dehydrogenase Complex Subunit D Succinate Ubiquinone Oxidoreductase Cytochrome B Small Subunit Succinate Ubiquinone Reductase Membrane Anchor Subunit |
| ENSMUSG00000000202 | ENSFM00440000236967 | Btb/Poz Domain Containing 17 Precursor Galectin 3 Binding |
| ENSMUSG00000000253 | ENSFM00250000002497 | Gmp Reductase EC_1.7.1.7 Guanosine 5' Monophosphate Oxidoreductase Guanosine Monophosphate Reductase |
| ENSMUSG00000000263 | ENSFM00400000131715 | Glycine Receptor Subunit Precursor Glycine Receptor Kda Subunit |
| ENSMUSG00000000276 | ENSFM00730001522404 | Diacylglycerol Kinase Epsilon Dag Kinase Epsilon EC_2.7.1.107 Diglyceride Kinase Epsilon Dgk Epsilon |
| ENSMUSG00000000290 | ENSFM00570000890148 | Unknown |
| ENSMUSG00000000290 | ENSFM00700001407845 | Unknown |
| ENSMUSG00000000290 | ENSFM00730001521210 | Integrin Beta Precursor Subunit Beta Subunit Beta Antigen |
| ENSMUSG00000000296 | ENSFM00810002041019 | Tumor D53 Tumor D52 1 |
| ENSMUSG00000000305 | ENSFM00390000130953 | Unknown |
| ENSMUSG00000000305 | ENSFM00730001521172 | Cadherin Precursor Cadherin Cadherin |
| ENSMUSG00000000308 | ENSFM00570000898238 | Unknown |
| ENSMUSG00000000308 | ENSFM00570000900073 | Unknown |
| ENSMUSG00000000308 | ENSFM00800002003437 | Creatine Kinase U Type Mitochondrial Precursor EC_2.7.3.2 Acidic Type Mitochondrial Creatine Kinase Mia Ck Ubiquitous Mitochondrial Creatine Kinase U Mtck |
| ENSMUSG00000000325 | ENSFM00570000877506 | Unknown |
| ENSMUSG00000000325 | ENSFM00730001521847 | Catenin Delta 1 Cadherin Associated Src Substrate Cas P120 Catenin P120 Ctn P120 Cas |
| ENSMUSG00000000326 | ENSFM00500000271337 | Catechol O Methyltransferase EC_2.1.1.6 |
| ENSMUSG00000000339 | ENSFM00570000884353 | Unknown |
| ENSMUSG00000000339 | ENSFM00250000005769 | Rna 3' Terminal Phosphate Cyclase Rna Cyclase Rna 3' Phosphate Cyclase EC_6.5.1.4 Rna Terminal Phosphate Cyclase Domain Containing 1 |
| ENSMUSG00000000340 | ENSFM00800002024492 | Unknown |
| ENSMUSG00000000340 | ENSFM00800002024493 | Unknown |
| ENSMUSG00000000340 | ENSFM00250000003682 | Lipoamide Acyltransferase Component Of Branched Chain Alpha Keto Acid Dehydrogenase Complex Mitochondrial Precursor EC_2.3.1.168 Branched Chain Alpha Keto Acid Dehydrogenase Complex Component E2 Bckad E2 BCKADE2 Dihydrolipoamide Acetyltransferase Componen |
| ENSMUSG00000000374 | ENSFM00730001522435 | Trafficking Particle Complex Subunit 10 Trafficking Particle Complex Subunit TMEM1 Transport Particle Subunit TMEM1 Trapp Subunit TMEM1 |
| ENSMUSG00000000378 | ENSFM00500000271756 | Malcavernin Cerebral Cavernous Malformations 2 |
| ENSMUSG00000000399 | ENSFM00250000004385 | Nadh Dehydrogenase 1 Alpha Subcomplex Subunit 9 Mitochondrial Precursor Complex I 39KD Ci 39KD Nadh Ubiquinone Oxidoreductase 39 Kda Subunit |
| ENSMUSG00000000416 | ENSFM00600000921238 | Cortactin Binding 2 CORTBP2 |
| ENSMUSG00000000531 | ENSFM00500000272853 | General Receptor Phosphoinositides 1 Associated Scaffold GRP1 Associated Scaffold |
| ENSMUSG00000000538 | ENSFM00250000001136 | TOM1 2 Target Of Myb 2 |
| ENSMUSG00000000560 | ENSFM00800002009455 | Unknown |
| ENSMUSG00000000560 | ENSFM00800002003385 | Gamma Aminobutyric Acid Receptor Subunit Alpha Precursor Gaba A Receptor Subunit Alpha |
| ENSMUSG00000000563 | ENSFM00570000877616 | Unknown |
| ENSMUSG00000000563 | ENSFM00250000004100 | Atp Synthase F 0 Complex Subunit B1 Mitochondrial Precursor Atp Synthase Subunit B Atpase Subunit B |
| ENSMUSG00000000568 | ENSFM00610000971968 | Unknown |
| ENSMUSG00000000568 | ENSFM00800002023620 | Unknown |
| ENSMUSG00000000568 | ENSFM00500000269788 | Heterogeneous Nuclear Ribonucleoprotein D Hnrnp D Hnrnp Dl |
| ENSMUSG00000000594 | ENSFM00250000006403 | Ganglioside GM2 Activator Precursor Cerebroside Sulfate Activator GM2 Ap Sphingolipid Activator 3 Sap 3 |
| ENSMUSG00000000600 | ENSFM00250000003734 | Krev Interaction Trapped 1 Krev Interaction Trapped 1 Cerebral Cavernous Malformations 1 |
| ENSMUSG00000000605 | ENSFM00280000058697 | H + /Cl Exchange Transporter 5 Chloride Channel 5 Clc 5 Chloride Transporter Clc 5 |
| ENSMUSG00000000627 | ENSFM00480000262746 | Semaphorin Precursor |
| ENSMUSG00000000628 | ENSFM00610000970732 | Unknown |
| ENSMUSG00000000628 | ENSFM00250000000532 | Hexokinase EC_2.7.1.1 Hexokinase Type Hk |
| ENSMUSG00000000631 | ENSFM00610000973552 | Unknown |
| ENSMUSG00000000631 | ENSFM00400000131775 | Unconventional Myosin Xviiia Molecule Associated With JAK3 N Terminus Majn Myosin Containing A Pdz Domain |
| ENSMUSG00000000631 | ENSFM00810002043502 | Unconventional Myosin Xviiia |
| ENSMUSG00000000711 | ENSFM00500000269759 | Ras Related Rab |
| ENSMUSG00000000738 | ENSFM00800002003722 | Paraplegin EC_3.4.24.- |
| ENSMUSG00000000740 | ENSFM00500000270284 | 60S Ribosomal L13 |
| ENSMUSG00000000743 | ENSFM00600000921337 | Charged Multivesicular Body 1B Chromatin Modifying 1B CHMP1B |
| ENSMUSG00000000759 | ENSFM00610000967253 | Unknown |
| ENSMUSG00000000759 | ENSFM00610000969293 | Unknown |
| ENSMUSG00000000759 | ENSFM00250000005014 | Gamma Tubulin Complex Component 3 Gamma Ring Complex |
| ENSMUSG00000000787 | ENSFM00770001847530 | Atp Dependent Rna Helicase DDX3Y EC_3.6.4.13 Dead Box 3 Y Chromosomal |
| ENSMUSG00000000805 | ENSFM00570000889881 | Unknown |
| ENSMUSG00000000805 | ENSFM00670001235760 | Carbonic Anhydrase 4 Precursor EC_4.2.1.1 Carbonate Dehydratase Iv Carbonic Anhydrase Iv Ca Iv |
| ENSMUSG00000000826 | ENSFM00500000270505 | Dnaj Homolog Subfamily C Member 5B Cysteine String Beta Csp Beta |
| ENSMUSG00000000827 | ENSFM00740001613634 | Unknown |
| ENSMUSG00000000827 | ENSFM00500000270547 | Tumor D54 Tumor D52 2 |
| ENSMUSG00000000838 | ENSFM00250000000920 | Fragile X Mental Retardation 1 |
| ENSMUSG00000000881 | ENSFM00250000000275 | Disks Large Homolog Synapse Associated Sap |
| ENSMUSG00000000915 | ENSFM00250000001308 | Huntingtin Interacting 1 Hip 1 Huntingtin Interacting I Hip I |
| ENSMUSG00000001016 | ENSFM00250000003528 | Interleukin Enhancer Binding Factor 2 |
| ENSMUSG00000001018 | ENSFM00740001603935 | Unknown |
| ENSMUSG00000001018 | ENSFM00740001607673 | Unknown |
| ENSMUSG00000001018 | ENSFM00500000273533 | Snare Associated Snapin Biogenesis Of Lysosome Related Organelles Complex 1 Subunit 7 Bloc 1 Subunit 7 Synaptosomal Associated 25 Binding Snap Associated |
| ENSMUSG00000001052 | ENSFM00750001632389 | Transport SEC24A SEC24 Related A |
| ENSMUSG00000001062 | ENSFM00740001589527 | VPS9 Domain Containing 1 |
| ENSMUSG00000001089 | ENSFM00610000966722 | Unknown |
| ENSMUSG00000001089 | ENSFM00610000973587 | Unknown |
| ENSMUSG00000001089 | ENSFM00800002003954 | Leucine Zipper 1 Leucine Zipper Motif Containing |
| ENSMUSG00000001120 | ENSFM00250000000747 | Poly Rc Binding Alpha Heterogeneous Nuclear Ribonucleoprotein Hnrnp |
| ENSMUSG00000001127 | ENSFM00290000065522 | Serine/Threonine Kinase EC_2.7.11.1 Proto Oncogene Raf |
| ENSMUSG00000001138 | ENSFM00250000000938 | Metal Transporter Ancient Conserved Domain Containing Cyclin |
| ENSMUSG00000001143 | ENSFM00680001314100 | Unknown |
| ENSMUSG00000001143 | ENSFM00260000050644 | Vesicular Integral Membrane VIP36 Precursor Lectin Mannose Binding 2 Vesicular Integral Membrane 36 VIP36 |
| ENSMUSG00000001173 | ENSFM00500000270009 | Type Ii Inositol 1 4 5 Trisphosphate 5 Phosphatase Precursor EC_3.1.3.36 Inositol Polyphosphate 5 Phosphatase Phosphoinositide 5 Phosphatase 5PTASE |
| ENSMUSG00000001211 | ENSFM00570000880145 | Unknown |
| ENSMUSG00000001211 | ENSFM00570000888711 | Unknown |
| ENSMUSG00000001211 | ENSFM00250000002628 | 1 Acyl Sn Glycerol 3 Phosphate Acyltransferase Delta EC_2.3.1.51 1 Acylglycerol 3 Phosphate O Acyltransferase 4 1 Agp Acyltransferase 4 1 Agpat 4 Lysophosphatidic Acid Acyltransferase Delta Lpaat Delta |
| ENSMUSG00000001227 | ENSFM00730001521376 | Semaphorin Precursor Semaphorin Sema |
| ENSMUSG00000001229 | ENSFM00250000001158 | Dipeptidyl Peptidase 9 DP9 EC_3.4.14.5 Dipeptidyl Peptidase Ix Dpp Ix Dipeptidyl Peptidase 9 DPLP9 |
| ENSMUSG00000001270 | ENSFM00800002003408 | Creatine Kinase M Type EC_2.7.3.2 Creatine Kinase M Chain M Ck |
| ENSMUSG00000001289 | ENSFM00610000956140 | Unknown |
| ENSMUSG00000001289 | ENSFM00610000960672 | Unknown |
| ENSMUSG00000001289 | ENSFM00610000960673 | Unknown |
| ENSMUSG00000001289 | ENSFM00500000272753 | Prefoldin Subunit 5 C Myc Binding Mm 1 Myc Modulator 1 |
| ENSMUSG00000001323 | ENSFM00500000271489 | Serine Racemase EC_4.3.1.17 EC_4.3.1.- 18 EC_5.1.-.- 1 18 D Serine Ammonia Lyase D Serine Dehydratase L Serine Ammonia Lyase L Serine Dehydratase |
| ENSMUSG00000001376 | ENSFM00250000002464 | Coiled Coil Domain Containing 132 |
| ENSMUSG00000001416 | ENSFM00610000969837 | Unknown |
| ENSMUSG00000001416 | ENSFM00500000270039 | T Complex 1 Subunit Gamma Tcp 1 Gamma Cct Gamma |
| ENSMUSG00000001424 | ENSFM00610000970527 | Unknown |
| ENSMUSG00000001424 | ENSFM00250000002294 | Staphylococcal Nuclease Domain Containing 1.100 Kda Coactivator P100 Co Activator |
| ENSMUSG00000001440 | ENSFM00250000005282 | Importin Subunit Beta 1 Karyopherin Subunit Beta 1 Nuclear Factor P97 Pore Targeting Complex 97 Kda Subunit PTAC97 |
| ENSMUSG00000001441 | ENSFM00610000955086 | Unknown |
| ENSMUSG00000001441 | ENSFM00610000973558 | Unknown |
| ENSMUSG00000001441 | ENSFM00760001714762 | Puromycin Sensitive Aminopeptidase Psa EC_3.4.11.14 Cytosol Alanyl Aminopeptidase Aap S |
| ENSMUSG00000001467 | ENSFM00800002003871 | Lanosterol 14 Alpha Demethylase Ldm EC_1.14.13.70 Cypli Cytochrome P450 51A1 Cytochrome P450 14DM Cytochrome Dm Cytochrome P450LI Sterol 14 Alpha Demethylase |
| ENSMUSG00000001473 | ENSFM00250000000193 | Tubulin Beta Chain |
| ENSMUSG00000001525 | ENSFM00640001119932 | Unknown |
| ENSMUSG00000001525 | ENSFM00250000000193 | Tubulin Beta Chain |
| ENSMUSG00000001552 | ENSFM00250000001012 | Catenin Beta Beta Catenin |
| ENSMUSG00000001576 | ENSFM00610000960834 | Unknown |
| ENSMUSG00000001576 | ENSFM00250000005380 | Endoplasmic Reticulum Golgi Intermediate Compartment 1 Er Golgi Intermediate Compartment 32 Kda Ergic 32 |
| ENSMUSG00000001642 | ENSFM00730001521171 | Reductase Aldo Keto Reductase Family 1 Member |
| ENSMUSG00000001666 | ENSFM00640001114985 | Unknown |
| ENSMUSG00000001666 | ENSFM00670001236245 | D Dopachrome Decarboxylase EC_4.1.1.84 D Dopachrome Tautomerase |
| ENSMUSG00000001687 | ENSFM00500000272867 | Ubiquitin 3 Precursor Membrane Anchored Ubiquitin Fold Mub |
| ENSMUSG00000001707 | ENSFM00250000008520 | Eukaryotic Translation Elongation Factor 1 Epsilon 1 Elongation Factor P18 Multisynthase Complex Auxiliary Component P18 |
| ENSMUSG00000001755 | ENSFM00440000236932 | Bifunctional Coenzyme A Synthase Coa Synthase Nbp Pov 2 |
| ENSMUSG00000001773 | ENSFM00730001521644 | 2 EC_3.4.17.21 Glutamate Carboxypeptidase Ii N Acetylated Alpha Linked Acidic Dipeptidase Naaladase |
| ENSMUSG00000001783 | ENSFM00250000005120 | Trna Splicing Ligase Rtcb Homolog |
| ENSMUSG00000001794 | ENSFM00250000000112 | Calpain Subunit Calcium Activated Neutral Proteinase Canp Calpain Subunit |
| ENSMUSG00000001829 | ENSFM00430000230197 | Caseinolytic Peptidase B Homolog Suppressor Of Potassium Transport Defect 3 |
| ENSMUSG00000001833 | ENSFM00760001714685 | Septin 7 CDC10 Homolog |
| ENSMUSG00000001847 | ENSFM00730001521186 | Ras Related C3 Botulinum Toxin Substrate Precursor P21 |
| ENSMUSG00000001891 | ENSFM00800002003558 | Utp Glucose 1 Phosphate Uridylyltransferase EC_2.7.7.9 Udp Glucose Pyrophosphorylase Udpgp Ugpase |
| ENSMUSG00000001924 | ENSFM00250000000763 | Ubiquitin Modifier Activating Enzyme Ubiquitin Activating Enzyme E1 |
| ENSMUSG00000001964 | ENSFM00570000893147 | Unknown |
| ENSMUSG00000001964 | ENSFM00250000010495 | Emerin |
| ENSMUSG00000001985 | ENSFM00720001492013 | Glutamate Receptor Ionotropic Kainate Precursor Glutamate Receptor |
| ENSMUSG00000001986 | ENSFM00720001492020 | Glutamate Receptor Precursor Glur Ampa Selective Glutamate Receptor Glur Glutamate Receptor Ionotropic Ampa |
| ENSMUSG00000001995 | ENSFM00730001521477 | Signal Induced Proliferation Associated 1 SIPA1 |
| ENSMUSG00000002006 | ENSFM00250000001242 | E3 Ubiquitin Ligase PDZRN3 EC_6.3.2.- Pdz Domain Containing Ring Finger 3 Semaphorin Cytoplasmic Domain Associated 3 SEMACAP3 |
| ENSMUSG00000002010 | ENSFM00770001847549 | Isocitrate Dehydrogenase Gamma Mitochondrial Precursor EC_1.1.1.41 Isocitric Dehydrogenase Subunit Gamma Nad + Specific Icdh Subunit Gamma |
| ENSMUSG00000002014 | ENSFM00250000005113 | Translocon Associated Subunit Delta Precursor Trap Delta Signal Sequence Receptor Subunit Delta Ssr Delta |
| ENSMUSG00000002015 | ENSFM00670001235596 | B Cell Receptor Associated Bcr Associated |
| ENSMUSG00000002017 | ENSFM00250000002232 | FAM98A |
| ENSMUSG00000002064 | ENSFM00250000003961 | Stromal Cell Derived Factor 2 1 Precursor SDF2 1 |
| ENSMUSG00000002102 | ENSFM00550000743154 | 26S Protease Regulatory Subunit 6A 26S Proteasome Aaa Atpase Subunit RPT5 Proteasome 26S Subunit Atpase 3 Tat Binding Tbp |
| ENSMUSG00000002103 | ENSFM00800002003509 | Acid Phosphatase Precursor EC_3.1.3.2 |
| ENSMUSG00000002107 | ENSFM00720001497880 | Unknown |
| ENSMUSG00000002107 | ENSFM00730001521889 | Cugbp Elav Family Member 2 Celf 2 Bruno 3 Cug Triplet Repeat Rna Binding 2 Cug BP2 Cug Bp And Etr 3 Factor 2 Elav Type Rna Binding 3 Etr 3 Rna Binding Brunol 3 |
| ENSMUSG00000002332 | ENSFM00250000006111 | Dehydrogenase/Reductase Sdr Family Member 1 EC_1.1.-.- |
| ENSMUSG00000002341 | ENSFM00730001521436 | Core Precursor Chondroitin Sulfate Proteoglycan |
| ENSMUSG00000002346 | ENSFM00500000271356 | Mitochondrial Coenzyme A Transporter Sl Solute Carrier Family 25 Member 42 |
| ENSMUSG00000002379 | ENSFM00500000275281 | Nadh Dehydrogenase 1 Alpha Subcomplex Subunit 11 Complex I B14 7 Ci B14 7 Nadh Ubiquinone Oxidoreductase Subunit B14 7 |
| ENSMUSG00000002395 | ENSFM00570000889168 | Unknown |
| ENSMUSG00000002395 | ENSFM00250000004790 | Vesicle Transport USE1 USE1 |
| ENSMUSG00000002416 | ENSFM00670001236841 | Nadh Dehydrogenase 1 Beta Subcomplex Subunit 2 Mitochondrial Precursor Complex I Aggg Ci Aggg Nadh Ubiquinone Oxidoreductase Aggg Subunit |
| ENSMUSG00000002458 | ENSFM00500000446065 | Unknown |
| ENSMUSG00000002458 | ENSFM00570000898922 | Unknown |
| ENSMUSG00000002458 | ENSFM00480000262761 | Regulator Of G Signaling |
| ENSMUSG00000002459 | ENSFM00610000972139 | Unknown |
| ENSMUSG00000002459 | ENSFM00480000262761 | Regulator Of G Signaling |
| ENSMUSG00000002475 | ENSFM00500000270392 | Abhydrolase Domain Containing EC_3.1.1.- Lung Alpha/Beta Hydrolase |
| ENSMUSG00000002496 | ENSFM00250000003183 | Tuberin Tuberous Sclerosis 2 |
| ENSMUSG00000002504 | ENSFM00740001589334 | Na + /H + Exchange Regulatory Cofactor Nhe RF1 Nherf 1 Ezrin Radixin Moesin Binding Phosphoprotein 50 EBP50 Regulatory Cofactor Of Na + /H + Exchanger Sodium Hydrogen Exchanger Regulatory Factor 1 Solute Carrier Family 9 A3 Regulatory Factor 1 |
| ENSMUSG00000002546 | ENSFM00500000270377 | Golgin Subfamily A Member 2.130 Kda Cis Golgi Matrix GM130 |
| ENSMUSG00000002660 | ENSFM00250000006881 | Atp Dependent Clp Protease Proteolytic Subunit Mitochondrial Precursor EC_3.4.21.92 Endopeptidase Clp |
| ENSMUSG00000002718 | ENSFM00610000960843 | Unknown |
| ENSMUSG00000002718 | ENSFM00250000005028 | Exportin 2 EXP2 Chromosome Segregation 1 Importin Alpha Re Exporter |
| ENSMUSG00000002731 | ENSFM00600000921352 | Risc Loading Complex Subunit TARBP2 |
| ENSMUSG00000002741 | ENSFM00810002040913 | Synaptobrevin Homolog YKT6 Precursor EC_2.3.1.- |
| ENSMUSG00000002771 | ENSFM00570000882063 | Unknown |
| ENSMUSG00000002771 | ENSFM00730001521500 | Glutamate Receptor Ionotropic Nmda Precursor Glutamate Receptor Subunit Epsilon N Methyl D Aspartate Receptor Subtype |
| ENSMUSG00000002808 | ENSFM00250000006488 | Mammalian Ependymin Related 1 Precursor Merp 1 |
| ENSMUSG00000002812 | ENSFM00730001522115 | Flightless 1 |
| ENSMUSG00000002944 | ENSFM00730001522348 | Platelet Glycoprotein 4 Glycoprotein Iiib Gpiiib Pas Iv Pas 4 Platelet Glycoprotein Iv Gpiv CD36 Antigen |
| ENSMUSG00000002948 | ENSFM00790001963074 | Dual Specificity Mitogen Activated Kinase Kinase 7 Map Kinase Kinase 7 Mapkk 7 EC_2.7.12.2 Jnk Activating Kinase 2 Mapk/Erk Kinase 7 Mek 7 C Jun N Terminal Kinase Kinase 2 Jnk Kinase 2 Jnkk 2 |
| ENSMUSG00000002949 | ENSFM00500000271055 | Mitochondrial Import Inner Membrane Translocase Subunit Precursor |
| ENSMUSG00000002957 | ENSFM00810002040615 | Ap 2 Complex Subunit Alpha 2.100 Kda Coated Vesicle C Adaptor Complex Ap 2 Subunit Alpha 2 Adaptor Related Complex 2 Subunit Alpha 2 Alpha Adaptin C ALPHA2 Adaptin Clathrin Assembly Complex 2 Alpha C Large Chain Plasma Membrane Adaptor HA2/AP2 Adaptin Alp |
| ENSMUSG00000002980 | ENSFM00570000897897 | Unknown |
| ENSMUSG00000002980 | ENSFM00500000273157 | Basal Cell Adhesion Molecule Precursor B Cam Cell Surface Glycoprotein Lutheran Antigen CD239 Antigen |
| ENSMUSG00000002981 | ENSFM00810002040917 | Cleft Lip And Palate Transmembrane 1 |
| ENSMUSG00000002984 | ENSFM00640001114360 | Unknown |
| ENSMUSG00000002984 | ENSFM00250000002521 | Mitochondrial Import Receptor Subunit TOM40 Homolog Translocase Of Outer Membrane 40 Kda Subunit Homolog |
| ENSMUSG00000002985 | ENSFM00640001113209 | Unknown |
| ENSMUSG00000002985 | ENSFM00500000271202 | Apolipoprotein E Precursor Apo E |
| ENSMUSG00000002997 | ENSFM00730001521993 | Camp Dependent Kinase Type Ii Regulatory Subunit |
| ENSMUSG00000003033 | ENSFM00730001521549 | Ap 1 Complex Subunit Mu 1 Ap Mu Chain Family Member MU1A Adaptor Complex Ap 1 Subunit Mu 1 Adaptor Related Complex 1 Subunit Mu 1 Clathrin Assembly Complex 1 Mu 1 Medium Chain 1 Golgi Adaptor HA1/AP1 Adaptin Mu 1 Subunit Mu Adaptin 1 MU1A Adaptin |
| ENSMUSG00000003037 | ENSFM00730001521184 | Ras Related Rab |
| ENSMUSG00000003062 | ENSFM00800002024367 | Unknown |
| ENSMUSG00000003062 | ENSFM00250000003670 | Star Related Lipid Transfer 3 Start Domain Containing 3 STARD3 |
| ENSMUSG00000003072 | ENSFM00500000272854 | Atp Synthase Subunit Delta Mitochondrial Precursor F Atpase Delta Subunit |
| ENSMUSG00000003099 | ENSFM00500000270236 | Serine/Threonine Phosphatase 5 PP5 EC_3.1.3.16 Phosphatase T Ppt |
| ENSMUSG00000003131 | ENSFM00250000004414 | Platelet Activating Factor Acetylhydrolase Ib Subunit Beta EC_3.1.1.47 Paf Acetylhydrolase 30 Kda Subunit Paf Ah 30 Kda Subunit Paf Ah Subunit Beta Pafah Subunit Beta |
| ENSMUSG00000003135 | ENSFM00250000006276 | CCR4 Not Transcription Complex Subunit 11 |
| ENSMUSG00000003153 | ENSFM00610000970890 | Unknown |
| ENSMUSG00000003153 | ENSFM00610000970892 | Unknown |
| ENSMUSG00000003153 | ENSFM00610000970893 | Unknown |
| ENSMUSG00000003153 | ENSFM00730001521168 | Solute Carrier Family 2 Facilitated Glucose Transporter Member Glucose Transporter Type Glut |
| ENSMUSG00000003161 | ENSFM00500000270522 | Sorcin |
| ENSMUSG00000003178 | ENSFM00250000000908 | Methionine Sulfoxide Oxidase EC_1.14.13.- Molecule Interacting With Casl Mical |
| ENSMUSG00000003200 | ENSFM00250000000986 | Endophilin SH3 Domain Containing GRB2 |
| ENSMUSG00000003269 | ENSFM00250000001040 | Cytohesin Ph SEC7 And Coiled Coil Domain Containing |
| ENSMUSG00000003279 | ENSFM00250000000718 | Disks Large Associated Dap Psd 95/SAP90 Binding SAP90/Psd 95 Associated |
| ENSMUSG00000003316 | ENSFM00250000002484 | Golgi Apparatus 1 Precursor E Selectin Ligand 1 Esl 1 Golgi Sialoglycoprotein Mg 160 |
| ENSMUSG00000003345 | ENSFM00730001521484 | Casein Kinase I Gamma Cki Gamma EC_2.7.11.1 |
| ENSMUSG00000003352 | ENSFM00250000000524 | Voltage Dependent L Type Calcium Channel Subunit Beta Calcium Channel Voltage Dependent Subunit Beta |
| ENSMUSG00000003363 | ENSFM00570000879838 | Unknown |
| ENSMUSG00000003363 | ENSFM00570000888995 | Unknown |
| ENSMUSG00000003363 | ENSFM00250000001612 | Phospholipase D3 Pld 3 EC_3.1.4.4 Choline Phosphatase 3 Phosphatidylcholine Hydrolyzing Phospholipase D3 |
| ENSMUSG00000003378 | ENSFM00720001492013 | Glutamate Receptor Ionotropic Kainate Precursor Glutamate Receptor |
| ENSMUSG00000003402 | ENSFM00500000271338 | Glucosidase 2 Subunit Beta Precursor 80K H Glucosidase Ii Subunit Beta Kinase C Substrate 60.1 Kda Heavy Chain Pkcsh |
| ENSMUSG00000003410 | ENSFM00250000000588 | Elav |
| ENSMUSG00000003411 | ENSFM00730001521441 | Ras Related Rab 3 |
| ENSMUSG00000003429 | ENSFM00800002003484 | 40S Ribosomal S11 |
| ENSMUSG00000003438 | ENSFM00500000271227 | Mitochondrial Import Inner Membrane Translocase Subunit TIM50 Precursor |
| ENSMUSG00000003452 | ENSFM00250000002108 | Bicaudal D Homolog Bic D |
| ENSMUSG00000003458 | ENSFM00570000898013 | Unknown |
| ENSMUSG00000003458 | ENSFM00250000005577 | Nicastrin Precursor |
| ENSMUSG00000003469 | ENSFM00250000003309 | Phytanoyl Coa Hydroxylase Interacting Phytanoyl Coa Hydroxylase Associated 1 Pahx AP1 PAHXAP1 |
| ENSMUSG00000003518 | ENSFM00660001182137 | Unknown |
| ENSMUSG00000003518 | ENSFM00500000272272 | Dual Specificity Phosphatase 3 EC_3.1.3.16 EC_3.1.3.- 48 Vaccinia H1 Related Phosphatase Vhr |
| ENSMUSG00000003526 | ENSFM00770001851658 | Uncharacterized |
| ENSMUSG00000003526 | ENSFM00570000881208 | Unknown |
| ENSMUSG00000003526 | ENSFM00250000001738 | Probable Proline Dehydrogenase 2 EC_1.5.5.2 Probable Proline Oxidase 2 |
| ENSMUSG00000003528 | ENSFM00730001522086 | Tricarboxylate Transport Protein Mitochondrial Precursor Citrate Transport Ctp Solute Carrier Family 25 Member 1 Tricarboxylate Carrier |
| ENSMUSG00000003573 | ENSFM00680001319354 | Unknown |
| ENSMUSG00000003573 | ENSFM00250000001140 | Homer Homolog Homer Vasp/Ena Related Gene Up Regulated During Seizure And Ltp Vesl |
| ENSMUSG00000003623 | ENSFM00730001522249 | Peroxisomal Carnitine O Octanoyltransferase Cot EC_2.3.1.137 |
| ENSMUSG00000003644 | ENSFM00570000851006 | Ribosomal S6 Kinase Alpha S6K Alpha EC_2.7.11.1.90 Kda Ribosomal S6 Kinase P90 Rsk Map Kinase Activated Kinase Mapk Activated Kinase Mapkap Kinase Mapkapk Ribosomal S6 Kinase Rsk |
| ENSMUSG00000003657 | ENSFM00500000270302 | Calbindin Calbindin D28 D 28K Vitamin D Dependent Calcium Binding Protein Avian Type |
| ENSMUSG00000003731 | ENSFM00810002040604 | Importin Subunit Alpha Karyopherin Subunit Alpha |
| ENSMUSG00000003779 | ENSFM00730001521759 | Kinesin KIF20B Kinesin Related Motor Interacting With PIN1 M Phase Phosphoprotein 1 MPP1 |
| ENSMUSG00000003808 | ENSFM00570000893661 | Unknown |
| ENSMUSG00000003808 | ENSFM00250000004351 | Phenylalanine Trna Ligase Alpha Subunit EC_6.1.1.20 Phenylalanyl Trna Synthetase Alpha Subunit Phers |
| ENSMUSG00000003809 | ENSFM00570000886815 | Unknown |
| ENSMUSG00000003809 | ENSFM00380000124741 | Glutaryl Coa Dehydrogenase Mitochondrial Precursor Gcd EC_1.3.8.6 |
| ENSMUSG00000003814 | ENSFM00500000269786 | Calreticulin Precursor |
| ENSMUSG00000003863 | ENSFM00250000000441 | Liprin Alpha Tyrosine Phosphatase Receptor Type F Polypeptide Interacting Alpha Ptprf Interacting Alpha |
| ENSMUSG00000003868 | ENSFM00730001521813 | Ruvb 2 EC_3.6.4.12 |
| ENSMUSG00000003872 | ENSFM00260000050595 | Lin 7 Homolog Lin Mammalian Lin Seven Mals Vertebrate Lin 7 Homolog Veli |
| ENSMUSG00000003923 | ENSFM00250000006597 | Transcription Factor A Mitochondrial Precursor Mttfa |
| ENSMUSG00000003934 | ENSFM00250000002014 | Ephrin B1 Precursor Elk Ligand Elk L Eph Related Receptor Tyrosine Kinase Ligand 2 Lerk 2 |
| ENSMUSG00000003955 | ENSFM00500000274855 | FAM162A E2 Induced Gene 5 Growth And Transformation Dependent Hgtd P |
| ENSMUSG00000003970 | ENSFM00250000002389 | 60S Ribosomal |
| ENSMUSG00000003974 | ENSFM00790001963004 | Metabotropic Glutamate Receptor Precursor |
| ENSMUSG00000004031 | ENSFM00250000001886 | Bmp/Retinoic Acid Inducible Neural Specific 1 Precursor Deleted In Bladder Cancer 1 |
| ENSMUSG00000004032 | ENSFM00610000969806 | Unknown |
| ENSMUSG00000004032 | ENSFM00610000973394 | Unknown |
| ENSMUSG00000004032 | ENSFM00500000269733 | Glutathione S Transferase EC_2.5.1.18 Gst Class Mu |
| ENSMUSG00000004069 | ENSFM00730001522148 | Dnaj Homolog Subfamily A Member 3 Mitochondrial Precursor Dnaj Tid 1 1 Tumorous Imaginal Discs TID56 Homolog |
| ENSMUSG00000004070 | ENSFM00570000893742 | Unknown |
| ENSMUSG00000004070 | ENSFM00250000002797 | Heme Oxygenase Ho EC_1.14.99.3 |
| ENSMUSG00000004071 | ENSFM00500000273378 | Cell Death Inducing P53 Target 1 Litaf |
| ENSMUSG00000004110 | ENSFM00740001589100 | Voltage Dependent Type Calcium Channel Subunit Alpha Brain Calcium Channel Calcium Channel L Type Alpha 1 Polypeptide Voltage Gated Calcium Channel Subunit Alpha CAV2 |
| ENSMUSG00000004113 | ENSFM00300000085455 | N Type Calcium Channel Pore Forming Subunit Alpha 1B |
| ENSMUSG00000004113 | ENSFM00700001391361 | Voltage N Type Calcium Channel |
| ENSMUSG00000004113 | ENSFM00740001589100 | Voltage Dependent Type Calcium Channel Subunit Alpha Brain Calcium Channel Calcium Channel L Type Alpha 1 Polypeptide Voltage Gated Calcium Channel Subunit Alpha CAV2 |
| ENSMUSG00000004207 | ENSFM00250000002893 | Proactivator Polypeptide Saposin A Saposin B Saposin B Saposin C Saposin D |
| ENSMUSG00000004264 | ENSFM00800002003689 | Prohibitin 2 |
| ENSMUSG00000004267 | ENSFM00730001521170 | Enolase EC_4.2.1.11 2 Phospho D Glycerate Hydro Lyase |
| ENSMUSG00000004285 | ENSFM00570000892542 | Unknown |
| ENSMUSG00000004285 | ENSFM00670001235732 | V Type Proton Atpase Subunit F V Atpase Subunit F V Atpase 14 Kda Subunit Vacuolar Proton Pump Subunit F |
| ENSMUSG00000004319 | ENSFM00280000058697 | H + /Cl Exchange Transporter 5 Chloride Channel 5 Clc 5 Chloride Transporter Clc 5 |
| ENSMUSG00000004364 | ENSFM00610000969310 | Unknown |
| ENSMUSG00000004364 | ENSFM00610000969311 | Unknown |
| ENSMUSG00000004364 | ENSFM00550000764342 | Cullin 3 |
| ENSMUSG00000004364 | ENSFM00730001521665 | Cullin 3 Cul 3 |
| ENSMUSG00000004366 | ENSFM00500000273500 | Somatostatin Precursor |
| ENSMUSG00000004383 | ENSFM00250000001914 | Glycosyltransferase LARGE2 EC_2.4.-.- Glycosyltransferase 1B |
| ENSMUSG00000004394 | ENSFM00500000270043 | Transmembrane EMP24 Domain Containing Precursor |
| ENSMUSG00000004451 | ENSFM00500000270233 | Ras Related Precursor |
| ENSMUSG00000004455 | ENSFM00800002023656 | Unknown |
| ENSMUSG00000004455 | ENSFM00800002023657 | Unknown |
| ENSMUSG00000004455 | ENSFM00800002023658 | Unknown |
| ENSMUSG00000004455 | ENSFM00420000140518 | Serine/Threonine Phosphatase PP1 Catalytic Subunit Pp EC_3.1.3.16 |
| ENSMUSG00000004460 | ENSFM00500000271339 | Dnaj Homolog Subfamily B Member 11 Precursor Er Associated Dnaj Er Associated HSP40 Co Chaperone Endoplasmic Reticulum Dna J Domain Containing 3 Er Resident ERDJ3 ERDJ3 ERJ3P |
| ENSMUSG00000004530 | ENSFM00610000967749 | Unknown |
| ENSMUSG00000004530 | ENSFM00730001521292 | Coronin |
| ENSMUSG00000004558 | ENSFM00250000000535 | N Myc Downstream Regulated Gene |
| ENSMUSG00000004565 | ENSFM00760001714681 | Neuropathy Target Esterase Sws |
| ENSMUSG00000004567 | ENSFM00250000001474 | Mucolipin |
| ENSMUSG00000004567 | ENSFM00590000917008 | Mucolipin 1 |
| ENSMUSG00000004610 | ENSFM00250000004626 | Electron Transfer Flavoprotein Subunit Beta Beta Etf |
| ENSMUSG00000004631 | ENSFM00250000002718 | Alpha Sarcoglycan Precursor Alpha Sg 50 Kda Dystrophin Associated Glycoprotein 50DAG Adhalin |
| ENSMUSG00000004637 | ENSFM00480000262794 | Ww Domain Containing Oxidoreductase EC_1.1.1.- |
| ENSMUSG00000004655 | ENSFM00750001632461 | Aquaporin 1 Aqp 1 Aquaporin Chip Water Channel Red Blood Cells And Kidney Proximal Tubule |
| ENSMUSG00000004748 | ENSFM00250000005790 | Mitochondrial Fission Process 1 Mitochondrial 18 Kda MTP18 |
| ENSMUSG00000004768 | ENSFM00700001403675 | Unknown |
| ENSMUSG00000004768 | ENSFM00400000131956 | Ras Related Rab 23 Precursor |
| ENSMUSG00000004789 | ENSFM00800002003585 | Dihydrolipoyllysine Residue Succinyltransferase Component Of 2 Oxoglutarate Dehydrogenase Complex Mitochondrial Precursor EC_2.3.1.61 2 Oxoglutarate Dehydrogenase Complex Component E2 Ogdc E2 Dihydrolipoamide Succinyltransferase Component Of 2 Oxoglutarat |
| ENSMUSG00000004843 | ENSFM00500000272414 | Charged Multivesicular Body 2B Chromatin Modifying 2B CHMP2B |
| ENSMUSG00000004849 | ENSFM00570000887691 | Unknown |
| ENSMUSG00000004849 | ENSFM00500000269841 | Ap 1 Complex Subunit Sigma Adaptor Complex Ap 1 Subunit Sigma Adaptor Related Complex 1 Subunit Sigma Clathrin Assembly Complex 1 Sigma Small Chain Golgi Adaptor HA1/AP1 Adaptin Sigma Subunit Sigma Subunit Of Ap 1 Clathrin Sigma Adaptin Adaptin |
| ENSMUSG00000004892 | ENSFM00730001521436 | Core Precursor Chondroitin Sulfate Proteoglycan |
| ENSMUSG00000004894 | ENSFM00590000918422 | Unknown |
| ENSMUSG00000004894 | ENSFM00730001521886 | Hyaluronan And Proteoglycan Link 1 Precursor Cartilage Linking 1 Cartilage Link Proteoglycan Link |
| ENSMUSG00000004902 | ENSFM00500000270170 | Mitochondrial Glutamate Carrier 1 Gc 1 Glutamate/H + Symporter 1 Solute Carrier Family 25 Member 22 |
| ENSMUSG00000004929 | ENSFM00760001714699 | Neurolysin Mitochondrial Precursor EC_3.4.24.16 Microsomal Endopeptidase Mep Mitochondrial Oligopeptidase M Neurotensin Endopeptidase |
| ENSMUSG00000004933 | ENSFM00730001522042 | Tyrosine Kinase Csk EC_2.7.10.2 C Src Kinase |
| ENSMUSG00000004936 | ENSFM00780001898380 | Dual Specificity Mitogen Activated Kinase Kinase 1 Map Kinase Kinase 1 Mapkk 1 EC_2.7.12.2 Erk Activator Kinase 1 Mapk/Erk Kinase 1 |
| ENSMUSG00000004937 | ENSFM00500000270552 | Small Glutamine Rich Tetratricopeptide Repeat Containing Sgt Small Glutamine Rich With Tetratricopeptide Repeats |
| ENSMUSG00000004951 | ENSFM00500000270924 | Heat Shock Beta 1 HSPB1 Heat Shock 27 Kda Hsp 27 |
| ENSMUSG00000004961 | ENSFM00720001492031 | Synaptotagmin |
| ENSMUSG00000004980 | ENSFM00270000056431 | Heterogeneous Nuclear Ribonucleoprotein |
| ENSMUSG00000005034 | ENSFM00790001962972 | Camp Dependent Kinase Catalytic Subunit Pka C EC_2.7.11.11 |
| ENSMUSG00000005069 | ENSFM00250000001819 | Peroxisomal Targeting Signal 1 Receptor PTS1 Receptor PTS1R PTS1 Bp Peroxin 5 Peroxisomal C Terminal Targeting Signal Import Receptor Peroxisome Receptor 1 |
| ENSMUSG00000005089 | ENSFM00800002003429 | Excitatory Amino Acid Transporter 2 Sodium Dependent Glutamate/Aspartate Transporter 2 Solute Carrier Family 1 Member 2 |
| ENSMUSG00000005103 | ENSFM00800002003651 | Wd Repeat Containing 1 Actin Interacting 1 |
| ENSMUSG00000005125 | ENSFM00610000969969 | Unknown |
| ENSMUSG00000005125 | ENSFM00610000969970 | Unknown |
| ENSMUSG00000005125 | ENSFM00610000969971 | Unknown |
| ENSMUSG00000005125 | ENSFM00610000973110 | Unknown |
| ENSMUSG00000005125 | ENSFM00610000973111 | Unknown |
| ENSMUSG00000005125 | ENSFM00250000000535 | N Myc Downstream Regulated Gene |
| ENSMUSG00000005161 | ENSFM00250000000757 | Thioredoxin Peroxidase Thioredoxin Dependent Peroxide Reductase |
| ENSMUSG00000005299 | ENSFM00810002057433 | Unknown |
| ENSMUSG00000005299 | ENSFM00810002057434 | Unknown |
| ENSMUSG00000005299 | ENSFM00740001589271 | LETM1 And Ef Hand Domain Containing 1 Mitochondrial Precursor Leucine Zipper Ef Hand Containing Transmembrane 1 |
| ENSMUSG00000005312 | ENSFM00250000001264 | Ubiquilin Linking Iap With Cytoskeleton Plic |
| ENSMUSG00000005338 | ENSFM00250000001126 | Cell Adhesion Molecule Precursor Immunoglobulin Superfamily Member Nectin Necl Synaptic Cell Adhesion Molecule |
| ENSMUSG00000005357 | ENSFM00800002003375 | Excitatory Amino Acid Transporter Sodium Dependent Glutamate/Aspartate Transporter Solute Carrier Family 1 Member |
| ENSMUSG00000005360 | ENSFM00800002003375 | Excitatory Amino Acid Transporter Sodium Dependent Glutamate/Aspartate Transporter Solute Carrier Family 1 Member |
| ENSMUSG00000005374 | ENSFM00570000897418 | Unknown |
| ENSMUSG00000005374 | ENSFM00810002057469 | Unknown |
| ENSMUSG00000005374 | ENSFM00250000006389 | Transducin Beta 2 |
| ENSMUSG00000005417 | ENSFM00600000921208 | Trio And F Actin Binding Protein Tara Trio Associated Repeat On Actin |
| ENSMUSG00000005447 | ENSFM00250000004414 | Platelet Activating Factor Acetylhydrolase Ib Subunit Beta EC_3.1.1.47 Paf Acetylhydrolase 30 Kda Subunit Paf Ah 30 Kda Subunit Paf Ah Subunit Beta Pafah Subunit Beta |
| ENSMUSG00000005469 | ENSFM00790001962972 | Camp Dependent Kinase Catalytic Subunit Pka C EC_2.7.11.11 |
| ENSMUSG00000005483 | ENSFM00440000236855 | Dnaj Homolog Subfamily B Member |
| ENSMUSG00000005510 | ENSFM00790001963038 | Nadh Dehydrogenase Iron Sulfur 3 Mitochondrial Precursor EC_1.6.5.3 EC_1.6.99.- 3 Complex I 30KD Ci 30KD Nadh Ubiquinone Oxidoreductase 30 Kda Subunit |
| ENSMUSG00000005514 | ENSFM00570000894700 | Unknown |
| ENSMUSG00000005514 | ENSFM00750001632390 | Nadph Cytochrome P450 Reductase Cpr P450R EC_1.6.2.4 |
| ENSMUSG00000005533 | ENSFM00730001521215 | Insulin Receptor Precursor EC_2.7.10.1 |
| ENSMUSG00000005575 | ENSFM00570000885916 | Unknown |
| ENSMUSG00000005575 | ENSFM00500000270983 | NEDD8 Conjugating Enzyme UBC12 EC_6.3.2.- NEDD8 Carrier NEDD8 Ligase Ubiquitin Conjugating Enzyme E2 M |
| ENSMUSG00000005580 | ENSFM00740001589278 | Adenylate Cyclase Type 9 EC_4.6.1.1 Atp Pyrophosphate Lyase 9 Adenylate Cyclase Type Ix Adenylyl Cyclase 9 |
| ENSMUSG00000005610 | ENSFM00590000918294 | Unknown |
| ENSMUSG00000005610 | ENSFM00320000100157 | Eukaryotic Translation Initiation Factor 4 Gamma 2 Eif 4 Gamma 2 Eif 4G 2 EIF4G 2 Apobec 1 Target 1 Translation Repressor NAT1 P97 |
| ENSMUSG00000005625 | ENSFM00810002040686 | 26S Proteasome Non Atpase Regulatory Subunit 4 26S Proteasome Regulatory Subunit RPN10 26S Proteasome Regulatory Subunit S5A Multiubiquitin Chain Binding |
| ENSMUSG00000005656 | ENSFM00250000002044 | Sorting Nexin |
| ENSMUSG00000005672 | ENSFM00730001521364 | Mast/Stem Cell Growth Factor Receptor Kit Precursor Scfr EC_2.7.10.1 Proto Oncogene C Kit Tyrosine Kinase Kit |
| ENSMUSG00000005674 | ENSFM00700001403680 | Unknown |
| ENSMUSG00000005674 | ENSFM00250000002521 | Mitochondrial Import Receptor Subunit TOM40 Homolog Translocase Of Outer Membrane 40 Kda Subunit Homolog |
| ENSMUSG00000005683 | ENSFM00250000002339 | Citrate Synthase Mitochondrial Precursor EC_2.3.3.1 Citrate Si Synthase |
| ENSMUSG00000005686 | ENSFM00770001847533 | Amp Deaminase EC_3.5.4.6 Amp Deaminase |
| ENSMUSG00000005716 | ENSFM00670001236518 | Parvalbumin Alpha |
| ENSMUSG00000005779 | ENSFM00800002003556 | Proteasome Subunit Beta Type 4 Precursor EC_3.4.25.1 Macropain Beta Chain Multicatalytic Endopeptidase Complex Beta Chain Proteasome Beta Chain Proteasome Chain 3 |
| ENSMUSG00000005803 | ENSFM00660001169747 | Unknown |
| ENSMUSG00000005803 | ENSFM00660001169748 | Unknown |
| ENSMUSG00000005803 | ENSFM00810002040854 | Sulfide:Quinone Oxidoreductase Mitochondrial Precursor Sqor EC_1.8.5.- |
| ENSMUSG00000005871 | ENSFM00610000963080 | Unknown |
| ENSMUSG00000005871 | ENSFM00610000970304 | Unknown |
| ENSMUSG00000005871 | ENSFM00250000001676 | Adenomatous Polyposis Coli |
| ENSMUSG00000005873 | ENSFM00500000270563 | Receptor Expression Enhancing Polyposis Locus 1 |
| ENSMUSG00000005882 | ENSFM00570000878392 | Unknown |
| ENSMUSG00000005882 | ENSFM00570000896684 | Unknown |
| ENSMUSG00000005882 | ENSFM00570000898900 | Unknown |
| ENSMUSG00000005882 | ENSFM00590000918369 | Unknown |
| ENSMUSG00000005882 | ENSFM00250000004739 | Ubiquinol Cytochrome C Reductase Complex Assembly Factor 1 Basic Fgf Repressed Zic Binding Ubiquinol Cytochrome C Reductase Complex Chaperone CBP3 Homolog |
| ENSMUSG00000005958 | ENSFM00590000918408 | Unknown |
| ENSMUSG00000005958 | ENSFM00730001521338 | Ephrin Type B Receptor Kinase |
| ENSMUSG00000005981 | ENSFM00720001503502 | Unknown |
| ENSMUSG00000005981 | ENSFM00730001522162 | Heat Shock 75 Kda Mitochondrial Precursor Hsp 75 Tnfr Associated 1 Tumor Necrosis Factor Type 1 Receptor Associated Trap 1 |
| ENSMUSG00000006010 | ENSFM00250000007657 | Odr 4 Homolog |
| ENSMUSG00000006024 | ENSFM00730001522002 | Soluble Nsf Attachment Snap N Ethylmaleimide Sensitive Factor Attachment |
| ENSMUSG00000006169 | ENSFM00500000270815 | Clathrin Interactor 1 Enthoprotin Epsin 4 Epsin Related Epsinr |
| ENSMUSG00000006205 | ENSFM00250000001030 | Serine Protease HTRA2 Mitochondrial |
| ENSMUSG00000006273 | ENSFM00810002040578 | V Type Proton Atpase Subunit B V Atpase Subunit B Endomembrane Proton Pump 58 Kda Subunit Vacuolar Proton Pump Subunit B |
| ENSMUSG00000006276 | ENSFM00250000001830 | Epidermal Growth Factor Receptor Substrate 15 1 EPS15 Related EPS15R |
| ENSMUSG00000006304 | ENSFM00760001763096 | Unknown |
| ENSMUSG00000006304 | ENSFM00800002003702 | Actin Related 2/3 Complex Subunit 2 ARP2/3 Complex 34 Kda Subunit P34 Arc |
| ENSMUSG00000006333 | ENSFM00500000295123 | Unknown |
| ENSMUSG00000006333 | ENSFM00570000893535 | Unknown |
| ENSMUSG00000006333 | ENSFM00250000003284 | 40S Ribosomal S9 |
| ENSMUSG00000006356 | ENSFM00740001589457 | Cysteine Rich Crp |
| ENSMUSG00000006373 | ENSFM00670001235707 | Membrane Associated Progesterone Receptor Component |
| ENSMUSG00000006390 | ENSFM00800002003497 | Elongation Of Very Long Chain Fatty Acids 7 EC_2.3.1.199 3 Keto Acyl Coa Synthase ELOVL7 Elovl Fatty Acid Elongase 7 Elovl Fa Elongase 7 Very Long Chain 3 Oxoacyl Coa Synthase 7 |
| ENSMUSG00000006412 | ENSFM00500000273880 | Prefoldin Subunit 2 |
| ENSMUSG00000006456 | ENSFM00350000105492 | Rna Binding 14 Rna Binding Motif 14 |
| ENSMUSG00000006464 | ENSFM00250000005034 | Bardet Biedl Syndrome 1 BBS2 2 |
| ENSMUSG00000006476 | ENSFM00570000884423 | Unknown |
| ENSMUSG00000006476 | ENSFM00250000002451 | Nmda Receptor Synaptonuclear Signaling And Neuronal Migration Factor Juxtasynaptic Attractor Of Caldendrin On Dendritic Boutons Jacob Nasal Embryonic Luteinizing Hormone Releasing Hormone Factor Nasal Embryonic Lhrh Factor |
| ENSMUSG00000006494 | ENSFM00500000269765 | Kinase Isozyme Mitochondrial Precursor EC_2.7.11.2 Pyruvate Dehydrogenase Kinase |
| ENSMUSG00000006498 | ENSFM00610000973070 | Unknown |
| ENSMUSG00000006498 | ENSFM00610000973071 | Unknown |
| ENSMUSG00000006498 | ENSFM00610000973072 | Unknown |
| ENSMUSG00000006498 | ENSFM00260000050389 | Polypyrimidine Tract Binding |
| ENSMUSG00000006522 | ENSFM00250000000536 | Inter Alpha Trypsin Inhibitor Heavy Chain Precursor Iti Heavy Chain Iti Inter Alpha Inhibitor Heavy Chain |
| ENSMUSG00000006527 | ENSFM00260000050568 | Scm With Four Mbt Domains |
| ENSMUSG00000006574 | ENSFM00730001521491 | Anion Exchange Ae Anion Exchanger Solute Carrier Family 4 Member |
| ENSMUSG00000006575 | ENSFM00250000002370 | Run Domain Containing |
| ENSMUSG00000006576 | ENSFM00570000880704 | Unknown |
| ENSMUSG00000006576 | ENSFM00730001521491 | Anion Exchange Ae Anion Exchanger Solute Carrier Family 4 Member |
| ENSMUSG00000006651 | ENSFM00250000000617 | Amyloid Beta A4 Precursor Abpp App Alzheimer Disease Amyloid A4 Homolog |
| ENSMUSG00000006699 | ENSFM00730001521235 | Cell Division Control 42 Homolog Precursor |
| ENSMUSG00000006717 | ENSFM00500000273552 | Acyl Coenzyme A Thioesterase 13 Acyl Coa Thioesterase 13 EC_3.1.2.- Thioesterase Superfamily Member 2 |
| ENSMUSG00000006731 | ENSFM00250000004460 | Beta 1 4 N Acetylgalactosaminyltransferase 1 EC_2.4.1.92 N Acetylneuraminyl Galactosylglucosylceramide GM2/GD2 Synthase Galnac T |
| ENSMUSG00000006740 | ENSFM00670001235340 | Kinesin Heavy Chain Kinesin Heavy Chain Kinesin Heavy Chain |
| ENSMUSG00000006782 | ENSFM00740001589364 | 2' 3' Cyclic Nucleotide 3' Phosphodiesterase Precursor Cnp Cnpase EC_3.1.4.37 |
| ENSMUSG00000006818 | ENSFM00780001898344 | Superoxide Dismutase Mitochondrial EC_1.15.1.1 |
| ENSMUSG00000006930 | ENSFM00500000273792 | Huntingtin Associated 1 Hap 1 |
| ENSMUSG00000006932 | ENSFM00570000883608 | Unknown |
| ENSMUSG00000006932 | ENSFM00610000969916 | Unknown |
| ENSMUSG00000006932 | ENSFM00250000001012 | Catenin Beta Beta Catenin |
| ENSMUSG00000006998 | ENSFM00610000967351 | Unknown |
| ENSMUSG00000006998 | ENSFM00250000002637 | 26S Proteasome Regulatory Subunit RPN1 Regulatory 26S Proteasome Subunit |
| ENSMUSG00000007021 | ENSFM00670001235552 | Synaptogyrin |
| ENSMUSG00000007029 | ENSFM00250000001468 | Valine Trna Ligase EC_6.1.1.9 Valyl Trna Synthetase Valrs |
| ENSMUSG00000007036 | ENSFM00640001113079 | Unknown |
| ENSMUSG00000007036 | ENSFM00250000005259 | Abhydrolase Domain Containing 16A Ec 3 Hla B Associated Transcript 5 |
| ENSMUSG00000007039 | ENSFM00250000003817 | N G N G Dimethylarginine Dimethylaminohydrolase 2 Ddah 2 Dimethylarginine Dimethylaminohydrolase 2 EC_3.5.3.18 Ddahii Dimethylargininase 2 |
| ENSMUSG00000007097 | ENSFM00650001139994 | Sodium/Potassium Transporting Atpase Subunit Alpha Na + /K + Atpase Alpha Subunit EC_3.6.3.9 Sodium Pump Subunit Alpha |
| ENSMUSG00000007107 | ENSFM00650001139994 | Sodium/Potassium Transporting Atpase Subunit Alpha Na + /K + Atpase Alpha Subunit EC_3.6.3.9 Sodium Pump Subunit Alpha |
| ENSMUSG00000007207 | ENSFM00250000000549 | Syntaxin |
| ENSMUSG00000007338 | ENSFM00500000273530 | 39S Ribosomal L49 Mitochondrial L49MT Mrp L49 |
| ENSMUSG00000007411 | ENSFM00730001521220 | Kinase EC_2.7.11.1 |
| ENSMUSG00000007458 | ENSFM00250000007261 | Cation Dependent Mannose 6 Phosphate Receptor Precursor Cd Man 6 P Receptor Cd Mpr 46 Kda Mannose 6 Phosphate Receptor Mpr 46 |
| ENSMUSG00000007476 | ENSFM00250000001418 | Volume Regulated Anion Channel Subunit Leucine Rich Repeat Containing |
| ENSMUSG00000007564 | ENSFM00640001117298 | Unknown |
| ENSMUSG00000007564 | ENSFM00250000001941 | Serine/Threonine Phosphatase 2A 65 Kda Regulatory Subunit A PP2A Subunit A PR65 PP2A Subunit A R1 |
| ENSMUSG00000007594 | ENSFM00730001521886 | Hyaluronan And Proteoglycan Link 1 Precursor Cartilage Linking 1 Cartilage Link Proteoglycan Link |
| ENSMUSG00000007617 | ENSFM00250000001140 | Homer Homolog Homer Vasp/Ena Related Gene Up Regulated During Seizure And Ltp Vesl |
| ENSMUSG00000007653 | ENSFM00400000131718 | Gamma Aminobutyric Acid Receptor Subunit Beta Precursor Gaba A Receptor Subunit Beta |
| ENSMUSG00000007659 | ENSFM00570000889665 | Unknown |
| ENSMUSG00000007659 | ENSFM00500000270604 | Bcl 2 1 BCL2 L 1 Apoptosis Regulator Bcl X |
| ENSMUSG00000007739 | ENSFM00640001120031 | Unknown |
| ENSMUSG00000007739 | ENSFM00640001120032 | Unknown |
| ENSMUSG00000007739 | ENSFM00730001521612 | T Complex 1 Subunit Delta Tcp 1 Delta Cct Delta |
| ENSMUSG00000007783 | ENSFM00500000269758 | Carnitine O Palmitoyltransferase 1 Muscle CPT1 M EC_2.3.1.21 Carnitine O Palmitoyltransferase I Muscle Cpt I Cpti M Carnitine Palmitoyltransferase 1B |
| ENSMUSG00000007815 | ENSFM00780001914034 | Unknown |
| ENSMUSG00000007815 | ENSFM00730001521223 | Rho Related Gtp Binding Precursor |
| ENSMUSG00000007891 | ENSFM00720001492011 | Precursor |
| ENSMUSG00000007892 | ENSFM00810002041089 | 60S Acidic Ribosomal P1 |
| ENSMUSG00000007944 | ENSFM00670001235990 | Tetratricopeptide Repeat Tpr Repeat |
| ENSMUSG00000008036 | ENSFM00670001235578 | Ap 2 Complex Subunit Sigma Adaptor Complex Ap 2 Subunit Sigma Adaptor Related Complex 2 Subunit Sigma Clathrin Assembly 2 Sigma Small Chain Clathrin Coat Assembly AP17 Clathrin Coat Associated AP17 Plasma Membrane Adaptor Ap 2.17 Kda SIGMA2 Adaptin |
| ENSMUSG00000008140 | ENSFM00250000006413 | Er Membrane Complex Subunit 10 Precursor |
| ENSMUSG00000008153 | ENSFM00250000001322 | Calsyntenin Precursor Alcadein Alc |
| ENSMUSG00000008435 | ENSFM00500000269664 | Retinol Dehydrogenase |
| ENSMUSG00000008475 | ENSFM00500000270526 | Unknown |
| ENSMUSG00000008475 | ENSFM00670001241191 | Actin Related 2/3 Complex Subunit 5 |
| ENSMUSG00000008489 | ENSFM00660001189998 | Unknown |
| ENSMUSG00000008489 | ENSFM00250000000588 | Elav |
| ENSMUSG00000008540 | ENSFM00570000879881 | Unknown |
| ENSMUSG00000008540 | ENSFM00500000272807 | Microsomal Glutathione S Transferase 1 Microsomal Gst 1 EC_2.5.1.18 Microsomal Gst I |
| ENSMUSG00000008540 | ENSFM00520000556550 | Microsomal Glutathione S Transferase 1 |
| ENSMUSG00000008683 | ENSFM00570000895097 | Unknown |
| ENSMUSG00000008683 | ENSFM00570000896056 | Unknown |
| ENSMUSG00000008683 | ENSFM00250000002123 | 40S Ribosomal |
| ENSMUSG00000008734 | ENSFM00670001235445 | G Coupled Receptor Family C Group 5 Member Retinoic Acid Induced Gene Raig |
| ENSMUSG00000008822 | ENSFM00500000296684 | Acylphosphatase 1 |
| ENSMUSG00000008822 | ENSFM00670001236427 | Acylphosphatase 1 EC_3.6.1.7 Acylphosphatase Organ Common Type Isozyme Acylphosphate Phosphohydrolase 1 |
| ENSMUSG00000008859 | ENSFM00500000270233 | Ras Related Precursor |
| ENSMUSG00000008892 | ENSFM00250000001189 | Voltage Dependent Anion Selective Channel Vdac Outer Mitochondrial Membrane Porin |
| ENSMUSG00000009013 | ENSFM00670001235517 | Dynein Light Chain Cytoplasmic 8 Kda Dynein Light Chain Dynein Light Chain LC8 Type |
| ENSMUSG00000009073 | ENSFM00610000969488 | Unknown |
| ENSMUSG00000009073 | ENSFM00610000973510 | Unknown |
| ENSMUSG00000009073 | ENSFM00750001632408 | Merlin Moesin Ezrin Radixin Neurofibromin 2 Schwannomin |
| ENSMUSG00000009079 | ENSFM00500000270534 | Rna Binding Ews |
| ENSMUSG00000009079 | ENSFM00500000311767 | Rna Binding Ews |
| ENSMUSG00000009090 | ENSFM00430000230089 | Ap 1 Complex Subunit Beta 1 Adaptor Complex Ap 1 Subunit Beta 1 Adaptor Related Complex 1 Subunit Beta 1 Beta 1 Adaptin Beta Adaptin 1 Clathrin Assembly Complex 1 Beta Large Chain Golgi Adaptor HA1/AP1 Adaptin Beta Subunit |
| ENSMUSG00000009112 | ENSFM00250000005222 | Bcl 2 13 BCL2 L 13 Bcl Rambo MIL1 |
| ENSMUSG00000009207 | ENSFM00570000889618 | Unknown |
| ENSMUSG00000009207 | ENSFM00250000003767 | Lunapark |
| ENSMUSG00000009216 | ENSFM00500000274513 | Unknown |
| ENSMUSG00000009394 | ENSFM00250000000771 | Synapsin Synapsin |
| ENSMUSG00000009418 | ENSFM00250000000514 | Neuron Navigator Pore Membrane And/Or Filament Interacting Steerin Unc 53 Homolog Un |
| ENSMUSG00000009535 | ENSFM00250000004534 | Cap Guanine N7 Methyltransferase EC_2.1.1.56 RG7MT1 Guanine N 7 Methyltransferase Cap Methyltransferase |
| ENSMUSG00000009549 | ENSFM00500000420639 | Signal Recognition Particle 14 Kda |
| ENSMUSG00000009549 | ENSFM00670001236179 | Signal Recognition Particle 14 Kda SRP14 |
| ENSMUSG00000009588 | ENSFM00250000002806 | Alpha N Acetylgalactosaminide Alpha 2 6 Sialyltransferase 2 EC_2.4.99.- Galnac Alpha 2 6 Sialyltransferase Ii ST6GALNAC Ii ST6GALNACII Sialyltransferase 7B SIAT7 B |
| ENSMUSG00000009647 | ENSFM00590000918380 | Unknown |
| ENSMUSG00000009647 | ENSFM00250000002939 | Calcium Uniporter Regulatory Subunit Mcub Mitochondrial Precursor Mcub Coiled Coil Domain Containing 109B |
| ENSMUSG00000009670 | ENSFM00250000004461 | Testis Expressed Sequence 11 |
| ENSMUSG00000009681 | ENSFM00250000000543 | Breakpoint Cluster Region |
| ENSMUSG00000009731 | ENSFM00300000079128 | Potassium Voltage Gated Channel Subfamily D Member Voltage Gated Potassium Channel Subunit KV4 |
| ENSMUSG00000009741 | ENSFM00250000001310 | Transcription Factor CP2 |
| ENSMUSG00000009863 | ENSFM00800002003507 | Succinate Dehydrogenase Iron Sulfur Subunit Mitochondrial Precursor EC_1.3.5.1 Iron Sulfur Subunit Of Complex Ii Ip |
| ENSMUSG00000009894 | ENSFM00570000876467 | Unknown |
| ENSMUSG00000009894 | ENSFM00570000891790 | Unknown |
| ENSMUSG00000009894 | ENSFM00570000894302 | Unknown |
| ENSMUSG00000009894 | ENSFM00250000004681 | Synaptosomal Associated 47 Snap 47 Synaptosomal Associated 47 Kda |
| ENSMUSG00000009927 | ENSFM00670001235896 | 40S Ribosomal S25 |
| ENSMUSG00000010025 | ENSFM00250000000672 | Aldehyde Dehydrogenase Aldehyde Dehydrogenase Aldehyde Dehydrogenase Family 3 Member |
| ENSMUSG00000010064 | ENSFM00250000000644 | Sodium Coupled Neutral Amino Acid Transporter Amino Acid Transporter Solute Carrier Family 38 Member System A Amino Acid Transporter 1 System N Amino Acid Transporter |
| ENSMUSG00000010066 | ENSFM00730001522093 | Voltage Dependent Calcium Channel Subunit Alpha 2/Delta 1 Precursor Voltage Gated Calcium Channel Subunit Alpha 2/Delta 1 |
| ENSMUSG00000010080 | ENSFM00500000269762 | Epsin Eps 15 Interacting |
| ENSMUSG00000010086 | ENSFM00250000007086 | Ring Finger 112 Brain Finger Zinc Finger 179 |
| ENSMUSG00000010095 | ENSFM00250000004066 | 4F2 Cell Surface Antigen Heavy Chain 4F2HC Solute Carrier Family 3 Member 2 CD98 Antigen |
| ENSMUSG00000010110 | ENSFM00660001179029 | Unknown |
| ENSMUSG00000010110 | ENSFM00660001179821 | Unknown |
| ENSMUSG00000010110 | ENSFM00660001179822 | Unknown |
| ENSMUSG00000010110 | ENSFM00760001714795 | Syntaxin 5 |
| ENSMUSG00000010277 | ENSFM00570000880907 | Unknown |
| ENSMUSG00000010277 | ENSFM00250000003975 | Precursor |
| ENSMUSG00000010376 | ENSFM00500000292193 | NEDD8 Precursor Neddylin Ubiquitin NEDD8 |
| ENSMUSG00000010392 | ENSFM00500000270907 | Golgi Snap Receptor Complex Member 1.28 Kda Golgi Snare 28 Kda Cis Golgi Snare P28 |
| ENSMUSG00000010660 | ENSFM00740001589144 | 1 Phosphatidylinositol 4 5 Bisphosphate Phosphodiesterase Delta EC_3.1.4.11 Phosphoinositide Phospholipase C Delta Phospholipase C Delta Plc Delta |
| ENSMUSG00000010663 | ENSFM00250000001297 | Fatty Acid Desaturase 2 EC_1.14.19.- Delta 6 Fatty Acid Desaturase Desaturase Delta 6 Desaturase |
| ENSMUSG00000010760 | ENSFM00500000282174 | Pleckstrin Homology Domain Family A Member 2 Imprinted In Placenta And Liver |
| ENSMUSG00000010803 | ENSFM00610000973527 | Unknown |
| ENSMUSG00000010803 | ENSFM00800002003385 | Gamma Aminobutyric Acid Receptor Subunit Alpha Precursor Gaba A Receptor Subunit Alpha |
| ENSMUSG00000010825 | ENSFM00500000273079 | Delphilin Glutamate Receptor Ionotropic Delta 2 Interacting 1 |
| ENSMUSG00000010914 | ENSFM00800002003776 | Pyruvate Dehydrogenase X Component Mitochondrial Precursor Dihydrolipoamide Dehydrogenase Binding Of Pyruvate Dehydrogenase Complex Lipoyl Containing Pyruvate Dehydrogenase Complex Component X |
| ENSMUSG00000010936 | ENSFM00760001714983 | VAC14 Homolog |
| ENSMUSG00000011589 | ENSFM00250000002725 | Fibronectin Type Iii And Spry Domain Containing 1 |
| ENSMUSG00000011751 | ENSFM00740001589095 | Spectrin Beta Chain Erythrocytic Beta Spectrin |
| ENSMUSG00000011752 | ENSFM00250000001043 | Phosphoglycerate Mutase EC_3.1.3.13 EC_5.4.2.- 11 EC_5.4.-.- 2 4 Bpg Dependent Pgam Phosphoglycerate Mutase Isozyme Pgam |
| ENSMUSG00000011832 | ENSFM00500000270092 | Ecotropic Viral Integration Site 5 |
| ENSMUSG00000011877 | ENSFM00250000001490 | Arf Gtpase Activating GIT1 Arf Gap GIT1 Cool Associated And Tyrosine Phosphorylated 1 Cat 1 CAT1 G Coupled Receptor Kinase Interactor 1 Grk Interacting 1 |
| ENSMUSG00000011884 | ENSFM00250000005320 | Glycolipid Transfer Gltp |
| ENSMUSG00000012848 | ENSFM00250000003056 | 40S Ribosomal S5 |
| ENSMUSG00000013033 | ENSFM00260000050328 | Latrophilin |
| ENSMUSG00000013160 | ENSFM00800002003469 | V Type Proton Atpase Subunit D V Atpase Subunit D Vacuolar Proton Pump Subunit D |
| ENSMUSG00000013236 | ENSFM00740001589097 | Receptor Type Tyrosine Phosphatase Precursor EC_3.1.3.48 |
| ENSMUSG00000013367 | ENSFM00290000065521 | Precursor |
| ENSMUSG00000013495 | ENSFM00250000006710 | Transmembrane 175 |
| ENSMUSG00000013523 | ENSFM00250000007762 | Breast Carcinoma Amplified Sequence 1 Amplified In Breast Cancer 1 |
| ENSMUSG00000013593 | ENSFM00250000004138 | Nadh Dehydrogenase Iron Sulfur 2 Mitochondrial Precursor EC_1.6.5.3 EC_1.6.99.- 3 Complex I 49KD Ci 49KD Nadh Ubiquinone Oxidoreductase 49 Kda Subunit |
| ENSMUSG00000013629 | ENSFM00670001235391 | Cad Glutamine Dependent Carbamoyl Phosphate Synthase EC_6.3.5.5 |
| ENSMUSG00000013629 | ENSFM00730001521216 | Carbamoyl Phosphate Synthase Mitochondrial Precursor EC_6.3.4.16 Carbamoyl Phosphate Synthetase I Cpsase I |
| ENSMUSG00000013662 | ENSFM00790001963005 | Atpase Family Aaa Domain Containing 1 EC_3.6.1.3 |
| ENSMUSG00000013698 | ENSFM00250000010619 | Astrocytic Phosphoprotein Pea 15.15 Kda Phosphoprotein Enriched In Astrocytes |
| ENSMUSG00000013701 | ENSFM00610000967248 | Unknown |
| ENSMUSG00000013701 | ENSFM00610000967249 | Unknown |
| ENSMUSG00000013701 | ENSFM00250000005363 | Mitochondrial Import Inner Membrane Translocase Subunit TIM23 |
| ENSMUSG00000013858 | ENSFM00250000003298 | Membralin Transmembrane 259 |
| ENSMUSG00000013878 | ENSFM00250000003576 | E3 Ubiquitin Ligase RNF170 EC_6.3.2.- Ring Finger 170 |
| ENSMUSG00000013921 | ENSFM00760001720057 | Unknown |
| ENSMUSG00000013921 | ENSFM00740001589331 | Cap Gly Domain Containing Linker 3 Cytoplasmic Linker 170 Related 59 Kda Clip 170 Related 59 Kda Clipr 59 |
| ENSMUSG00000014077 | ENSFM00570000879445 | Unknown |
| ENSMUSG00000014077 | ENSFM00500000270459 | Calcineurin B Homologous 1 Calcineurin B Calcium Binding Chp Calcium Binding P22 Ef Hand Calcium Binding Domain Containing P22 |
| ENSMUSG00000014226 | ENSFM00250000005060 | Calcyclin Binding Cacybp |
| ENSMUSG00000014294 | ENSFM00670001236219 | Nadh Dehydrogenase 1 Alpha Subcomplex Subunit 2 Complex I B8 Ci B8 Nadh Ubiquinone Oxidoreductase B8 Subunit |
| ENSMUSG00000014301 | ENSFM00570000887254 | Unknown |
| ENSMUSG00000014301 | ENSFM00670001236447 | Mitochondrial Import Inner Membrane Translocase Subunit TIM16 Mitochondria Associated Granulocyte Macrophage Csf Signaling Molecule Presequence Translocated Associated Motor Subunit PAM16 |
| ENSMUSG00000014313 | ENSFM00500000410038 | Unknown |
| ENSMUSG00000014313 | ENSFM00570000879209 | Unknown |
| ENSMUSG00000014313 | ENSFM00570000894887 | Unknown |
| ENSMUSG00000014353 | ENSFM00250000002118 | Transmembrane Precursor |
| ENSMUSG00000014361 | ENSFM00730001521921 | Tyrosine Kinase Precursor EC_2.7.10.1 |
| ENSMUSG00000014402 | ENSFM00500000270204 | Tumor Susceptibility Gene 101 Escrt I Complex Subunit TSG101 |
| ENSMUSG00000014601 | ENSFM00740001589092 | Adenosylhomocysteinase Adohcyase EC_3.3.1.1 S Adenosyl L Homocysteine Hydrolase 2 |
| ENSMUSG00000014602 | ENSFM00760001763121 | Unknown |
| ENSMUSG00000014602 | ENSFM00760001763122 | Unknown |
| ENSMUSG00000014602 | ENSFM00800002003381 | Kinesin |
| ENSMUSG00000014606 | ENSFM00570000892647 | Unknown |
| ENSMUSG00000014606 | ENSFM00770001847568 | Mitochondrial 2 Oxoglutarate/Malate Carrier Ogcp Solute Carrier Family 25 Member 11 |
| ENSMUSG00000014769 | ENSFM00800002003627 | Proteasome Subunit Beta Type 1 EC_3.4.25.1 Proteasome |
| ENSMUSG00000014846 | ENSFM00250000003009 | Tubulin Polymerization Promoting Family Member |
| ENSMUSG00000014867 | ENSFM00700001390183 | Surfeit Locus |
| ENSMUSG00000014932 | ENSFM00730001521177 | Tyrosine Kinase EC_2.7.10.2 |
| ENSMUSG00000014956 | ENSFM00810002057428 | Unknown |
| ENSMUSG00000014956 | ENSFM00420000140518 | Serine/Threonine Phosphatase PP1 Catalytic Subunit Pp EC_3.1.3.16 |
| ENSMUSG00000015002 | ENSFM00640001109165 | Unknown |
| ENSMUSG00000015002 | ENSFM00250000001283 | EFR3 Homolog |
| ENSMUSG00000015016 | ENSFM00800002003672 | Acyl Coa Synthetase Family Member 3 Mitochondrial Precursor EC_6.2.1.- |
| ENSMUSG00000015085 | ENSFM00250000001339 | Ectonucleoside Triphosphate Diphosphohydrolase Ntpdase EC_3.6.1.5 Ecto Atp Diphosphohydrolase Ecto Atpdase Ecto Atpase Ecto Apyrase |
| ENSMUSG00000015087 | ENSFM00250000002978 | Rab 6 Gtp Binding Parf Rab 1 RBEL1 |
| ENSMUSG00000015090 | ENSFM00500000272118 | Prostaglandin H2 D Isomerase Precursor EC_5.3.99.2 Glutathione Independent Pgd Synthase Lipocalin Type Prostaglandin D Synthase Prostaglandin D2 Synthase PGD2 Synthase Pgds PGDS2 |
| ENSMUSG00000015092 | ENSFM00670001235719 | Endothelial Differentiation Related Factor 1 Edf 1 Multiprotein Bridging Factor 1 MBF1 |
| ENSMUSG00000015094 | ENSFM00250000007990 | Neural Proliferation Differentiation And Control 1 Precursor Npdc 1 |
| ENSMUSG00000015112 | ENSFM00800002023336 | Unknown |
| ENSMUSG00000015112 | ENSFM00800002003431 | Calcium Binding Mitochondrial Carrier Mitochondrial Aspartate Glutamate Carrier Solute Carrier Family 25 Member |
| ENSMUSG00000015149 | ENSFM00810002040646 | Nad Dependent Deacetylase Sirtuin 2 EC_3.5.1.- Regulatory SIR2 Homolog 2 SIR2 2 |
| ENSMUSG00000015165 | ENSFM00260000050504 | Heterogeneous Nuclear Ribonucleoprotein L |
| ENSMUSG00000015214 | ENSFM00730001521553 | Myotubularin Phosphatidylinositol 3 5 Bisphosphate 3 Phosphatase EC_3.1.3.95 |
| ENSMUSG00000015222 | ENSFM00640001118144 | Unknown |
| ENSMUSG00000015222 | ENSFM00640001119229 | Unknown |
| ENSMUSG00000015222 | ENSFM00260000050441 | Microtubule Associated Tau Neurofibrillary Tangle Paired Helical Filament Tau Phf Tau |
| ENSMUSG00000015224 | ENSFM00400000131705 | Cytochrome P450 |
| ENSMUSG00000015243 | ENSFM00740001589079 | Atp Binding Cassette Sub Family A Member Atp Binding Cassette Transporter Atp Binding Cassette |
| ENSMUSG00000015247 | ENSFM00250000007263 | Nipsnap Homolog 3A NIPSNAP3A |
| ENSMUSG00000015290 | ENSFM00570000863849 | Unknown |
| ENSMUSG00000015290 | ENSFM00500000273690 | Ubiquitin 4A |
| ENSMUSG00000015291 | ENSFM00570000895605 | Unknown |
| ENSMUSG00000015291 | ENSFM00800002003449 | Rab Gdp Dissociation Inhibitor Alpha Rab Gdi Alpha Guanosine Diphosphate Dissociation Inhibitor 1 Gdi 1 |
| ENSMUSG00000015337 | ENSFM00790001963101 | Endonuclease G Mitochondrial Precursor Endo G EC_3.1.30.- |
| ENSMUSG00000015341 | ENSFM00250000003047 | Golgin Subfamily A Member 7 |
| ENSMUSG00000015363 | ENSFM00610000965646 | Unknown |
| ENSMUSG00000015363 | ENSFM00250000005731 | Trab Domain Containing |
| ENSMUSG00000015476 | ENSFM00600000921799 | Proline Rich Transmembrane 1 Dispanin Subfamily D Member 1 DSPD1 |
| ENSMUSG00000015488 | ENSFM00570000881220 | Unknown |
| ENSMUSG00000015488 | ENSFM00500000272815 | Calcium Channel Flower Homolog Calcium Channel Flower Domain Containing 1 |
| ENSMUSG00000015656 | ENSFM00730001521103 | Heat Shock Kda Heat Shock 70 Kda |
| ENSMUSG00000015668 | ENSFM00400000131930 | Pdz Domain Containing 11 |
| ENSMUSG00000015671 | ENSFM00800002003520 | Proteasome Subunit Alpha Type 2 EC_3.4.25.1 Macropain Subunit C3 Multicatalytic Endopeptidase Complex Subunit C3 Proteasome Component C3 |
| ENSMUSG00000015714 | ENSFM00730001521585 | Ceramide Synthase LAG1 Longevity Assurance Homolog |
| ENSMUSG00000015733 | ENSFM00250000001955 | F Actin Capping Subunit Alpha 2 Capz Alpha 2 |
| ENSMUSG00000015747 | ENSFM00250000005970 | Vacuolar Sorting Associated 45 |
| ENSMUSG00000015766 | ENSFM00570000899024 | Unknown |
| ENSMUSG00000015766 | ENSFM00500000270023 | Epidermal Growth Factor Receptor Kinase Substrate 8 EPS8 Epidermal Growth Factor Receptor Pathway Substrate 8 Related EPS8 Related |
| ENSMUSG00000015806 | ENSFM00250000004373 | Dihydropteridine Reductase EC_1.5.1.34 Hdhpr Quinoid Dihydropteridine Reductase |
| ENSMUSG00000015829 | ENSFM00740001589132 | Tenascin Precursor Tn |
| ENSMUSG00000015837 | ENSFM00250000004435 | Sequestosome 1 Ubiquitin Binding P62 |
| ENSMUSG00000015869 | ENSFM00730001521917 | Phosphoribosyl Pyrophosphate Synthase Associated Prpp Synthase Associated |
| ENSMUSG00000015889 | ENSFM00810002040760 | Leukotriene A 4 Hydrolase Lta 4 Hydrolase EC_3.3.2.6 Leukotriene A 4 Hydrolase |
| ENSMUSG00000015932 | ENSFM00500000270025 | Cofilin Cofilin Muscle |
| ENSMUSG00000015943 | ENSFM00500000273601 | Bola 1 |
| ENSMUSG00000016150 | ENSFM00250000000375 | Teneurin Ten Odd Oz/Ten M Homolog Tenascin Ten Teneurin Transmembrane |
| ENSMUSG00000016252 | ENSFM00500000444477 | Unknown |
| ENSMUSG00000016319 | ENSFM00250000000816 | Adp/Atp Translocase Adp Atp Carrier Adenine Nucleotide Translocator Ant Solute Carrier Family 25 Member |
| ENSMUSG00000016346 | ENSFM00250000000417 | Potassium Voltage Gated Channel Subfamily Kqt Member Kqt Potassium Channel Subunit Alpha Voltage Gated Potassium Channel Subunit KV7 |
| ENSMUSG00000016349 | ENSFM00800002003364 | Elongation Factor 1 Alpha Ef 1 Alpha Eukaryotic Elongation Factor 1 A EEF1A |
| ENSMUSG00000016427 | ENSFM00500000386654 | Unknown |
| ENSMUSG00000016481 | ENSFM00780001917710 | Unknown |
| ENSMUSG00000016481 | ENSFM00740001589381 | Complement Component Receptor 1 Precursor Complement |
| ENSMUSG00000016494 | ENSFM00780001915745 | Unknown |
| ENSMUSG00000016494 | ENSFM00250000005487 | Hematopoietic Progenitor Cell Antigen CD34 Precursor CD34 Antigen |
| ENSMUSG00000016495 | ENSFM00570000879077 | Unknown |
| ENSMUSG00000016495 | ENSFM00570000883039 | Unknown |
| ENSMUSG00000016495 | ENSFM00570000887956 | Unknown |
| ENSMUSG00000016495 | ENSFM00570000900159 | Unknown |
| ENSMUSG00000016495 | ENSFM00500000274177 | Plasminogen Receptor Kt Plg R Kt |
| ENSMUSG00000016534 | ENSFM00640001103074 | Lysosome Associated Membrane Glycoprotein 1 Precursor Lamp 1 Lysosome Associated Membrane 1 CD107 Antigen Family Member A CD107A Antigen |
| ENSMUSG00000016541 | ENSFM00250000007503 | Ataxin 10 Spinocerebellar Ataxia Type 10 |
| ENSMUSG00000016664 | ENSFM00250000001466 | Kinase C And Casein Kinase Substrate In Neurons |
| ENSMUSG00000016995 | ENSFM00740001589150 | Precursor |
| ENSMUSG00000017009 | ENSFM00500000273193 | Syndecan 4 Precursor SYND4 |
| ENSMUSG00000017132 | ENSFM00250000001040 | Cytohesin Ph SEC7 And Coiled Coil Domain Containing |
| ENSMUSG00000017144 | ENSFM00500000269995 | Rho Related Gtp Binding Precursor Rho Family Gtpase |
| ENSMUSG00000017167 | ENSFM00250000000380 | Contactin Associated 5 Precursor Cell Recognition Molecule CASPR5 |
| ENSMUSG00000017188 | ENSFM00670001236757 | Cytochrome C Oxidase Assembly Factor 3 Homolog Mitochondrial Coiled Coil Domain Containing 56 |
| ENSMUSG00000017221 | ENSFM00250000003614 | 26S Proteasome Non Atpase Regulatory Subunit 3 26S Proteasome Subunit 26S Proteasome Regulatory Subunit |
| ENSMUSG00000017286 | ENSFM00610000963542 | Unknown |
| ENSMUSG00000017286 | ENSFM00610000963543 | Unknown |
| ENSMUSG00000017286 | ENSFM00250000002922 | Glyoxalase Domain Containing 4 |
| ENSMUSG00000017288 | ENSFM00610000973548 | Unknown |
| ENSMUSG00000017288 | ENSFM00610000973549 | Unknown |
| ENSMUSG00000017288 | ENSFM00800002003735 | Vacuolar Sorting Associated 53 Homolog |
| ENSMUSG00000017291 | ENSFM00250000000892 | Serine/Threonine Kinase EC_2.7.11.1 Thousand And One Amino Acid |
| ENSMUSG00000017307 | ENSFM00640001115284 | Unknown |
| ENSMUSG00000017307 | ENSFM00500000271864 | Acyl Coenzyme A Thioesterase 8 Acyl Coa Thioesterase 8 EC_3.1.2.27 Choloyl Coenzyme A Thioesterase Acyl Coa Thioesterase Pte 2 Peroxisomal Acyl Coenzyme A Thioester Hydrolase 1 Pte 1 Peroxisomal Long Chain Acyl Coa Thioesterase 1 |
| ENSMUSG00000017314 | ENSFM00730001521911 | Maguk P55 Subfamily Member 2 Discs Large Homolog 2 MPP2 |
| ENSMUSG00000017390 | ENSFM00250000000488 | Fructose Bisphosphate Aldolase EC_4.1.2.13 Type Aldolase |
| ENSMUSG00000017404 | ENSFM00250000001949 | 60S Ribosomal L19 |
| ENSMUSG00000017412 | ENSFM00250000000524 | Voltage Dependent L Type Calcium Channel Subunit Beta Calcium Channel Voltage Dependent Subunit Beta |
| ENSMUSG00000017428 | ENSFM00640001114961 | Unknown |
| ENSMUSG00000017428 | ENSFM00640001114962 | Unknown |
| ENSMUSG00000017428 | ENSFM00250000003951 | 26S Proteasome Non Atpase Regulatory Subunit 26S Proteasome Regulatory Subunit RPN6 |
| ENSMUSG00000017631 | ENSFM00250000000543 | Breakpoint Cluster Region |
| ENSMUSG00000017670 | ENSFM00570000880327 | Unknown |
| ENSMUSG00000017670 | ENSFM00570000901128 | Unknown |
| ENSMUSG00000017670 | ENSFM00250000001494 | Engulfment And Cell Motility |
| ENSMUSG00000017686 | ENSFM00250000000940 | Mitochondrial Rho Gtpase 2 Miro 2 EC_3.6.5.- Ras Homolog Gene Family Member T2 |
| ENSMUSG00000017715 | ENSFM00750001632469 | Cdp Diacylglycerol Glycerol 3 Phosphate 3 Phosphatidyltransferase Mitochondrial Precursor EC_2.7.8.5 Phosphatidylglycerophosphate Synthase 1 Pgp Synthase 1 |
| ENSMUSG00000017721 | ENSFM00780001898466 | Gpi Transamidase Component Pig T Precursor Phosphatidylinositol Glycan Biosynthesis Class T |
| ENSMUSG00000017740 | ENSFM00810002057382 | Unknown |
| ENSMUSG00000017740 | ENSFM00260000050376 | Solute Carrier Family 12 Member Electroneutral Potassium Chloride Cotransporter K Cl Cotransporter |
| ENSMUSG00000017774 | ENSFM00500000314344 | Unknown |
| ENSMUSG00000017774 | ENSFM00800002003512 | Unconventional Myosin Ic Myosin I Beta Mmi Beta Mmib |
| ENSMUSG00000017776 | ENSFM00250000002274 | Adapter Molecule Crk Proto Oncogene C Crk P38 |
| ENSMUSG00000017778 | ENSFM00570000879128 | Unknown |
| ENSMUSG00000017778 | ENSFM00570000891961 | Unknown |
| ENSMUSG00000017778 | ENSFM00570000894444 | Unknown |
| ENSMUSG00000017781 | ENSFM00500000269976 | Phosphatidylinositol Transfer Alpha Pi Tp Alpha Ptdins Transfer Alpha Ptdinstp Alpha |
| ENSMUSG00000017802 | ENSFM00500000272621 | Unknown |
| ENSMUSG00000017831 | ENSFM00500000269759 | Ras Related Rab |
| ENSMUSG00000017837 | ENSFM00250000003149 | Nf Kappa B Inhibitor Interacting Ras 2 I Kappa B Interacting Ras 2 Kappa B Ras 2 Kappab RAS2 |
| ENSMUSG00000017843 | ENSFM00810002040632 | Serine/Threonine Phosphatase 2A 56 Kda Regulatory Subunit Gamma PP2A B Subunit B' PP2A B Subunit B' Gamma PP2A B Subunit B56 Gamma PP2A B Subunit PR61 Gamma PP2A B Subunit R5 Gamma |
| ENSMUSG00000017943 | ENSFM00250000004188 | Ganglioside Induced Differentiation Associated 1 GDAP1 |
| ENSMUSG00000018001 | ENSFM00570000899921 | Unknown |
| ENSMUSG00000018001 | ENSFM00660001188987 | Unknown |
| ENSMUSG00000018001 | ENSFM00660001188989 | Unknown |
| ENSMUSG00000018001 | ENSFM00250000001040 | Cytohesin Ph SEC7 And Coiled Coil Domain Containing |
| ENSMUSG00000018012 | ENSFM00730001521186 | Ras Related C3 Botulinum Toxin Substrate Precursor P21 |
| ENSMUSG00000018042 | ENSFM00730001521809 | Nadh Cytochrome B5 Reductase B5R EC_1.6.2.2 |
| ENSMUSG00000018076 | ENSFM00250000001693 | Mediator Of Rna Polymerase Ii Transcription Subunit 13 Thyroid Hormone Receptor Associated Thyroid Hormone Receptor Associated Complex 240 Kda Component |
| ENSMUSG00000018171 | ENSFM00570000898042 | Unknown |
| ENSMUSG00000018171 | ENSFM00250000003161 | Vacuole Membrane 1 Transmembrane 49 |
| ENSMUSG00000018189 | ENSFM00670001235447 | Ubiquitin Carboxyl Terminal Hydrolase Isozyme L5 Uch L5 EC_3.4.19.12 Ubiquitin C Terminal Hydrolase UCH37 Ubiquitin Thioesterase L5 |
| ENSMUSG00000018196 | ENSFM00760001723674 | Unknown |
| ENSMUSG00000018196 | ENSFM00500000272856 | Glutaredoxin 2 Mitochondrial Precursor |
| ENSMUSG00000018286 | ENSFM00810002040613 | Proteasome Subunit Beta Type 9 Precursor EC_3.4.25.1 Low Molecular Mass 2 Macropain Chain 7 Multicatalytic Endopeptidase Complex Chain 7 Proteasome Chain 7 Proteasome Subunit Beta 1I Really Interesting New Gene 12 |
| ENSMUSG00000018322 | ENSFM00740001589368 | Mitochondrial Import Receptor Subunit TOM34 Translocase Of Outer Membrane 34 Kda Subunit |
| ENSMUSG00000018326 | ENSFM00730001521154 | Unknown |
| ENSMUSG00000018340 | ENSFM00750001632387 | Annexin Annexin Annexin Calphobindin Ii Lipocortin |
| ENSMUSG00000018398 | ENSFM00590000918438 | Unknown |
| ENSMUSG00000018398 | ENSFM00350000105408 | Septin |
| ENSMUSG00000018411 | ENSFM00500000327757 | Unknown |
| ENSMUSG00000018411 | ENSFM00260000050441 | Microtubule Associated Tau Neurofibrillary Tangle Paired Helical Filament Tau Phf Tau |
| ENSMUSG00000018417 | ENSFM00800002003560 | Unconventional Myosin Ia Brush Border Myosin I Bbm I Bbmi Myosin I Heavy Chain Mihc |
| ENSMUSG00000018446 | ENSFM00250000004016 | Complement Component 1 Q Subcomponent Binding Protein Mitochondrial Precursor GC1Q R Glycoprotein GC1QBP C1QBP |
| ENSMUSG00000018459 | ENSFM00250000000477 | Solute Carrier Family 13 Member Na + /Dicarboxylate Cotransporter Nadc |
| ENSMUSG00000018470 | ENSFM00780001898357 | Voltage Gated Potassium Channel Subunit Beta K + Channel Subunit Beta Kv Beta |
| ENSMUSG00000018507 | ENSFM00500000269768 | Transient Receptor Potential Cation Channel Subfamily V Member 1 TRPV1 Osm 9 Trp Channel 1 OTRPC1 Vanilloid Receptor 1 |
| ENSMUSG00000018547 | ENSFM00260000050567 | Phosphatidylinositol 5 Phosphate 4 Kinase Type 2 EC_2.7.1.149 1 Phosphatidylinositol 5 Phosphate 4 Kinase 2 Diphosphoinositide Kinase 2 Phosphatidylinositol 5 Phosphate 4 Kinase Type Ii Pi 5 P 4 Kinase Type Ii PIP4KII Ptdins 5 P 4 Kinase 2 |
| ENSMUSG00000018574 | ENSFM00500000270065 | Very Long Chain Specific Acyl Coa Dehydrogenase Mitochondrial Precursor EC_1.3.8.9 |
| ENSMUSG00000018583 | ENSFM00250000002045 | Ras Gtpase Activating Binding 1 G3BP 1 EC_3.6.4.12 EC_3.6.4.- 13 Atp Dependent Helicase |
| ENSMUSG00000018593 | ENSFM00250000001867 | Sparc Precursor Basement Membrane 40 Bm 40 Osteonectin On Secreted Acidic And Rich In Cysteine |
| ENSMUSG00000018669 | ENSFM00570000878559 | Unknown |
| ENSMUSG00000018669 | ENSFM00250000006242 | CDK5 Regulatory Subunit Associated 3 |
| ENSMUSG00000018707 | ENSFM00730001521523 | Dynein Heavy Chain Cytoplasmic Dynein Heavy Chain Cytosolic |
| ENSMUSG00000018733 | ENSFM00570000889673 | Unknown |
| ENSMUSG00000018733 | ENSFM00660001177557 | Unknown |
| ENSMUSG00000018733 | ENSFM00250000006034 | Peroxisome Assembly 12 Peroxin 12 Peroxisome Assembly Factor 3 Paf 3 |
| ENSMUSG00000018736 | ENSFM00250000002568 | Nuclear Distribution Nude 1 |
| ENSMUSG00000018740 | ENSFM00400000131818 | Solute Carrier Family 25 Member |
| ENSMUSG00000018765 | ENSFM00250000000920 | Fragile X Mental Retardation 1 |
| ENSMUSG00000018796 | ENSFM00570000888598 | Unknown |
| ENSMUSG00000018796 | ENSFM00500000269666 | Long Chain Fatty Acid Coa Ligase EC_6.2.1.3 Long Chain Acyl Coa Synthetase Lacs |
| ENSMUSG00000018800 | ENSFM00570000851109 | Atp Binding Cassette Sub Family A Member |
| ENSMUSG00000018819 | ENSFM00500000273526 | Lymphocyte Specific Kda Phosphoprotein PP52 Lymphocyte Specific Antigen WP34 |
| ENSMUSG00000018830 | ENSFM00270000056424 | Myosin Cellular Myosin Heavy Chain Type Myosin Heavy Chain Myosin Heavy Chain Non Muscle Non Muscle Myosin Heavy Chain Nmmhc Non Muscle Myosin Heavy Chain Nmmhc Ii Nmmhc |
| ENSMUSG00000018848 | ENSFM00440000236919 | Arginine Trna Ligase Cytoplasmic EC_6.1.1.19 Arginyl Trna Synthetase Argrs |
| ENSMUSG00000018858 | ENSFM00570000886758 | Unknown |
| ENSMUSG00000018858 | ENSFM00500000273212 | Peptidyl Trna Hydrolase ICT1 Mitochondrial Precursor EC_3.1.1.29 39S Ribosomal L58 Mitochondrial Mrp L58 Immature Colon Carcinoma Transcript 1 |
| ENSMUSG00000018861 | ENSFM00250000003227 | Nadph:Adrenodoxin Oxidoreductase Mitochondrial Precursor Ar Adrenodoxin Reductase EC_1.18.1.6 Ferredoxin Nadp + Reductase Ferredoxin Reductase |
| ENSMUSG00000018909 | ENSFM00250000000572 | Arrestin |
| ENSMUSG00000018965 | ENSFM00730001521154 | Unknown |
| ENSMUSG00000019054 | ENSFM00600000921667 | Mitochondrial Fission 1 FIS1 Homolog Tetratricopeptide Repeat 11 Tpr Repeat 11 |
| ENSMUSG00000019066 | ENSFM00730001521441 | Ras Related Rab 3 |
| ENSMUSG00000019082 | ENSFM00720001503227 | Unknown |
| ENSMUSG00000019082 | ENSFM00720001503349 | Unknown |
| ENSMUSG00000019082 | ENSFM00720001503415 | Unknown |
| ENSMUSG00000019082 | ENSFM00740001599670 | Unknown |
| ENSMUSG00000019082 | ENSFM00740001613500 | Unknown |
| ENSMUSG00000019082 | ENSFM00810002056307 | Unknown |
| ENSMUSG00000019082 | ENSFM00810002057416 | Unknown |
| ENSMUSG00000019082 | ENSFM00500000270170 | Mitochondrial Glutamate Carrier 1 Gc 1 Glutamate/H + Symporter 1 Solute Carrier Family 25 Member 22 |
| ENSMUSG00000019087 | ENSFM00570000885080 | Unknown |
| ENSMUSG00000019087 | ENSFM00570000888862 | Unknown |
| ENSMUSG00000019087 | ENSFM00560000771081 | V Type Proton Atpase Subunit S1 Precursor V Atpase Subunit S1 V Atpase AC45 Subunit V Atpase S1 Accessory Vacuolar Proton Pump Subunit S1 |
| ENSMUSG00000019124 | ENSFM00570000885243 | Unknown |
| ENSMUSG00000019124 | ENSFM00250000002467 | Secernin |
| ENSMUSG00000019146 | ENSFM00250000002019 | Voltage Dependent Calcium Channel Gamma Subunit Neuronal Voltage Gated Calcium Channel Gamma Subunit Transmembrane Ampar Regulatory Gamma Tarp Gamma |
| ENSMUSG00000019173 | ENSFM00570000890095 | Unknown |
| ENSMUSG00000019173 | ENSFM00500000269759 | Ras Related Rab |
| ENSMUSG00000019179 | ENSFM00680001326152 | Unknown |
| ENSMUSG00000019179 | ENSFM00730001521810 | Malate Dehydrogenase Mitochondrial Precursor EC_1.1.1.37 |
| ENSMUSG00000019188 | ENSFM00250000002681 | Minor Histocompatibility Antigen H13 EC_3.4.23.- Presenilin 3 Signal Peptide Peptidase |
| ENSMUSG00000019194 | ENSFM00600000921511 | Sodium Channel Subunit Beta 1 Precursor |
| ENSMUSG00000019210 | ENSFM00250000002828 | V Type Proton Atpase Subunit E V Atpase Subunit E V Atpase Kda Subunit Vacuolar Proton Pump Subunit E |
| ENSMUSG00000019232 | ENSFM00750001632360 | Ethanolamine Phosphate Phospho Lyase EC_4.2.3.2 Alanine Glyoxylate Aminotransferase 2 1 |
| ENSMUSG00000019261 | ENSFM00250000000942 | Microtubule Associated Map |
| ENSMUSG00000019302 | ENSFM00570000881199 | Unknown |
| ENSMUSG00000019302 | ENSFM00250000000490 | V Type Proton Atpase 116 Kda Subunit A V Atpase 116 Kda Proton 116 Kda Subunit Vacuolar Proton Translocating Atpase 116 Kda Subunit A |
| ENSMUSG00000019370 | ENSFM00640001118108 | Unknown |
| ENSMUSG00000019370 | ENSFM00640001118555 | Unknown |
| ENSMUSG00000019370 | ENSFM00750001632311 | Calmodulin Cam |
| ENSMUSG00000019373 | ENSFM00570000892434 | Unknown |
| ENSMUSG00000019373 | ENSFM00250000006013 | COP9 Signalosome Complex Subunit 3 Signalosome Subunit 3 |
| ENSMUSG00000019428 | ENSFM00520000547997 | Unknown |
| ENSMUSG00000019428 | ENSFM00570000883795 | Unknown |
| ENSMUSG00000019428 | ENSFM00250000005307 | Peptidyl Prolyl Cis Trans Isomerase FKBP8 Ppiase FKBP8 EC_5.2.1.8.38 Kda FK506 Binding 38 Kda Fkbp Fkbp 38 FK506 Binding 8 Fkbp 8 FKBPR38 Rotamase |
| ENSMUSG00000019433 | ENSFM00250000002325 | Pdz Domain Containing |
| ENSMUSG00000019467 | ENSFM00740001589124 | EC_2.7.11.1 |
| ENSMUSG00000019471 | ENSFM00500000270663 | HSP90 Co Chaperone CDC37 HSP90 Chaperone Kinase Targeting Subunit P50CDC37 |
| ENSMUSG00000019478 | ENSFM00800002003435 | Ras Related Rab |
| ENSMUSG00000019494 | ENSFM00250000007478 | COP9 Signalosome Complex Subunit 6 Signalosome Subunit 6 |
| ENSMUSG00000019578 | ENSFM00250000005080 | Ubx Domain Containing 6 Ubx Domain Containing 1 |
| ENSMUSG00000019579 | ENSFM00670001236674 | Precursor Interleukin 25 Il 25 Stromal Cell Derived Growth Factor SF20 |
| ENSMUSG00000019763 | ENSFM00570000879481 | Unknown |
| ENSMUSG00000019763 | ENSFM00570000884368 | Unknown |
| ENSMUSG00000019763 | ENSFM00570000894366 | Unknown |
| ENSMUSG00000019763 | ENSFM00500000271332 | Required Meiotic Nuclear Division 1 Homolog Precursor |
| ENSMUSG00000019775 | ENSFM00480000262761 | Regulator Of G Signaling |
| ENSMUSG00000019790 | ENSFM00500000269888 | Syntaxin Binding 5 Lethal 2 Giant Larvae Homolog Tomosyn |
| ENSMUSG00000019791 | ENSFM00590000917183 | Unknown |
| ENSMUSG00000019791 | ENSFM00500000273620 | Histidine Triad Nucleotide Binding 3 Hint 3 Ec 3 |
| ENSMUSG00000019795 | ENSFM00590000917209 | Unknown |
| ENSMUSG00000019795 | ENSFM00590000917210 | Unknown |
| ENSMUSG00000019795 | ENSFM00590000918378 | Unknown |
| ENSMUSG00000019795 | ENSFM00250000003005 | L Isoaspartate D Aspartate O Methyltransferase Pimt EC_2.1.1.77 L Isoaspartyl Carboxyl Methyltransferase L Isoaspartyl/D Aspartyl Methyltransferase Beta Aspartate Methyltransferase |
| ENSMUSG00000019802 | ENSFM00250000003542 | Translocation SEC63 Homolog |
| ENSMUSG00000019804 | ENSFM00250000002957 | Sorting Nexin |
| ENSMUSG00000019809 | ENSFM00250000005471 | Peroxisomal Biogenesis Factor 3 Peroxin 3 Peroxisomal Assembly PEX3 |
| ENSMUSG00000019820 | ENSFM00250000000455 | Dystrophin |
| ENSMUSG00000019828 | ENSFM00730001521627 | Metabotropic Glutamate Receptor Precursor |
| ENSMUSG00000019831 | ENSFM00250000001269 | Wiskott Aldrich Syndrome Family Member 1 Wasp Family Member 1 Wave 1 |
| ENSMUSG00000019843 | ENSFM00570000890474 | Unknown |
| ENSMUSG00000019843 | ENSFM00730001521177 | Tyrosine Kinase EC_2.7.10.2 |
| ENSMUSG00000019849 | ENSFM00250000003032 | Prolyl Endopeptidase Pe EC_3.4.21.26 Post Proline Cleaving Enzyme |
| ENSMUSG00000019864 | ENSFM00500000271278 | Reticulon 4 Interacting 1 Mitochondrial Precursor |
| ENSMUSG00000019868 | ENSFM00570000866653 | Unknown |
| ENSMUSG00000019868 | ENSFM00570000882696 | Unknown |
| ENSMUSG00000019868 | ENSFM00250000005580 | Vacuolar Sorting Associated VTA1 Homolog SKD1 Binding 1 SBP1 |
| ENSMUSG00000019874 | ENSFM00800002003523 | Fatty Acid Binding Protein Heart Fatty Acid Binding 3 Heart Type Fatty Acid Binding H Fabp |
| ENSMUSG00000019877 | ENSFM00250000000772 | Serine Incorporator Tumor Differentially Expressed |
| ENSMUSG00000019906 | ENSFM00260000050595 | Lin 7 Homolog Lin Mammalian Lin Seven Mals Vertebrate Lin 7 Homolog Veli |
| ENSMUSG00000019907 | ENSFM00500000269806 | Phosphatase 1 Regulatory Subunit 12A Myosin Phosphatase Targeting Subunit 1 Myosin Phosphatase Target Subunit 1 Phosphatase Myosin Binding Subunit |
| ENSMUSG00000019923 | ENSFM00250000009507 | ZW10 Interactor ZW10 Interacting 1 Zwint 1 |
| ENSMUSG00000019943 | ENSFM00250000000445 | Plasma Membrane Calcium Transporting Atpase EC_3.6.3.8 Plasma Membrane Calcium Atpase Plasma Membrane Calcium Pump |
| ENSMUSG00000019951 | ENSFM00250000002015 | UHRF1 Binding 1 |
| ENSMUSG00000019961 | ENSFM00600000921391 | Lamina Associated Polypeptide 2 Thymopoietin Tp Thymopoietin Related Peptide Tprp |
| ENSMUSG00000019969 | ENSFM00250000001423 | Presenilin Ps EC_3.4.23.- |
| ENSMUSG00000019978 | ENSFM00800002003537 | Unknown |
| ENSMUSG00000019986 | ENSFM00250000003083 | Jouberin Abelson Helper Integration Site 1 Ahi 1 |
| ENSMUSG00000019996 | ENSFM00250000002902 | Ensconsin Epithelial Microtubule Associated Of 115 Kda E Map 115 Microtubule Associated 7 Map 7 |
| ENSMUSG00000019998 | ENSFM00500000270193 | Syntaxin |
| ENSMUSG00000020015 | ENSFM00740001589171 | Cyclin Dependent Kinase EC_2.7.11.22 Cell Division Kinase Pctaire Motif Kinase Serine/Threonine Kinase Pctaire |
| ENSMUSG00000020018 | ENSFM00670001236821 | Small Nuclear Ribonucleoprotein F Snrnp F Sm F Sm F Smf |
| ENSMUSG00000020022 | ENSFM00570000899974 | Unknown |
| ENSMUSG00000020022 | ENSFM00500000272434 | Nadh Dehydrogenase 1 Alpha Subcomplex Subunit Complex I B17 2 Ci B17 2 CIB17 2 Nadh Ubiquinone Oxidoreductase Subunit B17 2 |
| ENSMUSG00000020042 | ENSFM00250000001718 | Ankyrin Repeat And Btb/Poz Domain Containing BTBD11 Btb/Poz Domain Containing 11 |
| ENSMUSG00000020048 | ENSFM00760001714713 | Endoplasmin 94 Kda Glucose Regulated Grp 94 Heat Shock 90 Kda Beta Member 1 |
| ENSMUSG00000020078 | ENSFM00810002040678 | Vacuolar Sorting Associated Vesicle Sorting |
| ENSMUSG00000020083 | ENSFM00670001236755 | Uncharacterized |
| ENSMUSG00000020083 | ENSFM00570000887169 | Unknown |
| ENSMUSG00000020088 | ENSFM00800002003440 | Gtp Binding SAR1 |
| ENSMUSG00000020089 | ENSFM00730001521782 | Inorganic Pyrophosphatase EC_3.6.1.1 Pyrophosphate Phospho Hydrolase Ppase |
| ENSMUSG00000020097 | ENSFM00570000900525 | Unknown |
| ENSMUSG00000020097 | ENSFM00800002003569 | Sphingosine 1 Phosphate Lyase 1 S1PL Sp Lyase 1 Spl 1 EC_4.1.2.27 Sphingosine 1 Phosphate Aldolase |
| ENSMUSG00000020109 | ENSFM00250000001153 | Dnaj Homolog Subfamily Member |
| ENSMUSG00000020111 | ENSFM00610000967698 | Unknown |
| ENSMUSG00000020111 | ENSFM00260000050684 | Calcium Uptake 1 Mitochondrial Calcium Binding Atopy Related Autoantigen 1 Homolog |
| ENSMUSG00000020114 | ENSFM00250000001833 | Cullin Associated NEDD8 Dissociated Cullin Associated And Neddylation Dissociated Tbp Interacting Of 120 Kda Tbp Interacting P120 |
| ENSMUSG00000020115 | ENSFM00250000003276 | Inhibitor Of Nuclear Factor Kappa B Kinase Subunit Epsilon I Kappa B Kinase Epsilon Ikk E Ikk Epsilon Ikbke EC_2.7.11.10 Inducible I Kappa B Kinase Ikk I |
| ENSMUSG00000020122 | ENSFM00410000138465 | Receptor Tyrosine Kinase Erbb Precursor EC_2.7.10.1 Proto Oncogene C Erbb |
| ENSMUSG00000020132 | ENSFM00500000271226 | Ras Related Rab 21 |
| ENSMUSG00000020142 | ENSFM00760001714847 | Neutral Amino Acid Transporter B 0 Atb 0 Sodium Dependent Neutral Amino Acid Transporter Type 2 Solute Carrier Family 1 Member 5 |
| ENSMUSG00000020149 | ENSFM00670001238119 | Unknown |
| ENSMUSG00000020149 | ENSFM00730001521238 | Ras Related Rab |
| ENSMUSG00000020152 | ENSFM00730001521596 | Actin Related 2 Actin 2 |
| ENSMUSG00000020153 | ENSFM00250000003821 | Nadh Dehydrogenase Iron Sulfur 7 Mitochondrial Precursor EC_1.6.5.3 EC_1.6.99.- 3 Complex I 20KD Ci 20KD Nadh Ubiquinone Oxidoreductase 20 Kda Subunit |
| ENSMUSG00000020170 | ENSFM00250000003196 | Fibroblast Growth Factor Receptor Substrate 3 Fgfr Substrate 3 Fgfr Signaling Adaptor SNT2 SUC1 Associated Neurotrophic Factor Target 2 Snt 2 |
| ENSMUSG00000020183 | ENSFM00730001521968 | Carboxypeptidase D Precursor EC_3.4.17.22 Metallocarboxypeptidase D GP180 |
| ENSMUSG00000020189 | ENSFM00730001521733 | Oxysterol Binding Related 5 Orp 5 Osbp Related 5 Oxysterol Binding Homolog 1 |
| ENSMUSG00000020198 | ENSFM00250000001618 | Ap 3 Complex Subunit Delta 1 Ap 3 Complex Subunit Delta Adaptor Related Complex 3 Subunit Delta 1 Delta Adaptin |
| ENSMUSG00000020219 | ENSFM00670001236643 | Mitochondrial Import Inner Membrane Translocase Subunit TIM13 |
| ENSMUSG00000020220 | ENSFM00250000001900 | Vacuolar Sorting Associated 13D |
| ENSMUSG00000020238 | ENSFM00570000898047 | Unknown |
| ENSMUSG00000020238 | ENSFM00370000118641 | Nicalin Precursor Nicastrin |
| ENSMUSG00000020250 | ENSFM00730001521412 | Thioredoxin Reductase EC_1.8.1.9 Thioredoxin Reductase |
| ENSMUSG00000020263 | ENSFM00660001171072 | Unknown |
| ENSMUSG00000020263 | ENSFM00660001189703 | Unknown |
| ENSMUSG00000020263 | ENSFM00250000003219 | Dcc Interacting 13 DIP13 Adapter Containing Ph Domain Ptb Domain And Leucine Zipper Motif |
| ENSMUSG00000020265 | ENSFM00640001111891 | Unknown |
| ENSMUSG00000020265 | ENSFM00640001111892 | Unknown |
| ENSMUSG00000020265 | ENSFM00640001116302 | Unknown |
| ENSMUSG00000020265 | ENSFM00640001119856 | Unknown |
| ENSMUSG00000020265 | ENSFM00670001235669 | Small Ubiquitin Related Modifier Precursor Sumo |
| ENSMUSG00000020267 | ENSFM00520000548728 | Histidine Triad Nucleotide Binding 1 |
| ENSMUSG00000020267 | ENSFM00670001235733 | Histidine Triad Nucleotide Binding 1 Ec 3 Adenosine 5' Monophosphoramidase Kinase C Inhibitor 1 Kinase C Interacting 1 Pkci 1 |
| ENSMUSG00000020277 | ENSFM00730001521507 | Atp Dependent 6 Phosphofructokinase Type |
| ENSMUSG00000020279 | ENSFM00570000864111 | Unknown |
| ENSMUSG00000020279 | ENSFM00250000006177 | Interleukin 9 Receptor Precursor Il 9 Receptor Il 9R CD129 Antigen |
| ENSMUSG00000020290 | ENSFM00550000764870 | Unknown |
| ENSMUSG00000020290 | ENSFM00250000001829 | Exportin 1 Chromosome Region Maintenance 1 |
| ENSMUSG00000020297 | ENSFM00500000298059 | Unknown |
| ENSMUSG00000020297 | ENSFM00500000315845 | Unknown |
| ENSMUSG00000020297 | ENSFM00250000005181 | Neuron Specific Family Member |
| ENSMUSG00000020300 | ENSFM00250000000830 | Cytoplasmic Polyadenylation Element Binding |
| ENSMUSG00000020308 | ENSFM00250000008950 | Tubulin Polyglutamylase Complex Subunit 1 PGS1 |
| ENSMUSG00000020311 | ENSFM00610000962379 | Unknown |
| ENSMUSG00000020311 | ENSFM00250000003055 | Endoplasmic Reticulum Lectin 1 Precursor Er Lectin Erlectin |
| ENSMUSG00000020315 | ENSFM00740001589095 | Spectrin Beta Chain Erythrocytic Beta Spectrin |
| ENSMUSG00000020321 | ENSFM00730001521535 | Malate Dehydrogenase Cytoplasmic EC_1.1.1.37 Cytosolic Malate Dehydrogenase |
| ENSMUSG00000020331 | ENSFM00250000000931 | Potassium/Sodium Hyperpolarization Activated Cyclic Nucleotide Gated Channel |
| ENSMUSG00000020333 | ENSFM00500000269666 | Long Chain Fatty Acid Coa Ligase EC_6.2.1.3 Long Chain Acyl Coa Synthetase Lacs |
| ENSMUSG00000020334 | ENSFM00760001714753 | Solute Carrier Family 22 Member Organic Cation/Carnitine Transporter |
| ENSMUSG00000020340 | ENSFM00570000884032 | Unknown |
| ENSMUSG00000020340 | ENSFM00250000001390 | Cytoplasmic FMR1 Interacting |
| ENSMUSG00000020349 | ENSFM00730001521389 | Serine/Threonine Phosphatase Catalytic Subunit EC_3.1.3.16 |
| ENSMUSG00000020358 | ENSFM00500000269788 | Heterogeneous Nuclear Ribonucleoprotein D Hnrnp D Hnrnp Dl |
| ENSMUSG00000020361 | ENSFM00360000109870 | Unknown |
| ENSMUSG00000020368 | ENSFM00500000269826 | Precursor |
| ENSMUSG00000020372 | ENSFM00250000002295 | Guanine Nucleotide Binding Subunit Beta Receptor Activated C Kinase Receptor Of Activated Kinase C 1 RACK1 |
| ENSMUSG00000020396 | ENSFM00730001521354 | Neurofilament Polypeptide Nf Kda Neurofilament Neurofilament Triplet |
| ENSMUSG00000020402 | ENSFM00250000001189 | Voltage Dependent Anion Selective Channel Vdac Outer Mitochondrial Membrane Porin |
| ENSMUSG00000020428 | ENSFM00800002003422 | Gamma Aminobutyric Acid Receptor Subunit Alpha Precursor Gaba A Receptor Subunit Alpha |
| ENSMUSG00000020431 | ENSFM00730001521247 | Adenylate Cyclase Type EC_4.6.1.1 Atp Pyrophosphate Lyase Adenylate Cyclase Type Adenylyl Cyclase Ca 2+ Adenylyl Cyclase |
| ENSMUSG00000020436 | ENSFM00730001521448 | Gamma Aminobutyric Acid Receptor Subunit Gamma Precursor Gaba A Receptor Subunit Gamma |
| ENSMUSG00000020440 | ENSFM00730001521124 | Adp Ribosylation Factor |
| ENSMUSG00000020444 | ENSFM00640001120069 | Unknown |
| ENSMUSG00000020444 | ENSFM00400000131805 | Guanylate Kinase EC_2.7.4.8 Gmp Kinase |
| ENSMUSG00000020456 | ENSFM00750001632363 | 2 Oxoglutarate Dehydrogenase Mitochondrial Precursor 2 Oxoglutarate Dehydrogenase Complex Component E1 Ogdc E1 Alpha Ketoglutarate Dehydrogenase |
| ENSMUSG00000020458 | ENSFM00500000269689 | Reticulon |
| ENSMUSG00000020460 | ENSFM00500000270032 | Ubiquitin 40S Ribosomal S27A Precursor Ubiquitin Carboxyl Extension 80 |
| ENSMUSG00000020476 | ENSFM00480000262787 | Drebrin |
| ENSMUSG00000020483 | ENSFM00570000882728 | Unknown |
| ENSMUSG00000020483 | ENSFM00670001235517 | Dynein Light Chain Cytoplasmic 8 Kda Dynein Light Chain Dynein Light Chain LC8 Type |
| ENSMUSG00000020486 | ENSFM00800002003401 | Septin |
| ENSMUSG00000020514 | ENSFM00250000005774 | 39S Ribosomal L22 Mitochondrial Precursor L22MT Mrp L22 |
| ENSMUSG00000020524 | ENSFM00640001120061 | Unknown |
| ENSMUSG00000020524 | ENSFM00720001492020 | Glutamate Receptor Precursor Glur Ampa Selective Glutamate Receptor Glur Glutamate Receptor Ionotropic Ampa |
| ENSMUSG00000020528 | ENSFM00730001521917 | Phosphoribosyl Pyrophosphate Synthase Associated Prpp Synthase Associated |
| ENSMUSG00000020532 | ENSFM00780001898351 | Acetyl Coa Carboxylase 1 ACC1 EC_6.4.1.2 Acc Alpha |
| ENSMUSG00000020536 | ENSFM00500000270191 | Lethal 2 Giant Larvae |
| ENSMUSG00000020537 | ENSFM00730001521857 | Developmentally Regulated Gtp Binding 2 Drg 2 |
| ENSMUSG00000020570 | ENSFM00570000892194 | Unknown |
| ENSMUSG00000020570 | ENSFM00800002003944 | Synaptophysin 1 Pantophysin |
| ENSMUSG00000020571 | ENSFM00500000270568 | Disulfide Isomerase A6 Precursor EC_5.3.4.1 Thioredoxin Domain Containing 7 |
| ENSMUSG00000020576 | ENSFM00250000003713 | Neuroblastoma Amplified Sequence Neuroblastoma Amplified Gene |
| ENSMUSG00000020580 | ENSFM00730001521800 | Rho Associated Kinase 1 EC_2.7.11.1 Rho Associated Coiled Coil Containing Kinase 1 Rho Associated Coiled Coil Containing Kinase I Rock I P160 Rock 1 P160ROCK |
| ENSMUSG00000020585 | ENSFM00500000271417 | Lysosomal Associated Transmembrane 4A |
| ENSMUSG00000020589 | ENSFM00250000002173 | FAM49A |
| ENSMUSG00000020598 | ENSFM00390000126295 | Neural Cell Adhesion Molecule L1 L1 L1 |
| ENSMUSG00000020599 | ENSFM00730001522095 | Regulator Of G Signaling |
| ENSMUSG00000020611 | ENSFM00730001522052 | Guanine Nucleotide Binding Subunit Alpha G Alpha G Subunit Alpha |
| ENSMUSG00000020612 | ENSFM00730001521681 | Camp Dependent Kinase Type I Alpha Regulatory Camp Dependent Kinase Type I Alpha Regulatory Subunit N Terminally Processed |
| ENSMUSG00000020621 | ENSFM00500000269664 | Retinol Dehydrogenase |
| ENSMUSG00000020628 | ENSFM00250000002755 | Trafficking Particle Complex Subunit 12 Tetratricopeptide Repeat 15 Tpr Repeat 15 |
| ENSMUSG00000020635 | ENSFM00670001235580 | Peptidyl Prolyl Cis Trans Isomerase FKBP1B Ppiase FKBP1B EC_5.2.1.8 12.6 Kda FK506 Binding 12.6 Kda Fkbp Fkbp 12 6 FK506 Binding 1B Fkbp 1B Immunophilin FKBP12 6 Rotamase |
| ENSMUSG00000020640 | ENSFM00250000000789 | Intersectin |
| ENSMUSG00000020650 | ENSFM00670001235596 | B Cell Receptor Associated Bcr Associated |
| ENSMUSG00000020654 | ENSFM00730001521247 | Adenylate Cyclase Type EC_4.6.1.1 Atp Pyrophosphate Lyase Adenylate Cyclase Type Adenylyl Cyclase Ca 2+ Adenylyl Cyclase |
| ENSMUSG00000020658 | ENSFM00250000001283 | EFR3 Homolog |
| ENSMUSG00000020664 | ENSFM00800002003534 | Dihydrolipoyl Dehydrogenase Mitochondrial Precursor EC_1.8.1.4 Dihydrolipoamide Dehydrogenase |
| ENSMUSG00000020668 | ENSFM00800002003386 | Kinesin |
| ENSMUSG00000020671 | ENSFM00730001521184 | Ras Related Rab |
| ENSMUSG00000020681 | ENSFM00250000000787 | Angiotensin Converting Enzyme Precursor Ace Carboxypeptidase |
| ENSMUSG00000020689 | ENSFM00730001521296 | Integrin Beta Precursor |
| ENSMUSG00000020701 | ENSFM00250000000808 | Transmembrane Precursor |
| ENSMUSG00000020708 | ENSFM00730001521682 | 26S Protease Regulatory Subunit 8 26S Proteasome Aaa Atpase Subunit RPT6 Proteasome 26S Subunit Atpase 5 Proteasome Subunit P45 P45/Sug |
| ENSMUSG00000020716 | ENSFM00250000001252 | Neurofibromin Neurofibromatosis Related Nf 1 |
| ENSMUSG00000020719 | ENSFM00720001503540 | Unknown |
| ENSMUSG00000020719 | ENSFM00420000140507 | Probable Atp Dependent Rna Helicase DDX5 EC_3.6.4.13 Dead Box 5 |
| ENSMUSG00000020720 | ENSFM00250000004556 | 26S Proteasome Non Atpase Regulatory Subunit 12 26S Proteasome Regulatory Subunit RPN5 26S Proteasome Regulatory Subunit P55 |
| ENSMUSG00000020732 | ENSFM00730001521896 | Ras Related Rab |
| ENSMUSG00000020733 | ENSFM00740001589334 | Na + /H + Exchange Regulatory Cofactor Nhe RF1 Nherf 1 Ezrin Radixin Moesin Binding Phosphoprotein 50 EBP50 Regulatory Cofactor Of Na + /H + Exchanger Sodium Hydrogen Exchanger Regulatory Factor 1 Solute Carrier Family 9 A3 Regulatory Factor 1 |
| ENSMUSG00000020734 | ENSFM00730001521500 | Glutamate Receptor Ionotropic Nmda Precursor Glutamate Receptor Subunit Epsilon N Methyl D Aspartate Receptor Subtype |
| ENSMUSG00000020736 | ENSFM00250000006985 | 5' 3' Deoxyribonucleotidase Cytosolic Type EC_3.1.3.- Cytosolic 5' 3' Pyrimidine Nucleotidase Deoxy 5' Nucleotidase 1 Dnt 1 |
| ENSMUSG00000020745 | ENSFM00810002040664 | Lissencephaly 1 |
| ENSMUSG00000020777 | ENSFM00250000000722 | Peroxisomal Acyl Coenzyme A Oxidase 1 Aox EC_1.3.3.6 Palmitoyl Coa Oxidase |
| ENSMUSG00000020780 | ENSFM00250000003522 | Signal Recognition Particle Subunit SRP68 SRP68 Signal Recognition Particle 68 Kda |
| ENSMUSG00000020788 | ENSFM00500000269652 | Sarcoplasmic/Endoplasmic Reticulum Calcium Atpase Sr Ca 2+ Atpase EC_3.6.3.8 Calcium Pump Calcium Transporting Atpase Sarcoplasmic Reticulum Type Twitch Skeletal Muscle Endoplasmic Reticulum Class 1/2 Ca 2+ Atpase |
| ENSMUSG00000020792 | ENSFM00570000877843 | Unknown |
| ENSMUSG00000020792 | ENSFM00250000003124 | Exocyst Complex Component 7 Exocyst Complex Component EXO70 |
| ENSMUSG00000020802 | ENSFM00250000002926 | E2/E3 Hybrid Ubiquitin Ligase UBE2O EC_6.3.2.- EC_6.3.2.19 Ubiquitin Carrier O Ubiquitin Conjugating Enzyme E2 O Ubiquitin Conjugating Enzyme E2 Of 230 Kda Ubiquitin Conjugating Enzyme E2 230K Ubiquitin Ligase O |
| ENSMUSG00000020805 | ENSFM00250000000477 | Solute Carrier Family 13 Member Na + /Dicarboxylate Cotransporter Nadc |
| ENSMUSG00000020817 | ENSFM00250000001974 | Rab Gtpase Binding Effector 1 Rabaptin 5 Rabaptin 5ALPHA |
| ENSMUSG00000020827 | ENSFM00680001326720 | Unknown |
| ENSMUSG00000020827 | ENSFM00260000050335 | Kinase EC_2.7.11.1 Kinase Mapk/Erk Kinase Kinase Kinase Mek Kinase Kinase Mekkk Kinase |
| ENSMUSG00000020828 | ENSFM00680001318976 | Unknown |
| ENSMUSG00000020828 | ENSFM00740001589239 | Phospholipase D2 Pld 2 EC_3.1.4.4 Choline Phosphatase 2 Phosphatidylcholine Hydrolyzing Phospholipase D2 |
| ENSMUSG00000020834 | ENSFM00500000313432 | Unknown |
| ENSMUSG00000020834 | ENSFM00570000880913 | Unknown |
| ENSMUSG00000020834 | ENSFM00500000269664 | Retinol Dehydrogenase |
| ENSMUSG00000020838 | ENSFM00730001521312 | Sodium Dependent Transporter Transporter Solute Carrier Family 6 Member |
| ENSMUSG00000020840 | ENSFM00610000963462 | Unknown |
| ENSMUSG00000020840 | ENSFM00610000966005 | Unknown |
| ENSMUSG00000020840 | ENSFM00810002040919 | Bleomycin Hydrolase Bh Blm Hydrolase Bmh EC_3.4.22.40 |
| ENSMUSG00000020841 | ENSFM00730001521968 | Carboxypeptidase D Precursor EC_3.4.17.22 Metallocarboxypeptidase D GP180 |
| ENSMUSG00000020843 | ENSFM00570000888842 | Unknown |
| ENSMUSG00000020843 | ENSFM00610000966469 | Unknown |
| ENSMUSG00000020843 | ENSFM00500000272831 | Mitochondrial Import Inner Membrane Translocase Subunit TIM22 |
| ENSMUSG00000020849 | ENSFM00730001521388 | Unknown |
| ENSMUSG00000020859 | ENSFM00250000000947 | C Jun Amino Terminal Kinase Interacting Jip Jnk Interacting Jnk Mitogen Activated Kinase 8 Interacting |
| ENSMUSG00000020869 | ENSFM00250000006592 | Leucine Rich Repeat Containing 59 |
| ENSMUSG00000020882 | ENSFM00250000000524 | Voltage Dependent L Type Calcium Channel Subunit Beta Calcium Channel Voltage Dependent Subunit Beta |
| ENSMUSG00000020883 | ENSFM00500000270304 | F Box/Lrr Repeat 2 |
| ENSMUSG00000020886 | ENSFM00680001326715 | Unknown |
| ENSMUSG00000020886 | ENSFM00680001326716 | Unknown |
| ENSMUSG00000020886 | ENSFM00250000000275 | Disks Large Homolog Synapse Associated Sap |
| ENSMUSG00000020894 | ENSFM00670001235687 | Vesicle Associated Membrane 2 Vamp 2 Synaptobrevin 2 |
| ENSMUSG00000020900 | ENSFM00270000056424 | Myosin Cellular Myosin Heavy Chain Type Myosin Heavy Chain Myosin Heavy Chain Non Muscle Non Muscle Myosin Heavy Chain Nmmhc Non Muscle Myosin Heavy Chain Nmmhc Ii Nmmhc |
| ENSMUSG00000020903 | ENSFM00250000005899 | Syntaxin 8 |
| ENSMUSG00000020903 | ENSFM00500000303360 | Syntaxin 8 CRA_D |
| ENSMUSG00000020917 | ENSFM00250000001673 | Atp Citrate Synthase EC_2.3.3.8 Atp Citrate Pro S Lyase Citrate Cleavage Enzyme |
| ENSMUSG00000020922 | ENSFM00250000006283 | LSM12 Homolog |
| ENSMUSG00000020923 | ENSFM00740001589345 | Nucleolar Transcription Factor 1 Upstream Binding Factor 1 Ubf 1 |
| ENSMUSG00000020926 | ENSFM00410000138484 | Disintegrin And Metalloproteinase Domain Containing Precursor Adam Metalloproteinase Disintegrin And Cysteine Rich Mdc |
| ENSMUSG00000020932 | ENSFM00730001521291 | Vimentin |
| ENSMUSG00000020935 | ENSFM00500000272373 | Dephospho Coa Kinase Domain Containing |
| ENSMUSG00000020946 | ENSFM00250000004732 | Golgi Snap Receptor Complex Member 2.27 Kda Golgi Snare Membrin |
| ENSMUSG00000020949 | ENSFM00390000126407 | Peptidyl Prolyl Cis Trans Isomerase FKBP3 Ppiase FKBP3 EC_5.2.1.8.25 Kda FK506 Binding 25 Kda Fkbp Fkbp 25 FK506 Binding 3 Fkbp 3 Immunophilin FKBP25 Rapamycin Selective 25 Kda Immunophilin Rotamase |
| ENSMUSG00000020952 | ENSFM00790001963053 | SEC1 Family Domain Containing 1 SLY1 Homolog SLY1P Syntaxin Binding 1 2 |
| ENSMUSG00000020954 | ENSFM00250000001473 | Striatin |
| ENSMUSG00000020964 | ENSFM00760001714807 | Sel 1 Homolog Precursor Suppressor Of Lin 12 Sel |
| ENSMUSG00000020986 | ENSFM00810002040640 | Transport SEC23 Related |
| ENSMUSG00000020988 | ENSFM00250000003783 | L 2 Hydroxyglutarate Dehydrogenase Mitochondrial Precursor EC_1.1.99.2 |
| ENSMUSG00000020993 | ENSFM00670001235686 | Trafficking Particle Complex Subunit 6A Trapp Complex Subunit 6A |
| ENSMUSG00000021000 | ENSFM00660001182217 | Unknown |
| ENSMUSG00000021000 | ENSFM00250000002588 | Ctage Family Member Ctage |
| ENSMUSG00000021003 | ENSFM00250000005474 | Galactocerebrosidase Precursor Galcerase EC_3.2.1.46 Galactocerebroside Beta Galactosidase Galactosylceramidase Galactosylceramide Beta Galactosidase |
| ENSMUSG00000021018 | ENSFM00500000272402 | Dna Directed Rna Polymerases I Ii And Iii Subunit RPABC3 Rna Polymerases I Ii And Iii Subunit ABC3 Dna Directed Rna Polymerase Ii Subunit H RPB17 RPB8 Homolog |
| ENSMUSG00000021024 | ENSFM00590000918027 | Unknown |
| ENSMUSG00000021024 | ENSFM00420000140606 | Proteasome Subunit Alpha Type EC_3.4.25.1 Macropain Multicatalytic Endopeptidase Complex Proteasome |
| ENSMUSG00000021027 | ENSFM00250000000944 | Ral Gtpase Activating Subunit Alpha 1 Gap Related Interacting Partner To E12 Gripe Gtpase Activating Domain 1 Tuberin 1 P240 |
| ENSMUSG00000021033 | ENSFM00250000003545 | Maleylacetoacetate Isomerase Maai EC_5.2.1.2 GSTZ1 1 Glutathione S Transferase Zeta 1 EC_2.5.1.- 18 |
| ENSMUSG00000021037 | ENSFM00500000270381 | Activator Of 90 Kda Heat Shock Atpase Homolog |
| ENSMUSG00000021039 | ENSFM00810002040569 | Pre Processing 45 |
| ENSMUSG00000021044 | ENSFM00730001522023 | Uncharacterized Aarf Domain Containing Kinase EC_2.7.11.- |
| ENSMUSG00000021047 | ENSFM00740001589294 | Rna Binding Nova 1 Neuro Oncological Ventral Antigen 1 Ventral Neuron Specific 1 |
| ENSMUSG00000021048 | ENSFM00800002003403 | C 1 Tetrahydrofolate Synthase Cytoplasmic C1 Thf Synthase |
| ENSMUSG00000021051 | ENSFM00760001714676 | Serine/Threonine Phosphatase 2A 56 Kda Regulatory Subunit Epsilon PP2A B Subunit B' Epsilon PP2A B Subunit B56 Epsilon PP2A B Subunit PR61 Epsilon PP2A B Subunit R5 Epsilon |
| ENSMUSG00000021057 | ENSFM00250000007751 | A Kinase Anchor 5 Akap 5 A Kinase Anchor Kda Akap Camp Dependent Kinase Regulatory Subunit Ii High Affinity Binding |
| ENSMUSG00000021061 | ENSFM00740001589095 | Spectrin Beta Chain Erythrocytic Beta Spectrin |
| ENSMUSG00000021062 | ENSFM00750001632504 | Ras Related Rab 15 |
| ENSMUSG00000021065 | ENSFM00250000004295 | Alpha 1 6 Fucosyltransferase ALPHA1 6FUCT EC_2.4.1.68 Gdp L Fuc:N Acetyl Beta D Glucosaminide ALPHA1 6 Fucosyltransferase Gdp Fucose Glycoprotein Fucosyltransferase Glycoprotein 6 Alpha L Fucosyltransferase |
| ENSMUSG00000021066 | ENSFM00250000001436 | Atlastin EC_3.6.5.- |
| ENSMUSG00000021071 | ENSFM00300000079125 | E3 Ubiquitin Ligase TRIM9 EC_6.3.2.- Tripartite Motif Containing 9 |
| ENSMUSG00000021072 | ENSFM00670001235637 | Thioredoxin Related Transmembrane Precursor Thioredoxin Domain Containing |
| ENSMUSG00000021076 | ENSFM00250000004727 | Actin Related 10 Actin Related 11 |
| ENSMUSG00000021079 | ENSFM00670001236198 | Mitochondrial Import Inner Membrane Translocase Subunit TIM9 |
| ENSMUSG00000021087 | ENSFM00500000269689 | Reticulon |
| ENSMUSG00000021096 | ENSFM00730001521429 | Phosphatase 1A EC_3.1.3.16 Phosphatase 2C Alpha PP2C Alpha |
| ENSMUSG00000021097 | ENSFM00500000271703 | Calmin Calponin Transmembrane Domain |
| ENSMUSG00000021102 | ENSFM00670001235945 | Glutaredoxin Related 5 Mitochondrial Precursor Monothiol Glutaredoxin 5 |
| ENSMUSG00000021112 | ENSFM00760001714797 | Maguk P55 Subfamily Member 5 |
| ENSMUSG00000021114 | ENSFM00770001847582 | V Type Proton Atpase Subunit D V Atpase Subunit D Vacuolar Proton Pump Subunit D |
| ENSMUSG00000021116 | ENSFM00810002040882 | Eukaryotic Translation Initiation Factor 2 Subunit 1 Eukaryotic Translation Initiation Factor 2 Subunit Alpha Eif 2 Alpha Eif 2A Eif 2ALPHA |
| ENSMUSG00000021124 | ENSFM00590000917484 | Unknown |
| ENSMUSG00000021124 | ENSFM00590000917485 | Unknown |
| ENSMUSG00000021124 | ENSFM00250000006099 | Vesicle Transport Through Interaction With T Snares Homolog 1B Vesicle Transport V Snare VTI1 1 VTI1 RP1 |
| ENSMUSG00000021131 | ENSFM00670001235831 | Enhancer Of Rudimentary |
| ENSMUSG00000021139 | ENSFM00610000973484 | Unknown |
| ENSMUSG00000021139 | ENSFM00670001236361 | Synaptojanin 2 Binding |
| ENSMUSG00000021140 | ENSFM00250000001163 | Pecanex |
| ENSMUSG00000021147 | ENSFM00660001167690 | Unknown |
| ENSMUSG00000021147 | ENSFM00660001167691 | Unknown |
| ENSMUSG00000021147 | ENSFM00660001172867 | Unknown |
| ENSMUSG00000021147 | ENSFM00660001173980 | Unknown |
| ENSMUSG00000021147 | ENSFM00660001181865 | Unknown |
| ENSMUSG00000021147 | ENSFM00250000004113 | Wd Repeat Containing 37 |
| ENSMUSG00000021171 | ENSFM00250000000964 | Extended Synaptotagmin E |
| ENSMUSG00000021178 | ENSFM00730001521694 | 26S Protease Regulatory Subunit 4 26S Proteasome Aaa Atpase Subunit RPT2 Proteasome 26S Subunit Atpase 1 |
| ENSMUSG00000021188 | ENSFM00250000003391 | Thyroid Receptor Interacting 11 Tr Interacting 11 Trip 11 Clonal Evolution Related Gene On Chromosome 14 Golgi Associated Microtubule Binding 210 Gmap 210 TRIP230 |
| ENSMUSG00000021192 | ENSFM00250000005275 | Golgin Subfamily A Member 5 Golgin 84 |
| ENSMUSG00000021193 | ENSFM00250000003510 | Presequence Protease EC_3.4.24.- |
| ENSMUSG00000021196 | ENSFM00570000877873 | Unknown |
| ENSMUSG00000021196 | ENSFM00570000886442 | Unknown |
| ENSMUSG00000021196 | ENSFM00730001521507 | Atp Dependent 6 Phosphofructokinase Type |
| ENSMUSG00000021198 | ENSFM00680001316020 | Unknown |
| ENSMUSG00000021198 | ENSFM00250000002517 | Unc 79 Homolog |
| ENSMUSG00000021218 | ENSFM00800002003449 | Rab Gdp Dissociation Inhibitor Alpha Rab Gdi Alpha Guanosine Diphosphate Dissociation Inhibitor 1 Gdi 1 |
| ENSMUSG00000021219 | ENSFM00810002057142 | Unknown |
| ENSMUSG00000021219 | ENSFM00730001521808 | Regulator Of G Signaling |
| ENSMUSG00000021224 | ENSFM00570000870030 | Unknown |
| ENSMUSG00000021224 | ENSFM00570000884795 | Unknown |
| ENSMUSG00000021224 | ENSFM00250000001973 | Numb |
| ENSMUSG00000021238 | ENSFM00410000138519 | Methylmalonate Semialdehyde Dehydrogenase EC_1.2.1.18 EC_1.2.1.- 27 Aldehyde Dehydrogenase Family 6 Member A1 |
| ENSMUSG00000021241 | ENSFM00500000274462 | Iron Sulfur Cluster Assembly 2 Homolog Mitochondrial Precursor Hesb Domain Containing 1 |
| ENSMUSG00000021248 | ENSFM00500000271022 | Transmembrane EMP24 Domain Containing Precursor |
| ENSMUSG00000021252 | ENSFM00570000894602 | Unknown |
| ENSMUSG00000021252 | ENSFM00500000272892 | Probable Ergosterol Biosynthetic 28 |
| ENSMUSG00000021259 | ENSFM00380000124753 | Cholesterol 24 Hydroxylase CH24H EC_1.14.13.98 Cytochrome P450 46A1 |
| ENSMUSG00000021262 | ENSFM00740001589204 | Ena/Vasp Ena/Vasodilator Stimulated Phosphoprotein |
| ENSMUSG00000021266 | ENSFM00590000917504 | Unknown |
| ENSMUSG00000021266 | ENSFM00590000917505 | Unknown |
| ENSMUSG00000021266 | ENSFM00250000003193 | Tryptophan Trna Ligase Cytoplasmic EC_6.1.1.2 Tryptophanyl Trna Synthetase Trprs |
| ENSMUSG00000021270 | ENSFM00250000000292 | Heat Shock |
| ENSMUSG00000021277 | ENSFM00760001714794 | Tnf Receptor Associated Factor 3 EC_6.3.2.- CD40 Receptor Associated Factor 1 CRAF1 |
| ENSMUSG00000021279 | ENSFM00730001521452 | Serine/Threonine Kinase Mrck EC_2.7.11.1 CDC42 Binding Kinase Dmpk Myotonic Dystrophy Kinase Related CDC42 Binding Kinase Mrck Myotonic Dystrophy Kinase |
| ENSMUSG00000021282 | ENSFM00760001714733 | Eukaryotic Translation Initiation Factor 5 Eif 5 |
| ENSMUSG00000021288 | ENSFM00250000000749 | Kinesin Light Chain |
| ENSMUSG00000021290 | ENSFM00500000327578 | Unknown |
| ENSMUSG00000021301 | ENSFM00730001522025 | E3 Ubiquitin Ligase EC_6.3.2.- Hect C2 And Ww Domain Containing NEDD4 E3 Ubiquitin Ligase |
| ENSMUSG00000021303 | ENSFM00500000362850 | Unknown |
| ENSMUSG00000021313 | ENSFM00250000000332 | Ryanodine Receptor Ryr Muscle Calcium Release Channel Muscle Type Ryanodine Receptor Type Ryanodine Receptor |
| ENSMUSG00000021314 | ENSFM00250000000861 | Bridging Integrator |
| ENSMUSG00000021338 | ENSFM00610000954856 | Unknown |
| ENSMUSG00000021338 | ENSFM00610000963235 | Unknown |
| ENSMUSG00000021338 | ENSFM00610000973168 | Unknown |
| ENSMUSG00000021338 | ENSFM00260000050454 | Leucine Rich Repeat Containing |
| ENSMUSG00000021339 | ENSFM00250000006689 | Magnesium Transporter MRS2 Homolog Mitochondrial Precursor MRS2 |
| ENSMUSG00000021357 | ENSFM00250000004215 | Exocyst Complex Component 2 Exocyst Complex Component SEC5 |
| ENSMUSG00000021361 | ENSFM00670001236776 | Transmembrane 14C |
| ENSMUSG00000021371 | ENSFM00610000952921 | Mitochondrial Calcium Uniporter Regulator 1 Coiled Coil Domain Containing 90A Mitochondrial |
| ENSMUSG00000021373 | ENSFM00250000001899 | Adenylyl Cyclase Associated Cap |
| ENSMUSG00000021417 | ENSFM00500000270657 | Enoyl Coa Delta Isomerase 2 Mitochondrial Precursor EC_5.3.3.8 Delta 3 Delta 2 Enoyl Coa Isomerase D3 D2 Enoyl Coa Isomerase Dodecenoyl Coa Isomerase Peroxisomal 3 2 Trans Enoyl Coa Isomerase Peci |
| ENSMUSG00000021427 | ENSFM00250000002354 | Translocon Associated Subunit Alpha Precursor Trap Alpha Signal Sequence Receptor Subunit Alpha Ssr Alpha |
| ENSMUSG00000021451 | ENSFM00480000262746 | Semaphorin Precursor |
| ENSMUSG00000021460 | ENSFM00500000270222 | Methylglutaconyl Coa Hydratase Mitochondrial Precursor EC_4.2.1.18 Au Specific Rna Binding Enoyl Coa Hydratase Au Binding Coa Hydratase |
| ENSMUSG00000021468 | ENSFM00800002003789 | Serine Palmitoyltransferase 1 EC_2.3.1.50 Long Chain Base Biosynthesis 1 Lcb 1 Serine Palmitoyl Coa Transferase 1 Spt 1 SPT1 |
| ENSMUSG00000021474 | ENSFM00730001521561 | Sideroflexin |
| ENSMUSG00000021477 | ENSFM00800002003395 | Cathepsin L1 EC_3.4.22.15 Cathepsin L |
| ENSMUSG00000021484 | ENSFM00260000050644 | Vesicular Integral Membrane VIP36 Precursor Lectin Mannose Binding 2 Vesicular Integral Membrane 36 VIP36 |
| ENSMUSG00000021501 | ENSFM00250000007633 | Calcium Signal Modulating Cyclophilin Ligand Caml |
| ENSMUSG00000021518 | ENSFM00800002003785 | Phosphatidylserine Synthase 1 Pss 1 Ptdser Synthase 1 EC_2.7.8.29 Serine Exchange Enzyme I |
| ENSMUSG00000021520 | ENSFM00670001236244 | Cytochrome B C1 Complex Subunit 7 Complex Iii Subunit 7 Complex Iii Subunit Vii Ubiquinol Cytochrome C Reductase Complex 14 Kda |
| ENSMUSG00000021536 | ENSFM00730001521648 | Adenylate Cyclase Type EC_4.6.1.1 Atp Pyrophosphate Lyase Adenylate Cyclase Type Adenylyl Cyclase |
| ENSMUSG00000021546 | ENSFM00660001174969 | Unknown |
| ENSMUSG00000021546 | ENSFM00250000001382 | Heterogeneous Nuclear Ribonucleoprotein K Hnrnp K |
| ENSMUSG00000021552 | ENSFM00250000003710 | G Kinase Anchoring 1 |
| ENSMUSG00000021559 | ENSFM00730001521806 | Death Associated Kinase Dap Kinase EC_2.7.11.1 Dap Kinase |
| ENSMUSG00000021573 | ENSFM00250000003009 | Tubulin Polymerization Promoting Family Member |
| ENSMUSG00000021576 | ENSFM00500000271967 | Programmed Cell Death 6 Probable Calcium Binding Alg 2 |
| ENSMUSG00000021577 | ENSFM00800002003532 | Succinate Dehydrogenase Flavoprotein Subunit Mitochondrial Precursor EC_1.3.5.1 Flavoprotein Subunit Of Complex Ii Fp |
| ENSMUSG00000021578 | ENSFM00590000917390 | Unknown |
| ENSMUSG00000021578 | ENSFM00250000008534 | Coiled Coil Domain Containing 127 |
| ENSMUSG00000021583 | ENSFM00740001589214 | Endoplasmic Reticulum Aminopeptidase EC_3.4.11.- Derived Aminopeptidase |
| ENSMUSG00000021591 | ENSFM00670001236435 | Glutaredoxin 1 Thioltransferase 1 Ttase 1 |
| ENSMUSG00000021596 | ENSFM00250000001118 | Multiple C2 And Transmembrane Domain Containing |
| ENSMUSG00000021606 | ENSFM00570000892282 | Unknown |
| ENSMUSG00000021606 | ENSFM00670001236132 | Nadh Dehydrogenase Iron Sulfur 6 Mitochondrial Precursor Complex I 13KD A Ci 13KD A Nadh Ubiquinone Oxidoreductase 13 Kda A Subunit |
| ENSMUSG00000021608 | ENSFM00570000885386 | Unknown |
| ENSMUSG00000021608 | ENSFM00570000889498 | Unknown |
| ENSMUSG00000021608 | ENSFM00250000001465 | Lysophosphatidylcholine Acyltransferase Lpc Acyltransferase Lpcat Lysopc Acyltransferase EC_2.3.1.23 1 Acylglycerophosphocholine O Acyltransferase 1 Alkylglycerophosphocholine O Acetyltransferase 67 Acetyl Coa:Lyso Platelet Activating Factor Acetyltransfe |
| ENSMUSG00000021609 | ENSFM00730001521312 | Sodium Dependent Transporter Transporter Solute Carrier Family 6 Member |
| ENSMUSG00000021610 | ENSFM00740001589427 | Cleft Lip And Palate Transmembrane 1 CLPTM1 |
| ENSMUSG00000021613 | ENSFM00730001521886 | Hyaluronan And Proteoglycan Link 1 Precursor Cartilage Linking 1 Cartilage Link Proteoglycan Link |
| ENSMUSG00000021614 | ENSFM00730001521436 | Core Precursor Chondroitin Sulfate Proteoglycan |
| ENSMUSG00000021646 | ENSFM00450000242169 | Methylcrotonoyl Coa Carboxylase Beta Chain Mitochondrial Precursor Mccase Subunit Beta EC_6.4.1.4 3 Methylcrotonyl Coa Carboxylase 2 3 Methylcrotonyl Coa Carboxylase Non Biotin Containing Subunit 3 Methylcrotonyl Coa:Carbon Dioxide Ligase Subunit Beta |
| ENSMUSG00000021665 | ENSFM00730001521836 | Beta Hexosaminidase Subunit Alpha Precursor EC_3.2.1.52 Beta N Acetylhexosaminidase Subunit Alpha Hexosaminidase Subunit A N Acetyl Beta Glucosaminidase Subunit Alpha |
| ENSMUSG00000021670 | ENSFM00610000969599 | Unknown |
| ENSMUSG00000021670 | ENSFM00770001847546 | 3 Hydroxy 3 Methylglutaryl Coenzyme A Reductase Hmg Coa Reductase EC_1.1.1.34 |
| ENSMUSG00000021684 | ENSFM00280000058709 | High Affinity Camp Specific And Ibmx Insensitive 3' 5' Cyclic Phosphodiesterase 8B EC_3.1.4.53 Cell Proliferation Inducing Gene 22 |
| ENSMUSG00000021687 | ENSFM00670001235365 | Secretory Carrier Associated Membrane Secretory Carrier Membrane |
| ENSMUSG00000021693 | ENSFM00250000000916 | Kinesin |
| ENSMUSG00000021699 | ENSFM00640001105232 | Unknown |
| ENSMUSG00000021699 | ENSFM00400000131721 | Camp Specific 3' 5' Cyclic Phosphodiesterase EC_3.1.4.53 |
| ENSMUSG00000021700 | ENSFM00730001521441 | Ras Related Rab 3 |
| ENSMUSG00000021704 | ENSFM00250000002187 | Metaxin 1 Mitochondrial Outer Membrane Import Complex 1 |
| ENSMUSG00000021708 | ENSFM00250000001613 | Ras Specific Guanine Nucleotide Releasing Factor 2 Ras GRF2 Ras Guanine Nucleotide Exchange Factor 2 |
| ENSMUSG00000021709 | ENSFM00730001522141 | Leucine Rich Repeat Containing 7 Densin 180 Densin |
| ENSMUSG00000021710 | ENSFM00760001714699 | Neurolysin Mitochondrial Precursor EC_3.4.24.16 Microsomal Endopeptidase Mep Mitochondrial Oligopeptidase M Neurotensin Endopeptidase |
| ENSMUSG00000021712 | ENSFM00590000917832 | Unknown |
| ENSMUSG00000021712 | ENSFM00730001522472 | E3 Ubiquitin Ligase TRIM23 EC_6.3.2.- Adp Ribosylation Factor Domain Containing 1 Gtp Binding Ard 1 Tripartite Motif Containing 23 |
| ENSMUSG00000021719 | ENSFM00250000004063 | Regulator Of G Signaling 7 Binding R7 Family Binding |
| ENSMUSG00000021728 | ENSFM00500000273736 | Embigin Precursor |
| ENSMUSG00000021730 | ENSFM00250000000931 | Potassium/Sodium Hyperpolarization Activated Cyclic Nucleotide Gated Channel |
| ENSMUSG00000021731 | ENSFM00250000008011 | 28S Ribosomal S30 Mitochondrial Mrp S30 S30MT |
| ENSMUSG00000021733 | ENSFM00730001521473 | Sodium Bicarbonate Solute Carrier Family 4 Member |
| ENSMUSG00000021737 | ENSFM00760001714781 | 26S Proteasome Non Atpase Regulatory Subunit 6 26S Proteasome Regulatory Subunit RPN7 26S Proteasome Regulatory Subunit S10 P42A |
| ENSMUSG00000021745 | ENSFM00730001521864 | Receptor Type Tyrosine Phosphatase Precursor R Ptp EC_3.1.3.48 |
| ENSMUSG00000021748 | ENSFM00500000270150 | Pyruvate Dehydrogenase E1 Component Subunit Beta Mitochondrial Precursor PDHE1 B EC_1.2.4.1 |
| ENSMUSG00000021750 | ENSFM00500000273986 | Unknown |
| ENSMUSG00000021756 | ENSFM00740001609388 | Unknown |
| ENSMUSG00000021756 | ENSFM00740001609389 | Unknown |
| ENSMUSG00000021756 | ENSFM00500000270708 | Interleukin 6 Receptor Subunit Beta Precursor Il 6 Receptor Subunit Beta Il 6R Subunit Beta Il 6R Beta Il 6RB Interleukin 6 Signal Transducer Membrane Glycoprotein 130 GP130 Oncostatin M Receptor Subunit Alpha CD130 Antigen |
| ENSMUSG00000021764 | ENSFM00250000005730 | Nadh Dehydrogenase Iron Sulfur 4 Mitochondrial Precursor Complex I 18 Kda Ci 18 Kda Nadh Ubiquinone Oxidoreductase 18 Kda Subunit |
| ENSMUSG00000021771 | ENSFM00570000888306 | Unknown |
| ENSMUSG00000021771 | ENSFM00250000001189 | Voltage Dependent Anion Selective Channel Vdac Outer Mitochondrial Membrane Porin |
| ENSMUSG00000021772 | ENSFM00570000869836 | Unknown |
| ENSMUSG00000021772 | ENSFM00250000003149 | Nf Kappa B Inhibitor Interacting Ras 2 I Kappa B Interacting Ras 2 Kappa B Ras 2 Kappab RAS2 |
| ENSMUSG00000021773 | ENSFM00270000056553 | Catechol O Methyltransferase Domain Containing 1 EC_2.1.1.- |
| ENSMUSG00000021791 | ENSFM00670001244624 | DPY30 Domain Containing 2 |
| ENSMUSG00000021792 | ENSFM00570000881559 | Unknown |
| ENSMUSG00000021792 | ENSFM00250000005616 | Redox Regulatory FAM213A Peroxiredoxin 2 Activated In M Csf Stimulated Monocytes Pamm |
| ENSMUSG00000021794 | ENSFM00250000001520 | Glutamate Dehydrogenase Mitochondrial Gdh EC_1.4.1.3 |
| ENSMUSG00000021796 | ENSFM00610000972853 | Unknown |
| ENSMUSG00000021796 | ENSFM00730001521645 | Bone Morphogenetic Receptor Type 1A Precursor Bmp Type 1A Receptor Bmpr 1A EC_2.7.11.30 Activin Receptor Kinase 3 Alk 3 Receptor CD292 Antigen |
| ENSMUSG00000021807 | ENSFM00250000005286 | Homolog |
| ENSMUSG00000021814 | ENSFM00760001714601 | Annexin Annexin Annexin |
| ENSMUSG00000021816 | ENSFM00310000089007 | Serine/Threonine Phosphatase 2B Catalytic Subunit EC_3.1.3.16 Cam Prp Catalytic Subunit Calmodulin Dependent Calcineurin A Subunit |
| ENSMUSG00000021820 | ENSFM00250000000111 | Calcium/Calmodulin Dependent Kinase Type Ii Subunit Cam Kinase Ii Subunit Camk Ii Subunit EC_2.7.11.17 |
| ENSMUSG00000021823 | ENSFM00250000001750 | Vinculin |
| ENSMUSG00000021824 | ENSFM00440000236903 | Ap 3 Complex Subunit Mu Adaptor Related Complex 3 Subunit Mu Clathrin Assembly Assembly Complex 3 Mu Medium Chain Clathrin Coat Assembly AP47 Homolog Clathrin Coat Associated AP47 Homolog Golgi Adaptor Ap 1.47 Kda Homolog HA1.47 Kda Subunit Homolog Adapti |
| ENSMUSG00000021831 | ENSFM00730001521998 | ERO1 Alpha Precursor ERO1 L ERO1 L Alpha EC_1.8.4.- Endoplasmic Reticulum Oxidoreductin 1 Oxidoreductin 1 L Alpha |
| ENSMUSG00000021832 | ENSFM00720001492034 | 26S Protease Regulatory Subunit 10B 26S Proteasome Aaa Atpase Subunit RPT4 Proteasome 26S Subunit Atpase 6 Proteasome Subunit P42 |
| ENSMUSG00000021843 | ENSFM00250000002379 | Kinectin |
| ENSMUSG00000021866 | ENSFM00760001714601 | Annexin Annexin Annexin |
| ENSMUSG00000021868 | ENSFM00750001632315 | Peptidyl Prolyl Cis Trans Isomerase Ppiase EC_5.2.1.8 Cyclophilin Rotamase |
| ENSMUSG00000021870 | ENSFM00250000002303 | Sarcolemmal Membrane Associated Sarcolemmal Associated |
| ENSMUSG00000021877 | ENSFM00610000970080 | Unknown |
| ENSMUSG00000021877 | ENSFM00610000972841 | Unknown |
| ENSMUSG00000021877 | ENSFM00730001521124 | Adp Ribosylation Factor |
| ENSMUSG00000021884 | ENSFM00570000881762 | Unknown |
| ENSMUSG00000021884 | ENSFM00610000969634 | Unknown |
| ENSMUSG00000021884 | ENSFM00250000003931 | 2 Hydroxyacyl Coa Lyase 1 EC_4.1.-.- 2 Hydroxyphytanoyl Coa Lyase 2 Hpcl Phytanoyl Coa 2 Hydroxylase 2 |
| ENSMUSG00000021910 | ENSFM00610000966894 | Unknown |
| ENSMUSG00000021910 | ENSFM00610000972849 | Unknown |
| ENSMUSG00000021910 | ENSFM00250000002121 | Nischarin Imidazoline Receptor 1 I 1 IR1 Imidazoline Receptor 1 Imidazoline 1 Receptor I1R |
| ENSMUSG00000021913 | ENSFM00750001632363 | 2 Oxoglutarate Dehydrogenase Mitochondrial Precursor 2 Oxoglutarate Dehydrogenase Complex Component E1 Ogdc E1 Alpha Ketoglutarate Dehydrogenase |
| ENSMUSG00000021917 | ENSFM00760001723268 | Unknown |
| ENSMUSG00000021917 | ENSFM00500000275173 | Signal Peptidase Complex Subunit 1 EC_3.4.-.- Microsomal Signal Peptidase 12 Kda Subunit Spase 12 Kda Subunit |
| ENSMUSG00000021929 | ENSFM00750001632344 | Importin Subunit Alpha 3 Importin Alpha Q1 QIP1 Karyopherin Subunit Alpha 4 |
| ENSMUSG00000021930 | ENSFM00250000004208 | Spry Domain Containing 7 Chronic Lymphocytic Leukemia Deletion Region Gene 6 Homolog Cll Deletion Region Gene 6 Homolog |
| ENSMUSG00000021938 | ENSFM00610000969895 | Unknown |
| ENSMUSG00000021938 | ENSFM00250000000671 | Paraspeckle Component 1 |
| ENSMUSG00000021939 | ENSFM00290000065537 | Cathepsin B Precursor EC_3.4.22.1 |
| ENSMUSG00000021948 | ENSFM00750001632332 | Kinase C Delta Type EC_2.7.11.13 Npkc Delta |
| ENSMUSG00000021957 | ENSFM00590000917972 | Unknown |
| ENSMUSG00000021957 | ENSFM00250000001413 | Transketolase EC_2.2.1.1 |
| ENSMUSG00000021969 | ENSFM00250000001255 | Palmitoyltransferase EC_2.3.1.225 Zinc Finger Dhhc Domain Containing Dhhc |
| ENSMUSG00000021983 | ENSFM00570000880471 | Unknown |
| ENSMUSG00000021983 | ENSFM00570000897944 | Unknown |
| ENSMUSG00000021983 | ENSFM00770001847537 | Phospholipid Transporting Atpase EC_3.6.3.1 Atpase Class I Type 8A Member P4 Atpase Flippase Complex Alpha Subunit At |
| ENSMUSG00000021986 | ENSFM00250000005156 | Apc Membrane Recruitment 2 AMER2 FAM123A |
| ENSMUSG00000021991 | ENSFM00730001521792 | Voltage Dependent Calcium Channel Subunit Alpha 2/Delta 3 Precursor Voltage Gated Calcium Channel Subunit Alpha 2/Delta 3 |
| ENSMUSG00000021994 | ENSFM00740001589193 | Wnt 5 |
| ENSMUSG00000022019 | ENSFM00250000004146 | Tudor Domain Containing 3 |
| ENSMUSG00000022024 | ENSFM00500000271163 | Suppressor Of G2 Allele Of SKP1 Homolog |
| ENSMUSG00000022037 | ENSFM00570000900344 | Unknown |
| ENSMUSG00000022037 | ENSFM00250000003194 | Clusterin Precursor |
| ENSMUSG00000022048 | ENSFM00250000000643 | Dihydropyrimidinase Related Drp Collapsin Response Mediator Crmp |
| ENSMUSG00000022052 | ENSFM00250000000829 | Serine/Threonine Phosphatase 2A 55 Kda Regulatory Subunit B PP2A Subunit B B55 PP2A Subunit B PR55 PP2A Subunit B R2 PP2A Subunit B |
| ENSMUSG00000022054 | ENSFM00730001521354 | Neurofilament Polypeptide Nf Kda Neurofilament Neurofilament Triplet |
| ENSMUSG00000022055 | ENSFM00730001521354 | Neurofilament Polypeptide Nf Kda Neurofilament Neurofilament Triplet |
| ENSMUSG00000022099 | ENSFM00500000271390 | Dematin Dematin Actin Binding Erythrocyte Membrane Band 4 9 |
| ENSMUSG00000022100 | ENSFM00250000001821 | Exportin 7 |
| ENSMUSG00000022103 | ENSFM00500000269996 | Gdnf Family Receptor Alpha Precursor Gdnf Receptor Alpha Gdnfr Alpha Gfr Alpha Tgf Beta Related Neurotrophic Factor Receptor |
| ENSMUSG00000022108 | ENSFM00600000921249 | Integral Membrane 2B Immature BRI2 IMBRI2 Transmembrane Bri Bri |
| ENSMUSG00000022110 | ENSFM00590000918377 | Unknown |
| ENSMUSG00000022110 | ENSFM00250000001376 | Succinyl Coa Ligase Subunit Beta Mitochondrial Precursor EC_6.2.1.5 Atp Specific Succinyl Coa Synthetase Subunit Beta Succinyl Coa Synthetase Beta A Chain Scs Betaa |
| ENSMUSG00000022112 | ENSFM00500000270283 | Glypican 3 Precursor Intestinal Oci 5 |
| ENSMUSG00000022114 | ENSFM00250000002344 | Sprouty Homolog Spry |
| ENSMUSG00000022132 | ENSFM00670001235591 | Claudin |
| ENSMUSG00000022136 | ENSFM00730001522230 | Dnaj Homolog Subfamily C Member 3 Precursor Interferon Induced Double Stranded Rna Activated Kinase Inhibitor Kinase Inhibitor Of 58 Kda Kinase Inhibitor P58 |
| ENSMUSG00000022150 | ENSFM00590000917915 | Unknown |
| ENSMUSG00000022150 | ENSFM00590000918392 | Unknown |
| ENSMUSG00000022150 | ENSFM00740001589270 | Disabled Homolog 2 Adaptor Molecule Disabled 2 Differentially Expressed In Ovarian Carcinoma 2 Doc 2 |
| ENSMUSG00000022159 | ENSFM00550000762928 | Unknown |
| ENSMUSG00000022159 | ENSFM00640001117456 | Unknown |
| ENSMUSG00000022159 | ENSFM00640001119687 | Unknown |
| ENSMUSG00000022159 | ENSFM00730001521469 | Ras Related Rab |
| ENSMUSG00000022174 | ENSFM00670001235800 | Dolichyl Diphosphooligosaccharide Glycosyltransferase Subunit DAD1 Oligosaccharyl Transferase Subunit DAD1 EC_2.4.99.18 Defender Against Cell Death 1 Dad 1 |
| ENSMUSG00000022176 | ENSFM00260000050685 | Gtp Binding Rem Rad And Gem Gtp Binding |
| ENSMUSG00000022180 | ENSFM00250000000331 | Amino Acid Transporter Solute Carrier Family 7 Member |
| ENSMUSG00000022186 | ENSFM00250000002519 | Succinyl Coa:3 Ketoacid Coenzyme A Transferase 1 Mitochondrial Precursor EC_2.8.3.5 3 Oxoacid Coa Transferase 1 Somatic Type Succinyl Coa:3 Oxoacid Coa Transferase Scot S |
| ENSMUSG00000022193 | ENSFM00320000100083 | Proteasome Subunit Beta Type Precursor EC_3.4.25.1 Macropain Multicatalytic Endopeptidase Complex Proteasome Proteasome Subunit |
| ENSMUSG00000022199 | ENSFM00650001140024 | Solute Carrier Family 22 Member 17 24P3 Receptor 24P3R Lipocalin 2 Receptor |
| ENSMUSG00000022208 | ENSFM00250000001356 | Junctophilin Jp Junctophilin |
| ENSMUSG00000022210 | ENSFM00500000270135 | Dehydrogenase/Reductase Sdr Family Member 4 EC_1.1.1.184 Nadph Dependent Carbonyl Reductase/Nadp Retinol Dehydrogenase Cr Phcr Nadph Dependent Retinol Dehydrogenase/Reductase Ndrd Peroxisomal Short Chain Alcohol Dehydrogenase Pscd |
| ENSMUSG00000022212 | ENSFM00800002003632 | Copine Copine |
| ENSMUSG00000022223 | ENSFM00250000006615 | Epimerase Family Sd EC_1.1.1.- Short Chain Dehydrogenase/Reductase Family 39U Member 1 |
| ENSMUSG00000022234 | ENSFM00730001521615 | T Complex 1 Subunit Epsilon Tcp 1 Epsilon Cct Epsilon |
| ENSMUSG00000022240 | ENSFM00730001521673 | Catenin Delta 2 |
| ENSMUSG00000022241 | ENSFM00250000001453 | Threonine Trna Ligase EC_6.1.1.3 Threonyl Trna Synthetase Thrrs Threonyl Trna Synthetase |
| ENSMUSG00000022246 | ENSFM00800002003877 | Ankycorbin Ankyrin Repeat And Coiled Coil Structure Containing Retinal Pigment Epithelial Cell Retinoic Acid Induced 14 |
| ENSMUSG00000022253 | ENSFM00760001763284 | Unknown |
| ENSMUSG00000022253 | ENSFM00250000003187 | Nad Kinase 2 Mitochondrial Precursor EC_2.7.1.23 Mitochondrial Nad Kinase Nad Kinase Domain Containing 1 Mitochondrial |
| ENSMUSG00000022255 | ENSFM00250000005409 | Lyric Lysine Rich CEACAM1 Co Isolated Metadherin Metastasis Adhesion |
| ENSMUSG00000022263 | ENSFM00740001589124 | EC_2.7.11.1 |
| ENSMUSG00000022265 | ENSFM00250000005694 | Progressive Ankylosis Homolog Ank |
| ENSMUSG00000022283 | ENSFM00570000851024 | Polyadenylate Binding Pabp Poly A Binding |
| ENSMUSG00000022285 | ENSFM00500000361339 | Unknown |
| ENSMUSG00000022285 | ENSFM00570000901593 | Unknown |
| ENSMUSG00000022285 | ENSFM00730001521154 | Unknown |
| ENSMUSG00000022295 | ENSFM00750001632375 | V Type Proton Atpase Subunit C V Atpase Subunit C Vacuolar Proton Pump Subunit C |
| ENSMUSG00000022307 | ENSFM00670001235411 | Oxidation Resistance 1 |
| ENSMUSG00000022321 | ENSFM00260000050333 | Cadherin Precursor |
| ENSMUSG00000022323 | ENSFM00500000272841 | Ribonuclease UK114 EC_3.1.-.- Kda Translational Inhibitor UK114 Antigen |
| ENSMUSG00000022337 | ENSFM00250000004805 | Er Membrane Complex Subunit 2 Tetratricopeptide Repeat 35 Tpr Repeat 35 |
| ENSMUSG00000022340 | ENSFM00250000002727 | Syntabulin Golgi Localized Syntaphilin Related Syntaxin 1 Binding |
| ENSMUSG00000022350 | ENSFM00250000005256 | Wash Complex Subunit Strumpellin |
| ENSMUSG00000022354 | ENSFM00250000004530 | Nadh Dehydrogenase 1 Beta Subcomplex Subunit 9 Complex I B22 Ci B22 Nadh Ubiquinone Oxidoreductase B22 Subunit |
| ENSMUSG00000022365 | ENSFM00500000270947 | Derlin 1 Degradation In Endoplasmic Reticulum 1 DER1 1 |
| ENSMUSG00000022376 | ENSFM00730001521247 | Adenylate Cyclase Type EC_4.6.1.1 Atp Pyrophosphate Lyase Adenylate Cyclase Type Adenylyl Cyclase Ca 2+ Adenylyl Cyclase |
| ENSMUSG00000022378 | ENSFM00250000002173 | FAM49A |
| ENSMUSG00000022390 | ENSFM00250000002307 | Zinc Finger Ccch Domain Containing |
| ENSMUSG00000022400 | ENSFM00670001235574 | Ring Box 1 |
| ENSMUSG00000022401 | ENSFM00610000960709 | Unknown |
| ENSMUSG00000022401 | ENSFM00800002003832 | Probable Xaa Pro Aminopeptidase 3 X Pro Aminopeptidase 3 EC_3.4.11.9 Aminopeptidase P3 APP3 |
| ENSMUSG00000022403 | ENSFM00610000960711 | Unknown |
| ENSMUSG00000022403 | ENSFM00610000973129 | Unknown |
| ENSMUSG00000022403 | ENSFM00250000002693 | HSC70 Interacting Hip Fa ST13 Homolog |
| ENSMUSG00000022407 | ENSFM00610000967132 | Unknown |
| ENSMUSG00000022407 | ENSFM00800002024349 | Unknown |
| ENSMUSG00000022407 | ENSFM00250000003164 | Adenylosuccinate Lyase Asl EC_4.3.2.2 Adenylosuccinase Asase |
| ENSMUSG00000022415 | ENSFM00570000881601 | Unknown |
| ENSMUSG00000022415 | ENSFM00670001235552 | Synaptogyrin |
| ENSMUSG00000022416 | ENSFM00590000917531 | Unknown |
| ENSMUSG00000022416 | ENSFM00740001589106 | Voltage Dependent T Type Calcium Channel Subunit Alpha Voltage Gated Calcium Channel Subunit Alpha CAV3 |
| ENSMUSG00000022416 | ENSFM00810002042227 | CACNA1I Splicing Variant |
| ENSMUSG00000022421 | ENSFM00500000269860 | Neuronal Pentraxin 2 Precursor NP2 Neuronal Pentraxin Ii Np Ii |
| ENSMUSG00000022427 | ENSFM00600000921823 | Mitochondrial Import Receptor Subunit TOM22 Homolog Translocase Of Outer Membrane 22 Kda Subunit Homolog |
| ENSMUSG00000022428 | ENSFM00600000921664 | Chibby Homolog 1 Cytosolic Leucine Rich Pigea 14 PKD2 Interactor Golgi And Endoplasmic Reticulum Associated 1 |
| ENSMUSG00000022433 | ENSFM00570000881021 | Unknown |
| ENSMUSG00000022433 | ENSFM00800002003414 | Casein Kinase I Delta Cki Delta Ckid EC_2.7.11.1 |
| ENSMUSG00000022436 | ENSFM00570000891648 | Unknown |
| ENSMUSG00000022436 | ENSFM00250000000900 | Rho Gtpase Activating Rho Type Gtpase Activating Rhogap Interacting With CIP4 Homologs Rich |
| ENSMUSG00000022436 | ENSFM00670001235619 | Pyridoxal Phosphate Phosphatase Plp Phosphatase EC_3.1.3.3 EC_3.1.3.- 74 Chronophin |
| ENSMUSG00000022437 | ENSFM00500000271410 | Sorting And Assembly Machinery Component 50 Homolog |
| ENSMUSG00000022443 | ENSFM00270000056424 | Myosin Cellular Myosin Heavy Chain Type Myosin Heavy Chain Myosin Heavy Chain Non Muscle Non Muscle Myosin Heavy Chain Nmmhc Non Muscle Myosin Heavy Chain Nmmhc Ii Nmmhc |
| ENSMUSG00000022450 | ENSFM00500000272594 | Nadh Dehydrogenase 1 Alpha Subcomplex Subunit 6 Complex I B14 Ci B14 Nadh Ubiquinone Oxidoreductase B14 Subunit |
| ENSMUSG00000022451 | ENSFM00500000270278 | Twinfilin |
| ENSMUSG00000022454 | ENSFM00250000001932 | Kinase C Binding NELL2 Precursor Nel 2 |
| ENSMUSG00000022456 | ENSFM00500000269805 | Septin |
| ENSMUSG00000022477 | ENSFM00250000002510 | Aconitate Hydratase Mitochondrial Precursor Aconitase EC_4.2.1.3 Citrate Hydro Lyase |
| ENSMUSG00000022489 | ENSFM00400000131765 | Calcium/Calmodulin Dependent 3' 5' Cyclic Nucleotide Phosphodiesterase Cam Pde EC_3.1.4.17 Kda Cam Pde |
| ENSMUSG00000022494 | ENSFM00500000276365 | Shisa Precursor |
| ENSMUSG00000022514 | ENSFM00500000270987 | Interleukin 1 Receptor Accessory Precursor Il 1 Receptor Accessory Il 1RACP |
| ENSMUSG00000022523 | ENSFM00500000269773 | Fibroblast Growth Factor Fgf Fibroblast Growth Factor Homologous Factor Fhf |
| ENSMUSG00000022540 | ENSFM00250000005331 | Rogdi Homolog |
| ENSMUSG00000022548 | ENSFM00670001235881 | Apolipoprotein D Precursor Apo D Apod |
| ENSMUSG00000022551 | ENSFM00250000003946 | Cytochrome C1 Heme Protein Mitochondrial Precursor Complex Iii Subunit 4 Complex Iii Subunit Iv Cytochrome B C1 Complex Subunit 4 Ubiquinol Cytochrome C Reductase Complex Cytochrome C1 Subunit Cytochrome C 1 |
| ENSMUSG00000022561 | ENSFM00610000965707 | Unknown |
| ENSMUSG00000022561 | ENSFM00610000965708 | Unknown |
| ENSMUSG00000022561 | ENSFM00610000973119 | Unknown |
| ENSMUSG00000022561 | ENSFM00500000272001 | Glycosylphosphatidylinositol Anchor Attachment 1 Gpi Anchor Attachment 1 GAA1 Homolog |
| ENSMUSG00000022565 | ENSFM00610000973117 | Unknown |
| ENSMUSG00000022565 | ENSFM00610000973118 | Unknown |
| ENSMUSG00000022565 | ENSFM00740001589199 | Plectin Pcn Pltn Plectin 1 |
| ENSMUSG00000022568 | ENSFM00570000899077 | Unknown |
| ENSMUSG00000022568 | ENSFM00730001522046 | Scribble |
| ENSMUSG00000022571 | ENSFM00500000269896 | Pyrroline 5 Carboxylate Reductase 3 P5C Reductase 3 P5CR 3 EC_1.5.1.2 Pyrroline 5 Carboxylate Reductase |
| ENSMUSG00000022577 | ENSFM00560000771895 | Lymphocyte Antigen 6H Precursor Ly 6H |
| ENSMUSG00000022602 | ENSFM00250000009482 | Activity Regulated Cytoskeleton Associated Arc/ARG3 1 Activity Regulated Gene 3 1 ARG3 1 |
| ENSMUSG00000022604 | ENSFM00250000003841 | Centrosomal Of 97 Kda CEP97 Leucine Rich Repeat And Iq Domain Containing 2 |
| ENSMUSG00000022620 | ENSFM00600000921289 | N Acetylgalactosamine 6 Sulfatase Precursor EC_3.1.6.4 Chondroitinsulfatase Chondroitinase Galactose 6 Sulfate Sulfatase N Acetylgalactosamine 6 Sulfate Sulfatase GALNAC6S Sulfatase |
| ENSMUSG00000022623 | ENSFM00250000000743 | SH3 And Multiple Ankyrin Repeat Domains Proline Rich Synapse Associated |
| ENSMUSG00000022623 | ENSFM00800002009948 | SHANK3E Splice Variant |
| ENSMUSG00000022629 | ENSFM00730001521483 | Kinesin |
| ENSMUSG00000022636 | ENSFM00500000270582 | CD166 Antigen Activated Leukocyte Cell Adhesion Molecule CD166 Antigen |
| ENSMUSG00000022656 | ENSFM00790001963937 | Unknown |
| ENSMUSG00000022656 | ENSFM00500000271628 | Nectin 3 Precursor Poliovirus Receptor Related 3 |
| ENSMUSG00000022658 | ENSFM00500000270015 | Transgelin |
| ENSMUSG00000022661 | ENSFM00250000008020 | Ox 2 Membrane Glycoprotein Precursor Mrc Ox 2 Antigen CD200 Antigen |
| ENSMUSG00000022663 | ENSFM00500000271371 | Ubiquitin Conjugating Enzyme ATG3 EC_6.3.2.- Autophagy Related 3 APG3 |
| ENSMUSG00000022671 | ENSFM00670001236572 | Mitotic Spindle Organizing Mitotic Spindle Organizing Associated With A Ring Of Gamma Tubulin |
| ENSMUSG00000022674 | ENSFM00520000550014 | Unknown |
| ENSMUSG00000022674 | ENSFM00250000001221 | Ubiquitin Conjugating Enzyme E2 Variant |
| ENSMUSG00000022698 | ENSFM00250000003966 | N Alpha Acetyltransferase 50 EC_2.3.1.- N Acetyltransferase Nate Catalytic Subunit |
| ENSMUSG00000022706 | ENSFM00500000274041 | 39S Ribosomal L40 Mitochondrial Precursor L40MT Mrp L40 Nuclear Localization Signal Containing Deleted In Velocardiofacial Syndrome |
| ENSMUSG00000022707 | ENSFM00250000003836 | 1 4 Alpha Glucan Branching Enzyme EC_2.4.1.18 Glycogen Branching Enzyme |
| ENSMUSG00000022710 | ENSFM00590000918024 | Ubiquitin Carboxyl Terminal Hydrolase 7 |
| ENSMUSG00000022710 | ENSFM00800002003516 | Ubiquitin Carboxyl Terminal Hydrolase 7 EC_3.4.19.12 Deubiquitinating Enzyme 7 Herpesvirus Associated Ubiquitin Specific Protease Ubiquitin Thioesterase 7 Ubiquitin Specific Processing Protease 7 |
| ENSMUSG00000022752 | ENSFM00710001441924 | Mitochondrial Import Receptor Subunit TOM70 Mitochondrial Precursor Proteins Import Receptor Translocase Of Outer Membrane 70 Kda Subunit |
| ENSMUSG00000022762 | ENSFM00250000000779 | Neural Cell Adhesion Molecule 1 Precursor N Cam 1 Ncam 1 |
| ENSMUSG00000022763 | ENSFM00800002003733 | Apoptosis Inducing Factor 3 Ec 1 Apoptosis Inducing Factor |
| ENSMUSG00000022765 | ENSFM00250000006834 | Synaptosomal Associated 29 Snap 29 Golgi Snare Of 32 Kda GS32 Soluble 29 Kda Nsf Attachment Vesicle Membrane Fusion Snap 29 |
| ENSMUSG00000022769 | ENSFM00250000003961 | Stromal Cell Derived Factor 2 1 Precursor SDF2 1 |
| ENSMUSG00000022770 | ENSFM00720001503508 | Unknown |
| ENSMUSG00000022770 | ENSFM00250000000275 | Disks Large Homolog Synapse Associated Sap |
| ENSMUSG00000022789 | ENSFM00780001898359 | Dynamin 1 EC_3.6.5.5 Dynamin Family Member Proline Rich Carboxyl Terminal Domain Less Dymple Dynamin Related 1 |
| ENSMUSG00000022792 | ENSFM00250000003377 | Tyrosine Trna Ligase Mitochondrial Precursor EC_6.1.1.1 Tyrosyl Trna Synthetase Tyrrs |
| ENSMUSG00000022797 | ENSFM00570000890137 | Unknown |
| ENSMUSG00000022797 | ENSFM00500000270210 | Transferrin Receptor 1 Tr Tfr TFR1 Trfr CD71 Antigen |
| ENSMUSG00000022799 | ENSFM00250000001205 | Rho Gtpase Activating Rho Type Gtpase Activating Gtpase Activating |
| ENSMUSG00000022801 | ENSFM00610000970063 | Unknown |
| ENSMUSG00000022801 | ENSFM00800002003638 | Leucine Rich Repeat And Calponin Homology Domain Containing |
| ENSMUSG00000022802 | ENSFM00250000004185 | Leishmanolysin Peptidase EC_3.4.24.- |
| ENSMUSG00000022808 | ENSFM00250000006922 | Sorting Nexin 4 |
| ENSMUSG00000022812 | ENSFM00810002040612 | Glycogen Synthase Kinase 3 Gsk 3 EC_2.7.11.26 Serine/Threonine Kinase EC_2.7.11.- 1 |
| ENSMUSG00000022817 | ENSFM00730001521296 | Integrin Beta Precursor |
| ENSMUSG00000022820 | ENSFM00500000272985 | Nadh Dehydrogenase 1 Beta Subcomplex Subunit 4 Complex I B15 Ci B15 Nadh Ubiquinone Oxidoreductase B15 Subunit |
| ENSMUSG00000022822 | ENSFM00730001521608 | Multidrug Resistance Associated Atp Binding Cassette Sub Family C Member |
| ENSMUSG00000022829 | ENSFM00500000269888 | Syntaxin Binding 5 Lethal 2 Giant Larvae Homolog Tomosyn |
| ENSMUSG00000022836 | ENSFM00740001589296 | Myosin Light Chain Kinase Smooth Muscle Mlck Smmlck EC_2.7.11.18 Telokin |
| ENSMUSG00000022840 | ENSFM00730001521247 | Adenylate Cyclase Type EC_4.6.1.1 Atp Pyrophosphate Lyase Adenylate Cyclase Type Adenylyl Cyclase Ca 2+ Adenylyl Cyclase |
| ENSMUSG00000022841 | ENSFM00730001521696 | Ap 2 Complex Subunit Mu Ap 2 Mu Chain Clathrin Assembly Complex 2 Mu Medium Chain Clathrin Coat Assembly AP50 Clathrin Coat Associated AP50 MU2 Adaptin Plasma Membrane Adaptor Ap Kda |
| ENSMUSG00000022843 | ENSFM00250000000612 | Chloride Channel Clc |
| ENSMUSG00000022848 | ENSFM00250000006697 | Disrupted In Renal Carcinoma 2 |
| ENSMUSG00000022861 | ENSFM00730001521692 | Diacylglycerol Kinase Dag Kinase EC_2.7.1.107 Kda Diacylglycerol Kinase Diglyceride Kinase Dgk |
| ENSMUSG00000022865 | ENSFM00500000271078 | Coxsackievirus And Adenovirus Receptor Precursor |
| ENSMUSG00000022883 | ENSFM00740001589121 | Roundabout Homolog Precursor |
| ENSMUSG00000022884 | ENSFM00730001521378 | Atp Dependent Rna Helicase EIF4A EC_3.6.4.13 Eukaryotic Initiation Factor 4A Eif 4A Translation Initiation Factor 1 |
| ENSMUSG00000022889 | ENSFM00250000006573 | 39S Ribosomal L39 Mitochondrial Mrp L39 |
| ENSMUSG00000022890 | ENSFM00670001236072 | Atp Synthase Coupling Factor 6 Mitochondrial Precursor Atpase Subunit F6 |
| ENSMUSG00000022892 | ENSFM00250000000617 | Amyloid Beta A4 Precursor Abpp App Alzheimer Disease Amyloid A4 Homolog |
| ENSMUSG00000022897 | ENSFM00730001521969 | Dual Specificity Tyrosine Phosphorylation Regulated Kinase Dual Specificity YAK1 Related Kinase Minibrain |
| ENSMUSG00000022956 | ENSFM00570000882034 | Unknown |
| ENSMUSG00000022956 | ENSFM00250000006301 | Downstream Neighbor Of Son |
| ENSMUSG00000022956 | ENSFM00670001235592 | Atp Synthase Subunit O Mitochondrial Precursor Oligomycin Sensitivity Conferral Oscp |
| ENSMUSG00000022973 | ENSFM00730001521871 | Synaptojanin EC_3.1.3.36 Synaptic Inositol 1 4 5 Trisphosphate 5 Phosphatase |
| ENSMUSG00000022982 | ENSFM00670001235331 | Superoxide Dismutase EC_1.15.1.1 |
| ENSMUSG00000022994 | ENSFM00730001521247 | Adenylate Cyclase Type EC_4.6.1.1 Atp Pyrophosphate Lyase Adenylate Cyclase Type Adenylyl Cyclase Ca 2+ Adenylyl Cyclase |
| ENSMUSG00000022995 | ENSFM00740001589204 | Ena/Vasp Ena/Vasodilator Stimulated Phosphoprotein |
| ENSMUSG00000023011 | ENSFM00500000269895 | Lifeguard 2 Fas Apoptotic Inhibitory Molecule 2 |
| ENSMUSG00000023017 | ENSFM00250000000608 | Acid Sensing Ion Channel Amiloride Sensitive Cation Channel |
| ENSMUSG00000023019 | ENSFM00800002003402 | Glycerol 3 Phosphate Dehydrogenase Cytoplasmic Gpd C Gpdh C EC_1.1.1.8 |
| ENSMUSG00000023020 | ENSFM00500000441205 | Unknown |
| ENSMUSG00000023022 | ENSFM00500000271499 | Lim Domain And Actin Binding 1 Epithelial Lost In Neoplasm |
| ENSMUSG00000023026 | ENSFM00250000000652 | Disco Interacting 2 Homolog DIP2 Homolog |
| ENSMUSG00000023032 | ENSFM00590000918402 | Unknown |
| ENSMUSG00000023032 | ENSFM00730001521473 | Sodium Bicarbonate Solute Carrier Family 4 Member |
| ENSMUSG00000023033 | ENSFM00550000769437 | Unknown |
| ENSMUSG00000023033 | ENSFM00740001589070 | Sodium Channel Type Subunit Alpha Sodium Channel Sodium Channel Type Subunit Alpha Voltage Gated Sodium Channel Subunit Alpha NAV1 |
| ENSMUSG00000023055 | ENSFM00500000271639 | Calcium Binding And Coiled Coil Domain Containing 1 |
| ENSMUSG00000023064 | ENSFM00670001236738 | Gamma Synuclein |
| ENSMUSG00000023089 | ENSFM00520000548784 | Unknown |
| ENSMUSG00000023089 | ENSFM00670001236297 | Nadh Dehydrogenase 1 Alpha Subcomplex Subunit 5 Complex I Subunit B13 Complex I 13KD B Ci 13KD B Nadh Ubiquinone Oxidoreductase 13 Kda B Subunit |
| ENSMUSG00000023110 | ENSFM00720001500980 | Unknown |
| ENSMUSG00000023110 | ENSFM00810002040716 | Arginine N Methyltransferase 5 EC_2.1.1.- Histone Arginine N Methyltransferase PRMT5 EC_2.1.1.125 SHK1 Kinase Binding 1 Homolog SKB1 Homolog |
| ENSMUSG00000023175 | ENSFM00610000952905 | Basigin Precursor CD147 Antigen |
| ENSMUSG00000023186 | ENSFM00570000893999 | Unknown |
| ENSMUSG00000023186 | ENSFM00260000050708 | Von Willebrand Factor A Domain Containing 5A Loss Of Heterozygosity 11 Chromosomal Region 2 Gene A |
| ENSMUSG00000023192 | ENSFM00790001963004 | Metabotropic Glutamate Receptor Precursor |
| ENSMUSG00000023216 | ENSFM00730001521321 | Glutamine Gamma Glutamyltransferase EC_2.3.2.13 Transglutaminase Tg Tgase Transglutaminase Tgase |
| ENSMUSG00000023262 | ENSFM00760001763418 | Unknown |
| ENSMUSG00000023262 | ENSFM00760001763419 | Unknown |
| ENSMUSG00000023262 | ENSFM00250000002657 | Aminoacylase Acy EC_3.5.1.14 N Acyl L Amino Acid Amidohydrolase |
| ENSMUSG00000023266 | ENSFM00570000896626 | Unknown |
| ENSMUSG00000023266 | ENSFM00250000003196 | Fibroblast Growth Factor Receptor Substrate 3 Fgfr Substrate 3 Fgfr Signaling Adaptor SNT2 SUC1 Associated Neurotrophic Factor Target 2 Snt 2 |
| ENSMUSG00000023277 | ENSFM00760001763416 | Unknown |
| ENSMUSG00000023277 | ENSFM00760001763417 | Unknown |
| ENSMUSG00000023277 | ENSFM00500000270278 | Twinfilin |
| ENSMUSG00000023307 | ENSFM00250000003363 | E3 Ubiquitin Ligase MARCH5 EC_6.3.2.- Membrane Associated Ring Finger 5 Membrane Associated Ring Ch V March V |
| ENSMUSG00000023328 | ENSFM00570000891640 | Unknown |
| ENSMUSG00000023328 | ENSFM00500000269646 | Acetylcholinesterase Precursor Ache EC_3.1.1.7 |
| ENSMUSG00000023353 | ENSFM00250000000604 | Arf Gap With Gtpase Ank Repeat And Ph Domain Containing Agap Centaurin Gamma |
| ENSMUSG00000023439 | ENSFM00250000000545 | Guanine Nucleotide Binding G I /G S /G T Subunit Beta Transducin Beta Chain |
| ENSMUSG00000023452 | ENSFM00570000893700 | Unknown |
| ENSMUSG00000023452 | ENSFM00800002023580 | Unknown |
| ENSMUSG00000023452 | ENSFM00600000921219 | Phosphatidylserine Decarboxylase Proenzyme EC_4.1.1.65 |
| ENSMUSG00000023456 | ENSFM00250000000728 | Triosephosphate Isomerase Tim EC_5.3.1.1 Triose Phosphate Isomerase |
| ENSMUSG00000023460 | ENSFM00720001492078 | Ras Related Rab 12 |
| ENSMUSG00000023484 | ENSFM00730001521291 | Vimentin |
| ENSMUSG00000023723 | ENSFM00500000272974 | 28S Ribosomal S23 Mitochondrial Mrp S23 S23MT |
| ENSMUSG00000023827 | ENSFM00250000002628 | 1 Acyl Sn Glycerol 3 Phosphate Acyltransferase Delta EC_2.3.1.51 1 Acylglycerol 3 Phosphate O Acyltransferase 4 1 Agp Acyltransferase 4 1 Agpat 4 Lysophosphatidic Acid Acyltransferase Delta Lpaat Delta |
| ENSMUSG00000023827 | ENSFM00500000294792 | 1 3 Phosphate Acyltransferase |
| ENSMUSG00000023830 | ENSFM00590000917226 | Unknown |
| ENSMUSG00000023830 | ENSFM00590000918426 | Unknown |
| ENSMUSG00000023830 | ENSFM00740001589412 | Cation Independent Mannose 6 Phosphate Receptor Precursor Ci Man 6 P Receptor Ci Mpr M6PR 300 Kda Mannose 6 Phosphate Receptor Mpr 300 Insulin Growth Factor 2 Receptor Insulin Growth Factor Ii Receptor Igf Ii Receptor M6P/IGF2 Receptor M6P/IGF2R CD222 A |
| ENSMUSG00000023832 | ENSFM00760001714915 | Acetyl Coa Acetyltransferase Cytosolic EC_2.3.1.9 Cytosolic Acetoacetyl Coa Thiolase |
| ENSMUSG00000023845 | ENSFM00740001589214 | Endoplasmic Reticulum Aminopeptidase EC_3.4.11.- Derived Aminopeptidase |
| ENSMUSG00000023861 | ENSFM00670001236064 | Mitochondrial Pyruvate Carrier 1 Brain 44 |
| ENSMUSG00000023868 | ENSFM00500000441667 | Unknown |
| ENSMUSG00000023868 | ENSFM00800002003845 | Camp And Camp Inhibited Cgmp 3' 5' Cyclic Phosphodiesterase 10A EC_3.1.4.17 EC_3.1.4.- 35 |
| ENSMUSG00000023912 | ENSFM00500000270868 | Mitochondrial Uncoupling 4 Ucp 4 Solute Carrier Family 25 Member 27 |
| ENSMUSG00000023915 | ENSFM00440000236917 | Tumor Necrosis Factor Receptor Superfamily Member 21 Precursor Death Receptor 6 CD358 Antigen |
| ENSMUSG00000023921 | ENSFM00250000004237 | Methylmalonyl Coa Mutase Mitochondrial Precursor Mcm EC_5.4.99.2 Methylmalonyl Coa Isomerase |
| ENSMUSG00000023932 | ENSFM00250000005324 | Pre Splicing Factor CEF1 |
| ENSMUSG00000023938 | ENSFM00250000001208 | Alanine Trna Ligase |
| ENSMUSG00000023939 | ENSFM00500000273213 | 39S Ribosomal L14 Mitochondrial Precursor L14MT Mrp L14 39S Ribosomal L32 Mitochondrial L32MT Mrp L32 |
| ENSMUSG00000023942 | ENSFM00610000970016 | Unknown |
| ENSMUSG00000023942 | ENSFM00610000973261 | Unknown |
| ENSMUSG00000023942 | ENSFM00610000973449 | Unknown |
| ENSMUSG00000023942 | ENSFM00500000270018 | Equilibrative Nucleoside Transporter Equilibrative Nitrobenzylmercaptopurine Riboside Nucleoside Transporter Equilibrative Nbmpr Nucleoside Transporter Nucleoside Transporter Type Solute Carrier Family 29 Member |
| ENSMUSG00000023944 | ENSFM00570000899544 | Unknown |
| ENSMUSG00000023944 | ENSFM00250000000292 | Heat Shock |
| ENSMUSG00000023960 | ENSFM00570000888029 | Unknown |
| ENSMUSG00000023960 | ENSFM00760001714771 | Bis 5' Adenosyl Triphosphatase ENPP4 Precursor EC_3.6.1.29 AP3A Hydrolase AP3AASE Ectonucleotide Pyrophosphatase/Phosphodiesterase Family Member 4 E Npp 4 Npp 4 |
| ENSMUSG00000023972 | ENSFM00260000050584 | Tyrosine Kinase Otk |
| ENSMUSG00000023973 | ENSFM00570000882944 | Unknown |
| ENSMUSG00000023973 | ENSFM00670001235696 | Canopy Homolog Precursor |
| ENSMUSG00000024008 | ENSFM00730001521728 | Copine Copine |
| ENSMUSG00000024012 | ENSFM00250000001870 | Mitochondrial Carrier Homolog |
| ENSMUSG00000024026 | ENSFM00500000271437 | Lactoylglutathione Lyase EC_4.4.1.5 Aldoketomutase Glyoxalase I Glx I Ketone Aldehyde Mutase Methylglyoxalase S D Lactoylglutathione Methylglyoxal Lyase |
| ENSMUSG00000024038 | ENSFM00760001752576 | Uncharacterized |
| ENSMUSG00000024038 | ENSFM00500000273812 | Nadh Dehydrogenase Flavoprotein 3 Mitochondrial |
| ENSMUSG00000024038 | ENSFM00500000301275 | Nadh Dehydrogenase Flavoprotein 3 Mitochondrial Precursor Complex I 9KD Ci 9KD Nadh Ubiquinone Oxidoreductase 9 Kda Subunit |
| ENSMUSG00000024044 | ENSFM00800002003400 | Unknown |
| ENSMUSG00000024065 | ENSFM00250000000766 | Eh Domain Containing |
| ENSMUSG00000024068 | ENSFM00800002003737 | Spastin |
| ENSMUSG00000024077 | ENSFM00250000001473 | Striatin |
| ENSMUSG00000024091 | ENSFM00250000001879 | Vesicle Associated Membrane Associated A Vamp A Vamp Associated A Vap A |
| ENSMUSG00000024096 | ENSFM00250000003792 | Rala Binding 1 RALBP1 Dinitrophenyl S Glutathione Atpase Dnp Sg Atpase Ral Interacting 1 |
| ENSMUSG00000024099 | ENSFM00570000901685 | Unknown |
| ENSMUSG00000024099 | ENSFM00700001404061 | Unknown |
| ENSMUSG00000024099 | ENSFM00250000003401 | Nadh Dehydrogenase Flavoprotein 2 Mitochondrial Precursor EC_1.6.5.3 EC_1.6.99.- 3 Nadh Ubiquinone Oxidoreductase 24 Kda Subunit |
| ENSMUSG00000024101 | ENSFM00250000002535 | Was Family Homolog |
| ENSMUSG00000024109 | ENSFM00740001589096 | Neurexin Precursor Neurexin |
| ENSMUSG00000024120 | ENSFM00250000002838 | Leucine Rich Ppr Motif Containing Protein Mitochondrial Precursor 130 Kda Leucine Rich Lrp 130 |
| ENSMUSG00000024121 | ENSFM00250000002076 | V Type Proton Atpase 16 Kda Proteolipid Subunit V Atpase 16 Kda Proteolipid Subunit Vacuolar Proton Pump 16 Kda Proteolipid Subunit |
| ENSMUSG00000024121 | ENSFM00570000891623 | V Type Proton Atpase 16 Kda Proteolipid Subunit |
| ENSMUSG00000024122 | ENSFM00570000891895 | Unknown |
| ENSMUSG00000024122 | ENSFM00800002003539 | 3 Phosphoinositide Dependent Kinase EC_2.7.11.1 |
| ENSMUSG00000024130 | ENSFM00740001589107 | Atp Binding Cassette Sub Family A Member 3 |
| ENSMUSG00000024132 | ENSFM00670001235679 | Enoyl Coa Delta Isomerase 1 Mitochondrial Precursor EC_5.3.3.8 3 2 Trans Enoyl Coa Isomerase Delta 3 Delta 2 Enoyl Coa Isomerase D3 D2 Enoyl Coa Isomerase Dodecenoyl Coa Isomerase |
| ENSMUSG00000024143 | ENSFM00730001521235 | Cell Division Control 42 Homolog Precursor |
| ENSMUSG00000024158 | ENSFM00730001521820 | Hydroxyacylglutathione Hydrolase Mitochondrial Precursor EC_3.1.2.6 Glyoxalase Ii Glx Ii |
| ENSMUSG00000024163 | ENSFM00250000000947 | C Jun Amino Terminal Kinase Interacting Jip Jnk Interacting Jnk Mitogen Activated Kinase 8 Interacting |
| ENSMUSG00000024187 | ENSFM00250000006994 | ITFG3 |
| ENSMUSG00000024191 | ENSFM00570000877984 | Unknown |
| ENSMUSG00000024191 | ENSFM00570000901053 | Unknown |
| ENSMUSG00000024191 | ENSFM00250000003982 | Vesicle Transport SEC20 |
| ENSMUSG00000024208 | ENSFM00670001236187 | Ubiquinol Cytochrome C Reductase Complex Assembly Factor 2 Precursor Mitochondrial Nucleoid Factor 1 Mitochondrial M19 |
| ENSMUSG00000024211 | ENSFM00570000893452 | Unknown |
| ENSMUSG00000024211 | ENSFM00250000000328 | Metabotropic Glutamate Receptor Precursor |
| ENSMUSG00000024213 | ENSFM00660001168570 | Unknown |
| ENSMUSG00000024213 | ENSFM00250000002183 | Diphosphoinositol Polyphosphate Phosphohydrolase Dipp EC_3.6.1.52 Diadenosine 5' 5''' P1 P6 Hexaphosphate Hydrolase EC_3.6.1.- Nucleoside Diphosphate Linked Moiety X Motif Nudix Motif |
| ENSMUSG00000024234 | ENSFM00250000005018 | Poly A Rna Polymerase Mitochondrial Precursor Pap EC_2.7.7.19 Pap Associated Domain Containing 1 Polynucleotide Adenylyltransferase |
| ENSMUSG00000024248 | ENSFM00500000274370 | Cytochrome C Oxidase Subunit 7A Related Protein Mitochondrial Precursor Cytochrome C Oxidase Subunit Viia Related |
| ENSMUSG00000024259 | ENSFM00250000005083 | Solute Carrier Family 25 Member 46 |
| ENSMUSG00000024261 | ENSFM00500000270270 | Synaptotagmin Synaptotagmin |
| ENSMUSG00000024270 | ENSFM00570000883974 | Unknown |
| ENSMUSG00000024270 | ENSFM00500000270550 | Zinc Transporter ZIP6 Precursor Solute Carrier Family 39 Member 6 Zrt And Irt 6 Zip 6 |
| ENSMUSG00000024277 | ENSFM00610000970007 | Unknown |
| ENSMUSG00000024277 | ENSFM00250000000867 | Microtubule Associated Rp/Eb Family Member |
| ENSMUSG00000024286 | ENSFM00250000001383 | Cyclin Y |
| ENSMUSG00000024300 | ENSFM00780001898391 | Unconventional Myosin Ie Unconventional Myosin 1E |
| ENSMUSG00000024302 | ENSFM00250000000811 | Dystrobrevin Beta Dtn B Beta Dystrobrevin |
| ENSMUSG00000024304 | ENSFM00730001521172 | Cadherin Precursor Cadherin Cadherin |
| ENSMUSG00000024309 | ENSFM00500000273478 | Prefoldin Subunit 6 |
| ENSMUSG00000024319 | ENSFM00500000271174 | Vacuolar Sorting Associated 52 Homolog SAC2 Suppressor Of Actin Mutations 2 |
| ENSMUSG00000024327 | ENSFM00500000271041 | Zinc Transporter Sl Histidine Rich Membrane KE4 Solute Carrier Family 39 Member 7 Zrt Irt 7 ZIP7 |
| ENSMUSG00000024347 | ENSFM00250000001069 | Ph And SEC7 Domain Containing 1 Exchange Factor ARF6 Pleckstrin Homology And SEC7 Domain Containing 1 |
| ENSMUSG00000024350 | ENSFM00250000001153 | Dnaj Homolog Subfamily Member |
| ENSMUSG00000024359 | ENSFM00730001521528 | Unknown |
| ENSMUSG00000024360 | ENSFM00680001326074 | Unknown |
| ENSMUSG00000024360 | ENSFM00250000005309 | Eukaryotic Peptide Chain Release Factor Subunit 1 Eukaryotic Release Factor 1 ERF1 |
| ENSMUSG00000024381 | ENSFM00250000000861 | Bridging Integrator |
| ENSMUSG00000024387 | ENSFM00500000464452 | Unknown |
| ENSMUSG00000024387 | ENSFM00800002003472 | Casein Kinase Ii Subunit Beta Ck Ii Beta |
| ENSMUSG00000024392 | ENSFM00250000003234 | Large Proline Rich BAG6 BCL2 Associated Athanogene 6 Hla B Associated Transcript 3 |
| ENSMUSG00000024393 | ENSFM00740001589307 | PRRC2A Hla B Associated Transcript 2 Proline Rich And Coiled Coil Containing 2A |
| ENSMUSG00000024403 | ENSFM00640001109543 | Unknown |
| ENSMUSG00000024403 | ENSFM00600000921884 | V Type Proton Atpase Subunit G 2 V Atpase Subunit G 2 Vacuolar Proton Pump Subunit G 2 |
| ENSMUSG00000024410 | ENSFM00250000005107 | Uncharacterized Colon Cancer Associated MIC1 Mic 1 |
| ENSMUSG00000024411 | ENSFM00730001522004 | Aquaporin |
| ENSMUSG00000024413 | ENSFM00780001898362 | Niemann Pick C1 Precursor |
| ENSMUSG00000024423 | ENSFM00500000271599 | Impact Imprinted And Ancient Gene Homolog |
| ENSMUSG00000024426 | ENSFM00570000879961 | Unknown |
| ENSMUSG00000024426 | ENSFM00250000002340 | Alpha Tubulin N Acetyltransferase 1 |
| ENSMUSG00000024429 | ENSFM00250000004865 | Guanine Nucleotide Binding 1 |
| ENSMUSG00000024462 | ENSFM00250000000928 | Gamma Aminobutyric Acid Type B Receptor Subunit 2 Precursor Gaba B Receptor 2 Gaba B R2 Gaba BR2 GABABR2 GB2 G Coupled Receptor 51 |
| ENSMUSG00000024479 | ENSFM00500000273729 | MAL2 |
| ENSMUSG00000024480 | ENSFM00500000270528 | Ap 3 Complex Subunit Sigma Ap 3 Complex Subunit Sigma Adaptor Related Complex 3 Subunit Sigma Sigma Adaptin Adaptin Sigma Adaptin |
| ENSMUSG00000024487 | ENSFM00250000003967 | YIPF5 YIP1 Family Member 5 |
| ENSMUSG00000024493 | ENSFM00250000002483 | Leucine Trna Ligase Cytoplasmic EC_6.1.1.4 Leucyl Trna Synthetase Leurs |
| ENSMUSG00000024501 | ENSFM00250000000643 | Dihydropyrimidinase Related Drp Collapsin Response Mediator Crmp |
| ENSMUSG00000024502 | ENSFM00250000001480 | Janus Kinase And Microtubule Interacting 1 Multiple Alpha Helices And Rna Linker 1 Marlin 1 |
| ENSMUSG00000024507 | ENSFM00500000270110 | Peroxisomal Multifunctional Enzyme Type 2 Mfe 2 17 Beta Hydroxysteroid Dehydrogenase 4 17 Beta Hsd 4 D Bifunctional Dbp Multifunctional 2 Mpf 2 |
| ENSMUSG00000024511 | ENSFM00730001521880 | Ras Related Rab |
| ENSMUSG00000024516 | ENSFM00790001962996 | Signal Peptidase Complex Catalytic Subunit SEC11A EC_3.4.21.89 Endopeptidase SP18 Microsomal Signal Peptidase 18 Kda Subunit Spase 18 Kda Subunit SEC11 Homolog A SEC11 1 SPC18 |
| ENSMUSG00000024524 | ENSFM00300000079126 | Guanine Nucleotide Binding G S Subunit Alpha Adenylate Cyclase Stimulating G Alpha |
| ENSMUSG00000024527 | ENSFM00800002003441 | AFG3 EC_3.4.24.- |
| ENSMUSG00000024533 | ENSFM00250000001244 | Spire Homolog |
| ENSMUSG00000024556 | ENSFM00250000000979 | Nadp Dependent Malic Enzyme Nadp Me EC_1.1.1.40 Malic Enzyme |
| ENSMUSG00000024566 | ENSFM00810002040637 | Probable Phospholipid Transporting Atpase EC_3.6.3.1 Atpase Class Ii Type |
| ENSMUSG00000024576 | ENSFM00770001847535 | Casein Kinase I Alpha Cki Alpha EC_2.7.11.1 CK1 |
| ENSMUSG00000024579 | ENSFM00780001901924 | Unknown |
| ENSMUSG00000024579 | ENSFM00250000003214 | Prenylcysteine Oxidase Precursor |
| ENSMUSG00000024581 | ENSFM00250000005767 | Gamma Soluble Nsf Attachment Snap Gamma N Ethylmaleimide Sensitive Factor Attachment Gamma |
| ENSMUSG00000024583 | ENSFM00500000271170 | Thioredoxin Thioredoxin Related |
| ENSMUSG00000024587 | ENSFM00500000270122 | Asparagine Trna Ligase Cytoplasmic EC_6.1.1.22 Asparaginyl Trna Synthetase Asnrs |
| ENSMUSG00000024588 | ENSFM00250000003660 | Ferrochelatase Mitochondrial Precursor EC_4.99.1.1 Heme Synthase Protoheme Ferro Lyase |
| ENSMUSG00000024589 | ENSFM00730001521822 | E3 Ubiquitin Ligase EC_6.3.2.- |
| ENSMUSG00000024590 | ENSFM00250000000425 | Lamin |
| ENSMUSG00000024597 | ENSFM00260000050342 | Solute Carrier Family 12 Member Bumetanide Sensitive Sodium Potassium Chloride Cotransporter Na K Cl Symporter |
| ENSMUSG00000024603 | ENSFM00250000004748 | Dynactin Subunit 4 Dynactin Subunit P62 |
| ENSMUSG00000024608 | ENSFM00250000003648 | 40S Ribosomal S14 |
| ENSMUSG00000024614 | ENSFM00250000006330 | Disulfide Isomerase TMX3 Precursor EC_5.3.4.1 Thioredoxin Domain Containing 10 Thioredoxin Related Transmembrane 3 |
| ENSMUSG00000024617 | ENSFM00500000347513 | Unknown |
| ENSMUSG00000024617 | ENSFM00250000000111 | Calcium/Calmodulin Dependent Kinase Type Ii Subunit Cam Kinase Ii Subunit Camk Ii Subunit EC_2.7.11.17 |
| ENSMUSG00000024620 | ENSFM00730001521867 | Platelet Derived Growth Factor Receptor Precursor Pdgf R Pdgfr EC_2.7.10.1 Platelet Derived Growth Factor Receptor Type Platelet Derived Growth Factor Receptor CD140 Antigen Family Member Platelet Derived Growth Factor Receptor Antigen |
| ENSMUSG00000024639 | ENSFM00730001521471 | Guanine Nucleotide Binding G Q Subunit Alpha Guanine Nucleotide Binding Alpha Q |
| ENSMUSG00000024640 | ENSFM00790001963003 | Phosphoserine Aminotransferase Psat EC_2.6.1.52 Endometrial Progesterone Induced Epip Phosphohydroxythreonine Aminotransferase |
| ENSMUSG00000024644 | ENSFM00800002003448 | Cytosolic Non Specific Dipeptidase EC_3.4.13.18 Cndp Dipeptidase 2 |
| ENSMUSG00000024646 | ENSFM00500000271733 | Cytochrome B5 |
| ENSMUSG00000024654 | ENSFM00500000271079 | Isoaspartyl Peptidase/L Asparaginase Precursor EC_3.4.19.5 EC_3.5.1.- 1 Asparaginase 1 Beta Aspartyl Peptidase Isoaspartyl Dipeptidase L Asparagine Amidohydrolase |
| ENSMUSG00000024661 | ENSFM00500000269681 | Ferritin Heavy Chain Ferritin H Subunit EC_1.16.3.1 |
| ENSMUSG00000024687 | ENSFM00500000269873 | Oxysterol Binding |
| ENSMUSG00000024736 | ENSFM00250000000808 | Transmembrane Precursor |
| ENSMUSG00000024740 | ENSFM00810002040719 | Dna Damage Binding 1 Damage Specific Dna Binding 1 |
| ENSMUSG00000024743 | ENSFM00740001589375 | Synaptotagmin 7 Synaptotagmin Vii Sytvii |
| ENSMUSG00000024758 | ENSFM00500000269689 | Reticulon |
| ENSMUSG00000024759 | ENSFM00250000001436 | Atlastin EC_3.6.5.- |
| ENSMUSG00000024767 | ENSFM00500000272123 | Ubiquitin Thioesterase OTUB1 EC_3.4.19.12 Deubiquitinating Enzyme OTUB1 Otu Domain Containing Ubiquitin Aldehyde Binding 1 Otubain 1 Ubiquitin Specific Processing Protease OTUB1 |
| ENSMUSG00000024772 | ENSFM00250000000766 | Eh Domain Containing |
| ENSMUSG00000024777 | ENSFM00760001714676 | Serine/Threonine Phosphatase 2A 56 Kda Regulatory Subunit Epsilon PP2A B Subunit B' Epsilon PP2A B Subunit B56 Epsilon PP2A B Subunit PR61 Epsilon PP2A B Subunit R5 Epsilon |
| ENSMUSG00000024782 | ENSFM00570000883900 | Unknown |
| ENSMUSG00000024782 | ENSFM00500000270040 | Adenylate Kinase 4 Mitochondrial |
| ENSMUSG00000024797 | ENSFM00590000917614 | Unknown |
| ENSMUSG00000024797 | ENSFM00250000003232 | Vacuolar Sorting Associated 51 Homolog Fat Free Homolog |
| ENSMUSG00000024812 | ENSFM00250000000691 | Tight Junction Zo Tight Junction Zona Occludens Zonula Occludens |
| ENSMUSG00000024835 | ENSFM00730001521292 | Coronin |
| ENSMUSG00000024844 | ENSFM00670001236579 | Barrier To Autointegration Factor Breakpoint Cluster Region 1 |
| ENSMUSG00000024851 | ENSFM00500000269931 | Membrane Associated Phosphatidylinositol Transfer Phosphatidylinositol Transfer Protein Membrane Associated Pitpnm PYK2 N Terminal Domain Interacting Receptor Nir |
| ENSMUSG00000024855 | ENSFM00250000001313 | Phosphofurin Acidic Cluster Sorting Pacs |
| ENSMUSG00000024858 | ENSFM00610000967883 | Unknown |
| ENSMUSG00000024858 | ENSFM00610000970067 | Unknown |
| ENSMUSG00000024858 | ENSFM00800002003453 | Beta Adrenergic Receptor Kinase 1 Beta Ark 1 EC_2.7.11.15 G Coupled Receptor Kinase 2 |
| ENSMUSG00000024862 | ENSFM00250000000749 | Kinesin Light Chain |
| ENSMUSG00000024870 | ENSFM00730001521238 | Ras Related Rab |
| ENSMUSG00000024883 | ENSFM00740001589251 | Ras And Rab Interactor Ras Interaction/Interference |
| ENSMUSG00000024885 | ENSFM00250000000672 | Aldehyde Dehydrogenase Aldehyde Dehydrogenase Aldehyde Dehydrogenase Family 3 Member |
| ENSMUSG00000024887 | ENSFM00810002040830 | Neutral Ceramidase N Cdase Ncdase EC_3.5.1.23 Acylsphingosine Deacylase 2 N Acylsphingosine Amidohydrolase 2 |
| ENSMUSG00000024891 | ENSFM00500000270018 | Equilibrative Nucleoside Transporter Equilibrative Nitrobenzylmercaptopurine Riboside Nucleoside Transporter Equilibrative Nbmpr Nucleoside Transporter Nucleoside Transporter Type Solute Carrier Family 29 Member |
| ENSMUSG00000024892 | ENSFM00720001492032 | Pyruvate Carboxylase EC_6.4.1.1 Pyruvic Carboxylase Pcb |
| ENSMUSG00000024900 | ENSFM00500000269758 | Carnitine O Palmitoyltransferase 1 Muscle CPT1 M EC_2.3.1.21 Carnitine O Palmitoyltransferase I Muscle Cpt I Cpti M Carnitine Palmitoyltransferase 1B |
| ENSMUSG00000024902 | ENSFM00500000272206 | 39S Ribosomal L11 Mitochondrial Precursor L11MT Mrp L11 |
| ENSMUSG00000024911 | ENSFM00250000004633 | Acidic Fibroblast Growth Factor Intracellular Binding Afgf Intracellular Binding Fgf 1 Intracellular Binding |
| ENSMUSG00000024924 | ENSFM00740001589117 | Low Density Lipoprotein Receptor Receptor |
| ENSMUSG00000024935 | ENSFM00800002003375 | Excitatory Amino Acid Transporter Sodium Dependent Glutamate/Aspartate Transporter Solute Carrier Family 1 Member |
| ENSMUSG00000024953 | ENSFM00670001235781 | Peroxiredoxin 5 Mitochondrial Precursor EC_1.11.1.15 Antioxidant Enzyme B166 AOEB166 Plp Peroxiredoxin V Prx V Peroxisomal Antioxidant Enzyme Thioredoxin Peroxidase PMP20 Thioredoxin Reductase |
| ENSMUSG00000024960 | ENSFM00800002003424 | 1 Phosphatidylinositol 4 5 Bisphosphate Phosphodiesterase Beta EC_3.1.4.11 Phosphoinositide Phospholipase C Beta Phospholipase C Beta Plc Beta |
| ENSMUSG00000024966 | ENSFM00810002040902 | Stress Induced Phosphoprotein 1 STI1 HSC70/HSP90 Organizing Hop |
| ENSMUSG00000024969 | ENSFM00610000972490 | Unknown |
| ENSMUSG00000024969 | ENSFM00610000972491 | Unknown |
| ENSMUSG00000024969 | ENSFM00730001521220 | Kinase EC_2.7.11.1 |
| ENSMUSG00000024974 | ENSFM00310000089021 | Structural Maintenance Of Chromosomes 3 Smc 3 Smc 3 Chondroitin Sulfate Proteoglycan 6 |
| ENSMUSG00000024976 | ENSFM00500000270686 | Leucine Rich Repeat Soc 2 Homolog Sur 8 Homolog Soc 2 Homolog |
| ENSMUSG00000024981 | ENSFM00500000269666 | Long Chain Fatty Acid Coa Ligase EC_6.2.1.3 Long Chain Acyl Coa Synthetase Lacs |
| ENSMUSG00000024983 | ENSFM00500000272468 | Vesicle Transport Through Interaction With T Snares Homolog 1A Vesicle Transport V Snare VTI1 2 VTI1 RP2 |
| ENSMUSG00000024991 | ENSFM00800002003567 | Eukaryotic Translation Initiation Factor 3 Subunit A |
| ENSMUSG00000024997 | ENSFM00250000000757 | Thioredoxin Peroxidase Thioredoxin Dependent Peroxide Reductase |
| ENSMUSG00000025006 | ENSFM00670001235644 | Sorbin And SH3 Domain Containing 1 Ponsin SH3 Domain 5 S C Cbl Associated Cap |
| ENSMUSG00000025007 | ENSFM00660001173763 | Unknown |
| ENSMUSG00000025007 | ENSFM00660001182007 | Unknown |
| ENSMUSG00000025007 | ENSFM00760001714924 | Delta 1 Pyrroline 5 Carboxylate Synthase P5CS Aldehyde Dehydrogenase Family 18 Member A1 |
| ENSMUSG00000025016 | ENSFM00730001521961 | Transmembrane 9 Superfamily Member Precursor |
| ENSMUSG00000025020 | ENSFM00250000000421 | Slit Homolog Precursor Slit |
| ENSMUSG00000025026 | ENSFM00250000000681 | Adducin |
| ENSMUSG00000025027 | ENSFM00720001503450 | Unknown |
| ENSMUSG00000025027 | ENSFM00800002003425 | Xaa Pro Aminopeptidase 1 EC_3.4.11.9 Aminoacylproline Aminopeptidase Cytosolic Aminopeptidase P Soluble Aminopeptidase P Samp X Pro Aminopeptidase 1 X Prolyl Aminopeptidase 1 Soluble |
| ENSMUSG00000025037 | ENSFM00250000002249 | Amine Oxidase EC_1.4.3.4 Monoamine Oxidase Type Mao |
| ENSMUSG00000025040 | ENSFM00670001235667 | FUN14 Domain Containing |
| ENSMUSG00000025059 | ENSFM00780001898348 | Glycerol Kinase Gk Glycerokinase EC_2.7.1.30 Atp:Glycerol 3 Phosphotransferase Glycerol Kinase Testis Specific |
| ENSMUSG00000025060 | ENSFM00250000000882 | Serine/Threonine Kinase EC_2.7.11.1 |
| ENSMUSG00000025085 | ENSFM00250000000459 | Actin Binding Lim Ablim Actin Binding Lim Family Member |
| ENSMUSG00000025092 | ENSFM00250000005151 | Unknown |
| ENSMUSG00000025094 | ENSFM00250000000980 | Synaptic Vesicular Amine Transporter Monoamine Transporter Solute Carrier Family 18 Member 2 Vesicular Amine Transporter 2 VAT2 |
| ENSMUSG00000025130 | ENSFM00730001521641 | Disulfide Isomerase Precursor Pdi EC_5.3.4.1 Prolyl 4 Hydroxylase Subunit Beta |
| ENSMUSG00000025132 | ENSFM00250000002426 | Rho Gdp Dissociation Inhibitor Rho Gdi Rho Gdi |
| ENSMUSG00000025137 | ENSFM00780001898417 | Ethanolamine Phosphate Cytidylyltransferase EC_2.7.7.14 Ctp:Phosphoethanolamine Cytidylyltransferase Phosphorylethanolamine Transferase |
| ENSMUSG00000025139 | ENSFM00250000004428 | Toll Interacting |
| ENSMUSG00000025151 | ENSFM00500000269827 | Melanoma Associated Antigen Mage Antigen |
| ENSMUSG00000025153 | ENSFM00510000502777 | Fatty Acid Synthase EC_2.3.1.85 S Acetyltransferase EC_2.3.1.- 38 |
| ENSMUSG00000025156 | ENSFM00570000893319 | Unknown |
| ENSMUSG00000025156 | ENSFM00250000004184 | COP9 Signalosome Complex Subunit 1 SGN1 Signalosome Subunit 1 G Pathway Suppressor 1 Gps 1 JAB1 Containing Signalosome Subunit 1 |
| ENSMUSG00000025162 | ENSFM00570000883886 | Unknown |
| ENSMUSG00000025162 | ENSFM00800002003414 | Casein Kinase I Delta Cki Delta Ckid EC_2.7.11.1 |
| ENSMUSG00000025173 | ENSFM00250000002405 | Wd Repeat Domain Phosphoinositide Interacting 3 Wipi 3 Wd Repeat Containing 45 WDR45 Wd Repeat Containing 45B |
| ENSMUSG00000025178 | ENSFM00250000002398 | Phosphatidylinositol 4 Kinase Type 2 EC_2.7.1.67 Phosphatidylinositol 4 Kinase Type Ii |
| ENSMUSG00000025189 | ENSFM00250000000938 | Metal Transporter Ancient Conserved Domain Containing Cyclin |
| ENSMUSG00000025190 | ENSFM00780001898358 | Aspartate Aminotransferase Cytoplasmic Caspat EC_2.6.1.1 EC_2.6.1.- 3 Cysteine Aminotransferase Cytoplasmic Cysteine Transaminase Cytoplasmic Ccat Glutamate Oxaloacetate Transaminase 1 Transaminase A |
| ENSMUSG00000025198 | ENSFM00610000961458 | Unknown |
| ENSMUSG00000025198 | ENSFM00610000961459 | Unknown |
| ENSMUSG00000025198 | ENSFM00610000972581 | Unknown |
| ENSMUSG00000025198 | ENSFM00250000003616 | Erlin 2 Endoplasmic Reticulum Lipid Raft Associated 2 Stomatin Prohibitin Flotillin Hflc/K Domain Containing 2 Spfh Domain Containing 2 |
| ENSMUSG00000025204 | ENSFM00610000972583 | Unknown |
| ENSMUSG00000025204 | ENSFM00250000006874 | Nadh Dehydrogenase 1 Beta Subcomplex Subunit 8 Mitochondrial Precursor Complex I Ashi Ci Ashi Nadh Ubiquinone Oxidoreductase Ashi Subunit |
| ENSMUSG00000025208 | ENSFM00250000003611 | 39S Ribosomal L43 Mitochondrial Precursor L43MT Mrp L43 Mitochondrial Ribosomal BMRP36A |
| ENSMUSG00000025212 | ENSFM00730001521561 | Sideroflexin |
| ENSMUSG00000025228 | ENSFM00730001521541 | Actin |
| ENSMUSG00000025232 | ENSFM00730001521836 | Beta Hexosaminidase Subunit Alpha Precursor EC_3.2.1.52 Beta N Acetylhexosaminidase Subunit Alpha Hexosaminidase Subunit A N Acetyl Beta Glucosaminidase Subunit Alpha |
| ENSMUSG00000025236 | ENSFM00250000002925 | Adp Dependent Glucokinase Precursor Adp Gk Adpgk EC_2.7.1.147 |
| ENSMUSG00000025240 | ENSFM00770001847625 | Phosphatidylinositide Phosphatase SAC1 EC_3.1.3.- Suppressor Of Actin Mutations 1 |
| ENSMUSG00000025241 | ENSFM00250000002726 | Fyve And Coiled Coil Domain Containing 1 |
| ENSMUSG00000025257 | ENSFM00500000273611 | RIB43A With Coiled Coils 1 |
| ENSMUSG00000025260 | ENSFM00500000270523 | 3 Hydroxyacyl Coa Dehydrogenase Type 2 EC_1.1.1.35 17 Beta Hydroxysteroid Dehydrogenase 10 17 Beta Hsd 10 EC_1.1.1.- 51 3 Hydroxy 2 Methylbutyryl Coa Dehydrogenase EC_1.1.-.- 1 178 3 Hydroxyacyl Coa Dehydrogenase Type Ii Endoplasmic Reticulum Associated A |
| ENSMUSG00000025261 | ENSFM00570000886279 | Unknown |
| ENSMUSG00000025261 | ENSFM00570000892564 | Unknown |
| ENSMUSG00000025261 | ENSFM00760001714810 | E3 Ubiquitin Ligase HUWE1 EC_6.3.2.- Hect Uba And Wwe Domain Containing 1 Upstream Regulatory Element Binding 1 Ure B1 Ure Binding 1 |
| ENSMUSG00000025262 | ENSFM00250000002493 | Constitutive Coactivator Of Ppar Gamma 1 Oxidative Stress Associated Src Activator FAM120A |
| ENSMUSG00000025277 | ENSFM00250000005911 | Monoacylglycerol Lipase ABHD6 EC_3.1.1.23 2 Arachidonoylglycerol Hydrolase Abhydrolase Domain Containing 6 |
| ENSMUSG00000025287 | ENSFM00720001503361 | Unknown |
| ENSMUSG00000025287 | ENSFM00250000003102 | Acyl Coenzyme A Thioesterase Mitochondrial Precursor Acyl Coa Thioesterase EC_3.1.2.- Mitochondrial 48 Kda Acyl Coa Thioester Hydrolase Mt ACT48 |
| ENSMUSG00000025289 | ENSFM00250000000757 | Thioredoxin Peroxidase Thioredoxin Dependent Peroxide Reductase |
| ENSMUSG00000025290 | ENSFM00500000270090 | 40S Ribosomal S24 |
| ENSMUSG00000025314 | ENSFM00310000089006 | Receptor Type Tyrosine Phosphatase Precursor Tyrosine Phosphatase R Ptp EC_3.1.3.48 Tyrosine Phosphatase Ptp |
| ENSMUSG00000025318 | ENSFM00250000001356 | Junctophilin Jp Junctophilin |
| ENSMUSG00000025321 | ENSFM00730001521296 | Integrin Beta Precursor |
| ENSMUSG00000025326 | ENSFM00790001977124 | Unknown |
| ENSMUSG00000025326 | ENSFM00810002057405 | Unknown |
| ENSMUSG00000025326 | ENSFM00810002057406 | Unknown |
| ENSMUSG00000025326 | ENSFM00760001714658 | Ubiquitin Ligase E3A EC_6.3.2.- Oncogenic Associated E6 Ap |
| ENSMUSG00000025364 | ENSFM00250000003247 | Proliferation Associated 2G4 |
| ENSMUSG00000025366 | ENSFM00250000000964 | Extended Synaptotagmin E |
| ENSMUSG00000025370 | ENSFM00260000050333 | Cadherin Precursor |
| ENSMUSG00000025371 | ENSFM00570000879244 | Unknown |
| ENSMUSG00000025371 | ENSFM00570000880208 | Unknown |
| ENSMUSG00000025371 | ENSFM00570000900828 | Unknown |
| ENSMUSG00000025371 | ENSFM00670001235835 | Charged Multivesicular Body 6 Chromatin Modifying 6 |
| ENSMUSG00000025372 | ENSFM00250000000906 | Brain Specific Angiogenesis Inhibitor 1 Associated 2 Bai Associated 2 BAI1 Associated 2 Insulin Receptor Substrate Of 53 Kda IRSP53 Insulin Receptor Substrate P53 |
| ENSMUSG00000025375 | ENSFM00670001235536 | Serine/Threonine Kinase EC_2.7.11.1 Apoptosis Associated Tyrosine Kinase Brain Kinase Lemur Tyrosine Kinase |
| ENSMUSG00000025381 | ENSFM00500000272939 | Canopy Homolog 2 Precursor Mir Interacting Saposin Secreted ZSIG9 Transmembrane 4 |
| ENSMUSG00000025393 | ENSFM00790001962974 | Atp Synthase Subunit Beta Mitochondrial EC_3.6.3.14 |
| ENSMUSG00000025403 | ENSFM00730001521489 | Serine Hydroxymethyltransferase Shmt EC_2.1.2.1 Glycine Hydroxymethyltransferase Serine Methylase |
| ENSMUSG00000025410 | ENSFM00250000005336 | Dynactin Subunit 2 |
| ENSMUSG00000025421 | ENSFM00570000899207 | Unknown |
| ENSMUSG00000025421 | ENSFM00500000271192 | Haloacid Dehalogenase Hydrolase Domain Containing 2 |
| ENSMUSG00000025422 | ENSFM00250000000604 | Arf Gap With Gtpase Ank Repeat And Ph Domain Containing Agap Centaurin Gamma |
| ENSMUSG00000025428 | ENSFM00570000888005 | Unknown |
| ENSMUSG00000025428 | ENSFM00810002040641 | Atp Synthase Subunit Alpha Mitochondrial Precursor |
| ENSMUSG00000025453 | ENSFM00250000002458 | Nad P Transhydrogenase Mitochondrial Precursor EC_1.6.1.2 Nicotinamide Nucleotide Transhydrogenase Pyridine Nucleotide Transhydrogenase |
| ENSMUSG00000025465 | ENSFM00500000270475 | Enoyl Coa Hydratase Mitochondrial Precursor EC_4.2.1.17 Enoyl Coa Hydratase 1 Short Chain Enoyl Coa Hydratase Sceh |
| ENSMUSG00000025466 | ENSFM00250000007872 | Fucose Mutarotase EC_5.1.3.29 |
| ENSMUSG00000025474 | ENSFM00800002003795 | Gamma Tubulin Complex Component 2 Gamma Ring Complex Kda |
| ENSMUSG00000025475 | ENSFM00250000001368 | G Coupled Receptor 124 Precursor Tumor Endothelial Marker 5 |
| ENSMUSG00000025477 | ENSFM00250000002520 | Type I Inositol 1 4 5 Trisphosphate 5 Phosphatase 5PTASE EC_3.1.3.56 |
| ENSMUSG00000025478 | ENSFM00250000000643 | Dihydropyrimidinase Related Drp Collapsin Response Mediator Crmp |
| ENSMUSG00000025485 | ENSFM00250000001722 | Synembryn Ric |
| ENSMUSG00000025487 | ENSFM00610000962626 | Unknown |
| ENSMUSG00000025487 | ENSFM00610000962627 | Unknown |
| ENSMUSG00000025487 | ENSFM00780001898464 | 26S Proteasome Non Atpase Regulatory Subunit 13 26S Proteasome Regulatory Subunit RPN9 26S Proteasome Regulatory Subunit S11 26S Proteasome Regulatory Subunit P40 5 |
| ENSMUSG00000025499 | ENSFM00730001521569 | Ras Precursor |
| ENSMUSG00000025503 | ENSFM00810002040751 | Transaldolase EC_2.2.1.2 |
| ENSMUSG00000025508 | ENSFM00600000922276 | 60S Acidic Ribosomal P2 |
| ENSMUSG00000025512 | ENSFM00250000002832 | Chitinase Domain Containing 1 Precursor |
| ENSMUSG00000025521 | ENSFM00250000007004 | Transmembrane 192 |
| ENSMUSG00000025525 | ENSFM00500000271869 | Micos Complex Subunit MIC27 Precursor Apolipoprotein O FAM121A |
| ENSMUSG00000025544 | ENSFM00610000972920 | Unknown |
| ENSMUSG00000025544 | ENSFM00780001898383 | Transmembrane 9 Superfamily Member 2 Precursor |
| ENSMUSG00000025545 | ENSFM00250000006225 | Citrate Lyase Subunit Beta Protein Mitochondrial Precursor Citrate Lyase Beta EC_4.1.-.- |
| ENSMUSG00000025555 | ENSFM00270000056477 | Ferm Rhogef And Pleckstrin Domain Containing |
| ENSMUSG00000025558 | ENSFM00500000269738 | Dedicator Of Cytokinesis CDC42 Guanine Nucleotide Exchange Factor Zizimin |
| ENSMUSG00000025579 | ENSFM00730001521472 | Lysosomal Alpha Glucosidase Precursor EC_3.2.1.20 Acid Maltase |
| ENSMUSG00000025580 | ENSFM00730001521529 | Atp Dependent Rna Helicase |
| ENSMUSG00000025582 | ENSFM00500000269860 | Neuronal Pentraxin 2 Precursor NP2 Neuronal Pentraxin Ii Np Ii |
| ENSMUSG00000025584 | ENSFM00280000058709 | High Affinity Camp Specific And Ibmx Insensitive 3' 5' Cyclic Phosphodiesterase 8B EC_3.1.4.53 Cell Proliferation Inducing Gene 22 |
| ENSMUSG00000025613 | ENSFM00660001189898 | Unknown |
| ENSMUSG00000025613 | ENSFM00800002003473 | T Complex 1 Subunit Theta Tcp 1 Theta Cct Theta |
| ENSMUSG00000025630 | ENSFM00250000001478 | Hypoxanthine Guanine Phosphoribosyltransferase Hgprt Hgprtase EC_2.4.2.8 |
| ENSMUSG00000025645 | ENSFM00250000006098 | Coiled Coil Domain Containing 51 |
| ENSMUSG00000025651 | ENSFM00780001917798 | Unknown |
| ENSMUSG00000025651 | ENSFM00780001917799 | Unknown |
| ENSMUSG00000025651 | ENSFM00780001898353 | Cytochrome B C1 Complex Subunit 1 Mitochondrial Precursor Complex Iii Subunit 1 Core I Ubiquinol Cytochrome C Reductase Complex Core 1 |
| ENSMUSG00000025656 | ENSFM00720001501783 | Unknown |
| ENSMUSG00000025656 | ENSFM00800002023268 | Unknown |
| ENSMUSG00000025656 | ENSFM00800002023269 | Unknown |
| ENSMUSG00000025656 | ENSFM00250000001570 | Rho Guanine Nucleotide Exchange Factor 9 Collybistin Rac/CDC42 Guanine Nucleotide Exchange Factor 9 |
| ENSMUSG00000025658 | ENSFM00500000270149 | Connector Enhancer Of Kinase Suppressor Of Ras Connector Enhancer Of Ksr Cnk Homolog |
| ENSMUSG00000025666 | ENSFM00500000273439 | Transmembrane 47 Transmembrane 4 Superfamily Member 10 |
| ENSMUSG00000025724 | ENSFM00790001962996 | Signal Peptidase Complex Catalytic Subunit SEC11A EC_3.4.21.89 Endopeptidase SP18 Microsomal Signal Peptidase 18 Kda Subunit Spase 18 Kda Subunit SEC11 Homolog A SEC11 1 SPC18 |
| ENSMUSG00000025731 | ENSFM00630001052488 | Unknown |
| ENSMUSG00000025731 | ENSFM00500000272527 | Homolog |
| ENSMUSG00000025733 | ENSFM00250000000940 | Mitochondrial Rho Gtpase 2 Miro 2 EC_3.6.5.- Ras Homolog Gene Family Member T2 |
| ENSMUSG00000025738 | ENSFM00250000003830 | F Box/Lrr Repeat 16 F Box And Leucine Rich Repeat 16 |
| ENSMUSG00000025739 | ENSFM00500000299734 | Unknown |
| ENSMUSG00000025743 | ENSFM00250000006742 | Syndecan 3 |
| ENSMUSG00000025745 | ENSFM00800002023575 | Unknown |
| ENSMUSG00000025745 | ENSFM00760001714774 | Trifunctional Enzyme Subunit Alpha Mitochondrial Precursor 78 Kda Gastrin Binding Tp Alpha |
| ENSMUSG00000025757 | ENSFM00360000109870 | Unknown |
| ENSMUSG00000025777 | ENSFM00760001743276 | Unknown |
| ENSMUSG00000025777 | ENSFM00250000004188 | Ganglioside Induced Differentiation Associated 1 GDAP1 |
| ENSMUSG00000025781 | ENSFM00750001632374 | Atp Synthase Subunit Gamma Mitochondrial F Atpase Gamma Subunit |
| ENSMUSG00000025791 | ENSFM00760001714662 | Phosphoglucomutase 1 Pgm 1 EC_5.4.2.2 Glucose Phosphomutase 1 |
| ENSMUSG00000025792 | ENSFM00760001714704 | 39S Ribosomal L12 Mitochondrial Precursor L12MT Mrp L12 |
| ENSMUSG00000025793 | ENSFM00500000271438 | Hepatocyte Growth Factor Regulated Tyrosine Kinase Substrate |
| ENSMUSG00000025794 | ENSFM00500000270978 | 60S Ribosomal L14 |
| ENSMUSG00000025809 | ENSFM00570000898867 | Unknown |
| ENSMUSG00000025809 | ENSFM00730001521210 | Integrin Beta Precursor Subunit Beta Subunit Beta Antigen |
| ENSMUSG00000025810 | ENSFM00250000000469 | Neuropilin Precursor Vascular Endothelial Cell Growth Factor 165 Receptor |
| ENSMUSG00000025813 | ENSFM00250000001140 | Homer Homolog Homer Vasp/Ena Related Gene Up Regulated During Seizure And Ltp Vesl |
| ENSMUSG00000025816 | ENSFM00250000001640 | Transport SEC61 Subunit Alpha |
| ENSMUSG00000025823 | ENSFM00800002003822 | Disulfide Isomerase A4 Precursor EC_5.3.4.1 Endoplasmic Reticulum Resident 70 Er 70 ERP70 Endoplasmic Reticulum Resident 72 Er 72 Erp 72 ERP72 |
| ENSMUSG00000025855 | ENSFM00570000895275 | Unknown |
| ENSMUSG00000025855 | ENSFM00730001521681 | Camp Dependent Kinase Type I Alpha Regulatory Camp Dependent Kinase Type I Alpha Regulatory Subunit N Terminally Processed |
| ENSMUSG00000025858 | ENSFM00250000005809 | Golgi To Er Traffic 4 Homolog |
| ENSMUSG00000025873 | ENSFM00570000883441 | Unknown |
| ENSMUSG00000025873 | ENSFM00570000886274 | Unknown |
| ENSMUSG00000025873 | ENSFM00570000898587 | Unknown |
| ENSMUSG00000025873 | ENSFM00250000005012 | Fas Associated Factor 2 Ubx Domain Containing 8 |
| ENSMUSG00000025889 | ENSFM00600000921478 | Alpha Synuclein |
| ENSMUSG00000025892 | ENSFM00720001492020 | Glutamate Receptor Precursor Glur Ampa Selective Glutamate Receptor Glur Glutamate Receptor Ionotropic Ampa |
| ENSMUSG00000025903 | ENSFM00500000270136 | Acyl Thioesterase 1 Apt 1 EC_3.1.2.- Lysophospholipase 1 Lysophospholipase I Lpl I Lysopla I |
| ENSMUSG00000025907 | ENSFM00590000917586 | Unknown |
| ENSMUSG00000025907 | ENSFM00250000002936 | RB1 Inducible Coiled Coil 1 Fak Family Kinase Interacting Of 200 Kda FIP200 |
| ENSMUSG00000025950 | ENSFM00760001763093 | Unknown |
| ENSMUSG00000025950 | ENSFM00770001847524 | Isocitrate Dehydrogenase Cytoplasmic Idh EC_1.1.1.42 Cytosolic Nadp Isocitrate Dehydrogenase Idp Nadp + Specific Icdh Oxalosuccinate Decarboxylase |
| ENSMUSG00000025964 | ENSFM00410000138484 | Disintegrin And Metalloproteinase Domain Containing Precursor Adam Metalloproteinase Disintegrin And Cysteine Rich Mdc |
| ENSMUSG00000025967 | ENSFM00640001117281 | Unknown |
| ENSMUSG00000025967 | ENSFM00640001117282 | Unknown |
| ENSMUSG00000025967 | ENSFM00700001403676 | Unknown |
| ENSMUSG00000025967 | ENSFM00700001407061 | Unknown |
| ENSMUSG00000025967 | ENSFM00700001407062 | Unknown |
| ENSMUSG00000025967 | ENSFM00730001521581 | Elongation Factor 1 Beta Ef 1 Beta |
| ENSMUSG00000025968 | ENSFM00760001763089 | Unknown |
| ENSMUSG00000025968 | ENSFM00760001763090 | Unknown |
| ENSMUSG00000025968 | ENSFM00760001763091 | Unknown |
| ENSMUSG00000025968 | ENSFM00250000003275 | Nadh Ubiquinone Oxidoreductase 75 Kda Subunit Mitochondrial Precursor EC_1.6.5.3 EC_1.6.99.- 3 Complex I 75KD Ci 75KD |
| ENSMUSG00000025969 | ENSFM00250000000469 | Neuropilin Precursor Vascular Endothelial Cell Growth Factor 165 Receptor |
| ENSMUSG00000025971 | ENSFM00250000009079 | Uncharacterized Mitochondrial Precursor |
| ENSMUSG00000025978 | ENSFM00700001401514 | Unknown |
| ENSMUSG00000025978 | ENSFM00390000126424 | Raftlin 2 Raft Linking 2 |
| ENSMUSG00000025979 | ENSFM00590000916954 | Unknown |
| ENSMUSG00000025979 | ENSFM00250000004752 | Mob Phocein Class Ii MMOB1 MOB1 Homolog 3 MOB3 Mps One Binder Kinase Activator 3 Preimplantation 3 |
| ENSMUSG00000025980 | ENSFM00760001714648 | 60 Kda Heat Shock Protein Mitochondrial 60 Kda Chaperonin Chaperonin 60 CPN60 Heat Shock 60 Hsp 60 HSP60 Mitochondrial Matrix P1 |
| ENSMUSG00000025986 | ENSFM00500000270550 | Zinc Transporter ZIP6 Precursor Solute Carrier Family 39 Member 6 Zrt And Irt 6 Zip 6 |
| ENSMUSG00000026000 | ENSFM00250000002547 | Lanc 1.40 Kda Erythrocyte Membrane P40 |
| ENSMUSG00000026003 | ENSFM00730001522380 | Long Chain Specific Acyl Coa Dehydrogenase Mitochondrial Precursor Lcad EC_1.3.8.8 |
| ENSMUSG00000026014 | ENSFM00250000001459 | Amyloid Beta A4 Precursor Binding Family B Member 1 Interacting APBB1 Interacting 1 |
| ENSMUSG00000026024 | ENSFM00760001751453 | Unknown |
| ENSMUSG00000026024 | ENSFM00250000002591 | Alsin Amyotrophic Lateral Sclerosis 2 |
| ENSMUSG00000026032 | ENSFM00810002044181 | Nadh Dehydrogenase 1 Beta Subcomplex Subunit 3 Complex I B12 Ci B12 Nadh Ubiquinone Oxidoreductase B12 Subunit |
| ENSMUSG00000026036 | ENSFM00500000270871 | Gtp Cyclohydrolase 1 Type 2 Ni EC_3.5.4.16 Gtp Cyclohydrolase I NIF3 1 |
| ENSMUSG00000026064 | ENSFM00810002057371 | Unknown |
| ENSMUSG00000026064 | ENSFM00250000001523 | Tyrosine Phosphatase Type Iva 1 Precursor EC_3.1.3.48 Tyrosine Phosphatase 4A1 Tyrosine Phosphatase Of Regenerating Liver 1 Prl 1 |
| ENSMUSG00000026074 | ENSFM00260000050335 | Kinase EC_2.7.11.1 Kinase Mapk/Erk Kinase Kinase Kinase Mek Kinase Kinase Mekkk Kinase |
| ENSMUSG00000026090 | ENSFM00250000019314 | Uncharacterized |
| ENSMUSG00000026090 | ENSFM00590000918345 | Unknown |
| ENSMUSG00000026102 | ENSFM00590000917084 | Unknown |
| ENSMUSG00000026102 | ENSFM00660001167674 | Unknown |
| ENSMUSG00000026102 | ENSFM00250000006580 | Inositol Polyphosphate 1 Phosphatase Ipp Ippase EC_3.1.3.57 |
| ENSMUSG00000026103 | ENSFM00250000001281 | Glutaminase Isoform Mitochondrial Precursor Gls EC_3.5.1.2 Glutaminase L Glutamine Amidohydrolase |
| ENSMUSG00000026109 | ENSFM00250000001843 | Tomoregulin 1 Precursor Tr 1 Transmembrane With Egf And One Follistatin Domain |
| ENSMUSG00000026113 | ENSFM00250000001432 | Type Inositol 3 4 Bisphosphate 4 Phosphatase EC_3.1.3.66 Inositol Polyphosphate 4 Phosphatase Type |
| ENSMUSG00000026150 | ENSFM00250000003442 | Mitochondrial Fission Factor |
| ENSMUSG00000026155 | ENSFM00600000921242 | Stromal Membrane Associated 2 Stromal Membrane Associated 1 |
| ENSMUSG00000026159 | ENSFM00650001140030 | Arf Gap Domain And Fg Repeat Containing 1 Hiv 1 Rev Binding Nucleoporin Rip |
| ENSMUSG00000026163 | ENSFM00250000003911 | A Kinase Anchor Sphkap SPHK1 Interactor And Akap Domain Containing Sphingosine Kinase Type 1 Interacting |
| ENSMUSG00000026172 | ENSFM00500000270206 | Mitochondrial Chaperone BCS1 BCS1 |
| ENSMUSG00000026176 | ENSFM00730001521778 | Carboxy Terminal Domain Rna Polymerase Ii Polypeptide A Small Phosphatase Nuclear Lim Interactor Interacting Factor Nli Interacting Factor Small C Terminal Domain Phosphatase Small Ctd Phosphatase |
| ENSMUSG00000026179 | ENSFM00500000272376 | Probable Hydrolase Pnkd Ec 3 Myofibrillogenesis Regulator 1 Mr 1 Paroxysmal Nonkinesiogenic Dyskinesia |
| ENSMUSG00000026181 | ENSFM00260000050737 | Phosphatase 1E EC_3.1.3.16 Ca 2+ /Calmodulin Dependent Kinase Phosphatase N Camkp N Camkp Nucleus Camkn Partner Of Pix 1 Partner Of Pix Alpha Partner Of Pixa |
| ENSMUSG00000026192 | ENSFM00250000003624 | Bifunctional Purine Biosynthesis Phosphoribosylaminoimidazolecarboxamide Formyltransferase EC_2.1.2.3 5 Aminoimidazole 4 Carboxamide Ribonucleotide Formyltransferase Aicar Transformylase |
| ENSMUSG00000026198 | ENSFM00590000918350 | Unknown |
| ENSMUSG00000026198 | ENSFM00730001521565 | Atp Binding Cassette Sub Family B Member 6 Mitochondrial Ubiquitously Expressed Mammalian Abc Half Transporter |
| ENSMUSG00000026202 | ENSFM00760001757230 | Unknown |
| ENSMUSG00000026202 | ENSFM00250000000217 | Tubulin Alpha Chain |
| ENSMUSG00000026203 | ENSFM00760001751325 | Unknown |
| ENSMUSG00000026203 | ENSFM00570000851144 | Dnaj Homolog Subfamily B Member |
| ENSMUSG00000026203 | ENSFM00760001751344 | Dnaj Homolog Subfamily B Member 2 |
| ENSMUSG00000026204 | ENSFM00760001762510 | Unknown |
| ENSMUSG00000026204 | ENSFM00260000050522 | Receptor Type Tyrosine Phosphatase Precursor R Ptp |
| ENSMUSG00000026207 | ENSFM00570000901221 | Unknown |
| ENSMUSG00000026207 | ENSFM00740001589455 | Striated Muscle Specific Serine/Threonine Kinase EC_2.7.11.1 Aortic Preferentially Expressed 1 Apeg 1 |
| ENSMUSG00000026209 | ENSFM00760001763105 | Unknown |
| ENSMUSG00000026209 | ENSFM00250000004224 | Aspartyl Aminopeptidase EC_3.4.11.21 |
| ENSMUSG00000026213 | ENSFM00250000004028 | Serine/Threonine Kinase 11 Interacting |
| ENSMUSG00000026223 | ENSFM00600000921249 | Integral Membrane 2B Immature BRI2 IMBRI2 Transmembrane Bri Bri |
| ENSMUSG00000026229 | ENSFM00570000891016 | Unknown |
| ENSMUSG00000026229 | ENSFM00250000002209 | 26S Proteasome Non Atpase Regulatory Subunit 1 26S Proteasome Regulatory Subunit RPN2 26S Proteasome Regulatory Subunit S1 |
| ENSMUSG00000026234 | ENSFM00760001751711 | Unknown |
| ENSMUSG00000026234 | ENSFM00500000270420 | Nucleolin |
| ENSMUSG00000026235 | ENSFM00760001763106 | Unknown |
| ENSMUSG00000026235 | ENSFM00730001521142 | Ephrin Type A Receptor Precursor EC_2.7.10.1 Kinase |
| ENSMUSG00000026238 | ENSFM00760001748815 | Unknown |
| ENSMUSG00000026238 | ENSFM00760001748816 | Prothymosin Alpha |
| ENSMUSG00000026238 | ENSFM00810002041148 | Prothymosin Alpha |
| ENSMUSG00000026245 | ENSFM00760001763107 | Unknown |
| ENSMUSG00000026245 | ENSFM00250000004384 | Phenylalanine Trna Ligase Beta Subunit EC_6.1.1.20 Phenylalanyl Trna Synthetase Beta Subunit Phers |
| ENSMUSG00000026247 | ENSFM00800002003370 | Endothelin Converting Enzyme Ece EC_3.4.24.71 |
| ENSMUSG00000026249 | ENSFM00740001589252 | Plasminogen Activator Inhibitor 1 Precursor Pai Pai 1 Endothelial Plasminogen Activator Inhibitor Serpin E1 |
| ENSMUSG00000026259 | ENSFM00740001589196 | Ephexin 1 Eph Interacting Exchange Neuronal Guanine Nucleotide Exchange Factor |
| ENSMUSG00000026260 | ENSFM00760001749841 | Unknown |
| ENSMUSG00000026260 | ENSFM00250000002870 | Nadh Dehydrogenase 1 Alpha Subcomplex Subunit 10 Mitochondrial Precursor Complex I 42KD Ci 42KD Nadh Ubiquinone Oxidoreductase 42 Kda Subunit |
| ENSMUSG00000026275 | ENSFM00810002040799 | Phosphatase 1 Regulatory Subunit 7 Phosphatase 1 Regulatory Subunit 22 |
| ENSMUSG00000026276 | ENSFM00570000881519 | Unknown |
| ENSMUSG00000026276 | ENSFM00640001119291 | Unknown |
| ENSMUSG00000026276 | ENSFM00800002003496 | Septin 2 |
| ENSMUSG00000026312 | ENSFM00570000892693 | Unknown |
| ENSMUSG00000026312 | ENSFM00700001406263 | Unknown |
| ENSMUSG00000026312 | ENSFM00760001759622 | Unknown |
| ENSMUSG00000026312 | ENSFM00260000050333 | Cadherin Precursor |
| ENSMUSG00000026319 | ENSFM00760001747407 | Unknown |
| ENSMUSG00000026319 | ENSFM00250000004714 | Lish Domain And Heat Repeat Containing |
| ENSMUSG00000026335 | ENSFM00250000001444 | Peptidyl Glycine Alpha Amidating Monooxygenase Precursor Pam |
| ENSMUSG00000026341 | ENSFM00760001763132 | Unknown |
| ENSMUSG00000026341 | ENSFM00760001763133 | Unknown |
| ENSMUSG00000026341 | ENSFM00800002003421 | Actin Related 3 Actin 3 |
| ENSMUSG00000026344 | ENSFM00590000917131 | Unknown |
| ENSMUSG00000026344 | ENSFM00250000006765 | LY6/Plaur Domain Containing 1 Precursor |
| ENSMUSG00000026347 | ENSFM00250000007006 | Transmembrane 163 Synaptic Vesicle Membrane Of 31 Kda |
| ENSMUSG00000026353 | ENSFM00250000005136 | Ubx Domain Containing 4 Ubx Domain Containing 2 |
| ENSMUSG00000026356 | ENSFM00250000003932 | Aspartate Trna Ligase Cytoplasmic EC_6.1.1.12 Aspartyl Trna Synthetase Asprs |
| ENSMUSG00000026356 | ENSFM00500000299788 | Aspartate Trna Ligase Cytoplasmic |
| ENSMUSG00000026374 | ENSFM00250000005250 | Translin EC_3.1.-.- Component 3 Of Promoter Of Risc C3PO |
| ENSMUSG00000026385 | ENSFM00500000413119 | Unknown |
| ENSMUSG00000026385 | ENSFM00700001403865 | Unknown |
| ENSMUSG00000026385 | ENSFM00670001239746 | Acyl Coa Binding Acbp Diazepam Binding Inhibitor Dbi Endozepine Ep |
| ENSMUSG00000026389 | ENSFM00250000001178 | Metalloreductase EC_1.16.1.- Six Transmembrane Epithelial Antigen Of Prostate |
| ENSMUSG00000026395 | ENSFM00780001917658 | Unknown |
| ENSMUSG00000026395 | ENSFM00740001589127 | Receptor Type Tyrosine Phosphatase Precursor Tyrosine Phosphatase R Ptp EC_3.1.3.48 |
| ENSMUSG00000026411 | ENSFM00670001235681 | Transmembrane Precursor |
| ENSMUSG00000026421 | ENSFM00740001589223 | Cysteine And Glycine Rich Cysteine Rich |
| ENSMUSG00000026425 | ENSFM00760001756891 | Unknown |
| ENSMUSG00000026425 | ENSFM00250000000639 | Slit Robo Rho Gtpase Activating Rho Gtpase Activating |
| ENSMUSG00000026426 | ENSFM00410000138507 | Adp Ribosylation Factor Small G Indispensable Equal Chromosome Segregation |
| ENSMUSG00000026437 | ENSFM00760001763136 | Unknown |
| ENSMUSG00000026437 | ENSFM00740001589171 | Cyclin Dependent Kinase EC_2.7.11.22 Cell Division Kinase Pctaire Motif Kinase Serine/Threonine Kinase Pctaire |
| ENSMUSG00000026442 | ENSFM00390000126295 | Neural Cell Adhesion Molecule L1 L1 L1 |
| ENSMUSG00000026452 | ENSFM00760001754333 | Unknown |
| ENSMUSG00000026452 | ENSFM00720001492031 | Synaptotagmin |
| ENSMUSG00000026456 | ENSFM00640001112518 | Unknown |
| ENSMUSG00000026456 | ENSFM00640001119305 | Unknown |
| ENSMUSG00000026456 | ENSFM00730001521809 | Nadh Cytochrome B5 Reductase B5R EC_1.6.2.2 |
| ENSMUSG00000026457 | ENSFM00730001521852 | Receptor |
| ENSMUSG00000026458 | ENSFM00250000000441 | Liprin Alpha Tyrosine Phosphatase Receptor Type F Polypeptide Interacting Alpha Ptprf Interacting Alpha |
| ENSMUSG00000026463 | ENSFM00610000972342 | Unknown |
| ENSMUSG00000026463 | ENSFM00250000000445 | Plasma Membrane Calcium Transporting Atpase EC_3.6.3.8 Plasma Membrane Calcium Atpase Plasma Membrane Calcium Pump |
| ENSMUSG00000026469 | ENSFM00500000270654 | Xenotropic And Polytropic Retrovirus Receptor 1 |
| ENSMUSG00000026470 | ENSFM00760001723625 | Uncharacterized |
| ENSMUSG00000026470 | ENSFM00250000002600 | Syntaxin |
| ENSMUSG00000026473 | ENSFM00750001632335 | Glutamine Synthetase Gs EC_6.3.1.2 Glutamate Decarboxylase EC_4.1.1.- 15 Glutamate Ammonia Ligase |
| ENSMUSG00000026490 | ENSFM00730001521452 | Serine/Threonine Kinase Mrck EC_2.7.11.1 CDC42 Binding Kinase Dmpk Myotonic Dystrophy Kinase Related CDC42 Binding Kinase Mrck Myotonic Dystrophy Kinase |
| ENSMUSG00000026499 | ENSFM00500000270849 | Golgi Resident GCP60 Acyl Coa Binding Domain Containing 3 Golgi Complex Associated 1 GOCAP1 Golgi Phosphoprotein 1 GOLPH1 Pbr And Pka Associated 7 Peripheral Benzodiazepine Receptor Associated PAP7 |
| ENSMUSG00000026510 | ENSFM00250000001053 | Apoptosis Stimulating Of P53 2 Tumor Suppressor P53 Binding 2 53BP2 P53 Binding 2 P53BP2 |
| ENSMUSG00000026520 | ENSFM00500000269896 | Pyrroline 5 Carboxylate Reductase 3 P5C Reductase 3 P5CR 3 EC_1.5.1.2 Pyrroline 5 Carboxylate Reductase |
| ENSMUSG00000026526 | ENSFM00660001189496 | Unknown |
| ENSMUSG00000026526 | ENSFM00250000003390 | Fumarate Hydratase Mitochondrial Precursor Fumarase EC_4.2.1.2 |
| ENSMUSG00000026527 | ENSFM00780001917687 | Unknown |
| ENSMUSG00000026527 | ENSFM00730001521808 | Regulator Of G Signaling |
| ENSMUSG00000026532 | ENSFM00740001589109 | Spectrin Alpha Chain Non Erythrocytic 1 Alpha Ii Spectrin Fodrin Alpha Chain |
| ENSMUSG00000026553 | ENSFM00570000887400 | Unknown |
| ENSMUSG00000026553 | ENSFM00260000050647 | Coatomer Subunit Alpha Alpha Coat Alpha Cop Hep Cop Hepcop |
| ENSMUSG00000026556 | ENSFM00250000002390 | Vang 2 Van Gogh |
| ENSMUSG00000026568 | ENSFM00780001917677 | Unknown |
| ENSMUSG00000026568 | ENSFM00500000273639 | Mitochondrial Pyruvate Carrier 2 Brain 44 |
| ENSMUSG00000026576 | ENSFM00500000270479 | Sodium/Potassium Transporting Atpase Subunit Beta 1 Sodium/Potassium Dependent Atpase Subunit Beta 1 |
| ENSMUSG00000026585 | ENSFM00250000003018 | Kinesin Associated 3 Kap 3 KAP3 |
| ENSMUSG00000026587 | ENSFM00780001911402 | Unknown |
| ENSMUSG00000026587 | ENSFM00250000001299 | Astrotactin Precursor |
| ENSMUSG00000026596 | ENSFM00730001521872 | Tyrosine Kinase EC_2.7.10.2 Abelson Murine Leukemia Viral Oncogene Homolog Abelson Tyrosine Kinase |
| ENSMUSG00000026605 | ENSFM00610000969319 | Unknown |
| ENSMUSG00000026605 | ENSFM00250000003245 | Centromere F Precursor Cenp F Ah Antigen Kinetochore Cenpf Mitosin |
| ENSMUSG00000026615 | ENSFM00800002003460 | Bifunctional Glutamate/Proline Trna Ligase Bifunctional Aminoacyl Trna Synthetase |
| ENSMUSG00000026617 | ENSFM00690001359175 | Unknown |
| ENSMUSG00000026617 | ENSFM00250000005952 | 3' 2' 5' Bisphosphate Nucleotidase 1 EC_3.1.3.7 Bisphosphate 3' Nucleotidase 1 Pap Inositol 1 4 Phosphatase Pip |
| ENSMUSG00000026618 | ENSFM00500000270016 | Isoleucine Trna Ligase Mitochondrial EC_6.1.1.5 Isoleucyl Trna Synthetase Ilers |
| ENSMUSG00000026620 | ENSFM00730001521220 | Kinase EC_2.7.11.1 |
| ENSMUSG00000026623 | ENSFM00570000895571 | Unknown |
| ENSMUSG00000026623 | ENSFM00250000004110 | Acyl Coa:Lysophosphatidylglycerol Acyltransferase 1 EC_2.3.1.- |
| ENSMUSG00000026626 | ENSFM00780001917704 | Unknown |
| ENSMUSG00000026626 | ENSFM00760001714676 | Serine/Threonine Phosphatase 2A 56 Kda Regulatory Subunit Epsilon PP2A B Subunit B' Epsilon PP2A B Subunit B56 Epsilon PP2A B Subunit PR61 Epsilon PP2A B Subunit R5 Epsilon |
| ENSMUSG00000026640 | ENSFM00570000890425 | Unknown |
| ENSMUSG00000026640 | ENSFM00730001521329 | Plexin Precursor |
| ENSMUSG00000026657 | ENSFM00250000001583 | Ferm Domain Containing 4B GRP1 Binding GRSP1 |
| ENSMUSG00000026687 | ENSFM00730001521985 | 4 Trimethylaminobutyraldehyde Dehydrogenase Tmabadh EC_1.2.1.47 Aldehyde Dehydrogenase Family 9 Member A1 EC_1.2.1.- 3 |
| ENSMUSG00000026688 | ENSFM00670001235926 | Microsomal Glutathione S Transferase 3 Microsomal Gst 3 EC_2.5.1.18 Microsomal Gst Iii |
| ENSMUSG00000026696 | ENSFM00780001910349 | Unknown |
| ENSMUSG00000026696 | ENSFM00500000273038 | Vesicle Associated Membrane 4 Vamp 4 |
| ENSMUSG00000026701 | ENSFM00780001917673 | Unknown |
| ENSMUSG00000026701 | ENSFM00730001522010 | Peroxiredoxin 6 EC_1.11.1.15 1 Cys Peroxiredoxin 1 Cys Prx Acidic Calcium Independent Phospholipase A2 AIPLA2 EC_3.1.1.- Antioxidant 2 Non Selenium Glutathione Peroxidase Nsgpx EC_1.11.-.- 1 9 |
| ENSMUSG00000026718 | ENSFM00780001917563 | Unknown |
| ENSMUSG00000026718 | ENSFM00250000002061 | Signal Transducing Adapter Molecule Stam |
| ENSMUSG00000026728 | ENSFM00730001521291 | Vimentin |
| ENSMUSG00000026737 | ENSFM00570000888534 | Unknown |
| ENSMUSG00000026737 | ENSFM00260000050567 | Phosphatidylinositol 5 Phosphate 4 Kinase Type 2 EC_2.7.1.149 1 Phosphatidylinositol 5 Phosphate 4 Kinase 2 Diphosphoinositide Kinase 2 Phosphatidylinositol 5 Phosphate 4 Kinase Type Ii Pi 5 P 4 Kinase Type Ii PIP4KII Ptdins 5 P 4 Kinase 2 |
| ENSMUSG00000026755 | ENSFM00500000270526 | Unknown |
| ENSMUSG00000026764 | ENSFM00670001235340 | Kinesin Heavy Chain Kinesin Heavy Chain Kinesin Heavy Chain |
| ENSMUSG00000026775 | ENSFM00800002003559 | Atp Dependent Zinc Metalloprotease Ym EC_3.4.24.- Atp Dependent Metalloprotease FTSH1 Meg 4 YME1 1 |
| ENSMUSG00000026782 | ENSFM00250000000845 | Abl Interactor 1 Abelson Interactor 1 Abi 1 EPS8 SH3 Domain Binding EPS8 Binding Spectrin SH3 Domain Binding 1 |
| ENSMUSG00000026787 | ENSFM00800002003398 | Glutamate Decarboxylase 1 EC_4.1.1.15.67 Kda Glutamic Acid Decarboxylase Gad 67 Glutamate Decarboxylase 67 Kda |
| ENSMUSG00000026797 | ENSFM00250000001101 | Syntaxin Binding 1 Unc 18 Homolog 1 UNC18 1 Unc 18 Homolog A Unc 18A |
| ENSMUSG00000026817 | ENSFM00740001589162 | Adenylate Kinase Isoenzyme 1 |
| ENSMUSG00000026819 | ENSFM00500000269757 | Calcium Binding Mitochondrial Carrier Scamc Small Calcium Binding Mitochondrial Carrier Solute Carrier Family 25 Member |
| ENSMUSG00000026820 | ENSFM00780001917901 | Unknown |
| ENSMUSG00000026820 | ENSFM00250000004186 | Prostaglandin E Synthase 2 |
| ENSMUSG00000026825 | ENSFM00570000892896 | Unknown |
| ENSMUSG00000026825 | ENSFM00520000517792 | Dynamin EC_3.6.5.5 |
| ENSMUSG00000026827 | ENSFM00250000001725 | Glycerol 3 Phosphate Dehydrogenase Mitochondrial Precursor Gpd M Gpdh M EC_1.1.5.3 |
| ENSMUSG00000026830 | ENSFM00250000009524 | Ermin Juxtanodin Jn |
| ENSMUSG00000026833 | ENSFM00320000100099 | Noelin Precursor Neuronal Olfactomedin Related Er Localized Olfactomedin 1 |
| ENSMUSG00000026841 | ENSFM00730001521406 | Fibrinogen C Domain Containing 1 |
| ENSMUSG00000026853 | ENSFM00570000890585 | Unknown |
| ENSMUSG00000026853 | ENSFM00570000894957 | Unknown |
| ENSMUSG00000026853 | ENSFM00570000900479 | Unknown |
| ENSMUSG00000026853 | ENSFM00610000954683 | Unknown |
| ENSMUSG00000026853 | ENSFM00730001521465 | Choline O Acetyltransferase Choactase Chat Choline Acetylase EC_2.3.1.6 |
| ENSMUSG00000026858 | ENSFM00660001175733 | Unknown |
| ENSMUSG00000026858 | ENSFM00250000002334 | FAM73B |
| ENSMUSG00000026860 | ENSFM00250000002047 | Endophilin SH3 Domain Containing GRB2 |
| ENSMUSG00000026864 | ENSFM00780001917908 | Unknown |
| ENSMUSG00000026864 | ENSFM00800002003443 | 78 Kda Glucose Regulated Grp 78 Heat Shock 70 Kda 5 Immunoglobulin Heavy Chain Binding Bip |
| ENSMUSG00000026869 | ENSFM00250000006842 | 26S Proteasome Non Atpase Regulatory Subunit 5 26S Protease Subunit S5 Basic 26S Proteasome Subunit S5B |
| ENSMUSG00000026878 | ENSFM00800002003610 | Ras Related Rab 14 |
| ENSMUSG00000026879 | ENSFM00760001714632 | Gelsolin Actin Depolymerizing Factor Adf |
| ENSMUSG00000026880 | ENSFM00250000000738 | Stomatin |
| ENSMUSG00000026883 | ENSFM00570000864156 | Unknown |
| ENSMUSG00000026883 | ENSFM00570000890343 | Unknown |
| ENSMUSG00000026883 | ENSFM00250000000610 | Ras Gtpase Activating |
| ENSMUSG00000026887 | ENSFM00590000917495 | Unknown |
| ENSMUSG00000026887 | ENSFM00250000005296 | Ribosome Recycling Factor Mitochondrial Precursor Rrf Ribosome Releasing Factor Mitochondrial |
| ENSMUSG00000026895 | ENSFM00250000005294 | Nadh Dehydrogenase 1 Alpha Subcomplex Subunit 8 Complex I 19KD Ci 19KD Complex I Pgiv Ci Pgiv Nadh Ubiquinone Oxidoreductase 19 Kda Subunit |
| ENSMUSG00000026904 | ENSFM00730001521473 | Sodium Bicarbonate Solute Carrier Family 4 Member |
| ENSMUSG00000026914 | ENSFM00250000004350 | 26S Proteasome Non Atpase Regulatory Subunit 14 EC_3.4.19.- 26S Proteasome Regulatory Subunit RPN11 |
| ENSMUSG00000026925 | ENSFM00420000140651 | 72 Kda Inositol Polyphosphate 5 Phosphatase Precursor EC_3.1.3.36 |
| ENSMUSG00000026926 | ENSFM00500000270616 | Mitochondrial Processing Peptidase Subunit Alpha Precursor EC_3.4.24.64 Alpha Mpp |
| ENSMUSG00000026930 | ENSFM00570000888385 | Unknown |
| ENSMUSG00000026930 | ENSFM00250000001225 | G Signaling Modulator 1 Activator Of G Signaling 3 |
| ENSMUSG00000026944 | ENSFM00740001589079 | Atp Binding Cassette Sub Family A Member Atp Binding Cassette Transporter Atp Binding Cassette |
| ENSMUSG00000026959 | ENSFM00730001522127 | Glutamate Receptor Subunit 1 |
| ENSMUSG00000026971 | ENSFM00730001521296 | Integrin Beta Precursor |
| ENSMUSG00000026991 | ENSFM00740001610501 | Unknown |
| ENSMUSG00000026991 | ENSFM00730001521673 | Catenin Delta 2 |
| ENSMUSG00000027002 | ENSFM00250000001880 | Nck Associated 1 1 Membrane Associated Hem |
| ENSMUSG00000027006 | ENSFM00250000002411 | Dnaj Homolog Subfamily C Member 10 Precursor EC_1.8.4.- Endoplasmic Reticulum Dna J Domain Containing 5 Er Resident ERDJ5 ERDJ5 |
| ENSMUSG00000027010 | ENSFM00550000747100 | Calcium Binding Mitochondrial Carrier ARALAR1 |
| ENSMUSG00000027010 | ENSFM00800002003431 | Calcium Binding Mitochondrial Carrier Mitochondrial Aspartate Glutamate Carrier Solute Carrier Family 25 Member |
| ENSMUSG00000027012 | ENSFM00250000000887 | Cytoplasmic Dynein 1 Intermediate Chain 2 Cytoplasmic Dynein Intermediate Chain 2 Dynein Intermediate Chain 2 Cytosolic Dh Ic 2 |
| ENSMUSG00000027015 | ENSFM00670001235694 | Cytochrome B Ascorbate Dependent 3 Ec 1 Cytochrome B561 Family Member A3 |
| ENSMUSG00000027022 | ENSFM00250000001847 | Xin Actin Binding Repeat Containing 2 Beta Xin Cardiomyopathy Associated 3 |
| ENSMUSG00000027075 | ENSFM00500000270086 | Large Neutral Amino Acids Transporter Small Subunit 4 L Type Amino Acid Transporter 4 Solute Carrier Family 43 Member 2 |
| ENSMUSG00000027076 | ENSFM00670001236523 | Mitochondrial Import Inner Membrane Translocase Subunit TIM10 |
| ENSMUSG00000027087 | ENSFM00570000898230 | Unknown |
| ENSMUSG00000027087 | ENSFM00500000269717 | Integrin Alpha Precursor Receptor Subunit Alpha |
| ENSMUSG00000027099 | ENSFM00250000005082 | Metaxin 2 Mitochondrial Outer Membrane Import Complex 2 |
| ENSMUSG00000027108 | ENSFM00250000002536 | Obg Atpase 1 |
| ENSMUSG00000027111 | ENSFM00400000131750 | Integrin Alpha Precursor CD49 Antigen Family Member Vla Antigen |
| ENSMUSG00000027130 | ENSFM00260000050376 | Solute Carrier Family 12 Member Electroneutral Potassium Chloride Cotransporter K Cl Cotransporter |
| ENSMUSG00000027131 | ENSFM00670001235866 | Er Membrane Complex Subunit 4 Transmembrane 85 |
| ENSMUSG00000027134 | ENSFM00250000001465 | Lysophosphatidylcholine Acyltransferase Lpc Acyltransferase Lpcat Lysopc Acyltransferase EC_2.3.1.23 1 Acylglycerophosphocholine O Acyltransferase 1 Alkylglycerophosphocholine O Acetyltransferase 67 Acetyl Coa:Lyso Platelet Activating Factor Acetyltransfe |
| ENSMUSG00000027162 | ENSFM00260000050595 | Lin 7 Homolog Lin Mammalian Lin Seven Mals Vertebrate Lin 7 Homolog Veli |
| ENSMUSG00000027163 | ENSFM00250000007011 | Comm Domain Containing 9 |
| ENSMUSG00000027184 | ENSFM00670001263995 | Unknown |
| ENSMUSG00000027184 | ENSFM00470000251449 | Caprin 1 Cytoplasmic Activation And Proliferation Associated 1 Gpi Anchored Membrane 1 Gpi Anchored P137 Gpi P137 P137GPI Membrane Component Chromosome 11 Surface Marker 1 Rna Granule 105 |
| ENSMUSG00000027187 | ENSFM00760001714613 | Catalase EC_1.11.1.6 |
| ENSMUSG00000027195 | ENSFM00670001235389 | Estradiol 17 Beta Dehydrogenase 12 EC_1.1.1.62 17 Beta Hydroxysteroid Dehydrogenase 12 17 Beta Hsd 12 |
| ENSMUSG00000027206 | ENSFM00250000004729 | COP9 Signalosome Complex Subunit 2 Signalosome Subunit 2 |
| ENSMUSG00000027215 | ENSFM00610000956195 | Unknown |
| ENSMUSG00000027215 | ENSFM00500000271329 | CD82 Antigen C33 Antigen IA4 Inducible Membrane R2 Metastasis Suppressor Kangai 1 CD82 Antigen |
| ENSMUSG00000027220 | ENSFM00400000131978 | Synaptotagmin 13 Synaptotagmin Xiii Sytxiii |
| ENSMUSG00000027233 | ENSFM00600000921370 | PAT1 Homolog 1 PAT1 1 PAT1 Homolog B PAT1B |
| ENSMUSG00000027247 | ENSFM00430000230144 | Rho Gtpase Activating Rho Type Gtpase Activating |
| ENSMUSG00000027248 | ENSFM00800002003471 | Disulfide Isomerase A3 Precursor EC_5.3.4.1.58 Kda Glucose Regulated 58 Kda Microsomal P58 Disulfide Isomerase Er 60 Endoplasmic Reticulum Resident 57 Er 57 ERP57 Endoplasmic Reticulum Resident 60 Er 60 ERP60 |
| ENSMUSG00000027254 | ENSFM00250000000942 | Microtubule Associated Map |
| ENSMUSG00000027255 | ENSFM00250000002206 | Adp Ribosylation Factor Gtpase Activating 2 Arf Gap 2 Zinc Finger 289 |
| ENSMUSG00000027257 | ENSFM00570000890106 | Unknown |
| ENSMUSG00000027257 | ENSFM00250000001466 | Kinase C And Casein Kinase Substrate In Neurons |
| ENSMUSG00000027270 | ENSFM00250000007191 | Lysosome Associated Membrane Glycoprotein 5 Precursor Brain And Dendritic Cell Associated Lamp Brain Associated Lamp Bad Lamp Lysosome Associated Membrane 5 Lamp 5 |
| ENSMUSG00000027273 | ENSFM00250000001228 | Synaptosomal Associated 25 Snap 25 Synaptosomal Associated 25 Kda |
| ENSMUSG00000027282 | ENSFM00570000874766 | Unknown |
| ENSMUSG00000027282 | ENSFM00250000001870 | Mitochondrial Carrier Homolog |
| ENSMUSG00000027286 | ENSFM00350000105477 | Leucine Rich Repeat Containing 57 |
| ENSMUSG00000027287 | ENSFM00250000001228 | Synaptosomal Associated 25 Snap 25 Synaptosomal Associated 25 Kda |
| ENSMUSG00000027291 | ENSFM00250000002875 | VAM6/VPS39 |
| ENSMUSG00000027293 | ENSFM00250000000766 | Eh Domain Containing |
| ENSMUSG00000027296 | ENSFM00250000001260 | Inositol Trisphosphate 3 Kinase EC_2.7.1.127 Inositol 1 4 5 Trisphosphate 3 Kinase IP3 3 Kinase IP3K Insp 3 Kinase |
| ENSMUSG00000027303 | ENSFM00740001589127 | Receptor Type Tyrosine Phosphatase Precursor Tyrosine Phosphatase R Ptp EC_3.1.3.48 |
| ENSMUSG00000027312 | ENSFM00250000000720 | Precursor |
| ENSMUSG00000027340 | ENSFM00570000877032 | Unknown |
| ENSMUSG00000027340 | ENSFM00250000000925 | Solute Carrier Family 23 Member Na + /L Ascorbic Acid Transporter Sodium Dependent Vitamin C Transporter Yolk Sac Permease Molecule |
| ENSMUSG00000027347 | ENSFM00250000001366 | Ras Guanyl Releasing Calcium And Dag Regulated Guanine Nucleotide Exchange Factor Caldag |
| ENSMUSG00000027349 | ENSFM00250000002232 | FAM98A |
| ENSMUSG00000027350 | ENSFM00250000007047 | Secretogranin 1 Precursor Chromogranin B Cgb Secretogranin I Sgi |
| ENSMUSG00000027351 | ENSFM00250000001487 | Sprouty Related EVH1 Domain Containing Spred |
| ENSMUSG00000027363 | ENSFM00730001521839 | Ubiquitin Carboxyl Terminal Hydrolase 8 EC_3.4.19.12 Deubiquitinating Enzyme 8 Ubiquitin Isopeptidase Y Ubiquitin Thioesterase 8 Ubiquitin Specific Processing Protease 8 |
| ENSMUSG00000027371 | ENSFM00500000271766 | Fumarylacetoacetate Hydrolase Domain Containing Ec 3 |
| ENSMUSG00000027397 | ENSFM00570000878959 | Unknown |
| ENSMUSG00000027397 | ENSFM00250000001093 | Sodium Dependent Phosphate Transporter Solute Carrier Family 20 Member |
| ENSMUSG00000027406 | ENSFM00740001613626 | IDH3B |
| ENSMUSG00000027406 | ENSFM00770001847589 | Isocitrate Dehydrogenase Subunit Beta Mitochondrial Precursor EC_1.1.1.41 Isocitric Dehydrogenase Subunit Beta Nad + Specific Icdh Subunit Beta |
| ENSMUSG00000027411 | ENSFM00250000004293 | Vacuolar Sorting Associated 16 |
| ENSMUSG00000027419 | ENSFM00760001714725 | Neuroendocrine Convertase 2 Precursor Nec 2 EC_3.4.21.94 KEX2 Endoprotease 2 Prohormone Convertase 2 Proprotein Convertase 2 PC2 |
| ENSMUSG00000027422 | ENSFM00250000002798 | Ribosome Binding Ribosome Receptor Rrp |
| ENSMUSG00000027423 | ENSFM00250000002044 | Sorting Nexin |
| ENSMUSG00000027429 | ENSFM00570000879929 | Unknown |
| ENSMUSG00000027429 | ENSFM00810002040640 | Transport SEC23 Related |
| ENSMUSG00000027430 | ENSFM00500000272679 | D Tyrosyl Trna Tyr Deacylase 1 EC_3.1.-.- Dna Unwinding Element Binding B Due B |
| ENSMUSG00000027438 | ENSFM00730001522002 | Soluble Nsf Attachment Snap N Ethylmaleimide Sensitive Factor Attachment |
| ENSMUSG00000027447 | ENSFM00670001237088 | Cystatin C Precursor Cystatin 3 |
| ENSMUSG00000027455 | ENSFM00500000270274 | Ubx Domain Containing 2B NSFL1 Cofactor P37 P97 Cofactor P37 |
| ENSMUSG00000027457 | ENSFM00720001500049 | Unknown |
| ENSMUSG00000027457 | ENSFM00720001503569 | Unknown |
| ENSMUSG00000027457 | ENSFM00250000002727 | Syntabulin Golgi Localized Syntaphilin Related Syntaxin 1 Binding |
| ENSMUSG00000027479 | ENSFM00250000000867 | Microtubule Associated Rp/Eb Family Member |
| ENSMUSG00000027488 | ENSFM00250000002085 | 1 Syntrophin 59 Kda Dystrophin Associated A1 Component 1 Syntrophin |
| ENSMUSG00000027499 | ENSFM00500000291172 | Unknown |
| ENSMUSG00000027499 | ENSFM00500000298869 | Unknown |
| ENSMUSG00000027500 | ENSFM00670001235346 | Stathmin |
| ENSMUSG00000027506 | ENSFM00570000888554 | Unknown |
| ENSMUSG00000027506 | ENSFM00570000899195 | Unknown |
| ENSMUSG00000027506 | ENSFM00810002040979 | Tumor D52 |
| ENSMUSG00000027508 | ENSFM00250000008582 | Phosphoprotein Associated With Glycosphingolipid Enriched Microdomains 1 Csk Binding Transmembrane Phosphoprotein Cbp |
| ENSMUSG00000027519 | ENSFM00570000886275 | Unknown |
| ENSMUSG00000027519 | ENSFM00730001522125 | Ras Related Rab |
| ENSMUSG00000027522 | ENSFM00570000882040 | Unknown |
| ENSMUSG00000027522 | ENSFM00500000271003 | Syntaxin 16 |
| ENSMUSG00000027523 | ENSFM00570000878388 | Unknown |
| ENSMUSG00000027523 | ENSFM00570000892390 | Unknown |
| ENSMUSG00000027523 | ENSFM00300000079126 | Guanine Nucleotide Binding G S Subunit Alpha Adenylate Cyclase Stimulating G Alpha |
| ENSMUSG00000027523 | ENSFM00600000922010 | Neuroendocrine Secretory 55 Precursor NESP55 |
| ENSMUSG00000027523 | ENSFM00740001591176 | Alex Alternative Gene Product Encoded By Xl Exon |
| ENSMUSG00000027531 | ENSFM00570000900355 | Unknown |
| ENSMUSG00000027531 | ENSFM00500000270262 | Inositol Monophosphatase 1 Imp 1 Impase 1 EC_3.1.3.25 |
| ENSMUSG00000027533 | ENSFM00500000273330 | Fatty Acid Binding Protein Epidermal Epidermal Type Fatty Acid Binding E Fabp Fatty Acid Binding 5 Psoriasis Associated Fatty Acid Binding Homolog Pa Fabp |
| ENSMUSG00000027534 | ENSFM00250000003292 | Sorting Nexin 16 |
| ENSMUSG00000027540 | ENSFM00270000056472 | Tyrosine Phosphatase Non Receptor Type 1 EC_3.1.3.48 Tyrosine Phosphatase 1B Ptp 1B |
| ENSMUSG00000027546 | ENSFM00570000890220 | Unknown |
| ENSMUSG00000027546 | ENSFM00810002040637 | Probable Phospholipid Transporting Atpase EC_3.6.3.1 Atpase Class Ii Type |
| ENSMUSG00000027562 | ENSFM00450000242133 | Carbonic Anhydrase EC_4.2.1.1 Carbonate Dehydratase Carbonic Anhydrase Ca |
| ENSMUSG00000027566 | ENSFM00810002040638 | Proteasome Subunit Alpha Type 7 EC_3.4.25.1 |
| ENSMUSG00000027573 | ENSFM00250000005529 | Glucose Induced Degradation 8 Homolog |
| ENSMUSG00000027575 | ENSFM00250000003758 | Adp Ribosylation Factor Gtpase Activating 1 Arf Gap 1 Adp Ribosylation Factor 1 Gtpase Activating ARF1 Gap ARF1 Directed Gtpase Activating |
| ENSMUSG00000027581 | ENSFM00670001235346 | Stathmin |
| ENSMUSG00000027599 | ENSFM00250000005628 | Armadillo Repeat Containing 1 |
| ENSMUSG00000027602 | ENSFM00500000270555 | Microtubule Associated Proteins 1A/1B Light Chain 3A Precursor Autophagy Related LC3 A Autophagy Related Ubiquitin Modifier LC3 A MAP1 Light Chain 3 1 MAP1A/MAP1B Light Chain 3 A MAP1A/MAP1B LC3 A Microtubule Associated 1 Light Chain 3 Alpha |
| ENSMUSG00000027603 | ENSFM00730001522526 | Gamma Glutamyltransferase 7 Precursor Ggt 7 EC_2.3.2.2 Gamma Glutamyltransferase 3 Gamma Glutamyltranspeptidase 7 Glutathione Hydrolase 7 EC_3.4.19.- 13 |
| ENSMUSG00000027618 | ENSFM00500000270207 | Cysteine Desulfurase Mitochondrial Precursor EC_2.8.1.7 |
| ENSMUSG00000027618 | ENSFM00740001613632 | Cysteine Desulfurase Mitochondrial |
| ENSMUSG00000027624 | ENSFM00800002003400 | Unknown |
| ENSMUSG00000027634 | ENSFM00250000000535 | N Myc Downstream Regulated Gene |
| ENSMUSG00000027642 | ENSFM00250000002871 | Dolichyl Diphosphooligosaccharide Glycosyltransferase Subunit 2 Precursor EC_2.4.99.18 Dolichyl Diphosphooligosaccharide Glycosyltransferase 63 Kda Subunit Ribophorin Ii Rpn Ii Ribophorin 2 |
| ENSMUSG00000027646 | ENSFM00730001521177 | Tyrosine Kinase EC_2.7.10.2 |
| ENSMUSG00000027652 | ENSFM00570000876486 | Unknown |
| ENSMUSG00000027652 | ENSFM00250000002523 | Ral Gtpase Activating Subunit Beta P170 |
| ENSMUSG00000027668 | ENSFM00250000001660 | Transmembrane Gtpase |
| ENSMUSG00000027669 | ENSFM00740001613509 | Unknown |
| ENSMUSG00000027669 | ENSFM00780001917804 | Unknown |
| ENSMUSG00000027669 | ENSFM00250000000545 | Guanine Nucleotide Binding G I /G S /G T Subunit Beta Transducin Beta Chain |
| ENSMUSG00000027673 | ENSFM00570000869305 | Unknown |
| ENSMUSG00000027673 | ENSFM00570000881141 | Unknown |
| ENSMUSG00000027673 | ENSFM00250000005807 | Nadh Dehydrogenase 1 Beta Subcomplex Subunit 5 Mitochondrial Precursor Complex I Sgdh Ci Sgdh Nadh Ubiquinone Oxidoreductase Sgdh Subunit |
| ENSMUSG00000027674 | ENSFM00250000001819 | Peroxisomal Targeting Signal 1 Receptor PTS1 Receptor PTS1R PTS1 Bp Peroxin 5 Peroxisomal C Terminal Targeting Signal Import Receptor Peroxisome Receptor 1 |
| ENSMUSG00000027679 | ENSFM00500000273595 | Mitochondrial Import Inner Membrane Translocase Subunit TIM14 Dnaj Homolog Subfamily C Member 19 |
| ENSMUSG00000027680 | ENSFM00800002020852 | Unknown |
| ENSMUSG00000027680 | ENSFM00800002023406 | Unknown |
| ENSMUSG00000027680 | ENSFM00250000000920 | Fragile X Mental Retardation 1 |
| ENSMUSG00000027692 | ENSFM00260000050335 | Kinase EC_2.7.11.1 Kinase Mapk/Erk Kinase Kinase Kinase Mek Kinase Kinase Mekkk Kinase |
| ENSMUSG00000027695 | ENSFM00740001589239 | Phospholipase D2 Pld 2 EC_3.1.4.4 Choline Phosphatase 2 Phosphatidylcholine Hydrolyzing Phospholipase D2 |
| ENSMUSG00000027698 | ENSFM00570000884077 | Unknown |
| ENSMUSG00000027698 | ENSFM00500000270047 | Neutral Cholesterol Ester Hydrolase 1 Nceh EC_3.1.1.- Arylacetamide Deacetylase 1 |
| ENSMUSG00000027709 | ENSFM00800002024464 | Unknown |
| ENSMUSG00000027709 | ENSFM00800002003420 | Methylcrotonoyl Coa Carboxylase Subunit Alpha Mitochondrial Precursor Mccase Subunit Alpha EC_6.4.1.4 3 Methylcrotonyl Coa Carboxylase 1 3 Methylcrotonyl Coa Carboxylase Biotin Containing Subunit 3 Methylcrotonyl Coa:Carbon Dioxide Ligase Subunit Alpha |
| ENSMUSG00000027710 | ENSFM00500000270065 | Very Long Chain Specific Acyl Coa Dehydrogenase Mitochondrial Precursor EC_1.3.8.9 |
| ENSMUSG00000027712 | ENSFM00750001632387 | Annexin Annexin Annexin Calphobindin Ii Lipocortin |
| ENSMUSG00000027737 | ENSFM00250000000331 | Amino Acid Transporter Solute Carrier Family 7 Member |
| ENSMUSG00000027739 | ENSFM00780001913222 | Unknown |
| ENSMUSG00000027739 | ENSFM00500000270315 | Ras Related Rab 33 |
| ENSMUSG00000027746 | ENSFM00520000531347 | Unknown |
| ENSMUSG00000027746 | ENSFM00570000863230 | Unknown |
| ENSMUSG00000027746 | ENSFM00670001236771 | Ubiquitin Fold Modifier 1 Precursor |
| ENSMUSG00000027774 | ENSFM00730001521750 | Elongation Factor G Mitochondrial |
| ENSMUSG00000027797 | ENSFM00700001407794 | Unknown |
| ENSMUSG00000027797 | ENSFM00400000131747 | Serine/Threonine Kinase EC_2.7.11.1 Doublecortin And Cam Kinase Doublecortin Kinase |
| ENSMUSG00000027799 | ENSFM00730001521650 | Neurobeachin |
| ENSMUSG00000027804 | ENSFM00780001913202 | Unknown |
| ENSMUSG00000027804 | ENSFM00730001521742 | Peptidyl Prolyl Cis Trans Isomerase D Ppiase D EC_5.2.1.8 Rotamase D |
| ENSMUSG00000027805 | ENSFM00670001235608 | Profilin Profilin |
| ENSMUSG00000027809 | ENSFM00250000005227 | Electron Transfer Flavoprotein Ubiquinone Oxidoreductase Mitochondrial Precursor Etf Qo Etf Ubiquinone Oxidoreductase EC_1.5.5.1 Electron Transferring Flavoprotein Dehydrogenase Etf Dehydrogenase |
| ENSMUSG00000027810 | ENSFM00570000894590 | Unknown |
| ENSMUSG00000027810 | ENSFM00570000901911 | Unknown |
| ENSMUSG00000027810 | ENSFM00780001898479 | Eukaryotic Translation Initiation Factor 2A Eif 2A |
| ENSMUSG00000027827 | ENSFM00780001898357 | Voltage Gated Potassium Channel Subunit Beta K + Channel Subunit Beta Kv Beta |
| ENSMUSG00000027828 | ENSFM00250000005568 | Translocon Associated Subunit Gamma Trap Gamma Signal Sequence Receptor Subunit Gamma Ssr Gamma |
| ENSMUSG00000027835 | ENSFM00590000916990 | Unknown |
| ENSMUSG00000027835 | ENSFM00250000004991 | Programmed Cell Death 10 |
| ENSMUSG00000027852 | ENSFM00570000878487 | Unknown |
| ENSMUSG00000027852 | ENSFM00800002017607 | Unknown |
| ENSMUSG00000027852 | ENSFM00730001521569 | Ras Precursor |
| ENSMUSG00000027858 | ENSFM00800002003930 | Tetraspanin 2 Tspan 2 |
| ENSMUSG00000027864 | ENSFM00570000851634 | Prostaglandin F2 Receptor Negative Regulator Precursor Prostaglandin F2 Alpha Receptor Regulatory Prostaglandin F2 Alpha Receptor Associated CD315 Antigen |
| ENSMUSG00000027879 | ENSFM00570000899167 | Unknown |
| ENSMUSG00000027879 | ENSFM00800002017956 | Unknown |
| ENSMUSG00000027879 | ENSFM00500000271789 | Vesicle Trafficking SEC22B Er Golgi Snare Of 24 Kda Ers 24 ERS24 SEC22 Vesicle Trafficking Homolog B |
| ENSMUSG00000027882 | ENSFM00800002010420 | Unknown |
| ENSMUSG00000027882 | ENSFM00250000001101 | Syntaxin Binding 1 Unc 18 Homolog 1 UNC18 1 Unc 18 Homolog A Unc 18A |
| ENSMUSG00000027884 | ENSFM00790001965928 | Unknown |
| ENSMUSG00000027884 | ENSFM00250000004452 | Chloride Channel Clic 1 Precursor Mid 1 Related Chloride Channel 1 |
| ENSMUSG00000027889 | ENSFM00770001847533 | Amp Deaminase EC_3.5.4.6 Amp Deaminase |
| ENSMUSG00000027893 | ENSFM00740001589092 | Adenosylhomocysteinase Adohcyase EC_3.3.1.1 S Adenosyl L Homocysteine Hydrolase 2 |
| ENSMUSG00000027894 | ENSFM00730001521463 | Sodium Dependent Neutral Amino Acid Transporter B 0 AT2 Sodium And Chloride Dependent Neurotransmitter Transporter NTT73 Solute Carrier Family 6 Member 15 Transporter V7 3 |
| ENSMUSG00000027935 | ENSFM00730001521184 | Ras Related Rab |
| ENSMUSG00000027940 | ENSFM00570000886978 | Unknown |
| ENSMUSG00000027940 | ENSFM00570000890222 | Unknown |
| ENSMUSG00000027940 | ENSFM00250000000220 | Tropomyosin Alpha 1 Chain Alpha Tropomyosin Tropomyosin 1 |
| ENSMUSG00000027942 | ENSFM00250000003053 | Uncharacterized |
| ENSMUSG00000027942 | ENSFM00670001253279 | Uncharacterized Homolog |
| ENSMUSG00000027942 | ENSFM00590000916953 | Unknown |
| ENSMUSG00000027962 | ENSFM00250000004468 | Vascular Cell Adhesion 1 Precursor V Cam 1 Vcam 1 CD106 Antigen |
| ENSMUSG00000027963 | ENSFM00500000271408 | Exostosin 2 EC_2.4.1.223 Alpha 1 4 N Acetylhexosaminyltransferase EXTL2 Alpha Galnact EXTL2 Ext Related 2 Glucuronyl Galactosyl Proteoglycan 4 Alpha N Acetylglucosaminyltransferase |
| ENSMUSG00000027965 | ENSFM00320000100099 | Noelin Precursor Neuronal Olfactomedin Related Er Localized Olfactomedin 1 |
| ENSMUSG00000027984 | ENSFM00730001522238 | Hydroxyacyl Coenzyme A Dehydrogenase Mitochondrial Precursor Hcdh EC_1.1.1.35 Medium And Short Chain L 3 Hydroxyacyl Coenzyme A Dehydrogenase Short Chain 3 Hydroxyacyl Coa Dehydrogenase |
| ENSMUSG00000027993 | ENSFM00570000883591 | Unknown |
| ENSMUSG00000027993 | ENSFM00750001632380 | Tripartite Motif Containing 2 EC_6.3.2.- E3 Ubiquitin Ligase TRIM2 |
| ENSMUSG00000028001 | ENSFM00250000004159 | Pre Splicing Factor PRP46 Pre Processing 46 |
| ENSMUSG00000028005 | ENSFM00730001521369 | Guanylate Cyclase Soluble Subunit Beta 1 Gcs Beta 1 EC_4.6.1.2 Guanylate Cyclase Soluble Subunit Beta 3 Gcs Beta 3 Soluble Guanylate Cyclase Small Subunit |
| ENSMUSG00000028013 | ENSFM00730001521782 | Inorganic Pyrophosphatase EC_3.6.1.1 Pyrophosphate Phospho Hydrolase Ppase |
| ENSMUSG00000028020 | ENSFM00570000886172 | Unknown |
| ENSMUSG00000028020 | ENSFM00570000893633 | Unknown |
| ENSMUSG00000028020 | ENSFM00400000131715 | Glycine Receptor Subunit Precursor Glycine Receptor Kda Subunit |
| ENSMUSG00000028029 | ENSFM00800002011480 | Unknown |
| ENSMUSG00000028029 | ENSFM00800002018118 | Unknown |
| ENSMUSG00000028029 | ENSFM00800002018119 | Unknown |
| ENSMUSG00000028029 | ENSFM00500000270832 | Aminoacyl Trna Synthase Complex Interacting Multifunctional 1 Multisynthase Complex Auxiliary Component P43 |
| ENSMUSG00000028033 | ENSFM00250000000417 | Potassium Voltage Gated Channel Subfamily Kqt Member Kqt Potassium Channel Subunit Alpha Voltage Gated Potassium Channel Subunit KV7 |
| ENSMUSG00000028035 | ENSFM00800002024518 | Unknown |
| ENSMUSG00000028035 | ENSFM00440000236855 | Dnaj Homolog Subfamily B Member |
| ENSMUSG00000028039 | ENSFM00500000271936 | Ephrin A3 Precursor EHK1 Ligand EHK1 L Eph Related Receptor Tyrosine Kinase Ligand 3 Lerk 3 |
| ENSMUSG00000028048 | ENSFM00250000003010 | Glucosylceramidase Precursor EC_3.2.1.45 Acid Beta Glucosidase Beta Glucocerebrosidase D Glucosyl N Acylsphingosine Glucohydrolase |
| ENSMUSG00000028049 | ENSFM00670001235365 | Secretory Carrier Associated Membrane Secretory Carrier Membrane |
| ENSMUSG00000028051 | ENSFM00250000000931 | Potassium/Sodium Hyperpolarization Activated Cyclic Nucleotide Gated Channel |
| ENSMUSG00000028059 | ENSFM00250000000618 | Rho Guanine Nucleotide Exchange Factor 2 Guanine Nucleotide Exchange Factor H1 Gef H1 |
| ENSMUSG00000028062 | ENSFM00520000554191 | Unknown |
| ENSMUSG00000028062 | ENSFM00670001235984 | Ragulator Complex LAMTOR2 Late Endosomal/Lysosomal Adaptor And Mapk And Mtor Activator 2 Roadblock Domain Containing 3 |
| ENSMUSG00000028063 | ENSFM00250000000425 | Lamin |
| ENSMUSG00000028064 | ENSFM00570000895576 | Unknown |
| ENSMUSG00000028064 | ENSFM00570000899480 | Unknown |
| ENSMUSG00000028064 | ENSFM00740001613578 | Unknown |
| ENSMUSG00000028064 | ENSFM00480000262746 | Semaphorin Precursor |
| ENSMUSG00000028070 | ENSFM00810002040729 | Nad P H Hydrate Epimerase |
| ENSMUSG00000028078 | ENSFM00400000131747 | Serine/Threonine Kinase EC_2.7.11.1 Doublecortin And Cam Kinase Doublecortin Kinase |
| ENSMUSG00000028081 | ENSFM00250000001462 | 40S Ribosomal S3A |
| ENSMUSG00000028093 | ENSFM00250000006412 | Lysophosphatidic Acid Phosphatase Type 6 Precursor EC_3.1.3.2 Acid Phosphatase 6 Lysophosphatidic Acid Phosphatase 1 PACPL1 |
| ENSMUSG00000028102 | ENSFM00570000895017 | Unknown |
| ENSMUSG00000028102 | ENSFM00670001235735 | Peroxisomal Membrane Peroxin Peroxisomal Biogenesis Factor PEX11 Homolog PEX11 |
| ENSMUSG00000028126 | ENSFM00730001521611 | Phosphatidylinositol 4 Phosphate 5 Kinase Type 1 Beta Pi Beta Ptdins 4 P 5 Kinase 1 Beta EC_2.7.1.68 Phosphatidylinositol 4 Phosphate 5 Kinase Type I Phosphatidylinositol 4 Phosphate 5 Kinase Beta |
| ENSMUSG00000028127 | ENSFM00800002024500 | Unknown |
| ENSMUSG00000028127 | ENSFM00800002003649 | Atp Binding Cassette Sub Family D Member Kda Peroxisomal Membrane PMP70 |
| ENSMUSG00000028128 | ENSFM00250000004048 | Tissue Factor Precursor Tf Coagulation Factor Iii CD142 Antigen |
| ENSMUSG00000028128 | ENSFM00680001306906 | Coagulation Factor Iii |
| ENSMUSG00000028134 | ENSFM00800002024494 | Unknown |
| ENSMUSG00000028134 | ENSFM00800002024495 | Unknown |
| ENSMUSG00000028134 | ENSFM00260000050389 | Polypyrimidine Tract Binding |
| ENSMUSG00000028136 | ENSFM00800002024472 | Unknown |
| ENSMUSG00000028136 | ENSFM00250000003280 | Sorting Nexin 27 |
| ENSMUSG00000028138 | ENSFM00800002024507 | Unknown |
| ENSMUSG00000028138 | ENSFM00800002024508 | Unknown |
| ENSMUSG00000028138 | ENSFM00250000000515 | Alcohol Dehydrogenase EC_1.1.1.1 Alcohol Dehydrogenase |
| ENSMUSG00000028145 | ENSFM00500000273840 | Acyl Coenzyme A Thioesterase THEM4 Precursor Acyl Coa Thioesterase THEM4 EC_3.1.2.2 Thioesterase Superfamily Member 4 |
| ENSMUSG00000028149 | ENSFM00800002017420 | Unknown |
| ENSMUSG00000028149 | ENSFM00250000002946 | RAP1 Gtpase Gdp Dissociation Stimulator 1 Exchange Factor Smggds Smg Gds Smg P21 Stimulatory Gdp/Gtp Exchange |
| ENSMUSG00000028161 | ENSFM00310000089007 | Serine/Threonine Phosphatase 2B Catalytic Subunit EC_3.1.3.16 Cam Prp Catalytic Subunit Calmodulin Dependent Calcineurin A Subunit |
| ENSMUSG00000028165 | ENSFM00600000921602 | Cdgsh Iron Sulfur Domain Containing 2 |
| ENSMUSG00000028176 | ENSFM00730001522141 | Leucine Rich Repeat Containing 7 Densin 180 Densin |
| ENSMUSG00000028184 | ENSFM00800002024517 | Unknown |
| ENSMUSG00000028184 | ENSFM00260000050328 | Latrophilin |
| ENSMUSG00000028194 | ENSFM00250000003817 | N G N G Dimethylarginine Dimethylaminohydrolase 2 Ddah 2 Dimethylarginine Dimethylaminohydrolase 2 EC_3.5.3.18 Ddahii Dimethylargininase 2 |
| ENSMUSG00000028207 | ENSFM00570000897930 | Unknown |
| ENSMUSG00000028207 | ENSFM00500000270363 | Aspartyl/Asparaginyl Beta Hydroxylase EC_1.14.11.16 |
| ENSMUSG00000028221 | ENSFM00250000004019 | Type Phosphatidylinositol 4 5 Bisphosphate 4 Phosphatase Type 2 Ptdins 4 5 P2 4 Ptase EC_3.1.3.78 Ptdins 4 5 P2 4 Ptase Ii Transmembrane 55A |
| ENSMUSG00000028222 | ENSFM00500000270302 | Calbindin Calbindin D28 D 28K Vitamin D Dependent Calcium Binding Protein Avian Type |
| ENSMUSG00000028223 | ENSFM00500000271494 | 2 4 Dienoyl Coa Reductase Mitochondrial Precursor EC_1.3.1.34 2 4 Dienoyl Coa Reductase |
| ENSMUSG00000028229 | ENSFM00500000271951 | Regulator Of Microtubule Dynamics 1 Rmd 1 FAM82B |
| ENSMUSG00000028232 | ENSFM00570000894942 | Unknown |
| ENSMUSG00000028232 | ENSFM00250000003959 | Transmembrane 68 |
| ENSMUSG00000028234 | ENSFM00670001235553 | 40S Ribosomal S20 |
| ENSMUSG00000028247 | ENSFM00500000271624 | Hexaprenyldihydroxybenzoate Methyltransferase Mitochondrial Precursor EC_2.1.1.114 2 Polyprenyl 6 Hydroxyphenol Methylase EC_2.1.1.- 222 3 4 Dihydroxy 5 Hexaprenylbenzoate Methyltransferase Dhhb Methyltransferase Dhhb Mt Dhhb Mtase 3 Demethylubiquinone 3 |
| ENSMUSG00000028249 | ENSFM00250000003553 | Syntenin Syndecan Binding |
| ENSMUSG00000028261 | ENSFM00500000273749 | Nadh Dehydrogenase 1 Alpha Subcomplex Assembly Factor 4 Hormone Regulated Proliferation Associated Of 20 Kda |
| ENSMUSG00000028273 | ENSFM00740001589114 | Pdz And Lim Domain |
| ENSMUSG00000028277 | ENSFM00670001235639 | Ubiquitin Conjugating Enzyme E2 J1 EC_6.3.2.19 Non Canonical Ubiquitin Conjugating Enzyme 1 Ncube 1 |
| ENSMUSG00000028292 | ENSFM00440000236914 | Arginine Trna Ligase EC_6.1.1.19 Arginyl Trna Synthetase Argrs |
| ENSMUSG00000028328 | ENSFM00640001116861 | Unknown |
| ENSMUSG00000028328 | ENSFM00500000269813 | Tropomodulin Tropomodulin Tmod |
| ENSMUSG00000028334 | ENSFM00250000006043 | Sialic Acid Synthase N Acetylneuraminate Synthase EC_2.5.1.56 N Acetylneuraminate 9 Phosphate Synthase EC_2.5.1.- 57 N Acetylneuraminic Acid Phosphate Synthase N Acetylneuraminic Acid Synthase |
| ENSMUSG00000028337 | ENSFM00730001521970 | Coronin |
| ENSMUSG00000028343 | ENSFM00250000004114 | Endoplasmic Reticulum Resident 44 Precursor Er 44 ERP44 Thioredoxin Domain Containing 4 |
| ENSMUSG00000028347 | ENSFM00250000001843 | Tomoregulin 1 Precursor Tr 1 Transmembrane With Egf And One Follistatin Domain |
| ENSMUSG00000028351 | ENSFM00250000001886 | Bmp/Retinoic Acid Inducible Neural Specific 1 Precursor Deleted In Bladder Cancer 1 |
| ENSMUSG00000028354 | ENSFM00250000001548 | Formin |
| ENSMUSG00000028364 | ENSFM00740001589132 | Tenascin Precursor Tn |
| ENSMUSG00000028367 | ENSFM00670001236147 | Thioredoxin Trx |
| ENSMUSG00000028373 | ENSFM00250000001299 | Astrotactin Precursor |
| ENSMUSG00000028383 | ENSFM00500000270535 | Hydroxysteroid Dehydrogenase 2 Ec 1 |
| ENSMUSG00000028385 | ENSFM00250000003712 | Sorting Nexin |
| ENSMUSG00000028393 | ENSFM00250000005243 | Delta Aminolevulinic Acid Dehydratase Aladh EC_4.2.1.24 Porphobilinogen Synthase |
| ENSMUSG00000028399 | ENSFM00640001120160 | Unknown |
| ENSMUSG00000028399 | ENSFM00640001120161 | Unknown |
| ENSMUSG00000028399 | ENSFM00740001589097 | Receptor Type Tyrosine Phosphatase Precursor EC_3.1.3.48 |
| ENSMUSG00000028402 | ENSFM00740001589189 | Multiple Pdz Domain Multi Pdz Domain 1 |
| ENSMUSG00000028405 | ENSFM00250000001292 | Cytoplasmic Aconitate Hydratase Aconitase EC_4.2.1.3 Citrate Hydro Lyase Iron Responsive Element Binding 1 Ire Bp 1 |
| ENSMUSG00000028410 | ENSFM00760001714614 | Dnaj Homolog Subfamily A Member Precursor |
| ENSMUSG00000028412 | ENSFM00500000270045 | Choline Transporter Solute Carrier Family 44 Member |
| ENSMUSG00000028434 | ENSFM00730001521622 | Band 4 1 |
| ENSMUSG00000028439 | ENSFM00250000007224 | Unknown |
| ENSMUSG00000028443 | ENSFM00500000272560 | Bis 5' Nucleosyl Tetraphosphatase EC_3.6.1.17 Diadenosine 5' 5''' P1 P4 Tetraphosphate Asymmetrical Hydrolase AP4A Hydrolase AP4AASE Diadenosine Tetraphosphatase Nucleoside Diphosphate Linked Moiety X Motif 2 Nudix Motif 2 |
| ENSMUSG00000028444 | ENSFM00570000894523 | Unknown |
| ENSMUSG00000028444 | ENSFM00250000003929 | Ciliary Neurotrophic Factor Receptor Subunit Alpha Precursor Cntf Receptor Subunit Alpha Cntfr Alpha |
| ENSMUSG00000028447 | ENSFM00250000007388 | Dynactin Subunit 3 |
| ENSMUSG00000028452 | ENSFM00750001632324 | Transitional Endoplasmic Reticulum Atpase Ter Atpase EC_3.6.4.6 15S Mg 2+ Atpase P97 Subunit Valosin Containing Vcp |
| ENSMUSG00000028455 | ENSFM00260000050877 | Stomatin 2 Mitochondrial Precursor Slp 2 |
| ENSMUSG00000028456 | ENSFM00250000000599 | Unc 13 Homolog MUNC13 |
| ENSMUSG00000028465 | ENSFM00500000330223 | Unknown |
| ENSMUSG00000028465 | ENSFM00250000001103 | Talin |
| ENSMUSG00000028470 | ENSFM00670001236173 | Histidine Triad Nucleotide Binding 2 Mitochondrial Precursor Hint 2 Ec 3 Hint 3 |
| ENSMUSG00000028478 | ENSFM00250000001836 | Clathrin Light Chain |
| ENSMUSG00000028480 | ENSFM00500000271973 | Golgi Associated Plant Pathogenesis Related 1 Gapr 1 Golgi Associated Pr 1 Glioma Pathogenesis Related 2 Glipr 2 |
| ENSMUSG00000028484 | ENSFM00570000899071 | Unknown |
| ENSMUSG00000028484 | ENSFM00500000271092 | PC4 And SFRS1 Interacting Lens Epithelium Derived Growth Factor |
| ENSMUSG00000028488 | ENSFM00250000000986 | Endophilin SH3 Domain Containing GRB2 |
| ENSMUSG00000028495 | ENSFM00250000001600 | 40S Ribosomal S6 |
| ENSMUSG00000028517 | ENSFM00260000050433 | Lipid Phosphate Phosphohydrolase EC_3.1.3.4 PAP2 Phosphatidate Phosphohydrolase Type Phosphatidic Acid Phosphatase Pap |
| ENSMUSG00000028519 | ENSFM00740001589270 | Disabled Homolog 2 Adaptor Molecule Disabled 2 Differentially Expressed In Ovarian Carcinoma 2 Doc 2 |
| ENSMUSG00000028524 | ENSFM00570000890494 | Unknown |
| ENSMUSG00000028524 | ENSFM00600000921280 | Fch Domain Only |
| ENSMUSG00000028525 | ENSFM00640001116163 | Unknown |
| ENSMUSG00000028525 | ENSFM00400000131721 | Camp Specific 3' 5' Cyclic Phosphodiesterase EC_3.1.4.53 |
| ENSMUSG00000028527 | ENSFM00500000270040 | Adenylate Kinase 4 Mitochondrial |
| ENSMUSG00000028528 | ENSFM00250000001678 | Tyrosine Phosphatase Auxilin EC_3.1.3.48 Dnaj Homolog Subfamily C Member 6 |
| ENSMUSG00000028530 | ENSFM00570000894607 | Unknown |
| ENSMUSG00000028530 | ENSFM00250000000777 | Tyrosine Kinase EC_2.7.10.2 Janus Kinase Jak |
| ENSMUSG00000028542 | ENSFM00550000769437 | Unknown |
| ENSMUSG00000028542 | ENSFM00610000973577 | Unknown |
| ENSMUSG00000028542 | ENSFM00730001521205 | Sodium And Chloride Dependent Glycine Transporter Glyt Solute Carrier Family 6 Member |
| ENSMUSG00000028546 | ENSFM00500000353790 | Unknown |
| ENSMUSG00000028546 | ENSFM00250000000588 | Elav |
| ENSMUSG00000028552 | ENSFM00250000001830 | Epidermal Growth Factor Receptor Substrate 15 1 EPS15 Related EPS15R |
| ENSMUSG00000028552 | ENSFM00720001494248 | Epidermal Growth Factor Receptor Substrate 15 |
| ENSMUSG00000028556 | ENSFM00500000269778 | Dedicator Of Cytokinesis |
| ENSMUSG00000028567 | ENSFM00320000100209 | Thioredoxin Domain Containing 12 Precursor EC_1.8.4.2 Endoplasmic Reticulum Resident 19 Er 19 ERP19 Thioredoxin P19 |
| ENSMUSG00000028603 | ENSFM00250000003026 | Non Specific Lipid Transfer Nsl Tp EC_2.3.1.176 Propanoyl Coa C Acyltransferase Scp Chi Scpx Sterol Carrier 2 Scp 2 Sterol Carrier X Scp X |
| ENSMUSG00000028607 | ENSFM00730001521962 | Carnitine O Palmitoyltransferase 2 Mitochondrial Precursor EC_2.3.1.21 Carnitine Palmitoyltransferase Ii Cpt Ii |
| ENSMUSG00000028614 | ENSFM00680001326889 | Unknown |
| ENSMUSG00000028614 | ENSFM00250000005168 | Nucleoporin NDC1 Transmembrane 48 |
| ENSMUSG00000028622 | ENSFM00250000006448 | 39S Ribosomal L37 Mitochondrial Precursor L37MT Mrp L37 |
| ENSMUSG00000028639 | ENSFM00670001235454 | Nuclease Sensitive Element Binding 1 Ccaat Binding Transcription Factor I Subunit A Cbf A Enhancer Factor I Subunit A Efi A Y Box Transcription Factor Y Box Binding 1 Yb 1 |
| ENSMUSG00000028645 | ENSFM00730001521168 | Solute Carrier Family 2 Facilitated Glucose Transporter Member Glucose Transporter Type Glut |
| ENSMUSG00000028648 | ENSFM00670001236390 | Nadh Dehydrogenase Iron Sulfur 5 Complex I 15 Kda Ci 15 Kda Nadh Ubiquinone Oxidoreductase 15 Kda Subunit |
| ENSMUSG00000028656 | ENSFM00250000001899 | Adenylyl Cyclase Associated Cap |
| ENSMUSG00000028657 | ENSFM00640001104958 | Unknown |
| ENSMUSG00000028657 | ENSFM00760001714836 | Palmitoyl Thioesterase 1 Precursor Ppt 1 EC_3.1.2.22 Palmitoyl Hydrolase 1 |
| ENSMUSG00000028664 | ENSFM00730001521338 | Ephrin Type B Receptor Kinase |
| ENSMUSG00000028672 | ENSFM00250000002331 | Hydroxymethylglutaryl Coa Lyase Mitochondrial Precursor Hl Hmg Coa Lyase EC_4.1.3.4 3 Hydroxy 3 Methylglutarate Coa Lyase |
| ENSMUSG00000028684 | ENSFM00250000004160 | Uroporphyrinogen Decarboxylase Upd Uro D EC_4.1.1.37 |
| ENSMUSG00000028691 | ENSFM00250000000757 | Thioredoxin Peroxidase Thioredoxin Dependent Peroxide Reductase |
| ENSMUSG00000028692 | ENSFM00730001521171 | Reductase Aldo Keto Reductase Family 1 Member |
| ENSMUSG00000028719 | ENSFM00570000884722 | Unknown |
| ENSMUSG00000028719 | ENSFM00570000897069 | Unknown |
| ENSMUSG00000028719 | ENSFM00770001847583 | Ump Cmp Kinase |
| ENSMUSG00000028737 | ENSFM00800002003550 | Delta 1 Pyrroline 5 Carboxylate Dehydrogenase Mitochondrial Precursor P5C Dehydrogenase EC_1.2.1.88 Aldehyde Dehydrogenase Family 4 Member A1 L Glutamate Gamma Semialdehyde Dehydrogenase |
| ENSMUSG00000028743 | ENSFM00760001715034 | Aflatoxin B1 Aldehyde Reductase Member 2 EC_1.1.1.- N11 Succinic Semialdehyde Reductase Ssa Reductase |
| ENSMUSG00000028745 | ENSFM00250000003778 | F Actin Capping Subunit Beta |
| ENSMUSG00000028753 | ENSFM00740001589353 | Von Willebrand Factor A Domain Containing |
| ENSMUSG00000028757 | ENSFM00250000004027 | Dolichyl Diphosphooligosaccharide Glycosyltransferase 48 Kda Subunit Precursor Ddost 48 Kda Subunit Oligosaccharyl Transferase 48 Kda Subunit EC_2.4.99.18 |
| ENSMUSG00000028759 | ENSFM00250000005391 | Heterochromatin 1 Binding 3 |
| ENSMUSG00000028760 | ENSFM00250000000634 | Eukaryotic Translation Initiation Factor 4 Gamma 3 Eif 4 Gamma 3 Eif 4G 3 EIF4G 3 Eif 4 Gamma Ii EIF4GII |
| ENSMUSG00000028766 | ENSFM00250000000564 | Alkaline Phosphatase Precursor EC_3.1.3.1 Alkaline Phosphatase |
| ENSMUSG00000028773 | ENSFM00800002003523 | Fatty Acid Binding Protein Heart Fatty Acid Binding 3 Heart Type Fatty Acid Binding H Fabp |
| ENSMUSG00000028782 | ENSFM00250000000737 | Brain Specific Angiogenesis Inhibitor Precursor |
| ENSMUSG00000028785 | ENSFM00570000880762 | Unknown |
| ENSMUSG00000028785 | ENSFM00760001714608 | Unknown |
| ENSMUSG00000028788 | ENSFM00250000001523 | Tyrosine Phosphatase Type Iva 1 Precursor EC_3.1.3.48 Tyrosine Phosphatase 4A1 Tyrosine Phosphatase Of Regenerating Liver 1 Prl 1 |
| ENSMUSG00000028790 | ENSFM00400000131768 | Kh Domain Containing Rna Binding Signal Transduction Associated SAM68 Mammalian Slm |
| ENSMUSG00000028792 | ENSFM00570000888519 | Unknown |
| ENSMUSG00000028792 | ENSFM00810002040800 | Adenylate Kinase |
| ENSMUSG00000028804 | ENSFM00740001589158 | Cub And Sushi Domain Containing Cub And Sushi Multiple Domains |
| ENSMUSG00000028811 | ENSFM00570000885949 | Unknown |
| ENSMUSG00000028811 | ENSFM00500000270186 | Tyrosine Trna Ligase Cytoplasmic EC_6.1.1.1 Tyrosyl Trna Synthetase Tyrrs |
| ENSMUSG00000028820 | ENSFM00250000000671 | Paraspeckle Component 1 |
| ENSMUSG00000028826 | ENSFM00590000917330 | Unknown |
| ENSMUSG00000028826 | ENSFM00250000002769 | Macoilin Transmembrane |
| ENSMUSG00000028830 | ENSFM00250000002086 | Dyslexia Associated |
| ENSMUSG00000028832 | ENSFM00800002003794 | Stathmin |
| ENSMUSG00000028833 | ENSFM00250000005281 | Neurochondrin |
| ENSMUSG00000028837 | ENSFM00750001632406 | Proteasome Subunit Beta Type 2 EC_3.4.25.1 Macropain Subunit C7 I Multicatalytic Endopeptidase Complex Subunit C7 I Proteasome Component C7 I |
| ENSMUSG00000028843 | ENSFM00470000254285 | Unknown |
| ENSMUSG00000028843 | ENSFM00670001236675 | SH3 Domain Binding Glutamic Acid Rich 3 |
| ENSMUSG00000028847 | ENSFM00250000005557 | Trafficking Particle Complex Subunit 3 |
| ENSMUSG00000028849 | ENSFM00250000004934 | MAP7 Domain Containing 1 |
| ENSMUSG00000028851 | ENSFM00670001235568 | Nuclear Migration Nudc Nuclear Distribution C Homolog |
| ENSMUSG00000028854 | ENSFM00800002003389 | Sodium/Hydrogen Exchanger 1 Na + /H + Exchanger 1 Nhe 1 Solute Carrier Family 9 Member 1 |
| ENSMUSG00000028857 | ENSFM00570000878386 | Unknown |
| ENSMUSG00000028857 | ENSFM00570000888828 | Unknown |
| ENSMUSG00000028857 | ENSFM00570000900673 | Unknown |
| ENSMUSG00000028857 | ENSFM00250000005108 | Transmembrane 222 |
| ENSMUSG00000028868 | ENSFM00250000001269 | Wiskott Aldrich Syndrome Family Member 1 Wasp Family Member 1 Wave 1 |
| ENSMUSG00000028879 | ENSFM00500000270193 | Syntaxin |
| ENSMUSG00000028909 | ENSFM00310000088999 | Receptor Type Tyrosine Phosphatase Precursor R Ptp EC_3.1.3.48 Receptor Type Tyrosine Phosphatase |
| ENSMUSG00000028910 | ENSFM00800002003721 | Trans 2 Enoyl Coa Reductase Mitochondrial Precursor EC_1.3.1.38 |
| ENSMUSG00000028927 | ENSFM00250000001768 | Arginine Deiminase Type EC_3.5.3.15 Peptidylarginine Deiminase Arginine Deiminase Type |
| ENSMUSG00000028931 | ENSFM00590000917429 | Unknown |
| ENSMUSG00000028931 | ENSFM00780001898357 | Voltage Gated Potassium Channel Subunit Beta K + Channel Subunit Beta Kv Beta |
| ENSMUSG00000028932 | ENSFM00730001521776 | 26S Protease Regulatory Subunit 7 26S Proteasome Aaa Atpase Subunit RPT1 Proteasome 26S Subunit Atpase 2 |
| ENSMUSG00000028936 | ENSFM00600000921476 | 60S Ribosomal L22 |
| ENSMUSG00000028937 | ENSFM00250000005415 | Cytosolic Acyl Coenzyme A Thioester Hydrolase EC_3.1.2.2 Acyl Coa Thioesterase 7 Brain Acyl Coa Hydrolase Bach Cte Iia Cte Ii Long Chain Acyl Coa Thioester Hydrolase |
| ENSMUSG00000028944 | ENSFM00570000892600 | Unknown |
| ENSMUSG00000028944 | ENSFM00250000001075 | 5' Amp Activated Kinase Subunit Gamma Ampk Ampk Subunit Gamma |
| ENSMUSG00000028945 | ENSFM00810002040784 | Gtpase RHEBL1 Precursor Ras Homolog Enriched In Brain 1 Rheb 1 |
| ENSMUSG00000028953 | ENSFM00730001521705 | Atp Binding Cassette Sub Family F Member 2 |
| ENSMUSG00000028955 | ENSFM00670001236814 | Vesicle Associated Membrane 3 Vamp 3 Synaptobrevin 3 |
| ENSMUSG00000028958 | ENSFM00500000273949 | Transmembrane And Ubiquitin Domain Containing 1 Hepatocyte Odd Shuttling |
| ENSMUSG00000028961 | ENSFM00540000717911 | 6 Phosphogluconate Dehydrogenase Decarboxylating EC_1.1.1.44 |
| ENSMUSG00000028964 | ENSFM00670001235693 | Dj 1 Precursor EC_3.4.-.- Parkinson Disease 7 Homolog |
| ENSMUSG00000028969 | ENSFM00730001521798 | Negative Regulator Of The Pho System Serine/Threonine Kinase PHO85 |
| ENSMUSG00000028973 | ENSFM00800002023566 | Unknown |
| ENSMUSG00000028973 | ENSFM00800002003665 | Atp Binding Cassette Sub Family B Member 8 Mitochondrial Precursor |
| ENSMUSG00000028975 | ENSFM00570000893486 | Unknown |
| ENSMUSG00000028975 | ENSFM00250000007000 | Peroxisomal Membrane PEX14 PTS1 Receptor Docking Peroxin 14 Peroxisomal Membrane Anchor PEX14 |
| ENSMUSG00000028991 | ENSFM00260000050608 | Serine/Threonine Kinase EC_2.7.11.1 Rapamycin 1 Target Of Rapamycin 1 |
| ENSMUSG00000028995 | ENSFM00250000003532 | Hyccin Down Regulated By CTNNB1 A FAM126A |
| ENSMUSG00000029016 | ENSFM00570000895521 | Unknown |
| ENSMUSG00000029016 | ENSFM00740001589241 | H + /Cl Exchange Transporter 7 Chloride Channel 7 Alpha Subunit Chloride Channel 7 Clc 7 |
| ENSMUSG00000029020 | ENSFM00250000001660 | Transmembrane Gtpase |
| ENSMUSG00000029028 | ENSFM00610000961855 | Unknown |
| ENSMUSG00000029028 | ENSFM00250000006095 | Leucine Rich Repeat Containing 47 |
| ENSMUSG00000029030 | ENSFM00250000004936 | Tumor P63 Regulated Gene 1 Mossy Fiber Terminal Associated Vertebrate Specific Presynaptic Protein FAM79A |
| ENSMUSG00000029033 | ENSFM00250000000926 | Arf Gap With Coiled Coil Ank Repeat And Ph Domain Containing Centaurin Beta Cnt |
| ENSMUSG00000029036 | ENSFM00740001603337 | Unknown |
| ENSMUSG00000029036 | ENSFM00250000002488 | Atpase Family Aaa Domain Containing 3 |
| ENSMUSG00000029048 | ENSFM00250000005251 | RER1 |
| ENSMUSG00000029055 | ENSFM00570000893447 | Unknown |
| ENSMUSG00000029055 | ENSFM00740001598015 | Unknown |
| ENSMUSG00000029055 | ENSFM00770001847600 | 1 Phosphatidylinositol 4 5 Bisphosphate Phosphodiesterase Eta 2 EC_3.1.4.11 Phosphoinositide Phospholipase C Eta 2 Phosphoinositide Phospholipase C 4 Plc L4 Phospholipase C 4 Phospholipase C Eta 2 Plc ETA2 |
| ENSMUSG00000029059 | ENSFM00250000007632 | Prostamide/Prostaglandin F Synthase Prostamide/Pg F Synthase Prostamide/Pgf Synthase EC_1.11.1.20 |
| ENSMUSG00000029064 | ENSFM00250000000545 | Guanine Nucleotide Binding G I /G S /G T Subunit Beta Transducin Beta Chain |
| ENSMUSG00000029084 | ENSFM00500000272366 | Adp Ribosyl Cyclase/Cyclic Adp Ribose Hydrolase 1 EC_3.2.2.6 2' Phospho Adp Ribosyl Cyclase 2' Phospho Adp Ribosyl Cyclase/2' Phospho Cyclic Adp Ribose Transferase EC_2.4.99.- 20 2' Phospho Cyclic Adp Ribose Transferase Adp Ribosyl Cyclase 1 Adprc 1 Cycli |
| ENSMUSG00000029086 | ENSFM00800002020433 | Unknown |
| ENSMUSG00000029086 | ENSFM00250000001124 | Prominin 2 Precursor Prom 2 Prominin 2 |
| ENSMUSG00000029088 | ENSFM00410000138464 | Kv Channel Interacting A Type Potassium Channel Modulatory Potassium Channel Interacting |
| ENSMUSG00000029093 | ENSFM00250000000793 | VPS10 Domain Containing Receptor Precursor |
| ENSMUSG00000029095 | ENSFM00810002057443 | Unknown |
| ENSMUSG00000029095 | ENSFM00250000000459 | Actin Binding Lim Ablim Actin Binding Lim Family Member |
| ENSMUSG00000029098 | ENSFM00570000879084 | Unknown |
| ENSMUSG00000029098 | ENSFM00730001522151 | Peroxisomal Acyl Coenzyme A Oxidase 3 EC_1.3.3.6 Branched Chain Acyl Coa Oxidase Brcacox Pristanoyl Coa Oxidase |
| ENSMUSG00000029101 | ENSFM00570000887105 | Unknown |
| ENSMUSG00000029101 | ENSFM00810002056023 | Unknown |
| ENSMUSG00000029101 | ENSFM00250000000973 | Regulator Of G Signaling |
| ENSMUSG00000029103 | ENSFM00250000004723 | Alpha 2 Macroglobulin Receptor Associated Precursor Alpha 2 Mrap Low Density Lipoprotein Receptor Related Associated 1 Rap |
| ENSMUSG00000029104 | ENSFM00250000003199 | Huntingtin Huntington Disease Homolog Hd Homolog |
| ENSMUSG00000029106 | ENSFM00250000000681 | Adducin |
| ENSMUSG00000029108 | ENSFM00760001714757 | Protocadherin Precursor Protocadherin |
| ENSMUSG00000029112 | ENSFM00500000277777 | NK1 Transcription Factor Related 1 Homeobox 153 Hpx 153 Homeobox Sax 2 Nkx 1 1 |
| ENSMUSG00000029120 | ENSFM00810002057415 | Unknown |
| ENSMUSG00000029120 | ENSFM00250000000829 | Serine/Threonine Phosphatase 2A 55 Kda Regulatory Subunit B PP2A Subunit B B55 PP2A Subunit B PR55 PP2A Subunit B R2 PP2A Subunit B |
| ENSMUSG00000029121 | ENSFM00250000000643 | Dihydropyrimidinase Related Drp Collapsin Response Mediator Crmp |
| ENSMUSG00000029125 | ENSFM00250000005997 | Syntaxin 18 |
| ENSMUSG00000029126 | ENSFM00810002055513 | Unknown |
| ENSMUSG00000029126 | ENSFM00810002055514 | Unknown |
| ENSMUSG00000029126 | ENSFM00250000005181 | Neuron Specific Family Member |
| ENSMUSG00000029131 | ENSFM00800002023572 | Unknown |
| ENSMUSG00000029131 | ENSFM00800002023573 | Unknown |
| ENSMUSG00000029131 | ENSFM00570000851144 | Dnaj Homolog Subfamily B Member |
| ENSMUSG00000029151 | ENSFM00810002057419 | Unknown |
| ENSMUSG00000029151 | ENSFM00810002057420 | Unknown |
| ENSMUSG00000029151 | ENSFM00250000001206 | Zinc Transporter Znt Solute Carrier Family 30 Member |
| ENSMUSG00000029152 | ENSFM00810002056010 | Unknown |
| ENSMUSG00000029152 | ENSFM00810002057449 | Unknown |
| ENSMUSG00000029152 | ENSFM00500000272374 | Ocia Domain Containing 1 |
| ENSMUSG00000029153 | ENSFM00810002056007 | Unknown |
| ENSMUSG00000029153 | ENSFM00500000274958 | Ocia Domain Containing 2 |
| ENSMUSG00000029166 | ENSFM00250000000867 | Microtubule Associated Rp/Eb Family Member |
| ENSMUSG00000029168 | ENSFM00250000000643 | Dihydropyrimidinase Related Drp Collapsin Response Mediator Crmp |
| ENSMUSG00000029169 | ENSFM00810002040596 | Pre Splicing Factor Atp Dependent Rna Helicase EC_3.6.4.13 Deah Box |
| ENSMUSG00000029198 | ENSFM00500000272223 | Grpe Homolog 1 Mitochondrial Precursor Mt Grpe#1 |
| ENSMUSG00000029211 | ENSFM00800002023605 | Unknown |
| ENSMUSG00000029211 | ENSFM00800002023606 | Unknown |
| ENSMUSG00000029211 | ENSFM00800002003422 | Gamma Aminobutyric Acid Receptor Subunit Alpha Precursor Gaba A Receptor Subunit Alpha |
| ENSMUSG00000029212 | ENSFM00400000131718 | Gamma Aminobutyric Acid Receptor Subunit Beta Precursor Gaba A Receptor Subunit Beta |
| ENSMUSG00000029219 | ENSFM00800002003969 | Sodium/Bile Acid Cotransporter 4 Na + /Bile Acid Cotransporter 4 Solute Carrier Family 10 Member 4 |
| ENSMUSG00000029221 | ENSFM00810002055818 | Unknown |
| ENSMUSG00000029221 | ENSFM00810002057448 | Unknown |
| ENSMUSG00000029221 | ENSFM00250000002884 | Zinc Transporter 9 Znt 9 Solute Carrier Family 30 Member 9 |
| ENSMUSG00000029223 | ENSFM00550000743168 | Ubiquitin Carboxyl Terminal Hydrolase Isozyme L1 Precursor Uch L1 EC_3.4.19.12 Ec 6 Ubiquitin Thioesterase L1 |
