Automated design of genomic Southern blot probes
Mike DR Croning, David G Fricker, Noboru H Komiyama and Seth GN Grant
Design Case - ProbeDesign24 - back to list
Summary
./analyse_probe_search_results : Mon Sep 5 11:45:28 2005
Connected to host: host, as user: user Database : gene_targeting_production Fetched probe design: ProbeDesign24 assembly : NCBIM33 bias : 5prime chromosome : 12 id : 1 strand : 1 ------------- Design window length: 2001bp All jobs for probe design are successful - total: 41 Fetched conf : Exonerate mouse_NCBI33 (id: 1) Genome file(s) at: /blastdb/Mouse/NCBIM33/genome/masked_dusted Fetched 405 putative probes to analyse Selection criteria to rank probes: Minimum score ratio : 10 (i.e. self-hit must score at least 10 times higher than the next best hit) Maximum percent repetitive bases: 5 Favour probes to the 5prime end of design window ------------------------ Putative probes without hits : 0 Putative probes with single_self_hit : 405/405 Putative probes exceeding selection criteria: 252/405 PASSED (GREEN) AND FAILED PUTATIVE PROBES IN THE DESIGN WINDOW --------------------------------------------------------------
Image
Unique (yellow), passed (green) and failed putative probes in the design window
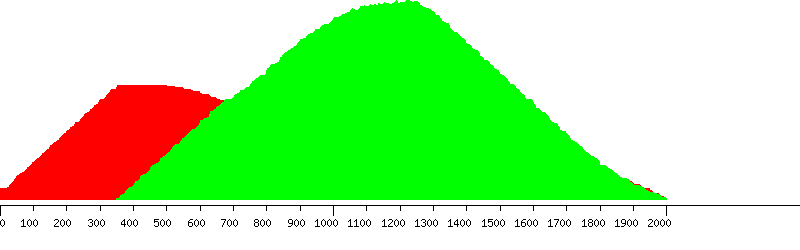
Unique putative probes
No probes
Putative probes
| Putative probe ID | Length | Score ratio | Actual | %rep DNA | Distance | Overall |
|---|---|---|---|---|---|---|
| 15 | 1000 | 32.9 | 5000 | - | 700 | 32.9 |
| 16 | 1000 | 32.9 | 5000 | - | 750 | 32.9 |
| 17 | 1000 | 32.9 | 5000 | - | 800 | 32.9 |
| 18 | 1000 | 32.9 | 5000 | - | 850 | 32.9 |
| 19 | 1000 | 32.9 | 5000 | - | 900 | 32.9 |
| 20 | 1000 | 32.9 | 5000 | - | 950 | 32.9 |
| 21 | 1000 | 32.9 | 5000 | - | 1000 | 32.9 |
| 37 | 950 | 31.2 | 4750 | - | 705 | 31.2 |
| 38 | 950 | 31.2 | 4750 | - | 752 | 31.2 |
| 39 | 950 | 31.2 | 4750 | - | 799 | 31.2 |
| 40 | 950 | 31.2 | 4750 | - | 846 | 31.2 |
| 41 | 950 | 31.2 | 4750 | - | 893 | 31.2 |
| 42 | 950 | 31.2 | 4750 | - | 940 | 31.2 |
| 43 | 950 | 31.2 | 4750 | - | 987 | 31.2 |
| 44 | 950 | 31.2 | 4750 | - | 1034 | 31.2 |
| 9 | 1000 | 29.9 | 5000 | - | 400 | 29.9 |
