Automated design of genomic Southern blot probes
Mike DR Croning, David G Fricker, Noboru H Komiyama and Seth GN Grant
Design Case - ProbeDesign53 - back to list
Summary
./analyse_probe_search_results : Mon Sep 26 11:57:28 2005
Connected to host: host, as user: user Database : southern_blot_designs Fetched probe design: ProbeDesign53 assembly : NCBIM34 bias : 3prime chromosome : X id : 38 strand : 1 ------------- Design window length: 6001bp All jobs for probe design are successful - total: 108 Fetched conf : Exonerate mouse_NCBIM34 (id: 1) Genome file(s) at: /blastdb/Mouse/NCBIM34/softmasked_dusted Fetched 2146 putative probes to analyse Selection criteria to rank probes: Minimum score ratio : 10 (i.e. self-hit must score at least 10 times higher than the next best hit) Maximum percent repetitive bases: 5 Favour probes to the 3prime end of design window ------------------------ Putative probes without hits : 0 Putative probes with single_self_hit : 2146/2146 Putative probes exceeding selection criteria: 55/2146 PASSED (GREEN) AND FAILED PUTATIVE PROBES IN THE DESIGN WINDOW --------------------------------------------------------------
Image
Unique (yellow), passed (green) and failed putative probes in the design window
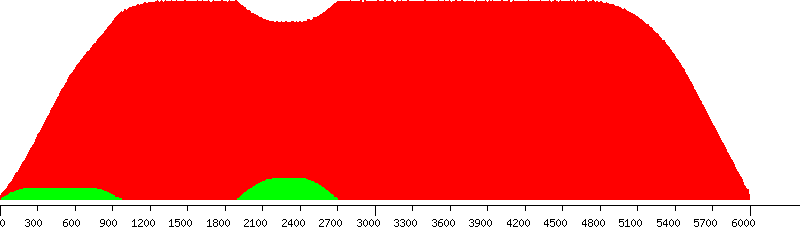
Unique putative probes
No probes
Putative probes
| Putative probe ID | Length | Score ratio | Actual | %rep DNA | Distance | Overall |
|---|---|---|---|---|---|---|
| 41296 | 700 | 18.6 | 3500 | - | 3325 | 18.6 |
| 41297 | 700 | 18.6 | 3500 | - | 3360 | 18.6 |
| 41457 | 650 | 17.3 | 3250 | - | 3328 | 17.3 |
| 41458 | 650 | 17.3 | 3250 | - | 3360 | 17.3 |
| 41459 | 650 | 17.3 | 3250 | - | 3392 | 17.3 |
| 41149 | 750 | 19.9 | 3750 | 1.5 | 3330 | 17.0 |
| 41631 | 600 | 16.0 | 3000 | - | 3300 | 16.0 |
| 41632 | 600 | 16.0 | 3000 | - | 3330 | 16.0 |
| 41633 | 600 | 16.0 | 3000 | - | 3360 | 16.0 |
| 41634 | 600 | 16.0 | 3000 | - | 3390 | 16.0 |
| 41635 | 600 | 16.0 | 3000 | - | 3420 | 16.0 |
| 41636 | 600 | 16.0 | 3000 | - | 3450 | 16.0 |
| 41460 | 650 | 17.3 | 3250 | 0.8 | 3424 | 15.7 |
| 41825 | 550 | 14.6 | 2750 | - | 3321 | 14.6 |
| 41826 | 550 | 14.6 | 2750 | - | 3348 | 14.6 |
| 41827 | 550 | 14.6 | 2750 | - | 3375 | 14.6 |
| 41828 | 550 | 14.6 | 2750 | - | 3402 | 14.6 |
| 41829 | 550 | 14.6 | 2750 | - | 3429 | 14.6 |
| 41830 | 550 | 14.6 | 2750 | - | 3456 | 14.6 |
| 41831 | 550 | 14.6 | 2750 | - | 3483 | 14.6 |
| 41832 | 550 | 14.6 | 2750 | - | 3510 | 14.6 |
| 42036 | 500 | 13.3 | 2500 | - | 3300 | 13.3 |
| 42037 | 500 | 13.3 | 2500 | - | 3325 | 13.3 |
| 42038 | 500 | 13.3 | 2500 | - | 3350 | 13.3 |
| 42039 | 500 | 13.3 | 2500 | - | 3375 | 13.3 |
| 42040 | 500 | 13.3 | 2500 | - | 3400 | 13.3 |
| 42041 | 500 | 13.3 | 2500 | - | 3425 | 13.3 |
| 42042 | 500 | 13.3 | 2500 | - | 3450 | 13.3 |
| 42043 | 500 | 13.3 | 2500 | - | 3475 | 13.3 |
| 42044 | 500 | 13.3 | 2500 | - | 3500 | 13.3 |
| 42045 | 500 | 13.3 | 2500 | - | 3525 | 13.3 |
| 42046 | 500 | 13.3 | 2500 | - | 3550 | 13.3 |
| 40926 | 850 | 12.5 | 4250 | - | 5082 | 12.5 |
| 41055 | 800 | 11.8 | 4000 | - | 5080 | 11.8 |
| 41056 | 800 | 11.8 | 4000 | - | 5120 | 11.8 |
| 41298 | 700 | 18.6 | 3500 | 3.7 | 3395 | 11.2 |
| 41196 | 750 | 11.0 | 3750 | - | 5069 | 11.0 |
| 41197 | 750 | 11.0 | 3750 | - | 5106 | 11.0 |
| 41198 | 750 | 11.0 | 3750 | - | 5143 | 11.0 |
| 41199 | 750 | 11.0 | 3750 | - | 5180 | 11.0 |
| 42047 | 500 | 13.3 | 2500 | 1.2 | 3575 | 10.9 |
| 40803 | 900 | 10.3 | 4500 | - | 5040 | 10.3 |
| 41346 | 700 | 10.3 | 3500 | - | 5075 | 10.3 |
| 41347 | 700 | 10.3 | 3500 | - | 5110 | 10.3 |
| 41348 | 700 | 10.3 | 3500 | - | 5145 | 10.3 |
| 41349 | 700 | 10.3 | 3500 | - | 5180 | 10.3 |
| 41350 | 700 | 10.3 | 3500 | - | 5215 | 10.3 |
| 41351 | 700 | 10.3 | 3500 | 1.0 | 5250 | 8.3 |
| 41833 | 550 | 14.6 | 2750 | 3.3 | 3537 | 8.1 |
| 41057 | 800 | 11.8 | 4000 | 2.1 | 5160 | 7.5 |
| 40927 | 850 | 12.5 | 4250 | 3.6 | 5124 | 5.2 |
| 41200 | 750 | 11.0 | 3750 | 3.2 | 5217 | 4.6 |
| 40804 | 900 | 13.2 | 4500 | 4.7 | 5085 | 3.9 |
| 40690 | 950 | 10.1 | 4750 | 3.8 | 5029 | 2.5 |
Overall score = score_ratio - 2 * repetitive DNA content
Redundant probes
| Putative probe ID | Length | Score ratio | Actual | %rep DNA | Distance | Overall |
|---|---|---|---|---|---|---|
| 41637 | 600 | 16.0 | 3000 | 1.8 | 3480 | 12.3 |
Overall score = score_ratio - 2 * repetitive DNA content