| ENSMUSG00000029231 | ENSFM00730001521867 | Platelet Derived Growth Factor Receptor Precursor Pdgf R Pdgfr EC_2.7.10.1 Platelet Derived Growth Factor Receptor Type Platelet Derived Growth Factor Receptor CD140 Antigen Family Member Platelet Derived Growth Factor Receptor Antigen |
| ENSMUSG00000029234 | ENSFM00500000271648 | Transmembrane 165 Precursor Transmembrane Tparl |
| ENSMUSG00000029247 | ENSFM00250000003230 | Multifunctional Phosphoribosylaminoimidazole Succinocarboxamide Synthase EC_6.3.2.6 Saicar Synthetase |
| ENSMUSG00000029247 | ENSFM00570000894579 | Multifunctional ADE2 |
| ENSMUSG00000029247 | ENSFM00570000901681 | Multifunctional ADE2 |
| ENSMUSG00000029291 | ENSFM00250000001058 | Run And Fyve Domain Containing |
| ENSMUSG00000029309 | ENSFM00250000001867 | Sparc Precursor Basement Membrane 40 Bm 40 Osteonectin On Secreted Acidic And Rich In Cysteine |
| ENSMUSG00000029311 | ENSFM00500000269766 | Dehydrogenase |
| ENSMUSG00000029330 | ENSFM00750001632370 | Phosphatidate Cytidylyltransferase 2 EC_2.7.7.41 Cdp Dag Synthase 2 Cdp Dg Synthase 2 Cdp Diacylglycerol Synthase 2 Cds 2 Cdp Diglyceride Pyrophosphorylase 2 Cdp Diglyceride Synthase 2 Ctp:Phosphatidate Cytidylyltransferase 2 |
| ENSMUSG00000029334 | ENSFM00400000131755 | Cgmp Dependent Kinase Cgk EC_2.7.11.12 Cgmp Dependent Kinase |
| ENSMUSG00000029348 | ENSFM00800002016787 | Unknown |
| ENSMUSG00000029348 | ENSFM00800002016788 | Unknown |
| ENSMUSG00000029348 | ENSFM00800002023638 | Unknown |
| ENSMUSG00000029348 | ENSFM00250000005876 | Aspartate Beta Hydroxylase Domain Containing EC_1.14.11.- |
| ENSMUSG00000029359 | ENSFM00250000006459 | Calcineurin B Homologous 3 Tescalcin |
| ENSMUSG00000029361 | ENSFM00720001501689 | Unknown |
| ENSMUSG00000029361 | ENSFM00470000251434 | Nitric Oxide Synthase EC_1.14.13.39 Nos Nos Type Peptidyl Cysteine S Nitrosylase |
| ENSMUSG00000029368 | ENSFM00250000001052 | Precursor |
| ENSMUSG00000029390 | ENSFM00800002023673 | Unknown |
| ENSMUSG00000029390 | ENSFM00500000270937 | Transmembrane EMP24 Domain Containing 2 Precursor Copi Coated Vesicle Membrane P24 P24 Family Beta 1 P24BETA1 |
| ENSMUSG00000029405 | ENSFM00250000002045 | Ras Gtpase Activating Binding 1 G3BP 1 EC_3.6.4.12 EC_3.6.4.- 13 Atp Dependent Helicase |
| ENSMUSG00000029406 | ENSFM00590000917808 | Unknown |
| ENSMUSG00000029406 | ENSFM00500000269931 | Membrane Associated Phosphatidylinositol Transfer Phosphatidylinositol Transfer Protein Membrane Associated Pitpnm PYK2 N Terminal Domain Interacting Receptor Nir |
| ENSMUSG00000029407 | ENSFM00810002056372 | Unknown |
| ENSMUSG00000029407 | ENSFM00500000270803 | General Vesicular Transport Factor P115 USO1 Homolog Transcytosis Associated Tap Vesicle Docking |
| ENSMUSG00000029408 | ENSFM00730001521297 | Antigen Peptide Transporter 1 APT1 Atp Binding Cassette Sub Family B Member 2 Peptide Transporter TAP1 |
| ENSMUSG00000029419 | ENSFM00500000276193 | Uncharacterized |
| ENSMUSG00000029419 | ENSFM00760001758450 | Unknown |
| ENSMUSG00000029420 | ENSFM00740001589344 | Peripheral Type Benzodiazepine Receptor Associated 1 Prax 1 Rims Binding 1 Rim BP1 |
| ENSMUSG00000029426 | ENSFM00800002003823 | Lysosome Membrane 2.85 Kda Lysosomal Membrane Sialoglycoprotein LGP85 CD36 Antigen 2 Lysosome Membrane Ii Limp Ii Scavenger Receptor Class B Member 2 CD36 Antigen |
| ENSMUSG00000029428 | ENSFM00800002023682 | Unknown |
| ENSMUSG00000029428 | ENSFM00250000000549 | Syntaxin |
| ENSMUSG00000029430 | ENSFM00250000002338 | Gtp Binding Nuclear Ran Gtpase Ran Ras Related Nuclear |
| ENSMUSG00000029432 | ENSFM00250000003191 | Nipsnap Homolog 2 NIPSNAP2 Glioblastoma Amplified Sequence |
| ENSMUSG00000029433 | ENSFM00800002023665 | Unknown |
| ENSMUSG00000029433 | ENSFM00250000004873 | Diablo Homolog Mitochondrial |
| ENSMUSG00000029434 | ENSFM00540000717941 | Vacuolar Sorting Associated |
| ENSMUSG00000029436 | ENSFM00760001714959 | Matrix Metalloproteinase 17 Precursor Mmp 17 EC_3.4.24.- Membrane Type Matrix Metalloproteinase 4 Mt Mmp 4 MTMMP4 Membrane Type 4 Matrix Metalloproteinase MT4 Mmp MT4MMP |
| ENSMUSG00000029449 | ENSFM00800002003736 | Rho Related Gtp Binding Precursor |
| ENSMUSG00000029455 | ENSFM00730001521185 | Dehydrogenase Aldehyde Dehydrogenase Family 1 Member Dehydrogenase |
| ENSMUSG00000029462 | ENSFM00520000542186 | Unknown |
| ENSMUSG00000029462 | ENSFM00570000870191 | Unknown |
| ENSMUSG00000029462 | ENSFM00570000892720 | Unknown |
| ENSMUSG00000029462 | ENSFM00250000005075 | Vacuolar Sorting Associated 29 Vesicle Sorting 29 |
| ENSMUSG00000029465 | ENSFM00500000270831 | Actin Related 2/3 Complex Subunit 3 ARP2/3 Complex 21 Kda Subunit P21 Arc |
| ENSMUSG00000029467 | ENSFM00500000269652 | Sarcoplasmic/Endoplasmic Reticulum Calcium Atpase Sr Ca 2+ Atpase EC_3.6.3.8 Calcium Pump Calcium Transporting Atpase Sarcoplasmic Reticulum Type Twitch Skeletal Muscle Endoplasmic Reticulum Class 1/2 Ca 2+ Atpase |
| ENSMUSG00000029471 | ENSFM00800002023662 | Unknown |
| ENSMUSG00000029471 | ENSFM00800002023663 | Unknown |
| ENSMUSG00000029471 | ENSFM00760001714763 | Calcium/Calmodulin Dependent Kinase Kinase 1 Cam Kk 1 Cam Kinase Kinase 1 Camkk 1 EC_2.7.11.17 Cam Kinase Iv Kinase Calcium/Calmodulin Dependent Kinase Kinase Alpha Cam Kk Alpha Cam Kinase Kinase Alpha Camkk Alpha |
| ENSMUSG00000029500 | ENSFM00250000004793 | Serine/Threonine Phosphatase PGAM5 Mitochondrial |
| ENSMUSG00000029502 | ENSFM00250000007244 | Golgin Subfamily A Member 3 Golgin 160 |
| ENSMUSG00000029516 | ENSFM00660001188751 | Unknown |
| ENSMUSG00000029516 | ENSFM00520000517879 | Citron Rho Interacting Kinase EC_2.7.11.1 Serine/Threonine Kinase 21 |
| ENSMUSG00000029518 | ENSFM00720001503436 | Unknown |
| ENSMUSG00000029518 | ENSFM00750001632414 | Ras Related Rab 35 |
| ENSMUSG00000029571 | ENSFM00570000885262 | Unknown |
| ENSMUSG00000029571 | ENSFM00600000921356 | Transmembrane |
| ENSMUSG00000029575 | ENSFM00250000007426 | Cob I Yrinic Acid A C Diamide Adenosyltransferase Mitochondrial Precursor EC_2.5.1.17 Cob I Alamin Adenosyltransferase Methylmalonic Aciduria Type B |
| ENSMUSG00000029580 | ENSFM00730001521060 | Actin |
| ENSMUSG00000029581 | ENSFM00250000001972 | Fascin |
| ENSMUSG00000029592 | ENSFM00250000005542 | Ubiquitin Carboxyl Terminal Hydrolase 30 EC_3.4.19.12 Deubiquitinating Enzyme 30 Ubiquitin Thioesterase 30 Ubiquitin Specific Processing Protease 30 Ub Specific Protease 30 |
| ENSMUSG00000029602 | ENSFM00570000879652 | Unknown |
| ENSMUSG00000029602 | ENSFM00760001714919 | Ras Gtpase Activating 4 Calcium Promoted Ras Inactivator Ras P21 Activator 4 Rasgap Activating 2 |
| ENSMUSG00000029608 | ENSFM00810002057459 | Unknown |
| ENSMUSG00000029608 | ENSFM00260000050490 | Double C2 Domain Containing DOC2 |
| ENSMUSG00000029610 | ENSFM00250000006128 | Aminoacyl Trna Synthase Complex Interacting Multifunctional 2 Multisynthase Complex Auxiliary Component P38 Jtv 1 |
| ENSMUSG00000029614 | ENSFM00810002057460 | Unknown |
| ENSMUSG00000029614 | ENSFM00500000270597 | 60S Ribosomal L6 |
| ENSMUSG00000029616 | ENSFM00500000326708 | Unknown |
| ENSMUSG00000029616 | ENSFM00570000865410 | Unknown |
| ENSMUSG00000029616 | ENSFM00570000885583 | Unknown |
| ENSMUSG00000029616 | ENSFM00250000008476 | Endoplasmic Reticulum Resident 29 Precursor ERP29 Endoplasmic Reticulum Resident 31 ERP31 |
| ENSMUSG00000029617 | ENSFM00250000003646 | Vacuolar Fusion CCZ1 Homolog |
| ENSMUSG00000029621 | ENSFM00760001714703 | Actin Related 2/3 Complex Subunit 1B ARP2/3 Complex 41 Kda Subunit P41 Arc |
| ENSMUSG00000029623 | ENSFM00600000921691 | 28 Kda Heat And Acid Stable Phosphoprotein Pdgf Associated Pap Pdgfa Associated 1 PAP1 |
| ENSMUSG00000029632 | ENSFM00670001238510 | Cytochrome C Oxidase Subunit NDUFA4 |
| ENSMUSG00000029636 | ENSFM00250000001269 | Wiskott Aldrich Syndrome Family Member 1 Wasp Family Member 1 Wave 1 |
| ENSMUSG00000029638 | ENSFM00250000002371 | Induced |
| ENSMUSG00000029651 | ENSFM00570000887300 | Unknown |
| ENSMUSG00000029651 | ENSFM00570000894628 | Unknown |
| ENSMUSG00000029651 | ENSFM00500000271268 | Microtubule Associated Tumor Suppressor Candidate 2 Cardiac Zipper Microtubule Plus End Tracking TIP150 Tracking Of 150 Kda |
| ENSMUSG00000029657 | ENSFM00360000109870 | Unknown |
| ENSMUSG00000029684 | ENSFM00250000001784 | Wiskott Aldrich Syndrome Wasp |
| ENSMUSG00000029686 | ENSFM00570000896172 | Unknown |
| ENSMUSG00000029686 | ENSFM00510000502743 | Cullin |
| ENSMUSG00000029713 | ENSFM00250000000545 | Guanine Nucleotide Binding G I /G S /G T Subunit Beta Transducin Beta Chain |
| ENSMUSG00000029714 | ENSFM00740001589295 | Perq Amino Acid Rich With Gyf Domain Containing 2 Trinucleotide Repeat Containing Gene 15 |
| ENSMUSG00000029723 | ENSFM00250000008373 | TSC22 Domain Family 4 TSC22 Related Inducible Leucine Zipper 2 Thg |
| ENSMUSG00000029723 | ENSFM00500000295696 | TSC22 Domain Family 4 |
| ENSMUSG00000029757 | ENSFM00570000882077 | Unknown |
| ENSMUSG00000029757 | ENSFM00250000000887 | Cytoplasmic Dynein 1 Intermediate Chain 2 Cytoplasmic Dynein Intermediate Chain 2 Dynein Intermediate Chain 2 Cytosolic Dh Ic 2 |
| ENSMUSG00000029761 | ENSFM00250000002835 | Caldesmon Cdm |
| ENSMUSG00000029763 | ENSFM00250000002477 | Exocyst Complex Component 4 Exocyst Complex Component SEC8 |
| ENSMUSG00000029765 | ENSFM00730001521329 | Plexin Precursor |
| ENSMUSG00000029769 | ENSFM00250000006497 | Coiled Coil Domain Containing 136 |
| ENSMUSG00000029772 | ENSFM00560000818040 | Unknown |
| ENSMUSG00000029772 | ENSFM00660001186554 | Unknown |
| ENSMUSG00000029772 | ENSFM00660001186556 | Unknown |
| ENSMUSG00000029772 | ENSFM00740001589092 | Adenosylhomocysteinase Adohcyase EC_3.3.1.1 S Adenosyl L Homocysteine Hydrolase 2 |
| ENSMUSG00000029776 | ENSFM00780001898427 | 3 Hydroxyisobutyrate Dehydrogenase Mitochondrial Hibadh EC_1.1.1.31 |
| ENSMUSG00000029777 | ENSFM00800002003540 | Glycine Trna Ligase EC_6.1.1.14 Diadenosine Tetraphosphate Synthetase Ap 4 A Synthetase Glycyl Trna Synthetase Glyrs |
| ENSMUSG00000029778 | ENSFM00610000970654 | Unknown |
| ENSMUSG00000029778 | ENSFM00730001521191 | Pituitary Adenylate Cyclase Activating Polypeptide Type Receptor Pacap Type Receptor Pacap R Pacap |
| ENSMUSG00000029802 | ENSFM00570000902030 | Unknown |
| ENSMUSG00000029802 | ENSFM00730001521925 | Atp Binding Cassette Sub Family G Member 2 Urate Exporter CD338 Antigen |
| ENSMUSG00000029840 | ENSFM00810002055357 | Unknown |
| ENSMUSG00000029840 | ENSFM00500000273644 | Myotrophin |
| ENSMUSG00000029861 | ENSFM00500000271560 | Unknown |
| ENSMUSG00000029864 | ENSFM00250000004030 | Glutathione S Transferase Kappa 1 EC_2.5.1.18 Gst 13 13 Gst Class Kappa GSTK1 1 Glutathione S Transferase Subunit 13 |
| ENSMUSG00000029911 | ENSFM00520000555852 | Unknown |
| ENSMUSG00000029911 | ENSFM00250000006324 | Single Stranded Dna Binding Protein Mitochondrial Precursor Mt Ssb Mtssb |
| ENSMUSG00000029916 | ENSFM00810002055665 | Unknown |
| ENSMUSG00000029916 | ENSFM00250000006088 | Acylglycerol Kinase Mitochondrial Precursor EC_2.7.1.107 EC_2.7.1.- 94 Multiple Substrate Lipid Kinase Mulk Multi Substrate Lipid Kinase |
| ENSMUSG00000029994 | ENSFM00760001714627 | Annexin Annexin Annexin |
| ENSMUSG00000029998 | ENSFM00250000003214 | Prenylcysteine Oxidase Precursor |
| ENSMUSG00000030000 | ENSFM00250000000681 | Adducin |
| ENSMUSG00000030007 | ENSFM00500000269991 | T Complex 1 Subunit Eta Tcp 1 Eta Cct Eta |
| ENSMUSG00000030020 | ENSFM00730001521542 | Prickle |
| ENSMUSG00000030029 | ENSFM00250000001262 | Leucine Rich Repeats And Immunoglobulin Domains Precursor Lig |
| ENSMUSG00000030036 | ENSFM00760001714954 | Mannosyl Oligosaccharide Glucosidase EC_3.2.1.106 Processing Glucosidase I |
| ENSMUSG00000030037 | ENSFM00500000276686 | 39S Ribosomal L53 Mitochondrial Precursor L53MT Mrp L53 |
| ENSMUSG00000030058 | ENSFM00800002003465 | Coatomer Subunit Gamma Gamma Coat Gamma Cop |
| ENSMUSG00000030062 | ENSFM00250000002976 | Dolichyl Diphosphooligosaccharide Glycosyltransferase Subunit 1 Precursor EC_2.4.99.18 Dolichyl Diphosphooligosaccharide Glycosyltransferase 67 Kda Subunit Ribophorin I Rpn I Ribophorin 1 |
| ENSMUSG00000030075 | ENSFM00390000126303 | Contactin Precursor Brain Derived Immunoglobulin Superfamily Big |
| ENSMUSG00000030077 | ENSFM00390000126295 | Neural Cell Adhesion Molecule L1 L1 L1 |
| ENSMUSG00000030084 | ENSFM00730001521329 | Plexin Precursor |
| ENSMUSG00000030086 | ENSFM00500000273806 | Micos Complex Subunit MIC25 Coiled Coil Helix Coiled Coil Helix Domain Containing |
| ENSMUSG00000030088 | ENSFM00730001521877 | Cytosolic 10 Formyltetrahydrofolate Dehydrogenase 10 Fthfdh Fdh EC_1.5.1.6 Aldehyde Dehydrogenase Family 1 Member L1 |
| ENSMUSG00000030092 | ENSFM00590000918177 | Unknown |
| ENSMUSG00000030092 | ENSFM00390000126303 | Contactin Precursor Brain Derived Immunoglobulin Superfamily Big |
| ENSMUSG00000030095 | ENSFM00250000004628 | Transmembrane 43 |
| ENSMUSG00000030098 | ENSFM00590000917517 | Unknown |
| ENSMUSG00000030098 | ENSFM00740001589268 | Glutamate Receptor Interacting |
| ENSMUSG00000030102 | ENSFM00250000000547 | Inositol 1 4 5 Trisphosphate Receptor Type IP3 Receptor IP3R Ins Type Inositol 1 4 5 Trisphosphate Receptor Type INSP3 Receptor |
| ENSMUSG00000030105 | ENSFM00410000138507 | Adp Ribosylation Factor Small G Indispensable Equal Chromosome Segregation |
| ENSMUSG00000030120 | ENSFM00500000271997 | Myeloid Leukemia Factor 2 Myelodysplasia Myeloid Leukemia Factor 2 |
| ENSMUSG00000030122 | ENSFM00810002042509 | Parathymosin |
| ENSMUSG00000030123 | ENSFM00800002003838 | Plexin C1 Precursor Virus Encoded Semaphorin Receptor CD232 Antigen |
| ENSMUSG00000030126 | ENSFM00640001113413 | Unknown |
| ENSMUSG00000030126 | ENSFM00640001116667 | Unknown |
| ENSMUSG00000030126 | ENSFM00250000001324 | Transmembrane And Coiled Coil Domains |
| ENSMUSG00000030127 | ENSFM00570000882296 | Unknown |
| ENSMUSG00000030127 | ENSFM00250000003382 | COP9 Signalosome Complex Subunit 7A SGN7A Signalosome Subunit 7A JAB1 Containing Signalosome Subunit 7A |
| ENSMUSG00000030134 | ENSFM00250000001384 | Ras Gef Domain Containing Family Member |
| ENSMUSG00000030137 | ENSFM00250000000217 | Tubulin Alpha Chain |
| ENSMUSG00000030161 | ENSFM00500000270406 | Gamma Aminobutyric Acid Receptor Associated 1 Precursor Gaba A Receptor Associated 1 |
| ENSMUSG00000030172 | ENSFM00250000000753 | Elks/RAB6 Interacting/Cast Family Member 1 Erc 1 RAB6 Interacting 2 |
| ENSMUSG00000030207 | ENSFM00800002003960 | Uncharacterized |
| ENSMUSG00000030207 | ENSFM00590000918208 | Unknown |
| ENSMUSG00000030207 | ENSFM00590000918209 | Unknown |
| ENSMUSG00000030209 | ENSFM00760001754226 | Unknown |
| ENSMUSG00000030209 | ENSFM00760001754227 | Unknown |
| ENSMUSG00000030209 | ENSFM00730001521500 | Glutamate Receptor Ionotropic Nmda Precursor Glutamate Receptor Subunit Epsilon N Methyl D Aspartate Receptor Subtype |
| ENSMUSG00000030224 | ENSFM00810002040832 | Serine Threonine Kinase Receptor Associated |
| ENSMUSG00000030231 | ENSFM00800002003674 | Pleckstrin Homology Domain Containing Family A Member 6 Ph Domain Containing Family A Member 6 Phosphoinositol 3 Phosphate Binding 3 Pepp 3 |
| ENSMUSG00000030245 | ENSFM00600000921609 | Vesicle Transport GOT1B Golgi Transport 1 Homolog B |
| ENSMUSG00000030246 | ENSFM00250000000475 | L Lactate Dehydrogenase Chain Ldh EC_1.1.1.27 |
| ENSMUSG00000030257 | ENSFM00610000970807 | Unknown |
| ENSMUSG00000030257 | ENSFM00250000000639 | Slit Robo Rho Gtpase Activating Rho Gtpase Activating |
| ENSMUSG00000030265 | ENSFM00570000883366 | Unknown |
| ENSMUSG00000030265 | ENSFM00570000889271 | Unknown |
| ENSMUSG00000030265 | ENSFM00730001521569 | Ras Precursor |
| ENSMUSG00000030268 | ENSFM00570000894976 | Unknown |
| ENSMUSG00000030268 | ENSFM00730001521772 | Branched Chain Amino Acid Aminotransferase EC_2.6.1.42 |
| ENSMUSG00000030270 | ENSFM00720001503378 | Unknown |
| ENSMUSG00000030270 | ENSFM00730001521728 | Copine Copine |
| ENSMUSG00000030279 | ENSFM00250000002423 | C2 Domain Containing 5.138 Kda C2 Domain Containing Phosphoprotein |
| ENSMUSG00000030282 | ENSFM00250000005763 | N Acylneuraminate Cytidylyltransferase EC_2.7.7.43 Cmp N Acetylneuraminic Acid Synthase Cmp Neunac Synthase |
| ENSMUSG00000030284 | ENSFM00250000002332 | Cysteine Rich With Egf Domain Precursor |
| ENSMUSG00000030286 | ENSFM00500000270967 | Er Membrane Complex Subunit 3 Transmembrane 111 |
| ENSMUSG00000030287 | ENSFM00250000000547 | Inositol 1 4 5 Trisphosphate Receptor Type IP3 Receptor IP3R Ins Type Inositol 1 4 5 Trisphosphate Receptor Type INSP3 Receptor |
| ENSMUSG00000030302 | ENSFM00250000000445 | Plasma Membrane Calcium Transporting Atpase EC_3.6.3.8 Plasma Membrane Calcium Atpase Plasma Membrane Calcium Pump |
| ENSMUSG00000030307 | ENSFM00730001521104 | Sodium And Chloride Dependent Transporter Solute Carrier Family 6 Member |
| ENSMUSG00000030310 | ENSFM00800002003411 | Sodium And Chloride Dependent Gaba Transporter 1 Gat 1 Solute Carrier Family 6 Member 1 |
| ENSMUSG00000030316 | ENSFM00500000271930 | Phosphatidate Cytidylyltransferase Mitochondrial Precursor EC_2.7.7.41 Cdp Diacylglycerol Synthase Cdp Dag Synthase Mitochondrial Translocator Assembly And Maintenance 41 Homolog TAM41 |
| ENSMUSG00000030327 | ENSFM00250000002415 | Adaptin Ear Binding Coat Associated 1 Necap Endocytosis Associated 1 Necap 1 |
| ENSMUSG00000030337 | ENSFM00670001235687 | Vesicle Associated Membrane 2 Vamp 2 Synaptobrevin 2 |
| ENSMUSG00000030342 | ENSFM00800002003594 | CD9 Antigen CD9 Antigen |
| ENSMUSG00000030350 | ENSFM00730001521302 | Arginine N Methyltransferase 1 EC_2.1.1.- Histone Arginine N Methyltransferase PRMT1 EC_2.1.1.125 |
| ENSMUSG00000030352 | ENSFM00570000884895 | Unknown |
| ENSMUSG00000030352 | ENSFM00810002057403 | Unknown |
| ENSMUSG00000030352 | ENSFM00500000270370 | Tetraspanin 9 Tspan 9 |
| ENSMUSG00000030357 | ENSFM00500000270053 | Peptidyl Prolyl Cis Trans Isomerase FKBP5 Ppiase FKBP5 EC_5.2.1.8.51 Kda FK506 Binding 51 Kda Fkbp Fkbp 51 FK506 Binding 5 Fkbp 5 Rotamase |
| ENSMUSG00000030359 | ENSFM00730001521356 | Precursor |
| ENSMUSG00000030374 | ENSFM00740001613490 | Unknown |
| ENSMUSG00000030374 | ENSFM00740001613491 | Unknown |
| ENSMUSG00000030374 | ENSFM00250000001473 | Striatin |
| ENSMUSG00000030376 | ENSFM00710001441664 | Sodium/Calcium Exchanger 1 Precursor Na + /Ca 2+ Exchange 1 Solute Carrier Family 8 Member 1 |
| ENSMUSG00000030397 | ENSFM00730001521220 | Kinase EC_2.7.11.1 |
| ENSMUSG00000030403 | ENSFM00800002003977 | Vasodilator Stimulated Phosphoprotein Vasp |
| ENSMUSG00000030410 | ENSFM00250000001646 | Dystrophia Myotonica Wd Repeat Containing Dystrophia Myotonica Containing Wd Repeat Motif Protein Dmr N9 |
| ENSMUSG00000030428 | ENSFM00250000001068 | Tweety Homolog |
| ENSMUSG00000030428 | ENSFM00570000864998 | Tweety Homolog |
| ENSMUSG00000030432 | ENSFM00570000887315 | Unknown |
| ENSMUSG00000030432 | ENSFM00500000271918 | 60S Ribosomal L28 |
| ENSMUSG00000030447 | ENSFM00250000001390 | Cytoplasmic FMR1 Interacting |
| ENSMUSG00000030465 | ENSFM00250000001069 | Ph And SEC7 Domain Containing 1 Exchange Factor ARF6 Pleckstrin Homology And SEC7 Domain Containing 1 |
| ENSMUSG00000030500 | ENSFM00730001521551 | Vesicular Glutamate Transporter Solute Carrier Family 17 Member |
| ENSMUSG00000030515 | ENSFM00250000001453 | Threonine Trna Ligase EC_6.1.1.3 Threonyl Trna Synthetase Thrrs Threonyl Trna Synthetase |
| ENSMUSG00000030516 | ENSFM00250000000691 | Tight Junction Zo Tight Junction Zona Occludens Zonula Occludens |
| ENSMUSG00000030532 | ENSFM00250000006266 | Guanosine 3' 5' Bis Diphosphate 3' Pyrophosphohydrolase MESH1 EC_3.1.7.2 Hd Domain Containing 3 Metazoan Spot Homolog 1 MESH1 Penta Phosphate Guanosine 3' Pyrophosphohydrolase Ppgpp Ase |
| ENSMUSG00000030539 | ENSFM00480000262746 | Semaphorin Precursor |
| ENSMUSG00000030541 | ENSFM00770001847524 | Isocitrate Dehydrogenase Cytoplasmic Idh EC_1.1.1.42 Cytosolic Nadp Isocitrate Dehydrogenase Idp Nadp + Specific Icdh Oxalosuccinate Decarboxylase |
| ENSMUSG00000030583 | ENSFM00720001502782 | Unknown |
| ENSMUSG00000030583 | ENSFM00720001502783 | Unknown |
| ENSMUSG00000030583 | ENSFM00720001503388 | Unknown |
| ENSMUSG00000030583 | ENSFM00730001521477 | Signal Induced Proliferation Associated 1 SIPA1 |
| ENSMUSG00000030600 | ENSFM00260000050443 | Leucine Rich Repeat And Fibronectin Type Iii Domain Containing Precursor Synaptic Adhesion Molecule |
| ENSMUSG00000030602 | ENSFM00250000002132 | Smaug Homolog 1 Smaug 1 Sterile Alpha Motif Domain Containing 4A |
| ENSMUSG00000030602 | ENSFM00730001521739 | Serine/Threonine Kinase Pak EC_2.7.11.1 P21 Activated Kinase Pak |
| ENSMUSG00000030603 | ENSFM00730001521737 | 26S Protease Regulatory Subunit 6B 26S Proteasome Aaa Atpase Subunit RPT3 Proteasome 26S Subunit Atpase 4 |
| ENSMUSG00000030605 | ENSFM00500000270078 | Lactadherin Precursor Mfgm Milk Fat Globule Egf Factor 8 Mfg E8 SED1 |
| ENSMUSG00000030607 | ENSFM00730001521436 | Core Precursor Chondroitin Sulfate Proteoglycan |
| ENSMUSG00000030612 | ENSFM00250000006279 | 39S Ribosomal L46 Mitochondrial Precursor L46MT Mrp L46 |
| ENSMUSG00000030613 | ENSFM00610000952921 | Mitochondrial Calcium Uniporter Regulator 1 Coiled Coil Domain Containing 90A Mitochondrial |
| ENSMUSG00000030615 | ENSFM00450000242207 | Transmembrane 126A |
| ENSMUSG00000030621 | ENSFM00250000000979 | Nadp Dependent Malic Enzyme Nadp Me EC_1.1.1.40 Malic Enzyme |
| ENSMUSG00000030630 | ENSFM00250000004343 | Fumarylacetoacetase Faa EC_3.7.1.2 |
| ENSMUSG00000030643 | ENSFM00730001522236 | Ras Related Rab 30 Precursor |
| ENSMUSG00000030647 | ENSFM00670001236356 | Nadh Dehydrogenase 1 Subunit C2 Complex I B14 5B Ci B14 5B Nadh Ubiquinone Oxidoreductase Subunit B14 5B |
| ENSMUSG00000030652 | ENSFM00250000004792 | Ubiquinone Biosynthesis COQ7 Homolog Coenzyme Q Biosynthesis 7 Homolog Timing Clk 1 Homolog |
| ENSMUSG00000030653 | ENSFM00730001522357 | Cgmp Dependent 3' 5' Cyclic Phosphodiesterase EC_3.1.4.17 Cyclic Gmp Stimulated Phosphodiesterase Cgs Pde Cgspde |
| ENSMUSG00000030659 | ENSFM00720001497502 | Unknown |
| ENSMUSG00000030659 | ENSFM00250000002033 | Nucleobindin Precursor |
| ENSMUSG00000030662 | ENSFM00800002003590 | Importin 5 IMP5 Importin Subunit Beta 3 Karyopherin Beta 3 Ran Binding 5 RANBP5 |
| ENSMUSG00000030682 | ENSFM00500000271211 | Cdp Diacylglycerol Inositol 3 Phosphatidyltransferase EC_2.7.8.11 Phosphatidylinositol Synthase Pi Synthase Ptdins Synthase |
| ENSMUSG00000030683 | ENSFM00570000889526 | Unknown |
| ENSMUSG00000030683 | ENSFM00250000001049 | Seizure 6 Precursor |
| ENSMUSG00000030695 | ENSFM00250000000488 | Fructose Bisphosphate Aldolase EC_4.1.2.13 Type Aldolase |
| ENSMUSG00000030701 | ENSFM00500000271631 | Pleckstrin Homology Domain Containing Family B Member 1 Ph Domain Containing Family B Member 1 Evectin 1 Ph Domain Containing In Retina 1 PHRET1 Pleckstrin Homology Domain Retinal 1 |
| ENSMUSG00000030704 | ENSFM00460000244935 | Ras Related Rab 6 |
| ENSMUSG00000030706 | ENSFM00570000888665 | Unknown |
| ENSMUSG00000030706 | ENSFM00250000005488 | 39S Ribosomal L48 Mitochondrial Precursor L48MT Mrp L48 |
| ENSMUSG00000030706 | ENSFM00250000019723 | 39S Ribosomal L48 Mitochondrial |
| ENSMUSG00000030707 | ENSFM00730001521292 | Coronin |
| ENSMUSG00000030718 | ENSFM00760001714910 | Phosphatase Methylesterase 1 Pme 1 EC_3.1.1.89 |
| ENSMUSG00000030729 | ENSFM00250000002020 | Phosphoglucomutase |
| ENSMUSG00000030731 | ENSFM00570000885121 | Unknown |
| ENSMUSG00000030731 | ENSFM00570000888896 | Unknown |
| ENSMUSG00000030731 | ENSFM00720001492041 | Synaptotagmin Synaptotagmin |
| ENSMUSG00000030739 | ENSFM00270000056424 | Myosin Cellular Myosin Heavy Chain Type Myosin Heavy Chain Myosin Heavy Chain Non Muscle Non Muscle Myosin Heavy Chain Nmmhc Non Muscle Myosin Heavy Chain Nmmhc Ii Nmmhc |
| ENSMUSG00000030744 | ENSFM00250000001742 | 40S Ribosomal S3 EC_4.2.99.18 |
| ENSMUSG00000030751 | ENSFM00400000131829 | Proteasome Subunit Alpha Type EC_3.4.25.1 Macropain Subunit Multicatalytic Endopeptidase Complex Subunit Proteasome Component |
| ENSMUSG00000030754 | ENSFM00250000004388 | Coatomer Subunit Beta Beta Coat Beta Cop |
| ENSMUSG00000030759 | ENSFM00570000889091 | Unknown |
| ENSMUSG00000030759 | ENSFM00250000000622 | Fatty Acyl Coa Reductase 1 EC_1.2.1.- N2 Male Sterility Domain Containing 2 |
| ENSMUSG00000030774 | ENSFM00730001521340 | Serine/Threonine Kinase Pak EC_2.7.11.1 Pak P21 Activated Kinase Pak |
| ENSMUSG00000030786 | ENSFM00670001256242 | Unknown |
| ENSMUSG00000030786 | ENSFM00730001521375 | Integrin Alpha Precursor CD11 Antigen Family Member Glycoprotein Alpha Leukocyte Antigen |
| ENSMUSG00000030795 | ENSFM00600000921301 | Rna Binding |
| ENSMUSG00000030795 | ENSFM00600000925489 | Fus |
| ENSMUSG00000030802 | ENSFM00500000271321 | Kinase Mitochondrial Precursor EC_2.7.11.4 Branched Chain Alpha Ketoacid Dehydrogenase Kinase Bckd Kinase Bckdhkin |
| ENSMUSG00000030805 | ENSFM00570000893393 | Unknown |
| ENSMUSG00000030805 | ENSFM00250000000549 | Syntaxin |
| ENSMUSG00000030806 | ENSFM00250000000549 | Syntaxin |
| ENSMUSG00000030824 | ENSFM00250000002033 | Nucleobindin Precursor |
| ENSMUSG00000030835 | ENSFM00250000002391 | Nodal Modulator Precursor |
| ENSMUSG00000030842 | ENSFM00250000006855 | Ragulator Complex LAMTOR1 Late Endosomal/Lysosomal Adaptor And Mapk And Mtor Activator 1 |
| ENSMUSG00000030854 | ENSFM00280000058726 | Tyrosine Phosphatase Non Receptor Type EC_3.1.3.48 Tyrosine Phosphatase Tyrosine Phosphatase |
| ENSMUSG00000030861 | ENSFM00730001521755 | Short/Branched Chain Specific Acyl Coa Dehydrogenase Mitochondrial Precursor Sbcad EC_1.3.8.5 2 Methyl Branched Chain Acyl Coa Dehydrogenase 2 Mebcad 2 Methylbutyryl Coenzyme A Dehydrogenase 2 Methylbutyryl Coa Dehydrogenase |
| ENSMUSG00000030868 | ENSFM00660001178171 | Unknown |
| ENSMUSG00000030868 | ENSFM00660001188113 | Unknown |
| ENSMUSG00000030868 | ENSFM00250000005726 | Dynactin Subunit 5 Dynactin Subunit P25 |
| ENSMUSG00000030869 | ENSFM00670001235829 | Acyl Carrier Protein Mitochondrial Precursor Acp Nadh Ubiquinone Oxidoreductase 9.6 Kda Subunit |
| ENSMUSG00000030881 | ENSFM00570000876081 | Unknown |
| ENSMUSG00000030881 | ENSFM00570000882500 | Unknown |
| ENSMUSG00000030881 | ENSFM00250000002848 | Arfaptin Adp Ribosylation Factor Interacting |
| ENSMUSG00000030884 | ENSFM00250000003169 | Cytochrome B C1 Complex Subunit 2 Mitochondrial Precursor Complex Iii Subunit 2 Core Ii Ubiquinol Cytochrome C Reductase Complex Core 2 |
| ENSMUSG00000030905 | ENSFM00250000008871 | Ketimine Reductase Mu Crystallin EC_1.5.1.25 Nadp Regulated Thyroid Hormone Binding |
| ENSMUSG00000030934 | ENSFM00790001963008 | Ornithine Aminotransferase Mitochondrial Precursor EC_2.6.1.13 Ornithine Oxo Acid Aminotransferase |
| ENSMUSG00000030987 | ENSFM00250000001930 | Stromal Interaction Molecule Precursor |
| ENSMUSG00000031007 | ENSFM00250000004412 | Renin Receptor Precursor Atpase H + Transporting Lysosomal Accessory 2 Atpase H + Transporting Lysosomal Interacting 2 Renin/Prorenin Receptor Vacuolar Atp Synthase Membrane Sector Associated M8 9 V Atpase M8 9 Subunit |
| ENSMUSG00000031010 | ENSFM00250000001518 | Probable Ubiquitin Carboxyl Terminal Hydrolase Faf EC_3.4.19.12 Deubiquitinating Enzyme Faf Fat Facets Related Linked Ubiquitin Thioesterase Faf Ubiquitin Specific Protease 9 Chromosome Ubiquitin Specific Processing Protease Faf |
| ENSMUSG00000031012 | ENSFM00730001521718 | 55 Kda Erythrocyte Membrane P55 Membrane Protein Palmitoylated 1 |
| ENSMUSG00000031021 | ENSFM00570000882023 | Unknown |
| ENSMUSG00000031021 | ENSFM00670001235681 | Transmembrane Precursor |
| ENSMUSG00000031029 | ENSFM00250000003700 | Eukaryotic Translation Initiation Factor 3 Subunit F |
| ENSMUSG00000031059 | ENSFM00250000007177 | Nadh Dehydrogenase 1 Beta Subcomplex Subunit 11 Mitochondrial Precursor Complex I Esss Ci Esss Nadh Ubiquinone Oxidoreductase Esss Subunit |
| ENSMUSG00000031066 | ENSFM00570000851025 | Ubiquitin Carboxyl Terminal Hydrolase EC_3.4.19.12 Deubiquitinating Enzyme Ubiquitin Thioesterase Ubiquitin Specific Processing Protease |
| ENSMUSG00000031068 | ENSFM00740001589388 | Glutaredoxin 3 Pkc Interacting Cousin Of Thioredoxin Picot Pkc Theta Interacting Pkcq Interacting Thioredoxin 2 |
| ENSMUSG00000031078 | ENSFM00600000921221 | Substrate |
| ENSMUSG00000031095 | ENSFM00730001521495 | Cullin Cul |
| ENSMUSG00000031105 | ENSFM00570000894083 | Unknown |
| ENSMUSG00000031105 | ENSFM00500000270162 | Kidney Mitochondrial Carrier 1 Solute Carrier Family 25 Member 30 |
| ENSMUSG00000031119 | ENSFM00500000269807 | Glypican Secreted Glypican |
| ENSMUSG00000031134 | ENSFM00500000270455 | Rna Binding Motif Protein X Heterogeneous Nuclear Ribonucleoprotein G Hnrnp G |
| ENSMUSG00000031143 | ENSFM00250000005354 | Coiled Coil Domain Containing 22 |
| ENSMUSG00000031144 | ENSFM00500000270007 | Synaptophysin |
| ENSMUSG00000031146 | ENSFM00500000274188 | Proteolipid 2 |
| ENSMUSG00000031149 | ENSFM00670001235692 | PRA1 Family 3 Adp Ribosylation Factor 6 Interacting 5 Arl 6 Interacting 5 Aip 5 |
| ENSMUSG00000031150 | ENSFM00570000882243 | Unknown |
| ENSMUSG00000031150 | ENSFM00250000009538 | Coiled Coil Domain Containing 120 |
| ENSMUSG00000031153 | ENSFM00250000004894 | GRIP1 Associated 1 Grasp 1 |
| ENSMUSG00000031158 | ENSFM00500000270373 | Mitochondrial Import Inner Membrane Translocase Subunit TIM17 |
| ENSMUSG00000031167 | ENSFM00600000921729 | Rna Binding 3 Rna Binding Motif 3 |
| ENSMUSG00000031168 | ENSFM00500000272667 | 3 Beta Hydroxysteroid Delta 8 Delta 7 Isomerase EC_5.3.3.5 Cholestenol Delta Isomerase Delta 8 Delta 7 Sterol Isomerase D8 D7 Sterol Isomerase Emopamil Binding |
| ENSMUSG00000031198 | ENSFM00670001235667 | FUN14 Domain Containing |
| ENSMUSG00000031202 | ENSFM00500000270441 | Ras Related Rab |
| ENSMUSG00000031207 | ENSFM00250000000517 | Moesin |
| ENSMUSG00000031217 | ENSFM00250000002014 | Ephrin B1 Precursor Elk Ligand Elk L Eph Related Receptor Tyrosine Kinase Ligand 2 Lerk 2 |
| ENSMUSG00000031278 | ENSFM00730001521866 | Long Chain Fatty Acid Coa Ligase EC_6.2.1.3 Long Chain Acyl Coa Synthetase Lacs |
| ENSMUSG00000031292 | ENSFM00740001589470 | Cyclin Dependent Kinase 5 EC_2.7.11.22 |
| ENSMUSG00000031299 | ENSFM00750001632368 | Pyruvate Dehydrogenase E1 Component Subunit Alpha Somatic Form Mitochondrial Precursor EC_1.2.4.1 PDHE1 A Type I |
| ENSMUSG00000031302 | ENSFM00380000124679 | Neuroligin Precursor |
| ENSMUSG00000031311 | ENSFM00250000000671 | Paraspeckle Component 1 |
| ENSMUSG00000031320 | ENSFM00250000001458 | 40S Ribosomal S4 |
| ENSMUSG00000031333 | ENSFM00730001521676 | Iron Sulfur Clusters Transporter ATM1 Mitochondrial Precursor |
| ENSMUSG00000031342 | ENSFM00660001189588 | Unknown |
| ENSMUSG00000031342 | ENSFM00670001264730 | Unknown |
| ENSMUSG00000031342 | ENSFM00250000000909 | Myelin Proteolipid Plp Lipophilin |
| ENSMUSG00000031343 | ENSFM00800002003385 | Gamma Aminobutyric Acid Receptor Subunit Alpha Precursor Gaba A Receptor Subunit Alpha |
| ENSMUSG00000031347 | ENSFM00750001632381 | Centrin |
| ENSMUSG00000031349 | ENSFM00500000271873 | Sterol 4 Alpha Carboxylate 3 Dehydrogenase Decarboxylating EC_1.1.1.170 |
| ENSMUSG00000031367 | ENSFM00500000269841 | Ap 1 Complex Subunit Sigma Adaptor Complex Ap 1 Subunit Sigma Adaptor Related Complex 1 Subunit Sigma Clathrin Assembly Complex 1 Sigma Small Chain Golgi Adaptor HA1/AP1 Adaptin Sigma Subunit Sigma Subunit Of Ap 1 Clathrin Sigma Adaptin Adaptin |
| ENSMUSG00000031376 | ENSFM00250000000445 | Plasma Membrane Calcium Transporting Atpase EC_3.6.3.8 Plasma Membrane Calcium Atpase Plasma Membrane Calcium Pump |
| ENSMUSG00000031386 | ENSFM00250000001451 | Host Cell Factor 1 Hcf Hcf 1 C1 Factor |
| ENSMUSG00000031391 | ENSFM00390000126295 | Neural Cell Adhesion Molecule L1 L1 L1 |
| ENSMUSG00000031397 | ENSFM00250000001413 | Transketolase EC_2.2.1.1 |
| ENSMUSG00000031400 | ENSFM00760001714666 | Glucose 6 Phosphate 1 Dehydrogenase G6PD EC_1.1.1.49 |
| ENSMUSG00000031402 | ENSFM00570000899672 | Unknown |
| ENSMUSG00000031402 | ENSFM00730001521718 | 55 Kda Erythrocyte Membrane P55 Membrane Protein Palmitoylated 1 |
| ENSMUSG00000031425 | ENSFM00250000000909 | Myelin Proteolipid Plp Lipophilin |
| ENSMUSG00000031442 | ENSFM00570000895390 | Unknown |
| ENSMUSG00000031442 | ENSFM00250000000478 | Guanine Nucleotide Exchange Factor Dbs Dbl'S Big Sister MCF2 Transforming Sequence |
| ENSMUSG00000031446 | ENSFM00570000894978 | Unknown |
| ENSMUSG00000031446 | ENSFM00730001521495 | Cullin Cul |
| ENSMUSG00000031447 | ENSFM00640001103074 | Lysosome Associated Membrane Glycoprotein 1 Precursor Lamp 1 Lysosome Associated Membrane 1 CD107 Antigen Family Member A CD107A Antigen |
| ENSMUSG00000031453 | ENSFM00570000892435 | Unknown |
| ENSMUSG00000031453 | ENSFM00760001714814 | Ras Gtpase Activating 3 GAP1 IP4BP Ins P4 Binding |
| ENSMUSG00000031467 | ENSFM00250000007868 | 1 Acyl Sn Glycerol 3 Phosphate Acyltransferase Epsilon EC_2.3.1.51 1 Acylglycerol 3 Phosphate O Acyltransferase 5 1 Agp Acyltransferase 5 1 Agpat 5 Lysophosphatidic Acid Acyltransferase Epsilon Lpaat Epsilon |
| ENSMUSG00000031483 | ENSFM00250000003616 | Erlin 2 Endoplasmic Reticulum Lipid Raft Associated 2 Stomatin Prohibitin Flotillin Hflc/K Domain Containing 2 Spfh Domain Containing 2 |
| ENSMUSG00000031505 | ENSFM00680001319081 | Unknown |
| ENSMUSG00000031505 | ENSFM00810002040702 | Atp Dependent S Nad P H Hydrate Dehydratase |
| ENSMUSG00000031511 | ENSFM00250000001135 | Rho Guanine Nucleotide Exchange Factor 6 Rac/CDC42 Guanine Nucleotide Exchange Factor 6 |
| ENSMUSG00000031517 | ENSFM00250000000909 | Myelin Proteolipid Plp Lipophilin |
| ENSMUSG00000031539 | ENSFM00440000236903 | Ap 3 Complex Subunit Mu Adaptor Related Complex 3 Subunit Mu Clathrin Assembly Assembly Complex 3 Mu Medium Chain Clathrin Coat Assembly AP47 Homolog Clathrin Coat Associated AP47 Homolog Golgi Adaptor Ap 1.47 Kda Homolog HA1.47 Kda Subunit Homolog Adapti |
| ENSMUSG00000031555 | ENSFM00800002003698 | Disintegrin And Metalloproteinase Domain Containing 9 Precursor Adam 9 EC_3.4.24.- Meltrin Gamma Metalloprotease/Disintegrin/Cysteine Rich 9 Myeloma Cell Metalloproteinase |
| ENSMUSG00000031561 | ENSFM00500000315657 | Unknown |
| ENSMUSG00000031561 | ENSFM00250000000375 | Teneurin Ten Odd Oz/Ten M Homolog Tenascin Ten Teneurin Transmembrane |
| ENSMUSG00000031584 | ENSFM00730001521746 | Glutathione Reductase Gr Grase EC_1.8.1.7 |
| ENSMUSG00000031591 | ENSFM00570000901226 | Unknown |
| ENSMUSG00000031591 | ENSFM00500000270714 | Acid Ceramidase Precursor Ac Acdase Acid Cdase EC_3.5.1.23 Acylsphingosine Deacylase N Acylsphingosine Amidohydrolase |
| ENSMUSG00000031592 | ENSFM00250000002581 | Pericentriolar Material 1 Pcm 1 |
| ENSMUSG00000031596 | ENSFM00570000892478 | Unknown |
| ENSMUSG00000031596 | ENSFM00250000000444 | Cationic Amino Acid Transporter 2 Cat 2 CAT2 Low Affinity Cationic Amino Acid Transporter 2 Solute Carrier Family 7 Member 2 |
| ENSMUSG00000031626 | ENSFM00570000879880 | Unknown |
| ENSMUSG00000031626 | ENSFM00570000883394 | Unknown |
| ENSMUSG00000031626 | ENSFM00570000887049 | Unknown |
| ENSMUSG00000031626 | ENSFM00570000891654 | Unknown |
| ENSMUSG00000031626 | ENSFM00570000895888 | Unknown |
| ENSMUSG00000031626 | ENSFM00570000900335 | Unknown |
| ENSMUSG00000031626 | ENSFM00550000743175 | Sorbin And SH3 Domain Containing 2 Arg/Abl Interacting 2 ARGBP2 |
| ENSMUSG00000031633 | ENSFM00250000000816 | Adp/Atp Translocase Adp Atp Carrier Adenine Nucleotide Translocator Ant Solute Carrier Family 25 Member |
| ENSMUSG00000031634 | ENSFM00500000271480 | UFM1 Specific Protease 2 UFSP2 EC_3.4.22.- |
| ENSMUSG00000031641 | ENSFM00500000272596 | Carbonyl Reductase Family Member 4 Ec 1.3.-.- Oxoacyl Reductase EC_1.1.1.- Quinone Reductase CBR4 |
| ENSMUSG00000031654 | ENSFM00250000002369 | Cerebellin Precursor |
| ENSMUSG00000031672 | ENSFM00730001521567 | Aspartate Aminotransferase Mitochondrial Precursor Maspat EC_2.6.1.1 EC_2.6.1.- 7 Fatty Acid Binding Fabp 1 Glutamate Oxaloacetate Transaminase 2 Kynurenine Aminotransferase 4 Kynurenine Aminotransferase Iv Kynurenine Oxoglutarate Transaminase 4 Kynurenin |
| ENSMUSG00000031673 | ENSFM00800002003474 | Cadherin Precursor |
| ENSMUSG00000031696 | ENSFM00250000003527 | Vacuolar Sorting Associated 35 Maternal Embryonic 3 Vesicle Sorting 35 |
| ENSMUSG00000031701 | ENSFM00760001714614 | Dnaj Homolog Subfamily A Member Precursor |
| ENSMUSG00000031703 | ENSFM00250000004403 | T Cell Immunomodulatory Tip Integrin Alpha Fg Gap Repeat Containing 1 |
| ENSMUSG00000031708 | ENSFM00250000002457 | Very Long Chain Enoyl Coa Reductase EC_1.3.1.93 Synaptic Glycoprotein SC2 Trans 2 3 Enoyl Coa Reductase Ter |
| ENSMUSG00000031729 | ENSFM00250000004551 | IST1 Homolog |
| ENSMUSG00000031730 | ENSFM00570000885026 | Unknown |
| ENSMUSG00000031730 | ENSFM00250000003878 | Dihydroorotate Dehydrogenase Quinone Mitochondrial Precursor Dhodehase EC_1.3.5.2 Dihydroorotate Oxidase |
| ENSMUSG00000031731 | ENSFM00680001326191 | Unknown |
| ENSMUSG00000031731 | ENSFM00810002040662 | Ap 1 Complex Subunit Gamma 1 Adaptor Complex Ap 1 Subunit Gamma 1 Adaptor Related Complex 1 Subunit Gamma 1 Clathrin Assembly Complex 1 Gamma 1 Large Chain Gamma Adaptin GAMMA1 Adaptin Golgi Adaptor HA1/AP1 Adaptin Subunit Gamma 1 |
| ENSMUSG00000031748 | ENSFM00570000894903 | Unknown |
| ENSMUSG00000031748 | ENSFM00410000138458 | Guanine Nucleotide Binding Subunit |
| ENSMUSG00000031751 | ENSFM00250000003925 | E3 Ubiquitin Ligase Amfr EC_6.3.2.- Autocrine Motility Factor Receptor Amf Receptor |
| ENSMUSG00000031754 | ENSFM00250000006035 | Cleavage And Polyadenylation Specificity Factor Subunit 5 Nucleoside Diphosphate Linked Moiety X Motif 21 Nudix Motif 21 |
| ENSMUSG00000031772 | ENSFM00250000000380 | Contactin Associated 5 Precursor Cell Recognition Molecule CASPR5 |
| ENSMUSG00000031775 | ENSFM00500000273560 | Plasmolipin Plasma Membrane Proteolipid |
| ENSMUSG00000031778 | ENSFM00570000876509 | Unknown |
| ENSMUSG00000031778 | ENSFM00570000877880 | Unknown |
| ENSMUSG00000031778 | ENSFM00250000008357 | Fractalkine Precursor C X3 C Motif Chemokine 1 CX3C Membrane Anchored Chemokine Neurotactin Small Inducible Cytokine D1 |
| ENSMUSG00000031782 | ENSFM00500000272185 | Ubiquinone Biosynthesis COQ9 Mitochondrial Precursor |
| ENSMUSG00000031785 | ENSFM00640001103085 | G Coupled Receptor 56 GPR56 N Terminal Fragment GPR56 Nt GPR56 N GPR56 Extracellular Subunit GPR56 Subunit Alpha |
| ENSMUSG00000031791 | ENSFM00570000851205 | Trimeric Intracellular Cation Channel Type A Tric A Trica Transmembrane 38A |
| ENSMUSG00000031807 | ENSFM00500000272386 | 6 Phosphogluconolactonase 6PGL EC_3.1.1.31 |
| ENSMUSG00000031808 | ENSFM00730001521827 | Long Chain Fatty Acid Transport 4 Fatp 4 Fatty Acid Transport 4 EC_6.2.1.- Solute Carrier Family 27 Member 4 |
| ENSMUSG00000031812 | ENSFM00700001400708 | Unknown |
| ENSMUSG00000031812 | ENSFM00500000270555 | Microtubule Associated Proteins 1A/1B Light Chain 3A Precursor Autophagy Related LC3 A Autophagy Related Ubiquitin Modifier LC3 A MAP1 Light Chain 3 1 MAP1A/MAP1B Light Chain 3 A MAP1A/MAP1B LC3 A Microtubule Associated 1 Light Chain 3 Alpha |
| ENSMUSG00000031818 | ENSFM00700001407963 | Unknown |
| ENSMUSG00000031818 | ENSFM00500000271199 | Cytochrome C Oxidase Subunit 4 1 Mitochondrial Cytochrome C Oxidase Polypeptide Iv Cytochrome C Oxidase Subunit Iv 1 Cox Iv 1 |
| ENSMUSG00000031819 | ENSFM00700001400973 | Unknown |
| ENSMUSG00000031819 | ENSFM00500000272397 | Er Membrane Complex Subunit 8 Neighbor Of COX4 |
| ENSMUSG00000031824 | ENSFM00250000006640 | Uncharacterized |
| ENSMUSG00000031824 | ENSFM00570000882328 | Unknown |
| ENSMUSG00000031827 | ENSFM00500000273700 | Coactosin |
| ENSMUSG00000031833 | ENSFM00250000000518 | Microtubule Associated Serine/Threonine Kinase EC_2.7.11.1 |
| ENSMUSG00000031837 | ENSFM00250000001762 | N Terminal Ef Hand Calcium Binding Ef Hand Calcium Binding Neuronal Calcium Binding |
| ENSMUSG00000031839 | ENSFM00500000401249 | Unknown |
| ENSMUSG00000031840 | ENSFM00570000900716 | Unknown |
| ENSMUSG00000031840 | ENSFM00730001521441 | Ras Related Rab 3 |
| ENSMUSG00000031841 | ENSFM00730001521172 | Cadherin Precursor Cadherin Cadherin |
| ENSMUSG00000031862 | ENSFM00500000270146 | Manganese Transporting Atpase EC_3.6.3.- |
| ENSMUSG00000031865 | ENSFM00760001714751 | Dynactin Subunit 1.150 Kda Dynein Associated Polypeptide Dap 150 Dp 150 Glued |
| ENSMUSG00000031904 | ENSFM00250000000331 | Amino Acid Transporter Solute Carrier Family 7 Member |
| ENSMUSG00000031906 | ENSFM00250000006694 | Sphingomyelin Phosphodiesterase 3 EC_3.1.4.12 Neutral Sphingomyelinase 2 Nsmase 2 NSMASE2 Neutral Sphingomyelinase Ii |
| ENSMUSG00000031913 | ENSFM00810002040610 | Vacuolar Sorting Associated EC_3.6.4.6 |
| ENSMUSG00000031918 | ENSFM00570000883574 | Unknown |
| ENSMUSG00000031918 | ENSFM00570000888483 | Unknown |
| ENSMUSG00000031918 | ENSFM00730001521553 | Myotubularin Phosphatidylinositol 3 5 Bisphosphate 3 Phosphatase EC_3.1.3.95 |
| ENSMUSG00000031924 | ENSFM00500000273474 | Cytochrome B5 Type B Precursor Cytochrome B5 Outer Mitochondrial Membrane |
| ENSMUSG00000031930 | ENSFM00730001521655 | NEDD4 E3 Ubiquitin Ligase EC_6.3.2.- Ww Domain Containing |
| ENSMUSG00000031948 | ENSFM00250000002843 | Lysine Trna Ligase EC_6.1.1.6 Lysyl Trna Synthetase Lysrs |
| ENSMUSG00000031950 | ENSFM00500000272384 | Gamma Aminobutyric Acid Receptor Associated 2 Precursor Gaba A Receptor Associated 2 Ganglioside Expression Factor 2 Gef 2 General Transport Factor P16 Golgi Associated Atpase Enhancer Of 16 Kda Gate 16 MAP1 Light Chain 3 Related |
| ENSMUSG00000031960 | ENSFM00570000894312 | Unknown |
| ENSMUSG00000031960 | ENSFM00250000001208 | Alanine Trna Ligase |
| ENSMUSG00000031969 | ENSFM00570000879755 | Unknown |
| ENSMUSG00000031969 | ENSFM00730001522325 | Isobutyryl Coa Dehydrogenase Mitochondrial Precursor EC_1.3.99.- Acyl Coa Dehydrogenase Family Member 8 Acad 8 |
| ENSMUSG00000031971 | ENSFM00250000006451 | Centriole Cilia And Spindle Associated |
| ENSMUSG00000031972 | ENSFM00800002003372 | Actin Muscle Precursor Alpha Actin |
| ENSMUSG00000031974 | ENSFM00730001521332 | Atp Mitochondrial Precursor Abc Transporter |
| ENSMUSG00000031983 | ENSFM00250000010056 | Uncharacterized |
| ENSMUSG00000031985 | ENSFM00250000004371 | Dihydroxyacetone Phosphate Acyltransferase Dap At Dhap At EC_2.3.1.42 Acyl Coa:Dihydroxyacetonephosphateacyltransferase Glycerone Phosphate O Acyltransferase |
| ENSMUSG00000031988 | ENSFM00810002040678 | Vacuolar Sorting Associated Vesicle Sorting |
| ENSMUSG00000031990 | ENSFM00500000272551 | Junctional Adhesion Molecule C Precursor Jam C Junctional Adhesion Molecule 3 Jam 3 |
| ENSMUSG00000031996 | ENSFM00250000000617 | Amyloid Beta A4 Precursor Abpp App Alzheimer Disease Amyloid A4 Homolog |
| ENSMUSG00000032011 | ENSFM00250000009003 | Thy 1 Membrane Glycoprotein Precursor Thy 1 Antigen CD90 Antigen |
| ENSMUSG00000032012 | ENSFM00500000270858 | Nectin 1 Precursor Herpes Virus Entry Mediator C Herpesvirus Entry Mediator C Hvec Poliovirus Receptor Related 1 CD111 Antigen |
| ENSMUSG00000032020 | ENSFM00570000885990 | Unknown |
| ENSMUSG00000032020 | ENSFM00660001189901 | Unknown |
| ENSMUSG00000032020 | ENSFM00250000002027 | Ubiquitin Associated And SH3 Domain Containing B EC_3.1.3.48 Cbl Interacting P70 Suppressor Of T Cell Receptor Signaling 1 Sts 1 T Cell Ubiquitin Ligand 2 Tula 2 Tyrosine Phosphatase STS1/TULA2 |
| ENSMUSG00000032024 | ENSFM00500000272636 | Cxadr Membrane Precursor Adipocyte Adhesion Molecule Coxsackie And Adenovirus Receptor Membrane Car Membrane |
| ENSMUSG00000032026 | ENSFM00500000271661 | Oligoribonuclease Mitochondrial Precursor EC_3.1.-.- Rna Exonuclease 2 Homolog Small Fragment Nuclease |
| ENSMUSG00000032030 | ENSFM00570000899340 | Unknown |
| ENSMUSG00000032030 | ENSFM00730001522203 | Cullin 5 Cul 5 Vasopressin Activated Calcium Mobilizing Receptor 1 Vacm 1 |
| ENSMUSG00000032036 | ENSFM00760001763379 | Unknown |
| ENSMUSG00000032036 | ENSFM00250000000717 | Kin Of Irre Precursor Kin Of Irregular Chiasm Nephrin |
| ENSMUSG00000032040 | ENSFM00500000271098 | M7GPPPX Diphosphatase EC_3.6.1.59 Dcs 1 Decapping Scavenger Enzyme Hint Related 7MEGMP Directed Hydrolase Histidine Triad Nucleotide Binding 5 Histidine Triad Member 5 Hint 5 Scavenger Decapping Enzyme Dcps |
| ENSMUSG00000032042 | ENSFM00790001963023 | Signal Recognition Particle Receptor Subunit Alpha Sr Alpha Docking Alpha Dp Alpha |
| ENSMUSG00000032046 | ENSFM00570000877269 | Unknown |
| ENSMUSG00000032046 | ENSFM00570000892528 | Unknown |
| ENSMUSG00000032046 | ENSFM00250000002071 | Monoacylglycerol Lipase ABHD12 EC_3.1.1.23 2 Arachidonoylglycerol Hydrolase Abhydrolase Domain Containing 12 |
| ENSMUSG00000032047 | ENSFM00730001521421 | Acetyl Coa Acetyltransferase Mitochondrial Precursor EC_2.3.1.9 Acetoacetyl Coa Thiolase |
| ENSMUSG00000032050 | ENSFM00250000000517 | Moesin |
| ENSMUSG00000032060 | ENSFM00670001235476 | Alpha Crystallin Chain |
| ENSMUSG00000032076 | ENSFM00250000001126 | Cell Adhesion Molecule Precursor Immunoglobulin Superfamily Member Nectin Necl Synaptic Cell Adhesion Molecule |
| ENSMUSG00000032081 | ENSFM00570000880516 | Unknown |
| ENSMUSG00000032081 | ENSFM00600000926313 | Apolipoprotein C Iii Precursor Apo Ciii Apoc Iii Apolipoprotein C3 |
| ENSMUSG00000032083 | ENSFM00470000251509 | Apolipoprotein A I Precursor Apo Ai Apoa I Apolipoprotein A1 |
| ENSMUSG00000032096 | ENSFM00810002040812 | Coatomer Subunit Delta Delta Coat Delta Cop |
| ENSMUSG00000032097 | ENSFM00540000717952 | Atp Dependent Rna Helicase EC_3.6.4.13 |
| ENSMUSG00000032109 | ENSFM00800002003951 | Nlr Family Member X1 Precursor |
| ENSMUSG00000032112 | ENSFM00500000271162 | Trafficking Particle Complex Subunit 4 |
| ENSMUSG00000032115 | ENSFM00780001898382 | Hypoxia Up Regulated 1 Precursor |
| ENSMUSG00000032116 | ENSFM00570000897300 | Unknown |
| ENSMUSG00000032116 | ENSFM00250000001541 | Dolichyl Diphosphooligosaccharide Glycosyltransferase Subunit Oligosaccharyl Transferase Subunit STT3 EC_2.4.99.18 |
| ENSMUSG00000032120 | ENSFM00260000050940 | C2 Domain Containing 2 Transmembrane 24 |
| ENSMUSG00000032127 | ENSFM00250000005369 | E3 Ubiquitin Ligase PEP5 |
| ENSMUSG00000032135 | ENSFM00570000885775 | Unknown |
| ENSMUSG00000032135 | ENSFM00500000271112 | Cell Surface Glycoprotein MUC18 Precursor Melanoma Cell Adhesion Molecule Melanoma Associated Antigen MUC18 CD146 Antigen |
| ENSMUSG00000032171 | ENSFM00670001235859 | Peptidyl Prolyl Cis Trans Isomerase Nima Interacting 1 EC_5.2.1.8 Peptidyl Prolyl Cis Trans Isomerase PIN1 Ppiase PIN1 |
| ENSMUSG00000032172 | ENSFM00320000100099 | Noelin Precursor Neuronal Olfactomedin Related Er Localized Olfactomedin 1 |
| ENSMUSG00000032174 | ENSFM00250000002453 | Intercellular Adhesion Molecule 1 Precursor Icam 1 CD54 Antigen |
| ENSMUSG00000032186 | ENSFM00500000269813 | Tropomodulin Tropomodulin Tmod |
| ENSMUSG00000032192 | ENSFM00770001847629 | Guanine Nucleotide Binding Subunit Beta 5 GBETA5 Transducin Beta Chain 5 |
| ENSMUSG00000032216 | ENSFM00740001613563 | Unknown |
| ENSMUSG00000032216 | ENSFM00730001521822 | E3 Ubiquitin Ligase EC_6.3.2.- |
| ENSMUSG00000032220 | ENSFM00780001898391 | Unconventional Myosin Ie Unconventional Myosin 1E |
| ENSMUSG00000032224 | ENSFM00570000901107 | Unknown |
| ENSMUSG00000032224 | ENSFM00480000262849 | FAM81A |
| ENSMUSG00000032231 | ENSFM00730001521439 | Annexin Annexin Annexin Calpactin Calpactin Chromobindin Lipocortin I |
| ENSMUSG00000032261 | ENSFM00760001743793 | Unknown |
| ENSMUSG00000032261 | ENSFM00670001236492 | SH3 Domain Binding Glutamic Acid Rich 2 |
| ENSMUSG00000032264 | ENSFM00250000006432 | Centromere/Kinetochore ZW10 Homolog |
| ENSMUSG00000032279 | ENSFM00810002040683 | Isocitrate Dehydrogenase Subunit Alpha Mitochondrial Precursor EC_1.1.1.41 Isocitric Dehydrogenase Subunit Alpha Nad + Specific Icdh Subunit Alpha |
| ENSMUSG00000032281 | ENSFM00570000877931 | Unknown |
| ENSMUSG00000032281 | ENSFM00260000050566 | Long Chain Fatty Acid Coa Ligase EC_6.2.1.3 Acyl Coa Synthetase Bubblegum Family Member |
| ENSMUSG00000032281 | ENSFM00670001256713 | Long Chain Fatty Acid Coa Ligase ACSBG1 |
| ENSMUSG00000032285 | ENSFM00760001714614 | Dnaj Homolog Subfamily A Member Precursor |
| ENSMUSG00000032290 | ENSFM00500000271161 | Tyrosine Phosphatase Non Receptor Type 9 EC_3.1.3.48 Tyrosine Phosphatase MEG2 Ptpase MEG2 |
| ENSMUSG00000032294 | ENSFM00250000000989 | Pyruvate Kinase Pkm EC_2.7.1.40 Pyruvate Kinase Muscle Isozyme |
| ENSMUSG00000032295 | ENSFM00590000917310 | Unknown |
| ENSMUSG00000032295 | ENSFM00590000917311 | Unknown |
| ENSMUSG00000032295 | ENSFM00590000917313 | Unknown |
| ENSMUSG00000032295 | ENSFM00590000918415 | Unknown |
| ENSMUSG00000032295 | ENSFM00250000003741 | Alpha Mannosidase 2C1 EC_3.2.1.24 Alpha D Mannoside Mannohydrolase Mannosidase Alpha Class 2C Member 1 |
| ENSMUSG00000032301 | ENSFM00500000270239 | Proteasome Subunit Alpha Type EC_3.4.25.1 Macropain Subunit Multicatalytic Endopeptidase Complex Subunit Proteasome Component Subunit |
| ENSMUSG00000032313 | ENSFM00250000007298 | Transmembrane |
| ENSMUSG00000032314 | ENSFM00250000004026 | Electron Transfer Flavoprotein Subunit Alpha Mitochondrial Precursor Alpha Etf |
| ENSMUSG00000032320 | ENSFM00570000891778 | Unknown |
| ENSMUSG00000032320 | ENSFM00500000271265 | Reticulocalbin 2 Precursor Taipoxin Associated Calcium Binding 49 Tcbp 49 |
| ENSMUSG00000032328 | ENSFM00500000270036 | Cell Cycle Control 50A P4 Atpase Flippase Complex Beta Subunit TMEM30A Transmembrane 30A |
| ENSMUSG00000032330 | ENSFM00670001243313 | Cytochrome C Oxidase Subunit 7A2 Mitochondrial Precursor Cytochrome C Oxidase Subunit Viia Liver/Heart Cytochrome C Oxidase Subunit Viia L |
| ENSMUSG00000032336 | ENSFM00660001189906 | Unknown |
| ENSMUSG00000032336 | ENSFM00610000952905 | Basigin Precursor CD147 Antigen |
| ENSMUSG00000032338 | ENSFM00250000000931 | Potassium/Sodium Hyperpolarization Activated Cyclic Nucleotide Gated Channel |
| ENSMUSG00000032340 | ENSFM00260000050557 | Netrin Receptor Dcc Precursor Tumor Suppressor Dcc |
| ENSMUSG00000032348 | ENSFM00270000056439 | Glutathione S Transferase EC_2.5.1.18 Gst Class Alpha Member Glutathione S Transferase |
| ENSMUSG00000032349 | ENSFM00570000895578 | Unknown |
| ENSMUSG00000032349 | ENSFM00760001714825 | Elongation Of Very Long Chain Fatty Acids 5 EC_2.3.1.199 3 Keto Acyl Coa Synthase ELOVL5 Elovl Fatty Acid Elongase 5 Elovl Fa Elongase 5 Very Long Chain 3 Oxoacyl Coa Synthase 5 |
| ENSMUSG00000032355 | ENSFM00740001605474 | Unknown |
| ENSMUSG00000032355 | ENSFM00740001589545 | Muscular Lmna Interacting Muscular Enriched A Type Laminin Interacting |
| ENSMUSG00000032356 | ENSFM00250000001613 | Ras Specific Guanine Nucleotide Releasing Factor 2 Ras GRF2 Ras Guanine Nucleotide Exchange Factor 2 |
| ENSMUSG00000032356 | ENSFM00500000289379 | Ras Specific Guanine Nucleotide Releasing Factor 1 CRA_B |
| ENSMUSG00000032366 | ENSFM00720001503517 | Unknown |
| ENSMUSG00000032366 | ENSFM00250000000220 | Tropomyosin Alpha 1 Chain Alpha Tropomyosin Tropomyosin 1 |
| ENSMUSG00000032370 | ENSFM00250000007515 | Serine Beta Lactamase Lactb Mitochondrial Precursor EC_3.4.-.- |
| ENSMUSG00000032382 | ENSFM00730001521918 | Sorting Nexin |
| ENSMUSG00000032383 | ENSFM00500000269861 | Peptidyl Prolyl Cis Trans Isomerase B Precursor Ppiase B EC_5.2.1.8 Rotamase B |
| ENSMUSG00000032384 | ENSFM00570000899402 | Unknown |
| ENSMUSG00000032384 | ENSFM00730001521484 | Casein Kinase I Gamma Cki Gamma EC_2.7.11.1 |
| ENSMUSG00000032399 | ENSFM00250000002321 | 60S Ribosomal L4 |
| ENSMUSG00000032412 | ENSFM00670001235362 | Sodium/Potassium Transporting Atpase Subunit Beta Sodium/Potassium Dependent Atpase Subunit Beta |
| ENSMUSG00000032418 | ENSFM00250000000979 | Nadp Dependent Malic Enzyme Nadp Me EC_1.1.1.40 Malic Enzyme |
| ENSMUSG00000032420 | ENSFM00250000001233 | 5' Nucleotidase Precursor 5' Nt EC_3.1.3.5 Ecto 5' Nucleotidase |
| ENSMUSG00000032423 | ENSFM00640001116175 | Unknown |
| ENSMUSG00000032423 | ENSFM00730001521753 | Heterogeneous Nuclear Ribonucleoprotein Q Hnrnp Q Glycine And Tyrosine Rich Rna Binding Gry Rbp NS1 Associated 1 Synaptotagmin Binding Cytoplasmic Rna Interacting |
| ENSMUSG00000032435 | ENSFM00250000001777 | Cytoplasmic Dynein 1 Light Intermediate Chain 1 Dynein Light Chain A Dlc A Dynein Light Intermediate Chain 1 Cytosolic |
| ENSMUSG00000032437 | ENSFM00250000001541 | Dolichyl Diphosphooligosaccharide Glycosyltransferase Subunit Oligosaccharyl Transferase Subunit STT3 EC_2.4.99.18 |
| ENSMUSG00000032458 | ENSFM00260000050742 | Coatomer Subunit Beta' Beta' Coat Beta' Cop |
| ENSMUSG00000032459 | ENSFM00250000006229 | 28S Ribosomal S22 Mitochondrial Mrp S22 S22MT |
| ENSMUSG00000032462 | ENSFM00570000900907 | Unknown |
| ENSMUSG00000032462 | ENSFM00730001521385 | Phosphatidylinositol 4 5 Bisphosphate 3 Kinase Catalytic Subunit PI3 Kinase Subunit PI3K Ptdins 3 Kinase Subunit EC_2.7.1.153 Phosphatidylinositol 4 5 Bisphosphate 3 Kinase 110 Kda Catalytic Subunit Ptdins 3 Kinase Subunit P110 |
| ENSMUSG00000032468 | ENSFM00250000004654 | Armadillo Repeat Containing 8 |
| ENSMUSG00000032470 | ENSFM00770001847543 | Ras Related R Ras |
| ENSMUSG00000032479 | ENSFM00800002020841 | Unknown |
| ENSMUSG00000032479 | ENSFM00260000050563 | Microtubule Associated 4 Map 4 |
| ENSMUSG00000032480 | ENSFM00800002017283 | Unknown |
| ENSMUSG00000032480 | ENSFM00800002024439 | Unknown |
| ENSMUSG00000032480 | ENSFM00800002024440 | Unknown |
| ENSMUSG00000032480 | ENSFM00800002024441 | Unknown |
| ENSMUSG00000032480 | ENSFM00800002024442 | Unknown |
| ENSMUSG00000032480 | ENSFM00260000050346 | Atp Dependent Rna Helicase EC_3.6.4.13 Deah Box |
| ENSMUSG00000032482 | ENSFM00760001715141 | Chondroitin Sulfate Proteoglycan 5 Precursor Acidic Leucine Rich Egf Domain Containing Brain Neuroglycan C |
| ENSMUSG00000032504 | ENSFM00500000270726 | Programmed Cell Death 6 Interacting Alg 2 Interacting 1 Alg 2 Interacting X |
| ENSMUSG00000032512 | ENSFM00460000244957 | Wd Repeat Containing 48 |
| ENSMUSG00000032517 | ENSFM00500000311584 | Unknown |
| ENSMUSG00000032517 | ENSFM00600000921972 | Myelin Associated Oligodendrocyte Basic |
| ENSMUSG00000032518 | ENSFM00250000001034 | 40S Ribosomal Sa |
| ENSMUSG00000032523 | ENSFM00260000051120 | Cysteine N Palmitoyltransferase Hhat Glycerol Uptake/Transporter Homolog Hedgehog Acyltransferase |
| ENSMUSG00000032527 | ENSFM00450000242182 | Propionyl Coa Carboxylase Beta Chain Mitochondrial Precursor Pccase Subunit Beta EC_6.4.1.3 Propanoyl Coa:Carbon Dioxide Ligase Subunit Beta |
| ENSMUSG00000032549 | ENSFM00460000244935 | Ras Related Rab 6 |
| ENSMUSG00000032557 | ENSFM00250000004803 | Ubiquitin Modifier Activating Enzyme 5 Ubiquitin Activating Enzyme 5 |
| ENSMUSG00000032558 | ENSFM00250000004673 | Nephrocystin 3 |
| ENSMUSG00000032560 | ENSFM00760001750153 | Unknown |
| ENSMUSG00000032560 | ENSFM00250000002783 | Dnaj Homolog Subfamily C Member 13 Required Receptor Mediated Endocytosis 8 Rme 8 |
| ENSMUSG00000032562 | ENSFM00780001917784 | Unknown |
| ENSMUSG00000032562 | ENSFM00780001917785 | Unknown |
| ENSMUSG00000032562 | ENSFM00410000138458 | Guanine Nucleotide Binding Subunit |
| ENSMUSG00000032564 | ENSFM00800002003632 | Copine Copine |
| ENSMUSG00000032570 | ENSFM00810002040624 | Calcium Transporting Atpase Type 2C Member Atpase EC_3.6.3.8 Secretory Pathway Ca 2+ Atpase |
| ENSMUSG00000032571 | ENSFM00800002003738 | Phosphoinositide 3 Kinase Regulatory Subunit 4 PI3 Kinase Regulatory Subunit 4 EC_2.7.11.1 |
| ENSMUSG00000032575 | ENSFM00590000917349 | Unknown |
| ENSMUSG00000032575 | ENSFM00670001235628 | Mesencephalic Astrocyte Derived Neurotrophic Factor Homolog Precursor Manf/Cdnf |
| ENSMUSG00000032583 | ENSFM00770001847570 | Vacuolar Fusion MON1 |
| ENSMUSG00000032589 | ENSFM00600000921250 | Piccolo Aczonin |
| ENSMUSG00000032590 | ENSFM00250000003584 | Acylamino Acid Releasing Enzyme Aare EC_3.4.19.1 Acyl Peptide Hydrolase Aph Acylaminoacyl Peptidase |
| ENSMUSG00000032598 | ENSFM00780001908780 | Unknown |
| ENSMUSG00000032598 | ENSFM00780001908781 | Unknown |
| ENSMUSG00000032598 | ENSFM00780001917797 | Unknown |
| ENSMUSG00000032598 | ENSFM00250000006300 | Nck Interacting With SH3 Domain 54 Kda Vaca Interacting Kda Interacting 90 Kda SH3 Interacting With Nck SH3 Adapter SPIN90 Wasp Interacting SH3 Domain Wish Wiskott Aldrich Syndrome |
| ENSMUSG00000032601 | ENSFM00780001917795 | Unknown |
| ENSMUSG00000032601 | ENSFM00730001521993 | Camp Dependent Kinase Type Ii Regulatory Subunit |
| ENSMUSG00000032602 | ENSFM00760001714734 | Mitochondrial Carnitine/Acylcarnitine Carrier Carnitine/Acylcarnitine Translocase Cac Solute Carrier Family 25 Member 20 |
| ENSMUSG00000032604 | ENSFM00780001898374 | Glutamine Trna Ligase EC_6.1.1.18 Glutaminyl Trna Synthetase Glnrs |
| ENSMUSG00000032606 | ENSFM00250000009545 | Nicolin 1 Tubulin Polyglutamylase Complex Subunit 5 PGS5 |
| ENSMUSG00000032615 | ENSFM00250000006985 | 5' 3' Deoxyribonucleotidase Cytosolic Type EC_3.1.3.- Cytosolic 5' 3' Pyrimidine Nucleotidase Deoxy 5' Nucleotidase 1 Dnt 1 |
| ENSMUSG00000032625 | ENSFM00740001589337 | Thrombospondin Type 1 Domain Containing Precursor |
| ENSMUSG00000032648 | ENSFM00250000001172 | Glycogen Phosphorylase Form EC_2.4.1.1 |
| ENSMUSG00000032666 | ENSFM00500000274384 | Uncharacterized |
| ENSMUSG00000032666 | ENSFM00760001721091 | Unknown |
| ENSMUSG00000032667 | ENSFM00250000001840 | Serum Paraoxonase/Arylesterase 2 Pon 2 EC_3.1.1.2 Aromatic Esterase 2 A Esterase 2 Serum Aryldialkylphosphatase 2 |
| ENSMUSG00000032705 | ENSFM00250000005784 | Exonuclease 3' 5' Domain Containing 2 Exonuclease 3' 5' Domain Containing 2 |
| ENSMUSG00000032735 | ENSFM00250000000459 | Actin Binding Lim Ablim Actin Binding Lim Family Member |
| ENSMUSG00000032740 | ENSFM00250000000961 | Girdin Akt Phosphorylation Enhancer Ape Coiled Coil Domain Containing 88A G Alpha Interacting Vesicle Associated Giv Girders Of Actin Filament Hook Related 1 HKRP1 |
| ENSMUSG00000032757 | ENSFM00570000852347 | BET1 Homolog Golgi Vesicular Membrane Trafficking P18 |
| ENSMUSG00000032763 | ENSFM00250000005381 | Acetolactate Synthase EC_2.2.1.- Ilvb |
| ENSMUSG00000032773 | ENSFM00660001189510 | Unknown |
| ENSMUSG00000032773 | ENSFM00800002003412 | Muscarinic Acetylcholine Receptor |
| ENSMUSG00000032788 | ENSFM00500000270834 | Pyridoxal Kinase EC_2.7.1.35 Pyridoxine Kinase |
| ENSMUSG00000032796 | ENSFM00740001589230 | Laminin Subunit Alpha 2 Precursor Laminin M Chain Laminin 12 Subunit Alpha Laminin 2 Subunit Alpha Laminin 4 Subunit Alpha Merosin Heavy Chain |
| ENSMUSG00000032803 | ENSFM00500000272616 | CDV3 Homolog |
| ENSMUSG00000032816 | ENSFM00740001589250 | Immunoglobulin Superfamily Dcc Subclass Member Precursor |
| ENSMUSG00000032826 | ENSFM00600000932983 | Uncharacterized |
| ENSMUSG00000032826 | ENSFM00720001501960 | Unknown |
| ENSMUSG00000032826 | ENSFM00500000269628 | Ankyrin Ank Ankyrin |
| ENSMUSG00000032826 | ENSFM00720001502851 | Ankyrin 2 |
| ENSMUSG00000032827 | ENSFM00250000001450 | Neurabin 1 Neurabin I Neural Tissue Specific F Actin Binding I Phosphatase 1 Regulatory Subunit 9A |
| ENSMUSG00000032842 | ENSFM00740001589321 | Multidrug Resistance Associated 7 Atp Binding Cassette Sub Family C Member 10 |
| ENSMUSG00000032854 | ENSFM00730001522351 | 2 Hydroxyacylsphingosine 1 Beta Galactosyltransferase Precursor EC_2.4.1.45 Ceramide Udp Galactosyltransferase Cerebroside Synthase Udp Galactose Ceramide Galactosyltransferase |
| ENSMUSG00000032878 | ENSFM00740001589413 | Coiled Coil Domain Containing |
| ENSMUSG00000032883 | ENSFM00730001521866 | Long Chain Fatty Acid Coa Ligase EC_6.2.1.3 Long Chain Acyl Coa Synthetase Lacs |
| ENSMUSG00000032890 | ENSFM00250000000629 | Regulating Synaptic Membrane Exocytosis Rab 3 Interacting Molecule Rim |
| ENSMUSG00000032902 | ENSFM00730001521446 | Monocarboxylate Transporter Mct Solute Carrier Family 16 Member |
| ENSMUSG00000032911 | ENSFM00570000851341 | Chondroitin Sulfate Proteoglycan 4 Precursor Chondroitin Sulfate Proteoglycan NG2 |
| ENSMUSG00000032932 | ENSFM00730001522410 | Heat Shock 70 Kda 13 Precursor Microsomal Stress 70 Atpase Core Stress 70 Chaperone Microsome Associated 60 Kda |
| ENSMUSG00000032936 | ENSFM00780001915889 | Unknown |
| ENSMUSG00000032936 | ENSFM00780001917789 | Unknown |
| ENSMUSG00000032936 | ENSFM00800002003895 | Cam Kinase Vesicle Associated |
| ENSMUSG00000032946 | ENSFM00500000318755 | Unknown |
| ENSMUSG00000032946 | ENSFM00500000386335 | Unknown |
| ENSMUSG00000032946 | ENSFM00570000900297 | Unknown |
| ENSMUSG00000032946 | ENSFM00250000001366 | Ras Guanyl Releasing Calcium And Dag Regulated Guanine Nucleotide Exchange Factor Caldag |
| ENSMUSG00000032959 | ENSFM00670001235483 | Phosphatidylethanolamine Binding 1 Pebp 1 Hcnppp |
| ENSMUSG00000032966 | ENSFM00500000293807 | Peptidyl Prolyl Cis Trans Isomerase |
| ENSMUSG00000032966 | ENSFM00670001235580 | Peptidyl Prolyl Cis Trans Isomerase FKBP1B Ppiase FKBP1B EC_5.2.1.8 12.6 Kda FK506 Binding 12.6 Kda Fkbp Fkbp 12 6 FK506 Binding 1B Fkbp 1B Immunophilin FKBP12 6 Rotamase |
| ENSMUSG00000033004 | ENSFM00250000001622 | E3 Ubiquitin Ligase MYCBP2 EC_6.3.2.- Myc Binding 2 Pam/Highwire/Rpm 1 Protein Associated With Myc |
| ENSMUSG00000033047 | ENSFM00250000003239 | Eukaryotic Translation Initiation Factor 3 Subunit L |
| ENSMUSG00000033059 | ENSFM00250000001172 | Glycogen Phosphorylase Form EC_2.4.1.1 |
| ENSMUSG00000033060 | ENSFM00250000003075 | Lim Domain Only 7 Lmo 7 F Box Only 20 Lomp |
| ENSMUSG00000033065 | ENSFM00730001521507 | Atp Dependent 6 Phosphofructokinase Type |
| ENSMUSG00000033066 | ENSFM00250000005898 | Growth Arrest Specific 7 Gas 7 |
| ENSMUSG00000033068 | ENSFM00250000002349 | Ectonucleoside Triphosphate Diphosphohydrolase 5 Precursor Ntpdase 5 EC_3.6.1.6 Guanosine Diphosphatase ENTPD5 Gdpase ENTPD5 EC_3.6.1.- 42 Uridine Diphosphatase ENTPD5 Udpase ENTPD5 |
| ENSMUSG00000033096 | ENSFM00440000236938 | Adipocyte Plasma Membrane Associated |
| ENSMUSG00000033124 | ENSFM00760001760759 | Unknown |
| ENSMUSG00000033124 | ENSFM00730001522009 | Autophagy Related APG9 |
| ENSMUSG00000033152 | ENSFM00250000007351 | Podocalyxin 2 Precursor Endoglycan |
| ENSMUSG00000033161 | ENSFM00650001139994 | Sodium/Potassium Transporting Atpase Subunit Alpha Na + /K + Atpase Alpha Subunit EC_3.6.3.9 Sodium Pump Subunit Alpha |
| ENSMUSG00000033174 | ENSFM00250000006453 | Monoglyceride Lipase Mgl EC_3.1.1.23 Monoacylglycerol Lipase Magl |
| ENSMUSG00000033184 | ENSFM00500000270192 | Transmembrane EMP24 Domain Containing Precursor P24 Family Gamma |
| ENSMUSG00000033196 | ENSFM00250000000024 | Myosin Myosin Heavy Chain Myosin Heavy Chain Muscle |
| ENSMUSG00000033208 | ENSFM00670001236994 | S100 B S 100 Beta Chain S 100 Subunit Beta S100 Calcium Binding B |
| ENSMUSG00000033272 | ENSFM00670001240014 | Uncharacterized |
| ENSMUSG00000033272 | ENSFM00250000007335 | Probable Udp Sugar Transporter Sl Solute Carrier Family 35 Member A4 |
| ENSMUSG00000033295 | ENSFM00740001589097 | Receptor Type Tyrosine Phosphatase Precursor EC_3.1.3.48 |
| ENSMUSG00000033307 | ENSFM00670001235997 | Macrophage Migration Inhibitory Factor Mif EC_5.3.2.1 L Dopachrome Isomerase L Dopachrome Tautomerase EC_5.3.3.- 12 Phenylpyruvate Tautomerase |
| ENSMUSG00000033335 | ENSFM00640001119884 | Unknown |
| ENSMUSG00000033335 | ENSFM00520000517792 | Dynamin EC_3.6.5.5 |
| ENSMUSG00000033352 | ENSFM00810002045789 | Unknown |
| ENSMUSG00000033352 | ENSFM00790001963039 | Dual Specificity Mitogen Activated Kinase Kinase 4 Map Kinase Kinase 4 Mapkk 4 EC_2.7.12.2 Jnk Activating Kinase 1 Mapk/Erk Kinase 4 Mek 4 Sapk/Erk Kinase 1 SEK1 |
| ENSMUSG00000033382 | ENSFM00250000003061 | Trafficking Particle Complex Subunit 8 TRS85 Homolog |
| ENSMUSG00000033389 | ENSFM00250000000900 | Rho Gtpase Activating Rho Type Gtpase Activating Rhogap Interacting With CIP4 Homologs Rich |
| ENSMUSG00000033392 | ENSFM00250000000820 | Clip Associating Cytoplasmic Linker Associated |
| ENSMUSG00000033400 | ENSFM00810002040809 | Glycogen Debranching Enzyme Glycogen Debrancher |
| ENSMUSG00000033419 | ENSFM00250000000859 | Clathrin Assembly |
| ENSMUSG00000033436 | ENSFM00250000001585 | Armadillo Repeat Containing X Linked |
| ENSMUSG00000033475 | ENSFM00500000297210 | Unknown |
| ENSMUSG00000033488 | ENSFM00780001915243 | Unknown |
| ENSMUSG00000033488 | ENSFM00500000276691 | Quinone Oxidoreductase 2 Ec 1 |
| ENSMUSG00000033530 | ENSFM00250000002093 | Tetratricopeptide Repeat Tpr Repeat |
| ENSMUSG00000033569 | ENSFM00250000000737 | Brain Specific Angiogenesis Inhibitor Precursor |
| ENSMUSG00000033577 | ENSFM00730001522029 | Unconventional Myosin Vi Unconventional Myosin 6 |
| ENSMUSG00000033578 | ENSFM00250000006851 | Transmembrane 35 |
| ENSMUSG00000033594 | ENSFM00740001589644 | Spermatogenesis Associated 2 SPATA2 |
| ENSMUSG00000033595 | ENSFM00250000002312 | Leucine Rich Repeat Lgi Family Member Precursor Leucine Rich Glioma Inactivated |
| ENSMUSG00000033597 | ENSFM00250000001241 | Caskin 1 Cask Interacting 1 |
| ENSMUSG00000033615 | ENSFM00570000891587 | Unknown |
| ENSMUSG00000033615 | ENSFM00670001235939 | Complexin Complexin Cpx Synaphin |
| ENSMUSG00000033624 | ENSFM00800002003582 | Pyruvate Dehydrogenase Phosphatase Regulatory Subunit Mitochondrial Precursor Pdpr |
| ENSMUSG00000033628 | ENSFM00410000138522 | Phosphatidylinositol 3 Kinase Catalytic Subunit Type 3 PI3 Kinase Type 3 PI3K Type 3 Ptdins 3 Kinase Type 3 EC_2.7.1.137 Phosphoinositide 3 Kinase Class 3 |
| ENSMUSG00000033629 | ENSFM00500000271426 | Very Long Chain 3R 3 Hydroxyacyl Coa Dehydratase 3 EC_4.2.1.134 3 Hydroxyacyl Coa Dehydratase 3 HACD3 Tyrosine Phosphatase PTPLAD1 Tyrosine Phosphatase A Domain Containing 1 |
| ENSMUSG00000033676 | ENSFM00400000131718 | Gamma Aminobutyric Acid Receptor Subunit Beta Precursor Gaba A Receptor Subunit Beta |
| ENSMUSG00000033697 | ENSFM00660001189878 | Unknown |
| ENSMUSG00000033697 | ENSFM00660001189879 | Unknown |
| ENSMUSG00000033697 | ENSFM00660001189880 | Unknown |
| ENSMUSG00000033697 | ENSFM00660001189881 | Unknown |
| ENSMUSG00000033697 | ENSFM00250000002627 | Rho Gtpase Activating 39 |
| ENSMUSG00000033717 | ENSFM00800002003463 | Alpha 2A Adrenergic Receptor Alpha 2A Adrenoreceptor Alpha 2A Adrenoceptor Alpha 2AAR |
| ENSMUSG00000033720 | ENSFM00470000258106 | Unknown |
| ENSMUSG00000033720 | ENSFM00740001589514 | Sideroflexin 5 |
| ENSMUSG00000033735 | ENSFM00250000004781 | Sepiapterin Reductase Spr EC_1.1.1.153 |
| ENSMUSG00000033751 | ENSFM00500000275503 | Growth Arrest And Dna Damage Inducible Proteins Interacting 1 39S Ribosomal L59 Mitochondrial Mrp L59 Papillomavirus L2 Interacting Nuclear 1 Plinp 1 |
| ENSMUSG00000033763 | ENSFM00250000002052 | Metastasis Suppressor 1 Missing In Metastasis |
| ENSMUSG00000033768 | ENSFM00740001589096 | Neurexin Precursor Neurexin |
| ENSMUSG00000033769 | ENSFM00590000917621 | Unknown |
| ENSMUSG00000033769 | ENSFM00250000001326 | Exocyst Complex Component 6 Exocyst Complex Component SEC15A SEC15 1 |
| ENSMUSG00000033793 | ENSFM00780001913229 | Unknown |
| ENSMUSG00000033793 | ENSFM00250000003378 | V Type Proton Atpase Subunit H V Atpase Subunit H Vacuolar Proton Pump Subunit H |
| ENSMUSG00000033808 | ENSFM00250000002118 | Transmembrane Precursor |
| ENSMUSG00000033819 | ENSFM00590000918398 | Unknown |
| ENSMUSG00000033819 | ENSFM00500000270382 | Phosphatase 1 Regulatory Inhibitor Subunit 16B Precursor Caax Box Timap Tgf Beta Inhibited Membrane Associated |
| ENSMUSG00000033860 | ENSFM00800002003901 | Fibrinogen Gamma Chain Precursor |
| ENSMUSG00000033900 | ENSFM00780001912614 | Unknown |
| ENSMUSG00000033900 | ENSFM00250000007724 | Microtubule Associated 9 Aster Associated |
| ENSMUSG00000033916 | ENSFM00600000921262 | Charged Multivesicular Body 2A Chromatin Modifying 2A CHMP2A |
| ENSMUSG00000033917 | ENSFM00250000004887 | Glycerophosphodiester Phosphodiesterase 1 EC_3.1.4.44 Membrane Interacting Of RGS16 |
| ENSMUSG00000033938 | ENSFM00670001236404 | Nadh Dehydrogenase 1 Beta Subcomplex Subunit 7 Complex I B18 Ci B18 Nadh Ubiquinone Oxidoreductase B18 Subunit |
| ENSMUSG00000033940 | ENSFM00500000311475 | Unknown |
| ENSMUSG00000033953 | ENSFM00500000269882 | Calcineurin Subunit B Regulatory Subunit Phosphatase Regulatory Subunit |
| ENSMUSG00000033960 | ENSFM00790001963123 | Junctional Associated With Coronary Artery Disease Jcad |
| ENSMUSG00000033981 | ENSFM00720001492020 | Glutamate Receptor Precursor Glur Ampa Selective Glutamate Receptor Glur Glutamate Receptor Ionotropic Ampa |
| ENSMUSG00000033987 | ENSFM00730001521362 | Dynein Heavy Chain Axonemal Axonemal Beta Dynein Heavy Chain Ciliary Dynein Heavy Chain |
| ENSMUSG00000033998 | ENSFM00600000921306 | Potassium Channel Subfamily K Member 1 Inward Rectifying Potassium Channel Twik 1 |
| ENSMUSG00000034000 | ENSFM00250000002795 | Sialidase 3 EC_3.2.1.18 Ganglioside Sialidase Membrane Sialidase N Acetyl Alpha Neuraminidase 3 |
| ENSMUSG00000034024 | ENSFM00500000270021 | T Complex 1 Subunit Beta Tcp 1 Beta Cct Beta |
| ENSMUSG00000034040 | ENSFM00500000269893 | Polypeptide N Acetylgalactosaminyltransferase EC_2.4.1.41 Polypeptide Galnac Transferase Galnac T Pp Gantase Udp Acetylgalactosaminyltransferase Udp Galnac:Polypeptide N Acetylgalactosaminyltransferase |
| ENSMUSG00000034064 | ENSFM00250000005054 | O Glucosyltransferase 1 Precursor EC_2.4.1.- O Glucosyltransferase Rumi Homolog O Xylosyltransferase EC_2.4.2.26 |
| ENSMUSG00000034075 | ENSFM00500000270072 | Palmitoyltransferase ZDHHC5 EC_2.3.1.225 Zinc Finger Dhhc Domain Containing 5 Dhhc 5 |
| ENSMUSG00000034088 | ENSFM00760001763124 | Unknown |
| ENSMUSG00000034088 | ENSFM00760001763125 | Unknown |
| ENSMUSG00000034088 | ENSFM00740001589253 | Vigilin High Density Lipoprotein Binding Hdl Binding |
| ENSMUSG00000034135 | ENSFM00570000893436 | Unknown |
| ENSMUSG00000034135 | ENSFM00730001521674 | Serine/Threonine Kinase EC_2.7.11.1 Salt Inducible Kinase Sik Serine/Threonine Kinase SNF1 Kinase |
| ENSMUSG00000034145 | ENSFM00250000001207 | Transmembrane |
| ENSMUSG00000034152 | ENSFM00250000001333 | Exocyst Complex Component 3 Exocyst Complex Component SEC6 |
| ENSMUSG00000034160 | ENSFM00250000002216 | Udp N Acetylglucosamine Peptide N Acetylglucosaminyltransferase 110 Kda Subunit EC_2.4.1.255 O Glcnac Transferase Subunit P110 O Linked N Acetylglucosamine Transferase 110 Kda Subunit Ogt |
| ENSMUSG00000034171 | ENSFM00500000270531 | Fatty Acid Amide Hydrolase 1 EC_3.5.1.99 Anandamide Amidohydrolase 1 Oleamide Hydrolase 1 |
| ENSMUSG00000034173 | ENSFM00500000270351 | Zinc Finger Bed Domain Containing |
| ENSMUSG00000034173 | ENSFM00600000921422 | Coiled Coil Helix Coiled Coil Helix Domain Containing Mitochondrial Precursor |
| ENSMUSG00000034187 | ENSFM00640001116846 | Unknown |
| ENSMUSG00000034187 | ENSFM00800002003482 | Vesicle Fusing Atpase EC_3.6.4.6 N Ethylmaleimide Sensitive Fusion Nem Sensitive Fusion Vesicular Fusion Nsf |
| ENSMUSG00000034189 | ENSFM00670001235389 | Estradiol 17 Beta Dehydrogenase 12 EC_1.1.1.62 17 Beta Hydroxysteroid Dehydrogenase 12 17 Beta Hsd 12 |
| ENSMUSG00000034201 | ENSFM00500000270880 | GAS2 GAS2 Related On Chromosome Growth Arrest Specific 2 |
| ENSMUSG00000034216 | ENSFM00250000004354 | Vacuolar Sorting Associated 18 |
| ENSMUSG00000034220 | ENSFM00500000269807 | Glypican Secreted Glypican |
| ENSMUSG00000034243 | ENSFM00250000005925 | Golgin Subfamily B Member 1.372 Kda Golgi Complex Associated GCP372 Giantin Macrogolgin |
| ENSMUSG00000034243 | ENSFM00470000252488 | GOLGB1 |
| ENSMUSG00000034245 | ENSFM00250000007111 | Histone Deacetylase 11 HD11 EC_3.5.1.98 |
| ENSMUSG00000034254 | ENSFM00640001113149 | Unknown |
| ENSMUSG00000034254 | ENSFM00640001117076 | Unknown |
| ENSMUSG00000034254 | ENSFM00500000270410 | 1 Acyl Sn Glycerol 3 Phosphate Acyltransferase Alpha Precursor EC_2.3.1.51 1 Acylglycerol 3 Phosphate O Acyltransferase 1 1 Agp Acyltransferase 1 1 Agpat 1 Lysophosphatidic Acid Acyltransferase Alpha Lpaat Alpha |
| ENSMUSG00000034275 | ENSFM00250000001385 | Turtle Homolog A Precursor Dendrite Arborization And Synapse Maturation 1 Immunoglobulin Superfamily Member 9A IGSF9A |
| ENSMUSG00000034285 | ENSFM00570000884536 | Unknown |
| ENSMUSG00000034285 | ENSFM00250000003191 | Nipsnap Homolog 2 NIPSNAP2 Glioblastoma Amplified Sequence |
| ENSMUSG00000034312 | ENSFM00250000001463 | Iq Motif And SEC7 Domain Containing |
| ENSMUSG00000034336 | ENSFM00730001521354 | Neurofilament Polypeptide Nf Kda Neurofilament Neurofilament Triplet |
| ENSMUSG00000034341 | ENSFM00500000271593 | Ww Domain Binding 2 Wbp 2 |
| ENSMUSG00000034361 | ENSFM00800002003432 | Copine Copine |
| ENSMUSG00000034390 | ENSFM00250000004890 | C Maf Inducing C Mip |
| ENSMUSG00000034412 | ENSFM00260000050553 | TBC1 Domain Family Member |
| ENSMUSG00000034432 | ENSFM00760001745951 | Unknown |
| ENSMUSG00000034432 | ENSFM00760001763114 | Unknown |
| ENSMUSG00000034432 | ENSFM00250000006097 | COP9 Signalosome Complex Subunit 8 SGN8 Signalosome Subunit 8 COP9 Homolog JAB1 Containing Signalosome Subunit 8 |
| ENSMUSG00000034471 | ENSFM00250000001241 | Caskin 1 Cask Interacting 1 |
| ENSMUSG00000034484 | ENSFM00730001521918 | Sorting Nexin |
| ENSMUSG00000034485 | ENSFM00800002003915 | Uveal Autoantigen With Coiled Coil Domains And Ankyrin Repeats |
| ENSMUSG00000034488 | ENSFM00500000270078 | Lactadherin Precursor Mfgm Milk Fat Globule Egf Factor 8 Mfg E8 SED1 |
| ENSMUSG00000034560 | ENSFM00250000004568 | Wash Complex Subunit 7 |
| ENSMUSG00000034566 | ENSFM00500000271216 | Atp Synthase Subunit D Mitochondrial Atpase Subunit D |
| ENSMUSG00000034574 | ENSFM00350000105429 | Disheveled Associated Activator Of Morphogenesis |
| ENSMUSG00000034593 | ENSFM00570000901355 | Unknown |
| ENSMUSG00000034593 | ENSFM00750001632326 | Unconventional Myosin Va Dilute Myosin Heavy Chain Non Muscle |
| ENSMUSG00000034602 | ENSFM00730001522201 | Homolog |
| ENSMUSG00000034613 | ENSFM00250000002013 | Phosphatase EC_3.1.3.16 |
| ENSMUSG00000034648 | ENSFM00250000001289 | Leucine Rich Repeat Neuronal Precursor Neuronal Leucine Rich Repeat Nlrr |
| ENSMUSG00000034656 | ENSFM00740001589100 | Voltage Dependent Type Calcium Channel Subunit Alpha Brain Calcium Channel Calcium Channel L Type Alpha 1 Polypeptide Voltage Gated Calcium Channel Subunit Alpha CAV2 |
| ENSMUSG00000034659 | ENSFM00570000883846 | Unknown |
| ENSMUSG00000034659 | ENSFM00570000897938 | Unknown |
| ENSMUSG00000034659 | ENSFM00250000010758 | Transmembrane 109 Precursor Mitsugumin 23 MG23 |
| ENSMUSG00000034675 | ENSFM00480000262790 | Drebrin Developmentally Regulated Brain |
| ENSMUSG00000034685 | ENSFM00740001589308 | Precursor |
| ENSMUSG00000034686 | ENSFM00250000010083 | Proline Rich 7 Synaptic Proline Rich Membrane |
| ENSMUSG00000034709 | ENSFM00250000003688 | Phosphatase 1 Regulatory Subunit 21 Coiled Coil Domain Containing 128 Klraq Motif Containing 1 |
| ENSMUSG00000034723 | ENSFM00670001235637 | Thioredoxin Related Transmembrane Precursor Thioredoxin Domain Containing |
| ENSMUSG00000034729 | ENSFM00570000888237 | Unknown |
| ENSMUSG00000034729 | ENSFM00250000004828 | 28S Ribosomal S10 Mitochondrial Mrp S10 S10MT |
| ENSMUSG00000034730 | ENSFM00250000000737 | Brain Specific Angiogenesis Inhibitor Precursor |
| ENSMUSG00000034751 | ENSFM00250000000518 | Microtubule Associated Serine/Threonine Kinase EC_2.7.11.1 |
| ENSMUSG00000034751 | ENSFM00670001238533 | Microtubule Associated Serine/Threonine Kinase 4 |
| ENSMUSG00000034781 | ENSFM00730001521471 | Guanine Nucleotide Binding G Q Subunit Alpha Guanine Nucleotide Binding Alpha Q |
| ENSMUSG00000034786 | ENSFM00640001113161 | Unknown |
| ENSMUSG00000034786 | ENSFM00640001113162 | Unknown |
| ENSMUSG00000034786 | ENSFM00600000921937 | G Signaling Modulator 3 |
| ENSMUSG00000034789 | ENSFM00500000272409 | Ras Related Rab 24 |
| ENSMUSG00000034799 | ENSFM00660001189536 | Unknown |
| ENSMUSG00000034799 | ENSFM00250000000599 | Unc 13 Homolog MUNC13 |
| ENSMUSG00000034807 | ENSFM00250000001576 | Procollagen Galactosyltransferase Precursor EC_2.4.1.50 Collagen Beta 1 O Galactosyltransferase Glycosyltransferase 25 Family Member Hydroxylysine Galactosyltransferase |
| ENSMUSG00000034813 | ENSFM00740001589268 | Glutamate Receptor Interacting |
| ENSMUSG00000034820 | ENSFM00500000272686 | Cleavage And Polyadenylation Specificity Factor Subunit 7 |
| ENSMUSG00000034853 | ENSFM00590000916928 | Unknown |
| ENSMUSG00000034853 | ENSFM00250000003357 | Acyl Coenzyme A Thioesterase 12 Acyl Coa Thioesterase 12 EC_3.1.2.1 Acyl Coa Thioester Hydrolase 12 Cytoplasmic Acetyl Coa Hydrolase 1 Cach 1 1 |
| ENSMUSG00000034863 | ENSFM00800002003835 | Anoctamin 8 Transmembrane 16H |
| ENSMUSG00000034868 | ENSFM00730001521616 | Myosin Regulatory Light Myosin Regulatory Light Chain 2 Smooth Muscle Myosin Regulatory Light Chain |
| ENSMUSG00000034891 | ENSFM00600000921732 | Beta Synuclein Phosphoneuroprotein 14 Pnp 14 |
| ENSMUSG00000034892 | ENSFM00500000277386 | Unknown |
| ENSMUSG00000034902 | ENSFM00590000917942 | Unknown |
| ENSMUSG00000034902 | ENSFM00590000918386 | Unknown |
| ENSMUSG00000034902 | ENSFM00590000918387 | Unknown |
| ENSMUSG00000034902 | ENSFM00590000918388 | Phosphatidylinositol 4 Phosphate 5 Kinase Type 1 Gamma |
| ENSMUSG00000034902 | ENSFM00730001521611 | Phosphatidylinositol 4 Phosphate 5 Kinase Type 1 Beta Pi Beta Ptdins 4 P 5 Kinase 1 Beta EC_2.7.1.68 Phosphatidylinositol 4 Phosphate 5 Kinase Type I Phosphatidylinositol 4 Phosphate 5 Kinase Beta |
| ENSMUSG00000034912 | ENSFM00740001599945 | Unknown |
| ENSMUSG00000034912 | ENSFM00250000002191 | Mam Domain Containing Glycosylphosphatidylinositol Anchor 2 Precursor Mam Domain Containing 1 |
| ENSMUSG00000034958 | ENSFM00250000001236 | Ambiguous |
| ENSMUSG00000034958 | ENSFM00570000895557 | Unknown |
| ENSMUSG00000034974 | ENSFM00730001521806 | Death Associated Kinase Dap Kinase EC_2.7.11.1 Dap Kinase |
| ENSMUSG00000034981 | ENSFM00250000008366 | Prostate Androgen Regulated Mucin 1 Precursor Parm 1 |
| ENSMUSG00000034993 | ENSFM00500000270300 | Synaptic Vesicle Membrane Vat 1 Ec 1 |
| ENSMUSG00000034994 | ENSFM00760001714607 | Elongation Factor 2 Ef 2 |
| ENSMUSG00000035067 | ENSFM00500000269993 | Xk Related |
| ENSMUSG00000035067 | ENSFM00670001258781 | Xk Related |
| ENSMUSG00000035104 | ENSFM00570000899708 | Unknown |
| ENSMUSG00000035104 | ENSFM00600000921769 | Eva 1 Homolog A FAM176A Transmembrane 166 |
| ENSMUSG00000035133 | ENSFM00250000002482 | Rho Gtpase Activating 35 Glucocorticoid Receptor Dna Binding Factor 1 |
| ENSMUSG00000035150 | ENSFM00250000002233 | Eukaryotic Translation Initiation Factor 2 Subunit 3 Eukaryotic Translation Initiation Factor 2 Subunit Gamma Eif 2 Gamma |
| ENSMUSG00000035152 | ENSFM00570000901915 | Unknown |
| ENSMUSG00000035152 | ENSFM00430000230089 | Ap 1 Complex Subunit Beta 1 Adaptor Complex Ap 1 Subunit Beta 1 Adaptor Related Complex 1 Subunit Beta 1 Beta 1 Adaptin Beta Adaptin 1 Clathrin Assembly Complex 1 Beta Large Chain Golgi Adaptor HA1/AP1 Adaptin Beta Subunit |
| ENSMUSG00000035168 | ENSFM00250000001143 | Tetratricopeptide Repeat Ankyrin Repeat And Coiled Coil Domain Containing |
| ENSMUSG00000035199 | ENSFM00670001235692 | PRA1 Family 3 Adp Ribosylation Factor 6 Interacting 5 Arl 6 Interacting 5 Aip 5 |
| ENSMUSG00000035202 | ENSFM00250000002298 | Leucine Trna Ligase Mitochondrial Precursor EC_6.1.1.4 Leucyl Trna Synthetase Leurs |
| ENSMUSG00000035203 | ENSFM00570000899105 | Unknown |
| ENSMUSG00000035203 | ENSFM00500000269762 | Epsin Eps 15 Interacting |
| ENSMUSG00000035226 | ENSFM00250000000629 | Regulating Synaptic Membrane Exocytosis Rab 3 Interacting Molecule Rim |
| ENSMUSG00000035227 | ENSFM00250000005526 | Signal Peptidase Complex Subunit 2 EC_3.4.-.- Microsomal Signal Peptidase 25 Kda Subunit Spase 25 Kda Subunit |
| ENSMUSG00000035232 | ENSFM00500000269765 | Kinase Isozyme Mitochondrial Precursor EC_2.7.11.2 Pyruvate Dehydrogenase Kinase |
| ENSMUSG00000035236 | ENSFM00570000877479 | Unknown |
| ENSMUSG00000035236 | ENSFM00570000898233 | Unknown |
| ENSMUSG00000035236 | ENSFM00250000003803 | Scai Suppressor Of Cancer Cell Invasion |
| ENSMUSG00000035246 | ENSFM00250000002167 | Choline Phosphate Cytidylyltransferase A EC_2.7.7.15 Cct Alpha Ctp:Phosphocholine Cytidylyltransferase A Cct A Ct A Phosphorylcholine Transferase A |
| ENSMUSG00000035274 | ENSFM00500000273235 | Trophoblast Glycoprotein Precursor 5T4 Oncofetal Trophoblast Glycoprotein 5T4 Oncotrophoblast Glycoprotein Wnt Activated Inhibitory Factor 1 WAIF1 |
| ENSMUSG00000035284 | ENSFM00250000000977 | Vacuolar Sorting Associated 13A Chorea Acanthocytosis Chorein |
| ENSMUSG00000035285 | ENSFM00600000921959 | N Acetyltransferase 14 EC_2.3.1.- |
| ENSMUSG00000035297 | ENSFM00570000892552 | Unknown |
| ENSMUSG00000035297 | ENSFM00250000005111 | COP9 Signalosome Complex Subunit 4 Signalosome Subunit 4 |
| ENSMUSG00000035325 | ENSFM00760001714697 | Transport SEC31A SEC31 1 SEC31 Related A |
| ENSMUSG00000035376 | ENSFM00500000270421 | Very Long Chain 3R 3 Hydroxyacyl Coa Dehydratase 2 EC_4.2.1.134 3 Hydroxyacyl Coa Dehydratase 2 HACD2 Tyrosine Phosphatase Member B |
| ENSMUSG00000035383 | ENSFM00500000273166 | Pro Mch Precursor |
| ENSMUSG00000035390 | ENSFM00740001589247 | Serine/Threonine Kinase BRSK2 EC_2.7.11.1 EC_2.7.11.- 26 Brain Specific Serine/Threonine Kinase 2 Br Serine/Threonine Kinase 2 Serine/Threonine Kinase Sad A |
| ENSMUSG00000035392 | ENSFM00570000876791 | Unknown |
| ENSMUSG00000035392 | ENSFM00570000883859 | Unknown |
| ENSMUSG00000035392 | ENSFM00570000888231 | Unknown |
| ENSMUSG00000035392 | ENSFM00570000888409 | Unknown |
| ENSMUSG00000035392 | ENSFM00250000001555 | Denn Domain Containing Connecdenn |
| ENSMUSG00000035395 | ENSFM00250000004211 | DDB1 And CUL4 Associated Factor 8 Wd Repeat Containing |
| ENSMUSG00000035441 | ENSFM00770001847571 | Unconventional Myosin |
| ENSMUSG00000035469 | ENSFM00550000769437 | Unknown |
| ENSMUSG00000035469 | ENSFM00640001117070 | Unknown |
| ENSMUSG00000035469 | ENSFM00640001117071 | Unknown |
| ENSMUSG00000035469 | ENSFM00640001119698 | Unknown |
| ENSMUSG00000035469 | ENSFM00260000050642 | RCC1 And Btb Domain Containing 2 Chromosome Condensation 1 Regulator Of Chromosome Condensation And Btb Domain Containing 2 |
| ENSMUSG00000035513 | ENSFM00260000050653 | Netrin Precursor Laminet |
| ENSMUSG00000035547 | ENSFM00570000885619 | Unknown |
| ENSMUSG00000035547 | ENSFM00570000889572 | Unknown |
| ENSMUSG00000035547 | ENSFM00730001522149 | Calpain |
| ENSMUSG00000035561 | ENSFM00730001521185 | Dehydrogenase Aldehyde Dehydrogenase Family 1 Member Dehydrogenase |
| ENSMUSG00000035566 | ENSFM00780001898335 | Protocadherin Precursor |
| ENSMUSG00000035596 | ENSFM00570000898185 | Unknown |
| ENSMUSG00000035596 | ENSFM00250000003877 | Lysophospholipid Acyltransferase 7 Lplat 7 EC_2.3.1.- Bladder And Breast Carcinoma Overexpressed Gene 1 Leukocyte Receptor Cluster Member 4 Membrane Bound O Acyltransferase Domain Containing 7 O Acyltransferase Domain Containing 7 Mboa 7 |
| ENSMUSG00000035637 | ENSFM00570000878149 | Unknown |
| ENSMUSG00000035637 | ENSFM00500000270235 | Glyoxylate Reductase/Hydroxypyruvate Reductase EC_1.1.1.79 EC_1.1.1.- 81 |
| ENSMUSG00000035640 | ENSFM00570000895905 | Unknown |
| ENSMUSG00000035640 | ENSFM00610000973073 | Unknown |
| ENSMUSG00000035640 | ENSFM00250000006978 | Dos |
| ENSMUSG00000035653 | ENSFM00260000050443 | Leucine Rich Repeat And Fibronectin Type Iii Domain Containing Precursor Synaptic Adhesion Molecule |
| ENSMUSG00000035674 | ENSFM00500000447488 | Unknown |
| ENSMUSG00000035674 | ENSFM00570000878851 | Unknown |
| ENSMUSG00000035674 | ENSFM00570000896115 | Unknown |
| ENSMUSG00000035674 | ENSFM00670001244910 | Nadh Dehydrogenase 1 Alpha Subcomplex Subunit 3 Complex I B9 Ci B9 Nadh Ubiquinone Oxidoreductase B9 Subunit |
| ENSMUSG00000035722 | ENSFM00740001589079 | Atp Binding Cassette Sub Family A Member Atp Binding Cassette Transporter Atp Binding Cassette |
| ENSMUSG00000035735 | ENSFM00250000001790 | SN1 Specific Diacylglycerol Lipase Alpha Dgl Alpha EC_3.1.1.- Neural Stem Cell Derived Dendrite Regulator |
| ENSMUSG00000035770 | ENSFM00250000001777 | Cytoplasmic Dynein 1 Light Intermediate Chain 1 Dynein Light Chain A Dlc A Dynein Light Intermediate Chain 1 Cytosolic |
| ENSMUSG00000035772 | ENSFM00250000006868 | 28S Ribosomal S2 Mitochondrial Mrp S2 S2MT |
| ENSMUSG00000035776 | ENSFM00500000272479 | CD99 Antigen 2 Precursor MIC2 1 CD99 Antigen |
| ENSMUSG00000035798 | ENSFM00500000270112 | Palmitoyltransferase EC_2.3.1.225 Huntingtin Interacting 14 Zinc Finger Dhhc Domain Containing Dhhc |
| ENSMUSG00000035805 | ENSFM00250000007537 | Membrane MLC1 |
| ENSMUSG00000035835 | ENSFM00610000968417 | Unknown |
| ENSMUSG00000035835 | ENSFM00500000270520 | Lipid Phosphate Phosphatase Related Type 4 EC_3.1.3.4 Brain Specific Phosphatidic Acid Phosphatase 1 Plasticity Related Gene 1 Prg 1 |
| ENSMUSG00000035863 | ENSFM00500000271464 | Paralemmin 1 Precursor Paralemmin |
| ENSMUSG00000035864 | ENSFM00720001492031 | Synaptotagmin |
| ENSMUSG00000035890 | ENSFM00620000999434 | E3 Ubiquitin Ligase RNF126 |
| ENSMUSG00000035900 | ENSFM00250000002904 | Gram Domain Containing 4 |
| ENSMUSG00000035936 | ENSFM00500000269985 | Succinate Semialdehyde Dehydrogenase Mitochondrial Precursor EC_1.2.1.24 Aldehyde Dehydrogenase Family 5 Member A1 Nad + Dependent Succinic Semialdehyde Dehydrogenase |
| ENSMUSG00000035953 | ENSFM00250000004019 | Type Phosphatidylinositol 4 5 Bisphosphate 4 Phosphatase Type 2 Ptdins 4 5 P2 4 Ptase EC_3.1.3.78 Ptdins 4 5 P2 4 Ptase Ii Transmembrane 55A |
| ENSMUSG00000035954 | ENSFM00760001714721 | Dedicator Of Cytokinesis 3 Modifier Of Cell Adhesion Presenilin Binding Pbp |
| ENSMUSG00000035960 | ENSFM00570000889367 | Unknown |
| ENSMUSG00000035960 | ENSFM00250000004097 | Dna Apurinic Or Apyrimidinic Site Lyase EC_3.1.-.- EC_4.2.99.18 Apex Nuclease Apen Apurinic Apyrimidinic Endonuclease 1 Ap Endonuclease 1 Ref 1 Redox Factor 1 |
| ENSMUSG00000036026 | ENSFM00250000001207 | Transmembrane |
| ENSMUSG00000036046 | ENSFM00250000003605 | Uncharacterized |
| ENSMUSG00000036052 | ENSFM00440000236855 | Dnaj Homolog Subfamily B Member |
| ENSMUSG00000036053 | ENSFM00350000105407 | Formin |
| ENSMUSG00000036062 | ENSFM00250000007322 | Ambiguous |
| ENSMUSG00000036062 | ENSFM00720001501746 | Unknown |
| ENSMUSG00000036062 | ENSFM00720001501747 | Unknown |
| ENSMUSG00000036062 | ENSFM00720001503558 | Unknown |
| ENSMUSG00000036087 | ENSFM00260000050845 | Slain Motif Containing |
| ENSMUSG00000036095 | ENSFM00730001521692 | Diacylglycerol Kinase Dag Kinase EC_2.7.1.107 Kda Diacylglycerol Kinase Diglyceride Kinase Dgk |
| ENSMUSG00000036103 | ENSFM00250000004242 | Collectin 12 Collectin Placenta 1 Cl P1 Scavenger Receptor |
| ENSMUSG00000036104 | ENSFM00250000004358 | RAB3 Gtpase Activating Catalytic Subunit RAB3 Gtpase Activating 130 Kda Subunit RAB3 Gap P130 RAB3 Gap |
| ENSMUSG00000036138 | ENSFM00500000269858 | 3 Ketoacyl Coa Thiolase Peroxisomal Precursor EC_2.3.1.16 Acetyl Coa Acyltransferase Beta Ketothiolase Peroxisomal 3 Oxoacyl Coa Thiolase |
| ENSMUSG00000036199 | ENSFM00500000273406 | Nadh Dehydrogenase 1 Alpha Subcomplex Subunit 13 Cell Death Regulatory Grim 19 Complex I B16 6 Ci B16 6 Gene Associated With Retinoic Interferon Induced Mortality 19 Grim 19 Nadh Ubiquinone Oxidoreductase B16 6 Subunit |
| ENSMUSG00000036257 | ENSFM00570000879553 | Unknown |
| ENSMUSG00000036257 | ENSFM00250000002364 | Calcium Independent Phospholipase A2 Gamma EC_3.1.1.5 Intracellular Membrane Associated Calcium Independent Phospholipase A2 Gamma IPLA2 Gamma Patatin Phospholipase Domain Containing 8 |
| ENSMUSG00000036278 | ENSFM00670001235583 | O Acetyl Adp Ribose Deacetylase EC_3.2.2.- EC_3.5.-.- 1 Macro Domain Containing Hydrolase |
| ENSMUSG00000036298 | ENSFM00500000270995 | Proton Myo Inositol Cotransporter H + Myo Inositol Cotransporter Hmit H + Myo Inositol Symporter Solute Carrier Family 2 Member 13 |
| ENSMUSG00000036306 | ENSFM00650001140033 | Leucine Zipper Tumor Suppressor 2 |
| ENSMUSG00000036309 | ENSFM00610000956029 | Unknown |
| ENSMUSG00000036309 | ENSFM00610000969567 | Unknown |
| ENSMUSG00000036309 | ENSFM00670001235396 | E3 Ubiquitin Ligase Complex Scf Subunit Sconc Sulfur Controller C Sulfur Metabolite Repression Control C |
| ENSMUSG00000036333 | ENSFM00250000001605 | Kinase D Interacting Substrate Of 220 Kda Ankyrin Repeat Rich Membrane Spanning |
| ENSMUSG00000036353 | ENSFM00440000236882 | P2Y Purinoceptor |
| ENSMUSG00000036357 | ENSFM00740001589582 | Probable G Coupled Receptor 101 |
| ENSMUSG00000036371 | ENSFM00500000270904 | Plasminogen Activator Inhibitor 1 Rna Binding PAI1 Rna Binding 1 Pai RBP1 SERPINE1 Binding 1 |
| ENSMUSG00000036372 | ENSFM00500000276946 | Unknown |
| ENSMUSG00000036372 | ENSFM00500000423017 | Unknown |
| ENSMUSG00000036377 | ENSFM00250000006430 | Uncharacterized |
| ENSMUSG00000036377 | ENSFM00520000559661 | Unknown |
| ENSMUSG00000036377 | ENSFM00570000896173 | Unknown |
| ENSMUSG00000036377 | ENSFM00570000899386 | Unknown |
| ENSMUSG00000036402 | ENSFM00500000279757 | Unknown |
| ENSMUSG00000036422 | ENSFM00790001962997 | Protocadherin Precursor |
| ENSMUSG00000036427 | ENSFM00250000000760 | Glucose 6 Phosphate Isomerase Gpi EC_5.3.1.9 Phosphoglucose Isomerase Pgi Phosphohexose Isomerase Phi |
| ENSMUSG00000036435 | ENSFM00250000004597 | Exocyst Complex Component 1 Exocyst Complex Component SEC3 |
| ENSMUSG00000036452 | ENSFM00250000000574 | Rho Gtpase Activating Rho Type Gtpase Activating |
| ENSMUSG00000036452 | ENSFM00760001762720 | Gene |
| ENSMUSG00000036473 | ENSFM00270000056490 | TBC1 Domain Family Member 24 |
| ENSMUSG00000036473 | ENSFM00570000851049 | Netrin Precursor |
| ENSMUSG00000036499 | ENSFM00430000230229 | Early Endosome Antigen 1 |
| ENSMUSG00000036504 | ENSFM00670001236604 | 14 Kda Phosphohistidine Phosphatase EC_3.1.3.- Phosphohistidine Phosphatase 1 |
| ENSMUSG00000036513 | ENSFM00590000916015 | Unknown |
| ENSMUSG00000036513 | ENSFM00250000006343 | Comm Domain Containing 2 |
| ENSMUSG00000036529 | ENSFM00250000000945 | Myotubularin Related Set Binding Factor |
| ENSMUSG00000036550 | ENSFM00780001898388 | CCR4 Not Transcription Complex Subunit 1 CCR4 Associated Factor 1 |
| ENSMUSG00000036560 | ENSFM00610000969178 | Unknown |
| ENSMUSG00000036560 | ENSFM00250000002312 | Leucine Rich Repeat Lgi Family Member Precursor Leucine Rich Glioma Inactivated |
| ENSMUSG00000036564 | ENSFM00610000972759 | Unknown |
| ENSMUSG00000036564 | ENSFM00250000000535 | N Myc Downstream Regulated Gene |
| ENSMUSG00000036565 | ENSFM00250000001068 | Tweety Homolog |
| ENSMUSG00000036568 | ENSFM00250000009373 | GLTSCR1 |
| ENSMUSG00000036570 | ENSFM00810002042307 | Phospholemman Precursor Fxyd Domain Containing Ion Transport Regulator 1 |
| ENSMUSG00000036578 | ENSFM00670001247403 | Fxyd Domain Containing Ion Transport Regulator 7 |
| ENSMUSG00000036585 | ENSFM00520000548481 | Unknown |
| ENSMUSG00000036585 | ENSFM00570000878929 | Unknown |
| ENSMUSG00000036585 | ENSFM00570000891312 | Unknown |
| ENSMUSG00000036585 | ENSFM00570000902091 | Unknown |
| ENSMUSG00000036585 | ENSFM00500000271792 | Fibroblast Growth Factor 1 Fgf 1 Acidic Fibroblast Growth Factor Afgf Heparin Binding Growth Factor 1 Hbgf 1 |
| ENSMUSG00000036591 | ENSFM00640001105460 | Unknown |
| ENSMUSG00000036591 | ENSFM00250000001623 | Rho Gtpase Activating 21 Rho Type Gtpase Activating 21 |
| ENSMUSG00000036606 | ENSFM00730001521536 | Plexin Precursor |
| ENSMUSG00000036611 | ENSFM00250000006173 | Endonuclease/Exonuclease/Phosphatase Family Domain Containing 1 |
| ENSMUSG00000036617 | ENSFM00250000001584 | Src Kinase Signaling Inhibitor 1 Snap 25 Interacting Snip P130CAS Associated P140CAP |
| ENSMUSG00000036622 | ENSFM00250000000568 | Probable Cation Transporting Atpase EC_3.6.3.- |
| ENSMUSG00000036634 | ENSFM00760001747992 | Unknown |
| ENSMUSG00000036634 | ENSFM00320000100165 | Myelin Associated Glycoprotein Precursor Siglec 4A |
| ENSMUSG00000036636 | ENSFM00740001589241 | H + /Cl Exchange Transporter 7 Chloride Channel 7 Alpha Subunit Chloride Channel 7 Clc 7 |
| ENSMUSG00000036686 | ENSFM00730001522120 | Coiled Coil And C2 Domain Containing Five Prime Repressor Element Under Dual Repression Binding Fre Under Dual Repression Binding Freud |
| ENSMUSG00000036698 | ENSFM00250000000492 | Argonaute |
| ENSMUSG00000036707 | ENSFM00570000901084 | Unknown |
| ENSMUSG00000036707 | ENSFM00250000002178 | Calcium Binding 39 MO25 |
| ENSMUSG00000036712 | ENSFM00250000002039 | Ubiquitin Carboxyl Terminal Hydrolase Cyld EC_3.4.19.12 Deubiquitinating Enzyme Cyld Ubiquitin Thioesterase Cyld Ubiquitin Specific Processing Protease Cyld |
| ENSMUSG00000036751 | ENSFM00670001236790 | Cytochrome C Oxidase Subunit 6B1 Cytochrome C Oxidase Subunit Vib 1 Cox Vib 1 |
| ENSMUSG00000036752 | ENSFM00250000000193 | Tubulin Beta Chain |
| ENSMUSG00000036766 | ENSFM00760001753778 | Unknown |
| ENSMUSG00000036766 | ENSFM00540000718025 | Delta And Notch Epidermal Growth Factor Related Receptor Precursor |
| ENSMUSG00000036781 | ENSFM00570000892545 | Unknown |
| ENSMUSG00000036781 | ENSFM00570000893201 | Unknown |
| ENSMUSG00000036781 | ENSFM00570000892970 | 40S Ribosomal S27 |
| ENSMUSG00000036781 | ENSFM00670001235597 | 40S Ribosomal S27 |
| ENSMUSG00000036880 | ENSFM00760001714736 | 3 Ketoacyl Coa Thiolase Mitochondrial EC_2.3.1.16 Acetyl Coa Acyltransferase Beta Ketothiolase Mitochondrial 3 Oxoacyl Coa Thiolase |
| ENSMUSG00000036882 | ENSFM00250000001205 | Rho Gtpase Activating Rho Type Gtpase Activating Gtpase Activating |
| ENSMUSG00000036894 | ENSFM00800002003504 | Ras Related Rap Precursor |
| ENSMUSG00000036896 | ENSFM00680001303632 | Complement C1Q Subcomponent Subunit Precursor |
| ENSMUSG00000036902 | ENSFM00250000001916 | Neuropilin And Tolloid Precursor Brain Specific Transmembrane Containing 2 Cub And 1 Ldl Receptor Class A Domains |
| ENSMUSG00000036905 | ENSFM00680001303632 | Complement C1Q Subcomponent Subunit Precursor |
| ENSMUSG00000036932 | ENSFM00250000002966 | Apoptosis Inducing Factor 1 Mitochondrial Precursor EC_1.1.1.- Programmed Cell Death 8 |
| ENSMUSG00000036943 | ENSFM00730001521184 | Ras Related Rab |
| ENSMUSG00000036948 | ENSFM00250000006401 | Uncharacterized |
| ENSMUSG00000036948 | ENSFM00590000916964 | Unknown |
| ENSMUSG00000036957 | ENSFM00260000050443 | Leucine Rich Repeat And Fibronectin Type Iii Domain Containing Precursor Synaptic Adhesion Molecule |
| ENSMUSG00000036989 | ENSFM00750001632380 | Tripartite Motif Containing 2 EC_6.3.2.- E3 Ubiquitin Ligase TRIM2 |
| ENSMUSG00000036992 | ENSFM00500000270737 | NTF2 Related Export |
| ENSMUSG00000037012 | ENSFM00570000888918 | Unknown |
| ENSMUSG00000037012 | ENSFM00250000000532 | Hexokinase EC_2.7.1.1 Hexokinase Type Hk |
| ENSMUSG00000037012 | ENSFM00570000883017 | Hexokinase 1 |
| ENSMUSG00000037022 | ENSFM00250000008244 | Methylmalonic Aciduria Type A Homolog Mitochondrial Precursor EC_3.6.-.- Methylmalonic Aciduria Type A 1 |
| ENSMUSG00000037062 | ENSFM00250000002047 | Endophilin SH3 Domain Containing GRB2 |
| ENSMUSG00000037072 | ENSFM00500000274344 | Unknown |
| ENSMUSG00000037072 | ENSFM00800002015848 | Unknown |
| ENSMUSG00000037126 | ENSFM00250000001069 | Ph And SEC7 Domain Containing 1 Exchange Factor ARF6 Pleckstrin Homology And SEC7 Domain Containing 1 |
| ENSMUSG00000037149 | ENSFM00250000003062 | Atp Dependent Rna Helicase DDX1 EC_3.6.4.13 Dead Box 1 |
| ENSMUSG00000037172 | ENSFM00250000006734 | Lchn |
| ENSMUSG00000037217 | ENSFM00250000000771 | Synapsin Synapsin |
| ENSMUSG00000037234 | ENSFM00250000001958 | Hook |
| ENSMUSG00000037236 | ENSFM00760001750219 | Unknown |
| ENSMUSG00000037236 | ENSFM00760001750220 | Unknown |
| ENSMUSG00000037236 | ENSFM00760001762716 | Unknown |
| ENSMUSG00000037236 | ENSFM00250000005840 | Matrin 3 |
| ENSMUSG00000037242 | ENSFM00250000000982 | Chloride Intracellular Channel |
| ENSMUSG00000037259 | ENSFM00250000004649 | Double Zinc Ribbon And Ankyrin Repeat Containing 1 |
| ENSMUSG00000037270 | ENSFM00380000124692 | Uncharacterized Fragile Site Associated |
| ENSMUSG00000037331 | ENSFM00250000001222 | La Related 1 La Ribonucleoprotein Domain Family Member 1 |
| ENSMUSG00000037341 | ENSFM00730001521509 | Sodium/Hydrogen Exchanger Na + /H + Exchanger Nhe Solute Carrier Family 9 Member |
| ENSMUSG00000037344 | ENSFM00570000878459 | Unknown |
| ENSMUSG00000037344 | ENSFM00800002003687 | Solute Carrier Family 12 Member 9 Cation Chloride Cotransporter Interacting 1 Potassium Chloride Transporter 9 |
| ENSMUSG00000037351 | ENSFM00730001521541 | Actin |
| ENSMUSG00000037353 | ENSFM00250000006470 | LETM1 Domain Containing 1 |
| ENSMUSG00000037373 | ENSFM00810002057430 | Unknown |
| ENSMUSG00000037373 | ENSFM00810002057431 | Unknown |
| ENSMUSG00000037373 | ENSFM00250000001273 | C Terminal Binding |
| ENSMUSG00000037386 | ENSFM00250000000629 | Regulating Synaptic Membrane Exocytosis Rab 3 Interacting Molecule Rim |
| ENSMUSG00000037419 | ENSFM00500000273498 | Endonuclease Domain Containing 1 Precursor EC_3.1.30.- |
| ENSMUSG00000037428 | ENSFM00250000010086 | Neurosecretory Vgf Neuroendocrine Regulatory Peptide 1 Nerp 1 |
| ENSMUSG00000037434 | ENSFM00710001441877 | Zinc Transporter 1 Znt 1 Solute Carrier Family 30 Member 1 |
| ENSMUSG00000037470 | ENSFM00640001117168 | Unknown |
| ENSMUSG00000037470 | ENSFM00640001119182 | Unknown |
| ENSMUSG00000037470 | ENSFM00800002003477 | Udp Glucose:Glycoprotein Glucosyltransferase 1 Precursor UGT1 EC_2.4.1.- Udp Glc:Glycoprotein Glucosyltransferase Udp Glucose Ceramide Glucosyltransferase 1 |
| ENSMUSG00000037499 | ENSFM00480000262834 | Neudesin Precursor Neuron Derived Neurotrophic Factor SCIRP10 Related Spinal Cord Injury Related 10 |
| ENSMUSG00000037514 | ENSFM00740001613629 | Unknown |
| ENSMUSG00000037514 | ENSFM00730001521878 | Pantothenate Kinase EC_2.7.1.33 Pantothenic Acid Kinase |
| ENSMUSG00000037519 | ENSFM00720001499800 | Unknown |
| ENSMUSG00000037519 | ENSFM00250000000441 | Liprin Alpha Tyrosine Phosphatase Receptor Type F Polypeptide Interacting Alpha Ptprf Interacting Alpha |
| ENSMUSG00000037523 | ENSFM00720001500067 | Unknown |
| ENSMUSG00000037523 | ENSFM00250000008057 | Mitochondrial Antiviral Signaling Mavs Card Adapter Inducing Interferon Beta Cardif Interferon Beta Promoter Stimulator 1 Ips 1 Virus Induced Signaling Adapter Visa |
| ENSMUSG00000037563 | ENSFM00250000002859 | 40S Ribosomal S16 |
| ENSMUSG00000037601 | ENSFM00250000000895 | Nucleoside Diphosphate Kinase Ndk Ndp Kinase EC_2.7.4.6 |
| ENSMUSG00000037601 | ENSFM00500000303122 | Nucleoside Diphosphate Kinase A |
| ENSMUSG00000037605 | ENSFM00570000897271 | Unknown |
| ENSMUSG00000037605 | ENSFM00260000050328 | Latrophilin |
| ENSMUSG00000037625 | ENSFM00260000051131 | Claudin 11 |
| ENSMUSG00000037653 | ENSFM00260000050545 | Btb/Poz Domain Containing |
| ENSMUSG00000037656 | ENSFM00250000001093 | Sodium Dependent Phosphate Transporter Solute Carrier Family 20 Member |
| ENSMUSG00000037669 | ENSFM00250000005114 | Unknown |
| ENSMUSG00000037679 | ENSFM00320000100171 | Inverted Formin 2 |
| ENSMUSG00000037685 | ENSFM00570000877289 | Phospholipid Transporting Atpase Ia |
| ENSMUSG00000037685 | ENSFM00770001847537 | Phospholipid Transporting Atpase EC_3.6.3.1 Atpase Class I Type 8A Member P4 Atpase Flippase Complex Alpha Subunit At |
| ENSMUSG00000037703 | ENSFM00650001140033 | Leucine Zipper Tumor Suppressor 2 |
| ENSMUSG00000037706 | ENSFM00800002003853 | CD81 Antigen 26 Kda Cell Surface Tapa 1 Target Of The Antiproliferative Antibody 1 CD81 Antigen |
| ENSMUSG00000037710 | ENSFM00500000274698 | Cdgsh Iron Sulfur Domain Containing 1 Mitoneet |
| ENSMUSG00000037720 | ENSFM00590000917165 | Unknown |
| ENSMUSG00000037720 | ENSFM00590000918332 | Unknown |
| ENSMUSG00000037720 | ENSFM00810002055293 | Unknown |
| ENSMUSG00000037720 | ENSFM00250000005590 | Transmembrane 33 |
| ENSMUSG00000037736 | ENSFM00570000885356 | Unknown |
| ENSMUSG00000037736 | ENSFM00570000898739 | Unknown |
| ENSMUSG00000037736 | ENSFM00810002057321 | Unknown |
| ENSMUSG00000037736 | ENSFM00250000003304 | Lim And Calponin Homology Domains Containing 1 |
| ENSMUSG00000037740 | ENSFM00500000275166 | 28S Ribosomal S26 Mitochondrial Precursor Mrp S26 S26MT |
| ENSMUSG00000037742 | ENSFM00800002003364 | Elongation Factor 1 Alpha Ef 1 Alpha Eukaryotic Elongation Factor 1 A EEF1A |
| ENSMUSG00000037747 | ENSFM00590000917567 | Unknown |
| ENSMUSG00000037747 | ENSFM00250000003309 | Phytanoyl Coa Hydroxylase Interacting Phytanoyl Coa Hydroxylase Associated 1 Pahx AP1 PAHXAP1 |
| ENSMUSG00000037754 | ENSFM00500000270382 | Phosphatase 1 Regulatory Inhibitor Subunit 16B Precursor Caax Box Timap Tgf Beta Inhibited Membrane Associated |
| ENSMUSG00000037771 | ENSFM00250000004214 | Vesicular Inhibitory Amino Acid Transporter Gaba And Glycine Transporter Solute Carrier Family 32 Member 1 Vesicular Gaba Transporter |
| ENSMUSG00000037772 | ENSFM00500000273802 | 39S Ribosomal L23 Mitochondrial L23MT Mrp L23 L23 Mitochondrial Related |
| ENSMUSG00000037805 | ENSFM00570000901645 | Unknown |
| ENSMUSG00000037805 | ENSFM00790001962982 | 60S Ribosomal L10A |
| ENSMUSG00000037815 | ENSFM00250000001018 | Catenin Alpha 2 Alpha N Catenin |
| ENSMUSG00000037851 | ENSFM00610000969945 | Unknown |
| ENSMUSG00000037851 | ENSFM00500000270033 | Isoleucine Trna Ligase Cytoplasmic EC_6.1.1.5 Isoleucyl Trna Synthetase Ilers |
| ENSMUSG00000037852 | ENSFM00730001521835 | Carboxypeptidase E Precursor Cpe EC_3.4.17.10 Carboxypeptidase H Cph Enkephalin Convertase Prohormone Processing Carboxypeptidase |
| ENSMUSG00000037855 | ENSFM00250000005396 | Unknown |
| ENSMUSG00000037902 | ENSFM00500000269963 | Tyrosine Phosphatase Non Receptor Type Substrate 1 Precursor Shp Substrate 1 Shps 1 Brain Ig Molecule With Tyrosine Based Activation Motifs Bit CD172 Antigen Family Member A Inhibitory Receptor Shps 1 Signal Regulatory Alpha 1 Sirp Alpha 1 CD172A Antige |
| ENSMUSG00000037905 | ENSFM00800002014565 | Unknown |
| ENSMUSG00000037905 | ENSFM00250000006424 | BRI3 Binding I3 Binding |
| ENSMUSG00000037916 | ENSFM00250000003345 | Nadh Dehydrogenase Flavoprotein 1 Mitochondrial Precursor EC_1.6.5.3 EC_1.6.99.- 3 Complex I 51KD Ci 51KD Nadh Ubiquinone Oxidoreductase 51 Kda Subunit |
| ENSMUSG00000037949 | ENSFM00800002003506 | Anoctamin 10 Transmembrane 16K |
| ENSMUSG00000037957 | ENSFM00250000001646 | Dystrophia Myotonica Wd Repeat Containing Dystrophia Myotonica Containing Wd Repeat Motif Protein Dmr N9 |
| ENSMUSG00000037972 | ENSFM00670001236736 | Stannin |
| ENSMUSG00000037977 | ENSFM00500000274877 | Uncharacterized |
| ENSMUSG00000037979 | ENSFM00570000895056 | Unknown |
| ENSMUSG00000037979 | ENSFM00250000009967 | Coiled Coil Domain Containing 92 |
| ENSMUSG00000037989 | ENSFM00250000000621 | Serine/Threonine Kinase EC_2.7.11.1 Kinase Lysine Deficient Kinase With No Lysine |
| ENSMUSG00000037996 | ENSFM00730001521845 | Sodium/Potassium/Calcium Exchanger 1 Na + /K + /Ca 2+ Exchange 1 Retinal Rod Na Ca+K Exchanger Solute Carrier Family 24 Member 1 |
| ENSMUSG00000038013 | ENSFM00500000273445 | Was/Wasl Interacting Family Member 2 Wasp Interacting Related Wip Related |
| ENSMUSG00000038014 | ENSFM00250000002493 | Constitutive Coactivator Of Ppar Gamma 1 Oxidative Stress Associated Src Activator FAM120A |
| ENSMUSG00000038028 | ENSFM00810002057401 | Unknown |
| ENSMUSG00000038028 | ENSFM00250000007002 | Fructose 2 6 Bisphosphatase Tigar EC_3.1.3.46 TP53 Induced Glycolysis And Apoptosis Regulator |
| ENSMUSG00000038034 | ENSFM00740001589327 | Immunoglobulin Superfamily Member Precursor Glu Trp Ile Ewi Motif Containing Ewi Antigen |
| ENSMUSG00000038077 | ENSFM00290000065519 | Potassium Voltage Gated Channel Subfamily A Member Voltage Gated Potassium Channel Subunit KV1 |
| ENSMUSG00000038079 | ENSFM00250000005158 | Transmembrane Amyotrophic Lateral Sclerosis 2 Chromosomal Region Candidate Gene 4 |
| ENSMUSG00000038084 | ENSFM00250000001929 | Dynamin 120 Kda Protein Mitochondrial Precursor EC_3.6.5.5 Optic Atrophy 1 |
| ENSMUSG00000038094 | ENSFM00720001502981 | Unknown |
| ENSMUSG00000038094 | ENSFM00720001503205 | Unknown |
| ENSMUSG00000038094 | ENSFM00250000000568 | Probable Cation Transporting Atpase EC_3.6.3.- |
| ENSMUSG00000038121 | ENSFM00250000007801 | Unknown |
| ENSMUSG00000038150 | ENSFM00500000270327 | ORM1 |
| ENSMUSG00000038173 | ENSFM00570000884802 | Unknown |
| ENSMUSG00000038173 | ENSFM00730001522540 | Ectonucleotide Pyrophosphatase/Phosphodiesterase Family Member 6 Precursor E Npp 6 Npp 6 EC_3.1.4.- EC_3.1.4.38 Choline Specific Glycerophosphodiester Phosphodiesterase Glycerophosphocholine Cholinephosphodiesterase Gpc Cpde |
| ENSMUSG00000038174 | ENSFM00670001258143 | Unknown |
| ENSMUSG00000038174 | ENSFM00250000003532 | Hyccin Down Regulated By CTNNB1 A FAM126A |
| ENSMUSG00000038178 | ENSFM00500000270086 | Large Neutral Amino Acids Transporter Small Subunit 4 L Type Amino Acid Transporter 4 Solute Carrier Family 43 Member 2 |
| ENSMUSG00000038264 | ENSFM00730001522534 | Semaphorin 7A Precursor Semaphorin K1 Sema K1 Semaphorin L Sema L CD108 Antigen |
| ENSMUSG00000038267 | ENSFM00650001140024 | Solute Carrier Family 22 Member 17 24P3 Receptor 24P3R Lipocalin 2 Receptor |
| ENSMUSG00000038286 | ENSFM00250000004835 | Valacyclovir Hydrolase Precursor Vacvase Valacyclovirase EC_3.1.-.- Biphenyl Hydrolase |
| ENSMUSG00000038291 | ENSFM00660001189527 | Unknown |
| ENSMUSG00000038291 | ENSFM00250000003977 | Sorting Nexin 25 |
| ENSMUSG00000038366 | ENSFM00470000259183 | Unknown |
| ENSMUSG00000038366 | ENSFM00570000883595 | Unknown |
| ENSMUSG00000038366 | ENSFM00570000888630 | Unknown |
| ENSMUSG00000038366 | ENSFM00570000892904 | Unknown |
| ENSMUSG00000038366 | ENSFM00570000895214 | Unknown |
| ENSMUSG00000038366 | ENSFM00570000895866 | Unknown |
| ENSMUSG00000038366 | ENSFM00610000965408 | Unknown |
| ENSMUSG00000038366 | ENSFM00500000270806 | Lim And SH3 Domain 1 Lasp 1 |
| ENSMUSG00000038370 | ENSFM00500000346194 | Unknown |
| ENSMUSG00000038387 | ENSFM00770001847543 | Ras Related R Ras |
| ENSMUSG00000038388 | ENSFM00610000970622 | Unknown |
| ENSMUSG00000038388 | ENSFM00730001521911 | Maguk P55 Subfamily Member 2 Discs Large Homolog 2 MPP2 |
| ENSMUSG00000038390 | ENSFM00250000004703 | Probable G Coupled Receptor 153 G Coupled Receptor PGR1 |
| ENSMUSG00000038412 | ENSFM00670001236403 | HIG1 Domain Family Member 1A Mitochondrial Hypoxia Inducible Gene 1 |
| ENSMUSG00000038422 | ENSFM00500000273549 | Haloacid Dehalogenase Hydrolase Domain Containing 3 |
| ENSMUSG00000038429 | ENSFM00760001714687 | Ubiquitin Carboxyl Terminal Hydrolase 13 EC_3.4.19.12 Deubiquitinating Enzyme 13 Ubiquitin Thioesterase 13 Ubiquitin Specific Processing Protease 13 |
| ENSMUSG00000038453 | ENSFM00250000001584 | Src Kinase Signaling Inhibitor 1 Snap 25 Interacting Snip P130CAS Associated P140CAP |
| ENSMUSG00000038462 | ENSFM00770001847545 | Cytochrome B C1 Complex Subunit Rieske Mitochondrial Precursor EC_1.10.2.2 Complex Iii Subunit 5 Cytochrome B C1 Complex Subunit 5 Rieske Iron Sulfur Risp Ubiquinol Cytochrome C Reductase Iron Sulfur Subunit |
| ENSMUSG00000038467 | ENSFM00600000921240 | Charged Multivesicular Body Chromatin Modifying |
| ENSMUSG00000038486 | ENSFM00250000000744 | Synaptic Vesicle Glycoprotein |
| ENSMUSG00000038498 | ENSFM00250000004508 | Cation Channel Sperm Associated 1 CATSPER1 |
| ENSMUSG00000038503 | ENSFM00570000888493 | Unknown |
| ENSMUSG00000038503 | ENSFM00800002003934 | Ldlr Chaperone Mesd Precursor Mesoderm Development Candidate 2 Mesoderm Development |
| ENSMUSG00000038510 | ENSFM00810002040861 | Ribosome Production Factor 2 Homolog Brix Domain Containing 1 Ribosome Biogenesis RPF2 Homolog |
| ENSMUSG00000038517 | ENSFM00250000006948 | Tank Binding Kinase 1 Binding 1 TBK1 Binding 1 |
| ENSMUSG00000038520 | ENSFM00730001521999 | TBC1 Domain Family Member 15 Gtpase Activating RAB7 Gap RAB7 RAB7 Gap |
| ENSMUSG00000038525 | ENSFM00250000001585 | Armadillo Repeat Containing X Linked |
| ENSMUSG00000038526 | ENSFM00800002003797 | Carbonic Anhydrase 12 Precursor EC_4.2.1.1 Carbonate Dehydratase Xii Carbonic Anhydrase Xii Ca Xii |
| ENSMUSG00000038555 | ENSFM00500000269981 | Receptor Expression Enhancing |
| ENSMUSG00000038587 | ENSFM00250000003924 | A Kinase Anchor 12 Akap 12 |
| ENSMUSG00000038602 | ENSFM00250000002791 | Solute Carrier Family 35 Member |
| ENSMUSG00000038608 | ENSFM00500000269738 | Dedicator Of Cytokinesis CDC42 Guanine Nucleotide Exchange Factor Zizimin |
| ENSMUSG00000038619 | ENSFM00500000271209 | Alpha Endosulfine |
| ENSMUSG00000038633 | ENSFM00760001714747 | Sphingolipid Delta 4 Desaturase DES1 EC_1.14.-.- Degenerative Spermatocyte Homolog 1 |
| ENSMUSG00000038650 | ENSFM00740001589564 | Ribonuclease Inhibitor Ribonuclease/Angiogenin Inhibitor 1 |
| ENSMUSG00000038665 | ENSFM00730001521936 | Diacylglycerol Kinase Zeta Dag Kinase Zeta EC_2.7.1.107 Diglyceride Kinase Zeta Dgk Zeta |
| ENSMUSG00000038690 | ENSFM00590000916152 | Unknown |
| ENSMUSG00000038690 | ENSFM00590000918343 | Unknown |
| ENSMUSG00000038690 | ENSFM00780001899296 | Atp Synthase Subunit F Mitochondrial |
| ENSMUSG00000038717 | ENSFM00670001236183 | Atp Synthase Subunit G Mitochondrial Atpase Subunit G |
| ENSMUSG00000038740 | ENSFM00500000272007 | Multivesicular Body Subunit 12B Escrt I Complex Subunit MVB12B FAM125B |
| ENSMUSG00000038776 | ENSFM00500000270247 | Epoxide Hydrolase 1 EC_3.3.2.9 Epoxide Hydratase Microsomal Epoxide Hydrolase |
| ENSMUSG00000038812 | ENSFM00500000274587 | Multifunctional Methyltransferase Subunit TRM112 Trna Methyltransferase 112 Homolog |
| ENSMUSG00000038828 | ENSFM00250000004458 | Transmembrane 214 |
| ENSMUSG00000038845 | ENSFM00790001963030 | Prohibitin |
| ENSMUSG00000038900 | ENSFM00780001917905 | Unknown |
| ENSMUSG00000038900 | ENSFM00250000002131 | 60S Ribosomal L12 |
| ENSMUSG00000038910 | ENSFM00770001847576 | Inactive Phospholipase C 2 Plc L 2 Plc L2 Phospholipase C L2 Phospholipase C Epsilon 2 Plc Epsilon 2 |
| ENSMUSG00000038916 | ENSFM00250000001168 | Soga Family Member |
| ENSMUSG00000038936 | ENSFM00250000003507 | Saccharopine Dehydrogenase Oxidoreductase Ec 1 |
| ENSMUSG00000038949 | ENSFM00250000005660 | Consortin |
| ENSMUSG00000038965 | ENSFM00500000270436 | Ubiquitin Conjugating Enzyme E2 L3 EC_6.3.2.19 Ubiquitin Carrier L3 Ubiquitin Ligase L3 |
| ENSMUSG00000038967 | ENSFM00500000269765 | Kinase Isozyme Mitochondrial Precursor EC_2.7.11.2 Pyruvate Dehydrogenase Kinase |
| ENSMUSG00000038970 | ENSFM00670001235536 | Serine/Threonine Kinase EC_2.7.11.1 Apoptosis Associated Tyrosine Kinase Brain Kinase Lemur Tyrosine Kinase |
| ENSMUSG00000038976 | ENSFM00250000001450 | Neurabin 1 Neurabin I Neural Tissue Specific F Actin Binding I Phosphatase 1 Regulatory Subunit 9A |
| ENSMUSG00000038991 | ENSFM00670001235739 | Thioredoxin Domain Containing 5 Precursor Endoplasmic Reticulum Resident 46 Er 46 ERP46 Thioredoxin P46 |
| ENSMUSG00000039016 | ENSFM00670001236920 | Mitochondrial Import Inner Membrane Translocase Subunit TIM8 B Deafness Dystonia 2 |
| ENSMUSG00000039047 | ENSFM00800002020638 | Unknown |
| ENSMUSG00000039047 | ENSFM00800002024521 | Unknown |
| ENSMUSG00000039047 | ENSFM00810002040906 | Gpi Anchor Transamidase Precursor Gpi Transamidase Ec 3 Phosphatidylinositol Glycan Biosynthesis Class K Pig K |
| ENSMUSG00000039058 | ENSFM00740001589162 | Adenylate Kinase Isoenzyme 1 |
| ENSMUSG00000039062 | ENSFM00250000000130 | Aminopeptidase N Ap N EC_3.4.11.2 Microsomal Aminopeptidase |
| ENSMUSG00000039067 | ENSFM00250000004823 | 26S Proteasome Non Atpase Regulatory Subunit 7 26S Proteasome Regulatory Subunit RPN8 |
| ENSMUSG00000039103 | ENSFM00800002024520 | Unknown |
| ENSMUSG00000039103 | ENSFM00250000002729 | Nexilin F Actin Binding |
| ENSMUSG00000039105 | ENSFM00670001236060 | V Type Proton Atpase Subunit G 1 V Atpase Subunit G 1 V Atpase 13 Kda Subunit 1 Vacuolar Proton Pump Subunit G 1 Vacuolar Proton Pump Subunit M16 |
| ENSMUSG00000039114 | ENSFM00500000274208 | Neuritin Precursor |
| ENSMUSG00000039156 | ENSFM00250000001930 | Stromal Interaction Molecule Precursor |
| ENSMUSG00000039163 | ENSFM00500000274574 | Cox Assembly Mitochondrial Homolog CMC1P |
| ENSMUSG00000039166 | ENSFM00660001179988 | Unknown |
| ENSMUSG00000039166 | ENSFM00660001189627 | Unknown |
| ENSMUSG00000039166 | ENSFM00500000273030 | A Kinase Anchor 7 Gamma Akap 7 Gamma A Kinase Anchor 18 Akap 18 Kinase A Anchoring 7 Gamma PRKA7 Gamma |
| ENSMUSG00000039166 | ENSFM00500000311253 | A Kinase Anchor 7 Alpha |
| ENSMUSG00000039166 | ENSFM00700001393728 | A Kinase Anchor 7 Alpha Akap 7 Alpha A Kinase Anchor 18 Akap 18 A Kinase Anchor 9 Kda Akap 9 Kinase A Anchoring 7 Alpha PRKA7 Alpha |
| ENSMUSG00000039221 | ENSFM00780001917087 | Unknown |
| ENSMUSG00000039221 | ENSFM00600000921635 | 60S Ribosomal L22 1 |
| ENSMUSG00000039252 | ENSFM00250000002312 | Leucine Rich Repeat Lgi Family Member Precursor Leucine Rich Glioma Inactivated |
| ENSMUSG00000039254 | ENSFM00570000880296 | Unknown |
| ENSMUSG00000039254 | ENSFM00780001917899 | Unknown |
| ENSMUSG00000039254 | ENSFM00730001522352 | O 1 EC_2.4.1.109 Dolichyl Phosphate Mannose Mannosyltransferase 1 |
| ENSMUSG00000039278 | ENSFM00600000922075 | Prosaas Precursor Proprotein Convertase Subtilisin/Kexin Type 1 Inhibitor Proprotein Convertase 1 Inhibitor Pro Saas |
| ENSMUSG00000039313 | ENSFM00500000272805 | Protein DD1 |
| ENSMUSG00000039316 | ENSFM00390000126378 | Raftlin Raft Linking |
| ENSMUSG00000039318 | ENSFM00250000002965 | RAB3 Gtpase Activating Non Catalytic Subunit RAB3 Gtpase Activating 150 Kda Subunit RAB3 Gap P150 RAB3 GAP150 RAB3 Gap Regulatory Subunit |
| ENSMUSG00000039361 | ENSFM00250000000859 | Clathrin Assembly |
| ENSMUSG00000039367 | ENSFM00770001847547 | Transport SEC24 Related |
| ENSMUSG00000039385 | ENSFM00260000050333 | Cadherin Precursor |
| ENSMUSG00000039414 | ENSFM00250000001534 | Heat Repeat Containing |
| ENSMUSG00000039419 | ENSFM00250000000380 | Contactin Associated 5 Precursor Cell Recognition Molecule CASPR5 |
| ENSMUSG00000039428 | ENSFM00670001235695 | Transmembrane 135 Peroxisomal Membrane 52 PMP52 |
| ENSMUSG00000039457 | ENSFM00500000270485 | Envoplakin 210 Kda Cornified Envelope Precursor P210 |
| ENSMUSG00000039474 | ENSFM00280000058784 | Wolframin |
| ENSMUSG00000039478 | ENSFM00260000050554 | Calcium Uptake 2 Mitochondrial Precursor Ef Hand Domain Containing Family Member A1 |
| ENSMUSG00000039488 | ENSFM00390000126303 | Contactin Precursor Brain Derived Immunoglobulin Superfamily Big |
| ENSMUSG00000039515 | ENSFM00550000750340 | Unknown |
| ENSMUSG00000039515 | ENSFM00570000898338 | Unknown |
| ENSMUSG00000039515 | ENSFM00500000286252 | Serine/Threonine Phosphatase 2A Activator |
| ENSMUSG00000039515 | ENSFM00760001714775 | Serine/Threonine Phosphatase 2A Activator EC_5.2.1.8 PP2A Subunit B' PR53 Phosphotyrosyl Phosphatase Activator Ptpa Serine/Threonine Phosphatase 2A Regulatory Subunit 4 Serine/Threonine Phosphatase 2A Regulatory Subunit B' |
| ENSMUSG00000039530 | ENSFM00250000002565 | Magnesium Transporter 1 Precursor MAGT1 |
| ENSMUSG00000039536 | ENSFM00250000001250 | Double Stranded Rna Binding Staufen Homolog |
| ENSMUSG00000039542 | ENSFM00780001917411 | Unknown |
| ENSMUSG00000039542 | ENSFM00250000000779 | Neural Cell Adhesion Molecule 1 Precursor N Cam 1 Ncam 1 |
| ENSMUSG00000039546 | ENSFM00570000900895 | Unknown |
| ENSMUSG00000039546 | ENSFM00250000009368 | Adherens Junction Associated 1 |
| ENSMUSG00000039585 | ENSFM00510000502753 | Unconventional Myosin Unconventional Myosin |
| ENSMUSG00000039640 | ENSFM00760001714704 | 39S Ribosomal L12 Mitochondrial Precursor L12MT Mrp L12 |
| ENSMUSG00000039652 | ENSFM00250000000830 | Cytoplasmic Polyadenylation Element Binding |
| ENSMUSG00000039682 | ENSFM00250000002996 | Cytosol Aminopeptidase EC_3.4.11.1 Leucine Aminopeptidase 3 Lap 3 Leucyl Aminopeptidase Proline Aminopeptidase EC_3.4.11.- 5 Prolyl Aminopeptidase |
| ENSMUSG00000039701 | ENSFM00250000002075 | Inactive Ubiquitin Carboxyl Terminal Hydrolase Inactive Ubiquitin Specific Peptidase |
| ENSMUSG00000039704 | ENSFM00250000005238 | LMBR1 Domain Containing 2 |
| ENSMUSG00000039716 | ENSFM00760001714721 | Dedicator Of Cytokinesis 3 Modifier Of Cell Adhesion Presenilin Binding Pbp |
| ENSMUSG00000039728 | ENSFM00730001521205 | Sodium And Chloride Dependent Glycine Transporter Glyt Solute Carrier Family 6 Member |
| ENSMUSG00000039735 | ENSFM00250000000770 | Formin Binding 1 |
| ENSMUSG00000039740 | ENSFM00800002003805 | Alpha 1 3/1 6 Mannosyltransferase ALG2 EC_2.4.1.132 EC_2.4.1.- 257 Asparagine Linked Glycosylation 2 Homolog Gdp Man:Man 1 Glcnac 2 Pp Dol Alpha 1 3 Mannosyltransferase Gdp Man:Man 1 Glcnac 2 Pp Dolichol Mannosyltransferase Gdp Man:Man 2 Glcnac 2 Pp Dol A |
| ENSMUSG00000039768 | ENSFM00250000002491 | Dnaj Homolog Subfamily C Member 11 |
| ENSMUSG00000039809 | ENSFM00250000000928 | Gamma Aminobutyric Acid Type B Receptor Subunit 2 Precursor Gaba B Receptor 2 Gaba B R2 Gaba BR2 GABABR2 GB2 G Coupled Receptor 51 |
| ENSMUSG00000039824 | ENSFM00250000000438 | Myosin Light Chain Myosin Light Chain Myosin Light Chain |
| ENSMUSG00000039828 | ENSFM00780001898455 | Wd Repeat Containing 70 |
| ENSMUSG00000039838 | ENSFM00500000271895 | Proton Associated Sugar Transporter A Past A Deleted In Neuroblastoma 5 Dnb 5 Solute Carrier Family 45 Member 1 |
| ENSMUSG00000039913 | ENSFM00730001521739 | Serine/Threonine Kinase Pak EC_2.7.11.1 P21 Activated Kinase Pak |
| ENSMUSG00000039929 | ENSFM00250000007283 | Nucleolar Pre Ribosomal Associated 1 URB1 Ribosome Biogenesis 1 Homolog |
| ENSMUSG00000039943 | ENSFM00800002003599 | 1 Phosphatidylinositol 4 5 Bisphosphate Phosphodiesterase Beta 4 EC_3.1.4.11 Phosphoinositide Phospholipase C Beta 4 Phospholipase C Beta 4 Plc Beta 4 |
| ENSMUSG00000039952 | ENSFM00780001915552 | Unknown |
| ENSMUSG00000039952 | ENSFM00780001917791 | Unknown |
| ENSMUSG00000039952 | ENSFM00250000004650 | Dystroglycan Precursor Dystrophin Associated Glycoprotein 1 |
| ENSMUSG00000039953 | ENSFM00250000001322 | Calsyntenin Precursor Alcadein Alc |
| ENSMUSG00000039959 | ENSFM00250000001308 | Huntingtin Interacting 1 Hip 1 Huntingtin Interacting I Hip I |
| ENSMUSG00000040003 | ENSFM00800002020509 | Unknown |
| ENSMUSG00000040003 | ENSFM00250000000552 | Membrane Associated Guanylate Kinase Ww And Pdz Domain Containing Membrane Associated Guanylate Kinase Inverted Magi |
| ENSMUSG00000040009 | ENSFM00410000138458 | Guanine Nucleotide Binding Subunit |
| ENSMUSG00000040010 | ENSFM00250000000331 | Amino Acid Transporter Solute Carrier Family 7 Member |
| ENSMUSG00000040022 | ENSFM00500000271919 | RAB11 Family Interacting 2 RAB11 FIP2 |
| ENSMUSG00000040028 | ENSFM00250000000588 | Elav |
| ENSMUSG00000040035 | ENSFM00250000002345 | Dispatched |
| ENSMUSG00000040037 | ENSFM00290000065521 | Precursor |
| ENSMUSG00000040048 | ENSFM00250000006353 | Nadh Dehydrogenase 1 Beta Subcomplex Subunit 10 Complex I Pdsw Ci Pdsw Nadh Ubiquinone Oxidoreductase Pdsw Subunit |
| ENSMUSG00000040055 | ENSFM00740001589167 | Gap Junction Beta Connexin |
| ENSMUSG00000040105 | ENSFM00250000003664 | Probable Lipid Phosphate Phosphatase PPAPDC3 EC_3.1.3.- Phosphatidic Acid Phosphatase Type 2 Domain Containing 3 |
| ENSMUSG00000040111 | ENSFM00500000270030 | Gram Domain Containing |
| ENSMUSG00000040112 | ENSFM00250000004577 | 28S Ribosomal S35 Mitochondrial Precursor Mrp S35 S35MT 28S Ribosomal S28 Mitochondrial Mrp S28 S28MT |
| ENSMUSG00000040118 | ENSFM00670001245035 | Uncharacterized |
| ENSMUSG00000040118 | ENSFM00730001522093 | Voltage Dependent Calcium Channel Subunit Alpha 2/Delta 1 Precursor Voltage Gated Calcium Channel Subunit Alpha 2/Delta 1 |
| ENSMUSG00000040147 | ENSFM00640001119741 | Unknown |
| ENSMUSG00000040147 | ENSFM00250000002249 | Amine Oxidase EC_1.4.3.4 Monoamine Oxidase Type Mao |
| ENSMUSG00000040188 | ENSFM00670001235365 | Secretory Carrier Associated Membrane Secretory Carrier Membrane |
| ENSMUSG00000040225 | ENSFM00500000270319 | PRRC2C BAT2 Domain Containing 1 Hla B Associated Transcript 2 2 Proline Rich And Coiled Coil Containing 2C |
| ENSMUSG00000040236 | ENSFM00500000273083 | Trafficking Particle Complex Subunit 5 |
| ENSMUSG00000040242 | ENSFM00800002003783 | Suppressor Of Ikbke 1 Suppressor Of Ikk Epsilon |
| ENSMUSG00000040249 | ENSFM00570000898112 | Unknown |
| ENSMUSG00000040249 | ENSFM00740001589098 | Density Lipoprotein Receptor Related Precursor Lrp |
| ENSMUSG00000040260 | ENSFM00350000105429 | Disheveled Associated Activator Of Morphogenesis |
| ENSMUSG00000040265 | ENSFM00520000517792 | Dynamin EC_3.6.5.5 |
| ENSMUSG00000040269 | ENSFM00670001236327 | 28S Ribosomal S28 Mitochondrial Precursor Mrp S28 S28MT |
| ENSMUSG00000040276 | ENSFM00250000001466 | Kinase C And Casein Kinase Substrate In Neurons |
| ENSMUSG00000040351 | ENSFM00400000131980 | Ankyrin Repeat And Ibr Domain Containing 1 |
| ENSMUSG00000040354 | ENSFM00250000004570 | Methionine Trna Ligase Cytoplasmic EC_6.1.1.10 Methionyl Trna Synthetase Metrs |
| ENSMUSG00000040359 | ENSFM00250000003769 | E3 UFM1 Ligase 1 EC_6.3.2.- |
| ENSMUSG00000040380 | ENSFM00250000002369 | Cerebellin Precursor |
| ENSMUSG00000040385 | ENSFM00420000140518 | Serine/Threonine Phosphatase PP1 Catalytic Subunit Pp EC_3.1.3.16 |
| ENSMUSG00000040389 | ENSFM00800002017904 | Unknown |
| ENSMUSG00000040389 | ENSFM00250000002280 | Wd Repeat Containing 47 Neuronal Enriched Map Interacting Nemitin |
| ENSMUSG00000040412 | ENSFM00740001613423 | Unknown |
| ENSMUSG00000040412 | ENSFM00250000001885 | Precursor Estrogen Induced Gene 121 |
| ENSMUSG00000040420 | ENSFM00260000050333 | Cadherin Precursor |
| ENSMUSG00000040430 | ENSFM00500000270877 | Cytoplasmic Phosphatidylinositol Transfer 1 Mammalian Rdgb Homolog Beta M Rdgb Beta Mrdgbbeta Retinal Degeneration B Homolog Beta Rdgbbeta |
| ENSMUSG00000040462 | ENSFM00250000005852 | Os 9 Precursor |
| ENSMUSG00000040466 | ENSFM00250000005350 | Flavin Reductase Nadph Fr EC_1.5.1.30 Biliverdin Reductase B Bvr B EC_1.3.1.- 24 Biliverdin Ix Beta Reductase Green Heme Binding Ghbp Nadph Dependent Diaphorase Nadph Flavin Reductase Flr |
| ENSMUSG00000040472 | ENSFM00500000271139 | Geranylgeranyl Transferase Type 2 Subunit Alpha EC_2.5.1.60 Geranylgeranyl Transferase Type Ii Subunit Alpha Rab Geranyl Geranyltransferase Subunit Alpha Rab Gg Transferase Alpha Rab Ggtase Alpha Rab Geranylgeranyltransferase Subunit Alpha |
| ENSMUSG00000040479 | ENSFM00570000877778 | Unknown |
| ENSMUSG00000040479 | ENSFM00570000883510 | Unknown |
| ENSMUSG00000040479 | ENSFM00730001521936 | Diacylglycerol Kinase Zeta Dag Kinase Zeta EC_2.7.1.107 Diglyceride Kinase Zeta Dgk Zeta |
| ENSMUSG00000040490 | ENSFM00260000050443 | Leucine Rich Repeat And Fibronectin Type Iii Domain Containing Precursor Synaptic Adhesion Molecule |
| ENSMUSG00000040495 | ENSFM00800002003483 | Muscarinic Acetylcholine Receptor |
| ENSMUSG00000040521 | ENSFM00250000005091 | Elongation Factor Ts Mitochondrial |
| ENSMUSG00000040548 | ENSFM00560000830190 | Unknown |
| ENSMUSG00000040548 | ENSFM00250000002486 | Testis Expressed Sequence 2 |
| ENSMUSG00000040549 | ENSFM00760001714778 | Cytoskeleton Associated 5 |
| ENSMUSG00000040560 | ENSFM00250000001912 | Wd Repeat Containing 7 Tgf Beta Resistance Associated Trag |
| ENSMUSG00000040563 | ENSFM00500000270116 | Lipid Phosphate Phosphatase Related Type Plasticity Related Gene Prg |
| ENSMUSG00000040584 | ENSFM00730001521159 | Multidrug Resistance EC_3.6.3.44 Atp Binding Cassette Sub Family B Member P Glycoprotein |
| ENSMUSG00000040606 | ENSFM00250000003884 | Kazrin |
| ENSMUSG00000040612 | ENSFM00740001589513 | Immunoglobulin Domain Containing Receptor 2 Precursor |
| ENSMUSG00000040618 | ENSFM00730001521206 | Phosphoenolpyruvate Carboxykinase Pepck EC_4.1.1.32 |
| ENSMUSG00000040620 | ENSFM00680001306470 | Unknown |
| ENSMUSG00000040620 | ENSFM00680001326723 | Unknown |
| ENSMUSG00000040620 | ENSFM00680001326724 | Unknown |
| ENSMUSG00000040620 | ENSFM00800002003714 | Atp Dependent Rna Helicase DHX33 EC_3.6.4.13 Deah Box 33 |
| ENSMUSG00000040624 | ENSFM00380000124706 | Pleckstrin Homology Domain Containing Family G Member Ph Domain Containing Family G Member |
| ENSMUSG00000040640 | ENSFM00250000000753 | Elks/RAB6 Interacting/Cast Family Member 1 Erc 1 RAB6 Interacting 2 |
| ENSMUSG00000040659 | ENSFM00250000003493 | Ef Hand Domain Containing Swiprosin |
| ENSMUSG00000040675 | ENSFM00800002003403 | C 1 Tetrahydrofolate Synthase Cytoplasmic C1 Thf Synthase |
| ENSMUSG00000040687 | ENSFM00250000001066 | Map Kinase Activating Death Domain RAB3 Exchange Factor |
| ENSMUSG00000040697 | ENSFM00250000004546 | Dnaj Homolog Subfamily C Member 16 Precursor |
| ENSMUSG00000040699 | ENSFM00500000273568 | Lim Domain Containing 2 |
| ENSMUSG00000040722 | ENSFM00740001589424 | Secretory Carrier Associated Membrane Secretory Carrier Membrane |
| ENSMUSG00000040724 | ENSFM00290000065519 | Potassium Voltage Gated Channel Subfamily A Member Voltage Gated Potassium Channel Subunit KV1 |
| ENSMUSG00000040731 | ENSFM00500000271538 | Eukaryotic Translation Initiation Factor 4H Eif 4H Williams Beuren Syndrome Chromosomal Region 1 |
| ENSMUSG00000040752 | ENSFM00500000316024 | Unknown |
| ENSMUSG00000040752 | ENSFM00250000000024 | Myosin Myosin Heavy Chain Myosin Heavy Chain Muscle |
| ENSMUSG00000040759 | ENSFM00500000273888 | Cklf Marvel Transmembrane Domain Containing 5 Chemokine Factor Superfamily Member 5 |
| ENSMUSG00000040760 | ENSFM00250000003219 | Dcc Interacting 13 DIP13 Adapter Containing Ph Domain Ptb Domain And Leucine Zipper Motif |
| ENSMUSG00000040794 | ENSFM00260000051148 | Complement C1Q Tumor Necrosis Factor Related 4 Precursor |
| ENSMUSG00000040797 | ENSFM00250000001463 | Iq Motif And SEC7 Domain Containing |
| ENSMUSG00000040813 | ENSFM00250000008293 | Testis Expressed Sequence 264 Precursor Secreted ZSIG11 |
| ENSMUSG00000040860 | ENSFM00570000880210 | Unknown |
| ENSMUSG00000040860 | ENSFM00550000743177 | Ciliary Rootlet Coiled Coil |
| ENSMUSG00000040875 | ENSFM00720001502110 | Unknown |
| ENSMUSG00000040875 | ENSFM00720001502111 | Unknown |
| ENSMUSG00000040875 | ENSFM00780001898376 | Oxysterol Binding Related 11 Orp 11 Osbp Related 11 |
| ENSMUSG00000040883 | ENSFM00500000275174 | Transmembrane 205 |
| ENSMUSG00000040888 | ENSFM00500000272782 | Fad Linked Sulfhydryl Oxidase Alr EC_1.8.3.2 Augmenter Of Liver Regeneration |
| ENSMUSG00000040896 | ENSFM00300000079128 | Potassium Voltage Gated Channel Subfamily D Member Voltage Gated Potassium Channel Subunit KV4 |
| ENSMUSG00000040907 | ENSFM00650001139994 | Sodium/Potassium Transporting Atpase Subunit Alpha Na + /K + Atpase Alpha Subunit EC_3.6.3.9 Sodium Pump Subunit Alpha |
| ENSMUSG00000040952 | ENSFM00720001499506 | Unknown |
| ENSMUSG00000040952 | ENSFM00670001235598 | 40S Ribosomal S19 |
| ENSMUSG00000040972 | ENSFM00250000006695 | Immunoglobulin Superfamily Member 21 Precursor IGSF21 |
| ENSMUSG00000040990 | ENSFM00600000921328 | SH3 Domain Containing Kinase Binding 1 Of Kinase |
| ENSMUSG00000041020 | ENSFM00500000375694 | Unknown |
| ENSMUSG00000041020 | ENSFM00250000005199 | MAP7 Domain Containing 2 |
| ENSMUSG00000041028 | ENSFM00250000004335 | Growth Hormone Inducible Transmembrane Precursor Mitochondrial Morphology And Cristae Structure 1 MICS1 |
| ENSMUSG00000041035 | ENSFM00250000005262 | Dpcd |
| ENSMUSG00000041073 | ENSFM00660001189960 | Unknown |
| ENSMUSG00000041073 | ENSFM00500000270490 | Nascent Polypeptide Associated Complex Subunit Alpha Nac Alpha Alpha Nac |
| ENSMUSG00000041078 | ENSFM00720001492054 | Glutamate Receptor Ionotropic Delta Precursor Glur Delta Subunit |
| ENSMUSG00000041112 | ENSFM00250000001494 | Engulfment And Cell Motility |
| ENSMUSG00000041115 | ENSFM00250000001463 | Iq Motif And SEC7 Domain Containing |
| ENSMUSG00000041119 | ENSFM00570000881490 | Unknown |
| ENSMUSG00000041119 | ENSFM00570000881576 | Unknown |
| ENSMUSG00000041119 | ENSFM00570000882033 | Unknown |
| ENSMUSG00000041119 | ENSFM00570000887766 | Unknown |
| ENSMUSG00000041119 | ENSFM00570000889256 | Unknown |
| ENSMUSG00000041119 | ENSFM00570000900811 | Unknown |
| ENSMUSG00000041119 | ENSFM00400000131822 | High Affinity Cgmp Specific 3' 5' Cyclic Phosphodiesterase 9A EC_3.1.4.35 |
| ENSMUSG00000041133 | ENSFM00720001501832 | Unknown |
| ENSMUSG00000041133 | ENSFM00790001962984 | Structural Maintenance Of Chromosomes 1A Smc 1A Smc 1A |
| ENSMUSG00000041168 | ENSFM00810002040688 | Lon Protease Homolog Mitochondrial |
| ENSMUSG00000041205 | ENSFM00250000010765 | MAP6 Domain Containing 1.21 Kda Stop SL21 |
| ENSMUSG00000041236 | ENSFM00250000002575 | Vacuolar Sorting Associated 41 |
| ENSMUSG00000041245 | ENSFM00250000000621 | Serine/Threonine Kinase EC_2.7.11.1 Kinase Lysine Deficient Kinase With No Lysine |
| ENSMUSG00000041278 | ENSFM00260000051026 | Tetratricopeptide Repeat 1 Tpr Repeat 1 |
| ENSMUSG00000041313 | ENSFM00250000000444 | Cationic Amino Acid Transporter 2 Cat 2 CAT2 Low Affinity Cationic Amino Acid Transporter 2 Solute Carrier Family 7 Member 2 |
| ENSMUSG00000041329 | ENSFM00670001235362 | Sodium/Potassium Transporting Atpase Subunit Beta Sodium/Potassium Dependent Atpase Subunit Beta |
| ENSMUSG00000041351 | ENSFM00800002003399 | Ubiquitin Carboxyl Terminal Hydrolase 48 EC_3.4.19.12 Deubiquitinating Enzyme 48 Ubiquitin Thioesterase 48 Ubiquitin Specific Processing Protease 48 |
| ENSMUSG00000041417 | ENSFM00250000000988 | Phosphatidylinositol 3 Kinase Regulatory Subunit Alpha PI3 Kinase Regulatory Subunit Alpha PI3K Regulatory Subunit Alpha Ptdins 3 Kinase Regulatory Subunit Alpha Phosphatidylinositol 3 Kinase 85 Kda Regulatory Subunit Alpha PI3 Kinase Subunit P85 Alpha Pt |
| ENSMUSG00000041426 | ENSFM00790001963058 | 3 Hydroxyisobutyryl Coa Hydrolase Mitochondrial Precursor EC_3.1.2.4 3 Hydroxyisobutyryl Coenzyme A Hydrolase Hib Coa Hydrolase Hibyl Coa H |
| ENSMUSG00000041439 | ENSFM00500000271322 | Major Facilitator Superfamily Domain Containing 6 Macrophage Mhc Class I Receptor 2 |
| ENSMUSG00000041444 | ENSFM00250000001205 | Rho Gtpase Activating Rho Type Gtpase Activating Gtpase Activating |
| ENSMUSG00000041453 | ENSFM00250000000614 | 60S Ribosomal L21 |
| ENSMUSG00000041459 | ENSFM00760001750470 | Unknown |
| ENSMUSG00000041459 | ENSFM00760001763533 | Unknown |
| ENSMUSG00000041459 | ENSFM00760001763651 | Unknown |
| ENSMUSG00000041459 | ENSFM00760001763652 | Unknown |
| ENSMUSG00000041459 | ENSFM00250000001956 | Tar Dna Binding 43 Tdp 43 |
| ENSMUSG00000041530 | ENSFM00250000000492 | Argonaute |
| ENSMUSG00000041556 | ENSFM00500000270014 | F Box Only 6 F Box That Recognizes Sugar Chains 2 |
| ENSMUSG00000041570 | ENSFM00600000921199 | Calmodulin Regulated Spectrin Associated |
| ENSMUSG00000041592 | ENSFM00260000050437 | Sidekick Precursor |
| ENSMUSG00000041598 | ENSFM00250000006209 | CDC42 Effector 4 Binder Of Rho Gtpases 4 |
| ENSMUSG00000041607 | ENSFM00470000260897 | Unknown |
| ENSMUSG00000041607 | ENSFM00720001502028 | Unknown |
| ENSMUSG00000041607 | ENSFM00250000002814 | Myelin Basic Mbp |
| ENSMUSG00000041608 | ENSFM00250000001339 | Ectonucleoside Triphosphate Diphosphohydrolase Ntpdase EC_3.6.1.5 Ecto Atp Diphosphohydrolase Ecto Atpdase Ecto Atpase Ecto Apyrase |
| ENSMUSG00000041609 | ENSFM00570000884207 | Unknown |
| ENSMUSG00000041609 | ENSFM00500000272101 | Bicaudal D Related 1 Bicd Related 1 Bicdr 1 Coiled Coil Domain Containing 64A |
| ENSMUSG00000041624 | ENSFM00730001521851 | Guanylate Cyclase Soluble Subunit Alpha 3 Gcs Alpha 3 EC_4.6.1.2 Gcs Alpha 1 Soluble Guanylate Cyclase Large Subunit |
| ENSMUSG00000041632 | ENSFM00250000006668 | 28S Ribosomal S27 Mitochondrial Mrp S27 S27MT |
| ENSMUSG00000041638 | ENSFM00250000002277 | Translational Activator GCN1 |
| ENSMUSG00000041650 | ENSFM00660001189568 | Unknown |
| ENSMUSG00000041650 | ENSFM00800002003706 | Propionyl Coa Carboxylase Alpha Chain Mitochondrial Precursor Pccase Subunit Alpha EC_6.4.1.3 Propanoyl Coa:Carbon Dioxide Ligase Subunit Alpha |
| ENSMUSG00000041670 | ENSFM00250000000629 | Regulating Synaptic Membrane Exocytosis Rab 3 Interacting Molecule Rim |
| ENSMUSG00000041685 | ENSFM00600000921280 | Fch Domain Only |
| ENSMUSG00000041688 | ENSFM00250000001614 | Angiomotin |
| ENSMUSG00000041697 | ENSFM00670001236141 | Cytochrome C Oxidase Subunit 6A1 Mitochondrial Precursor Cytochrome C Oxidase Polypeptide Via Liver |
| ENSMUSG00000041720 | ENSFM00570000876532 | Unknown |
| ENSMUSG00000041720 | ENSFM00760001714819 | Phosphatidylinositol 4 Kinase Alpha PI4 Kinase Alpha PI4K Alpha Ptdins 4 Kinase Alpha EC_2.7.1.67 |
| ENSMUSG00000041729 | ENSFM00730001521970 | Coronin |
| ENSMUSG00000041757 | ENSFM00800002003674 | Pleckstrin Homology Domain Containing Family A Member 6 Ph Domain Containing Family A Member 6 Phosphoinositol 3 Phosphate Binding 3 Pepp 3 |
| ENSMUSG00000041763 | ENSFM00250000003674 | Tripeptidyl Peptidase 2 Tpp 2 EC_3.4.14.10 Tripeptidyl Aminopeptidase Tripeptidyl Peptidase Ii Tpp Ii |
| ENSMUSG00000041771 | ENSFM00730001521994 | Sodium/Potassium/Calcium Exchanger 3 Precursor Na + /K + /Ca 2+ Exchange 3 Solute Carrier Family 24 Member 3 |
| ENSMUSG00000041794 | ENSFM00500000271516 | Rab Effector Myrip Exophilin 8 Myosin Viia And Rab Interacting Synaptotagmin Lacking C2 Domains C SLAC2 C Slp Homolog Lacking C2 Domains C |
| ENSMUSG00000041797 | ENSFM00570000851109 | Atp Binding Cassette Sub Family A Member |
| ENSMUSG00000041836 | ENSFM00740001589127 | Receptor Type Tyrosine Phosphatase Precursor Tyrosine Phosphatase R Ptp EC_3.1.3.48 |
| ENSMUSG00000041881 | ENSFM00640001119910 | Unknown |
| ENSMUSG00000041881 | ENSFM00500000274400 | Nadh Dehydrogenase 1 Alpha Subcomplex Subunit 7 Complex I B14 5A Ci B14 5A Nadh Ubiquinone Oxidoreductase Subunit B14 5A |
| ENSMUSG00000041889 | ENSFM00500000274793 | Shisa 4 Precursor Transmembrane 58 |
| ENSMUSG00000041912 | ENSFM00800002016924 | Unknown |
| ENSMUSG00000041912 | ENSFM00250000006342 | Tudor And Kh Domain Containing Tudor Domain Containing 2 |
| ENSMUSG00000041926 | ENSFM00790001963080 | Aminopeptidase B Ap B EC_3.4.11.6 Arginine Aminopeptidase Arginyl Aminopeptidase Cytosol Aminopeptidase Iv |
| ENSMUSG00000041936 | ENSFM00760001714908 | Agrin N Terminal 110 Kda Subunit |
| ENSMUSG00000041957 | ENSFM00730001522005 | Plakophilin 1 Band 6 B6P |
| ENSMUSG00000041958 | ENSFM00500000271389 | Gpi Transamidase Component Pig S Phosphatidylinositol Glycan Biosynthesis Class S |
| ENSMUSG00000041977 | ENSFM00600000921196 | Rho Guanine Nucleotide Exchange Factor |
| ENSMUSG00000041986 | ENSFM00250000003015 | Elmo Domain Containing |
| ENSMUSG00000042035 | ENSFM00740001589327 | Immunoglobulin Superfamily Member Precursor Glu Trp Ile Ewi Motif Containing Ewi Antigen |
| ENSMUSG00000042066 | ENSFM00250000001324 | Transmembrane And Coiled Coil Domains |
| ENSMUSG00000042078 | ENSFM00250000002557 | Synaptic Vesicle 2 Related SV2 Related |
| ENSMUSG00000042096 | ENSFM00670001235511 | D Amino Acid Oxidase Daao Damox Dao EC_1.4.3.3 |
| ENSMUSG00000042165 | ENSFM00250000003585 | Proteasomal Ubiquitin Receptor Adhesion Regulating Molecule 1 Arm 1 |
| ENSMUSG00000042215 | ENSFM00760001743808 | Unknown |
| ENSMUSG00000042215 | ENSFM00260000051255 | Bag Family Molecular Chaperone Regulator 2 Bag 2 Bcl 2 Associated Athanogene 2 |
| ENSMUSG00000042228 | ENSFM00730001521707 | Tyrosine Kinase EC_2.7.10.2 |
| ENSMUSG00000042251 | ENSFM00500000271359 | Probable Carboxypeptidase P Precursor EC_3.4.17.- Peptidase M20 Domain Containing 1 |
| ENSMUSG00000042272 | ENSFM00250000002761 | SEC14 Domain And Spectrin Repeat Containing 1 |
| ENSMUSG00000042275 | ENSFM00250000004827 | Pelota Homolog EC_3.1.-.- |
| ENSMUSG00000042302 | ENSFM00740001589265 | Eh Domain Binding 1 |
| ENSMUSG00000042331 | ENSFM00250000001872 | Cytospin A SPECC1 Sperm Antigen With Calponin Homology And Coiled Coil Domains 1 |
| ENSMUSG00000042345 | ENSFM00250000002027 | Ubiquitin Associated And SH3 Domain Containing B EC_3.1.3.48 Cbl Interacting P70 Suppressor Of T Cell Receptor Signaling 1 Sts 1 T Cell Ubiquitin Ligand 2 Tula 2 Tyrosine Phosphatase STS1/TULA2 |
| ENSMUSG00000042348 | ENSFM00420000140675 | Adp Ribosylation Factor 15 Adp Ribosylation Factor Related 2 Arf Related 2 |
| ENSMUSG00000042363 | ENSFM00500000272380 | Galectin Related Lectin Galactoside Binding |
| ENSMUSG00000042388 | ENSFM00250000000718 | Disks Large Associated Dap Psd 95/SAP90 Binding SAP90/Psd 95 Associated |
| ENSMUSG00000042401 | ENSFM00250000002903 | Cartilage Acidic 1 Precursor 68 Kda Chondrocyte Expressed Cep 68 Aspic |
| ENSMUSG00000042404 | ENSFM00250000001005 | Denn Domain Containing |
| ENSMUSG00000042410 | ENSFM00660001180466 | Unknown |
| ENSMUSG00000042410 | ENSFM00660001190018 | Unknown |
| ENSMUSG00000042410 | ENSFM00250000004718 | Alkyldihydroxyacetonephosphate Synthase Alkyl Dhap Synthase EC_2.5.1.26 Alkylglycerone Phosphate Synthase |
| ENSMUSG00000042425 | ENSFM00250000001492 | Ferm And Pdz Domain Containing 4 Pdz Domain Containing 10 Psd 95 Interacting Regulator Of Spine Morphogenesis Preso |
| ENSMUSG00000042426 | ENSFM00260000050346 | Atp Dependent Rna Helicase EC_3.6.4.13 Deah Box |
| ENSMUSG00000042444 | ENSFM00250000002444 | FAM63A |
| ENSMUSG00000042447 | ENSFM00500000271280 | Wd Repeat Containing Mio |
| ENSMUSG00000042492 | ENSFM00260000050553 | TBC1 Domain Family Member |
| ENSMUSG00000042520 | ENSFM00800002020257 | Unknown |
| ENSMUSG00000042520 | ENSFM00800002020258 | Unknown |
| ENSMUSG00000042520 | ENSFM00600000921193 | Ubiquitin Associated 2 |
| ENSMUSG00000042532 | ENSFM00250000003047 | Golgin Subfamily A Member 7 |
| ENSMUSG00000042535 | ENSFM00250000001372 | Gtp Binding |
| ENSMUSG00000042569 | ENSFM00560000771026 | Dehydrogenase/Reductase Sdr Family Member EC_1.1.-.- |
| ENSMUSG00000042604 | ENSFM00290000065519 | Potassium Voltage Gated Channel Subfamily A Member Voltage Gated Potassium Channel Subunit KV1 |
| ENSMUSG00000042605 | ENSFM00590000917643 | Unknown |
| ENSMUSG00000042605 | ENSFM00590000917644 | Unknown |
| ENSMUSG00000042605 | ENSFM00800002017351 | Unknown |
| ENSMUSG00000042605 | ENSFM00600000921253 | Ataxin 2 Spinocerebellar Ataxia Type 2 |
| ENSMUSG00000042644 | ENSFM00250000000547 | Inositol 1 4 5 Trisphosphate Receptor Type IP3 Receptor IP3R Ins Type Inositol 1 4 5 Trisphosphate Receptor Type INSP3 Receptor |
| ENSMUSG00000042647 | ENSFM00500000333101 | Uncharacterized |
| ENSMUSG00000042647 | ENSFM00800002023650 | Unknown |
| ENSMUSG00000042647 | ENSFM00780001898384 | Acyl Coa Dehydrogenase Family Member Acad EC_1.3.99.- |
| ENSMUSG00000042662 | ENSFM00740001598893 | Unknown |
| ENSMUSG00000042662 | ENSFM00670001235708 | Dual Specificity Phosphatase EC_3.1.3.16 EC_3.1.3.- 48 |
| ENSMUSG00000042662 | ENSFM00670001254208 | DUSP15 |
| ENSMUSG00000042671 | ENSFM00670001235642 | Regulator Of G Signaling |
| ENSMUSG00000042682 | ENSFM00670001236822 | Selenoprotein K Selk |
| ENSMUSG00000042686 | ENSFM00250000001356 | Junctophilin Jp Junctophilin |
| ENSMUSG00000042699 | ENSFM00760001763154 | Unknown |
| ENSMUSG00000042699 | ENSFM00800002003717 | Atp Dependent Rna Helicase A EC_3.6.4.13 Deah Box 9 Nuclear Dna Helicase Ii Ndh Ii |
| ENSMUSG00000042700 | ENSFM00730001521477 | Signal Induced Proliferation Associated 1 SIPA1 |
| ENSMUSG00000042705 | ENSFM00250000008194 | Comm Domain Containing 10 |
| ENSMUSG00000042734 | ENSFM00670001235990 | Tetratricopeptide Repeat Tpr Repeat |
| ENSMUSG00000042744 | ENSFM00500000270499 | Probable E3 Ubiquitin Ligase HECTD4 EC_6.3.2.- Hect Domain Containing 4 |
| ENSMUSG00000042766 | ENSFM00570000879224 | Unknown |
| ENSMUSG00000042766 | ENSFM00400000131833 | Tripartite Motif Containing 46 Gene Y Geney Tripartite Fibronectin Type Iii And C Terminal Spry Motif |
| ENSMUSG00000042770 | ENSFM00250000006764 | Heme Binding 1 |
| ENSMUSG00000042787 | ENSFM00500000272927 | Nuclease Exog Mitochondrial Precursor EC_3.1.30.- Endonuclease G 1 Endo G 1 |
| ENSMUSG00000042790 | ENSFM00590000917795 | Unknown |
| ENSMUSG00000042790 | ENSFM00590000917796 | Unknown |
| ENSMUSG00000042790 | ENSFM00250000005433 | Ring Finger 214 |
| ENSMUSG00000042807 | ENSFM00730001522025 | E3 Ubiquitin Ligase EC_6.3.2.- Hect C2 And Ww Domain Containing NEDD4 E3 Ubiquitin Ligase |
| ENSMUSG00000042846 | ENSFM00400000131776 | Leucine Rich Repeat Transmembrane Neuronal Precursor |
| ENSMUSG00000042870 | ENSFM00250000001136 | TOM1 2 Target Of Myb 2 |
| ENSMUSG00000042873 | ENSFM00250000002406 | Lipoma Hmgic Fusion Partner |
| ENSMUSG00000042901 | ENSFM00780001917699 | Unknown |
| ENSMUSG00000042901 | ENSFM00250000003456 | Axin Interactor Dorsalization Associated Axin Interaction Partner And Dorsalization Antagonist |
| ENSMUSG00000042992 | ENSFM00250000006838 | Loss Of Heterozygosity 12 Chromosomal Region 1 |
| ENSMUSG00000043004 | ENSFM00500000277611 | Unknown |
| ENSMUSG00000043079 | ENSFM00590000918103 | Unknown |
| ENSMUSG00000043079 | ENSFM00250000008348 | Synaptopodin |
| ENSMUSG00000043091 | ENSFM00250000000217 | Tubulin Alpha Chain |
| ENSMUSG00000043192 | ENSFM00250000000760 | Glucose 6 Phosphate Isomerase Gpi EC_5.3.1.9 Phosphoglucose Isomerase Pgi Phosphohexose Isomerase Phi |
| ENSMUSG00000043284 | ENSFM00250000007165 | Transmembrane 11 Mitochondrial |
| ENSMUSG00000043301 | ENSFM00730001521214 | G Activated Inward Rectifier Potassium Channel Girk Inward Rectifier K + Channel KIR3 Potassium Channel Inwardly Rectifying Subfamily J Member |
| ENSMUSG00000043384 | ENSFM00600000921594 | G Coupled Receptor Associated Sorting Gasp |
| ENSMUSG00000043445 | ENSFM00670001235619 | Pyridoxal Phosphate Phosphatase Plp Phosphatase EC_3.1.3.3 EC_3.1.3.- 74 Chronophin |
| ENSMUSG00000043448 | ENSFM00730001521860 | Gap Junction Gamma 1 Connexin 45 CX45 Gap Junction Alpha 7 |
| ENSMUSG00000043460 | ENSFM00250000002984 | Phosphatase 1 Regulatory Subunit 29 Precursor Extracellular Leucine Rich Repeat And Fibronectin Type Iii Domain Containing 2 Leucine Rich Repeat And Fibronectin Type Iii Domain Containing 6 Leucine Rich Repeat Containing 62 |
| ENSMUSG00000043463 | ENSFM00750001632398 | Ras Related Rab |
| ENSMUSG00000043542 | ENSFM00500000272347 | Zinc Finger C2HC Domain Containing 1A |
| ENSMUSG00000043557 | ENSFM00250000002191 | Mam Domain Containing Glycosylphosphatidylinositol Anchor 2 Precursor Mam Domain Containing 1 |
| ENSMUSG00000043572 | ENSFM00500000270765 | Probable Proline Trna Ligase Mitochondrial Precursor EC_6.1.1.15 Prolyl Trna Synthetase Prors |
| ENSMUSG00000043629 | ENSFM00500000286919 | Small Membrane A Kinase Anchor Small Membrane Akap Smakap |
| ENSMUSG00000043670 | ENSFM00500000270725 | Gtp Binding Di Precursor Distinct Subgroup Of The Ras Family Member |
| ENSMUSG00000043716 | ENSFM00250000000871 | 60S Ribosomal L7 |
| ENSMUSG00000043807 | ENSFM00250000009121 | Lymphocyte Antigen 6 Complex Locus G5B Precursor |
| ENSMUSG00000043811 | ENSFM00250000002242 | Reticulon 4 Receptor Precursor Nogo Receptor Nogo 66 Receptor Homolog Nogo 66 Receptor Related |
| ENSMUSG00000043843 | ENSFM00250000005962 | Transmembrane 145 |
| ENSMUSG00000043940 | ENSFM00780001898367 | Wd Repeat And Fyve Domain Containing |
| ENSMUSG00000043991 | ENSFM00250000002171 | Transcriptional Activator Pur Beta Purine Rich Element Binding B |
| ENSMUSG00000044018 | ENSFM00250000009476 | 39S Ribosomal L50 Mitochondrial L50MT Mrp L50 |
| ENSMUSG00000044024 | ENSFM00250000009583 | Relt 2 |
| ENSMUSG00000044060 | ENSFM00690001356664 | Btb/Poz Domain Containing 8 |
| ENSMUSG00000044080 | ENSFM00500000444029 | Unknown |
| ENSMUSG00000044080 | ENSFM00670001236593 | S100 A1 S 100 Alpha Chain S 100 Subunit Alpha S100 Calcium Binding A1 |
| ENSMUSG00000044147 | ENSFM00750001632347 | Adp Ribosylation Factor |
| ENSMUSG00000044216 | ENSFM00730001521253 | Inward Rectifier Potassium Channel Inward Rectifier K + Channel KIR2 Irk Potassium Channel Inwardly Rectifying Subfamily J Member |
| ENSMUSG00000044252 | ENSFM00570000883987 | Unknown |
| ENSMUSG00000044252 | ENSFM00570000900012 | Unknown |
| ENSMUSG00000044252 | ENSFM00500000269986 | Oxysterol Binding Related 1 Orp 1 Osbp Related 1 |
| ENSMUSG00000044287 | ENSFM00600000921973 | Neuritin Precursor |
| ENSMUSG00000044288 | ENSFM00250000000847 | Cannabinoid Receptor 1 Cb R CB1 |
| ENSMUSG00000044352 | ENSFM00500000274456 | Ankyrin Repeat Domain Containing Sowaha Precursor Ankyrin Repeat Domain Containing 43 Sosondowah Homolog A |
| ENSMUSG00000044424 | ENSFM00250000002224 | 40S Ribosomal S7 |
| ENSMUSG00000044433 | ENSFM00600000921199 | Calmodulin Regulated Spectrin Associated |
| ENSMUSG00000044447 | ENSFM00760001714647 | Dedicator Of Cytokinesis |
| ENSMUSG00000044629 | ENSFM00250000008302 | CB1 Cannabinoid Receptor Interacting 1 Crip 1 |
| ENSMUSG00000044667 | ENSFM00500000270520 | Lipid Phosphate Phosphatase Related Type 4 EC_3.1.3.4 Brain Specific Phosphatidic Acid Phosphatase 1 Plasticity Related Gene 1 Prg 1 |
| ENSMUSG00000044708 | ENSFM00730001521400 | Atp Sensitive Inward Rectifier Potassium Channel Atp Potassium Channel K Inward Rectifier K + Channel 1 Potassium Channel Inwardly Rectifying Subfamily J Member |
| ENSMUSG00000044734 | ENSFM00500000269596 | Serpin |
| ENSMUSG00000044847 | ENSFM00250000007949 | U7 Snrna Associated Sm LSM11 |
| ENSMUSG00000044894 | ENSFM00500000446966 | Unknown |
| ENSMUSG00000044894 | ENSFM00500000290955 | Cytochrome B C1 Complex Subunit 8 Complex Iii Subunit 8 Complex Iii Subunit Viii Ubiquinol Cytochrome C Reductase Complex 9.5 Kda Ubiquinol Cytochrome C Reductase Complex Ubiquinone Binding Qp C |
| ENSMUSG00000044986 | ENSFM00500000270326 | Thiosulfate Sulfurtransferase EC_2.8.1.1 Rhodanese |
| ENSMUSG00000045007 | ENSFM00420000140599 | Tubulin Gamma Chain Gamma Tubulin |
| ENSMUSG00000045009 | ENSFM00250000007114 | Proline Rich Transmembrane 3 Precursor |
| ENSMUSG00000045038 | ENSFM00750001632342 | Kinase C EC_2.7.11.13 |
| ENSMUSG00000045045 | ENSFM00260000050443 | Leucine Rich Repeat And Fibronectin Type Iii Domain Containing Precursor Synaptic Adhesion Molecule |
| ENSMUSG00000045083 | ENSFM00250000001884 | Leucine Rich Repeat And Immunoglobulin Domain Containing Nogo Receptor Interacting Precursor Leucine Rich Repeat Neuronal |
| ENSMUSG00000045092 | ENSFM00730001521347 | Sphingosine 1 Phosphate Receptor S1P Receptor Endothelial Differentiation G Coupled Receptor Sphingosine 1 Phosphate Receptor Edg S1P Receptor Edg |
| ENSMUSG00000045095 | ENSFM00250000000552 | Membrane Associated Guanylate Kinase Ww And Pdz Domain Containing Membrane Associated Guanylate Kinase Inverted Magi |
| ENSMUSG00000045103 | ENSFM00250000000455 | Dystrophin |
| ENSMUSG00000045114 | ENSFM00810002055295 | Unknown |
| ENSMUSG00000045114 | ENSFM00810002057413 | Unknown |
| ENSMUSG00000045114 | ENSFM00810002057414 | Unknown |
| ENSMUSG00000045114 | ENSFM00500000274820 | Proline Rich Transmembrane 2 Dispanin Subfamily B Member 3 DSPB3 |
| ENSMUSG00000045128 | ENSFM00760001714679 | 60S Ribosomal L18A |
| ENSMUSG00000045136 | ENSFM00250000000193 | Tubulin Beta Chain |
| ENSMUSG00000045176 | ENSFM00500000274101 | Uncharacterized |
| ENSMUSG00000045193 | ENSFM00600000921577 | Cold Inducible Rna Binding Glycine Rich Rna Binding Cirp |
| ENSMUSG00000045210 | ENSFM00250000008023 | Deubiquitinating VCIP135 EC_3.4.19.12 Valosin Containing P97/P47 Complex Interacting 1 Valosin Containing P97/P47 Complex Interacting P135 |
| ENSMUSG00000045211 | ENSFM00260000051397 | 8 Oxo Dgdp Phosphatase NUDT18 EC_3.6.1.58 2 Hydroxy Dadp Phosphatase 7 8 Dihydro 8 Oxoguanine Phosphatase Mutt Homolog 3 Nucleoside Diphosphate Linked Moiety X Motif 18 Nudix Motif 18 |
| ENSMUSG00000045302 | ENSFM00810002055838 | Unknown |
| ENSMUSG00000045302 | ENSFM00250000005591 | Prolactin Regulatory Element Binding Mammalian Guanine Nucleotide Exchange Factor MSEC12 |
| ENSMUSG00000045316 | ENSFM00670001235551 | Acylpyruvase FAHD1 Mitochondrial Precursor EC_3.7.1.5 Fumarylacetoacetate Hydrolase Domain Containing 1 |
| ENSMUSG00000045348 | ENSFM00250000007087 | Neuronal Tyrosine Phosphorylated Phosphoinositide 3 Kinase Adapter 1 |
| ENSMUSG00000045411 | ENSFM00570000879994 | Unknown |
| ENSMUSG00000045414 | ENSFM00250000009380 | Deleted In Autism 1 Precursor Golgi Of 49 Kda GOPRO49 Hypoxia And Akt Induced Stem Cell Factor Hasf |
| ENSMUSG00000045441 | ENSFM00250000008609 | G Regulated Inducer Of Neurite Outgrowth 3 GRIN3 |
| ENSMUSG00000045455 | ENSFM00670001236601 | Mitochondrial Import Inner Membrane Translocase Subunit TIM8 A Deafness Dystonia 1 |
| ENSMUSG00000045589 | ENSFM00570000876542 | Unknown |
| ENSMUSG00000045589 | ENSFM00250000008517 | Domon Domain Containing FRRS1L Ferric Chelate Reductase 1 |
| ENSMUSG00000045659 | ENSFM00800002003882 | Pleckstrin Homology Domain Containing Family A Member 7 Ph Domain Containing Family A Member 7 Heart Adapter 1 |
| ENSMUSG00000045671 | ENSFM00250000001487 | Sprouty Related EVH1 Domain Containing Spred |
| ENSMUSG00000045731 | ENSFM00670001235427 | Prepronociceptin Neuropeptide 1 |
| ENSMUSG00000045763 | ENSFM00800002007336 | Brain Acid Soluble 1.22 Kda Neuronal Tissue Enriched Acidic Neuronal Axonal Membrane Nap 22 |
| ENSMUSG00000045912 | ENSFM00500000273557 | C2 Calcium Dependent Domain Containing 4C Nuclear Localized Factor 3 FAM148C |
| ENSMUSG00000045962 | ENSFM00590000917927 | Unknown |
| ENSMUSG00000045962 | ENSFM00590000917928 | Unknown |
| ENSMUSG00000045962 | ENSFM00590000918190 | Unknown |
| ENSMUSG00000045962 | ENSFM00250000000621 | Serine/Threonine Kinase EC_2.7.11.1 Kinase Lysine Deficient Kinase With No Lysine |
| ENSMUSG00000045967 | ENSFM00250000001603 | Probable G Coupled Receptor Precursor |
| ENSMUSG00000045973 | ENSFM00500000271232 | Solute Carrier Family 25 Member 51 Mitochondrial Carrier Triple Repeat 1 |
| ENSMUSG00000045983 | ENSFM00570000887850 | Unknown |
| ENSMUSG00000045983 | ENSFM00250000000634 | Eukaryotic Translation Initiation Factor 4 Gamma 3 Eif 4 Gamma 3 Eif 4G 3 EIF4G 3 Eif 4 Gamma Ii EIF4GII |
| ENSMUSG00000046032 | ENSFM00250000002957 | Sorting Nexin |
| ENSMUSG00000046079 | ENSFM00570000894492 | Unknown |
| ENSMUSG00000046079 | ENSFM00570000896228 | Unknown |
| ENSMUSG00000046079 | ENSFM00250000001418 | Volume Regulated Anion Channel Subunit Leucine Rich Repeat Containing |
| ENSMUSG00000046093 | ENSFM00570000890067 | Unknown |
| ENSMUSG00000046093 | ENSFM00760001714608 | Unknown |
| ENSMUSG00000046159 | ENSFM00800002003412 | Muscarinic Acetylcholine Receptor |
| ENSMUSG00000046182 | ENSFM00670001235575 | Germ Cell Specific Gene 1 GSG1 |
| ENSMUSG00000046230 | ENSFM00250000000977 | Vacuolar Sorting Associated 13A Chorea Acanthocytosis Chorein |
| ENSMUSG00000046240 | ENSFM00250000007963 | Hepatocyte Cell Adhesion Molecule Precursor Hepacam |
| ENSMUSG00000046312 | ENSFM00570000883301 | Unknown |
| ENSMUSG00000046312 | ENSFM00540000717964 | Uncharacterized Family 31 Glucosidase EC_3.2.1.- |
| ENSMUSG00000046314 | ENSFM00570000892739 | Unknown |
| ENSMUSG00000046314 | ENSFM00500000272618 | Syntaxin Binding 6 |
| ENSMUSG00000046324 | ENSFM00250000001559 | Endoplasmic Reticulum Metallopeptidase 1 |
| ENSMUSG00000046329 | ENSFM00610000973458 | Unknown |
| ENSMUSG00000046329 | ENSFM00500000269757 | Calcium Binding Mitochondrial Carrier Scamc Small Calcium Binding Mitochondrial Carrier Solute Carrier Family 25 Member |
| ENSMUSG00000046330 | ENSFM00670001236020 | 60S Ribosomal L37A |
| ENSMUSG00000046364 | ENSFM00640001118765 | Unknown |
| ENSMUSG00000046364 | ENSFM00780001898350 | 60S Ribosomal L27A |
| ENSMUSG00000046417 | ENSFM00250000005430 | Leucine Rich Repeat Containing Leucine Rich Repeat Containing |
| ENSMUSG00000046434 | ENSFM00270000056431 | Heterogeneous Nuclear Ribonucleoprotein |
| ENSMUSG00000046480 | ENSFM00500000274501 | Sodium Channel Subunit Beta 4 Precursor |
| ENSMUSG00000046573 | ENSFM00670001238243 | Lyr Motif Containing 4 |
| ENSMUSG00000046598 | ENSFM00500000271183 | D Beta Hydroxybutyrate Dehydrogenase Mitochondrial Bdh EC_1.1.1.30 3 Hydroxybutyrate Dehydrogenase |
| ENSMUSG00000046671 | ENSFM00250000006972 | Mitochondrial Fission Regulator 1 |
| ENSMUSG00000046691 | ENSFM00250000008429 | Chromosome Transmission Fidelity 8 Homolog 2 |
| ENSMUSG00000046691 | ENSFM00500000283580 | Chromosome Transmission Fidelity 8 Homolog |
| ENSMUSG00000046699 | ENSFM00250000001398 | Slit And Ntrk Precursor |
| ENSMUSG00000046707 | ENSFM00250000001591 | Casein Kinase Ii Subunit Ck Ii EC_2.7.11.1 |
| ENSMUSG00000046709 | ENSFM00460000244930 | Mitogen Activated Kinase Map Kinase Mapk EC_2.7.11.24 Stress Activated Kinase C Jun N Terminal Kinase |
| ENSMUSG00000046798 | ENSFM00570000889082 | Unknown |
| ENSMUSG00000046798 | ENSFM00400000131954 | Claudin 12 |
| ENSMUSG00000046841 | ENSFM00250000007369 | Cytoskeleton Associated 4.63 Kda Cytoskeleton Linking Membrane Climp 63 P63 |
| ENSMUSG00000046844 | ENSFM00500000270300 | Synaptic Vesicle Membrane Vat 1 Ec 1 |
| ENSMUSG00000046879 | ENSFM00250000001767 | Interferon Inducible Gtpase 5 EC_3.6.5.- Immunity Related Gtpase Cinema 1 |
| ENSMUSG00000046985 | ENSFM00800002017596 | Unknown |
| ENSMUSG00000046985 | ENSFM00800002023590 | Unknown |
| ENSMUSG00000046985 | ENSFM00500000271643 | Transmembrane Anterior Posterior Transformation 1 |
| ENSMUSG00000047013 | ENSFM00250000006718 | F Box Only 41 |
| ENSMUSG00000047037 | ENSFM00730001521603 | Magnesium Transporter Non Imprinted In Prader Willi/Angelman Syndrome Region |
| ENSMUSG00000047085 | ENSFM00260000050662 | Leucine Rich Repeat Containing Precursor Netrin Ligand Ngl |
| ENSMUSG00000047126 | ENSFM00250000001393 | Clathrin Heavy Chain |
| ENSMUSG00000047187 | ENSFM00730001521469 | Ras Related Rab |
| ENSMUSG00000047215 | ENSFM00800002021381 | Unknown |
| ENSMUSG00000047215 | ENSFM00750001632341 | 60S Ribosomal L9 |
| ENSMUSG00000047250 | ENSFM00250000001175 | Prostaglandin G/H Synthase 2 Precursor EC_1.14.99.1 Cyclooxygenase 2 Cox 2 Phs Ii Prostaglandin H2 Synthase 2 Pgh Synthase 2 Pghs 2 Prostaglandin Endoperoxide Synthase 2 |
| ENSMUSG00000047260 | ENSFM00670001236443 | Er Membrane Complex Subunit 6 Transmembrane 93 |
| ENSMUSG00000047261 | ENSFM00250000005389 | Neuromodulin Axonal Membrane Gap 43 Growth Associated 43 |
| ENSMUSG00000047368 | ENSFM00250000001156 | Alpha/Beta Hydrolase Domain Containing Ec 3 |
| ENSMUSG00000047379 | ENSFM00250000008535 | Beta 1 4 Glucuronyltransferase 1 |
| ENSMUSG00000047414 | ENSFM00250000003081 | Leucine Rich Repeat Transmembrane Precursor Fibronectin Domain Containing Leucine Rich Transmembrane |
| ENSMUSG00000047454 | ENSFM00790001963049 | Molybdopterin Adenylyltransferase Mpt Adenylyltransferase EC_2.7.7.75 Domain G |
| ENSMUSG00000047459 | ENSFM00740001589456 | Dynein Light Chain Roadblock Type Dynein Light Chain Cytoplasmic |
| ENSMUSG00000047495 | ENSFM00250000000718 | Disks Large Associated Dap Psd 95/SAP90 Binding SAP90/Psd 95 Associated |
| ENSMUSG00000047507 | ENSFM00720001499239 | Unknown |
| ENSMUSG00000047507 | ENSFM00250000001781 | Unc 13 Homolog D MUNC13 4 |
| ENSMUSG00000047547 | ENSFM00570000887013 | Unknown |
| ENSMUSG00000047547 | ENSFM00250000001836 | Clathrin Light Chain |
| ENSMUSG00000047557 | ENSFM00250000009454 | Latexin Endogenous Carboxypeptidase Inhibitor Eci Tissue Carboxypeptidase Inhibitor Tci |
| ENSMUSG00000047675 | ENSFM00800002003452 | 40S Ribosomal S8 |
| ENSMUSG00000047804 | ENSFM00740001589182 | Serine/Threonine Kinase EC_2.7.11.1 Unc 51 Kinase |
| ENSMUSG00000047842 | ENSFM00500000270725 | Gtp Binding Di Precursor Distinct Subgroup Of The Ras Family Member |
| ENSMUSG00000047866 | ENSFM00730001521929 | Lon Protease Homolog 2 Peroxisomal |
| ENSMUSG00000047879 | ENSFM00780001898425 | Ubiquitin Carboxyl Terminal Hydrolase 14 EC_3.4.19.12 Deubiquitinating Enzyme 14 Ubiquitin Thioesterase 14 Ubiquitin Specific Processing Protease 14 |
| ENSMUSG00000047921 | ENSFM00250000002395 | Trafficking Particle Complex Subunit 9 Nik And Ikbkb Binding |
| ENSMUSG00000047945 | ENSFM00600000921868 | Marcks Related Marcks 1 Macrophage Myristoylated Alanine Rich C Kinase Substrate Mac Marcks Macmarcks |
| ENSMUSG00000047959 | ENSFM00290000065519 | Potassium Voltage Gated Channel Subfamily A Member Voltage Gated Potassium Channel Subunit KV1 |
| ENSMUSG00000047976 | ENSFM00290000065519 | Potassium Voltage Gated Channel Subfamily A Member Voltage Gated Potassium Channel Subunit KV1 |
| ENSMUSG00000047986 | ENSFM00250000010790 | Paralemmin 3 Precursor |
| ENSMUSG00000048000 | ENSFM00640001110751 | Unknown |
| ENSMUSG00000048000 | ENSFM00640001118145 | Unknown |
| ENSMUSG00000048000 | ENSFM00740001589295 | Perq Amino Acid Rich With Gyf Domain Containing 2 Trinucleotide Repeat Containing Gene 15 |
| ENSMUSG00000048022 | ENSFM00500000277126 | Transmembrane 229A |
| ENSMUSG00000048040 | ENSFM00250000004611 | Signal Peptidase Complex Subunit 3 EC_3.4.-.- Microsomal Signal Peptidase 22/23 Kda Subunit SPC22/23 Spase 22/23 Kda Subunit |
| ENSMUSG00000048076 | ENSFM00730001521124 | Adp Ribosylation Factor |
| ENSMUSG00000048078 | ENSFM00250000000375 | Teneurin Ten Odd Oz/Ten M Homolog Tenascin Ten Teneurin Transmembrane |
| ENSMUSG00000048087 | ENSFM00730001521588 | Adenosylhomocysteinase Adohcyase EC_3.3.1.1 S Adenosyl L Homocysteine Hydrolase |
| ENSMUSG00000048120 | ENSFM00570000895392 | Unknown |
| ENSMUSG00000048120 | ENSFM00250000001339 | Ectonucleoside Triphosphate Diphosphohydrolase Ntpdase EC_3.6.1.5 Ecto Atp Diphosphohydrolase Ecto Atpdase Ecto Atpase Ecto Apyrase |
| ENSMUSG00000048279 | ENSFM00740001589315 | Sacsin Dnaj Homolog Subfamily C Member 29 DNAJC29 |
| ENSMUSG00000048304 | ENSFM00250000001398 | Slit And Ntrk Precursor |
| ENSMUSG00000048307 | ENSFM00250000008243 | Ankyrin Repeat Domain Containing 46 Ankyrin Repeat Small Ank S |
| ENSMUSG00000048351 | ENSFM00250000006626 | Cytochrome C Oxidase Assembly Factor Beta Lactamase Hcp SEL1 Repeat Containing |
| ENSMUSG00000048388 | ENSFM00740001589308 | Precursor |
| ENSMUSG00000048429 | ENSFM00500000274239 | Uncharacterized |
| ENSMUSG00000048537 | ENSFM00570000895969 | Unknown |
| ENSMUSG00000048537 | ENSFM00600000921202 | Pleckstrin Homology Domain Family B Member LL5 |
| ENSMUSG00000048550 | ENSFM00570000886184 | Unknown |
| ENSMUSG00000048550 | ENSFM00770001847663 | Threonine Synthase 1 TSH1 |
| ENSMUSG00000048578 | ENSFM00250000005592 | Malectin Precursor |
| ENSMUSG00000048707 | ENSFM00250000006760 | Taperin |
| ENSMUSG00000048755 | ENSFM00250000005002 | Malonyl Coa Acyl Carrier Transacylase Mitochondrial Precursor Mct EC_2.3.1.39 Malonyltransferase |
| ENSMUSG00000048758 | ENSFM00600000921342 | 60S Ribosomal L29 |
| ENSMUSG00000048787 | ENSFM00570000894687 | Unknown |
| ENSMUSG00000048787 | ENSFM00500000271989 | DCN1 3 DCUN1 Domain Containing 3 Defective In Cullin Neddylation 1 3 |
| ENSMUSG00000048832 | ENSFM00500000274002 | Vacuolar Sorting Associated 37C Escrt I Complex Subunit VPS37C |
| ENSMUSG00000048988 | ENSFM00250000002984 | Phosphatase 1 Regulatory Subunit 29 Precursor Extracellular Leucine Rich Repeat And Fibronectin Type Iii Domain Containing 2 Leucine Rich Repeat And Fibronectin Type Iii Domain Containing 6 Leucine Rich Repeat Containing 62 |
| ENSMUSG00000049044 | ENSFM00250000000899 | Rap Guanine Nucleotide Exchange Factor Exchange Factor Directly Activated By Camp Exchange Directly Activated By Camp Epac Camp Regulated Guanine Nucleotide Exchange Factor Camp |
| ENSMUSG00000049047 | ENSFM00250000001585 | Armadillo Repeat Containing X Linked |
| ENSMUSG00000049076 | ENSFM00250000000926 | Arf Gap With Coiled Coil Ank Repeat And Ph Domain Containing Centaurin Beta Cnt |
| ENSMUSG00000049100 | ENSFM00780001898335 | Protocadherin Precursor |
| ENSMUSG00000049119 | ENSFM00500000272914 | Unknown |
| ENSMUSG00000049184 | ENSFM00250000002171 | Transcriptional Activator Pur Beta Purine Rich Element Binding B |
| ENSMUSG00000049225 | ENSFM00250000002844 | Phosphatase 1 Mitochondrial Precursor Pdp 1 EC_3.1.3.43 Phosphatase 2C Pyruvate Dehydrogenase Phosphatase Catalytic Subunit 1 Pdpc 1 |
| ENSMUSG00000049230 | ENSFM00740001589263 | Myelin Expression Factor 2 Mef 2 Myef 2 |
| ENSMUSG00000049252 | ENSFM00740001589098 | Density Lipoprotein Receptor Related Precursor Lrp |
| ENSMUSG00000049281 | ENSFM00500000271888 | Sodium Channel Subunit Beta 3 Precursor |
| ENSMUSG00000049287 | ENSFM00500000273104 | Transferase CAF17 Mitochondrial Precursor EC_2.1.-.- Iron Sulfur Cluster Assembly Factor Homolog |
| ENSMUSG00000049303 | ENSFM00610000969294 | Unknown |
| ENSMUSG00000049303 | ENSFM00500000272992 | Synaptotagmin 12 Synaptotagmin Xii Sytxii |
| ENSMUSG00000049313 | ENSFM00570000851314 | Sortilin Related Receptor Low Density Lipoprotein Receptor Relative With 11 Ligand Binding Repeats Ldlr Relative With 11 Ligand Binding Repeats LR11 Sorla 1 Sorting Related Receptor Containing Ldlr Class A Repeats |
| ENSMUSG00000049336 | ENSFM00250000000375 | Teneurin Ten Odd Oz/Ten M Homolog Tenascin Ten Teneurin Transmembrane |
| ENSMUSG00000049339 | ENSFM00500000272612 | Unknown |
| ENSMUSG00000049339 | ENSFM00760001751343 | Unknown |
| ENSMUSG00000049339 | ENSFM00760001763101 | Unknown |
| ENSMUSG00000049339 | ENSFM00810002043394 | Unknown |
| ENSMUSG00000049354 | ENSFM00500000271100 | DDB1 And CUL4 Associated Factor 7 Wd Repeat Containing 68 Wd Repeat Containing AN11 Homolog |
| ENSMUSG00000049511 | ENSFM00800002003603 | 5 Hydroxytryptamine Receptor 1B 5 Ht 1B 5 HT1B Serotonin Receptor 1B |
| ENSMUSG00000049517 | ENSFM00250000002784 | 40S Ribosomal S23 |
| ENSMUSG00000049521 | ENSFM00250000009952 | CDC42 Effector 1 Binder Of Rho Gtpases 5 |
| ENSMUSG00000049550 | ENSFM00250000000764 | Cap Gly Domain Containing Linker 2 Cytoplasmic Linker 115 Clip 115 Cytoplasmic Linker 2 |
| ENSMUSG00000049556 | ENSFM00250000001884 | Leucine Rich Repeat And Immunoglobulin Domain Containing Nogo Receptor Interacting Precursor Leucine Rich Repeat Neuronal |
| ENSMUSG00000049583 | ENSFM00730001521627 | Metabotropic Glutamate Receptor Precursor |
| ENSMUSG00000049612 | ENSFM00250000008621 | Oligodendrocyte Myelin Glycoprotein Precursor |
| ENSMUSG00000049760 | ENSFM00250000011790 | QIL1 |
| ENSMUSG00000049775 | ENSFM00500000306692 | Unknown |
| ENSMUSG00000049775 | ENSFM00550000746767 | Unknown |
| ENSMUSG00000049792 | ENSFM00590000917510 | Unknown |
| ENSMUSG00000049792 | ENSFM00590000917511 | Unknown |
| ENSMUSG00000049792 | ENSFM00250000007925 | Bag Family Molecular Chaperone Regulator 5 Bag 5 Bcl 2 Associated Athanogene 5 |
| ENSMUSG00000049804 | ENSFM00810002041142 | Armadillo Repeat Containing X Linked 4 |
| ENSMUSG00000049807 | ENSFM00250000001623 | Rho Gtpase Activating 21 Rho Type Gtpase Activating 21 |
| ENSMUSG00000049858 | ENSFM00500000270791 | Sulfite Oxidase Mitochondrial Precursor EC_1.8.3.1 |
| ENSMUSG00000049940 | ENSFM00670001235707 | Membrane Associated Progesterone Receptor Component |
| ENSMUSG00000050029 | ENSFM00800002003504 | Ras Related Rap Precursor |
| ENSMUSG00000050043 | ENSFM00570000891191 | Unknown |
| ENSMUSG00000050043 | ENSFM00250000004757 | Thioredoxin Related Transmembrane 2 Precursor Thioredoxin Domain Containing 14 |
| ENSMUSG00000050079 | ENSFM00250000004877 | Ring Finger And Spry Domain Containing 1 Precursor |
| ENSMUSG00000050121 | ENSFM00600000921671 | Opalin Oligodendrocytic Myelin Paranodal And Inner Loop Transmembrane 10 |
| ENSMUSG00000050132 | ENSFM00250000003414 | Sterile Alpha And Tir Motif Containing Tir 1 Homolog |
| ENSMUSG00000050148 | ENSFM00250000001264 | Ubiquilin Linking Iap With Cytoskeleton Plic |
| ENSMUSG00000050211 | ENSFM00540000717923 | Cytosolic Phospholipase A2 CPLA2 EC_3.1.1.4 Phospholipase A2 Group |
| ENSMUSG00000050321 | ENSFM00250000001916 | Neuropilin And Tolloid Precursor Brain Specific Transmembrane Containing 2 Cub And 1 Ldl Receptor Class A Domains |
| ENSMUSG00000050323 | ENSFM00500000273764 | Nadh Dehydrogenase Ubiquinone Complex I Assembly Factor 6 Precursor |
| ENSMUSG00000050357 | ENSFM00260000050454 | Leucine Rich Repeat Containing |
| ENSMUSG00000050379 | ENSFM00350000105408 | Septin |
| ENSMUSG00000050390 | ENSFM00250000008611 | Uncharacterized |
| ENSMUSG00000050390 | ENSFM00570000882097 | Unknown |
| ENSMUSG00000050390 | ENSFM00570000887662 | Unknown |
| ENSMUSG00000050490 | ENSFM00500000270276 | Proteasome Subunit Alpha Type 5 EC_3.4.25.1 Macropain Multicatalytic Endopeptidase Complex Proteasome |
| ENSMUSG00000050530 | ENSFM00740001589308 | Precursor |
| ENSMUSG00000050552 | ENSFM00670001237240 | Ragulator Complex LAMTOR4 Late Endosomal/Lysosomal Adaptor And Mapk And Mtor Activator 4 |
| ENSMUSG00000050556 | ENSFM00270000056440 | Potassium Voltage Gated Channel Subfamily Member Voltage Gated Potassium Channel Subunit |
| ENSMUSG00000050587 | ENSFM00610000973611 | Unknown |
| ENSMUSG00000050587 | ENSFM00260000050662 | Leucine Rich Repeat Containing Precursor Netrin Ligand Ngl |
| ENSMUSG00000050621 | ENSFM00670001235597 | 40S Ribosomal S27 |
| ENSMUSG00000050627 | ENSFM00800002003402 | Glycerol 3 Phosphate Dehydrogenase Cytoplasmic Gpd C Gpdh C EC_1.1.1.8 |
| ENSMUSG00000050663 | ENSFM00250000000130 | Aminopeptidase N Ap N EC_3.4.11.2 Microsomal Aminopeptidase |
| ENSMUSG00000050697 | ENSFM00760001714720 | 5' Amp Activated Kinase Catalytic Subunit Alpha 1 Ampk Subunit Alpha 1 EC_2.7.11.1 Acetyl Coa Carboxylase Kinase Acaca Kinase EC_2.7.11.- 27 Hydroxymethylglutaryl Coa Reductase Kinase Hmgcr Kinase EC_2.7.-.- 11 31 Tau Kinase PRKAA1 Ec 2.7.11.26 |
| ENSMUSG00000050705 | ENSFM00500000272346 | Uncharacterized |
| ENSMUSG00000050705 | ENSFM00570000890740 | Unknown |
| ENSMUSG00000050711 | ENSFM00250000004240 | Secretogranin 2 Precursor Secretogranin Ii Sgii |
| ENSMUSG00000050732 | ENSFM00670001236743 | Vesicle Associated Membrane 8 Vamp 8 |
| ENSMUSG00000050812 | ENSFM00250000003543 | Proteasome ECM29 |
| ENSMUSG00000050840 | ENSFM00260000050333 | Cadherin Precursor |
| ENSMUSG00000050856 | ENSFM00500000427096 | Unknown |
| ENSMUSG00000050856 | ENSFM00520000547368 | Unknown |
| ENSMUSG00000050875 | ENSFM00600000921820 | Unknown |
| ENSMUSG00000050875 | ENSFM00660001160931 | Unknown |
| ENSMUSG00000050890 | ENSFM00250000003729 | Serine/Threonine Kinase EC_2.7.11.1 PDLIM1 Interacting Kinase 1 |
| ENSMUSG00000050896 | ENSFM00250000002242 | Reticulon 4 Receptor Precursor Nogo Receptor Nogo 66 Receptor Homolog Nogo 66 Receptor Related |
| ENSMUSG00000050947 | ENSFM00380000124725 | Amphoterin Induced Precursor Amigo Alivin |
| ENSMUSG00000050953 | ENSFM00760001714761 | Gap Junction Alpha 1 Connexin 43 CX43 |
| ENSMUSG00000050965 | ENSFM00520000517802 | Kinase C Type Pkc EC_2.7.11.13 |
| ENSMUSG00000051007 | ENSFM00250000005901 | Parkinson Disease 7 Domain Containing 1 Precursor |
| ENSMUSG00000051065 | ENSFM00250000008630 | M Mab 21 Domain Containing 2 |
| ENSMUSG00000051067 | ENSFM00250000001884 | Leucine Rich Repeat And Immunoglobulin Domain Containing Nogo Receptor Interacting Precursor Leucine Rich Repeat Neuronal |
| ENSMUSG00000051098 | ENSFM00250000007523 | Metallo Beta Lactamase Domain Containing 2 Ec 3 |
| ENSMUSG00000051111 | ENSFM00250000000744 | Synaptic Vesicle Glycoprotein |
| ENSMUSG00000051154 | ENSFM00250000008895 | Comm Domain Containing 3 |
| ENSMUSG00000051177 | ENSFM00810002057518 | Unknown |
| ENSMUSG00000051177 | ENSFM00800002003424 | 1 Phosphatidylinositol 4 5 Bisphosphate Phosphodiesterase Beta EC_3.1.4.11 Phosphoinositide Phospholipase C Beta Phospholipase C Beta Plc Beta |
| ENSMUSG00000051242 | ENSFM00730001521419 | Protocadherin Beta Precursor Pcdh Beta |
| ENSMUSG00000051243 | ENSFM00260000051182 | Immunoglobulin Superfamily Containing Leucine Rich Repeat Precursor |
| ENSMUSG00000051323 | ENSFM00760001762688 | Unknown |
| ENSMUSG00000051323 | ENSFM00780001898335 | Protocadherin Precursor |
| ENSMUSG00000051331 | ENSFM00500000269632 | Voltage Dependent L Type Calcium Channel Subunit Alpha Calcium Channel L Type Alpha 1 Polypeptide Voltage Gated Calcium Channel Subunit Alpha CAV1 |
| ENSMUSG00000051343 | ENSFM00500000272387 | RAB11 Family Interacting 5 RAB11 FIP5 RAB11 Interacting RIP11 |
| ENSMUSG00000051355 | ENSFM00590000916064 | Unknown |
| ENSMUSG00000051355 | ENSFM00250000007033 | Comm Domain Containing 1 |
| ENSMUSG00000051359 | ENSFM00760001714608 | Unknown |
| ENSMUSG00000051375 | ENSFM00780001917497 | Unknown |
| ENSMUSG00000051375 | ENSFM00780001917498 | Unknown |
| ENSMUSG00000051375 | ENSFM00760001714757 | Protocadherin Precursor Protocadherin |
| ENSMUSG00000051379 | ENSFM00250000003081 | Leucine Rich Repeat Transmembrane Precursor Fibronectin Domain Containing Leucine Rich Transmembrane |
| ENSMUSG00000051391 | ENSFM00730001521154 | Unknown |
| ENSMUSG00000051401 | ENSFM00260000050545 | Btb/Poz Domain Containing |
| ENSMUSG00000051412 | ENSFM00250000005040 | Vesicle Associated Membrane 7 Vamp 7 Synaptobrevin 1 |
| ENSMUSG00000051483 | ENSFM00250000002301 | Carbonyl Reductase EC_1.1.1.- 197 20 Beta Hydroxysteroid Dehydrogenase Nadph Dependent Carbonyl Reductase 1 Prostaglandin 9 Ketoreductase Prostaglandin E 2 9 Reductase EC_1.1.-.- 1 189 |
| ENSMUSG00000051615 | ENSFM00800002003504 | Ras Related Rap Precursor |
| ENSMUSG00000051675 | ENSFM00250000008223 | E3 Ubiquitin Ligase TRIM32 EC_6.3.2.- Tripartite Motif Containing 32 |
| ENSMUSG00000051695 | ENSFM00250000000747 | Poly Rc Binding Alpha Heterogeneous Nuclear Ribonucleoprotein Hnrnp |
| ENSMUSG00000051747 | ENSFM00670001267275 | Uncharacterized |
| ENSMUSG00000051747 | ENSFM00570000894573 | Unknown |
| ENSMUSG00000051747 | ENSFM00740001589077 | Titin EC_2.7.11.1 Connectin |
| ENSMUSG00000051790 | ENSFM00380000124679 | Neuroligin Precursor |
| ENSMUSG00000051839 | ENSFM00260000053088 | Glycophorin A CD235A Antigen |
| ENSMUSG00000051934 | ENSFM00520000517852 | Spermatogenesis Associated Serine Rich 2 Serine Rich Spermatocytes And Round Spermatid 59 Kda P59SCR |
| ENSMUSG00000051951 | ENSFM00500000269993 | Xk Related |
| ENSMUSG00000052026 | ENSFM00730001521205 | Sodium And Chloride Dependent Glycine Transporter Glyt Solute Carrier Family 6 Member |
| ENSMUSG00000052033 | ENSFM00610000954462 | Unknown |
| ENSMUSG00000052033 | ENSFM00610000955092 | Unknown |
| ENSMUSG00000052033 | ENSFM00610000962596 | Unknown |
| ENSMUSG00000052033 | ENSFM00610000973625 | Unknown |
| ENSMUSG00000052033 | ENSFM00500000273754 | Prefoldin Subunit 4 |
| ENSMUSG00000052087 | ENSFM00570000895016 | Unknown |
| ENSMUSG00000052087 | ENSFM00250000000973 | Regulator Of G Signaling |
| ENSMUSG00000052146 | ENSFM00670001235534 | 40S Ribosomal S10 |
| ENSMUSG00000052214 | ENSFM00600000921688 | Optic Atrophy 3 |
| ENSMUSG00000052298 | ENSFM00570000885284 | Unknown |
| ENSMUSG00000052298 | ENSFM00670001236658 | CDC42 Small Effector 2 |
| ENSMUSG00000052337 | ENSFM00250000002778 | Micos Complex Subunit MIC60 Precursor Mitochondrial Inner Membrane Mitofilin |
| ENSMUSG00000052372 | ENSFM00500000270404 | Interleukin 1 Receptor Accessory 1 Precursor Il 1 Rapl 1 Il 1RAPL 1 IL1RAPL 1 X Linked Interleukin 1 Receptor Accessory 1 |
| ENSMUSG00000052373 | ENSFM00760001714665 | Maguk P55 Subfamily Member |
| ENSMUSG00000052374 | ENSFM00440000236866 | Alpha Actinin Alpha Actinin F Actin Cross Linking |
| ENSMUSG00000052387 | ENSFM00730001521288 | Transient Receptor Potential Cation Channel Subfamily M Member Long Transient Receptor Potential Channel |
| ENSMUSG00000052397 | ENSFM00250000000517 | Moesin |
| ENSMUSG00000052428 | ENSFM00780001909040 | Unknown |
| ENSMUSG00000052428 | ENSFM00250000005494 | Transmembrane And Coiled Coil 1 |
| ENSMUSG00000052429 | ENSFM00570000898596 | Unknown |
| ENSMUSG00000052429 | ENSFM00730001521302 | Arginine N Methyltransferase 1 EC_2.1.1.- Histone Arginine N Methyltransferase PRMT1 EC_2.1.1.125 |
| ENSMUSG00000052456 | ENSFM00250000003927 | Atpase GET3 |
| ENSMUSG00000052459 | ENSFM00570000885853 | Unknown |
| ENSMUSG00000052459 | ENSFM00250000002169 | V Type Proton Atpase Catalytic Subunit A V Atpase Subunit A EC_3.6.3.14 V Atpase 69 Kda Subunit Vacuolar Proton Pump Subunit Alpha |
| ENSMUSG00000052516 | ENSFM00740001589121 | Roundabout Homolog Precursor |
| ENSMUSG00000052539 | ENSFM00250000000552 | Membrane Associated Guanylate Kinase Ww And Pdz Domain Containing Membrane Associated Guanylate Kinase Inverted Magi |
| ENSMUSG00000052572 | ENSFM00250000000275 | Disks Large Homolog Synapse Associated Sap |
| ENSMUSG00000052581 | ENSFM00570000896064 | Unknown |
| ENSMUSG00000052581 | ENSFM00400000131776 | Leucine Rich Repeat Transmembrane Neuronal Precursor |
| ENSMUSG00000052681 | ENSFM00730001521249 | Ras Related Rap 1 |
| ENSMUSG00000052698 | ENSFM00250000001103 | Talin |
| ENSMUSG00000052727 | ENSFM00250000000942 | Microtubule Associated Map |
| ENSMUSG00000052738 | ENSFM00640001118348 | Unknown |
| ENSMUSG00000052738 | ENSFM00250000003768 | Succinyl Coa Ligase Subunit Alpha Mitochondrial Precursor EC_6.2.1.4 EC_6.2.1.- 5 Succinyl Coa Synthetase Subunit Alpha Scs Alpha |
| ENSMUSG00000052852 | ENSFM00500000269981 | Receptor Expression Enhancing |
| ENSMUSG00000052889 | ENSFM00520000517802 | Kinase C Type Pkc EC_2.7.11.13 |
| ENSMUSG00000052981 | ENSFM00760001715179 | Ubiquitin Conjugating Enzyme E2Q EC_6.3.2.19 |
| ENSMUSG00000053024 | ENSFM00760001763137 | Unknown |
| ENSMUSG00000053024 | ENSFM00760001763138 | Unknown |
| ENSMUSG00000053024 | ENSFM00800002003683 | Contactin 1 Precursor Neural Cell Surface F3 |
| ENSMUSG00000053025 | ENSFM00250000000744 | Synaptic Vesicle Glycoprotein |
| ENSMUSG00000053046 | ENSFM00640001118862 | Unknown |
| ENSMUSG00000053046 | ENSFM00740001589247 | Serine/Threonine Kinase BRSK2 EC_2.7.11.1 EC_2.7.11.- 26 Brain Specific Serine/Threonine Kinase 2 Br Serine/Threonine Kinase 2 Serine/Threonine Kinase Sad A |
| ENSMUSG00000053062 | ENSFM00500000272329 | Junctional Adhesion Molecule B Precursor Jam B Junctional Adhesion Molecule 2 Jam 2 Vascular Endothelial Junction Associated Molecule Ve Jam CD322 Antigen |
| ENSMUSG00000053119 | ENSFM00500000271306 | Charged Multivesicular Body 3 Chromatin Modifying 3 Vacuolar Sorting Associated 24 |
| ENSMUSG00000053141 | ENSFM00310000088999 | Receptor Type Tyrosine Phosphatase Precursor R Ptp EC_3.1.3.48 Receptor Type Tyrosine Phosphatase |
| ENSMUSG00000053253 | ENSFM00600000921294 | NEDD4 Family Interacting NEDD4 Ww Domain Binding |
| ENSMUSG00000053279 | ENSFM00730001521185 | Dehydrogenase Aldehyde Dehydrogenase Family 1 Member Dehydrogenase |
| ENSMUSG00000053289 | ENSFM00730001521664 | Atp Dependent Rna Helicase EC_3.6.4.13 |
| ENSMUSG00000053291 | ENSFM00800002003435 | Ras Related Rab |
| ENSMUSG00000053317 | ENSFM00570000900708 | Unknown |
| ENSMUSG00000053317 | ENSFM00670001236538 | Transport SEC61 Subunit Beta |
| ENSMUSG00000053329 | ENSFM00250000005161 | ES1 Mitochondrial Precursor |
| ENSMUSG00000053395 | ENSFM00250000002019 | Voltage Dependent Calcium Channel Gamma Subunit Neuronal Voltage Gated Calcium Channel Gamma Subunit Transmembrane Ampar Regulatory Gamma Tarp Gamma |
| ENSMUSG00000053398 | ENSFM00500000270390 | D 3 Phosphoglycerate Dehydrogenase 3 Pgdh EC_1.1.1.95 |
| ENSMUSG00000053519 | ENSFM00410000138464 | Kv Channel Interacting A Type Potassium Channel Modulatory Potassium Channel Interacting |
| ENSMUSG00000053550 | ENSFM00500000276971 | Shisa 7 Precursor Shisa 6 |
| ENSMUSG00000053580 | ENSFM00610000973561 | Unknown |
| ENSMUSG00000053580 | ENSFM00250000001143 | Tetratricopeptide Repeat Ankyrin Repeat And Coiled Coil Domain Containing |
| ENSMUSG00000053580 | ENSFM00670001247752 | TANC2 |
| ENSMUSG00000053617 | ENSFM00740001589343 | SH3 And Px Domain Containing 2A Five SH3 Domain Containing SH3 Multiple Domains 1 Tyrosine Kinase Substrate With Five SH3 Domains |
| ENSMUSG00000053644 | ENSFM00420000140646 | Alpha Aminoadipic Semialdehyde Dehydrogenase Precursor Alpha Aasa Dehydrogenase EC_1.2.1.31 Aldehyde Dehydrogenase Family 7 Member A1 EC_1.2.1.- 3 Antiquitin 1 Betaine Aldehyde Dehydrogenase EC_1.2.-.- 1 8 DELTA1 Piperideine 6 Carboxylate Dehydrogenase P6 |
| ENSMUSG00000053646 | ENSFM00730001521536 | Plexin Precursor |
| ENSMUSG00000053693 | ENSFM00250000000518 | Microtubule Associated Serine/Threonine Kinase EC_2.7.11.1 |
| ENSMUSG00000053702 | ENSFM00500000270806 | Lim And SH3 Domain 1 Lasp 1 |
| ENSMUSG00000053702 | ENSFM00740001589165 | Nebulin Related Anchoring N Rap |
| ENSMUSG00000053768 | ENSFM00500000272284 | Micos Complex Subunit MIC19 Coiled Coil Helix Coiled Coil Helix Domain Containing 3 |
| ENSMUSG00000053799 | ENSFM00250000001326 | Exocyst Complex Component 6 Exocyst Complex Component SEC15A SEC15 1 |
| ENSMUSG00000053819 | ENSFM00250000000111 | Calcium/Calmodulin Dependent Kinase Type Ii Subunit Cam Kinase Ii Subunit Camk Ii Subunit EC_2.7.11.17 |
| ENSMUSG00000053825 | ENSFM00250000000441 | Liprin Alpha Tyrosine Phosphatase Receptor Type F Polypeptide Interacting Alpha Ptprf Interacting Alpha |
| ENSMUSG00000053898 | ENSFM00260000050929 | Delta 3 5 Delta 2 4 Dienoyl Coa Isomerase Mitochondrial Precursor EC_5.3.3.- |
| ENSMUSG00000053907 | ENSFM00810002040570 | S Adenosylmethionine Synthase Adomet Synthase EC_2.5.1.6 Methionine Adenosyltransferase Mat |
| ENSMUSG00000053930 | ENSFM00500000272417 | Shisa 6 Homolog Precursor |
| ENSMUSG00000053931 | ENSFM00800002024498 | Unknown |
| ENSMUSG00000053931 | ENSFM00800002024499 | Unknown |
| ENSMUSG00000053931 | ENSFM00500000269939 | Calponin Calponin Smooth Muscle |
| ENSMUSG00000053963 | ENSFM00500000275444 | Stum Homolog |
| ENSMUSG00000054027 | ENSFM00250000001969 | 5' Nucleotidase Domain Containing EC_3.1.3.- |
| ENSMUSG00000054091 | ENSFM00500000446102 | Unknown |
| ENSMUSG00000054099 | ENSFM00800002023532 | Unknown |
| ENSMUSG00000054099 | ENSFM00250000002826 | Solute Carrier Family 25 Member |
| ENSMUSG00000054252 | ENSFM00570000898156 | Unknown |
| ENSMUSG00000054252 | ENSFM00250000000093 | Fibroblast Growth Factor Receptor Precursor Fgfr EC_2.7.10.1 |
| ENSMUSG00000054277 | ENSFM00250000002206 | Adp Ribosylation Factor Gtpase Activating 2 Arf Gap 2 Zinc Finger 289 |
| ENSMUSG00000054312 | ENSFM00570000884963 | Unknown |
| ENSMUSG00000054312 | ENSFM00670001236631 | 28S Ribosomal S21 Mitochondrial Mrp S21 S21MT |
| ENSMUSG00000054364 | ENSFM00730001521223 | Rho Related Gtp Binding Precursor |
| ENSMUSG00000054423 | ENSFM00250000000780 | Calcium Dependent Secretion Activator 1 Calcium Dependent Activator Secretion 1 Caps 1 |
| ENSMUSG00000054453 | ENSFM00730001522102 | Synaptotagmin 4 Exophilin 2 Granuphilin |
| ENSMUSG00000054455 | ENSFM00250000001879 | Vesicle Associated Membrane Associated A Vamp A Vamp Associated A Vap A |
| ENSMUSG00000054459 | ENSFM00760001714608 | Unknown |
| ENSMUSG00000054469 | ENSFM00570000878308 | Unknown |
| ENSMUSG00000054469 | ENSFM00500000270992 | Lysocardiolipin Acyltransferase 1 EC_2.3.1.- EC_2.3.1.51 Acyl Coa:Lysocardiolipin Acyltransferase 1 |
| ENSMUSG00000054477 | ENSFM00250000000796 | Small Conductance Calcium Activated Potassium Channel Skca KCA2 |
| ENSMUSG00000054619 | ENSFM00250000004491 | Methyltransferase Precursor EC_2.1.1.- |
| ENSMUSG00000054640 | ENSFM00710001441664 | Sodium/Calcium Exchanger 1 Precursor Na + /Ca 2+ Exchange 1 Solute Carrier Family 8 Member 1 |
| ENSMUSG00000054693 | ENSFM00390000126345 | Disintegrin And Metalloproteinase Domain Containing 10 Precursor Adam 10 EC_3.4.24.81 Kuzbanian Homolog Mammalian Disintegrin Metalloprotease CD156C Antigen |
| ENSMUSG00000054720 | ENSFM00570000878992 | Unknown |
| ENSMUSG00000054720 | ENSFM00250000001418 | Volume Regulated Anion Channel Subunit Leucine Rich Repeat Containing |
| ENSMUSG00000054728 | ENSFM00760001714738 | Phosphatase And Actin Regulator |
| ENSMUSG00000054733 | ENSFM00500000271413 | Mitochondrial Peptide Methionine Sulfoxide Reductase Precursor EC_1.8.4.11 Peptide Methionine S S Oxide Reductase Peptide Met O Reductase Methionine S Oxide Reductase |
| ENSMUSG00000054793 | ENSFM00250000001126 | Cell Adhesion Molecule Precursor Immunoglobulin Superfamily Member Nectin Necl Synaptic Cell Adhesion Molecule |
| ENSMUSG00000054808 | ENSFM00440000236866 | Alpha Actinin Alpha Actinin F Actin Cross Linking |
| ENSMUSG00000054814 | ENSFM00810002057450 | Unknown |
| ENSMUSG00000054814 | ENSFM00760001714668 | Ubiquitin Carboxyl Terminal Hydrolase 12 EC_3.4.19.12 Deubiquitinating Enzyme 12 Ubiquitin Thioesterase 12 Ubiquitin Specific Processing Protease 12 |
| ENSMUSG00000054874 | ENSFM00250000001163 | Pecanex |
| ENSMUSG00000054901 | ENSFM00250000009469 | Rho Guanine Nucleotide Exchange Factor 33 |
| ENSMUSG00000054942 | ENSFM00250000002334 | FAM73B |
| ENSMUSG00000054976 | ENSFM00250000005153 | Neuronal Tyrosine Phosphorylated Phosphoinositide 3 Kinase Adapter 2 |
| ENSMUSG00000055003 | ENSFM00570000880212 | Unknown |
| ENSMUSG00000055003 | ENSFM00670001235765 | Leucine Rich Repeat And Transmembrane Domain Containing Precursor |
| ENSMUSG00000055013 | ENSFM00250000000604 | Arf Gap With Gtpase Ank Repeat And Ph Domain Containing Agap Centaurin Gamma |
| ENSMUSG00000055022 | ENSFM00800002003683 | Contactin 1 Precursor Neural Cell Surface F3 |
| ENSMUSG00000055065 | ENSFM00420000140507 | Probable Atp Dependent Rna Helicase DDX5 EC_3.6.4.13 Dead Box 5 |
| ENSMUSG00000055069 | ENSFM00500000270441 | Ras Related Rab |
| ENSMUSG00000055194 | ENSFM00730001521060 | Actin |
| ENSMUSG00000055254 | ENSFM00730001521357 | Nt 3 Growth Factor Receptor Precursor EC_2.7.10.1 Neurotrophic Tyrosine Kinase Receptor Type 3 |
| ENSMUSG00000055333 | ENSFM00570000851039 | Protocadherin Fat Precursor Fat Tumor Suppressor Homolog |
| ENSMUSG00000055407 | ENSFM00250000004498 | Microtubule Associated 6 Map 6 Stable Tubule Only Polypeptide Stop |
| ENSMUSG00000055421 | ENSFM00780001908951 | Unknown |
| ENSMUSG00000055421 | ENSFM00760001714657 | Protocadherin 11 X Linked Precursor Protocadherin 11 Protocadherin On The X Chromosome Pcdh X |
| ENSMUSG00000055447 | ENSFM00250000005008 | Leukocyte Surface Antigen CD47 Precursor Integrin Associated Iap CD47 Antigen |
| ENSMUSG00000055531 | ENSFM00660001169823 | Unknown |
| ENSMUSG00000055531 | ENSFM00660001189768 | Unknown |
| ENSMUSG00000055531 | ENSFM00600000921459 | Cleavage And Polyadenylation Specificity Factor Subunit 6 |
| ENSMUSG00000055567 | ENSFM00470000251461 | Unc 80 |
| ENSMUSG00000055723 | ENSFM00770001847543 | Ras Related R Ras |
| ENSMUSG00000055762 | ENSFM00730001521722 | Elongation Factor 1 Delta Ef 1 Delta |
| ENSMUSG00000055782 | ENSFM00800002003396 | Atp Binding Cassette Sub Family D Member Adrenoleukodystrophy |
| ENSMUSG00000055805 | ENSFM00350000105407 | Formin |
| ENSMUSG00000055839 | ENSFM00500000273664 | Transcription Elongation Factor B Polypeptide 2 Elongin 18 Kda Subunit Elongin B Elob Rna Polymerase Ii Transcription Factor Siii Subunit B Siii P18 |
| ENSMUSG00000055943 | ENSFM00500000273154 | Er Membrane Complex Subunit 7 Precursor |
| ENSMUSG00000055963 | ENSFM00810002042668 | Triple Qxxk/R Motif Containing Triple Repetitive Sequence Of Qxxk/R Homolog |
| ENSMUSG00000056004 | ENSFM00570000891130 | Unknown |
| ENSMUSG00000056004 | ENSFM00250000001885 | Precursor Estrogen Induced Gene 121 |
| ENSMUSG00000056050 | ENSFM00800002008628 | Unknown |
| ENSMUSG00000056050 | ENSFM00500000270934 | Melanoma Inhibitory Activity 3 Precursor Transport And Golgi Organization 1 |
| ENSMUSG00000056073 | ENSFM00720001492013 | Glutamate Receptor Ionotropic Kainate Precursor Glutamate Receptor |
| ENSMUSG00000056158 | ENSFM00400000131811 | Carbonic Anhydrase Related 11 Precursor Ca Rp Xi Ca Xi Carp Xi |
| ENSMUSG00000056158 | ENSFM00500000308824 | Carbonic Anhydrase Related 10 |
| ENSMUSG00000056201 | ENSFM00500000270025 | Cofilin Cofilin Muscle |
| ENSMUSG00000056209 | ENSFM00500000274611 | Nucleoplasmin 3 |
| ENSMUSG00000056222 | ENSFM00250000001454 | Testican Precursor Sparc/Osteonectin Cwcv And Kazal Domains Proteoglycan |
| ENSMUSG00000056367 | ENSFM00800002003421 | Actin Related 3 Actin 3 |
| ENSMUSG00000056413 | ENSFM00250000002436 | Arf Gap With Dual Ph Domain Containing 2 Centaurin Alpha 2 Cnt A2 |
| ENSMUSG00000056429 | ENSFM00500000276364 | Trans Golgi Network Integral Membrane Precursor |
| ENSMUSG00000056515 | ENSFM00730001522125 | Ras Related Rab |
| ENSMUSG00000056553 | ENSFM00260000050522 | Receptor Type Tyrosine Phosphatase Precursor R Ptp |
| ENSMUSG00000056602 | ENSFM00250000000754 | Furry |
| ENSMUSG00000056629 | ENSFM00500000273014 | Peptidyl Prolyl Cis Trans Isomerase FKBP2 Precursor Ppiase FKBP2 EC_5.2.1.8.13 Kda FK506 Binding 13 Kda Fkbp Fkbp 13 FK506 Binding 2 Fkbp 2 Immunophilin FKBP13 Rotamase |
| ENSMUSG00000056665 | ENSFM00560000771357 | THEM6 Precursor |
| ENSMUSG00000056755 | ENSFM00640001118416 | Unknown |
| ENSMUSG00000056755 | ENSFM00640001118417 | Unknown |
| ENSMUSG00000056755 | ENSFM00250000000328 | Metabotropic Glutamate Receptor Precursor |
| ENSMUSG00000056812 | ENSFM00800002003921 | Sia Alpha 2 3 Gal Beta 1 4 Glcnac R:Alpha 2 8 Sialyltransferase EC_2.4.99.- Alpha 2 8 Sialyltransferase 8C Alpha 2 8 Sialyltransferase Iii ST8 Alpha N Acetyl Neuraminide Alpha 2 8 Sialyltransferase 3 Sialyltransferase 8C SIAT8 C Sialyltransferase ST8SIA I |
| ENSMUSG00000056851 | ENSFM00250000000747 | Poly Rc Binding Alpha Heterogeneous Nuclear Ribonucleoprotein Hnrnp |
| ENSMUSG00000056856 | ENSFM00610000971715 | Unknown |
| ENSMUSG00000056856 | ENSFM00250000001480 | Janus Kinase And Microtubule Interacting 1 Multiple Alpha Helices And Rna Linker 1 Marlin 1 |
| ENSMUSG00000056966 | ENSFM00500000274805 | Gap Junction Gamma 3 Gap Junction Epsilon 1 |
| ENSMUSG00000057113 | ENSFM00500000270251 | Nucleophosmin Npm Nucleolar Phosphoprotein B23 Nucleolar NO38 Numatrin |
| ENSMUSG00000057177 | ENSFM00810002040612 | Glycogen Synthase Kinase 3 Gsk 3 EC_2.7.11.26 Serine/Threonine Kinase EC_2.7.11.- 1 |
| ENSMUSG00000057182 | ENSFM00740001589070 | Sodium Channel Type Subunit Alpha Sodium Channel Sodium Channel Type Subunit Alpha Voltage Gated Sodium Channel Subunit Alpha NAV1 |
| ENSMUSG00000057193 | ENSFM00500000269830 | Choline Transporter Solute Carrier Family 44 Member |
| ENSMUSG00000057230 | ENSFM00730001521956 | AP2 Associated Kinase 1 EC_2.7.11.1 Adaptor Associated Kinase 1 |
| ENSMUSG00000057262 | ENSFM00250000001504 | 60S Ribosomal L15 |
| ENSMUSG00000057322 | ENSFM00500000287342 | Unknown |
| ENSMUSG00000057335 | ENSFM00740001589275 | Unknown |
| ENSMUSG00000057378 | ENSFM00250000000332 | Ryanodine Receptor Ryr Muscle Calcium Release Channel Muscle Type Ryanodine Receptor Type Ryanodine Receptor |
| ENSMUSG00000057497 | ENSFM00500000273742 | Unknown |
| ENSMUSG00000057614 | ENSFM00410000138458 | Guanine Nucleotide Binding Subunit |
| ENSMUSG00000057666 | ENSFM00720001501961 | Unknown |
| ENSMUSG00000057666 | ENSFM00720001503163 | Unknown |
| ENSMUSG00000057666 | ENSFM00730001521081 | Glyceraldehyde 3 Phosphate Dehydrogenase Gapdh EC_1.2.1.12 Peptidyl Cysteine S Nitrosylase Gapdh EC_2.6.99.- |
| ENSMUSG00000057716 | ENSFM00800002003927 | Transmembrane 178B Precursor |
| ENSMUSG00000057738 | ENSFM00740001589109 | Spectrin Alpha Chain Non Erythrocytic 1 Alpha Ii Spectrin Fodrin Alpha Chain |
| ENSMUSG00000057841 | ENSFM00500000270098 | 60S Ribosomal L32 |
| ENSMUSG00000057863 | ENSFM00670001235763 | 60S Ribosomal L36 |
| ENSMUSG00000057880 | ENSFM00810002040728 | 4 Aminobutyrate Aminotransferase Mitochondrial Precursor EC_2.6.1.19 S 3 Amino 2 Methylpropionate Transaminase EC_2.6.1.- 22 Gaba Aminotransferase Gaba At Gamma Amino N Butyrate Transaminase Gaba Transaminase Gaba T L Aibat |
| ENSMUSG00000057897 | ENSFM00250000000111 | Calcium/Calmodulin Dependent Kinase Type Ii Subunit Cam Kinase Ii Subunit Camk Ii Subunit EC_2.7.11.17 |
| ENSMUSG00000057914 | ENSFM00780001917886 | Unknown |
| ENSMUSG00000057914 | ENSFM00250000000524 | Voltage Dependent L Type Calcium Channel Subunit Beta Calcium Channel Voltage Dependent Subunit Beta |
| ENSMUSG00000058013 | ENSFM00350000105408 | Septin |
| ENSMUSG00000058064 | ENSFM00250000002087 | 60S Ribosomal L11 |
| ENSMUSG00000058070 | ENSFM00520000517805 | Echinoderm Microtubule Associated Emap |
| ENSMUSG00000058076 | ENSFM00570000876575 | Unknown |
| ENSMUSG00000058076 | ENSFM00500000272004 | Succinate Dehydrogenase Cytochrome B560 Subunit Mitochondrial Precursor Integral Membrane Cii 3 Qps 1 QPS1 Succinate Dehydrogenase Complex Subunit C |
| ENSMUSG00000058135 | ENSFM00500000269733 | Glutathione S Transferase EC_2.5.1.18 Gst Class Mu |
| ENSMUSG00000058153 | ENSFM00250000001049 | Seizure 6 Precursor |
| ENSMUSG00000058207 | ENSFM00250000000235 | Precursor |
| ENSMUSG00000058254 | ENSFM00500000270260 | Tetraspanin 7 Tspan 7 Transmembrane 4 Superfamily Member 2 CD231 Antigen |
| ENSMUSG00000058301 | ENSFM00500000270457 | Atp Dependent Helicase Nonsense 1 Up Frameshift Suppressor 1 |
| ENSMUSG00000058325 | ENSFM00760001714647 | Dedicator Of Cytokinesis |
| ENSMUSG00000058420 | ENSFM00500000272887 | Synaptotagmin 17 Synaptotagmin Xvii Sytxvii |
| ENSMUSG00000058441 | ENSFM00590000917448 | Unknown |
| ENSMUSG00000058441 | ENSFM00250000007933 | Pannexin 2 |
| ENSMUSG00000058443 | ENSFM00800002003415 | 60S Ribosomal L10 |
| ENSMUSG00000058546 | ENSFM00670001235314 | 60S Ribosomal |
| ENSMUSG00000058558 | ENSFM00670001235299 | 60S Ribosomal L5 |
| ENSMUSG00000058569 | ENSFM00500000270043 | Transmembrane EMP24 Domain Containing Precursor |
| ENSMUSG00000058587 | ENSFM00500000269813 | Tropomodulin Tropomodulin Tmod |
| ENSMUSG00000058589 | ENSFM00720001498764 | Unknown |
| ENSMUSG00000058589 | ENSFM00720001498765 | Unknown |
| ENSMUSG00000058589 | ENSFM00720001501008 | Unknown |
| ENSMUSG00000058589 | ENSFM00720001503484 | Unknown |
| ENSMUSG00000058589 | ENSFM00720001503485 | Unknown |
| ENSMUSG00000058589 | ENSFM00250000001010 | Ankyrin Repeat And Sterile Alpha Motif Domain Containing 1B Amyloid Beta Intracellular Domain Associated 1 Aida 1 E2A PBX1 Associated Eb 1 |
| ENSMUSG00000058600 | ENSFM00670001235478 | 60S Ribosomal L30 |
| ENSMUSG00000058624 | ENSFM00250000005049 | Guanine Deaminase Guanase Guanine Aminase EC_3.5.4.3 Guanine Aminohydrolase Gah |
| ENSMUSG00000058655 | ENSFM00500000270799 | Eukaryotic Translation Initiation Factor 4B Eif 4B |
| ENSMUSG00000058672 | ENSFM00250000000193 | Tubulin Beta Chain |
| ENSMUSG00000058715 | ENSFM00780001911544 | Unknown |
| ENSMUSG00000058715 | ENSFM00670001245974 | High Affinity Immunoglobulin Epsilon Receptor Subunit Gamma Precursor Fc Receptor Gamma Chain Fcrgamma Fc Epsilon Ri Gamma Ige Fc Receptor Subunit Gamma Fceri Gamma |
| ENSMUSG00000058793 | ENSFM00660001190026 | Unknown |
| ENSMUSG00000058793 | ENSFM00660001190027 | Unknown |
| ENSMUSG00000058793 | ENSFM00750001632370 | Phosphatidate Cytidylyltransferase 2 EC_2.7.7.41 Cdp Dag Synthase 2 Cdp Dg Synthase 2 Cdp Diacylglycerol Synthase 2 Cds 2 Cdp Diglyceride Pyrophosphorylase 2 Cdp Diglyceride Synthase 2 Ctp:Phosphatidate Cytidylyltransferase 2 |
| ENSMUSG00000058799 | ENSFM00500000269862 | Nucleosome Assembly 1 |
| ENSMUSG00000058835 | ENSFM00250000000845 | Abl Interactor 1 Abelson Interactor 1 Abi 1 EPS8 SH3 Domain Binding EPS8 Binding Spectrin SH3 Domain Binding 1 |
| ENSMUSG00000058897 | ENSFM00500000271217 | Collagen Alpha 1 Xxv Chain Clac P |
| ENSMUSG00000058927 | ENSFM00670001235376 | Cytochrome C |
| ENSMUSG00000058966 | ENSFM00250000003740 | FAM57B |
| ENSMUSG00000058975 | ENSFM00260000050384 | Potassium Voltage Gated Channel Subfamily C Member Voltage Gated Potassium Channel KV3 |
| ENSMUSG00000058997 | ENSFM00580000902189 | Von Willebrand Factor A Domain Containing 8 |
| ENSMUSG00000059003 | ENSFM00730001521500 | Glutamate Receptor Ionotropic Nmda Precursor Glutamate Receptor Subunit Epsilon N Methyl D Aspartate Receptor Subtype |
| ENSMUSG00000059005 | ENSFM00270000056431 | Heterogeneous Nuclear Ribonucleoprotein |
| ENSMUSG00000059040 | ENSFM00730001521170 | Enolase EC_4.2.1.11 2 Phospho D Glycerate Hydro Lyase |
| ENSMUSG00000059070 | ENSFM00640001117531 | Unknown |
| ENSMUSG00000059070 | ENSFM00640001118658 | Unknown |
| ENSMUSG00000059070 | ENSFM00810002040668 | 60S Ribosomal L18 |
| ENSMUSG00000059119 | ENSFM00500000269862 | Nucleosome Assembly 1 |
| ENSMUSG00000059146 | ENSFM00730001521357 | Nt 3 Growth Factor Receptor Precursor EC_2.7.10.1 Neurotrophic Tyrosine Kinase Receptor Type 3 |
| ENSMUSG00000059173 | ENSFM00400000131765 | Calcium/Calmodulin Dependent 3' 5' Cyclic Nucleotide Phosphodiesterase Cam Pde EC_3.1.4.17 Kda Cam Pde |
| ENSMUSG00000059208 | ENSFM00570000887653 | Unknown |
| ENSMUSG00000059208 | ENSFM00740001589263 | Myelin Expression Factor 2 Mef 2 Myef 2 |
| ENSMUSG00000059213 | ENSFM00250000009854 | Dendrin |
| ENSMUSG00000059248 | ENSFM00570000901008 | Unknown |
| ENSMUSG00000059248 | ENSFM00500000269805 | Septin |
| ENSMUSG00000059291 | ENSFM00250000002087 | 60S Ribosomal L11 |
| ENSMUSG00000059316 | ENSFM00730001521827 | Long Chain Fatty Acid Transport 4 Fatp 4 Fatty Acid Transport 4 EC_6.2.1.- Solute Carrier Family 27 Member 4 |
| ENSMUSG00000059323 | ENSFM00610000967091 | Unknown |
| ENSMUSG00000059323 | ENSFM00610000967178 | Unknown |
| ENSMUSG00000059323 | ENSFM00610000973122 | Unknown |
| ENSMUSG00000059323 | ENSFM00250000004206 | Tonsoku Inhibitor Of Kappa B Related I Kappa B Related Ikappabr Nf Kappa B Inhibitor 2 Nuclear Factor Of Kappa Light Polypeptide Gene Enhancer In B Cells Inhibitor 2 |
| ENSMUSG00000059323 | ENSFM00500000271517 | Vacuolar Sorting Associated 28 Homolog Escrt I Complex Subunit VPS28 |
| ENSMUSG00000059336 | ENSFM00250000001770 | Urea Transporter 1 Solute Carrier Family 14 Member 1 Urea Transporter B Ut B Urea Transporter Erythrocyte |
| ENSMUSG00000059409 | ENSFM00810002040632 | Serine/Threonine Phosphatase 2A 56 Kda Regulatory Subunit Gamma PP2A B Subunit B' PP2A B Subunit B' Gamma PP2A B Subunit B56 Gamma PP2A B Subunit PR61 Gamma PP2A B Subunit R5 Gamma |
| ENSMUSG00000059439 | ENSFM00810002045978 | Unknown |
| ENSMUSG00000059439 | ENSFM00250000002665 | Breast Carcinoma Amplified Sequence 3 |
| ENSMUSG00000059447 | ENSFM00800002023576 | Unknown |
| ENSMUSG00000059447 | ENSFM00440000236928 | Trifunctional Enzyme Subunit Beta Mitochondrial Precursor Tp Beta |
| ENSMUSG00000059495 | ENSFM00600000921196 | Rho Guanine Nucleotide Exchange Factor |
| ENSMUSG00000059534 | ENSFM00500000442182 | Unknown |
| ENSMUSG00000059554 | ENSFM00640001110165 | Unknown |
| ENSMUSG00000059554 | ENSFM00500000272544 | Coiled Coil Domain Containing 28A CCRL1AP |
| ENSMUSG00000059602 | ENSFM00250000000771 | Synapsin Synapsin |
| ENSMUSG00000059714 | ENSFM00640001119931 | Unknown |
| ENSMUSG00000059714 | ENSFM00780001898418 | Flotillin 1 |
| ENSMUSG00000059734 | ENSFM00250000005068 | Nadh Dehydrogenase Iron Sulfur 8 Mitochondrial Precursor EC_1.6.5.3 EC_1.6.99.- 3 Complex I 23KD Ci 23KD Nadh Ubiquinone Oxidoreductase 23 Kda Subunit |
| ENSMUSG00000059742 | ENSFM00730001521633 | Potassium Voltage Gated Channel Subfamily H Member 2 Ether A Go Go Related Gene Potassium Channel 1 Erg 1 Eag Related 1 Ether A Go Go Related 1 Voltage Gated Potassium Channel Subunit KV11 1 |
| ENSMUSG00000059743 | ENSFM00800002003519 | Farnesyl Pyrophosphate Synthase Fpp Synthase Fps EC_2.5.1.10 2E 6E Farnesyl Diphosphate Synthase Cholesterol Regulated 39 Kda Cr 39 Dimethylallyltranstransferase EC_2.5.1.- 1 Farnesyl Diphosphate Synthase Geranyltranstransferase |
| ENSMUSG00000059775 | ENSFM00670001235586 | 40S Ribosomal S26 |
| ENSMUSG00000059811 | ENSFM00250000001436 | Atlastin EC_3.6.5.- |
| ENSMUSG00000059857 | ENSFM00260000050653 | Netrin Precursor Laminet |
| ENSMUSG00000059890 | ENSFM00570000887673 | Unknown |
| ENSMUSG00000059890 | ENSFM00730001522369 | Ubiquitin Conjugation Factor E4 A |
| ENSMUSG00000059923 | ENSFM00570000883977 | Unknown |
| ENSMUSG00000059923 | ENSFM00500000270097 | Growth Factor Receptor Bound 2 Adapter GRB2 SH2/SH3 Adapter GRB2 |
| ENSMUSG00000059970 | ENSFM00730001521103 | Heat Shock Kda Heat Shock 70 Kda |
| ENSMUSG00000059974 | ENSFM00290000065521 | Precursor |
| ENSMUSG00000059981 | ENSFM00250000000892 | Serine/Threonine Kinase EC_2.7.11.1 Thousand And One Amino Acid |
| ENSMUSG00000060002 | ENSFM00570000892250 | Unknown |
| ENSMUSG00000060002 | ENSFM00500000270126 | Cholinephosphotransferase 1 EC_2.7.8.2 Diacylglycerol Cholinephosphotransferase 1 |
| ENSMUSG00000060012 | ENSFM00730001521492 | Kinesin |
| ENSMUSG00000060036 | ENSFM00810002040577 | 60S Ribosomal L3 |
| ENSMUSG00000060073 | ENSFM00590000916022 | Unknown |
| ENSMUSG00000060073 | ENSFM00590000917798 | Unknown |
| ENSMUSG00000060073 | ENSFM00590000918013 | Unknown |
| ENSMUSG00000060073 | ENSFM00590000918014 | Unknown |
| ENSMUSG00000060073 | ENSFM00810002040700 | Proteasome Subunit Alpha Type 3 EC_3.4.25.1 Macropain Subunit C8 Multicatalytic Endopeptidase Complex Subunit C8 Proteasome Component C8 |
| ENSMUSG00000060081 | ENSFM00670001235241 | Histone H2A |
| ENSMUSG00000060090 | ENSFM00250000005619 | XRP2 |
| ENSMUSG00000060126 | ENSFM00500000270782 | Translationally Controlled Tumor Tctp |
| ENSMUSG00000060147 | ENSFM00610000961279 | Unknown |
| ENSMUSG00000060147 | ENSFM00610000969846 | Unknown |
| ENSMUSG00000060147 | ENSFM00610000973173 | Unknown |
| ENSMUSG00000060147 | ENSFM00500000269596 | Serpin |
| ENSMUSG00000060166 | ENSFM00500000270072 | Palmitoyltransferase ZDHHC5 EC_2.3.1.225 Zinc Finger Dhhc Domain Containing 5 Dhhc 5 |
| ENSMUSG00000060227 | ENSFM00250000003320 | CASC4 Cancer Susceptibility Candidate Gene 4 |
| ENSMUSG00000060240 | ENSFM00600000922041 | Cell Cycle Exit And Neuronal Differentiation 1 |
| ENSMUSG00000060261 | ENSFM00810002049264 | Unknown |
| ENSMUSG00000060261 | ENSFM00810002049698 | Unknown |
| ENSMUSG00000060261 | ENSFM00760001714782 | General Transcription Factor Ii I Repeat Domain Containing 1 GTF2I Repeat Domain Containing 1 |
| ENSMUSG00000060275 | ENSFM00600000921191 | Pro Neuregulin 1 Membrane Bound Precursor Pro NRG1 |
| ENSMUSG00000060279 | ENSFM00610000970149 | Unknown |
| ENSMUSG00000060279 | ENSFM00610000971381 | Unknown |
| ENSMUSG00000060279 | ENSFM00810002040615 | Ap 2 Complex Subunit Alpha 2.100 Kda Coated Vesicle C Adaptor Complex Ap 2 Subunit Alpha 2 Adaptor Related Complex 2 Subunit Alpha 2 Alpha Adaptin C ALPHA2 Adaptin Clathrin Assembly Complex 2 Alpha C Large Chain Plasma Membrane Adaptor HA2/AP2 Adaptin Alp |
| ENSMUSG00000060373 | ENSFM00810002040643 | Heterogeneous Nuclear Ribonucleoprotein C Hnrnp C |
| ENSMUSG00000060429 | ENSFM00250000002085 | 1 Syntrophin 59 Kda Dystrophin Associated A1 Component 1 Syntrophin |
| ENSMUSG00000060534 | ENSFM00260000050557 | Netrin Receptor Dcc Precursor Tumor Suppressor Dcc |
| ENSMUSG00000060636 | ENSFM00500000271015 | 60S Ribosomal L35A |
| ENSMUSG00000060675 | ENSFM00500000271831 | Hras Suppressor 3 HRSL3 EC_3.1.1.32 EC_3.1.1.- 4 Group Xvi Phospholipase A1/A2 H Rev 107 |
| ENSMUSG00000060681 | ENSFM00730001521509 | Sodium/Hydrogen Exchanger Na + /H + Exchanger Nhe Solute Carrier Family 9 Member |
| ENSMUSG00000060743 | ENSFM00590000918360 | Unknown |
| ENSMUSG00000060743 | ENSFM00590000918361 | Unknown |
| ENSMUSG00000060743 | ENSFM00250000000259 | Histone H3 |
| ENSMUSG00000060780 | ENSFM00400000131776 | Leucine Rich Repeat Transmembrane Neuronal Precursor |
| ENSMUSG00000060803 | ENSFM00500000270850 | Glutathione S Transferase P EC_2.5.1.18 Gst Class Pi |
| ENSMUSG00000060843 | ENSFM00250000001018 | Catenin Alpha 2 Alpha N Catenin |
| ENSMUSG00000060882 | ENSFM00300000079128 | Potassium Voltage Gated Channel Subfamily D Member Voltage Gated Potassium Channel Subunit KV4 |
| ENSMUSG00000060904 | ENSFM00610000973080 | Unknown |
| ENSMUSG00000060904 | ENSFM00730001521760 | Adp Ribosylation Factor 1 |
| ENSMUSG00000060923 | ENSFM00670001236363 | Acylphosphatase 2 EC_3.6.1.7 Acylphosphatase Muscle Type Isozyme Acylphosphate Phosphohydrolase 2 |
| ENSMUSG00000060924 | ENSFM00740001589158 | Cub And Sushi Domain Containing Cub And Sushi Multiple Domains |
| ENSMUSG00000060938 | ENSFM00500000270069 | 60S Ribosomal L26 |
| ENSMUSG00000060961 | ENSFM00730001521721 | Electrogenic Sodium Bicarbonate Cotransporter 1 Sodium Bicarbonate Cotransporter Na + /HCO3 Cotransporter Solute Carrier Family 4 Member 4 |
| ENSMUSG00000060961 | ENSFM00800002006266 | Electrogenic Sodium Bicarbonate Cotransporter 1 |
| ENSMUSG00000060992 | ENSFM00500000270533 | Coatomer Subunit Zeta Zeta Coat Zeta Cop |
| ENSMUSG00000061080 | ENSFM00760001748822 | Unknown |
| ENSMUSG00000061080 | ENSFM00760001763360 | Unknown |
| ENSMUSG00000061080 | ENSFM00760001763361 | Unknown |
| ENSMUSG00000061080 | ENSFM00760001763362 | Unknown |
| ENSMUSG00000061080 | ENSFM00290000065521 | Precursor |
| ENSMUSG00000061086 | ENSFM00570000895631 | Unknown |
| ENSMUSG00000061086 | ENSFM00570000897194 | Unknown |
| ENSMUSG00000061086 | ENSFM00250000000438 | Myosin Light Chain Myosin Light Chain Myosin Light Chain |
| ENSMUSG00000061111 | ENSFM00670001237224 | Unknown |
| ENSMUSG00000061244 | ENSFM00590000918374 | Unknown |
| ENSMUSG00000061244 | ENSFM00250000005479 | Exocyst Complex Component Exocyst Complex Component SEC10 |
| ENSMUSG00000061273 | ENSFM00500000273449 | Membrane Magnesium Transporter 1 Precursor Er Membrane Complex Subunit 5 Transmembrane 32 |
| ENSMUSG00000061455 | ENSFM00250000004622 | Syntaxin 17 |
| ENSMUSG00000061461 | ENSFM00570000899182 | Unknown |
| ENSMUSG00000061474 | ENSFM00670001236924 | 28S Ribosomal S36 Mitochondrial Mrp S36 S36MT |
| ENSMUSG00000061518 | ENSFM00780001901644 | Unknown |
| ENSMUSG00000061518 | ENSFM00780001912206 | Unknown |
| ENSMUSG00000061518 | ENSFM00670001235955 | Cytochrome C Oxidase Subunit 5B Mitochondrial Precursor Cytochrome C Oxidase Polypeptide Vb |
| ENSMUSG00000061576 | ENSFM00570000878930 | Unknown |
| ENSMUSG00000061576 | ENSFM00750001632364 | Dipeptidyl Aminopeptidase 6 Dppx Dipeptidyl Aminopeptidase Related Dipeptidyl Peptidase 6 Dipeptidyl Peptidase Iv Dipeptidyl Peptidase Vi Dpp Vi |
| ENSMUSG00000061601 | ENSFM00600000921250 | Piccolo Aczonin |
| ENSMUSG00000061666 | ENSFM00250000003307 | Glycerophosphodiester Phosphodiesterase Domain Containing EC_3.1.-.- |
| ENSMUSG00000061689 | ENSFM00660001182988 | Unknown |
| ENSMUSG00000061689 | ENSFM00660001190031 | Unknown |
| ENSMUSG00000061689 | ENSFM00250000000718 | Disks Large Associated Dap Psd 95/SAP90 Binding SAP90/Psd 95 Associated |
| ENSMUSG00000061718 | ENSFM00500000273519 | Phosphatase 1 Regulatory Subunit 1B Darpp 32 Dopamine And Camp Regulated Neuronal Phosphoprotein |
| ENSMUSG00000061740 | ENSFM00770001847552 | Cytochrome P450 |
| ENSMUSG00000061751 | ENSFM00740001589124 | EC_2.7.11.1 |
| ENSMUSG00000061758 | ENSFM00730001521171 | Reductase Aldo Keto Reductase Family 1 Member |
| ENSMUSG00000061787 | ENSFM00780001898364 | 40S Ribosomal S17 |
| ENSMUSG00000061808 | ENSFM00500000271252 | Transthyretin Precursor Prealbumin |
| ENSMUSG00000061816 | ENSFM00250000000438 | Myosin Light Chain Myosin Light Chain Myosin Light Chain |
| ENSMUSG00000061904 | ENSFM00760001714669 | Phosphate Carrier Protein Mitochondrial Precursor Phosphate Transport Ptp Solute Carrier Family 25 Member 3 |
| ENSMUSG00000061981 | ENSFM00780001898447 | Flotillin 2 |
| ENSMUSG00000062006 | ENSFM00800002015933 | Unknown |
| ENSMUSG00000062006 | ENSFM00500000271952 | 60S Ribosomal L34 |
| ENSMUSG00000062014 | ENSFM00500000270656 | Glia Maturation Factor Gmf |
| ENSMUSG00000062044 | ENSFM00570000890117 | Unknown |
| ENSMUSG00000062044 | ENSFM00670001235536 | Serine/Threonine Kinase EC_2.7.11.1 Apoptosis Associated Tyrosine Kinase Brain Kinase Lemur Tyrosine Kinase |
| ENSMUSG00000062070 | ENSFM00790001962976 | Phosphoglycerate Kinase EC_2.7.2.3 |
| ENSMUSG00000062075 | ENSFM00250000000425 | Lamin |
| ENSMUSG00000062078 | ENSFM00400000131794 | Quaking |
| ENSMUSG00000062110 | ENSFM00810002057151 | Unknown |
| ENSMUSG00000062110 | ENSFM00250000004937 | SEC1 Family Domain Containing 2 Syntaxin Binding 1 1 |
| ENSMUSG00000062127 | ENSFM00600000921238 | Cortactin Binding 2 CORTBP2 |
| ENSMUSG00000062151 | ENSFM00250000000599 | Unc 13 Homolog MUNC13 |
| ENSMUSG00000062190 | ENSFM00250000002547 | Lanc 1.40 Kda Erythrocyte Membrane P40 |
| ENSMUSG00000062209 | ENSFM00410000138465 | Receptor Tyrosine Kinase Erbb Precursor EC_2.7.10.1 Proto Oncogene C Erbb |
| ENSMUSG00000062232 | ENSFM00250000001338 | Rap Guanine Nucleotide Exchange Factor 2 Cyclic Nucleotide Ras Gef Cnrasgef Neural Rap Guanine Nucleotide Exchange Nrap Gep Pdz Domain Containing Guanine Nucleotide Exchange Factor 1 Pdz GEF1 Ra Gef 1 Ras/RAP1 Associating Gef 1 |
| ENSMUSG00000062234 | ENSFM00800002023625 | Unknown |
| ENSMUSG00000062234 | ENSFM00800002023626 | Unknown |
| ENSMUSG00000062234 | ENSFM00250000001678 | Tyrosine Phosphatase Auxilin EC_3.1.3.48 Dnaj Homolog Subfamily C Member 6 |
| ENSMUSG00000062257 | ENSFM00290000065521 | Precursor |
| ENSMUSG00000062328 | ENSFM00250000001117 | 60S Ribosomal L17 |
| ENSMUSG00000062373 | ENSFM00290000065595 | Transmembrane 65 |
| ENSMUSG00000062376 | ENSFM00670001236522 | Unknown |
| ENSMUSG00000062380 | ENSFM00250000000193 | Tubulin Beta Chain |
| ENSMUSG00000062382 | ENSFM00730001522100 | Ferritin Light Chain Ferritin L Subunit |
| ENSMUSG00000062444 | ENSFM00480000262766 | Ap 3 Complex Subunit Beta 1 Adaptor Complex Ap 3 Subunit Beta 1 Adaptor Related Complex 3 Subunit Beta 1 Beta 3A Adaptin Clathrin Assembly Complex 3 Beta 1 Large Chain |
| ENSMUSG00000062580 | ENSFM00570000894972 | Unknown |
| ENSMUSG00000062580 | ENSFM00500000270373 | Mitochondrial Import Inner Membrane Translocase Subunit TIM17 |
| ENSMUSG00000062591 | ENSFM00250000000193 | Tubulin Beta Chain |
| ENSMUSG00000062604 | ENSFM00800002023563 | Unknown |
| ENSMUSG00000062604 | ENSFM00800002023564 | Unknown |
| ENSMUSG00000062604 | ENSFM00500000269720 | Srsf Kinase EC_2.7.11.1 Kinase Serine/Arginine Rich Specific Kinase Sr Specific Kinase |
| ENSMUSG00000062647 | ENSFM00750001632337 | 60S Ribosomal L7A |
| ENSMUSG00000062661 | ENSFM00780001917898 | Unknown |
| ENSMUSG00000062661 | ENSFM00760001714671 | Neuronal Calcium Sensor 1 Ncs 1 |
| ENSMUSG00000062762 | ENSFM00740001613556 | Unknown |
| ENSMUSG00000062762 | ENSFM00250000005747 | Etoposide Induced 2 4 P53 Induced Gene 8 |
| ENSMUSG00000062785 | ENSFM00260000050384 | Potassium Voltage Gated Channel Subfamily C Member Voltage Gated Potassium Channel KV3 |
| ENSMUSG00000062825 | ENSFM00730001521060 | Actin |
| ENSMUSG00000062908 | ENSFM00570000896410 | Medium Chain Specific Acyl Coa Dehydrogenase Mitochondrial |
| ENSMUSG00000062908 | ENSFM00730001521951 | Medium Chain Specific Acyl Coa Dehydrogenase Mitochondrial Precursor Mcad EC_1.3.8.7 |
| ENSMUSG00000062929 | ENSFM00500000270025 | Cofilin Cofilin Muscle |
| ENSMUSG00000062949 | ENSFM00730001521628 | Phospholipid Transporting Atpase EC_3.6.3.1 Atpase Class Vi Type P4 Atpase Flippase Complex Alpha Subunit |
| ENSMUSG00000062961 | ENSFM00260000052939 | Coiled Coil Domain Containing 177 |
| ENSMUSG00000062963 | ENSFM00250000007775 | Ubiquitin Fold Modifier Conjugating Enzyme 1 UFM1 Conjugating Enzyme 1 |
| ENSMUSG00000063052 | ENSFM00570000879628 | Unknown |
| ENSMUSG00000063052 | ENSFM00570000893042 | Unknown |
| ENSMUSG00000063052 | ENSFM00800002024524 | Unknown |
| ENSMUSG00000063052 | ENSFM00440000236958 | Leucine Rich Repeat Containing 40 |
| ENSMUSG00000063063 | ENSFM00250000001018 | Catenin Alpha 2 Alpha N Catenin |
| ENSMUSG00000063065 | ENSFM00730001521833 | Mitogen Activated Kinase 1 Map Kinase 1 Mapk 1 EC_2.7.11.24 ERT1 Extracellular Signal Regulated Kinase 2 Erk 2 Mitogen Activated Kinase 2 Map Kinase 2 Mapk 2 |
| ENSMUSG00000063142 | ENSFM00500000288687 | Uncharacterized |
| ENSMUSG00000063142 | ENSFM00250000000903 | Calcium Activated Potassium Channel Subunit Alpha 1 Bk Channel Bkca Alpha Calcium Activated Potassium Channel Subfamily M Subunit Alpha 1 K Vca Alpha KCA1 1 Maxi K Channel Maxik Slo Alpha SLO1 Slowpoke Homolog Slo Homolog |
| ENSMUSG00000063146 | ENSFM00250000000764 | Cap Gly Domain Containing Linker 2 Cytoplasmic Linker 115 Clip 115 Cytoplasmic Linker 2 |
| ENSMUSG00000063160 | ENSFM00250000001973 | Numb |
| ENSMUSG00000063229 | ENSFM00250000000475 | L Lactate Dehydrogenase Chain Ldh EC_1.1.1.27 |
| ENSMUSG00000063239 | ENSFM00250000000328 | Metabotropic Glutamate Receptor Precursor |
| ENSMUSG00000063297 | ENSFM00250000005652 | Leucine Zipper 2 Precursor |
| ENSMUSG00000063317 | ENSFM00260000050633 | Ubiquitin Carboxyl Terminal Hydrolase 43 EC_3.4.19.12 Deubiquitinating Enzyme 43 Ubiquitin Thioesterase 43 Ubiquitin Specific Processing Protease 43 |
| ENSMUSG00000063358 | ENSFM00730001521833 | Mitogen Activated Kinase 1 Map Kinase 1 Mapk 1 EC_2.7.11.24 ERT1 Extracellular Signal Regulated Kinase 2 Erk 2 Mitogen Activated Kinase 2 Map Kinase 2 Mapk 2 |
| ENSMUSG00000063410 | ENSFM00810002040592 | Serine/Threonine Kinase EC_2.7.11.1 Mammalian STE20 Kinase Mst STE20 Kinase Serine/Threonine Kinase |
| ENSMUSG00000063446 | ENSFM00500000270116 | Lipid Phosphate Phosphatase Related Type Plasticity Related Gene Prg |
| ENSMUSG00000063455 | ENSFM00600000921514 | Ambiguous |
| ENSMUSG00000063457 | ENSFM00250000003189 | 40S Ribosomal S15 |
| ENSMUSG00000063646 | ENSFM00640001112222 | Unknown |
| ENSMUSG00000063646 | ENSFM00250000001480 | Janus Kinase And Microtubule Interacting 1 Multiple Alpha Helices And Rna Linker 1 Marlin 1 |
| ENSMUSG00000063696 | ENSFM00250000001360 | 60S Acidic Ribosomal P0 60S Ribosomal L10E |
| ENSMUSG00000063801 | ENSFM00500000270528 | Ap 3 Complex Subunit Sigma Ap 3 Complex Subunit Sigma Adaptor Related Complex 3 Subunit Sigma Sigma Adaptin Adaptin Sigma Adaptin |
| ENSMUSG00000063856 | ENSFM00500000270307 | Glutathione Peroxidase 2 Gpx 2 Gshpx 2 EC_1.11.1.9 Glutathione Peroxidase Gastrointestinal Gpx Gi Gshpx Gi |
| ENSMUSG00000063882 | ENSFM00500000356682 | Unknown |
| ENSMUSG00000063882 | ENSFM00810002043076 | Cytochrome B C1 Complex Subunit 6 Mitochondrial Precursor Complex Iii Subunit 6 Complex Iii Subunit Viii Cytochrome C1 Non Heme 11 Kda Mitochondrial Hinge Ubiquinol Cytochrome C Reductase Complex 11 Kda |
| ENSMUSG00000063887 | ENSFM00380000124679 | Neuroligin Precursor |
| ENSMUSG00000063904 | ENSFM00810002040563 | Dipeptidyl Peptidase 3 EC_3.4.14.4 Dipeptidyl Aminopeptidase Iii Dipeptidyl Arylamidase Iii Dipeptidyl Peptidase Iii Dpp Iii |
| ENSMUSG00000064030 | ENSFM00500000273807 | Partner Of Y14 And Mago Wibg Homolog |
| ENSMUSG00000064068 | ENSFM00640001115744 | Unknown |
| ENSMUSG00000064068 | ENSFM00640001116925 | Unknown |
| ENSMUSG00000064068 | ENSFM00640001119895 | Unknown |
| ENSMUSG00000064068 | ENSFM00640001119896 | Unknown |
| ENSMUSG00000064068 | ENSFM00250000002187 | Metaxin 1 Mitochondrial Outer Membrane Import Complex 1 |
| ENSMUSG00000064105 | ENSFM00250000000938 | Metal Transporter Ancient Conserved Domain Containing Cyclin |
| ENSMUSG00000064115 | ENSFM00570000887569 | Unknown |
| ENSMUSG00000064115 | ENSFM00250000001126 | Cell Adhesion Molecule Precursor Immunoglobulin Superfamily Member Nectin Necl Synaptic Cell Adhesion Molecule |
| ENSMUSG00000064125 | ENSFM00660001189520 | Unknown |
| ENSMUSG00000064125 | ENSFM00250000009146 | Proline Rich 36 |
| ENSMUSG00000064293 | ENSFM00390000126303 | Contactin Precursor Brain Derived Immunoglobulin Superfamily Big |
| ENSMUSG00000064302 | ENSFM00760001753540 | Unknown |
| ENSMUSG00000064302 | ENSFM00250000000820 | Clip Associating Cytoplasmic Linker Associated |
| ENSMUSG00000064329 | ENSFM00740001589070 | Sodium Channel Type Subunit Alpha Sodium Channel Sodium Channel Type Subunit Alpha Voltage Gated Sodium Channel Subunit Alpha NAV1 |
| ENSMUSG00000064341 | ENSFM00250000000180 | Nadh Ubiquinone Oxidoreductase Chain 1 EC_1.6.5.3 Nadh Dehydrogenase Subunit 1 |
| ENSMUSG00000064345 | ENSFM00750001632314 | Nadh Ubiquinone Oxidoreductase Chain 2 EC_1.6.5.3 Nadh Dehydrogenase Subunit 2 |
| ENSMUSG00000064351 | ENSFM00730001521132 | Cytochrome C Oxidase Subunit 1 EC_1.9.3.1 Cytochrome C Oxidase Polypeptide I |
| ENSMUSG00000064354 | ENSFM00250000000189 | Cytochrome C Oxidase Subunit 2 Cytochrome C Oxidase Polypeptide Ii |
| ENSMUSG00000064356 | ENSFM00500000344754 | Unknown |
| ENSMUSG00000064358 | ENSFM00270000056420 | Cytochrome C Oxidase Subunit 3 Cytochrome C Oxidase Polypeptide Iii |
| ENSMUSG00000064363 | ENSFM00250000000110 | Nadh Ubiquinone Oxidoreductase Chain 4 EC_1.6.5.3 Nadh Dehydrogenase Subunit 4 |
| ENSMUSG00000064367 | ENSFM00730001521259 | Nadh Ubiquinone Oxidoreductase Chain 5 EC_1.6.5.3 Nadh Dehydrogenase Subunit 5 |
| ENSMUSG00000065954 | ENSFM00740001589245 | Transforming Acidic Coiled Coil Containing |
| ENSMUSG00000066036 | ENSFM00250000001486 | E3 Ubiquitin Ligase UBR4 EC_6.3.2.- N Recognin 4 Zinc Finger UBR1 Type 1 |
| ENSMUSG00000066129 | ENSFM00250000004496 | Very Kind Kind Domain Containing 1 Kinase Non Catalytic C Lobe Domain Containing 1 Ras Gef Domain Containing Family Member 2 |
| ENSMUSG00000066151 | ENSFM00250000004912 | Unknown |
| ENSMUSG00000066189 | ENSFM00250000002019 | Voltage Dependent Calcium Channel Gamma Subunit Neuronal Voltage Gated Calcium Channel Gamma Subunit Transmembrane Ampar Regulatory Gamma Tarp Gamma |
| ENSMUSG00000066232 | ENSFM00760001714748 | Importin 7 IMP7 Ran Binding 7 RANBP7 |
| ENSMUSG00000066235 | ENSFM00250000007877 | O Linked Mannose Beta 1 4 N Acetylglucosaminyltransferase 2 POMGNT2 EC_2.4.1.- |
| ENSMUSG00000066278 | ENSFM00500000273456 | Vacuolar Sorting Associated 37B Escrt I Complex Subunit VPS37B |
| ENSMUSG00000066324 | ENSFM00250000004572 | Inositol Monophosphatase 3 Imp 3 Impase 3 EC_3.1.3.25 EC_3.1.3.- 7 Golgi 3 Prime Phosphoadenosine 5 Prime Phosphate 3 Prime Phosphatase Golgi Resident Pap Phosphatase Gpapp Inositol Monophosphatase Domain Containing 1 Inositol 1 Or 4 Monophosphatase 3 Myo |
| ENSMUSG00000066392 | ENSFM00740001589096 | Neurexin Precursor Neurexin |
| ENSMUSG00000066438 | ENSFM00570000882117 | Unknown |
| ENSMUSG00000066438 | ENSFM00430000230337 | Pleckstrin Homology Domain Containing Family D Member 1 Ph Domain Containing Family D Member 1 |
| ENSMUSG00000066441 | ENSFM00590000918434 | Unknown |
| ENSMUSG00000066441 | ENSFM00500000269664 | Retinol Dehydrogenase |
| ENSMUSG00000066487 | ENSFM00250000000709 | 40S Ribosomal S2 |
| ENSMUSG00000066621 | ENSFM00740001589473 | Tectonin Beta Propeller Repeat Containing |
| ENSMUSG00000066705 | ENSFM00670001236586 | Fxyd Domain Containing Ion Transport Regulator 6 Precursor Phosphohippolin |
| ENSMUSG00000066839 | ENSFM00250000005244 | Evolutionarily Conserved Signaling Intermediate In Toll Pathway Mitochondrial Precursor |
| ENSMUSG00000067038 | ENSFM00500000270361 | 40S Ribosomal S12 |
| ENSMUSG00000067194 | ENSFM00250000002567 | Eukaryotic Translation Initiation Factor 1A Eif 1A Eukaryotic Translation Initiation Factor 4C Eif 4C |
| ENSMUSG00000067242 | ENSFM00800002009181 | Unknown |
| ENSMUSG00000067242 | ENSFM00250000002312 | Leucine Rich Repeat Lgi Family Member Precursor Leucine Rich Glioma Inactivated |
| ENSMUSG00000067274 | ENSFM00250000001360 | 60S Acidic Ribosomal P0 60S Ribosomal L10E |
| ENSMUSG00000067288 | ENSFM00500000284355 | Unknown |
| ENSMUSG00000067288 | ENSFM00640001110125 | Unknown |
| ENSMUSG00000067336 | ENSFM00730001522171 | Anti Muellerian Hormone Type 2 Receptor Precursor EC_2.7.11.30 Anti Muellerian Hormone Type Ii Receptor Amh Type Ii Receptor Mis Type Ii Receptor Misrii Mrii |
| ENSMUSG00000067629 | ENSFM00810002057507 | Unknown |
| ENSMUSG00000067629 | ENSFM00250000000610 | Ras Gtpase Activating |
| ENSMUSG00000067713 | ENSFM00250000001075 | 5' Amp Activated Kinase Subunit Gamma Ampk Ampk Subunit Gamma |
| ENSMUSG00000067722 | ENSFM00720001492156 | CCSMST1 Precursor |
| ENSMUSG00000067889 | ENSFM00740001589095 | Spectrin Beta Chain Erythrocytic Beta Spectrin |
| ENSMUSG00000068015 | ENSFM00800002003728 | Leucine Rich Repeat And Calponin Homology Domain Containing 1 Calponin Homology Domain Containing 1 |
| ENSMUSG00000068036 | ENSFM00250000001587 | Afadin Af 6 |
| ENSMUSG00000068039 | ENSFM00500000270058 | T Complex 1 Subunit Alpha Tcp 1 Alpha Cct Alpha |
| ENSMUSG00000068040 | ENSFM00780001898361 | Transmembrane 9 Superfamily Member Precursor |
| ENSMUSG00000068184 | ENSFM00670001236509 | Mimitin Mitochondrial Precursor Myc Induced Mitochondrial Mmtn Nadh Dehydrogenase 1 Alpha Subcomplex Assembly Factor 2 NDUFA12 |
| ENSMUSG00000068290 | ENSFM00570000879313 | Unknown |
| ENSMUSG00000068290 | ENSFM00570000896336 | Unknown |
| ENSMUSG00000068290 | ENSFM00600000921555 | Ddrgk Domain Containing 1 |
| ENSMUSG00000068329 | ENSFM00250000001030 | Serine Protease HTRA2 Mitochondrial |
| ENSMUSG00000068373 | ENSFM00250000005157 | Ambiguous |
| ENSMUSG00000068566 | ENSFM00500000271004 | Myeloid Associated Differentiation Marker |
| ENSMUSG00000068735 | ENSFM00500000272770 | Tumor P53 Inducible 11 P53 Induced Gene 11 |
| ENSMUSG00000068739 | ENSFM00770001847577 | Serine Trna Ligase Cytoplasmic EC_6.1.1.11 Seryl Trna Synthetase Serrs Seryl Trna Ser/Sec Synthetase |
| ENSMUSG00000068740 | ENSFM00560000770988 | Cadherin Egf Lag Seven Pass G Type Receptor Multiple Epidermal Growth Factor Domains Multiple Egf Domains |
| ENSMUSG00000068747 | ENSFM00500000271702 | Sortilin Precursor Neurotensin Receptor 3 NTR3 |
| ENSMUSG00000068748 | ENSFM00730001521864 | Receptor Type Tyrosine Phosphatase Precursor R Ptp EC_3.1.3.48 |
| ENSMUSG00000068749 | ENSFM00800002024487 | Unknown |
| ENSMUSG00000068749 | ENSFM00500000270276 | Proteasome Subunit Alpha Type 5 EC_3.4.25.1 Macropain Multicatalytic Endopeptidase Complex Proteasome |
| ENSMUSG00000068798 | ENSFM00730001521249 | Ras Related Rap 1 |
| ENSMUSG00000068823 | ENSFM00250000002562 | Cold Shock Domain Containing E1 |
| ENSMUSG00000068923 | ENSFM00500000270270 | Synaptotagmin Synaptotagmin |
| ENSMUSG00000069045 | ENSFM00760001763663 | Unknown |
| ENSMUSG00000069045 | ENSFM00770001847530 | Atp Dependent Rna Helicase DDX3Y EC_3.6.4.13 Dead Box 3 Y Chromosomal |
| ENSMUSG00000069072 | ENSFM00800002003868 | Probable Cationic Amino Acid Transporter Solute Carrier Family 7 Member 14 |
| ENSMUSG00000069117 | ENSFM00250000002236 | 40S Ribosomal S18 |
| ENSMUSG00000069227 | ENSFM00250000008086 | G Regulated Inducer Of Neurite Outgrowth 1 GRIN1 |
| ENSMUSG00000069266 | ENSFM00500000269649 | Histone H4 |
| ENSMUSG00000069308 | ENSFM00500000269608 | Histone H2B |
| ENSMUSG00000069539 | ENSFM00760001714811 | SCY1 2 Coated Vesicle Associated Kinase Of 104 Kda |
| ENSMUSG00000069601 | ENSFM00500000269628 | Ankyrin Ank Ankyrin |
| ENSMUSG00000069662 | ENSFM00800002005729 | Myristoylated Alanine Rich C Kinase Substrate Marcks |
| ENSMUSG00000069744 | ENSFM00250000003368 | Proteasome Subunit Beta Type 3 EC_3.4.25.1 Proteasome Chain 13 Proteasome Component C10 Ii Proteasome Theta Chain |
| ENSMUSG00000069769 | ENSFM00500000270111 | Rna Binding Musashi Homolog |
| ENSMUSG00000069833 | ENSFM00740001589261 | Associated |
| ENSMUSG00000069844 | ENSFM00500000270642 | Homolog Mitochondrial Precursor |
| ENSMUSG00000069919 | ENSFM00500000269637 | Hemoglobin Subunit Alpha Alpha Globin Hemoglobin Alpha Chain |
| ENSMUSG00000070047 | ENSFM00760001763202 | Unknown |
| ENSMUSG00000070047 | ENSFM00570000851039 | Protocadherin Fat Precursor Fat Tumor Suppressor Homolog |
| ENSMUSG00000070283 | ENSFM00780001908782 | Unknown |
| ENSMUSG00000070283 | ENSFM00500000273238 | Nadh Dehydrogenase 1 Alpha Subcomplex Assembly Factor 3 |
| ENSMUSG00000070287 | ENSFM00500000272943 | Solute Carrier Family 35 Member G2 Transmembrane 22 |
| ENSMUSG00000070304 | ENSFM00780001898523 | Sodium Channel Subunit Beta 2 Precursor |
| ENSMUSG00000070372 | ENSFM00250000001955 | F Actin Capping Subunit Alpha 2 Capz Alpha 2 |
| ENSMUSG00000070394 | ENSFM00670001236968 | Transmembrane 256 Precursor |
| ENSMUSG00000070436 | ENSFM00310000089046 | Serpin H1 Precursor 47 Kda Heat Shock Collagen Binding Colligin |
| ENSMUSG00000070462 | ENSFM00250000008923 | Mesoderm Development Candidate 1 |
| ENSMUSG00000070498 | ENSFM00250000000808 | Transmembrane Precursor |
| ENSMUSG00000070509 | ENSFM00250000001343 | Rgm Domain Family Member |
| ENSMUSG00000070565 | ENSFM00250000000610 | Ras Gtpase Activating |
| ENSMUSG00000070570 | ENSFM00730001521551 | Vesicular Glutamate Transporter Solute Carrier Family 17 Member |
| ENSMUSG00000070639 | ENSFM00250000001418 | Volume Regulated Anion Channel Subunit Leucine Rich Repeat Containing |
| ENSMUSG00000070695 | ENSFM00250000000380 | Contactin Associated 5 Precursor Cell Recognition Molecule CASPR5 |
| ENSMUSG00000070730 | ENSFM00570000889560 | Unknown |
| ENSMUSG00000070730 | ENSFM00570000894879 | Unknown |
| ENSMUSG00000070730 | ENSFM00570000901405 | Regulator Of Microtubule Dynamics 3 |
| ENSMUSG00000070730 | ENSFM00670001235671 | Regulator Of Microtubule Dynamics Rmd Fa |
| ENSMUSG00000070871 | ENSFM00250000001383 | Cyclin Y |
| ENSMUSG00000070880 | ENSFM00570000877622 | Unknown |
| ENSMUSG00000070880 | ENSFM00800002003398 | Glutamate Decarboxylase 1 EC_4.1.1.15.67 Kda Glutamic Acid Decarboxylase Gad 67 Glutamate Decarboxylase 67 Kda |
| ENSMUSG00000071014 | ENSFM00600000921719 | Nadh Dehydrogenase 1 Beta Subcomplex Subunit 6 Complex I B17 Ci B17 Nadh Ubiquinone Oxidoreductase B17 Subunit |
| ENSMUSG00000071014 | ENSFM00770001850404 | Nadh Dehydrogenase 1 Beta Subcomplex Subunit 6 |
| ENSMUSG00000071073 | ENSFM00250000009938 | Leucine Rich Repeat Containing 73 |
| ENSMUSG00000071074 | ENSFM00250000005379 | YIPF3 YIP1 Family Member 3 |
| ENSMUSG00000071172 | ENSFM00500000272005 | Serine/Arginine Rich Splicing Factor 3 Pre Splicing Factor SRP20 Splicing Factor Arginine/Serine Rich 3 |
| ENSMUSG00000071379 | ENSFM00760001714608 | Unknown |
| ENSMUSG00000071415 | ENSFM00570000883216 | Unknown |
| ENSMUSG00000071415 | ENSFM00250000003000 | 60S Ribosomal L23 |
| ENSMUSG00000071424 | ENSFM00720001492054 | Glutamate Receptor Ionotropic Delta Precursor Glur Delta Subunit |
| ENSMUSG00000071519 | ENSFM00730001521212 | Trypsin Precursor EC_3.4.21.4 |
| ENSMUSG00000071528 | ENSFM00500000445520 | Unknown |
| ENSMUSG00000071531 | ENSFM00280000058989 | G Regulated Inducer Of Neurite Outgrowth 2 GRIN2 |
| ENSMUSG00000071644 | ENSFM00250000001516 | Elongation Factor 1 Gamma Ef 1 Gamma Eef 1B Gamma |
| ENSMUSG00000071649 | ENSFM00250000001080 | Galactosylgalactosylxylosylprotein 3 Beta Glucuronosyltransferase EC_2.4.1.135 Beta 1 3 Glucuronyltransferase Glucuronosyltransferase Glcat Udp Beta 1 3 Glucuronyltransferase Glcuat |
| ENSMUSG00000071650 | ENSFM00810002040602 | Neutral Alpha Glucosidase Ab Precursor EC_3.2.1.84 Alpha Glucosidase 2 Glucosidase Ii Subunit Alpha |
| ENSMUSG00000071654 | ENSFM00500000295485 | Ubiquinol Cytochrome C Reductase Complex Assembly Factor 3 |
| ENSMUSG00000071658 | ENSFM00500000277869 | Unknown |
| ENSMUSG00000071660 | ENSFM00600000927711 | Tetratricopeptide Repeat 9C |
| ENSMUSG00000071660 | ENSFM00670001236445 | Tetratricopeptide Repeat 9C Tpr Repeat 9C |
| ENSMUSG00000071711 | ENSFM00500000270326 | Thiosulfate Sulfurtransferase EC_2.8.1.1 Rhodanese |
| ENSMUSG00000071862 | ENSFM00400000131776 | Leucine Rich Repeat Transmembrane Neuronal Precursor |
| ENSMUSG00000071866 | ENSFM00750001632321 | Peptidyl Prolyl Cis Trans Isomerase A Ppiase A EC_5.2.1.8 Cyclophilin A Cyclosporin A Binding Rotamase A |
| ENSMUSG00000072214 | ENSFM00800002003401 | Septin |
| ENSMUSG00000072235 | ENSFM00250000000217 | Tubulin Alpha Chain |
| ENSMUSG00000072582 | ENSFM00500000273926 | Peptidyl Trna Hydrolase 2 Mitochondrial Precursor Pth 2 EC_3.1.1.29 Bcl 2 Inhibitor Of Transcription |
| ENSMUSG00000072772 | ENSFM00500000274497 | C10 |
| ENSMUSG00000072812 | ENSFM00740001589261 | Associated |
| ENSMUSG00000072825 | ENSFM00740001589275 | Unknown |
| ENSMUSG00000073002 | ENSFM00600000925372 | Vesicle Associated Membrane 5 Vamp 5 Myobrevin |
| ENSMUSG00000073131 | ENSFM00670001237448 | Vacuolar Atpase Assembly Integral Membrane VMA21 |
| ENSMUSG00000073422 | ENSFM00500000270902 | Estradiol 17 Beta Dehydrogenase 8 EC_1.1.1.62 17 Beta Hydroxysteroid Dehydrogenase 8 17 Beta Hsd 8 3 Oxoacyl Reductase EC_1.1.1.- Testosterone 17 Beta Dehydrogenase 8 EC_1.1.-.- 1 239 |
| ENSMUSG00000073435 | ENSFM00250000000895 | Nucleoside Diphosphate Kinase Ndk Ndp Kinase EC_2.7.4.6 |
| ENSMUSG00000073481 | ENSFM00500000270661 | Mitochondrial Amidoxime Reducing Component 2 Precursor MARC2 Ec 1 Molybdenum Cofactor Sulfurase C Terminal Domain Containing 2 Mosc Domain Containing 2 Moco Sulfurase C Terminal Domain Containing 2 |
| ENSMUSG00000073557 | ENSFM00500000269806 | Phosphatase 1 Regulatory Subunit 12A Myosin Phosphatase Targeting Subunit 1 Myosin Phosphatase Target Subunit 1 Phosphatase Myosin Binding Subunit |
| ENSMUSG00000073563 | ENSFM00730001521484 | Casein Kinase I Gamma Cki Gamma EC_2.7.11.1 |
| ENSMUSG00000073639 | ENSFM00500000270602 | Ras Related Rab 18 |
| ENSMUSG00000073640 | ENSFM00670001235486 | 60S Ribosomal L27 |
| ENSMUSG00000073643 | ENSFM00570000880478 | Unknown |
| ENSMUSG00000073643 | ENSFM00250000002558 | Wd Repeat And Fyve Domain Containing WD40 And Fyve Domain Containing |
| ENSMUSG00000073676 | ENSFM00670001235755 | 10 Kda Heat Shock Protein Mitochondrial HSP10.10 Kda Chaperonin Chaperonin 10 CPN10 |
| ENSMUSG00000073678 | ENSFM00460000244973 | Gpi Inositol Deacylase EC_3.1.-.- Post Gpi Attachment To Proteins Factor 1 |
| ENSMUSG00000073702 | ENSFM00780001913412 | Unknown |
| ENSMUSG00000073702 | ENSFM00670001235422 | 60S Ribosomal L31 |
| ENSMUSG00000073725 | ENSFM00250000004677 | Probable Lysosomal Cobalamin Transporter LMBR1 Domain Containing 1 |
| ENSMUSG00000073805 | ENSFM00250000008262 | Unknown |
| ENSMUSG00000073838 | ENSFM00260000050713 | Elongation Factor Tu Mitochondrial Precursor |
| ENSMUSG00000073940 | ENSFM00250000000136 | Hemoglobin Subunit Beta Beta Globin Hemoglobin Beta Chain |
| ENSMUSG00000073982 | ENSFM00730001521186 | Ras Related C3 Botulinum Toxin Substrate Precursor P21 |
| ENSMUSG00000074030 | ENSFM00250000004839 | Exocyst Complex Component 8 Exocyst Complex 84 Kda Subunit |
| ENSMUSG00000074064 | ENSFM00250000006084 | Malonyl Coa Decarboxylase Mitochondrial Precursor Mcd EC_4.1.1.9 |
| ENSMUSG00000074093 | ENSFM00680001307767 | Unknown |
| ENSMUSG00000074129 | ENSFM00790001962987 | 60S Ribosomal L13A |
| ENSMUSG00000074211 | ENSFM00670001237409 | Succinate Dehydrogenase Assembly Factor 1 Mitochondrial |
| ENSMUSG00000074212 | ENSFM00250000001153 | Dnaj Homolog Subfamily Member |
| ENSMUSG00000074305 | ENSFM00760001742183 | Unknown |
| ENSMUSG00000074305 | ENSFM00740001589410 | Pseudopodium Enriched Atypical Kinase 1 EC_2.7.10.2 Sugen Kinase 269 Tyrosine Kinase SGK269 |
| ENSMUSG00000074457 | ENSFM00570000882450 | Unknown |
| ENSMUSG00000074457 | ENSFM00600000921934 | S100 A16 S100 Calcium Binding A16 |
| ENSMUSG00000074505 | ENSFM00570000851039 | Protocadherin Fat Precursor Fat Tumor Suppressor Homolog |
| ENSMUSG00000074657 | ENSFM00670001235340 | Kinesin Heavy Chain Kinesin Heavy Chain Kinesin Heavy Chain |
| ENSMUSG00000074698 | ENSFM00570000890230 | Unknown |
| ENSMUSG00000074698 | ENSFM00250000001591 | Casein Kinase Ii Subunit Ck Ii EC_2.7.11.1 |
| ENSMUSG00000074736 | ENSFM00570000890894 | Unknown |
| ENSMUSG00000074736 | ENSFM00670001235828 | Synapse Differentiation Inducing Gene 1 Capucin Dispanin Subfamily C Member 1 DSPC1 Transmembrane 90A |
| ENSMUSG00000074746 | ENSFM00250000005716 | Pdz Domain Containing 8 Sarcoma Antigen Ny Sar 84/Ny Sar 104 |
| ENSMUSG00000074781 | ENSFM00500000270143 | Ubiquitin Conjugating Enzyme E2 N EC_6.3.2.19 Ubiquitin Carrier N Ubiquitin Ligase N |
| ENSMUSG00000074785 | ENSFM00700001407985 | Unknown |
| ENSMUSG00000074785 | ENSFM00800002003838 | Plexin C1 Precursor Virus Encoded Semaphorin Receptor CD232 Antigen |
| ENSMUSG00000074793 | ENSFM00250000005151 | Unknown |
| ENSMUSG00000074793 | ENSFM00720001503567 | Unknown |
| ENSMUSG00000074797 | ENSFM00670001235533 | Inosine Triphosphate Pyrophosphatase |
| ENSMUSG00000074886 | ENSFM00730001521304 | G Coupled Receptor Kinase G Coupled Receptor Kinase |
| ENSMUSG00000074923 | ENSFM00730001521739 | Serine/Threonine Kinase Pak EC_2.7.11.1 P21 Activated Kinase Pak |
| ENSMUSG00000075229 | ENSFM00500000273461 | Coiled Coil Domain Containing 58 |
| ENSMUSG00000075318 | ENSFM00740001589070 | Sodium Channel Type Subunit Alpha Sodium Channel Sodium Channel Type Subunit Alpha Voltage Gated Sodium Channel Subunit Alpha NAV1 |
| ENSMUSG00000075402 | ENSFM00250000000206 | Keratin Type Ii Cytoskeletal Cytokeratin Ck Keratin Type Ii Keratin |
| ENSMUSG00000075415 | ENSFM00250000000770 | Formin Binding 1 |
| ENSMUSG00000075478 | ENSFM00250000001398 | Slit And Ntrk Precursor |
| ENSMUSG00000075605 | ENSFM00670001246027 | Secreted Ly 6/Upar Related 2 |
| ENSMUSG00000075700 | ENSFM00250000003123 | Selenoprotein Precursor |
| ENSMUSG00000075701 | ENSFM00500000273466 | Selenoprotein S Sels Vcp Interacting Membrane |
| ENSMUSG00000075702 | ENSFM00670001237106 | Selenoprotein M Precursor Selm |
| ENSMUSG00000075703 | ENSFM00570000881232 | Unknown |
| ENSMUSG00000075703 | ENSFM00250000002697 | Ethanolaminephosphotransferase 1 EC_2.7.8.1 Selenoprotein I Seli |
| ENSMUSG00000075704 | ENSFM00730001521412 | Thioredoxin Reductase EC_1.8.1.9 Thioredoxin Reductase |
| ENSMUSG00000075706 | ENSFM00500000272603 | Phospholipid Hydroperoxide Glutathione Peroxidase Mitochondrial Precursor Phgpx EC_1.11.1.12 Glutathione Peroxidase 4 Gpx 4 Gshpx 4 |
| ENSMUSG00000076432 | ENSFM00730001521154 | Unknown |
| ENSMUSG00000076435 | ENSFM00760001714805 | Acyl Coa Synthetase Family Member 2 Mitochondrial Precursor EC_6.2.1.- |
| ENSMUSG00000076439 | ENSFM00380000124712 | Myelin Oligodendrocyte Glycoprotein Precursor |
| ENSMUSG00000076617 | ENSFM00740001589433 | Ig Chain C Region |
| ENSMUSG00000077450 | ENSFM00500000269744 | Ras Related Rab Precursor |
| ENSMUSG00000078137 | ENSFM00500000295837 | Ankyrin Repeat Domain Containing 63 |
| ENSMUSG00000078139 | ENSFM00500000272843 | Iron Sulfur Cluster Assembly 1 Homolog Mitochondrial Precursor Hesb Domain Containing 2 Iron Sulfur Assembly Isca |
| ENSMUSG00000078193 | ENSFM00500000271690 | 60S Ribosomal L35 |
| ENSMUSG00000078249 | ENSFM00680001326820 | Unknown |
| ENSMUSG00000078249 | ENSFM00810002041146 | High Mobility Group Hmg I/Hmg Y Hmg I Y High Mobility Group At Hook 1 High Mobility Group A1 |
| ENSMUSG00000078350 | ENSFM00720001501322 | Unknown |
| ENSMUSG00000078350 | ENSFM00720001501323 | Unknown |
| ENSMUSG00000078350 | ENSFM00720001501324 | Unknown |
| ENSMUSG00000078350 | ENSFM00720001503264 | Unknown |
| ENSMUSG00000078441 | ENSFM00740001589424 | Secretory Carrier Associated Membrane Secretory Carrier Membrane |
| ENSMUSG00000078517 | ENSFM00500000270724 | Er Membrane Complex Subunit 1 Precursor |
| ENSMUSG00000078532 | ENSFM00250000002642 | Sodium/Potassium Transporting Atpase Subunit Beta 1 Interacting Na + /K + Transporting Atpase Subunit Beta 1 Interacting |
| ENSMUSG00000078622 | ENSFM00250000004158 | Coiled Coil Domain Containing 47 Precursor |
| ENSMUSG00000078656 | ENSFM00500000271998 | Vacuolar Sorting Associated 25 Escrt Ii Complex Subunit VPS25 |
| ENSMUSG00000078695 | ENSFM00500000292816 | Cdgsh Iron Sulfur Domain Containing 3 Mitochondrial Precursor |
| ENSMUSG00000078713 | ENSFM00500000331103 | Unknown |
| ENSMUSG00000078713 | ENSFM00500000343387 | Unknown |
| ENSMUSG00000078713 | ENSFM00500000394168 | Unknown |
| ENSMUSG00000078713 | ENSFM00500000447447 | Unknown |
| ENSMUSG00000078794 | ENSFM00390000126901 | Dapper Homolog 3 Dapper Antagonist Of Catenin 3 |
| ENSMUSG00000078812 | ENSFM00680001318390 | Unknown |
| ENSMUSG00000078812 | ENSFM00250000002695 | Eukaryotic Translation Initiation Factor 5A Eif 5A Eif Eukaryotic Initiation Factor 5A |
| ENSMUSG00000078816 | ENSFM00520000517802 | Kinase C Type Pkc EC_2.7.11.13 |
| ENSMUSG00000078908 | ENSFM00770001847570 | Vacuolar Fusion MON1 |
| ENSMUSG00000078919 | ENSFM00250000004763 | Dolichol Phosphate Mannosyltransferase Subunit 1 EC_2.4.1.83 Dolichol Phosphate Mannose Synthase Subunit 1 Dpm Synthase Subunit 1 Dolichyl Phosphate Beta D Mannosyltransferase Subunit 1 Mannose P Dolichol Synthase Subunit 1 Mpd Synthase Subunit 1 |
| ENSMUSG00000078923 | ENSFM00250000001221 | Ubiquitin Conjugating Enzyme E2 Variant |
| ENSMUSG00000078974 | ENSFM00500000275476 | Unknown |
| ENSMUSG00000078974 | ENSFM00700001395304 | CRA_B |
| ENSMUSG00000079037 | ENSFM00250000000454 | Major Prion Precursor Prp PRP27 30 PRP33 35C CD230 Antigen |
| ENSMUSG00000079055 | ENSFM00710001441664 | Sodium/Calcium Exchanger 1 Precursor Na + /Ca 2+ Exchange 1 Solute Carrier Family 8 Member 1 |
| ENSMUSG00000079104 | ENSFM00250000000896 | Ribose Phosphate Pyrophosphokinase EC_2.7.6.1 Phosphoribosyl Pyrophosphate Synthase |
| ENSMUSG00000079108 | ENSFM00250000002862 | Signal Recognition Particle 54 Kda SRP54 |
| ENSMUSG00000079316 | ENSFM00570000876450 | Unknown |
| ENSMUSG00000079316 | ENSFM00750001632398 | Ras Related Rab |
| ENSMUSG00000079426 | ENSFM00250000003587 | Actin Related 2/3 Complex Subunit 4 ARP2/3 Complex 20 Kda P20 Arc |
| ENSMUSG00000079477 | ENSFM00390000126319 | Ras Related Rab |
| ENSMUSG00000079480 | ENSFM00670001235905 | Peptidyl Prolyl Cis Trans Isomerase Nima Interacting 4 EC_5.2.1.8 Parvulin 14 PAR14 Peptidyl Prolyl Cis Trans Isomerase PIN4 Ppiase PIN4 Rotamase PIN4 |
| ENSMUSG00000079481 | ENSFM00400000132091 | Nhs 2 |
| ENSMUSG00000079508 | ENSFM00500000272483 | Apolipoprotein O FAM121B |
| ENSMUSG00000079598 | ENSFM00500000286464 | C Type Lectin Domain Family 2 Member L |
| ENSMUSG00000079658 | ENSFM00760001743923 | Unknown |
| ENSMUSG00000079658 | ENSFM00500000272323 | Transcription Elongation Factor B Polypeptide 1 Elongin 15 Kda Subunit Elongin C Eloc Rna Polymerase Ii Transcription Factor Siii Subunit C Siii P15 Stromal Membrane Associated SMAP1B Homolog |
| ENSMUSG00000079677 | ENSFM00670001235975 | Adrenodoxin Protein Mitochondrial Precursor Ferredoxin 1 |
| ENSMUSG00000084845 | ENSFM00500000314906 | Transmembrane 240 |
| ENSMUSG00000086040 | ENSFM00600000921808 | Was/Wasl Interacting Family Member 3 Corticosteroids And Regional Expression 16 |
| ENSMUSG00000086784 | ENSFM00670001235660 | Isochorismatase Domain Containing |
| ENSMUSG00000087260 | ENSFM00670001236711 | Ragulator Complex LAMTOR5 Late Endosomal/Lysosomal Adaptor And Mapk And Mtor Activator 5 |
| ENSMUSG00000087408 | ENSFM00440000237016 | Ceramide Synthase 1 CERS1 LAG1 Longevity Assurance Homolog 1 Longevity Assurance Gene 1 Homolog 1 Uog 1 |
| ENSMUSG00000087408 | ENSFM00800002004042 | Embryonic Growth/Differentiation Factor 1 Precursor Gdf 1 |
| ENSMUSG00000087651 | ENSFM00610000968876 | Unknown |
| ENSMUSG00000089774 | ENSFM00250000000584 | Cotransporter Na + Cotransporter Solute Carrier Family 5 Member |
| ENSMUSG00000089847 | ENSFM00470000258105 | Unknown |
| ENSMUSG00000089847 | ENSFM00570000883024 | Unknown |
| ENSMUSG00000089847 | ENSFM00570000890077 | Unknown |
| ENSMUSG00000089847 | ENSFM00670001236558 | Mitochondrial Import Inner Membrane Translocase Subunit TIM10 B Mitochondrial Import Inner Membrane Translocase Subunit TIM9 B TIMM10B TIM10B |
| ENSMUSG00000089945 | ENSFM00740001589417 | A Kinase Anchor 2 Akap 2 Akap Kl Kinase A Anchoring 2 PRKA2 |
| ENSMUSG00000090053 | ENSFM00740001589417 | A Kinase Anchor 2 Akap 2 Akap Kl Kinase A Anchoring 2 PRKA2 |
| ENSMUSG00000090071 | ENSFM00250000002836 | Cyclin Dependent Kinase 5 Activator 1 Precursor CDK5 Activator 1 Cyclin Dependent Kinase 5 Regulatory Subunit 1 Tpkii Regulatory Subunit |
| ENSMUSG00000090150 | ENSFM00760001763413 | Unknown |
| ENSMUSG00000090150 | ENSFM00780001898384 | Acyl Coa Dehydrogenase Family Member Acad EC_1.3.99.- |
| ENSMUSG00000090523 | ENSFM00610000965985 | Unknown |
| ENSMUSG00000090523 | ENSFM00640001115365 | Unknown |
| ENSMUSG00000090523 | ENSFM00560000828284 | Glycophorin C CD236 Antigen |
| ENSMUSG00000090639 | ENSFM00250000000343 | Precursor |
| ENSMUSG00000090841 | ENSFM00250000000438 | Myosin Light Chain Myosin Light Chain Myosin Light Chain |
| ENSMUSG00000090862 | ENSFM00250000003348 | 40S Ribosomal S13 |
| ENSMUSG00000091228 | ENSFM00250000000895 | Nucleoside Diphosphate Kinase Ndk Ndp Kinase EC_2.7.4.6 |
| ENSMUSG00000091780 | ENSFM00500000270642 | Homolog Mitochondrial Precursor |
| ENSMUSG00000091971 | ENSFM00730001521103 | Heat Shock Kda Heat Shock 70 Kda |
| ENSMUSG00000092305 | ENSFM00250000000896 | Ribose Phosphate Pyrophosphokinase EC_2.7.6.1 Phosphoribosyl Pyrophosphate Synthase |
| ENSMUSG00000093904 | ENSFM00500000272385 | Mitochondrial Import Receptor Subunit TOM20 Homolog Mitochondrial 20 Kda Outer Membrane Outer Mitochondrial Membrane Receptor TOM20 |
| ENSMUSG00000093930 | ENSFM00770001847558 | Hydroxymethylglutaryl Coa Synthase Hmg Coa Synthase EC_2.3.3.10 3 Hydroxy 3 Methylglutaryl Coenzyme A Synthase |
| ENSMUSG00000094103 | ENSFM00500000273173 | Unknown |
| ENSMUSG00000094483 | ENSFM00250000002171 | Transcriptional Activator Pur Beta Purine Rich Element Binding B |
| ENSMUSG00000094626 | ENSFM00250000010793 | Cat Eye Syndrome Critical Region 6 |
| ENSMUSG00000094685 | ENSFM00250000006602 | Ragulator Complex LAMTOR3 Late Endosomal/Lysosomal Adaptor And Mapk And Mtor Activator 3 Mitogen Activated Kinase Scaffold 1 |
| ENSMUSG00000096054 | ENSFM00250000000542 | Nesprin 2 Nuclear Envelope Spectrin Repeat 2 Nucleus And Actin Connecting Element Protein Nuance Synaptic Nuclear Envelope 2 Syne 2 |
| ENSMUSG00000096188 | ENSFM00260000051403 | Cklf Marvel Transmembrane Domain Containing 4 Chemokine Factor Superfamily Member 4 |
| ENSMUSG00000098112 | ENSFM00250000000861 | Bridging Integrator |
| ENSMUSG00000098274 | ENSFM00670001235623 | 60S Ribosomal L24 |
| ENSMUSG00000098557 | ENSFM00260000050545 | Btb/Poz Domain Containing |
| ENSMUSG00000100679 | ENSFM00500000270327 | ORM1 |
| ENSMUSG00000100725 | ENSFM00760001763162 | Unknown |
| ENSMUSG00000100725 | ENSFM00250000001411 | Heterogeneous Nuclear Ribonucleoprotein U Scaffold Attachment Factor Saf |
| ENSMUSG00000102226 | ENSFM00260000050975 | Brain Enriched Guanylate Kinase Associated |
| ENSMUSG00000102543 | ENSFM00780001917496 | Unknown |
| ENSMUSG00000102543 | ENSFM00730001521313 | Protocadherin Gamma Precursor Pcdh Gamma |
| ENSMUSG00000102543 | ENSFM00780001898335 | Protocadherin Precursor |
| ENSMUSG00000102697 | ENSFM00780001898335 | Protocadherin Precursor |
| ENSMUSG00000102918 | ENSFM00780001917495 | Unknown |
| ENSMUSG00000102918 | ENSFM00730001521313 | Protocadherin Gamma Precursor Pcdh Gamma |
| ENSMUSG00000103888 | ENSFM00740001589112 | Microtubule Actin Cross Linking Factor 1 Actin Cross Linking Family 7 |
| ENSMUSG00000106379 | ENSFM00680001311925 | Unknown |
| ENSMUSG00000106379 | ENSFM00250000002406 | Lipoma Hmgic Fusion Partner |
| ENSMUSG00000107283 | ENSFM00500000272108 | MPV17 |
Supplementary Table 4b. Ensemble Families from mouse postsynaptic density proteins
For each mouse postsynaptic density protein its corresponding Ensembl Family code and descriptor are given.
