Automated design of genomic Southern blot probes
Mike DR Croning, David G Fricker, Noboru H Komiyama and Seth GN Grant
Design Case - ProbeDesign119 - back to list
Summary
./analyse_probe_search_results : Wed Jun 28 17:02:22 2006
Connected to host: host, as user: user Database : southern_blot_designs Fetched probe design: ProbeDesign119 assembly : NCBIM36 bias : 3prime chromosome : 7 id : 113 strand : 1 ------------- Design window length: 2501bp All jobs for probe design are successful - total: 35 Fetched conf : Exonerate mouse_NCBIM36 (id: 2) Genome file(s) at: /blastdb/Mouse/NCBIM36/genome/softmasked_dusted Fetched 700 putative probes to analyse Selection criteria to rank probes: Minimum score ratio : 10 (i.e. self-hit must score at least 10 times higher than the next best hit) Maximum percent repetitive bases: 5 Favour probes to the 3prime end of design window ------------------------ Putative probes without hits : 0 Putative probes with single_self_hit : 700/700 Putative probes that are unique : 29/700 Putative probes exceeding selection criteria : 54/700 (both unique and not unique) Putative probes with successfully-picked primers: 54/54 Identified 14 'redundant' probes exceeding selection criteria (but wholly contained by other probes, and higher repetitive DNA content)
Image
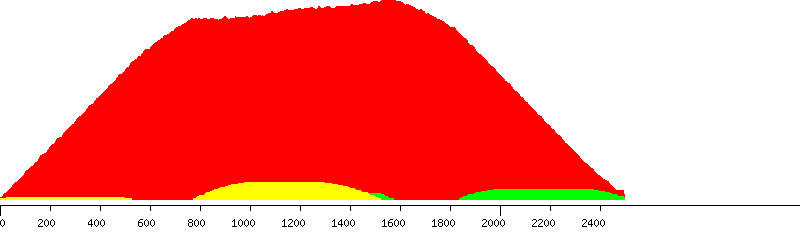
Unique (yellow), passed (green) and failed putative probes in the design window
Unique putative probes
| Putative probe ID | Length | Actual | %rep DNA | Distance |
|---|---|---|---|---|
| 106773 | 700 | 3500 | - | 980 |
| 106828 | 650 | 3250 | - | 992 |
| 106829 | 650 | 3250 | - | 1024 |
| 106888 | 600 | 3000 | - | 990 |
| 106889 | 600 | 3000 | - | 1020 |
| 106890 | 600 | 3000 | - | 1050 |
| 106891 | 600 | 3000 | - | 1080 |
| 106956 | 550 | 2750 | - | 999 |
| 106957 | 550 | 2750 | - | 1026 |
| 106958 | 550 | 2750 | - | 1053 |
| 106959 | 550 | 2750 | - | 1080 |
| 106960 | 550 | 2750 | - | 1107 |
| 106961 | 550 | 2750 | - | 1134 |
| 107032 | 500 | 2500 | - | 1000 |
| 107033 | 500 | 2500 | - | 1025 |
| 107034 | 500 | 2500 | - | 1050 |
| 107035 | 500 | 2500 | - | 1075 |
| 107036 | 500 | 2500 | - | 1100 |
| 107037 | 500 | 2500 | - | 1125 |
| 107038 | 500 | 2500 | - | 1150 |
| 107039 | 500 | 2500 | - | 1175 |
| 107040 | 500 | 2500 | - | 1200 |
| 106830 | 650 | 3250 | 0.6 | 1056 |
| 106892 | 600 | 3000 | 1.3 | 1110 |
| 106962 | 550 | 2750 | 1.6 | 1161 |
| 106774 | 700 | 3500 | 1.9 | 1015 |
| 107041 | 500 | 2500 | 4.6 | 1225 |
| 107071 | 500 | 2500 | 18.8 | 1975 |
| 107072 | 500 | 2500 | 18.8 | 2000 |
Putative probes
| Putative probe ID | Length | Score ratio | Actual | %rep DNA | Distance | Overall |
|---|---|---|---|---|---|---|
| 106723 | 750 | 22.1 | 3750 | 1.3 | 962 | 19.4 |
| 106827 | 650 | 19.1 | 3250 | 0.3 | 960 | 18.5 |
| 106887 | 600 | 17.6 | 3000 | 0.3 | 960 | 17.0 |
| 106955 | 550 | 16.7 | 2750 | - | 972 | 16.7 |
| 107031 | 500 | 15.7 | 2500 | - | 975 | 15.7 |
| 106772 | 700 | 20.1 | 3500 | 2.4 | 945 | 15.3 |
| 106797 | 650 | 20.6 | 3250 | 3.4 | 0 | 13.8 |
| 106722 | 750 | 21.6 | 3750 | 4.9 | 925 | 11.7 |
| 106857 | 600 | 19.0 | 3000 | 3.7 | 60 | 11.7 |
| 106954 | 550 | 15.8 | 2750 | 3.1 | 945 | 9.6 |
| 107030 | 500 | 14.4 | 2500 | 2.4 | 950 | 9.6 |
Overall score = score_ratio - 2 * repetitive DNA content
Redundant probes
| Putative probe ID | Length | Score ratio | Actual | %rep DNA | Distance | Overall |
|---|---|---|---|---|---|---|
| 106855 | 600 | 19.0 | 3000 | 3.7 | 0 | 11.7 |
| 106856 | 600 | 19.0 | 3000 | 3.7 | 30 | 11.7 |
| 106919 | 550 | 17.4 | 2750 | 4.0 | 0 | 9.4 |
| 106920 | 550 | 17.4 | 2750 | 4.0 | 27 | 9.4 |
| 106921 | 550 | 17.4 | 2750 | 4.0 | 54 | 9.4 |
| 106922 | 550 | 17.4 | 2750 | 4.0 | 81 | 9.4 |
| 106923 | 550 | 17.4 | 2750 | 4.0 | 108 | 9.4 |
| 106992 | 500 | 15.8 | 2500 | 4.4 | 0 | 7.0 |
| 106993 | 500 | 15.8 | 2500 | 4.4 | 25 | 7.0 |
| 106994 | 500 | 15.8 | 2500 | 4.4 | 50 | 7.0 |
| 106995 | 500 | 15.8 | 2500 | 4.4 | 75 | 7.0 |
| 106996 | 500 | 15.8 | 2500 | 4.4 | 100 | 7.0 |
| 106997 | 500 | 15.8 | 2500 | 4.4 | 125 | 7.0 |
| 106998 | 500 | 15.8 | 2500 | 4.4 | 150 | 7.0 |
Overall score = score_ratio - 2 * repetitive DNA content
