Automated design of genomic Southern blot probes
Mike DR Croning, David G Fricker, Noboru H Komiyama and Seth GN Grant
Design Case - ProbeDesign43 - back to list
Summary
./analyse_probe_search_results : Tue Sep 13 14:38:24 2005
Connected to host: host, as user: user Database : southern_blot_designs Fetched probe design: ProbeDesign43 assembly : NCBIM34 bias : 3prime chromosome : X id : 25 strand : 1 ------------- Design window length: 3001bp All jobs for probe design are successful - total: 46 Fetched conf : Exonerate mouse_NCBIM34 (id: 1) Genome file(s) at: /blastdb/Mouse/NCBIM34/softmasked_dusted Fetched 907 putative probes to analyse Selection criteria to rank probes: Minimum score ratio : 10 (i.e. self-hit must score at least 10 times higher than the next best hit) Maximum percent repetitive bases: 5 Favour probes to the 3prime end of design window ------------------------ Putative probes without hits : 0 Putative probes with single_self_hit : 907/907 Putative probes exceeding selection criteria: 214/907 PASSED (GREEN) AND FAILED PUTATIVE PROBES IN THE DESIGN WINDOW --------------------------------------------------------------
Image
Unique (yellow), passed (green) and failed putative probes in the design window
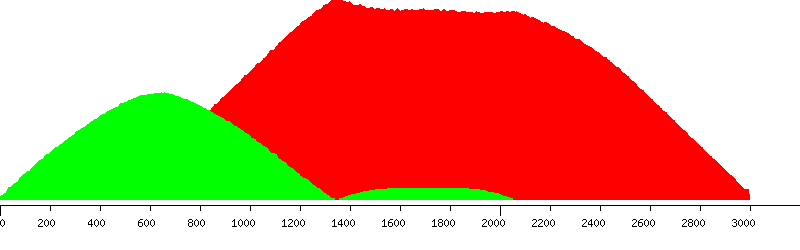
Unique putative probes
No probes
Putative probes
| Putative probe ID | Length | Score ratio | Actual | %rep DNA | Distance | Overall |
|---|---|---|---|---|---|---|
| 27287 | 1150 | 34.6 | 5750 | 1.0 | 1710 | 32.7 |
| 27358 | 1050 | 33.2 | 5250 | 0.3 | 1716 | 32.7 |
| 27321 | 1100 | 34.8 | 5500 | 1.5 | 1705 | 31.9 |
| 27398 | 1000 | 31.6 | 5000 | - | 1750 | 31.6 |
| 27322 | 1100 | 33.1 | 5500 | 0.9 | 1760 | 31.3 |
| 27225 | 1250 | 31.7 | 6250 | 0.8 | 1736 | 30.1 |
| 27441 | 950 | 30.1 | 4750 | - | 1739 | 30.1 |
| 27442 | 950 | 30.1 | 4750 | - | 1786 | 30.1 |
| 27359 | 1050 | 31.6 | 5250 | 1.0 | 1768 | 29.7 |
| 27399 | 1000 | 31.6 | 5000 | 1.0 | 1800 | 29.6 |
| 27397 | 1000 | 31.6 | 5000 | 1.1 | 1700 | 29.4 |
| 27255 | 1200 | 30.5 | 6000 | 0.8 | 1740 | 28.8 |
| 27256 | 1200 | 30.5 | 6000 | 0.8 | 1800 | 28.8 |
| 27487 | 900 | 28.5 | 4500 | - | 1755 | 28.5 |
| 27488 | 900 | 28.5 | 4500 | - | 1800 | 28.5 |
| 27489 | 900 | 28.5 | 4500 | - | 1845 | 28.5 |
| 27486 | 900 | 28.5 | 4500 | 0.1 | 1710 | 28.3 |
| 27196 | 1300 | 33.0 | 6500 | 2.4 | 1690 | 28.2 |
| 27400 | 1000 | 30.1 | 5000 | 1.0 | 1850 | 28.1 |
| 27443 | 950 | 30.1 | 4750 | 1.1 | 1833 | 28.0 |