| Ensembl Gene Id | Ensembl Protein Family Id(S) | Ensembl Family Description |
|---|---|---|
| ENSMUSG00000000001 | ENSFM00410000138458 | Guanine Nucleotide Binding Subunit |
| ENSMUSG00000000149 | ENSFM00730001522052 | Guanine Nucleotide Binding Subunit Alpha G Alpha G Subunit Alpha |
| ENSMUSG00000000202 | ENSFM00440000236967 | Btb/Poz Domain Containing 17 Precursor Galectin 3 Binding |
| ENSMUSG00000000263 | ENSFM00400000131715 | Glycine Receptor Subunit Precursor Glycine Receptor Kda Subunit |
| ENSMUSG00000000276 | ENSFM00730001522404 | Diacylglycerol Kinase Epsilon Dag Kinase Epsilon EC_2.7.1.107 Diglyceride Kinase Epsilon Dgk Epsilon |
| ENSMUSG00000000296 | ENSFM00810002041019 | Tumor D53 Tumor D52 1 |
| ENSMUSG00000000305 | ENSFM00390000130953 | Unknown |
| ENSMUSG00000000305 | ENSFM00730001521172 | Cadherin Precursor Cadherin Cadherin |
| ENSMUSG00000000308 | ENSFM00570000898238 | Unknown |
| ENSMUSG00000000308 | ENSFM00570000900073 | Unknown |
| ENSMUSG00000000308 | ENSFM00800002003437 | Creatine Kinase U Type Mitochondrial Precursor EC_2.7.3.2 Acidic Type Mitochondrial Creatine Kinase Mia Ck Ubiquitous Mitochondrial Creatine Kinase U Mtck |
| ENSMUSG00000000325 | ENSFM00570000877506 | Unknown |
| ENSMUSG00000000325 | ENSFM00730001521847 | Catenin Delta 1 Cadherin Associated Src Substrate Cas P120 Catenin P120 Ctn P120 Cas |
| ENSMUSG00000000339 | ENSFM00570000884353 | Unknown |
| ENSMUSG00000000339 | ENSFM00250000005769 | Rna 3' Terminal Phosphate Cyclase Rna Cyclase Rna 3' Phosphate Cyclase EC_6.5.1.4 Rna Terminal Phosphate Cyclase Domain Containing 1 |
| ENSMUSG00000000340 | ENSFM00800002024492 | Unknown |
| ENSMUSG00000000340 | ENSFM00800002024493 | Unknown |
| ENSMUSG00000000340 | ENSFM00250000003682 | Lipoamide Acyltransferase Component Of Branched Chain Alpha Keto Acid Dehydrogenase Complex Mitochondrial Precursor EC_2.3.1.168 Branched Chain Alpha Keto Acid Dehydrogenase Complex Component E2 Bckad E2 BCKADE2 Dihydrolipoamide Acetyltransferase Componen |
| ENSMUSG00000000374 | ENSFM00730001522435 | Trafficking Particle Complex Subunit 10 Trafficking Particle Complex Subunit TMEM1 Transport Particle Subunit TMEM1 Trapp Subunit TMEM1 |
| ENSMUSG00000000378 | ENSFM00500000271756 | Malcavernin Cerebral Cavernous Malformations 2 |
| ENSMUSG00000000399 | ENSFM00250000004385 | Nadh Dehydrogenase 1 Alpha Subcomplex Subunit 9 Mitochondrial Precursor Complex I 39KD Ci 39KD Nadh Ubiquinone Oxidoreductase 39 Kda Subunit |
| ENSMUSG00000000416 | ENSFM00600000921238 | Cortactin Binding 2 CORTBP2 |
| ENSMUSG00000000531 | ENSFM00500000272853 | General Receptor Phosphoinositides 1 Associated Scaffold GRP1 Associated Scaffold |
| ENSMUSG00000000538 | ENSFM00250000001136 | TOM1 2 Target Of Myb 2 |
| ENSMUSG00000000560 | ENSFM00800002009455 | Unknown |
| ENSMUSG00000000560 | ENSFM00800002003385 | Gamma Aminobutyric Acid Receptor Subunit Alpha Precursor Gaba A Receptor Subunit Alpha |
| ENSMUSG00000000568 | ENSFM00610000971968 | Unknown |
| ENSMUSG00000000568 | ENSFM00800002023620 | Unknown |
| ENSMUSG00000000568 | ENSFM00500000269788 | Heterogeneous Nuclear Ribonucleoprotein D Hnrnp D Hnrnp Dl |
| ENSMUSG00000000600 | ENSFM00250000003734 | Krev Interaction Trapped 1 Krev Interaction Trapped 1 Cerebral Cavernous Malformations 1 |
| ENSMUSG00000000627 | ENSFM00480000262746 | Semaphorin Precursor |
| ENSMUSG00000000628 | ENSFM00610000970732 | Unknown |
| ENSMUSG00000000628 | ENSFM00250000000532 | Hexokinase EC_2.7.1.1 Hexokinase Type Hk |
| ENSMUSG00000000631 | ENSFM00610000973552 | Unknown |
| ENSMUSG00000000631 | ENSFM00810002043502 | Unconventional Myosin Xviiia |
| ENSMUSG00000000631 | ENSFM00400000131775 | Unconventional Myosin Xviiia Molecule Associated With JAK3 N Terminus Majn Myosin Containing A Pdz Domain |
| ENSMUSG00000000740 | ENSFM00500000270284 | 60S Ribosomal L13 |
| ENSMUSG00000000743 | ENSFM00600000921337 | Charged Multivesicular Body 1B Chromatin Modifying 1B CHMP1B |
| ENSMUSG00000000759 | ENSFM00610000967253 | Unknown |
| ENSMUSG00000000759 | ENSFM00610000969293 | Unknown |
| ENSMUSG00000000759 | ENSFM00250000005014 | Gamma Tubulin Complex Component 3 Gamma Ring Complex |
| ENSMUSG00000000787 | ENSFM00770001847530 | Atp Dependent Rna Helicase DDX3Y EC_3.6.4.13 Dead Box 3 Y Chromosomal |
| ENSMUSG00000000805 | ENSFM00570000889881 | Unknown |
| ENSMUSG00000000805 | ENSFM00670001235760 | Carbonic Anhydrase 4 Precursor EC_4.2.1.1 Carbonate Dehydratase Iv Carbonic Anhydrase Iv Ca Iv |
| ENSMUSG00000000827 | ENSFM00740001613634 | Unknown |
| ENSMUSG00000000827 | ENSFM00500000270547 | Tumor D54 Tumor D52 2 |
| ENSMUSG00000000838 | ENSFM00250000000920 | Fragile X Mental Retardation 1 |
| ENSMUSG00000000881 | ENSFM00250000000275 | Disks Large Homolog Synapse Associated Sap |
| ENSMUSG00000000915 | ENSFM00250000001308 | Huntingtin Interacting 1 Hip 1 Huntingtin Interacting I Hip I |
| ENSMUSG00000001016 | ENSFM00250000003528 | Interleukin Enhancer Binding Factor 2 |
| ENSMUSG00000001052 | ENSFM00750001632389 | Transport SEC24A SEC24 Related A |
| ENSMUSG00000001062 | ENSFM00740001589527 | VPS9 Domain Containing 1 |
| ENSMUSG00000001089 | ENSFM00610000966722 | Unknown |
| ENSMUSG00000001089 | ENSFM00610000973587 | Unknown |
| ENSMUSG00000001089 | ENSFM00800002003954 | Leucine Zipper 1 Leucine Zipper Motif Containing |
| ENSMUSG00000001120 | ENSFM00250000000747 | Poly Rc Binding Alpha Heterogeneous Nuclear Ribonucleoprotein Hnrnp |
| ENSMUSG00000001127 | ENSFM00290000065522 | Serine/Threonine Kinase EC_2.7.11.1 Proto Oncogene Raf |
| ENSMUSG00000001229 | ENSFM00250000001158 | Dipeptidyl Peptidase 9 DP9 EC_3.4.14.5 Dipeptidyl Peptidase Ix Dpp Ix Dipeptidyl Peptidase 9 DPLP9 |
| ENSMUSG00000001323 | ENSFM00500000271489 | Serine Racemase EC_4.3.1.17 EC_4.3.1.- 18 EC_5.1.-.- 1 18 D Serine Ammonia Lyase D Serine Dehydratase L Serine Ammonia Lyase L Serine Dehydratase |
| ENSMUSG00000001416 | ENSFM00610000969837 | Unknown |
| ENSMUSG00000001416 | ENSFM00500000270039 | T Complex 1 Subunit Gamma Tcp 1 Gamma Cct Gamma |
| ENSMUSG00000001424 | ENSFM00610000970527 | Unknown |
| ENSMUSG00000001424 | ENSFM00250000002294 | Staphylococcal Nuclease Domain Containing 1.100 Kda Coactivator P100 Co Activator |
| ENSMUSG00000001473 | ENSFM00250000000193 | Tubulin Beta Chain |
| ENSMUSG00000001525 | ENSFM00640001119932 | Unknown |
| ENSMUSG00000001525 | ENSFM00250000000193 | Tubulin Beta Chain |
| ENSMUSG00000001552 | ENSFM00250000001012 | Catenin Beta Beta Catenin |
| ENSMUSG00000001687 | ENSFM00500000272867 | Ubiquitin 3 Precursor Membrane Anchored Ubiquitin Fold Mub |
| ENSMUSG00000001707 | ENSFM00250000008520 | Eukaryotic Translation Elongation Factor 1 Epsilon 1 Elongation Factor P18 Multisynthase Complex Auxiliary Component P18 |
| ENSMUSG00000001783 | ENSFM00250000005120 | Trna Splicing Ligase Rtcb Homolog |
| ENSMUSG00000001794 | ENSFM00250000000112 | Calpain Subunit Calcium Activated Neutral Proteinase Canp Calpain Subunit |
| ENSMUSG00000001829 | ENSFM00430000230197 | Caseinolytic Peptidase B Homolog Suppressor Of Potassium Transport Defect 3 |
| ENSMUSG00000001833 | ENSFM00760001714685 | Septin 7 CDC10 Homolog |
| ENSMUSG00000001847 | ENSFM00730001521186 | Ras Related C3 Botulinum Toxin Substrate Precursor P21 |
| ENSMUSG00000001964 | ENSFM00570000893147 | Unknown |
| ENSMUSG00000001964 | ENSFM00250000010495 | Emerin |
| ENSMUSG00000001985 | ENSFM00720001492013 | Glutamate Receptor Ionotropic Kainate Precursor Glutamate Receptor |
| ENSMUSG00000001986 | ENSFM00720001492020 | Glutamate Receptor Precursor Glur Ampa Selective Glutamate Receptor Glur Glutamate Receptor Ionotropic Ampa |
| ENSMUSG00000001995 | ENSFM00730001521477 | Signal Induced Proliferation Associated 1 SIPA1 |
| ENSMUSG00000002006 | ENSFM00250000001242 | E3 Ubiquitin Ligase PDZRN3 EC_6.3.2.- Pdz Domain Containing Ring Finger 3 Semaphorin Cytoplasmic Domain Associated 3 SEMACAP3 |
| ENSMUSG00000002010 | ENSFM00770001847549 | Isocitrate Dehydrogenase Gamma Mitochondrial Precursor EC_1.1.1.41 Isocitric Dehydrogenase Subunit Gamma Nad + Specific Icdh Subunit Gamma |
| ENSMUSG00000002014 | ENSFM00250000005113 | Translocon Associated Subunit Delta Precursor Trap Delta Signal Sequence Receptor Subunit Delta Ssr Delta |
| ENSMUSG00000002015 | ENSFM00670001235596 | B Cell Receptor Associated Bcr Associated |
| ENSMUSG00000002017 | ENSFM00250000002232 | FAM98A |
| ENSMUSG00000002064 | ENSFM00250000003961 | Stromal Cell Derived Factor 2 1 Precursor SDF2 1 |
| ENSMUSG00000002102 | ENSFM00550000743154 | 26S Protease Regulatory Subunit 6A 26S Proteasome Aaa Atpase Subunit RPT5 Proteasome 26S Subunit Atpase 3 Tat Binding Tbp |
| ENSMUSG00000002107 | ENSFM00720001497880 | Unknown |
| ENSMUSG00000002107 | ENSFM00730001521889 | Cugbp Elav Family Member 2 Celf 2 Bruno 3 Cug Triplet Repeat Rna Binding 2 Cug BP2 Cug Bp And Etr 3 Factor 2 Elav Type Rna Binding 3 Etr 3 Rna Binding Brunol 3 |
| ENSMUSG00000002341 | ENSFM00730001521436 | Core Precursor Chondroitin Sulfate Proteoglycan |
| ENSMUSG00000002346 | ENSFM00500000271356 | Mitochondrial Coenzyme A Transporter Sl Solute Carrier Family 25 Member 42 |
| ENSMUSG00000002416 | ENSFM00670001236841 | Nadh Dehydrogenase 1 Beta Subcomplex Subunit 2 Mitochondrial Precursor Complex I Aggg Ci Aggg Nadh Ubiquinone Oxidoreductase Aggg Subunit |
| ENSMUSG00000002458 | ENSFM00500000446065 | Unknown |
| ENSMUSG00000002458 | ENSFM00570000898922 | Unknown |
| ENSMUSG00000002458 | ENSFM00480000262761 | Regulator Of G Signaling |
| ENSMUSG00000002459 | ENSFM00610000972139 | Unknown |
| ENSMUSG00000002459 | ENSFM00480000262761 | Regulator Of G Signaling |
| ENSMUSG00000002496 | ENSFM00250000003183 | Tuberin Tuberous Sclerosis 2 |
| ENSMUSG00000002504 | ENSFM00740001589334 | Na + /H + Exchange Regulatory Cofactor Nhe RF1 Nherf 1 Ezrin Radixin Moesin Binding Phosphoprotein 50 EBP50 Regulatory Cofactor Of Na + /H + Exchanger Sodium Hydrogen Exchanger Regulatory Factor 1 Solute Carrier Family 9 A3 Regulatory Factor 1 |
| ENSMUSG00000002731 | ENSFM00600000921352 | Risc Loading Complex Subunit TARBP2 |
| ENSMUSG00000002741 | ENSFM00810002040913 | Synaptobrevin Homolog YKT6 Precursor EC_2.3.1.- |
| ENSMUSG00000002771 | ENSFM00570000882063 | Unknown |
| ENSMUSG00000002771 | ENSFM00730001521500 | Glutamate Receptor Ionotropic Nmda Precursor Glutamate Receptor Subunit Epsilon N Methyl D Aspartate Receptor Subtype |
| ENSMUSG00000002808 | ENSFM00250000006488 | Mammalian Ependymin Related 1 Precursor Merp 1 |
| ENSMUSG00000002812 | ENSFM00730001522115 | Flightless 1 |
| ENSMUSG00000002948 | ENSFM00790001963074 | Dual Specificity Mitogen Activated Kinase Kinase 7 Map Kinase Kinase 7 Mapkk 7 EC_2.7.12.2 Jnk Activating Kinase 2 Mapk/Erk Kinase 7 Mek 7 C Jun N Terminal Kinase Kinase 2 Jnk Kinase 2 Jnkk 2 |
| ENSMUSG00000002949 | ENSFM00500000271055 | Mitochondrial Import Inner Membrane Translocase Subunit Precursor |
| ENSMUSG00000002957 | ENSFM00810002040615 | Ap 2 Complex Subunit Alpha 2.100 Kda Coated Vesicle C Adaptor Complex Ap 2 Subunit Alpha 2 Adaptor Related Complex 2 Subunit Alpha 2 Alpha Adaptin C ALPHA2 Adaptin Clathrin Assembly Complex 2 Alpha C Large Chain Plasma Membrane Adaptor HA2/AP2 Adaptin Alp |
| ENSMUSG00000002984 | ENSFM00640001114360 | Unknown |
| ENSMUSG00000002984 | ENSFM00250000002521 | Mitochondrial Import Receptor Subunit TOM40 Homolog Translocase Of Outer Membrane 40 Kda Subunit Homolog |
| ENSMUSG00000002997 | ENSFM00730001521993 | Camp Dependent Kinase Type Ii Regulatory Subunit |
| ENSMUSG00000003033 | ENSFM00730001521549 | Ap 1 Complex Subunit Mu 1 Ap Mu Chain Family Member MU1A Adaptor Complex Ap 1 Subunit Mu 1 Adaptor Related Complex 1 Subunit Mu 1 Clathrin Assembly Complex 1 Mu 1 Medium Chain 1 Golgi Adaptor HA1/AP1 Adaptin Mu 1 Subunit Mu Adaptin 1 MU1A Adaptin |
| ENSMUSG00000003037 | ENSFM00730001521184 | Ras Related Rab |
| ENSMUSG00000003131 | ENSFM00250000004414 | Platelet Activating Factor Acetylhydrolase Ib Subunit Beta EC_3.1.1.47 Paf Acetylhydrolase 30 Kda Subunit Paf Ah 30 Kda Subunit Paf Ah Subunit Beta Pafah Subunit Beta |
| ENSMUSG00000003178 | ENSFM00250000000908 | Methionine Sulfoxide Oxidase EC_1.14.13.- Molecule Interacting With Casl Mical |
| ENSMUSG00000003269 | ENSFM00250000001040 | Cytohesin Ph SEC7 And Coiled Coil Domain Containing |
| ENSMUSG00000003279 | ENSFM00250000000718 | Disks Large Associated Dap Psd 95/SAP90 Binding SAP90/Psd 95 Associated |
| ENSMUSG00000003345 | ENSFM00730001521484 | Casein Kinase I Gamma Cki Gamma EC_2.7.11.1 |
| ENSMUSG00000003352 | ENSFM00250000000524 | Voltage Dependent L Type Calcium Channel Subunit Beta Calcium Channel Voltage Dependent Subunit Beta |
| ENSMUSG00000003363 | ENSFM00570000879838 | Unknown |
| ENSMUSG00000003363 | ENSFM00570000888995 | Unknown |
| ENSMUSG00000003363 | ENSFM00250000001612 | Phospholipase D3 Pld 3 EC_3.1.4.4 Choline Phosphatase 3 Phosphatidylcholine Hydrolyzing Phospholipase D3 |
| ENSMUSG00000003378 | ENSFM00720001492013 | Glutamate Receptor Ionotropic Kainate Precursor Glutamate Receptor |
| ENSMUSG00000003410 | ENSFM00250000000588 | Elav |
| ENSMUSG00000003429 | ENSFM00800002003484 | 40S Ribosomal S11 |
| ENSMUSG00000003469 | ENSFM00250000003309 | Phytanoyl Coa Hydroxylase Interacting Phytanoyl Coa Hydroxylase Associated 1 Pahx AP1 PAHXAP1 |
| ENSMUSG00000003518 | ENSFM00660001182137 | Unknown |
| ENSMUSG00000003518 | ENSFM00500000272272 | Dual Specificity Phosphatase 3 EC_3.1.3.16 EC_3.1.3.- 48 Vaccinia H1 Related Phosphatase Vhr |
| ENSMUSG00000003528 | ENSFM00730001522086 | Tricarboxylate Transport Protein Mitochondrial Precursor Citrate Transport Ctp Solute Carrier Family 25 Member 1 Tricarboxylate Carrier |
| ENSMUSG00000003573 | ENSFM00680001319354 | Unknown |
| ENSMUSG00000003573 | ENSFM00250000001140 | Homer Homolog Homer Vasp/Ena Related Gene Up Regulated During Seizure And Ltp Vesl |
| ENSMUSG00000003644 | ENSFM00570000851006 | Ribosomal S6 Kinase Alpha S6K Alpha EC_2.7.11.1.90 Kda Ribosomal S6 Kinase P90 Rsk Map Kinase Activated Kinase Mapk Activated Kinase Mapkap Kinase Mapkapk Ribosomal S6 Kinase Rsk |
| ENSMUSG00000003779 | ENSFM00730001521759 | Kinesin KIF20B Kinesin Related Motor Interacting With PIN1 M Phase Phosphoprotein 1 MPP1 |
| ENSMUSG00000003808 | ENSFM00570000893661 | Unknown |
| ENSMUSG00000003808 | ENSFM00250000004351 | Phenylalanine Trna Ligase Alpha Subunit EC_6.1.1.20 Phenylalanyl Trna Synthetase Alpha Subunit Phers |
| ENSMUSG00000003809 | ENSFM00570000886815 | Unknown |
| ENSMUSG00000003809 | ENSFM00380000124741 | Glutaryl Coa Dehydrogenase Mitochondrial Precursor Gcd EC_1.3.8.6 |
| ENSMUSG00000003863 | ENSFM00250000000441 | Liprin Alpha Tyrosine Phosphatase Receptor Type F Polypeptide Interacting Alpha Ptprf Interacting Alpha |
| ENSMUSG00000003872 | ENSFM00260000050595 | Lin 7 Homolog Lin Mammalian Lin Seven Mals Vertebrate Lin 7 Homolog Veli |
| ENSMUSG00000003923 | ENSFM00250000006597 | Transcription Factor A Mitochondrial Precursor Mttfa |
| ENSMUSG00000003955 | ENSFM00500000274855 | FAM162A E2 Induced Gene 5 Growth And Transformation Dependent Hgtd P |
| ENSMUSG00000003970 | ENSFM00250000002389 | 60S Ribosomal |
| ENSMUSG00000003974 | ENSFM00790001963004 | Metabotropic Glutamate Receptor Precursor |
| ENSMUSG00000004031 | ENSFM00250000001886 | Bmp/Retinoic Acid Inducible Neural Specific 1 Precursor Deleted In Bladder Cancer 1 |
| ENSMUSG00000004069 | ENSFM00730001522148 | Dnaj Homolog Subfamily A Member 3 Mitochondrial Precursor Dnaj Tid 1 1 Tumorous Imaginal Discs TID56 Homolog |
| ENSMUSG00000004113 | ENSFM00300000085455 | N Type Calcium Channel Pore Forming Subunit Alpha 1B |
| ENSMUSG00000004113 | ENSFM00740001589100 | Voltage Dependent Type Calcium Channel Subunit Alpha Brain Calcium Channel Calcium Channel L Type Alpha 1 Polypeptide Voltage Gated Calcium Channel Subunit Alpha CAV2 |
| ENSMUSG00000004113 | ENSFM00700001391361 | Voltage N Type Calcium Channel |
| ENSMUSG00000004264 | ENSFM00800002003689 | Prohibitin 2 |
| ENSMUSG00000004285 | ENSFM00570000892542 | Unknown |
| ENSMUSG00000004285 | ENSFM00670001235732 | V Type Proton Atpase Subunit F V Atpase Subunit F V Atpase 14 Kda Subunit Vacuolar Proton Pump Subunit F |
| ENSMUSG00000004364 | ENSFM00610000969310 | Unknown |
| ENSMUSG00000004364 | ENSFM00610000969311 | Unknown |
| ENSMUSG00000004364 | ENSFM00550000764342 | Cullin 3 |
| ENSMUSG00000004364 | ENSFM00730001521665 | Cullin 3 Cul 3 |
| ENSMUSG00000004366 | ENSFM00500000273500 | Somatostatin Precursor |
| ENSMUSG00000004451 | ENSFM00500000270233 | Ras Related Precursor |
| ENSMUSG00000004455 | ENSFM00800002023656 | Unknown |
| ENSMUSG00000004455 | ENSFM00800002023657 | Unknown |
| ENSMUSG00000004455 | ENSFM00800002023658 | Unknown |
| ENSMUSG00000004455 | ENSFM00420000140518 | Serine/Threonine Phosphatase PP1 Catalytic Subunit Pp EC_3.1.3.16 |
| ENSMUSG00000004460 | ENSFM00500000271339 | Dnaj Homolog Subfamily B Member 11 Precursor Er Associated Dnaj Er Associated HSP40 Co Chaperone Endoplasmic Reticulum Dna J Domain Containing 3 Er Resident ERDJ3 ERDJ3 ERJ3P |
| ENSMUSG00000004530 | ENSFM00610000967749 | Unknown |
| ENSMUSG00000004530 | ENSFM00730001521292 | Coronin |
| ENSMUSG00000004565 | ENSFM00760001714681 | Neuropathy Target Esterase Sws |
| ENSMUSG00000004631 | ENSFM00250000002718 | Alpha Sarcoglycan Precursor Alpha Sg 50 Kda Dystrophin Associated Glycoprotein 50DAG Adhalin |
| ENSMUSG00000004637 | ENSFM00480000262794 | Ww Domain Containing Oxidoreductase EC_1.1.1.- |
| ENSMUSG00000004655 | ENSFM00750001632461 | Aquaporin 1 Aqp 1 Aquaporin Chip Water Channel Red Blood Cells And Kidney Proximal Tubule |
| ENSMUSG00000004789 | ENSFM00800002003585 | Dihydrolipoyllysine Residue Succinyltransferase Component Of 2 Oxoglutarate Dehydrogenase Complex Mitochondrial Precursor EC_2.3.1.61 2 Oxoglutarate Dehydrogenase Complex Component E2 Ogdc E2 Dihydrolipoamide Succinyltransferase Component Of 2 Oxoglutarat |
| ENSMUSG00000004843 | ENSFM00500000272414 | Charged Multivesicular Body 2B Chromatin Modifying 2B CHMP2B |
| ENSMUSG00000004849 | ENSFM00570000887691 | Unknown |
| ENSMUSG00000004849 | ENSFM00500000269841 | Ap 1 Complex Subunit Sigma Adaptor Complex Ap 1 Subunit Sigma Adaptor Related Complex 1 Subunit Sigma Clathrin Assembly Complex 1 Sigma Small Chain Golgi Adaptor HA1/AP1 Adaptin Sigma Subunit Sigma Subunit Of Ap 1 Clathrin Sigma Adaptin Adaptin |
| ENSMUSG00000004892 | ENSFM00730001521436 | Core Precursor Chondroitin Sulfate Proteoglycan |
| ENSMUSG00000004894 | ENSFM00590000918422 | Unknown |
| ENSMUSG00000004894 | ENSFM00730001521886 | Hyaluronan And Proteoglycan Link 1 Precursor Cartilage Linking 1 Cartilage Link Proteoglycan Link |
| ENSMUSG00000004902 | ENSFM00500000270170 | Mitochondrial Glutamate Carrier 1 Gc 1 Glutamate/H + Symporter 1 Solute Carrier Family 25 Member 22 |
| ENSMUSG00000004933 | ENSFM00730001522042 | Tyrosine Kinase Csk EC_2.7.10.2 C Src Kinase |
| ENSMUSG00000004936 | ENSFM00780001898380 | Dual Specificity Mitogen Activated Kinase Kinase 1 Map Kinase Kinase 1 Mapkk 1 EC_2.7.12.2 Erk Activator Kinase 1 Mapk/Erk Kinase 1 |
| ENSMUSG00000004951 | ENSFM00500000270924 | Heat Shock Beta 1 HSPB1 Heat Shock 27 Kda Hsp 27 |
| ENSMUSG00000004961 | ENSFM00720001492031 | Synaptotagmin |
| ENSMUSG00000004980 | ENSFM00270000056431 | Heterogeneous Nuclear Ribonucleoprotein |
| ENSMUSG00000005034 | ENSFM00790001962972 | Camp Dependent Kinase Catalytic Subunit Pka C EC_2.7.11.11 |
| ENSMUSG00000005069 | ENSFM00250000001819 | Peroxisomal Targeting Signal 1 Receptor PTS1 Receptor PTS1R PTS1 Bp Peroxin 5 Peroxisomal C Terminal Targeting Signal Import Receptor Peroxisome Receptor 1 |
| ENSMUSG00000005161 | ENSFM00250000000757 | Thioredoxin Peroxidase Thioredoxin Dependent Peroxide Reductase |
| ENSMUSG00000005360 | ENSFM00800002003375 | Excitatory Amino Acid Transporter Sodium Dependent Glutamate/Aspartate Transporter Solute Carrier Family 1 Member |
| ENSMUSG00000005374 | ENSFM00570000897418 | Unknown |
| ENSMUSG00000005374 | ENSFM00810002057469 | Unknown |
| ENSMUSG00000005374 | ENSFM00250000006389 | Transducin Beta 2 |
| ENSMUSG00000005417 | ENSFM00600000921208 | Trio And F Actin Binding Protein Tara Trio Associated Repeat On Actin |
| ENSMUSG00000005469 | ENSFM00790001962972 | Camp Dependent Kinase Catalytic Subunit Pka C EC_2.7.11.11 |
| ENSMUSG00000005483 | ENSFM00440000236855 | Dnaj Homolog Subfamily B Member |
| ENSMUSG00000005510 | ENSFM00790001963038 | Nadh Dehydrogenase Iron Sulfur 3 Mitochondrial Precursor EC_1.6.5.3 EC_1.6.99.- 3 Complex I 30KD Ci 30KD Nadh Ubiquinone Oxidoreductase 30 Kda Subunit |
| ENSMUSG00000005610 | ENSFM00590000918294 | Unknown |
| ENSMUSG00000005610 | ENSFM00320000100157 | Eukaryotic Translation Initiation Factor 4 Gamma 2 Eif 4 Gamma 2 Eif 4G 2 EIF4G 2 Apobec 1 Target 1 Translation Repressor NAT1 P97 |
| ENSMUSG00000005674 | ENSFM00700001403680 | Unknown |
| ENSMUSG00000005674 | ENSFM00250000002521 | Mitochondrial Import Receptor Subunit TOM40 Homolog Translocase Of Outer Membrane 40 Kda Subunit Homolog |
| ENSMUSG00000005686 | ENSFM00770001847533 | Amp Deaminase EC_3.5.4.6 Amp Deaminase |
| ENSMUSG00000005779 | ENSFM00800002003556 | Proteasome Subunit Beta Type 4 Precursor EC_3.4.25.1 Macropain Beta Chain Multicatalytic Endopeptidase Complex Beta Chain Proteasome Beta Chain Proteasome Chain 3 |
| ENSMUSG00000005803 | ENSFM00660001169747 | Unknown |
| ENSMUSG00000005803 | ENSFM00660001169748 | Unknown |
| ENSMUSG00000005803 | ENSFM00810002040854 | Sulfide:Quinone Oxidoreductase Mitochondrial Precursor Sqor EC_1.8.5.- |
| ENSMUSG00000005871 | ENSFM00610000963080 | Unknown |
| ENSMUSG00000005871 | ENSFM00610000970304 | Unknown |
| ENSMUSG00000005871 | ENSFM00250000001676 | Adenomatous Polyposis Coli |
| ENSMUSG00000006169 | ENSFM00500000270815 | Clathrin Interactor 1 Enthoprotin Epsin 4 Epsin Related Epsinr |
| ENSMUSG00000006205 | ENSFM00250000001030 | Serine Protease HTRA2 Mitochondrial |
| ENSMUSG00000006273 | ENSFM00810002040578 | V Type Proton Atpase Subunit B V Atpase Subunit B Endomembrane Proton Pump 58 Kda Subunit Vacuolar Proton Pump Subunit B |
| ENSMUSG00000006276 | ENSFM00250000001830 | Epidermal Growth Factor Receptor Substrate 15 1 EPS15 Related EPS15R |
| ENSMUSG00000006304 | ENSFM00760001763096 | Unknown |
| ENSMUSG00000006304 | ENSFM00800002003702 | Actin Related 2/3 Complex Subunit 2 ARP2/3 Complex 34 Kda Subunit P34 Arc |
| ENSMUSG00000006333 | ENSFM00500000295123 | Unknown |
| ENSMUSG00000006333 | ENSFM00570000893535 | Unknown |
| ENSMUSG00000006333 | ENSFM00250000003284 | 40S Ribosomal S9 |
| ENSMUSG00000006356 | ENSFM00740001589457 | Cysteine Rich Crp |
| ENSMUSG00000006456 | ENSFM00350000105492 | Rna Binding 14 Rna Binding Motif 14 |
| ENSMUSG00000006464 | ENSFM00250000005034 | Bardet Biedl Syndrome 1 BBS2 2 |
| ENSMUSG00000006476 | ENSFM00570000884423 | Unknown |
| ENSMUSG00000006476 | ENSFM00250000002451 | Nmda Receptor Synaptonuclear Signaling And Neuronal Migration Factor Juxtasynaptic Attractor Of Caldendrin On Dendritic Boutons Jacob Nasal Embryonic Luteinizing Hormone Releasing Hormone Factor Nasal Embryonic Lhrh Factor |
| ENSMUSG00000006498 | ENSFM00610000973070 | Unknown |
| ENSMUSG00000006498 | ENSFM00610000973071 | Unknown |
| ENSMUSG00000006498 | ENSFM00610000973072 | Unknown |
| ENSMUSG00000006498 | ENSFM00260000050389 | Polypyrimidine Tract Binding |
| ENSMUSG00000006522 | ENSFM00250000000536 | Inter Alpha Trypsin Inhibitor Heavy Chain Precursor Iti Heavy Chain Iti Inter Alpha Inhibitor Heavy Chain |
| ENSMUSG00000006527 | ENSFM00260000050568 | Scm With Four Mbt Domains |
| ENSMUSG00000006574 | ENSFM00730001521491 | Anion Exchange Ae Anion Exchanger Solute Carrier Family 4 Member |
| ENSMUSG00000006575 | ENSFM00250000002370 | Run Domain Containing |
| ENSMUSG00000006731 | ENSFM00250000004460 | Beta 1 4 N Acetylgalactosaminyltransferase 1 EC_2.4.1.92 N Acetylneuraminyl Galactosylglucosylceramide GM2/GD2 Synthase Galnac T |
| ENSMUSG00000006740 | ENSFM00670001235340 | Kinesin Heavy Chain Kinesin Heavy Chain Kinesin Heavy Chain |
| ENSMUSG00000006782 | ENSFM00740001589364 | 2' 3' Cyclic Nucleotide 3' Phosphodiesterase Precursor Cnp Cnpase EC_3.1.4.37 |
| ENSMUSG00000006930 | ENSFM00500000273792 | Huntingtin Associated 1 Hap 1 |
| ENSMUSG00000006932 | ENSFM00570000883608 | Unknown |
| ENSMUSG00000006932 | ENSFM00610000969916 | Unknown |
| ENSMUSG00000006932 | ENSFM00250000001012 | Catenin Beta Beta Catenin |
| ENSMUSG00000007021 | ENSFM00670001235552 | Synaptogyrin |
| ENSMUSG00000007029 | ENSFM00250000001468 | Valine Trna Ligase EC_6.1.1.9 Valyl Trna Synthetase Valrs |
| ENSMUSG00000007036 | ENSFM00640001113079 | Unknown |
| ENSMUSG00000007036 | ENSFM00250000005259 | Abhydrolase Domain Containing 16A Ec 3 Hla B Associated Transcript 5 |
| ENSMUSG00000007411 | ENSFM00730001521220 | Kinase EC_2.7.11.1 |
| ENSMUSG00000007476 | ENSFM00250000001418 | Volume Regulated Anion Channel Subunit Leucine Rich Repeat Containing |
| ENSMUSG00000007564 | ENSFM00640001117298 | Unknown |
| ENSMUSG00000007564 | ENSFM00250000001941 | Serine/Threonine Phosphatase 2A 65 Kda Regulatory Subunit A PP2A Subunit A PR65 PP2A Subunit A R1 |
| ENSMUSG00000007594 | ENSFM00730001521886 | Hyaluronan And Proteoglycan Link 1 Precursor Cartilage Linking 1 Cartilage Link Proteoglycan Link |
| ENSMUSG00000007617 | ENSFM00250000001140 | Homer Homolog Homer Vasp/Ena Related Gene Up Regulated During Seizure And Ltp Vesl |
| ENSMUSG00000007653 | ENSFM00400000131718 | Gamma Aminobutyric Acid Receptor Subunit Beta Precursor Gaba A Receptor Subunit Beta |
| ENSMUSG00000007739 | ENSFM00640001120031 | Unknown |
| ENSMUSG00000007739 | ENSFM00640001120032 | Unknown |
| ENSMUSG00000007739 | ENSFM00730001521612 | T Complex 1 Subunit Delta Tcp 1 Delta Cct Delta |
| ENSMUSG00000007892 | ENSFM00810002041089 | 60S Acidic Ribosomal P1 |
| ENSMUSG00000007944 | ENSFM00670001235990 | Tetratricopeptide Repeat Tpr Repeat |
| ENSMUSG00000008036 | ENSFM00670001235578 | Ap 2 Complex Subunit Sigma Adaptor Complex Ap 2 Subunit Sigma Adaptor Related Complex 2 Subunit Sigma Clathrin Assembly 2 Sigma Small Chain Clathrin Coat Assembly AP17 Clathrin Coat Associated AP17 Plasma Membrane Adaptor Ap 2.17 Kda SIGMA2 Adaptin |
| ENSMUSG00000008140 | ENSFM00250000006413 | Er Membrane Complex Subunit 10 Precursor |
| ENSMUSG00000008435 | ENSFM00500000269664 | Retinol Dehydrogenase |
| ENSMUSG00000008489 | ENSFM00660001189998 | Unknown |
| ENSMUSG00000008489 | ENSFM00250000000588 | Elav |
| ENSMUSG00000008683 | ENSFM00570000895097 | Unknown |
| ENSMUSG00000008683 | ENSFM00570000896056 | Unknown |
| ENSMUSG00000008683 | ENSFM00250000002123 | 40S Ribosomal |
| ENSMUSG00000008734 | ENSFM00670001235445 | G Coupled Receptor Family C Group 5 Member Retinoic Acid Induced Gene Raig |
| ENSMUSG00000008859 | ENSFM00500000270233 | Ras Related Precursor |
| ENSMUSG00000008892 | ENSFM00250000001189 | Voltage Dependent Anion Selective Channel Vdac Outer Mitochondrial Membrane Porin |
| ENSMUSG00000009013 | ENSFM00670001235517 | Dynein Light Chain Cytoplasmic 8 Kda Dynein Light Chain Dynein Light Chain LC8 Type |
| ENSMUSG00000009073 | ENSFM00610000969488 | Unknown |
| ENSMUSG00000009073 | ENSFM00610000973510 | Unknown |
| ENSMUSG00000009073 | ENSFM00750001632408 | Merlin Moesin Ezrin Radixin Neurofibromin 2 Schwannomin |
| ENSMUSG00000009079 | ENSFM00500000270534 | Rna Binding Ews |
| ENSMUSG00000009079 | ENSFM00500000311767 | Rna Binding Ews |
| ENSMUSG00000009090 | ENSFM00430000230089 | Ap 1 Complex Subunit Beta 1 Adaptor Complex Ap 1 Subunit Beta 1 Adaptor Related Complex 1 Subunit Beta 1 Beta 1 Adaptin Beta Adaptin 1 Clathrin Assembly Complex 1 Beta Large Chain Golgi Adaptor HA1/AP1 Adaptin Beta Subunit |
| ENSMUSG00000009216 | ENSFM00500000274513 | Unknown |
| ENSMUSG00000009394 | ENSFM00250000000771 | Synapsin Synapsin |
| ENSMUSG00000009418 | ENSFM00250000000514 | Neuron Navigator Pore Membrane And/Or Filament Interacting Steerin Unc 53 Homolog Un |
| ENSMUSG00000009535 | ENSFM00250000004534 | Cap Guanine N7 Methyltransferase EC_2.1.1.56 RG7MT1 Guanine N 7 Methyltransferase Cap Methyltransferase |
| ENSMUSG00000009549 | ENSFM00500000420639 | Signal Recognition Particle 14 Kda |
| ENSMUSG00000009549 | ENSFM00670001236179 | Signal Recognition Particle 14 Kda SRP14 |
| ENSMUSG00000009647 | ENSFM00590000918380 | Unknown |
| ENSMUSG00000009647 | ENSFM00250000002939 | Calcium Uniporter Regulatory Subunit Mcub Mitochondrial Precursor Mcub Coiled Coil Domain Containing 109B |
| ENSMUSG00000009670 | ENSFM00250000004461 | Testis Expressed Sequence 11 |
| ENSMUSG00000009681 | ENSFM00250000000543 | Breakpoint Cluster Region |
| ENSMUSG00000009731 | ENSFM00300000079128 | Potassium Voltage Gated Channel Subfamily D Member Voltage Gated Potassium Channel Subunit KV4 |
| ENSMUSG00000009741 | ENSFM00250000001310 | Transcription Factor CP2 |
| ENSMUSG00000009894 | ENSFM00570000876467 | Unknown |
| ENSMUSG00000009894 | ENSFM00570000891790 | Unknown |
| ENSMUSG00000009894 | ENSFM00570000894302 | Unknown |
| ENSMUSG00000009894 | ENSFM00250000004681 | Synaptosomal Associated 47 Snap 47 Synaptosomal Associated 47 Kda |
| ENSMUSG00000009927 | ENSFM00670001235896 | 40S Ribosomal S25 |
| ENSMUSG00000010025 | ENSFM00250000000672 | Aldehyde Dehydrogenase Aldehyde Dehydrogenase Aldehyde Dehydrogenase Family 3 Member |
| ENSMUSG00000010066 | ENSFM00730001522093 | Voltage Dependent Calcium Channel Subunit Alpha 2/Delta 1 Precursor Voltage Gated Calcium Channel Subunit Alpha 2/Delta 1 |
| ENSMUSG00000010080 | ENSFM00500000269762 | Epsin Eps 15 Interacting |
| ENSMUSG00000010086 | ENSFM00250000007086 | Ring Finger 112 Brain Finger Zinc Finger 179 |
| ENSMUSG00000010110 | ENSFM00660001179029 | Unknown |
| ENSMUSG00000010110 | ENSFM00660001179821 | Unknown |
| ENSMUSG00000010110 | ENSFM00660001179822 | Unknown |
| ENSMUSG00000010110 | ENSFM00760001714795 | Syntaxin 5 |
| ENSMUSG00000010376 | ENSFM00500000292193 | NEDD8 Precursor Neddylin Ubiquitin NEDD8 |
| ENSMUSG00000010392 | ENSFM00500000270907 | Golgi Snap Receptor Complex Member 1.28 Kda Golgi Snare 28 Kda Cis Golgi Snare P28 |
| ENSMUSG00000010803 | ENSFM00610000973527 | Unknown |
| ENSMUSG00000010803 | ENSFM00800002003385 | Gamma Aminobutyric Acid Receptor Subunit Alpha Precursor Gaba A Receptor Subunit Alpha |
| ENSMUSG00000010825 | ENSFM00500000273079 | Delphilin Glutamate Receptor Ionotropic Delta 2 Interacting 1 |
| ENSMUSG00000010914 | ENSFM00800002003776 | Pyruvate Dehydrogenase X Component Mitochondrial Precursor Dihydrolipoamide Dehydrogenase Binding Of Pyruvate Dehydrogenase Complex Lipoyl Containing Pyruvate Dehydrogenase Complex Component X |
| ENSMUSG00000010936 | ENSFM00760001714983 | VAC14 Homolog |
| ENSMUSG00000011589 | ENSFM00250000002725 | Fibronectin Type Iii And Spry Domain Containing 1 |
| ENSMUSG00000011751 | ENSFM00740001589095 | Spectrin Beta Chain Erythrocytic Beta Spectrin |
| ENSMUSG00000011752 | ENSFM00250000001043 | Phosphoglycerate Mutase EC_3.1.3.13 EC_5.4.2.- 11 EC_5.4.-.- 2 4 Bpg Dependent Pgam Phosphoglycerate Mutase Isozyme Pgam |
| ENSMUSG00000011832 | ENSFM00500000270092 | Ecotropic Viral Integration Site 5 |
| ENSMUSG00000011877 | ENSFM00250000001490 | Arf Gtpase Activating GIT1 Arf Gap GIT1 Cool Associated And Tyrosine Phosphorylated 1 Cat 1 CAT1 G Coupled Receptor Kinase Interactor 1 Grk Interacting 1 |
| ENSMUSG00000011884 | ENSFM00250000005320 | Glycolipid Transfer Gltp |
| ENSMUSG00000012848 | ENSFM00250000003056 | 40S Ribosomal S5 |
| ENSMUSG00000013033 | ENSFM00260000050328 | Latrophilin |
| ENSMUSG00000013160 | ENSFM00800002003469 | V Type Proton Atpase Subunit D V Atpase Subunit D Vacuolar Proton Pump Subunit D |
| ENSMUSG00000013236 | ENSFM00740001589097 | Receptor Type Tyrosine Phosphatase Precursor EC_3.1.3.48 |
| ENSMUSG00000013367 | ENSFM00290000065521 | Precursor |
| ENSMUSG00000013523 | ENSFM00250000007762 | Breast Carcinoma Amplified Sequence 1 Amplified In Breast Cancer 1 |
| ENSMUSG00000013662 | ENSFM00790001963005 | Atpase Family Aaa Domain Containing 1 EC_3.6.1.3 |
| ENSMUSG00000013698 | ENSFM00250000010619 | Astrocytic Phosphoprotein Pea 15.15 Kda Phosphoprotein Enriched In Astrocytes |
| ENSMUSG00000013921 | ENSFM00760001720057 | Unknown |
| ENSMUSG00000013921 | ENSFM00740001589331 | Cap Gly Domain Containing Linker 3 Cytoplasmic Linker 170 Related 59 Kda Clip 170 Related 59 Kda Clipr 59 |
| ENSMUSG00000014077 | ENSFM00570000879445 | Unknown |
| ENSMUSG00000014077 | ENSFM00500000270459 | Calcineurin B Homologous 1 Calcineurin B Calcium Binding Chp Calcium Binding P22 Ef Hand Calcium Binding Domain Containing P22 |
| ENSMUSG00000014294 | ENSFM00670001236219 | Nadh Dehydrogenase 1 Alpha Subcomplex Subunit 2 Complex I B8 Ci B8 Nadh Ubiquinone Oxidoreductase B8 Subunit |
| ENSMUSG00000014301 | ENSFM00570000887254 | Unknown |
| ENSMUSG00000014301 | ENSFM00670001236447 | Mitochondrial Import Inner Membrane Translocase Subunit TIM16 Mitochondria Associated Granulocyte Macrophage Csf Signaling Molecule Presequence Translocated Associated Motor Subunit PAM16 |
| ENSMUSG00000014353 | ENSFM00250000002118 | Transmembrane Precursor |
| ENSMUSG00000014402 | ENSFM00500000270204 | Tumor Susceptibility Gene 101 Escrt I Complex Subunit TSG101 |
| ENSMUSG00000014606 | ENSFM00570000892647 | Unknown |
| ENSMUSG00000014606 | ENSFM00770001847568 | Mitochondrial 2 Oxoglutarate/Malate Carrier Ogcp Solute Carrier Family 25 Member 11 |
| ENSMUSG00000014769 | ENSFM00800002003627 | Proteasome Subunit Beta Type 1 EC_3.4.25.1 Proteasome |
| ENSMUSG00000014932 | ENSFM00730001521177 | Tyrosine Kinase EC_2.7.10.2 |
| ENSMUSG00000014956 | ENSFM00810002057428 | Unknown |
| ENSMUSG00000014956 | ENSFM00420000140518 | Serine/Threonine Phosphatase PP1 Catalytic Subunit Pp EC_3.1.3.16 |
| ENSMUSG00000015002 | ENSFM00640001109165 | Unknown |
| ENSMUSG00000015002 | ENSFM00250000001283 | EFR3 Homolog |
| ENSMUSG00000015092 | ENSFM00670001235719 | Endothelial Differentiation Related Factor 1 Edf 1 Multiprotein Bridging Factor 1 MBF1 |
| ENSMUSG00000015165 | ENSFM00260000050504 | Heterogeneous Nuclear Ribonucleoprotein L |
| ENSMUSG00000015214 | ENSFM00730001521553 | Myotubularin Phosphatidylinositol 3 5 Bisphosphate 3 Phosphatase EC_3.1.3.95 |
| ENSMUSG00000015290 | ENSFM00570000863849 | Unknown |
| ENSMUSG00000015290 | ENSFM00500000273690 | Ubiquitin 4A |
| ENSMUSG00000015337 | ENSFM00790001963101 | Endonuclease G Mitochondrial Precursor Endo G EC_3.1.30.- |
| ENSMUSG00000015341 | ENSFM00250000003047 | Golgin Subfamily A Member 7 |
| ENSMUSG00000015476 | ENSFM00600000921799 | Proline Rich Transmembrane 1 Dispanin Subfamily D Member 1 DSPD1 |
| ENSMUSG00000015656 | ENSFM00730001521103 | Heat Shock Kda Heat Shock 70 Kda |
| ENSMUSG00000015668 | ENSFM00400000131930 | Pdz Domain Containing 11 |
| ENSMUSG00000015733 | ENSFM00250000001955 | F Actin Capping Subunit Alpha 2 Capz Alpha 2 |
| ENSMUSG00000015747 | ENSFM00250000005970 | Vacuolar Sorting Associated 45 |
| ENSMUSG00000015766 | ENSFM00570000899024 | Unknown |
| ENSMUSG00000015766 | ENSFM00500000270023 | Epidermal Growth Factor Receptor Kinase Substrate 8 EPS8 Epidermal Growth Factor Receptor Pathway Substrate 8 Related EPS8 Related |
| ENSMUSG00000015829 | ENSFM00740001589132 | Tenascin Precursor Tn |
| ENSMUSG00000015837 | ENSFM00250000004435 | Sequestosome 1 Ubiquitin Binding P62 |
| ENSMUSG00000015869 | ENSFM00730001521917 | Phosphoribosyl Pyrophosphate Synthase Associated Prpp Synthase Associated |
| ENSMUSG00000015943 | ENSFM00500000273601 | Bola 1 |
| ENSMUSG00000016319 | ENSFM00250000000816 | Adp/Atp Translocase Adp Atp Carrier Adenine Nucleotide Translocator Ant Solute Carrier Family 25 Member |
| ENSMUSG00000016346 | ENSFM00250000000417 | Potassium Voltage Gated Channel Subfamily Kqt Member Kqt Potassium Channel Subunit Alpha Voltage Gated Potassium Channel Subunit KV7 |
| ENSMUSG00000016349 | ENSFM00800002003364 | Elongation Factor 1 Alpha Ef 1 Alpha Eukaryotic Elongation Factor 1 A EEF1A |
| ENSMUSG00000016427 | ENSFM00500000386654 | Unknown |
| ENSMUSG00000016495 | ENSFM00570000879077 | Unknown |
| ENSMUSG00000016495 | ENSFM00570000883039 | Unknown |
| ENSMUSG00000016495 | ENSFM00570000887956 | Unknown |
| ENSMUSG00000016495 | ENSFM00570000900159 | Unknown |
| ENSMUSG00000016495 | ENSFM00500000274177 | Plasminogen Receptor Kt Plg R Kt |
| ENSMUSG00000016541 | ENSFM00250000007503 | Ataxin 10 Spinocerebellar Ataxia Type 10 |
| ENSMUSG00000016995 | ENSFM00740001589150 | Precursor |
| ENSMUSG00000017132 | ENSFM00250000001040 | Cytohesin Ph SEC7 And Coiled Coil Domain Containing |
| ENSMUSG00000017167 | ENSFM00250000000380 | Contactin Associated 5 Precursor Cell Recognition Molecule CASPR5 |
| ENSMUSG00000017188 | ENSFM00670001236757 | Cytochrome C Oxidase Assembly Factor 3 Homolog Mitochondrial Coiled Coil Domain Containing 56 |
| ENSMUSG00000017221 | ENSFM00250000003614 | 26S Proteasome Non Atpase Regulatory Subunit 3 26S Proteasome Subunit 26S Proteasome Regulatory Subunit |
| ENSMUSG00000017288 | ENSFM00610000973548 | Unknown |
| ENSMUSG00000017288 | ENSFM00610000973549 | Unknown |
| ENSMUSG00000017288 | ENSFM00800002003735 | Vacuolar Sorting Associated 53 Homolog |
| ENSMUSG00000017291 | ENSFM00250000000892 | Serine/Threonine Kinase EC_2.7.11.1 Thousand And One Amino Acid |
| ENSMUSG00000017307 | ENSFM00640001115284 | Unknown |
| ENSMUSG00000017307 | ENSFM00500000271864 | Acyl Coenzyme A Thioesterase 8 Acyl Coa Thioesterase 8 EC_3.1.2.27 Choloyl Coenzyme A Thioesterase Acyl Coa Thioesterase Pte 2 Peroxisomal Acyl Coenzyme A Thioester Hydrolase 1 Pte 1 Peroxisomal Long Chain Acyl Coa Thioesterase 1 |
| ENSMUSG00000017314 | ENSFM00730001521911 | Maguk P55 Subfamily Member 2 Discs Large Homolog 2 MPP2 |
| ENSMUSG00000017404 | ENSFM00250000001949 | 60S Ribosomal L19 |
| ENSMUSG00000017412 | ENSFM00250000000524 | Voltage Dependent L Type Calcium Channel Subunit Beta Calcium Channel Voltage Dependent Subunit Beta |
| ENSMUSG00000017428 | ENSFM00640001114961 | Unknown |
| ENSMUSG00000017428 | ENSFM00640001114962 | Unknown |
| ENSMUSG00000017428 | ENSFM00250000003951 | 26S Proteasome Non Atpase Regulatory Subunit 26S Proteasome Regulatory Subunit RPN6 |
| ENSMUSG00000017670 | ENSFM00570000880327 | Unknown |
| ENSMUSG00000017670 | ENSFM00570000901128 | Unknown |
| ENSMUSG00000017670 | ENSFM00250000001494 | Engulfment And Cell Motility |
| ENSMUSG00000017686 | ENSFM00250000000940 | Mitochondrial Rho Gtpase 2 Miro 2 EC_3.6.5.- Ras Homolog Gene Family Member T2 |
| ENSMUSG00000017715 | ENSFM00750001632469 | Cdp Diacylglycerol Glycerol 3 Phosphate 3 Phosphatidyltransferase Mitochondrial Precursor EC_2.7.8.5 Phosphatidylglycerophosphate Synthase 1 Pgp Synthase 1 |
| ENSMUSG00000017721 | ENSFM00780001898466 | Gpi Transamidase Component Pig T Precursor Phosphatidylinositol Glycan Biosynthesis Class T |
| ENSMUSG00000017774 | ENSFM00500000314344 | Unknown |
| ENSMUSG00000017774 | ENSFM00800002003512 | Unconventional Myosin Ic Myosin I Beta Mmi Beta Mmib |
| ENSMUSG00000017776 | ENSFM00250000002274 | Adapter Molecule Crk Proto Oncogene C Crk P38 |
| ENSMUSG00000017781 | ENSFM00500000269976 | Phosphatidylinositol Transfer Alpha Pi Tp Alpha Ptdins Transfer Alpha Ptdinstp Alpha |
| ENSMUSG00000017837 | ENSFM00250000003149 | Nf Kappa B Inhibitor Interacting Ras 2 I Kappa B Interacting Ras 2 Kappa B Ras 2 Kappab RAS2 |
| ENSMUSG00000017843 | ENSFM00810002040632 | Serine/Threonine Phosphatase 2A 56 Kda Regulatory Subunit Gamma PP2A B Subunit B' PP2A B Subunit B' Gamma PP2A B Subunit B56 Gamma PP2A B Subunit PR61 Gamma PP2A B Subunit R5 Gamma |
| ENSMUSG00000017943 | ENSFM00250000004188 | Ganglioside Induced Differentiation Associated 1 GDAP1 |
| ENSMUSG00000018001 | ENSFM00570000899921 | Unknown |
| ENSMUSG00000018001 | ENSFM00660001188987 | Unknown |
| ENSMUSG00000018001 | ENSFM00660001188989 | Unknown |
| ENSMUSG00000018001 | ENSFM00250000001040 | Cytohesin Ph SEC7 And Coiled Coil Domain Containing |
| ENSMUSG00000018012 | ENSFM00730001521186 | Ras Related C3 Botulinum Toxin Substrate Precursor P21 |
| ENSMUSG00000018076 | ENSFM00250000001693 | Mediator Of Rna Polymerase Ii Transcription Subunit 13 Thyroid Hormone Receptor Associated Thyroid Hormone Receptor Associated Complex 240 Kda Component |
| ENSMUSG00000018286 | ENSFM00810002040613 | Proteasome Subunit Beta Type 9 Precursor EC_3.4.25.1 Low Molecular Mass 2 Macropain Chain 7 Multicatalytic Endopeptidase Complex Chain 7 Proteasome Chain 7 Proteasome Subunit Beta 1I Really Interesting New Gene 12 |
| ENSMUSG00000018326 | ENSFM00730001521154 | Unknown |
| ENSMUSG00000018398 | ENSFM00590000918438 | Unknown |
| ENSMUSG00000018398 | ENSFM00350000105408 | Septin |
| ENSMUSG00000018411 | ENSFM00500000327757 | Unknown |
| ENSMUSG00000018411 | ENSFM00260000050441 | Microtubule Associated Tau Neurofibrillary Tangle Paired Helical Filament Tau Phf Tau |
| ENSMUSG00000018417 | ENSFM00800002003560 | Unconventional Myosin Ia Brush Border Myosin I Bbm I Bbmi Myosin I Heavy Chain Mihc |
| ENSMUSG00000018446 | ENSFM00250000004016 | Complement Component 1 Q Subcomponent Binding Protein Mitochondrial Precursor GC1Q R Glycoprotein GC1QBP C1QBP |
| ENSMUSG00000018507 | ENSFM00500000269768 | Transient Receptor Potential Cation Channel Subfamily V Member 1 TRPV1 Osm 9 Trp Channel 1 OTRPC1 Vanilloid Receptor 1 |
| ENSMUSG00000018547 | ENSFM00260000050567 | Phosphatidylinositol 5 Phosphate 4 Kinase Type 2 EC_2.7.1.149 1 Phosphatidylinositol 5 Phosphate 4 Kinase 2 Diphosphoinositide Kinase 2 Phosphatidylinositol 5 Phosphate 4 Kinase Type Ii Pi 5 P 4 Kinase Type Ii PIP4KII Ptdins 5 P 4 Kinase 2 |
| ENSMUSG00000018583 | ENSFM00250000002045 | Ras Gtpase Activating Binding 1 G3BP 1 EC_3.6.4.12 EC_3.6.4.- 13 Atp Dependent Helicase |
| ENSMUSG00000018593 | ENSFM00250000001867 | Sparc Precursor Basement Membrane 40 Bm 40 Osteonectin On Secreted Acidic And Rich In Cysteine |
| ENSMUSG00000018669 | ENSFM00570000878559 | Unknown |
| ENSMUSG00000018669 | ENSFM00250000006242 | CDK5 Regulatory Subunit Associated 3 |
| ENSMUSG00000018733 | ENSFM00570000889673 | Unknown |
| ENSMUSG00000018733 | ENSFM00660001177557 | Unknown |
| ENSMUSG00000018733 | ENSFM00250000006034 | Peroxisome Assembly 12 Peroxin 12 Peroxisome Assembly Factor 3 Paf 3 |
| ENSMUSG00000018736 | ENSFM00250000002568 | Nuclear Distribution Nude 1 |
| ENSMUSG00000018765 | ENSFM00250000000920 | Fragile X Mental Retardation 1 |
| ENSMUSG00000018819 | ENSFM00500000273526 | Lymphocyte Specific Kda Phosphoprotein PP52 Lymphocyte Specific Antigen WP34 |
| ENSMUSG00000018830 | ENSFM00270000056424 | Myosin Cellular Myosin Heavy Chain Type Myosin Heavy Chain Myosin Heavy Chain Non Muscle Non Muscle Myosin Heavy Chain Nmmhc Non Muscle Myosin Heavy Chain Nmmhc Ii Nmmhc |
| ENSMUSG00000018858 | ENSFM00570000886758 | Unknown |
| ENSMUSG00000018858 | ENSFM00500000273212 | Peptidyl Trna Hydrolase ICT1 Mitochondrial Precursor EC_3.1.1.29 39S Ribosomal L58 Mitochondrial Mrp L58 Immature Colon Carcinoma Transcript 1 |
| ENSMUSG00000018965 | ENSFM00730001521154 | Unknown |
| ENSMUSG00000019066 | ENSFM00730001521441 | Ras Related Rab 3 |
| ENSMUSG00000019082 | ENSFM00720001503227 | Unknown |
| ENSMUSG00000019082 | ENSFM00720001503349 | Unknown |
| ENSMUSG00000019082 | ENSFM00720001503415 | Unknown |
| ENSMUSG00000019082 | ENSFM00740001599670 | Unknown |
| ENSMUSG00000019082 | ENSFM00740001613500 | Unknown |
| ENSMUSG00000019082 | ENSFM00810002056307 | Unknown |
| ENSMUSG00000019082 | ENSFM00810002057416 | Unknown |
| ENSMUSG00000019082 | ENSFM00500000270170 | Mitochondrial Glutamate Carrier 1 Gc 1 Glutamate/H + Symporter 1 Solute Carrier Family 25 Member 22 |
| ENSMUSG00000019087 | ENSFM00570000885080 | Unknown |
| ENSMUSG00000019087 | ENSFM00570000888862 | Unknown |
| ENSMUSG00000019087 | ENSFM00560000771081 | V Type Proton Atpase Subunit S1 Precursor V Atpase Subunit S1 V Atpase AC45 Subunit V Atpase S1 Accessory Vacuolar Proton Pump Subunit S1 |
| ENSMUSG00000019124 | ENSFM00570000885243 | Unknown |
| ENSMUSG00000019124 | ENSFM00250000002467 | Secernin |
| ENSMUSG00000019146 | ENSFM00250000002019 | Voltage Dependent Calcium Channel Gamma Subunit Neuronal Voltage Gated Calcium Channel Gamma Subunit Transmembrane Ampar Regulatory Gamma Tarp Gamma |
| ENSMUSG00000019210 | ENSFM00250000002828 | V Type Proton Atpase Subunit E V Atpase Subunit E V Atpase Kda Subunit Vacuolar Proton Pump Subunit E |
| ENSMUSG00000019261 | ENSFM00250000000942 | Microtubule Associated Map |
| ENSMUSG00000019302 | ENSFM00570000881199 | Unknown |
| ENSMUSG00000019302 | ENSFM00250000000490 | V Type Proton Atpase 116 Kda Subunit A V Atpase 116 Kda Proton 116 Kda Subunit Vacuolar Proton Translocating Atpase 116 Kda Subunit A |
| ENSMUSG00000019370 | ENSFM00640001118108 | Unknown |
| ENSMUSG00000019370 | ENSFM00640001118555 | Unknown |
| ENSMUSG00000019370 | ENSFM00750001632311 | Calmodulin Cam |
| ENSMUSG00000019373 | ENSFM00570000892434 | Unknown |
| ENSMUSG00000019373 | ENSFM00250000006013 | COP9 Signalosome Complex Subunit 3 Signalosome Subunit 3 |
| ENSMUSG00000019428 | ENSFM00520000547997 | Unknown |
| ENSMUSG00000019428 | ENSFM00570000883795 | Unknown |
| ENSMUSG00000019428 | ENSFM00250000005307 | Peptidyl Prolyl Cis Trans Isomerase FKBP8 Ppiase FKBP8 EC_5.2.1.8.38 Kda FK506 Binding 38 Kda Fkbp Fkbp 38 FK506 Binding 8 Fkbp 8 FKBPR38 Rotamase |
| ENSMUSG00000019433 | ENSFM00250000002325 | Pdz Domain Containing |
| ENSMUSG00000019467 | ENSFM00740001589124 | EC_2.7.11.1 |
| ENSMUSG00000019578 | ENSFM00250000005080 | Ubx Domain Containing 6 Ubx Domain Containing 1 |
| ENSMUSG00000019763 | ENSFM00570000879481 | Unknown |
| ENSMUSG00000019763 | ENSFM00570000884368 | Unknown |
| ENSMUSG00000019763 | ENSFM00570000894366 | Unknown |
| ENSMUSG00000019763 | ENSFM00500000271332 | Required Meiotic Nuclear Division 1 Homolog Precursor |
| ENSMUSG00000019775 | ENSFM00480000262761 | Regulator Of G Signaling |
| ENSMUSG00000019795 | ENSFM00590000917209 | Unknown |
| ENSMUSG00000019795 | ENSFM00590000917210 | Unknown |
| ENSMUSG00000019795 | ENSFM00590000918378 | Unknown |
| ENSMUSG00000019795 | ENSFM00250000003005 | L Isoaspartate D Aspartate O Methyltransferase Pimt EC_2.1.1.77 L Isoaspartyl Carboxyl Methyltransferase L Isoaspartyl/D Aspartyl Methyltransferase Beta Aspartate Methyltransferase |
| ENSMUSG00000019809 | ENSFM00250000005471 | Peroxisomal Biogenesis Factor 3 Peroxin 3 Peroxisomal Assembly PEX3 |
| ENSMUSG00000019820 | ENSFM00250000000455 | Dystrophin |
| ENSMUSG00000019831 | ENSFM00250000001269 | Wiskott Aldrich Syndrome Family Member 1 Wasp Family Member 1 Wave 1 |
| ENSMUSG00000019843 | ENSFM00570000890474 | Unknown |
| ENSMUSG00000019843 | ENSFM00730001521177 | Tyrosine Kinase EC_2.7.10.2 |
| ENSMUSG00000019868 | ENSFM00570000866653 | Unknown |
| ENSMUSG00000019868 | ENSFM00570000882696 | Unknown |
| ENSMUSG00000019868 | ENSFM00250000005580 | Vacuolar Sorting Associated VTA1 Homolog SKD1 Binding 1 SBP1 |
| ENSMUSG00000019906 | ENSFM00260000050595 | Lin 7 Homolog Lin Mammalian Lin Seven Mals Vertebrate Lin 7 Homolog Veli |
| ENSMUSG00000019907 | ENSFM00500000269806 | Phosphatase 1 Regulatory Subunit 12A Myosin Phosphatase Targeting Subunit 1 Myosin Phosphatase Target Subunit 1 Phosphatase Myosin Binding Subunit |
| ENSMUSG00000019923 | ENSFM00250000009507 | ZW10 Interactor ZW10 Interacting 1 Zwint 1 |
| ENSMUSG00000019961 | ENSFM00600000921391 | Lamina Associated Polypeptide 2 Thymopoietin Tp Thymopoietin Related Peptide Tprp |
| ENSMUSG00000019969 | ENSFM00250000001423 | Presenilin Ps EC_3.4.23.- |
| ENSMUSG00000019986 | ENSFM00250000003083 | Jouberin Abelson Helper Integration Site 1 Ahi 1 |
| ENSMUSG00000019996 | ENSFM00250000002902 | Ensconsin Epithelial Microtubule Associated Of 115 Kda E Map 115 Microtubule Associated 7 Map 7 |
| ENSMUSG00000019998 | ENSFM00500000270193 | Syntaxin |
| ENSMUSG00000020015 | ENSFM00740001589171 | Cyclin Dependent Kinase EC_2.7.11.22 Cell Division Kinase Pctaire Motif Kinase Serine/Threonine Kinase Pctaire |
| ENSMUSG00000020022 | ENSFM00570000899974 | Unknown |
| ENSMUSG00000020022 | ENSFM00500000272434 | Nadh Dehydrogenase 1 Alpha Subcomplex Subunit Complex I B17 2 Ci B17 2 CIB17 2 Nadh Ubiquinone Oxidoreductase Subunit B17 2 |
| ENSMUSG00000020042 | ENSFM00250000001718 | Ankyrin Repeat And Btb/Poz Domain Containing BTBD11 Btb/Poz Domain Containing 11 |
| ENSMUSG00000020088 | ENSFM00800002003440 | Gtp Binding SAR1 |
| ENSMUSG00000020097 | ENSFM00570000900525 | Unknown |
| ENSMUSG00000020097 | ENSFM00800002003569 | Sphingosine 1 Phosphate Lyase 1 S1PL Sp Lyase 1 Spl 1 EC_4.1.2.27 Sphingosine 1 Phosphate Aldolase |
| ENSMUSG00000020109 | ENSFM00250000001153 | Dnaj Homolog Subfamily Member |
| ENSMUSG00000020111 | ENSFM00610000967698 | Unknown |
| ENSMUSG00000020111 | ENSFM00260000050684 | Calcium Uptake 1 Mitochondrial Calcium Binding Atopy Related Autoantigen 1 Homolog |
| ENSMUSG00000020115 | ENSFM00250000003276 | Inhibitor Of Nuclear Factor Kappa B Kinase Subunit Epsilon I Kappa B Kinase Epsilon Ikk E Ikk Epsilon Ikbke EC_2.7.11.10 Inducible I Kappa B Kinase Ikk I |
| ENSMUSG00000020132 | ENSFM00500000271226 | Ras Related Rab 21 |
| ENSMUSG00000020152 | ENSFM00730001521596 | Actin Related 2 Actin 2 |
| ENSMUSG00000020153 | ENSFM00250000003821 | Nadh Dehydrogenase Iron Sulfur 7 Mitochondrial Precursor EC_1.6.5.3 EC_1.6.99.- 3 Complex I 20KD Ci 20KD Nadh Ubiquinone Oxidoreductase 20 Kda Subunit |
| ENSMUSG00000020170 | ENSFM00250000003196 | Fibroblast Growth Factor Receptor Substrate 3 Fgfr Substrate 3 Fgfr Signaling Adaptor SNT2 SUC1 Associated Neurotrophic Factor Target 2 Snt 2 |
| ENSMUSG00000020183 | ENSFM00730001521968 | Carboxypeptidase D Precursor EC_3.4.17.22 Metallocarboxypeptidase D GP180 |
| ENSMUSG00000020198 | ENSFM00250000001618 | Ap 3 Complex Subunit Delta 1 Ap 3 Complex Subunit Delta Adaptor Related Complex 3 Subunit Delta 1 Delta Adaptin |
| ENSMUSG00000020219 | ENSFM00670001236643 | Mitochondrial Import Inner Membrane Translocase Subunit TIM13 |
| ENSMUSG00000020238 | ENSFM00570000898047 | Unknown |
| ENSMUSG00000020238 | ENSFM00370000118641 | Nicalin Precursor Nicastrin |
| ENSMUSG00000020263 | ENSFM00660001171072 | Unknown |
| ENSMUSG00000020263 | ENSFM00660001189703 | Unknown |
| ENSMUSG00000020263 | ENSFM00250000003219 | Dcc Interacting 13 DIP13 Adapter Containing Ph Domain Ptb Domain And Leucine Zipper Motif |
| ENSMUSG00000020267 | ENSFM00520000548728 | Histidine Triad Nucleotide Binding 1 |
| ENSMUSG00000020267 | ENSFM00670001235733 | Histidine Triad Nucleotide Binding 1 Ec 3 Adenosine 5' Monophosphoramidase Kinase C Inhibitor 1 Kinase C Interacting 1 Pkci 1 |
| ENSMUSG00000020277 | ENSFM00730001521507 | Atp Dependent 6 Phosphofructokinase Type |
| ENSMUSG00000020279 | ENSFM00570000864111 | Unknown |
| ENSMUSG00000020279 | ENSFM00250000006177 | Interleukin 9 Receptor Precursor Il 9 Receptor Il 9R CD129 Antigen |
| ENSMUSG00000020297 | ENSFM00500000298059 | Unknown |
| ENSMUSG00000020297 | ENSFM00500000315845 | Unknown |
| ENSMUSG00000020297 | ENSFM00250000005181 | Neuron Specific Family Member |
| ENSMUSG00000020300 | ENSFM00250000000830 | Cytoplasmic Polyadenylation Element Binding |
| ENSMUSG00000020308 | ENSFM00250000008950 | Tubulin Polyglutamylase Complex Subunit 1 PGS1 |
| ENSMUSG00000020315 | ENSFM00740001589095 | Spectrin Beta Chain Erythrocytic Beta Spectrin |
| ENSMUSG00000020340 | ENSFM00570000884032 | Unknown |
| ENSMUSG00000020340 | ENSFM00250000001390 | Cytoplasmic FMR1 Interacting |
| ENSMUSG00000020349 | ENSFM00730001521389 | Serine/Threonine Phosphatase Catalytic Subunit EC_3.1.3.16 |
| ENSMUSG00000020358 | ENSFM00500000269788 | Heterogeneous Nuclear Ribonucleoprotein D Hnrnp D Hnrnp Dl |
| ENSMUSG00000020396 | ENSFM00730001521354 | Neurofilament Polypeptide Nf Kda Neurofilament Neurofilament Triplet |
| ENSMUSG00000020402 | ENSFM00250000001189 | Voltage Dependent Anion Selective Channel Vdac Outer Mitochondrial Membrane Porin |
| ENSMUSG00000020428 | ENSFM00800002003422 | Gamma Aminobutyric Acid Receptor Subunit Alpha Precursor Gaba A Receptor Subunit Alpha |
| ENSMUSG00000020431 | ENSFM00730001521247 | Adenylate Cyclase Type EC_4.6.1.1 Atp Pyrophosphate Lyase Adenylate Cyclase Type Adenylyl Cyclase Ca 2+ Adenylyl Cyclase |
| ENSMUSG00000020436 | ENSFM00730001521448 | Gamma Aminobutyric Acid Receptor Subunit Gamma Precursor Gaba A Receptor Subunit Gamma |
| ENSMUSG00000020440 | ENSFM00730001521124 | Adp Ribosylation Factor |
| ENSMUSG00000020444 | ENSFM00640001120069 | Unknown |
| ENSMUSG00000020444 | ENSFM00400000131805 | Guanylate Kinase EC_2.7.4.8 Gmp Kinase |
| ENSMUSG00000020456 | ENSFM00750001632363 | 2 Oxoglutarate Dehydrogenase Mitochondrial Precursor 2 Oxoglutarate Dehydrogenase Complex Component E1 Ogdc E1 Alpha Ketoglutarate Dehydrogenase |
| ENSMUSG00000020460 | ENSFM00500000270032 | Ubiquitin 40S Ribosomal S27A Precursor Ubiquitin Carboxyl Extension 80 |
| ENSMUSG00000020476 | ENSFM00480000262787 | Drebrin |
| ENSMUSG00000020483 | ENSFM00570000882728 | Unknown |
| ENSMUSG00000020483 | ENSFM00670001235517 | Dynein Light Chain Cytoplasmic 8 Kda Dynein Light Chain Dynein Light Chain LC8 Type |
| ENSMUSG00000020486 | ENSFM00800002003401 | Septin |
| ENSMUSG00000020514 | ENSFM00250000005774 | 39S Ribosomal L22 Mitochondrial Precursor L22MT Mrp L22 |
| ENSMUSG00000020524 | ENSFM00640001120061 | Unknown |
| ENSMUSG00000020524 | ENSFM00720001492020 | Glutamate Receptor Precursor Glur Ampa Selective Glutamate Receptor Glur Glutamate Receptor Ionotropic Ampa |
| ENSMUSG00000020528 | ENSFM00730001521917 | Phosphoribosyl Pyrophosphate Synthase Associated Prpp Synthase Associated |
| ENSMUSG00000020537 | ENSFM00730001521857 | Developmentally Regulated Gtp Binding 2 Drg 2 |
| ENSMUSG00000020585 | ENSFM00500000271417 | Lysosomal Associated Transmembrane 4A |
| ENSMUSG00000020599 | ENSFM00730001522095 | Regulator Of G Signaling |
| ENSMUSG00000020612 | ENSFM00730001521681 | Camp Dependent Kinase Type I Alpha Regulatory Camp Dependent Kinase Type I Alpha Regulatory Subunit N Terminally Processed |
| ENSMUSG00000020635 | ENSFM00670001235580 | Peptidyl Prolyl Cis Trans Isomerase FKBP1B Ppiase FKBP1B EC_5.2.1.8 12.6 Kda FK506 Binding 12.6 Kda Fkbp Fkbp 12 6 FK506 Binding 1B Fkbp 1B Immunophilin FKBP12 6 Rotamase |
| ENSMUSG00000020640 | ENSFM00250000000789 | Intersectin |
| ENSMUSG00000020650 | ENSFM00670001235596 | B Cell Receptor Associated Bcr Associated |
| ENSMUSG00000020658 | ENSFM00250000001283 | EFR3 Homolog |
| ENSMUSG00000020664 | ENSFM00800002003534 | Dihydrolipoyl Dehydrogenase Mitochondrial Precursor EC_1.8.1.4 Dihydrolipoamide Dehydrogenase |
| ENSMUSG00000020668 | ENSFM00800002003386 | Kinesin |
| ENSMUSG00000020708 | ENSFM00730001521682 | 26S Protease Regulatory Subunit 8 26S Proteasome Aaa Atpase Subunit RPT6 Proteasome 26S Subunit Atpase 5 Proteasome Subunit P45 P45/Sug |
| ENSMUSG00000020716 | ENSFM00250000001252 | Neurofibromin Neurofibromatosis Related Nf 1 |
| ENSMUSG00000020719 | ENSFM00720001503540 | Unknown |
| ENSMUSG00000020719 | ENSFM00420000140507 | Probable Atp Dependent Rna Helicase DDX5 EC_3.6.4.13 Dead Box 5 |
| ENSMUSG00000020720 | ENSFM00250000004556 | 26S Proteasome Non Atpase Regulatory Subunit 12 26S Proteasome Regulatory Subunit RPN5 26S Proteasome Regulatory Subunit P55 |
| ENSMUSG00000020734 | ENSFM00730001521500 | Glutamate Receptor Ionotropic Nmda Precursor Glutamate Receptor Subunit Epsilon N Methyl D Aspartate Receptor Subtype |
| ENSMUSG00000020745 | ENSFM00810002040664 | Lissencephaly 1 |
| ENSMUSG00000020780 | ENSFM00250000003522 | Signal Recognition Particle Subunit SRP68 SRP68 Signal Recognition Particle 68 Kda |
| ENSMUSG00000020792 | ENSFM00570000877843 | Unknown |
| ENSMUSG00000020792 | ENSFM00250000003124 | Exocyst Complex Component 7 Exocyst Complex Component EXO70 |
| ENSMUSG00000020817 | ENSFM00250000001974 | Rab Gtpase Binding Effector 1 Rabaptin 5 Rabaptin 5ALPHA |
| ENSMUSG00000020827 | ENSFM00680001326720 | Unknown |
| ENSMUSG00000020827 | ENSFM00260000050335 | Kinase EC_2.7.11.1 Kinase Mapk/Erk Kinase Kinase Kinase Mek Kinase Kinase Mekkk Kinase |
| ENSMUSG00000020834 | ENSFM00500000313432 | Unknown |
| ENSMUSG00000020834 | ENSFM00570000880913 | Unknown |
| ENSMUSG00000020834 | ENSFM00500000269664 | Retinol Dehydrogenase |
| ENSMUSG00000020843 | ENSFM00570000888842 | Unknown |
| ENSMUSG00000020843 | ENSFM00610000966469 | Unknown |
| ENSMUSG00000020843 | ENSFM00500000272831 | Mitochondrial Import Inner Membrane Translocase Subunit TIM22 |
| ENSMUSG00000020849 | ENSFM00730001521388 | Unknown |
| ENSMUSG00000020869 | ENSFM00250000006592 | Leucine Rich Repeat Containing 59 |
| ENSMUSG00000020882 | ENSFM00250000000524 | Voltage Dependent L Type Calcium Channel Subunit Beta Calcium Channel Voltage Dependent Subunit Beta |
| ENSMUSG00000020886 | ENSFM00680001326715 | Unknown |
| ENSMUSG00000020886 | ENSFM00680001326716 | Unknown |
| ENSMUSG00000020886 | ENSFM00250000000275 | Disks Large Homolog Synapse Associated Sap |
| ENSMUSG00000020894 | ENSFM00670001235687 | Vesicle Associated Membrane 2 Vamp 2 Synaptobrevin 2 |
| ENSMUSG00000020900 | ENSFM00270000056424 | Myosin Cellular Myosin Heavy Chain Type Myosin Heavy Chain Myosin Heavy Chain Non Muscle Non Muscle Myosin Heavy Chain Nmmhc Non Muscle Myosin Heavy Chain Nmmhc Ii Nmmhc |
| ENSMUSG00000020903 | ENSFM00250000005899 | Syntaxin 8 |
| ENSMUSG00000020903 | ENSFM00500000303360 | Syntaxin 8 CRA_D |
| ENSMUSG00000020923 | ENSFM00740001589345 | Nucleolar Transcription Factor 1 Upstream Binding Factor 1 Ubf 1 |
| ENSMUSG00000020926 | ENSFM00410000138484 | Disintegrin And Metalloproteinase Domain Containing Precursor Adam Metalloproteinase Disintegrin And Cysteine Rich Mdc |
| ENSMUSG00000020932 | ENSFM00730001521291 | Vimentin |
| ENSMUSG00000020935 | ENSFM00500000272373 | Dephospho Coa Kinase Domain Containing |
| ENSMUSG00000020954 | ENSFM00250000001473 | Striatin |
| ENSMUSG00000020986 | ENSFM00810002040640 | Transport SEC23 Related |
| ENSMUSG00000020993 | ENSFM00670001235686 | Trafficking Particle Complex Subunit 6A Trapp Complex Subunit 6A |
| ENSMUSG00000021003 | ENSFM00250000005474 | Galactocerebrosidase Precursor Galcerase EC_3.2.1.46 Galactocerebroside Beta Galactosidase Galactosylceramidase Galactosylceramide Beta Galactosidase |
| ENSMUSG00000021024 | ENSFM00590000918027 | Unknown |
| ENSMUSG00000021024 | ENSFM00420000140606 | Proteasome Subunit Alpha Type EC_3.4.25.1 Macropain Multicatalytic Endopeptidase Complex Proteasome |
| ENSMUSG00000021027 | ENSFM00250000000944 | Ral Gtpase Activating Subunit Alpha 1 Gap Related Interacting Partner To E12 Gripe Gtpase Activating Domain 1 Tuberin 1 P240 |
| ENSMUSG00000021037 | ENSFM00500000270381 | Activator Of 90 Kda Heat Shock Atpase Homolog |
| ENSMUSG00000021039 | ENSFM00810002040569 | Pre Processing 45 |
| ENSMUSG00000021047 | ENSFM00740001589294 | Rna Binding Nova 1 Neuro Oncological Ventral Antigen 1 Ventral Neuron Specific 1 |
| ENSMUSG00000021048 | ENSFM00800002003403 | C 1 Tetrahydrofolate Synthase Cytoplasmic C1 Thf Synthase |
| ENSMUSG00000021051 | ENSFM00760001714676 | Serine/Threonine Phosphatase 2A 56 Kda Regulatory Subunit Epsilon PP2A B Subunit B' Epsilon PP2A B Subunit B56 Epsilon PP2A B Subunit PR61 Epsilon PP2A B Subunit R5 Epsilon |
| ENSMUSG00000021057 | ENSFM00250000007751 | A Kinase Anchor 5 Akap 5 A Kinase Anchor Kda Akap Camp Dependent Kinase Regulatory Subunit Ii High Affinity Binding |
| ENSMUSG00000021061 | ENSFM00740001589095 | Spectrin Beta Chain Erythrocytic Beta Spectrin |
| ENSMUSG00000021062 | ENSFM00750001632504 | Ras Related Rab 15 |
| ENSMUSG00000021065 | ENSFM00250000004295 | Alpha 1 6 Fucosyltransferase ALPHA1 6FUCT EC_2.4.1.68 Gdp L Fuc:N Acetyl Beta D Glucosaminide ALPHA1 6 Fucosyltransferase Gdp Fucose Glycoprotein Fucosyltransferase Glycoprotein 6 Alpha L Fucosyltransferase |
| ENSMUSG00000021071 | ENSFM00300000079125 | E3 Ubiquitin Ligase TRIM9 EC_6.3.2.- Tripartite Motif Containing 9 |
| ENSMUSG00000021076 | ENSFM00250000004727 | Actin Related 10 Actin Related 11 |
| ENSMUSG00000021097 | ENSFM00500000271703 | Calmin Calponin Transmembrane Domain |
| ENSMUSG00000021112 | ENSFM00760001714797 | Maguk P55 Subfamily Member 5 |
| ENSMUSG00000021114 | ENSFM00770001847582 | V Type Proton Atpase Subunit D V Atpase Subunit D Vacuolar Proton Pump Subunit D |
| ENSMUSG00000021124 | ENSFM00590000917484 | Unknown |
| ENSMUSG00000021124 | ENSFM00590000917485 | Unknown |
| ENSMUSG00000021124 | ENSFM00250000006099 | Vesicle Transport Through Interaction With T Snares Homolog 1B Vesicle Transport V Snare VTI1 1 VTI1 RP1 |
| ENSMUSG00000021131 | ENSFM00670001235831 | Enhancer Of Rudimentary |
| ENSMUSG00000021139 | ENSFM00610000973484 | Unknown |
| ENSMUSG00000021139 | ENSFM00670001236361 | Synaptojanin 2 Binding |
| ENSMUSG00000021140 | ENSFM00250000001163 | Pecanex |
| ENSMUSG00000021147 | ENSFM00660001167690 | Unknown |
| ENSMUSG00000021147 | ENSFM00660001167691 | Unknown |
| ENSMUSG00000021147 | ENSFM00660001172867 | Unknown |
| ENSMUSG00000021147 | ENSFM00660001173980 | Unknown |
| ENSMUSG00000021147 | ENSFM00660001181865 | Unknown |
| ENSMUSG00000021147 | ENSFM00250000004113 | Wd Repeat Containing 37 |
| ENSMUSG00000021171 | ENSFM00250000000964 | Extended Synaptotagmin E |
| ENSMUSG00000021188 | ENSFM00250000003391 | Thyroid Receptor Interacting 11 Tr Interacting 11 Trip 11 Clonal Evolution Related Gene On Chromosome 14 Golgi Associated Microtubule Binding 210 Gmap 210 TRIP230 |
| ENSMUSG00000021196 | ENSFM00570000877873 | Unknown |
| ENSMUSG00000021196 | ENSFM00570000886442 | Unknown |
| ENSMUSG00000021196 | ENSFM00730001521507 | Atp Dependent 6 Phosphofructokinase Type |
| ENSMUSG00000021198 | ENSFM00680001316020 | Unknown |
| ENSMUSG00000021198 | ENSFM00250000002517 | Unc 79 Homolog |
| ENSMUSG00000021219 | ENSFM00810002057142 | Unknown |
| ENSMUSG00000021219 | ENSFM00730001521808 | Regulator Of G Signaling |
| ENSMUSG00000021224 | ENSFM00570000870030 | Unknown |
| ENSMUSG00000021224 | ENSFM00570000884795 | Unknown |
| ENSMUSG00000021224 | ENSFM00250000001973 | Numb |
| ENSMUSG00000021248 | ENSFM00500000271022 | Transmembrane EMP24 Domain Containing Precursor |
| ENSMUSG00000021259 | ENSFM00380000124753 | Cholesterol 24 Hydroxylase CH24H EC_1.14.13.98 Cytochrome P450 46A1 |
| ENSMUSG00000021262 | ENSFM00740001589204 | Ena/Vasp Ena/Vasodilator Stimulated Phosphoprotein |
| ENSMUSG00000021277 | ENSFM00760001714794 | Tnf Receptor Associated Factor 3 EC_6.3.2.- CD40 Receptor Associated Factor 1 CRAF1 |
| ENSMUSG00000021279 | ENSFM00730001521452 | Serine/Threonine Kinase Mrck EC_2.7.11.1 CDC42 Binding Kinase Dmpk Myotonic Dystrophy Kinase Related CDC42 Binding Kinase Mrck Myotonic Dystrophy Kinase |
| ENSMUSG00000021288 | ENSFM00250000000749 | Kinesin Light Chain |
| ENSMUSG00000021290 | ENSFM00500000327578 | Unknown |
| ENSMUSG00000021303 | ENSFM00500000362850 | Unknown |
| ENSMUSG00000021313 | ENSFM00250000000332 | Ryanodine Receptor Ryr Muscle Calcium Release Channel Muscle Type Ryanodine Receptor Type Ryanodine Receptor |
| ENSMUSG00000021339 | ENSFM00250000006689 | Magnesium Transporter MRS2 Homolog Mitochondrial Precursor MRS2 |
| ENSMUSG00000021357 | ENSFM00250000004215 | Exocyst Complex Component 2 Exocyst Complex Component SEC5 |
| ENSMUSG00000021373 | ENSFM00250000001899 | Adenylyl Cyclase Associated Cap |
| ENSMUSG00000021451 | ENSFM00480000262746 | Semaphorin Precursor |
| ENSMUSG00000021460 | ENSFM00500000270222 | Methylglutaconyl Coa Hydratase Mitochondrial Precursor EC_4.2.1.18 Au Specific Rna Binding Enoyl Coa Hydratase Au Binding Coa Hydratase |
| ENSMUSG00000021518 | ENSFM00800002003785 | Phosphatidylserine Synthase 1 Pss 1 Ptdser Synthase 1 EC_2.7.8.29 Serine Exchange Enzyme I |
| ENSMUSG00000021536 | ENSFM00730001521648 | Adenylate Cyclase Type EC_4.6.1.1 Atp Pyrophosphate Lyase Adenylate Cyclase Type Adenylyl Cyclase |
| ENSMUSG00000021546 | ENSFM00660001174969 | Unknown |
| ENSMUSG00000021546 | ENSFM00250000001382 | Heterogeneous Nuclear Ribonucleoprotein K Hnrnp K |
| ENSMUSG00000021552 | ENSFM00250000003710 | G Kinase Anchoring 1 |
| ENSMUSG00000021559 | ENSFM00730001521806 | Death Associated Kinase Dap Kinase EC_2.7.11.1 Dap Kinase |
| ENSMUSG00000021573 | ENSFM00250000003009 | Tubulin Polymerization Promoting Family Member |
| ENSMUSG00000021576 | ENSFM00500000271967 | Programmed Cell Death 6 Probable Calcium Binding Alg 2 |
| ENSMUSG00000021578 | ENSFM00590000917390 | Unknown |
| ENSMUSG00000021578 | ENSFM00250000008534 | Coiled Coil Domain Containing 127 |
| ENSMUSG00000021591 | ENSFM00670001236435 | Glutaredoxin 1 Thioltransferase 1 Ttase 1 |
| ENSMUSG00000021606 | ENSFM00570000892282 | Unknown |
| ENSMUSG00000021606 | ENSFM00670001236132 | Nadh Dehydrogenase Iron Sulfur 6 Mitochondrial Precursor Complex I 13KD A Ci 13KD A Nadh Ubiquinone Oxidoreductase 13 Kda A Subunit |
| ENSMUSG00000021613 | ENSFM00730001521886 | Hyaluronan And Proteoglycan Link 1 Precursor Cartilage Linking 1 Cartilage Link Proteoglycan Link |
| ENSMUSG00000021614 | ENSFM00730001521436 | Core Precursor Chondroitin Sulfate Proteoglycan |
| ENSMUSG00000021646 | ENSFM00450000242169 | Methylcrotonoyl Coa Carboxylase Beta Chain Mitochondrial Precursor Mccase Subunit Beta EC_6.4.1.4 3 Methylcrotonyl Coa Carboxylase 2 3 Methylcrotonyl Coa Carboxylase Non Biotin Containing Subunit 3 Methylcrotonyl Coa:Carbon Dioxide Ligase Subunit Beta |
| ENSMUSG00000021665 | ENSFM00730001521836 | Beta Hexosaminidase Subunit Alpha Precursor EC_3.2.1.52 Beta N Acetylhexosaminidase Subunit Alpha Hexosaminidase Subunit A N Acetyl Beta Glucosaminidase Subunit Alpha |
| ENSMUSG00000021684 | ENSFM00280000058709 | High Affinity Camp Specific And Ibmx Insensitive 3' 5' Cyclic Phosphodiesterase 8B EC_3.1.4.53 Cell Proliferation Inducing Gene 22 |
| ENSMUSG00000021693 | ENSFM00250000000916 | Kinesin |
| ENSMUSG00000021699 | ENSFM00640001105232 | Unknown |
| ENSMUSG00000021699 | ENSFM00400000131721 | Camp Specific 3' 5' Cyclic Phosphodiesterase EC_3.1.4.53 |
| ENSMUSG00000021704 | ENSFM00250000002187 | Metaxin 1 Mitochondrial Outer Membrane Import Complex 1 |
| ENSMUSG00000021708 | ENSFM00250000001613 | Ras Specific Guanine Nucleotide Releasing Factor 2 Ras GRF2 Ras Guanine Nucleotide Exchange Factor 2 |
| ENSMUSG00000021709 | ENSFM00730001522141 | Leucine Rich Repeat Containing 7 Densin 180 Densin |
| ENSMUSG00000021712 | ENSFM00590000917832 | Unknown |
| ENSMUSG00000021712 | ENSFM00730001522472 | E3 Ubiquitin Ligase TRIM23 EC_6.3.2.- Adp Ribosylation Factor Domain Containing 1 Gtp Binding Ard 1 Tripartite Motif Containing 23 |
| ENSMUSG00000021719 | ENSFM00250000004063 | Regulator Of G Signaling 7 Binding R7 Family Binding |
| ENSMUSG00000021731 | ENSFM00250000008011 | 28S Ribosomal S30 Mitochondrial Mrp S30 S30MT |
| ENSMUSG00000021737 | ENSFM00760001714781 | 26S Proteasome Non Atpase Regulatory Subunit 6 26S Proteasome Regulatory Subunit RPN7 26S Proteasome Regulatory Subunit S10 P42A |
| ENSMUSG00000021745 | ENSFM00730001521864 | Receptor Type Tyrosine Phosphatase Precursor R Ptp EC_3.1.3.48 |
| ENSMUSG00000021748 | ENSFM00500000270150 | Pyruvate Dehydrogenase E1 Component Subunit Beta Mitochondrial Precursor PDHE1 B EC_1.2.4.1 |
| ENSMUSG00000021750 | ENSFM00500000273986 | Unknown |
| ENSMUSG00000021771 | ENSFM00570000888306 | Unknown |
| ENSMUSG00000021771 | ENSFM00250000001189 | Voltage Dependent Anion Selective Channel Vdac Outer Mitochondrial Membrane Porin |
| ENSMUSG00000021772 | ENSFM00570000869836 | Unknown |
| ENSMUSG00000021772 | ENSFM00250000003149 | Nf Kappa B Inhibitor Interacting Ras 2 I Kappa B Interacting Ras 2 Kappa B Ras 2 Kappab RAS2 |
| ENSMUSG00000021773 | ENSFM00270000056553 | Catechol O Methyltransferase Domain Containing 1 EC_2.1.1.- |
| ENSMUSG00000021791 | ENSFM00670001244624 | DPY30 Domain Containing 2 |
| ENSMUSG00000021792 | ENSFM00570000881559 | Unknown |
| ENSMUSG00000021792 | ENSFM00250000005616 | Redox Regulatory FAM213A Peroxiredoxin 2 Activated In M Csf Stimulated Monocytes Pamm |
| ENSMUSG00000021794 | ENSFM00250000001520 | Glutamate Dehydrogenase Mitochondrial Gdh EC_1.4.1.3 |
| ENSMUSG00000021807 | ENSFM00250000005286 | Homolog |
| ENSMUSG00000021814 | ENSFM00760001714601 | Annexin Annexin Annexin |
| ENSMUSG00000021820 | ENSFM00250000000111 | Calcium/Calmodulin Dependent Kinase Type Ii Subunit Cam Kinase Ii Subunit Camk Ii Subunit EC_2.7.11.17 |
| ENSMUSG00000021823 | ENSFM00250000001750 | Vinculin |
| ENSMUSG00000021824 | ENSFM00440000236903 | Ap 3 Complex Subunit Mu Adaptor Related Complex 3 Subunit Mu Clathrin Assembly Assembly Complex 3 Mu Medium Chain Clathrin Coat Assembly AP47 Homolog Clathrin Coat Associated AP47 Homolog Golgi Adaptor Ap 1.47 Kda Homolog HA1.47 Kda Subunit Homolog Adapti |
| ENSMUSG00000021832 | ENSFM00720001492034 | 26S Protease Regulatory Subunit 10B 26S Proteasome Aaa Atpase Subunit RPT4 Proteasome 26S Subunit Atpase 6 Proteasome Subunit P42 |
| ENSMUSG00000021843 | ENSFM00250000002379 | Kinectin |
| ENSMUSG00000021866 | ENSFM00760001714601 | Annexin Annexin Annexin |
| ENSMUSG00000021870 | ENSFM00250000002303 | Sarcolemmal Membrane Associated Sarcolemmal Associated |
| ENSMUSG00000021910 | ENSFM00610000966894 | Unknown |
| ENSMUSG00000021910 | ENSFM00610000972849 | Unknown |
| ENSMUSG00000021910 | ENSFM00250000002121 | Nischarin Imidazoline Receptor 1 I 1 IR1 Imidazoline Receptor 1 Imidazoline 1 Receptor I1R |
| ENSMUSG00000021913 | ENSFM00750001632363 | 2 Oxoglutarate Dehydrogenase Mitochondrial Precursor 2 Oxoglutarate Dehydrogenase Complex Component E1 Ogdc E1 Alpha Ketoglutarate Dehydrogenase |
| ENSMUSG00000021917 | ENSFM00760001723268 | Unknown |
| ENSMUSG00000021917 | ENSFM00500000275173 | Signal Peptidase Complex Subunit 1 EC_3.4.-.- Microsomal Signal Peptidase 12 Kda Subunit Spase 12 Kda Subunit |
| ENSMUSG00000021930 | ENSFM00250000004208 | Spry Domain Containing 7 Chronic Lymphocytic Leukemia Deletion Region Gene 6 Homolog Cll Deletion Region Gene 6 Homolog |
| ENSMUSG00000021938 | ENSFM00610000969895 | Unknown |
| ENSMUSG00000021938 | ENSFM00250000000671 | Paraspeckle Component 1 |
| ENSMUSG00000021986 | ENSFM00250000005156 | Apc Membrane Recruitment 2 AMER2 FAM123A |
| ENSMUSG00000021991 | ENSFM00730001521792 | Voltage Dependent Calcium Channel Subunit Alpha 2/Delta 3 Precursor Voltage Gated Calcium Channel Subunit Alpha 2/Delta 3 |
| ENSMUSG00000021994 | ENSFM00740001589193 | Wnt 5 |
| ENSMUSG00000022019 | ENSFM00250000004146 | Tudor Domain Containing 3 |
| ENSMUSG00000022024 | ENSFM00500000271163 | Suppressor Of G2 Allele Of SKP1 Homolog |
| ENSMUSG00000022037 | ENSFM00570000900344 | Unknown |
| ENSMUSG00000022037 | ENSFM00250000003194 | Clusterin Precursor |
| ENSMUSG00000022052 | ENSFM00250000000829 | Serine/Threonine Phosphatase 2A 55 Kda Regulatory Subunit B PP2A Subunit B B55 PP2A Subunit B PR55 PP2A Subunit B R2 PP2A Subunit B |
| ENSMUSG00000022054 | ENSFM00730001521354 | Neurofilament Polypeptide Nf Kda Neurofilament Neurofilament Triplet |
| ENSMUSG00000022055 | ENSFM00730001521354 | Neurofilament Polypeptide Nf Kda Neurofilament Neurofilament Triplet |
| ENSMUSG00000022099 | ENSFM00500000271390 | Dematin Dematin Actin Binding Erythrocyte Membrane Band 4 9 |
| ENSMUSG00000022103 | ENSFM00500000269996 | Gdnf Family Receptor Alpha Precursor Gdnf Receptor Alpha Gdnfr Alpha Gfr Alpha Tgf Beta Related Neurotrophic Factor Receptor |
| ENSMUSG00000022108 | ENSFM00600000921249 | Integral Membrane 2B Immature BRI2 IMBRI2 Transmembrane Bri Bri |
| ENSMUSG00000022112 | ENSFM00500000270283 | Glypican 3 Precursor Intestinal Oci 5 |
| ENSMUSG00000022114 | ENSFM00250000002344 | Sprouty Homolog Spry |
| ENSMUSG00000022132 | ENSFM00670001235591 | Claudin |
| ENSMUSG00000022150 | ENSFM00590000917915 | Unknown |
| ENSMUSG00000022150 | ENSFM00590000918392 | Unknown |
| ENSMUSG00000022150 | ENSFM00740001589270 | Disabled Homolog 2 Adaptor Molecule Disabled 2 Differentially Expressed In Ovarian Carcinoma 2 Doc 2 |
| ENSMUSG00000022176 | ENSFM00260000050685 | Gtp Binding Rem Rad And Gem Gtp Binding |
| ENSMUSG00000022208 | ENSFM00250000001356 | Junctophilin Jp Junctophilin |
| ENSMUSG00000022210 | ENSFM00500000270135 | Dehydrogenase/Reductase Sdr Family Member 4 EC_1.1.1.184 Nadph Dependent Carbonyl Reductase/Nadp Retinol Dehydrogenase Cr Phcr Nadph Dependent Retinol Dehydrogenase/Reductase Ndrd Peroxisomal Short Chain Alcohol Dehydrogenase Pscd |
| ENSMUSG00000022212 | ENSFM00800002003632 | Copine Copine |
| ENSMUSG00000022234 | ENSFM00730001521615 | T Complex 1 Subunit Epsilon Tcp 1 Epsilon Cct Epsilon |
| ENSMUSG00000022240 | ENSFM00730001521673 | Catenin Delta 2 |
| ENSMUSG00000022246 | ENSFM00800002003877 | Ankycorbin Ankyrin Repeat And Coiled Coil Structure Containing Retinal Pigment Epithelial Cell Retinoic Acid Induced 14 |
| ENSMUSG00000022255 | ENSFM00250000005409 | Lyric Lysine Rich CEACAM1 Co Isolated Metadherin Metastasis Adhesion |
| ENSMUSG00000022263 | ENSFM00740001589124 | EC_2.7.11.1 |
| ENSMUSG00000022265 | ENSFM00250000005694 | Progressive Ankylosis Homolog Ank |
| ENSMUSG00000022283 | ENSFM00570000851024 | Polyadenylate Binding Pabp Poly A Binding |
| ENSMUSG00000022285 | ENSFM00500000361339 | Unknown |
| ENSMUSG00000022285 | ENSFM00570000901593 | Unknown |
| ENSMUSG00000022285 | ENSFM00730001521154 | Unknown |
| ENSMUSG00000022295 | ENSFM00750001632375 | V Type Proton Atpase Subunit C V Atpase Subunit C Vacuolar Proton Pump Subunit C |
| ENSMUSG00000022321 | ENSFM00260000050333 | Cadherin Precursor |
| ENSMUSG00000022337 | ENSFM00250000004805 | Er Membrane Complex Subunit 2 Tetratricopeptide Repeat 35 Tpr Repeat 35 |
| ENSMUSG00000022340 | ENSFM00250000002727 | Syntabulin Golgi Localized Syntaphilin Related Syntaxin 1 Binding |
| ENSMUSG00000022350 | ENSFM00250000005256 | Wash Complex Subunit Strumpellin |
| ENSMUSG00000022354 | ENSFM00250000004530 | Nadh Dehydrogenase 1 Beta Subcomplex Subunit 9 Complex I B22 Ci B22 Nadh Ubiquinone Oxidoreductase B22 Subunit |
| ENSMUSG00000022390 | ENSFM00250000002307 | Zinc Finger Ccch Domain Containing |
| ENSMUSG00000022401 | ENSFM00610000960709 | Unknown |
| ENSMUSG00000022401 | ENSFM00800002003832 | Probable Xaa Pro Aminopeptidase 3 X Pro Aminopeptidase 3 EC_3.4.11.9 Aminopeptidase P3 APP3 |
| ENSMUSG00000022427 | ENSFM00600000921823 | Mitochondrial Import Receptor Subunit TOM22 Homolog Translocase Of Outer Membrane 22 Kda Subunit Homolog |
| ENSMUSG00000022428 | ENSFM00600000921664 | Chibby Homolog 1 Cytosolic Leucine Rich Pigea 14 PKD2 Interactor Golgi And Endoplasmic Reticulum Associated 1 |
| ENSMUSG00000022433 | ENSFM00570000881021 | Unknown |
| ENSMUSG00000022433 | ENSFM00800002003414 | Casein Kinase I Delta Cki Delta Ckid EC_2.7.11.1 |
| ENSMUSG00000022437 | ENSFM00500000271410 | Sorting And Assembly Machinery Component 50 Homolog |
| ENSMUSG00000022443 | ENSFM00270000056424 | Myosin Cellular Myosin Heavy Chain Type Myosin Heavy Chain Myosin Heavy Chain Non Muscle Non Muscle Myosin Heavy Chain Nmmhc Non Muscle Myosin Heavy Chain Nmmhc Ii Nmmhc |
| ENSMUSG00000022450 | ENSFM00500000272594 | Nadh Dehydrogenase 1 Alpha Subcomplex Subunit 6 Complex I B14 Ci B14 Nadh Ubiquinone Oxidoreductase B14 Subunit |
| ENSMUSG00000022451 | ENSFM00500000270278 | Twinfilin |
| ENSMUSG00000022456 | ENSFM00500000269805 | Septin |
| ENSMUSG00000022489 | ENSFM00400000131765 | Calcium/Calmodulin Dependent 3' 5' Cyclic Nucleotide Phosphodiesterase Cam Pde EC_3.1.4.17 Kda Cam Pde |
| ENSMUSG00000022523 | ENSFM00500000269773 | Fibroblast Growth Factor Fgf Fibroblast Growth Factor Homologous Factor Fhf |
| ENSMUSG00000022540 | ENSFM00250000005331 | Rogdi Homolog |
| ENSMUSG00000022551 | ENSFM00250000003946 | Cytochrome C1 Heme Protein Mitochondrial Precursor Complex Iii Subunit 4 Complex Iii Subunit Iv Cytochrome B C1 Complex Subunit 4 Ubiquinol Cytochrome C Reductase Complex Cytochrome C1 Subunit Cytochrome C 1 |
| ENSMUSG00000022565 | ENSFM00610000973117 | Unknown |
| ENSMUSG00000022565 | ENSFM00610000973118 | Unknown |
| ENSMUSG00000022565 | ENSFM00740001589199 | Plectin Pcn Pltn Plectin 1 |
| ENSMUSG00000022568 | ENSFM00570000899077 | Unknown |
| ENSMUSG00000022568 | ENSFM00730001522046 | Scribble |
| ENSMUSG00000022571 | ENSFM00500000269896 | Pyrroline 5 Carboxylate Reductase 3 P5C Reductase 3 P5CR 3 EC_1.5.1.2 Pyrroline 5 Carboxylate Reductase |
| ENSMUSG00000022602 | ENSFM00250000009482 | Activity Regulated Cytoskeleton Associated Arc/ARG3 1 Activity Regulated Gene 3 1 ARG3 1 |
| ENSMUSG00000022604 | ENSFM00250000003841 | Centrosomal Of 97 Kda CEP97 Leucine Rich Repeat And Iq Domain Containing 2 |
| ENSMUSG00000022623 | ENSFM00250000000743 | SH3 And Multiple Ankyrin Repeat Domains Proline Rich Synapse Associated |
| ENSMUSG00000022623 | ENSFM00800002009948 | SHANK3E Splice Variant |
| ENSMUSG00000022658 | ENSFM00500000270015 | Transgelin |
| ENSMUSG00000022671 | ENSFM00670001236572 | Mitotic Spindle Organizing Mitotic Spindle Organizing Associated With A Ring Of Gamma Tubulin |
| ENSMUSG00000022674 | ENSFM00520000550014 | Unknown |
| ENSMUSG00000022674 | ENSFM00250000001221 | Ubiquitin Conjugating Enzyme E2 Variant |
| ENSMUSG00000022706 | ENSFM00500000274041 | 39S Ribosomal L40 Mitochondrial Precursor L40MT Mrp L40 Nuclear Localization Signal Containing Deleted In Velocardiofacial Syndrome |
| ENSMUSG00000022752 | ENSFM00710001441924 | Mitochondrial Import Receptor Subunit TOM70 Mitochondrial Precursor Proteins Import Receptor Translocase Of Outer Membrane 70 Kda Subunit |
| ENSMUSG00000022763 | ENSFM00800002003733 | Apoptosis Inducing Factor 3 Ec 1 Apoptosis Inducing Factor |
| ENSMUSG00000022765 | ENSFM00250000006834 | Synaptosomal Associated 29 Snap 29 Golgi Snare Of 32 Kda GS32 Soluble 29 Kda Nsf Attachment Vesicle Membrane Fusion Snap 29 |
| ENSMUSG00000022770 | ENSFM00720001503508 | Unknown |
| ENSMUSG00000022770 | ENSFM00250000000275 | Disks Large Homolog Synapse Associated Sap |
| ENSMUSG00000022789 | ENSFM00780001898359 | Dynamin 1 EC_3.6.5.5 Dynamin Family Member Proline Rich Carboxyl Terminal Domain Less Dymple Dynamin Related 1 |
| ENSMUSG00000022792 | ENSFM00250000003377 | Tyrosine Trna Ligase Mitochondrial Precursor EC_6.1.1.1 Tyrosyl Trna Synthetase Tyrrs |
| ENSMUSG00000022799 | ENSFM00250000001205 | Rho Gtpase Activating Rho Type Gtpase Activating Gtpase Activating |
| ENSMUSG00000022801 | ENSFM00610000970063 | Unknown |
| ENSMUSG00000022801 | ENSFM00800002003638 | Leucine Rich Repeat And Calponin Homology Domain Containing |
| ENSMUSG00000022802 | ENSFM00250000004185 | Leishmanolysin Peptidase EC_3.4.24.- |
| ENSMUSG00000022812 | ENSFM00810002040612 | Glycogen Synthase Kinase 3 Gsk 3 EC_2.7.11.26 Serine/Threonine Kinase EC_2.7.11.- 1 |
| ENSMUSG00000022820 | ENSFM00500000272985 | Nadh Dehydrogenase 1 Beta Subcomplex Subunit 4 Complex I B15 Ci B15 Nadh Ubiquinone Oxidoreductase B15 Subunit |
| ENSMUSG00000022836 | ENSFM00740001589296 | Myosin Light Chain Kinase Smooth Muscle Mlck Smmlck EC_2.7.11.18 Telokin |
| ENSMUSG00000022841 | ENSFM00730001521696 | Ap 2 Complex Subunit Mu Ap 2 Mu Chain Clathrin Assembly Complex 2 Mu Medium Chain Clathrin Coat Assembly AP50 Clathrin Coat Associated AP50 MU2 Adaptin Plasma Membrane Adaptor Ap Kda |
| ENSMUSG00000022843 | ENSFM00250000000612 | Chloride Channel Clc |
| ENSMUSG00000022861 | ENSFM00730001521692 | Diacylglycerol Kinase Dag Kinase EC_2.7.1.107 Kda Diacylglycerol Kinase Diglyceride Kinase Dgk |
| ENSMUSG00000022883 | ENSFM00740001589121 | Roundabout Homolog Precursor |
| ENSMUSG00000022884 | ENSFM00730001521378 | Atp Dependent Rna Helicase EIF4A EC_3.6.4.13 Eukaryotic Initiation Factor 4A Eif 4A Translation Initiation Factor 1 |
| ENSMUSG00000022897 | ENSFM00730001521969 | Dual Specificity Tyrosine Phosphorylation Regulated Kinase Dual Specificity YAK1 Related Kinase Minibrain |
| ENSMUSG00000022956 | ENSFM00570000882034 | Unknown |
| ENSMUSG00000022956 | ENSFM00670001235592 | Atp Synthase Subunit O Mitochondrial Precursor Oligomycin Sensitivity Conferral Oscp |
| ENSMUSG00000022956 | ENSFM00250000006301 | Downstream Neighbor Of Son |
| ENSMUSG00000022994 | ENSFM00730001521247 | Adenylate Cyclase Type EC_4.6.1.1 Atp Pyrophosphate Lyase Adenylate Cyclase Type Adenylyl Cyclase Ca 2+ Adenylyl Cyclase |
| ENSMUSG00000022995 | ENSFM00740001589204 | Ena/Vasp Ena/Vasodilator Stimulated Phosphoprotein |
| ENSMUSG00000023011 | ENSFM00500000269895 | Lifeguard 2 Fas Apoptotic Inhibitory Molecule 2 |
| ENSMUSG00000023017 | ENSFM00250000000608 | Acid Sensing Ion Channel Amiloride Sensitive Cation Channel |
| ENSMUSG00000023020 | ENSFM00500000441205 | Unknown |
| ENSMUSG00000023022 | ENSFM00500000271499 | Lim Domain And Actin Binding 1 Epithelial Lost In Neoplasm |
| ENSMUSG00000023026 | ENSFM00250000000652 | Disco Interacting 2 Homolog DIP2 Homolog |
| ENSMUSG00000023055 | ENSFM00500000271639 | Calcium Binding And Coiled Coil Domain Containing 1 |
| ENSMUSG00000023192 | ENSFM00790001963004 | Metabotropic Glutamate Receptor Precursor |
| ENSMUSG00000023216 | ENSFM00730001521321 | Glutamine Gamma Glutamyltransferase EC_2.3.2.13 Transglutaminase Tg Tgase Transglutaminase Tgase |
| ENSMUSG00000023266 | ENSFM00570000896626 | Unknown |
| ENSMUSG00000023266 | ENSFM00250000003196 | Fibroblast Growth Factor Receptor Substrate 3 Fgfr Substrate 3 Fgfr Signaling Adaptor SNT2 SUC1 Associated Neurotrophic Factor Target 2 Snt 2 |
| ENSMUSG00000023277 | ENSFM00760001763416 | Unknown |
| ENSMUSG00000023277 | ENSFM00760001763417 | Unknown |
| ENSMUSG00000023277 | ENSFM00500000270278 | Twinfilin |
| ENSMUSG00000023307 | ENSFM00250000003363 | E3 Ubiquitin Ligase MARCH5 EC_6.3.2.- Membrane Associated Ring Finger 5 Membrane Associated Ring Ch V March V |
| ENSMUSG00000023353 | ENSFM00250000000604 | Arf Gap With Gtpase Ank Repeat And Ph Domain Containing Agap Centaurin Gamma |
| ENSMUSG00000023439 | ENSFM00250000000545 | Guanine Nucleotide Binding G I /G S /G T Subunit Beta Transducin Beta Chain |
| ENSMUSG00000023452 | ENSFM00570000893700 | Unknown |
| ENSMUSG00000023452 | ENSFM00800002023580 | Unknown |
| ENSMUSG00000023452 | ENSFM00600000921219 | Phosphatidylserine Decarboxylase Proenzyme EC_4.1.1.65 |
| ENSMUSG00000023456 | ENSFM00250000000728 | Triosephosphate Isomerase Tim EC_5.3.1.1 Triose Phosphate Isomerase |
| ENSMUSG00000023460 | ENSFM00720001492078 | Ras Related Rab 12 |
| ENSMUSG00000023484 | ENSFM00730001521291 | Vimentin |
| ENSMUSG00000023723 | ENSFM00500000272974 | 28S Ribosomal S23 Mitochondrial Mrp S23 S23MT |
| ENSMUSG00000023912 | ENSFM00500000270868 | Mitochondrial Uncoupling 4 Ucp 4 Solute Carrier Family 25 Member 27 |
| ENSMUSG00000023932 | ENSFM00250000005324 | Pre Splicing Factor CEF1 |
| ENSMUSG00000023939 | ENSFM00500000273213 | 39S Ribosomal L14 Mitochondrial Precursor L14MT Mrp L14 39S Ribosomal L32 Mitochondrial L32MT Mrp L32 |
| ENSMUSG00000023960 | ENSFM00570000888029 | Unknown |
| ENSMUSG00000023960 | ENSFM00760001714771 | Bis 5' Adenosyl Triphosphatase ENPP4 Precursor EC_3.6.1.29 AP3A Hydrolase AP3AASE Ectonucleotide Pyrophosphatase/Phosphodiesterase Family Member 4 E Npp 4 Npp 4 |
| ENSMUSG00000024008 | ENSFM00730001521728 | Copine Copine |
| ENSMUSG00000024012 | ENSFM00250000001870 | Mitochondrial Carrier Homolog |
| ENSMUSG00000024038 | ENSFM00760001752576 | Uncharacterized |
| ENSMUSG00000024038 | ENSFM00500000273812 | Nadh Dehydrogenase Flavoprotein 3 Mitochondrial |
| ENSMUSG00000024038 | ENSFM00500000301275 | Nadh Dehydrogenase Flavoprotein 3 Mitochondrial Precursor Complex I 9KD Ci 9KD Nadh Ubiquinone Oxidoreductase 9 Kda Subunit |
| ENSMUSG00000024044 | ENSFM00800002003400 | Unknown |
| ENSMUSG00000024065 | ENSFM00250000000766 | Eh Domain Containing |
| ENSMUSG00000024068 | ENSFM00800002003737 | Spastin |
| ENSMUSG00000024077 | ENSFM00250000001473 | Striatin |
| ENSMUSG00000024091 | ENSFM00250000001879 | Vesicle Associated Membrane Associated A Vamp A Vamp Associated A Vap A |
| ENSMUSG00000024096 | ENSFM00250000003792 | Rala Binding 1 RALBP1 Dinitrophenyl S Glutathione Atpase Dnp Sg Atpase Ral Interacting 1 |
| ENSMUSG00000024099 | ENSFM00570000901685 | Unknown |
| ENSMUSG00000024099 | ENSFM00700001404061 | Unknown |
| ENSMUSG00000024099 | ENSFM00250000003401 | Nadh Dehydrogenase Flavoprotein 2 Mitochondrial Precursor EC_1.6.5.3 EC_1.6.99.- 3 Nadh Ubiquinone Oxidoreductase 24 Kda Subunit |
| ENSMUSG00000024101 | ENSFM00250000002535 | Was Family Homolog |
| ENSMUSG00000024121 | ENSFM00570000891623 | V Type Proton Atpase 16 Kda Proteolipid Subunit |
| ENSMUSG00000024121 | ENSFM00250000002076 | V Type Proton Atpase 16 Kda Proteolipid Subunit V Atpase 16 Kda Proteolipid Subunit Vacuolar Proton Pump 16 Kda Proteolipid Subunit |
| ENSMUSG00000024122 | ENSFM00570000891895 | Unknown |
| ENSMUSG00000024122 | ENSFM00800002003539 | 3 Phosphoinositide Dependent Kinase EC_2.7.11.1 |
| ENSMUSG00000024143 | ENSFM00730001521235 | Cell Division Control 42 Homolog Precursor |
| ENSMUSG00000024163 | ENSFM00250000000947 | C Jun Amino Terminal Kinase Interacting Jip Jnk Interacting Jnk Mitogen Activated Kinase 8 Interacting |
| ENSMUSG00000024187 | ENSFM00250000006994 | ITFG3 |
| ENSMUSG00000024208 | ENSFM00670001236187 | Ubiquinol Cytochrome C Reductase Complex Assembly Factor 2 Precursor Mitochondrial Nucleoid Factor 1 Mitochondrial M19 |
| ENSMUSG00000024211 | ENSFM00570000893452 | Unknown |
| ENSMUSG00000024211 | ENSFM00250000000328 | Metabotropic Glutamate Receptor Precursor |
| ENSMUSG00000024234 | ENSFM00250000005018 | Poly A Rna Polymerase Mitochondrial Precursor Pap EC_2.7.7.19 Pap Associated Domain Containing 1 Polynucleotide Adenylyltransferase |
| ENSMUSG00000024248 | ENSFM00500000274370 | Cytochrome C Oxidase Subunit 7A Related Protein Mitochondrial Precursor Cytochrome C Oxidase Subunit Viia Related |
| ENSMUSG00000024259 | ENSFM00250000005083 | Solute Carrier Family 25 Member 46 |
| ENSMUSG00000024270 | ENSFM00570000883974 | Unknown |
| ENSMUSG00000024270 | ENSFM00500000270550 | Zinc Transporter ZIP6 Precursor Solute Carrier Family 39 Member 6 Zrt And Irt 6 Zip 6 |
| ENSMUSG00000024277 | ENSFM00610000970007 | Unknown |
| ENSMUSG00000024277 | ENSFM00250000000867 | Microtubule Associated Rp/Eb Family Member |
| ENSMUSG00000024286 | ENSFM00250000001383 | Cyclin Y |
| ENSMUSG00000024300 | ENSFM00780001898391 | Unconventional Myosin Ie Unconventional Myosin 1E |
| ENSMUSG00000024304 | ENSFM00730001521172 | Cadherin Precursor Cadherin Cadherin |
| ENSMUSG00000024319 | ENSFM00500000271174 | Vacuolar Sorting Associated 52 Homolog SAC2 Suppressor Of Actin Mutations 2 |
| ENSMUSG00000024327 | ENSFM00500000271041 | Zinc Transporter Sl Histidine Rich Membrane KE4 Solute Carrier Family 39 Member 7 Zrt Irt 7 ZIP7 |
| ENSMUSG00000024350 | ENSFM00250000001153 | Dnaj Homolog Subfamily Member |
| ENSMUSG00000024360 | ENSFM00680001326074 | Unknown |
| ENSMUSG00000024360 | ENSFM00250000005309 | Eukaryotic Peptide Chain Release Factor Subunit 1 Eukaryotic Release Factor 1 ERF1 |
| ENSMUSG00000024387 | ENSFM00500000464452 | Unknown |
| ENSMUSG00000024387 | ENSFM00800002003472 | Casein Kinase Ii Subunit Beta Ck Ii Beta |
| ENSMUSG00000024393 | ENSFM00740001589307 | PRRC2A Hla B Associated Transcript 2 Proline Rich And Coiled Coil Containing 2A |
| ENSMUSG00000024403 | ENSFM00640001109543 | Unknown |
| ENSMUSG00000024403 | ENSFM00600000921884 | V Type Proton Atpase Subunit G 2 V Atpase Subunit G 2 Vacuolar Proton Pump Subunit G 2 |
| ENSMUSG00000024410 | ENSFM00250000005107 | Uncharacterized Colon Cancer Associated MIC1 Mic 1 |
| ENSMUSG00000024426 | ENSFM00570000879961 | Unknown |
| ENSMUSG00000024426 | ENSFM00250000002340 | Alpha Tubulin N Acetyltransferase 1 |
| ENSMUSG00000024462 | ENSFM00250000000928 | Gamma Aminobutyric Acid Type B Receptor Subunit 2 Precursor Gaba B Receptor 2 Gaba B R2 Gaba BR2 GABABR2 GB2 G Coupled Receptor 51 |
| ENSMUSG00000024479 | ENSFM00500000273729 | MAL2 |
| ENSMUSG00000024480 | ENSFM00500000270528 | Ap 3 Complex Subunit Sigma Ap 3 Complex Subunit Sigma Adaptor Related Complex 3 Subunit Sigma Sigma Adaptin Adaptin Sigma Adaptin |
| ENSMUSG00000024493 | ENSFM00250000002483 | Leucine Trna Ligase Cytoplasmic EC_6.1.1.4 Leucyl Trna Synthetase Leurs |
| ENSMUSG00000024502 | ENSFM00250000001480 | Janus Kinase And Microtubule Interacting 1 Multiple Alpha Helices And Rna Linker 1 Marlin 1 |
| ENSMUSG00000024507 | ENSFM00500000270110 | Peroxisomal Multifunctional Enzyme Type 2 Mfe 2 17 Beta Hydroxysteroid Dehydrogenase 4 17 Beta Hsd 4 D Bifunctional Dbp Multifunctional 2 Mpf 2 |
| ENSMUSG00000024516 | ENSFM00790001962996 | Signal Peptidase Complex Catalytic Subunit SEC11A EC_3.4.21.89 Endopeptidase SP18 Microsomal Signal Peptidase 18 Kda Subunit Spase 18 Kda Subunit SEC11 Homolog A SEC11 1 SPC18 |
| ENSMUSG00000024524 | ENSFM00300000079126 | Guanine Nucleotide Binding G S Subunit Alpha Adenylate Cyclase Stimulating G Alpha |
| ENSMUSG00000024527 | ENSFM00800002003441 | AFG3 EC_3.4.24.- |
| ENSMUSG00000024533 | ENSFM00250000001244 | Spire Homolog |
| ENSMUSG00000024576 | ENSFM00770001847535 | Casein Kinase I Alpha Cki Alpha EC_2.7.11.1 CK1 |
| ENSMUSG00000024583 | ENSFM00500000271170 | Thioredoxin Thioredoxin Related |
| ENSMUSG00000024588 | ENSFM00250000003660 | Ferrochelatase Mitochondrial Precursor EC_4.99.1.1 Heme Synthase Protoheme Ferro Lyase |
| ENSMUSG00000024589 | ENSFM00730001521822 | E3 Ubiquitin Ligase EC_6.3.2.- |
| ENSMUSG00000024590 | ENSFM00250000000425 | Lamin |
| ENSMUSG00000024597 | ENSFM00260000050342 | Solute Carrier Family 12 Member Bumetanide Sensitive Sodium Potassium Chloride Cotransporter Na K Cl Symporter |
| ENSMUSG00000024603 | ENSFM00250000004748 | Dynactin Subunit 4 Dynactin Subunit P62 |
| ENSMUSG00000024608 | ENSFM00250000003648 | 40S Ribosomal S14 |
| ENSMUSG00000024617 | ENSFM00500000347513 | Unknown |
| ENSMUSG00000024617 | ENSFM00250000000111 | Calcium/Calmodulin Dependent Kinase Type Ii Subunit Cam Kinase Ii Subunit Camk Ii Subunit EC_2.7.11.17 |
| ENSMUSG00000024639 | ENSFM00730001521471 | Guanine Nucleotide Binding G Q Subunit Alpha Guanine Nucleotide Binding Alpha Q |
| ENSMUSG00000024661 | ENSFM00500000269681 | Ferritin Heavy Chain Ferritin H Subunit EC_1.16.3.1 |
| ENSMUSG00000024740 | ENSFM00810002040719 | Dna Damage Binding 1 Damage Specific Dna Binding 1 |
| ENSMUSG00000024743 | ENSFM00740001589375 | Synaptotagmin 7 Synaptotagmin Vii Sytvii |
| ENSMUSG00000024767 | ENSFM00500000272123 | Ubiquitin Thioesterase OTUB1 EC_3.4.19.12 Deubiquitinating Enzyme OTUB1 Otu Domain Containing Ubiquitin Aldehyde Binding 1 Otubain 1 Ubiquitin Specific Processing Protease OTUB1 |
| ENSMUSG00000024772 | ENSFM00250000000766 | Eh Domain Containing |
| ENSMUSG00000024777 | ENSFM00760001714676 | Serine/Threonine Phosphatase 2A 56 Kda Regulatory Subunit Epsilon PP2A B Subunit B' Epsilon PP2A B Subunit B56 Epsilon PP2A B Subunit PR61 Epsilon PP2A B Subunit R5 Epsilon |
| ENSMUSG00000024782 | ENSFM00570000883900 | Unknown |
| ENSMUSG00000024782 | ENSFM00500000270040 | Adenylate Kinase 4 Mitochondrial |
| ENSMUSG00000024797 | ENSFM00590000917614 | Unknown |
| ENSMUSG00000024797 | ENSFM00250000003232 | Vacuolar Sorting Associated 51 Homolog Fat Free Homolog |
| ENSMUSG00000024812 | ENSFM00250000000691 | Tight Junction Zo Tight Junction Zona Occludens Zonula Occludens |
| ENSMUSG00000024835 | ENSFM00730001521292 | Coronin |
| ENSMUSG00000024844 | ENSFM00670001236579 | Barrier To Autointegration Factor Breakpoint Cluster Region 1 |
| ENSMUSG00000024855 | ENSFM00250000001313 | Phosphofurin Acidic Cluster Sorting Pacs |
| ENSMUSG00000024858 | ENSFM00610000967883 | Unknown |
| ENSMUSG00000024858 | ENSFM00610000970067 | Unknown |
| ENSMUSG00000024858 | ENSFM00800002003453 | Beta Adrenergic Receptor Kinase 1 Beta Ark 1 EC_2.7.11.15 G Coupled Receptor Kinase 2 |
| ENSMUSG00000024862 | ENSFM00250000000749 | Kinesin Light Chain |
| ENSMUSG00000024883 | ENSFM00740001589251 | Ras And Rab Interactor Ras Interaction/Interference |
| ENSMUSG00000024885 | ENSFM00250000000672 | Aldehyde Dehydrogenase Aldehyde Dehydrogenase Aldehyde Dehydrogenase Family 3 Member |
| ENSMUSG00000024900 | ENSFM00500000269758 | Carnitine O Palmitoyltransferase 1 Muscle CPT1 M EC_2.3.1.21 Carnitine O Palmitoyltransferase I Muscle Cpt I Cpti M Carnitine Palmitoyltransferase 1B |
| ENSMUSG00000024902 | ENSFM00500000272206 | 39S Ribosomal L11 Mitochondrial Precursor L11MT Mrp L11 |
| ENSMUSG00000024911 | ENSFM00250000004633 | Acidic Fibroblast Growth Factor Intracellular Binding Afgf Intracellular Binding Fgf 1 Intracellular Binding |
| ENSMUSG00000024960 | ENSFM00800002003424 | 1 Phosphatidylinositol 4 5 Bisphosphate Phosphodiesterase Beta EC_3.1.4.11 Phosphoinositide Phospholipase C Beta Phospholipase C Beta Plc Beta |
| ENSMUSG00000024969 | ENSFM00610000972490 | Unknown |
| ENSMUSG00000024969 | ENSFM00610000972491 | Unknown |
| ENSMUSG00000024969 | ENSFM00730001521220 | Kinase EC_2.7.11.1 |
| ENSMUSG00000024974 | ENSFM00310000089021 | Structural Maintenance Of Chromosomes 3 Smc 3 Smc 3 Chondroitin Sulfate Proteoglycan 6 |
| ENSMUSG00000024976 | ENSFM00500000270686 | Leucine Rich Repeat Soc 2 Homolog Sur 8 Homolog Soc 2 Homolog |
| ENSMUSG00000024983 | ENSFM00500000272468 | Vesicle Transport Through Interaction With T Snares Homolog 1A Vesicle Transport V Snare VTI1 2 VTI1 RP2 |
| ENSMUSG00000025006 | ENSFM00670001235644 | Sorbin And SH3 Domain Containing 1 Ponsin SH3 Domain 5 S C Cbl Associated Cap |
| ENSMUSG00000025026 | ENSFM00250000000681 | Adducin |
| ENSMUSG00000025040 | ENSFM00670001235667 | FUN14 Domain Containing |
| ENSMUSG00000025059 | ENSFM00780001898348 | Glycerol Kinase Gk Glycerokinase EC_2.7.1.30 Atp:Glycerol 3 Phosphotransferase Glycerol Kinase Testis Specific |
| ENSMUSG00000025085 | ENSFM00250000000459 | Actin Binding Lim Ablim Actin Binding Lim Family Member |
| ENSMUSG00000025092 | ENSFM00250000005151 | Unknown |
| ENSMUSG00000025132 | ENSFM00250000002426 | Rho Gdp Dissociation Inhibitor Rho Gdi Rho Gdi |
| ENSMUSG00000025137 | ENSFM00780001898417 | Ethanolamine Phosphate Cytidylyltransferase EC_2.7.7.14 Ctp:Phosphoethanolamine Cytidylyltransferase Phosphorylethanolamine Transferase |
| ENSMUSG00000025139 | ENSFM00250000004428 | Toll Interacting |
| ENSMUSG00000025151 | ENSFM00500000269827 | Melanoma Associated Antigen Mage Antigen |
| ENSMUSG00000025162 | ENSFM00570000883886 | Unknown |
| ENSMUSG00000025162 | ENSFM00800002003414 | Casein Kinase I Delta Cki Delta Ckid EC_2.7.11.1 |
| ENSMUSG00000025198 | ENSFM00610000961458 | Unknown |
| ENSMUSG00000025198 | ENSFM00610000961459 | Unknown |
| ENSMUSG00000025198 | ENSFM00610000972581 | Unknown |
| ENSMUSG00000025198 | ENSFM00250000003616 | Erlin 2 Endoplasmic Reticulum Lipid Raft Associated 2 Stomatin Prohibitin Flotillin Hflc/K Domain Containing 2 Spfh Domain Containing 2 |
| ENSMUSG00000025204 | ENSFM00610000972583 | Unknown |
| ENSMUSG00000025204 | ENSFM00250000006874 | Nadh Dehydrogenase 1 Beta Subcomplex Subunit 8 Mitochondrial Precursor Complex I Ashi Ci Ashi Nadh Ubiquinone Oxidoreductase Ashi Subunit |
| ENSMUSG00000025208 | ENSFM00250000003611 | 39S Ribosomal L43 Mitochondrial Precursor L43MT Mrp L43 Mitochondrial Ribosomal BMRP36A |
| ENSMUSG00000025212 | ENSFM00730001521561 | Sideroflexin |
| ENSMUSG00000025228 | ENSFM00730001521541 | Actin |
| ENSMUSG00000025240 | ENSFM00770001847625 | Phosphatidylinositide Phosphatase SAC1 EC_3.1.3.- Suppressor Of Actin Mutations 1 |
| ENSMUSG00000025241 | ENSFM00250000002726 | Fyve And Coiled Coil Domain Containing 1 |
| ENSMUSG00000025257 | ENSFM00500000273611 | RIB43A With Coiled Coils 1 |
| ENSMUSG00000025260 | ENSFM00500000270523 | 3 Hydroxyacyl Coa Dehydrogenase Type 2 EC_1.1.1.35 17 Beta Hydroxysteroid Dehydrogenase 10 17 Beta Hsd 10 EC_1.1.1.- 51 3 Hydroxy 2 Methylbutyryl Coa Dehydrogenase EC_1.1.-.- 1 178 3 Hydroxyacyl Coa Dehydrogenase Type Ii Endoplasmic Reticulum Associated A |
| ENSMUSG00000025262 | ENSFM00250000002493 | Constitutive Coactivator Of Ppar Gamma 1 Oxidative Stress Associated Src Activator FAM120A |
| ENSMUSG00000025277 | ENSFM00250000005911 | Monoacylglycerol Lipase ABHD6 EC_3.1.1.23 2 Arachidonoylglycerol Hydrolase Abhydrolase Domain Containing 6 |
| ENSMUSG00000025287 | ENSFM00720001503361 | Unknown |
| ENSMUSG00000025287 | ENSFM00250000003102 | Acyl Coenzyme A Thioesterase Mitochondrial Precursor Acyl Coa Thioesterase EC_3.1.2.- Mitochondrial 48 Kda Acyl Coa Thioester Hydrolase Mt ACT48 |
| ENSMUSG00000025289 | ENSFM00250000000757 | Thioredoxin Peroxidase Thioredoxin Dependent Peroxide Reductase |
| ENSMUSG00000025290 | ENSFM00500000270090 | 40S Ribosomal S24 |
| ENSMUSG00000025318 | ENSFM00250000001356 | Junctophilin Jp Junctophilin |
| ENSMUSG00000025364 | ENSFM00250000003247 | Proliferation Associated 2G4 |
| ENSMUSG00000025370 | ENSFM00260000050333 | Cadherin Precursor |
| ENSMUSG00000025372 | ENSFM00250000000906 | Brain Specific Angiogenesis Inhibitor 1 Associated 2 Bai Associated 2 BAI1 Associated 2 Insulin Receptor Substrate Of 53 Kda IRSP53 Insulin Receptor Substrate P53 |
| ENSMUSG00000025403 | ENSFM00730001521489 | Serine Hydroxymethyltransferase Shmt EC_2.1.2.1 Glycine Hydroxymethyltransferase Serine Methylase |
| ENSMUSG00000025410 | ENSFM00250000005336 | Dynactin Subunit 2 |
| ENSMUSG00000025422 | ENSFM00250000000604 | Arf Gap With Gtpase Ank Repeat And Ph Domain Containing Agap Centaurin Gamma |
| ENSMUSG00000025465 | ENSFM00500000270475 | Enoyl Coa Hydratase Mitochondrial Precursor EC_4.2.1.17 Enoyl Coa Hydratase 1 Short Chain Enoyl Coa Hydratase Sceh |
| ENSMUSG00000025475 | ENSFM00250000001368 | G Coupled Receptor 124 Precursor Tumor Endothelial Marker 5 |
| ENSMUSG00000025477 | ENSFM00250000002520 | Type I Inositol 1 4 5 Trisphosphate 5 Phosphatase 5PTASE EC_3.1.3.56 |
| ENSMUSG00000025485 | ENSFM00250000001722 | Synembryn Ric |
| ENSMUSG00000025487 | ENSFM00610000962626 | Unknown |
| ENSMUSG00000025487 | ENSFM00610000962627 | Unknown |
| ENSMUSG00000025487 | ENSFM00780001898464 | 26S Proteasome Non Atpase Regulatory Subunit 13 26S Proteasome Regulatory Subunit RPN9 26S Proteasome Regulatory Subunit S11 26S Proteasome Regulatory Subunit P40 5 |
| ENSMUSG00000025499 | ENSFM00730001521569 | Ras Precursor |
| ENSMUSG00000025508 | ENSFM00600000922276 | 60S Acidic Ribosomal P2 |
| ENSMUSG00000025525 | ENSFM00500000271869 | Micos Complex Subunit MIC27 Precursor Apolipoprotein O FAM121A |
| ENSMUSG00000025555 | ENSFM00270000056477 | Ferm Rhogef And Pleckstrin Domain Containing |
| ENSMUSG00000025558 | ENSFM00500000269738 | Dedicator Of Cytokinesis CDC42 Guanine Nucleotide Exchange Factor Zizimin |
| ENSMUSG00000025580 | ENSFM00730001521529 | Atp Dependent Rna Helicase |
| ENSMUSG00000025582 | ENSFM00500000269860 | Neuronal Pentraxin 2 Precursor NP2 Neuronal Pentraxin Ii Np Ii |
| ENSMUSG00000025584 | ENSFM00280000058709 | High Affinity Camp Specific And Ibmx Insensitive 3' 5' Cyclic Phosphodiesterase 8B EC_3.1.4.53 Cell Proliferation Inducing Gene 22 |
| ENSMUSG00000025613 | ENSFM00660001189898 | Unknown |
| ENSMUSG00000025613 | ENSFM00800002003473 | T Complex 1 Subunit Theta Tcp 1 Theta Cct Theta |
| ENSMUSG00000025645 | ENSFM00250000006098 | Coiled Coil Domain Containing 51 |
| ENSMUSG00000025656 | ENSFM00720001501783 | Unknown |
| ENSMUSG00000025656 | ENSFM00800002023268 | Unknown |
| ENSMUSG00000025656 | ENSFM00800002023269 | Unknown |
| ENSMUSG00000025656 | ENSFM00250000001570 | Rho Guanine Nucleotide Exchange Factor 9 Collybistin Rac/CDC42 Guanine Nucleotide Exchange Factor 9 |
| ENSMUSG00000025658 | ENSFM00500000270149 | Connector Enhancer Of Kinase Suppressor Of Ras Connector Enhancer Of Ksr Cnk Homolog |
| ENSMUSG00000025666 | ENSFM00500000273439 | Transmembrane 47 Transmembrane 4 Superfamily Member 10 |
| ENSMUSG00000025724 | ENSFM00790001962996 | Signal Peptidase Complex Catalytic Subunit SEC11A EC_3.4.21.89 Endopeptidase SP18 Microsomal Signal Peptidase 18 Kda Subunit Spase 18 Kda Subunit SEC11 Homolog A SEC11 1 SPC18 |
| ENSMUSG00000025739 | ENSFM00500000299734 | Unknown |
| ENSMUSG00000025743 | ENSFM00250000006742 | Syndecan 3 |
| ENSMUSG00000025745 | ENSFM00800002023575 | Unknown |
| ENSMUSG00000025745 | ENSFM00760001714774 | Trifunctional Enzyme Subunit Alpha Mitochondrial Precursor 78 Kda Gastrin Binding Tp Alpha |
| ENSMUSG00000025777 | ENSFM00760001743276 | Unknown |
| ENSMUSG00000025777 | ENSFM00250000004188 | Ganglioside Induced Differentiation Associated 1 GDAP1 |
| ENSMUSG00000025792 | ENSFM00760001714704 | 39S Ribosomal L12 Mitochondrial Precursor L12MT Mrp L12 |
| ENSMUSG00000025794 | ENSFM00500000270978 | 60S Ribosomal L14 |
| ENSMUSG00000025813 | ENSFM00250000001140 | Homer Homolog Homer Vasp/Ena Related Gene Up Regulated During Seizure And Ltp Vesl |
| ENSMUSG00000025816 | ENSFM00250000001640 | Transport SEC61 Subunit Alpha |
| ENSMUSG00000025855 | ENSFM00570000895275 | Unknown |
| ENSMUSG00000025855 | ENSFM00730001521681 | Camp Dependent Kinase Type I Alpha Regulatory Camp Dependent Kinase Type I Alpha Regulatory Subunit N Terminally Processed |
| ENSMUSG00000025858 | ENSFM00250000005809 | Golgi To Er Traffic 4 Homolog |
| ENSMUSG00000025892 | ENSFM00720001492020 | Glutamate Receptor Precursor Glur Ampa Selective Glutamate Receptor Glur Glutamate Receptor Ionotropic Ampa |
| ENSMUSG00000025964 | ENSFM00410000138484 | Disintegrin And Metalloproteinase Domain Containing Precursor Adam Metalloproteinase Disintegrin And Cysteine Rich Mdc |
| ENSMUSG00000025979 | ENSFM00590000916954 | Unknown |
| ENSMUSG00000025979 | ENSFM00250000004752 | Mob Phocein Class Ii MMOB1 MOB1 Homolog 3 MOB3 Mps One Binder Kinase Activator 3 Preimplantation 3 |
| ENSMUSG00000026000 | ENSFM00250000002547 | Lanc 1.40 Kda Erythrocyte Membrane P40 |
| ENSMUSG00000026014 | ENSFM00250000001459 | Amyloid Beta A4 Precursor Binding Family B Member 1 Interacting APBB1 Interacting 1 |
| ENSMUSG00000026032 | ENSFM00810002044181 | Nadh Dehydrogenase 1 Beta Subcomplex Subunit 3 Complex I B12 Ci B12 Nadh Ubiquinone Oxidoreductase B12 Subunit |
| ENSMUSG00000026074 | ENSFM00260000050335 | Kinase EC_2.7.11.1 Kinase Mapk/Erk Kinase Kinase Kinase Mek Kinase Kinase Mekkk Kinase |
| ENSMUSG00000026090 | ENSFM00250000019314 | Uncharacterized |
| ENSMUSG00000026090 | ENSFM00590000918345 | Unknown |
| ENSMUSG00000026109 | ENSFM00250000001843 | Tomoregulin 1 Precursor Tr 1 Transmembrane With Egf And One Follistatin Domain |
| ENSMUSG00000026113 | ENSFM00250000001432 | Type Inositol 3 4 Bisphosphate 4 Phosphatase EC_3.1.3.66 Inositol Polyphosphate 4 Phosphatase Type |
| ENSMUSG00000026150 | ENSFM00250000003442 | Mitochondrial Fission Factor |
| ENSMUSG00000026155 | ENSFM00600000921242 | Stromal Membrane Associated 2 Stromal Membrane Associated 1 |
| ENSMUSG00000026159 | ENSFM00650001140030 | Arf Gap Domain And Fg Repeat Containing 1 Hiv 1 Rev Binding Nucleoporin Rip |
| ENSMUSG00000026163 | ENSFM00250000003911 | A Kinase Anchor Sphkap SPHK1 Interactor And Akap Domain Containing Sphingosine Kinase Type 1 Interacting |
| ENSMUSG00000026179 | ENSFM00500000272376 | Probable Hydrolase Pnkd Ec 3 Myofibrillogenesis Regulator 1 Mr 1 Paroxysmal Nonkinesiogenic Dyskinesia |
| ENSMUSG00000026181 | ENSFM00260000050737 | Phosphatase 1E EC_3.1.3.16 Ca 2+ /Calmodulin Dependent Kinase Phosphatase N Camkp N Camkp Nucleus Camkn Partner Of Pix 1 Partner Of Pix Alpha Partner Of Pixa |
| ENSMUSG00000026202 | ENSFM00760001757230 | Unknown |
| ENSMUSG00000026202 | ENSFM00250000000217 | Tubulin Alpha Chain |
| ENSMUSG00000026203 | ENSFM00760001751325 | Unknown |
| ENSMUSG00000026203 | ENSFM00570000851144 | Dnaj Homolog Subfamily B Member |
| ENSMUSG00000026203 | ENSFM00760001751344 | Dnaj Homolog Subfamily B Member 2 |
| ENSMUSG00000026207 | ENSFM00570000901221 | Unknown |
| ENSMUSG00000026207 | ENSFM00740001589455 | Striated Muscle Specific Serine/Threonine Kinase EC_2.7.11.1 Aortic Preferentially Expressed 1 Apeg 1 |
| ENSMUSG00000026223 | ENSFM00600000921249 | Integral Membrane 2B Immature BRI2 IMBRI2 Transmembrane Bri Bri |
| ENSMUSG00000026234 | ENSFM00760001751711 | Unknown |
| ENSMUSG00000026234 | ENSFM00500000270420 | Nucleolin |
| ENSMUSG00000026235 | ENSFM00760001763106 | Unknown |
| ENSMUSG00000026235 | ENSFM00730001521142 | Ephrin Type A Receptor Precursor EC_2.7.10.1 Kinase |
| ENSMUSG00000026245 | ENSFM00760001763107 | Unknown |
| ENSMUSG00000026245 | ENSFM00250000004384 | Phenylalanine Trna Ligase Beta Subunit EC_6.1.1.20 Phenylalanyl Trna Synthetase Beta Subunit Phers |
| ENSMUSG00000026247 | ENSFM00800002003370 | Endothelin Converting Enzyme Ece EC_3.4.24.71 |
| ENSMUSG00000026249 | ENSFM00740001589252 | Plasminogen Activator Inhibitor 1 Precursor Pai Pai 1 Endothelial Plasminogen Activator Inhibitor Serpin E1 |
| ENSMUSG00000026259 | ENSFM00740001589196 | Ephexin 1 Eph Interacting Exchange Neuronal Guanine Nucleotide Exchange Factor |
| ENSMUSG00000026276 | ENSFM00570000881519 | Unknown |
| ENSMUSG00000026276 | ENSFM00640001119291 | Unknown |
| ENSMUSG00000026276 | ENSFM00800002003496 | Septin 2 |
| ENSMUSG00000026312 | ENSFM00570000892693 | Unknown |
| ENSMUSG00000026312 | ENSFM00700001406263 | Unknown |
| ENSMUSG00000026312 | ENSFM00760001759622 | Unknown |
| ENSMUSG00000026312 | ENSFM00260000050333 | Cadherin Precursor |
| ENSMUSG00000026319 | ENSFM00760001747407 | Unknown |
| ENSMUSG00000026319 | ENSFM00250000004714 | Lish Domain And Heat Repeat Containing |
| ENSMUSG00000026335 | ENSFM00250000001444 | Peptidyl Glycine Alpha Amidating Monooxygenase Precursor Pam |
| ENSMUSG00000026341 | ENSFM00760001763132 | Unknown |
| ENSMUSG00000026341 | ENSFM00760001763133 | Unknown |
| ENSMUSG00000026341 | ENSFM00800002003421 | Actin Related 3 Actin 3 |
| ENSMUSG00000026344 | ENSFM00590000917131 | Unknown |
| ENSMUSG00000026344 | ENSFM00250000006765 | LY6/Plaur Domain Containing 1 Precursor |
| ENSMUSG00000026347 | ENSFM00250000007006 | Transmembrane 163 Synaptic Vesicle Membrane Of 31 Kda |
| ENSMUSG00000026353 | ENSFM00250000005136 | Ubx Domain Containing 4 Ubx Domain Containing 2 |
| ENSMUSG00000026356 | ENSFM00500000299788 | Aspartate Trna Ligase Cytoplasmic |
| ENSMUSG00000026356 | ENSFM00250000003932 | Aspartate Trna Ligase Cytoplasmic EC_6.1.1.12 Aspartyl Trna Synthetase Asprs |
| ENSMUSG00000026421 | ENSFM00740001589223 | Cysteine And Glycine Rich Cysteine Rich |
| ENSMUSG00000026425 | ENSFM00760001756891 | Unknown |
| ENSMUSG00000026425 | ENSFM00250000000639 | Slit Robo Rho Gtpase Activating Rho Gtpase Activating |
| ENSMUSG00000026426 | ENSFM00410000138507 | Adp Ribosylation Factor Small G Indispensable Equal Chromosome Segregation |
| ENSMUSG00000026437 | ENSFM00760001763136 | Unknown |
| ENSMUSG00000026437 | ENSFM00740001589171 | Cyclin Dependent Kinase EC_2.7.11.22 Cell Division Kinase Pctaire Motif Kinase Serine/Threonine Kinase Pctaire |
| ENSMUSG00000026458 | ENSFM00250000000441 | Liprin Alpha Tyrosine Phosphatase Receptor Type F Polypeptide Interacting Alpha Ptprf Interacting Alpha |
| ENSMUSG00000026470 | ENSFM00760001723625 | Uncharacterized |
| ENSMUSG00000026470 | ENSFM00250000002600 | Syntaxin |
| ENSMUSG00000026490 | ENSFM00730001521452 | Serine/Threonine Kinase Mrck EC_2.7.11.1 CDC42 Binding Kinase Dmpk Myotonic Dystrophy Kinase Related CDC42 Binding Kinase Mrck Myotonic Dystrophy Kinase |
| ENSMUSG00000026499 | ENSFM00500000270849 | Golgi Resident GCP60 Acyl Coa Binding Domain Containing 3 Golgi Complex Associated 1 GOCAP1 Golgi Phosphoprotein 1 GOLPH1 Pbr And Pka Associated 7 Peripheral Benzodiazepine Receptor Associated PAP7 |
| ENSMUSG00000026510 | ENSFM00250000001053 | Apoptosis Stimulating Of P53 2 Tumor Suppressor P53 Binding 2 53BP2 P53 Binding 2 P53BP2 |
| ENSMUSG00000026520 | ENSFM00500000269896 | Pyrroline 5 Carboxylate Reductase 3 P5C Reductase 3 P5CR 3 EC_1.5.1.2 Pyrroline 5 Carboxylate Reductase |
| ENSMUSG00000026527 | ENSFM00780001917687 | Unknown |
| ENSMUSG00000026527 | ENSFM00730001521808 | Regulator Of G Signaling |
| ENSMUSG00000026532 | ENSFM00740001589109 | Spectrin Alpha Chain Non Erythrocytic 1 Alpha Ii Spectrin Fodrin Alpha Chain |
| ENSMUSG00000026553 | ENSFM00570000887400 | Unknown |
| ENSMUSG00000026553 | ENSFM00260000050647 | Coatomer Subunit Alpha Alpha Coat Alpha Cop Hep Cop Hepcop |
| ENSMUSG00000026556 | ENSFM00250000002390 | Vang 2 Van Gogh |
| ENSMUSG00000026568 | ENSFM00780001917677 | Unknown |
| ENSMUSG00000026568 | ENSFM00500000273639 | Mitochondrial Pyruvate Carrier 2 Brain 44 |
| ENSMUSG00000026585 | ENSFM00250000003018 | Kinesin Associated 3 Kap 3 KAP3 |
| ENSMUSG00000026587 | ENSFM00780001911402 | Unknown |
| ENSMUSG00000026587 | ENSFM00250000001299 | Astrotactin Precursor |
| ENSMUSG00000026596 | ENSFM00730001521872 | Tyrosine Kinase EC_2.7.10.2 Abelson Murine Leukemia Viral Oncogene Homolog Abelson Tyrosine Kinase |
| ENSMUSG00000026620 | ENSFM00730001521220 | Kinase EC_2.7.11.1 |
| ENSMUSG00000026623 | ENSFM00570000895571 | Unknown |
| ENSMUSG00000026623 | ENSFM00250000004110 | Acyl Coa:Lysophosphatidylglycerol Acyltransferase 1 EC_2.3.1.- |
| ENSMUSG00000026657 | ENSFM00250000001583 | Ferm Domain Containing 4B GRP1 Binding GRSP1 |
| ENSMUSG00000026688 | ENSFM00670001235926 | Microsomal Glutathione S Transferase 3 Microsomal Gst 3 EC_2.5.1.18 Microsomal Gst Iii |
| ENSMUSG00000026696 | ENSFM00780001910349 | Unknown |
| ENSMUSG00000026696 | ENSFM00500000273038 | Vesicle Associated Membrane 4 Vamp 4 |
| ENSMUSG00000026728 | ENSFM00730001521291 | Vimentin |
| ENSMUSG00000026737 | ENSFM00570000888534 | Unknown |
| ENSMUSG00000026737 | ENSFM00260000050567 | Phosphatidylinositol 5 Phosphate 4 Kinase Type 2 EC_2.7.1.149 1 Phosphatidylinositol 5 Phosphate 4 Kinase 2 Diphosphoinositide Kinase 2 Phosphatidylinositol 5 Phosphate 4 Kinase Type Ii Pi 5 P 4 Kinase Type Ii PIP4KII Ptdins 5 P 4 Kinase 2 |
| ENSMUSG00000026755 | ENSFM00500000270526 | Unknown |
| ENSMUSG00000026764 | ENSFM00670001235340 | Kinesin Heavy Chain Kinesin Heavy Chain Kinesin Heavy Chain |
| ENSMUSG00000026775 | ENSFM00800002003559 | Atp Dependent Zinc Metalloprotease Ym EC_3.4.24.- Atp Dependent Metalloprotease FTSH1 Meg 4 YME1 1 |
| ENSMUSG00000026782 | ENSFM00250000000845 | Abl Interactor 1 Abelson Interactor 1 Abi 1 EPS8 SH3 Domain Binding EPS8 Binding Spectrin SH3 Domain Binding 1 |
| ENSMUSG00000026817 | ENSFM00740001589162 | Adenylate Kinase Isoenzyme 1 |
| ENSMUSG00000026825 | ENSFM00570000892896 | Unknown |
| ENSMUSG00000026825 | ENSFM00520000517792 | Dynamin EC_3.6.5.5 |
| ENSMUSG00000026830 | ENSFM00250000009524 | Ermin Juxtanodin Jn |
| ENSMUSG00000026833 | ENSFM00320000100099 | Noelin Precursor Neuronal Olfactomedin Related Er Localized Olfactomedin 1 |
| ENSMUSG00000026841 | ENSFM00730001521406 | Fibrinogen C Domain Containing 1 |
| ENSMUSG00000026879 | ENSFM00760001714632 | Gelsolin Actin Depolymerizing Factor Adf |
| ENSMUSG00000026880 | ENSFM00250000000738 | Stomatin |
| ENSMUSG00000026883 | ENSFM00570000864156 | Unknown |
| ENSMUSG00000026883 | ENSFM00570000890343 | Unknown |
| ENSMUSG00000026883 | ENSFM00250000000610 | Ras Gtpase Activating |
| ENSMUSG00000026887 | ENSFM00590000917495 | Unknown |
| ENSMUSG00000026887 | ENSFM00250000005296 | Ribosome Recycling Factor Mitochondrial Precursor Rrf Ribosome Releasing Factor Mitochondrial |
| ENSMUSG00000026895 | ENSFM00250000005294 | Nadh Dehydrogenase 1 Alpha Subcomplex Subunit 8 Complex I 19KD Ci 19KD Complex I Pgiv Ci Pgiv Nadh Ubiquinone Oxidoreductase 19 Kda Subunit |
| ENSMUSG00000026914 | ENSFM00250000004350 | 26S Proteasome Non Atpase Regulatory Subunit 14 EC_3.4.19.- 26S Proteasome Regulatory Subunit RPN11 |
| ENSMUSG00000026925 | ENSFM00420000140651 | 72 Kda Inositol Polyphosphate 5 Phosphatase Precursor EC_3.1.3.36 |
| ENSMUSG00000026930 | ENSFM00570000888385 | Unknown |
| ENSMUSG00000026930 | ENSFM00250000001225 | G Signaling Modulator 1 Activator Of G Signaling 3 |
| ENSMUSG00000026959 | ENSFM00730001522127 | Glutamate Receptor Subunit 1 |
| ENSMUSG00000026991 | ENSFM00740001610501 | Unknown |
| ENSMUSG00000026991 | ENSFM00730001521673 | Catenin Delta 2 |
| ENSMUSG00000027002 | ENSFM00250000001880 | Nck Associated 1 1 Membrane Associated Hem |
| ENSMUSG00000027006 | ENSFM00250000002411 | Dnaj Homolog Subfamily C Member 10 Precursor EC_1.8.4.- Endoplasmic Reticulum Dna J Domain Containing 5 Er Resident ERDJ5 ERDJ5 |
| ENSMUSG00000027010 | ENSFM00550000747100 | Calcium Binding Mitochondrial Carrier ARALAR1 |
| ENSMUSG00000027010 | ENSFM00800002003431 | Calcium Binding Mitochondrial Carrier Mitochondrial Aspartate Glutamate Carrier Solute Carrier Family 25 Member |
| ENSMUSG00000027012 | ENSFM00250000000887 | Cytoplasmic Dynein 1 Intermediate Chain 2 Cytoplasmic Dynein Intermediate Chain 2 Dynein Intermediate Chain 2 Cytosolic Dh Ic 2 |
| ENSMUSG00000027015 | ENSFM00670001235694 | Cytochrome B Ascorbate Dependent 3 Ec 1 Cytochrome B561 Family Member A3 |
| ENSMUSG00000027022 | ENSFM00250000001847 | Xin Actin Binding Repeat Containing 2 Beta Xin Cardiomyopathy Associated 3 |
| ENSMUSG00000027075 | ENSFM00500000270086 | Large Neutral Amino Acids Transporter Small Subunit 4 L Type Amino Acid Transporter 4 Solute Carrier Family 43 Member 2 |
| ENSMUSG00000027099 | ENSFM00250000005082 | Metaxin 2 Mitochondrial Outer Membrane Import Complex 2 |
| ENSMUSG00000027131 | ENSFM00670001235866 | Er Membrane Complex Subunit 4 Transmembrane 85 |
| ENSMUSG00000027162 | ENSFM00260000050595 | Lin 7 Homolog Lin Mammalian Lin Seven Mals Vertebrate Lin 7 Homolog Veli |
| ENSMUSG00000027163 | ENSFM00250000007011 | Comm Domain Containing 9 |
| ENSMUSG00000027184 | ENSFM00670001263995 | Unknown |
| ENSMUSG00000027184 | ENSFM00470000251449 | Caprin 1 Cytoplasmic Activation And Proliferation Associated 1 Gpi Anchored Membrane 1 Gpi Anchored P137 Gpi P137 P137GPI Membrane Component Chromosome 11 Surface Marker 1 Rna Granule 105 |
| ENSMUSG00000027195 | ENSFM00670001235389 | Estradiol 17 Beta Dehydrogenase 12 EC_1.1.1.62 17 Beta Hydroxysteroid Dehydrogenase 12 17 Beta Hsd 12 |
| ENSMUSG00000027220 | ENSFM00400000131978 | Synaptotagmin 13 Synaptotagmin Xiii Sytxiii |
| ENSMUSG00000027233 | ENSFM00600000921370 | PAT1 Homolog 1 PAT1 1 PAT1 Homolog B PAT1B |
| ENSMUSG00000027254 | ENSFM00250000000942 | Microtubule Associated Map |
| ENSMUSG00000027255 | ENSFM00250000002206 | Adp Ribosylation Factor Gtpase Activating 2 Arf Gap 2 Zinc Finger 289 |
| ENSMUSG00000027257 | ENSFM00570000890106 | Unknown |
| ENSMUSG00000027257 | ENSFM00250000001466 | Kinase C And Casein Kinase Substrate In Neurons |
| ENSMUSG00000027273 | ENSFM00250000001228 | Synaptosomal Associated 25 Snap 25 Synaptosomal Associated 25 Kda |
| ENSMUSG00000027282 | ENSFM00570000874766 | Unknown |
| ENSMUSG00000027282 | ENSFM00250000001870 | Mitochondrial Carrier Homolog |
| ENSMUSG00000027286 | ENSFM00350000105477 | Leucine Rich Repeat Containing 57 |
| ENSMUSG00000027287 | ENSFM00250000001228 | Synaptosomal Associated 25 Snap 25 Synaptosomal Associated 25 Kda |
| ENSMUSG00000027291 | ENSFM00250000002875 | VAM6/VPS39 |
| ENSMUSG00000027296 | ENSFM00250000001260 | Inositol Trisphosphate 3 Kinase EC_2.7.1.127 Inositol 1 4 5 Trisphosphate 3 Kinase IP3 3 Kinase IP3K Insp 3 Kinase |
| ENSMUSG00000027347 | ENSFM00250000001366 | Ras Guanyl Releasing Calcium And Dag Regulated Guanine Nucleotide Exchange Factor Caldag |
| ENSMUSG00000027349 | ENSFM00250000002232 | FAM98A |
| ENSMUSG00000027350 | ENSFM00250000007047 | Secretogranin 1 Precursor Chromogranin B Cgb Secretogranin I Sgi |
| ENSMUSG00000027351 | ENSFM00250000001487 | Sprouty Related EVH1 Domain Containing Spred |
| ENSMUSG00000027363 | ENSFM00730001521839 | Ubiquitin Carboxyl Terminal Hydrolase 8 EC_3.4.19.12 Deubiquitinating Enzyme 8 Ubiquitin Isopeptidase Y Ubiquitin Thioesterase 8 Ubiquitin Specific Processing Protease 8 |
| ENSMUSG00000027411 | ENSFM00250000004293 | Vacuolar Sorting Associated 16 |
| ENSMUSG00000027422 | ENSFM00250000002798 | Ribosome Binding Ribosome Receptor Rrp |
| ENSMUSG00000027429 | ENSFM00570000879929 | Unknown |
| ENSMUSG00000027429 | ENSFM00810002040640 | Transport SEC23 Related |
| ENSMUSG00000027447 | ENSFM00670001237088 | Cystatin C Precursor Cystatin 3 |
| ENSMUSG00000027457 | ENSFM00720001500049 | Unknown |
| ENSMUSG00000027457 | ENSFM00720001503569 | Unknown |
| ENSMUSG00000027457 | ENSFM00250000002727 | Syntabulin Golgi Localized Syntaphilin Related Syntaxin 1 Binding |
| ENSMUSG00000027479 | ENSFM00250000000867 | Microtubule Associated Rp/Eb Family Member |
| ENSMUSG00000027506 | ENSFM00570000888554 | Unknown |
| ENSMUSG00000027506 | ENSFM00570000899195 | Unknown |
| ENSMUSG00000027506 | ENSFM00810002040979 | Tumor D52 |
| ENSMUSG00000027508 | ENSFM00250000008582 | Phosphoprotein Associated With Glycosphingolipid Enriched Microdomains 1 Csk Binding Transmembrane Phosphoprotein Cbp |
| ENSMUSG00000027519 | ENSFM00570000886275 | Unknown |
| ENSMUSG00000027519 | ENSFM00730001522125 | Ras Related Rab |
| ENSMUSG00000027522 | ENSFM00570000882040 | Unknown |
| ENSMUSG00000027522 | ENSFM00500000271003 | Syntaxin 16 |
| ENSMUSG00000027523 | ENSFM00570000878388 | Unknown |
| ENSMUSG00000027523 | ENSFM00570000892390 | Unknown |
| ENSMUSG00000027523 | ENSFM00740001591176 | Alex Alternative Gene Product Encoded By Xl Exon |
| ENSMUSG00000027523 | ENSFM00300000079126 | Guanine Nucleotide Binding G S Subunit Alpha Adenylate Cyclase Stimulating G Alpha |
| ENSMUSG00000027523 | ENSFM00600000922010 | Neuroendocrine Secretory 55 Precursor NESP55 |
| ENSMUSG00000027546 | ENSFM00570000890220 | Unknown |
| ENSMUSG00000027546 | ENSFM00810002040637 | Probable Phospholipid Transporting Atpase EC_3.6.3.1 Atpase Class Ii Type |
| ENSMUSG00000027566 | ENSFM00810002040638 | Proteasome Subunit Alpha Type 7 EC_3.4.25.1 |
| ENSMUSG00000027573 | ENSFM00250000005529 | Glucose Induced Degradation 8 Homolog |
| ENSMUSG00000027575 | ENSFM00250000003758 | Adp Ribosylation Factor Gtpase Activating 1 Arf Gap 1 Adp Ribosylation Factor 1 Gtpase Activating ARF1 Gap ARF1 Directed Gtpase Activating |
| ENSMUSG00000027581 | ENSFM00670001235346 | Stathmin |
| ENSMUSG00000027599 | ENSFM00250000005628 | Armadillo Repeat Containing 1 |
| ENSMUSG00000027602 | ENSFM00500000270555 | Microtubule Associated Proteins 1A/1B Light Chain 3A Precursor Autophagy Related LC3 A Autophagy Related Ubiquitin Modifier LC3 A MAP1 Light Chain 3 1 MAP1A/MAP1B Light Chain 3 A MAP1A/MAP1B LC3 A Microtubule Associated 1 Light Chain 3 Alpha |
| ENSMUSG00000027618 | ENSFM00740001613632 | Cysteine Desulfurase Mitochondrial |
| ENSMUSG00000027618 | ENSFM00500000270207 | Cysteine Desulfurase Mitochondrial Precursor EC_2.8.1.7 |
| ENSMUSG00000027624 | ENSFM00800002003400 | Unknown |
| ENSMUSG00000027642 | ENSFM00250000002871 | Dolichyl Diphosphooligosaccharide Glycosyltransferase Subunit 2 Precursor EC_2.4.99.18 Dolichyl Diphosphooligosaccharide Glycosyltransferase 63 Kda Subunit Ribophorin Ii Rpn Ii Ribophorin 2 |
| ENSMUSG00000027646 | ENSFM00730001521177 | Tyrosine Kinase EC_2.7.10.2 |
| ENSMUSG00000027652 | ENSFM00570000876486 | Unknown |
| ENSMUSG00000027652 | ENSFM00250000002523 | Ral Gtpase Activating Subunit Beta P170 |
| ENSMUSG00000027669 | ENSFM00740001613509 | Unknown |
| ENSMUSG00000027669 | ENSFM00780001917804 | Unknown |
| ENSMUSG00000027669 | ENSFM00250000000545 | Guanine Nucleotide Binding G I /G S /G T Subunit Beta Transducin Beta Chain |
| ENSMUSG00000027673 | ENSFM00570000869305 | Unknown |
| ENSMUSG00000027673 | ENSFM00570000881141 | Unknown |
| ENSMUSG00000027673 | ENSFM00250000005807 | Nadh Dehydrogenase 1 Beta Subcomplex Subunit 5 Mitochondrial Precursor Complex I Sgdh Ci Sgdh Nadh Ubiquinone Oxidoreductase Sgdh Subunit |
| ENSMUSG00000027679 | ENSFM00500000273595 | Mitochondrial Import Inner Membrane Translocase Subunit TIM14 Dnaj Homolog Subfamily C Member 19 |
| ENSMUSG00000027680 | ENSFM00800002020852 | Unknown |
| ENSMUSG00000027680 | ENSFM00800002023406 | Unknown |
| ENSMUSG00000027680 | ENSFM00250000000920 | Fragile X Mental Retardation 1 |
| ENSMUSG00000027692 | ENSFM00260000050335 | Kinase EC_2.7.11.1 Kinase Mapk/Erk Kinase Kinase Kinase Mek Kinase Kinase Mekkk Kinase |
| ENSMUSG00000027695 | ENSFM00740001589239 | Phospholipase D2 Pld 2 EC_3.1.4.4 Choline Phosphatase 2 Phosphatidylcholine Hydrolyzing Phospholipase D2 |
| ENSMUSG00000027709 | ENSFM00800002024464 | Unknown |
| ENSMUSG00000027709 | ENSFM00800002003420 | Methylcrotonoyl Coa Carboxylase Subunit Alpha Mitochondrial Precursor Mccase Subunit Alpha EC_6.4.1.4 3 Methylcrotonyl Coa Carboxylase 1 3 Methylcrotonyl Coa Carboxylase Biotin Containing Subunit 3 Methylcrotonyl Coa:Carbon Dioxide Ligase Subunit Alpha |
| ENSMUSG00000027739 | ENSFM00780001913222 | Unknown |
| ENSMUSG00000027739 | ENSFM00500000270315 | Ras Related Rab 33 |
| ENSMUSG00000027797 | ENSFM00700001407794 | Unknown |
| ENSMUSG00000027797 | ENSFM00400000131747 | Serine/Threonine Kinase EC_2.7.11.1 Doublecortin And Cam Kinase Doublecortin Kinase |
| ENSMUSG00000027804 | ENSFM00780001913202 | Unknown |
| ENSMUSG00000027804 | ENSFM00730001521742 | Peptidyl Prolyl Cis Trans Isomerase D Ppiase D EC_5.2.1.8 Rotamase D |
| ENSMUSG00000027810 | ENSFM00570000894590 | Unknown |
| ENSMUSG00000027810 | ENSFM00570000901911 | Unknown |
| ENSMUSG00000027810 | ENSFM00780001898479 | Eukaryotic Translation Initiation Factor 2A Eif 2A |
| ENSMUSG00000027827 | ENSFM00780001898357 | Voltage Gated Potassium Channel Subunit Beta K + Channel Subunit Beta Kv Beta |
| ENSMUSG00000027828 | ENSFM00250000005568 | Translocon Associated Subunit Gamma Trap Gamma Signal Sequence Receptor Subunit Gamma Ssr Gamma |
| ENSMUSG00000027835 | ENSFM00590000916990 | Unknown |
| ENSMUSG00000027835 | ENSFM00250000004991 | Programmed Cell Death 10 |
| ENSMUSG00000027858 | ENSFM00800002003930 | Tetraspanin 2 Tspan 2 |
| ENSMUSG00000027879 | ENSFM00570000899167 | Unknown |
| ENSMUSG00000027879 | ENSFM00800002017956 | Unknown |
| ENSMUSG00000027879 | ENSFM00500000271789 | Vesicle Trafficking SEC22B Er Golgi Snare Of 24 Kda Ers 24 ERS24 SEC22 Vesicle Trafficking Homolog B |
| ENSMUSG00000027889 | ENSFM00770001847533 | Amp Deaminase EC_3.5.4.6 Amp Deaminase |
| ENSMUSG00000027935 | ENSFM00730001521184 | Ras Related Rab |
| ENSMUSG00000027940 | ENSFM00570000886978 | Unknown |
| ENSMUSG00000027940 | ENSFM00570000890222 | Unknown |
| ENSMUSG00000027940 | ENSFM00250000000220 | Tropomyosin Alpha 1 Chain Alpha Tropomyosin Tropomyosin 1 |
| ENSMUSG00000027963 | ENSFM00500000271408 | Exostosin 2 EC_2.4.1.223 Alpha 1 4 N Acetylhexosaminyltransferase EXTL2 Alpha Galnact EXTL2 Ext Related 2 Glucuronyl Galactosyl Proteoglycan 4 Alpha N Acetylglucosaminyltransferase |
| ENSMUSG00000027965 | ENSFM00320000100099 | Noelin Precursor Neuronal Olfactomedin Related Er Localized Olfactomedin 1 |
| ENSMUSG00000027993 | ENSFM00570000883591 | Unknown |
| ENSMUSG00000027993 | ENSFM00750001632380 | Tripartite Motif Containing 2 EC_6.3.2.- E3 Ubiquitin Ligase TRIM2 |
| ENSMUSG00000028001 | ENSFM00250000004159 | Pre Splicing Factor PRP46 Pre Processing 46 |
| ENSMUSG00000028005 | ENSFM00730001521369 | Guanylate Cyclase Soluble Subunit Beta 1 Gcs Beta 1 EC_4.6.1.2 Guanylate Cyclase Soluble Subunit Beta 3 Gcs Beta 3 Soluble Guanylate Cyclase Small Subunit |
| ENSMUSG00000028020 | ENSFM00570000886172 | Unknown |
| ENSMUSG00000028020 | ENSFM00570000893633 | Unknown |
| ENSMUSG00000028020 | ENSFM00400000131715 | Glycine Receptor Subunit Precursor Glycine Receptor Kda Subunit |
| ENSMUSG00000028029 | ENSFM00800002011480 | Unknown |
| ENSMUSG00000028029 | ENSFM00800002018118 | Unknown |
| ENSMUSG00000028029 | ENSFM00800002018119 | Unknown |
| ENSMUSG00000028029 | ENSFM00500000270832 | Aminoacyl Trna Synthase Complex Interacting Multifunctional 1 Multisynthase Complex Auxiliary Component P43 |
| ENSMUSG00000028035 | ENSFM00800002024518 | Unknown |
| ENSMUSG00000028035 | ENSFM00440000236855 | Dnaj Homolog Subfamily B Member |
| ENSMUSG00000028039 | ENSFM00500000271936 | Ephrin A3 Precursor EHK1 Ligand EHK1 L Eph Related Receptor Tyrosine Kinase Ligand 3 Lerk 3 |
| ENSMUSG00000028059 | ENSFM00250000000618 | Rho Guanine Nucleotide Exchange Factor 2 Guanine Nucleotide Exchange Factor H1 Gef H1 |
| ENSMUSG00000028063 | ENSFM00250000000425 | Lamin |
| ENSMUSG00000028078 | ENSFM00400000131747 | Serine/Threonine Kinase EC_2.7.11.1 Doublecortin And Cam Kinase Doublecortin Kinase |
| ENSMUSG00000028081 | ENSFM00250000001462 | 40S Ribosomal S3A |
| ENSMUSG00000028102 | ENSFM00570000895017 | Unknown |
| ENSMUSG00000028102 | ENSFM00670001235735 | Peroxisomal Membrane Peroxin Peroxisomal Biogenesis Factor PEX11 Homolog PEX11 |
| ENSMUSG00000028126 | ENSFM00730001521611 | Phosphatidylinositol 4 Phosphate 5 Kinase Type 1 Beta Pi Beta Ptdins 4 P 5 Kinase 1 Beta EC_2.7.1.68 Phosphatidylinositol 4 Phosphate 5 Kinase Type I Phosphatidylinositol 4 Phosphate 5 Kinase Beta |
| ENSMUSG00000028127 | ENSFM00800002024500 | Unknown |
| ENSMUSG00000028127 | ENSFM00800002003649 | Atp Binding Cassette Sub Family D Member Kda Peroxisomal Membrane PMP70 |
| ENSMUSG00000028128 | ENSFM00680001306906 | Coagulation Factor Iii |
| ENSMUSG00000028128 | ENSFM00250000004048 | Tissue Factor Precursor Tf Coagulation Factor Iii CD142 Antigen |
| ENSMUSG00000028134 | ENSFM00800002024494 | Unknown |
| ENSMUSG00000028134 | ENSFM00800002024495 | Unknown |
| ENSMUSG00000028134 | ENSFM00260000050389 | Polypyrimidine Tract Binding |
| ENSMUSG00000028136 | ENSFM00800002024472 | Unknown |
| ENSMUSG00000028136 | ENSFM00250000003280 | Sorting Nexin 27 |
| ENSMUSG00000028176 | ENSFM00730001522141 | Leucine Rich Repeat Containing 7 Densin 180 Densin |
| ENSMUSG00000028184 | ENSFM00800002024517 | Unknown |
| ENSMUSG00000028184 | ENSFM00260000050328 | Latrophilin |
| ENSMUSG00000028207 | ENSFM00570000897930 | Unknown |
| ENSMUSG00000028207 | ENSFM00500000270363 | Aspartyl/Asparaginyl Beta Hydroxylase EC_1.14.11.16 |
| ENSMUSG00000028221 | ENSFM00250000004019 | Type Phosphatidylinositol 4 5 Bisphosphate 4 Phosphatase Type 2 Ptdins 4 5 P2 4 Ptase EC_3.1.3.78 Ptdins 4 5 P2 4 Ptase Ii Transmembrane 55A |
| ENSMUSG00000028223 | ENSFM00500000271494 | 2 4 Dienoyl Coa Reductase Mitochondrial Precursor EC_1.3.1.34 2 4 Dienoyl Coa Reductase |
| ENSMUSG00000028232 | ENSFM00570000894942 | Unknown |
| ENSMUSG00000028232 | ENSFM00250000003959 | Transmembrane 68 |
| ENSMUSG00000028234 | ENSFM00670001235553 | 40S Ribosomal S20 |
| ENSMUSG00000028249 | ENSFM00250000003553 | Syntenin Syndecan Binding |
| ENSMUSG00000028261 | ENSFM00500000273749 | Nadh Dehydrogenase 1 Alpha Subcomplex Assembly Factor 4 Hormone Regulated Proliferation Associated Of 20 Kda |
| ENSMUSG00000028292 | ENSFM00440000236914 | Arginine Trna Ligase EC_6.1.1.19 Arginyl Trna Synthetase Argrs |
| ENSMUSG00000028328 | ENSFM00640001116861 | Unknown |
| ENSMUSG00000028328 | ENSFM00500000269813 | Tropomodulin Tropomodulin Tmod |
| ENSMUSG00000028337 | ENSFM00730001521970 | Coronin |
| ENSMUSG00000028347 | ENSFM00250000001843 | Tomoregulin 1 Precursor Tr 1 Transmembrane With Egf And One Follistatin Domain |
| ENSMUSG00000028351 | ENSFM00250000001886 | Bmp/Retinoic Acid Inducible Neural Specific 1 Precursor Deleted In Bladder Cancer 1 |
| ENSMUSG00000028354 | ENSFM00250000001548 | Formin |
| ENSMUSG00000028364 | ENSFM00740001589132 | Tenascin Precursor Tn |
| ENSMUSG00000028373 | ENSFM00250000001299 | Astrotactin Precursor |
| ENSMUSG00000028383 | ENSFM00500000270535 | Hydroxysteroid Dehydrogenase 2 Ec 1 |
| ENSMUSG00000028399 | ENSFM00640001120160 | Unknown |
| ENSMUSG00000028399 | ENSFM00640001120161 | Unknown |
| ENSMUSG00000028399 | ENSFM00740001589097 | Receptor Type Tyrosine Phosphatase Precursor EC_3.1.3.48 |
| ENSMUSG00000028402 | ENSFM00740001589189 | Multiple Pdz Domain Multi Pdz Domain 1 |
| ENSMUSG00000028410 | ENSFM00760001714614 | Dnaj Homolog Subfamily A Member Precursor |
| ENSMUSG00000028434 | ENSFM00730001521622 | Band 4 1 |
| ENSMUSG00000028444 | ENSFM00570000894523 | Unknown |
| ENSMUSG00000028444 | ENSFM00250000003929 | Ciliary Neurotrophic Factor Receptor Subunit Alpha Precursor Cntf Receptor Subunit Alpha Cntfr Alpha |
| ENSMUSG00000028447 | ENSFM00250000007388 | Dynactin Subunit 3 |
| ENSMUSG00000028455 | ENSFM00260000050877 | Stomatin 2 Mitochondrial Precursor Slp 2 |
| ENSMUSG00000028456 | ENSFM00250000000599 | Unc 13 Homolog MUNC13 |
| ENSMUSG00000028478 | ENSFM00250000001836 | Clathrin Light Chain |
| ENSMUSG00000028480 | ENSFM00500000271973 | Golgi Associated Plant Pathogenesis Related 1 Gapr 1 Golgi Associated Pr 1 Glioma Pathogenesis Related 2 Glipr 2 |
| ENSMUSG00000028484 | ENSFM00570000899071 | Unknown |
| ENSMUSG00000028484 | ENSFM00500000271092 | PC4 And SFRS1 Interacting Lens Epithelium Derived Growth Factor |
| ENSMUSG00000028495 | ENSFM00250000001600 | 40S Ribosomal S6 |
| ENSMUSG00000028517 | ENSFM00260000050433 | Lipid Phosphate Phosphohydrolase EC_3.1.3.4 PAP2 Phosphatidate Phosphohydrolase Type Phosphatidic Acid Phosphatase Pap |
| ENSMUSG00000028519 | ENSFM00740001589270 | Disabled Homolog 2 Adaptor Molecule Disabled 2 Differentially Expressed In Ovarian Carcinoma 2 Doc 2 |
| ENSMUSG00000028524 | ENSFM00570000890494 | Unknown |
| ENSMUSG00000028524 | ENSFM00600000921280 | Fch Domain Only |
| ENSMUSG00000028525 | ENSFM00640001116163 | Unknown |
| ENSMUSG00000028525 | ENSFM00400000131721 | Camp Specific 3' 5' Cyclic Phosphodiesterase EC_3.1.4.53 |
| ENSMUSG00000028527 | ENSFM00500000270040 | Adenylate Kinase 4 Mitochondrial |
| ENSMUSG00000028530 | ENSFM00570000894607 | Unknown |
| ENSMUSG00000028530 | ENSFM00250000000777 | Tyrosine Kinase EC_2.7.10.2 Janus Kinase Jak |
| ENSMUSG00000028546 | ENSFM00500000353790 | Unknown |
| ENSMUSG00000028546 | ENSFM00250000000588 | Elav |
| ENSMUSG00000028556 | ENSFM00500000269778 | Dedicator Of Cytokinesis |
| ENSMUSG00000028567 | ENSFM00320000100209 | Thioredoxin Domain Containing 12 Precursor EC_1.8.4.2 Endoplasmic Reticulum Resident 19 Er 19 ERP19 Thioredoxin P19 |
| ENSMUSG00000028639 | ENSFM00670001235454 | Nuclease Sensitive Element Binding 1 Ccaat Binding Transcription Factor I Subunit A Cbf A Enhancer Factor I Subunit A Efi A Y Box Transcription Factor Y Box Binding 1 Yb 1 |
| ENSMUSG00000028645 | ENSFM00730001521168 | Solute Carrier Family 2 Facilitated Glucose Transporter Member Glucose Transporter Type Glut |
| ENSMUSG00000028657 | ENSFM00640001104958 | Unknown |
| ENSMUSG00000028657 | ENSFM00760001714836 | Palmitoyl Thioesterase 1 Precursor Ppt 1 EC_3.1.2.22 Palmitoyl Hydrolase 1 |
| ENSMUSG00000028691 | ENSFM00250000000757 | Thioredoxin Peroxidase Thioredoxin Dependent Peroxide Reductase |
| ENSMUSG00000028719 | ENSFM00570000884722 | Unknown |
| ENSMUSG00000028719 | ENSFM00570000897069 | Unknown |
| ENSMUSG00000028719 | ENSFM00770001847583 | Ump Cmp Kinase |
| ENSMUSG00000028745 | ENSFM00250000003778 | F Actin Capping Subunit Beta |
| ENSMUSG00000028757 | ENSFM00250000004027 | Dolichyl Diphosphooligosaccharide Glycosyltransferase 48 Kda Subunit Precursor Ddost 48 Kda Subunit Oligosaccharyl Transferase 48 Kda Subunit EC_2.4.99.18 |
| ENSMUSG00000028759 | ENSFM00250000005391 | Heterochromatin 1 Binding 3 |
| ENSMUSG00000028766 | ENSFM00250000000564 | Alkaline Phosphatase Precursor EC_3.1.3.1 Alkaline Phosphatase |
| ENSMUSG00000028782 | ENSFM00250000000737 | Brain Specific Angiogenesis Inhibitor Precursor |
| ENSMUSG00000028785 | ENSFM00570000880762 | Unknown |
| ENSMUSG00000028785 | ENSFM00760001714608 | Unknown |
| ENSMUSG00000028790 | ENSFM00400000131768 | Kh Domain Containing Rna Binding Signal Transduction Associated SAM68 Mammalian Slm |
| ENSMUSG00000028811 | ENSFM00570000885949 | Unknown |
| ENSMUSG00000028811 | ENSFM00500000270186 | Tyrosine Trna Ligase Cytoplasmic EC_6.1.1.1 Tyrosyl Trna Synthetase Tyrrs |
| ENSMUSG00000028820 | ENSFM00250000000671 | Paraspeckle Component 1 |
| ENSMUSG00000028826 | ENSFM00590000917330 | Unknown |
| ENSMUSG00000028826 | ENSFM00250000002769 | Macoilin Transmembrane |
| ENSMUSG00000028847 | ENSFM00250000005557 | Trafficking Particle Complex Subunit 3 |
| ENSMUSG00000028849 | ENSFM00250000004934 | MAP7 Domain Containing 1 |
| ENSMUSG00000028868 | ENSFM00250000001269 | Wiskott Aldrich Syndrome Family Member 1 Wasp Family Member 1 Wave 1 |
| ENSMUSG00000028879 | ENSFM00500000270193 | Syntaxin |
| ENSMUSG00000028909 | ENSFM00310000088999 | Receptor Type Tyrosine Phosphatase Precursor R Ptp EC_3.1.3.48 Receptor Type Tyrosine Phosphatase |
| ENSMUSG00000028932 | ENSFM00730001521776 | 26S Protease Regulatory Subunit 7 26S Proteasome Aaa Atpase Subunit RPT1 Proteasome 26S Subunit Atpase 2 |
| ENSMUSG00000028936 | ENSFM00600000921476 | 60S Ribosomal L22 |
| ENSMUSG00000028945 | ENSFM00810002040784 | Gtpase RHEBL1 Precursor Ras Homolog Enriched In Brain 1 Rheb 1 |
| ENSMUSG00000028953 | ENSFM00730001521705 | Atp Binding Cassette Sub Family F Member 2 |
| ENSMUSG00000028955 | ENSFM00670001236814 | Vesicle Associated Membrane 3 Vamp 3 Synaptobrevin 3 |
| ENSMUSG00000028969 | ENSFM00730001521798 | Negative Regulator Of The Pho System Serine/Threonine Kinase PHO85 |
| ENSMUSG00000028973 | ENSFM00800002023566 | Unknown |
| ENSMUSG00000028973 | ENSFM00800002003665 | Atp Binding Cassette Sub Family B Member 8 Mitochondrial Precursor |
| ENSMUSG00000028975 | ENSFM00570000893486 | Unknown |
| ENSMUSG00000028975 | ENSFM00250000007000 | Peroxisomal Membrane PEX14 PTS1 Receptor Docking Peroxin 14 Peroxisomal Membrane Anchor PEX14 |
| ENSMUSG00000028991 | ENSFM00260000050608 | Serine/Threonine Kinase EC_2.7.11.1 Rapamycin 1 Target Of Rapamycin 1 |
| ENSMUSG00000028995 | ENSFM00250000003532 | Hyccin Down Regulated By CTNNB1 A FAM126A |
| ENSMUSG00000029016 | ENSFM00570000895521 | Unknown |
| ENSMUSG00000029016 | ENSFM00740001589241 | H + /Cl Exchange Transporter 7 Chloride Channel 7 Alpha Subunit Chloride Channel 7 Clc 7 |
| ENSMUSG00000029036 | ENSFM00740001603337 | Unknown |
| ENSMUSG00000029036 | ENSFM00250000002488 | Atpase Family Aaa Domain Containing 3 |
| ENSMUSG00000029055 | ENSFM00570000893447 | Unknown |
| ENSMUSG00000029055 | ENSFM00740001598015 | Unknown |
| ENSMUSG00000029055 | ENSFM00770001847600 | 1 Phosphatidylinositol 4 5 Bisphosphate Phosphodiesterase Eta 2 EC_3.1.4.11 Phosphoinositide Phospholipase C Eta 2 Phosphoinositide Phospholipase C 4 Plc L4 Phospholipase C 4 Phospholipase C Eta 2 Plc ETA2 |
| ENSMUSG00000029059 | ENSFM00250000007632 | Prostamide/Prostaglandin F Synthase Prostamide/Pg F Synthase Prostamide/Pgf Synthase EC_1.11.1.20 |
| ENSMUSG00000029088 | ENSFM00410000138464 | Kv Channel Interacting A Type Potassium Channel Modulatory Potassium Channel Interacting |
| ENSMUSG00000029093 | ENSFM00250000000793 | VPS10 Domain Containing Receptor Precursor |
| ENSMUSG00000029095 | ENSFM00810002057443 | Unknown |
| ENSMUSG00000029095 | ENSFM00250000000459 | Actin Binding Lim Ablim Actin Binding Lim Family Member |
| ENSMUSG00000029106 | ENSFM00250000000681 | Adducin |
| ENSMUSG00000029120 | ENSFM00810002057415 | Unknown |
| ENSMUSG00000029120 | ENSFM00250000000829 | Serine/Threonine Phosphatase 2A 55 Kda Regulatory Subunit B PP2A Subunit B B55 PP2A Subunit B PR55 PP2A Subunit B R2 PP2A Subunit B |
| ENSMUSG00000029125 | ENSFM00250000005997 | Syntaxin 18 |
| ENSMUSG00000029126 | ENSFM00810002055513 | Unknown |
| ENSMUSG00000029126 | ENSFM00810002055514 | Unknown |
| ENSMUSG00000029126 | ENSFM00250000005181 | Neuron Specific Family Member |
| ENSMUSG00000029131 | ENSFM00800002023572 | Unknown |
| ENSMUSG00000029131 | ENSFM00800002023573 | Unknown |
| ENSMUSG00000029131 | ENSFM00570000851144 | Dnaj Homolog Subfamily B Member |
| ENSMUSG00000029152 | ENSFM00810002056010 | Unknown |
| ENSMUSG00000029152 | ENSFM00810002057449 | Unknown |
| ENSMUSG00000029152 | ENSFM00500000272374 | Ocia Domain Containing 1 |
| ENSMUSG00000029166 | ENSFM00250000000867 | Microtubule Associated Rp/Eb Family Member |
| ENSMUSG00000029169 | ENSFM00810002040596 | Pre Splicing Factor Atp Dependent Rna Helicase EC_3.6.4.13 Deah Box |
| ENSMUSG00000029211 | ENSFM00800002023605 | Unknown |
| ENSMUSG00000029211 | ENSFM00800002023606 | Unknown |
| ENSMUSG00000029211 | ENSFM00800002003422 | Gamma Aminobutyric Acid Receptor Subunit Alpha Precursor Gaba A Receptor Subunit Alpha |
| ENSMUSG00000029212 | ENSFM00400000131718 | Gamma Aminobutyric Acid Receptor Subunit Beta Precursor Gaba A Receptor Subunit Beta |
| ENSMUSG00000029247 | ENSFM00570000894579 | Multifunctional ADE2 |
| ENSMUSG00000029247 | ENSFM00570000901681 | Multifunctional ADE2 |
| ENSMUSG00000029247 | ENSFM00250000003230 | Multifunctional Phosphoribosylaminoimidazole Succinocarboxamide Synthase EC_6.3.2.6 Saicar Synthetase |
| ENSMUSG00000029291 | ENSFM00250000001058 | Run And Fyve Domain Containing |
| ENSMUSG00000029334 | ENSFM00400000131755 | Cgmp Dependent Kinase Cgk EC_2.7.11.12 Cgmp Dependent Kinase |
| ENSMUSG00000029348 | ENSFM00800002016787 | Unknown |
| ENSMUSG00000029348 | ENSFM00800002016788 | Unknown |
| ENSMUSG00000029348 | ENSFM00800002023638 | Unknown |
| ENSMUSG00000029348 | ENSFM00250000005876 | Aspartate Beta Hydroxylase Domain Containing EC_1.14.11.- |
| ENSMUSG00000029405 | ENSFM00250000002045 | Ras Gtpase Activating Binding 1 G3BP 1 EC_3.6.4.12 EC_3.6.4.- 13 Atp Dependent Helicase |
| ENSMUSG00000029406 | ENSFM00590000917808 | Unknown |
| ENSMUSG00000029406 | ENSFM00500000269931 | Membrane Associated Phosphatidylinositol Transfer Phosphatidylinositol Transfer Protein Membrane Associated Pitpnm PYK2 N Terminal Domain Interacting Receptor Nir |
| ENSMUSG00000029419 | ENSFM00500000276193 | Uncharacterized |
| ENSMUSG00000029419 | ENSFM00760001758450 | Unknown |
| ENSMUSG00000029420 | ENSFM00740001589344 | Peripheral Type Benzodiazepine Receptor Associated 1 Prax 1 Rims Binding 1 Rim BP1 |
| ENSMUSG00000029430 | ENSFM00250000002338 | Gtp Binding Nuclear Ran Gtpase Ran Ras Related Nuclear |
| ENSMUSG00000029432 | ENSFM00250000003191 | Nipsnap Homolog 2 NIPSNAP2 Glioblastoma Amplified Sequence |
| ENSMUSG00000029434 | ENSFM00540000717941 | Vacuolar Sorting Associated |
| ENSMUSG00000029436 | ENSFM00760001714959 | Matrix Metalloproteinase 17 Precursor Mmp 17 EC_3.4.24.- Membrane Type Matrix Metalloproteinase 4 Mt Mmp 4 MTMMP4 Membrane Type 4 Matrix Metalloproteinase MT4 Mmp MT4MMP |
| ENSMUSG00000029449 | ENSFM00800002003736 | Rho Related Gtp Binding Precursor |
| ENSMUSG00000029462 | ENSFM00520000542186 | Unknown |
| ENSMUSG00000029462 | ENSFM00570000870191 | Unknown |
| ENSMUSG00000029462 | ENSFM00570000892720 | Unknown |
| ENSMUSG00000029462 | ENSFM00250000005075 | Vacuolar Sorting Associated 29 Vesicle Sorting 29 |
| ENSMUSG00000029465 | ENSFM00500000270831 | Actin Related 2/3 Complex Subunit 3 ARP2/3 Complex 21 Kda Subunit P21 Arc |
| ENSMUSG00000029471 | ENSFM00800002023662 | Unknown |
| ENSMUSG00000029471 | ENSFM00800002023663 | Unknown |
| ENSMUSG00000029471 | ENSFM00760001714763 | Calcium/Calmodulin Dependent Kinase Kinase 1 Cam Kk 1 Cam Kinase Kinase 1 Camkk 1 EC_2.7.11.17 Cam Kinase Iv Kinase Calcium/Calmodulin Dependent Kinase Kinase Alpha Cam Kk Alpha Cam Kinase Kinase Alpha Camkk Alpha |
| ENSMUSG00000029500 | ENSFM00250000004793 | Serine/Threonine Phosphatase PGAM5 Mitochondrial |
| ENSMUSG00000029516 | ENSFM00660001188751 | Unknown |
| ENSMUSG00000029516 | ENSFM00520000517879 | Citron Rho Interacting Kinase EC_2.7.11.1 Serine/Threonine Kinase 21 |
| ENSMUSG00000029518 | ENSFM00720001503436 | Unknown |
| ENSMUSG00000029518 | ENSFM00750001632414 | Ras Related Rab 35 |
| ENSMUSG00000029580 | ENSFM00730001521060 | Actin |
| ENSMUSG00000029592 | ENSFM00250000005542 | Ubiquitin Carboxyl Terminal Hydrolase 30 EC_3.4.19.12 Deubiquitinating Enzyme 30 Ubiquitin Thioesterase 30 Ubiquitin Specific Processing Protease 30 Ub Specific Protease 30 |
| ENSMUSG00000029602 | ENSFM00570000879652 | Unknown |
| ENSMUSG00000029602 | ENSFM00760001714919 | Ras Gtpase Activating 4 Calcium Promoted Ras Inactivator Ras P21 Activator 4 Rasgap Activating 2 |
| ENSMUSG00000029608 | ENSFM00810002057459 | Unknown |
| ENSMUSG00000029608 | ENSFM00260000050490 | Double C2 Domain Containing DOC2 |
| ENSMUSG00000029610 | ENSFM00250000006128 | Aminoacyl Trna Synthase Complex Interacting Multifunctional 2 Multisynthase Complex Auxiliary Component P38 Jtv 1 |
| ENSMUSG00000029614 | ENSFM00810002057460 | Unknown |
| ENSMUSG00000029614 | ENSFM00500000270597 | 60S Ribosomal L6 |
| ENSMUSG00000029617 | ENSFM00250000003646 | Vacuolar Fusion CCZ1 Homolog |
| ENSMUSG00000029621 | ENSFM00760001714703 | Actin Related 2/3 Complex Subunit 1B ARP2/3 Complex 41 Kda Subunit P41 Arc |
| ENSMUSG00000029623 | ENSFM00600000921691 | 28 Kda Heat And Acid Stable Phosphoprotein Pdgf Associated Pap Pdgfa Associated 1 PAP1 |
| ENSMUSG00000029632 | ENSFM00670001238510 | Cytochrome C Oxidase Subunit NDUFA4 |
| ENSMUSG00000029636 | ENSFM00250000001269 | Wiskott Aldrich Syndrome Family Member 1 Wasp Family Member 1 Wave 1 |
| ENSMUSG00000029651 | ENSFM00570000887300 | Unknown |
| ENSMUSG00000029651 | ENSFM00570000894628 | Unknown |
| ENSMUSG00000029651 | ENSFM00500000271268 | Microtubule Associated Tumor Suppressor Candidate 2 Cardiac Zipper Microtubule Plus End Tracking TIP150 Tracking Of 150 Kda |
| ENSMUSG00000029684 | ENSFM00250000001784 | Wiskott Aldrich Syndrome Wasp |
| ENSMUSG00000029686 | ENSFM00570000896172 | Unknown |
| ENSMUSG00000029686 | ENSFM00510000502743 | Cullin |
| ENSMUSG00000029713 | ENSFM00250000000545 | Guanine Nucleotide Binding G I /G S /G T Subunit Beta Transducin Beta Chain |
| ENSMUSG00000029714 | ENSFM00740001589295 | Perq Amino Acid Rich With Gyf Domain Containing 2 Trinucleotide Repeat Containing Gene 15 |
| ENSMUSG00000029723 | ENSFM00500000295696 | TSC22 Domain Family 4 |
| ENSMUSG00000029723 | ENSFM00250000008373 | TSC22 Domain Family 4 TSC22 Related Inducible Leucine Zipper 2 Thg |
| ENSMUSG00000029757 | ENSFM00570000882077 | Unknown |
| ENSMUSG00000029757 | ENSFM00250000000887 | Cytoplasmic Dynein 1 Intermediate Chain 2 Cytoplasmic Dynein Intermediate Chain 2 Dynein Intermediate Chain 2 Cytosolic Dh Ic 2 |
| ENSMUSG00000029761 | ENSFM00250000002835 | Caldesmon Cdm |
| ENSMUSG00000029763 | ENSFM00250000002477 | Exocyst Complex Component 4 Exocyst Complex Component SEC8 |
| ENSMUSG00000029769 | ENSFM00250000006497 | Coiled Coil Domain Containing 136 |
| ENSMUSG00000029840 | ENSFM00810002055357 | Unknown |
| ENSMUSG00000029840 | ENSFM00500000273644 | Myotrophin |
| ENSMUSG00000029861 | ENSFM00500000271560 | Unknown |
| ENSMUSG00000029911 | ENSFM00520000555852 | Unknown |
| ENSMUSG00000029911 | ENSFM00250000006324 | Single Stranded Dna Binding Protein Mitochondrial Precursor Mt Ssb Mtssb |
| ENSMUSG00000029916 | ENSFM00810002055665 | Unknown |
| ENSMUSG00000029916 | ENSFM00250000006088 | Acylglycerol Kinase Mitochondrial Precursor EC_2.7.1.107 EC_2.7.1.- 94 Multiple Substrate Lipid Kinase Mulk Multi Substrate Lipid Kinase |
| ENSMUSG00000029994 | ENSFM00760001714627 | Annexin Annexin Annexin |
| ENSMUSG00000030000 | ENSFM00250000000681 | Adducin |
| ENSMUSG00000030007 | ENSFM00500000269991 | T Complex 1 Subunit Eta Tcp 1 Eta Cct Eta |
| ENSMUSG00000030020 | ENSFM00730001521542 | Prickle |
| ENSMUSG00000030036 | ENSFM00760001714954 | Mannosyl Oligosaccharide Glucosidase EC_3.2.1.106 Processing Glucosidase I |
| ENSMUSG00000030037 | ENSFM00500000276686 | 39S Ribosomal L53 Mitochondrial Precursor L53MT Mrp L53 |
| ENSMUSG00000030058 | ENSFM00800002003465 | Coatomer Subunit Gamma Gamma Coat Gamma Cop |
| ENSMUSG00000030062 | ENSFM00250000002976 | Dolichyl Diphosphooligosaccharide Glycosyltransferase Subunit 1 Precursor EC_2.4.99.18 Dolichyl Diphosphooligosaccharide Glycosyltransferase 67 Kda Subunit Ribophorin I Rpn I Ribophorin 1 |
| ENSMUSG00000030075 | ENSFM00390000126303 | Contactin Precursor Brain Derived Immunoglobulin Superfamily Big |
| ENSMUSG00000030086 | ENSFM00500000273806 | Micos Complex Subunit MIC25 Coiled Coil Helix Coiled Coil Helix Domain Containing |
| ENSMUSG00000030092 | ENSFM00590000918177 | Unknown |
| ENSMUSG00000030092 | ENSFM00390000126303 | Contactin Precursor Brain Derived Immunoglobulin Superfamily Big |
| ENSMUSG00000030098 | ENSFM00590000917517 | Unknown |
| ENSMUSG00000030098 | ENSFM00740001589268 | Glutamate Receptor Interacting |
| ENSMUSG00000030102 | ENSFM00250000000547 | Inositol 1 4 5 Trisphosphate Receptor Type IP3 Receptor IP3R Ins Type Inositol 1 4 5 Trisphosphate Receptor Type INSP3 Receptor |
| ENSMUSG00000030120 | ENSFM00500000271997 | Myeloid Leukemia Factor 2 Myelodysplasia Myeloid Leukemia Factor 2 |
| ENSMUSG00000030127 | ENSFM00570000882296 | Unknown |
| ENSMUSG00000030127 | ENSFM00250000003382 | COP9 Signalosome Complex Subunit 7A SGN7A Signalosome Subunit 7A JAB1 Containing Signalosome Subunit 7A |
| ENSMUSG00000030134 | ENSFM00250000001384 | Ras Gef Domain Containing Family Member |
| ENSMUSG00000030137 | ENSFM00250000000217 | Tubulin Alpha Chain |
| ENSMUSG00000030161 | ENSFM00500000270406 | Gamma Aminobutyric Acid Receptor Associated 1 Precursor Gaba A Receptor Associated 1 |
| ENSMUSG00000030172 | ENSFM00250000000753 | Elks/RAB6 Interacting/Cast Family Member 1 Erc 1 RAB6 Interacting 2 |
| ENSMUSG00000030209 | ENSFM00760001754226 | Unknown |
| ENSMUSG00000030209 | ENSFM00760001754227 | Unknown |
| ENSMUSG00000030209 | ENSFM00730001521500 | Glutamate Receptor Ionotropic Nmda Precursor Glutamate Receptor Subunit Epsilon N Methyl D Aspartate Receptor Subtype |
| ENSMUSG00000030224 | ENSFM00810002040832 | Serine Threonine Kinase Receptor Associated |
| ENSMUSG00000030231 | ENSFM00800002003674 | Pleckstrin Homology Domain Containing Family A Member 6 Ph Domain Containing Family A Member 6 Phosphoinositol 3 Phosphate Binding 3 Pepp 3 |
| ENSMUSG00000030257 | ENSFM00610000970807 | Unknown |
| ENSMUSG00000030257 | ENSFM00250000000639 | Slit Robo Rho Gtpase Activating Rho Gtpase Activating |
| ENSMUSG00000030265 | ENSFM00570000883366 | Unknown |
| ENSMUSG00000030265 | ENSFM00570000889271 | Unknown |
| ENSMUSG00000030265 | ENSFM00730001521569 | Ras Precursor |
| ENSMUSG00000030270 | ENSFM00720001503378 | Unknown |
| ENSMUSG00000030270 | ENSFM00730001521728 | Copine Copine |
| ENSMUSG00000030279 | ENSFM00250000002423 | C2 Domain Containing 5.138 Kda C2 Domain Containing Phosphoprotein |
| ENSMUSG00000030282 | ENSFM00250000005763 | N Acylneuraminate Cytidylyltransferase EC_2.7.7.43 Cmp N Acetylneuraminic Acid Synthase Cmp Neunac Synthase |
| ENSMUSG00000030286 | ENSFM00500000270967 | Er Membrane Complex Subunit 3 Transmembrane 111 |
| ENSMUSG00000030287 | ENSFM00250000000547 | Inositol 1 4 5 Trisphosphate Receptor Type IP3 Receptor IP3R Ins Type Inositol 1 4 5 Trisphosphate Receptor Type INSP3 Receptor |
| ENSMUSG00000030316 | ENSFM00500000271930 | Phosphatidate Cytidylyltransferase Mitochondrial Precursor EC_2.7.7.41 Cdp Diacylglycerol Synthase Cdp Dag Synthase Mitochondrial Translocator Assembly And Maintenance 41 Homolog TAM41 |
| ENSMUSG00000030327 | ENSFM00250000002415 | Adaptin Ear Binding Coat Associated 1 Necap Endocytosis Associated 1 Necap 1 |
| ENSMUSG00000030337 | ENSFM00670001235687 | Vesicle Associated Membrane 2 Vamp 2 Synaptobrevin 2 |
| ENSMUSG00000030350 | ENSFM00730001521302 | Arginine N Methyltransferase 1 EC_2.1.1.- Histone Arginine N Methyltransferase PRMT1 EC_2.1.1.125 |
| ENSMUSG00000030359 | ENSFM00730001521356 | Precursor |
| ENSMUSG00000030374 | ENSFM00740001613490 | Unknown |
| ENSMUSG00000030374 | ENSFM00740001613491 | Unknown |
| ENSMUSG00000030374 | ENSFM00250000001473 | Striatin |
| ENSMUSG00000030397 | ENSFM00730001521220 | Kinase EC_2.7.11.1 |
| ENSMUSG00000030403 | ENSFM00800002003977 | Vasodilator Stimulated Phosphoprotein Vasp |
| ENSMUSG00000030410 | ENSFM00250000001646 | Dystrophia Myotonica Wd Repeat Containing Dystrophia Myotonica Containing Wd Repeat Motif Protein Dmr N9 |
| ENSMUSG00000030432 | ENSFM00570000887315 | Unknown |
| ENSMUSG00000030432 | ENSFM00500000271918 | 60S Ribosomal L28 |
| ENSMUSG00000030447 | ENSFM00250000001390 | Cytoplasmic FMR1 Interacting |
| ENSMUSG00000030465 | ENSFM00250000001069 | Ph And SEC7 Domain Containing 1 Exchange Factor ARF6 Pleckstrin Homology And SEC7 Domain Containing 1 |
| ENSMUSG00000030516 | ENSFM00250000000691 | Tight Junction Zo Tight Junction Zona Occludens Zonula Occludens |
| ENSMUSG00000030583 | ENSFM00720001502782 | Unknown |
| ENSMUSG00000030583 | ENSFM00720001502783 | Unknown |
| ENSMUSG00000030583 | ENSFM00720001503388 | Unknown |
| ENSMUSG00000030583 | ENSFM00730001521477 | Signal Induced Proliferation Associated 1 SIPA1 |
| ENSMUSG00000030600 | ENSFM00260000050443 | Leucine Rich Repeat And Fibronectin Type Iii Domain Containing Precursor Synaptic Adhesion Molecule |
| ENSMUSG00000030602 | ENSFM00730001521739 | Serine/Threonine Kinase Pak EC_2.7.11.1 P21 Activated Kinase Pak |
| ENSMUSG00000030602 | ENSFM00250000002132 | Smaug Homolog 1 Smaug 1 Sterile Alpha Motif Domain Containing 4A |
| ENSMUSG00000030603 | ENSFM00730001521737 | 26S Protease Regulatory Subunit 6B 26S Proteasome Aaa Atpase Subunit RPT3 Proteasome 26S Subunit Atpase 4 |
| ENSMUSG00000030605 | ENSFM00500000270078 | Lactadherin Precursor Mfgm Milk Fat Globule Egf Factor 8 Mfg E8 SED1 |
| ENSMUSG00000030607 | ENSFM00730001521436 | Core Precursor Chondroitin Sulfate Proteoglycan |
| ENSMUSG00000030615 | ENSFM00450000242207 | Transmembrane 126A |
| ENSMUSG00000030647 | ENSFM00670001236356 | Nadh Dehydrogenase 1 Subunit C2 Complex I B14 5B Ci B14 5B Nadh Ubiquinone Oxidoreductase Subunit B14 5B |
| ENSMUSG00000030653 | ENSFM00730001522357 | Cgmp Dependent 3' 5' Cyclic Phosphodiesterase EC_3.1.4.17 Cyclic Gmp Stimulated Phosphodiesterase Cgs Pde Cgspde |
| ENSMUSG00000030682 | ENSFM00500000271211 | Cdp Diacylglycerol Inositol 3 Phosphatidyltransferase EC_2.7.8.11 Phosphatidylinositol Synthase Pi Synthase Ptdins Synthase |
| ENSMUSG00000030695 | ENSFM00250000000488 | Fructose Bisphosphate Aldolase EC_4.1.2.13 Type Aldolase |
| ENSMUSG00000030701 | ENSFM00500000271631 | Pleckstrin Homology Domain Containing Family B Member 1 Ph Domain Containing Family B Member 1 Evectin 1 Ph Domain Containing In Retina 1 PHRET1 Pleckstrin Homology Domain Retinal 1 |
| ENSMUSG00000030718 | ENSFM00760001714910 | Phosphatase Methylesterase 1 Pme 1 EC_3.1.1.89 |
| ENSMUSG00000030731 | ENSFM00570000885121 | Unknown |
| ENSMUSG00000030731 | ENSFM00570000888896 | Unknown |
| ENSMUSG00000030731 | ENSFM00720001492041 | Synaptotagmin Synaptotagmin |
| ENSMUSG00000030739 | ENSFM00270000056424 | Myosin Cellular Myosin Heavy Chain Type Myosin Heavy Chain Myosin Heavy Chain Non Muscle Non Muscle Myosin Heavy Chain Nmmhc Non Muscle Myosin Heavy Chain Nmmhc Ii Nmmhc |
| ENSMUSG00000030744 | ENSFM00250000001742 | 40S Ribosomal S3 EC_4.2.99.18 |
| ENSMUSG00000030759 | ENSFM00570000889091 | Unknown |
| ENSMUSG00000030759 | ENSFM00250000000622 | Fatty Acyl Coa Reductase 1 EC_1.2.1.- N2 Male Sterility Domain Containing 2 |
| ENSMUSG00000030774 | ENSFM00730001521340 | Serine/Threonine Kinase Pak EC_2.7.11.1 Pak P21 Activated Kinase Pak |
| ENSMUSG00000030795 | ENSFM00600000925489 | Fus |
| ENSMUSG00000030795 | ENSFM00600000921301 | Rna Binding |
| ENSMUSG00000030802 | ENSFM00500000271321 | Kinase Mitochondrial Precursor EC_2.7.11.4 Branched Chain Alpha Ketoacid Dehydrogenase Kinase Bckd Kinase Bckdhkin |
| ENSMUSG00000030842 | ENSFM00250000006855 | Ragulator Complex LAMTOR1 Late Endosomal/Lysosomal Adaptor And Mapk And Mtor Activator 1 |
| ENSMUSG00000030861 | ENSFM00730001521755 | Short/Branched Chain Specific Acyl Coa Dehydrogenase Mitochondrial Precursor Sbcad EC_1.3.8.5 2 Methyl Branched Chain Acyl Coa Dehydrogenase 2 Mebcad 2 Methylbutyryl Coenzyme A Dehydrogenase 2 Methylbutyryl Coa Dehydrogenase |
| ENSMUSG00000030868 | ENSFM00660001178171 | Unknown |
| ENSMUSG00000030868 | ENSFM00660001188113 | Unknown |
| ENSMUSG00000030868 | ENSFM00250000005726 | Dynactin Subunit 5 Dynactin Subunit P25 |
| ENSMUSG00000031007 | ENSFM00250000004412 | Renin Receptor Precursor Atpase H + Transporting Lysosomal Accessory 2 Atpase H + Transporting Lysosomal Interacting 2 Renin/Prorenin Receptor Vacuolar Atp Synthase Membrane Sector Associated M8 9 V Atpase M8 9 Subunit |
| ENSMUSG00000031012 | ENSFM00730001521718 | 55 Kda Erythrocyte Membrane P55 Membrane Protein Palmitoylated 1 |
| ENSMUSG00000031021 | ENSFM00570000882023 | Unknown |
| ENSMUSG00000031021 | ENSFM00670001235681 | Transmembrane Precursor |
| ENSMUSG00000031029 | ENSFM00250000003700 | Eukaryotic Translation Initiation Factor 3 Subunit F |
| ENSMUSG00000031059 | ENSFM00250000007177 | Nadh Dehydrogenase 1 Beta Subcomplex Subunit 11 Mitochondrial Precursor Complex I Esss Ci Esss Nadh Ubiquinone Oxidoreductase Esss Subunit |
| ENSMUSG00000031068 | ENSFM00740001589388 | Glutaredoxin 3 Pkc Interacting Cousin Of Thioredoxin Picot Pkc Theta Interacting Pkcq Interacting Thioredoxin 2 |
| ENSMUSG00000031078 | ENSFM00600000921221 | Substrate |
| ENSMUSG00000031134 | ENSFM00500000270455 | Rna Binding Motif Protein X Heterogeneous Nuclear Ribonucleoprotein G Hnrnp G |
| ENSMUSG00000031150 | ENSFM00570000882243 | Unknown |
| ENSMUSG00000031150 | ENSFM00250000009538 | Coiled Coil Domain Containing 120 |
| ENSMUSG00000031153 | ENSFM00250000004894 | GRIP1 Associated 1 Grasp 1 |
| ENSMUSG00000031158 | ENSFM00500000270373 | Mitochondrial Import Inner Membrane Translocase Subunit TIM17 |
| ENSMUSG00000031167 | ENSFM00600000921729 | Rna Binding 3 Rna Binding Motif 3 |
| ENSMUSG00000031198 | ENSFM00670001235667 | FUN14 Domain Containing |
| ENSMUSG00000031278 | ENSFM00730001521866 | Long Chain Fatty Acid Coa Ligase EC_6.2.1.3 Long Chain Acyl Coa Synthetase Lacs |
| ENSMUSG00000031292 | ENSFM00740001589470 | Cyclin Dependent Kinase 5 EC_2.7.11.22 |
| ENSMUSG00000031299 | ENSFM00750001632368 | Pyruvate Dehydrogenase E1 Component Subunit Alpha Somatic Form Mitochondrial Precursor EC_1.2.4.1 PDHE1 A Type I |
| ENSMUSG00000031302 | ENSFM00380000124679 | Neuroligin Precursor |
| ENSMUSG00000031311 | ENSFM00250000000671 | Paraspeckle Component 1 |
| ENSMUSG00000031320 | ENSFM00250000001458 | 40S Ribosomal S4 |
| ENSMUSG00000031333 | ENSFM00730001521676 | Iron Sulfur Clusters Transporter ATM1 Mitochondrial Precursor |
| ENSMUSG00000031343 | ENSFM00800002003385 | Gamma Aminobutyric Acid Receptor Subunit Alpha Precursor Gaba A Receptor Subunit Alpha |
| ENSMUSG00000031347 | ENSFM00750001632381 | Centrin |
| ENSMUSG00000031367 | ENSFM00500000269841 | Ap 1 Complex Subunit Sigma Adaptor Complex Ap 1 Subunit Sigma Adaptor Related Complex 1 Subunit Sigma Clathrin Assembly Complex 1 Sigma Small Chain Golgi Adaptor HA1/AP1 Adaptin Sigma Subunit Sigma Subunit Of Ap 1 Clathrin Sigma Adaptin Adaptin |
| ENSMUSG00000031386 | ENSFM00250000001451 | Host Cell Factor 1 Hcf Hcf 1 C1 Factor |
| ENSMUSG00000031391 | ENSFM00390000126295 | Neural Cell Adhesion Molecule L1 L1 L1 |
| ENSMUSG00000031402 | ENSFM00570000899672 | Unknown |
| ENSMUSG00000031402 | ENSFM00730001521718 | 55 Kda Erythrocyte Membrane P55 Membrane Protein Palmitoylated 1 |
| ENSMUSG00000031442 | ENSFM00570000895390 | Unknown |
| ENSMUSG00000031442 | ENSFM00250000000478 | Guanine Nucleotide Exchange Factor Dbs Dbl'S Big Sister MCF2 Transforming Sequence |
| ENSMUSG00000031446 | ENSFM00570000894978 | Unknown |
| ENSMUSG00000031446 | ENSFM00730001521495 | Cullin Cul |
| ENSMUSG00000031453 | ENSFM00570000892435 | Unknown |
| ENSMUSG00000031453 | ENSFM00760001714814 | Ras Gtpase Activating 3 GAP1 IP4BP Ins P4 Binding |
| ENSMUSG00000031467 | ENSFM00250000007868 | 1 Acyl Sn Glycerol 3 Phosphate Acyltransferase Epsilon EC_2.3.1.51 1 Acylglycerol 3 Phosphate O Acyltransferase 5 1 Agp Acyltransferase 5 1 Agpat 5 Lysophosphatidic Acid Acyltransferase Epsilon Lpaat Epsilon |
| ENSMUSG00000031483 | ENSFM00250000003616 | Erlin 2 Endoplasmic Reticulum Lipid Raft Associated 2 Stomatin Prohibitin Flotillin Hflc/K Domain Containing 2 Spfh Domain Containing 2 |
| ENSMUSG00000031511 | ENSFM00250000001135 | Rho Guanine Nucleotide Exchange Factor 6 Rac/CDC42 Guanine Nucleotide Exchange Factor 6 |
| ENSMUSG00000031539 | ENSFM00440000236903 | Ap 3 Complex Subunit Mu Adaptor Related Complex 3 Subunit Mu Clathrin Assembly Assembly Complex 3 Mu Medium Chain Clathrin Coat Assembly AP47 Homolog Clathrin Coat Associated AP47 Homolog Golgi Adaptor Ap 1.47 Kda Homolog HA1.47 Kda Subunit Homolog Adapti |
| ENSMUSG00000031561 | ENSFM00500000315657 | Unknown |
| ENSMUSG00000031561 | ENSFM00250000000375 | Teneurin Ten Odd Oz/Ten M Homolog Tenascin Ten Teneurin Transmembrane |
| ENSMUSG00000031591 | ENSFM00570000901226 | Unknown |
| ENSMUSG00000031591 | ENSFM00500000270714 | Acid Ceramidase Precursor Ac Acdase Acid Cdase EC_3.5.1.23 Acylsphingosine Deacylase N Acylsphingosine Amidohydrolase |
| ENSMUSG00000031592 | ENSFM00250000002581 | Pericentriolar Material 1 Pcm 1 |
| ENSMUSG00000031626 | ENSFM00570000879880 | Unknown |
| ENSMUSG00000031626 | ENSFM00570000883394 | Unknown |
| ENSMUSG00000031626 | ENSFM00570000887049 | Unknown |
| ENSMUSG00000031626 | ENSFM00570000891654 | Unknown |
| ENSMUSG00000031626 | ENSFM00570000895888 | Unknown |
| ENSMUSG00000031626 | ENSFM00570000900335 | Unknown |
| ENSMUSG00000031626 | ENSFM00550000743175 | Sorbin And SH3 Domain Containing 2 Arg/Abl Interacting 2 ARGBP2 |
| ENSMUSG00000031633 | ENSFM00250000000816 | Adp/Atp Translocase Adp Atp Carrier Adenine Nucleotide Translocator Ant Solute Carrier Family 25 Member |
| ENSMUSG00000031654 | ENSFM00250000002369 | Cerebellin Precursor |
| ENSMUSG00000031673 | ENSFM00800002003474 | Cadherin Precursor |
| ENSMUSG00000031701 | ENSFM00760001714614 | Dnaj Homolog Subfamily A Member Precursor |
| ENSMUSG00000031703 | ENSFM00250000004403 | T Cell Immunomodulatory Tip Integrin Alpha Fg Gap Repeat Containing 1 |
| ENSMUSG00000031708 | ENSFM00250000002457 | Very Long Chain Enoyl Coa Reductase EC_1.3.1.93 Synaptic Glycoprotein SC2 Trans 2 3 Enoyl Coa Reductase Ter |
| ENSMUSG00000031729 | ENSFM00250000004551 | IST1 Homolog |
| ENSMUSG00000031748 | ENSFM00570000894903 | Unknown |
| ENSMUSG00000031748 | ENSFM00410000138458 | Guanine Nucleotide Binding Subunit |
| ENSMUSG00000031754 | ENSFM00250000006035 | Cleavage And Polyadenylation Specificity Factor Subunit 5 Nucleoside Diphosphate Linked Moiety X Motif 21 Nudix Motif 21 |
| ENSMUSG00000031775 | ENSFM00500000273560 | Plasmolipin Plasma Membrane Proteolipid |
| ENSMUSG00000031808 | ENSFM00730001521827 | Long Chain Fatty Acid Transport 4 Fatp 4 Fatty Acid Transport 4 EC_6.2.1.- Solute Carrier Family 27 Member 4 |
| ENSMUSG00000031812 | ENSFM00700001400708 | Unknown |
| ENSMUSG00000031812 | ENSFM00500000270555 | Microtubule Associated Proteins 1A/1B Light Chain 3A Precursor Autophagy Related LC3 A Autophagy Related Ubiquitin Modifier LC3 A MAP1 Light Chain 3 1 MAP1A/MAP1B Light Chain 3 A MAP1A/MAP1B LC3 A Microtubule Associated 1 Light Chain 3 Alpha |
| ENSMUSG00000031818 | ENSFM00700001407963 | Unknown |
| ENSMUSG00000031818 | ENSFM00500000271199 | Cytochrome C Oxidase Subunit 4 1 Mitochondrial Cytochrome C Oxidase Polypeptide Iv Cytochrome C Oxidase Subunit Iv 1 Cox Iv 1 |
| ENSMUSG00000031819 | ENSFM00700001400973 | Unknown |
| ENSMUSG00000031819 | ENSFM00500000272397 | Er Membrane Complex Subunit 8 Neighbor Of COX4 |
| ENSMUSG00000031833 | ENSFM00250000000518 | Microtubule Associated Serine/Threonine Kinase EC_2.7.11.1 |
| ENSMUSG00000031837 | ENSFM00250000001762 | N Terminal Ef Hand Calcium Binding Ef Hand Calcium Binding Neuronal Calcium Binding |
| ENSMUSG00000031839 | ENSFM00500000401249 | Unknown |
| ENSMUSG00000031841 | ENSFM00730001521172 | Cadherin Precursor Cadherin Cadherin |
| ENSMUSG00000031913 | ENSFM00810002040610 | Vacuolar Sorting Associated EC_3.6.4.6 |
| ENSMUSG00000031918 | ENSFM00570000883574 | Unknown |
| ENSMUSG00000031918 | ENSFM00570000888483 | Unknown |
| ENSMUSG00000031918 | ENSFM00730001521553 | Myotubularin Phosphatidylinositol 3 5 Bisphosphate 3 Phosphatase EC_3.1.3.95 |
| ENSMUSG00000031924 | ENSFM00500000273474 | Cytochrome B5 Type B Precursor Cytochrome B5 Outer Mitochondrial Membrane |
| ENSMUSG00000031930 | ENSFM00730001521655 | NEDD4 E3 Ubiquitin Ligase EC_6.3.2.- Ww Domain Containing |
| ENSMUSG00000031950 | ENSFM00500000272384 | Gamma Aminobutyric Acid Receptor Associated 2 Precursor Gaba A Receptor Associated 2 Ganglioside Expression Factor 2 Gef 2 General Transport Factor P16 Golgi Associated Atpase Enhancer Of 16 Kda Gate 16 MAP1 Light Chain 3 Related |
| ENSMUSG00000031969 | ENSFM00570000879755 | Unknown |
| ENSMUSG00000031969 | ENSFM00730001522325 | Isobutyryl Coa Dehydrogenase Mitochondrial Precursor EC_1.3.99.- Acyl Coa Dehydrogenase Family Member 8 Acad 8 |
| ENSMUSG00000031971 | ENSFM00250000006451 | Centriole Cilia And Spindle Associated |
| ENSMUSG00000031972 | ENSFM00800002003372 | Actin Muscle Precursor Alpha Actin |
| ENSMUSG00000031983 | ENSFM00250000010056 | Uncharacterized |
| ENSMUSG00000031985 | ENSFM00250000004371 | Dihydroxyacetone Phosphate Acyltransferase Dap At Dhap At EC_2.3.1.42 Acyl Coa:Dihydroxyacetonephosphateacyltransferase Glycerone Phosphate O Acyltransferase |
| ENSMUSG00000031988 | ENSFM00810002040678 | Vacuolar Sorting Associated Vesicle Sorting |
| ENSMUSG00000031990 | ENSFM00500000272551 | Junctional Adhesion Molecule C Precursor Jam C Junctional Adhesion Molecule 3 Jam 3 |
| ENSMUSG00000032011 | ENSFM00250000009003 | Thy 1 Membrane Glycoprotein Precursor Thy 1 Antigen CD90 Antigen |
| ENSMUSG00000032030 | ENSFM00570000899340 | Unknown |
| ENSMUSG00000032030 | ENSFM00730001522203 | Cullin 5 Cul 5 Vasopressin Activated Calcium Mobilizing Receptor 1 Vacm 1 |
| ENSMUSG00000032036 | ENSFM00760001763379 | Unknown |
| ENSMUSG00000032036 | ENSFM00250000000717 | Kin Of Irre Precursor Kin Of Irregular Chiasm Nephrin |
| ENSMUSG00000032046 | ENSFM00570000877269 | Unknown |
| ENSMUSG00000032046 | ENSFM00570000892528 | Unknown |
| ENSMUSG00000032046 | ENSFM00250000002071 | Monoacylglycerol Lipase ABHD12 EC_3.1.1.23 2 Arachidonoylglycerol Hydrolase Abhydrolase Domain Containing 12 |
| ENSMUSG00000032047 | ENSFM00730001521421 | Acetyl Coa Acetyltransferase Mitochondrial Precursor EC_2.3.1.9 Acetoacetyl Coa Thiolase |
| ENSMUSG00000032060 | ENSFM00670001235476 | Alpha Crystallin Chain |
| ENSMUSG00000032081 | ENSFM00570000880516 | Unknown |
| ENSMUSG00000032081 | ENSFM00600000926313 | Apolipoprotein C Iii Precursor Apo Ciii Apoc Iii Apolipoprotein C3 |
| ENSMUSG00000032096 | ENSFM00810002040812 | Coatomer Subunit Delta Delta Coat Delta Cop |
| ENSMUSG00000032097 | ENSFM00540000717952 | Atp Dependent Rna Helicase EC_3.6.4.13 |
| ENSMUSG00000032116 | ENSFM00570000897300 | Unknown |
| ENSMUSG00000032116 | ENSFM00250000001541 | Dolichyl Diphosphooligosaccharide Glycosyltransferase Subunit Oligosaccharyl Transferase Subunit STT3 EC_2.4.99.18 |
| ENSMUSG00000032127 | ENSFM00250000005369 | E3 Ubiquitin Ligase PEP5 |
| ENSMUSG00000032171 | ENSFM00670001235859 | Peptidyl Prolyl Cis Trans Isomerase Nima Interacting 1 EC_5.2.1.8 Peptidyl Prolyl Cis Trans Isomerase PIN1 Ppiase PIN1 |
| ENSMUSG00000032172 | ENSFM00320000100099 | Noelin Precursor Neuronal Olfactomedin Related Er Localized Olfactomedin 1 |
| ENSMUSG00000032186 | ENSFM00500000269813 | Tropomodulin Tropomodulin Tmod |
| ENSMUSG00000032192 | ENSFM00770001847629 | Guanine Nucleotide Binding Subunit Beta 5 GBETA5 Transducin Beta Chain 5 |
| ENSMUSG00000032220 | ENSFM00780001898391 | Unconventional Myosin Ie Unconventional Myosin 1E |
| ENSMUSG00000032224 | ENSFM00570000901107 | Unknown |
| ENSMUSG00000032224 | ENSFM00480000262849 | FAM81A |
| ENSMUSG00000032231 | ENSFM00730001521439 | Annexin Annexin Annexin Calpactin Calpactin Chromobindin Lipocortin I |
| ENSMUSG00000032261 | ENSFM00760001743793 | Unknown |
| ENSMUSG00000032261 | ENSFM00670001236492 | SH3 Domain Binding Glutamic Acid Rich 2 |
| ENSMUSG00000032264 | ENSFM00250000006432 | Centromere/Kinetochore ZW10 Homolog |
| ENSMUSG00000032295 | ENSFM00590000917310 | Unknown |
| ENSMUSG00000032295 | ENSFM00590000917311 | Unknown |
| ENSMUSG00000032295 | ENSFM00590000917313 | Unknown |
| ENSMUSG00000032295 | ENSFM00590000918415 | Unknown |
| ENSMUSG00000032295 | ENSFM00250000003741 | Alpha Mannosidase 2C1 EC_3.2.1.24 Alpha D Mannoside Mannohydrolase Mannosidase Alpha Class 2C Member 1 |
| ENSMUSG00000032320 | ENSFM00570000891778 | Unknown |
| ENSMUSG00000032320 | ENSFM00500000271265 | Reticulocalbin 2 Precursor Taipoxin Associated Calcium Binding 49 Tcbp 49 |
| ENSMUSG00000032328 | ENSFM00500000270036 | Cell Cycle Control 50A P4 Atpase Flippase Complex Beta Subunit TMEM30A Transmembrane 30A |
| ENSMUSG00000032348 | ENSFM00270000056439 | Glutathione S Transferase EC_2.5.1.18 Gst Class Alpha Member Glutathione S Transferase |
| ENSMUSG00000032355 | ENSFM00740001605474 | Unknown |
| ENSMUSG00000032355 | ENSFM00740001589545 | Muscular Lmna Interacting Muscular Enriched A Type Laminin Interacting |
| ENSMUSG00000032356 | ENSFM00500000289379 | Ras Specific Guanine Nucleotide Releasing Factor 1 CRA_B |
| ENSMUSG00000032356 | ENSFM00250000001613 | Ras Specific Guanine Nucleotide Releasing Factor 2 Ras GRF2 Ras Guanine Nucleotide Exchange Factor 2 |
| ENSMUSG00000032366 | ENSFM00720001503517 | Unknown |
| ENSMUSG00000032366 | ENSFM00250000000220 | Tropomyosin Alpha 1 Chain Alpha Tropomyosin Tropomyosin 1 |
| ENSMUSG00000032370 | ENSFM00250000007515 | Serine Beta Lactamase Lactb Mitochondrial Precursor EC_3.4.-.- |
| ENSMUSG00000032383 | ENSFM00500000269861 | Peptidyl Prolyl Cis Trans Isomerase B Precursor Ppiase B EC_5.2.1.8 Rotamase B |
| ENSMUSG00000032384 | ENSFM00570000899402 | Unknown |
| ENSMUSG00000032384 | ENSFM00730001521484 | Casein Kinase I Gamma Cki Gamma EC_2.7.11.1 |
| ENSMUSG00000032399 | ENSFM00250000002321 | 60S Ribosomal L4 |
| ENSMUSG00000032420 | ENSFM00250000001233 | 5' Nucleotidase Precursor 5' Nt EC_3.1.3.5 Ecto 5' Nucleotidase |
| ENSMUSG00000032423 | ENSFM00640001116175 | Unknown |
| ENSMUSG00000032423 | ENSFM00730001521753 | Heterogeneous Nuclear Ribonucleoprotein Q Hnrnp Q Glycine And Tyrosine Rich Rna Binding Gry Rbp NS1 Associated 1 Synaptotagmin Binding Cytoplasmic Rna Interacting |
| ENSMUSG00000032435 | ENSFM00250000001777 | Cytoplasmic Dynein 1 Light Intermediate Chain 1 Dynein Light Chain A Dlc A Dynein Light Intermediate Chain 1 Cytosolic |
| ENSMUSG00000032437 | ENSFM00250000001541 | Dolichyl Diphosphooligosaccharide Glycosyltransferase Subunit Oligosaccharyl Transferase Subunit STT3 EC_2.4.99.18 |
| ENSMUSG00000032459 | ENSFM00250000006229 | 28S Ribosomal S22 Mitochondrial Mrp S22 S22MT |
| ENSMUSG00000032470 | ENSFM00770001847543 | Ras Related R Ras |
| ENSMUSG00000032480 | ENSFM00800002017283 | Unknown |
| ENSMUSG00000032480 | ENSFM00800002024439 | Unknown |
| ENSMUSG00000032480 | ENSFM00800002024440 | Unknown |
| ENSMUSG00000032480 | ENSFM00800002024441 | Unknown |
| ENSMUSG00000032480 | ENSFM00800002024442 | Unknown |
| ENSMUSG00000032480 | ENSFM00260000050346 | Atp Dependent Rna Helicase EC_3.6.4.13 Deah Box |
| ENSMUSG00000032512 | ENSFM00460000244957 | Wd Repeat Containing 48 |
| ENSMUSG00000032517 | ENSFM00500000311584 | Unknown |
| ENSMUSG00000032517 | ENSFM00600000921972 | Myelin Associated Oligodendrocyte Basic |
| ENSMUSG00000032562 | ENSFM00780001917784 | Unknown |
| ENSMUSG00000032562 | ENSFM00780001917785 | Unknown |
| ENSMUSG00000032562 | ENSFM00410000138458 | Guanine Nucleotide Binding Subunit |
| ENSMUSG00000032564 | ENSFM00800002003632 | Copine Copine |
| ENSMUSG00000032571 | ENSFM00800002003738 | Phosphoinositide 3 Kinase Regulatory Subunit 4 PI3 Kinase Regulatory Subunit 4 EC_2.7.11.1 |
| ENSMUSG00000032583 | ENSFM00770001847570 | Vacuolar Fusion MON1 |
| ENSMUSG00000032589 | ENSFM00600000921250 | Piccolo Aczonin |
| ENSMUSG00000032598 | ENSFM00780001908780 | Unknown |
| ENSMUSG00000032598 | ENSFM00780001908781 | Unknown |
| ENSMUSG00000032598 | ENSFM00780001917797 | Unknown |
| ENSMUSG00000032598 | ENSFM00250000006300 | Nck Interacting With SH3 Domain 54 Kda Vaca Interacting Kda Interacting 90 Kda SH3 Interacting With Nck SH3 Adapter SPIN90 Wasp Interacting SH3 Domain Wish Wiskott Aldrich Syndrome |
| ENSMUSG00000032601 | ENSFM00780001917795 | Unknown |
| ENSMUSG00000032601 | ENSFM00730001521993 | Camp Dependent Kinase Type Ii Regulatory Subunit |
| ENSMUSG00000032604 | ENSFM00780001898374 | Glutamine Trna Ligase EC_6.1.1.18 Glutaminyl Trna Synthetase Glnrs |
| ENSMUSG00000032606 | ENSFM00250000009545 | Nicolin 1 Tubulin Polyglutamylase Complex Subunit 5 PGS5 |
| ENSMUSG00000032666 | ENSFM00500000274384 | Uncharacterized |
| ENSMUSG00000032666 | ENSFM00760001721091 | Unknown |
| ENSMUSG00000032705 | ENSFM00250000005784 | Exonuclease 3' 5' Domain Containing 2 Exonuclease 3' 5' Domain Containing 2 |
| ENSMUSG00000032735 | ENSFM00250000000459 | Actin Binding Lim Ablim Actin Binding Lim Family Member |
| ENSMUSG00000032740 | ENSFM00250000000961 | Girdin Akt Phosphorylation Enhancer Ape Coiled Coil Domain Containing 88A G Alpha Interacting Vesicle Associated Giv Girders Of Actin Filament Hook Related 1 HKRP1 |
| ENSMUSG00000032763 | ENSFM00250000005381 | Acetolactate Synthase EC_2.2.1.- Ilvb |
| ENSMUSG00000032773 | ENSFM00660001189510 | Unknown |
| ENSMUSG00000032773 | ENSFM00800002003412 | Muscarinic Acetylcholine Receptor |
| ENSMUSG00000032826 | ENSFM00600000932983 | Uncharacterized |
| ENSMUSG00000032826 | ENSFM00720001501960 | Unknown |
| ENSMUSG00000032826 | ENSFM00720001502851 | Ankyrin 2 |
| ENSMUSG00000032826 | ENSFM00500000269628 | Ankyrin Ank Ankyrin |
| ENSMUSG00000032827 | ENSFM00250000001450 | Neurabin 1 Neurabin I Neural Tissue Specific F Actin Binding I Phosphatase 1 Regulatory Subunit 9A |
| ENSMUSG00000032854 | ENSFM00730001522351 | 2 Hydroxyacylsphingosine 1 Beta Galactosyltransferase Precursor EC_2.4.1.45 Ceramide Udp Galactosyltransferase Cerebroside Synthase Udp Galactose Ceramide Galactosyltransferase |
| ENSMUSG00000032878 | ENSFM00740001589413 | Coiled Coil Domain Containing |
| ENSMUSG00000032883 | ENSFM00730001521866 | Long Chain Fatty Acid Coa Ligase EC_6.2.1.3 Long Chain Acyl Coa Synthetase Lacs |
| ENSMUSG00000032890 | ENSFM00250000000629 | Regulating Synaptic Membrane Exocytosis Rab 3 Interacting Molecule Rim |
| ENSMUSG00000032936 | ENSFM00780001915889 | Unknown |
| ENSMUSG00000032936 | ENSFM00780001917789 | Unknown |
| ENSMUSG00000032936 | ENSFM00800002003895 | Cam Kinase Vesicle Associated |
| ENSMUSG00000032946 | ENSFM00500000318755 | Unknown |
| ENSMUSG00000032946 | ENSFM00500000386335 | Unknown |
| ENSMUSG00000032946 | ENSFM00570000900297 | Unknown |
| ENSMUSG00000032946 | ENSFM00250000001366 | Ras Guanyl Releasing Calcium And Dag Regulated Guanine Nucleotide Exchange Factor Caldag |
| ENSMUSG00000033004 | ENSFM00250000001622 | E3 Ubiquitin Ligase MYCBP2 EC_6.3.2.- Myc Binding 2 Pam/Highwire/Rpm 1 Protein Associated With Myc |
| ENSMUSG00000033047 | ENSFM00250000003239 | Eukaryotic Translation Initiation Factor 3 Subunit L |
| ENSMUSG00000033060 | ENSFM00250000003075 | Lim Domain Only 7 Lmo 7 F Box Only 20 Lomp |
| ENSMUSG00000033065 | ENSFM00730001521507 | Atp Dependent 6 Phosphofructokinase Type |
| ENSMUSG00000033066 | ENSFM00250000005898 | Growth Arrest Specific 7 Gas 7 |
| ENSMUSG00000033068 | ENSFM00250000002349 | Ectonucleoside Triphosphate Diphosphohydrolase 5 Precursor Ntpdase 5 EC_3.6.1.6 Guanosine Diphosphatase ENTPD5 Gdpase ENTPD5 EC_3.6.1.- 42 Uridine Diphosphatase ENTPD5 Udpase ENTPD5 |
| ENSMUSG00000033335 | ENSFM00640001119884 | Unknown |
| ENSMUSG00000033335 | ENSFM00520000517792 | Dynamin EC_3.6.5.5 |
| ENSMUSG00000033352 | ENSFM00810002045789 | Unknown |
| ENSMUSG00000033352 | ENSFM00790001963039 | Dual Specificity Mitogen Activated Kinase Kinase 4 Map Kinase Kinase 4 Mapkk 4 EC_2.7.12.2 Jnk Activating Kinase 1 Mapk/Erk Kinase 4 Mek 4 Sapk/Erk Kinase 1 SEK1 |
| ENSMUSG00000033382 | ENSFM00250000003061 | Trafficking Particle Complex Subunit 8 TRS85 Homolog |
| ENSMUSG00000033389 | ENSFM00250000000900 | Rho Gtpase Activating Rho Type Gtpase Activating Rhogap Interacting With CIP4 Homologs Rich |
| ENSMUSG00000033392 | ENSFM00250000000820 | Clip Associating Cytoplasmic Linker Associated |
| ENSMUSG00000033436 | ENSFM00250000001585 | Armadillo Repeat Containing X Linked |
| ENSMUSG00000033475 | ENSFM00500000297210 | Unknown |
| ENSMUSG00000033530 | ENSFM00250000002093 | Tetratricopeptide Repeat Tpr Repeat |
| ENSMUSG00000033569 | ENSFM00250000000737 | Brain Specific Angiogenesis Inhibitor Precursor |
| ENSMUSG00000033577 | ENSFM00730001522029 | Unconventional Myosin Vi Unconventional Myosin 6 |
| ENSMUSG00000033578 | ENSFM00250000006851 | Transmembrane 35 |
| ENSMUSG00000033594 | ENSFM00740001589644 | Spermatogenesis Associated 2 SPATA2 |
| ENSMUSG00000033595 | ENSFM00250000002312 | Leucine Rich Repeat Lgi Family Member Precursor Leucine Rich Glioma Inactivated |
| ENSMUSG00000033597 | ENSFM00250000001241 | Caskin 1 Cask Interacting 1 |
| ENSMUSG00000033629 | ENSFM00500000271426 | Very Long Chain 3R 3 Hydroxyacyl Coa Dehydratase 3 EC_4.2.1.134 3 Hydroxyacyl Coa Dehydratase 3 HACD3 Tyrosine Phosphatase PTPLAD1 Tyrosine Phosphatase A Domain Containing 1 |
| ENSMUSG00000033676 | ENSFM00400000131718 | Gamma Aminobutyric Acid Receptor Subunit Beta Precursor Gaba A Receptor Subunit Beta |
| ENSMUSG00000033697 | ENSFM00660001189878 | Unknown |
| ENSMUSG00000033697 | ENSFM00660001189879 | Unknown |
| ENSMUSG00000033697 | ENSFM00660001189880 | Unknown |
| ENSMUSG00000033697 | ENSFM00660001189881 | Unknown |
| ENSMUSG00000033697 | ENSFM00250000002627 | Rho Gtpase Activating 39 |
| ENSMUSG00000033717 | ENSFM00800002003463 | Alpha 2A Adrenergic Receptor Alpha 2A Adrenoreceptor Alpha 2A Adrenoceptor Alpha 2AAR |
| ENSMUSG00000033751 | ENSFM00500000275503 | Growth Arrest And Dna Damage Inducible Proteins Interacting 1 39S Ribosomal L59 Mitochondrial Mrp L59 Papillomavirus L2 Interacting Nuclear 1 Plinp 1 |
| ENSMUSG00000033763 | ENSFM00250000002052 | Metastasis Suppressor 1 Missing In Metastasis |
| ENSMUSG00000033769 | ENSFM00590000917621 | Unknown |
| ENSMUSG00000033769 | ENSFM00250000001326 | Exocyst Complex Component 6 Exocyst Complex Component SEC15A SEC15 1 |
| ENSMUSG00000033793 | ENSFM00780001913229 | Unknown |
| ENSMUSG00000033793 | ENSFM00250000003378 | V Type Proton Atpase Subunit H V Atpase Subunit H Vacuolar Proton Pump Subunit H |
| ENSMUSG00000033860 | ENSFM00800002003901 | Fibrinogen Gamma Chain Precursor |
| ENSMUSG00000033900 | ENSFM00780001912614 | Unknown |
| ENSMUSG00000033900 | ENSFM00250000007724 | Microtubule Associated 9 Aster Associated |
| ENSMUSG00000033916 | ENSFM00600000921262 | Charged Multivesicular Body 2A Chromatin Modifying 2A CHMP2A |
| ENSMUSG00000033938 | ENSFM00670001236404 | Nadh Dehydrogenase 1 Beta Subcomplex Subunit 7 Complex I B18 Ci B18 Nadh Ubiquinone Oxidoreductase B18 Subunit |
| ENSMUSG00000033940 | ENSFM00500000311475 | Unknown |
| ENSMUSG00000033960 | ENSFM00790001963123 | Junctional Associated With Coronary Artery Disease Jcad |
| ENSMUSG00000033981 | ENSFM00720001492020 | Glutamate Receptor Precursor Glur Ampa Selective Glutamate Receptor Glur Glutamate Receptor Ionotropic Ampa |
| ENSMUSG00000034000 | ENSFM00250000002795 | Sialidase 3 EC_3.2.1.18 Ganglioside Sialidase Membrane Sialidase N Acetyl Alpha Neuraminidase 3 |
| ENSMUSG00000034024 | ENSFM00500000270021 | T Complex 1 Subunit Beta Tcp 1 Beta Cct Beta |
| ENSMUSG00000034040 | ENSFM00500000269893 | Polypeptide N Acetylgalactosaminyltransferase EC_2.4.1.41 Polypeptide Galnac Transferase Galnac T Pp Gantase Udp Acetylgalactosaminyltransferase Udp Galnac:Polypeptide N Acetylgalactosaminyltransferase |
| ENSMUSG00000034075 | ENSFM00500000270072 | Palmitoyltransferase ZDHHC5 EC_2.3.1.225 Zinc Finger Dhhc Domain Containing 5 Dhhc 5 |
| ENSMUSG00000034135 | ENSFM00570000893436 | Unknown |
| ENSMUSG00000034135 | ENSFM00730001521674 | Serine/Threonine Kinase EC_2.7.11.1 Salt Inducible Kinase Sik Serine/Threonine Kinase SNF1 Kinase |
| ENSMUSG00000034152 | ENSFM00250000001333 | Exocyst Complex Component 3 Exocyst Complex Component SEC6 |
| ENSMUSG00000034160 | ENSFM00250000002216 | Udp N Acetylglucosamine Peptide N Acetylglucosaminyltransferase 110 Kda Subunit EC_2.4.1.255 O Glcnac Transferase Subunit P110 O Linked N Acetylglucosamine Transferase 110 Kda Subunit Ogt |
| ENSMUSG00000034171 | ENSFM00500000270531 | Fatty Acid Amide Hydrolase 1 EC_3.5.1.99 Anandamide Amidohydrolase 1 Oleamide Hydrolase 1 |
| ENSMUSG00000034173 | ENSFM00600000921422 | Coiled Coil Helix Coiled Coil Helix Domain Containing Mitochondrial Precursor |
| ENSMUSG00000034173 | ENSFM00500000270351 | Zinc Finger Bed Domain Containing |
| ENSMUSG00000034189 | ENSFM00670001235389 | Estradiol 17 Beta Dehydrogenase 12 EC_1.1.1.62 17 Beta Hydroxysteroid Dehydrogenase 12 17 Beta Hsd 12 |
| ENSMUSG00000034201 | ENSFM00500000270880 | GAS2 GAS2 Related On Chromosome Growth Arrest Specific 2 |
| ENSMUSG00000034216 | ENSFM00250000004354 | Vacuolar Sorting Associated 18 |
| ENSMUSG00000034220 | ENSFM00500000269807 | Glypican Secreted Glypican |
| ENSMUSG00000034243 | ENSFM00470000252488 | GOLGB1 |
| ENSMUSG00000034243 | ENSFM00250000005925 | Golgin Subfamily B Member 1.372 Kda Golgi Complex Associated GCP372 Giantin Macrogolgin |
| ENSMUSG00000034245 | ENSFM00250000007111 | Histone Deacetylase 11 HD11 EC_3.5.1.98 |
| ENSMUSG00000034254 | ENSFM00640001113149 | Unknown |
| ENSMUSG00000034254 | ENSFM00640001117076 | Unknown |
| ENSMUSG00000034254 | ENSFM00500000270410 | 1 Acyl Sn Glycerol 3 Phosphate Acyltransferase Alpha Precursor EC_2.3.1.51 1 Acylglycerol 3 Phosphate O Acyltransferase 1 1 Agp Acyltransferase 1 1 Agpat 1 Lysophosphatidic Acid Acyltransferase Alpha Lpaat Alpha |
| ENSMUSG00000034275 | ENSFM00250000001385 | Turtle Homolog A Precursor Dendrite Arborization And Synapse Maturation 1 Immunoglobulin Superfamily Member 9A IGSF9A |
| ENSMUSG00000034285 | ENSFM00570000884536 | Unknown |
| ENSMUSG00000034285 | ENSFM00250000003191 | Nipsnap Homolog 2 NIPSNAP2 Glioblastoma Amplified Sequence |
| ENSMUSG00000034312 | ENSFM00250000001463 | Iq Motif And SEC7 Domain Containing |
| ENSMUSG00000034336 | ENSFM00730001521354 | Neurofilament Polypeptide Nf Kda Neurofilament Neurofilament Triplet |
| ENSMUSG00000034361 | ENSFM00800002003432 | Copine Copine |
| ENSMUSG00000034390 | ENSFM00250000004890 | C Maf Inducing C Mip |
| ENSMUSG00000034412 | ENSFM00260000050553 | TBC1 Domain Family Member |
| ENSMUSG00000034432 | ENSFM00760001745951 | Unknown |
| ENSMUSG00000034432 | ENSFM00760001763114 | Unknown |
| ENSMUSG00000034432 | ENSFM00250000006097 | COP9 Signalosome Complex Subunit 8 SGN8 Signalosome Subunit 8 COP9 Homolog JAB1 Containing Signalosome Subunit 8 |
| ENSMUSG00000034471 | ENSFM00250000001241 | Caskin 1 Cask Interacting 1 |
| ENSMUSG00000034485 | ENSFM00800002003915 | Uveal Autoantigen With Coiled Coil Domains And Ankyrin Repeats |
| ENSMUSG00000034488 | ENSFM00500000270078 | Lactadherin Precursor Mfgm Milk Fat Globule Egf Factor 8 Mfg E8 SED1 |
| ENSMUSG00000034560 | ENSFM00250000004568 | Wash Complex Subunit 7 |
| ENSMUSG00000034566 | ENSFM00500000271216 | Atp Synthase Subunit D Mitochondrial Atpase Subunit D |
| ENSMUSG00000034574 | ENSFM00350000105429 | Disheveled Associated Activator Of Morphogenesis |
| ENSMUSG00000034593 | ENSFM00570000901355 | Unknown |
| ENSMUSG00000034593 | ENSFM00750001632326 | Unconventional Myosin Va Dilute Myosin Heavy Chain Non Muscle |
| ENSMUSG00000034613 | ENSFM00250000002013 | Phosphatase EC_3.1.3.16 |
| ENSMUSG00000034656 | ENSFM00740001589100 | Voltage Dependent Type Calcium Channel Subunit Alpha Brain Calcium Channel Calcium Channel L Type Alpha 1 Polypeptide Voltage Gated Calcium Channel Subunit Alpha CAV2 |
| ENSMUSG00000034659 | ENSFM00570000883846 | Unknown |
| ENSMUSG00000034659 | ENSFM00570000897938 | Unknown |
| ENSMUSG00000034659 | ENSFM00250000010758 | Transmembrane 109 Precursor Mitsugumin 23 MG23 |
| ENSMUSG00000034675 | ENSFM00480000262790 | Drebrin Developmentally Regulated Brain |
| ENSMUSG00000034685 | ENSFM00740001589308 | Precursor |
| ENSMUSG00000034686 | ENSFM00250000010083 | Proline Rich 7 Synaptic Proline Rich Membrane |
| ENSMUSG00000034729 | ENSFM00570000888237 | Unknown |
| ENSMUSG00000034729 | ENSFM00250000004828 | 28S Ribosomal S10 Mitochondrial Mrp S10 S10MT |
| ENSMUSG00000034730 | ENSFM00250000000737 | Brain Specific Angiogenesis Inhibitor Precursor |
| ENSMUSG00000034751 | ENSFM00670001238533 | Microtubule Associated Serine/Threonine Kinase 4 |
| ENSMUSG00000034751 | ENSFM00250000000518 | Microtubule Associated Serine/Threonine Kinase EC_2.7.11.1 |
| ENSMUSG00000034786 | ENSFM00640001113161 | Unknown |
| ENSMUSG00000034786 | ENSFM00640001113162 | Unknown |
| ENSMUSG00000034786 | ENSFM00600000921937 | G Signaling Modulator 3 |
| ENSMUSG00000034799 | ENSFM00660001189536 | Unknown |
| ENSMUSG00000034799 | ENSFM00250000000599 | Unc 13 Homolog MUNC13 |
| ENSMUSG00000034807 | ENSFM00250000001576 | Procollagen Galactosyltransferase Precursor EC_2.4.1.50 Collagen Beta 1 O Galactosyltransferase Glycosyltransferase 25 Family Member Hydroxylysine Galactosyltransferase |
| ENSMUSG00000034813 | ENSFM00740001589268 | Glutamate Receptor Interacting |
| ENSMUSG00000034820 | ENSFM00500000272686 | Cleavage And Polyadenylation Specificity Factor Subunit 7 |
| ENSMUSG00000034868 | ENSFM00730001521616 | Myosin Regulatory Light Myosin Regulatory Light Chain 2 Smooth Muscle Myosin Regulatory Light Chain |
| ENSMUSG00000034892 | ENSFM00500000277386 | Unknown |
| ENSMUSG00000034902 | ENSFM00590000917942 | Unknown |
| ENSMUSG00000034902 | ENSFM00590000918386 | Unknown |
| ENSMUSG00000034902 | ENSFM00590000918387 | Unknown |
| ENSMUSG00000034902 | ENSFM00730001521611 | Phosphatidylinositol 4 Phosphate 5 Kinase Type 1 Beta Pi Beta Ptdins 4 P 5 Kinase 1 Beta EC_2.7.1.68 Phosphatidylinositol 4 Phosphate 5 Kinase Type I Phosphatidylinositol 4 Phosphate 5 Kinase Beta |
| ENSMUSG00000034902 | ENSFM00590000918388 | Phosphatidylinositol 4 Phosphate 5 Kinase Type 1 Gamma |
| ENSMUSG00000034912 | ENSFM00740001599945 | Unknown |
| ENSMUSG00000034912 | ENSFM00250000002191 | Mam Domain Containing Glycosylphosphatidylinositol Anchor 2 Precursor Mam Domain Containing 1 |
| ENSMUSG00000034958 | ENSFM00250000001236 | Ambiguous |
| ENSMUSG00000034958 | ENSFM00570000895557 | Unknown |
| ENSMUSG00000034974 | ENSFM00730001521806 | Death Associated Kinase Dap Kinase EC_2.7.11.1 Dap Kinase |
| ENSMUSG00000035067 | ENSFM00500000269993 | Xk Related |
| ENSMUSG00000035067 | ENSFM00670001258781 | Xk Related |
| ENSMUSG00000035104 | ENSFM00570000899708 | Unknown |
| ENSMUSG00000035104 | ENSFM00600000921769 | Eva 1 Homolog A FAM176A Transmembrane 166 |
| ENSMUSG00000035150 | ENSFM00250000002233 | Eukaryotic Translation Initiation Factor 2 Subunit 3 Eukaryotic Translation Initiation Factor 2 Subunit Gamma Eif 2 Gamma |
| ENSMUSG00000035152 | ENSFM00570000901915 | Unknown |
| ENSMUSG00000035152 | ENSFM00430000230089 | Ap 1 Complex Subunit Beta 1 Adaptor Complex Ap 1 Subunit Beta 1 Adaptor Related Complex 1 Subunit Beta 1 Beta 1 Adaptin Beta Adaptin 1 Clathrin Assembly Complex 1 Beta Large Chain Golgi Adaptor HA1/AP1 Adaptin Beta Subunit |
| ENSMUSG00000035168 | ENSFM00250000001143 | Tetratricopeptide Repeat Ankyrin Repeat And Coiled Coil Domain Containing |
| ENSMUSG00000035226 | ENSFM00250000000629 | Regulating Synaptic Membrane Exocytosis Rab 3 Interacting Molecule Rim |
| ENSMUSG00000035227 | ENSFM00250000005526 | Signal Peptidase Complex Subunit 2 EC_3.4.-.- Microsomal Signal Peptidase 25 Kda Subunit Spase 25 Kda Subunit |
| ENSMUSG00000035232 | ENSFM00500000269765 | Kinase Isozyme Mitochondrial Precursor EC_2.7.11.2 Pyruvate Dehydrogenase Kinase |
| ENSMUSG00000035236 | ENSFM00570000877479 | Unknown |
| ENSMUSG00000035236 | ENSFM00570000898233 | Unknown |
| ENSMUSG00000035236 | ENSFM00250000003803 | Scai Suppressor Of Cancer Cell Invasion |
| ENSMUSG00000035285 | ENSFM00600000921959 | N Acetyltransferase 14 EC_2.3.1.- |
| ENSMUSG00000035297 | ENSFM00570000892552 | Unknown |
| ENSMUSG00000035297 | ENSFM00250000005111 | COP9 Signalosome Complex Subunit 4 Signalosome Subunit 4 |
| ENSMUSG00000035376 | ENSFM00500000270421 | Very Long Chain 3R 3 Hydroxyacyl Coa Dehydratase 2 EC_4.2.1.134 3 Hydroxyacyl Coa Dehydratase 2 HACD2 Tyrosine Phosphatase Member B |
| ENSMUSG00000035383 | ENSFM00500000273166 | Pro Mch Precursor |
| ENSMUSG00000035390 | ENSFM00740001589247 | Serine/Threonine Kinase BRSK2 EC_2.7.11.1 EC_2.7.11.- 26 Brain Specific Serine/Threonine Kinase 2 Br Serine/Threonine Kinase 2 Serine/Threonine Kinase Sad A |
| ENSMUSG00000035392 | ENSFM00570000876791 | Unknown |
| ENSMUSG00000035392 | ENSFM00570000883859 | Unknown |
| ENSMUSG00000035392 | ENSFM00570000888231 | Unknown |
| ENSMUSG00000035392 | ENSFM00570000888409 | Unknown |
| ENSMUSG00000035392 | ENSFM00250000001555 | Denn Domain Containing Connecdenn |
| ENSMUSG00000035395 | ENSFM00250000004211 | DDB1 And CUL4 Associated Factor 8 Wd Repeat Containing |
| ENSMUSG00000035441 | ENSFM00770001847571 | Unconventional Myosin |
| ENSMUSG00000035469 | ENSFM00550000769437 | Unknown |
| ENSMUSG00000035469 | ENSFM00640001117070 | Unknown |
| ENSMUSG00000035469 | ENSFM00640001117071 | Unknown |
| ENSMUSG00000035469 | ENSFM00640001119698 | Unknown |
| ENSMUSG00000035469 | ENSFM00260000050642 | RCC1 And Btb Domain Containing 2 Chromosome Condensation 1 Regulator Of Chromosome Condensation And Btb Domain Containing 2 |
| ENSMUSG00000035513 | ENSFM00260000050653 | Netrin Precursor Laminet |
| ENSMUSG00000035547 | ENSFM00570000885619 | Unknown |
| ENSMUSG00000035547 | ENSFM00570000889572 | Unknown |
| ENSMUSG00000035547 | ENSFM00730001522149 | Calpain |
| ENSMUSG00000035735 | ENSFM00250000001790 | SN1 Specific Diacylglycerol Lipase Alpha Dgl Alpha EC_3.1.1.- Neural Stem Cell Derived Dendrite Regulator |
| ENSMUSG00000035770 | ENSFM00250000001777 | Cytoplasmic Dynein 1 Light Intermediate Chain 1 Dynein Light Chain A Dlc A Dynein Light Intermediate Chain 1 Cytosolic |
| ENSMUSG00000035772 | ENSFM00250000006868 | 28S Ribosomal S2 Mitochondrial Mrp S2 S2MT |
| ENSMUSG00000035805 | ENSFM00250000007537 | Membrane MLC1 |
| ENSMUSG00000035835 | ENSFM00610000968417 | Unknown |
| ENSMUSG00000035835 | ENSFM00500000270520 | Lipid Phosphate Phosphatase Related Type 4 EC_3.1.3.4 Brain Specific Phosphatidic Acid Phosphatase 1 Plasticity Related Gene 1 Prg 1 |
| ENSMUSG00000035863 | ENSFM00500000271464 | Paralemmin 1 Precursor Paralemmin |
| ENSMUSG00000035864 | ENSFM00720001492031 | Synaptotagmin |
| ENSMUSG00000035900 | ENSFM00250000002904 | Gram Domain Containing 4 |
| ENSMUSG00000035954 | ENSFM00760001714721 | Dedicator Of Cytokinesis 3 Modifier Of Cell Adhesion Presenilin Binding Pbp |
| ENSMUSG00000036026 | ENSFM00250000001207 | Transmembrane |
| ENSMUSG00000036046 | ENSFM00250000003605 | Uncharacterized |
| ENSMUSG00000036052 | ENSFM00440000236855 | Dnaj Homolog Subfamily B Member |
| ENSMUSG00000036053 | ENSFM00350000105407 | Formin |
| ENSMUSG00000036087 | ENSFM00260000050845 | Slain Motif Containing |
| ENSMUSG00000036095 | ENSFM00730001521692 | Diacylglycerol Kinase Dag Kinase EC_2.7.1.107 Kda Diacylglycerol Kinase Diglyceride Kinase Dgk |
| ENSMUSG00000036103 | ENSFM00250000004242 | Collectin 12 Collectin Placenta 1 Cl P1 Scavenger Receptor |
| ENSMUSG00000036199 | ENSFM00500000273406 | Nadh Dehydrogenase 1 Alpha Subcomplex Subunit 13 Cell Death Regulatory Grim 19 Complex I B16 6 Ci B16 6 Gene Associated With Retinoic Interferon Induced Mortality 19 Grim 19 Nadh Ubiquinone Oxidoreductase B16 6 Subunit |
| ENSMUSG00000036306 | ENSFM00650001140033 | Leucine Zipper Tumor Suppressor 2 |
| ENSMUSG00000036309 | ENSFM00610000956029 | Unknown |
| ENSMUSG00000036309 | ENSFM00610000969567 | Unknown |
| ENSMUSG00000036309 | ENSFM00670001235396 | E3 Ubiquitin Ligase Complex Scf Subunit Sconc Sulfur Controller C Sulfur Metabolite Repression Control C |
| ENSMUSG00000036371 | ENSFM00500000270904 | Plasminogen Activator Inhibitor 1 Rna Binding PAI1 Rna Binding 1 Pai RBP1 SERPINE1 Binding 1 |
| ENSMUSG00000036372 | ENSFM00500000276946 | Unknown |
| ENSMUSG00000036372 | ENSFM00500000423017 | Unknown |
| ENSMUSG00000036377 | ENSFM00250000006430 | Uncharacterized |
| ENSMUSG00000036377 | ENSFM00520000559661 | Unknown |
| ENSMUSG00000036377 | ENSFM00570000896173 | Unknown |
| ENSMUSG00000036377 | ENSFM00570000899386 | Unknown |
| ENSMUSG00000036402 | ENSFM00500000279757 | Unknown |
| ENSMUSG00000036435 | ENSFM00250000004597 | Exocyst Complex Component 1 Exocyst Complex Component SEC3 |
| ENSMUSG00000036452 | ENSFM00760001762720 | Gene |
| ENSMUSG00000036452 | ENSFM00250000000574 | Rho Gtpase Activating Rho Type Gtpase Activating |
| ENSMUSG00000036473 | ENSFM00570000851049 | Netrin Precursor |
| ENSMUSG00000036473 | ENSFM00270000056490 | TBC1 Domain Family Member 24 |
| ENSMUSG00000036550 | ENSFM00780001898388 | CCR4 Not Transcription Complex Subunit 1 CCR4 Associated Factor 1 |
| ENSMUSG00000036560 | ENSFM00610000969178 | Unknown |
| ENSMUSG00000036560 | ENSFM00250000002312 | Leucine Rich Repeat Lgi Family Member Precursor Leucine Rich Glioma Inactivated |
| ENSMUSG00000036568 | ENSFM00250000009373 | GLTSCR1 |
| ENSMUSG00000036570 | ENSFM00810002042307 | Phospholemman Precursor Fxyd Domain Containing Ion Transport Regulator 1 |
| ENSMUSG00000036585 | ENSFM00520000548481 | Unknown |
| ENSMUSG00000036585 | ENSFM00570000878929 | Unknown |
| ENSMUSG00000036585 | ENSFM00570000891312 | Unknown |
| ENSMUSG00000036585 | ENSFM00570000902091 | Unknown |
| ENSMUSG00000036585 | ENSFM00500000271792 | Fibroblast Growth Factor 1 Fgf 1 Acidic Fibroblast Growth Factor Afgf Heparin Binding Growth Factor 1 Hbgf 1 |
| ENSMUSG00000036591 | ENSFM00640001105460 | Unknown |
| ENSMUSG00000036591 | ENSFM00250000001623 | Rho Gtpase Activating 21 Rho Type Gtpase Activating 21 |
| ENSMUSG00000036611 | ENSFM00250000006173 | Endonuclease/Exonuclease/Phosphatase Family Domain Containing 1 |
| ENSMUSG00000036617 | ENSFM00250000001584 | Src Kinase Signaling Inhibitor 1 Snap 25 Interacting Snip P130CAS Associated P140CAP |
| ENSMUSG00000036686 | ENSFM00730001522120 | Coiled Coil And C2 Domain Containing Five Prime Repressor Element Under Dual Repression Binding Fre Under Dual Repression Binding Freud |
| ENSMUSG00000036698 | ENSFM00250000000492 | Argonaute |
| ENSMUSG00000036707 | ENSFM00570000901084 | Unknown |
| ENSMUSG00000036707 | ENSFM00250000002178 | Calcium Binding 39 MO25 |
| ENSMUSG00000036712 | ENSFM00250000002039 | Ubiquitin Carboxyl Terminal Hydrolase Cyld EC_3.4.19.12 Deubiquitinating Enzyme Cyld Ubiquitin Thioesterase Cyld Ubiquitin Specific Processing Protease Cyld |
| ENSMUSG00000036752 | ENSFM00250000000193 | Tubulin Beta Chain |
| ENSMUSG00000036781 | ENSFM00570000892545 | Unknown |
| ENSMUSG00000036781 | ENSFM00570000893201 | Unknown |
| ENSMUSG00000036781 | ENSFM00670001235597 | 40S Ribosomal S27 |
| ENSMUSG00000036781 | ENSFM00570000892970 | 40S Ribosomal S27 |
| ENSMUSG00000036880 | ENSFM00760001714736 | 3 Ketoacyl Coa Thiolase Mitochondrial EC_2.3.1.16 Acetyl Coa Acyltransferase Beta Ketothiolase Mitochondrial 3 Oxoacyl Coa Thiolase |
| ENSMUSG00000036882 | ENSFM00250000001205 | Rho Gtpase Activating Rho Type Gtpase Activating Gtpase Activating |
| ENSMUSG00000036896 | ENSFM00680001303632 | Complement C1Q Subcomponent Subunit Precursor |
| ENSMUSG00000036902 | ENSFM00250000001916 | Neuropilin And Tolloid Precursor Brain Specific Transmembrane Containing 2 Cub And 1 Ldl Receptor Class A Domains |
| ENSMUSG00000036905 | ENSFM00680001303632 | Complement C1Q Subcomponent Subunit Precursor |
| ENSMUSG00000036932 | ENSFM00250000002966 | Apoptosis Inducing Factor 1 Mitochondrial Precursor EC_1.1.1.- Programmed Cell Death 8 |
| ENSMUSG00000036943 | ENSFM00730001521184 | Ras Related Rab |
| ENSMUSG00000036948 | ENSFM00250000006401 | Uncharacterized |
| ENSMUSG00000036948 | ENSFM00590000916964 | Unknown |
| ENSMUSG00000036957 | ENSFM00260000050443 | Leucine Rich Repeat And Fibronectin Type Iii Domain Containing Precursor Synaptic Adhesion Molecule |
| ENSMUSG00000036989 | ENSFM00750001632380 | Tripartite Motif Containing 2 EC_6.3.2.- E3 Ubiquitin Ligase TRIM2 |
| ENSMUSG00000036992 | ENSFM00500000270737 | NTF2 Related Export |
| ENSMUSG00000037012 | ENSFM00570000888918 | Unknown |
| ENSMUSG00000037012 | ENSFM00570000883017 | Hexokinase 1 |
| ENSMUSG00000037012 | ENSFM00250000000532 | Hexokinase EC_2.7.1.1 Hexokinase Type Hk |
| ENSMUSG00000037022 | ENSFM00250000008244 | Methylmalonic Aciduria Type A Homolog Mitochondrial Precursor EC_3.6.-.- Methylmalonic Aciduria Type A 1 |
| ENSMUSG00000037126 | ENSFM00250000001069 | Ph And SEC7 Domain Containing 1 Exchange Factor ARF6 Pleckstrin Homology And SEC7 Domain Containing 1 |
| ENSMUSG00000037149 | ENSFM00250000003062 | Atp Dependent Rna Helicase DDX1 EC_3.6.4.13 Dead Box 1 |
| ENSMUSG00000037217 | ENSFM00250000000771 | Synapsin Synapsin |
| ENSMUSG00000037234 | ENSFM00250000001958 | Hook |
| ENSMUSG00000037236 | ENSFM00760001750219 | Unknown |
| ENSMUSG00000037236 | ENSFM00760001750220 | Unknown |
| ENSMUSG00000037236 | ENSFM00760001762716 | Unknown |
| ENSMUSG00000037236 | ENSFM00250000005840 | Matrin 3 |
| ENSMUSG00000037259 | ENSFM00250000004649 | Double Zinc Ribbon And Ankyrin Repeat Containing 1 |
| ENSMUSG00000037331 | ENSFM00250000001222 | La Related 1 La Ribonucleoprotein Domain Family Member 1 |
| ENSMUSG00000037351 | ENSFM00730001521541 | Actin |
| ENSMUSG00000037353 | ENSFM00250000006470 | LETM1 Domain Containing 1 |
| ENSMUSG00000037373 | ENSFM00810002057430 | Unknown |
| ENSMUSG00000037373 | ENSFM00810002057431 | Unknown |
| ENSMUSG00000037373 | ENSFM00250000001273 | C Terminal Binding |
| ENSMUSG00000037386 | ENSFM00250000000629 | Regulating Synaptic Membrane Exocytosis Rab 3 Interacting Molecule Rim |
| ENSMUSG00000037428 | ENSFM00250000010086 | Neurosecretory Vgf Neuroendocrine Regulatory Peptide 1 Nerp 1 |
| ENSMUSG00000037519 | ENSFM00720001499800 | Unknown |
| ENSMUSG00000037519 | ENSFM00250000000441 | Liprin Alpha Tyrosine Phosphatase Receptor Type F Polypeptide Interacting Alpha Ptprf Interacting Alpha |
| ENSMUSG00000037563 | ENSFM00250000002859 | 40S Ribosomal S16 |
| ENSMUSG00000037605 | ENSFM00570000897271 | Unknown |
| ENSMUSG00000037605 | ENSFM00260000050328 | Latrophilin |
| ENSMUSG00000037625 | ENSFM00260000051131 | Claudin 11 |
| ENSMUSG00000037653 | ENSFM00260000050545 | Btb/Poz Domain Containing |
| ENSMUSG00000037703 | ENSFM00650001140033 | Leucine Zipper Tumor Suppressor 2 |
| ENSMUSG00000037706 | ENSFM00800002003853 | CD81 Antigen 26 Kda Cell Surface Tapa 1 Target Of The Antiproliferative Antibody 1 CD81 Antigen |
| ENSMUSG00000037710 | ENSFM00500000274698 | Cdgsh Iron Sulfur Domain Containing 1 Mitoneet |
| ENSMUSG00000037720 | ENSFM00590000917165 | Unknown |
| ENSMUSG00000037720 | ENSFM00590000918332 | Unknown |
| ENSMUSG00000037720 | ENSFM00810002055293 | Unknown |
| ENSMUSG00000037720 | ENSFM00250000005590 | Transmembrane 33 |
| ENSMUSG00000037736 | ENSFM00570000885356 | Unknown |
| ENSMUSG00000037736 | ENSFM00570000898739 | Unknown |
| ENSMUSG00000037736 | ENSFM00810002057321 | Unknown |
| ENSMUSG00000037736 | ENSFM00250000003304 | Lim And Calponin Homology Domains Containing 1 |
| ENSMUSG00000037742 | ENSFM00800002003364 | Elongation Factor 1 Alpha Ef 1 Alpha Eukaryotic Elongation Factor 1 A EEF1A |
| ENSMUSG00000037754 | ENSFM00500000270382 | Phosphatase 1 Regulatory Inhibitor Subunit 16B Precursor Caax Box Timap Tgf Beta Inhibited Membrane Associated |
| ENSMUSG00000037772 | ENSFM00500000273802 | 39S Ribosomal L23 Mitochondrial L23MT Mrp L23 L23 Mitochondrial Related |
| ENSMUSG00000037805 | ENSFM00570000901645 | Unknown |
| ENSMUSG00000037805 | ENSFM00790001962982 | 60S Ribosomal L10A |
| ENSMUSG00000037815 | ENSFM00250000001018 | Catenin Alpha 2 Alpha N Catenin |
| ENSMUSG00000037851 | ENSFM00610000969945 | Unknown |
| ENSMUSG00000037851 | ENSFM00500000270033 | Isoleucine Trna Ligase Cytoplasmic EC_6.1.1.5 Isoleucyl Trna Synthetase Ilers |
| ENSMUSG00000037855 | ENSFM00250000005396 | Unknown |
| ENSMUSG00000037905 | ENSFM00800002014565 | Unknown |
| ENSMUSG00000037905 | ENSFM00250000006424 | BRI3 Binding I3 Binding |
| ENSMUSG00000037957 | ENSFM00250000001646 | Dystrophia Myotonica Wd Repeat Containing Dystrophia Myotonica Containing Wd Repeat Motif Protein Dmr N9 |
| ENSMUSG00000037979 | ENSFM00570000895056 | Unknown |
| ENSMUSG00000037979 | ENSFM00250000009967 | Coiled Coil Domain Containing 92 |
| ENSMUSG00000037989 | ENSFM00250000000621 | Serine/Threonine Kinase EC_2.7.11.1 Kinase Lysine Deficient Kinase With No Lysine |
| ENSMUSG00000038013 | ENSFM00500000273445 | Was/Wasl Interacting Family Member 2 Wasp Interacting Related Wip Related |
| ENSMUSG00000038014 | ENSFM00250000002493 | Constitutive Coactivator Of Ppar Gamma 1 Oxidative Stress Associated Src Activator FAM120A |
| ENSMUSG00000038028 | ENSFM00810002057401 | Unknown |
| ENSMUSG00000038028 | ENSFM00250000007002 | Fructose 2 6 Bisphosphatase Tigar EC_3.1.3.46 TP53 Induced Glycolysis And Apoptosis Regulator |
| ENSMUSG00000038034 | ENSFM00740001589327 | Immunoglobulin Superfamily Member Precursor Glu Trp Ile Ewi Motif Containing Ewi Antigen |
| ENSMUSG00000038077 | ENSFM00290000065519 | Potassium Voltage Gated Channel Subfamily A Member Voltage Gated Potassium Channel Subunit KV1 |
| ENSMUSG00000038079 | ENSFM00250000005158 | Transmembrane Amyotrophic Lateral Sclerosis 2 Chromosomal Region Candidate Gene 4 |
| ENSMUSG00000038173 | ENSFM00570000884802 | Unknown |
| ENSMUSG00000038173 | ENSFM00730001522540 | Ectonucleotide Pyrophosphatase/Phosphodiesterase Family Member 6 Precursor E Npp 6 Npp 6 EC_3.1.4.- EC_3.1.4.38 Choline Specific Glycerophosphodiester Phosphodiesterase Glycerophosphocholine Cholinephosphodiesterase Gpc Cpde |
| ENSMUSG00000038174 | ENSFM00670001258143 | Unknown |
| ENSMUSG00000038174 | ENSFM00250000003532 | Hyccin Down Regulated By CTNNB1 A FAM126A |
| ENSMUSG00000038264 | ENSFM00730001522534 | Semaphorin 7A Precursor Semaphorin K1 Sema K1 Semaphorin L Sema L CD108 Antigen |
| ENSMUSG00000038366 | ENSFM00470000259183 | Unknown |
| ENSMUSG00000038366 | ENSFM00570000883595 | Unknown |
| ENSMUSG00000038366 | ENSFM00570000888630 | Unknown |
| ENSMUSG00000038366 | ENSFM00570000892904 | Unknown |
| ENSMUSG00000038366 | ENSFM00570000895214 | Unknown |
| ENSMUSG00000038366 | ENSFM00570000895866 | Unknown |
| ENSMUSG00000038366 | ENSFM00610000965408 | Unknown |
| ENSMUSG00000038366 | ENSFM00500000270806 | Lim And SH3 Domain 1 Lasp 1 |
| ENSMUSG00000038387 | ENSFM00770001847543 | Ras Related R Ras |
| ENSMUSG00000038390 | ENSFM00250000004703 | Probable G Coupled Receptor 153 G Coupled Receptor PGR1 |
| ENSMUSG00000038453 | ENSFM00250000001584 | Src Kinase Signaling Inhibitor 1 Snap 25 Interacting Snip P130CAS Associated P140CAP |
| ENSMUSG00000038467 | ENSFM00600000921240 | Charged Multivesicular Body Chromatin Modifying |
| ENSMUSG00000038510 | ENSFM00810002040861 | Ribosome Production Factor 2 Homolog Brix Domain Containing 1 Ribosome Biogenesis RPF2 Homolog |
| ENSMUSG00000038517 | ENSFM00250000006948 | Tank Binding Kinase 1 Binding 1 TBK1 Binding 1 |
| ENSMUSG00000038525 | ENSFM00250000001585 | Armadillo Repeat Containing X Linked |
| ENSMUSG00000038555 | ENSFM00500000269981 | Receptor Expression Enhancing |
| ENSMUSG00000038608 | ENSFM00500000269738 | Dedicator Of Cytokinesis CDC42 Guanine Nucleotide Exchange Factor Zizimin |
| ENSMUSG00000038665 | ENSFM00730001521936 | Diacylglycerol Kinase Zeta Dag Kinase Zeta EC_2.7.1.107 Diglyceride Kinase Zeta Dgk Zeta |
| ENSMUSG00000038740 | ENSFM00500000272007 | Multivesicular Body Subunit 12B Escrt I Complex Subunit MVB12B FAM125B |
| ENSMUSG00000038828 | ENSFM00250000004458 | Transmembrane 214 |
| ENSMUSG00000038845 | ENSFM00790001963030 | Prohibitin |
| ENSMUSG00000038900 | ENSFM00780001917905 | Unknown |
| ENSMUSG00000038900 | ENSFM00250000002131 | 60S Ribosomal L12 |
| ENSMUSG00000038910 | ENSFM00770001847576 | Inactive Phospholipase C 2 Plc L 2 Plc L2 Phospholipase C L2 Phospholipase C Epsilon 2 Plc Epsilon 2 |
| ENSMUSG00000038916 | ENSFM00250000001168 | Soga Family Member |
| ENSMUSG00000038936 | ENSFM00250000003507 | Saccharopine Dehydrogenase Oxidoreductase Ec 1 |
| ENSMUSG00000038970 | ENSFM00670001235536 | Serine/Threonine Kinase EC_2.7.11.1 Apoptosis Associated Tyrosine Kinase Brain Kinase Lemur Tyrosine Kinase |
| ENSMUSG00000038976 | ENSFM00250000001450 | Neurabin 1 Neurabin I Neural Tissue Specific F Actin Binding I Phosphatase 1 Regulatory Subunit 9A |
| ENSMUSG00000039058 | ENSFM00740001589162 | Adenylate Kinase Isoenzyme 1 |
| ENSMUSG00000039062 | ENSFM00250000000130 | Aminopeptidase N Ap N EC_3.4.11.2 Microsomal Aminopeptidase |
| ENSMUSG00000039067 | ENSFM00250000004823 | 26S Proteasome Non Atpase Regulatory Subunit 7 26S Proteasome Regulatory Subunit RPN8 |
| ENSMUSG00000039103 | ENSFM00800002024520 | Unknown |
| ENSMUSG00000039103 | ENSFM00250000002729 | Nexilin F Actin Binding |
| ENSMUSG00000039105 | ENSFM00670001236060 | V Type Proton Atpase Subunit G 1 V Atpase Subunit G 1 V Atpase 13 Kda Subunit 1 Vacuolar Proton Pump Subunit G 1 Vacuolar Proton Pump Subunit M16 |
| ENSMUSG00000039114 | ENSFM00500000274208 | Neuritin Precursor |
| ENSMUSG00000039156 | ENSFM00250000001930 | Stromal Interaction Molecule Precursor |
| ENSMUSG00000039221 | ENSFM00780001917087 | Unknown |
| ENSMUSG00000039221 | ENSFM00600000921635 | 60S Ribosomal L22 1 |
| ENSMUSG00000039252 | ENSFM00250000002312 | Leucine Rich Repeat Lgi Family Member Precursor Leucine Rich Glioma Inactivated |
| ENSMUSG00000039254 | ENSFM00570000880296 | Unknown |
| ENSMUSG00000039254 | ENSFM00780001917899 | Unknown |
| ENSMUSG00000039254 | ENSFM00730001522352 | O 1 EC_2.4.1.109 Dolichyl Phosphate Mannose Mannosyltransferase 1 |
| ENSMUSG00000039316 | ENSFM00390000126378 | Raftlin Raft Linking |
| ENSMUSG00000039367 | ENSFM00770001847547 | Transport SEC24 Related |
| ENSMUSG00000039385 | ENSFM00260000050333 | Cadherin Precursor |
| ENSMUSG00000039457 | ENSFM00500000270485 | Envoplakin 210 Kda Cornified Envelope Precursor P210 |
| ENSMUSG00000039474 | ENSFM00280000058784 | Wolframin |
| ENSMUSG00000039478 | ENSFM00260000050554 | Calcium Uptake 2 Mitochondrial Precursor Ef Hand Domain Containing Family Member A1 |
| ENSMUSG00000039488 | ENSFM00390000126303 | Contactin Precursor Brain Derived Immunoglobulin Superfamily Big |
| ENSMUSG00000039515 | ENSFM00550000750340 | Unknown |
| ENSMUSG00000039515 | ENSFM00570000898338 | Unknown |
| ENSMUSG00000039515 | ENSFM00500000286252 | Serine/Threonine Phosphatase 2A Activator |
| ENSMUSG00000039515 | ENSFM00760001714775 | Serine/Threonine Phosphatase 2A Activator EC_5.2.1.8 PP2A Subunit B' PR53 Phosphotyrosyl Phosphatase Activator Ptpa Serine/Threonine Phosphatase 2A Regulatory Subunit 4 Serine/Threonine Phosphatase 2A Regulatory Subunit B' |
| ENSMUSG00000039530 | ENSFM00250000002565 | Magnesium Transporter 1 Precursor MAGT1 |
| ENSMUSG00000039536 | ENSFM00250000001250 | Double Stranded Rna Binding Staufen Homolog |
| ENSMUSG00000039546 | ENSFM00570000900895 | Unknown |
| ENSMUSG00000039546 | ENSFM00250000009368 | Adherens Junction Associated 1 |
| ENSMUSG00000039585 | ENSFM00510000502753 | Unconventional Myosin Unconventional Myosin |
| ENSMUSG00000039640 | ENSFM00760001714704 | 39S Ribosomal L12 Mitochondrial Precursor L12MT Mrp L12 |
| ENSMUSG00000039652 | ENSFM00250000000830 | Cytoplasmic Polyadenylation Element Binding |
| ENSMUSG00000039701 | ENSFM00250000002075 | Inactive Ubiquitin Carboxyl Terminal Hydrolase Inactive Ubiquitin Specific Peptidase |
| ENSMUSG00000039716 | ENSFM00760001714721 | Dedicator Of Cytokinesis 3 Modifier Of Cell Adhesion Presenilin Binding Pbp |
| ENSMUSG00000039740 | ENSFM00800002003805 | Alpha 1 3/1 6 Mannosyltransferase ALG2 EC_2.4.1.132 EC_2.4.1.- 257 Asparagine Linked Glycosylation 2 Homolog Gdp Man:Man 1 Glcnac 2 Pp Dol Alpha 1 3 Mannosyltransferase Gdp Man:Man 1 Glcnac 2 Pp Dolichol Mannosyltransferase Gdp Man:Man 2 Glcnac 2 Pp Dol A |
| ENSMUSG00000039768 | ENSFM00250000002491 | Dnaj Homolog Subfamily C Member 11 |
| ENSMUSG00000039809 | ENSFM00250000000928 | Gamma Aminobutyric Acid Type B Receptor Subunit 2 Precursor Gaba B Receptor 2 Gaba B R2 Gaba BR2 GABABR2 GB2 G Coupled Receptor 51 |
| ENSMUSG00000039824 | ENSFM00250000000438 | Myosin Light Chain Myosin Light Chain Myosin Light Chain |
| ENSMUSG00000039913 | ENSFM00730001521739 | Serine/Threonine Kinase Pak EC_2.7.11.1 P21 Activated Kinase Pak |
| ENSMUSG00000039929 | ENSFM00250000007283 | Nucleolar Pre Ribosomal Associated 1 URB1 Ribosome Biogenesis 1 Homolog |
| ENSMUSG00000039943 | ENSFM00800002003599 | 1 Phosphatidylinositol 4 5 Bisphosphate Phosphodiesterase Beta 4 EC_3.1.4.11 Phosphoinositide Phospholipase C Beta 4 Phospholipase C Beta 4 Plc Beta 4 |
| ENSMUSG00000039959 | ENSFM00250000001308 | Huntingtin Interacting 1 Hip 1 Huntingtin Interacting I Hip I |
| ENSMUSG00000040003 | ENSFM00800002020509 | Unknown |
| ENSMUSG00000040003 | ENSFM00250000000552 | Membrane Associated Guanylate Kinase Ww And Pdz Domain Containing Membrane Associated Guanylate Kinase Inverted Magi |
| ENSMUSG00000040009 | ENSFM00410000138458 | Guanine Nucleotide Binding Subunit |
| ENSMUSG00000040022 | ENSFM00500000271919 | RAB11 Family Interacting 2 RAB11 FIP2 |
| ENSMUSG00000040037 | ENSFM00290000065521 | Precursor |
| ENSMUSG00000040048 | ENSFM00250000006353 | Nadh Dehydrogenase 1 Beta Subcomplex Subunit 10 Complex I Pdsw Ci Pdsw Nadh Ubiquinone Oxidoreductase Pdsw Subunit |
| ENSMUSG00000040055 | ENSFM00740001589167 | Gap Junction Beta Connexin |
| ENSMUSG00000040105 | ENSFM00250000003664 | Probable Lipid Phosphate Phosphatase PPAPDC3 EC_3.1.3.- Phosphatidic Acid Phosphatase Type 2 Domain Containing 3 |
| ENSMUSG00000040118 | ENSFM00670001245035 | Uncharacterized |
| ENSMUSG00000040118 | ENSFM00730001522093 | Voltage Dependent Calcium Channel Subunit Alpha 2/Delta 1 Precursor Voltage Gated Calcium Channel Subunit Alpha 2/Delta 1 |
| ENSMUSG00000040225 | ENSFM00500000270319 | PRRC2C BAT2 Domain Containing 1 Hla B Associated Transcript 2 2 Proline Rich And Coiled Coil Containing 2C |
| ENSMUSG00000040236 | ENSFM00500000273083 | Trafficking Particle Complex Subunit 5 |
| ENSMUSG00000040242 | ENSFM00800002003783 | Suppressor Of Ikbke 1 Suppressor Of Ikk Epsilon |
| ENSMUSG00000040260 | ENSFM00350000105429 | Disheveled Associated Activator Of Morphogenesis |
| ENSMUSG00000040265 | ENSFM00520000517792 | Dynamin EC_3.6.5.5 |
| ENSMUSG00000040269 | ENSFM00670001236327 | 28S Ribosomal S28 Mitochondrial Precursor Mrp S28 S28MT |
| ENSMUSG00000040351 | ENSFM00400000131980 | Ankyrin Repeat And Ibr Domain Containing 1 |
| ENSMUSG00000040359 | ENSFM00250000003769 | E3 UFM1 Ligase 1 EC_6.3.2.- |
| ENSMUSG00000040380 | ENSFM00250000002369 | Cerebellin Precursor |
| ENSMUSG00000040385 | ENSFM00420000140518 | Serine/Threonine Phosphatase PP1 Catalytic Subunit Pp EC_3.1.3.16 |
| ENSMUSG00000040389 | ENSFM00800002017904 | Unknown |
| ENSMUSG00000040389 | ENSFM00250000002280 | Wd Repeat Containing 47 Neuronal Enriched Map Interacting Nemitin |
| ENSMUSG00000040420 | ENSFM00260000050333 | Cadherin Precursor |
| ENSMUSG00000040472 | ENSFM00500000271139 | Geranylgeranyl Transferase Type 2 Subunit Alpha EC_2.5.1.60 Geranylgeranyl Transferase Type Ii Subunit Alpha Rab Geranyl Geranyltransferase Subunit Alpha Rab Gg Transferase Alpha Rab Ggtase Alpha Rab Geranylgeranyltransferase Subunit Alpha |
| ENSMUSG00000040479 | ENSFM00570000877778 | Unknown |
| ENSMUSG00000040479 | ENSFM00570000883510 | Unknown |
| ENSMUSG00000040479 | ENSFM00730001521936 | Diacylglycerol Kinase Zeta Dag Kinase Zeta EC_2.7.1.107 Diglyceride Kinase Zeta Dgk Zeta |
| ENSMUSG00000040495 | ENSFM00800002003483 | Muscarinic Acetylcholine Receptor |
| ENSMUSG00000040549 | ENSFM00760001714778 | Cytoskeleton Associated 5 |
| ENSMUSG00000040560 | ENSFM00250000001912 | Wd Repeat Containing 7 Tgf Beta Resistance Associated Trag |
| ENSMUSG00000040563 | ENSFM00500000270116 | Lipid Phosphate Phosphatase Related Type Plasticity Related Gene Prg |
| ENSMUSG00000040584 | ENSFM00730001521159 | Multidrug Resistance EC_3.6.3.44 Atp Binding Cassette Sub Family B Member P Glycoprotein |
| ENSMUSG00000040606 | ENSFM00250000003884 | Kazrin |
| ENSMUSG00000040612 | ENSFM00740001589513 | Immunoglobulin Domain Containing Receptor 2 Precursor |
| ENSMUSG00000040620 | ENSFM00680001306470 | Unknown |
| ENSMUSG00000040620 | ENSFM00680001326723 | Unknown |
| ENSMUSG00000040620 | ENSFM00680001326724 | Unknown |
| ENSMUSG00000040620 | ENSFM00800002003714 | Atp Dependent Rna Helicase DHX33 EC_3.6.4.13 Deah Box 33 |
| ENSMUSG00000040624 | ENSFM00380000124706 | Pleckstrin Homology Domain Containing Family G Member Ph Domain Containing Family G Member |
| ENSMUSG00000040640 | ENSFM00250000000753 | Elks/RAB6 Interacting/Cast Family Member 1 Erc 1 RAB6 Interacting 2 |
| ENSMUSG00000040697 | ENSFM00250000004546 | Dnaj Homolog Subfamily C Member 16 Precursor |
| ENSMUSG00000040699 | ENSFM00500000273568 | Lim Domain Containing 2 |
| ENSMUSG00000040722 | ENSFM00740001589424 | Secretory Carrier Associated Membrane Secretory Carrier Membrane |
| ENSMUSG00000040724 | ENSFM00290000065519 | Potassium Voltage Gated Channel Subfamily A Member Voltage Gated Potassium Channel Subunit KV1 |
| ENSMUSG00000040794 | ENSFM00260000051148 | Complement C1Q Tumor Necrosis Factor Related 4 Precursor |
| ENSMUSG00000040797 | ENSFM00250000001463 | Iq Motif And SEC7 Domain Containing |
| ENSMUSG00000040860 | ENSFM00570000880210 | Unknown |
| ENSMUSG00000040860 | ENSFM00550000743177 | Ciliary Rootlet Coiled Coil |
| ENSMUSG00000040896 | ENSFM00300000079128 | Potassium Voltage Gated Channel Subfamily D Member Voltage Gated Potassium Channel Subunit KV4 |
| ENSMUSG00000040952 | ENSFM00720001499506 | Unknown |
| ENSMUSG00000040952 | ENSFM00670001235598 | 40S Ribosomal S19 |
| ENSMUSG00000040972 | ENSFM00250000006695 | Immunoglobulin Superfamily Member 21 Precursor IGSF21 |
| ENSMUSG00000040990 | ENSFM00600000921328 | SH3 Domain Containing Kinase Binding 1 Of Kinase |
| ENSMUSG00000041020 | ENSFM00500000375694 | Unknown |
| ENSMUSG00000041020 | ENSFM00250000005199 | MAP7 Domain Containing 2 |
| ENSMUSG00000041112 | ENSFM00250000001494 | Engulfment And Cell Motility |
| ENSMUSG00000041115 | ENSFM00250000001463 | Iq Motif And SEC7 Domain Containing |
| ENSMUSG00000041119 | ENSFM00570000881490 | Unknown |
| ENSMUSG00000041119 | ENSFM00570000881576 | Unknown |
| ENSMUSG00000041119 | ENSFM00570000882033 | Unknown |
| ENSMUSG00000041119 | ENSFM00570000887766 | Unknown |
| ENSMUSG00000041119 | ENSFM00570000889256 | Unknown |
| ENSMUSG00000041119 | ENSFM00570000900811 | Unknown |
| ENSMUSG00000041119 | ENSFM00400000131822 | High Affinity Cgmp Specific 3' 5' Cyclic Phosphodiesterase 9A EC_3.1.4.35 |
| ENSMUSG00000041133 | ENSFM00720001501832 | Unknown |
| ENSMUSG00000041133 | ENSFM00790001962984 | Structural Maintenance Of Chromosomes 1A Smc 1A Smc 1A |
| ENSMUSG00000041205 | ENSFM00250000010765 | MAP6 Domain Containing 1.21 Kda Stop SL21 |
| ENSMUSG00000041236 | ENSFM00250000002575 | Vacuolar Sorting Associated 41 |
| ENSMUSG00000041351 | ENSFM00800002003399 | Ubiquitin Carboxyl Terminal Hydrolase 48 EC_3.4.19.12 Deubiquitinating Enzyme 48 Ubiquitin Thioesterase 48 Ubiquitin Specific Processing Protease 48 |
| ENSMUSG00000041417 | ENSFM00250000000988 | Phosphatidylinositol 3 Kinase Regulatory Subunit Alpha PI3 Kinase Regulatory Subunit Alpha PI3K Regulatory Subunit Alpha Ptdins 3 Kinase Regulatory Subunit Alpha Phosphatidylinositol 3 Kinase 85 Kda Regulatory Subunit Alpha PI3 Kinase Subunit P85 Alpha Pt |
| ENSMUSG00000041444 | ENSFM00250000001205 | Rho Gtpase Activating Rho Type Gtpase Activating Gtpase Activating |
| ENSMUSG00000041453 | ENSFM00250000000614 | 60S Ribosomal L21 |
| ENSMUSG00000041459 | ENSFM00760001750470 | Unknown |
| ENSMUSG00000041459 | ENSFM00760001763533 | Unknown |
| ENSMUSG00000041459 | ENSFM00760001763651 | Unknown |
| ENSMUSG00000041459 | ENSFM00760001763652 | Unknown |
| ENSMUSG00000041459 | ENSFM00250000001956 | Tar Dna Binding 43 Tdp 43 |
| ENSMUSG00000041530 | ENSFM00250000000492 | Argonaute |
| ENSMUSG00000041556 | ENSFM00500000270014 | F Box Only 6 F Box That Recognizes Sugar Chains 2 |
| ENSMUSG00000041570 | ENSFM00600000921199 | Calmodulin Regulated Spectrin Associated |
| ENSMUSG00000041598 | ENSFM00250000006209 | CDC42 Effector 4 Binder Of Rho Gtpases 4 |
| ENSMUSG00000041607 | ENSFM00470000260897 | Unknown |
| ENSMUSG00000041607 | ENSFM00720001502028 | Unknown |
| ENSMUSG00000041607 | ENSFM00250000002814 | Myelin Basic Mbp |
| ENSMUSG00000041609 | ENSFM00570000884207 | Unknown |
| ENSMUSG00000041609 | ENSFM00500000272101 | Bicaudal D Related 1 Bicd Related 1 Bicdr 1 Coiled Coil Domain Containing 64A |
| ENSMUSG00000041624 | ENSFM00730001521851 | Guanylate Cyclase Soluble Subunit Alpha 3 Gcs Alpha 3 EC_4.6.1.2 Gcs Alpha 1 Soluble Guanylate Cyclase Large Subunit |
| ENSMUSG00000041632 | ENSFM00250000006668 | 28S Ribosomal S27 Mitochondrial Mrp S27 S27MT |
| ENSMUSG00000041638 | ENSFM00250000002277 | Translational Activator GCN1 |
| ENSMUSG00000041670 | ENSFM00250000000629 | Regulating Synaptic Membrane Exocytosis Rab 3 Interacting Molecule Rim |
| ENSMUSG00000041685 | ENSFM00600000921280 | Fch Domain Only |
| ENSMUSG00000041688 | ENSFM00250000001614 | Angiomotin |
| ENSMUSG00000041697 | ENSFM00670001236141 | Cytochrome C Oxidase Subunit 6A1 Mitochondrial Precursor Cytochrome C Oxidase Polypeptide Via Liver |
| ENSMUSG00000041720 | ENSFM00570000876532 | Unknown |
| ENSMUSG00000041720 | ENSFM00760001714819 | Phosphatidylinositol 4 Kinase Alpha PI4 Kinase Alpha PI4K Alpha Ptdins 4 Kinase Alpha EC_2.7.1.67 |
| ENSMUSG00000041729 | ENSFM00730001521970 | Coronin |
| ENSMUSG00000041757 | ENSFM00800002003674 | Pleckstrin Homology Domain Containing Family A Member 6 Ph Domain Containing Family A Member 6 Phosphoinositol 3 Phosphate Binding 3 Pepp 3 |
| ENSMUSG00000041881 | ENSFM00640001119910 | Unknown |
| ENSMUSG00000041881 | ENSFM00500000274400 | Nadh Dehydrogenase 1 Alpha Subcomplex Subunit 7 Complex I B14 5A Ci B14 5A Nadh Ubiquinone Oxidoreductase Subunit B14 5A |
| ENSMUSG00000041912 | ENSFM00800002016924 | Unknown |
| ENSMUSG00000041912 | ENSFM00250000006342 | Tudor And Kh Domain Containing Tudor Domain Containing 2 |
| ENSMUSG00000041957 | ENSFM00730001522005 | Plakophilin 1 Band 6 B6P |
| ENSMUSG00000041958 | ENSFM00500000271389 | Gpi Transamidase Component Pig S Phosphatidylinositol Glycan Biosynthesis Class S |
| ENSMUSG00000041977 | ENSFM00600000921196 | Rho Guanine Nucleotide Exchange Factor |
| ENSMUSG00000041986 | ENSFM00250000003015 | Elmo Domain Containing |
| ENSMUSG00000042066 | ENSFM00250000001324 | Transmembrane And Coiled Coil Domains |
| ENSMUSG00000042096 | ENSFM00670001235511 | D Amino Acid Oxidase Daao Damox Dao EC_1.4.3.3 |
| ENSMUSG00000042215 | ENSFM00760001743808 | Unknown |
| ENSMUSG00000042215 | ENSFM00260000051255 | Bag Family Molecular Chaperone Regulator 2 Bag 2 Bcl 2 Associated Athanogene 2 |
| ENSMUSG00000042228 | ENSFM00730001521707 | Tyrosine Kinase EC_2.7.10.2 |
| ENSMUSG00000042272 | ENSFM00250000002761 | SEC14 Domain And Spectrin Repeat Containing 1 |
| ENSMUSG00000042275 | ENSFM00250000004827 | Pelota Homolog EC_3.1.-.- |
| ENSMUSG00000042331 | ENSFM00250000001872 | Cytospin A SPECC1 Sperm Antigen With Calponin Homology And Coiled Coil Domains 1 |
| ENSMUSG00000042388 | ENSFM00250000000718 | Disks Large Associated Dap Psd 95/SAP90 Binding SAP90/Psd 95 Associated |
| ENSMUSG00000042401 | ENSFM00250000002903 | Cartilage Acidic 1 Precursor 68 Kda Chondrocyte Expressed Cep 68 Aspic |
| ENSMUSG00000042425 | ENSFM00250000001492 | Ferm And Pdz Domain Containing 4 Pdz Domain Containing 10 Psd 95 Interacting Regulator Of Spine Morphogenesis Preso |
| ENSMUSG00000042447 | ENSFM00500000271280 | Wd Repeat Containing Mio |
| ENSMUSG00000042492 | ENSFM00260000050553 | TBC1 Domain Family Member |
| ENSMUSG00000042520 | ENSFM00800002020257 | Unknown |
| ENSMUSG00000042520 | ENSFM00800002020258 | Unknown |
| ENSMUSG00000042520 | ENSFM00600000921193 | Ubiquitin Associated 2 |
| ENSMUSG00000042532 | ENSFM00250000003047 | Golgin Subfamily A Member 7 |
| ENSMUSG00000042535 | ENSFM00250000001372 | Gtp Binding |
| ENSMUSG00000042569 | ENSFM00560000771026 | Dehydrogenase/Reductase Sdr Family Member EC_1.1.-.- |
| ENSMUSG00000042604 | ENSFM00290000065519 | Potassium Voltage Gated Channel Subfamily A Member Voltage Gated Potassium Channel Subunit KV1 |
| ENSMUSG00000042605 | ENSFM00590000917643 | Unknown |
| ENSMUSG00000042605 | ENSFM00590000917644 | Unknown |
| ENSMUSG00000042605 | ENSFM00800002017351 | Unknown |
| ENSMUSG00000042605 | ENSFM00600000921253 | Ataxin 2 Spinocerebellar Ataxia Type 2 |
| ENSMUSG00000042647 | ENSFM00500000333101 | Uncharacterized |
| ENSMUSG00000042647 | ENSFM00800002023650 | Unknown |
| ENSMUSG00000042647 | ENSFM00780001898384 | Acyl Coa Dehydrogenase Family Member Acad EC_1.3.99.- |
| ENSMUSG00000042662 | ENSFM00740001598893 | Unknown |
| ENSMUSG00000042662 | ENSFM00670001235708 | Dual Specificity Phosphatase EC_3.1.3.16 EC_3.1.3.- 48 |
| ENSMUSG00000042662 | ENSFM00670001254208 | DUSP15 |
| ENSMUSG00000042671 | ENSFM00670001235642 | Regulator Of G Signaling |
| ENSMUSG00000042682 | ENSFM00670001236822 | Selenoprotein K Selk |
| ENSMUSG00000042686 | ENSFM00250000001356 | Junctophilin Jp Junctophilin |
| ENSMUSG00000042699 | ENSFM00760001763154 | Unknown |
| ENSMUSG00000042699 | ENSFM00800002003717 | Atp Dependent Rna Helicase A EC_3.6.4.13 Deah Box 9 Nuclear Dna Helicase Ii Ndh Ii |
| ENSMUSG00000042700 | ENSFM00730001521477 | Signal Induced Proliferation Associated 1 SIPA1 |
| ENSMUSG00000042705 | ENSFM00250000008194 | Comm Domain Containing 10 |
| ENSMUSG00000042734 | ENSFM00670001235990 | Tetratricopeptide Repeat Tpr Repeat |
| ENSMUSG00000042744 | ENSFM00500000270499 | Probable E3 Ubiquitin Ligase HECTD4 EC_6.3.2.- Hect Domain Containing 4 |
| ENSMUSG00000042766 | ENSFM00570000879224 | Unknown |
| ENSMUSG00000042766 | ENSFM00400000131833 | Tripartite Motif Containing 46 Gene Y Geney Tripartite Fibronectin Type Iii And C Terminal Spry Motif |
| ENSMUSG00000042787 | ENSFM00500000272927 | Nuclease Exog Mitochondrial Precursor EC_3.1.30.- Endonuclease G 1 Endo G 1 |
| ENSMUSG00000042790 | ENSFM00590000917795 | Unknown |
| ENSMUSG00000042790 | ENSFM00590000917796 | Unknown |
| ENSMUSG00000042790 | ENSFM00250000005433 | Ring Finger 214 |
| ENSMUSG00000042807 | ENSFM00730001522025 | E3 Ubiquitin Ligase EC_6.3.2.- Hect C2 And Ww Domain Containing NEDD4 E3 Ubiquitin Ligase |
| ENSMUSG00000042846 | ENSFM00400000131776 | Leucine Rich Repeat Transmembrane Neuronal Precursor |
| ENSMUSG00000042870 | ENSFM00250000001136 | TOM1 2 Target Of Myb 2 |
| ENSMUSG00000042873 | ENSFM00250000002406 | Lipoma Hmgic Fusion Partner |
| ENSMUSG00000043004 | ENSFM00500000277611 | Unknown |
| ENSMUSG00000043079 | ENSFM00590000918103 | Unknown |
| ENSMUSG00000043079 | ENSFM00250000008348 | Synaptopodin |
| ENSMUSG00000043091 | ENSFM00250000000217 | Tubulin Alpha Chain |
| ENSMUSG00000043284 | ENSFM00250000007165 | Transmembrane 11 Mitochondrial |
| ENSMUSG00000043384 | ENSFM00600000921594 | G Coupled Receptor Associated Sorting Gasp |
| ENSMUSG00000043448 | ENSFM00730001521860 | Gap Junction Gamma 1 Connexin 45 CX45 Gap Junction Alpha 7 |
| ENSMUSG00000043460 | ENSFM00250000002984 | Phosphatase 1 Regulatory Subunit 29 Precursor Extracellular Leucine Rich Repeat And Fibronectin Type Iii Domain Containing 2 Leucine Rich Repeat And Fibronectin Type Iii Domain Containing 6 Leucine Rich Repeat Containing 62 |
| ENSMUSG00000043542 | ENSFM00500000272347 | Zinc Finger C2HC Domain Containing 1A |
| ENSMUSG00000043557 | ENSFM00250000002191 | Mam Domain Containing Glycosylphosphatidylinositol Anchor 2 Precursor Mam Domain Containing 1 |
| ENSMUSG00000043629 | ENSFM00500000286919 | Small Membrane A Kinase Anchor Small Membrane Akap Smakap |
| ENSMUSG00000043670 | ENSFM00500000270725 | Gtp Binding Di Precursor Distinct Subgroup Of The Ras Family Member |
| ENSMUSG00000043716 | ENSFM00250000000871 | 60S Ribosomal L7 |
| ENSMUSG00000043807 | ENSFM00250000009121 | Lymphocyte Antigen 6 Complex Locus G5B Precursor |
| ENSMUSG00000043811 | ENSFM00250000002242 | Reticulon 4 Receptor Precursor Nogo Receptor Nogo 66 Receptor Homolog Nogo 66 Receptor Related |
| ENSMUSG00000043991 | ENSFM00250000002171 | Transcriptional Activator Pur Beta Purine Rich Element Binding B |
| ENSMUSG00000044018 | ENSFM00250000009476 | 39S Ribosomal L50 Mitochondrial L50MT Mrp L50 |
| ENSMUSG00000044060 | ENSFM00690001356664 | Btb/Poz Domain Containing 8 |
| ENSMUSG00000044147 | ENSFM00750001632347 | Adp Ribosylation Factor |
| ENSMUSG00000044216 | ENSFM00730001521253 | Inward Rectifier Potassium Channel Inward Rectifier K + Channel KIR2 Irk Potassium Channel Inwardly Rectifying Subfamily J Member |
| ENSMUSG00000044252 | ENSFM00570000883987 | Unknown |
| ENSMUSG00000044252 | ENSFM00570000900012 | Unknown |
| ENSMUSG00000044252 | ENSFM00500000269986 | Oxysterol Binding Related 1 Orp 1 Osbp Related 1 |
| ENSMUSG00000044287 | ENSFM00600000921973 | Neuritin Precursor |
| ENSMUSG00000044352 | ENSFM00500000274456 | Ankyrin Repeat Domain Containing Sowaha Precursor Ankyrin Repeat Domain Containing 43 Sosondowah Homolog A |
| ENSMUSG00000044424 | ENSFM00250000002224 | 40S Ribosomal S7 |
| ENSMUSG00000044433 | ENSFM00600000921199 | Calmodulin Regulated Spectrin Associated |
| ENSMUSG00000044447 | ENSFM00760001714647 | Dedicator Of Cytokinesis |
| ENSMUSG00000044667 | ENSFM00500000270520 | Lipid Phosphate Phosphatase Related Type 4 EC_3.1.3.4 Brain Specific Phosphatidic Acid Phosphatase 1 Plasticity Related Gene 1 Prg 1 |
| ENSMUSG00000044847 | ENSFM00250000007949 | U7 Snrna Associated Sm LSM11 |
| ENSMUSG00000044894 | ENSFM00500000446966 | Unknown |
| ENSMUSG00000044894 | ENSFM00500000290955 | Cytochrome B C1 Complex Subunit 8 Complex Iii Subunit 8 Complex Iii Subunit Viii Ubiquinol Cytochrome C Reductase Complex 9.5 Kda Ubiquinol Cytochrome C Reductase Complex Ubiquinone Binding Qp C |
| ENSMUSG00000045007 | ENSFM00420000140599 | Tubulin Gamma Chain Gamma Tubulin |
| ENSMUSG00000045045 | ENSFM00260000050443 | Leucine Rich Repeat And Fibronectin Type Iii Domain Containing Precursor Synaptic Adhesion Molecule |
| ENSMUSG00000045092 | ENSFM00730001521347 | Sphingosine 1 Phosphate Receptor S1P Receptor Endothelial Differentiation G Coupled Receptor Sphingosine 1 Phosphate Receptor Edg S1P Receptor Edg |
| ENSMUSG00000045095 | ENSFM00250000000552 | Membrane Associated Guanylate Kinase Ww And Pdz Domain Containing Membrane Associated Guanylate Kinase Inverted Magi |
| ENSMUSG00000045128 | ENSFM00760001714679 | 60S Ribosomal L18A |
| ENSMUSG00000045136 | ENSFM00250000000193 | Tubulin Beta Chain |
| ENSMUSG00000045193 | ENSFM00600000921577 | Cold Inducible Rna Binding Glycine Rich Rna Binding Cirp |
| ENSMUSG00000045211 | ENSFM00260000051397 | 8 Oxo Dgdp Phosphatase NUDT18 EC_3.6.1.58 2 Hydroxy Dadp Phosphatase 7 8 Dihydro 8 Oxoguanine Phosphatase Mutt Homolog 3 Nucleoside Diphosphate Linked Moiety X Motif 18 Nudix Motif 18 |
| ENSMUSG00000045302 | ENSFM00810002055838 | Unknown |
| ENSMUSG00000045302 | ENSFM00250000005591 | Prolactin Regulatory Element Binding Mammalian Guanine Nucleotide Exchange Factor MSEC12 |
| ENSMUSG00000045316 | ENSFM00670001235551 | Acylpyruvase FAHD1 Mitochondrial Precursor EC_3.7.1.5 Fumarylacetoacetate Hydrolase Domain Containing 1 |
| ENSMUSG00000045348 | ENSFM00250000007087 | Neuronal Tyrosine Phosphorylated Phosphoinositide 3 Kinase Adapter 1 |
| ENSMUSG00000045411 | ENSFM00570000879994 | Unknown |
| ENSMUSG00000045414 | ENSFM00250000009380 | Deleted In Autism 1 Precursor Golgi Of 49 Kda GOPRO49 Hypoxia And Akt Induced Stem Cell Factor Hasf |
| ENSMUSG00000045441 | ENSFM00250000008609 | G Regulated Inducer Of Neurite Outgrowth 3 GRIN3 |
| ENSMUSG00000045455 | ENSFM00670001236601 | Mitochondrial Import Inner Membrane Translocase Subunit TIM8 A Deafness Dystonia 1 |
| ENSMUSG00000045589 | ENSFM00570000876542 | Unknown |
| ENSMUSG00000045589 | ENSFM00250000008517 | Domon Domain Containing FRRS1L Ferric Chelate Reductase 1 |
| ENSMUSG00000045659 | ENSFM00800002003882 | Pleckstrin Homology Domain Containing Family A Member 7 Ph Domain Containing Family A Member 7 Heart Adapter 1 |
| ENSMUSG00000045671 | ENSFM00250000001487 | Sprouty Related EVH1 Domain Containing Spred |
| ENSMUSG00000045731 | ENSFM00670001235427 | Prepronociceptin Neuropeptide 1 |
| ENSMUSG00000045763 | ENSFM00800002007336 | Brain Acid Soluble 1.22 Kda Neuronal Tissue Enriched Acidic Neuronal Axonal Membrane Nap 22 |
| ENSMUSG00000045912 | ENSFM00500000273557 | C2 Calcium Dependent Domain Containing 4C Nuclear Localized Factor 3 FAM148C |
| ENSMUSG00000045967 | ENSFM00250000001603 | Probable G Coupled Receptor Precursor |
| ENSMUSG00000045983 | ENSFM00570000887850 | Unknown |
| ENSMUSG00000045983 | ENSFM00250000000634 | Eukaryotic Translation Initiation Factor 4 Gamma 3 Eif 4 Gamma 3 Eif 4G 3 EIF4G 3 Eif 4 Gamma Ii EIF4GII |
| ENSMUSG00000046079 | ENSFM00570000894492 | Unknown |
| ENSMUSG00000046079 | ENSFM00570000896228 | Unknown |
| ENSMUSG00000046079 | ENSFM00250000001418 | Volume Regulated Anion Channel Subunit Leucine Rich Repeat Containing |
| ENSMUSG00000046093 | ENSFM00570000890067 | Unknown |
| ENSMUSG00000046093 | ENSFM00760001714608 | Unknown |
| ENSMUSG00000046159 | ENSFM00800002003412 | Muscarinic Acetylcholine Receptor |
| ENSMUSG00000046182 | ENSFM00670001235575 | Germ Cell Specific Gene 1 GSG1 |
| ENSMUSG00000046240 | ENSFM00250000007963 | Hepatocyte Cell Adhesion Molecule Precursor Hepacam |
| ENSMUSG00000046312 | ENSFM00570000883301 | Unknown |
| ENSMUSG00000046312 | ENSFM00540000717964 | Uncharacterized Family 31 Glucosidase EC_3.2.1.- |
| ENSMUSG00000046314 | ENSFM00570000892739 | Unknown |
| ENSMUSG00000046314 | ENSFM00500000272618 | Syntaxin Binding 6 |
| ENSMUSG00000046330 | ENSFM00670001236020 | 60S Ribosomal L37A |
| ENSMUSG00000046364 | ENSFM00640001118765 | Unknown |
| ENSMUSG00000046364 | ENSFM00780001898350 | 60S Ribosomal L27A |
| ENSMUSG00000046417 | ENSFM00250000005430 | Leucine Rich Repeat Containing Leucine Rich Repeat Containing |
| ENSMUSG00000046434 | ENSFM00270000056431 | Heterogeneous Nuclear Ribonucleoprotein |
| ENSMUSG00000046671 | ENSFM00250000006972 | Mitochondrial Fission Regulator 1 |
| ENSMUSG00000046691 | ENSFM00500000283580 | Chromosome Transmission Fidelity 8 Homolog |
| ENSMUSG00000046691 | ENSFM00250000008429 | Chromosome Transmission Fidelity 8 Homolog 2 |
| ENSMUSG00000046707 | ENSFM00250000001591 | Casein Kinase Ii Subunit Ck Ii EC_2.7.11.1 |
| ENSMUSG00000046709 | ENSFM00460000244930 | Mitogen Activated Kinase Map Kinase Mapk EC_2.7.11.24 Stress Activated Kinase C Jun N Terminal Kinase |
| ENSMUSG00000046798 | ENSFM00570000889082 | Unknown |
| ENSMUSG00000046798 | ENSFM00400000131954 | Claudin 12 |
| ENSMUSG00000046841 | ENSFM00250000007369 | Cytoskeleton Associated 4.63 Kda Cytoskeleton Linking Membrane Climp 63 P63 |
| ENSMUSG00000046879 | ENSFM00250000001767 | Interferon Inducible Gtpase 5 EC_3.6.5.- Immunity Related Gtpase Cinema 1 |
| ENSMUSG00000047013 | ENSFM00250000006718 | F Box Only 41 |
| ENSMUSG00000047037 | ENSFM00730001521603 | Magnesium Transporter Non Imprinted In Prader Willi/Angelman Syndrome Region |
| ENSMUSG00000047215 | ENSFM00800002021381 | Unknown |
| ENSMUSG00000047215 | ENSFM00750001632341 | 60S Ribosomal L9 |
| ENSMUSG00000047261 | ENSFM00250000005389 | Neuromodulin Axonal Membrane Gap 43 Growth Associated 43 |
| ENSMUSG00000047414 | ENSFM00250000003081 | Leucine Rich Repeat Transmembrane Precursor Fibronectin Domain Containing Leucine Rich Transmembrane |
| ENSMUSG00000047454 | ENSFM00790001963049 | Molybdopterin Adenylyltransferase Mpt Adenylyltransferase EC_2.7.7.75 Domain G |
| ENSMUSG00000047495 | ENSFM00250000000718 | Disks Large Associated Dap Psd 95/SAP90 Binding SAP90/Psd 95 Associated |
| ENSMUSG00000047507 | ENSFM00720001499239 | Unknown |
| ENSMUSG00000047507 | ENSFM00250000001781 | Unc 13 Homolog D MUNC13 4 |
| ENSMUSG00000047547 | ENSFM00570000887013 | Unknown |
| ENSMUSG00000047547 | ENSFM00250000001836 | Clathrin Light Chain |
| ENSMUSG00000047675 | ENSFM00800002003452 | 40S Ribosomal S8 |
| ENSMUSG00000047842 | ENSFM00500000270725 | Gtp Binding Di Precursor Distinct Subgroup Of The Ras Family Member |
| ENSMUSG00000047866 | ENSFM00730001521929 | Lon Protease Homolog 2 Peroxisomal |
| ENSMUSG00000047921 | ENSFM00250000002395 | Trafficking Particle Complex Subunit 9 Nik And Ikbkb Binding |
| ENSMUSG00000047959 | ENSFM00290000065519 | Potassium Voltage Gated Channel Subfamily A Member Voltage Gated Potassium Channel Subunit KV1 |
| ENSMUSG00000047976 | ENSFM00290000065519 | Potassium Voltage Gated Channel Subfamily A Member Voltage Gated Potassium Channel Subunit KV1 |
| ENSMUSG00000047986 | ENSFM00250000010790 | Paralemmin 3 Precursor |
| ENSMUSG00000048000 | ENSFM00640001110751 | Unknown |
| ENSMUSG00000048000 | ENSFM00640001118145 | Unknown |
| ENSMUSG00000048000 | ENSFM00740001589295 | Perq Amino Acid Rich With Gyf Domain Containing 2 Trinucleotide Repeat Containing Gene 15 |
| ENSMUSG00000048076 | ENSFM00730001521124 | Adp Ribosylation Factor |
| ENSMUSG00000048279 | ENSFM00740001589315 | Sacsin Dnaj Homolog Subfamily C Member 29 DNAJC29 |
| ENSMUSG00000048304 | ENSFM00250000001398 | Slit And Ntrk Precursor |
| ENSMUSG00000048388 | ENSFM00740001589308 | Precursor |
| ENSMUSG00000048537 | ENSFM00570000895969 | Unknown |
| ENSMUSG00000048537 | ENSFM00600000921202 | Pleckstrin Homology Domain Family B Member LL5 |
| ENSMUSG00000048578 | ENSFM00250000005592 | Malectin Precursor |
| ENSMUSG00000048707 | ENSFM00250000006760 | Taperin |
| ENSMUSG00000048758 | ENSFM00600000921342 | 60S Ribosomal L29 |
| ENSMUSG00000048832 | ENSFM00500000274002 | Vacuolar Sorting Associated 37C Escrt I Complex Subunit VPS37C |
| ENSMUSG00000048988 | ENSFM00250000002984 | Phosphatase 1 Regulatory Subunit 29 Precursor Extracellular Leucine Rich Repeat And Fibronectin Type Iii Domain Containing 2 Leucine Rich Repeat And Fibronectin Type Iii Domain Containing 6 Leucine Rich Repeat Containing 62 |
| ENSMUSG00000049044 | ENSFM00250000000899 | Rap Guanine Nucleotide Exchange Factor Exchange Factor Directly Activated By Camp Exchange Directly Activated By Camp Epac Camp Regulated Guanine Nucleotide Exchange Factor Camp |
| ENSMUSG00000049076 | ENSFM00250000000926 | Arf Gap With Coiled Coil Ank Repeat And Ph Domain Containing Centaurin Beta Cnt |
| ENSMUSG00000049100 | ENSFM00780001898335 | Protocadherin Precursor |
| ENSMUSG00000049119 | ENSFM00500000272914 | Unknown |
| ENSMUSG00000049184 | ENSFM00250000002171 | Transcriptional Activator Pur Beta Purine Rich Element Binding B |
| ENSMUSG00000049230 | ENSFM00740001589263 | Myelin Expression Factor 2 Mef 2 Myef 2 |
| ENSMUSG00000049336 | ENSFM00250000000375 | Teneurin Ten Odd Oz/Ten M Homolog Tenascin Ten Teneurin Transmembrane |
| ENSMUSG00000049354 | ENSFM00500000271100 | DDB1 And CUL4 Associated Factor 7 Wd Repeat Containing 68 Wd Repeat Containing AN11 Homolog |
| ENSMUSG00000049517 | ENSFM00250000002784 | 40S Ribosomal S23 |
| ENSMUSG00000049521 | ENSFM00250000009952 | CDC42 Effector 1 Binder Of Rho Gtpases 5 |
| ENSMUSG00000049550 | ENSFM00250000000764 | Cap Gly Domain Containing Linker 2 Cytoplasmic Linker 115 Clip 115 Cytoplasmic Linker 2 |
| ENSMUSG00000049583 | ENSFM00730001521627 | Metabotropic Glutamate Receptor Precursor |
| ENSMUSG00000049612 | ENSFM00250000008621 | Oligodendrocyte Myelin Glycoprotein Precursor |
| ENSMUSG00000049760 | ENSFM00250000011790 | QIL1 |
| ENSMUSG00000049775 | ENSFM00500000306692 | Unknown |
| ENSMUSG00000049775 | ENSFM00550000746767 | Unknown |
| ENSMUSG00000049792 | ENSFM00590000917510 | Unknown |
| ENSMUSG00000049792 | ENSFM00590000917511 | Unknown |
| ENSMUSG00000049792 | ENSFM00250000007925 | Bag Family Molecular Chaperone Regulator 5 Bag 5 Bcl 2 Associated Athanogene 5 |
| ENSMUSG00000049804 | ENSFM00810002041142 | Armadillo Repeat Containing X Linked 4 |
| ENSMUSG00000049807 | ENSFM00250000001623 | Rho Gtpase Activating 21 Rho Type Gtpase Activating 21 |
| ENSMUSG00000050079 | ENSFM00250000004877 | Ring Finger And Spry Domain Containing 1 Precursor |
| ENSMUSG00000050132 | ENSFM00250000003414 | Sterile Alpha And Tir Motif Containing Tir 1 Homolog |
| ENSMUSG00000050211 | ENSFM00540000717923 | Cytosolic Phospholipase A2 CPLA2 EC_3.1.1.4 Phospholipase A2 Group |
| ENSMUSG00000050321 | ENSFM00250000001916 | Neuropilin And Tolloid Precursor Brain Specific Transmembrane Containing 2 Cub And 1 Ldl Receptor Class A Domains |
| ENSMUSG00000050323 | ENSFM00500000273764 | Nadh Dehydrogenase Ubiquinone Complex I Assembly Factor 6 Precursor |
| ENSMUSG00000050357 | ENSFM00260000050454 | Leucine Rich Repeat Containing |
| ENSMUSG00000050379 | ENSFM00350000105408 | Septin |
| ENSMUSG00000050390 | ENSFM00250000008611 | Uncharacterized |
| ENSMUSG00000050390 | ENSFM00570000882097 | Unknown |
| ENSMUSG00000050390 | ENSFM00570000887662 | Unknown |
| ENSMUSG00000050490 | ENSFM00500000270276 | Proteasome Subunit Alpha Type 5 EC_3.4.25.1 Macropain Multicatalytic Endopeptidase Complex Proteasome |
| ENSMUSG00000050530 | ENSFM00740001589308 | Precursor |
| ENSMUSG00000050556 | ENSFM00270000056440 | Potassium Voltage Gated Channel Subfamily Member Voltage Gated Potassium Channel Subunit |
| ENSMUSG00000050587 | ENSFM00610000973611 | Unknown |
| ENSMUSG00000050587 | ENSFM00260000050662 | Leucine Rich Repeat Containing Precursor Netrin Ligand Ngl |
| ENSMUSG00000050621 | ENSFM00670001235597 | 40S Ribosomal S27 |
| ENSMUSG00000050663 | ENSFM00250000000130 | Aminopeptidase N Ap N EC_3.4.11.2 Microsomal Aminopeptidase |
| ENSMUSG00000050711 | ENSFM00250000004240 | Secretogranin 2 Precursor Secretogranin Ii Sgii |
| ENSMUSG00000050840 | ENSFM00260000050333 | Cadherin Precursor |
| ENSMUSG00000050856 | ENSFM00500000427096 | Unknown |
| ENSMUSG00000050856 | ENSFM00520000547368 | Unknown |
| ENSMUSG00000050890 | ENSFM00250000003729 | Serine/Threonine Kinase EC_2.7.11.1 PDLIM1 Interacting Kinase 1 |
| ENSMUSG00000050896 | ENSFM00250000002242 | Reticulon 4 Receptor Precursor Nogo Receptor Nogo 66 Receptor Homolog Nogo 66 Receptor Related |
| ENSMUSG00000050953 | ENSFM00760001714761 | Gap Junction Alpha 1 Connexin 43 CX43 |
| ENSMUSG00000050965 | ENSFM00520000517802 | Kinase C Type Pkc EC_2.7.11.13 |
| ENSMUSG00000051067 | ENSFM00250000001884 | Leucine Rich Repeat And Immunoglobulin Domain Containing Nogo Receptor Interacting Precursor Leucine Rich Repeat Neuronal |
| ENSMUSG00000051154 | ENSFM00250000008895 | Comm Domain Containing 3 |
| ENSMUSG00000051242 | ENSFM00730001521419 | Protocadherin Beta Precursor Pcdh Beta |
| ENSMUSG00000051343 | ENSFM00500000272387 | RAB11 Family Interacting 5 RAB11 FIP5 RAB11 Interacting RIP11 |
| ENSMUSG00000051355 | ENSFM00590000916064 | Unknown |
| ENSMUSG00000051355 | ENSFM00250000007033 | Comm Domain Containing 1 |
| ENSMUSG00000051379 | ENSFM00250000003081 | Leucine Rich Repeat Transmembrane Precursor Fibronectin Domain Containing Leucine Rich Transmembrane |
| ENSMUSG00000051391 | ENSFM00730001521154 | Unknown |
| ENSMUSG00000051401 | ENSFM00260000050545 | Btb/Poz Domain Containing |
| ENSMUSG00000051412 | ENSFM00250000005040 | Vesicle Associated Membrane 7 Vamp 7 Synaptobrevin 1 |
| ENSMUSG00000051483 | ENSFM00250000002301 | Carbonyl Reductase EC_1.1.1.- 197 20 Beta Hydroxysteroid Dehydrogenase Nadph Dependent Carbonyl Reductase 1 Prostaglandin 9 Ketoreductase Prostaglandin E 2 9 Reductase EC_1.1.-.- 1 189 |
| ENSMUSG00000051695 | ENSFM00250000000747 | Poly Rc Binding Alpha Heterogeneous Nuclear Ribonucleoprotein Hnrnp |
| ENSMUSG00000051747 | ENSFM00670001267275 | Uncharacterized |
| ENSMUSG00000051747 | ENSFM00570000894573 | Unknown |
| ENSMUSG00000051747 | ENSFM00740001589077 | Titin EC_2.7.11.1 Connectin |
| ENSMUSG00000051790 | ENSFM00380000124679 | Neuroligin Precursor |
| ENSMUSG00000051839 | ENSFM00260000053088 | Glycophorin A CD235A Antigen |
| ENSMUSG00000051934 | ENSFM00520000517852 | Spermatogenesis Associated Serine Rich 2 Serine Rich Spermatocytes And Round Spermatid 59 Kda P59SCR |
| ENSMUSG00000051951 | ENSFM00500000269993 | Xk Related |
| ENSMUSG00000052146 | ENSFM00670001235534 | 40S Ribosomal S10 |
| ENSMUSG00000052214 | ENSFM00600000921688 | Optic Atrophy 3 |
| ENSMUSG00000052298 | ENSFM00570000885284 | Unknown |
| ENSMUSG00000052298 | ENSFM00670001236658 | CDC42 Small Effector 2 |
| ENSMUSG00000052337 | ENSFM00250000002778 | Micos Complex Subunit MIC60 Precursor Mitochondrial Inner Membrane Mitofilin |
| ENSMUSG00000052372 | ENSFM00500000270404 | Interleukin 1 Receptor Accessory 1 Precursor Il 1 Rapl 1 Il 1RAPL 1 IL1RAPL 1 X Linked Interleukin 1 Receptor Accessory 1 |
| ENSMUSG00000052373 | ENSFM00760001714665 | Maguk P55 Subfamily Member |
| ENSMUSG00000052374 | ENSFM00440000236866 | Alpha Actinin Alpha Actinin F Actin Cross Linking |
| ENSMUSG00000052428 | ENSFM00780001909040 | Unknown |
| ENSMUSG00000052428 | ENSFM00250000005494 | Transmembrane And Coiled Coil 1 |
| ENSMUSG00000052429 | ENSFM00570000898596 | Unknown |
| ENSMUSG00000052429 | ENSFM00730001521302 | Arginine N Methyltransferase 1 EC_2.1.1.- Histone Arginine N Methyltransferase PRMT1 EC_2.1.1.125 |
| ENSMUSG00000052459 | ENSFM00570000885853 | Unknown |
| ENSMUSG00000052459 | ENSFM00250000002169 | V Type Proton Atpase Catalytic Subunit A V Atpase Subunit A EC_3.6.3.14 V Atpase 69 Kda Subunit Vacuolar Proton Pump Subunit Alpha |
| ENSMUSG00000052516 | ENSFM00740001589121 | Roundabout Homolog Precursor |
| ENSMUSG00000052539 | ENSFM00250000000552 | Membrane Associated Guanylate Kinase Ww And Pdz Domain Containing Membrane Associated Guanylate Kinase Inverted Magi |
| ENSMUSG00000052572 | ENSFM00250000000275 | Disks Large Homolog Synapse Associated Sap |
| ENSMUSG00000052581 | ENSFM00570000896064 | Unknown |
| ENSMUSG00000052581 | ENSFM00400000131776 | Leucine Rich Repeat Transmembrane Neuronal Precursor |
| ENSMUSG00000052681 | ENSFM00730001521249 | Ras Related Rap 1 |
| ENSMUSG00000052727 | ENSFM00250000000942 | Microtubule Associated Map |
| ENSMUSG00000052738 | ENSFM00640001118348 | Unknown |
| ENSMUSG00000052738 | ENSFM00250000003768 | Succinyl Coa Ligase Subunit Alpha Mitochondrial Precursor EC_6.2.1.4 EC_6.2.1.- 5 Succinyl Coa Synthetase Subunit Alpha Scs Alpha |
| ENSMUSG00000052889 | ENSFM00520000517802 | Kinase C Type Pkc EC_2.7.11.13 |
| ENSMUSG00000052981 | ENSFM00760001715179 | Ubiquitin Conjugating Enzyme E2Q EC_6.3.2.19 |
| ENSMUSG00000053024 | ENSFM00760001763137 | Unknown |
| ENSMUSG00000053024 | ENSFM00760001763138 | Unknown |
| ENSMUSG00000053024 | ENSFM00800002003683 | Contactin 1 Precursor Neural Cell Surface F3 |
| ENSMUSG00000053046 | ENSFM00640001118862 | Unknown |
| ENSMUSG00000053046 | ENSFM00740001589247 | Serine/Threonine Kinase BRSK2 EC_2.7.11.1 EC_2.7.11.- 26 Brain Specific Serine/Threonine Kinase 2 Br Serine/Threonine Kinase 2 Serine/Threonine Kinase Sad A |
| ENSMUSG00000053119 | ENSFM00500000271306 | Charged Multivesicular Body 3 Chromatin Modifying 3 Vacuolar Sorting Associated 24 |
| ENSMUSG00000053141 | ENSFM00310000088999 | Receptor Type Tyrosine Phosphatase Precursor R Ptp EC_3.1.3.48 Receptor Type Tyrosine Phosphatase |
| ENSMUSG00000053289 | ENSFM00730001521664 | Atp Dependent Rna Helicase EC_3.6.4.13 |
| ENSMUSG00000053317 | ENSFM00570000900708 | Unknown |
| ENSMUSG00000053317 | ENSFM00670001236538 | Transport SEC61 Subunit Beta |