| 10 | 1000 | 29.9 | 5000 | - | 450 | 29.9 |
| 11 | 1000 | 29.9 | 5000 | - | 500 | 29.9 |
| 12 | 1000 | 29.9 | 5000 | - | 550 | 29.9 |
| 13 | 1000 | 29.9 | 5000 | - | 600 | 29.9 |
| 14 | 1000 | 29.9 | 5000 | - | 650 | 29.9 |
| 60 | 900 | 29.6 | 4500 | - | 675 | 29.6 |
| 61 | 900 | 29.6 | 4500 | - | 720 | 29.6 |
| 62 | 900 | 29.6 | 4500 | - | 765 | 29.6 |
| 63 | 900 | 29.6 | 4500 | - | 810 | 29.6 |
| 64 | 900 | 29.6 | 4500 | - | 855 | 29.6 |
| 65 | 900 | 29.6 | 4500 | - | 900 | 29.6 |
| 66 | 900 | 29.6 | 4500 | - | 945 | 29.6 |
| 67 | 900 | 29.6 | 4500 | - | 990 | 29.6 |
| 68 | 900 | 29.6 | 4500 | - | 1035 | 29.6 |
| 69 | 900 | 29.6 | 4500 | - | 1080 | 29.6 |
| 31 | 950 | 28.4 | 4750 | - | 423 | 28.4 |
| 32 | 950 | 28.4 | 4750 | - | 470 | 28.4 |
| 33 | 950 | 28.4 | 4750 | - | 517 | 28.4 |
| 34 | 950 | 28.4 | 4750 | - | 564 | 28.4 |
| 35 | 950 | 28.4 | 4750 | - | 611 | 28.4 |
| 36 | 950 | 28.4 | 4750 | - | 658 | 28.4 |
| 86 | 850 | 28.0 | 4250 | - | 672 | 28.0 |
| 87 | 850 | 28.0 | 4250 | - | 714 | 28.0 |
| 88 | 850 | 28.0 | 4250 | - | 756 | 28.0 |
| 89 | 850 | 28.0 | 4250 | - | 798 | 28.0 |
| 90 | 850 | 28.0 | 4250 | - | 840 | 28.0 |
| 91 | 850 | 28.0 | 4250 | - | 882 | 28.0 |
| 92 | 850 | 28.0 | 4250 | - | 924 | 28.0 |
| 93 | 850 | 28.0 | 4250 | - | 966 | 28.0 |
| 94 | 850 | 28.0 | 4250 | - | 1008 | 28.0 |
| 95 | 850 | 28.0 | 4250 | - | 1050 | 28.0 |
| 96 | 850 | 28.0 | 4250 | - | 1092 | 28.0 |
| 97 | 850 | 28.0 | 4250 | - | 1134 | 28.0 |
| 54 | 900 | 26.9 | 4500 | - | 405 | 26.9 |
| 55 | 900 | 26.9 | 4500 | - | 450 | 26.9 |
| 56 | 900 | 26.9 | 4500 | - | 495 | 26.9 |
| 57 | 900 | 26.9 | 4500 | - | 540 | 26.9 |
| 58 | 900 | 26.9 | 4500 | - | 585 | 26.9 |
| 59 | 900 | 26.9 | 4500 | - | 630 | 26.9 |
| 115 | 800 | 26.3 | 4000 | - | 680 | 26.3 |
| 116 | 800 | 26.3 | 4000 | - | 720 | 26.3 |
| 117 | 800 | 26.3 | 4000 | - | 760 | 26.3 |
| 118 | 800 | 26.3 | 4000 | - | 800 | 26.3 |
| 119 | 800 | 26.3 | 4000 | - | 840 | 26.3 |
| 120 | 800 | 26.3 | 4000 | - | 880 | 26.3 |
| 121 | 800 | 26.3 | 4000 | - | 920 | 26.3 |
| 122 | 800 | 26.3 | 4000 | - | 960 | 26.3 |
| 123 | 800 | 26.3 | 4000 | - | 1000 | 26.3 |
| 124 | 800 | 26.3 | 4000 | - | 1040 | 26.3 |