| 27288 | 1150 | 29.2 | 5750 | 0.9 | 1767 | 27.4 |
| 27289 | 1150 | 29.2 | 5750 | 0.9 | 1824 | 27.4 |
| 27536 | 850 | 26.9 | 4250 | - | 1722 | 26.9 |
| 27537 | 850 | 26.9 | 4250 | - | 1764 | 26.9 |
| 27538 | 850 | 26.9 | 4250 | - | 1806 | 26.9 |
| 27539 | 850 | 26.9 | 4250 | - | 1848 | 26.9 |
| 27540 | 850 | 26.9 | 4250 | - | 1890 | 26.9 |
| 27444 | 950 | 28.6 | 4750 | 1.1 | 1880 | 26.5 |
| 27490 | 900 | 28.5 | 4500 | 1.1 | 1890 | 26.3 |
| 27323 | 1100 | 27.9 | 5500 | 0.9 | 1815 | 26.1 |
| 27324 | 1100 | 27.9 | 5500 | 0.9 | 1870 | 26.1 |
| 27440 | 950 | 30.1 | 4750 | 2.0 | 1692 | 26.1 |
| 27590 | 800 | 25.3 | 4000 | - | 1720 | 25.3 |
| 27591 | 800 | 25.3 | 4000 | - | 1760 | 25.3 |
| 27592 | 800 | 25.3 | 4000 | - | 1800 | 25.3 |
| 27593 | 800 | 25.3 | 4000 | - | 1840 | 25.3 |
| 27594 | 800 | 25.3 | 4000 | - | 1880 | 25.3 |
| 27595 | 800 | 25.3 | 4000 | - | 1920 | 25.3 |
| 27596 | 800 | 25.3 | 4000 | - | 1960 | 25.3 |
| 27491 | 900 | 27.1 | 4500 | 1.1 | 1935 | 24.9 |
| 27360 | 1050 | 26.6 | 5250 | 1.0 | 1820 | 24.7 |
| 27361 | 1050 | 26.6 | 5250 | 1.0 | 1872 | 24.7 |
| 27362 | 1050 | 26.6 | 5250 | 1.0 | 1924 | 24.7 |
| 27541 | 850 | 26.9 | 4250 | 1.2 | 1932 | 24.5 |
| 27357 | 1050 | 33.2 | 5250 | 4.5 | 1664 | 24.3 |
| 27224 | 1250 | 31.7 | 6250 | 3.8 | 1674 | 24.2 |
| 27650 | 750 | 23.7 | 3750 | - | 1739 | 23.7 |
| 27651 | 750 | 23.7 | 3750 | - | 1776 | 23.7 |
| 27652 | 750 | 23.7 | 3750 | - | 1813 | 23.7 |
| 27653 | 750 | 23.7 | 3750 | - | 1850 | 23.7 |
| 27654 | 750 | 23.7 | 3750 | - | 1887 | 23.7 |
| 27655 | 750 | 23.7 | 3750 | - | 1924 | 23.7 |
| 27656 | 750 | 23.7 | 3750 | - | 1961 | 23.7 |
| 27657 | 750 | 23.7 | 3750 | - | 1998 | 23.7 |
| 27254 | 1200 | 30.5 | 6000 | 3.4 | 1680 | 23.6 |
| 27401 | 1000 | 25.4 | 5000 | 1.0 | 1900 | 23.4 |
| 27402 | 1000 | 25.4 | 5000 | 1.0 | 1950 | 23.4 |
| 27542 | 850 | 25.6 | 4250 | 1.2 | 1974 | 23.2 |
| 27543 | 850 | 25.6 | 4250 | 1.2 | 2016 | 23.2 |
| 27597 | 800 | 25.3 | 4000 | 1.2 | 2000 | 22.8 |
| 27713 | 700 | 22.2 | 3500 | - | 1715 | 22.2 |
| 27714 | 700 | 22.2 | 3500 | - | 1750 | 22.2 |
| 27715 | 700 | 22.2 | 3500 | - | 1785 | 22.2 |
| 27716 | 700 | 22.2 | 3500 | - | 1820 | 22.2 |
| 27717 | 700 | 22.2 | 3500 | - | 1855 | 22.2 |
| 27718 | 700 | 22.2 | 3500 | - | 1890 | 22.2 |
| 27719 | 700 | 22.2 | 3500 | - | 1925 | 22.2 |
| 27720 | 700 | 22.2 | 3500 | - | 1960 | 22.2 |
| 27721 | 700 | 22.2 | 3500 | - | 1995 | 22.2 |
| 27722 | 700 | 22.2 | 3500 | - | 2030 | 22.2 |