| ENSMUSG00000053329 | ENSFM00250000005161 | ES1 Mitochondrial Precursor |
| ENSMUSG00000053395 | ENSFM00250000002019 | Voltage Dependent Calcium Channel Gamma Subunit Neuronal Voltage Gated Calcium Channel Gamma Subunit Transmembrane Ampar Regulatory Gamma Tarp Gamma |
| ENSMUSG00000053519 | ENSFM00410000138464 | Kv Channel Interacting A Type Potassium Channel Modulatory Potassium Channel Interacting |
| ENSMUSG00000053550 | ENSFM00500000276971 | Shisa 7 Precursor Shisa 6 |
| ENSMUSG00000053580 | ENSFM00610000973561 | Unknown |
| ENSMUSG00000053580 | ENSFM00670001247752 | TANC2 |
| ENSMUSG00000053580 | ENSFM00250000001143 | Tetratricopeptide Repeat Ankyrin Repeat And Coiled Coil Domain Containing |
| ENSMUSG00000053617 | ENSFM00740001589343 | SH3 And Px Domain Containing 2A Five SH3 Domain Containing SH3 Multiple Domains 1 Tyrosine Kinase Substrate With Five SH3 Domains |
| ENSMUSG00000053693 | ENSFM00250000000518 | Microtubule Associated Serine/Threonine Kinase EC_2.7.11.1 |
| ENSMUSG00000053702 | ENSFM00500000270806 | Lim And SH3 Domain 1 Lasp 1 |
| ENSMUSG00000053702 | ENSFM00740001589165 | Nebulin Related Anchoring N Rap |
| ENSMUSG00000053768 | ENSFM00500000272284 | Micos Complex Subunit MIC19 Coiled Coil Helix Coiled Coil Helix Domain Containing 3 |
| ENSMUSG00000053799 | ENSFM00250000001326 | Exocyst Complex Component 6 Exocyst Complex Component SEC15A SEC15 1 |
| ENSMUSG00000053819 | ENSFM00250000000111 | Calcium/Calmodulin Dependent Kinase Type Ii Subunit Cam Kinase Ii Subunit Camk Ii Subunit EC_2.7.11.17 |
| ENSMUSG00000053825 | ENSFM00250000000441 | Liprin Alpha Tyrosine Phosphatase Receptor Type F Polypeptide Interacting Alpha Ptprf Interacting Alpha |
| ENSMUSG00000053930 | ENSFM00500000272417 | Shisa 6 Homolog Precursor |
| ENSMUSG00000053931 | ENSFM00800002024498 | Unknown |
| ENSMUSG00000053931 | ENSFM00800002024499 | Unknown |
| ENSMUSG00000053931 | ENSFM00500000269939 | Calponin Calponin Smooth Muscle |
| ENSMUSG00000053963 | ENSFM00500000275444 | Stum Homolog |
| ENSMUSG00000054099 | ENSFM00800002023532 | Unknown |
| ENSMUSG00000054099 | ENSFM00250000002826 | Solute Carrier Family 25 Member |
| ENSMUSG00000054277 | ENSFM00250000002206 | Adp Ribosylation Factor Gtpase Activating 2 Arf Gap 2 Zinc Finger 289 |
| ENSMUSG00000054312 | ENSFM00570000884963 | Unknown |
| ENSMUSG00000054312 | ENSFM00670001236631 | 28S Ribosomal S21 Mitochondrial Mrp S21 S21MT |
| ENSMUSG00000054453 | ENSFM00730001522102 | Synaptotagmin 4 Exophilin 2 Granuphilin |
| ENSMUSG00000054455 | ENSFM00250000001879 | Vesicle Associated Membrane Associated A Vamp A Vamp Associated A Vap A |
| ENSMUSG00000054459 | ENSFM00760001714608 | Unknown |
| ENSMUSG00000054477 | ENSFM00250000000796 | Small Conductance Calcium Activated Potassium Channel Skca KCA2 |
| ENSMUSG00000054720 | ENSFM00570000878992 | Unknown |
| ENSMUSG00000054720 | ENSFM00250000001418 | Volume Regulated Anion Channel Subunit Leucine Rich Repeat Containing |
| ENSMUSG00000054728 | ENSFM00760001714738 | Phosphatase And Actin Regulator |
| ENSMUSG00000054808 | ENSFM00440000236866 | Alpha Actinin Alpha Actinin F Actin Cross Linking |
| ENSMUSG00000054814 | ENSFM00810002057450 | Unknown |
| ENSMUSG00000054814 | ENSFM00760001714668 | Ubiquitin Carboxyl Terminal Hydrolase 12 EC_3.4.19.12 Deubiquitinating Enzyme 12 Ubiquitin Thioesterase 12 Ubiquitin Specific Processing Protease 12 |
| ENSMUSG00000054874 | ENSFM00250000001163 | Pecanex |
| ENSMUSG00000054901 | ENSFM00250000009469 | Rho Guanine Nucleotide Exchange Factor 33 |
| ENSMUSG00000054942 | ENSFM00250000002334 | FAM73B |
| ENSMUSG00000054976 | ENSFM00250000005153 | Neuronal Tyrosine Phosphorylated Phosphoinositide 3 Kinase Adapter 2 |
| ENSMUSG00000055003 | ENSFM00570000880212 | Unknown |
| ENSMUSG00000055003 | ENSFM00670001235765 | Leucine Rich Repeat And Transmembrane Domain Containing Precursor |
| ENSMUSG00000055013 | ENSFM00250000000604 | Arf Gap With Gtpase Ank Repeat And Ph Domain Containing Agap Centaurin Gamma |
| ENSMUSG00000055022 | ENSFM00800002003683 | Contactin 1 Precursor Neural Cell Surface F3 |
| ENSMUSG00000055065 | ENSFM00420000140507 | Probable Atp Dependent Rna Helicase DDX5 EC_3.6.4.13 Dead Box 5 |
| ENSMUSG00000055194 | ENSFM00730001521060 | Actin |
| ENSMUSG00000055333 | ENSFM00570000851039 | Protocadherin Fat Precursor Fat Tumor Suppressor Homolog |
| ENSMUSG00000055407 | ENSFM00250000004498 | Microtubule Associated 6 Map 6 Stable Tubule Only Polypeptide Stop |
| ENSMUSG00000055447 | ENSFM00250000005008 | Leukocyte Surface Antigen CD47 Precursor Integrin Associated Iap CD47 Antigen |
| ENSMUSG00000055531 | ENSFM00660001169823 | Unknown |
| ENSMUSG00000055531 | ENSFM00660001189768 | Unknown |
| ENSMUSG00000055531 | ENSFM00600000921459 | Cleavage And Polyadenylation Specificity Factor Subunit 6 |
| ENSMUSG00000055567 | ENSFM00470000251461 | Unc 80 |
| ENSMUSG00000055723 | ENSFM00770001847543 | Ras Related R Ras |
| ENSMUSG00000055762 | ENSFM00730001521722 | Elongation Factor 1 Delta Ef 1 Delta |
| ENSMUSG00000055782 | ENSFM00800002003396 | Atp Binding Cassette Sub Family D Member Adrenoleukodystrophy |
| ENSMUSG00000055805 | ENSFM00350000105407 | Formin |
| ENSMUSG00000055839 | ENSFM00500000273664 | Transcription Elongation Factor B Polypeptide 2 Elongin 18 Kda Subunit Elongin B Elob Rna Polymerase Ii Transcription Factor Siii Subunit B Siii P18 |
| ENSMUSG00000055943 | ENSFM00500000273154 | Er Membrane Complex Subunit 7 Precursor |
| ENSMUSG00000055963 | ENSFM00810002042668 | Triple Qxxk/R Motif Containing Triple Repetitive Sequence Of Qxxk/R Homolog |
| ENSMUSG00000056073 | ENSFM00720001492013 | Glutamate Receptor Ionotropic Kainate Precursor Glutamate Receptor |
| ENSMUSG00000056158 | ENSFM00500000308824 | Carbonic Anhydrase Related 10 |
| ENSMUSG00000056158 | ENSFM00400000131811 | Carbonic Anhydrase Related 11 Precursor Ca Rp Xi Ca Xi Carp Xi |
| ENSMUSG00000056201 | ENSFM00500000270025 | Cofilin Cofilin Muscle |
| ENSMUSG00000056209 | ENSFM00500000274611 | Nucleoplasmin 3 |
| ENSMUSG00000056367 | ENSFM00800002003421 | Actin Related 3 Actin 3 |
| ENSMUSG00000056413 | ENSFM00250000002436 | Arf Gap With Dual Ph Domain Containing 2 Centaurin Alpha 2 Cnt A2 |
| ENSMUSG00000056553 | ENSFM00260000050522 | Receptor Type Tyrosine Phosphatase Precursor R Ptp |
| ENSMUSG00000056602 | ENSFM00250000000754 | Furry |
| ENSMUSG00000056665 | ENSFM00560000771357 | THEM6 Precursor |
| ENSMUSG00000056755 | ENSFM00640001118416 | Unknown |
| ENSMUSG00000056755 | ENSFM00640001118417 | Unknown |
| ENSMUSG00000056755 | ENSFM00250000000328 | Metabotropic Glutamate Receptor Precursor |
| ENSMUSG00000056812 | ENSFM00800002003921 | Sia Alpha 2 3 Gal Beta 1 4 Glcnac R:Alpha 2 8 Sialyltransferase EC_2.4.99.- Alpha 2 8 Sialyltransferase 8C Alpha 2 8 Sialyltransferase Iii ST8 Alpha N Acetyl Neuraminide Alpha 2 8 Sialyltransferase 3 Sialyltransferase 8C SIAT8 C Sialyltransferase ST8SIA I |
| ENSMUSG00000056851 | ENSFM00250000000747 | Poly Rc Binding Alpha Heterogeneous Nuclear Ribonucleoprotein Hnrnp |
| ENSMUSG00000056856 | ENSFM00610000971715 | Unknown |
| ENSMUSG00000056856 | ENSFM00250000001480 | Janus Kinase And Microtubule Interacting 1 Multiple Alpha Helices And Rna Linker 1 Marlin 1 |
| ENSMUSG00000057113 | ENSFM00500000270251 | Nucleophosmin Npm Nucleolar Phosphoprotein B23 Nucleolar NO38 Numatrin |
| ENSMUSG00000057230 | ENSFM00730001521956 | AP2 Associated Kinase 1 EC_2.7.11.1 Adaptor Associated Kinase 1 |
| ENSMUSG00000057262 | ENSFM00250000001504 | 60S Ribosomal L15 |
| ENSMUSG00000057322 | ENSFM00500000287342 | Unknown |
| ENSMUSG00000057335 | ENSFM00740001589275 | Unknown |
| ENSMUSG00000057378 | ENSFM00250000000332 | Ryanodine Receptor Ryr Muscle Calcium Release Channel Muscle Type Ryanodine Receptor Type Ryanodine Receptor |
| ENSMUSG00000057614 | ENSFM00410000138458 | Guanine Nucleotide Binding Subunit |
| ENSMUSG00000057666 | ENSFM00720001501961 | Unknown |
| ENSMUSG00000057666 | ENSFM00720001503163 | Unknown |
| ENSMUSG00000057666 | ENSFM00730001521081 | Glyceraldehyde 3 Phosphate Dehydrogenase Gapdh EC_1.2.1.12 Peptidyl Cysteine S Nitrosylase Gapdh EC_2.6.99.- |
| ENSMUSG00000057716 | ENSFM00800002003927 | Transmembrane 178B Precursor |
| ENSMUSG00000057738 | ENSFM00740001589109 | Spectrin Alpha Chain Non Erythrocytic 1 Alpha Ii Spectrin Fodrin Alpha Chain |
| ENSMUSG00000057841 | ENSFM00500000270098 | 60S Ribosomal L32 |
| ENSMUSG00000057863 | ENSFM00670001235763 | 60S Ribosomal L36 |
| ENSMUSG00000057897 | ENSFM00250000000111 | Calcium/Calmodulin Dependent Kinase Type Ii Subunit Cam Kinase Ii Subunit Camk Ii Subunit EC_2.7.11.17 |
| ENSMUSG00000057914 | ENSFM00780001917886 | Unknown |
| ENSMUSG00000057914 | ENSFM00250000000524 | Voltage Dependent L Type Calcium Channel Subunit Beta Calcium Channel Voltage Dependent Subunit Beta |
| ENSMUSG00000058013 | ENSFM00350000105408 | Septin |
| ENSMUSG00000058064 | ENSFM00250000002087 | 60S Ribosomal L11 |
| ENSMUSG00000058301 | ENSFM00500000270457 | Atp Dependent Helicase Nonsense 1 Up Frameshift Suppressor 1 |
| ENSMUSG00000058325 | ENSFM00760001714647 | Dedicator Of Cytokinesis |
| ENSMUSG00000058420 | ENSFM00500000272887 | Synaptotagmin 17 Synaptotagmin Xvii Sytxvii |
| ENSMUSG00000058441 | ENSFM00590000917448 | Unknown |
| ENSMUSG00000058441 | ENSFM00250000007933 | Pannexin 2 |
| ENSMUSG00000058443 | ENSFM00800002003415 | 60S Ribosomal L10 |
| ENSMUSG00000058546 | ENSFM00670001235314 | 60S Ribosomal |
| ENSMUSG00000058558 | ENSFM00670001235299 | 60S Ribosomal L5 |
| ENSMUSG00000058569 | ENSFM00500000270043 | Transmembrane EMP24 Domain Containing Precursor |
| ENSMUSG00000058587 | ENSFM00500000269813 | Tropomodulin Tropomodulin Tmod |
| ENSMUSG00000058589 | ENSFM00720001498764 | Unknown |
| ENSMUSG00000058589 | ENSFM00720001498765 | Unknown |
| ENSMUSG00000058589 | ENSFM00720001501008 | Unknown |
| ENSMUSG00000058589 | ENSFM00720001503484 | Unknown |
| ENSMUSG00000058589 | ENSFM00720001503485 | Unknown |
| ENSMUSG00000058589 | ENSFM00250000001010 | Ankyrin Repeat And Sterile Alpha Motif Domain Containing 1B Amyloid Beta Intracellular Domain Associated 1 Aida 1 E2A PBX1 Associated Eb 1 |
| ENSMUSG00000058600 | ENSFM00670001235478 | 60S Ribosomal L30 |
| ENSMUSG00000058655 | ENSFM00500000270799 | Eukaryotic Translation Initiation Factor 4B Eif 4B |
| ENSMUSG00000058672 | ENSFM00250000000193 | Tubulin Beta Chain |
| ENSMUSG00000058715 | ENSFM00780001911544 | Unknown |
| ENSMUSG00000058715 | ENSFM00670001245974 | High Affinity Immunoglobulin Epsilon Receptor Subunit Gamma Precursor Fc Receptor Gamma Chain Fcrgamma Fc Epsilon Ri Gamma Ige Fc Receptor Subunit Gamma Fceri Gamma |
| ENSMUSG00000058799 | ENSFM00500000269862 | Nucleosome Assembly 1 |
| ENSMUSG00000058835 | ENSFM00250000000845 | Abl Interactor 1 Abelson Interactor 1 Abi 1 EPS8 SH3 Domain Binding EPS8 Binding Spectrin SH3 Domain Binding 1 |
| ENSMUSG00000058897 | ENSFM00500000271217 | Collagen Alpha 1 Xxv Chain Clac P |
| ENSMUSG00000058966 | ENSFM00250000003740 | FAM57B |
| ENSMUSG00000058997 | ENSFM00580000902189 | Von Willebrand Factor A Domain Containing 8 |
| ENSMUSG00000059003 | ENSFM00730001521500 | Glutamate Receptor Ionotropic Nmda Precursor Glutamate Receptor Subunit Epsilon N Methyl D Aspartate Receptor Subtype |
| ENSMUSG00000059005 | ENSFM00270000056431 | Heterogeneous Nuclear Ribonucleoprotein |
| ENSMUSG00000059070 | ENSFM00640001117531 | Unknown |
| ENSMUSG00000059070 | ENSFM00640001118658 | Unknown |
| ENSMUSG00000059070 | ENSFM00810002040668 | 60S Ribosomal L18 |
| ENSMUSG00000059119 | ENSFM00500000269862 | Nucleosome Assembly 1 |
| ENSMUSG00000059208 | ENSFM00570000887653 | Unknown |
| ENSMUSG00000059208 | ENSFM00740001589263 | Myelin Expression Factor 2 Mef 2 Myef 2 |
| ENSMUSG00000059213 | ENSFM00250000009854 | Dendrin |
| ENSMUSG00000059248 | ENSFM00570000901008 | Unknown |
| ENSMUSG00000059248 | ENSFM00500000269805 | Septin |
| ENSMUSG00000059291 | ENSFM00250000002087 | 60S Ribosomal L11 |
| ENSMUSG00000059316 | ENSFM00730001521827 | Long Chain Fatty Acid Transport 4 Fatp 4 Fatty Acid Transport 4 EC_6.2.1.- Solute Carrier Family 27 Member 4 |
| ENSMUSG00000059323 | ENSFM00610000967091 | Unknown |
| ENSMUSG00000059323 | ENSFM00610000967178 | Unknown |
| ENSMUSG00000059323 | ENSFM00610000973122 | Unknown |
| ENSMUSG00000059323 | ENSFM00250000004206 | Tonsoku Inhibitor Of Kappa B Related I Kappa B Related Ikappabr Nf Kappa B Inhibitor 2 Nuclear Factor Of Kappa Light Polypeptide Gene Enhancer In B Cells Inhibitor 2 |
| ENSMUSG00000059323 | ENSFM00500000271517 | Vacuolar Sorting Associated 28 Homolog Escrt I Complex Subunit VPS28 |
| ENSMUSG00000059336 | ENSFM00250000001770 | Urea Transporter 1 Solute Carrier Family 14 Member 1 Urea Transporter B Ut B Urea Transporter Erythrocyte |
| ENSMUSG00000059439 | ENSFM00810002045978 | Unknown |
| ENSMUSG00000059439 | ENSFM00250000002665 | Breast Carcinoma Amplified Sequence 3 |
| ENSMUSG00000059447 | ENSFM00800002023576 | Unknown |
| ENSMUSG00000059447 | ENSFM00440000236928 | Trifunctional Enzyme Subunit Beta Mitochondrial Precursor Tp Beta |
| ENSMUSG00000059554 | ENSFM00640001110165 | Unknown |
| ENSMUSG00000059554 | ENSFM00500000272544 | Coiled Coil Domain Containing 28A CCRL1AP |
| ENSMUSG00000059602 | ENSFM00250000000771 | Synapsin Synapsin |
| ENSMUSG00000059714 | ENSFM00640001119931 | Unknown |
| ENSMUSG00000059714 | ENSFM00780001898418 | Flotillin 1 |
| ENSMUSG00000059775 | ENSFM00670001235586 | 40S Ribosomal S26 |
| ENSMUSG00000059857 | ENSFM00260000050653 | Netrin Precursor Laminet |
| ENSMUSG00000059970 | ENSFM00730001521103 | Heat Shock Kda Heat Shock 70 Kda |
| ENSMUSG00000059974 | ENSFM00290000065521 | Precursor |
| ENSMUSG00000059981 | ENSFM00250000000892 | Serine/Threonine Kinase EC_2.7.11.1 Thousand And One Amino Acid |
| ENSMUSG00000060036 | ENSFM00810002040577 | 60S Ribosomal L3 |
| ENSMUSG00000060073 | ENSFM00590000916022 | Unknown |
| ENSMUSG00000060073 | ENSFM00590000917798 | Unknown |
| ENSMUSG00000060073 | ENSFM00590000918013 | Unknown |
| ENSMUSG00000060073 | ENSFM00590000918014 | Unknown |
| ENSMUSG00000060073 | ENSFM00810002040700 | Proteasome Subunit Alpha Type 3 EC_3.4.25.1 Macropain Subunit C8 Multicatalytic Endopeptidase Complex Subunit C8 Proteasome Component C8 |
| ENSMUSG00000060081 | ENSFM00670001235241 | Histone H2A |
| ENSMUSG00000060090 | ENSFM00250000005619 | XRP2 |
| ENSMUSG00000060166 | ENSFM00500000270072 | Palmitoyltransferase ZDHHC5 EC_2.3.1.225 Zinc Finger Dhhc Domain Containing 5 Dhhc 5 |
| ENSMUSG00000060227 | ENSFM00250000003320 | CASC4 Cancer Susceptibility Candidate Gene 4 |
| ENSMUSG00000060240 | ENSFM00600000922041 | Cell Cycle Exit And Neuronal Differentiation 1 |
| ENSMUSG00000060261 | ENSFM00810002049264 | Unknown |
| ENSMUSG00000060261 | ENSFM00810002049698 | Unknown |
| ENSMUSG00000060261 | ENSFM00760001714782 | General Transcription Factor Ii I Repeat Domain Containing 1 GTF2I Repeat Domain Containing 1 |
| ENSMUSG00000060275 | ENSFM00600000921191 | Pro Neuregulin 1 Membrane Bound Precursor Pro NRG1 |
| ENSMUSG00000060279 | ENSFM00610000970149 | Unknown |
| ENSMUSG00000060279 | ENSFM00610000971381 | Unknown |
| ENSMUSG00000060279 | ENSFM00810002040615 | Ap 2 Complex Subunit Alpha 2.100 Kda Coated Vesicle C Adaptor Complex Ap 2 Subunit Alpha 2 Adaptor Related Complex 2 Subunit Alpha 2 Alpha Adaptin C ALPHA2 Adaptin Clathrin Assembly Complex 2 Alpha C Large Chain Plasma Membrane Adaptor HA2/AP2 Adaptin Alp |
| ENSMUSG00000060373 | ENSFM00810002040643 | Heterogeneous Nuclear Ribonucleoprotein C Hnrnp C |
| ENSMUSG00000060636 | ENSFM00500000271015 | 60S Ribosomal L35A |
| ENSMUSG00000060743 | ENSFM00590000918360 | Unknown |
| ENSMUSG00000060743 | ENSFM00590000918361 | Unknown |
| ENSMUSG00000060743 | ENSFM00250000000259 | Histone H3 |
| ENSMUSG00000060780 | ENSFM00400000131776 | Leucine Rich Repeat Transmembrane Neuronal Precursor |
| ENSMUSG00000060843 | ENSFM00250000001018 | Catenin Alpha 2 Alpha N Catenin |
| ENSMUSG00000060882 | ENSFM00300000079128 | Potassium Voltage Gated Channel Subfamily D Member Voltage Gated Potassium Channel Subunit KV4 |
| ENSMUSG00000060938 | ENSFM00500000270069 | 60S Ribosomal L26 |
| ENSMUSG00000060992 | ENSFM00500000270533 | Coatomer Subunit Zeta Zeta Coat Zeta Cop |
| ENSMUSG00000061080 | ENSFM00760001748822 | Unknown |
| ENSMUSG00000061080 | ENSFM00760001763360 | Unknown |
| ENSMUSG00000061080 | ENSFM00760001763361 | Unknown |
| ENSMUSG00000061080 | ENSFM00760001763362 | Unknown |
| ENSMUSG00000061080 | ENSFM00290000065521 | Precursor |
| ENSMUSG00000061086 | ENSFM00570000895631 | Unknown |
| ENSMUSG00000061086 | ENSFM00570000897194 | Unknown |
| ENSMUSG00000061086 | ENSFM00250000000438 | Myosin Light Chain Myosin Light Chain Myosin Light Chain |
| ENSMUSG00000061111 | ENSFM00670001237224 | Unknown |
| ENSMUSG00000061244 | ENSFM00590000918374 | Unknown |
| ENSMUSG00000061244 | ENSFM00250000005479 | Exocyst Complex Component Exocyst Complex Component SEC10 |
| ENSMUSG00000061273 | ENSFM00500000273449 | Membrane Magnesium Transporter 1 Precursor Er Membrane Complex Subunit 5 Transmembrane 32 |
| ENSMUSG00000061455 | ENSFM00250000004622 | Syntaxin 17 |
| ENSMUSG00000061461 | ENSFM00570000899182 | Unknown |
| ENSMUSG00000061474 | ENSFM00670001236924 | 28S Ribosomal S36 Mitochondrial Mrp S36 S36MT |
| ENSMUSG00000061601 | ENSFM00600000921250 | Piccolo Aczonin |
| ENSMUSG00000061689 | ENSFM00660001182988 | Unknown |
| ENSMUSG00000061689 | ENSFM00660001190031 | Unknown |
| ENSMUSG00000061689 | ENSFM00250000000718 | Disks Large Associated Dap Psd 95/SAP90 Binding SAP90/Psd 95 Associated |
| ENSMUSG00000061751 | ENSFM00740001589124 | EC_2.7.11.1 |
| ENSMUSG00000061758 | ENSFM00730001521171 | Reductase Aldo Keto Reductase Family 1 Member |
| ENSMUSG00000061787 | ENSFM00780001898364 | 40S Ribosomal S17 |
| ENSMUSG00000061816 | ENSFM00250000000438 | Myosin Light Chain Myosin Light Chain Myosin Light Chain |
| ENSMUSG00000061981 | ENSFM00780001898447 | Flotillin 2 |
| ENSMUSG00000062006 | ENSFM00800002015933 | Unknown |
| ENSMUSG00000062006 | ENSFM00500000271952 | 60S Ribosomal L34 |
| ENSMUSG00000062014 | ENSFM00500000270656 | Glia Maturation Factor Gmf |
| ENSMUSG00000062044 | ENSFM00570000890117 | Unknown |
| ENSMUSG00000062044 | ENSFM00670001235536 | Serine/Threonine Kinase EC_2.7.11.1 Apoptosis Associated Tyrosine Kinase Brain Kinase Lemur Tyrosine Kinase |
| ENSMUSG00000062075 | ENSFM00250000000425 | Lamin |
| ENSMUSG00000062078 | ENSFM00400000131794 | Quaking |
| ENSMUSG00000062110 | ENSFM00810002057151 | Unknown |
| ENSMUSG00000062110 | ENSFM00250000004937 | SEC1 Family Domain Containing 2 Syntaxin Binding 1 1 |
| ENSMUSG00000062127 | ENSFM00600000921238 | Cortactin Binding 2 CORTBP2 |
| ENSMUSG00000062151 | ENSFM00250000000599 | Unc 13 Homolog MUNC13 |
| ENSMUSG00000062190 | ENSFM00250000002547 | Lanc 1.40 Kda Erythrocyte Membrane P40 |
| ENSMUSG00000062232 | ENSFM00250000001338 | Rap Guanine Nucleotide Exchange Factor 2 Cyclic Nucleotide Ras Gef Cnrasgef Neural Rap Guanine Nucleotide Exchange Nrap Gep Pdz Domain Containing Guanine Nucleotide Exchange Factor 1 Pdz GEF1 Ra Gef 1 Ras/RAP1 Associating Gef 1 |
| ENSMUSG00000062234 | ENSFM00800002023625 | Unknown |
| ENSMUSG00000062234 | ENSFM00800002023626 | Unknown |
| ENSMUSG00000062234 | ENSFM00250000001678 | Tyrosine Phosphatase Auxilin EC_3.1.3.48 Dnaj Homolog Subfamily C Member 6 |
| ENSMUSG00000062257 | ENSFM00290000065521 | Precursor |
| ENSMUSG00000062328 | ENSFM00250000001117 | 60S Ribosomal L17 |
| ENSMUSG00000062380 | ENSFM00250000000193 | Tubulin Beta Chain |
| ENSMUSG00000062444 | ENSFM00480000262766 | Ap 3 Complex Subunit Beta 1 Adaptor Complex Ap 3 Subunit Beta 1 Adaptor Related Complex 3 Subunit Beta 1 Beta 3A Adaptin Clathrin Assembly Complex 3 Beta 1 Large Chain |
| ENSMUSG00000062591 | ENSFM00250000000193 | Tubulin Beta Chain |
| ENSMUSG00000062604 | ENSFM00800002023563 | Unknown |
| ENSMUSG00000062604 | ENSFM00800002023564 | Unknown |
| ENSMUSG00000062604 | ENSFM00500000269720 | Srsf Kinase EC_2.7.11.1 Kinase Serine/Arginine Rich Specific Kinase Sr Specific Kinase |
| ENSMUSG00000062647 | ENSFM00750001632337 | 60S Ribosomal L7A |
| ENSMUSG00000062661 | ENSFM00780001917898 | Unknown |
| ENSMUSG00000062661 | ENSFM00760001714671 | Neuronal Calcium Sensor 1 Ncs 1 |
| ENSMUSG00000062785 | ENSFM00260000050384 | Potassium Voltage Gated Channel Subfamily C Member Voltage Gated Potassium Channel KV3 |
| ENSMUSG00000062825 | ENSFM00730001521060 | Actin |
| ENSMUSG00000062929 | ENSFM00500000270025 | Cofilin Cofilin Muscle |
| ENSMUSG00000062961 | ENSFM00260000052939 | Coiled Coil Domain Containing 177 |
| ENSMUSG00000062963 | ENSFM00250000007775 | Ubiquitin Fold Modifier Conjugating Enzyme 1 UFM1 Conjugating Enzyme 1 |
| ENSMUSG00000063052 | ENSFM00570000879628 | Unknown |
| ENSMUSG00000063052 | ENSFM00570000893042 | Unknown |
| ENSMUSG00000063052 | ENSFM00800002024524 | Unknown |
| ENSMUSG00000063052 | ENSFM00440000236958 | Leucine Rich Repeat Containing 40 |
| ENSMUSG00000063063 | ENSFM00250000001018 | Catenin Alpha 2 Alpha N Catenin |
| ENSMUSG00000063065 | ENSFM00730001521833 | Mitogen Activated Kinase 1 Map Kinase 1 Mapk 1 EC_2.7.11.24 ERT1 Extracellular Signal Regulated Kinase 2 Erk 2 Mitogen Activated Kinase 2 Map Kinase 2 Mapk 2 |
| ENSMUSG00000063146 | ENSFM00250000000764 | Cap Gly Domain Containing Linker 2 Cytoplasmic Linker 115 Clip 115 Cytoplasmic Linker 2 |
| ENSMUSG00000063160 | ENSFM00250000001973 | Numb |
| ENSMUSG00000063239 | ENSFM00250000000328 | Metabotropic Glutamate Receptor Precursor |
| ENSMUSG00000063297 | ENSFM00250000005652 | Leucine Zipper 2 Precursor |
| ENSMUSG00000063317 | ENSFM00260000050633 | Ubiquitin Carboxyl Terminal Hydrolase 43 EC_3.4.19.12 Deubiquitinating Enzyme 43 Ubiquitin Thioesterase 43 Ubiquitin Specific Processing Protease 43 |
| ENSMUSG00000063358 | ENSFM00730001521833 | Mitogen Activated Kinase 1 Map Kinase 1 Mapk 1 EC_2.7.11.24 ERT1 Extracellular Signal Regulated Kinase 2 Erk 2 Mitogen Activated Kinase 2 Map Kinase 2 Mapk 2 |
| ENSMUSG00000063410 | ENSFM00810002040592 | Serine/Threonine Kinase EC_2.7.11.1 Mammalian STE20 Kinase Mst STE20 Kinase Serine/Threonine Kinase |
| ENSMUSG00000063455 | ENSFM00600000921514 | Ambiguous |
| ENSMUSG00000063457 | ENSFM00250000003189 | 40S Ribosomal S15 |
| ENSMUSG00000063646 | ENSFM00640001112222 | Unknown |
| ENSMUSG00000063646 | ENSFM00250000001480 | Janus Kinase And Microtubule Interacting 1 Multiple Alpha Helices And Rna Linker 1 Marlin 1 |
| ENSMUSG00000063696 | ENSFM00250000001360 | 60S Acidic Ribosomal P0 60S Ribosomal L10E |
| ENSMUSG00000063801 | ENSFM00500000270528 | Ap 3 Complex Subunit Sigma Ap 3 Complex Subunit Sigma Adaptor Related Complex 3 Subunit Sigma Sigma Adaptin Adaptin Sigma Adaptin |
| ENSMUSG00000063856 | ENSFM00500000270307 | Glutathione Peroxidase 2 Gpx 2 Gshpx 2 EC_1.11.1.9 Glutathione Peroxidase Gastrointestinal Gpx Gi Gshpx Gi |
| ENSMUSG00000064030 | ENSFM00500000273807 | Partner Of Y14 And Mago Wibg Homolog |
| ENSMUSG00000064068 | ENSFM00640001115744 | Unknown |
| ENSMUSG00000064068 | ENSFM00640001116925 | Unknown |
| ENSMUSG00000064068 | ENSFM00640001119895 | Unknown |
| ENSMUSG00000064068 | ENSFM00640001119896 | Unknown |
| ENSMUSG00000064068 | ENSFM00250000002187 | Metaxin 1 Mitochondrial Outer Membrane Import Complex 1 |
| ENSMUSG00000064125 | ENSFM00660001189520 | Unknown |
| ENSMUSG00000064125 | ENSFM00250000009146 | Proline Rich 36 |
| ENSMUSG00000064293 | ENSFM00390000126303 | Contactin Precursor Brain Derived Immunoglobulin Superfamily Big |
| ENSMUSG00000064302 | ENSFM00760001753540 | Unknown |
| ENSMUSG00000064302 | ENSFM00250000000820 | Clip Associating Cytoplasmic Linker Associated |
| ENSMUSG00000064354 | ENSFM00250000000189 | Cytochrome C Oxidase Subunit 2 Cytochrome C Oxidase Polypeptide Ii |
| ENSMUSG00000064356 | ENSFM00500000344754 | Unknown |
| ENSMUSG00000065954 | ENSFM00740001589245 | Transforming Acidic Coiled Coil Containing |
| ENSMUSG00000066036 | ENSFM00250000001486 | E3 Ubiquitin Ligase UBR4 EC_6.3.2.- N Recognin 4 Zinc Finger UBR1 Type 1 |
| ENSMUSG00000066129 | ENSFM00250000004496 | Very Kind Kind Domain Containing 1 Kinase Non Catalytic C Lobe Domain Containing 1 Ras Gef Domain Containing Family Member 2 |
| ENSMUSG00000066189 | ENSFM00250000002019 | Voltage Dependent Calcium Channel Gamma Subunit Neuronal Voltage Gated Calcium Channel Gamma Subunit Transmembrane Ampar Regulatory Gamma Tarp Gamma |
| ENSMUSG00000066232 | ENSFM00760001714748 | Importin 7 IMP7 Ran Binding 7 RANBP7 |
| ENSMUSG00000066235 | ENSFM00250000007877 | O Linked Mannose Beta 1 4 N Acetylglucosaminyltransferase 2 POMGNT2 EC_2.4.1.- |
| ENSMUSG00000066278 | ENSFM00500000273456 | Vacuolar Sorting Associated 37B Escrt I Complex Subunit VPS37B |
| ENSMUSG00000066392 | ENSFM00740001589096 | Neurexin Precursor Neurexin |
| ENSMUSG00000066438 | ENSFM00570000882117 | Unknown |
| ENSMUSG00000066438 | ENSFM00430000230337 | Pleckstrin Homology Domain Containing Family D Member 1 Ph Domain Containing Family D Member 1 |
| ENSMUSG00000066487 | ENSFM00250000000709 | 40S Ribosomal S2 |
| ENSMUSG00000066839 | ENSFM00250000005244 | Evolutionarily Conserved Signaling Intermediate In Toll Pathway Mitochondrial Precursor |
| ENSMUSG00000067038 | ENSFM00500000270361 | 40S Ribosomal S12 |
| ENSMUSG00000067242 | ENSFM00800002009181 | Unknown |
| ENSMUSG00000067242 | ENSFM00250000002312 | Leucine Rich Repeat Lgi Family Member Precursor Leucine Rich Glioma Inactivated |
| ENSMUSG00000067274 | ENSFM00250000001360 | 60S Acidic Ribosomal P0 60S Ribosomal L10E |
| ENSMUSG00000067288 | ENSFM00500000284355 | Unknown |
| ENSMUSG00000067288 | ENSFM00640001110125 | Unknown |
| ENSMUSG00000067629 | ENSFM00810002057507 | Unknown |
| ENSMUSG00000067629 | ENSFM00250000000610 | Ras Gtpase Activating |
| ENSMUSG00000067889 | ENSFM00740001589095 | Spectrin Beta Chain Erythrocytic Beta Spectrin |
| ENSMUSG00000068015 | ENSFM00800002003728 | Leucine Rich Repeat And Calponin Homology Domain Containing 1 Calponin Homology Domain Containing 1 |
| ENSMUSG00000068036 | ENSFM00250000001587 | Afadin Af 6 |
| ENSMUSG00000068039 | ENSFM00500000270058 | T Complex 1 Subunit Alpha Tcp 1 Alpha Cct Alpha |
| ENSMUSG00000068290 | ENSFM00570000879313 | Unknown |
| ENSMUSG00000068290 | ENSFM00570000896336 | Unknown |
| ENSMUSG00000068290 | ENSFM00600000921555 | Ddrgk Domain Containing 1 |
| ENSMUSG00000068373 | ENSFM00250000005157 | Ambiguous |
| ENSMUSG00000068566 | ENSFM00500000271004 | Myeloid Associated Differentiation Marker |
| ENSMUSG00000068735 | ENSFM00500000272770 | Tumor P53 Inducible 11 P53 Induced Gene 11 |
| ENSMUSG00000068739 | ENSFM00770001847577 | Serine Trna Ligase Cytoplasmic EC_6.1.1.11 Seryl Trna Synthetase Serrs Seryl Trna Ser/Sec Synthetase |
| ENSMUSG00000068748 | ENSFM00730001521864 | Receptor Type Tyrosine Phosphatase Precursor R Ptp EC_3.1.3.48 |
| ENSMUSG00000068749 | ENSFM00800002024487 | Unknown |
| ENSMUSG00000068749 | ENSFM00500000270276 | Proteasome Subunit Alpha Type 5 EC_3.4.25.1 Macropain Multicatalytic Endopeptidase Complex Proteasome |
| ENSMUSG00000068798 | ENSFM00730001521249 | Ras Related Rap 1 |
| ENSMUSG00000068823 | ENSFM00250000002562 | Cold Shock Domain Containing E1 |
| ENSMUSG00000068923 | ENSFM00500000270270 | Synaptotagmin Synaptotagmin |
| ENSMUSG00000069045 | ENSFM00760001763663 | Unknown |
| ENSMUSG00000069045 | ENSFM00770001847530 | Atp Dependent Rna Helicase DDX3Y EC_3.6.4.13 Dead Box 3 Y Chromosomal |
| ENSMUSG00000069117 | ENSFM00250000002236 | 40S Ribosomal S18 |
| ENSMUSG00000069227 | ENSFM00250000008086 | G Regulated Inducer Of Neurite Outgrowth 1 GRIN1 |
| ENSMUSG00000069266 | ENSFM00500000269649 | Histone H4 |
| ENSMUSG00000069308 | ENSFM00500000269608 | Histone H2B |
| ENSMUSG00000069601 | ENSFM00500000269628 | Ankyrin Ank Ankyrin |
| ENSMUSG00000069769 | ENSFM00500000270111 | Rna Binding Musashi Homolog |
| ENSMUSG00000069833 | ENSFM00740001589261 | Associated |
| ENSMUSG00000070047 | ENSFM00760001763202 | Unknown |
| ENSMUSG00000070047 | ENSFM00570000851039 | Protocadherin Fat Precursor Fat Tumor Suppressor Homolog |
| ENSMUSG00000070283 | ENSFM00780001908782 | Unknown |
| ENSMUSG00000070283 | ENSFM00500000273238 | Nadh Dehydrogenase 1 Alpha Subcomplex Assembly Factor 3 |
| ENSMUSG00000070372 | ENSFM00250000001955 | F Actin Capping Subunit Alpha 2 Capz Alpha 2 |
| ENSMUSG00000070394 | ENSFM00670001236968 | Transmembrane 256 Precursor |
| ENSMUSG00000070436 | ENSFM00310000089046 | Serpin H1 Precursor 47 Kda Heat Shock Collagen Binding Colligin |
| ENSMUSG00000070462 | ENSFM00250000008923 | Mesoderm Development Candidate 1 |
| ENSMUSG00000070498 | ENSFM00250000000808 | Transmembrane Precursor |
| ENSMUSG00000070509 | ENSFM00250000001343 | Rgm Domain Family Member |
| ENSMUSG00000070565 | ENSFM00250000000610 | Ras Gtpase Activating |
| ENSMUSG00000070639 | ENSFM00250000001418 | Volume Regulated Anion Channel Subunit Leucine Rich Repeat Containing |
| ENSMUSG00000070730 | ENSFM00570000889560 | Unknown |
| ENSMUSG00000070730 | ENSFM00570000894879 | Unknown |
| ENSMUSG00000070730 | ENSFM00570000901405 | Regulator Of Microtubule Dynamics 3 |
| ENSMUSG00000070730 | ENSFM00670001235671 | Regulator Of Microtubule Dynamics Rmd Fa |
| ENSMUSG00000071014 | ENSFM00770001850404 | Nadh Dehydrogenase 1 Beta Subcomplex Subunit 6 |
| ENSMUSG00000071014 | ENSFM00600000921719 | Nadh Dehydrogenase 1 Beta Subcomplex Subunit 6 Complex I B17 Ci B17 Nadh Ubiquinone Oxidoreductase B17 Subunit |
| ENSMUSG00000071073 | ENSFM00250000009938 | Leucine Rich Repeat Containing 73 |
| ENSMUSG00000071172 | ENSFM00500000272005 | Serine/Arginine Rich Splicing Factor 3 Pre Splicing Factor SRP20 Splicing Factor Arginine/Serine Rich 3 |
| ENSMUSG00000071379 | ENSFM00760001714608 | Unknown |
| ENSMUSG00000071415 | ENSFM00570000883216 | Unknown |
| ENSMUSG00000071415 | ENSFM00250000003000 | 60S Ribosomal L23 |
| ENSMUSG00000071424 | ENSFM00720001492054 | Glutamate Receptor Ionotropic Delta Precursor Glur Delta Subunit |
| ENSMUSG00000071519 | ENSFM00730001521212 | Trypsin Precursor EC_3.4.21.4 |
| ENSMUSG00000071528 | ENSFM00500000445520 | Unknown |
| ENSMUSG00000071531 | ENSFM00280000058989 | G Regulated Inducer Of Neurite Outgrowth 2 GRIN2 |
| ENSMUSG00000071644 | ENSFM00250000001516 | Elongation Factor 1 Gamma Ef 1 Gamma Eef 1B Gamma |
| ENSMUSG00000071649 | ENSFM00250000001080 | Galactosylgalactosylxylosylprotein 3 Beta Glucuronosyltransferase EC_2.4.1.135 Beta 1 3 Glucuronyltransferase Glucuronosyltransferase Glcat Udp Beta 1 3 Glucuronyltransferase Glcuat |
| ENSMUSG00000071660 | ENSFM00600000927711 | Tetratricopeptide Repeat 9C |
| ENSMUSG00000071660 | ENSFM00670001236445 | Tetratricopeptide Repeat 9C Tpr Repeat 9C |
| ENSMUSG00000071862 | ENSFM00400000131776 | Leucine Rich Repeat Transmembrane Neuronal Precursor |
| ENSMUSG00000072214 | ENSFM00800002003401 | Septin |
| ENSMUSG00000072582 | ENSFM00500000273926 | Peptidyl Trna Hydrolase 2 Mitochondrial Precursor Pth 2 EC_3.1.1.29 Bcl 2 Inhibitor Of Transcription |
| ENSMUSG00000072772 | ENSFM00500000274497 | C10 |
| ENSMUSG00000072812 | ENSFM00740001589261 | Associated |
| ENSMUSG00000072825 | ENSFM00740001589275 | Unknown |
| ENSMUSG00000073002 | ENSFM00600000925372 | Vesicle Associated Membrane 5 Vamp 5 Myobrevin |
| ENSMUSG00000073435 | ENSFM00250000000895 | Nucleoside Diphosphate Kinase Ndk Ndp Kinase EC_2.7.4.6 |
| ENSMUSG00000073481 | ENSFM00500000270661 | Mitochondrial Amidoxime Reducing Component 2 Precursor MARC2 Ec 1 Molybdenum Cofactor Sulfurase C Terminal Domain Containing 2 Mosc Domain Containing 2 Moco Sulfurase C Terminal Domain Containing 2 |
| ENSMUSG00000073557 | ENSFM00500000269806 | Phosphatase 1 Regulatory Subunit 12A Myosin Phosphatase Targeting Subunit 1 Myosin Phosphatase Target Subunit 1 Phosphatase Myosin Binding Subunit |
| ENSMUSG00000073563 | ENSFM00730001521484 | Casein Kinase I Gamma Cki Gamma EC_2.7.11.1 |
| ENSMUSG00000073640 | ENSFM00670001235486 | 60S Ribosomal L27 |
| ENSMUSG00000073678 | ENSFM00460000244973 | Gpi Inositol Deacylase EC_3.1.-.- Post Gpi Attachment To Proteins Factor 1 |
| ENSMUSG00000073702 | ENSFM00780001913412 | Unknown |
| ENSMUSG00000073702 | ENSFM00670001235422 | 60S Ribosomal L31 |
| ENSMUSG00000073805 | ENSFM00250000008262 | Unknown |
| ENSMUSG00000073838 | ENSFM00260000050713 | Elongation Factor Tu Mitochondrial Precursor |
| ENSMUSG00000073982 | ENSFM00730001521186 | Ras Related C3 Botulinum Toxin Substrate Precursor P21 |
| ENSMUSG00000074030 | ENSFM00250000004839 | Exocyst Complex Component 8 Exocyst Complex 84 Kda Subunit |
| ENSMUSG00000074064 | ENSFM00250000006084 | Malonyl Coa Decarboxylase Mitochondrial Precursor Mcd EC_4.1.1.9 |
| ENSMUSG00000074129 | ENSFM00790001962987 | 60S Ribosomal L13A |
| ENSMUSG00000074212 | ENSFM00250000001153 | Dnaj Homolog Subfamily Member |
| ENSMUSG00000074305 | ENSFM00760001742183 | Unknown |
| ENSMUSG00000074305 | ENSFM00740001589410 | Pseudopodium Enriched Atypical Kinase 1 EC_2.7.10.2 Sugen Kinase 269 Tyrosine Kinase SGK269 |
| ENSMUSG00000074457 | ENSFM00570000882450 | Unknown |
| ENSMUSG00000074457 | ENSFM00600000921934 | S100 A16 S100 Calcium Binding A16 |
| ENSMUSG00000074505 | ENSFM00570000851039 | Protocadherin Fat Precursor Fat Tumor Suppressor Homolog |
| ENSMUSG00000074698 | ENSFM00570000890230 | Unknown |
| ENSMUSG00000074698 | ENSFM00250000001591 | Casein Kinase Ii Subunit Ck Ii EC_2.7.11.1 |
| ENSMUSG00000074736 | ENSFM00570000890894 | Unknown |
| ENSMUSG00000074736 | ENSFM00670001235828 | Synapse Differentiation Inducing Gene 1 Capucin Dispanin Subfamily C Member 1 DSPC1 Transmembrane 90A |
| ENSMUSG00000074781 | ENSFM00500000270143 | Ubiquitin Conjugating Enzyme E2 N EC_6.3.2.19 Ubiquitin Carrier N Ubiquitin Ligase N |
| ENSMUSG00000074793 | ENSFM00250000005151 | Unknown |
| ENSMUSG00000074793 | ENSFM00720001503567 | Unknown |
| ENSMUSG00000074886 | ENSFM00730001521304 | G Coupled Receptor Kinase G Coupled Receptor Kinase |
| ENSMUSG00000074923 | ENSFM00730001521739 | Serine/Threonine Kinase Pak EC_2.7.11.1 P21 Activated Kinase Pak |
| ENSMUSG00000075402 | ENSFM00250000000206 | Keratin Type Ii Cytoskeletal Cytokeratin Ck Keratin Type Ii Keratin |
| ENSMUSG00000075701 | ENSFM00500000273466 | Selenoprotein S Sels Vcp Interacting Membrane |
| ENSMUSG00000075706 | ENSFM00500000272603 | Phospholipid Hydroperoxide Glutathione Peroxidase Mitochondrial Precursor Phgpx EC_1.11.1.12 Glutathione Peroxidase 4 Gpx 4 Gshpx 4 |
| ENSMUSG00000076432 | ENSFM00730001521154 | Unknown |
| ENSMUSG00000076439 | ENSFM00380000124712 | Myelin Oligodendrocyte Glycoprotein Precursor |
| ENSMUSG00000076617 | ENSFM00740001589433 | Ig Chain C Region |
| ENSMUSG00000078137 | ENSFM00500000295837 | Ankyrin Repeat Domain Containing 63 |
| ENSMUSG00000078139 | ENSFM00500000272843 | Iron Sulfur Cluster Assembly 1 Homolog Mitochondrial Precursor Hesb Domain Containing 2 Iron Sulfur Assembly Isca |
| ENSMUSG00000078193 | ENSFM00500000271690 | 60S Ribosomal L35 |
| ENSMUSG00000078249 | ENSFM00680001326820 | Unknown |
| ENSMUSG00000078249 | ENSFM00810002041146 | High Mobility Group Hmg I/Hmg Y Hmg I Y High Mobility Group At Hook 1 High Mobility Group A1 |
| ENSMUSG00000078350 | ENSFM00720001501322 | Unknown |
| ENSMUSG00000078350 | ENSFM00720001501323 | Unknown |
| ENSMUSG00000078350 | ENSFM00720001501324 | Unknown |
| ENSMUSG00000078350 | ENSFM00720001503264 | Unknown |
| ENSMUSG00000078517 | ENSFM00500000270724 | Er Membrane Complex Subunit 1 Precursor |
| ENSMUSG00000078532 | ENSFM00250000002642 | Sodium/Potassium Transporting Atpase Subunit Beta 1 Interacting Na + /K + Transporting Atpase Subunit Beta 1 Interacting |
| ENSMUSG00000078622 | ENSFM00250000004158 | Coiled Coil Domain Containing 47 Precursor |
| ENSMUSG00000078656 | ENSFM00500000271998 | Vacuolar Sorting Associated 25 Escrt Ii Complex Subunit VPS25 |
| ENSMUSG00000078695 | ENSFM00500000292816 | Cdgsh Iron Sulfur Domain Containing 3 Mitochondrial Precursor |
| ENSMUSG00000078713 | ENSFM00500000331103 | Unknown |
| ENSMUSG00000078713 | ENSFM00500000343387 | Unknown |
| ENSMUSG00000078713 | ENSFM00500000394168 | Unknown |
| ENSMUSG00000078713 | ENSFM00500000447447 | Unknown |
| ENSMUSG00000078794 | ENSFM00390000126901 | Dapper Homolog 3 Dapper Antagonist Of Catenin 3 |
| ENSMUSG00000078816 | ENSFM00520000517802 | Kinase C Type Pkc EC_2.7.11.13 |
| ENSMUSG00000078908 | ENSFM00770001847570 | Vacuolar Fusion MON1 |
| ENSMUSG00000078919 | ENSFM00250000004763 | Dolichol Phosphate Mannosyltransferase Subunit 1 EC_2.4.1.83 Dolichol Phosphate Mannose Synthase Subunit 1 Dpm Synthase Subunit 1 Dolichyl Phosphate Beta D Mannosyltransferase Subunit 1 Mannose P Dolichol Synthase Subunit 1 Mpd Synthase Subunit 1 |
| ENSMUSG00000078923 | ENSFM00250000001221 | Ubiquitin Conjugating Enzyme E2 Variant |
| ENSMUSG00000079104 | ENSFM00250000000896 | Ribose Phosphate Pyrophosphokinase EC_2.7.6.1 Phosphoribosyl Pyrophosphate Synthase |
| ENSMUSG00000079108 | ENSFM00250000002862 | Signal Recognition Particle 54 Kda SRP54 |
| ENSMUSG00000079426 | ENSFM00250000003587 | Actin Related 2/3 Complex Subunit 4 ARP2/3 Complex 20 Kda P20 Arc |
| ENSMUSG00000079481 | ENSFM00400000132091 | Nhs 2 |
| ENSMUSG00000079658 | ENSFM00760001743923 | Unknown |
| ENSMUSG00000079658 | ENSFM00500000272323 | Transcription Elongation Factor B Polypeptide 1 Elongin 15 Kda Subunit Elongin C Eloc Rna Polymerase Ii Transcription Factor Siii Subunit C Siii P15 Stromal Membrane Associated SMAP1B Homolog |
| ENSMUSG00000084845 | ENSFM00500000314906 | Transmembrane 240 |
| ENSMUSG00000086040 | ENSFM00600000921808 | Was/Wasl Interacting Family Member 3 Corticosteroids And Regional Expression 16 |
| ENSMUSG00000087260 | ENSFM00670001236711 | Ragulator Complex LAMTOR5 Late Endosomal/Lysosomal Adaptor And Mapk And Mtor Activator 5 |
| ENSMUSG00000087651 | ENSFM00610000968876 | Unknown |
| ENSMUSG00000089945 | ENSFM00740001589417 | A Kinase Anchor 2 Akap 2 Akap Kl Kinase A Anchoring 2 PRKA2 |
| ENSMUSG00000090053 | ENSFM00740001589417 | A Kinase Anchor 2 Akap 2 Akap Kl Kinase A Anchoring 2 PRKA2 |
| ENSMUSG00000090071 | ENSFM00250000002836 | Cyclin Dependent Kinase 5 Activator 1 Precursor CDK5 Activator 1 Cyclin Dependent Kinase 5 Regulatory Subunit 1 Tpkii Regulatory Subunit |
| ENSMUSG00000090150 | ENSFM00760001763413 | Unknown |
| ENSMUSG00000090150 | ENSFM00780001898384 | Acyl Coa Dehydrogenase Family Member Acad EC_1.3.99.- |
| ENSMUSG00000090523 | ENSFM00610000965985 | Unknown |
| ENSMUSG00000090523 | ENSFM00640001115365 | Unknown |
| ENSMUSG00000090523 | ENSFM00560000828284 | Glycophorin C CD236 Antigen |
| ENSMUSG00000090639 | ENSFM00250000000343 | Precursor |
| ENSMUSG00000090841 | ENSFM00250000000438 | Myosin Light Chain Myosin Light Chain Myosin Light Chain |
| ENSMUSG00000090862 | ENSFM00250000003348 | 40S Ribosomal S13 |
| ENSMUSG00000091228 | ENSFM00250000000895 | Nucleoside Diphosphate Kinase Ndk Ndp Kinase EC_2.7.4.6 |
| ENSMUSG00000091780 | ENSFM00500000270642 | Homolog Mitochondrial Precursor |
| ENSMUSG00000091971 | ENSFM00730001521103 | Heat Shock Kda Heat Shock 70 Kda |
| ENSMUSG00000092305 | ENSFM00250000000896 | Ribose Phosphate Pyrophosphokinase EC_2.7.6.1 Phosphoribosyl Pyrophosphate Synthase |
| ENSMUSG00000093904 | ENSFM00500000272385 | Mitochondrial Import Receptor Subunit TOM20 Homolog Mitochondrial 20 Kda Outer Membrane Outer Mitochondrial Membrane Receptor TOM20 |
| ENSMUSG00000094483 | ENSFM00250000002171 | Transcriptional Activator Pur Beta Purine Rich Element Binding B |
| ENSMUSG00000094626 | ENSFM00250000010793 | Cat Eye Syndrome Critical Region 6 |
| ENSMUSG00000094685 | ENSFM00250000006602 | Ragulator Complex LAMTOR3 Late Endosomal/Lysosomal Adaptor And Mapk And Mtor Activator 3 Mitogen Activated Kinase Scaffold 1 |
| ENSMUSG00000096054 | ENSFM00250000000542 | Nesprin 2 Nuclear Envelope Spectrin Repeat 2 Nucleus And Actin Connecting Element Protein Nuance Synaptic Nuclear Envelope 2 Syne 2 |
| ENSMUSG00000098274 | ENSFM00670001235623 | 60S Ribosomal L24 |
| ENSMUSG00000098557 | ENSFM00260000050545 | Btb/Poz Domain Containing |
| ENSMUSG00000100725 | ENSFM00760001763162 | Unknown |
| ENSMUSG00000100725 | ENSFM00250000001411 | Heterogeneous Nuclear Ribonucleoprotein U Scaffold Attachment Factor Saf |
| ENSMUSG00000102226 | ENSFM00260000050975 | Brain Enriched Guanylate Kinase Associated |
| ENSMUSG00000102543 | ENSFM00780001917496 | Unknown |
| ENSMUSG00000102543 | ENSFM00730001521313 | Protocadherin Gamma Precursor Pcdh Gamma |
| ENSMUSG00000102543 | ENSFM00780001898335 | Protocadherin Precursor |
| ENSMUSG00000102697 | ENSFM00780001898335 | Protocadherin Precursor |
| ENSMUSG00000103888 | ENSFM00740001589112 | Microtubule Actin Cross Linking Factor 1 Actin Cross Linking Family 7 |
| ENSMUSG00000106379 | ENSFM00680001311925 | Unknown |
| ENSMUSG00000106379 | ENSFM00250000002406 | Lipoma Hmgic Fusion Partner |
| Voltage Dependent Calcium Channel Subunit Delta 1 | ||
| Nociceptin Orphanin Fq Ppnoc | ||
| Molybdopterin Molybdenumtransferase Mpt Mo Transferase EC_2.10.1.- 1 Domain E | ||
| Light | ||
| Cyclin Dependent Kinase 5 Activator 1 P25 P25 Tau Kinase Ii |
Supplementary Table 4c. Ensemble Families from zebrafish synaptosomal proteins
For each zebrafish synaptosomal protein its corresponding Ensembl Family code and descriptor are given.
| Ensembl Gene Id | Ensembl Protein Family Id(S) | Ensembl Family Description |
|---|---|---|
| ENSDARG00000000068 | ENSFM00740001589334 | Na + /H + Exchange Regulatory Cofactor Nhe RF1 Nherf 1 Ezrin Radixin Moesin Binding Phosphoprotein 50 EBP50 Regulatory Cofactor Of Na + /H + Exchanger Sodium Hydrogen Exchanger Regulatory Factor 1 Solute Carrier Family 9 A3 Regulatory Factor 1 |
| ENSDARG00000000086 | ENSFM00250000000789 | Intersectin |
| ENSDARG00000000103 | ENSFM00270000056424 | Myosin Cellular Myosin Heavy Chain Type Myosin Heavy Chain Myosin Heavy Chain Non Muscle Non Muscle Myosin Heavy Chain Nmmhc Non Muscle Myosin Heavy Chain Nmmhc Ii Nmmhc |
| ENSDARG00000000183 | ENSFM00440000236900 | Tyrosine Phosphatase Non Receptor Type EC_3.1.3.48 Tyrosine Phosphatase |
| ENSDARG00000000189 | ENSFM00730001521376 | Semaphorin Precursor Semaphorin Sema |
| ENSDARG00000000370 | ENSFM00740001589124 | EC_2.7.11.1 |
| ENSDARG00000000472 | ENSFM00800002003683 | Contactin 1 Precursor Neural Cell Surface F3 |
| ENSDARG00000000503 | ENSFM00250000000549 | Syntaxin |
| ENSDARG00000000551 | ENSFM00760001714847 | Neutral Amino Acid Transporter B 0 Atb 0 Sodium Dependent Neutral Amino Acid Transporter Type 2 Solute Carrier Family 1 Member 5 |
| ENSDARG00000000563 | ENSFM00800002009114 | Unknown |
| ENSDARG00000000563 | ENSFM00740001589077 | Titin EC_2.7.11.1 Connectin |
| ENSDARG00000000730 | ENSFM00730001521104 | Sodium And Chloride Dependent Transporter Solute Carrier Family 6 Member |
| ENSDARG00000000779 | ENSFM00790001963053 | SEC1 Family Domain Containing 1 SLY1 Homolog SLY1P Syntaxin Binding 1 2 |
| ENSDARG00000000861 | ENSFM00730001522046 | Scribble |
| ENSDARG00000001014 | ENSFM00270000056424 | Myosin Cellular Myosin Heavy Chain Type Myosin Heavy Chain Myosin Heavy Chain Non Muscle Non Muscle Myosin Heavy Chain Nmmhc Non Muscle Myosin Heavy Chain Nmmhc Ii Nmmhc |
| ENSDARG00000001127 | ENSFM00730001521551 | Vesicular Glutamate Transporter Solute Carrier Family 17 Member |
| ENSDARG00000001154 | ENSFM00740001589344 | Peripheral Type Benzodiazepine Receptor Associated 1 Prax 1 Rims Binding 1 Rim BP1 |
| ENSDARG00000001220 | ENSFM00250000001622 | E3 Ubiquitin Ligase MYCBP2 EC_6.3.2.- Myc Binding 2 Pam/Highwire/Rpm 1 Protein Associated With Myc |
| ENSDARG00000001241 | ENSFM00250000002682 | Poly U Binding Splicing Factor Kda Poly U Binding Splicing Factor |
| ENSDARG00000001437 | ENSFM00730001521168 | Solute Carrier Family 2 Facilitated Glucose Transporter Member Glucose Transporter Type Glut |
| ENSDARG00000001557 | ENSFM00740001589368 | Mitochondrial Import Receptor Subunit TOM34 Translocase Of Outer Membrane 34 Kda Subunit |
| ENSDARG00000001559 | ENSFM00740001589158 | Cub And Sushi Domain Containing Cub And Sushi Multiple Domains |
| ENSDARG00000001578 | ENSFM00500000270475 | Enoyl Coa Hydratase Mitochondrial Precursor EC_4.2.1.17 Enoyl Coa Hydratase 1 Short Chain Enoyl Coa Hydratase Sceh |
| ENSDARG00000001676 | ENSFM00250000000909 | Myelin Proteolipid Plp Lipophilin |
| ENSDARG00000001710 | ENSFM00780001898418 | Flotillin 1 |
| ENSDARG00000001729 | ENSFM00250000001473 | Striatin |
| ENSDARG00000001733 | ENSFM00250000004323 | Ptb Domain Containing Engulfment Adapter 1 Cell Death 6 Homolog Ptb Domain Adapter Ced 6 |
| ENSDARG00000001769 | ENSFM00740001589127 | Receptor Type Tyrosine Phosphatase Precursor Tyrosine Phosphatase R Ptp EC_3.1.3.48 |
| ENSDARG00000001788 | ENSFM00670001235592 | Atp Synthase Subunit O Mitochondrial Precursor Oligomycin Sensitivity Conferral Oscp |
| ENSDARG00000001835 | ENSFM00250000001647 | Sodium Leak Channel Non Selective Voltage Gated Channel 1 |
| ENSDARG00000001870 | ENSFM00650001139994 | Sodium/Potassium Transporting Atpase Subunit Alpha Na + /K + Atpase Alpha Subunit EC_3.6.3.9 Sodium Pump Subunit Alpha |
| ENSDARG00000001873 | ENSFM00500000270390 | D 3 Phosphoglycerate Dehydrogenase 3 Pgdh EC_1.1.1.95 |
| ENSDARG00000001881 | ENSFM00250000000524 | Voltage Dependent L Type Calcium Channel Subunit Beta Calcium Channel Voltage Dependent Subunit Beta |
| ENSDARG00000001890 | ENSFM00250000001541 | Dolichyl Diphosphooligosaccharide Glycosyltransferase Subunit Oligosaccharyl Transferase Subunit STT3 EC_2.4.99.18 |
| ENSDARG00000001939 | ENSFM00250000001080 | Galactosylgalactosylxylosylprotein 3 Beta Glucuronosyltransferase EC_2.4.1.135 Beta 1 3 Glucuronyltransferase Glucuronosyltransferase Glcat Udp Beta 1 3 Glucuronyltransferase Glcuat |
| ENSDARG00000001950 | ENSFM00740001589162 | Adenylate Kinase Isoenzyme 1 |
| ENSDARG00000001994 | ENSFM00250000001101 | Syntaxin Binding 1 Unc 18 Homolog 1 UNC18 1 Unc 18 Homolog A Unc 18A |
| ENSDARG00000002021 | ENSFM00250000001172 | Glycogen Phosphorylase Form EC_2.4.1.1 |
| ENSDARG00000002026 | ENSFM00400000131794 | Quaking |
| ENSDARG00000002082 | ENSFM00250000001591 | Casein Kinase Ii Subunit Ck Ii EC_2.7.11.1 |
| ENSDARG00000002127 | ENSFM00250000002061 | Signal Transducing Adapter Molecule Stam |
| ENSDARG00000002167 | ENSFM00250000000524 | Voltage Dependent L Type Calcium Channel Subunit Beta Calcium Channel Voltage Dependent Subunit Beta |
| ENSDARG00000002184 | ENSFM00250000004174 | Immunoglobulin Binding 1 ALPHA4 Phosphoprotein CD79A Binding 1 Phosphatase 2/4/6 Regulatory Subunit |
| ENSDARG00000002230 | ENSFM00500000270007 | Synaptophysin |
| ENSDARG00000002240 | ENSFM00810002040613 | Proteasome Subunit Beta Type 9 Precursor EC_3.4.25.1 Low Molecular Mass 2 Macropain Chain 7 Multicatalytic Endopeptidase Complex Chain 7 Proteasome Chain 7 Proteasome Subunit Beta 1I Really Interesting New Gene 12 |
| ENSDARG00000002249 | ENSFM00800002003855 | Thromboxane A Synthase Txa Synthase Txs EC_5.3.99.5 Cytochrome P450 5A1 |
| ENSDARG00000002255 | ENSFM00800002003400 | Unknown |
| ENSDARG00000002255 | ENSFM00800002024337 | Unknown |
| ENSDARG00000002305 | ENSFM00250000000979 | Nadp Dependent Malic Enzyme Nadp Me EC_1.1.1.40 Malic Enzyme |
| ENSDARG00000002467 | ENSFM00500000269664 | Retinol Dehydrogenase |
| ENSDARG00000002523 | ENSFM00500000270535 | Hydroxysteroid Dehydrogenase 2 Ec 1 |
| ENSDARG00000002536 | ENSFM00780001898361 | Transmembrane 9 Superfamily Member Precursor |
| ENSDARG00000002538 | ENSFM00250000004547 | WD40 Repeat Containing SMU1 Smu 1 Suppressor Of Mec 8 And Unc 52 Homolog |
| ENSDARG00000002564 | ENSFM00670001235552 | Synaptogyrin |
| ENSDARG00000002571 | ENSFM00500000269813 | Tropomodulin Tropomodulin Tmod |
| ENSDARG00000002587 | ENSFM00250000000643 | Dihydropyrimidinase Related Drp Collapsin Response Mediator Crmp |
| ENSDARG00000002591 | ENSFM00730001521814 | Ruvb 1 EC_3.6.4.12 |
| ENSDARG00000002600 | ENSFM00250000000295 | Proprotein Convertase Subtilisin/Kexin Type Precursor EC_3.4.21.- Convertase |
| ENSDARG00000002632 | ENSFM00760001714601 | Annexin Annexin Annexin |
| ENSDARG00000002642 | ENSFM00250000000986 | Endophilin SH3 Domain Containing GRB2 |
| ENSDARG00000002656 | ENSFM00500000269888 | Syntaxin Binding 5 Lethal 2 Giant Larvae Homolog Tomosyn |
| ENSDARG00000002659 | ENSFM00250000000867 | Microtubule Associated Rp/Eb Family Member |
| ENSDARG00000002690 | ENSFM00670001236256 | Guanine Nucleotide Exchange Factor MSS4 Rab Interacting Factor |
| ENSDARG00000002710 | ENSFM00500000270420 | Nucleolin |
| ENSDARG00000002768 | ENSFM00670001235702 | Parvalbumin Beta |
| ENSDARG00000002790 | ENSFM00730001521696 | Ap 2 Complex Subunit Mu Ap 2 Mu Chain Clathrin Assembly Complex 2 Mu Medium Chain Clathrin Coat Assembly AP50 Clathrin Coat Associated AP50 MU2 Adaptin Plasma Membrane Adaptor Ap Kda |
| ENSDARG00000002791 | ENSFM00650001139994 | Sodium/Potassium Transporting Atpase Subunit Alpha Na + /K + Atpase Alpha Subunit EC_3.6.3.9 Sodium Pump Subunit Alpha |
| ENSDARG00000002816 | ENSFM00250000001613 | Ras Specific Guanine Nucleotide Releasing Factor 2 Ras GRF2 Ras Guanine Nucleotide Exchange Factor 2 |
| ENSDARG00000002840 | ENSFM00500000269652 | Sarcoplasmic/Endoplasmic Reticulum Calcium Atpase Sr Ca 2+ Atpase EC_3.6.3.8 Calcium Pump Calcium Transporting Atpase Sarcoplasmic Reticulum Type Twitch Skeletal Muscle Endoplasmic Reticulum Class 1/2 Ca 2+ Atpase |
| ENSDARG00000002877 | ENSFM00250000003177 | RALBP1 Associated Eps Domain Containing 1 RALBP1 Interacting 1 |
| ENSDARG00000002880 | ENSFM00250000002464 | Coiled Coil Domain Containing 132 |
| ENSDARG00000002949 | ENSFM00770001847529 | Serine/Threonine Phosphatase 6 Catalytic Subunit PP6C EC_3.1.3.16 Phosphatase |
| ENSDARG00000002956 | ENSFM00730001521375 | Integrin Alpha Precursor CD11 Antigen Family Member Glycoprotein Alpha Leukocyte Antigen |
| ENSDARG00000002991 | ENSFM00250000002276 | Testis Specific Gene 10 |
| ENSDARG00000003008 | ENSFM00750001632342 | Kinase C EC_2.7.11.13 |
| ENSDARG00000003020 | ENSFM00260000050662 | Leucine Rich Repeat Containing Precursor Netrin Ligand Ngl |
| ENSDARG00000003032 | ENSFM00730001521378 | Atp Dependent Rna Helicase EIF4A EC_3.6.4.13 Eukaryotic Initiation Factor 4A Eif 4A Translation Initiation Factor 1 |
| ENSDARG00000003035 | ENSFM00730001521528 | Unknown |
| ENSDARG00000003046 | ENSFM00550000743175 | Sorbin And SH3 Domain Containing 2 Arg/Abl Interacting 2 ARGBP2 |
| ENSDARG00000003087 | ENSFM00250000005080 | Ubx Domain Containing 6 Ubx Domain Containing 1 |
| ENSDARG00000003127 | ENSFM00250000004463 | Monooxygenase P33MONOX Ec 1 |
| ENSDARG00000003146 | ENSFM00730001521354 | Neurofilament Polypeptide Nf Kda Neurofilament Neurofilament Triplet |
| ENSDARG00000003151 | ENSFM00670001235396 | E3 Ubiquitin Ligase Complex Scf Subunit Sconc Sulfur Controller C Sulfur Metabolite Repression Control C |
| ENSDARG00000003169 | ENSFM00250000000552 | Membrane Associated Guanylate Kinase Ww And Pdz Domain Containing Membrane Associated Guanylate Kinase Inverted Magi |
| ENSDARG00000003189 | ENSFM00250000002209 | 26S Proteasome Non Atpase Regulatory Subunit 1 26S Proteasome Regulatory Subunit RPN2 26S Proteasome Regulatory Subunit S1 |
| ENSDARG00000003216 | ENSFM00730001521439 | Annexin Annexin Annexin Calpactin Calpactin Chromobindin Lipocortin I |
| ENSDARG00000003307 | ENSFM00500000271003 | Syntaxin 16 |
| ENSDARG00000003308 | ENSFM00250000004805 | Er Membrane Complex Subunit 2 Tetratricopeptide Repeat 35 Tpr Repeat 35 |
| ENSDARG00000003320 | ENSFM00250000003694 | Unknown |
| ENSDARG00000003375 | ENSFM00630001060922 | Unknown |
| ENSDARG00000003375 | ENSFM00630001070087 | Unknown |
| ENSDARG00000003380 | ENSFM00250000000775 | Peroxisomal N 1 Acetyl Spermine/Spermidine Oxidase EC_1.5.3.13 Polyamine Oxidase |
| ENSDARG00000003403 | ENSFM00250000000375 | Teneurin Ten Odd Oz/Ten M Homolog Tenascin Ten Teneurin Transmembrane |
| ENSDARG00000003444 | ENSFM00500000271510 | Dehydrogenase/Reductase Sdr Family Member 7 Precursor EC_1.1.-.- Retinal Short Chain Dehydrogenase/Reductase 4 RETSDR4 |
| ENSDARG00000003449 | ENSFM00800002003845 | Camp And Camp Inhibited Cgmp 3' 5' Cyclic Phosphodiesterase 10A EC_3.1.4.17 EC_3.1.4.- 35 |
| ENSDARG00000003462 | ENSFM00250000003660 | Ferrochelatase Mitochondrial Precursor EC_4.99.1.1 Heme Synthase Protoheme Ferro Lyase |
| ENSDARG00000003486 | ENSFM00420000140518 | Serine/Threonine Phosphatase PP1 Catalytic Subunit Pp EC_3.1.3.16 |
| ENSDARG00000003495 | ENSFM00250000001066 | Map Kinase Activating Death Domain RAB3 Exchange Factor |
| ENSDARG00000003512 | ENSFM00250000001010 | Ankyrin Repeat And Sterile Alpha Motif Domain Containing 1B Amyloid Beta Intracellular Domain Associated 1 Aida 1 E2A PBX1 Associated Eb 1 |
| ENSDARG00000003517 | ENSFM00610000973509 | Unknown |
| ENSDARG00000003517 | ENSFM00500000270262 | Inositol Monophosphatase 1 Imp 1 Impase 1 EC_3.1.3.25 |
| ENSDARG00000003519 | ENSFM00500000272306 | Adenine Phosphoribosyltransferase Aprt EC_2.4.2.7 |
| ENSDARG00000003526 | ENSFM00500000270276 | Proteasome Subunit Alpha Type 5 EC_3.4.25.1 Macropain Multicatalytic Endopeptidase Complex Proteasome |
| ENSDARG00000003544 | ENSFM00250000000663 | Double Stranded Rna Specific Editase EC_3.5.-.- Rna Adenosine Deaminase Rna Editing Deaminase Rna Editing Enzyme Dsrna Adenosine Deaminase |
| ENSDARG00000003570 | ENSFM00760001714713 | Endoplasmin 94 Kda Glucose Regulated Grp 94 Heat Shock 90 Kda Beta Member 1 |
| ENSDARG00000003592 | ENSFM00700001390183 | Surfeit Locus |
| ENSDARG00000003599 | ENSFM00810002040577 | 60S Ribosomal L3 |
| ENSDARG00000003641 | ENSFM00600000921432 | Tfg Trk Fused Gene |
| ENSDARG00000003655 | ENSFM00500000272495 | Transmembrane 59 Precursor Brain Specific Membrane Anchored |
| ENSDARG00000003675 | ENSFM00250000003625 | Lambda Crystallin EC_1.1.1.45 L Gulonate 3 Dehydrogenase GUL3DH |
| ENSDARG00000003693 | ENSFM00770001847539 | Histidine Trna Ligase EC_6.1.1.21 Histidyl Trna Synthetase Hisrs |
| ENSDARG00000003695 | ENSFM00250000001189 | Voltage Dependent Anion Selective Channel Vdac Outer Mitochondrial Membrane Porin |
| ENSDARG00000003754 | ENSFM00800002003594 | CD9 Antigen CD9 Antigen |
| ENSDARG00000003776 | ENSFM00260000050567 | Phosphatidylinositol 5 Phosphate 4 Kinase Type 2 EC_2.7.1.149 1 Phosphatidylinositol 5 Phosphate 4 Kinase 2 Diphosphoinositide Kinase 2 Phosphatidylinositol 5 Phosphate 4 Kinase Type Ii Pi 5 P 4 Kinase Type Ii PIP4KII Ptdins 5 P 4 Kinase 2 |
| ENSDARG00000003779 | ENSFM00730001521673 | Catenin Delta 2 |
| ENSDARG00000003795 | ENSFM00770001847524 | Isocitrate Dehydrogenase Cytoplasmic Idh EC_1.1.1.42 Cytosolic Nadp Isocitrate Dehydrogenase Idp Nadp + Specific Icdh Oxalosuccinate Decarboxylase |
| ENSDARG00000003811 | ENSFM00730001521536 | Plexin Precursor |
| ENSDARG00000003829 | ENSFM00730001521512 | Polypeptide N Acetylgalactosaminyltransferase EC_2.4.1.41 Polypeptide Galnac Transferase Galnac Pp Gantase Udp Acetylgalactosaminyltransferase Udp Galnac:Polypeptide N Acetylgalactosaminyltransferase |
| ENSDARG00000003835 | ENSFM00250000000738 | Stomatin |
| ENSDARG00000003854 | ENSFM00500000269666 | Long Chain Fatty Acid Coa Ligase EC_6.2.1.3 Long Chain Acyl Coa Synthetase Lacs |
| ENSDARG00000003866 | ENSFM00780001898383 | Transmembrane 9 Superfamily Member 2 Precursor |
| ENSDARG00000003869 | ENSFM00500000271494 | 2 4 Dienoyl Coa Reductase Mitochondrial Precursor EC_1.3.1.34 2 4 Dienoyl Coa Reductase |
| ENSDARG00000003931 | ENSFM00800002003448 | Cytosolic Non Specific Dipeptidase EC_3.4.13.18 Cndp Dipeptidase 2 |
| ENSDARG00000003940 | ENSFM00610000967182 | Unknown |
| ENSDARG00000003940 | ENSFM00250000006972 | Mitochondrial Fission Regulator 1 |
| ENSDARG00000003952 | ENSFM00670001235608 | Profilin Profilin |
| ENSDARG00000003961 | ENSFM00730001522506 | Poly Synthase 3 Padprt 3 |
| ENSDARG00000003963 | ENSFM00500000270778 | Tnf Receptor Associated Factor 4 Cysteine Rich Associated Ring And Traf Domains 1 |
| ENSDARG00000003994 | ENSFM00720001492041 | Synaptotagmin Synaptotagmin |
| ENSDARG00000003998 | ENSFM00250000003309 | Phytanoyl Coa Hydroxylase Interacting Phytanoyl Coa Hydroxylase Associated 1 Pahx AP1 PAHXAP1 |
| ENSDARG00000004017 | ENSFM00740001589368 | Mitochondrial Import Receptor Subunit TOM34 Translocase Of Outer Membrane 34 Kda Subunit |
| ENSDARG00000004055 | ENSFM00250000002015 | UHRF1 Binding 1 |
| ENSDARG00000004078 | ENSFM00730001521866 | Long Chain Fatty Acid Coa Ligase EC_6.2.1.3 Long Chain Acyl Coa Synthetase Lacs |
| ENSDARG00000004151 | ENSFM00400000131956 | Ras Related Rab 23 Precursor |
| ENSDARG00000004173 | ENSFM00260000050647 | Coatomer Subunit Alpha Alpha Coat Alpha Cop Hep Cop Hepcop |
| ENSDARG00000004174 | ENSFM00780001898388 | CCR4 Not Transcription Complex Subunit 1 CCR4 Associated Factor 1 |
| ENSDARG00000004261 | ENSFM00500000270043 | Transmembrane EMP24 Domain Containing Precursor |
| ENSDARG00000004301 | ENSFM00730001521186 | Ras Related C3 Botulinum Toxin Substrate Precursor P21 |
| ENSDARG00000004305 | ENSFM00250000002390 | Vang 2 Van Gogh |
| ENSDARG00000004336 | ENSFM00250000002477 | Exocyst Complex Component 4 Exocyst Complex Component SEC8 |
| ENSDARG00000004386 | ENSFM00250000001885 | Precursor Estrogen Induced Gene 121 |
| ENSDARG00000004406 | ENSFM00250000004781 | Sepiapterin Reductase Spr EC_1.1.1.153 |
| ENSDARG00000004445 | ENSFM00730001521627 | Metabotropic Glutamate Receptor Precursor |
| ENSDARG00000004470 | ENSFM00500000271338 | Glucosidase 2 Subunit Beta Precursor 80K H Glucosidase Ii Subunit Beta Kinase C Substrate 60.1 Kda Heavy Chain Pkcsh |
| ENSDARG00000004527 | ENSFM00670001235905 | Peptidyl Prolyl Cis Trans Isomerase Nima Interacting 4 EC_5.2.1.8 Parvulin 14 PAR14 Peptidyl Prolyl Cis Trans Isomerase PIN4 Ppiase PIN4 Rotamase PIN4 |
| ENSDARG00000004561 | ENSFM00520000517802 | Kinase C Type Pkc EC_2.7.11.13 |
| ENSDARG00000004581 | ENSFM00760001714807 | Sel 1 Homolog Precursor Suppressor Of Lin 12 Sel |
| ENSDARG00000004597 | ENSFM00260000050662 | Leucine Rich Repeat Containing Precursor Netrin Ligand Ngl |
| ENSDARG00000004621 | ENSFM00250000000909 | Myelin Proteolipid Plp Lipophilin |
| ENSDARG00000004634 | ENSFM00650001148889 | Unknown |
| ENSDARG00000004634 | ENSFM00500000269873 | Oxysterol Binding |
| ENSDARG00000004635 | ENSFM00730001521142 | Ephrin Type A Receptor Precursor EC_2.7.10.1 Kinase |
| ENSDARG00000004648 | ENSFM00260000050545 | Btb/Poz Domain Containing |
| ENSDARG00000004680 | ENSFM00570000851144 | Dnaj Homolog Subfamily B Member |
| ENSDARG00000004687 | ENSFM00500000269858 | 3 Ketoacyl Coa Thiolase Peroxisomal Precursor EC_2.3.1.16 Acetyl Coa Acyltransferase Beta Ketothiolase Peroxisomal 3 Oxoacyl Coa Thiolase |
| ENSDARG00000004754 | ENSFM00360000109870 | Unknown |
| ENSDARG00000004796 | ENSFM00250000001774 | E3 Ubiquitin Ligase MGRN1 EC_6.3.2.- Mahogunin Ring Finger 1 |
| ENSDARG00000004830 | ENSFM00630001070744 | Unknown |
| ENSDARG00000004830 | ENSFM00780001898447 | Flotillin 2 |
| ENSDARG00000004836 | ENSFM00500000270505 | Dnaj Homolog Subfamily C Member 5B Cysteine String Beta Csp Beta |
| ENSDARG00000004870 | ENSFM00630001071866 | Unknown |
| ENSDARG00000004870 | ENSFM00250000001423 | Presenilin Ps EC_3.4.23.- |
| ENSDARG00000004906 | ENSFM00810002040902 | Stress Induced Phosphoprotein 1 STI1 HSC70/HSP90 Organizing Hop |
| ENSDARG00000004930 | ENSFM00610000965711 | Unknown |
| ENSDARG00000004930 | ENSFM00250000003075 | Lim Domain Only 7 Lmo 7 F Box Only 20 Lomp |
| ENSDARG00000004931 | ENSFM00710001441664 | Sodium/Calcium Exchanger 1 Precursor Na + /Ca 2+ Exchange 1 Solute Carrier Family 8 Member 1 |
| ENSDARG00000004964 | ENSFM00730001521393 | Cytochrome P450 Lauric Acid Omega Hydroxylase EC_1.14.15.3 |
| ENSDARG00000004988 | ENSFM00310000089007 | Serine/Threonine Phosphatase 2B Catalytic Subunit EC_3.1.3.16 Cam Prp Catalytic Subunit Calmodulin Dependent Calcineurin A Subunit |
| ENSDARG00000005085 | ENSFM00630001072006 | Unknown |
| ENSDARG00000005085 | ENSFM00500000272604 | Gamma Glutamylcyclotransferase EC_2.3.2.4 |
| ENSDARG00000005122 | ENSFM00500000269652 | Sarcoplasmic/Endoplasmic Reticulum Calcium Atpase Sr Ca 2+ Atpase EC_3.6.3.8 Calcium Pump Calcium Transporting Atpase Sarcoplasmic Reticulum Type Twitch Skeletal Muscle Endoplasmic Reticulum Class 1/2 Ca 2+ Atpase |
| ENSDARG00000005134 | ENSFM00250000003951 | 26S Proteasome Non Atpase Regulatory Subunit 26S Proteasome Regulatory Subunit RPN6 |
| ENSDARG00000005141 | ENSFM00800002003895 | Cam Kinase Vesicle Associated |
| ENSDARG00000005159 | ENSFM00250000001040 | Cytohesin Ph SEC7 And Coiled Coil Domain Containing |
| ENSDARG00000005162 | ENSFM00610000969203 | Unknown |
| ENSDARG00000005162 | ENSFM00250000000220 | Tropomyosin Alpha 1 Chain Alpha Tropomyosin Tropomyosin 1 |
| ENSDARG00000005163 | ENSFM00250000001283 | EFR3 Homolog |
| ENSDARG00000005191 | ENSFM00730001521588 | Adenosylhomocysteinase Adohcyase EC_3.3.1.1 S Adenosyl L Homocysteine Hydrolase |
| ENSDARG00000005210 | ENSFM00730001521970 | Coronin |
| ENSDARG00000005220 | ENSFM00500000271967 | Programmed Cell Death 6 Probable Calcium Binding Alg 2 |
| ENSDARG00000005251 | ENSFM00810002040663 | Ribulose Phosphate 3 Epimerase EC_5.1.3.1 5 Phosphate Epimerase |
| ENSDARG00000005350 | ENSFM00250000000652 | Disco Interacting 2 Homolog DIP2 Homolog |
| ENSDARG00000005359 | ENSFM00250000001376 | Succinyl Coa Ligase Subunit Beta Mitochondrial Precursor EC_6.2.1.5 Atp Specific Succinyl Coa Synthetase Subunit Beta Succinyl Coa Synthetase Beta A Chain Scs Betaa |
| ENSDARG00000005372 | ENSFM00770001847668 | Calcium/Calmodulin Dependent Kinase Type Iv Camk Iv EC_2.7.11.17 Cam Kinase Gr |
| ENSDARG00000005397 | ENSFM00750001632380 | Tripartite Motif Containing 2 EC_6.3.2.- E3 Ubiquitin Ligase TRIM2 |
| ENSDARG00000005423 | ENSFM00250000001043 | Phosphoglycerate Mutase EC_3.1.3.13 EC_5.4.2.- 11 EC_5.4.-.- 2 4 Bpg Dependent Pgam Phosphoglycerate Mutase Isozyme Pgam |
| ENSDARG00000005425 | ENSFM00750001642432 | Unknown |
| ENSDARG00000005425 | ENSFM00250000002558 | Wd Repeat And Fyve Domain Containing WD40 And Fyve Domain Containing |
| ENSDARG00000005451 | ENSFM00800002003449 | Rab Gdp Dissociation Inhibitor Alpha Rab Gdi Alpha Guanosine Diphosphate Dissociation Inhibitor 1 Gdi 1 |
| ENSDARG00000005479 | ENSFM00250000000375 | Teneurin Ten Odd Oz/Ten M Homolog Tenascin Ten Teneurin Transmembrane |
| ENSDARG00000005482 | ENSFM00250000001338 | Rap Guanine Nucleotide Exchange Factor 2 Cyclic Nucleotide Ras Gef Cnrasgef Neural Rap Guanine Nucleotide Exchange Nrap Gep Pdz Domain Containing Guanine Nucleotide Exchange Factor 1 Pdz GEF1 Ra Gef 1 Ras/RAP1 Associating Gef 1 |
| ENSDARG00000005513 | ENSFM00500000270490 | Nascent Polypeptide Associated Complex Subunit Alpha Nac Alpha Alpha Nac |
| ENSDARG00000005547 | ENSFM00250000003702 | RAD50 Interacting 1 RAD50 Interactor 1 Rint 1 |
| ENSDARG00000005560 | ENSFM00730001521154 | Unknown |
| ENSDARG00000005643 | ENSFM00730001522241 | 2 Amino 3 Ketobutyrate Coenzyme A Ligase Mitochondrial Precursor Akb Ligase EC_2.3.1.29 Aminoacetone Synthase Glycine Acetyltransferase |
| ENSDARG00000005645 | ENSFM00740001589121 | Roundabout Homolog Precursor |
| ENSDARG00000005690 | ENSFM00500000269757 | Calcium Binding Mitochondrial Carrier Scamc Small Calcium Binding Mitochondrial Carrier Solute Carrier Family 25 Member |
| ENSDARG00000005699 | ENSFM00760001714673 | Atp Dependent Rna Helicase DDX25 EC_3.6.4.13 Dead Box 25 Rna Helicase |
| ENSDARG00000005739 | ENSFM00250000000909 | Myelin Proteolipid Plp Lipophilin |
| ENSDARG00000005749 | ENSFM00250000001833 | Cullin Associated NEDD8 Dissociated Cullin Associated And Neddylation Dissociated Tbp Interacting Of 120 Kda Tbp Interacting P120 |
| ENSDARG00000005754 | ENSFM00740001589097 | Receptor Type Tyrosine Phosphatase Precursor EC_3.1.3.48 |
| ENSDARG00000005774 | ENSFM00770001847530 | Atp Dependent Rna Helicase DDX3Y EC_3.6.4.13 Dead Box 3 Y Chromosomal |
| ENSDARG00000005775 | ENSFM00740001589070 | Sodium Channel Type Subunit Alpha Sodium Channel Sodium Channel Type Subunit Alpha Voltage Gated Sodium Channel Subunit Alpha NAV1 |
| ENSDARG00000005791 | ENSFM00500000271918 | 60S Ribosomal L28 |
| ENSDARG00000005840 | ENSFM00730001521567 | Aspartate Aminotransferase Mitochondrial Precursor Maspat EC_2.6.1.1 EC_2.6.1.- 7 Fatty Acid Binding Fabp 1 Glutamate Oxaloacetate Transaminase 2 Kynurenine Aminotransferase 4 Kynurenine Aminotransferase Iv Kynurenine Oxoglutarate Transaminase 4 Kynurenin |
| ENSDARG00000005853 | ENSFM00250000000816 | Adp/Atp Translocase Adp Atp Carrier Adenine Nucleotide Translocator Ant Solute Carrier Family 25 Member |
| ENSDARG00000005861 | ENSFM00250000000944 | Ral Gtpase Activating Subunit Alpha 1 Gap Related Interacting Partner To E12 Gripe Gtpase Activating Domain 1 Tuberin 1 P240 |
| ENSDARG00000005891 | ENSFM00650001147970 | Unknown |
| ENSDARG00000005891 | ENSFM00730001521809 | Nadh Cytochrome B5 Reductase B5R EC_1.6.2.2 |
| ENSDARG00000005897 | ENSFM00250000005338 | Deoxyribose Phosphate Aldolase Dera EC_4.1.2.4 |
| ENSDARG00000005924 | ENSFM00250000000235 | Precursor |
| ENSDARG00000005926 | ENSFM00810002040800 | Adenylate Kinase |
| ENSDARG00000005941 | ENSFM00730001521177 | Tyrosine Kinase EC_2.7.10.2 |
| ENSDARG00000006019 | ENSFM00250000001413 | Transketolase EC_2.2.1.1 |
| ENSDARG00000006025 | ENSFM00500000272770 | Tumor P53 Inducible 11 P53 Induced Gene 11 |
| ENSDARG00000006029 | ENSFM00810002040760 | Leukotriene A 4 Hydrolase Lta 4 Hydrolase EC_3.3.2.6 Leukotriene A 4 Hydrolase |
| ENSDARG00000006031 | ENSFM00810002040728 | 4 Aminobutyrate Aminotransferase Mitochondrial Precursor EC_2.6.1.19 S 3 Amino 2 Methylpropionate Transaminase EC_2.6.1.- 22 Gaba Aminotransferase Gaba At Gamma Amino N Butyrate Transaminase Gaba Transaminase Gaba T L Aibat |
| ENSDARG00000006040 | ENSFM00250000005150 | Cytochrome P450 20A1 EC_1.14.-.- |
| ENSDARG00000006062 | ENSFM00250000003470 | A Kinase Anchor 1 Mitochondrial Precursor A Kinase Anchor Kda Akap Dual Specificity A Kinase Anchoring 1 D Akap 1 Kinase A Anchoring 1 PRKA1 Spermatid A Kinase Anchor 84 S AKAP84 |
| ENSDARG00000006094 | ENSFM00740001589412 | Cation Independent Mannose 6 Phosphate Receptor Precursor Ci Man 6 P Receptor Ci Mpr M6PR 300 Kda Mannose 6 Phosphate Receptor Mpr 300 Insulin Growth Factor 2 Receptor Insulin Growth Factor Ii Receptor Igf Ii Receptor M6P/IGF2 Receptor M6P/IGF2R CD222 A |
| ENSDARG00000006125 | ENSFM00800002003414 | Casein Kinase I Delta Cki Delta Ckid EC_2.7.11.1 |
| ENSDARG00000006128 | ENSFM00740001589275 | Unknown |
| ENSDARG00000006225 | ENSFM00810002040601 | Spliceosome Rna Helicase DDX39B EC_3.6.4.13.56 Kda U2AF65 Associated Dead Box UAP56 |
| ENSDARG00000006240 | ENSFM00730001521827 | Long Chain Fatty Acid Transport 4 Fatp 4 Fatty Acid Transport 4 EC_6.2.1.- Solute Carrier Family 27 Member 4 |
| ENSDARG00000006257 | ENSFM00740001589117 | Low Density Lipoprotein Receptor Receptor |
| ENSDARG00000006260 | ENSFM00250000000217 | Tubulin Alpha Chain |
| ENSDARG00000006266 | ENSFM00500000269705 | Signal Transducer And Activator Of Transcription |
| ENSDARG00000006290 | ENSFM00670001238662 | Nadh Dehydrogenase Iron Sulfur 5 |
| ENSDARG00000006299 | ENSFM00250000001135 | Rho Guanine Nucleotide Exchange Factor 6 Rac/CDC42 Guanine Nucleotide Exchange Factor 6 |
| ENSDARG00000006314 | ENSFM00500000269717 | Integrin Alpha Precursor Receptor Subunit Alpha |
| ENSDARG00000006316 | ENSFM00670001235314 | 60S Ribosomal |
| ENSDARG00000006356 | ENSFM00800002003873 | Vesicular Acetylcholine Transporter Vacht Solute Carrier Family 18 Member 3 |
| ENSDARG00000006370 | ENSFM00250000003378 | V Type Proton Atpase Subunit H V Atpase Subunit H Vacuolar Proton Pump Subunit H |
| ENSDARG00000006383 | ENSFM00500000269888 | Syntaxin Binding 5 Lethal 2 Giant Larvae Homolog Tomosyn |
| ENSDARG00000006396 | ENSFM00390000126295 | Neural Cell Adhesion Molecule L1 L1 L1 |
| ENSDARG00000006399 | ENSFM00730001521388 | Unknown |
| ENSDARG00000006413 | ENSFM00500000276121 | Unknown |
| ENSDARG00000006497 | ENSFM00630001071959 | Unknown |
| ENSDARG00000006497 | ENSFM00500000269689 | Reticulon |
| ENSDARG00000006546 | ENSFM00500000270040 | Adenylate Kinase 4 Mitochondrial |
| ENSDARG00000006553 | ENSFM00770001847543 | Ras Related R Ras |
| ENSDARG00000006585 | ENSFM00250000001646 | Dystrophia Myotonica Wd Repeat Containing Dystrophia Myotonica Containing Wd Repeat Motif Protein Dmr N9 |
| ENSDARG00000006601 | ENSFM00250000003884 | Kazrin |
| ENSDARG00000006609 | ENSFM00780001898380 | Dual Specificity Mitogen Activated Kinase Kinase 1 Map Kinase Kinase 1 Mapkk 1 EC_2.7.12.2 Erk Activator Kinase 1 Mapk/Erk Kinase 1 |
| ENSDARG00000006617 | ENSFM00250000005767 | Gamma Soluble Nsf Attachment Snap Gamma N Ethylmaleimide Sensitive Factor Attachment Gamma |
| ENSDARG00000006639 | ENSFM00250000003767 | Lunapark |
| ENSDARG00000006642 | ENSFM00730001521857 | Developmentally Regulated Gtp Binding 2 Drg 2 |
| ENSDARG00000006691 | ENSFM00250000002131 | 60S Ribosomal L12 |
| ENSDARG00000006712 | ENSFM00500000273445 | Was/Wasl Interacting Family Member 2 Wasp Interacting Related Wip Related |
| ENSDARG00000006766 | ENSFM00250000002294 | Staphylococcal Nuclease Domain Containing 1.100 Kda Coactivator P100 Co Activator |
| ENSDARG00000006797 | ENSFM00250000004579 | 5' Nucleotidase Domain Containing 1 EC_3.1.3.- |
| ENSDARG00000006832 | ENSFM00500000269893 | Polypeptide N Acetylgalactosaminyltransferase EC_2.4.1.41 Polypeptide Galnac Transferase Galnac T Pp Gantase Udp Acetylgalactosaminyltransferase Udp Galnac:Polypeptide N Acetylgalactosaminyltransferase |
| ENSDARG00000006849 | ENSFM00250000000608 | Acid Sensing Ion Channel Amiloride Sensitive Cation Channel |
| ENSDARG00000006863 | ENSFM00360000109952 | Coiled Coil Domain Containing 108 |
| ENSDARG00000006900 | ENSFM00760001714645 | Inosine 5' Monophosphate Dehydrogenase |
| ENSDARG00000006915 | ENSFM00650001147969 | Unknown |
| ENSDARG00000006915 | ENSFM00410000138507 | Adp Ribosylation Factor Small G Indispensable Equal Chromosome Segregation |
| ENSDARG00000006923 | ENSFM00740001589100 | Voltage Dependent Type Calcium Channel Subunit Alpha Brain Calcium Channel Calcium Channel L Type Alpha 1 Polypeptide Voltage Gated Calcium Channel Subunit Alpha CAV2 |
| ENSDARG00000006963 | ENSFM00250000005028 | Exportin 2 EXP2 Chromosome Segregation 1 Importin Alpha Re Exporter |
| ENSDARG00000007025 | ENSFM00670001236445 | Tetratricopeptide Repeat 9C Tpr Repeat 9C |
| ENSDARG00000007034 | ENSFM00250000001382 | Heterogeneous Nuclear Ribonucleoprotein K Hnrnp K |
| ENSDARG00000007034 | ENSFM00540000725842 | Hnrpkl |
| ENSDARG00000007098 | ENSFM00600000921249 | Integral Membrane 2B Immature BRI2 IMBRI2 Transmembrane Bri Bri |
| ENSDARG00000007127 | ENSFM00760001714915 | Acetyl Coa Acetyltransferase Cytosolic EC_2.3.1.9 Cytosolic Acetoacetyl Coa Thiolase |
| ENSDARG00000007136 | ENSFM00500000272182 | SH3 Domain Binding 5 SH3BP 5 |
| ENSDARG00000007141 | ENSFM00550000743154 | 26S Protease Regulatory Subunit 6A 26S Proteasome Aaa Atpase Subunit RPT5 Proteasome 26S Subunit Atpase 3 Tat Binding Tbp |
| ENSDARG00000007149 | ENSFM00390000126295 | Neural Cell Adhesion Molecule L1 L1 L1 |
| ENSDARG00000007172 | ENSFM00730001521329 | Plexin Precursor |
| ENSDARG00000007195 | ENSFM00790001963004 | Metabotropic Glutamate Receptor Precursor |
| ENSDARG00000007196 | ENSFM00780001898474 | Export Factor RAE1 Homolog Associated Mrnp 41 |
| ENSDARG00000007216 | ENSFM00250000004383 | Translation Initiation Factor RLI1 Atp Binding Cassette Sub Family E Member RLI1 Rnase L Inhibitor |
| ENSDARG00000007219 | ENSFM00440000236866 | Alpha Actinin Alpha Actinin F Actin Cross Linking |
| ENSDARG00000007220 | ENSFM00250000000779 | Neural Cell Adhesion Molecule 1 Precursor N Cam 1 Ncam 1 |
| ENSDARG00000007244 | ENSFM00800002003509 | Acid Phosphatase Precursor EC_3.1.3.2 |
| ENSDARG00000007247 | ENSFM00250000001722 | Synembryn Ric |
| ENSDARG00000007250 | ENSFM00250000004790 | Vesicle Transport USE1 USE1 |
| ENSDARG00000007257 | ENSFM00500000269759 | Ras Related Rab |
| ENSDARG00000007264 | ENSFM00250000003191 | Nipsnap Homolog 2 NIPSNAP2 Glioblastoma Amplified Sequence |
| ENSDARG00000007275 | ENSFM00720001492013 | Glutamate Receptor Ionotropic Kainate Precursor Glutamate Receptor |
| ENSDARG00000007294 | ENSFM00250000002510 | Aconitate Hydratase Mitochondrial Precursor Aconitase EC_4.2.1.3 Citrate Hydro Lyase |
| ENSDARG00000007320 | ENSFM00250000000871 | 60S Ribosomal L7 |
| ENSDARG00000007323 | ENSFM00630001070600 | Unknown |
| ENSDARG00000007323 | ENSFM00600000921240 | Charged Multivesicular Body Chromatin Modifying |
| ENSDARG00000007359 | ENSFM00500000273690 | Ubiquitin 4A |
| ENSDARG00000007370 | ENSFM00730001521574 | Fer 1 |
| ENSDARG00000007385 | ENSFM00500000269991 | T Complex 1 Subunit Eta Tcp 1 Eta Cct Eta |
| ENSDARG00000007412 | ENSFM00730001521168 | Solute Carrier Family 2 Facilitated Glucose Transporter Member Glucose Transporter Type Glut |
| ENSDARG00000007480 | ENSFM00250000000935 | Retinoid Isomerohydrolase EC_3.1.1.64 All Trans Retinyl Palmitate Hydrolase Retinal Pigment Epithelium Specific 65 Kda Retinol Isomerase |
| ENSDARG00000007496 | ENSFM00250000003769 | E3 UFM1 Ligase 1 EC_6.3.2.- |
| ENSDARG00000007526 | ENSFM00250000004138 | Nadh Dehydrogenase Iron Sulfur 2 Mitochondrial Precursor EC_1.6.5.3 EC_1.6.99.- 3 Complex I 49KD Ci 49KD Nadh Ubiquinone Oxidoreductase 49 Kda Subunit |
| ENSDARG00000007654 | ENSFM00800002003482 | Vesicle Fusing Atpase EC_3.6.4.6 N Ethylmaleimide Sensitive Fusion Nem Sensitive Fusion Vesicular Fusion Nsf |
| ENSDARG00000007663 | ENSFM00250000000861 | Bridging Integrator |
| ENSDARG00000007678 | ENSFM00250000001068 | Tweety Homolog |
| ENSDARG00000007697 | ENSFM00800002003523 | Fatty Acid Binding Protein Heart Fatty Acid Binding 3 Heart Type Fatty Acid Binding H Fabp |
| ENSDARG00000007720 | ENSFM00670001235917 | Activated Rna Polymerase Ii Transcriptional Coactivator P15 Positive Cofactor 4 PC4 SUB1 Homolog P14 |
| ENSDARG00000007723 | ENSFM00250000002014 | Ephrin B1 Precursor Elk Ligand Elk L Eph Related Receptor Tyrosine Kinase Ligand 2 Lerk 2 |
| ENSDARG00000007745 | ENSFM00770001847545 | Cytochrome B C1 Complex Subunit Rieske Mitochondrial Precursor EC_1.10.2.2 Complex Iii Subunit 5 Cytochrome B C1 Complex Subunit 5 Rieske Iron Sulfur Risp Ubiquinol Cytochrome C Reductase Iron Sulfur Subunit |
| ENSDARG00000007753 | ENSFM00800002003432 | Copine Copine |
| ENSDARG00000007786 | ENSFM00250000004757 | Thioredoxin Related Transmembrane 2 Precursor Thioredoxin Domain Containing 14 |
| ENSDARG00000007791 | ENSFM00250000001941 | Serine/Threonine Phosphatase 2A 65 Kda Regulatory Subunit A PP2A Subunit A PR65 PP2A Subunit A R1 |
| ENSDARG00000007825 | ENSFM00780001898380 | Dual Specificity Mitogen Activated Kinase Kinase 1 Map Kinase Kinase 1 Mapkk 1 EC_2.7.12.2 Erk Activator Kinase 1 Mapk/Erk Kinase 1 |
| ENSDARG00000007869 | ENSFM00250000000766 | Eh Domain Containing |
| ENSDARG00000007916 | ENSFM00800002003832 | Probable Xaa Pro Aminopeptidase 3 X Pro Aminopeptidase 3 EC_3.4.11.9 Aminopeptidase P3 APP3 |
| ENSDARG00000007955 | ENSFM00500000270033 | Isoleucine Trna Ligase Cytoplasmic EC_6.1.1.5 Isoleucyl Trna Synthetase Ilers |
| ENSDARG00000007959 | ENSFM00780001898427 | 3 Hydroxyisobutyrate Dehydrogenase Mitochondrial Hibadh EC_1.1.1.31 |
| ENSDARG00000007960 | ENSFM00500000269788 | Heterogeneous Nuclear Ribonucleoprotein D Hnrnp D Hnrnp Dl |
| ENSDARG00000007976 | ENSFM00800002006688 | Unknown |
| ENSDARG00000007989 | ENSFM00670001265228 | Neural Cell Expressed Developmentally Down Regulated 8 |
| ENSDARG00000008026 | ENSFM00250000002649 | Niban |
| ENSDARG00000008030 | ENSFM00730001521616 | Myosin Regulatory Light Myosin Regulatory Light Chain 2 Smooth Muscle Myosin Regulatory Light Chain |
| ENSDARG00000008100 | ENSFM00250000000331 | Amino Acid Transporter Solute Carrier Family 7 Member |
| ENSDARG00000008107 | ENSFM00730001521177 | Tyrosine Kinase EC_2.7.10.2 |
| ENSDARG00000008142 | ENSFM00250000001101 | Syntaxin Binding 1 Unc 18 Homolog 1 UNC18 1 Unc 18 Homolog A Unc 18A |
| ENSDARG00000008155 | ENSFM00250000003170 | Spermine Synthase Spmsy EC_2.5.1.22 Spermidine Aminopropyltransferase |
| ENSDARG00000008200 | ENSFM00800002003558 | Utp Glucose 1 Phosphate Uridylyltransferase EC_2.7.7.9 Udp Glucose Pyrophosphorylase Udpgp Ugpase |
| ENSDARG00000008224 | ENSFM00250000003527 | Vacuolar Sorting Associated 35 Maternal Embryonic 3 Vesicle Sorting 35 |
| ENSDARG00000008237 | ENSFM00770001847577 | Serine Trna Ligase Cytoplasmic EC_6.1.1.11 Seryl Trna Synthetase Serrs Seryl Trna Ser/Sec Synthetase |
| ENSDARG00000008243 | ENSFM00800002003473 | T Complex 1 Subunit Theta Tcp 1 Theta Cct Theta |
| ENSDARG00000008249 | ENSFM00730001522293 | Patched Domain Containing |
| ENSDARG00000008329 | ENSFM00250000000608 | Acid Sensing Ion Channel Amiloride Sensitive Cation Channel |
| ENSDARG00000008370 | ENSFM00800002003414 | Casein Kinase I Delta Cki Delta Ckid EC_2.7.11.1 |
| ENSDARG00000008383 | ENSFM00760001714703 | Actin Related 2/3 Complex Subunit 1B ARP2/3 Complex 41 Kda Subunit P41 Arc |
| ENSDARG00000008398 | ENSFM00500000269632 | Voltage Dependent L Type Calcium Channel Subunit Alpha Calcium Channel L Type Alpha 1 Polypeptide Voltage Gated Calcium Channel Subunit Alpha CAV1 |
| ENSDARG00000008407 | ENSFM00500000270260 | Tetraspanin 7 Tspan 7 Transmembrane 4 Superfamily Member 2 CD231 Antigen |
| ENSDARG00000008434 | ENSFM00500000270604 | Bcl 2 1 BCL2 L 1 Apoptosis Regulator Bcl X |
| ENSDARG00000008447 | ENSFM00500000270053 | Peptidyl Prolyl Cis Trans Isomerase FKBP5 Ppiase FKBP5 EC_5.2.1.8.51 Kda FK506 Binding 51 Kda Fkbp Fkbp 51 FK506 Binding 5 Fkbp 5 Rotamase |
| ENSDARG00000008457 | ENSFM00630001071945 | Unknown |
| ENSDARG00000008457 | ENSFM00250000002702 | Fatty Acid Amide Hydrolase 2 EC_3.5.1.99 |
| ENSDARG00000008487 | ENSFM00250000000455 | Dystrophin |
| ENSDARG00000008491 | ENSFM00500000269664 | Retinol Dehydrogenase |
| ENSDARG00000008494 | ENSFM00250000000438 | Myosin Light Chain Myosin Light Chain Myosin Light Chain |
| ENSDARG00000008568 | ENSFM00500000269757 | Calcium Binding Mitochondrial Carrier Scamc Small Calcium Binding Mitochondrial Carrier Solute Carrier Family 25 Member |
| ENSDARG00000008573 | ENSFM00800002003872 | Glycosaminoglycan Xylosylkinase EC_2.7.1.- Xylose Kinase |
| ENSDARG00000008593 | ENSFM00250000003713 | Neuroblastoma Amplified Sequence Neuroblastoma Amplified Gene |
| ENSDARG00000008623 | ENSFM00250000006013 | COP9 Signalosome Complex Subunit 3 Signalosome Subunit 3 |
| ENSDARG00000008660 | ENSFM00730001521292 | Coronin |
| ENSDARG00000008678 | ENSFM00250000002957 | Sorting Nexin |
| ENSDARG00000008723 | ENSFM00520000517802 | Kinase C Type Pkc EC_2.7.11.13 |
| ENSDARG00000008735 | ENSFM00630001059629 | Unknown |
| ENSDARG00000008735 | ENSFM00250000004412 | Renin Receptor Precursor Atpase H + Transporting Lysosomal Accessory 2 Atpase H + Transporting Lysosomal Interacting 2 Renin/Prorenin Receptor Vacuolar Atp Synthase Membrane Sector Associated M8 9 V Atpase M8 9 Subunit |
| ENSDARG00000008748 | ENSFM00500000270143 | Ubiquitin Conjugating Enzyme E2 N EC_6.3.2.19 Ubiquitin Carrier N Ubiquitin Ligase N |
| ENSDARG00000008765 | ENSFM00260000050657 | Transmembrane EMP24 Domain Containing Precursor P24 Family Gamma |
| ENSDARG00000008785 | ENSFM00800002003534 | Dihydrolipoyl Dehydrogenase Mitochondrial Precursor EC_1.8.1.4 Dihydrolipoamide Dehydrogenase |
| ENSDARG00000008790 | ENSFM00800002003421 | Actin Related 3 Actin 3 |
| ENSDARG00000008803 | ENSFM00600000929886 | Myristoylated Alanine Rich Kinase C Substrate B |
| ENSDARG00000008808 | ENSFM00520000517805 | Echinoderm Microtubule Associated Emap |
| ENSDARG00000008816 | ENSFM00250000001520 | Glutamate Dehydrogenase Mitochondrial Gdh EC_1.4.1.3 |
| ENSDARG00000008867 | ENSFM00730001521249 | Ras Related Rap 1 |
| ENSDARG00000008884 | ENSFM00250000001478 | Hypoxanthine Guanine Phosphoribosyltransferase Hgprt Hgprtase EC_2.4.2.8 |
| ENSDARG00000008936 | ENSFM00790001962996 | Signal Peptidase Complex Catalytic Subunit SEC11A EC_3.4.21.89 Endopeptidase SP18 Microsomal Signal Peptidase 18 Kda Subunit Spase 18 Kda Subunit SEC11 Homolog A SEC11 1 SPC18 |
| ENSDARG00000008969 | ENSFM00500000270953 | Fibrinogen Beta Chain Fibrinopeptide B |
| ENSDARG00000008982 | ENSFM00250000002399 | Calsequestrin Precursor Calsequestrin Muscle |
| ENSDARG00000008983 | ENSFM00250000002047 | Endophilin SH3 Domain Containing GRB2 |
| ENSDARG00000009001 | ENSFM00500000270568 | Disulfide Isomerase A6 Precursor EC_5.3.4.1 Thioredoxin Domain Containing 7 |
| ENSDARG00000009018 | ENSFM00280000058667 | Ammonium Transporter Rh Type B Rhesus Blood Group Family Type B Glycoprotein Rh Family Type B Glycoprotein Rh Type B Glycoprotein |
| ENSDARG00000009020 | ENSFM00250000000847 | Cannabinoid Receptor 1 Cb R CB1 |
| ENSDARG00000009026 | ENSFM00500000269628 | Ankyrin Ank Ankyrin |
| ENSDARG00000009031 | ENSFM00260000050335 | Kinase EC_2.7.11.1 Kinase Mapk/Erk Kinase Kinase Kinase Mek Kinase Kinase Mekkk Kinase |
| ENSDARG00000009087 | ENSFM00630001072007 | Unknown |
| ENSDARG00000009087 | ENSFM00500000283613 | Chain |
| ENSDARG00000009208 | ENSFM00750001632332 | Kinase C Delta Type EC_2.7.11.13 Npkc Delta |
| ENSDARG00000009209 | ENSFM00730001521509 | Sodium/Hydrogen Exchanger Na + /H + Exchanger Nhe Solute Carrier Family 9 Member |
| ENSDARG00000009212 | ENSFM00750001632315 | Peptidyl Prolyl Cis Trans Isomerase Ppiase EC_5.2.1.8 Cyclophilin Rotamase |
| ENSDARG00000009217 | ENSFM00250000001136 | TOM1 2 Target Of Myb 2 |
| ENSDARG00000009250 | ENSFM00250000004626 | Electron Transfer Flavoprotein Subunit Beta Beta Etf |
| ENSDARG00000009252 | ENSFM00810002040954 | N Acyl Phosphatidylethanolamine Hydrolyzing Phospholipase D N Acyl Phosphatidylethanolamine Phospholipase D Nape Pld Nape Hydrolyzing Phospholipase D EC_3.1.4.54 |
| ENSDARG00000009281 | ENSFM00520000517792 | Dynamin EC_3.6.5.5 |
| ENSDARG00000009285 | ENSFM00250000001504 | 60S Ribosomal L15 |
| ENSDARG00000009285 | ENSFM00730001521550 | Ubiquitin Conjugating Enzyme E2 EC_6.3.2.19 Ubiquitin Carrier Ubiquitin Ligase |
| ENSDARG00000009315 | ENSFM00500000269826 | Precursor |
| ENSDARG00000009342 | ENSFM00670001235739 | Thioredoxin Domain Containing 5 Precursor Endoplasmic Reticulum Resident 46 Er 46 ERP46 Thioredoxin P46 |
| ENSDARG00000009350 | ENSFM00500000271371 | Ubiquitin Conjugating Enzyme ATG3 EC_6.3.2.- Autophagy Related 3 APG3 |
| ENSDARG00000009372 | ENSFM00500000270725 | Gtp Binding Di Precursor Distinct Subgroup Of The Ras Family Member |
| ENSDARG00000009401 | ENSFM00730001521436 | Core Precursor Chondroitin Sulfate Proteoglycan |
| ENSDARG00000009499 | ENSFM00250000000542 | Nesprin 2 Nuclear Envelope Spectrin Repeat 2 Nucleus And Actin Connecting Element Protein Nuance Synaptic Nuclear Envelope 2 Syne 2 |
| ENSDARG00000009549 | ENSFM00250000001486 | E3 Ubiquitin Ligase UBR4 EC_6.3.2.- N Recognin 4 Zinc Finger UBR1 Type 1 |
| ENSDARG00000009553 | ENSFM00500000437551 | Unknown |
| ENSDARG00000009567 | ENSFM00740001589455 | Striated Muscle Specific Serine/Threonine Kinase EC_2.7.11.1 Aortic Preferentially Expressed 1 Apeg 1 |
| ENSDARG00000009640 | ENSFM00800002003627 | Proteasome Subunit Beta Type 1 EC_3.4.25.1 Proteasome |
| ENSDARG00000009677 | ENSFM00250000000275 | Disks Large Homolog Synapse Associated Sap |
| ENSDARG00000009685 | ENSFM00800002009095 | Unknown |
| ENSDARG00000009685 | ENSFM00730001521782 | Inorganic Pyrophosphatase EC_3.6.1.1 Pyrophosphate Phospho Hydrolase Ppase |
| ENSDARG00000009689 | ENSFM00350000105429 | Disheveled Associated Activator Of Morphogenesis |
| ENSDARG00000009693 | ENSFM00500000270191 | Lethal 2 Giant Larvae |
| ENSDARG00000009724 | ENSFM00250000002871 | Dolichyl Diphosphooligosaccharide Glycosyltransferase Subunit 2 Precursor EC_2.4.99.18 Dolichyl Diphosphooligosaccharide Glycosyltransferase 63 Kda Subunit Ribophorin Ii Rpn Ii Ribophorin 2 |
| ENSDARG00000009738 | ENSFM00250000000524 | Voltage Dependent L Type Calcium Channel Subunit Beta Calcium Channel Voltage Dependent Subunit Beta |
| ENSDARG00000009740 | ENSFM00810002040799 | Phosphatase 1 Regulatory Subunit 7 Phosphatase 1 Regulatory Subunit 22 |
| ENSDARG00000009771 | ENSFM00760001714910 | Phosphatase Methylesterase 1 Pme 1 EC_3.1.1.89 |
| ENSDARG00000009782 | ENSFM00270000056424 | Myosin Cellular Myosin Heavy Chain Type Myosin Heavy Chain Myosin Heavy Chain Non Muscle Non Muscle Myosin Heavy Chain Nmmhc Non Muscle Myosin Heavy Chain Nmmhc Ii Nmmhc |
| ENSDARG00000009830 | ENSFM00740001589253 | Vigilin High Density Lipoprotein Binding Hdl Binding |
| ENSDARG00000009837 | ENSFM00250000006088 | Acylglycerol Kinase Mitochondrial Precursor EC_2.7.1.107 EC_2.7.1.- 94 Multiple Substrate Lipid Kinase Mulk Multi Substrate Lipid Kinase |
| ENSDARG00000009930 | ENSFM00250000001126 | Cell Adhesion Molecule Precursor Immunoglobulin Superfamily Member Nectin Necl Synaptic Cell Adhesion Molecule |
| ENSDARG00000009941 | ENSFM00250000000753 | Elks/RAB6 Interacting/Cast Family Member 1 Erc 1 RAB6 Interacting 2 |
| ENSDARG00000009978 | ENSFM00670001247480 | S100 |
| ENSDARG00000010002 | ENSFM00730001521471 | Guanine Nucleotide Binding G Q Subunit Alpha Guanine Nucleotide Binding Alpha Q |
| ENSDARG00000010008 | ENSFM00730001521291 | Vimentin |
| ENSDARG00000010021 | ENSFM00500000272223 | Grpe Homolog 1 Mitochondrial Precursor Mt Grpe#1 |
| ENSDARG00000010042 | ENSFM00520000517792 | Dynamin EC_3.6.5.5 |
| ENSDARG00000010052 | ENSFM00250000000535 | N Myc Downstream Regulated Gene |
| ENSDARG00000010070 | ENSFM00610000971869 | Unknown |
| ENSDARG00000010070 | ENSFM00800002003698 | Disintegrin And Metalloproteinase Domain Containing 9 Precursor Adam 9 EC_3.4.24.- Meltrin Gamma Metalloprotease/Disintegrin/Cysteine Rich 9 Myeloma Cell Metalloproteinase |
| ENSDARG00000010078 | ENSFM00250000005326 | Nuclear Pore Complex NUP133.133 Kda Nucleoporin Nucleoporin NUP133 |
| ENSDARG00000010096 | ENSFM00800002003375 | Excitatory Amino Acid Transporter Sodium Dependent Glutamate/Aspartate Transporter Solute Carrier Family 1 Member |
| ENSDARG00000010108 | ENSFM00250000006424 | BRI3 Binding I3 Binding |
| ENSDARG00000010113 | ENSFM00250000006874 | Nadh Dehydrogenase 1 Beta Subcomplex Subunit 8 Mitochondrial Precursor Complex I Ashi Ci Ashi Nadh Ubiquinone Oxidoreductase Ashi Subunit |
| ENSDARG00000010130 | ENSFM00500000270338 | Acid Sphingomyelinase Phosphodiesterase Precursor Asm Phosphodiesterase EC_3.1.4.- |
| ENSDARG00000010144 | ENSFM00500000270520 | Lipid Phosphate Phosphatase Related Type 4 EC_3.1.3.4 Brain Specific Phosphatidic Acid Phosphatase 1 Plasticity Related Gene 1 Prg 1 |
| ENSDARG00000010149 | ENSFM00810002040641 | Atp Synthase Subunit Alpha Mitochondrial Precursor |
| ENSDARG00000010155 | ENSFM00250000000845 | Abl Interactor 1 Abelson Interactor 1 Abi 1 EPS8 SH3 Domain Binding EPS8 Binding Spectrin SH3 Domain Binding 1 |
| ENSDARG00000010160 | ENSFM00250000002123 | 40S Ribosomal |
| ENSDARG00000010194 | ENSFM00670001281972 | Unknown |
| ENSDARG00000010194 | ENSFM00500000270799 | Eukaryotic Translation Initiation Factor 4B Eif 4B |
| ENSDARG00000010246 | ENSFM00730001521302 | Arginine N Methyltransferase 1 EC_2.1.1.- Histone Arginine N Methyltransferase PRMT1 EC_2.1.1.125 |
| ENSDARG00000010250 | ENSFM00250000003453 | Nad P H Dehydrogenase 1 EC_1.6.5.2 Azoreductase Dt Diaphorase Dtd Menadione Reductase Nad P H:Quinone Oxidoreductase 1 Phylloquinone Reductase Quinone Reductase 1 QR1 |
| ENSDARG00000010279 | ENSFM00670001235365 | Secretory Carrier Associated Membrane Secretory Carrier Membrane |
| ENSDARG00000010280 | ENSFM00610000970574 | Unknown |
| ENSDARG00000010280 | ENSFM00250000000820 | Clip Associating Cytoplasmic Linker Associated |
| ENSDARG00000010312 | ENSFM00750001632325 | Transmembrane 4 L6 Family Member |
| ENSDARG00000010316 | ENSFM00780001898374 | Glutamine Trna Ligase EC_6.1.1.18 Glutaminyl Trna Synthetase Glnrs |
| ENSDARG00000010411 | ENSFM00500000269762 | Epsin Eps 15 Interacting |
| ENSDARG00000010442 | ENSFM00260000051011 | Ring Finger 11 |
| ENSDARG00000010445 | ENSFM00250000005731 | Trab Domain Containing |
| ENSDARG00000010472 | ENSFM00650001139994 | Sodium/Potassium Transporting Atpase Subunit Alpha Na + /K + Atpase Alpha Subunit EC_3.6.3.9 Sodium Pump Subunit Alpha |
| ENSDARG00000010516 | ENSFM00250000000614 | 60S Ribosomal L21 |
| ENSDARG00000010531 | ENSFM00740001589424 | Secretory Carrier Associated Membrane Secretory Carrier Membrane |
| ENSDARG00000010553 | ENSFM00500000273449 | Membrane Magnesium Transporter 1 Precursor Er Membrane Complex Subunit 5 Transmembrane 32 |
| ENSDARG00000010601 | ENSFM00260000050612 | Myotubularin Related |
| ENSDARG00000010610 | ENSFM00250000003414 | Sterile Alpha And Tir Motif Containing Tir 1 Homolog |
| ENSDARG00000010721 | ENSFM00350000105408 | Septin |
| ENSDARG00000010728 | ENSFM00760001714632 | Gelsolin Actin Depolymerizing Factor Adf |
| ENSDARG00000010745 | ENSFM00760001714614 | Dnaj Homolog Subfamily A Member Precursor |
| ENSDARG00000010764 | ENSFM00500000272331 | Dehydrogenase/Reductase Sdr Family Member 12 EC_1.1.-.- |
| ENSDARG00000010767 | ENSFM00730001521142 | Ephrin Type A Receptor Precursor EC_2.7.10.1 Kinase |
| ENSDARG00000010773 | ENSFM00760001714681 | Neuropathy Target Esterase Sws |
| ENSDARG00000010784 | ENSFM00500000269806 | Phosphatase 1 Regulatory Subunit 12A Myosin Phosphatase Targeting Subunit 1 Myosin Phosphatase Target Subunit 1 Phosphatase Myosin Binding Subunit |
| ENSDARG00000010789 | ENSFM00250000001140 | Homer Homolog Homer Vasp/Ena Related Gene Up Regulated During Seizure And Ltp Vesl |
| ENSDARG00000010816 | ENSFM00610000973508 | Unknown |
| ENSDARG00000010816 | ENSFM00800002003868 | Probable Cationic Amino Acid Transporter Solute Carrier Family 7 Member 14 |
| ENSDARG00000010844 | ENSFM00730001521569 | Ras Precursor |
| ENSDARG00000010936 | ENSFM00730001521159 | Multidrug Resistance EC_3.6.3.44 Atp Binding Cassette Sub Family B Member P Glycoprotein |
| ENSDARG00000010941 | ENSFM00250000007877 | O Linked Mannose Beta 1 4 N Acetylglucosaminyltransferase 2 POMGNT2 EC_2.4.1.- |
| ENSDARG00000010953 | ENSFM00250000002568 | Nuclear Distribution Nude 1 |
| ENSDARG00000010957 | ENSFM00730001521911 | Maguk P55 Subfamily Member 2 Discs Large Homolog 2 MPP2 |
| ENSDARG00000010965 | ENSFM00810002040638 | Proteasome Subunit Alpha Type 7 EC_3.4.25.1 |
| ENSDARG00000011065 | ENSFM00250000000111 | Calcium/Calmodulin Dependent Kinase Type Ii Subunit Cam Kinase Ii Subunit Camk Ii Subunit EC_2.7.11.17 |
| ENSDARG00000011141 | ENSFM00250000000643 | Dihydropyrimidinase Related Drp Collapsin Response Mediator Crmp |
| ENSDARG00000011146 | ENSFM00670001275535 | Unknown |
| ENSDARG00000011146 | ENSFM00670001236244 | Cytochrome B C1 Complex Subunit 7 Complex Iii Subunit 7 Complex Iii Subunit Vii Ubiquinol Cytochrome C Reductase Complex 14 Kda |
| ENSDARG00000011166 | ENSFM00450000242133 | Carbonic Anhydrase EC_4.2.1.1 Carbonate Dehydratase Carbonic Anhydrase Ca |
| ENSDARG00000011170 | ENSFM00250000000535 | N Myc Downstream Regulated Gene |
| ENSDARG00000011171 | ENSFM00250000000375 | Teneurin Ten Odd Oz/Ten M Homolog Tenascin Ten Teneurin Transmembrane |
| ENSDARG00000011175 | ENSFM00770001847582 | V Type Proton Atpase Subunit D V Atpase Subunit D Vacuolar Proton Pump Subunit D |
| ENSDARG00000011196 | ENSFM00250000002491 | Dnaj Homolog Subfamily C Member 11 |
| ENSDARG00000011201 | ENSFM00500000374444 | RPLP2L |
| ENSDARG00000011208 | ENSFM00670001235955 | Cytochrome C Oxidase Subunit 5B Mitochondrial Precursor Cytochrome C Oxidase Polypeptide Vb |
| ENSDARG00000011272 | ENSFM00670001236187 | Ubiquinol Cytochrome C Reductase Complex Assembly Factor 2 Precursor Mitochondrial Nucleoid Factor 1 Mitochondrial M19 |
| ENSDARG00000011301 | ENSFM00250000003349 | NEDD8 Activating Enzyme E1 Regulatory Subunit Amyloid Beta Precursor Binding 1.59 Kda App BP1 Amyloid Binding 1 |
| ENSDARG00000011312 | ENSFM00730001521973 | Serine/Threonine Kinase 4 EC_2.7.11.1 |
| ENSDARG00000011333 | ENSFM00250000001205 | Rho Gtpase Activating Rho Type Gtpase Activating Gtpase Activating |
| ENSDARG00000011334 | ENSFM00760001714608 | Unknown |
| ENSDARG00000011370 | ENSFM00730001521177 | Tyrosine Kinase EC_2.7.10.2 |
| ENSDARG00000011405 | ENSFM00250000003284 | 40S Ribosomal S9 |
| ENSDARG00000011418 | ENSFM00500000271373 | Sigma Non Opioid Intracellular Receptor 1 Sigma 1 Type Opioid Receptor SIGMA1 Receptor SIGMA1R |
| ENSDARG00000011459 | ENSFM00760001714632 | Gelsolin Actin Depolymerizing Factor Adf |
| ENSDARG00000011462 | ENSFM00500000272123 | Ubiquitin Thioesterase OTUB1 EC_3.4.19.12 Deubiquitinating Enzyme OTUB1 Otu Domain Containing Ubiquitin Aldehyde Binding 1 Otubain 1 Ubiquitin Specific Processing Protease OTUB1 |
| ENSDARG00000011487 | ENSFM00730001521471 | Guanine Nucleotide Binding G Q Subunit Alpha Guanine Nucleotide Binding Alpha Q |
| ENSDARG00000011488 | ENSFM00810002040646 | Nad Dependent Deacetylase Sirtuin 2 EC_3.5.1.- Regulatory SIR2 Homolog 2 SIR2 2 |
| ENSDARG00000011553 | ENSFM00250000004100 | Atp Synthase F 0 Complex Subunit B1 Mitochondrial Precursor Atp Synthase Subunit B Atpase Subunit B |
| ENSDARG00000011564 | ENSFM00250000000671 | Paraspeckle Component 1 |
| ENSDARG00000011602 | ENSFM00250000006430 | Uncharacterized |
| ENSDARG00000011602 | ENSFM00730001542826 | Unknown |
| ENSDARG00000011611 | ENSFM00730001521541 | Actin |
| ENSDARG00000011640 | ENSFM00720001492031 | Synaptotagmin |
| ENSDARG00000011661 | ENSFM00500000270278 | Twinfilin |
| ENSDARG00000011665 | ENSFM00250000000488 | Fructose Bisphosphate Aldolase EC_4.1.2.13 Type Aldolase |
| ENSDARG00000011770 | ENSFM00500000272331 | Dehydrogenase/Reductase Sdr Family Member 12 EC_1.1.-.- |
| ENSDARG00000011777 | ENSFM00600000921221 | Substrate |
| ENSDARG00000011841 | ENSFM00670001238040 | Atp Synthase Subunit G |
| ENSDARG00000011855 | ENSFM00730001521956 | AP2 Associated Kinase 1 EC_2.7.11.1 Adaptor Associated Kinase 1 |
| ENSDARG00000011862 | ENSFM00730001521354 | Neurofilament Polypeptide Nf Kda Neurofilament Neurofilament Triplet |
| ENSDARG00000011863 | ENSFM00260000050584 | Tyrosine Kinase Otk |
| ENSDARG00000011897 | ENSFM00500000270204 | Tumor Susceptibility Gene 101 Escrt I Complex Subunit TSG101 |
| ENSDARG00000011909 | ENSFM00250000000547 | Inositol 1 4 5 Trisphosphate Receptor Type IP3 Receptor IP3R Ins Type Inositol 1 4 5 Trisphosphate Receptor Type INSP3 Receptor |
| ENSDARG00000011921 | ENSFM00500000271170 | Thioredoxin Thioredoxin Related |
| ENSDARG00000011929 | ENSFM00250000000909 | Myelin Proteolipid Plp Lipophilin |
| ENSDARG00000011934 | ENSFM00250000001801 | Glycogenin 1 Gn 1 GN1 EC_2.4.1.186 |
| ENSDARG00000011945 | ENSFM00250000005741 | Solute Carrier Family 35 Member E1 |
| ENSDARG00000011948 | ENSFM00730001521215 | Insulin Receptor Precursor EC_2.7.10.1 |
| ENSDARG00000012019 | ENSFM00400000131715 | Glycine Receptor Subunit Precursor Glycine Receptor Kda Subunit |
| ENSDARG00000012021 | ENSFM00250000005493 | 3 Ketodihydrosphingosine Reductase Precursor Kds Reductase EC_1.1.1.102 3 Dehydrosphinganine Reductase Follicular Variant Translocation 1 Fvt 1 |
| ENSDARG00000012072 | ENSFM00250000005226 | Atp Synthase Subunit S Mitochondrial Precursor Atp Synthase Coupling Factor B Mitochondrial Atp Synthase Regulatory Component Factor B |
| ENSDARG00000012076 | ENSFM00560000771472 | Apolipoprotein A I Precursor Apo Ai Apoa I Apolipoprotein A1 |
| ENSDARG00000012144 | ENSFM00500000273154 | Er Membrane Complex Subunit 7 Precursor |
| ENSDARG00000012177 | ENSFM00800002003435 | Ras Related Rab |
| ENSDARG00000012194 | ENSFM00250000003026 | Non Specific Lipid Transfer Nsl Tp EC_2.3.1.176 Propanoyl Coa C Acyltransferase Scp Chi Scpx Sterol Carrier 2 Scp 2 Sterol Carrier X Scp X |
| ENSDARG00000012204 | ENSFM00740001589171 | Cyclin Dependent Kinase EC_2.7.11.22 Cell Division Kinase Pctaire Motif Kinase Serine/Threonine Kinase Pctaire |
| ENSDARG00000012248 | ENSFM00250000001343 | Rgm Domain Family Member |
| ENSDARG00000012275 | ENSFM00760001714943 | Katanin P60 Atpase Containing Subunit A 2 |
| ENSDARG00000012281 | ENSFM00730001521354 | Neurofilament Polypeptide Nf Kda Neurofilament Neurofilament Triplet |
| ENSDARG00000012295 | ENSFM00500000270206 | Mitochondrial Chaperone BCS1 BCS1 |
| ENSDARG00000012367 | ENSFM00400000131833 | Tripartite Motif Containing 46 Gene Y Geney Tripartite Fibronectin Type Iii And C Terminal Spry Motif |
| ENSDARG00000012378 | ENSFM00250000003167 | N Alpha Acetyltransferase 35 Natc Auxiliary Subunit Embryonic Growth Associated MAK10 Homolog |
| ENSDARG00000012381 | ENSFM00730001521103 | Heat Shock Kda Heat Shock 70 Kda |
| ENSDARG00000012387 | ENSFM00750001632368 | Pyruvate Dehydrogenase E1 Component Subunit Alpha Somatic Form Mitochondrial Precursor EC_1.2.4.1 PDHE1 A Type I |
| ENSDARG00000012388 | ENSFM00500000271199 | Cytochrome C Oxidase Subunit 4 1 Mitochondrial Cytochrome C Oxidase Polypeptide Iv Cytochrome C Oxidase Subunit Iv 1 Cox Iv 1 |
| ENSDARG00000012426 | ENSFM00730001521354 | Neurofilament Polypeptide Nf Kda Neurofilament Neurofilament Triplet |
| ENSDARG00000012513 | ENSFM00250000003553 | Syntenin Syndecan Binding |
| ENSDARG00000012519 | ENSFM00250000001451 | Host Cell Factor 1 Hcf Hcf 1 C1 Factor |
| ENSDARG00000012534 | ENSFM00730001521104 | Sodium And Chloride Dependent Transporter Solute Carrier Family 6 Member |
| ENSDARG00000012555 | ENSFM00740001589162 | Adenylate Kinase Isoenzyme 1 |
| ENSDARG00000012586 | ENSFM00350000105407 | Formin |
| ENSDARG00000012588 | ENSFM00800002003785 | Phosphatidylserine Synthase 1 Pss 1 Ptdser Synthase 1 EC_2.7.8.29 Serine Exchange Enzyme I |
| ENSDARG00000012609 | ENSFM00290000065616 | Hemopexin Precursor |
| ENSDARG00000012682 | ENSFM00670001235608 | Profilin Profilin |
| ENSDARG00000012684 | ENSFM00250000000445 | Plasma Membrane Calcium Transporting Atpase EC_3.6.3.8 Plasma Membrane Calcium Atpase Plasma Membrane Calcium Pump |
| ENSDARG00000012694 | ENSFM00250000000285 | Complement C3 Complement C3 Beta Chain |
| ENSDARG00000012801 | ENSFM00730001521507 | Atp Dependent 6 Phosphofructokinase Type |
| ENSDARG00000012818 | ENSFM00250000001591 | Casein Kinase Ii Subunit Ck Ii EC_2.7.11.1 |
| ENSDARG00000012823 | ENSFM00250000000718 | Disks Large Associated Dap Psd 95/SAP90 Binding SAP90/Psd 95 Associated |
| ENSDARG00000012840 | ENSFM00250000002920 | Alpha 1 2 Mannosyltransferase ALG9 EC_2.4.1.259 EC_2.4.1.- 261 Asparagine Linked Glycosylation 9 Homolog Disrupted In Bipolar Disorder 1 Dol P Man:Man 6 Glcnac 2 Pp Dol Alpha 1 2 Mannosyltransferase Dol P Man:Man 8 Glcnac 2 Pp Dol Alpha 1 2 Mannosyltransf |
| ENSDARG00000012866 | ENSFM00250000000859 | Clathrin Assembly |
| ENSDARG00000012871 | ENSFM00250000005785 | Probable Aminopeptidase NPEPL1 EC_3.4.11.- Aminopeptidase 1 |
| ENSDARG00000012874 | ENSFM00250000001228 | Synaptosomal Associated 25 Snap 25 Synaptosomal Associated 25 Kda |
| ENSDARG00000012881 | ENSFM00730001521491 | Anion Exchange Ae Anion Exchanger Solute Carrier Family 4 Member |
| ENSDARG00000012915 | ENSFM00800002024121 | Unknown |
| ENSDARG00000012915 | ENSFM00260000050929 | Delta 3 5 Delta 2 4 Dienoyl Coa Isomerase Mitochondrial Precursor EC_5.3.3.- |
| ENSDARG00000012927 | ENSFM00250000000929 | Transcription Elongation Factor A Transcription Elongation Factor S Ii Transcription Elongation Factor Tfiis |
| ENSDARG00000012932 | ENSFM00670001235672 | Methionine Adenosyltransferase 2 Subunit Beta Methionine Adenosyltransferase Ii Beta Mat Ii Beta |
| ENSDARG00000012942 | ENSFM00730001521296 | Integrin Beta Precursor |
| ENSDARG00000012944 | ENSFM00250000000024 | Myosin Myosin Heavy Chain Myosin Heavy Chain Muscle |
| ENSDARG00000012953 | ENSFM00730001521660 | Mob Kinase Activator 1A MOB1 Homolog 1B Mps One Binder Kinase Activator 1B |
| ENSDARG00000012982 | ENSFM00750001657117 | Unknown |
| ENSDARG00000012982 | ENSFM00250000001252 | Neurofibromin Neurofibromatosis Related Nf 1 |
| ENSDARG00000012987 | ENSFM00250000000760 | Glucose 6 Phosphate Isomerase Gpi EC_5.3.1.9 Phosphoglucose Isomerase Pgi Phosphohexose Isomerase Phi |
| ENSDARG00000013000 | ENSFM00250000000441 | Liprin Alpha Tyrosine Phosphatase Receptor Type F Polypeptide Interacting Alpha Ptprf Interacting Alpha |
| ENSDARG00000013005 | ENSFM00290000065521 | Precursor |
| ENSDARG00000013020 | ENSFM00250000000811 | Dystrobrevin Beta Dtn B Beta Dystrobrevin |
| ENSDARG00000013044 | ENSFM00250000003401 | Nadh Dehydrogenase Flavoprotein 2 Mitochondrial Precursor EC_1.6.5.3 EC_1.6.99.- 3 Nadh Ubiquinone Oxidoreductase 24 Kda Subunit |
| ENSDARG00000013063 | ENSFM00610000962512 | Unknown |
| ENSDARG00000013063 | ENSFM00250000002519 | Succinyl Coa:3 Ketoacid Coenzyme A Transferase 1 Mitochondrial Precursor EC_2.8.3.5 3 Oxoacid Coa Transferase 1 Somatic Type Succinyl Coa:3 Oxoacid Coa Transferase Scot S |
| ENSDARG00000013075 | ENSFM00500000272206 | 39S Ribosomal L11 Mitochondrial Precursor L11MT Mrp L11 |
| ENSDARG00000013078 | ENSFM00730001521154 | Unknown |
| ENSDARG00000013079 | ENSFM00800002003795 | Gamma Tubulin Complex Component 2 Gamma Ring Complex Kda |
| ENSDARG00000013087 | ENSFM00250000000535 | N Myc Downstream Regulated Gene |
| ENSDARG00000013110 | ENSFM00500000271390 | Dematin Dematin Actin Binding Erythrocyte Membrane Band 4 9 |
| ENSDARG00000013128 | ENSFM00500000269765 | Kinase Isozyme Mitochondrial Precursor EC_2.7.11.2 Pyruvate Dehydrogenase Kinase |
| ENSDARG00000013144 | ENSFM00500000270479 | Sodium/Potassium Transporting Atpase Subunit Beta 1 Sodium/Potassium Dependent Atpase Subunit Beta 1 |
| ENSDARG00000013161 | ENSFM00730001521569 | Ras Precursor |
| ENSDARG00000013236 | ENSFM00500000270882 | Delta 24 Sterol Reductase Precursor EC_1.3.1.72 24 Dehydrocholesterol Reductase 3 Beta Hydroxysterol Delta 24 Reductase |
| ENSDARG00000013250 | ENSFM00250000001453 | Threonine Trna Ligase EC_6.1.1.3 Threonyl Trna Synthetase Thrrs Threonyl Trna Synthetase |
| ENSDARG00000013295 | ENSFM00730001521168 | Solute Carrier Family 2 Facilitated Glucose Transporter Member Glucose Transporter Type Glut |
| ENSDARG00000013307 | ENSFM00250000001949 | 60S Ribosomal L19 |
| ENSDARG00000013317 | ENSFM00250000001172 | Glycogen Phosphorylase Form EC_2.4.1.1 |
| ENSDARG00000013333 | ENSFM00250000002870 | Nadh Dehydrogenase 1 Alpha Subcomplex Subunit 10 Mitochondrial Precursor Complex I 42KD Ci 42KD Nadh Ubiquinone Oxidoreductase 42 Kda Subunit |
| ENSDARG00000013335 | ENSFM00750001632387 | Annexin Annexin Annexin Calphobindin Ii Lipocortin |
| ENSDARG00000013351 | ENSFM00610000972707 | Unknown |
| ENSDARG00000013351 | ENSFM00600000926155 | Inducible Rna Binding |
| ENSDARG00000013360 | ENSFM00250000000986 | Endophilin SH3 Domain Containing GRB2 |
| ENSDARG00000013371 | ENSFM00670001235660 | Isochorismatase Domain Containing |
| ENSDARG00000013414 | ENSFM00260000050595 | Lin 7 Homolog Lin Mammalian Lin Seven Mals Vertebrate Lin 7 Homolog Veli |
| ENSDARG00000013430 | ENSFM00250000004735 | Betaine Homocysteine S Methyltransferase |
| ENSDARG00000013443 | ENSFM00810002040578 | V Type Proton Atpase Subunit B V Atpase Subunit B Endomembrane Proton Pump 58 Kda Subunit Vacuolar Proton Pump Subunit B |
| ENSDARG00000013475 | ENSFM00730001521612 | T Complex 1 Subunit Delta Tcp 1 Delta Cct Delta |
| ENSDARG00000013500 | ENSFM00740001589079 | Atp Binding Cassette Sub Family A Member Atp Binding Cassette Transporter Atp Binding Cassette |
| ENSDARG00000013500 | ENSFM00770001864617 | Si:Dkey 266H7 1 |
| ENSDARG00000013542 | ENSFM00710001459591 | Unknown |
| ENSDARG00000013542 | ENSFM00250000004110 | Acyl Coa:Lysophosphatidylglycerol Acyltransferase 1 EC_2.3.1.- |
| ENSDARG00000013561 | ENSFM00760001714662 | Phosphoglucomutase 1 Pgm 1 EC_5.4.2.2 Glucose Phosphomutase 1 |
| ENSDARG00000013582 | ENSFM00250000001591 | Casein Kinase Ii Subunit Ck Ii EC_2.7.11.1 |
| ENSDARG00000013613 | ENSFM00760001714601 | Annexin Annexin Annexin |
| ENSDARG00000013623 | ENSFM00250000001189 | Voltage Dependent Anion Selective Channel Vdac Outer Mitochondrial Membrane Porin |
| ENSDARG00000013647 | ENSFM00390000126303 | Contactin Precursor Brain Derived Immunoglobulin Superfamily Big |
| ENSDARG00000013655 | ENSFM00500000270547 | Tumor D54 Tumor D52 2 |
| ENSDARG00000013659 | ENSFM00250000006516 | BRO1 Domain Containing Brox Precursor BRO1 Domain And Caax Motif Containing |
| ENSDARG00000013669 | ENSFM00730001522002 | Soluble Nsf Attachment Snap N Ethylmaleimide Sensitive Factor Attachment |
| ENSDARG00000013670 | ENSFM00780001898382 | Hypoxia Up Regulated 1 Precursor |
| ENSDARG00000013708 | ENSFM00250000001518 | Probable Ubiquitin Carboxyl Terminal Hydrolase Faf EC_3.4.19.12 Deubiquitinating Enzyme Faf Fat Facets Related Linked Ubiquitin Thioesterase Faf Ubiquitin Specific Protease 9 Chromosome Ubiquitin Specific Processing Protease Faf |
| ENSDARG00000013730 | ENSFM00730001521721 | Electrogenic Sodium Bicarbonate Cotransporter 1 Sodium Bicarbonate Cotransporter Na + /HCO3 Cotransporter Solute Carrier Family 4 Member 4 |
| ENSDARG00000013732 | ENSFM00250000005580 | Vacuolar Sorting Associated VTA1 Homolog SKD1 Binding 1 SBP1 |
| ENSDARG00000013741 | ENSFM00250000002547 | Lanc 1.40 Kda Erythrocyte Membrane P40 |
| ENSDARG00000013750 | ENSFM00730001521170 | Enolase EC_4.2.1.11 2 Phospho D Glycerate Hydro Lyase |
| ENSDARG00000013763 | ENSFM00250000002798 | Ribosome Binding Ribosome Receptor Rrp |
| ENSDARG00000013776 | ENSFM00760001714699 | Neurolysin Mitochondrial Precursor EC_3.4.24.16 Microsomal Endopeptidase Mep Mitochondrial Oligopeptidase M Neurotensin Endopeptidase |
| ENSDARG00000013804 | ENSFM00250000000112 | Calpain Subunit Calcium Activated Neutral Proteinase Canp Calpain Subunit |
| ENSDARG00000013841 | ENSFM00730001521872 | Tyrosine Kinase EC_2.7.10.2 Abelson Murine Leukemia Viral Oncogene Homolog Abelson Tyrosine Kinase |
| ENSDARG00000013843 | ENSFM00800002003401 | Septin |
| ENSDARG00000013860 | ENSFM00500000274425 | Phosphatidylglycerophosphatase And Tyrosine Phosphatase 1 Precursor EC_3.1.3.27 Pten Phosphatase Phosphoinositide Lipid Phosphatase Tyrosine Phosphatase Mitochondrial 1 EC_3.1.3.- 16 EC_3.1.-.- 3 48 |
| ENSDARG00000013861 | ENSFM00250000005628 | Armadillo Repeat Containing 1 |
| ENSDARG00000013892 | ENSFM00570000851314 | Sortilin Related Receptor Low Density Lipoprotein Receptor Relative With 11 Ligand Binding Repeats Ldlr Relative With 11 Ligand Binding Repeats LR11 Sorla 1 Sorting Related Receptor Containing Ldlr Class A Repeats |
| ENSDARG00000013931 | ENSFM00250000004630 | Eukaryotic Translation Initiation Factor 3 Subunit M |
| ENSDARG00000013938 | ENSFM00250000003368 | Proteasome Subunit Beta Type 3 EC_3.4.25.1 Proteasome Chain 13 Proteasome Component C10 Ii Proteasome Theta Chain |
| ENSDARG00000013950 | ENSFM00500000276040 | Peptide Deformylase Mitochondrial Precursor EC_3.5.1.88 Polypeptide Deformylase |
| ENSDARG00000013961 | ENSFM00500000271463 | High Affinity Copper Uptake 1 Copper Transporter 1 Solute Carrier Family 31 Member 1 |
| ENSDARG00000013966 | ENSFM00420000140606 | Proteasome Subunit Alpha Type EC_3.4.25.1 Macropain Multicatalytic Endopeptidase Complex Proteasome |
| ENSDARG00000013976 | ENSFM00760001714601 | Annexin Annexin Annexin |
| ENSDARG00000014013 | ENSFM00730001522072 | Lamin B Receptor Integral Nuclear Envelope Inner Membrane |
| ENSDARG00000014015 | ENSFM00800002003471 | Disulfide Isomerase A3 Precursor EC_5.3.4.1.58 Kda Glucose Regulated 58 Kda Microsomal P58 Disulfide Isomerase Er 60 Endoplasmic Reticulum Resident 57 Er 57 ERP57 Endoplasmic Reticulum Resident 60 Er 60 ERP60 |
| ENSDARG00000014031 | ENSFM00730001521139 | Multidrug Resistance Associated 1 Atp Binding Cassette Sub Family C Member 1 Leukotriene C 4 Transporter LTC4 Transporter |
| ENSDARG00000014053 | ENSFM00610000967352 | Unknown |
| ENSDARG00000014053 | ENSFM00320000100099 | Noelin Precursor Neuronal Olfactomedin Related Er Localized Olfactomedin 1 |
| ENSDARG00000014068 | ENSFM00250000001043 | Phosphoglycerate Mutase EC_3.1.3.13 EC_5.4.2.- 11 EC_5.4.-.- 2 4 Bpg Dependent Pgam Phosphoglycerate Mutase Isozyme Pgam |
| ENSDARG00000014081 | ENSFM00500000270682 | Unknown |
| ENSDARG00000014113 | ENSFM00250000001784 | Wiskott Aldrich Syndrome Wasp |
| ENSDARG00000014137 | ENSFM00250000000859 | Clathrin Assembly |
| ENSDARG00000014139 | ENSFM00250000003679 | Signal Recognition Particle Subunit SRP72 SRP72 Signal Recognition Particle 72 Kda |
| ENSDARG00000014165 | ENSFM00250000005568 | Translocon Associated Subunit Gamma Trap Gamma Signal Sequence Receptor Subunit Gamma Ssr Gamma |
| ENSDARG00000014169 | ENSFM00720001492031 | Synaptotagmin |
| ENSDARG00000014215 | ENSFM00730001521172 | Cadherin Precursor Cadherin Cadherin |
| ENSDARG00000014222 | ENSFM00250000001591 | Casein Kinase Ii Subunit Ck Ii EC_2.7.11.1 |
| ENSDARG00000014230 | ENSFM00800002003585 | Dihydrolipoyllysine Residue Succinyltransferase Component Of 2 Oxoglutarate Dehydrogenase Complex Mitochondrial Precursor EC_2.3.1.61 2 Oxoglutarate Dehydrogenase Complex Component E2 Ogdc E2 Dihydrolipoamide Succinyltransferase Component Of 2 Oxoglutarat |
| ENSDARG00000014233 | ENSFM00350000105408 | Septin |
| ENSDARG00000014239 | ENSFM00250000000964 | Extended Synaptotagmin E |
| ENSDARG00000014273 | ENSFM00250000000111 | Calcium/Calmodulin Dependent Kinase Type Ii Subunit Cam Kinase Ii Subunit Camk Ii Subunit EC_2.7.11.17 |
| ENSDARG00000014280 | ENSFM00250000000718 | Disks Large Associated Dap Psd 95/SAP90 Binding SAP90/Psd 95 Associated |
| ENSDARG00000014287 | ENSFM00670001269577 | Unknown |
| ENSDARG00000014287 | ENSFM00730001521170 | Enolase EC_4.2.1.11 2 Phospho D Glycerate Hydro Lyase |
| ENSDARG00000014302 | ENSFM00250000001886 | Bmp/Retinoic Acid Inducible Neural Specific 1 Precursor Deleted In Bladder Cancer 1 |
| ENSDARG00000014303 | ENSFM00760001714983 | VAC14 Homolog |
| ENSDARG00000014313 | ENSFM00600000928429 | Atp Synthase Coupling Factor 6 Mitochondrial |
| ENSDARG00000014378 | ENSFM00260000050376 | Solute Carrier Family 12 Member Electroneutral Potassium Chloride Cotransporter K Cl Cotransporter |
| ENSDARG00000014420 | ENSFM00250000000588 | Elav |
| ENSDARG00000014462 | ENSFM00730001521441 | Ras Related Rab 3 |
| ENSDARG00000014517 | ENSFM00760001714687 | Ubiquitin Carboxyl Terminal Hydrolase 13 EC_3.4.19.12 Deubiquitinating Enzyme 13 Ubiquitin Thioesterase 13 Ubiquitin Specific Processing Protease 13 |
| ENSDARG00000014522 | ENSFM00260000050333 | Cadherin Precursor |
| ENSDARG00000014527 | ENSFM00500000269765 | Kinase Isozyme Mitochondrial Precursor EC_2.7.11.2 Pyruvate Dehydrogenase Kinase |
| ENSDARG00000014532 | ENSFM00250000003456 | Axin Interactor Dorsalization Associated Axin Interaction Partner And Dorsalization Antagonist |
| ENSDARG00000014536 | ENSFM00670001236109 | Prefoldin Subunit 1 |
| ENSDARG00000014571 | ENSFM00250000001012 | Catenin Beta Beta Catenin |
| ENSDARG00000014582 | ENSFM00250000001333 | Exocyst Complex Component 3 Exocyst Complex Component SEC6 |
| ENSDARG00000014591 | ENSFM00250000003528 | Interleukin Enhancer Binding Factor 2 |
| ENSDARG00000014634 | ENSFM00250000004850 | Membrane Bound Transcription Factor Site 1 Protease Precursor EC_3.4.21.112 Endopeptidase S1P Sterol Regulated Luminal Protease Subtilisin/Kexin Isozyme 1 Ski 1 |
| ENSDARG00000014646 | ENSFM00250000001706 | Amine Oxidase EC_1.4.3.21 Amine Oxidase Semicarbazide Sensitive Amine Oxidase Ssao |
| ENSDARG00000014651 | ENSFM00250000001422 | Supervillin Archvillin P205/P250 |
| ENSDARG00000014674 | ENSFM00730001521866 | Long Chain Fatty Acid Coa Ligase EC_6.2.1.3 Long Chain Acyl Coa Synthetase Lacs |
| ENSDARG00000014690 | ENSFM00250000001458 | 40S Ribosomal S4 |
| ENSDARG00000014717 | ENSFM00730001521523 | Dynein Heavy Chain Cytoplasmic Dynein Heavy Chain Cytosolic |
| ENSDARG00000014727 | ENSFM00250000000722 | Peroxisomal Acyl Coenzyme A Oxidase 1 Aox EC_1.3.3.6 Palmitoyl Coa Oxidase |
| ENSDARG00000014792 | ENSFM00260000050662 | Leucine Rich Repeat Containing Precursor Netrin Ligand Ngl |
| ENSDARG00000014793 | ENSFM00250000000766 | Eh Domain Containing |
| ENSDARG00000014794 | ENSFM00250000003169 | Cytochrome B C1 Complex Subunit 2 Mitochondrial Precursor Complex Iii Subunit 2 Core Ii Ubiquinol Cytochrome C Reductase Complex Core 2 |
| ENSDARG00000014804 | ENSFM00730001522093 | Voltage Dependent Calcium Channel Subunit Alpha 2/Delta 1 Precursor Voltage Gated Calcium Channel Subunit Alpha 2/Delta 1 |
| ENSDARG00000014806 | ENSFM00500000270421 | Very Long Chain 3R 3 Hydroxyacyl Coa Dehydratase 2 EC_4.2.1.134 3 Hydroxyacyl Coa Dehydratase 2 HACD2 Tyrosine Phosphatase Member B |
| ENSDARG00000014867 | ENSFM00250000002389 | 60S Ribosomal |
| ENSDARG00000014871 | ENSFM00670001235552 | Synaptogyrin |
| ENSDARG00000014875 | ENSFM00250000000845 | Abl Interactor 1 Abelson Interactor 1 Abi 1 EPS8 SH3 Domain Binding EPS8 Binding Spectrin SH3 Domain Binding 1 |
| ENSDARG00000014891 | ENSFM00740001589121 | Roundabout Homolog Precursor |
| ENSDARG00000014915 | ENSFM00670001235829 | Acyl Carrier Protein Mitochondrial Precursor Acp Nadh Ubiquinone Oxidoreductase 9.6 Kda Subunit |
| ENSDARG00000014941 | ENSFM00500000272173 | Ekc/Keops Complex Subunit Tprkb TP53RK Binding |
| ENSDARG00000014956 | ENSFM00250000004873 | Diablo Homolog Mitochondrial |
| ENSDARG00000014962 | ENSFM00310000089007 | Serine/Threonine Phosphatase 2B Catalytic Subunit EC_3.1.3.16 Cam Prp Catalytic Subunit Calmodulin Dependent Calcineurin A Subunit |
| ENSDARG00000014973 | ENSFM00260000050653 | Netrin Precursor Laminet |
| ENSDARG00000015002 | ENSFM00400000134066 | Cadherin 4 Retinal |
| ENSDARG00000015002 | ENSFM00730001521172 | Cadherin Precursor Cadherin Cadherin |
| ENSDARG00000015003 | ENSFM00250000000639 | Slit Robo Rho Gtpase Activating Rho Gtpase Activating |
| ENSDARG00000015006 | ENSFM00780001898359 | Dynamin 1 EC_3.6.5.5 Dynamin Family Member Proline Rich Carboxyl Terminal Domain Less Dymple Dynamin Related 1 |
| ENSDARG00000015025 | ENSFM00390000126295 | Neural Cell Adhesion Molecule L1 L1 L1 |
| ENSDARG00000015050 | ENSFM00750001632311 | Calmodulin Cam |
| ENSDARG00000015053 | ENSFM00740001589268 | Glutamate Receptor Interacting |
| ENSDARG00000015059 | ENSFM00350000105429 | Disheveled Associated Activator Of Morphogenesis |
| ENSDARG00000015065 | ENSFM00250000000285 | Complement C3 Complement C3 Beta Chain |
| ENSDARG00000015088 | ENSFM00500000271339 | Dnaj Homolog Subfamily B Member 11 Precursor Er Associated Dnaj Er Associated HSP40 Co Chaperone Endoplasmic Reticulum Dna J Domain Containing 3 Er Resident ERDJ3 ERDJ3 ERJ3P |
| ENSDARG00000015126 | ENSFM00250000002089 | Golgi Reassembly Stacking 1 Golgi Peripheral Membrane P65 Golgi Reassembly Stacking Of 65 Kda GRASP65 |
| ENSDARG00000015128 | ENSFM00670001235486 | 60S Ribosomal L27 |
| ENSDARG00000015174 | ENSFM00250000000490 | V Type Proton Atpase 116 Kda Subunit A V Atpase 116 Kda Proton 116 Kda Subunit Vacuolar Proton Translocating Atpase 116 Kda Subunit A |
| ENSDARG00000015184 | ENSFM00760001714665 | Maguk P55 Subfamily Member |
| ENSDARG00000015201 | ENSFM00250000003005 | L Isoaspartate D Aspartate O Methyltransferase Pimt EC_2.1.1.77 L Isoaspartyl Carboxyl Methyltransferase L Isoaspartyl/D Aspartyl Methyltransferase Beta Aspartate Methyltransferase |
| ENSDARG00000015236 | ENSFM00800002009595 | Pycrl |
| ENSDARG00000015236 | ENSFM00500000269896 | Pyrroline 5 Carboxylate Reductase 3 P5C Reductase 3 P5CR 3 EC_1.5.1.2 Pyrroline 5 Carboxylate Reductase |
| ENSDARG00000015239 | ENSFM00800002003664 | Pre Processing Factor 19 |
| ENSDARG00000015240 | ENSFM00740001589470 | Cyclin Dependent Kinase 5 EC_2.7.11.22 |
| ENSDARG00000015315 | ENSFM00730001521682 | 26S Protease Regulatory Subunit 8 26S Proteasome Aaa Atpase Subunit RPT6 Proteasome 26S Subunit Atpase 5 Proteasome Subunit P45 P45/Sug |
| ENSDARG00000015337 | ENSFM00500000271337 | Catechol O Methyltransferase EC_2.1.1.6 |
| ENSDARG00000015343 | ENSFM00540000717911 | 6 Phosphogluconate Dehydrogenase Decarboxylating EC_1.1.1.44 |
| ENSDARG00000015385 | ENSFM00790001963038 | Nadh Dehydrogenase Iron Sulfur 3 Mitochondrial Precursor EC_1.6.5.3 EC_1.6.99.- 3 Complex I 30KD Ci 30KD Nadh Ubiquinone Oxidoreductase 30 Kda Subunit |
| ENSDARG00000015392 | ENSFM00810002040596 | Pre Splicing Factor Atp Dependent Rna Helicase EC_3.6.4.13 Deah Box |
| ENSDARG00000015425 | ENSFM00730001521994 | Sodium/Potassium/Calcium Exchanger 3 Precursor Na + /K + /Ca 2+ Exchange 3 Solute Carrier Family 24 Member 3 |
| ENSDARG00000015449 | ENSFM00250000004295 | Alpha 1 6 Fucosyltransferase ALPHA1 6FUCT EC_2.4.1.68 Gdp L Fuc:N Acetyl Beta D Glucosaminide ALPHA1 6 Fucosyltransferase Gdp Fucose Glycoprotein Fucosyltransferase Glycoprotein 6 Alpha L Fucosyltransferase |
| ENSDARG00000015474 | ENSFM00760001714676 | Serine/Threonine Phosphatase 2A 56 Kda Regulatory Subunit Epsilon PP2A B Subunit B' Epsilon PP2A B Subunit B56 Epsilon PP2A B Subunit PR61 Epsilon PP2A B Subunit R5 Epsilon |
| ENSDARG00000015502 | ENSFM00390000126345 | Disintegrin And Metalloproteinase Domain Containing 10 Precursor Adam 10 EC_3.4.24.81 Kuzbanian Homolog Mammalian Disintegrin Metalloprotease CD156C Antigen |
| ENSDARG00000015524 | ENSFM00250000000896 | Ribose Phosphate Pyrophosphokinase EC_2.7.6.1 Phosphoribosyl Pyrophosphate Synthase |
| ENSDARG00000015531 | ENSFM00730001521473 | Sodium Bicarbonate Solute Carrier Family 4 Member |
| ENSDARG00000015537 | ENSFM00800002003398 | Glutamate Decarboxylase 1 EC_4.1.1.15.67 Kda Glutamic Acid Decarboxylase Gad 67 Glutamate Decarboxylase 67 Kda |
| ENSDARG00000015546 | ENSFM00250000000564 | Alkaline Phosphatase Precursor EC_3.1.3.1 Alkaline Phosphatase |
| ENSDARG00000015551 | ENSFM00500000269681 | Ferritin Heavy Chain Ferritin H Subunit EC_1.16.3.1 |
| ENSDARG00000015610 | ENSFM00420000140599 | Tubulin Gamma Chain Gamma Tubulin |
| ENSDARG00000015627 | ENSFM00730001521808 | Regulator Of G Signaling |
| ENSDARG00000015649 | ENSFM00800002003504 | Ras Related Rap Precursor |
| ENSDARG00000015681 | ENSFM00610000973032 | Unknown |
| ENSDARG00000015681 | ENSFM00810002040612 | Glycogen Synthase Kinase 3 Gsk 3 EC_2.7.11.26 Serine/Threonine Kinase EC_2.7.11.- 1 |
| ENSDARG00000015757 | ENSFM00250000004152 | Transmembrane |
| ENSDARG00000015790 | ENSFM00670001235362 | Sodium/Potassium Transporting Atpase Subunit Beta Sodium/Potassium Dependent Atpase Subunit Beta |
| ENSDARG00000015807 | ENSFM00730001522125 | Ras Related Rab |
| ENSDARG00000015823 | ENSFM00810002040678 | Vacuolar Sorting Associated Vesicle Sorting |
| ENSDARG00000015854 | ENSFM00730001521465 | Choline O Acetyltransferase Choactase Chat Choline Acetylase EC_2.3.1.6 |
| ENSDARG00000015856 | ENSFM00250000002826 | Solute Carrier Family 25 Member |
| ENSDARG00000015862 | ENSFM00670001235299 | 60S Ribosomal L5 |
| ENSDARG00000015866 | ENSFM00500000286061 | 14 Kda Apolipoprotein |
| ENSDARG00000015891 | ENSFM00740001589127 | Receptor Type Tyrosine Phosphatase Precursor Tyrosine Phosphatase R Ptp EC_3.1.3.48 |
| ENSDARG00000015918 | ENSFM00810002040672 | Dihydrolipoyllysine Residue Acetyltransferase Component Of Pyruvate Dehydrogenase Complex Mitochondrial EC_2.3.1.12 Dihydrolipoamide Acetyltransferase Component Of Pyruvate Dehydrogenase Complex Pyruvate Dehydrogenase Complex Component E2 Pdc E2 PDCE2 |
| ENSDARG00000015921 | ENSFM00800002003812 | Periodic Tryptophan 1 Homolog |
| ENSDARG00000015931 | ENSFM00250000000859 | Clathrin Assembly |
| ENSDARG00000015947 | ENSFM00740001589150 | Precursor |
| ENSDARG00000015978 | ENSFM00670001235955 | Cytochrome C Oxidase Subunit 5B Mitochondrial Precursor Cytochrome C Oxidase Polypeptide Vb |
| ENSDARG00000016016 | ENSFM00480000262787 | Drebrin |
| ENSDARG00000016058 | ENSFM00250000004633 | Acidic Fibroblast Growth Factor Intracellular Binding Afgf Intracellular Binding Fgf 1 Intracellular Binding |
| ENSDARG00000016059 | ENSFM00500000269759 | Ras Related Rab |
| ENSDARG00000016074 | ENSFM00500000270893 | Quinone Oxidoreductase EC_1.6.5.5 Nadph:Quinone Reductase Zeta Crystallin |
| ENSDARG00000016128 | ENSFM00440000236903 | Ap 3 Complex Subunit Mu Adaptor Related Complex 3 Subunit Mu Clathrin Assembly Assembly Complex 3 Mu Medium Chain Clathrin Coat Assembly AP47 Homolog Clathrin Coat Associated AP47 Homolog Golgi Adaptor Ap 1.47 Kda Homolog HA1.47 Kda Subunit Homolog Adapti |
| ENSDARG00000016141 | ENSFM00730001521312 | Sodium Dependent Transporter Transporter Solute Carrier Family 6 Member |
| ENSDARG00000016153 | ENSFM00250000004650 | Dystroglycan Precursor Dystrophin Associated Glycoprotein 1 |
| ENSDARG00000016173 | ENSFM00500000270039 | T Complex 1 Subunit Gamma Tcp 1 Gamma Cct Gamma |
| ENSDARG00000016231 | ENSFM00670001235596 | B Cell Receptor Associated Bcr Associated |
| ENSDARG00000016238 | ENSFM00670001235692 | PRA1 Family 3 Adp Ribosylation Factor 6 Interacting 5 Arl 6 Interacting 5 Aip 5 |
| ENSDARG00000016239 | ENSFM00780001898464 | 26S Proteasome Non Atpase Regulatory Subunit 13 26S Proteasome Regulatory Subunit RPN9 26S Proteasome Regulatory Subunit S11 26S Proteasome Regulatory Subunit P40 5 |
| ENSDARG00000016253 | ENSFM00500000270233 | Ras Related Precursor |
| ENSDARG00000016255 | ENSFM00600000921240 | Charged Multivesicular Body Chromatin Modifying |
| ENSDARG00000016263 | ENSFM00500000270072 | Palmitoyltransferase ZDHHC5 EC_2.3.1.225 Zinc Finger Dhhc Domain Containing 5 Dhhc 5 |
| ENSDARG00000016301 | ENSFM00250000000193 | Tubulin Beta Chain |
| ENSDARG00000016302 | ENSFM00500000270457 | Atp Dependent Helicase Nonsense 1 Up Frameshift Suppressor 1 |
| ENSDARG00000016307 | ENSFM00250000006279 | 39S Ribosomal L46 Mitochondrial Precursor L46MT Mrp L46 |
| ENSDARG00000016319 | ENSFM00740001589515 | Complement Component C9 Precursor |
| ENSDARG00000016348 | ENSFM00250000000652 | Disco Interacting 2 Homolog DIP2 Homolog |
| ENSDARG00000016350 | ENSFM00250000001899 | Adenylyl Cyclase Associated Cap |
| ENSDARG00000016357 | ENSFM00250000000577 | Dimethylaniline Monooxygenase EC_1.14.13.8 Dimethylaniline Oxidase Flavin Containing Monooxygenase Fmo |
| ENSDARG00000016396 | ENSFM00250000007224 | Unknown |
| ENSDARG00000016396 | ENSFM00610000967087 | Unknown |
| ENSDARG00000016415 | ENSFM00800002003741 | Probable 2 Oxoglutarate Dehydrogenase E1 Component DHKTD1 Mitochondrial Precursor EC_1.2.4.2 Dehydrogenase E1 And Transketolase Domain Containing 1 |
| ENSDARG00000016429 | ENSFM00270000056477 | Ferm Rhogef And Pleckstrin Domain Containing |
| ENSDARG00000016463 | ENSFM00670001235686 | Trafficking Particle Complex Subunit 6A Trapp Complex Subunit 6A |
| ENSDARG00000016464 | ENSFM00730001521452 | Serine/Threonine Kinase Mrck EC_2.7.11.1 CDC42 Binding Kinase Dmpk Myotonic Dystrophy Kinase Related CDC42 Binding Kinase Mrck Myotonic Dystrophy Kinase |
| ENSDARG00000016470 | ENSFM00750001632387 | Annexin Annexin Annexin Calphobindin Ii Lipocortin |
| ENSDARG00000016480 | ENSFM00730001521551 | Vesicular Glutamate Transporter Solute Carrier Family 17 Member |
| ENSDARG00000016545 | ENSFM00730001521465 | Choline O Acetyltransferase Choactase Chat Choline Acetylase EC_2.3.1.6 |
| ENSDARG00000016551 | ENSFM00250000001463 | Iq Motif And SEC7 Domain Containing |
| ENSDARG00000016584 | ENSFM00730001521808 | Regulator Of G Signaling |
| ENSDARG00000016598 | ENSFM00800002003437 | Creatine Kinase U Type Mitochondrial Precursor EC_2.7.3.2 Acidic Type Mitochondrial Creatine Kinase Mia Ck Ubiquitous Mitochondrial Creatine Kinase U Mtck |
| ENSDARG00000016611 | ENSFM00250000002467 | Secernin |
| ENSDARG00000016667 | ENSFM00250000000928 | Gamma Aminobutyric Acid Type B Receptor Subunit 2 Precursor Gaba B Receptor 2 Gaba B R2 Gaba BR2 GABABR2 GB2 G Coupled Receptor 51 |
| ENSDARG00000016676 | ENSFM00410000138458 | Guanine Nucleotide Binding Subunit |
| ENSDARG00000016687 | ENSFM00500000270065 | Very Long Chain Specific Acyl Coa Dehydrogenase Mitochondrial Precursor EC_1.3.8.9 |
| ENSDARG00000016706 | ENSFM00250000003624 | Bifunctional Purine Biosynthesis Phosphoribosylaminoimidazolecarboxamide Formyltransferase EC_2.1.2.3 5 Aminoimidazole 4 Carboxamide Ribonucleotide Formyltransferase Aicar Transformylase |
| ENSDARG00000016721 | ENSFM00800002003532 | Succinate Dehydrogenase Flavoprotein Subunit Mitochondrial Precursor EC_1.3.5.1 Flavoprotein Subunit Of Complex Ii Fp |
| ENSDARG00000016733 | ENSFM00790001963003 | Phosphoserine Aminotransferase Psat EC_2.6.1.52 Endometrial Progesterone Induced Epip Phosphohydroxythreonine Aminotransferase |
| ENSDARG00000016739 | ENSFM00260000050662 | Leucine Rich Repeat Containing Precursor Netrin Ligand Ngl |
| ENSDARG00000016745 | ENSFM00500000271904 | Solute Carrier Family 35 Member F6 Precursor ANT2 Binding ANT2BP Transport And Golgi Organization 9 Homolog |
| ENSDARG00000016754 | ENSFM00670001238431 | Tubulin Specific Chaperone A |
| ENSDARG00000016765 | ENSFM00670001235365 | Secretory Carrier Associated Membrane Secretory Carrier Membrane |
| ENSDARG00000016771 | ENSFM00250000000343 | Precursor |
| ENSDARG00000016782 | ENSFM00760001714810 | E3 Ubiquitin Ligase HUWE1 EC_6.3.2.- Hect Uba And Wwe Domain Containing 1 Upstream Regulatory Element Binding 1 Ure B1 Ure Binding 1 |
| ENSDARG00000016809 | ENSFM00790001962972 | Camp Dependent Kinase Catalytic Subunit Pka C EC_2.7.11.11 |
| ENSDARG00000016825 | ENSFM00250000000813 | Vitellogenin Precursor |
| ENSDARG00000016837 | ENSFM00500000271973 | Golgi Associated Plant Pathogenesis Related 1 Gapr 1 Golgi Associated Pr 1 Glioma Pathogenesis Related 2 Glipr 2 |
| ENSDARG00000016856 | ENSFM00250000001840 | Serum Paraoxonase/Arylesterase 2 Pon 2 EC_3.1.1.2 Aromatic Esterase 2 A Esterase 2 Serum Aryldialkylphosphatase 2 |
| ENSDARG00000016864 | ENSFM00250000004384 | Phenylalanine Trna Ligase Beta Subunit EC_6.1.1.20 Phenylalanyl Trna Synthetase Beta Subunit Phers |
| ENSDARG00000016889 | ENSFM00810002040848 | Eukaryotic Translation Initiation Factor 3 Subunit G |
| ENSDARG00000016904 | ENSFM00500000271321 | Kinase Mitochondrial Precursor EC_2.7.11.4 Branched Chain Alpha Ketoacid Dehydrogenase Kinase Bckd Kinase Bckdhkin |
| ENSDARG00000016939 | ENSFM00730001521210 | Integrin Beta Precursor Subunit Beta Subunit Beta Antigen |
| ENSDARG00000016968 | ENSFM00250000004458 | Transmembrane 214 |
| ENSDARG00000016977 | ENSFM00250000003280 | Sorting Nexin 27 |
| ENSDARG00000017014 | ENSFM00730001521388 | Unknown |
| ENSDARG00000017034 | ENSFM00810002040854 | Sulfide:Quinone Oxidoreductase Mitochondrial Precursor Sqor EC_1.8.5.- |
| ENSDARG00000017047 | ENSFM00730001521827 | Long Chain Fatty Acid Transport 4 Fatp 4 Fatty Acid Transport 4 EC_6.2.1.- Solute Carrier Family 27 Member 4 |
| ENSDARG00000017049 | ENSFM00250000003164 | Adenylosuccinate Lyase Asl EC_4.3.2.2 Adenylosuccinase Asase |
| ENSDARG00000017105 | ENSFM00740001589204 | Ena/Vasp Ena/Vasodilator Stimulated Phosphoprotein |
| ENSDARG00000017126 | ENSFM00250000005381 | Acetolactate Synthase EC_2.2.1.- Ilvb |
| ENSDARG00000017140 | ENSFM00800002003496 | Septin 2 |
| ENSDARG00000017162 | ENSFM00800002003386 | Kinesin |
| ENSDARG00000017180 | ENSFM00780001898362 | Niemann Pick C1 Precursor |
| ENSDARG00000017211 | ENSFM00730001521536 | Plexin Precursor |
| ENSDARG00000017217 | ENSFM00500000272427 | Immunoglobulin Superfamily Member 11 Precursor IGSF11 Brain And Testis Specific Immunoglobulin Superfamily Bt Igsf |
| ENSDARG00000017219 | ENSFM00570000851024 | Polyadenylate Binding Pabp Poly A Binding |
| ENSDARG00000017235 | ENSFM00250000002695 | Eukaryotic Translation Initiation Factor 5A Eif 5A Eif Eukaryotic Initiation Factor 5A |
| ENSDARG00000017261 | ENSFM00250000003307 | Glycerophosphodiester Phosphodiesterase Domain Containing EC_3.1.-.- |
| ENSDARG00000017299 | ENSFM00540000718180 | Fatty Acid Binding |
| ENSDARG00000017312 | ENSFM00250000005238 | LMBR1 Domain Containing 2 |
| ENSDARG00000017316 | ENSFM00250000007925 | Bag Family Molecular Chaperone Regulator 5 Bag 5 Bcl 2 Associated Athanogene 5 |
| ENSDARG00000017338 | ENSFM00250000001605 | Kinase D Interacting Substrate Of 220 Kda Ankyrin Repeat Rich Membrane Spanning |
| ENSDARG00000017354 | ENSFM00730001521142 | Ephrin Type A Receptor Precursor EC_2.7.10.1 Kinase |
| ENSDARG00000017388 | ENSFM00500000270299 | Glutathione S Transferase Theta EC_2.5.1.18 Gst Class Theta |
| ENSDARG00000017422 | ENSFM00440000236938 | Adipocyte Plasma Membrane Associated |
| ENSDARG00000017431 | ENSFM00250000002444 | FAM63A |
| ENSDARG00000017441 | ENSFM00610000972103 | Unknown |
| ENSDARG00000017441 | ENSFM00250000000438 | Myosin Light Chain Myosin Light Chain Myosin Light Chain |
| ENSDARG00000017466 | ENSFM00250000000779 | Neural Cell Adhesion Molecule 1 Precursor N Cam 1 Ncam 1 |
| ENSDARG00000017571 | ENSFM00450000242169 | Methylcrotonoyl Coa Carboxylase Beta Chain Mitochondrial Precursor Mccase Subunit Beta EC_6.4.1.4 3 Methylcrotonyl Coa Carboxylase 2 3 Methylcrotonyl Coa Carboxylase Non Biotin Containing Subunit 3 Methylcrotonyl Coa:Carbon Dioxide Ligase Subunit Beta |
| ENSDARG00000017590 | ENSFM00810002040842 | Omega Amidase NIT2 EC_3.5.1.3 Nitrilase Homolog 2 |
| ENSDARG00000017591 | ENSFM00570000851039 | Protocadherin Fat Precursor Fat Tumor Suppressor Homolog |
| ENSDARG00000017612 | ENSFM00500000271344 | Pyridoxine 5' Phosphate Oxidase EC_1.4.3.5 Pyridoxamine Phosphate Oxidase |
| ENSDARG00000017624 | ENSFM00740001589131 | Keratin Type Ii Cytoskeletal 8 Cytokeratin 8 Ck 8 Keratin 8 K8 Type Ii Keratin KB8 |
| ENSDARG00000017658 | ENSFM00500000270423 | Mannose 1 Phosphate Guanyltransferase EC_2.7.7.13 Gdp Mannose Pyrophosphorylase Gtp Mannose 1 Phosphate Guanylyltransferase |
| ENSDARG00000017661 | ENSFM00290000065522 | Serine/Threonine Kinase EC_2.7.11.1 Proto Oncogene Raf |
| ENSDARG00000017673 | ENSFM00740001589294 | Rna Binding Nova 1 Neuro Oncological Ventral Antigen 1 Ventral Neuron Specific 1 |
| ENSDARG00000017728 | ENSFM00800002003689 | Prohibitin 2 |
| ENSDARG00000017740 | ENSFM00250000003542 | Translocation SEC63 Homolog |
| ENSDARG00000017741 | ENSFM00250000002045 | Ras Gtpase Activating Binding 1 G3BP 1 EC_3.6.4.12 EC_3.6.4.- 13 Atp Dependent Helicase |
| ENSDARG00000017742 | ENSFM00250000000328 | Metabotropic Glutamate Receptor Precursor |
| ENSDARG00000017772 | ENSFM00610000972252 | Unknown |
| ENSDARG00000017772 | ENSFM00610000972253 | Unknown |
| ENSDARG00000017772 | ENSFM00730001521535 | Malate Dehydrogenase Cytoplasmic EC_1.1.1.37 Cytosolic Malate Dehydrogenase |
| ENSDARG00000017781 | ENSFM00500000270523 | 3 Hydroxyacyl Coa Dehydrogenase Type 2 EC_1.1.1.35 17 Beta Hydroxysteroid Dehydrogenase 10 17 Beta Hsd 10 EC_1.1.1.- 51 3 Hydroxy 2 Methylbutyryl Coa Dehydrogenase EC_1.1.-.- 1 178 3 Hydroxyacyl Coa Dehydrogenase Type Ii Endoplasmic Reticulum Associated A |
| ENSDARG00000017785 | ENSFM00800002009756 | Unknown |
| ENSDARG00000017785 | ENSFM00250000000906 | Brain Specific Angiogenesis Inhibitor 1 Associated 2 Bai Associated 2 BAI1 Associated 2 Insulin Receptor Substrate Of 53 Kda IRSP53 Insulin Receptor Substrate P53 |
| ENSDARG00000017803 | ENSFM00810002040612 | Glycogen Synthase Kinase 3 Gsk 3 EC_2.7.11.26 Serine/Threonine Kinase EC_2.7.11.- 1 |
| ENSDARG00000017813 | ENSFM00500000271389 | Gpi Transamidase Component Pig S Phosphatidylinositol Glycan Biosynthesis Class S |
| ENSDARG00000017841 | ENSFM00250000001833 | Cullin Associated NEDD8 Dissociated Cullin Associated And Neddylation Dissociated Tbp Interacting Of 120 Kda Tbp Interacting P120 |
| ENSDARG00000017842 | ENSFM00500000271553 | E3 Ubiquitin Ligase Synoviolin Precursor EC_6.3.2.- Synovial Apoptosis Inhibitor 1 |
| ENSDARG00000017844 | ENSFM00500000270533 | Coatomer Subunit Zeta Zeta Coat Zeta Cop |
| ENSDARG00000017891 | ENSFM00500000270058 | T Complex 1 Subunit Alpha Tcp 1 Alpha Cct Alpha |
| ENSDARG00000017901 | ENSFM00610000960380 | Uncharacterized |
| ENSDARG00000017901 | ENSFM00250000001103 | Talin |
| ENSDARG00000017986 | ENSFM00250000001900 | Vacuolar Sorting Associated 13D |
| ENSDARG00000018000 | ENSFM00250000001310 | Transcription Factor CP2 |
| ENSDARG00000018002 | ENSFM00670001235679 | Enoyl Coa Delta Isomerase 1 Mitochondrial Precursor EC_5.3.3.8 3 2 Trans Enoyl Coa Isomerase Delta 3 Delta 2 Enoyl Coa Isomerase D3 D2 Enoyl Coa Isomerase Dodecenoyl Coa Isomerase |
| ENSDARG00000018032 | ENSFM00740001589070 | Sodium Channel Type Subunit Alpha Sodium Channel Sodium Channel Type Subunit Alpha Voltage Gated Sodium Channel Subunit Alpha NAV1 |
| ENSDARG00000018065 | ENSFM00290000065521 | Precursor |
| ENSDARG00000018069 | ENSFM00500000269664 | Retinol Dehydrogenase |
| ENSDARG00000018110 | ENSFM00730001521739 | Serine/Threonine Kinase Pak EC_2.7.11.1 P21 Activated Kinase Pak |
| ENSDARG00000018124 | ENSFM00250000003614 | 26S Proteasome Non Atpase Regulatory Subunit 3 26S Proteasome Subunit 26S Proteasome Regulatory Subunit |
| ENSDARG00000018146 | ENSFM00500000270307 | Glutathione Peroxidase 2 Gpx 2 Gshpx 2 EC_1.11.1.9 Glutathione Peroxidase Gastrointestinal Gpx Gi Gshpx Gi |
| ENSDARG00000018162 | ENSFM00770001847643 | Alpha Catulin Alpha Catenin Related Acrp Catenin Alpha 1 |
| ENSDARG00000018174 | ENSFM00410000138458 | Guanine Nucleotide Binding Subunit |
| ENSDARG00000018190 | ENSFM00250000003927 | Atpase GET3 |
| ENSDARG00000018259 | ENSFM00650001139994 | Sodium/Potassium Transporting Atpase Subunit Alpha Na + /K + Atpase Alpha Subunit EC_3.6.3.9 Sodium Pump Subunit Alpha |
| ENSDARG00000018270 | ENSFM00320000100099 | Noelin Precursor Neuronal Olfactomedin Related Er Localized Olfactomedin 1 |
| ENSDARG00000018285 | ENSFM00800002003539 | 3 Phosphoinositide Dependent Kinase EC_2.7.11.1 |
| ENSDARG00000018334 | ENSFM00500000271690 | 60S Ribosomal L35 |
| ENSDARG00000018397 | ENSFM00760001714608 | Unknown |
| ENSDARG00000018404 | ENSFM00560000771030 | Keratin Type I Cytoskeletal 18 Cytokeratin 18 Ck 18 Keratin 18 K18 |
| ENSDARG00000018426 | ENSFM00420000140646 | Alpha Aminoadipic Semialdehyde Dehydrogenase Precursor Alpha Aasa Dehydrogenase EC_1.2.1.31 Aldehyde Dehydrogenase Family 7 Member A1 EC_1.2.1.- 3 Antiquitin 1 Betaine Aldehyde Dehydrogenase EC_1.2.-.- 1 8 DELTA1 Piperideine 6 Carboxylate Dehydrogenase P6 |
| ENSDARG00000018432 | ENSFM00250000007261 | Cation Dependent Mannose 6 Phosphate Receptor Precursor Cd Man 6 P Receptor Cd Mpr 46 Kda Mannose 6 Phosphate Receptor Mpr 46 |
| ENSDARG00000018491 | ENSFM00800002003822 | Disulfide Isomerase A4 Precursor EC_5.3.4.1 Endoplasmic Reticulum Resident 70 Er 70 ERP70 Endoplasmic Reticulum Resident 72 Er 72 Erp 72 ERP72 |
| ENSDARG00000018542 | ENSFM00730001521886 | Hyaluronan And Proteoglycan Link 1 Precursor Cartilage Linking 1 Cartilage Link Proteoglycan Link |
| ENSDARG00000018562 | ENSFM00500000271207 | Coatomer Subunit Epsilon Epsilon Coat Epsilon Cop |
| ENSDARG00000018600 | ENSFM00250000003890 | All Trans Retinol 13 14 Reductase Precursor EC_1.3.99.23 All Trans 13 14 Dihydroretinol Saturase Retsat Ppar Alpha Regulated And Starvation Induced Gene |
| ENSDARG00000018602 | ENSFM00500000269759 | Ras Related Rab |
| ENSDARG00000018618 | ENSFM00600000921459 | Cleavage And Polyadenylation Specificity Factor Subunit 6 |
| ENSDARG00000018637 | ENSFM00500000275476 | Unknown |
| ENSDARG00000018653 | ENSFM00250000005415 | Cytosolic Acyl Coenzyme A Thioester Hydrolase EC_3.1.2.2 Acyl Coa Thioesterase 7 Brain Acyl Coa Hydrolase Bach Cte Iia Cte Ii Long Chain Acyl Coa Thioester Hydrolase |
| ENSDARG00000018693 | ENSFM00730001521172 | Cadherin Precursor Cadherin Cadherin |
| ENSDARG00000018716 | ENSFM00320000100117 | Diacylglycerol Kinase Eta |
| ENSDARG00000018738 | ENSFM00250000006432 | Centromere/Kinetochore ZW10 Homolog |
| ENSDARG00000018743 | ENSFM00740001589424 | Secretory Carrier Associated Membrane Secretory Carrier Membrane |
| ENSDARG00000018817 | ENSFM00730001521380 | Brain Derived Neurotrophic Factor Bdnf |
| ENSDARG00000018817 | ENSFM00790001962973 | Brain Derived Neurotrophic Factor Precursor Bdnf |
| ENSDARG00000018820 | ENSFM00740001589123 | Filamin Fln Abp 280 Actin Binding Filamin |
| ENSDARG00000018903 | ENSFM00250000006128 | Aminoacyl Trna Synthase Complex Interacting Multifunctional 2 Multisynthase Complex Auxiliary Component P38 Jtv 1 |
| ENSDARG00000018923 | ENSFM00570000851039 | Protocadherin Fat Precursor Fat Tumor Suppressor Homolog |
| ENSDARG00000018966 | ENSFM00730001521809 | Nadh Cytochrome B5 Reductase B5R EC_1.6.2.2 |
| ENSDARG00000018967 | ENSFM00250000000928 | Gamma Aminobutyric Acid Type B Receptor Subunit 2 Precursor Gaba B Receptor 2 Gaba B R2 Gaba BR2 GABABR2 GB2 G Coupled Receptor 51 |
| ENSDARG00000018973 | ENSFM00810002040917 | Cleft Lip And Palate Transmembrane 1 |
| ENSDARG00000018989 | ENSFM00360000109870 | Unknown |
| ENSDARG00000018997 | ENSFM00670001235939 | Complexin Complexin Cpx Synaphin |
| ENSDARG00000019000 | ENSFM00310000089021 | Structural Maintenance Of Chromosomes 3 Smc 3 Smc 3 Chondroitin Sulfate Proteoglycan 6 |
| ENSDARG00000019008 | ENSFM00250000004114 | Endoplasmic Reticulum Resident 44 Precursor Er 44 ERP44 Thioredoxin Domain Containing 4 |
| ENSDARG00000019060 | ENSFM00250000002957 | Sorting Nexin |
| ENSDARG00000019062 | ENSFM00500000270526 | Unknown |
| ENSDARG00000019098 | ENSFM00500000271329 | CD82 Antigen C33 Antigen IA4 Inducible Membrane R2 Metastasis Suppressor Kangai 1 CD82 Antigen |
| ENSDARG00000019144 | ENSFM00800002003435 | Ras Related Rab |
| ENSDARG00000019181 | ENSFM00250000001034 | 40S Ribosomal Sa |
| ENSDARG00000019191 | ENSFM00500000269805 | Septin |
| ENSDARG00000019205 | ENSFM00250000002493 | Constitutive Coactivator Of Ppar Gamma 1 Oxidative Stress Associated Src Activator FAM120A |
| ENSDARG00000019230 | ENSFM00750001632337 | 60S Ribosomal L7A |
| ENSDARG00000019231 | ENSFM00740001589109 | Spectrin Alpha Chain Non Erythrocytic 1 Alpha Ii Spectrin Fodrin Alpha Chain |
| ENSDARG00000019239 | ENSFM00510000502743 | Cullin |
| ENSDARG00000019260 | ENSFM00500000269693 | Precursor EC_1.1.-.- 3 Alpha Hydroxysteroid 3 Alpha |
| ENSDARG00000019278 | ENSFM00250000000545 | Guanine Nucleotide Binding G I /G S /G T Subunit Beta Transducin Beta Chain |
| ENSDARG00000019280 | ENSFM00250000002483 | Leucine Trna Ligase Cytoplasmic EC_6.1.1.4 Leucyl Trna Synthetase Leurs |
| ENSDARG00000019326 | ENSFM00730001521917 | Phosphoribosyl Pyrophosphate Synthase Associated Prpp Synthase Associated |
| ENSDARG00000019328 | ENSFM00730001521329 | Plexin Precursor |
| ENSDARG00000019332 | ENSFM00250000011695 | Uncharacterized |
| ENSDARG00000019332 | ENSFM00670001238241 | Nadh Dehydrogenase 1 Beta Subcomplex Subunit 4 |
| ENSDARG00000019341 | ENSFM00500000269807 | Glypican Secreted Glypican |
| ENSDARG00000019345 | ENSFM00610000958444 | Unknown |
| ENSDARG00000019345 | ENSFM00730001521220 | Kinase EC_2.7.11.1 |
| ENSDARG00000019355 | ENSFM00250000006626 | Cytochrome C Oxidase Assembly Factor Beta Lactamase Hcp SEL1 Repeat Containing |
| ENSDARG00000019371 | ENSFM00440000236870 | Vascular Endothelial Growth Factor Receptor Precursor Vegfr EC_2.7.10.1 Fms Tyrosine Kinase Flt Tyrosine Kinase Receptor |
| ENSDARG00000019383 | ENSFM00730001521452 | Serine/Threonine Kinase Mrck EC_2.7.11.1 CDC42 Binding Kinase Dmpk Myotonic Dystrophy Kinase Related CDC42 Binding Kinase Mrck Myotonic Dystrophy Kinase |
| ENSDARG00000019398 | ENSFM00420000140606 | Proteasome Subunit Alpha Type EC_3.4.25.1 Macropain Multicatalytic Endopeptidase Complex Proteasome |
| ENSDARG00000019404 | ENSFM00500000272854 | Atp Synthase Subunit Delta Mitochondrial Precursor F Atpase Delta Subunit |
| ENSDARG00000019426 | ENSFM00740001589124 | EC_2.7.11.1 |
| ENSDARG00000019444 | ENSFM00250000005113 | Translocon Associated Subunit Delta Precursor Trap Delta Signal Sequence Receptor Subunit Delta Ssr Delta |
| ENSDARG00000019451 | ENSFM00760001714725 | Neuroendocrine Convertase 2 Precursor Nec 2 EC_3.4.21.94 KEX2 Endoprotease 2 Prohormone Convertase 2 Proprotein Convertase 2 PC2 |
| ENSDARG00000019496 | ENSFM00250000003999 | S Formylglutathione Hydrolase Fgh EC_3.1.2.12 Esterase D |
| ENSDARG00000019526 | ENSFM00650001140030 | Arf Gap Domain And Fg Repeat Containing 1 Hiv 1 Rev Binding Nucleoporin Rip |
| ENSDARG00000019529 | ENSFM00250000001234 | Poly Synthase 1 |
| ENSDARG00000019532 | ENSFM00250000001297 | Fatty Acid Desaturase 2 EC_1.14.19.- Delta 6 Fatty Acid Desaturase Desaturase Delta 6 Desaturase |
| ENSDARG00000019549 | ENSFM00750001632370 | Phosphatidate Cytidylyltransferase 2 EC_2.7.7.41 Cdp Dag Synthase 2 Cdp Dg Synthase 2 Cdp Diacylglycerol Synthase 2 Cds 2 Cdp Diglyceride Pyrophosphorylase 2 Cdp Diglyceride Synthase 2 Ctp:Phosphatidate Cytidylyltransferase 2 |
| ENSDARG00000019556 | ENSFM00740001589241 | H + /Cl Exchange Transporter 7 Chloride Channel 7 Alpha Subunit Chloride Channel 7 Clc 7 |
| ENSDARG00000019581 | ENSFM00500000270803 | General Vesicular Transport Factor P115 USO1 Homolog Transcytosis Associated Tap Vesicle Docking |
| ENSDARG00000019596 | ENSFM00730001521888 | Nardilysin Precursor EC_3.4.24.61 N Arginine Dibasic Convertase Nrd Convertase Nrd C |
| ENSDARG00000019644 | ENSFM00250000000475 | L Lactate Dehydrogenase Chain Ldh EC_1.1.1.27 |
| ENSDARG00000019651 | ENSFM00800002003401 | Septin |
| ENSDARG00000019702 | ENSFM00250000000488 | Fructose Bisphosphate Aldolase EC_4.1.2.13 Type Aldolase |
| ENSDARG00000019713 | ENSFM00400000131714 | Solute Carrier Family 22 Member Organic Anion Transporter |
| ENSDARG00000019732 | ENSFM00250000001870 | Mitochondrial Carrier Homolog |
| ENSDARG00000019743 | ENSFM00760001714751 | Dynactin Subunit 1.150 Kda Dynein Associated Polypeptide Dap 150 Dp 150 Glued |
| ENSDARG00000019778 | ENSFM00250000001600 | 40S Ribosomal S6 |
| ENSDARG00000019856 | ENSFM00650001139994 | Sodium/Potassium Transporting Atpase Subunit Alpha Na + /K + Atpase Alpha Subunit EC_3.6.3.9 Sodium Pump Subunit Alpha |
| ENSDARG00000019881 | ENSFM00610000952905 | Basigin Precursor CD147 Antigen |
| ENSDARG00000019924 | ENSFM00770001847583 | Ump Cmp Kinase |
| ENSDARG00000019945 | ENSFM00740001589097 | Receptor Type Tyrosine Phosphatase Precursor EC_3.1.3.48 |
| ENSDARG00000019951 | ENSFM00250000004532 | Translocation SEC62 Translocation 1 Tp 1 |
| ENSDARG00000019963 | ENSFM00730001521561 | Sideroflexin |
| ENSDARG00000020007 | ENSFM00250000000184 | Collagen Alpha I Chain Precursor Alpha Type I Collagen |
| ENSDARG00000020008 | ENSFM00750001632324 | Transitional Endoplasmic Reticulum Atpase Ter Atpase EC_3.6.4.6 15S Mg 2+ Atpase P97 Subunit Valosin Containing Vcp |
| ENSDARG00000020054 | ENSFM00250000000203 | Aldehyde Oxidase EC_1.2.3.1 Azaheterocycle Hydroxylase EC_1.17.3.- |
| ENSDARG00000020057 | ENSFM00730001522171 | Anti Muellerian Hormone Type 2 Receptor Precursor EC_2.7.11.30 Anti Muellerian Hormone Type Ii Receptor Amh Type Ii Receptor Mis Type Ii Receptor Misrii Mrii |
| ENSDARG00000020101 | ENSFM00730001521776 | 26S Protease Regulatory Subunit 7 26S Proteasome Aaa Atpase Subunit RPT1 Proteasome 26S Subunit Atpase 2 |
| ENSDARG00000020102 | ENSFM00260000050545 | Btb/Poz Domain Containing |
| ENSDARG00000020123 | ENSFM00780001898387 | Chaperone Activity Of BC1 Complex Mitochondrial Precursor Chaperone ABC1 EC_2.7.11.- Aarf Domain Containing Kinase 3 |
| ENSDARG00000020128 | ENSFM00730001522125 | Ras Related Rab |
| ENSDARG00000020134 | ENSFM00730001521477 | Signal Induced Proliferation Associated 1 SIPA1 |
| ENSDARG00000020149 | ENSFM00500000272795 | Acyl Coenzyme A Oxidase Acyl Coa Oxidase EC_1.3.3.- |
| ENSDARG00000020164 | ENSFM00250000002014 | Ephrin B1 Precursor Elk Ligand Elk L Eph Related Receptor Tyrosine Kinase Ligand 2 Lerk 2 |
| ENSDARG00000020231 | ENSFM00250000000867 | Microtubule Associated Rp/Eb Family Member |
| ENSDARG00000020261 | ENSFM00730001521469 | Ras Related Rab |
| ENSDARG00000020328 | ENSFM00800002003674 | Pleckstrin Homology Domain Containing Family A Member 6 Ph Domain Containing Family A Member 6 Phosphoinositol 3 Phosphate Binding 3 Pepp 3 |
| ENSDARG00000020345 | ENSFM00250000000820 | Clip Associating Cytoplasmic Linker Associated |
| ENSDARG00000020387 | ENSFM00250000007263 | Nipsnap Homolog 3A NIPSNAP3A |
| ENSDARG00000020405 | ENSFM00730001522002 | Soluble Nsf Attachment Snap N Ethylmaleimide Sensitive Factor Attachment |
| ENSDARG00000020443 | ENSFM00500000269993 | Xk Related |
| ENSDARG00000020449 | ENSFM00250000005379 | YIPF3 YIP1 Family Member 3 |
| ENSDARG00000020450 | ENSFM00250000002019 | Voltage Dependent Calcium Channel Gamma Subunit Neuronal Voltage Gated Calcium Channel Gamma Subunit Transmembrane Ampar Regulatory Gamma Tarp Gamma |
| ENSDARG00000020454 | ENSFM00250000004187 | 26S Proteasome Non Atpase Regulatory Subunit 8 26S Proteasome Regulatory Subunit RPN12 26S Proteasome Regulatory Subunit S14 |
| ENSDARG00000020482 | ENSFM00250000000671 | Paraspeckle Component 1 |
| ENSDARG00000020493 | ENSFM00250000002312 | Leucine Rich Repeat Lgi Family Member Precursor Leucine Rich Glioma Inactivated |
| ENSDARG00000020497 | ENSFM00390000126319 | Ras Related Rab |
| ENSDARG00000020521 | ENSFM00250000001326 | Exocyst Complex Component 6 Exocyst Complex Component SEC15A SEC15 1 |
| ENSDARG00000020529 | ENSFM00630001070444 | Unknown |
| ENSDARG00000020529 | ENSFM00250000003931 | 2 Hydroxyacyl Coa Lyase 1 EC_4.1.-.- 2 Hydroxyphytanoyl Coa Lyase 2 Hpcl Phytanoyl Coa 2 Hydroxylase 2 |
| ENSDARG00000020573 | ENSFM00770001847530 | Atp Dependent Rna Helicase DDX3Y EC_3.6.4.13 Dead Box 3 Y Chromosomal |
| ENSDARG00000020607 | ENSFM00500000270967 | Er Membrane Complex Subunit 3 Transmembrane 111 |
| ENSDARG00000020609 | ENSFM00250000001228 | Synaptosomal Associated 25 Snap 25 Synaptosomal Associated 25 Kda |
| ENSDARG00000020618 | ENSFM00250000005161 | ES1 Mitochondrial Precursor |
| ENSDARG00000020621 | ENSFM00430000230089 | Ap 1 Complex Subunit Beta 1 Adaptor Complex Ap 1 Subunit Beta 1 Adaptor Related Complex 1 Subunit Beta 1 Beta 1 Adaptin Beta Adaptin 1 Clathrin Assembly Complex 1 Beta Large Chain Golgi Adaptor HA1/AP1 Adaptin Beta Subunit |
| ENSDARG00000020645 | ENSFM00250000000444 | Cationic Amino Acid Transporter 2 Cat 2 CAT2 Low Affinity Cationic Amino Acid Transporter 2 Solute Carrier Family 7 Member 2 |
| ENSDARG00000020676 | ENSFM00810002040563 | Dipeptidyl Peptidase 3 EC_3.4.14.4 Dipeptidyl Aminopeptidase Iii Dipeptidyl Arylamidase Iii Dipeptidyl Peptidase Iii Dpp Iii |
| ENSDARG00000020679 | ENSFM00250000006881 | Atp Dependent Clp Protease Proteolytic Subunit Mitochondrial Precursor EC_3.4.21.92 Endopeptidase Clp |
| ENSDARG00000020718 | ENSFM00500000270170 | Mitochondrial Glutamate Carrier 1 Gc 1 Glutamate/H + Symporter 1 Solute Carrier Family 25 Member 22 |
| ENSDARG00000020741 | ENSFM00250000004159 | Pre Splicing Factor PRP46 Pre Processing 46 |
| ENSDARG00000020764 | ENSFM00250000002303 | Sarcolemmal Membrane Associated Sarcolemmal Associated |
| ENSDARG00000020771 | ENSFM00740001589132 | Tenascin Precursor Tn |
| ENSDARG00000020795 | ENSFM00730001521186 | Ras Related C3 Botulinum Toxin Substrate Precursor P21 |
| ENSDARG00000020798 | ENSFM00250000002415 | Adaptin Ear Binding Coat Associated 1 Necap Endocytosis Associated 1 Necap 1 |
| ENSDARG00000020850 | ENSFM00800002003364 | Elongation Factor 1 Alpha Ef 1 Alpha Eukaryotic Elongation Factor 1 A EEF1A |
| ENSDARG00000020871 | ENSFM00730001521864 | Receptor Type Tyrosine Phosphatase Precursor R Ptp EC_3.1.3.48 |
| ENSDARG00000020890 | ENSFM00500000269813 | Tropomodulin Tropomodulin Tmod |
| ENSDARG00000020895 | ENSFM00250000006518 | Bag Family Molecular Chaperone Regulator 1 Bag 1 Bcl 2 Associated Athanogene 1 |
| ENSDARG00000020953 | ENSFM00570000851144 | Dnaj Homolog Subfamily B Member |
| ENSDARG00000020960 | ENSFM00500000271438 | Hepatocyte Growth Factor Regulated Tyrosine Kinase Substrate |
| ENSDARG00000020982 | ENSFM00730001521542 | Prickle |
| ENSDARG00000021048 | ENSFM00740001589427 | Cleft Lip And Palate Transmembrane 1 CLPTM1 |
| ENSDARG00000021124 | ENSFM00500000270025 | Cofilin Cofilin Muscle |
| ENSDARG00000021135 | ENSFM00500000270135 | Dehydrogenase/Reductase Sdr Family Member 4 EC_1.1.1.184 Nadph Dependent Carbonyl Reductase/Nadp Retinol Dehydrogenase Cr Phcr Nadph Dependent Retinol Dehydrogenase/Reductase Ndrd Peroxisomal Short Chain Alcohol Dehydrogenase Pscd |
| ENSDARG00000021143 | ENSFM00500000269689 | Reticulon |
| ENSDARG00000021149 | ENSFM00250000002301 | Carbonyl Reductase EC_1.1.1.- 197 20 Beta Hydroxysteroid Dehydrogenase Nadph Dependent Carbonyl Reductase 1 Prostaglandin 9 Ketoreductase Prostaglandin E 2 9 Reductase EC_1.1.-.- 1 189 |
| ENSDARG00000021193 | ENSFM00610000967750 | Unknown |
| ENSDARG00000021193 | ENSFM00730001521292 | Coronin |
| ENSDARG00000021200 | ENSFM00500000270806 | Lim And SH3 Domain 1 Lasp 1 |
| ENSDARG00000021220 | ENSFM00250000002331 | Hydroxymethylglutaryl Coa Lyase Mitochondrial Precursor Hl Hmg Coa Lyase EC_4.1.3.4 3 Hydroxy 3 Methylglutarate Coa Lyase |
| ENSDARG00000021242 | ENSFM00250000005419 | Major Vault Mvp |
| ENSDARG00000021252 | ENSFM00350000105437 | T Complex 1 Subunit Zeta Tcp 1 Zeta Cct Zeta |
| ENSDARG00000021257 | ENSFM00670001275463 | Unknown |
| ENSDARG00000021257 | ENSFM00670001275464 | Unknown |
| ENSDARG00000021257 | ENSFM00670001281310 | Unknown |
| ENSDARG00000021257 | ENSFM00250000003134 | Eukaryotic Translation Initiation Factor 3 Subunit D |
| ENSDARG00000021287 | ENSFM00390000126319 | Ras Related Rab |
| ENSDARG00000021309 | ENSFM00730001521223 | Rho Related Gtp Binding Precursor |
| ENSDARG00000021346 | ENSFM00500000270150 | Pyruvate Dehydrogenase E1 Component Subunit Beta Mitochondrial Precursor PDHE1 B EC_1.2.4.1 |
| ENSDARG00000021351 | ENSFM00730001521354 | Neurofilament Polypeptide Nf Kda Neurofilament Neurofilament Triplet |
| ENSDARG00000021352 | ENSFM00720001492020 | Glutamate Receptor Precursor Glur Ampa Selective Glutamate Receptor Glur Glutamate Receptor Ionotropic Ampa |
| ENSDARG00000021366 | ENSFM00250000001433 | Fructose 1 6 Bisphosphatase 1 Fbpase 1 EC_3.1.3.11 D Fructose 1 6 Bisphosphate 1 Phosphohydrolase 1 Liver Fbpase |
| ENSDARG00000021404 | ENSFM00760001714944 | NFU1 Iron Sulfur Cluster Scaffold Homolog Mitochondrial |
| ENSDARG00000021442 | ENSFM00800002003474 | Cadherin Precursor |
| ENSDARG00000021539 | ENSFM00730001521338 | Ephrin Type B Receptor Kinase |
| ENSDARG00000021556 | ENSFM00500000272867 | Ubiquitin 3 Precursor Membrane Anchored Ubiquitin Fold Mub |
| ENSDARG00000021564 | ENSFM00250000001189 | Voltage Dependent Anion Selective Channel Vdac Outer Mitochondrial Membrane Porin |
| ENSDARG00000021582 | ENSFM00800002017697 | Unknown |
| ENSDARG00000021582 | ENSFM00740001589456 | Dynein Light Chain Roadblock Type Dynein Light Chain Cytoplasmic |
| ENSDARG00000021584 | ENSFM00390000126303 | Contactin Precursor Brain Derived Immunoglobulin Superfamily Big |
| ENSDARG00000021590 | ENSFM00250000000552 | Membrane Associated Guanylate Kinase Ww And Pdz Domain Containing Membrane Associated Guanylate Kinase Inverted Magi |
| ENSDARG00000021595 | ENSFM00250000002406 | Lipoma Hmgic Fusion Partner |
| ENSDARG00000021607 | ENSFM00290000065521 | Precursor |
| ENSDARG00000021611 | ENSFM00610000972000 | Unknown |
| ENSDARG00000021611 | ENSFM00250000000621 | Serine/Threonine Kinase EC_2.7.11.1 Kinase Lysine Deficient Kinase With No Lysine |
| ENSDARG00000021647 | ENSFM00410000138458 | Guanine Nucleotide Binding Subunit |
| ENSDARG00000021669 | ENSFM00250000001640 | Transport SEC61 Subunit Alpha |
| ENSDARG00000021681 | ENSFM00500000271079 | Isoaspartyl Peptidase/L Asparaginase Precursor EC_3.4.19.5 EC_3.5.1.- 1 Asparaginase 1 Beta Aspartyl Peptidase Isoaspartyl Dipeptidase L Asparagine Amidohydrolase |
| ENSDARG00000021808 | ENSFM00630001072000 | Unknown |
| ENSDARG00000021808 | ENSFM00760001714660 | Glycylpeptide N Tetradecanoyltransferase 1 EC_2.3.1.97 |
| ENSDARG00000021838 | ENSFM00250000002784 | 40S Ribosomal S23 |
| ENSDARG00000021859 | ENSFM00740001589214 | Endoplasmic Reticulum Aminopeptidase EC_3.4.11.- Derived Aminopeptidase |
| ENSDARG00000021864 | ENSFM00610000961726 | Unknown |
| ENSDARG00000021864 | ENSFM00500000308224 | Ribosomal P1 |
| ENSDARG00000021948 | ENSFM00740001589132 | Tenascin Precursor Tn |
| ENSDARG00000021973 | ENSFM00250000002693 | HSC70 Interacting Hip Fa ST13 Homolog |
| ENSDARG00000021984 | ENSFM00670001239126 | Nadh Dehydrogenase 1 Alpha Subcomplex Subunit 2 |
| ENSDARG00000021987 | ENSFM00740001589199 | Plectin Pcn Pltn Plectin 1 |
| ENSDARG00000021991 | ENSFM00250000003616 | Erlin 2 Endoplasmic Reticulum Lipid Raft Associated 2 Stomatin Prohibitin Flotillin Hflc/K Domain Containing 2 Spfh Domain Containing 2 |
| ENSDARG00000021996 | ENSFM00250000000829 | Serine/Threonine Phosphatase 2A 55 Kda Regulatory Subunit B PP2A Subunit B B55 PP2A Subunit B PR55 PP2A Subunit B R2 PP2A Subunit B |
| ENSDARG00000022045 | ENSFM00250000000942 | Microtubule Associated Map |
| ENSDARG00000022129 | ENSFM00610000973231 | Unknown |
| ENSDARG00000022129 | ENSFM00350000105447 | Rna Binding Rna Binding Motif Rna Binding Motif |
| ENSDARG00000022165 | ENSFM00500000272807 | Microsomal Glutathione S Transferase 1 Microsomal Gst 1 EC_2.5.1.18 Microsomal Gst I |
| ENSDARG00000022176 | ENSFM00290000065521 | Precursor |
| ENSDARG00000022183 | ENSFM00500000270301 | Glutathione S Transferase Omega 1 Gsto 1 EC_2.5.1.18 Glutathione S Transferase Omega 1 1 Gsto 1 1 Glutathione Dependent Dehydroascorbate Reductase EC_1.8.5.- 1 Monomethylarsonic Acid Reductase Mma V Reductase EC_1.20.-.- 4 2 S Phenacyl Glutathione Reducta |
| ENSDARG00000022254 | ENSFM00520000517802 | Kinase C Type Pkc EC_2.7.11.13 |
| ENSDARG00000022303 | ENSFM00500000314819 | Hypoxia Induced Gene 1 |
| ENSDARG00000022315 | ENSFM00670001236060 | V Type Proton Atpase Subunit G 1 V Atpase Subunit G 1 V Atpase 13 Kda Subunit 1 Vacuolar Proton Pump Subunit G 1 Vacuolar Proton Pump Subunit M16 |
| ENSDARG00000022437 | ENSFM00800002003853 | CD81 Antigen 26 Kda Cell Surface Tapa 1 Target Of The Antiproliferative Antibody 1 CD81 Antigen |
| ENSDARG00000022438 | ENSFM00670001237914 | Cytochrome C Oxidase Subunit 6A Mitochondrial |
| ENSDARG00000022456 | ENSFM00730001521170 | Enolase EC_4.2.1.11 2 Phospho D Glycerate Hydro Lyase |
| ENSDARG00000022509 | ENSFM00500000271199 | Cytochrome C Oxidase Subunit 4 1 Mitochondrial Cytochrome C Oxidase Polypeptide Iv Cytochrome C Oxidase Subunit Iv 1 Cox Iv 1 |
| ENSDARG00000022531 | ENSFM00570000851049 | Netrin Precursor |
| ENSDARG00000022599 | ENSFM00250000004224 | Aspartyl Aminopeptidase EC_3.4.11.21 |
| ENSDARG00000022659 | ENSFM00250000006922 | Sorting Nexin 4 |
| ENSDARG00000022668 | ENSFM00500000270097 | Growth Factor Receptor Bound 2 Adapter GRB2 SH2/SH3 Adapter GRB2 |
| ENSDARG00000022684 | ENSFM00670001235580 | Peptidyl Prolyl Cis Trans Isomerase FKBP1B Ppiase FKBP1B EC_5.2.1.8 12.6 Kda FK506 Binding 12.6 Kda Fkbp Fkbp 12 6 FK506 Binding 1B Fkbp 1B Immunophilin FKBP12 6 Rotamase |
| ENSDARG00000022763 | ENSFM00610000963421 | Unknown |
| ENSDARG00000022763 | ENSFM00760001714608 | Unknown |
| ENSDARG00000022767 | ENSFM00740001589094 | Apolipoprotein B 100 Precursor Apo B 100 |
| ENSDARG00000022788 | ENSFM00250000003382 | COP9 Signalosome Complex Subunit 7A SGN7A Signalosome Subunit 7A JAB1 Containing Signalosome Subunit 7A |
| ENSDARG00000022971 | ENSFM00730001521142 | Ephrin Type A Receptor Precursor EC_2.7.10.1 Kinase |
| ENSDARG00000022983 | ENSFM00500000272544 | Coiled Coil Domain Containing 28A CCRL1AP |
| ENSDARG00000023151 | ENSFM00500000269770 | Mitochondrial Brown Fat Uncoupling 1 Ucp 1 Solute Carrier Family 25 Member 7 Thermogenin |
| ENSDARG00000023181 | ENSFM00500000330711 | Unknown |
| ENSDARG00000023190 | ENSFM00750001632344 | Importin Subunit Alpha 3 Importin Alpha Q1 QIP1 Karyopherin Subunit Alpha 4 |
| ENSDARG00000023220 | ENSFM00250000003123 | Selenoprotein Precursor |
| ENSDARG00000023228 | ENSFM00760001714608 | Unknown |
| ENSDARG00000023267 | ENSFM00790001963005 | Atpase Family Aaa Domain Containing 1 EC_3.6.1.3 |
| ENSDARG00000023279 | ENSFM00810002040686 | 26S Proteasome Non Atpase Regulatory Subunit 4 26S Proteasome Regulatory Subunit RPN10 26S Proteasome Regulatory Subunit S5A Multiubiquitin Chain Binding |
| ENSDARG00000023290 | ENSFM00800002003523 | Fatty Acid Binding Protein Heart Fatty Acid Binding 3 Heart Type Fatty Acid Binding H Fabp |
| ENSDARG00000023298 | ENSFM00670001235597 | 40S Ribosomal S27 |
| ENSDARG00000023299 | ENSFM00670001235480 | Unknown |
| ENSDARG00000023309 | ENSFM00260000051295 | Spry Domain Containing 4 |
| ENSDARG00000023323 | ENSFM00730001521154 | Unknown |
| ENSDARG00000023443 | ENSFM00250000000691 | Tight Junction Zo Tight Junction Zona Occludens Zonula Occludens |
| ENSDARG00000023445 | ENSFM00250000000445 | Plasma Membrane Calcium Transporting Atpase EC_3.6.3.8 Plasma Membrane Calcium Atpase Plasma Membrane Calcium Pump |
| ENSDARG00000023472 | ENSFM00250000001012 | Catenin Beta Beta Catenin |
| ENSDARG00000023536 | ENSFM00250000002458 | Nad P Transhydrogenase Mitochondrial Precursor EC_1.6.1.2 Nicotinamide Nucleotide Transhydrogenase Pyridine Nucleotide Transhydrogenase |
| ENSDARG00000023542 | ENSFM00260000050333 | Cadherin Precursor |
| ENSDARG00000023583 | ENSFM00500000272185 | Ubiquinone Biosynthesis COQ9 Mitochondrial Precursor |
| ENSDARG00000023600 | ENSFM00250000000986 | Endophilin SH3 Domain Containing GRB2 |
| ENSDARG00000023648 | ENSFM00540000729047 | Unknown |
| ENSDARG00000023648 | ENSFM00770001847549 | Isocitrate Dehydrogenase Gamma Mitochondrial Precursor EC_1.1.1.41 Isocitric Dehydrogenase Subunit Gamma Nad + Specific Icdh Subunit Gamma |
| ENSDARG00000023672 | ENSFM00670001238234 | Mitochondrial Import Inner Membrane Translocase Subunit TIM8 A |
| ENSDARG00000023712 | ENSFM00250000002249 | Amine Oxidase EC_1.4.3.4 Monoamine Oxidase Type Mao |
| ENSDARG00000023713 | ENSFM00750001632461 | Aquaporin 1 Aqp 1 Aquaporin Chip Water Channel Red Blood Cells And Kidney Proximal Tubule |
| ENSDARG00000023771 | ENSFM00400000131718 | Gamma Aminobutyric Acid Receptor Subunit Beta Precursor Gaba A Receptor Subunit Beta |
| ENSDARG00000023797 | ENSFM00250000000332 | Ryanodine Receptor Ryr Muscle Calcium Release Channel Muscle Type Ryanodine Receptor Type Ryanodine Receptor |
| ENSDARG00000023914 | ENSFM00730001521220 | Kinase EC_2.7.11.1 |
| ENSDARG00000023920 | ENSFM00500000270191 | Lethal 2 Giant Larvae |
| ENSDARG00000023940 | ENSFM00250000001269 | Wiskott Aldrich Syndrome Family Member 1 Wasp Family Member 1 Wave 1 |
| ENSDARG00000023967 | ENSFM00750001632375 | V Type Proton Atpase Subunit C V Atpase Subunit C Vacuolar Proton Pump Subunit C |
| ENSDARG00000023988 | ENSFM00250000004748 | Dynactin Subunit 4 Dynactin Subunit P62 |
| ENSDARG00000024017 | ENSFM00250000002191 | Mam Domain Containing Glycosylphosphatidylinositol Anchor 2 Precursor Mam Domain Containing 1 |
| ENSDARG00000024116 | ENSFM00500000297266 | Uncharacterized |
| ENSDARG00000024317 | ENSFM00730001522162 | Heat Shock 75 Kda Mitochondrial Precursor Hsp 75 Tnfr Associated 1 Tumor Necrosis Factor Type 1 Receptor Associated Trap 1 |
| ENSDARG00000024324 | ENSFM00430000230144 | Rho Gtpase Activating Rho Type Gtpase Activating |
| ENSDARG00000024433 | ENSFM00670001235702 | Parvalbumin Beta |
| ENSDARG00000024443 | ENSFM00250000003409 | Glucosamine 6 Phosphate Isomerase EC_3.5.99.6 Glucosamine 6 Phosphate Deaminase Gnpda GLCN6P Deaminase |
| ENSDARG00000024478 | ENSFM00500000273639 | Mitochondrial Pyruvate Carrier 2 Brain 44 |
| ENSDARG00000024479 | ENSFM00810002040906 | Gpi Anchor Transamidase Precursor Gpi Transamidase Ec 3 Phosphatidylinositol Glycan Biosynthesis Class K Pig K |
| ENSDARG00000024564 | ENSFM00250000003507 | Saccharopine Dehydrogenase Oxidoreductase Ec 1 |
| ENSDARG00000024642 | ENSFM00730001521611 | Phosphatidylinositol 4 Phosphate 5 Kinase Type 1 Beta Pi Beta Ptdins 4 P 5 Kinase 1 Beta EC_2.7.1.68 Phosphatidylinositol 4 Phosphate 5 Kinase Type I Phosphatidylinositol 4 Phosphate 5 Kinase Beta |
| ENSDARG00000024694 | ENSFM00800002003560 | Unconventional Myosin Ia Brush Border Myosin I Bbm I Bbmi Myosin I Heavy Chain Mihc |
| ENSDARG00000024717 | ENSFM00250000002879 | Selenium Binding |
| ENSDARG00000024744 | ENSFM00750001632364 | Dipeptidyl Aminopeptidase 6 Dppx Dipeptidyl Aminopeptidase Related Dipeptidyl Peptidase 6 Dipeptidyl Peptidase Iv Dipeptidyl Peptidase Vi Dpp Vi |
| ENSDARG00000024746 | ENSFM00250000000292 | Heat Shock |
| ENSDARG00000024785 | ENSFM00250000001018 | Catenin Alpha 2 Alpha N Catenin |
| ENSDARG00000024846 | ENSFM00250000006097 | COP9 Signalosome Complex Subunit 8 SGN8 Signalosome Subunit 8 COP9 Homolog JAB1 Containing Signalosome Subunit 8 |
| ENSDARG00000024874 | ENSFM00760001714721 | Dedicator Of Cytokinesis 3 Modifier Of Cell Adhesion Presenilin Binding Pbp |
| ENSDARG00000024877 | ENSFM00730001522225 | Prostaglandin Reductase 1 Prg 1 EC_1.3.1.- 15 Oxoprostaglandin 13 Reductase EC_1.3.1.48 Nadp Dependent Leukotriene B4 12 Hydroxydehydrogenase EC_1.3.-.- 1 74 |
| ENSDARG00000024954 | ENSFM00500000270043 | Transmembrane EMP24 Domain Containing Precursor |
| ENSDARG00000024966 | ENSFM00730001521220 | Kinase EC_2.7.11.1 |
| ENSDARG00000025011 | ENSFM00730001521871 | Synaptojanin EC_3.1.3.36 Synaptic Inositol 1 4 5 Trisphosphate 5 Phosphatase |
| ENSDARG00000025012 | ENSFM00250000000728 | Triosephosphate Isomerase Tim EC_5.3.1.1 Triose Phosphate Isomerase |
| ENSDARG00000025034 | ENSFM00670001235552 | Synaptogyrin |
| ENSDARG00000025073 | ENSFM00630001070331 | Unknown |
| ENSDARG00000025073 | ENSFM00760001714679 | 60S Ribosomal L18A |
| ENSDARG00000025074 | ENSFM00760001714704 | 39S Ribosomal L12 Mitochondrial Precursor L12MT Mrp L12 |
| ENSDARG00000025091 | ENSFM00710001464690 | Unknown |
| ENSDARG00000025091 | ENSFM00250000000517 | Moesin |
| ENSDARG00000025106 | ENSFM00310000089007 | Serine/Threonine Phosphatase 2B Catalytic Subunit EC_3.1.3.16 Cam Prp Catalytic Subunit Calmodulin Dependent Calcineurin A Subunit |
| ENSDARG00000025108 | ENSFM00250000000552 | Membrane Associated Guanylate Kinase Ww And Pdz Domain Containing Membrane Associated Guanylate Kinase Inverted Magi |
| ENSDARG00000025189 | ENSFM00730001521728 | Copine Copine |
| ENSDARG00000025206 | ENSFM00720001492031 | Synaptotagmin |
| ENSDARG00000025254 | ENSFM00670001238517 | S100 |
| ENSDARG00000025269 | ENSFM00500000270726 | Programmed Cell Death 6 Interacting Alg 2 Interacting 1 Alg 2 Interacting X |
| ENSDARG00000025299 | ENSFM00500000270370 | Tetraspanin 9 Tspan 9 |
| ENSDARG00000025301 | ENSFM00730001521291 | Vimentin |
| ENSDARG00000025302 | ENSFM00250000002896 | Dixin Coiled Coil DIX1 Coiled Coil DIX1 Dix Domain Containing 1 |
| ENSDARG00000025319 | ENSFM00730001521177 | Tyrosine Kinase EC_2.7.10.2 |
| ENSDARG00000025338 | ENSFM00730001521820 | Hydroxyacylglutathione Hydrolase Mitochondrial Precursor EC_3.1.2.6 Glyoxalase Ii Glx Ii |
| ENSDARG00000025350 | ENSFM00250000000757 | Thioredoxin Peroxidase Thioredoxin Dependent Peroxide Reductase |
| ENSDARG00000025375 | ENSFM00770001847524 | Isocitrate Dehydrogenase Cytoplasmic Idh EC_1.1.1.42 Cytosolic Nadp Isocitrate Dehydrogenase Idp Nadp + Specific Icdh Oxalosuccinate Decarboxylase |
| ENSDARG00000025391 | ENSFM00500000273880 | Prefoldin Subunit 2 |
| ENSDARG00000025500 | ENSFM00250000002187 | Metaxin 1 Mitochondrial Outer Membrane Import Complex 1 |
| ENSDARG00000025555 | ENSFM00610000961155 | Unknown |
| ENSDARG00000025555 | ENSFM00500000270327 | ORM1 |
| ENSDARG00000025559 | ENSFM00250000005998 | Peroxisomal Membrane 11C Peroxin 11C Peroxisomal Biogenesis Factor 11C PEX11 Homolog Gamma PEX11 Gamma |
| ENSDARG00000025566 | ENSFM00610000973264 | Unknown |
| ENSDARG00000025566 | ENSFM00760001714669 | Phosphate Carrier Protein Mitochondrial Precursor Phosphate Transport Ptp Solute Carrier Family 25 Member 3 |
| ENSDARG00000025581 | ENSFM00800002003415 | 60S Ribosomal L10 |
| ENSDARG00000025679 | ENSFM00610000973476 | Unknown |
| ENSDARG00000025679 | ENSFM00500000271337 | Catechol O Methyltransferase EC_2.1.1.6 |
| ENSDARG00000025693 | ENSFM00670001235681 | Transmembrane Precursor |
| ENSDARG00000025728 | ENSFM00730001522127 | Glutamate Receptor Subunit 1 |
| ENSDARG00000025850 | ENSFM00670001236070 | 40S Ribosomal S21 |
| ENSDARG00000025859 | ENSFM00250000004656 | Lipase Maturation Factor 2 |
| ENSDARG00000025889 | ENSFM00780001898425 | Ubiquitin Carboxyl Terminal Hydrolase 14 EC_3.4.19.12 Deubiquitinating Enzyme 14 Ubiquitin Thioesterase 14 Ubiquitin Specific Processing Protease 14 |
| ENSDARG00000025903 | ENSFM00740001589285 | Galectin Gal |
| ENSDARG00000025904 | ENSFM00250000002457 | Very Long Chain Enoyl Coa Reductase EC_1.3.1.93 Synaptic Glycoprotein SC2 Trans 2 3 Enoyl Coa Reductase Ter |
| ENSDARG00000025953 | ENSFM00610000970041 | Unknown |
| ENSDARG00000025953 | ENSFM00730001521235 | Cell Division Control 42 Homolog Precursor |
| ENSDARG00000025972 | ENSFM00500000271084 | Malignant T Cell Amplified Sequence 1 Mct |
| ENSDARG00000025974 | ENSFM00250000000552 | Membrane Associated Guanylate Kinase Ww And Pdz Domain Containing Membrane Associated Guanylate Kinase Inverted Magi |
| ENSDARG00000026068 | ENSFM00500000273083 | Trafficking Particle Complex Subunit 5 |
| ENSDARG00000026072 | ENSFM00560000771298 | Programmed Cell Death 5 Tf 1 Cell Apoptosis Related 19 TFAR19 |
| ENSDARG00000026137 | ENSFM00740001589514 | Sideroflexin 5 |
| ENSDARG00000026322 | ENSFM00500000269664 | Retinol Dehydrogenase |
| ENSDARG00000026333 | ENSFM00500000269993 | Xk Related |
| ENSDARG00000026369 | ENSFM00610000964944 | Unknown |
| ENSDARG00000026376 | ENSFM00250000001292 | Cytoplasmic Aconitate Hydratase Aconitase EC_4.2.1.3 Citrate Hydro Lyase Iron Responsive Element Binding 1 Ire Bp 1 |
| ENSDARG00000026484 | ENSFM00750001632504 | Ras Related Rab 15 |
| ENSDARG00000026499 | ENSFM00260000050737 | Phosphatase 1E EC_3.1.3.16 Ca 2+ /Calmodulin Dependent Kinase Phosphatase N Camkp N Camkp Nucleus Camkn Partner Of Pix 1 Partner Of Pix Alpha Partner Of Pixa |
| ENSDARG00000026531 | ENSFM00500000270582 | CD166 Antigen Activated Leukocyte Cell Adhesion Molecule CD166 Antigen |
| ENSDARG00000026595 | ENSFM00810002040664 | Lissencephaly 1 |
| ENSDARG00000026629 | ENSFM00610000961238 | Gdp Mannose 4 6 Dehydratase |
| ENSDARG00000026629 | ENSFM00250000003487 | Gdp Mannose 4 6 Dehydratase EC_4.2.1.47 Gdp D Mannose Dehydratase Gmd |
| ENSDARG00000026630 | ENSFM00730001521220 | Kinase EC_2.7.11.1 |
| ENSDARG00000026654 | ENSFM00500000270907 | Golgi Snap Receptor Complex Member 1.28 Kda Golgi Snare 28 Kda Cis Golgi Snare P28 |
| ENSDARG00000026680 | ENSFM00500000270162 | Kidney Mitochondrial Carrier 1 Solute Carrier Family 25 Member 30 |
| ENSDARG00000026712 | ENSFM00500000269759 | Ras Related Rab |
| ENSDARG00000026726 | ENSFM00730001521439 | Annexin Annexin Annexin Calpactin Calpactin Chromobindin Lipocortin I |
| ENSDARG00000026762 | ENSFM00800002020379 | Unknown |
| ENSDARG00000026762 | ENSFM00250000003532 | Hyccin Down Regulated By CTNNB1 A FAM126A |
| ENSDARG00000026784 | ENSFM00740001589121 | Roundabout Homolog Precursor |
| ENSDARG00000026796 | ENSFM00730001521627 | Metabotropic Glutamate Receptor Precursor |
| ENSDARG00000026801 | ENSFM00250000001250 | Double Stranded Rna Binding Staufen Homolog |
| ENSDARG00000026811 | ENSFM00500000270921 | Exostosin 3 EC_2.4.1.223 Glucuronyl Galactosyl Proteoglycan 4 Alpha N Acetylglucosaminyltransferase Multiple Exostosis 3 |
| ENSDARG00000026829 | ENSFM00500000273700 | Coactosin |
| ENSDARG00000026835 | ENSFM00260000050648 | Mitochondrial Folate Transporter/Carrier Solute Carrier Family 25 Member 32 |
| ENSDARG00000026845 | ENSFM00730001521223 | Rho Related Gtp Binding Precursor |
| ENSDARG00000026871 | ENSFM00550000743168 | Ubiquitin Carboxyl Terminal Hydrolase Isozyme L1 Precursor Uch L1 EC_3.4.19.12 Ec 6 Ubiquitin Thioesterase L1 |
| ENSDARG00000026882 | ENSFM00500000271464 | Paralemmin 1 Precursor Paralemmin |
| ENSDARG00000026902 | ENSFM00250000007609 | Quinone Oxidoreductase 1 Quinone Oxidoreductase Homolog 1 Qoh 1 Zeta Crystallin Homolog |
| ENSDARG00000026908 | ENSFM00500000270937 | Transmembrane EMP24 Domain Containing 2 Precursor Copi Coated Vesicle Membrane P24 P24 Family Beta 1 P24BETA1 |
| ENSDARG00000026967 | ENSFM00670001235578 | Ap 2 Complex Subunit Sigma Adaptor Complex Ap 2 Subunit Sigma Adaptor Related Complex 2 Subunit Sigma Clathrin Assembly 2 Sigma Small Chain Clathrin Coat Assembly AP17 Clathrin Coat Associated AP17 Plasma Membrane Adaptor Ap 2.17 Kda SIGMA2 Adaptin |
| ENSDARG00000027041 | ENSFM00780001898335 | Protocadherin Precursor |
| ENSDARG00000027065 | ENSFM00250000000644 | Sodium Coupled Neutral Amino Acid Transporter Amino Acid Transporter Solute Carrier Family 38 Member System A Amino Acid Transporter 1 System N Amino Acid Transporter |
| ENSDARG00000027099 | ENSFM00610000973063 | Unknown |
| ENSDARG00000027099 | ENSFM00730001521737 | 26S Protease Regulatory Subunit 6B 26S Proteasome Aaa Atpase Subunit RPT3 Proteasome 26S Subunit Atpase 4 |
| ENSDARG00000027141 | ENSFM00500000270436 | Ubiquitin Conjugating Enzyme E2 L3 EC_6.3.2.19 Ubiquitin Carrier L3 Ubiquitin Ligase L3 |
| ENSDARG00000027192 | ENSFM00250000000931 | Potassium/Sodium Hyperpolarization Activated Cyclic Nucleotide Gated Channel |
| ENSDARG00000027200 | ENSFM00500000272384 | Gamma Aminobutyric Acid Receptor Associated 2 Precursor Gaba A Receptor Associated 2 Ganglioside Expression Factor 2 Gef 2 General Transport Factor P16 Golgi Associated Atpase Enhancer Of 16 Kda Gate 16 MAP1 Light Chain 3 Related |
| ENSDARG00000027236 | ENSFM00500000271931 | Retinoschisin Precursor X Linked Juvenile Retinoschisis |
| ENSDARG00000027279 | ENSFM00250000001973 | Numb |
| ENSDARG00000027290 | ENSFM00250000000469 | Neuropilin Precursor Vascular Endothelial Cell Growth Factor 165 Receptor |
| ENSDARG00000027322 | ENSFM00260000050595 | Lin 7 Homolog Lin Mammalian Lin Seven Mals Vertebrate Lin 7 Homolog Veli |
| ENSDARG00000027355 | ENSFM00250000000816 | Adp/Atp Translocase Adp Atp Carrier Adenine Nucleotide Translocator Ant Solute Carrier Family 25 Member |
| ENSDARG00000027394 | ENSFM00800002003927 | Transmembrane 178B Precursor |
| ENSDARG00000027397 | ENSFM00250000002390 | Vang 2 Van Gogh |
| ENSDARG00000027418 | ENSFM00500000271162 | Trafficking Particle Complex Subunit 4 |
| ENSDARG00000027419 | ENSFM00800002003398 | Glutamate Decarboxylase 1 EC_4.1.1.15.67 Kda Glutamic Acid Decarboxylase Gad 67 Glutamate Decarboxylase 67 Kda |
| ENSDARG00000027552 | ENSFM00630001065056 | Unknown |
| ENSDARG00000027552 | ENSFM00730001521833 | Mitogen Activated Kinase 1 Map Kinase 1 Mapk 1 EC_2.7.11.24 ERT1 Extracellular Signal Regulated Kinase 2 Erk 2 Mitogen Activated Kinase 2 Map Kinase 2 Mapk 2 |
| ENSDARG00000027564 | ENSFM00730001521739 | Serine/Threonine Kinase Pak EC_2.7.11.1 P21 Activated Kinase Pak |
| ENSDARG00000027590 | ENSFM00800002003496 | Septin 2 |
| ENSDARG00000027602 | ENSFM00260000050443 | Leucine Rich Repeat And Fibronectin Type Iii Domain Containing Precursor Synaptic Adhesion Molecule |
| ENSDARG00000027618 | ENSFM00740001589268 | Glutamate Receptor Interacting |
| ENSDARG00000027734 | ENSFM00500000269839 | Serine/Arginine Rich Splicing Factor Pre Splicing Factor Splicing Factor Arginine/Serine Rich |
| ENSDARG00000027740 | ENSFM00670001235823 | Pituitary Adenylate Cyclase Activating Polypeptide Precursor Pacap |
| ENSDARG00000027828 | ENSFM00730001522127 | Glutamate Receptor Subunit 1 |
| ENSDARG00000027887 | ENSFM00610000966963 | Unknown |
| ENSDARG00000027887 | ENSFM00250000006728 | Zinc Finger Fyve Domain Containing 21 |
| ENSDARG00000027915 | ENSFM00500000272347 | Zinc Finger C2HC Domain Containing 1A |
| ENSDARG00000027963 | ENSFM00800002003895 | Cam Kinase Vesicle Associated |
| ENSDARG00000027984 | ENSFM00250000003545 | Maleylacetoacetate Isomerase Maai EC_5.2.1.2 GSTZ1 1 Glutathione S Transferase Zeta 1 EC_2.5.1.- 18 |
| ENSDARG00000028000 | ENSFM00730001521507 | Atp Dependent 6 Phosphofructokinase Type |
| ENSDARG00000028048 | ENSFM00670001235503 | Estradiol 17 Beta Dehydrogenase 1 EC_1.1.1.62 17 Beta Hydroxysteroid Dehydrogenase Type 1 17 Beta Hsd 1 |
| ENSDARG00000028066 | ENSFM00500000270725 | Gtp Binding Di Precursor Distinct Subgroup Of The Ras Family Member |
| ENSDARG00000028087 | ENSFM00730001521185 | Dehydrogenase Aldehyde Dehydrogenase Family 1 Member Dehydrogenase |
| ENSDARG00000028198 | ENSFM00250000001221 | Ubiquitin Conjugating Enzyme E2 Variant |
| ENSDARG00000028213 | ENSFM00800002010267 | Unknown |
| ENSDARG00000028213 | ENSFM00740001589077 | Titin EC_2.7.11.1 Connectin |
| ENSDARG00000028259 | ENSFM00250000000672 | Aldehyde Dehydrogenase Aldehyde Dehydrogenase Aldehyde Dehydrogenase Family 3 Member |
| ENSDARG00000028275 | ENSFM00730001521311 | Sulfotransferase S Sulfotransferase |
| ENSDARG00000028306 | ENSFM00730001521291 | Vimentin |
| ENSDARG00000028322 | ENSFM00250000007000 | Peroxisomal Membrane PEX14 PTS1 Receptor Docking Peroxin 14 Peroxisomal Membrane Anchor PEX14 |
| ENSDARG00000028327 | ENSFM00500000270656 | Glia Maturation Factor Gmf |
| ENSDARG00000028354 | ENSFM00500000272618 | Syntaxin Binding 6 |
| ENSDARG00000028386 | ENSFM00500000271809 | Oxidoreductase HTATIP2 EC_1.1.1.- |
| ENSDARG00000028393 | ENSFM00350000105429 | Disheveled Associated Activator Of Morphogenesis |
| ENSDARG00000028401 | ENSFM00730001521186 | Ras Related C3 Botulinum Toxin Substrate Precursor P21 |
| ENSDARG00000028533 | ENSFM00740001589112 | Microtubule Actin Cross Linking Factor 1 Actin Cross Linking Family 7 |
| ENSDARG00000028546 | ENSFM00250000003275 | Nadh Ubiquinone Oxidoreductase 75 Kda Subunit Mitochondrial Precursor EC_1.6.5.3 EC_1.6.99.- 3 Complex I 75KD Ci 75KD |
| ENSDARG00000028552 | ENSFM00500000270520 | Lipid Phosphate Phosphatase Related Type 4 EC_3.1.3.4 Brain Specific Phosphatidic Acid Phosphatase 1 Plasticity Related Gene 1 Prg 1 |
| ENSDARG00000028618 | ENSFM00560000771030 | Keratin Type I Cytoskeletal 18 Cytokeratin 18 Ck 18 Keratin 18 K18 |
| ENSDARG00000028628 | ENSFM00250000004188 | Ganglioside Induced Differentiation Associated 1 GDAP1 |
| ENSDARG00000028661 | ENSFM00250000003929 | Ciliary Neurotrophic Factor Receptor Subunit Alpha Precursor Cntf Receptor Subunit Alpha Cntfr Alpha |
| ENSDARG00000028748 | ENSFM00730001521961 | Transmembrane 9 Superfamily Member Precursor |
| ENSDARG00000028800 | ENSFM00250000001040 | Cytohesin Ph SEC7 And Coiled Coil Domain Containing |
| ENSDARG00000028889 | ENSFM00250000006353 | Nadh Dehydrogenase 1 Beta Subcomplex Subunit 10 Complex I Pdsw Ci Pdsw Nadh Ubiquinone Oxidoreductase Pdsw Subunit |
| ENSDARG00000028900 | ENSFM00500000270052 | Outer Dense Fiber Cenexin Outer Dense Fiber Of Sperm Tails 2 |
| ENSDARG00000028982 | ENSFM00800002003706 | Propionyl Coa Carboxylase Alpha Chain Mitochondrial Precursor Pccase Subunit Alpha EC_6.4.1.3 Propanoyl Coa:Carbon Dioxide Ligase Subunit Alpha |
| ENSDARG00000029011 | ENSFM00800002003425 | Xaa Pro Aminopeptidase 1 EC_3.4.11.9 Aminoacylproline Aminopeptidase Cytosolic Aminopeptidase P Soluble Aminopeptidase P Samp X Pro Aminopeptidase 1 X Prolyl Aminopeptidase 1 Soluble |
| ENSDARG00000029019 | ENSFM00800002003537 | Unknown |
| ENSDARG00000029064 | ENSFM00670001264207 | Ubiquinol Cytochrome C Reductase Complex Iii Subunit Vii |
| ENSDARG00000029114 | ENSFM00670001235536 | Serine/Threonine Kinase EC_2.7.11.1 Apoptosis Associated Tyrosine Kinase Brain Kinase Lemur Tyrosine Kinase |
| ENSDARG00000029133 | ENSFM00250000005014 | Gamma Tubulin Complex Component 3 Gamma Ring Complex |
| ENSDARG00000029146 | ENSFM00740001589098 | Density Lipoprotein Receptor Related Precursor Lrp |
| ENSDARG00000029150 | ENSFM00250000000292 | Heat Shock |
| ENSDARG00000029170 | ENSFM00250000000859 | Clathrin Assembly |
| ENSDARG00000029234 | ENSFM00500000269888 | Syntaxin Binding 5 Lethal 2 Giant Larvae Homolog Tomosyn |
| ENSDARG00000029239 | ENSFM00720001492041 | Synaptotagmin Synaptotagmin |
| ENSDARG00000029248 | ENSFM00250000001137 | Far Upstream Element Binding 2 Fuse Binding 2 Kh Type Splicing Regulatory Ksrp |
| ENSDARG00000029252 | ENSFM00500000270730 | Lupus La Homolog La Autoantigen Homolog La Ribonucleoprotein |
| ENSDARG00000029307 | ENSFM00800002024333 | Unknown |
| ENSDARG00000029307 | ENSFM00250000005107 | Uncharacterized Colon Cancer Associated MIC1 Mic 1 |
| ENSDARG00000029381 | ENSFM00250000000672 | Aldehyde Dehydrogenase Aldehyde Dehydrogenase Aldehyde Dehydrogenase Family 3 Member |
| ENSDARG00000029415 | ENSFM00250000005082 | Metaxin 2 Mitochondrial Outer Membrane Import Complex 2 |
| ENSDARG00000029463 | ENSFM00250000001190 | Arf Gap With SH3 Domain Ank Repeat And Ph Domain Containing 1.130 Kda Phosphatidylinositol 4 5 Bisphosphate Dependent ARF1 Gtpase Activating Adp Ribosylation Factor Directed Gtpase Activating 1 Arf Gtpase Activating 1 Development And Differentiation Enhan |
| ENSDARG00000029472 | ENSFM00250000001587 | Afadin Af 6 |
| ENSDARG00000029473 | ENSFM00500000269733 | Glutathione S Transferase EC_2.5.1.18 Gst Class Mu |
| ENSDARG00000029480 | ENSFM00250000001872 | Cytospin A SPECC1 Sperm Antigen With Calponin Homology And Coiled Coil Domains 1 |
| ENSDARG00000029500 | ENSFM00500000271952 | 60S Ribosomal L34 |
| ENSDARG00000029501 | ENSFM00810002040612 | Glycogen Synthase Kinase 3 Gsk 3 EC_2.7.11.26 Serine/Threonine Kinase EC_2.7.11.- 1 |
| ENSDARG00000029510 | ENSFM00500000270373 | Mitochondrial Import Inner Membrane Translocase Subunit TIM17 |
| ENSDARG00000029533 | ENSFM00810002040668 | 60S Ribosomal L18 |
| ENSDARG00000029609 | ENSFM00670001235583 | O Acetyl Adp Ribose Deacetylase EC_3.2.2.- EC_3.5.-.- 1 Macro Domain Containing Hydrolase |
| ENSDARG00000029639 | ENSFM00710001441924 | Mitochondrial Import Receptor Subunit TOM70 Mitochondrial Precursor Proteins Import Receptor Translocase Of Outer Membrane 70 Kda Subunit |
| ENSDARG00000029689 | ENSFM00250000001413 | Transketolase EC_2.2.1.1 |
| ENSDARG00000029692 | ENSFM00250000001058 | Run And Fyve Domain Containing |
| ENSDARG00000029722 | ENSFM00500000269645 | High Mobility Group B1 High Mobility Group 1 Hmg 1 |
| ENSDARG00000029729 | ENSFM00250000006225 | Citrate Lyase Subunit Beta Protein Mitochondrial Precursor Citrate Lyase Beta EC_4.1.-.- |
| ENSDARG00000029885 | ENSFM00460000244935 | Ras Related Rab 6 |
| ENSDARG00000029931 | ENSFM00500000270146 | Manganese Transporting Atpase EC_3.6.3.- |
| ENSDARG00000029952 | ENSFM00770001847533 | Amp Deaminase EC_3.5.4.6 Amp Deaminase |
| ENSDARG00000030038 | ENSFM00500000274497 | C10 |
| ENSDARG00000030053 | ENSFM00730001521722 | Elongation Factor 1 Delta Ef 1 Delta |
| ENSDARG00000030097 | ENSFM00250000003553 | Syntenin Syndecan Binding |
| ENSDARG00000030108 | ENSFM00730001543948 | Unknown |
| ENSDARG00000030108 | ENSFM00500000271789 | Vesicle Trafficking SEC22B Er Golgi Snare Of 24 Kda Ers 24 ERS24 SEC22 Vesicle Trafficking Homolog B |
| ENSDARG00000030154 | ENSFM00730001521739 | Serine/Threonine Kinase Pak EC_2.7.11.1 P21 Activated Kinase Pak |
| ENSDARG00000030177 | ENSFM00550000743168 | Ubiquitin Carboxyl Terminal Hydrolase Isozyme L1 Precursor Uch L1 EC_3.4.19.12 Ec 6 Ubiquitin Thioesterase L1 |
| ENSDARG00000030236 | ENSFM00500000270036 | Cell Cycle Control 50A P4 Atpase Flippase Complex Beta Subunit TMEM30A Transmembrane 30A |
| ENSDARG00000030237 | ENSFM00670001235707 | Membrane Associated Progesterone Receptor Component |
| ENSDARG00000030241 | ENSFM00610000955927 | Unknown |
| ENSDARG00000030241 | ENSFM00500000271934 | Prefoldin Subunit 3 Von Hippel Lindau Binding 1 Vbp 1 Vhl Binding 1 |
| ENSDARG00000030278 | ENSFM00810002040683 | Isocitrate Dehydrogenase Subunit Alpha Mitochondrial Precursor EC_1.1.1.41 Isocitric Dehydrogenase Subunit Alpha Nad + Specific Icdh Subunit Alpha |
| ENSDARG00000030376 | ENSFM00730001521500 | Glutamate Receptor Ionotropic Nmda Precursor Glutamate Receptor Subunit Epsilon N Methyl D Aspartate Receptor Subtype |
| ENSDARG00000030441 | ENSFM00730001521782 | Inorganic Pyrophosphatase EC_3.6.1.1 Pyrophosphate Phospho Hydrolase Ppase |
| ENSDARG00000030479 | ENSFM00750001659003 | Unknown |
| ENSDARG00000030479 | ENSFM00500000269645 | High Mobility Group B1 High Mobility Group 1 Hmg 1 |
| ENSDARG00000030490 | ENSFM00740001589095 | Spectrin Beta Chain Erythrocytic Beta Spectrin |
| ENSDARG00000030508 | ENSFM00250000007961 | Coiled Coil Domain Containing 134 Precursor |
| ENSDARG00000030514 | ENSFM00500000269666 | Long Chain Fatty Acid Coa Ligase EC_6.2.1.3 Long Chain Acyl Coa Synthetase Lacs |
| ENSDARG00000030598 | ENSFM00250000004448 | Nicotinamide Phosphoribosyltransferase Namprtase Nampt EC_2.4.2.12 Pre B Cell Colony Enhancing Factor 1 Homolog Pbef Visfatin |
| ENSDARG00000030602 | ENSFM00670001235598 | 40S Ribosomal S19 |
| ENSDARG00000030614 | ENSFM00720001492031 | Synaptotagmin |
| ENSDARG00000030641 | ENSFM00250000006470 | LETM1 Domain Containing 1 |
| ENSDARG00000030644 | ENSFM00410000138458 | Guanine Nucleotide Binding Subunit |
| ENSDARG00000030656 | ENSFM00500000269805 | Septin |
| ENSDARG00000030694 | ENSFM00250000002828 | V Type Proton Atpase Subunit E V Atpase Subunit E V Atpase Kda Subunit Vacuolar Proton Pump Subunit E |
| ENSDARG00000030765 | ENSFM00730001522238 | Hydroxyacyl Coenzyme A Dehydrogenase Mitochondrial Precursor Hcdh EC_1.1.1.35 Medium And Short Chain L 3 Hydroxyacyl Coenzyme A Dehydrogenase Short Chain 3 Hydroxyacyl Coa Dehydrogenase |
| ENSDARG00000030775 | ENSFM00250000005040 | Vesicle Associated Membrane 7 Vamp 7 Synaptobrevin 1 |
| ENSDARG00000030781 | ENSFM00730001522195 | Short Chain Specific Acyl Coa Dehydrogenase Mitochondrial Precursor Scad EC_1.3.8.1 Butyryl Coa Dehydrogenase |
| ENSDARG00000030786 | ENSFM00780001898394 | Mannose 6 Phosphate Isomerase EC_5.3.1.8 Phosphohexomutase Phosphomannose Isomerase Pmi |
| ENSDARG00000030832 | ENSFM00730001521834 | Fer 1 |
| ENSDARG00000030887 | ENSFM00500000273791 | Phd Finger 23 |
| ENSDARG00000030945 | ENSFM00250000003975 | Precursor |
| ENSDARG00000030949 | ENSFM00670001236233 | Signal Recognition Particle Receptor Subunit Beta Sr Beta |
| ENSDARG00000031020 | ENSFM00260000050567 | Phosphatidylinositol 5 Phosphate 4 Kinase Type 2 EC_2.7.1.149 1 Phosphatidylinositol 5 Phosphate 4 Kinase 2 Diphosphoinositide Kinase 2 Phosphatidylinositol 5 Phosphate 4 Kinase Type Ii Pi 5 P 4 Kinase Type Ii PIP4KII Ptdins 5 P 4 Kinase 2 |
| ENSDARG00000031049 | ENSFM00250000006695 | Immunoglobulin Superfamily Member 21 Precursor IGSF21 |
| ENSDARG00000031075 | ENSFM00250000001126 | Cell Adhesion Molecule Precursor Immunoglobulin Superfamily Member Nectin Necl Synaptic Cell Adhesion Molecule |
| ENSDARG00000031098 | ENSFM00500000271227 | Mitochondrial Import Inner Membrane Translocase Subunit TIM50 Precursor |
| ENSDARG00000031119 | ENSFM00600000938696 | Unknown |
| ENSDARG00000031119 | ENSFM00250000000906 | Brain Specific Angiogenesis Inhibitor 1 Associated 2 Bai Associated 2 BAI1 Associated 2 Insulin Receptor Substrate Of 53 Kda IRSP53 Insulin Receptor Substrate P53 |
| ENSDARG00000031136 | ENSFM00800002003555 | Dbh Monooxygenase Precursor EC_1.14.17.- |
| ENSDARG00000031164 | ENSFM00250000000217 | Tubulin Alpha Chain |
| ENSDARG00000031200 | ENSFM00810002040632 | Serine/Threonine Phosphatase 2A 56 Kda Regulatory Subunit Gamma PP2A B Subunit B' PP2A B Subunit B' Gamma PP2A B Subunit B56 Gamma PP2A B Subunit PR61 Gamma PP2A B Subunit R5 Gamma |
| ENSDARG00000031214 | ENSFM00810002040812 | Coatomer Subunit Delta Delta Coat Delta Cop |
| ENSDARG00000031283 | ENSFM00670001235687 | Vesicle Associated Membrane 2 Vamp 2 Synaptobrevin 2 |
| ENSDARG00000031302 | ENSFM00600000921242 | Stromal Membrane Associated 2 Stromal Membrane Associated 1 |
| ENSDARG00000031317 | ENSFM00670001246510 | Pancreatic Progenitor Cell Differentiation And Proliferation Factor B Exocrine Differentiation And Proliferation Factor B |
| ENSDARG00000031343 | ENSFM00460000244935 | Ras Related Rab 6 |
| ENSDARG00000031366 | ENSFM00500000271278 | Reticulon 4 Interacting 1 Mitochondrial Precursor |
| ENSDARG00000031388 | ENSFM00250000005336 | Dynactin Subunit 2 |
| ENSDARG00000031435 | ENSFM00670001237763 | Thioredoxin |
| ENSDARG00000031438 | ENSFM00760001714772 | Atp Sensitive Inward Rectifier Potassium Channel 11 Inward Rectifier K + Channel KIR6 2 Potassium Channel Inwardly Rectifying Subfamily J Member 11 |
| ENSDARG00000031495 | ENSFM00500000270208 | Set Phosphatase 2A Inhibitor I2PP2A I 2PP2A Template Activating Factor I Taf I |
| ENSDARG00000031506 | ENSFM00500000270147 | Feline Leukemia Virus Subgroup C Receptor Related |
| ENSDARG00000031511 | ENSFM00750001632406 | Proteasome Subunit Beta Type 2 EC_3.4.25.1 Macropain Subunit C7 I Multicatalytic Endopeptidase Complex Subunit C7 I Proteasome Component C7 I |
| ENSDARG00000031548 | ENSFM00730001521338 | Ephrin Type B Receptor Kinase |
| ENSDARG00000031560 | ENSFM00500000269720 | Srsf Kinase EC_2.7.11.1 Kinase Serine/Arginine Rich Specific Kinase Sr Specific Kinase |
| ENSDARG00000031712 | ENSFM00790001963004 | Metabotropic Glutamate Receptor Precursor |
| ENSDARG00000031720 | ENSFM00250000001322 | Calsyntenin Precursor Alcadein Alc |
| ENSDARG00000031720 | ENSFM00770001863632 | CLSTN1 |
| ENSDARG00000031795 | ENSFM00760001714769 | Atp Binding Cassette Sub Family F Member 1 Atp Binding Cassette 50 |
| ENSDARG00000031796 | ENSFM00500000269646 | Acetylcholinesterase Precursor Ache EC_3.1.1.7 |
| ENSDARG00000031886 | ENSFM00250000002602 | Intraflagellar Transport 140 Homolog Wd And Tetratricopeptide Repeats 2 |
| ENSDARG00000031890 | ENSFM00250000001656 | T Complex 11 |
| ENSDARG00000031981 | ENSFM00670001238103 | Pterin 4 Alpha Carbinolamine Dehydratase |
| ENSDARG00000031985 | ENSFM00760001714775 | Serine/Threonine Phosphatase 2A Activator EC_5.2.1.8 PP2A Subunit B' PR53 Phosphotyrosyl Phosphatase Activator Ptpa Serine/Threonine Phosphatase 2A Regulatory Subunit 4 Serine/Threonine Phosphatase 2A Regulatory Subunit B' |
| ENSDARG00000032013 | ENSFM00810002040664 | Lissencephaly 1 |
| ENSDARG00000032049 | ENSFM00740001589204 | Ena/Vasp Ena/Vasodilator Stimulated Phosphoprotein |
| ENSDARG00000032083 | ENSFM00250000000643 | Dihydropyrimidinase Related Drp Collapsin Response Mediator Crmp |
| ENSDARG00000032102 | ENSFM00250000000757 | Thioredoxin Peroxidase Thioredoxin Dependent Peroxide Reductase |
| ENSDARG00000032117 | ENSFM00250000003062 | Atp Dependent Rna Helicase DDX1 EC_3.6.4.13 Dead Box 1 |
| ENSDARG00000032155 | ENSFM00730001521429 | Phosphatase 1A EC_3.1.3.16 Phosphatase 2C Alpha PP2C Alpha |
| ENSDARG00000032157 | ENSFM00730001521304 | G Coupled Receptor Kinase G Coupled Receptor Kinase |
| ENSDARG00000032161 | ENSFM00250000000639 | Slit Robo Rho Gtpase Activating Rho Gtpase Activating |
| ENSDARG00000032188 | ENSFM00250000001418 | Volume Regulated Anion Channel Subunit Leucine Rich Repeat Containing |
| ENSDARG00000032199 | ENSFM00610000973096 | Unknown |
| ENSDARG00000032199 | ENSFM00500000270283 | Glypican 3 Precursor Intestinal Oci 5 |
| ENSDARG00000032206 | ENSFM00810002040651 | Cystathionine Gamma Lyase EC_4.4.1.1 Cysteine Sulfhydrase Gamma Cystathionase |
| ENSDARG00000032238 | ENSFM00520000517792 | Dynamin EC_3.6.5.5 |
| ENSDARG00000032326 | ENSFM00630001067617 | Unknown |
| ENSDARG00000032326 | ENSFM00800002003721 | Trans 2 Enoyl Coa Reductase Mitochondrial Precursor EC_1.3.1.38 |
| ENSDARG00000032340 | ENSFM00500000272115 | Serine Hydrolase EC_3.1.-.- |
| ENSDARG00000032430 | ENSFM00250000001941 | Serine/Threonine Phosphatase 2A 65 Kda Regulatory Subunit A PP2A Subunit A PR65 PP2A Subunit A R1 |
| ENSDARG00000032431 | ENSFM00500000272108 | MPV17 |
| ENSDARG00000032458 | ENSFM00730001521220 | Kinase EC_2.7.11.1 |
| ENSDARG00000032535 | ENSFM00670001235692 | PRA1 Family 3 Adp Ribosylation Factor 6 Interacting 5 Arl 6 Interacting 5 Aip 5 |
| ENSDARG00000032552 | ENSFM00250000003363 | E3 Ubiquitin Ligase MARCH5 EC_6.3.2.- Membrane Associated Ring Finger 5 Membrane Associated Ring Ch V March V |
| ENSDARG00000032565 | ENSFM00250000002019 | Voltage Dependent Calcium Channel Gamma Subunit Neuronal Voltage Gated Calcium Channel Gamma Subunit Transmembrane Ampar Regulatory Gamma Tarp Gamma |
| ENSDARG00000032575 | ENSFM00730001521154 | Unknown |
| ENSDARG00000032577 | ENSFM00740001589241 | H + /Cl Exchange Transporter 7 Chloride Channel 7 Alpha Subunit Chloride Channel 7 Clc 7 |
| ENSDARG00000032606 | ENSFM00350000105408 | Septin |
| ENSDARG00000032618 | ENSFM00500000272807 | Microsomal Glutathione S Transferase 1 Microsomal Gst 1 EC_2.5.1.18 Microsomal Gst I |
| ENSDARG00000032714 | ENSFM00720001492020 | Glutamate Receptor Precursor Glur Ampa Selective Glutamate Receptor Glur Glutamate Receptor Ionotropic Ampa |
| ENSDARG00000032725 | ENSFM00500000270032 | Ubiquitin 40S Ribosomal S27A Precursor Ubiquitin Carboxyl Extension 80 |
| ENSDARG00000032737 | ENSFM00720001492020 | Glutamate Receptor Precursor Glur Ampa Selective Glutamate Receptor Glur Glutamate Receptor Ionotropic Ampa |
| ENSDARG00000032799 | ENSFM00300000079128 | Potassium Voltage Gated Channel Subfamily D Member Voltage Gated Potassium Channel Subunit KV4 |
| ENSDARG00000032802 | ENSFM00250000002379 | Kinectin |
| ENSDARG00000032849 | ENSFM00250000000535 | N Myc Downstream Regulated Gene |
| ENSDARG00000032862 | ENSFM00730001521710 | Udp Glucuronosyltransferase Ugt 58 Precursor Udpgt 58 EC_2.4.1.17 |
| ENSDARG00000032865 | ENSFM00250000001466 | Kinase C And Casein Kinase Substrate In Neurons |
| ENSDARG00000032866 | ENSFM00670001235831 | Enhancer Of Rudimentary |
| ENSDARG00000032919 | ENSFM00500000272284 | Micos Complex Subunit MIC19 Coiled Coil Helix Coiled Coil Helix Domain Containing 3 |
| ENSDARG00000032970 | ENSFM00500000271199 | Cytochrome C Oxidase Subunit 4 1 Mitochondrial Cytochrome C Oxidase Polypeptide Iv Cytochrome C Oxidase Subunit Iv 1 Cox Iv 1 |
| ENSDARG00000032990 | ENSFM00730001521536 | Plexin Precursor |
| ENSDARG00000033012 | ENSFM00250000004492 | Pericentrin |
| ENSDARG00000033184 | ENSFM00730001521993 | Camp Dependent Kinase Type Ii Regulatory Subunit |
| ENSDARG00000033201 | ENSFM00710001456837 | Unknown |
| ENSDARG00000033201 | ENSFM00740001589457 | Cysteine Rich Crp |
| ENSDARG00000033259 | ENSFM00760001714930 | DIS3 Exonuclease 2 |
| ENSDARG00000033413 | ENSFM00250000000948 | Acyl Coenzyme A Thioesterase Acyl Coa Thioesterase EC_3.1.2.2 Acyl Coenzyme A Thioester Hydrolase Long Chain Acyl Coa |
| ENSDARG00000033437 | ENSFM00570000851006 | Ribosomal S6 Kinase Alpha S6K Alpha EC_2.7.11.1.90 Kda Ribosomal S6 Kinase P90 Rsk Map Kinase Activated Kinase Mapk Activated Kinase Mapkap Kinase Mapkapk Ribosomal S6 Kinase Rsk |
| ENSDARG00000033516 | ENSFM00250000001283 | EFR3 Homolog |
| ENSDARG00000033539 | ENSFM00250000003230 | Multifunctional Phosphoribosylaminoimidazole Succinocarboxamide Synthase EC_6.3.2.6 Saicar Synthetase |
| ENSDARG00000033567 | ENSFM00670001235580 | Peptidyl Prolyl Cis Trans Isomerase FKBP1B Ppiase FKBP1B EC_5.2.1.8 12.6 Kda FK506 Binding 12.6 Kda Fkbp Fkbp 12 6 FK506 Binding 1B Fkbp 1B Immunophilin FKBP12 6 Rotamase |
| ENSDARG00000033597 | ENSFM00250000003859 | Apoptosis Inhibitor 5 Api |
| ENSDARG00000033599 | ENSFM00800002003471 | Disulfide Isomerase A3 Precursor EC_5.3.4.1.58 Kda Glucose Regulated 58 Kda Microsomal P58 Disulfide Isomerase Er 60 Endoplasmic Reticulum Resident 57 Er 57 ERP57 Endoplasmic Reticulum Resident 60 Er 60 ERP60 |
| ENSDARG00000033604 | ENSFM00250000004723 | Alpha 2 Macroglobulin Receptor Associated Precursor Alpha 2 Mrap Low Density Lipoprotein Receptor Related Associated 1 Rap |
| ENSDARG00000033609 | ENSFM00500000270555 | Microtubule Associated Proteins 1A/1B Light Chain 3A Precursor Autophagy Related LC3 A Autophagy Related Ubiquitin Modifier LC3 A MAP1 Light Chain 3 1 MAP1A/MAP1B Light Chain 3 A MAP1A/MAP1B LC3 A Microtubule Associated 1 Light Chain 3 Alpha |
| ENSDARG00000033655 | ENSFM00800002003794 | Stathmin |
| ENSDARG00000033666 | ENSFM00250000002398 | Phosphatidylinositol 4 Kinase Type 2 EC_2.7.1.67 Phosphatidylinositol 4 Kinase Type Ii |
| ENSDARG00000033683 | ENSFM00250000000220 | Tropomyosin Alpha 1 Chain Alpha Tropomyosin Tropomyosin 1 |
| ENSDARG00000033738 | ENSFM00250000000438 | Myosin Light Chain Myosin Light Chain Myosin Light Chain |
| ENSDARG00000033789 | ENSFM00670001236404 | Nadh Dehydrogenase 1 Beta Subcomplex Subunit 7 Complex I B18 Ci B18 Nadh Ubiquinone Oxidoreductase B18 Subunit |
| ENSDARG00000033802 | ENSFM00720001492028 | Sterol 26 Hydroxylase Mitochondrial Precursor EC_1.14.13.15 5 Beta Cholestane 3 Alpha 7 Alpha 12 Alpha Triol 27 Hydroxylase Cytochrome P 450C27/25 Cytochrome P450 27 Sterol 27 Hydroxylase Vitamin D 3 25 Hydroxylase |
| ENSDARG00000033804 | ENSFM00250000003280 | Sorting Nexin 27 |
| ENSDARG00000033840 | ENSFM00570000851039 | Protocadherin Fat Precursor Fat Tumor Suppressor Homolog |
| ENSDARG00000033845 | ENSFM00250000001385 | Turtle Homolog A Precursor Dendrite Arborization And Synapse Maturation 1 Immunoglobulin Superfamily Member 9A IGSF9A |
| ENSDARG00000033899 | ENSFM00730001521696 | Ap 2 Complex Subunit Mu Ap 2 Mu Chain Clathrin Assembly Complex 2 Mu Medium Chain Clathrin Coat Assembly AP50 Clathrin Coat Associated AP50 MU2 Adaptin Plasma Membrane Adaptor Ap Kda |
| ENSDARG00000033978 | ENSFM00740001589502 | Nucleoredoxin EC_1.8.1.8 |
| ENSDARG00000033993 | ENSFM00250000001258 | Transporter 1 Solute Carrier Family 19 Member 1 |
| ENSDARG00000034007 | ENSFM00250000001124 | Prominin 2 Precursor Prom 2 Prominin 2 |
| ENSDARG00000034011 | ENSFM00800002003385 | Gamma Aminobutyric Acid Receptor Subunit Alpha Precursor Gaba A Receptor Subunit Alpha |
| ENSDARG00000034056 | ENSFM00610000971777 | Unknown |
| ENSDARG00000034056 | ENSFM00730001521484 | Casein Kinase I Gamma Cki Gamma EC_2.7.11.1 |
| ENSDARG00000034093 | ENSFM00400000131747 | Serine/Threonine Kinase EC_2.7.11.1 Doublecortin And Cam Kinase Doublecortin Kinase |
| ENSDARG00000034165 | ENSFM00250000001884 | Leucine Rich Repeat And Immunoglobulin Domain Containing Nogo Receptor Interacting Precursor Leucine Rich Repeat Neuronal |
| ENSDARG00000034189 | ENSFM00320000100094 | STE20/SPS1 Related Proline Alanine Rich Kinase Ste 20 Related Kinase EC_2.7.11.1 Serine/Threonine Kinase 39 |
| ENSDARG00000034195 | ENSFM00740001589149 | Dna Topoisomerase 2 EC_5.99.1.3 Dna Topoisomerase Ii |
| ENSDARG00000034210 | ENSFM00730001521806 | Death Associated Kinase Dap Kinase EC_2.7.11.1 Dap Kinase |
| ENSDARG00000034215 | ENSFM00500000270441 | Ras Related Rab |
| ENSDARG00000034240 | ENSFM00630001071983 | Unknown |
| ENSDARG00000034240 | ENSFM00250000001955 | F Actin Capping Subunit Alpha 2 Capz Alpha 2 |
| ENSDARG00000034270 | ENSFM00750001632363 | 2 Oxoglutarate Dehydrogenase Mitochondrial Precursor 2 Oxoglutarate Dehydrogenase Complex Component E1 Ogdc E1 Alpha Ketoglutarate Dehydrogenase |
| ENSDARG00000034313 | ENSFM00750001659041 | Unknown |
| ENSDARG00000034313 | ENSFM00500000270236 | Serine/Threonine Phosphatase 5 PP5 EC_3.1.3.16 Phosphatase T Ppt |
| ENSDARG00000034344 | ENSFM00780001898335 | Protocadherin Precursor |
| ENSDARG00000034368 | ENSFM00730001521836 | Beta Hexosaminidase Subunit Alpha Precursor EC_3.2.1.52 Beta N Acetylhexosaminidase Subunit Alpha Hexosaminidase Subunit A N Acetyl Beta Glucosaminidase Subunit Alpha |
| ENSDARG00000034396 | ENSFM00250000004570 | Methionine Trna Ligase Cytoplasmic EC_6.1.1.10 Methionyl Trna Synthetase Metrs |
| ENSDARG00000034423 | ENSFM00670001250790 | Gamma Synuclein |
| ENSDARG00000034424 | ENSFM00670001235362 | Sodium/Potassium Transporting Atpase Subunit Beta Sodium/Potassium Dependent Atpase Subunit Beta |
| ENSDARG00000034434 | ENSFM00730001521215 | Insulin Receptor Precursor EC_2.7.10.1 |
| ENSDARG00000034470 | ENSFM00250000000488 | Fructose Bisphosphate Aldolase EC_4.1.2.13 Type Aldolase |
| ENSDARG00000034473 | ENSFM00250000001068 | Tweety Homolog |
| ENSDARG00000034493 | ENSFM00730001521500 | Glutamate Receptor Ionotropic Nmda Precursor Glutamate Receptor Subunit Epsilon N Methyl D Aspartate Receptor Subtype |
| ENSDARG00000034522 | ENSFM00460000244935 | Ras Related Rab 6 |
| ENSDARG00000034534 | ENSFM00250000002169 | V Type Proton Atpase Catalytic Subunit A V Atpase Subunit A EC_3.6.3.14 V Atpase 69 Kda Subunit Vacuolar Proton Pump Subunit Alpha |
| ENSDARG00000034600 | ENSFM00500000271648 | Transmembrane 165 Precursor Transmembrane Tparl |
| ENSDARG00000034616 | ENSFM00500000271997 | Myeloid Leukemia Factor 2 Myelodysplasia Myeloid Leukemia Factor 2 |
| ENSDARG00000034650 | ENSFM00800002003523 | Fatty Acid Binding Protein Heart Fatty Acid Binding 3 Heart Type Fatty Acid Binding H Fabp |
| ENSDARG00000034705 | ENSFM00500000290457 | Parvalbumin 7 Parvalbumin Alpha Parvalbumin 4A |
| ENSDARG00000034777 | ENSFM00500000272779 | Thioredoxin Mitochondrial Precursor Mtrx Mt Trx Thioredoxin 2 |
| ENSDARG00000034823 | ENSFM00800002003465 | Coatomer Subunit Gamma Gamma Coat Gamma Cop |
| ENSDARG00000034826 | ENSFM00670001235693 | Dj 1 Precursor EC_3.4.-.- Parkinson Disease 7 Homolog |
| ENSDARG00000034883 | ENSFM00610000961514 | Unknown |
| ENSDARG00000034883 | ENSFM00500000271070 | Acyl Coa Binding Domain Containing 5 |
| ENSDARG00000034897 | ENSFM00630001060822 | Unknown |
| ENSDARG00000034897 | ENSFM00670001235534 | 40S Ribosomal S10 |
| ENSDARG00000035043 | ENSFM00500000272753 | Prefoldin Subunit 5 C Myc Binding Mm 1 Myc Modulator 1 |
| ENSDARG00000035054 | ENSFM00250000004991 | Programmed Cell Death 10 |
| ENSDARG00000035066 | ENSFM00740001589345 | Nucleolar Transcription Factor 1 Upstream Binding Factor 1 Ubf 1 |
| ENSDARG00000035131 | ENSFM00700001390183 | Surfeit Locus |
| ENSDARG00000035152 | ENSFM00430000230089 | Ap 1 Complex Subunit Beta 1 Adaptor Complex Ap 1 Subunit Beta 1 Adaptor Related Complex 1 Subunit Beta 1 Beta 1 Adaptin Beta Adaptin 1 Clathrin Assembly Complex 1 Beta Large Chain Golgi Adaptor HA1/AP1 Adaptin Beta Subunit |
| ENSDARG00000035167 | ENSFM00250000005488 | 39S Ribosomal L48 Mitochondrial Precursor L48MT Mrp L48 |
| ENSDARG00000035181 | ENSFM00250000005083 | Solute Carrier Family 25 Member 46 |
| ENSDARG00000035188 | ENSFM00800002003610 | Ras Related Rab 14 |
| ENSDARG00000035256 | ENSFM00760001714607 | Elongation Factor 2 Ef 2 |
| ENSDARG00000035282 | ENSFM00670001236443 | Er Membrane Complex Subunit 6 Transmembrane 93 |
| ENSDARG00000035329 | ENSFM00250000000112 | Calpain Subunit Calcium Activated Neutral Proteinase Canp Calpain Subunit |
| ENSDARG00000035338 | ENSFM00250000003009 | Tubulin Polymerization Promoting Family Member |
| ENSDARG00000035340 | ENSFM00730001522339 | Gap Junction Delta 2 Connexin Gap Junction Alpha 9 |
| ENSDARG00000035352 | ENSFM00250000004414 | Platelet Activating Factor Acetylhydrolase Ib Subunit Beta EC_3.1.1.47 Paf Acetylhydrolase 30 Kda Subunit Paf Ah 30 Kda Subunit Paf Ah Subunit Beta Pafah Subunit Beta |
| ENSDARG00000035357 | ENSFM00250000000545 | Guanine Nucleotide Binding G I /G S /G T Subunit Beta Transducin Beta Chain |
| ENSDARG00000035360 | ENSFM00260000050335 | Kinase EC_2.7.11.1 Kinase Mapk/Erk Kinase Kinase Kinase Mek Kinase Kinase Mekkk Kinase |
| ENSDARG00000035400 | ENSFM00500000270103 | Transcription Factor BTF3 Homolog 4 Basic Transcription Factor 3 4 |
| ENSDARG00000035433 | ENSFM00500000270526 | Unknown |
| ENSDARG00000035458 | ENSFM00500000269652 | Sarcoplasmic/Endoplasmic Reticulum Calcium Atpase Sr Ca 2+ Atpase EC_3.6.3.8 Calcium Pump Calcium Transporting Atpase Sarcoplasmic Reticulum Type Twitch Skeletal Muscle Endoplasmic Reticulum Class 1/2 Ca 2+ Atpase |
| ENSDARG00000035468 | ENSFM00500000269757 | Calcium Binding Mitochondrial Carrier Scamc Small Calcium Binding Mitochondrial Carrier Solute Carrier Family 25 Member |
| ENSDARG00000035470 | ENSFM00250000002047 | Endophilin SH3 Domain Containing GRB2 |
| ENSDARG00000035538 | ENSFM00250000000490 | V Type Proton Atpase 116 Kda Subunit A V Atpase 116 Kda Proton 116 Kda Subunit Vacuolar Proton Translocating Atpase 116 Kda Subunit A |
| ENSDARG00000035577 | ENSFM00750001632370 | Phosphatidate Cytidylyltransferase 2 EC_2.7.7.41 Cdp Dag Synthase 2 Cdp Dg Synthase 2 Cdp Diacylglycerol Synthase 2 Cds 2 Cdp Diglyceride Pyrophosphorylase 2 Cdp Diglyceride Synthase 2 Ctp:Phosphatidate Cytidylyltransferase 2 |
| ENSDARG00000035598 | ENSFM00730001521292 | Coronin |
| ENSDARG00000035603 | ENSFM00670001235511 | D Amino Acid Oxidase Daao Damox Dao EC_1.4.3.3 |
| ENSDARG00000035605 | ENSFM00250000007069 | Trichoplein Keratin Filament Binding Protein Tchp Mitochondrial With Oncostatic Activity Mitostatin |
| ENSDARG00000035608 | ENSFM00250000004793 | Serine/Threonine Phosphatase PGAM5 Mitochondrial |
| ENSDARG00000035618 | ENSFM00770001847616 | 2 Methoxy 6 Polyprenyl 1 4 Benzoquinol Methylase Mitochondrial Precursor EC_2.1.1.201 Ubiquinone Biosynthesis Methyltransferase COQ5 |
| ENSDARG00000035631 | ENSFM00250000003961 | Stromal Cell Derived Factor 2 1 Precursor SDF2 1 |
| ENSDARG00000035650 | ENSFM00740001589204 | Ena/Vasp Ena/Vasodilator Stimulated Phosphoprotein |
| ENSDARG00000035660 | ENSFM00250000002086 | Dyslexia Associated |
| ENSDARG00000035676 | ENSFM00250000001523 | Tyrosine Phosphatase Type Iva 1 Precursor EC_3.1.3.48 Tyrosine Phosphatase 4A1 Tyrosine Phosphatase Of Regenerating Liver 1 Prl 1 |
| ENSDARG00000035679 | ENSFM00800002003520 | Proteasome Subunit Alpha Type 2 EC_3.4.25.1 Macropain Subunit C3 Multicatalytic Endopeptidase Complex Subunit C3 Proteasome Component C3 |
| ENSDARG00000035692 | ENSFM00250000001462 | 40S Ribosomal S3A |
| ENSDARG00000035699 | ENSFM00670001236018 | Signal Recognition Particle 19 Kda SRP19 |
| ENSDARG00000035715 | ENSFM00500000304598 | Marcks 2 |
| ENSDARG00000035741 | ENSFM00800002023359 | Unknown |
| ENSDARG00000035741 | ENSFM00770001847568 | Mitochondrial 2 Oxoglutarate/Malate Carrier Ogcp Solute Carrier Family 25 Member 11 |
| ENSDARG00000035751 | ENSFM00760001714748 | Importin 7 IMP7 Ran Binding 7 RANBP7 |
| ENSDARG00000035763 | ENSFM00250000005997 | Syntaxin 18 |
| ENSDARG00000035808 | ENSFM00280000058697 | H + /Cl Exchange Transporter 5 Chloride Channel 5 Clc 5 Chloride Transporter Clc 5 |
| ENSDARG00000035860 | ENSFM00500000289305 | Unknown |
| ENSDARG00000035860 | ENSFM00750001641929 | Unknown |
| ENSDARG00000035868 | ENSFM00250000001244 | Spire Homolog |
| ENSDARG00000035871 | ENSFM00670001235478 | 60S Ribosomal L30 |
| ENSDARG00000035872 | ENSFM00670001235389 | Estradiol 17 Beta Dehydrogenase 12 EC_1.1.1.62 17 Beta Hydroxysteroid Dehydrogenase 12 17 Beta Hsd 12 |
| ENSDARG00000035880 | ENSFM00750001632375 | V Type Proton Atpase Subunit C V Atpase Subunit C Vacuolar Proton Pump Subunit C |
| ENSDARG00000035882 | ENSFM00500000272841 | Ribonuclease UK114 EC_3.1.-.- Kda Translational Inhibitor UK114 Antigen |
| ENSDARG00000035899 | ENSFM00250000001884 | Leucine Rich Repeat And Immunoglobulin Domain Containing Nogo Receptor Interacting Precursor Leucine Rich Repeat Neuronal |
| ENSDARG00000035907 | ENSFM00250000002173 | FAM49A |
| ENSDARG00000035913 | ENSFM00500000270186 | Tyrosine Trna Ligase Cytoplasmic EC_6.1.1.1 Tyrosyl Trna Synthetase Tyrrs |
| ENSDARG00000035949 | ENSFM00600000921356 | Transmembrane |
| ENSDARG00000035970 | ENSFM00260000050522 | Receptor Type Tyrosine Phosphatase Precursor R Ptp |
| ENSDARG00000035985 | ENSFM00250000007801 | Unknown |
| ENSDARG00000035993 | ENSFM00670001235669 | Small Ubiquitin Related Modifier Precursor Sumo |
| ENSDARG00000036031 | ENSFM00800002003401 | Septin |
| ENSDARG00000036056 | ENSFM00700001390294 | Inactive Serine/Threonine Kinase VRK3 Serine/Threonine Pseudokinase VRK3 Vaccinia Related Kinase 3 |
| ENSDARG00000036058 | ENSFM00410000138458 | Guanine Nucleotide Binding Subunit |
| ENSDARG00000036080 | ENSFM00800002003853 | CD81 Antigen 26 Kda Cell Surface Tapa 1 Target Of The Antiproliferative Antibody 1 CD81 Antigen |
| ENSDARG00000036104 | ENSFM00770001847571 | Unconventional Myosin |
| ENSDARG00000036109 | ENSFM00250000002187 | Metaxin 1 Mitochondrial Outer Membrane Import Complex 1 |
| ENSDARG00000036164 | ENSFM00290000065553 | Cysteine Trna Ligase EC_6.1.1.16 Cysteinyl Trna Synthetase Cysrs |
| ENSDARG00000036175 | ENSFM00760001714757 | Protocadherin Precursor Protocadherin |
| ENSDARG00000036179 | ENSFM00780001898391 | Unconventional Myosin Ie Unconventional Myosin 1E |
| ENSDARG00000036186 | ENSFM00630001068294 | Unknown |
| ENSDARG00000036186 | ENSFM00800002005438 | Unknown |
| ENSDARG00000036186 | ENSFM00800002005102 | Myelin Basic |
| ENSDARG00000036252 | ENSFM00770001847543 | Ras Related R Ras |
| ENSDARG00000036281 | ENSFM00730001522120 | Coiled Coil And C2 Domain Containing Five Prime Repressor Element Under Dual Repression Binding Fre Under Dual Repression Binding Freud |
| ENSDARG00000036295 | ENSFM00250000003047 | Golgin Subfamily A Member 7 |
| ENSDARG00000036298 | ENSFM00250000003348 | 40S Ribosomal S13 |
| ENSDARG00000036299 | ENSFM00250000002033 | Nucleobindin Precursor |
| ENSDARG00000036329 | ENSFM00610000954065 | Unknown |
| ENSDARG00000036335 | ENSFM00250000007309 | C1GALT1 Specific Chaperone 1 Core 1 BETA1 3 Galactosyltransferase 2 C1GAL T2 C1GALT2 Core 1 BETA3 Gal T2 Core 1 BETA3 Galactosyltransferase Specific Molecular Chaperone |
| ENSDARG00000036338 | ENSFM00250000005369 | E3 Ubiquitin Ligase PEP5 |
| ENSDARG00000036344 | ENSFM00500000270302 | Calbindin Calbindin D28 D 28K Vitamin D Dependent Calcium Binding Protein Avian Type |
| ENSDARG00000036375 | ENSFM00250000001390 | Cytoplasmic FMR1 Interacting |
| ENSDARG00000036383 | ENSFM00730001521994 | Sodium/Potassium/Calcium Exchanger 3 Precursor Na + /K + /Ca 2+ Exchange 3 Solute Carrier Family 24 Member 3 |
| ENSDARG00000036427 | ENSFM00250000004066 | 4F2 Cell Surface Antigen Heavy Chain 4F2HC Solute Carrier Family 3 Member 2 CD98 Antigen |
| ENSDARG00000036438 | ENSFM00250000003345 | Nadh Dehydrogenase Flavoprotein 1 Mitochondrial Precursor EC_1.6.5.3 EC_1.6.99.- 3 Complex I 51KD Ci 51KD Nadh Ubiquinone Oxidoreductase 51 Kda Subunit |
| ENSDARG00000036446 | ENSFM00730001521993 | Camp Dependent Kinase Type Ii Regulatory Subunit |
| ENSDARG00000036456 | ENSFM00760001714627 | Annexin Annexin Annexin |
| ENSDARG00000036501 | ENSFM00500000270441 | Ras Related Rab |
| ENSDARG00000036546 | ENSFM00500000270834 | Pyridoxal Kinase EC_2.7.1.35 Pyridoxine Kinase |
| ENSDARG00000036577 | ENSFM00250000002076 | V Type Proton Atpase 16 Kda Proteolipid Subunit V Atpase 16 Kda Proteolipid Subunit Vacuolar Proton Pump 16 Kda Proteolipid Subunit |
| ENSDARG00000036587 | ENSFM00250000002301 | Carbonyl Reductase EC_1.1.1.- 197 20 Beta Hydroxysteroid Dehydrogenase Nadph Dependent Carbonyl Reductase 1 Prostaglandin 9 Ketoreductase Prostaglandin E 2 9 Reductase EC_1.1.-.- 1 189 |
| ENSDARG00000036626 | ENSFM00730001521376 | Semaphorin Precursor Semaphorin Sema |
| ENSDARG00000036629 | ENSFM00250000003648 | 40S Ribosomal S14 |
| ENSDARG00000036658 | ENSFM00250000002178 | Calcium Binding 39 MO25 |
| ENSDARG00000036684 | ENSFM00760001715902 | Nadh Dehydrogenase 1 Alpha Subcomplex Subunit 7 |
| ENSDARG00000036721 | ENSFM00250000002521 | Mitochondrial Import Receptor Subunit TOM40 Homolog Translocase Of Outer Membrane 40 Kda Subunit Homolog |
| ENSDARG00000036774 | ENSFM00250000000973 | Regulator Of G Signaling |
| ENSDARG00000036809 | ENSFM00250000000130 | Aminopeptidase N Ap N EC_3.4.11.2 Microsomal Aminopeptidase |
| ENSDARG00000036820 | ENSFM00250000006453 | Monoglyceride Lipase Mgl EC_3.1.1.23 Monoacylglycerol Lipase Magl |
| ENSDARG00000036830 | ENSFM00670001270143 | Unknown |
| ENSDARG00000036830 | ENSFM00730001521138 | Keratin Type I Cytoskeletal Cytokeratin Ck Keratin |
| ENSDARG00000036840 | ENSFM00730001521138 | Keratin Type I Cytoskeletal Cytokeratin Ck Keratin |
| ENSDARG00000036863 | ENSFM00770001847571 | Unconventional Myosin |
| ENSDARG00000036875 | ENSFM00500000270361 | 40S Ribosomal S12 |
| ENSDARG00000036878 | ENSFM00670001240188 | SH3 Domain Binding Glutamic Acid Rich 2 |
| ENSDARG00000036888 | ENSFM00610000973299 | Unknown |
| ENSDARG00000036888 | ENSFM00610000973300 | Unknown |
| ENSDARG00000036911 | ENSFM00670001235365 | Secretory Carrier Associated Membrane Secretory Carrier Membrane |
| ENSDARG00000036985 | ENSFM00730001521536 | Plexin Precursor |
| ENSDARG00000036998 | ENSFM00730001521124 | Adp Ribosylation Factor |
| ENSDARG00000037000 | ENSFM00670001236165 | Peptidyl Prolyl Cis Trans Isomerase FKBP11 Precursor Ppiase FKBP11 EC_5.2.1.8.19 Kda FK506 Binding 19 Kda Fkbp Fkbp 19 FK506 Binding 11 Fkbp 11 Rotamase |
| ENSDARG00000037008 | ENSFM00250000004645 | Seipin Bernardinelli Seip Congenital Lipodystrophy Type 2 |
| ENSDARG00000037009 | ENSFM00670001238356 | Barrier To Autointegration Factor |
| ENSDARG00000037012 | ENSFM00250000004066 | 4F2 Cell Surface Antigen Heavy Chain 4F2HC Solute Carrier Family 3 Member 2 CD98 Antigen |
| ENSDARG00000037017 | ENSFM00760001714925 | Ubiquitin Conjugation Factor E4 B |
| ENSDARG00000037038 | ENSFM00720001492034 | 26S Protease Regulatory Subunit 10B 26S Proteasome Aaa Atpase Subunit RPT4 Proteasome 26S Subunit Atpase 6 Proteasome Subunit P42 |
| ENSDARG00000037057 | ENSFM00380000124741 | Glutaryl Coa Dehydrogenase Mitochondrial Precursor Gcd EC_1.3.8.6 |
| ENSDARG00000037059 | ENSFM00500000269830 | Choline Transporter Solute Carrier Family 44 Member |
| ENSDARG00000037071 | ENSFM00670001235586 | 40S Ribosomal S26 |
| ENSDARG00000037122 | ENSFM00250000000375 | Teneurin Ten Odd Oz/Ten M Homolog Tenascin Ten Teneurin Transmembrane |
| ENSDARG00000037145 | ENSFM00710001441664 | Sodium/Calcium Exchanger 1 Precursor Na + /Ca 2+ Exchange 1 Solute Carrier Family 8 Member 1 |
| ENSDARG00000037153 | ENSFM00560000771081 | V Type Proton Atpase Subunit S1 Precursor V Atpase Subunit S1 V Atpase AC45 Subunit V Atpase S1 Accessory Vacuolar Proton Pump Subunit S1 |
| ENSDARG00000037172 | ENSFM00600000921555 | Ddrgk Domain Containing 1 |
| ENSDARG00000037229 | ENSFM00670001235711 | Density Regulated Drp |
| ENSDARG00000037259 | ENSFM00540000718275 | Nadh Dehydrogenase 1 Beta Subcomplex 6 |
| ENSDARG00000037261 | ENSFM00500000270600 | S Methyl 5' Thioadenosine Phosphorylase |
| ENSDARG00000037281 | ENSFM00800002003901 | Fibrinogen Gamma Chain Precursor |
| ENSDARG00000037318 | ENSFM00250000004027 | Dolichyl Diphosphooligosaccharide Glycosyltransferase 48 Kda Subunit Precursor Ddost 48 Kda Subunit Oligosaccharyl Transferase 48 Kda Subunit EC_2.4.99.18 |
| ENSDARG00000037350 | ENSFM00750001632341 | 60S Ribosomal L9 |
| ENSDARG00000037373 | ENSFM00730001521338 | Ephrin Type B Receptor Kinase |
| ENSDARG00000037403 | ENSFM00730001521103 | Heat Shock Kda Heat Shock 70 Kda |
| ENSDARG00000037406 | ENSFM00250000003366 | Wd Repeat Containing 19 |
| ENSDARG00000037433 | ENSFM00250000000920 | Fragile X Mental Retardation 1 |
| ENSDARG00000037455 | ENSFM00730001522351 | 2 Hydroxyacylsphingosine 1 Beta Galactosyltransferase Precursor EC_2.4.1.45 Ceramide Udp Galactosyltransferase Cerebroside Synthase Udp Galactose Ceramide Galactosyltransferase |
| ENSDARG00000037478 | ENSFM00800002003474 | Cadherin Precursor |
| ENSDARG00000037484 | ENSFM00250000007004 | Transmembrane 192 |
| ENSDARG00000037488 | ENSFM00500000269826 | Precursor |
| ENSDARG00000037496 | ENSFM00720001492020 | Glutamate Receptor Precursor Glur Ampa Selective Glutamate Receptor Glur Glutamate Receptor Ionotropic Ampa |
| ENSDARG00000037498 | ENSFM00720001492020 | Glutamate Receptor Precursor Glur Ampa Selective Glutamate Receptor Glur Glutamate Receptor Ionotropic Ampa |
| ENSDARG00000037506 | ENSFM00250000000896 | Ribose Phosphate Pyrophosphokinase EC_2.7.6.1 Phosphoribosyl Pyrophosphate Synthase |
| ENSDARG00000037553 | ENSFM00500000270404 | Interleukin 1 Receptor Accessory 1 Precursor Il 1 Rapl 1 Il 1RAPL 1 IL1RAPL 1 X Linked Interleukin 1 Receptor Accessory 1 |
| ENSDARG00000037559 | ENSFM00250000000763 | Ubiquitin Modifier Activating Enzyme Ubiquitin Activating Enzyme E1 |
| ENSDARG00000037589 | ENSFM00250000005529 | Glucose Induced Degradation 8 Homolog |
| ENSDARG00000037593 | ENSFM00800002023350 | Unknown |
| ENSDARG00000037593 | ENSFM00730001521542 | Prickle |
| ENSDARG00000037679 | ENSFM00500000273639 | Mitochondrial Pyruvate Carrier 2 Brain 44 |
| ENSDARG00000037746 | ENSFM00730001521060 | Actin |
| ENSDARG00000037747 | ENSFM00250000001972 | Fascin |
| ENSDARG00000037783 | ENSFM00250000000442 | Coagulation Factor Precursor |
| ENSDARG00000037794 | ENSFM00500000269860 | Neuronal Pentraxin 2 Precursor NP2 Neuronal Pentraxin Ii Np Ii |
| ENSDARG00000037846 | ENSFM00250000002681 | Minor Histocompatibility Antigen H13 EC_3.4.23.- Presenilin 3 Signal Peptide Peptidase |
| ENSDARG00000037860 | ENSFM00670001238299 | Cytochrome C Oxidase Subunit Vib 1 |
| ENSDARG00000037870 | ENSFM00730001521060 | Actin |
| ENSDARG00000037884 | ENSFM00730001522236 | Ras Related Rab 30 Precursor |
| ENSDARG00000037905 | ENSFM00740001589100 | Voltage Dependent Type Calcium Channel Subunit Alpha Brain Calcium Channel Calcium Channel L Type Alpha 1 Polypeptide Voltage Gated Calcium Channel Subunit Alpha CAV2 |
| ENSDARG00000037909 | ENSFM00500000274239 | Uncharacterized |
| ENSDARG00000037921 | ENSFM00500000361151 | Unknown |
| ENSDARG00000037924 | ENSFM00730001522052 | Guanine Nucleotide Binding Subunit Alpha G Alpha G Subunit Alpha |
| ENSDARG00000037932 | ENSFM00260000050595 | Lin 7 Homolog Lin Mammalian Lin Seven Mals Vertebrate Lin 7 Homolog Veli |
| ENSDARG00000037935 | ENSFM00400000131967 | Aldehyde Dehydrogenase Family 16 Member A1 |
| ENSDARG00000037946 | ENSFM00500000271451 | Prolactin Precursor Prl |
| ENSDARG00000037962 | ENSFM00810002040603 | Proteasome Subunit Beta Type Precursor EC_3.4.25.1 Macropain Multicatalytic Endopeptidase Complex Proteasome Subunit |
| ENSDARG00000037967 | ENSFM00750001632414 | Ras Related Rab 35 |
| ENSDARG00000037980 | ENSFM00500000273664 | Transcription Elongation Factor B Polypeptide 2 Elongin 18 Kda Subunit Elongin B Elob Rna Polymerase Ii Transcription Factor Siii Subunit B Siii P18 |
| ENSDARG00000037997 | ENSFM00250000000193 | Tubulin Beta Chain |
| ENSDARG00000037998 | ENSFM00780001898418 | Flotillin 1 |
| ENSDARG00000038010 | ENSFM00730001521186 | Ras Related C3 Botulinum Toxin Substrate Precursor P21 |
| ENSDARG00000038028 | ENSFM00500000272594 | Nadh Dehydrogenase 1 Alpha Subcomplex Subunit 6 Complex I B14 Ci B14 Nadh Ubiquinone Oxidoreductase B14 Subunit |
| ENSDARG00000038030 | ENSFM00610000962968 | Unknown |
| ENSDARG00000038030 | ENSFM00670001235574 | Ring Box 1 |
| ENSDARG00000038059 | ENSFM00500000270097 | Growth Factor Receptor Bound 2 Adapter GRB2 SH2/SH3 Adapter GRB2 |
| ENSDARG00000038067 | ENSFM00750001632391 | E3 Ubiquitin Ligase SMURF2 EC_6.3.2.- Smad Ubiquitination Regulatory Factor 2 Smad Specific E3 Ubiquitin Ligase 2 |
| ENSDARG00000038068 | ENSFM00420000140507 | Probable Atp Dependent Rna Helicase DDX5 EC_3.6.4.13 Dead Box 5 |
| ENSDARG00000038075 | ENSFM00250000003946 | Cytochrome C1 Heme Protein Mitochondrial Precursor Complex Iii Subunit 4 Complex Iii Subunit Iv Cytochrome B C1 Complex Subunit 4 Ubiquinol Cytochrome C Reductase Complex Cytochrome C1 Subunit Cytochrome C 1 |
| ENSDARG00000038123 | ENSFM00730001521616 | Myosin Regulatory Light Myosin Regulatory Light Chain 2 Smooth Muscle Myosin Regulatory Light Chain |
| ENSDARG00000038207 | ENSFM00800002003550 | Delta 1 Pyrroline 5 Carboxylate Dehydrogenase Mitochondrial Precursor P5C Dehydrogenase EC_1.2.1.88 Aldehyde Dehydrogenase Family 4 Member A1 L Glutamate Gamma Semialdehyde Dehydrogenase |
| ENSDARG00000038225 | ENSFM00730001521569 | Ras Precursor |
| ENSDARG00000038228 | ENSFM00670001250817 | Selenoprotein W 2A |
| ENSDARG00000038270 | ENSFM00790001963096 | Gpi Ethanolamine Phosphate Transferase 2 Ec 2 GPI7 Homolog HGPI7 Phosphatidylinositol Glycan Biosynthesis Class G Pig G |
| ENSDARG00000038308 | ENSFM00810002040913 | Synaptobrevin Homolog YKT6 Precursor EC_2.3.1.- |
| ENSDARG00000038315 | ENSFM00670001235619 | Pyridoxal Phosphate Phosphatase Plp Phosphatase EC_3.1.3.3 EC_3.1.3.- 74 Chronophin |
| ENSDARG00000038414 | ENSFM00250000006614 | Beta 1 3 Galactosyltransferase 6 Beta 1 3 Galtase 6 BETA3GAL T6 BETA3GALT6 EC_2.4.1.134 Gag Galtii Galactosyltransferase Ii Galactosylxylosylprotein 3 Beta Galactosyltransferase Udp Gal:Betagal Beta 1 3 Galactosyltransferase Polypeptide 6 |
| ENSDARG00000038432 | ENSFM00250000004727 | Actin Related 10 Actin Related 11 |
| ENSDARG00000038445 | ENSFM00250000001244 | Spire Homolog |
| ENSDARG00000038446 | ENSFM00250000000469 | Neuropilin Precursor Vascular Endothelial Cell Growth Factor 165 Receptor |
| ENSDARG00000038446 | ENSFM00320000100837 | Soluble Neuropilin |
| ENSDARG00000038467 | ENSFM00740001589327 | Immunoglobulin Superfamily Member Precursor Glu Trp Ile Ewi Motif Containing Ewi Antigen |
| ENSDARG00000038518 | ENSFM00250000006834 | Synaptosomal Associated 29 Snap 29 Golgi Snare Of 32 Kda GS32 Soluble 29 Kda Nsf Attachment Vesicle Membrane Fusion Snap 29 |
| ENSDARG00000038543 | ENSFM00460000244957 | Wd Repeat Containing 48 |
| ENSDARG00000038563 | ENSFM00250000003510 | Presequence Protease EC_3.4.24.- |
| ENSDARG00000038577 | ENSFM00500000411071 | Unknown |
| ENSDARG00000038608 | ENSFM00500000272004 | Succinate Dehydrogenase Cytochrome B560 Subunit Mitochondrial Precursor Integral Membrane Cii 3 Qps 1 QPS1 Succinate Dehydrogenase Complex Subunit C |
| ENSDARG00000038609 | ENSFM00480000262818 | Myelin P0 Precursor Myelin Peripheral Mpp Myelin Zero |
| ENSDARG00000038615 | ENSFM00250000004019 | Type Phosphatidylinositol 4 5 Bisphosphate 4 Phosphatase Type 2 Ptdins 4 5 P2 4 Ptase EC_3.1.3.78 Ptdins 4 5 P2 4 Ptase Ii Transmembrane 55A |
| ENSDARG00000038618 | ENSFM00730001521962 | Carnitine O Palmitoyltransferase 2 Mitochondrial Precursor EC_2.3.1.21 Carnitine Palmitoyltransferase Ii Cpt Ii |
| ENSDARG00000038655 | ENSFM00250000009368 | Adherens Junction Associated 1 |
| ENSDARG00000038693 | ENSFM00500000386940 | Unknown |
| ENSDARG00000038694 | ENSFM00610000960810 | Unknown |
| ENSDARG00000038694 | ENSFM00500000270563 | Receptor Expression Enhancing Polyposis Locus 1 |
| ENSDARG00000038695 | ENSFM00250000000588 | Elav |
| ENSDARG00000038703 | ENSFM00250000000532 | Hexokinase EC_2.7.1.1 Hexokinase Type Hk |
| ENSDARG00000038731 | ENSFM00800002024004 | Unknown |
| ENSDARG00000038731 | ENSFM00730001521754 | Solute Carrier Family 25 Member |
| ENSDARG00000038768 | ENSFM00760001714704 | 39S Ribosomal L12 Mitochondrial Precursor L12MT Mrp L12 |
| ENSDARG00000038834 | ENSFM00250000005227 | Electron Transfer Flavoprotein Ubiquinone Oxidoreductase Mitochondrial Precursor Etf Qo Etf Ubiquinone Oxidoreductase EC_1.5.5.1 Electron Transferring Flavoprotein Dehydrogenase Etf Dehydrogenase |
| ENSDARG00000038835 | ENSFM00600000938248 | Unknown |
| ENSDARG00000038835 | ENSFM00730001521742 | Peptidyl Prolyl Cis Trans Isomerase D Ppiase D EC_5.2.1.8 Rotamase D |
| ENSDARG00000038845 | ENSFM00250000004441 | D Lactate Dehydrogenase Mitochondrial Precursor EC_1.1.2.4 D Lactate Ferricytochrome C Oxidoreductase D Lcr |
| ENSDARG00000038859 | ENSFM00480000262761 | Regulator Of G Signaling |
| ENSDARG00000038865 | ENSFM00730001522151 | Peroxisomal Acyl Coenzyme A Oxidase 3 EC_1.3.3.6 Branched Chain Acyl Coa Oxidase Brcacox Pristanoyl Coa Oxidase |
| ENSDARG00000038866 | ENSFM00250000003061 | Trafficking Particle Complex Subunit 8 TRS85 Homolog |
| ENSDARG00000038881 | ENSFM00760001714736 | 3 Ketoacyl Coa Thiolase Mitochondrial EC_2.3.1.16 Acetyl Coa Acyltransferase Beta Ketothiolase Mitochondrial 3 Oxoacyl Coa Thiolase |
| ENSDARG00000038900 | ENSFM00730001521951 | Medium Chain Specific Acyl Coa Dehydrogenase Mitochondrial Precursor Mcad EC_1.3.8.7 |
| ENSDARG00000038901 | ENSFM00800002006771 | Transmembrane 256 Precursor |
| ENSDARG00000038910 | ENSFM00450000242182 | Propionyl Coa Carboxylase Beta Chain Mitochondrial Precursor Pccase Subunit Beta EC_6.4.1.3 Propanoyl Coa:Carbon Dioxide Ligase Subunit Beta |
| ENSDARG00000038967 | ENSFM00730001521665 | Cullin 3 Cul 3 |
| ENSDARG00000038978 | ENSFM00440000236855 | Dnaj Homolog Subfamily B Member |
| ENSDARG00000038980 | ENSFM00320000100209 | Thioredoxin Domain Containing 12 Precursor EC_1.8.4.2 Endoplasmic Reticulum Resident 19 Er 19 ERP19 Thioredoxin P19 |
| ENSDARG00000038991 | ENSFM00800002003482 | Vesicle Fusing Atpase EC_3.6.4.6 N Ethylmaleimide Sensitive Fusion Nem Sensitive Fusion Vesicular Fusion Nsf |
| ENSDARG00000039005 | ENSFM00610000962953 | Unknown |
| ENSDARG00000039005 | ENSFM00500000272371 | Trafficking Particle Complex Subunit 1 |
| ENSDARG00000039007 | ENSFM00730001521170 | Enolase EC_4.2.1.11 2 Phospho D Glycerate Hydro Lyase |
| ENSDARG00000039022 | ENSFM00810002040592 | Serine/Threonine Kinase EC_2.7.11.1 Mammalian STE20 Kinase Mst STE20 Kinase Serine/Threonine Kinase |
| ENSDARG00000039034 | ENSFM00540000726513 | Marcks 1 |
| ENSDARG00000039093 | ENSFM00780001898358 | Aspartate Aminotransferase Cytoplasmic Caspat EC_2.6.1.1 EC_2.6.1.- 3 Cysteine Aminotransferase Cytoplasmic Cysteine Transaminase Cytoplasmic Ccat Glutamate Oxaloacetate Transaminase 1 Transaminase A |
| ENSDARG00000039136 | ENSFM00500000333809 | Cytochrome C Oxidase Assembly |
| ENSDARG00000039142 | ENSFM00500000270526 | Unknown |
| ENSDARG00000039182 | ENSFM00730001522093 | Voltage Dependent Calcium Channel Subunit Alpha 2/Delta 1 Precursor Voltage Gated Calcium Channel Subunit Alpha 2/Delta 1 |
| ENSDARG00000039203 | ENSFM00250000003442 | Mitochondrial Fission Factor |
| ENSDARG00000039225 | ENSFM00250000000977 | Vacuolar Sorting Associated 13A Chorea Acanthocytosis Chorein |
| ENSDARG00000039241 | ENSFM00520000517802 | Kinase C Type Pkc EC_2.7.11.13 |
| ENSDARG00000039268 | ENSFM00600000938103 | Unknown |
| ENSDARG00000039270 | ENSFM00250000006099 | Vesicle Transport Through Interaction With T Snares Homolog 1B Vesicle Transport V Snare VTI1 1 VTI1 RP1 |
| ENSDARG00000039272 | ENSFM00250000001554 | Plastin |
| ENSDARG00000039279 | ENSFM00250000005275 | Golgin Subfamily A Member 5 Golgin 84 |
| ENSDARG00000039343 | ENSFM00750001658617 | Unknown |
| ENSDARG00000039343 | ENSFM00740001589092 | Adenosylhomocysteinase Adohcyase EC_3.3.1.1 S Adenosyl L Homocysteine Hydrolase 2 |
| ENSDARG00000039345 | ENSFM00730001521812 | Developmentally Regulated Gtp Binding 1 Drg 1 |
| ENSDARG00000039346 | ENSFM00670001236297 | Nadh Dehydrogenase 1 Alpha Subcomplex Subunit 5 Complex I Subunit B13 Complex I 13KD B Ci 13KD B Nadh Ubiquinone Oxidoreductase 13 Kda B Subunit |
| ENSDARG00000039347 | ENSFM00500000270090 | 40S Ribosomal S24 |
| ENSDARG00000039350 | ENSFM00250000006324 | Single Stranded Dna Binding Protein Mitochondrial Precursor Mt Ssb Mtssb |
| ENSDARG00000039356 | ENSFM00440000236938 | Adipocyte Plasma Membrane Associated |
| ENSDARG00000039363 | ENSFM00250000001153 | Dnaj Homolog Subfamily Member |
| ENSDARG00000039378 | ENSFM00500000271265 | Reticulocalbin 2 Precursor Taipoxin Associated Calcium Binding 49 Tcbp 49 |
| ENSDARG00000039392 | ENSFM00250000000621 | Serine/Threonine Kinase EC_2.7.11.1 Kinase Lysine Deficient Kinase With No Lysine |
| ENSDARG00000039399 | ENSFM00250000001915 | Cytosolic Purine 5' Nucleotidase EC_3.1.3.5 Cytosolic 5' Nucleotidase Ii |
| ENSDARG00000039435 | ENSFM00480000262761 | Regulator Of G Signaling |
| ENSDARG00000039452 | ENSFM00250000000532 | Hexokinase EC_2.7.1.1 Hexokinase Type Hk |
| ENSDARG00000039489 | ENSFM00250000001490 | Arf Gtpase Activating GIT1 Arf Gap GIT1 Cool Associated And Tyrosine Phosphorylated 1 Cat 1 CAT1 G Coupled Receptor Kinase Interactor 1 Grk Interacting 1 |
| ENSDARG00000039500 | ENSFM00260000050443 | Leucine Rich Repeat And Fibronectin Type Iii Domain Containing Precursor Synaptic Adhesion Molecule |
| ENSDARG00000039502 | ENSFM00800002003364 | Elongation Factor 1 Alpha Ef 1 Alpha Eukaryotic Elongation Factor 1 A EEF1A |
| ENSDARG00000039522 | ENSFM00250000000193 | Tubulin Beta Chain |
| ENSDARG00000039578 | ENSFM00250000003247 | Proliferation Associated 2G4 |
| ENSDARG00000039647 | ENSFM00800002003411 | Sodium And Chloride Dependent Gaba Transporter 1 Gat 1 Solute Carrier Family 6 Member 1 |
| ENSDARG00000039666 | ENSFM00250000004363 | Gdp L Fucose Synthase EC_1.1.1.271 Gdp 4 Keto 6 Deoxy D Mannose 3 5 Epimerase 4 Reductase Fx Red Cell Nadp H Binding |
| ENSDARG00000039681 | ENSFM00500000271410 | Sorting And Assembly Machinery Component 50 Homolog |
| ENSDARG00000039724 | ENSFM00730001521733 | Oxysterol Binding Related 5 Orp 5 Osbp Related 5 Oxysterol Binding Homolog 1 |
| ENSDARG00000039729 | ENSFM00250000001190 | Arf Gap With SH3 Domain Ank Repeat And Ph Domain Containing 1.130 Kda Phosphatidylinositol 4 5 Bisphosphate Dependent ARF1 Gtpase Activating Adp Ribosylation Factor Directed Gtpase Activating 1 Arf Gtpase Activating 1 Development And Differentiation Enhan |
| ENSDARG00000039830 | ENSFM00500000366462 | Unknown |
| ENSDARG00000039872 | ENSFM00670001235984 | Ragulator Complex LAMTOR2 Late Endosomal/Lysosomal Adaptor And Mapk And Mtor Activator 2 Roadblock Domain Containing 3 |
| ENSDARG00000039882 | ENSFM00610000965597 | Unknown |
| ENSDARG00000039882 | ENSFM00500000270528 | Ap 3 Complex Subunit Sigma Ap 3 Complex Subunit Sigma Adaptor Related Complex 3 Subunit Sigma Sigma Adaptin Adaptin Sigma Adaptin |
| ENSDARG00000039887 | ENSFM00250000004016 | Complement Component 1 Q Subcomponent Binding Protein Mitochondrial Precursor GC1Q R Glycoprotein GC1QBP C1QBP |
| ENSDARG00000039901 | ENSFM00250000000986 | Endophilin SH3 Domain Containing GRB2 |
| ENSDARG00000039914 | ENSFM00730001521081 | Glyceraldehyde 3 Phosphate Dehydrogenase Gapdh EC_1.2.1.12 Peptidyl Cysteine S Nitrosylase Gapdh EC_2.6.99.- |
| ENSDARG00000039915 | ENSFM00670001239768 | Uncharacterized |
| ENSDARG00000039929 | ENSFM00800002003437 | Creatine Kinase U Type Mitochondrial Precursor EC_2.7.3.2 Acidic Type Mitochondrial Creatine Kinase Mia Ck Ubiquitous Mitochondrial Creatine Kinase U Mtck |
| ENSDARG00000039966 | ENSFM00250000001124 | Prominin 2 Precursor Prom 2 Prominin 2 |
| ENSDARG00000039974 | ENSFM00800002003877 | Ankycorbin Ankyrin Repeat And Coiled Coil Structure Containing Retinal Pigment Epithelial Cell Retinoic Acid Induced 14 |
| ENSDARG00000039980 | ENSFM00760001714836 | Palmitoyl Thioesterase 1 Precursor Ppt 1 EC_3.1.2.22 Palmitoyl Hydrolase 1 |
| ENSDARG00000040001 | ENSFM00250000000754 | Furry |
| ENSDARG00000040002 | ENSFM00760001714681 | Neuropathy Target Esterase Sws |
| ENSDARG00000040021 | ENSFM00250000007576 | Receptor Binding Cancer Antigen Expressed On Siso Cells Cancer Associated Surface Antigen RCAS1 Estrogen Receptor Binding Fragment Associated Gene 9 |
| ENSDARG00000040024 | ENSFM00800002003402 | Glycerol 3 Phosphate Dehydrogenase Cytoplasmic Gpd C Gpdh C EC_1.1.1.8 |
| ENSDARG00000040064 | ENSFM00250000006412 | Lysophosphatidic Acid Phosphatase Type 6 Precursor EC_3.1.3.2 Acid Phosphatase 6 Lysophosphatidic Acid Phosphatase 1 PACPL1 |
| ENSDARG00000040087 | ENSFM00760001715069 | Band 4 1 NBL4 |
| ENSDARG00000040156 | ENSFM00250000000328 | Metabotropic Glutamate Receptor Precursor |
| ENSDARG00000040158 | ENSFM00730001521235 | Cell Division Control 42 Homolog Precursor |
| ENSDARG00000040190 | ENSFM00250000004373 | Dihydropteridine Reductase EC_1.5.1.34 Hdhpr Quinoid Dihydropteridine Reductase |
| ENSDARG00000040245 | ENSFM00800002003590 | Importin 5 IMP5 Importin Subunit Beta 3 Karyopherin Beta 3 Ran Binding 5 RANBP5 |
| ENSDARG00000040246 | ENSFM00800002003504 | Ras Related Rap Precursor |
| ENSDARG00000040248 | ENSFM00730001521465 | Choline O Acetyltransferase Choactase Chat Choline Acetylase EC_2.3.1.6 |
| ENSDARG00000040255 | ENSFM00500000271283 | Bifunctional Epoxide Hydrolase 2 |
| ENSDARG00000040287 | ENSFM00730001521154 | Unknown |
| ENSDARG00000040291 | ENSFM00250000001126 | Cell Adhesion Molecule Precursor Immunoglobulin Superfamily Member Nectin Necl Synaptic Cell Adhesion Molecule |
| ENSDARG00000040295 | ENSFM00500000275143 | Apolipoprotein Eb Precursor Apo Eb |
| ENSDARG00000040309 | ENSFM00250000007262 | Px Domain Containing 1 |
| ENSDARG00000040334 | ENSFM00810002040570 | S Adenosylmethionine Synthase Adomet Synthase EC_2.5.1.6 Methionine Adenosyltransferase Mat |
| ENSDARG00000040352 | ENSFM00730001522249 | Peroxisomal Carnitine O Octanoyltransferase Cot EC_2.3.1.137 |
| ENSDARG00000040380 | ENSFM00250000000618 | Rho Guanine Nucleotide Exchange Factor 2 Guanine Nucleotide Exchange Factor H1 Gef H1 |
| ENSDARG00000040401 | ENSFM00760001714734 | Mitochondrial Carnitine/Acylcarnitine Carrier Carnitine/Acylcarnitine Translocase Cac Solute Carrier Family 25 Member 20 |
| ENSDARG00000040430 | ENSFM00500000269860 | Neuronal Pentraxin 2 Precursor NP2 Neuronal Pentraxin Ii Np Ii |
| ENSDARG00000040455 | ENSFM00500000271471 | Unc 50 Homolog |
| ENSDARG00000040463 | ENSFM00500000271232 | Solute Carrier Family 25 Member 51 Mitochondrial Carrier Triple Repeat 1 |
| ENSDARG00000040469 | ENSFM00730001522540 | Ectonucleotide Pyrophosphatase/Phosphodiesterase Family Member 6 Precursor E Npp 6 Npp 6 EC_3.1.4.- EC_3.1.4.38 Choline Specific Glycerophosphodiester Phosphodiesterase Glycerophosphocholine Cholinephosphodiesterase Gpc Cpde |
| ENSDARG00000040492 | ENSFM00690001369712 | Unknown |
| ENSDARG00000040492 | ENSFM00690001374215 | Unknown |
| ENSDARG00000040492 | ENSFM00800002003403 | C 1 Tetrahydrofolate Synthase Cytoplasmic C1 Thf Synthase |
| ENSDARG00000040528 | ENSFM00440000236993 | Galectin 3 Binding Precursor Lectin Galactoside Binding Soluble 3 Binding |
| ENSDARG00000040555 | ENSFM00250000004858 | 2 Oxoisovalerate Dehydrogenase Subunit Alpha Mitochondrial Precursor EC_1.2.4.4 Branched Chain Alpha Keto Acid Dehydrogenase E1 Component Alpha Chain BCKDE1A Bckdh E1 Alpha |
| ENSDARG00000040565 | ENSFM00800002003408 | Creatine Kinase M Type EC_2.7.3.2 Creatine Kinase M Chain M Ck |
| ENSDARG00000040582 | ENSFM00250000007733 | Unknown |
| ENSDARG00000040614 | ENSFM00250000002761 | SEC14 Domain And Spectrin Repeat Containing 1 |
| ENSDARG00000040650 | ENSFM00250000004184 | COP9 Signalosome Complex Subunit 1 SGN1 Signalosome Subunit 1 G Pathway Suppressor 1 Gps 1 JAB1 Containing Signalosome Subunit 1 |
| ENSDARG00000040674 | ENSFM00730001521778 | Carboxy Terminal Domain Rna Polymerase Ii Polypeptide A Small Phosphatase Nuclear Lim Interactor Interacting Factor Nli Interacting Factor Small C Terminal Domain Phosphatase Small Ctd Phosphatase |
| ENSDARG00000040705 | ENSFM00250000001281 | Glutaminase Isoform Mitochondrial Precursor Gls EC_3.5.1.2 Glutaminase L Glutamine Amidohydrolase |
| ENSDARG00000040778 | ENSFM00500000270233 | Ras Related Precursor |
| ENSDARG00000040803 | ENSFM00250000007515 | Serine Beta Lactamase Lactb Mitochondrial Precursor EC_3.4.-.- |
| ENSDARG00000040822 | ENSFM00670001235667 | FUN14 Domain Containing |
| ENSDARG00000040853 | ENSFM00500000270686 | Leucine Rich Repeat Soc 2 Homolog Sur 8 Homolog Soc 2 Homolog |
| ENSDARG00000040854 | ENSFM00500000270204 | Tumor Susceptibility Gene 101 Escrt I Complex Subunit TSG101 |
| ENSDARG00000040874 | ENSFM00610000969353 | Unknown |
| ENSDARG00000040874 | ENSFM00250000000681 | Adducin |
| ENSDARG00000040881 | ENSFM00740001589125 | Heterogeneous Nuclear Ribonucleoprotein Hnrnp |
| ENSDARG00000040890 | ENSFM00800002003519 | Farnesyl Pyrophosphate Synthase Fpp Synthase Fps EC_2.5.1.10 2E 6E Farnesyl Diphosphate Synthase Cholesterol Regulated 39 Kda Cr 39 Dimethylallyltranstransferase EC_2.5.1.- 1 Farnesyl Diphosphate Synthase Geranyltranstransferase |
| ENSDARG00000040926 | ENSFM00730001521440 | Coup Transcription Factor Coup Coup Transcription Factor Coup Tf Nuclear Receptor Subfamily 2 Group F Member |
| ENSDARG00000040942 | ENSFM00250000002129 | Purine Nucleoside Phosphorylase Pnp EC_2.4.2.1 Inosine Phosphorylase Inosine Guanosine Phosphorylase |
| ENSDARG00000040965 | ENSFM00250000001228 | Synaptosomal Associated 25 Snap 25 Synaptosomal Associated 25 Kda |
| ENSDARG00000040984 | ENSFM00730001522410 | Heat Shock 70 Kda 13 Precursor Microsomal Stress 70 Atpase Core Stress 70 Chaperone Microsome Associated 60 Kda |
| ENSDARG00000040988 | ENSFM00250000000728 | Triosephosphate Isomerase Tim EC_5.3.1.1 Triose Phosphate Isomerase |
| ENSDARG00000040991 | ENSFM00600000921238 | Cortactin Binding 2 CORTBP2 |
| ENSDARG00000041060 | ENSFM00740001589285 | Galectin Gal |
| ENSDARG00000041062 | ENSFM00500000270302 | Calbindin Calbindin D28 D 28K Vitamin D Dependent Calcium Binding Protein Avian Type |
| ENSDARG00000041065 | ENSFM00690001369041 | Unknown |
| ENSDARG00000041065 | ENSFM00500000270924 | Heat Shock Beta 1 HSPB1 Heat Shock 27 Kda Hsp 27 |
| ENSDARG00000041068 | ENSFM00730001521567 | Aspartate Aminotransferase Mitochondrial Precursor Maspat EC_2.6.1.1 EC_2.6.1.- 7 Fatty Acid Binding Fabp 1 Glutamate Oxaloacetate Transaminase 2 Kynurenine Aminotransferase 4 Kynurenine Aminotransferase Iv Kynurenine Oxoglutarate Transaminase 4 Kynurenin |
| ENSDARG00000041137 | ENSFM00500000269664 | Retinol Dehydrogenase |
| ENSDARG00000041150 | ENSFM00730001521551 | Vesicular Glutamate Transporter Solute Carrier Family 17 Member |
| ENSDARG00000041182 | ENSFM00250000002321 | 60S Ribosomal L4 |
| ENSDARG00000041232 | ENSFM00500000375647 | Unknown |
| ENSDARG00000041314 | ENSFM00250000004530 | Nadh Dehydrogenase 1 Beta Subcomplex Subunit 9 Complex I B22 Ci B22 Nadh Ubiquinone Oxidoreductase B22 Subunit |
| ENSDARG00000041332 | ENSFM00250000005590 | Transmembrane 33 |
| ENSDARG00000041358 | ENSFM00250000002312 | Leucine Rich Repeat Lgi Family Member Precursor Leucine Rich Glioma Inactivated |
| ENSDARG00000041363 | ENSFM00250000007388 | Dynactin Subunit 3 |
| ENSDARG00000041391 | ENSFM00500000271022 | Transmembrane EMP24 Domain Containing Precursor |
| ENSDARG00000041407 | ENSFM00670001235671 | Regulator Of Microtubule Dynamics Rmd Fa |
| ENSDARG00000041417 | ENSFM00560000771081 | V Type Proton Atpase Subunit S1 Precursor V Atpase Subunit S1 V Atpase AC45 Subunit V Atpase S1 Accessory Vacuolar Proton Pump Subunit S1 |
| ENSDARG00000041450 | ENSFM00500000269744 | Ras Related Rab Precursor |
| ENSDARG00000041595 | ENSFM00350000105388 | Carboxylesterase |
| ENSDARG00000041619 | ENSFM00800002020786 | Unknown |
| ENSDARG00000041619 | ENSFM00250000002295 | Guanine Nucleotide Binding Subunit Beta Receptor Activated C Kinase Receptor Of Activated Kinase C 1 RACK1 |
| ENSDARG00000041685 | ENSFM00730001521356 | Precursor |
| ENSDARG00000041703 | ENSFM00250000002798 | Ribosome Binding Ribosome Receptor Rrp |
| ENSDARG00000041720 | ENSFM00500000270526 | Unknown |
| ENSDARG00000041723 | ENSFM00250000000193 | Tubulin Beta Chain |
| ENSDARG00000041728 | ENSFM00730001521698 | Mannosyl Oligosaccharide 1 2 Alpha Mannosidase Ia EC_3.2.1.113 Man 9 Alpha Mannosidase Mannosidase Alpha Class 1A Member 1 Processing Alpha 1 2 Mannosidase Ia Alpha 1 2 Mannosidase Ia |
| ENSDARG00000041736 | ENSFM00670001235389 | Estradiol 17 Beta Dehydrogenase 12 EC_1.1.1.62 17 Beta Hydroxysteroid Dehydrogenase 12 17 Beta Hsd 12 |
| ENSDARG00000041799 | ENSFM00760001714761 | Gap Junction Alpha 1 Connexin 43 CX43 |
| ENSDARG00000041811 | ENSFM00670001235896 | 40S Ribosomal S25 |
| ENSDARG00000041830 | ENSFM00250000005250 | Translin EC_3.1.-.- Component 3 Of Promoter Of Risc C3PO |
| ENSDARG00000041875 | ENSFM00250000001221 | Ubiquitin Conjugating Enzyme E2 Variant |
| ENSDARG00000041895 | ENSFM00670001235391 | Cad Glutamine Dependent Carbamoyl Phosphate Synthase EC_6.3.5.5 |
| ENSDARG00000041895 | ENSFM00730001521216 | Carbamoyl Phosphate Synthase Mitochondrial Precursor EC_6.3.4.16 Carbamoyl Phosphate Synthetase I Cpsase I |
| ENSDARG00000041896 | ENSFM00500000270505 | Dnaj Homolog Subfamily C Member 5B Cysteine String Beta Csp Beta |
| ENSDARG00000041915 | ENSFM00500000275444 | Stum Homolog |
| ENSDARG00000041921 | ENSFM00500000408964 | Unknown |
| ENSDARG00000041921 | ENSFM00610000963039 | Unknown |
| ENSDARG00000041921 | ENSFM00800002014373 | Unknown |
| ENSDARG00000041926 | ENSFM00250000000275 | Disks Large Homolog Synapse Associated Sap |
| ENSDARG00000041974 | ENSFM00500000294166 | Unknown |
| ENSDARG00000041974 | ENSFM00610000955359 | Unknown |
| ENSDARG00000042069 | ENSFM00670001235483 | Phosphatidylethanolamine Binding 1 Pebp 1 Hcnppp |
| ENSDARG00000042114 | ENSFM00250000000861 | Bridging Integrator |
| ENSDARG00000042126 | ENSFM00250000001494 | Engulfment And Cell Motility |
| ENSDARG00000042128 | ENSFM00250000001466 | Kinase C And Casein Kinase Substrate In Neurons |
| ENSDARG00000042141 | ENSFM00730001522029 | Unconventional Myosin Vi Unconventional Myosin 6 |
| ENSDARG00000042249 | ENSFM00250000001583 | Ferm Domain Containing 4B GRP1 Binding GRSP1 |
| ENSDARG00000042252 | ENSFM00500000271538 | Eukaryotic Translation Initiation Factor 4H Eif 4H Williams Beuren Syndrome Chromosomal Region 1 |
| ENSDARG00000042277 | ENSFM00250000002014 | Ephrin B1 Precursor Elk Ligand Elk L Eph Related Receptor Tyrosine Kinase Ligand 2 Lerk 2 |
| ENSDARG00000042357 | ENSFM00500000269751 | Claudin |
| ENSDARG00000042418 | ENSFM00500000269893 | Polypeptide N Acetylgalactosaminyltransferase EC_2.4.1.41 Polypeptide Galnac Transferase Galnac T Pp Gantase Udp Acetylgalactosaminyltransferase Udp Galnac:Polypeptide N Acetylgalactosaminyltransferase |
| ENSDARG00000042469 | ENSFM00500000272434 | Nadh Dehydrogenase 1 Alpha Subcomplex Subunit Complex I B17 2 Ci B17 2 CIB17 2 Nadh Ubiquinone Oxidoreductase Subunit B17 2 |
| ENSDARG00000042509 | ENSFM00250000002922 | Glyoxalase Domain Containing 4 |
| ENSDARG00000042533 | ENSFM00500000269733 | Glutathione S Transferase EC_2.5.1.18 Gst Class Mu |
| ENSDARG00000042539 | ENSFM00730001521154 | Unknown |
| ENSDARG00000042566 | ENSFM00250000002224 | 40S Ribosomal S7 |
| ENSDARG00000042630 | ENSFM00500000273740 | Heme Binding 2 Soul |
| ENSDARG00000042641 | ENSFM00800002003871 | Lanosterol 14 Alpha Demethylase Ldm EC_1.14.13.70 Cypli Cytochrome P450 51A1 Cytochrome P450 14DM Cytochrome Dm Cytochrome P450LI Sterol 14 Alpha Demethylase |
| ENSDARG00000042644 | ENSFM00780001898344 | Superoxide Dismutase Mitochondrial EC_1.15.1.1 |
| ENSDARG00000042658 | ENSFM00730001522325 | Isobutyryl Coa Dehydrogenase Mitochondrial Precursor EC_1.3.99.- Acyl Coa Dehydrogenase Family Member 8 Acad 8 |
| ENSDARG00000042670 | ENSFM00250000001830 | Epidermal Growth Factor Receptor Substrate 15 1 EPS15 Related EPS15R |
| ENSDARG00000042677 | ENSFM00250000001126 | Cell Adhesion Molecule Precursor Immunoglobulin Superfamily Member Nectin Necl Synaptic Cell Adhesion Molecule |
| ENSDARG00000042690 | ENSFM00730001521347 | Sphingosine 1 Phosphate Receptor S1P Receptor Endothelial Differentiation G Coupled Receptor Sphingosine 1 Phosphate Receptor Edg S1P Receptor Edg |
| ENSDARG00000042708 | ENSFM00250000000217 | Tubulin Alpha Chain |
| ENSDARG00000042723 | ENSFM00500000269996 | Gdnf Family Receptor Alpha Precursor Gdnf Receptor Alpha Gdnfr Alpha Gfr Alpha Tgf Beta Related Neurotrophic Factor Receptor |
| ENSDARG00000042777 | ENSFM00500000282727 | Nadh Dehydrogenase 1 Alpha Subcomplex 11 |
| ENSDARG00000042780 | ENSFM00740001589094 | Apolipoprotein B 100 Precursor Apo B 100 |
| ENSDARG00000042803 | ENSFM00730001521441 | Ras Related Rab 3 |
| ENSDARG00000042821 | ENSFM00250000004718 | Alkyldihydroxyacetonephosphate Synthase Alkyl Dhap Synthase EC_2.5.1.26 Alkylglycerone Phosphate Synthase |