| 125 | 800 | 26.3 | 4000 | - | 1080 | 26.3 |
| 126 | 800 | 26.3 | 4000 | - | 1120 | 26.3 |
| 127 | 800 | 26.3 | 4000 | - | 1160 | 26.3 |
| 128 | 800 | 26.3 | 4000 | - | 1200 | 26.3 |
| 30 | 950 | 28.4 | 4750 | 1.3 | 376 | 25.9 |
| 80 | 850 | 25.4 | 4250 | - | 420 | 25.4 |
| 81 | 850 | 25.4 | 4250 | - | 462 | 25.4 |
| 82 | 850 | 25.4 | 4250 | - | 504 | 25.4 |
| 83 | 850 | 25.4 | 4250 | - | 546 | 25.4 |
| 84 | 850 | 25.4 | 4250 | - | 588 | 25.4 |
| 85 | 850 | 25.4 | 4250 | - | 630 | 25.4 |
| 147 | 750 | 24.7 | 3750 | - | 666 | 24.7 |
| 148 | 750 | 24.7 | 3750 | - | 703 | 24.7 |
| 149 | 750 | 24.7 | 3750 | - | 740 | 24.7 |
| 150 | 750 | 24.7 | 3750 | - | 777 | 24.7 |
| 151 | 750 | 24.7 | 3750 | - | 814 | 24.7 |
| 152 | 750 | 24.7 | 3750 | - | 851 | 24.7 |
| 153 | 750 | 24.7 | 3750 | - | 888 | 24.7 |
| 154 | 750 | 24.7 | 3750 | - | 925 | 24.7 |
| 155 | 750 | 24.7 | 3750 | - | 962 | 24.7 |
| 156 | 750 | 24.7 | 3750 | - | 999 | 24.7 |
| 157 | 750 | 24.7 | 3750 | - | 1036 | 24.7 |
| 158 | 750 | 24.7 | 3750 | - | 1073 | 24.7 |
| 159 | 750 | 24.7 | 3750 | - | 1110 | 24.7 |
| 160 | 750 | 24.7 | 3750 | - | 1147 | 24.7 |
| 161 | 750 | 24.7 | 3750 | - | 1184 | 24.7 |
| 162 | 750 | 24.7 | 3750 | - | 1221 | 24.7 |
| 108 | 800 | 24.0 | 4000 | - | 400 | 24.0 |
| 109 | 800 | 24.0 | 4000 | - | 440 | 24.0 |
| 110 | 800 | 24.0 | 4000 | - | 480 | 24.0 |
| 111 | 800 | 24.0 | 4000 | - | 520 | 24.0 |
| 112 | 800 | 24.0 | 4000 | - | 560 | 24.0 |
| 113 | 800 | 24.0 | 4000 | - | 600 | 24.0 |
| 114 | 800 | 24.0 | 4000 | - | 640 | 24.0 |
| 79 | 850 | 25.4 | 4250 | 1.2 | 378 | 23.1 |
| 182 | 700 | 23.0 | 3500 | - | 665 | 23.0 |
| 183 | 700 | 23.0 | 3500 | - | 700 | 23.0 |
| 184 | 700 | 23.0 | 3500 | - | 735 | 23.0 |
| 185 | 700 | 23.0 | 3500 | - | 770 | 23.0 |
| 186 | 700 | 23.0 | 3500 | - | 805 | 23.0 |
| 187 | 700 | 23.0 | 3500 | - | 840 | 23.0 |
| 188 | 700 | 23.0 | 3500 | - | 875 | 23.0 |
| 189 | 700 | 23.0 | 3500 | - | 910 | 23.0 |
| 190 | 700 | 23.0 | 3500 | - | 945 | 23.0 |
| 191 | 700 | 23.0 | 3500 | - | 980 | 23.0 |
| 192 | 700 | 23.0 | 3500 | - | 1015 | 23.0 |
| 193 | 700 | 23.0 | 3500 | - | 1050 | 23.0 |
| 194 | 700 | 23.0 | 3500 | - | 1085 | 23.0 |