| 27445 | 950 | 24.1 | 4750 | 1.1 | 1927 | 22.0 |
| 27446 | 950 | 24.1 | 4750 | 1.1 | 1974 | 22.0 |
| 27447 | 950 | 24.1 | 4750 | 1.1 | 2021 | 22.0 |
| 27598 | 800 | 24.1 | 4000 | 1.2 | 2040 | 21.6 |
| 27723 | 700 | 22.2 | 3500 | 0.3 | 2065 | 21.6 |
| 27649 | 750 | 23.7 | 3750 | 1.2 | 1702 | 21.3 |
| 27658 | 750 | 23.7 | 3750 | 1.3 | 2035 | 21.1 |
| 27492 | 900 | 22.8 | 4500 | 1.1 | 1980 | 20.6 |
| 27493 | 900 | 22.8 | 4500 | 1.1 | 2025 | 20.6 |
| 27494 | 900 | 22.8 | 4500 | 1.1 | 2070 | 20.6 |
| 27784 | 650 | 20.6 | 3250 | - | 1728 | 20.6 |
| 27785 | 650 | 20.6 | 3250 | - | 1760 | 20.6 |
| 27786 | 650 | 20.6 | 3250 | - | 1792 | 20.6 |
| 27787 | 650 | 20.6 | 3250 | - | 1824 | 20.6 |
| 27788 | 650 | 20.6 | 3250 | - | 1856 | 20.6 |
| 27789 | 650 | 20.6 | 3250 | - | 1888 | 20.6 |
| 27790 | 650 | 20.6 | 3250 | - | 1920 | 20.6 |
| 27791 | 650 | 20.6 | 3250 | - | 1952 | 20.6 |
| 27792 | 650 | 20.6 | 3250 | - | 1984 | 20.6 |
| 27793 | 650 | 20.6 | 3250 | - | 2016 | 20.6 |
| 27794 | 650 | 20.6 | 3250 | - | 2048 | 20.6 |
| 27795 | 650 | 20.6 | 3250 | - | 2080 | 20.6 |
| 27796 | 650 | 20.6 | 3250 | - | 2112 | 20.6 |
| 27659 | 750 | 22.6 | 3750 | 1.3 | 2072 | 19.9 |
| 27660 | 750 | 22.6 | 3750 | 1.3 | 2109 | 19.9 |
| 27535 | 850 | 26.9 | 4250 | 3.6 | 1680 | 19.6 |
| 27862 | 600 | 19.5 | 3000 | - | 1740 | 19.5 |
| 27724 | 700 | 22.2 | 3500 | 1.4 | 2100 | 19.3 |
| 27544 | 850 | 21.6 | 4250 | 1.2 | 2058 | 19.2 |
| 27545 | 850 | 21.6 | 4250 | 1.2 | 2100 | 19.2 |
| 27863 | 600 | 19.0 | 3000 | - | 1770 | 19.0 |
| 27864 | 600 | 19.0 | 3000 | - | 1800 | 19.0 |
| 27865 | 600 | 19.0 | 3000 | - | 1830 | 19.0 |
| 27866 | 600 | 19.0 | 3000 | - | 1860 | 19.0 |
| 27867 | 600 | 19.0 | 3000 | - | 1890 | 19.0 |
| 27868 | 600 | 19.0 | 3000 | - | 1920 | 19.0 |
| 27869 | 600 | 19.0 | 3000 | - | 1950 | 19.0 |
| 27870 | 600 | 19.0 | 3000 | - | 1980 | 19.0 |
| 27871 | 600 | 19.0 | 3000 | - | 2010 | 19.0 |
| 27872 | 600 | 19.0 | 3000 | - | 2040 | 19.0 |
| 27873 | 600 | 19.0 | 3000 | - | 2070 | 19.0 |
| 27874 | 600 | 19.0 | 3000 | - | 2100 | 19.0 |
| 27875 | 600 | 19.0 | 3000 | - | 2130 | 19.0 |
| 27876 | 600 | 19.0 | 3000 | - | 2160 | 19.0 |
| 27725 | 700 | 21.1 | 3500 | 1.4 | 2135 | 18.2 |
| 27599 | 800 | 20.3 | 4000 | 1.2 | 2080 | 17.8 |
| 27600 | 800 | 20.3 | 4000 | 1.2 | 2120 | 17.8 |
| 27601 | 800 | 20.3 | 4000 | 1.2 | 2160 | 17.8 |
| 27797 | 650 | 20.6 | 3250 | 1.5 | 2144 | 17.5 |
| 27952 | 550 | 17.4 | 2750 | - | 1809 | 17.4 |