| ENSDARG00000042823 | ENSFM00250000005526 | Signal Peptidase Complex Subunit 2 EC_3.4.-.- Microsomal Signal Peptidase 25 Kda Subunit Spase 25 Kda Subunit |
| ENSDARG00000042837 | ENSFM00670001235362 | Sodium/Potassium Transporting Atpase Subunit Beta Sodium/Potassium Dependent Atpase Subunit Beta |
| ENSDARG00000042848 | ENSFM00250000003005 | L Isoaspartate D Aspartate O Methyltransferase Pimt EC_2.1.1.77 L Isoaspartyl Carboxyl Methyltransferase L Isoaspartyl/D Aspartyl Methyltransferase Beta Aspartate Methyltransferase |
| ENSDARG00000042853 | ENSFM00730001521875 | Isovaleryl Coa Dehydrogenase Mitochondrial Precursor Ivd EC_1.3.8.4 |
| ENSDARG00000042854 | ENSFM00500000270247 | Epoxide Hydrolase 1 EC_3.3.2.9 Epoxide Hydratase Microsomal Epoxide Hydrolase |
| ENSDARG00000042872 | ENSFM00250000002881 | Ankyrin Repeat And Lem Domain Containing 2 Lem Domain Containing 4 |
| ENSDARG00000042872 | ENSFM00500000270072 | Palmitoyltransferase ZDHHC5 EC_2.3.1.225 Zinc Finger Dhhc Domain Containing 5 Dhhc 5 |
| ENSDARG00000042905 | ENSFM00610000960974 | Unknown |
| ENSDARG00000042905 | ENSFM00790001962982 | 60S Ribosomal L10A |
| ENSDARG00000042948 | ENSFM00500000270505 | Dnaj Homolog Subfamily C Member 5B Cysteine String Beta Csp Beta |
| ENSDARG00000042974 | ENSFM00500000270007 | Synaptophysin |
| ENSDARG00000042975 | ENSFM00500000270222 | Methylglutaconyl Coa Hydratase Mitochondrial Precursor EC_4.2.1.18 Au Specific Rna Binding Enoyl Coa Hydratase Au Binding Coa Hydratase |
| ENSDARG00000042978 | ENSFM00400000131705 | Cytochrome P450 |
| ENSDARG00000043010 | ENSFM00250000000111 | Calcium/Calmodulin Dependent Kinase Type Ii Subunit Cam Kinase Ii Subunit Camk Ii Subunit EC_2.7.11.17 |
| ENSDARG00000043012 | ENSFM00250000005767 | Gamma Soluble Nsf Attachment Snap Gamma N Ethylmaleimide Sensitive Factor Attachment Gamma |
| ENSDARG00000043019 | ENSFM00250000004597 | Exocyst Complex Component 1 Exocyst Complex Component SEC3 |
| ENSDARG00000043026 | ENSFM00670001235358 | Tau Tubulin Kinase EC_2.7.11.1 |
| ENSDARG00000043094 | ENSFM00300000079126 | Guanine Nucleotide Binding G S Subunit Alpha Adenylate Cyclase Stimulating G Alpha |
| ENSDARG00000043134 | ENSFM00250000001072 | Homeobox Cut 1 Ccaat Displacement Cdp Homeobox Cux 1 |
| ENSDARG00000043148 | ENSFM00800002003375 | Excitatory Amino Acid Transporter Sodium Dependent Glutamate/Aspartate Transporter Solute Carrier Family 1 Member |
| ENSDARG00000043180 | ENSFM00800002003402 | Glycerol 3 Phosphate Dehydrogenase Cytoplasmic Gpd C Gpdh C EC_1.1.1.8 |
| ENSDARG00000043241 | ENSFM00250000000572 | Arrestin |
| ENSDARG00000043250 | ENSFM00730001521429 | Phosphatase 1A EC_3.1.3.16 Phosphatase 2C Alpha PP2C Alpha |
| ENSDARG00000043257 | ENSFM00710001464696 | Unknown |
| ENSDARG00000043257 | ENSFM00800002003408 | Creatine Kinase M Type EC_2.7.3.2 Creatine Kinase M Chain M Ck |
| ENSDARG00000043313 | ENSFM00500000269628 | Ankyrin Ank Ankyrin |
| ENSDARG00000043357 | ENSFM00250000006389 | Transducin Beta 2 |
| ENSDARG00000043371 | ENSFM00730001521810 | Malate Dehydrogenase Mitochondrial Precursor EC_1.1.1.37 |
| ENSDARG00000043406 | ENSFM00710001441664 | Sodium/Calcium Exchanger 1 Precursor Na + /Ca 2+ Exchange 1 Solute Carrier Family 8 Member 1 |
| ENSDARG00000043410 | ENSFM00500000269993 | Xk Related |
| ENSDARG00000043416 | ENSFM00710001464693 | Unknown |
| ENSDARG00000043451 | ENSFM00250000002178 | Calcium Binding 39 MO25 |
| ENSDARG00000043453 | ENSFM00250000003056 | 40S Ribosomal S5 |
| ENSDARG00000043457 | ENSFM00610000973040 | Unknown |
| ENSDARG00000043457 | ENSFM00730001521081 | Glyceraldehyde 3 Phosphate Dehydrogenase Gapdh EC_1.2.1.12 Peptidyl Cysteine S Nitrosylase Gapdh EC_2.6.99.- |
| ENSDARG00000043460 | ENSFM00670001238056 | C Type Natriuretic Peptide Precursor C Type Natriuretic Peptide Cnp 22 |
| ENSDARG00000043465 | ENSFM00810002040578 | V Type Proton Atpase Subunit B V Atpase Subunit B Endomembrane Proton Pump 58 Kda Subunit Vacuolar Proton Pump Subunit B |
| ENSDARG00000043467 | ENSFM00540000720122 | Nadh Dehydrogenase 1 Beta Subcomplex Subunit 11 Mitochondrial |
| ENSDARG00000043474 | ENSFM00250000000445 | Plasma Membrane Calcium Transporting Atpase EC_3.6.3.8 Plasma Membrane Calcium Atpase Plasma Membrane Calcium Pump |
| ENSDARG00000043493 | ENSFM00250000001393 | Clathrin Heavy Chain |
| ENSDARG00000043497 | ENSFM00250000002467 | Secernin |
| ENSDARG00000043509 | ENSFM00250000002087 | 60S Ribosomal L11 |
| ENSDARG00000043510 | ENSFM00500000273038 | Vesicle Associated Membrane 4 Vamp 4 |
| ENSDARG00000043511 | ENSFM00730001522010 | Peroxiredoxin 6 EC_1.11.1.15 1 Cys Peroxiredoxin 1 Cys Prx Acidic Calcium Independent Phospholipase A2 AIPLA2 EC_3.1.1.- Antioxidant 2 Non Selenium Glutathione Peroxidase Nsgpx EC_1.11.-.- 1 9 |
| ENSDARG00000043514 | ENSFM00250000000577 | Dimethylaniline Monooxygenase EC_1.14.13.8 Dimethylaniline Oxidase Flavin Containing Monooxygenase Fmo |
| ENSDARG00000043518 | ENSFM00250000000577 | Dimethylaniline Monooxygenase EC_1.14.13.8 Dimethylaniline Oxidase Flavin Containing Monooxygenase Fmo |
| ENSDARG00000043527 | ENSFM00250000003664 | Probable Lipid Phosphate Phosphatase PPAPDC3 EC_3.1.3.- Phosphatidic Acid Phosphatase Type 2 Domain Containing 3 |
| ENSDARG00000043554 | ENSFM00500000274041 | 39S Ribosomal L40 Mitochondrial Precursor L40MT Mrp L40 Nuclear Localization Signal Containing Deleted In Velocardiofacial Syndrome |
| ENSDARG00000043555 | ENSFM00500000270036 | Cell Cycle Control 50A P4 Atpase Flippase Complex Beta Subunit TMEM30A Transmembrane 30A |
| ENSDARG00000043561 | ENSFM00730001521694 | 26S Protease Regulatory Subunit 4 26S Proteasome Aaa Atpase Subunit RPT2 Proteasome 26S Subunit Atpase 1 |
| ENSDARG00000043571 | ENSFM00250000000916 | Kinesin |
| ENSDARG00000043589 | ENSFM00670001235760 | Carbonic Anhydrase 4 Precursor EC_4.2.1.1 Carbonate Dehydratase Iv Carbonic Anhydrase Iv Ca Iv |
| ENSDARG00000043661 | ENSFM00250000000780 | Calcium Dependent Secretion Activator 1 Calcium Dependent Activator Secretion 1 Caps 1 |
| ENSDARG00000043665 | ENSFM00670001235945 | Glutaredoxin Related 5 Mitochondrial Precursor Monothiol Glutaredoxin 5 |
| ENSDARG00000043673 | ENSFM00260000050975 | Brain Enriched Guanylate Kinase Associated |
| ENSDARG00000043697 | ENSFM00730001521354 | Neurofilament Polypeptide Nf Kda Neurofilament Neurofilament Triplet |
| ENSDARG00000043732 | ENSFM00250000005111 | COP9 Signalosome Complex Subunit 4 Signalosome Subunit 4 |
| ENSDARG00000043746 | ENSFM00740001589096 | Neurexin Precursor Neurexin |
| ENSDARG00000043795 | ENSFM00250000002426 | Rho Gdp Dissociation Inhibitor Rho Gdi Rho Gdi |
| ENSDARG00000043835 | ENSFM00730001521441 | Ras Related Rab 3 |
| ENSDARG00000043842 | ENSFM00250000010056 | Uncharacterized |
| ENSDARG00000043843 | ENSFM00610000961323 | Unknown |
| ENSDARG00000043843 | ENSFM00500000273030 | A Kinase Anchor 7 Gamma Akap 7 Gamma A Kinase Anchor 18 Akap 18 Kinase A Anchoring 7 Gamma PRKA7 Gamma |
| ENSDARG00000043848 | ENSFM00670001235331 | Superoxide Dismutase EC_1.15.1.1 |
| ENSDARG00000043857 | ENSFM00670001236771 | Ubiquitin Fold Modifier 1 Precursor |
| ENSDARG00000043864 | ENSFM00610000952905 | Basigin Precursor CD147 Antigen |
| ENSDARG00000043865 | ENSFM00670001236198 | Mitochondrial Import Inner Membrane Translocase Subunit TIM9 |
| ENSDARG00000043938 | ENSFM00350000105477 | Leucine Rich Repeat Containing 57 |
| ENSDARG00000043970 | ENSFM00250000002838 | Leucine Rich Ppr Motif Containing Protein Mitochondrial Precursor 130 Kda Leucine Rich Lrp 130 |
| ENSDARG00000043976 | ENSFM00250000005309 | Eukaryotic Peptide Chain Release Factor Subunit 1 Eukaryotic Release Factor 1 ERF1 |
| ENSDARG00000044015 | ENSFM00500000269996 | Gdnf Family Receptor Alpha Precursor Gdnf Receptor Alpha Gdnfr Alpha Gfr Alpha Tgf Beta Related Neurotrophic Factor Receptor |
| ENSDARG00000044016 | ENSFM00730001522029 | Unconventional Myosin Vi Unconventional Myosin 6 |
| ENSDARG00000044053 | ENSFM00760001714608 | Unknown |
| ENSDARG00000044062 | ENSFM00250000001273 | C Terminal Binding |
| ENSDARG00000044090 | ENSFM00770001847613 | Caax Prenyl Protease 1 Homolog EC_3.4.24.84 Farnesylated Proteins Converting Enzyme 1 Face 1 Prenyl Specific Endoprotease 1 Zinc Metalloproteinase STE24 Homolog |
| ENSDARG00000044091 | ENSFM00500000269976 | Phosphatidylinositol Transfer Alpha Pi Tp Alpha Ptdins Transfer Alpha Ptdinstp Alpha |
| ENSDARG00000044092 | ENSFM00600000927268 | Atpase Inhibitor B Mitochondrial Precursor Inhibitor Of F 1 F O Atpase B If 1 B IF1 B |
| ENSDARG00000044093 | ENSFM00790001962987 | 60S Ribosomal L13A |
| ENSDARG00000044125 | ENSFM00670001237763 | Thioredoxin |
| ENSDARG00000044134 | ENSFM00500000270016 | Isoleucine Trna Ligase Mitochondrial EC_6.1.1.5 Isoleucyl Trna Synthetase Ilers |
| ENSDARG00000044142 | ENSFM00800002003390 | Acetyl Coenzyme A Synthetase EC_6.2.1.1 Acetate Coa Ligase Acyl Activating Enzyme |
| ENSDARG00000044153 | ENSFM00420000140518 | Serine/Threonine Phosphatase PP1 Catalytic Subunit Pp EC_3.1.3.16 |
| ENSDARG00000044161 | ENSFM00720001492054 | Glutamate Receptor Ionotropic Delta Precursor Glur Delta Subunit |
| ENSDARG00000044167 | ENSFM00250000001768 | Arginine Deiminase Type EC_3.5.3.15 Peptidylarginine Deiminase Arginine Deiminase Type |
| ENSDARG00000044182 | ENSFM00250000001250 | Double Stranded Rna Binding Staufen Homolog |
| ENSDARG00000044212 | ENSFM00560000788827 | Unknown |
| ENSDARG00000044216 | ENSFM00250000006452 | Alpha Ketoglutarate Dependent Dioxygenase Fto EC_1.14.11.- Fat Mass And Obesity Associated |
| ENSDARG00000044241 | ENSFM00500000271657 | Dna Damage Regulated Autophagy Modulator 2 Transmembrane 77 |
| ENSDARG00000044254 | ENSFM00760001714627 | Annexin Annexin Annexin |
| ENSDARG00000044281 | ENSFM00730001522141 | Leucine Rich Repeat Containing 7 Densin 180 Densin |
| ENSDARG00000044299 | ENSFM00250000000425 | Lamin |
| ENSDARG00000044339 | ENSFM00250000005619 | XRP2 |
| ENSDARG00000044341 | ENSFM00500000270226 | Carbohydrate Sulfotransferase EC_2.8.2.- Galactose/N Acetylglucosamine/N Acetylglucosamine 6 O Sulfotransferase N Acetylglucosamine 6 O Sulfotransferase GLCNAC6ST Sulfotransferase N Acetylglucosamine 6 O Sulfotransferase GLCNAC6ST GN6ST |
| ENSDARG00000044345 | ENSFM00250000001390 | Cytoplasmic FMR1 Interacting |
| ENSDARG00000044420 | ENSFM00500000273595 | Mitochondrial Import Inner Membrane Translocase Subunit TIM14 Dnaj Homolog Subfamily C Member 19 |
| ENSDARG00000044501 | ENSFM00730001521291 | Vimentin |
| ENSDARG00000044514 | ENSFM00250000002180 | Beta Galactoside Alpha 2 6 Sialyltransferase 2 Alpha 2 6 St 2 EC_2.4.99.1 Cmp N Acetylneuraminate Beta Galactosamide Alpha 2 6 Sialyltransferase 2 ST6GAL Ii ST6GALII Sialyltransferase 2 |
| ENSDARG00000044521 | ENSFM00730001521581 | Elongation Factor 1 Beta Ef 1 Beta |
| ENSDARG00000044540 | ENSFM00500000273014 | Peptidyl Prolyl Cis Trans Isomerase FKBP2 Precursor Ppiase FKBP2 EC_5.2.1.8.13 Kda FK506 Binding 13 Kda Fkbp Fkbp 13 FK506 Binding 2 Fkbp 2 Immunophilin FKBP13 Rotamase |
| ENSDARG00000044551 | ENSFM00730001522009 | Autophagy Related APG9 |
| ENSDARG00000044562 | ENSFM00670001235376 | Cytochrome C |
| ENSDARG00000044565 | ENSFM00250000002536 | Obg Atpase 1 |
| ENSDARG00000044573 | ENSFM00730001521235 | Cell Division Control 42 Homolog Precursor |
| ENSDARG00000044575 | ENSFM00250000001468 | Valine Trna Ligase EC_6.1.1.9 Valyl Trna Synthetase Valrs |
| ENSDARG00000044601 | ENSFM00500000269689 | Reticulon |
| ENSDARG00000044605 | ENSFM00500000270193 | Syntaxin |
| ENSDARG00000044626 | ENSFM00610000952921 | Mitochondrial Calcium Uniporter Regulator 1 Coiled Coil Domain Containing 90A Mitochondrial |
| ENSDARG00000044628 | ENSFM00500000272679 | D Tyrosyl Trna Tyr Deacylase 1 EC_3.1.-.- Dna Unwinding Element Binding B Due B |
| ENSDARG00000044669 | ENSFM00250000005391 | Heterochromatin 1 Binding 3 |
| ENSDARG00000044719 | ENSFM00250000007237 | Neurensin 1 Neuro P24 Vesicular Membrane Of 24 Kda Vesicular Membrane P24 |
| ENSDARG00000044751 | ENSFM00670001240292 | D Dopachrome |
| ENSDARG00000044753 | ENSFM00770001847589 | Isocitrate Dehydrogenase Subunit Beta Mitochondrial Precursor EC_1.1.1.41 Isocitric Dehydrogenase Subunit Beta Nad + Specific Icdh Subunit Beta |
| ENSDARG00000044760 | ENSFM00410000138458 | Guanine Nucleotide Binding Subunit |
| ENSDARG00000044874 | ENSFM00250000002627 | Rho Gtpase Activating 39 |
| ENSDARG00000044902 | ENSFM00250000000445 | Plasma Membrane Calcium Transporting Atpase EC_3.6.3.8 Plasma Membrane Calcium Atpase Plasma Membrane Calcium Pump |
| ENSDARG00000044914 | ENSFM00780001898454 | Succinyl Coa Ligase Subunit Beta Mitochondrial EC_6.2.1.4 Gtp Specific Succinyl Coa Synthetase Subunit Beta Succinyl Coa Synthetase Beta G Chain Scs Betag |
| ENSDARG00000044943 | ENSFM00760001714762 | Puromycin Sensitive Aminopeptidase Psa EC_3.4.11.14 Cytosol Alanyl Aminopeptidase Aap S |
| ENSDARG00000044968 | ENSFM00250000001750 | Vinculin |
| ENSDARG00000044972 | ENSFM00670001235596 | B Cell Receptor Associated Bcr Associated |
| ENSDARG00000044975 | ENSFM00730001521138 | Keratin Type I Cytoskeletal Cytokeratin Ck Keratin |
| ENSDARG00000045006 | ENSFM00730001521864 | Receptor Type Tyrosine Phosphatase Precursor R Ptp EC_3.1.3.48 |
| ENSDARG00000045014 | ENSFM00250000000217 | Tubulin Alpha Chain |
| ENSDARG00000045023 | ENSFM00250000002406 | Lipoma Hmgic Fusion Partner |
| ENSDARG00000045027 | ENSFM00750001632398 | Ras Related Rab |
| ENSDARG00000045036 | ENSFM00250000006209 | CDC42 Effector 4 Binder Of Rho Gtpases 4 |
| ENSDARG00000045064 | ENSFM00730001543875 | Unknown |
| ENSDARG00000045064 | ENSFM00250000000459 | Actin Binding Lim Ablim Actin Binding Lim Family Member |
| ENSDARG00000045065 | ENSFM00500000272468 | Vesicle Transport Through Interaction With T Snares Homolog 1A Vesicle Transport V Snare VTI1 2 VTI1 RP2 |
| ENSDARG00000045066 | ENSFM00250000001339 | Ectonucleoside Triphosphate Diphosphohydrolase Ntpdase EC_3.6.1.5 Ecto Atp Diphosphohydrolase Ecto Atpdase Ecto Atpase Ecto Apyrase |
| ENSDARG00000045087 | ENSFM00250000002836 | Cyclin Dependent Kinase 5 Activator 1 Precursor CDK5 Activator 1 Cyclin Dependent Kinase 5 Regulatory Subunit 1 Tpkii Regulatory Subunit |
| ENSDARG00000045132 | ENSFM00250000001189 | Voltage Dependent Anion Selective Channel Vdac Outer Mitochondrial Membrane Porin |
| ENSDARG00000045140 | ENSFM00500000271002 | Leucine Carboxyl Methyltransferase 1 EC_2.1.1.233 Leucine Carboxy Methyltransferase 1 |
| ENSDARG00000045145 | ENSFM00500000276365 | Shisa Precursor |
| ENSDARG00000045146 | ENSFM00800002024278 | Unknown |
| ENSDARG00000045146 | ENSFM00500000303715 | TOMM22 |
| ENSDARG00000045150 | ENSFM00800002003414 | Casein Kinase I Delta Cki Delta Ckid EC_2.7.11.1 |
| ENSDARG00000045156 | ENSFM00730001522095 | Regulator Of G Signaling |
| ENSDARG00000045200 | ENSFM00440000237023 | Dual Specificity Phosphatase 19 EC_3.1.3.16 EC_3.1.3.- 48 Phosphatase SKRP1 Stress Activated Kinase Pathway Regulating Phosphatase 1 |
| ENSDARG00000045228 | ENSFM00610000960150 | Unknown |
| ENSDARG00000045230 | ENSFM00670001239338 | Cytochrome C Oxidase Subunit Vib 1 |
| ENSDARG00000045249 | ENSFM00500000270871 | Gtp Cyclohydrolase 1 Type 2 Ni EC_3.5.4.16 Gtp Cyclohydrolase I NIF3 1 |
| ENSDARG00000045277 | ENSFM00500000269664 | Retinol Dehydrogenase |
| ENSDARG00000045294 | ENSFM00250000002047 | Endophilin SH3 Domain Containing GRB2 |
| ENSDARG00000045297 | ENSFM00800002003689 | Prohibitin 2 |
| ENSDARG00000045331 | ENSFM00730001521673 | Catenin Delta 2 |
| ENSDARG00000045351 | ENSFM00500000273461 | Coiled Coil Domain Containing 58 |
| ENSDARG00000045355 | ENSFM00500000272323 | Transcription Elongation Factor B Polypeptide 1 Elongin 15 Kda Subunit Elongin C Eloc Rna Polymerase Ii Transcription Factor Siii Subunit C Siii P15 Stromal Membrane Associated SMAP1B Homolog |
| ENSDARG00000045364 | ENSFM00250000005557 | Trafficking Particle Complex Subunit 3 |
| ENSDARG00000045367 | ENSFM00250000000217 | Tubulin Alpha Chain |
| ENSDARG00000045399 | ENSFM00730001521615 | T Complex 1 Subunit Epsilon Tcp 1 Epsilon Cct Epsilon |
| ENSDARG00000045411 | ENSFM00780001898384 | Acyl Coa Dehydrogenase Family Member Acad EC_1.3.99.- |
| ENSDARG00000045415 | ENSFM00300000079126 | Guanine Nucleotide Binding G S Subunit Alpha Adenylate Cyclase Stimulating G Alpha |
| ENSDARG00000045417 | ENSFM00610000971059 | Unknown |
| ENSDARG00000045417 | ENSFM00250000002173 | FAM49A |
| ENSDARG00000045420 | ENSFM00250000006488 | Mammalian Ependymin Related 1 Precursor Merp 1 |
| ENSDARG00000045421 | ENSFM00250000003670 | Star Related Lipid Transfer 3 Start Domain Containing 3 STARD3 |
| ENSDARG00000045447 | ENSFM00500000272943 | Solute Carrier Family 35 Member G2 Transmembrane 22 |
| ENSDARG00000045463 | ENSFM00500000270233 | Ras Related Precursor |
| ENSDARG00000045465 | ENSFM00250000002547 | Lanc 1.40 Kda Erythrocyte Membrane P40 |
| ENSDARG00000045487 | ENSFM00250000002859 | 40S Ribosomal S16 |
| ENSDARG00000045514 | ENSFM00750001632374 | Atp Synthase Subunit Gamma Mitochondrial F Atpase Gamma Subunit |
| ENSDARG00000045540 | ENSFM00250000001098 | Serine/Threonine Phosphatase 6 Regulatory Subunit Saps Domain Family Member |
| ENSDARG00000045542 | ENSFM00670001240165 | Ragulator Complex LAMTOR4 Late Endosomal/Lysosomal Adaptor And Mapk And Mtor Activator 4 |
| ENSDARG00000045543 | ENSFM00670001235732 | V Type Proton Atpase Subunit F V Atpase Subunit F V Atpase 14 Kda Subunit Vacuolar Proton Pump Subunit F |
| ENSDARG00000045555 | ENSFM00500000270642 | Homolog Mitochondrial Precursor |
| ENSDARG00000045567 | ENSFM00610000972962 | Unknown |
| ENSDARG00000045567 | ENSFM00670001236604 | 14 Kda Phosphohistidine Phosphatase EC_3.1.3.- Phosphohistidine Phosphatase 1 |
| ENSDARG00000045568 | ENSFM00730001521772 | Branched Chain Amino Acid Aminotransferase EC_2.6.1.42 |
| ENSDARG00000045618 | ENSFM00250000001836 | Clathrin Light Chain |
| ENSDARG00000045620 | ENSFM00250000006043 | Sialic Acid Synthase N Acetylneuraminate Synthase EC_2.5.1.56 N Acetylneuraminate 9 Phosphate Synthase EC_2.5.1.- 57 N Acetylneuraminic Acid Phosphate Synthase N Acetylneuraminic Acid Synthase |
| ENSDARG00000045628 | ENSFM00270000056513 | Ras Related Rab |
| ENSDARG00000045639 | ENSFM00250000000588 | Elav |
| ENSDARG00000045685 | ENSFM00800002003683 | Contactin 1 Precursor Neural Cell Surface F3 |
| ENSDARG00000045696 | ENSFM00500000273802 | 39S Ribosomal L23 Mitochondrial L23MT Mrp L23 L23 Mitochondrial Related |
| ENSDARG00000045706 | ENSFM00500000270370 | Tetraspanin 9 Tspan 9 |
| ENSDARG00000045748 | ENSFM00740001589370 | Stabilin 2 Precursor Fasciclin Egf Laminin Type Egf And Link Domain Containing Scavenger Receptor 2 Feel 2 Hyaluronan Receptor Endocytosis |
| ENSDARG00000045749 | ENSFM00250000001883 | Liprin Beta 2 Tyrosine Phosphatase Receptor Type F Polypeptide Interacting Binding 2 Ptprf Interacting Binding 2 |
| ENSDARG00000045799 | ENSFM00730001521436 | Core Precursor Chondroitin Sulfate Proteoglycan |
| ENSDARG00000045814 | ENSFM00500000271410 | Sorting And Assembly Machinery Component 50 Homolog |
| ENSDARG00000045832 | ENSFM00670001238501 | Cocaine And Amphetamine Regulated Transcript |
| ENSDARG00000045843 | ENSFM00250000004097 | Dna Apurinic Or Apyrimidinic Site Lyase EC_3.1.-.- EC_4.2.99.18 Apex Nuclease Apen Apurinic Apyrimidinic Endonuclease 1 Ap Endonuclease 1 Ref 1 Redox Factor 1 |
| ENSDARG00000045870 | ENSFM00800002003674 | Pleckstrin Homology Domain Containing Family A Member 6 Ph Domain Containing Family A Member 6 Phosphoinositol 3 Phosphate Binding 3 Pepp 3 |
| ENSDARG00000045877 | ENSFM00500000270143 | Ubiquitin Conjugating Enzyme E2 N EC_6.3.2.19 Ubiquitin Carrier N Ubiquitin Ligase N |
| ENSDARG00000045888 | ENSFM00730001521421 | Acetyl Coa Acetyltransferase Mitochondrial Precursor EC_2.3.1.9 Acetoacetyl Coa Thiolase |
| ENSDARG00000045893 | ENSFM00500000270346 | Btb/Poz Domain Containing KCTD15 Potassium Channel Tetramerization Domain Containing 15 |
| ENSDARG00000045926 | ENSFM00500000270567 | Cellular Retinoic Acid Binding 1 Cellular Retinoic Acid Binding I Crabp I |
| ENSDARG00000045928 | ENSFM00500000270239 | Proteasome Subunit Alpha Type EC_3.4.25.1 Macropain Subunit Multicatalytic Endopeptidase Complex Subunit Proteasome Component Subunit |
| ENSDARG00000045944 | ENSFM00800002003411 | Sodium And Chloride Dependent Gaba Transporter 1 Gat 1 Solute Carrier Family 6 Member 1 |
| ENSDARG00000045945 | ENSFM00250000000771 | Synapsin Synapsin |
| ENSDARG00000045976 | ENSFM00250000002455 | SID1 Transmembrane Family Member Precursor |
| ENSDARG00000045978 | ENSFM00670001237761 | Micos Complex Subunit MIC25A Coiled Coil Helix Coiled Coil Helix Domain Containing 6 |
| ENSDARG00000046022 | ENSFM00500000270192 | Transmembrane EMP24 Domain Containing Precursor P24 Family Gamma |
| ENSDARG00000046048 | ENSFM00250000001879 | Vesicle Associated Membrane Associated A Vamp A Vamp Associated A Vap A |
| ENSDARG00000046054 | ENSFM00730001521711 | Acyl Coa Synthetase Fatty Acid Transport Fatp Fatty Acid Coenzyme A Ligase Very Long Chain Solute Carrier Family 27 Member 2 Very Long Chain Coa |
| ENSDARG00000046106 | ENSFM00730001521184 | Ras Related Rab |
| ENSDARG00000046107 | ENSFM00250000001241 | Caskin 1 Cask Interacting 1 |
| ENSDARG00000046157 | ENSFM00750001651938 | Unknown |
| ENSDARG00000046157 | ENSFM00750001659008 | Unknown |
| ENSDARG00000046157 | ENSFM00780001898364 | 40S Ribosomal S17 |
| ENSDARG00000051756 | ENSFM00800002003776 | Pyruvate Dehydrogenase X Component Mitochondrial Precursor Dihydrolipoamide Dehydrogenase Binding Of Pyruvate Dehydrogenase Complex Lipoyl Containing Pyruvate Dehydrogenase Complex Component X |
| ENSDARG00000051783 | ENSFM00250000001360 | 60S Acidic Ribosomal P0 60S Ribosomal L10E |
| ENSDARG00000051814 | ENSFM00730001521864 | Receptor Type Tyrosine Phosphatase Precursor R Ptp EC_3.1.3.48 |
| ENSDARG00000051816 | ENSFM00740001589372 | Alpha Aminoadipic Semialdehyde Synthase Mitochondrial Precursor Lkr/Sdh |
| ENSDARG00000051824 | ENSFM00250000001843 | Tomoregulin 1 Precursor Tr 1 Transmembrane With Egf And One Follistatin Domain |
| ENSDARG00000051852 | ENSFM00260000050384 | Potassium Voltage Gated Channel Subfamily C Member Voltage Gated Potassium Channel KV3 |
| ENSDARG00000051874 | ENSFM00250000005421 | Stimulated By Retinoic Acid Gene 6 |
| ENSDARG00000051875 | ENSFM00260000051182 | Immunoglobulin Superfamily Containing Leucine Rich Repeat Precursor |
| ENSDARG00000051879 | ENSFM00730001521882 | Atp Binding Cassette Sub Family C Member Sulfonylurea Receptor |
| ENSDARG00000051888 | ENSFM00250000004551 | IST1 Homolog |
| ENSDARG00000051889 | ENSFM00250000003878 | Dihydroorotate Dehydrogenase Quinone Mitochondrial Precursor Dhodehase EC_1.3.5.2 Dihydroorotate Oxidase |
| ENSDARG00000051970 | ENSFM00250000004547 | WD40 Repeat Containing SMU1 Smu 1 Suppressor Of Mec 8 And Unc 52 Homolog |
| ENSDARG00000051986 | ENSFM00250000005068 | Nadh Dehydrogenase Iron Sulfur 8 Mitochondrial Precursor EC_1.6.5.3 EC_1.6.99.- 3 Complex I 23KD Ci 23KD Nadh Ubiquinone Oxidoreductase 23 Kda Subunit |
| ENSDARG00000052010 | ENSFM00500000272289 | Mitotic Spindle Associated Mmxd Complex Subunit MIP18 FAM96B |
| ENSDARG00000052020 | ENSFM00260000051403 | Cklf Marvel Transmembrane Domain Containing 4 Chemokine Factor Superfamily Member 4 |
| ENSDARG00000052030 | ENSFM00730001521171 | Reductase Aldo Keto Reductase Family 1 Member |
| ENSDARG00000052114 | ENSFM00500000273341 | Creb Regulated Transcription Coactivator 3 Transducer Of Regulated Camp Response Element Binding 3 Torc 3 Transducer Of Creb 3 |
| ENSDARG00000052178 | ENSFM00810002040882 | Eukaryotic Translation Initiation Factor 2 Subunit 1 Eukaryotic Translation Initiation Factor 2 Subunit Alpha Eif 2 Alpha Eif 2A Eif 2ALPHA |
| ENSDARG00000052207 | ENSFM00250000000285 | Complement C3 Complement C3 Beta Chain |
| ENSDARG00000052263 | ENSFM00730001521238 | Ras Related Rab |
| ENSDARG00000052304 | ENSFM00780001898353 | Cytochrome B C1 Complex Subunit 1 Mitochondrial Precursor Complex Iii Subunit 1 Core I Ubiquinol Cytochrome C Reductase Complex Core 1 |
| ENSDARG00000052334 | ENSFM00500000272374 | Ocia Domain Containing 1 |
| ENSDARG00000052374 | ENSFM00250000005012 | Fas Associated Factor 2 Ubx Domain Containing 8 |
| ENSDARG00000052377 | ENSFM00670001277927 | Unknown |
| ENSDARG00000052377 | ENSFM00250000004556 | 26S Proteasome Non Atpase Regulatory Subunit 12 26S Proteasome Regulatory Subunit RPN5 26S Proteasome Regulatory Subunit P55 |
| ENSDARG00000052379 | ENSFM00250000005354 | Coiled Coil Domain Containing 22 |
| ENSDARG00000052390 | ENSFM00750001657472 | Unknown |
| ENSDARG00000052390 | ENSFM00750001658547 | Unknown |
| ENSDARG00000052390 | ENSFM00500000272421 | TMED8 |
| ENSDARG00000052438 | ENSFM00730001521596 | Actin Related 2 Actin 2 |
| ENSDARG00000052462 | ENSFM00600000921219 | Phosphatidylserine Decarboxylase Proenzyme EC_4.1.1.65 |
| ENSDARG00000052518 | ENSFM00250000000549 | Syntaxin |
| ENSDARG00000052604 | ENSFM00250000000830 | Cytoplasmic Polyadenylation Element Binding |
| ENSDARG00000052606 | ENSFM00250000001158 | Dipeptidyl Peptidase 9 DP9 EC_3.4.14.5 Dipeptidyl Peptidase Ix Dpp Ix Dipeptidyl Peptidase 9 DPLP9 |
| ENSDARG00000052625 | ENSFM00670001235580 | Peptidyl Prolyl Cis Trans Isomerase FKBP1B Ppiase FKBP1B EC_5.2.1.8 12.6 Kda FK506 Binding 12.6 Kda Fkbp Fkbp 12 6 FK506 Binding 1B Fkbp 1B Immunophilin FKBP12 6 Rotamase |
| ENSDARG00000052642 | ENSFM00500000276365 | Shisa Precursor |
| ENSDARG00000052644 | ENSFM00610000965197 | Unknown |
| ENSDARG00000052644 | ENSFM00400000131811 | Carbonic Anhydrase Related 11 Precursor Ca Rp Xi Ca Xi Carp Xi |
| ENSDARG00000052673 | ENSFM00760001714685 | Septin 7 CDC10 Homolog |
| ENSDARG00000052674 | ENSFM00770001847535 | Casein Kinase I Alpha Cki Alpha EC_2.7.11.1 CK1 |
| ENSDARG00000052680 | ENSFM00500000313233 | Si:Dkey 182G1 2 |
| ENSDARG00000052702 | ENSFM00250000000112 | Calpain Subunit Calcium Activated Neutral Proteinase Canp Calpain Subunit |
| ENSDARG00000052703 | ENSFM00600000921602 | Cdgsh Iron Sulfur Domain Containing 2 |
| ENSDARG00000052712 | ENSFM00250000003768 | Succinyl Coa Ligase Subunit Alpha Mitochondrial Precursor EC_6.2.1.4 EC_6.2.1.- 5 Succinyl Coa Synthetase Subunit Alpha Scs Alpha |
| ENSDARG00000052713 | ENSFM00400000131776 | Leucine Rich Repeat Transmembrane Neuronal Precursor |
| ENSDARG00000052748 | ENSFM00250000000112 | Calpain Subunit Calcium Activated Neutral Proteinase Canp Calpain Subunit |
| ENSDARG00000052765 | ENSFM00720001492020 | Glutamate Receptor Precursor Glur Ampa Selective Glutamate Receptor Glur Glutamate Receptor Ionotropic Ampa |
| ENSDARG00000052782 | ENSFM00400000131715 | Glycine Receptor Subunit Precursor Glycine Receptor Kda Subunit |
| ENSDARG00000052840 | ENSFM00250000005730 | Nadh Dehydrogenase Iron Sulfur 4 Mitochondrial Precursor Complex I 18 Kda Ci 18 Kda Nadh Ubiquinone Oxidoreductase 18 Kda Subunit |
| ENSDARG00000052856 | ENSFM00400000131768 | Kh Domain Containing Rna Binding Signal Transduction Associated SAM68 Mammalian Slm |
| ENSDARG00000052859 | ENSFM00500000270459 | Calcineurin B Homologous 1 Calcineurin B Calcium Binding Chp Calcium Binding P22 Ef Hand Calcium Binding Domain Containing P22 |
| ENSDARG00000052866 | ENSFM00250000003199 | Huntingtin Huntington Disease Homolog Hd Homolog |
| ENSDARG00000052928 | ENSFM00750001632347 | Adp Ribosylation Factor |
| ENSDARG00000052950 | ENSFM00250000001623 | Rho Gtpase Activating 21 Rho Type Gtpase Activating 21 |
| ENSDARG00000052957 | ENSFM00740001589144 | 1 Phosphatidylinositol 4 5 Bisphosphate Phosphodiesterase Delta EC_3.1.4.11 Phosphoinositide Phospholipase C Delta Phospholipase C Delta Plc Delta |
| ENSDARG00000053003 | ENSFM00250000002791 | Solute Carrier Family 35 Member |
| ENSDARG00000053058 | ENSFM00800002003484 | 40S Ribosomal S11 |
| ENSDARG00000053066 | ENSFM00250000000193 | Tubulin Beta Chain |
| ENSDARG00000053070 | ENSFM00250000004732 | Golgi Snap Receptor Complex Member 2.27 Kda Golgi Snare Membrin |
| ENSDARG00000053113 | ENSFM00800002003595 | Secretory Phospholipase A2 Receptor Precursor PLA2 R PLA2R 180 Kda Secretory Phospholipase A2 Receptor M Type Receptor |
| ENSDARG00000053122 | ENSFM00500000270651 | Clavesin Retinaldehyde Binding 1 |
| ENSDARG00000053129 | ENSFM00670001235746 | Calcium Regulated Heat Stable 1 Calcium Regulated Heat Stable Of 24 Kda Crhsp 24 |
| ENSDARG00000053130 | ENSFM00500000386893 | Unknown |
| ENSDARG00000053138 | ENSFM00250000006095 | Leucine Rich Repeat Containing 47 |
| ENSDARG00000053194 | ENSFM00400000131930 | Pdz Domain Containing 11 |
| ENSDARG00000053196 | ENSFM00260000050608 | Serine/Threonine Kinase EC_2.7.11.1 Rapamycin 1 Target Of Rapamycin 1 |
| ENSDARG00000053205 | ENSFM00250000000441 | Liprin Alpha Tyrosine Phosphatase Receptor Type F Polypeptide Interacting Alpha Ptprf Interacting Alpha |
| ENSDARG00000053217 | ENSFM00750001646347 | Unknown |
| ENSDARG00000053217 | ENSFM00670001261071 | Cytochrome C Oxidase Subunit Viia 2 |
| ENSDARG00000053248 | ENSFM00730001521354 | Neurofilament Polypeptide Nf Kda Neurofilament Neurofilament Triplet |
| ENSDARG00000053262 | ENSFM00670001235362 | Sodium/Potassium Transporting Atpase Subunit Beta Sodium/Potassium Dependent Atpase Subunit Beta |
| ENSDARG00000053326 | ENSFM00730001521471 | Guanine Nucleotide Binding G Q Subunit Alpha Guanine Nucleotide Binding Alpha Q |
| ENSDARG00000053358 | ENSFM00600000936395 | Brain Acid Soluble 1 Homolog |
| ENSDARG00000053365 | ENSFM00670001235422 | 60S Ribosomal L31 |
| ENSDARG00000053395 | ENSFM00500000298957 | Uncharacterized |
| ENSDARG00000053395 | ENSFM00670001272366 | Unknown |
| ENSDARG00000053446 | ENSFM00670001235917 | Activated Rna Polymerase Ii Transcriptional Coactivator P15 Positive Cofactor 4 PC4 SUB1 Homolog P14 |
| ENSDARG00000053453 | ENSFM00730001521911 | Maguk P55 Subfamily Member 2 Discs Large Homolog 2 MPP2 |
| ENSDARG00000053454 | ENSFM00390000126303 | Contactin Precursor Brain Derived Immunoglobulin Superfamily Big |
| ENSDARG00000053456 | ENSFM00630001071936 | Unknown |
| ENSDARG00000053456 | ENSFM00780001898348 | Glycerol Kinase Gk Glycerokinase EC_2.7.1.30 Atp:Glycerol 3 Phosphotransferase Glycerol Kinase Testis Specific |
| ENSDARG00000053457 | ENSFM00250000003000 | 60S Ribosomal L23 |
| ENSDARG00000053467 | ENSFM00250000001372 | Gtp Binding |
| ENSDARG00000053468 | ENSFM00390000126345 | Disintegrin And Metalloproteinase Domain Containing 10 Precursor Adam 10 EC_3.4.24.81 Kuzbanian Homolog Mammalian Disintegrin Metalloprotease CD156C Antigen |
| ENSDARG00000053474 | ENSFM00250000006342 | Tudor And Kh Domain Containing Tudor Domain Containing 2 |
| ENSDARG00000053485 | ENSFM00410000138519 | Methylmalonate Semialdehyde Dehydrogenase EC_1.2.1.18 EC_1.2.1.- 27 Aldehyde Dehydrogenase Family 6 Member A1 |
| ENSDARG00000053493 | ENSFM00730001521185 | Dehydrogenase Aldehyde Dehydrogenase Family 1 Member Dehydrogenase |
| ENSDARG00000053517 | ENSFM00730001522226 | Echinoderm Microtubule Associated 6 Emap 6 Echinoderm Microtubule Associated 5 |
| ENSDARG00000053527 | ENSFM00500000272475 | Sorting Nexin 24 |
| ENSDARG00000053542 | ENSFM00260000050545 | Btb/Poz Domain Containing |
| ENSDARG00000053544 | ENSFM00360000109870 | Unknown |
| ENSDARG00000053547 | ENSFM00250000001480 | Janus Kinase And Microtubule Interacting 1 Multiple Alpha Helices And Rna Linker 1 Marlin 1 |
| ENSDARG00000053609 | ENSFM00250000005901 | Parkinson Disease 7 Domain Containing 1 Precursor |
| ENSDARG00000053617 | ENSFM00250000000111 | Calcium/Calmodulin Dependent Kinase Type Ii Subunit Cam Kinase Ii Subunit Camk Ii Subunit EC_2.7.11.17 |
| ENSDARG00000053652 | ENSFM00500000273764 | Nadh Dehydrogenase Ubiquinone Complex I Assembly Factor 6 Precursor |
| ENSDARG00000053656 | ENSFM00500000270136 | Acyl Thioesterase 1 Apt 1 EC_3.1.2.- Lysophospholipase 1 Lysophospholipase I Lpl I Lysopla I |
| ENSDARG00000053665 | ENSFM00730001521448 | Gamma Aminobutyric Acid Receptor Subunit Gamma Precursor Gaba A Receptor Subunit Gamma |
| ENSDARG00000053668 | ENSFM00250000000734 | Cohesin Subunit Sa SCC3 Homolog Stromal Antigen |
| ENSDARG00000053753 | ENSFM00250000003442 | Mitochondrial Fission Factor |
| ENSDARG00000053853 | ENSFM00250000000477 | Solute Carrier Family 13 Member Na + /Dicarboxylate Cotransporter Nadc |
| ENSDARG00000053956 | ENSFM00740001589095 | Spectrin Beta Chain Erythrocytic Beta Spectrin |
| ENSDARG00000053979 | ENSFM00600000921262 | Charged Multivesicular Body 2A Chromatin Modifying 2A CHMP2A |
| ENSDARG00000054007 | ENSFM00670001235869 | Phosphatase Inhibitor Phosphatase 1 Regulatory Subunit 2 Pseudogene |
| ENSDARG00000054032 | ENSFM00500000272409 | Ras Related Rab 24 |
| ENSDARG00000054172 | ENSFM00670001235707 | Membrane Associated Progesterone Receptor Component |
| ENSDARG00000054177 | ENSFM00250000001368 | G Coupled Receptor 124 Precursor Tumor Endothelial Marker 5 |
| ENSDARG00000054186 | ENSFM00760001714671 | Neuronal Calcium Sensor 1 Ncs 1 |
| ENSDARG00000054191 | ENSFM00790001962976 | Phosphoglycerate Kinase EC_2.7.2.3 |
| ENSDARG00000054208 | ENSFM00450000242164 | Phosphorylase B Kinase Gamma Catalytic Chain Skeletal Muscle/Heart EC_2.7.11.19 Phosphorylase Kinase Subunit Gamma 1 Serine/Threonine Kinase PHKG1 EC_2.7.11.- 1 EC_2.7.-.- 11 26 |
| ENSDARG00000054300 | ENSFM00250000006111 | Dehydrogenase/Reductase Sdr Family Member 1 EC_1.1.-.- |
| ENSDARG00000054319 | ENSFM00250000002519 | Succinyl Coa:3 Ketoacid Coenzyme A Transferase 1 Mitochondrial Precursor EC_2.8.3.5 3 Oxoacid Coa Transferase 1 Somatic Type Succinyl Coa:3 Oxoacid Coa Transferase Scot S |
| ENSDARG00000054320 | ENSFM00810002040615 | Ap 2 Complex Subunit Alpha 2.100 Kda Coated Vesicle C Adaptor Complex Ap 2 Subunit Alpha 2 Adaptor Related Complex 2 Subunit Alpha 2 Alpha Adaptin C ALPHA2 Adaptin Clathrin Assembly Complex 2 Alpha C Large Chain Plasma Membrane Adaptor HA2/AP2 Adaptin Alp |
| ENSDARG00000054337 | ENSFM00810002040662 | Ap 1 Complex Subunit Gamma 1 Adaptor Complex Ap 1 Subunit Gamma 1 Adaptor Related Complex 1 Subunit Gamma 1 Clathrin Assembly Complex 1 Gamma 1 Large Chain Gamma Adaptin GAMMA1 Adaptin Golgi Adaptor HA1/AP1 Adaptin Subunit Gamma 1 |
| ENSDARG00000054355 | ENSFM00500000271100 | DDB1 And CUL4 Associated Factor 7 Wd Repeat Containing 68 Wd Repeat Containing AN11 Homolog |
| ENSDARG00000054362 | ENSFM00250000004158 | Coiled Coil Domain Containing 47 Precursor |
| ENSDARG00000054400 | ENSFM00610000971754 | Unknown |
| ENSDARG00000054400 | ENSFM00450000242169 | Methylcrotonoyl Coa Carboxylase Beta Chain Mitochondrial Precursor Mccase Subunit Beta EC_6.4.1.4 3 Methylcrotonyl Coa Carboxylase 2 3 Methylcrotonyl Coa Carboxylase Non Biotin Containing Subunit 3 Methylcrotonyl Coa:Carbon Dioxide Ligase Subunit Beta |
| ENSDARG00000054420 | ENSFM00250000000935 | Retinoid Isomerohydrolase EC_3.1.1.64 All Trans Retinyl Palmitate Hydrolase Retinal Pigment Epithelium Specific 65 Kda Retinol Isomerase |
| ENSDARG00000054454 | ENSFM00730001521142 | Ephrin Type A Receptor Precursor EC_2.7.10.1 Kinase |
| ENSDARG00000054456 | ENSFM00740001589331 | Cap Gly Domain Containing Linker 3 Cytoplasmic Linker 170 Related 59 Kda Clip 170 Related 59 Kda Clipr 59 |
| ENSDARG00000054458 | ENSFM00800002014386 | Unknown |
| ENSDARG00000054458 | ENSFM00800002017278 | Unknown |
| ENSDARG00000054458 | ENSFM00250000002303 | Sarcolemmal Membrane Associated Sarcolemmal Associated |
| ENSDARG00000054530 | ENSFM00440000236919 | Arginine Trna Ligase Cytoplasmic EC_6.1.1.19 Arginyl Trna Synthetase Argrs |
| ENSDARG00000054578 | ENSFM00250000007414 | Adp Ribosylation Factor 6 Interacting 1 Arl 6 Interacting 1 Aip 1 |
| ENSDARG00000054610 | ENSFM00730001521292 | Coronin |
| ENSDARG00000054696 | ENSFM00800002003556 | Proteasome Subunit Beta Type 4 Precursor EC_3.4.25.1 Macropain Beta Chain Multicatalytic Endopeptidase Complex Beta Chain Proteasome Beta Chain Proteasome Chain 3 |
| ENSDARG00000054746 | ENSFM00800002003477 | Udp Glucose:Glycoprotein Glucosyltransferase 1 Precursor UGT1 EC_2.4.1.- Udp Glc:Glycoprotein Glucosyltransferase Udp Glucose Ceramide Glucosyltransferase 1 |
| ENSDARG00000054748 | ENSFM00250000005024 | Cue Domain Containing 1 |
| ENSDARG00000054793 | ENSFM00250000006413 | Er Membrane Complex Subunit 10 Precursor |
| ENSDARG00000054818 | ENSFM00500000270098 | 60S Ribosomal L32 |
| ENSDARG00000054833 | ENSFM00250000002033 | Nucleobindin Precursor |
| ENSDARG00000054849 | ENSFM00730001521772 | Branched Chain Amino Acid Aminotransferase EC_2.6.1.42 |
| ENSDARG00000054858 | ENSFM00250000001053 | Apoptosis Stimulating Of P53 2 Tumor Suppressor P53 Binding 2 53BP2 P53 Binding 2 P53BP2 |
| ENSDARG00000054864 | ENSFM00250000000617 | Amyloid Beta A4 Precursor Abpp App Alzheimer Disease Amyloid A4 Homolog |
| ENSDARG00000054867 | ENSFM00790001963058 | 3 Hydroxyisobutyryl Coa Hydrolase Mitochondrial Precursor EC_3.1.2.4 3 Hydroxyisobutyryl Coenzyme A Hydrolase Hib Coa Hydrolase Hibyl Coa H |
| ENSDARG00000054890 | ENSFM00250000006713 | Rpe Retinal G Coupled Receptor |
| ENSDARG00000054907 | ENSFM00670001237705 | Cytochrome C Oxidase Subunit Viia Related Protein Mitochondrial |
| ENSDARG00000054931 | ENSFM00760001714676 | Serine/Threonine Phosphatase 2A 56 Kda Regulatory Subunit Epsilon PP2A B Subunit B' Epsilon PP2A B Subunit B56 Epsilon PP2A B Subunit PR61 Epsilon PP2A B Subunit R5 Epsilon |
| ENSDARG00000054942 | ENSFM00670001237282 | Beta Galactoside Binding Lectin 14 Kda Lectin Electrolectin |
| ENSDARG00000055052 | ENSFM00800002005222 | Uncharacterized |
| ENSDARG00000055052 | ENSFM00260000050441 | Microtubule Associated Tau Neurofibrillary Tangle Paired Helical Filament Tau Phf Tau |
| ENSDARG00000055053 | ENSFM00250000000536 | Inter Alpha Trypsin Inhibitor Heavy Chain Precursor Iti Heavy Chain Iti Inter Alpha Inhibitor Heavy Chain |
| ENSDARG00000055064 | ENSFM00670001235781 | Peroxiredoxin 5 Mitochondrial Precursor EC_1.11.1.15 Antioxidant Enzyme B166 AOEB166 Plp Peroxiredoxin V Prx V Peroxisomal Antioxidant Enzyme Thioredoxin Peroxidase PMP20 Thioredoxin Reductase |
| ENSDARG00000055075 | ENSFM00730001543869 | Unknown |
| ENSDARG00000055075 | ENSFM00250000001422 | Supervillin Archvillin P205/P250 |
| ENSDARG00000055080 | ENSFM00350000105447 | Rna Binding Rna Binding Motif Rna Binding Motif |
| ENSDARG00000055092 | ENSFM00750001632390 | Nadph Cytochrome P450 Reductase Cpr P450R EC_1.6.2.4 |
| ENSDARG00000055101 | ENSFM00250000002797 | Heme Oxygenase Ho EC_1.14.99.3 |
| ENSDARG00000055108 | ENSFM00250000004887 | Glycerophosphodiester Phosphodiesterase 1 EC_3.1.4.44 Membrane Interacting Of RGS16 |
| ENSDARG00000055120 | ENSFM00290000065537 | Cathepsin B Precursor EC_3.4.22.1 |
| ENSDARG00000055133 | ENSFM00250000003245 | Centromere F Precursor Cenp F Ah Antigen Kinetochore Cenpf Mitosin |
| ENSDARG00000055154 | ENSFM00710001441664 | Sodium/Calcium Exchanger 1 Precursor Na + /Ca 2+ Exchange 1 Solute Carrier Family 8 Member 1 |
| ENSDARG00000055223 | ENSFM00250000000459 | Actin Binding Lim Ablim Actin Binding Lim Family Member |
| ENSDARG00000055225 | ENSFM00500000269778 | Dedicator Of Cytokinesis |
| ENSDARG00000055255 | ENSFM00500000269931 | Membrane Associated Phosphatidylinositol Transfer Phosphatidylinositol Transfer Protein Membrane Associated Pitpnm PYK2 N Terminal Domain Interacting Receptor Nir |
| ENSDARG00000055288 | ENSFM00500000270508 | Slowmo Homolog |
| ENSDARG00000055291 | ENSFM00630001071973 | Unknown |
| ENSDARG00000055291 | ENSFM00500000270602 | Ras Related Rab 18 |
| ENSDARG00000055302 | ENSFM00720001492054 | Glutamate Receptor Ionotropic Delta Precursor Glur Delta Subunit |
| ENSDARG00000055377 | ENSFM00770001847629 | Guanine Nucleotide Binding Subunit Beta 5 GBETA5 Transducin Beta Chain 5 |
| ENSDARG00000055455 | ENSFM00250000000909 | Myelin Proteolipid Plp Lipophilin |
| ENSDARG00000055459 | ENSFM00250000000718 | Disks Large Associated Dap Psd 95/SAP90 Binding SAP90/Psd 95 Associated |
| ENSDARG00000055510 | ENSFM00500000270081 | Yippee |
| ENSDARG00000055525 | ENSFM00500000272380 | Galectin Related Lectin Galactoside Binding |
| ENSDARG00000055543 | ENSFM00250000000617 | Amyloid Beta A4 Precursor Abpp App Alzheimer Disease Amyloid A4 Homolog |
| ENSDARG00000055565 | ENSFM00250000000524 | Voltage Dependent L Type Calcium Channel Subunit Beta Calcium Channel Voltage Dependent Subunit Beta |
| ENSDARG00000055569 | ENSFM00250000008610 | GH3 Domain Containing Precursor |
| ENSDARG00000055591 | ENSFM00250000006230 | Peroxisomal Sarcosine Oxidase Pso EC_1.5.3.1 EC_1.5.3.- 7 L Pipecolate Oxidase L Pipecolic Acid Oxidase |
| ENSDARG00000055607 | ENSFM00740001589340 | Cingulin |
| ENSDARG00000055610 | ENSFM00250000004215 | Exocyst Complex Component 2 Exocyst Complex Component SEC5 |
| ENSDARG00000055620 | ENSFM00500000270065 | Very Long Chain Specific Acyl Coa Dehydrogenase Mitochondrial Precursor EC_1.3.8.9 |
| ENSDARG00000055635 | ENSFM00250000002274 | Adapter Molecule Crk Proto Oncogene C Crk P38 |
| ENSDARG00000055639 | ENSFM00730001521813 | Ruvb 2 EC_3.6.4.12 |
| ENSDARG00000055648 | ENSFM00730001521968 | Carboxypeptidase D Precursor EC_3.4.17.22 Metallocarboxypeptidase D GP180 |
| ENSDARG00000055678 | ENSFM00250000003924 | A Kinase Anchor 12 Akap 12 |
| ENSDARG00000055708 | ENSFM00500000444882 | Translocase Of Inner Mitochondrial Membrane 8 Homolog B Yeast |
| ENSDARG00000055759 | ENSFM00250000003493 | Ef Hand Domain Containing Swiprosin |
| ENSDARG00000055791 | ENSFM00250000004066 | 4F2 Cell Surface Antigen Heavy Chain 4F2HC Solute Carrier Family 3 Member 2 CD98 Antigen |
| ENSDARG00000055809 | ENSFM00250000000813 | Vitellogenin Precursor |
| ENSDARG00000055825 | ENSFM00560000770988 | Cadherin Egf Lag Seven Pass G Type Receptor Multiple Epidermal Growth Factor Domains Multiple Egf Domains |
| ENSDARG00000055843 | ENSFM00260000050333 | Cadherin Precursor |
| ENSDARG00000055855 | ENSFM00260000050384 | Potassium Voltage Gated Channel Subfamily C Member Voltage Gated Potassium Channel KV3 |
| ENSDARG00000055857 | ENSFM00250000001352 | Dopey |
| ENSDARG00000055874 | ENSFM00730001521835 | Carboxypeptidase E Precursor Cpe EC_3.4.17.10 Carboxypeptidase H Cph Enkephalin Convertase Prohormone Processing Carboxypeptidase |
| ENSDARG00000055899 | ENSFM00250000008930 | Lzic Leucine Zipper And CTNNBIP1 Domain Containing Leucine Zipper And Icat Homologous Domain Containing |
| ENSDARG00000055901 | ENSFM00250000001178 | Metalloreductase EC_1.16.1.- Six Transmembrane Epithelial Antigen Of Prostate |
| ENSDARG00000055976 | ENSFM00500000271562 | Peroxisomal Trans 2 Enoyl Coa Reductase Terp EC_1.3.1.38 2 4 Dienoyl Coa Reductase |
| ENSDARG00000055996 | ENSFM00800002003452 | 40S Ribosomal S8 |
| ENSDARG00000056001 | ENSFM00250000000754 | Furry |
| ENSDARG00000056036 | ENSFM00250000001101 | Syntaxin Binding 1 Unc 18 Homolog 1 UNC18 1 Unc 18 Homolog A Unc 18A |
| ENSDARG00000056075 | ENSFM00780001898918 | Membrane Bound Complement Regulatory |
| ENSDARG00000056078 | ENSFM00390000126424 | Raftlin 2 Raft Linking 2 |
| ENSDARG00000056080 | ENSFM00250000002946 | RAP1 Gtpase Gdp Dissociation Stimulator 1 Exchange Factor Smggds Smg Gds Smg P21 Stimulatory Gdp/Gtp Exchange |
| ENSDARG00000056085 | ENSFM00250000004752 | Mob Phocein Class Ii MMOB1 MOB1 Homolog 3 MOB3 Mps One Binder Kinase Activator 3 Preimplantation 3 |
| ENSDARG00000056089 | ENSFM00250000001959 | Nuclear Receptor Binding |
| ENSDARG00000056090 | ENSFM00250000001955 | F Actin Capping Subunit Alpha 2 Capz Alpha 2 |
| ENSDARG00000056101 | ENSFM00300000079128 | Potassium Voltage Gated Channel Subfamily D Member Voltage Gated Potassium Channel Subunit KV4 |
| ENSDARG00000056102 | ENSFM00250000004210 | Udp Glucuronic Acid Decarboxylase 1 EC_4.1.1.35 Udp Glucuronate Decarboxylase 1 Uxs 1 |
| ENSDARG00000056108 | ENSFM00500000306879 | Cytochrome C Oxidase Subunit NDUFA4 |
| ENSDARG00000056112 | ENSFM00750001632433 | 1 Phosphatidylinositol 3 Phosphate 5 Kinase Phosphatidylinositol 3 Phosphate 5 Kinase EC_2.7.1.150 Fyve Finger Containing Phosphoinositide Kinase Pikfyve Phosphatidylinositol 3 Phosphate 5 Kinase Type Iii Pipkin Iii Type Iii Pip Kinase |
| ENSDARG00000056119 | ENSFM00250000001516 | Elongation Factor 1 Gamma Ef 1 Gamma Eef 1B Gamma |
| ENSDARG00000056121 | ENSFM00280000058758 | SERAC1 Serine Active Site Containing 1 |
| ENSDARG00000056122 | ENSFM00800002003449 | Rab Gdp Dissociation Inhibitor Alpha Rab Gdi Alpha Guanosine Diphosphate Dissociation Inhibitor 1 Gdi 1 |
| ENSDARG00000056160 | ENSFM00760001714648 | 60 Kda Heat Shock Protein Mitochondrial 60 Kda Chaperonin Chaperonin 60 CPN60 Heat Shock 60 Hsp 60 HSP60 Mitochondrial Matrix P1 |
| ENSDARG00000056163 | ENSFM00500000271232 | Solute Carrier Family 25 Member 51 Mitochondrial Carrier Triple Repeat 1 |
| ENSDARG00000056165 | ENSFM00500000272346 | Uncharacterized |
| ENSDARG00000056167 | ENSFM00670001235755 | 10 Kda Heat Shock Protein Mitochondrial HSP10.10 Kda Chaperonin Chaperonin 10 CPN10 |
| ENSDARG00000056181 | ENSFM00250000000779 | Neural Cell Adhesion Molecule 1 Precursor N Cam 1 Ncam 1 |
| ENSDARG00000056206 | ENSFM00250000000111 | Calcium/Calmodulin Dependent Kinase Type Ii Subunit Cam Kinase Ii Subunit Camk Ii Subunit EC_2.7.11.17 |
| ENSDARG00000056218 | ENSFM00260000050335 | Kinase EC_2.7.11.1 Kinase Mapk/Erk Kinase Kinase Kinase Mek Kinase Kinase Mekkk Kinase |
| ENSDARG00000056250 | ENSFM00250000000681 | Adducin |
| ENSDARG00000056252 | ENSFM00500000271702 | Sortilin Precursor Neurotensin Receptor 3 NTR3 |
| ENSDARG00000056262 | ENSFM00500000272943 | Solute Carrier Family 35 Member G2 Transmembrane 22 |
| ENSDARG00000056314 | ENSFM00730001521356 | Precursor |
| ENSDARG00000056328 | ENSFM00250000000940 | Mitochondrial Rho Gtpase 2 Miro 2 EC_3.6.5.- Ras Homolog Gene Family Member T2 |
| ENSDARG00000056331 | ENSFM00740001589092 | Adenosylhomocysteinase Adohcyase EC_3.3.1.1 S Adenosyl L Homocysteine Hydrolase 2 |
| ENSDARG00000056347 | ENSFM00730001521441 | Ras Related Rab 3 |
| ENSDARG00000056379 | ENSFM00670001237454 | C Type Lectin |
| ENSDARG00000056394 | ENSFM00250000001136 | TOM1 2 Target Of Myb 2 |
| ENSDARG00000056479 | ENSFM00730001521238 | Ras Related Rab |
| ENSDARG00000056481 | ENSFM00500000270300 | Synaptic Vesicle Membrane Vat 1 Ec 1 |
| ENSDARG00000056510 | ENSFM00250000004030 | Glutathione S Transferase Kappa 1 EC_2.5.1.18 Gst 13 13 Gst Class Kappa GSTK1 1 Glutathione S Transferase Subunit 13 |
| ENSDARG00000056557 | ENSFM00250000004388 | Coatomer Subunit Beta Beta Coat Beta Cop |
| ENSDARG00000056559 | ENSFM00250000003646 | Vacuolar Fusion CCZ1 Homolog |
| ENSDARG00000056566 | ENSFM00250000001762 | N Terminal Ef Hand Calcium Binding Ef Hand Calcium Binding Neuronal Calcium Binding |
| ENSDARG00000056583 | ENSFM00670001236132 | Nadh Dehydrogenase Iron Sulfur 6 Mitochondrial Precursor Complex I 13KD A Ci 13KD A Nadh Ubiquinone Oxidoreductase 13 Kda A Subunit |
| ENSDARG00000056603 | ENSFM00250000000478 | Guanine Nucleotide Exchange Factor Dbs Dbl'S Big Sister MCF2 Transforming Sequence |
| ENSDARG00000056609 | ENSFM00790001963005 | Atpase Family Aaa Domain Containing 1 EC_3.6.1.3 |
| ENSDARG00000056639 | ENSFM00260000050454 | Leucine Rich Repeat Containing |
| ENSDARG00000056643 | ENSFM00400000131714 | Solute Carrier Family 22 Member Organic Anion Transporter |
| ENSDARG00000056648 | ENSFM00780001898505 | Exostosin 2 EC_2.4.1.224 EC_2.4.1.- 225 Glucuronosyl N Acetylglucosaminyl Proteoglycan/N Acetylglucosaminyl Proteoglycan 4 Alpha N Acetylglucosaminyltransferase Multiple Exostoses 2 |
| ENSDARG00000056651 | ENSFM00500000269996 | Gdnf Family Receptor Alpha Precursor Gdnf Receptor Alpha Gdnfr Alpha Gfr Alpha Tgf Beta Related Neurotrophic Factor Receptor |
| ENSDARG00000056664 | ENSFM00250000002848 | Arfaptin Adp Ribosylation Factor Interacting |
| ENSDARG00000056672 | ENSFM00800002003665 | Atp Binding Cassette Sub Family B Member 8 Mitochondrial Precursor |
| ENSDARG00000056683 | ENSFM00730001521798 | Negative Regulator Of The Pho System Serine/Threonine Kinase PHO85 |
| ENSDARG00000056690 | ENSFM00730001521553 | Myotubularin Phosphatidylinositol 3 5 Bisphosphate 3 Phosphatase EC_3.1.3.95 |
| ENSDARG00000056722 | ENSFM00670001242505 | CD99 Antigen 2 Precursor CD99 Antigen |
| ENSDARG00000056725 | ENSFM00600000944062 | Unknown |
| ENSDARG00000056725 | ENSFM00500000269645 | High Mobility Group B1 High Mobility Group 1 Hmg 1 |
| ENSDARG00000056726 | ENSFM00250000004491 | Methyltransferase Precursor EC_2.1.1.- |
| ENSDARG00000056742 | ENSFM00250000000643 | Dihydropyrimidinase Related Drp Collapsin Response Mediator Crmp |
| ENSDARG00000056745 | ENSFM00250000001762 | N Terminal Ef Hand Calcium Binding Ef Hand Calcium Binding Neuronal Calcium Binding |
| ENSDARG00000056753 | ENSFM00760001714751 | Dynactin Subunit 1.150 Kda Dynein Associated Polypeptide Dap 150 Dp 150 Glued |
| ENSDARG00000056774 | ENSFM00250000003445 | Prosaposin Receptor Gp Precursor Endothelin B Receptor 2 Etbr Lp 2 G Coupled Receptor 37 1 |
| ENSDARG00000056797 | ENSFM00250000000829 | Serine/Threonine Phosphatase 2A 55 Kda Regulatory Subunit B PP2A Subunit B B55 PP2A Subunit B PR55 PP2A Subunit B R2 PP2A Subunit B |
| ENSDARG00000056803 | ENSFM00500000269841 | Ap 1 Complex Subunit Sigma Adaptor Complex Ap 1 Subunit Sigma Adaptor Related Complex 1 Subunit Sigma Clathrin Assembly Complex 1 Sigma Small Chain Golgi Adaptor HA1/AP1 Adaptin Sigma Subunit Sigma Subunit Of Ap 1 Clathrin Sigma Adaptin Adaptin |
| ENSDARG00000056831 | ENSFM00500000301701 | Unknown |
| ENSDARG00000056831 | ENSFM00800002013988 | Unknown |
| ENSDARG00000056833 | ENSFM00250000002557 | Synaptic Vesicle 2 Related SV2 Related |
| ENSDARG00000056874 | ENSFM00500000272583 | Lysozyme G EC_3.2.1.17 1 4 Beta N Acetylmuramidase |
| ENSDARG00000056877 | ENSFM00670001235687 | Vesicle Associated Membrane 2 Vamp 2 Synaptobrevin 2 |
| ENSDARG00000056892 | ENSFM00630001071998 | Unknown |
| ENSDARG00000056892 | ENSFM00730001521911 | Maguk P55 Subfamily Member 2 Discs Large Homolog 2 MPP2 |
| ENSDARG00000056910 | ENSFM00390000126295 | Neural Cell Adhesion Molecule L1 L1 L1 |
| ENSDARG00000056934 | ENSFM00250000007963 | Hepatocyte Cell Adhesion Molecule Precursor Hepacam |
| ENSDARG00000056978 | ENSFM00740001589271 | LETM1 And Ef Hand Domain Containing 1 Mitochondrial Precursor Leucine Zipper Ef Hand Containing Transmembrane 1 |
| ENSDARG00000056998 | ENSFM00250000000717 | Kin Of Irre Precursor Kin Of Irregular Chiasm Nephrin |
| ENSDARG00000057000 | ENSFM00800002003895 | Cam Kinase Vesicle Associated |
| ENSDARG00000057007 | ENSFM00250000001273 | C Terminal Binding |
| ENSDARG00000057011 | ENSFM00250000004763 | Dolichol Phosphate Mannosyltransferase Subunit 1 EC_2.4.1.83 Dolichol Phosphate Mannose Synthase Subunit 1 Dpm Synthase Subunit 1 Dolichyl Phosphate Beta D Mannosyltransferase Subunit 1 Mannose P Dolichol Synthase Subunit 1 Mpd Synthase Subunit 1 |
| ENSDARG00000057013 | ENSFM00250000001126 | Cell Adhesion Molecule Precursor Immunoglobulin Superfamily Member Nectin Necl Synaptic Cell Adhesion Molecule |
| ENSDARG00000057026 | ENSFM00250000002338 | Gtp Binding Nuclear Ran Gtpase Ran Ras Related Nuclear |
| ENSDARG00000057075 | ENSFM00250000006602 | Ragulator Complex LAMTOR3 Late Endosomal/Lysosomal Adaptor And Mapk And Mtor Activator 3 Mitogen Activated Kinase Scaffold 1 |
| ENSDARG00000057125 | ENSFM00740001589239 | Phospholipase D2 Pld 2 EC_3.1.4.4 Choline Phosphatase 2 Phosphatidylcholine Hydrolyzing Phospholipase D2 |
| ENSDARG00000057128 | ENSFM00760001714774 | Trifunctional Enzyme Subunit Alpha Mitochondrial Precursor 78 Kda Gastrin Binding Tp Alpha |
| ENSDARG00000057151 | ENSFM00250000003032 | Prolyl Endopeptidase Pe EC_3.4.21.26 Post Proline Cleaving Enzyme |
| ENSDARG00000057159 | ENSFM00440000236974 | Ankyrin Repeat Domain Containing 29 |
| ENSDARG00000057169 | ENSFM00740001589079 | Atp Binding Cassette Sub Family A Member Atp Binding Cassette Transporter Atp Binding Cassette |
| ENSDARG00000057239 | ENSFM00800002003538 | Tryptophan 5 Hydroxylase EC_1.14.16.4 Tryptophan 5 Monooxygenase |
| ENSDARG00000057255 | ENSFM00500000270724 | Er Membrane Complex Subunit 1 Precursor |
| ENSDARG00000057262 | ENSFM00720001492028 | Sterol 26 Hydroxylase Mitochondrial Precursor EC_1.14.13.15 5 Beta Cholestane 3 Alpha 7 Alpha 12 Alpha Triol 27 Hydroxylase Cytochrome P 450C27/25 Cytochrome P450 27 Sterol 27 Hydroxylase Vitamin D 3 25 Hydroxylase |
| ENSDARG00000057272 | ENSFM00250000002884 | Zinc Transporter 9 Znt 9 Solute Carrier Family 30 Member 9 |
| ENSDARG00000057286 | ENSFM00250000001889 | Differentially Expressed In Fdcp 6 |
| ENSDARG00000057287 | ENSFM00740001589430 | Graves Disease Carrier Gdc Mitochondrial Solute Carrier Homolog Solute Carrier Family 25 Member 16 |
| ENSDARG00000057303 | ENSFM00800002003793 | N Acetylgalactosaminyltransferase 7 EC_2.4.1.- Polypeptide Galnac Transferase 7 Galnac T7 Pp Gantase 7 Udp Acetylgalactosaminyltransferase 7 Udp Galnac:Polypeptide N Acetylgalactosaminyltransferase 7 |
| ENSDARG00000057368 | ENSFM00800002003452 | 40S Ribosomal S8 |
| ENSDARG00000057374 | ENSFM00730001521561 | Sideroflexin |
| ENSDARG00000057378 | ENSFM00610000963233 | Unknown |
| ENSDARG00000057378 | ENSFM00250000005616 | Redox Regulatory FAM213A Peroxiredoxin 2 Activated In M Csf Stimulated Monocytes Pamm |
| ENSDARG00000057394 | ENSFM00500000270315 | Ras Related Rab 33 |
| ENSDARG00000057414 | ENSFM00790001963030 | Prohibitin |
| ENSDARG00000057456 | ENSFM00310000089007 | Serine/Threonine Phosphatase 2B Catalytic Subunit EC_3.1.3.16 Cam Prp Catalytic Subunit Calmodulin Dependent Calcineurin A Subunit |
| ENSDARG00000057465 | ENSFM00780001898365 | Glutamine Fructose 6 Phosphate Aminotransferase 2 EC_2.6.1.16 D Fructose 6 Phosphate Amidotransferase 2 Glutamine:Fructose 6 Phosphate Amidotransferase 2 Gfat 2 GFAT2 Hexosephosphate Aminotransferase 2 |
| ENSDARG00000057556 | ENSFM00250000001117 | 60S Ribosomal L17 |
| ENSDARG00000057568 | ENSFM00730001521354 | Neurofilament Polypeptide Nf Kda Neurofilament Neurofilament Triplet |
| ENSDARG00000057598 | ENSFM00500000304120 | S100 Calcium Binding Protein Beta Neural |
| ENSDARG00000057624 | ENSFM00250000004521 | COP9 Signalosome Complex Subunit 5 EC_3.4.-.- |
| ENSDARG00000057661 | ENSFM00250000000488 | Fructose Bisphosphate Aldolase EC_4.1.2.13 Type Aldolase |
| ENSDARG00000057665 | ENSFM00600000926850 | Prion |
| ENSDARG00000057676 | ENSFM00250000003047 | Golgin Subfamily A Member 7 |
| ENSDARG00000057698 | ENSFM00720001492011 | Precursor |
| ENSDARG00000057826 | ENSFM00250000002895 | Occludin |
| ENSDARG00000057853 | ENSFM00250000002076 | V Type Proton Atpase 16 Kda Proteolipid Subunit V Atpase 16 Kda Proteolipid Subunit Vacuolar Proton Pump 16 Kda Proteolipid Subunit |
| ENSDARG00000057855 | ENSFM00250000005086 | 28S Ribosomal S31 Mitochondrial Precursor Mrp S31 S31MT |
| ENSDARG00000057857 | ENSFM00480000262814 | Adp Ribosylation Factor Related 1 Arf Related 1 |
| ENSDARG00000057867 | ENSFM00500000270806 | Lim And SH3 Domain 1 Lasp 1 |
| ENSDARG00000057881 | ENSFM00730001521354 | Neurofilament Polypeptide Nf Kda Neurofilament Neurofilament Triplet |
| ENSDARG00000057882 | ENSFM00500000270831 | Actin Related 2/3 Complex Subunit 3 ARP2/3 Complex 21 Kda Subunit P21 Arc |
| ENSDARG00000057912 | ENSFM00250000002567 | Eukaryotic Translation Initiation Factor 1A Eif 1A Eukaryotic Translation Initiation Factor 4C Eif 4C |
| ENSDARG00000057928 | ENSFM00780001898757 | Mitotic Spindle Organizing 2 Mitotic Spindle Organizing Associated With A Ring Of Gamma Tubulin 2 |
| ENSDARG00000057995 | ENSFM00420000140675 | Adp Ribosylation Factor 15 Adp Ribosylation Factor Related 2 Arf Related 2 |
| ENSDARG00000058022 | ENSFM00500000270327 | ORM1 |
| ENSDARG00000058041 | ENSFM00250000005294 | Nadh Dehydrogenase 1 Alpha Subcomplex Subunit 8 Complex I 19KD Ci 19KD Complex I Pgiv Ci Pgiv Nadh Ubiquinone Oxidoreductase 19 Kda Subunit |
| ENSDARG00000058044 | ENSFM00730001521238 | Ras Related Rab |
| ENSDARG00000058047 | ENSFM00610000965688 | Unknown |
| ENSDARG00000058047 | ENSFM00250000004414 | Platelet Activating Factor Acetylhydrolase Ib Subunit Beta EC_3.1.1.47 Paf Acetylhydrolase 30 Kda Subunit Paf Ah 30 Kda Subunit Paf Ah Subunit Beta Pafah Subunit Beta |
| ENSDARG00000058088 | ENSFM00250000002966 | Apoptosis Inducing Factor 1 Mitochondrial Precursor EC_1.1.1.- Programmed Cell Death 8 |
| ENSDARG00000058102 | ENSFM00800002003410 | Dehydrogenase Mitochondrial Precursor |
| ENSDARG00000058114 | ENSFM00250000004572 | Inositol Monophosphatase 3 Imp 3 Impase 3 EC_3.1.3.25 EC_3.1.3.- 7 Golgi 3 Prime Phosphoadenosine 5 Prime Phosphate 3 Prime Phosphatase Golgi Resident Pap Phosphatase Gpapp Inositol Monophosphatase Domain Containing 1 Inositol 1 Or 4 Monophosphatase 3 Myo |
| ENSDARG00000058115 | ENSFM00250000000093 | Fibroblast Growth Factor Receptor Precursor Fgfr EC_2.7.10.1 |
| ENSDARG00000058117 | ENSFM00250000001228 | Synaptosomal Associated 25 Snap 25 Synaptosomal Associated 25 Kda |
| ENSDARG00000058128 | ENSFM00250000000517 | Moesin |
| ENSDARG00000058162 | ENSFM00250000002167 | Choline Phosphate Cytidylyltransferase A EC_2.7.7.15 Cct Alpha Ctp:Phosphocholine Cytidylyltransferase A Cct A Ct A Phosphorylcholine Transferase A |
| ENSDARG00000058178 | ENSFM00250000001255 | Palmitoyltransferase EC_2.3.1.225 Zinc Finger Dhhc Domain Containing Dhhc |
| ENSDARG00000058202 | ENSFM00250000005766 | Peroxisomal Membrane PEX16 Peroxin 16 Peroxisomal Biogenesis Factor 16 |
| ENSDARG00000058203 | ENSFM00790001962984 | Structural Maintenance Of Chromosomes 1A Smc 1A Smc 1A |
| ENSDARG00000058207 | ENSFM00250000000845 | Abl Interactor 1 Abelson Interactor 1 Abi 1 EPS8 SH3 Domain Binding EPS8 Binding Spectrin SH3 Domain Binding 1 |
| ENSDARG00000058208 | ENSFM00500000270824 | S Adenosylmethionine Mitochondrial Carrier Mitochondrial S Adenosylmethionine Transporter Solute Carrier Family 25 Member 26 |
| ENSDARG00000058221 | ENSFM00250000004201 | Ceramide Glucosyltransferase EC_2.4.1.80 Udp Glucose Ceramide Glucosyltransferase |
| ENSDARG00000058225 | ENSFM00250000003587 | Actin Related 2/3 Complex Subunit 4 ARP2/3 Complex 20 Kda P20 Arc |
| ENSDARG00000058297 | ENSFM00670001236643 | Mitochondrial Import Inner Membrane Translocase Subunit TIM13 |
| ENSDARG00000058302 | ENSFM00670001236078 | SH3 Domain Binding Glutamic Acid Rich |
| ENSDARG00000058348 | ENSFM00760001714632 | Gelsolin Actin Depolymerizing Factor Adf |
| ENSDARG00000058352 | ENSFM00250000003102 | Acyl Coenzyme A Thioesterase Mitochondrial Precursor Acyl Coa Thioesterase EC_3.1.2.- Mitochondrial 48 Kda Acyl Coa Thioester Hydrolase Mt ACT48 |
| ENSDARG00000058354 | ENSFM00250000003123 | Selenoprotein Precursor |
| ENSDARG00000058358 | ENSFM00740001589131 | Keratin Type Ii Cytoskeletal 8 Cytokeratin 8 Ck 8 Keratin 8 K8 Type Ii Keratin KB8 |
| ENSDARG00000058367 | ENSFM00500000272855 | Alpha/Beta Hydrolase Domain Containing 14A Abhydrolase Domain Containing 14A Ec 3 |
| ENSDARG00000058371 | ENSFM00740001589131 | Keratin Type Ii Cytoskeletal 8 Cytokeratin 8 Ck 8 Keratin 8 K8 Type Ii Keratin KB8 |
| ENSDARG00000058394 | ENSFM00500000270015 | Transgelin |
| ENSDARG00000058419 | ENSFM00250000002277 | Translational Activator GCN1 |
| ENSDARG00000058421 | ENSFM00250000002312 | Leucine Rich Repeat Lgi Family Member Precursor Leucine Rich Glioma Inactivated |
| ENSDARG00000058425 | ENSFM00750001632414 | Ras Related Rab 35 |
| ENSDARG00000058444 | ENSFM00250000002044 | Sorting Nexin |
| ENSDARG00000058451 | ENSFM00500000270597 | 60S Ribosomal L6 |
| ENSDARG00000058463 | ENSFM00670001235829 | Acyl Carrier Protein Mitochondrial Precursor Acp Nadh Ubiquinone Oxidoreductase 9.6 Kda Subunit |
| ENSDARG00000058494 | ENSFM00730001522148 | Dnaj Homolog Subfamily A Member 3 Mitochondrial Precursor Dnaj Tid 1 1 Tumorous Imaginal Discs TID56 Homolog |
| ENSDARG00000058504 | ENSFM00500000269841 | Ap 1 Complex Subunit Sigma Adaptor Complex Ap 1 Subunit Sigma Adaptor Related Complex 1 Subunit Sigma Clathrin Assembly Complex 1 Sigma Small Chain Golgi Adaptor HA1/AP1 Adaptin Sigma Subunit Sigma Subunit Of Ap 1 Clathrin Sigma Adaptin Adaptin |
| ENSDARG00000058558 | ENSFM00650001140033 | Leucine Zipper Tumor Suppressor 2 |
| ENSDARG00000058574 | ENSFM00730001521925 | Atp Binding Cassette Sub Family G Member 2 Urate Exporter CD338 Antigen |
| ENSDARG00000058601 | ENSFM00250000004188 | Ganglioside Induced Differentiation Associated 1 GDAP1 |
| ENSDARG00000058644 | ENSFM00570000851144 | Dnaj Homolog Subfamily B Member |
| ENSDARG00000058646 | ENSFM00260000050522 | Receptor Type Tyrosine Phosphatase Precursor R Ptp |
| ENSDARG00000058649 | ENSFM00400000131833 | Tripartite Motif Containing 46 Gene Y Geney Tripartite Fibronectin Type Iii And C Terminal Spry Motif |
| ENSDARG00000058734 | ENSFM00250000000757 | Thioredoxin Peroxidase Thioredoxin Dependent Peroxide Reductase |
| ENSDARG00000058736 | ENSFM00800002003422 | Gamma Aminobutyric Acid Receptor Subunit Alpha Precursor Gaba A Receptor Subunit Alpha |
| ENSDARG00000058828 | ENSFM00250000006098 | Coiled Coil Domain Containing 51 |
| ENSDARG00000058848 | ENSFM00250000001474 | Mucolipin |
| ENSDARG00000058861 | ENSFM00370000118641 | Nicalin Precursor Nicastrin |
| ENSDARG00000058868 | ENSFM00250000001676 | Adenomatous Polyposis Coli |
| ENSDARG00000058892 | ENSFM00740001589275 | Unknown |
| ENSDARG00000058966 | ENSFM00500000269664 | Retinol Dehydrogenase |
| ENSDARG00000058969 | ENSFM00250000000380 | Contactin Associated 5 Precursor Cell Recognition Molecule CASPR5 |
| ENSDARG00000058984 | ENSFM00800002003492 | Uv Excision Repair RAD23 Homolog |
| ENSDARG00000059003 | ENSFM00740001589092 | Adenosylhomocysteinase Adohcyase EC_3.3.1.1 S Adenosyl L Homocysteine Hydrolase 2 |
| ENSDARG00000059035 | ENSFM00750001632390 | Nadph Cytochrome P450 Reductase Cpr P450R EC_1.6.2.4 |
| ENSDARG00000059048 | ENSFM00500000272563 | Myelin Zero 1 Precursor |
| ENSDARG00000059067 | ENSFM00250000001012 | Catenin Beta Beta Catenin |
| ENSDARG00000059070 | ENSFM00800002003540 | Glycine Trna Ligase EC_6.1.1.14 Diadenosine Tetraphosphate Synthetase Ap 4 A Synthetase Glycyl Trna Synthetase Glyrs |
| ENSDARG00000059083 | ENSFM00810002040632 | Serine/Threonine Phosphatase 2A 56 Kda Regulatory Subunit Gamma PP2A B Subunit B' PP2A B Subunit B' Gamma PP2A B Subunit B56 Gamma PP2A B Subunit PR61 Gamma PP2A B Subunit R5 Gamma |
| ENSDARG00000059093 | ENSFM00500000269628 | Ankyrin Ank Ankyrin |
| ENSDARG00000059125 | ENSFM00790001962972 | Camp Dependent Kinase Catalytic Subunit Pka C EC_2.7.11.11 |
| ENSDARG00000059128 | ENSFM00810002042428 | Cytochrome B C1 Complex Subunit 6 |
| ENSDARG00000059166 | ENSFM00800002003814 | Solute Carrier Family 22 Member 13 Organic Cation Transporter 3 Orctl 3 |
| ENSDARG00000059231 | ENSFM00750001632325 | Transmembrane 4 L6 Family Member |
| ENSDARG00000059234 | ENSFM00250000006668 | 28S Ribosomal S27 Mitochondrial Mrp S27 S27MT |
| ENSDARG00000059246 | ENSFM00500000269788 | Heterogeneous Nuclear Ribonucleoprotein D Hnrnp D Hnrnp Dl |
| ENSDARG00000059298 | ENSFM00670001239874 | Signal Peptidase Complex Subunit 1 |
| ENSDARG00000059304 | ENSFM00600000921422 | Coiled Coil Helix Coiled Coil Helix Domain Containing Mitochondrial Precursor |
| ENSDARG00000059311 | ENSFM00630001053079 | Unknown |
| ENSDARG00000059311 | ENSFM00250000000643 | Dihydropyrimidinase Related Drp Collapsin Response Mediator Crmp |
| ENSDARG00000059323 | ENSFM00250000002976 | Dolichyl Diphosphooligosaccharide Glycosyltransferase Subunit 1 Precursor EC_2.4.99.18 Dolichyl Diphosphooligosaccharide Glycosyltransferase 67 Kda Subunit Ribophorin I Rpn I Ribophorin 1 |
| ENSDARG00000059333 | ENSFM00610000965417 | Unknown |
| ENSDARG00000059333 | ENSFM00250000001584 | Src Kinase Signaling Inhibitor 1 Snap 25 Interacting Snip P130CAS Associated P140CAP |
| ENSDARG00000059340 | ENSFM00250000000718 | Disks Large Associated Dap Psd 95/SAP90 Binding SAP90/Psd 95 Associated |
| ENSDARG00000059349 | ENSFM00800002008304 | Unknown |
| ENSDARG00000059368 | ENSFM00720001492020 | Glutamate Receptor Precursor Glur Ampa Selective Glutamate Receptor Glur Glutamate Receptor Ionotropic Ampa |
| ENSDARG00000059388 | ENSFM00500000271183 | D Beta Hydroxybutyrate Dehydrogenase Mitochondrial Bdh EC_1.1.1.30 3 Hydroxybutyrate Dehydrogenase |
| ENSDARG00000059409 | ENSFM00800002003396 | Atp Binding Cassette Sub Family D Member Adrenoleukodystrophy |
| ENSDARG00000059472 | ENSFM00250000001205 | Rho Gtpase Activating Rho Type Gtpase Activating Gtpase Activating |
| ENSDARG00000059473 | ENSFM00670001235382 | Kn Motif And Ankyrin Repeat Domain Containing Ankyrin Repeat Domain Containing |
| ENSDARG00000059596 | ENSFM00250000000764 | Cap Gly Domain Containing Linker 2 Cytoplasmic Linker 115 Clip 115 Cytoplasmic Linker 2 |
| ENSDARG00000059600 | ENSFM00250000001974 | Rab Gtpase Binding Effector 1 Rabaptin 5 Rabaptin 5ALPHA |
| ENSDARG00000059601 | ENSFM00250000000942 | Microtubule Associated Map |
| ENSDARG00000059629 | ENSFM00250000005876 | Aspartate Beta Hydroxylase Domain Containing EC_1.14.11.- |
| ENSDARG00000059630 | ENSFM00250000005592 | Malectin Precursor |
| ENSDARG00000059680 | ENSFM00250000001972 | Fascin |
| ENSDARG00000059701 | ENSFM00730001522115 | Flightless 1 |
| ENSDARG00000059738 | ENSFM00740001589097 | Receptor Type Tyrosine Phosphatase Precursor EC_3.1.3.48 |
| ENSDARG00000059763 | ENSFM00400000131718 | Gamma Aminobutyric Acid Receptor Subunit Beta Precursor Gaba A Receptor Subunit Beta |
| ENSDARG00000059770 | ENSFM00630001072039 | Unknown |
| ENSDARG00000059770 | ENSFM00500000269758 | Carnitine O Palmitoyltransferase 1 Muscle CPT1 M EC_2.3.1.21 Carnitine O Palmitoyltransferase I Muscle Cpt I Cpti M Carnitine Palmitoyltransferase 1B |
| ENSDARG00000059773 | ENSFM00250000001584 | Src Kinase Signaling Inhibitor 1 Snap 25 Interacting Snip P130CAS Associated P140CAP |
| ENSDARG00000059775 | ENSFM00250000004214 | Vesicular Inhibitory Amino Acid Transporter Gaba And Glycine Transporter Solute Carrier Family 32 Member 1 Vesicular Gaba Transporter |
| ENSDARG00000059806 | ENSFM00770001847625 | Phosphatidylinositide Phosphatase SAC1 EC_3.1.3.- Suppressor Of Actin Mutations 1 |
| ENSDARG00000059809 | ENSFM00730001521718 | 55 Kda Erythrocyte Membrane P55 Membrane Protein Palmitoylated 1 |
| ENSDARG00000059826 | ENSFM00610000966260 | Unknown |
| ENSDARG00000059826 | ENSFM00250000002903 | Cartilage Acidic 1 Precursor 68 Kda Chondrocyte Expressed Cep 68 Aspic |
| ENSDARG00000059832 | ENSFM00250000000737 | Brain Specific Angiogenesis Inhibitor Precursor |
| ENSDARG00000059853 | ENSFM00500000440578 | Unknown |
| ENSDARG00000059857 | ENSFM00250000007418 | Biliverdin Reductase A Precursor Bvr A EC_1.3.1.24 Biliverdin Ix Alpha Reductase |
| ENSDARG00000059888 | ENSFM00250000000492 | Argonaute |
| ENSDARG00000059897 | ENSFM00730001521357 | Nt 3 Growth Factor Receptor Precursor EC_2.7.10.1 Neurotrophic Tyrosine Kinase Receptor Type 3 |
| ENSDARG00000059902 | ENSFM00250000004293 | Vacuolar Sorting Associated 16 |
| ENSDARG00000059933 | ENSFM00260000050433 | Lipid Phosphate Phosphohydrolase EC_3.1.3.4 PAP2 Phosphatidate Phosphohydrolase Type Phosphatidic Acid Phosphatase Pap |
| ENSDARG00000059939 | ENSFM00740001589270 | Disabled Homolog 2 Adaptor Molecule Disabled 2 Differentially Expressed In Ovarian Carcinoma 2 Doc 2 |
| ENSDARG00000059945 | ENSFM00250000000744 | Synaptic Vesicle Glycoprotein |
| ENSDARG00000059960 | ENSFM00770001847600 | 1 Phosphatidylinositol 4 5 Bisphosphate Phosphodiesterase Eta 2 EC_3.1.4.11 Phosphoinositide Phospholipase C Eta 2 Phosphoinositide Phospholipase C 4 Plc L4 Phospholipase C 4 Phospholipase C Eta 2 Plc ETA2 |
| ENSDARG00000059997 | ENSFM00250000000744 | Synaptic Vesicle Glycoprotein |
| ENSDARG00000060006 | ENSFM00730001521822 | E3 Ubiquitin Ligase EC_6.3.2.- |
| ENSDARG00000060036 | ENSFM00500000270832 | Aminoacyl Trna Synthase Complex Interacting Multifunctional 1 Multisynthase Complex Auxiliary Component P43 |
| ENSDARG00000060069 | ENSFM00670001235660 | Isochorismatase Domain Containing |
| ENSDARG00000060093 | ENSFM00730001521806 | Death Associated Kinase Dap Kinase EC_2.7.11.1 Dap Kinase |
| ENSDARG00000060101 | ENSFM00250000002755 | Trafficking Particle Complex Subunit 12 Tetratricopeptide Repeat 15 Tpr Repeat 15 |
| ENSDARG00000060112 | ENSFM00250000002727 | Syntabulin Golgi Localized Syntaphilin Related Syntaxin 1 Binding |
| ENSDARG00000060116 | ENSFM00400000131731 | Calcium/Calmodulin Dependent Kinase Type EC_2.7.11.17 Cam Kinase I Cam Kinase Cam Ki Camki Kinase |
| ENSDARG00000060123 | ENSFM00730001521864 | Receptor Type Tyrosine Phosphatase Precursor R Ptp EC_3.1.3.48 |
| ENSDARG00000060124 | ENSFM00500000270373 | Mitochondrial Import Inner Membrane Translocase Subunit TIM17 |
| ENSDARG00000060126 | ENSFM00630001071865 | Unknown |
| ENSDARG00000060149 | ENSFM00250000000459 | Actin Binding Lim Ablim Actin Binding Lim Family Member |
| ENSDARG00000060150 | ENSFM00670001276795 | Unknown |
| ENSDARG00000060150 | ENSFM00250000003951 | 26S Proteasome Non Atpase Regulatory Subunit 26S Proteasome Regulatory Subunit RPN6 |
| ENSDARG00000060196 | ENSFM00500000271837 | Phospholysine Phosphohistidine Inorganic Pyrophosphate Phosphatase EC_3.1.3.- EC_3.6.1.1 |
| ENSDARG00000060238 | ENSFM00800002003915 | Uveal Autoantigen With Coiled Coil Domains And Ankyrin Repeats |
| ENSDARG00000060252 | ENSFM00760001714954 | Mannosyl Oligosaccharide Glucosidase EC_3.2.1.106 Processing Glucosidase I |
| ENSDARG00000060288 | ENSFM00500000271153 | Proline Synthase Co Transcribed Bacterial Homolog |
| ENSDARG00000060303 | ENSFM00730001521473 | Sodium Bicarbonate Solute Carrier Family 4 Member |
| ENSDARG00000060306 | ENSFM00250000001310 | Transcription Factor CP2 |
| ENSDARG00000060309 | ENSFM00250000000639 | Slit Robo Rho Gtpase Activating Rho Gtpase Activating |
| ENSDARG00000060312 | ENSFM00250000001662 | Mitogen Activated Kinase Binding 1 |
| ENSDARG00000060323 | ENSFM00250000005479 | Exocyst Complex Component Exocyst Complex Component SEC10 |
| ENSDARG00000060326 | ENSFM00250000000942 | Microtubule Associated Map |
| ENSDARG00000060349 | ENSFM00250000001269 | Wiskott Aldrich Syndrome Family Member 1 Wasp Family Member 1 Wave 1 |
| ENSDARG00000060368 | ENSFM00250000000771 | Synapsin Synapsin |
| ENSDARG00000060372 | ENSFM00730001521329 | Plexin Precursor |
| ENSDARG00000060380 | ENSFM00800002003815 | Ubiquinone Biosynthesis Monooxygenase COQ6 EC_1.14.13.- Coenzyme Q10 Monooxygenase 6 |
| ENSDARG00000060383 | ENSFM00250000008244 | Methylmalonic Aciduria Type A Homolog Mitochondrial Precursor EC_3.6.-.- Methylmalonic Aciduria Type A 1 |
| ENSDARG00000060393 | ENSFM00730001521406 | Fibrinogen C Domain Containing 1 |
| ENSDARG00000060434 | ENSFM00250000000942 | Microtubule Associated Map |
| ENSDARG00000060452 | ENSFM00260000050437 | Sidekick Precursor |
| ENSDARG00000060469 | ENSFM00800002003738 | Phosphoinositide 3 Kinase Regulatory Subunit 4 PI3 Kinase Regulatory Subunit 4 EC_2.7.11.1 |
| ENSDARG00000060477 | ENSFM00500000270891 | Vacuolar Sorting Associated 8 Homolog |
| ENSDARG00000060481 | ENSFM00250000001436 | Atlastin EC_3.6.5.- |
| ENSDARG00000060484 | ENSFM00250000002697 | Ethanolaminephosphotransferase 1 EC_2.7.8.1 Selenoprotein I Seli |
| ENSDARG00000060494 | ENSFM00800002003460 | Bifunctional Glutamate/Proline Trna Ligase Bifunctional Aminoacyl Trna Synthetase |
| ENSDARG00000060504 | ENSFM00730001521507 | Atp Dependent 6 Phosphofructokinase Type |
| ENSDARG00000060532 | ENSFM00410000138484 | Disintegrin And Metalloproteinase Domain Containing Precursor Adam Metalloproteinase Disintegrin And Cysteine Rich Mdc |
| ENSDARG00000060539 | ENSFM00250000000743 | SH3 And Multiple Ankyrin Repeat Domains Proline Rich Synapse Associated |
| ENSDARG00000060554 | ENSFM00250000004237 | Methylmalonyl Coa Mutase Mitochondrial Precursor Mcm EC_5.4.99.2 Methylmalonyl Coa Isomerase |
| ENSDARG00000060594 | ENSFM00760001714774 | Trifunctional Enzyme Subunit Alpha Mitochondrial Precursor 78 Kda Gastrin Binding Tp Alpha |
| ENSDARG00000060596 | ENSFM00760001714720 | 5' Amp Activated Kinase Catalytic Subunit Alpha 1 Ampk Subunit Alpha 1 EC_2.7.11.1 Acetyl Coa Carboxylase Kinase Acaca Kinase EC_2.7.11.- 27 Hydroxymethylglutaryl Coa Reductase Kinase Hmgcr Kinase EC_2.7.-.- 11 31 Tau Kinase PRKAA1 Ec 2.7.11.26 |
| ENSDARG00000060610 | ENSFM00760001714757 | Protocadherin Precursor Protocadherin |
| ENSDARG00000060626 | ENSFM00730001521692 | Diacylglycerol Kinase Dag Kinase EC_2.7.1.107 Kda Diacylglycerol Kinase Diglyceride Kinase Dgk |
| ENSDARG00000060627 | ENSFM00250000001308 | Huntingtin Interacting 1 Hip 1 Huntingtin Interacting I Hip I |
| ENSDARG00000060683 | ENSFM00250000005832 | E3 Ubiquitin Ligase LRSAM1 EC_6.3.2.- Leucine Rich Repeat And Sterile Alpha Motif Containing 1 TSG101 Associated Ligase |
| ENSDARG00000060711 | ENSFM00250000000744 | Synaptic Vesicle Glycoprotein |
| ENSDARG00000060723 | ENSFM00250000001930 | Stromal Interaction Molecule Precursor |
| ENSDARG00000060725 | ENSFM00250000004546 | Dnaj Homolog Subfamily C Member 16 Precursor |
| ENSDARG00000060734 | ENSFM00250000003219 | Dcc Interacting 13 DIP13 Adapter Containing Ph Domain Ptb Domain And Leucine Zipper Motif |
| ENSDARG00000060796 | ENSFM00250000001093 | Sodium Dependent Phosphate Transporter Solute Carrier Family 20 Member |
| ENSDARG00000060797 | ENSFM00730001521507 | Atp Dependent 6 Phosphofructokinase Type |
| ENSDARG00000060805 | ENSFM00250000000942 | Microtubule Associated Map |
| ENSDARG00000060813 | ENSFM00800002003882 | Pleckstrin Homology Domain Containing Family A Member 7 Ph Domain Containing Family A Member 7 Heart Adapter 1 |
| ENSDARG00000060853 | ENSFM00250000001880 | Nck Associated 1 1 Membrane Associated Hem |
| ENSDARG00000060854 | ENSFM00250000002046 | SH3KBP1 Binding 1 |
| ENSDARG00000060865 | ENSFM00250000003803 | Scai Suppressor Of Cancer Cell Invasion |
| ENSDARG00000060978 | ENSFM00500000269652 | Sarcoplasmic/Endoplasmic Reticulum Calcium Atpase Sr Ca 2+ Atpase EC_3.6.3.8 Calcium Pump Calcium Transporting Atpase Sarcoplasmic Reticulum Type Twitch Skeletal Muscle Endoplasmic Reticulum Class 1/2 Ca 2+ Atpase |
| ENSDARG00000061013 | ENSFM00310000089031 | Rabankyrin 5 |
| ENSDARG00000061031 | ENSFM00540000718025 | Delta And Notch Epidermal Growth Factor Related Receptor Precursor |
| ENSDARG00000061042 | ENSFM00250000000928 | Gamma Aminobutyric Acid Type B Receptor Subunit 2 Precursor Gaba B Receptor 2 Gaba B R2 Gaba BR2 GABABR2 GB2 G Coupled Receptor 51 |
| ENSDARG00000061051 | ENSFM00250000001886 | Bmp/Retinoic Acid Inducible Neural Specific 1 Precursor Deleted In Bladder Cancer 1 |
| ENSDARG00000061066 | ENSFM00260000050940 | C2 Domain Containing 2 Transmembrane 24 |
| ENSDARG00000061093 | ENSFM00600000921202 | Pleckstrin Homology Domain Family B Member LL5 |
| ENSDARG00000061099 | ENSFM00390000126295 | Neural Cell Adhesion Molecule L1 L1 L1 |
| ENSDARG00000061100 | ENSFM00500000270122 | Asparagine Trna Ligase Cytoplasmic EC_6.1.1.22 Asparaginyl Trna Synthetase Asnrs |
| ENSDARG00000061121 | ENSFM00260000050328 | Latrophilin |
| ENSDARG00000061131 | ENSFM00730001521483 | Kinesin |
| ENSDARG00000061143 | ENSFM00500000271643 | Transmembrane Anterior Posterior Transformation 1 |
| ENSDARG00000061165 | ENSFM00730001521312 | Sodium Dependent Transporter Transporter Solute Carrier Family 6 Member |
| ENSDARG00000061180 | ENSFM00250000005970 | Vacuolar Sorting Associated 45 |
| ENSDARG00000061207 | ENSFM00250000002093 | Tetratricopeptide Repeat Tpr Repeat |
| ENSDARG00000061255 | ENSFM00500000272272 | Dual Specificity Phosphatase 3 EC_3.1.3.16 EC_3.1.3.- 48 Vaccinia H1 Related Phosphatase Vhr |
| ENSDARG00000061257 | ENSFM00250000007974 | Njmu R1 |
| ENSDARG00000061268 | ENSFM00250000000492 | Argonaute |
| ENSDARG00000061282 | ENSFM00250000002484 | Golgi Apparatus 1 Precursor E Selectin Ligand 1 Esl 1 Golgi Sialoglycoprotein Mg 160 |
| ENSDARG00000061294 | ENSFM00250000002482 | Rho Gtpase Activating 35 Glucocorticoid Receptor Dna Binding Factor 1 |
| ENSDARG00000061301 | ENSFM00250000002497 | Gmp Reductase EC_1.7.1.7 Guanosine 5' Monophosphate Oxidoreductase Guanosine Monophosphate Reductase |
| ENSDARG00000061304 | ENSFM00250000001450 | Neurabin 1 Neurabin I Neural Tissue Specific F Actin Binding I Phosphatase 1 Regulatory Subunit 9A |
| ENSDARG00000061335 | ENSFM00730001521200 | Polypeptide N Acetylgalactosaminyltransferase EC_2.4.1.41 Polypeptide Galnac Transferase Galnac Pp Gantase Udp Acetylgalactosaminyltransferase Udp Galnac:Polypeptide N Acetylgalactosaminyltransferase |
| ENSDARG00000061338 | ENSFM00540000717952 | Atp Dependent Rna Helicase EC_3.6.4.13 |
| ENSDARG00000061397 | ENSFM00610000972403 | Unknown |
| ENSDARG00000061397 | ENSFM00760001714822 | E3 Ubiquitin Ligase TRIP12 EC_6.3.2.- Thyroid Receptor Interacting 12 Tr Interacting 12 Trip 12 |
| ENSDARG00000061398 | ENSFM00730001521622 | Band 4 1 |
| ENSDARG00000061413 | ENSFM00250000002785 | Phospholipase DDHD2 EC_3.1.1.- Ddhd Domain Containing 2 Sam Wwe And Ddhd Domain Containing 1 |
| ENSDARG00000061421 | ENSFM00740001589247 | Serine/Threonine Kinase BRSK2 EC_2.7.11.1 EC_2.7.11.- 26 Brain Specific Serine/Threonine Kinase 2 Br Serine/Threonine Kinase 2 Serine/Threonine Kinase Sad A |
| ENSDARG00000061445 | ENSFM00730001521247 | Adenylate Cyclase Type EC_4.6.1.1 Atp Pyrophosphate Lyase Adenylate Cyclase Type Adenylyl Cyclase Ca 2+ Adenylyl Cyclase |
| ENSDARG00000061454 | ENSFM00740001589096 | Neurexin Precursor Neurexin |
| ENSDARG00000061466 | ENSFM00730001521728 | Copine Copine |
| ENSDARG00000061473 | ENSFM00250000006948 | Tank Binding Kinase 1 Binding 1 TBK1 Binding 1 |
| ENSDARG00000061479 | ENSFM00740001589337 | Thrombospondin Type 1 Domain Containing Precursor |
| ENSDARG00000061517 | ENSFM00740001589098 | Density Lipoprotein Receptor Related Precursor Lrp |
| ENSDARG00000061525 | ENSFM00250000000882 | Serine/Threonine Kinase EC_2.7.11.1 |
| ENSDARG00000061551 | ENSFM00800002003399 | Ubiquitin Carboxyl Terminal Hydrolase 48 EC_3.4.19.12 Deubiquitinating Enzyme 48 Ubiquitin Thioesterase 48 Ubiquitin Specific Processing Protease 48 |
| ENSDARG00000061579 | ENSFM00800002003512 | Unconventional Myosin Ic Myosin I Beta Mmi Beta Mmib |
| ENSDARG00000061591 | ENSFM00730001521332 | Atp Mitochondrial Precursor Abc Transporter |
| ENSDARG00000061603 | ENSFM00610000973095 | Unknown |
| ENSDARG00000061603 | ENSFM00550000743175 | Sorbin And SH3 Domain Containing 2 Arg/Abl Interacting 2 ARGBP2 |
| ENSDARG00000061635 | ENSFM00750001632326 | Unconventional Myosin Va Dilute Myosin Heavy Chain Non Muscle |
| ENSDARG00000061638 | ENSFM00500000272373 | Dephospho Coa Kinase Domain Containing |
| ENSDARG00000061644 | ENSFM00500000271209 | Alpha Endosulfine |
| ENSDARG00000061647 | ENSFM00740001589096 | Neurexin Precursor Neurexin |
| ENSDARG00000061660 | ENSFM00500000269758 | Carnitine O Palmitoyltransferase 1 Muscle CPT1 M EC_2.3.1.21 Carnitine O Palmitoyltransferase I Muscle Cpt I Cpti M Carnitine Palmitoyltransferase 1B |
| ENSDARG00000061688 | ENSFM00730001521847 | Catenin Delta 1 Cadherin Associated Src Substrate Cas P120 Catenin P120 Ctn P120 Cas |
| ENSDARG00000061692 | ENSFM00810002040729 | Nad P H Hydrate Epimerase |
| ENSDARG00000061699 | ENSFM00730001521477 | Signal Induced Proliferation Associated 1 SIPA1 |
| ENSDARG00000061713 | ENSFM00810002040979 | Tumor D52 |
| ENSDARG00000061718 | ENSFM00670001235606 | Zinc Finger |
| ENSDARG00000061719 | ENSFM00250000004568 | Wash Complex Subunit 7 |
| ENSDARG00000061728 | ENSFM00500000273552 | Acyl Coenzyme A Thioesterase 13 Acyl Coa Thioesterase 13 EC_3.1.2.- Thioesterase Superfamily Member 2 |
| ENSDARG00000061736 | ENSFM00500000269628 | Ankyrin Ank Ankyrin |
| ENSDARG00000061741 | ENSFM00250000000547 | Inositol 1 4 5 Trisphosphate Receptor Type IP3 Receptor IP3R Ins Type Inositol 1 4 5 Trisphosphate Receptor Type INSP3 Receptor |
| ENSDARG00000061746 | ENSFM00250000001570 | Rho Guanine Nucleotide Exchange Factor 9 Collybistin Rac/CDC42 Guanine Nucleotide Exchange Factor 9 |
| ENSDARG00000061761 | ENSFM00600000921688 | Optic Atrophy 3 |
| ENSDARG00000061764 | ENSFM00740001589261 | Associated |
| ENSDARG00000061778 | ENSFM00250000001548 | Formin |
| ENSDARG00000061817 | ENSFM00800002003381 | Kinesin |
| ENSDARG00000061829 | ENSFM00250000000599 | Unc 13 Homolog MUNC13 |
| ENSDARG00000061854 | ENSFM00260000050644 | Vesicular Integral Membrane VIP36 Precursor Lectin Mannose Binding 2 Vesicular Integral Membrane 36 VIP36 |
| ENSDARG00000061862 | ENSFM00400000131775 | Unconventional Myosin Xviiia Molecule Associated With JAK3 N Terminus Majn Myosin Containing A Pdz Domain |
| ENSDARG00000061865 | ENSFM00730001521664 | Atp Dependent Rna Helicase EC_3.6.4.13 |
| ENSDARG00000061951 | ENSFM00250000005925 | Golgin Subfamily B Member 1.372 Kda Golgi Complex Associated GCP372 Giantin Macrogolgin |
| ENSDARG00000061981 | ENSFM00250000002996 | Cytosol Aminopeptidase EC_3.4.11.1 Leucine Aminopeptidase 3 Lap 3 Leucyl Aminopeptidase Proline Aminopeptidase EC_3.4.11.- 5 Prolyl Aminopeptidase |
| ENSDARG00000062003 | ENSFM00750001632326 | Unconventional Myosin Va Dilute Myosin Heavy Chain Non Muscle |
| ENSDARG00000062008 | ENSFM00290000065564 | Heparan Sulfate 2 O Sulfotransferase 1 2 O Sulfotransferase 2OST EC_2.8.2.- |
| ENSDARG00000062013 | ENSFM00250000000130 | Aminopeptidase N Ap N EC_3.4.11.2 Microsomal Aminopeptidase |
| ENSDARG00000062016 | ENSFM00250000003232 | Vacuolar Sorting Associated 51 Homolog Fat Free Homolog |
| ENSDARG00000062025 | ENSFM00250000006593 | Oxygen Dependent Coproporphyrinogen Iii Oxidase Cox Coprogen Oxidase Coproporphyrinogenase EC_1.3.3.3 |
| ENSDARG00000062045 | ENSFM00500000270404 | Interleukin 1 Receptor Accessory 1 Precursor Il 1 Rapl 1 Il 1RAPL 1 IL1RAPL 1 X Linked Interleukin 1 Receptor Accessory 1 |
| ENSDARG00000062054 | ENSFM00500000269758 | Carnitine O Palmitoyltransferase 1 Muscle CPT1 M EC_2.3.1.21 Carnitine O Palmitoyltransferase I Muscle Cpt I Cpti M Carnitine Palmitoyltransferase 1B |
| ENSDARG00000062086 | ENSFM00250000005157 | Ambiguous |
| ENSDARG00000062086 | ENSFM00600000934186 | Si:Dkey 177F24 1 |
| ENSDARG00000062087 | ENSFM00500000270047 | Neutral Cholesterol Ester Hydrolase 1 Nceh EC_3.1.1.- Arylacetamide Deacetylase 1 |
| ENSDARG00000062089 | ENSFM00800002003477 | Udp Glucose:Glycoprotein Glucosyltransferase 1 Precursor UGT1 EC_2.4.1.- Udp Glc:Glycoprotein Glucosyltransferase Udp Glucose Ceramide Glucosyltransferase 1 |
| ENSDARG00000062096 | ENSFM00500000271359 | Probable Carboxypeptidase P Precursor EC_3.4.17.- Peptidase M20 Domain Containing 1 |
| ENSDARG00000062103 | ENSFM00800002003877 | Ankycorbin Ankyrin Repeat And Coiled Coil Structure Containing Retinal Pigment Epithelial Cell Retinoic Acid Induced 14 |
| ENSDARG00000062103 | ENSFM00800002003981 | Ankyrin Repeat Domain Containing 24 |
| ENSDARG00000062123 | ENSFM00250000000604 | Arf Gap With Gtpase Ank Repeat And Ph Domain Containing Agap Centaurin Gamma |
| ENSDARG00000062134 | ENSFM00780001898357 | Voltage Gated Potassium Channel Subunit Beta K + Channel Subunit Beta Kv Beta |
| ENSDARG00000062173 | ENSFM00600000921199 | Calmodulin Regulated Spectrin Associated |
| ENSDARG00000062174 | ENSFM00250000000524 | Voltage Dependent L Type Calcium Channel Subunit Beta Calcium Channel Voltage Dependent Subunit Beta |
| ENSDARG00000062208 | ENSFM00600000921799 | Proline Rich Transmembrane 1 Dispanin Subfamily D Member 1 DSPD1 |
| ENSDARG00000062217 | ENSFM00730001521214 | G Activated Inward Rectifier Potassium Channel Girk Inward Rectifier K + Channel KIR3 Potassium Channel Inwardly Rectifying Subfamily J Member |
| ENSDARG00000062220 | ENSFM00800002003882 | Pleckstrin Homology Domain Containing Family A Member 7 Ph Domain Containing Family A Member 7 Heart Adapter 1 |
| ENSDARG00000062237 | ENSFM00500000270207 | Cysteine Desulfurase Mitochondrial Precursor EC_2.8.1.7 |
| ENSDARG00000062272 | ENSFM00800002003441 | AFG3 EC_3.4.24.- |
| ENSDARG00000062305 | ENSFM00250000000629 | Regulating Synaptic Membrane Exocytosis Rab 3 Interacting Molecule Rim |
| ENSDARG00000062310 | ENSFM00810002040846 | Wd Repeat Containing 26 |
| ENSDARG00000062323 | ENSFM00410000138484 | Disintegrin And Metalloproteinase Domain Containing Precursor Adam Metalloproteinase Disintegrin And Cysteine Rich Mdc |
| ENSDARG00000062325 | ENSFM00250000000743 | SH3 And Multiple Ankyrin Repeat Domains Proline Rich Synapse Associated |
| ENSDARG00000062330 | ENSFM00380000124692 | Uncharacterized Fragile Site Associated |
| ENSDARG00000062335 | ENSFM00250000005546 | Uridine Diphosphate Glucose Pyrophosphatase Udpg Pyrophosphatase Ugppase EC_3.6.1.45 Nucleoside Diphosphate Linked Moiety X Motif 14 Nudix Motif 14 |
| ENSDARG00000062347 | ENSFM00500000271268 | Microtubule Associated Tumor Suppressor Candidate 2 Cardiac Zipper Microtubule Plus End Tracking TIP150 Tracking Of 150 Kda |
| ENSDARG00000062350 | ENSFM00500000294137 | Uncharacterized |
| ENSDARG00000062370 | ENSFM00250000005222 | Bcl 2 13 BCL2 L 13 Bcl Rambo MIL1 |
| ENSDARG00000062376 | ENSFM00380000124679 | Neuroligin Precursor |
| ENSDARG00000062396 | ENSFM00250000001010 | Ankyrin Repeat And Sterile Alpha Motif Domain Containing 1B Amyloid Beta Intracellular Domain Associated 1 Aida 1 E2A PBX1 Associated Eb 1 |
| ENSDARG00000062415 | ENSFM00730001521673 | Catenin Delta 2 |
| ENSDARG00000062430 | ENSFM00250000001725 | Glycerol 3 Phosphate Dehydrogenase Mitochondrial Precursor Gpd M Gpdh M EC_1.1.5.3 |
| ENSDARG00000062445 | ENSFM00250000000995 | E3 Sumo Ligase EC_6.3.2.- Inhibitor Of Activated Stat |
| ENSDARG00000062447 | ENSFM00730001522025 | E3 Ubiquitin Ligase EC_6.3.2.- Hect C2 And Ww Domain Containing NEDD4 E3 Ubiquitin Ligase |
| ENSDARG00000062477 | ENSFM00250000005157 | Ambiguous |
| ENSDARG00000062511 | ENSFM00250000007244 | Golgin Subfamily A Member 3 Golgin 160 |
| ENSDARG00000062521 | ENSFM00810002040637 | Probable Phospholipid Transporting Atpase EC_3.6.3.1 Atpase Class Ii Type |
| ENSDARG00000062531 | ENSFM00250000000947 | C Jun Amino Terminal Kinase Interacting Jip Jnk Interacting Jnk Mitogen Activated Kinase 8 Interacting |
| ENSDARG00000062542 | ENSFM00500000269773 | Fibroblast Growth Factor Fgf Fibroblast Growth Factor Homologous Factor Fhf |
| ENSDARG00000062558 | ENSFM00250000004877 | Ring Finger And Spry Domain Containing 1 Precursor |
| ENSDARG00000062561 | ENSFM00750001653065 | Unknown |
| ENSDARG00000062561 | ENSFM00730001522023 | Uncharacterized Aarf Domain Containing Kinase EC_2.7.11.- |
| ENSDARG00000062577 | ENSFM00250000002482 | Rho Gtpase Activating 35 Glucocorticoid Receptor Dna Binding Factor 1 |
| ENSDARG00000062590 | ENSFM00740001589112 | Microtubule Actin Cross Linking Factor 1 Actin Cross Linking Family 7 |
| ENSDARG00000062590 | ENSFM00740001589199 | Plectin Pcn Pltn Plectin 1 |
| ENSDARG00000062594 | ENSFM00250000004792 | Ubiquinone Biosynthesis COQ7 Homolog Coenzyme Q Biosynthesis 7 Homolog Timing Clk 1 Homolog |
| ENSDARG00000062612 | ENSFM00440000236925 | Atp Dependent Rna Helicase EC_3.6.4.13 |
| ENSDARG00000062633 | ENSFM00250000001126 | Cell Adhesion Molecule Precursor Immunoglobulin Superfamily Member Nectin Necl Synaptic Cell Adhesion Molecule |
| ENSDARG00000062667 | ENSFM00760001714665 | Maguk P55 Subfamily Member |
| ENSDARG00000062693 | ENSFM00740001589096 | Neurexin Precursor Neurexin |
| ENSDARG00000062696 | ENSFM00610000967131 | Unknown |
| ENSDARG00000062696 | ENSFM00730001521692 | Diacylglycerol Kinase Dag Kinase EC_2.7.1.107 Kda Diacylglycerol Kinase Diglyceride Kinase Dgk |
| ENSDARG00000062720 | ENSFM00760001714757 | Protocadherin Precursor Protocadherin |
| ENSDARG00000062744 | ENSFM00740001589070 | Sodium Channel Type Subunit Alpha Sodium Channel Sodium Channel Type Subunit Alpha Voltage Gated Sodium Channel Subunit Alpha NAV1 |
| ENSDARG00000062780 | ENSFM00800002003733 | Apoptosis Inducing Factor 3 Ec 1 Apoptosis Inducing Factor |
| ENSDARG00000062794 | ENSFM00400000131833 | Tripartite Motif Containing 46 Gene Y Geney Tripartite Fibronectin Type Iii And C Terminal Spry Motif |
| ENSDARG00000062795 | ENSFM00730001521676 | Iron Sulfur Clusters Transporter ATM1 Mitochondrial Precursor |
| ENSDARG00000062799 | ENSFM00250000000906 | Brain Specific Angiogenesis Inhibitor 1 Associated 2 Bai Associated 2 BAI1 Associated 2 Insulin Receptor Substrate Of 53 Kda IRSP53 Insulin Receptor Substrate P53 |
| ENSDARG00000062812 | ENSFM00250000001281 | Glutaminase Isoform Mitochondrial Precursor Gls EC_3.5.1.2 Glutaminase L Glutamine Amidohydrolase |
| ENSDARG00000062823 | ENSFM00760001714819 | Phosphatidylinositol 4 Kinase Alpha PI4 Kinase Alpha PI4K Alpha Ptdins 4 Kinase Alpha EC_2.7.1.67 |
| ENSDARG00000062831 | ENSFM00500000270303 | Nectin 2 Precursor Herpes Virus Entry Mediator B Herpesvirus Entry Mediator B Hveb Poliovirus Receptor Related 2 CD112 Antigen |
| ENSDARG00000062834 | ENSFM00250000005244 | Evolutionarily Conserved Signaling Intermediate In Toll Pathway Mitochondrial Precursor |
| ENSDARG00000062846 | ENSFM00500000270268 | Transmembrane And Tpr Repeat Containing |
| ENSDARG00000062854 | ENSFM00260000050437 | Sidekick Precursor |
| ENSDARG00000062868 | ENSFM00430000230229 | Early Endosome Antigen 1 |
| ENSDARG00000062942 | ENSFM00290000065519 | Potassium Voltage Gated Channel Subfamily A Member Voltage Gated Potassium Channel Subunit KV1 |
| ENSDARG00000062948 | ENSFM00250000001269 | Wiskott Aldrich Syndrome Family Member 1 Wasp Family Member 1 Wave 1 |
| ENSDARG00000062949 | ENSFM00250000001969 | 5' Nucleotidase Domain Containing EC_3.1.3.- |
| ENSDARG00000062951 | ENSFM00250000004834 | Tld Domain Containing 1 Tbc/Lysm Associated Domain Containing 1 |
| ENSDARG00000062956 | ENSFM00250000001790 | SN1 Specific Diacylglycerol Lipase Alpha Dgl Alpha EC_3.1.1.- Neural Stem Cell Derived Dendrite Regulator |
| ENSDARG00000062968 | ENSFM00250000000945 | Myotubularin Related Set Binding Factor |
| ENSDARG00000062972 | ENSFM00250000001308 | Huntingtin Interacting 1 Hip 1 Huntingtin Interacting I Hip I |
| ENSDARG00000062991 | ENSFM00250000000845 | Abl Interactor 1 Abelson Interactor 1 Abi 1 EPS8 SH3 Domain Binding EPS8 Binding Spectrin SH3 Domain Binding 1 |
| ENSDARG00000062997 | ENSFM00250000000550 | Hyaluronidase Precursor EC_3.2.1.35 Hyaluronoglucosaminidase |
| ENSDARG00000063001 | ENSFM00770001847537 | Phospholipid Transporting Atpase EC_3.6.3.1 Atpase Class I Type 8A Member P4 Atpase Flippase Complex Alpha Subunit At |
| ENSDARG00000063008 | ENSFM00600000921514 | Ambiguous |
| ENSDARG00000063026 | ENSFM00250000007537 | Membrane MLC1 |
| ENSDARG00000063036 | ENSFM00250000001069 | Ph And SEC7 Domain Containing 1 Exchange Factor ARF6 Pleckstrin Homology And SEC7 Domain Containing 1 |
| ENSDARG00000063040 | ENSFM00250000001718 | Ankyrin Repeat And Btb/Poz Domain Containing BTBD11 Btb/Poz Domain Containing 11 |
| ENSDARG00000063054 | ENSFM00250000000743 | SH3 And Multiple Ankyrin Repeat Domains Proline Rich Synapse Associated |
| ENSDARG00000063068 | ENSFM00250000000542 | Nesprin 2 Nuclear Envelope Spectrin Repeat 2 Nucleus And Actin Connecting Element Protein Nuance Synaptic Nuclear Envelope 2 Syne 2 |
| ENSDARG00000063069 | ENSFM00250000004894 | GRIP1 Associated 1 Grasp 1 |
| ENSDARG00000063079 | ENSFM00250000000492 | Argonaute |
| ENSDARG00000063100 | ENSFM00250000004350 | 26S Proteasome Non Atpase Regulatory Subunit 14 EC_3.4.19.- 26S Proteasome Regulatory Subunit RPN11 |
| ENSDARG00000063101 | ENSFM00500000270074 | Alpha Mannosidase 2 EC_3.2.1.114 Golgi Alpha Mannosidase Ii Aman Ii Man Ii Mannosidase Alpha Class 2A Member 1 Mannosyl Oligosaccharide 1 3 1 6 Alpha Mannosidase |
| ENSDARG00000063133 | ENSFM00730001521473 | Sodium Bicarbonate Solute Carrier Family 4 Member |
| ENSDARG00000063145 | ENSFM00250000006597 | Transcription Factor A Mitochondrial Precursor Mttfa |
| ENSDARG00000063149 | ENSFM00800002003912 | Transmembrane And Tpr Repeat Containing 1 |
| ENSDARG00000063150 | ENSFM00740001589096 | Neurexin Precursor Neurexin |
| ENSDARG00000063158 | ENSFM00730001521845 | Sodium/Potassium/Calcium Exchanger 1 Na + /K + /Ca 2+ Exchange 1 Retinal Rod Na Ca+K Exchanger Solute Carrier Family 24 Member 1 |
| ENSDARG00000063159 | ENSFM00780001898359 | Dynamin 1 EC_3.6.5.5 Dynamin Family Member Proline Rich Carboxyl Terminal Domain Less Dymple Dynamin Related 1 |
| ENSDARG00000063180 | ENSFM00760001714721 | Dedicator Of Cytokinesis 3 Modifier Of Cell Adhesion Presenilin Binding Pbp |
| ENSDARG00000063187 | ENSFM00250000007868 | 1 Acyl Sn Glycerol 3 Phosphate Acyltransferase Epsilon EC_2.3.1.51 1 Acylglycerol 3 Phosphate O Acyltransferase 5 1 Agp Acyltransferase 5 1 Agpat 5 Lysophosphatidic Acid Acyltransferase Epsilon Lpaat Epsilon |
| ENSDARG00000063197 | ENSFM00630001068445 | Unknown |
| ENSDARG00000063197 | ENSFM00500000270377 | Golgin Subfamily A Member 2.130 Kda Cis Golgi Matrix GM130 |
| ENSDARG00000063207 | ENSFM00410000138465 | Receptor Tyrosine Kinase Erbb Precursor EC_2.7.10.1 Proto Oncogene C Erbb |
| ENSDARG00000063211 | ENSFM00250000004839 | Exocyst Complex Component 8 Exocyst Complex 84 Kda Subunit |
| ENSDARG00000063253 | ENSFM00730001522025 | E3 Ubiquitin Ligase EC_6.3.2.- Hect C2 And Ww Domain Containing NEDD4 E3 Ubiquitin Ligase |
| ENSDARG00000063255 | ENSFM00250000001718 | Ankyrin Repeat And Btb/Poz Domain Containing BTBD11 Btb/Poz Domain Containing 11 |
| ENSDARG00000063264 | ENSFM00760001714657 | Protocadherin 11 X Linked Precursor Protocadherin 11 Protocadherin On The X Chromosome Pcdh X |
| ENSDARG00000063295 | ENSFM00270000056424 | Myosin Cellular Myosin Heavy Chain Type Myosin Heavy Chain Myosin Heavy Chain Non Muscle Non Muscle Myosin Heavy Chain Nmmhc Non Muscle Myosin Heavy Chain Nmmhc Ii Nmmhc |
| ENSDARG00000063297 | ENSFM00730001521565 | Atp Binding Cassette Sub Family B Member 6 Mitochondrial Ubiquitously Expressed Mammalian Abc Half Transporter |
| ENSDARG00000063299 | ENSFM00650001146246 | Unknown |
| ENSDARG00000063299 | ENSFM00600000921250 | Piccolo Aczonin |
| ENSDARG00000063310 | ENSFM00670001235411 | Oxidation Resistance 1 |
| ENSDARG00000063332 | ENSFM00250000000743 | SH3 And Multiple Ankyrin Repeat Domains Proline Rich Synapse Associated |
| ENSDARG00000063344 | ENSFM00500000307906 | Unknown |
| ENSDARG00000063352 | ENSFM00250000001432 | Type Inositol 3 4 Bisphosphate 4 Phosphatase EC_3.1.3.66 Inositol Polyphosphate 4 Phosphatase Type |
| ENSDARG00000063358 | ENSFM00260000050684 | Calcium Uptake 1 Mitochondrial Calcium Binding Atopy Related Autoantigen 1 Homolog |
| ENSDARG00000063361 | ENSFM00600000921514 | Ambiguous |
| ENSDARG00000063375 | ENSFM00250000006576 | Phosphotriesterase Related EC_3.1.-.- Parathion Hydrolase Related |
| ENSDARG00000063376 | ENSFM00250000002369 | Cerebellin Precursor |
| ENSDARG00000063390 | ENSFM00500000270303 | Nectin 2 Precursor Herpes Virus Entry Mediator B Herpesvirus Entry Mediator B Hveb Poliovirus Receptor Related 2 CD112 Antigen |
| ENSDARG00000063411 | ENSFM00250000009938 | Leucine Rich Repeat Containing 73 |
| ENSDARG00000063414 | ENSFM00250000007556 | Transmembrane 5 |
| ENSDARG00000063416 | ENSFM00310000088999 | Receptor Type Tyrosine Phosphatase Precursor R Ptp EC_3.1.3.48 Receptor Type Tyrosine Phosphatase |
| ENSDARG00000063419 | ENSFM00800002009474 | Unknown |
| ENSDARG00000063419 | ENSFM00730001522013 | Probable Atp Dependent Rna Helicase DHX35 EC_3.6.4.13 Deah Box 35 |
| ENSDARG00000063433 | ENSFM00250000000445 | Plasma Membrane Calcium Transporting Atpase EC_3.6.3.8 Plasma Membrane Calcium Atpase Plasma Membrane Calcium Pump |
| ENSDARG00000063436 | ENSFM00260000050490 | Double C2 Domain Containing DOC2 |
| ENSDARG00000063446 | ENSFM00500000273104 | Transferase CAF17 Mitochondrial Precursor EC_2.1.-.- Iron Sulfur Cluster Assembly Factor Homolog |
| ENSDARG00000063525 | ENSFM00250000002925 | Adp Dependent Glucokinase Precursor Adp Gk Adpgk EC_2.7.1.147 |
| ENSDARG00000063527 | ENSFM00250000001494 | Engulfment And Cell Motility |
| ENSDARG00000063544 | ENSFM00260000050567 | Phosphatidylinositol 5 Phosphate 4 Kinase Type 2 EC_2.7.1.149 1 Phosphatidylinositol 5 Phosphate 4 Kinase 2 Diphosphoinositide Kinase 2 Phosphatidylinositol 5 Phosphate 4 Kinase Type Ii Pi 5 P 4 Kinase Type Ii PIP4KII Ptdins 5 P 4 Kinase 2 |
| ENSDARG00000063568 | ENSFM00800002014767 | Unknown |
| ENSDARG00000063568 | ENSFM00740001589375 | Synaptotagmin 7 Synaptotagmin Vii Sytvii |
| ENSDARG00000063570 | ENSFM00730001521969 | Dual Specificity Tyrosine Phosphorylation Regulated Kinase Dual Specificity YAK1 Related Kinase Minibrain |
| ENSDARG00000063578 | ENSFM00730001521936 | Diacylglycerol Kinase Zeta Dag Kinase Zeta EC_2.7.1.107 Diglyceride Kinase Zeta Dgk Zeta |
| ENSDARG00000063588 | ENSFM00250000004803 | Ubiquitin Modifier Activating Enzyme 5 Ubiquitin Activating Enzyme 5 |
| ENSDARG00000063624 | ENSFM00730001521750 | Elongation Factor G Mitochondrial |
| ENSDARG00000063677 | ENSFM00250000001383 | Cyclin Y |
| ENSDARG00000063684 | ENSFM00250000002013 | Phosphatase EC_3.1.3.16 |
| ENSDARG00000063711 | ENSFM00750001632380 | Tripartite Motif Containing 2 EC_6.3.2.- E3 Ubiquitin Ligase TRIM2 |
| ENSDARG00000063713 | ENSFM00250000000610 | Ras Gtpase Activating |
| ENSDARG00000063719 | ENSFM00730001521839 | Ubiquitin Carboxyl Terminal Hydrolase 8 EC_3.4.19.12 Deubiquitinating Enzyme 8 Ubiquitin Isopeptidase Y Ubiquitin Thioesterase 8 Ubiquitin Specific Processing Protease 8 |
| ENSDARG00000063882 | ENSFM00500000391179 | Uncharacterized |
| ENSDARG00000063895 | ENSFM00800002003365 | Nadh Ubiquinone Oxidoreductase Chain 1 EC_1.6.5.3 Nadh Dehydrogenase Subunit 1 |
| ENSDARG00000063899 | ENSFM00420000140506 | Nadh Ubiquinone Oxidoreductase Chain 2 EC_1.6.5.3 Nadh Dehydrogenase Subunit 2 |
| ENSDARG00000063908 | ENSFM00350000105374 | Cytochrome C Oxidase Subunit 2 EC_1.9.3.1 Cytochrome C Oxidase Polypeptide Ii |
| ENSDARG00000063910 | ENSFM00500000363081 | Unknown |
| ENSDARG00000063911 | ENSFM00300000082445 | Atp Synthase Subunit A F Atpase 6 |
| ENSDARG00000063917 | ENSFM00730001521085 | Nadh Ubiquinone Oxidoreductase Chain 4 EC_1.6.5.3 Nadh Dehydrogenase Subunit 4 |
| ENSDARG00000063921 | ENSFM00760001714727 | Nadh Ubiquinone Oxidoreductase Chain 5 EC_1.6.5.3 Nadh Dehydrogenase Subunit 5 |
| ENSDARG00000063924 | ENSFM00810002040539 | Cytochrome B Complex Iii Subunit 3 Complex Iii Subunit Iii Cytochrome B C1 Complex Subunit 3 Ubiquinol Cytochrome C Reductase Complex Cytochrome B Subunit |
| ENSDARG00000067507 | ENSFM00260000050545 | Btb/Poz Domain Containing |
| ENSDARG00000067509 | ENSFM00730001521994 | Sodium/Potassium/Calcium Exchanger 3 Precursor Na + /K + /Ca 2+ Exchange 3 Solute Carrier Family 24 Member 3 |
| ENSDARG00000067567 | ENSFM00730001521104 | Sodium And Chloride Dependent Transporter Solute Carrier Family 6 Member |
| ENSDARG00000067591 | ENSFM00250000002171 | Transcriptional Activator Pur Beta Purine Rich Element Binding B |
| ENSDARG00000067593 | ENSFM00500000270642 | Homolog Mitochondrial Precursor |
| ENSDARG00000067605 | ENSFM00730001522002 | Soluble Nsf Attachment Snap N Ethylmaleimide Sensitive Factor Attachment |
| ENSDARG00000067626 | ENSFM00730001521154 | Unknown |
| ENSDARG00000067676 | ENSFM00770001847576 | Inactive Phospholipase C 2 Plc L 2 Plc L2 Phospholipase C L2 Phospholipase C Epsilon 2 Plc Epsilon 2 |
| ENSDARG00000067717 | ENSFM00500000360114 | Unknown |
| ENSDARG00000067717 | ENSFM00800002014652 | Unknown |
| ENSDARG00000067784 | ENSFM00800002003389 | Sodium/Hydrogen Exchanger 1 Na + /H + Exchanger 1 Nhe 1 Solute Carrier Family 9 Member 1 |
| ENSDARG00000067815 | ENSFM00250000001884 | Leucine Rich Repeat And Immunoglobulin Domain Containing Nogo Receptor Interacting Precursor Leucine Rich Repeat Neuronal |
| ENSDARG00000067824 | ENSFM00250000000380 | Contactin Associated 5 Precursor Cell Recognition Molecule CASPR5 |
| ENSDARG00000067920 | ENSFM00730001521184 | Ras Related Rab |
| ENSDARG00000067964 | ENSFM00730001521205 | Sodium And Chloride Dependent Glycine Transporter Glyt Solute Carrier Family 6 Member |
| ENSDARG00000068125 | ENSFM00760001714944 | NFU1 Iron Sulfur Cluster Scaffold Homolog Mitochondrial |
| ENSDARG00000068149 | ENSFM00250000004936 | Tumor P63 Regulated Gene 1 Mossy Fiber Terminal Associated Vertebrate Specific Presynaptic Protein FAM79A |
| ENSDARG00000068156 | ENSFM00500000323213 | CHCHD7 |
| ENSDARG00000068166 | ENSFM00250000001412 | Whirlin |
| ENSDARG00000068176 | ENSFM00250000004739 | Ubiquinol Cytochrome C Reductase Complex Assembly Factor 1 Basic Fgf Repressed Zic Binding Ubiquinol Cytochrome C Reductase Complex Chaperone CBP3 Homolog |
| ENSDARG00000068177 | ENSFM00730001521340 | Serine/Threonine Kinase Pak EC_2.7.11.1 Pak P21 Activated Kinase Pak |
| ENSDARG00000068187 | ENSFM00800002003722 | Paraplegin EC_3.4.24.- |
| ENSDARG00000068246 | ENSFM00800002003424 | 1 Phosphatidylinositol 4 5 Bisphosphate Phosphodiesterase Beta EC_3.1.4.11 Phosphoinositide Phospholipase C Beta Phospholipase C Beta Plc Beta |
| ENSDARG00000068262 | ENSFM00500000429078 | Unknown |
| ENSDARG00000068264 | ENSFM00500000270235 | Glyoxylate Reductase/Hydroxypyruvate Reductase EC_1.1.1.79 EC_1.1.1.- 81 |
| ENSDARG00000068289 | ENSFM00250000006253 | Eukaryotic Translation Initiation Factor 3 Subunit K |
| ENSDARG00000068305 | ENSFM00500000274118 | 28S Ribosomal S14 Mitochondrial Mrp S14 S14MT |
| ENSDARG00000068323 | ENSFM00250000001299 | Astrotactin Precursor |
| ENSDARG00000068404 | ENSFM00610000963188 | Unknown |
| ENSDARG00000068404 | ENSFM00500000272127 | Tubulin Folding Cofactor B Cytoskeleton Associated 1 Cytoskeleton Associated Ckapi Tubulin Specific Chaperone B |
| ENSDARG00000068415 | ENSFM00250000004186 | Prostaglandin E Synthase 2 |
| ENSDARG00000068421 | ENSFM00670001235990 | Tetratricopeptide Repeat Tpr Repeat |
| ENSDARG00000068480 | ENSFM00250000005199 | MAP7 Domain Containing 2 |
| ENSDARG00000068516 | ENSFM00610000973362 | Unknown |
| ENSDARG00000068516 | ENSFM00730001521886 | Hyaluronan And Proteoglycan Link 1 Precursor Cartilage Linking 1 Cartilage Link Proteoglycan Link |
| ENSDARG00000068624 | ENSFM00740001589095 | Spectrin Beta Chain Erythrocytic Beta Spectrin |
| ENSDARG00000068628 | ENSFM00730001521184 | Ras Related Rab |
| ENSDARG00000068653 | ENSFM00730001521186 | Ras Related C3 Botulinum Toxin Substrate Precursor P21 |
| ENSDARG00000068691 | ENSFM00500000273388 | Btb/Poz Domain Containing KCTD4 |
| ENSDARG00000068738 | ENSFM00670001235955 | Cytochrome C Oxidase Subunit 5B Mitochondrial Precursor Cytochrome C Oxidase Polypeptide Vb |
| ENSDARG00000068745 | ENSFM00780001898729 | Microtubule Associated |
| ENSDARG00000068787 | ENSFM00730001521463 | Sodium Dependent Neutral Amino Acid Transporter B 0 AT2 Sodium And Chloride Dependent Neurotransmitter Transporter NTT73 Solute Carrier Family 6 Member 15 Transporter V7 3 |
| ENSDARG00000068822 | ENSFM00250000002171 | Transcriptional Activator Pur Beta Purine Rich Element Binding B |
| ENSDARG00000068868 | ENSFM00500000269862 | Nucleosome Assembly 1 |
| ENSDARG00000068886 | ENSFM00500000273213 | 39S Ribosomal L14 Mitochondrial Precursor L14MT Mrp L14 39S Ribosomal L32 Mitochondrial L32MT Mrp L32 |
| ENSDARG00000068910 | ENSFM00470000251434 | Nitric Oxide Synthase EC_1.14.13.39 Nos Nos Type Peptidyl Cysteine S Nitrosylase |
| ENSDARG00000068940 | ENSFM00500000331301 | Unknown |
| ENSDARG00000068978 | ENSFM00500000271437 | Lactoylglutathione Lyase EC_4.4.1.5 Aldoketomutase Glyoxalase I Glx I Ketone Aldehyde Mutase Methylglyoxalase S D Lactoylglutathione Methylglyoxal Lyase |
| ENSDARG00000068981 | ENSFM00250000004289 | D Glucuronyl C5 Epimerase EC_5.1.3.17 Heparan Sulfate C5 Epimerase Hsepi Heparin/Heparan Sulfate:Glucuronic Acid C5 Epimerase Heparosan N Sulfate Glucuronate 5 Epimerase |
| ENSDARG00000068989 | ENSFM00800002003385 | Gamma Aminobutyric Acid Receptor Subunit Alpha Precursor Gaba A Receptor Subunit Alpha |
| ENSDARG00000068992 | ENSFM00730001521103 | Heat Shock Kda Heat Shock 70 Kda |
| ENSDARG00000069003 | ENSFM00250000003214 | Prenylcysteine Oxidase Precursor |
| ENSDARG00000069013 | ENSFM00250000000757 | Thioredoxin Peroxidase Thioredoxin Dependent Peroxide Reductase |
| ENSDARG00000069026 | ENSFM00730001521247 | Adenylate Cyclase Type EC_4.6.1.1 Atp Pyrophosphate Lyase Adenylate Cyclase Type Adenylyl Cyclase Ca 2+ Adenylyl Cyclase |
| ENSDARG00000069046 | ENSFM00730001521138 | Keratin Type I Cytoskeletal Cytokeratin Ck Keratin |
| ENSDARG00000069090 | ENSFM00800002003469 | V Type Proton Atpase Subunit D V Atpase Subunit D Vacuolar Proton Pump Subunit D |
| ENSDARG00000069099 | ENSFM00250000005494 | Transmembrane And Coiled Coil 1 |
| ENSDARG00000069100 | ENSFM00730001521985 | 4 Trimethylaminobutyraldehyde Dehydrogenase Tmabadh EC_1.2.1.47 Aldehyde Dehydrogenase Family 9 Member A1 EC_1.2.1.- 3 |
| ENSDARG00000069101 | ENSFM00730001522002 | Soluble Nsf Attachment Snap N Ethylmaleimide Sensitive Factor Attachment |
| ENSDARG00000069113 | ENSFM00480000262790 | Drebrin Developmentally Regulated Brain |
| ENSDARG00000069116 | ENSFM00670001239775 | Mitochondrial Import Inner Membrane Translocase Subunit TIM10 |
| ENSDARG00000069118 | ENSFM00760001714676 | Serine/Threonine Phosphatase 2A 56 Kda Regulatory Subunit Epsilon PP2A B Subunit B' Epsilon PP2A B Subunit B56 Epsilon PP2A B Subunit PR61 Epsilon PP2A B Subunit R5 Epsilon |
| ENSDARG00000069142 | ENSFM00250000001208 | Alanine Trna Ligase |
| ENSDARG00000069150 | ENSFM00730001522318 | Gtp Binding RIT1 Ras Expressed In Many Tissues Ras Without Caax 1 |
| ENSDARG00000069242 | ENSFM00260000050925 | Disulfide Isomerase A5 Precursor EC_5.3.4.1 |
| ENSDARG00000069254 | ENSFM00800002003483 | Muscarinic Acetylcholine Receptor |
| ENSDARG00000069269 | ENSFM00250000002834 | Wd Repeat Containing 35 |
| ENSDARG00000069293 | ENSFM00500000278690 | 252G14 4 |
| ENSDARG00000069313 | ENSFM00500000271370 | Mitochondrial Inner Membrane OXA1L Precursor Oxidase Assembly 1 OXA1 |
| ENSDARG00000069318 | ENSFM00250000001283 | EFR3 Homolog |
| ENSDARG00000069324 | ENSFM00800002003735 | Vacuolar Sorting Associated 53 Homolog |
| ENSDARG00000069339 | ENSFM00270000056490 | TBC1 Domain Family Member 24 |
| ENSDARG00000069356 | ENSFM00260000050328 | Latrophilin |
| ENSDARG00000069360 | ENSFM00500000269882 | Calcineurin Subunit B Regulatory Subunit Phosphatase Regulatory Subunit |
| ENSDARG00000069402 | ENSFM00260000050662 | Leucine Rich Repeat Containing Precursor Netrin Ligand Ngl |
| ENSDARG00000069420 | ENSFM00730001522472 | E3 Ubiquitin Ligase TRIM23 EC_6.3.2.- Adp Ribosylation Factor Domain Containing 1 Gtp Binding Ard 1 Tripartite Motif Containing 23 |
| ENSDARG00000069427 | ENSFM00250000002642 | Sodium/Potassium Transporting Atpase Subunit Beta 1 Interacting Na + /K + Transporting Atpase Subunit Beta 1 Interacting |
| ENSDARG00000069430 | ENSFM00500000272770 | Tumor P53 Inducible 11 P53 Induced Gene 11 |
| ENSDARG00000069446 | ENSFM00800002003860 | Dopamine Beta Hydroxylase EC_1.14.17.1 Dopamine Beta Monooxygenase |
| ENSDARG00000069467 | ENSFM00250000001385 | Turtle Homolog A Precursor Dendrite Arborization And Synapse Maturation 1 Immunoglobulin Superfamily Member 9A IGSF9A |
| ENSDARG00000069484 | ENSFM00250000000610 | Ras Gtpase Activating |
| ENSDARG00000069494 | ENSFM00500000295586 | Transmembrane 223 |
| ENSDARG00000069496 | ENSFM00760001714716 | Adp Ribosylation Factor 2 |
| ENSDARG00000069501 | ENSFM00500000272327 | Alpha/Beta Hydrolase Domain Containing 11 Abhydrolase Domain Containing 11 Ec 3 Williams Beuren Syndrome Chromosomal Region 21 |
| ENSDARG00000069521 | ENSFM00250000005075 | Vacuolar Sorting Associated 29 Vesicle Sorting 29 |
| ENSDARG00000069529 | ENSFM00670001235889 | Dual Specificity Phosphatase EC_3.1.3.16 EC_3.1.3.- 48 |
| ENSDARG00000069560 | ENSFM00730001521919 | Potassium Voltage Gated Channel Subfamily H Member Brain Specific Eag Channel Ether A Go Go Potassium Channel Elk Channel Voltage Gated Potassium Channel Subunit KV12 |
| ENSDARG00000069583 | ENSFM00800002003448 | Cytosolic Non Specific Dipeptidase EC_3.4.13.18 Cndp Dipeptidase 2 |
| ENSDARG00000069600 | ENSFM00250000001269 | Wiskott Aldrich Syndrome Family Member 1 Wasp Family Member 1 Wave 1 |
| ENSDARG00000069603 | ENSFM00250000000892 | Serine/Threonine Kinase EC_2.7.11.1 Thousand And One Amino Acid |
| ENSDARG00000069608 | ENSFM00740001589417 | A Kinase Anchor 2 Akap 2 Akap Kl Kinase A Anchoring 2 PRKA2 |
| ENSDARG00000069615 | ENSFM00800002003437 | Creatine Kinase U Type Mitochondrial Precursor EC_2.7.3.2 Acidic Type Mitochondrial Creatine Kinase Mia Ck Ubiquitous Mitochondrial Creatine Kinase U Mtck |
| ENSDARG00000069649 | ENSFM00500000289607 | Transmembrane 160 |
| ENSDARG00000069654 | ENSFM00250000001098 | Serine/Threonine Phosphatase 6 Regulatory Subunit Saps Domain Family Member |
| ENSDARG00000069671 | ENSFM00250000009079 | Uncharacterized Mitochondrial Precursor |
| ENSDARG00000069673 | ENSFM00250000002832 | Chitinase Domain Containing 1 Precursor |
| ENSDARG00000069734 | ENSFM00500000270525 | Hemoglobin Subunit Beta Beta Globin Hemoglobin Beta Chain |
| ENSDARG00000069735 | ENSFM00500000270029 | Hemoglobin Subunit Alpha Alpha Globin Hemoglobin Alpha Chain |
| ENSDARG00000069742 | ENSFM00730001522203 | Cullin 5 Cul 5 Vasopressin Activated Calcium Mobilizing Receptor 1 Vacm 1 |
| ENSDARG00000069748 | ENSFM00730001522149 | Calpain |
| ENSDARG00000069765 | ENSFM00250000000610 | Ras Gtpase Activating |
| ENSDARG00000069774 | ENSFM00780001898447 | Flotillin 2 |
| ENSDARG00000069832 | ENSFM00800002004131 | Sideroflexin 4 |
| ENSDARG00000069843 | ENSFM00260000050545 | Btb/Poz Domain Containing |
| ENSDARG00000069846 | ENSFM00250000005161 | ES1 Mitochondrial Precursor |
| ENSDARG00000069878 | ENSFM00670001235480 | Unknown |
| ENSDARG00000069940 | ENSFM00260000050433 | Lipid Phosphate Phosphohydrolase EC_3.1.3.4 PAP2 Phosphatidate Phosphohydrolase Type Phosphatidic Acid Phosphatase Pap |
| ENSDARG00000069941 | ENSFM00250000006394 | Tyrosyl Dna Phosphodiesterase 2 Tyr Dna Phosphodiesterase 2 EC_3.1.4.- 5' Tyrosyl Dna Phosphodiesterase 5' Tyr Dna Phosphodiesterase Traf And Tnf Receptor Associated |
| ENSDARG00000069951 | ENSFM00800002003364 | Elongation Factor 1 Alpha Ef 1 Alpha Eukaryotic Elongation Factor 1 A EEF1A |
| ENSDARG00000069970 | ENSFM00250000001884 | Leucine Rich Repeat And Immunoglobulin Domain Containing Nogo Receptor Interacting Precursor Leucine Rich Repeat Neuronal |
| ENSDARG00000069977 | ENSFM00250000003351 | MYG1 Mitochondrial Precursor |
| ENSDARG00000069980 | ENSFM00500000270950 | Ergic 53 Precursor Er Golgi Intermediate Compartment 53 Kda Lectin Mannose Binding 1 |
| ENSDARG00000069981 | ENSFM00760001715141 | Chondroitin Sulfate Proteoglycan 5 Precursor Acidic Leucine Rich Egf Domain Containing Brain Neuroglycan C |
| ENSDARG00000070029 | ENSFM00760001715025 | Peroxisomal Bifunctional Enzyme Pbe Pbfe |
| ENSDARG00000070043 | ENSFM00250000003932 | Aspartate Trna Ligase Cytoplasmic EC_6.1.1.12 Aspartyl Trna Synthetase Asprs |
| ENSDARG00000070074 | ENSFM00740001589117 | Low Density Lipoprotein Receptor Receptor |
| ENSDARG00000070080 | ENSFM00730001521650 | Neurobeachin |
| ENSDARG00000070083 | ENSFM00790001962974 | Atp Synthase Subunit Beta Mitochondrial EC_3.6.3.14 |
| ENSDARG00000070097 | ENSFM00500000273466 | Selenoprotein S Sels Vcp Interacting Membrane |
| ENSDARG00000070100 | ENSFM00740001589251 | Ras And Rab Interactor Ras Interaction/Interference |
| ENSDARG00000070124 | ENSFM00500000271580 | Tyrosine Phosphatase Non Receptor Type 23 EC_3.1.3.48 His Domain Containing Tyrosine Phosphatase Hd Ptp Tyrosine Phosphatase TD14 Ptp TD14 |
| ENSDARG00000070140 | ENSFM00250000003890 | All Trans Retinol 13 14 Reductase Precursor EC_1.3.99.23 All Trans 13 14 Dihydroretinol Saturase Retsat Ppar Alpha Regulated And Starvation Induced Gene |
| ENSDARG00000070145 | ENSFM00250000000552 | Membrane Associated Guanylate Kinase Ww And Pdz Domain Containing Membrane Associated Guanylate Kinase Inverted Magi |
| ENSDARG00000070173 | ENSFM00720001492020 | Glutamate Receptor Precursor Glur Ampa Selective Glutamate Receptor Glur Glutamate Receptor Ionotropic Ampa |
| ENSDARG00000070230 | ENSFM00730001521877 | Cytosolic 10 Formyltetrahydrofolate Dehydrogenase 10 Fthfdh Fdh EC_1.5.1.6 Aldehyde Dehydrogenase Family 1 Member L1 |
| ENSDARG00000070393 | ENSFM00250000002643 | Coenzyme Q Binding COQ10 Homolog Mitochondrial Precursor |
| ENSDARG00000070399 | ENSFM00800002003805 | Alpha 1 3/1 6 Mannosyltransferase ALG2 EC_2.4.1.132 EC_2.4.1.- 257 Asparagine Linked Glycosylation 2 Homolog Gdp Man:Man 1 Glcnac 2 Pp Dol Alpha 1 3 Mannosyltransferase Gdp Man:Man 1 Glcnac 2 Pp Dolichol Mannosyltransferase Gdp Man:Man 2 Glcnac 2 Pp Dol A |
| ENSDARG00000070433 | ENSFM00250000004354 | Vacuolar Sorting Associated 18 |
| ENSDARG00000070435 | ENSFM00250000001879 | Vesicle Associated Membrane Associated A Vamp A Vamp Associated A Vap A |
| ENSDARG00000070437 | ENSFM00600000921476 | 60S Ribosomal L22 |
| ENSDARG00000070441 | ENSFM00500000270112 | Palmitoyltransferase EC_2.3.1.225 Huntingtin Interacting 14 Zinc Finger Dhhc Domain Containing Dhhc |
| ENSDARG00000070442 | ENSFM00730001522534 | Semaphorin 7A Precursor Semaphorin K1 Sema K1 Semaphorin L Sema L CD108 Antigen |
| ENSDARG00000070448 | ENSFM00730001521304 | G Coupled Receptor Kinase G Coupled Receptor Kinase |
| ENSDARG00000070487 | ENSFM00500000270661 | Mitochondrial Amidoxime Reducing Component 2 Precursor MARC2 Ec 1 Molybdenum Cofactor Sulfurase C Terminal Domain Containing 2 Mosc Domain Containing 2 Moco Sulfurase C Terminal Domain Containing 2 |
| ENSDARG00000070491 | ENSFM00760001714608 | Unknown |
| ENSDARG00000070498 | ENSFM00700001391940 | Unknown |
| ENSDARG00000070498 | ENSFM00250000003309 | Phytanoyl Coa Hydroxylase Interacting Phytanoyl Coa Hydroxylase Associated 1 Pahx AP1 PAHXAP1 |
| ENSDARG00000070511 | ENSFM00720001492013 | Glutamate Receptor Ionotropic Kainate Precursor Glutamate Receptor |
| ENSDARG00000070543 | ENSFM00730001521500 | Glutamate Receptor Ionotropic Nmda Precursor Glutamate Receptor Subunit Epsilon N Methyl D Aspartate Receptor Subtype |
| ENSDARG00000070560 | ENSFM00500000269862 | Nucleosome Assembly 1 |
| ENSDARG00000070567 | ENSFM00250000000780 | Calcium Dependent Secretion Activator 1 Calcium Dependent Activator Secretion 1 Caps 1 |
| ENSDARG00000070573 | ENSFM00730001521833 | Mitogen Activated Kinase 1 Map Kinase 1 Mapk 1 EC_2.7.11.24 ERT1 Extracellular Signal Regulated Kinase 2 Erk 2 Mitogen Activated Kinase 2 Map Kinase 2 Mapk 2 |
| ENSDARG00000070575 | ENSFM00500000271560 | Unknown |
| ENSDARG00000070603 | ENSFM00250000005151 | Unknown |
| ENSDARG00000070620 | ENSFM00730001521500 | Glutamate Receptor Ionotropic Nmda Precursor Glutamate Receptor Subunit Epsilon N Methyl D Aspartate Receptor Subtype |
| ENSDARG00000070626 | ENSFM00250000002019 | Voltage Dependent Calcium Channel Gamma Subunit Neuronal Voltage Gated Calcium Channel Gamma Subunit Transmembrane Ampar Regulatory Gamma Tarp Gamma |
| ENSDARG00000070657 | ENSFM00250000003247 | Proliferation Associated 2G4 |
| ENSDARG00000070674 | ENSFM00760001714781 | 26S Proteasome Non Atpase Regulatory Subunit 6 26S Proteasome Regulatory Subunit RPN7 26S Proteasome Regulatory Subunit S10 P42A |
| ENSDARG00000070675 | ENSFM00250000001936 | Precursor |
| ENSDARG00000070682 | ENSFM00260000051009 | 45 Kda Calcium Binding Precursor CAB45 Stromal Cell Derived Factor 4 Sdf 4 |
| ENSDARG00000070686 | ENSFM00500000271211 | Cdp Diacylglycerol Inositol 3 Phosphatidyltransferase EC_2.7.8.11 Phosphatidylinositol Synthase Pi Synthase Ptdins Synthase |
| ENSDARG00000070688 | ENSFM00760001714608 | Unknown |
| ENSDARG00000070717 | ENSFM00500000270170 | Mitochondrial Glutamate Carrier 1 Gc 1 Glutamate/H + Symporter 1 Solute Carrier Family 25 Member 22 |
| ENSDARG00000070723 | ENSFM00250000004385 | Nadh Dehydrogenase 1 Alpha Subcomplex Subunit 9 Mitochondrial Precursor Complex I 39KD Ci 39KD Nadh Ubiquinone Oxidoreductase 39 Kda Subunit |
| ENSDARG00000070787 | ENSFM00250000001012 | Catenin Beta Beta Catenin |
| ENSDARG00000070801 | ENSFM00250000001929 | Dynamin 120 Kda Protein Mitochondrial Precursor EC_3.6.5.5 Optic Atrophy 1 |
| ENSDARG00000070822 | ENSFM00610000953338 | Unknown |
| ENSDARG00000070822 | ENSFM00800002024428 | Unknown |
| ENSDARG00000070824 | ENSFM00250000005807 | Nadh Dehydrogenase 1 Beta Subcomplex Subunit 5 Mitochondrial Precursor Complex I Sgdh Ci Sgdh Nadh Ubiquinone Oxidoreductase Sgdh Subunit |
| ENSDARG00000070849 | ENSFM00250000003189 | 40S Ribosomal S15 |
| ENSDARG00000070866 | ENSFM00250000007165 | Transmembrane 11 Mitochondrial |
| ENSDARG00000070919 | ENSFM00730001521728 | Copine Copine |
| ENSDARG00000070952 | ENSFM00500000271560 | Unknown |
| ENSDARG00000071004 | ENSFM00250000002071 | Monoacylglycerol Lipase ABHD12 EC_3.1.1.23 2 Arachidonoylglycerol Hydrolase Abhydrolase Domain Containing 12 |
| ENSDARG00000071014 | ENSFM00560000773950 | S100U |
| ENSDARG00000071017 | ENSFM00250000001233 | 5' Nucleotidase Precursor 5' Nt EC_3.1.3.5 Ecto 5' Nucleotidase |
| ENSDARG00000071031 | ENSFM00730001521918 | Sorting Nexin |
| ENSDARG00000071065 | ENSFM00760001714666 | Glucose 6 Phosphate 1 Dehydrogenase G6PD EC_1.1.1.49 |
| ENSDARG00000071076 | ENSFM00250000000475 | L Lactate Dehydrogenase Chain Ldh EC_1.1.1.27 |
| ENSDARG00000071192 | ENSFM00760001714761 | Gap Junction Alpha 1 Connexin 43 CX43 |
| ENSDARG00000071230 | ENSFM00260000050443 | Leucine Rich Repeat And Fibronectin Type Iii Domain Containing Precursor Synaptic Adhesion Molecule |
| ENSDARG00000071331 | ENSFM00250000000332 | Ryanodine Receptor Ryr Muscle Calcium Release Channel Muscle Type Ryanodine Receptor Type Ryanodine Receptor |
| ENSDARG00000071336 | ENSFM00670001235997 | Macrophage Migration Inhibitory Factor Mif EC_5.3.2.1 L Dopachrome Isomerase L Dopachrome Tautomerase EC_5.3.3.- 12 Phenylpyruvate Tautomerase |
| ENSDARG00000071353 | ENSFM00500000311464 | Uncharacterized |
| ENSDARG00000071357 | ENSFM00310000089004 | Mitogen Activated Kinase Kinase Kinase Kinase EC_2.7.11.1 Kinase Mapk/Erk Kinase Kinase Kinase Mek Kinase Kinase Mekkk |
| ENSDARG00000071374 | ENSFM00400000131776 | Leucine Rich Repeat Transmembrane Neuronal Precursor |
| ENSDARG00000071378 | ENSFM00610000964234 | Unknown |
| ENSDARG00000071378 | ENSFM00780001898920 | QIL1 |
| ENSDARG00000071395 | ENSFM00250000000111 | Calcium/Calmodulin Dependent Kinase Type Ii Subunit Cam Kinase Ii Subunit Camk Ii Subunit EC_2.7.11.17 |
| ENSDARG00000071396 | ENSFM00250000005363 | Mitochondrial Import Inner Membrane Translocase Subunit TIM23 |
| ENSDARG00000071409 | ENSFM00760001714815 | Adp Ribosylation Factor 3 |
| ENSDARG00000071424 | ENSFM00250000001618 | Ap 3 Complex Subunit Delta 1 Ap 3 Complex Subunit Delta Adaptor Related Complex 3 Subunit Delta 1 Delta Adaptin |
| ENSDARG00000071426 | ENSFM00250000006592 | Leucine Rich Repeat Containing 59 |
| ENSDARG00000071437 | ENSFM00800002009056 | Unknown |
| ENSDARG00000071437 | ENSFM00740001589127 | Receptor Type Tyrosine Phosphatase Precursor Tyrosine Phosphatase R Ptp EC_3.1.3.48 |
| ENSDARG00000071454 | ENSFM00500000273107 | Pentraxin Fusion Precursor |
| ENSDARG00000071493 | ENSFM00320000100099 | Noelin Precursor Neuronal Olfactomedin Related Er Localized Olfactomedin 1 |
| ENSDARG00000071524 | ENSFM00730001521215 | Insulin Receptor Precursor EC_2.7.10.1 |
| ENSDARG00000071551 | ENSFM00250000004190 | X Ray Repair Cross Complementing 6 EC_3.6.4.- EC_4.2.-.- 99 5' Deoxyribose 5 Phosphate Lyase KU70 5' Lyase KU70 Atp Dependent Dna Helicase 2 Subunit 1 Atp Dependent Dna Helicase Ii 70 Kda Subunit Ctc Box Binding Factor 75 Kda Subunit CTC75 Ctcbf Dna Repai |
| ENSDARG00000071566 | ENSFM00420000140518 | Serine/Threonine Phosphatase PP1 Catalytic Subunit Pp EC_3.1.3.16 |
| ENSDARG00000071573 | ENSFM00610000959757 | Unknown |
| ENSDARG00000071573 | ENSFM00670001236563 | U6 Snrna Associated Sm LSM5 |
| ENSDARG00000071594 | ENSFM00250000006057 | Mitochondrial Import Inner Membrane Translocase Subunit TIM21 Precursor TIM21 Protein Mitochondrial |
| ENSDARG00000071658 | ENSFM00730001521154 | Unknown |
| ENSDARG00000071662 | ENSFM00740001593996 | Unknown |
| ENSDARG00000071679 | ENSFM00670001236674 | Precursor Interleukin 25 Il 25 Stromal Cell Derived Growth Factor SF20 |
| ENSDARG00000071691 | ENSFM00250000003169 | Cytochrome B C1 Complex Subunit 2 Mitochondrial Precursor Complex Iii Subunit 2 Core Ii Ubiquinol Cytochrome C Reductase Complex Core 2 |
| ENSDARG00000071709 | ENSFM00250000001450 | Neurabin 1 Neurabin I Neural Tissue Specific F Actin Binding I Phosphatase 1 Regulatory Subunit 9A |
| ENSDARG00000071774 | ENSFM00250000008520 | Eukaryotic Translation Elongation Factor 1 Epsilon 1 Elongation Factor P18 Multisynthase Complex Auxiliary Component P18 |
| ENSDARG00000071881 | ENSFM00500000273845 | Adp Ribosylation Factor 9 |
| ENSDARG00000071906 | ENSFM00750001632389 | Transport SEC24A SEC24 Related A |
| ENSDARG00000073707 | ENSFM00500000270149 | Connector Enhancer Of Kinase Suppressor Of Ras Connector Enhancer Of Ksr Cnk Homolog |
| ENSDARG00000073710 | ENSFM00800002003516 | Ubiquitin Carboxyl Terminal Hydrolase 7 EC_3.4.19.12 Deubiquitinating Enzyme 7 Herpesvirus Associated Ubiquitin Specific Protease Ubiquitin Thioesterase 7 Ubiquitin Specific Processing Protease 7 |
| ENSDARG00000073713 | ENSFM00260000050653 | Netrin Precursor Laminet |
| ENSDARG00000073724 | ENSFM00800002003951 | Nlr Family Member X1 Precursor |
| ENSDARG00000073732 | ENSFM00270000056424 | Myosin Cellular Myosin Heavy Chain Type Myosin Heavy Chain Myosin Heavy Chain Non Muscle Non Muscle Myosin Heavy Chain Nmmhc Non Muscle Myosin Heavy Chain Nmmhc Ii Nmmhc |
| ENSDARG00000073741 | ENSFM00250000002946 | RAP1 Gtpase Gdp Dissociation Stimulator 1 Exchange Factor Smggds Smg Gds Smg P21 Stimulatory Gdp/Gtp Exchange |
| ENSDARG00000073752 | ENSFM00610000969046 | Unknown |
| ENSDARG00000073764 | ENSFM00720001492028 | Sterol 26 Hydroxylase Mitochondrial Precursor EC_1.14.13.15 5 Beta Cholestane 3 Alpha 7 Alpha 12 Alpha Triol 27 Hydroxylase Cytochrome P 450C27/25 Cytochrome P450 27 Sterol 27 Hydroxylase Vitamin D 3 25 Hydroxylase |
| ENSDARG00000073765 | ENSFM00730001522052 | Guanine Nucleotide Binding Subunit Alpha G Alpha G Subunit Alpha |
| ENSDARG00000073769 | ENSFM00250000000552 | Membrane Associated Guanylate Kinase Ww And Pdz Domain Containing Membrane Associated Guanylate Kinase Inverted Magi |
| ENSDARG00000073792 | ENSFM00780001898478 | Endoplasmic Reticulum Mannosyl Oligosaccharide 1 2 Alpha Mannosidase EC_3.2.1.113 Er Alpha 1 2 Mannosidase Er Mannosidase 1 ERMAN1 MAN9GLCNAC2 Specific Processing Alpha Mannosidase Mannosidase Alpha Class 1B Member 1 |
| ENSDARG00000073822 | ENSFM00250000001463 | Iq Motif And SEC7 Domain Containing |
| ENSDARG00000073824 | ENSFM00250000001613 | Ras Specific Guanine Nucleotide Releasing Factor 2 Ras GRF2 Ras Guanine Nucleotide Exchange Factor 2 |
| ENSDARG00000073841 | ENSFM00740001589186 | E3 Ubiquitin Ligase HERC2 EC_6.3.2.- Hect Domain And RCC1 Domain Containing 2 |
| ENSDARG00000073843 | ENSFM00510000502753 | Unconventional Myosin Unconventional Myosin |
| ENSDARG00000073848 | ENSFM00250000001135 | Rho Guanine Nucleotide Exchange Factor 6 Rac/CDC42 Guanine Nucleotide Exchange Factor 6 |
| ENSDARG00000073872 | ENSFM00250000002945 | Tyrosine Sulfotransferase EC_2.8.2.20 Tyrosylprotein Sulfotransferase Tpst |
| ENSDARG00000073883 | ENSFM00250000001322 | Calsyntenin Precursor Alcadein Alc |
| ENSDARG00000073896 | ENSFM00730001521860 | Gap Junction Gamma 1 Connexin 45 CX45 Gap Junction Alpha 7 |
| ENSDARG00000073902 | ENSFM00500000271161 | Tyrosine Phosphatase Non Receptor Type 9 EC_3.1.3.48 Tyrosine Phosphatase MEG2 Ptpase MEG2 |
| ENSDARG00000073905 | ENSFM00260000050708 | Von Willebrand Factor A Domain Containing 5A Loss Of Heterozygosity 11 Chromosomal Region 2 Gene A |
| ENSDARG00000073923 | ENSFM00790001963134 | Uncharacterized Aarf Domain Containing Kinase 2 EC_2.7.11.- |
| ENSDARG00000073931 | ENSFM00800002003852 | Ubiquitin Thioesterase Otulin EC_3.4.19.12 Deubiquitinating Enzyme Otulin Otu Domain Containing Deubiquitinase With Linear Linkage Specificity Ubiquitin Thioesterase Gumby |
| ENSDARG00000074041 | ENSFM00570000851109 | Atp Binding Cassette Sub Family A Member |
| ENSDARG00000074059 | ENSFM00360000109935 | Disks Large Homolog 5 Discs Large P Dlg Placenta And Prostate Dlg |
| ENSDARG00000074073 | ENSFM00250000004498 | Microtubule Associated 6 Map 6 Stable Tubule Only Polypeptide Stop |
| ENSDARG00000074084 | ENSFM00740001589092 | Adenosylhomocysteinase Adohcyase EC_3.3.1.1 S Adenosyl L Homocysteine Hydrolase 2 |
| ENSDARG00000074131 | ENSFM00670001235340 | Kinesin Heavy Chain Kinesin Heavy Chain Kinesin Heavy Chain |
| ENSDARG00000074149 | ENSFM00250000000547 | Inositol 1 4 5 Trisphosphate Receptor Type IP3 Receptor IP3R Ins Type Inositol 1 4 5 Trisphosphate Receptor Type INSP3 Receptor |
| ENSDARG00000074153 | ENSFM00250000001398 | Slit And Ntrk Precursor |
| ENSDARG00000074169 | ENSFM00600000921257 | Glycerol 3 Phosphate Acyltransferase 1 Mitochondrial Precursor Gpat 1 EC_2.3.1.15 |
| ENSDARG00000074183 | ENSFM00320000100117 | Diacylglycerol Kinase Eta |
| ENSDARG00000074201 | ENSFM00740001589123 | Filamin Fln Abp 280 Actin Binding Filamin |
| ENSDARG00000074235 | ENSFM00500000355992 | Uncharacterized |
| ENSDARG00000074242 | ENSFM00500000270904 | Plasminogen Activator Inhibitor 1 Rna Binding PAI1 Rna Binding 1 Pai RBP1 SERPINE1 Binding 1 |
| ENSDARG00000074246 | ENSFM00800002003610 | Ras Related Rab 14 |
| ENSDARG00000074255 | ENSFM00260000050554 | Calcium Uptake 2 Mitochondrial Precursor Ef Hand Domain Containing Family Member A1 |
| ENSDARG00000074271 | ENSFM00250000008577 | Tail Anchored Insertion Receptor Wrb Tryptophan Rich Basic Wrb |
| ENSDARG00000074275 | ENSFM00250000003304 | Lim And Calponin Homology Domains Containing 1 |
| ENSDARG00000074282 | ENSFM00730001521472 | Lysosomal Alpha Glucosidase Precursor EC_3.2.1.20 Acid Maltase |
| ENSDARG00000074289 | ENSFM00800002017874 | Unknown |
| ENSDARG00000074289 | ENSFM00250000000217 | Tubulin Alpha Chain |
| ENSDARG00000074308 | ENSFM00250000005430 | Leucine Rich Repeat Containing Leucine Rich Repeat Containing |
| ENSDARG00000074309 | ENSFM00250000000938 | Metal Transporter Ancient Conserved Domain Containing Cyclin |
| ENSDARG00000074320 | ENSFM00780001898357 | Voltage Gated Potassium Channel Subunit Beta K + Channel Subunit Beta Kv Beta |
| ENSDARG00000074328 | ENSFM00250000001217 | Amyloid Beta A4 Precursor Binding Family A Member Adapter Neuron Specific Neuronal MUNC18 1 Interacting Mint |
| ENSDARG00000074329 | ENSFM00730001521847 | Catenin Delta 1 Cadherin Associated Src Substrate Cas P120 Catenin P120 Ctn P120 Cas |
| ENSDARG00000074367 | ENSFM00760001714668 | Ubiquitin Carboxyl Terminal Hydrolase 12 EC_3.4.19.12 Deubiquitinating Enzyme 12 Ubiquitin Thioesterase 12 Ubiquitin Specific Processing Protease 12 |
| ENSDARG00000074372 | ENSFM00250000002984 | Phosphatase 1 Regulatory Subunit 29 Precursor Extracellular Leucine Rich Repeat And Fibronectin Type Iii Domain Containing 2 Leucine Rich Repeat And Fibronectin Type Iii Domain Containing 6 Leucine Rich Repeat Containing 62 |
| ENSDARG00000074376 | ENSFM00250000002191 | Mam Domain Containing Glycosylphosphatidylinositol Anchor 2 Precursor Mam Domain Containing 1 |
| ENSDARG00000074381 | ENSFM00270000056477 | Ferm Rhogef And Pleckstrin Domain Containing |
| ENSDARG00000074419 | ENSFM00250000000931 | Potassium/Sodium Hyperpolarization Activated Cyclic Nucleotide Gated Channel |
| ENSDARG00000074431 | ENSFM00810002040719 | Dna Damage Binding 1 Damage Specific Dna Binding 1 |
| ENSDARG00000074435 | ENSFM00250000005373 | Tetratricopeptide Repeat 19 Mitochondrial Precursor Tpr Repeat 19 |
| ENSDARG00000074443 | ENSFM00250000005898 | Growth Arrest Specific 7 Gas 7 |
| ENSDARG00000074465 | ENSFM00250000000977 | Vacuolar Sorting Associated 13A Chorea Acanthocytosis Chorein |
| ENSDARG00000074480 | ENSFM00500000270149 | Connector Enhancer Of Kinase Suppressor Of Ras Connector Enhancer Of Ksr Cnk Homolog |
| ENSDARG00000074490 | ENSFM00250000003320 | CASC4 Cancer Susceptibility Candidate Gene 4 |
| ENSDARG00000074507 | ENSFM00500000271951 | Regulator Of Microtubule Dynamics 1 Rmd 1 FAM82B |
| ENSDARG00000074521 | ENSFM00250000004498 | Microtubule Associated 6 Map 6 Stable Tubule Only Polypeptide Stop |
| ENSDARG00000074524 | ENSFM00250000000380 | Contactin Associated 5 Precursor Cell Recognition Molecule CASPR5 |
| ENSDARG00000074531 | ENSFM00250000000947 | C Jun Amino Terminal Kinase Interacting Jip Jnk Interacting Jnk Mitogen Activated Kinase 8 Interacting |
| ENSDARG00000074533 | ENSFM00500000271856 | Solute Carrier Family 25 Member 38 |
| ENSDARG00000074535 | ENSFM00250000001884 | Leucine Rich Repeat And Immunoglobulin Domain Containing Nogo Receptor Interacting Precursor Leucine Rich Repeat Neuronal |
| ENSDARG00000074552 | ENSFM00250000003821 | Nadh Dehydrogenase Iron Sulfur 7 Mitochondrial Precursor EC_1.6.5.3 EC_1.6.99.- 3 Complex I 20KD Ci 20KD Nadh Ubiquinone Oxidoreductase 20 Kda Subunit |
| ENSDARG00000074558 | ENSFM00250000000380 | Contactin Associated 5 Precursor Cell Recognition Molecule CASPR5 |
| ENSDARG00000074581 | ENSFM00250000000681 | Adducin |
| ENSDARG00000074583 | ENSFM00800002014678 | Unknown |
| ENSDARG00000074583 | ENSFM00720001492054 | Glutamate Receptor Ionotropic Delta Precursor Glur Delta Subunit |
| ENSDARG00000074611 | ENSFM00250000004113 | Wd Repeat Containing 37 |
| ENSDARG00000074617 | ENSFM00280000058784 | Wolframin |
| ENSDARG00000074671 | ENSFM00500000269860 | Neuronal Pentraxin 2 Precursor NP2 Neuronal Pentraxin Ii Np Ii |
| ENSDARG00000074680 | ENSFM00610000972522 | Unknown |
| ENSDARG00000074680 | ENSFM00250000000629 | Regulating Synaptic Membrane Exocytosis Rab 3 Interacting Molecule Rim |
| ENSDARG00000074690 | ENSFM00250000002013 | Phosphatase EC_3.1.3.16 |
| ENSDARG00000074723 | ENSFM00720001492050 | Pleckstrin Homology Domain Containing Family H Member 3 Precursor Ph Domain Containing Family H Member 3 |
| ENSDARG00000074727 | ENSFM00250000002411 | Dnaj Homolog Subfamily C Member 10 Precursor EC_1.8.4.- Endoplasmic Reticulum Dna J Domain Containing 5 Er Resident ERDJ5 ERDJ5 |
| ENSDARG00000074746 | ENSFM00300000079128 | Potassium Voltage Gated Channel Subfamily D Member Voltage Gated Potassium Channel Subunit KV4 |
| ENSDARG00000074777 | ENSFM00500000269628 | Ankyrin Ank Ankyrin |
| ENSDARG00000074791 | ENSFM00250000000672 | Aldehyde Dehydrogenase Aldehyde Dehydrogenase Aldehyde Dehydrogenase Family 3 Member |
| ENSDARG00000074844 | ENSFM00800002004636 | Cdna Cssl: |
| ENSDARG00000074865 | ENSFM00250000001492 | Ferm And Pdz Domain Containing 4 Pdz Domain Containing 10 Psd 95 Interacting Regulator Of Spine Morphogenesis Preso |
| ENSDARG00000074866 | ENSFM00280000058726 | Tyrosine Phosphatase Non Receptor Type EC_3.1.3.48 Tyrosine Phosphatase Tyrosine Phosphatase |
| ENSDARG00000074876 | ENSFM00800002003396 | Atp Binding Cassette Sub Family D Member Adrenoleukodystrophy |
| ENSDARG00000074894 | ENSFM00540000717964 | Uncharacterized Family 31 Glucosidase EC_3.2.1.- |
| ENSDARG00000074899 | ENSFM00250000000892 | Serine/Threonine Kinase EC_2.7.11.1 Thousand And One Amino Acid |
| ENSDARG00000075007 | ENSFM00500000270226 | Carbohydrate Sulfotransferase EC_2.8.2.- Galactose/N Acetylglucosamine/N Acetylglucosamine 6 O Sulfotransferase N Acetylglucosamine 6 O Sulfotransferase GLCNAC6ST Sulfotransferase N Acetylglucosamine 6 O Sulfotransferase GLCNAC6ST GN6ST |
| ENSDARG00000075016 | ENSFM00740001589094 | Apolipoprotein B 100 Precursor Apo B 100 |
| ENSDARG00000075041 | ENSFM00350000105407 | Formin |
| ENSDARG00000075083 | ENSFM00250000002645 | Dna Dependent Kinase Catalytic Subunit Dna Pk Catalytic Subunit Dna Pkcs EC_2.7.11.1 |
| ENSDARG00000075098 | ENSFM00250000004932 | Wd Repeat Containing 17 |
| ENSDARG00000075100 | ENSFM00800002020697 | Unknown |
| ENSDARG00000075100 | ENSFM00260000050557 | Netrin Receptor Dcc Precursor Tumor Suppressor Dcc |
| ENSDARG00000075110 | ENSFM00250000000610 | Ras Gtpase Activating |
| ENSDARG00000075132 | ENSFM00250000003390 | Fumarate Hydratase Mitochondrial Precursor Fumarase EC_4.2.1.2 |
| ENSDARG00000075133 | ENSFM00250000000737 | Brain Specific Angiogenesis Inhibitor Precursor |
| ENSDARG00000075141 | ENSFM00670001235445 | G Coupled Receptor Family C Group 5 Member Retinoic Acid Induced Gene Raig |
| ENSDARG00000075159 | ENSFM00250000000343 | Precursor |
| ENSDARG00000075180 | ENSFM00670001238331 | Transmembrane 14C |
| ENSDARG00000075189 | ENSFM00250000000380 | Contactin Associated 5 Precursor Cell Recognition Molecule CASPR5 |
| ENSDARG00000075192 | ENSFM00800002003559 | Atp Dependent Zinc Metalloprotease Ym EC_3.4.24.- Atp Dependent Metalloprotease FTSH1 Meg 4 YME1 1 |
| ENSDARG00000075201 | ENSFM00250000001432 | Type Inositol 3 4 Bisphosphate 4 Phosphatase EC_3.1.3.66 Inositol Polyphosphate 4 Phosphatase Type |
| ENSDARG00000075245 | ENSFM00250000005384 | Wd Repeat Containing 11 Bromodomain And Wd Repeat Containing 2 |
| ENSDARG00000075249 | ENSFM00740001589308 | Precursor |
| ENSDARG00000075285 | ENSFM00770001847543 | Ras Related R Ras |
| ENSDARG00000075327 | ENSFM00500000269738 | Dedicator Of Cytokinesis CDC42 Guanine Nucleotide Exchange Factor Zizimin |
| ENSDARG00000075362 | ENSFM00250000001412 | Whirlin |
| ENSDARG00000075376 | ENSFM00250000001203 | Small G Signaling Modulator Run And TBC1 Domain Containing |
| ENSDARG00000075382 | ENSFM00730001521509 | Sodium/Hydrogen Exchanger Na + /H + Exchanger Nhe Solute Carrier Family 9 Member |
| ENSDARG00000075445 | ENSFM00320000100083 | Proteasome Subunit Beta Type Precursor EC_3.4.25.1 Macropain Multicatalytic Endopeptidase Complex Proteasome Proteasome Subunit |
| ENSDARG00000075455 | ENSFM00250000001168 | Soga Family Member |
| ENSDARG00000075491 | ENSFM00500000270149 | Connector Enhancer Of Kinase Suppressor Of Ras Connector Enhancer Of Ksr Cnk Homolog |
| ENSDARG00000075505 | ENSFM00570000851634 | Prostaglandin F2 Receptor Negative Regulator Precursor Prostaglandin F2 Alpha Receptor Regulatory Prostaglandin F2 Alpha Receptor Associated CD315 Antigen |
| ENSDARG00000075536 | ENSFM00800002003432 | Copine Copine |
| ENSDARG00000075539 | ENSFM00250000002727 | Syntabulin Golgi Localized Syntaphilin Related Syntaxin 1 Binding |
| ENSDARG00000075556 | ENSFM00800002003954 | Leucine Zipper 1 Leucine Zipper Motif Containing |
| ENSDARG00000075580 | ENSFM00800002003728 | Leucine Rich Repeat And Calponin Homology Domain Containing 1 Calponin Homology Domain Containing 1 |
| ENSDARG00000075584 | ENSFM00250000004403 | T Cell Immunomodulatory Tip Integrin Alpha Fg Gap Repeat Containing 1 |
| ENSDARG00000075625 | ENSFM00800002011902 | Unknown |
| ENSDARG00000075625 | ENSFM00250000001262 | Leucine Rich Repeats And Immunoglobulin Domains Precursor Lig |
| ENSDARG00000075673 | ENSFM00250000001623 | Rho Gtpase Activating 21 Rho Type Gtpase Activating 21 |
| ENSDARG00000075677 | ENSFM00500000272736 | 39S Ribosomal L17 Mitochondrial Precursor L17MT Mrp L17 |
| ENSDARG00000075709 | ENSFM00540000734587 | Uncharacterized |
| ENSDARG00000075726 | ENSFM00560000771026 | Dehydrogenase/Reductase Sdr Family Member EC_1.1.-.- |
| ENSDARG00000075733 | ENSFM00730001521785 | Zyxin |
| ENSDARG00000075752 | ENSFM00400000131775 | Unconventional Myosin Xviiia Molecule Associated With JAK3 N Terminus Majn Myosin Containing A Pdz Domain |
| ENSDARG00000075754 | ENSFM00250000005225 | Methylthioribose 1 Phosphate Isomerase |
| ENSDARG00000075758 | ENSFM00730001521154 | Unknown |
| ENSDARG00000075768 | ENSFM00800002003507 | Succinate Dehydrogenase Iron Sulfur Subunit Mitochondrial Precursor EC_1.3.5.1 Iron Sulfur Subunit Of Complex Ii Ip |
| ENSDARG00000075793 | ENSFM00610000970147 | Unknown |
| ENSDARG00000075793 | ENSFM00250000001999 | Phosphatidylinositol 3 4 5 Trisphosphate Dependent Rac Exchanger 2 P REX2 Ptdins 3 4 5 Dependent Rac Exchanger 2 Dep Domain Containing 2 |
| ENSDARG00000075806 | ENSFM00250000000717 | Kin Of Irre Precursor Kin Of Irregular Chiasm Nephrin |
| ENSDARG00000075815 | ENSFM00260000050376 | Solute Carrier Family 12 Member Electroneutral Potassium Chloride Cotransporter K Cl Cotransporter |
| ENSDARG00000075830 | ENSFM00720001492041 | Synaptotagmin Synaptotagmin |
| ENSDARG00000075831 | ENSFM00250000000331 | Amino Acid Transporter Solute Carrier Family 7 Member |
| ENSDARG00000075853 | ENSFM00600000921328 | SH3 Domain Containing Kinase Binding 1 Of Kinase |
| ENSDARG00000075857 | ENSFM00750001632469 | Cdp Diacylglycerol Glycerol 3 Phosphate 3 Phosphatidyltransferase Mitochondrial Precursor EC_2.7.8.5 Phosphatidylglycerophosphate Synthase 1 Pgp Synthase 1 |
| ENSDARG00000075892 | ENSFM00250000003234 | Large Proline Rich BAG6 BCL2 Associated Athanogene 6 Hla B Associated Transcript 3 |
| ENSDARG00000075899 | ENSFM00260000050328 | Latrophilin |
| ENSDARG00000075924 | ENSFM00250000000899 | Rap Guanine Nucleotide Exchange Factor Exchange Factor Directly Activated By Camp Exchange Directly Activated By Camp Epac Camp Regulated Guanine Nucleotide Exchange Factor Camp |
| ENSDARG00000075933 | ENSFM00790001963059 | Cytochrome C Oxidase Assembly COX15 Homolog |
| ENSDARG00000075949 | ENSFM00250000000930 | Serine/Threonine Kinase EC_2.7.11.13 |
| ENSDARG00000075963 | ENSFM00500000269621 | Mhc Class I Antigen |
| ENSDARG00000075989 | ENSFM00800002003702 | Actin Related 2/3 Complex Subunit 2 ARP2/3 Complex 34 Kda Subunit P34 Arc |
| ENSDARG00000076001 | ENSFM00730001521611 | Phosphatidylinositol 4 Phosphate 5 Kinase Type 1 Beta Pi Beta Ptdins 4 P 5 Kinase 1 Beta EC_2.7.1.68 Phosphatidylinositol 4 Phosphate 5 Kinase Type I Phosphatidylinositol 4 Phosphate 5 Kinase Beta |
| ENSDARG00000076014 | ENSFM00250000000753 | Elks/RAB6 Interacting/Cast Family Member 1 Erc 1 RAB6 Interacting 2 |
| ENSDARG00000076027 | ENSFM00670001235340 | Kinesin Heavy Chain Kinesin Heavy Chain Kinesin Heavy Chain |
| ENSDARG00000076030 | ENSFM00250000000524 | Voltage Dependent L Type Calcium Channel Subunit Beta Calcium Channel Voltage Dependent Subunit Beta |
| ENSDARG00000076052 | ENSFM00250000001049 | Seizure 6 Precursor |
| ENSDARG00000076058 | ENSFM00250000003010 | Glucosylceramidase Precursor EC_3.2.1.45 Acid Beta Glucosidase Beta Glucocerebrosidase D Glucosyl N Acylsphingosine Glucohydrolase |
| ENSDARG00000076070 | ENSFM00250000000718 | Disks Large Associated Dap Psd 95/SAP90 Binding SAP90/Psd 95 Associated |
| ENSDARG00000076103 | ENSFM00500000273079 | Delphilin Glutamate Receptor Ionotropic Delta 2 Interacting 1 |
| ENSDARG00000076117 | ENSFM00500000273097 | Star Related Lipid Transfer 5 Start Domain Containing 5 STARD5 |
| ENSDARG00000076128 | ENSFM00730001521681 | Camp Dependent Kinase Type I Alpha Regulatory Camp Dependent Kinase Type I Alpha Regulatory Subunit N Terminally Processed |
| ENSDARG00000076155 | ENSFM00250000005018 | Poly A Rna Polymerase Mitochondrial Precursor Pap EC_2.7.7.19 Pap Associated Domain Containing 1 Polynucleotide Adenylyltransferase |
| ENSDARG00000076174 | ENSFM00750001632380 | Tripartite Motif Containing 2 EC_6.3.2.- E3 Ubiquitin Ligase TRIM2 |
| ENSDARG00000076179 | ENSFM00740001589196 | Ephexin 1 Eph Interacting Exchange Neuronal Guanine Nucleotide Exchange Factor |
| ENSDARG00000076213 | ENSFM00760001714721 | Dedicator Of Cytokinesis 3 Modifier Of Cell Adhesion Presenilin Binding Pbp |
| ENSDARG00000076268 | ENSFM00500000276193 | Uncharacterized |
| ENSDARG00000076290 | ENSFM00500000269786 | Calreticulin Precursor |
| ENSDARG00000076302 | ENSFM00500000270006 | E3 Ubiquitin Ligase EC_6.3.2.- Deltex |
| ENSDARG00000076310 | ENSFM00250000006718 | F Box Only 41 |
| ENSDARG00000076318 | ENSFM00250000002169 | V Type Proton Atpase Catalytic Subunit A V Atpase Subunit A EC_3.6.3.14 V Atpase 69 Kda Subunit Vacuolar Proton Pump Subunit Alpha |
| ENSDARG00000076324 | ENSFM00670001235536 | Serine/Threonine Kinase EC_2.7.11.1 Apoptosis Associated Tyrosine Kinase Brain Kinase Lemur Tyrosine Kinase |
| ENSDARG00000076334 | ENSFM00250000003611 | 39S Ribosomal L43 Mitochondrial Precursor L43MT Mrp L43 Mitochondrial Ribosomal BMRP36A |
| ENSDARG00000076364 | ENSFM00800002003400 | Unknown |
| ENSDARG00000076376 | ENSFM00250000006710 | Transmembrane 175 |
| ENSDARG00000076381 | ENSFM00730001522086 | Tricarboxylate Transport Protein Mitochondrial Precursor Citrate Transport Ctp Solute Carrier Family 25 Member 1 Tricarboxylate Carrier |
| ENSDARG00000076393 | ENSFM00290000065595 | Transmembrane 65 |
| ENSDARG00000076401 | ENSFM00250000002019 | Voltage Dependent Calcium Channel Gamma Subunit Neuronal Voltage Gated Calcium Channel Gamma Subunit Transmembrane Ampar Regulatory Gamma Tarp Gamma |
| ENSDARG00000076464 | ENSFM00250000006855 | Ragulator Complex LAMTOR1 Late Endosomal/Lysosomal Adaptor And Mapk And Mtor Activator 1 |
| ENSDARG00000076486 | ENSFM00500000271298 | Low Molecular Weight Phosphotyrosine Phosphatase Lmw Ptp Lmw Ptpase EC_3.1.3.48 Low Molecular Weight Cytosolic Acid Phosphatase EC_3.1.3.- 2 |
| ENSDARG00000076508 | ENSFM00250000000328 | Metabotropic Glutamate Receptor Precursor |
| ENSDARG00000076544 | ENSFM00500000269985 | Succinate Semialdehyde Dehydrogenase Mitochondrial Precursor EC_1.2.1.24 Aldehyde Dehydrogenase Family 5 Member A1 Nad + Dependent Succinic Semialdehyde Dehydrogenase |
| ENSDARG00000076568 | ENSFM00630001066521 | Unknown |
| ENSDARG00000076568 | ENSFM00670001236538 | Transport SEC61 Subunit Beta |
| ENSDARG00000076602 | ENSFM00750001659061 | Unknown |
| ENSDARG00000076602 | ENSFM00670001235488 | Thymus Specific Serine Protease Precursor EC_3.4.-.- Serine Protease 16 |
| ENSDARG00000076673 | ENSFM00740001589199 | Plectin Pcn Pltn Plectin 1 |
| ENSDARG00000076697 | ENSFM00730001521477 | Signal Induced Proliferation Associated 1 SIPA1 |
| ENSDARG00000076701 | ENSFM00250000002796 | Beta 1 4 N Acetylgalactosaminyltransferase 3 BETA4GALNAC T3 BETA4GALNACT3 EC_2.4.1.244 Beta 1 4 N Acetylgalactosaminyltransferase Iii N Acetyl Beta Glucosaminyl Glycoprotein 4 Beta N Acetylgalactosaminyltransferase 2 Ngalnac T2 |
| ENSDARG00000076702 | ENSFM00250000001069 | Ph And SEC7 Domain Containing 1 Exchange Factor ARF6 Pleckstrin Homology And SEC7 Domain Containing 1 |
| ENSDARG00000076718 | ENSFM00600000921352 | Risc Loading Complex Subunit TARBP2 |
| ENSDARG00000076724 | ENSFM00760001714819 | Phosphatidylinositol 4 Kinase Alpha PI4 Kinase Alpha PI4K Alpha Ptdins 4 Kinase Alpha EC_2.7.1.67 |
| ENSDARG00000076742 | ENSFM00250000001040 | Cytohesin Ph SEC7 And Coiled Coil Domain Containing |
| ENSDARG00000076754 | ENSFM00730001521509 | Sodium/Hydrogen Exchanger Na + /H + Exchanger Nhe Solute Carrier Family 9 Member |
| ENSDARG00000076757 | ENSFM00730001521338 | Ephrin Type B Receptor Kinase |
| ENSDARG00000076780 | ENSFM00730001521755 | Short/Branched Chain Specific Acyl Coa Dehydrogenase Mitochondrial Precursor Sbcad EC_1.3.8.5 2 Methyl Branched Chain Acyl Coa Dehydrogenase 2 Mebcad 2 Methylbutyryl Coenzyme A Dehydrogenase 2 Methylbutyryl Coa Dehydrogenase |
| ENSDARG00000076796 | ENSFM00250000000275 | Disks Large Homolog Synapse Associated Sap |
| ENSDARG00000076804 | ENSFM00250000001068 | Tweety Homolog |
| ENSDARG00000076815 | ENSFM00800002003567 | Eukaryotic Translation Initiation Factor 3 Subunit A |
| ENSDARG00000076826 | ENSFM00750001632364 | Dipeptidyl Aminopeptidase 6 Dppx Dipeptidyl Aminopeptidase Related Dipeptidyl Peptidase 6 Dipeptidyl Peptidase Iv Dipeptidyl Peptidase Vi Dpp Vi |
| ENSDARG00000076829 | ENSFM00400000131980 | Ankyrin Repeat And Ibr Domain Containing 1 |
| ENSDARG00000076833 | ENSFM00500000270479 | Sodium/Potassium Transporting Atpase Subunit Beta 1 Sodium/Potassium Dependent Atpase Subunit Beta 1 |
| ENSDARG00000076836 | ENSFM00500000272603 | Phospholipid Hydroperoxide Glutathione Peroxidase Mitochondrial Precursor Phgpx EC_1.11.1.12 Glutathione Peroxidase 4 Gpx 4 Gshpx 4 |
| ENSDARG00000076873 | ENSFM00500000276193 | Uncharacterized |
| ENSDARG00000076888 | ENSFM00810002040602 | Neutral Alpha Glucosidase Ab Precursor EC_3.2.1.84 Alpha Glucosidase 2 Glucosidase Ii Subunit Alpha |
| ENSDARG00000076889 | ENSFM00250000000944 | Ral Gtpase Activating Subunit Alpha 1 Gap Related Interacting Partner To E12 Gripe Gtpase Activating Domain 1 Tuberin 1 P240 |
| ENSDARG00000076923 | ENSFM00250000003884 | Kazrin |
| ENSDARG00000076962 | ENSFM00750001656366 | Unknown |
| ENSDARG00000076962 | ENSFM00250000001697 | Glycerophosphodiester Phosphodiesterase Domain Containing EC_3.1.-.- Glycerophosphodiester Phosphodiesterase |
| ENSDARG00000076970 | ENSFM00500000272331 | Dehydrogenase/Reductase Sdr Family Member 12 EC_1.1.-.- |
| ENSDARG00000077002 | ENSFM00740001589327 | Immunoglobulin Superfamily Member Precursor Glu Trp Ile Ewi Motif Containing Ewi Antigen |
| ENSDARG00000077004 | ENSFM00730001521877 | Cytosolic 10 Formyltetrahydrofolate Dehydrogenase 10 Fthfdh Fdh EC_1.5.1.6 Aldehyde Dehydrogenase Family 1 Member L1 |
| ENSDARG00000077023 | ENSFM00780001898335 | Protocadherin Precursor |
| ENSDARG00000077045 | ENSFM00250000006800 | Neuroendocrine 7B2 Precursor Secretogranin V Secretogranin 5 Secretory Granule Endocrine I |
| ENSDARG00000077047 | ENSFM00260000050522 | Receptor Type Tyrosine Phosphatase Precursor R Ptp |
| ENSDARG00000077053 | ENSFM00250000000441 | Liprin Alpha Tyrosine Phosphatase Receptor Type F Polypeptide Interacting Alpha Ptprf Interacting Alpha |
| ENSDARG00000077080 | ENSFM00670001235445 | G Coupled Receptor Family C Group 5 Member Retinoic Acid Induced Gene Raig |
| ENSDARG00000077087 | ENSFM00800002010337 | Unknown |
| ENSDARG00000077121 | ENSFM00250000002048 | Cytochrome P450 EC_1.14.-.- |
| ENSDARG00000077124 | ENSFM00740001589247 | Serine/Threonine Kinase BRSK2 EC_2.7.11.1 EC_2.7.11.- 26 Brain Specific Serine/Threonine Kinase 2 Br Serine/Threonine Kinase 2 Serine/Threonine Kinase Sad A |
| ENSDARG00000077134 | ENSFM00250000001603 | Probable G Coupled Receptor Precursor |
| ENSDARG00000077145 | ENSFM00730001521247 | Adenylate Cyclase Type EC_4.6.1.1 Atp Pyrophosphate Lyase Adenylate Cyclase Type Adenylyl Cyclase Ca 2+ Adenylyl Cyclase |
| ENSDARG00000077174 | ENSFM00250000001659 | Sulfhydryl Oxidase Precursor EC_1.8.3.2 Quiescin Q6 |
| ENSDARG00000077177 | ENSFM00790001963039 | Dual Specificity Mitogen Activated Kinase Kinase 4 Map Kinase Kinase 4 Mapkk 4 EC_2.7.12.2 Jnk Activating Kinase 1 Mapk/Erk Kinase 4 Mek 4 Sapk/Erk Kinase 1 SEK1 |
| ENSDARG00000077228 | ENSFM00730001521357 | Nt 3 Growth Factor Receptor Precursor EC_2.7.10.1 Neurotrophic Tyrosine Kinase Receptor Type 3 |
| ENSDARG00000077229 | ENSFM00800002003835 | Anoctamin 8 Transmembrane 16H |
| ENSDARG00000077291 | ENSFM00250000000709 | 40S Ribosomal S2 |
| ENSDARG00000077327 | ENSFM00800002003383 | Anoctamin 7 New Gene Expressed In Prostate Transmembrane 16G |
| ENSDARG00000077329 | ENSFM00380000124679 | Neuroligin Precursor |
| ENSDARG00000077339 | ENSFM00500000273926 | Peptidyl Trna Hydrolase 2 Mitochondrial Precursor Pth 2 EC_3.1.1.29 Bcl 2 Inhibitor Of Transcription |
| ENSDARG00000077343 | ENSFM00780001898335 | Protocadherin Precursor |
| ENSDARG00000077349 | ENSFM00250000000793 | VPS10 Domain Containing Receptor Precursor |
| ENSDARG00000077367 | ENSFM00260000050653 | Netrin Precursor Laminet |
| ENSDARG00000077368 | ENSFM00810002048698 | Unknown |
| ENSDARG00000077368 | ENSFM00250000004892 | Zinc Transporter 6 Znt 6 Solute Carrier Family 30 Member 6 |
| ENSDARG00000077372 | ENSFM00500000270210 | Transferrin Receptor 1 Tr Tfr TFR1 Trfr CD71 Antigen |
| ENSDARG00000077383 | ENSFM00760001714601 | Annexin Annexin Annexin |
| ENSDARG00000077385 | ENSFM00480000262761 | Regulator Of G Signaling |
| ENSDARG00000077465 | ENSFM00250000000793 | VPS10 Domain Containing Receptor Precursor |
| ENSDARG00000077468 | ENSFM00500000269855 | Phospholipid Scramblase Pl Scramblase Ca 2+ Dependent Phospholipid Scramblase |
| ENSDARG00000077492 | ENSFM00770001847537 | Phospholipid Transporting Atpase EC_3.6.3.1 Atpase Class I Type 8A Member P4 Atpase Flippase Complex Alpha Subunit At |
| ENSDARG00000077506 | ENSFM00750001659004 | Unknown |
| ENSDARG00000077506 | ENSFM00800002009548 | Unknown |
| ENSDARG00000077506 | ENSFM00250000000691 | Tight Junction Zo Tight Junction Zona Occludens Zonula Occludens |
| ENSDARG00000077515 | ENSFM00740001589271 | LETM1 And Ef Hand Domain Containing 1 Mitochondrial Precursor Leucine Zipper Ef Hand Containing Transmembrane 1 |
| ENSDARG00000077533 | ENSFM00250000003700 | Eukaryotic Translation Initiation Factor 3 Subunit F |
| ENSDARG00000077545 | ENSFM00740001589247 | Serine/Threonine Kinase BRSK2 EC_2.7.11.1 EC_2.7.11.- 26 Brain Specific Serine/Threonine Kinase 2 Br Serine/Threonine Kinase 2 Serine/Threonine Kinase Sad A |
| ENSDARG00000077549 | ENSFM00500000271132 | Apoptosis Inducing Factor 2 Ec 1 |
| ENSDARG00000077553 | ENSFM00800002003504 | Ras Related Rap Precursor |
| ENSDARG00000077560 | ENSFM00730001521500 | Glutamate Receptor Ionotropic Nmda Precursor Glutamate Receptor Subunit Epsilon N Methyl D Aspartate Receptor Subtype |
| ENSDARG00000077582 | ENSFM00500000269628 | Ankyrin Ank Ankyrin |
| ENSDARG00000077596 | ENSFM00250000007322 | Ambiguous |
| ENSDARG00000077609 | ENSFM00250000002984 | Phosphatase 1 Regulatory Subunit 29 Precursor Extracellular Leucine Rich Repeat And Fibronectin Type Iii Domain Containing 2 Leucine Rich Repeat And Fibronectin Type Iii Domain Containing 6 Leucine Rich Repeat Containing 62 |
| ENSDARG00000077633 | ENSFM00250000000552 | Membrane Associated Guanylate Kinase Ww And Pdz Domain Containing Membrane Associated Guanylate Kinase Inverted Magi |
| ENSDARG00000077654 | ENSFM00250000000328 | Metabotropic Glutamate Receptor Precursor |
| ENSDARG00000077656 | ENSFM00570000851482 | Brefeldin A Inhibited Guanine Nucleotide Exchange 3 |
| ENSDARG00000077686 | ENSFM00730001521956 | AP2 Associated Kinase 1 EC_2.7.11.1 Adaptor Associated Kinase 1 |
| ENSDARG00000077709 | ENSFM00250000001463 | Iq Motif And SEC7 Domain Containing |
| ENSDARG00000077710 | ENSFM00380000124679 | Neuroligin Precursor |
| ENSDARG00000077715 | ENSFM00720001492013 | Glutamate Receptor Ionotropic Kainate Precursor Glutamate Receptor |
| ENSDARG00000077732 | ENSFM00500000272450 | Tho Complex Subunit 4 THO4 Aly/Ref Export Factor |
| ENSDARG00000077736 | ENSFM00670001235939 | Complexin Complexin Cpx Synaphin |
| ENSDARG00000077740 | ENSFM00250000004863 | Beta 1 3 Glucosyltransferase BETA3GLC T EC_2.4.1.- Beta 3 Glycosyltransferase |
| ENSDARG00000077764 | ENSFM00500000270531 | Fatty Acid Amide Hydrolase 1 EC_3.5.1.99 Anandamide Amidohydrolase 1 Oleamide Hydrolase 1 |
| ENSDARG00000077765 | ENSFM00730001521860 | Gap Junction Gamma 1 Connexin 45 CX45 Gap Junction Alpha 7 |
| ENSDARG00000077776 | ENSFM00800002003472 | Casein Kinase Ii Subunit Beta Ck Ii Beta |
| ENSDARG00000077777 | ENSFM00540000728697 | Unknown |
| ENSDARG00000077847 | ENSFM00320000100099 | Noelin Precursor Neuronal Olfactomedin Related Er Localized Olfactomedin 1 |
| ENSDARG00000077920 | ENSFM00740001589158 | Cub And Sushi Domain Containing Cub And Sushi Multiple Domains |
| ENSDARG00000077940 | ENSFM00500000275011 | V Set And Transmembrane Domain Containing 2B Precursor |
| ENSDARG00000077945 | ENSFM00250000007963 | Hepatocyte Cell Adhesion Molecule Precursor Hepacam |
| ENSDARG00000077946 | ENSFM00250000001546 | Swi/Snf Complex Subunit SMARCC2 BRG1 Associated Factor 170 BAF170 Swi/Snf Complex 170 Kda Subunit Swi/Snf Related Matrix Associated Actin Dependent Regulator Of Chromatin Subfamily C Member 2 |
| ENSDARG00000077967 | ENSFM00690001369327 | Unknown |
| ENSDARG00000077967 | ENSFM00800002005976 | Unknown |
| ENSDARG00000077967 | ENSFM00800002006897 | Unknown |
| ENSDARG00000077996 | ENSFM00800002003474 | Cadherin Precursor |
| ENSDARG00000078016 | ENSFM00610000968796 | Unknown |
| ENSDARG00000078016 | ENSFM00500000270006 | E3 Ubiquitin Ligase EC_6.3.2.- Deltex |
| ENSDARG00000078060 | ENSFM00740001589375 | Synaptotagmin 7 Synaptotagmin Vii Sytvii |
| ENSDARG00000078073 | ENSFM00500000270979 | Adp Sugar Pyrophosphatase EC_3.6.1.13 8 Oxo Dgdp Phosphatase EC_3.6.1.- 58 Nucleoside Diphosphate Linked Moiety X Motif 5 Nudix Motif 5 |
| ENSDARG00000078088 | ENSFM00570000851266 | Cadherin |
| ENSDARG00000078094 | ENSFM00250000004656 | Lipase Maturation Factor 2 |
| ENSDARG00000078102 | ENSFM00250000001069 | Ph And SEC7 Domain Containing 1 Exchange Factor ARF6 Pleckstrin Homology And SEC7 Domain Containing 1 |
| ENSDARG00000078109 | ENSFM00760001714668 | Ubiquitin Carboxyl Terminal Hydrolase 12 EC_3.4.19.12 Deubiquitinating Enzyme 12 Ubiquitin Thioesterase 12 Ubiquitin Specific Processing Protease 12 |
| ENSDARG00000078113 | ENSFM00540000737558 | Unknown |
| ENSDARG00000078125 | ENSFM00800002004549 | Unknown |
| ENSDARG00000078136 | ENSFM00250000002280 | Wd Repeat Containing 47 Neuronal Enriched Map Interacting Nemitin |
| ENSDARG00000078139 | ENSFM00250000003219 | Dcc Interacting 13 DIP13 Adapter Containing Ph Domain Ptb Domain And Leucine Zipper Motif |
| ENSDARG00000078141 | ENSFM00250000002984 | Phosphatase 1 Regulatory Subunit 29 Precursor Extracellular Leucine Rich Repeat And Fibronectin Type Iii Domain Containing 2 Leucine Rich Repeat And Fibronectin Type Iii Domain Containing 6 Leucine Rich Repeat Containing 62 |
| ENSDARG00000078149 | ENSFM00730001521500 | Glutamate Receptor Ionotropic Nmda Precursor Glutamate Receptor Subunit Epsilon N Methyl D Aspartate Receptor Subtype |
| ENSDARG00000078169 | ENSFM00730001522093 | Voltage Dependent Calcium Channel Subunit Alpha 2/Delta 1 Precursor Voltage Gated Calcium Channel Subunit Alpha 2/Delta 1 |
| ENSDARG00000078172 | ENSFM00570000851634 | Prostaglandin F2 Receptor Negative Regulator Precursor Prostaglandin F2 Alpha Receptor Regulatory Prostaglandin F2 Alpha Receptor Associated CD315 Antigen |
| ENSDARG00000078185 | ENSFM00250000001313 | Phosphofurin Acidic Cluster Sorting Pacs |
| ENSDARG00000078187 | ENSFM00260000050376 | Solute Carrier Family 12 Member Electroneutral Potassium Chloride Cotransporter K Cl Cotransporter |
| ENSDARG00000078215 | ENSFM00250000005337 | Rabenosyn 5 |
| ENSDARG00000078216 | ENSFM00250000001830 | Epidermal Growth Factor Receptor Substrate 15 1 EPS15 Related EPS15R |
| ENSDARG00000078227 | ENSFM00570000851341 | Chondroitin Sulfate Proteoglycan 4 Precursor Chondroitin Sulfate Proteoglycan NG2 |
| ENSDARG00000078233 | ENSFM00800002024108 | Unknown |
| ENSDARG00000078233 | ENSFM00730001521847 | Catenin Delta 1 Cadherin Associated Src Substrate Cas P120 Catenin P120 Ctn P120 Cas |
| ENSDARG00000078258 | ENSFM00760001714625 | Gamma Precursor Ggt EC_2.3.2.2 Gamma Glutamyltransferase 1 Glutathione Hydrolase EC_3.4.19.- 13 Leukotriene C4 Hydrolase EC_3.4.-.- 19 14 |
| ENSDARG00000078284 | ENSFM00760001714816 | Phosphorylase B Kinase Regulatory Subunit Beta Phosphorylase Kinase Subunit Beta |
| ENSDARG00000078302 | ENSFM00250000001886 | Bmp/Retinoic Acid Inducible Neural Specific 1 Precursor Deleted In Bladder Cancer 1 |
| ENSDARG00000078317 | ENSFM00500000271927 | Dual Specificity Phosphatase 10 EC_3.1.3.16 EC_3.1.3.- 48 Mitogen Activated Kinase Phosphatase 5 Map Kinase Phosphatase 5 Mkp 5 |
| ENSDARG00000078322 | ENSFM00740001589200 | Collagen Alpha 1 Chain |
| ENSDARG00000078327 | ENSFM00740001589275 | Unknown |
| ENSDARG00000078328 | ENSFM00670001235664 | Vesicle Trafficking SEC22A SEC22 Vesicle Trafficking Homolog A SEC22 Vesicle Trafficking 2 |
| ENSDARG00000078362 | ENSFM00740001589132 | Tenascin Precursor Tn |
| ENSDARG00000078381 | ENSFM00250000003391 | Thyroid Receptor Interacting 11 Tr Interacting 11 Trip 11 Clonal Evolution Related Gene On Chromosome 14 Golgi Associated Microtubule Binding 210 Gmap 210 TRIP230 |
| ENSDARG00000078386 | ENSFM00250000000887 | Cytoplasmic Dynein 1 Intermediate Chain 2 Cytoplasmic Dynein Intermediate Chain 2 Dynein Intermediate Chain 2 Cytosolic Dh Ic 2 |
| ENSDARG00000078399 | ENSFM00500000269666 | Long Chain Fatty Acid Coa Ligase EC_6.2.1.3 Long Chain Acyl Coa Synthetase Lacs |
| ENSDARG00000078425 | ENSFM00790001963008 | Ornithine Aminotransferase Mitochondrial Precursor EC_2.6.1.13 Ornithine Oxo Acid Aminotransferase |
| ENSDARG00000078452 | ENSFM00250000002797 | Heme Oxygenase Ho EC_1.14.99.3 |
| ENSDARG00000078485 | ENSFM00250000002727 | Syntabulin Golgi Localized Syntaphilin Related Syntaxin 1 Binding |
| ENSDARG00000078512 | ENSFM00780001898351 | Acetyl Coa Carboxylase 1 ACC1 EC_6.4.1.2 Acc Alpha |
| ENSDARG00000078529 | ENSFM00250000000737 | Brain Specific Angiogenesis Inhibitor Precursor |
| ENSDARG00000078592 | ENSFM00250000002391 | Nodal Modulator Precursor |
| ENSDARG00000078593 | ENSFM00580000902189 | Von Willebrand Factor A Domain Containing 8 |
| ENSDARG00000078599 | ENSFM00260000050554 | Calcium Uptake 2 Mitochondrial Precursor Ef Hand Domain Containing Family Member A1 |
| ENSDARG00000078603 | ENSFM00800002003512 | Unconventional Myosin Ic Myosin I Beta Mmi Beta Mmib |
| ENSDARG00000078604 | ENSFM00260000050553 | TBC1 Domain Family Member |
| ENSDARG00000078624 | ENSFM00250000001570 | Rho Guanine Nucleotide Exchange Factor 9 Collybistin Rac/CDC42 Guanine Nucleotide Exchange Factor 9 |
| ENSDARG00000078645 | ENSFM00670001280708 | Unknown |
| ENSDARG00000078645 | ENSFM00760001714763 | Calcium/Calmodulin Dependent Kinase Kinase 1 Cam Kk 1 Cam Kinase Kinase 1 Camkk 1 EC_2.7.11.17 Cam Kinase Iv Kinase Calcium/Calmodulin Dependent Kinase Kinase Alpha Cam Kk Alpha Cam Kinase Kinase Alpha Camkk Alpha |
| ENSDARG00000078675 | ENSFM00500000269778 | Dedicator Of Cytokinesis |
| ENSDARG00000078686 | ENSFM00410000138507 | Adp Ribosylation Factor Small G Indispensable Equal Chromosome Segregation |
| ENSDARG00000078688 | ENSFM00250000003196 | Fibroblast Growth Factor Receptor Substrate 3 Fgfr Substrate 3 Fgfr Signaling Adaptor SNT2 SUC1 Associated Neurotrophic Factor Target 2 Snt 2 |
| ENSDARG00000078703 | ENSFM00500000270363 | Aspartyl/Asparaginyl Beta Hydroxylase EC_1.14.11.16 |
| ENSDARG00000078734 | ENSFM00780001898391 | Unconventional Myosin Ie Unconventional Myosin 1E |
| ENSDARG00000078745 | ENSFM00250000002364 | Calcium Independent Phospholipase A2 Gamma EC_3.1.1.5 Intracellular Membrane Associated Calcium Independent Phospholipase A2 Gamma IPLA2 Gamma Patatin Phospholipase Domain Containing 8 |
| ENSDARG00000078751 | ENSFM00250000003674 | Tripeptidyl Peptidase 2 Tpp 2 EC_3.4.14.10 Tripeptidyl Aminopeptidase Tripeptidyl Peptidase Ii Tpp Ii |
| ENSDARG00000078759 | ENSFM00250000002526 | Dna Replication Atp Dependent Helicase/Nuclease DNA2 Dna Replication Atp Dependent Helicase Homolog |
| ENSDARG00000078760 | ENSFM00730001521792 | Voltage Dependent Calcium Channel Subunit Alpha 2/Delta 3 Precursor Voltage Gated Calcium Channel Subunit Alpha 2/Delta 3 |
| ENSDARG00000078761 | ENSFM00670001235671 | Regulator Of Microtubule Dynamics Rmd Fa |
| ENSDARG00000078770 | ENSFM00250000004371 | Dihydroxyacetone Phosphate Acyltransferase Dap At Dhap At EC_2.3.1.42 Acyl Coa:Dihydroxyacetonephosphateacyltransferase Glycerone Phosphate O Acyltransferase |
| ENSDARG00000078815 | ENSFM00730001521304 | G Coupled Receptor Kinase G Coupled Receptor Kinase |
| ENSDARG00000078839 | ENSFM00400000131776 | Leucine Rich Repeat Transmembrane Neuronal Precursor |
| ENSDARG00000078842 | ENSFM00250000000591 | Tensin |
| ENSDARG00000078854 | ENSFM00250000002565 | Magnesium Transporter 1 Precursor MAGT1 |
| ENSDARG00000078856 | ENSFM00650001140024 | Solute Carrier Family 22 Member 17 24P3 Receptor 24P3R Lipocalin 2 Receptor |
| ENSDARG00000078866 | ENSFM00260000050437 | Sidekick Precursor |
| ENSDARG00000078890 | ENSFM00780001898367 | Wd Repeat And Fyve Domain Containing |
| ENSDARG00000078891 | ENSFM00250000006401 | Uncharacterized |
| ENSDARG00000078898 | ENSFM00760001714757 | Protocadherin Precursor Protocadherin |
| ENSDARG00000078901 | ENSFM00250000001010 | Ankyrin Repeat And Sterile Alpha Motif Domain Containing 1B Amyloid Beta Intracellular Domain Associated 1 Aida 1 E2A PBX1 Associated Eb 1 |
| ENSDARG00000078902 | ENSFM00250000000629 | Regulating Synaptic Membrane Exocytosis Rab 3 Interacting Molecule Rim |
| ENSDARG00000078908 | ENSFM00250000006718 | F Box Only 41 |
| ENSDARG00000078929 | ENSFM00250000005259 | Abhydrolase Domain Containing 16A Ec 3 Hla B Associated Transcript 5 |
| ENSDARG00000078998 | ENSFM00250000002406 | Lipoma Hmgic Fusion Partner |
| ENSDARG00000079004 | ENSFM00250000001143 | Tetratricopeptide Repeat Ankyrin Repeat And Coiled Coil Domain Containing |
| ENSDARG00000079067 | ENSFM00500000271304 | MMS19 Nucleotide Excision Repair Homolog MMS19 |
| ENSDARG00000079078 | ENSFM00500000270029 | Hemoglobin Subunit Alpha Alpha Globin Hemoglobin Alpha Chain |
| ENSDARG00000079097 | ENSFM00250000001143 | Tetratricopeptide Repeat Ankyrin Repeat And Coiled Coil Domain Containing |
| ENSDARG00000079161 | ENSFM00600000921250 | Piccolo Aczonin |
| ENSDARG00000079166 | ENSFM00250000000787 | Angiotensin Converting Enzyme Precursor Ace Carboxypeptidase |
| ENSDARG00000079202 | ENSFM00250000001234 | Poly Synthase 1 |
| ENSDARG00000079204 | ENSFM00410000138484 | Disintegrin And Metalloproteinase Domain Containing Precursor Adam Metalloproteinase Disintegrin And Cysteine Rich Mdc |
| ENSDARG00000079219 | ENSFM00250000001660 | Transmembrane Gtpase |
| ENSDARG00000079249 | ENSFM00750001632363 | 2 Oxoglutarate Dehydrogenase Mitochondrial Precursor 2 Oxoglutarate Dehydrogenase Complex Component E1 Ogdc E1 Alpha Ketoglutarate Dehydrogenase |
| ENSDARG00000079251 | ENSFM00380000124679 | Neuroligin Precursor |
| ENSDARG00000079264 | ENSFM00250000001781 | Unc 13 Homolog D MUNC13 4 |
| ENSDARG00000079295 | ENSFM00740001589100 | Voltage Dependent Type Calcium Channel Subunit Alpha Brain Calcium Channel Calcium Channel L Type Alpha 1 Polypeptide Voltage Gated Calcium Channel Subunit Alpha CAV2 |
| ENSDARG00000079308 | ENSFM00250000001107 | Precursor Torsin Family Member A |
| ENSDARG00000079348 | ENSFM00730001521500 | Glutamate Receptor Ionotropic Nmda Precursor Glutamate Receptor Subunit Epsilon N Methyl D Aspartate Receptor Subtype |
| ENSDARG00000079366 | ENSFM00250000001450 | Neurabin 1 Neurabin I Neural Tissue Specific F Actin Binding I Phosphatase 1 Regulatory Subunit 9A |
| ENSDARG00000079374 | ENSFM00250000000691 | Tight Junction Zo Tight Junction Zona Occludens Zonula Occludens |
| ENSDARG00000079378 | ENSFM00600000921202 | Pleckstrin Homology Domain Family B Member LL5 |
| ENSDARG00000079388 | ENSFM00760001714908 | Agrin N Terminal 110 Kda Subunit |
| ENSDARG00000079396 | ENSFM00260000050443 | Leucine Rich Repeat And Fibronectin Type Iii Domain Containing Precursor Synaptic Adhesion Molecule |
| ENSDARG00000079438 | ENSFM00780001898562 | Plakophilin 3 |
| ENSDARG00000079440 | ENSFM00730001521970 | Coronin |
| ENSDARG00000079455 | ENSFM00380000124679 | Neuroligin Precursor |
| ENSDARG00000079475 | ENSFM00250000006300 | Nck Interacting With SH3 Domain 54 Kda Vaca Interacting Kda Interacting 90 Kda SH3 Interacting With Nck SH3 Adapter SPIN90 Wasp Interacting SH3 Domain Wish Wiskott Aldrich Syndrome |
| ENSDARG00000079483 | ENSFM00250000000764 | Cap Gly Domain Containing Linker 2 Cytoplasmic Linker 115 Clip 115 Cytoplasmic Linker 2 |
| ENSDARG00000079484 | ENSFM00250000001245 | Potassium Channel Subfamily T Member Sequence Activated Potassium Channel |
| ENSDARG00000079491 | ENSFM00290000065519 | Potassium Voltage Gated Channel Subfamily A Member Voltage Gated Potassium Channel Subunit KV1 |
| ENSDARG00000079504 | ENSFM00250000001660 | Transmembrane Gtpase |
| ENSDARG00000079516 | ENSFM00670001243324 | Cdgsh Domain Containing 1 |
| ENSDARG00000079556 | ENSFM00500000273557 | C2 Calcium Dependent Domain Containing 4C Nuclear Localized Factor 3 FAM148C |
| ENSDARG00000079572 | ENSFM00740001589144 | 1 Phosphatidylinositol 4 5 Bisphosphate Phosphodiesterase Delta EC_3.1.4.11 Phosphoinositide Phospholipase C Delta Phospholipase C Delta Plc Delta |
| ENSDARG00000079586 | ENSFM00400000131718 | Gamma Aminobutyric Acid Receptor Subunit Beta Precursor Gaba A Receptor Subunit Beta |
| ENSDARG00000079610 | ENSFM00250000001853 | A Kinase Anchor 9 Akap 9 A Kinase Anchor Kda Akap 120 Kinase A Anchoring 9 PRKA9 |
| ENSDARG00000079611 | ENSFM00480000262746 | Semaphorin Precursor |
| ENSDARG00000079645 | ENSFM00800002004636 | Cdna Cssl: |
| ENSDARG00000079651 | ENSFM00800002003441 | AFG3 EC_3.4.24.- |
| ENSDARG00000079656 | ENSFM00740001589308 | Precursor |
| ENSDARG00000079665 | ENSFM00250000001603 | Probable G Coupled Receptor Precursor |
| ENSDARG00000079670 | ENSFM00730001522141 | Leucine Rich Repeat Containing 7 Densin 180 Densin |
| ENSDARG00000079671 | ENSFM00500000270520 | Lipid Phosphate Phosphatase Related Type 4 EC_3.1.3.4 Brain Specific Phosphatidic Acid Phosphatase 1 Plasticity Related Gene 1 Prg 1 |
| ENSDARG00000079725 | ENSFM00800002003717 | Atp Dependent Rna Helicase A EC_3.6.4.13 Deah Box 9 Nuclear Dna Helicase Ii Ndh Ii |
| ENSDARG00000079771 | ENSFM00250000010619 | Astrocytic Phosphoprotein Pea 15.15 Kda Phosphoprotein Enriched In Astrocytes |
| ENSDARG00000079780 | ENSFM00250000001411 | Heterogeneous Nuclear Ribonucleoprotein U Scaffold Attachment Factor Saf |
| ENSDARG00000079781 | ENSFM00250000001398 | Slit And Ntrk Precursor |
| ENSDARG00000079805 | ENSFM00500000270015 | Transgelin |
| ENSDARG00000079823 | ENSFM00540000732848 | Unknown |
| ENSDARG00000079824 | ENSFM00740001597625 | Ninjurin 2 |
| ENSDARG00000079840 | ENSFM00250000000903 | Calcium Activated Potassium Channel Subunit Alpha 1 Bk Channel Bkca Alpha Calcium Activated Potassium Channel Subfamily M Subunit Alpha 1 K Vca Alpha KCA1 1 Maxi K Channel Maxik Slo Alpha SLO1 Slowpoke Homolog Slo Homolog |
| ENSDARG00000079847 | ENSFM00500000269762 | Epsin Eps 15 Interacting |
| ENSDARG00000079850 | ENSFM00740001589084 | Precursor |
| ENSDARG00000079858 | ENSFM00250000007006 | Transmembrane 163 Synaptic Vesicle Membrane Of 31 Kda |
| ENSDARG00000079872 | ENSFM00250000000899 | Rap Guanine Nucleotide Exchange Factor Exchange Factor Directly Activated By Camp Exchange Directly Activated By Camp Epac Camp Regulated Guanine Nucleotide Exchange Factor Camp |
| ENSDARG00000079881 | ENSFM00250000005995 | T Cell Activation Inhibitor Mitochondrial Tolerance Associated Gene 1 Toag 1 |
| ENSDARG00000079891 | ENSFM00250000001678 | Tyrosine Phosphatase Auxilin EC_3.1.3.48 Dnaj Homolog Subfamily C Member 6 |
| ENSDARG00000079892 | ENSFM00730001521536 | Plexin Precursor |
| ENSDARG00000079932 | ENSFM00250000006073 | Aminopeptidase 4 EC_3.4.11.- |
| ENSDARG00000079949 | ENSFM00250000002938 | Fact Complex Subunit SPT16 Facilitates Chromatin Transcription Complex Subunit SPT16 |
| ENSDARG00000079985 | ENSFM00500000274257 | Nuclear Receptor Interacting 2 |
| ENSDARG00000080000 | ENSFM00730001522086 | Tricarboxylate Transport Protein Mitochondrial Precursor Citrate Transport Ctp Solute Carrier Family 25 Member 1 Tricarboxylate Carrier |
| ENSDARG00000080010 | ENSFM00250000000515 | Alcohol Dehydrogenase EC_1.1.1.1 Alcohol Dehydrogenase |
| ENSDARG00000086034 | ENSFM00500000270858 | Nectin 1 Precursor Herpes Virus Entry Mediator C Herpesvirus Entry Mediator C Hvec Poliovirus Receptor Related 1 CD111 Antigen |
| ENSDARG00000086037 | ENSFM00500000272551 | Junctional Adhesion Molecule C Precursor Jam C Junctional Adhesion Molecule 3 Jam 3 |
| ENSDARG00000086104 | ENSFM00610000955325 | Unknown |
| ENSDARG00000086104 | ENSFM00730001521392 | Solute Carrier Organic Anion Transporter Family Member Organic Anion Transporting Polypeptide Organic Anion Solute Carrier Family 21 Member |
| ENSDARG00000086207 | ENSFM00730001521500 | Glutamate Receptor Ionotropic Nmda Precursor Glutamate Receptor Subunit Epsilon N Methyl D Aspartate Receptor Subtype |
| ENSDARG00000086523 | ENSFM00250000003616 | Erlin 2 Endoplasmic Reticulum Lipid Raft Associated 2 Stomatin Prohibitin Flotillin Hflc/K Domain Containing 2 Spfh Domain Containing 2 |
| ENSDARG00000086573 | ENSFM00250000008621 | Oligodendrocyte Myelin Glycoprotein Precursor |
| ENSDARG00000086618 | ENSFM00810002040700 | Proteasome Subunit Alpha Type 3 EC_3.4.25.1 Macropain Subunit C8 Multicatalytic Endopeptidase Complex Subunit C8 Proteasome Component C8 |
| ENSDARG00000086740 | ENSFM00250000007215 | Phytanoyl Coa Dioxygenase Peroxisomal Precursor EC_1.14.11.18 Phytanic Acid Oxidase Phytanoyl Coa Alpha Hydroxylase Phyh |
| ENSDARG00000086746 | ENSFM00250000001738 | Probable Proline Dehydrogenase 2 EC_1.5.5.2 Probable Proline Oxidase 2 |
| ENSDARG00000086756 | ENSFM00500000274428 | Regulator Of G Signaling 9 Binding RGS9 |
| ENSDARG00000086790 | ENSFM00730001521369 | Guanylate Cyclase Soluble Subunit Beta 1 Gcs Beta 1 EC_4.6.1.2 Guanylate Cyclase Soluble Subunit Beta 3 Gcs Beta 3 Soluble Guanylate Cyclase Small Subunit |
| ENSDARG00000086848 | ENSFM00250000002488 | Atpase Family Aaa Domain Containing 3 |
| ENSDARG00000086866 | ENSFM00780001898365 | Glutamine Fructose 6 Phosphate Aminotransferase 2 EC_2.6.1.16 D Fructose 6 Phosphate Amidotransferase 2 Glutamine:Fructose 6 Phosphate Amidotransferase 2 Gfat 2 GFAT2 Hexosephosphate Aminotransferase 2 |
| ENSDARG00000086870 | ENSFM00260000050342 | Solute Carrier Family 12 Member Bumetanide Sensitive Sodium Potassium Chloride Cotransporter Na K Cl Symporter |
| ENSDARG00000086969 | ENSFM00780001898335 | Protocadherin Precursor |
| ENSDARG00000086990 | ENSFM00800002009421 | Unknown |
| ENSDARG00000086990 | ENSFM00260000050454 | Leucine Rich Repeat Containing |
| ENSDARG00000087165 | ENSFM00600000942006 | Unknown |
| ENSDARG00000087206 | ENSFM00500000270021 | T Complex 1 Subunit Beta Tcp 1 Beta Cct Beta |
| ENSDARG00000087229 | ENSFM00250000003124 | Exocyst Complex Component 7 Exocyst Complex Component EXO70 |
| ENSDARG00000087243 | ENSFM00390000126319 | Ras Related Rab |
| ENSDARG00000087247 | ENSFM00780001898357 | Voltage Gated Potassium Channel Subunit Beta K + Channel Subunit Beta Kv Beta |
| ENSDARG00000087260 | ENSFM00250000002052 | Metastasis Suppressor 1 Missing In Metastasis |
| ENSDARG00000087346 | ENSFM00730001521249 | Ras Related Rap 1 |
| ENSDARG00000087364 | ENSFM00600000944558 | Unknown |
| ENSDARG00000087402 | ENSFM00250000000220 | Tropomyosin Alpha 1 Chain Alpha Tropomyosin Tropomyosin 1 |
| ENSDARG00000087456 | ENSFM00600000941833 | Unknown |
| ENSDARG00000087538 | ENSFM00800002003514 | Kinesin KIF3A Microtubule Plus End Directed Kinesin Motor 3A |
| ENSDARG00000087573 | ENSFM00250000007111 | Histone Deacetylase 11 HD11 EC_3.5.1.98 |
| ENSDARG00000087599 | ENSFM00250000000375 | Teneurin Ten Odd Oz/Ten M Homolog Tenascin Ten Teneurin Transmembrane |
| ENSDARG00000087616 | ENSFM00750001658357 | Unknown |
| ENSDARG00000087616 | ENSFM00800002006620 | Unknown |
| ENSDARG00000087616 | ENSFM00260000050441 | Microtubule Associated Tau Neurofibrillary Tangle Paired Helical Filament Tau Phf Tau |
| ENSDARG00000087640 | ENSFM00500000270274 | Ubx Domain Containing 2B NSFL1 Cofactor P37 P97 Cofactor P37 |
| ENSDARG00000087647 | ENSFM00760001714685 | Septin 7 CDC10 Homolog |
| ENSDARG00000087657 | ENSFM00630001067970 | Unknown |
| ENSDARG00000087657 | ENSFM00510000502777 | Fatty Acid Synthase EC_2.3.1.85 S Acetyltransferase EC_2.3.1.- 38 |
| ENSDARG00000087704 | ENSFM00500000269996 | Gdnf Family Receptor Alpha Precursor Gdnf Receptor Alpha Gdnfr Alpha Gfr Alpha Tgf Beta Related Neurotrophic Factor Receptor |
| ENSDARG00000087762 | ENSFM00730001521880 | Ras Related Rab |
| ENSDARG00000087822 | ENSFM00250000000604 | Arf Gap With Gtpase Ank Repeat And Ph Domain Containing Agap Centaurin Gamma |
| ENSDARG00000087843 | ENSFM00800002003683 | Contactin 1 Precursor Neural Cell Surface F3 |
| ENSDARG00000087897 | ENSFM00400000131810 | Core Histone Macro H2A 1 Histone Macro M H2A Y H2A/Y |
| ENSDARG00000087897 | ENSFM00550000756898 | Histone H2A |
| ENSDARG00000087916 | ENSFM00610000971995 | Unknown |
| ENSDARG00000087916 | ENSFM00500000314906 | Transmembrane 240 |
| ENSDARG00000087981 | ENSFM00730001521104 | Sodium And Chloride Dependent Transporter Solute Carrier Family 6 Member |
| ENSDARG00000088013 | ENSFM00720001492028 | Sterol 26 Hydroxylase Mitochondrial Precursor EC_1.14.13.15 5 Beta Cholestane 3 Alpha 7 Alpha 12 Alpha Triol 27 Hydroxylase Cytochrome P 450C27/25 Cytochrome P450 27 Sterol 27 Hydroxylase Vitamin D 3 25 Hydroxylase |
| ENSDARG00000088030 | ENSFM00670001273033 | Unknown |
| ENSDARG00000088030 | ENSFM00670001273034 | Unknown |
| ENSDARG00000088030 | ENSFM00500000271015 | 60S Ribosomal L35A |
| ENSDARG00000088073 | ENSFM00500000272914 | Unknown |
| ENSDARG00000088091 | ENSFM00600000941716 | Profilin |
| ENSDARG00000088100 | ENSFM00410000138464 | Kv Channel Interacting A Type Potassium Channel Modulatory Potassium Channel Interacting |
| ENSDARG00000088117 | ENSFM00250000004295 | Alpha 1 6 Fucosyltransferase ALPHA1 6FUCT EC_2.4.1.68 Gdp L Fuc:N Acetyl Beta D Glucosaminide ALPHA1 6 Fucosyltransferase Gdp Fucose Glycoprotein Fucosyltransferase Glycoprotein 6 Alpha L Fucosyltransferase |
| ENSDARG00000088136 | ENSFM00780001898335 | Protocadherin Precursor |
| ENSDARG00000088168 | ENSFM00250000000459 | Actin Binding Lim Ablim Actin Binding Lim Family Member |
| ENSDARG00000088227 | ENSFM00250000007006 | Transmembrane 163 Synaptic Vesicle Membrane Of 31 Kda |
| ENSDARG00000088247 | ENSFM00600000942312 | Unknown |
| ENSDARG00000088276 | ENSFM00500000269608 | Histone H2B |
| ENSDARG00000088277 | ENSFM00250000008968 | Sushi Domain Containing 5 Precursor |
| ENSDARG00000088293 | ENSFM00260000050490 | Double C2 Domain Containing DOC2 |
| ENSDARG00000088357 | ENSFM00730001522380 | Long Chain Specific Acyl Coa Dehydrogenase Mitochondrial Precursor Lcad EC_1.3.8.8 |
| ENSDARG00000088383 | ENSFM00500000272199 | Cytochrome C Oxidase Subunit 5A Mitochondrial Precursor Cytochrome C Oxidase Polypeptide Va |
| ENSDARG00000088439 | ENSFM00250000006403 | Ganglioside GM2 Activator Precursor Cerebroside Sulfate Activator GM2 Ap Sphingolipid Activator 3 Sap 3 |
| ENSDARG00000088466 | ENSFM00250000006330 | Disulfide Isomerase TMX3 Precursor EC_5.3.4.1 Thioredoxin Domain Containing 10 Thioredoxin Related Transmembrane 3 |
| ENSDARG00000088634 | ENSFM00730001521247 | Adenylate Cyclase Type EC_4.6.1.1 Atp Pyrophosphate Lyase Adenylate Cyclase Type Adenylyl Cyclase Ca 2+ Adenylyl Cyclase |
| ENSDARG00000088663 | ENSFM00260000050644 | Vesicular Integral Membrane VIP36 Precursor Lectin Mannose Binding 2 Vesicular Integral Membrane 36 VIP36 |
| ENSDARG00000088699 | ENSFM00250000005716 | Pdz Domain Containing 8 Sarcoma Antigen Ny Sar 84/Ny Sar 104 |
| ENSDARG00000088825 | ENSFM00520000517879 | Citron Rho Interacting Kinase EC_2.7.11.1 Serine/Threonine Kinase 21 |
| ENSDARG00000088862 | ENSFM00500000272618 | Syntaxin Binding 6 |
| ENSDARG00000088882 | ENSFM00600000941712 | Unknown |
| ENSDARG00000088899 | ENSFM00250000002523 | Ral Gtpase Activating Subunit Beta P170 |
| ENSDARG00000088959 | ENSFM00500000270834 | Pyridoxal Kinase EC_2.7.1.35 Pyridoxine Kinase |
| ENSDARG00000089066 | ENSFM00400000132091 | Nhs 2 |
| ENSDARG00000089076 | ENSFM00790001963049 | Molybdopterin Adenylyltransferase Mpt Adenylyltransferase EC_2.7.7.75 Domain G |
| ENSDARG00000089190 | ENSFM00250000001143 | Tetratricopeptide Repeat Ankyrin Repeat And Coiled Coil Domain Containing |
| ENSDARG00000089255 | ENSFM00600000940244 | Unknown |
| ENSDARG00000089292 | ENSFM00260000050328 | Latrophilin |
| ENSDARG00000089314 | ENSFM00260000050441 | Microtubule Associated Tau Neurofibrillary Tangle Paired Helical Filament Tau Phf Tau |
| ENSDARG00000089413 | ENSFM00600000939910 | Unknown |
| ENSDARG00000089413 | ENSFM00630001069159 | Unknown |
| ENSDARG00000089413 | ENSFM00630001071995 | Unknown |
| ENSDARG00000089418 | ENSFM00600000944882 | Unknown |
| ENSDARG00000089444 | ENSFM00250000008621 | Oligodendrocyte Myelin Glycoprotein Precursor |
| ENSDARG00000089477 | ENSFM00710001450268 | Unknown |
| ENSDARG00000089477 | ENSFM00800002014124 | Unknown |
| ENSDARG00000089486 | ENSFM00670001235924 | Complexin 3 Precursor Complexin Iii Cpx Iii |
| ENSDARG00000089562 | ENSFM00780001898335 | Protocadherin Precursor |
| ENSDARG00000089643 | ENSFM00500000271112 | Cell Surface Glycoprotein MUC18 Precursor Melanoma Cell Adhesion Molecule Melanoma Associated Antigen MUC18 CD146 Antigen |
| ENSDARG00000089681 | ENSFM00500000270103 | Transcription Factor BTF3 Homolog 4 Basic Transcription Factor 3 4 |
| ENSDARG00000089720 | ENSFM00600000944776 | Unknown |
| ENSDARG00000089856 | ENSFM00520000517879 | Citron Rho Interacting Kinase EC_2.7.11.1 Serine/Threonine Kinase 21 |
| ENSDARG00000089913 | ENSFM00740001589106 | Voltage Dependent T Type Calcium Channel Subunit Alpha Voltage Gated Calcium Channel Subunit Alpha CAV3 |
| ENSDARG00000089924 | ENSFM00730001521185 | Dehydrogenase Aldehyde Dehydrogenase Family 1 Member Dehydrogenase |
| ENSDARG00000089957 | ENSFM00250000002312 | Leucine Rich Repeat Lgi Family Member Precursor Leucine Rich Glioma Inactivated |
| ENSDARG00000089976 | ENSFM00800002016195 | Unknown |
| ENSDARG00000089976 | ENSFM00250000004611 | Signal Peptidase Complex Subunit 3 EC_3.4.-.- Microsomal Signal Peptidase 22/23 Kda Subunit SPC22/23 Spase 22/23 Kda Subunit |
| ENSDARG00000090035 | ENSFM00800002023314 | Unknown |
| ENSDARG00000090035 | ENSFM00250000002242 | Reticulon 4 Receptor Precursor Nogo Receptor Nogo 66 Receptor Homolog Nogo 66 Receptor Related |
| ENSDARG00000090036 | ENSFM00250000007281 | Rho Related Btb Domain Containing 3 EC_3.6.1.- |
| ENSDARG00000090062 | ENSFM00730001521186 | Ras Related C3 Botulinum Toxin Substrate Precursor P21 |
| ENSDARG00000090086 | ENSFM00800002024011 | Unknown |
| ENSDARG00000090086 | ENSFM00500000269744 | Ras Related Rab Precursor |
| ENSDARG00000090106 | ENSFM00730001521551 | Vesicular Glutamate Transporter Solute Carrier Family 17 Member |
| ENSDARG00000090145 | ENSFM00500000314906 | Transmembrane 240 |
| ENSDARG00000090149 | ENSFM00500000269649 | Histone H4 |
| ENSDARG00000090183 | ENSFM00250000002127 | Gtpase Activating And VPS9 Domain Containing 1 |
| ENSDARG00000090190 | ENSFM00500000273157 | Basal Cell Adhesion Molecule Precursor B Cam Cell Surface Glycoprotein Lutheran Antigen CD239 Antigen |
| ENSDARG00000090190 | ENSFM00500000271112 | Cell Surface Glycoprotein MUC18 Precursor Melanoma Cell Adhesion Molecule Melanoma Associated Antigen MUC18 CD146 Antigen |
| ENSDARG00000090194 | ENSFM00670001240264 | Hepatitis B Virus X Interacting |
| ENSDARG00000090195 | ENSFM00250000002187 | Metaxin 1 Mitochondrial Outer Membrane Import Complex 1 |
| ENSDARG00000090213 | ENSFM00810002040784 | Gtpase RHEBL1 Precursor Ras Homolog Enriched In Brain 1 Rheb 1 |
| ENSDARG00000090228 | ENSFM00270000056439 | Glutathione S Transferase EC_2.5.1.18 Gst Class Alpha Member Glutathione S Transferase |
| ENSDARG00000090286 | ENSFM00250000000235 | Precursor |
| ENSDARG00000090348 | ENSFM00780001898335 | Protocadherin Precursor |
| ENSDARG00000090389 | ENSFM00670001252646 | Unknown |
| ENSDARG00000090389 | ENSFM00800002015471 | Unknown |
| ENSDARG00000090444 | ENSFM00600000940278 | Unknown |
| ENSDARG00000090444 | ENSFM00610000973229 | Unknown |
| ENSDARG00000090444 | ENSFM00610000973230 | Unknown |
| ENSDARG00000090462 | ENSFM00500000273357 | 39S Ribosomal L20 Mitochondrial Precursor L20MT Mrp L20 |
| ENSDARG00000090481 | ENSFM00670001239693 | Tumor Suppressor Candidate 5 Homolog Dispanin Subfamily B Member 1 DSPB1 |
| ENSDARG00000090585 | ENSFM00500000269807 | Glypican Secreted Glypican |
| ENSDARG00000090615 | ENSFM00600000940243 | Unknown |
| ENSDARG00000090624 | ENSFM00260000050328 | Latrophilin |
| ENSDARG00000090654 | ENSFM00250000001678 | Tyrosine Phosphatase Auxilin EC_3.1.3.48 Dnaj Homolog Subfamily C Member 6 |
| ENSDARG00000090716 | ENSFM00250000001393 | Clathrin Heavy Chain |
| ENSDARG00000090815 | ENSFM00730001521400 | Atp Sensitive Inward Rectifier Potassium Channel Atp Potassium Channel K Inward Rectifier K + Channel 1 Potassium Channel Inwardly Rectifying Subfamily J Member |
| ENSDARG00000090850 | ENSFM00250000000235 | Precursor |
| ENSDARG00000090883 | ENSFM00800002003385 | Gamma Aminobutyric Acid Receptor Subunit Alpha Precursor Gaba A Receptor Subunit Alpha |
| ENSDARG00000090962 | ENSFM00250000037376 | N Acetyltransferase 14 EC_2.3.1.- |
| ENSDARG00000090982 | ENSFM00500000270045 | Choline Transporter Solute Carrier Family 44 Member |
| ENSDARG00000090988 | ENSFM00250000002483 | Leucine Trna Ligase Cytoplasmic EC_6.1.1.4 Leucyl Trna Synthetase Leurs |
| ENSDARG00000091042 | ENSFM00740001589315 | Sacsin Dnaj Homolog Subfamily C Member 29 DNAJC29 |
| ENSDARG00000091130 | ENSFM00250000000417 | Potassium Voltage Gated Channel Subfamily Kqt Member Kqt Potassium Channel Subunit Alpha Voltage Gated Potassium Channel Subunit KV7 |
| ENSDARG00000091136 | ENSFM00250000000235 | Precursor |
| ENSDARG00000091138 | ENSFM00800002003421 | Actin Related 3 Actin 3 |
| ENSDARG00000091269 | ENSFM00740001591432 | Si:Dkey 182G1 3 |
| ENSDARG00000091293 | ENSFM00250000001467 | Dmx 1 X 1 |
| ENSDARG00000091348 | ENSFM00500000269689 | Reticulon |
| ENSDARG00000091433 | ENSFM00250000001156 | Alpha/Beta Hydrolase Domain Containing Ec 3 |
| ENSDARG00000091457 | ENSFM00250000001269 | Wiskott Aldrich Syndrome Family Member 1 Wasp Family Member 1 Wave 1 |
| ENSDARG00000091525 | ENSFM00250000000259 | Histone H3 |
| ENSDARG00000091618 | ENSFM00250000001393 | Clathrin Heavy Chain |
| ENSDARG00000091655 | ENSFM00500000271167 | Glycine Cleavage System H Protein Mitochondrial Precursor Lipoic Acid Containing |
| ENSDARG00000091683 | ENSFM00250000008302 | CB1 Cannabinoid Receptor Interacting 1 Crip 1 |
| ENSDARG00000091777 | ENSFM00250000005977 | Target Of Rapamycin Complex 2 Subunit MAPKAP1 TORC2 Subunit MAPKAP1 Mitogen Activated Kinase 2 Associated 1 Stress Activated Map Kinase Interacting 1 Sapk Interacting 1 |
| ENSDARG00000091801 | ENSFM00500000269596 | Serpin |
| ENSDARG00000092112 | ENSFM00250000002521 | Mitochondrial Import Receptor Subunit TOM40 Homolog Translocase Of Outer Membrane 40 Kda Subunit Homolog |
| ENSDARG00000092124 | ENSFM00610000964585 | Unknown |
| ENSDARG00000092126 | ENSFM00250000000813 | Vitellogenin Precursor |
| ENSDARG00000092154 | ENSFM00250000008293 | Testis Expressed Sequence 264 Precursor Secreted ZSIG11 |
| ENSDARG00000092155 | ENSFM00670001265093 | Apolipoprotein C Ii |
| ENSDARG00000092170 | ENSFM00670001255935 | Unknown |
| ENSDARG00000092233 | ENSFM00250000000813 | Vitellogenin Precursor |
| ENSDARG00000092376 | ENSFM00250000000718 | Disks Large Associated Dap Psd 95/SAP90 Binding SAP90/Psd 95 Associated |
| ENSDARG00000092419 | ENSFM00250000000813 | Vitellogenin Precursor |
| ENSDARG00000092470 | ENSFM00500000273656 | Thrombomodulin Tm CD141 Antigen |
| ENSDARG00000092517 | ENSFM00610000964217 | Unknown |
| ENSDARG00000092553 | ENSFM00250000000816 | Adp/Atp Translocase Adp Atp Carrier Adenine Nucleotide Translocator Ant Solute Carrier Family 25 Member |
| ENSDARG00000092610 | ENSFM00640001103074 | Lysosome Associated Membrane Glycoprotein 1 Precursor Lamp 1 Lysosome Associated Membrane 1 CD107 Antigen Family Member A CD107A Antigen |
| ENSDARG00000092624 | ENSFM00740001589095 | Spectrin Beta Chain Erythrocytic Beta Spectrin |
| ENSDARG00000092644 | ENSFM00250000000492 | Argonaute |
| ENSDARG00000092677 | ENSFM00610000963214 | Unknown |
| ENSDARG00000092693 | ENSFM00500000270782 | Translationally Controlled Tumor Tctp |
| ENSDARG00000092746 | ENSFM00610000964941 | Unknown |
| ENSDARG00000092787 | ENSFM00610000965952 | Unknown |
| ENSDARG00000092798 | ENSFM00500000269861 | Peptidyl Prolyl Cis Trans Isomerase B Precursor Ppiase B EC_5.2.1.8 Rotamase B |
| ENSDARG00000092856 | ENSFM00610000967994 | Unknown |
| ENSDARG00000092873 | ENSFM00710001464695 | Unknown |
| ENSDARG00000092948 | ENSFM00730001521471 | Guanine Nucleotide Binding G Q Subunit Alpha Guanine Nucleotide Binding Alpha Q |
| ENSDARG00000092970 | ENSFM00610000972992 | Unknown |
| ENSDARG00000092976 | ENSFM00250000002249 | Amine Oxidase EC_1.4.3.4 Monoamine Oxidase Type Mao |
| ENSDARG00000093091 | ENSFM00250000001463 | Iq Motif And SEC7 Domain Containing |
| ENSDARG00000093185 | ENSFM00730001521851 | Guanylate Cyclase Soluble Subunit Alpha 3 Gcs Alpha 3 EC_4.6.1.2 Gcs Alpha 1 Soluble Guanylate Cyclase Large Subunit |
| ENSDARG00000093195 | ENSFM00500000269608 | Histone H2B |
| ENSDARG00000093359 | ENSFM00500000270275 | Phosphatidylinositol 4 5 Bisphosphate 5 Phosphatase A EC_3.1.3.56 Inositol Polyphosphate 5 Phosphatase J |
| ENSDARG00000093374 | ENSFM00610000973108 | Unknown |
| ENSDARG00000093374 | ENSFM00800002003537 | Unknown |
| ENSDARG00000093382 | ENSFM00610000969748 | Unknown |
| ENSDARG00000093382 | ENSFM00610000973342 | Unknown |
| ENSDARG00000093382 | ENSFM00610000973343 | Unknown |
| ENSDARG00000093413 | ENSFM00500000270078 | Lactadherin Precursor Mfgm Milk Fat Globule Egf Factor 8 Mfg E8 SED1 |
| ENSDARG00000093448 | ENSFM00670001236064 | Mitochondrial Pyruvate Carrier 1 Brain 44 |
| ENSDARG00000093945 | ENSFM00540000740857 | Vacuolar Atpase Assembly Integral Membrane VMA21 |
| ENSDARG00000094052 | ENSFM00250000000892 | Serine/Threonine Kinase EC_2.7.11.1 Thousand And One Amino Acid |
| ENSDARG00000094074 | ENSFM00610000963788 | Si:Dkey 81H8 1 |
| ENSDARG00000094133 | ENSFM00610000958535 | Unknown |
| ENSDARG00000094258 | ENSFM00250000007011 | Comm Domain Containing 9 |
| ENSDARG00000094512 | ENSFM00800002003799 | Gamma Aminobutyric Acid Receptor Subunit Pi Precursor Gaba A Receptor Subunit Pi |
| ENSDARG00000094526 | ENSFM00450000242166 | Kinase Suppressor Of Ras |
| ENSDARG00000094577 | ENSFM00670001236658 | CDC42 Small Effector 2 |
| ENSDARG00000094590 | ENSFM00250000002423 | C2 Domain Containing 5.138 Kda C2 Domain Containing Phosphoprotein |
| ENSDARG00000094695 | ENSFM00750001641915 | Unknown |
| ENSDARG00000094704 | ENSFM00500000271823 | Apoptosis Regulator Bcl 2 |
| ENSDARG00000094730 | ENSFM00610000964945 | Unknown |
| ENSDARG00000094730 | ENSFM00500000321096 | ACBD7 |
| ENSDARG00000095005 | ENSFM00500000270056 | Uncharacterized |
| ENSDARG00000095034 | ENSFM00250000004260 | Uncharacterized |
| ENSDARG00000095170 | ENSFM00500000270615 | Leucine Rich Repeat Flightless Interacting Lrr Flii Interacting |
| ENSDARG00000095459 | ENSFM00610000964147 | Unknown |
| ENSDARG00000095464 | ENSFM00500000270299 | Glutathione S Transferase Theta EC_2.5.1.18 Gst Class Theta |
| ENSDARG00000095577 | ENSFM00610000961526 | Unknown |
| ENSDARG00000095577 | ENSFM00670001235694 | Cytochrome B Ascorbate Dependent 3 Ec 1 Cytochrome B561 Family Member A3 |
| ENSDARG00000095579 | ENSFM00600000921667 | Mitochondrial Fission 1 FIS1 Homolog Tetratricopeptide Repeat 11 Tpr Repeat 11 |
| ENSDARG00000095603 | ENSFM00500000273079 | Delphilin Glutamate Receptor Ionotropic Delta 2 Interacting 1 |
| ENSDARG00000095751 | ENSFM00250000005636 | Precursor |
| ENSDARG00000095848 | ENSFM00250000000198 | Atrial Natriuretic Peptide Receptor Precursor EC_4.6.1.2 Atrial Natriuretic Peptide Receptor Type Anp Anpr Npr Guanylate Cyclase Gc |
| ENSDARG00000095897 | ENSFM00630001065265 | Unknown |
| ENSDARG00000096003 | ENSFM00650001147076 | Unknown |
| ENSDARG00000096249 | ENSFM00670001282088 | Unknown |
| ENSDARG00000096249 | ENSFM00730001521549 | Ap 1 Complex Subunit Mu 1 Ap Mu Chain Family Member MU1A Adaptor Complex Ap 1 Subunit Mu 1 Adaptor Related Complex 1 Subunit Mu 1 Clathrin Assembly Complex 1 Mu 1 Medium Chain 1 Golgi Adaptor HA1/AP1 Adaptin Mu 1 Subunit Mu Adaptin 1 MU1A Adaptin |
| ENSDARG00000096271 | ENSFM00740001589308 | Precursor |
| ENSDARG00000096346 | ENSFM00670001276890 | Unknown |
| ENSDARG00000096355 | ENSFM00560000771464 | Ig Mu Chain C Region Membrane Bound Form |
| ENSDARG00000096368 | ENSFM00250000002795 | Sialidase 3 EC_3.2.1.18 Ganglioside Sialidase Membrane Sialidase N Acetyl Alpha Neuraminidase 3 |
| ENSDARG00000096454 | ENSFM00730001521549 | Ap 1 Complex Subunit Mu 1 Ap Mu Chain Family Member MU1A Adaptor Complex Ap 1 Subunit Mu 1 Adaptor Related Complex 1 Subunit Mu 1 Clathrin Assembly Complex 1 Mu 1 Medium Chain 1 Golgi Adaptor HA1/AP1 Adaptin Mu 1 Subunit Mu Adaptin 1 MU1A Adaptin |
| ENSDARG00000096546 | ENSFM00250000000469 | Neuropilin Precursor Vascular Endothelial Cell Growth Factor 165 Receptor |
| ENSDARG00000096622 | ENSFM00730001521501 | Egf Module Containing Mucin Hormone Receptor Precursor Egf Module Receptor |
| ENSDARG00000096651 | ENSFM00710001457231 | Unknown |
| ENSDARG00000096651 | ENSFM00250000002158 | Precursor Lung Seven Transmembrane Receptor |
| ENSDARG00000096661 | ENSFM00710001456770 | Unknown |
| ENSDARG00000096687 | ENSFM00730001521808 | Regulator Of G Signaling |
| ENSDARG00000096829 | ENSFM00800002009624 | Unknown |
| ENSDARG00000096829 | ENSFM00250000005350 | Flavin Reductase Nadph Fr EC_1.5.1.30 Biliverdin Reductase B Bvr B EC_1.3.1.- 24 Biliverdin Ix Beta Reductase Green Heme Binding Ghbp Nadph Dependent Diaphorase Nadph Flavin Reductase Flr |
| ENSDARG00000096867 | ENSFM00500000271703 | Calmin Calponin Transmembrane Domain |
| ENSDARG00000097011 | ENSFM00500000270029 | Hemoglobin Subunit Alpha Alpha Globin Hemoglobin Alpha Chain |
| ENSDARG00000097110 | ENSFM00750001649403 | Unknown |
| ENSDARG00000097110 | ENSFM00750001649404 | Unknown |
| ENSDARG00000097170 | ENSFM00730001521583 | Protocadherin Alpha Precursor Pcdh Alpha |
| ENSDARG00000097170 | ENSFM00780001898335 | Protocadherin Precursor |
| ENSDARG00000097238 | ENSFM00500000270525 | Hemoglobin Subunit Beta Beta Globin Hemoglobin Beta Chain |
| ENSDARG00000097245 | ENSFM00250000001168 | Soga Family Member |
| ENSDARG00000097351 | ENSFM00740001589123 | Filamin Fln Abp 280 Actin Binding Filamin |
| ENSDARG00000097491 | ENSFM00730001521346 | Udp Glucuronosyltransferase 1 Precursor Udpgt 1 UGT1 UGT1 EC_2.4.1.17 Udp Glucuronosyltransferase |
| ENSDARG00000097528 | ENSFM00750001638793 | Unknown |
| ENSDARG00000097572 | ENSFM00310000088999 | Receptor Type Tyrosine Phosphatase Precursor R Ptp EC_3.1.3.48 Receptor Type Tyrosine Phosphatase |
| ENSDARG00000097576 | ENSFM00670001235687 | Vesicle Associated Membrane 2 Vamp 2 Synaptobrevin 2 |
| ENSDARG00000097660 | ENSFM00500000271336 | Trafficking Particle Complex Subunit 2 |
| ENSDARG00000097797 | ENSFM00750001646006 | Unknown |
| ENSDARG00000097797 | ENSFM00750001658958 | Unknown |
| ENSDARG00000097827 | ENSFM00740001589098 | Density Lipoprotein Receptor Related Precursor Lrp |
| ENSDARG00000097855 | ENSFM00550000764011 | Small Membrane A Kinase Anchor Small Membrane Akap Smakap |
| ENSDARG00000097897 | ENSFM00600000921280 | Fch Domain Only |
| ENSDARG00000098057 | ENSFM00800002020364 | Unknown |
| ENSDARG00000098057 | ENSFM00810002040701 | Down Syndrome Cell Adhesion Molecule Precursor |
| ENSDARG00000098075 | ENSFM00670001240600 | SH3 Domain Binding Glutamic Acid Rich 3 |
| ENSDARG00000098118 | ENSFM00730001522435 | Trafficking Particle Complex Subunit 10 Trafficking Particle Complex Subunit TMEM1 Transport Particle Subunit TMEM1 Trapp Subunit TMEM1 |
| ENSDARG00000098140 | ENSFM00800002003462 | Ankyrin Repeat Domain Containing |
| ENSDARG00000098161 | ENSFM00390000126303 | Contactin Precursor Brain Derived Immunoglobulin Superfamily Big |
| ENSDARG00000098226 | ENSFM00250000006694 | Sphingomyelin Phosphodiesterase 3 EC_3.1.4.12 Neutral Sphingomyelinase 2 Nsmase 2 NSMASE2 Neutral Sphingomyelinase Ii |
| ENSDARG00000098290 | ENSFM00470000251461 | Unc 80 |
| ENSDARG00000098312 | ENSFM00800002003823 | Lysosome Membrane 2.85 Kda Lysosomal Membrane Sialoglycoprotein LGP85 CD36 Antigen 2 Lysosome Membrane Ii Limp Ii Scavenger Receptor Class B Member 2 CD36 Antigen |
| ENSDARG00000098317 | ENSFM00250000001777 | Cytoplasmic Dynein 1 Light Intermediate Chain 1 Dynein Light Chain A Dlc A Dynein Light Intermediate Chain 1 Cytosolic |
| ENSDARG00000098320 | ENSFM00730001521627 | Metabotropic Glutamate Receptor Precursor |
| ENSDARG00000098344 | ENSFM00500000270602 | Ras Related Rab 18 |
| ENSDARG00000098355 | ENSFM00500000271216 | Atp Synthase Subunit D Mitochondrial Atpase Subunit D |
| ENSDARG00000098367 | ENSFM00250000002862 | Signal Recognition Particle 54 Kda SRP54 |
| ENSDARG00000098368 | ENSFM00250000000617 | Amyloid Beta A4 Precursor Abpp App Alzheimer Disease Amyloid A4 Homolog |
| ENSDARG00000098374 | ENSFM00740001589123 | Filamin Fln Abp 280 Actin Binding Filamin |
| ENSDARG00000098401 | ENSFM00800002016163 | Unknown |
| ENSDARG00000098417 | ENSFM00250000000771 | Synapsin Synapsin |
| ENSDARG00000098458 | ENSFM00670001236020 | 60S Ribosomal L37A |
| ENSDARG00000098465 | ENSFM00800002018337 | Unknown |
| ENSDARG00000098486 | ENSFM00250000004428 | Toll Interacting |
| ENSDARG00000098497 | ENSFM00730001521569 | Ras Precursor |
| ENSDARG00000098523 | ENSFM00250000000779 | Neural Cell Adhesion Molecule 1 Precursor N Cam 1 Ncam 1 |
| ENSDARG00000098584 | ENSFM00800002013985 | Unknown |
| ENSDARG00000098584 | ENSFM00800002013986 | Unknown |
| ENSDARG00000098589 | ENSFM00500000271733 | Cytochrome B5 |
| ENSDARG00000098591 | ENSFM00250000000193 | Tubulin Beta Chain |
| ENSDARG00000098599 | ENSFM00250000001467 | Dmx 1 X 1 |
| ENSDARG00000098612 | ENSFM00800002003483 | Muscarinic Acetylcholine Receptor |
| ENSDARG00000098639 | ENSFM00500000269896 | Pyrroline 5 Carboxylate Reductase 3 P5C Reductase 3 P5CR 3 EC_1.5.1.2 Pyrroline 5 Carboxylate Reductase |
| ENSDARG00000098644 | ENSFM00250000001313 | Phosphofurin Acidic Cluster Sorting Pacs |
| ENSDARG00000098645 | ENSFM00500000271083 | Regucalcin Rc Gluconolactonase Gnl EC_3.1.1.17 Senescence Marker 30 Smp 30 |
| ENSDARG00000098654 | ENSFM00390000126378 | Raftlin Raft Linking |
| ENSDARG00000098749 | ENSFM00250000002042 | Tubulin Specific Chaperone D Beta Tubulin Cofactor D Tubulin Folding Cofactor D |
| ENSDARG00000098751 | ENSFM00650001140033 | Leucine Zipper Tumor Suppressor 2 |
| ENSDARG00000098753 | ENSFM00250000001494 | Engulfment And Cell Motility |
| ENSDARG00000098766 | ENSFM00720001492032 | Pyruvate Carboxylase EC_6.4.1.1 Pyruvic Carboxylase Pcb |
| ENSDARG00000098782 | ENSFM00730001521465 | Choline O Acetyltransferase Choactase Chat Choline Acetylase EC_2.3.1.6 |
| ENSDARG00000098785 | ENSFM00740001589388 | Glutaredoxin 3 Pkc Interacting Cousin Of Thioredoxin Picot Pkc Theta Interacting Pkcq Interacting Thioredoxin 2 |
| ENSDARG00000098788 | ENSFM00780001898335 | Protocadherin Precursor |
| ENSDARG00000098809 | ENSFM00250000000859 | Clathrin Assembly |
| ENSDARG00000098813 | ENSFM00500000270193 | Syntaxin |
| ENSDARG00000098825 | ENSFM00250000004351 | Phenylalanine Trna Ligase Alpha Subunit EC_6.1.1.20 Phenylalanyl Trna Synthetase Alpha Subunit Phers |
| ENSDARG00000098831 | ENSFM00380000124741 | Glutaryl Coa Dehydrogenase Mitochondrial Precursor Gcd EC_1.3.8.6 |
| ENSDARG00000098860 | ENSFM00250000003740 | FAM57B |
| ENSDARG00000098874 | ENSFM00250000001326 | Exocyst Complex Component 6 Exocyst Complex Component SEC15A SEC15 1 |
| ENSDARG00000098880 | ENSFM00800002018422 | Unknown |
| ENSDARG00000098880 | ENSFM00600000921250 | Piccolo Aczonin |
| ENSDARG00000098936 | ENSFM00670001235340 | Kinesin Heavy Chain Kinesin Heavy Chain Kinesin Heavy Chain |
| ENSDARG00000098967 | ENSFM00500000269829 | Rab Gtpase Activating 1 |
| ENSDARG00000098969 | ENSFM00740001589472 | Reelin EC_3.4.21.- |
| ENSDARG00000098983 | ENSFM00250000004335 | Growth Hormone Inducible Transmembrane Precursor Mitochondrial Morphology And Cristae Structure 1 MICS1 |
| ENSDARG00000099031 | ENSFM00500000274501 | Sodium Channel Subunit Beta 4 Precursor |
| ENSDARG00000099039 | ENSFM00250000000747 | Poly Rc Binding Alpha Heterogeneous Nuclear Ribonucleoprotein Hnrnp |
| ENSDARG00000099056 | ENSFM00760001714736 | 3 Ketoacyl Coa Thiolase Mitochondrial EC_2.3.1.16 Acetyl Coa Acyltransferase Beta Ketothiolase Mitochondrial 3 Oxoacyl Coa Thiolase |
| ENSDARG00000099079 | ENSFM00250000001673 | Atp Citrate Synthase EC_2.3.3.8 Atp Citrate Pro S Lyase Citrate Cleavage Enzyme |
| ENSDARG00000099096 | ENSFM00400000131718 | Gamma Aminobutyric Acid Receptor Subunit Beta Precursor Gaba A Receptor Subunit Beta |
| ENSDARG00000099104 | ENSFM00670001235623 | 60S Ribosomal L24 |
| ENSDARG00000099116 | ENSFM00250000006339 | N Acetyl D Glucosamine Kinase N Acetylglucosamine Kinase EC_2.7.1.59 Glcnac Kinase |
| ENSDARG00000099118 | ENSFM00250000002395 | Trafficking Particle Complex Subunit 9 Nik And Ikbkb Binding |
| ENSDARG00000099143 | ENSFM00500000271023 | Farnesyltransferase/Geranylgeranyltransferase Type 1 Subunit Alpha EC_2.5.1.58 EC_2.5.1.- 59 Caax Farnesyltransferase Subunit Alpha Ftase Alpha Ras Proteins Prenyltransferase Subunit Alpha Type I Geranyl Geranyltransferase Subunit Alpha Ggtase I Alpha |
| ENSDARG00000099175 | ENSFM00500000269645 | High Mobility Group B1 High Mobility Group 1 Hmg 1 |
| ENSDARG00000099184 | ENSFM00770001847556 | Dual Specificity Mitogen Activated Kinase Kinase 6 Map Kinase Kinase 6 Mapkk 6 EC_2.7.12.2 Mapk/Erk Kinase 6 Mek 6 |
| ENSDARG00000099190 | ENSFM00250000008534 | Coiled Coil Domain Containing 127 |
| ENSDARG00000099199 | ENSFM00730001521655 | NEDD4 E3 Ubiquitin Ligase EC_6.3.2.- Ww Domain Containing |
| ENSDARG00000099203 | ENSFM00670001235362 | Sodium/Potassium Transporting Atpase Subunit Beta Sodium/Potassium Dependent Atpase Subunit Beta |
| ENSDARG00000099214 | ENSFM00500000270313 | Histone H2A |
| ENSDARG00000099226 | ENSFM00420000140518 | Serine/Threonine Phosphatase PP1 Catalytic Subunit Pp EC_3.1.3.16 |
| ENSDARG00000099236 | ENSFM00350000105408 | Septin |
| ENSDARG00000099241 | ENSFM00730001521389 | Serine/Threonine Phosphatase Catalytic Subunit EC_3.1.3.16 |
| ENSDARG00000099258 | ENSFM00500000271174 | Vacuolar Sorting Associated 52 Homolog SAC2 Suppressor Of Actin Mutations 2 |
| ENSDARG00000099301 | ENSFM00500000270663 | HSP90 Co Chaperone CDC37 HSP90 Chaperone Kinase Targeting Subunit P50CDC37 |
| ENSDARG00000099306 | ENSFM00500000270014 | F Box Only 6 F Box That Recognizes Sugar Chains 2 |
| ENSDARG00000099312 | ENSFM00670001240176 | Hematological And Neurological Expressed 1 |
| ENSDARG00000099321 | ENSFM00440000236967 | Btb/Poz Domain Containing 17 Precursor Galectin 3 Binding |
| ENSDARG00000099323 | ENSFM00250000000275 | Disks Large Homolog Synapse Associated Sap |
| ENSDARG00000099339 | ENSFM00800002020482 | Unknown |
| ENSDARG00000099339 | ENSFM00800002020483 | Unknown |
| ENSDARG00000099339 | ENSFM00250000001466 | Kinase C And Casein Kinase Substrate In Neurons |
| ENSDARG00000099353 | ENSFM00800002003431 | Calcium Binding Mitochondrial Carrier Mitochondrial Aspartate Glutamate Carrier Solute Carrier Family 25 Member |
| ENSDARG00000099368 | ENSFM00500000271356 | Mitochondrial Coenzyme A Transporter Sl Solute Carrier Family 25 Member 42 |
| ENSDARG00000099380 | ENSFM00500000270284 | 60S Ribosomal L13 |
| ENSDARG00000099386 | ENSFM00400000131776 | Leucine Rich Repeat Transmembrane Neuronal Precursor |
| ENSDARG00000099412 | ENSFM00730001521436 | Core Precursor Chondroitin Sulfate Proteoglycan |
| ENSDARG00000099478 | ENSFM00290000065564 | Heparan Sulfate 2 O Sulfotransferase 1 2 O Sulfotransferase 2OST EC_2.8.2.- |
| ENSDARG00000099511 | ENSFM00800002004636 | Cdna Cssl: |
| ENSDARG00000099524 | ENSFM00250000003836 | 1 4 Alpha Glucan Branching Enzyme EC_2.4.1.18 Glycogen Branching Enzyme |
| ENSDARG00000099568 | ENSFM00250000000938 | Metal Transporter Ancient Conserved Domain Containing Cyclin |
| ENSDARG00000099579 | ENSFM00760001714924 | Delta 1 Pyrroline 5 Carboxylate Synthase P5CS Aldehyde Dehydrogenase Family 18 Member A1 |
| ENSDARG00000099614 | ENSFM00250000002171 | Transcriptional Activator Pur Beta Purine Rich Element Binding B |
| ENSDARG00000099617 | ENSFM00250000006615 | Epimerase Family Sd EC_1.1.1.- Short Chain Dehydrogenase/Reductase Family 39U Member 1 |
| ENSDARG00000099624 | ENSFM00600000921337 | Charged Multivesicular Body 1B Chromatin Modifying 1B CHMP1B |
| ENSDARG00000099648 | ENSFM00500000269666 | Long Chain Fatty Acid Coa Ligase EC_6.2.1.3 Long Chain Acyl Coa Synthetase Lacs |
| ENSDARG00000099653 | ENSFM00250000002637 | 26S Proteasome Regulatory Subunit RPN1 Regulatory 26S Proteasome Subunit |
| ENSDARG00000099663 | ENSFM00500000272199 | Cytochrome C Oxidase Subunit 5A Mitochondrial Precursor Cytochrome C Oxidase Polypeptide Va |
| ENSDARG00000099664 | ENSFM00500000274344 | Unknown |
| ENSDARG00000099685 | ENSFM00770001847629 | Guanine Nucleotide Binding Subunit Beta 5 GBETA5 Transducin Beta Chain 5 |
| ENSDARG00000099690 | ENSFM00740001589204 | Ena/Vasp Ena/Vasodilator Stimulated Phosphoprotein |
| ENSDARG00000099692 | ENSFM00250000001383 | Cyclin Y |
| ENSDARG00000099720 | ENSFM00740001589204 | Ena/Vasp Ena/Vasodilator Stimulated Phosphoprotein |
| ENSDARG00000099729 | ENSFM00800002024092 | Unknown |
| ENSDARG00000099729 | ENSFM00780001898335 | Protocadherin Precursor |
| ENSDARG00000099730 | ENSFM00800002024340 | Unknown |
| ENSDARG00000099730 | ENSFM00800002024341 | Unknown |
| ENSDARG00000099730 | ENSFM00250000000989 | Pyruvate Kinase Pkm EC_2.7.1.40 Pyruvate Kinase Muscle Isozyme |
| ENSDARG00000099740 | ENSFM00250000000749 | Kinesin Light Chain |
| ENSDARG00000099744 | ENSFM00600000930021 | Neuromodulin Axonal Membrane Gap 43 Growth Associated 43 |
| ENSDARG00000099755 | ENSFM00730001521507 | Atp Dependent 6 Phosphofructokinase Type |
| ENSDARG00000099757 | ENSFM00250000002280 | Wd Repeat Containing 47 Neuronal Enriched Map Interacting Nemitin |
| ENSDARG00000099766 | ENSFM00730001521616 | Myosin Regulatory Light Myosin Regulatory Light Chain 2 Smooth Muscle Myosin Regulatory Light Chain |
| ENSDARG00000099774 | ENSFM00670001238567 | Uncharacterized |
| ENSDARG00000099776 | ENSFM00750001632335 | Glutamine Synthetase Gs EC_6.3.1.2 Glutamate Decarboxylase EC_4.1.1.- 15 Glutamate Ammonia Ligase |
| ENSDARG00000099783 | ENSFM00780001898335 | Protocadherin Precursor |
| ENSDARG00000099786 | ENSFM00440000236866 | Alpha Actinin Alpha Actinin F Actin Cross Linking |
| ENSDARG00000099793 | ENSFM00800002024303 | Unknown |
| ENSDARG00000099793 | ENSFM00760001715141 | Chondroitin Sulfate Proteoglycan 5 Precursor Acidic Leucine Rich Egf Domain Containing Brain Neuroglycan C |
| ENSDARG00000099799 | ENSFM00800002011864 | Unknown |
| ENSDARG00000099799 | ENSFM00500000339659 | Cdna Cssl: |
| ENSDARG00000099802 | ENSFM00250000002129 | Purine Nucleoside Phosphorylase Pnp EC_2.4.2.1 Inosine Phosphorylase Inosine Guanosine Phosphorylase |
| ENSDARG00000099844 | ENSFM00410000138458 | Guanine Nucleotide Binding Subunit |
| ENSDARG00000099850 | ENSFM00250000001463 | Iq Motif And SEC7 Domain Containing |
| ENSDARG00000099874 | ENSFM00250000000604 | Arf Gap With Gtpase Ank Repeat And Ph Domain Containing Agap Centaurin Gamma |
| ENSDARG00000099908 | ENSFM00250000005790 | Mitochondrial Fission Process 1 Mitochondrial 18 Kda MTP18 |
| ENSDARG00000099943 | ENSFM00250000000867 | Microtubule Associated Rp/Eb Family Member |
| ENSDARG00000099961 | ENSFM00670001235622 | BCL2/Adenovirus E1B 19 Kda Interacting 3 |
| ENSDARG00000099995 | ENSFM00250000001062 | Amyloid Beta A4 Precursor Binding Family B Member |
| ENSDARG00000099996 | ENSFM00250000000940 | Mitochondrial Rho Gtpase 2 Miro 2 EC_3.6.5.- Ras Homolog Gene Family Member T2 |
| ENSDARG00000100003 | ENSFM00750001632335 | Glutamine Synthetase Gs EC_6.3.1.2 Glutamate Decarboxylase EC_4.1.1.- 15 Glutamate Ammonia Ligase |
| ENSDARG00000100052 | ENSFM00540000738060 | Uncharacterized |
| ENSDARG00000100075 | ENSFM00730001521925 | Atp Binding Cassette Sub Family G Member 2 Urate Exporter CD338 Antigen |
| ENSDARG00000100089 | ENSFM00250000000111 | Calcium/Calmodulin Dependent Kinase Type Ii Subunit Cam Kinase Ii Subunit Camk Ii Subunit EC_2.7.11.17 |
| ENSDARG00000100115 | ENSFM00730001521131 | Dynein Heavy Chain Axonemal Axonemal Beta Dynein Heavy Chain Ciliary Dynein Heavy Chain |
| ENSDARG00000100119 | ENSFM00730001521718 | 55 Kda Erythrocyte Membrane P55 Membrane Protein Palmitoylated 1 |
| ENSDARG00000100145 | ENSFM00520000517792 | Dynamin EC_3.6.5.5 |
| ENSDARG00000100152 | ENSFM00800002017761 | Unknown |
| ENSDARG00000100157 | ENSFM00500000271930 | Phosphatidate Cytidylyltransferase Mitochondrial Precursor EC_2.7.7.41 Cdp Diacylglycerol Synthase Cdp Dag Synthase Mitochondrial Translocator Assembly And Maintenance 41 Homolog TAM41 |
| ENSDARG00000100170 | ENSFM00250000001444 | Peptidyl Glycine Alpha Amidating Monooxygenase Precursor Pam |
| ENSDARG00000100177 | ENSFM00730001521896 | Ras Related Rab |
| ENSDARG00000100252 | ENSFM00250000003778 | F Actin Capping Subunit Beta |
| ENSDARG00000100267 | ENSFM00250000000328 | Metabotropic Glutamate Receptor Precursor |
| ENSDARG00000100274 | ENSFM00250000001912 | Wd Repeat Containing 7 Tgf Beta Resistance Associated Trag |
| ENSDARG00000100304 | ENSFM00750001632424 | Choline Dehydrogenase Mitochondrial Precursor Cdh Chd EC_1.1.99.1 |
| ENSDARG00000100313 | ENSFM00730001521611 | Phosphatidylinositol 4 Phosphate 5 Kinase Type 1 Beta Pi Beta Ptdins 4 P 5 Kinase 1 Beta EC_2.7.1.68 Phosphatidylinositol 4 Phosphate 5 Kinase Type I Phosphatidylinositol 4 Phosphate 5 Kinase Beta |
| ENSDARG00000100349 | ENSFM00790001962972 | Camp Dependent Kinase Catalytic Subunit Pka C EC_2.7.11.11 |
| ENSDARG00000100371 | ENSFM00760001714607 | Elongation Factor 2 Ef 2 |
| ENSDARG00000100372 | ENSFM00250000005243 | Delta Aminolevulinic Acid Dehydratase Aladh EC_4.2.1.24 Porphobilinogen Synthase |
| ENSDARG00000100374 | ENSFM00730001521412 | Thioredoxin Reductase EC_1.8.1.9 Thioredoxin Reductase |
| ENSDARG00000100392 | ENSFM00250000002236 | 40S Ribosomal S18 |
| ENSDARG00000100392 | ENSFM00670001276328 | 40S Ribosomal S18 |
| ENSDARG00000100394 | ENSFM00740001589214 | Endoplasmic Reticulum Aminopeptidase EC_3.4.11.- Derived Aminopeptidase |
| ENSDARG00000100400 | ENSFM00250000003532 | Hyccin Down Regulated By CTNNB1 A FAM126A |
| ENSDARG00000100409 | ENSFM00250000007633 | Calcium Signal Modulating Cyclophilin Ligand Caml |
| ENSDARG00000100422 | ENSFM00790001963124 | Receptor Type Tyrosine Phosphatase O Precursor R Ptp O EC_3.1.3.48 Glomerular Epithelial 1 Tyrosine Phosphatase U2 Ptp U2 Ptpase U2 |
| ENSDARG00000100435 | ENSFM00500000271789 | Vesicle Trafficking SEC22B Er Golgi Snare Of 24 Kda Ers 24 ERS24 SEC22 Vesicle Trafficking Homolog B |
| ENSDARG00000100510 | ENSFM00800002003421 | Actin Related 3 Actin 3 |
| ENSDARG00000100513 | ENSFM00770001848133 | 40S Ribosomal S27 |
| ENSDARG00000100531 | ENSFM00500000272278 | Alpha/Beta Hydrolase Domain Containing 14B Abhydrolase Domain Containing 14B Ec 3 CCG1 Interacting Factor B |
| ENSDARG00000100562 | ENSFM00670001240058 | Syndecan |
| ENSDARG00000100585 | ENSFM00670001267729 | Cytochrome C Oxidase Assembly Factor 3 Homolog Mitochondrial Coiled Coil Domain Containing 56 |
| ENSDARG00000100588 | ENSFM00670001235763 | 60S Ribosomal L36 |
| ENSDARG00000100592 | ENSFM00710001441664 | Sodium/Calcium Exchanger 1 Precursor Na + /Ca 2+ Exchange 1 Solute Carrier Family 8 Member 1 |
| ENSDARG00000100594 | ENSFM00250000001049 | Seizure 6 Precursor |
| ENSDARG00000100620 | ENSFM00250000000892 | Serine/Threonine Kinase EC_2.7.11.1 Thousand And One Amino Acid |
| ENSDARG00000100679 | ENSFM00800002003638 | Leucine Rich Repeat And Calponin Homology Domain Containing |
| ENSDARG00000100718 | ENSFM00250000000979 | Nadp Dependent Malic Enzyme Nadp Me EC_1.1.1.40 Malic Enzyme |
| ENSDARG00000100725 | ENSFM00730001521338 | Ephrin Type B Receptor Kinase |
| ENSDARG00000100729 | ENSFM00250000001103 | Talin |
| ENSDARG00000100731 | ENSFM00730001521711 | Acyl Coa Synthetase Fatty Acid Transport Fatp Fatty Acid Coenzyme A Ligase Very Long Chain Solute Carrier Family 27 Member 2 Very Long Chain Coa |
| ENSDARG00000100742 | ENSFM00500000270563 | Receptor Expression Enhancing Polyposis Locus 1 |
| ENSDARG00000100786 | ENSFM00800002003599 | 1 Phosphatidylinositol 4 5 Bisphosphate Phosphodiesterase Beta 4 EC_3.1.4.11 Phosphoinositide Phospholipase C Beta 4 Phospholipase C Beta 4 Plc Beta 4 |
| ENSDARG00000100789 | ENSFM00500000274177 | Plasminogen Receptor Kt Plg R Kt |
| ENSDARG00000100823 | ENSFM00670001257472 | Selenoprotein K Selk |
| ENSDARG00000100829 | ENSFM00780001898335 | Protocadherin Precursor |
| ENSDARG00000100851 | ENSFM00790001963049 | Molybdopterin Adenylyltransferase Mpt Adenylyltransferase EC_2.7.7.75 Domain G |
| ENSDARG00000100869 | ENSFM00740001589278 | Adenylate Cyclase Type 9 EC_4.6.1.1 Atp Pyrophosphate Lyase 9 Adenylate Cyclase Type Ix Adenylyl Cyclase 9 |
| ENSDARG00000100894 | ENSFM00800002023443 | Unknown |
| ENSDARG00000100897 | ENSFM00730001521867 | Platelet Derived Growth Factor Receptor Precursor Pdgf R Pdgfr EC_2.7.10.1 Platelet Derived Growth Factor Receptor Type Platelet Derived Growth Factor Receptor CD140 Antigen Family Member Platelet Derived Growth Factor Receptor Antigen |
| ENSDARG00000100971 | ENSFM00610000965976 | Fxyd Domain Containing Ion Transport Regulator 6 |
| ENSDARG00000100972 | ENSFM00270000056424 | Myosin Cellular Myosin Heavy Chain Type Myosin Heavy Chain Myosin Heavy Chain Non Muscle Non Muscle Myosin Heavy Chain Nmmhc Non Muscle Myosin Heavy Chain Nmmhc Ii Nmmhc |
| ENSDARG00000100990 | ENSFM00250000000895 | Nucleoside Diphosphate Kinase Ndk Ndp Kinase EC_2.7.4.6 |
| ENSDARG00000101047 | ENSFM00800002017597 | Unknown |
| ENSDARG00000101047 | ENSFM00250000005120 | Trna Splicing Ligase Rtcb Homolog |
| ENSDARG00000101074 | ENSFM00250000001520 | Glutamate Dehydrogenase Mitochondrial Gdh EC_1.4.1.3 |
| ENSDARG00000101079 | ENSFM00500000270499 | Probable E3 Ubiquitin Ligase HECTD4 EC_6.3.2.- Hect Domain Containing 4 |
| ENSDARG00000101082 | ENSFM00250000003239 | Eukaryotic Translation Initiation Factor 3 Subunit L |
| ENSDARG00000101127 | ENSFM00500000270555 | Microtubule Associated Proteins 1A/1B Light Chain 3A Precursor Autophagy Related LC3 A Autophagy Related Ubiquitin Modifier LC3 A MAP1 Light Chain 3 1 MAP1A/MAP1B Light Chain 3 A MAP1A/MAP1B LC3 A Microtubule Associated 1 Light Chain 3 Alpha |
| ENSDARG00000101130 | ENSFM00780001898387 | Chaperone Activity Of BC1 Complex Mitochondrial Precursor Chaperone ABC1 EC_2.7.11.- Aarf Domain Containing Kinase 3 |
| ENSDARG00000101139 | ENSFM00500000270410 | 1 Acyl Sn Glycerol 3 Phosphate Acyltransferase Alpha Precursor EC_2.3.1.51 1 Acylglycerol 3 Phosphate O Acyltransferase 1 1 Agp Acyltransferase 1 1 Agpat 1 Lysophosphatidic Acid Acyltransferase Alpha Lpaat Alpha |
| ENSDARG00000101175 | ENSFM00250000002939 | Calcium Uniporter Regulatory Subunit Mcub Mitochondrial Precursor Mcub Coiled Coil Domain Containing 109B |
| ENSDARG00000101199 | ENSFM00250000003212 | Retinol Binding 4 Plasma Retinol Binding Prbp |
| ENSDARG00000101210 | ENSFM00250000002484 | Golgi Apparatus 1 Precursor E Selectin Ligand 1 Esl 1 Golgi Sialoglycoprotein Mg 160 |
| ENSDARG00000101232 | ENSFM00780001898335 | Protocadherin Precursor |
| ENSDARG00000101233 | ENSFM00250000000599 | Unc 13 Homolog MUNC13 |
| ENSDARG00000101239 | ENSFM00500000270110 | Peroxisomal Multifunctional Enzyme Type 2 Mfe 2 17 Beta Hydroxysteroid Dehydrogenase 4 17 Beta Hsd 4 D Bifunctional Dbp Multifunctional 2 Mpf 2 |
| ENSDARG00000101251 | ENSFM00250000000475 | L Lactate Dehydrogenase Chain Ldh EC_1.1.1.27 |
| ENSDARG00000101289 | ENSFM00500000270018 | Equilibrative Nucleoside Transporter Equilibrative Nitrobenzylmercaptopurine Riboside Nucleoside Transporter Equilibrative Nbmpr Nucleoside Transporter Nucleoside Transporter Type Solute Carrier Family 29 Member |
| ENSDARG00000101297 | ENSFM00250000007298 | Transmembrane |
| ENSDARG00000101317 | ENSFM00500000269895 | Lifeguard 2 Fas Apoptotic Inhibitory Molecule 2 |
| ENSDARG00000101322 | ENSFM00500000270210 | Transferrin Receptor 1 Tr Tfr TFR1 Trfr CD71 Antigen |
| ENSDARG00000101324 | ENSFM00560000771472 | Apolipoprotein A I Precursor Apo Ai Apoa I Apolipoprotein A1 |
| ENSDARG00000101334 | ENSFM00260000050761 | Dipeptidyl Peptidase 1 Precursor EC_3.4.14.1 Cathepsin C Cathepsin J Dipeptidyl Peptidase I Dpp I Dppi Dipeptidyl Transferase |
| ENSDARG00000101380 | ENSFM00670001235476 | Alpha Crystallin Chain |
| ENSDARG00000101406 | ENSFM00500000312340 | Ribosomal Protein Large P2 |
| ENSDARG00000101443 | ENSFM00250000004714 | Lish Domain And Heat Repeat Containing |
| ENSDARG00000101510 | ENSFM00750001632381 | Centrin |
| ENSDARG00000101512 | ENSFM00670001235637 | Thioredoxin Related Transmembrane Precursor Thioredoxin Domain Containing |
| ENSDARG00000101560 | ENSFM00400000131829 | Proteasome Subunit Alpha Type EC_3.4.25.1 Macropain Subunit Multicatalytic Endopeptidase Complex Subunit Proteasome Component |
| ENSDARG00000101561 | ENSFM00250000003682 | Lipoamide Acyltransferase Component Of Branched Chain Alpha Keto Acid Dehydrogenase Complex Mitochondrial Precursor EC_2.3.1.168 Branched Chain Alpha Keto Acid Dehydrogenase Complex Component E2 Bckad E2 BCKADE2 Dihydrolipoamide Acetyltransferase Componen |
| ENSDARG00000101606 | ENSFM00250000000629 | Regulating Synaptic Membrane Exocytosis Rab 3 Interacting Molecule Rim |
| ENSDARG00000101619 | ENSFM00800002013928 | Unknown |
| ENSDARG00000101624 | ENSFM00250000000425 | Lamin |
| ENSDARG00000101627 | ENSFM00250000002354 | Translocon Associated Subunit Alpha Precursor Trap Alpha Signal Sequence Receptor Subunit Alpha Ssr Alpha |
| ENSDARG00000101631 | ENSFM00250000004026 | Electron Transfer Flavoprotein Subunit Alpha Mitochondrial Precursor Alpha Etf |
| ENSDARG00000101669 | ENSFM00800002020440 | Unknown |
| ENSDARG00000101669 | ENSFM00250000003015 | Elmo Domain Containing |
| ENSDARG00000101687 | ENSFM00260000050563 | Microtubule Associated 4 Map 4 |
| ENSDARG00000101713 | ENSFM00780001898523 | Sodium Channel Subunit Beta 2 Precursor |
| ENSDARG00000101719 | ENSFM00250000003384 | Poly Adp Ribose Glycohydrolase EC_3.2.1.143 |
| ENSDARG00000101726 | ENSFM00350000105388 | Carboxylesterase |
| ENSDARG00000101755 | ENSFM00730001521681 | Camp Dependent Kinase Type I Alpha Regulatory Camp Dependent Kinase Type I Alpha Regulatory Subunit N Terminally Processed |
| ENSDARG00000101759 | ENSFM00250000001140 | Homer Homolog Homer Vasp/Ena Related Gene Up Regulated During Seizure And Ltp Vesl |
| ENSDARG00000101766 | ENSFM00510000505850 | Prothymosin Alpha B |
| ENSDARG00000101776 | ENSFM00500000272992 | Synaptotagmin 12 Synaptotagmin Xii Sytxii |
| ENSDARG00000101793 | ENSFM00250000001660 | Transmembrane Gtpase |
| ENSDARG00000101804 | ENSFM00800002022006 | Unknown |
| ENSDARG00000101812 | ENSFM00250000001584 | Src Kinase Signaling Inhibitor 1 Snap 25 Interacting Snip P130CAS Associated P140CAP |
| ENSDARG00000101858 | ENSFM00740001589112 | Microtubule Actin Cross Linking Factor 1 Actin Cross Linking Family 7 |
| ENSDARG00000101869 | ENSFM00250000000552 | Membrane Associated Guanylate Kinase Ww And Pdz Domain Containing Membrane Associated Guanylate Kinase Inverted Magi |
| ENSDARG00000101915 | ENSFM00670001235859 | Peptidyl Prolyl Cis Trans Isomerase Nima Interacting 1 EC_5.2.1.8 Peptidyl Prolyl Cis Trans Isomerase PIN1 Ppiase PIN1 |
| ENSDARG00000101949 | ENSFM00250000001973 | Numb |
| ENSDARG00000102009 | ENSFM00250000000829 | Serine/Threonine Phosphatase 2A 55 Kda Regulatory Subunit B PP2A Subunit B B55 PP2A Subunit B PR55 PP2A Subunit B R2 PP2A Subunit B |
| ENSDARG00000102024 | ENSFM00670001235835 | Charged Multivesicular Body 6 Chromatin Modifying 6 |
| ENSDARG00000102064 | ENSFM00290000065519 | Potassium Voltage Gated Channel Subfamily A Member Voltage Gated Potassium Channel Subunit KV1 |
| ENSDARG00000102067 | ENSFM00730001521627 | Metabotropic Glutamate Receptor Precursor |
| ENSDARG00000102078 | ENSFM00750001632414 | Ras Related Rab 35 |
| ENSDARG00000102081 | ENSFM00600000921208 | Trio And F Actin Binding Protein Tara Trio Associated Repeat On Actin |
| ENSDARG00000102100 | ENSFM00800002003635 | Macrophage Mannose Receptor 1 Precursor Mmr CD206 Antigen |
| ENSDARG00000102115 | ENSFM00670001238685 | Nadh Dehydrogenase 1 Subunit C2 |
| ENSDARG00000102125 | ENSFM00250000001463 | Iq Motif And SEC7 Domain Containing |
| ENSDARG00000102142 | ENSFM00800002003370 | Endothelin Converting Enzyme Ece EC_3.4.24.71 |
| ENSDARG00000102153 | ENSFM00250000000469 | Neuropilin Precursor Vascular Endothelial Cell Growth Factor 165 Receptor |
| ENSDARG00000102184 | ENSFM00250000001057 | E3 Ubiquitin Ligase EC_6.3.2.- Mind Bomb Homolog |
| ENSDARG00000102191 | ENSFM00730001521728 | Copine Copine |
| ENSDARG00000102200 | ENSFM00500000270074 | Alpha Mannosidase 2 EC_3.2.1.114 Golgi Alpha Mannosidase Ii Aman Ii Man Ii Mannosidase Alpha Class 2A Member 1 Mannosyl Oligosaccharide 1 3 1 6 Alpha Mannosidase |
| ENSDARG00000102216 | ENSFM00250000000275 | Disks Large Homolog Synapse Associated Sap |
| ENSDARG00000102249 | ENSFM00730001522144 | Xaa Pro Dipeptidase X Pro Dipeptidase EC_3.4.13.9 Imidodipeptidase Peptidase D Proline Dipeptidase Prolidase |
| ENSDARG00000102254 | ENSFM00500000269896 | Pyrroline 5 Carboxylate Reductase 3 P5C Reductase 3 P5CR 3 EC_1.5.1.2 Pyrroline 5 Carboxylate Reductase |
| ENSDARG00000102295 | ENSFM00730001522148 | Dnaj Homolog Subfamily A Member 3 Mitochondrial Precursor Dnaj Tid 1 1 Tumorous Imaginal Discs TID56 Homolog |
| ENSDARG00000102317 | ENSFM00500000270069 | 60S Ribosomal L26 |
| ENSDARG00000102335 | ENSFM00800002003409 | Anoctamin Transmembrane |
| ENSDARG00000102356 | ENSFM00250000003026 | Non Specific Lipid Transfer Nsl Tp EC_2.3.1.176 Propanoyl Coa C Acyltransferase Scp Chi Scpx Sterol Carrier 2 Scp 2 Sterol Carrier X Scp X |
| ENSDARG00000102362 | ENSFM00800002003431 | Calcium Binding Mitochondrial Carrier Mitochondrial Aspartate Glutamate Carrier Solute Carrier Family 25 Member |
| ENSDARG00000102365 | ENSFM00250000003294 | Transcription Elongation Factor B Polypeptide 3 Elongin 110 Kda Subunit Elongin A Eloa Rna Polymerase Ii Transcription Factor Siii Subunit A1 Siii P110 |
| ENSDARG00000102376 | ENSFM00250000002019 | Voltage Dependent Calcium Channel Gamma Subunit Neuronal Voltage Gated Calcium Channel Gamma Subunit Transmembrane Ampar Regulatory Gamma Tarp Gamma |
| ENSDARG00000102381 | ENSFM00500000269830 | Choline Transporter Solute Carrier Family 44 Member |
| ENSDARG00000102387 | ENSFM00500000270245 | Zinc Transporter ZIP14 Precursor Solute Carrier Family 39 Member 14 Zrt And Irt 14 Zip 14 |
| ENSDARG00000102394 | ENSFM00810002040662 | Ap 1 Complex Subunit Gamma 1 Adaptor Complex Ap 1 Subunit Gamma 1 Adaptor Related Complex 1 Subunit Gamma 1 Clathrin Assembly Complex 1 Gamma 1 Large Chain Gamma Adaptin GAMMA1 Adaptin Golgi Adaptor HA1/AP1 Adaptin Subunit Gamma 1 |
| ENSDARG00000102412 | ENSFM00800002017135 | Unknown |
| ENSDARG00000102412 | ENSFM00800002021480 | Unknown |
| ENSDARG00000102412 | ENSFM00500000270657 | Enoyl Coa Delta Isomerase 2 Mitochondrial Precursor EC_5.3.3.8 Delta 3 Delta 2 Enoyl Coa Isomerase D3 D2 Enoyl Coa Isomerase Dodecenoyl Coa Isomerase Peroxisomal 3 2 Trans Enoyl Coa Isomerase Peci |
| ENSDARG00000102415 | ENSFM00760001714632 | Gelsolin Actin Depolymerizing Factor Adf |
| ENSDARG00000102417 | ENSFM00250000004823 | 26S Proteasome Non Atpase Regulatory Subunit 7 26S Proteasome Regulatory Subunit RPN8 |
| ENSDARG00000102424 | ENSFM00250000000900 | Rho Gtpase Activating Rho Type Gtpase Activating Rhogap Interacting With CIP4 Homologs Rich |
| ENSDARG00000102441 | ENSFM00250000001018 | Catenin Alpha 2 Alpha N Catenin |
| ENSDARG00000102453 | ENSFM00800002003429 | Excitatory Amino Acid Transporter 2 Sodium Dependent Glutamate/Aspartate Transporter 2 Solute Carrier Family 1 Member 2 |
| ENSDARG00000102458 | ENSFM00250000001308 | Huntingtin Interacting 1 Hip 1 Huntingtin Interacting I Hip I |
| ENSDARG00000102472 | ENSFM00440000236967 | Btb/Poz Domain Containing 17 Precursor Galectin 3 Binding |
| ENSDARG00000102532 | ENSFM00250000001140 | Homer Homolog Homer Vasp/Ena Related Gene Up Regulated During Seizure And Ltp Vesl |
| ENSDARG00000102538 | ENSFM00800002003420 | Methylcrotonoyl Coa Carboxylase Subunit Alpha Mitochondrial Precursor Mccase Subunit Alpha EC_6.4.1.4 3 Methylcrotonyl Coa Carboxylase 1 3 Methylcrotonyl Coa Carboxylase Biotin Containing Subunit 3 Methylcrotonyl Coa:Carbon Dioxide Ligase Subunit Alpha |
| ENSDARG00000102584 | ENSFM00800002003632 | Copine Copine |
| ENSDARG00000102587 | ENSFM00500000271280 | Wd Repeat Containing Mio |
| ENSDARG00000102607 | ENSFM00500000270616 | Mitochondrial Processing Peptidase Subunit Alpha Precursor EC_3.4.24.64 Alpha Mpp |
| ENSDARG00000102626 | ENSFM00740001589159 | Extracellular Matrix Precursor |
| ENSDARG00000102640 | ENSFM00800002003471 | Disulfide Isomerase A3 Precursor EC_5.3.4.1.58 Kda Glucose Regulated 58 Kda Microsomal P58 Disulfide Isomerase Er 60 Endoplasmic Reticulum Resident 57 Er 57 ERP57 Endoplasmic Reticulum Resident 60 Er 60 ERP60 |
| ENSDARG00000102643 | ENSFM00600000921337 | Charged Multivesicular Body 1B Chromatin Modifying 1B CHMP1B |
| ENSDARG00000102674 | ENSFM00760001714778 | Cytoskeleton Associated 5 |
| ENSDARG00000102684 | ENSFM00730001522046 | Scribble |
| ENSDARG00000102690 | ENSFM00250000000629 | Regulating Synaptic Membrane Exocytosis Rab 3 Interacting Molecule Rim |
| ENSDARG00000102696 | ENSFM00250000002491 | Dnaj Homolog Subfamily C Member 11 |
| ENSDARG00000102730 | ENSFM00460000244930 | Mitogen Activated Kinase Map Kinase Mapk EC_2.7.11.24 Stress Activated Kinase C Jun N Terminal Kinase |
| ENSDARG00000102744 | ENSFM00670001235926 | Microsomal Glutathione S Transferase 3 Microsomal Gst 3 EC_2.5.1.18 Microsomal Gst Iii |
| ENSDARG00000102755 | ENSFM00730001522226 | Echinoderm Microtubule Associated 6 Emap 6 Echinoderm Microtubule Associated 5 |
| ENSDARG00000102765 | ENSFM00810002040688 | Lon Protease Homolog Mitochondrial |
| ENSDARG00000102780 | ENSFM00520000517802 | Kinase C Type Pkc EC_2.7.11.13 |
| ENSDARG00000102790 | ENSFM00250000002597 | Polyribonucleotide Nucleotidyltransferase 1 Mitochondrial Precursor EC_2.7.7.8 3' 5' Rna Exonuclease OLD35 Pnpase Old 35 Polynucleotide Phosphorylase 1 Pnpase 1 Polynucleotide Phosphorylase |
| ENSDARG00000102793 | ENSFM00500000271937 | Monocarboxylate Transporter 7 Mct 7 Monocarboxylate Transporter 6 Mct 6 Solute Carrier Family 16 Member 6 |
| ENSDARG00000102808 | ENSFM00500000269786 | Calreticulin Precursor |
| ENSDARG00000102813 | ENSFM00800002020563 | Unknown |
| ENSDARG00000102822 | ENSFM00670001236447 | Mitochondrial Import Inner Membrane Translocase Subunit TIM16 Mitochondria Associated Granulocyte Macrophage Csf Signaling Molecule Presequence Translocated Associated Motor Subunit PAM16 |
| ENSDARG00000102824 | ENSFM00780001898335 | Protocadherin Precursor |
| ENSDARG00000102825 | ENSFM00320000100099 | Noelin Precursor Neuronal Olfactomedin Related Er Localized Olfactomedin 1 |
| ENSDARG00000102845 | ENSFM00400000131747 | Serine/Threonine Kinase EC_2.7.11.1 Doublecortin And Cam Kinase Doublecortin Kinase |
| ENSDARG00000102855 | ENSFM00260000050557 | Netrin Receptor Dcc Precursor Tumor Suppressor Dcc |
| ENSDARG00000102874 | ENSFM00250000002778 | Micos Complex Subunit MIC60 Precursor Mitochondrial Inner Membrane Mitofilin |
| ENSDARG00000102877 | ENSFM00250000004835 | Valacyclovir Hydrolase Precursor Vacvase Valacyclovirase EC_3.1.-.- Biphenyl Hydrolase |
| ENSDARG00000102878 | ENSFM00250000000867 | Microtubule Associated Rp/Eb Family Member |
| ENSDARG00000102883 | ENSFM00740001589095 | Spectrin Beta Chain Erythrocytic Beta Spectrin |
| ENSDARG00000102889 | ENSFM00760001714685 | Septin 7 CDC10 Homolog |
| ENSDARG00000102975 | ENSFM00250000005181 | Neuron Specific Family Member |
| ENSDARG00000103007 | ENSFM00250000001742 | 40S Ribosomal S3 EC_4.2.99.18 |
| ENSDARG00000103032 | ENSFM00800002003504 | Ras Related Rap Precursor |
| ENSDARG00000103052 | ENSFM00250000003214 | Prenylcysteine Oxidase Precursor |
| ENSDARG00000103069 | ENSFM00290000065521 | Precursor |
| ENSDARG00000103083 | ENSFM00810002040719 | Dna Damage Binding 1 Damage Specific Dna Binding 1 |
| ENSDARG00000103095 | ENSFM00250000003276 | Inhibitor Of Nuclear Factor Kappa B Kinase Subunit Epsilon I Kappa B Kinase Epsilon Ikk E Ikk Epsilon Ikbke EC_2.7.11.10 Inducible I Kappa B Kinase Ikk I |
| ENSDARG00000103109 | ENSFM00800002024380 | Unknown |
| ENSDARG00000103109 | ENSFM00390000126295 | Neural Cell Adhesion Molecule L1 L1 L1 |
| ENSDARG00000103123 | ENSFM00250000007087 | Neuronal Tyrosine Phosphorylated Phosphoinositide 3 Kinase Adapter 1 |
| ENSDARG00000103135 | ENSFM00260000050877 | Stomatin 2 Mitochondrial Precursor Slp 2 |
| ENSDARG00000103144 | ENSFM00250000003688 | Phosphatase 1 Regulatory Subunit 21 Coiled Coil Domain Containing 128 Klraq Motif Containing 1 |
| ENSDARG00000103173 | ENSFM00800002015497 | Unknown |
| ENSDARG00000103173 | ENSFM00250000005899 | Syntaxin 8 |
| ENSDARG00000103190 | ENSFM00250000002637 | 26S Proteasome Regulatory Subunit RPN1 Regulatory 26S Proteasome Subunit |
| ENSDARG00000103200 | ENSFM00250000002158 | Precursor Lung Seven Transmembrane Receptor |
| ENSDARG00000103203 | ENSFM00800002022009 | Unknown |
| ENSDARG00000103203 | ENSFM00500000271517 | Vacuolar Sorting Associated 28 Homolog Escrt I Complex Subunit VPS28 |
| ENSDARG00000103256 | ENSFM00780001898335 | Protocadherin Precursor |
| ENSDARG00000103284 | ENSFM00250000005577 | Nicastrin Precursor |
| ENSDARG00000103288 | ENSFM00800002003582 | Pyruvate Dehydrogenase Phosphatase Regulatory Subunit Mitochondrial Precursor Pdpr |
| ENSDARG00000103298 | ENSFM00500000270247 | Epoxide Hydrolase 1 EC_3.3.2.9 Epoxide Hydratase Microsomal Epoxide Hydrolase |
| ENSDARG00000103313 | ENSFM00250000000940 | Mitochondrial Rho Gtpase 2 Miro 2 EC_3.6.5.- Ras Homolog Gene Family Member T2 |
| ENSDARG00000103332 | ENSFM00730001521400 | Atp Sensitive Inward Rectifier Potassium Channel Atp Potassium Channel K Inward Rectifier K + Channel 1 Potassium Channel Inwardly Rectifying Subfamily J Member |
| ENSDARG00000103333 | ENSFM00250000000906 | Brain Specific Angiogenesis Inhibitor 1 Associated 2 Bai Associated 2 BAI1 Associated 2 Insulin Receptor Substrate Of 53 Kda IRSP53 Insulin Receptor Substrate P53 |
| ENSDARG00000103359 | ENSFM00250000001398 | Slit And Ntrk Precursor |
| ENSDARG00000103364 | ENSFM00250000002339 | Citrate Synthase Mitochondrial Precursor EC_2.3.3.1 Citrate Si Synthase |
| ENSDARG00000103369 | ENSFM00810002040751 | Transaldolase EC_2.2.1.2 |
| ENSDARG00000103391 | ENSFM00750001632398 | Ras Related Rab |
| ENSDARG00000103394 | ENSFM00670001235340 | Kinesin Heavy Chain Kinesin Heavy Chain Kinesin Heavy Chain |
| ENSDARG00000103403 | ENSFM00800002003440 | Gtp Binding SAR1 |
| ENSDARG00000103404 | ENSFM00670001235447 | Ubiquitin Carboxyl Terminal Hydrolase Isozyme L5 Uch L5 EC_3.4.19.12 Ubiquitin C Terminal Hydrolase UCH37 Ubiquitin Thioesterase L5 |
| ENSDARG00000103428 | ENSFM00750001632363 | 2 Oxoglutarate Dehydrogenase Mitochondrial Precursor 2 Oxoglutarate Dehydrogenase Complex Component E1 Ogdc E1 Alpha Ketoglutarate Dehydrogenase |
| ENSDARG00000103433 | ENSFM00500000270978 | 60S Ribosomal L14 |
| ENSDARG00000103435 | ENSFM00670001235644 | Sorbin And SH3 Domain Containing 1 Ponsin SH3 Domain 5 S C Cbl Associated Cap |
| ENSDARG00000103453 | ENSFM00250000006331 | Zinc Finger 1 |
| ENSDARG00000103457 | ENSFM00250000003963 | Spartin |
| ENSDARG00000103479 | ENSFM00740001589097 | Receptor Type Tyrosine Phosphatase Precursor EC_3.1.3.48 |
| ENSDARG00000103490 | ENSFM00250000000643 | Dihydropyrimidinase Related Drp Collapsin Response Mediator Crmp |
| ENSDARG00000103498 | ENSFM00500000273787 | Ependymin Precursor Epd |
| ENSDARG00000103503 | ENSFM00500000270667 | Diacylglycerol O Acyltransferase 1 EC_2.3.1.20 Acyl Coa Retinol O Fatty Acyltransferase Arat Retinol O Fatty Acyltransferase EC_2.3.1.- 76 Diglyceride Acyltransferase |
| ENSDARG00000103516 | ENSFM00770001847547 | Transport SEC24 Related |
| ENSDARG00000103521 | ENSFM00250000002018 | Calcyphosin |
| ENSDARG00000103529 | ENSFM00730001521918 | Sorting Nexin |
| ENSDARG00000103551 | ENSFM00730001522194 | E3 Ubiquitin Ligase RNF213 EC_6.3.2.- |
| ENSDARG00000103563 | ENSFM00250000002234 | Intraflagellar Transport 122 Homolog Wd Repeat Containing 10 |
| ENSDARG00000103672 | ENSFM00600000929849 | Uncharacterized |
| ENSDARG00000103714 | ENSFM00460000244935 | Ras Related Rab 6 |
| ENSDARG00000103740 | ENSFM00670001235667 | FUN14 Domain Containing |
| ENSDARG00000103770 | ENSFM00250000004612 | Armadillo Repeat Containing 6 |
| ENSDARG00000103774 | ENSFM00250000003304 | Lim And Calponin Homology Domains Containing 1 |
| ENSDARG00000103791 | ENSFM00250000000895 | Nucleoside Diphosphate Kinase Ndk Ndp Kinase EC_2.7.4.6 |
| ENSDARG00000103799 | ENSFM00250000002843 | Lysine Trna Ligase EC_6.1.1.6 Lysyl Trna Synthetase Lysrs |
| ENSDARG00000103800 | ENSFM00730001521681 | Camp Dependent Kinase Type I Alpha Regulatory Camp Dependent Kinase Type I Alpha Regulatory Subunit N Terminally Processed |
| ENSDARG00000103811 | ENSFM00810002040809 | Glycogen Debranching Enzyme Glycogen Debrancher |
| ENSDARG00000103824 | ENSFM00670001235733 | Histidine Triad Nucleotide Binding 1 Ec 3 Adenosine 5' Monophosphoramidase Kinase C Inhibitor 1 Kinase C Interacting 1 Pkci 1 |
| ENSDARG00000103846 | ENSFM00800002003443 | 78 Kda Glucose Regulated Grp 78 Heat Shock 70 Kda 5 Immunoglobulin Heavy Chain Binding Bip |
| ENSDARG00000103847 | ENSFM00740001589278 | Adenylate Cyclase Type 9 EC_4.6.1.1 Atp Pyrophosphate Lyase 9 Adenylate Cyclase Type Ix Adenylyl Cyclase 9 |
| ENSDARG00000103900 | ENSFM00500000269805 | Septin |
| ENSDARG00000103902 | ENSFM00250000002568 | Nuclear Distribution Nude 1 |
| ENSDARG00000103914 | ENSFM00560000771357 | THEM6 Precursor |
| ENSDARG00000103937 | ENSFM00800002023711 | Unknown |
| ENSDARG00000103937 | ENSFM00250000000535 | N Myc Downstream Regulated Gene |
| ENSDARG00000103959 | ENSFM00730001521340 | Serine/Threonine Kinase Pak EC_2.7.11.1 Pak P21 Activated Kinase Pak |
| ENSDARG00000103962 | ENSFM00250000000681 | Adducin |
| ENSDARG00000103979 | ENSFM00500000269786 | Calreticulin Precursor |
| ENSDARG00000103987 | ENSFM00500000272336 | Calcium And Integrin Binding 1 Cib Calmyrin Dna Pkcs Interacting Kinase Interacting Kip |
| ENSDARG00000103993 | ENSFM00500000313527 | Secretogranin Ii |
| ENSDARG00000103994 | ENSFM00750001632315 | Peptidyl Prolyl Cis Trans Isomerase Ppiase EC_5.2.1.8 Cyclophilin Rotamase |
| ENSDARG00000104014 | ENSFM00730001521220 | Kinase EC_2.7.11.1 |
| ENSDARG00000104018 | ENSFM00500000273644 | Myotrophin |
| ENSDARG00000104023 | ENSFM00800002024129 | Unknown |
| ENSDARG00000104023 | ENSFM00320000100165 | Myelin Associated Glycoprotein Precursor Siglec 4A |
| ENSDARG00000104065 | ENSFM00740001589073 | Zinc Finger |
| ENSDARG00000104066 | ENSFM00760001714614 | Dnaj Homolog Subfamily A Member Precursor |
| ENSDARG00000104068 | ENSFM00500000270850 | Glutathione S Transferase P EC_2.5.1.18 Gst Class Pi |
| ENSDARG00000104085 | ENSFM00800002003649 | Atp Binding Cassette Sub Family D Member Kda Peroxisomal Membrane PMP70 |
| ENSDARG00000104086 | ENSFM00250000002627 | Rho Gtpase Activating 39 |
| ENSDARG00000104094 | ENSFM00800002003453 | Beta Adrenergic Receptor Kinase 1 Beta Ark 1 EC_2.7.11.15 G Coupled Receptor Kinase 2 |
| ENSDARG00000104101 | ENSFM00800002023681 | Unknown |
| ENSDARG00000104101 | ENSFM00740001589344 | Peripheral Type Benzodiazepine Receptor Associated 1 Prax 1 Rims Binding 1 Rim BP1 |
| ENSDARG00000104106 | ENSFM00500000270247 | Epoxide Hydrolase 1 EC_3.3.2.9 Epoxide Hydratase Microsomal Epoxide Hydrolase |
| ENSDARG00000104111 | ENSFM00250000008011 | 28S Ribosomal S30 Mitochondrial Mrp S30 S30MT |
| ENSDARG00000104119 | ENSFM00740001589124 | EC_2.7.11.1 |
| ENSDARG00000104139 | ENSFM00650001139994 | Sodium/Potassium Transporting Atpase Subunit Alpha Na + /K + Atpase Alpha Subunit EC_3.6.3.9 Sodium Pump Subunit Alpha |
| ENSDARG00000104157 | ENSFM00800002024283 | Unknown |
| ENSDARG00000104172 | ENSFM00250000004873 | Diablo Homolog Mitochondrial |
| ENSDARG00000104173 | ENSFM00260000050713 | Elongation Factor Tu Mitochondrial Precursor |
| ENSDARG00000104179 | ENSFM00500000271919 | RAB11 Family Interacting 2 RAB11 FIP2 |
| ENSDARG00000104225 | ENSFM00250000002568 | Nuclear Distribution Nude 1 |
| ENSDARG00000104230 | ENSFM00810002040640 | Transport SEC23 Related |
| ENSDARG00000104235 | ENSFM00750001632326 | Unconventional Myosin Va Dilute Myosin Heavy Chain Non Muscle |
| ENSDARG00000104264 | ENSFM00500000269993 | Xk Related |
| ENSDARG00000104267 | ENSFM00800002015296 | Unknown |
| ENSDARG00000104267 | ENSFM00760001719894 | Osteonectin |
| ENSDARG00000104267 | ENSFM00250000001947 | Transforming Growth Factor Beta Induced Ig H3 Precursor Beta Ig H3 Kerato Epithelin Rgd Containing Collagen Associated Rgd Cap |
| ENSDARG00000104279 | ENSFM00250000000617 | Amyloid Beta A4 Precursor Abpp App Alzheimer Disease Amyloid A4 Homolog |
| ENSDARG00000104282 | ENSFM00260000050557 | Netrin Receptor Dcc Precursor Tumor Suppressor Dcc |
| ENSDARG00000104283 | ENSFM00730001521452 | Serine/Threonine Kinase Mrck EC_2.7.11.1 CDC42 Binding Kinase Dmpk Myotonic Dystrophy Kinase Related CDC42 Binding Kinase Mrck Myotonic Dystrophy Kinase |
| ENSDARG00000104342 | ENSFM00730001521484 | Casein Kinase I Gamma Cki Gamma EC_2.7.11.1 |
| ENSDARG00000104343 | ENSFM00500000269861 | Peptidyl Prolyl Cis Trans Isomerase B Precursor Ppiase B EC_5.2.1.8 Rotamase B |
| ENSDARG00000104358 | ENSFM00250000000820 | Clip Associating Cytoplasmic Linker Associated |
| ENSDARG00000104375 | ENSFM00750001632389 | Transport SEC24A SEC24 Related A |
| ENSDARG00000104396 | ENSFM00390000126422 | E3 Ubiquitin/ISG15 Ligase TRIM25 EC_6.3.2.19 EC_6.3.2.- N3 Estrogen Responsive Finger Tripartite Motif Containing 25 Ubiquitin/ISG15 Conjugating Enzyme TRIM25 Zinc Finger 147 |
| ENSDARG00000104401 | ENSFM00800002003559 | Atp Dependent Zinc Metalloprotease Ym EC_3.4.24.- Atp Dependent Metalloprotease FTSH1 Meg 4 YME1 1 |
| ENSDARG00000104413 | ENSFM00250000005331 | Rogdi Homolog |
| ENSDARG00000104414 | ENSFM00730001521489 | Serine Hydroxymethyltransferase Shmt EC_2.1.2.1 Glycine Hydroxymethyltransferase Serine Methylase |
| ENSDARG00000104431 | ENSFM00800002003375 | Excitatory Amino Acid Transporter Sodium Dependent Glutamate/Aspartate Transporter Solute Carrier Family 1 Member |
| ENSDARG00000104439 | ENSFM00250000006965 | Claudin Domain Containing 1 |
| ENSDARG00000104458 | ENSFM00670001241589 | Ribonuclease Kappa B Rnase K B Rnase Kappa B EC_3.1.-.- |
| ENSDARG00000104480 | ENSFM00250000000931 | Potassium/Sodium Hyperpolarization Activated Cyclic Nucleotide Gated Channel |
| ENSDARG00000104483 | ENSFM00250000002325 | Pdz Domain Containing |
| ENSDARG00000104517 | ENSFM00800002016273 | Unknown |
| ENSDARG00000104537 | ENSFM00800002014398 | Unknown |
| ENSDARG00000104549 | ENSFM00250000000900 | Rho Gtpase Activating Rho Type Gtpase Activating Rhogap Interacting With CIP4 Homologs Rich |
| ENSDARG00000104581 | ENSFM00250000001136 | TOM1 2 Target Of Myb 2 |
| ENSDARG00000104615 | ENSFM00730001521674 | Serine/Threonine Kinase EC_2.7.11.1 Salt Inducible Kinase Sik Serine/Threonine Kinase SNF1 Kinase |
| ENSDARG00000104664 | ENSFM00400000131747 | Serine/Threonine Kinase EC_2.7.11.1 Doublecortin And Cam Kinase Doublecortin Kinase |
| ENSDARG00000104665 | ENSFM00250000000717 | Kin Of Irre Precursor Kin Of Irregular Chiasm Nephrin |
| ENSDARG00000104668 | ENSFM00800002022057 | Unknown |
| ENSDARG00000104674 | ENSFM00500000270021 | T Complex 1 Subunit Beta Tcp 1 Beta Cct Beta |
| ENSDARG00000104696 | ENSFM00500000272483 | Apolipoprotein O FAM121B |
| ENSDARG00000104701 | ENSFM00800002024297 | Unknown |
| ENSDARG00000104701 | ENSFM00250000004934 | MAP7 Domain Containing 1 |
| ENSDARG00000104702 | ENSFM00760001714613 | Catalase EC_1.11.1.6 |
| ENSDARG00000104732 | ENSFM00250000006084 | Malonyl Coa Decarboxylase Mitochondrial Precursor Mcd EC_4.1.1.9 |
| ENSDARG00000104767 | ENSFM00750001632364 | Dipeptidyl Aminopeptidase 6 Dppx Dipeptidyl Aminopeptidase Related Dipeptidyl Peptidase 6 Dipeptidyl Peptidase Iv Dipeptidyl Peptidase Vi Dpp Vi |
| ENSDARG00000104771 | ENSFM00500000271055 | Mitochondrial Import Inner Membrane Translocase Subunit Precursor |
| ENSDARG00000104786 | ENSFM00380000124679 | Neuroligin Precursor |
| ENSDARG00000104793 | ENSFM00730001522404 | Diacylglycerol Kinase Epsilon Dag Kinase Epsilon EC_2.7.1.107 Diglyceride Kinase Epsilon Dgk Epsilon |
| ENSDARG00000104801 | ENSFM00800002024325 | Unknown |
| ENSDARG00000104801 | ENSFM00250000000193 | Tubulin Beta Chain |
| ENSDARG00000104820 | ENSFM00250000001069 | Ph And SEC7 Domain Containing 1 Exchange Factor ARF6 Pleckstrin Homology And SEC7 Domain Containing 1 |
| ENSDARG00000104826 | ENSFM00780001898335 | Protocadherin Precursor |
| ENSDARG00000104889 | ENSFM00250000005282 | Importin Subunit Beta 1 Karyopherin Subunit Beta 1 Nuclear Factor P97 Pore Targeting Complex 97 Kda Subunit PTAC97 |
| ENSDARG00000104934 | ENSFM00250000001103 | Talin |
| ENSDARG00000104937 | ENSFM00740001589097 | Receptor Type Tyrosine Phosphatase Precursor EC_3.1.3.48 |
| ENSDARG00000104945 | ENSFM00670001239308 | Beta Synuclein |
| ENSDARG00000104953 | ENSFM00250000001541 | Dolichyl Diphosphooligosaccharide Glycosyltransferase Subunit Oligosaccharyl Transferase Subunit STT3 EC_2.4.99.18 |
| ENSDARG00000104958 | ENSFM00250000005444 | Kelch Domain Containing |
| ENSDARG00000105003 | ENSFM00600000921511 | Sodium Channel Subunit Beta 1 Precursor |
| ENSDARG00000105009 | ENSFM00250000003817 | N G N G Dimethylarginine Dimethylaminohydrolase 2 Ddah 2 Dimethylarginine Dimethylaminohydrolase 2 EC_3.5.3.18 Ddahii Dimethylargininase 2 |
| ENSDARG00000105029 | ENSFM00250000003009 | Tubulin Polymerization Promoting Family Member |
| ENSDARG00000105088 | ENSFM00800002024124 | Unknown |
| ENSDARG00000105088 | ENSFM00250000000375 | Teneurin Ten Odd Oz/Ten M Homolog Tenascin Ten Teneurin Transmembrane |
| ENSDARG00000105099 | ENSFM00250000003047 | Golgin Subfamily A Member 7 |
| ENSDARG00000105115 | ENSFM00250000000608 | Acid Sensing Ion Channel Amiloride Sensitive Cation Channel |
| ENSDARG00000105116 | ENSFM00730001521641 | Disulfide Isomerase Precursor Pdi EC_5.3.4.1 Prolyl 4 Hydroxylase Subunit Beta |
| ENSDARG00000105142 | ENSFM00250000000490 | V Type Proton Atpase 116 Kda Subunit A V Atpase 116 Kda Proton 116 Kda Subunit Vacuolar Proton Translocating Atpase 116 Kda Subunit A |
| ENSDARG00000105159 | ENSFM00760001714682 | Phosphorylase B Kinase Regulatory Subunit Alpha Phosphorylase Kinase Subunit |
| ENSDARG00000105201 | ENSFM00730001521191 | Pituitary Adenylate Cyclase Activating Polypeptide Type Receptor Pacap Type Receptor Pacap R Pacap |
| ENSDARG00000105209 | ENSFM00600000921208 | Trio And F Actin Binding Protein Tara Trio Associated Repeat On Actin |
| ENSDARG00000105243 | ENSFM00800002017484 | Unknown |
| ENSDARG00000105243 | ENSFM00800002017485 | Unknown |
| ENSDARG00000105243 | ENSFM00250000002216 | Udp N Acetylglucosamine Peptide N Acetylglucosaminyltransferase 110 Kda Subunit EC_2.4.1.255 O Glcnac Transferase Subunit P110 O Linked N Acetylglucosamine Transferase 110 Kda Subunit Ogt |
| ENSDARG00000105296 | ENSFM00480000262766 | Ap 3 Complex Subunit Beta 1 Adaptor Complex Ap 3 Subunit Beta 1 Adaptor Related Complex 3 Subunit Beta 1 Beta 3A Adaptin Clathrin Assembly Complex 3 Beta 1 Large Chain |
Supplementary Table 4d. Ensemble Families from mouse postsynaptic density proteins
For each zebrafish postsynaptic density protein its corresponding Ensembl Family code and descriptor are given.