| 195 | 700 | 23.0 | 3500 | - | 1120 | 23.0 |
| 196 | 700 | 23.0 | 3500 | - | 1155 | 23.0 |
| 197 | 700 | 23.0 | 3500 | - | 1190 | 23.0 |
| 198 | 700 | 23.0 | 3500 | - | 1225 | 23.0 |
| 140 | 750 | 22.5 | 3750 | - | 407 | 22.5 |
| 141 | 750 | 22.5 | 3750 | - | 444 | 22.5 |
| 142 | 750 | 22.5 | 3750 | - | 481 | 22.5 |
| 143 | 750 | 22.5 | 3750 | - | 518 | 22.5 |
| 144 | 750 | 22.5 | 3750 | - | 555 | 22.5 |
| 145 | 750 | 22.5 | 3750 | - | 592 | 22.5 |
| 146 | 750 | 22.5 | 3750 | - | 629 | 22.5 |
| 222 | 650 | 21.4 | 3250 | - | 672 | 21.4 |
| 223 | 650 | 21.4 | 3250 | - | 704 | 21.4 |
| 224 | 650 | 21.4 | 3250 | - | 736 | 21.4 |
| 225 | 650 | 21.4 | 3250 | - | 768 | 21.4 |
| 226 | 650 | 21.4 | 3250 | - | 800 | 21.4 |
| 227 | 650 | 21.4 | 3250 | - | 832 | 21.4 |
| 228 | 650 | 21.4 | 3250 | - | 864 | 21.4 |
| 229 | 650 | 21.4 | 3250 | - | 896 | 21.4 |
| 230 | 650 | 21.4 | 3250 | - | 928 | 21.4 |
| 231 | 650 | 21.4 | 3250 | - | 960 | 21.4 |
| 232 | 650 | 21.4 | 3250 | - | 992 | 21.4 |
| 233 | 650 | 21.4 | 3250 | - | 1024 | 21.4 |
| 234 | 650 | 21.4 | 3250 | - | 1056 | 21.4 |
| 235 | 650 | 21.4 | 3250 | - | 1088 | 21.4 |
| 236 | 650 | 21.4 | 3250 | - | 1120 | 21.4 |
| 237 | 650 | 21.4 | 3250 | - | 1152 | 21.4 |
| 238 | 650 | 21.4 | 3250 | - | 1184 | 21.4 |
| 239 | 650 | 21.4 | 3250 | - | 1216 | 21.4 |
| 175 | 700 | 21.0 | 3500 | - | 420 | 21.0 |
| 176 | 700 | 21.0 | 3500 | - | 455 | 21.0 |
| 177 | 700 | 21.0 | 3500 | - | 490 | 21.0 |
| 178 | 700 | 21.0 | 3500 | - | 525 | 21.0 |
| 179 | 700 | 21.0 | 3500 | - | 560 | 21.0 |
| 180 | 700 | 21.0 | 3500 | - | 595 | 21.0 |
| 181 | 700 | 21.0 | 3500 | - | 630 | 21.0 |
| 53 | 900 | 26.9 | 4500 | 3.1 | 360 | 20.7 |
| 8 | 1000 | 27.8 | 5000 | 3.8 | 350 | 20.2 |
| 174 | 700 | 21.0 | 3500 | 0.4 | 385 | 20.1 |
| 268 | 600 | 19.7 | 3000 | - | 720 | 19.7 |
| 269 | 600 | 19.7 | 3000 | - | 750 | 19.7 |
| 270 | 600 | 19.7 | 3000 | - | 780 | 19.7 |
| 271 | 600 | 19.7 | 3000 | - | 810 | 19.7 |
| 272 | 600 | 19.7 | 3000 | - | 840 | 19.7 |
| 273 | 600 | 19.7 | 3000 | - | 870 | 19.7 |
| 274 | 600 | 19.7 | 3000 | - | 900 | 19.7 |
| 275 | 600 | 19.7 | 3000 | - | 930 | 19.7 |
| 276 | 600 | 19.7 | 3000 | - | 960 | 19.7 |
| 277 | 600 | 19.7 | 3000 | - | 990 | 19.7 |