| 27953 | 550 | 17.4 | 2750 | - | 1836 | 17.4 |
| 27954 | 550 | 17.4 | 2750 | - | 1863 | 17.4 |
| 27955 | 550 | 17.4 | 2750 | - | 1890 | 17.4 |
| 27956 | 550 | 17.4 | 2750 | - | 1917 | 17.4 |
| 27957 | 550 | 17.4 | 2750 | - | 1944 | 17.4 |
| 27958 | 550 | 17.4 | 2750 | - | 1971 | 17.4 |
| 27959 | 550 | 17.4 | 2750 | - | 1998 | 17.4 |
| 27960 | 550 | 17.4 | 2750 | - | 2025 | 17.4 |
| 27961 | 550 | 17.4 | 2750 | - | 2052 | 17.4 |
| 27962 | 550 | 17.4 | 2750 | - | 2079 | 17.4 |
| 27963 | 550 | 17.4 | 2750 | - | 2106 | 17.4 |
| 27964 | 550 | 17.4 | 2750 | - | 2133 | 17.4 |
| 27965 | 550 | 17.4 | 2750 | - | 2160 | 17.4 |
| 27966 | 550 | 17.4 | 2750 | - | 2187 | 17.4 |
| 27967 | 550 | 17.4 | 2750 | 0.2 | 2214 | 17.0 |
| 27798 | 650 | 19.6 | 3250 | 1.5 | 2176 | 16.5 |
| 27799 | 650 | 19.6 | 3250 | 1.5 | 2208 | 16.5 |
| 27661 | 750 | 19.0 | 3750 | 1.3 | 2146 | 16.4 |
| 27662 | 750 | 19.0 | 3750 | 1.3 | 2183 | 16.4 |
| 27663 | 750 | 19.0 | 3750 | 1.3 | 2220 | 16.4 |
| 27783 | 650 | 20.7 | 3250 | 2.3 | 1696 | 16.1 |
| 28050 | 500 | 15.8 | 2500 | - | 1850 | 15.8 |
| 28051 | 500 | 15.8 | 2500 | - | 1875 | 15.8 |
| 28052 | 500 | 15.8 | 2500 | - | 1900 | 15.8 |
| 28053 | 500 | 15.8 | 2500 | - | 1925 | 15.8 |
| 28054 | 500 | 15.8 | 2500 | - | 1950 | 15.8 |
| 28055 | 500 | 15.8 | 2500 | - | 1975 | 15.8 |
| 28056 | 500 | 15.8 | 2500 | - | 2000 | 15.8 |
| 28057 | 500 | 15.8 | 2500 | - | 2025 | 15.8 |
| 28058 | 500 | 15.8 | 2500 | - | 2050 | 15.8 |
| 28059 | 500 | 15.8 | 2500 | - | 2075 | 15.8 |
| 28060 | 500 | 15.8 | 2500 | - | 2100 | 15.8 |
| 28061 | 500 | 15.8 | 2500 | - | 2125 | 15.8 |
| 28062 | 500 | 15.8 | 2500 | - | 2150 | 15.8 |
| 28063 | 500 | 15.8 | 2500 | - | 2175 | 15.8 |
| 28064 | 500 | 15.8 | 2500 | - | 2200 | 15.8 |
| 28065 | 500 | 15.8 | 2500 | - | 2225 | 15.8 |
| 28066 | 500 | 15.8 | 2500 | - | 2250 | 15.8 |
| 27877 | 600 | 19.0 | 3000 | 1.7 | 2190 | 15.7 |
| 27726 | 700 | 17.8 | 3500 | 1.4 | 2170 | 14.9 |
| 27727 | 700 | 17.8 | 3500 | 1.4 | 2205 | 14.9 |
| 27728 | 700 | 17.8 | 3500 | 1.4 | 2240 | 14.9 |
| 27729 | 700 | 17.8 | 3500 | 1.4 | 2275 | 14.9 |
| 27878 | 600 | 18.1 | 3000 | 1.7 | 2220 | 14.7 |
| 27879 | 600 | 18.1 | 3000 | 1.7 | 2250 | 14.7 |
| 27837 | 600 | 17.5 | 3000 | 1.7 | 990 | 14.2 |
| 27838 | 600 | 17.5 | 3000 | 1.7 | 1020 | 14.2 |
| 27968 | 550 | 17.4 | 2750 | 1.8 | 2241 | 13.8 |
| 27800 | 650 | 16.5 | 3250 | 1.5 | 2240 | 13.4 |
| 27801 | 650 | 16.5 | 3250 | 1.5 | 2272 | 13.4 |