| Ensembl Gene Id | Ensembl Protein Family Id(S) | Ensembl Family Description |
|---|---|---|
| ENSDARG00000000103 | ENSFM00270000056424 | Myosin Cellular Myosin Heavy Chain Type Myosin Heavy Chain Myosin Heavy Chain Non Muscle Non Muscle Myosin Heavy Chain Nmmhc Non Muscle Myosin Heavy Chain Nmmhc Ii Nmmhc |
| ENSDARG00000000189 | ENSFM00730001521376 | Semaphorin Precursor Semaphorin Sema |
| ENSDARG00000000370 | ENSFM00740001589124 | EC_2.7.11.1 |
| ENSDARG00000000472 | ENSFM00800002003683 | Contactin 1 Precursor Neural Cell Surface F3 |
| ENSDARG00000000861 | ENSFM00730001522046 | Scribble |
| ENSDARG00000001014 | ENSFM00270000056424 | Myosin Cellular Myosin Heavy Chain Type Myosin Heavy Chain Myosin Heavy Chain Non Muscle Non Muscle Myosin Heavy Chain Nmmhc Non Muscle Myosin Heavy Chain Nmmhc Ii Nmmhc |
| ENSDARG00000001154 | ENSFM00740001589344 | Peripheral Type Benzodiazepine Receptor Associated 1 Prax 1 Rims Binding 1 Rim BP1 |
| ENSDARG00000001220 | ENSFM00250000001622 | E3 Ubiquitin Ligase MYCBP2 EC_6.3.2.- Myc Binding 2 Pam/Highwire/Rpm 1 Protein Associated With Myc |
| ENSDARG00000001241 | ENSFM00250000002682 | Poly U Binding Splicing Factor Kda Poly U Binding Splicing Factor |
| ENSDARG00000001578 | ENSFM00500000270475 | Enoyl Coa Hydratase Mitochondrial Precursor EC_4.2.1.17 Enoyl Coa Hydratase 1 Short Chain Enoyl Coa Hydratase Sceh |
| ENSDARG00000001710 | ENSFM00780001898418 | Flotillin 1 |
| ENSDARG00000001733 | ENSFM00250000004323 | Ptb Domain Containing Engulfment Adapter 1 Cell Death 6 Homolog Ptb Domain Adapter Ced 6 |
| ENSDARG00000001835 | ENSFM00250000001647 | Sodium Leak Channel Non Selective Voltage Gated Channel 1 |
| ENSDARG00000001873 | ENSFM00500000270390 | D 3 Phosphoglycerate Dehydrogenase 3 Pgdh EC_1.1.1.95 |
| ENSDARG00000001881 | ENSFM00250000000524 | Voltage Dependent L Type Calcium Channel Subunit Beta Calcium Channel Voltage Dependent Subunit Beta |
| ENSDARG00000001939 | ENSFM00250000001080 | Galactosylgalactosylxylosylprotein 3 Beta Glucuronosyltransferase EC_2.4.1.135 Beta 1 3 Glucuronyltransferase Glucuronosyltransferase Glcat Udp Beta 1 3 Glucuronyltransferase Glcuat |
| ENSDARG00000002026 | ENSFM00400000131794 | Quaking |
| ENSDARG00000002082 | ENSFM00250000001591 | Casein Kinase Ii Subunit Ck Ii EC_2.7.11.1 |
| ENSDARG00000002167 | ENSFM00250000000524 | Voltage Dependent L Type Calcium Channel Subunit Beta Calcium Channel Voltage Dependent Subunit Beta |
| ENSDARG00000002538 | ENSFM00250000004547 | WD40 Repeat Containing SMU1 Smu 1 Suppressor Of Mec 8 And Unc 52 Homolog [Contains WD40 Repeat Containing SMU1 N Terminally Processed] |
| ENSDARG00000002571 | ENSFM00500000269813 | Tropomodulin Tropomodulin Tmod |
| ENSDARG00000002591 | ENSFM00730001521814 | Ruvb 1 EC_3.6.4.12 |
| ENSDARG00000002710 | ENSFM00500000270420 | Nucleolin |
| ENSDARG00000002790 | ENSFM00730001521696 | Ap 2 Complex Subunit Mu Ap 2 Mu Chain Clathrin Assembly Complex 2 Mu Medium Chain Clathrin Coat Assembly AP50 Clathrin Coat Associated AP50 MU2 Adaptin Plasma Membrane Adaptor Ap Kda |
| ENSDARG00000002816 | ENSFM00250000001613 | Ras Specific Guanine Nucleotide Releasing Factor 2 Ras GRF2 Ras Guanine Nucleotide Exchange Factor 2 |
| ENSDARG00000002991 | ENSFM00250000002276 | Testis Specific Gene 10 |
| ENSDARG00000003020 | ENSFM00260000050662 | Leucine Rich Repeat Containing Precursor Netrin Ligand Ngl |
| ENSDARG00000003046 | ENSFM00550000743175 | Sorbin And SH3 Domain Containing 2 Arg/Abl Interacting 2 ARGBP2 |
| ENSDARG00000003146 | ENSFM00730001521354 | Neurofilament Polypeptide Nf Kda Neurofilament Neurofilament Triplet |
| ENSDARG00000003151 | ENSFM00670001235396 | E3 Ubiquitin Ligase Complex Scf Subunit Sconc Sulfur Controller C Sulfur Metabolite Repression Control C |
| ENSDARG00000003169 | ENSFM00250000000552 | Membrane Associated Guanylate Kinase Ww And Pdz Domain Containing Membrane Associated Guanylate Kinase Inverted Magi |
| ENSDARG00000003308 | ENSFM00250000004805 | Er Membrane Complex Subunit 2 Tetratricopeptide Repeat 35 Tpr Repeat 35 |
| ENSDARG00000003320 | ENSFM00250000003694 | Unknown |
| ENSDARG00000003375 | ENSFM00630001060922 | Unknown |
| ENSDARG00000003375 | ENSFM00630001070087 | Unknown |
| ENSDARG00000003486 | ENSFM00420000140518 | Serine/Threonine Phosphatase PP1 Catalytic Subunit Pp EC_3.1.3.16 |
| ENSDARG00000003512 | ENSFM00250000001010 | Ankyrin Repeat And Sterile Alpha Motif Domain Containing 1B Amyloid Beta Intracellular Domain Associated 1 Aida 1 E2A PBX1 Associated Eb 1 |
| ENSDARG00000003544 | ENSFM00250000000663 | Double Stranded Rna Specific Editase EC_3.5.-.- Rna Adenosine Deaminase Rna Editing Deaminase Rna Editing Enzyme Dsrna Adenosine Deaminase |
| ENSDARG00000003599 | ENSFM00810002040577 | 60S Ribosomal L3 |
| ENSDARG00000003754 | ENSFM00800002003594 | CD9 Antigen CD9 Antigen |
| ENSDARG00000003779 | ENSFM00730001521673 | Catenin Delta 2 |
| ENSDARG00000003829 | ENSFM00730001521512 | Polypeptide N Acetylgalactosaminyltransferase EC_2.4.1.41 Polypeptide Galnac Transferase Galnac Pp Gantase Udp Acetylgalactosaminyltransferase Udp Galnac:Polypeptide N Acetylgalactosaminyltransferase |
| ENSDARG00000003869 | ENSFM00500000271494 | 2 4 Dienoyl Coa Reductase Mitochondrial Precursor EC_1.3.1.34 2 4 Dienoyl Coa Reductase [Nadph] 4 Enoyl Coa Reductase [Nadph] |
| ENSDARG00000003940 | ENSFM00610000967182 | Unknown |
| ENSDARG00000003940 | ENSFM00250000006972 | Mitochondrial Fission Regulator 1 |
| ENSDARG00000003961 | ENSFM00730001522506 | Poly [Adp Ribose] Polymerase 3 Parp 3 Hparp 3 EC_2.4.2.30 Adp Ribosyltransferase Diphtheria Toxin 3 ARTD3 IRT1 Nad + Adp Ribosyltransferase 3 Adprt 3 Poly[Adp Ribose] Synthase 3 Padprt 3 |
| ENSDARG00000003963 | ENSFM00500000270778 | Tnf Receptor Associated Factor 4 Cysteine Rich Associated Ring And Traf Domains 1 |
| ENSDARG00000003994 | ENSFM00720001492041 | Synaptotagmin Synaptotagmin |
| ENSDARG00000004017 | ENSFM00740001589368 | Mitochondrial Import Receptor Subunit TOM34 Translocase Of Outer Membrane 34 Kda Subunit |
| ENSDARG00000004174 | ENSFM00780001898388 | CCR4 Not Transcription Complex Subunit 1 CCR4 Associated Factor 1 |
| ENSDARG00000004301 | ENSFM00730001521186 | Ras Related C3 Botulinum Toxin Substrate Precursor P21 |
| ENSDARG00000004305 | ENSFM00250000002390 | Vang 2 Van Gogh |
| ENSDARG00000004336 | ENSFM00250000002477 | Exocyst Complex Component 4 Exocyst Complex Component SEC8 |
| ENSDARG00000004445 | ENSFM00730001521627 | Metabotropic Glutamate Receptor Precursor |
| ENSDARG00000004648 | ENSFM00260000050545 | Btb/Poz Domain Containing |
| ENSDARG00000004680 | ENSFM00570000851144 | Dnaj Homolog Subfamily B Member |
| ENSDARG00000004830 | ENSFM00630001070744 | Unknown |
| ENSDARG00000004830 | ENSFM00780001898447 | Flotillin 2 |
| ENSDARG00000005159 | ENSFM00250000001040 | Cytohesin Ph SEC7 And Coiled Coil Domain Containing |
| ENSDARG00000005162 | ENSFM00610000969203 | Unknown |
| ENSDARG00000005162 | ENSFM00250000000220 | Tropomyosin Alpha 1 Chain Alpha Tropomyosin Tropomyosin 1 |
| ENSDARG00000005163 | ENSFM00250000001283 | EFR3 Homolog |
| ENSDARG00000005210 | ENSFM00730001521970 | Coronin |
| ENSDARG00000005220 | ENSFM00500000271967 | Programmed Cell Death 6 Probable Calcium Binding Alg 2 |
| ENSDARG00000005350 | ENSFM00250000000652 | Disco Interacting 2 Homolog DIP2 Homolog |
| ENSDARG00000005397 | ENSFM00750001632380 | Tripartite Motif Containing 2 EC_6.3.2.- E3 Ubiquitin Ligase TRIM2 |
| ENSDARG00000005560 | ENSFM00730001521154 | Unknown |
| ENSDARG00000005774 | ENSFM00770001847530 | Atp Dependent Rna Helicase DDX3Y EC_3.6.4.13 Dead Box 3 Y Chromosomal |
| ENSDARG00000005791 | ENSFM00500000271918 | 60S Ribosomal L28 |
| ENSDARG00000005853 | ENSFM00250000000816 | Adp/Atp Translocase Adp Atp Carrier Adenine Nucleotide Translocator Ant Solute Carrier Family 25 Member [Contains Adp/Atp Translocase N Terminally Processed] |
| ENSDARG00000005861 | ENSFM00250000000944 | Ral Gtpase Activating Subunit Alpha 1 Gap Related Interacting Partner To E12 Gripe Gtpase Activating Domain 1 Tuberin 1 P240 |
| ENSDARG00000005897 | ENSFM00250000005338 | Deoxyribose Phosphate Aldolase Dera EC_4.1.2.4 |
| ENSDARG00000005924 | ENSFM00250000000235 | Precursor |
| ENSDARG00000005941 | ENSFM00730001521177 | Tyrosine Kinase EC_2.7.10.2 |
| ENSDARG00000006128 | ENSFM00740001589275 | Unknown |
| ENSDARG00000006225 | ENSFM00810002040601 | Spliceosome Rna Helicase DDX39B EC_3.6.4.13.56 Kda U2AF65 Associated Dead Box UAP56 |
| ENSDARG00000006240 | ENSFM00730001521827 | Long Chain Fatty Acid Transport 4 Fatp 4 Fatty Acid Transport 4 EC_6.2.1.- Solute Carrier Family 27 Member 4 |
| ENSDARG00000006260 | ENSFM00250000000217 | Tubulin Alpha Chain |
| ENSDARG00000006290 | ENSFM00670001238662 | Nadh Dehydrogenase Iron Sulfur 5 |
| ENSDARG00000006299 | ENSFM00250000001135 | Rho Guanine Nucleotide Exchange Factor 6 Rac/CDC42 Guanine Nucleotide Exchange Factor 6 |
| ENSDARG00000006316 | ENSFM00670001235314 | 60S Ribosomal |
| ENSDARG00000006370 | ENSFM00250000003378 | V Type Proton Atpase Subunit H V Atpase Subunit H Vacuolar Proton Pump Subunit H |
| ENSDARG00000006413 | ENSFM00500000276121 | Unknown |
| ENSDARG00000006546 | ENSFM00500000270040 | Adenylate Kinase 4 Mitochondrial |
| ENSDARG00000006585 | ENSFM00250000001646 | Dystrophia Myotonica Wd Repeat Containing Dystrophia Myotonica Containing Wd Repeat Motif Protein Dmr N9 |
| ENSDARG00000006601 | ENSFM00250000003884 | Kazrin |
| ENSDARG00000006609 | ENSFM00780001898380 | Dual Specificity Mitogen Activated Kinase Kinase 1 Map Kinase Kinase 1 Mapkk 1 EC_2.7.12.2 Erk Activator Kinase 1 Mapk/Erk Kinase 1 |
| ENSDARG00000006691 | ENSFM00250000002131 | 60S Ribosomal L12 |
| ENSDARG00000006712 | ENSFM00500000273445 | Was/Wasl Interacting Family Member 2 Wasp Interacting Related Wip Related |
| ENSDARG00000006832 | ENSFM00500000269893 | Polypeptide N Acetylgalactosaminyltransferase EC_2.4.1.41 Polypeptide Galnac Transferase Galnac T Pp Gantase Udp Acetylgalactosaminyltransferase Udp Galnac:Polypeptide N Acetylgalactosaminyltransferase |
| ENSDARG00000006900 | ENSFM00760001714645 | Inosine 5' Monophosphate Dehydrogenase |
| ENSDARG00000006923 | ENSFM00740001589100 | Voltage Dependent Type Calcium Channel Subunit Alpha Brain Calcium Channel Calcium Channel L Type Alpha 1 Polypeptide Voltage Gated Calcium Channel Subunit Alpha CAV2 |
| ENSDARG00000007034 | ENSFM00250000001382 | Heterogeneous Nuclear Ribonucleoprotein K Hnrnp K |
| ENSDARG00000007034 | ENSFM00540000725842 | Hnrpkl |
| ENSDARG00000007098 | ENSFM00600000921249 | Integral Membrane 2B Immature BRI2 IMBRI2 Transmembrane Bri Bri [Contains BRI2 Membrane Form Mature BRI2 MBRI2 |
| ENSDARG00000007195 | ENSFM00790001963004 | Metabotropic Glutamate Receptor Precursor |
| ENSDARG00000007196 | ENSFM00780001898474 | Export Factor RAE1 Homolog Associated Mrnp 41 |
| ENSDARG00000007216 | ENSFM00250000004383 | Translation Initiation Factor RLI1 Atp Binding Cassette Sub Family E Member RLI1 Rnase L Inhibitor |
| ENSDARG00000007219 | ENSFM00440000236866 | Alpha Actinin Alpha Actinin F Actin Cross Linking |
| ENSDARG00000007264 | ENSFM00250000003191 | Nipsnap Homolog 2 NIPSNAP2 Glioblastoma Amplified Sequence |
| ENSDARG00000007320 | ENSFM00250000000871 | 60S Ribosomal L7 |
| ENSDARG00000007526 | ENSFM00250000004138 | Nadh Dehydrogenase [Ubiquinone] Iron Sulfur 2 Mitochondrial Precursor EC_1.6.5.3 EC_1.6.99.- 3 Complex I 49KD Ci 49KD Nadh Ubiquinone Oxidoreductase 49 Kda Subunit |
| ENSDARG00000007723 | ENSFM00250000002014 | Ephrin B1 Precursor Elk Ligand Elk L Eph Related Receptor Tyrosine Kinase Ligand 2 Lerk 2 |
| ENSDARG00000007745 | ENSFM00770001847545 | Cytochrome B C1 Complex Subunit Rieske Mitochondrial Precursor EC_1.10.2.2 Complex Iii Subunit 5 Cytochrome B C1 Complex Subunit 5 Rieske Iron Sulfur Risp Ubiquinol Cytochrome C Reductase Iron Sulfur Subunit [Contains Cytochrome B C1 Complex Subunit 11 Co |
| ENSDARG00000007753 | ENSFM00800002003432 | Copine Copine |
| ENSDARG00000007791 | ENSFM00250000001941 | Serine/Threonine Phosphatase 2A 65 Kda Regulatory Subunit A PP2A Subunit A PR65 PP2A Subunit A R1 |
| ENSDARG00000007825 | ENSFM00780001898380 | Dual Specificity Mitogen Activated Kinase Kinase 1 Map Kinase Kinase 1 Mapkk 1 EC_2.7.12.2 Erk Activator Kinase 1 Mapk/Erk Kinase 1 |
| ENSDARG00000007916 | ENSFM00800002003832 | Probable Xaa Pro Aminopeptidase 3 X Pro Aminopeptidase 3 EC_3.4.11.9 Aminopeptidase P3 APP3 |
| ENSDARG00000007955 | ENSFM00500000270033 | Isoleucine Trna Ligase Cytoplasmic EC_6.1.1.5 Isoleucyl Trna Synthetase Ilers |
| ENSDARG00000007989 | ENSFM00670001265228 | Neural Cell Expressed Developmentally Down Regulated 8 |
| ENSDARG00000008030 | ENSFM00730001521616 | Myosin Regulatory Light Myosin Regulatory Light Chain 2 Smooth Muscle Myosin Regulatory Light Chain |
| ENSDARG00000008107 | ENSFM00730001521177 | Tyrosine Kinase EC_2.7.10.2 |
| ENSDARG00000008249 | ENSFM00730001522293 | Patched Domain Containing |
| ENSDARG00000008329 | ENSFM00250000000608 | Acid Sensing Ion Channel Amiloride Sensitive Cation Channel |
| ENSDARG00000008370 | ENSFM00800002003414 | Casein Kinase I Delta Cki Delta Ckid EC_2.7.11.1 |
| ENSDARG00000008383 | ENSFM00760001714703 | Actin Related 2/3 Complex Subunit 1B ARP2/3 Complex 41 Kda Subunit P41 Arc |
| ENSDARG00000008487 | ENSFM00250000000455 | Dystrophin |
| ENSDARG00000008491 | ENSFM00500000269664 | Retinol Dehydrogenase |
| ENSDARG00000008494 | ENSFM00250000000438 | Myosin Light Chain Myosin Light Chain Myosin Light Chain |
| ENSDARG00000008573 | ENSFM00800002003872 | Glycosaminoglycan Xylosylkinase EC_2.7.1.- Xylose Kinase |
| ENSDARG00000008660 | ENSFM00730001521292 | Coronin |
| ENSDARG00000008735 | ENSFM00630001059629 | Unknown |
| ENSDARG00000008735 | ENSFM00250000004412 | Renin Receptor Precursor Atpase H + Transporting Lysosomal Accessory 2 Atpase H + Transporting Lysosomal Interacting 2 Renin/Prorenin Receptor Vacuolar Atp Synthase Membrane Sector Associated M8 9 V Atpase M8 9 Subunit |
| ENSDARG00000008765 | ENSFM00260000050657 | Transmembrane EMP24 Domain Containing Precursor P24 Family Gamma |
| ENSDARG00000008785 | ENSFM00800002003534 | Dihydrolipoyl Dehydrogenase Mitochondrial Precursor EC_1.8.1.4 Dihydrolipoamide Dehydrogenase |
| ENSDARG00000008790 | ENSFM00800002003421 | Actin Related 3 Actin 3 |
| ENSDARG00000008816 | ENSFM00250000001520 | Glutamate Dehydrogenase Mitochondrial Gdh EC_1.4.1.3 |
| ENSDARG00000008969 | ENSFM00500000270953 | Fibrinogen Beta Chain Fibrinopeptide B |
| ENSDARG00000008982 | ENSFM00250000002399 | Calsequestrin Precursor Calsequestrin Muscle |
| ENSDARG00000009018 | ENSFM00280000058667 | Ammonium Transporter Rh Type B Rhesus Blood Group Family Type B Glycoprotein Rh Family Type B Glycoprotein Rh Type B Glycoprotein |
| ENSDARG00000009026 | ENSFM00500000269628 | Ankyrin Ank Ankyrin |
| ENSDARG00000009031 | ENSFM00260000050335 | Kinase EC_2.7.11.1 Kinase Mapk/Erk Kinase Kinase Kinase Mek Kinase Kinase Mekkk Kinase |
| ENSDARG00000009208 | ENSFM00750001632332 | Kinase C Delta Type EC_2.7.11.13 Npkc Delta [Contains Kinase C Delta Type Regulatory Subunit |
| ENSDARG00000009285 | ENSFM00250000001504 | 60S Ribosomal L15 |
| ENSDARG00000009285 | ENSFM00730001521550 | Ubiquitin Conjugating Enzyme E2 EC_6.3.2.19 Ubiquitin Carrier Ubiquitin Ligase |
| ENSDARG00000009401 | ENSFM00730001521436 | Core Precursor Chondroitin Sulfate Proteoglycan |
| ENSDARG00000009499 | ENSFM00250000000542 | Nesprin 2 Nuclear Envelope Spectrin Repeat 2 Nucleus And Actin Connecting Element Protein Nuance Synaptic Nuclear Envelope 2 Syne 2 |
| ENSDARG00000009553 | ENSFM00500000437551 | Unknown |
| ENSDARG00000009567 | ENSFM00740001589455 | Striated Muscle Specific Serine/Threonine Kinase EC_2.7.11.1 Aortic Preferentially Expressed 1 Apeg 1 |
| ENSDARG00000009677 | ENSFM00250000000275 | Disks Large Homolog Synapse Associated Sap |
| ENSDARG00000009689 | ENSFM00350000105429 | Disheveled Associated Activator Of Morphogenesis |
| ENSDARG00000009738 | ENSFM00250000000524 | Voltage Dependent L Type Calcium Channel Subunit Beta Calcium Channel Voltage Dependent Subunit Beta |
| ENSDARG00000009782 | ENSFM00270000056424 | Myosin Cellular Myosin Heavy Chain Type Myosin Heavy Chain Myosin Heavy Chain Non Muscle Non Muscle Myosin Heavy Chain Nmmhc Non Muscle Myosin Heavy Chain Nmmhc Ii Nmmhc |
| ENSDARG00000009941 | ENSFM00250000000753 | Elks/RAB6 Interacting/Cast Family Member 1 Erc 1 RAB6 Interacting 2 |
| ENSDARG00000009978 | ENSFM00670001247480 | S100 |
| ENSDARG00000010002 | ENSFM00730001521471 | Guanine Nucleotide Binding G Q Subunit Alpha Guanine Nucleotide Binding Alpha Q |
| ENSDARG00000010008 | ENSFM00730001521291 | Vimentin |
| ENSDARG00000010078 | ENSFM00250000005326 | Nuclear Pore Complex NUP133.133 Kda Nucleoporin Nucleoporin NUP133 |
| ENSDARG00000010108 | ENSFM00250000006424 | BRI3 Binding I3 Binding |
| ENSDARG00000010113 | ENSFM00250000006874 | Nadh Dehydrogenase [Ubiquinone] 1 Beta Subcomplex Subunit 8 Mitochondrial Precursor Complex I Ashi Ci Ashi Nadh Ubiquinone Oxidoreductase Ashi Subunit |
| ENSDARG00000010130 | ENSFM00500000270338 | Acid Sphingomyelinase Phosphodiesterase Precursor Asm Phosphodiesterase EC_3.1.4.- |
| ENSDARG00000010144 | ENSFM00500000270520 | Lipid Phosphate Phosphatase Related Type 4 EC_3.1.3.4 Brain Specific Phosphatidic Acid Phosphatase 1 Plasticity Related Gene 1 Prg 1 |
| ENSDARG00000010155 | ENSFM00250000000845 | Abl Interactor 1 Abelson Interactor 1 Abi 1 EPS8 SH3 Domain Binding EPS8 Binding Spectrin SH3 Domain Binding 1 |
| ENSDARG00000010160 | ENSFM00250000002123 | 40S Ribosomal |
| ENSDARG00000010280 | ENSFM00610000970574 | Unknown |
| ENSDARG00000010280 | ENSFM00250000000820 | Clip Associating Cytoplasmic Linker Associated |
| ENSDARG00000010316 | ENSFM00780001898374 | Glutamine Trna Ligase EC_6.1.1.18 Glutaminyl Trna Synthetase Glnrs |
| ENSDARG00000010516 | ENSFM00250000000614 | 60S Ribosomal L21 |
| ENSDARG00000010553 | ENSFM00500000273449 | Membrane Magnesium Transporter 1 Precursor Er Membrane Complex Subunit 5 Transmembrane 32 |
| ENSDARG00000010610 | ENSFM00250000003414 | Sterile Alpha And Tir Motif Containing Tir 1 Homolog |
| ENSDARG00000010721 | ENSFM00350000105408 | Septin |
| ENSDARG00000010728 | ENSFM00760001714632 | Gelsolin Actin Depolymerizing Factor Adf |
| ENSDARG00000010745 | ENSFM00760001714614 | Dnaj Homolog Subfamily A Member Precursor |
| ENSDARG00000010764 | ENSFM00500000272331 | Dehydrogenase/Reductase Sdr Family Member 12 EC_1.1.-.- |
| ENSDARG00000010784 | ENSFM00500000269806 | Phosphatase 1 Regulatory Subunit 12A Myosin Phosphatase Targeting Subunit 1 Myosin Phosphatase Target Subunit 1 Phosphatase Myosin Binding Subunit |
| ENSDARG00000010789 | ENSFM00250000001140 | Homer Homolog Homer Vasp/Ena Related Gene Up Regulated During Seizure And Ltp Vesl |
| ENSDARG00000010844 | ENSFM00730001521569 | Ras Precursor |
| ENSDARG00000010941 | ENSFM00250000007877 | O Linked Mannose Beta 1 4 N Acetylglucosaminyltransferase 2 POMGNT2 EC_2.4.1.- |
| ENSDARG00000010953 | ENSFM00250000002568 | Nuclear Distribution Nude 1 |
| ENSDARG00000011065 | ENSFM00250000000111 | Calcium/Calmodulin Dependent Kinase Type Ii Subunit Cam Kinase Ii Subunit Camk Ii Subunit EC_2.7.11.17 |
| ENSDARG00000011171 | ENSFM00250000000375 | Teneurin Ten Odd Oz/Ten M Homolog Tenascin Ten Teneurin Transmembrane [Contains Ten Intracellular Domain Ten Icd ] |
| ENSDARG00000011175 | ENSFM00770001847582 | V Type Proton Atpase Subunit D V Atpase Subunit D Vacuolar Proton Pump Subunit D |
| ENSDARG00000011196 | ENSFM00250000002491 | Dnaj Homolog Subfamily C Member 11 |
| ENSDARG00000011201 | ENSFM00500000374444 | RPLP2L |
| ENSDARG00000011333 | ENSFM00250000001205 | Rho Gtpase Activating Rho Type Gtpase Activating Gtpase Activating |
| ENSDARG00000011370 | ENSFM00730001521177 | Tyrosine Kinase EC_2.7.10.2 |
| ENSDARG00000011405 | ENSFM00250000003284 | 40S Ribosomal S9 |
| ENSDARG00000011459 | ENSFM00760001714632 | Gelsolin Actin Depolymerizing Factor Adf |
| ENSDARG00000011564 | ENSFM00250000000671 | Paraspeckle Component 1 |
| ENSDARG00000011602 | ENSFM00250000006430 | Uncharacterized |
| ENSDARG00000011602 | ENSFM00730001542826 | Unknown |
| ENSDARG00000011611 | ENSFM00730001521541 | Actin |
| ENSDARG00000011661 | ENSFM00500000270278 | Twinfilin |
| ENSDARG00000011665 | ENSFM00250000000488 | Fructose Bisphosphate Aldolase EC_4.1.2.13 Type Aldolase |
| ENSDARG00000011777 | ENSFM00600000921221 | Substrate |
| ENSDARG00000011855 | ENSFM00730001521956 | AP2 Associated Kinase 1 EC_2.7.11.1 Adaptor Associated Kinase 1 |
| ENSDARG00000011862 | ENSFM00730001521354 | Neurofilament Polypeptide Nf Kda Neurofilament Neurofilament Triplet |
| ENSDARG00000011909 | ENSFM00250000000547 | Inositol 1 4 5 Trisphosphate Receptor Type IP3 Receptor IP3R Ins Type Inositol 1 4 5 Trisphosphate Receptor Type INSP3 Receptor |
| ENSDARG00000011929 | ENSFM00250000000909 | Myelin Proteolipid Plp Lipophilin |
| ENSDARG00000012019 | ENSFM00400000131715 | Glycine Receptor Subunit Precursor Glycine Receptor Kda Subunit |
| ENSDARG00000012144 | ENSFM00500000273154 | Er Membrane Complex Subunit 7 Precursor |
| ENSDARG00000012204 | ENSFM00740001589171 | Cyclin Dependent Kinase EC_2.7.11.22 Cell Division Kinase Pctaire Motif Kinase Serine/Threonine Kinase Pctaire |
| ENSDARG00000012248 | ENSFM00250000001343 | Rgm Domain Family Member |
| ENSDARG00000012281 | ENSFM00730001521354 | Neurofilament Polypeptide Nf Kda Neurofilament Neurofilament Triplet |
| ENSDARG00000012367 | ENSFM00400000131833 | Tripartite Motif Containing 46 Gene Y Geney Tripartite Fibronectin Type Iii And C Terminal Spry Motif |
| ENSDARG00000012378 | ENSFM00250000003167 | N Alpha Acetyltransferase 35 Natc Auxiliary Subunit Embryonic Growth Associated MAK10 Homolog |
| ENSDARG00000012381 | ENSFM00730001521103 | Heat Shock Kda Heat Shock 70 Kda |
| ENSDARG00000012387 | ENSFM00750001632368 | Pyruvate Dehydrogenase E1 Component Subunit Alpha Somatic Form Mitochondrial Precursor EC_1.2.4.1 PDHE1 A Type I |
| ENSDARG00000012388 | ENSFM00500000271199 | Cytochrome C Oxidase Subunit 4 1 Mitochondrial Cytochrome C Oxidase Polypeptide Iv Cytochrome C Oxidase Subunit Iv 1 Cox Iv 1 |
| ENSDARG00000012426 | ENSFM00730001521354 | Neurofilament Polypeptide Nf Kda Neurofilament Neurofilament Triplet |
| ENSDARG00000012513 | ENSFM00250000003553 | Syntenin Syndecan Binding |
| ENSDARG00000012519 | ENSFM00250000001451 | Host Cell Factor 1 Hcf Hcf 1 C1 Factor [Contains Hcf N Terminal Chain 1 |
| ENSDARG00000012555 | ENSFM00740001589162 | Adenylate Kinase Isoenzyme 1 |
| ENSDARG00000012586 | ENSFM00350000105407 | Formin |
| ENSDARG00000012801 | ENSFM00730001521507 | Atp Dependent 6 Phosphofructokinase Type |
| ENSDARG00000012818 | ENSFM00250000001591 | Casein Kinase Ii Subunit Ck Ii EC_2.7.11.1 |
| ENSDARG00000012823 | ENSFM00250000000718 | Disks Large Associated Dap Psd 95/SAP90 Binding SAP90/Psd 95 Associated |
| ENSDARG00000012881 | ENSFM00730001521491 | Anion Exchange Ae Anion Exchanger Solute Carrier Family 4 Member |
| ENSDARG00000012944 | ENSFM00250000000024 | Myosin Myosin Heavy Chain Myosin Heavy Chain Muscle |
| ENSDARG00000012982 | ENSFM00750001657117 | Unknown |
| ENSDARG00000012982 | ENSFM00250000001252 | Neurofibromin Neurofibromatosis Related Nf 1 |
| ENSDARG00000013000 | ENSFM00250000000441 | Liprin Alpha Tyrosine Phosphatase Receptor Type F Polypeptide Interacting Alpha Ptprf Interacting Alpha |
| ENSDARG00000013005 | ENSFM00290000065521 | Precursor |
| ENSDARG00000013020 | ENSFM00250000000811 | Dystrobrevin Beta Dtn B Beta Dystrobrevin |
| ENSDARG00000013044 | ENSFM00250000003401 | Nadh Dehydrogenase [Ubiquinone] Flavoprotein 2 Mitochondrial Precursor EC_1.6.5.3 EC_1.6.99.- 3 Nadh Ubiquinone Oxidoreductase 24 Kda Subunit |
| ENSDARG00000013078 | ENSFM00730001521154 | Unknown |
| ENSDARG00000013079 | ENSFM00800002003795 | Gamma Tubulin Complex Component 2 Gamma Ring Complex Kda |
| ENSDARG00000013110 | ENSFM00500000271390 | Dematin Dematin Actin Binding Erythrocyte Membrane Band 4 9 |
| ENSDARG00000013128 | ENSFM00500000269765 | [Pyruvate Dehydrogenase Acetyl Transferring ] Kinase Isozyme Mitochondrial Precursor EC_2.7.11.2 Pyruvate Dehydrogenase Kinase |
| ENSDARG00000013161 | ENSFM00730001521569 | Ras Precursor |
| ENSDARG00000013236 | ENSFM00500000270882 | Delta 24 Sterol Reductase Precursor EC_1.3.1.72 24 Dehydrocholesterol Reductase 3 Beta Hydroxysterol Delta 24 Reductase |
| ENSDARG00000013250 | ENSFM00250000001453 | Threonine Trna Ligase EC_6.1.1.3 Threonyl Trna Synthetase Thrrs Threonyl Trna Synthetase |
| ENSDARG00000013307 | ENSFM00250000001949 | 60S Ribosomal L19 |
| ENSDARG00000013351 | ENSFM00610000972707 | Unknown |
| ENSDARG00000013351 | ENSFM00600000926155 | Inducible Rna Binding |
| ENSDARG00000013414 | ENSFM00260000050595 | Lin 7 Homolog Lin Mammalian Lin Seven Mals Vertebrate Lin 7 Homolog Veli |
| ENSDARG00000013443 | ENSFM00810002040578 | V Type Proton Atpase Subunit B V Atpase Subunit B Endomembrane Proton Pump 58 Kda Subunit Vacuolar Proton Pump Subunit B |
| ENSDARG00000013542 | ENSFM00710001459591 | Unknown |
| ENSDARG00000013542 | ENSFM00250000004110 | Acyl Coa:Lysophosphatidylglycerol Acyltransferase 1 EC_2.3.1.- |
| ENSDARG00000013582 | ENSFM00250000001591 | Casein Kinase Ii Subunit Ck Ii EC_2.7.11.1 |
| ENSDARG00000013647 | ENSFM00390000126303 | Contactin Precursor Brain Derived Immunoglobulin Superfamily Big |
| ENSDARG00000013741 | ENSFM00250000002547 | Lanc 1.40 Kda Erythrocyte Membrane P40 |
| ENSDARG00000013763 | ENSFM00250000002798 | Ribosome Binding Ribosome Receptor Rrp |
| ENSDARG00000013841 | ENSFM00730001521872 | Tyrosine Kinase EC_2.7.10.2 Abelson Murine Leukemia Viral Oncogene Homolog Abelson Tyrosine Kinase |
| ENSDARG00000013843 | ENSFM00800002003401 | Septin |
| ENSDARG00000014013 | ENSFM00730001522072 | Lamin B Receptor Integral Nuclear Envelope Inner Membrane |
| ENSDARG00000014053 | ENSFM00610000967352 | Unknown |
| ENSDARG00000014053 | ENSFM00320000100099 | Noelin Precursor Neuronal Olfactomedin Related Er Localized Olfactomedin 1 |
| ENSDARG00000014113 | ENSFM00250000001784 | Wiskott Aldrich Syndrome Wasp |
| ENSDARG00000014139 | ENSFM00250000003679 | Signal Recognition Particle Subunit SRP72 SRP72 Signal Recognition Particle 72 Kda |
| ENSDARG00000014165 | ENSFM00250000005568 | Translocon Associated Subunit Gamma Trap Gamma Signal Sequence Receptor Subunit Gamma Ssr Gamma |
| ENSDARG00000014215 | ENSFM00730001521172 | Cadherin Precursor Cadherin Cadherin |
| ENSDARG00000014222 | ENSFM00250000001591 | Casein Kinase Ii Subunit Ck Ii EC_2.7.11.1 |
| ENSDARG00000014230 | ENSFM00800002003585 | Dihydrolipoyllysine Residue Succinyltransferase Component Of 2 Oxoglutarate Dehydrogenase Complex Mitochondrial Precursor EC_2.3.1.61 2 Oxoglutarate Dehydrogenase Complex Component E2 Ogdc E2 Dihydrolipoamide Succinyltransferase Component Of 2 Oxoglutarat |
| ENSDARG00000014233 | ENSFM00350000105408 | Septin |
| ENSDARG00000014273 | ENSFM00250000000111 | Calcium/Calmodulin Dependent Kinase Type Ii Subunit Cam Kinase Ii Subunit Camk Ii Subunit EC_2.7.11.17 |
| ENSDARG00000014280 | ENSFM00250000000718 | Disks Large Associated Dap Psd 95/SAP90 Binding SAP90/Psd 95 Associated |
| ENSDARG00000014302 | ENSFM00250000001886 | Bmp/Retinoic Acid Inducible Neural Specific 1 Precursor Deleted In Bladder Cancer 1 |
| ENSDARG00000014522 | ENSFM00260000050333 | Cadherin Precursor |
| ENSDARG00000014527 | ENSFM00500000269765 | [Pyruvate Dehydrogenase Acetyl Transferring ] Kinase Isozyme Mitochondrial Precursor EC_2.7.11.2 Pyruvate Dehydrogenase Kinase |
| ENSDARG00000014571 | ENSFM00250000001012 | Catenin Beta Beta Catenin |
| ENSDARG00000014582 | ENSFM00250000001333 | Exocyst Complex Component 3 Exocyst Complex Component SEC6 |
| ENSDARG00000014591 | ENSFM00250000003528 | Interleukin Enhancer Binding Factor 2 |
| ENSDARG00000014634 | ENSFM00250000004850 | Membrane Bound Transcription Factor Site 1 Protease Precursor EC_3.4.21.112 Endopeptidase S1P Sterol Regulated Luminal Protease Subtilisin/Kexin Isozyme 1 Ski 1 |
| ENSDARG00000014651 | ENSFM00250000001422 | Supervillin Archvillin P205/P250 |
| ENSDARG00000014690 | ENSFM00250000001458 | 40S Ribosomal S4 |
| ENSDARG00000014793 | ENSFM00250000000766 | Eh Domain Containing |
| ENSDARG00000014804 | ENSFM00730001522093 | Voltage Dependent Calcium Channel Subunit Alpha 2/Delta 1 Precursor Voltage Gated Calcium Channel Subunit Alpha 2/Delta 1 [Contains Voltage Dependent Calcium Channel Subunit Alpha 2 1 |
| ENSDARG00000014867 | ENSFM00250000002389 | 60S Ribosomal |
| ENSDARG00000014875 | ENSFM00250000000845 | Abl Interactor 1 Abelson Interactor 1 Abi 1 EPS8 SH3 Domain Binding EPS8 Binding Spectrin SH3 Domain Binding 1 |
| ENSDARG00000014891 | ENSFM00740001589121 | Roundabout Homolog Precursor |
| ENSDARG00000014915 | ENSFM00670001235829 | Acyl Carrier Protein Mitochondrial Precursor Acp Nadh Ubiquinone Oxidoreductase 9.6 Kda Subunit |
| ENSDARG00000014973 | ENSFM00260000050653 | Netrin Precursor Laminet |
| ENSDARG00000015002 | ENSFM00400000134066 | Cadherin 4 Retinal |
| ENSDARG00000015002 | ENSFM00730001521172 | Cadherin Precursor Cadherin Cadherin |
| ENSDARG00000015050 | ENSFM00750001632311 | Calmodulin Cam |
| ENSDARG00000015053 | ENSFM00740001589268 | Glutamate Receptor Interacting |
| ENSDARG00000015059 | ENSFM00350000105429 | Disheveled Associated Activator Of Morphogenesis |
| ENSDARG00000015065 | ENSFM00250000000285 | Complement C3 Complement C3 Beta Chain |
| ENSDARG00000015128 | ENSFM00670001235486 | 60S Ribosomal L27 |
| ENSDARG00000015174 | ENSFM00250000000490 | V Type Proton Atpase 116 Kda Subunit A V Atpase 116 Kda Proton 116 Kda Subunit Vacuolar Proton Translocating Atpase 116 Kda Subunit A |
| ENSDARG00000015184 | ENSFM00760001714665 | Maguk P55 Subfamily Member |
| ENSDARG00000015236 | ENSFM00800002009595 | Pycrl |
| ENSDARG00000015236 | ENSFM00500000269896 | Pyrroline 5 Carboxylate Reductase 3 P5C Reductase 3 P5CR 3 EC_1.5.1.2 Pyrroline 5 Carboxylate Reductase |
| ENSDARG00000015239 | ENSFM00800002003664 | Pre Processing Factor 19 |
| ENSDARG00000015240 | ENSFM00740001589470 | Cyclin Dependent Kinase 5 EC_2.7.11.22 |
| ENSDARG00000015385 | ENSFM00790001963038 | Nadh Dehydrogenase [Ubiquinone] Iron Sulfur 3 Mitochondrial Precursor EC_1.6.5.3 EC_1.6.99.- 3 Complex I 30KD Ci 30KD Nadh Ubiquinone Oxidoreductase 30 Kda Subunit |
| ENSDARG00000015392 | ENSFM00810002040596 | Pre Splicing Factor Atp Dependent Rna Helicase EC_3.6.4.13 Deah Box |
| ENSDARG00000015449 | ENSFM00250000004295 | Alpha 1 6 Fucosyltransferase ALPHA1 6FUCT EC_2.4.1.68 Gdp L Fuc:N Acetyl Beta D Glucosaminide ALPHA1 6 Fucosyltransferase Gdp Fucose Glycoprotein Fucosyltransferase Glycoprotein 6 Alpha L Fucosyltransferase |
| ENSDARG00000015546 | ENSFM00250000000564 | Alkaline Phosphatase Precursor EC_3.1.3.1 Alkaline Phosphatase |
| ENSDARG00000015610 | ENSFM00420000140599 | Tubulin Gamma Chain Gamma Tubulin |
| ENSDARG00000015649 | ENSFM00800002003504 | Ras Related Rap Precursor |
| ENSDARG00000015681 | ENSFM00610000973032 | Unknown |
| ENSDARG00000015681 | ENSFM00810002040612 | Glycogen Synthase Kinase 3 Gsk 3 EC_2.7.11.26 Serine/Threonine Kinase EC_2.7.11.- 1 |
| ENSDARG00000015862 | ENSFM00670001235299 | 60S Ribosomal L5 |
| ENSDARG00000015918 | ENSFM00810002040672 | Dihydrolipoyllysine Residue Acetyltransferase Component Of Pyruvate Dehydrogenase Complex Mitochondrial EC_2.3.1.12 Dihydrolipoamide Acetyltransferase Component Of Pyruvate Dehydrogenase Complex Pyruvate Dehydrogenase Complex Component E2 Pdc E2 PDCE2 |
| ENSDARG00000015921 | ENSFM00800002003812 | Periodic Tryptophan 1 Homolog |
| ENSDARG00000015947 | ENSFM00740001589150 | Precursor |
| ENSDARG00000016016 | ENSFM00480000262787 | Drebrin |
| ENSDARG00000016128 | ENSFM00440000236903 | Ap 3 Complex Subunit Mu Adaptor Related Complex 3 Subunit Mu Clathrin Assembly Assembly Complex 3 Mu Medium Chain Clathrin Coat Assembly AP47 Homolog Clathrin Coat Associated AP47 Homolog Golgi Adaptor Ap 1.47 Kda Homolog HA1.47 Kda Subunit Homolog Adapti |
| ENSDARG00000016255 | ENSFM00600000921240 | Charged Multivesicular Body Chromatin Modifying |
| ENSDARG00000016263 | ENSFM00500000270072 | Palmitoyltransferase ZDHHC5 EC_2.3.1.225 Zinc Finger Dhhc Domain Containing 5 Dhhc 5 |
| ENSDARG00000016301 | ENSFM00250000000193 | Tubulin Beta Chain |
| ENSDARG00000016302 | ENSFM00500000270457 | Atp Dependent Helicase Nonsense 1 Up Frameshift Suppressor 1 |
| ENSDARG00000016307 | ENSFM00250000006279 | 39S Ribosomal L46 Mitochondrial Precursor L46MT Mrp L46 |
| ENSDARG00000016348 | ENSFM00250000000652 | Disco Interacting 2 Homolog DIP2 Homolog |
| ENSDARG00000016463 | ENSFM00670001235686 | Trafficking Particle Complex Subunit 6A Trapp Complex Subunit 6A |
| ENSDARG00000016464 | ENSFM00730001521452 | Serine/Threonine Kinase Mrck EC_2.7.11.1 CDC42 Binding Kinase Dmpk Myotonic Dystrophy Kinase Related CDC42 Binding Kinase Mrck Myotonic Dystrophy Kinase |
| ENSDARG00000016545 | ENSFM00730001521465 | Choline O Acetyltransferase Choactase Chat Choline Acetylase EC_2.3.1.6 |
| ENSDARG00000016551 | ENSFM00250000001463 | Iq Motif And SEC7 Domain Containing |
| ENSDARG00000016584 | ENSFM00730001521808 | Regulator Of G Signaling |
| ENSDARG00000016598 | ENSFM00800002003437 | Creatine Kinase U Type Mitochondrial Precursor EC_2.7.3.2 Acidic Type Mitochondrial Creatine Kinase Mia Ck Ubiquitous Mitochondrial Creatine Kinase U Mtck |
| ENSDARG00000016667 | ENSFM00250000000928 | Gamma Aminobutyric Acid Type B Receptor Subunit 2 Precursor Gaba B Receptor 2 Gaba B R2 Gaba BR2 GABABR2 GB2 G Coupled Receptor 51 |
| ENSDARG00000016676 | ENSFM00410000138458 | Guanine Nucleotide Binding Subunit |
| ENSDARG00000016825 | ENSFM00250000000813 | Vitellogenin Precursor [Contains Lipovitellin I Lvi |
| ENSDARG00000016837 | ENSFM00500000271973 | Golgi Associated Plant Pathogenesis Related 1 Gapr 1 Golgi Associated Pr 1 Glioma Pathogenesis Related 2 Glipr 2 |
| ENSDARG00000016904 | ENSFM00500000271321 | [3 Methyl 2 Oxobutanoate Dehydrogenase [Lipoamide]] Kinase Mitochondrial Precursor EC_2.7.11.4 Branched Chain Alpha Ketoacid Dehydrogenase Kinase Bckd Kinase Bckdhkin |
| ENSDARG00000016968 | ENSFM00250000004458 | Transmembrane 214 |
| ENSDARG00000017034 | ENSFM00810002040854 | Sulfide:Quinone Oxidoreductase Mitochondrial Precursor Sqor EC_1.8.5.- |
| ENSDARG00000017105 | ENSFM00740001589204 | Ena/Vasp Ena/Vasodilator Stimulated Phosphoprotein |
| ENSDARG00000017126 | ENSFM00250000005381 | Acetolactate Synthase EC_2.2.1.- Ilvb |
| ENSDARG00000017140 | ENSFM00800002003496 | Septin 2 |
| ENSDARG00000017162 | ENSFM00800002003386 | Kinesin |
| ENSDARG00000017219 | ENSFM00570000851024 | Polyadenylate Binding Pabp Poly A Binding |
| ENSDARG00000017316 | ENSFM00250000007925 | Bag Family Molecular Chaperone Regulator 5 Bag 5 Bcl 2 Associated Athanogene 5 |
| ENSDARG00000017441 | ENSFM00610000972103 | Unknown |
| ENSDARG00000017441 | ENSFM00250000000438 | Myosin Light Chain Myosin Light Chain Myosin Light Chain |
| ENSDARG00000017624 | ENSFM00740001589131 | Keratin Type Ii Cytoskeletal 8 Cytokeratin 8 Ck 8 Keratin 8 K8 Type Ii Keratin KB8 |
| ENSDARG00000017728 | ENSFM00800002003689 | Prohibitin 2 |
| ENSDARG00000017742 | ENSFM00250000000328 | Metabotropic Glutamate Receptor Precursor |
| ENSDARG00000017781 | ENSFM00500000270523 | 3 Hydroxyacyl Coa Dehydrogenase Type 2 EC_1.1.1.35 17 Beta Hydroxysteroid Dehydrogenase 10 17 Beta Hsd 10 EC_1.1.1.- 51 3 Hydroxy 2 Methylbutyryl Coa Dehydrogenase EC_1.1.-.- 1 178 3 Hydroxyacyl Coa Dehydrogenase Type Ii Endoplasmic Reticulum Associated A |
| ENSDARG00000017785 | ENSFM00800002009756 | Unknown |
| ENSDARG00000017785 | ENSFM00250000000906 | Brain Specific Angiogenesis Inhibitor 1 Associated 2 Bai Associated 2 BAI1 Associated 2 Insulin Receptor Substrate Of 53 Kda IRSP53 Insulin Receptor Substrate P53 |
| ENSDARG00000017803 | ENSFM00810002040612 | Glycogen Synthase Kinase 3 Gsk 3 EC_2.7.11.26 Serine/Threonine Kinase EC_2.7.11.- 1 |
| ENSDARG00000017986 | ENSFM00250000001900 | Vacuolar Sorting Associated 13D |
| ENSDARG00000018000 | ENSFM00250000001310 | Transcription Factor CP2 |
| ENSDARG00000018065 | ENSFM00290000065521 | Precursor |
| ENSDARG00000018110 | ENSFM00730001521739 | Serine/Threonine Kinase Pak EC_2.7.11.1 P21 Activated Kinase Pak |
| ENSDARG00000018146 | ENSFM00500000270307 | Glutathione Peroxidase 2 Gpx 2 Gshpx 2 EC_1.11.1.9 Glutathione Peroxidase Gastrointestinal Gpx Gi Gshpx Gi |
| ENSDARG00000018162 | ENSFM00770001847643 | Alpha Catulin Alpha Catenin Related Acrp Catenin Alpha 1 |
| ENSDARG00000018174 | ENSFM00410000138458 | Guanine Nucleotide Binding Subunit |
| ENSDARG00000018270 | ENSFM00320000100099 | Noelin Precursor Neuronal Olfactomedin Related Er Localized Olfactomedin 1 |
| ENSDARG00000018285 | ENSFM00800002003539 | 3 Phosphoinositide Dependent Kinase EC_2.7.11.1 |
| ENSDARG00000018334 | ENSFM00500000271690 | 60S Ribosomal L35 |
| ENSDARG00000018404 | ENSFM00560000771030 | Keratin Type I Cytoskeletal 18 Cytokeratin 18 Ck 18 Keratin 18 K18 |
| ENSDARG00000018542 | ENSFM00730001521886 | Hyaluronan And Proteoglycan Link 1 Precursor Cartilage Linking 1 Cartilage Link Proteoglycan Link |
| ENSDARG00000018618 | ENSFM00600000921459 | Cleavage And Polyadenylation Specificity Factor Subunit 6 |
| ENSDARG00000018693 | ENSFM00730001521172 | Cadherin Precursor Cadherin Cadherin |
| ENSDARG00000018820 | ENSFM00740001589123 | Filamin Fln Abp 280 Actin Binding Filamin |
| ENSDARG00000018903 | ENSFM00250000006128 | Aminoacyl Trna Synthase Complex Interacting Multifunctional 2 Multisynthase Complex Auxiliary Component P38 Jtv 1 |
| ENSDARG00000018967 | ENSFM00250000000928 | Gamma Aminobutyric Acid Type B Receptor Subunit 2 Precursor Gaba B Receptor 2 Gaba B R2 Gaba BR2 GABABR2 GB2 G Coupled Receptor 51 |
| ENSDARG00000019000 | ENSFM00310000089021 | Structural Maintenance Of Chromosomes 3 Smc 3 Smc 3 Chondroitin Sulfate Proteoglycan 6 |
| ENSDARG00000019062 | ENSFM00500000270526 | Unknown |
| ENSDARG00000019191 | ENSFM00500000269805 | Septin |
| ENSDARG00000019205 | ENSFM00250000002493 | Constitutive Coactivator Of Ppar Gamma 1 Oxidative Stress Associated Src Activator FAM120A |
| ENSDARG00000019230 | ENSFM00750001632337 | 60S Ribosomal L7A |
| ENSDARG00000019231 | ENSFM00740001589109 | Spectrin Alpha Chain Non Erythrocytic 1 Alpha Ii Spectrin Fodrin Alpha Chain |
| ENSDARG00000019239 | ENSFM00510000502743 | Cullin |
| ENSDARG00000019341 | ENSFM00500000269807 | Glypican Secreted Glypican |
| ENSDARG00000019345 | ENSFM00610000958444 | Unknown |
| ENSDARG00000019345 | ENSFM00730001521220 | Kinase EC_2.7.11.1 |
| ENSDARG00000019371 | ENSFM00440000236870 | Vascular Endothelial Growth Factor Receptor Precursor Vegfr EC_2.7.10.1 Fms Tyrosine Kinase Flt Tyrosine Kinase Receptor |
| ENSDARG00000019383 | ENSFM00730001521452 | Serine/Threonine Kinase Mrck EC_2.7.11.1 CDC42 Binding Kinase Dmpk Myotonic Dystrophy Kinase Related CDC42 Binding Kinase Mrck Myotonic Dystrophy Kinase |
| ENSDARG00000019426 | ENSFM00740001589124 | EC_2.7.11.1 |
| ENSDARG00000019529 | ENSFM00250000001234 | Poly [Adp Ribose] Polymerase 1 Parp 1 EC_2.4.2.30 Adp Ribosyltransferase Diphtheria Toxin 1 ARTD1 Nad + Adp Ribosyltransferase 1 Adprt 1 Poly[Adp Ribose] Synthase 1 |
| ENSDARG00000019651 | ENSFM00800002003401 | Septin |
| ENSDARG00000019743 | ENSFM00760001714751 | Dynactin Subunit 1.150 Kda Dynein Associated Polypeptide Dap 150 Dp 150 Glued |
| ENSDARG00000019778 | ENSFM00250000001600 | 40S Ribosomal S6 |
| ENSDARG00000019945 | ENSFM00740001589097 | Receptor Type Tyrosine Phosphatase Precursor EC_3.1.3.48 |
| ENSDARG00000020007 | ENSFM00250000000184 | Collagen Alpha I Chain Precursor Alpha Type I Collagen |
| ENSDARG00000020102 | ENSFM00260000050545 | Btb/Poz Domain Containing |
| ENSDARG00000020134 | ENSFM00730001521477 | Signal Induced Proliferation Associated 1 SIPA1 |
| ENSDARG00000020149 | ENSFM00500000272795 | Acyl Coenzyme A Oxidase Acyl Coa Oxidase EC_1.3.3.- |
| ENSDARG00000020328 | ENSFM00800002003674 | Pleckstrin Homology Domain Containing Family A Member 6 Ph Domain Containing Family A Member 6 Phosphoinositol 3 Phosphate Binding 3 Pepp 3 |
| ENSDARG00000020345 | ENSFM00250000000820 | Clip Associating Cytoplasmic Linker Associated |
| ENSDARG00000020443 | ENSFM00500000269993 | Xk Related |
| ENSDARG00000020450 | ENSFM00250000002019 | Voltage Dependent Calcium Channel Gamma Subunit Neuronal Voltage Gated Calcium Channel Gamma Subunit Transmembrane Ampar Regulatory Gamma Tarp Gamma |
| ENSDARG00000020482 | ENSFM00250000000671 | Paraspeckle Component 1 |
| ENSDARG00000020493 | ENSFM00250000002312 | Leucine Rich Repeat Lgi Family Member Precursor Leucine Rich Glioma Inactivated |
| ENSDARG00000020521 | ENSFM00250000001326 | Exocyst Complex Component 6 Exocyst Complex Component SEC15A SEC15 1 |
| ENSDARG00000020573 | ENSFM00770001847530 | Atp Dependent Rna Helicase DDX3Y EC_3.6.4.13 Dead Box 3 Y Chromosomal |
| ENSDARG00000020609 | ENSFM00250000001228 | Synaptosomal Associated 25 Snap 25 Synaptosomal Associated 25 Kda |
| ENSDARG00000020621 | ENSFM00430000230089 | Ap 1 Complex Subunit Beta 1 Adaptor Complex Ap 1 Subunit Beta 1 Adaptor Related Complex 1 Subunit Beta 1 Beta 1 Adaptin Beta Adaptin 1 Clathrin Assembly Complex 1 Beta Large Chain Golgi Adaptor HA1/AP1 Adaptin Beta Subunit |
| ENSDARG00000020718 | ENSFM00500000270170 | Mitochondrial Glutamate Carrier 1 Gc 1 Glutamate/H + Symporter 1 Solute Carrier Family 25 Member 22 |
| ENSDARG00000020741 | ENSFM00250000004159 | Pre Splicing Factor PRP46 Pre Processing 46 |
| ENSDARG00000020771 | ENSFM00740001589132 | Tenascin Precursor Tn |
| ENSDARG00000020795 | ENSFM00730001521186 | Ras Related C3 Botulinum Toxin Substrate Precursor P21 |
| ENSDARG00000020850 | ENSFM00800002003364 | Elongation Factor 1 Alpha Ef 1 Alpha Eukaryotic Elongation Factor 1 A EEF1A |
| ENSDARG00000020890 | ENSFM00500000269813 | Tropomodulin Tropomodulin Tmod |
| ENSDARG00000020953 | ENSFM00570000851144 | Dnaj Homolog Subfamily B Member |
| ENSDARG00000020982 | ENSFM00730001521542 | Prickle |
| ENSDARG00000021048 | ENSFM00740001589427 | Cleft Lip And Palate Transmembrane 1 CLPTM1 |
| ENSDARG00000021135 | ENSFM00500000270135 | Dehydrogenase/Reductase Sdr Family Member 4 EC_1.1.1.184 Nadph Dependent Carbonyl Reductase/Nadp Retinol Dehydrogenase Cr Phcr Nadph Dependent Retinol Dehydrogenase/Reductase Ndrd Peroxisomal Short Chain Alcohol Dehydrogenase Pscd |
| ENSDARG00000021193 | ENSFM00610000967750 | Unknown |
| ENSDARG00000021193 | ENSFM00730001521292 | Coronin |
| ENSDARG00000021200 | ENSFM00500000270806 | Lim And SH3 Domain 1 Lasp 1 |
| ENSDARG00000021309 | ENSFM00730001521223 | Rho Related Gtp Binding Precursor |
| ENSDARG00000021351 | ENSFM00730001521354 | Neurofilament Polypeptide Nf Kda Neurofilament Neurofilament Triplet |
| ENSDARG00000021352 | ENSFM00720001492020 | Glutamate Receptor Precursor Glur Ampa Selective Glutamate Receptor Glur Glutamate Receptor Ionotropic Ampa |
| ENSDARG00000021366 | ENSFM00250000001433 | Fructose 1 6 Bisphosphatase 1 Fbpase 1 EC_3.1.3.11 D Fructose 1 6 Bisphosphate 1 Phosphohydrolase 1 Liver Fbpase |
| ENSDARG00000021442 | ENSFM00800002003474 | Cadherin Precursor |
| ENSDARG00000021584 | ENSFM00390000126303 | Contactin Precursor Brain Derived Immunoglobulin Superfamily Big |
| ENSDARG00000021590 | ENSFM00250000000552 | Membrane Associated Guanylate Kinase Ww And Pdz Domain Containing Membrane Associated Guanylate Kinase Inverted Magi |
| ENSDARG00000021595 | ENSFM00250000002406 | Lipoma Hmgic Fusion Partner |
| ENSDARG00000021607 | ENSFM00290000065521 | Precursor |
| ENSDARG00000021838 | ENSFM00250000002784 | 40S Ribosomal S23 |
| ENSDARG00000021864 | ENSFM00610000961726 | Unknown |
| ENSDARG00000021864 | ENSFM00500000308224 | Ribosomal P1 |
| ENSDARG00000021948 | ENSFM00740001589132 | Tenascin Precursor Tn |
| ENSDARG00000021984 | ENSFM00670001239126 | Nadh Dehydrogenase [Ubiquinone] 1 Alpha Subcomplex Subunit 2 |
| ENSDARG00000021987 | ENSFM00740001589199 | Plectin Pcn Pltn Plectin 1 |
| ENSDARG00000022045 | ENSFM00250000000942 | Microtubule Associated Map [Contains Heavy Chain |
| ENSDARG00000022129 | ENSFM00610000973231 | Unknown |
| ENSDARG00000022129 | ENSFM00350000105447 | Rna Binding Rna Binding Motif Rna Binding Motif |
| ENSDARG00000022176 | ENSFM00290000065521 | Precursor |
| ENSDARG00000022315 | ENSFM00670001236060 | V Type Proton Atpase Subunit G 1 V Atpase Subunit G 1 V Atpase 13 Kda Subunit 1 Vacuolar Proton Pump Subunit G 1 Vacuolar Proton Pump Subunit M16 |
| ENSDARG00000022438 | ENSFM00670001237914 | Cytochrome C Oxidase Subunit 6A Mitochondrial |
| ENSDARG00000022509 | ENSFM00500000271199 | Cytochrome C Oxidase Subunit 4 1 Mitochondrial Cytochrome C Oxidase Polypeptide Iv Cytochrome C Oxidase Subunit Iv 1 Cox Iv 1 |
| ENSDARG00000022668 | ENSFM00500000270097 | Growth Factor Receptor Bound 2 Adapter GRB2 SH2/SH3 Adapter GRB2 |
| ENSDARG00000023267 | ENSFM00790001963005 | Atpase Family Aaa Domain Containing 1 EC_3.6.1.3 |
| ENSDARG00000023298 | ENSFM00670001235597 | 40S Ribosomal S27 |
| ENSDARG00000023299 | ENSFM00670001235480 | Unknown |
| ENSDARG00000023443 | ENSFM00250000000691 | Tight Junction Zo Tight Junction Zona Occludens Zonula Occludens |
| ENSDARG00000023472 | ENSFM00250000001012 | Catenin Beta Beta Catenin |
| ENSDARG00000023542 | ENSFM00260000050333 | Cadherin Precursor |
| ENSDARG00000023672 | ENSFM00670001238234 | Mitochondrial Import Inner Membrane Translocase Subunit TIM8 A |
| ENSDARG00000023713 | ENSFM00750001632461 | Aquaporin 1 Aqp 1 Aquaporin Chip Water Channel Red Blood Cells And Kidney Proximal Tubule |
| ENSDARG00000023771 | ENSFM00400000131718 | Gamma Aminobutyric Acid Receptor Subunit Beta Precursor Gaba A Receptor Subunit Beta |
| ENSDARG00000023797 | ENSFM00250000000332 | Ryanodine Receptor Ryr Muscle Calcium Release Channel Muscle Type Ryanodine Receptor Type Ryanodine Receptor |
| ENSDARG00000023914 | ENSFM00730001521220 | Kinase EC_2.7.11.1 |
| ENSDARG00000023940 | ENSFM00250000001269 | Wiskott Aldrich Syndrome Family Member 1 Wasp Family Member 1 Wave 1 |
| ENSDARG00000023967 | ENSFM00750001632375 | V Type Proton Atpase Subunit C V Atpase Subunit C Vacuolar Proton Pump Subunit C |
| ENSDARG00000024017 | ENSFM00250000002191 | Mam Domain Containing Glycosylphosphatidylinositol Anchor 2 Precursor Mam Domain Containing 1 |
| ENSDARG00000024478 | ENSFM00500000273639 | Mitochondrial Pyruvate Carrier 2 Brain 44 |
| ENSDARG00000024642 | ENSFM00730001521611 | Phosphatidylinositol 4 Phosphate 5 Kinase Type 1 Beta Pi Beta Ptdins 4 P 5 Kinase 1 Beta EC_2.7.1.68 Phosphatidylinositol 4 Phosphate 5 Kinase Type I Phosphatidylinositol 4 Phosphate 5 Kinase Beta |
| ENSDARG00000024694 | ENSFM00800002003560 | Unconventional Myosin Ia Brush Border Myosin I Bbm I Bbmi Myosin I Heavy Chain Mihc |
| ENSDARG00000024785 | ENSFM00250000001018 | Catenin Alpha 2 Alpha N Catenin |
| ENSDARG00000024874 | ENSFM00760001714721 | Dedicator Of Cytokinesis 3 Modifier Of Cell Adhesion Presenilin Binding Pbp |
| ENSDARG00000024954 | ENSFM00500000270043 | Transmembrane EMP24 Domain Containing Precursor |
| ENSDARG00000024966 | ENSFM00730001521220 | Kinase EC_2.7.11.1 |
| ENSDARG00000025073 | ENSFM00630001070331 | Unknown |
| ENSDARG00000025073 | ENSFM00760001714679 | 60S Ribosomal L18A |
| ENSDARG00000025074 | ENSFM00760001714704 | 39S Ribosomal L12 Mitochondrial Precursor L12MT Mrp L12 |
| ENSDARG00000025108 | ENSFM00250000000552 | Membrane Associated Guanylate Kinase Ww And Pdz Domain Containing Membrane Associated Guanylate Kinase Inverted Magi |
| ENSDARG00000025189 | ENSFM00730001521728 | Copine Copine |
| ENSDARG00000025301 | ENSFM00730001521291 | Vimentin |
| ENSDARG00000025319 | ENSFM00730001521177 | Tyrosine Kinase EC_2.7.10.2 |
| ENSDARG00000025350 | ENSFM00250000000757 | Thioredoxin Peroxidase Thioredoxin Dependent Peroxide Reductase |
| ENSDARG00000025581 | ENSFM00800002003415 | 60S Ribosomal L10 |
| ENSDARG00000025728 | ENSFM00730001522127 | Glutamate [Nmda] Receptor Subunit 1 |
| ENSDARG00000025850 | ENSFM00670001236070 | 40S Ribosomal S21 |
| ENSDARG00000025903 | ENSFM00740001589285 | Galectin Gal |
| ENSDARG00000025904 | ENSFM00250000002457 | Very Long Chain Enoyl Coa Reductase EC_1.3.1.93 Synaptic Glycoprotein SC2 Trans 2 3 Enoyl Coa Reductase Ter |
| ENSDARG00000025953 | ENSFM00610000970041 | Unknown |
| ENSDARG00000025953 | ENSFM00730001521235 | Cell Division Control 42 Homolog Precursor |
| ENSDARG00000025974 | ENSFM00250000000552 | Membrane Associated Guanylate Kinase Ww And Pdz Domain Containing Membrane Associated Guanylate Kinase Inverted Magi |
| ENSDARG00000026068 | ENSFM00500000273083 | Trafficking Particle Complex Subunit 5 |
| ENSDARG00000026333 | ENSFM00500000269993 | Xk Related |
| ENSDARG00000026484 | ENSFM00750001632504 | Ras Related Rab 15 |
| ENSDARG00000026595 | ENSFM00810002040664 | Lissencephaly 1 |
| ENSDARG00000026630 | ENSFM00730001521220 | Kinase EC_2.7.11.1 |
| ENSDARG00000026680 | ENSFM00500000270162 | Kidney Mitochondrial Carrier 1 Solute Carrier Family 25 Member 30 |
| ENSDARG00000026762 | ENSFM00800002020379 | Unknown |
| ENSDARG00000026762 | ENSFM00250000003532 | Hyccin Down Regulated By CTNNB1 A FAM126A |
| ENSDARG00000026801 | ENSFM00250000001250 | Double Stranded Rna Binding Staufen Homolog |
| ENSDARG00000026811 | ENSFM00500000270921 | Exostosin 3 EC_2.4.1.223 Glucuronyl Galactosyl Proteoglycan 4 Alpha N Acetylglucosaminyltransferase Multiple Exostosis 3 |
| ENSDARG00000026845 | ENSFM00730001521223 | Rho Related Gtp Binding Precursor |
| ENSDARG00000026882 | ENSFM00500000271464 | Paralemmin 1 Precursor Paralemmin |
| ENSDARG00000026967 | ENSFM00670001235578 | Ap 2 Complex Subunit Sigma Adaptor Complex Ap 2 Subunit Sigma Adaptor Related Complex 2 Subunit Sigma Clathrin Assembly 2 Sigma Small Chain Clathrin Coat Assembly AP17 Clathrin Coat Associated AP17 Plasma Membrane Adaptor Ap 2.17 Kda SIGMA2 Adaptin |
| ENSDARG00000027200 | ENSFM00500000272384 | Gamma Aminobutyric Acid Receptor Associated 2 Precursor Gaba A Receptor Associated 2 Ganglioside Expression Factor 2 Gef 2 General Transport Factor P16 Golgi Associated Atpase Enhancer Of 16 Kda Gate 16 MAP1 Light Chain 3 Related |
| ENSDARG00000027279 | ENSFM00250000001973 | Numb |
| ENSDARG00000027322 | ENSFM00260000050595 | Lin 7 Homolog Lin Mammalian Lin Seven Mals Vertebrate Lin 7 Homolog Veli |
| ENSDARG00000027355 | ENSFM00250000000816 | Adp/Atp Translocase Adp Atp Carrier Adenine Nucleotide Translocator Ant Solute Carrier Family 25 Member [Contains Adp/Atp Translocase N Terminally Processed] |
| ENSDARG00000027397 | ENSFM00250000002390 | Vang 2 Van Gogh |
| ENSDARG00000027564 | ENSFM00730001521739 | Serine/Threonine Kinase Pak EC_2.7.11.1 P21 Activated Kinase Pak |
| ENSDARG00000027590 | ENSFM00800002003496 | Septin 2 |
| ENSDARG00000027602 | ENSFM00260000050443 | Leucine Rich Repeat And Fibronectin Type Iii Domain Containing Precursor Synaptic Adhesion Molecule |
| ENSDARG00000027618 | ENSFM00740001589268 | Glutamate Receptor Interacting |
| ENSDARG00000027734 | ENSFM00500000269839 | Serine/Arginine Rich Splicing Factor Pre Splicing Factor Splicing Factor Arginine/Serine Rich |
| ENSDARG00000027740 | ENSFM00670001235823 | Pituitary Adenylate Cyclase Activating Polypeptide Precursor Pacap [Contains Pacap Related Peptide Prp 48 |
| ENSDARG00000027828 | ENSFM00730001522127 | Glutamate [Nmda] Receptor Subunit 1 |
| ENSDARG00000027915 | ENSFM00500000272347 | Zinc Finger C2HC Domain Containing 1A |
| ENSDARG00000027963 | ENSFM00800002003895 | Cam Kinase Vesicle Associated |
| ENSDARG00000028000 | ENSFM00730001521507 | Atp Dependent 6 Phosphofructokinase Type |
| ENSDARG00000028259 | ENSFM00250000000672 | Aldehyde Dehydrogenase Aldehyde Dehydrogenase Aldehyde Dehydrogenase Family 3 Member |
| ENSDARG00000028306 | ENSFM00730001521291 | Vimentin |
| ENSDARG00000028354 | ENSFM00500000272618 | Syntaxin Binding 6 |
| ENSDARG00000028393 | ENSFM00350000105429 | Disheveled Associated Activator Of Morphogenesis |
| ENSDARG00000028401 | ENSFM00730001521186 | Ras Related C3 Botulinum Toxin Substrate Precursor P21 |
| ENSDARG00000028533 | ENSFM00740001589112 | Microtubule Actin Cross Linking Factor 1 Actin Cross Linking Family 7 |
| ENSDARG00000028552 | ENSFM00500000270520 | Lipid Phosphate Phosphatase Related Type 4 EC_3.1.3.4 Brain Specific Phosphatidic Acid Phosphatase 1 Plasticity Related Gene 1 Prg 1 |
| ENSDARG00000028618 | ENSFM00560000771030 | Keratin Type I Cytoskeletal 18 Cytokeratin 18 Ck 18 Keratin 18 K18 |
| ENSDARG00000028748 | ENSFM00730001521961 | Transmembrane 9 Superfamily Member Precursor |
| ENSDARG00000028800 | ENSFM00250000001040 | Cytohesin Ph SEC7 And Coiled Coil Domain Containing |
| ENSDARG00000028900 | ENSFM00500000270052 | Outer Dense Fiber Cenexin Outer Dense Fiber Of Sperm Tails 2 |
| ENSDARG00000029019 | ENSFM00800002003537 | Unknown |
| ENSDARG00000029064 | ENSFM00670001264207 | Ubiquinol Cytochrome C Reductase Complex Iii Subunit Vii |
| ENSDARG00000029114 | ENSFM00670001235536 | Serine/Threonine Kinase EC_2.7.11.1 Apoptosis Associated Tyrosine Kinase Brain Kinase Lemur Tyrosine Kinase |
| ENSDARG00000029133 | ENSFM00250000005014 | Gamma Tubulin Complex Component 3 Gamma Ring Complex |
| ENSDARG00000029234 | ENSFM00500000269888 | Syntaxin Binding 5 Lethal 2 Giant Larvae Homolog Tomosyn |
| ENSDARG00000029239 | ENSFM00720001492041 | Synaptotagmin Synaptotagmin |
| ENSDARG00000029252 | ENSFM00500000270730 | Lupus La Homolog La Autoantigen Homolog La Ribonucleoprotein |
| ENSDARG00000029307 | ENSFM00800002024333 | Unknown |
| ENSDARG00000029307 | ENSFM00250000005107 | Uncharacterized Colon Cancer Associated MIC1 Mic 1 |
| ENSDARG00000029472 | ENSFM00250000001587 | Afadin Af 6 |
| ENSDARG00000029480 | ENSFM00250000001872 | Cytospin A SPECC1 Sperm Antigen With Calponin Homology And Coiled Coil Domains 1 |
| ENSDARG00000029500 | ENSFM00500000271952 | 60S Ribosomal L34 |
| ENSDARG00000029533 | ENSFM00810002040668 | 60S Ribosomal L18 |
| ENSDARG00000029689 | ENSFM00250000001413 | Transketolase EC_2.2.1.1 |
| ENSDARG00000029692 | ENSFM00250000001058 | Run And Fyve Domain Containing |
| ENSDARG00000029931 | ENSFM00500000270146 | Manganese Transporting Atpase EC_3.6.3.- |
| ENSDARG00000030038 | ENSFM00500000274497 | C10 |
| ENSDARG00000030097 | ENSFM00250000003553 | Syntenin Syndecan Binding |
| ENSDARG00000030154 | ENSFM00730001521739 | Serine/Threonine Kinase Pak EC_2.7.11.1 P21 Activated Kinase Pak |
| ENSDARG00000030376 | ENSFM00730001521500 | Glutamate Receptor Ionotropic Nmda Precursor Glutamate [Nmda] Receptor Subunit Epsilon N Methyl D Aspartate Receptor Subtype |
| ENSDARG00000030479 | ENSFM00750001659003 | Unknown |
| ENSDARG00000030479 | ENSFM00500000269645 | High Mobility Group B1 High Mobility Group 1 Hmg 1 |
| ENSDARG00000030490 | ENSFM00740001589095 | Spectrin Beta Chain Erythrocytic Beta Spectrin |
| ENSDARG00000030602 | ENSFM00670001235598 | 40S Ribosomal S19 |
| ENSDARG00000030641 | ENSFM00250000006470 | LETM1 Domain Containing 1 |
| ENSDARG00000030644 | ENSFM00410000138458 | Guanine Nucleotide Binding Subunit |
| ENSDARG00000030656 | ENSFM00500000269805 | Septin |
| ENSDARG00000030694 | ENSFM00250000002828 | V Type Proton Atpase Subunit E V Atpase Subunit E V Atpase Kda Subunit Vacuolar Proton Pump Subunit E |
| ENSDARG00000030887 | ENSFM00500000273791 | Phd Finger 23 |
| ENSDARG00000031020 | ENSFM00260000050567 | Phosphatidylinositol 5 Phosphate 4 Kinase Type 2 EC_2.7.1.149 1 Phosphatidylinositol 5 Phosphate 4 Kinase 2 Diphosphoinositide Kinase 2 Phosphatidylinositol 5 Phosphate 4 Kinase Type Ii Pi 5 P 4 Kinase Type Ii PIP4KII Ptdins 5 P 4 Kinase 2 |
| ENSDARG00000031049 | ENSFM00250000006695 | Immunoglobulin Superfamily Member 21 Precursor IGSF21 |
| ENSDARG00000031119 | ENSFM00600000938696 | Unknown |
| ENSDARG00000031119 | ENSFM00250000000906 | Brain Specific Angiogenesis Inhibitor 1 Associated 2 Bai Associated 2 BAI1 Associated 2 Insulin Receptor Substrate Of 53 Kda IRSP53 Insulin Receptor Substrate P53 |
| ENSDARG00000031136 | ENSFM00800002003555 | Dbh Monooxygenase Precursor EC_1.14.17.- |
| ENSDARG00000031164 | ENSFM00250000000217 | Tubulin Alpha Chain |
| ENSDARG00000031200 | ENSFM00810002040632 | Serine/Threonine Phosphatase 2A 56 Kda Regulatory Subunit Gamma PP2A B Subunit B' PP2A B Subunit B' Gamma PP2A B Subunit B56 Gamma PP2A B Subunit PR61 Gamma PP2A B Subunit R5 Gamma |
| ENSDARG00000031214 | ENSFM00810002040812 | Coatomer Subunit Delta Delta Coat Delta Cop |
| ENSDARG00000031388 | ENSFM00250000005336 | Dynactin Subunit 2 |
| ENSDARG00000031560 | ENSFM00500000269720 | Srsf Kinase EC_2.7.11.1 Kinase Serine/Arginine Rich Specific Kinase Sr Specific Kinase |
| ENSDARG00000031796 | ENSFM00500000269646 | Acetylcholinesterase Precursor Ache EC_3.1.1.7 |
| ENSDARG00000031890 | ENSFM00250000001656 | T Complex 11 |
| ENSDARG00000032049 | ENSFM00740001589204 | Ena/Vasp Ena/Vasodilator Stimulated Phosphoprotein |
| ENSDARG00000032102 | ENSFM00250000000757 | Thioredoxin Peroxidase Thioredoxin Dependent Peroxide Reductase |
| ENSDARG00000032117 | ENSFM00250000003062 | Atp Dependent Rna Helicase DDX1 EC_3.6.4.13 Dead Box 1 |
| ENSDARG00000032161 | ENSFM00250000000639 | Slit Robo Rho Gtpase Activating Rho Gtpase Activating |
| ENSDARG00000032188 | ENSFM00250000001418 | Volume Regulated Anion Channel Subunit Leucine Rich Repeat Containing |
| ENSDARG00000032199 | ENSFM00610000973096 | Unknown |
| ENSDARG00000032199 | ENSFM00500000270283 | Glypican 3 Precursor Intestinal Oci 5 |
| ENSDARG00000032238 | ENSFM00520000517792 | Dynamin EC_3.6.5.5 |
| ENSDARG00000032430 | ENSFM00250000001941 | Serine/Threonine Phosphatase 2A 65 Kda Regulatory Subunit A PP2A Subunit A PR65 PP2A Subunit A R1 |
| ENSDARG00000032458 | ENSFM00730001521220 | Kinase EC_2.7.11.1 |
| ENSDARG00000032565 | ENSFM00250000002019 | Voltage Dependent Calcium Channel Gamma Subunit Neuronal Voltage Gated Calcium Channel Gamma Subunit Transmembrane Ampar Regulatory Gamma Tarp Gamma |
| ENSDARG00000032575 | ENSFM00730001521154 | Unknown |
| ENSDARG00000032577 | ENSFM00740001589241 | H + /Cl Exchange Transporter 7 Chloride Channel 7 Alpha Subunit Chloride Channel 7 Clc 7 |
| ENSDARG00000032606 | ENSFM00350000105408 | Septin |
| ENSDARG00000032714 | ENSFM00720001492020 | Glutamate Receptor Precursor Glur Ampa Selective Glutamate Receptor Glur Glutamate Receptor Ionotropic Ampa |
| ENSDARG00000032725 | ENSFM00500000270032 | Ubiquitin 40S Ribosomal S27A Precursor Ubiquitin Carboxyl Extension 80 [Contains Ubiquitin |
| ENSDARG00000032737 | ENSFM00720001492020 | Glutamate Receptor Precursor Glur Ampa Selective Glutamate Receptor Glur Glutamate Receptor Ionotropic Ampa |
| ENSDARG00000032799 | ENSFM00300000079128 | Potassium Voltage Gated Channel Subfamily D Member Voltage Gated Potassium Channel Subunit KV4 |
| ENSDARG00000032866 | ENSFM00670001235831 | Enhancer Of Rudimentary |
| ENSDARG00000032919 | ENSFM00500000272284 | Micos Complex Subunit MIC19 Coiled Coil Helix Coiled Coil Helix Domain Containing 3 |
| ENSDARG00000032970 | ENSFM00500000271199 | Cytochrome C Oxidase Subunit 4 1 Mitochondrial Cytochrome C Oxidase Polypeptide Iv Cytochrome C Oxidase Subunit Iv 1 Cox Iv 1 |
| ENSDARG00000033201 | ENSFM00710001456837 | Unknown |
| ENSDARG00000033201 | ENSFM00740001589457 | Cysteine Rich Crp |
| ENSDARG00000033516 | ENSFM00250000001283 | EFR3 Homolog |
| ENSDARG00000033609 | ENSFM00500000270555 | Microtubule Associated Proteins 1A/1B Light Chain 3A Precursor Autophagy Related LC3 A Autophagy Related Ubiquitin Modifier LC3 A MAP1 Light Chain 3 1 MAP1A/MAP1B Light Chain 3 A MAP1A/MAP1B LC3 A Microtubule Associated 1 Light Chain 3 Alpha |
| ENSDARG00000033683 | ENSFM00250000000220 | Tropomyosin Alpha 1 Chain Alpha Tropomyosin Tropomyosin 1 |
| ENSDARG00000033738 | ENSFM00250000000438 | Myosin Light Chain Myosin Light Chain Myosin Light Chain |
| ENSDARG00000033845 | ENSFM00250000001385 | Turtle Homolog A Precursor Dendrite Arborization And Synapse Maturation 1 Immunoglobulin Superfamily Member 9A IGSF9A |
| ENSDARG00000033899 | ENSFM00730001521696 | Ap 2 Complex Subunit Mu Ap 2 Mu Chain Clathrin Assembly Complex 2 Mu Medium Chain Clathrin Coat Assembly AP50 Clathrin Coat Associated AP50 MU2 Adaptin Plasma Membrane Adaptor Ap Kda |
| ENSDARG00000034056 | ENSFM00610000971777 | Unknown |
| ENSDARG00000034056 | ENSFM00730001521484 | Casein Kinase I Gamma Cki Gamma EC_2.7.11.1 |
| ENSDARG00000034093 | ENSFM00400000131747 | Serine/Threonine Kinase EC_2.7.11.1 Doublecortin And Cam Kinase Doublecortin Kinase |
| ENSDARG00000034165 | ENSFM00250000001884 | Leucine Rich Repeat And Immunoglobulin Domain Containing Nogo Receptor Interacting Precursor Leucine Rich Repeat Neuronal |
| ENSDARG00000034195 | ENSFM00740001589149 | Dna Topoisomerase 2 EC_5.99.1.3 Dna Topoisomerase Ii |
| ENSDARG00000034240 | ENSFM00630001071983 | Unknown |
| ENSDARG00000034240 | ENSFM00250000001955 | F Actin Capping Subunit Alpha 2 Capz Alpha 2 |
| ENSDARG00000034270 | ENSFM00750001632363 | 2 Oxoglutarate Dehydrogenase Mitochondrial Precursor 2 Oxoglutarate Dehydrogenase Complex Component E1 Ogdc E1 Alpha Ketoglutarate Dehydrogenase |
| ENSDARG00000034344 | ENSFM00780001898335 | Protocadherin Precursor |
| ENSDARG00000034396 | ENSFM00250000004570 | Methionine Trna Ligase Cytoplasmic EC_6.1.1.10 Methionyl Trna Synthetase Metrs |
| ENSDARG00000034470 | ENSFM00250000000488 | Fructose Bisphosphate Aldolase EC_4.1.2.13 Type Aldolase |
| ENSDARG00000034493 | ENSFM00730001521500 | Glutamate Receptor Ionotropic Nmda Precursor Glutamate [Nmda] Receptor Subunit Epsilon N Methyl D Aspartate Receptor Subtype |
| ENSDARG00000034616 | ENSFM00500000271997 | Myeloid Leukemia Factor 2 Myelodysplasia Myeloid Leukemia Factor 2 |
| ENSDARG00000034883 | ENSFM00610000961514 | Unknown |
| ENSDARG00000034883 | ENSFM00500000271070 | Acyl Coa Binding Domain Containing 5 |
| ENSDARG00000034897 | ENSFM00630001060822 | Unknown |
| ENSDARG00000034897 | ENSFM00670001235534 | 40S Ribosomal S10 |
| ENSDARG00000035066 | ENSFM00740001589345 | Nucleolar Transcription Factor 1 Upstream Binding Factor 1 Ubf 1 |
| ENSDARG00000035152 | ENSFM00430000230089 | Ap 1 Complex Subunit Beta 1 Adaptor Complex Ap 1 Subunit Beta 1 Adaptor Related Complex 1 Subunit Beta 1 Beta 1 Adaptin Beta Adaptin 1 Clathrin Assembly Complex 1 Beta Large Chain Golgi Adaptor HA1/AP1 Adaptin Beta Subunit |
| ENSDARG00000035256 | ENSFM00760001714607 | Elongation Factor 2 Ef 2 |
| ENSDARG00000035282 | ENSFM00670001236443 | Er Membrane Complex Subunit 6 Transmembrane 93 |
| ENSDARG00000035340 | ENSFM00730001522339 | Gap Junction Delta 2 Connexin Gap Junction Alpha 9 |
| ENSDARG00000035357 | ENSFM00250000000545 | Guanine Nucleotide Binding G I /G S /G T Subunit Beta Transducin Beta Chain |
| ENSDARG00000035360 | ENSFM00260000050335 | Kinase EC_2.7.11.1 Kinase Mapk/Erk Kinase Kinase Kinase Mek Kinase Kinase Mekkk Kinase |
| ENSDARG00000035433 | ENSFM00500000270526 | Unknown |
| ENSDARG00000035538 | ENSFM00250000000490 | V Type Proton Atpase 116 Kda Subunit A V Atpase 116 Kda Proton 116 Kda Subunit Vacuolar Proton Translocating Atpase 116 Kda Subunit A |
| ENSDARG00000035598 | ENSFM00730001521292 | Coronin |
| ENSDARG00000035605 | ENSFM00250000007069 | Trichoplein Keratin Filament Binding Protein Tchp Mitochondrial With Oncostatic Activity Mitostatin |
| ENSDARG00000035608 | ENSFM00250000004793 | Serine/Threonine Phosphatase PGAM5 Mitochondrial |
| ENSDARG00000035650 | ENSFM00740001589204 | Ena/Vasp Ena/Vasodilator Stimulated Phosphoprotein |
| ENSDARG00000035692 | ENSFM00250000001462 | 40S Ribosomal S3A |
| ENSDARG00000035699 | ENSFM00670001236018 | Signal Recognition Particle 19 Kda SRP19 |
| ENSDARG00000035741 | ENSFM00800002023359 | Unknown |
| ENSDARG00000035741 | ENSFM00770001847568 | Mitochondrial 2 Oxoglutarate/Malate Carrier Ogcp Solute Carrier Family 25 Member 11 |
| ENSDARG00000035763 | ENSFM00250000005997 | Syntaxin 18 |
| ENSDARG00000035860 | ENSFM00500000289305 | Unknown |
| ENSDARG00000035860 | ENSFM00750001641929 | Unknown |
| ENSDARG00000035868 | ENSFM00250000001244 | Spire Homolog |
| ENSDARG00000035871 | ENSFM00670001235478 | 60S Ribosomal L30 |
| ENSDARG00000035872 | ENSFM00670001235389 | Estradiol 17 Beta Dehydrogenase 12 EC_1.1.1.62 17 Beta Hydroxysteroid Dehydrogenase 12 17 Beta Hsd 12 |
| ENSDARG00000035970 | ENSFM00260000050522 | Receptor Type Tyrosine Phosphatase Precursor R Ptp |
| ENSDARG00000035985 | ENSFM00250000007801 | Unknown |
| ENSDARG00000036031 | ENSFM00800002003401 | Septin |
| ENSDARG00000036056 | ENSFM00700001390294 | Inactive Serine/Threonine Kinase VRK3 Serine/Threonine Pseudokinase VRK3 Vaccinia Related Kinase 3 |
| ENSDARG00000036058 | ENSFM00410000138458 | Guanine Nucleotide Binding Subunit |
| ENSDARG00000036104 | ENSFM00770001847571 | Unconventional Myosin |
| ENSDARG00000036164 | ENSFM00290000065553 | Cysteine Trna Ligase EC_6.1.1.16 Cysteinyl Trna Synthetase Cysrs |
| ENSDARG00000036175 | ENSFM00760001714757 | Protocadherin Precursor Protocadherin |
| ENSDARG00000036179 | ENSFM00780001898391 | Unconventional Myosin Ie Unconventional Myosin 1E |
| ENSDARG00000036186 | ENSFM00630001068294 | Unknown |
| ENSDARG00000036186 | ENSFM00800002005438 | Unknown |
| ENSDARG00000036186 | ENSFM00800002005102 | Myelin Basic |
| ENSDARG00000036281 | ENSFM00730001522120 | Coiled Coil And C2 Domain Containing Five Prime Repressor Element Under Dual Repression Binding Fre Under Dual Repression Binding Freud |
| ENSDARG00000036295 | ENSFM00250000003047 | Golgin Subfamily A Member 7 |
| ENSDARG00000036298 | ENSFM00250000003348 | 40S Ribosomal S13 |
| ENSDARG00000036329 | ENSFM00610000954065 | Unknown |
| ENSDARG00000036338 | ENSFM00250000005369 | E3 Ubiquitin Ligase PEP5 |
| ENSDARG00000036375 | ENSFM00250000001390 | Cytoplasmic FMR1 Interacting |
| ENSDARG00000036577 | ENSFM00250000002076 | V Type Proton Atpase 16 Kda Proteolipid Subunit V Atpase 16 Kda Proteolipid Subunit Vacuolar Proton Pump 16 Kda Proteolipid Subunit |
| ENSDARG00000036626 | ENSFM00730001521376 | Semaphorin Precursor Semaphorin Sema |
| ENSDARG00000036629 | ENSFM00250000003648 | 40S Ribosomal S14 |
| ENSDARG00000036684 | ENSFM00760001715902 | Nadh Dehydrogenase 1 Alpha Subcomplex Subunit 7 |
| ENSDARG00000036774 | ENSFM00250000000973 | Regulator Of G Signaling |
| ENSDARG00000036830 | ENSFM00670001270143 | Unknown |
| ENSDARG00000036830 | ENSFM00730001521138 | Keratin Type I Cytoskeletal Cytokeratin Ck Keratin |
| ENSDARG00000036840 | ENSFM00730001521138 | Keratin Type I Cytoskeletal Cytokeratin Ck Keratin |
| ENSDARG00000036863 | ENSFM00770001847571 | Unconventional Myosin |
| ENSDARG00000036875 | ENSFM00500000270361 | 40S Ribosomal S12 |
| ENSDARG00000036878 | ENSFM00670001240188 | SH3 Domain Binding Glutamic Acid Rich 2 |
| ENSDARG00000036888 | ENSFM00610000973299 | Unknown |
| ENSDARG00000036888 | ENSFM00610000973300 | Unknown |
| ENSDARG00000037009 | ENSFM00670001238356 | Barrier To Autointegration Factor |
| ENSDARG00000037038 | ENSFM00720001492034 | 26S Protease Regulatory Subunit 10B 26S Proteasome Aaa Atpase Subunit RPT4 Proteasome 26S Subunit Atpase 6 Proteasome Subunit P42 |
| ENSDARG00000037057 | ENSFM00380000124741 | Glutaryl Coa Dehydrogenase Mitochondrial Precursor Gcd EC_1.3.8.6 |
| ENSDARG00000037071 | ENSFM00670001235586 | 40S Ribosomal S26 |
| ENSDARG00000037122 | ENSFM00250000000375 | Teneurin Ten Odd Oz/Ten M Homolog Tenascin Ten Teneurin Transmembrane [Contains Ten Intracellular Domain Ten Icd ] |
| ENSDARG00000037153 | ENSFM00560000771081 | V Type Proton Atpase Subunit S1 Precursor V Atpase Subunit S1 V Atpase AC45 Subunit V Atpase S1 Accessory Vacuolar Proton Pump Subunit S1 |
| ENSDARG00000037259 | ENSFM00540000718275 | Nadh Dehydrogenase 1 Beta Subcomplex 6 |
| ENSDARG00000037281 | ENSFM00800002003901 | Fibrinogen Gamma Chain Precursor |
| ENSDARG00000037318 | ENSFM00250000004027 | Dolichyl Diphosphooligosaccharide Glycosyltransferase 48 Kda Subunit Precursor Ddost 48 Kda Subunit Oligosaccharyl Transferase 48 Kda Subunit EC_2.4.99.18 |
| ENSDARG00000037350 | ENSFM00750001632341 | 60S Ribosomal L9 |
| ENSDARG00000037403 | ENSFM00730001521103 | Heat Shock Kda Heat Shock 70 Kda |
| ENSDARG00000037433 | ENSFM00250000000920 | Fragile X Mental Retardation 1 |
| ENSDARG00000037478 | ENSFM00800002003474 | Cadherin Precursor |
| ENSDARG00000037496 | ENSFM00720001492020 | Glutamate Receptor Precursor Glur Ampa Selective Glutamate Receptor Glur Glutamate Receptor Ionotropic Ampa |
| ENSDARG00000037498 | ENSFM00720001492020 | Glutamate Receptor Precursor Glur Ampa Selective Glutamate Receptor Glur Glutamate Receptor Ionotropic Ampa |
| ENSDARG00000037553 | ENSFM00500000270404 | Interleukin 1 Receptor Accessory 1 Precursor Il 1 Rapl 1 Il 1RAPL 1 IL1RAPL 1 X Linked Interleukin 1 Receptor Accessory 1 |
| ENSDARG00000037589 | ENSFM00250000005529 | Glucose Induced Degradation 8 Homolog |
| ENSDARG00000037593 | ENSFM00800002023350 | Unknown |
| ENSDARG00000037593 | ENSFM00730001521542 | Prickle |
| ENSDARG00000037679 | ENSFM00500000273639 | Mitochondrial Pyruvate Carrier 2 Brain 44 |
| ENSDARG00000037746 | ENSFM00730001521060 | Actin |
| ENSDARG00000037783 | ENSFM00250000000442 | Coagulation Factor Precursor [Contains Factor Light Chain |
| ENSDARG00000037794 | ENSFM00500000269860 | Neuronal Pentraxin 2 Precursor NP2 Neuronal Pentraxin Ii Np Ii |
| ENSDARG00000037870 | ENSFM00730001521060 | Actin |
| ENSDARG00000037905 | ENSFM00740001589100 | Voltage Dependent Type Calcium Channel Subunit Alpha Brain Calcium Channel Calcium Channel L Type Alpha 1 Polypeptide Voltage Gated Calcium Channel Subunit Alpha CAV2 |
| ENSDARG00000037921 | ENSFM00500000361151 | Unknown |
| ENSDARG00000037932 | ENSFM00260000050595 | Lin 7 Homolog Lin Mammalian Lin Seven Mals Vertebrate Lin 7 Homolog Veli |
| ENSDARG00000037935 | ENSFM00400000131967 | Aldehyde Dehydrogenase Family 16 Member A1 |
| ENSDARG00000037946 | ENSFM00500000271451 | Prolactin Precursor Prl |
| ENSDARG00000037980 | ENSFM00500000273664 | Transcription Elongation Factor B Polypeptide 2 Elongin 18 Kda Subunit Elongin B Elob Rna Polymerase Ii Transcription Factor Siii Subunit B Siii P18 |
| ENSDARG00000037997 | ENSFM00250000000193 | Tubulin Beta Chain |
| ENSDARG00000037998 | ENSFM00780001898418 | Flotillin 1 |
| ENSDARG00000038010 | ENSFM00730001521186 | Ras Related C3 Botulinum Toxin Substrate Precursor P21 |
| ENSDARG00000038028 | ENSFM00500000272594 | Nadh Dehydrogenase [Ubiquinone] 1 Alpha Subcomplex Subunit 6 Complex I B14 Ci B14 Nadh Ubiquinone Oxidoreductase B14 Subunit |
| ENSDARG00000038030 | ENSFM00610000962968 | Unknown |
| ENSDARG00000038030 | ENSFM00670001235574 | Ring Box 1 |
| ENSDARG00000038059 | ENSFM00500000270097 | Growth Factor Receptor Bound 2 Adapter GRB2 SH2/SH3 Adapter GRB2 |
| ENSDARG00000038068 | ENSFM00420000140507 | Probable Atp Dependent Rna Helicase DDX5 EC_3.6.4.13 Dead Box 5 |
| ENSDARG00000038075 | ENSFM00250000003946 | Cytochrome C1 Heme Protein Mitochondrial Precursor Complex Iii Subunit 4 Complex Iii Subunit Iv Cytochrome B C1 Complex Subunit 4 Ubiquinol Cytochrome C Reductase Complex Cytochrome C1 Subunit Cytochrome C 1 |
| ENSDARG00000038123 | ENSFM00730001521616 | Myosin Regulatory Light Myosin Regulatory Light Chain 2 Smooth Muscle Myosin Regulatory Light Chain |
| ENSDARG00000038308 | ENSFM00810002040913 | Synaptobrevin Homolog YKT6 Precursor EC_2.3.1.- |
| ENSDARG00000038315 | ENSFM00670001235619 | Pyridoxal Phosphate Phosphatase Plp Phosphatase EC_3.1.3.3 EC_3.1.3.- 74 Chronophin |
| ENSDARG00000038445 | ENSFM00250000001244 | Spire Homolog |
| ENSDARG00000038543 | ENSFM00460000244957 | Wd Repeat Containing 48 |
| ENSDARG00000038577 | ENSFM00500000411071 | Unknown |
| ENSDARG00000038609 | ENSFM00480000262818 | Myelin P0 Precursor Myelin Peripheral Mpp Myelin Zero |
| ENSDARG00000038655 | ENSFM00250000009368 | Adherens Junction Associated 1 |
| ENSDARG00000038693 | ENSFM00500000386940 | Unknown |
| ENSDARG00000038695 | ENSFM00250000000588 | Elav |
| ENSDARG00000038703 | ENSFM00250000000532 | Hexokinase EC_2.7.1.1 Hexokinase Type Hk |
| ENSDARG00000038768 | ENSFM00760001714704 | 39S Ribosomal L12 Mitochondrial Precursor L12MT Mrp L12 |
| ENSDARG00000038859 | ENSFM00480000262761 | Regulator Of G Signaling |
| ENSDARG00000038967 | ENSFM00730001521665 | Cullin 3 Cul 3 |
| ENSDARG00000038978 | ENSFM00440000236855 | Dnaj Homolog Subfamily B Member |
| ENSDARG00000039005 | ENSFM00610000962953 | Unknown |
| ENSDARG00000039005 | ENSFM00500000272371 | Trafficking Particle Complex Subunit 1 |
| ENSDARG00000039136 | ENSFM00500000333809 | Cytochrome C Oxidase Assembly |
| ENSDARG00000039142 | ENSFM00500000270526 | Unknown |
| ENSDARG00000039182 | ENSFM00730001522093 | Voltage Dependent Calcium Channel Subunit Alpha 2/Delta 1 Precursor Voltage Gated Calcium Channel Subunit Alpha 2/Delta 1 [Contains Voltage Dependent Calcium Channel Subunit Alpha 2 1 |
| ENSDARG00000039268 | ENSFM00600000938103 | Unknown |
| ENSDARG00000039270 | ENSFM00250000006099 | Vesicle Transport Through Interaction With T Snares Homolog 1B Vesicle Transport V Snare VTI1 1 VTI1 RP1 |
| ENSDARG00000039272 | ENSFM00250000001554 | Plastin |
| ENSDARG00000039346 | ENSFM00670001236297 | Nadh Dehydrogenase [Ubiquinone] 1 Alpha Subcomplex Subunit 5 Complex I Subunit B13 Complex I 13KD B Ci 13KD B Nadh Ubiquinone Oxidoreductase 13 Kda B Subunit |
| ENSDARG00000039347 | ENSFM00500000270090 | 40S Ribosomal S24 |
| ENSDARG00000039350 | ENSFM00250000006324 | Single Stranded Dna Binding Protein Mitochondrial Precursor Mt Ssb Mtssb |
| ENSDARG00000039363 | ENSFM00250000001153 | Dnaj Homolog Subfamily Member |
| ENSDARG00000039392 | ENSFM00250000000621 | Serine/Threonine Kinase EC_2.7.11.1 Kinase Lysine Deficient Kinase With No Lysine |
| ENSDARG00000039435 | ENSFM00480000262761 | Regulator Of G Signaling |
| ENSDARG00000039452 | ENSFM00250000000532 | Hexokinase EC_2.7.1.1 Hexokinase Type Hk |
| ENSDARG00000039489 | ENSFM00250000001490 | Arf Gtpase Activating GIT1 Arf Gap GIT1 Cool Associated And Tyrosine Phosphorylated 1 Cat 1 CAT1 G Coupled Receptor Kinase Interactor 1 Grk Interacting 1 |
| ENSDARG00000039502 | ENSFM00800002003364 | Elongation Factor 1 Alpha Ef 1 Alpha Eukaryotic Elongation Factor 1 A EEF1A |
| ENSDARG00000039522 | ENSFM00250000000193 | Tubulin Beta Chain |
| ENSDARG00000039578 | ENSFM00250000003247 | Proliferation Associated 2G4 |
| ENSDARG00000039724 | ENSFM00730001521733 | Oxysterol Binding Related 5 Orp 5 Osbp Related 5 Oxysterol Binding Homolog 1 |
| ENSDARG00000039887 | ENSFM00250000004016 | Complement Component 1 Q Subcomponent Binding Protein Mitochondrial Precursor GC1Q R Glycoprotein GC1QBP C1QBP |
| ENSDARG00000039929 | ENSFM00800002003437 | Creatine Kinase U Type Mitochondrial Precursor EC_2.7.3.2 Acidic Type Mitochondrial Creatine Kinase Mia Ck Ubiquitous Mitochondrial Creatine Kinase U Mtck |
| ENSDARG00000039974 | ENSFM00800002003877 | Ankycorbin Ankyrin Repeat And Coiled Coil Structure Containing Retinal Pigment Epithelial Cell Retinoic Acid Induced 14 |
| ENSDARG00000040001 | ENSFM00250000000754 | Furry |
| ENSDARG00000040002 | ENSFM00760001714681 | Neuropathy Target Esterase Sws |
| ENSDARG00000040087 | ENSFM00760001715069 | Band 4 1 NBL4 |
| ENSDARG00000040156 | ENSFM00250000000328 | Metabotropic Glutamate Receptor Precursor |
| ENSDARG00000040287 | ENSFM00730001521154 | Unknown |
| ENSDARG00000040380 | ENSFM00250000000618 | Rho Guanine Nucleotide Exchange Factor 2 Guanine Nucleotide Exchange Factor H1 Gef H1 |
| ENSDARG00000040455 | ENSFM00500000271471 | Unc 50 Homolog |
| ENSDARG00000040469 | ENSFM00730001522540 | Ectonucleotide Pyrophosphatase/Phosphodiesterase Family Member 6 Precursor E Npp 6 Npp 6 EC_3.1.4.- EC_3.1.4.38 Choline Specific Glycerophosphodiester Phosphodiesterase Glycerophosphocholine Cholinephosphodiesterase Gpc Cpde |
| ENSDARG00000040492 | ENSFM00690001369712 | Unknown |
| ENSDARG00000040492 | ENSFM00690001374215 | Unknown |
| ENSDARG00000040492 | ENSFM00800002003403 | C 1 Tetrahydrofolate Synthase Cytoplasmic C1 Thf Synthase [Includes Methylenetetrahydrofolate Dehydrogenase EC_1.5.1.5 |
| ENSDARG00000040528 | ENSFM00440000236993 | Galectin 3 Binding Precursor Lectin Galactoside Binding Soluble 3 Binding |
| ENSDARG00000040582 | ENSFM00250000007733 | Unknown |
| ENSDARG00000040614 | ENSFM00250000002761 | SEC14 Domain And Spectrin Repeat Containing 1 |
| ENSDARG00000040778 | ENSFM00500000270233 | Ras Related Precursor |
| ENSDARG00000040803 | ENSFM00250000007515 | Serine Beta Lactamase Lactb Mitochondrial Precursor EC_3.4.-.- |
| ENSDARG00000040822 | ENSFM00670001235667 | FUN14 Domain Containing |
| ENSDARG00000040853 | ENSFM00500000270686 | Leucine Rich Repeat Soc 2 Homolog Sur 8 Homolog Soc 2 Homolog |
| ENSDARG00000040874 | ENSFM00610000969353 | Unknown |
| ENSDARG00000040874 | ENSFM00250000000681 | Adducin |
| ENSDARG00000040881 | ENSFM00740001589125 | Heterogeneous Nuclear Ribonucleoprotein Hnrnp [Contains Heterogeneous Nuclear Ribonucleoprotein N Terminally Processed] |
| ENSDARG00000040926 | ENSFM00730001521440 | Coup Transcription Factor Coup Coup Transcription Factor Coup Tf Nuclear Receptor Subfamily 2 Group F Member |
| ENSDARG00000040984 | ENSFM00730001522410 | Heat Shock 70 Kda 13 Precursor Microsomal Stress 70 Atpase Core Stress 70 Chaperone Microsome Associated 60 Kda |
| ENSDARG00000040991 | ENSFM00600000921238 | Cortactin Binding 2 CORTBP2 |
| ENSDARG00000041060 | ENSFM00740001589285 | Galectin Gal |
| ENSDARG00000041182 | ENSFM00250000002321 | 60S Ribosomal L4 |
| ENSDARG00000041232 | ENSFM00500000375647 | Unknown |
| ENSDARG00000041314 | ENSFM00250000004530 | Nadh Dehydrogenase [Ubiquinone] 1 Beta Subcomplex Subunit 9 Complex I B22 Ci B22 Nadh Ubiquinone Oxidoreductase B22 Subunit |
| ENSDARG00000041358 | ENSFM00250000002312 | Leucine Rich Repeat Lgi Family Member Precursor Leucine Rich Glioma Inactivated |
| ENSDARG00000041363 | ENSFM00250000007388 | Dynactin Subunit 3 |
| ENSDARG00000041391 | ENSFM00500000271022 | Transmembrane EMP24 Domain Containing Precursor |
| ENSDARG00000041417 | ENSFM00560000771081 | V Type Proton Atpase Subunit S1 Precursor V Atpase Subunit S1 V Atpase AC45 Subunit V Atpase S1 Accessory Vacuolar Proton Pump Subunit S1 |
| ENSDARG00000041703 | ENSFM00250000002798 | Ribosome Binding Ribosome Receptor Rrp |
| ENSDARG00000041720 | ENSFM00500000270526 | Unknown |
| ENSDARG00000041723 | ENSFM00250000000193 | Tubulin Beta Chain |
| ENSDARG00000041728 | ENSFM00730001521698 | Mannosyl Oligosaccharide 1 2 Alpha Mannosidase Ia EC_3.2.1.113 Man 9 Alpha Mannosidase Mannosidase Alpha Class 1A Member 1 Processing Alpha 1 2 Mannosidase Ia Alpha 1 2 Mannosidase Ia |
| ENSDARG00000041799 | ENSFM00760001714761 | Gap Junction Alpha 1 Connexin 43 CX43 |
| ENSDARG00000041811 | ENSFM00670001235896 | 40S Ribosomal S25 |
| ENSDARG00000041921 | ENSFM00500000408964 | Unknown |
| ENSDARG00000041921 | ENSFM00610000963039 | Unknown |
| ENSDARG00000041921 | ENSFM00800002014373 | Unknown |
| ENSDARG00000041926 | ENSFM00250000000275 | Disks Large Homolog Synapse Associated Sap |
| ENSDARG00000041974 | ENSFM00500000294166 | Unknown |
| ENSDARG00000041974 | ENSFM00610000955359 | Unknown |
| ENSDARG00000042126 | ENSFM00250000001494 | Engulfment And Cell Motility |
| ENSDARG00000042141 | ENSFM00730001522029 | Unconventional Myosin Vi Unconventional Myosin 6 |
| ENSDARG00000042249 | ENSFM00250000001583 | Ferm Domain Containing 4B GRP1 Binding GRSP1 |
| ENSDARG00000042357 | ENSFM00500000269751 | Claudin |
| ENSDARG00000042418 | ENSFM00500000269893 | Polypeptide N Acetylgalactosaminyltransferase EC_2.4.1.41 Polypeptide Galnac Transferase Galnac T Pp Gantase Udp Acetylgalactosaminyltransferase Udp Galnac:Polypeptide N Acetylgalactosaminyltransferase |
| ENSDARG00000042469 | ENSFM00500000272434 | Nadh Dehydrogenase [Ubiquinone] 1 Alpha Subcomplex Subunit Complex I B17 2 Ci B17 2 CIB17 2 Nadh Ubiquinone Oxidoreductase Subunit B17 2 |
| ENSDARG00000042566 | ENSFM00250000002224 | 40S Ribosomal S7 |
| ENSDARG00000042670 | ENSFM00250000001830 | Epidermal Growth Factor Receptor Substrate 15 1 EPS15 Related EPS15R |
| ENSDARG00000042690 | ENSFM00730001521347 | Sphingosine 1 Phosphate Receptor S1P Receptor Endothelial Differentiation G Coupled Receptor Sphingosine 1 Phosphate Receptor Edg S1P Receptor Edg |
| ENSDARG00000042708 | ENSFM00250000000217 | Tubulin Alpha Chain |
| ENSDARG00000042723 | ENSFM00500000269996 | Gdnf Family Receptor Alpha Precursor Gdnf Receptor Alpha Gdnfr Alpha Gfr Alpha Tgf Beta Related Neurotrophic Factor Receptor |
| ENSDARG00000042777 | ENSFM00500000282727 | Nadh Dehydrogenase 1 Alpha Subcomplex 11 |
| ENSDARG00000042803 | ENSFM00730001521441 | Ras Related Rab 3 |
| ENSDARG00000042821 | ENSFM00250000004718 | Alkyldihydroxyacetonephosphate Synthase Alkyl Dhap Synthase EC_2.5.1.26 Alkylglycerone Phosphate Synthase |
| ENSDARG00000042905 | ENSFM00610000960974 | Unknown |
| ENSDARG00000042905 | ENSFM00790001962982 | 60S Ribosomal L10A |
| ENSDARG00000043010 | ENSFM00250000000111 | Calcium/Calmodulin Dependent Kinase Type Ii Subunit Cam Kinase Ii Subunit Camk Ii Subunit EC_2.7.11.17 |
| ENSDARG00000043019 | ENSFM00250000004597 | Exocyst Complex Component 1 Exocyst Complex Component SEC3 |
| ENSDARG00000043026 | ENSFM00670001235358 | Tau Tubulin Kinase EC_2.7.11.1 |
| ENSDARG00000043094 | ENSFM00300000079126 | Guanine Nucleotide Binding G S Subunit Alpha Adenylate Cyclase Stimulating G Alpha |
| ENSDARG00000043134 | ENSFM00250000001072 | Homeobox Cut 1 Ccaat Displacement Cdp Homeobox Cux 1 |
| ENSDARG00000043313 | ENSFM00500000269628 | Ankyrin Ank Ankyrin |
| ENSDARG00000043357 | ENSFM00250000006389 | Transducin Beta 2 |
| ENSDARG00000043410 | ENSFM00500000269993 | Xk Related |
| ENSDARG00000043453 | ENSFM00250000003056 | 40S Ribosomal S5[Contains 40S Ribosomal S5 N Terminally Processed] |
| ENSDARG00000043465 | ENSFM00810002040578 | V Type Proton Atpase Subunit B V Atpase Subunit B Endomembrane Proton Pump 58 Kda Subunit Vacuolar Proton Pump Subunit B |
| ENSDARG00000043467 | ENSFM00540000720122 | Nadh Dehydrogenase 1 Beta Subcomplex Subunit 11 Mitochondrial |
| ENSDARG00000043509 | ENSFM00250000002087 | 60S Ribosomal L11 |
| ENSDARG00000043527 | ENSFM00250000003664 | Probable Lipid Phosphate Phosphatase PPAPDC3 EC_3.1.3.- Phosphatidic Acid Phosphatase Type 2 Domain Containing 3 |
| ENSDARG00000043554 | ENSFM00500000274041 | 39S Ribosomal L40 Mitochondrial Precursor L40MT Mrp L40 Nuclear Localization Signal Containing Deleted In Velocardiofacial Syndrome |
| ENSDARG00000043571 | ENSFM00250000000916 | Kinesin |
| ENSDARG00000043589 | ENSFM00670001235760 | Carbonic Anhydrase 4 Precursor EC_4.2.1.1 Carbonate Dehydratase Iv Carbonic Anhydrase Iv Ca Iv |
| ENSDARG00000043673 | ENSFM00260000050975 | Brain Enriched Guanylate Kinase Associated |
| ENSDARG00000043697 | ENSFM00730001521354 | Neurofilament Polypeptide Nf Kda Neurofilament Neurofilament Triplet |
| ENSDARG00000043746 | ENSFM00740001589096 | Neurexin Precursor Neurexin |
| ENSDARG00000043842 | ENSFM00250000010056 | Uncharacterized |
| ENSDARG00000043864 | ENSFM00610000952905 | Basigin Precursor CD147 Antigen |
| ENSDARG00000043938 | ENSFM00350000105477 | Leucine Rich Repeat Containing 57 |
| ENSDARG00000043976 | ENSFM00250000005309 | Eukaryotic Peptide Chain Release Factor Subunit 1 Eukaryotic Release Factor 1 ERF1 |
| ENSDARG00000044015 | ENSFM00500000269996 | Gdnf Family Receptor Alpha Precursor Gdnf Receptor Alpha Gdnfr Alpha Gfr Alpha Tgf Beta Related Neurotrophic Factor Receptor |
| ENSDARG00000044016 | ENSFM00730001522029 | Unconventional Myosin Vi Unconventional Myosin 6 |
| ENSDARG00000044062 | ENSFM00250000001273 | C Terminal Binding |
| ENSDARG00000044093 | ENSFM00790001962987 | 60S Ribosomal L13A |
| ENSDARG00000044153 | ENSFM00420000140518 | Serine/Threonine Phosphatase PP1 Catalytic Subunit Pp EC_3.1.3.16 |
| ENSDARG00000044182 | ENSFM00250000001250 | Double Stranded Rna Binding Staufen Homolog |
| ENSDARG00000044281 | ENSFM00730001522141 | Leucine Rich Repeat Containing 7 Densin 180 Densin |
| ENSDARG00000044299 | ENSFM00250000000425 | Lamin |
| ENSDARG00000044339 | ENSFM00250000005619 | XRP2 |
| ENSDARG00000044341 | ENSFM00500000270226 | Carbohydrate Sulfotransferase EC_2.8.2.- Galactose/N Acetylglucosamine/N Acetylglucosamine 6 O Sulfotransferase N Acetylglucosamine 6 O Sulfotransferase GLCNAC6ST Sulfotransferase N Acetylglucosamine 6 O Sulfotransferase GLCNAC6ST GN6ST |
| ENSDARG00000044345 | ENSFM00250000001390 | Cytoplasmic FMR1 Interacting |
| ENSDARG00000044501 | ENSFM00730001521291 | Vimentin |
| ENSDARG00000044575 | ENSFM00250000001468 | Valine Trna Ligase EC_6.1.1.9 Valyl Trna Synthetase Valrs |
| ENSDARG00000044669 | ENSFM00250000005391 | Heterochromatin 1 Binding 3 |
| ENSDARG00000044760 | ENSFM00410000138458 | Guanine Nucleotide Binding Subunit |
| ENSDARG00000044874 | ENSFM00250000002627 | Rho Gtpase Activating 39 |
| ENSDARG00000044968 | ENSFM00250000001750 | Vinculin |
| ENSDARG00000044972 | ENSFM00670001235596 | B Cell Receptor Associated Bcr Associated |
| ENSDARG00000044975 | ENSFM00730001521138 | Keratin Type I Cytoskeletal Cytokeratin Ck Keratin |
| ENSDARG00000045006 | ENSFM00730001521864 | Receptor Type Tyrosine Phosphatase Precursor R Ptp EC_3.1.3.48 |
| ENSDARG00000045014 | ENSFM00250000000217 | Tubulin Alpha Chain |
| ENSDARG00000045023 | ENSFM00250000002406 | Lipoma Hmgic Fusion Partner |
| ENSDARG00000045064 | ENSFM00730001543875 | Unknown |
| ENSDARG00000045064 | ENSFM00250000000459 | Actin Binding Lim Ablim Actin Binding Lim Family Member |
| ENSDARG00000045087 | ENSFM00250000002836 | Cyclin Dependent Kinase 5 Activator 1 Precursor CDK5 Activator 1 Cyclin Dependent Kinase 5 Regulatory Subunit 1 Tpkii Regulatory Subunit [Contains Cyclin Dependent Kinase 5 Activator 1 P35 P35 |
| ENSDARG00000045145 | ENSFM00500000276365 | Shisa Precursor |
| ENSDARG00000045150 | ENSFM00800002003414 | Casein Kinase I Delta Cki Delta Ckid EC_2.7.11.1 |
| ENSDARG00000045156 | ENSFM00730001522095 | Regulator Of G Signaling |
| ENSDARG00000045228 | ENSFM00610000960150 | Unknown |
| ENSDARG00000045297 | ENSFM00800002003689 | Prohibitin 2 |
| ENSDARG00000045331 | ENSFM00730001521673 | Catenin Delta 2 |
| ENSDARG00000045355 | ENSFM00500000272323 | Transcription Elongation Factor B Polypeptide 1 Elongin 15 Kda Subunit Elongin C Eloc Rna Polymerase Ii Transcription Factor Siii Subunit C Siii P15 Stromal Membrane Associated SMAP1B Homolog |
| ENSDARG00000045364 | ENSFM00250000005557 | Trafficking Particle Complex Subunit 3 |
| ENSDARG00000045367 | ENSFM00250000000217 | Tubulin Alpha Chain |
| ENSDARG00000045411 | ENSFM00780001898384 | Acyl Coa Dehydrogenase Family Member Acad EC_1.3.99.- |
| ENSDARG00000045415 | ENSFM00300000079126 | Guanine Nucleotide Binding G S Subunit Alpha Adenylate Cyclase Stimulating G Alpha |
| ENSDARG00000045463 | ENSFM00500000270233 | Ras Related Precursor |
| ENSDARG00000045465 | ENSFM00250000002547 | Lanc 1.40 Kda Erythrocyte Membrane P40 |
| ENSDARG00000045487 | ENSFM00250000002859 | 40S Ribosomal S16 |
| ENSDARG00000045540 | ENSFM00250000001098 | Serine/Threonine Phosphatase 6 Regulatory Subunit Saps Domain Family Member |
| ENSDARG00000045543 | ENSFM00670001235732 | V Type Proton Atpase Subunit F V Atpase Subunit F V Atpase 14 Kda Subunit Vacuolar Proton Pump Subunit F |
| ENSDARG00000045639 | ENSFM00250000000588 | Elav |
| ENSDARG00000045685 | ENSFM00800002003683 | Contactin 1 Precursor Neural Cell Surface F3 |
| ENSDARG00000045696 | ENSFM00500000273802 | 39S Ribosomal L23 Mitochondrial L23MT Mrp L23 L23 Mitochondrial Related |
| ENSDARG00000045749 | ENSFM00250000001883 | Liprin Beta 2 Tyrosine Phosphatase Receptor Type F Polypeptide Interacting Binding 2 Ptprf Interacting Binding 2 |
| ENSDARG00000045799 | ENSFM00730001521436 | Core Precursor Chondroitin Sulfate Proteoglycan |
| ENSDARG00000045832 | ENSFM00670001238501 | Cocaine And Amphetamine Regulated Transcript |
| ENSDARG00000045870 | ENSFM00800002003674 | Pleckstrin Homology Domain Containing Family A Member 6 Ph Domain Containing Family A Member 6 Phosphoinositol 3 Phosphate Binding 3 Pepp 3 |
| ENSDARG00000045877 | ENSFM00500000270143 | Ubiquitin Conjugating Enzyme E2 N EC_6.3.2.19 Ubiquitin Carrier N Ubiquitin Ligase N |
| ENSDARG00000045893 | ENSFM00500000270346 | Btb/Poz Domain Containing KCTD15 Potassium Channel Tetramerization Domain Containing 15 |
| ENSDARG00000045945 | ENSFM00250000000771 | Synapsin Synapsin |
| ENSDARG00000045976 | ENSFM00250000002455 | SID1 Transmembrane Family Member Precursor |
| ENSDARG00000046022 | ENSFM00500000270192 | Transmembrane EMP24 Domain Containing Precursor P24 Family Gamma |
| ENSDARG00000046048 | ENSFM00250000001879 | Vesicle Associated Membrane Associated A Vamp A Vamp Associated A Vap A |
| ENSDARG00000046107 | ENSFM00250000001241 | Caskin 1 Cask Interacting 1 |
| ENSDARG00000046157 | ENSFM00750001651938 | Unknown |
| ENSDARG00000046157 | ENSFM00750001659008 | Unknown |
| ENSDARG00000046157 | ENSFM00780001898364 | 40S Ribosomal S17 |
| ENSDARG00000051756 | ENSFM00800002003776 | Pyruvate Dehydrogenase X Component Mitochondrial Precursor Dihydrolipoamide Dehydrogenase Binding Of Pyruvate Dehydrogenase Complex Lipoyl Containing Pyruvate Dehydrogenase Complex Component X |
| ENSDARG00000051783 | ENSFM00250000001360 | 60S Acidic Ribosomal P0 60S Ribosomal L10E |
| ENSDARG00000051814 | ENSFM00730001521864 | Receptor Type Tyrosine Phosphatase Precursor R Ptp EC_3.1.3.48 |
| ENSDARG00000051824 | ENSFM00250000001843 | Tomoregulin 1 Precursor Tr 1 Transmembrane With Egf And One Follistatin Domain |
| ENSDARG00000051888 | ENSFM00250000004551 | IST1 Homolog |
| ENSDARG00000051970 | ENSFM00250000004547 | WD40 Repeat Containing SMU1 Smu 1 Suppressor Of Mec 8 And Unc 52 Homolog [Contains WD40 Repeat Containing SMU1 N Terminally Processed] |
| ENSDARG00000052020 | ENSFM00260000051403 | Cklf Marvel Transmembrane Domain Containing 4 Chemokine Factor Superfamily Member 4 |
| ENSDARG00000052114 | ENSFM00500000273341 | Creb Regulated Transcription Coactivator 3 Transducer Of Regulated Camp Response Element Binding 3 Torc 3 Transducer Of Creb 3 |
| ENSDARG00000052379 | ENSFM00250000005354 | Coiled Coil Domain Containing 22 |
| ENSDARG00000052438 | ENSFM00730001521596 | Actin Related 2 Actin 2 |
| ENSDARG00000052462 | ENSFM00600000921219 | Phosphatidylserine Decarboxylase Proenzyme EC_4.1.1.65 [Contains Phosphatidylserine Decarboxylase Alpha Chain |
| ENSDARG00000052604 | ENSFM00250000000830 | Cytoplasmic Polyadenylation Element Binding |
| ENSDARG00000052625 | ENSFM00670001235580 | Peptidyl Prolyl Cis Trans Isomerase FKBP1B Ppiase FKBP1B EC_5.2.1.8 12.6 Kda FK506 Binding 12.6 Kda Fkbp Fkbp 12 6 FK506 Binding 1B Fkbp 1B Immunophilin FKBP12 6 Rotamase |
| ENSDARG00000052642 | ENSFM00500000276365 | Shisa Precursor |
| ENSDARG00000052644 | ENSFM00610000965197 | Unknown |
| ENSDARG00000052644 | ENSFM00400000131811 | Carbonic Anhydrase Related 11 Precursor Ca Rp Xi Ca Xi Carp Xi |
| ENSDARG00000052673 | ENSFM00760001714685 | Septin 7 CDC10 Homolog |
| ENSDARG00000052674 | ENSFM00770001847535 | Casein Kinase I Alpha Cki Alpha EC_2.7.11.1 CK1 |
| ENSDARG00000052703 | ENSFM00600000921602 | Cdgsh Iron Sulfur Domain Containing 2 |
| ENSDARG00000052713 | ENSFM00400000131776 | Leucine Rich Repeat Transmembrane Neuronal Precursor |
| ENSDARG00000052765 | ENSFM00720001492020 | Glutamate Receptor Precursor Glur Ampa Selective Glutamate Receptor Glur Glutamate Receptor Ionotropic Ampa |
| ENSDARG00000052782 | ENSFM00400000131715 | Glycine Receptor Subunit Precursor Glycine Receptor Kda Subunit |
| ENSDARG00000052840 | ENSFM00250000005730 | Nadh Dehydrogenase [Ubiquinone] Iron Sulfur 4 Mitochondrial Precursor Complex I 18 Kda Ci 18 Kda Nadh Ubiquinone Oxidoreductase 18 Kda Subunit |
| ENSDARG00000052856 | ENSFM00400000131768 | Kh Domain Containing Rna Binding Signal Transduction Associated SAM68 Mammalian Slm |
| ENSDARG00000052950 | ENSFM00250000001623 | Rho Gtpase Activating 21 Rho Type Gtpase Activating 21 |
| ENSDARG00000052957 | ENSFM00740001589144 | 1 Phosphatidylinositol 4 5 Bisphosphate Phosphodiesterase Delta EC_3.1.4.11 Phosphoinositide Phospholipase C Delta Phospholipase C Delta Plc Delta |
| ENSDARG00000053058 | ENSFM00800002003484 | 40S Ribosomal S11 |
| ENSDARG00000053066 | ENSFM00250000000193 | Tubulin Beta Chain |
| ENSDARG00000053070 | ENSFM00250000004732 | Golgi Snap Receptor Complex Member 2.27 Kda Golgi Snare Membrin |
| ENSDARG00000053194 | ENSFM00400000131930 | Pdz Domain Containing 11 |
| ENSDARG00000053196 | ENSFM00260000050608 | Serine/Threonine Kinase EC_2.7.11.1 Rapamycin 1 Target Of Rapamycin 1 |
| ENSDARG00000053205 | ENSFM00250000000441 | Liprin Alpha Tyrosine Phosphatase Receptor Type F Polypeptide Interacting Alpha Ptprf Interacting Alpha |
| ENSDARG00000053217 | ENSFM00750001646347 | Unknown |
| ENSDARG00000053217 | ENSFM00670001261071 | Cytochrome C Oxidase Subunit Viia 2 |
| ENSDARG00000053248 | ENSFM00730001521354 | Neurofilament Polypeptide Nf Kda Neurofilament Neurofilament Triplet |
| ENSDARG00000053358 | ENSFM00600000936395 | Brain Acid Soluble 1 Homolog |
| ENSDARG00000053365 | ENSFM00670001235422 | 60S Ribosomal L31 |
| ENSDARG00000053453 | ENSFM00730001521911 | Maguk P55 Subfamily Member 2 Discs Large Homolog 2 MPP2 |
| ENSDARG00000053454 | ENSFM00390000126303 | Contactin Precursor Brain Derived Immunoglobulin Superfamily Big |
| ENSDARG00000053457 | ENSFM00250000003000 | 60S Ribosomal L23 |
| ENSDARG00000053467 | ENSFM00250000001372 | Gtp Binding |
| ENSDARG00000053517 | ENSFM00730001522226 | Echinoderm Microtubule Associated 6 Emap 6 Echinoderm Microtubule Associated 5 |
| ENSDARG00000053527 | ENSFM00500000272475 | Sorting Nexin 24 |
| ENSDARG00000053547 | ENSFM00250000001480 | Janus Kinase And Microtubule Interacting 1 Multiple Alpha Helices And Rna Linker 1 Marlin 1 |
| ENSDARG00000053609 | ENSFM00250000005901 | Parkinson Disease 7 Domain Containing 1 Precursor |
| ENSDARG00000053617 | ENSFM00250000000111 | Calcium/Calmodulin Dependent Kinase Type Ii Subunit Cam Kinase Ii Subunit Camk Ii Subunit EC_2.7.11.17 |
| ENSDARG00000053652 | ENSFM00500000273764 | Nadh Dehydrogenase Ubiquinone Complex I Assembly Factor 6 Precursor |
| ENSDARG00000053665 | ENSFM00730001521448 | Gamma Aminobutyric Acid Receptor Subunit Gamma Precursor Gaba A Receptor Subunit Gamma |
| ENSDARG00000053668 | ENSFM00250000000734 | Cohesin Subunit Sa SCC3 Homolog Stromal Antigen |
| ENSDARG00000053956 | ENSFM00740001589095 | Spectrin Beta Chain Erythrocytic Beta Spectrin |
| ENSDARG00000053979 | ENSFM00600000921262 | Charged Multivesicular Body 2A Chromatin Modifying 2A CHMP2A |
| ENSDARG00000054177 | ENSFM00250000001368 | G Coupled Receptor 124 Precursor Tumor Endothelial Marker 5 |
| ENSDARG00000054208 | ENSFM00450000242164 | Phosphorylase B Kinase Gamma Catalytic Chain Skeletal Muscle/Heart EC_2.7.11.19 Phosphorylase Kinase Subunit Gamma 1 Serine/Threonine Kinase PHKG1 EC_2.7.11.- 1 EC_2.7.-.- 11 26 |
| ENSDARG00000054320 | ENSFM00810002040615 | Ap 2 Complex Subunit Alpha 2.100 Kda Coated Vesicle C Adaptor Complex Ap 2 Subunit Alpha 2 Adaptor Related Complex 2 Subunit Alpha 2 Alpha Adaptin C ALPHA2 Adaptin Clathrin Assembly Complex 2 Alpha C Large Chain Plasma Membrane Adaptor HA2/AP2 Adaptin Alp |
| ENSDARG00000054337 | ENSFM00810002040662 | Ap 1 Complex Subunit Gamma 1 Adaptor Complex Ap 1 Subunit Gamma 1 Adaptor Related Complex 1 Subunit Gamma 1 Clathrin Assembly Complex 1 Gamma 1 Large Chain Gamma Adaptin GAMMA1 Adaptin Golgi Adaptor HA1/AP1 Adaptin Subunit Gamma 1 |
| ENSDARG00000054355 | ENSFM00500000271100 | DDB1 And CUL4 Associated Factor 7 Wd Repeat Containing 68 Wd Repeat Containing AN11 Homolog |
| ENSDARG00000054456 | ENSFM00740001589331 | Cap Gly Domain Containing Linker 3 Cytoplasmic Linker 170 Related 59 Kda Clip 170 Related 59 Kda Clipr 59 |
| ENSDARG00000054530 | ENSFM00440000236919 | Arginine Trna Ligase Cytoplasmic EC_6.1.1.19 Arginyl Trna Synthetase Argrs |
| ENSDARG00000054610 | ENSFM00730001521292 | Coronin |
| ENSDARG00000054748 | ENSFM00250000005024 | Cue Domain Containing 1 |
| ENSDARG00000054818 | ENSFM00500000270098 | 60S Ribosomal L32 |
| ENSDARG00000054907 | ENSFM00670001237705 | Cytochrome C Oxidase Subunit Viia Related Protein Mitochondrial |
| ENSDARG00000054931 | ENSFM00760001714676 | Serine/Threonine Phosphatase 2A 56 Kda Regulatory Subunit Epsilon PP2A B Subunit B' Epsilon PP2A B Subunit B56 Epsilon PP2A B Subunit PR61 Epsilon PP2A B Subunit R5 Epsilon |
| ENSDARG00000055052 | ENSFM00800002005222 | Uncharacterized |
| ENSDARG00000055052 | ENSFM00260000050441 | Microtubule Associated Tau Neurofibrillary Tangle Paired Helical Filament Tau Phf Tau |
| ENSDARG00000055053 | ENSFM00250000000536 | Inter Alpha Trypsin Inhibitor Heavy Chain Precursor Iti Heavy Chain Iti Inter Alpha Inhibitor Heavy Chain |
| ENSDARG00000055064 | ENSFM00670001235781 | Peroxiredoxin 5 Mitochondrial Precursor EC_1.11.1.15 Antioxidant Enzyme B166 AOEB166 Plp Peroxiredoxin V Prx V Peroxisomal Antioxidant Enzyme Thioredoxin Peroxidase PMP20 Thioredoxin Reductase |
| ENSDARG00000055075 | ENSFM00730001543869 | Unknown |
| ENSDARG00000055075 | ENSFM00250000001422 | Supervillin Archvillin P205/P250 |
| ENSDARG00000055080 | ENSFM00350000105447 | Rna Binding Rna Binding Motif Rna Binding Motif |
| ENSDARG00000055133 | ENSFM00250000003245 | Centromere F Precursor Cenp F Ah Antigen Kinetochore Cenpf Mitosin |
| ENSDARG00000055223 | ENSFM00250000000459 | Actin Binding Lim Ablim Actin Binding Lim Family Member |
| ENSDARG00000055225 | ENSFM00500000269778 | Dedicator Of Cytokinesis |
| ENSDARG00000055302 | ENSFM00720001492054 | Glutamate Receptor Ionotropic Delta Precursor Glur Delta Subunit |
| ENSDARG00000055377 | ENSFM00770001847629 | Guanine Nucleotide Binding Subunit Beta 5 GBETA5 Transducin Beta Chain 5 |
| ENSDARG00000055459 | ENSFM00250000000718 | Disks Large Associated Dap Psd 95/SAP90 Binding SAP90/Psd 95 Associated |
| ENSDARG00000055510 | ENSFM00500000270081 | Yippee |
| ENSDARG00000055565 | ENSFM00250000000524 | Voltage Dependent L Type Calcium Channel Subunit Beta Calcium Channel Voltage Dependent Subunit Beta |
| ENSDARG00000055569 | ENSFM00250000008610 | GH3 Domain Containing Precursor |
| ENSDARG00000055607 | ENSFM00740001589340 | Cingulin |
| ENSDARG00000055610 | ENSFM00250000004215 | Exocyst Complex Component 2 Exocyst Complex Component SEC5 |
| ENSDARG00000055639 | ENSFM00730001521813 | Ruvb 2 EC_3.6.4.12 |
| ENSDARG00000055843 | ENSFM00260000050333 | Cadherin Precursor |
| ENSDARG00000055857 | ENSFM00250000001352 | Dopey |
| ENSDARG00000055976 | ENSFM00500000271562 | Peroxisomal Trans 2 Enoyl Coa Reductase Terp EC_1.3.1.38 2 4 Dienoyl Coa Reductase |
| ENSDARG00000055996 | ENSFM00800002003452 | 40S Ribosomal S8 |
| ENSDARG00000056001 | ENSFM00250000000754 | Furry |
| ENSDARG00000056075 | ENSFM00780001898918 | Membrane Bound Complement Regulatory |
| ENSDARG00000056078 | ENSFM00390000126424 | Raftlin 2 Raft Linking 2 |
| ENSDARG00000056085 | ENSFM00250000004752 | Mob Phocein Class Ii MMOB1 MOB1 Homolog 3 MOB3 Mps One Binder Kinase Activator 3 Preimplantation 3 |
| ENSDARG00000056090 | ENSFM00250000001955 | F Actin Capping Subunit Alpha 2 Capz Alpha 2 |
| ENSDARG00000056101 | ENSFM00300000079128 | Potassium Voltage Gated Channel Subfamily D Member Voltage Gated Potassium Channel Subunit KV4 |
| ENSDARG00000056119 | ENSFM00250000001516 | Elongation Factor 1 Gamma Ef 1 Gamma Eef 1B Gamma |
| ENSDARG00000056121 | ENSFM00280000058758 | SERAC1 Serine Active Site Containing 1 |
| ENSDARG00000056206 | ENSFM00250000000111 | Calcium/Calmodulin Dependent Kinase Type Ii Subunit Cam Kinase Ii Subunit Camk Ii Subunit EC_2.7.11.17 |
| ENSDARG00000056218 | ENSFM00260000050335 | Kinase EC_2.7.11.1 Kinase Mapk/Erk Kinase Kinase Kinase Mek Kinase Kinase Mekkk Kinase |
| ENSDARG00000056250 | ENSFM00250000000681 | Adducin |
| ENSDARG00000056347 | ENSFM00730001521441 | Ras Related Rab 3 |
| ENSDARG00000056394 | ENSFM00250000001136 | TOM1 2 Target Of Myb 2 |
| ENSDARG00000056559 | ENSFM00250000003646 | Vacuolar Fusion CCZ1 Homolog |
| ENSDARG00000056583 | ENSFM00670001236132 | Nadh Dehydrogenase [Ubiquinone] Iron Sulfur 6 Mitochondrial Precursor Complex I 13KD A Ci 13KD A Nadh Ubiquinone Oxidoreductase 13 Kda A Subunit |
| ENSDARG00000056603 | ENSFM00250000000478 | Guanine Nucleotide Exchange Factor Dbs Dbl'S Big Sister MCF2 Transforming Sequence |
| ENSDARG00000056648 | ENSFM00780001898505 | Exostosin 2 EC_2.4.1.224 EC_2.4.1.- 225 Glucuronosyl N Acetylglucosaminyl Proteoglycan/N Acetylglucosaminyl Proteoglycan 4 Alpha N Acetylglucosaminyltransferase Multiple Exostoses 2 |
| ENSDARG00000056683 | ENSFM00730001521798 | Negative Regulator Of The Pho System Serine/Threonine Kinase PHO85 |
| ENSDARG00000056725 | ENSFM00600000944062 | Unknown |
| ENSDARG00000056725 | ENSFM00500000269645 | High Mobility Group B1 High Mobility Group 1 Hmg 1 |
| ENSDARG00000056745 | ENSFM00250000001762 | N Terminal Ef Hand Calcium Binding Ef Hand Calcium Binding Neuronal Calcium Binding |
| ENSDARG00000056797 | ENSFM00250000000829 | Serine/Threonine Phosphatase 2A 55 Kda Regulatory Subunit B PP2A Subunit B B55 PP2A Subunit B PR55 PP2A Subunit B R2 PP2A Subunit B |
| ENSDARG00000056803 | ENSFM00500000269841 | Ap 1 Complex Subunit Sigma Adaptor Complex Ap 1 Subunit Sigma Adaptor Related Complex 1 Subunit Sigma Clathrin Assembly Complex 1 Sigma Small Chain Golgi Adaptor HA1/AP1 Adaptin Sigma Subunit Sigma Subunit Of Ap 1 Clathrin Sigma Adaptin Adaptin |
| ENSDARG00000056831 | ENSFM00500000301701 | Unknown |
| ENSDARG00000056831 | ENSFM00800002013988 | Unknown |
| ENSDARG00000056892 | ENSFM00630001071998 | Unknown |
| ENSDARG00000056892 | ENSFM00730001521911 | Maguk P55 Subfamily Member 2 Discs Large Homolog 2 MPP2 |
| ENSDARG00000056910 | ENSFM00390000126295 | Neural Cell Adhesion Molecule L1 L1 L1 |
| ENSDARG00000056934 | ENSFM00250000007963 | Hepatocyte Cell Adhesion Molecule Precursor Hepacam |
| ENSDARG00000057000 | ENSFM00800002003895 | Cam Kinase Vesicle Associated |
| ENSDARG00000057007 | ENSFM00250000001273 | C Terminal Binding |
| ENSDARG00000057011 | ENSFM00250000004763 | Dolichol Phosphate Mannosyltransferase Subunit 1 EC_2.4.1.83 Dolichol Phosphate Mannose Synthase Subunit 1 Dpm Synthase Subunit 1 Dolichyl Phosphate Beta D Mannosyltransferase Subunit 1 Mannose P Dolichol Synthase Subunit 1 Mpd Synthase Subunit 1 |
| ENSDARG00000057026 | ENSFM00250000002338 | Gtp Binding Nuclear Ran Gtpase Ran Ras Related Nuclear |
| ENSDARG00000057125 | ENSFM00740001589239 | Phospholipase D2 Pld 2 EC_3.1.4.4 Choline Phosphatase 2 Phosphatidylcholine Hydrolyzing Phospholipase D2 |
| ENSDARG00000057128 | ENSFM00760001714774 | Trifunctional Enzyme Subunit Alpha Mitochondrial Precursor 78 Kda Gastrin Binding Tp Alpha [Includes Long Chain Enoyl Coa Hydratase EC_4.2.1.17 |
| ENSDARG00000057159 | ENSFM00440000236974 | Ankyrin Repeat Domain Containing 29 |
| ENSDARG00000057239 | ENSFM00800002003538 | Tryptophan 5 Hydroxylase EC_1.14.16.4 Tryptophan 5 Monooxygenase |
| ENSDARG00000057287 | ENSFM00740001589430 | Graves Disease Carrier Gdc Mitochondrial Solute Carrier Homolog Solute Carrier Family 25 Member 16 |
| ENSDARG00000057368 | ENSFM00800002003452 | 40S Ribosomal S8 |
| ENSDARG00000057414 | ENSFM00790001963030 | Prohibitin |
| ENSDARG00000057556 | ENSFM00250000001117 | 60S Ribosomal L17 |
| ENSDARG00000057568 | ENSFM00730001521354 | Neurofilament Polypeptide Nf Kda Neurofilament Neurofilament Triplet |
| ENSDARG00000057661 | ENSFM00250000000488 | Fructose Bisphosphate Aldolase EC_4.1.2.13 Type Aldolase |
| ENSDARG00000057665 | ENSFM00600000926850 | Prion |
| ENSDARG00000057676 | ENSFM00250000003047 | Golgin Subfamily A Member 7 |
| ENSDARG00000057826 | ENSFM00250000002895 | Occludin |
| ENSDARG00000057853 | ENSFM00250000002076 | V Type Proton Atpase 16 Kda Proteolipid Subunit V Atpase 16 Kda Proteolipid Subunit Vacuolar Proton Pump 16 Kda Proteolipid Subunit |
| ENSDARG00000057855 | ENSFM00250000005086 | 28S Ribosomal S31 Mitochondrial Precursor Mrp S31 S31MT |
| ENSDARG00000057867 | ENSFM00500000270806 | Lim And SH3 Domain 1 Lasp 1 |
| ENSDARG00000057881 | ENSFM00730001521354 | Neurofilament Polypeptide Nf Kda Neurofilament Neurofilament Triplet |
| ENSDARG00000057882 | ENSFM00500000270831 | Actin Related 2/3 Complex Subunit 3 ARP2/3 Complex 21 Kda Subunit P21 Arc |
| ENSDARG00000058102 | ENSFM00800002003410 | Dehydrogenase Mitochondrial Precursor |
| ENSDARG00000058117 | ENSFM00250000001228 | Synaptosomal Associated 25 Snap 25 Synaptosomal Associated 25 Kda |
| ENSDARG00000058162 | ENSFM00250000002167 | Choline Phosphate Cytidylyltransferase A EC_2.7.7.15 Cct Alpha Ctp:Phosphocholine Cytidylyltransferase A Cct A Ct A Phosphorylcholine Transferase A |
| ENSDARG00000058202 | ENSFM00250000005766 | Peroxisomal Membrane PEX16 Peroxin 16 Peroxisomal Biogenesis Factor 16 |
| ENSDARG00000058203 | ENSFM00790001962984 | Structural Maintenance Of Chromosomes 1A Smc 1A Smc 1A |
| ENSDARG00000058207 | ENSFM00250000000845 | Abl Interactor 1 Abelson Interactor 1 Abi 1 EPS8 SH3 Domain Binding EPS8 Binding Spectrin SH3 Domain Binding 1 |
| ENSDARG00000058225 | ENSFM00250000003587 | Actin Related 2/3 Complex Subunit 4 ARP2/3 Complex 20 Kda P20 Arc |
| ENSDARG00000058297 | ENSFM00670001236643 | Mitochondrial Import Inner Membrane Translocase Subunit TIM13 |
| ENSDARG00000058348 | ENSFM00760001714632 | Gelsolin Actin Depolymerizing Factor Adf |
| ENSDARG00000058352 | ENSFM00250000003102 | Acyl Coenzyme A Thioesterase Mitochondrial Precursor Acyl Coa Thioesterase EC_3.1.2.- Mitochondrial 48 Kda Acyl Coa Thioester Hydrolase Mt ACT48 |
| ENSDARG00000058358 | ENSFM00740001589131 | Keratin Type Ii Cytoskeletal 8 Cytokeratin 8 Ck 8 Keratin 8 K8 Type Ii Keratin KB8 |
| ENSDARG00000058371 | ENSFM00740001589131 | Keratin Type Ii Cytoskeletal 8 Cytokeratin 8 Ck 8 Keratin 8 K8 Type Ii Keratin KB8 |
| ENSDARG00000058421 | ENSFM00250000002312 | Leucine Rich Repeat Lgi Family Member Precursor Leucine Rich Glioma Inactivated |
| ENSDARG00000058425 | ENSFM00750001632414 | Ras Related Rab 35 |
| ENSDARG00000058451 | ENSFM00500000270597 | 60S Ribosomal L6 |
| ENSDARG00000058463 | ENSFM00670001235829 | Acyl Carrier Protein Mitochondrial Precursor Acp Nadh Ubiquinone Oxidoreductase 9.6 Kda Subunit |
| ENSDARG00000058494 | ENSFM00730001522148 | Dnaj Homolog Subfamily A Member 3 Mitochondrial Precursor Dnaj Tid 1 1 Tumorous Imaginal Discs TID56 Homolog |
| ENSDARG00000058504 | ENSFM00500000269841 | Ap 1 Complex Subunit Sigma Adaptor Complex Ap 1 Subunit Sigma Adaptor Related Complex 1 Subunit Sigma Clathrin Assembly Complex 1 Sigma Small Chain Golgi Adaptor HA1/AP1 Adaptin Sigma Subunit Sigma Subunit Of Ap 1 Clathrin Sigma Adaptin Adaptin |
| ENSDARG00000058558 | ENSFM00650001140033 | Leucine Zipper Tumor Suppressor 2 |
| ENSDARG00000058644 | ENSFM00570000851144 | Dnaj Homolog Subfamily B Member |
| ENSDARG00000058646 | ENSFM00260000050522 | Receptor Type Tyrosine Phosphatase Precursor R Ptp |
| ENSDARG00000058649 | ENSFM00400000131833 | Tripartite Motif Containing 46 Gene Y Geney Tripartite Fibronectin Type Iii And C Terminal Spry Motif |
| ENSDARG00000058734 | ENSFM00250000000757 | Thioredoxin Peroxidase Thioredoxin Dependent Peroxide Reductase |
| ENSDARG00000058868 | ENSFM00250000001676 | Adenomatous Polyposis Coli |
| ENSDARG00000058892 | ENSFM00740001589275 | Unknown |
| ENSDARG00000059003 | ENSFM00740001589092 | Adenosylhomocysteinase Adohcyase EC_3.3.1.1 S Adenosyl L Homocysteine Hydrolase 2 |
| ENSDARG00000059067 | ENSFM00250000001012 | Catenin Beta Beta Catenin |
| ENSDARG00000059083 | ENSFM00810002040632 | Serine/Threonine Phosphatase 2A 56 Kda Regulatory Subunit Gamma PP2A B Subunit B' PP2A B Subunit B' Gamma PP2A B Subunit B56 Gamma PP2A B Subunit PR61 Gamma PP2A B Subunit R5 Gamma |
| ENSDARG00000059231 | ENSFM00750001632325 | Transmembrane 4 L6 Family Member |
| ENSDARG00000059333 | ENSFM00610000965417 | Unknown |
| ENSDARG00000059333 | ENSFM00250000001584 | Src Kinase Signaling Inhibitor 1 Snap 25 Interacting Snip P130CAS Associated P140CAP |
| ENSDARG00000059340 | ENSFM00250000000718 | Disks Large Associated Dap Psd 95/SAP90 Binding SAP90/Psd 95 Associated |
| ENSDARG00000059349 | ENSFM00800002008304 | Unknown |
| ENSDARG00000059368 | ENSFM00720001492020 | Glutamate Receptor Precursor Glur Ampa Selective Glutamate Receptor Glur Glutamate Receptor Ionotropic Ampa |
| ENSDARG00000059409 | ENSFM00800002003396 | Atp Binding Cassette Sub Family D Member Adrenoleukodystrophy |
| ENSDARG00000059472 | ENSFM00250000001205 | Rho Gtpase Activating Rho Type Gtpase Activating Gtpase Activating |
| ENSDARG00000059596 | ENSFM00250000000764 | Cap Gly Domain Containing Linker 2 Cytoplasmic Linker 115 Clip 115 Cytoplasmic Linker 2 |
| ENSDARG00000059601 | ENSFM00250000000942 | Microtubule Associated Map [Contains Heavy Chain |
| ENSDARG00000059629 | ENSFM00250000005876 | Aspartate Beta Hydroxylase Domain Containing EC_1.14.11.- |
| ENSDARG00000059701 | ENSFM00730001522115 | Flightless 1 |
| ENSDARG00000059738 | ENSFM00740001589097 | Receptor Type Tyrosine Phosphatase Precursor EC_3.1.3.48 |
| ENSDARG00000059763 | ENSFM00400000131718 | Gamma Aminobutyric Acid Receptor Subunit Beta Precursor Gaba A Receptor Subunit Beta |
| ENSDARG00000059770 | ENSFM00630001072039 | Unknown |
| ENSDARG00000059770 | ENSFM00500000269758 | Carnitine O Palmitoyltransferase 1 Muscle CPT1 M EC_2.3.1.21 Carnitine O Palmitoyltransferase I Muscle Cpt I Cpti M Carnitine Palmitoyltransferase 1B |
| ENSDARG00000059773 | ENSFM00250000001584 | Src Kinase Signaling Inhibitor 1 Snap 25 Interacting Snip P130CAS Associated P140CAP |
| ENSDARG00000059806 | ENSFM00770001847625 | Phosphatidylinositide Phosphatase SAC1 EC_3.1.3.- Suppressor Of Actin Mutations 1 |
| ENSDARG00000059809 | ENSFM00730001521718 | 55 Kda Erythrocyte Membrane P55 Membrane Protein Palmitoylated 1 |
| ENSDARG00000059826 | ENSFM00610000966260 | Unknown |
| ENSDARG00000059826 | ENSFM00250000002903 | Cartilage Acidic 1 Precursor 68 Kda Chondrocyte Expressed Cep 68 Aspic |
| ENSDARG00000059832 | ENSFM00250000000737 | Brain Specific Angiogenesis Inhibitor Precursor |
| ENSDARG00000059853 | ENSFM00500000440578 | Unknown |
| ENSDARG00000060006 | ENSFM00730001521822 | E3 Ubiquitin Ligase EC_6.3.2.- |
| ENSDARG00000060036 | ENSFM00500000270832 | Aminoacyl Trna Synthase Complex Interacting Multifunctional 1 Multisynthase Complex Auxiliary Component P43 [Contains Endothelial Monocyte Activating Polypeptide 2 Emap 2 Endothelial Monocyte Activating Polypeptide Ii Emap Ii Small Inducible Cytokine Subf |
| ENSDARG00000060093 | ENSFM00730001521806 | Death Associated Kinase Dap Kinase EC_2.7.11.1 Dap Kinase |
| ENSDARG00000060112 | ENSFM00250000002727 | Syntabulin Golgi Localized Syntaphilin Related Syntaxin 1 Binding |
| ENSDARG00000060149 | ENSFM00250000000459 | Actin Binding Lim Ablim Actin Binding Lim Family Member |
| ENSDARG00000060238 | ENSFM00800002003915 | Uveal Autoantigen With Coiled Coil Domains And Ankyrin Repeats |
| ENSDARG00000060252 | ENSFM00760001714954 | Mannosyl Oligosaccharide Glucosidase EC_3.2.1.106 Processing Glucosidase I |
| ENSDARG00000060306 | ENSFM00250000001310 | Transcription Factor CP2 |
| ENSDARG00000060312 | ENSFM00250000001662 | Mitogen Activated Kinase Binding 1 |
| ENSDARG00000060323 | ENSFM00250000005479 | Exocyst Complex Component Exocyst Complex Component SEC10 |
| ENSDARG00000060326 | ENSFM00250000000942 | Microtubule Associated Map [Contains Heavy Chain |
| ENSDARG00000060349 | ENSFM00250000001269 | Wiskott Aldrich Syndrome Family Member 1 Wasp Family Member 1 Wave 1 |
| ENSDARG00000060368 | ENSFM00250000000771 | Synapsin Synapsin |
| ENSDARG00000060383 | ENSFM00250000008244 | Methylmalonic Aciduria Type A Homolog Mitochondrial Precursor EC_3.6.-.- Methylmalonic Aciduria Type A 1 |
| ENSDARG00000060434 | ENSFM00250000000942 | Microtubule Associated Map [Contains Heavy Chain |
| ENSDARG00000060494 | ENSFM00800002003460 | Bifunctional Glutamate/Proline Trna Ligase Bifunctional Aminoacyl Trna Synthetase [Includes Glutamate Trna Ligase EC_6.1.1.17 Glutamyl Trna Synthetase Glurs |
| ENSDARG00000060504 | ENSFM00730001521507 | Atp Dependent 6 Phosphofructokinase Type |
| ENSDARG00000060532 | ENSFM00410000138484 | Disintegrin And Metalloproteinase Domain Containing Precursor Adam Metalloproteinase Disintegrin And Cysteine Rich Mdc |
| ENSDARG00000060539 | ENSFM00250000000743 | SH3 And Multiple Ankyrin Repeat Domains Proline Rich Synapse Associated |
| ENSDARG00000060594 | ENSFM00760001714774 | Trifunctional Enzyme Subunit Alpha Mitochondrial Precursor 78 Kda Gastrin Binding Tp Alpha [Includes Long Chain Enoyl Coa Hydratase EC_4.2.1.17 |
| ENSDARG00000060725 | ENSFM00250000004546 | Dnaj Homolog Subfamily C Member 16 Precursor |
| ENSDARG00000060734 | ENSFM00250000003219 | Dcc Interacting 13 DIP13 Adapter Containing Ph Domain Ptb Domain And Leucine Zipper Motif |
| ENSDARG00000060797 | ENSFM00730001521507 | Atp Dependent 6 Phosphofructokinase Type |
| ENSDARG00000060805 | ENSFM00250000000942 | Microtubule Associated Map [Contains Heavy Chain |
| ENSDARG00000060813 | ENSFM00800002003882 | Pleckstrin Homology Domain Containing Family A Member 7 Ph Domain Containing Family A Member 7 Heart Adapter 1 |
| ENSDARG00000060853 | ENSFM00250000001880 | Nck Associated 1 1 Membrane Associated Hem |
| ENSDARG00000060854 | ENSFM00250000002046 | SH3KBP1 Binding 1 |
| ENSDARG00000060865 | ENSFM00250000003803 | Scai Suppressor Of Cancer Cell Invasion |
| ENSDARG00000061042 | ENSFM00250000000928 | Gamma Aminobutyric Acid Type B Receptor Subunit 2 Precursor Gaba B Receptor 2 Gaba B R2 Gaba BR2 GABABR2 GB2 G Coupled Receptor 51 |
| ENSDARG00000061066 | ENSFM00260000050940 | C2 Domain Containing 2 Transmembrane 24 |
| ENSDARG00000061093 | ENSFM00600000921202 | Pleckstrin Homology Domain Family B Member LL5 |
| ENSDARG00000061207 | ENSFM00250000002093 | Tetratricopeptide Repeat Tpr Repeat |
| ENSDARG00000061257 | ENSFM00250000007974 | Njmu R1 |
| ENSDARG00000061294 | ENSFM00250000002482 | Rho Gtpase Activating 35 Glucocorticoid Receptor Dna Binding Factor 1 |
| ENSDARG00000061304 | ENSFM00250000001450 | Neurabin 1 Neurabin I Neural Tissue Specific F Actin Binding I Phosphatase 1 Regulatory Subunit 9A |
| ENSDARG00000061338 | ENSFM00540000717952 | Atp Dependent Rna Helicase EC_3.6.4.13 |
| ENSDARG00000061397 | ENSFM00610000972403 | Unknown |
| ENSDARG00000061397 | ENSFM00760001714822 | E3 Ubiquitin Ligase TRIP12 EC_6.3.2.- Thyroid Receptor Interacting 12 Tr Interacting 12 Trip 12 |
| ENSDARG00000061398 | ENSFM00730001521622 | Band 4 1 |
| ENSDARG00000061421 | ENSFM00740001589247 | Serine/Threonine Kinase BRSK2 EC_2.7.11.1 EC_2.7.11.- 26 Brain Specific Serine/Threonine Kinase 2 Br Serine/Threonine Kinase 2 Serine/Threonine Kinase Sad A |
| ENSDARG00000061466 | ENSFM00730001521728 | Copine Copine |
| ENSDARG00000061473 | ENSFM00250000006948 | Tank Binding Kinase 1 Binding 1 TBK1 Binding 1 |
| ENSDARG00000061579 | ENSFM00800002003512 | Unconventional Myosin Ic Myosin I Beta Mmi Beta Mmib |
| ENSDARG00000061603 | ENSFM00610000973095 | Unknown |
| ENSDARG00000061603 | ENSFM00550000743175 | Sorbin And SH3 Domain Containing 2 Arg/Abl Interacting 2 ARGBP2 |
| ENSDARG00000061635 | ENSFM00750001632326 | Unconventional Myosin Va Dilute Myosin Heavy Chain Non Muscle |
| ENSDARG00000061638 | ENSFM00500000272373 | Dephospho Coa Kinase Domain Containing |
| ENSDARG00000061647 | ENSFM00740001589096 | Neurexin Precursor Neurexin |
| ENSDARG00000061660 | ENSFM00500000269758 | Carnitine O Palmitoyltransferase 1 Muscle CPT1 M EC_2.3.1.21 Carnitine O Palmitoyltransferase I Muscle Cpt I Cpti M Carnitine Palmitoyltransferase 1B |
| ENSDARG00000061688 | ENSFM00730001521847 | Catenin Delta 1 Cadherin Associated Src Substrate Cas P120 Catenin P120 Ctn P120 Cas |
| ENSDARG00000061692 | ENSFM00810002040729 | Nad P H Hydrate Epimerase |
| ENSDARG00000061699 | ENSFM00730001521477 | Signal Induced Proliferation Associated 1 SIPA1 |
| ENSDARG00000061718 | ENSFM00670001235606 | Zinc Finger |
| ENSDARG00000061719 | ENSFM00250000004568 | Wash Complex Subunit 7 |
| ENSDARG00000061736 | ENSFM00500000269628 | Ankyrin Ank Ankyrin |
| ENSDARG00000061741 | ENSFM00250000000547 | Inositol 1 4 5 Trisphosphate Receptor Type IP3 Receptor IP3R Ins Type Inositol 1 4 5 Trisphosphate Receptor Type INSP3 Receptor |
| ENSDARG00000061746 | ENSFM00250000001570 | Rho Guanine Nucleotide Exchange Factor 9 Collybistin Rac/CDC42 Guanine Nucleotide Exchange Factor 9 |
| ENSDARG00000061761 | ENSFM00600000921688 | Optic Atrophy 3 |
| ENSDARG00000061764 | ENSFM00740001589261 | Associated |
| ENSDARG00000061778 | ENSFM00250000001548 | Formin |
| ENSDARG00000061829 | ENSFM00250000000599 | Unc 13 Homolog MUNC13 |
| ENSDARG00000061862 | ENSFM00400000131775 | Unconventional Myosin Xviiia Molecule Associated With JAK3 N Terminus Majn Myosin Containing A Pdz Domain |
| ENSDARG00000061865 | ENSFM00730001521664 | Atp Dependent Rna Helicase EC_3.6.4.13 |
| ENSDARG00000062003 | ENSFM00750001632326 | Unconventional Myosin Va Dilute Myosin Heavy Chain Non Muscle |
| ENSDARG00000062008 | ENSFM00290000065564 | Heparan Sulfate 2 O Sulfotransferase 1 2 O Sulfotransferase 2OST EC_2.8.2.- |
| ENSDARG00000062016 | ENSFM00250000003232 | Vacuolar Sorting Associated 51 Homolog Fat Free Homolog |
| ENSDARG00000062045 | ENSFM00500000270404 | Interleukin 1 Receptor Accessory 1 Precursor Il 1 Rapl 1 Il 1RAPL 1 IL1RAPL 1 X Linked Interleukin 1 Receptor Accessory 1 |
| ENSDARG00000062054 | ENSFM00500000269758 | Carnitine O Palmitoyltransferase 1 Muscle CPT1 M EC_2.3.1.21 Carnitine O Palmitoyltransferase I Muscle Cpt I Cpti M Carnitine Palmitoyltransferase 1B |
| ENSDARG00000062086 | ENSFM00250000005157 | Ambiguous |
| ENSDARG00000062089 | ENSFM00800002003477 | Udp Glucose:Glycoprotein Glucosyltransferase 1 Precursor UGT1 EC_2.4.1.- Udp Glc:Glycoprotein Glucosyltransferase Udp Glucose Ceramide Glucosyltransferase 1 |
| ENSDARG00000062103 | ENSFM00800002003877 | Ankycorbin Ankyrin Repeat And Coiled Coil Structure Containing Retinal Pigment Epithelial Cell Retinoic Acid Induced 14 |
| ENSDARG00000062103 | ENSFM00800002003981 | Ankyrin Repeat Domain Containing 24 |
| ENSDARG00000062123 | ENSFM00250000000604 | Arf Gap With Gtpase Ank Repeat And Ph Domain Containing Agap Centaurin Gamma |
| ENSDARG00000062173 | ENSFM00600000921199 | Calmodulin Regulated Spectrin Associated |
| ENSDARG00000062174 | ENSFM00250000000524 | Voltage Dependent L Type Calcium Channel Subunit Beta Calcium Channel Voltage Dependent Subunit Beta |
| ENSDARG00000062220 | ENSFM00800002003882 | Pleckstrin Homology Domain Containing Family A Member 7 Ph Domain Containing Family A Member 7 Heart Adapter 1 |
| ENSDARG00000062237 | ENSFM00500000270207 | Cysteine Desulfurase Mitochondrial Precursor EC_2.8.1.7 |
| ENSDARG00000062305 | ENSFM00250000000629 | Regulating Synaptic Membrane Exocytosis Rab 3 Interacting Molecule Rim |
| ENSDARG00000062310 | ENSFM00810002040846 | Wd Repeat Containing 26 |
| ENSDARG00000062323 | ENSFM00410000138484 | Disintegrin And Metalloproteinase Domain Containing Precursor Adam Metalloproteinase Disintegrin And Cysteine Rich Mdc |
| ENSDARG00000062325 | ENSFM00250000000743 | SH3 And Multiple Ankyrin Repeat Domains Proline Rich Synapse Associated |
| ENSDARG00000062347 | ENSFM00500000271268 | Microtubule Associated Tumor Suppressor Candidate 2 Cardiac Zipper Microtubule Plus End Tracking TIP150 Tracking Of 150 Kda |
| ENSDARG00000062350 | ENSFM00500000294137 | Uncharacterized |
| ENSDARG00000062376 | ENSFM00380000124679 | Neuroligin Precursor |
| ENSDARG00000062396 | ENSFM00250000001010 | Ankyrin Repeat And Sterile Alpha Motif Domain Containing 1B Amyloid Beta Intracellular Domain Associated 1 Aida 1 E2A PBX1 Associated Eb 1 |
| ENSDARG00000062415 | ENSFM00730001521673 | Catenin Delta 2 |
| ENSDARG00000062447 | ENSFM00730001522025 | E3 Ubiquitin Ligase EC_6.3.2.- Hect C2 And Ww Domain Containing NEDD4 E3 Ubiquitin Ligase |
| ENSDARG00000062477 | ENSFM00250000005157 | Ambiguous |
| ENSDARG00000062511 | ENSFM00250000007244 | Golgin Subfamily A Member 3 Golgin 160 |
| ENSDARG00000062521 | ENSFM00810002040637 | Probable Phospholipid Transporting Atpase EC_3.6.3.1 Atpase Class Ii Type |
| ENSDARG00000062558 | ENSFM00250000004877 | Ring Finger And Spry Domain Containing 1 Precursor |
| ENSDARG00000062561 | ENSFM00750001653065 | Unknown |
| ENSDARG00000062561 | ENSFM00730001522023 | Uncharacterized Aarf Domain Containing Kinase EC_2.7.11.- |
| ENSDARG00000062590 | ENSFM00740001589112 | Microtubule Actin Cross Linking Factor 1 Actin Cross Linking Family 7 |
| ENSDARG00000062590 | ENSFM00740001589199 | Plectin Pcn Pltn Plectin 1 |
| ENSDARG00000062612 | ENSFM00440000236925 | Atp Dependent Rna Helicase EC_3.6.4.13 |
| ENSDARG00000062667 | ENSFM00760001714665 | Maguk P55 Subfamily Member |
| ENSDARG00000062693 | ENSFM00740001589096 | Neurexin Precursor Neurexin |
| ENSDARG00000062696 | ENSFM00610000967131 | Unknown |
| ENSDARG00000062696 | ENSFM00730001521692 | Diacylglycerol Kinase Dag Kinase EC_2.7.1.107 Kda Diacylglycerol Kinase Diglyceride Kinase Dgk |
| ENSDARG00000062780 | ENSFM00800002003733 | Apoptosis Inducing Factor 3 Ec 1 Apoptosis Inducing Factor |
| ENSDARG00000062794 | ENSFM00400000131833 | Tripartite Motif Containing 46 Gene Y Geney Tripartite Fibronectin Type Iii And C Terminal Spry Motif |
| ENSDARG00000062799 | ENSFM00250000000906 | Brain Specific Angiogenesis Inhibitor 1 Associated 2 Bai Associated 2 BAI1 Associated 2 Insulin Receptor Substrate Of 53 Kda IRSP53 Insulin Receptor Substrate P53 |
| ENSDARG00000062823 | ENSFM00760001714819 | Phosphatidylinositol 4 Kinase Alpha PI4 Kinase Alpha PI4K Alpha Ptdins 4 Kinase Alpha EC_2.7.1.67 |
| ENSDARG00000062854 | ENSFM00260000050437 | Sidekick Precursor |
| ENSDARG00000062948 | ENSFM00250000001269 | Wiskott Aldrich Syndrome Family Member 1 Wasp Family Member 1 Wave 1 |
| ENSDARG00000062951 | ENSFM00250000004834 | Tld Domain Containing 1 Tbc/Lysm Associated Domain Containing 1 |
| ENSDARG00000062956 | ENSFM00250000001790 | SN1 Specific Diacylglycerol Lipase Alpha Dgl Alpha EC_3.1.1.- Neural Stem Cell Derived Dendrite Regulator |
| ENSDARG00000062991 | ENSFM00250000000845 | Abl Interactor 1 Abelson Interactor 1 Abi 1 EPS8 SH3 Domain Binding EPS8 Binding Spectrin SH3 Domain Binding 1 |
| ENSDARG00000062997 | ENSFM00250000000550 | Hyaluronidase Precursor EC_3.2.1.35 Hyaluronoglucosaminidase |
| ENSDARG00000063008 | ENSFM00600000921514 | Ambiguous |
| ENSDARG00000063026 | ENSFM00250000007537 | Membrane MLC1 |
| ENSDARG00000063036 | ENSFM00250000001069 | Ph And SEC7 Domain Containing 1 Exchange Factor ARF6 Pleckstrin Homology And SEC7 Domain Containing 1 |
| ENSDARG00000063040 | ENSFM00250000001718 | Ankyrin Repeat And Btb/Poz Domain Containing BTBD11 Btb/Poz Domain Containing 11 |
| ENSDARG00000063054 | ENSFM00250000000743 | SH3 And Multiple Ankyrin Repeat Domains Proline Rich Synapse Associated |
| ENSDARG00000063068 | ENSFM00250000000542 | Nesprin 2 Nuclear Envelope Spectrin Repeat 2 Nucleus And Actin Connecting Element Protein Nuance Synaptic Nuclear Envelope 2 Syne 2 |
| ENSDARG00000063079 | ENSFM00250000000492 | Argonaute |
| ENSDARG00000063101 | ENSFM00500000270074 | Alpha Mannosidase 2 EC_3.2.1.114 Golgi Alpha Mannosidase Ii Aman Ii Man Ii Mannosidase Alpha Class 2A Member 1 Mannosyl Oligosaccharide 1 3 1 6 Alpha Mannosidase |
| ENSDARG00000063145 | ENSFM00250000006597 | Transcription Factor A Mitochondrial Precursor Mttfa |
| ENSDARG00000063149 | ENSFM00800002003912 | Transmembrane And Tpr Repeat Containing 1 |
| ENSDARG00000063150 | ENSFM00740001589096 | Neurexin Precursor Neurexin |
| ENSDARG00000063180 | ENSFM00760001714721 | Dedicator Of Cytokinesis 3 Modifier Of Cell Adhesion Presenilin Binding Pbp |
| ENSDARG00000063207 | ENSFM00410000138465 | Receptor Tyrosine Kinase Erbb Precursor EC_2.7.10.1 Proto Oncogene C Erbb |
| ENSDARG00000063211 | ENSFM00250000004839 | Exocyst Complex Component 8 Exocyst Complex 84 Kda Subunit |
| ENSDARG00000063253 | ENSFM00730001522025 | E3 Ubiquitin Ligase EC_6.3.2.- Hect C2 And Ww Domain Containing NEDD4 E3 Ubiquitin Ligase |
| ENSDARG00000063255 | ENSFM00250000001718 | Ankyrin Repeat And Btb/Poz Domain Containing BTBD11 Btb/Poz Domain Containing 11 |
| ENSDARG00000063264 | ENSFM00760001714657 | Protocadherin 11 X Linked Precursor Protocadherin 11 Protocadherin On The X Chromosome Pcdh X |
| ENSDARG00000063295 | ENSFM00270000056424 | Myosin Cellular Myosin Heavy Chain Type Myosin Heavy Chain Myosin Heavy Chain Non Muscle Non Muscle Myosin Heavy Chain Nmmhc Non Muscle Myosin Heavy Chain Nmmhc Ii Nmmhc |
| ENSDARG00000063299 | ENSFM00650001146246 | Unknown |
| ENSDARG00000063299 | ENSFM00600000921250 | Piccolo Aczonin |
| ENSDARG00000063332 | ENSFM00250000000743 | SH3 And Multiple Ankyrin Repeat Domains Proline Rich Synapse Associated |
| ENSDARG00000063352 | ENSFM00250000001432 | Type Inositol 3 4 Bisphosphate 4 Phosphatase EC_3.1.3.66 Inositol Polyphosphate 4 Phosphatase Type |
| ENSDARG00000063358 | ENSFM00260000050684 | Calcium Uptake 1 Mitochondrial Calcium Binding Atopy Related Autoantigen 1 Homolog |
| ENSDARG00000063361 | ENSFM00600000921514 | Ambiguous |
| ENSDARG00000063411 | ENSFM00250000009938 | Leucine Rich Repeat Containing 73 |
| ENSDARG00000063414 | ENSFM00250000007556 | Transmembrane 5 |
| ENSDARG00000063419 | ENSFM00800002009474 | Unknown |
| ENSDARG00000063419 | ENSFM00730001522013 | Probable Atp Dependent Rna Helicase DHX35 EC_3.6.4.13 Deah Box 35 |
| ENSDARG00000063527 | ENSFM00250000001494 | Engulfment And Cell Motility |
| ENSDARG00000063568 | ENSFM00800002014767 | Unknown |
| ENSDARG00000063568 | ENSFM00740001589375 | Synaptotagmin 7 Synaptotagmin Vii Sytvii |
| ENSDARG00000063570 | ENSFM00730001521969 | Dual Specificity Tyrosine Phosphorylation Regulated Kinase Dual Specificity YAK1 Related Kinase Minibrain |
| ENSDARG00000063578 | ENSFM00730001521936 | Diacylglycerol Kinase Zeta Dag Kinase Zeta EC_2.7.1.107 Diglyceride Kinase Zeta Dgk Zeta |
| ENSDARG00000063677 | ENSFM00250000001383 | Cyclin Y |
| ENSDARG00000063711 | ENSFM00750001632380 | Tripartite Motif Containing 2 EC_6.3.2.- E3 Ubiquitin Ligase TRIM2 |
| ENSDARG00000063713 | ENSFM00250000000610 | Ras Gtpase Activating |
| ENSDARG00000063899 | ENSFM00420000140506 | Nadh Ubiquinone Oxidoreductase Chain 2 EC_1.6.5.3 Nadh Dehydrogenase Subunit 2 |
| ENSDARG00000063911 | ENSFM00300000082445 | Atp Synthase Subunit A F Atpase 6 |
| ENSDARG00000067507 | ENSFM00260000050545 | Btb/Poz Domain Containing |
| ENSDARG00000067591 | ENSFM00250000002171 | Transcriptional Activator Pur Beta Purine Rich Element Binding B |
| ENSDARG00000067815 | ENSFM00250000001884 | Leucine Rich Repeat And Immunoglobulin Domain Containing Nogo Receptor Interacting Precursor Leucine Rich Repeat Neuronal |
| ENSDARG00000068166 | ENSFM00250000001412 | Whirlin |
| ENSDARG00000068289 | ENSFM00250000006253 | Eukaryotic Translation Initiation Factor 3 Subunit K |
| ENSDARG00000068305 | ENSFM00500000274118 | 28S Ribosomal S14 Mitochondrial Mrp S14 S14MT |
| ENSDARG00000068323 | ENSFM00250000001299 | Astrotactin Precursor |
| ENSDARG00000068480 | ENSFM00250000005199 | MAP7 Domain Containing 2 |
| ENSDARG00000068516 | ENSFM00610000973362 | Unknown |
| ENSDARG00000068516 | ENSFM00730001521886 | Hyaluronan And Proteoglycan Link 1 Precursor Cartilage Linking 1 Cartilage Link Proteoglycan Link |
| ENSDARG00000068624 | ENSFM00740001589095 | Spectrin Beta Chain Erythrocytic Beta Spectrin |
| ENSDARG00000068745 | ENSFM00780001898729 | Microtubule Associated |
| ENSDARG00000068822 | ENSFM00250000002171 | Transcriptional Activator Pur Beta Purine Rich Element Binding B |
| ENSDARG00000068981 | ENSFM00250000004289 | D Glucuronyl C5 Epimerase EC_5.1.3.17 Heparan Sulfate C5 Epimerase Hsepi Heparin/Heparan Sulfate:Glucuronic Acid C5 Epimerase Heparosan N Sulfate Glucuronate 5 Epimerase |
| ENSDARG00000068989 | ENSFM00800002003385 | Gamma Aminobutyric Acid Receptor Subunit Alpha Precursor Gaba A Receptor Subunit Alpha |
| ENSDARG00000068992 | ENSFM00730001521103 | Heat Shock Kda Heat Shock 70 Kda |
| ENSDARG00000069026 | ENSFM00730001521247 | Adenylate Cyclase Type EC_4.6.1.1 Atp Pyrophosphate Lyase Adenylate Cyclase Type Adenylyl Cyclase Ca 2+ Adenylyl Cyclase |
| ENSDARG00000069046 | ENSFM00730001521138 | Keratin Type I Cytoskeletal Cytokeratin Ck Keratin |
| ENSDARG00000069090 | ENSFM00800002003469 | V Type Proton Atpase Subunit D V Atpase Subunit D Vacuolar Proton Pump Subunit D |
| ENSDARG00000069099 | ENSFM00250000005494 | Transmembrane And Coiled Coil 1 |
| ENSDARG00000069113 | ENSFM00480000262790 | Drebrin Developmentally Regulated Brain |
| ENSDARG00000069118 | ENSFM00760001714676 | Serine/Threonine Phosphatase 2A 56 Kda Regulatory Subunit Epsilon PP2A B Subunit B' Epsilon PP2A B Subunit B56 Epsilon PP2A B Subunit PR61 Epsilon PP2A B Subunit R5 Epsilon |
| ENSDARG00000069150 | ENSFM00730001522318 | Gtp Binding RIT1 Ras Expressed In Many Tissues Ras Without Caax 1 |
| ENSDARG00000069269 | ENSFM00250000002834 | Wd Repeat Containing 35 |
| ENSDARG00000069313 | ENSFM00500000271370 | Mitochondrial Inner Membrane OXA1L Precursor Oxidase Assembly 1 OXA1 |
| ENSDARG00000069318 | ENSFM00250000001283 | EFR3 Homolog |
| ENSDARG00000069324 | ENSFM00800002003735 | Vacuolar Sorting Associated 53 Homolog |
| ENSDARG00000069360 | ENSFM00500000269882 | Calcineurin Subunit B Regulatory Subunit Phosphatase Regulatory Subunit |
| ENSDARG00000069402 | ENSFM00260000050662 | Leucine Rich Repeat Containing Precursor Netrin Ligand Ngl |
| ENSDARG00000069420 | ENSFM00730001522472 | E3 Ubiquitin Ligase TRIM23 EC_6.3.2.- Adp Ribosylation Factor Domain Containing 1 Gtp Binding Ard 1 Tripartite Motif Containing 23 |
| ENSDARG00000069427 | ENSFM00250000002642 | Sodium/Potassium Transporting Atpase Subunit Beta 1 Interacting Na + /K + Transporting Atpase Subunit Beta 1 Interacting |
| ENSDARG00000069467 | ENSFM00250000001385 | Turtle Homolog A Precursor Dendrite Arborization And Synapse Maturation 1 Immunoglobulin Superfamily Member 9A IGSF9A |
| ENSDARG00000069484 | ENSFM00250000000610 | Ras Gtpase Activating |
| ENSDARG00000069494 | ENSFM00500000295586 | Transmembrane 223 |
| ENSDARG00000069529 | ENSFM00670001235889 | Dual Specificity Phosphatase EC_3.1.3.16 EC_3.1.3.- 48 |
| ENSDARG00000069600 | ENSFM00250000001269 | Wiskott Aldrich Syndrome Family Member 1 Wasp Family Member 1 Wave 1 |
| ENSDARG00000069603 | ENSFM00250000000892 | Serine/Threonine Kinase EC_2.7.11.1 Thousand And One Amino Acid |
| ENSDARG00000069608 | ENSFM00740001589417 | A Kinase Anchor 2 Akap 2 Akap Kl Kinase A Anchoring 2 PRKA2 |
| ENSDARG00000069615 | ENSFM00800002003437 | Creatine Kinase U Type Mitochondrial Precursor EC_2.7.3.2 Acidic Type Mitochondrial Creatine Kinase Mia Ck Ubiquitous Mitochondrial Creatine Kinase U Mtck |
| ENSDARG00000069649 | ENSFM00500000289607 | Transmembrane 160 |
| ENSDARG00000069742 | ENSFM00730001522203 | Cullin 5 Cul 5 Vasopressin Activated Calcium Mobilizing Receptor 1 Vacm 1 |
| ENSDARG00000069765 | ENSFM00250000000610 | Ras Gtpase Activating |
| ENSDARG00000069774 | ENSFM00780001898447 | Flotillin 2 |
| ENSDARG00000069843 | ENSFM00260000050545 | Btb/Poz Domain Containing |
| ENSDARG00000069878 | ENSFM00670001235480 | Unknown |
| ENSDARG00000069951 | ENSFM00800002003364 | Elongation Factor 1 Alpha Ef 1 Alpha Eukaryotic Elongation Factor 1 A EEF1A |
| ENSDARG00000069970 | ENSFM00250000001884 | Leucine Rich Repeat And Immunoglobulin Domain Containing Nogo Receptor Interacting Precursor Leucine Rich Repeat Neuronal |
| ENSDARG00000070029 | ENSFM00760001715025 | Peroxisomal Bifunctional Enzyme Pbe Pbfe [Includes Enoyl Coa Hydratase/3 2 Trans Enoyl Coa Isomerase EC_4.2.1.17 EC_5.3.3.- 8 |
| ENSDARG00000070043 | ENSFM00250000003932 | Aspartate Trna Ligase Cytoplasmic EC_6.1.1.12 Aspartyl Trna Synthetase Asprs |
| ENSDARG00000070100 | ENSFM00740001589251 | Ras And Rab Interactor Ras Interaction/Interference |
| ENSDARG00000070145 | ENSFM00250000000552 | Membrane Associated Guanylate Kinase Ww And Pdz Domain Containing Membrane Associated Guanylate Kinase Inverted Magi |
| ENSDARG00000070173 | ENSFM00720001492020 | Glutamate Receptor Precursor Glur Ampa Selective Glutamate Receptor Glur Glutamate Receptor Ionotropic Ampa |
| ENSDARG00000070393 | ENSFM00250000002643 | Coenzyme Q Binding COQ10 Homolog Mitochondrial Precursor |
| ENSDARG00000070435 | ENSFM00250000001879 | Vesicle Associated Membrane Associated A Vamp A Vamp Associated A Vap A |
| ENSDARG00000070437 | ENSFM00600000921476 | 60S Ribosomal L22 |
| ENSDARG00000070441 | ENSFM00500000270112 | Palmitoyltransferase EC_2.3.1.225 Huntingtin Interacting 14 Zinc Finger Dhhc Domain Containing Dhhc |
| ENSDARG00000070442 | ENSFM00730001522534 | Semaphorin 7A Precursor Semaphorin K1 Sema K1 Semaphorin L Sema L CD108 Antigen |
| ENSDARG00000070448 | ENSFM00730001521304 | G Coupled Receptor Kinase G Coupled Receptor Kinase |
| ENSDARG00000070543 | ENSFM00730001521500 | Glutamate Receptor Ionotropic Nmda Precursor Glutamate [Nmda] Receptor Subunit Epsilon N Methyl D Aspartate Receptor Subtype |
| ENSDARG00000070575 | ENSFM00500000271560 | Unknown |
| ENSDARG00000070603 | ENSFM00250000005151 | Unknown |
| ENSDARG00000070620 | ENSFM00730001521500 | Glutamate Receptor Ionotropic Nmda Precursor Glutamate [Nmda] Receptor Subunit Epsilon N Methyl D Aspartate Receptor Subtype |
| ENSDARG00000070626 | ENSFM00250000002019 | Voltage Dependent Calcium Channel Gamma Subunit Neuronal Voltage Gated Calcium Channel Gamma Subunit Transmembrane Ampar Regulatory Gamma Tarp Gamma |
| ENSDARG00000070717 | ENSFM00500000270170 | Mitochondrial Glutamate Carrier 1 Gc 1 Glutamate/H + Symporter 1 Solute Carrier Family 25 Member 22 |
| ENSDARG00000070787 | ENSFM00250000001012 | Catenin Beta Beta Catenin |
| ENSDARG00000070824 | ENSFM00250000005807 | Nadh Dehydrogenase [Ubiquinone] 1 Beta Subcomplex Subunit 5 Mitochondrial Precursor Complex I Sgdh Ci Sgdh Nadh Ubiquinone Oxidoreductase Sgdh Subunit |
| ENSDARG00000070849 | ENSFM00250000003189 | 40S Ribosomal S15 |
| ENSDARG00000070919 | ENSFM00730001521728 | Copine Copine |
| ENSDARG00000070952 | ENSFM00500000271560 | Unknown |
| ENSDARG00000071014 | ENSFM00560000773950 | S100U |
| ENSDARG00000071017 | ENSFM00250000001233 | 5' Nucleotidase Precursor 5' Nt EC_3.1.3.5 Ecto 5' Nucleotidase |
| ENSDARG00000071031 | ENSFM00730001521918 | Sorting Nexin |
| ENSDARG00000071192 | ENSFM00760001714761 | Gap Junction Alpha 1 Connexin 43 CX43 |
| ENSDARG00000071230 | ENSFM00260000050443 | Leucine Rich Repeat And Fibronectin Type Iii Domain Containing Precursor Synaptic Adhesion Molecule |
| ENSDARG00000071331 | ENSFM00250000000332 | Ryanodine Receptor Ryr Muscle Calcium Release Channel Muscle Type Ryanodine Receptor Type Ryanodine Receptor |
| ENSDARG00000071353 | ENSFM00500000311464 | Uncharacterized |
| ENSDARG00000071357 | ENSFM00310000089004 | Mitogen Activated Kinase Kinase Kinase Kinase EC_2.7.11.1 Kinase Mapk/Erk Kinase Kinase Kinase Mek Kinase Kinase Mekkk |
| ENSDARG00000071395 | ENSFM00250000000111 | Calcium/Calmodulin Dependent Kinase Type Ii Subunit Cam Kinase Ii Subunit Camk Ii Subunit EC_2.7.11.17 |
| ENSDARG00000071493 | ENSFM00320000100099 | Noelin Precursor Neuronal Olfactomedin Related Er Localized Olfactomedin 1 |
| ENSDARG00000071551 | ENSFM00250000004190 | X Ray Repair Cross Complementing 6 EC_3.6.4.- EC_4.2.-.- 99 5' Deoxyribose 5 Phosphate Lyase KU70 5' Lyase KU70 Atp Dependent Dna Helicase 2 Subunit 1 Atp Dependent Dna Helicase Ii 70 Kda Subunit Ctc Box Binding Factor 75 Kda Subunit CTC75 Ctcbf Dna Repai |
| ENSDARG00000071566 | ENSFM00420000140518 | Serine/Threonine Phosphatase PP1 Catalytic Subunit Pp EC_3.1.3.16 |
| ENSDARG00000071573 | ENSFM00610000959757 | Unknown |
| ENSDARG00000071573 | ENSFM00670001236563 | U6 Snrna Associated Sm LSM5 |
| ENSDARG00000071594 | ENSFM00250000006057 | Mitochondrial Import Inner Membrane Translocase Subunit TIM21 Precursor TIM21 Protein Mitochondrial |
| ENSDARG00000071709 | ENSFM00250000001450 | Neurabin 1 Neurabin I Neural Tissue Specific F Actin Binding I Phosphatase 1 Regulatory Subunit 9A |
| ENSDARG00000071774 | ENSFM00250000008520 | Eukaryotic Translation Elongation Factor 1 Epsilon 1 Elongation Factor P18 Multisynthase Complex Auxiliary Component P18 |
| ENSDARG00000071906 | ENSFM00750001632389 | Transport SEC24A SEC24 Related A |
| ENSDARG00000073707 | ENSFM00500000270149 | Connector Enhancer Of Kinase Suppressor Of Ras Connector Enhancer Of Ksr Cnk Homolog |
| ENSDARG00000073713 | ENSFM00260000050653 | Netrin Precursor Laminet |
| ENSDARG00000073724 | ENSFM00800002003951 | Nlr Family Member X1 Precursor |
| ENSDARG00000073732 | ENSFM00270000056424 | Myosin Cellular Myosin Heavy Chain Type Myosin Heavy Chain Myosin Heavy Chain Non Muscle Non Muscle Myosin Heavy Chain Nmmhc Non Muscle Myosin Heavy Chain Nmmhc Ii Nmmhc |
| ENSDARG00000073752 | ENSFM00610000969046 | Unknown |
| ENSDARG00000073769 | ENSFM00250000000552 | Membrane Associated Guanylate Kinase Ww And Pdz Domain Containing Membrane Associated Guanylate Kinase Inverted Magi |
| ENSDARG00000073792 | ENSFM00780001898478 | Endoplasmic Reticulum Mannosyl Oligosaccharide 1 2 Alpha Mannosidase EC_3.2.1.113 Er Alpha 1 2 Mannosidase Er Mannosidase 1 ERMAN1 MAN9GLCNAC2 Specific Processing Alpha Mannosidase Mannosidase Alpha Class 1B Member 1 |
| ENSDARG00000073822 | ENSFM00250000001463 | Iq Motif And SEC7 Domain Containing |
| ENSDARG00000073824 | ENSFM00250000001613 | Ras Specific Guanine Nucleotide Releasing Factor 2 Ras GRF2 Ras Guanine Nucleotide Exchange Factor 2 |
| ENSDARG00000073841 | ENSFM00740001589186 | E3 Ubiquitin Ligase HERC2 EC_6.3.2.- Hect Domain And RCC1 Domain Containing 2 |
| ENSDARG00000073843 | ENSFM00510000502753 | Unconventional Myosin Unconventional Myosin |
| ENSDARG00000073872 | ENSFM00250000002945 | Tyrosine Sulfotransferase EC_2.8.2.20 Tyrosylprotein Sulfotransferase Tpst |
| ENSDARG00000073896 | ENSFM00730001521860 | Gap Junction Gamma 1 Connexin 45 CX45 Gap Junction Alpha 7 |
| ENSDARG00000074059 | ENSFM00360000109935 | Disks Large Homolog 5 Discs Large P Dlg Placenta And Prostate Dlg |
| ENSDARG00000074073 | ENSFM00250000004498 | Microtubule Associated 6 Map 6 Stable Tubule Only Polypeptide Stop |
| ENSDARG00000074149 | ENSFM00250000000547 | Inositol 1 4 5 Trisphosphate Receptor Type IP3 Receptor IP3R Ins Type Inositol 1 4 5 Trisphosphate Receptor Type INSP3 Receptor |
| ENSDARG00000074153 | ENSFM00250000001398 | Slit And Ntrk Precursor |
| ENSDARG00000074242 | ENSFM00500000270904 | Plasminogen Activator Inhibitor 1 Rna Binding PAI1 Rna Binding 1 Pai RBP1 SERPINE1 Binding 1 |
| ENSDARG00000074255 | ENSFM00260000050554 | Calcium Uptake 2 Mitochondrial Precursor Ef Hand Domain Containing Family Member A1 |
| ENSDARG00000074275 | ENSFM00250000003304 | Lim And Calponin Homology Domains Containing 1 |
| ENSDARG00000074289 | ENSFM00800002017874 | Unknown |
| ENSDARG00000074289 | ENSFM00250000000217 | Tubulin Alpha Chain |
| ENSDARG00000074308 | ENSFM00250000005430 | Leucine Rich Repeat Containing Leucine Rich Repeat Containing |
| ENSDARG00000074320 | ENSFM00780001898357 | Voltage Gated Potassium Channel Subunit Beta K + Channel Subunit Beta Kv Beta |
| ENSDARG00000074329 | ENSFM00730001521847 | Catenin Delta 1 Cadherin Associated Src Substrate Cas P120 Catenin P120 Ctn P120 Cas |
| ENSDARG00000074367 | ENSFM00760001714668 | Ubiquitin Carboxyl Terminal Hydrolase 12 EC_3.4.19.12 Deubiquitinating Enzyme 12 Ubiquitin Thioesterase 12 Ubiquitin Specific Processing Protease 12 |
| ENSDARG00000074372 | ENSFM00250000002984 | Phosphatase 1 Regulatory Subunit 29 Precursor Extracellular Leucine Rich Repeat And Fibronectin Type Iii Domain Containing 2 Leucine Rich Repeat And Fibronectin Type Iii Domain Containing 6 Leucine Rich Repeat Containing 62 |
| ENSDARG00000074376 | ENSFM00250000002191 | Mam Domain Containing Glycosylphosphatidylinositol Anchor 2 Precursor Mam Domain Containing 1 |
| ENSDARG00000074381 | ENSFM00270000056477 | Ferm Rhogef And Pleckstrin Domain Containing |
| ENSDARG00000074480 | ENSFM00500000270149 | Connector Enhancer Of Kinase Suppressor Of Ras Connector Enhancer Of Ksr Cnk Homolog |
| ENSDARG00000074490 | ENSFM00250000003320 | CASC4 Cancer Susceptibility Candidate Gene 4 |
| ENSDARG00000074521 | ENSFM00250000004498 | Microtubule Associated 6 Map 6 Stable Tubule Only Polypeptide Stop |
| ENSDARG00000074524 | ENSFM00250000000380 | Contactin Associated 5 Precursor Cell Recognition Molecule CASPR5 |
| ENSDARG00000074533 | ENSFM00500000271856 | Solute Carrier Family 25 Member 38 |
| ENSDARG00000074558 | ENSFM00250000000380 | Contactin Associated 5 Precursor Cell Recognition Molecule CASPR5 |
| ENSDARG00000074581 | ENSFM00250000000681 | Adducin |
| ENSDARG00000074611 | ENSFM00250000004113 | Wd Repeat Containing 37 |
| ENSDARG00000074680 | ENSFM00610000972522 | Unknown |
| ENSDARG00000074680 | ENSFM00250000000629 | Regulating Synaptic Membrane Exocytosis Rab 3 Interacting Molecule Rim |
| ENSDARG00000074723 | ENSFM00720001492050 | Pleckstrin Homology Domain Containing Family H Member 3 Precursor Ph Domain Containing Family H Member 3 |
| ENSDARG00000074727 | ENSFM00250000002411 | Dnaj Homolog Subfamily C Member 10 Precursor EC_1.8.4.- Endoplasmic Reticulum Dna J Domain Containing 5 Er Resident ERDJ5 ERDJ5 |
| ENSDARG00000074746 | ENSFM00300000079128 | Potassium Voltage Gated Channel Subfamily D Member Voltage Gated Potassium Channel Subunit KV4 |
| ENSDARG00000074791 | ENSFM00250000000672 | Aldehyde Dehydrogenase Aldehyde Dehydrogenase Aldehyde Dehydrogenase Family 3 Member |
| ENSDARG00000074865 | ENSFM00250000001492 | Ferm And Pdz Domain Containing 4 Pdz Domain Containing 10 Psd 95 Interacting Regulator Of Spine Morphogenesis Preso |
| ENSDARG00000074876 | ENSFM00800002003396 | Atp Binding Cassette Sub Family D Member Adrenoleukodystrophy |
| ENSDARG00000074899 | ENSFM00250000000892 | Serine/Threonine Kinase EC_2.7.11.1 Thousand And One Amino Acid |
| ENSDARG00000075007 | ENSFM00500000270226 | Carbohydrate Sulfotransferase EC_2.8.2.- Galactose/N Acetylglucosamine/N Acetylglucosamine 6 O Sulfotransferase N Acetylglucosamine 6 O Sulfotransferase GLCNAC6ST Sulfotransferase N Acetylglucosamine 6 O Sulfotransferase GLCNAC6ST GN6ST |
| ENSDARG00000075041 | ENSFM00350000105407 | Formin |
| ENSDARG00000075083 | ENSFM00250000002645 | Dna Dependent Kinase Catalytic Subunit Dna Pk Catalytic Subunit Dna Pkcs EC_2.7.11.1 |
| ENSDARG00000075100 | ENSFM00800002020697 | Unknown |
| ENSDARG00000075100 | ENSFM00260000050557 | Netrin Receptor Dcc Precursor Tumor Suppressor Dcc |
| ENSDARG00000075110 | ENSFM00250000000610 | Ras Gtpase Activating |
| ENSDARG00000075133 | ENSFM00250000000737 | Brain Specific Angiogenesis Inhibitor Precursor |
| ENSDARG00000075141 | ENSFM00670001235445 | G Coupled Receptor Family C Group 5 Member Retinoic Acid Induced Gene Raig |
| ENSDARG00000075159 | ENSFM00250000000343 | Precursor |
| ENSDARG00000075192 | ENSFM00800002003559 | Atp Dependent Zinc Metalloprotease Ym EC_3.4.24.- Atp Dependent Metalloprotease FTSH1 Meg 4 YME1 1 |
| ENSDARG00000075245 | ENSFM00250000005384 | Wd Repeat Containing 11 Bromodomain And Wd Repeat Containing 2 |
| ENSDARG00000075327 | ENSFM00500000269738 | Dedicator Of Cytokinesis CDC42 Guanine Nucleotide Exchange Factor Zizimin |
| ENSDARG00000075362 | ENSFM00250000001412 | Whirlin |
| ENSDARG00000075455 | ENSFM00250000001168 | Soga Family Member |
| ENSDARG00000075491 | ENSFM00500000270149 | Connector Enhancer Of Kinase Suppressor Of Ras Connector Enhancer Of Ksr Cnk Homolog |
| ENSDARG00000075539 | ENSFM00250000002727 | Syntabulin Golgi Localized Syntaphilin Related Syntaxin 1 Binding |
| ENSDARG00000075556 | ENSFM00800002003954 | Leucine Zipper 1 Leucine Zipper Motif Containing |
| ENSDARG00000075580 | ENSFM00800002003728 | Leucine Rich Repeat And Calponin Homology Domain Containing 1 Calponin Homology Domain Containing 1 |
| ENSDARG00000075673 | ENSFM00250000001623 | Rho Gtpase Activating 21 Rho Type Gtpase Activating 21 |
| ENSDARG00000075677 | ENSFM00500000272736 | 39S Ribosomal L17 Mitochondrial Precursor L17MT Mrp L17 |
| ENSDARG00000075709 | ENSFM00540000734587 | Uncharacterized |
| ENSDARG00000075733 | ENSFM00730001521785 | Zyxin |
| ENSDARG00000075752 | ENSFM00400000131775 | Unconventional Myosin Xviiia Molecule Associated With JAK3 N Terminus Majn Myosin Containing A Pdz Domain |
| ENSDARG00000075758 | ENSFM00730001521154 | Unknown |
| ENSDARG00000075793 | ENSFM00610000970147 | Unknown |
| ENSDARG00000075793 | ENSFM00250000001999 | Phosphatidylinositol 3 4 5 Trisphosphate Dependent Rac Exchanger 2 P REX2 Ptdins 3 4 5 Dependent Rac Exchanger 2 Dep Domain Containing 2 |
| ENSDARG00000075806 | ENSFM00250000000717 | Kin Of Irre Precursor Kin Of Irregular Chiasm Nephrin |
| ENSDARG00000075830 | ENSFM00720001492041 | Synaptotagmin Synaptotagmin |
| ENSDARG00000075899 | ENSFM00260000050328 | Latrophilin |
| ENSDARG00000075924 | ENSFM00250000000899 | Rap Guanine Nucleotide Exchange Factor Exchange Factor Directly Activated By Camp Exchange Directly Activated By Camp Epac Camp Regulated Guanine Nucleotide Exchange Factor Camp |
| ENSDARG00000075949 | ENSFM00250000000930 | Serine/Threonine Kinase EC_2.7.11.13 |
| ENSDARG00000075989 | ENSFM00800002003702 | Actin Related 2/3 Complex Subunit 2 ARP2/3 Complex 34 Kda Subunit P34 Arc |
| ENSDARG00000076014 | ENSFM00250000000753 | Elks/RAB6 Interacting/Cast Family Member 1 Erc 1 RAB6 Interacting 2 |
| ENSDARG00000076030 | ENSFM00250000000524 | Voltage Dependent L Type Calcium Channel Subunit Beta Calcium Channel Voltage Dependent Subunit Beta |
| ENSDARG00000076070 | ENSFM00250000000718 | Disks Large Associated Dap Psd 95/SAP90 Binding SAP90/Psd 95 Associated |
| ENSDARG00000076103 | ENSFM00500000273079 | Delphilin Glutamate Receptor Ionotropic Delta 2 Interacting 1 |
| ENSDARG00000076128 | ENSFM00730001521681 | Camp Dependent Kinase Type I Alpha Regulatory Camp Dependent Kinase Type I Alpha Regulatory Subunit N Terminally Processed] |
| ENSDARG00000076155 | ENSFM00250000005018 | Poly A Rna Polymerase Mitochondrial Precursor Pap EC_2.7.7.19 Pap Associated Domain Containing 1 Polynucleotide Adenylyltransferase |
| ENSDARG00000076174 | ENSFM00750001632380 | Tripartite Motif Containing 2 EC_6.3.2.- E3 Ubiquitin Ligase TRIM2 |
| ENSDARG00000076213 | ENSFM00760001714721 | Dedicator Of Cytokinesis 3 Modifier Of Cell Adhesion Presenilin Binding Pbp |
| ENSDARG00000076268 | ENSFM00500000276193 | Uncharacterized |
| ENSDARG00000076302 | ENSFM00500000270006 | E3 Ubiquitin Ligase EC_6.3.2.- Deltex |
| ENSDARG00000076310 | ENSFM00250000006718 | F Box Only 41 |
| ENSDARG00000076334 | ENSFM00250000003611 | 39S Ribosomal L43 Mitochondrial Precursor L43MT Mrp L43 Mitochondrial Ribosomal BMRP36A |
| ENSDARG00000076381 | ENSFM00730001522086 | Tricarboxylate Transport Protein Mitochondrial Precursor Citrate Transport Ctp Solute Carrier Family 25 Member 1 Tricarboxylate Carrier |
| ENSDARG00000076393 | ENSFM00290000065595 | Transmembrane 65 |
| ENSDARG00000076401 | ENSFM00250000002019 | Voltage Dependent Calcium Channel Gamma Subunit Neuronal Voltage Gated Calcium Channel Gamma Subunit Transmembrane Ampar Regulatory Gamma Tarp Gamma |
| ENSDARG00000076508 | ENSFM00250000000328 | Metabotropic Glutamate Receptor Precursor |
| ENSDARG00000076673 | ENSFM00740001589199 | Plectin Pcn Pltn Plectin 1 |
| ENSDARG00000076697 | ENSFM00730001521477 | Signal Induced Proliferation Associated 1 SIPA1 |
| ENSDARG00000076701 | ENSFM00250000002796 | Beta 1 4 N Acetylgalactosaminyltransferase 3 BETA4GALNAC T3 BETA4GALNACT3 EC_2.4.1.244 Beta 1 4 N Acetylgalactosaminyltransferase Iii N Acetyl Beta Glucosaminyl Glycoprotein 4 Beta N Acetylgalactosaminyltransferase 2 Ngalnac T2 |
| ENSDARG00000076702 | ENSFM00250000001069 | Ph And SEC7 Domain Containing 1 Exchange Factor ARF6 Pleckstrin Homology And SEC7 Domain Containing 1 |
| ENSDARG00000076724 | ENSFM00760001714819 | Phosphatidylinositol 4 Kinase Alpha PI4 Kinase Alpha PI4K Alpha Ptdins 4 Kinase Alpha EC_2.7.1.67 |
| ENSDARG00000076742 | ENSFM00250000001040 | Cytohesin Ph SEC7 And Coiled Coil Domain Containing |
| ENSDARG00000076796 | ENSFM00250000000275 | Disks Large Homolog Synapse Associated Sap |
| ENSDARG00000076815 | ENSFM00800002003567 | Eukaryotic Translation Initiation Factor 3 Subunit A |
| ENSDARG00000076829 | ENSFM00400000131980 | Ankyrin Repeat And Ibr Domain Containing 1 |
| ENSDARG00000076873 | ENSFM00500000276193 | Uncharacterized |
| ENSDARG00000076889 | ENSFM00250000000944 | Ral Gtpase Activating Subunit Alpha 1 Gap Related Interacting Partner To E12 Gripe Gtpase Activating Domain 1 Tuberin 1 P240 |
| ENSDARG00000076923 | ENSFM00250000003884 | Kazrin |
| ENSDARG00000076970 | ENSFM00500000272331 | Dehydrogenase/Reductase Sdr Family Member 12 EC_1.1.-.- |
| ENSDARG00000077023 | ENSFM00780001898335 | Protocadherin Precursor |
| ENSDARG00000077047 | ENSFM00260000050522 | Receptor Type Tyrosine Phosphatase Precursor R Ptp |
| ENSDARG00000077053 | ENSFM00250000000441 | Liprin Alpha Tyrosine Phosphatase Receptor Type F Polypeptide Interacting Alpha Ptprf Interacting Alpha |
| ENSDARG00000077080 | ENSFM00670001235445 | G Coupled Receptor Family C Group 5 Member Retinoic Acid Induced Gene Raig |
| ENSDARG00000077124 | ENSFM00740001589247 | Serine/Threonine Kinase BRSK2 EC_2.7.11.1 EC_2.7.11.- 26 Brain Specific Serine/Threonine Kinase 2 Br Serine/Threonine Kinase 2 Serine/Threonine Kinase Sad A |
| ENSDARG00000077134 | ENSFM00250000001603 | Probable G Coupled Receptor Precursor |
| ENSDARG00000077145 | ENSFM00730001521247 | Adenylate Cyclase Type EC_4.6.1.1 Atp Pyrophosphate Lyase Adenylate Cyclase Type Adenylyl Cyclase Ca 2+ Adenylyl Cyclase |
| ENSDARG00000077174 | ENSFM00250000001659 | Sulfhydryl Oxidase Precursor EC_1.8.3.2 Quiescin Q6 |
| ENSDARG00000077228 | ENSFM00730001521357 | Nt 3 Growth Factor Receptor Precursor EC_2.7.10.1 Neurotrophic Tyrosine Kinase Receptor Type 3 |
| ENSDARG00000077291 | ENSFM00250000000709 | 40S Ribosomal S2 |
| ENSDARG00000077329 | ENSFM00380000124679 | Neuroligin Precursor |
| ENSDARG00000077339 | ENSFM00500000273926 | Peptidyl Trna Hydrolase 2 Mitochondrial Precursor Pth 2 EC_3.1.1.29 Bcl 2 Inhibitor Of Transcription |
| ENSDARG00000077343 | ENSFM00780001898335 | Protocadherin Precursor |
| ENSDARG00000077349 | ENSFM00250000000793 | VPS10 Domain Containing Receptor Precursor |
| ENSDARG00000077367 | ENSFM00260000050653 | Netrin Precursor Laminet |
| ENSDARG00000077368 | ENSFM00810002048698 | Unknown |
| ENSDARG00000077368 | ENSFM00250000004892 | Zinc Transporter 6 Znt 6 Solute Carrier Family 30 Member 6 |
| ENSDARG00000077385 | ENSFM00480000262761 | Regulator Of G Signaling |
| ENSDARG00000077506 | ENSFM00750001659004 | Unknown |
| ENSDARG00000077506 | ENSFM00800002009548 | Unknown |
| ENSDARG00000077506 | ENSFM00250000000691 | Tight Junction Zo Tight Junction Zona Occludens Zonula Occludens |
| ENSDARG00000077515 | ENSFM00740001589271 | LETM1 And Ef Hand Domain Containing 1 Mitochondrial Precursor Leucine Zipper Ef Hand Containing Transmembrane 1 |
| ENSDARG00000077545 | ENSFM00740001589247 | Serine/Threonine Kinase BRSK2 EC_2.7.11.1 EC_2.7.11.- 26 Brain Specific Serine/Threonine Kinase 2 Br Serine/Threonine Kinase 2 Serine/Threonine Kinase Sad A |
| ENSDARG00000077560 | ENSFM00730001521500 | Glutamate Receptor Ionotropic Nmda Precursor Glutamate [Nmda] Receptor Subunit Epsilon N Methyl D Aspartate Receptor Subtype |
| ENSDARG00000077582 | ENSFM00500000269628 | Ankyrin Ank Ankyrin |
| ENSDARG00000077609 | ENSFM00250000002984 | Phosphatase 1 Regulatory Subunit 29 Precursor Extracellular Leucine Rich Repeat And Fibronectin Type Iii Domain Containing 2 Leucine Rich Repeat And Fibronectin Type Iii Domain Containing 6 Leucine Rich Repeat Containing 62 |
| ENSDARG00000077633 | ENSFM00250000000552 | Membrane Associated Guanylate Kinase Ww And Pdz Domain Containing Membrane Associated Guanylate Kinase Inverted Magi |
| ENSDARG00000077654 | ENSFM00250000000328 | Metabotropic Glutamate Receptor Precursor |
| ENSDARG00000077709 | ENSFM00250000001463 | Iq Motif And SEC7 Domain Containing |
| ENSDARG00000077732 | ENSFM00500000272450 | Tho Complex Subunit 4 THO4 Aly/Ref Export Factor |
| ENSDARG00000077765 | ENSFM00730001521860 | Gap Junction Gamma 1 Connexin 45 CX45 Gap Junction Alpha 7 |
| ENSDARG00000077776 | ENSFM00800002003472 | Casein Kinase Ii Subunit Beta Ck Ii Beta |
| ENSDARG00000077847 | ENSFM00320000100099 | Noelin Precursor Neuronal Olfactomedin Related Er Localized Olfactomedin 1 |
| ENSDARG00000077946 | ENSFM00250000001546 | Swi/Snf Complex Subunit SMARCC2 BRG1 Associated Factor 170 BAF170 Swi/Snf Complex 170 Kda Subunit Swi/Snf Related Matrix Associated Actin Dependent Regulator Of Chromatin Subfamily C Member 2 |
| ENSDARG00000077996 | ENSFM00800002003474 | Cadherin Precursor |
| ENSDARG00000078016 | ENSFM00610000968796 | Unknown |
| ENSDARG00000078016 | ENSFM00500000270006 | E3 Ubiquitin Ligase EC_6.3.2.- Deltex |
| ENSDARG00000078060 | ENSFM00740001589375 | Synaptotagmin 7 Synaptotagmin Vii Sytvii |
| ENSDARG00000078102 | ENSFM00250000001069 | Ph And SEC7 Domain Containing 1 Exchange Factor ARF6 Pleckstrin Homology And SEC7 Domain Containing 1 |
| ENSDARG00000078125 | ENSFM00800002004549 | Unknown |
| ENSDARG00000078136 | ENSFM00250000002280 | Wd Repeat Containing 47 Neuronal Enriched Map Interacting Nemitin |
| ENSDARG00000078139 | ENSFM00250000003219 | Dcc Interacting 13 DIP13 Adapter Containing Ph Domain Ptb Domain And Leucine Zipper Motif |
| ENSDARG00000078141 | ENSFM00250000002984 | Phosphatase 1 Regulatory Subunit 29 Precursor Extracellular Leucine Rich Repeat And Fibronectin Type Iii Domain Containing 2 Leucine Rich Repeat And Fibronectin Type Iii Domain Containing 6 Leucine Rich Repeat Containing 62 |
| ENSDARG00000078149 | ENSFM00730001521500 | Glutamate Receptor Ionotropic Nmda Precursor Glutamate [Nmda] Receptor Subunit Epsilon N Methyl D Aspartate Receptor Subtype |
| ENSDARG00000078169 | ENSFM00730001522093 | Voltage Dependent Calcium Channel Subunit Alpha 2/Delta 1 Precursor Voltage Gated Calcium Channel Subunit Alpha 2/Delta 1 [Contains Voltage Dependent Calcium Channel Subunit Alpha 2 1 |
| ENSDARG00000078216 | ENSFM00250000001830 | Epidermal Growth Factor Receptor Substrate 15 1 EPS15 Related EPS15R |
| ENSDARG00000078233 | ENSFM00800002024108 | Unknown |
| ENSDARG00000078233 | ENSFM00730001521847 | Catenin Delta 1 Cadherin Associated Src Substrate Cas P120 Catenin P120 Ctn P120 Cas |
| ENSDARG00000078302 | ENSFM00250000001886 | Bmp/Retinoic Acid Inducible Neural Specific 1 Precursor Deleted In Bladder Cancer 1 |
| ENSDARG00000078317 | ENSFM00500000271927 | Dual Specificity Phosphatase 10 EC_3.1.3.16 EC_3.1.3.- 48 Mitogen Activated Kinase Phosphatase 5 Map Kinase Phosphatase 5 Mkp 5 |
| ENSDARG00000078322 | ENSFM00740001589200 | Collagen Alpha 1 Chain |
| ENSDARG00000078327 | ENSFM00740001589275 | Unknown |
| ENSDARG00000078362 | ENSFM00740001589132 | Tenascin Precursor Tn |
| ENSDARG00000078485 | ENSFM00250000002727 | Syntabulin Golgi Localized Syntaphilin Related Syntaxin 1 Binding |
| ENSDARG00000078529 | ENSFM00250000000737 | Brain Specific Angiogenesis Inhibitor Precursor |
| ENSDARG00000078599 | ENSFM00260000050554 | Calcium Uptake 2 Mitochondrial Precursor Ef Hand Domain Containing Family Member A1 |
| ENSDARG00000078603 | ENSFM00800002003512 | Unconventional Myosin Ic Myosin I Beta Mmi Beta Mmib |
| ENSDARG00000078624 | ENSFM00250000001570 | Rho Guanine Nucleotide Exchange Factor 9 Collybistin Rac/CDC42 Guanine Nucleotide Exchange Factor 9 |
| ENSDARG00000078675 | ENSFM00500000269778 | Dedicator Of Cytokinesis |
| ENSDARG00000078688 | ENSFM00250000003196 | Fibroblast Growth Factor Receptor Substrate 3 Fgfr Substrate 3 Fgfr Signaling Adaptor SNT2 SUC1 Associated Neurotrophic Factor Target 2 Snt 2 |
| ENSDARG00000078703 | ENSFM00500000270363 | Aspartyl/Asparaginyl Beta Hydroxylase EC_1.14.11.16 |
| ENSDARG00000078734 | ENSFM00780001898391 | Unconventional Myosin Ie Unconventional Myosin 1E |
| ENSDARG00000078745 | ENSFM00250000002364 | Calcium Independent Phospholipase A2 Gamma EC_3.1.1.5 Intracellular Membrane Associated Calcium Independent Phospholipase A2 Gamma IPLA2 Gamma Patatin Phospholipase Domain Containing 8 |
| ENSDARG00000078759 | ENSFM00250000002526 | Dna Replication Atp Dependent Helicase/Nuclease DNA2 Dna Replication Atp Dependent Helicase Homolog [Includes Dna Replication Nuclease DNA2 EC_3.1.-.- |
| ENSDARG00000078760 | ENSFM00730001521792 | Voltage Dependent Calcium Channel Subunit Alpha 2/Delta 3 Precursor Voltage Gated Calcium Channel Subunit Alpha 2/Delta 3 [Contains Voltage Dependent Calcium Channel Subunit Alpha 2 3 |
| ENSDARG00000078770 | ENSFM00250000004371 | Dihydroxyacetone Phosphate Acyltransferase Dap At Dhap At EC_2.3.1.42 Acyl Coa:Dihydroxyacetonephosphateacyltransferase Glycerone Phosphate O Acyltransferase |
| ENSDARG00000078839 | ENSFM00400000131776 | Leucine Rich Repeat Transmembrane Neuronal Precursor |
| ENSDARG00000078854 | ENSFM00250000002565 | Magnesium Transporter 1 Precursor MAGT1 |
| ENSDARG00000078891 | ENSFM00250000006401 | Uncharacterized |
| ENSDARG00000078901 | ENSFM00250000001010 | Ankyrin Repeat And Sterile Alpha Motif Domain Containing 1B Amyloid Beta Intracellular Domain Associated 1 Aida 1 E2A PBX1 Associated Eb 1 |
| ENSDARG00000078902 | ENSFM00250000000629 | Regulating Synaptic Membrane Exocytosis Rab 3 Interacting Molecule Rim |
| ENSDARG00000078908 | ENSFM00250000006718 | F Box Only 41 |
| ENSDARG00000078998 | ENSFM00250000002406 | Lipoma Hmgic Fusion Partner |
| ENSDARG00000079004 | ENSFM00250000001143 | Tetratricopeptide Repeat Ankyrin Repeat And Coiled Coil Domain Containing |
| ENSDARG00000079097 | ENSFM00250000001143 | Tetratricopeptide Repeat Ankyrin Repeat And Coiled Coil Domain Containing |
| ENSDARG00000079161 | ENSFM00600000921250 | Piccolo Aczonin |
| ENSDARG00000079202 | ENSFM00250000001234 | Poly [Adp Ribose] Polymerase 1 Parp 1 EC_2.4.2.30 Adp Ribosyltransferase Diphtheria Toxin 1 ARTD1 Nad + Adp Ribosyltransferase 1 Adprt 1 Poly[Adp Ribose] Synthase 1 |
| ENSDARG00000079204 | ENSFM00410000138484 | Disintegrin And Metalloproteinase Domain Containing Precursor Adam Metalloproteinase Disintegrin And Cysteine Rich Mdc |
| ENSDARG00000079249 | ENSFM00750001632363 | 2 Oxoglutarate Dehydrogenase Mitochondrial Precursor 2 Oxoglutarate Dehydrogenase Complex Component E1 Ogdc E1 Alpha Ketoglutarate Dehydrogenase |
| ENSDARG00000079251 | ENSFM00380000124679 | Neuroligin Precursor |
| ENSDARG00000079295 | ENSFM00740001589100 | Voltage Dependent Type Calcium Channel Subunit Alpha Brain Calcium Channel Calcium Channel L Type Alpha 1 Polypeptide Voltage Gated Calcium Channel Subunit Alpha CAV2 |
| ENSDARG00000079348 | ENSFM00730001521500 | Glutamate Receptor Ionotropic Nmda Precursor Glutamate [Nmda] Receptor Subunit Epsilon N Methyl D Aspartate Receptor Subtype |
| ENSDARG00000079366 | ENSFM00250000001450 | Neurabin 1 Neurabin I Neural Tissue Specific F Actin Binding I Phosphatase 1 Regulatory Subunit 9A |
| ENSDARG00000079374 | ENSFM00250000000691 | Tight Junction Zo Tight Junction Zona Occludens Zonula Occludens |
| ENSDARG00000079378 | ENSFM00600000921202 | Pleckstrin Homology Domain Family B Member LL5 |
| ENSDARG00000079388 | ENSFM00760001714908 | Agrin N Terminal 110 Kda Subunit |
| ENSDARG00000079396 | ENSFM00260000050443 | Leucine Rich Repeat And Fibronectin Type Iii Domain Containing Precursor Synaptic Adhesion Molecule |
| ENSDARG00000079438 | ENSFM00780001898562 | Plakophilin 3 |
| ENSDARG00000079440 | ENSFM00730001521970 | Coronin |
| ENSDARG00000079475 | ENSFM00250000006300 | Nck Interacting With SH3 Domain 54 Kda Vaca Interacting Kda Interacting 90 Kda SH3 Interacting With Nck SH3 Adapter SPIN90 Wasp Interacting SH3 Domain Wish Wiskott Aldrich Syndrome |
| ENSDARG00000079491 | ENSFM00290000065519 | Potassium Voltage Gated Channel Subfamily A Member Voltage Gated Potassium Channel Subunit KV1 |
| ENSDARG00000079556 | ENSFM00500000273557 | C2 Calcium Dependent Domain Containing 4C Nuclear Localized Factor 3 FAM148C |
| ENSDARG00000079586 | ENSFM00400000131718 | Gamma Aminobutyric Acid Receptor Subunit Beta Precursor Gaba A Receptor Subunit Beta |
| ENSDARG00000079610 | ENSFM00250000001853 | A Kinase Anchor 9 Akap 9 A Kinase Anchor Kda Akap 120 Kinase A Anchoring 9 PRKA9 |
| ENSDARG00000079611 | ENSFM00480000262746 | Semaphorin Precursor |
| ENSDARG00000079645 | ENSFM00800002004636 | Cdna Cssl: |
| ENSDARG00000079665 | ENSFM00250000001603 | Probable G Coupled Receptor Precursor |
| ENSDARG00000079670 | ENSFM00730001522141 | Leucine Rich Repeat Containing 7 Densin 180 Densin |
| ENSDARG00000079671 | ENSFM00500000270520 | Lipid Phosphate Phosphatase Related Type 4 EC_3.1.3.4 Brain Specific Phosphatidic Acid Phosphatase 1 Plasticity Related Gene 1 Prg 1 |
| ENSDARG00000079725 | ENSFM00800002003717 | Atp Dependent Rna Helicase A EC_3.6.4.13 Deah Box 9 Nuclear Dna Helicase Ii Ndh Ii |
| ENSDARG00000079780 | ENSFM00250000001411 | Heterogeneous Nuclear Ribonucleoprotein U Scaffold Attachment Factor Saf |
| ENSDARG00000079781 | ENSFM00250000001398 | Slit And Ntrk Precursor |
| ENSDARG00000079805 | ENSFM00500000270015 | Transgelin |
| ENSDARG00000079823 | ENSFM00540000732848 | Unknown |
| ENSDARG00000079824 | ENSFM00740001597625 | Ninjurin 2 |
| ENSDARG00000079840 | ENSFM00250000000903 | Calcium Activated Potassium Channel Subunit Alpha 1 Bk Channel Bkca Alpha Calcium Activated Potassium Channel Subfamily M Subunit Alpha 1 K Vca Alpha KCA1 1 Maxi K Channel Maxik Slo Alpha SLO1 Slowpoke Homolog Slo Homolog |
| ENSDARG00000079872 | ENSFM00250000000899 | Rap Guanine Nucleotide Exchange Factor Exchange Factor Directly Activated By Camp Exchange Directly Activated By Camp Epac Camp Regulated Guanine Nucleotide Exchange Factor Camp |
| ENSDARG00000079881 | ENSFM00250000005995 | T Cell Activation Inhibitor Mitochondrial Tolerance Associated Gene 1 Toag 1 |
| ENSDARG00000079949 | ENSFM00250000002938 | Fact Complex Subunit SPT16 Facilitates Chromatin Transcription Complex Subunit SPT16 |
| ENSDARG00000080000 | ENSFM00730001522086 | Tricarboxylate Transport Protein Mitochondrial Precursor Citrate Transport Ctp Solute Carrier Family 25 Member 1 Tricarboxylate Carrier |
| ENSDARG00000086104 | ENSFM00610000955325 | Unknown |
| ENSDARG00000086104 | ENSFM00730001521392 | Solute Carrier Organic Anion Transporter Family Member Organic Anion Transporting Polypeptide Organic Anion Solute Carrier Family 21 Member |
| ENSDARG00000086207 | ENSFM00730001521500 | Glutamate Receptor Ionotropic Nmda Precursor Glutamate [Nmda] Receptor Subunit Epsilon N Methyl D Aspartate Receptor Subtype |
| ENSDARG00000086573 | ENSFM00250000008621 | Oligodendrocyte Myelin Glycoprotein Precursor |
| ENSDARG00000086756 | ENSFM00500000274428 | Regulator Of G Signaling 9 Binding RGS9 |
| ENSDARG00000086790 | ENSFM00730001521369 | Guanylate Cyclase Soluble Subunit Beta 1 Gcs Beta 1 EC_4.6.1.2 Guanylate Cyclase Soluble Subunit Beta 3 Gcs Beta 3 Soluble Guanylate Cyclase Small Subunit |
| ENSDARG00000086848 | ENSFM00250000002488 | Atpase Family Aaa Domain Containing 3 |
| ENSDARG00000086969 | ENSFM00780001898335 | Protocadherin Precursor |
| ENSDARG00000086990 | ENSFM00800002009421 | Unknown |
| ENSDARG00000086990 | ENSFM00260000050454 | Leucine Rich Repeat Containing |
| ENSDARG00000087229 | ENSFM00250000003124 | Exocyst Complex Component 7 Exocyst Complex Component EXO70 |
| ENSDARG00000087247 | ENSFM00780001898357 | Voltage Gated Potassium Channel Subunit Beta K + Channel Subunit Beta Kv Beta |
| ENSDARG00000087346 | ENSFM00730001521249 | Ras Related Rap 1 |
| ENSDARG00000087364 | ENSFM00600000944558 | Unknown |
| ENSDARG00000087402 | ENSFM00250000000220 | Tropomyosin Alpha 1 Chain Alpha Tropomyosin Tropomyosin 1 |
| ENSDARG00000087456 | ENSFM00600000941833 | Unknown |
| ENSDARG00000087538 | ENSFM00800002003514 | Kinesin KIF3A Microtubule Plus End Directed Kinesin Motor 3A |
| ENSDARG00000087573 | ENSFM00250000007111 | Histone Deacetylase 11 HD11 EC_3.5.1.98 |
| ENSDARG00000087616 | ENSFM00750001658357 | Unknown |
| ENSDARG00000087616 | ENSFM00800002006620 | Unknown |
| ENSDARG00000087616 | ENSFM00260000050441 | Microtubule Associated Tau Neurofibrillary Tangle Paired Helical Filament Tau Phf Tau |
| ENSDARG00000087647 | ENSFM00760001714685 | Septin 7 CDC10 Homolog |
| ENSDARG00000087704 | ENSFM00500000269996 | Gdnf Family Receptor Alpha Precursor Gdnf Receptor Alpha Gdnfr Alpha Gfr Alpha Tgf Beta Related Neurotrophic Factor Receptor |
| ENSDARG00000087822 | ENSFM00250000000604 | Arf Gap With Gtpase Ank Repeat And Ph Domain Containing Agap Centaurin Gamma |
| ENSDARG00000087843 | ENSFM00800002003683 | Contactin 1 Precursor Neural Cell Surface F3 |
| ENSDARG00000087897 | ENSFM00400000131810 | Core Histone Macro H2A 1 Histone Macro M H2A Y H2A/Y |
| ENSDARG00000087897 | ENSFM00550000756898 | Histone H2A |
| ENSDARG00000087916 | ENSFM00610000971995 | Unknown |
| ENSDARG00000087916 | ENSFM00500000314906 | Transmembrane 240 |
| ENSDARG00000088030 | ENSFM00670001273033 | Unknown |
| ENSDARG00000088030 | ENSFM00670001273034 | Unknown |
| ENSDARG00000088030 | ENSFM00500000271015 | 60S Ribosomal L35A |
| ENSDARG00000088073 | ENSFM00500000272914 | Unknown |
| ENSDARG00000088100 | ENSFM00410000138464 | Kv Channel Interacting A Type Potassium Channel Modulatory Potassium Channel Interacting |
| ENSDARG00000088117 | ENSFM00250000004295 | Alpha 1 6 Fucosyltransferase ALPHA1 6FUCT EC_2.4.1.68 Gdp L Fuc:N Acetyl Beta D Glucosaminide ALPHA1 6 Fucosyltransferase Gdp Fucose Glycoprotein Fucosyltransferase Glycoprotein 6 Alpha L Fucosyltransferase |
| ENSDARG00000088136 | ENSFM00780001898335 | Protocadherin Precursor |
| ENSDARG00000088168 | ENSFM00250000000459 | Actin Binding Lim Ablim Actin Binding Lim Family Member |
| ENSDARG00000088276 | ENSFM00500000269608 | Histone H2B |
| ENSDARG00000088293 | ENSFM00260000050490 | Double C2 Domain Containing DOC2 |
| ENSDARG00000088383 | ENSFM00500000272199 | Cytochrome C Oxidase Subunit 5A Mitochondrial Precursor Cytochrome C Oxidase Polypeptide Va |
| ENSDARG00000088634 | ENSFM00730001521247 | Adenylate Cyclase Type EC_4.6.1.1 Atp Pyrophosphate Lyase Adenylate Cyclase Type Adenylyl Cyclase Ca 2+ Adenylyl Cyclase |
| ENSDARG00000088825 | ENSFM00520000517879 | Citron Rho Interacting Kinase EC_2.7.11.1 Serine/Threonine Kinase 21 |
| ENSDARG00000088882 | ENSFM00600000941712 | Unknown |
| ENSDARG00000088899 | ENSFM00250000002523 | Ral Gtpase Activating Subunit Beta P170 |
| ENSDARG00000089066 | ENSFM00400000132091 | Nhs 2 |
| ENSDARG00000089076 | ENSFM00790001963049 | Molybdopterin Adenylyltransferase Mpt Adenylyltransferase EC_2.7.7.75 Domain G |
| ENSDARG00000089190 | ENSFM00250000001143 | Tetratricopeptide Repeat Ankyrin Repeat And Coiled Coil Domain Containing |
| ENSDARG00000089255 | ENSFM00600000940244 | Unknown |
| ENSDARG00000089292 | ENSFM00260000050328 | Latrophilin |
| ENSDARG00000089314 | ENSFM00260000050441 | Microtubule Associated Tau Neurofibrillary Tangle Paired Helical Filament Tau Phf Tau |
| ENSDARG00000089413 | ENSFM00600000939910 | Unknown |
| ENSDARG00000089413 | ENSFM00630001069159 | Unknown |
| ENSDARG00000089413 | ENSFM00630001071995 | Unknown |
| ENSDARG00000089418 | ENSFM00600000944882 | Unknown |
| ENSDARG00000089444 | ENSFM00250000008621 | Oligodendrocyte Myelin Glycoprotein Precursor |
| ENSDARG00000089562 | ENSFM00780001898335 | Protocadherin Precursor |
| ENSDARG00000089856 | ENSFM00520000517879 | Citron Rho Interacting Kinase EC_2.7.11.1 Serine/Threonine Kinase 21 |
| ENSDARG00000089957 | ENSFM00250000002312 | Leucine Rich Repeat Lgi Family Member Precursor Leucine Rich Glioma Inactivated |
| ENSDARG00000090035 | ENSFM00800002023314 | Unknown |
| ENSDARG00000090035 | ENSFM00250000002242 | Reticulon 4 Receptor Precursor Nogo Receptor Nogo 66 Receptor Homolog Nogo 66 Receptor Related |
| ENSDARG00000090036 | ENSFM00250000007281 | Rho Related Btb Domain Containing 3 EC_3.6.1.- |
| ENSDARG00000090062 | ENSFM00730001521186 | Ras Related C3 Botulinum Toxin Substrate Precursor P21 |
| ENSDARG00000090145 | ENSFM00500000314906 | Transmembrane 240 |
| ENSDARG00000090149 | ENSFM00500000269649 | Histone H4 |
| ENSDARG00000090462 | ENSFM00500000273357 | 39S Ribosomal L20 Mitochondrial Precursor L20MT Mrp L20 |
| ENSDARG00000090585 | ENSFM00500000269807 | Glypican Secreted Glypican |
| ENSDARG00000090615 | ENSFM00600000940243 | Unknown |
| ENSDARG00000090654 | ENSFM00250000001678 | Tyrosine Phosphatase Auxilin EC_3.1.3.48 Dnaj Homolog Subfamily C Member 6 |
| ENSDARG00000090815 | ENSFM00730001521400 | Atp Sensitive Inward Rectifier Potassium Channel Atp Potassium Channel K Inward Rectifier K + Channel 1 Potassium Channel Inwardly Rectifying Subfamily J Member |
| ENSDARG00000090883 | ENSFM00800002003385 | Gamma Aminobutyric Acid Receptor Subunit Alpha Precursor Gaba A Receptor Subunit Alpha |
| ENSDARG00000091042 | ENSFM00740001589315 | Sacsin Dnaj Homolog Subfamily C Member 29 DNAJC29 |
| ENSDARG00000091138 | ENSFM00800002003421 | Actin Related 3 Actin 3 |
| ENSDARG00000091293 | ENSFM00250000001467 | Dmx 1 X 1 |
| ENSDARG00000091457 | ENSFM00250000001269 | Wiskott Aldrich Syndrome Family Member 1 Wasp Family Member 1 Wave 1 |
| ENSDARG00000091525 | ENSFM00250000000259 | Histone H3 |
| ENSDARG00000091777 | ENSFM00250000005977 | Target Of Rapamycin Complex 2 Subunit MAPKAP1 TORC2 Subunit MAPKAP1 Mitogen Activated Kinase 2 Associated 1 Stress Activated Map Kinase Interacting 1 Sapk Interacting 1 |
| ENSDARG00000092126 | ENSFM00250000000813 | Vitellogenin Precursor [Contains Lipovitellin I Lvi |
| ENSDARG00000092376 | ENSFM00250000000718 | Disks Large Associated Dap Psd 95/SAP90 Binding SAP90/Psd 95 Associated |
| ENSDARG00000092517 | ENSFM00610000964217 | Unknown |
| ENSDARG00000092553 | ENSFM00250000000816 | Adp/Atp Translocase Adp Atp Carrier Adenine Nucleotide Translocator Ant Solute Carrier Family 25 Member [Contains Adp/Atp Translocase N Terminally Processed] |
| ENSDARG00000092624 | ENSFM00740001589095 | Spectrin Beta Chain Erythrocytic Beta Spectrin |
| ENSDARG00000092644 | ENSFM00250000000492 | Argonaute |
| ENSDARG00000092746 | ENSFM00610000964941 | Unknown |
| ENSDARG00000092873 | ENSFM00710001464695 | Unknown |
| ENSDARG00000092948 | ENSFM00730001521471 | Guanine Nucleotide Binding G Q Subunit Alpha Guanine Nucleotide Binding Alpha Q |
| ENSDARG00000093091 | ENSFM00250000001463 | Iq Motif And SEC7 Domain Containing |
| ENSDARG00000093195 | ENSFM00500000269608 | Histone H2B |
| ENSDARG00000093382 | ENSFM00610000969748 | Unknown |
| ENSDARG00000093382 | ENSFM00610000973342 | Unknown |
| ENSDARG00000093382 | ENSFM00610000973343 | Unknown |
| ENSDARG00000093413 | ENSFM00500000270078 | Lactadherin Precursor Mfgm Milk Fat Globule Egf Factor 8 Mfg E8 SED1 |
| ENSDARG00000094526 | ENSFM00450000242166 | Kinase Suppressor Of Ras |
| ENSDARG00000094590 | ENSFM00250000002423 | C2 Domain Containing 5.138 Kda C2 Domain Containing Phosphoprotein |
| ENSDARG00000094695 | ENSFM00750001641915 | Unknown |
| ENSDARG00000095005 | ENSFM00500000270056 | Uncharacterized |
| ENSDARG00000095034 | ENSFM00250000004260 | Uncharacterized |
| ENSDARG00000095170 | ENSFM00500000270615 | Leucine Rich Repeat Flightless Interacting Lrr Flii Interacting |
| ENSDARG00000095459 | ENSFM00610000964147 | Unknown |
| ENSDARG00000095603 | ENSFM00500000273079 | Delphilin Glutamate Receptor Ionotropic Delta 2 Interacting 1 |
| ENSDARG00000096249 | ENSFM00670001282088 | Unknown |
| ENSDARG00000096249 | ENSFM00730001521549 | Ap 1 Complex Subunit Mu 1 Ap Mu Chain Family Member MU1A Adaptor Complex Ap 1 Subunit Mu 1 Adaptor Related Complex 1 Subunit Mu 1 Clathrin Assembly Complex 1 Mu 1 Medium Chain 1 Golgi Adaptor HA1/AP1 Adaptin Mu 1 Subunit Mu Adaptin 1 MU1A Adaptin |
| ENSDARG00000096355 | ENSFM00560000771464 | Ig Mu Chain C Region Membrane Bound Form |
| ENSDARG00000096368 | ENSFM00250000002795 | Sialidase 3 EC_3.2.1.18 Ganglioside Sialidase Membrane Sialidase N Acetyl Alpha Neuraminidase 3 |
| ENSDARG00000096454 | ENSFM00730001521549 | Ap 1 Complex Subunit Mu 1 Ap Mu Chain Family Member MU1A Adaptor Complex Ap 1 Subunit Mu 1 Adaptor Related Complex 1 Subunit Mu 1 Clathrin Assembly Complex 1 Mu 1 Medium Chain 1 Golgi Adaptor HA1/AP1 Adaptin Mu 1 Subunit Mu Adaptin 1 MU1A Adaptin |
| ENSDARG00000096687 | ENSFM00730001521808 | Regulator Of G Signaling |
| ENSDARG00000096867 | ENSFM00500000271703 | Calmin Calponin Transmembrane Domain |
| ENSDARG00000097170 | ENSFM00730001521583 | Protocadherin Alpha Precursor Pcdh Alpha |
| ENSDARG00000097170 | ENSFM00780001898335 | Protocadherin Precursor |
| ENSDARG00000097245 | ENSFM00250000001168 | Soga Family Member |
| ENSDARG00000097351 | ENSFM00740001589123 | Filamin Fln Abp 280 Actin Binding Filamin |
| ENSDARG00000097528 | ENSFM00750001638793 | Unknown |
| ENSDARG00000097572 | ENSFM00310000088999 | Receptor Type Tyrosine Phosphatase Precursor R Ptp EC_3.1.3.48 Receptor Type Tyrosine Phosphatase |
| ENSDARG00000097660 | ENSFM00500000271336 | Trafficking Particle Complex Subunit 2 |
| ENSDARG00000097855 | ENSFM00550000764011 | Small Membrane A Kinase Anchor Small Membrane Akap Smakap |
| ENSDARG00000097897 | ENSFM00600000921280 | Fch Domain Only |
| ENSDARG00000098057 | ENSFM00800002020364 | Unknown |
| ENSDARG00000098057 | ENSFM00810002040701 | Down Syndrome Cell Adhesion Molecule Precursor |
| ENSDARG00000098118 | ENSFM00730001522435 | Trafficking Particle Complex Subunit 10 Trafficking Particle Complex Subunit TMEM1 Transport Particle Subunit TMEM1 Trapp Subunit TMEM1 |
| ENSDARG00000098161 | ENSFM00390000126303 | Contactin Precursor Brain Derived Immunoglobulin Superfamily Big |
| ENSDARG00000098290 | ENSFM00470000251461 | Unc 80 |
| ENSDARG00000098367 | ENSFM00250000002862 | Signal Recognition Particle 54 Kda SRP54 |
| ENSDARG00000098374 | ENSFM00740001589123 | Filamin Fln Abp 280 Actin Binding Filamin |
| ENSDARG00000098417 | ENSFM00250000000771 | Synapsin Synapsin |
| ENSDARG00000098458 | ENSFM00670001236020 | 60S Ribosomal L37A |
| ENSDARG00000098486 | ENSFM00250000004428 | Toll Interacting |
| ENSDARG00000098584 | ENSFM00800002013985 | Unknown |
| ENSDARG00000098584 | ENSFM00800002013986 | Unknown |
| ENSDARG00000098591 | ENSFM00250000000193 | Tubulin Beta Chain |
| ENSDARG00000098599 | ENSFM00250000001467 | Dmx 1 X 1 |
| ENSDARG00000098639 | ENSFM00500000269896 | Pyrroline 5 Carboxylate Reductase 3 P5C Reductase 3 P5CR 3 EC_1.5.1.2 Pyrroline 5 Carboxylate Reductase |
| ENSDARG00000098654 | ENSFM00390000126378 | Raftlin Raft Linking |
| ENSDARG00000098751 | ENSFM00650001140033 | Leucine Zipper Tumor Suppressor 2 |
| ENSDARG00000098753 | ENSFM00250000001494 | Engulfment And Cell Motility |
| ENSDARG00000098782 | ENSFM00730001521465 | Choline O Acetyltransferase Choactase Chat Choline Acetylase EC_2.3.1.6 |
| ENSDARG00000098831 | ENSFM00380000124741 | Glutaryl Coa Dehydrogenase Mitochondrial Precursor Gcd EC_1.3.8.6 |
| ENSDARG00000098860 | ENSFM00250000003740 | FAM57B |
| ENSDARG00000098874 | ENSFM00250000001326 | Exocyst Complex Component 6 Exocyst Complex Component SEC15A SEC15 1 |
| ENSDARG00000098880 | ENSFM00800002018422 | Unknown |
| ENSDARG00000098880 | ENSFM00600000921250 | Piccolo Aczonin |
| ENSDARG00000099096 | ENSFM00400000131718 | Gamma Aminobutyric Acid Receptor Subunit Beta Precursor Gaba A Receptor Subunit Beta |
| ENSDARG00000099104 | ENSFM00670001235623 | 60S Ribosomal L24 |
| ENSDARG00000099118 | ENSFM00250000002395 | Trafficking Particle Complex Subunit 9 Nik And Ikbkb Binding |
| ENSDARG00000099175 | ENSFM00500000269645 | High Mobility Group B1 High Mobility Group 1 Hmg 1 |
| ENSDARG00000099184 | ENSFM00770001847556 | Dual Specificity Mitogen Activated Kinase Kinase 6 Map Kinase Kinase 6 Mapkk 6 EC_2.7.12.2 Mapk/Erk Kinase 6 Mek 6 |
| ENSDARG00000099214 | ENSFM00500000270313 | Histone H2A |
| ENSDARG00000099226 | ENSFM00420000140518 | Serine/Threonine Phosphatase PP1 Catalytic Subunit Pp EC_3.1.3.16 |
| ENSDARG00000099236 | ENSFM00350000105408 | Septin |
| ENSDARG00000099306 | ENSFM00500000270014 | F Box Only 6 F Box That Recognizes Sugar Chains 2 |
| ENSDARG00000099321 | ENSFM00440000236967 | Btb/Poz Domain Containing 17 Precursor Galectin 3 Binding |
| ENSDARG00000099323 | ENSFM00250000000275 | Disks Large Homolog Synapse Associated Sap |
| ENSDARG00000099339 | ENSFM00800002020482 | Unknown |
| ENSDARG00000099339 | ENSFM00800002020483 | Unknown |
| ENSDARG00000099339 | ENSFM00250000001466 | Kinase C And Casein Kinase Substrate In Neurons |
| ENSDARG00000099368 | ENSFM00500000271356 | Mitochondrial Coenzyme A Transporter Sl Solute Carrier Family 25 Member 42 |
| ENSDARG00000099380 | ENSFM00500000270284 | 60S Ribosomal L13 |
| ENSDARG00000099386 | ENSFM00400000131776 | Leucine Rich Repeat Transmembrane Neuronal Precursor |
| ENSDARG00000099478 | ENSFM00290000065564 | Heparan Sulfate 2 O Sulfotransferase 1 2 O Sulfotransferase 2OST EC_2.8.2.- |
| ENSDARG00000099579 | ENSFM00760001714924 | Delta 1 Pyrroline 5 Carboxylate Synthase P5CS Aldehyde Dehydrogenase Family 18 Member A1 [Includes Glutamate 5 Kinase Gk EC_2.7.2.11 Gamma Glutamyl Kinase |
| ENSDARG00000099614 | ENSFM00250000002171 | Transcriptional Activator Pur Beta Purine Rich Element Binding B |
| ENSDARG00000099624 | ENSFM00600000921337 | Charged Multivesicular Body 1B Chromatin Modifying 1B CHMP1B |
| ENSDARG00000099664 | ENSFM00500000274344 | Unknown |
| ENSDARG00000099685 | ENSFM00770001847629 | Guanine Nucleotide Binding Subunit Beta 5 GBETA5 Transducin Beta Chain 5 |
| ENSDARG00000099690 | ENSFM00740001589204 | Ena/Vasp Ena/Vasodilator Stimulated Phosphoprotein |
| ENSDARG00000099720 | ENSFM00740001589204 | Ena/Vasp Ena/Vasodilator Stimulated Phosphoprotein |
| ENSDARG00000099729 | ENSFM00800002024092 | Unknown |
| ENSDARG00000099729 | ENSFM00780001898335 | Protocadherin Precursor |
| ENSDARG00000099730 | ENSFM00800002024340 | Unknown |
| ENSDARG00000099730 | ENSFM00800002024341 | Unknown |
| ENSDARG00000099730 | ENSFM00250000000989 | Pyruvate Kinase Pkm EC_2.7.1.40 Pyruvate Kinase Muscle Isozyme |
| ENSDARG00000099744 | ENSFM00600000930021 | Neuromodulin Axonal Membrane Gap 43 Growth Associated 43 |
| ENSDARG00000099755 | ENSFM00730001521507 | Atp Dependent 6 Phosphofructokinase Type |
| ENSDARG00000099766 | ENSFM00730001521616 | Myosin Regulatory Light Myosin Regulatory Light Chain 2 Smooth Muscle Myosin Regulatory Light Chain |
| ENSDARG00000099783 | ENSFM00780001898335 | Protocadherin Precursor |
| ENSDARG00000099786 | ENSFM00440000236866 | Alpha Actinin Alpha Actinin F Actin Cross Linking |
| ENSDARG00000099844 | ENSFM00410000138458 | Guanine Nucleotide Binding Subunit |
| ENSDARG00000099850 | ENSFM00250000001463 | Iq Motif And SEC7 Domain Containing |
| ENSDARG00000099874 | ENSFM00250000000604 | Arf Gap With Gtpase Ank Repeat And Ph Domain Containing Agap Centaurin Gamma |
| ENSDARG00000099996 | ENSFM00250000000940 | Mitochondrial Rho Gtpase 2 Miro 2 EC_3.6.5.- Ras Homolog Gene Family Member T2 |
| ENSDARG00000100052 | ENSFM00540000738060 | Uncharacterized |
| ENSDARG00000100089 | ENSFM00250000000111 | Calcium/Calmodulin Dependent Kinase Type Ii Subunit Cam Kinase Ii Subunit Camk Ii Subunit EC_2.7.11.17 |
| ENSDARG00000100115 | ENSFM00730001521131 | Dynein Heavy Chain Axonemal Axonemal Beta Dynein Heavy Chain Ciliary Dynein Heavy Chain |
| ENSDARG00000100119 | ENSFM00730001521718 | 55 Kda Erythrocyte Membrane P55 Membrane Protein Palmitoylated 1 |
| ENSDARG00000100152 | ENSFM00800002017761 | Unknown |
| ENSDARG00000100170 | ENSFM00250000001444 | Peptidyl Glycine Alpha Amidating Monooxygenase Precursor Pam [Includes Peptidylglycine Alpha Hydroxylating Monooxygenase Phm EC_1.14.17.3 |
| ENSDARG00000100252 | ENSFM00250000003778 | F Actin Capping Subunit Beta |
| ENSDARG00000100267 | ENSFM00250000000328 | Metabotropic Glutamate Receptor Precursor |
| ENSDARG00000100274 | ENSFM00250000001912 | Wd Repeat Containing 7 Tgf Beta Resistance Associated Trag |
| ENSDARG00000100392 | ENSFM00250000002236 | 40S Ribosomal S18 |
| ENSDARG00000100392 | ENSFM00670001276328 | 40S Ribosomal S18 |
| ENSDARG00000100400 | ENSFM00250000003532 | Hyccin Down Regulated By CTNNB1 A FAM126A |
| ENSDARG00000100435 | ENSFM00500000271789 | Vesicle Trafficking SEC22B Er Golgi Snare Of 24 Kda Ers 24 ERS24 SEC22 Vesicle Trafficking Homolog B |
| ENSDARG00000100510 | ENSFM00800002003421 | Actin Related 3 Actin 3 |
| ENSDARG00000100513 | ENSFM00770001848133 | 40S Ribosomal S27 |
| ENSDARG00000100562 | ENSFM00670001240058 | Syndecan |
| ENSDARG00000100588 | ENSFM00670001235763 | 60S Ribosomal L36 |
| ENSDARG00000100620 | ENSFM00250000000892 | Serine/Threonine Kinase EC_2.7.11.1 Thousand And One Amino Acid |
| ENSDARG00000100679 | ENSFM00800002003638 | Leucine Rich Repeat And Calponin Homology Domain Containing |
| ENSDARG00000100729 | ENSFM00250000001103 | Talin |
| ENSDARG00000100789 | ENSFM00500000274177 | Plasminogen Receptor Kt Plg R Kt |
| ENSDARG00000100829 | ENSFM00780001898335 | Protocadherin Precursor |
| ENSDARG00000100851 | ENSFM00790001963049 | Molybdopterin Adenylyltransferase Mpt Adenylyltransferase EC_2.7.7.75 Domain G |
| ENSDARG00000100972 | ENSFM00270000056424 | Myosin Cellular Myosin Heavy Chain Type Myosin Heavy Chain Myosin Heavy Chain Non Muscle Non Muscle Myosin Heavy Chain Nmmhc Non Muscle Myosin Heavy Chain Nmmhc Ii Nmmhc |
| ENSDARG00000101047 | ENSFM00800002017597 | Unknown |
| ENSDARG00000101047 | ENSFM00250000005120 | Trna Splicing Ligase Rtcb Homolog |
| ENSDARG00000101074 | ENSFM00250000001520 | Glutamate Dehydrogenase Mitochondrial Gdh EC_1.4.1.3 |
| ENSDARG00000101079 | ENSFM00500000270499 | Probable E3 Ubiquitin Ligase HECTD4 EC_6.3.2.- Hect Domain Containing 4 |
| ENSDARG00000101082 | ENSFM00250000003239 | Eukaryotic Translation Initiation Factor 3 Subunit L |
| ENSDARG00000101127 | ENSFM00500000270555 | Microtubule Associated Proteins 1A/1B Light Chain 3A Precursor Autophagy Related LC3 A Autophagy Related Ubiquitin Modifier LC3 A MAP1 Light Chain 3 1 MAP1A/MAP1B Light Chain 3 A MAP1A/MAP1B LC3 A Microtubule Associated 1 Light Chain 3 Alpha |
| ENSDARG00000101139 | ENSFM00500000270410 | 1 Acyl Sn Glycerol 3 Phosphate Acyltransferase Alpha Precursor EC_2.3.1.51 1 Acylglycerol 3 Phosphate O Acyltransferase 1 1 Agp Acyltransferase 1 1 Agpat 1 Lysophosphatidic Acid Acyltransferase Alpha Lpaat Alpha |
| ENSDARG00000101233 | ENSFM00250000000599 | Unc 13 Homolog MUNC13 |
| ENSDARG00000101380 | ENSFM00670001235476 | Alpha Crystallin Chain |
| ENSDARG00000101406 | ENSFM00500000312340 | Ribosomal Protein Large P2 |
| ENSDARG00000101510 | ENSFM00750001632381 | Centrin |
| ENSDARG00000101561 | ENSFM00250000003682 | Lipoamide Acyltransferase Component Of Branched Chain Alpha Keto Acid Dehydrogenase Complex Mitochondrial Precursor EC_2.3.1.168 Branched Chain Alpha Keto Acid Dehydrogenase Complex Component E2 Bckad E2 BCKADE2 Dihydrolipoamide Acetyltransferase Componen |
| ENSDARG00000101606 | ENSFM00250000000629 | Regulating Synaptic Membrane Exocytosis Rab 3 Interacting Molecule Rim |
| ENSDARG00000101624 | ENSFM00250000000425 | Lamin |
| ENSDARG00000101687 | ENSFM00260000050563 | Microtubule Associated 4 Map 4 |
| ENSDARG00000101719 | ENSFM00250000003384 | Poly Adp Ribose Glycohydrolase EC_3.2.1.143 |
| ENSDARG00000101755 | ENSFM00730001521681 | Camp Dependent Kinase Type I Alpha Regulatory Camp Dependent Kinase Type I Alpha Regulatory Subunit N Terminally Processed] |
| ENSDARG00000101759 | ENSFM00250000001140 | Homer Homolog Homer Vasp/Ena Related Gene Up Regulated During Seizure And Ltp Vesl |
| ENSDARG00000101812 | ENSFM00250000001584 | Src Kinase Signaling Inhibitor 1 Snap 25 Interacting Snip P130CAS Associated P140CAP |
| ENSDARG00000101858 | ENSFM00740001589112 | Microtubule Actin Cross Linking Factor 1 Actin Cross Linking Family 7 |
| ENSDARG00000101869 | ENSFM00250000000552 | Membrane Associated Guanylate Kinase Ww And Pdz Domain Containing Membrane Associated Guanylate Kinase Inverted Magi |
| ENSDARG00000102024 | ENSFM00670001235835 | Charged Multivesicular Body 6 Chromatin Modifying 6 |
| ENSDARG00000102067 | ENSFM00730001521627 | Metabotropic Glutamate Receptor Precursor |
| ENSDARG00000102078 | ENSFM00750001632414 | Ras Related Rab 35 |
| ENSDARG00000102081 | ENSFM00600000921208 | Trio And F Actin Binding Protein Tara Trio Associated Repeat On Actin |
| ENSDARG00000102115 | ENSFM00670001238685 | Nadh Dehydrogenase [Ubiquinone] 1 Subunit C2 |
| ENSDARG00000102125 | ENSFM00250000001463 | Iq Motif And SEC7 Domain Containing |
| ENSDARG00000102184 | ENSFM00250000001057 | E3 Ubiquitin Ligase EC_6.3.2.- Mind Bomb Homolog |
| ENSDARG00000102191 | ENSFM00730001521728 | Copine Copine |
| ENSDARG00000102216 | ENSFM00250000000275 | Disks Large Homolog Synapse Associated Sap |
| ENSDARG00000102254 | ENSFM00500000269896 | Pyrroline 5 Carboxylate Reductase 3 P5C Reductase 3 P5CR 3 EC_1.5.1.2 Pyrroline 5 Carboxylate Reductase |
| ENSDARG00000102295 | ENSFM00730001522148 | Dnaj Homolog Subfamily A Member 3 Mitochondrial Precursor Dnaj Tid 1 1 Tumorous Imaginal Discs TID56 Homolog |
| ENSDARG00000102317 | ENSFM00500000270069 | 60S Ribosomal L26 |
| ENSDARG00000102365 | ENSFM00250000003294 | Transcription Elongation Factor B Polypeptide 3 Elongin 110 Kda Subunit Elongin A Eloa Rna Polymerase Ii Transcription Factor Siii Subunit A1 Siii P110 |
| ENSDARG00000102376 | ENSFM00250000002019 | Voltage Dependent Calcium Channel Gamma Subunit Neuronal Voltage Gated Calcium Channel Gamma Subunit Transmembrane Ampar Regulatory Gamma Tarp Gamma |
| ENSDARG00000102381 | ENSFM00500000269830 | Choline Transporter Solute Carrier Family 44 Member |
| ENSDARG00000102394 | ENSFM00810002040662 | Ap 1 Complex Subunit Gamma 1 Adaptor Complex Ap 1 Subunit Gamma 1 Adaptor Related Complex 1 Subunit Gamma 1 Clathrin Assembly Complex 1 Gamma 1 Large Chain Gamma Adaptin GAMMA1 Adaptin Golgi Adaptor HA1/AP1 Adaptin Subunit Gamma 1 |
| ENSDARG00000102441 | ENSFM00250000001018 | Catenin Alpha 2 Alpha N Catenin |
| ENSDARG00000102472 | ENSFM00440000236967 | Btb/Poz Domain Containing 17 Precursor Galectin 3 Binding |
| ENSDARG00000102532 | ENSFM00250000001140 | Homer Homolog Homer Vasp/Ena Related Gene Up Regulated During Seizure And Ltp Vesl |
| ENSDARG00000102584 | ENSFM00800002003632 | Copine Copine |
| ENSDARG00000102587 | ENSFM00500000271280 | Wd Repeat Containing Mio |
| ENSDARG00000102626 | ENSFM00740001589159 | Extracellular Matrix Precursor |
| ENSDARG00000102643 | ENSFM00600000921337 | Charged Multivesicular Body 1B Chromatin Modifying 1B CHMP1B |
| ENSDARG00000102674 | ENSFM00760001714778 | Cytoskeleton Associated 5 |
| ENSDARG00000102684 | ENSFM00730001522046 | Scribble |
| ENSDARG00000102690 | ENSFM00250000000629 | Regulating Synaptic Membrane Exocytosis Rab 3 Interacting Molecule Rim |
| ENSDARG00000102730 | ENSFM00460000244930 | Mitogen Activated Kinase Map Kinase Mapk EC_2.7.11.24 Stress Activated Kinase C Jun N Terminal Kinase |
| ENSDARG00000102744 | ENSFM00670001235926 | Microsomal Glutathione S Transferase 3 Microsomal Gst 3 EC_2.5.1.18 Microsomal Gst Iii |
| ENSDARG00000102755 | ENSFM00730001522226 | Echinoderm Microtubule Associated 6 Emap 6 Echinoderm Microtubule Associated 5 |
| ENSDARG00000102780 | ENSFM00520000517802 | Kinase C Type Pkc EC_2.7.11.13 |
| ENSDARG00000102790 | ENSFM00250000002597 | Polyribonucleotide Nucleotidyltransferase 1 Mitochondrial Precursor EC_2.7.7.8 3' 5' Rna Exonuclease OLD35 Pnpase Old 35 Polynucleotide Phosphorylase 1 Pnpase 1 Polynucleotide Phosphorylase |
| ENSDARG00000102813 | ENSFM00800002020563 | Unknown |
| ENSDARG00000102822 | ENSFM00670001236447 | Mitochondrial Import Inner Membrane Translocase Subunit TIM16 Mitochondria Associated Granulocyte Macrophage Csf Signaling Molecule Presequence Translocated Associated Motor Subunit PAM16 |
| ENSDARG00000102824 | ENSFM00780001898335 | Protocadherin Precursor |
| ENSDARG00000102825 | ENSFM00320000100099 | Noelin Precursor Neuronal Olfactomedin Related Er Localized Olfactomedin 1 |
| ENSDARG00000102845 | ENSFM00400000131747 | Serine/Threonine Kinase EC_2.7.11.1 Doublecortin And Cam Kinase Doublecortin Kinase |
| ENSDARG00000102855 | ENSFM00260000050557 | Netrin Receptor Dcc Precursor Tumor Suppressor Dcc |
| ENSDARG00000102883 | ENSFM00740001589095 | Spectrin Beta Chain Erythrocytic Beta Spectrin |
| ENSDARG00000102889 | ENSFM00760001714685 | Septin 7 CDC10 Homolog |
| ENSDARG00000103007 | ENSFM00250000001742 | 40S Ribosomal S3 EC_4.2.99.18 |
| ENSDARG00000103032 | ENSFM00800002003504 | Ras Related Rap Precursor |
| ENSDARG00000103069 | ENSFM00290000065521 | Precursor |
| ENSDARG00000103083 | ENSFM00810002040719 | Dna Damage Binding 1 Damage Specific Dna Binding 1 |
| ENSDARG00000103095 | ENSFM00250000003276 | Inhibitor Of Nuclear Factor Kappa B Kinase Subunit Epsilon I Kappa B Kinase Epsilon Ikk E Ikk Epsilon Ikbke EC_2.7.11.10 Inducible I Kappa B Kinase Ikk I |
| ENSDARG00000103123 | ENSFM00250000007087 | Neuronal Tyrosine Phosphorylated Phosphoinositide 3 Kinase Adapter 1 |
| ENSDARG00000103144 | ENSFM00250000003688 | Phosphatase 1 Regulatory Subunit 21 Coiled Coil Domain Containing 128 Klraq Motif Containing 1 |
| ENSDARG00000103203 | ENSFM00800002022009 | Unknown |
| ENSDARG00000103203 | ENSFM00500000271517 | Vacuolar Sorting Associated 28 Homolog Escrt I Complex Subunit VPS28 |
| ENSDARG00000103256 | ENSFM00780001898335 | Protocadherin Precursor |
| ENSDARG00000103332 | ENSFM00730001521400 | Atp Sensitive Inward Rectifier Potassium Channel Atp Potassium Channel K Inward Rectifier K + Channel 1 Potassium Channel Inwardly Rectifying Subfamily J Member |
| ENSDARG00000103333 | ENSFM00250000000906 | Brain Specific Angiogenesis Inhibitor 1 Associated 2 Bai Associated 2 BAI1 Associated 2 Insulin Receptor Substrate Of 53 Kda IRSP53 Insulin Receptor Substrate P53 |
| ENSDARG00000103359 | ENSFM00250000001398 | Slit And Ntrk Precursor |
| ENSDARG00000103428 | ENSFM00750001632363 | 2 Oxoglutarate Dehydrogenase Mitochondrial Precursor 2 Oxoglutarate Dehydrogenase Complex Component E1 Ogdc E1 Alpha Ketoglutarate Dehydrogenase |
| ENSDARG00000103433 | ENSFM00500000270978 | 60S Ribosomal L14 |
| ENSDARG00000103435 | ENSFM00670001235644 | Sorbin And SH3 Domain Containing 1 Ponsin SH3 Domain 5 S C Cbl Associated Cap |
| ENSDARG00000103479 | ENSFM00740001589097 | Receptor Type Tyrosine Phosphatase Precursor EC_3.1.3.48 |
| ENSDARG00000103521 | ENSFM00250000002018 | Calcyphosin |
| ENSDARG00000103563 | ENSFM00250000002234 | Intraflagellar Transport 122 Homolog Wd Repeat Containing 10 |
| ENSDARG00000103774 | ENSFM00250000003304 | Lim And Calponin Homology Domains Containing 1 |
| ENSDARG00000103791 | ENSFM00250000000895 | Nucleoside Diphosphate Kinase Ndk Ndp Kinase EC_2.7.4.6 |
| ENSDARG00000103799 | ENSFM00250000002843 | Lysine Trna Ligase EC_6.1.1.6 Lysyl Trna Synthetase Lysrs |
| ENSDARG00000103800 | ENSFM00730001521681 | Camp Dependent Kinase Type I Alpha Regulatory Camp Dependent Kinase Type I Alpha Regulatory Subunit N Terminally Processed] |
| ENSDARG00000103900 | ENSFM00500000269805 | Septin |
| ENSDARG00000103902 | ENSFM00250000002568 | Nuclear Distribution Nude 1 |
| ENSDARG00000103937 | ENSFM00800002023711 | Unknown |
| ENSDARG00000103937 | ENSFM00250000000535 | N Myc Downstream Regulated Gene |
| ENSDARG00000103962 | ENSFM00250000000681 | Adducin |
| ENSDARG00000103987 | ENSFM00500000272336 | Calcium And Integrin Binding 1 Cib Calmyrin Dna Pkcs Interacting Kinase Interacting Kip |
| ENSDARG00000103993 | ENSFM00500000313527 | Secretogranin Ii |
| ENSDARG00000104014 | ENSFM00730001521220 | Kinase EC_2.7.11.1 |
| ENSDARG00000104023 | ENSFM00800002024129 | Unknown |
| ENSDARG00000104023 | ENSFM00320000100165 | Myelin Associated Glycoprotein Precursor Siglec 4A |
| ENSDARG00000104066 | ENSFM00760001714614 | Dnaj Homolog Subfamily A Member Precursor |
| ENSDARG00000104085 | ENSFM00800002003649 | Atp Binding Cassette Sub Family D Member Kda Peroxisomal Membrane PMP70 |
| ENSDARG00000104086 | ENSFM00250000002627 | Rho Gtpase Activating 39 |
| ENSDARG00000104101 | ENSFM00800002023681 | Unknown |
| ENSDARG00000104101 | ENSFM00740001589344 | Peripheral Type Benzodiazepine Receptor Associated 1 Prax 1 Rims Binding 1 Rim BP1 |
| ENSDARG00000104111 | ENSFM00250000008011 | 28S Ribosomal S30 Mitochondrial Mrp S30 S30MT |
| ENSDARG00000104119 | ENSFM00740001589124 | EC_2.7.11.1 |
| ENSDARG00000104139 | ENSFM00650001139994 | Sodium/Potassium Transporting Atpase Subunit Alpha Na + /K + Atpase Alpha Subunit EC_3.6.3.9 Sodium Pump Subunit Alpha |
| ENSDARG00000104157 | ENSFM00800002024283 | Unknown |
| ENSDARG00000104173 | ENSFM00260000050713 | Elongation Factor Tu Mitochondrial Precursor |
| ENSDARG00000104225 | ENSFM00250000002568 | Nuclear Distribution Nude 1 |
| ENSDARG00000104230 | ENSFM00810002040640 | Transport SEC23 Related |
| ENSDARG00000104235 | ENSFM00750001632326 | Unconventional Myosin Va Dilute Myosin Heavy Chain Non Muscle |
| ENSDARG00000104264 | ENSFM00500000269993 | Xk Related |
| ENSDARG00000104267 | ENSFM00800002015296 | Unknown |
| ENSDARG00000104267 | ENSFM00760001719894 | Osteonectin |
| ENSDARG00000104267 | ENSFM00250000001947 | Transforming Growth Factor Beta Induced Ig H3 Precursor Beta Ig H3 Kerato Epithelin Rgd Containing Collagen Associated Rgd Cap |
| ENSDARG00000104282 | ENSFM00260000050557 | Netrin Receptor Dcc Precursor Tumor Suppressor Dcc |
| ENSDARG00000104342 | ENSFM00730001521484 | Casein Kinase I Gamma Cki Gamma EC_2.7.11.1 |
| ENSDARG00000104358 | ENSFM00250000000820 | Clip Associating Cytoplasmic Linker Associated |
| ENSDARG00000104375 | ENSFM00750001632389 | Transport SEC24A SEC24 Related A |
| ENSDARG00000104396 | ENSFM00390000126422 | E3 Ubiquitin/ISG15 Ligase TRIM25 EC_6.3.2.19 EC_6.3.2.- N3 Estrogen Responsive Finger Tripartite Motif Containing 25 Ubiquitin/ISG15 Conjugating Enzyme TRIM25 Zinc Finger 147 |
| ENSDARG00000104401 | ENSFM00800002003559 | Atp Dependent Zinc Metalloprotease Ym EC_3.4.24.- Atp Dependent Metalloprotease FTSH1 Meg 4 YME1 1 |
| ENSDARG00000104413 | ENSFM00250000005331 | Rogdi Homolog |
| ENSDARG00000104414 | ENSFM00730001521489 | Serine Hydroxymethyltransferase Shmt EC_2.1.2.1 Glycine Hydroxymethyltransferase Serine Methylase |
| ENSDARG00000104439 | ENSFM00250000006965 | Claudin Domain Containing 1 |
| ENSDARG00000104458 | ENSFM00670001241589 | Ribonuclease Kappa B Rnase K B Rnase Kappa B EC_3.1.-.- |
| ENSDARG00000104483 | ENSFM00250000002325 | Pdz Domain Containing |
| ENSDARG00000104517 | ENSFM00800002016273 | Unknown |
| ENSDARG00000104549 | ENSFM00250000000900 | Rho Gtpase Activating Rho Type Gtpase Activating Rhogap Interacting With CIP4 Homologs Rich |
| ENSDARG00000104581 | ENSFM00250000001136 | TOM1 2 Target Of Myb 2 |
| ENSDARG00000104615 | ENSFM00730001521674 | Serine/Threonine Kinase EC_2.7.11.1 Salt Inducible Kinase Sik Serine/Threonine Kinase SNF1 Kinase |
| ENSDARG00000104664 | ENSFM00400000131747 | Serine/Threonine Kinase EC_2.7.11.1 Doublecortin And Cam Kinase Doublecortin Kinase |
| ENSDARG00000104668 | ENSFM00800002022057 | Unknown |
| ENSDARG00000104701 | ENSFM00800002024297 | Unknown |
| ENSDARG00000104701 | ENSFM00250000004934 | MAP7 Domain Containing 1 |
| ENSDARG00000104820 | ENSFM00250000001069 | Ph And SEC7 Domain Containing 1 Exchange Factor ARF6 Pleckstrin Homology And SEC7 Domain Containing 1 |
| ENSDARG00000104937 | ENSFM00740001589097 | Receptor Type Tyrosine Phosphatase Precursor EC_3.1.3.48 |
| ENSDARG00000104958 | ENSFM00250000005444 | Kelch Domain Containing |
| ENSDARG00000105099 | ENSFM00250000003047 | Golgin Subfamily A Member 7 |
| ENSDARG00000105115 | ENSFM00250000000608 | Acid Sensing Ion Channel Amiloride Sensitive Cation Channel |
| ENSDARG00000105142 | ENSFM00250000000490 | V Type Proton Atpase 116 Kda Subunit A V Atpase 116 Kda Proton 116 Kda Subunit Vacuolar Proton Translocating Atpase 116 Kda Subunit A |
| ENSDARG00000105159 | ENSFM00760001714682 | Phosphorylase B Kinase Regulatory Subunit Alpha Phosphorylase Kinase Subunit |
| ENSDARG00000105209 | ENSFM00600000921208 | Trio And F Actin Binding Protein Tara Trio Associated Repeat On Actin |
| ENSDARG00000105243 | ENSFM00800002017484 | Unknown |
| ENSDARG00000105243 | ENSFM00800002017485 | Unknown |
| ENSDARG00000105243 | ENSFM00250000002216 | Udp N Acetylglucosamine Peptide N Acetylglucosaminyltransferase 110 Kda Subunit EC_2.4.1.255 O Glcnac Transferase Subunit P110 O Linked N Acetylglucosamine Transferase 110 Kda Subunit Ogt |
Supplementary Table 5. Domains in mouse and zebrafish synaptic proteins
Domain types as described by the PFAM database (pfam.xfam.org) in mouse and zebrafish proteins. The count of domain types is given for mouse PSD proteins (M.PSD), mouse SYN proteins (M.SYN), mouse Genome (M.Genome), mouse brain (M.brain), zebrafish PSD proteins (Z.PSD), zebrafish SYN proteins (Z.SYN), zebrafish Genome (Z.Genome), zebrafish brain (Z.brain).