| 278 | 600 | 19.7 | 3000 | - | 1020 | 19.7 |
| 279 | 600 | 19.7 | 3000 | - | 1050 | 19.7 |
| 280 | 600 | 19.7 | 3000 | - | 1080 | 19.7 |
| 281 | 600 | 19.7 | 3000 | - | 1110 | 19.7 |
| 282 | 600 | 19.7 | 3000 | - | 1140 | 19.7 |
| 283 | 600 | 19.7 | 3000 | - | 1170 | 19.7 |
| 284 | 600 | 19.7 | 3000 | - | 1200 | 19.7 |
| 285 | 600 | 19.7 | 3000 | - | 1230 | 19.7 |
| 214 | 650 | 19.5 | 3250 | - | 416 | 19.5 |
| 215 | 650 | 19.5 | 3250 | - | 448 | 19.5 |
| 216 | 650 | 19.5 | 3250 | - | 480 | 19.5 |
| 217 | 650 | 19.5 | 3250 | - | 512 | 19.5 |
| 218 | 650 | 19.5 | 3250 | - | 544 | 19.5 |
| 219 | 650 | 19.5 | 3250 | - | 576 | 19.5 |
| 220 | 650 | 19.5 | 3250 | - | 608 | 19.5 |
| 221 | 650 | 19.5 | 3250 | - | 640 | 19.5 |
| 213 | 650 | 19.5 | 3250 | 0.6 | 384 | 18.2 |
| 337 | 550 | 18.2 | 2750 | - | 1242 | 18.2 |
| 319 | 550 | 18.1 | 2750 | - | 756 | 18.1 |
| 320 | 550 | 18.1 | 2750 | - | 783 | 18.1 |
| 321 | 550 | 18.1 | 2750 | - | 810 | 18.1 |
| 322 | 550 | 18.1 | 2750 | - | 837 | 18.1 |
| 323 | 550 | 18.1 | 2750 | - | 864 | 18.1 |
| 324 | 550 | 18.1 | 2750 | - | 891 | 18.1 |
| 325 | 550 | 18.1 | 2750 | - | 918 | 18.1 |
| 326 | 550 | 18.1 | 2750 | - | 945 | 18.1 |
| 327 | 550 | 18.1 | 2750 | - | 972 | 18.1 |
| 328 | 550 | 18.1 | 2750 | - | 999 | 18.1 |
| 329 | 550 | 18.1 | 2750 | - | 1026 | 18.1 |
| 330 | 550 | 18.1 | 2750 | - | 1053 | 18.1 |
| 331 | 550 | 18.1 | 2750 | - | 1080 | 18.1 |
| 332 | 550 | 18.1 | 2750 | - | 1107 | 18.1 |
| 333 | 550 | 18.1 | 2750 | - | 1134 | 18.1 |
| 334 | 550 | 18.1 | 2750 | - | 1161 | 18.1 |
| 335 | 550 | 18.1 | 2750 | - | 1188 | 18.1 |
| 336 | 550 | 18.1 | 2750 | - | 1215 | 18.1 |
| 257 | 600 | 18.0 | 3000 | - | 390 | 18.0 |
| 258 | 600 | 18.0 | 3000 | - | 420 | 18.0 |
| 259 | 600 | 18.0 | 3000 | - | 450 | 18.0 |
| 260 | 600 | 18.0 | 3000 | - | 480 | 18.0 |
| 261 | 600 | 18.0 | 3000 | - | 510 | 18.0 |
| 262 | 600 | 18.0 | 3000 | - | 540 | 18.0 |
| 263 | 600 | 18.0 | 3000 | - | 570 | 18.0 |
| 264 | 600 | 18.0 | 3000 | - | 600 | 18.0 |
| 265 | 600 | 18.0 | 3000 | - | 630 | 18.0 |
| 266 | 600 | 18.0 | 3000 | - | 660 | 18.0 |
| 139 | 750 | 22.5 | 3750 | 2.4 | 370 | 17.7 |