| 27802 | 650 | 16.5 | 3250 | 1.5 | 2304 | 13.4 |
| 27803 | 650 | 16.5 | 3250 | 1.5 | 2336 | 13.4 |
| 27969 | 550 | 16.6 | 2750 | 1.8 | 2268 | 12.9 |
| 27970 | 550 | 16.6 | 2750 | 1.8 | 2295 | 12.9 |
| 27922 | 550 | 16.1 | 2750 | 1.8 | 999 | 12.4 |
| 27923 | 550 | 16.1 | 2750 | 1.8 | 1026 | 12.4 |
| 27924 | 550 | 16.1 | 2750 | 1.8 | 1053 | 12.4 |
| 28068 | 500 | 16.2 | 2500 | 2.0 | 2300 | 12.2 |
| 27760 | 650 | 19.0 | 3250 | 3.5 | 960 | 11.9 |
| 27880 | 600 | 15.2 | 3000 | 1.7 | 2280 | 11.9 |
| 27881 | 600 | 15.2 | 3000 | 1.7 | 2310 | 11.9 |
| 27882 | 600 | 15.2 | 3000 | 1.7 | 2340 | 11.9 |
| 27883 | 600 | 15.2 | 3000 | 1.7 | 2370 | 11.9 |
| 28015 | 500 | 14.6 | 2500 | 1.4 | 975 | 11.8 |
| 28069 | 500 | 15.1 | 2500 | 2.0 | 2325 | 11.1 |
| 28070 | 500 | 15.1 | 2500 | 2.0 | 2350 | 11.1 |
| 28016 | 500 | 14.6 | 2500 | 2.0 | 1000 | 10.6 |
| 28017 | 500 | 14.6 | 2500 | 2.0 | 1025 | 10.6 |
| 28018 | 500 | 14.6 | 2500 | 2.0 | 1050 | 10.6 |
| 28019 | 500 | 14.6 | 2500 | 2.0 | 1075 | 10.6 |
| 28020 | 500 | 14.6 | 2500 | 2.0 | 1100 | 10.6 |
| 27971 | 550 | 14.0 | 2750 | 1.8 | 2322 | 10.3 |
| 27972 | 550 | 14.0 | 2750 | 1.8 | 2349 | 10.3 |
| 27973 | 550 | 14.0 | 2750 | 1.8 | 2376 | 10.3 |
| 27974 | 550 | 14.0 | 2750 | 1.8 | 2403 | 10.3 |
| 27975 | 550 | 14.0 | 2750 | 1.8 | 2430 | 10.3 |
| 27761 | 650 | 19.0 | 3250 | 4.6 | 992 | 9.8 |
| 27925 | 550 | 16.1 | 2750 | 3.3 | 1080 | 9.5 |
| 28021 | 500 | 14.6 | 2500 | 2.6 | 1125 | 9.4 |
| 27921 | 550 | 16.1 | 2750 | 3.5 | 972 | 9.2 |
| 28071 | 500 | 12.7 | 2500 | 2.0 | 2375 | 8.7 |
| 28072 | 500 | 12.7 | 2500 | 2.0 | 2400 | 8.7 |
| 28073 | 500 | 12.7 | 2500 | 2.0 | 2425 | 8.7 |
| 28074 | 500 | 12.7 | 2500 | 2.0 | 2450 | 8.7 |
| 28075 | 500 | 12.7 | 2500 | 2.0 | 2475 | 8.7 |
| 28014 | 500 | 14.6 | 2500 | 4.6 | 950 | 5.4 |
Overall score = score_ratio - 2 * repetitive DNA content
Redundant probes
| Putative probe ID | Length | Score ratio | Actual | %rep DNA | Distance | Overall |
|---|---|---|---|---|---|---|
| 27403 | 1000 | 25.4 | 5000 | 1.0 | 2000 | 23.4 |
| 27546 | 850 | 21.6 | 4250 | 1.2 | 2142 | 19.2 |
| 27602 | 800 | 20.3 | 4000 | 1.2 | 2200 | 17.8 |
| 27589 | 800 | 25.3 | 4000 | 3.9 | 1680 | 17.6 |
| 27712 | 700 | 22.2 | 3500 | 4.4 | 1680 | 13.3 |
| 27884 | 600 | 15.2 | 3000 | 1.7 | 2400 | 11.9 |
| 27836 | 600 | 17.5 | 3000 | 3.8 | 960 | 9.9 |
| 28076 | 500 | 12.7 | 2500 | 2.0 | 2500 | 8.7 |
Overall score = score_ratio - 2 * repetitive DNA content