| Pfam | M.PSD | M.SYN | M.Genome | M.Brain | Z.PSD | Z.SYN | Z.Genome | Z.Brain | Description | Count |
|---|---|---|---|---|---|---|---|---|---|---|
| PF00001 | 6 | 10 | 383 | 19 | 1 | 6 | 645 | 3 | 7 transmembrane receptor (rhodopsin family) | 1073 |
| PF00002 | 7 | 9 | 44 | 2 | 6 | 11 | 67 | 1 | 7 transmembrane receptor (Secretin family) | 147 |
| PF00003 | 10 | 11 | 143 | 4 | 15 | 18 | 61 | 2 | 7 transmembrane sweet-taste receptor of 3 GCPR | 264 |
| PF00004 | 12 | 16 | 43 | 15 | 4 | 19 | 45 | 12 | ATPase family associated with various cellular activities (AAA) | 166 |
| PF00005 | 6 | 17 | 52 | 12 | 3 | 16 | 52 | 3 | ABC transporter | 161 |
| PF00006 | 2 | 4 | 5 | 4 | 2 | 6 | 6 | 6 | "ATP synthase alpha/beta family, nucleotide-binding domain" | 35 |
| PF00007 | 0 | 0 | 9 | 1 | 0 | 0 | 12 | 0 | Cystine-knot domain | 22 |
| PF00008 | 7 | 14 | 60 | 7 | 6 | 13 | 69 | 5 | EGF-like domain | 181 |
| PF00009 | 5 | 7 | 20 | 8 | 6 | 7 | 23 | 10 | Elongation factor Tu GTP binding domain | 86 |
| PF00010 | 0 | 0 | 103 | 5 | 0 | 0 | 118 | 0 | Helix-loop-helix DNA-binding domain | 226 |
| PF00011 | 2 | 2 | 10 | 3 | 1 | 1 | 11 | 3 | Hsp20/alpha crystallin family | 33 |
| PF00012 | 3 | 10 | 13 | 5 | 4 | 10 | 18 | 8 | Hsp70 protein | 71 |
| PF00013 | 11 | 11 | 36 | 10 | 4 | 10 | 37 | 14 | KH domain | 133 |
| PF00014 | 0 | 1 | 19 | 1 | 0 | 3 | 22 | 2 | Kunitz/Bovine pancreatic trypsin inhibitor domain | 48 |
| PF00017 | 8 | 8 | 97 | 16 | 7 | 9 | 114 | 5 | SH2 domain | 264 |
| PF00018 | 29 | 30 | 94 | 23 | 25 | 30 | 104 | 13 | SH3 domain | 348 |
| PF00019 | 0 | 1 | 36 | 0 | 0 | 0 | 43 | 0 | Transforming growth factor beta like domain | 80 |
| PF00020 | 0 | 0 | 18 | 1 | 0 | 0 | 22 | 0 | TNFR/NGFR cysteine-rich region | 41 |
| PF00021 | 0 | 2 | 31 | 2 | 2 | 2 | 7 | 2 | u-PAR/Ly-6 domain | 48 |
| PF00022 | 10 | 10 | 26 | 9 | 7 | 7 | 20 | 8 | Actin | 97 |
| PF00023 | 10 | 9 | 70 | 12 | 13 | 14 | 74 | 5 | Ankyrin repeat | 207 |
| PF00024 | 0 | 0 | 5 | 0 | 0 | 0 | 2 | 1 | PAN domain | 8 |
| PF00025 | 6 | 10 | 30 | 15 | 1 | 11 | 37 | 12 | ADP-ribosylation factor family | 122 |
| PF00026 | 0 | 1 | 9 | 1 | 0 | 1 | 4 | 1 | Eukaryotic aspartyl protease | 17 |
| PF00027 | 8 | 13 | 32 | 8 | 6 | 14 | 44 | 2 | Cyclic nucleotide-binding domain | 127 |
| PF00028 | 17 | 21 | 97 | 6 | 22 | 38 | 87 | 6 | Cadherin domain | 294 |
| PF00029 | 3 | 4 | 20 | 3 | 5 | 2 | 41 | 1 | Connexin | 79 |
| PF00030 | 0 | 0 | 16 | 1 | 0 | 0 | 60 | 19 | Beta/Gamma crystallin | 96 |
| PF00031 | 1 | 1 | 27 | 3 | 0 | 3 | 13 | 8 | Cystatin domain | 56 |
| PF00032 | 0 | 0 | 0 | 1 | 0 | 1 | 1 | 1 | Cytochrome b(C-terminal)/b6/petD | 4 |
| PF00034 | 0 | 1 | 2 | 0 | 0 | 1 | 2 | 1 | Cytochrome c | 7 |
| PF00035 | 3 | 0 | 15 | 5 | 4 | 4 | 16 | 6 | Double-stranded RNA binding motif | 53 |
| PF00036 | 4 | 5 | 23 | 7 | 1 | 7 | 23 | 7 | EF hand | 77 |
| PF00037 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 4Fe-4S binding domain | 6 |
| PF00038 | 11 | 9 | 71 | 9 | 25 | 22 | 58 | 30 | Intermediate filament protein | 235 |
| PF00039 | 0 | 0 | 4 | 0 | 0 | 0 | 2 | 0 | Fibronectin type I domain | 6 |
| PF00040 | 0 | 2 | 12 | 2 | 0 | 4 | 10 | 1 | Fibronectin type II domain | 31 |
| PF00041 | 30 | 46 | 141 | 18 | 30 | 64 | 170 | 21 | Fibronectin type III domain | 520 |
| PF00042 | 0 | 2 | 13 | 5 | 0 | 5 | 21 | 6 | Globin | 52 |
| PF00043 | 4 | 8 | 24 | 9 | 4 | 11 | 16 | 13 | "Glutathione S-transferase, C-terminal domain" | 89 |
| PF00044 | 1 | 1 | 2 | 0 | 0 | 2 | 2 | 2 | "Glyceraldehyde 3-phosphate dehydrogenase, NAD binding domain" | 10 |
| PF00045 | 1 | 0 | 22 | 0 | 0 | 0 | 24 | 1 | Hemopexin | 48 |
| PF00046 | 0 | 2 | 236 | 13 | 0 | 0 | 241 | 0 | Homeobox domain | 492 |
| PF00047 | 2 | 7 | 30 | 3 | 2 | 3 | 6 | 1 | Immunoglobulin domain | 54 |
| PF00048 | 0 | 1 | 41 | 1 | 0 | 0 | 35 | 0 | "Small cytokines (intecrine/chemokine), interleukin-8 like" | 78 |
| PF00049 | 0 | 0 | 9 | 1 | 0 | 1 | 10 | 1 | Insulin/IGF/Relaxin family | 22 |
| PF00050 | 1 | 3 | 18 | 1 | 2 | 2 | 12 | 1 | Kazal-type serine protease inhibitor domain | 40 |
| PF00051 | 0 | 0 | 15 | 0 | 0 | 0 | 14 | 2 | Kringle domain | 31 |
| PF00052 | 0 | 1 | 8 | 0 | 0 | 0 | 5 | 3 | Laminin B (Domain IV) | 17 |
| PF00053 | 3 | 5 | 32 | 2 | 4 | 6 | 38 | 6 | Laminin EGF-like (Domains III and V) | 96 |
| PF00054 | 0 | 3 | 11 | 2 | 1 | 1 | 9 | 3 | Laminin G domain | 30 |
| PF00055 | 3 | 3 | 15 | 1 | 3 | 4 | 15 | 2 | Laminin N-terminal (Domain VI) | 46 |
| PF00056 | 0 | 4 | 6 | 3 | 0 | 5 | 5 | 4 | "lactate/malate dehydrogenase, NAD binding domain" | 27 |
| PF00057 | 2 | 4 | 41 | 2 | 0 | 7 | 49 | 6 | Low-density lipoprotein receptor domain class A | 111 |
| PF00058 | 0 | 4 | 12 | 1 | 0 | 6 | 19 | 3 | Low-density lipoprotein receptor repeat class B | 45 |
| PF00059 | 5 | 6 | 109 | 2 | 2 | 7 | 148 | 3 | Lectin C-type domain | 282 |
| PF00060 | 13 | 11 | 17 | 6 | 19 | 21 | 30 | 7 | Ligand-gated ion channel | 124 |
| PF00061 | 0 | 4 | 59 | 7 | 1 | 9 | 25 | 9 | Lipocalin / cytosolic fatty-acid binding protein family | 114 |
| PF00062 | 0 | 0 | 9 | 1 | 0 | 0 | 1 | 1 | C-type lysozyme/alpha-lactalbumin family | 12 |
| PF00063 | 13 | 11 | 37 | 5 | 23 | 16 | 54 | 16 | Myosin head (motor domain) | 175 |
| PF00066 | 0 | 0 | 7 | 0 | 0 | 0 | 7 | 0 | LNR domain | 14 |
| PF00067 | 1 | 4 | 99 | 6 | 0 | 10 | 68 | 5 | Cytochrome P450 | 193 |
| PF00068 | 0 | 0 | 8 | 0 | 0 | 0 | 2 | 0 | Phospholipase A2 | 10 |
| PF00069 | 77 | 69 | 363 | 98 | 72 | 103 | 690 | 47 | Protein kinase domain | 1519 |
| PF00070 | 3 | 6 | 7 | 4 | 2 | 5 | 9 | 3 | Pyridine nucleotide-disulphide oxidoreductase | 39 |
| PF00071 | 33 | 70 | 130 | 56 | 25 | 83 | 163 | 57 | Ras family | 617 |
| PF00072 | 2 | 1 | 2 | 1 | 0 | 0 | 2 | 0 | Response regulator receiver domain | 8 |
| PF00074 | 0 | 0 | 19 | 0 | 0 | 0 | 3 | 0 | Pancreatic ribonuclease | 22 |
| PF00075 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 0 | RNase H | 3 |
| PF00076 | 32 | 22 | 169 | 61 | 13 | 18 | 169 | 64 | "RNA recognition motif. (a.k.a. RRM, RBD, or RNP domain)" | 548 |
| PF00078 | 0 | 0 | 1 | 0 | 0 | 0 | 9 | 0 | Reverse transcriptase (RNA-dependent DNA polymerase) | 10 |
| PF00079 | 2 | 5 | 62 | 11 | 1 | 5 | 29 | 11 | Serpin (serine protease inhibitor) | 126 |
| PF00080 | 0 | 1 | 3 | 3 | 0 | 1 | 5 | 1 | Copper/zinc superoxide dismutase (SODC) | 14 |
| PF00081 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | "Iron/manganese superoxide dismutases, alpha-hairpin domain" | 6 |
| PF00082 | 0 | 2 | 11 | 4 | 1 | 3 | 11 | 1 | Subtilase family | 33 |
| PF00083 | 1 | 10 | 39 | 4 | 0 | 11 | 44 | 6 | Sugar (and other) transporter | 115 |
| PF00084 | 5 | 10 | 51 | 3 | 5 | 11 | 47 | 5 | Sushi domain (SCR repeat) | 137 |
| PF00085 | 5 | 16 | 29 | 13 | 3 | 19 | 27 | 10 | Thioredoxin | 122 |
| PF00086 | 0 | 1 | 17 | 3 | 0 | 1 | 19 | 2 | Thyroglobulin type-1 repeat | 43 |
| PF00088 | 0 | 1 | 7 | 0 | 0 | 1 | 10 | 0 | Trefoil (P-type) domain | 19 |
| PF00089 | 1 | 2 | 152 | 3 | 2 | 2 | 129 | 15 | Trypsin | 306 |
| PF00090 | 3 | 4 | 58 | 5 | 3 | 4 | 67 | 2 | Thrombospondin type 1 domain | 146 |
| PF00091 | 11 | 10 | 20 | 7 | 13 | 12 | 21 | 13 | "Tubulin/FtsZ family, GTPase domain" | 107 |
| PF00092 | 4 | 4 | 42 | 5 | 6 | 5 | 54 | 12 | von Willebrand factor type A domain | 132 |
| PF00093 | 0 | 1 | 27 | 4 | 0 | 0 | 29 | 3 | von Willebrand factor type C domain | 64 |
| PF00094 | 0 | 0 | 14 | 0 | 0 | 1 | 17 | 1 | von Willebrand factor type D domain | 33 |
| PF00095 | 0 | 0 | 19 | 0 | 0 | 0 | 8 | 0 | WAP-type (Whey Acidic Protein) 'four-disulfide core' | 27 |
| PF00096 | 0 | 0 | 247 | 17 | 0 | 0 | 254 | 2 | "Zinc finger, C2H2 type" | 520 |
| PF00097 | 4 | 4 | 56 | 6 | 3 | 1 | 60 | 0 | "Zinc finger, C3HC4 type (RING finger)" | 134 |
| PF00098 | 0 | 0 | 19 | 3 | 1 | 0 | 24 | 4 | Zinc knuckle | 51 |
| PF00100 | 0 | 0 | 17 | 1 | 0 | 0 | 54 | 1 | Zona pellucida-like domain | 73 |
| PF00102 | 7 | 16 | 39 | 7 | 10 | 23 | 43 | 4 | Protein-tyrosine phosphatase | 149 |
| PF00103 | 0 | 0 | 27 | 2 | 1 | 1 | 5 | 0 | Somatotropin hormone family | 36 |
| PF00104 | 0 | 0 | 49 | 2 | 1 | 0 | 64 | 1 | Ligand-binding domain of nuclear hormone receptor | 117 |
| PF00105 | 0 | 0 | 47 | 2 | 1 | 0 | 61 | 1 | "Zinc finger, C4 type (two domains)" | 112 |
| PF00106 | 10 | 19 | 51 | 18 | 6 | 28 | 75 | 24 | short chain dehydrogenase | 231 |
| PF00107 | 0 | 5 | 15 | 9 | 0 | 6 | 14 | 6 | Zinc-binding dehydrogenase | 55 |
| PF00108 | 3 | 6 | 8 | 4 | 0 | 6 | 5 | 5 | "Thiolase, N-terminal domain" | 37 |
| PF00109 | 0 | 1 | 2 | 2 | 0 | 1 | 2 | 0 | "Beta-ketoacyl synthase, N-terminal domain" | 8 |
| PF00110 | 1 | 0 | 19 | 1 | 0 | 0 | 24 | 0 | wnt family | 45 |
| PF00111 | 0 | 1 | 7 | 2 | 0 | 1 | 6 | 0 | 2Fe-2S iron-sulfur cluster binding domain | 17 |
| PF00112 | 0 | 2 | 23 | 6 | 0 | 2 | 26 | 4 | Papain family cysteine protease | 63 |
| PF00113 | 0 | 2 | 4 | 1 | 0 | 4 | 5 | 5 | "Enolase, C-terminal TIM barrel domain" | 21 |
| PF00115 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 1 | Cytochrome C and Quinol oxidase polypeptide I | 3 |
| PF00116 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 1 | "Cytochrome C oxidase subunit II, periplasmic domain" | 6 |
| PF00117 | 0 | 1 | 5 | 1 | 0 | 1 | 4 | 1 | Glutamine amidotransferase class-I | 13 |
| PF00118 | 7 | 8 | 14 | 6 | 0 | 11 | 13 | 9 | TCP-1/cpn60 chaperonin family | 68 |
| PF00119 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | ATP synthase A chain | 5 |
| PF00120 | 0 | 1 | 2 | 0 | 0 | 2 | 2 | 1 | "Glutamine synthetase, catalytic domain" | 8 |
| PF00121 | 1 | 1 | 1 | 0 | 0 | 2 | 2 | 2 | Triosephosphate isomerase | 9 |
| PF00122 | 1 | 19 | 36 | 8 | 3 | 19 | 37 | 18 | E1-E2 ATPase | 141 |
| PF00123 | 0 | 0 | 6 | 0 | 2 | 1 | 7 | 1 | Peptide hormone | 17 |
| PF00125 | 4 | 4 | 93 | 5 | 6 | 5 | 91 | 8 | Core histone H2A/H2B/H3/H4 | 216 |
| PF00128 | 0 | 2 | 7 | 1 | 0 | 4 | 9 | 4 | "Alpha amylase, catalytic domain" | 27 |
| PF00129 | 0 | 0 | 32 | 0 | 0 | 1 | 12 | 1 | "Class I Histocompatibility antigen, domains alpha 1 and 2" | 46 |
| PF00130 | 17 | 14 | 49 | 13 | 7 | 20 | 54 | 11 | Phorbol esters/diacylglycerol binding domain (C1 domain) | 185 |
| PF00131 | 0 | 0 | 4 | 3 | 0 | 0 | 1 | 0 | Metallothionein | 8 |
| PF00132 | 1 | 1 | 5 | 3 | 0 | 1 | 5 | 1 | Bacterial transferase hexapeptide (six repeats) | 17 |
| PF00133 | 3 | 5 | 6 | 4 | 2 | 3 | 7 | 4 | "tRNA synthetases class I (I, L, M and V)" | 34 |
| PF00134 | 1 | 2 | 30 | 2 | 1 | 2 | 28 | 0 | "Cyclin, N-terminal domain" | 66 |
| PF00135 | 2 | 4 | 24 | 4 | 4 | 9 | 15 | 6 | Carboxylesterase family | 68 |
| PF00136 | 0 | 0 | 4 | 0 | 0 | 0 | 4 | 0 | DNA polymerase family B | 8 |
| PF00137 | 1 | 1 | 5 | 1 | 2 | 2 | 6 | 2 | ATP synthase subunit C | 20 |
| PF00143 | 0 | 0 | 30 | 0 | 0 | 0 | 3 | 0 | Interferon alpha/beta domain | 33 |
| PF00144 | 1 | 1 | 2 | 1 | 1 | 1 | 3 | 0 | Beta-lactamase | 10 |
| PF00145 | 0 | 0 | 4 | 1 | 0 | 0 | 8 | 0 | C-5 cytosine-specific DNA methylase | 13 |
| PF00146 | 0 | 1 | 0 | 0 | 0 | 1 | 1 | 1 | NADH dehydrogenase | 4 |
| PF00147 | 5 | 2 | 23 | 2 | 6 | 6 | 44 | 8 | "Fibrinogen beta and gamma chains, C-terminal globular domain" | 96 |
| PF00149 | 5 | 8 | 28 | 16 | 6 | 11 | 26 | 10 | Calcineurin-like phosphoesterase | 110 |
| PF00151 | 0 | 0 | 8 | 0 | 0 | 0 | 7 | 0 | Lipase | 15 |
| PF00152 | 1 | 3 | 5 | 2 | 2 | 3 | 4 | 2 | "tRNA synthetases class II (D, K and N)" | 22 |
| PF00153 | 14 | 22 | 52 | 24 | 13 | 29 | 58 | 16 | Mitochondrial carrier protein | 228 |
| PF00155 | 0 | 3 | 17 | 7 | 0 | 4 | 17 | 6 | Aminotransferase class I and II | 54 |
| PF00156 | 0 | 1 | 4 | 2 | 0 | 2 | 8 | 4 | Phosphoribosyl transferase domain | 21 |
| PF00157 | 0 | 0 | 13 | 0 | 0 | 0 | 17 | 0 | Pou domain - N-terminal to homeobox domain | 30 |
| PF00159 | 0 | 0 | 3 | 0 | 0 | 0 | 3 | 0 | Pancreatic hormone peptide | 6 |
| PF00160 | 2 | 4 | 17 | 10 | 0 | 5 | 17 | 5 | Cyclophilin type peptidyl-prolyl cis-trans isomerase/CLD | 60 |
| PF00162 | 0 | 1 | 2 | 0 | 0 | 1 | 1 | 1 | Phosphoglycerate kinase | 6 |
| PF00163 | 1 | 1 | 2 | 0 | 1 | 1 | 1 | 1 | Ribosomal protein S4/S9 N-terminal domain | 8 |
| PF00164 | 1 | 0 | 2 | 1 | 1 | 1 | 2 | 1 | Ribosomal protein S12/S23 | 9 |
| PF00166 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | Chaperonin 10 Kd subunit | 5 |
| PF00167 | 2 | 1 | 22 | 2 | 0 | 1 | 25 | 2 | Fibroblast growth factor | 55 |
| PF00168 | 47 | 46 | 117 | 32 | 32 | 53 | 145 | 26 | C2 domain | 498 |
| PF00169 | 34 | 36 | 170 | 39 | 29 | 37 | 196 | 16 | PH domain | 557 |
| PF00170 | 0 | 0 | 29 | 1 | 0 | 0 | 31 | 1 | bZIP transcription factor | 62 |
| PF00171 | 2 | 12 | 21 | 7 | 4 | 15 | 20 | 13 | Aldehyde dehydrogenase family | 94 |
| PF00173 | 1 | 7 | 15 | 4 | 1 | 5 | 9 | 3 | Cytochrome b5-like Heme/Steroid binding domain | 45 |
| PF00174 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | Oxidoreductase molybdopterin binding domain | 2 |
| PF00175 | 0 | 4 | 12 | 4 | 0 | 5 | 12 | 2 | Oxidoreductase NAD-binding domain | 39 |
| PF00176 | 0 | 0 | 31 | 4 | 0 | 0 | 29 | 4 | SNF2 family N-terminal domain | 68 |
| PF00177 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | Ribosomal protein S7p/S5e | 9 |
| PF00178 | 0 | 0 | 26 | 0 | 0 | 0 | 26 | 0 | Ets-domain | 52 |
| PF00179 | 4 | 7 | 40 | 10 | 2 | 5 | 40 | 10 | Ubiquitin-conjugating enzyme | 118 |
| PF00180 | 1 | 5 | 6 | 3 | 0 | 5 | 5 | 5 | Isocitrate/isopropylmalate dehydrogenase | 30 |
| PF00181 | 1 | 1 | 2 | 2 | 1 | 1 | 2 | 1 | "Ribosomal Proteins L2, RNA binding domain" | 11 |
| PF00183 | 0 | 4 | 4 | 2 | 0 | 4 | 5 | 4 | Hsp90 protein | 23 |
| PF00184 | 0 | 0 | 2 | 1 | 0 | 0 | 2 | 0 | "Neurohypophysial hormones, C-terminal Domain" | 5 |
| PF00185 | 0 | 1 | 2 | 0 | 0 | 1 | 2 | 0 | "Aspartate/ornithine carbamoyltransferase, Asp/Orn binding domain" | 6 |
| PF00186 | 0 | 0 | 1 | 1 | 0 | 0 | 2 | 0 | Dihydrofolate reductase | 4 |
| PF00188 | 1 | 1 | 13 | 0 | 1 | 1 | 11 | 1 | Cysteine-rich secretory protein family | 29 |
| PF00189 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | "Ribosomal protein S3, C-terminal domain" | 6 |
| PF00191 | 4 | 5 | 11 | 2 | 0 | 10 | 13 | 11 | Annexin | 56 |
| PF00193 | 7 | 6 | 14 | 2 | 4 | 6 | 16 | 2 | Extracellular link domain | 57 |
| PF00194 | 4 | 6 | 18 | 4 | 5 | 8 | 22 | 4 | Eukaryotic-type carbonic anhydrase | 71 |
| PF00198 | 3 | 4 | 4 | 2 | 4 | 4 | 3 | 3 | 2-oxoacid dehydrogenases acyltransferase (catalytic domain) | 27 |
| PF00199 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | Catalase | 3 |
| PF00200 | 2 | 4 | 35 | 2 | 3 | 6 | 17 | 3 | Disintegrin | 72 |
| PF00201 | 1 | 1 | 20 | 1 | 0 | 3 | 24 | 2 | UDP-glucoronosyl and UDP-glucosyl transferase | 52 |
| PF00202 | 0 | 3 | 5 | 4 | 0 | 2 | 4 | 2 | Aminotransferase class-III | 20 |
| PF00203 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | Ribosomal protein S19 | 7 |
| PF00204 | 0 | 0 | 2 | 0 | 1 | 0 | 2 | 0 | DNA gyrase B | 5 |
| PF00205 | 1 | 2 | 2 | 2 | 1 | 2 | 2 | 0 | "Thiamine pyrophosphate enzyme, central domain" | 12 |
| PF00206 | 0 | 2 | 3 | 1 | 0 | 2 | 3 | 3 | Lyase | 14 |
| PF00207 | 1 | 1 | 9 | 0 | 1 | 4 | 24 | 7 | Alpha-2-macroglobulin family | 47 |
| PF00208 | 1 | 1 | 1 | 0 | 2 | 2 | 1 | 1 | Glutamate/Leucine/Phenylalanine/Valine dehydrogenase | 9 |
| PF00209 | 0 | 8 | 20 | 3 | 0 | 10 | 34 | 6 | Sodium:neurotransmitter symporter family | 81 |
| PF00210 | 1 | 2 | 11 | 1 | 0 | 1 | 9 | 2 | Ferritin-like domain | 27 |
| PF00211 | 5 | 9 | 20 | 3 | 4 | 8 | 26 | 1 | Adenylate and Guanylate cyclase catalytic domain | 76 |
| PF00212 | 0 | 0 | 3 | 0 | 0 | 1 | 7 | 0 | Atrial natriuretic peptide | 11 |
| PF00213 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | ATP synthase delta (OSCP) subunit | 7 |
| PF00214 | 0 | 0 | 5 | 0 | 0 | 0 | 6 | 0 | Calcitonin / CGRP / IAPP family | 11 |
| PF00215 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | Orotidine 5'-phosphate decarboxylase / HUMPS family | 4 |
| PF00217 | 1 | 2 | 4 | 3 | 3 | 5 | 7 | 6 | "ATP:guanido phosphotransferase, C-terminal catalytic domain" | 31 |
| PF00219 | 1 | 1 | 17 | 2 | 0 | 0 | 19 | 0 | Insulin-like growth factor binding protein | 40 |
| PF00220 | 0 | 0 | 2 | 1 | 0 | 0 | 2 | 0 | "Neurohypophysial hormones, N-terminal Domain" | 5 |
| PF00221 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Aromatic amino acid lyase | 2 |
| PF00224 | 0 | 1 | 2 | 0 | 1 | 1 | 1 | 0 | "Pyruvate kinase, barrel domain" | 6 |
| PF00225 | 5 | 7 | 41 | 3 | 3 | 8 | 49 | 3 | Kinesin motor domain | 119 |
| PF00226 | 17 | 20 | 49 | 16 | 13 | 19 | 50 | 6 | DnaJ domain | 190 |
| PF00227 | 8 | 13 | 19 | 8 | 0 | 15 | 22 | 15 | Proteasome subunit | 100 |
| PF00229 | 0 | 0 | 19 | 2 | 0 | 0 | 17 | 0 | TNF(Tumour Necrosis Factor) family | 38 |
| PF00230 | 1 | 2 | 12 | 0 | 1 | 1 | 15 | 0 | Major intrinsic protein | 32 |
| PF00231 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | ATP synthase | 6 |
| PF00232 | 0 | 0 | 4 | 0 | 0 | 0 | 7 | 0 | Glycosyl hydrolase family 1 | 11 |
| PF00233 | 7 | 6 | 21 | 8 | 0 | 1 | 24 | 1 | 3'5'-cyclic nucleotide phosphodiesterase | 68 |
| PF00235 | 0 | 1 | 4 | 1 | 0 | 3 | 4 | 3 | Profilin | 16 |
| PF00236 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Glycoprotein hormone | 3 |
| PF00237 | 2 | 2 | 2 | 1 | 1 | 1 | 2 | 1 | Ribosomal protein L22p/L17e | 12 |
| PF00238 | 2 | 1 | 2 | 1 | 1 | 2 | 2 | 1 | Ribosomal protein L14p/L23e | 12 |
| PF00240 | 5 | 9 | 43 | 9 | 5 | 9 | 35 | 12 | Ubiquitin family | 127 |
| PF00241 | 7 | 9 | 9 | 5 | 4 | 7 | 12 | 7 | Cofilin/tropomyosin-type actin-binding protein | 60 |
| PF00243 | 0 | 0 | 4 | 0 | 0 | 1 | 4 | 0 | Nerve growth factor family | 9 |
| PF00244 | 6 | 6 | 7 | 5 | 5 | 11 | 11 | 11 | 14-3-3 protein | 62 |
| PF00245 | 1 | 1 | 4 | 0 | 1 | 1 | 3 | 1 | Alkaline phosphatase | 12 |
| PF00246 | 1 | 3 | 21 | 2 | 0 | 2 | 21 | 4 | Zinc carboxypeptidase | 54 |
| PF00248 | 2 | 7 | 21 | 3 | 2 | 4 | 14 | 5 | Aldo/keto reductase family | 58 |
| PF00249 | 0 | 0 | 20 | 2 | 1 | 1 | 23 | 3 | Myb-like DNA-binding domain | 50 |
| PF00250 | 0 | 0 | 44 | 2 | 0 | 0 | 62 | 1 | Fork head domain | 109 |
| PF00252 | 1 | 1 | 3 | 0 | 1 | 1 | 2 | 1 | Ribosomal protein L16p/L10e | 10 |
| PF00253 | 1 | 1 | 2 | 0 | 2 | 2 | 2 | 1 | Ribosomal protein S14p/S29e | 11 |
| PF00254 | 2 | 7 | 16 | 5 | 1 | 5 | 16 | 6 | FKBP-type peptidyl-prolyl cis-trans isomerase | 58 |
| PF00255 | 2 | 2 | 8 | 1 | 1 | 2 | 8 | 2 | Glutathione peroxidase | 26 |
| PF00258 | 0 | 2 | 7 | 2 | 0 | 3 | 8 | 0 | Flavodoxin | 22 |
| PF00260 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Protamine P1 | 1 |
| PF00261 | 2 | 2 | 4 | 2 | 3 | 3 | 5 | 3 | Tropomyosin | 24 |
| PF00262 | 0 | 2 | 5 | 2 | 0 | 5 | 4 | 3 | Calreticulin family | 21 |
| PF00264 | 0 | 0 | 3 | 0 | 0 | 0 | 4 | 0 | Common central domain of tyrosinase | 7 |
| PF00265 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Thymidine kinase | 1 |
| PF00266 | 1 | 2 | 6 | 2 | 1 | 2 | 5 | 3 | Aminotransferase class-V | 22 |
| PF00268 | 0 | 0 | 2 | 0 | 0 | 0 | 3 | 0 | "Ribonucleotide reductase, small chain" | 5 |
| PF00270 | 13 | 10 | 70 | 19 | 10 | 12 | 54 | 16 | DEAD/DEAH box helicase | 204 |
| PF00271 | 13 | 10 | 105 | 25 | 11 | 13 | 88 | 20 | Helicase conserved C-terminal domain | 285 |
| PF00273 | 0 | 1 | 5 | 1 | 0 | 0 | 1 | 1 | Serum albumin family | 9 |
| PF00274 | 1 | 2 | 5 | 4 | 3 | 4 | 5 | 5 | Fructose-bisphosphate aldolase class-I | 29 |
| PF00276 | 2 | 2 | 2 | 0 | 2 | 2 | 2 | 1 | Ribosomal protein L23 | 13 |
| PF00277 | 0 | 0 | 4 | 0 | 0 | 0 | 1 | 0 | Serum amyloid A protein | 5 |
| PF00278 | 0 | 0 | 4 | 0 | 0 | 0 | 3 | 0 | "Pyridoxal-dependent decarboxylase, C-terminal sheet domain" | 7 |
| PF00281 | 2 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | Ribosomal protein L5 | 8 |
| PF00282 | 1 | 3 | 8 | 6 | 0 | 2 | 8 | 3 | Pyridoxal-dependent decarboxylase conserved domain | 31 |
| PF00285 | 0 | 2 | 3 | 2 | 0 | 2 | 1 | 0 | Citrate synthase | 10 |
| PF00287 | 0 | 3 | 5 | 3 | 0 | 7 | 6 | 6 | Sodium / potassium ATPase beta chain | 30 |
| PF00288 | 0 | 0 | 5 | 4 | 0 | 0 | 5 | 1 | GHMP kinases N terminal domain | 15 |
| PF00289 | 1 | 5 | 7 | 1 | 0 | 5 | 6 | 2 | "Carbamoyl-phosphate synthase L chain, N-terminal domain" | 27 |
| PF00291 | 1 | 2 | 6 | 4 | 0 | 0 | 6 | 2 | Pyridoxal-phosphate dependent enzyme | 21 |
| PF00292 | 0 | 0 | 9 | 0 | 0 | 0 | 12 | 0 | 'Paired box' domain | 21 |
| PF00293 | 1 | 3 | 22 | 6 | 0 | 2 | 19 | 5 | NUDIX domain | 58 |
| PF00294 | 0 | 1 | 4 | 2 | 0 | 2 | 7 | 2 | pfkB family carbohydrate kinase | 18 |
| PF00297 | 1 | 1 | 3 | 1 | 1 | 1 | 1 | 1 | Ribosomal protein L3 | 10 |
| PF00298 | 2 | 2 | 2 | 1 | 1 | 2 | 2 | 1 | "Ribosomal protein L11, RNA binding domain" | 13 |
| PF00300 | 3 | 5 | 11 | 5 | 1 | 3 | 16 | 5 | Histidine phosphatase superfamily (branch 1) | 49 |
| PF00303 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Thymidylate synthase | 2 |
| PF00305 | 0 | 0 | 7 | 0 | 0 | 0 | 7 | 0 | Lipoxygenase | 14 |
| PF00306 | 2 | 4 | 5 | 4 | 2 | 6 | 6 | 6 | "ATP synthase alpha/beta chain, C terminal domain" | 35 |
| PF00307 | 23 | 22 | 65 | 22 | 22 | 26 | 85 | 21 | Calponin homology (CH) domain | 286 |
| PF00310 | 0 | 0 | 3 | 1 | 0 | 1 | 3 | 1 | Glutamine amidotransferases class-II | 9 |
| PF00312 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | Ribosomal protein S15 | 9 |
| PF00313 | 2 | 1 | 8 | 2 | 0 | 1 | 7 | 1 | 'Cold-shock' DNA-binding domain | 22 |
| PF00316 | 0 | 0 | 2 | 0 | 1 | 0 | 3 | 3 | Fructose-1-6-bisphosphatase | 9 |
| PF00317 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | "Ribonucleotide reductase, all-alpha domain" | 2 |
| PF00318 | 1 | 1 | 2 | 1 | 0 | 1 | 1 | 1 | Ribosomal protein S2 | 8 |
| PF00319 | 0 | 0 | 6 | 1 | 0 | 0 | 7 | 0 | SRF-type transcription factor (DNA-binding and dimerisation domain) | 14 |
| PF00320 | 0 | 0 | 14 | 3 | 0 | 0 | 15 | 1 | GATA zinc finger | 33 |
| PF00322 | 0 | 0 | 3 | 0 | 0 | 0 | 4 | 0 | Endothelin family | 7 |
| PF00323 | 0 | 0 | 16 | 0 | 0 | 0 | 0 | 0 | Mammalian defensin | 16 |
| PF00324 | 1 | 4 | 9 | 0 | 0 | 4 | 12 | 2 | Amino acid permease | 32 |
| PF00326 | 1 | 3 | 9 | 6 | 0 | 5 | 9 | 4 | Prolyl oligopeptidase family | 37 |
| PF00327 | 1 | 1 | 3 | 1 | 1 | 1 | 3 | 1 | Ribosomal protein L30p/L7e | 12 |
| PF00328 | 0 | 2 | 7 | 3 | 0 | 2 | 8 | 0 | Histidine phosphatase superfamily (branch 2) | 22 |
| PF00329 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | "Respiratory-chain NADH dehydrogenase, 30 Kd subunit" | 7 |
| PF00330 | 0 | 2 | 3 | 1 | 0 | 2 | 3 | 2 | Aconitase family (aconitate hydratase) | 13 |
| PF00333 | 1 | 1 | 2 | 1 | 1 | 1 | 2 | 1 | "Ribosomal protein S5, N-terminal domain" | 10 |
| PF00334 | 3 | 4 | 9 | 2 | 2 | 3 | 6 | 1 | Nucleoside diphosphate kinase | 30 |
| PF00335 | 2 | 6 | 31 | 11 | 1 | 7 | 49 | 5 | Tetraspanin family | 112 |
| PF00337 | 0 | 1 | 11 | 2 | 2 | 4 | 18 | 7 | Galactoside-binding lectin | 45 |
| PF00338 | 2 | 3 | 3 | 0 | 0 | 1 | 2 | 1 | Ribosomal protein S10p/S20e | 12 |
| PF00339 | 0 | 1 | 10 | 2 | 0 | 1 | 17 | 0 | "Arrestin (or S-antigen), N-terminal domain" | 31 |
| PF00340 | 0 | 0 | 9 | 1 | 0 | 0 | 2 | 0 | Interleukin-1 / 18 | 12 |
| PF00341 | 0 | 0 | 9 | 1 | 0 | 0 | 11 | 0 | PDGF/VEGF domain | 21 |
| PF00342 | 0 | 2 | 1 | 0 | 0 | 1 | 1 | 1 | Phosphoglucose isomerase | 6 |
| PF00343 | 0 | 2 | 3 | 2 | 0 | 2 | 4 | 4 | Carbohydrate phosphorylase | 17 |
| PF00344 | 1 | 0 | 2 | 1 | 0 | 1 | 2 | 1 | SecY translocase | 8 |
| PF00346 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | "Respiratory-chain NADH dehydrogenase, 49 Kd subunit" | 7 |
| PF00347 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | Ribosomal protein L6 | 7 |
| PF00348 | 0 | 1 | 4 | 1 | 0 | 1 | 4 | 2 | Polyprenyl synthetase | 13 |
| PF00349 | 2 | 2 | 5 | 1 | 2 | 2 | 3 | 2 | Hexokinase | 19 |
| PF00350 | 6 | 10 | 13 | 6 | 2 | 11 | 35 | 12 | Dynamin family | 95 |
| PF00351 | 0 | 0 | 4 | 1 | 1 | 1 | 6 | 0 | Biopterin-dependent aromatic amino acid hydroxylase | 13 |
| PF00352 | 0 | 0 | 3 | 0 | 0 | 0 | 3 | 0 | "Transcription factor TFIID (or TATA-binding protein, TBP)" | 6 |
| PF00354 | 1 | 2 | 11 | 3 | 1 | 4 | 21 | 6 | Pentaxin family | 49 |
| PF00355 | 1 | 2 | 3 | 1 | 2 | 2 | 7 | 1 | Rieske [2Fe-2S] domain | 19 |
| PF00357 | 0 | 3 | 10 | 0 | 0 | 1 | 3 | 1 | Integrin alpha cytoplasmic region | 18 |
| PF00359 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | "Phosphoenolpyruvate-dependent sugar phosphotransferase system, EIIA 2" | 2 |
| PF00361 | 0 | 3 | 0 | 1 | 1 | 3 | 3 | 3 | "NADH-Ubiquinone/plastoquinone (complex I), various chains" | 14 |
| PF00362 | 0 | 6 | 9 | 1 | 0 | 2 | 11 | 1 | "Integrin, beta chain" | 30 |
| PF00363 | 0 | 0 | 4 | 0 | 0 | 0 | 0 | 0 | Casein | 4 |
| PF00364 | 4 | 8 | 9 | 3 | 4 | 8 | 7 | 5 | Biotin-requiring enzyme | 48 |
| PF00365 | 3 | 3 | 3 | 2 | 5 | 5 | 5 | 4 | Phosphofructokinase | 30 |
| PF00366 | 1 | 1 | 2 | 2 | 1 | 1 | 3 | 1 | Ribosomal protein S17 | 12 |
| PF00368 | 0 | 1 | 1 | 0 | 0 | 0 | 1 | 0 | Hydroxymethylglutaryl-coenzyme A reductase | 3 |
| PF00370 | 1 | 1 | 7 | 1 | 0 | 1 | 4 | 0 | "FGGY family of carbohydrate kinases, N-terminal domain" | 15 |
| PF00373 | 7 | 9 | 42 | 9 | 8 | 16 | 59 | 9 | FERM central domain | 159 |
| PF00375 | 1 | 5 | 7 | 5 | 0 | 5 | 8 | 2 | Sodium:dicarboxylate symporter family | 33 |
| PF00377 | 0 | 1 | 3 | 1 | 0 | 0 | 0 | 0 | Prion/Doppel alpha-helical domain | 5 |
| PF00378 | 3 | 7 | 14 | 5 | 4 | 9 | 10 | 7 | Enoyl-CoA hydratase/isomerase family | 59 |
| PF00380 | 1 | 1 | 2 | 0 | 1 | 1 | 2 | 1 | Ribosomal protein S9/S16 | 9 |
| PF00382 | 0 | 0 | 2 | 1 | 0 | 0 | 2 | 0 | Transcription factor TFIIB repeat | 5 |
| PF00383 | 0 | 0 | 5 | 1 | 0 | 0 | 6 | 0 | Cytidine and deoxycytidylate deaminase zinc-binding region | 12 |
| PF00384 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | Molybdopterin oxidoreductase | 5 |
| PF00385 | 0 | 0 | 23 | 3 | 0 | 0 | 23 | 1 | Chromo (CHRromatin Organisation MOdifier) domain | 50 |
| PF00386 | 5 | 3 | 29 | 6 | 0 | 1 | 55 | 1 | C1q domain | 100 |
| PF00387 | 4 | 5 | 15 | 2 | 1 | 6 | 17 | 1 | "Phosphatidylinositol-specific phospholipase C, Y domain" | 51 |
| PF00388 | 4 | 5 | 18 | 2 | 1 | 6 | 25 | 3 | "Phosphatidylinositol-specific phospholipase C, X domain" | 64 |
| PF00389 | 1 | 3 | 4 | 2 | 3 | 4 | 7 | 5 | "D-isomer specific 2-hydroxyacid dehydrogenase, catalytic domain" | 29 |
| PF00390 | 0 | 3 | 3 | 1 | 0 | 2 | 2 | 1 | "Malic enzyme, N-terminal domain" | 12 |
| PF00393 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | "6-phosphogluconate dehydrogenase, C-terminal domain" | 5 |
| PF00394 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Multicopper oxidase | 1 |
| PF00396 | 0 | 0 | 1 | 0 | 0 | 0 | 4 | 0 | Granulin | 5 |
| PF00397 | 12 | 12 | 43 | 18 | 15 | 15 | 38 | 4 | WW domain | 157 |
| PF00398 | 0 | 0 | 3 | 1 | 0 | 0 | 3 | 0 | Ribosomal RNA adenine dimethylase | 7 |
| PF00400 | 30 | 43 | 233 | 75 | 30 | 43 | 219 | 29 | "WD domain, G-beta repeat" | 702 |
| PF00402 | 2 | 2 | 6 | 5 | 1 | 2 | 7 | 6 | Calponin family repeat | 31 |
| PF00403 | 0 | 0 | 4 | 1 | 0 | 0 | 4 | 2 | Heavy-metal-associated domain | 11 |
| PF00405 | 1 | 1 | 4 | 2 | 1 | 2 | 4 | 1 | Transferrin | 16 |
| PF00406 | 5 | 6 | 8 | 4 | 2 | 5 | 9 | 7 | Adenylate kinase | 46 |
| PF00408 | 0 | 2 | 4 | 1 | 0 | 1 | 4 | 3 | "Phosphoglucomutase/phosphomannomutase, C-terminal domain" | 15 |
| PF00410 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | Ribosomal protein S8 | 7 |
| PF00411 | 1 | 1 | 2 | 0 | 1 | 1 | 2 | 1 | Ribosomal protein S11 | 9 |
| PF00412 | 13 | 13 | 71 | 20 | 12 | 9 | 91 | 16 | LIM domain | 245 |
| PF00413 | 1 | 0 | 21 | 0 | 0 | 0 | 21 | 0 | Matrixin | 43 |
| PF00414 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | Neuraxin and MAP1B repeat | 3 |
| PF00415 | 2 | 2 | 19 | 6 | 2 | 1 | 22 | 2 | Regulator of chromosome condensation (RCC1) repeat | 56 |
| PF00416 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 0 | Ribosomal protein S13/S18 | 5 |
| PF00418 | 1 | 3 | 3 | 0 | 5 | 5 | 4 | 4 | "Tau and MAP protein, tubulin-binding repeat" | 25 |
| PF00420 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | NADH-ubiquinone/plastoquinone oxidoreductase chain 4L | 2 |
| PF00428 | 4 | 4 | 3 | 2 | 4 | 4 | 3 | 3 | 60s Acidic ribosomal protein | 27 |
| PF00429 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | ENV polyprotein (coat polyprotein) | 2 |
| PF00431 | 2 | 7 | 49 | 3 | 0 | 8 | 49 | 2 | CUB domain | 120 |
| PF00432 | 0 | 0 | 4 | 2 | 0 | 0 | 3 | 0 | Prenyltransferase and squalene oxidase repeat | 9 |
| PF00433 | 6 | 7 | 33 | 12 | 3 | 10 | 38 | 10 | Protein kinase C terminal domain | 119 |
| PF00435 | 13 | 13 | 22 | 3 | 17 | 15 | 23 | 7 | Spectrin repeat | 113 |
| PF00436 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | Single-strand binding protein family | 6 |
| PF00438 | 0 | 1 | 2 | 1 | 0 | 1 | 4 | 1 | "S-adenosylmethionine synthetase, N-terminal domain" | 10 |
| PF00439 | 0 | 0 | 37 | 5 | 0 | 0 | 38 | 3 | Bromodomain | 83 |
| PF00441 | 5 | 10 | 14 | 4 | 4 | 13 | 13 | 9 | "Acyl-CoA dehydrogenase, C-terminal domain" | 72 |
| PF00443 | 6 | 8 | 55 | 14 | 1 | 6 | 46 | 5 | Ubiquitin carboxyl-terminal hydrolase | 141 |
| PF00444 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Ribosomal protein L36 | 2 |
| PF00445 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | Ribonuclease T2 family | 4 |
| PF00446 | 0 | 0 | 1 | 0 | 0 | 0 | 2 | 0 | Gonadotropin-releasing hormone | 3 |
| PF00447 | 0 | 0 | 6 | 0 | 0 | 0 | 4 | 0 | HSF-type DNA-binding | 10 |
| PF00448 | 1 | 1 | 3 | 1 | 1 | 1 | 1 | 0 | "SRP54-type protein, GTPase domain" | 9 |
| PF00450 | 0 | 0 | 3 | 1 | 0 | 0 | 3 | 0 | Serine carboxypeptidase | 7 |
| PF00452 | 0 | 2 | 13 | 2 | 0 | 3 | 13 | 1 | "Apoptosis regulator proteins, Bcl-2 family" | 34 |
| PF00453 | 0 | 0 | 1 | 0 | 1 | 1 | 1 | 0 | Ribosomal protein L20 | 4 |
| PF00454 | 2 | 5 | 17 | 3 | 3 | 4 | 18 | 1 | Phosphatidylinositol 3- and 4-kinase | 53 |
| PF00456 | 0 | 2 | 3 | 0 | 1 | 2 | 2 | 2 | "Transketolase, thiamine diphosphate binding domain" | 12 |
| PF00458 | 0 | 4 | 5 | 3 | 2 | 4 | 4 | 4 | WHEP-TRS domain | 26 |
| PF00459 | 0 | 4 | 5 | 4 | 0 | 2 | 5 | 3 | Inositol monophosphatase family | 23 |
| PF00462 | 2 | 4 | 6 | 3 | 0 | 4 | 3 | 2 | Glutaredoxin | 24 |
| PF00464 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 0 | Serine hydroxymethyltransferase | 8 |
| PF00465 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Iron-containing alcohol dehydrogenase | 2 |
| PF00466 | 2 | 2 | 2 | 2 | 1 | 1 | 3 | 1 | Ribosomal protein L10 | 14 |
| PF00467 | 3 | 2 | 6 | 2 | 3 | 3 | 5 | 2 | KOW motif | 26 |
| PF00468 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Ribosomal protein L34 | 2 |
| PF00472 | 1 | 1 | 4 | 3 | 0 | 0 | 4 | 0 | RF-1 domain | 13 |
| PF00473 | 0 | 0 | 2 | 0 | 0 | 0 | 3 | 0 | Corticotropin-releasing factor family | 5 |
| PF00474 | 0 | 1 | 14 | 0 | 0 | 0 | 13 | 0 | Sodium:solute symporter family | 28 |
| PF00476 | 0 | 0 | 3 | 0 | 0 | 0 | 2 | 0 | DNA polymerase family A | 5 |
| PF00478 | 0 | 1 | 4 | 2 | 1 | 2 | 5 | 3 | IMP dehydrogenase / GMP reductase domain | 18 |
| PF00479 | 0 | 1 | 3 | 2 | 0 | 1 | 2 | 1 | "Glucose-6-phosphate dehydrogenase, NAD binding domain" | 10 |
| PF00480 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | ROK family | 2 |
| PF00481 | 2 | 3 | 19 | 9 | 0 | 5 | 23 | 6 | Protein phosphatase 2C | 67 |
| PF00483 | 0 | 0 | 3 | 3 | 0 | 1 | 7 | 1 | Nucleotidyl transferase | 15 |
| PF00485 | 0 | 0 | 3 | 1 | 0 | 0 | 6 | 0 | Phosphoribulokinase / Uridine kinase family | 10 |
| PF00487 | 0 | 2 | 12 | 0 | 0 | 1 | 6 | 0 | Fatty acid desaturase | 21 |
| PF00488 | 0 | 0 | 5 | 0 | 0 | 0 | 4 | 0 | MutS domain V | 9 |
| PF00489 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | Interleukin-6/G-CSF/MGF family | 3 |
| PF00490 | 0 | 1 | 1 | 1 | 0 | 1 | 0 | 0 | Delta-aminolevulinic acid dehydratase | 4 |
| PF00491 | 0 | 0 | 3 | 0 | 0 | 0 | 3 | 2 | Arginase family | 8 |
| PF00493 | 0 | 0 | 9 | 0 | 0 | 0 | 8 | 0 | MCM2/3/5 family | 17 |
| PF00494 | 1 | 1 | 2 | 2 | 1 | 1 | 1 | 0 | Squalene/phytoene synthase | 9 |
| PF00497 | 7 | 6 | 10 | 4 | 17 | 17 | 22 | 5 | "Bacterial extracellular solute-binding proteins, family 3" | 88 |
| PF00498 | 4 | 6 | 26 | 4 | 4 | 5 | 32 | 2 | FHA domain | 83 |
| PF00499 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | NADH-ubiquinone/plastoquinone oxidoreductase chain 6 | 1 |
| PF00501 | 5 | 11 | 29 | 10 | 3 | 13 | 30 | 7 | AMP-binding enzyme | 108 |
| PF00503 | 9 | 11 | 16 | 9 | 10 | 15 | 27 | 10 | G-protein alpha subunit | 107 |
| PF00505 | 2 | 1 | 47 | 7 | 5 | 6 | 53 | 8 | HMG (high mobility group) box | 129 |
| PF00507 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | "NADH-ubiquinone/plastoquinone oxidoreductase, chain 3" | 2 |
| PF00510 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | Cytochrome c oxidase subunit III | 2 |
| PF00514 | 8 | 12 | 28 | 11 | 12 | 15 | 31 | 8 | Armadillo/beta-catenin-like repeat | 125 |
| PF00515 | 7 | 15 | 56 | 18 | 5 | 13 | 46 | 11 | Tetratricopeptide repeat | 171 |
| PF00520 | 18 | 35 | 105 | 14 | 13 | 25 | 116 | 6 | Ion transport protein | 332 |
| PF00521 | 0 | 0 | 2 | 0 | 1 | 0 | 2 | 0 | "DNA gyrase/topoisomerase IV, subunit A" | 5 |
| PF00525 | 1 | 1 | 3 | 1 | 1 | 0 | 2 | 2 | "Alpha crystallin A chain, N terminal" | 11 |
| PF00530 | 0 | 0 | 22 | 1 | 1 | 1 | 31 | 1 | Scavenger receptor cysteine-rich domain | 57 |
| PF00531 | 3 | 3 | 34 | 8 | 5 | 7 | 32 | 4 | Death domain | 96 |
| PF00533 | 0 | 0 | 16 | 1 | 1 | 1 | 14 | 1 | BRCA1 C Terminus (BRCT) domain | 34 |
| PF00534 | 1 | 1 | 5 | 3 | 0 | 1 | 5 | 0 | Glycosyl transferases group 1 | 16 |
| PF00535 | 3 | 3 | 25 | 5 | 4 | 6 | 25 | 0 | Glycosyl transferase family 2 | 71 |
| PF00536 | 12 | 11 | 61 | 12 | 16 | 22 | 74 | 7 | SAM domain (Sterile alpha motif) | 215 |
| PF00538 | 1 | 0 | 12 | 5 | 3 | 3 | 19 | 16 | linker histone H1 and H5 family | 59 |
| PF00542 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | Ribosomal protein L7/L12 C-terminal domain | 8 |
| PF00549 | 1 | 3 | 4 | 2 | 0 | 4 | 4 | 3 | CoA-ligase | 21 |
| PF00550 | 0 | 3 | 5 | 2 | 2 | 4 | 5 | 3 | Phosphopantetheine attachment site | 24 |
| PF00551 | 0 | 1 | 4 | 0 | 0 | 2 | 4 | 2 | Formyl transferase | 13 |
| PF00554 | 0 | 0 | 10 | 0 | 0 | 0 | 9 | 0 | Rel homology domain (RHD) | 19 |
| PF00557 | 2 | 2 | 9 | 5 | 3 | 5 | 9 | 5 | Metallopeptidase family M24 | 40 |
| PF00560 | 13 | 18 | 92 | 20 | 15 | 20 | 99 | 3 | Leucine Rich Repeat | 280 |
| PF00561 | 0 | 1 | 9 | 2 | 0 | 2 | 4 | 1 | alpha/beta hydrolase fold | 19 |
| PF00562 | 0 | 0 | 3 | 0 | 0 | 0 | 2 | 0 | "RNA polymerase Rpb2, domain 6" | 5 |
| PF00564 | 1 | 1 | 14 | 3 | 0 | 1 | 15 | 0 | PB1 domain | 35 |
| PF00565 | 1 | 1 | 1 | 1 | 1 | 2 | 2 | 0 | Staphylococcal nuclease homologue | 9 |
| PF00566 | 4 | 4 | 39 | 10 | 0 | 4 | 43 | 2 | Rab-GTPase-TBC domain | 106 |
| PF00567 | 2 | 2 | 11 | 1 | 0 | 3 | 14 | 1 | Tudor domain | 34 |
| PF00568 | 9 | 8 | 11 | 6 | 8 | 8 | 12 | 0 | WH1 domain | 62 |
| PF00569 | 2 | 4 | 17 | 3 | 4 | 1 | 12 | 2 | "Zinc finger, ZZ type" | 45 |
| PF00570 | 0 | 0 | 3 | 1 | 0 | 0 | 2 | 0 | HRDC domain | 6 |
| PF00571 | 2 | 10 | 19 | 6 | 2 | 6 | 22 | 2 | CBS domain | 69 |
| PF00572 | 1 | 1 | 2 | 1 | 1 | 1 | 2 | 1 | Ribosomal protein L13 | 10 |
| PF00573 | 1 | 1 | 2 | 1 | 1 | 1 | 2 | 1 | Ribosomal protein L4/L1 family | 10 |
| PF00574 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | Clp protease | 5 |
| PF00575 | 0 | 1 | 8 | 1 | 0 | 1 | 8 | 0 | S1 RNA binding domain | 19 |
| PF00576 | 0 | 1 | 2 | 1 | 0 | 0 | 2 | 0 | HIUase/Transthyretin family | 6 |
| PF00578 | 3 | 5 | 6 | 5 | 3 | 6 | 6 | 6 | AhpC/TSA family | 40 |
| PF00579 | 2 | 3 | 4 | 4 | 0 | 1 | 3 | 1 | tRNA synthetases class I (W and Y) | 18 |
| PF00580 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | UvrD/REP helicase N-terminal domain | 2 |
| PF00581 | 1 | 2 | 23 | 8 | 0 | 0 | 16 | 1 | Rhodanese-like domain | 51 |
| PF00583 | 1 | 1 | 24 | 7 | 0 | 0 | 19 | 2 | Acetyltransferase (GNAT) family | 54 |
| PF00584 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | 0 | SecE/Sec61-gamma subunits of protein translocation complex | 4 |
| PF00586 | 0 | 0 | 4 | 1 | 0 | 0 | 3 | 1 | "AIR synthase related protein, N-terminal domain" | 9 |
| PF00587 | 1 | 6 | 8 | 3 | 2 | 4 | 7 | 4 | "tRNA synthetase class II core domain (G, H, P, S and T)" | 35 |
| PF00588 | 0 | 0 | 3 | 0 | 0 | 0 | 5 | 0 | SpoU rRNA Methylase family | 8 |
| PF00590 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Tetrapyrrole (Corrin/Porphyrin) Methylases | 1 |
| PF00591 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | "Glycosyl transferase family, a/b domain" | 1 |
| PF00594 | 0 | 0 | 16 | 1 | 1 | 1 | 19 | 2 | Vitamin K-dependent carboxylation/gamma-carboxyglutamic (GLA) domain | 40 |
| PF00595 | 53 | 44 | 132 | 43 | 60 | 62 | 169 | 16 | PDZ domain (Also known as DHR or GLGF) | 579 |
| PF00596 | 3 | 2 | 4 | 3 | 4 | 4 | 4 | 3 | Class II Aldolase and Adducin N-terminal domain | 27 |
| PF00605 | 0 | 0 | 9 | 1 | 0 | 0 | 12 | 0 | Interferon regulatory factor transcription factor | 22 |
| PF00609 | 5 | 4 | 9 | 3 | 2 | 6 | 8 | 2 | Diacylglycerol kinase accessory domain | 39 |
| PF00610 | 4 | 4 | 20 | 4 | 6 | 8 | 26 | 1 | "Domain found in Dishevelled, Egl-10, and Pleckstrin (DEP)" | 73 |
| PF00611 | 5 | 8 | 20 | 6 | 2 | 7 | 24 | 5 | "Fes/CIP4, and EFC/F-BAR homology domain" | 77 |
| PF00612 | 9 | 9 | 46 | 9 | 11 | 8 | 43 | 5 | IQ calmodulin-binding motif | 140 |
| PF00613 | 1 | 3 | 8 | 1 | 1 | 1 | 10 | 0 | "Phosphoinositide 3-kinase family, accessory domain (PIK domain)" | 25 |
| PF00614 | 1 | 1 | 3 | 1 | 1 | 0 | 4 | 0 | Phospholipase D Active site motif | 11 |
| PF00615 | 9 | 12 | 33 | 12 | 8 | 12 | 33 | 1 | Regulator of G protein signaling domain | 120 |
| PF00616 | 6 | 5 | 14 | 4 | 5 | 6 | 16 | 4 | GTPase-activator protein for Ras-like GTPase | 60 |
| PF00617 | 8 | 4 | 25 | 6 | 4 | 5 | 29 | 0 | RasGEF domain | 81 |
| PF00618 | 8 | 4 | 28 | 4 | 4 | 5 | 23 | 0 | RasGEF N-terminal motif | 76 |
| PF00619 | 0 | 0 | 25 | 4 | 1 | 0 | 26 | 3 | Caspase recruitment domain | 59 |
| PF00620 | 14 | 14 | 66 | 15 | 10 | 12 | 80 | 6 | RhoGAP domain | 217 |
| PF00621 | 16 | 16 | 66 | 7 | 12 | 15 | 81 | 4 | RhoGEF domain | 217 |
| PF00622 | 8 | 6 | 79 | 11 | 7 | 9 | 237 | 9 | SPRY domain | 366 |
| PF00623 | 0 | 0 | 3 | 1 | 0 | 0 | 0 | 0 | "RNA polymerase Rpb1, domain 2" | 4 |
| PF00625 | 19 | 15 | 25 | 13 | 28 | 25 | 38 | 6 | Guanylate kinase | 169 |
| PF00626 | 6 | 5 | 14 | 7 | 8 | 10 | 14 | 4 | Gelsolin repeat | 68 |
| PF00627 | 3 | 11 | 30 | 8 | 2 | 6 | 26 | 8 | UBA/TS-N domain | 94 |
| PF00628 | 0 | 0 | 63 | 9 | 1 | 0 | 62 | 2 | PHD-finger | 137 |
| PF00629 | 4 | 4 | 17 | 3 | 2 | 6 | 21 | 0 | MAM domain | 57 |
| PF00630 | 3 | 3 | 11 | 2 | 7 | 7 | 14 | 3 | Filamin/ABP280 repeat | 50 |
| PF00631 | 7 | 8 | 16 | 10 | 9 | 12 | 19 | 6 | GGL domain | 87 |
| PF00632 | 4 | 7 | 27 | 10 | 6 | 8 | 31 | 4 | HECT-domain (ubiquitin-transferase) | 97 |
| PF00633 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | Helix-hairpin-helix motif | 4 |
| PF00634 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | BRCA2 repeat | 2 |
| PF00635 | 2 | 2 | 6 | 2 | 2 | 2 | 5 | 2 | MSP (Major sperm protein) domain | 23 |
| PF00636 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | Ribonuclease III domain | 4 |
| PF00637 | 4 | 5 | 6 | 3 | 1 | 6 | 9 | 2 | Region in Clathrin and VPS | 36 |
| PF00638 | 0 | 0 | 4 | 0 | 0 | 0 | 6 | 1 | RanBP1 domain | 11 |
| PF00639 | 1 | 1 | 2 | 1 | 0 | 1 | 0 | 0 | PPIC-type PPIASE domain | 6 |
| PF00640 | 6 | 6 | 27 | 9 | 7 | 11 | 33 | 2 | Phosphotyrosine interaction domain (PTB/PID) | 101 |
| PF00641 | 2 | 0 | 16 | 6 | 0 | 0 | 18 | 3 | Zn-finger in Ran binding protein and others | 45 |
| PF00642 | 1 | 0 | 38 | 5 | 0 | 0 | 31 | 1 | Zinc finger C-x8-C-x5-C-x3-H type (and similar) | 76 |
| PF00643 | 5 | 6 | 64 | 9 | 8 | 6 | 146 | 3 | B-box zinc finger | 247 |
| PF00644 | 0 | 0 | 15 | 2 | 3 | 2 | 19 | 2 | Poly(ADP-ribose) polymerase catalytic domain | 43 |
| PF00645 | 0 | 0 | 2 | 1 | 1 | 1 | 2 | 1 | Poly(ADP-ribose) polymerase and DNA-Ligase Zn-finger region | 8 |
| PF00646 | 1 | 1 | 19 | 3 | 0 | 0 | 26 | 1 | F-box domain | 51 |
| PF00647 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | "Elongation factor 1 gamma, conserved domain" | 8 |
| PF00648 | 1 | 1 | 13 | 4 | 0 | 3 | 21 | 3 | Calpain family cysteine protease | 46 |
| PF00650 | 0 | 1 | 12 | 4 | 0 | 2 | 9 | 1 | CRAL/TRIO domain | 29 |
| PF00651 | 3 | 2 | 154 | 10 | 7 | 7 | 130 | 1 | BTB/POZ domain | 314 |
| PF00652 | 1 | 1 | 20 | 3 | 3 | 5 | 21 | 1 | Ricin-type beta-trefoil lectin domain | 55 |
| PF00653 | 0 | 0 | 10 | 1 | 0 | 0 | 6 | 0 | Inhibitor of Apoptosis domain | 17 |
| PF00654 | 2 | 5 | 9 | 4 | 1 | 3 | 11 | 0 | Voltage gated chloride channel | 35 |
| PF00656 | 0 | 0 | 12 | 1 | 0 | 0 | 22 | 1 | Caspase domain | 36 |
| PF00657 | 0 | 0 | 3 | 1 | 0 | 0 | 3 | 0 | GDSL-like Lipase/Acylhydrolase | 7 |
| PF00658 | 1 | 1 | 6 | 0 | 1 | 1 | 5 | 3 | "Poly-adenylate binding protein, unique domain" | 18 |
| PF00659 | 0 | 0 | 5 | 1 | 0 | 0 | 5 | 0 | POLO box duplicated region | 11 |
| PF00662 | 0 | 1 | 0 | 0 | 0 | 1 | 1 | 1 | "NADH-Ubiquinone oxidoreductase (complex I), chain 5 N-terminus" | 4 |
| PF00664 | 3 | 8 | 23 | 4 | 0 | 7 | 27 | 1 | ABC transporter transmembrane region | 73 |
| PF00665 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | Integrase core domain | 2 |
| PF00666 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | Cathelicidin | 2 |
| PF00667 | 0 | 2 | 6 | 2 | 0 | 3 | 7 | 0 | FAD binding domain | 20 |
| PF00670 | 0 | 3 | 3 | 1 | 0 | 3 | 4 | 3 | "S-adenosyl-L-homocysteine hydrolase, NAD binding domain" | 17 |
| PF00673 | 2 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | ribosomal L5P family C-terminus | 8 |
| PF00675 | 0 | 4 | 6 | 3 | 0 | 6 | 8 | 5 | Insulinase (Peptidase family M16) | 32 |
| PF00676 | 3 | 3 | 6 | 3 | 4 | 6 | 6 | 5 | Dehydrogenase E1 component | 36 |
| PF00679 | 0 | 2 | 6 | 3 | 1 | 3 | 8 | 3 | Elongation factor G C-terminus | 26 |
| PF00681 | 2 | 2 | 6 | 0 | 3 | 2 | 6 | 5 | Plectin repeat | 26 |
| PF00682 | 0 | 2 | 3 | 1 | 0 | 2 | 3 | 2 | HMGL-like | 13 |
| PF00683 | 0 | 0 | 6 | 0 | 0 | 0 | 5 | 0 | TB domain | 11 |
| PF00684 | 3 | 4 | 4 | 4 | 4 | 4 | 3 | 1 | DnaJ central domain | 27 |
| PF00685 | 0 | 0 | 41 | 1 | 2 | 2 | 41 | 4 | Sulfotransferase domain | 91 |
| PF00686 | 0 | 0 | 3 | 1 | 0 | 0 | 2 | 0 | Starch binding domain | 6 |
| PF00687 | 1 | 1 | 2 | 1 | 1 | 1 | 2 | 1 | Ribosomal protein L1p/L10e family | 10 |
| PF00688 | 0 | 1 | 24 | 0 | 0 | 0 | 33 | 0 | TGF-beta propeptide | 58 |
| PF00689 | 0 | 11 | 16 | 5 | 1 | 15 | 22 | 15 | "Cation transporting ATPase, C-terminus" | 85 |
| PF00690 | 0 | 12 | 18 | 5 | 1 | 15 | 23 | 15 | "Cation transporter/ATPase, N-terminus" | 89 |
| PF00692 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | dUTPase | 2 |
| PF00694 | 0 | 2 | 3 | 1 | 0 | 2 | 3 | 2 | Aconitase C-terminal domain | 13 |
| PF00696 | 0 | 1 | 1 | 0 | 1 | 1 | 0 | 0 | Amino acid kinase family | 4 |
| PF00698 | 0 | 2 | 2 | 2 | 0 | 1 | 4 | 0 | Acyl transferase domain | 11 |
| PF00701 | 0 | 0 | 2 | 1 | 0 | 0 | 2 | 0 | Dihydrodipicolinate synthetase family | 5 |
| PF00702 | 1 | 13 | 20 | 6 | 3 | 17 | 25 | 16 | haloacid dehalogenase-like hydrolase | 101 |
| PF00703 | 0 | 0 | 1 | 0 | 0 | 0 | 2 | 0 | Glycosyl hydrolases family 2 | 3 |
| PF00704 | 0 | 1 | 9 | 2 | 0 | 1 | 6 | 0 | Glycosyl hydrolases family 18 | 19 |
| PF00705 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | "Proliferating cell nuclear antigen, N-terminal domain" | 3 |
| PF00707 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | "Translation initiation factor IF-3, C-terminal domain" | 3 |
| PF00708 | 0 | 2 | 2 | 2 | 0 | 0 | 1 | 0 | Acylphosphatase | 7 |
| PF00709 | 0 | 0 | 2 | 2 | 0 | 0 | 2 | 1 | Adenylosuccinate synthetase | 7 |
| PF00710 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Asparaginase | 2 |
| PF00711 | 0 | 0 | 23 | 0 | 0 | 0 | 0 | 0 | Beta defensin | 23 |
| PF00714 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Interferon gamma | 2 |
| PF00715 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Interleukin 2 | 1 |
| PF00717 | 2 | 2 | 3 | 1 | 0 | 1 | 3 | 1 | Peptidase S24-like | 13 |
| PF00719 | 0 | 2 | 2 | 2 | 0 | 2 | 2 | 2 | Inorganic pyrophosphatase | 12 |
| PF00723 | 0 | 0 | 3 | 2 | 1 | 2 | 2 | 1 | Glycosyl hydrolases family 15 | 11 |
| PF00725 | 1 | 2 | 4 | 4 | 3 | 5 | 5 | 4 | "3-hydroxyacyl-CoA dehydrogenase, C-terminal domain" | 28 |
| PF00726 | 0 | 0 | 4 | 0 | 0 | 0 | 2 | 0 | Interleukin 10 | 6 |
| PF00727 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Interleukin 4 | 2 |
| PF00728 | 1 | 2 | 3 | 1 | 0 | 1 | 4 | 1 | "Glycosyl hydrolase family 20, catalytic domain" | 13 |
| PF00730 | 0 | 0 | 4 | 0 | 0 | 0 | 3 | 0 | HhH-GPD superfamily base excision DNA repair protein | 7 |
| PF00731 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | AIR carboxylase | 6 |
| PF00732 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | GMC oxidoreductase | 2 |
| PF00733 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | Asparagine synthase | 4 |
| PF00735 | 9 | 9 | 13 | 7 | 15 | 15 | 20 | 9 | Septin | 97 |
| PF00736 | 1 | 2 | 2 | 2 | 0 | 2 | 2 | 1 | EF-1 guanine nucleotide exchange domain | 12 |
| PF00743 | 0 | 0 | 9 | 1 | 0 | 3 | 5 | 1 | Flavin-binding monooxygenase-like | 19 |
| PF00748 | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 0 | Calpain inhibitor | 3 |
| PF00749 | 1 | 2 | 3 | 3 | 2 | 2 | 2 | 2 | "tRNA synthetases class I (E and Q), catalytic domain" | 17 |
| PF00750 | 1 | 1 | 2 | 1 | 1 | 1 | 2 | 1 | tRNA synthetases class I (R) | 10 |
| PF00751 | 0 | 0 | 7 | 0 | 0 | 0 | 5 | 0 | DM DNA binding domain | 12 |
| PF00752 | 0 | 0 | 4 | 1 | 0 | 0 | 4 | 0 | XPG N-terminal domain | 9 |
| PF00753 | 1 | 2 | 10 | 7 | 0 | 1 | 8 | 2 | Metallo-beta-lactamase superfamily | 31 |
| PF00754 | 3 | 8 | 24 | 3 | 3 | 11 | 29 | 3 | F5/8 type C domain | 84 |
| PF00755 | 1 | 5 | 7 | 3 | 5 | 8 | 11 | 5 | Choline/Carnitine o-acyltransferase | 45 |
| PF00756 | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 1 | Putative esterase | 4 |
| PF00757 | 0 | 3 | 7 | 1 | 1 | 4 | 12 | 0 | Furin-like cysteine rich region | 28 |
| PF00758 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | Erythropoietin/thrombopoietin | 4 |
| PF00762 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | Ferrochelatase | 7 |
| PF00763 | 1 | 2 | 4 | 2 | 1 | 1 | 4 | 1 | "Tetrahydrofolate dehydrogenase/cyclohydrolase, catalytic domain" | 16 |
| PF00764 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Arginosuccinate synthase | 1 |
| PF00766 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | Electron transfer flavoprotein FAD-binding domain | 3 |
| PF00769 | 1 | 3 | 4 | 1 | 0 | 2 | 6 | 3 | Ezrin/radixin/moesin family | 20 |
| PF00773 | 0 | 0 | 4 | 1 | 0 | 1 | 5 | 0 | RNB domain | 11 |
| PF00777 | 1 | 2 | 20 | 1 | 0 | 1 | 24 | 0 | Glycosyltransferase family 29 (sialyltransferase) | 49 |
| PF00778 | 0 | 0 | 6 | 1 | 0 | 1 | 7 | 0 | DIX domain | 15 |
| PF00779 | 2 | 1 | 8 | 1 | 0 | 0 | 5 | 0 | BTK motif | 17 |
| PF00780 | 7 | 7 | 15 | 6 | 7 | 8 | 15 | 1 | CNH domain | 66 |
| PF00781 | 6 | 5 | 14 | 6 | 2 | 7 | 13 | 2 | Diacylglycerol kinase catalytic domain | 55 |
| PF00782 | 2 | 3 | 38 | 13 | 2 | 5 | 58 | 1 | "Dual specificity phosphatase, catalytic domain" | 122 |
| PF00784 | 1 | 1 | 8 | 1 | 2 | 2 | 14 | 0 | MyTH4 domain | 29 |
| PF00786 | 8 | 6 | 14 | 9 | 3 | 5 | 18 | 1 | P21-Rho-binding domain | 64 |
| PF00787 | 5 | 14 | 46 | 12 | 4 | 9 | 51 | 6 | PX domain | 147 |
| PF00788 | 6 | 5 | 36 | 11 | 2 | 5 | 46 | 1 | Ras association (RalGDS/AF-6) domain | 112 |
| PF00789 | 2 | 4 | 13 | 5 | 0 | 3 | 10 | 2 | UBX domain | 39 |
| PF00790 | 2 | 4 | 9 | 2 | 2 | 5 | 6 | 4 | VHS domain | 34 |
| PF00791 | 3 | 3 | 11 | 2 | 6 | 8 | 15 | 4 | ZU5 domain | 52 |
| PF00792 | 0 | 2 | 7 | 1 | 0 | 0 | 9 | 0 | Phosphoinositide 3-kinase C2 | 19 |
| PF00794 | 0 | 1 | 6 | 0 | 0 | 0 | 8 | 0 | "PI3-kinase family, ras-binding domain" | 15 |
| PF00795 | 0 | 0 | 7 | 2 | 0 | 1 | 3 | 1 | Carbon-nitrogen hydrolase | 14 |
| PF00797 | 0 | 0 | 3 | 0 | 0 | 0 | 7 | 1 | N-acetyltransferase | 11 |
| PF00801 | 1 | 1 | 6 | 2 | 1 | 1 | 6 | 0 | PKD domain | 18 |
| PF00804 | 2 | 6 | 9 | 3 | 0 | 3 | 14 | 2 | Syntaxin | 39 |
| PF00805 | 0 | 0 | 1 | 0 | 0 | 0 | 2 | 0 | Pentapeptide repeats (8 copies) | 3 |
| PF00806 | 0 | 0 | 2 | 1 | 0 | 0 | 1 | 0 | Pumilio-family RNA binding repeat | 4 |
| PF00808 | 0 | 0 | 7 | 2 | 0 | 0 | 8 | 1 | Histone-like transcription factor (CBF/NF-Y) and archaeal histone | 18 |
| PF00809 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Pterin binding enzyme | 1 |
| PF00810 | 0 | 0 | 3 | 0 | 0 | 0 | 3 | 0 | ER lumen protein retaining receptor | 6 |
| PF00811 | 1 | 1 | 1 | 1 | 0 | 2 | 3 | 2 | Ependymin | 11 |
| PF00812 | 1 | 3 | 8 | 5 | 1 | 3 | 11 | 0 | Ephrin | 32 |
| PF00814 | 0 | 1 | 2 | 0 | 0 | 0 | 2 | 2 | Glycoprotease family | 7 |
| PF00817 | 0 | 0 | 4 | 1 | 0 | 0 | 4 | 0 | impB/mucB/samB family | 9 |
| PF00821 | 0 | 1 | 2 | 1 | 0 | 0 | 2 | 1 | Phosphoenolpyruvate carboxykinase | 7 |
| PF00822 | 5 | 5 | 41 | 4 | 6 | 6 | 59 | 2 | PMP-22/EMP/MP20/Claudin family | 128 |
| PF00827 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | Ribosomal L15 | 8 |
| PF00828 | 2 | 2 | 3 | 0 | 1 | 1 | 1 | 1 | Ribosomal protein L18e/L15 | 11 |
| PF00829 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Ribosomal prokaryotic L21 protein | 2 |
| PF00830 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Ribosomal L28 family | 1 |
| PF00831 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | Ribosomal L29 protein | 7 |
| PF00832 | 0 | 0 | 2 | 1 | 0 | 0 | 1 | 0 | Ribosomal L39 protein | 4 |
| PF00833 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | Ribosomal S17 | 7 |
| PF00834 | 0 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | Ribulose-phosphate 3 epimerase family | 5 |
| PF00835 | 3 | 3 | 3 | 3 | 2 | 4 | 5 | 2 | SNAP-25 family | 25 |
| PF00836 | 1 | 3 | 4 | 1 | 0 | 1 | 7 | 1 | Stathmin family | 18 |
| PF00837 | 0 | 0 | 3 | 0 | 0 | 0 | 2 | 0 | Iodothyronine deiodinase | 5 |
| PF00838 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | Translationally controlled tumour protein | 5 |
| PF00839 | 0 | 1 | 1 | 1 | 0 | 2 | 1 | 0 | Cysteine rich repeat | 6 |
| PF00841 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Sperm histone P2 | 1 |
| PF00849 | 0 | 0 | 4 | 2 | 0 | 0 | 3 | 0 | RNA pseudouridylate synthase | 9 |
| PF00850 | 1 | 1 | 10 | 3 | 1 | 1 | 8 | 1 | Histone deacetylase domain | 26 |
| PF00852 | 0 | 0 | 5 | 2 | 0 | 0 | 13 | 0 | Glycosyltransferase family 10 (fucosyltransferase) | 20 |
| PF00853 | 0 | 0 | 3 | 0 | 0 | 0 | 4 | 0 | Runt domain | 7 |
| PF00854 | 0 | 0 | 4 | 0 | 0 | 0 | 3 | 0 | POT family | 7 |
| PF00855 | 1 | 1 | 22 | 5 | 0 | 0 | 17 | 2 | PWWP domain | 48 |
| PF00856 | 0 | 0 | 40 | 6 | 0 | 0 | 43 | 1 | SET domain | 90 |
| PF00857 | 0 | 1 | 3 | 1 | 0 | 2 | 2 | 2 | Isochorismatase family | 11 |
| PF00858 | 1 | 0 | 8 | 2 | 2 | 3 | 4 | 0 | Amiloride-sensitive sodium channel | 20 |
| PF00859 | 0 | 0 | 4 | 3 | 0 | 0 | 4 | 0 | CTF/NF-I family transcription modulation region | 11 |
| PF00860 | 0 | 1 | 4 | 1 | 0 | 0 | 4 | 0 | Permease family | 10 |
| PF00861 | 1 | 1 | 1 | 1 | 1 | 1 | 3 | 1 | Ribosomal L18p/L5e family | 10 |
| PF00864 | 0 | 0 | 7 | 0 | 0 | 0 | 9 | 0 | ATP P2X receptor | 16 |
| PF00865 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Osteopontin | 1 |
| PF00867 | 0 | 0 | 4 | 1 | 0 | 0 | 4 | 0 | XPG I-region | 9 |
| PF00868 | 1 | 1 | 8 | 0 | 0 | 0 | 9 | 2 | Transglutaminase family | 21 |
| PF00870 | 0 | 0 | 3 | 0 | 0 | 0 | 3 | 0 | P53 DNA-binding domain | 6 |
| PF00875 | 0 | 0 | 2 | 1 | 0 | 0 | 8 | 0 | DNA photolyase | 11 |
| PF00876 | 1 | 1 | 3 | 0 | 0 | 0 | 4 | 0 | Innexin | 9 |
| PF00877 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | NlpC/P60 family | 1 |
| PF00878 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | Cation-independent mannose-6-phosphate receptor repeat | 5 |
| PF00879 | 0 | 0 | 28 | 0 | 0 | 0 | 0 | 0 | Defensin propeptide | 28 |
| PF00880 | 2 | 2 | 4 | 2 | 2 | 2 | 4 | 2 | Nebulin repeat | 20 |
| PF00881 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Nitroreductase family | 2 |
| PF00882 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Zinc dependent phospholipase C | 1 |
| PF00883 | 0 | 1 | 2 | 0 | 0 | 3 | 3 | 2 | "Cytosol aminopeptidase family, catalytic domain" | 11 |
| PF00884 | 0 | 1 | 12 | 1 | 0 | 0 | 15 | 0 | Sulfatase | 29 |
| PF00886 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | Ribosomal protein S16 | 2 |
| PF00887 | 1 | 3 | 8 | 2 | 1 | 4 | 8 | 2 | Acyl CoA binding protein | 29 |
| PF00888 | 4 | 5 | 10 | 4 | 3 | 3 | 9 | 5 | Cullin family | 43 |
| PF00889 | 0 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | Elongation factor TS | 5 |
| PF00890 | 0 | 1 | 1 | 1 | 0 | 2 | 2 | 2 | FAD binding domain | 9 |
| PF00891 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 1 | O-methyltransferase | 3 |
| PF00892 | 0 | 1 | 8 | 0 | 0 | 3 | 7 | 0 | EamA-like transporter family | 19 |
| PF00895 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 1 | ATP synthase protein 8 | 6 |
| PF00899 | 0 | 2 | 11 | 5 | 0 | 3 | 8 | 4 | ThiF family | 33 |
| PF00900 | 1 | 0 | 1 | 0 | 1 | 1 | 1 | 1 | Ribosomal family S4e | 6 |
| PF00903 | 0 | 1 | 3 | 1 | 0 | 1 | 3 | 2 | Glyoxalase/Bleomycin resistance protein/Dioxygenase superfamily | 11 |
| PF00907 | 0 | 0 | 17 | 1 | 0 | 0 | 22 | 0 | T-box | 40 |
| PF00909 | 0 | 0 | 4 | 0 | 1 | 1 | 6 | 0 | Ammonium Transporter Family | 12 |
| PF00916 | 0 | 0 | 10 | 0 | 0 | 0 | 12 | 0 | Sulfate transporter family | 22 |
| PF00917 | 1 | 2 | 18 | 1 | 1 | 2 | 16 | 1 | MATH domain | 42 |
| PF00918 | 0 | 0 | 2 | 1 | 1 | 1 | 1 | 0 | Gastrin/cholecystokinin family | 6 |
| PF00919 | 0 | 0 | 2 | 2 | 0 | 0 | 1 | 0 | Uncharacterized protein family UPF0004 | 5 |
| PF00923 | 0 | 1 | 1 | 1 | 0 | 1 | 0 | 0 | Transaldolase | 4 |
| PF00927 | 1 | 1 | 8 | 0 | 0 | 0 | 9 | 2 | "Transglutaminase family, C-terminal ig like domain" | 21 |
| PF00928 | 5 | 4 | 10 | 5 | 6 | 6 | 12 | 4 | Adaptor complexes medium subunit family | 52 |
| PF00929 | 0 | 1 | 12 | 2 | 0 | 0 | 9 | 0 | Exonuclease | 24 |
| PF00930 | 1 | 1 | 6 | 4 | 0 | 4 | 5 | 2 | Dipeptidyl peptidase IV (DPP IV) N-terminal region | 23 |
| PF00931 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | NB-ARC domain | 2 |
| PF00932 | 3 | 2 | 5 | 1 | 2 | 2 | 3 | 2 | Lamin Tail Domain | 20 |
| PF00935 | 0 | 0 | 2 | 1 | 0 | 0 | 1 | 1 | Ribosomal protein L44 | 5 |
| PF00939 | 0 | 2 | 5 | 0 | 0 | 1 | 6 | 0 | Sodium:sulfate symporter transmembrane region | 14 |
| PF00940 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | DNA-dependent RNA polymerase | 2 |
| PF00941 | 0 | 0 | 5 | 0 | 0 | 1 | 3 | 0 | FAD binding domain in molybdopterin dehydrogenase | 9 |
| PF00953 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Glycosyl transferase family 4 | 2 |
| PF00955 | 1 | 6 | 10 | 0 | 1 | 5 | 12 | 4 | HCO3- transporter family | 39 |
| PF00956 | 2 | 2 | 11 | 4 | 0 | 3 | 5 | 3 | Nucleosome assembly protein (NAP) | 30 |
| PF00957 | 9 | 10 | 9 | 3 | 2 | 10 | 10 | 3 | Synaptobrevin | 56 |
| PF00958 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | GMP synthase C terminal domain | 4 |
| PF00962 | 2 | 2 | 5 | 2 | 0 | 1 | 8 | 1 | Adenosine/AMP deaminase | 21 |
| PF00965 | 0 | 0 | 4 | 0 | 0 | 0 | 3 | 0 | Tissue inhibitor of metalloproteinase | 7 |
| PF00969 | 0 | 0 | 6 | 0 | 0 | 0 | 6 | 1 | "Class II histocompatibility antigen, beta domain" | 13 |
| PF00970 | 0 | 2 | 5 | 2 | 0 | 2 | 4 | 2 | Oxidoreductase FAD-binding domain | 17 |
| PF00975 | 0 | 1 | 2 | 1 | 0 | 1 | 2 | 0 | Thioesterase domain | 7 |
| PF00976 | 0 | 0 | 1 | 1 | 0 | 0 | 2 | 0 | Corticotropin ACTH domain | 4 |
| PF00984 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | "UDP-glucose/GDP-mannose dehydrogenase family, central domain" | 4 |
| PF00988 | 0 | 1 | 2 | 0 | 0 | 1 | 2 | 0 | "Carbamoyl-phosphate synthase small chain, CPSase domain" | 6 |
| PF00989 | 0 | 0 | 18 | 2 | 0 | 0 | 20 | 0 | PAS fold | 40 |
| PF00992 | 0 | 0 | 6 | 0 | 0 | 0 | 19 | 4 | Troponin | 29 |
| PF00993 | 0 | 0 | 3 | 0 | 0 | 0 | 4 | 1 | "Class II histocompatibility antigen, alpha domain" | 8 |
| PF00994 | 1 | 1 | 2 | 1 | 2 | 2 | 2 | 1 | Probable molybdopterin binding domain | 12 |
| PF00995 | 3 | 6 | 8 | 6 | 0 | 5 | 9 | 2 | Sec1 family | 39 |
| PF00996 | 0 | 2 | 4 | 2 | 0 | 2 | 3 | 2 | GDP dissociation inhibitor | 15 |
| PF00997 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Kappa casein | 1 |
| PF00999 | 0 | 3 | 13 | 3 | 0 | 4 | 11 | 0 | Sodium/hydrogen exchanger family | 34 |
| PF01000 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | RNA polymerase Rpb3/RpoA insert domain | 4 |
| PF01007 | 1 | 2 | 15 | 4 | 2 | 4 | 27 | 0 | Inward rectifier potassium channel | 55 |
| PF01008 | 0 | 0 | 4 | 3 | 0 | 1 | 4 | 2 | Initiation factor 2 subunit family | 14 |
| PF01011 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | PQQ enzyme repeat | 1 |
| PF01012 | 0 | 2 | 2 | 1 | 0 | 2 | 1 | 1 | Electron transfer flavoprotein domain | 9 |
| PF01014 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Uricase | 2 |
| PF01015 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | Ribosomal S3Ae family | 7 |
| PF01016 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Ribosomal L27 protein | 3 |
| PF01017 | 0 | 0 | 7 | 1 | 0 | 1 | 8 | 0 | "STAT protein, all-alpha domain" | 17 |
| PF01018 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | GTP1/OBG | 4 |
| PF01019 | 0 | 1 | 4 | 1 | 0 | 1 | 10 | 1 | Gamma-glutamyltranspeptidase | 18 |
| PF01020 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 0 | Ribosomal L40e family | 3 |
| PF01023 | 1 | 3 | 23 | 6 | 1 | 4 | 11 | 5 | S-100/ICaBP type calcium binding domain | 54 |
| PF01025 | 0 | 1 | 2 | 1 | 0 | 1 | 2 | 2 | GrpE | 9 |
| PF01026 | 0 | 0 | 3 | 2 | 0 | 0 | 4 | 0 | TatD related DNase | 9 |
| PF01027 | 1 | 2 | 7 | 2 | 0 | 1 | 8 | 0 | Inhibitor of apoptosis-promoting Bax1 | 21 |
| PF01028 | 0 | 0 | 2 | 0 | 0 | 0 | 4 | 1 | "Eukaryotic DNA topoisomerase I, catalytic core" | 7 |
| PF01030 | 0 | 3 | 7 | 1 | 1 | 4 | 12 | 0 | Receptor L domain | 28 |
| PF01031 | 4 | 4 | 4 | 3 | 1 | 6 | 14 | 7 | Dynamin central region | 43 |
| PF01033 | 0 | 0 | 8 | 0 | 0 | 0 | 12 | 0 | Somatomedin B domain | 20 |
| PF01034 | 2 | 5 | 7 | 2 | 5 | 6 | 8 | 2 | Syndecan domain | 37 |
| PF01035 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | "6-O-methylguanine DNA methyltransferase, DNA binding domain" | 2 |
| PF01039 | 1 | 3 | 4 | 1 | 0 | 4 | 5 | 3 | Carboxyl transferase domain | 21 |
| PF01040 | 0 | 0 | 3 | 0 | 0 | 0 | 3 | 0 | UbiA prenyltransferase family | 6 |
| PF01042 | 0 | 1 | 1 | 0 | 0 | 1 | 2 | 1 | Endoribonuclease L-PSP | 6 |
| PF01044 | 4 | 3 | 5 | 2 | 4 | 4 | 3 | 2 | Vinculin family | 27 |
| PF01048 | 0 | 0 | 5 | 2 | 0 | 3 | 7 | 2 | Phosphorylase superfamily | 19 |
| PF01049 | 9 | 5 | 26 | 1 | 8 | 10 | 26 | 3 | Cadherin cytoplasmic region | 88 |
| PF01053 | 0 | 0 | 1 | 1 | 0 | 1 | 2 | 1 | Cys/Met metabolism PLP-dependent enzyme | 6 |
| PF01055 | 1 | 2 | 6 | 2 | 0 | 3 | 7 | 1 | Glycosyl hydrolases family 31 | 22 |
| PF01056 | 0 | 0 | 5 | 0 | 0 | 0 | 6 | 0 | Myc amino-terminal region | 11 |
| PF01058 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | "NADH ubiquinone oxidoreductase, 20 Kd subunit" | 6 |
| PF01059 | 0 | 1 | 0 | 1 | 0 | 1 | 1 | 1 | "NADH-ubiquinone oxidoreductase chain 4, amino terminus" | 5 |
| PF01061 | 0 | 1 | 6 | 3 | 0 | 2 | 8 | 0 | ABC-2 type transporter | 20 |
| PF01062 | 0 | 0 | 3 | 0 | 0 | 0 | 3 | 0 | "Bestrophin, RFP-TM, chloride channel" | 6 |
| PF01063 | 0 | 1 | 2 | 2 | 0 | 2 | 2 | 2 | Aminotransferase class IV | 11 |
| PF01064 | 0 | 2 | 12 | 6 | 0 | 1 | 10 | 0 | Activin types I and II receptor domain | 31 |
| PF01066 | 1 | 3 | 5 | 3 | 0 | 2 | 8 | 1 | CDP-alcohol phosphatidyltransferase | 23 |
| PF01067 | 1 | 1 | 12 | 3 | 0 | 3 | 19 | 3 | "Calpain large subunit, domain III" | 42 |
| PF01068 | 0 | 0 | 2 | 0 | 0 | 0 | 3 | 0 | ATP dependent DNA ligase domain | 5 |
| PF01070 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | FMN-dependent dehydrogenase | 4 |
| PF01071 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | "Phosphoribosylglycinamide synthetase, ATP-grasp (A) domain" | 1 |
| PF01073 | 0 | 1 | 10 | 2 | 0 | 0 | 5 | 0 | 3-beta hydroxysteroid dehydrogenase/isomerase family | 18 |
| PF01074 | 1 | 1 | 5 | 1 | 1 | 2 | 4 | 0 | Glycosyl hydrolases family 38 N-terminal domain | 15 |
| PF01079 | 0 | 0 | 3 | 0 | 0 | 0 | 5 | 0 | Hint module | 8 |
| PF01080 | 1 | 1 | 2 | 0 | 0 | 1 | 2 | 0 | Presenilin | 7 |
| PF01082 | 1 | 1 | 4 | 0 | 2 | 2 | 3 | 0 | "Copper type II ascorbate-dependent monooxygenase, N-terminal domain" | 13 |
| PF01084 | 0 | 0 | 3 | 2 | 0 | 0 | 3 | 0 | Ribosomal protein S18 | 8 |
| PF01085 | 0 | 0 | 3 | 0 | 0 | 0 | 5 | 0 | Hedgehog amino-terminal signalling domain | 8 |
| PF01086 | 2 | 2 | 2 | 2 | 0 | 1 | 2 | 2 | Clathrin light chain | 13 |
| PF01087 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | "Galactose-1-phosphate uridyl transferase, N-terminal domain" | 2 |
| PF01088 | 0 | 2 | 5 | 3 | 0 | 3 | 3 | 2 | "Ubiquitin carboxyl-terminal hydrolase, family 1" | 18 |
| PF01090 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | Ribosomal protein S19e | 7 |
| PF01091 | 0 | 0 | 2 | 1 | 0 | 0 | 2 | 1 | "PTN/MK heparin-binding protein family, C-terminal domain" | 6 |
| PF01092 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | Ribosomal protein S6e | 7 |
| PF01093 | 1 | 1 | 1 | 0 | 0 | 0 | 2 | 0 | Clusterin | 5 |
| PF01094 | 21 | 20 | 159 | 9 | 29 | 33 | 93 | 7 | Receptor family ligand binding region | 371 |
| PF01096 | 0 | 0 | 6 | 2 | 0 | 1 | 6 | 1 | Transcription factor S-II (TFIIS) | 16 |
| PF01099 | 0 | 0 | 11 | 0 | 0 | 0 | 0 | 0 | Uteroglobin family | 11 |
| PF01101 | 0 | 0 | 4 | 1 | 0 | 0 | 3 | 3 | HMG14 and HMG17 | 11 |
| PF01102 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | Glycophorin A | 3 |
| PF01103 | 1 | 1 | 1 | 1 | 0 | 2 | 2 | 2 | Surface antigen | 10 |
| PF01105 | 2 | 5 | 10 | 2 | 4 | 6 | 11 | 2 | emp24/gp25L/p24 family/GOLD | 42 |
| PF01106 | 0 | 0 | 1 | 1 | 0 | 2 | 2 | 2 | NifU-like domain | 8 |
| PF01108 | 1 | 1 | 12 | 0 | 0 | 1 | 12 | 0 | Tissue factor | 27 |
| PF01109 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Granulocyte-macrophage colony-stimulating factor | 1 |
| PF01110 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Ciliary neurotrophic factor | 2 |
| PF01111 | 0 | 0 | 3 | 0 | 0 | 0 | 2 | 0 | Cyclin-dependent kinase regulatory subunit | 5 |
| PF01112 | 0 | 1 | 3 | 2 | 0 | 1 | 6 | 1 | Asparaginase | 14 |
| PF01114 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | "Colipase, N-terminal domain" | 1 |
| PF01115 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 0 | "F-actin capping protein, beta subunit" | 5 |
| PF01119 | 0 | 0 | 4 | 1 | 0 | 0 | 4 | 0 | "DNA mismatch repair protein, C-terminal domain" | 9 |
| PF01120 | 0 | 0 | 2 | 0 | 0 | 0 | 3 | 0 | Alpha-L-fucosidase | 5 |
| PF01121 | 1 | 2 | 2 | 2 | 1 | 1 | 2 | 0 | Dephospho-CoA kinase | 11 |
| PF01122 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 0 | Eukaryotic cobalamin-binding protein | 3 |
| PF01124 | 1 | 2 | 6 | 2 | 1 | 3 | 9 | 2 | MAPEG family | 26 |
| PF01125 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | G10 protein | 2 |
| PF01126 | 0 | 1 | 2 | 1 | 0 | 2 | 3 | 0 | Heme oxygenase | 9 |
| PF01127 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | Succinate dehydrogenase/Fumarate reductase transmembrane subunit | 6 |
| PF01128 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase | 2 |
| PF01129 | 0 | 0 | 5 | 0 | 0 | 0 | 8 | 0 | NAD:arginine ADP-ribosyltransferase | 13 |
| PF01130 | 0 | 2 | 3 | 1 | 0 | 1 | 2 | 0 | CD36 family | 9 |
| PF01131 | 0 | 0 | 2 | 1 | 0 | 0 | 2 | 1 | DNA topoisomerase | 6 |
| PF01133 | 1 | 0 | 1 | 0 | 1 | 0 | 1 | 1 | Enhancer of rudimentary | 5 |
| PF01134 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Glucose inhibited division protein A | 2 |
| PF01135 | 1 | 1 | 3 | 1 | 0 | 2 | 4 | 2 | Protein-L-isoaspartate(D-aspartate) O-methyltransferase (PCMT) | 14 |
| PF01137 | 1 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | RNA 3'-terminal phosphate cyclase | 5 |
| PF01138 | 0 | 0 | 6 | 1 | 1 | 0 | 4 | 0 | "3' exoribonuclease family, domain 1" | 12 |
| PF01139 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | tRNA-splicing ligase RtcB | 6 |
| PF01142 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | tRNA pseudouridine synthase D (TruD) | 4 |
| PF01144 | 0 | 1 | 3 | 1 | 0 | 2 | 2 | 1 | Coenzyme A transferase | 10 |
| PF01145 | 8 | 8 | 11 | 7 | 7 | 11 | 16 | 10 | SPFH domain / Band 7 family | 78 |
| PF01146 | 0 | 0 | 3 | 1 | 0 | 0 | 4 | 1 | Caveolin | 9 |
| PF01148 | 0 | 2 | 2 | 2 | 0 | 2 | 2 | 1 | Cytidylyltransferase family | 11 |
| PF01149 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | Formamidopyrimidine-DNA glycosylase N-terminal domain | 4 |
| PF01150 | 1 | 4 | 8 | 2 | 0 | 1 | 10 | 0 | GDA1/CD39 (nucleoside phosphatase) family | 26 |
| PF01151 | 0 | 2 | 7 | 1 | 0 | 0 | 10 | 1 | GNS1/SUR4 family | 21 |
| PF01153 | 2 | 3 | 6 | 4 | 3 | 3 | 7 | 1 | Glypican | 29 |
| PF01154 | 0 | 1 | 2 | 0 | 0 | 0 | 0 | 0 | Hydroxymethylglutaryl-coenzyme A synthase N terminal | 3 |
| PF01156 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 1 | Inosine-uridine preferring nucleoside hydrolase | 3 |
| PF01157 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | Ribosomal protein L21e | 7 |
| PF01158 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 0 | Ribosomal protein L36e | 5 |
| PF01159 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | Ribosomal protein L6e | 8 |
| PF01160 | 1 | 0 | 3 | 1 | 0 | 0 | 5 | 0 | Vertebrate endogenous opioids neuropeptide | 10 |
| PF01161 | 0 | 1 | 4 | 1 | 0 | 1 | 2 | 1 | Phosphatidylethanolamine-binding protein | 10 |
| PF01163 | 0 | 0 | 3 | 0 | 0 | 0 | 3 | 0 | RIO1 family | 6 |
| PF01165 | 1 | 1 | 1 | 0 | 0 | 0 | 1 | 0 | Ribosomal protein S21 | 4 |
| PF01166 | 1 | 1 | 4 | 1 | 0 | 0 | 4 | 0 | TSC-22/dip/bun family | 11 |
| PF01167 | 0 | 0 | 5 | 1 | 0 | 0 | 7 | 0 | Tub family | 13 |
| PF01168 | 0 | 0 | 1 | 1 | 0 | 1 | 2 | 2 | "Alanine racemase, N-terminal domain" | 7 |
| PF01169 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | Uncharacterized protein family UPF0016 | 5 |
| PF01170 | 0 | 0 | 3 | 1 | 0 | 0 | 3 | 0 | Putative RNA methylase family UPF0020 | 7 |
| PF01171 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | PP-loop family | 3 |
| PF01172 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Shwachman-Bodian-Diamond syndrome (SBDS) protein | 3 |
| PF01175 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Urocanase | 2 |
| PF01176 | 0 | 1 | 9 | 1 | 0 | 1 | 3 | 2 | Translation initiation factor 1A / IF-1 | 17 |
| PF01179 | 0 | 0 | 6 | 1 | 0 | 1 | 2 | 2 | "Copper amine oxidase, enzyme domain" | 12 |
| PF01180 | 0 | 1 | 2 | 2 | 0 | 1 | 3 | 1 | Dihydroorotate dehydrogenase | 10 |
| PF01182 | 0 | 1 | 4 | 4 | 0 | 1 | 4 | 2 | Glucosamine-6-phosphate isomerases/6-phosphogluconolactonase | 16 |
| PF01186 | 0 | 0 | 5 | 0 | 0 | 0 | 9 | 0 | Lysyl oxidase | 14 |
| PF01187 | 0 | 2 | 2 | 2 | 0 | 2 | 2 | 2 | Macrophage migration inhibitory factor (MIF) | 12 |
| PF01188 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | "Mandelate racemase / muconate lactonizing enzyme, C-terminal domain" | 2 |
| PF01189 | 0 | 0 | 7 | 2 | 0 | 0 | 5 | 0 | NOL1/NOP2/sun family | 14 |
| PF01191 | 0 | 0 | 1 | 1 | 0 | 0 | 2 | 0 | "RNA polymerase Rpb5, C-terminal domain" | 4 |
| PF01192 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | RNA polymerase Rpb6 | 2 |
| PF01193 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | RNA polymerase Rpb3/Rpb11 dimerisation domain | 4 |
| PF01194 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 0 | RNA polymerases N / 8 kDa subunit | 3 |
| PF01195 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | Peptidyl-tRNA hydrolase | 2 |
| PF01196 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | Ribosomal protein L17 | 5 |
| PF01198 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | Ribosomal protein L31e | 7 |
| PF01199 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | Ribosomal protein L34e | 7 |
| PF01200 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | Ribosomal protein S28e | 7 |
| PF01201 | 1 | 1 | 2 | 0 | 2 | 2 | 3 | 2 | Ribosomal protein S8e | 13 |
| PF01202 | 0 | 1 | 2 | 2 | 0 | 0 | 0 | 0 | Shikimate kinase | 5 |
| PF01204 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Trehalase | 2 |
| PF01205 | 0 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | Uncharacterized protein family UPF0029 | 4 |
| PF01207 | 0 | 0 | 4 | 2 | 0 | 0 | 3 | 0 | Dihydrouridine synthase (Dus) | 9 |
| PF01208 | 0 | 1 | 1 | 0 | 0 | 0 | 1 | 1 | Uroporphyrinogen decarboxylase (URO-D) | 4 |
| PF01209 | 0 | 0 | 1 | 1 | 0 | 1 | 1 | 0 | ubiE/COQ5 methyltransferase family | 4 |
| PF01210 | 0 | 2 | 2 | 1 | 0 | 2 | 4 | 3 | NAD-dependent glycerol-3-phosphate dehydrogenase N-terminus | 14 |
| PF01212 | 0 | 0 | 2 | 1 | 0 | 0 | 1 | 0 | Beta-eliminating lyase | 4 |
| PF01213 | 1 | 2 | 2 | 2 | 0 | 1 | 1 | 1 | Adenylate cyclase associated (CAP) N terminal | 10 |
| PF01214 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | Casein kinase II regulatory subunit | 7 |
| PF01215 | 0 | 1 | 1 | 0 | 0 | 3 | 3 | 3 | Cytochrome c oxidase subunit Vb | 11 |
| PF01216 | 0 | 0 | 2 | 0 | 1 | 1 | 3 | 2 | Calsequestrin | 9 |
| PF01217 | 11 | 9 | 16 | 9 | 9 | 11 | 19 | 7 | Clathrin adaptor complex small chain | 91 |
| PF01218 | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 1 | Coproporphyrinogen III oxidase | 4 |
| PF01221 | 2 | 2 | 4 | 2 | 0 | 0 | 6 | 2 | Dynein light chain type 1 | 18 |
| PF01222 | 0 | 0 | 3 | 0 | 1 | 0 | 3 | 1 | Ergosterol biosynthesis ERG4/ERG24 family | 8 |
| PF01223 | 2 | 1 | 5 | 2 | 1 | 1 | 14 | 2 | DNA/RNA non-specific endonuclease | 28 |
| PF01227 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | GTP cyclohydrolase I | 2 |
| PF01229 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Glycosyl hydrolases family 39 | 2 |
| PF01230 | 1 | 2 | 3 | 2 | 0 | 1 | 2 | 1 | HIT domain | 12 |
| PF01231 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 0 | "Indoleamine 2,3-dioxygenase" | 3 |
| PF01233 | 0 | 0 | 2 | 1 | 0 | 1 | 3 | 1 | "Myristoyl-CoA:protein N-myristoyltransferase, N-terminal domain" | 8 |
| PF01234 | 0 | 0 | 3 | 0 | 0 | 0 | 1 | 0 | NNMT/PNMT/TEMT family | 4 |
| PF01237 | 1 | 3 | 11 | 6 | 1 | 2 | 15 | 3 | Oxysterol-binding protein | 42 |
| PF01238 | 0 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | Phosphomannose isomerase type I | 5 |
| PF01239 | 1 | 0 | 3 | 2 | 0 | 1 | 1 | 1 | Protein prenyltransferase alpha subunit repeat | 9 |
| PF01242 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | 6-pyruvoyl tetrahydropterin synthase | 3 |
| PF01243 | 0 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | Pyridoxamine 5'-phosphate oxidase | 5 |
| PF01244 | 0 | 0 | 3 | 0 | 0 | 0 | 2 | 0 | Membrane dipeptidase (Peptidase family M19) | 5 |
| PF01245 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Ribosomal protein L19 | 3 |
| PF01246 | 1 | 1 | 2 | 0 | 1 | 1 | 1 | 0 | Ribosomal protein L24e | 7 |
| PF01247 | 1 | 0 | 1 | 0 | 1 | 1 | 1 | 1 | Ribosomal protein L35Ae | 6 |
| PF01248 | 3 | 3 | 11 | 3 | 5 | 5 | 14 | 6 | Ribosomal protein L7Ae/L30e/S12e/Gadd45 family | 50 |
| PF01249 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | Ribosomal protein S21e | 6 |
| PF01250 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Ribosomal protein S6 | 2 |
| PF01251 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | Ribosomal protein S7e | 8 |
| PF01253 | 0 | 0 | 4 | 2 | 0 | 1 | 4 | 2 | Translation initiation factor SUI1 | 13 |
| PF01254 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Nuclear transition protein 2 | 1 |
| PF01255 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | Putative undecaprenyl diphosphate synthase | 4 |
| PF01256 | 0 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | Carbohydrate kinase | 4 |
| PF01257 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | Thioredoxin-like [2Fe-2S] ferredoxin | 7 |
| PF01259 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | SAICAR synthetase | 6 |
| PF01261 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 1 | Xylose isomerase-like TIM barrel | 3 |
| PF01262 | 0 | 1 | 1 | 1 | 0 | 2 | 3 | 3 | "Alanine dehydrogenase/PNT, C-terminal domain" | 11 |
| PF01263 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 1 | Aldose 1-epimerase | 3 |
| PF01265 | 0 | 0 | 1 | 0 | 0 | 0 | 2 | 0 | Cytochrome c/c1 heme lyase | 3 |
| PF01266 | 1 | 4 | 9 | 3 | 1 | 5 | 9 | 1 | FAD dependent oxidoreductase | 33 |
| PF01267 | 2 | 2 | 3 | 1 | 2 | 2 | 2 | 2 | F-actin capping protein alpha subunit | 16 |
| PF01268 | 1 | 2 | 2 | 1 | 1 | 1 | 3 | 1 | Formate--tetrahydrofolate ligase | 12 |
| PF01269 | 0 | 0 | 2 | 1 | 0 | 0 | 1 | 1 | Fibrillarin | 5 |
| PF01271 | 2 | 1 | 3 | 3 | 2 | 1 | 2 | 1 | Granin (chromogranin or secretogranin) | 15 |
| PF01273 | 0 | 0 | 17 | 1 | 0 | 0 | 1 | 1 | "LBP / BPI / CETP family, N-terminal domain" | 20 |
| PF01274 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | Malate synthase | 1 |
| PF01275 | 0 | 3 | 3 | 1 | 1 | 5 | 5 | 5 | Myelin proteolipid protein (PLP or lipophilin) | 23 |
| PF01279 | 0 | 0 | 2 | 0 | 0 | 0 | 4 | 0 | Parathyroid hormone family | 6 |
| PF01280 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | Ribosomal protein L19e | 8 |
| PF01281 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | "Ribosomal protein L9, N-terminal domain" | 3 |
| PF01282 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | Ribosomal protein S24e | 7 |
| PF01283 | 1 | 1 | 1 | 0 | 1 | 1 | 2 | 1 | Ribosomal protein S26e | 8 |
| PF01284 | 4 | 9 | 24 | 7 | 2 | 8 | 31 | 6 | Membrane-associating domain | 91 |
| PF01285 | 0 | 0 | 4 | 0 | 0 | 0 | 5 | 0 | TEA/ATTS domain family | 9 |
| PF01286 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | XPA protein N-terminal | 2 |
| PF01287 | 0 | 1 | 2 | 1 | 0 | 1 | 2 | 2 | "Eukaryotic elongation factor 5A hypusine, DNA-binding OB fold" | 9 |
| PF01290 | 1 | 0 | 6 | 2 | 0 | 2 | 3 | 2 | Thymosin beta-4 family | 16 |
| PF01291 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | LIF / OSM family | 2 |
| PF01294 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 0 | Ribosomal protein L13e | 5 |
| PF01296 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | Galanin | 4 |
| PF01299 | 0 | 3 | 6 | 2 | 0 | 2 | 5 | 1 | Lysosome-associated membrane glycoprotein (Lamp) | 19 |
| PF01300 | 0 | 0 | 1 | 0 | 0 | 0 | 2 | 0 | Telomere recombination | 3 |
| PF01301 | 0 | 0 | 3 | 1 | 0 | 0 | 4 | 1 | Glycosyl hydrolases family 35 | 9 |
| PF01302 | 4 | 5 | 10 | 4 | 3 | 6 | 16 | 4 | CAP-Gly domain | 52 |
| PF01315 | 0 | 0 | 5 | 0 | 0 | 1 | 3 | 0 | "Aldehyde oxidase and xanthine dehydrogenase, a/b hammerhead domain" | 9 |
| PF01321 | 0 | 1 | 2 | 0 | 0 | 1 | 2 | 1 | Creatinase/Prolidase N-terminal domain | 7 |
| PF01323 | 0 | 1 | 1 | 1 | 0 | 1 | 4 | 1 | DSBA-like thioredoxin domain | 9 |
| PF01327 | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 0 | Polypeptide deformylase | 3 |
| PF01329 | 0 | 0 | 2 | 1 | 0 | 1 | 1 | 0 | Pterin 4 alpha carbinolamine dehydratase | 5 |
| PF01331 | 0 | 0 | 1 | 1 | 0 | 0 | 2 | 0 | "mRNA capping enzyme, catalytic domain" | 4 |
| PF01335 | 1 | 1 | 6 | 2 | 0 | 1 | 8 | 1 | Death effector domain | 20 |
| PF01336 | 1 | 3 | 9 | 5 | 2 | 3 | 9 | 3 | OB-fold nucleic acid binding domain | 35 |
| PF01342 | 0 | 0 | 7 | 1 | 0 | 0 | 9 | 0 | SAND domain | 17 |
| PF01344 | 1 | 2 | 59 | 4 | 2 | 2 | 61 | 2 | Kelch motif | 133 |
| PF01347 | 0 | 0 | 2 | 0 | 2 | 7 | 13 | 9 | Lipoprotein amino terminal region | 33 |
| PF01351 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Ribonuclease HII | 2 |
| PF01352 | 0 | 0 | 260 | 4 | 0 | 0 | 0 | 0 | KRAB box | 264 |
| PF01363 | 2 | 5 | 30 | 5 | 0 | 8 | 22 | 3 | FYVE zinc finger | 75 |
| PF01365 | 4 | 4 | 6 | 0 | 5 | 5 | 8 | 2 | RIH domain | 34 |
| PF01368 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | DHH family | 4 |
| PF01369 | 8 | 7 | 15 | 1 | 12 | 10 | 17 | 4 | Sec7 domain | 74 |
| PF01370 | 0 | 1 | 7 | 4 | 0 | 3 | 9 | 2 | NAD dependent epimerase/dehydratase family | 26 |
| PF01379 | 0 | 0 | 1 | 0 | 0 | 0 | 2 | 0 | "Porphobilinogen deaminase, dipyromethane cofactor binding domain" | 3 |
| PF01380 | 0 | 0 | 2 | 0 | 0 | 1 | 2 | 1 | SIS domain | 6 |
| PF01381 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | Helix-turn-helix | 5 |
| PF01382 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | Avidin family | 1 |
| PF01384 | 0 | 2 | 2 | 1 | 0 | 1 | 3 | 0 | Phosphate transporter family | 9 |
| PF01387 | 0 | 3 | 3 | 2 | 0 | 2 | 1 | 1 | Synuclein | 12 |
| PF01388 | 0 | 0 | 15 | 0 | 0 | 0 | 15 | 0 | ARID/BRIGHT DNA binding domain | 30 |
| PF01390 | 0 | 1 | 21 | 0 | 1 | 1 | 24 | 1 | SEA domain | 49 |
| PF01391 | 4 | 1 | 79 | 4 | 2 | 1 | 97 | 9 | Collagen triple helix repeat (20 copies) | 197 |
| PF01392 | 0 | 0 | 23 | 1 | 0 | 0 | 25 | 0 | Fz domain | 49 |
| PF01393 | 0 | 0 | 4 | 2 | 0 | 0 | 5 | 1 | Chromo shadow domain | 12 |
| PF01394 | 0 | 1 | 1 | 0 | 0 | 3 | 3 | 2 | Clathrin propeller repeat | 10 |
| PF01396 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 1 | Topoisomerase DNA binding C4 zinc finger | 3 |
| PF01398 | 3 | 3 | 12 | 8 | 0 | 4 | 10 | 4 | JAB1/Mov34/MPN/PAD-1 ubiquitin protease | 44 |
| PF01399 | 8 | 11 | 16 | 11 | 1 | 11 | 16 | 12 | PCI domain | 86 |
| PF01400 | 0 | 0 | 6 | 0 | 0 | 0 | 17 | 1 | Astacin (Peptidase family M12A) | 24 |
| PF01401 | 0 | 1 | 3 | 1 | 0 | 1 | 2 | 1 | Angiotensin-converting enzyme | 9 |
| PF01403 | 3 | 12 | 30 | 3 | 4 | 11 | 47 | 6 | Sema domain | 116 |
| PF01404 | 1 | 3 | 14 | 0 | 0 | 9 | 16 | 0 | Ephrin receptor ligand binding domain | 43 |
| PF01406 | 0 | 0 | 2 | 1 | 1 | 1 | 2 | 1 | tRNA synthetases class I (C) catalytic domain | 8 |
| PF01408 | 0 | 0 | 4 | 2 | 0 | 1 | 6 | 1 | "Oxidoreductase family, NAD-binding Rossmann fold" | 14 |
| PF01409 | 1 | 1 | 2 | 1 | 0 | 1 | 1 | 0 | tRNA synthetases class II core domain (F) | 7 |
| PF01410 | 0 | 0 | 11 | 1 | 1 | 0 | 15 | 4 | Fibrillar collagen C-terminal domain | 32 |
| PF01411 | 0 | 2 | 3 | 3 | 0 | 1 | 2 | 1 | tRNA synthetases class II (A) | 12 |
| PF01412 | 11 | 11 | 23 | 12 | 5 | 8 | 29 | 2 | Putative GTPase activating protein for Arf | 101 |
| PF01413 | 0 | 0 | 6 | 0 | 0 | 0 | 5 | 0 | C-terminal tandem repeated domain in type 4 procollagen | 11 |
| PF01414 | 0 | 0 | 4 | 1 | 0 | 0 | 10 | 0 | Delta serrate ligand | 15 |
| PF01415 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | Interleukin 7/9 family | 2 |
| PF01416 | 0 | 0 | 3 | 0 | 0 | 0 | 3 | 0 | tRNA pseudouridine synthase | 6 |
| PF01417 | 2 | 3 | 6 | 1 | 0 | 2 | 8 | 2 | ENTH domain | 24 |
| PF01419 | 0 | 0 | 5 | 0 | 0 | 0 | 0 | 0 | Jacalin-like lectin domain | 5 |
| PF01421 | 2 | 3 | 52 | 3 | 3 | 4 | 27 | 2 | Reprolysin (M12B) family zinc metalloprotease | 96 |
| PF01422 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 0 | NF-X1 type zinc finger | 3 |
| PF01423 | 1 | 2 | 19 | 7 | 1 | 1 | 15 | 10 | LSM domain | 56 |
| PF01424 | 0 | 0 | 11 | 1 | 0 | 0 | 4 | 1 | R3H domain | 17 |
| PF01425 | 1 | 1 | 2 | 2 | 0 | 2 | 4 | 0 | Amidase | 12 |
| PF01426 | 0 | 0 | 11 | 2 | 0 | 0 | 12 | 1 | BAH domain | 26 |
| PF01428 | 0 | 0 | 8 | 2 | 0 | 0 | 7 | 0 | AN1-like Zinc finger | 17 |
| PF01429 | 0 | 0 | 9 | 3 | 0 | 0 | 8 | 1 | Methyl-CpG binding domain | 21 |
| PF01431 | 1 | 1 | 7 | 1 | 0 | 1 | 3 | 0 | Peptidase family M13 | 14 |
| PF01432 | 0 | 2 | 3 | 1 | 0 | 1 | 4 | 1 | Peptidase family M3 | 12 |
| PF01433 | 2 | 7 | 12 | 2 | 0 | 6 | 13 | 3 | Peptidase family M1 | 45 |
| PF01434 | 2 | 3 | 4 | 2 | 2 | 5 | 4 | 1 | Peptidase family M41 | 23 |
| PF01435 | 0 | 0 | 2 | 2 | 0 | 1 | 2 | 1 | Peptidase family M48 | 8 |
| PF01436 | 3 | 4 | 8 | 4 | 5 | 5 | 7 | 2 | NHL repeat | 38 |
| PF01437 | 3 | 14 | 28 | 5 | 4 | 11 | 43 | 6 | Plexin repeat | 114 |
| PF01442 | 0 | 2 | 4 | 1 | 0 | 5 | 5 | 3 | Apolipoprotein A1/A4/E domain | 20 |
| PF01448 | 0 | 0 | 12 | 2 | 0 | 0 | 17 | 1 | ELM2 domain | 32 |
| PF01451 | 0 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | Low molecular weight phosphotyrosine protein phosphatase | 5 |
| PF01454 | 1 | 0 | 30 | 2 | 0 | 0 | 1 | 0 | MAGE family | 34 |
| PF01457 | 1 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Leishmanolysin | 4 |
| PF01459 | 5 | 4 | 5 | 3 | 0 | 6 | 6 | 5 | Eukaryotic porin | 34 |
| PF01462 | 6 | 9 | 41 | 11 | 3 | 6 | 40 | 2 | Leucine rich repeat N-terminal domain | 118 |
| PF01463 | 0 | 2 | 7 | 0 | 0 | 1 | 5 | 0 | Leucine rich repeat C-terminal domain | 15 |
| PF01464 | 0 | 0 | 1 | 0 | 0 | 1 | 2 | 1 | Transglycosylase SLT domain | 5 |
| PF01465 | 0 | 0 | 4 | 2 | 0 | 0 | 3 | 0 | GRIP domain | 9 |
| PF01466 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | "Skp1 family, dimerisation domain" | 7 |
| PF01467 | 1 | 2 | 7 | 4 | 1 | 1 | 8 | 1 | Cytidylyltransferase | 25 |
| PF01470 | 0 | 0 | 2 | 1 | 0 | 0 | 2 | 0 | Pyroglutamyl peptidase | 5 |
| PF01471 | 1 | 0 | 19 | 0 | 0 | 0 | 17 | 0 | Putative peptidoglycan binding domain | 37 |
| PF01472 | 0 | 0 | 3 | 1 | 0 | 1 | 3 | 1 | PUA domain | 9 |
| PF01476 | 0 | 1 | 4 | 2 | 0 | 1 | 5 | 2 | LysM domain | 15 |
| PF01477 | 0 | 0 | 20 | 1 | 0 | 0 | 18 | 0 | PLAT/LH2 domain | 39 |
| PF01479 | 2 | 1 | 3 | 0 | 1 | 1 | 2 | 1 | S4 domain | 11 |
| PF01480 | 0 | 0 | 5 | 1 | 0 | 0 | 5 | 0 | PWI domain | 11 |
| PF01483 | 0 | 1 | 7 | 2 | 0 | 2 | 7 | 0 | Proprotein convertase P-domain | 19 |
| PF01485 | 1 | 1 | 13 | 1 | 1 | 1 | 22 | 0 | IBR domain | 40 |
| PF01490 | 0 | 2 | 17 | 3 | 0 | 2 | 18 | 3 | Transmembrane amino acid transporter protein | 45 |
| PF01491 | 0 | 0 | 1 | 1 | 1 | 0 | 2 | 0 | Frataxin-like domain | 5 |
| PF01494 | 0 | 0 | 5 | 1 | 0 | 1 | 9 | 2 | FAD binding domain | 18 |
| PF01496 | 1 | 1 | 3 | 1 | 3 | 3 | 5 | 1 | V-type ATPase 116kDa subunit family | 18 |
| PF01498 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | Transposase | 2 |
| PF01500 | 0 | 0 | 7 | 0 | 0 | 0 | 0 | 0 | "Keratin, high sulfur B2 protein" | 7 |
| PF01501 | 0 | 1 | 9 | 1 | 1 | 3 | 13 | 3 | Glycosyl transferase family 8 | 31 |
| PF01504 | 4 | 3 | 8 | 4 | 2 | 7 | 10 | 4 | Phosphatidylinositol-4-phosphate 5-Kinase | 42 |
| PF01505 | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 1 | Major Vault Protein repeat | 4 |
| PF01507 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Phosphoadenosine phosphosulfate reductase family | 3 |
| PF01509 | 0 | 0 | 3 | 1 | 0 | 0 | 3 | 0 | TruB family pseudouridylate synthase (N terminal domain) | 7 |
| PF01510 | 0 | 0 | 4 | 1 | 0 | 0 | 2 | 0 | N-acetylmuramoyl-L-alanine amidase | 7 |
| PF01512 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | Respiratory-chain NADH dehydrogenase 51 Kd subunit | 6 |
| PF01513 | 0 | 1 | 1 | 0 | 0 | 0 | 2 | 0 | ATP-NAD kinase | 4 |
| PF01521 | 1 | 1 | 2 | 0 | 0 | 0 | 2 | 0 | Iron-sulphur cluster biosynthesis | 6 |
| PF01529 | 2 | 3 | 24 | 2 | 2 | 4 | 29 | 1 | DHHC palmitoyltransferase | 67 |
| PF01530 | 0 | 0 | 5 | 0 | 0 | 0 | 8 | 0 | "Zinc finger, C2HC type" | 13 |
| PF01531 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | Glycosyl transferase family 11 | 3 |
| PF01532 | 0 | 0 | 7 | 2 | 2 | 2 | 8 | 0 | Glycosyl hydrolase family 47 | 21 |
| PF01534 | 0 | 0 | 11 | 1 | 0 | 0 | 12 | 0 | Frizzled/Smoothened family membrane region | 24 |
| PF01535 | 0 | 1 | 1 | 0 | 0 | 1 | 3 | 1 | PPR repeat | 7 |
| PF01536 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 0 | Adenosylmethionine decarboxylase | 3 |
| PF01541 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | GIY-YIG catalytic domain | 1 |
| PF01545 | 0 | 3 | 10 | 2 | 0 | 3 | 10 | 0 | Cation efflux family | 28 |
| PF01546 | 0 | 3 | 4 | 3 | 0 | 3 | 5 | 2 | Peptidase family M20/M25/M40 | 20 |
| PF01549 | 0 | 0 | 1 | 0 | 0 | 0 | 3 | 0 | ShK domain-like | 4 |
| PF01551 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Peptidase family M23 | 1 |
| PF01553 | 5 | 10 | 18 | 8 | 3 | 4 | 14 | 2 | Acyltransferase | 64 |
| PF01554 | 0 | 0 | 2 | 0 | 0 | 0 | 5 | 0 | MatE | 7 |
| PF01556 | 7 | 6 | 9 | 8 | 5 | 6 | 10 | 1 | DnaJ C terminal domain | 52 |
| PF01557 | 1 | 3 | 3 | 2 | 0 | 0 | 1 | 0 | Fumarylacetoacetate (FAA) hydrolase family | 10 |
| PF01562 | 2 | 4 | 53 | 3 | 3 | 6 | 28 | 3 | Reprolysin family propeptide | 102 |
| PF01564 | 0 | 0 | 3 | 3 | 0 | 1 | 3 | 2 | Spermine/spermidine synthase | 12 |
| PF01565 | 0 | 1 | 5 | 1 | 2 | 3 | 4 | 0 | FAD binding domain | 16 |
| PF01566 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 0 | Natural resistance-associated macrophage protein | 3 |
| PF01569 | 5 | 4 | 18 | 5 | 4 | 6 | 23 | 0 | PAP2 superfamily | 65 |
| PF01571 | 0 | 2 | 5 | 3 | 1 | 3 | 4 | 3 | Aminomethyltransferase folate-binding domain | 21 |
| PF01575 | 1 | 1 | 2 | 1 | 0 | 1 | 0 | 0 | MaoC like domain | 6 |
| PF01576 | 5 | 7 | 18 | 2 | 10 | 8 | 23 | 11 | Myosin tail | 84 |
| PF01582 | 1 | 2 | 22 | 3 | 2 | 1 | 27 | 1 | TIR domain | 59 |
| PF01583 | 0 | 0 | 2 | 0 | 0 | 0 | 3 | 0 | Adenylylsulphate kinase | 5 |
| PF01585 | 0 | 0 | 20 | 5 | 0 | 0 | 13 | 0 | G-patch domain | 38 |
| PF01586 | 0 | 0 | 4 | 0 | 0 | 0 | 4 | 0 | Myogenic Basic domain | 8 |
| PF01588 | 2 | 2 | 2 | 1 | 1 | 2 | 3 | 2 | Putative tRNA binding domain | 15 |
| PF01590 | 1 | 2 | 7 | 1 | 0 | 1 | 8 | 1 | GAF domain | 21 |
| PF01591 | 0 | 0 | 4 | 0 | 0 | 0 | 7 | 0 | 6-phosphofructo-2-kinase | 11 |
| PF01592 | 0 | 0 | 1 | 1 | 0 | 0 | 2 | 1 | NifU-like N terminal domain | 5 |
| PF01593 | 0 | 2 | 10 | 2 | 0 | 3 | 9 | 1 | Flavin containing amine oxidoreductase | 27 |
| PF01594 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Domain of unknown function DUF20 | 2 |
| PF01595 | 0 | 3 | 4 | 0 | 0 | 2 | 4 | 0 | Domain of unknown function DUF21 | 13 |
| PF01596 | 1 | 2 | 3 | 1 | 0 | 2 | 4 | 2 | O-methyltransferase | 15 |
| PF01597 | 0 | 0 | 2 | 1 | 0 | 1 | 2 | 0 | Glycine cleavage H-protein | 6 |
| PF01599 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | Ribosomal protein S27a | 7 |
| PF01602 | 7 | 9 | 13 | 7 | 5 | 9 | 8 | 6 | Adaptin N terminal region | 64 |
| PF01603 | 3 | 4 | 5 | 4 | 4 | 4 | 7 | 4 | Protein phosphatase 2A regulatory B subunit (B56 family) | 35 |
| PF01607 | 0 | 0 | 2 | 0 | 0 | 0 | 3 | 0 | Chitin binding Peritrophin-A domain | 5 |
| PF01608 | 2 | 4 | 4 | 1 | 1 | 5 | 3 | 2 | I/LWEQ domain | 22 |
| PF01612 | 1 | 1 | 4 | 1 | 0 | 0 | 3 | 0 | 3'-5' exonuclease | 10 |
| PF01619 | 0 | 1 | 2 | 0 | 0 | 1 | 3 | 0 | Proline dehydrogenase | 7 |
| PF01624 | 0 | 0 | 3 | 0 | 0 | 0 | 2 | 0 | MutS domain I | 5 |
| PF01625 | 0 | 1 | 1 | 1 | 0 | 0 | 2 | 0 | Peptide methionine sulfoxide reductase | 5 |
| PF01630 | 0 | 0 | 7 | 0 | 1 | 0 | 8 | 0 | Hyaluronidase | 16 |
| PF01632 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Ribosomal protein L35 | 2 |
| PF01633 | 0 | 0 | 4 | 2 | 0 | 0 | 5 | 0 | Choline/ethanolamine kinase | 11 |
| PF01636 | 1 | 1 | 3 | 0 | 1 | 1 | 3 | 0 | Phosphotransferase enzyme family | 10 |
| PF01641 | 0 | 0 | 3 | 2 | 0 | 0 | 4 | 0 | SelR domain | 9 |
| PF01642 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | Methylmalonyl-CoA mutase | 5 |
| PF01648 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 4'-phosphopantetheinyl transferase superfamily | 2 |
| PF01650 | 0 | 1 | 2 | 1 | 0 | 1 | 8 | 0 | Peptidase C13 family | 13 |
| PF01652 | 0 | 0 | 4 | 3 | 0 | 0 | 7 | 3 | Eukaryotic initiation factor 4E | 17 |
| PF01655 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | Ribosomal protein L32 | 8 |
| PF01658 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | Myo-inositol-1-phosphate synthase | 2 |
| PF01661 | 0 | 1 | 8 | 3 | 1 | 1 | 7 | 3 | Macro domain | 24 |
| PF01663 | 2 | 1 | 10 | 2 | 1 | 2 | 11 | 0 | Type I phosphodiesterase / nucleotide pyrophosphatase | 29 |
| PF01667 | 2 | 1 | 3 | 1 | 2 | 2 | 2 | 2 | Ribosomal protein S27 | 15 |
| PF01669 | 1 | 1 | 1 | 0 | 2 | 2 | 2 | 2 | Myelin basic protein | 11 |
| PF01687 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Riboflavin kinase | 3 |
| PF01693 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Caulimovirus viroplasmin | 2 |
| PF01694 | 0 | 0 | 9 | 2 | 0 | 0 | 9 | 0 | Rhomboid family | 20 |
| PF01697 | 0 | 0 | 0 | 0 | 0 | 0 | 5 | 0 | Glycosyltransferase family 92 | 5 |
| PF01699 | 0 | 5 | 9 | 3 | 0 | 9 | 17 | 2 | Sodium/calcium exchanger protein | 45 |
| PF01702 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | Queuine tRNA-ribosyltransferase | 4 |
| PF01704 | 0 | 1 | 3 | 2 | 0 | 1 | 4 | 3 | UTP--glucose-1-phosphate uridylyltransferase | 14 |
| PF01709 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Transcriptional regulator | 3 |
| PF01712 | 0 | 1 | 4 | 1 | 0 | 1 | 5 | 1 | Deoxynucleoside kinase | 13 |
| PF01713 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Smr domain | 2 |
| PF01715 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | IPP transferase | 2 |
| PF01722 | 1 | 1 | 3 | 3 | 0 | 1 | 1 | 0 | BolA-like protein | 10 |
| PF01725 | 0 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | Ham1 family | 4 |
| PF01728 | 0 | 0 | 5 | 1 | 0 | 0 | 4 | 0 | FtsJ-like methyltransferase | 10 |
| PF01729 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | "Quinolinate phosphoribosyl transferase, C-terminal domain" | 1 |
| PF01731 | 0 | 1 | 3 | 2 | 0 | 1 | 4 | 0 | Arylesterase | 11 |
| PF01733 | 0 | 2 | 4 | 1 | 0 | 1 | 5 | 0 | Nucleoside transporter | 13 |
| PF01734 | 1 | 2 | 8 | 1 | 2 | 3 | 7 | 1 | Patatin-like phospholipase | 25 |
| PF01735 | 1 | 1 | 6 | 1 | 0 | 0 | 7 | 1 | Lysophospholipase catalytic domain | 17 |
| PF01738 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | Dienelactone hydrolase family | 4 |
| PF01739 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | "CheR methyltransferase, SAM binding domain" | 1 |
| PF01740 | 0 | 0 | 10 | 0 | 0 | 0 | 12 | 0 | STAS domain | 22 |
| PF01743 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Poly A polymerase head domain | 3 |
| PF01746 | 0 | 0 | 3 | 1 | 0 | 0 | 3 | 0 | tRNA (Guanine-1)-methyltransferase | 7 |
| PF01747 | 0 | 0 | 2 | 0 | 0 | 0 | 3 | 0 | ATP-sulfurylase | 5 |
| PF01749 | 0 | 2 | 6 | 3 | 0 | 1 | 7 | 2 | Importin beta binding domain | 21 |
| PF01751 | 0 | 0 | 4 | 1 | 1 | 0 | 4 | 1 | Toprim domain | 11 |
| PF01753 | 0 | 0 | 21 | 2 | 1 | 1 | 25 | 2 | MYND finger | 52 |
| PF01754 | 0 | 0 | 5 | 2 | 0 | 0 | 8 | 0 | A20-like zinc finger | 15 |
| PF01755 | 1 | 0 | 3 | 0 | 0 | 0 | 5 | 0 | Glycosyltransferase family 25 (LPS biosynthesis protein) | 9 |
| PF01756 | 0 | 2 | 4 | 0 | 1 | 3 | 3 | 2 | Acyl-CoA oxidase | 15 |
| PF01757 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Acyltransferase family | 2 |
| PF01758 | 0 | 1 | 7 | 0 | 0 | 0 | 3 | 0 | Sodium Bile acid symporter family | 11 |
| PF01759 | 1 | 1 | 17 | 0 | 1 | 3 | 27 | 4 | UNC-6/NTR/C345C module | 54 |
| PF01761 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 3-dehydroquinate synthase | 1 |
| PF01762 | 0 | 0 | 15 | 1 | 0 | 1 | 26 | 0 | Galactosyltransferase | 43 |
| PF01764 | 1 | 1 | 2 | 1 | 1 | 1 | 2 | 0 | Lipase (class 3) | 9 |
| PF01765 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | Ribosome recycling factor | 5 |
| PF01769 | 0 | 0 | 3 | 0 | 0 | 0 | 4 | 0 | Divalent cation transporter | 7 |
| PF01770 | 0 | 0 | 3 | 0 | 0 | 1 | 4 | 0 | Reduced folate carrier | 8 |
| PF01773 | 0 | 0 | 4 | 0 | 0 | 0 | 1 | 0 | Na+ dependent nucleoside transporter N-terminus | 5 |
| PF01775 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | Ribosomal L18ae/LX protein domain | 8 |
| PF01776 | 2 | 2 | 2 | 2 | 1 | 1 | 2 | 1 | Ribosomal L22e protein family | 13 |
| PF01777 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | Ribosomal L27e protein family | 7 |
| PF01778 | 1 | 1 | 2 | 1 | 1 | 1 | 2 | 1 | Ribosomal L28e protein family | 10 |
| PF01779 | 1 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Ribosomal L29e protein family | 3 |
| PF01780 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | Ribosomal L37ae protein family | 5 |
| PF01781 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | Ribosomal L38e protein family | 7 |
| PF01783 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Ribosomal L32p protein family | 2 |
| PF01784 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | NIF3 (NGG1p interacting factor 3) | 5 |
| PF01791 | 0 | 0 | 1 | 0 | 1 | 1 | 1 | 1 | DeoC/LacD family aldolase | 5 |
| PF01794 | 0 | 1 | 10 | 1 | 0 | 1 | 7 | 0 | Ferric reductase like transmembrane component | 20 |
| PF01795 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | MraW methylase family | 3 |
| PF01798 | 0 | 0 | 3 | 0 | 0 | 0 | 2 | 1 | Putative snoRNA binding domain | 6 |
| PF01799 | 0 | 0 | 5 | 0 | 0 | 1 | 3 | 0 | [2Fe-2S] binding domain | 9 |
| PF01803 | 0 | 0 | 2 | 1 | 0 | 0 | 3 | 0 | LIM-domain binding protein | 6 |
| PF01805 | 0 | 0 | 6 | 1 | 0 | 0 | 6 | 2 | Surp module | 15 |
| PF01808 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | AICARFT/IMPCHase bienzyme | 5 |
| PF01812 | 0 | 0 | 3 | 0 | 0 | 0 | 1 | 0 | 5-formyltetrahydrofolate cyclo-ligase family | 4 |
| PF01813 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | ATP synthase subunit D | 8 |
| PF01814 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Hemerythrin HHE cation binding domain | 2 |
| PF01821 | 0 | 0 | 6 | 0 | 1 | 2 | 10 | 4 | Anaphylotoxin-like domain | 23 |
| PF01822 | 0 | 0 | 5 | 0 | 0 | 0 | 3 | 0 | WSC domain | 8 |
| PF01823 | 3 | 3 | 12 | 2 | 2 | 4 | 23 | 3 | MAC/Perforin domain | 52 |
| PF01825 | 6 | 8 | 30 | 2 | 5 | 10 | 50 | 1 | Latrophilin/CL-1-like GPS domain | 112 |
| PF01826 | 0 | 0 | 11 | 0 | 0 | 0 | 11 | 0 | Trypsin Inhibitor like cysteine rich domain | 22 |
| PF01833 | 1 | 8 | 24 | 3 | 1 | 8 | 28 | 5 | IPT/TIG domain | 78 |
| PF01834 | 0 | 0 | 3 | 0 | 0 | 0 | 2 | 0 | XRCC1 N terminal domain | 5 |
| PF01835 | 1 | 1 | 10 | 0 | 1 | 4 | 23 | 7 | MG2 domain | 47 |
| PF01839 | 0 | 2 | 11 | 0 | 1 | 1 | 9 | 1 | FG-GAP repeat | 25 |
| PF01840 | 0 | 0 | 7 | 0 | 0 | 0 | 0 | 0 | TCL1/MTCP1 family | 7 |
| PF01841 | 1 | 1 | 10 | 1 | 0 | 0 | 11 | 3 | Transglutaminase-like superfamily | 27 |
| PF01842 | 0 | 0 | 3 | 0 | 1 | 1 | 4 | 0 | ACT domain | 9 |
| PF01843 | 2 | 2 | 6 | 2 | 4 | 2 | 9 | 1 | DIL domain | 28 |
| PF01844 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | HNH endonuclease | 2 |
| PF01846 | 0 | 1 | 6 | 2 | 1 | 1 | 5 | 0 | FF domain | 16 |
| PF01847 | 0 | 0 | 1 | 0 | 0 | 0 | 2 | 0 | von Hippel-Lindau disease tumour suppressor protein | 3 |
| PF01849 | 0 | 1 | 4 | 1 | 0 | 3 | 4 | 3 | NAC domain | 16 |
| PF01851 | 0 | 2 | 3 | 2 | 0 | 2 | 1 | 1 | Proteasome/cyclosome repeat | 11 |
| PF01852 | 0 | 1 | 14 | 6 | 0 | 1 | 17 | 0 | START domain | 39 |
| PF01853 | 0 | 0 | 5 | 1 | 0 | 0 | 7 | 0 | MOZ/SAS family | 13 |
| PF01857 | 0 | 0 | 3 | 0 | 0 | 0 | 3 | 0 | Retinoblastoma-associated protein B domain | 6 |
| PF01858 | 0 | 0 | 3 | 0 | 0 | 0 | 3 | 0 | Retinoblastoma-associated protein A domain | 6 |
| PF01866 | 0 | 0 | 2 | 1 | 0 | 0 | 2 | 0 | Putative diphthamide synthesis protein | 5 |
| PF01868 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | Domain of unknown function UPF0086 | 2 |
| PF01869 | 0 | 0 | 1 | 1 | 0 | 1 | 0 | 0 | BadF/BadG/BcrA/BcrD ATPase family | 3 |
| PF01871 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 0 | AMMECR1 | 3 |
| PF01873 | 0 | 1 | 2 | 2 | 0 | 0 | 2 | 2 | Domain found in IF2B/IF5 | 9 |
| PF01875 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Memo-like protein | 3 |
| PF01876 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | RNase P subunit p30 | 3 |
| PF01878 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | EVE domain | 3 |
| PF01883 | 0 | 0 | 2 | 1 | 0 | 1 | 1 | 0 | Domain of unknown function DUF59 | 5 |
| PF01885 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | "RNA 2'-phosphotransferase, Tpt1 / KptA family" | 2 |
| PF01896 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | Eukaryotic and archaeal DNA primase small subunit | 4 |
| PF01900 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | Rpp14/Pop5 family | 4 |
| PF01902 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | ATP-binding region | 2 |
| PF01907 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Ribosomal protein L37e | 2 |
| PF01909 | 0 | 0 | 11 | 0 | 0 | 0 | 4 | 0 | Nucleotidyltransferase domain | 15 |
| PF01912 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | eIF-6 family | 3 |
| PF01916 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Deoxyhypusine synthase | 2 |
| PF01918 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | Alba | 4 |
| PF01920 | 0 | 3 | 5 | 3 | 0 | 2 | 5 | 3 | Prefoldin subunit | 21 |
| PF01922 | 0 | 0 | 1 | 0 | 1 | 1 | 1 | 1 | SRP19 protein | 5 |
| PF01923 | 0 | 1 | 1 | 0 | 0 | 0 | 1 | 0 | Cobalamin adenosyltransferase | 3 |
| PF01926 | 1 | 3 | 16 | 7 | 0 | 3 | 21 | 1 | 50S ribosome-binding GTPase | 52 |
| PF01927 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | Mut7-C RNAse domain | 1 |
| PF01928 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | CYTH domain | 3 |
| PF01929 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 0 | Ribosomal protein L14 | 5 |
| PF01931 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Protein of unknown function DUF84 | 2 |
| PF01936 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | NYN domain | 1 |
| PF01937 | 0 | 0 | 2 | 1 | 0 | 0 | 1 | 0 | Protein of unknown function DUF89 | 4 |
| PF01938 | 0 | 0 | 2 | 2 | 0 | 0 | 1 | 0 | TRAM domain | 5 |
| PF01940 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Integral membrane protein DUF92 | 1 |
| PF01946 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Thi4 family | 1 |
| PF01951 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | Archease protein family (MTH1598/TM1083) | 4 |
| PF01956 | 2 | 2 | 2 | 2 | 1 | 2 | 3 | 2 | Integral membrane protein DUF106 | 16 |
| PF01958 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Domain of unknown function DUF108 | 3 |
| PF01963 | 0 | 1 | 2 | 0 | 0 | 1 | 3 | 0 | TraB family | 7 |
| PF01965 | 1 | 3 | 3 | 2 | 1 | 4 | 6 | 6 | DJ-1/PfpI family | 26 |
| PF01966 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | HD domain | 3 |
| PF01967 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | MoaC family | 1 |
| PF01968 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | Hydantoinase/oxoprolinase | 4 |
| PF01974 | 0 | 0 | 2 | 1 | 0 | 0 | 2 | 0 | "tRNA intron endonuclease, catalytic C-terminal domain" | 5 |
| PF01979 | 0 | 6 | 8 | 4 | 0 | 7 | 9 | 5 | Amidohydrolase family | 39 |
| PF01980 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Uncharacterised protein family UPF0066 | 2 |
| PF01981 | 1 | 1 | 2 | 0 | 1 | 2 | 3 | 2 | Peptidyl-tRNA hydrolase PTH2 | 12 |
| PF01984 | 0 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | Double-stranded DNA-binding domain | 5 |
| PF01990 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | ATP synthase (F/14-kDa) subunit | 8 |
| PF01991 | 1 | 1 | 2 | 1 | 1 | 1 | 2 | 1 | ATP synthase (E/31 kDa) subunit | 10 |
| PF01992 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | ATP synthase (C/AC39) subunit | 9 |
| PF01997 | 0 | 1 | 2 | 2 | 0 | 1 | 2 | 2 | Translin family | 10 |
| PF02002 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | TFIIE alpha subunit | 3 |
| PF02005 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | "N2,N2-dimethylguanosine tRNA methyltransferase" | 4 |
| PF02008 | 0 | 0 | 12 | 0 | 0 | 0 | 14 | 0 | CXXC zinc finger domain | 26 |
| PF02010 | 0 | 1 | 5 | 0 | 0 | 1 | 3 | 0 | REJ domain | 10 |
| PF02014 | 0 | 0 | 3 | 1 | 0 | 0 | 12 | 0 | Reeler domain | 16 |
| PF02017 | 0 | 0 | 5 | 0 | 0 | 0 | 4 | 0 | CIDE-N domain | 9 |
| PF02019 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | WIF domain | 4 |
| PF02020 | 2 | 3 | 7 | 2 | 0 | 0 | 8 | 3 | eIF4-gamma/eIF5/eIF2-epsilon | 25 |
| PF02023 | 0 | 0 | 41 | 1 | 0 | 0 | 0 | 0 | SCAN domain | 42 |
| PF02024 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Leptin | 2 |
| PF02025 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Interleukin 5 | 1 |
| PF02026 | 2 | 1 | 3 | 0 | 2 | 2 | 4 | 1 | RyR domain | 15 |
| PF02029 | 2 | 0 | 2 | 1 | 0 | 0 | 2 | 1 | Caldesmon | 8 |
| PF02033 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Ribosome-binding factor A | 2 |
| PF02036 | 2 | 3 | 5 | 2 | 0 | 4 | 3 | 2 | SCP-2 sterol transfer family | 21 |
| PF02037 | 1 | 1 | 19 | 7 | 2 | 2 | 23 | 9 | SAP domain | 64 |
| PF02038 | 1 | 3 | 7 | 3 | 0 | 2 | 3 | 0 | ATP1G1/PLM/MAT8 family | 19 |
| PF02044 | 0 | 0 | 2 | 0 | 0 | 0 | 3 | 0 | Bombesin-like peptide | 5 |
| PF02045 | 0 | 0 | 1 | 0 | 0 | 0 | 2 | 0 | CCAAT-binding transcription factor (CBF-B/NF-YA) subunit B | 3 |
| PF02046 | 1 | 1 | 2 | 2 | 1 | 1 | 2 | 1 | Cytochrome c oxidase subunit VIa | 11 |
| PF02055 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | O-Glycosyl hydrolase family 30 | 5 |
| PF02057 | 1 | 1 | 1 | 0 | 0 | 0 | 2 | 0 | Glycosyl hydrolase family 59 | 5 |
| PF02058 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | Guanylin precursor | 2 |
| PF02059 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Interleukin-3 | 1 |
| PF02060 | 0 | 0 | 5 | 0 | 0 | 0 | 2 | 0 | Slow voltage-gated potassium channel | 7 |
| PF02063 | 0 | 2 | 2 | 2 | 0 | 3 | 4 | 3 | MARCKS family | 16 |
| PF02064 | 1 | 1 | 2 | 0 | 0 | 0 | 0 | 0 | MAS20 protein import receptor | 4 |
| PF02065 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 0 | Melibiase | 3 |
| PF02070 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Neuromedin U | 2 |
| PF02072 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Prepro-orexin | 2 |
| PF02077 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | SURF4 family | 6 |
| PF02078 | 3 | 3 | 3 | 2 | 2 | 2 | 2 | 2 | "Synapsin, N-terminal domain" | 19 |
| PF02079 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Nuclear transition protein 1 | 1 |
| PF02083 | 0 | 0 | 2 | 0 | 0 | 0 | 3 | 0 | Urotensin II | 5 |
| PF02089 | 1 | 1 | 5 | 0 | 0 | 1 | 6 | 1 | Palmitoyl protein thioesterase | 15 |
| PF02096 | 0 | 0 | 2 | 1 | 1 | 1 | 1 | 0 | 60Kd inner membrane protein | 6 |
| PF02099 | 0 | 0 | 3 | 3 | 0 | 0 | 2 | 0 | Josephin | 8 |
| PF02100 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | Ornithine decarboxylase antizyme | 4 |
| PF02101 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Ocular albinism type 1 protein | 2 |
| PF02104 | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 0 | SURF1 family | 3 |
| PF02106 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Fanconi anaemia group C protein | 2 |
| PF02109 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | DAD family | 2 |
| PF02114 | 0 | 0 | 4 | 1 | 0 | 0 | 3 | 0 | Phosducin | 8 |
| PF02115 | 1 | 1 | 3 | 2 | 0 | 1 | 2 | 2 | RHO protein GDP dissociation inhibitor | 12 |
| PF02121 | 2 | 4 | 5 | 5 | 0 | 1 | 5 | 2 | Phosphatidylinositol transfer protein | 24 |
| PF02126 | 0 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | Phosphotriesterase family | 5 |
| PF02127 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | Aminopeptidase I zinc metalloprotease (M18) | 5 |
| PF02130 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Uncharacterized protein family UPF0054 | 2 |
| PF02134 | 0 | 1 | 6 | 3 | 0 | 1 | 4 | 1 | Repeat in ubiquitin-activating (UBA) protein | 16 |
| PF02135 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 0 | TAZ zinc finger | 3 |
| PF02136 | 3 | 1 | 9 | 4 | 0 | 1 | 8 | 3 | Nuclear transport factor 2 (NTF2) domain | 29 |
| PF02137 | 0 | 0 | 6 | 1 | 1 | 1 | 6 | 2 | Adenosine-deaminase (editase) domain | 17 |
| PF02138 | 0 | 2 | 9 | 2 | 0 | 2 | 7 | 0 | Beige/BEACH domain | 22 |
| PF02140 | 3 | 3 | 4 | 1 | 2 | 4 | 43 | 1 | Galactose binding lectin domain | 61 |
| PF02141 | 1 | 4 | 16 | 4 | 0 | 2 | 14 | 0 | DENN (AEX-3) domain | 41 |
| PF02142 | 0 | 2 | 3 | 0 | 0 | 2 | 3 | 1 | MGS-like domain | 11 |
| PF02144 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Repair protein Rad1/Rec1/Rad17 | 2 |
| PF02145 | 6 | 4 | 10 | 2 | 5 | 4 | 12 | 1 | Rap/ran-GAP | 44 |
| PF02146 | 0 | 1 | 7 | 2 | 0 | 1 | 7 | 2 | Sir2 family | 20 |
| PF02148 | 0 | 1 | 14 | 4 | 0 | 1 | 13 | 1 | Zn-finger in ubiquitin-hydrolases and other protein | 34 |
| PF02149 | 4 | 4 | 5 | 1 | 6 | 5 | 7 | 2 | Kinase associated domain 1 | 34 |
| PF02150 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | RNA polymerases M/15 Kd subunit | 4 |
| PF02151 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | UvrB/uvrC motif | 1 |
| PF02155 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Glucocorticoid receptor | 2 |
| PF02157 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | Mannose-6-phosphate receptor | 6 |
| PF02158 | 1 | 0 | 2 | 1 | 0 | 0 | 2 | 0 | Neuregulin family | 6 |
| PF02159 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Oestrogen receptor | 2 |
| PF02161 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Progesterone receptor | 1 |
| PF02163 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Peptidase family M50 | 2 |
| PF02165 | 0 | 0 | 1 | 0 | 0 | 0 | 2 | 0 | Wilm's tumour protein | 3 |
| PF02166 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Androgen receptor | 2 |
| PF02167 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | Cytochrome C1 family | 8 |
| PF02170 | 2 | 1 | 8 | 0 | 2 | 3 | 7 | 3 | PAZ domain | 26 |
| PF02171 | 2 | 1 | 7 | 0 | 2 | 3 | 6 | 3 | Piwi domain | 24 |
| PF02172 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 0 | KIX domain | 3 |
| PF02173 | 0 | 0 | 3 | 0 | 0 | 0 | 4 | 1 | pKID domain | 8 |
| PF02174 | 2 | 4 | 15 | 2 | 2 | 3 | 14 | 1 | PTB domain (IRS-1 type) | 43 |
| PF02176 | 1 | 1 | 7 | 1 | 1 | 1 | 11 | 0 | TRAF-type zinc finger | 23 |
| PF02177 | 0 | 3 | 3 | 2 | 0 | 4 | 2 | 0 | Amyloid A4 N-terminal heparin-binding | 14 |
| PF02178 | 1 | 0 | 4 | 2 | 0 | 0 | 4 | 1 | AT hook motif | 12 |
| PF02179 | 2 | 2 | 5 | 3 | 2 | 2 | 4 | 2 | BAG domain | 22 |
| PF02180 | 0 | 1 | 3 | 0 | 0 | 2 | 3 | 0 | Bcl-2 homology region 4 | 9 |
| PF02181 | 6 | 6 | 15 | 6 | 7 | 7 | 21 | 0 | Formin Homology 2 Domain | 68 |
| PF02182 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | SAD/SRA domain | 2 |
| PF02184 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | HAT (Half-A-TPR) repeat | 2 |
| PF02185 | 0 | 0 | 7 | 1 | 0 | 0 | 9 | 2 | Hr1 repeat | 19 |
| PF02186 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | TFIIE beta subunit core domain | 2 |
| PF02187 | 1 | 0 | 7 | 2 | 2 | 2 | 7 | 1 | Growth-Arrest-Specific Protein 2 Domain | 22 |
| PF02188 | 3 | 5 | 7 | 4 | 2 | 1 | 9 | 1 | GoLoco motif | 32 |
| PF02189 | 1 | 1 | 7 | 0 | 0 | 0 | 0 | 0 | Immunoreceptor tyrosine-based activation motif | 9 |
| PF02190 | 1 | 2 | 6 | 1 | 0 | 1 | 4 | 0 | ATP-dependent protease La (LON) domain | 15 |
| PF02191 | 6 | 6 | 13 | 2 | 7 | 10 | 22 | 1 | Olfactomedin-like domain | 67 |
| PF02192 | 0 | 1 | 3 | 0 | 0 | 0 | 4 | 0 | "PI3-kinase family, p85-binding domain" | 8 |
| PF02194 | 0 | 1 | 4 | 1 | 0 | 0 | 4 | 0 | PXA domain | 10 |
| PF02195 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | ParB-like nuclease domain | 2 |
| PF02196 | 1 | 2 | 6 | 2 | 1 | 2 | 12 | 1 | Raf-like Ras-binding domain | 27 |
| PF02197 | 4 | 4 | 7 | 2 | 2 | 4 | 5 | 1 | Regulatory subunit of type II PKA R-subunit | 29 |
| PF02198 | 0 | 0 | 10 | 0 | 0 | 0 | 9 | 0 | Sterile alpha motif (SAM)/Pointed domain | 19 |
| PF02199 | 0 | 1 | 3 | 1 | 0 | 0 | 2 | 1 | Saposin A-type domain | 8 |
| PF02201 | 0 | 0 | 5 | 0 | 0 | 0 | 6 | 0 | SWIB/MDM2 domain | 11 |
| PF02204 | 2 | 3 | 10 | 3 | 1 | 2 | 12 | 0 | Vacuolar sorting protein 9 (VPS9) domain | 33 |
| PF02205 | 6 | 6 | 15 | 6 | 6 | 6 | 15 | 2 | WH2 motif | 62 |
| PF02207 | 1 | 1 | 7 | 1 | 0 | 1 | 6 | 0 | Putative zinc finger in N-recognin (UBR box) | 17 |
| PF02208 | 2 | 1 | 3 | 1 | 3 | 2 | 3 | 0 | Sorbin homologous domain | 15 |
| PF02209 | 4 | 3 | 8 | 3 | 6 | 3 | 9 | 2 | Villin headpiece domain | 38 |
| PF02210 | 5 | 15 | 41 | 5 | 6 | 16 | 50 | 9 | Laminin G domain | 147 |
| PF02212 | 4 | 4 | 4 | 3 | 1 | 5 | 14 | 7 | Dynamin GTPase effector domain | 42 |
| PF02213 | 2 | 2 | 3 | 1 | 0 | 0 | 3 | 0 | GYF domain | 11 |
| PF02214 | 13 | 14 | 51 | 10 | 10 | 15 | 53 | 4 | BTB/POZ domain | 170 |
| PF02218 | 1 | 1 | 2 | 0 | 1 | 1 | 2 | 1 | Repeat in HS1/Cortactin | 9 |
| PF02219 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Methylenetetrahydrofolate reductase | 2 |
| PF02221 | 0 | 1 | 4 | 1 | 0 | 1 | 4 | 1 | ML domain | 12 |
| PF02223 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | Thymidylate kinase | 4 |
| PF02225 | 0 | 2 | 18 | 3 | 0 | 2 | 11 | 0 | PA domain | 36 |
| PF02229 | 0 | 0 | 1 | 1 | 0 | 2 | 2 | 2 | Transcriptional Coactivator p15 (PC4) | 8 |
| PF02230 | 0 | 1 | 3 | 1 | 0 | 1 | 2 | 1 | Phospholipase/Carboxylesterase | 9 |
| PF02233 | 0 | 1 | 0 | 0 | 0 | 1 | 3 | 3 | NAD(P) transhydrogenase beta subunit | 8 |
| PF02234 | 0 | 0 | 3 | 1 | 0 | 0 | 4 | 0 | Cyclin-dependent kinase inhibitor | 8 |
| PF02237 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | Biotin protein ligase C terminal domain | 2 |
| PF02238 | 1 | 2 | 3 | 2 | 2 | 2 | 4 | 3 | Cytochrome c oxidase subunit VIIa | 19 |
| PF02244 | 0 | 0 | 8 | 1 | 0 | 0 | 7 | 3 | Carboxypeptidase activation peptide | 19 |
| PF02245 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Methylpurine-DNA glycosylase (MPG) | 2 |
| PF02251 | 0 | 0 | 3 | 3 | 0 | 0 | 3 | 3 | Proteasome activator pa28 alpha subunit | 12 |
| PF02252 | 0 | 0 | 3 | 3 | 0 | 0 | 3 | 3 | Proteasome activator pa28 beta subunit | 12 |
| PF02254 | 0 | 1 | 0 | 0 | 1 | 1 | 1 | 0 | TrkA-N domain | 4 |
| PF02256 | 0 | 0 | 2 | 1 | 0 | 0 | 2 | 0 | Iron hydrogenase small subunit | 5 |
| PF02257 | 0 | 0 | 9 | 0 | 0 | 0 | 9 | 0 | RFX DNA-binding domain | 18 |
| PF02258 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | Shiga-like toxin beta subunit | 2 |
| PF02259 | 1 | 1 | 5 | 0 | 2 | 2 | 5 | 0 | FAT domain | 16 |
| PF02260 | 1 | 1 | 5 | 0 | 2 | 2 | 5 | 0 | FATC domain | 16 |
| PF02262 | 0 | 0 | 3 | 0 | 0 | 0 | 3 | 0 | CBL proto-oncogene N-terminal domain 1 | 6 |
| PF02263 | 1 | 3 | 13 | 5 | 0 | 1 | 11 | 1 | "Guanylate-binding protein, N-terminal domain" | 35 |
| PF02267 | 0 | 1 | 2 | 1 | 0 | 0 | 0 | 0 | ADP-ribosyl cyclase | 4 |
| PF02268 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | "Transcription initiation factor IIA, gamma subunit, helical domain" | 2 |
| PF02269 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 0 | "Transcription initiation factor IID, 18kD subunit" | 3 |
| PF02270 | 0 | 0 | 1 | 1 | 0 | 0 | 2 | 0 | "Transcription initiation factor IIF, beta subunit" | 4 |
| PF02271 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | Ubiquinol-cytochrome C reductase complex 14kD subunit | 6 |
| PF02272 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | DHHA1 domain | 6 |
| PF02274 | 0 | 2 | 3 | 3 | 0 | 1 | 1 | 0 | Amidinotransferase | 10 |
| PF02275 | 1 | 1 | 2 | 1 | 0 | 0 | 2 | 0 | "Linear amide C-N hydrolases, choloylglycine hydrolase family" | 7 |
| PF02284 | 0 | 1 | 1 | 0 | 1 | 2 | 1 | 0 | Cytochrome c oxidase subunit Va | 6 |
| PF02285 | 0 | 0 | 3 | 0 | 0 | 0 | 3 | 0 | Cytochrome oxidase c subunit VIII | 6 |
| PF02290 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | Signal recognition particle 14kD protein | 3 |
| PF02291 | 0 | 0 | 3 | 1 | 0 | 0 | 1 | 0 | "Transcription initiation factor IID, 31kD subunit" | 5 |
| PF02295 | 0 | 0 | 2 | 1 | 0 | 0 | 2 | 1 | Adenosine deaminase z-alpha domain | 6 |
| PF02296 | 2 | 2 | 2 | 2 | 1 | 1 | 1 | 1 | "Alpha adaptin AP2, C-terminal domain" | 12 |
| PF02297 | 0 | 1 | 3 | 1 | 0 | 3 | 3 | 2 | Cytochrome oxidase c subunit VIb | 13 |
| PF02301 | 0 | 0 | 4 | 1 | 0 | 0 | 4 | 0 | HORMA domain | 9 |
| PF02310 | 0 | 1 | 2 | 0 | 0 | 1 | 1 | 1 | B12 binding domain | 6 |
| PF02312 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Core binding factor beta subunit | 2 |
| PF02318 | 5 | 4 | 10 | 5 | 2 | 3 | 12 | 0 | FYVE-type zinc finger | 41 |
| PF02319 | 0 | 0 | 10 | 0 | 0 | 0 | 9 | 0 | E2F/DP family winged-helix DNA-binding domain | 19 |
| PF02320 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | Ubiquinol-cytochrome C reductase hinge protein | 6 |
| PF02330 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | Mitochondrial glycoprotein | 7 |
| PF02338 | 0 | 1 | 12 | 3 | 0 | 0 | 12 | 0 | OTU-like cysteine protease | 28 |
| PF02344 | 0 | 0 | 1 | 0 | 0 | 0 | 2 | 0 | Myc leucine zipper domain | 3 |
| PF02347 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Glycine cleavage system P-protein | 3 |
| PF02348 | 1 | 1 | 1 | 0 | 0 | 0 | 1 | 1 | Cytidylyltransferase | 5 |
| PF02350 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | UDP-N-acetylglucosamine 2-epimerase | 2 |
| PF02351 | 1 | 0 | 6 | 1 | 3 | 3 | 10 | 0 | GDNF/GAS1 domain | 24 |
| PF02354 | 3 | 3 | 2 | 1 | 2 | 5 | 6 | 1 | Latrophilin Cytoplasmic C-terminal region | 23 |
| PF02359 | 0 | 2 | 2 | 0 | 0 | 3 | 4 | 3 | "Cell division protein 48 (CDC48), N-terminal domain" | 14 |
| PF02366 | 1 | 1 | 2 | 2 | 0 | 0 | 2 | 0 | Dolichyl-phosphate-mannose-protein mannosyltransferase | 8 |
| PF02368 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Bacterial Ig-like domain (group 2) | 3 |
| PF02372 | 0 | 0 | 2 | 0 | 0 | 0 | 3 | 0 | Interleukin 15 | 5 |
| PF02373 | 0 | 0 | 24 | 3 | 0 | 0 | 16 | 1 | "JmjC domain, hydroxylase" | 44 |
| PF02374 | 0 | 1 | 2 | 2 | 0 | 1 | 1 | 1 | Anion-transporting ATPase | 8 |
| PF02375 | 0 | 0 | 9 | 0 | 0 | 0 | 8 | 0 | jmjN domain | 17 |
| PF02376 | 0 | 0 | 7 | 1 | 0 | 0 | 10 | 0 | CUT domain | 18 |
| PF02377 | 0 | 0 | 3 | 0 | 0 | 0 | 4 | 0 | Dishevelled specific domain | 7 |
| PF02383 | 1 | 2 | 5 | 2 | 1 | 2 | 4 | 2 | SacI homology domain | 19 |
| PF02389 | 0 | 0 | 4 | 0 | 0 | 0 | 0 | 0 | Cornifin (SPRR) family | 4 |
| PF02390 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Putative methyltransferase | 3 |
| PF02391 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | MoaE protein | 2 |
| PF02394 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | Interleukin-1 propeptide | 2 |
| PF02403 | 1 | 1 | 2 | 0 | 0 | 1 | 1 | 1 | Seryl-tRNA synthetase N-terminal domain | 7 |
| PF02404 | 0 | 0 | 1 | 0 | 0 | 0 | 2 | 0 | Stem cell factor | 3 |
| PF02410 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Oligomerisation domain | 2 |
| PF02412 | 0 | 0 | 5 | 1 | 0 | 0 | 6 | 1 | Thrombospondin type 3 repeat | 13 |
| PF02423 | 0 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | Ornithine cyclodeaminase/mu-crystallin family | 4 |
| PF02434 | 0 | 0 | 5 | 1 | 0 | 1 | 9 | 0 | Fringe-like | 16 |
| PF02436 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | Conserved carboxylase domain | 5 |
| PF02437 | 0 | 0 | 7 | 0 | 0 | 0 | 11 | 0 | SKI/SNO/DAC family | 18 |
| PF02441 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Flavoprotein | 3 |
| PF02450 | 0 | 0 | 2 | 1 | 0 | 0 | 0 | 0 | Lecithin:cholesterol acyltransferase | 3 |
| PF02453 | 0 | 4 | 7 | 4 | 0 | 5 | 8 | 5 | Reticulon | 33 |
| PF02460 | 0 | 1 | 9 | 0 | 1 | 1 | 10 | 0 | Patched family | 22 |
| PF02463 | 2 | 0 | 7 | 3 | 2 | 2 | 8 | 2 | RecF/RecN/SMC N terminal domain | 26 |
| PF02466 | 2 | 5 | 7 | 2 | 1 | 4 | 6 | 1 | Tim17/Tim22/Tim23/Pmp24 family | 28 |
| PF02469 | 0 | 0 | 4 | 0 | 1 | 1 | 4 | 2 | Fasciclin domain | 12 |
| PF02475 | 0 | 0 | 2 | 1 | 0 | 0 | 2 | 0 | Met-10+ like-protein | 5 |
| PF02485 | 0 | 0 | 7 | 0 | 0 | 0 | 7 | 0 | Core-2/I-Branching enzyme | 14 |
| PF02487 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | CLN3 protein | 2 |
| PF02492 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | "CobW/HypB/UreG, nucleotide-binding domain" | 2 |
| PF02493 | 3 | 3 | 14 | 2 | 0 | 0 | 13 | 0 | MORN repeat | 35 |
| PF02494 | 0 | 0 | 3 | 0 | 0 | 0 | 3 | 0 | HYR domain | 6 |
| PF02513 | 0 | 0 | 5 | 1 | 0 | 0 | 3 | 0 | Spin/Ssty Family | 9 |
| PF02515 | 0 | 0 | 2 | 1 | 0 | 0 | 1 | 0 | CoA-transferase family III | 4 |
| PF02516 | 2 | 2 | 2 | 2 | 0 | 2 | 1 | 0 | Oligosaccharyl transferase STT3 subunit | 11 |
| PF02517 | 0 | 0 | 1 | 0 | 0 | 0 | 2 | 0 | CAAX protease self-immunity | 3 |
| PF02518 | 2 | 6 | 11 | 4 | 4 | 5 | 11 | 6 | "Histidine kinase-, DNA gyrase B-, and HSP90-like ATPase" | 49 |
| PF02525 | 0 | 0 | 2 | 0 | 0 | 1 | 1 | 1 | Flavodoxin-like fold | 5 |
| PF02535 | 2 | 3 | 14 | 5 | 0 | 1 | 8 | 0 | ZIP Zinc transporter | 33 |
| PF02536 | 0 | 0 | 5 | 1 | 0 | 0 | 4 | 0 | mTERF | 10 |
| PF02538 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | Hydantoinase B/oxoprolinase | 4 |
| PF02540 | 0 | 0 | 2 | 1 | 0 | 0 | 1 | 1 | NAD synthase | 5 |
| PF02544 | 1 | 1 | 5 | 1 | 1 | 1 | 7 | 1 | 3-oxo-5-alpha-steroid 4-dehydrogenase | 18 |
| PF02545 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | Maf-like protein | 1 |
| PF02567 | 0 | 0 | 1 | 0 | 0 | 0 | 2 | 0 | Phenazine biosynthesis-like protein | 3 |
| PF02574 | 0 | 0 | 3 | 0 | 0 | 1 | 2 | 1 | Homocysteine S-methyltransferase | 7 |
| PF02578 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Multi-copper polyphenol oxidoreductase laccase | 2 |
| PF02580 | 0 | 1 | 2 | 2 | 0 | 1 | 2 | 0 | D-Tyr-tRNA(Tyr) deacylase | 8 |
| PF02582 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | "Uncharacterised ACR, YagE family COG1723" | 5 |
| PF02585 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | GlcNAc-PI de-N-acetylase | 2 |
| PF02586 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | "Uncharacterised ACR, COG2135" | 2 |
| PF02594 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | "Uncharacterised ACR, YggU family COG1872" | 2 |
| PF02597 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | ThiS family | 2 |
| PF02598 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Putative RNA methyltransferase | 2 |
| PF02602 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Uroporphyrinogen-III synthase HemD | 3 |
| PF02607 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | B12 binding domain | 1 |
| PF02617 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | ATP-dependent Clp protease adaptor protein ClpS | 4 |
| PF02627 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | Carboxymuconolactone decarboxylase family | 1 |
| PF02628 | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 0 | Cytochrome oxidase assembly protein | 3 |
| PF02629 | 1 | 2 | 2 | 1 | 0 | 2 | 2 | 1 | CoA binding domain | 11 |
| PF02630 | 1 | 2 | 2 | 0 | 0 | 2 | 2 | 1 | SCO1/SenC | 10 |
| PF02636 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Putative S-adenosyl-L-methionine-dependent methyltransferase | 2 |
| PF02637 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | GatB domain | 3 |
| PF02661 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Fic/DOC family | 2 |
| PF02666 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | Phosphatidylserine decarboxylase | 6 |
| PF02668 | 0 | 0 | 1 | 2 | 0 | 0 | 2 | 0 | "Taurine catabolism dioxygenase TauD, TfdA family" | 5 |
| PF02671 | 0 | 0 | 3 | 0 | 0 | 0 | 5 | 0 | Paired amphipathic helix repeat | 8 |
| PF02676 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Methyltransferase TYW3 | 2 |
| PF02678 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Pirin | 3 |
| PF02686 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Glu-tRNAGln amidotransferase C subunit | 2 |
| PF02690 | 0 | 0 | 3 | 0 | 0 | 0 | 4 | 0 | Na+/Pi-cotransporter | 7 |
| PF02696 | 0 | 0 | 1 | 0 | 0 | 0 | 2 | 0 | "Uncharacterized ACR, YdiU/UPF0061 family" | 3 |
| PF02709 | 0 | 0 | 11 | 0 | 0 | 0 | 9 | 1 | N-terminal domain of galactosyltransferase | 21 |
| PF02714 | 1 | 2 | 3 | 1 | 0 | 0 | 2 | 0 | Domain of unknown function DUF221 | 9 |
| PF02724 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | CDC45-like protein | 2 |
| PF02727 | 0 | 0 | 6 | 1 | 0 | 1 | 2 | 2 | "Copper amine oxidase, N2 domain" | 12 |
| PF02728 | 0 | 0 | 6 | 1 | 0 | 1 | 2 | 2 | "Copper amine oxidase, N3 domain" | 12 |
| PF02729 | 0 | 1 | 2 | 0 | 0 | 1 | 2 | 0 | "Aspartate/ornithine carbamoyltransferase, carbamoyl-P binding domain" | 6 |
| PF02731 | 1 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | SKIP/SNW domain | 3 |
| PF02732 | 0 | 0 | 5 | 0 | 0 | 0 | 4 | 0 | ERCC4 domain | 9 |
| PF02733 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Dak1 domain | 2 |
| PF02734 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | DAK2 domain | 2 |
| PF02735 | 0 | 0 | 2 | 0 | 1 | 1 | 1 | 0 | Ku70/Ku80 beta-barrel domain | 5 |
| PF02736 | 3 | 5 | 13 | 1 | 7 | 5 | 18 | 10 | Myosin N-terminal SH3-like domain | 62 |
| PF02737 | 1 | 2 | 4 | 4 | 3 | 5 | 6 | 4 | "3-hydroxyacyl-CoA dehydrogenase, NAD binding domain" | 29 |
| PF02738 | 0 | 0 | 5 | 0 | 0 | 1 | 3 | 0 | Molybdopterin-binding domain of aldehyde dehydrogenase | 9 |
| PF02740 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | "Colipase, C-terminal domain" | 1 |
| PF02743 | 3 | 3 | 4 | 2 | 3 | 3 | 6 | 1 | Cache domain | 25 |
| PF02744 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | "Galactose-1-phosphate uridyl transferase, C-terminal domain" | 2 |
| PF02746 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | "Mandelate racemase / muconate lactonizing enzyme, N-terminal domain" | 2 |
| PF02747 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | "Proliferating cell nuclear antigen, C-terminal domain" | 3 |
| PF02749 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | "Quinolinate phosphoribosyl transferase, N-terminal domain" | 1 |
| PF02750 | 3 | 3 | 3 | 2 | 2 | 2 | 2 | 2 | "Synapsin, ATP binding domain" | 19 |
| PF02751 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | "Transcription initiation factor IIA, gamma subunit" | 2 |
| PF02752 | 0 | 1 | 10 | 2 | 0 | 1 | 16 | 0 | "Arrestin (or S-antigen), C-terminal domain" | 30 |
| PF02755 | 1 | 1 | 7 | 0 | 0 | 0 | 11 | 0 | RPEL repeat | 20 |
| PF02758 | 0 | 0 | 26 | 2 | 0 | 0 | 13 | 0 | PAAD/DAPIN/Pyrin domain | 41 |
| PF02759 | 3 | 2 | 19 | 6 | 2 | 3 | 20 | 2 | RUN domain | 57 |
| PF02760 | 0 | 0 | 10 | 1 | 0 | 0 | 0 | 0 | HIN-200/IF120x domain | 11 |
| PF02761 | 0 | 0 | 3 | 0 | 0 | 0 | 3 | 0 | "CBL proto-oncogene N-terminus, EF hand-like domain" | 6 |
| PF02762 | 0 | 0 | 3 | 0 | 0 | 0 | 3 | 0 | "CBL proto-oncogene N-terminus, SH2-like domain" | 6 |
| PF02765 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 0 | Telomeric single stranded DNA binding POT1/CDC13 | 3 |
| PF02769 | 0 | 0 | 4 | 1 | 0 | 0 | 3 | 1 | "AIR synthase related protein, C-terminal domain" | 9 |
| PF02770 | 5 | 11 | 15 | 4 | 4 | 14 | 14 | 10 | "Acyl-CoA dehydrogenase, middle domain" | 77 |
| PF02771 | 5 | 9 | 12 | 4 | 3 | 11 | 11 | 8 | "Acyl-CoA dehydrogenase, N-terminal domain" | 63 |
| PF02772 | 0 | 1 | 2 | 1 | 0 | 1 | 4 | 1 | "S-adenosylmethionine synthetase, central domain" | 10 |
| PF02773 | 0 | 1 | 2 | 1 | 0 | 1 | 4 | 1 | "S-adenosylmethionine synthetase, C-terminal domain" | 10 |
| PF02775 | 1 | 2 | 2 | 2 | 1 | 2 | 2 | 0 | "Thiamine pyrophosphate enzyme, C-terminal TPP binding domain" | 12 |
| PF02776 | 1 | 2 | 2 | 2 | 1 | 2 | 2 | 0 | "Thiamine pyrophosphate enzyme, N-terminal TPP binding domain" | 12 |
| PF02777 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | "Iron/manganese superoxide dismutases, C-terminal domain" | 6 |
| PF02778 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | "tRNA intron endonuclease, N-terminal domain" | 2 |
| PF02779 | 3 | 5 | 8 | 4 | 4 | 7 | 7 | 7 | "Transketolase, pyrimidine binding domain" | 45 |
| PF02780 | 1 | 3 | 5 | 2 | 1 | 3 | 4 | 4 | "Transketolase, C-terminal domain" | 23 |
| PF02781 | 0 | 1 | 3 | 2 | 0 | 1 | 2 | 1 | "Glucose-6-phosphate dehydrogenase, C-terminal domain" | 10 |
| PF02782 | 1 | 1 | 6 | 0 | 0 | 1 | 4 | 0 | "FGGY family of carbohydrate kinases, C-terminal domain" | 13 |
| PF02784 | 0 | 0 | 4 | 0 | 0 | 0 | 3 | 0 | "Pyridoxal-dependent decarboxylase, pyridoxal binding domain" | 7 |
| PF02785 | 1 | 4 | 5 | 1 | 0 | 4 | 4 | 2 | Biotin carboxylase C-terminal domain | 21 |
| PF02786 | 1 | 5 | 7 | 1 | 0 | 5 | 6 | 2 | "Carbamoyl-phosphate synthase L chain, ATP binding domain" | 27 |
| PF02787 | 0 | 1 | 2 | 0 | 0 | 1 | 2 | 0 | "Carbamoyl-phosphate synthetase large chain, oligomerisation domain" | 6 |
| PF02789 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | "Cytosol aminopeptidase family, N-terminal domain" | 5 |
| PF02790 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 1 | "Cytochrome C oxidase subunit II, transmembrane domain" | 6 |
| PF02791 | 0 | 0 | 4 | 0 | 0 | 0 | 4 | 0 | DDT domain | 8 |
| PF02792 | 0 | 0 | 2 | 1 | 0 | 0 | 1 | 1 | Mago nashi protein | 5 |
| PF02793 | 6 | 7 | 24 | 1 | 4 | 7 | 25 | 1 | Hormone receptor domain | 75 |
| PF02798 | 3 | 6 | 18 | 4 | 1 | 7 | 10 | 8 | "Glutathione S-transferase, N-terminal domain" | 57 |
| PF02799 | 0 | 0 | 2 | 1 | 0 | 1 | 3 | 1 | "Myristoyl-CoA:protein N-myristoyltransferase, C-terminal domain" | 8 |
| PF02800 | 1 | 1 | 2 | 0 | 0 | 2 | 2 | 2 | "Glyceraldehyde 3-phosphate dehydrogenase, C-terminal domain" | 10 |
| PF02801 | 0 | 1 | 2 | 2 | 0 | 1 | 2 | 0 | "Beta-ketoacyl synthase, C-terminal domain" | 8 |
| PF02803 | 3 | 6 | 8 | 4 | 0 | 6 | 5 | 5 | "Thiolase, C-terminal domain" | 37 |
| PF02806 | 0 | 1 | 5 | 0 | 0 | 1 | 4 | 1 | "Alpha amylase, C-terminal all-beta domain" | 12 |
| PF02807 | 1 | 2 | 4 | 3 | 2 | 4 | 6 | 5 | "ATP:guanido phosphotransferase, N-terminal domain" | 27 |
| PF02809 | 2 | 5 | 10 | 5 | 0 | 2 | 6 | 1 | Ubiquitin interaction motif | 31 |
| PF02812 | 1 | 1 | 1 | 0 | 2 | 2 | 1 | 1 | "Glu/Leu/Phe/Val dehydrogenase, dimerisation domain" | 9 |
| PF02815 | 6 | 7 | 10 | 3 | 5 | 6 | 12 | 2 | MIR domain | 51 |
| PF02816 | 0 | 0 | 6 | 0 | 0 | 0 | 5 | 0 | Alpha-kinase family | 11 |
| PF02817 | 2 | 3 | 3 | 1 | 3 | 3 | 2 | 2 | e3 binding domain | 19 |
| PF02818 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | PPAK motif | 3 |
| PF02820 | 1 | 1 | 8 | 1 | 0 | 0 | 9 | 0 | mbt repeat | 20 |
| PF02822 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Antistasin family | 3 |
| PF02823 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | "ATP synthase, Delta/Epsilon chain, beta-sandwich domain" | 5 |
| PF02824 | 1 | 3 | 5 | 3 | 1 | 3 | 3 | 1 | TGS domain | 20 |
| PF02825 | 0 | 1 | 8 | 0 | 4 | 4 | 13 | 1 | WWE domain | 31 |
| PF02826 | 1 | 3 | 4 | 2 | 3 | 4 | 7 | 5 | "D-isomer specific 2-hydroxyacid dehydrogenase, NAD binding domain" | 29 |
| PF02827 | 0 | 1 | 3 | 1 | 0 | 0 | 2 | 0 | cAMP-dependent protein kinase inhibitor | 7 |
| PF02828 | 7 | 6 | 10 | 6 | 8 | 9 | 15 | 4 | L27 domain | 65 |
| PF02833 | 0 | 0 | 2 | 1 | 0 | 0 | 1 | 1 | DHHA2 domain | 5 |
| PF02836 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | "Glycosyl hydrolases family 2, TIM barrel domain" | 4 |
| PF02837 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | "Glycosyl hydrolases family 2, sugar binding domain" | 4 |
| PF02840 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Prp18 domain | 2 |
| PF02841 | 0 | 2 | 11 | 5 | 0 | 0 | 8 | 0 | "Guanylate-binding protein, C-terminal domain" | 26 |
| PF02843 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | "Phosphoribosylglycinamide synthetase, C domain" | 1 |
| PF02844 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | "Phosphoribosylglycinamide synthetase, N domain" | 1 |
| PF02845 | 1 | 2 | 8 | 2 | 2 | 1 | 8 | 0 | CUE domain | 24 |
| PF02847 | 2 | 2 | 6 | 0 | 0 | 0 | 9 | 2 | MA3 domain | 21 |
| PF02852 | 1 | 4 | 4 | 2 | 1 | 2 | 4 | 1 | "Pyridine nucleotide-disulphide oxidoreductase, dimerisation domain" | 19 |
| PF02854 | 2 | 2 | 10 | 2 | 0 | 0 | 9 | 1 | MIF4G domain | 26 |
| PF02862 | 1 | 2 | 6 | 3 | 0 | 2 | 6 | 1 | DDHD domain | 21 |
| PF02864 | 0 | 0 | 7 | 1 | 0 | 1 | 8 | 0 | "STAT protein, DNA binding domain" | 17 |
| PF02865 | 0 | 0 | 7 | 1 | 0 | 1 | 8 | 0 | "STAT protein, protein interaction domain" | 17 |
| PF02866 | 0 | 4 | 7 | 3 | 0 | 5 | 6 | 4 | "lactate/malate dehydrogenase, alpha/beta C-terminal domain" | 29 |
| PF02867 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | "Ribonucleotide reductase, barrel domain" | 2 |
| PF02870 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | "6-O-methylguanine DNA methyltransferase, ribonuclease-like domain" | 2 |
| PF02872 | 1 | 1 | 2 | 1 | 1 | 0 | 1 | 0 | "5'-nucleotidase, C-terminal domain" | 7 |
| PF02874 | 2 | 4 | 5 | 4 | 2 | 6 | 6 | 6 | "ATP synthase alpha/beta family, beta-barrel domain" | 35 |
| PF02877 | 0 | 0 | 3 | 2 | 3 | 2 | 3 | 2 | "Poly(ADP-ribose) polymerase, regulatory domain" | 15 |
| PF02878 | 0 | 2 | 5 | 1 | 0 | 1 | 5 | 4 | "Phosphoglucomutase/phosphomannomutase, alpha/beta/alpha domain I" | 18 |
| PF02879 | 0 | 2 | 5 | 1 | 0 | 1 | 5 | 4 | "Phosphoglucomutase/phosphomannomutase, alpha/beta/alpha domain II" | 18 |
| PF02880 | 0 | 2 | 4 | 1 | 0 | 1 | 4 | 3 | "Phosphoglucomutase/phosphomannomutase, alpha/beta/alpha domain III" | 15 |
| PF02881 | 1 | 1 | 3 | 1 | 1 | 1 | 1 | 0 | "SRP54-type protein, helical bundle domain" | 9 |
| PF02882 | 1 | 2 | 4 | 2 | 1 | 1 | 4 | 1 | "Tetrahydrofolate dehydrogenase/cyclohydrolase, NAD(P)-binding domain" | 16 |
| PF02883 | 4 | 5 | 9 | 5 | 5 | 5 | 5 | 4 | Adaptin C-terminal domain | 42 |
| PF02885 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | "Glycosyl transferase family, helical bundle domain" | 1 |
| PF02886 | 0 | 0 | 9 | 0 | 0 | 0 | 1 | 1 | "LBP / BPI / CETP family, C-terminal domain" | 11 |
| PF02887 | 0 | 1 | 2 | 0 | 1 | 1 | 1 | 0 | "Pyruvate kinase, alpha/beta domain" | 6 |
| PF02888 | 1 | 1 | 3 | 0 | 0 | 0 | 5 | 0 | Calmodulin binding domain | 10 |
| PF02889 | 0 | 1 | 4 | 0 | 0 | 1 | 3 | 2 | Sec63 Brl domain | 11 |
| PF02891 | 0 | 0 | 6 | 2 | 0 | 1 | 6 | 0 | MIZ/SP-RING zinc finger | 15 |
| PF02892 | 0 | 0 | 3 | 0 | 0 | 0 | 9 | 1 | BED zinc finger | 13 |
| PF02893 | 3 | 6 | 17 | 3 | 0 | 2 | 17 | 0 | GRAM domain | 48 |
| PF02894 | 0 | 0 | 2 | 2 | 0 | 0 | 2 | 0 | "Oxidoreductase family, C-terminal alpha/beta domain" | 6 |
| PF02897 | 0 | 1 | 2 | 2 | 0 | 1 | 2 | 1 | "Prolyl oligopeptidase, N-terminal beta-propeller domain" | 9 |
| PF02898 | 0 | 1 | 3 | 1 | 0 | 1 | 3 | 0 | "Nitric oxide synthase, oxygenase domain" | 9 |
| PF02902 | 0 | 0 | 10 | 3 | 0 | 0 | 11 | 1 | "Ulp1 protease family, C-terminal catalytic domain" | 25 |
| PF02906 | 0 | 0 | 2 | 1 | 0 | 0 | 2 | 0 | "Iron only hydrogenase large subunit, C-terminal domain" | 5 |
| PF02910 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | Fumarate reductase flavoprotein C-term | 6 |
| PF02911 | 0 | 1 | 3 | 0 | 0 | 2 | 3 | 2 | "Formyl transferase, C-terminal domain" | 11 |
| PF02913 | 0 | 1 | 3 | 1 | 1 | 2 | 3 | 0 | "FAD linked oxidases, C-terminal domain" | 11 |
| PF02919 | 0 | 0 | 2 | 0 | 0 | 0 | 3 | 1 | "Eukaryotic DNA topoisomerase I, DNA binding fragment" | 6 |
| PF02920 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | DNA binding domain of tn916 integrase | 2 |
| PF02921 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | Ubiquinol cytochrome reductase transmembrane region | 7 |
| PF02922 | 0 | 1 | 0 | 0 | 0 | 1 | 1 | 0 | Carbohydrate-binding module 48 (Isoamylase N-terminal domain) | 3 |
| PF02926 | 0 | 0 | 3 | 2 | 0 | 0 | 2 | 0 | THUMP domain | 7 |
| PF02928 | 0 | 0 | 5 | 0 | 0 | 0 | 5 | 0 | C5HC2 zinc finger | 10 |
| PF02931 | 11 | 10 | 42 | 11 | 9 | 10 | 59 | 2 | Neurotransmitter-gated ion-channel ligand binding domain | 154 |
| PF02932 | 11 | 10 | 42 | 11 | 8 | 10 | 58 | 2 | Neurotransmitter-gated ion-channel transmembrane region | 152 |
| PF02933 | 0 | 2 | 2 | 0 | 0 | 3 | 4 | 3 | "Cell division protein 48 (CDC48), domain 2" | 14 |
| PF02934 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | GatB/GatE catalytic domain | 3 |
| PF02935 | 0 | 1 | 1 | 1 | 0 | 1 | 0 | 0 | Cytochrome c oxidase subunit VIIc | 4 |
| PF02936 | 1 | 1 | 2 | 2 | 3 | 3 | 3 | 3 | Cytochrome c oxidase subunit IV | 18 |
| PF02937 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | Cytochrome c oxidase subunit VIc | 7 |
| PF02938 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | GAD domain | 3 |
| PF02939 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | UcrQ family | 8 |
| PF02944 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | BESS motif | 2 |
| PF02946 | 1 | 1 | 3 | 1 | 0 | 0 | 2 | 0 | GTF2I-like repeat | 8 |
| PF02947 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | flt3 ligand | 1 |
| PF02948 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Amelogenin | 1 |
| PF02953 | 2 | 6 | 7 | 6 | 2 | 5 | 6 | 3 | Tim10/DDP family zinc finger | 37 |
| PF02961 | 1 | 1 | 2 | 1 | 1 | 1 | 2 | 1 | Barrier to autointegration factor | 10 |
| PF02965 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | "Vitamin B12 dependent methionine synthase, activation domain" | 1 |
| PF02966 | 0 | 0 | 2 | 1 | 0 | 0 | 2 | 0 | Mitosis protein DIM1 | 5 |
| PF02969 | 0 | 0 | 2 | 1 | 0 | 0 | 1 | 0 | TATA box binding protein associated factor (TAF) | 4 |
| PF02970 | 0 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | Tubulin binding cofactor A | 5 |
| PF02971 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Formiminotransferase domain | 2 |
| PF02978 | 1 | 0 | 2 | 0 | 1 | 1 | 0 | 0 | Signal peptide binding domain | 5 |
| PF02984 | 0 | 0 | 15 | 1 | 0 | 0 | 13 | 0 | "Cyclin, C-terminal domain" | 29 |
| PF02985 | 1 | 4 | 15 | 1 | 2 | 4 | 12 | 3 | HEAT repeat | 42 |
| PF02988 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Phospholipase A2 inhibitor | 1 |
| PF02990 | 0 | 3 | 5 | 4 | 1 | 3 | 5 | 0 | Endomembrane protein 70 | 21 |
| PF02991 | 4 | 3 | 5 | 2 | 3 | 3 | 6 | 3 | Autophagy protein Atg8 ubiquitin like | 29 |
| PF02994 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | L1 transposable element | 1 |
| PF02996 | 0 | 1 | 4 | 1 | 0 | 2 | 4 | 2 | Prefoldin subunit | 14 |
| PF03002 | 1 | 0 | 2 | 1 | 0 | 0 | 5 | 0 | Somatostatin/Cortistatin family | 9 |
| PF03006 | 0 | 1 | 11 | 2 | 0 | 0 | 15 | 1 | Haemolysin-III related | 30 |
| PF03009 | 0 | 2 | 7 | 3 | 0 | 3 | 9 | 0 | Glycerophosphoryl diester phosphodiesterase family | 24 |
| PF03015 | 1 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | Male sterility protein | 5 |
| PF03016 | 0 | 0 | 4 | 0 | 3 | 3 | 5 | 0 | Exostosin family | 15 |
| PF03020 | 2 | 1 | 7 | 2 | 2 | 2 | 8 | 2 | LEM domain | 26 |
| PF03024 | 0 | 0 | 7 | 0 | 0 | 0 | 3 | 0 | Folate receptor family | 10 |
| PF03028 | 0 | 2 | 13 | 1 | 1 | 2 | 12 | 1 | Dynein heavy chain and region D6 of dynein motor | 32 |
| PF03029 | 0 | 0 | 3 | 0 | 0 | 0 | 2 | 0 | Conserved hypothetical ATP binding protein | 5 |
| PF03031 | 0 | 2 | 8 | 3 | 0 | 2 | 12 | 1 | NLI interacting factor-like phosphatase | 28 |
| PF03034 | 1 | 1 | 2 | 1 | 0 | 1 | 1 | 1 | Phosphatidyl serine synthase | 8 |
| PF03036 | 0 | 0 | 4 | 1 | 0 | 0 | 4 | 0 | Perilipin family | 9 |
| PF03039 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Interleukin-12 alpha subunit | 2 |
| PF03045 | 0 | 0 | 7 | 0 | 0 | 0 | 4 | 0 | DAN domain | 11 |
| PF03051 | 0 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | Peptidase C1-like family | 5 |
| PF03054 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | tRNA methyl transferase | 3 |
| PF03055 | 0 | 0 | 3 | 1 | 0 | 2 | 6 | 0 | Retinal pigment epithelial membrane protein | 12 |
| PF03061 | 0 | 4 | 7 | 3 | 0 | 2 | 6 | 2 | Thioesterase superfamily | 24 |
| PF03062 | 0 | 2 | 11 | 3 | 0 | 1 | 14 | 0 | "MBOAT, membrane-bound O-acyltransferase family" | 31 |
| PF03066 | 2 | 1 | 3 | 2 | 0 | 0 | 4 | 2 | Nucleoplasmin | 14 |
| PF03068 | 0 | 1 | 5 | 0 | 0 | 1 | 1 | 1 | Protein-arginine deiminase (PAD) | 9 |
| PF03071 | 0 | 0 | 2 | 0 | 0 | 0 | 3 | 0 | GNT-I family | 5 |
| PF03073 | 0 | 0 | 2 | 1 | 0 | 0 | 1 | 0 | TspO/MBR family | 4 |
| PF03074 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | Glutamate-cysteine ligase | 4 |
| PF03079 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | ARD/ARD' family | 2 |
| PF03081 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | Exo70 exocyst complex subunit | 7 |
| PF03083 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Sugar efflux transporter for intercellular exchange | 2 |
| PF03088 | 0 | 1 | 1 | 0 | 0 | 2 | 2 | 1 | Strictosidine synthase | 7 |
| PF03089 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Recombination activating protein 2 | 2 |
| PF03091 | 0 | 0 | 2 | 1 | 0 | 0 | 1 | 0 | CutA1 divalent ion tolerance protein | 4 |
| PF03095 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | Phosphotyrosyl phosphate activator (PTPA) protein | 7 |
| PF03096 | 0 | 4 | 4 | 4 | 1 | 5 | 5 | 4 | Ndr family | 27 |
| PF03097 | 0 | 1 | 5 | 2 | 0 | 3 | 5 | 2 | BRO1-like domain | 18 |
| PF03098 | 0 | 1 | 9 | 0 | 0 | 0 | 7 | 0 | Animal haem peroxidase | 17 |
| PF03099 | 0 | 0 | 3 | 2 | 0 | 0 | 2 | 0 | Biotin/lipoate A/B protein ligase family | 7 |
| PF03102 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | NeuB family | 6 |
| PF03104 | 0 | 0 | 4 | 0 | 0 | 0 | 4 | 0 | "DNA polymerase family B, exonuclease domain" | 8 |
| PF03105 | 0 | 1 | 1 | 1 | 0 | 0 | 2 | 0 | SPX domain | 5 |
| PF03109 | 0 | 1 | 5 | 3 | 1 | 4 | 5 | 1 | ABC1 family | 20 |
| PF03114 | 1 | 10 | 15 | 8 | 1 | 11 | 22 | 8 | BAR domain | 76 |
| PF03121 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Herpesviridae UL52/UL70 DNA primase | 2 |
| PF03124 | 0 | 1 | 1 | 1 | 0 | 0 | 2 | 0 | EXS family | 5 |
| PF03126 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Plus-3 domain | 3 |
| PF03127 | 2 | 2 | 6 | 2 | 2 | 3 | 3 | 2 | GAT domain | 22 |
| PF03128 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | CXCXC repeat | 2 |
| PF03129 | 0 | 5 | 9 | 4 | 2 | 4 | 8 | 4 | Anticodon binding domain | 36 |
| PF03130 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | PBS lyase HEAT-like repeat | 2 |
| PF03131 | 0 | 0 | 13 | 0 | 0 | 0 | 17 | 0 | bZIP Maf transcription factor | 30 |
| PF03133 | 0 | 0 | 14 | 1 | 0 | 0 | 10 | 0 | Tubulin-tyrosine ligase family | 25 |
| PF03134 | 1 | 3 | 6 | 2 | 0 | 2 | 6 | 1 | "TB2/DP1, HVA22 family" | 21 |
| PF03137 | 0 | 0 | 15 | 2 | 1 | 1 | 8 | 0 | Organic Anion Transporter Polypeptide (OATP) family | 27 |
| PF03142 | 0 | 0 | 1 | 0 | 0 | 0 | 4 | 0 | Chitin synthase | 5 |
| PF03143 | 4 | 4 | 8 | 3 | 5 | 4 | 10 | 5 | Elongation factor Tu C-terminal domain | 43 |
| PF03144 | 5 | 7 | 18 | 7 | 6 | 7 | 22 | 10 | Elongation factor Tu domain 2 | 82 |
| PF03145 | 0 | 0 | 4 | 0 | 0 | 0 | 2 | 0 | Seven in absentia protein family | 6 |
| PF03146 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | Agrin NtA domain | 6 |
| PF03147 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | Ferredoxin-fold anticodon binding domain | 4 |
| PF03148 | 0 | 0 | 6 | 1 | 0 | 0 | 4 | 0 | Tektin family | 11 |
| PF03151 | 0 | 0 | 8 | 0 | 0 | 1 | 7 | 0 | Triose-phosphate Transporter family | 16 |
| PF03152 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Ubiquitin fusion degradation protein UFD1 | 2 |
| PF03153 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | "Transcription factor IIA, alpha/beta subunit" | 4 |
| PF03154 | 0 | 0 | 2 | 0 | 0 | 0 | 3 | 0 | Atrophin-1 family | 5 |
| PF03155 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | "ALG6, ALG8 glycosyltransferase family" | 4 |
| PF03157 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | High molecular weight glutenin subunit | 3 |
| PF03159 | 0 | 0 | 2 | 1 | 0 | 0 | 1 | 0 | XRN 5'-3' exonuclease N-terminus | 4 |
| PF03160 | 0 | 3 | 9 | 1 | 1 | 5 | 13 | 2 | Calx-beta domain | 34 |
| PF03164 | 2 | 2 | 2 | 2 | 0 | 0 | 3 | 0 | Trafficking protein Mon1 | 11 |
| PF03165 | 0 | 0 | 12 | 6 | 0 | 0 | 17 | 0 | MH1 domain | 35 |
| PF03166 | 0 | 0 | 8 | 3 | 0 | 0 | 13 | 0 | MH2 domain | 24 |
| PF03167 | 0 | 0 | 3 | 0 | 0 | 0 | 4 | 0 | Uracil DNA glycosylase superfamily | 7 |
| PF03171 | 0 | 0 | 7 | 1 | 0 | 0 | 10 | 0 | 2OG-Fe(II) oxygenase superfamily | 18 |
| PF03172 | 0 | 0 | 7 | 0 | 0 | 0 | 3 | 0 | Sp100 domain | 10 |
| PF03177 | 0 | 0 | 2 | 1 | 1 | 1 | 2 | 0 | Non-repetitive/WGA-negative nucleoporin C-terminal | 7 |
| PF03178 | 1 | 1 | 3 | 2 | 0 | 1 | 2 | 1 | CPSF A subunit region | 11 |
| PF03179 | 2 | 2 | 3 | 2 | 1 | 1 | 1 | 1 | Vacuolar (H+)-ATPase G subunit | 13 |
| PF03184 | 0 | 0 | 8 | 1 | 0 | 0 | 2 | 0 | DDE superfamily endonuclease | 11 |
| PF03185 | 0 | 0 | 4 | 1 | 0 | 0 | 3 | 0 | "Calcium-activated potassium channel, beta subunit" | 8 |
| PF03188 | 1 | 1 | 5 | 0 | 0 | 1 | 7 | 0 | Eukaryotic cytochrome b561 | 15 |
| PF03189 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | Otopetrin | 4 |
| PF03190 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | "Protein of unknown function, DUF255" | 2 |
| PF03194 | 0 | 0 | 3 | 2 | 0 | 0 | 3 | 1 | LUC7 N_terminus | 9 |
| PF03199 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Eukaryotic glutathione synthase | 3 |
| PF03200 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | Mannosyl oligosaccharide glucosidase | 7 |
| PF03205 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | Molybdopterin guanine dinucleotide synthesis protein B | 4 |
| PF03208 | 0 | 2 | 3 | 2 | 0 | 2 | 4 | 1 | PRA1 family protein | 14 |
| PF03215 | 0 | 0 | 3 | 0 | 0 | 0 | 2 | 0 | Rad17 cell cycle checkpoint protein | 5 |
| PF03221 | 0 | 0 | 8 | 1 | 0 | 0 | 2 | 0 | Tc5 transposase DNA-binding domain | 11 |
| PF03223 | 1 | 1 | 2 | 1 | 1 | 2 | 3 | 2 | V-ATPase subunit C | 13 |
| PF03224 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | V-ATPase subunit H | 9 |
| PF03226 | 0 | 0 | 7 | 0 | 1 | 1 | 7 | 0 | "Yippee zinc-binding/DNA-binding /Mis18, centromere assembly" | 16 |
| PF03227 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 1 | Gamma interferon inducible lysosomal thiol reductase (GILT) | 3 |
| PF03232 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | Ubiquinone biosynthesis protein COQ7 | 6 |
| PF03234 | 0 | 1 | 1 | 1 | 0 | 1 | 0 | 0 | Cdc37 N terminal kinase binding | 4 |
| PF03247 | 0 | 2 | 2 | 0 | 0 | 1 | 2 | 1 | Prothymosin/parathymosin family | 8 |
| PF03248 | 0 | 1 | 1 | 0 | 0 | 0 | 1 | 0 | Rer1 family | 3 |
| PF03250 | 3 | 3 | 7 | 0 | 2 | 2 | 7 | 2 | Tropomodulin | 26 |
| PF03253 | 1 | 1 | 2 | 0 | 0 | 0 | 1 | 0 | Urea transporter | 5 |
| PF03256 | 0 | 0 | 6 | 1 | 1 | 0 | 4 | 1 | "Anaphase-promoting complex, subunit 10 (APC10)" | 13 |
| PF03259 | 0 | 2 | 3 | 0 | 0 | 2 | 3 | 1 | Roadblock/LC7 domain | 11 |
| PF03261 | 1 | 1 | 2 | 1 | 1 | 1 | 5 | 0 | Cyclin-dependent kinase 5 activator protein | 12 |
| PF03265 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | Deoxyribonuclease II | 4 |
| PF03266 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | NTPase | 4 |
| PF03271 | 3 | 3 | 3 | 2 | 0 | 4 | 3 | 2 | EB1-like C-terminal motif | 20 |
| PF03281 | 0 | 1 | 12 | 3 | 0 | 0 | 11 | 0 | Mab-21 protein | 27 |
| PF03283 | 0 | 0 | 1 | 0 | 0 | 0 | 3 | 0 | Pectinacetylesterase | 4 |
| PF03285 | 3 | 3 | 3 | 1 | 1 | 2 | 3 | 2 | Paralemmin | 18 |
| PF03291 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | mRNA capping enzyme | 6 |
| PF03297 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | S25 ribosomal protein | 7 |
| PF03298 | 0 | 0 | 2 | 0 | 0 | 0 | 3 | 1 | Stanniocalcin family | 6 |
| PF03299 | 0 | 0 | 5 | 0 | 0 | 0 | 6 | 0 | Transcription factor AP-2 | 11 |
| PF03301 | 0 | 0 | 1 | 0 | 0 | 0 | 2 | 0 | "Tryptophan 2,3-dioxygenase" | 3 |
| PF03308 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | ArgK protein | 7 |
| PF03311 | 0 | 0 | 4 | 1 | 0 | 0 | 4 | 0 | Cornichon protein | 9 |
| PF03318 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | Clostridium epsilon toxin ETX/Bacillus mosquitocidal toxin MTX2 | 2 |
| PF03321 | 0 | 0 | 1 | 0 | 1 | 1 | 1 | 0 | GH3 auxin-responsive promoter | 4 |
| PF03328 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | HpcH/HpaI aldolase/citrate lyase family | 6 |
| PF03332 | 0 | 0 | 2 | 1 | 0 | 0 | 2 | 1 | Eukaryotic phosphomannomutase | 6 |
| PF03343 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 1 | SART-1 family | 3 |
| PF03344 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Daxx Family | 2 |
| PF03345 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | Oligosaccharyltransferase 48 kDa subunit beta | 7 |
| PF03348 | 0 | 1 | 5 | 2 | 0 | 0 | 6 | 0 | Serine incorporator (Serinc) | 14 |
| PF03351 | 1 | 1 | 5 | 1 | 1 | 1 | 6 | 0 | DOMON domain | 16 |
| PF03357 | 5 | 6 | 10 | 4 | 5 | 4 | 10 | 2 | Snf7 | 46 |
| PF03359 | 4 | 4 | 5 | 3 | 6 | 6 | 9 | 1 | Guanylate-kinase-associated protein (GKAP) protein | 38 |
| PF03360 | 1 | 1 | 3 | 1 | 1 | 1 | 4 | 0 | Glycosyltransferase family 43 | 12 |
| PF03364 | 0 | 0 | 2 | 0 | 1 | 0 | 3 | 0 | Polyketide cyclase / dehydrase and lipid transport | 6 |
| PF03366 | 0 | 0 | 4 | 0 | 0 | 0 | 5 | 0 | YEATS family | 9 |
| PF03367 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | ZPR1 zinc-finger domain | 3 |
| PF03368 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Dicer dimerisation domain | 2 |
| PF03370 | 0 | 0 | 7 | 1 | 0 | 0 | 9 | 0 | Putative phosphatase regulatory subunit | 17 |
| PF03371 | 0 | 0 | 2 | 1 | 0 | 0 | 2 | 0 | PRP38 family | 5 |
| PF03372 | 2 | 5 | 27 | 9 | 0 | 5 | 30 | 3 | Endonuclease/Exonuclease/phosphatase family | 81 |
| PF03378 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | "CAS/CSE protein, C-terminus" | 6 |
| PF03381 | 1 | 1 | 3 | 1 | 0 | 2 | 4 | 1 | LEM3 (ligand-effect modulator 3) family / CDC50 family | 13 |
| PF03388 | 0 | 2 | 4 | 3 | 0 | 3 | 3 | 1 | Legume-like lectin family | 16 |
| PF03398 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | Regulator of Vps4 activity in the MVB pathway | 8 |
| PF03399 | 0 | 0 | 3 | 1 | 0 | 0 | 3 | 0 | SAC3/GANP/Nin1/mts3/eIF-3 p25 family | 7 |
| PF03402 | 0 | 0 | 195 | 0 | 0 | 0 | 0 | 0 | "Vomeronasal organ pheromone receptor family, V1R" | 195 |
| PF03403 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 0 | "Platelet-activating factor acetylhydrolase, isoform II" | 3 |
| PF03404 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | Mo-co oxidoreductase dimerisation domain | 2 |
| PF03414 | 0 | 0 | 7 | 0 | 0 | 0 | 10 | 0 | Glycosyltransferase family 6 | 17 |
| PF03416 | 0 | 0 | 3 | 1 | 0 | 0 | 5 | 0 | Peptidase family C54 | 9 |
| PF03435 | 1 | 1 | 2 | 2 | 0 | 2 | 4 | 2 | Saccharopine dehydrogenase | 14 |
| PF03439 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | "Early transcription elongation factor of RNA pol II, NGN section" | 3 |
| PF03441 | 0 | 0 | 2 | 1 | 0 | 0 | 8 | 0 | FAD binding domain of DNA photolyase | 11 |
| PF03446 | 0 | 2 | 3 | 2 | 0 | 2 | 2 | 2 | NAD binding domain of 6-phosphogluconate dehydrogenase | 13 |
| PF03447 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | "Homoserine dehydrogenase, NAD binding domain" | 3 |
| PF03450 | 0 | 0 | 5 | 0 | 0 | 1 | 3 | 0 | CO dehydrogenase flavoprotein C-terminal domain | 9 |
| PF03451 | 0 | 1 | 5 | 2 | 2 | 1 | 4 | 1 | HELP motif | 16 |
| PF03453 | 1 | 1 | 1 | 0 | 2 | 2 | 1 | 1 | MoeA N-terminal region (domain I and II) | 9 |
| PF03454 | 1 | 1 | 1 | 0 | 2 | 2 | 1 | 1 | MoeA C-terminal region (domain IV) | 9 |
| PF03455 | 1 | 4 | 16 | 4 | 0 | 2 | 14 | 0 | dDENN domain | 41 |
| PF03456 | 1 | 4 | 15 | 4 | 0 | 2 | 14 | 0 | uDENN domain | 40 |
| PF03462 | 0 | 0 | 2 | 2 | 0 | 0 | 2 | 0 | PCRF domain | 6 |
| PF03463 | 2 | 1 | 2 | 0 | 1 | 1 | 2 | 2 | eRF1 domain 1 | 11 |
| PF03464 | 2 | 1 | 2 | 0 | 1 | 1 | 2 | 2 | eRF1 domain 2 | 11 |
| PF03465 | 2 | 1 | 2 | 0 | 1 | 1 | 2 | 2 | eRF1 domain 3 | 11 |
| PF03467 | 0 | 0 | 2 | 1 | 0 | 0 | 2 | 0 | Smg-4/UPF3 family | 5 |
| PF03473 | 1 | 1 | 3 | 1 | 0 | 1 | 1 | 0 | MOSC domain | 8 |
| PF03474 | 0 | 0 | 3 | 0 | 0 | 0 | 2 | 0 | DMRTA motif | 5 |
| PF03476 | 1 | 1 | 3 | 1 | 0 | 1 | 1 | 0 | MOSC N-terminal beta barrel domain | 8 |
| PF03477 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | ATP cone domain | 2 |
| PF03483 | 1 | 2 | 2 | 2 | 0 | 2 | 2 | 1 | B3/4 domain | 12 |
| PF03484 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | tRNA synthetase B5 domain | 7 |
| PF03485 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | Arginyl tRNA synthetase N terminal domain | 7 |
| PF03487 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Interleukin-13 | 1 |
| PF03489 | 0 | 1 | 4 | 1 | 0 | 0 | 5 | 1 | "Saposin-like type B, region 2" | 12 |
| PF03491 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | "Serotonin (5-HT) neurotransmitter transporter, N-terminus" | 2 |
| PF03493 | 0 | 1 | 3 | 0 | 1 | 2 | 3 | 1 | Calcium-activated BK potassium channel alpha subunit | 11 |
| PF03494 | 0 | 1 | 1 | 1 | 0 | 2 | 1 | 0 | Beta-amyloid peptide (beta-APP) | 6 |
| PF03501 | 2 | 2 | 2 | 0 | 2 | 2 | 2 | 2 | Plectin/S10 domain | 14 |
| PF03508 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | Gap junction alpha-1 protein (Cx43) | 7 |
| PF03509 | 0 | 0 | 1 | 0 | 0 | 0 | 2 | 0 | Gap junction alpha-8 protein (Cx50) | 3 |
| PF03511 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Fanconi anaemia group A protein | 1 |
| PF03517 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Regulator of volume decrease after cellular swelling | 1 |
| PF03520 | 1 | 2 | 5 | 0 | 0 | 1 | 7 | 0 | KCNQ voltage-gated potassium channel | 16 |
| PF03521 | 1 | 1 | 2 | 1 | 0 | 0 | 3 | 0 | Kv2 voltage-gated K+ channel | 8 |
| PF03522 | 0 | 1 | 2 | 0 | 0 | 1 | 1 | 0 | K-Cl Co-transporter type 1 (KCC1) | 5 |
| PF03523 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Macrophage scavenger receptor | 1 |
| PF03528 | 1 | 1 | 2 | 0 | 0 | 1 | 2 | 0 | Rabaptin | 7 |
| PF03529 | 0 | 0 | 3 | 0 | 0 | 0 | 5 | 0 | Otx1 transcription factor | 8 |
| PF03530 | 1 | 1 | 3 | 0 | 0 | 0 | 5 | 0 | Calcium-activated SK potassium channel | 10 |
| PF03531 | 0 | 0 | 1 | 0 | 0 | 0 | 2 | 1 | Structure-specific recognition protein (SSRP1) | 4 |
| PF03533 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | SPO11 homologue | 1 |
| PF03535 | 0 | 0 | 2 | 1 | 0 | 0 | 2 | 0 | Paxillin family | 5 |
| PF03540 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Transcription initiation factor TFIID 23-30kDa subunit | 2 |
| PF03542 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | Tuberin | 3 |
| PF03546 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | Treacher Collins syndrome protein Treacle | 2 |
| PF03547 | 0 | 0 | 1 | 0 | 0 | 0 | 2 | 0 | Membrane transport protein | 3 |
| PF03556 | 0 | 1 | 5 | 1 | 0 | 0 | 4 | 0 | Cullin binding | 11 |
| PF03561 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Allantoicase repeat | 2 |
| PF03567 | 0 | 0 | 12 | 1 | 2 | 1 | 19 | 0 | Sulfotransferase family | 35 |
| PF03568 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Peptidase family C50 | 2 |
| PF03571 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | Peptidase family M49 | 5 |
| PF03572 | 0 | 0 | 1 | 0 | 0 | 0 | 2 | 0 | Peptidase family S41 | 3 |
| PF03575 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | Peptidase family S51 | 1 |
| PF03577 | 1 | 1 | 3 | 3 | 0 | 2 | 2 | 2 | Peptidase family C69 | 14 |
| PF03587 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | EMG1/NEP1 methyltransferase | 3 |
| PF03600 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Citrate transporter | 2 |
| PF03604 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | "DNA directed RNA polymerase, 7 kDa subunit" | 2 |
| PF03607 | 2 | 1 | 8 | 2 | 3 | 3 | 7 | 0 | Doublecortin | 26 |
| PF03615 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 0 | GCM motif protein | 3 |
| PF03619 | 0 | 0 | 4 | 0 | 0 | 0 | 6 | 0 | Organic solute transporter Ostalpha | 10 |
| PF03623 | 0 | 0 | 2 | 0 | 0 | 0 | 3 | 0 | Focal adhesion targeting region | 5 |
| PF03629 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Domain of unknown function (DUF303) | 2 |
| PF03630 | 0 | 1 | 4 | 1 | 0 | 0 | 3 | 0 | Fumble | 9 |
| PF03632 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Glycosyl hydrolase family 65 central catalytic domain | 3 |
| PF03635 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | Vacuolar protein sorting-associated protein 35 | 6 |
| PF03637 | 1 | 1 | 7 | 0 | 1 | 2 | 8 | 1 | Mob1/phocein family | 21 |
| PF03638 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | "Tesmin/TSO1-like CXC domain, cysteine-rich domain" | 4 |
| PF03643 | 1 | 2 | 3 | 2 | 0 | 1 | 4 | 2 | Vacuolar protein sorting-associated protein 26 | 15 |
| PF03644 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Glycosyl hydrolase family 85 | 2 |
| PF03645 | 0 | 0 | 10 | 2 | 0 | 0 | 4 | 1 | Tctex-1 family | 17 |
| PF03647 | 0 | 1 | 2 | 0 | 0 | 1 | 3 | 0 | Transmembrane proteins 14C | 7 |
| PF03650 | 1 | 2 | 2 | 2 | 2 | 3 | 3 | 3 | Uncharacterised protein family (UPF0041) | 18 |
| PF03656 | 1 | 1 | 1 | 2 | 1 | 1 | 0 | 0 | Pam16 | 7 |
| PF03657 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Uncharacterised protein family (UPF0113) | 2 |
| PF03660 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | PHF5-like protein | 3 |
| PF03661 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | Uncharacterised protein family (UPF0121) | 7 |
| PF03662 | 0 | 0 | 2 | 1 | 0 | 0 | 2 | 0 | "Glycosyl hydrolase family 79, N-terminal domain" | 5 |
| PF03665 | 1 | 1 | 2 | 2 | 0 | 0 | 2 | 0 | Uncharacterised protein family (UPF0172) | 8 |
| PF03666 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Nitrogen Permease regulator of amino acid transport activity 3 | 2 |
| PF03669 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | Uncharacterised protein family (UPF0139) | 1 |
| PF03670 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Uncharacterised protein family (UPF0184) | 2 |
| PF03671 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | Ubiquitin fold modifier 1 protein | 6 |
| PF03676 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Uncharacterised protein family (UPF0183) | 2 |
| PF03690 | 0 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | Uncharacterised protein family (UPF0160) | 5 |
| PF03694 | 0 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | Erg28 like protein | 4 |
| PF03700 | 0 | 2 | 2 | 1 | 1 | 2 | 1 | 1 | "Sorting nexin, N-terminal domain" | 10 |
| PF03712 | 1 | 1 | 4 | 0 | 2 | 2 | 3 | 0 | "Copper type II ascorbate-dependent monooxygenase, C-terminal domain" | 13 |
| PF03715 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Noc2p family | 2 |
| PF03719 | 1 | 1 | 2 | 1 | 1 | 1 | 2 | 1 | "Ribosomal protein S5, C-terminal domain" | 10 |
| PF03720 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | "UDP-glucose/GDP-mannose dehydrogenase family, UDP binding domain" | 4 |
| PF03721 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | "UDP-glucose/GDP-mannose dehydrogenase family, NAD binding domain" | 4 |
| PF03725 | 0 | 0 | 6 | 1 | 0 | 0 | 3 | 0 | "3' exoribonuclease family, domain 2" | 10 |
| PF03726 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | "Polyribonucleotide nucleotidyltransferase, RNA binding domain" | 1 |
| PF03727 | 2 | 2 | 5 | 1 | 2 | 2 | 3 | 2 | Hexokinase | 19 |
| PF03730 | 0 | 0 | 2 | 0 | 1 | 1 | 1 | 0 | Ku70/Ku80 C-terminal arm | 5 |
| PF03731 | 0 | 0 | 2 | 0 | 1 | 1 | 1 | 0 | Ku70/Ku80 N-terminal alpha/beta domain | 5 |
| PF03732 | 0 | 0 | 5 | 0 | 0 | 0 | 2 | 0 | Retrotransposon gag protein | 7 |
| PF03733 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | Domain of unknown function (DUF307) | 2 |
| PF03735 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | ENT domain | 2 |
| PF03736 | 4 | 3 | 5 | 4 | 4 | 4 | 7 | 0 | EPTP domain | 31 |
| PF03747 | 0 | 0 | 3 | 1 | 0 | 0 | 3 | 2 | ADP-ribosylglycohydrolase | 9 |
| PF03762 | 0 | 0 | 1 | 0 | 0 | 0 | 2 | 0 | Vitelline membrane outer layer protein I (VOMI) | 3 |
| PF03764 | 0 | 2 | 4 | 2 | 1 | 3 | 6 | 3 | "Elongation factor G, domain IV" | 21 |
| PF03765 | 0 | 0 | 10 | 4 | 0 | 2 | 7 | 1 | "CRAL/TRIO, N-terminal domain" | 24 |
| PF03770 | 1 | 1 | 7 | 2 | 0 | 0 | 10 | 0 | Inositol polyphosphate kinase | 21 |
| PF03781 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | Sulfatase-modifying factor enzyme 1 | 4 |
| PF03782 | 0 | 0 | 3 | 0 | 0 | 0 | 3 | 0 | AMOP domain | 6 |
| PF03792 | 0 | 0 | 4 | 3 | 0 | 0 | 3 | 0 | PBC domain | 10 |
| PF03798 | 1 | 3 | 16 | 7 | 1 | 1 | 20 | 0 | TLC domain | 49 |
| PF03800 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Nuf2 family | 2 |
| PF03801 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | HEC/Ndc80p family | 2 |
| PF03803 | 0 | 0 | 6 | 1 | 0 | 1 | 7 | 1 | Scramblase | 16 |
| PF03807 | 2 | 3 | 7 | 3 | 3 | 4 | 5 | 1 | NADP oxidoreductase coenzyme F420-dependent | 28 |
| PF03810 | 1 | 5 | 14 | 6 | 0 | 3 | 14 | 4 | Importin-beta N-terminal domain | 47 |
| PF03813 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Nrap protein | 2 |
| PF03815 | 0 | 0 | 6 | 1 | 0 | 0 | 7 | 1 | LCCL domain | 15 |
| PF03819 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | MazG nucleotide pyrophosphohydrolase domain | 2 |
| PF03820 | 1 | 3 | 5 | 3 | 0 | 4 | 5 | 4 | Tricarboxylate carrier | 25 |
| PF03821 | 1 | 0 | 3 | 0 | 0 | 0 | 2 | 0 | Golgi 4-transmembrane spanning transporter | 6 |
| PF03823 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Neurokinin B | 1 |
| PF03826 | 0 | 0 | 14 | 0 | 0 | 0 | 20 | 0 | OAR domain | 34 |
| PF03827 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Orexin receptor type 2 | 2 |
| PF03828 | 1 | 1 | 7 | 2 | 1 | 1 | 6 | 0 | Cid1 family poly A polymerase | 19 |
| PF03832 | 1 | 2 | 2 | 1 | 0 | 1 | 1 | 1 | WSK motif | 9 |
| PF03834 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Binding domain of DNA repair protein Ercc1 (rad10/Swi10) | 2 |
| PF03835 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Rad4 transglutaminase-like domain | 2 |
| PF03836 | 0 | 0 | 3 | 0 | 0 | 0 | 2 | 1 | RasGAP C-terminus | 6 |
| PF03839 | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 0 | Translocation protein Sec62 | 3 |
| PF03847 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Transcription initiation factor TFIID subunit A | 1 |
| PF03849 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Transcription factor Tfb2 | 3 |
| PF03850 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Transcription factor Tfb4 | 2 |
| PF03853 | 0 | 1 | 2 | 2 | 1 | 1 | 3 | 2 | YjeF-related protein N-terminus | 12 |
| PF03857 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | Colicin immunity protein | 3 |
| PF03859 | 0 | 0 | 2 | 0 | 0 | 0 | 3 | 0 | CG-1 domain | 5 |
| PF03868 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | "Ribosomal protein L6, N-terminal domain" | 8 |
| PF03870 | 0 | 1 | 1 | 0 | 0 | 0 | 1 | 0 | RNA polymerase Rpb8 | 3 |
| PF03871 | 0 | 0 | 1 | 1 | 0 | 0 | 2 | 0 | "RNA polymerase Rpb5, N-terminal domain" | 4 |
| PF03874 | 0 | 0 | 2 | 1 | 0 | 0 | 2 | 0 | RNA polymerase Rpb4 | 5 |
| PF03876 | 0 | 0 | 3 | 1 | 0 | 0 | 2 | 0 | SHS2 domain found in N terminus of Rpb7p/Rpc25p/MJ0397 | 6 |
| PF03878 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | YIF1 | 4 |
| PF03879 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | Cgr1 family | 1 |
| PF03881 | 0 | 0 | 2 | 1 | 0 | 0 | 0 | 0 | Fructosamine kinase | 3 |
| PF03896 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | "Translocon-associated protein (TRAP), alpha subunit" | 3 |
| PF03900 | 0 | 0 | 1 | 0 | 0 | 0 | 2 | 0 | "Porphobilinogen deaminase, C-terminal domain" | 3 |
| PF03901 | 0 | 0 | 4 | 1 | 0 | 1 | 2 | 0 | Alg9-like mannosyltransferase family | 8 |
| PF03908 | 0 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | Sec20 | 4 |
| PF03909 | 0 | 0 | 3 | 2 | 0 | 0 | 3 | 0 | BSD domain | 8 |
| PF03911 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | Sec61beta family | 6 |
| PF03914 | 0 | 0 | 3 | 0 | 0 | 0 | 3 | 0 | CBF/Mak21 family | 6 |
| PF03915 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 0 | Actin interacting protein 3 | 14 |
| PF03917 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | "Eukaryotic glutathione synthase, ATP binding domain" | 3 |
| PF03919 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | "mRNA capping enzyme, C-terminal domain" | 3 |
| PF03920 | 0 | 0 | 5 | 2 | 0 | 0 | 7 | 0 | Groucho/TLE N-terminal Q-rich domain | 14 |
| PF03921 | 0 | 1 | 5 | 0 | 0 | 2 | 4 | 0 | "Intercellular adhesion molecule (ICAM), N-terminal domain" | 12 |
| PF03931 | 2 | 2 | 2 | 1 | 2 | 2 | 3 | 2 | "Skp1 family, tetramerisation domain" | 16 |
| PF03932 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | CutC family | 3 |
| PF03937 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Flavinator of succinate dehydrogenase | 3 |
| PF03939 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | "Ribosomal protein L23, N-terminal domain" | 7 |
| PF03941 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | "Inner centromere protein, ARK binding region" | 1 |
| PF03942 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 0 | DTW domain | 3 |
| PF03943 | 0 | 0 | 4 | 0 | 0 | 0 | 2 | 0 | TAP C-terminal domain | 6 |
| PF03946 | 2 | 2 | 2 | 1 | 1 | 2 | 2 | 1 | "Ribosomal protein L11, N-terminal domain" | 13 |
| PF03947 | 1 | 1 | 2 | 2 | 1 | 1 | 2 | 1 | "Ribosomal Proteins L2, C-terminal domain" | 11 |
| PF03949 | 0 | 3 | 3 | 1 | 0 | 2 | 2 | 1 | "Malic enzyme, NAD binding domain" | 12 |
| PF03950 | 1 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | "tRNA synthetases class I (E and Q), anti-codon binding domain" | 15 |
| PF03951 | 0 | 1 | 2 | 0 | 0 | 2 | 2 | 1 | "Glutamine synthetase, beta-Grasp domain" | 8 |
| PF03952 | 0 | 2 | 4 | 1 | 0 | 4 | 5 | 5 | "Enolase, N-terminal domain" | 21 |
| PF03953 | 11 | 10 | 19 | 7 | 13 | 12 | 20 | 13 | Tubulin C-terminal domain | 105 |
| PF03954 | 0 | 0 | 4 | 0 | 0 | 0 | 0 | 0 | "Hepatic lectin, N-terminal domain" | 4 |
| PF03957 | 0 | 0 | 3 | 0 | 0 | 0 | 3 | 0 | Jun-like transcription factor | 6 |
| PF03959 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Serine hydrolase (FSH1) | 1 |
| PF03962 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Mnd1 family | 2 |
| PF03966 | 0 | 1 | 2 | 1 | 0 | 0 | 2 | 0 | Trm112p-like protein | 6 |
| PF03969 | 0 | 0 | 1 | 0 | 0 | 0 | 2 | 0 | AFG1-like ATPase | 3 |
| PF03972 | 0 | 0 | 1 | 0 | 0 | 0 | 2 | 0 | MmgE/PrpD family | 3 |
| PF03980 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Nnf1 | 2 |
| PF03981 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | Ubiquinol-cytochrome C chaperone | 5 |
| PF03982 | 0 | 0 | 6 | 0 | 0 | 0 | 4 | 2 | Diacylglycerol acyltransferase | 12 |
| PF03985 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Paf1 | 1 |
| PF03986 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | "Autophagocytosis associated protein (Atg3), N-terminal domain" | 6 |
| PF03987 | 0 | 1 | 2 | 1 | 0 | 1 | 1 | 1 | "Autophagocytosis associated protein, active-site domain" | 7 |
| PF03992 | 1 | 1 | 3 | 2 | 1 | 2 | 3 | 1 | Antibiotic biosynthesis monooxygenase | 14 |
| PF03997 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 0 | VPS28 protein | 5 |
| PF03998 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Utp11 protein | 2 |
| PF03999 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | Microtubule associated protein (MAP65/ASE1 family) | 4 |
| PF04000 | 0 | 0 | 3 | 0 | 0 | 0 | 3 | 0 | Sas10/Utp3/C1D family | 6 |
| PF04003 | 0 | 0 | 3 | 0 | 0 | 0 | 3 | 0 | Dip2/Utp12 Family | 6 |
| PF04004 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Leo1-like protein | 3 |
| PF04005 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | Hus1-like protein | 2 |
| PF04006 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Mpp10 protein | 2 |
| PF04030 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | "D-arabinono-1,4-lactone oxidase" | 1 |
| PF04031 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Las1-like | 2 |
| PF04032 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | RNAse P Rpr2/Rpp21/SNM1 subunit domain | 2 |
| PF04034 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Domain of unknown function (DUF367) | 2 |
| PF04037 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 1 | Domain of unknown function (DUF382) | 3 |
| PF04042 | 0 | 0 | 3 | 0 | 0 | 0 | 2 | 1 | DNA polymerase alpha/epsilon subunit B | 6 |
| PF04045 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | "Arp2/3 complex, 34 kD subunit p34-Arc" | 8 |
| PF04046 | 0 | 0 | 1 | 0 | 0 | 0 | 2 | 1 | PSP | 4 |
| PF04048 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | Sec8 exocyst complex component specific domain | 7 |
| PF04049 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Anaphase promoting complex subunit 8 / Cdc23 | 2 |
| PF04050 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | Up-frameshift suppressor 2 | 2 |
| PF04051 | 3 | 3 | 5 | 2 | 3 | 3 | 4 | 2 | Transport protein particle (TRAPP) component | 25 |
| PF04053 | 1 | 2 | 2 | 1 | 0 | 1 | 2 | 2 | Coatomer WD associated region | 11 |
| PF04054 | 1 | 0 | 1 | 0 | 1 | 1 | 1 | 0 | "CCR4-Not complex component, Not1" | 5 |
| PF04055 | 0 | 0 | 8 | 4 | 0 | 0 | 7 | 0 | Radical SAM superfamily | 19 |
| PF04056 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Ssl1-like | 2 |
| PF04057 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | "Replication factor-A protein 1, N-terminal domain" | 4 |
| PF04061 | 0 | 2 | 3 | 0 | 0 | 2 | 3 | 0 | ORMDL family | 10 |
| PF04062 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | ARP2/3 complex ARPC3 (21 kDa) subunit | 7 |
| PF04063 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Domain of unknown function (DUF383) | 1 |
| PF04064 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Domain of unknown function (DUF384) | 1 |
| PF04065 | 0 | 0 | 1 | 1 | 0 | 0 | 2 | 0 | "Not1 N-terminal domain, CCR4-Not complex component" | 4 |
| PF04068 | 0 | 0 | 2 | 1 | 1 | 1 | 2 | 1 | "Possible Fer4-like domain in RNase L inhibitor, RLI" | 8 |
| PF04072 | 0 | 0 | 2 | 0 | 0 | 1 | 2 | 0 | Leucine carboxyl methyltransferase | 5 |
| PF04073 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | Aminoacyl-tRNA editing domain | 2 |
| PF04078 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | "Cell differentiation family, Rcd1-like" | 4 |
| PF04080 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Per1-like | 2 |
| PF04081 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | "DNA polymerase delta, subunit 4" | 2 |
| PF04083 | 0 | 0 | 6 | 0 | 0 | 0 | 1 | 0 | Partial alpha/beta-hydrolase lipase region | 7 |
| PF04084 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | Origin recognition complex subunit 2 | 2 |
| PF04086 | 0 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | "Signal recognition particle, alpha subunit, N-terminal" | 4 |
| PF04088 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | "Peroxin 13, N-terminal region" | 2 |
| PF04089 | 2 | 2 | 10 | 1 | 1 | 1 | 6 | 0 | BRICHOS domain | 23 |
| PF04091 | 2 | 1 | 2 | 1 | 2 | 2 | 1 | 0 | Exocyst complex subunit Sec15-like | 11 |
| PF04095 | 0 | 0 | 2 | 1 | 0 | 1 | 5 | 1 | Nicotinate phosphoribosyltransferase (NAPRTase) family | 10 |
| PF04096 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Nucleoporin autopeptidase | 2 |
| PF04097 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | Nup93/Nic96 | 4 |
| PF04098 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 0 | Rad52/22 family double-strand break repair protein | 3 |
| PF04099 | 0 | 1 | 2 | 2 | 1 | 2 | 2 | 1 | Sybindin-like family | 11 |
| PF04100 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | "Vps53-like, N-terminal" | 6 |
| PF04101 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 0 | Glycosyltransferase family 28 C-terminal domain | 3 |
| PF04103 | 0 | 0 | 25 | 1 | 1 | 1 | 34 | 0 | CD20-like family | 62 |
| PF04104 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | "Eukaryotic and archaeal DNA primase, large subunit" | 2 |
| PF04106 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Autophagy protein Apg5 | 3 |
| PF04109 | 0 | 1 | 2 | 0 | 0 | 1 | 2 | 0 | Autophagy protein Apg9 | 6 |
| PF04110 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Ubiquitin-like autophagy protein Apg12 | 2 |
| PF04111 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Autophagy protein Apg6 | 2 |
| PF04112 | 0 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | "Mak10 subunit, NatC N(alpha)-terminal acetyltransferase" | 3 |
| PF04113 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | "Gpi16 subunit, GPI transamidase component" | 5 |
| PF04114 | 0 | 1 | 1 | 0 | 0 | 0 | 1 | 0 | "Gaa1-like, GPI transamidase component" | 3 |
| PF04116 | 0 | 0 | 5 | 0 | 0 | 0 | 10 | 0 | Fatty acid hydroxylase superfamily | 15 |
| PF04117 | 0 | 1 | 3 | 1 | 0 | 1 | 4 | 1 | Mpv17 / PMP22 family | 11 |
| PF04118 | 0 | 0 | 2 | 1 | 1 | 1 | 2 | 1 | "Dopey, N-terminal" | 8 |
| PF04121 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Nuclear pore protein 84 / 107 | 3 |
| PF04124 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Dor1-like family | 2 |
| PF04127 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | DNA / pantothenate metabolism flavoprotein | 4 |
| PF04129 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | Vps52 / Sac2 family | 4 |
| PF04130 | 1 | 1 | 5 | 2 | 2 | 2 | 4 | 0 | Spc97 / Spc98 family | 17 |
| PF04133 | 0 | 0 | 2 | 2 | 0 | 0 | 1 | 0 | Vacuolar protein sorting 55 | 5 |
| PF04134 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | "Protein of unknown function, DUF393" | 1 |
| PF04135 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | "Nucleolar RNA-binding protein, Nop10p family" | 2 |
| PF04136 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Sec34-like family | 1 |
| PF04137 | 0 | 1 | 2 | 2 | 0 | 0 | 1 | 0 | Endoplasmic Reticulum Oxidoreductin 1 (ERO1) | 6 |
| PF04139 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | Rad9 | 4 |
| PF04140 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Isoprenylcysteine carboxyl methyltransferase (ICMT) family | 2 |
| PF04142 | 0 | 1 | 5 | 1 | 0 | 0 | 6 | 0 | Nucleotide-sugar transporter | 13 |
| PF04144 | 1 | 5 | 5 | 3 | 0 | 5 | 7 | 3 | SCAMP family | 29 |
| PF04145 | 0 | 0 | 2 | 0 | 0 | 1 | 2 | 0 | Ctr copper transporter family | 5 |
| PF04146 | 0 | 0 | 5 | 1 | 0 | 0 | 4 | 0 | YT521-B-like domain | 10 |
| PF04147 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Nop14-like family | 1 |
| PF04148 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Transmembrane adaptor Erv26 | 2 |
| PF04152 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Mre11 DNA-binding presumed domain | 1 |
| PF04153 | 0 | 0 | 2 | 2 | 0 | 0 | 3 | 0 | NOT2 / NOT3 / NOT5 family | 7 |
| PF04157 | 0 | 0 | 2 | 2 | 0 | 0 | 2 | 0 | EAP30/Vps36 family | 6 |
| PF04158 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Sof1-like domain | 2 |
| PF04161 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Arv1-like family | 2 |
| PF04176 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | TIP41-like family | 3 |
| PF04177 | 0 | 0 | 2 | 1 | 0 | 1 | 1 | 1 | TAP42-like family | 6 |
| PF04178 | 0 | 1 | 6 | 2 | 0 | 0 | 7 | 0 | Got1/Sft2-like family | 16 |
| PF04180 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Low temperature viability protein | 2 |
| PF04181 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Rtr1/RPAP2 family | 2 |
| PF04182 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | B-block binding subunit of TFIIIC | 1 |
| PF04184 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | ST7 protein | 4 |
| PF04188 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Mannosyltransferase (PIG-V)) | 1 |
| PF04189 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Gcd10p family | 3 |
| PF04190 | 1 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Protein of unknown function (DUF410) | 4 |
| PF04191 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Phospholipid methyltransferase | 1 |
| PF04192 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Utp21 specific WD40 associated putative domain | 2 |
| PF04193 | 0 | 0 | 5 | 0 | 0 | 0 | 7 | 0 | PQ loop repeat | 12 |
| PF04194 | 0 | 0 | 2 | 1 | 0 | 0 | 2 | 0 | "Programmed cell death protein 2, C-terminal putative domain" | 5 |
| PF04201 | 3 | 1 | 4 | 0 | 0 | 2 | 4 | 2 | Tumour protein D52 family | 16 |
| PF04209 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | "homogentisate 1,2-dioxygenase" | 2 |
| PF04212 | 2 | 0 | 9 | 4 | 0 | 1 | 8 | 1 | MIT (microtubule interacting and transport) domain | 25 |
| PF04218 | 0 | 0 | 6 | 0 | 0 | 0 | 0 | 0 | CENP-B N-terminal DNA-binding domain | 6 |
| PF04227 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | Indigoidine synthase A like protein | 1 |
| PF04241 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | Protein of unknown function (DUF423) | 7 |
| PF04253 | 0 | 2 | 5 | 1 | 0 | 2 | 3 | 0 | Transferrin receptor-like dimerisation domain | 13 |
| PF04258 | 0 | 1 | 5 | 0 | 0 | 1 | 3 | 0 | Signal peptide peptidase | 10 |
| PF04263 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | "Thiamin pyrophosphokinase, catalytic domain" | 3 |
| PF04265 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | "Thiamin pyrophosphokinase, vitamin B1 binding domain" | 3 |
| PF04266 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | ASCH domain | 2 |
| PF04272 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Phospholamban | 2 |
| PF04275 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Phosphomevalonate kinase | 3 |
| PF04280 | 1 | 1 | 2 | 1 | 0 | 1 | 1 | 0 | Tim44-like domain | 7 |
| PF04281 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | Mitochondrial import receptor subunit Tom22 | 7 |
| PF04300 | 1 | 1 | 6 | 2 | 1 | 1 | 11 | 1 | F-box associated region | 24 |
| PF04305 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | Protein of unknown function (DUF455) | 1 |
| PF04321 | 0 | 0 | 1 | 1 | 0 | 1 | 1 | 0 | RmlD substrate binding domain | 4 |
| PF04326 | 0 | 0 | 9 | 0 | 0 | 0 | 0 | 0 | Divergent AAA domain | 9 |
| PF04360 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Serglycin | 2 |
| PF04366 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Family of unknown function (DUF500) | 2 |
| PF04376 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | "Arginine-tRNA-protein transferase, N terminus" | 1 |
| PF04377 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | "Arginine-tRNA-protein transferase, C terminus" | 1 |
| PF04379 | 0 | 0 | 2 | 2 | 0 | 0 | 2 | 0 | Protein of unknown function (DUF525) | 6 |
| PF04382 | 2 | 3 | 4 | 1 | 1 | 4 | 4 | 2 | SAB domain | 21 |
| PF04387 | 2 | 1 | 4 | 1 | 0 | 1 | 2 | 1 | "Protein tyrosine phosphatase-like protein, PTPLA" | 12 |
| PF04388 | 0 | 0 | 1 | 1 | 0 | 0 | 2 | 0 | Hamartin protein | 4 |
| PF04389 | 1 | 4 | 10 | 4 | 0 | 3 | 8 | 0 | Peptidase family M28 | 30 |
| PF04402 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Protein of unknown function (DUF541) | 2 |
| PF04406 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Type IIB DNA topoisomerase | 2 |
| PF04408 | 4 | 2 | 17 | 5 | 4 | 4 | 15 | 2 | Helicase associated domain (HA2) | 53 |
| PF04410 | 0 | 0 | 2 | 1 | 0 | 0 | 2 | 1 | Gar1/Naf1 RNA binding region | 6 |
| PF04418 | 0 | 0 | 1 | 1 | 0 | 1 | 0 | 0 | Domain of unknown function (DUF543) | 3 |
| PF04419 | 0 | 0 | 3 | 0 | 0 | 0 | 4 | 0 | 4F5 protein family | 7 |
| PF04420 | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 0 | CHD5-like protein | 3 |
| PF04421 | 0 | 0 | 1 | 1 | 0 | 1 | 1 | 0 | Mss4 protein | 4 |
| PF04423 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Rad50 zinc hook motif | 2 |
| PF04424 | 0 | 1 | 2 | 1 | 0 | 1 | 2 | 0 | Protein of unknown function (DUF544) | 7 |
| PF04427 | 1 | 1 | 5 | 0 | 0 | 0 | 4 | 0 | Brix domain | 11 |
| PF04430 | 1 | 1 | 2 | 2 | 0 | 0 | 0 | 0 | Protein of unknown function (DUF498/DUF598) | 6 |
| PF04433 | 0 | 0 | 6 | 1 | 1 | 1 | 5 | 1 | SWIRM domain | 15 |
| PF04434 | 0 | 0 | 5 | 0 | 0 | 0 | 2 | 0 | SWIM zinc finger | 7 |
| PF04437 | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 0 | RINT-1 / TIP-1 family | 3 |
| PF04438 | 0 | 0 | 6 | 0 | 0 | 0 | 6 | 0 | HIT zinc finger | 12 |
| PF04440 | 0 | 0 | 3 | 0 | 0 | 0 | 4 | 0 | Dysbindin (Dystrobrevin binding protein 1) | 7 |
| PF04442 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Cytochrome c oxidase assembly protein CtaG/Cox11 | 3 |
| PF04446 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | tRNAHis guanylyltransferase | 2 |
| PF04487 | 0 | 0 | 3 | 0 | 0 | 0 | 3 | 0 | CITED | 6 |
| PF04488 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | Glycosyltransferase sugar-binding region containing DXD motif | 2 |
| PF04493 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Endonuclease V | 1 |
| PF04494 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | WD40 associated region in TFIID subunit | 4 |
| PF04495 | 0 | 0 | 2 | 2 | 0 | 1 | 2 | 0 | GRASP55/65 PDZ-like domain | 7 |
| PF04499 | 0 | 0 | 3 | 1 | 1 | 2 | 3 | 2 | SIT4 phosphatase-associated protein | 12 |
| PF04500 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | FLYWCH zinc finger domain | 2 |
| PF04502 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | Family of unknown function (DUF572) | 4 |
| PF04503 | 0 | 0 | 3 | 0 | 0 | 0 | 5 | 0 | "Single-stranded DNA binding protein, SSDP" | 8 |
| PF04505 | 2 | 3 | 14 | 0 | 0 | 2 | 10 | 1 | Interferon-induced transmembrane protein | 32 |
| PF04506 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Rft protein | 2 |
| PF04511 | 0 | 1 | 3 | 0 | 0 | 0 | 3 | 0 | Der1-like family | 7 |
| PF04515 | 0 | 2 | 5 | 1 | 1 | 2 | 5 | 2 | Plasma-membrane choline transporter | 18 |
| PF04516 | 1 | 0 | 6 | 2 | 2 | 1 | 7 | 0 | CP2 transcription factor | 19 |
| PF04538 | 0 | 0 | 12 | 2 | 0 | 0 | 0 | 0 | Brain expressed X-linked like family | 14 |
| PF04547 | 0 | 2 | 10 | 2 | 0 | 3 | 12 | 0 | Calcium-activated chloride channel | 29 |
| PF04548 | 0 | 0 | 9 | 0 | 0 | 3 | 85 | 0 | AIG1 family | 97 |
| PF04549 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | CD47 transmembrane region | 4 |
| PF04553 | 0 | 0 | 2 | 0 | 0 | 0 | 3 | 0 | "Tis11B like protein, N terminus" | 5 |
| PF04557 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | "Glutaminyl-tRNA synthetase, non-specific RNA binding region part 2" | 8 |
| PF04558 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | "Glutaminyl-tRNA synthetase, non-specific RNA binding region part 1" | 8 |
| PF04560 | 0 | 0 | 3 | 0 | 0 | 0 | 2 | 0 | "RNA polymerase Rpb2, domain 7" | 5 |
| PF04561 | 0 | 0 | 3 | 0 | 0 | 0 | 2 | 0 | "RNA polymerase Rpb2, domain 2" | 5 |
| PF04563 | 0 | 0 | 3 | 0 | 0 | 0 | 2 | 0 | RNA polymerase beta subunit | 5 |
| PF04564 | 1 | 2 | 6 | 3 | 2 | 3 | 7 | 1 | U-box domain | 25 |
| PF04565 | 0 | 0 | 3 | 0 | 0 | 0 | 2 | 0 | "RNA polymerase Rpb2, domain 3" | 5 |
| PF04566 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 0 | "RNA polymerase Rpb2, domain 4" | 3 |
| PF04567 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 0 | "RNA polymerase Rpb2, domain 5" | 3 |
| PF04568 | 0 | 0 | 1 | 0 | 0 | 1 | 2 | 2 | "Mitochondrial ATPase inhibitor, IATP" | 6 |
| PF04571 | 0 | 0 | 3 | 1 | 0 | 0 | 3 | 0 | "lipin, N-terminal conserved region" | 7 |
| PF04572 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | "Alpha 1,4-glycosyltransferase conserved region" | 2 |
| PF04573 | 0 | 1 | 3 | 0 | 0 | 1 | 1 | 1 | Signal peptidase subunit | 7 |
| PF04577 | 1 | 1 | 2 | 1 | 1 | 1 | 2 | 0 | Protein of unknown function (DUF563) | 9 |
| PF04579 | 0 | 0 | 3 | 2 | 0 | 0 | 0 | 0 | "Keratin, high-sulphur matrix protein" | 5 |
| PF04587 | 0 | 1 | 1 | 1 | 0 | 1 | 2 | 0 | ADP-specific Phosphofructokinase/Glucokinase conserved region | 6 |
| PF04588 | 0 | 1 | 5 | 1 | 1 | 2 | 2 | 0 | Hypoxia induced protein conserved region | 12 |
| PF04589 | 0 | 0 | 3 | 0 | 0 | 0 | 3 | 0 | RFX1 transcription activation region | 6 |
| PF04592 | 0 | 0 | 1 | 0 | 0 | 0 | 2 | 1 | "Selenoprotein P, N terminal region" | 4 |
| PF04593 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 1 | "Selenoprotein P, C terminal region" | 3 |
| PF04597 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | Ribophorin I | 7 |
| PF04598 | 0 | 0 | 10 | 2 | 0 | 0 | 3 | 0 | Gasdermin family | 15 |
| PF04603 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | Ran-interacting Mog1 protein | 2 |
| PF04614 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Pex19 protein family | 2 |
| PF04615 | 0 | 0 | 2 | 1 | 0 | 0 | 1 | 0 | Utp14 protein | 4 |
| PF04617 | 0 | 0 | 4 | 0 | 0 | 0 | 4 | 0 | Hox9 activation region | 8 |
| PF04621 | 0 | 0 | 3 | 0 | 0 | 0 | 3 | 0 | PEA3 subfamily ETS-domain transcription factor N terminal domain | 6 |
| PF04622 | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 1 | ERG2 and Sigma1 receptor like protein | 4 |
| PF04627 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | Mitochondrial ATP synthase epsilon chain | 6 |
| PF04628 | 0 | 0 | 3 | 2 | 1 | 1 | 1 | 0 | "Sedlin, N-terminal conserved region" | 8 |
| PF04629 | 0 | 0 | 2 | 1 | 0 | 0 | 2 | 0 | "Islet cell autoantigen ICA69, C-terminal domain" | 5 |
| PF04636 | 0 | 0 | 3 | 0 | 0 | 0 | 3 | 0 | PA26 p53-induced protein (sestrin) | 6 |
| PF04641 | 0 | 0 | 2 | 2 | 0 | 0 | 3 | 0 | Rtf2 RING-finger | 7 |
| PF04643 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Motilin/ghrelin-associated peptide | 2 |
| PF04644 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Motilin/ghrelin | 1 |
| PF04652 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | Vta1 like | 7 |
| PF04658 | 0 | 0 | 3 | 1 | 0 | 0 | 1 | 0 | TAFII55 protein conserved region | 5 |
| PF04664 | 0 | 0 | 2 | 1 | 0 | 0 | 3 | 0 | Opioid growth factor receptor (OGFr) conserved region | 6 |
| PF04666 | 0 | 0 | 6 | 0 | 0 | 0 | 4 | 0 | N-Acetylglucosaminyltransferase-IV (GnT-IV) conserved region | 10 |
| PF04667 | 0 | 1 | 2 | 2 | 0 | 1 | 4 | 0 | cAMP-regulated phosphoprotein/endosulfine conserved region | 10 |
| PF04668 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | Twisted gastrulation (Tsg) protein conserved region | 2 |
| PF04669 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Polysaccharide biosynthesis | 3 |
| PF04670 | 0 | 0 | 4 | 2 | 0 | 0 | 2 | 0 | Gtr1/RagA G protein conserved region | 8 |
| PF04675 | 0 | 0 | 2 | 0 | 0 | 0 | 3 | 0 | DNA ligase N terminus | 5 |
| PF04676 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | Protein similar to CwfJ C-terminus 2 | 4 |
| PF04677 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | Protein similar to CwfJ C-terminus 1 | 4 |
| PF04678 | 1 | 1 | 2 | 1 | 0 | 1 | 0 | 0 | "Protein of unknown function, DUF607" | 6 |
| PF04679 | 0 | 0 | 2 | 0 | 0 | 0 | 3 | 0 | ATP dependent DNA ligase C terminal region | 5 |
| PF04683 | 0 | 1 | 1 | 0 | 0 | 0 | 1 | 0 | Proteasome complex subunit Rpn13 ubiquitin receptor | 3 |
| PF04685 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | "Protein of unknown function, DUF608" | 2 |
| PF04691 | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 1 | Apolipoprotein C-I (ApoC-1) | 4 |
| PF04692 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | "Platelet-derived growth factor, N terminal region" | 4 |
| PF04695 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | Peroxisomal membrane anchor protein (Pex14p) conserved region | 6 |
| PF04696 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | pinin/SDK/memA/ protein conserved region | 3 |
| PF04697 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | pinin/SDK conserved region | 3 |
| PF04698 | 0 | 1 | 2 | 1 | 0 | 0 | 4 | 0 | Rab effector MyRIP/melanophilin C-terminus | 8 |
| PF04699 | 1 | 2 | 2 | 2 | 4 | 4 | 4 | 3 | ARP2/3 complex 16 kDa subunit (p16-Arc) | 22 |
| PF04704 | 0 | 0 | 4 | 0 | 0 | 0 | 2 | 0 | Zfx / Zfy transcription activation region | 6 |
| PF04706 | 0 | 0 | 4 | 2 | 0 | 0 | 4 | 0 | Dickkopf N-terminal cysteine-rich region | 10 |
| PF04707 | 0 | 0 | 6 | 2 | 0 | 1 | 3 | 0 | PRELI-like family | 12 |
| PF04709 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | "Anti-Mullerian hormone, N terminal region" | 2 |
| PF04710 | 0 | 0 | 3 | 0 | 0 | 0 | 5 | 0 | Pellino | 8 |
| PF04711 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Apolipoprotein A-II (ApoA-II) | 1 |
| PF04712 | 0 | 0 | 3 | 0 | 0 | 0 | 2 | 0 | Radial spokehead-like protein | 5 |
| PF04714 | 0 | 0 | 3 | 0 | 0 | 0 | 2 | 1 | "BCL7, N-terminal conserver region" | 6 |
| PF04716 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | ETC complex I subunit conserved region | 6 |
| PF04718 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | Mitochondrial ATP synthase g subunit | 5 |
| PF04719 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | hTAFII28-like protein conserved region | 2 |
| PF04721 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Domain of unknown function (DUF750) | 3 |
| PF04722 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Ssu72-like protein | 2 |
| PF04724 | 0 | 0 | 1 | 0 | 0 | 0 | 2 | 0 | Glycosyltransferase family 17 | 3 |
| PF04727 | 3 | 3 | 6 | 2 | 3 | 3 | 4 | 1 | ELMO/CED-12 family | 25 |
| PF04729 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 0 | ASF1 like histone chaperone | 3 |
| PF04731 | 0 | 0 | 3 | 0 | 0 | 0 | 2 | 0 | Caudal like protein activation region | 5 |
| PF04732 | 6 | 5 | 7 | 4 | 8 | 8 | 12 | 8 | Intermediate filament head (DNA binding) region | 58 |
| PF04733 | 0 | 0 | 1 | 1 | 0 | 1 | 1 | 0 | Coatomer epsilon subunit | 4 |
| PF04734 | 0 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | Neutral/alkaline non-lysosomal ceramidase | 4 |
| PF04739 | 0 | 0 | 2 | 2 | 0 | 0 | 3 | 0 | "5'-AMP-activated protein kinase beta subunit, interation domain" | 7 |
| PF04749 | 0 | 0 | 3 | 0 | 0 | 1 | 11 | 0 | PLAC8 family | 15 |
| PF04750 | 0 | 0 | 2 | 0 | 0 | 0 | 3 | 0 | FAR-17a/AIG1-like protein | 5 |
| PF04752 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | ChaC-like protein | 4 |
| PF04756 | 1 | 1 | 2 | 1 | 1 | 1 | 2 | 0 | OST3 / OST6 family | 9 |
| PF04757 | 1 | 1 | 3 | 1 | 0 | 0 | 3 | 0 | Pex2 / Pex12 amino terminal region | 9 |
| PF04758 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | Ribosomal protein S30 | 8 |
| PF04762 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | IKI3 family | 3 |
| PF04768 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Protein of unknown function (DUF619) | 2 |
| PF04774 | 1 | 0 | 2 | 0 | 1 | 1 | 3 | 2 | Hyaluronan / mRNA binding family | 10 |
| PF04775 | 0 | 0 | 9 | 2 | 0 | 1 | 9 | 1 | Acyl-CoA thioester hydrolase/BAAT N-terminal region | 22 |
| PF04777 | 0 | 1 | 3 | 3 | 1 | 1 | 3 | 0 | Erv1 / Alr family | 12 |
| PF04784 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | "Protein of unknown function, DUF547" | 1 |
| PF04790 | 0 | 0 | 4 | 1 | 0 | 0 | 2 | 0 | Sarcoglycan complex subunit protein | 7 |
| PF04791 | 0 | 2 | 4 | 2 | 0 | 1 | 3 | 0 | LMBR1-like membrane protein | 12 |
| PF04794 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | YdjC-like protein | 3 |
| PF04795 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | PAPA-1-like conserved region | 2 |
| PF04799 | 0 | 2 | 2 | 0 | 0 | 2 | 2 | 0 | fzo-like conserved region | 8 |
| PF04800 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | ETC complex I subunit conserved region | 6 |
| PF04801 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Sin-like protein conserved region | 3 |
| PF04802 | 0 | 0 | 4 | 1 | 0 | 0 | 1 | 0 | Component of IIS longevity pathway SMK-1 | 6 |
| PF04803 | 0 | 0 | 41 | 0 | 0 | 0 | 1 | 0 | Cor1/Xlr/Xmr conserved region | 42 |
| PF04810 | 4 | 3 | 6 | 3 | 3 | 4 | 4 | 0 | Sec23/Sec24 zinc finger | 27 |
| PF04811 | 4 | 3 | 6 | 3 | 3 | 4 | 4 | 0 | Sec23/Sec24 trunk domain | 27 |
| PF04812 | 0 | 0 | 2 | 0 | 0 | 0 | 3 | 0 | "Hepatocyte nuclear factor 1 (HNF-1), beta isoform C terminus" | 5 |
| PF04813 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | "Hepatocyte nuclear factor 1 (HNF-1), alpha isoform C terminus" | 2 |
| PF04814 | 0 | 0 | 3 | 1 | 0 | 0 | 6 | 0 | "Hepatocyte nuclear factor 1 (HNF-1), N terminus" | 10 |
| PF04815 | 4 | 3 | 6 | 3 | 3 | 4 | 4 | 0 | Sec23/Sec24 helical domain | 27 |
| PF04818 | 0 | 0 | 7 | 2 | 0 | 0 | 6 | 1 | RNA polymerase II-binding domain. | 16 |
| PF04819 | 0 | 0 | 6 | 0 | 0 | 0 | 2 | 0 | Family of unknown function (DUF716) | 8 |
| PF04821 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Timeless protein | 2 |
| PF04822 | 0 | 0 | 43 | 0 | 1 | 0 | 2 | 1 | Takusan | 47 |
| PF04824 | 0 | 0 | 3 | 0 | 0 | 0 | 3 | 0 | Conserved region of Rad21 / Rec8 like protein | 6 |
| PF04825 | 0 | 0 | 3 | 0 | 0 | 0 | 5 | 0 | N terminus of Rad21 / Rec8 like protein | 8 |
| PF04826 | 4 | 3 | 11 | 1 | 0 | 0 | 1 | 0 | Armadillo-like | 20 |
| PF04828 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Glutathione-dependent formaldehyde-activating enzyme | 2 |
| PF04831 | 0 | 0 | 3 | 0 | 0 | 0 | 3 | 0 | Popeye protein conserved region | 6 |
| PF04832 | 0 | 1 | 2 | 1 | 0 | 1 | 4 | 1 | SOUL heme-binding protein | 10 |
| PF04836 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | Interferon-related protein conserved region | 4 |
| PF04840 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 0 | "Vps16, C-terminal region" | 5 |
| PF04841 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 0 | "Vps16, N-terminal region" | 5 |
| PF04845 | 3 | 3 | 3 | 2 | 3 | 3 | 2 | 1 | PurA ssDNA and RNA-binding protein | 20 |
| PF04847 | 0 | 0 | 3 | 1 | 0 | 0 | 3 | 0 | Calcipressin | 7 |
| PF04849 | 2 | 0 | 4 | 1 | 0 | 0 | 2 | 0 | HAP1 N-terminal conserved region | 9 |
| PF04851 | 0 | 0 | 4 | 1 | 0 | 0 | 3 | 0 | "Type III restriction enzyme, res subunit" | 8 |
| PF04855 | 0 | 0 | 1 | 1 | 0 | 0 | 2 | 0 | SNF5 / SMARCB1 / INI1 | 4 |
| PF04856 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Securin sister-chromatid separation inhibitor | 1 |
| PF04857 | 0 | 0 | 5 | 1 | 0 | 0 | 3 | 0 | CAF1 family ribonuclease | 9 |
| PF04858 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | TH1 protein | 3 |
| PF04868 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 0 | "Retinal cGMP phosphodiesterase, gamma subunit" | 3 |
| PF04869 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | "Uso1 / p115 like vesicle tethering protein, head region" | 6 |
| PF04871 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | "Uso1 / p115 like vesicle tethering protein, C terminal region" | 6 |
| PF04873 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Ethylene insensitive 3 | 1 |
| PF04874 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Mak16 protein C-terminal region | 2 |
| PF04880 | 1 | 1 | 2 | 1 | 3 | 3 | 1 | 0 | "NUDE protein, C-terminal conserved region" | 12 |
| PF04882 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | Peroxin-3 | 5 |
| PF04884 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Vitamin B6 photo-protection and homoeostasis | 2 |
| PF04886 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | PT repeat | 1 |
| PF04889 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Cwf15/Cwc15 cell cycle control protein | 2 |
| PF04893 | 0 | 2 | 7 | 1 | 0 | 0 | 5 | 0 | Yip1 domain | 15 |
| PF04900 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | Fcf1 | 2 |
| PF04901 | 0 | 0 | 3 | 0 | 0 | 0 | 3 | 0 | Receptor activity modifying family | 6 |
| PF04902 | 0 | 0 | 1 | 0 | 0 | 0 | 2 | 0 | Conserved region in Nab1 | 3 |
| PF04904 | 0 | 0 | 2 | 0 | 0 | 0 | 3 | 0 | NAB conserved region 1 (NCD1) | 5 |
| PF04905 | 0 | 0 | 2 | 0 | 0 | 0 | 3 | 0 | NAB conserved region 2 (NCD2) | 5 |
| PF04906 | 0 | 2 | 3 | 2 | 0 | 3 | 5 | 1 | Tweety | 16 |
| PF04908 | 1 | 2 | 4 | 3 | 1 | 3 | 4 | 2 | "SH3-binding, glutamic acid-rich protein" | 20 |
| PF04909 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Amidohydrolase | 2 |
| PF04910 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Transcriptional repressor TCF25 | 2 |
| PF04912 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | Dynamitin | 8 |
| PF04916 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | Phospholipase B | 4 |
| PF04921 | 0 | 0 | 2 | 1 | 0 | 0 | 1 | 0 | "XAP5, circadian clock regulator" | 4 |
| PF04922 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | DIE2/ALG10 family | 3 |
| PF04923 | 0 | 0 | 2 | 0 | 1 | 1 | 1 | 0 | Ninjurin | 5 |
| PF04925 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | SHQ1 protein | 2 |
| PF04926 | 0 | 0 | 3 | 0 | 0 | 0 | 2 | 0 | Poly(A) polymerase predicted RNA binding domain | 5 |
| PF04928 | 0 | 0 | 3 | 0 | 0 | 0 | 2 | 0 | Poly(A) polymerase central domain | 5 |
| PF04930 | 2 | 2 | 2 | 1 | 1 | 2 | 1 | 1 | FUN14 family | 12 |
| PF04931 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | DNA polymerase phi | 3 |
| PF04934 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | MED6 mediator sub complex component | 3 |
| PF04935 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Surfeit locus protein 6 | 2 |
| PF04938 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Survival motor neuron (SMN) interacting protein 1 (SIP1) | 2 |
| PF04939 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Ribosome biogenesis regulatory protein (RRS1) | 2 |
| PF04950 | 0 | 0 | 2 | 1 | 0 | 0 | 2 | 0 | Protein of unknown function (DUF663) | 5 |
| PF04952 | 0 | 0 | 2 | 1 | 0 | 0 | 3 | 0 | Succinylglutamate desuccinylase / Aspartoacylase family | 6 |
| PF04959 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 1 | Arsenite-resistance protein 2 | 3 |
| PF04960 | 0 | 1 | 2 | 2 | 0 | 2 | 5 | 2 | Glutaminase | 14 |
| PF04961 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Formiminotransferase-cyclodeaminase | 2 |
| PF04968 | 0 | 0 | 2 | 1 | 0 | 0 | 3 | 0 | CHORD | 6 |
| PF04969 | 2 | 4 | 15 | 6 | 0 | 0 | 13 | 2 | CS domain | 42 |
| PF04970 | 0 | 1 | 7 | 0 | 0 | 0 | 11 | 0 | Lecithin retinol acyltransferase | 19 |
| PF04979 | 0 | 0 | 2 | 0 | 0 | 1 | 1 | 0 | Protein phosphatase inhibitor 2 (IPP-2) | 4 |
| PF04981 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | NMD3 family | 3 |
| PF04983 | 0 | 0 | 3 | 1 | 0 | 0 | 0 | 0 | "RNA polymerase Rpb1, domain 3" | 4 |
| PF04987 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Phosphatidylinositolglycan class N (PIG-N) | 2 |
| PF04988 | 0 | 0 | 3 | 1 | 0 | 0 | 0 | 0 | A-kinase anchoring protein 95 (AKAP95) | 4 |
| PF04990 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | "RNA polymerase Rpb1, domain 7" | 2 |
| PF04991 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 0 | LicD family | 3 |
| PF04992 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | "RNA polymerase Rpb1, domain 6" | 2 |
| PF04997 | 0 | 0 | 3 | 1 | 0 | 0 | 0 | 0 | "RNA polymerase Rpb1, domain 1" | 4 |
| PF04998 | 0 | 0 | 3 | 1 | 0 | 0 | 0 | 0 | "RNA polymerase Rpb1, domain 5" | 4 |
| PF05000 | 0 | 0 | 3 | 1 | 0 | 0 | 0 | 0 | "RNA polymerase Rpb1, domain 4" | 4 |
| PF05001 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | RNA polymerase Rpb1 C-terminal repeat | 2 |
| PF05002 | 1 | 2 | 2 | 1 | 0 | 0 | 1 | 0 | SGS domain | 7 |
| PF05004 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | Interferon-related developmental regulator (IFRD) | 4 |
| PF05005 | 0 | 1 | 1 | 0 | 0 | 1 | 2 | 2 | Janus/Ocnus family (Ocnus) | 7 |
| PF05007 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Mannosyltransferase (PIG-M) | 2 |
| PF05008 | 2 | 2 | 2 | 1 | 1 | 2 | 2 | 0 | Vesicle transport v-SNARE protein N-terminus | 12 |
| PF05010 | 1 | 1 | 3 | 0 | 0 | 0 | 3 | 0 | Transforming acidic coiled-coil-containing protein (TACC) | 8 |
| PF05011 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | "Lariat debranching enzyme, C-terminal domain" | 3 |
| PF05014 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Nucleoside 2-deoxyribosyltransferase | 2 |
| PF05018 | 0 | 0 | 3 | 1 | 0 | 0 | 2 | 0 | Protein of unknown function (DUF667) | 6 |
| PF05019 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | Coenzyme Q (ubiquinone) biosynthesis protein Coq4 | 2 |
| PF05020 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | "NPL4 family, putative zinc binding region" | 1 |
| PF05021 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | NPL4 family | 1 |
| PF05022 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | "SRP40, C-terminal domain" | 2 |
| PF05024 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | N-acetylglucosaminyl transferase component (Gpi1) | 3 |
| PF05025 | 0 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | RbsD / FucU transport protein family | 4 |
| PF05026 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | "Dcp2, box A domain" | 1 |
| PF05028 | 0 | 0 | 1 | 0 | 1 | 1 | 1 | 1 | Poly (ADP-ribose) glycohydrolase (PARG) | 5 |
| PF05029 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Timeless protein C terminal region | 2 |
| PF05030 | 0 | 0 | 3 | 0 | 0 | 0 | 1 | 0 | SSXT protein (N-terminal region) | 4 |
| PF05033 | 0 | 0 | 7 | 3 | 0 | 0 | 6 | 0 | Pre-SET motif | 16 |
| PF05038 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Cytochrome Cytochrome b558 alpha-subunit | 2 |
| PF05039 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | Agouti protein | 4 |
| PF05041 | 2 | 0 | 4 | 1 | 0 | 0 | 4 | 0 | Pecanex protein (C-terminus) | 11 |
| PF05044 | 0 | 0 | 2 | 0 | 0 | 0 | 3 | 0 | Homeo-prospero domain | 5 |
| PF05046 | 0 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | Mitochondrial large subunit ribosomal protein (Img2) | 4 |
| PF05047 | 2 | 2 | 3 | 3 | 2 | 2 | 3 | 1 | Mitochondrial ribosomal protein L51 / S25 / CI-B8 domain | 18 |
| PF05048 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Periplasmic copper-binding protein (NosD) | 2 |
| PF05049 | 1 | 1 | 15 | 0 | 0 | 1 | 10 | 0 | Interferon-inducible GTPase (IIGP) | 28 |
| PF05051 | 0 | 0 | 2 | 0 | 0 | 0 | 3 | 0 | Cytochrome C oxidase copper chaperone (COX17) | 5 |
| PF05053 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Menin | 2 |
| PF05057 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 0 | Putative serine esterase (DUF676) | 3 |
| PF05060 | 0 | 0 | 1 | 1 | 0 | 0 | 2 | 0 | N-acetylglucosaminyltransferase II (MGAT2) | 4 |
| PF05063 | 0 | 0 | 3 | 0 | 0 | 0 | 3 | 0 | MT-A70 | 6 |
| PF05064 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 0 | Nsp1-like C-terminal region | 3 |
| PF05066 | 0 | 0 | 2 | 0 | 0 | 0 | 3 | 0 | "HB1, ASXL, restriction endonuclease HTH domain" | 5 |
| PF05071 | 1 | 2 | 2 | 1 | 1 | 2 | 3 | 1 | NADH ubiquinone oxidoreductase subunit NDUFA12 | 13 |
| PF05076 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Suppressor of fused protein (SUFU) | 1 |
| PF05083 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | LST-1 protein | 1 |
| PF05089 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Alpha-N-acetylglucosaminidase (NAGLU) tim-barrel domain | 2 |
| PF05090 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 1 | Vitamin K-dependent gamma-carboxylase | 3 |
| PF05091 | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 1 | Eukaryotic translation initiation factor 3 subunit 7 (eIF-3) | 4 |
| PF05093 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | "Cytokine-induced anti-apoptosis inhibitor 1, Fe-S biogenesis" | 4 |
| PF05104 | 2 | 1 | 2 | 0 | 2 | 2 | 2 | 0 | Ribosome receptor lysine/proline rich region | 11 |
| PF05110 | 0 | 0 | 4 | 1 | 0 | 0 | 3 | 0 | AF-4 proto-oncoprotein | 8 |
| PF05111 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Ameloblastin precursor (Amelin) | 1 |
| PF05118 | 2 | 2 | 3 | 2 | 2 | 2 | 3 | 0 | Aspartyl/Asparaginyl beta-hydroxylase | 16 |
| PF05127 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Helicase | 3 |
| PF05129 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Transcription elongation factor Elf1 like | 1 |
| PF05131 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | Pep3/Vps18/deep orange family | 6 |
| PF05132 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | RNA polymerase III RPC4 | 2 |
| PF05147 | 2 | 2 | 3 | 2 | 2 | 2 | 2 | 1 | Lanthionine synthetase C-like protein | 16 |
| PF05148 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Hypothetical methyltransferase | 3 |
| PF05153 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Family of unknown function (DUF706) | 2 |
| PF05154 | 0 | 0 | 4 | 1 | 0 | 0 | 4 | 0 | TM2 domain | 9 |
| PF05158 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | RNA polymerase Rpc34 subunit | 2 |
| PF05160 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | DSS1/SEM1 family | 2 |
| PF05161 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 1 | MOFRL family | 3 |
| PF05162 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Ribosomal protein L41 | 1 |
| PF05168 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | HEPN domain | 6 |
| PF05172 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Nup53/35/40-type RNA recognition motif | 3 |
| PF05175 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | Methyltransferase small domain | 4 |
| PF05178 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | KRI1-like family | 2 |
| PF05179 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | "RNA pol II accessory factor, Cdc73 family" | 3 |
| PF05180 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | DNL zinc finger | 2 |
| PF05181 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | XPA protein C-terminus | 2 |
| PF05182 | 0 | 0 | 1 | 0 | 0 | 0 | 2 | 0 | Fip1 motif | 3 |
| PF05184 | 0 | 1 | 3 | 1 | 0 | 0 | 5 | 1 | "Saposin-like type B, region 1" | 11 |
| PF05185 | 0 | 1 | 3 | 1 | 0 | 0 | 4 | 0 | PRMT5 arginine-N-methyltransferase | 9 |
| PF05186 | 1 | 1 | 5 | 1 | 0 | 0 | 5 | 1 | Dpy-30 motif | 14 |
| PF05187 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | Electron transfer flavoprotein-ubiquinone oxidoreductase | 5 |
| PF05188 | 0 | 0 | 4 | 0 | 0 | 0 | 3 | 0 | MutS domain II | 7 |
| PF05189 | 1 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | "RNA 3'-terminal phosphate cyclase (RTC), insert domain" | 5 |
| PF05190 | 0 | 0 | 5 | 0 | 0 | 0 | 4 | 0 | MutS family domain IV | 9 |
| PF05191 | 2 | 3 | 3 | 3 | 1 | 2 | 3 | 3 | "Adenylate kinase, active site lid" | 20 |
| PF05192 | 0 | 0 | 5 | 0 | 0 | 0 | 4 | 0 | MutS domain III | 9 |
| PF05193 | 0 | 4 | 6 | 3 | 0 | 6 | 8 | 5 | Peptidase M16 inactive domain | 32 |
| PF05195 | 1 | 0 | 2 | 2 | 1 | 1 | 1 | 0 | "Aminopeptidase P, N-terminal domain" | 8 |
| PF05196 | 0 | 0 | 2 | 1 | 0 | 0 | 2 | 1 | "PTN/MK heparin-binding protein family, N-terminal domain" | 6 |
| PF05197 | 0 | 1 | 2 | 1 | 0 | 0 | 1 | 0 | TRIC channel | 5 |
| PF05198 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | "Translation initiation factor IF-3, N-terminal domain" | 3 |
| PF05199 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | GMC oxidoreductase | 2 |
| PF05205 | 0 | 0 | 2 | 1 | 0 | 0 | 1 | 0 | COMPASS (Complex proteins associated with Set1p) component shg1 | 4 |
| PF05206 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Methyltransferase TRM13 | 1 |
| PF05207 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | CSL zinc finger | 4 |
| PF05208 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | ALG3 protein | 2 |
| PF05210 | 3 | 3 | 6 | 3 | 0 | 0 | 5 | 0 | Sprouty protein (Spry) | 20 |
| PF05216 | 0 | 0 | 1 | 0 | 1 | 1 | 1 | 0 | UNC-50 family | 4 |
| PF05217 | 2 | 2 | 4 | 1 | 3 | 3 | 4 | 2 | STOP protein | 21 |
| PF05219 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | DREV methyltransferase | 2 |
| PF05221 | 0 | 3 | 3 | 1 | 0 | 3 | 4 | 3 | S-adenosyl-L-homocysteine hydrolase | 17 |
| PF05222 | 0 | 1 | 1 | 1 | 0 | 2 | 3 | 3 | "Alanine dehydrogenase/PNT, N-terminal domain" | 11 |
| PF05224 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 0 | NDT80 / PhoG like DNA-binding family | 3 |
| PF05225 | 0 | 0 | 2 | 0 | 0 | 0 | 3 | 0 | "helix-turn-helix, Psq domain" | 5 |
| PF05236 | 0 | 0 | 2 | 0 | 0 | 0 | 3 | 0 | Transcription initiation factor TFIID component TAF4 family | 5 |
| PF05237 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | MoeZ/MoeB domain | 3 |
| PF05238 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Kinetochore protein CHL4 like | 2 |
| PF05240 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | APOBEC-like C-terminal domain | 2 |
| PF05241 | 0 | 1 | 2 | 0 | 0 | 0 | 2 | 0 | Emopamil binding protein | 5 |
| PF05242 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Glycosylation-dependent cell adhesion molecule 1 (GlyCAM-1) | 1 |
| PF05250 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Uncharacterised protein family (UPF0193) | 2 |
| PF05251 | 1 | 1 | 1 | 0 | 0 | 0 | 1 | 0 | Uncharacterised protein family (UPF0197) | 4 |
| PF05253 | 0 | 0 | 6 | 0 | 0 | 0 | 2 | 0 | U11-48K-like CHHC zinc finger | 8 |
| PF05254 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Uncharacterised protein family (UPF0203) | 3 |
| PF05255 | 0 | 0 | 2 | 0 | 0 | 1 | 1 | 1 | Uncharacterised protein family (UPF0220) | 5 |
| PF05270 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | Alpha-L-arabinofuranosidase B (ABFB) | 4 |
| PF05276 | 0 | 0 | 2 | 0 | 0 | 1 | 3 | 0 | SH3 domain-binding protein 5 (SH3BP5) | 6 |
| PF05277 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Protein of unknown function (DUF726) | 2 |
| PF05279 | 1 | 1 | 1 | 1 | 1 | 1 | 3 | 0 | Aspartyl beta-hydroxylase N-terminal region | 9 |
| PF05281 | 0 | 0 | 1 | 1 | 0 | 1 | 2 | 1 | Neuroendocrine protein 7B2 precursor (Secretogranin V) | 6 |
| PF05282 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | AAR2 protein | 2 |
| PF05283 | 0 | 0 | 3 | 0 | 0 | 0 | 2 | 0 | Multi-glycosylated core protein 24 (MGC-24) | 5 |
| PF05285 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | SDA1 | 1 |
| PF05287 | 0 | 0 | 10 | 0 | 0 | 0 | 0 | 0 | PMG protein | 10 |
| PF05291 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Bystin | 2 |
| PF05292 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 0 | Malonyl-CoA decarboxylase (MCD) | 5 |
| PF05296 | 0 | 0 | 69 | 0 | 0 | 0 | 4 | 0 | Mammalian taste receptor protein (TAS2R) | 73 |
| PF05300 | 2 | 2 | 2 | 1 | 1 | 2 | 4 | 3 | Protein of unknown function (DUF737) | 17 |
| PF05301 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | Touch receptor neuron protein Mec-17 | 5 |
| PF05303 | 0 | 0 | 1 | 1 | 0 | 0 | 2 | 0 | Protein of unknown function (DUF727) | 4 |
| PF05308 | 1 | 1 | 3 | 1 | 1 | 1 | 3 | 0 | Mitochondrial fission regulator | 11 |
| PF05324 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Sperm antigen HE2 | 1 |
| PF05326 | 0 | 0 | 5 | 0 | 0 | 0 | 0 | 0 | Seminal vesicle autoantigen (SVA) | 5 |
| PF05327 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | RNA polymerase I specific transcription initiation factor RRN3 | 1 |
| PF05328 | 0 | 1 | 1 | 0 | 0 | 0 | 2 | 1 | CybS | 5 |
| PF05334 | 0 | 0 | 2 | 1 | 0 | 0 | 2 | 2 | Protein of unknown function (DUF719) | 7 |
| PF05337 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Macrophage colony stimulating factor-1 (CSF-1) | 1 |
| PF05346 | 0 | 1 | 1 | 0 | 0 | 1 | 2 | 0 | Eukaryotic membrane protein family | 5 |
| PF05347 | 2 | 4 | 9 | 4 | 2 | 2 | 7 | 2 | Complex 1 protein (LYR family) | 32 |
| PF05348 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Proteasome maturation factor UMP1 | 2 |
| PF05349 | 0 | 0 | 3 | 0 | 0 | 0 | 1 | 0 | "GATA-type transcription activator, N-terminal" | 4 |
| PF05350 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 0 | Glycogen synthase kinase-3 binding | 3 |
| PF05351 | 0 | 0 | 3 | 2 | 0 | 0 | 5 | 0 | "GMP-PDE, delta subunit" | 10 |
| PF05355 | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 0 | Apolipoprotein C-II | 3 |
| PF05361 | 0 | 0 | 5 | 2 | 0 | 0 | 5 | 0 | PKC-activated protein phosphatase-1 inhibitor | 12 |
| PF05362 | 1 | 2 | 2 | 0 | 0 | 1 | 0 | 0 | Lon protease (S16) C-terminal proteolytic domain | 6 |
| PF05365 | 0 | 1 | 1 | 0 | 1 | 1 | 0 | 0 | "Ubiquinol-cytochrome C reductase, UQCRX/QCR9 like" | 4 |
| PF05366 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Sarcolipin | 1 |
| PF05368 | 1 | 1 | 2 | 1 | 0 | 0 | 1 | 0 | NmrA-like family | 6 |
| PF05375 | 0 | 0 | 1 | 0 | 0 | 0 | 3 | 0 | Pacifastin inhibitor (LCMII) | 4 |
| PF05378 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | Hydantoinase/oxoprolinase N-terminal region | 4 |
| PF05383 | 1 | 1 | 7 | 2 | 1 | 1 | 8 | 1 | La domain | 22 |
| PF05386 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | TEP1 N-terminal domain | 1 |
| PF05391 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | Lsm interaction motif | 2 |
| PF05392 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | Cytochrome C oxidase chain VIIB | 2 |
| PF05395 | 0 | 1 | 3 | 2 | 0 | 0 | 2 | 0 | Protein phosphatase inhibitor 1/DARPP-32 | 8 |
| PF05399 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | Ectropic viral integration site 2A protein (EVI2A) | 2 |
| PF05404 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | "Translocon-associated protein, delta subunit precursor (TRAP-delta)" | 7 |
| PF05405 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | Mitochondrial ATP synthase B chain precursor (ATP-synt_B) | 6 |
| PF05406 | 0 | 0 | 3 | 2 | 3 | 2 | 3 | 2 | WGR domain | 15 |
| PF05422 | 0 | 0 | 1 | 0 | 1 | 1 | 1 | 0 | Stress-activated map kinase interacting protein 1 (SIN1) | 4 |
| PF05427 | 1 | 1 | 1 | 0 | 0 | 1 | 2 | 0 | Acidic fibroblast growth factor binding (FIBP) | 6 |
| PF05428 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Corticotropin-releasing factor binding protein (CRF-BP) | 3 |
| PF05432 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Bone sialoprotein II (BSP-II) | 1 |
| PF05434 | 1 | 2 | 2 | 1 | 0 | 1 | 2 | 0 | TMEM9 | 9 |
| PF05438 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Thyrotropin-releasing hormone (TRH) | 2 |
| PF05439 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Jumping translocation breakpoint protein (JTB) | 2 |
| PF05450 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | Nicastrin | 3 |
| PF05454 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | Dystroglycan (Dystrophin-associated glycoprotein 1) | 5 |
| PF05456 | 0 | 0 | 3 | 1 | 0 | 0 | 5 | 1 | Eukaryotic translation initiation factor 4E binding protein (EIF4EBP) | 10 |
| PF05458 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Cd27 binding protein (Siva) | 1 |
| PF05460 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Origin recognition complex subunit 6 (ORC6) | 2 |
| PF05461 | 0 | 0 | 13 | 0 | 0 | 0 | 10 | 0 | Apolipoprotein L | 23 |
| PF05462 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | Slime mold cyclic AMP receptor | 1 |
| PF05463 | 0 | 0 | 2 | 0 | 0 | 0 | 3 | 0 | Sclerostin (SOST) | 5 |
| PF05466 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | Brain acid soluble protein 1 (BASP1 protein) | 8 |
| PF05470 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 1 | Eukaryotic translation initiation factor 3 subunit 8 N-terminus | 3 |
| PF05474 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | Semenogelin | 2 |
| PF05477 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Surfeit locus protein 2 (SURF2) | 2 |
| PF05478 | 0 | 1 | 2 | 0 | 0 | 2 | 3 | 1 | Prominin | 9 |
| PF05483 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Synaptonemal complex protein 1 (SCP-1) | 2 |
| PF05485 | 0 | 0 | 7 | 1 | 0 | 0 | 32 | 0 | THAP domain | 40 |
| PF05486 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Signal recognition particle 9 kDa protein (SRP9) | 3 |
| PF05493 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 0 | ATP synthase subunit H | 3 |
| PF05495 | 0 | 0 | 1 | 1 | 0 | 0 | 2 | 0 | CHY zinc finger | 4 |
| PF05499 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | DNA methyltransferase 1-associated protein 1 (DMAP1) | 2 |
| PF05502 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | Dynactin p62 family | 6 |
| PF05507 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | Microfibril-associated glycoprotein (MAGP) | 4 |
| PF05510 | 1 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | Sarcoglycan alpha/epsilon | 5 |
| PF05511 | 0 | 1 | 1 | 1 | 0 | 2 | 3 | 2 | Mitochondrial ATP synthase coupling factor 6 | 10 |
| PF05517 | 1 | 2 | 3 | 2 | 0 | 2 | 2 | 2 | p25-alpha | 14 |
| PF05527 | 0 | 0 | 4 | 1 | 0 | 0 | 4 | 1 | Domain of unknown function (DUF758) | 10 |
| PF05529 | 2 | 2 | 2 | 2 | 1 | 2 | 2 | 1 | B-cell receptor-associated protein 31-like | 14 |
| PF05536 | 0 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | Neurochondrin | 3 |
| PF05544 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | Proline racemase | 2 |
| PF05556 | 0 | 0 | 3 | 0 | 0 | 0 | 5 | 0 | Calcineurin-binding protein (Calsarcin) | 8 |
| PF05557 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Mitotic checkpoint protein | 3 |
| PF05571 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 0 | Protein of unknown function (DUF766) | 3 |
| PF05572 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | Pregnancy-associated plasma protein-A | 4 |
| PF05577 | 0 | 0 | 3 | 1 | 0 | 1 | 3 | 0 | Serine carboxypeptidase S28 | 8 |
| PF05586 | 0 | 0 | 2 | 1 | 0 | 0 | 5 | 0 | Anthrax receptor C-terminus region | 8 |
| PF05587 | 0 | 0 | 3 | 1 | 0 | 0 | 5 | 0 | Anthrax receptor extracellular domain | 9 |
| PF05593 | 0 | 1 | 1 | 0 | 2 | 4 | 3 | 0 | RHS Repeat | 11 |
| PF05600 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | Protein of unknown function (DUF773) | 5 |
| PF05602 | 0 | 2 | 2 | 2 | 1 | 2 | 2 | 0 | Cleft lip and palate transmembrane protein 1 (CLPTM1) | 11 |
| PF05603 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Protein of unknown function (DUF775) | 1 |
| PF05604 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Protein of unknown function (DUF776) | 2 |
| PF05605 | 0 | 0 | 8 | 3 | 0 | 0 | 6 | 0 | "Drought induced 19 protein (Di19), zinc-binding" | 17 |
| PF05608 | 0 | 1 | 1 | 0 | 0 | 0 | 1 | 0 | Protein of unknown function (DUF778) | 3 |
| PF05609 | 0 | 0 | 2 | 1 | 0 | 0 | 2 | 0 | Lamina-associated polypeptide 1C (LAP1C) | 5 |
| PF05612 | 0 | 0 | 1 | 0 | 0 | 1 | 2 | 2 | Mouse protein of unknown function (DUF781) | 6 |
| PF05615 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Tho complex subunit 7 | 2 |
| PF05620 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Protein of unknown function (DUF788) | 2 |
| PF05622 | 2 | 2 | 6 | 1 | 0 | 0 | 5 | 0 | HOOK protein | 16 |
| PF05624 | 1 | 1 | 3 | 0 | 0 | 0 | 2 | 0 | Lipolysis stimulated receptor (LSR) | 7 |
| PF05625 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | PAXNEB protein | 2 |
| PF05631 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Protein of unknown function (DUF791) | 2 |
| PF05640 | 1 | 0 | 3 | 0 | 1 | 0 | 4 | 0 | "Na,K-Atpase Interacting protein" | 9 |
| PF05641 | 3 | 3 | 3 | 1 | 1 | 1 | 3 | 1 | Agenet domain | 16 |
| PF05644 | 1 | 1 | 1 | 1 | 0 | 2 | 2 | 0 | Mitochondrial and peroxisomal fission factor Mff | 8 |
| PF05645 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | RNA polymerase III subunit RPC82 | 2 |
| PF05648 | 1 | 1 | 3 | 0 | 0 | 1 | 3 | 0 | Peroxisomal biogenesis factor 11 (PEX11) | 9 |
| PF05649 | 1 | 1 | 7 | 1 | 0 | 1 | 4 | 0 | Peptidase family M13 | 15 |
| PF05652 | 0 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | Scavenger mRNA decapping enzyme (DcpS) N-terminal | 4 |
| PF05653 | 1 | 1 | 6 | 0 | 0 | 0 | 5 | 0 | Magnesium transporter NIPA | 13 |
| PF05667 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | Protein of unknown function (DUF812) | 6 |
| PF05669 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | SOH1 | 2 |
| PF05670 | 0 | 0 | 2 | 1 | 0 | 0 | 1 | 0 | Domain of unknown function (DUF814) | 4 |
| PF05672 | 3 | 2 | 3 | 1 | 2 | 2 | 5 | 0 | MAP7 (E-MAP-115) family | 18 |
| PF05676 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | NADH-ubiquinone oxidoreductase B18 subunit (NDUFB7) | 6 |
| PF05679 | 0 | 0 | 8 | 2 | 1 | 1 | 12 | 0 | Chondroitin N-acetylgalactosaminyltransferase | 24 |
| PF05680 | 1 | 1 | 1 | 0 | 0 | 2 | 2 | 2 | ATP synthase E chain | 9 |
| PF05686 | 0 | 1 | 3 | 1 | 0 | 0 | 3 | 0 | Glycosyl transferase family 90 | 8 |
| PF05693 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 1 | Glycogen synthase | 5 |
| PF05694 | 0 | 0 | 2 | 0 | 0 | 1 | 1 | 1 | 56kDa selenium binding protein (SBP56) | 5 |
| PF05699 | 0 | 0 | 5 | 0 | 0 | 0 | 4 | 1 | hAT family C-terminal dimerisation region | 10 |
| PF05700 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Breast carcinoma amplified sequence 2 (BCAS2) | 3 |
| PF05705 | 0 | 0 | 1 | 0 | 0 | 0 | 2 | 0 | Eukaryotic protein of unknown function (DUF829) | 3 |
| PF05706 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Cyclin-dependent kinase inhibitor 3 (CDKN3) | 2 |
| PF05712 | 0 | 0 | 4 | 1 | 0 | 0 | 1 | 0 | MRG | 6 |
| PF05715 | 2 | 2 | 2 | 0 | 2 | 2 | 1 | 1 | Piccolo Zn-finger | 12 |
| PF05716 | 1 | 1 | 3 | 0 | 0 | 0 | 2 | 0 | A-kinase anchor protein 110 kDa (AKAP 110) | 7 |
| PF05719 | 0 | 0 | 2 | 1 | 0 | 0 | 3 | 0 | Golgi phosphoprotein 3 (GPP34) | 6 |
| PF05721 | 0 | 0 | 2 | 1 | 0 | 1 | 3 | 0 | Phytanoyl-CoA dioxygenase (PhyH) | 7 |
| PF05724 | 0 | 0 | 1 | 0 | 0 | 0 | 2 | 0 | Thiopurine S-methyltransferase (TPMT) | 3 |
| PF05726 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Pirin C-terminal cupin domain | 3 |
| PF05729 | 0 | 2 | 33 | 0 | 1 | 2 | 146 | 4 | NACHT domain | 188 |
| PF05731 | 0 | 0 | 2 | 1 | 0 | 0 | 2 | 0 | TROVE domain | 5 |
| PF05735 | 0 | 0 | 5 | 1 | 0 | 0 | 6 | 1 | Thrombospondin C-terminal region | 13 |
| PF05739 | 10 | 15 | 19 | 9 | 2 | 12 | 27 | 6 | SNARE domain | 100 |
| PF05741 | 0 | 0 | 3 | 0 | 0 | 0 | 2 | 0 | Nanos RNA binding domain | 5 |
| PF05742 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | NRDE protein | 2 |
| PF05743 | 1 | 1 | 2 | 1 | 0 | 2 | 5 | 0 | UEV domain | 12 |
| PF05746 | 1 | 1 | 3 | 1 | 1 | 1 | 3 | 1 | DALR anticodon binding domain | 12 |
| PF05753 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Translocon-associated protein beta (TRAPB) | 2 |
| PF05760 | 0 | 0 | 3 | 0 | 0 | 0 | 3 | 0 | Immediate early response protein (IER) | 6 |
| PF05761 | 0 | 1 | 4 | 1 | 0 | 3 | 6 | 3 | 5' nucleotidase family | 18 |
| PF05764 | 0 | 0 | 1 | 0 | 0 | 0 | 2 | 0 | YL1 nuclear protein | 3 |
| PF05768 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Glutaredoxin-like domain (DUF836) | 2 |
| PF05769 | 1 | 1 | 2 | 1 | 0 | 0 | 1 | 0 | Protein of unknown function (DUF837) | 6 |
| PF05770 | 0 | 0 | 1 | 1 | 0 | 0 | 2 | 0 | "Inositol 1, 3, 4-trisphosphate 5/6-kinase" | 4 |
| PF05773 | 0 | 1 | 12 | 4 | 0 | 0 | 10 | 1 | RWD domain | 28 |
| PF05778 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Apolipoprotein CIII (Apo-CIII) | 2 |
| PF05781 | 0 | 0 | 2 | 0 | 0 | 0 | 4 | 0 | MRVI1 protein | 6 |
| PF05782 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Extracellular matrix protein 1 (ECM1) | 1 |
| PF05783 | 2 | 2 | 3 | 3 | 0 | 1 | 2 | 1 | Dynein light intermediate chain (DLIC) | 14 |
| PF05786 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Condensin complex subunit 2 | 2 |
| PF05790 | 0 | 1 | 3 | 0 | 0 | 0 | 0 | 0 | Immunoglobulin C2-set domain | 4 |
| PF05793 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | "Transcription initiation factor IIF, alpha subunit (TFIIF-alpha)" | 2 |
| PF05794 | 0 | 0 | 4 | 1 | 1 | 1 | 2 | 0 | T-complex protein 11 | 9 |
| PF05804 | 1 | 1 | 1 | 1 | 0 | 0 | 3 | 0 | Kinesin-associated protein (KAP) | 7 |
| PF05805 | 0 | 0 | 5 | 0 | 0 | 0 | 6 | 0 | L6 membrane protein | 11 |
| PF05806 | 0 | 0 | 1 | 0 | 0 | 0 | 3 | 0 | Noggin | 4 |
| PF05808 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Podoplanin | 2 |
| PF05811 | 0 | 1 | 1 | 1 | 0 | 1 | 0 | 0 | Eukaryotic protein of unknown function (DUF842) | 4 |
| PF05817 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | Oligosaccharyltransferase subunit Ribophorin II | 7 |
| PF05821 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | NADH-ubiquinone oxidoreductase ASHI subunit (CI-ASHI or NDUFB8) | 8 |
| PF05822 | 0 | 0 | 2 | 1 | 0 | 0 | 1 | 0 | Pyrimidine 5'-nucleotidase (UMPH-1) | 4 |
| PF05824 | 1 | 0 | 1 | 0 | 0 | 0 | 2 | 0 | Pro-melanin-concentrating hormone (Pro-MCH) | 4 |
| PF05825 | 0 | 0 | 2 | 0 | 0 | 0 | 4 | 0 | Beta-microseminoprotein (PSP-94) | 6 |
| PF05826 | 0 | 0 | 1 | 0 | 0 | 0 | 2 | 0 | Phospholipase A2 | 3 |
| PF05827 | 1 | 1 | 1 | 1 | 2 | 2 | 4 | 1 | Vacuolar ATP synthase subunit S1 (ATP6S1) | 13 |
| PF05832 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | Eukaryotic protein of unknown function (DUF846) | 4 |
| PF05833 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | Fibronectin-binding protein A N-terminus (FbpA) | 2 |
| PF05835 | 0 | 1 | 4 | 1 | 0 | 3 | 9 | 4 | Synaphin protein | 22 |
| PF05837 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Centromere protein H (CENP-H) | 2 |
| PF05839 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Apc13p protein | 2 |
| PF05843 | 0 | 0 | 2 | 1 | 0 | 0 | 1 | 0 | Suppressor of forked protein (Suf) | 4 |
| PF05856 | 1 | 1 | 1 | 0 | 1 | 1 | 2 | 2 | ARP2/3 complex 20 kDa subunit (ARPC4) | 9 |
| PF05859 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Mis12 protein | 2 |
| PF05871 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | ESCRT-II complex subunit | 5 |
| PF05873 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | "ATP synthase D chain, mitochondrial (ATP5H)" | 4 |
| PF05875 | 0 | 0 | 3 | 0 | 0 | 0 | 3 | 0 | Ceramidase | 6 |
| PF05879 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | Root hair defective 3 GTP-binding protein (RHD3) | 3 |
| PF05881 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 0 | "2',3'-cyclic nucleotide 3'-phosphodiesterase (CNP or CNPase)" | 5 |
| PF05889 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Soluble liver antigen/liver pancreas antigen (SLA/LP autoantigen) | 3 |
| PF05890 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Eukaryotic rRNA processing protein EBP2 | 3 |
| PF05891 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | AdoMet dependent proline di-methyltransferase | 4 |
| PF05902 | 2 | 3 | 4 | 1 | 0 | 3 | 4 | 3 | 4.1 protein C-terminal domain (CTD) | 20 |
| PF05903 | 0 | 0 | 2 | 2 | 0 | 0 | 3 | 0 | PPPDE putative peptidase domain | 7 |
| PF05907 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Eukaryotic protein of unknown function (DUF866) | 3 |
| PF05914 | 1 | 1 | 2 | 0 | 0 | 0 | 2 | 0 | RIB43A | 6 |
| PF05915 | 0 | 0 | 1 | 0 | 0 | 0 | 3 | 0 | Eukaryotic protein of unknown function (DUF872) | 4 |
| PF05916 | 0 | 0 | 3 | 0 | 0 | 0 | 3 | 0 | GINS complex protein | 6 |
| PF05918 | 0 | 0 | 1 | 1 | 0 | 1 | 1 | 0 | Apoptosis inhibitory protein 5 (API5) | 4 |
| PF05920 | 0 | 0 | 17 | 2 | 0 | 0 | 18 | 0 | Homeobox KN domain | 37 |
| PF05922 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Peptidase inhibitor I9 | 2 |
| PF05923 | 1 | 1 | 2 | 0 | 1 | 1 | 2 | 0 | APC cysteine-rich region | 8 |
| PF05924 | 1 | 1 | 2 | 0 | 1 | 1 | 2 | 0 | SAMP Motif | 8 |
| PF05934 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | Mid-1-related chloride channel (MCLC) | 2 |
| PF05937 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | EB-1 Binding Domain | 6 |
| PF05956 | 1 | 1 | 2 | 0 | 1 | 1 | 2 | 0 | APC basic domain | 8 |
| PF05958 | 0 | 0 | 1 | 0 | 0 | 0 | 2 | 0 | tRNA (Uracil-5-)-methyltransferase | 3 |
| PF05964 | 0 | 0 | 5 | 0 | 0 | 0 | 7 | 1 | F/Y-rich N-terminus | 13 |
| PF05965 | 0 | 0 | 5 | 0 | 0 | 0 | 7 | 1 | F/Y rich C-terminus | 13 |
| PF05970 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | PIF1-like helicase | 2 |
| PF05971 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Protein of unknown function (DUF890) | 2 |
| PF05972 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | APC 15 residue motif | 6 |
| PF05978 | 0 | 0 | 4 | 0 | 0 | 0 | 2 | 0 | Ion channel regulatory protein UNC-93 | 6 |
| PF05983 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | MED7 protein | 2 |
| PF05986 | 0 | 0 | 24 | 1 | 0 | 0 | 20 | 0 | ADAM-TS Spacer 1 | 45 |
| PF05994 | 2 | 2 | 2 | 0 | 2 | 2 | 2 | 2 | Cytoplasmic Fragile-X interacting family | 14 |
| PF05995 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Cysteine dioxygenase type I | 1 |
| PF05997 | 0 | 0 | 2 | 1 | 0 | 0 | 0 | 0 | "Nucleolar protein,Nop52" | 3 |
| PF06001 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 0 | Domain of Unknown Function (DUF902) | 3 |
| PF06003 | 1 | 0 | 3 | 1 | 0 | 0 | 3 | 0 | Survival motor neuron protein (SMN) | 8 |
| PF06008 | 0 | 1 | 5 | 0 | 0 | 0 | 4 | 1 | Laminin Domain I | 11 |
| PF06009 | 0 | 1 | 5 | 0 | 1 | 1 | 5 | 2 | Laminin Domain II | 15 |
| PF06012 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | Domain of Unknown Function (DUF908) | 5 |
| PF06017 | 5 | 3 | 8 | 2 | 7 | 5 | 10 | 2 | Myosin tail | 42 |
| PF06021 | 0 | 0 | 4 | 0 | 0 | 0 | 2 | 0 | Aralkyl acyl-CoA:amino acid N-acyltransferase | 6 |
| PF06025 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | Domain of Unknown Function (DUF913) | 5 |
| PF06026 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Ribose 5-phosphate isomerase A (phosphoriboisomerase A) | 2 |
| PF06027 | 0 | 1 | 3 | 1 | 0 | 2 | 4 | 0 | Eukaryotic protein of unknown function (DUF914) | 11 |
| PF06031 | 0 | 0 | 5 | 0 | 0 | 0 | 6 | 0 | SERTA motif | 11 |
| PF06046 | 1 | 1 | 4 | 2 | 1 | 2 | 7 | 1 | Exocyst complex component Sec6 | 19 |
| PF06047 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 0 | Ras-induced vulval development antagonist | 3 |
| PF06049 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Coagulation Factor V LSPD Repeat | 1 |
| PF06052 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 1 | 3-hydroxyanthranilic acid dioxygenase | 3 |
| PF06058 | 0 | 0 | 2 | 1 | 0 | 0 | 1 | 0 | Dcp1-like decapping family | 4 |
| PF06060 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | Pre-pro-megakaryocyte potentiating factor precursor (Mesothelin) | 4 |
| PF06068 | 0 | 1 | 2 | 2 | 2 | 2 | 2 | 1 | TIP49 C-terminus | 12 |
| PF06071 | 0 | 1 | 2 | 2 | 0 | 1 | 1 | 1 | Protein of unknown function (DUF933) | 8 |
| PF06079 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Apyrase | 2 |
| PF06080 | 0 | 1 | 1 | 1 | 0 | 0 | 2 | 0 | Protein of unknown function (DUF938) | 5 |
| PF06083 | 0 | 0 | 5 | 0 | 0 | 0 | 6 | 0 | Interleukin-17 | 11 |
| PF06087 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Tyrosyl-DNA phosphodiesterase | 2 |
| PF06090 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Inositol-pentakisphosphate 2-kinase | 2 |
| PF06093 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | Spt4/RpoE2 zinc finger | 2 |
| PF06094 | 0 | 0 | 1 | 1 | 0 | 0 | 3 | 0 | AIG2-like family | 5 |
| PF06098 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 0 | Radial spoke protein 3 | 3 |
| PF06102 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Domain of unknown function (DUF947) | 2 |
| PF06105 | 0 | 0 | 3 | 0 | 0 | 0 | 1 | 0 | Aph-1 protein | 4 |
| PF06110 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | Eukaryotic protein of unknown function (DUF953) | 4 |
| PF06113 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Brain and reproductive organ-expressed protein (BRE) | 3 |
| PF06119 | 0 | 0 | 4 | 0 | 0 | 0 | 12 | 1 | Nidogen-like | 17 |
| PF06121 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Domain of Unknown Function (DUF959) | 1 |
| PF06140 | 0 | 0 | 3 | 0 | 0 | 0 | 8 | 0 | Interferon-induced 6-16 family | 11 |
| PF06148 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | "COG (conserved oligomeric Golgi) complex component, COG2" | 2 |
| PF06155 | 0 | 0 | 1 | 2 | 0 | 0 | 2 | 0 | Protein of unknown function (DUF971) | 5 |
| PF06159 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Protein of unknown function (DUF974) | 2 |
| PF06179 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Surfeit locus protein 5 subunit 22 of Mediator complex | 2 |
| PF06189 | 0 | 0 | 2 | 0 | 0 | 0 | 4 | 0 | 5'-nucleotidase | 6 |
| PF06193 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Orthopoxvirus A5L protein-like | 2 |
| PF06201 | 1 | 1 | 2 | 0 | 0 | 1 | 1 | 1 | PITH domain | 7 |
| PF06202 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | "Amylo-alpha-1,6-glucosidase" | 3 |
| PF06209 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Cofactor of BRCA1 (COBRA1) | 3 |
| PF06211 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | BMP and activin membrane-bound inhibitor (BAMBI) N-terminal domain | 2 |
| PF06212 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | GRIM-19 protein | 6 |
| PF06214 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Signaling lymphocytic activation molecule (SLAM) protein | 1 |
| PF06218 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Nitrogen permease regulator 2 | 2 |
| PF06220 | 0 | 0 | 3 | 1 | 0 | 0 | 3 | 0 | U1 zinc finger | 7 |
| PF06221 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | "Putative zinc finger motif, C2HC5-type" | 1 |
| PF06229 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | FRG1-like family | 3 |
| PF06237 | 0 | 0 | 2 | 0 | 0 | 0 | 3 | 0 | Protein of unknown function (DUF1011) | 5 |
| PF06239 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | Evolutionarily conserved signalling intermediate in Toll pathway | 6 |
| PF06244 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | Protein of unknown function (DUF1014) | 4 |
| PF06246 | 0 | 0 | 1 | 0 | 0 | 0 | 2 | 0 | Isy1-like splicing family | 3 |
| PF06248 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | Centromere/kinetochore Zw10 | 6 |
| PF06268 | 0 | 1 | 3 | 1 | 0 | 2 | 4 | 2 | Fascin domain | 13 |
| PF06271 | 0 | 0 | 1 | 0 | 0 | 0 | 2 | 0 | RDD family | 3 |
| PF06278 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Protein of unknown function (DUF1032) | 2 |
| PF06292 | 3 | 2 | 5 | 0 | 1 | 3 | 5 | 2 | Domain of Unknown Function (DUF1041) | 21 |
| PF06293 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 1 | Lipopolysaccharide kinase (Kdo/WaaP) family | 4 |
| PF06294 | 0 | 0 | 4 | 0 | 0 | 0 | 2 | 0 | Domain of Unknown Function (DUF1042) | 6 |
| PF06297 | 1 | 1 | 8 | 2 | 2 | 1 | 7 | 1 | PET Domain | 23 |
| PF06309 | 0 | 0 | 5 | 1 | 0 | 1 | 8 | 0 | Torsin | 15 |
| PF06311 | 2 | 2 | 2 | 1 | 1 | 2 | 1 | 0 | NUMB domain | 11 |
| PF06312 | 0 | 0 | 8 | 0 | 0 | 0 | 12 | 0 | Neurexophilin | 20 |
| PF06320 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | GCN5-like protein 1 (GCN5L1) | 2 |
| PF06325 | 1 | 1 | 6 | 1 | 0 | 0 | 5 | 0 | Ribosomal protein L11 methyltransferase (PrmA) | 14 |
| PF06327 | 2 | 4 | 6 | 1 | 0 | 1 | 9 | 0 | Domain of Unknown Function (DUF1053) | 23 |
| PF06328 | 0 | 1 | 4 | 0 | 0 | 0 | 3 | 0 | Ig-like C2-type domain | 8 |
| PF06331 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Transcription factor TFIIH complex subunit Tfb5 | 2 |
| PF06333 | 1 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | Mediator complex subunit 13 C-terminal | 5 |
| PF06337 | 0 | 1 | 6 | 1 | 0 | 0 | 5 | 0 | DUSP domain | 13 |
| PF06345 | 0 | 0 | 3 | 1 | 0 | 0 | 1 | 0 | DRF Autoregulatory Domain | 5 |
| PF06346 | 1 | 1 | 3 | 0 | 0 | 0 | 0 | 0 | Formin Homology Region 1 | 5 |
| PF06350 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Hormone-sensitive lipase (HSL) N-terminus | 3 |
| PF06365 | 0 | 2 | 4 | 2 | 0 | 0 | 2 | 0 | CD34/Podocalyxin family | 10 |
| PF06367 | 4 | 5 | 9 | 5 | 5 | 5 | 9 | 0 | Diaphanous FH3 Domain | 42 |
| PF06369 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | Sea anemone cytotoxic protein | 1 |
| PF06371 | 4 | 5 | 9 | 5 | 5 | 5 | 9 | 0 | Diaphanous GTPase-binding Domain | 42 |
| PF06372 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Gemin6 protein | 1 |
| PF06373 | 0 | 0 | 1 | 0 | 1 | 1 | 4 | 0 | Cocaine and amphetamine regulated transcript protein (CART) | 7 |
| PF06374 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | NADH-ubiquinone oxidoreductase subunit b14.5b (NDUFC2) | 6 |
| PF06375 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | Bovine leukaemia virus receptor (BLVR) | 7 |
| PF06384 | 0 | 0 | 2 | 2 | 0 | 1 | 2 | 1 | Beta-catenin-interacting protein ICAT | 8 |
| PF06387 | 2 | 2 | 3 | 3 | 0 | 1 | 1 | 0 | D1 dopamine receptor-interacting protein (calcyon) | 12 |
| PF06388 | 1 | 1 | 2 | 1 | 0 | 1 | 1 | 1 | Protein of unknown function (DUF1075) | 8 |
| PF06390 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | Neuroendocrine-specific golgi protein P55 (NESP55) | 4 |
| PF06391 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | CDK-activating kinase assembly factor MAT1 | 3 |
| PF06393 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | BH3 interacting domain (BID) | 3 |
| PF06396 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | "Angiotensin II, type I receptor-associated protein (AGTRAP)" | 2 |
| PF06398 | 0 | 1 | 2 | 0 | 0 | 0 | 2 | 0 | Integral peroxisomal membrane peroxin | 5 |
| PF06399 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | GTP cyclohydrolase I feedback regulatory protein (GFRP) | 1 |
| PF06400 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | "Alpha-2-macroglobulin RAP, N-terminal domain" | 5 |
| PF06401 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | "Alpha-2-macroglobulin RAP, C-terminal domain" | 5 |
| PF06413 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Neugrin | 2 |
| PF06417 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | Protein of unknown function (DUF1077) | 6 |
| PF06418 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 0 | CTP synthase N-terminus | 3 |
| PF06419 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | Conserved oligomeric complex COG6 | 2 |
| PF06421 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | GTP-binding protein LepA C-terminus | 3 |
| PF06423 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | GWT1 | 2 |
| PF06424 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | "PRP1 splicing factor, N-terminal" | 3 |
| PF06427 | 0 | 1 | 2 | 0 | 1 | 2 | 2 | 2 | UDP-glucose:Glycoprotein Glucosyltransferase | 10 |
| PF06428 | 0 | 0 | 2 | 1 | 0 | 0 | 1 | 0 | GDP/GTP exchange factor Sec2p | 4 |
| PF06432 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Phosphatidylinositol N-acetylglucosaminyltransferase | 2 |
| PF06441 | 0 | 1 | 1 | 1 | 0 | 2 | 1 | 1 | Epoxide hydrolase N terminus | 7 |
| PF06444 | 0 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | NADH dehydrogenase subunit 2 C-terminus | 5 |
| PF06445 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | GyrI-like small molecule binding domain | 2 |
| PF06446 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | Hepcidin | 2 |
| PF06448 | 0 | 0 | 1 | 0 | 0 | 3 | 3 | 3 | Domain of Unknown Function (DUF1081) | 10 |
| PF06455 | 0 | 1 | 0 | 0 | 0 | 1 | 1 | 1 | NADH dehydrogenase subunit 5 C-terminus | 4 |
| PF06456 | 0 | 1 | 5 | 1 | 0 | 1 | 6 | 0 | Arfaptin-like domain | 14 |
| PF06459 | 2 | 1 | 3 | 0 | 2 | 2 | 4 | 1 | Ryanodine Receptor TM 4-6 | 15 |
| PF06461 | 0 | 0 | 3 | 1 | 0 | 0 | 3 | 0 | Domain of Unknown Function (DUF1086) | 7 |
| PF06462 | 0 | 1 | 2 | 0 | 0 | 0 | 7 | 1 | Propeller | 11 |
| PF06463 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Molybdenum Cofactor Synthesis C | 2 |
| PF06464 | 1 | 1 | 5 | 1 | 2 | 2 | 7 | 0 | DMAP1-binding Domain | 19 |
| PF06465 | 0 | 0 | 3 | 1 | 0 | 0 | 3 | 0 | Domain of Unknown Function (DUF1087) | 7 |
| PF06466 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 0 | PCAF (P300/CBP-associated factor) N-terminal domain | 3 |
| PF06467 | 0 | 0 | 7 | 1 | 0 | 0 | 3 | 0 | MYM-type Zinc finger with FCS sequence motif | 11 |
| PF06468 | 0 | 0 | 2 | 1 | 0 | 0 | 4 | 0 | Spondin_N | 7 |
| PF06469 | 0 | 1 | 2 | 1 | 0 | 1 | 2 | 0 | Domain of Unknown Function (DUF1088) | 7 |
| PF06470 | 2 | 0 | 6 | 3 | 2 | 2 | 6 | 2 | SMC proteins Flexible Hinge Domain | 23 |
| PF06472 | 2 | 2 | 4 | 0 | 3 | 3 | 4 | 1 | ABC transporter transmembrane region 2 | 19 |
| PF06473 | 0 | 0 | 2 | 0 | 0 | 1 | 5 | 0 | FGF binding protein 1 (FGF-BP1) | 8 |
| PF06479 | 0 | 0 | 3 | 1 | 0 | 0 | 3 | 0 | Ribonuclease 2-5A | 7 |
| PF06480 | 1 | 2 | 3 | 1 | 0 | 3 | 3 | 1 | FtsH Extracellular | 14 |
| PF06482 | 0 | 0 | 2 | 0 | 0 | 0 | 3 | 0 | Collagenase NC10 and Endostatin | 5 |
| PF06484 | 2 | 4 | 4 | 0 | 2 | 6 | 7 | 0 | Teneurin Intracellular Region | 25 |
| PF06487 | 0 | 0 | 2 | 2 | 0 | 0 | 1 | 1 | Sin3 associated polypeptide p18 (SAP18) | 6 |
| PF06495 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | Fruit fly transformer protein | 2 |
| PF06512 | 0 | 4 | 10 | 0 | 0 | 3 | 6 | 0 | Sodium ion transport-associated | 23 |
| PF06522 | 1 | 1 | 3 | 1 | 0 | 1 | 3 | 1 | NADH-ubiquinone reductase complex 1 MLRQ subunit | 11 |
| PF06524 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | NOA36 protein | 3 |
| PF06529 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Vertebrate interleukin-3 regulated transcription factor | 2 |
| PF06534 | 1 | 1 | 3 | 1 | 1 | 1 | 4 | 0 | Repulsive guidance molecule (RGM) C-terminus | 12 |
| PF06535 | 1 | 1 | 3 | 1 | 1 | 1 | 4 | 0 | Repulsive guidance molecule (RGM) N-terminus | 12 |
| PF06540 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Galanin message associated peptide (GMAP) | 2 |
| PF06541 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Protein of unknown function (DUF1113) | 2 |
| PF06544 | 0 | 0 | 4 | 1 | 0 | 0 | 2 | 0 | Protein of unknown function (DUF1115) | 7 |
| PF06546 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | Vertebrate heat shock transcription factor | 4 |
| PF06553 | 0 | 0 | 2 | 0 | 0 | 1 | 4 | 0 | BNIP3 | 7 |
| PF06554 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Olfactory marker protein | 2 |
| PF06565 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 1 | Repeat of unknown function (DUF1126) | 5 |
| PF06566 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | Chondroitin sulphate attachment domain | 2 |
| PF06567 | 0 | 1 | 1 | 0 | 0 | 2 | 1 | 1 | Neural chondroitin sulphate proteoglycan cytoplasmic domain | 6 |
| PF06573 | 0 | 0 | 2 | 1 | 0 | 0 | 1 | 0 | Churchill protein | 4 |
| PF06581 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Mad1 and Cdc20-bound-Mad2 binding | 1 |
| PF06583 | 0 | 2 | 2 | 0 | 2 | 2 | 1 | 0 | Neogenin C-terminus | 9 |
| PF06584 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | DIRP | 2 |
| PF06588 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Muskelin N-terminus | 3 |
| PF06602 | 2 | 3 | 14 | 6 | 0 | 3 | 15 | 0 | Myotubularin-like phosphatase domain | 43 |
| PF06607 | 0 | 0 | 3 | 1 | 0 | 0 | 2 | 0 | Prokineticin | 6 |
| PF06608 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Protein of unknown function (DUF1143) | 1 |
| PF06614 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | Neuromodulin | 4 |
| PF06617 | 0 | 0 | 3 | 1 | 0 | 0 | 1 | 0 | M-phase inducer phosphatase | 5 |
| PF06621 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | Single-minded protein C-terminus | 4 |
| PF06623 | 0 | 0 | 4 | 0 | 0 | 0 | 0 | 0 | MHC_I C-terminus | 4 |
| PF06624 | 0 | 0 | 2 | 0 | 0 | 0 | 3 | 0 | Ribosome associated membrane protein RAMP4 | 5 |
| PF06625 | 1 | 0 | 2 | 2 | 1 | 1 | 6 | 1 | Protein of unknown function (DUF1151) | 14 |
| PF06628 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | Catalase-related immune-responsive | 3 |
| PF06631 | 2 | 2 | 3 | 1 | 0 | 2 | 2 | 0 | Protein of unknown function (DUF1154) | 12 |
| PF06632 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | DNA double-strand break repair and V(D)J recombination protein XRCC4 | 2 |
| PF06637 | 0 | 0 | 1 | 0 | 0 | 0 | 2 | 0 | PV-1 protein (PLVAP) | 3 |
| PF06638 | 1 | 0 | 2 | 1 | 2 | 2 | 2 | 0 | Strabismus protein | 10 |
| PF06644 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | ATP11 protein | 3 |
| PF06645 | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 0 | Microsomal signal peptidase 12 kDa subunit (SPC12) | 5 |
| PF06650 | 0 | 3 | 4 | 0 | 1 | 3 | 3 | 0 | Protein of unknown function (DUF1162) | 14 |
| PF06657 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | Centrosome microtubule-binding domain of Cep57 | 4 |
| PF06658 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Protein of unknown function (DUF1168) | 2 |
| PF06662 | 0 | 0 | 1 | 0 | 1 | 1 | 2 | 0 | D-glucuronyl C5-epimerase C-terminus | 5 |
| PF06663 | 1 | 1 | 2 | 1 | 3 | 2 | 3 | 1 | Protein of unknown function (DUF1170) | 14 |
| PF06664 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | Wnt-binding factor required for Wnt secretion | 4 |
| PF06668 | 1 | 1 | 5 | 0 | 1 | 0 | 5 | 0 | Inter-alpha-trypsin inhibitor heavy chain C-terminus | 13 |
| PF06677 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Sjogren's syndrome/scleroderma autoantigen 1 (Autoantigen p27) | 3 |
| PF06679 | 0 | 0 | 3 | 0 | 0 | 0 | 1 | 0 | Protein of unknown function (DUF1180) | 4 |
| PF06682 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Protein of unknown function (DUF1183) | 3 |
| PF06699 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | GPI biosynthesis protein family Pig-F | 2 |
| PF06701 | 0 | 0 | 4 | 1 | 2 | 0 | 2 | 1 | Mib_herc2 | 10 |
| PF06702 | 0 | 0 | 4 | 1 | 1 | 1 | 5 | 0 | Protein of unknown function (DUF1193) | 12 |
| PF06703 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 0 | Microsomal signal peptidase 25 kDa subunit (SPC25) | 5 |
| PF06726 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Bladder cancer-related protein BC10 | 2 |
| PF06728 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | GPI transamidase subunit PIG-U | 2 |
| PF06729 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | "Kinetochore component, CENP-R" | 2 |
| PF06730 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 0 | FAM92 protein | 3 |
| PF06732 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Pescadillo N-terminus | 2 |
| PF06733 | 0 | 0 | 4 | 0 | 0 | 0 | 4 | 0 | DEAD_2 | 8 |
| PF06736 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | 0 | Protein of unknown function (DUF1211) | 4 |
| PF06741 | 1 | 0 | 2 | 1 | 0 | 0 | 2 | 1 | LsmAD domain | 7 |
| PF06743 | 0 | 0 | 7 | 2 | 0 | 0 | 4 | 0 | "FAST kinase-like protein, subdomain 1" | 13 |
| PF06745 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | KaiC | 1 |
| PF06747 | 2 | 2 | 8 | 5 | 0 | 3 | 6 | 2 | CHCH domain | 28 |
| PF06752 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | Enhancer of Polycomb C-terminus | 4 |
| PF06762 | 0 | 0 | 2 | 0 | 0 | 2 | 2 | 1 | Lipase maturation factor | 7 |
| PF06773 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Bim protein N-terminus | 1 |
| PF06775 | 0 | 0 | 1 | 0 | 0 | 1 | 2 | 0 | Putative adipose-regulatory protein (Seipin) | 4 |
| PF06777 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Protein of unknown function (DUF1227) | 2 |
| PF06783 | 0 | 0 | 1 | 0 | 1 | 1 | 1 | 1 | Uncharacterised protein family (UPF0239) | 5 |
| PF06784 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | Uncharacterised protein family (UPF0240) | 7 |
| PF06789 | 0 | 2 | 2 | 0 | 0 | 0 | 1 | 0 | Uncharacterised protein family (UPF0258) | 5 |
| PF06803 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | Protein of unknown function (DUF1232) | 2 |
| PF06807 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | Pre-mRNA cleavage complex II protein Clp1 | 4 |
| PF06809 | 0 | 1 | 1 | 0 | 0 | 0 | 2 | 0 | Neural proliferation differentiation control-1 protein (NPDC1) | 4 |
| PF06814 | 1 | 1 | 4 | 0 | 0 | 2 | 2 | 0 | Lung seven transmembrane receptor | 10 |
| PF06816 | 0 | 0 | 3 | 0 | 0 | 0 | 3 | 0 | NOTCH protein | 6 |
| PF06818 | 2 | 2 | 4 | 0 | 2 | 2 | 5 | 0 | Fez1 | 17 |
| PF06821 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | Serine hydrolase | 4 |
| PF06825 | 1 | 1 | 2 | 1 | 1 | 0 | 3 | 0 | Heat shock factor binding protein 1 | 9 |
| PF06827 | 0 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | Zinc finger found in FPG and IleRS | 3 |
| PF06831 | 0 | 0 | 3 | 0 | 0 | 0 | 2 | 0 | Formamidopyrimidine-DNA glycosylase H2TH domain | 5 |
| PF06839 | 0 | 0 | 7 | 0 | 0 | 0 | 3 | 1 | GRF zinc finger | 11 |
| PF06840 | 1 | 1 | 1 | 1 | 0 | 1 | 2 | 0 | Protein of unknown function (DUF1241) | 7 |
| PF06842 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | Protein of unknown function (DUF1242) | 4 |
| PF06858 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Nucleolar GTP-binding protein 1 (NOG1) | 3 |
| PF06859 | 0 | 0 | 2 | 0 | 0 | 0 | 3 | 0 | Bicoid-interacting protein 3 (Bin3) | 5 |
| PF06862 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Protein of unknown function (DUF1253) | 2 |
| PF06870 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | A49-like RNA polymerase I associated factor | 2 |
| PF06875 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 0 | Plethodontid receptivity factor PRF | 3 |
| PF06881 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | RNA polymerase II transcription factor SIII (Elongin) subunit A | 2 |
| PF06883 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | "RNA polymerase I, Rpa2 specific domain" | 2 |
| PF06886 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Targeting protein for Xklp2 (TPX2) | 2 |
| PF06888 | 0 | 0 | 2 | 1 | 0 | 0 | 1 | 0 | Putative Phosphatase | 4 |
| PF06905 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | Fas apoptotic inhibitory molecule (FAIM1) | 4 |
| PF06907 | 0 | 1 | 2 | 1 | 0 | 0 | 1 | 0 | Latexin | 5 |
| PF06910 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Male enhanced antigen 1 (MEA1) | 2 |
| PF06911 | 0 | 0 | 1 | 1 | 0 | 1 | 1 | 0 | Senescence-associated protein | 4 |
| PF06916 | 0 | 1 | 2 | 1 | 1 | 2 | 3 | 1 | Protein of unknown function (DUF1279) | 11 |
| PF06920 | 7 | 5 | 10 | 0 | 6 | 5 | 10 | 0 | Dedicator of cytokinesis | 43 |
| PF06936 | 1 | 1 | 2 | 0 | 0 | 1 | 1 | 1 | Selenoprotein S (SelS) | 7 |
| PF06937 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | EURL protein | 2 |
| PF06941 | 0 | 2 | 2 | 2 | 0 | 0 | 0 | 0 | "5' nucleotidase, deoxy (Pyrimidine), cytosolic type C protein (NT5C)" | 6 |
| PF06951 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | Group XII secretory phospholipase A2 precursor (PLA2G12) | 4 |
| PF06954 | 0 | 0 | 4 | 0 | 0 | 0 | 0 | 0 | Resistin | 4 |
| PF06957 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | Coatomer (COPI) alpha subunit C-terminus | 6 |
| PF06959 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | RecQ helicase protein-like 5 (RecQ5) | 1 |
| PF06963 | 0 | 0 | 1 | 0 | 0 | 0 | 2 | 0 | Ferroportin1 (FPN1) | 3 |
| PF06966 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | Protein of unknown function (DUF1295) | 1 |
| PF06969 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | HemN C-terminal domain | 2 |
| PF06978 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | Ribonucleases P/MRP protein subunit POP1 | 2 |
| PF06979 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Protein of unknown function (DUF1301) | 2 |
| PF06984 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Mitochondrial 39-S ribosomal protein L47 (MRP-L47) | 3 |
| PF06989 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | BAALC N-terminus | 2 |
| PF06990 | 0 | 0 | 6 | 0 | 0 | 0 | 5 | 0 | Galactose-3-O-sulfotransferase | 11 |
| PF06991 | 0 | 0 | 2 | 2 | 0 | 0 | 1 | 0 | "Splicing factor, Prp19-binding domain" | 5 |
| PF06994 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Involucrin | 1 |
| PF07000 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Protein of unknown function (DUF1308) | 2 |
| PF07001 | 2 | 2 | 3 | 1 | 0 | 0 | 3 | 0 | BAT2 N-terminus | 11 |
| PF07002 | 5 | 2 | 9 | 5 | 6 | 6 | 5 | 2 | Copine | 40 |
| PF07004 | 0 | 0 | 10 | 1 | 0 | 0 | 5 | 0 | Sperm-tail PG-rich repeat | 16 |
| PF07010 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Endomucin | 1 |
| PF07019 | 0 | 1 | 2 | 1 | 1 | 1 | 2 | 0 | Rab5-interacting protein (Rab5ip) | 8 |
| PF07034 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Origin recognition complex (ORC) subunit 3 N-terminus | 2 |
| PF07035 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | Colon cancer-associated protein Mic1-like | 7 |
| PF07039 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | SGF29 tudor-like domain | 2 |
| PF07047 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | Optic atrophy 3 protein (OPA3) | 7 |
| PF07051 | 1 | 2 | 2 | 2 | 0 | 1 | 3 | 1 | Ovarian carcinoma immunoreactive antigen (OCIA) | 12 |
| PF07052 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Hepatocellular carcinoma-associated antigen 59 | 2 |
| PF07061 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Swi5 | 3 |
| PF07064 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | RIC1 | 1 |
| PF07065 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | D123 | 3 |
| PF07074 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | "Translocon-associated protein, gamma subunit (TRAP-gamma)" | 7 |
| PF07078 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | Forty-two-three protein | 2 |
| PF07084 | 0 | 0 | 2 | 2 | 0 | 0 | 3 | 0 | Thyroid hormone-inducible hepatic protein Spot 14 | 7 |
| PF07086 | 0 | 0 | 1 | 0 | 0 | 0 | 2 | 0 | Protein of unknown function (DUF1352) | 3 |
| PF07092 | 0 | 1 | 3 | 1 | 0 | 1 | 3 | 0 | Protein of unknown function (DUF1356) | 9 |
| PF07093 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | SGT1 protein | 3 |
| PF07096 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Protein of unknown function (DUF1358) | 2 |
| PF07106 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Tat binding protein 1(TBP-1)-interacting protein (TBPIP) | 3 |
| PF07111 | 0 | 0 | 1 | 0 | 0 | 0 | 2 | 0 | Alpha helical coiled-coil rod protein (HCR) | 3 |
| PF07114 | 1 | 1 | 2 | 0 | 0 | 0 | 1 | 0 | Protein of unknown function (DUF1370) | 5 |
| PF07139 | 1 | 1 | 2 | 1 | 0 | 0 | 0 | 0 | Protein of unknown function (DUF1387) | 5 |
| PF07140 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Interferon gamma receptor (IFNGR1) | 1 |
| PF07142 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | Repeat of unknown function (DUF1388) | 3 |
| PF07145 | 1 | 0 | 8 | 2 | 0 | 0 | 4 | 1 | Ataxin-2 C-terminal region | 16 |
| PF07147 | 1 | 1 | 2 | 2 | 1 | 1 | 1 | 0 | Mitochondrial 28S ribosomal protein S30 (PDCD9) | 9 |
| PF07156 | 0 | 2 | 2 | 2 | 0 | 2 | 1 | 0 | Prenylcysteine lyase | 9 |
| PF07159 | 0 | 2 | 2 | 2 | 1 | 3 | 5 | 3 | Protein of unknown function (DUF1394) | 18 |
| PF07160 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Protein of unknown function (DUF1395) | 2 |
| PF07162 | 0 | 0 | 3 | 0 | 0 | 0 | 3 | 0 | "Ciliary basal body-associated, B9 protein" | 6 |
| PF07163 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Pex26 protein | 2 |
| PF07175 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Osteoregulin | 1 |
| PF07177 | 0 | 0 | 5 | 1 | 0 | 0 | 5 | 0 | Neuralized | 11 |
| PF07189 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Splicing factor 3B subunit 10 (SF3b10) | 3 |
| PF07192 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | SNURF/RPN4 protein | 2 |
| PF07200 | 2 | 1 | 4 | 3 | 0 | 0 | 4 | 0 | Modifier of rudimentary (Mod(r)) protein | 14 |
| PF07202 | 0 | 0 | 4 | 0 | 0 | 0 | 2 | 0 | T-complex protein 10 C-terminus | 6 |
| PF07213 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | DAP10 membrane protein | 2 |
| PF07221 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 1 | N-acylglucosamine 2-epimerase (GlcNAc 2-epimerase) | 3 |
| PF07222 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Proacrosin binding protein sp32 | 1 |
| PF07225 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | NADH-ubiquinone oxidoreductase B15 subunit (NDUFB4) | 7 |
| PF07228 | 0 | 0 | 1 | 0 | 0 | 0 | 2 | 0 | Stage II sporulation protein E (SpoIIE) | 3 |
| PF07258 | 4 | 5 | 10 | 5 | 0 | 1 | 9 | 1 | HCaRG protein | 35 |
| PF07259 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | ProSAAS precursor | 2 |
| PF07260 | 1 | 1 | 1 | 1 | 0 | 0 | 2 | 0 | Progressive ankylosis protein (ANKH) | 6 |
| PF07263 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Dentin matrix protein 1 (DMP1) | 1 |
| PF07264 | 0 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | Etoposide-induced protein 2.4 (EI24) | 4 |
| PF07270 | 0 | 0 | 7 | 0 | 0 | 0 | 0 | 0 | Protein of unknown function (DUF1438) | 7 |
| PF07281 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | Insulin-induced protein (INSIG) | 4 |
| PF07286 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Protein of unknown function (DUF1445) | 3 |
| PF07289 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Protein of unknown function (DUF1448) | 2 |
| PF07292 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 1 | Nmi/IFP 35 domain (NID) | 4 |
| PF07297 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Dolichol phosphate-mannose biosynthesis regulatory protein (DPM2) | 2 |
| PF07303 | 0 | 0 | 6 | 0 | 1 | 1 | 10 | 0 | Occludin homology domain | 18 |
| PF07304 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Steroid receptor RNA activator (SRA1) | 3 |
| PF07324 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | DiGeorge syndrome critical region 6 (DGCR6) protein | 2 |
| PF07326 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Protein of unknown function (DUF1466) | 1 |
| PF07334 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | Interferon-induced 35 kDa protein (IFP 35) N-terminus | 2 |
| PF07347 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | NADH:ubiquinone oxidoreductase subunit B14.5a (Complex I-B14.5a) | 8 |
| PF07353 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Uroplakin II | 1 |
| PF07354 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | Zona-pellucida-binding protein (Sp38) | 2 |
| PF07359 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Liver-expressed antimicrobial peptide 2 precursor (LEAP-2) | 2 |
| PF07386 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | Protein of unknown function (DUF1499) | 1 |
| PF07392 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Cyclin-dependent kinase inhibitor 2a p19Arf N-terminus | 1 |
| PF07393 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | Exocyst complex component Sec10 | 7 |
| PF07400 | 0 | 0 | 1 | 0 | 0 | 0 | 2 | 0 | Interleukin 11 | 3 |
| PF07406 | 0 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | NICE-3 protein | 4 |
| PF07412 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 0 | Geminin | 3 |
| PF07421 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Neurotensin/neuromedin N precursor | 3 |
| PF07426 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | Dynactin subunit p22 | 7 |
| PF07443 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | HepA-related protein (HARP) | 1 |
| PF07448 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Secreted phosphoprotein 24 (Spp-24) | 2 |
| PF07452 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | CHRD domain | 2 |
| PF07469 | 0 | 0 | 3 | 1 | 0 | 0 | 1 | 0 | Domain of unknown function (DUF1518) | 5 |
| PF07474 | 0 | 0 | 3 | 0 | 0 | 0 | 5 | 1 | G2F domain | 9 |
| PF07479 | 0 | 2 | 2 | 1 | 0 | 2 | 4 | 3 | NAD-dependent glycerol-3-phosphate dehydrogenase C-terminus | 14 |
| PF07491 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Protein phosphatase inhibitor | 3 |
| PF07496 | 0 | 0 | 6 | 0 | 0 | 0 | 3 | 0 | CW-type Zinc Finger | 9 |
| PF07500 | 0 | 0 | 7 | 3 | 0 | 1 | 6 | 1 | "Transcription factor S-II (TFIIS), central domain" | 18 |
| PF07502 | 0 | 0 | 4 | 0 | 0 | 0 | 3 | 0 | MANEC domain | 7 |
| PF07521 | 0 | 0 | 3 | 3 | 0 | 0 | 3 | 0 | RNA-metabolising metallo-beta-lactamase | 9 |
| PF07522 | 0 | 0 | 3 | 0 | 0 | 0 | 3 | 0 | DNA repair metallo-beta-lactamase | 6 |
| PF07524 | 0 | 0 | 3 | 0 | 0 | 0 | 3 | 0 | Bromodomain associated | 6 |
| PF07525 | 0 | 0 | 35 | 3 | 0 | 0 | 43 | 0 | SOCS box | 81 |
| PF07527 | 0 | 0 | 10 | 0 | 0 | 0 | 16 | 0 | Hairy Orange | 26 |
| PF07528 | 1 | 0 | 5 | 3 | 1 | 1 | 5 | 2 | DZF domain | 18 |
| PF07529 | 0 | 0 | 3 | 0 | 0 | 0 | 3 | 1 | HSA | 7 |
| PF07531 | 0 | 0 | 5 | 0 | 0 | 0 | 6 | 0 | NHR1 homology to TAF | 11 |
| PF07533 | 0 | 0 | 6 | 0 | 0 | 0 | 6 | 1 | BRK domain | 13 |
| PF07534 | 1 | 2 | 6 | 1 | 1 | 3 | 14 | 3 | TLD | 31 |
| PF07535 | 0 | 0 | 1 | 0 | 0 | 0 | 2 | 0 | DBF zinc finger | 3 |
| PF07539 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Down-regulated in metastasis | 1 |
| PF07540 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Nucleolar complex-associated protein | 2 |
| PF07541 | 0 | 1 | 1 | 1 | 0 | 1 | 2 | 0 | Eukaryotic translation initiation factor 2 alpha subunit | 6 |
| PF07542 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | ATP12 chaperone protein | 3 |
| PF07544 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | RNA polymerase II transcription mediator complex subunit 9 | 3 |
| PF07545 | 0 | 0 | 3 | 0 | 0 | 0 | 4 | 0 | Vestigial/Tondu family | 7 |
| PF07546 | 0 | 0 | 12 | 1 | 0 | 0 | 8 | 0 | EMI domain | 21 |
| PF07555 | 0 | 0 | 1 | 1 | 0 | 0 | 2 | 0 | beta-N-acetylglucosaminidase | 4 |
| PF07557 | 0 | 0 | 1 | 0 | 0 | 0 | 2 | 1 | Shugoshin C terminus | 4 |
| PF07558 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Shugoshin N-terminal coiled-coil region | 1 |
| PF07562 | 6 | 7 | 133 | 2 | 8 | 11 | 53 | 1 | Nine Cysteines Domain of family 3 GPCR | 221 |
| PF07565 | 1 | 6 | 9 | 0 | 1 | 5 | 11 | 4 | Band 3 cytoplasmic domain | 37 |
| PF07569 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | TUP1-like enhancer of split | 2 |
| PF07571 | 0 | 0 | 2 | 1 | 0 | 0 | 1 | 0 | Protein of unknown function (DUF1546) | 4 |
| PF07572 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | Bucentaur or craniofacial development | 4 |
| PF07574 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Nse1 non-SMC component of SMC5-6 complex | 1 |
| PF07575 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | Nup85 Nucleoporin | 2 |
| PF07576 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | BRCA1-associated protein 2 | 3 |
| PF07593 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | ASPIC and UnbV | 7 |
| PF07645 | 3 | 8 | 73 | 11 | 0 | 8 | 85 | 6 | Calcium-binding EGF domain | 194 |
| PF07646 | 0 | 0 | 5 | 1 | 0 | 0 | 1 | 0 | Kelch motif | 7 |
| PF07647 | 10 | 11 | 38 | 10 | 11 | 12 | 35 | 4 | SAM domain (Sterile alpha motif) | 131 |
| PF07648 | 3 | 5 | 37 | 5 | 3 | 3 | 31 | 1 | Kazal-type serine protease inhibitor domain | 88 |
| PF07650 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | KH domain | 7 |
| PF07651 | 2 | 4 | 4 | 0 | 0 | 8 | 6 | 2 | ANTH domain | 26 |
| PF07653 | 18 | 16 | 55 | 12 | 19 | 19 | 64 | 4 | Variant SH3 domain | 207 |
| PF07654 | 1 | 2 | 52 | 0 | 1 | 3 | 54 | 5 | Immunoglobulin C1-set domain | 118 |
| PF07657 | 0 | 0 | 5 | 1 | 0 | 0 | 10 | 0 | N terminus of Notch ligand | 16 |
| PF07662 | 0 | 0 | 4 | 0 | 0 | 0 | 1 | 0 | Na+ dependent nucleoside transporter C-terminus | 5 |
| PF07670 | 0 | 0 | 4 | 0 | 0 | 0 | 1 | 0 | Nucleoside recognition | 5 |
| PF07676 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | WD40-like Beta Propeller Repeat | 3 |
| PF07677 | 1 | 1 | 9 | 0 | 1 | 4 | 24 | 7 | A-macroglobulin receptor | 47 |
| PF07678 | 1 | 1 | 9 | 0 | 1 | 4 | 24 | 7 | A-macroglobulin complement component | 47 |
| PF07679 | 32 | 59 | 166 | 33 | 47 | 83 | 209 | 28 | Immunoglobulin I-set domain | 657 |
| PF07683 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Cobalamin synthesis protein cobW C-terminal domain | 2 |
| PF07684 | 0 | 0 | 4 | 0 | 0 | 0 | 4 | 0 | NOTCH protein | 8 |
| PF07686 | 14 | 29 | 178 | 17 | 15 | 40 | 370 | 17 | Immunoglobulin V-set domain | 680 |
| PF07687 | 0 | 3 | 5 | 3 | 0 | 3 | 5 | 2 | Peptidase dimerisation domain | 21 |
| PF07690 | 1 | 11 | 65 | 9 | 0 | 11 | 82 | 7 | Major Facilitator Superfamily | 186 |
| PF07691 | 0 | 0 | 2 | 0 | 1 | 1 | 5 | 0 | PA14 domain | 9 |
| PF07693 | 0 | 1 | 2 | 0 | 0 | 1 | 3 | 0 | KAP family P-loop domain | 7 |
| PF07699 | 0 | 2 | 10 | 0 | 0 | 3 | 10 | 2 | GCC2 and GCC3 | 27 |
| PF07700 | 2 | 2 | 4 | 0 | 1 | 1 | 2 | 1 | Heme NO binding | 13 |
| PF07701 | 2 | 2 | 5 | 0 | 1 | 2 | 8 | 1 | Heme NO binding associated | 21 |
| PF07703 | 1 | 1 | 9 | 0 | 1 | 4 | 24 | 7 | Alpha-2-macroglobulin family N-terminal region | 47 |
| PF07706 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Aminotransferase ubiquitination site | 1 |
| PF07707 | 1 | 1 | 56 | 5 | 3 | 3 | 60 | 1 | BTB And C-terminal Kelch | 130 |
| PF07710 | 0 | 0 | 3 | 0 | 0 | 0 | 3 | 0 | P53 tetramerisation motif | 6 |
| PF07711 | 1 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | "Rab geranylgeranyl transferase alpha-subunit, insert domain" | 5 |
| PF07713 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Protein of unknown function (DUF1604) | 2 |
| PF07714 | 11 | 24 | 120 | 18 | 10 | 29 | 142 | 3 | Protein tyrosine kinase | 357 |
| PF07716 | 0 | 0 | 10 | 0 | 0 | 0 | 20 | 0 | Basic region leucine zipper | 30 |
| PF07717 | 4 | 2 | 16 | 5 | 4 | 4 | 15 | 2 | Oligonucleotide/oligosaccharide-binding (OB)-fold | 52 |
| PF07718 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | Coatomer beta C-terminal region | 6 |
| PF07719 | 7 | 8 | 43 | 11 | 4 | 5 | 37 | 4 | Tetratricopeptide repeat | 119 |
| PF07722 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Peptidase C26 | 2 |
| PF07723 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Leucine Rich Repeat | 1 |
| PF07724 | 1 | 1 | 2 | 1 | 0 | 0 | 3 | 0 | AAA domain (Cdc48 subfamily) | 8 |
| PF07728 | 1 | 3 | 13 | 1 | 1 | 3 | 11 | 1 | AAA domain (dynein-related subfamily) | 34 |
| PF07731 | 0 | 0 | 4 | 0 | 1 | 2 | 3 | 2 | Multicopper oxidase | 12 |
| PF07732 | 0 | 0 | 5 | 0 | 1 | 2 | 4 | 2 | Multicopper oxidase | 14 |
| PF07738 | 0 | 0 | 7 | 1 | 0 | 0 | 5 | 0 | Sad1 / UNC-like C-terminal | 13 |
| PF07741 | 0 | 0 | 1 | 0 | 0 | 0 | 2 | 0 | Brf1-like TBP-binding domain | 3 |
| PF07742 | 0 | 0 | 8 | 0 | 0 | 0 | 8 | 0 | BTG family | 16 |
| PF07743 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | HSCB C-terminal oligomerisation domain | 2 |
| PF07744 | 0 | 0 | 5 | 2 | 0 | 0 | 7 | 0 | SPOC domain | 14 |
| PF07748 | 1 | 1 | 5 | 1 | 1 | 2 | 4 | 0 | Glycosyl hydrolases family 38 C-terminal domain | 15 |
| PF07749 | 0 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | "Endoplasmic reticulum protein ERp29, C-terminal domain" | 3 |
| PF07757 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Predicted AdoMet-dependent methyltransferase | 2 |
| PF07763 | 0 | 0 | 2 | 1 | 0 | 0 | 3 | 0 | FEZ-like protein | 6 |
| PF07766 | 1 | 1 | 3 | 2 | 2 | 3 | 3 | 1 | LETM1-like protein | 16 |
| PF07767 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Nop53 (60S ribosomal biogenesis) | 1 |
| PF07773 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | Protein of unknown function (DUF1619) | 4 |
| PF07774 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | Protein of unknown function (DUF1620) | 7 |
| PF07776 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | Zinc-finger associated domain (zf-AD) | 2 |
| PF07778 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Mis6 | 2 |
| PF07779 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 10 TM Acyl Transferase domain found in Cas1p | 2 |
| PF07780 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Spb1 C-terminal domain | 1 |
| PF07782 | 0 | 0 | 3 | 0 | 0 | 0 | 4 | 0 | DC-STAMP-like protein | 7 |
| PF07786 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Protein of unknown function (DUF1624) | 2 |
| PF07787 | 0 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | Protein of unknown function (DUF1625) | 3 |
| PF07798 | 0 | 2 | 2 | 0 | 0 | 1 | 2 | 0 | Protein of unknown function (DUF1640) | 7 |
| PF07803 | 1 | 0 | 2 | 0 | 0 | 0 | 5 | 0 | GSG1-like protein | 8 |
| PF07807 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | RED-like protein C-terminal region | 3 |
| PF07808 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | RED-like protein N-terminal region | 3 |
| PF07809 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 0 | RTP801 C-terminal region | 3 |
| PF07810 | 0 | 0 | 8 | 1 | 0 | 0 | 7 | 0 | TMC domain | 16 |
| PF07814 | 0 | 0 | 1 | 0 | 0 | 0 | 2 | 0 | Wings apart-like protein regulation of heterochromatin | 3 |
| PF07815 | 2 | 2 | 3 | 3 | 6 | 6 | 6 | 2 | Abl-interactor HHR | 30 |
| PF07817 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | GLE1-like protein | 2 |
| PF07818 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | HCNGP-like protein | 2 |
| PF07819 | 1 | 1 | 2 | 0 | 1 | 0 | 2 | 0 | PGAP1-like protein | 7 |
| PF07830 | 0 | 1 | 3 | 2 | 0 | 2 | 6 | 2 | "Protein serine/threonine phosphatase 2C, C-terminal domain" | 16 |
| PF07831 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Pyrimidine nucleoside phosphorylase C-terminal domain | 1 |
| PF07834 | 0 | 0 | 1 | 0 | 0 | 0 | 2 | 0 | RanGAP1 C-terminal domain | 3 |
| PF07837 | 0 | 0 | 1 | 0 | 0 | 0 | 2 | 0 | "Formiminotransferase domain, N-terminal subdomain" | 3 |
| PF07842 | 0 | 0 | 3 | 2 | 0 | 0 | 3 | 0 | GC-rich sequence DNA-binding factor-like protein | 8 |
| PF07847 | 0 | 0 | 1 | 1 | 0 | 0 | 2 | 0 | Protein of unknown function (DUF1637) | 4 |
| PF07850 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | Renin receptor-like protein | 7 |
| PF07851 | 0 | 0 | 2 | 1 | 0 | 0 | 3 | 0 | TMPIT-like protein | 6 |
| PF07855 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Protein of unknown function (DUF1649) | 2 |
| PF07856 | 0 | 0 | 3 | 0 | 0 | 0 | 3 | 0 | Mediator of CRAC channel activity | 6 |
| PF07857 | 0 | 0 | 1 | 0 | 0 | 0 | 2 | 0 | CEO family (DUF1632) | 3 |
| PF07859 | 0 | 1 | 15 | 1 | 0 | 1 | 7 | 0 | alpha/beta hydrolase fold | 25 |
| PF07884 | 0 | 0 | 2 | 2 | 0 | 0 | 2 | 0 | Vitamin K epoxide reductase family | 6 |
| PF07885 | 1 | 2 | 19 | 0 | 0 | 1 | 27 | 1 | Ion channel | 51 |
| PF07888 | 1 | 1 | 3 | 0 | 0 | 0 | 5 | 0 | Calcium binding and coiled-coil domain (CALCOCO1) like | 10 |
| PF07890 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Rrp15p | 2 |
| PF07894 | 0 | 0 | 8 | 0 | 0 | 0 | 9 | 0 | Protein of unknown function (DUF1669) | 17 |
| PF07896 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Protein of unknown function (DUF1674) | 2 |
| PF07904 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Chromatin modification-related protein EAF7 | 2 |
| PF07910 | 0 | 1 | 3 | 2 | 0 | 0 | 3 | 0 | Peptidase family C78 | 9 |
| PF07912 | 0 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | "ERp29, N-terminal domain" | 3 |
| PF07915 | 0 | 2 | 2 | 0 | 0 | 0 | 2 | 0 | Glucosidase II beta subunit-like protein | 6 |
| PF07919 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | "Gryzun, putative trafficking through Golgi" | 2 |
| PF07923 | 0 | 1 | 2 | 1 | 1 | 1 | 2 | 0 | N1221-like protein | 8 |
| PF07926 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | TPR/MLP1/MLP2-like protein | 3 |
| PF07928 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Vps54-like protein | 1 |
| PF07933 | 1 | 1 | 2 | 1 | 0 | 1 | 2 | 2 | Protein of unknown function (DUF1681) | 10 |
| PF07934 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | "8-oxoguanine DNA glycosylase, N-terminal domain" | 2 |
| PF07941 | 1 | 1 | 1 | 0 | 0 | 0 | 1 | 0 | Potassium channel Kv1.4 tandem inactivation domain | 4 |