| 107 | 800 | 24.0 | 4000 | 3.5 | 360 | 17.0 |
| 306 | 550 | 16.5 | 2750 | - | 405 | 16.5 |
| 307 | 550 | 16.5 | 2750 | - | 432 | 16.5 |
| 308 | 550 | 16.5 | 2750 | - | 459 | 16.5 |
| 309 | 550 | 16.5 | 2750 | - | 486 | 16.5 |
| 310 | 550 | 16.5 | 2750 | - | 513 | 16.5 |
| 311 | 550 | 16.5 | 2750 | - | 540 | 16.5 |
| 312 | 550 | 16.5 | 2750 | - | 567 | 16.5 |
| 313 | 550 | 16.5 | 2750 | - | 594 | 16.5 |
| 314 | 550 | 16.5 | 2750 | - | 621 | 16.5 |
| 315 | 550 | 16.5 | 2750 | - | 648 | 16.5 |
| 377 | 500 | 16.4 | 2500 | - | 800 | 16.4 |
| 378 | 500 | 16.4 | 2500 | - | 825 | 16.4 |
| 379 | 500 | 16.4 | 2500 | - | 850 | 16.4 |
| 380 | 500 | 16.4 | 2500 | - | 875 | 16.4 |
| 381 | 500 | 16.4 | 2500 | - | 900 | 16.4 |
| 382 | 500 | 16.4 | 2500 | - | 925 | 16.4 |
| 383 | 500 | 16.4 | 2500 | - | 950 | 16.4 |
| 384 | 500 | 16.4 | 2500 | - | 975 | 16.4 |
| 385 | 500 | 16.4 | 2500 | - | 1000 | 16.4 |
| 386 | 500 | 16.4 | 2500 | - | 1025 | 16.4 |
| 387 | 500 | 16.4 | 2500 | - | 1050 | 16.4 |
| 388 | 500 | 16.4 | 2500 | - | 1075 | 16.4 |
| 389 | 500 | 16.4 | 2500 | - | 1100 | 16.4 |
| 390 | 500 | 16.4 | 2500 | - | 1125 | 16.4 |
| 391 | 500 | 16.4 | 2500 | - | 1150 | 16.4 |
| 392 | 500 | 16.4 | 2500 | - | 1175 | 16.4 |
| 393 | 500 | 16.4 | 2500 | - | 1200 | 16.4 |
| 394 | 500 | 16.4 | 2500 | - | 1225 | 16.4 |
| 361 | 500 | 15.0 | 2500 | - | 400 | 15.0 |
| 362 | 500 | 15.0 | 2500 | - | 425 | 15.0 |
| 363 | 500 | 15.0 | 2500 | - | 450 | 15.0 |
| 364 | 500 | 15.0 | 2500 | - | 475 | 15.0 |
| 365 | 500 | 15.0 | 2500 | - | 500 | 15.0 |
| 366 | 500 | 15.0 | 2500 | - | 525 | 15.0 |
| 367 | 500 | 15.0 | 2500 | - | 550 | 15.0 |
| 368 | 500 | 15.0 | 2500 | - | 575 | 15.0 |
| 369 | 500 | 15.0 | 2500 | - | 600 | 15.0 |
| 370 | 500 | 15.0 | 2500 | - | 625 | 15.0 |
| 371 | 500 | 15.0 | 2500 | - | 650 | 15.0 |
| 305 | 550 | 16.5 | 2750 | 1.8 | 378 | 12.8 |
| 360 | 500 | 15.0 | 2500 | 2.6 | 375 | 9.8 |
Overall score = score_ratio - 2 * repetitive DNA content
Redundant probes
| Putative probe ID | Length | Score ratio | Actual | %rep DNA | Distance | Overall |
|---|---|---|---|---|---|---|
| 256 | 600 | 18.0 | 3000 | 4.7 | 360 | 8.6 |
Overall score = score_ratio - 2 * repetitive DNA content
