An Integrative Neuroscience Program Linking Mouse Genes to Cognition and Disease
Seth G. N. Grant
(Taken from Behavioural Genetics in the Post Genomic Era, Edited by Robert Plomin, John C Defries, Ian W Craig and Peter McGuffin, American Psychological Association 2002. ISBN 1-55798-926-5) PDF version
- Introduction
- General Strategy & Outline
- Connecting the Molecular Mechanisms of Learning
- A Multilayer Organisation
- Layer 1: Identification of Genes Encoding Assemblies
- Layer 2: Genomics
- Layer 3: Functional Genomics - Experimental Neuroscience
- Layer 4: Informatics
- Structural Issues for a Large Multidisciplinary Program
- Conclusions
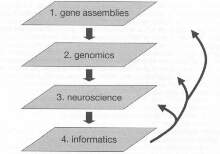
A Multilayer Organisation
The G2C can be organised into four layers (see figure 1). These layers are briefly summarized here and discussed in more detail later.
The entry point for the strategy (Layer 1) is molecular information derived from basic science studies. Strong emphasis is placed on the value of genetically modifiable organisms with nervous systems (invertebrates: fruit fly, Drosophila; worm, Caenorhabditis elegans; vertebrates: mouse, Mus musculus; zebra fish, Danio rerio). Through the use of genetic screens and mutations, these organisms have generated lists of proteins that are involved with various phenotypes. Compiling the set of genes that are involved in a common phenotype (e.g. learning) or involved in a multiprotein complex, or some other ways of classifying sets, produces useful information for a human genotyping study. A prototype for this set is that derived from the molecular studies of the multiprotein complexes (Hebbosomes) underlying acquisition of learning (10862698).
Layer 2 of the G2C takes forward the candidate genes from Layer 1 into human genotyping. Using genome sequencing technology, human single-nucleotide polymorphisms (SNPs) can be determined for all genes in the set and DNAs from relevant humans genotyped. Given the rapid pace of the SNP identification and characterization, information covering the first phase of this should be available in the public domain in the very near future.
Layer 3 of the G2C is aimed at validating the biological significance of variant alleles found in humans. Here functional assays are required, and mouse ES cell technology again is used to provide several complementary in vivo and in vitro approaches. One could assemble a wide range of molecular and neuroscience methods in a highly interactive research program. These neurobiological studies can e linked to human neurobiological studies, thus providing a broad framework of connections at many levels of analysis.
There will be an important role for informatics at all stages of the G2C, and Layer 4 is the platform for this technology. This will include access to existing databases as well as generating new databases. These databases and links should generate a novel and valuable resource for the scientific and medical community.
Layer 1: Identification of Genes Encoding Assemblies
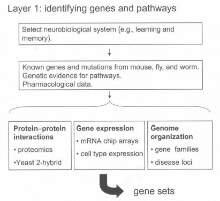
Figure 2: This layer requires bioinformatics and expertise of sciences within the area of basic biology.
Sets of genes are defined using several sources of information (see figure 2). This layer requires bioinformatics and expertise of sciences within the area of basic biology. Types of molecular information that will be used to select genes include the following: (a) mutant phenotypes of mice and other genetic organisms, (b) knowledge of molecular pathways, (c) protein interaction networks obtained from proteomic and yeast 2-hybrid screens, and (d) gene families, chromosomal organisation, and syntenic regions between human and mouse. This prioritization of genes will provide the information for Layer 2.
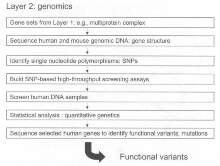
Layer 2: Genomics
The overall goal of Layer 2 is to identify variant structures in specific human genes, which are candidates for detailed functional testing (see figure 3). The basic gene structure for those loci that have been selected and prioritized according to Layer 1 of the G2C will be determined for human and mouse using available finished sequence from the various genome sequencing projects (see www.sanger.ac.uk for information on genome projects). The comparative gene structure of mouse and human serves several purposes. First, it provides a basis for comparing gene structure and assigning intron/exon and other regulatory features to the sequence. (11465055). This information is useful in designing genotyping strategies, including those involving SNP detection. A second reason for obtaining mouse sequence is that in Layer 3 of the G2C this information is useful as a guide for construction of gene-targeting vectors for engineering specific mutations into the mouse.
A major collaborative international effort is underway to identify SNP in the human genome. This SNP Consortium (10757651; 11029002) aims to generate sufficient numbers of SNPs that can then be used in high-thoroughput genotyping assays (11256593; 11172497; 11258600). Statistical analysis (11258193; 11474211) of SNP frequency in populations is used to implicate a gene in the phenotype relevant to the human DNAs. The identification of statistical association will motivate resequencing of the alleles in affected individuals to identify potential functional variants. Sequence information may predict the nature of the functional impairment, such as premature termination condons, and these putative functional variants will be tested in Layer 3.
There are potentially interesting features of a genotyping strategy based on genes encoding proteins known to be components of pathways. As has been shown in model genetic organisms, construction of compound mutations allows ne to examine the functional relationship between the two genes. Epistatic interactions between genes (traditionally defined as the presence of one allele at one locus preventing the expression of an allele at a different locus) is a feature of genes encoding proteins in common pathways. By extension, it may be that some diseases manifest symptoms only if a pathway is debilitated, and this may require the presence of two affected genes. Thus, statistical analysis of the set of genes in Layer 1 may show that sets of SNPs identifying particular variant genes will detect these genes. The effects of these variant genes alone may not give statistical significance association with the disease, although the subsets of genes may do so.