| PF07942 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | N2227-like protein | 1 |
| PF07946 | 1 | 1 | 1 | 1 | 0 | 1 | 2 | 0 | Protein of unknown function (DUF1682) | 7 |
| PF07947 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | YhhN-like protein | 4 |
| PF07959 | 0 | 0 | 2 | 1 | 0 | 0 | 2 | 0 | L-fucokinase | 5 |
| PF07962 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Replication Fork Protection Component Swi3 | 2 |
| PF07965 | 0 | 5 | 6 | 1 | 0 | 2 | 7 | 1 | Integrin beta tail domain | 22 |
| PF07966 | 0 | 1 | 6 | 1 | 0 | 1 | 3 | 1 | A1 Propeptide | 13 |
| PF07967 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | C3HC zinc finger-like | 2 |
| PF07970 | 0 | 1 | 3 | 0 | 0 | 0 | 3 | 0 | Endoplasmic reticulum vesicle transporter | 7 |
| PF07973 | 0 | 4 | 6 | 4 | 1 | 2 | 4 | 2 | Threonyl and Alanyl tRNA synthetase second additional domain | 23 |
| PF07974 | 2 | 4 | 12 | 2 | 3 | 3 | 13 | 2 | EGF-like domain | 41 |
| PF07975 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | TFIIH C1-like domain | 2 |
| PF07978 | 2 | 3 | 4 | 1 | 1 | 2 | 3 | 1 | NIPSNAP | 17 |
| PF07984 | 0 | 0 | 4 | 0 | 0 | 0 | 6 | 0 | Domain of unknown function (DUF1693) | 10 |
| PF07985 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | SRR1 | 2 |
| PF07986 | 1 | 1 | 3 | 1 | 1 | 1 | 3 | 0 | Tubulin binding cofactor C | 11 |
| PF07988 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | LMSTEN motif | 4 |
| PF07989 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | Microtubule associated | 4 |
| PF07992 | 4 | 7 | 9 | 4 | 3 | 6 | 11 | 4 | Pyridine nucleotide-disulphide oxidoreductase | 48 |
| PF07993 | 1 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | Male sterility protein | 5 |
| PF07994 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | Myo-inositol-1-phosphate synthase | 2 |
| PF07995 | 0 | 0 | 3 | 0 | 0 | 0 | 4 | 0 | Glucose / Sorbosone dehydrogenase | 7 |
| PF07998 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Peptidase family M54 | 3 |
| PF08005 | 1 | 1 | 5 | 0 | 1 | 1 | 6 | 0 | PHR domain | 15 |
| PF08007 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | Cupin superfamily protein | 4 |
| PF08014 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | Domain of unknown function (DUF1704) | 4 |
| PF08016 | 0 | 1 | 10 | 1 | 0 | 1 | 7 | 0 | Polycystin cation channel | 20 |
| PF08022 | 0 | 0 | 6 | 1 | 0 | 0 | 4 | 0 | FAD-binding domain | 11 |
| PF08030 | 0 | 0 | 6 | 1 | 0 | 0 | 4 | 0 | Ferric reductase NAD binding domain | 11 |
| PF08032 | 0 | 0 | 2 | 0 | 0 | 0 | 3 | 0 | RNA 2'-O ribose methyltransferase substrate binding | 5 |
| PF08033 | 4 | 3 | 6 | 3 | 3 | 4 | 4 | 0 | Sec23/Sec24 beta-sandwich domain | 27 |
| PF08035 | 0 | 0 | 1 | 1 | 0 | 0 | 2 | 0 | Opioids neuropeptide | 4 |
| PF08038 | 0 | 0 | 1 | 1 | 0 | 1 | 1 | 0 | TOM7 family | 4 |
| PF08039 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | Mitochondrial proteolipid | 7 |
| PF08040 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | MNLL subunit | 4 |
| PF08043 | 1 | 1 | 2 | 0 | 0 | 0 | 3 | 0 | Xin repeat | 7 |
| PF08059 | 0 | 1 | 4 | 1 | 0 | 1 | 3 | 1 | SEP domain | 11 |
| PF08060 | 0 | 0 | 3 | 0 | 0 | 0 | 2 | 1 | NOSIC (NUC001) domain | 6 |
| PF08061 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | P68HR (NUC004) repeat | 4 |
| PF08062 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | P120R (NUC006) repeat | 1 |
| PF08063 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | PADR1 (NUC008) domain | 6 |
| PF08064 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | UME (NUC010) domain | 2 |
| PF08065 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | K167R (NUC007) repeat | 2 |
| PF08066 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | PMC2NT (NUC016) domain | 3 |
| PF08067 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 2 | ROKNT (NUC014) domain | 10 |
| PF08068 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | DKCLD (NUC011) domain | 2 |
| PF08069 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | Ribosomal S13/S15 N-terminal domain | 7 |
| PF08070 | 0 | 0 | 2 | 0 | 1 | 0 | 1 | 0 | DTHCT (NUC029) region | 4 |
| PF08071 | 1 | 0 | 1 | 0 | 1 | 1 | 1 | 1 | RS4NT (NUC023) domain | 6 |
| PF08072 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | BDHCT (NUC031) domain | 2 |
| PF08073 | 0 | 0 | 3 | 1 | 0 | 0 | 3 | 0 | CHDNT (NUC034) domain | 7 |
| PF08074 | 0 | 0 | 3 | 1 | 0 | 0 | 3 | 0 | CHDCT2 (NUC038) domain | 7 |
| PF08075 | 3 | 3 | 3 | 3 | 2 | 2 | 3 | 2 | NOPS (NUC059) domain | 21 |
| PF08079 | 1 | 1 | 2 | 0 | 1 | 1 | 2 | 1 | Ribosomal L30 N-terminal domain | 9 |
| PF08080 | 0 | 0 | 3 | 3 | 1 | 0 | 2 | 0 | RNPHF zinc finger | 9 |
| PF08081 | 1 | 0 | 3 | 2 | 0 | 0 | 1 | 1 | RBM1CTR (NUC064) family | 8 |
| PF08082 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 1 | "PRO8NT (NUC069), PrP8 N-terminal domain" | 3 |
| PF08083 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 1 | PROCN (NUC071) domain | 3 |
| PF08084 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 1 | PROCT (NUC072) domain | 3 |
| PF08122 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | NADH-ubiquinone oxidoreductase B12 subunit family | 8 |
| PF08123 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Histone methylation protein DOT1 | 2 |
| PF08127 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | Peptidase family C1 propeptide | 6 |
| PF08142 | 0 | 0 | 2 | 1 | 0 | 0 | 2 | 0 | AARP2CN (NUC121) domain | 5 |
| PF08143 | 2 | 2 | 2 | 2 | 0 | 2 | 3 | 3 | CBFNT (NUC161) domain | 16 |
| PF08144 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | CPL (NUC119) domain | 2 |
| PF08145 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | BOP1NT (NUC169) domain | 2 |
| PF08146 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | BP28CT (NUC211) domain | 2 |
| PF08147 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | DBP10CT (NUC160) domain | 1 |
| PF08148 | 0 | 0 | 2 | 1 | 0 | 0 | 2 | 0 | DSHCT (NUC185) domain | 5 |
| PF08149 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | BING4CT (NUC141) domain | 2 |
| PF08150 | 0 | 0 | 3 | 1 | 0 | 2 | 7 | 1 | FerB (NUC096) domain | 14 |
| PF08151 | 0 | 0 | 3 | 1 | 0 | 2 | 7 | 1 | FerI (NUC094) domain | 14 |
| PF08152 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 0 | GUCT (NUC152) domain | 3 |
| PF08153 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | NGP1NT (NUC091) domain | 1 |
| PF08154 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | NLE (NUC135) domain | 4 |
| PF08155 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | NOGCT (NUC087) domain | 3 |
| PF08156 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 1 | NOP5NT (NUC127) domain | 4 |
| PF08157 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | NUC129 domain | 3 |
| PF08158 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | NUC130/3NT domain | 1 |
| PF08159 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | NUC153 domain | 4 |
| PF08161 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | NUC173 domain | 2 |
| PF08163 | 0 | 0 | 1 | 0 | 1 | 1 | 1 | 0 | NUC194 domain | 4 |
| PF08164 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | "Apoptosis-antagonizing transcription factor, C-terminal" | 2 |
| PF08165 | 0 | 0 | 2 | 1 | 0 | 1 | 3 | 0 | FerA (NUC095) domain | 7 |
| PF08166 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | NUC202 domain | 2 |
| PF08167 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | rRNA processing/ribosome biogenesis | 2 |
| PF08168 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | NUC205 domain | 1 |
| PF08169 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | RBB1NT (NUC162) domain | 4 |
| PF08170 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | POPLD (NUC188) domain | 2 |
| PF08172 | 0 | 0 | 1 | 0 | 1 | 1 | 1 | 0 | CASP C terminal | 4 |
| PF08174 | 0 | 0 | 3 | 0 | 0 | 0 | 4 | 0 | Cell division protein anillin | 7 |
| PF08190 | 0 | 0 | 5 | 1 | 0 | 0 | 3 | 0 | pre-RNA processing PIH1/Nop17 | 9 |
| PF08198 | 1 | 0 | 1 | 1 | 1 | 1 | 3 | 1 | Thymopoietin protein | 9 |
| PF08202 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Mis12-Mtw1 protein family | 1 |
| PF08204 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | CD47 immunoglobulin-like domain | 4 |
| PF08205 | 2 | 13 | 51 | 10 | 2 | 16 | 44 | 8 | CD80-like C2-set immunoglobulin domain | 146 |
| PF08208 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | DNA-directed RNA polymerase I subunit RPA34.5 | 2 |
| PF08209 | 0 | 0 | 1 | 1 | 0 | 0 | 2 | 0 | Sgf11 (transcriptional regulation protein) | 4 |
| PF08210 | 0 | 0 | 5 | 0 | 0 | 0 | 3 | 0 | APOBEC-like N-terminal domain | 8 |
| PF08212 | 0 | 1 | 1 | 1 | 0 | 0 | 3 | 0 | Lipocalin-like domain | 6 |
| PF08213 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Mitochondrial domain of unknown function (DUF1713) | 2 |
| PF08214 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 0 | Histone acetylation protein | 3 |
| PF08216 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | "Catenin-beta-like, Arm-motif containing nuclear" | 2 |
| PF08217 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | Fungal domain of unknown function (DUF1712) | 7 |
| PF08221 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | RNA polymerase III subunit RPC82 helix-turn-helix domain | 2 |
| PF08227 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | DASH complex subunit Hsk3 like | 2 |
| PF08231 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | SYF2 splicing factor | 2 |
| PF08232 | 3 | 3 | 3 | 1 | 0 | 1 | 1 | 0 | Striatin family | 12 |
| PF08234 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Chromosome segregation protein Spc25 | 2 |
| PF08235 | 1 | 2 | 6 | 3 | 0 | 1 | 4 | 0 | LNS2 (Lipin/Ned1/Smp2) | 17 |
| PF08236 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | SRI (Set2 Rpb1 interacting) domain | 2 |
| PF08238 | 0 | 2 | 7 | 2 | 0 | 2 | 6 | 0 | Sel1 repeat | 19 |
| PF08239 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | Bacterial SH3 domain | 6 |
| PF08240 | 0 | 6 | 14 | 8 | 0 | 6 | 12 | 7 | Alcohol dehydrogenase GroES-like domain | 53 |
| PF08241 | 0 | 1 | 13 | 4 | 0 | 1 | 12 | 2 | Methyltransferase domain | 33 |
| PF08242 | 0 | 1 | 2 | 1 | 0 | 0 | 1 | 0 | Methyltransferase domain | 5 |
| PF08243 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | SPT2 chromatin protein | 2 |
| PF08245 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | Mur ligase middle domain | 1 |
| PF08246 | 0 | 1 | 19 | 2 | 0 | 0 | 27 | 3 | Cathepsin propeptide inhibitor domain (I29) | 52 |
| PF08264 | 3 | 5 | 6 | 4 | 2 | 3 | 5 | 4 | Anticodon-binding domain of tRNA | 32 |
| PF08265 | 0 | 0 | 2 | 0 | 0 | 0 | 3 | 0 | YL1 nuclear protein C-terminal domain | 5 |
| PF08266 | 4 | 10 | 49 | 3 | 12 | 20 | 35 | 1 | Cadherin-like | 134 |
| PF08271 | 0 | 0 | 4 | 2 | 0 | 0 | 4 | 0 | TFIIB zinc-binding | 10 |
| PF08279 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | HTH domain | 4 |
| PF08285 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Dolichol-phosphate mannosyltransferase subunit 3 (DPM3) | 3 |
| PF08286 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Spc24 subunit of Ndc80 | 2 |
| PF08288 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | PIGA (GPI anchor biosynthesis) | 2 |
| PF08292 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | RNA polymerase III subunit Rpc25 | 1 |
| PF08293 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Mitochondrial ribosomal subunit S27 | 2 |
| PF08294 | 0 | 0 | 1 | 0 | 1 | 1 | 1 | 0 | TIM21 | 4 |
| PF08295 | 0 | 0 | 2 | 0 | 0 | 0 | 3 | 0 | Sin3 family co-repressor | 5 |
| PF08311 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | Mad3/BUB1 homology region 1 | 4 |
| PF08312 | 0 | 0 | 3 | 0 | 0 | 0 | 3 | 0 | cwf21 domain | 6 |
| PF08313 | 0 | 0 | 4 | 1 | 0 | 0 | 5 | 0 | "SCA7, zinc-binding domain" | 10 |
| PF08314 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | 0 | Secretory pathway protein Sec39 | 4 |
| PF08315 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | cwf18 pre-mRNA splicing factor | 2 |
| PF08318 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | COG4 transport protein | 3 |
| PF08320 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | PIG-X / PBN1 | 2 |
| PF08321 | 0 | 1 | 3 | 0 | 0 | 1 | 2 | 1 | PPP5 TPR repeat region | 8 |
| PF08324 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 1 | PUL domain | 3 |
| PF08326 | 0 | 1 | 2 | 0 | 0 | 1 | 2 | 0 | "Acetyl-CoA carboxylase, central region" | 6 |
| PF08327 | 1 | 1 | 2 | 1 | 0 | 0 | 1 | 0 | Activator of Hsp90 ATPase homolog 1-like protein | 6 |
| PF08332 | 4 | 2 | 4 | 3 | 7 | 7 | 6 | 6 | Calcium/calmodulin dependent protein kinase II Association | 39 |
| PF08336 | 0 | 0 | 3 | 1 | 0 | 0 | 5 | 0 | "Prolyl 4-Hydroxylase alpha-subunit, N-terminal region" | 9 |
| PF08337 | 0 | 7 | 9 | 2 | 1 | 8 | 9 | 4 | Plexin cytoplasmic RasGAP domain | 40 |
| PF08338 | 0 | 1 | 1 | 1 | 0 | 1 | 0 | 0 | Domain of unknown function (DUF1731) | 4 |
| PF08344 | 0 | 0 | 8 | 0 | 0 | 0 | 10 | 1 | Transient receptor ion channel II | 19 |
| PF08347 | 0 | 0 | 4 | 0 | 0 | 0 | 5 | 0 | N-terminal CTNNB1 binding | 9 |
| PF08351 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Domain of unknown function (DUF1726) | 3 |
| PF08355 | 1 | 2 | 2 | 0 | 1 | 3 | 1 | 0 | EF hand associated | 10 |
| PF08356 | 1 | 2 | 2 | 0 | 1 | 3 | 1 | 0 | EF hand associated | 10 |
| PF08357 | 0 | 0 | 6 | 1 | 0 | 0 | 5 | 0 | SEFIR domain | 12 |
| PF08365 | 0 | 0 | 1 | 0 | 0 | 0 | 2 | 0 | Insulin-like growth factor II E-peptide | 3 |
| PF08366 | 0 | 3 | 4 | 2 | 1 | 5 | 5 | 0 | LLGL2 | 20 |
| PF08367 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | Peptidase M16C associated | 6 |
| PF08368 | 0 | 0 | 7 | 2 | 0 | 0 | 3 | 0 | "FAST kinase-like protein, subdomain 2" | 12 |
| PF08372 | 0 | 1 | 2 | 0 | 0 | 0 | 5 | 0 | Plant phosphoribosyltransferase C-terminal | 8 |
| PF08373 | 0 | 0 | 6 | 2 | 0 | 0 | 4 | 0 | RAP domain | 12 |
| PF08374 | 0 | 3 | 4 | 1 | 2 | 5 | 5 | 0 | Protocadherin | 20 |
| PF08375 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | Proteasome regulatory subunit C-terminal | 6 |
| PF08377 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | MAP2/Tau projection domain | 6 |
| PF08378 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | Nuclease-related domain | 1 |
| PF08383 | 0 | 0 | 4 | 0 | 0 | 0 | 5 | 0 | Maf N-terminal region | 9 |
| PF08384 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | "Pro-opiomelanocortin, N-terminal region" | 3 |
| PF08385 | 0 | 2 | 9 | 0 | 0 | 1 | 6 | 2 | "Dynein heavy chain, N-terminal region 1" | 20 |
| PF08389 | 0 | 1 | 5 | 1 | 0 | 0 | 6 | 3 | Exportin 1-like protein | 16 |
| PF08390 | 0 | 0 | 3 | 2 | 0 | 0 | 3 | 0 | TRAM1-like protein | 8 |
| PF08391 | 0 | 0 | 12 | 0 | 0 | 0 | 1 | 0 | "Ly49-like protein, N-terminal region" | 13 |
| PF08393 | 0 | 2 | 13 | 1 | 1 | 2 | 13 | 2 | "Dynein heavy chain, N-terminal region 2" | 34 |
| PF08397 | 3 | 3 | 6 | 3 | 4 | 3 | 9 | 2 | IRSp53/MIM homology domain | 33 |
| PF08399 | 3 | 3 | 4 | 2 | 5 | 5 | 8 | 2 | VWA N-terminal | 32 |
| PF08403 | 1 | 1 | 3 | 0 | 0 | 1 | 5 | 0 | Amino acid permease N-terminal | 11 |
| PF08409 | 0 | 0 | 4 | 0 | 1 | 2 | 5 | 0 | Domain of unknown function (DUF1736) | 12 |
| PF08412 | 0 | 4 | 4 | 3 | 0 | 3 | 5 | 1 | Ion transport protein N-terminal | 20 |
| PF08416 | 1 | 1 | 8 | 3 | 0 | 1 | 7 | 0 | Phosphotyrosine-binding domain | 21 |
| PF08418 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 1 | DNA polymerase alpha subunit B N-terminal | 3 |
| PF08423 | 0 | 0 | 7 | 0 | 0 | 0 | 7 | 0 | Rad51 | 14 |
| PF08424 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | "NRDE-2, necessary for RNA interference" | 2 |
| PF08427 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Domain of unknown function (DUF1741) | 1 |
| PF08429 | 0 | 0 | 4 | 0 | 0 | 0 | 3 | 0 | PLU-1-like protein | 7 |
| PF08430 | 0 | 0 | 3 | 0 | 0 | 0 | 2 | 0 | Forkhead N-terminal region | 5 |
| PF08433 | 0 | 0 | 2 | 1 | 0 | 0 | 1 | 0 | Chromatin associated protein KTI12 | 4 |
| PF08434 | 0 | 0 | 7 | 0 | 0 | 0 | 2 | 0 | Calcium-activated chloride channel | 9 |
| PF08441 | 0 | 3 | 18 | 1 | 0 | 2 | 19 | 2 | Integrin alpha | 45 |
| PF08442 | 0 | 2 | 3 | 2 | 0 | 3 | 3 | 2 | ATP-grasp domain | 15 |
| PF08443 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 0 | RimK-like ATP-grasp domain | 3 |
| PF08444 | 0 | 0 | 4 | 0 | 0 | 0 | 0 | 0 | "Aralkyl acyl-CoA:amino acid N-acyltransferase, C-terminal region" | 4 |
| PF08445 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | FR47-like protein | 2 |
| PF08447 | 0 | 0 | 6 | 0 | 0 | 0 | 8 | 0 | PAS fold | 14 |
| PF08449 | 0 | 0 | 6 | 1 | 0 | 0 | 5 | 0 | UAA transporter family | 12 |
| PF08450 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | SMP-30/Gluconolaconase/LRE-like region | 2 |
| PF08451 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | Adenosine/AMP deaminase N-terminal | 2 |
| PF08454 | 4 | 4 | 6 | 0 | 5 | 5 | 8 | 2 | RyR and IP3R Homology associated | 34 |
| PF08457 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | Sfi1 spindle body protein | 1 |
| PF08466 | 0 | 0 | 2 | 1 | 0 | 0 | 3 | 0 | Inward rectifier potassium channel N-terminal | 6 |
| PF08473 | 2 | 2 | 2 | 0 | 2 | 2 | 2 | 1 | Neuronal voltage-dependent calcium channel alpha 2acd | 13 |
| PF08474 | 0 | 0 | 3 | 0 | 0 | 0 | 2 | 0 | Myelin transcription factor 1 | 5 |
| PF08477 | 4 | 6 | 10 | 2 | 4 | 6 | 9 | 0 | Miro-like protein | 41 |
| PF08487 | 1 | 2 | 8 | 2 | 1 | 1 | 9 | 1 | Vault protein inter-alpha-trypsin domain | 25 |
| PF08490 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Domain of unknown function (DUF1744) | 2 |
| PF08491 | 0 | 0 | 1 | 0 | 0 | 0 | 2 | 0 | Squalene epoxidase | 3 |
| PF08492 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | SRP72 RNA-binding domain | 6 |
| PF08499 | 1 | 2 | 3 | 2 | 0 | 0 | 2 | 0 | 3'5'-cyclic nucleotide phosphodiesterase N-terminal | 10 |
| PF08504 | 0 | 0 | 3 | 0 | 0 | 0 | 3 | 0 | Runx inhibition domain | 6 |
| PF08506 | 1 | 2 | 3 | 2 | 0 | 2 | 3 | 2 | Cse1 | 15 |
| PF08510 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | PIG-P | 2 |
| PF08511 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | COQ9 | 6 |
| PF08512 | 0 | 0 | 2 | 0 | 1 | 1 | 3 | 2 | Histone chaperone Rttp106-like | 9 |
| PF08513 | 2 | 1 | 7 | 4 | 2 | 3 | 10 | 2 | LisH | 31 |
| PF08514 | 0 | 0 | 3 | 1 | 1 | 0 | 5 | 0 | STAG domain | 10 |
| PF08515 | 0 | 1 | 7 | 3 | 0 | 0 | 8 | 0 | Transforming growth factor beta type I GS-motif | 19 |
| PF08516 | 2 | 3 | 32 | 2 | 3 | 4 | 12 | 2 | ADAM cysteine-rich | 60 |
| PF08517 | 0 | 0 | 3 | 0 | 0 | 0 | 4 | 0 | Ataxin-1 and HBP1 module (AXH) | 7 |
| PF08518 | 1 | 1 | 2 | 0 | 1 | 1 | 3 | 0 | Spa2 homology domain (SHD) of GIT | 9 |
| PF08519 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Replication factor RFC1 C terminal domain | 2 |
| PF08523 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | Multiprotein bridging factor 1 | 5 |
| PF08524 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | rRNA processing | 1 |
| PF08526 | 0 | 1 | 5 | 0 | 0 | 1 | 1 | 1 | Protein-arginine deiminase (PAD) N-terminal domain | 9 |
| PF08527 | 0 | 1 | 5 | 0 | 0 | 1 | 1 | 1 | Protein-arginine deiminase (PAD) middle domain | 9 |
| PF08534 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | Redoxin | 7 |
| PF08539 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | HbrB-like | 4 |
| PF08540 | 0 | 1 | 2 | 0 | 0 | 0 | 0 | 0 | Hydroxymethylglutaryl-coenzyme A synthase C terminal | 3 |
| PF08542 | 0 | 0 | 4 | 0 | 0 | 0 | 4 | 0 | Replication factor C C-terminal domain | 8 |
| PF08544 | 0 | 0 | 4 | 4 | 0 | 0 | 5 | 1 | GHMP kinases C terminal | 14 |
| PF08547 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Complex I intermediate-associated protein 30 (CIA30) | 3 |
| PF08551 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Eukaryotic integral membrane protein (DUF1751) | 2 |
| PF08555 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Eukaryotic family of unknown function (DUF1754) | 2 |
| PF08557 | 0 | 1 | 3 | 0 | 0 | 0 | 2 | 0 | Sphingolipid Delta4-desaturase (DES) | 6 |
| PF08558 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | Telomere repeat binding factor (TRF) | 4 |
| PF08561 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Mitochondrial ribosomal protein L37 | 3 |
| PF08562 | 0 | 0 | 4 | 0 | 0 | 0 | 0 | 0 | Crisp | 4 |
| PF08563 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | P53 transactivation motif | 1 |
| PF08564 | 0 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | Cdc37 C terminal domain | 4 |
| PF08565 | 0 | 1 | 2 | 2 | 0 | 1 | 1 | 0 | Cdc37 Hsp90 binding domain | 7 |
| PF08567 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | "TFIIH p62 subunit, N-terminal domain" | 2 |
| PF08568 | 0 | 0 | 1 | 1 | 0 | 0 | 2 | 1 | "Uncharacterised protein family, YAP/Alf4/glomulin" | 5 |
| PF08569 | 1 | 1 | 2 | 1 | 0 | 2 | 3 | 2 | Mo25-like | 12 |
| PF08571 | 0 | 1 | 2 | 0 | 0 | 0 | 1 | 1 | Yos1-like | 5 |
| PF08572 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | pre-mRNA processing factor 3 (PRP3) | 3 |
| PF08573 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | DNA repair protein endonuclease SAE2/CtIP C-terminus | 3 |
| PF08574 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Protein of unknown function (DUF1762) | 2 |
| PF08577 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | PI31 proteasome regulator | 2 |
| PF08581 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | Tup N-terminal | 1 |
| PF08583 | 0 | 1 | 2 | 1 | 0 | 1 | 1 | 0 | Cytochrome c oxidase biogenesis protein Cmc1 like | 6 |
| PF08584 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Ribonuclease P 40kDa (Rpp40) subunit | 2 |
| PF08585 | 1 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | Domain of unknown function (DUF1767) | 5 |
| PF08590 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Domain of unknown function (DUF1771) | 2 |
| PF08596 | 0 | 3 | 3 | 2 | 1 | 3 | 3 | 0 | "Lethal giant larvae(Lgl) like, C-terminal" | 15 |
| PF08597 | 0 | 0 | 2 | 0 | 0 | 0 | 3 | 1 | Translation initiation factor eIF3 subunit | 6 |
| PF08598 | 0 | 0 | 3 | 0 | 0 | 0 | 4 | 0 | Sds3-like | 7 |
| PF08599 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | DNA damage repair protein Nbs1 | 2 |
| PF08600 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Rsm1-like | 2 |
| PF08603 | 1 | 2 | 2 | 2 | 0 | 1 | 1 | 1 | Adenylate cyclase associated (CAP) C terminal | 10 |
| PF08604 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Nucleoporin Nup153-like | 1 |
| PF08606 | 0 | 0 | 1 | 0 | 1 | 1 | 1 | 1 | Prp19/Pso4-like | 5 |
| PF08608 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Wyosine base formation | 2 |
| PF08610 | 0 | 0 | 1 | 0 | 1 | 1 | 1 | 0 | Peroxisomal membrane protein (Pex16) | 4 |
| PF08612 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | TATA-binding related factor (TRF) of subunit 20 of Mediator complex | 2 |
| PF08613 | 0 | 0 | 1 | 0 | 0 | 0 | 2 | 0 | Cyclin | 3 |
| PF08614 | 0 | 0 | 2 | 1 | 0 | 0 | 1 | 0 | Autophagy protein 16 (ATG16) | 4 |
| PF08615 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Ribonuclease H2 non-catalytic subunit (Ylr154p-like) | 3 |
| PF08616 | 0 | 0 | 3 | 0 | 0 | 0 | 2 | 0 | Stabilization of polarity axis | 5 |
| PF08617 | 0 | 0 | 1 | 1 | 0 | 1 | 1 | 0 | Kinase binding protein CGI-121 | 4 |
| PF08620 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | "RPAP1-like, C-terminal" | 2 |
| PF08621 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | "RPAP1-like, N-terminal" | 2 |
| PF08623 | 0 | 1 | 2 | 0 | 0 | 2 | 2 | 1 | TATA-binding protein interacting (TIP20) | 8 |
| PF08625 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Utp13 specific WD40 associated domain | 2 |
| PF08626 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 0 | "Transport protein Trs120 or TRAPPC9, TRAPP II complex subunit" | 5 |
| PF08628 | 0 | 1 | 4 | 1 | 0 | 0 | 4 | 0 | Sorting nexin C terminal | 10 |
| PF08629 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | PDE8 phosphodiesterase | 2 |
| PF08631 | 1 | 1 | 1 | 0 | 0 | 0 | 1 | 0 | Meiosis protein SPO22/ZIP4 like | 4 |
| PF08638 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Mediator complex subunit MED14 | 2 |
| PF08640 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | U3 small nucleolar RNA-associated protein 6 | 1 |
| PF08641 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Kinetochore protein Mis14 like | 2 |
| PF08644 | 0 | 0 | 1 | 0 | 1 | 1 | 1 | 1 | FACT complex subunit (SPT16/CDC68) | 5 |
| PF08645 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Polynucleotide kinase 3 phosphatase | 1 |
| PF08646 | 0 | 0 | 2 | 1 | 0 | 0 | 2 | 1 | Replication factor-A C terminal domain | 6 |
| PF08647 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | BRE1 E3 ubiquitin ligase | 1 |
| PF08648 | 0 | 0 | 1 | 0 | 0 | 0 | 2 | 0 | Protein of unknown function (DUF1777) | 3 |
| PF08652 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | RAI1 like PD-(D/E)XK nuclease | 2 |
| PF08658 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Rad54 N terminal | 2 |
| PF08659 | 0 | 1 | 1 | 1 | 0 | 1 | 2 | 0 | KR domain | 6 |
| PF08660 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Oligosaccharide biosynthesis protein Alg14 like | 1 |
| PF08661 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Replication factor A protein 3 | 3 |
| PF08662 | 1 | 0 | 2 | 0 | 0 | 0 | 3 | 1 | Eukaryotic translation initiation factor eIF2A | 7 |
| PF08666 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | SAF domain | 6 |
| PF08669 | 0 | 2 | 5 | 3 | 1 | 3 | 4 | 3 | Glycine cleavage T-protein C-terminal barrel domain | 21 |
| PF08672 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Anaphase promoting complex (APC) subunit 2 | 1 |
| PF08674 | 0 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | Acetylcholinesterase tetramerisation domain | 8 |
| PF08675 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | RNA binding domain | 1 |
| PF08676 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | MutL C terminal dimerisation domain | 4 |
| PF08683 | 2 | 2 | 3 | 0 | 1 | 1 | 6 | 0 | Microtubule-binding calmodulin-regulated spectrin-associated | 15 |
| PF08685 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | GON domain | 2 |
| PF08686 | 0 | 0 | 14 | 2 | 0 | 0 | 10 | 0 | PLAC (protease and lacunin) domain | 26 |
| PF08687 | 0 | 0 | 4 | 0 | 0 | 0 | 4 | 0 | Apx/Shroom domain ASD2 | 8 |
| PF08688 | 0 | 0 | 3 | 0 | 0 | 0 | 3 | 0 | Apx/Shroom domain ASD1 | 6 |
| PF08694 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | Ubiquitin-fold modifier-conjugating enzyme 1 | 6 |
| PF08695 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 0 | Cytochrome oxidase complex assembly protein 1 | 3 |
| PF08696 | 0 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | DNA replication factor Dna2 | 3 |
| PF08698 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Fcf2 pre-rRNA processing | 2 |
| PF08699 | 2 | 1 | 4 | 0 | 2 | 3 | 4 | 2 | Domain of unknown function (DUF1785) | 18 |
| PF08700 | 2 | 2 | 3 | 1 | 2 | 2 | 2 | 0 | Vps51/Vps67 | 14 |
| PF08701 | 0 | 0 | 1 | 0 | 0 | 0 | 2 | 0 | GNL3L/Grn1 putative GTPase | 3 |
| PF08702 | 2 | 0 | 3 | 1 | 3 | 2 | 3 | 3 | Fibrinogen alpha/beta chain family | 17 |
| PF08703 | 1 | 1 | 3 | 0 | 0 | 1 | 2 | 0 | PLC-beta C terminal | 8 |
| PF08704 | 0 | 0 | 1 | 0 | 0 | 0 | 2 | 0 | tRNA methyltransferase complex GCD14 subunit | 3 |
| PF08709 | 4 | 4 | 6 | 0 | 5 | 5 | 8 | 2 | "Inositol 1,4,5-trisphosphate/ryanodine receptor" | 34 |
| PF08711 | 0 | 0 | 9 | 4 | 1 | 1 | 8 | 2 | TFIIS helical bundle-like domain | 25 |
| PF08712 | 0 | 0 | 1 | 1 | 0 | 2 | 2 | 2 | Scaffold protein Nfu/NifU N terminal | 8 |
| PF08718 | 1 | 1 | 4 | 1 | 0 | 0 | 6 | 1 | Glycolipid transfer protein (GLTP) | 14 |
| PF08725 | 0 | 5 | 6 | 1 | 0 | 2 | 8 | 1 | Integrin beta cytoplasmic domain | 23 |
| PF08726 | 4 | 4 | 6 | 2 | 2 | 2 | 5 | 4 | Ca2+ insensitive EF hand | 29 |
| PF08729 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | HPC2 and ubinuclein domain | 4 |
| PF08736 | 4 | 4 | 13 | 1 | 4 | 9 | 17 | 4 | FERM adjacent (FA) | 56 |
| PF08737 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Rgp1 | 2 |
| PF08740 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | BCS1 N terminal | 5 |
| PF08742 | 0 | 0 | 10 | 0 | 0 | 0 | 13 | 0 | C8 domain | 23 |
| PF08743 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Nse4 C-terminal | 2 |
| PF08746 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | RING-like domain | 1 |
| PF08752 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | Coatomer gamma subunit appendage platform subdomain | 7 |
| PF08755 | 0 | 0 | 1 | 1 | 0 | 0 | 2 | 0 | Hemimethylated DNA-binding protein YccV like | 4 |
| PF08758 | 3 | 2 | 7 | 0 | 2 | 2 | 3 | 1 | Cadherin prodomain like | 20 |
| PF08763 | 2 | 4 | 6 | 2 | 3 | 4 | 11 | 0 | Voltage gated calcium channel IQ domain | 32 |
| PF08766 | 0 | 0 | 4 | 1 | 0 | 0 | 5 | 1 | DEK C terminal domain | 11 |
| PF08767 | 0 | 1 | 2 | 0 | 0 | 0 | 3 | 2 | CRM1 C terminal | 8 |
| PF08768 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Domain of unknown function (DUF1794) | 2 |
| PF08771 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | Rapamycin binding domain | 6 |
| PF08772 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Nin one binding (NOB1) Zn-ribbon like | 3 |
| PF08773 | 0 | 0 | 1 | 1 | 0 | 1 | 0 | 0 | Cathepsin C exclusion domain | 3 |
| PF08774 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | VRR-NUC domain | 2 |
| PF08776 | 3 | 2 | 3 | 0 | 5 | 5 | 3 | 0 | VASP tetramerisation domain | 21 |
| PF08777 | 0 | 0 | 2 | 1 | 1 | 1 | 2 | 1 | RNA binding motif | 8 |
| PF08778 | 0 | 0 | 2 | 0 | 0 | 0 | 5 | 0 | HIF-1 alpha C terminal transactivation domain | 7 |
| PF08781 | 0 | 0 | 2 | 0 | 0 | 0 | 3 | 0 | Transcription factor DP | 5 |
| PF08782 | 0 | 0 | 4 | 0 | 0 | 0 | 7 | 0 | c-SKI Smad4 binding domain | 11 |
| PF08783 | 0 | 0 | 1 | 0 | 0 | 0 | 43 | 0 | DWNN domain | 44 |
| PF08784 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Replication protein A C terminal | 2 |
| PF08785 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Ku C terminal domain like | 1 |
| PF08788 | 0 | 0 | 3 | 0 | 0 | 0 | 3 | 0 | NHR2 domain like | 6 |
| PF08790 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | LYAR-type C2HC zinc finger | 1 |
| PF08797 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | HIRAN domain | 2 |
| PF08799 | 0 | 0 | 2 | 1 | 0 | 0 | 2 | 0 | pre-mRNA processing factor 4 (PRP4) like | 5 |
| PF08801 | 0 | 0 | 2 | 1 | 1 | 1 | 2 | 0 | Nup133 N terminal like | 7 |
| PF08806 | 0 | 2 | 2 | 0 | 1 | 1 | 1 | 0 | Sep15/SelM redox domain | 7 |
| PF08815 | 0 | 0 | 3 | 1 | 0 | 0 | 3 | 0 | Nuclear receptor coactivator | 7 |
| PF08824 | 0 | 0 | 3 | 0 | 0 | 0 | 3 | 0 | Serine rich protein interaction domain | 6 |
| PF08825 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | E2 binding domain | 2 |
| PF08826 | 2 | 2 | 4 | 1 | 2 | 3 | 2 | 0 | DMPK coiled coil domain like | 16 |
| PF08831 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Class II MHC-associated invariant chain trimerisation domain | 1 |
| PF08832 | 0 | 0 | 3 | 1 | 0 | 0 | 3 | 0 | Steroid receptor coactivator | 7 |
| PF08833 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | Axin beta-catenin binding domain | 2 |
| PF08839 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | DNA replication factor CDT1 like | 2 |
| PF08840 | 0 | 0 | 9 | 2 | 0 | 1 | 9 | 1 | BAAT / Acyl-CoA thioester hydrolase C terminal | 22 |
| PF08880 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 1 | QLQ | 5 |
| PF08910 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | 0 | Aida N-terminus | 4 |
| PF08911 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | NUP50 (Nucleoporin 50 kDa) | 2 |
| PF08912 | 0 | 1 | 2 | 0 | 0 | 0 | 4 | 1 | Rho Binding | 8 |
| PF08913 | 0 | 2 | 2 | 1 | 1 | 2 | 1 | 1 | Vinculin Binding Site | 10 |
| PF08914 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | Rap1 Myb domain | 2 |
| PF08916 | 0 | 0 | 3 | 0 | 0 | 0 | 3 | 0 | Phenylalanine zipper | 6 |
| PF08917 | 0 | 0 | 1 | 0 | 0 | 0 | 3 | 0 | Transforming growth factor beta receptor 2 ectodomain | 4 |
| PF08919 | 1 | 1 | 2 | 0 | 1 | 1 | 2 | 0 | F-actin binding | 8 |
| PF08920 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 1 | Splicing factor 3B subunit 1 | 3 |
| PF08923 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 0 | Mitogen-activated protein kinase kinase 1 interacting | 5 |
| PF08925 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | Domain of Unknown Function (DUF1907) | 2 |
| PF08926 | 3 | 0 | 4 | 1 | 0 | 0 | 5 | 1 | Domain of unknown function (DUF1908) | 14 |
| PF08934 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 0 | Rb C-terminal domain | 3 |
| PF08938 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | HBS1 N-terminus | 2 |
| PF08939 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Domain of unknown function (DUF1917) | 3 |
| PF08941 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | USP8 interacting | 3 |
| PF08944 | 0 | 0 | 1 | 0 | 0 | 0 | 3 | 0 | "NADPH oxidase subunit p47Phox, C terminal domain" | 4 |
| PF08945 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | "Bcl-x interacting, BH3 domain" | 2 |
| PF08947 | 0 | 0 | 3 | 2 | 0 | 0 | 4 | 0 | BPS (Between PH and SH2) | 9 |
| PF08952 | 0 | 1 | 2 | 0 | 0 | 1 | 1 | 1 | Domain of unknown function (DUF1866) | 6 |
| PF08953 | 4 | 5 | 6 | 6 | 6 | 6 | 8 | 3 | Domain of unknown function (DUF1899) | 44 |
| PF08954 | 4 | 5 | 7 | 7 | 6 | 6 | 9 | 3 | Domain of unknown function (DUF1900) | 47 |
| PF08961 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Domain of unknown function (DUF1875) | 3 |
| PF08969 | 1 | 0 | 3 | 2 | 0 | 1 | 4 | 1 | USP8 dimerisation domain | 12 |
| PF08976 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Domain of unknown function (DUF1880) | 1 |
| PF08983 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | Domain of unknown function (DUF1856) | 4 |
| PF08991 | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 0 | Domain of unknown function (DUF1903) | 3 |
| PF08996 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | DNA Polymerase alpha zinc finger | 2 |
| PF08997 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | "Ubiquinol-cytochrome C reductase complex, 6.4kD protein" | 2 |
| PF08999 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | "Surfactant protein C, N terminal propeptide" | 1 |
| PF09004 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Domain of unknown function (DUF1891) | 1 |
| PF09005 | 0 | 0 | 3 | 1 | 0 | 1 | 3 | 3 | Domain of unknown function (DUF1897) | 11 |
| PF09006 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Lung surfactant protein D coiled-coil trimerisation | 1 |
| PF09007 | 1 | 2 | 2 | 1 | 0 | 1 | 3 | 1 | "EBP50, C-terminal" | 11 |
| PF09011 | 2 | 1 | 8 | 2 | 4 | 5 | 7 | 5 | HMG-box domain | 34 |
| PF09014 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Beta-2-glycoprotein-1 fifth domain | 2 |
| PF09026 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Centromere protein B dimerisation domain | 1 |
| PF09027 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | GTPase binding | 4 |
| PF09029 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | 5-aminolevulinate synthase presequence | 4 |
| PF09030 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | Creb binding | 2 |
| PF09032 | 0 | 1 | 1 | 0 | 0 | 0 | 1 | 0 | "Siah interacting protein, N terminal" | 3 |
| PF09033 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | "DNA Fragmentation factor 45kDa, C terminal domain" | 2 |
| PF09034 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | "TRADD, N-terminal domain" | 2 |
| PF09036 | 1 | 1 | 1 | 0 | 0 | 0 | 3 | 0 | Bcr-Abl oncoprotein oligomerisation domain | 6 |
| PF09038 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Tumour suppressor p53-binding protein-1 Tudor | 2 |
| PF09040 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | "Gastric H+/K+-ATPase, N terminal domain" | 2 |
| PF09041 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Aurora-A binding | 1 |
| PF09042 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | Titin Z | 3 |
| PF09045 | 1 | 1 | 2 | 2 | 0 | 0 | 2 | 0 | L27_2 | 8 |
| PF09047 | 0 | 0 | 1 | 0 | 0 | 0 | 2 | 0 | MEF2 binding | 3 |
| PF09049 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | Stannin transmembrane | 2 |
| PF09050 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | Stannin unstructured linker | 2 |
| PF09051 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | Stannin cytoplasmic | 2 |
| PF09057 | 0 | 1 | 1 | 0 | 0 | 2 | 1 | 0 | Second Mitochondria-derived Activator of Caspases | 5 |
| PF09058 | 2 | 2 | 2 | 2 | 3 | 3 | 2 | 1 | L27_1 | 17 |
| PF09060 | 1 | 0 | 1 | 1 | 0 | 0 | 2 | 0 | L27_N | 5 |
| PF09064 | 0 | 0 | 1 | 1 | 0 | 1 | 1 | 0 | "Thrombomodulin like fifth domain, EGF-like" | 4 |
| PF09066 | 2 | 2 | 3 | 1 | 2 | 2 | 3 | 2 | "Beta2-adaptin appendage, C-terminal sub-domain" | 17 |
| PF09067 | 0 | 0 | 5 | 0 | 0 | 0 | 8 | 0 | "Erythropoietin receptor, ligand binding" | 13 |
| PF09068 | 1 | 3 | 6 | 0 | 2 | 1 | 5 | 1 | EF hand | 19 |
| PF09069 | 1 | 3 | 6 | 0 | 2 | 1 | 5 | 1 | EF-hand | 19 |
| PF09070 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 1 | PFU (PLAA family ubiquitin binding) | 3 |
| PF09072 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Translation machinery associated TMA7 | 2 |
| PF09073 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | BUD22 | 2 |
| PF09079 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | "CDC6, C terminal" | 4 |
| PF09085 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | "Adhesion molecule, immunoglobulin-like" | 1 |
| PF09088 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | MIF4G like | 1 |
| PF09090 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | MIF4G like | 1 |
| PF09103 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | "BRCA2, oligonucleotide/oligosaccharide-binding, domain 1" | 2 |
| PF09104 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | "BRCA2, oligonucleotide/oligosaccharide-binding, domain 3" | 2 |
| PF09110 | 0 | 0 | 2 | 1 | 0 | 0 | 2 | 1 | HAND | 6 |
| PF09111 | 0 | 0 | 2 | 1 | 0 | 0 | 2 | 1 | SLIDE | 6 |
| PF09112 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | "Peptide-N-glycosidase F, N terminal" | 1 |
| PF09113 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | "Peptide-N-glycosidase F, C terminal" | 1 |
| PF09121 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Tower | 2 |
| PF09127 | 0 | 2 | 4 | 0 | 0 | 1 | 4 | 2 | "Leukotriene A4 hydrolase, C-terminal" | 13 |
| PF09128 | 1 | 2 | 3 | 1 | 0 | 0 | 3 | 0 | Regulator of G protein signalling-like domain | 10 |
| PF09133 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | SANTA (SANT Associated) | 2 |
| PF09138 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Urm1 (Ubiquitin related modifier) | 2 |
| PF09139 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | Mitochondrial matrix Mmp37 | 4 |
| PF09140 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | ATPase MipZ | 1 |
| PF09141 | 0 | 2 | 2 | 1 | 1 | 3 | 1 | 1 | "Talin, middle domain" | 11 |
| PF09162 | 0 | 0 | 4 | 0 | 0 | 0 | 2 | 0 | "Tap, RNA-binding" | 6 |
| PF09164 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | "Vitamin D binding protein, domain III" | 1 |
| PF09165 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | "Ubiquinol-cytochrome c reductase 8 kDa, N-terminal" | 7 |
| PF09166 | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 1 | "Biliverdin reductase, catalytic" | 4 |
| PF09169 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | "BRCA2, helical" | 2 |
| PF09170 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | "CST, Suppressor of cdc thirteen homolog, complex subunit STN1" | 3 |
| PF09172 | 0 | 0 | 1 | 0 | 2 | 7 | 12 | 9 | Domain of unknown function (DUF1943) | 31 |
| PF09173 | 1 | 1 | 2 | 1 | 0 | 0 | 1 | 1 | "Initiation factor eIF2 gamma, C terminal" | 7 |
| PF09174 | 0 | 0 | 1 | 0 | 0 | 0 | 2 | 0 | Maf1 regulator | 3 |
| PF09175 | 0 | 0 | 0 | 0 | 2 | 4 | 8 | 6 | Domain of unknown function (DUF1944) | 20 |
| PF09177 | 1 | 1 | 1 | 1 | 0 | 0 | 2 | 0 | "Syntaxin 6, N-terminal" | 6 |
| PF09180 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | "Prolyl-tRNA synthetase, C-terminal" | 7 |
| PF09184 | 0 | 0 | 1 | 1 | 0 | 0 | 2 | 0 | PPP4R2 | 4 |
| PF09191 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | "CD4, extracellular" | 1 |
| PF09193 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | "Cholecystokinin A receptor, N-terminal" | 1 |
| PF09202 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | "Rio2, N-terminal" | 2 |
| PF09229 | 1 | 1 | 2 | 1 | 0 | 0 | 1 | 0 | "Activator of Hsp90 ATPase, N-terminal" | 6 |
| PF09230 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | DNA fragmentation factor 40 kDa | 2 |
| PF09238 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | "Interleukin-4 receptor alpha chain, N-terminal" | 1 |
| PF09240 | 0 | 1 | 11 | 0 | 0 | 0 | 3 | 0 | "Interleukin-6 receptor alpha chain, binding" | 15 |
| PF09243 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Mitochondrial small ribosomal subunit Rsm22 | 1 |
| PF09247 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | TATA box-binding protein binding | 2 |
| PF09252 | 0 | 0 | 11 | 0 | 0 | 0 | 0 | 0 | Allergen Fel d I-B chain | 11 |
| PF09256 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | "BAFF-R, TALL-1 binding" | 1 |
| PF09257 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | "BCMA, TALL-1 binding" | 1 |
| PF09258 | 1 | 0 | 5 | 1 | 2 | 2 | 4 | 0 | Glycosyl transferase family 64 domain | 15 |
| PF09261 | 1 | 1 | 5 | 1 | 1 | 2 | 4 | 0 | "Alpha mannosidase, middle domain" | 15 |
| PF09262 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | "Peroxisome biogenesis factor 1, N-terminal" | 1 |
| PF09263 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | "Peroxisome biogenesis factor 1, N-terminal" | 1 |
| PF09268 | 0 | 1 | 1 | 0 | 0 | 3 | 3 | 2 | "Clathrin, heavy-chain linker" | 10 |
| PF09270 | 0 | 0 | 2 | 1 | 0 | 0 | 3 | 0 | Beta-trefoil DNA-binding domain | 6 |
| PF09271 | 0 | 0 | 2 | 1 | 0 | 0 | 3 | 0 | "LAG1, DNA binding" | 6 |
| PF09272 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | "Hepsin, SRCR" | 2 |
| PF09273 | 0 | 0 | 3 | 0 | 0 | 0 | 3 | 0 | Rubisco LSMT substrate-binding | 6 |
| PF09279 | 4 | 5 | 15 | 2 | 1 | 6 | 16 | 1 | "Phosphoinositide-specific phospholipase C, efhand-like" | 50 |
| PF09280 | 0 | 0 | 2 | 0 | 0 | 1 | 3 | 3 | XPC-binding domain | 9 |
| PF09282 | 1 | 1 | 1 | 0 | 0 | 0 | 1 | 0 | Mago binding | 4 |
| PF09286 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | "Pro-kumamolisin, activation domain" | 3 |
| PF09289 | 1 | 2 | 5 | 1 | 0 | 0 | 7 | 0 | Follistatin/Osteonectin-like EGF domain | 16 |
| PF09292 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | "Endonuclease VIII-like 1, DNA bind" | 2 |
| PF09294 | 1 | 1 | 12 | 0 | 0 | 1 | 10 | 0 | "Interferon-alpha/beta receptor, fibronectin type III" | 25 |
| PF09296 | 0 | 0 | 2 | 1 | 0 | 0 | 1 | 0 | NADH pyrophosphatase-like rudimentary NUDIX domain | 4 |
| PF09297 | 0 | 0 | 2 | 1 | 0 | 0 | 1 | 0 | NADH pyrophosphatase zinc ribbon domain | 4 |
| PF09298 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | Fumarylacetoacetase N-terminal | 2 |
| PF09302 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | XLF (XRCC4-like factor) | 1 |
| PF09303 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | "KCNMB2, ball and chain domain" | 2 |
| PF09305 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | "TACI, cysteine-rich domain" | 1 |
| PF09307 | 0 | 0 | 1 | 0 | 0 | 1 | 2 | 0 | "CLIP, MHC2 interacting" | 4 |
| PF09309 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | "FCP1, C-terminal" | 3 |
| PF09310 | 0 | 0 | 1 | 0 | 0 | 0 | 3 | 0 | "POU domain, class 2, associating factor 1" | 4 |
| PF09311 | 3 | 3 | 6 | 1 | 0 | 2 | 5 | 1 | Rabaptin-like protein | 21 |
| PF09315 | 0 | 0 | 7 | 0 | 0 | 0 | 1 | 0 | Domain of unknown function (DUF1973) | 8 |
| PF09316 | 0 | 0 | 3 | 0 | 0 | 0 | 3 | 1 | "C-myb, C-terminal" | 7 |
| PF09320 | 3 | 2 | 3 | 0 | 1 | 1 | 4 | 0 | Domain of unknown function (DUF1977) | 14 |
| PF09324 | 0 | 1 | 4 | 1 | 0 | 1 | 3 | 0 | Domain of unknown function (DUF1981) | 10 |
| PF09325 | 0 | 4 | 5 | 3 | 1 | 3 | 5 | 3 | Vps5 C terminal like | 24 |
| PF09326 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | Domain of unknown function (DUF1982) | 5 |
| PF09329 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Primase zinc finger | 2 |
| PF09332 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Mcm10 replication factor | 2 |
| PF09333 | 0 | 2 | 5 | 0 | 0 | 1 | 1 | 0 | ATG C terminal domain | 9 |
| PF09334 | 0 | 1 | 2 | 0 | 1 | 2 | 3 | 2 | tRNA synthetases class I (M) | 11 |
| PF09335 | 0 | 0 | 3 | 0 | 0 | 0 | 4 | 0 | SNARE associated Golgi protein | 7 |
| PF09336 | 1 | 1 | 6 | 3 | 0 | 1 | 5 | 2 | Vps4 C terminal oligomerisation domain | 19 |
| PF09340 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | Histone acetyltransferase subunit NuA4 | 2 |
| PF09341 | 0 | 0 | 4 | 1 | 0 | 0 | 0 | 0 | Transcription factor Pcc1 | 5 |
| PF09346 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | SMI1 / KNR4 family (SUKH-1) | 3 |
| PF09349 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | OHCU decarboxylase | 2 |
| PF09350 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Domain of unknown function (DUF1992) | 2 |
| PF09354 | 0 | 0 | 3 | 0 | 0 | 0 | 2 | 0 | HNF3 C-terminal domain | 5 |
| PF09358 | 0 | 1 | 4 | 2 | 0 | 1 | 3 | 1 | Ubiquitin-activating enzyme e1 C-terminal domain | 12 |
| PF09360 | 2 | 2 | 3 | 2 | 1 | 3 | 3 | 3 | Iron-binding zinc finger CDGSH type | 19 |
| PF09368 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Sas10 C-terminal domain | 2 |
| PF09371 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Tex-like protein N-terminal domain | 2 |
| PF09377 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | SBDS protein C-terminal domain | 3 |
| PF09379 | 7 | 9 | 30 | 6 | 7 | 15 | 44 | 9 | FERM N-terminal domain | 127 |
| PF09380 | 6 | 7 | 24 | 4 | 5 | 12 | 33 | 8 | FERM C-terminal PH-like domain | 99 |
| PF09382 | 0 | 0 | 3 | 0 | 0 | 0 | 2 | 0 | RQC domain | 5 |
| PF09384 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | UTP15 C terminal | 2 |
| PF09387 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Mitochondrial RNA binding protein MRP | 3 |
| PF09396 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 1 | Thrombin light chain | 3 |
| PF09398 | 0 | 0 | 2 | 1 | 1 | 0 | 2 | 0 | FOP N terminal dimerisation domain | 6 |
| PF09402 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 0 | Man1-Src1p-C-terminal domain | 3 |
| PF09404 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Eukaryotic protein of unknown function (DUF2003) | 2 |
| PF09405 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | CASC3/Barentsz eIF4AIII binding | 2 |
| PF09409 | 1 | 1 | 3 | 2 | 0 | 1 | 3 | 1 | PUB domain | 12 |
| PF09412 | 0 | 0 | 1 | 0 | 0 | 0 | 4 | 0 | Endoribonuclease XendoU | 5 |
| PF09415 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | CENP-S associating Centromere protein X | 2 |
| PF09416 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | RNA helicase (UPF2 interacting domain) | 7 |
| PF09420 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Ribosome biogenesis protein Nop16 | 3 |
| PF09422 | 1 | 1 | 3 | 2 | 0 | 0 | 3 | 0 | WTX protein | 10 |
| PF09429 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | WW domain binding protein 11 | 1 |
| PF09430 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | Protein of unknown function (DUF2012) | 8 |
| PF09431 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | Protein of unknown function (DUF2013) | 6 |
| PF09439 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | Signal recognition particle receptor beta subunit | 7 |
| PF09440 | 0 | 0 | 1 | 1 | 0 | 0 | 2 | 1 | eIF3 subunit 6 N terminal domain | 5 |
| PF09443 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 0 | Cripto_Frl-1_Cryptic (CFC) | 3 |
| PF09445 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | RNA cap guanine-N2 methyltransferase | 2 |
| PF09446 | 0 | 1 | 2 | 1 | 0 | 1 | 1 | 0 | VMA21-like domain | 6 |
| PF09453 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | HIRA B motif | 2 |
| PF09454 | 1 | 1 | 1 | 1 | 0 | 2 | 2 | 0 | Vps23 core domain | 8 |
| PF09457 | 3 | 3 | 6 | 1 | 1 | 2 | 4 | 0 | FIP domain | 20 |
| PF09465 | 0 | 0 | 1 | 0 | 1 | 0 | 1 | 1 | Lamin-B receptor of TUDOR domain | 4 |
| PF09468 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Ydr279p protein family (RNase H2 complex component) | 2 |
| PF09469 | 0 | 0 | 2 | 0 | 0 | 0 | 3 | 0 | Cordon-bleu ubiquitin-like domain | 5 |
| PF09470 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Telethonin protein | 2 |
| PF09494 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Slx4 endonuclease | 2 |
| PF09495 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Protein of unknown function (DUF2462) | 2 |
| PF09496 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Cenp-O kinetochore centromere component | 2 |
| PF09497 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Transcription mediator complex subunit Med12 | 1 |
| PF09507 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | DNA polymerase subunit Cdc27 | 3 |
| PF09514 | 0 | 0 | 11 | 0 | 0 | 0 | 0 | 0 | SSXRD motif | 11 |
| PF09531 | 0 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | Nucleoporin protein Ndc1-Nup | 4 |
| PF09532 | 0 | 0 | 3 | 3 | 0 | 0 | 3 | 1 | FDF domain | 10 |
| PF09596 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | MamL-1 domain | 4 |
| PF09606 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 0 | ARC105 or Med15 subunit of Mediator complex non-fungal | 3 |
| PF09607 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Brinker DNA-binding domain | 1 |
| PF09631 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Sen15 protein | 2 |
| PF09637 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Med18 protein | 2 |
| PF09649 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Histone chaperone domain CHZ | 2 |
| PF09666 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Sororin protein | 2 |
| PF09667 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Domain of unknown function (DUF2028) | 2 |
| PF09668 | 0 | 0 | 4 | 1 | 0 | 1 | 2 | 0 | Aspartyl protease | 8 |
| PF09696 | 1 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Ctf8 | 3 |
| PF09724 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Uncharacterized conserved protein (DUF2036) | 2 |
| PF09725 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Folate-sensitive fragile site protein Fra10Ac1 | 1 |
| PF09726 | 1 | 0 | 1 | 1 | 0 | 0 | 3 | 0 | Transmembrane protein | 6 |
| PF09727 | 2 | 1 | 4 | 0 | 1 | 1 | 5 | 0 | Cortactin-binding protein-2 | 14 |
| PF09728 | 0 | 0 | 3 | 0 | 0 | 0 | 4 | 0 | Myosin-like coiled-coil protein | 7 |
| PF09730 | 0 | 1 | 2 | 0 | 0 | 0 | 3 | 0 | Microtubule-associated protein Bicaudal-D | 6 |
| PF09731 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 0 | Mitochondrial inner membrane protein | 5 |
| PF09732 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Cactus-binding C-terminus of cactin protein | 2 |
| PF09733 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | VEFS-Box of polycomb protein | 2 |
| PF09734 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | RNA polymerase III transcription factor (TF)IIIC subunit | 3 |
| PF09735 | 1 | 1 | 2 | 1 | 1 | 1 | 2 | 1 | Membrane-associated apoptosis protein | 10 |
| PF09736 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Pre-mRNA-splicing factor of RES complex | 2 |
| PF09737 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | De-etiolated protein 1 Det1 | 1 |
| PF09738 | 0 | 0 | 2 | 1 | 1 | 0 | 3 | 0 | Double stranded RNA binding protein (DUF2051) | 7 |
| PF09739 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Mini-chromosome maintenance replisome factor | 3 |
| PF09740 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Uncharacterized conserved protein (DUF2043) | 1 |
| PF09741 | 1 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Uncharacterized conserved protein (DUF2045) | 3 |
| PF09742 | 0 | 0 | 1 | 1 | 0 | 0 | 2 | 0 | Dyggve-Melchior-Clausen syndrome protein | 4 |
| PF09743 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 0 | Uncharacterized conserved protein (DUF2042) | 5 |
| PF09744 | 1 | 2 | 4 | 2 | 0 | 2 | 5 | 1 | JNK_SAPK-associated protein-1 | 17 |
| PF09745 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Coiled-coil domain-containing protein 55 (DUF2040) | 2 |
| PF09746 | 0 | 1 | 1 | 0 | 0 | 0 | 1 | 0 | Tumour-associated protein | 3 |
| PF09747 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Coiled-coil domain containing protein (DUF2052) | 3 |
| PF09748 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Transcription factor subunit Med10 of Mediator complex | 2 |
| PF09749 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Uncharacterised conserved protein | 2 |
| PF09750 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | Alternative splicing regulator | 4 |
| PF09751 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Nuclear protein Es2 | 2 |
| PF09752 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Uncharacterized conserved protein (DUF2048) | 1 |
| PF09753 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | 0 | Membrane fusion protein Use1 | 4 |
| PF09754 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | PAC2 family | 3 |
| PF09755 | 0 | 0 | 1 | 0 | 0 | 0 | 2 | 0 | Uncharacterized conserved protein H4 (DUF2046) | 3 |
| PF09756 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | DDRGK domain | 3 |
| PF09757 | 0 | 0 | 1 | 0 | 0 | 0 | 2 | 0 | Arb2 domain | 3 |
| PF09758 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Uncharacterised conserved protein | 2 |
| PF09759 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | Spinocerebellar ataxia type 10 protein domain | 5 |
| PF09762 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Coiled-coil domain-containing protein (DUF2037) | 3 |
| PF09763 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | Exocyst complex component Sec3 | 7 |
| PF09764 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | N-terminal glutamine amidase | 2 |
| PF09765 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | WD-repeat region | 2 |
| PF09766 | 0 | 0 | 1 | 1 | 0 | 0 | 2 | 0 | Fms-interacting protein | 4 |
| PF09767 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Predicted membrane protein (DUF2053) | 2 |
| PF09768 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Peptidase M76 family | 2 |
| PF09769 | 1 | 2 | 2 | 2 | 0 | 3 | 2 | 1 | Apolipoprotein O | 13 |
| PF09770 | 1 | 0 | 2 | 0 | 0 | 0 | 2 | 1 | Topoisomerase II-associated protein PAT1 | 6 |
| PF09771 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Transmembrane protein 188 | 2 |
| PF09772 | 0 | 0 | 1 | 0 | 0 | 0 | 3 | 0 | Transmembrane protein 26 | 4 |
| PF09773 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 1 | Meckelin (Transmembrane protein 67) | 3 |
| PF09774 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | Caffeine-induced death protein 2 | 5 |
| PF09775 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Keratinocyte-associated protein 2 | 2 |
| PF09776 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Mitochondrial ribosomal protein L55 | 2 |
| PF09777 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Osteopetrosis-associated transmembrane protein 1 precursor | 2 |
| PF09778 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Guanylylate cyclase | 2 |
| PF09779 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Ima1 N-terminal domain | 2 |
| PF09781 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | "NADH:ubiquinone oxidoreductase, NDUFB5/SGDH subunit" | 8 |
| PF09782 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | "NADH:ubiquinone oxidoreductase, NDUFB6/B17 subunit" | 8 |
| PF09783 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Vacuolar import and degradation protein | 2 |
| PF09785 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Prp31 C terminal domain | 2 |
| PF09786 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | "Cytochrome B561, N terminal" | 2 |
| PF09787 | 0 | 1 | 2 | 1 | 0 | 1 | 1 | 0 | Golgin subfamily A member 5 | 6 |
| PF09788 | 1 | 2 | 2 | 1 | 0 | 1 | 3 | 0 | Transmembrane protein 55A | 10 |
| PF09789 | 0 | 0 | 1 | 0 | 0 | 0 | 2 | 0 | Uncharacterized coiled-coil protein (DUF2353) | 3 |
| PF09790 | 2 | 2 | 2 | 1 | 2 | 2 | 1 | 0 | Hyccin | 12 |
| PF09791 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | "Oxidoreductase-like protein, N-terminal" | 2 |
| PF09793 | 0 | 1 | 1 | 1 | 0 | 0 | 2 | 0 | Anticodon-binding domain | 5 |
| PF09794 | 0 | 0 | 3 | 0 | 0 | 0 | 3 | 1 | Transport protein Avl9 | 7 |
| PF09797 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | N-acetyltransferase B complex (NatB) non catalytic subunit | 3 |
| PF09799 | 0 | 0 | 3 | 0 | 0 | 0 | 3 | 0 | Predicted membrane protein | 6 |
| PF09801 | 0 | 0 | 1 | 0 | 0 | 0 | 2 | 0 | Integral membrane protein S linking to the trans Golgi network | 3 |
| PF09803 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Uncharacterized conserved protein (DUF2346) | 2 |
| PF09804 | 0 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | Uncharacterized conserved protein (DUF2347) | 4 |
| PF09805 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Nucleolar protein 12 (25kDa) | 2 |
| PF09806 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | Cyclin-dependent kinase 2-associated protein | 2 |
| PF09807 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Uncharacterized conserved protein (DUF2348) | 3 |
| PF09808 | 0 | 0 | 1 | 0 | 0 | 0 | 2 | 0 | "Small nuclear RNA activating complex (SNAPc), subunit SNAP43" | 3 |
| PF09809 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Mitochondrial ribosomal protein L27 | 2 |
| PF09810 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Exonuclease V - a 5' deoxyribonuclease | 2 |
| PF09811 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 0 | "Essential protein Yae1, N terminal" | 3 |
| PF09812 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | Mitochondrial ribosomal protein L28 | 7 |
| PF09813 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | Coiled-coil domain-containing protein 56 | 6 |
| PF09814 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | HECT-like Ubiquitin-conjugating enzyme (E2)-binding | 2 |
| PF09815 | 2 | 2 | 8 | 0 | 4 | 4 | 12 | 0 | XK-related protein | 32 |
| PF09816 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | RNA polymerase II transcription elongation factor | 4 |
| PF09817 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Uncharacterized conserved protein (DUF2352) | 2 |
| PF09822 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | ABC-type uncharacterized transport system | 2 |
| PF09848 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | Uncharacterized conserved protein (DUF2075) | 3 |
| PF09992 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Predicted periplasmic protein (DUF2233) | 2 |
| PF10018 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Vitamin-D-receptor interacting Mediator subunit 4 | 2 |
| PF10033 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Autophagy-related protein 13 | 3 |
| PF10034 | 0 | 0 | 4 | 0 | 0 | 0 | 2 | 0 | Q-cell neuroblast polarisation | 6 |
| PF10036 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | Putative carnitine deficiency-associated protein | 6 |
| PF10037 | 1 | 1 | 2 | 0 | 0 | 1 | 1 | 0 | Mitochondrial 28S ribosomal protein S27 | 6 |
| PF10044 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Retinal tissue protein | 2 |
| PF10046 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Biogenesis of lysosome-related organelles complex-1 subunit 2 | 2 |
| PF10058 | 0 | 1 | 1 | 1 | 0 | 1 | 2 | 0 | Predicted integral membrane metal-binding protein (DUF2296) | 6 |
| PF10075 | 1 | 1 | 3 | 1 | 1 | 3 | 3 | 3 | "COP9 signalosome, subunit CSN8" | 16 |
| PF10146 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Zinc finger-containing protein | 3 |
| PF10147 | 1 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Growth arrest and DNA-damage-inducible proteins-interacting protein 1 | 4 |
| PF10148 | 0 | 0 | 1 | 0 | 0 | 0 | 2 | 0 | Schwannomin-interacting protein 1 | 3 |
| PF10149 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Transmembrane protein 231 | 2 |
| PF10151 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | Uncharacterised conserved protein (DUF2359) | 6 |
| PF10152 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Predicted coiled-coil domain-containing protein (DUF2360) | 2 |
| PF10154 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Uncharacterized conserved protein (DUF2362) | 3 |
| PF10155 | 0 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | Uncharacterized conserved protein (DUF2363) | 4 |
| PF10156 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Subunit 17 of Mediator complex | 2 |
| PF10157 | 0 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | Uncharacterized conserved protein (DUF2365) | 4 |
| PF10158 | 0 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | Tumour suppressor protein | 4 |
| PF10159 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Kinase phosphorylation protein | 2 |
| PF10160 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Predicted membrane protein | 2 |
| PF10161 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Putative mitochondrial precursor protein | 2 |
| PF10162 | 0 | 0 | 3 | 0 | 0 | 0 | 3 | 0 | G8 domain | 6 |
| PF10163 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Transcription factor e(y)2 | 3 |
| PF10164 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Uncharacterized conserved protein (DUF2367) | 2 |
| PF10165 | 1 | 1 | 2 | 2 | 0 | 1 | 4 | 1 | Guanine nucleotide exchange factor synembryn | 12 |
| PF10166 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | Uncharacterised conserved protein (DUF2368) | 6 |
| PF10167 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Uncharacterised conserved protein | 2 |
| PF10168 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Nuclear pore component | 3 |
| PF10169 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Learning-associated protein | 2 |
| PF10170 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Cysteine-rich domain | 2 |
| PF10171 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | Uncharacterised conserved protein (DUF2366) | 5 |
| PF10172 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Det1 complexing ubiquitin ligase | 2 |
| PF10174 | 2 | 2 | 2 | 1 | 2 | 2 | 3 | 1 | RIM-binding protein of the cytomatrix active zone | 15 |
| PF10175 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | M-phase phosphoprotein 6 | 2 |
| PF10176 | 0 | 1 | 2 | 0 | 0 | 0 | 2 | 0 | Protein of unknown function (DUF2370) | 5 |
| PF10177 | 0 | 0 | 3 | 0 | 0 | 0 | 1 | 0 | Uncharacterised conserved protein (DUF2371) | 4 |
| PF10178 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Uncharacterised conserved protein (DUF2372) | 1 |
| PF10179 | 0 | 0 | 1 | 0 | 0 | 0 | 2 | 0 | Uncharacterised conserved protein (DUF2369) | 3 |
| PF10180 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Uncharacterised conserved protein (DUF2373) | 2 |
| PF10181 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | "GPI-GlcNAc transferase complex, PIG-H component" | 2 |
| PF10183 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | ESSS subunit of NADH:ubiquinone oxidoreductase (complex I) | 9 |
| PF10184 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | 0 | Uncharacterized conserved protein (DUF2358) | 4 |
| PF10185 | 0 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | Chaperone for wingless signalling and trafficking of LDL receptor | 5 |
| PF10186 | 0 | 0 | 2 | 1 | 0 | 0 | 2 | 0 | UV radiation resistance protein and autophagy-related subunit 14 | 5 |
| PF10187 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | N-terminal domain of NEFA-interacting nuclear protein NIP30 | 2 |
| PF10188 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | Organic solute transport protein 1 | 2 |
| PF10189 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Conserved protein (DUF2356) | 2 |
| PF10190 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | Putative transmembrane protein 170 | 4 |
| PF10191 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Golgi complex component 7 (COG7) | 2 |
| PF10192 | 0 | 1 | 2 | 0 | 0 | 0 | 1 | 0 | Rhodopsin-like GPCR transmembrane domain | 4 |
| PF10193 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Telomere length regulation protein | 2 |
| PF10195 | 0 | 0 | 2 | 1 | 0 | 0 | 1 | 0 | DNA-binding nuclear phosphoprotein p8 | 4 |
| PF10197 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | N-terminal domain of CBF1 interacting co-repressor CIR | 4 |
| PF10198 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 1 | Histone acetyltransferases subunit 3 | 3 |
| PF10199 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Alpha and gamma adaptin binding protein p34 | 2 |
| PF10200 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | "NADH:ubiquinone oxidoreductase, NDUFS5-15kDa" | 7 |
| PF10203 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Cytochrome c oxidase assembly protein PET191 | 3 |
| PF10204 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 0 | Dual oxidase maturation factor | 3 |
| PF10205 | 0 | 1 | 1 | 0 | 1 | 1 | 0 | 0 | Predicted coiled-coil domain-containing protein | 4 |
| PF10206 | 0 | 1 | 1 | 0 | 0 | 1 | 2 | 1 | "Mitochondrial F1F0-ATP synthase, subunit f" | 6 |
| PF10208 | 0 | 1 | 2 | 0 | 0 | 0 | 2 | 1 | Degradation arginine-rich protein for mis-folding | 6 |
| PF10209 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Uncharacterized conserved protein (DUF2340) | 2 |
| PF10210 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Mitochondrial 28S ribosomal protein S32 | 3 |
| PF10211 | 0 | 0 | 1 | 0 | 0 | 0 | 2 | 0 | Axonemal dynein light chain | 3 |
| PF10212 | 0 | 1 | 1 | 0 | 1 | 1 | 0 | 0 | Predicted coiled-coil domain-containing protein | 4 |
| PF10213 | 0 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | Mitochondrial ribosomal subunit protein | 4 |
| PF10215 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Oligosaccaryltransferase | 2 |
| PF10217 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Uncharacterized conserved protein (DUF2039) | 2 |
| PF10218 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Uncharacterized conserved protein (DUF2054) | 2 |
| PF10220 | 0 | 0 | 2 | 2 | 0 | 0 | 2 | 0 | Uncharacterized conserved protein (DUF2146) | 6 |
| PF10221 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Cell cycle and development regulator | 2 |
| PF10222 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Uncharacterized conserved protein (DUF2152) | 2 |
| PF10223 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | Uncharacterized conserved protein (DUF2181) | 4 |
| PF10224 | 0 | 0 | 1 | 0 | 0 | 0 | 2 | 0 | Predicted coiled-coil protein (DUF2205) | 3 |
| PF10225 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | Uncharacterized conserved protein (DUF2215) | 4 |
| PF10226 | 1 | 0 | 3 | 0 | 0 | 0 | 5 | 0 | Uncharacterized conserved proteins (DUF2216) | 9 |
| PF10228 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Uncharacterised conserved protein (DUF2228) | 3 |
| PF10229 | 0 | 0 | 1 | 0 | 0 | 0 | 2 | 0 | Uncharacterized conserved protein (DUF2246) | 3 |
| PF10230 | 0 | 1 | 1 | 0 | 0 | 0 | 1 | 0 | Uncharacterised conserved protein (DUF2305) | 3 |
| PF10231 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Uncharacterised conserved protein (DUF2315) | 2 |
| PF10232 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Mediator of RNA polymerase II transcription complex subunit 8 | 1 |
| PF10233 | 0 | 1 | 1 | 0 | 0 | 0 | 1 | 0 | Uncharacterized conserved protein CG6151-P | 3 |
| PF10234 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Clusterin-associated protein-1 | 2 |
| PF10235 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Microtubule-associated protein CRIPT | 1 |
| PF10236 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Mitochondrial ribosomal death-associated protein 3 | 3 |
| PF10237 | 0 | 0 | 2 | 1 | 0 | 0 | 1 | 0 | Probable N6-adenine methyltransferase | 4 |
| PF10238 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | E2F-associated phosphoprotein | 2 |
| PF10239 | 2 | 1 | 3 | 1 | 0 | 0 | 3 | 1 | Protein of unknown function (DUF2465) | 11 |
| PF10240 | 1 | 1 | 2 | 2 | 0 | 0 | 2 | 0 | Protein of unknown function (DUF2464) | 8 |
| PF10241 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Uncharacterized conserved protein | 2 |
| PF10242 | 2 | 1 | 6 | 1 | 3 | 2 | 8 | 0 | Lipoma HMGIC fusion partner-like protein | 23 |
| PF10243 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Microtubule-binding protein MIP-T3 | 2 |
| PF10244 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Mitochondrial ribosomal subunit | 3 |
| PF10245 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | Mitochondrial 28S ribosomal protein S22 | 5 |
| PF10246 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | Mitochondrial ribosomal protein MRP-S35 | 5 |
| PF10247 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | Reactive mitochondrial oxygen species modulator 1 | 4 |
| PF10248 | 1 | 1 | 2 | 1 | 1 | 1 | 2 | 0 | Myelodysplasia-myeloid leukemia factor 1-interacting protein | 9 |
| PF10249 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | NADH-ubiquinone oxidoreductase subunit 10 | 7 |
| PF10250 | 0 | 0 | 2 | 2 | 0 | 0 | 2 | 0 | GDP-fucose protein O-fucosyltransferase | 6 |
| PF10251 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Presenilin enhancer-2 subunit of gamma secretase | 2 |
| PF10252 | 1 | 1 | 1 | 1 | 0 | 0 | 2 | 0 | Casein kinase substrate phosphoprotein PP28 | 6 |
| PF10253 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | "Mitotic checkpoint regulator, MAD2B-interacting" | 2 |
| PF10254 | 1 | 1 | 2 | 1 | 0 | 2 | 2 | 0 | PACS-1 cytosolic sorting protein | 9 |
| PF10255 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 0 | RNA polymerase I-associated factor PAF67 | 5 |
| PF10256 | 2 | 2 | 4 | 1 | 3 | 3 | 4 | 0 | Golgin subfamily A member 7/ERF4 family | 19 |
| PF10257 | 0 | 0 | 4 | 0 | 0 | 0 | 4 | 0 | Retinoic acid induced 16-like protein | 8 |
| PF10258 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | PHAX RNA-binding domain | 2 |
| PF10259 | 1 | 1 | 0 | 0 | 1 | 1 | 0 | 0 | Rogdi leucine zipper containing protein | 4 |
| PF10260 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Uncharacterized conserved domain (SAYSvFN) | 3 |
| PF10261 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | Inositol phospholipid synthesis and fat-storage-inducing TM | 4 |
| PF10262 | 0 | 1 | 5 | 1 | 0 | 3 | 7 | 1 | Rdx family | 18 |
| PF10263 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | SprT-like family | 1 |
| PF10264 | 0 | 0 | 2 | 0 | 0 | 0 | 3 | 0 | Winged helix Storkhead-box1 domain | 5 |
| PF10265 | 1 | 2 | 2 | 1 | 0 | 0 | 2 | 0 | Uncharacterized conserved protein (DUF2217) | 8 |
| PF10266 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | Hereditary spastic paraplegia protein strumpellin | 4 |
| PF10267 | 2 | 3 | 5 | 2 | 0 | 0 | 4 | 0 | Predicted transmembrane and coiled-coil 2 protein | 16 |
| PF10268 | 0 | 0 | 2 | 1 | 0 | 0 | 2 | 0 | Predicted transmembrane protein 161AB | 5 |
| PF10269 | 0 | 0 | 4 | 0 | 0 | 0 | 1 | 0 | Transmembrane Fragile-X-F protein | 5 |
| PF10270 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 0 | Membrane magnesium transporter | 8 |
| PF10271 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 0 | Putative transmembrane protein | 3 |
| PF10272 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Putative transmembrane protein precursor | 2 |
| PF10273 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Pre-rRNA-processing protein TSR2 | 3 |
| PF10274 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 0 | Parkin co-regulated protein | 3 |
| PF10275 | 1 | 1 | 3 | 1 | 0 | 1 | 2 | 2 | Peptidase C65 Otubain | 11 |
| PF10276 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | Zinc-finger domain | 7 |
| PF10277 | 0 | 0 | 6 | 0 | 0 | 1 | 10 | 0 | Frag1/DRAM/Sfk1 family | 17 |
| PF10278 | 0 | 0 | 1 | 1 | 0 | 0 | 2 | 0 | Mediator of RNA pol II transcription subunit 19 | 4 |
| PF10280 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Mediator complex protein | 2 |
| PF10283 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Zinc-finger (CX5CX6HX5H) motif | 2 |
| PF10288 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Protein of unknown function (DUF2392) | 3 |
| PF10291 | 2 | 1 | 3 | 0 | 1 | 1 | 3 | 0 | Muniscin C-terminal mu homology domain | 11 |
| PF10293 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | Domain of unknown function (DUF2405) | 2 |
| PF10294 | 0 | 0 | 9 | 0 | 0 | 0 | 8 | 0 | Putative methyltransferase | 17 |
| PF10296 | 0 | 1 | 1 | 0 | 0 | 1 | 3 | 0 | Putative integral membrane protein conserved region (DUF2404) | 6 |
| PF10300 | 0 | 0 | 4 | 1 | 0 | 0 | 2 | 0 | Protein of unknown function (DUF3808) | 7 |
| PF10304 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Domain of unknown function (DUF2411) | 2 |
| PF10309 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Protein of unknown function (DUF2414) | 2 |
| PF10312 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Conserved mid region of cactin | 2 |
| PF10320 | 0 | 0 | 38 | 0 | 0 | 0 | 0 | 0 | Serpentine type 7TM GPCR chemoreceptor Srsx | 38 |
| PF10341 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | "Shelterin complex subunit, TPP1/ACD" | 2 |
| PF10343 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Protein of unknown function (DUF2419) | 2 |
| PF10344 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | 0 | Mitochondrial protein from FMP27 | 4 |
| PF10345 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Cohesin loading factor | 2 |
| PF10347 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | 0 | RNA pol II promoter Fmp27 protein domain | 4 |
| PF10349 | 0 | 1 | 2 | 0 | 0 | 0 | 2 | 0 | WW-domain ligand protein | 5 |
| PF10350 | 0 | 0 | 1 | 0 | 0 | 0 | 2 | 0 | Putative death-receptor fusion protein (DUF2428) | 3 |
| PF10351 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | 0 | Golgi-body localisation protein domain | 4 |
| PF10354 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Domain of unknown function (DUF2431) | 2 |
| PF10357 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Domain of Kin17 curved DNA-binding protein | 2 |
| PF10358 | 0 | 1 | 4 | 0 | 0 | 0 | 5 | 0 | N-terminal C2 in EEIG1 and EHBP1 proteins | 10 |
| PF10359 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | RNA pol II promoter Fmp27 protein domain | 2 |
| PF10363 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 0 | Protein of unknown function (DUF2435) | 3 |
| PF10366 | 1 | 1 | 2 | 2 | 0 | 0 | 3 | 0 | Vacuolar sorting protein 39 domain 1 | 9 |
| PF10367 | 1 | 1 | 2 | 2 | 0 | 0 | 4 | 0 | Vacuolar sorting protein 39 domain 2 | 10 |
| PF10373 | 0 | 0 | 3 | 1 | 0 | 0 | 2 | 0 | Est1 DNA/RNA binding domain | 6 |
| PF10374 | 0 | 0 | 3 | 1 | 0 | 0 | 2 | 0 | Telomerase activating protein Est1 | 6 |
| PF10376 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Double-strand recombination repair protein | 3 |
| PF10377 | 0 | 1 | 1 | 0 | 0 | 0 | 1 | 0 | Autophagy-related protein 11 | 3 |
| PF10381 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | Autophagocytosis associated protein C-terminal | 6 |
| PF10382 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Protein of unknown function (DUF2439) | 1 |
| PF10384 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Centromere protein Scm3 | 1 |
| PF10390 | 0 | 0 | 3 | 0 | 0 | 0 | 3 | 0 | RNA polymerase II elongation factor ELL | 6 |
| PF10391 | 0 | 0 | 4 | 0 | 0 | 0 | 3 | 0 | Fingers domain of DNA polymerase lambda | 7 |
| PF10392 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Golgi transport complex subunit 5 | 2 |
| PF10393 | 1 | 0 | 4 | 0 | 1 | 0 | 4 | 1 | Trimeric coiled-coil oligomerisation domain of matrilin | 11 |
| PF10394 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Histone acetyl transferase HAT1 N-terminus | 2 |
| PF10396 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | GTP-binding protein TrmE N-terminus | 3 |
| PF10397 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | Adenylosuccinate lyase C-terminus | 6 |
| PF10401 | 0 | 0 | 7 | 1 | 0 | 0 | 8 | 0 | Interferon-regulatory factor 3 | 16 |
| PF10403 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Rad4 beta-hairpin domain 1 | 2 |
| PF10404 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Rad4 beta-hairpin domain 2 | 2 |
| PF10405 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Rad4 beta-hairpin domain 3 | 2 |
| PF10406 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Transcription factor TFIID complex subunit 8 C-term | 2 |
| PF10408 | 0 | 1 | 2 | 1 | 0 | 1 | 2 | 0 | Ubiquitin elongating factor core | 7 |
| PF10409 | 1 | 2 | 7 | 2 | 1 | 3 | 11 | 1 | C2 domain of PTEN tumour-suppressor protein | 28 |
| PF10413 | 0 | 0 | 1 | 0 | 0 | 0 | 6 | 0 | Amino terminal of the G-protein receptor rhodopsin | 7 |
| PF10415 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | Fumarase C C-terminus | 5 |
| PF10417 | 3 | 5 | 6 | 5 | 3 | 5 | 5 | 5 | C-terminal domain of 1-Cys peroxiredoxin | 37 |
| PF10419 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | TFIIIC subunit | 2 |
| PF10420 | 0 | 0 | 1 | 0 | 0 | 0 | 3 | 0 | Cytokine interleukin-12p40 C-terminus | 4 |
| PF10421 | 0 | 0 | 11 | 0 | 0 | 0 | 0 | 0 | "2'-5'-oligoadenylate synthetase 1, domain 2, C-terminus" | 11 |
| PF10426 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Recombination-activating protein 1 zinc-finger domain | 1 |
| PF10427 | 0 | 0 | 3 | 0 | 0 | 0 | 3 | 0 | Argonaute hook | 6 |
| PF10430 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Tie-2 Ig-like domain 1 | 2 |
| PF10431 | 1 | 1 | 2 | 1 | 0 | 0 | 3 | 0 | "C-terminal, D2-small domain, of ClpB protein" | 8 |
| PF10433 | 1 | 1 | 3 | 2 | 1 | 1 | 1 | 0 | Mono-functional DNA-alkylating methyl methanesulfonate N-term | 10 |
| PF10436 | 2 | 4 | 5 | 2 | 3 | 3 | 6 | 3 | Mitochondrial branched-chain alpha-ketoacid dehydrogenase kinase | 28 |
| PF10441 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Urb2/Npa2 family | 2 |
| PF10442 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | FIST C domain | 3 |
| PF10444 | 0 | 0 | 1 | 0 | 0 | 0 | 2 | 0 | Nbl1 / Borealin N terminal | 3 |
| PF10447 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Exosome component EXOSC1/CSL4 | 2 |
| PF10451 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | Telomere regulation protein Stn1 | 2 |
| PF10453 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Nuclear fragile X mental retardation-interacting protein 1 (NUFIP1) | 1 |
| PF10456 | 0 | 0 | 3 | 1 | 0 | 0 | 5 | 0 | WASP-binding domain of Sorting nexin protein | 9 |
| PF10457 | 0 | 1 | 2 | 1 | 0 | 1 | 2 | 0 | Cholesterol-capturing domain | 7 |
| PF10461 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Peptidase S68 | 1 |
| PF10469 | 0 | 1 | 3 | 1 | 0 | 1 | 3 | 0 | AKAP7 2'5' RNA ligase-like domain | 9 |
| PF10470 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | 0 | PKA-RI-RII subunit binding domain of A-kinase anchor protein | 4 |
| PF10471 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Anaphase-promoting complex APC subunit 1 | 2 |
| PF10472 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | eIF2-alpha phosphatase phosphorylation constitutive repressor | 1 |
| PF10473 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | Leucine-rich repeats of kinetochore protein Cenp-F/LEK1 | 5 |
| PF10474 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | Protein of unknown function C-terminus (DUF2451) | 5 |
| PF10475 | 0 | 1 | 2 | 0 | 0 | 1 | 2 | 1 | Protein of unknown function N-terminal domain (DUF2450) | 7 |
| PF10476 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Protein of unknown function C-terminus (DUF2448) | 2 |
| PF10477 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 1 | Nucleocytoplasmic shuttling protein for mRNA cap-binding EIF4E | 3 |
| PF10479 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | 0 | Fragile site-associated protein C-terminus | 4 |
| PF10480 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Beta-1 integrin binding protein | 2 |
| PF10481 | 0 | 1 | 1 | 0 | 1 | 1 | 2 | 0 | Cenp-F N-terminal domain | 6 |
| PF10482 | 0 | 0 | 2 | 1 | 0 | 0 | 2 | 0 | Tumour-suppressor protein CtIP N-terminal domain | 5 |
| PF10483 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Elongator subunit Iki1 | 3 |
| PF10484 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | Mitochondrial ribosomal protein S23 | 5 |
| PF10486 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 0 | Phosphoinositide 3-kinase gamma adapter protein p101 subunit | 3 |
| PF10487 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Nucleoporin subcomplex protein binding to Pom34 | 3 |
| PF10488 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | Phosphatase-1 catalytic subunit binding region | 4 |
| PF10490 | 0 | 1 | 1 | 0 | 1 | 1 | 2 | 0 | Rb-binding domain of kinetochore protein Cenp-F/LEK1 | 6 |
| PF10491 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | NLS-binding and DNA-binding and dimerisation domains of Nrf1 | 3 |
| PF10492 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Nrf1 activator activation site binding domain | 3 |
| PF10493 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Rough deal protein C-terminal region | 1 |
| PF10494 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Serine-threonine protein kinase 19 | 3 |
| PF10495 | 0 | 0 | 2 | 0 | 1 | 2 | 2 | 1 | Pericentrin-AKAP-450 domain of centrosomal targeting protein | 8 |
| PF10496 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | SNARE-complex protein Syntaxin-18 N-terminus | 7 |
| PF10497 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | Zinc-finger domain of monoamine-oxidase A repressor R1 | 4 |
| PF10498 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Intra-flagellar transport protein 57 | 3 |
| PF10500 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Nuclear RNA-splicing-associated protein | 1 |
| PF10501 | 1 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | Ribosomal subunit 39S | 3 |
| PF10502 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | "Signal peptidase, peptidase S26" | 1 |
| PF10504 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Protein of unknown function (DUF2452) | 3 |
| PF10505 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | NMDA receptor-regulated gene protein 2 C-terminus | 2 |
| PF10506 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 0 | PDZ domain of MCC-2 bdg protein for Usher syndrome | 3 |
| PF10507 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | Protein of unknown function (DUF2453) | 6 |
| PF10508 | 0 | 1 | 1 | 0 | 0 | 0 | 1 | 0 | Proteasome non-ATPase 26S subunit | 3 |
| PF10509 | 0 | 0 | 2 | 2 | 0 | 0 | 2 | 1 | Galactokinase galactose-binding signature | 7 |
| PF10510 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | Phosphatidylinositol-glycan biosynthesis class S protein | 6 |
| PF10512 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Cell division cycle-associated protein 8 | 2 |
| PF10513 | 0 | 0 | 8 | 1 | 0 | 0 | 9 | 1 | Enhancer of polycomb-like | 19 |
| PF10514 | 0 | 0 | 1 | 1 | 0 | 0 | 2 | 0 | "Pro-apoptotic Bcl-2 protein, BAD" | 4 |
| PF10515 | 0 | 3 | 3 | 2 | 0 | 4 | 2 | 0 | beta-amyloid precursor protein C-terminus | 14 |
| PF10516 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | SHNi-TPR | 4 |
| PF10520 | 0 | 0 | 1 | 0 | 0 | 0 | 2 | 0 | Kua-ubiquitin conjugating enzyme hybrid localisation domain | 3 |
| PF10521 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Protein of unknown function (DUF2454) | 2 |
| PF10522 | 1 | 3 | 6 | 1 | 0 | 0 | 0 | 0 | RII binding domain | 11 |
| PF10523 | 0 | 0 | 8 | 0 | 0 | 0 | 10 | 0 | BEN domain | 18 |
| PF10524 | 0 | 0 | 4 | 3 | 0 | 0 | 4 | 0 | Nuclear factor I protein pre-N-terminus | 11 |
| PF10525 | 0 | 0 | 2 | 1 | 0 | 0 | 3 | 0 | Engrailed homeobox C-terminal signature domain | 6 |
| PF10529 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Histidine-rich Calcium-binding repeat region | 1 |
| PF10531 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | SLBB domain | 6 |
| PF10534 | 1 | 1 | 3 | 1 | 3 | 2 | 4 | 1 | Connector enhancer of kinase suppressor of ras | 16 |
| PF10537 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | ATP-utilising chromatin assembly and remodelling N-terminal | 2 |
| PF10540 | 4 | 1 | 5 | 0 | 2 | 3 | 4 | 1 | Munc13 (mammalian uncoordinated) homology domain | 20 |
| PF10541 | 1 | 1 | 4 | 0 | 2 | 2 | 6 | 0 | Nuclear envelope localisation domain | 16 |
| PF10545 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | Alcohol dehydrogenase transcription factor Myb/SANT-like | 2 |
| PF10551 | 0 | 0 | 1 | 0 | 0 | 0 | 3 | 0 | MULE transposase domain | 4 |
| PF10557 | 4 | 5 | 6 | 4 | 3 | 3 | 7 | 5 | Cullin protein neddylation domain | 37 |
| PF10558 | 0 | 1 | 1 | 1 | 0 | 1 | 0 | 0 | Mitochondrial 18 KDa protein (MTP18) | 4 |
| PF10559 | 1 | 0 | 2 | 1 | 0 | 1 | 2 | 1 | Plug domain of Sec61p | 8 |
| PF10561 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Uncharacterised protein family UPF0565 | 2 |
| PF10562 | 1 | 0 | 1 | 1 | 2 | 2 | 2 | 0 | Calmodulin-binding domain C0 of NMDA receptor NR1 subunit | 9 |
| PF10565 | 3 | 2 | 3 | 0 | 6 | 4 | 6 | 0 | N-methyl D-aspartate receptor 2B3 C-terminus | 24 |
| PF10568 | 3 | 3 | 3 | 1 | 0 | 3 | 3 | 2 | Outer mitochondrial membrane transport complex protein | 18 |
| PF10569 | 1 | 1 | 8 | 0 | 1 | 4 | 23 | 6 | Alpha-macro-globulin thiol-ester bond-forming region | 44 |
| PF10570 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Myelin-PO cytoplasmic C-term p65 binding region | 1 |
| PF10571 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Uncharacterised protein family UPF0547 | 2 |
| PF10572 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | Uncharacterised protein family UPF0556 | 5 |
| PF10573 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Uncharacterised protein family UPF0561 | 2 |
| PF10574 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | Uncharacterised protein family UPF0552 | 4 |
| PF10576 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Iron-sulfur binding domain of endonuclease III | 1 |
| PF10577 | 3 | 3 | 4 | 2 | 0 | 3 | 5 | 1 | Uncharacterised protein family UPF0560 | 21 |
| PF10578 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Seminal vesicle protein repeat | 1 |
| PF10579 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Rapsyn N-terminal myristoylation and linker region | 2 |
| PF10580 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | Gap junction protein N-terminal region | 6 |
| PF10581 | 3 | 3 | 3 | 2 | 3 | 3 | 2 | 2 | Synapsin N-terminal | 21 |
| PF10582 | 3 | 4 | 20 | 3 | 5 | 2 | 40 | 1 | Gap junction channel protein cysteine-rich domain | 78 |
| PF10583 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Involucrin of squamous epithelia N-terminus | 1 |
| PF10584 | 5 | 7 | 8 | 4 | 0 | 8 | 9 | 8 | Proteasome subunit A N-terminal signature | 49 |
| PF10585 | 0 | 1 | 6 | 3 | 0 | 1 | 4 | 1 | Ubiquitin-activating enzyme active site | 16 |
| PF10587 | 1 | 2 | 2 | 2 | 0 | 2 | 2 | 1 | Eukaryotic elongation factor 1 beta central acidic region | 12 |
| PF10588 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | NADH-ubiquinone oxidoreductase-G iron-sulfur binding region | 5 |
| PF10589 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | NADH-ubiquinone oxidoreductase-F iron-sulfur binding region | 6 |
| PF10590 | 0 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | Pyridoxine 5'-phosphate oxidase C-terminal dimerisation region | 5 |
| PF10591 | 1 | 3 | 7 | 3 | 0 | 0 | 7 | 0 | Secreted protein acidic and rich in cysteine Ca binding region | 21 |
| PF10595 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | Uncharacterised protein family UPF0564 | 4 |
| PF10596 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 1 | U6-snRNA interacting domain of PrP8 | 3 |
| PF10597 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 1 | U5-snRNA binding site 2 of PrP8 | 3 |
| PF10598 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 1 | RNA recognition motif of the spliceosomal PrP8 | 3 |
| PF10599 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Retro-transposon transporting motif | 1 |
| PF10600 | 4 | 3 | 4 | 3 | 5 | 5 | 4 | 2 | PDZ-associated domain of NMDA receptors | 30 |
| PF10601 | 0 | 1 | 2 | 0 | 0 | 0 | 7 | 0 | LITAF-like zinc ribbon domain | 10 |
| PF10602 | 1 | 2 | 2 | 2 | 0 | 2 | 2 | 1 | 26S proteasome subunit RPN7 | 12 |
| PF10606 | 1 | 2 | 2 | 1 | 2 | 4 | 2 | 0 | Homer-binding domain of metabotropic glutamate receptor | 14 |
| PF10607 | 1 | 0 | 6 | 3 | 1 | 0 | 6 | 1 | CTLH/CRA C-terminal to LisH motif domain | 18 |
| PF10608 | 4 | 3 | 4 | 3 | 5 | 5 | 3 | 2 | Polyubiquitination (PEST) N-terminal domain of MAGUK | 29 |
| PF10609 | 0 | 0 | 3 | 2 | 0 | 0 | 3 | 0 | ParA/MinD ATPase like | 8 |
| PF10613 | 7 | 6 | 8 | 3 | 4 | 6 | 10 | 3 | Ligated ion channel L-glutamate- and glycine-binding site | 47 |
| PF10629 | 0 | 0 | 3 | 0 | 0 | 0 | 1 | 0 | Protein of unknown function (DUF2475) | 4 |
| PF10630 | 0 | 0 | 4 | 0 | 0 | 0 | 0 | 0 | Protein of unknown function (DUF2476) | 4 |
| PF10631 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Protein of unknown function (DUF2477) | 1 |
| PF10637 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Oxoglutarate and iron-dependent oxygenase degradation C-term | 3 |
| PF10639 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Uncharacterised protein family UPF0546 | 2 |
| PF10644 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Misato Segment II tubulin-like domain | 2 |
| PF10650 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Putative zinc-finger domain | 1 |
| PF10660 | 1 | 2 | 2 | 2 | 1 | 1 | 1 | 1 | Iron-containing outer mitochondrial membrane protein N-terminus | 11 |
| PF10744 | 0 | 0 | 1 | 0 | 0 | 0 | 2 | 0 | Mediator of RNA polymerase II transcription subunit 1 | 3 |
| PF10780 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | 39S ribosomal protein L53/MRP-L53 | 5 |
| PF10856 | 0 | 0 | 1 | 0 | 0 | 0 | 2 | 0 | Protein of unknown function (DUF2678) | 3 |
| PF10873 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Protein of unknown function (DUF2668) | 1 |
| PF10914 | 0 | 0 | 3 | 0 | 0 | 0 | 2 | 0 | Protein of unknown function (DUF2781) | 5 |
| PF10937 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | Protein of unknown function (DUF2638) | 4 |
| PF10961 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | Protein of unknown function (DUF2763) | 4 |
| PF10996 | 0 | 0 | 4 | 3 | 0 | 0 | 4 | 0 | Beta-Casp domain | 11 |
| PF11002 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | RFPL defining motif (RDM) | 1 |
| PF11027 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Protein of unknown function (DUF2615) | 2 |
| PF11028 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Protein of unknown function (DUF2723) | 2 |
| PF11029 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | DAZ associated protein 2 (DAZAP2) | 2 |
| PF11032 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 1 | Apolipoprotein M (ApoM) | 3 |
| PF11035 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Small nuclear RNA activating complex subunit 2-like | 2 |
| PF11037 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Insulin-resistance promoting peptide in skeletal muscle | 2 |
| PF11057 | 0 | 0 | 3 | 0 | 0 | 0 | 1 | 0 | Cortexin of kidney | 4 |
| PF11069 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Protein of unknown function (DUF2870) | 2 |
| PF11092 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Neuronal protein 3.1 (p311) | 1 |
| PF11095 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Gem-associated protein 7 (Gemin7) | 2 |
| PF11107 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Fanconi anemia group F protein (FANCF) | 2 |
| PF11109 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Orexigenic neuropeptide Qrfp/P518 | 1 |
| PF11111 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Centromere protein M (CENP-M) | 2 |
| PF11176 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Protein of unknown function (DUF2962) | 3 |
| PF11182 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | Alginate O-acetyl transferase AlgF | 1 |
| PF11221 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Subunit 21 of Mediator complex | 2 |
| PF11229 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Protein of unknown function (DUF3028) | 2 |
| PF11232 | 0 | 0 | 2 | 1 | 0 | 0 | 1 | 0 | Mediator complex subunit 25 PTOV activation and synapsin 2 | 4 |
| PF11235 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Mediator complex subunit 25 synapsin 1 | 3 |
| PF11244 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | Mediator complex subunit 25 C-terminal NR box-containing | 2 |
| PF11261 | 0 | 0 | 3 | 2 | 0 | 0 | 3 | 0 | Interferon regulatory factor 2-binding protein zinc finger | 8 |
| PF11262 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 1 | Transcription factor/nuclear export subunit protein 2 | 4 |
| PF11265 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Mediator complex subunit 25 von Willebrand factor type A | 3 |
| PF11277 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Mediator complex subunit 24 N-terminal | 1 |
| PF11290 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Protein of unknown function (DUF3090) | 1 |
| PF11303 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | Protein of unknown function (DUF3105) | 1 |
| PF11315 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Mediator complex subunit 30 | 1 |
| PF11326 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Protein of unknown function (DUF3128) | 2 |
| PF11357 | 0 | 0 | 3 | 0 | 0 | 0 | 1 | 0 | Cell cycle regulatory protein | 4 |
| PF11365 | 1 | 1 | 3 | 1 | 2 | 2 | 6 | 1 | Protein of unknown function (DUF3166) | 17 |
| PF11380 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Protein of unknown function (DUF3184) | 2 |
| PF11404 | 0 | 0 | 1 | 1 | 0 | 1 | 2 | 0 | Potassium voltage-gated channel | 5 |
| PF11409 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Smad anchor for receptor activation (SARA) | 2 |
| PF11411 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | DNA ligase IV | 2 |
| PF11413 | 0 | 0 | 3 | 0 | 0 | 0 | 5 | 0 | Hypoxia-inducible factor-1 | 8 |
| PF11414 | 1 | 1 | 4 | 0 | 1 | 1 | 3 | 0 | Adenomatous polyposis coli tumour suppressor protein | 11 |
| PF11461 | 0 | 0 | 3 | 1 | 0 | 0 | 2 | 0 | Rab interacting lysosomal protein | 6 |
| PF11464 | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 0 | Rabenosyn Rab binding domain | 3 |
| PF11465 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Natural killer cell receptor 2B4 | 1 |
| PF11466 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | Prion-like protein Doppel | 2 |
| PF11467 | 1 | 1 | 2 | 1 | 0 | 0 | 1 | 0 | Lens epithelium-derived growth factor (LEDGF) | 6 |
| PF11470 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | GLUT4 regulating protein TUG | 2 |
| PF11488 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 0 | Transcriptional regulatory protein LGE1 | 3 |
| PF11502 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | B-cell lymphoma 9 protein | 4 |
| PF11510 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | Fanconi Anaemia group E protein FANCE | 1 |
| PF11515 | 0 | 0 | 3 | 0 | 1 | 0 | 1 | 1 | Mouse development and cellular proliferation protein Cullin-7 | 6 |
| PF11521 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | C-terminal general transcription factor TFIIE alpha | 3 |
| PF11527 | 0 | 0 | 2 | 2 | 0 | 0 | 2 | 0 | The ARF-like 2 binding protein BART | 6 |
| PF11531 | 0 | 0 | 1 | 1 | 0 | 0 | 2 | 0 | Coactivator-associated arginine methyltransferase 1 N terminal | 4 |
| PF11532 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | Heterogeneous nuclear ribonucleoprotein M | 6 |
| PF11538 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Snurportin1 | 2 |
| PF11540 | 2 | 2 | 2 | 2 | 0 | 1 | 2 | 0 | Cytoplasmic dynein 1 intermediate chain 2 | 11 |
| PF11543 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Nuclear pore localisation protein NPL4 | 1 |
| PF11547 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | E3 ubiquitin ligase EDD | 2 |
| PF11548 | 1 | 2 | 2 | 0 | 3 | 3 | 3 | 1 | Protein-tyrosine phosphatase receptor IA-2 | 15 |
| PF11555 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 0 | EGFR receptor inhibitor Mig-6 | 3 |
| PF11559 | 0 | 0 | 1 | 0 | 0 | 0 | 2 | 0 | Afadin- and alpha -actinin-Binding | 3 |
| PF11560 | 1 | 0 | 2 | 1 | 0 | 0 | 0 | 0 | Lamina-associated polypeptide 2 alpha | 4 |
| PF11566 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 0 | PI31 proteasome regulator N-terminal | 3 |
| PF11568 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Mediator complex subunit 29 | 3 |
| PF11569 | 0 | 0 | 3 | 0 | 0 | 0 | 1 | 0 | "Homeodomain leucine-zipper encoding, Homez" | 4 |
| PF11571 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Mediator complex subunit 27 | 3 |
| PF11573 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Mediator complex subunit 23 | 2 |
| PF11577 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | NF-kappa-B essential modulator NEMO | 4 |
| PF11587 | 0 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | Major prion protein bPrPp - N terminal | 3 |
| PF11594 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Mediator complex subunit 28 | 3 |
| PF11597 | 1 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | Mediator complex subunit 13 N-terminal | 5 |
| PF11598 | 0 | 0 | 3 | 1 | 0 | 0 | 3 | 0 | Cartilage oligomeric matrix protein | 7 |
| PF11600 | 1 | 1 | 2 | 1 | 0 | 0 | 1 | 0 | "Chromatin assembly factor 1 complex p150 subunit, N-terminal" | 6 |
| PF11601 | 3 | 3 | 3 | 2 | 3 | 3 | 3 | 1 | Shal-type voltage-gated potassium channels | 21 |
| PF11605 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Vacuolar protein sorting protein 36 Vps36 | 3 |
| PF11608 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Limkain b1 | 1 |
| PF11613 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 0 | "Agonist of corticotropin releasing factor R2, Urocortin-2" | 3 |
| PF11616 | 0 | 0 | 2 | 1 | 0 | 0 | 2 | 0 | WD repeat binding protein EZH2 | 5 |
| PF11618 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | Protein of unknown function (DUF3250) | 4 |
| PF11620 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | GA-binding protein alpha chain | 2 |
| PF11626 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | TRF2-interacting telomeric protein/Rap1 - C terminal domain | 2 |
| PF11627 | 3 | 3 | 3 | 1 | 0 | 0 | 2 | 2 | Nuclear factor hnRNPA1 | 14 |
| PF11628 | 1 | 1 | 2 | 0 | 0 | 0 | 1 | 0 | T-cell surface glycoprotein CD3 zeta chain | 5 |
| PF11629 | 0 | 0 | 2 | 0 | 0 | 1 | 1 | 0 | C terminal SARAH domain of Mst1 | 4 |
| PF11635 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Mediator complex subunit 16 | 2 |
| PF11636 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Troponin I residues 1-32 | 1 |
| PF11640 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Telomere-length maintenance and DNA damage repair | 2 |
| PF11648 | 0 | 0 | 3 | 0 | 0 | 0 | 2 | 0 | C-terminal domain of RIG-I | 5 |
| PF11652 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | Protein of unknown function (DUF3259) | 4 |
| PF11669 | 1 | 0 | 4 | 1 | 0 | 0 | 3 | 0 | WW domain-binding protein 1 | 9 |
| PF11698 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | V-ATPase subunit H | 8 |
| PF11699 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Mif2/CENP-C like | 2 |
| PF11701 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 0 | Myosin-binding striated muscle assembly central | 3 |
| PF11704 | 0 | 0 | 2 | 1 | 0 | 0 | 3 | 0 | Vesicle coat protein involved in Golgi to plasma membrane transport | 6 |
| PF11705 | 0 | 0 | 2 | 0 | 0 | 0 | 3 | 0 | DNA-directed RNA polymerase III subunit Rpc31 | 5 |
| PF11707 | 1 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Ribosome 60S biogenesis N-terminal | 3 |
| PF11708 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Pre-mRNA splicing Prp18-interacting factor | 2 |
| PF11712 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Endoplasmic reticulum-based factor for assembly of V-ATPase | 3 |
| PF11715 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Nucleoporin Nup120/160 | 3 |
| PF11717 | 0 | 0 | 6 | 2 | 0 | 0 | 6 | 0 | RNA binding activity-knot of a chromodomain | 14 |
| PF11718 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Pre-mRNA 3'-end-processing endonuclease polyadenylation factor C-term | 3 |
| PF11719 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | DNA replication and checkpoint protein | 2 |
| PF11721 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | Di-glucose binding within endoplasmic reticulum | 7 |
| PF11722 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | CCCH zinc finger in TRM13 protein | 1 |
| PF11732 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 1 | Transcription- and export-related complex subunit | 4 |
| PF11759 | 0 | 0 | 11 | 0 | 0 | 0 | 0 | 0 | Keratin-associated matrix | 11 |
| PF11764 | 0 | 0 | 2 | 1 | 0 | 0 | 2 | 0 | COMPASS (Complex proteins associated with Set1p) component N | 5 |
| PF11768 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Protein of unknown function (DUF3312) | 2 |
| PF11769 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | Protein of unknown function (DUF3313) | 3 |
| PF11770 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | GRB2-binding adapter (GAPT) | 1 |
| PF11771 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Protein of unknown function (DUF3314) | 2 |
| PF11779 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | Protein of unknown function (DUF3317) | 4 |
| PF11781 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | RNA polymerase I-specific transcription initiation factor Rrn7 | 2 |
| PF11788 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 39S mitochondrial ribosomal protein L46 | 6 |
| PF11789 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Zinc-finger of the MIZ type in Nse subunit | 2 |
| PF11793 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | FANCL C-terminal domain | 4 |
| PF11798 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | IMS family HHH motif | 2 |
| PF11799 | 0 | 0 | 4 | 1 | 0 | 0 | 4 | 0 | impB/mucB/samB family C-terminal domain | 9 |
| PF11801 | 2 | 2 | 2 | 0 | 0 | 3 | 3 | 2 | Tom37 C-terminal domain | 14 |
| PF11802 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Centromere-associated protein K | 2 |
| PF11803 | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 0 | UDP-glucuronate decarboxylase N-terminal | 3 |
| PF11816 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | Domain of unknown function (DUF3337) | 7 |
| PF11817 | 1 | 1 | 2 | 0 | 0 | 0 | 1 | 0 | Foie gras liver health family 1 | 5 |
| PF11819 | 2 | 0 | 4 | 1 | 1 | 1 | 7 | 1 | Domain of unknown function (DUF3338) | 17 |
| PF11822 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Domain of unknown function (DUF3342) | 3 |
| PF11825 | 0 | 0 | 3 | 1 | 0 | 0 | 5 | 0 | Nuclear/hormone receptor activator site AF-1 | 9 |
| PF11830 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | Domain of unknown function (DUF3350) | 4 |
| PF11831 | 1 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | pre-mRNA splicing factor component | 3 |
| PF11834 | 0 | 0 | 1 | 0 | 0 | 0 | 2 | 0 | Domain of unknown function (DUF3354) | 3 |
| PF11838 | 2 | 5 | 7 | 2 | 0 | 5 | 10 | 1 | ERAP1-like C-terminal domain | 32 |
| PF11841 | 2 | 2 | 3 | 2 | 3 | 2 | 2 | 1 | Domain of unknown function (DUF3361) | 17 |
| PF11851 | 0 | 0 | 4 | 0 | 0 | 0 | 4 | 0 | Domain of unknown function (DUF3371) | 8 |
| PF11857 | 0 | 0 | 4 | 0 | 0 | 0 | 6 | 0 | Domain of unknown function (DUF3377) | 10 |
| PF11861 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Domain of unknown function (DUF3381) | 1 |
| PF11864 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | Domain of unknown function (DUF3384) | 3 |
| PF11865 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | Domain of unknown function (DUF3385) | 6 |
| PF11875 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 0 | Domain of unknown function (DUF3395) | 8 |
| PF11878 | 3 | 2 | 6 | 0 | 3 | 3 | 6 | 0 | Domain of unknown function (DUF3398) | 23 |
| PF11879 | 3 | 3 | 3 | 2 | 3 | 3 | 3 | 1 | Domain of unknown function (DUF3399) | 21 |
| PF11881 | 3 | 1 | 3 | 1 | 2 | 1 | 4 | 0 | Domain of unknown function (DUF3401) | 15 |
| PF11882 | 0 | 1 | 2 | 1 | 1 | 1 | 2 | 0 | Domain of unknown function (DUF3402) | 8 |
| PF11894 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Protein of unknown function (DUF3414) | 2 |
| PF11904 | 0 | 0 | 4 | 2 | 0 | 1 | 2 | 1 | GPCR-chaperone | 10 |
| PF11911 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Protein of unknown function (DUF3429) | 2 |
| PF11914 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Domain of unknown function (DUF3432) | 2 |
| PF11916 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | Vacuolar protein 14 C-terminal Fig4p binding | 7 |
| PF11918 | 0 | 0 | 1 | 0 | 0 | 0 | 2 | 0 | Domain of unknown function (DUF3436) | 3 |
| PF11919 | 0 | 0 | 1 | 0 | 0 | 0 | 2 | 0 | Domain of unknown function (DUF3437) | 3 |
| PF11923 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | Domain of unknown function (DUF3441) | 2 |
| PF11928 | 0 | 0 | 3 | 1 | 0 | 0 | 4 | 0 | Domain of unknown function (DUF3446) | 8 |
| PF11931 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | Domain of unknown function (DUF3449) | 2 |
| PF11933 | 0 | 4 | 6 | 0 | 0 | 3 | 5 | 0 | Domain of unknown function (DUF3451) | 18 |
| PF11934 | 0 | 0 | 3 | 0 | 0 | 0 | 3 | 0 | Domain of unknown function (DUF3452) | 6 |
| PF11935 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Domain of unknown function (DUF3453) | 2 |
| PF11936 | 0 | 0 | 3 | 0 | 0 | 0 | 3 | 0 | Domain of unknown function (DUF3454) | 6 |
| PF11938 | 0 | 3 | 7 | 5 | 0 | 0 | 6 | 1 | TLR4 regulator and MIR-interacting MSAP | 22 |
| PF11942 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | "Spt5 transcription elongation factor, acidic N-terminal" | 3 |
| PF11945 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | WAHD domain of WASH complex | 5 |
| PF11952 | 0 | 0 | 2 | 1 | 0 | 1 | 2 | 0 | Protein of unknown function (DUF3469) | 6 |
| PF11956 | 1 | 1 | 2 | 0 | 0 | 1 | 3 | 0 | Ankyrin-G binding motif of KCNQ2-3 | 8 |
| PF11957 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | THO complex subunit 1 transcription elongation factor | 2 |
| PF11968 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Putative methyltransferase (DUF3321) | 1 |
| PF11969 | 0 | 2 | 3 | 2 | 0 | 0 | 4 | 0 | Scavenger mRNA decapping enzyme C-term binding | 11 |
| PF11971 | 1 | 1 | 6 | 0 | 1 | 1 | 4 | 0 | CAMSAP CH domain | 14 |
| PF11976 | 0 | 1 | 4 | 1 | 0 | 1 | 4 | 1 | Ubiquitin-2 like Rad60 SUMO-like | 12 |
| PF11977 | 0 | 0 | 8 | 2 | 0 | 0 | 4 | 0 | Zc3h12a-like Ribonuclease NYN domain | 14 |
| PF11978 | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 1 | Shoulder domain | 4 |
| PF11979 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 0 | Domain of unknown function (DUF3480) | 3 |
| PF11980 | 0 | 2 | 2 | 0 | 0 | 4 | 3 | 0 | Domain of unknown function (DUF3481) | 11 |
| PF11987 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 1 | Translation-initiation factor 2 | 5 |
| PF12001 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | Domain of unknown function (DUF3496) | 3 |
| PF12002 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | MgsA AAA+ ATPase C terminal | 2 |
| PF12003 | 6 | 7 | 10 | 1 | 5 | 9 | 13 | 1 | Domain of unknown function (DUF3497) | 52 |
| PF12004 | 3 | 3 | 3 | 0 | 4 | 4 | 5 | 2 | Domain of unknown function (DUF3498) | 24 |
| PF12009 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Telomerase ribonucleoprotein complex - RNA binding domain | 2 |
| PF12012 | 0 | 0 | 6 | 1 | 0 | 0 | 7 | 0 | Domain of unknown function (DUF3504) | 14 |
| PF12016 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | Stonin 2 | 2 |
| PF12017 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | Transposase protein | 1 |
| PF12018 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Domain of unknown function (DUF3508) | 2 |
| PF12020 | 0 | 0 | 5 | 1 | 0 | 0 | 9 | 0 | TAFA family | 15 |
| PF12022 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Domain of unknown function (DUF3510) | 2 |
| PF12024 | 0 | 0 | 2 | 1 | 0 | 0 | 2 | 0 | Domain of unknown function (DUF3512) | 5 |
| PF12026 | 0 | 0 | 4 | 0 | 0 | 0 | 4 | 0 | Domain of unknown function (DUF3513) | 8 |
| PF12031 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 0 | Domain of unknown function (DUF3518) | 3 |
| PF12036 | 0 | 0 | 3 | 0 | 0 | 0 | 2 | 0 | Protein of unknown function (DUF3522) | 5 |
| PF12037 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | Domain of unknown function (DUF3523) | 8 |
| PF12038 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Domain of unknown function (DUF3524) | 2 |
| PF12045 | 0 | 0 | 3 | 0 | 0 | 0 | 3 | 0 | Protein of unknown function (DUF3528) | 6 |
| PF12047 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Cytosine specific DNA methyltransferase replication foci domain | 2 |
| PF12052 | 4 | 3 | 4 | 1 | 5 | 5 | 5 | 0 | Voltage gated calcium channel subunit beta domain 4Aa N terminal | 27 |
| PF12053 | 0 | 0 | 2 | 0 | 0 | 0 | 4 | 0 | Domain of unknown function (DUF3534) | 6 |
| PF12054 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Domain of unknown function (DUF3535) | 2 |
| PF12057 | 0 | 1 | 1 | 1 | 0 | 1 | 2 | 1 | Domain of unknown function (DUF3538) | 7 |
| PF12062 | 0 | 0 | 4 | 0 | 0 | 0 | 4 | 0 | heparan sulfate-N-deacetylase | 8 |
| PF12063 | 0 | 0 | 2 | 0 | 0 | 0 | 3 | 0 | Domain of unknown function (DUF3543) | 5 |
| PF12064 | 0 | 0 | 1 | 0 | 0 | 0 | 2 | 1 | Domain of unknown function (DUF3544) | 4 |
| PF12066 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 1 | Domain of unknown function (DUF3546) | 3 |
| PF12067 | 0 | 0 | 3 | 0 | 0 | 0 | 2 | 0 | Sox C-terminal transactivation domain | 5 |
| PF12068 | 0 | 1 | 2 | 1 | 0 | 0 | 2 | 0 | Domain of unknown function (DUF3548) | 6 |
| PF12070 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | Protein of unknown function (DUF3550/UPF0682) | 6 |
| PF12074 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | Domain of unknown function (DUF3554) | 6 |
| PF12075 | 0 | 0 | 4 | 2 | 0 | 1 | 5 | 0 | KN motif | 12 |
| PF12090 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Spt20 family | 2 |
| PF12104 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | T cell CD4 receptor C terminal region | 1 |
| PF12108 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | Splicing factor SF3a60 binding domain | 2 |
| PF12109 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | CXCR4 Chemokine receptor N terminal | 1 |
| PF12110 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | Nuclear protein 96 | 1 |
| PF12114 | 0 | 0 | 3 | 0 | 0 | 0 | 4 | 0 | Period protein 2/3C-terminal region | 7 |
| PF12125 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 0 | D domain of beta-TrCP | 3 |
| PF12126 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | Protein of unknown function (DUF3583) | 2 |
| PF12129 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 0 | Male germ-cell putative homeodomain transcription factor | 3 |
| PF12130 | 0 | 1 | 7 | 2 | 0 | 0 | 13 | 1 | Protein of unknown function (DUF3585) | 24 |
| PF12134 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 1 | PRP8 domain IV core | 3 |
| PF12140 | 1 | 1 | 5 | 1 | 0 | 0 | 4 | 0 | Protein of unknown function (DUF3588) | 12 |
| PF12144 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Eukaryotic Mediator 12 catenin-binding domain | 1 |
| PF12145 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Eukaryotic Mediator 12 subunit domain | 1 |
| PF12146 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | Putative lysophospholipase | 6 |
| PF12148 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | Protein of unknown function (DUF3590) | 2 |
| PF12153 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | LPS binding domain of CAP18 (C terminal) | 1 |
| PF12157 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | Protein of unknown function (DUF3591) | 2 |
| PF12160 | 1 | 0 | 1 | 0 | 1 | 0 | 1 | 1 | Fibrinogen alpha C domain | 5 |
| PF12162 | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 0 | STAT1 TAZ2 binding domain | 3 |
| PF12166 | 0 | 0 | 1 | 0 | 0 | 0 | 2 | 0 | Protein of unknown function (DUF3595) | 3 |
| PF12171 | 0 | 0 | 13 | 0 | 0 | 0 | 12 | 0 | Zinc-finger double-stranded RNA-binding | 25 |
| PF12177 | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 0 | Prohormone convertase enzyme | 3 |
| PF12178 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Chromosome passenger complex (CPC) protein INCENP N terminal | 1 |
| PF12179 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 0 | I-kappa-kinase-beta NEMO binding domain | 3 |
| PF12180 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 0 | TSG101 and ALIX binding domain of CEP55 | 3 |
| PF12185 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Nup358/RanBP2 E3 ligase domain | 2 |
| PF12188 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Signal transducer and activator of transcription 2 C terminal | 1 |
| PF12191 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Tumour necrosis factor receptor stn_TNFRSF12A_TNFR domain | 1 |
| PF12196 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | FHA Ki67 binding domain of hNIFK | 3 |
| PF12201 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | "Bcl2-interacting killer, BH3-domain containing" | 1 |
| PF12202 | 1 | 3 | 6 | 2 | 1 | 3 | 6 | 2 | Oxidative-stress-responsive kinase 1 C terminal | 24 |
| PF12203 | 0 | 0 | 3 | 0 | 0 | 0 | 2 | 0 | Glutamine rich N terminal domain of histone deacetylase 4 | 5 |
| PF12205 | 1 | 1 | 2 | 0 | 1 | 1 | 3 | 0 | G protein-coupled receptor kinase-interacting protein 1 C term | 9 |
| PF12210 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | Hepatocyte growth factor-regulated tyrosine kinase substrate | 5 |
| PF12213 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | DNA polymerases epsilon N terminal | 1 |
| PF12214 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Cell cycle regulated microtubule associated protein | 2 |
| PF12215 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | beta-Glucocerebrosidase 2 N terminal | 2 |
| PF12220 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 1 | U1 small nuclear ribonucleoprotein of 70kDa MW N terminal | 3 |
| PF12230 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | Pre-mRNA splicing factor PRP21 like protein | 4 |
| PF12231 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 1 | Rap1-interacting factor 1 N terminal | 3 |
| PF12232 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | Myogenic determination factor 5 | 4 |
| PF12234 | 0 | 0 | 2 | 0 | 2 | 2 | 1 | 1 | RAVE protein 1 C terminal | 8 |
| PF12235 | 3 | 3 | 3 | 1 | 1 | 1 | 3 | 1 | Fragile X-related 1 protein C terminal | 16 |
| PF12237 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | Phosphorylated CTD interacting factor 1 WW domain | 2 |
| PF12240 | 1 | 1 | 3 | 1 | 0 | 0 | 4 | 0 | Angiomotin C terminal | 10 |
| PF12248 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | Farnesoic acid 0-methyl transferase | 1 |
| PF12251 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | snRNA-activating protein of 50kDa MW C terminal | 1 |
| PF12253 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Chromatin assembly factor 1 subunit A | 2 |
| PF12254 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | DNA polymerase alpha subunit p180 N terminal | 2 |
| PF12257 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Protein of unknown function (DUF3608) | 2 |
| PF12258 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Microcephalin protein | 1 |
| PF12260 | 1 | 0 | 6 | 1 | 0 | 0 | 7 | 0 | Protein-kinase domain of FAM69 | 15 |
| PF12265 | 0 | 0 | 3 | 1 | 0 | 0 | 3 | 1 | Histone-binding protein RBBP4 or subunit C of CAF1 complex | 8 |
| PF12269 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | CpG binding protein zinc finger C terminal domain | 2 |
| PF12280 | 0 | 0 | 2 | 1 | 0 | 1 | 2 | 0 | Brain specific membrane anchored protein | 6 |
| PF12284 | 0 | 0 | 3 | 0 | 0 | 0 | 4 | 0 | Hox protein A13 N terminal | 7 |
| PF12287 | 1 | 1 | 2 | 2 | 0 | 0 | 2 | 1 | Cytoplasmic activation/proliferation-associated protein-1 C term | 9 |
| PF12295 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Symplekin tight junction protein C terminal | 2 |
| PF12297 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Ellis van Creveld protein 2 like protein | 1 |
| PF12301 | 0 | 1 | 1 | 1 | 0 | 2 | 3 | 0 | CD99 antigen like protein 2 | 8 |
| PF12304 | 0 | 0 | 2 | 0 | 0 | 0 | 3 | 0 | Beta-casein like protein | 5 |
| PF12307 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Protein of unknown function (DUF3631) | 1 |
| PF12308 | 3 | 3 | 3 | 1 | 5 | 5 | 5 | 0 | Neurogenesis glycoprotein | 25 |
| PF12309 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | KIF-1 binding protein C terminal | 3 |
| PF12310 | 0 | 0 | 3 | 0 | 0 | 0 | 3 | 0 | Transcription factor protein N terminal | 6 |
| PF12316 | 0 | 0 | 3 | 0 | 0 | 0 | 4 | 0 | Segment polarity protein dishevelled (Dsh) C terminal | 7 |
| PF12317 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Intraflagellar transport complex B protein 46 C terminal | 3 |
| PF12325 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | TATA element modulatory factor 1 TATA binding | 2 |
| PF12328 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Rpp20 subunit of nuclear RNase MRP and P | 1 |
| PF12329 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | TATA element modulatory factor 1 DNA binding | 2 |
| PF12330 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Domain of unknown function (DUF3635) | 2 |
| PF12333 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Rix1 complex component involved in 60S ribosome maturation | 2 |
| PF12335 | 0 | 1 | 2 | 0 | 0 | 1 | 3 | 1 | Myotubularin protein | 8 |
| PF12336 | 0 | 0 | 5 | 0 | 0 | 0 | 8 | 0 | SOX transcription factor | 13 |
| PF12341 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | Protein of unknown function (DUF3639) | 1 |
| PF12346 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Holliday junction recognition protein-associated repeat | 1 |
| PF12347 | 0 | 0 | 4 | 1 | 0 | 0 | 5 | 0 | Holliday junction regulator protein family C-terminal repeat | 10 |
| PF12348 | 3 | 3 | 3 | 0 | 4 | 3 | 2 | 1 | CLASP N terminal | 19 |
| PF12349 | 0 | 3 | 7 | 2 | 0 | 0 | 5 | 0 | Sterol-sensing domain of SREBP cleavage-activation | 17 |
| PF12352 | 4 | 5 | 5 | 3 | 2 | 4 | 4 | 0 | Snare region anchored in the vesicle membrane C-terminus | 27 |
| PF12353 | 0 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | Eukaryotic translation initiation factor 3 subunit G | 5 |
| PF12356 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Protein of unknown function (DUF3643) | 2 |
| PF12360 | 0 | 0 | 2 | 0 | 0 | 0 | 3 | 0 | Paired box protein 7 | 5 |
| PF12366 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Cancer susceptibility candidate 1 | 2 |
| PF12369 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Gonadotropin hormone receptor transmembrane region | 2 |
| PF12371 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 0 | Protein of unknown function (DUF3651) | 3 |
| PF12372 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | Huntingtin protein region | 6 |
| PF12374 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Double-sex mab3 related transcription factor 1 | 2 |
| PF12394 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 0 | Protein of unknown function (DUF3657) | 3 |
| PF12397 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | U3 small nucleolar RNA-associated protein 10 | 2 |
| PF12400 | 0 | 0 | 1 | 0 | 0 | 0 | 2 | 0 | Vaculolar membrane protein | 3 |
| PF12402 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | NocA-like zinc-finger protein 1 | 4 |
| PF12403 | 0 | 0 | 3 | 0 | 0 | 0 | 4 | 0 | Paired-box protein 2 C terminal | 7 |
| PF12409 | 0 | 2 | 4 | 0 | 0 | 0 | 2 | 0 | P5-type ATPase cation transporter | 8 |
| PF12413 | 0 | 0 | 3 | 0 | 0 | 0 | 4 | 0 | Homeobox protein distal-less-like N terminal | 7 |
| PF12414 | 0 | 0 | 3 | 0 | 0 | 0 | 5 | 0 | Calcitonin gene-related peptide regulator C terminal | 8 |
| PF12416 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Cep120 protein | 2 |
| PF12417 | 0 | 0 | 6 | 0 | 0 | 0 | 0 | 0 | Zinc finger protein | 6 |
| PF12422 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Condensin II non structural maintenance of chromosomes subunit | 2 |
| PF12423 | 0 | 2 | 4 | 1 | 0 | 1 | 7 | 0 | Kinesin protein 1B | 15 |
| PF12424 | 0 | 4 | 4 | 1 | 0 | 5 | 6 | 5 | Plasma membrane calcium transporter ATPase C terminal | 25 |
| PF12430 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Abscisic acid G-protein coupled receptor | 2 |
| PF12432 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Protein of unknown function (DUF3677) | 1 |
| PF12436 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | ICP0-binding domain of Ubiquitin-specific protease 7 | 5 |
| PF12440 | 0 | 0 | 14 | 0 | 0 | 0 | 0 | 0 | Melanoma associated antigen family N terminal | 14 |
| PF12443 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | AT-hook-containing transcription factor | 2 |
| PF12444 | 0 | 0 | 3 | 1 | 0 | 0 | 4 | 0 | Sox developmental protein N terminal | 8 |
| PF12448 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | Kinesin associated protein | 4 |
| PF12451 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | Vacuolar protein sorting protein 11 C terminal | 6 |
| PF12453 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | Protein tyrosine phosphatase N terminal | 2 |
| PF12455 | 0 | 1 | 1 | 0 | 1 | 2 | 2 | 2 | Dynein associated protein | 9 |
| PF12456 | 0 | 1 | 3 | 2 | 0 | 1 | 3 | 2 | Inositol phosphatase | 12 |
| PF12457 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Tuftelin interacting protein N terminal | 3 |
| PF12460 | 0 | 0 | 1 | 1 | 0 | 1 | 1 | 0 | RNAPII transcription regulator C-terminal | 4 |
| PF12463 | 0 | 0 | 1 | 1 | 0 | 0 | 2 | 0 | Protein of unknown function (DUF3689) | 4 |
| PF12465 | 0 | 0 | 2 | 1 | 0 | 1 | 3 | 1 | Proteasome beta subunits C terminal | 8 |
| PF12470 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Suppressor of Fused Gli/Ci N terminal binding domain | 1 |
| PF12473 | 0 | 2 | 6 | 2 | 0 | 2 | 7 | 0 | Kinesin protein | 19 |
| PF12474 | 0 | 1 | 2 | 0 | 0 | 1 | 3 | 1 | Polo kinase kinase | 8 |
| PF12478 | 1 | 1 | 2 | 0 | 0 | 0 | 3 | 2 | Ubiquitin-associated protein 2 | 9 |
| PF12480 | 0 | 0 | 8 | 0 | 0 | 0 | 0 | 0 | Protein of unknown function (DUF3699) | 8 |
| PF12483 | 0 | 0 | 1 | 0 | 0 | 1 | 3 | 1 | E3 Ubiquitin ligase | 6 |
| PF12485 | 0 | 0 | 3 | 0 | 0 | 0 | 5 | 0 | Lymphocyte signaling adaptor protein | 8 |
| PF12489 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Nuclear coactivator | 2 |
| PF12490 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | Breast carcinoma amplified sequence 3 | 5 |
| PF12491 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Apolipoprotein B100 C terminal | 1 |
| PF12494 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Protein of unknown function (DUF3695) | 2 |
| PF12496 | 1 | 1 | 4 | 1 | 0 | 0 | 5 | 1 | Bcl2-/adenovirus E1B nineteen kDa-interacting protein 2 | 13 |
| PF12497 | 0 | 0 | 1 | 0 | 0 | 0 | 2 | 0 | Estrogen receptor beta | 3 |
| PF12509 | 0 | 0 | 3 | 0 | 0 | 0 | 3 | 0 | Protein of unknown function (DUF3715) | 6 |
| PF12510 | 0 | 0 | 1 | 1 | 0 | 0 | 2 | 0 | Smoothelin cytoskeleton protein | 4 |
| PF12513 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Mitochondrial degradasome RNA helicase subunit C terminal | 3 |
| PF12516 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | Protein of unknown function (DUF3719) | 4 |
| PF12529 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | Xylosyltransferase C terminal | 4 |
| PF12530 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Protein of unknown function (DUF3730) | 2 |
| PF12533 | 0 | 0 | 4 | 1 | 0 | 0 | 5 | 0 | Neuronal helix-loop-helix transcription factor | 10 |
| PF12534 | 4 | 4 | 6 | 2 | 1 | 1 | 4 | 0 | Leucine-rich repeat containing protein 8 | 22 |
| PF12537 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Protein of unknown function (DUF3735) | 2 |
| PF12540 | 0 | 0 | 1 | 0 | 0 | 0 | 2 | 0 | Protein of unknown function (DUF3736) | 3 |
| PF12542 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Pre-mRNA splicing factor | 2 |
| PF12547 | 0 | 0 | 1 | 0 | 0 | 0 | 2 | 0 | Capicua transcriptional repressor modulator | 3 |
| PF12548 | 0 | 0 | 2 | 0 | 0 | 0 | 3 | 0 | Sulfatase protein | 5 |
| PF12549 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Tyrosine hydroxylase N terminal | 3 |
| PF12554 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Mitotic-spindle organizing gamma-tubulin ring associated | 2 |
| PF12567 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | Leukocyte receptor CD45 | 2 |
| PF12569 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 1 | NMDA receptor-regulated protein 1 | 5 |
| PF12572 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Protein of unknown function (DUF3752) | 2 |
| PF12576 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Protein of unknown function (DUF3754) | 1 |
| PF12577 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | PPAR gamma N-terminal region | 2 |
| PF12578 | 0 | 0 | 3 | 1 | 0 | 1 | 3 | 0 | Myotubularin-associated protein | 8 |
| PF12580 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | Tripeptidyl peptidase II | 6 |
| PF12583 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | Tripeptidyl peptidase II N terminal | 3 |
| PF12584 | 1 | 1 | 1 | 0 | 1 | 0 | 0 | 0 | "Trafficking protein particle complex subunit 10, TRAPPC10" | 4 |
| PF12589 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Methyltransferase involved in Williams-Beuren syndrome | 2 |
| PF12595 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | Rhomboid serine protease | 4 |
| PF12597 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | Protein of unknown function (DUF3767) | 2 |
| PF12598 | 0 | 0 | 2 | 0 | 0 | 0 | 3 | 0 | T-box transcription factor | 5 |
| PF12605 | 3 | 3 | 3 | 1 | 2 | 2 | 2 | 0 | Casein kinase 1 gamma C terminal | 16 |
| PF12606 | 0 | 1 | 3 | 1 | 0 | 0 | 2 | 0 | Tumour necrosis factor receptor superfamily member 19 | 7 |
| PF12610 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | Suppressor of cytokine signalling | 4 |
| PF12612 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | Tubulin folding cofactor D C terminal | 2 |
| PF12618 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 0 | Protein of unknown function (DUF3776) | 3 |
| PF12619 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Mini-chromosome maintenance protein 2 | 1 |
| PF12624 | 0 | 4 | 8 | 1 | 1 | 4 | 6 | 0 | "N-terminal region of Chorein, a TM vesicle-mediated sorter" | 24 |
| PF12627 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Probable RNA and SrmB- binding site of polymerase A | 3 |
| PF12631 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | "Catalytic cysteine-containing C-terminus of GTPase, MnmE" | 3 |
| PF12632 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Mysoin-binding motif of peroxisomes | 3 |
| PF12640 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | UPF0489 domain | 2 |
| PF12656 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | DExH-box splicing factor binding site | 3 |
| PF12657 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Transcription factor IIIC subunit delta N-term | 2 |
| PF12660 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Putative zinc-finger of transcription factor IIIC complex | 2 |
| PF12661 | 5 | 9 | 33 | 4 | 5 | 10 | 36 | 4 | Human growth factor-like EGF | 106 |
| PF12662 | 0 | 1 | 15 | 2 | 0 | 2 | 21 | 4 | Complement Clr-like EGF-like | 45 |
| PF12678 | 0 | 1 | 2 | 1 | 1 | 1 | 2 | 1 | RING-H2 zinc finger | 9 |
| PF12681 | 0 | 1 | 2 | 1 | 0 | 1 | 2 | 1 | Glyoxalase-like domain | 8 |
| PF12689 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Acid Phosphatase | 3 |
| PF12695 | 2 | 3 | 12 | 5 | 0 | 5 | 12 | 0 | Alpha/beta hydrolase family | 39 |
| PF12697 | 2 | 4 | 18 | 5 | 0 | 5 | 16 | 5 | Alpha/beta hydrolase family | 55 |
| PF12698 | 0 | 6 | 15 | 1 | 0 | 3 | 9 | 0 | ABC-2 family transporter protein | 34 |
| PF12701 | 0 | 0 | 3 | 3 | 0 | 0 | 3 | 1 | Scd6-like Sm domain | 10 |
| PF12705 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | PD-(D/E)XK nuclease superfamily | 2 |
| PF12706 | 0 | 0 | 6 | 3 | 0 | 1 | 6 | 0 | Beta-lactamase superfamily domain | 16 |
| PF12710 | 0 | 6 | 17 | 2 | 0 | 1 | 13 | 1 | haloacid dehalogenase-like hydrolase | 40 |
| PF12711 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Kinesin motor | 2 |
| PF12714 | 0 | 0 | 4 | 0 | 0 | 0 | 5 | 0 | TILa domain | 9 |
| PF12717 | 0 | 0 | 3 | 1 | 0 | 0 | 2 | 0 | non-SMC mitotic condensation complex subunit 1 | 6 |
| PF12718 | 1 | 1 | 2 | 2 | 1 | 1 | 1 | 1 | Tropomyosin like | 10 |
| PF12719 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | "Nuclear condensing complex subunits, C-term domain" | 2 |
| PF12721 | 0 | 0 | 4 | 1 | 0 | 0 | 1 | 0 | RIP homotypic interaction motif | 6 |
| PF12722 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | High-temperature-induced dauer-formation protein | 2 |
| PF12726 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | SEN1 N terminal | 1 |
| PF12733 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Cadherin-like beta sandwich domain | 2 |
| PF12736 | 0 | 0 | 5 | 1 | 0 | 0 | 4 | 0 | "Cell-cycle sustaining, positive selection," | 10 |
| PF12738 | 0 | 0 | 6 | 1 | 0 | 0 | 5 | 0 | twin BRCT domain | 12 |
| PF12739 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 0 | ER-Golgi trafficking TRAPP I complex 85 kDa subunit | 5 |
| PF12743 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Oestrogen-type nuclear receptor final C-terminal | 1 |
| PF12745 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Anticodon binding domain of tRNAs | 1 |
| PF12752 | 0 | 0 | 4 | 1 | 0 | 0 | 2 | 0 | SUZ domain | 7 |
| PF12755 | 1 | 1 | 2 | 1 | 0 | 1 | 3 | 1 | Vacuolar 14 Fab1-binding region | 10 |
| PF12756 | 0 | 0 | 10 | 0 | 0 | 0 | 8 | 0 | C2H2 type zinc-finger (2 copies) | 18 |
| PF12762 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | ISXO2-like transposase domain | 3 |
| PF12763 | 4 | 6 | 10 | 3 | 3 | 6 | 12 | 7 | Cytoskeletal-regulatory complex EF hand | 51 |
| PF12765 | 0 | 0 | 3 | 0 | 0 | 0 | 1 | 0 | HEAT repeat associated with sister chromatid cohesion | 4 |
| PF12767 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | "Transcriptional regulator of RNA polII, SAGA, subunit" | 1 |
| PF12769 | 0 | 0 | 0 | 0 | 0 | 1 | 2 | 2 | Domain of unknown function (DUF3814) | 5 |
| PF12770 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | CHAT domain | 1 |
| PF12772 | 0 | 0 | 1 | 0 | 0 | 0 | 2 | 0 | Growth hormone receptor binding | 3 |
| PF12773 | 1 | 1 | 1 | 0 | 0 | 0 | 1 | 0 | Double zinc ribbon | 4 |
| PF12774 | 0 | 2 | 13 | 1 | 1 | 2 | 13 | 2 | Hydrolytic ATP binding site of dynein motor region D1 | 34 |
| PF12775 | 0 | 2 | 13 | 1 | 1 | 2 | 12 | 1 | P-loop containing dynein motor region D3 | 32 |
| PF12777 | 0 | 2 | 13 | 1 | 1 | 2 | 12 | 1 | Microtubule-binding stalk of dynein motor | 32 |
| PF12780 | 0 | 2 | 13 | 0 | 1 | 1 | 11 | 0 | P-loop containing dynein motor region D4 | 28 |
| PF12781 | 0 | 2 | 13 | 0 | 1 | 2 | 11 | 1 | ATP-binding dynein motor region D5 | 30 |
| PF12783 | 0 | 1 | 4 | 1 | 0 | 0 | 3 | 0 | Guanine nucleotide exchange factor in Golgi transport N-terminal | 9 |
| PF12796 | 35 | 30 | 213 | 31 | 33 | 36 | 225 | 11 | Ankyrin repeats (3 copies) | 614 |
| PF12798 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 4Fe-4S binding domain | 1 |
| PF12799 | 13 | 16 | 57 | 15 | 12 | 14 | 50 | 5 | Leucine Rich repeats (2 copies) | 182 |
| PF12807 | 0 | 0 | 1 | 1 | 0 | 0 | 3 | 0 | Translation initiation factor eIF3 subunit 135 | 5 |
| PF12810 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | Glycine rich protein | 4 |
| PF12811 | 0 | 1 | 1 | 1 | 0 | 1 | 0 | 0 | Bax inhibitor 1 like | 4 |
| PF12813 | 0 | 0 | 1 | 1 | 0 | 0 | 2 | 0 | XPG domain containing | 4 |
| PF12814 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | Meiotic cell cortex C-terminal pleckstrin homology | 3 |
| PF12815 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Spt5 C-terminal nonapeptide repeat binding Spt4 | 3 |
| PF12816 | 0 | 0 | 1 | 1 | 0 | 1 | 1 | 0 | Golgi CORVET complex core vacuolar protein 8 | 4 |
| PF12820 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Serine-rich domain associated with BRCT | 1 |
| PF12826 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Helix-hairpin-helix motif | 2 |
| PF12830 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Sister chromatid cohesion C-terminus | 2 |
| PF12832 | 0 | 1 | 2 | 1 | 0 | 0 | 3 | 0 | MFS_1 like family | 7 |
| PF12836 | 1 | 1 | 4 | 0 | 0 | 0 | 2 | 0 | Helix-hairpin-helix motif | 8 |
| PF12838 | 0 | 1 | 1 | 1 | 0 | 1 | 2 | 1 | 4Fe-4S dicluster domain | 7 |
| PF12842 | 1 | 0 | 1 | 0 | 1 | 1 | 1 | 0 | Domain of unknown function (DUF3819) | 5 |
| PF12845 | 1 | 0 | 3 | 0 | 1 | 1 | 1 | 0 | TBD domain | 7 |
| PF12847 | 0 | 0 | 3 | 2 | 0 | 0 | 4 | 0 | Methyltransferase domain | 9 |
| PF12848 | 1 | 0 | 3 | 3 | 0 | 0 | 2 | 0 | ABC transporter | 9 |
| PF12850 | 1 | 1 | 1 | 1 | 0 | 1 | 2 | 1 | Calcineurin-like phosphoesterase superfamily domain | 8 |
| PF12851 | 0 | 0 | 3 | 0 | 0 | 0 | 3 | 0 | Oxygenase domain of the 2OGFeDO superfamily | 6 |
| PF12859 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Anaphase-promoting complex subunit 1 | 2 |
| PF12861 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | Anaphase-promoting complex subunit 11 RING-H2 finger | 2 |
| PF12862 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Anaphase-promoting complex subunit 5 | 3 |
| PF12871 | 0 | 0 | 1 | 0 | 1 | 1 | 1 | 0 | Pre-mRNA-splicing factor 38-associated hydrophilic C-term | 4 |
| PF12872 | 0 | 0 | 3 | 0 | 0 | 0 | 4 | 0 | OST-HTH/LOTUS domain | 7 |
| PF12874 | 0 | 0 | 22 | 3 | 0 | 0 | 37 | 1 | Zinc-finger of C2H2 type | 63 |
| PF12877 | 2 | 3 | 3 | 0 | 4 | 4 | 4 | 0 | Domain of unknown function (DUF3827) | 20 |
| PF12881 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | NUT protein N terminus | 2 |
| PF12882 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | NUT protein C terminal | 2 |
| PF12884 | 0 | 0 | 3 | 2 | 0 | 0 | 3 | 0 | "Transducer of regulated CREB activity, N terminus" | 8 |
| PF12885 | 0 | 0 | 3 | 2 | 1 | 0 | 4 | 0 | Transducer of regulated CREB activity middle domain | 10 |
| PF12886 | 0 | 0 | 3 | 2 | 1 | 0 | 3 | 0 | "Transducer of regulated CREB activity, C terminus" | 9 |
| PF12894 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Anaphase-promoting complex subunit 4 WD40 domain | 3 |
| PF12895 | 0 | 0 | 2 | 2 | 0 | 0 | 2 | 0 | "Anaphase-promoting complex, cyclosome, subunit 3" | 6 |
| PF12896 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | "Anaphase-promoting complex, cyclosome, subunit 4" | 3 |
| PF12901 | 1 | 1 | 3 | 1 | 0 | 0 | 4 | 0 | SUZ-C motif | 10 |
| PF12906 | 1 | 1 | 11 | 0 | 0 | 1 | 13 | 1 | RING-variant domain | 28 |
| PF12907 | 0 | 0 | 1 | 0 | 0 | 0 | 2 | 0 | Zinc-binding | 3 |
| PF12922 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | "non-SMC mitotic condensation complex subunit 1, N-term" | 2 |
| PF12923 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Ribosomal RNA-processing protein 7 (RRP7) | 1 |
| PF12924 | 0 | 3 | 3 | 2 | 0 | 4 | 2 | 0 | "Copper-binding of amyloid precursor, CuBD" | 14 |
| PF12925 | 0 | 3 | 3 | 2 | 0 | 4 | 2 | 0 | E2 domain of amyloid precursor protein | 14 |
| PF12926 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 0 | Mitotic-spindle organizing gamma-tubulin ring associated | 5 |
| PF12928 | 0 | 0 | 1 | 0 | 0 | 1 | 2 | 0 | tRNA-splicing endonuclease subunit sen54 N-term | 4 |
| PF12931 | 0 | 1 | 4 | 0 | 0 | 0 | 1 | 0 | Sec23-binding domain of Sec16 | 6 |
| PF12932 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | Vesicle coat trafficking protein Sec16 mid-region | 2 |
| PF12933 | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 1 | FTO catalytic domain | 4 |
| PF12934 | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 1 | FTO C-terminal domain | 4 |
| PF12936 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | KRI1-like family C-terminal | 2 |
| PF12937 | 1 | 2 | 54 | 7 | 2 | 2 | 42 | 2 | F-box-like | 112 |
| PF12938 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | M domain of GW182 | 2 |
| PF12940 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Recombination-activation protein 1 (RAG1) | 2 |
| PF12947 | 0 | 1 | 10 | 1 | 0 | 1 | 15 | 2 | EGF domain | 30 |
| PF12971 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Alpha-N-acetylglucosaminidase (NAGLU) N-terminal domain | 2 |
| PF12972 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Alpha-N-acetylglucosaminidase (NAGLU) C-terminal domain | 2 |
| PF12998 | 0 | 0 | 5 | 0 | 0 | 0 | 5 | 0 | Inhibitor of growth proteins N-terminal histone-binding | 10 |
| PF12999 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | Glucosidase II beta subunit-like | 6 |
| PF13000 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Acetyl-coenzyme A transporter 1 | 2 |
| PF13001 | 0 | 1 | 1 | 0 | 0 | 0 | 1 | 0 | Proteasome stabiliser | 3 |
| PF13003 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Ribosomal protein L1 | 2 |
| PF13012 | 3 | 3 | 4 | 3 | 0 | 3 | 2 | 2 | Maintenance of mitochondrial structure and function | 20 |
| PF13014 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | KH domain | 1 |
| PF13015 | 0 | 1 | 2 | 1 | 0 | 1 | 2 | 1 | Glucosidase II beta subunit-like protein | 8 |
| PF13017 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | piRNA pathway germ-plasm component | 1 |
| PF13019 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 1 | Telomere stability and silencing | 3 |
| PF13020 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | Domain of unknown function (DUF3883) | 1 |
| PF13023 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | HD domain | 2 |
| PF13041 | 0 | 0 | 2 | 1 | 0 | 0 | 1 | 0 | PPR repeat family | 4 |
| PF13083 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | KH domain | 8 |
| PF13085 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 2Fe-2S iron-sulfur cluster binding domain | 6 |
| PF13086 | 1 | 1 | 11 | 0 | 2 | 1 | 12 | 1 | AAA domain | 29 |
| PF13087 | 1 | 1 | 11 | 0 | 2 | 1 | 12 | 1 | AAA domain | 29 |
| PF13088 | 1 | 0 | 4 | 4 | 1 | 0 | 7 | 1 | BNR repeat-like domain | 18 |
| PF13091 | 2 | 2 | 4 | 2 | 1 | 1 | 7 | 0 | PLD-like domain | 19 |
| PF13092 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Kinetochore complex Sim4 subunit Fta1 | 2 |
| PF13094 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | "CENP-Q, a CENPA-CAD centromere complex subunit" | 1 |
| PF13096 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | "CENP-A-nucleosome distal (CAD) centromere subunit, CENP-P" | 2 |
| PF13097 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | CENP-A nucleosome associated complex (NAC) subunit | 2 |
| PF13147 | 0 | 1 | 3 | 1 | 0 | 0 | 1 | 0 | Amidohydrolase | 6 |
| PF13167 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | GTP-binding GTPase N-terminal | 1 |
| PF13173 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | AAA domain | 2 |
| PF13174 | 1 | 1 | 5 | 3 | 2 | 2 | 5 | 1 | Tetratricopeptide repeat | 20 |
| PF13176 | 3 | 4 | 9 | 2 | 0 | 1 | 5 | 0 | Tetratricopeptide repeat | 24 |
| PF13177 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | "DNA polymerase III, delta subunit" | 2 |
| PF13180 | 0 | 2 | 5 | 2 | 0 | 1 | 31 | 0 | PDZ domain | 41 |
| PF13181 | 2 | 2 | 14 | 3 | 2 | 4 | 17 | 2 | Tetratricopeptide repeat | 46 |
| PF13185 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | GAF domain | 1 |
| PF13191 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | AAA ATPase domain | 4 |
| PF13193 | 2 | 4 | 16 | 5 | 1 | 6 | 16 | 3 | AMP-binding enzyme C-terminal domain | 53 |
| PF13202 | 3 | 4 | 21 | 8 | 2 | 6 | 18 | 5 | EF hand | 67 |
| PF13207 | 0 | 0 | 3 | 0 | 0 | 0 | 3 | 1 | AAA domain | 7 |
| PF13229 | 0 | 0 | 5 | 0 | 0 | 0 | 3 | 0 | Right handed beta helix region | 8 |
| PF13231 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Dolichyl-phosphate-mannose-protein mannosyltransferase | 1 |
| PF13232 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | Complex1_LYR-like | 2 |
| PF13233 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Complex1_LYR-like | 2 |
| PF13234 | 0 | 0 | 2 | 1 | 0 | 0 | 2 | 0 | rRNA-processing arch domain | 5 |
| PF13236 | 0 | 0 | 1 | 1 | 0 | 0 | 3 | 0 | Clustered mitochondria | 5 |
| PF13238 | 0 | 0 | 3 | 0 | 0 | 0 | 3 | 0 | AAA domain | 6 |
| PF13242 | 0 | 3 | 4 | 2 | 1 | 2 | 5 | 4 | HAD-hyrolase-like | 21 |
| PF13246 | 0 | 1 | 4 | 1 | 0 | 1 | 3 | 1 | Putative hydrolase of sodium-potassium ATPase alpha subunit | 11 |
| PF13248 | 0 | 0 | 1 | 0 | 1 | 1 | 1 | 1 | zinc-ribbon domain | 5 |
| PF13249 | 0 | 0 | 4 | 2 | 0 | 0 | 3 | 0 | Prenyltransferase-like | 9 |
| PF13251 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Domain of unknown function (DUF4042) | 2 |
| PF13270 | 1 | 1 | 2 | 2 | 0 | 1 | 2 | 0 | Domain of unknown function (DUF4061) | 9 |
| PF13271 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Domain of unknown function (DUF4062) | 2 |
| PF13279 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 0 | Thioesterase-like superfamily | 5 |
| PF13281 | 0 | 0 | 3 | 0 | 0 | 0 | 2 | 0 | Domain of unknown function (DUF4071) | 5 |
| PF13289 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 0 | SIR2-like domain | 3 |
| PF13290 | 1 | 1 | 1 | 0 | 0 | 0 | 1 | 0 | Chitobiase/beta-hexosaminidase C-terminal domain | 4 |
| PF13293 | 0 | 0 | 3 | 0 | 0 | 0 | 2 | 0 | Domain of unknown function (DUF4074) | 5 |
| PF13297 | 0 | 0 | 2 | 1 | 0 | 0 | 1 | 1 | Telomere stability C-terminal | 5 |
| PF13299 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Cleavage and polyadenylation factor 2 C-terminal | 3 |
| PF13300 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Domain of unknown function (DUF4078) | 2 |
| PF13302 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Acetyltransferase (GNAT) domain | 2 |
| PF13306 | 1 | 3 | 9 | 3 | 1 | 1 | 14 | 0 | Leucine rich repeats (6 copies) | 32 |
| PF13307 | 0 | 0 | 4 | 0 | 0 | 0 | 4 | 0 | Helicase C-terminal domain | 8 |
| PF13324 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | Grap2 and cyclin-D-interacting | 4 |
| PF13325 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | N-terminal region of micro-spherule protein | 2 |
| PF13328 | 0 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | HD domain | 4 |
| PF13330 | 0 | 0 | 5 | 0 | 0 | 0 | 5 | 0 | Mucin-2 protein WxxW repeating region | 10 |
| PF13339 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Apoptosis antagonizing transcription factor | 2 |
| PF13341 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | RAG2 PHD domain | 2 |
| PF13344 | 0 | 3 | 5 | 2 | 1 | 2 | 5 | 4 | Haloacid dehalogenase-like hydrolase | 22 |
| PF13345 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Domain of unknown function (DUF4098) | 2 |
| PF13347 | 0 | 0 | 7 | 0 | 0 | 0 | 10 | 0 | MFS/sugar transport protein | 17 |
| PF13353 | 0 | 0 | 2 | 0 | 0 | 0 | 3 | 0 | 4Fe-4S single cluster domain | 5 |
| PF13358 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 0 | DDE superfamily endonuclease | 3 |
| PF13359 | 0 | 0 | 1 | 0 | 0 | 0 | 24 | 0 | DDE superfamily endonuclease | 25 |
| PF13360 | 1 | 1 | 2 | 1 | 0 | 1 | 2 | 1 | PQQ-like domain | 9 |
| PF13361 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 0 | UvrD-like helicase C-terminal domain | 3 |
| PF13364 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | Beta-galactosidase jelly roll domain | 1 |
| PF13365 | 1 | 2 | 6 | 1 | 0 | 0 | 23 | 0 | Trypsin-like peptidase domain | 33 |
| PF13369 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Transglutaminase-like superfamily | 3 |
| PF13371 | 1 | 1 | 10 | 2 | 1 | 3 | 7 | 1 | Tetratricopeptide repeat | 26 |
| PF13374 | 3 | 4 | 11 | 4 | 0 | 1 | 8 | 1 | Tetratricopeptide repeat | 32 |
| PF13378 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | Enolase C-terminal domain-like | 2 |
| PF13383 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Methyltransferase domain | 2 |
| PF13384 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | Homeodomain-like domain | 2 |
| PF13385 | 0 | 3 | 14 | 3 | 0 | 3 | 14 | 0 | Concanavalin A-like lectin/glucanases superfamily | 37 |
| PF13392 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | HNH endonuclease | 1 |
| PF13393 | 0 | 0 | 3 | 1 | 0 | 1 | 1 | 1 | Histidyl-tRNA synthetase | 7 |
| PF13394 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 4Fe-4S single cluster domain | 1 |
| PF13401 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | AAA domain | 2 |
| PF13402 | 0 | 0 | 3 | 1 | 0 | 0 | 3 | 0 | Peptidase M60-like family | 7 |
| PF13405 | 11 | 15 | 31 | 9 | 5 | 10 | 28 | 11 | EF-hand domain | 120 |
| PF13409 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | "Glutathione S-transferase, N-terminal domain" | 2 |
| PF13410 | 3 | 4 | 7 | 5 | 0 | 2 | 7 | 4 | "Glutathione S-transferase, C-terminal domain" | 32 |
| PF13414 | 11 | 20 | 76 | 20 | 8 | 16 | 73 | 10 | TPR repeat | 234 |
| PF13415 | 1 | 1 | 12 | 5 | 1 | 1 | 10 | 2 | "Galactose oxidase, central domain" | 33 |
| PF13417 | 2 | 5 | 14 | 9 | 0 | 5 | 11 | 8 | "Glutathione S-transferase, N-terminal domain" | 54 |
| PF13418 | 0 | 1 | 11 | 4 | 0 | 0 | 15 | 0 | "Galactose oxidase, central domain" | 31 |
| PF13419 | 0 | 1 | 7 | 4 | 0 | 1 | 5 | 2 | Haloacid dehalogenase-like hydrolase | 20 |
| PF13423 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Ubiquitin carboxyl-terminal hydrolase | 2 |
| PF13424 | 3 | 4 | 17 | 5 | 0 | 2 | 19 | 1 | Tetratricopeptide repeat | 51 |
| PF13426 | 2 | 2 | 11 | 2 | 0 | 1 | 13 | 0 | PAS domain | 31 |
| PF13428 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | Tetratricopeptide repeat | 1 |
| PF13429 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Tetratricopeptide repeat | 1 |
| PF13431 | 0 | 0 | 1 | 0 | 0 | 0 | 2 | 0 | Tetratricopeptide repeat | 3 |
| PF13432 | 2 | 2 | 5 | 1 | 0 | 0 | 2 | 0 | Tetratricopeptide repeat | 12 |
| PF13445 | 0 | 1 | 4 | 0 | 1 | 0 | 4 | 0 | RING-type zinc-finger | 10 |
| PF13450 | 0 | 3 | 7 | 3 | 0 | 4 | 6 | 3 | NAD(P)-binding Rossmann-like domain | 26 |
| PF13452 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 0 | N-terminal half of MaoC dehydratase | 5 |
| PF13460 | 0 | 1 | 2 | 0 | 0 | 4 | 3 | 2 | NADH(P)-binding | 12 |
| PF13465 | 0 | 0 | 496 | 24 | 0 | 1 | 600 | 2 | Zinc-finger double domain | 1123 |
| PF13469 | 0 | 0 | 2 | 0 | 1 | 1 | 2 | 0 | Sulfotransferase family | 6 |
| PF13472 | 1 | 2 | 2 | 1 | 0 | 2 | 2 | 2 | GDSL-like Lipase/Acylhydrolase family | 12 |
| PF13476 | 0 | 0 | 1 | 0 | 0 | 0 | 2 | 0 | AAA domain | 3 |
| PF13481 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | AAA domain | 1 |
| PF13483 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Beta-lactamase superfamily domain | 2 |
| PF13489 | 0 | 1 | 5 | 3 | 0 | 0 | 6 | 1 | Methyltransferase domain | 16 |
| PF13499 | 20 | 30 | 101 | 31 | 17 | 41 | 126 | 32 | EF-hand domain pair | 398 |
| PF13504 | 5 | 7 | 24 | 6 | 2 | 2 | 21 | 1 | Leucine rich repeat | 68 |
| PF13506 | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 0 | Glycosyl transferase family 21 | 3 |
| PF13507 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | CobB/CobQ-like glutamine amidotransferase domain | 2 |
| PF13508 | 0 | 0 | 3 | 1 | 0 | 0 | 2 | 0 | Acetyltransferase (GNAT) domain | 6 |
| PF13510 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 2Fe-2S iron-sulfur cluster binding domain | 5 |
| PF13513 | 1 | 3 | 10 | 2 | 0 | 2 | 6 | 0 | HEAT-like repeat | 24 |
| PF13516 | 6 | 10 | 66 | 10 | 5 | 9 | 184 | 5 | Leucine Rich repeat | 295 |
| PF13517 | 1 | 2 | 7 | 2 | 1 | 3 | 9 | 1 | "Repeat domain in Vibrio, Colwellia, Bradyrhizobium and Shewanella" | 26 |
| PF13519 | 2 | 3 | 14 | 4 | 0 | 2 | 11 | 0 | von Willebrand factor type A domain | 36 |
| PF13520 | 0 | 7 | 16 | 5 | 0 | 4 | 18 | 2 | Amino acid permease | 52 |
| PF13522 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | Glutamine amidotransferase domain | 1 |
| PF13532 | 0 | 0 | 7 | 1 | 0 | 0 | 8 | 0 | 2OG-Fe(II) oxygenase superfamily | 16 |
| PF13534 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 4Fe-4S dicluster domain | 6 |
| PF13535 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | ATP-grasp domain | 1 |
| PF13536 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Multidrug resistance efflux transporter | 2 |
| PF13537 | 0 | 0 | 2 | 0 | 0 | 0 | 3 | 0 | Glutamine amidotransferase domain | 5 |
| PF13538 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | UvrD-like helicase C-terminal domain | 2 |
| PF13540 | 1 | 1 | 5 | 0 | 1 | 1 | 2 | 0 | Regulator of chromosome condensation (RCC1) repeat | 11 |
| PF13543 | 0 | 0 | 1 | 0 | 1 | 1 | 4 | 0 | SAM like domain present in kinase suppressor RAS 1 | 7 |
| PF13553 | 0 | 0 | 2 | 0 | 0 | 0 | 6 | 1 | Function to find | 9 |
| PF13558 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | "Putative exonuclease SbcCD, C subunit" | 2 |
| PF13561 | 2 | 2 | 8 | 1 | 3 | 3 | 7 | 3 | Enoyl-(Acyl carrier protein) reductase | 29 |
| PF13564 | 1 | 1 | 2 | 1 | 0 | 0 | 1 | 0 | DoxX-like family | 6 |
| PF13570 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | PQQ-like domain | 2 |
| PF13574 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | Metallo-peptidase family M12B Reprolysin-like | 5 |
| PF13579 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | Glycosyl transferase 4-like domain | 6 |
| PF13589 | 0 | 2 | 10 | 3 | 0 | 3 | 10 | 2 | "Histidine kinase-, DNA gyrase B-, and HSP90-like ATPase" | 30 |
| PF13593 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | SBF-like CPA transporter family (DUF4137) | 1 |
| PF13594 | 0 | 2 | 2 | 1 | 0 | 2 | 2 | 1 | Amidohydrolase | 10 |
| PF13599 | 0 | 3 | 3 | 3 | 0 | 3 | 4 | 3 | Pentapeptide repeats (9 copies) | 19 |
| PF13602 | 0 | 3 | 3 | 0 | 0 | 2 | 2 | 2 | Zinc-binding dehydrogenase | 12 |
| PF13603 | 0 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | "Leucyl-tRNA synthetase, Domain 2" | 4 |
| PF13604 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | AAA domain | 2 |
| PF13606 | 0 | 0 | 5 | 1 | 0 | 0 | 3 | 0 | Ankyrin repeat | 9 |
| PF13613 | 0 | 0 | 0 | 0 | 0 | 0 | 9 | 0 | Helix-turn-helix of DDE superfamily endonuclease | 9 |
| PF13614 | 0 | 0 | 2 | 1 | 0 | 0 | 2 | 0 | AAA domain | 5 |
| PF13615 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Putative alanine racemase | 3 |
| PF13616 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | PPIC-type PPIASE domain | 5 |
| PF13620 | 1 | 4 | 10 | 1 | 0 | 3 | 11 | 2 | Carboxypeptidase regulatory-like domain | 32 |
| PF13621 | 0 | 0 | 8 | 2 | 0 | 0 | 7 | 0 | Cupin-like domain | 17 |
| PF13622 | 1 | 1 | 1 | 0 | 0 | 0 | 1 | 0 | Thioesterase-like superfamily | 4 |
| PF13625 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Helicase conserved C-terminal domain | 3 |
| PF13631 | 0 | 0 | 0 | 1 | 0 | 1 | 1 | 1 | Cytochrome b(N-terminal)/b6/petB | 4 |
| PF13634 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | Nucleoporin FG repeat region | 1 |
| PF13637 | 6 | 5 | 34 | 0 | 4 | 4 | 30 | 2 | Ankyrin repeats (many copies) | 85 |
| PF13638 | 0 | 0 | 4 | 1 | 0 | 0 | 3 | 0 | PIN domain | 8 |
| PF13639 | 1 | 3 | 70 | 13 | 4 | 8 | 72 | 1 | Ring finger domain | 172 |
| PF13640 | 0 | 0 | 9 | 2 | 0 | 0 | 8 | 0 | 2OG-Fe(II) oxygenase superfamily | 19 |
| PF13641 | 0 | 0 | 2 | 0 | 0 | 0 | 3 | 0 | Glycosyltransferase like family 2 | 5 |
| PF13646 | 3 | 5 | 15 | 4 | 3 | 6 | 12 | 5 | HEAT repeats | 53 |
| PF13650 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Aspartyl protease | 2 |
| PF13656 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | RNA polymerase Rpb3/Rpb11 dimerisation domain | 4 |
| PF13659 | 0 | 0 | 3 | 1 | 0 | 0 | 2 | 0 | Methyltransferase domain | 6 |
| PF13660 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 1 | Domain of unknown function (DUF4147) | 3 |
| PF13661 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 2OG-Fe(II) oxygenase superfamily | 1 |
| PF13664 | 0 | 1 | 1 | 0 | 0 | 0 | 1 | 0 | Domain of unknown function (DUF4149) | 3 |
| PF13669 | 0 | 0 | 1 | 1 | 0 | 0 | 3 | 1 | Glyoxalase/Bleomycin resistance protein/Dioxygenase superfamily | 6 |
| PF13671 | 2 | 2 | 9 | 2 | 1 | 1 | 7 | 4 | AAA domain | 28 |
| PF13673 | 0 | 0 | 1 | 0 | 0 | 1 | 2 | 0 | Acetyltransferase (GNAT) domain | 4 |
| PF13676 | 1 | 1 | 3 | 1 | 1 | 1 | 2 | 0 | TIR domain | 10 |
| PF13679 | 0 | 0 | 3 | 0 | 0 | 0 | 3 | 0 | Methyltransferase domain | 6 |
| PF13688 | 0 | 1 | 2 | 0 | 0 | 2 | 5 | 1 | Metallo-peptidase family M12 | 11 |
| PF13691 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | tRNase Z endonuclease | 3 |
| PF13692 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Glycosyl transferases group 1 | 2 |
| PF13695 | 0 | 0 | 6 | 0 | 0 | 0 | 3 | 1 | Zinc-binding domain | 10 |
| PF13696 | 0 | 0 | 1 | 0 | 0 | 0 | 2 | 0 | Zinc knuckle | 3 |
| PF13704 | 1 | 0 | 3 | 0 | 0 | 0 | 5 | 0 | Glycosyl transferase family 2 | 9 |
| PF13705 | 0 | 0 | 2 | 0 | 0 | 0 | 4 | 0 | TRC8 N-terminal domain | 6 |
| PF13716 | 6 | 6 | 14 | 4 | 5 | 5 | 18 | 4 | Divergent CRAL/TRIO domain | 62 |
| PF13718 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | GNAT acetyltransferase 2 | 3 |
| PF13725 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Possible tRNA binding domain | 3 |
| PF13733 | 0 | 0 | 7 | 0 | 0 | 0 | 7 | 1 | N-terminal region of glycosyl transferase group 7 | 15 |
| PF13738 | 0 | 0 | 3 | 0 | 0 | 0 | 3 | 0 | Pyridine nucleotide-disulphide oxidoreductase | 6 |
| PF13757 | 0 | 1 | 3 | 2 | 0 | 0 | 2 | 0 | Vault protein inter-alpha-trypsin domain | 8 |
| PF13764 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 0 | E3 ubiquitin-protein ligase UBR4 | 5 |
| PF13765 | 0 | 0 | 35 | 1 | 1 | 3 | 218 | 3 | SPRY-associated domain | 261 |
| PF13766 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 2-enoyl-CoA Hydratase C-terminal region | 6 |
| PF13768 | 0 | 2 | 8 | 3 | 1 | 2 | 10 | 1 | von Willebrand factor type A domain | 27 |
| PF13771 | 0 | 0 | 16 | 0 | 0 | 0 | 13 | 2 | PHD-like zinc-binding domain | 31 |
| PF13772 | 0 | 0 | 1 | 1 | 0 | 1 | 2 | 1 | AIG2-like family | 6 |
| PF13774 | 3 | 3 | 4 | 1 | 2 | 5 | 5 | 1 | Regulated-SNARE-like domain | 24 |
| PF13775 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Domain of unknown function (DUF4171) | 2 |
| PF13778 | 0 | 0 | 3 | 0 | 0 | 0 | 5 | 0 | Domain of unknown function (DUF4174) | 8 |
| PF13792 | 0 | 0 | 10 | 0 | 0 | 0 | 12 | 0 | Sulfate transporter N-terminal domain with GLY motif | 22 |
| PF13793 | 4 | 3 | 6 | 2 | 0 | 3 | 3 | 3 | N-terminal domain of ribose phosphate pyrophosphokinase | 24 |
| PF13802 | 0 | 2 | 4 | 1 | 0 | 2 | 5 | 1 | Galactose mutarotase-like | 15 |
| PF13806 | 0 | 0 | 1 | 0 | 0 | 0 | 3 | 0 | Rieske-like [2Fe-2S] domain | 4 |
| PF13812 | 0 | 1 | 3 | 1 | 0 | 0 | 1 | 0 | Pentatricopeptide repeat domain | 6 |
| PF13815 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | Iguana/Dzip1-like DAZ-interacting protein N-terminal | 4 |
| PF13820 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Putative nucleic acid-binding region | 2 |
| PF13821 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Domain of unknown function (DUF4187) | 2 |
| PF13831 | 0 | 0 | 11 | 1 | 0 | 0 | 12 | 1 | PHD-finger | 25 |
| PF13832 | 0 | 0 | 12 | 1 | 0 | 0 | 12 | 1 | PHD-zinc-finger like domain | 26 |
| PF13833 | 8 | 10 | 39 | 7 | 9 | 14 | 53 | 13 | EF-hand domain pair | 153 |
| PF13836 | 0 | 0 | 3 | 1 | 0 | 0 | 0 | 0 | Domain of unknown function (DUF4195) | 4 |
| PF13837 | 0 | 0 | 6 | 0 | 0 | 1 | 17 | 1 | Myb/SANT-like DNA-binding domain | 25 |
| PF13838 | 0 | 1 | 2 | 0 | 0 | 3 | 3 | 2 | Clathrin-H-link | 11 |
| PF13839 | 0 | 0 | 4 | 0 | 0 | 0 | 4 | 0 | GDSL/SGNH-like Acyl-Esterase family found in Pmr5 and Cas1p | 8 |
| PF13840 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 0 | ACT domain | 3 |
| PF13841 | 0 | 0 | 19 | 0 | 0 | 0 | 1 | 0 | Beta defensin | 20 |
| PF13842 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | DDE_Tnp_1-like zinc-ribbon | 1 |
| PF13843 | 0 | 0 | 2 | 1 | 0 | 0 | 7 | 0 | Transposase IS4 | 10 |
| PF13844 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 0 | Glycosyl transferase family 41 | 5 |
| PF13846 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Domain of unknown function (DUF4196) | 1 |
| PF13847 | 1 | 1 | 7 | 2 | 0 | 1 | 8 | 4 | Methyltransferase domain | 24 |
| PF13848 | 0 | 5 | 10 | 4 | 0 | 6 | 8 | 3 | Thioredoxin-like domain | 36 |
| PF13850 | 0 | 1 | 3 | 0 | 0 | 0 | 3 | 0 | Endoplasmic Reticulum-Golgi Intermediate Compartment (ERGIC) | 7 |
| PF13851 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Growth-arrest specific micro-tubule binding | 1 |
| PF13853 | 0 | 0 | 1030 | 3 | 0 | 0 | 19 | 0 | Olfactory receptor | 1052 |
| PF13854 | 0 | 0 | 6 | 1 | 1 | 1 | 6 | 2 | Kelch motif | 17 |
| PF13855 | 38 | 45 | 171 | 47 | 33 | 43 | 184 | 8 | Leucine rich repeat | 569 |
| PF13857 | 3 | 1 | 19 | 5 | 3 | 2 | 10 | 0 | Ankyrin repeats (many copies) | 43 |
| PF13862 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | p21-C-terminal region-binding protein | 2 |
| PF13863 | 0 | 0 | 4 | 0 | 0 | 0 | 3 | 0 | Domain of unknown function (DUF4200) | 7 |
| PF13864 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | Calmodulin-binding | 4 |
| PF13865 | 0 | 0 | 3 | 2 | 1 | 0 | 3 | 1 | C-terminal duplication domain of Friend of PRMT1 | 10 |
| PF13866 | 0 | 0 | 2 | 1 | 0 | 0 | 1 | 0 | SAP30 zinc-finger | 4 |
| PF13867 | 0 | 0 | 2 | 1 | 0 | 0 | 1 | 0 | Sin3 binding region of histone deacetylase complex subunit SAP30 | 4 |
| PF13868 | 0 | 0 | 5 | 2 | 1 | 0 | 4 | 0 | "Tumour suppressor, Mitostatin" | 12 |
| PF13869 | 1 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | Nucleotide hydrolase | 5 |
| PF13870 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | Domain of unknown function (DUF4201) | 4 |
| PF13871 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 0 | Helicase_C-like | 3 |
| PF13872 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 0 | P-loop containing NTP hydrolase pore-1 | 3 |
| PF13873 | 0 | 0 | 6 | 0 | 0 | 0 | 5 | 0 | Myb/SANT-like DNA-binding domain | 11 |
| PF13874 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | Nucleoporin complex subunit 54 | 2 |
| PF13877 | 0 | 0 | 3 | 1 | 0 | 0 | 2 | 0 | Potential Monad-binding region of RPAP3 | 6 |
| PF13878 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | zinc-finger of acetyl-transferase ESCO | 4 |
| PF13879 | 0 | 0 | 3 | 0 | 0 | 0 | 2 | 0 | KIAA1430 homologue | 5 |
| PF13880 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | ESCO1/2 acetyl-transferase | 4 |
| PF13881 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | Ubiquitin-2 like Rad60 SUMO-like | 6 |
| PF13882 | 1 | 4 | 4 | 1 | 1 | 5 | 6 | 4 | Bravo-like intracellular region | 26 |
| PF13883 | 0 | 0 | 2 | 1 | 0 | 0 | 1 | 0 | Pyridoxamine 5'-phosphate oxidase | 4 |
| PF13884 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 0 | Chaperone of endosialidase | 3 |
| PF13885 | 0 | 0 | 39 | 0 | 0 | 0 | 0 | 0 | "Keratin, high sulfur B2 protein" | 39 |
| PF13886 | 0 | 0 | 3 | 0 | 0 | 0 | 3 | 0 | Domain of unknown function (DUF4203) | 6 |
| PF13887 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 0 | Myelin gene regulatory factor -C-terminal domain 1 | 3 |
| PF13888 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 0 | Myelin gene regulatory factor C-terminal domain 2 | 3 |
| PF13889 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | Chromosome segregation during meiosis | 4 |
| PF13890 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | Rab3 GTPase-activating protein catalytic subunit | 2 |
| PF13891 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | Potential DNA-binding domain | 4 |
| PF13892 | 0 | 0 | 3 | 1 | 0 | 0 | 2 | 0 | DNA-binding domain | 6 |
| PF13893 | 4 | 2 | 31 | 7 | 1 | 2 | 28 | 8 | "RNA recognition motif. (a.k.a. RRM, RBD, or RNP domain)" | 83 |
| PF13894 | 0 | 0 | 134 | 10 | 0 | 0 | 150 | 1 | C2H2-type zinc finger | 295 |
| PF13895 | 20 | 51 | 155 | 26 | 25 | 56 | 242 | 20 | Immunoglobulin domain | 595 |
| PF13896 | 0 | 2 | 3 | 0 | 0 | 0 | 3 | 0 | Glycosyl-transferase for dystroglycan | 8 |
| PF13897 | 1 | 1 | 2 | 1 | 0 | 1 | 4 | 0 | Golgi-dynamics membrane-trafficking | 10 |
| PF13898 | 0 | 0 | 2 | 0 | 0 | 0 | 3 | 0 | Domain of unknown function (DUF4205) | 5 |
| PF13899 | 1 | 1 | 4 | 2 | 0 | 1 | 4 | 0 | Thioredoxin-like | 13 |
| PF13901 | 0 | 0 | 5 | 2 | 0 | 0 | 4 | 0 | Domain of unknown function (DUF4206) | 11 |
| PF13902 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | R3H-associated N-terminal domain | 2 |
| PF13903 | 2 | 2 | 14 | 0 | 1 | 1 | 20 | 0 | PMP-22/EMP/MP20/Claudin tight junction | 40 |
| PF13904 | 1 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | Domain of unknown function (DUF4207) | 3 |
| PF13905 | 0 | 0 | 4 | 2 | 0 | 1 | 4 | 0 | Thioredoxin-like | 11 |
| PF13906 | 0 | 3 | 6 | 1 | 0 | 2 | 7 | 0 | C-terminus of AA_permease | 19 |
| PF13907 | 0 | 0 | 3 | 0 | 0 | 0 | 1 | 0 | Domain of unknown function (DUF4208) | 4 |
| PF13908 | 2 | 4 | 9 | 1 | 4 | 3 | 12 | 0 | Wnt and FGF inhibitory regulator | 35 |
| PF13909 | 0 | 0 | 17 | 0 | 0 | 0 | 9 | 0 | C2H2-type zinc-finger domain | 26 |
| PF13910 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Domain of unknown function (DUF4209) | 1 |
| PF13911 | 2 | 2 | 3 | 2 | 0 | 1 | 4 | 2 | AhpC/TSA antioxidant enzyme | 16 |
| PF13912 | 0 | 0 | 101 | 7 | 0 | 1 | 226 | 1 | C2H2-type zinc finger | 336 |
| PF13913 | 1 | 1 | 5 | 1 | 1 | 1 | 4 | 1 | zinc-finger of a C2HC-type | 15 |
| PF13914 | 1 | 0 | 2 | 0 | 0 | 0 | 1 | 0 | Phostensin PP1-binding and SH3-binding region | 4 |
| PF13915 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | Domain of unknown function (DUF4210) | 4 |
| PF13916 | 1 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | "PP1-regulatory protein, Phostensin N-terminal" | 3 |
| PF13917 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | Zinc knuckle | 4 |
| PF13918 | 1 | 1 | 3 | 1 | 0 | 0 | 3 | 0 | PLD-like domain | 9 |
| PF13919 | 0 | 0 | 3 | 0 | 0 | 0 | 2 | 0 | Asx homology domain | 5 |
| PF13920 | 1 | 0 | 34 | 7 | 2 | 2 | 36 | 1 | "Zinc finger, C3HC4 type (RING finger)" | 83 |
| PF13921 | 1 | 0 | 7 | 0 | 0 | 0 | 11 | 1 | Myb-like DNA-binding domain | 20 |
| PF13922 | 0 | 0 | 3 | 0 | 0 | 0 | 2 | 0 | "PHD domain of transcriptional enhancer, Asx" | 5 |
| PF13923 | 2 | 2 | 19 | 3 | 1 | 1 | 34 | 0 | "Zinc finger, C3HC4 type (RING finger)" | 62 |
| PF13925 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | con80 domain of Katanin | 4 |
| PF13926 | 0 | 0 | 3 | 0 | 0 | 0 | 3 | 0 | Domain of unknown function (DUF4211) | 6 |
| PF13927 | 3 | 9 | 45 | 7 | 3 | 12 | 83 | 5 | Immunoglobulin domain | 167 |
| PF13931 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 1 | Kinesin-associated microtubule-binding | 3 |
| PF13932 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | GidA associated domain 3 | 2 |
| PF13934 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Nuclear pore complex assembly | 2 |
| PF13948 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | Domain of unknown function (DUF4215) | 3 |
| PF13949 | 0 | 1 | 2 | 0 | 0 | 2 | 2 | 1 | ALIX V-shaped domain binding to HIV | 8 |
| PF13950 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | UDP-glucose 4-epimerase C-term subunit | 3 |
| PF13959 | 1 | 0 | 4 | 1 | 1 | 1 | 4 | 0 | Domain of unknown function (DUF4217) | 12 |
| PF13964 | 0 | 0 | 11 | 2 | 1 | 1 | 12 | 0 | Kelch motif | 27 |
| PF13965 | 0 | 0 | 2 | 0 | 1 | 0 | 1 | 0 | dsRNA-gated channel SID-1 | 4 |
| PF13966 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | zinc-binding in reverse transcriptase | 1 |
| PF13967 | 1 | 2 | 3 | 1 | 0 | 0 | 2 | 0 | "Late exocytosis, associated with Golgi transport" | 9 |
| PF13971 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Meiosis-specific protein Mei4 | 2 |
| PF13975 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | gag-polyprotein putative aspartyl protease | 1 |
| PF14008 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Iron/zinc purple acid phosphatase-like protein C | 1 |
| PF14031 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | Putative serine dehydratase domain | 2 |
| PF14046 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Nuclear receptor repeat | 1 |
| PF14047 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | Dppa2/4 conserved region | 3 |
| PF14048 | 0 | 0 | 4 | 1 | 0 | 0 | 2 | 0 | C-terminal domain of methyl-CpG binding protein 2 and 3 | 7 |
| PF14049 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Dppa2/4 conserved region in higher vertebrates | 1 |
| PF14050 | 0 | 1 | 2 | 2 | 0 | 0 | 0 | 0 | N-terminal conserved domain of Nudc. | 5 |
| PF14051 | 0 | 0 | 3 | 2 | 0 | 0 | 3 | 0 | N-terminal domain of DPF2/REQ. | 8 |
| PF14061 | 0 | 0 | 3 | 0 | 0 | 0 | 3 | 0 | Polycomb-like MTF2 factor 2 | 6 |
| PF14073 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | Centrosome localisation domain of Cep57 | 4 |
| PF14075 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | Ubinuclein conserved middle domain | 4 |
| PF14077 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Alternative WD40 repeat motif | 2 |
| PF14138 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | Cytochrome c oxidase assembly protein COX16 | 7 |
| PF14160 | 1 | 0 | 4 | 1 | 1 | 0 | 4 | 0 | Centrosome-associated C terminus | 11 |
| PF14161 | 1 | 0 | 4 | 1 | 1 | 0 | 4 | 0 | Centrosome-associated N terminus | 11 |
| PF14186 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | 0 | Cytoskeletal adhesion | 4 |
| PF14204 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | Ribosomal L18 C-terminal region | 9 |
| PF14222 | 1 | 1 | 2 | 0 | 2 | 2 | 3 | 0 | Cell morphogenesis N-terminal | 11 |
| PF14225 | 1 | 1 | 2 | 0 | 2 | 2 | 3 | 0 | Cell morphogenesis C-terminal | 11 |
| PF14226 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | non-haem dioxygenase in morphine synthesis N-terminal | 1 |
| PF14228 | 1 | 1 | 2 | 0 | 2 | 2 | 3 | 0 | Cell morphogenesis central region | 11 |
| PF14237 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | Domain of unknown function (DUF4339) | 2 |
| PF14259 | 7 | 5 | 40 | 14 | 5 | 2 | 37 | 9 | "RNA recognition motif (a.k.a. RRM, RBD, or RNP domain)" | 119 |
| PF14260 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | C4-type zinc-finger of DNA polymerase delta | 4 |
| PF14291 | 1 | 1 | 3 | 0 | 0 | 0 | 2 | 0 | Domain of unknown function (DUF4371) | 7 |
| PF14304 | 0 | 0 | 2 | 1 | 0 | 0 | 1 | 0 | Transcription termination and cleavage factor C-terminal | 4 |
| PF14306 | 0 | 0 | 2 | 0 | 0 | 0 | 3 | 0 | PUA-like domain | 5 |
| PF14324 | 0 | 0 | 4 | 1 | 0 | 1 | 5 | 0 | PINIT domain | 11 |
| PF14327 | 0 | 0 | 2 | 1 | 0 | 0 | 1 | 0 | Hinge domain of cleavage stimulation factor subunit 2 | 4 |
| PF14336 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Domain of unknown function (DUF4392) | 3 |
| PF14360 | 0 | 0 | 3 | 0 | 0 | 0 | 5 | 0 | PAP2 superfamily C-terminal | 8 |
| PF14369 | 0 | 1 | 2 | 2 | 0 | 0 | 1 | 0 | zinc-finger | 6 |
| PF14370 | 0 | 0 | 2 | 0 | 0 | 0 | 4 | 1 | C-terminal topoisomerase domain | 7 |
| PF14374 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 60S ribosomal protein L4 C-terminal domain | 8 |
| PF14377 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | Domain of unknown function (DUF4414) | 5 |
| PF14381 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Ethylene-responsive protein kinase Le-CTR1 | 2 |
| PF14382 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | Exosome complex exonuclease RRP4 N-terminal region | 4 |
| PF14392 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 2 | Zinc knuckle | 6 |
| PF14396 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | "Cystic fibrosis TM conductance regulator (CFTR), regulator domain" | 2 |
| PF14413 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Thg1 C terminal domain | 2 |
| PF14429 | 7 | 5 | 10 | 0 | 6 | 5 | 10 | 0 | C2 domain in Dock180 and Zizimin proteins | 43 |
| PF14438 | 1 | 0 | 2 | 1 | 0 | 0 | 2 | 1 | Ataxin 2 SM domain | 7 |
| PF14443 | 0 | 0 | 2 | 1 | 0 | 0 | 2 | 0 | DBC1 | 5 |
| PF14444 | 0 | 0 | 2 | 1 | 0 | 0 | 2 | 0 | S1-like | 5 |
| PF14465 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | NFRKB Winged Helix-like | 2 |
| PF14469 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 28 kDa A-kinase anchor | 1 |
| PF14473 | 0 | 0 | 2 | 0 | 0 | 0 | 4 | 0 | RD3 protein | 6 |
| PF14478 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Domain of unknown function (DUF4430) | 2 |
| PF14484 | 0 | 0 | 2 | 0 | 0 | 1 | 128 | 2 | Fish-specific NACHT associated domain | 133 |
| PF14492 | 0 | 2 | 5 | 2 | 1 | 3 | 7 | 3 | "Elongation Factor G, domain II" | 23 |
| PF14493 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Helix-turn-helix domain | 1 |
| PF14497 | 3 | 5 | 6 | 2 | 1 | 4 | 6 | 4 | "Glutathione S-transferase, C-terminal domain" | 31 |
| PF14500 | 0 | 0 | 1 | 1 | 0 | 1 | 1 | 0 | Dos2-interacting transcription regulator of RNA-Pol-II | 4 |
| PF14513 | 2 | 2 | 3 | 1 | 1 | 2 | 3 | 1 | Diacylglycerol kinase N-terminus | 15 |
| PF14520 | 0 | 0 | 2 | 0 | 0 | 0 | 3 | 0 | Helix-hairpin-helix domain | 5 |
| PF14523 | 2 | 2 | 3 | 1 | 1 | 3 | 3 | 1 | Syntaxin-like protein | 16 |
| PF14529 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | Endonuclease-reverse transcriptase | 2 |
| PF14533 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | Ubiquitin-specific protease C-terminal | 5 |
| PF14538 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Raptor N-terminal CASPase like domain | 1 |
| PF14541 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Xylanase inhibitor C-terminal | 1 |
| PF14542 | 0 | 0 | 1 | 0 | 0 | 0 | 2 | 0 | GCN5-related N-acetyl-transferase | 3 |
| PF14545 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 0 | "Dof, BCAP, and BANK (DBB) motif," | 3 |
| PF14551 | 0 | 0 | 7 | 0 | 0 | 0 | 8 | 0 | MCM N-terminal domain | 15 |
| PF14554 | 0 | 0 | 2 | 1 | 0 | 0 | 1 | 0 | VEGF heparin-binding domain | 4 |
| PF14555 | 0 | 2 | 11 | 4 | 0 | 2 | 11 | 1 | UBA-like domain | 31 |
| PF14559 | 1 | 1 | 7 | 3 | 1 | 1 | 3 | 1 | Tetratricopeptide repeat | 18 |
| PF14560 | 0 | 0 | 4 | 2 | 0 | 1 | 3 | 1 | Ubiquitin-like domain | 11 |
| PF14565 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | Interleukin 22 IL-10-related T-cell-derived-inducible factor | 2 |
| PF14566 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Inositol hexakisphosphate | 2 |
| PF14570 | 0 | 0 | 1 | 0 | 0 | 0 | 2 | 0 | RING/Ubox like zinc-binding domain | 3 |
| PF14572 | 4 | 3 | 6 | 2 | 0 | 3 | 3 | 3 | Phosphoribosyl synthetase-associated domain | 24 |
| PF14575 | 1 | 3 | 14 | 0 | 0 | 10 | 17 | 1 | Ephrin type-A receptor 2 transmembrane domain | 46 |
| PF14578 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 1 | Elongation factor Tu domain 4 | 3 |
| PF14580 | 1 | 2 | 15 | 5 | 0 | 1 | 10 | 4 | Leucine-rich repeat | 38 |
| PF14586 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | "Class I Histocompatibility antigen, NKG2D ligand, domains 1 and 2" | 2 |
| PF14593 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 0 | PH domain | 8 |
| PF14596 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | STAT6 C-terminal | 1 |
| PF14598 | 0 | 0 | 15 | 2 | 0 | 0 | 20 | 0 | PAS domain | 37 |
| PF14599 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Zinc-ribbon | 3 |
| PF14603 | 0 | 0 | 3 | 0 | 0 | 0 | 3 | 0 | Helically-extended SH3 domain | 6 |
| PF14604 | 22 | 25 | 88 | 31 | 24 | 32 | 100 | 17 | Variant SH3 domain | 339 |
| PF14608 | 0 | 0 | 2 | 1 | 0 | 0 | 1 | 0 | Zinc finger C-x8-C-x5-C-x3-H type | 4 |
| PF14609 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | gamma-Tubulin ring complex non-core subunit mod21 | 1 |
| PF14617 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | U3-containing 90S pre-ribosomal complex subunit | 2 |
| PF14619 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 1 | "Snf2-ATP coupling, chromatin remodelling complex" | 5 |
| PF14621 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | RFX5 DNA-binding domain | 1 |
| PF14622 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Ribonuclease-III-like | 2 |
| PF14629 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Origin recognition complex (ORC) subunit 4 C-terminus | 1 |
| PF14630 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Origin recognition complex (ORC) subunit 5 C-terminus | 2 |
| PF14631 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Fanconi anaemia protein FancD2 nuclease | 2 |
| PF14632 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Acidic N-terminal SPT6 | 2 |
| PF14633 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | SH2 domain | 2 |
| PF14634 | 2 | 2 | 9 | 3 | 2 | 2 | 6 | 1 | zinc-RING finger domain | 27 |
| PF14635 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Helix-hairpin-helix motif | 2 |
| PF14636 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 0 | Folliculin-interacting protein N-terminus | 3 |
| PF14637 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 0 | Folliculin-interacting protein middle domain | 3 |
| PF14638 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 0 | Folliculin-interacting protein C-terminus | 3 |
| PF14639 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Holliday-junction resolvase-like of SPT6 | 2 |
| PF14640 | 0 | 0 | 1 | 0 | 1 | 1 | 1 | 0 | Transmembrane protein 223 | 4 |
| PF14641 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Helix-turn-helix DNA-binding domain of SPT6 | 2 |
| PF14642 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | FAM47 family | 3 |
| PF14643 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Domain of unknown function (DUF4455) | 2 |
| PF14644 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Domain of unknown function (DUF4456) | 2 |
| PF14645 | 1 | 0 | 2 | 0 | 0 | 0 | 1 | 0 | Chibby family | 4 |
| PF14646 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | MYCBP-associated protein family | 2 |
| PF14647 | 0 | 0 | 1 | 0 | 1 | 1 | 1 | 1 | FAM91 N-terminus | 5 |
| PF14648 | 0 | 0 | 1 | 0 | 1 | 1 | 1 | 1 | FAM91 C-terminus | 5 |
| PF14649 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Spatacsin C-terminus | 2 |
| PF14650 | 0 | 0 | 8 | 0 | 0 | 0 | 0 | 0 | FAM75 family | 8 |
| PF14651 | 0 | 0 | 2 | 0 | 0 | 0 | 5 | 2 | Lipocalin / cytosolic fatty-acid binding protein family | 9 |
| PF14652 | 0 | 0 | 1 | 0 | 0 | 0 | 3 | 0 | Domain of unknown function (DUF4457) | 4 |
| PF14653 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Insulin growth factor-like family | 1 |
| PF14654 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | "Mucin, catalytic, TM and cytoplasmic tail region" | 1 |
| PF14655 | 0 | 1 | 1 | 0 | 0 | 0 | 1 | 0 | Rab3 GTPase-activating protein regulatory subunit N-terminus | 3 |
| PF14656 | 0 | 1 | 1 | 0 | 0 | 0 | 1 | 0 | Rab3 GTPase-activating protein regulatory subunit C-terminus | 3 |
| PF14658 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | EF-hand domain | 2 |
| PF14661 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | HAUS augmin-like complex subunit 6 N-terminus | 1 |
| PF14662 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Coiled-coil region of CCDC155 | 2 |
| PF14663 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Rapamycin-insensitive companion of mTOR RasGEF_N domain | 2 |
| PF14664 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | "Rapamycin-insensitive companion of mTOR, N-term" | 2 |
| PF14665 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | "Rapamycin-insensitive companion of mTOR, phosphorylation-site" | 2 |
| PF14666 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | "Rapamycin-insensitive companion of mTOR, middle domain" | 2 |
| PF14668 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | "Rapamycin-insensitive companion of mTOR, domain 5" | 2 |
| PF14669 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Putative aspartate racemase | 1 |
| PF14670 | 3 | 5 | 41 | 3 | 1 | 4 | 41 | 4 | Coagulation Factor Xa inhibitory site | 102 |
| PF14671 | 0 | 0 | 2 | 0 | 0 | 0 | 3 | 0 | "Dual specificity protein phosphatase, N-terminal half" | 5 |
| PF14672 | 0 | 0 | 20 | 0 | 0 | 0 | 0 | 0 | Late cornified envelope | 20 |
| PF14674 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | FANCI solenoid 1 cap | 2 |
| PF14675 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | FANCI solenoid 1 | 2 |
| PF14676 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | FANCI solenoid 2 | 2 |
| PF14677 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | FANCI solenoid 3 | 2 |
| PF14678 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | FANCI solenoid 4 | 2 |
| PF14679 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | FANCI helical domain 1 | 2 |
| PF14680 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | FANCI helical domain 2 | 2 |
| PF14681 | 0 | 0 | 2 | 0 | 0 | 0 | 4 | 0 | Uracil phosphoribosyltransferase | 6 |
| PF14687 | 0 | 0 | 1 | 0 | 1 | 1 | 1 | 0 | Domain of unknown function (DUF4460) | 4 |
| PF14688 | 0 | 0 | 1 | 0 | 1 | 1 | 1 | 0 | Domain of unknown function (DUF4461) | 4 |
| PF14691 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | "Dihydroprymidine dehydrogenase domain II, 4Fe-4S cluster" | 3 |
| PF14694 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Lines N-terminus | 2 |
| PF14695 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Lines C-terminus | 2 |
| PF14697 | 0 | 0 | 1 | 1 | 0 | 0 | 2 | 0 | 4Fe-4S dicluster domain | 4 |
| PF14698 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 1 | Argininosuccinate lyase C-terminal | 3 |
| PF14699 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | N-terminal domain from the human glycogen debranching enzyme | 3 |
| PF14700 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | DNA-directed RNA polymerase N-terminal | 2 |
| PF14701 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | glucanotransferase domain of human glycogen debranching enzyme | 3 |
| PF14702 | 0 | 1 | 2 | 0 | 0 | 1 | 0 | 0 | central domain of human glycogen debranching enzyme | 4 |
| PF14703 | 1 | 2 | 3 | 1 | 0 | 0 | 2 | 0 | Domain of unknown function (DUF4463) | 9 |
| PF14704 | 0 | 0 | 1 | 0 | 0 | 0 | 3 | 0 | Dermatopontin | 4 |
| PF14705 | 0 | 0 | 2 | 0 | 0 | 0 | 4 | 0 | Costars | 6 |
| PF14707 | 0 | 1 | 3 | 0 | 0 | 0 | 4 | 0 | C-terminal region of aryl-sulfatase | 8 |
| PF14709 | 0 | 0 | 4 | 0 | 0 | 0 | 3 | 0 | double strand RNA binding domain from DEAD END PROTEIN 1 | 7 |
| PF14712 | 0 | 1 | 2 | 2 | 0 | 0 | 2 | 0 | Snapin/Pallidin | 7 |
| PF14713 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Domain of unknown function (DUF4464) | 2 |
| PF14716 | 0 | 0 | 4 | 0 | 0 | 0 | 3 | 0 | Helix-hairpin-helix domain | 7 |
| PF14719 | 0 | 0 | 3 | 0 | 0 | 0 | 2 | 0 | Phosphotyrosine interaction domain (PTB/PID) | 5 |
| PF14721 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | "Apoptosis-inducing factor, mitochondrion-associated, C-term" | 7 |
| PF14722 | 0 | 0 | 3 | 0 | 0 | 0 | 3 | 0 | Ki-ras-induced actin-interacting protein-IP3R-interacting domain | 6 |
| PF14723 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | Sperm-specific antigen 2 C-terminus | 4 |
| PF14724 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Mitochondrial-associated sphingomyelin phosphodiesterase | 2 |
| PF14726 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | "Rotatin, an armadillo repeat protein, centriole functioning" | 2 |
| PF14727 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | PTHB1 N-terminus | 3 |
| PF14728 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | PTHB1 C-terminus | 3 |
| PF14732 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | Ubiquitin/SUMO-activating enzyme ubiquitin-like domain | 2 |
| PF14735 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | HAUS augmin-like complex subunit 4 | 2 |
| PF14736 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Protein N-terminal asparagine amidohydrolase | 1 |
| PF14737 | 0 | 0 | 1 | 0 | 0 | 0 | 2 | 0 | Domain of unknown function (DUF4470) | 3 |
| PF14738 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | "Solute carrier (proton/amino acid symporter), TRAMD3 or PAT1" | 2 |
| PF14739 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Domain of unknown function (DUF4472) | 2 |
| PF14740 | 0 | 0 | 1 | 0 | 0 | 0 | 2 | 0 | Domain of unknown function (DUF4471) | 3 |
| PF14744 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | WASH complex subunit 7 | 6 |
| PF14745 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | "WASH complex subunit 7, N-terminal" | 6 |
| PF14746 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | "WASH complex subunit 7, C-terminal" | 6 |
| PF14748 | 2 | 2 | 3 | 3 | 3 | 3 | 1 | 1 | Pyrroline-5-carboxylate reductase dimerisation | 18 |
| PF14749 | 0 | 1 | 2 | 0 | 0 | 1 | 1 | 1 | Acyl-coenzyme A oxidase N-terminal | 6 |
| PF14750 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Integrator complex subunit 2 | 2 |
| PF14752 | 0 | 0 | 2 | 0 | 0 | 1 | 2 | 0 | Retinol binding protein receptor | 5 |
| PF14753 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 0 | Domain of unknown function (DUF4475) | 3 |
| PF14759 | 1 | 1 | 1 | 0 | 1 | 1 | 3 | 0 | Reductase C-terminal | 8 |
| PF14761 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Hermansky-Pudlak syndrome 3 | 2 |
| PF14762 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | "Hermansky-Pudlak syndrome 3, middle region" | 2 |
| PF14763 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | "Hermansky-Pudlak syndrome 3, C-terminal" | 2 |
| PF14764 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | "AP-5 complex subunit, vesicle trafficking" | 2 |
| PF14765 | 0 | 1 | 1 | 1 | 0 | 1 | 2 | 0 | Polyketide synthase dehydratase | 6 |
| PF14766 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Replication protein A interacting N-terminal | 3 |
| PF14767 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Replication protein A interacting middle | 3 |
| PF14768 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Replication protein A interacting C-terminal | 3 |
| PF14769 | 0 | 0 | 3 | 0 | 0 | 0 | 3 | 0 | Flagellar C1a complex subunit C1a-32 | 6 |
| PF14770 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Transmembrane protein 18 | 2 |
| PF14771 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Domain of unknown function (DUF4476) | 2 |
| PF14772 | 0 | 0 | 1 | 0 | 0 | 0 | 2 | 0 | Sperm tail | 3 |
| PF14773 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | "Helicase-associated putative binding domain, C-terminal" | 2 |
| PF14774 | 0 | 1 | 2 | 0 | 0 | 0 | 1 | 0 | FAM177 family | 4 |
| PF14775 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | Sperm tail C-terminal domain | 1 |
| PF14776 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Cation-channel complex subunit UNC-79 | 2 |
| PF14777 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Cilia BBSome complex subunit 10 | 2 |
| PF14778 | 0 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | Olfactory receptor 4-like | 4 |
| PF14779 | 1 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Ciliary BBSome complex subunit 1 | 3 |
| PF14780 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 1 | Domain of unknown function (DUF4477) | 3 |
| PF14781 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | "Ciliary BBSome complex subunit 2, N-terminal" | 3 |
| PF14782 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | "Ciliary BBSome complex subunit 2, C-terminal" | 3 |
| PF14783 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | "Ciliary BBSome complex subunit 2, middle region" | 3 |
| PF14784 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | C-terminal domain of the ECSIT protein | 6 |
| PF14788 | 0 | 1 | 3 | 1 | 1 | 2 | 6 | 0 | EF hand | 14 |
| PF14791 | 0 | 0 | 4 | 0 | 0 | 0 | 2 | 0 | DNA polymerase beta thumb | 6 |
| PF14792 | 0 | 0 | 4 | 0 | 0 | 0 | 2 | 0 | DNA polymerase beta palm | 6 |
| PF14796 | 1 | 1 | 2 | 0 | 0 | 1 | 0 | 0 | Clathrin-adaptor complex-3 beta-1 subunit C-terminal | 5 |
| PF14797 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | "Serine-rich region of AP3B1, clathrin-adaptor complex" | 1 |
| PF14798 | 0 | 0 | 5 | 0 | 0 | 0 | 9 | 0 | Calcium homeostasis modulator | 14 |
| PF14799 | 1 | 1 | 2 | 0 | 0 | 0 | 2 | 0 | FAM195 family | 6 |
| PF14800 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Domain of unknown function (DUF4481) | 2 |
| PF14802 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | 0 | TMEM192 family | 4 |
| PF14806 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | Coatomer beta subunit appendage platform | 6 |
| PF14807 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Adaptin AP4 complex epsilon appendage platform | 1 |
| PF14808 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | TMEM164 family | 1 |
| PF14811 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Protein of unknown function TPD sequence-motif | 2 |
| PF14813 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | NADH dehydrogenase 1 beta subcomplex subunit 2 | 5 |
| PF14815 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | NUDIX domain | 2 |
| PF14816 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 0 | "Family of unknown function, FAM178" | 3 |
| PF14817 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | HAUS augmin-like complex subunit 5 | 2 |
| PF14818 | 0 | 0 | 3 | 1 | 0 | 0 | 3 | 0 | Domain of unknown function (DUF4482) | 7 |
| PF14820 | 0 | 0 | 9 | 0 | 0 | 0 | 0 | 0 | Small proline-rich 2 | 9 |
| PF14821 | 0 | 1 | 2 | 2 | 0 | 0 | 1 | 0 | Threonine synthase N terminus | 6 |
| PF14822 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | Vasohibin | 4 |
| PF14825 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Domain of unknown function (DUF4483) | 2 |
| PF14826 | 0 | 0 | 1 | 0 | 1 | 1 | 1 | 1 | FACT complex subunit SPT16 N-terminal lobe domain | 5 |
| PF14828 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Amnionless | 2 |
| PF14833 | 0 | 1 | 2 | 2 | 0 | 1 | 1 | 1 | NAD-binding of NADP-dependent 3-hydroxyisobutyrate dehydrogenase | 8 |
| PF14835 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | zf-RING of BARD1-type protein | 1 |
| PF14836 | 0 | 1 | 5 | 1 | 0 | 0 | 3 | 0 | Ubiquitin-like domain | 10 |
| PF14837 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Integrator complex subunit 5 N-terminus | 3 |
| PF14838 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Integrator complex subunit 5 C-terminus | 3 |
| PF14839 | 0 | 0 | 2 | 1 | 0 | 0 | 3 | 0 | DOR family | 6 |
| PF14843 | 0 | 2 | 6 | 2 | 1 | 1 | 10 | 0 | Growth factor receptor domain IV | 22 |
| PF14844 | 0 | 2 | 7 | 2 | 0 | 2 | 6 | 0 | PH domain associated with Beige/BEACH | 19 |
| PF14845 | 1 | 2 | 2 | 1 | 0 | 1 | 2 | 1 | beta-acetyl hexosaminidase like | 10 |
| PF14846 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Domain of unknown function (DUF4485) | 2 |
| PF14851 | 1 | 2 | 4 | 1 | 0 | 0 | 3 | 0 | FAM176 family | 11 |
| PF14852 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | Fis1 N-terminal tetratricopeptide repeat | 5 |
| PF14853 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | Fis1 C-terminal tetratricopeptide repeat | 5 |
| PF14854 | 0 | 0 | 3 | 1 | 0 | 0 | 2 | 0 | Leucine rich adaptor protein | 6 |
| PF14857 | 0 | 0 | 2 | 0 | 0 | 0 | 5 | 0 | TMEM151 family | 7 |
| PF14862 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | Big defensin | 1 |
| PF14863 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | Alkyl sulfatase dimerisation | 1 |
| PF14868 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Domain of unknown function (DUF4487) | 2 |
| PF14874 | 0 | 0 | 3 | 1 | 0 | 1 | 1 | 0 | Flagellar-associated PapD-like | 6 |
| PF14875 | 0 | 0 | 2 | 1 | 0 | 0 | 2 | 0 | "N-term cysteine-rich ER, FAM69" | 5 |
| PF14878 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Death-like domain of SPT6 | 2 |
| PF14880 | 1 | 0 | 1 | 0 | 0 | 1 | 1 | 0 | Cytochrome oxidase c assembly | 4 |
| PF14881 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Tubulin domain | 2 |
| PF14886 | 0 | 0 | 1 | 0 | 0 | 0 | 2 | 0 | FAM183A and FAM183B related | 3 |
| PF14887 | 1 | 0 | 1 | 0 | 1 | 1 | 2 | 0 | HMG (high mobility group) box 5 | 6 |
| PF14892 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 0 | Domain of unknown function (DUF4490) | 3 |
| PF14893 | 0 | 0 | 10 | 1 | 0 | 0 | 0 | 0 | PNMA | 11 |
| PF14895 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Protein phosphatase 1 inhibitor | 1 |
| PF14901 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Cleavage inducing molecular chaperone | 1 |
| PF14904 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Family of unknown function | 2 |
| PF14906 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Domain of unknown function (DUF4495) | 2 |
| PF14908 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Domain of unknown function (DUF4496) | 1 |
| PF14909 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | Spermatogenesis-assoc protein 6 | 4 |
| PF14910 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | "S-phase genomic integrity recombination mediator, N-terminal" | 2 |
| PF14911 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | "S-phase genomic integrity recombination mediator, C-terminal" | 2 |
| PF14912 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 0 | Testicular haploid expressed repeat | 3 |
| PF14913 | 0 | 1 | 1 | 0 | 0 | 0 | 1 | 0 | DPCD protein family | 3 |
| PF14914 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | LRRC37A/B like protein 1 C-terminal domain | 1 |
| PF14915 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | CCDC144C protein coiled-coil region | 1 |
| PF14916 | 1 | 0 | 3 | 1 | 0 | 0 | 4 | 0 | Coiled-coil domain of unknown function | 9 |
| PF14917 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | "Coiled coil protein 74, C terminal" | 2 |
| PF14918 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | MDM2-binding | 2 |
| PF14919 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | MDM2-binding | 2 |
| PF14920 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | MDM2-binding | 2 |
| PF14921 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Adenomatosis polyposis coli down-regulated 1 | 2 |
| PF14922 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | Protein of unknown function | 2 |
| PF14923 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Coiled-coil protein 142 | 1 |
| PF14924 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Protein of unknown function (DUF4497) | 1 |
| PF14925 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Domain of unknown function | 1 |
| PF14926 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Domain of unknown function (DUF4498) | 3 |
| PF14927 | 0 | 0 | 4 | 0 | 0 | 1 | 5 | 0 | Neurensin | 10 |
| PF14929 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | TAF RNA Polymerase I subunit A | 1 |
| PF14931 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | "Intraflagellar transport complex B, subunit 20" | 3 |
| PF14932 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | HAUS augmin-like complex subunit 3 | 2 |
| PF14933 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | CEP19-like protein | 2 |
| PF14934 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | Domain of unknown function (DUF4499) | 3 |
| PF14935 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Transmembrane protein 138 | 2 |
| PF14936 | 1 | 1 | 1 | 1 | 0 | 2 | 2 | 0 | Tumour protein p53-inducible protein 11 | 8 |
| PF14937 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Domain of unknown function (DUF4500) | 2 |
| PF14938 | 0 | 3 | 3 | 3 | 0 | 6 | 7 | 6 | "Soluble NSF attachment protein, SNAP" | 28 |
| PF14939 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | "DDB1-and CUL4-substrate receptor 15, WD repeat" | 2 |
| PF14940 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | Transmembrane 219 | 4 |
| PF14941 | 0 | 0 | 1 | 0 | 0 | 0 | 2 | 0 | "Transcriptional regulator, Out at first" | 3 |
| PF14942 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | "Organelle biogenesis, Muted-like protein" | 1 |
| PF14943 | 0 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | Mitochondrial ribosome subunit S26 | 4 |
| PF14944 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | Tongue Cancer Chemotherapy Resistant Protein 1 | 2 |
| PF14945 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | "Normal lung function maintenance, Low in Lung Cancer 1 protein" | 2 |
| PF14946 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Domain of unknown function (DUF4501) | 2 |
| PF14948 | 0 | 1 | 2 | 0 | 4 | 4 | 4 | 1 | RESP18 domain | 16 |
| PF14949 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 0 | ARF7 effector protein C-terminus | 3 |
| PF14950 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Domain of unknown function (DUF4502) | 1 |
| PF14951 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Domain of unknown function (DUF4503) | 1 |
| PF14952 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | "Putative treble-clef, zinc-finger, Zn-binding" | 2 |
| PF14953 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Domain of unknown function (DUF4504) | 2 |
| PF14954 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 1 | Limb expression 1 | 5 |
| PF14955 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | Mitochondrial ribosome subunit S24 | 2 |
| PF14956 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Domain of unknown function (DUF4505) | 2 |
| PF14957 | 2 | 2 | 6 | 3 | 0 | 1 | 7 | 0 | Cdc42 effector | 21 |
| PF14958 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Domain of unknown function (DUF4506) | 2 |
| PF14959 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | gamma-Secretase-activating protein C-term | 2 |
| PF14960 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | ATP synthase regulation | 6 |
| PF14961 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Broad-minded protein | 2 |
| PF14962 | 1 | 1 | 2 | 1 | 1 | 2 | 2 | 1 | Mitochondria Localisation Sequence | 11 |
| PF14963 | 0 | 1 | 1 | 1 | 0 | 1 | 0 | 0 | Calcium signal-modulating cyclophilin ligand | 4 |
| PF14964 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Domain of unknown function (DUF4507) | 3 |
| PF14965 | 2 | 2 | 2 | 1 | 1 | 1 | 2 | 1 | Negative regulator of p53/TP53 | 12 |
| PF14966 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | DNA repair REX1-B | 2 |
| PF14967 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | FAM70 protein | 4 |
| PF14968 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Coiled coil protein 84 | 2 |
| PF14969 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Domain of unknown function (DUF4508) | 2 |
| PF14970 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Domain of unknown function (DUF4509) | 2 |
| PF14971 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Domain of unknown function (DUF4510) | 2 |
| PF14972 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | Mitochondrial morphogenesis regulator | 7 |
| PF14973 | 0 | 0 | 1 | 0 | 0 | 0 | 3 | 0 | TERF1-interacting nuclear factor 2 N-terminus | 4 |
| PF14974 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | Domain of unknown function (DUF4511) | 7 |
| PF14975 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Domain of unknown function (DUF4512) | 1 |
| PF14976 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | FAM72 protein | 2 |
| PF14977 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | FAM194 protein | 2 |
| PF14978 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Mitochondrial ribosome protein 63 | 2 |
| PF14979 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | Transmembrane 52 | 3 |
| PF14980 | 0 | 0 | 1 | 0 | 1 | 1 | 1 | 0 | TIP39 peptide | 4 |
| PF14981 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | FAM165 family | 1 |
| PF14982 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | UPF0731 family | 2 |
| PF14983 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Domain of unknown function (DUF4513) | 1 |
| PF14984 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | CD24 protein | 1 |
| PF14985 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | TM140 protein family | 1 |
| PF14986 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Domain of unknown function (DUF4514) | 1 |
| PF14987 | 0 | 1 | 1 | 0 | 0 | 0 | 1 | 1 | NADH dehydrogenase 1 alpha subcomplex subunit 3 | 4 |
| PF14988 | 0 | 0 | 6 | 0 | 0 | 0 | 2 | 1 | Domain of unknown function (DUF4515) | 9 |
| PF14989 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Coiled-coil domain containing 32 | 2 |
| PF14990 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Domain of unknown function (DUF4516) | 2 |
| PF14991 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Protein melan-A | 1 |
| PF14992 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | TMCO5 family | 3 |
| PF14993 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Neuropeptide S precursor protein | 1 |
| PF14994 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Testis-specific gene 13 protein | 1 |
| PF14995 | 0 | 0 | 1 | 0 | 0 | 0 | 2 | 0 | Transmembrane protein | 3 |
| PF14996 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Retinal Maintenance | 1 |
| PF14997 | 1 | 1 | 2 | 0 | 0 | 0 | 5 | 0 | CECR6/TMEM121 family | 9 |
| PF14998 | 0 | 0 | 3 | 0 | 0 | 0 | 3 | 0 | Transcription Regulator | 6 |
| PF14999 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | "Shadow of prion protein, neuroprotective" | 1 |
| PF15000 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Tumour suppressor candidate 2 | 3 |
| PF15001 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | AP-5 complex subunit sigma-1 | 2 |
| PF15002 | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 0 | "ERK and JNK pathways, inhibitor" | 3 |
| PF15003 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | HAUS augmin-like complex subunit 2 | 2 |
| PF15004 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Myeloma-overexpressed-like | 3 |
| PF15005 | 0 | 0 | 4 | 0 | 0 | 0 | 0 | 0 | Izumo sperm-egg fusion | 4 |
| PF15006 | 0 | 0 | 1 | 1 | 0 | 0 | 2 | 2 | Domain of unknown function (DUF4517) | 6 |
| PF15007 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | "Centrosomal spindle body, CEP44" | 2 |
| PF15008 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Domain of unknown function (DUF4518) | 2 |
| PF15009 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Transmembrane protein 173 | 2 |
| PF15010 | 1 | 1 | 3 | 0 | 2 | 2 | 4 | 0 | Putative cell signalling | 13 |
| PF15011 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | Casein Kinase 2 substrate | 1 |
| PF15012 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Domain of unknown function (DUF4519) | 2 |
| PF15013 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | CCSMST1 family | 2 |
| PF15014 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Ceroid-lipofuscinosis neuronal protein 5 | 2 |
| PF15015 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | "Spermatogenesis-associated, N-terminal" | 1 |
| PF15016 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Domain of unknown function (DUF4520) | 2 |
| PF15017 | 0 | 0 | 3 | 2 | 0 | 0 | 2 | 0 | Drug resistance and apoptosis regulator | 7 |
| PF15018 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | TRP-interacting helix | 2 |
| PF15019 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | C9orf72-like protein family | 3 |
| PF15020 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | Cation channel sperm-associated protein subunit delta | 2 |
| PF15021 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Protein of unknown function (DUF4521) | 1 |
| PF15022 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Protein of unknown function (DUF4522) | 1 |
| PF15023 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Protein of unknown function (DUF4523) | 1 |
| PF15024 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 0 | Glycosyltransferase family 18 | 3 |
| PF15025 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Domain of unknown function (DUF4524) | 2 |
| PF15027 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | Domain of unknown function (DUF4525) | 4 |
| PF15029 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Protein of unknown function (DUF4526) | 2 |
| PF15031 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Domain of unknown function (DUF4528) | 2 |
| PF15032 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Protein of unknown function (DUF4529) | 2 |
| PF15033 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Kinocilin protein | 1 |
| PF15034 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | KRTAP type 7 family | 1 |
| PF15035 | 1 | 1 | 2 | 0 | 1 | 0 | 2 | 0 | "Ciliary rootlet component, centrosome cohesion" | 7 |
| PF15036 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Interleukin 34 | 3 |
| PF15037 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 0 | Interleukin-17 receptor extracellular region | 3 |
| PF15038 | 0 | 0 | 1 | 0 | 1 | 1 | 1 | 0 | Jiraiya | 4 |
| PF15039 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Domain of unknown function (DUF4530) | 1 |
| PF15040 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Humanin family | 1 |
| PF15041 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Domain of unknown function (DUF4531) | 1 |
| PF15042 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Late cornified envelope-like proline-rich protein 1 | 1 |
| PF15043 | 0 | 1 | 1 | 0 | 0 | 1 | 2 | 1 | CB1 cannabinoid receptor-interacting protein 1 | 6 |
| PF15044 | 0 | 0 | 1 | 1 | 0 | 0 | 3 | 0 | "Mitochondrial function, CLU-N-term" | 5 |
| PF15045 | 0 | 0 | 2 | 1 | 0 | 0 | 2 | 0 | "Clathrin-binding box of Aftiphilin, vesicle trafficking" | 5 |
| PF15046 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Protein of unknown function (DUF4532) | 1 |
| PF15047 | 0 | 0 | 2 | 1 | 0 | 0 | 0 | 0 | Protein of unknown function (DUF4533) | 3 |
| PF15048 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | Organic solute transporter subunit beta protein | 2 |
| PF15049 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Protein of unknown function (DUF4534) | 1 |
| PF15050 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | SCIMP protein | 1 |
| PF15051 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | FAM198 protein | 4 |
| PF15052 | 0 | 0 | 1 | 0 | 0 | 0 | 2 | 0 | TMEM169 protein family | 3 |
| PF15053 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | Mjmu-R1-like protein family | 5 |
| PF15054 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | Domain of unknown function (DUF4535) | 2 |
| PF15055 | 0 | 0 | 1 | 0 | 0 | 1 | 2 | 0 | Domain of unknown function (DUF4536) | 4 |
| PF15056 | 2 | 1 | 2 | 1 | 1 | 0 | 4 | 0 | Neuritin protein family | 11 |
| PF15057 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Domain of unknown function (DUF4537) | 1 |
| PF15058 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | Speriolin N terminus | 2 |
| PF15059 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | Speriolin C-terminus | 2 |
| PF15060 | 0 | 0 | 1 | 0 | 0 | 1 | 2 | 0 | Differentiation and proliferation regulator | 4 |
| PF15061 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | Domain of unknown function (DUF4538) | 3 |
| PF15062 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Haemopoietic lineage transmembrane helix | 2 |
| PF15063 | 0 | 0 | 2 | 0 | 0 | 0 | 3 | 0 | Thyroid cancer protein 1 | 5 |
| PF15064 | 0 | 0 | 2 | 1 | 0 | 0 | 0 | 0 | Cation channel sperm-associated protein subunit gamma | 3 |
| PF15065 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | "Lysosomal transcription factor, NCU-G1" | 3 |
| PF15066 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Cancer-associated gene protein 1 family | 1 |
| PF15067 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | FAM124 family | 4 |
| PF15068 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | FAM101 family | 4 |
| PF15069 | 1 | 1 | 2 | 2 | 0 | 0 | 3 | 0 | FAM163 family | 9 |
| PF15070 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | 0 | Putative golgin subfamily A member 2-like protein 5 | 4 |
| PF15071 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | "Transmembrane family 220, helix" | 2 |
| PF15072 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Domain of unknown function (DUF4539) | 2 |
| PF15073 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Domain of unknown function (DUF4540) | 1 |
| PF15074 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Domain of unknown function (DUF4541) | 2 |
| PF15075 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Domain of unknown function (DUF4542) | 1 |
| PF15076 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Domain of unknown function (DUF4543) | 1 |
| PF15077 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Domain of unknown function (DUF4544) | 1 |
| PF15078 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Domain of unknown function (DUF4545) | 1 |
| PF15079 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Domain of unknown function (DUF4546) | 1 |
| PF15080 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Domain of unknown function (DUF4547) | 1 |
| PF15081 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Domain of unknown function (DUF4548) | 1 |
| PF15082 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | Domain of unknown function (DUF4549) | 1 |
| PF15083 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | Colipase-like | 2 |
| PF15084 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | Domain of unknown function (DUF4550) | 1 |
| PF15085 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Neuropeptide FF | 2 |
| PF15086 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Uncharacterised protein family UPF0542 | 2 |
| PF15087 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Protein of unknown function (DUF4551) | 2 |
| PF15088 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | "NADH dehydrogenase [ubiquinone] 1 subunit C1, mitochondrial" | 1 |
| PF15090 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | Domain of unknown function (DUF4553) | 1 |
| PF15091 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Domain of unknown function (DUF4554) | 2 |
| PF15092 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Uncharacterised protein family UPF0728 | 2 |
| PF15093 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Domain of unknown function (DUF4555) | 1 |
| PF15094 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Domain of unknown function (DUF4556) | 2 |
| PF15095 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Interleukin 33 | 1 |
| PF15096 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | G6B family | 1 |
| PF15097 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Immunoglobulin J chain | 1 |
| PF15098 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | TMEM89 protein family | 1 |
| PF15099 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 0 | Phosphoinositide-interacting protein family | 3 |
| PF15100 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | TMEM187 protein family | 1 |
| PF15101 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Domain of unknown function (DUF4557) | 2 |
| PF15102 | 0 | 0 | 1 | 0 | 0 | 0 | 2 | 0 | TMEM154 protein family | 3 |
| PF15103 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | G0/G1 switch protein 2 | 2 |
| PF15104 | 0 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | Domain of unknown function (DUF4558) | 3 |
| PF15107 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | FAM216B protein family | 2 |
| PF15108 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Voltage-dependent calcium channel gamma-like subunit protein family | 2 |
| PF15109 | 0 | 0 | 1 | 0 | 0 | 0 | 2 | 0 | TMEM125 protein family | 3 |
| PF15110 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | TMEM141 protein family | 1 |
| PF15111 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | TMEM101 protein family | 1 |
| PF15112 | 0 | 0 | 1 | 1 | 1 | 1 | 2 | 0 | Domain of unknown function (DUF4559) | 6 |
| PF15113 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | TMEM117 protein family | 2 |
| PF15114 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | Uncharacterised protein family UPF0640 | 1 |
| PF15115 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 0 | Domain of unknown function with conserved HDNR motif | 3 |
| PF15116 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | CAMPATH-1 antigen | 1 |
| PF15117 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Uncharacterised protein family UPF0697 | 2 |
| PF15118 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | Domain of unknown function (DUF4560) | 2 |
| PF15119 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Apolipoprotein C4 | 1 |
| PF15120 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Domain of unknown function (DUF4561) | 2 |
| PF15121 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | TMEM71 protein family | 2 |
| PF15122 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | TMEM206 protein family | 2 |
| PF15123 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Domain of unknown function (DUF4562) | 2 |
| PF15124 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Domain of unknown function (DUF4563) | 1 |
| PF15125 | 0 | 0 | 1 | 0 | 0 | 0 | 3 | 0 | TMEM238 protein family | 4 |
| PF15127 | 1 | 0 | 1 | 0 | 1 | 1 | 1 | 0 | Protein of unknown function (DUF4565) | 5 |
| PF15128 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | T-cell leukemia translocation-altered | 2 |
| PF15129 | 0 | 0 | 2 | 0 | 0 | 0 | 3 | 0 | FAM150 family | 5 |
| PF15130 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Domain of unknown function (DUF4566) | 2 |
| PF15132 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Domain of unknown function (DUF4568) | 1 |
| PF15133 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | Domain of unknown function (DUF4569) | 2 |
| PF15134 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Domain of unknown function (DUF4570) | 2 |
| PF15135 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Uncharacterised protein UPF0515 | 3 |
| PF15136 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Uncharacterised protein family UPF0449 | 1 |
| PF15137 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Domain of unknown function (DUF4571) | 1 |
| PF15138 | 0 | 0 | 1 | 0 | 0 | 0 | 2 | 0 | Syncollin | 3 |
| PF15139 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Domain of unknown function (DUF4572) | 1 |
| PF15140 | 0 | 0 | 1 | 1 | 0 | 0 | 2 | 0 | Domain of unknown function (DUF4573) | 4 |
| PF15141 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | Domain of unknown function (DUF4574) | 6 |
| PF15142 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | INCA1 | 1 |
| PF15144 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Domain of unknown function (DUF4576) | 1 |
| PF15145 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Domain of unknown function (DUF4577) | 1 |
| PF15146 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Fanconi anemia-associated | 3 |
| PF15147 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Domain of unknown function (DUF4578) | 1 |
| PF15148 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 0 | Apolipoprotein F | 3 |
| PF15149 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Cation channel sperm-associated protein subunit beta protein family | 1 |
| PF15150 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Phorbol-12-myristate-13-acetate-induced | 2 |
| PF15151 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Response gene to complement 32 protein family | 2 |
| PF15152 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 0 | Kisspeptin | 3 |
| PF15153 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Cytokine-like protein 1 | 2 |
| PF15155 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | MORF4 family-associated protein1 | 1 |
| PF15156 | 0 | 0 | 1 | 0 | 0 | 0 | 2 | 0 | Ceroid-lipofuscinosis neuronal protein 6 | 3 |
| PF15157 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | IQ-like | 2 |
| PF15159 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Phosphatidylinositol N-acetylglucosaminyltransferase subunit Y | 2 |
| PF15160 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Spermatogenesis-associated serine-rich protein 1 | 1 |
| PF15161 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Neuropeptide-like | 2 |
| PF15162 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Domain of unknown function (DUF4580) | 2 |
| PF15163 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Meiosis-expressed | 2 |
| PF15164 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Williams-Beuren syndrome chromosomal region 28 protein homologue | 1 |
| PF15165 | 0 | 0 | 1 | 0 | 0 | 0 | 2 | 0 | Meiotic recombination protein REC114-like | 3 |
| PF15167 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Domain of unknown function (DUF4581) | 2 |
| PF15168 | 1 | 1 | 1 | 0 | 0 | 0 | 1 | 0 | Triple QxxK/R motif-containing protein family | 4 |
| PF15169 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Domain of unknown function (DUF4564) | 2 |
| PF15170 | 0 | 0 | 2 | 2 | 0 | 0 | 3 | 0 | Calcium/calmodulin-dependent protein kinase II inhibitor | 7 |
| PF15171 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | "Neuropeptide secretory protein family, NPQ, spexin" | 1 |
| PF15172 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Prolactin-releasing peptide | 2 |
| PF15173 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | FAM180 family | 1 |
| PF15175 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Spermatogenesis-associated protein 24 | 1 |
| PF15176 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | Leucine-rich repeat family 19 TM domain | 4 |
| PF15177 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | Interleukin-28A | 2 |
| PF15178 | 1 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Mitochondrial import receptor subunit TOM5 homolog | 4 |
| PF15179 | 0 | 0 | 1 | 0 | 0 | 0 | 2 | 0 | Myc target protein 1 | 3 |
| PF15180 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 0 | Neuropeptides B and W | 3 |
| PF15181 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Spermatid-specific manchette-related protein 1 | 1 |
| PF15182 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Otospiralin | 2 |
| PF15183 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | Melanocortin-2 receptor accessory protein family | 4 |
| PF15184 | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 0 | Mitochondrial import receptor subunit TOM6 homolog | 5 |
| PF15185 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | "Bcl-2-modifying factor, apoptosis" | 2 |
| PF15186 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | Testis-expressed sequence 13 protein family | 3 |
| PF15187 | 0 | 0 | 1 | 0 | 0 | 0 | 2 | 0 | Oesophageal cancer-related gene 4 | 3 |
| PF15188 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Coiled-coil domain-containing protein 167 | 2 |
| PF15189 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Domain of unknown function (DUF4582) | 2 |
| PF15190 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Domain of unknown function (DUF4583) | 2 |
| PF15191 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Synaptonemal complex central element protein 3 | 2 |
| PF15192 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | TMEM213 family | 1 |
| PF15193 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | FAM24 family | 2 |
| PF15194 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | TMEM191C family | 1 |
| PF15195 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | TMEM210 family | 1 |
| PF15196 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Activator of apoptosis harakiri | 1 |
| PF15198 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Dexamethasone-induced | 2 |
| PF15200 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Keratinocyte differentiation-associated | 1 |
| PF15201 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Progressive rod-cone degeneration | 1 |
| PF15202 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Adipogenin | 1 |
| PF15203 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | TMEM95 family | 1 |
| PF15205 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | Placenta-specific protein 9 | 2 |
| PF15206 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | FAM209 family | 1 |
| PF15207 | 1 | 0 | 1 | 0 | 2 | 2 | 2 | 0 | TMEM240 family | 8 |
| PF15208 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Rab15 effector | 1 |
| PF15209 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Interleukin 31 | 1 |
| PF15210 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Surfactant-associated protein 2 | 1 |
| PF15212 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | "Spermatogenesis-associated protein 19, mitochondrial" | 1 |
| PF15213 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | CMT1A duplicated region transcript 4 protein | 1 |
| PF15214 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Peroxisomal testis-specific protein 1 | 1 |
| PF15216 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Thymic stromal lymphopoietin | 1 |
| PF15217 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | TSC21 family | 1 |
| PF15218 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Spermatogenesis-associated protein 25 | 1 |
| PF15219 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Testis-expressed 12 | 1 |
| PF15220 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Hypoxia-inducible lipid droplet-associated | 1 |
| PF15221 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Lens epithelial cell protein LEP503 | 2 |
| PF15222 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Kidney androgen-regulated | 1 |
| PF15223 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | Domain of unknown function (DUF4584) | 2 |
| PF15224 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | Scrapie-responsive protein 1 | 2 |
| PF15226 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | HCF-1 beta-propeller-interacting protein family | 1 |
| PF15227 | 0 | 0 | 33 | 2 | 1 | 1 | 92 | 2 | "zinc finger of C3HC4-type, RING" | 131 |
| PF15228 | 0 | 0 | 2 | 0 | 0 | 0 | 3 | 1 | Death-associated protein | 6 |
| PF15229 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 0 | POM121 family | 3 |
| PF15230 | 0 | 0 | 2 | 1 | 0 | 0 | 1 | 0 | Serine/arginine repetitive matrix protein C-terminus | 4 |
| PF15232 | 0 | 0 | 1 | 0 | 0 | 0 | 3 | 1 | Domain of unknown function (DUF4585) | 5 |
| PF15233 | 0 | 0 | 2 | 1 | 0 | 0 | 1 | 0 | Synaptonemal complex central element protein 1 | 4 |
| PF15234 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Linker for activation of T-cells | 1 |
| PF15235 | 3 | 2 | 3 | 1 | 2 | 2 | 2 | 0 | G protein-regulated inducer of neurite outgrowth C-terminus | 15 |
| PF15236 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | Coiled-coil domain-containing protein 66 | 2 |
| PF15237 | 0 | 0 | 4 | 3 | 0 | 0 | 6 | 2 | PTRF/SDPR family | 15 |
| PF15238 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | FAM181 | 1 |
| PF15239 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Domain of unknown function (DUF4586) | 2 |
| PF15240 | 0 | 0 | 4 | 0 | 0 | 0 | 0 | 0 | Proline-rich | 4 |
| PF15241 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | Cylicin N-terminus | 2 |
| PF15242 | 0 | 0 | 3 | 1 | 0 | 0 | 2 | 0 | Family of FAM53 | 6 |
| PF15243 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Anaphase-promoting complex subunit 15 | 1 |
| PF15244 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Hydroxy-steroid dehydrogenase | 2 |
| PF15245 | 0 | 0 | 1 | 0 | 0 | 0 | 2 | 0 | Transcription cofactor vestigial-like protein 4 | 3 |
| PF15246 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 0 | "Nck-associated protein 5, Peripheral clock protein" | 3 |
| PF15247 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Histone RNA hairpin-binding protein RNA-binding domain | 2 |
| PF15248 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 0 | Domain of unknown function (DUF4587) | 3 |
| PF15249 | 1 | 1 | 2 | 0 | 0 | 0 | 3 | 0 | Glioma tumor suppressor candidate region | 7 |
| PF15250 | 1 | 2 | 2 | 1 | 2 | 2 | 2 | 0 | Raftlin | 12 |
| PF15251 | 0 | 0 | 1 | 0 | 0 | 0 | 2 | 0 | Domain of unknown function (DUF4588) | 3 |
| PF15252 | 0 | 0 | 2 | 1 | 0 | 0 | 1 | 0 | Domain of unknown function (DUF4589) | 4 |
| PF15253 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | SCL-interrupting locus protein N-terminus | 2 |
| PF15254 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Coiled-coil domain-containing protein 14 | 1 |
| PF15255 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | "WASH complex subunit CAP-Z interacting, central region" | 4 |
| PF15256 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 0 | SPATIAL | 3 |
| PF15257 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Domain of unknown function (DUF4590) | 2 |
| PF15258 | 0 | 0 | 2 | 0 | 0 | 0 | 4 | 0 | Protein family of FAM222A | 6 |
| PF15259 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 0 | G-2 and S-phase expressed 1 | 3 |
| PF15260 | 0 | 1 | 2 | 0 | 0 | 1 | 2 | 0 | Protein family FAM219A | 6 |
| PF15261 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Domain of unknown function (DUF4591) | 2 |
| PF15262 | 2 | 1 | 2 | 0 | 1 | 1 | 4 | 0 | Domain of unknown function (DUF4592) | 11 |
| PF15264 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Tumour suppressing sub-chromosomal transferable candidate 4 | 2 |
| PF15265 | 1 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | FAM196 family | 3 |
| PF15266 | 0 | 0 | 2 | 1 | 0 | 0 | 2 | 0 | Domain of unknown function (DUF4594) | 5 |
| PF15268 | 1 | 0 | 3 | 0 | 1 | 1 | 3 | 0 | Dapper | 9 |
| PF15269 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Zinc-finger | 2 |
| PF15273 | 1 | 1 | 3 | 0 | 1 | 1 | 5 | 0 | NHS-like | 12 |
| PF15274 | 1 | 1 | 1 | 0 | 0 | 0 | 1 | 0 | Muscular LMNA-interacting protein | 4 |
| PF15275 | 0 | 0 | 3 | 1 | 0 | 0 | 5 | 0 | PEHE domain | 9 |
| PF15276 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 1 | Protein phosphatase 1 binding | 5 |
| PF15277 | 2 | 2 | 2 | 0 | 2 | 3 | 4 | 2 | Exocyst complex component SEC3 N-terminal PIP2 binding PH | 17 |
| PF15279 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | Sine oculis-binding protein | 4 |
| PF15280 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Protein aurora borealis N-terminus | 2 |
| PF15281 | 0 | 1 | 1 | 1 | 0 | 0 | 2 | 0 | Consortin C-terminus | 5 |
| PF15282 | 0 | 0 | 1 | 0 | 0 | 1 | 2 | 1 | BMP-2-inducible protein kinase C-terminus | 5 |
| PF15285 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | "Beclin-1 BH3 domain, Bcl-2-interacting" | 2 |
| PF15287 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | KRBA1 family repeat | 1 |
| PF15288 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Zinc knuckle | 3 |
| PF15289 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Regulatory factor X-associated C-terminal binding domain | 2 |
| PF15290 | 2 | 2 | 2 | 0 | 3 | 2 | 3 | 0 | Golgi-localised syntaxin-1-binding clamp | 14 |
| PF15292 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Treslin N-terminus | 1 |
| PF15293 | 0 | 0 | 1 | 0 | 0 | 0 | 2 | 0 | Nuclear fragile X mental retardation-interacting protein 2 | 3 |
| PF15294 | 1 | 0 | 2 | 0 | 0 | 0 | 1 | 0 | Leucine zipper | 4 |
| PF15295 | 0 | 0 | 1 | 0 | 0 | 0 | 2 | 0 | Coiled-coil domain-containing protein 50 N-terminus | 3 |
| PF15296 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Codanin-1 C-terminus | 2 |
| PF15297 | 0 | 0 | 2 | 1 | 0 | 0 | 1 | 0 | Cytoskeleton-associated protein 2 C-terminus | 4 |
| PF15298 | 1 | 0 | 2 | 0 | 1 | 1 | 1 | 0 | AJAP1/PANP C-terminus | 6 |
| PF15299 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Amyotrophic lateral sclerosis 2 chromosomal region candidate gene 8 | 2 |
| PF15300 | 0 | 0 | 3 | 0 | 0 | 0 | 2 | 0 | INTS6/SAGE1/DDX26B/CT45 C-terminus | 5 |
| PF15301 | 1 | 1 | 2 | 0 | 0 | 0 | 3 | 0 | SLAIN motif-containing family | 7 |
| PF15302 | 0 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | P33 mono-oxygenase | 5 |
| PF15303 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | E3 ubiquitin-protein ligase Arkadia N-terminus | 4 |
| PF15304 | 1 | 1 | 2 | 0 | 3 | 2 | 4 | 1 | A-kinase anchor protein 2 C-terminus | 14 |
| PF15305 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Intraflagellar transport protein 43 | 1 |
| PF15306 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | LIN37 | 2 |
| PF15307 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | Sperm acrosome-associated protein 7 | 2 |
| PF15308 | 2 | 2 | 2 | 0 | 3 | 2 | 4 | 0 | CEP170 C-terminus | 15 |
| PF15309 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 0 | ALMS motif | 3 |
| PF15310 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Vitamin A-deficiency (VAD) rat model signalling | 1 |
| PF15311 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Hydrolethalus syndrome protein 1 C-terminus | 1 |
| PF15312 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Junctional sarcoplasmic reticulum protein | 2 |
| PF15313 | 0 | 0 | 2 | 1 | 0 | 0 | 1 | 0 | Hexamethylene bis-acetamide-inducible protein | 4 |
| PF15314 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | "Proline-rich acidic protein 1, pregnancy-specific uterine" | 1 |
| PF15315 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Facioscapulohumeral muscular dystrophy candidate 2 | 1 |
| PF15316 | 0 | 0 | 3 | 0 | 0 | 0 | 3 | 0 | MyoD family inhibitor | 6 |
| PF15317 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | "Cardiac transcription factor regulator, Developmental protein" | 4 |
| PF15318 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Putative Bcl-2 like protein of testis | 1 |
| PF15319 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | "RAD9, RAD1, HUS1-interacting nuclear orphan protein" | 1 |
| PF15320 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | "mRNA cap methylation, RNMT-activating mini protein" | 2 |
| PF15321 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | ATPase family AAA domain containing 4 | 4 |
| PF15322 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | "Protein missing in infertile sperm 1, putative" | 2 |
| PF15323 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Developmental protein | 3 |
| PF15324 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Hedgehog signalling target | 2 |
| PF15325 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Modulator of retrovirus infection | 1 |
| PF15326 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Testis expressed sequence 15 | 1 |
| PF15327 | 0 | 0 | 2 | 1 | 0 | 0 | 2 | 0 | Tankyrase binding protein C terminal domain | 5 |
| PF15328 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | Putative GRINL1B complex locus protein 2 | 4 |
| PF15330 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | "SHP2-interacting transmembrane adaptor protein, SIT" | 3 |
| PF15331 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Cellular tumour antigen p53-inducible 5 | 1 |
| PF15332 | 0 | 0 | 1 | 0 | 1 | 1 | 1 | 0 | Lck-interacting transmembrane adapter 1 | 4 |
| PF15333 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | TATA box-binding protein-associated factor 1D | 1 |
| PF15334 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Aurora kinase A and ninein interacting protein | 1 |
| PF15335 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Caspase activity and apoptosis inhibitor 1 | 2 |
| PF15336 | 0 | 0 | 2 | 0 | 0 | 0 | 4 | 0 | Autism susceptibility gene 2 protein | 6 |
| PF15337 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 0 | Vascular protein family Vasculin-like 1 | 3 |
| PF15339 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Acrosome formation-associated factor | 1 |
| PF15340 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Cooperator of PRMT5 family | 1 |
| PF15341 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Ribosome biogenesis protein SLX9 | 2 |
| PF15342 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | FAM212 family | 4 |
| PF15343 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Decidual protein induced by progesterone family | 2 |
| PF15344 | 0 | 0 | 2 | 1 | 0 | 0 | 2 | 0 | FAM217 family | 5 |
| PF15345 | 0 | 0 | 1 | 0 | 0 | 0 | 2 | 0 | Transmembrane protein 51 | 3 |
| PF15346 | 0 | 0 | 1 | 0 | 0 | 0 | 2 | 0 | Arginine and glutamate-rich 1 | 3 |
| PF15347 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | Phosphoprotein associated with glycosphingolipid-enriched | 6 |
| PF15348 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Gemini of Cajal bodies-associated protein 8 | 2 |
| PF15350 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Ewing's tumour-associated antigen 1 homologue | 2 |
| PF15351 | 1 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Junctional protein associated with coronary artery disease | 4 |
| PF15352 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Susceptibility to monomelic amyotrophy | 2 |
| PF15353 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Headcase protein family homologue | 2 |
| PF15355 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 1 | "Stretch-responsive small skeletal muscle X protein, Chisel" | 3 |
| PF15356 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Psoriasis susceptibility locus 2 | 1 |
| PF15358 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Testis-specific serine kinase substrate | 1 |
| PF15359 | 0 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | Carnitine deficiency-associated protein 3 | 5 |
| PF15360 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | APJ endogenous ligand | 2 |
| PF15361 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Resistance to inhibitors of cholinesterase homologue 3 | 2 |
| PF15362 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Enamelin | 1 |
| PF15363 | 2 | 2 | 2 | 0 | 0 | 1 | 1 | 0 | Domain of unknown function (DUF4596) | 8 |
| PF15364 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | PAXIP1-associated-protein-1 C term PTIP binding protein | 2 |
| PF15365 | 0 | 0 | 2 | 1 | 0 | 0 | 2 | 0 | Proline-rich nuclear receptor coactivator | 5 |
| PF15366 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Domain of unknown function (DUF4597) | 1 |
| PF15367 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Calcium-binding and spermatid-specific protein 1 | 1 |
| PF15368 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | Spermatogenesis family BioT2 | 2 |
| PF15369 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | Uncharacterised protein KIAA1328 | 1 |
| PF15370 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Domain of unknown function (DUF4598) | 2 |
| PF15371 | 0 | 0 | 5 | 0 | 0 | 0 | 0 | 0 | Domain of unknown function (DUF4599) | 5 |
| PF15372 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Domain of unknown function (DUF4600) | 2 |
| PF15373 | 0 | 0 | 1 | 0 | 0 | 0 | 2 | 0 | Domain of unknown function (DUF4601) | 3 |
| PF15374 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 0 | Coiled-coil domain-containing protein 71L | 3 |
| PF15375 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Domain of unknown function (DUF4602) | 2 |
| PF15376 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | Domain of unknown function (DUF4603) | 1 |
| PF15377 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Domain of unknown function (DUF4604) | 2 |
| PF15378 | 0 | 1 | 2 | 1 | 0 | 0 | 1 | 0 | Domain of unknown function (DUF4605) | 5 |
| PF15379 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Domain of unknown function (DUF4606) | 1 |
| PF15380 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Domain of unknown function (DUF4607) | 1 |
| PF15382 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Domain of unknown function (DUF4609) | 1 |
| PF15383 | 1 | 0 | 1 | 0 | 0 | 0 | 2 | 0 | Transmembrane protein 237 | 4 |
| PF15384 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Domain of unknown function (DUF4610) | 2 |
| PF15385 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | Specifically androgen-regulated gene protein | 4 |
| PF15386 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 0 | Drosophila Tantalus-like | 3 |
| PF15387 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 0 | Domain of unknown function (DUF4611) | 3 |
| PF15388 | 0 | 1 | 3 | 1 | 0 | 0 | 5 | 0 | Protein Family FAM117 | 10 |
| PF15389 | 1 | 1 | 1 | 0 | 0 | 0 | 1 | 0 | Domain of unknown function (DUF4612) | 4 |
| PF15390 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Domain of unknown function (DUF4613) | 2 |
| PF15391 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Domain of unknown function (DUF4614) | 2 |
| PF15392 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Joubert syndrome-associated | 2 |
| PF15393 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | Domain of unknown function (DUF4615) | 1 |
| PF15394 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Domain of unknown function (DUF4616) | 1 |
| PF15395 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Domain of unknown function (DUF4617) | 1 |
| PF15396 | 0 | 0 | 1 | 0 | 0 | 0 | 2 | 0 | Protein Family FAM60A | 3 |
| PF15397 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Domain of unknown function (DUF4618) | 2 |
| PF15398 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Domain of unknown function (DUF4619) | 1 |
| PF15400 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Testis-expressed sequence 33 protein family | 1 |
| PF15405 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Pleckstrin homology domain | 2 |
| PF15409 | 0 | 1 | 4 | 3 | 0 | 0 | 5 | 0 | Pleckstrin homology domain | 13 |
| PF15410 | 8 | 8 | 10 | 1 | 11 | 10 | 11 | 2 | Pleckstrin homology domain | 61 |
| PF15412 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Binding domain of Nse4/EID3 to Nse3-MAGE | 2 |
| PF15413 | 1 | 1 | 2 | 1 | 0 | 0 | 1 | 0 | Pleckstrin homology domain | 6 |
| PF15423 | 0 | 0 | 2 | 1 | 0 | 0 | 0 | 0 | FLYWCH-type zinc finger-containing protein | 3 |
| PF15424 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Odontogenic ameloblast-associated family | 1 |
| PF15427 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | S100P-binding protein | 1 |
| PF15429 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Domain of unknown function (DUF4628) | 2 |
| PF15431 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Transmembrane protein 190 | 1 |
| PF15433 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | Mitochondrial 28S ribosomal protein S31 | 5 |
| PF15434 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 0 | Family 104 | 3 |
| PF15435 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | UNC119-binding protein C5orf30 homologue | 2 |
| PF15439 | 2 | 1 | 3 | 1 | 1 | 0 | 3 | 0 | Neuronal tyrosine-phosphorylated phosphoinositide-3-kinase adapter | 11 |
| PF15440 | 0 | 0 | 3 | 1 | 0 | 0 | 1 | 0 | THRAP3/BCLAF1 family | 5 |
| PF15441 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Rho guanine nucleotide exchange factor 5/35 | 1 |
| PF15442 | 0 | 0 | 5 | 0 | 0 | 0 | 0 | 0 | Domain of unknown function (DUF4629) | 5 |
| PF15443 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | Domain of unknown function (DUF4630) | 2 |
| PF15449 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Retinal protein | 2 |
| PF15450 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Domain of unknown function (DUF4631) | 1 |
| PF15452 | 2 | 1 | 2 | 1 | 1 | 0 | 2 | 0 | Neuronal tyrosine-phosphorylated phosphoinositide-3-kinase adapter | 9 |
| PF15453 | 0 | 0 | 1 | 0 | 1 | 1 | 2 | 0 | Protein incorporated later into Tight Junctions | 5 |
| PF15454 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | Late endosomal/lysosomal adaptor and MAPK and MTOR activator | 6 |
| PF15455 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Proline-rich 19 | 2 |
| PF15462 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | "Bartter syndrome, infantile, with sensorineural deafness (Barttin)" | 2 |
| PF15464 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Domain of unknown function (DUF4633) | 1 |
| PF15465 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Domain of unknown function (DUF4634) | 1 |
| PF15466 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Domain of unknown function (DUF4635) | 1 |
| PF15467 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | Secretogranin-3 | 3 |
| PF15468 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Domain of unknown function (DUF4636) | 1 |
| PF15469 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | Exocyst complex component Sec5 | 7 |
| PF15470 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Domain of unknown function (DUF4637) | 1 |
| PF15471 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Transmembrane protein family 171 | 1 |
| PF15472 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Domain of unknown function (DUF4638) | 1 |
| PF15473 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 1 | "PEST, proteolytic signal-containing nuclear protein family" | 3 |
| PF15475 | 0 | 0 | 1 | 1 | 1 | 1 | 3 | 1 | "Transmembrane protein C12orf23, UPF0444" | 8 |
| PF15476 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Histone deacetylase complex subunit SAP25 | 1 |
| PF15477 | 0 | 0 | 3 | 1 | 0 | 0 | 3 | 0 | Small acidic protein family | 7 |
| PF15478 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Family of unknown function with LKAAEAR motif | 1 |
| PF15479 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Domain of unknown function (DUF4639) | 1 |
| PF15480 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Domain of unknown function (DUF4640) | 1 |
| PF15482 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Coiled-coil domain-containing glutamate-rich protein family 1 | 2 |
| PF15483 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Domain of unknown function (DUF4641) | 1 |
| PF15484 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Domain of unknown function (DUF4642) | 1 |
| PF15485 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | Domain of unknown function (DUF4643) | 1 |
| PF15486 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Domain of unknown function (DUF4644) | 1 |
| PF15487 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | FAM220 family | 1 |
| PF15488 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Domain of unknown function (DUF4645) | 1 |
| PF15489 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | "CST, telomere maintenance, complex subunit CTC1" | 3 |
| PF15490 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | "Telomere-capping, CST complex subunit" | 2 |
| PF15492 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | 0 | "Neuroblastoma-amplified sequence, N terminal" | 4 |
| PF15494 | 0 | 0 | 6 | 0 | 0 | 0 | 6 | 0 | Scavenger receptor cysteine-rich domain | 12 |
| PF15497 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | "snRNA-activating protein complex subunit 19, SNAPc subunit 19" | 2 |
| PF15498 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | "Nephrin and CD2AP-binding protein, Dendrin" | 2 |
| PF15499 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | "Ubiquitin-specific peptidase-like, SUMO isopeptidase" | 2 |
| PF15501 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Nuclear protein MDM1 | 2 |
| PF15502 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 0 | M-phase-specific PLK1-interacting protein | 3 |
| PF15503 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Protein phosphatase 1 regulatory subunit 35 C-terminus | 2 |
| PF15504 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Domain of unknown function (DUF4647) | 1 |
| PF15505 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | Domain of unknown function (DUF4648) | 2 |
| PF15506 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 0 | OCC1 family | 5 |
| PF15508 | 1 | 1 | 2 | 1 | 0 | 0 | 2 | 0 | beta subunit of N-acylethanolamine-hydrolyzing acid amidase | 7 |
| PF15509 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Domain of unknown function (DUF4650) | 1 |
| PF15510 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Centromere kinetochore component W | 1 |
| PF15511 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Centromere kinetochore component CENP-T | 2 |
| PF15512 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | "Chromatin assembly factor complex 1 subunit p60, C-terminal" | 2 |
| PF15519 | 0 | 0 | 1 | 0 | 0 | 0 | 2 | 0 | linker between RRM2 and RRM3 domains in RBM39 protein | 3 |
| PF15539 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | "CAF1 complex subunit p150, region binding to CAF1-p60 at C-term" | 2 |
| PF15546 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Domain of unknown function (DUF4653) | 1 |
| PF15547 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Domain of unknown function (DUF4654) | 1 |
| PF15549 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | PGC7/Stella/Dppa3 domain | 2 |
| PF15550 | 0 | 0 | 1 | 0 | 0 | 0 | 2 | 0 | Draxin | 3 |
| PF15551 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Domain of unknown function (DUF4656) | 2 |
| PF15552 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Domain of unknown function (DUF4657) | 1 |
| PF15553 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | Testis-expressed protein 19 | 2 |
| PF15554 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | FSIP1 family | 2 |
| PF15555 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Domain of unknown function (DUF4658) | 1 |
| PF15556 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | ZW10 interactor | 4 |
| PF15557 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | "CAF1 complex subunit p150, region binding to PCNA" | 2 |
| PF15558 | 1 | 1 | 2 | 0 | 0 | 0 | 1 | 0 | Domain of unknown function (DUF4659) | 5 |
| PF15559 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | Domain of unknown function (DUF4660) | 2 |
| PF15576 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Domain of unknown function (DUF4661) | 2 |
| PF15612 | 0 | 0 | 5 | 0 | 0 | 0 | 2 | 0 | "WSTF, HB1, Itc1p, MBD9 motif 1" | 7 |
| PF15613 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | "WSTF, HB1, Itc1p, MBD9 motif 2" | 4 |
| PF15614 | 0 | 0 | 3 | 0 | 0 | 0 | 2 | 0 | "WSTF, HB1, Itc1p, MBD9 motif 3" | 5 |
| PF15619 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 0 | Ciliary protein causing Leber congenital amaurosis disease | 3 |
| PF15620 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Centromere assembly component CENP-C middle DNMT3B-binding region | 1 |
| PF15621 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | Proline-rich submaxillary gland androgen-regulated family | 3 |
| PF15622 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | Kinetochore assembly subunit CENP-C N-terminal | 1 |
| PF15625 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | CC2D2A N-terminal C2 domain | 3 |
| PF15627 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | CEP76 C2 domain | 3 |
| PF15630 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | Kinetochore component CENP-S | 2 |
| PF15636 | 2 | 4 | 4 | 0 | 2 | 6 | 5 | 0 | GHH signature containing HNH/Endo VII superfamily nuclease toxin | 23 |
Supplementary Table 6a. GO analysis of mouse specific PSD proteins.
Biological Process and Cellular Component terms enriched amongst PSD proteins absent from the zebrafish synapse. For each enriched term the following information is given: number of genes in the genome annotated for that term, observed number of PSD proteins annotated for that term, expected number of PSD proteins annotated for that term, fold enrichment, (+) over-represented or (-) under-represented, Binomial statistics p-value for the enrichment of that term amongst mouse PSD proteins absent from zebrafish synapse.
| GO biological process | Annotated in Mouse Genome | observed | expected | Fold Enrichment | +/- | P value |
|---|---|---|---|---|---|---|
| regulation of cation transmembrane transport | 159 | 21 | 4.92 | 4.27 | + | 3.80E-04 |
| regulation of exocytosis | 165 | 21 | 5.10 | 4.12 | + | 6.97E-04 |
| establishment of protein localization to membrane | 148 | 18 | 4.58 | 3.93 | + | 1.07E-02 |
| regulation of ion transmembrane transporter activity | 167 | 20 | 5.16 | 3.87 | + | 3.57E-03 |
| protein localization to plasma membrane | 142 | 17 | 4.39 | 3.87 | + | 2.55E-02 |
| protein localization to cell periphery | 145 | 17 | 4.48 | 3.79 | + | 3.34E-02 |
| regulation of transmembrane transporter activity | 172 | 20 | 5.32 | 3.76 | + | 5.59E-03 |
| vesicle localization | 164 | 19 | 5.07 | 3.75 | + | 1.11E-02 |
| regulation of transporter activity | 182 | 21 | 5.63 | 3.73 | + | 3.37E-03 |
| regulation of synapse structure or activity | 237 | 26 | 7.33 | 3.55 | + | 3.96E-04 |
| endosomal transport | 192 | 21 | 5.94 | 3.54 | + | 7.79E-03 |
| exocytosis | 211 | 23 | 6.52 | 3.53 | + | 2.56E-03 |
| small GTPase mediated signal transduction | 302 | 30 | 9.34 | 3.21 | + | 3.29E-04 |
| organelle localization | 339 | 33 | 10.48 | 3.15 | + | 1.12E-04 |
| regulation of actin filament-based process | 302 | 29 | 9.34 | 3.11 | + | 1.10E-03 |
| regulation of vesicle-mediated transport | 396 | 38 | 12.24 | 3.10 | + | 1.33E-05 |
| protein localization to membrane | 271 | 26 | 8.38 | 3.10 | + | 4.89E-03 |
| microtubule-based process | 485 | 46 | 14.99 | 3.07 | + | 3.68E-07 |
| negative regulation of organelle organization | 260 | 24 | 8.04 | 2.99 | + | 2.49E-02 |
| establishment of organelle localization | 263 | 24 | 8.13 | 2.95 | + | 3.01E-02 |
| positive regulation of cell projection organization | 307 | 28 | 9.49 | 2.95 | + | 4.91E-03 |
| single-organism cellular localization | 669 | 60 | 20.68 | 2.90 | + | 3.77E-09 |
| regulation of cytoskeleton organization | 382 | 34 | 11.81 | 2.88 | + | 5.59E-04 |
| endomembrane system organization | 378 | 33 | 11.69 | 2.82 | + | 1.33E-03 |
| cytoskeleton organization | 779 | 68 | 24.08 | 2.82 | + | 3.32E-10 |
| microtubule cytoskeleton organization | 333 | 29 | 10.29 | 2.82 | + | 7.72E-03 |
| vesicle-mediated transport | 878 | 76 | 27.14 | 2.80 | + | 1.44E-11 |
| regulation of cellular component size | 289 | 25 | 8.93 | 2.80 | + | 4.73E-02 |
| actin cytoskeleton organization | 365 | 31 | 11.28 | 2.75 | + | 5.54E-03 |
| regulation of cell morphogenesis | 471 | 40 | 14.56 | 2.75 | + | 1.39E-04 |
| regulation of metal ion transport | 330 | 28 | 10.20 | 2.74 | + | 1.92E-02 |
| actin filament-based process | 389 | 33 | 12.03 | 2.74 | + | 2.51E-03 |
| protein transport | 983 | 83 | 30.39 | 2.73 | + | 2.55E-12 |
| secretion by cell | 359 | 30 | 11.10 | 2.70 | + | 1.15E-02 |
| negative regulation of cellular component organization | 517 | 43 | 15.98 | 2.69 | + | 7.31E-05 |
| establishment of protein localization | 1071 | 89 | 33.11 | 2.69 | + | 4.92E-13 |
| membrane organization | 603 | 50 | 18.64 | 2.68 | + | 4.92E-06 |
| intracellular protein transport | 538 | 44 | 16.63 | 2.65 | + | 7.95E-05 |
| regulation of cellular component biogenesis | 637 | 52 | 19.69 | 2.64 | + | 3.76E-06 |
| regulation of cell projection organization | 508 | 40 | 15.70 | 2.55 | + | 9.91E-04 |
| regulation of neuron projection development | 400 | 31 | 12.37 | 2.51 | + | 3.48E-02 |
| single-organism intracellular transport | 856 | 65 | 26.46 | 2.46 | + | 4.59E-07 |
| protein localization | 1464 | 111 | 45.26 | 2.45 | + | 2.99E-14 |
| cytoplasmic transport | 515 | 39 | 15.92 | 2.45 | + | 3.70E-03 |
| single-organism membrane organization | 534 | 40 | 16.51 | 2.42 | + | 3.46E-03 |
| neuron projection development | 506 | 37 | 15.64 | 2.37 | + | 1.61E-02 |
| intracellular transport | 1026 | 75 | 31.72 | 2.36 | + | 8.01E-08 |
| cellular protein localization | 956 | 69 | 29.55 | 2.33 | + | 9.81E-07 |
| cellular macromolecule localization | 963 | 69 | 29.77 | 2.32 | + | 1.33E-06 |
| establishment of localization in cell | 1386 | 99 | 42.85 | 2.31 | + | 1.02E-10 |
| cellular localization | 1757 | 121 | 54.32 | 2.23 | + | 9.66E-13 |
| macromolecule localization | 1728 | 117 | 53.42 | 2.19 | + | 1.12E-11 |
| cell projection organization | 846 | 56 | 26.15 | 2.14 | + | 1.02E-03 |
| regulation of organelle organization | 1021 | 67 | 31.56 | 2.12 | + | 7.82E-05 |
| protein complex subunit organization | 1146 | 75 | 35.43 | 2.12 | + | 1.08E-05 |
| positive regulation of cellular component organization | 1142 | 74 | 35.30 | 2.10 | + | 2.12E-05 |
| regulation of nervous system development | 781 | 50 | 24.14 | 2.07 | + | 1.25E-02 |
| regulation of cellular component organization | 2095 | 132 | 64.76 | 2.04 | + | 2.84E-11 |
| generation of neurons | 1291 | 81 | 39.91 | 2.03 | + | 1.49E-05 |
| neuron differentiation | 821 | 50 | 25.38 | 1.97 | + | 4.81E-02 |
| organic substance transport | 1552 | 94 | 47.98 | 1.96 | + | 3.84E-06 |
| neurogenesis | 1372 | 83 | 42.41 | 1.96 | + | 4.73E-05 |
| single-organism localization | 2664 | 156 | 82.35 | 1.89 | + | 2.39E-11 |
| single-organism transport | 2467 | 143 | 76.26 | 1.88 | + | 1.06E-09 |
| transport | 3170 | 183 | 98.00 | 1.87 | + | 1.45E-13 |
| single-organism organelle organization | 1767 | 102 | 54.62 | 1.87 | + | 8.22E-06 |
| establishment of localization | 3295 | 190 | 101.86 | 1.87 | + | 2.99E-14 |
| cellular component assembly | 1581 | 91 | 48.88 | 1.86 | + | 8.49E-05 |
| nervous system development | 1800 | 103 | 55.65 | 1.85 | + | 1.06E-05 |
| organelle organization | 2494 | 142 | 77.10 | 1.84 | + | 5.03E-09 |
| cellular component organization | 4121 | 228 | 127.40 | 1.79 | + | 3.45E-16 |
| intracellular signal transduction | 1213 | 67 | 37.50 | 1.79 | + | 3.36E-02 |
| positive regulation of molecular function | 1327 | 73 | 41.02 | 1.78 | + | 1.41E-02 |
| macromolecular complex subunit organization | 1737 | 95 | 53.70 | 1.77 | + | 4.31E-04 |
| localization | 4100 | 221 | 126.75 | 1.74 | + | 3.97E-14 |
| cellular component biogenesis | 1748 | 94 | 54.04 | 1.74 | + | 1.09E-03 |
| cellular component organization or biogenesis | 4260 | 229 | 131.69 | 1.74 | + | 9.43E-15 |
| regulation of localization | 2246 | 119 | 69.43 | 1.71 | + | 3.97E-05 |
| regulation of transport | 1676 | 87 | 51.81 | 1.68 | + | 1.40E-02 |
| regulation of biological quality | 2643 | 134 | 81.71 | 1.64 | + | 5.17E-05 |
| regulation of signaling | 2486 | 125 | 76.85 | 1.63 | + | 3.01E-04 |
| regulation of molecular function | 2260 | 113 | 69.87 | 1.62 | + | 2.05E-03 |
| regulation of cell communication | 2620 | 131 | 80.99 | 1.62 | + | 1.81E-04 |
| cell differentiation | 3118 | 149 | 96.39 | 1.55 | + | 2.71E-04 |
| cellular developmental process | 3284 | 154 | 101.52 | 1.52 | + | 4.94E-04 |
| system development | 3499 | 163 | 108.17 | 1.51 | + | 2.74E-04 |
| anatomical structure development | 4183 | 190 | 129.31 | 1.47 | + | 8.34E-05 |
| single-organism developmental process | 4666 | 209 | 144.24 | 1.45 | + | 3.25E-05 |
| developmental process | 4693 | 210 | 145.08 | 1.45 | + | 3.17E-05 |
| multicellular organismal development | 4036 | 178 | 124.77 | 1.43 | + | 2.30E-03 |
| negative regulation of cellular process | 3811 | 166 | 117.81 | 1.41 | + | 1.45E-02 |
| negative regulation of biological process | 4133 | 176 | 127.77 | 1.38 | + | 2.58E-02 |
| positive regulation of biological process | 4910 | 205 | 151.79 | 1.35 | + | 1.06E-02 |
| single-organism cellular process | 10788 | 420 | 333.50 | 1.26 | + | 1.90E-07 |
| single-organism process | 12188 | 474 | 376.78 | 1.26 | + | 2.15E-10 |
| cellular process | 13249 | 502 | 409.58 | 1.23 | + | 1.07E-09 |
| biological regulation | 10699 | 399 | 330.75 | 1.21 | + | 8.89E-04 |
| regulation of cellular process | 9809 | 364 | 303.24 | 1.20 | + | 1.55E-02 |
| regulation of biological process | 10256 | 379 | 317.05 | 1.20 | + | 1.04E-02 |
| biological_process | 20668 | 668 | 638.93 | 1.05 | + | 1.46E-02 |
| organic cyclic compound biosynthetic process | 2284 | 38 | 70.61 | .54 | - | 4.88E-02 |
| G-protein coupled receptor signaling pathway | 1870 | 23 | 57.81 | .40 | - | 4.73E-04 |
| sensory perception | 1571 | 18 | 48.57 | .37 | - | 1.60E-03 |
| detection of stimulus | 1279 | 5 | 39.54 | < 0.2 | - | 1.86E-08 |
| sensory perception of chemical stimulus | 1230 | 4 | 38.02 | < 0.2 | - | 9.19E-09 |
| detection of stimulus involved in sensory perception | 1186 | 2 | 36.66 | < 0.2 | - | 2.54E-10 |
| sensory perception of smell | 1107 | 1 | 34.22 | < 0.2 | - | 1.57E-10 |
| detection of chemical stimulus | 1154 | 1 | 35.67 | < 0.2 | - | 3.54E-11 |
| GO cellular component complete | Annotated in Mouse Genome | observed | expected | Fold Enrichment | +/- | P value |
|---|---|---|---|---|---|---|
| ESCRT I complex | 9 | 5 | .28 | > 5 | + | 1.26E-02 |
| HOPS complex | 11 | 5 | .34 | > 5 | + | 3.26E-02 |
| ESCRT complex | 20 | 9 | .62 | > 5 | + | 2.31E-05 |
| peroxisomal part | 63 | 10 | 1.95 | > 5 | + | 4.14E-02 |
| microbody part | 63 | 10 | 1.95 | > 5 | + | 4.14E-02 |
| cortical cytoskeleton | 84 | 13 | 2.60 | > 5 | + | 3.85E-03 |
| postsynaptic density | 198 | 26 | 6.12 | 4.25 | + | 1.76E-06 |
| postsynaptic membrane | 212 | 27 | 6.55 | 4.12 | + | 1.62E-06 |
| postsynapse | 394 | 48 | 12.18 | 3.94 | + | 3.19E-12 |
| synaptic membrane | 259 | 31 | 8.01 | 3.87 | + | 4.33E-07 |
| endosome membrane | 154 | 18 | 4.76 | 3.78 | + | 2.87E-03 |
| cell cortex part | 120 | 14 | 3.71 | 3.77 | + | 3.73E-02 |
| coated vesicle | 129 | 15 | 3.99 | 3.76 | + | 2.05E-02 |
| extrinsic component of plasma membrane | 139 | 16 | 4.30 | 3.72 | + | 1.23E-02 |
| early endosome | 194 | 22 | 6.00 | 3.67 | + | 3.75E-04 |
| endosomal part | 166 | 18 | 5.13 | 3.51 | + | 7.89E-03 |
| microtubule | 356 | 38 | 11.01 | 3.45 | + | 1.14E-07 |
| synapse part | 591 | 58 | 18.27 | 3.17 | + | 3.85E-11 |
| ion channel complex | 257 | 25 | 7.94 | 3.15 | + | 9.77E-04 |
| synapse | 777 | 75 | 24.02 | 3.12 | + | 1.21E-14 |
| transporter complex | 284 | 27 | 8.78 | 3.08 | + | 5.55E-04 |
| myelin sheath | 200 | 19 | 6.18 | 3.07 | + | 2.71E-02 |
| transmembrane transporter complex | 280 | 26 | 8.66 | 3.00 | + | 1.38E-03 |
| cell cortex | 205 | 19 | 6.34 | 3.00 | + | 3.76E-02 |
| dendrite | 516 | 47 | 15.95 | 2.95 | + | 1.31E-07 |
| cell junction | 1114 | 100 | 34.44 | 2.90 | + | 3.63E-18 |
| adherens junction | 410 | 36 | 12.67 | 2.84 | + | 5.02E-05 |
| extrinsic component of membrane | 255 | 22 | 7.88 | 2.79 | + | 2.71E-02 |
| anchoring junction | 425 | 36 | 13.14 | 2.74 | + | 1.19E-04 |
| somatodendritic compartment | 786 | 65 | 24.30 | 2.68 | + | 2.04E-09 |
| focal adhesion | 349 | 28 | 10.79 | 2.60 | + | 8.35E-03 |
| membrane protein complex | 968 | 77 | 29.92 | 2.57 | + | 1.07E-10 |
| cell-substrate adherens junction | 354 | 28 | 10.94 | 2.56 | + | 1.08E-02 |
| cell leading edge | 305 | 24 | 9.43 | 2.55 | + | 4.98E-02 |
| cell-substrate junction | 359 | 28 | 11.10 | 2.52 | + | 1.38E-02 |
| neuron projection | 1059 | 82 | 32.74 | 2.50 | + | 6.35E-11 |
| neuron part | 1323 | 102 | 40.90 | 2.49 | + | 4.46E-14 |
| actin cytoskeleton | 406 | 31 | 12.55 | 2.47 | + | 7.25E-03 |
| endosome | 686 | 52 | 21.21 | 2.45 | + | 6.82E-06 |
| cell-cell junction | 383 | 28 | 11.84 | 2.36 | + | 4.27E-02 |
| perinuclear region of cytoplasm | 538 | 39 | 16.63 | 2.34 | + | 1.64E-03 |
| cell projection | 1928 | 138 | 59.60 | 2.32 | + | 2.32E-17 |
| cell body | 636 | 45 | 19.66 | 2.29 | + | 4.89E-04 |
| centrosome | 458 | 32 | 14.16 | 2.26 | + | 2.90E-02 |
| plasma membrane protein complex | 447 | 31 | 13.82 | 2.24 | + | 4.43E-02 |
| whole membrane | 1559 | 108 | 48.19 | 2.24 | + | 6.06E-12 |
| neuronal cell body | 570 | 39 | 17.62 | 2.21 | + | 6.24E-03 |
| cell projection part | 930 | 63 | 28.75 | 2.19 | + | 1.10E-05 |
| ribonucleoprotein complex | 631 | 42 | 19.51 | 2.15 | + | 5.30E-03 |
| cytoskeletal part | 1228 | 81 | 37.96 | 2.13 | + | 2.40E-07 |
| cytoskeleton | 1801 | 117 | 55.68 | 2.10 | + | 2.99E-11 |
| microtubule cytoskeleton | 947 | 61 | 29.28 | 2.08 | + | 1.09E-04 |
| membrane region | 1047 | 67 | 32.37 | 2.07 | + | 3.14E-05 |
| plasma membrane region | 847 | 54 | 26.18 | 2.06 | + | 8.37E-04 |
| bounding membrane of organelle | 1144 | 72 | 35.37 | 2.04 | + | 1.76E-05 |
| organelle membrane | 1693 | 103 | 52.34 | 1.97 | + | 6.01E-08 |
| cytoplasmic membrane-bounded vesicle | 726 | 44 | 22.44 | 1.96 | + | 3.02E-02 |
| cytoplasmic vesicle | 1028 | 61 | 31.78 | 1.92 | + | 1.61E-03 |
| organelle envelope | 863 | 51 | 26.68 | 1.91 | + | 1.35E-02 |
| envelope | 866 | 51 | 26.77 | 1.91 | + | 1.48E-02 |
| extracellular vesicle | 2580 | 146 | 79.76 | 1.83 | + | 5.35E-10 |
| extracellular exosome | 2567 | 145 | 79.36 | 1.83 | + | 7.57E-10 |
| extracellular organelle | 2586 | 146 | 79.94 | 1.83 | + | 6.38E-10 |
| extracellular membrane-bounded organelle | 2573 | 145 | 79.54 | 1.82 | + | 9.02E-10 |
| membrane-bounded vesicle | 3118 | 175 | 96.39 | 1.82 | + | 2.14E-12 |
| vesicle | 3396 | 190 | 104.98 | 1.81 | + | 1.04E-13 |
| intracellular non-membrane-bounded organelle | 3397 | 190 | 105.01 | 1.81 | + | 1.07E-13 |
| non-membrane-bounded organelle | 3397 | 190 | 105.01 | 1.81 | + | 1.07E-13 |
| mitochondrion | 1693 | 94 | 52.34 | 1.80 | + | 4.00E-05 |
| endomembrane system | 3183 | 176 | 98.40 | 1.79 | + | 6.81E-12 |
| cytosol | 1522 | 82 | 47.05 | 1.74 | + | 1.09E-03 |
| cytoplasmic part | 6482 | 346 | 200.38 | 1.73 | + | 2.69E-28 |
| macromolecular complex | 4629 | 246 | 143.10 | 1.72 | + | 1.41E-16 |
| protein complex | 4016 | 210 | 124.15 | 1.69 | + | 1.75E-12 |
| cell periphery | 4568 | 236 | 141.22 | 1.67 | + | 4.08E-14 |
| plasma membrane | 4463 | 224 | 137.97 | 1.62 | + | 1.07E-11 |
| cytoplasm | 9660 | 477 | 298.63 | 1.60 | + | 1.05E-39 |
| extracellular region part | 3567 | 170 | 110.27 | 1.54 | + | 3.77E-06 |
| organelle part | 6035 | 286 | 186.57 | 1.53 | + | 2.59E-13 |
| intracellular organelle part | 5840 | 274 | 180.54 | 1.52 | + | 7.19E-12 |
| membrane | 8967 | 404 | 277.21 | 1.46 | + | 2.10E-19 |
| extracellular region | 4113 | 179 | 127.15 | 1.41 | + | 7.91E-04 |
| intracellular part | 12214 | 526 | 377.58 | 1.39 | + | 9.80E-29 |
| intracellular | 12408 | 532 | 383.58 | 1.39 | + | 4.99E-29 |
| intracellular organelle | 10618 | 450 | 328.24 | 1.37 | + | 9.18E-18 |
| organelle | 11628 | 481 | 359.47 | 1.34 | + | 4.20E-18 |
| cell | 14515 | 594 | 448.72 | 1.32 | + | 8.43E-33 |
| cell part | 14485 | 592 | 447.79 | 1.32 | + | 3.90E-32 |
| membrane-bounded organelle | 10749 | 434 | 332.29 | 1.31 | + | 5.81E-12 |
| intracellular membrane-bounded organelle | 9638 | 388 | 297.95 | 1.30 | + | 4.56E-09 |
| cellular_component | 20887 | 680 | 645.70 | 1.05 | + | 2.78E-07 |
| Unclassified | 1433 | 14 | 44.30 | .32 | - | 0.00E00 |
Supplementary Table 6b. GO analysis of zebrafish specific PSD proteins.
Biological Process and Cellular Component terms enriched amongst PSD proteins absent from the mouse synapse. For each enriched term the following information is given: number of genes in the genome annotated for that term, observed number of PSD proteins annotated for that term, expected number of PSD proteins annotated for that term, fold enrichment, (+) over-represented or (-) under-represented, Binomial statistics p-value for the enrichment of that term amongst zebrafish PSD proteins absent from mouse synapse.
| GO biological process | Annotated in Zebrafish Genome | observed | expected | Fold Enrichment | +/- | P value |
|---|---|---|---|---|---|---|
| protein polymerization | 42 | 10 | .69 | > 5 | + | 8.78E-06 |
| cellular protein complex assembly | 131 | 15 | 2.15 | > 5 | + | 2.23E-05 |
| protein complex assembly | 288 | 25 | 4.72 | > 5 | + | 8.41E-08 |
| protein complex biogenesis | 288 | 25 | 4.72 | > 5 | + | 8.41E-08 |
| cellular macromolecular complex assembly | 270 | 22 | 4.43 | 4.97 | + | 4.10E-06 |
| protein complex subunit organization | 452 | 32 | 7.42 | 4.32 | + | 2.68E-08 |
| macromolecular complex assembly | 400 | 28 | 6.56 | 4.27 | + | 7.04E-07 |
| cellular component assembly | 710 | 41 | 11.65 | 3.52 | + | 1.75E-08 |
| cellular component biogenesis | 825 | 45 | 13.53 | 3.32 | + | 1.01E-08 |
| macromolecular complex subunit organization | 762 | 39 | 12.50 | 3.12 | + | 1.81E-06 |
| cellular component organization | 2017 | 74 | 33.09 | 2.24 | + | 1.87E-07 |
| cellular component organization or biogenesis | 2128 | 78 | 34.91 | 2.23 | + | 5.33E-08 |
| single-organism process | 8573 | 185 | 140.65 | 1.32 | + | 1.12E-02 |
| cellular process | 9313 | 195 | 152.79 | 1.28 | + | 3.91E-02 |
| biological_process | 14385 | 287 | 236.00 | 1.22 | + | 5.14E-04 |
| GO cellular component complete | Annotated in Zebrafish Genome | observed | expected | Fold Enrichment | +/- | P value |
|---|---|---|---|---|---|---|
| intermediate filament | 54 | 10 | .89 | > 5 | + | 1.93E-05 |
| intermediate filament cytoskeleton | 55 | 10 | .90 | > 5 | + | 2.29E-05 |
| myosin complex | 61 | 7 | 1.00 | > 5 | + | 4.64E-02 |
| actin cytoskeleton | 162 | 14 | 2.66 | > 5 | + | 4.26E-04 |
| cytoskeletal part | 496 | 34 | 8.14 | 4.18 | + | 2.97E-09 |
| cytoskeleton | 671 | 45 | 11.01 | 4.09 | + | 1.88E-12 |
| intracellular non-membrane-bounded organelle | 1277 | 64 | 20.95 | 3.05 | + | 1.73E-12 |
| non-membrane-bounded organelle | 1277 | 64 | 20.95 | 3.05 | + | 1.73E-12 |
| ribonucleoprotein complex | 402 | 19 | 6.60 | 2.88 | + | 2.93E-02 |
| macromolecular complex | 2412 | 90 | 39.57 | 2.27 | + | 7.68E-11 |
| intracellular organelle part | 2346 | 87 | 38.49 | 2.26 | + | 3.06E-10 |
| organelle part | 2389 | 87 | 39.19 | 2.22 | + | 8.14E-10 |
| protein complex | 2042 | 73 | 33.50 | 2.18 | + | 1.73E-07 |
| cytoplasm | 3962 | 96 | 65.00 | 1.48 | + | 2.61E-02 |
| intracellular organelle | 5188 | 122 | 85.11 | 1.43 | + | 6.48E-03 |
| organelle | 5237 | 123 | 85.92 | 1.43 | + | 6.21E-03 |
| intracellular part | 6645 | 151 | 109.02 | 1.39 | + | 2.18E-03 |
| intracellular | 7002 | 159 | 114.87 | 1.38 | + | 1.01E-03 |
| cell | 8569 | 180 | 140.58 | 1.28 | + | 2.12E-02 |
| cell part | 8569 | 180 | 140.58 | 1.28 | + | 2.12E-02 |
| cellular_component | 13404 | 269 | 219.90 | 1.22 | + | 4.22E-04 |
Supplementary Table 7a. Mouse SYN orthologues in zebrafish SYN.
For each mouse synaptosomal protein with an orthologue in the zebrafish synaptosomal fraction the following information is given: mouse ENSEBLE ID, zebrafish orthologue ENSEBLE ID in SYN, % of protein identity between mouse and zebrafish proteins and orthology type.
Supplementary Table 7b. Mouse PSD orthologues in zebrafish PSD.
For each mouse postsynaptic density protein with an orthologue in the zebrafish postsynaptic density fraction the following information is given: mouse ENSEBLE ID, zebrafish orthologue ENSEBLE ID in PSD, % of protein identity between mouse and zebrafish proteins and orthology type.
Supplementary Table 7c. Zebrafish SYN orthologues in Mouse SYN.
For each zebrafish synaptosomal protein with an orthologue in the mouse synaptosomal fraction the following information is given: zebrafish ENSEBLE ID, mouse orthologue ENSEBLE ID in SYN, % of protein identity between mouse and zebrafish proteins and orthology type.
Supplementary Table 7d. Zebrafish PSD orthologues in mouse PSD.
For each mouse postsynaptic density protein with an orthologue in the zebrafish postsynaptic density fraction the following information is given: zebrafish ENSEBLE ID, mouse orthologue ENSEBLE ID in PSD, % of protein identity between mouse and zebrafish proteins and orthology type.