Layer 3: Functional Genomics - Experimental Neuroscience
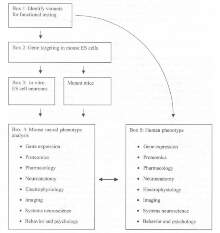
An output from Layer 2 will be variants in the sequence of a human gene. In addition to the statistical analysis used to make a case that a variant gene may be at the basis of some altered phenotype in humans, there is a need to generate biological data showing this variant has function consequences. The simplest way forward may be to use some king of specific in vitro assay that is sensitive to the function of the protein involved. The G2C includes this aspect; however, it proposes to use wider, integrative program of study where the variant is tested in sets of assays relevant to the cells on one hand and the cognitive processes on the other - in other words, many assays at the molecular, cellular, and animal level (see figure 4).
Studying gene function in the nervous system requires general tools applicable to neurons and glia. This in contrast to some areas of cell biology, such as DNA replication or growth control, which can be studied in generic cells. Moreover, in the context of heritable differences in gene structure and the implications for behaviour, it is ultimately necessary to study the gene in the context of the whole animal. Gene targeting in mouse provides an ideal way to bridge the gap between cell biology in cultured neurons and biology of the whole animal. This is because of the pluripotential nature of ES cells and thus the ability to derive cells and animals from the same genetically modified cell. Layer 3 outlines some of the applications of mouse gene targeting and the analysis of mice.
Gene targeting in mouse ES cells (Figure 4 Box 2) is ideally suited for studies of human gene function in complex organs such as the brain because almost any type of gene or chromosomal engineering is feasible in ES cells. The following are some of the relevant technologies:
- Gene knock out - complete disruption of expression (10840733; 11236657; 9735384; 11385465).
- Point mutation and other fine mutations (9735384); this may be particularly useful for introducing SNPs into mouse genes.
- Larger sequence modification, including "humanization' or substitution of human wild-type or mutant genes for mouse genes; this uses techniques of chromosomal engineering <% 11377795 %>.
- Conditional gene modification; these methods allow the desired genetic modification to be "active" or "inactive" in a desired cell (neurone or specific neuronal population) at a specific time during the lifespan of the animal <% 11434315 11084322 %>. For example, a gene that regulates synaptic plasticity, which may be encoded in almost all neurons, can be inactivated in a set of neurons in a selected brain region (e.g., hippocampus) and the effects on cognitive functions assessed.
- Rescue of knock-out allele with mutant or variant genes (Kojuma et al, 1997).
- Insertion of reporter constructs to monitor gene expression (9853749) and subcellular localization of proteins (e.g., Green Fluorescent Protein technology).
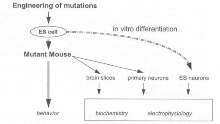
Although mutant mice are useful, there are some neuronal phenotypes that can be studied in neurons grown in culture (see figure 4, Box 4). A major limitation to the study of synapse function has been the lack of clonal cell lines that form synapses with the properties of central nervous system synapses. 'Very recently, it was found that totipotent murine ES cells can be induced to differentiate in culture into neurones (embryonic stem cell neurons; ESNs) comparable with those prepared from neonatal cortex ( (9777634). Importantly, the ESNs display the ability to form functional synapses. Combining gene targeting with ESN technology allows the creation of mutant neurons in vitro. This opens the possibility toward various in vitro screens in mutant neurons (see figure 5).
The phenotype of the cells, animal tissues, and whole animal can be systematically studied in a variety of studies ranging from the molecular to the psychological (see example list in Figure 4, Box 5). It is unnecessary here to break this list into further detail but rather to draw attention to the value of multiple lines of experimental analysis. The first advantage of testing a variant allele in multiple assays is that it makes it more likely that a phenotype can be identified. A greater challenge is to understand why a variant allele may be involved with the human phenotype. Here it is necessary to have some information on the brain at many levels. For example, if one were to only examine synapse function, one may overlook some other critical role in, say, glial function. The advantage of the mouse is that it is possible to explore many levels using ethically acceptable approaches, unlike humans, for which it is not possible to perform similarly invasive procedures. Thus, it is necessary to compare and contrast at those levels where it is possible-the phenotype of mouse and human (see figure 4).
Comparison of mouse and human phenotypes can be pursued on two levels: (a) comparing the mouse and human where each carries a mutation in the same gene and (b) comparing similar phenotypes where the genetic basis in humans is unknown. As illustrated in Boxes 4 and 5 of Figure 4, it would be important to have detailed annotation of phenotype information, assembled in appropriate databases, so that genotype information could be used to ascribe gene function to a phenotype. Developing "neuroscience phenotyping" assays for comparison of humans and mouse is an area that needs further development. Many tests have been developed for rats and are readily transferable to mice. This program of research could promote further efforts to improve and find new ways to examine neurological phenotypes in mice.
Layer 4: Informatics
This broadly integrated program places emphasis on the need for user mobility between datasets as well as storage and recall of information. Linking the datasets together is perhaps one of the most difficult challenges, and a fluid interface would be extremely important. Here are some examples of questions that a well-designed informatic interface may be able to handle:
- List all genes that are encoded in a particular region of chromosome 6 and expressed in the hippocampus. Further sort those genes into those that are known to be important for development or synaptic plasticity of the hippocampus.
- Identify the regions of the human brain that are altered in functional magnetic resonance imaging studies in various genetic diseases, and contrast the regions with the known gene expression profiles and the biochemical function of these genes.
- Catalog the multiprotein complexes involved with synaptic signaling, and list the corresponding human syndromes involving those genes.
- List the genotyping assays that could be used to differentially diagnose chosen psychiatric disorders.
- List the human polymorphisms that result in altered expression of neuronal membrane proteins, and link this to drugs known to modulate those proteins.
Although some aspects of these questions could be answered today, the amount of labor involved with the current data mining tools is enormous. In principle, it should be possible to have answers to these questions in just hours with appropriately designed databases and search engines.